- 1Wildlife and Ecology Group, School of Natural Sciences, Massey University, Palmerston North, New Zealand
- 2Wildbase Research, Massey University, Palmerston North, New Zealand
- 3School of Fundamental Sciences, Massey University, Palmerston North, New Zealand
Reinforcement translocations are increasingly utilised in conservation with the goal of achieving genetic rescue. However, concerns regarding undesirable results, such as genetic homogenisation or replacement, are widespread. One factor influencing translocation outcomes is the rate at which the resident and the introduced individuals interbreed. Consequently, post-release mate choice is a key behaviour to consider in conservation planning. Here we studied mating, and its consequences for genomic admixture, in the North Island brown kiwi Apteryx mantelli population on Ponui Island which was founded by two translocation events over 50 years ago. The two source populations used are now recognised as belonging to two separate management units between which birds differ in size and are genetically differentiated. We examined the correlation between male and female morphometrics for 17 known pairs and quantified the relatedness of 20 pairs from this admixed population. In addition, we compared the genetic similarity and makeup of 106 Ponui Island birds, including 23 known pairs, to birds representing the source populations for the original translocations. We found no evidence for size-assortative mating. On the contrary, genomic SNP data suggested that kiwi of one feather did not flock together, meaning that mate choice resulted in pairing between individuals that were less related than expected by random chance. Furthermore, the birds in the current Ponui Island population were found to fall along a gradient of genomic composition consistent with non-clustered representation of the two parental genomes. These findings indicate potential for successful genetic rescue in future Apteryx reinforcement translocations, a potential that is currently under utilised due to restrictive translocation policies. In light of our findings, we suggest that reconsideration of these policies could render great benefits for the future diversity of this iconic genus in New Zealand.
Introduction
Translocations are increasingly utilised in conservation management to reintroduce species within their former ranges, introduce them to new sites predicted to be suitable for them, or reinforce already existing populations (IUCN/SSC, 2013). Foreseeing the outcome of such interventions remains challenging, partly due to the complexity of the interface between genotype, phenotype, environment, and post translocation behaviour (Renan et al., 2018; Undin et al., 2021b). Post translocation breeding behaviour, such as mating system, and mate choice, warrant special attention, for instance, thanks to its direct link to the post translocation effective population size (Ne; Anthony and Blumstein, 2000).
Mate choice will directly impact reinforcement translocation outcomes. These translocations commonly involve the movement of individuals from a genetically distinct source into a target population. When such interventions result in a fitness increase and/or increased population growth rate, it is referred to as genetic rescue (Ingvarsson, 2001; Hedrick et al., 2011; Whiteley et al., 2015; Bell et al., 2019). Support is growing for genetic rescue being an effective conservation tool, especially for target populations showing symptoms of inbreeding depression (Weeks et al., 2011; Frankham, 2015; Whiteley et al., 2015; Bell et al., 2019; Ralls et al., 2020). Yet, several recent publications argue that genetic rescue has not been attempted as often as it should (Frankham, 2015; Ralls et al., 2018, 2020). One reason for hesitation toward this deliberate admixture of individuals, is the perceived high risk of causing outbreeding depression and loss of unique genetic diversity through homogenisation or genetic replacement (Love Stowell et al., 2017; vonHoldt et al., 2018; Ralls et al., 2020). A fundamental factor determining whether a translocation results in rescue, homogenisation, or replacement is whether, and at what rate, introduced, and original individuals interbreed (Vila et al., 2003; Hedrick and Fredrickson, 2010; Adams et al., 2011; Weiser et al., 2013; Bateson et al., 2014; Mussmann et al., 2017; Thavornkanlapachai et al., 2019). Consequently, post translocation mate choice, and, specifically, the presence of assortative mating, has the potential to greatly affect the outcome of this type of management (Bradley et al., 2014; Engler et al., 2019).
Assortative mating is defined as a positive or negative correlation between phenotypic or genetic traits of paired males and females (Jiang et al., 2013). Some level of positive assortative preference (“like mates alike”) is fundamental in biology since this is a key mechanism for generating new as well as preserving existing species, subspecies, and ecotypes (Verzijden et al., 2005; Kopp et al., 2017; Schumer et al., 2017; Janicke et al., 2019). However, the results of positive assortative mating are context- and situation-dependent (Schumer et al., 2017). For instance, in birds as well as other groups, this behaviour has been shown to maintain sharp morphological separation in hybrid zones (Semenov et al., 2017) but has also been suggested to explain cases of instant speciation through hybridisation (Melo et al., 2009; Hermansen et al., 2011). This mating strategy has been linked to a preference for matching plumage colouration or pattern in several bird species (Andersson et al., 1998; Masello and Quillfeldt, 2003; Semenov et al., 2017; Sun et al., 2019). Yet, the most discussed and studied type of positive assortative mating is size matching between males and females, generally resulting in big individuals mating with other big individuals and small with small (Delestrade, 2001; Helfenstein et al., 2004; Jiang et al., 2013; Ippi et al., 2018; Janicke et al., 2019).
The opposite of positive assortative mating is negative assortative mating, also known as disassortative mating. This mating behaviour results in pairing between individuals less genetically and/or phenotypically similar than expected by chance, and/or between individuals with dissimilar or “opposite” traits (Jiang et al., 2013). Several studies of birds have found strong evidence for disassortative mating being a key mechanism for ensuring high fitness of offspring, for instance, since it manifests as incest-avoidance, reduced risk of inbreeding, and/or as a way of matching beneficial genotypes (Walters et al., 1988; Nelson-Flower, 2009; Nelson-Flower et al., 2012; Riehl and Stern, 2015; Riehl, 2017). Most research on this topic has focused on the major histocompatibility complex (MHC; Brown, 1998; Bonneaud et al., 2006; Kamiya et al., 2014; Santos et al., 2016; O'Connor et al., 2019). Growing evidence suggests a strong heterozygotic advantage for these immune system genes, resulting in fitness benefits for pairs with differing or otherwise compatible MHC profiles (Strandh et al., 2012; Kamiya et al., 2014; Brambilla et al., 2018; Hoover et al., 2018). However, disassortative mating has also been linked to other traits. A famous example is the white-throated sparrow (Zonotrichia albicollis). Within this species, mating almost exclusively occurs between individuals of the opposite of the two prevalent colour morphs (Hedrick et al., 2018).
Whether positive or negative, assortative mating is by necessity linked to mechanisms for detecting compatibility, phenotypic similarity, and/or genetic kinship (Kopp et al., 2017). The mechanisms behind such kin-recognition vary greatly between species but can be based on visual (Walters and Garcia, 2016; Sun et al., 2019), audible (Humphries, 2013), and/or scent cues (Zelano and Edwards, 2002; Bonadonna and Sanz-Aguilar, 2012; Strandh et al., 2012; Hoover et al., 2018), and the ability to detect (dis)similarity can be learnt or innate (Riehl and Stern, 2015). For many species, the mechanism(s) for kin-recognition remain unknown but, there is, for instance, growing support for MHC linked mate choice being based on scent cues in birds as well as mammals (Zelano and Edwards, 2002; Bonadonna and Sanz-Aguilar, 2012; Strandh et al., 2012). More specifically, Zelano and Edwards (2002) speculated that the most likely birds to evolve the ability to make active mate choice based on MHC identity would (1) lack the opportunity for kin-recognition based on social recognition, for instance, due to having precocial young leaving the parents at an early stage, (2) have at least one parent whose only or main contribution to the offspring is genetic material, (3) be long-lived, (4) be monogamous, and (5) have the highly developed olfactory sense. Part of the reasoning behind this is that a long life-span and high partner fidelity generates selection for mechanisms for identifying optimal partners (Kvarnemo, 2018).
One group matching all five of these criteria is Apteryx or kiwi birds; in particular, the focus species of this paper North Island brown kiwi, Apteryx mantelli (1) has super-precocial chicks, allowing conservationists to collect eggs and hatch them in captivity without the presence of a parent (Colbourne et al., 2005), (2) has male-only incubation (Colbourne, 2002), (3) can live for at least 40 years (Barlow, 2011; Robertson and de Monchy, 2012), (4) shows high levels of pair fidelity and monogamy (Taborsky and Taborsky, 1999), and (5) uses scent as their primary sense (Cunningham et al., 2009; Castro et al., 2010).
Here we exploit research opportunities provided by the unique high-density A. mantelli population on Ponui Island that has been thoroughly researched for the last 17 years. This population is the result of two translocation events over 50 years ago. The origin of the translocated birds was Hauturu-o-Toi (also known as Little Barrier Island) and the Waipoua Forest (Figure 1). The birds at these sites now belong to two of the four management units recognised within A. mantelli, the Northland and Western management units, respectively (Craig et al., 2011; Scrimgeour and Pickett, 2011; Germano et al., 2018). This division into management units followed the identification of four genetically separate taxa (lineages) within A. mantelli (Northland, Western, Eastern, and Coromandel; Weir et al., 2016). The study by Weir et al. (2016) found a Pairwise Hudson's Fst of 0.139 (95% CI 0.120–0.158) between the Northland and Western taxa, and dated the split between these management units, and thus between the parental populations of Ponui Island, to between 100,000 and 220,000 years ago. Whilst local adaptations might have evolved over this period of temporal divergence, the extent and nature of any local adaptation remains unknown (Undin et al., 2021a).
