
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 11 March 2025
Sec. Antibiotic Resistance and New Antimicrobial drugs
Volume 15 - 2025 | https://doi.org/10.3389/fcimb.2025.1551240
This article is part of the Research Topic Deciphering Antimicrobial Resistance: Genetic Insights and Perspectives View all 3 articles
Background: Bacterial resistance to aminoglycoside antimicrobials is becoming increasingly severe due to their use as commonly prescribed antibiotics. The discovery of new molecular mechanisms of aminoglycoside resistance is critical for the effective treatment of bacterial infections.
Methods: Bacteria in goose feces were isolated by plate streaking. The identification and characterization of a novel resistance gene from the bacterial genome involved various techniques, including molecular cloning, drug susceptibility testing, protein expression and purification, and enzyme kinetic analysis. Additionally, whole-genome sequencing and phylogenetic studies were performed.
Results: Brucella intermedia DW0551, isolated from goose feces, was resistant to 35 antibiotics, and the minimum inhibitory concentration (MIC) was particularly high for most aminoglycoside antibiotics. The novel aminoglycoside resistance gene aac(6’)-Iaq encoded by B. intermedia DW0551 conferred resistance to netilmicin, sisomicin, amikacin, kanamycin, gentamicin, tobramycin, and ribostamycin. The amino acid sequence of AAC(6’)-Iaq shared the highest identity (52.63%) with the functionally characterized aminoglycoside acetyltransferase AAC(6’)-If. AAC(6’)-Iaq contained all the conserved sites of the acetyltransferase family NAT_SF. The enzyme exhibited strong affinity and catalytic activity toward netilmicin and sisomicin. The mobile genetic element (MGE) was not found in the flanking regions of the aac(6’)-Iaq and aac(6’)-Iaq-like genes.
Conclusion: In this work, a novel aminoglycoside acetyltransferase gene, designated aac(6’)-Iaq, which conferred resistance to a variety of aminoglycoside antimicrobials, was identified in an animal Brucella intermedia isolate. Identification of new antibiotic resistance mechanisms in bacteria isolated from animals could aid in the treatment of animal and human infectious diseases caused by related bacterial species.
Aminoglycosides are antimicrobial molecules that are widely used in clinical practice and are usually used in combination with β-lactam drugs for the treatment of infections caused by Gram-positive and Gram-negative bacteria (Abdul-Aziz et al., 2020). Aminoglycoside resistance genes confer drug resistance to bacteria, encoding mainly aminoglycoside resistance proteins, namely, aminoglycoside acetyltransferases (AACs), aminoglycoside nucleotidyltransferases (ANTs) and aminoglycoside phosphotransferases (APHs), which acetylate, adenylate and phosphorylate drugs at specific sites, respectively (Ramirez and Tolmasky, 2010). The AAC(6’) enzyme, the most common aminoglycoside N-acetyltransferase, which modifies the 6’ site of aminoglycosides, such as kanamycin, tobramycin, gentamicin and amikacin, and usually confers resistance to these drugs (Ramirez and Tolmasky, 2010).
To date, 68 genes of the aac(6’) gene family have been included in the Comprehensive Antibiotic Resistance Database (CARD) (Alcock et al., 2023). These genes are located on chromosomal and plasmid DNA and are generally related to mobile genetic elements (MGEs), such as transposons and integrons (Alcock et al., 2023). Proteins encoded by the aac(6’) genes usually exhibit resistance to more than one aminoglycoside antibiotic. For example, AAC(6’)-Ia shows resistance to tobramycin, gentamicin, amikacin, isepamicin and netilmicin (Shaw et al., 1992), while AAC(6’)-Iy shows resistance to tobramycin, amikacin and netilmicin (Magnet et al., 1999).
Brucella intermedia (formerly known as Ochrobactrum intermedium, type strain LMG 3301 = NCTC 12171 = CNS 2-75) was first isolated from human blood in 1988 (Holmes et al., 1988) and was first described and named in 1998 (Velasco et al., 1998). These bacteria are Gram-negative facultative coccobacilli and are usually present as circular, low-convexity and smooth colonies approximately 1 mm in diameter. These bacteria generally have two chromosomes, with the larger one being responsible for activities related to functions of bacterial life and the smaller one encoding genes related to virulence. Brucella spp. were once thought to cause disease only in immunosuppressed populations, but more recent studies have indicated their role as opportunistic pathogens causing a wide range of diseases (Ryan and Pembroke, 2020). For example, they can lead to bacteremia in individuals with malignant tumors (Apisarnthanarak et al., 2005; Kassab et al., 2021), inflammation of the respiratory and cardiovascular systems (Hirai et al., 2016; Bharucha et al., 2017), and endophthalmitis in otherwise healthy individuals (Jacobs et al., 2013). These infections have also been reported to be associated with outbreaks of nosocomial P. aeruginosa infections (Sekiguchi et al., 2007). The emergence of multidrug-resistant B. intermedia has attracted increasing attention in recent years due to the increase in its pathogenicity (Ryan and Pembroke, 2020; Sheng et al., 2023). However, the mechanism underlying the resistance to aminoglycosides has rarely been reported (Lu et al., 2021).
In one of our recent projects, to investigate the antimicrobial resistance mechanisms of animal, human and environmental bacteria, we isolated hundreds of bacteria from fecal samples. The antimicrobial resistance profiles of these strains were examined, and their genomes were sequenced. In this work, the resistance phenotype and genotype of a microbe, designated B. intermedia DW0551 isolated from a goose, were characterized, and a novel chromosomal aminoglycoside resistance gene designated aac(6’)-Iaq was identified from this bacterium.
A total of 576 strains of bacteria were isolated from samples collected from sewage and domestic fowl and livestock in animal farms in Wenzhou, Zhejiang Province, China. For all of them, the minimum inhibitory concentrations (MICs) of numerous antimicrobial agents were tested, and the genomes were sequenced. The isolates were initially identified at the species level using 16S rRNA gene homology and average nucleotide identity (ANI) analyses (Konstantinidis and Tiedje, 2005). One of them, designated B. intermedia DW0551 herein, was isolated from goose feces. Table 1 lists the strains and plasmids used in this study.
According to the M100 performance standards for antimicrobial susceptibility testing by the Clinical and Laboratory Standards Institute (CLSI) (Lewis, 2022), the MICs were determined by the agar dilution method using Mueller–Hinton (MH) agar plates with twofold serial dilutions of antibiotics (Crump et al., 2015). After 16-20 h of incubation at 37°C, the MIC results were interpreted according to M100 performance standards and the guidelines of the European Committee on Antimicrobial Susceptibility Testing (v12.0) (EUCAST, 2022). A total of 43 antimicrobial molecules were tested (Table 2), all of which were purchased from hospitals or pharmaceutical companies. Pseudomonas aeruginosa ATCC27853, also a Gram-negative bacillus as B. intermedia DW0551, was used for quality control. The MIC results of the quality control strain remained within the predetermined limits, indicating the credible test results.
The genomic DNA of the bacterium was extracted using the Universal Genomic DNA Purification Mini Spin Kit (Beyotime Biotechnology Co., Ltd., Shanghai, China). For analysis of the resistance genes in 576 bacteria, the genome of each bacterium was sequenced on the Illumina NovaSeq 6000 platform (Shanghai Personal Biotechnology Co., Ltd., Shanghai, China). The Illumina short reads of all 576 bacterial genomes were pooled together and assembled by SKESA v2.4.0 (Souvorov et al., 2018). The antibiotic resistance-related genes were annotated by Prokka (v.1.14.6) (Seemann, 2014) against the CARD (Alcock et al., 2020) and ResFinder database (Bortolaia et al., 2020). For whole-genome sequencing of Brucella intermedia DW0551, the PacBio Sequel II (Shanghai Personal Biotechnology Co., Ltd., Shanghai, China) sequencing platform was subsequently used. The long reads from PacBio Sequel II and the short reads from Illumina NovaSeq 6000 were hybrid assembled in a short-read-first manner using Unicycler (v0.4.8) (Wick et al., 2017) and then polished by Pilon (v 1.23) (Walker et al., 2014). The optimization of the assembly results was performed using Racon (v1.4.13) (Huang et al., 2022) and Pilon (v1.23) (Walker et al., 2014) and was assessed using QUAST (v5.0.2-fb0b821) (Gurevich et al., 2013). Putative proteins were annotated against the NCBI nonredundant protein database using DIAMOND (v2.0.14) (Buchfink et al., 2021). The average nucleotide identity (ANI) was calculated with FastANI (Zong, 2020). Multiple sequence alignment was performed with MAFFT (v7.407) (Katoh and Standley, 2013). IQ-TREE v2.2.2.3 was used to select the model that minimized the BIC score to construct a phylogenetic tree using the log-likelihood method with the Bayesian information criterion (BIC) (Lorah and Womack, 2019). The protein domains of AAC(6’)-Iaq were predicted using CD-search (Marchler-Bauer and Bryant, 2004). Genetic context analysis of aac(6’)-Iaq and other related sequences was carried out using clinker (v.0.0.25) (Gilchrist and Chooi, 2021).
