
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 13 March 2025
Sec. Antibiotic Resistance and New Antimicrobial drugs
Volume 15 - 2025 | https://doi.org/10.3389/fcimb.2025.1544757
 Jing Wang1,2†
Jing Wang1,2† Zi-Han Dong1†
Zi-Han Dong1† Xian-Yuan Zhou3†
Xian-Yuan Zhou3† Qin-Chun Ma1
Qin-Chun Ma1 Zhen-Yu Wang1
Zhen-Yu Wang1 Dachuan Lin4
Dachuan Lin4 Ying-Feng Huang3
Ying-Feng Huang3 Chi Zhang3
Chi Zhang3 Xinan Jiao1
Xinan Jiao1 Deng Li3*
Deng Li3* Qiuchun Li1*
Qiuchun Li1*Salmonellosis, caused by non-typhoidal Salmonella, is a common foodborne gastrointestinal infection. Third-generation cephalosporins are recommended as the first-line treatment for Salmonella infections. Our study aimed to investigate the molecular epidemiology, antimicrobial resistance, and the transmission of extended-spectrum β-lactamases (ESBL) genes in 96 clinical Salmonella isolates collected between 2020 and 2022 at a tertiary hospital in Shenzhen, China. We performed antimicrobial susceptibility testing and whole-genome sequencing to identify serotypes, multilocus sequence typing, antimicrobial resistance genes in these isolates, and the genetic structures of the blaCTX-M/blaCMY genes. Seventeen Salmonella serotypes were identified, with S. 4,[5],12:i:- (37.5%) being the most common, followed by S. Enteritidis (15.63%), S. Typhimurium (14.58%), S. London (7.29%), and S. Rissen (5.21%). MLST analysis revealed 19 distinct sequence types (STs), with ST34 being the most prevalent (36.46%), followed by ST11 (15.63%) and ST19 (13.54%). Antimicrobial resistance testing showed those isolates had high levels of resistance to ampicillin (72.92%) and tetracycline (71.88%), with 70.83% of isolates as multidrug-resistant (MDR). Three blaCTX-M genes (blaCTX-M-14, blaCTX-M-55, and blaCTX-M-65) and blaCMY-2 were identified among 18 cefotaxime-resistant strains, of which one and 12 isolates successfully transferred blaCMY or blaCTX-M to E. coli C600 via conjugation, respectively. The blaCTX-M/blaCMY-2-carrying contigs in nine Salmonella isolates ranged from 2,156 to 164,862 bp, were located either on the chromosome (n=1) or plasmids (IncI1, IncK1, IncA/C) (n=9), and the blaCTX-M/blaCMY-2 genes were associated with ISEcp1. Our study demonstrates the diversity of MDR Salmonella serotypes in clinical isolates, and highlights the role of plasmids and mobile genetic elements in the horizontal transfer of blaCTX-M/blaCMY, emphasizing the need for continuous surveillance of Salmonella in clinical samples.
Non-typhoidal Salmonella is a leading cause of foodborne illness globally, causing salmonellosis, which typically presents with symptoms such as diarrhea, fever, abdominal cramps, and vomiting (CDC, 2019; EFSA and ECDC, 2023). In severe cases, particularly in immunocompromised individuals, Salmonella infection can lead to life-threatening complications (Ruiz et al., 2004). These infections are primarily caused by the consumption of Salmonella-contaminated food, particularly raw or undercooked poultry, eggs, and beef (EFSA and ECDC, 2023). To date, 2,659 Salmonella serovars have been identified (Monte and Sellera, 2020), with Salmonella enterica serovar Typhimurium (including its monophasic variant) and S. Enteritidis being the most common serotypes in human infections (EFSA and ECDC, 2023).
In recent decades, antimicrobial resistance has become a significant challenge in treating Salmonella infections (CDC, 2019; EFSA and ECDC, 2023). Third-generation cephalosporins are considered first-line antibiotics for treating Salmonella infection, but the isolates acquiring genes to produce extended-spectrum β-lactamases (ESBLs) - enzymes can confer the bacterial resistance to a broad range of β-lactam antibiotics, including penicillins and cephalosporins, which has significantly restricted treatment options (Ruiz et al., 2004; Crump et al., 2015; Bevan et al., 2017). The global prevalence of ESBL-producing Salmonella, particularly strains encoding CTX-M/CMY β-lactamases, continues to rise, presenting an increasing public health concern worldwide (Sun et al., 2022; Wang et al., 2020). Therefore, monitoring these Salmonella strains is crucial for understanding their prevalence and transmission characteristics. This study aims to investigate the molecular epidemiology and antimicrobial resistance characteristics of Salmonella isolates, with a particular focus on the prevalence and transmission of ESBL-producing strains in a tertiary hospital in Shenzhen, China.