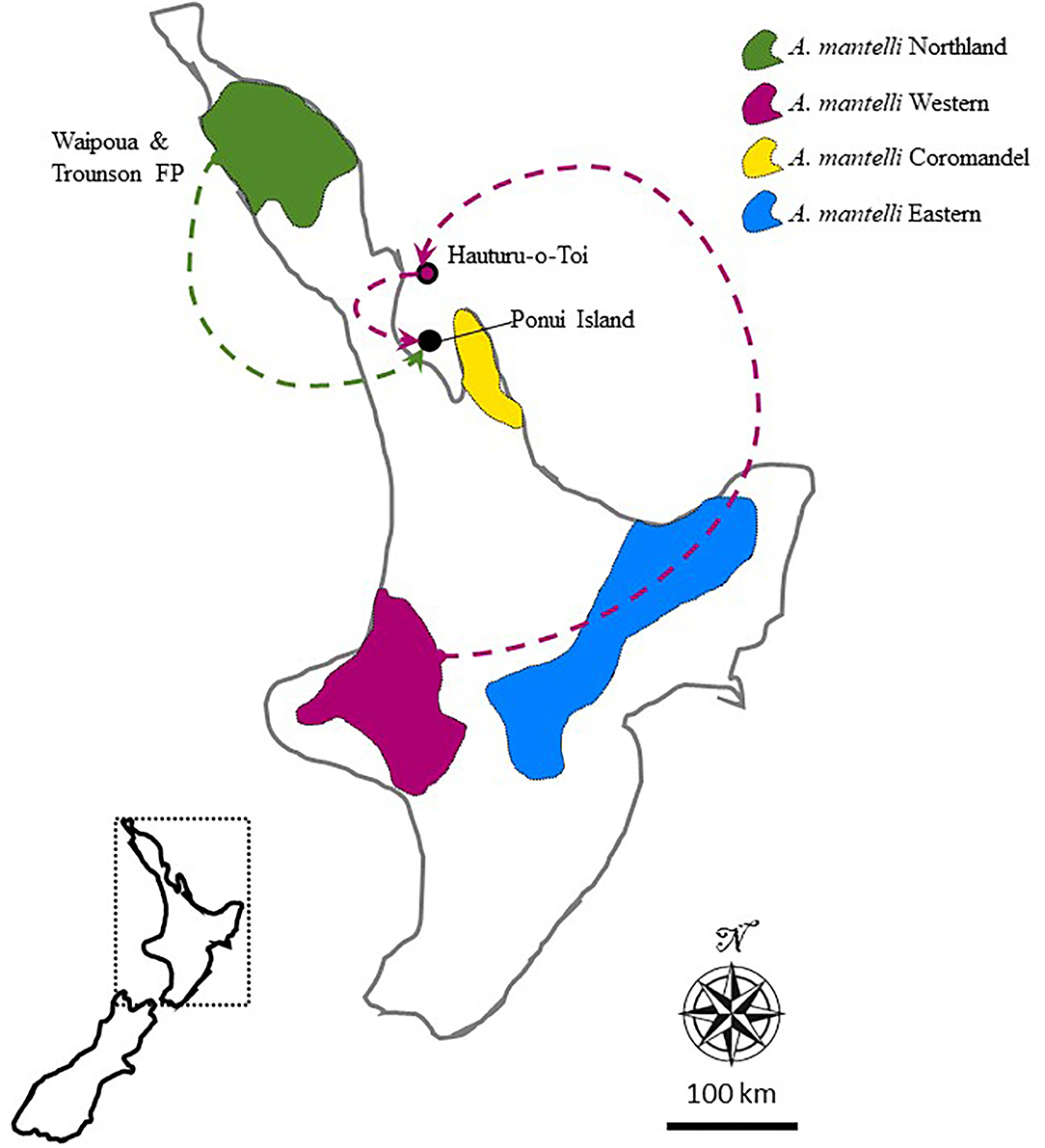
Figure 1. Map of New Zealand's North Island illustrating the approximate distribution of the four management units (also referred to as taxa) recognised within this species. The location of Ponui Island, its parental populations, and the origin of the Hauturu birds are indicated. Modified from Undin et al. (2021b).
The unique features of A. mantelli, the island's long history of continuous research, and the population's multi-origin background make the A. mantelli on Ponui Island ideal for research of behaviour after translocation into a novel environment and, in particular, of mate choice. We examine size matching and genetic (dis)similarity of 17 to 23 Ponui Island pairs by utilising morphometric data collected over 17 years combined with high-density SNP data obtained through genotype-by-sequencing (GBS; a method for reduced representation sequencing; Elshire et al., 2011). We then utilise the SNP data to infer breeding behaviour over time on a population level by investigating whether or not there is evidence for cryptic clusters within the population. Finally, we discuss what the results suggest about A. mantelli mate choice and the plausible implications of this behaviour for future conservation efforts. We focus on reinforcement translocations which are aimed at achieving genetic rescue in this species. However, discussion of our findings is applicable for all species in need of augmented gene flow.
Materials and Methods
Study Sites, Blood Sampling, and Morphometric Measurements
Ponui Island is 1770 ha, and its kiwi density is estimated to one bird per ha. Over the last 17 years, at any given time between 30 and 50 adult A. mantelli on the island have been tagged with radio transmitters. Tracking these birds with transmitters, combined with detailed studies of their habitat use, foraging, and breeding (partly using camera traps) has provided extensive data on individual features as well as on mating and pair identities. The datasets utilised herein contained morphometric data from 53 birds from Ponui Island including 17 pairs, and blood samples from 117 birds including 23 pairs (see further below). Most of these birds—paired birds as well as reference birds—were first caught as adults (>4 years old) over 10 years ago (Table 1). However, the true age was only known for the 12 reference birds that were caught as juveniles between 2004 and 2011. Based on the population being founded in 1964, and an estimated generation time of 8–15 years, the analysed birds can be expected to belong to between the 4th and 6th generation born on the island. However, the oldest known A. mantelli individual was over 40 years old and with a maximum lifespan of 80 to 100 years suggested for Apteryx, it is possible that some of the originally released birds and/or their offspring may still be alive. Blood samples have been collected in four separate cohorts: in 2004, 2006–2008, 2010, and 2017–2019. When multiple samples were available from the same bird, the newest was used to increase the likelihood of high quality. While paired and reference birds had a similar age spread, newer blood samples tended to be more available for the paired birds (Table 1).
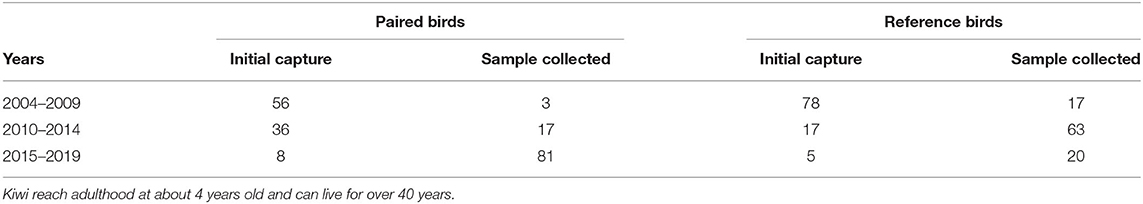
Table 1. Number of birds and samples collected at three-time intervals for paired and reference Ponui Island birds in this study, illustrating that the majority of birds were initially caught at least 10 years ago.
The Ponui Island population was founded through two translocations in 1964 of eight birds from the Waipoua Forest (now belonging to the A. mantelli Northland management unit) and six birds from Hauturu-o-Toi (Hauturu; Colbourne, 2005). The Hauturu birds are usually considered as belonging to the A. mantelli Western management unit, but some claim their origin is hybrid or unclear (Burbidge et al., 2003; Colbourne, 2005; Scrimgeour and Pickett, 2011; Germano et al., 2018).
Unfortunately, data could not be collected directly from the Waipoua Forest due to arduous terrain and a substantial reduction in population density since the early 1990s (Craig et al., 2011). Fortunately, we were able to source both blood samples (n = 20) and morphometric data (n = 15 adults) from the Trounson Kauri Park (hereafter Trounson). Trounson is about 10 km in a straight line, south of the Waipoua Forest, which is within the known dispersal distance of this species (Forbes, 2009). Banded birds have also been observed moving between the two areas on several occasions (Meduna and Donovan pers. comm. 2020) suggesting high potential for sufficient gene flow between the areas to consider them as representing one gene pool.
Morphometric measurements were also collected from 16 adult kiwi on Hauturu-o-Toi. Five additional Hauturu morphometric samples and 13 blood samples for genetic analyses were collected from the Remutaka Forest and the Pūkaha National Wildlife Centre (also known as Mt Bruce) from birds that were translocated to these sites from Hauturu in 2010 and 2008, respectively. Throughout this text, these birds will jointly be referred to as Hauturu birds to reduce confusion.
Blood sampling and morphometric measurements of the birds were conducted in accordance with the Kiwi Best Practise Manual (Robertson and Colbourne, 2017), the Massey University Animal Ethics Committee (MUAEC) permits 06/05, 07/144, 16/92, and 18/83, and the Department of Conservation Wildlife permits AK-14969-RES, AK-21519-FAU, 50249-FAU and 70875-RES. Samples from the parental populations were collected in 2020 and 2021. For birds fitted with transmitters (most of the Ponui Island birds analysed, all the studied pairs, and most of the Hauturu birds from the Remutaka Forest and the Pūkaha National Wildlife Centre), blood sampling, and measuring occurred together with the annual transmitter replacement. A licenced kiwi dog assisted with catching the remaining birds, which were caught specifically for this study. The density of kiwi has been estimated to be around one kiwi per three ha both in Trounson and on Hauturu (in 2007 and 2008, respectively; Holzapfel et al., 2008). Age of the birds was mostly unknown, hence individuals were considered adults if they were known to have bred or, when the breeding status was unknown, based on their sex, size, and weight combined; females were considered adult if weighing >2000 g, or >1700 g if having a tarsus width (TW) >11 mm or a bill >113 mm; males were considered adults if weighting >1700 g, or >1400 g if having a tarsus length (TL) >90 mm or a bill >90 mm.
Size Assortative Mating
Bill length, tarsus length, and the ratio between these measurements were used to explore differences in size distribution among adult birds from Hauturu, Trounson, and Ponui Island. Linear modelling (lm) in R (R core team version 3.6.2) was used for this analysis. To analyse size assortative mating, Pearson correlations between female and male morphometric values were investigated. The measurements analysed were TL, TW, bill length, weight to TL ratio, bill length to TL ratio, and body condition was used for 17 Ponui Island pairs. Body condition was calculated based on weight and TW following Taborsky and Taborsky (1999).
DNA Purification, Sequencing, and SNP Calling
DNA was extracted from 10 to 50 μl thawed A. mantelli blood using a High Pure PCR Template Preparation Kit (Roche, Basel, Switzerland). The manufacturer's instructions were followed with the exception that the DNA was eluted twice using 50 μl of elution buffer for each centrifugation round. The DNA extraction success and quality were validated using agarose gel electrophoresis (1% (w/v) agarose in 1x TAE buffer) and the concentration of DNA was measured with a Qubit 2.0 fluorometer using the dsDNA High Sensitivity assay (Life Technologies, CA, USA).
Pair-ended Genotype-by-sequencing (GBS) library, sequencing preparation, and associated quality cheques were done by The Elshire Group Limited. GBS libraries were constructed using 100 ng of genomic DNA, 1.44 ng of adapters, the restriction enzyme EcoT22i, and 18 PCR cycles, and otherwise following the protocol presented in Elshire et al. (2011). For this study, sequencing of 150 unique birds across three 96-well plates were analysed. Sample location within plates was randomised and each plate contained one positive and one negative control. Sequencing was performed on an Illumina HiSeq XTen with 2 × 150bp paired-end reads.
The previously published A. mantelli genome (Le Duc et al., 2015) was deemed suboptimal as a reference genome for our study based on being (1) highly fragmented and (2) a composite genome of multiple individuals representing two separate management units within the species. Raw short-read data generated by Le Duc et al. (2015) was accessed from the European Nucleotide Archive (Study Assession RJEB6383) and the ERR519283_1.fastq.gz date file was used to re-assemble a reference genome based on a single individual. Raw reads were aggressively trimmed using trim_galore (https://github.com/FelixKrueger/TrimGalore) before assembly with Meraculous 2.2.5.1 (Chapman et al., 2011) using a kmer size of 35 and heterozygous mode 1. The resulting reference genome utilised for our analyses was haploid, with heterozygous regions collapsed into a single haplotype chosen at random between the two possible sequences.