The ORF of aac(6’)-Iaq (447 bp) with its upstream promoter region predicted by BPROM (www.softberry.com) (Supplementary Figure S1) was amplified by PCR with primers designed by SnapGene v6.0 (www.snapgene.com) (Supplementary Table S1). The PCR products (575 bp) were subsequently ligated into the T-Vector pMD™19 using T4 ligase (Takara Biomedical Technology Co., Ltd.). The recombinant plasmid pMD19-T-aac(6’)-Iaq was subsequently transformed into competent E. coli DH5α cells, after which the recombinant strain DH5α(pMD19-T-aac(6’)-Iaq) was selected on LB agar plates supplemented with 100 µg/mL ampicillin. Finally, the cloned fragment [aac(6’)-Iaq with its upstream promoter region] was verified by Sanger sequencing (Shanghai Sunny Biotechnology Co., Ltd., Shanghai, China).
The PCR-amplified ORF of the aac(6’)-Iaq gene was inserted into the pCold I vector (Table 1 and Supplementary Table S1), and the recombinant plasmid pCold I-aac(6’)-Iaq was subsequently transformed into E. coli BL21. The recombinant BL21(pColdI-aac(6’)-Iaq) was cultured in 100 mL of LB to an OD600 of 0.6 in a shaker at a constant temperature of 37°C. After cooling, 1 mM isopropyl-β-D-thiogalactopyranoside was added, and the mixture was shaken at a constant temperature of 16°C for 18 h. Bacteria were then collected by centrifugation at 10,000 × g for 10 min at 4°C and lysed with 4 mL of nondenaturing lysis solution. Subsequently, the lysed bacteria were subjected to 10 min of ultrasonic lysis. After 30 min of centrifugation (10,000 × g) at 4°C, the supernatant containing the recombinant protein was obtained, and the protein was purified using the Beyotime His-tag Protein Purification Kit (Beyotime Biotechnology Co., Ltd., Shanghai, China). The His tag of the recombinant protein was excised using Thrombin (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) at 4°C for 72 h. The AAC(6’)-Iaq protein was then concentrated using an Amicon Ultra15 centrifugal filter equipped with an Ultracel-10 membrane (Casanovas et al., 2017) and stored at -20°C with 50% glycerol.
The kinetic parameters of AAC(6’)-Iaq were measured as reported previously with slight modifications (Vetting et al., 2008). The AAC(6’)-Iaq activity was determined by measuring the NTB ions produced from the interaction of the acetylation reaction product CoA-SH (containing sulfhydryl groups) with DTNB (CoA-SH + DTNB → TNB-). The total reaction volume was 200 µL, and the pH ranged from 7-7.5. The mixture contained 2 mM DTNB, 1 mM EDTA, 100 mM CoA-SH, 25 mM MES, 5-500 µg/mL aminoglycoside substrate dissolved using sterile double-distilled water and 2 µg of purified AAC(6’)-Iaq protein (Magnet et al., 2001). The reaction was monitored at 412 nm using Synergy Neo2 (BioTek Instruments Inc., VT, United States) at a constant temperature of 37°C for 10 min at 4-second intervals. The AAC(6’)-Iaq protein in the reaction system was replaced with an equal volume of double distilled water as a control for each assay, and the experiments were repeated three times for each aminoglycoside substrate. The steady-state kinetic parameters (kcat and Km) were determined by nonlinear regression of the initial reaction rates with the Michaelis-Menten equation in Prism (v9.4.0) software (GraphPad Software, CA, United States). Another AAC(6’) variant [AAC(6′)-Va] was analyzed together as a positive control (Zhang et al., 2023).
Reverse transcription quantitative PCR (RT-qPCR) was performed according to previous methods with slight changes (Rocha et al., 2020). Bacteria were cultured in LB broth until the OD600 reached 0.5, and for the experimental group, 1/4 MIC ribostamycin (1,024 μg/mL) was then added. The bacteria were further incubated for 2, 4, or 8 h, followed by RNA extraction with RNAprep Pure Cell/Bacteria Kit (Tiangen, Beijing, China) and quantification with a DS-11+ Spectrophotometer (DeNovix, Delaware, United States). The DNA-free RNA extract was verified by PCR of the Brucella intermedia 16S rRNA gene. cDNA was synthesized for each sample using HiScript III RT SuperMix for qPCR (Vazyme Biotech, Nanjing, China). RT-qPCR was performed on a CFX96™ Touch Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, United States), and the increase in real-time fluorescence was monitored by using SYBR qPCR Master Mix (Vazyme Biotech, Nanjing, China). The housekeeping 16S rRNA gene was utilized as a reference gene for relevant quantification via the CT method (Schmittgen and Livak, 2008). Comparisons of expression levels were performed using a t-test to evaluate the effects of aminoglycosides, and p ≤ 0.05 was considered to indicate statistical significance.
The nucleotide sequences of the B. intermedia DW0551 genome have been submitted to GenBank under accession number CP131474 for chromosome_1, CP131475 for chromosome_2, CP131476 for pDW0551, and OR395485 for the aac(6’)-Iaq gene.
As mentioned above, to explore the resistance mechanisms of the bacteria against antimicrobial molecules, we sequenced 576 bacteria isolated from sewage, domestic fowl and livestock feces. Annotation of the pooled genomic sequences of these bacteria via Illumina sequencing demonstrated that they encoded putative resistance genes against various classes of antimicrobial agents, including resistance genes for aminoglycosides, β-lactams, fluoroquinolones, tetracyclines, phenicols, and glycopeptides. In this work, we focused on identifying new resistance mechanisms for aminoglycosides. Of the potential aminoglycoside antibiotic resistance genes annotated from the pooled genomic sequences of 576 bacteria, ten [aph(3’)-Ia-, ant(9)-Ia-, aadA5-, aph(6)-Ic-, aph(6)-Ic-, aac(3)-IIIb-, aac(6’)-If-, aac(6’)-Iaa-, aph(6)-Id-, and aac(2’)-IIb-like genes sharing < 80% amino acid sequence identity with the functionally characterized aminoglycoside resistance genes] were cloned, and their resistance functions were determined by MIC tests of several aminoglycoside antimicrobials. Finally, we found that one of these genes, an aac(6’)-If homolog [eventually designated aac(6’)-Iaq in this work] carried by the isolate DW0551 isolated from a goose, conferred resistance to several aminoglycosides.
To analyze the structure of the novel resistance gene-related sequence, the whole genome of DW0551, which consists of two chromosomes and one plasmid (designated pDW0551), was sequenced. The larger chromosome (chromosome_1) is approximately 2.67 Mb in size, encoding 2,787 open reading frames (ORFs) with an average length of 847 bp, and the other (chromosome_2) is approximately 1.97 Mb in length, encoding 2,034 ORFs with an average length of 871 bp (Figure 1). The plasmid is 221.64 kb in size and encodes 250 ORFs (Table 3). Species identification analysis of DW0551 revealed that it shared the highest genome-wide ANI (97.83%) with the type strain Brucella intermedia 34576_H01 (GenBank assembly accession: GCA_900454225.1) in the NCBI nucleotide database, and 16S rRNA gene homology analysis revealed that the 16S rRNA gene of DW0551 shared the highest similarity (99.66%) with that of Brucella intermedia O. intermedium_CIP_105838 (GCA_012103055.1). According to the criteria for classifying a bacterium as a certain species (a threshold of ≥ 95% ANI was set to classify a bacterium as a certain species) (Richter and Rosselló-Móra, 2009), the isolate DW0551 belonged to the species Brucella intermedia and was thus designated Brucella intermedia DW0551.