From March 2020 to November 2022, 96 Salmonella strains were isolated from non-repetitive samples, including feces (n=93), blood (n=2), and purulent secretion (n=1), obtained from 87 different patients for routine diagnostics at a tertiary hospital in Shenzhen, China. The collected samples (2-3 g for feces; 1-3 mL for other liquid samples) were inoculated onto Salmonella Shigella (SS) agar plates using a standardized streaking technique. The plates were then incubated aerobically at 37°C for 24 hours. Following incubation, a single presumptive Salmonella colony was selected from each plate and subjected to species identification using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Colonies confirmed as Salmonella spp. were subsequently streaked onto xylose lysine deoxycholate (XLD) agar plates for purification.
All Salmonella isolates were tested for susceptibility to colistin using the ISO-standard broth microdilution method, recommended by the joint CLSI-EUCAST Polymyxin Breakpoints Working Group (https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/General_documents/Recommendations_for_MIC_determination_of_colistin_March_2016.pdf). In addition, susceptibility to 12 other antimicrobial agents, including ampicillin, cefotaxime, meropenem, gentamicin, amikacin, streptomycin, tetracycline, chloramphenicol, nalidixic acid, ciprofloxacin, fosfomycin, and sulfamethoxazole/trimethoprim, was assessed using the agar dilution method following the guidelines of the Clinical and Laboratory Standards Institute (CLSI) M07 (CLSI, 2012). Escherichia coli ATCC 25922 was used as the quality control strain. Results were interpreted according to the 32nd edition of the CLSI M100 (CLSI, 2022). The interpretation of streptomycin (>16 mg/L) was based on the epidemiological cut-off value for Salmonella enterica established by the European Committee on Antimicrobial Susceptibility Testing (EUCAST; www.eucast.org).
Genomic DNA from all Salmonella isolates was extracted using the TIANamp Bacteria DNA Kit (Tiangen, Beijing, China) and sequenced on the Illumina NovaSeq platform. The genomic libraries were prepared using the NEB NEXT Ultra DNA Library Prep Kit for Illumina (New England Biolabs, USA), and sequencing was performed to generate 150 bp paired-end reads. For each Salmonella isolates subjected to WGS, a minimum coverage depth of 100× was achieved. Raw reads with less than 90% Q30 bases were trimmed and filtered using the NGSQC Toolkit v2.3.3, and assembled into contigs using SPAdes 3.8.2 (Bankevich et al., 2012). Serotypes were determined using the Salmonella In Silico Typing Resource (SISTR) (Yoshida et al., 2016). Genomic sequences were subjected to multilocus sequence typing (MLST) analysis using MLST 2.0 (https://cge.food.dtu.dk/services/MLST/). Additionally, antimicrobial resistance genes and mutations were identified using ResFinder and PointFinder, respectively (http://genepi.food.dtu.dk/resfinder). Contigs carrying blaCTX-M/blaCMY genes were retrieved from the draft genomes and analyzed using the ISfinder platform (https://www-is.biotoul.fr) to identify insertion sequences, PlasmidFinder 2.1 (https://cge.food.dtu.dk/services/PlasmidFinder/) to detect plasmid replicons, and BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) for sequence homology and annotation.
Conjugation experiments were performed using cefotaxime-resistant Salmonella strains as donors and E. coli C600 (which exhibits high-level streptomycin resistance) as the recipient. Donor and recipient strains were inoculated in 2 mL of LB broth and incubated at 37°C, 180 rpm for 4 hours. The cultures were then mixed in a 1:4 ratio (v/v) and incubated at 37°C for 12 h. Transconjugants were selected on MacConkey agar plates containing cefotaxime (2 mg/mL) and streptomycin (3000 mg/mL) and confirmed by PCR detection of blaCTX-M or blaCMY (Liu et al., 2007). All experiments were conducted in triplicate.