Processing of raw read data, including filtering, trimming, alignment, and SNP calling was conducted by Tea Break Bioinformatics. The 1,538,639,658 raw sequencing reads were de-multiplexed using Axe (axe-demux; Murray and Borevitz, 2018), adapters and barcodes were trimmed using the batch_trim.pl script (https://github.com/Lanilen/GBS-PreProcess) using default parameters. Forward and reverse reads were pair-matched and aligned to the reference genome using Bowtie 2 (Langmead and Salzberg, 2012) using default parameters.
SNP-calling was conducted in STACKS 2.5 (Catchen et al., 2013) using the populations program set for the EcoT22i enzyme, bootstrapping, and site merging. Initially, this was done for each plate separately. The graphical output from Kinship-using-GBS-with-Depth-adjustment program (KGD; Dodds et al., 2015) and Tensorflow Projector (https://projector.tensorflow.org/) were then used to verify the absence of bias or batch effects after which Stacks 2.5 was rerun for the combined dataset of all three plates using the following command line: –vcf -r 0.1 –min-maf 0.1 -e ecoT22I –ordered-export –bootstrap –merge-sites –genepop –structure –fasta-loci –fasta-samples –fasta-samples-raw –write-single-snp. These settings allowed for up to 90% missing data per locus (-r 0.1) to maximise the number of individuals included in the resulting dataset. SNPs derived from different cut sites that had overlapping read coverage were combined into single loci (–merge-sites). To ensure robust SNP identification and to restrict noise, the minimum minor allele frequency was set to 10% (min-maf 0.1). The output was filtered to only include the first variable site per locus (–write-single-snp) resulting in 51691 SNPs that were utilised for analyses with an average depth of 24 reads (with a standard deviation of 12), and an average call rate of 0.6 (with a standard deviation of 0.07). The generated output provided as text files formatted for genepop (see below), and fasta files for loci and samples (–genepop –fasta-loci –fasta-samples).
Relatedness of Paired Birds
For relatedness analyses, known first-degree relatives (known offspring and siblings) were excluded and these analyses contained 20 known pairs and a total of 74 Ponui Island individuals. Unscaled pairwise relatedness values were derived using KGD (Dodds et al., 2015). This matrix of relatedness values is based on pairwise proportional allelic similarity but accounts for read depth (including missing data) which can lead to values >1 and below zero. These values were used to evaluate whether paired individuals were more or less related than expected under a scenario of random mating in two ways. Test one explored whether the relatedness of paired individuals to their partner was higher or lower than the average relatedness of the paired individuals to all other Ponui Island birds in the data set. This approach provided an indication of whether paired individuals were more closely related than expected by chance (a prediction of assortative mating) or more distantly related than expected by chance (a prediction of disassortative mating). To account for possible bias caused by cryptic clusters (in particular if a few paired individuals belonged to a cluster very distantly related to the rest), test one was complemented with test two. Test two involved ranking the relatedness of the female to the male and vice versa for each pair and then comparing these results to expectations based on a computer simulation of ranked relatedness under a scenario of random mating. A ranking of 1 suggested that the partner of a paired bird was the least related individual within the dataset to that paired bird and a ranking of 73 that the partner was the most related in the dataset Random mating was simulated using 10*40 randomly drawn ranks between 1 and 73. T-tests were used for both these tests.
In addition, the average relatedness of the male and female in each pair on Ponui Island (n = 23 Ponui Island pairs) to the birds from Hauturu, was used to categorise pairs as being “similar” or “different” in their genetic makeup). Based on the average relatedness to the Hauturu birds for 106 Ponui Island, the relatedness space was split into four quadrants to categorise pairs. Birds were classified as “similar” when both male and female were found to be less related or both more related to Hauturu than the average value. Birds were classified as “different” when the male was more and the female less related than the average or vice versa.
Genetic Differentiation
Lastly, we examined genetic differentiation of the Hauturu, Trounson, and Ponui Island populations. Using the R package genepop (Rousset, 2008) we calculated pairwise Fst values between the parental populations and between each parental population and the population on Ponui Island. Low Fst values between the parental populations and the current population on Ponui Island would be expected if allelic variation from both parental genomes remains present on the island. Since presence of admixture on Ponui Island would be a crucial step in achieving genetic rescue (Weiser et al., 2013), the relatedness values derived from KGD were then used to calculate the average relatedness to the Hauturu and the Trounson birds for each of the Ponui Island birds in the sample (n = 106 Ponui Island birds). These two factors of relatedness were then linearly normalised to span from 1 to 0; i.e., the Ponui Island birds most related to Trounson had a value of 1 and the bird least related a value of 0, and the same was done for the relatedness of Ponui Island birds to Hauturu birds. The relatedness of each Ponui Island bird to Hauturu was then plotted against the same bird's relatedness to Trounson to explore potential skews toward either parental population and/or patterns of discontinuous spread (or clustering) along the relatedness gradient suggestive of positive assortative mating over time having generated cryptic subpopulations on the island.
Results
Morphology
For bill length and bill to tarsus ratio, the birds from Hauturu were found to be significantly smaller compared to birds from Trounson, while the distribution of individual morphometric values of the Ponui Island birds spanned the range of both parental populations (Figures 2A,C; Bill: F = 8.00, df = 2, p = 0.001; Bill/TL: F = 11.10, df = 2, p < 0.001). A bimodal distribution for both these characteristics was due to the size difference between the sexes (Figures 2A,B; Robertson and Colbourne, 2017); accounting for this difference, the trait distributions within the Ponui Island population was continuous. The difference in tarsus length was not found to be statistically significant (TL: F = 2.60, df = 2, p = 0.08).
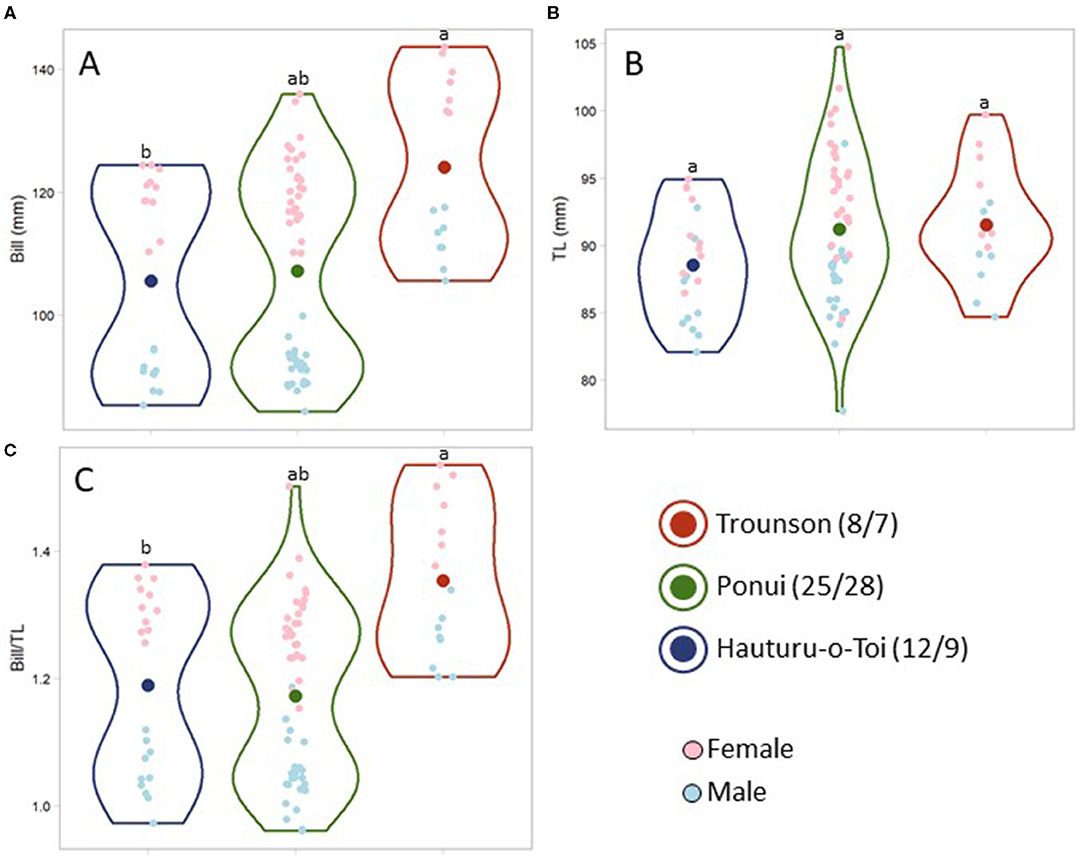
Figure 2. Violin plots comparing morphometrics of adult A. mantelli from Hauturu-o-Toi, Ponui Island, and Trounson Kauri Park, specifically bill length (A), tarsus length (B), and the ratio between the two (C). Different lowercase letters indicate significant difference. Small dots represent each data point and dot colour indicates the sex. Large dots represent the average size or ratio estimated for each population. The shape of the violin outline indicates the distribution of the data points. Sample size for males/females is indicated in the legend for each population.
None of the analysed morphometric characteristics (bill length, weight, tarsus length, tarsus width, bill to tarsus and weight to tarsus ratio, or body condition) showed any support for size assortative mating among the Ponui Island pairs (n = 17 pairs; Figure 3).