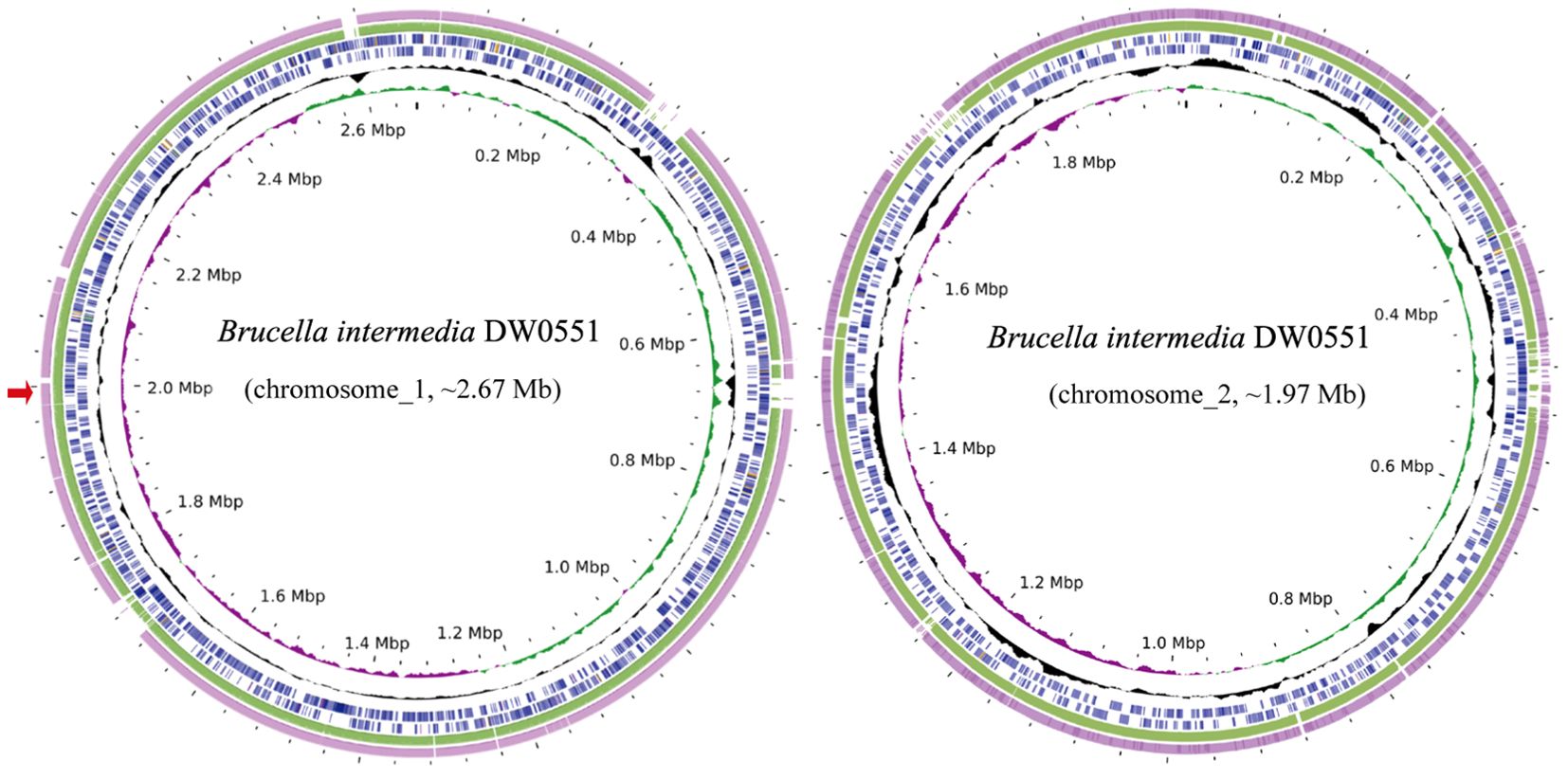
Figure 1. Genome maps of B intermedia DW0551 and its closest relatives. The circles, from inside to outside, represent the GC skew, GC content, and genes encoded in the forward and reverse strands of chromosome_1 (chromosome_2) of B intermedia DW0551, B intermedia ZJ499 chromosome 1 (chromosome 2) (CP061039.1), and B intermedia SG. G2 chromosome 1 (chromosome 2) (CP106662.1). The red arrow on the left represents the location of the novel resistance gene aac(6’)-Iaq.
A total of 59 Brucella intermedia genomes were present in the NCBI genome database, the genome sizes of which ranged from 3.94 Mb (GCA_001637305.1) to 5.39 Mb (GCA_028621395.1). Only four strains had complete genomes, namely, B. intermedia ZJ499 (GCA_014495725.1), B. intermedia SG. G2 (GCA_025490555.1), B. intermedia ZL (GCA_029834515.1) and B. intermedia TSBOI (GCA_029855085.1). The remaining genomes were incomplete draft genomes. The four complete genomes each contained two chromosomes, and only B. intermedia SG. G2 had a plasmid, which was 176.21 kb in length, approximately 45 kb smaller than the plasmid pDW0551 in this study. When searching for similar plasmid sequences in the NCBI nucleotide database, no sequence that shared an identity of more than 10% with pDW0551 was found.
The results of antibiotic susceptibility testing revealed that DW0551 had high MIC levels of ≥ 16 µg/mL to 81.40% (35/43) of the antimicrobial agents tested. The MICs for aminoglycosides were especially high. Among the 10 aminoglycosides tested, except for a relatively low MIC value of 16 µg/mL for neomycin, DW0551 exhibited high MICs for the other 9 antimicrobial agents, with MIC ≥ 512 µg/mL for sisomicin and ≥ 64 µg/mL for netilmicin (Table 2). Compared to the recombinant carrying aac(6’)-Iaq DH5α (pMD19-T-aac(6’)-Iaq), the wild strain DW0551 showed higher MIC levels to aminoglycosides tested. This variation may be due to the presence of other resistance mechanisms, such as efflux pumps, modifying enzymes, target bypass, different intrinsic resistance mechanisms between different bacterial species and so on.
Considering the resistance genotype of the genome, a total of 106 drug resistance-related genes that shared amino acid identities ≥ 30% with functionally characterized resistance genes were predicted (Supplementary Table S2). Among them, 8 were putative aminoglycoside resistance genes encoding aminoglycoside-modifying enzymes, of which both aac(6’)-Il (Bunny et al., 1995) and ant(2’’)-Ia (Cox et al., 2015) shared amino acid identities of 100% with the functionally characterized resistance genes, while the remaining six (including the novel resistance gene aac(6’)-Iaq of this work) shared amino acid identities ranging from 44.1% to 73.2% with the functionally characterized genes.
To determine the drug resistance function of the aac(6’)-Iaq gene, the ORF of the aac(6’)-Iaq gene with its promoter region was cloned (the aac(6’)-Iaq gene and its flanking regions, including the proposed -10 and -35 regions of the promoter, are shown in Supplementary Figure S1), and the recombinant strain DH5α(pMD19-T-aac(6’)-Iaq) exhibited resistance to numerous aminoglycosides, including ribostamycin, netilmicin, sisomicin, tobramycin, gentamicin, kanamycin, and amikacin, with 128-, 64-, 64-, 32-, 8-, 8-, and 8-fold higher MIC levels, respectively, than that of the control strain DH5α(pMD19-T) (Table 2). However, the MICs of spectinomycin and streptomycin did not vary.