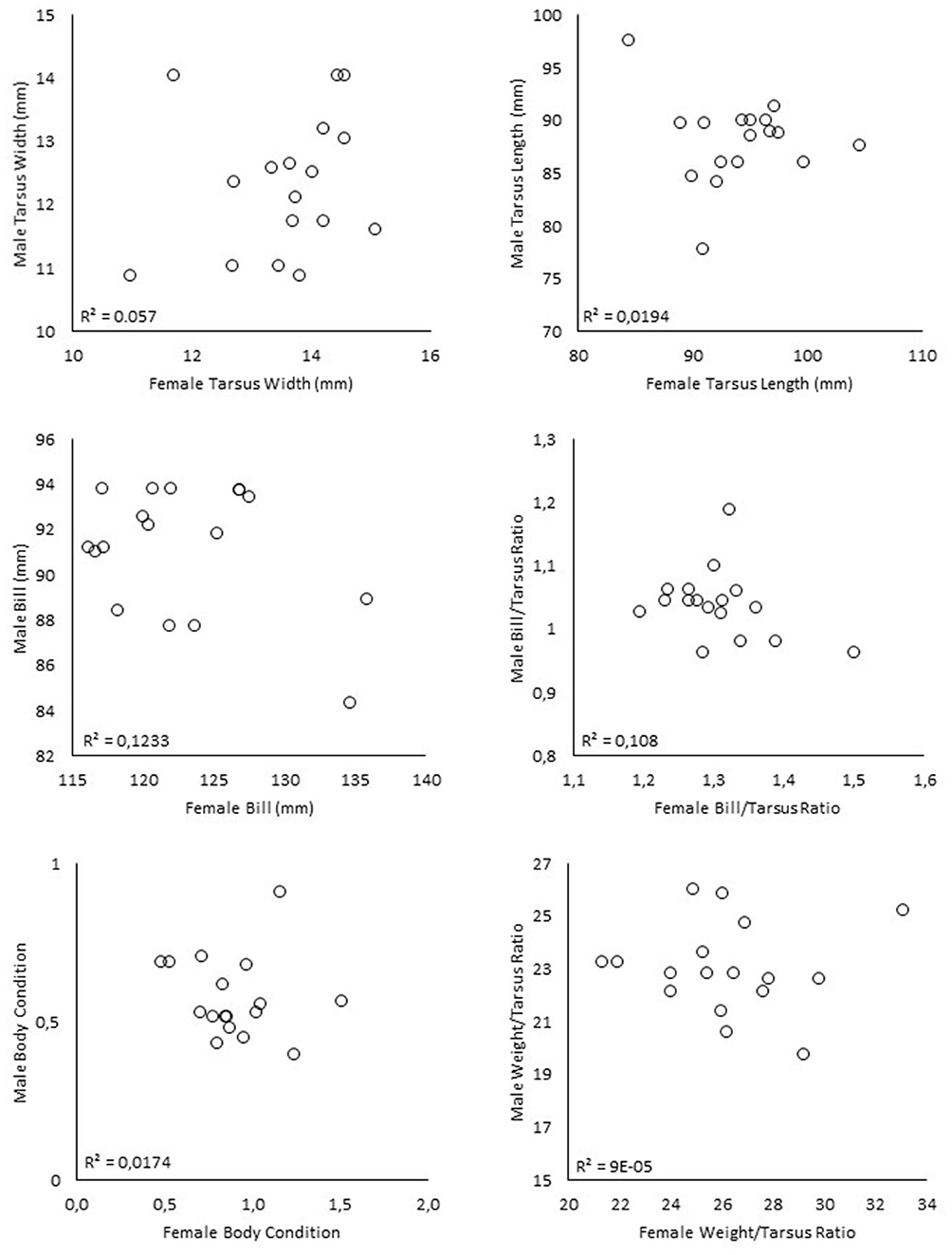
Figure 3. Correlations between morphometric measurements of paired male and female A. mantelli from Ponui Island. The unit of weight is grams. Bill, tarsus length, and tarsus width are given in millimetres. Body condition is based on a combination of weight and tarsus width (Taborsky and Taborsky, 1999). None of the correlations were found to be significant. R2 values refer to linear regression.
Relatedness of Partners
Overall, the allelic similarity within pairs was found to be significantly lower compared to the paired individuals' average relatedness to all other Ponui Island birds in the dataset (0.057 vs 0.011, respectively; t = −4.06, df = 39, p-value < 0.001; n = 20 pairs and 74 birds overall; Figure 4A). Looking at individuals, 75% of the paired birds had a lower value of allelic similarity to their partner compared to their average similarity to the full dataset. The results based on ranked relatedness further supported that pairs were less related than expected under random mating (t = 2.43, df = 73.25, p-value = 0.018; Figure 4B). On average across paired birds, 69% of the birds in the full data set had a higher ranked relatedness than the partner of that individual. In five cases, the partner was in fact one of the top three least related individuals in the data set. However, there was large variation, and one pair was found to have an allelic similarity close to that of first-degree relatives.
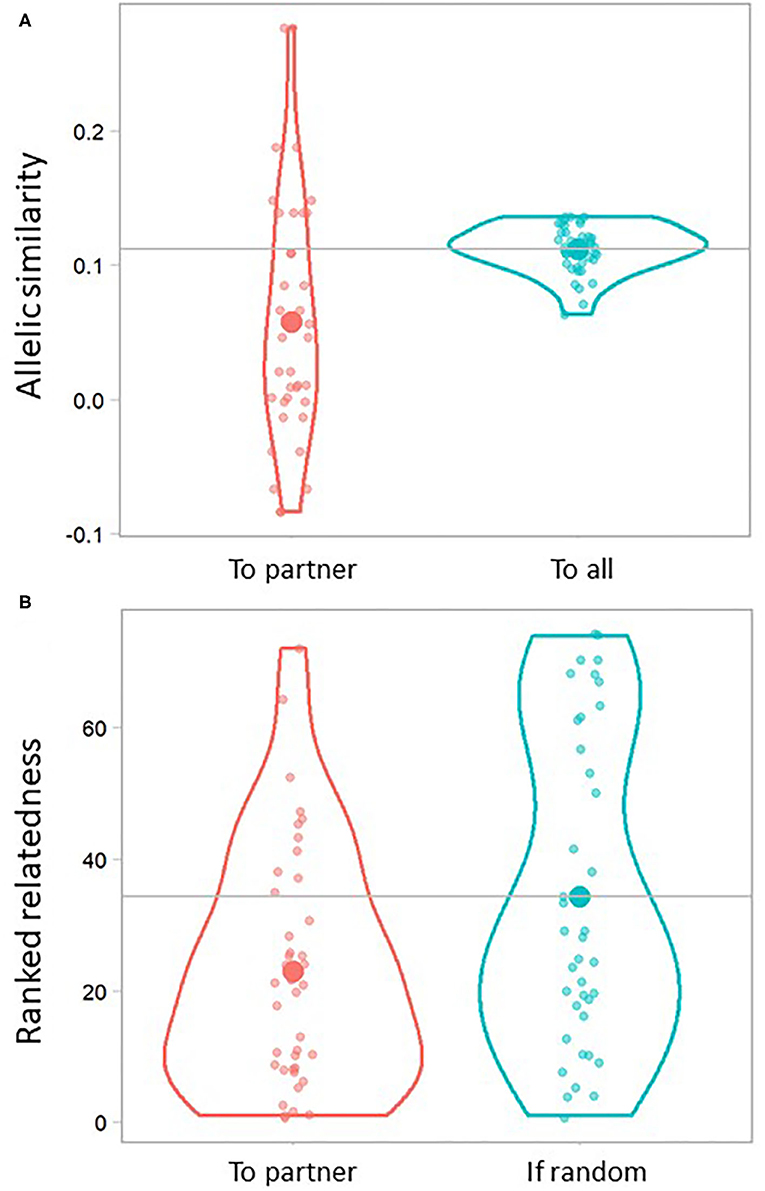
Figure 4. Violin plots illustrating relatedness of paired individuals to their partner (n = 40 datapoints representing 20 pairs). (A) Pairwise relatedness based on allelic similarity between each paired bird and its partner (left) and the average pairwise relatedness based on allelic similarity of each paired bird to all other birds in the entire sample set (right; n = 74). (B) Ranked (or relative) relatedness of each paired bird to its partner (left) compared to one example iteration of 40 randomly generated rank values. A ranking of 1 indicates that the partner of a paired bird was the least related individual in the sample set, while a ranking of 73 indicates that the partner was the most related bird in the data set. Small dots represent each data point, and the shape of the violin outline represents their distribution. Large dots represent the average. Horizontal grey lines indicate average allelic similarity and ranked relatedness, respectively, expected under the null hypothesis of random mating.
Based on the paired individuals' average relatedness to Hauturu birds, an equal number of pairs could be classified as similar (both male and female over or under the average relatedness to Hauturu) and different (one bird less and one more related to Hauturu than average; 10 pairs vs. 10 pairs; Figure 5A). The remaining three pairs were placed in neither category due to either or both partners falling on the threshold value (0.07; Figure 5A).
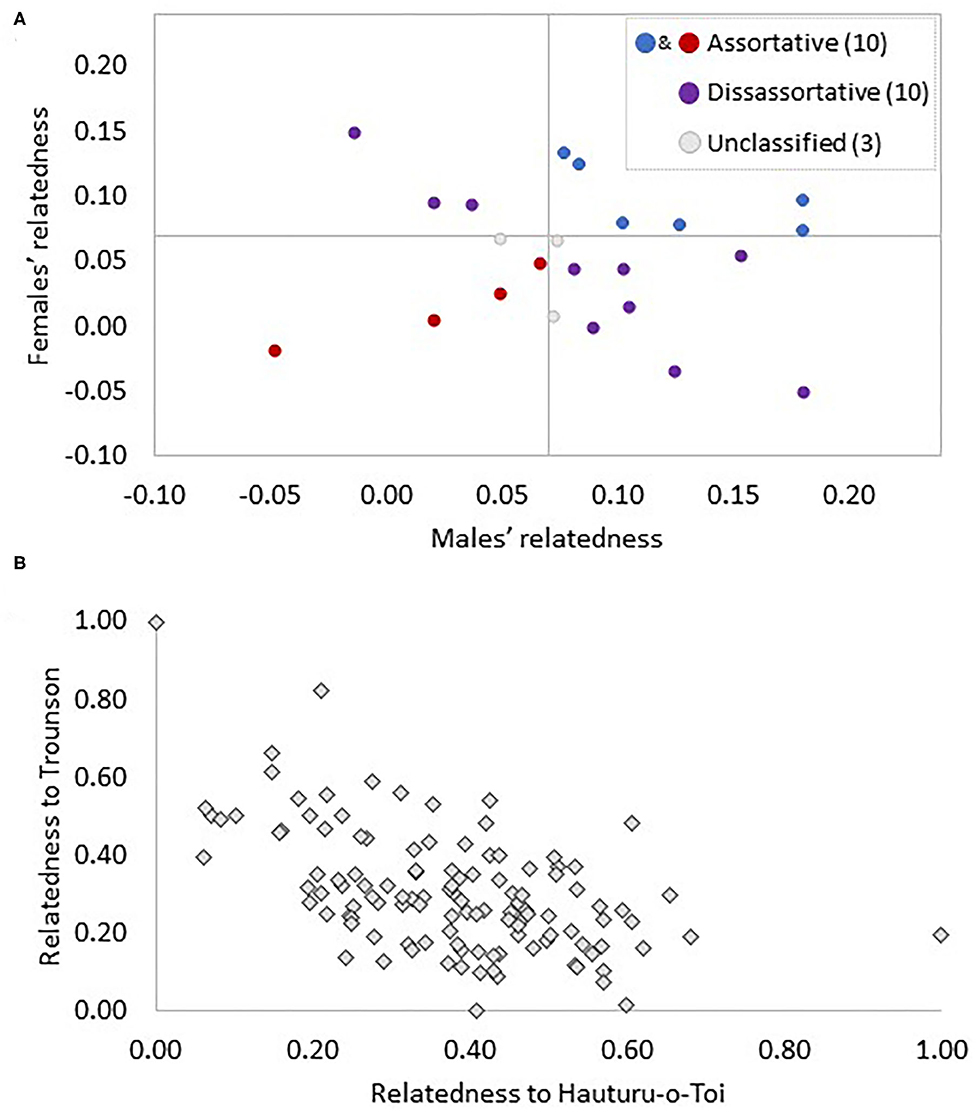
Figure 5. Illustration of the genetic makeup of Ponui Island pairs (A) and the Ponui Island population overall (B). In panel (A) 23 Ponui Island pairs are represented by one dot each and grey lines divide the graph into quadrants using the average relatedness of 106 Ponui Island birds to the Hauturu birds as a threshold value. In pairs falling in the lower left quadrant (red dots), both the male and female are less related to Hauturu than the average Ponui Island bird and, thus, are classified as “assortative” or “similar”. Pairs in the upper right quadrant (blue) are also “assortative” or “similar” (both less related to Hauturu than the average Ponui Island bird), while the top left and bottom right quadrant contains pairs where the male and the female fall on opposite sides of the threshold and are, hence, referred to as “disassortative” or “different” pairs (purple). Three pairs fall on the threshold value and remain unclassified (grey). In panel (B) each of the 106 Ponui Island birds is represented by a diamond indicating their normalised relatedness to Hauturu and Trounson birds. Relatedness values were spread uniformly across the gradient, i.e., the scatter plot indicates no genetic clustering suggestive of positive assortative mating.
Genetic Differentiation and Evidence for Admixture
The pairwise Fst value between Trounson and Hauturu was found to be 0.159 while that between Ponui Island and Trounson was 0.101 and between Ponui Island and Hauturu was 0.092. These values indicate less genetic differentiation between both source populations and the Ponui Island population compared to between the two source populations. A strong negative correlation in relatedness for Ponui Island birds to Hauturu and Trounson was observed. The relatedness values formed a continuous gradient of allelic similarity. Thus, there was no indication of the Ponui Island birds clustering into cryptic subpopulations with individuals either mostly related to Hauturu or to Trounson birds (Figure 5B). These results are consistent with a lack of assortative mate choice over time, generating repeated mixing and backcrossing and resulting in a genetic structure on Ponui Island expected in a hybrid swarm (Hwang et al., 2011).
Discussion
Taken together we found no evidence for positive assortative mating in A. mantelli. This finding was supported both by morphometric data, which indicated no size assortative mating, and genetic data, which suggested an equal likelihood of pairing with an individual of similar or dissimilar overall genetic makeup. The Ponui Island population originated from two genetically distinct taxa (Colbourne, 2005; Weir et al., 2016; Germano et al., 2018). Our morphometric sample set from Hauturu and Trounson suggested that these populations differ morphologically with Hauturu individuals being smaller. If size assortative mating occurred with limited admixture after the translocations to Ponui Island, we would thus expect to observe (cryptic) sub-populations on the island. However, we found no evidence for this. Morphological variation of individual Ponui Island birds spanned that of both parental populations. Furthermore, our genetic analyses indicated high level of shared diversity between both Ponui Island and both two founder genomes (based on Fst values) and a continuum of relatedness without clustering between the Ponui Island birds and the parental populations (based on allelic similarity). These results are expected if there is little genetic, preferential, or behavioural barriers to admixture following multi-origin translocations. Based on this, we suggest that the extant population in 2019—close to six decades after the translocation—has retained allelic variants from both parental populations, and that the population today constitutes a successful hybrid swarm in which there is large individual variation in genomic makeup and no evidence for lingering cryptic separation into a Hauturu and a Waipoua cluster.
In fact, pairwise allelic similarity and ranked relatedness suggested that paired individuals on Ponui were, on average, less related to each other than expected by chance. This disassortative mating is interesting for two reasons. Firstly, it is indicative of a post translocation behaviour that has the potential to increase the speed of admixture between genetically distinct groups, and this rate of admixture is directly linked to the potential for genetic rescue. Mate choice and mating system will drastically impact translocation outcomes (Anthony and Blumstein, 2000; Renan et al., 2018; Zecherle et al., 2021). Promiscuity, polygamy, and/or consecutive alteration of breeding units will all potentially increase the speed of admixture and reduce the long-term impact of initial mating after translocation events (Sander et al., 1997). However, A. mantelli has been described as monogamous with relatively high pair fidelity, thus, initial pairing post translocation will have comparatively larger impact on admixture and thus on the chances of achieving genetic rescue (Taborsky and Taborsky, 1999; Colbourne, 2002). It is possible that the initial scenario on Ponui Island following the translocations in 1964 was artificially bimodal with birds from the two source locations representing two strict groups. Unfortunately, important information of sex distribution and post translocation survival in the two cohorts is missing. However, the lack of genetic clustering on Ponui today indicates that paring between individuals with different degrees of mixed ancestry has happened over time. Thus, the lower-than-expected relatedness between birds in current pairs, cannot be explained by random chance impacting a small initial population. Furthermore, the population of Apteryx on Ponui Island today represents one of very few Apteryx populations that exhibit what is thought to resemble kiwi densities found before the introduction of exotic predators (Potter, 1990; McLennan and Potter, 1992; Miles et al., 1997; Craig et al., 2011; Germano et al., 2018) and, consequently, a comparatively “natural” frequency of mate choice opportunity suggesting that disassortative mate choice might have been more pronounced in the past.
The second reason the lower-than-expected relatedness between paired individuals is interesting is that it is suggestive of kin-recognition in A. mantelli, and that this mechanism is being utilised to reduce inbreeding and increase diversity. Such disassortative mating based on phenotype has been found to be rare (Jiang et al., 2013), and A. mantelli fit the profile of a bird expected to exhibit mate choice linked to MHC compatibility exceptionally well, not the least thanks to its well-developed sense of smell (Zelano and Edwards, 2002; Cunningham et al., 2009; Castro et al., 2010; Corfield et al., 2014; Le Duc et al., 2015). In fact, when comparing across Aves, Zelenitsky et al. (2011) noted that two extant groups stand out due to the large size of their olfaction bulb and inferred positive selection for this part of the brain compared to their sister clades: Apteryx and the order Procellariiformes (the petrels and allies, including albatrosses and shearwaters). Furthermore, Zelano and Edwards (2002), in studies of Procellariiformes have, indeed, found olfaction-based kin-recognition in European storm petrels, Hydrobates pelagicus, and blue petrels, Halobaena caerulea (Bonadonna et al., 2004; Bonadonna and Sanz-Aguilar, 2012) as well as MCH-based disassortative mating in Leach's storm-petrel (Oceanodroma leucorhoa) and H. caerulea (Strandh et al., 2012; Hoover et al., 2018). Studies of MHC diversity and structure in Apteryx have so far focused on the two species with the most drastic bottleneck history: A. rowi and A. owenii (Binney, 2007; Miller et al., 2011). The MHC diversity in A. owenii was found to be extremely low (Binney, 2007; Miller et al., 2011). If MHC profiles are important for Apteryx mate choice as well as fitness, this low diversity could provide an additional hurdle to A. owenii recovery. On the contrary, MHC diversity in A. rowi was found to be higher than expected based on their small current population size and recent tight bottleneck (Binney, 2007). This finding could suggest that a population reduction to about 150 birds (as in A. rowi) was still sufficient for mate choice to keep MHC diversity high, while a population minimum as low as five breeding birds (as in A. owenii) left little or no variation for mate choice to act upon. In view of these findings, we suggest that further studies into MHC diversity, compatibility, detectability, and preference in Apteryx should receive high priority. A first step would be to further examine the genetic dissimilarity between partners and investigate if it is predominantly linked to and/or exaggerated in certain regions of the genome (Beaumont and Balding, 2004; Fitzpatrick et al., 2010; Keller et al., 2013). Such studies would not only increase our understanding of Apteryx breeding but also take us closer to understanding the significance and nature of the genetic differences observed between Apteryx taxa today (see Undin et al., 2021a).
Hesitation toward translocations between genetically distinct lineages (populations, subspecies, or species) remains widespread. This is despite the fact that an increasing number of empirical studies, theoretical studies, and meta-studies suggests that admixture reduces genetic load and symptoms of inbreeding depression, increases heterozygosity and genetic diversity, boosts resilience and recovery rates, and can introduce favourable genetic variation that can increase the ability to cope with change especially in a situation of environmental change (Frankham et al., 2011; Weeks et al., 2011; Frankham, 2015; Jamieson, 2015; Whiteley et al., 2015; Ralls et al., 2018; Zecherle et al., 2021). An interesting parallel to the A. mantelli population on Ponui Island is the population of burrowing bettongs (or boodie, Bettongia lesuer) in Matuwa (Western Australia). This population was established in 2010 through the translocations of individuals from Barrow Island and Dryandra (Thavornkanlapachai et al., 2019). Similar to the case of Ponui Island, the Barrow Island and Dryandra populations represent two distinct taxa within B. lesuer (sometimes referred to as different subspecies; Thavornkanlapachai et al., 2019). A second similarity is that the Dryandra population, like Hauturu, was the result of a previous translocation, in this case from Dorre Island. Thirdly, these taxa show marked differences in phenotype, with the Barrow Island bettongs being substantially smaller (Short and Turner, 1999; Donaldson et al., 2017). When exploring the results of this multi-origin introduction, Thavornkanlapachai et al. (2019) and Rick et al. (2019) discovered that the first breeding at Matuwa happened almost exclusively within the two taxa. However, thanks to their low-fidelity breeding system, over half of the offspring in the Matuwa population were found to be of mixed origin after three years (Thavornkanlapachai et al., 2019). Despite their differences, the A. mantelli on Ponui Island and the B. lesuer in Matuwa can both be seen as examples of (1) that neither genetic nor phenotypic differences need to be hurdles for generating successful and genetically diverse populations through active augmented geneflow, translocations, and admixture, and (2) that breeding behaviour post translocations is directly influencing outcomes (Frankham, 2015; Frankham et al., 2017; vonHoldt et al., 2018; Rick et al., 2019; Hoffmann et al., 2020). Further studies are needed into the mechanisms of genetic rescue and outbreeding depression to increase the predictability of translocation outcomes (Undin et al., 2021b). In relation to behaviour, particular focus is needed to elucidate the effects of source site identity, translocation timing, number of individuals released, as well as population density for post translocation on mate choice and thus on the rate of admixture (see further Zecherle et al., 2021).