The aac(6’)-Iaq gene encoded in the chromosome 1 (Figure 1) is 447 bp in length and encodes a protein of 148 amino acids with a theoretical pI value of 4.64. To investigate the possible induction of aac(6’)-Iaq expression by antibiotics, we performed RT-qPCR experiments to compare the relative expression levels of the genes in the presence and absence of antibiotics. After induction by ribostamycin for 8 h, the expression of the aac(6’)-Iaq gene in the ribostamycin-treated group increased approximately 3-fold in comparison to that in the control group (P < 0.05) (Figure 2). When analyzing the relationship of aac(6’)-Iaq with functionally characterized proteins, a total of 4 functionally characterized resistance genes (all aac(6’)-I genes) with aa identities greater than 50% were found in the public antibiotic resistance gene database CARD. These genes were aac(6’)-If (Ploy et al., 1994), aac(6’)-Iaa (Salipante and Hall, 2003), aac(6’)-Iy (Magnet et al., 1999) and aac(6’)-Ic (Shaw et al., 1992), and the proteins they encoded shared amino acid identities of 52.63%, 52.05%, 51.37% and 50.74%, respectively, with that encoded by aac(6’)-Iaq. Evolutionary analysis of all the members of the functionally characterized AAC(6’) family revealed that they were roughly clustered into 4 clusters (C1-C4) (Figure 3; Supplementary Table S3). The AAC(6’) genes that shared higher identities with aac(6’)-Iaq were clustered together.
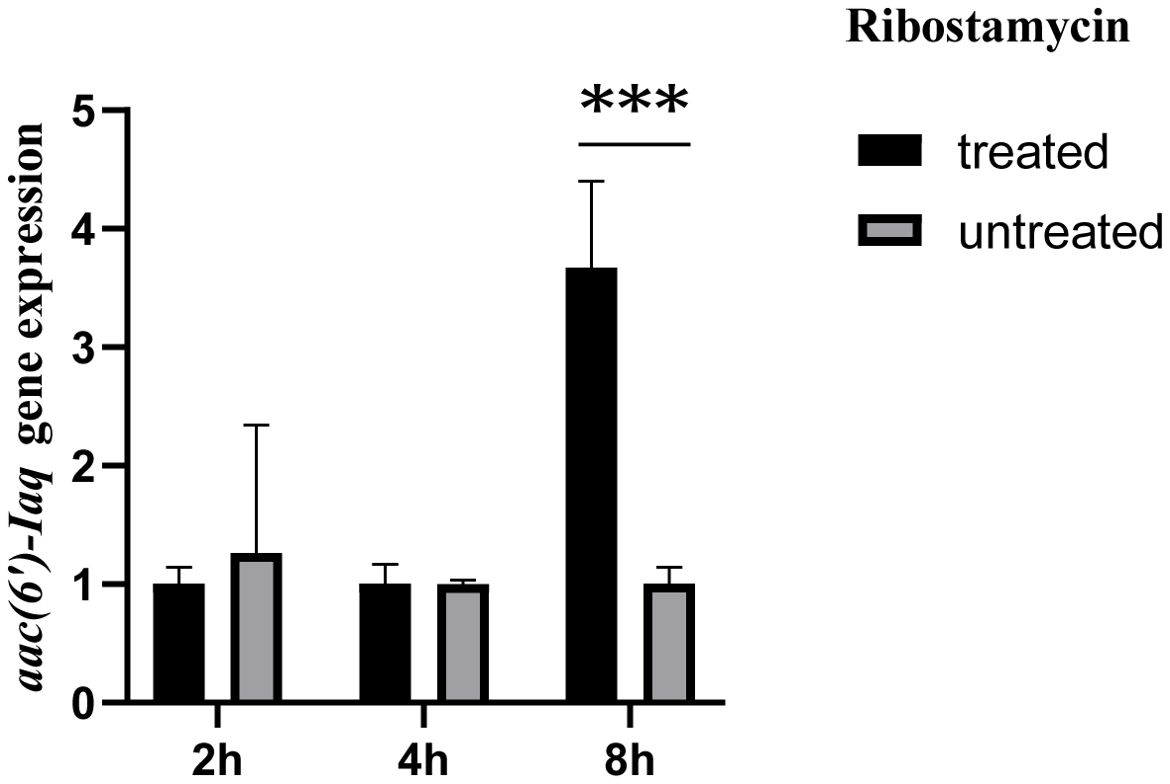
Figure 2. Comparison of the expression levels of the aac(6’)-Iaq gene in the recombinant strains treated with or without 1024 µg/mL ribostamycin. The bars indicate the means ± standard errors, and the experiments were conducted in triplicate. A statistically significant difference was observed between the treated and untreated groups at 8 h. “***”: Significant difference, P < 0.05.
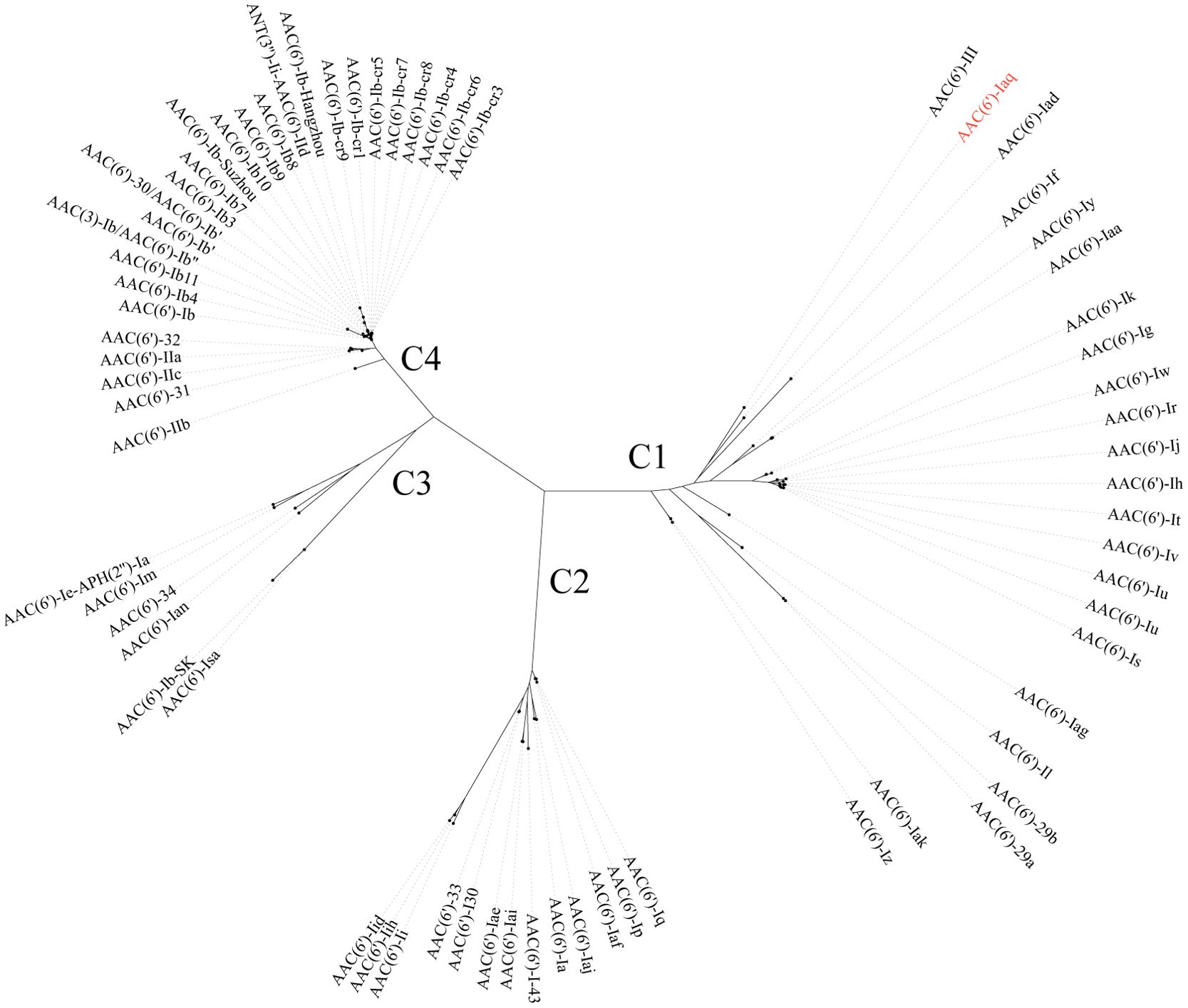
Figure 3. A phylogenetic tree showing the relationship of AAC(6’)-Iaq with other functionally characterized AAC(6’)s; C1-C4 refer to clusters 1-4. AAC(6’)-Iaq is highlighted in red. The accession numbers of these genes are listed in Supplementary Table S3.