Conclusion
Our results suggest that A. mantelli do not mate assortatively with genetically or morphologically similar mates. In fact, pairs were on average found to be less related than expected. This behaviour has the potential to work as a counterforce that reduces inbreeding in general (Walters et al., 1988; Nelson-Flower, 2009; Nelson-Flower et al., 2012; Riehl and Stern, 2015; Riehl, 2017), and, importantly, it suggests that interbreeding between resident and introduced kiwi after reinforcement translocations is likely. This creates potential for genetic rescue. Currently, a discussion is ongoing about the potential impact of inbreeding on Apteryx fitness and viability (Weiser et al., 2013; Innes et al., 2015, 2016; Germano et al., 2018). At the same time, policy around translocations is restrictive, strongly recommending minimisation of the geographic as well as the genetic distance between target and source populations (Craig et al., 2011; Scrimgeour and Pickett, 2011; Innes et al., 2016; Kiwis for Kiwi, 2016; Germano et al., 2018; Taylor et al., 2021). Arguments for this policy include that mixing populations that are too different risks resulting in outbreeding depression, loss of local adaptation, and overall loss of diversity. We recognise that mixing could result in undesirable outcomes and that this depends on the nature of the difference between source and target population (Undin et al., 2021a,b). However, our results presented herein indicate that genomic admixture can be a steady but slow process that retains diversity from both parental populations and that the Ponui Island population represents a successful balancing of the two key conservation goals of introducing genetic material without eradicating local diversity or homogenising population differences. Further support that there is currently underutilised potential for genetic rescue in Apteryx is the astonishing success of the Ponui Island population. This hybrid population has, on average, exhibited 9% annual population growth for over 50 years and is now one of the densest in the country despite the presence of some species of introduced predators and competitors (Cunningham and Castro, 2011; Strang, 2018). Additional study of this population should be prioritised to elucidate the particular factors that generated this success to enable practitioners to optimise strategy for future translocations.
We agree with other recent publications stressing that behaviour in the context of conservation needs more attention (Caro and Sherman, 2011; Renan et al., 2018; Zecherle et al., 2021). Consequently, we hope that this case study can contribute to the ongoing discussions concerning (1) attempting genetic rescue, (2) identifying suitable source populations for translocations, (3) the usefulness of genomics and epigenomics to take behavioural research to a population level and add a temporal scale, (4) how knowledge of social behaviour post translocations is important for appropriate conservation management design, and (5) how conservation management plans would benefit from incorporating the opportunity to express the full range of natural behaviours as an explicit goal and a part of how conservation success is defined. For instance, the presence of active mate choice in kiwi raises concern that, for the vast majority of Apteryx, the opportunity for mate choice today is substantially restricted either due to low population densities, small population sizes, reduced connectivity, and/or artificial settings such as so-called kohanga kiwi sites (sites where predators have been excluded, and the goal is to breed kiwi and harvest juveniles for introduction, reintroduction, and/or reinforcement translocations; Innes et al., 2016).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: Dryad https://datadryad.org/stash/share/4VAEtkWXTdMm7vlU5jG5qDr_iBNyPZoT1l8C-xpRu-E. The following data files associated with this publication is available via Dryad. (1) Morphometric data compared between adults of three populations; (2) Morphometric data compared among 17 pairs of birds; (3) A relatedness matrix including all 150 individuals analysed; (4) Relatedness values and ranked relatedness values among 20 analysed pairs. Requests for raw reads from the underlying GBS sequence library should be made to Richard Witehira: cmljaGFyZHdpdGVoaXJhQHh0cmEuY28ubno= or Andre Witehira: YW5kcmUud2l0ZWhpcmFAZ21haWwuY29t. Permission would then be sought from the kaitiaki (guardians) representing hapu (sub-tribes) that affiliate with the areas of sample collection. Kiwi are taonga (treasures) to the indigenous Māori people of Aotearoa New Zealand. All individuals, samples, or genomic data obtained from taonga species have whakapapa (genealogy, connections, and belonging) and are considered taonga in their own right. Tikanga Māori (Māori customary practices) determines their use.
Ethics Statement
The animal study was reviewed and approved by Massey University Animal Ethics Committee (MUAEC).
Author Contributions
MU performed the literature search, analysed the data, and wrote the first draft of the manuscript. IC is the leader of the long-term Ponui Island Kiwi Research and led the fieldwork and sample collection for this study. All authors jointly developed the concept for the article, contributed to the interpretation of the data, commented on previous versions of the manuscript, and approved the final manuscript.
Acknowledgements
First, we want to acknowledge the people of the land, in particular Te Patukeha, Ngati Kuta Hapū and Richard Witehira—without you, this project would never have started. We thank the Chamberlin family for allowing us to have a program of research to study the Ponui kiwi for the last 21 years.
Funding
Funding for the many projects on Ponui Island over the years was provided by an anonymous donor, Kiwis for kiwi, Forest and Bird, and Massey University Research Fund. We are also very grateful to Bernard Sabrair and Elka Gouzer-Waechter who funded part of this research through the Massey Foundation. Kiwi Rescue's MBIE Program C09X1609, the New Zealand Marsden Fund MAU1707 and the Ngā Pae o te Māramatanga Kia Ārohi Kia Mārama grant id 18SCO17 provided specific funding for the last 4 years.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adams, J. R., Vucetich, L. M., Hedrick, P. W., Peterson, R. O., and Vucetich, J. A. (2011). Genomic sweep and potential genetic rescue during limiting environmental conditions in an isolated wolf population. Proc. Royal Soc.f London B: Biol. Sci. 278, 3336–3344. doi: 10.1098/rspb.2011.0261
Andersson, S., Örnborg, J., and Andersson, M. (1998). Ultraviolet sexual dimorphism and assortative mating in blue tits. Proc. Royal Soc.f London B: Biol. Sci. 265, 445–450. doi: 10.1098/rspb.1998.0315
Anthony, L. L., and Blumstein, D. T. (2000). Integrating behaviour into wildlife conservation: the multiple ways that behaviour can reduce Ne. Biol. Conserv. 95, 303–315. doi: 10.1016/S0006-3207(00)00037-9
Bateson, Z. W., Dunn, P. O., Hull, S. D., Henschen, A. E., Johnson, J. A., and Whittingham, L. A. (2014). Genetic restoration of a threatened population of greater prairie-chickens. Biol. Conserv. 174, 12–19. doi: 10.1016/j.biocon.2014.03.008
Beaumont, M. A., and Balding, D. J. (2004). Identifying adaptive genetic divergence among populations from genome scans. Mol. Ecol. 13, 969–980. doi: 10.1111/j.1365-294X.2004.02125.x
Bell, D. A., Robinson, Z. L., Funk, W. C., Fitzpatrick, S. W., Allendorf, F. W., Tallmon, D. A., et al. (2019). The exciting potential and remaining uncertainties of genetic rescue. Trends Ecol. Evol. 34, 1070–1079. doi: 10.1016/j.tree.2019.06.006
Binney, B. M. (2007). The Major Histocompatibility Complex (MHC) of the kiwi (Apteryx spp.). Auckland: Master of Science in Molecular Bioscience, Massey University.
Bonadonna, F., and Sanz-Aguilar, A. (2012). Kin recognition and inbreeding avoidance in wild birds: the first evidence for individual kin-related odour recognition. Anim. Behav. 84, 509–513. doi: 10.1016/j.anbehav.2012.06.014
Bonadonna, F., Villafane, M., Bajzak, C., and Jouventin, P. (2004). Recognition of burrow's olfactory signature in blue petrels, Halobaena caerulea: an efficient discrimination mechanism in the dark. Anim. Behav. 67, 893–898. doi: 10.1016/j.anbehav.2003.08.013
Bonneaud, C., Chastel, O., Federici, P., Westerdahl, H., and Sorci, G. (2006). Complex MHC-based mate choice in a wild passerine. Proc. Royal Soc. London B: Biol. Sci. 273, 1111–1116. doi: 10.1098/rspb.2005.3325
Bradley, D. W., Molles, L. E., and Waas, J. R. (2014). Post-translocation assortative pairing and social implications for the conservation of an endangered songbird. Anim. Conserv. 17, 197–203. doi: 10.1111/acv.12083
Brambilla, A., Keller, L., Bassano, B., and Grossen, C. (2018). Heterozygosity–fitness correlation at the major histocompatibility complex despite low variation in alpine ibex (Capra ibex). Evol. Appl. 11, 631–644. doi: 10.1111/eva.12575
Brown, J. L. (1998). The new heterozygosity theory of mate choice and the MHC. Genetica 104, 215–221. doi: 10.1023/A:1026409220292
Burbidge, M. L., Colbourne, R. M., Robertson, H. A., and Baker, A. J. (2003). Molecular and other biological evidence supports the recognition of at least three species of brown kiwi. Conserv. Gene.s 4, 167–177. doi: 10.1023/A:1023386506067
Caro, T., and Sherman, P. W. (2011). Endangered species and a threatened discipline: behavioural ecology. Trends Ecol. Evol. 26, 111–118. doi: 10.1016/j.tree.2010.12.008
Castro, I., Cunningham, S. J., Gsell, A. C., Jaffe, K., Cabrera, A., and Liendo, C. (2010). Olfaction in birds: a closer look at the kiwi (Apterygidae). J. Avian Biol. 41, 213–218. doi: 10.1111/j.1600-048X.2010.05010.x
Catchen, J., Hohenlohe, P. A., Bassham, S., Amores, A., and Cresko, W. A. (2013). Stacks: an analysis tool set for population genomics. Mol. Ecol. 22, 3124–3140. doi: 10.1111/mec.12354
Chapman, J. A., Ho, I., Sunkara, S., Luo, S., Schroth, G. P., and Rokhsar, D. S. (2011). Meraculous: de novo genome assembly with short paired-end reads. PLoS ONE 6:e23501. doi: 10.1371/journal.pone.0023501
Colbourne, R. (2002). Incubation behaviour and egg physiology of kiwi (Apteryx spp.) in natural habitats. NZ J. Ecol. 129–138. https://www.jstor.org/stable/24055315
Colbourne, R. (2005). Kiwi (Apteryx spp.) on offshore New Zealand islands. Dept. Conserv. Res. Develop. Series 208:24.
Colbourne, R., Bassett, S., Billing, T., McCormick, H., McLennan, J., Nelson, A., et al. (2005). The development of Operation Nest Egg as a tool in the conservation management of kiwi. Dept. Conserv. Res. Develop. Series 259:24.
Corfield, J. R., Eisthen, H. L., Iwaniuk, A. N., and Parsons, S. (2014). Anatomical specializations for enhanced olfactory sensitivity in kiwi, Apteryx mantelli. Brain Behav. Evol. 84, 214–226. doi: 10.1159/000365564
Craig, E., Gardiner, C., Renwick, N., and Sporle, W. (2011). Taxon plan for Northland brown kiwi (Apteryx mantelli).