To compare the resistance spectra of strains harboring different aac(6’) genes, the resistance phenotypes of 13 aac(6’) genes from different evolutionary clusters were analyzed; four of these genes were most closely related to aac(6’)-Iaq (no resistance profile was available for aac(6’)-If) in cluster 1, and the remaining 9 aac(6’) genes were randomly selected from the other 3 clusters (Supplementary Table S4). The novel aminoglycoside resistance gene aac(6’)-Iaq conferred resistance to all seven aminoglycosides detected; however, for the other 13 aac(6’) genes, among the 4 to 7 antimicrobial agents tested, each showed susceptibility to 1 or 2 antirobial agents. Among the three antimicrobial agents tested, all the genes conferred resistance to tobramycin, and most (12/14) conferred resistance to amikacin; however, only a small portion (4/14) of the genes conferred resistance to gentamicin. All of the genes tested conferred resistance to the antimicrobial molecules tested, except for aac(6’)-Iaf (Kitao et al., 2009) and aac(6’)-Isa (Hamano et al., 2004), both of which showed susceptibility to sisomicin. The aac(6’) genes exhibited different resistance spectra even though they were phylogenetically closely related.
When analyzing the essential functional residues of the protein, multiple sequence alignment of AAC(6’)-Iaq with the functionally characterized proteins of the AAC(6’)-I proteins was performed (Marchler-Bauer et al., 2017). The coenzyme A chemical binding pocket of the conserved protein domain family NAT_SF (PSSM-Id: 173926), consisting of 5 residues (I84, Y85, V86, A96 and R97) (Rojas et al., 1999), was present in AAC(6’)-Iaq (I84-Y85-V86-X-X-X-X-X-X-X-X-X-A96-R97) (Figure 4). The conserved protein domain family NAT_SF belongs to the cl17182 superfamily (domain architecture ID 10456837, a family of proteins containing various enzymes with catalytic acyl-transfer substrates) (Wybenga-Groot et al., 1999).
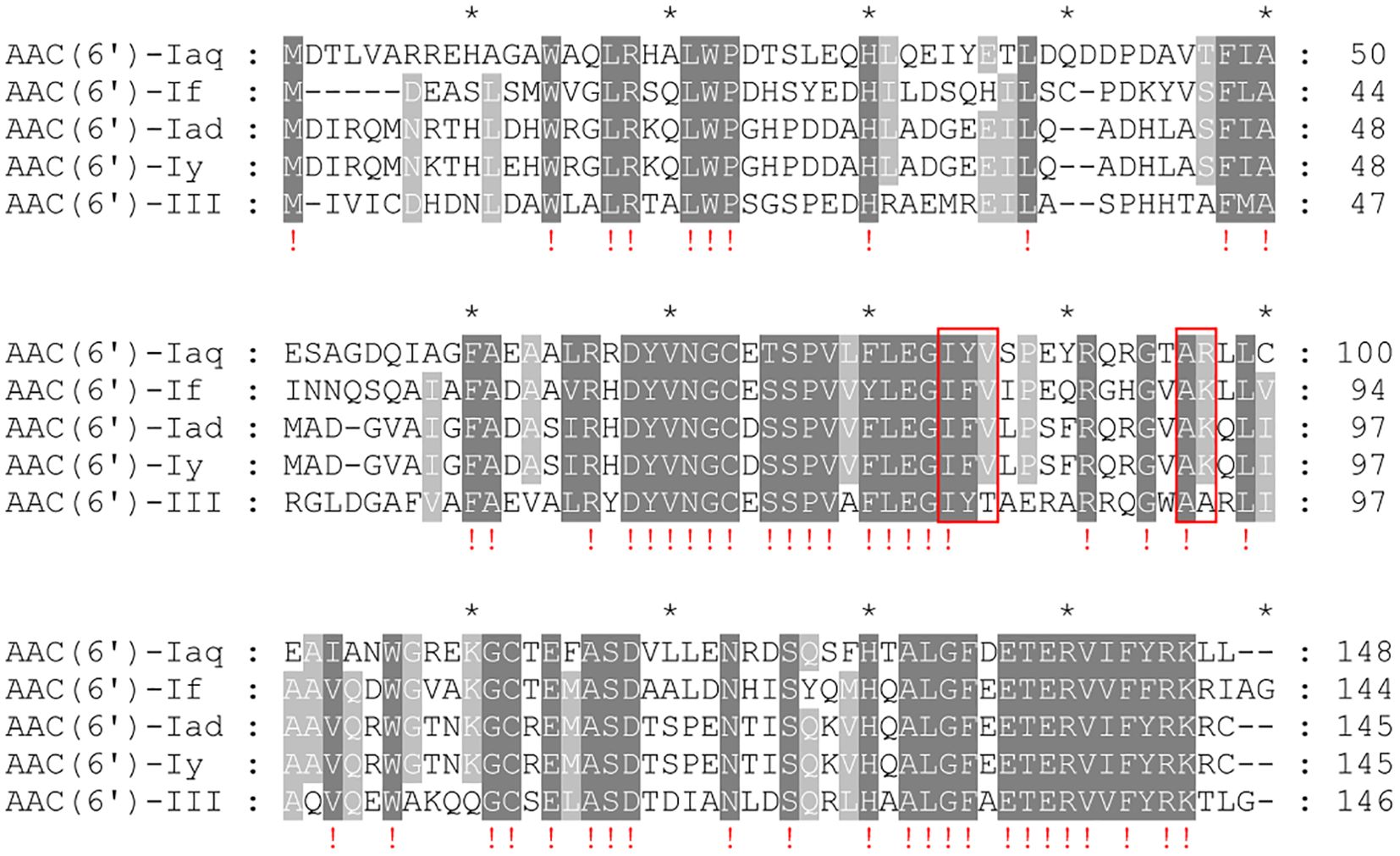
Figure 4. Multiple sequence alignment of AAC(6’)-Iaq with other close relatives of the C1 cluster. The numbers on the right represent the corresponding amino acid sequence length. Exclamation marks indicate fully conserved residues; gaps are represented using hyphens. The red frames represent the residues of the conserved protein domain family NAT_SF. “*” is a location marker. Dark gray indicates conserved sites, and light gray indicates relatively conserved sites.
The results of the acetyltransferase activity and enzyme kinetic parameter analyses of AAC(6’)-Iaq demonstrated that, consistent with the MIC results, AAC(6’)-Iaq was able to acetylate ribostamycin, netilmicin, sisomicin, kanamycin and amikacin. No acetyltransferase activity could be detected for streptomycin. According to the Michaelis-Menten constant, this enzyme had the highest affinities and catalytic efficiencies for netilmicin [Km of 3.83 ± 0.34 µM and kcat/Km of (9.08 ± 1.83) × 104 M-1/s-1] and sisomicin [Km of 4.33 ± 1.32 µM and kcat/Km of (6.13 ± 2.26) × 104 M-1/s-1] (Table 4).
In addition to streptomycin, which has a hydroxyl group at the 6’-position, AAC(6’)-Iaq exhibited acetyltransferase activity toward five other aminoglycosides harboring an amino group at the 6’ position, and this regiospecific acetyltransferase transfer is consistent with previous findings (Magnet et al., 2001). Among these aminoglycoside substrates, amikacin and kanamycin were poor substrates, exhibiting lower affinities (with Km values of 14.08 ± 0.76 µM and 13.34 ± 9.07 µM, respectively); this difference is thought to be related to the fact that the substituent at position 1 of ring I of amikacin and kanamycin is a hydroxyl group rather than an amino group (Tada et al., 2016).
AAC(6’)-Iaq exhibits a higher affinity and a higher catalytic efficiency for netilmicin than several other AAC(6’) enzymes reported previously. The Km values for netilmicin of AAC(6’)-Ial, AAC(6)-Iy, and AAC(6’)-Ic were 23 ± 6, 8 ± 1 and 20 ± 5 µM, respectively (Magnet et al., 2001; Tada et al., 2016), while the kcat/Km values for netilmicin of AAC(6’)-Ial, AAC(6’)-Iap and AAC(6’)-III [AAC(6’)-Ic] were 4.1×104 M-1/s-1, 2.2 × 104 M-1/s-1, and 2.9×104 M-1/s-1, respectively (Kim et al., 2007; Tada et al., 2016); however, AAC(6’)-Ib (Vetting et al., 2008) had a much greater catalytic efficiency for netilmicin than the other AAC(6’)-I proteins, showing a kcat/Km value of (2.0 ± 0.5) × 106 M-1/s-1.
To analyze the genetic context of the aac(6’)-Iaq-encoding region, a sequence approximately 20 kb in length with aac(6’)-Iaq at the center was used as a query to search the nonredundant nucleotide database of the NCBI. A total of 18 sequences with > 80% nucleotide identity were retrieved, and all of them were from the genus Brucella/Ochrobactrum. Of these 18 sequences, the four sequences with the highest identities (> 96.0%) were from Brucella intermedia (CP123056.1, identity 98.29%, CP061039.1, identity 97.20%, CP122438.1, identity 97.04%, and CP106662.1, identity 96.98%), and all of them contained an aac(6’)-Iaq-like gene (an aac(6’)-Iaq-like gene is a gene other than aac(6’)-Iaq according to the public database and the protein it encoded shares aa identity of ≥ 80% with AAC(6’)-Iaq). The remaining 14 sequences showed identities ranging from 82.24% to 85.64% and were all free of an aac(6’)-Iaq-like gene. Comparative structural analysis of 10 sequences, including four sequences that shared identities higher than 96.0% and five sequences that shared lower identities with the sequence of interest in this work, was performed, and the results revealed that they had similar genetic contexts in terms of gene content and gene order. Except for the aac(6’)-Iaq-like gene, at most one or two predicted ORFs (such as vapC, mazE, fbcH, rnasel, yijF, rpmH or a hypothetical protein-encoding gene) differed between them (Figure 5). No MGE was found within the 20 kb sequences. However, the mechanism underlying its appearance in Brucella intermedia chromosomes remains to be further studied.
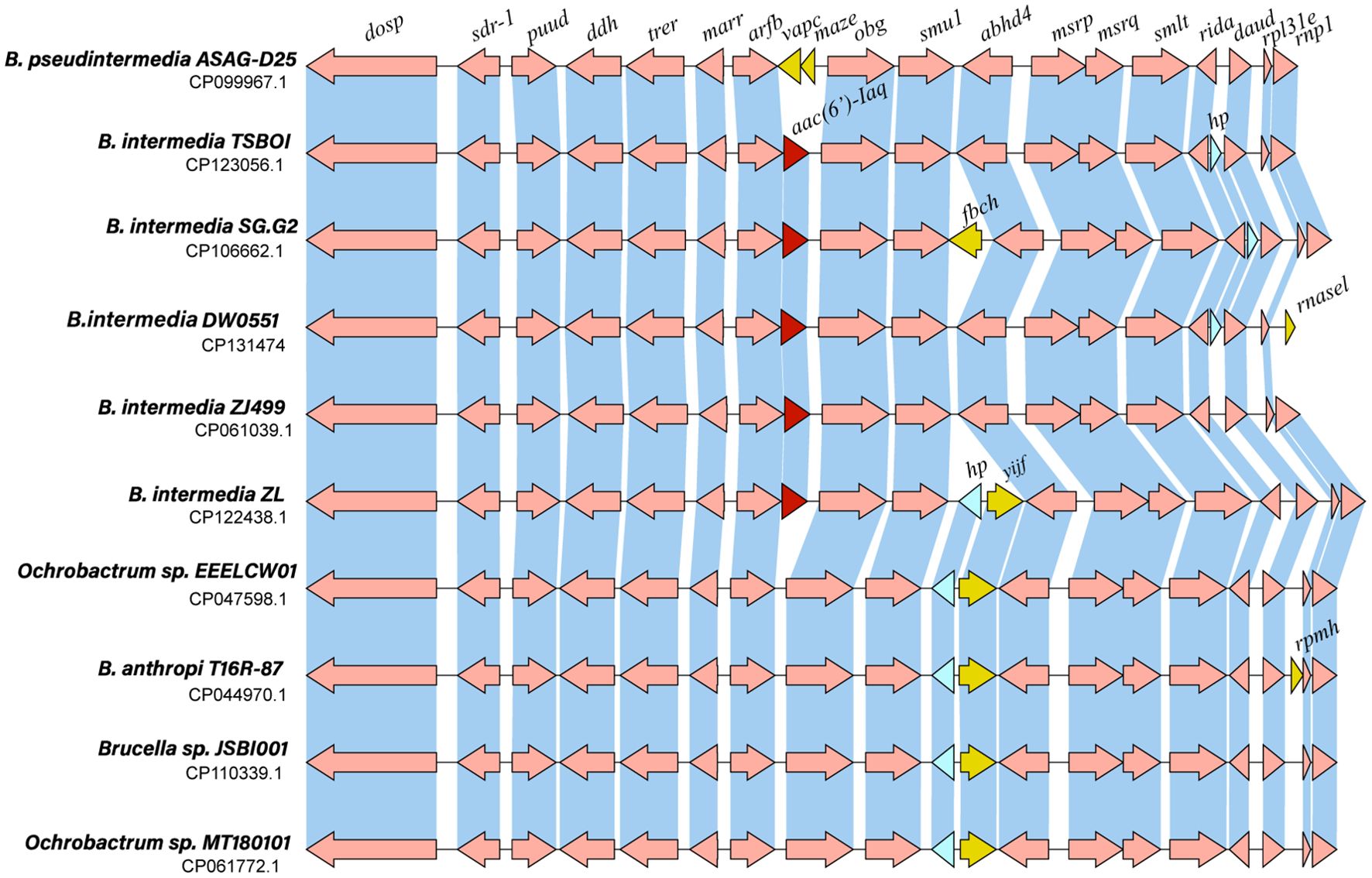
Figure 5. Genetic context of aac(6’)-Iaq and other related sequences. Regions with ≥ 80% amino acid identity are in blue. hp: hypothetical protein. The aac(6’)-Iaq and aac(6’)-Iaq-like genes (similarity > 95.0%) are shown in red. The genes specific to each sequence are shown in yellow, and the hypothetical protein-encoding genes are shown in blue.
To analyze the distribution of the aac(6’)-Iaq(-like) genes, the aa sequence of AAC(6’)-Iaq was used as a query to search the NCBI nonredundant nucleotide database, and approximately one hundred sequences with amino acid similarities greater than 50.0% were retrieved. Of these sequences, only four (mentioned above) had similarities > 95.0% and they were from the same species as AAC(6’)-Iaq, Brucella intermedia. As mentioned above, there are four complete Brucella intermedia genomes in the NCBI genome database, and the sequences carrying the AAC(6’)-Iaq(-like) proteins are from them, respectively: CP106662.1 (identity 97.3% chromosome of SG.G2), isolated from houseplant; CP122438.1 (identity 96.62%, chromosome of ZL), isolated from environment; CP061039.1 (identity 96.62%, chromosome of ZJ499), isolated from human sputum; CP123056.1 (identity 95.95%, chromosome of TSBOI), isolated from soil, and all the others had similarities < 60% with AAC(6’)-Iaq; these similar sequences were from bacteria of different families, such as one (CP120373.1, identity 54.60%) from Ensifer garamanticus LMG 24692, or from different classes, such as one (CP116005.1, identity 59.59%) from Sphingosinicella microcystinivorans DMF-3. The genetic context of these sequences was entirely different from that of the sequence in this work (Brucella intermedia DW0551). To analyze the evolutionary relationships between these genes, additional aac(6’)-Iaq-like genes with higher identities would be included.