Cunningham, S. J., and Castro, I. (2011). The secret life of wild brown kiwi: studying behaviour of a cryptic species by direct observation. NZ J. Ecol. 35, 209–219. https://www.jstor.org/stable/24060731
Cunningham, S. J., Castro, I., and Potter, M. A. (2009). The relative importance of olfaction and remote touch in prey detection by North Island brown kiwis. Anim. Behav. 78, 899–905. doi: 10.1016/j.anbehav.2009.07.015
Delestrade, A. (2001). Sexual size dimorphism and positive assortative mating in alpine choughs (Pyrrhocorax graculus). Auk 118, 553–556. doi: 10.1093/auk/118.2.553
Dodds, K. G., McEwan, J. C., Brauning, R., Anderson, R. M., van Stijn, T. C., Kristjánsson, T., et al. (2015). Construction of relatedness matrices using genotyping-by-sequencing data. BMC Genom. 16:1047. doi: 10.1186/s12864-015-2252-3
Donaldson, F., Bencini, R., Morris, K., Teale, R., Wale, C. H., How, R. A., et al. (2017). The burrowing bettongs of Barrow Island: demographic and genetic insights into a threatened macropod. Aust. J. Zool. 65, 257–272. doi: 10.1071/ZO17049
Elshire, R. J., Glaubitz, J. C., Sun, Q., Poland, J. A., Kawamoto, K., Buckler, E. S., et al. (2011). A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6:e19379. doi: 10.1371/journal.pone.0019379
Engler, J., O., Sacher, T., Coppack, T., and Bairlein, F. (2019). Assortative mating frames establishment in a young island bird population. R. Soc. Open Sci. 6:190050. doi: 10.1098/rsos.190050
Fitzpatrick, B. M., Johnson, J. R., Kump, D. K., Smith, J. J., Voss, S. R., and Shaffer, H. B. (2010). Rapid spread of invasive genes into a threatened native species. Proc. Nat. Acad. Sci. 107, 3606–3610. doi: 10.1073/pnas.0911802107
Forbes, Y. (2009). Natal Dispersal, Habitat Selection and Mortality of North Island brown kiwi (Apteryx mantelli) at the Moehau Kiwi Sanctuary, Coromandel. Auckland: Master of Applied Science in Environmental Studies, Auckland University of Technology.
Frankham, R. (2015). Genetic rescue of small inbred populations: meta-analysis reveals large and consistent benefits of gene flow. Mol. Ecol. 24, 2610–2618. doi: 10.1111/mec.13139
Frankham, R., Ballou, J. D., Eldridge, M. D., Lacy, R. C., Ralls, K., Dudash, M. R., et al. (2011). Predicting the probability of outbreeding depression. Conserv. Biol. 25, 465–475. doi: 10.1111/j.1523-1739.2011.01662.x
Frankham, R., Ballou, J. D., Ralls, K., Eldridge, M., Dudash, M. R., Fenster, C. B., et al. (2017). Genetic Management of Fragmented Animal and Plant Populations. Don Mills, ON: Oxford University Press.
Germano, J., Barlow, S., Castro, I., Colbourne, R., Cox, M., Gillies, C., et al. (2018). Kiwi Recovery Plan 2018–2028 Mahere Whakaora Kiwi 2018–2028.
Hedrick, P. W., Adams, J. R., and Vucetich, J. A. (2011). Reevaluating and broadening the definition of genetic rescue. Conserv. Biol. 25, 1069–1070. doi: 10.1111/j.1523-1739.2011.01751.x
Hedrick, P. W., and Fredrickson, R. (2010). Genetic rescue guidelines with examples from Mexican wolves and Florida panthers. Conserv. Gene. 11, 615–626. doi: 10.1007/s10592-009-9999-5
Hedrick, P. W., Tuttle, E. M., and Gonser, R. A. (2018). Negative-assortative mating in the white-throated sparrow. J. Hered. 109, 223–231. doi: 10.1093/jhered/esx086
Helfenstein, F., Danchin, E., and Wagner, R. H. (2004). Assortative mating and sexual size dimorphism in black-legged kittiwakes. Waterbirds 27, 350–355. doi: 10.1675/1524-4695(2004)027[0350:AMASSD]2.0.CO;2
Hermansen, J. S., Saether, S. A., Elgvin, T. O., Borge, T., Hjelle, E., and Sætre, G. P. (2011). Hybrid speciation in sparrows I: phenotypic intermediacy, genetic admixture and barriers to gene flow. Mol. Ecol. 20, 3812–3822. doi: 10.1111/j.1365-294X.2011.05183.x
Hoffmann, A. A., Miller, A. D., and Weeks, A. R. (2020). Genetic mixing for population management: from genetic rescue to provenancing. Evol. Appl. 14, 634–652. doi: 10.1111/eva.13154
Holzapfel, S., Robertson, H., and McLennan, J. A. (2008). Kiwi (Apteryx spp.) recovery plan 2008–2018. Threat. Species Recov. Plan 60, 1–73.
Hoover, B., Alcaide, M., Jennings, S., Sin, S. Y. W., Edwards, S. V., and Nevitt, G. A. (2018). Ecology can inform genetics: disassortative mating contributes to MHC polymorphism in Leach's storm-petrels (Oceanodroma leucorhoa). Mol. Ecol. 27, 3371–3385. doi: 10.1111/mec.14801
Humphries, D. J. (2013). The Mechanisms and Function of Social Recognition in the Cooperatively Breeding Southern Pied Babbler, Turdoides bicolor (Doctorate). Sydney, NSW: Degree in Biology, Macquarie University.
Hwang, A., Northrup, S., Alexander, J., Vo, K., and Edmands, S. (2011). Long-term experimental hybrid swarms between moderately incompatible Tigriopus californicus populations: hybrid inferiority in early generations yields to hybrid superiority in later generations. Conserv. Gene. 12, 895–909. doi: 10.1007/s10592-011-0193-1
Ingvarsson, P. K. (2001). Restoration of genetic variation lost–the genetic rescue hypothesis. Trends Ecol. Evol. 16, 62–63. doi: 10.1016/S0169-5347(00)02065-6
Innes, J., Eppink, F., Andreson, D., and Robertson, H. A. (2016). Roles for 'kohanga' in Kiwi Conservation a Review. Lincoin: Landcare Research/Manaaki Whenua Contract Report LC2504 for the Department of Conservation.
Innes, J., Eppink, F. V., and Robertson, H. A. (2015). Saving a National Icon: Preliminary Estimation of the Additional Cost of Achieving kiwi Population Stability or 2% Auckland, NZ: Landcare Research Manaaki Whenua.
Ippi, S., Cerón, G., Alvarez, L. M., Aráoz, R., and Blendinger, P. G. (2018). Relationships among territory size, body size, and food availability in a specialist river duck. Emu 118, 293–303. doi: 10.1080/01584197.2018.1438848
IUCN/SSC (2013). Guidelines for Reintroductions and Other Conservation translocations. Version 1.0. IUCN Species Survival Commission.
Jamieson, I. G. (2015). Significance of population genetics for managing small natural and reintroduced populations in New Zealand. N. Z. J. Ecol. 39, 1–18. https://www.jstor.org/stable/26198690
Janicke, T., Marie-Orleach, L., Aubier, T. G., Perrier, C., and Morrow, E. H. (2019). Assortative mating in animals and its role for speciation. Am. Nat. 194, 865–875. doi: 10.1086/705825
Jiang, Y., Bolnick, D. I., and Kirkpatrick, M. (2013). Assortative mating in animals. Am. Nat. 181, E125–E138. doi: 10.1086/670160
Kamiya, T., O'Dwyer, K., Westerdahl, H., Senior, A., and Nakagawa, S. (2014). A quantitative review of MHC-based mating preference: the role of diversity and dissimilarity. Mol. Ecol. 23, 5151–5163. doi: 10.1111/mec.12934
Keller, I., Wagner, C., Greuter, L., Mwaiko, S., Selz, O., Sivasundar, A., et al. (2013). Population genomic signatures of divergent adaptation, gene flow and hybrid speciation in the rapid radiation of Lake Victoria cichlid fishes. Mol. Ecol. 22, 2848–2863. doi: 10.1111/mec.12083
Kiwis for Kiwi (2016). “Target 2% working with new zealanders to grow kiwi populations,” in Kiwis for Kiwi Investment. Auckland: Kiwis.
Kopp, M., Servedio, M. R., Mendelson, T. C., Safran, R. J., Rodríguez, R. L., Hauber, M. E., et al. (2017). Mechanisms of assortative mating in speciation with gene flow: connecting theory and empirical research. Am. Nat. 191, 1–20. doi: 10.1086/694889
Kvarnemo, C. (2018). Why do some animals mate with one partner rather than many? a review of causes and consequences of monogamy. Biol. Rev. 93, 1795–1812. doi: 10.1111/brv.12421
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Le Duc, D., Renaud, G., Krishnan, A., Almén, M. S., Huynen, L., Prohaska, S. J., et al. (2015). Kiwi genome provides insights into evolution of a nocturnal lifestyle. Genome Biol. 16, 147–162. doi: 10.1186/s13059-015-0711-4
Love Stowell, S. M., Pinzone, C. A., and Martin, A. P. (2017). Overcoming barriers to active interventions for genetic diversity. Biodivers. Conserv. 26, 1753–1765. doi: 10.1007/s10531-017-1330-z
Masello, J. F., and Quillfeldt, P. (2003). Body size, body condition and ornamental feathers of burrowing parrots: variation between years and sexes, assortative mating and influences on breeding success. Emu 103, 149–161. doi: 10.1071/MU02036
McLennan, J., and Potter, M. (1992). Distribution, population changes and management of brown kiwi in Hawke's Bay. N. Z. J. Ecol. 16, 91–102. doi: 10.1111/24053603
Melo, M. C., Salazar, C., Jiggins, C. D., and Linares, M. (2009). Assortative mating preferences among hybrids offers a route to hybrid speciation. Evol: Int. J. Organ. Evol. 63, 1660–1665. doi: 10.1111/j.1558-5646.2009.00633.x
Miles, J. R. G., Potter, M. A., and Fordham, R. A. (1997). Northern brown kiwi (Apteryx australis mantelli) in Tongariro National Park and Tongariro Forest: ecology and threats. (0478018991). Wellington: Department of Conservation Wellington.
Miller, H. C., Bowker-Wright, G., Kharkrang, M., and Ramstad, K. (2011). Characterisation of class II B MHC genes from a ratite bird, the little spotted kiwi (Apteryx owenii). Immunogenetics 63, 223–233. doi: 10.1007/s00251-010-0503-7
Murray, K. D., and Borevitz, J. O. (2018). Axe: rapid, competitive sequence read demultiplexing using a trie. Bioinformatics 34, 3924–3925. doi: 10.1093/bioinformatics/bty432
Mussmann, S. M., Douglas, M. R., Anthonysamy, W. J., Davis, M. A., Simpson, S. A., Louis, W., et al. (2017). Genetic rescue, the greater prairie chicken and the problem of conservation reliance in the Anthropocene. R. Soc. Open Sci. 4:160736. doi: 10.1098/rsos.160736
Nelson-Flower, M. J. (2009). Kinship and its Consequences in the Cooperatively Breeding Southern Pied Babbler Turdoides Bicolor (Doctorate Degree). Cape Town: University of Cape Town.