In this study, a novel aminoglycoside resistance gene, designated aac(6’)-Iaq, encoded on the chromosome of a Brucella intermedia isolate from goose feces, was identified. AAC(6’)-Iaq shares < 60% amino acid identity with all functionally characterized aminoglycoside resistance gene encoded proteins and shows resistance to several aminoglycosides including ribostamycin, netilmicin, sisomicin, tobramycin, gentamicin, kanamycin, and amikacin. AAC(6’)-Iaq exhibited acetylation activity toward the aminoglycoside substrates analyzed. The discovery of novel resistance mechanisms in animal bacteria might also aid in the treatment of microbial infections in animals and humans.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
NL: Formal Analysis, Investigation, Methodology, Software, Writing – original draft. WX: Data curation, Formal Analysis, Investigation, Writing – original draft. DH: Conceptualization, Project administration, Supervision, Writing – original draft. CL: Formal Analysis, Investigation, Resources, Writing – original draft. JL: Conceptualization, Data curation, Validation, Writing – original draft. MZ: Data curation, Methodology, Resources, Writing – review & editing. QB: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. WP: Funding acquisition, Resources, Writing – review & editing.
This study used strains isolated from animals in animal farms in Wenzhou, China. The owners of the farms were informed in writing of the study and provided approval for the sampling of animals. The studies involving human participants and animals were reviewed and approved by the Animal Welfare and Ethics Committee of Wenzhou Medical University, Zhejiang Province, China (protocol number: wydw2021-0323).
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by Zhejiang Provincial Natural Science Foundation of China (QN25H190009), the Science & Technology Project of Jinhua City, China (2024-4-030, 2022-2-013), the Science & Technology Project of Taizhou City, China (21ywb126), and the Science & Technology Project of Wenzhou City, China (N20210001).
The authors would like to acknowledge the teachers and scientists of the Science and Technology Platforms of Wenzhou Medical University who helped with the analysis of the enzyme kinetic parameters.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1551240/full#supplementary-material
Supplementary Figure 1 | The aac(6’)-Iaq gene and its flanking regions. The underlined regions are the -10 and -35 regions of the proposed promoter. The aac(6’)-Iaq gene is shaded gray.
Supplementary Table 1 | Primers used in this study.
Supplementary Table 2 | 106 resistance genes predicted from the DW0551 genome.
Supplementary Table 3 | Sequences used to reconstruct the phylogenetic tree.
Supplementary Table 4 | Resistance phenotypes conferred by aac(6’) genes belonging to different phylogenetic clusters.
Abdul-Aziz, M. H., Alffenaar, J. C., Bassetti, M., Bracht, H., Dimopoulos, G., Marriott, D., et al. (2020). Antimicrobial therapeutic drug monitoring in critically ill adult patients: a Position Paper(). Intensive Care Med. 46, 1127–1153. doi: 10.1007/s00134-020-06050-1
Alcock, B. P., Huynh, W., Chalil, R., Smith, K. W., Raphenya, A. R., Wlodarski, M. A., et al. (2023). CARD 2023: expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 51, D690–d699. doi: 10.1093/nar/gkac920
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Apisarnthanarak, A., Kiratisin, P., Mundy, L. M. (2005). Evaluation of Ochrobactrum intermedium bacteremia in a patient with bladder cancer. Diagn. Microbiol. Infect. Dis. 53, 153–155. doi: 10.1016/j.diagmicrobio.2005.05.014
Bharucha, T., Sharma, D., Sharma, H., Kandil, H., Collier, S. (2017). Ochromobactrum intermedium: an emerging opportunistic pathogen-case of recurrent bacteraemia associated with infective endocarditis in a haemodialysis patient. New Microbes New Infections 15, 14–15. doi: 10.1016/j.nmni.2016.09.016
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrobial Chemotherapy 75, 3491–3500. doi: 10.1093/jac/dkaa345
Buchfink, B., Reuter, K., Drost, H.-G. (2021). Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 18, 366–368. doi: 10.1038/s41592-021-01101-x
Bunny, K. L., Hall, R. M., Stokes, H. W. (1995). New mobile gene cassettes containing an aminoglycoside resistance gene, aacA7, and a chloramphenicol resistance gene, catB3, in an integron in pBWH301. Antimicrobial Agents Chemotherapy 39, 686–693. doi: 10.1128/AAC.39.3.686
Casanovas, A., Pinto-Llorente, R., Carrascal, M., Abian, J. (2017). Large-scale filter-aided sample preparation method for the analysis of the ubiquitinome. Analytical Chem. 89, 3840–3846. doi: 10.1021/acs.analchem.6b04804
Cox, G., Stogios, P. J., Savchenko, A., Wright, G. D. (2015). Structural and molecular basis for resistance to aminoglycoside antibiotics by the adenylyltransferase ANT(2″)-Ia. MBio 6, e02180–14. doi: 10.1128/mBio.02180-14
Crump, J. A., Sjölund-Karlsson, M., Gordon, M. A., Parry, C. M. (2015). Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive salmonella infections. Clin. Microbiol. Rev. 28, 901–937. doi: 10.1128/CMR.00002-15
EUCAST (2022). Breakpoint tables for interpretation of MICs and zone diameters. Version 12.0. Available online at: http://www.eucast.org (Accessed October 25, 2022).
Gilchrist, C. L. M., Chooi, Y.-H. (2021). clinker & clustermap.js: automatic generation of gene cluster comparison figures. Bioinf. (Oxford England) 37, 2473–2475. doi: 10.1093/bioinformatics/btab007
Gurevich, A., Saveliev, V., Vyahhi, N., Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinf. (Oxford England) 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Hamano, Y., Hoshino, Y., Nakamori, S., Takagi, H. (2004). Overexpression and characterization of an aminoglycoside 6’-N-acetyltransferase with broad specificity from an epsilon-poly-L-lysine producer, Streptomyces albulus IFO14147. J. Biochem. 136, 517–524. doi: 10.1093/jb/mvh146
Hirai, J., Yamagishi, Y., Sakanashi, D., Koizumi, Y., Suematsu, H., Mikamo, H. (2016). A case of bacteremia caused by ochrobacterium intermedium]. Kansenshogaku zasshi. J. Japanese Assoc. For Infect. Dis. 90, 129–133. doi: 10.11150/kansenshogakuzasshi.90.129
Holmes, B., Popoff, M., Kiredjian, M., Kersters, K. (1988). Ochrobactrum anthropi gen. nov., sp. nov. from Human Clinical Specimens and Previously Known as Group Vd. Int. J. Syst. Evol. Microbiol. 38, 406–416. doi: 10.1099/00207713-38-4-406
Huang, N., Nie, F., Ni, P., Gao, X., Luo, F., Wang, J. (2022). BlockPolish: accurate polishing of long-read assembly via block divide-and-conquer. Briefings In Bioinf. 23, bbab406. doi: 10.1093/bib/bbab405
Jacobs, D. J., Grube, T. J., Flynn, H. W., Greven, C. M., Pathengay, A., Miller, D., et al. (2013). Intravitreal moxifloxacin in the management of Ochrobactrum intermedium endophthalmitis due to metallic intraocular foreign body. Clin. Ophthalmol. (Auckland N.Z.) 7, 1727–1730. doi: 10.2147/OPTH.S44212
Kassab, I., Sarsam, N., Affas, S., Ayas, M., Baang, J. H. (2021). A case of ochrobactrum intermedium bacteremia secondary to cholangitis with a literature review. Cureus 13, e14648. doi: 10.7759/cureus.14648
Katoh, K., Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kim, C., Villegas-Estrada, A., Hesek, D., Mobashery, S. (2007). Mechanistic characterization of the bifunctional aminoglycoside-modifying enzyme AAC(3)-Ib/AAC(6’)-Ib’ from Pseudomonas aeruginosa. Biochemistry 46, 5270–5282. doi: 10.1021/bi700111z
Kitao, T., Miyoshi-Akiyama, T., Kirikae, T. (2009). AAC(6’)-Iaf, a novel aminoglycoside 6’-N-acetyltransferase from multidrug-resistant Pseudomonas aeruginosa clinical isolates. Antimicrobial Agents Chemotherapy 53, 2327–2334. doi: 10.1128/AAC.01360-08
Konstantinidis, K. T., Tiedje, J. M. (2005). Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. United States America 102, 2567–2572.