Nelson-Flower, M. J., Hockey, P. A. R., O'Ryan, C., and Ridley, A. R. (2012). Inbreeding avoidance mechanisms: dispersal dynamics in cooperatively breeding southern pied babblers. J. Anim. Ecol. 81, 876–883. doi: 10.1111/j.1365-2656.2012.01983.x
O'Connor, E. A., Westerdahl, H., Burri, R., and Edwards, S. V. (2019). Avian MHC evolution in the era of genomics: phase 1.0. Cells 8:1152. doi: 10.3390/cells8101152
Potter, M. (1990). Movement of North Island brown kiwi (Apteryx australis mantelli) between forest remnants. N. Z. J. Ecol. 14, 17–24. doi: 10.1111/24053307
Ralls, K., Ballou, J. D., Dudash, M. R., Eldridge, M. D. B., Fenster, C. B., Lacy, R. C., et al. (2018). Call for a paradigm shift in the genetic management of fragmented populations. Conserv. Lett. 11:e12412. doi: 10.1111/conl.12412
Ralls, K., Sunnucks, P., Lacy, R. C., and Frankham, R. (2020). Genetic rescue: a critique of the evidence supports maximizing genetic diversity rather than minimizing the introduction of putatively harmful genetic variation. Biol. Conserv. 251:108784. doi: 10.1016/j.biocon.2020.108784
Renan, S., Speyer, E., Ben-Nun, T., Ziv, A., Greenbaum, G., Templeton, A. R., et al. (2018). Fission-fusion social structure of a reintroduced ungulate: Implications for conservation. Biol. Conserv. 222, 261–267. doi: 10.1016/j.biocon.2018.04.013
Rick, K., Ottewell, K., Lohr, C., Thavornkanlapachai, R., Byrne, M., and Kennington, W. J. (2019). Population genomics of Bettongia lesueur: admixing increases genetic diversity with no evidence of outbreeding depression. Genes 10:851. doi: 10.3390/genes10110851
Riehl, C. (2017). Kinship and incest avoidance drive patterns of reproductive skew in cooperatively breeding birds. Am. Nat. 190, 774–785. doi: 10.1086/694411
Riehl, C., and Stern, C. A. (2015). How cooperatively breeding birds identify relatives and avoid incest: new insights into dispersal and kin recognition. Bioessays 37, 1303–1308. doi: 10.1002/bies.201500120
Robertson, H. A., and Colbourne, R. (2017). Kiwi Best Practice Manual. Wellington: Terrestrial Ecosystems Unit, Department of Conservation.
Robertson, H. A., and de Monchy, P. J. (2012). Varied success from the landscape-scale management of kiwi Apteryx spp. in five sanctuaries in New Zealand. Bird Conserv. Int. 22, 429–444. doi: 10.1017/S0959270912000044
Rousset, F. (2008). genepop'007: a complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 8, 103–106. doi: 10.1111/j.1471-8286.2007.01931.x
Sander, U., Short, J., and Turner, B. (1997). Social organisation and warren use of the burrowing bettong, Bettongia lesueur (Macropodoidea: Potoroidae). Wildlife Res. 24, 143–157. doi: 10.1071/WR96021
Santos, P. S. C., Courtiol, A., Heidel, A. J., Höner, O. P., Heckmann, I., Nagy, M., et al. (2016). MHC-dependent mate choice is linked to a trace-amine-associated receptor gene in a mammal [Article]. Sci. Rep. 6:38490. doi: 10.1038/srep38490
Schumer, M., Powell, D. L., Delclós, P. J., Squire, M., Cui, R., Andolfatto, P., et al. (2017). Assortative mating and persistent reproductive isolation in hybrids. Proc. Nat. Acad. Sci. 114, 10936–10941. doi: 10.1073/pnas.1711238114
Scrimgeour, J., and Pickett, A. J. (2011). Taxon Plan for Western Brown Kiwi (Apteryx mantelli) Strategic Plan for the Recovery of Western Brown Kiwi for the Period 2011 – 2021 and Beyond, Including Key Actions Required for Their Recovery. Whanganui: Department of Conservation Tongariro Whanganui Taranaki Conservancy.
Semenov, G. A., Scordato, E. S. C., Khaydarov, D. R., Smith, C.hris, C., R., Kane, N. C., et al. (2017). Effects of assortative mate choice on the genomic and morphological structure of a hybrid zone between two bird subspecies. Mol. Ecol. 26, 6430–6444. doi: 10.1111/mec.14376
Short, J., and Turner, B. (1999). Ecology of burrowing bettongs, Bettongia lesueur (Marsupialia: Potoroidae), on Dorre and Bernier Islands, Western Australia. Wildlife Res. 26, 651–669. doi: 10.1071/WR98039
Strandh, M., Westerdahl, H., Pontarp, M., Canbäck, B., Dubois, M.-P., Miquel, C., et al. (2012). Major histocompatibility complex class II compatibility, but not class I, predicts mate choice in a bird with highly developed olfaction. Proc. Royal Soc. London B: Biol. Sci. 279, 4457–4463. doi: 10.1098/rspb.2012.1562
Strang, K. E. (2018). The Ecology of Feral Cats (Felis catus) on a New Zealand Offshore Island: Considerations for Management (Doctorate Degree in Ecology). Palmerston North: Massey University.
Sun, L., Zhou, T., Stone, G. N., Wan, Q. H., and Fang, S. G. (2019). Seeing-good-gene-based mate choice: From genes to behavioural preferences. J. Anim. Ecol. 88, 1708–1719. doi: 10.1111/1365-2656.13071
Taborsky, B., and Taborsky, M. (1999). The mating system and stability of pairs in kiwi Apteryx spp. J. Avian Biol. 30, 143–151. doi: 10.2307/3677123
Taylor, H. R., Robertson, H., Carter, A., and Ramstad, K. M. (2021). The conservation management implications of isolation by distance and high genetic diversity in Great Spotted Kiwi (Apteryx haastii). Emu-Austral Ornithol. 1–13. doi: 10.1080/01584197.2021.1888126
Thavornkanlapachai, R., Mills, H. R., Ottewell, K., Dunlop, J., Sims, C., Morris, K., et al. (2019). Mixing genetically and morphologically distinct populations in translocations: asymmetrical introgression in a newly established population of the boodie (Bettongia lesueur). Genes 10, 729–747. doi: 10.3390/genes10090729
Undin, M., Hills, S. F. K., Lockhart, P. J., and Castro, I. (2021a). Gaps in genetic knowledge affect conservation management of kiwi (Apteryx) species. Ibis, 1–20. doi: 10.1111/ibi.12951
Undin, M., Lockhart, P. J., Hills, S. F. K., and Castro, I. (2021b). Genetic rescue and the plight of Ponui hybrids [Review]. Front. Conserv. Sci. 1–8. doi: 10.3389/fcosc.2020.622191
Verzijden, M. N., Lachlan, R. F., and Servedio, M. R. (2005). Female mate-choice bhaviour and sympatric speciation. Evolution 59, 2097–2108. doi: 10.1111/j.0014-3820.2005.tb00920.x
Vila, C., Sundqvist, A. K., Flagstad, Ø., Seddon, J., Kojola, I., Casulli, A., et al. (2003). Rescue of a severely bottlenecked wolf (Canis lupus) population by a single immigrant. Proc. Royal Soc. London B: Biol. Sci. 270, 91–97. doi: 10.1098/rspb.2002.2184
vonHoldt, B. M., Brzeski, K. E., Wilcove, D. S., and Rutledge, L. Y. (2018). Redefining the role of admixture and genomics in species conservation. Conserv. Lett. 11:e12371. doi: 10.1111/conl.12371
Walters, J. R., Doerr, P. D., and Carter Iii, J. H. (1988). The cooperative breeding system of the red-cockaded woodpecker. Ethology 78, 275–305. doi: 10.1111/j.1439-0310.1988.tb00239.x
Walters, J. R., and Garcia, V. (2016). “Red-cockaded woodpeckers: alternative pathways to breeding success,” in Cooperative Breeding in Vertebrates: Studies of Ecology, Evolution, and Behavior, eds W. Koenig and J. Dickinson (New York, NY:Cambridge University Press), 58–76.
Weeks, A. R., Sgro, C. M., Young, A. G., Frankham, R., Mitchell, N. J., Miller, K. A., et al. (2011). Assessing the benefits and risks of translocations in changing environments: a genetic perspective. Evol. Appl. 4, 709–725. doi: 10.1111/j.1752-4571.2011.00192.x
Weir, J. T., Haddrath, O., Robertson, H. A., Colbourne, R. M., and Baker, A. J. (2016). Explosive ice age diversification of kiwi. Proc. Nat. Acad. Sci. 113, E5580–E5587. doi: 10.1073/pnas.1603795113
Weiser, E. L., Grueber, C. E., and Jamieson, I. G. (2013). Simulating retention of rare alleles in small populations to assess management options for species with different life histories. Conserv. Bio. 27, 335–344. doi: 10.1111/cobi.12011
Whiteley, A. R., Fitzpatrick, S. W., Funk, W. C., and Tallmon, D. A. (2015). Genetic rescue to the rescue. Trends Ecol. Evol. 30, 42–49. doi: 10.1016/j.tree.2014.10.009
Zecherle, L. J., Nichols, H. J., Bar-David, S., Brown, R. P., Hipperson, H., Horsburgh, G. J., et al. (2021). Subspecies hybridization as a potential conservation tool in species reintroductions. Evol. Appl. 14, 1216–1224. doi: 10.1111/eva.13191
Zelano, B., and Edwards, S. V. (2002). An MHC component to kin recognition and mate choice in birds: predictions, progress, and prospects. Am. Nat. 160, S225–S237. doi: 10.1086/342897
Keywords: aves, assortative mating, breeding behaviour, conservation management, hybridisation, mate choice, translocation
Citation: Undin M, Lockhart PJ, Hills SFK, Armstrong DP and Castro I (2021) Mixed Mating in a Multi-Origin Population Suggests High Potential for Genetic Rescue in North Island Brown Kiwi, Apteryx mantelli. Front. Conserv. Sci. 2:702128. doi: 10.3389/fcosc.2021.702128
Received: 29 April 2021; Accepted: 15 July 2021;
Published: 10 August 2021.
Edited by:
Oded Berger-Tal, Ben-Gurion University of the Negev, IsraelReviewed by:
Shirli Bar-David, Ben-Gurion University of the Negev, IsraelJordan Bemmels, University of Toronto Scarborough, Canada
Carolyn Hogg, The University of Sydney, Australia
Copyright © 2021 Undin, Lockhart, Hills, Armstrong and Castro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Malin Undin, bWFsaW4udW5kaW5AZ21haWwuY29t
 Malin Undin
Malin Undin Peter J. Lockhart
Peter J. Lockhart Simon F. K. Hills
Simon F. K. Hills Doug P. Armstrong
Doug P. Armstrong Isabel Castro
Isabel Castro