Lewis, J. S., II (2022). M100 performance standards for antimicrobial susceptibility testing (West Valley Road, Suite 2500 Wayne, PA 19087, USA: Clinical and Laboratory Standards Institute), 402. P., FIDSA.
Lorah, J., Womack, A. (2019). Value of sample size for computation of the Bayesian information criterion (BIC) in multilevel modeling. Behav. Res. Methods 51, 440–450. doi: 10.3758/s13428-018-1188-3
Lu, W., Li, K., Huang, J., Sun, Z., Li, A., Liu, H., et al. (2021). Identification and characteristics of a novel aminoglycoside phosphotransferase, APH(3’)-IId, from an MDR clinical isolate of Brucella intermedia. J. Antimicrobial Chemotherapy 76, 2787–2794. doi: 10.1093/jac/dkab272
Magnet, S., Courvalin, P., Lambert, T. (1999). Activation of the cryptic aac(6’)-Iy aminoglycoside resistance gene of Salmonella by a chromosomal deletion generating a transcriptional fusion. J. Bacteriol 181, 6650–6655. doi: 10.1128/jb.181.21.6650-6655.1999
Magnet, S., Lambert, T., Courvalin, P., Blanchard, J. S. (2001). Kinetic and mutagenic characterization of the chromosomally encoded Salmonella enterica AAC(6’)-Iy aminoglycoside N-acetyltransferase. Biochemistry 40, 3700–3709. doi: 10.1021/bi002736e
Marchler-Bauer, A., Bo, Y., Han, L., He, J., Lanczycki, C. J., Lu, S., et al. (2017). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200–D203. doi: 10.1093/nar/gkw1129
Marchler-Bauer, A., Bryant, S. H. (2004). CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32, W327–W331. doi: 10.1093/nar/gkh454
Ploy, M. C., Giamarellou, H., Bourlioux, P., Courvalin, P., Lambert, T. (1994). Detection of aac(6’)-I genes in amikacin-resistant Acinetobacter spp. by PCR. Antimicrobial Agents Chemotherapy 38, 2925–2928. doi: 10.1128/AAC.38.12.2925
Ramirez, M. S., Tolmasky, M. E. (2010). Aminoglycoside modifying enzymes. Drug Resist. Update 13, 151–171. doi: 10.1016/j.drup.2010.08.003
Richter, M., Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. United States America 106, 19126–19131. doi: 10.1073/pnas.0906412106
Rocha, D. J. P. G., Castro, T. L. P., Aguiar, E. R. G. R., Pacheco, L. G. C. (2020). Gene expression analysis in bacteria by RT-qPCR. Methods In Mol. Biol. (Clifton N.J.) 2065, 119–137. doi: 10.1007/978-1-4939-9833-3_10
Rojas, J. R., Trievel, R. C., Zhou, J., Mo, Y., Li, X., Berger, S. L., et al. (1999). Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature 401, 93–98. doi: 10.1038/43487
Ryan, M. P., Pembroke, J. T. (2020). The genus ochrobactrum as major opportunistic pathogens. Microorganisms 8, 1797. doi: 10.3390/microorganisms8111797
Salipante, S. J., Hall, B. G. (2003). Determining the limits of the evolutionary potential of an antibiotic resistance gene. Mol. Biol. Evol. 20, 653–659. doi: 10.1093/molbev/msg074
Schmittgen, T. D., Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinf. (Oxford England) 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Sekiguchi, J.-I., Asagi, T., Miyoshi-Akiyama, T., Kasai, A., Mizuguchi, Y., Araake, M., et al. (2007). Outbreaks of multidrug-resistant Pseudomonas aeruginosa in community hospitals in Japan. J. Clin. Microbiol. 45, 979–989. doi: 10.1128/JCM.01772-06
Shaw, K. J., Rather, P. N., Sabatelli, F. J., Mann, P., Munayyer, H., Mierzwa, R., et al. (1992). Characterization of the chromosomal aac(6’)-Ic gene from Serratia marcescens. Antimicrob. Agents Chemother. 36, 1447–1455. doi: 10.1128/aac.36.7.1447
Sheng, X., Lu, W., Li, A., Lu, J., Song, C., Xu, J., et al. (2023). ANT(9)-ic, a novel chromosomally encoded aminoglycoside nucleotidyltransferase from brucella intermedia. Microbiol. Spectr. 11, e0062023. doi: 10.1128/spectrum.00620-23
Souvorov, A., Agarwala, R., Lipman, D. J. (2018). SKESA: strategic k-mer extension for scrupulous assemblies. Genome Biol. 19, 153. doi: 10.1186/s13059-018-1540-z
Tada, T., Miyoshi-Akiyama, T., Shimada, K., Dahal, R. K., Mishra, S. K., Ohara, H., et al. (2016). A novel 6’-N-aminoglycoside acetyltransferase, AAC(6’)-ial, from a clinical isolate of serratia marcescens. Microbial Drug Resistance (Larchmont N.Y.) 22, 103–108. doi: 10.1089/mdr.2015.0126
Velasco, J., Romero, C., López-Goñi, I., Leiva, J., Díaz, R., Moriyón, I. (1998). Evaluation of the relatedness of Brucella spp. and Ochrobactrum anthropi and description of Ochrobactrum intermedium sp. nov., a new species with a closer relationship to Brucella spp. Int. J. Systematic Bacteriology 48 Pt 3, 759–768. doi: 10.1099/00207713-48-3-759
Vetting, M. W., Park, C. H., Hegde, S. S., Jacoby, G. A., Hooper, D. C., Blanchard, J. S. (2008). Mechanistic and structural analysis of aminoglycoside N-acetyltransferase AAC(6’)-Ib and its bifunctional, fluoroquinolone-active AAC(6’)-Ib-cr variant. Biochemistry 47, 9825–9835. doi: 10.1021/bi800664x
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PloS One 9, e112963. doi: 10.1371/journal.pone.0112963
Wick, R. R., Judd, L. M., Gorrie, C. L., Holt, K. E. (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PloS Comput. Biol. 13, e1005595. doi: 10.1371/journal.pcbi.1005595
Wybenga-Groot, L. E., Draker, K., Wright, G. D., Berghuis, A. M. (1999). Crystal structure of an aminoglycoside 6’-N-acetyltransferase: defining the GCN5-related N-acetyltransferase superfamily fold. Structure (London England: 1993) 7, 497–507. doi: 10.1016/S0969-2126(99)80066-5
Zhang, G., Zhang, L., Sha, Y., Chen, Q., Lin, N., Zhao, J., et al. (2023). Identification and characterization of a novel 6’-N-aminoglycoside acetyltransferase AAC(6’)-Va from a clinical isolate of Aeromonas hydrophila. Front. In Microbiol. 14. doi: 10.3389/fmicb.2023.1229593
Keywords: Brucella intermedia, resistance mechanism, aminoglycoside acetyltransferase, aac(6’)-Iaq, kinetic parameter
Citation: Lin N, Xu W, Huang D, Liu C, Lu J, Zhu M, Bao Q and Pan W (2025) aac(6’)-Iaq, a novel aminoglycoside acetyltransferase gene identified from an animal isolate Brucella intermedia DW0551. Front. Cell. Infect. Microbiol. 15:1551240. doi: 10.3389/fcimb.2025.1551240
Received: 25 December 2024; Accepted: 18 February 2025;
Published: 11 March 2025.
Edited by:
Massimiliano Lucidi, Roma Tre University, ItalyReviewed by:
Peibo Yuan, Southern Medical University, ChinaCopyright © 2025 Lin, Xu, Huang, Liu, Lu, Zhu, Bao and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mei Zhu, emh1bWVpX2RAMTYzLmNvbQ==; Qiyu Bao, YmFvcXlAZ2Vub21pY3MuY24=; Wei Pan, MjU2NTg1MDdAcXEuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.