The whole genome sequences of the Salmonella isolates have been deposited in GenBank under the accession number: PRJEB83553.
This study analyzed 96 Salmonella isolates obtained from fecal (96.88%, n=93), blood (2.08%, 2/96), and pus samples (1.04%, 1/96) from 87 patients. Of these, 29 isolates were collected in 2020, 38 in 2021, and 29 in 2022, representing 30.21%, 39.58%, and 30.21%, respectively. As shown in Figure 1, the highest detection rate occurred in July (17.71%, n=17), while the lowest was observed in January (2.08%, n=2). The patient cohort comprised 53 males and 34 females, with a male-to-female ratio of 1.56:1. The age of the patients ranged from 24 days to 80 years, with 77.01% (n=67) aged ≤ 5 years, 20.69% (n=18) between 6 and 59 years old, and 2.30% (n=2) ≥ 60 years.
A total of 17 different Salmonella serotypes were identified among the 96 isolates (Figure 2). The most common serotype was S. Typhimurium monophasic variant S. 4,[5],12:i:- (37.50%, n=36), followed by S. Enteritidis (15.63%, n=15), S. Typhimurium (14.58%, n=14), S. London (7.29%, n=7), and S. Rissen (5.21%, n=5). S. Give and S. Goldcoast each accounted for 3.13% (n=3), while S. Paratyphi B, S. Agona, and S. Weltevreden each had two isolates. Additionally, one isolate was identified for each of the following serovars: S. Sandiego, S. Anatum, S. Saintpaul, S. Corvallis, S. Stanley, S. Virchow, and S. Newport.

Figure 2. The prevalence of individual serovar with their sequence type (ST) detected in this study.
The 96 Salmonella isolates were classified into 19 sequence types (STs) using in silico MLST (Figure 2). The most prevalent ST was ST34 (36.46%, n=35), followed by ST11 (15.63%, n=15), ST19 (13.54%, n=13), ST155 (7.29%, n=7), and ST469 (5.21%, n=5). A novel ST, named ST10830, was identified in two S. Typhimurium isolates and differed from ST34 in the purE locus.
Comparison of MLST and serotyping revealed that each ST generally corresponds to a single serotype, with the exceptions of ST19 and ST34. Specifically, ST19 included two isolates of S. 4,[5],12:i:- and 11 isolates of S. Typhimurium, while ST34 comprised 34 isolates of S. 4,[5],12:i:- and one of S. Typhimurium. Additionally, three serovars were associated with multiple STs: 36 S. 4,[5],12:i:-isolates included both ST19 and ST34; 14 S. Typhimurium isolates included ST19, ST34 and ST10830; and two S. Paratyphi B isolates were classified as ST43 and ST86.
Antimicrobial susceptibility testing of the 96 Salmonella isolates revealed high resistance rates to ampicillin (72.92%, n=70) and tetracycline (71.88%, n=69), followed by streptomycin (48.96%, n=47), sulfamethoxazole/trimethoprim (44.79%, n=43), and chloramphenicol (43.75%, n=42). Additionally, gentamicin resistance was observed in 15 (15.63%) isolates. One S. 4,[5],12:i:- isolate (1.04%) from a one-year-old male infant showed resistance to amikacin and fosfomycin, while four S. Enteritidis isolates (4.17%) were resistant to colistin. All isolates were susceptible to meropenem. Additionally, 18 isolates (18.75%) exhibited resistance to cefotaxime, while 28 (29.17%) and three (3.13%) isolates were resistant to nalidixic acid and ciprofloxacin, respectively. Particularly, only one isolate (1.04%) exhibited co-resistance to third-generation cephalosporin (cefotaxime) and fluoroquinolone (ciprofloxacin), which are considered first-line treatments for salmonellosis.
Of the 96 Salmonella isolates, seven (7.29%) were pan-susceptible, exhibiting susceptibility to all tested antimicrobial agents, while 68 isolates (70.83%) were resistant to at least three antimicrobial classes and thus classified as multidrug-resistant (MDR). Among the 89 isolates resistant to at least one antimicrobial agent, 34 distinct antimicrobial resistance patterns were identified. The most common pattern was resistance to ampicillin-tetracycline-chloramphenicol-sulfamethoxazole/trimethoprim, observed in 13 isolates (13.54%) (Supplementary Table S1).
To investigate the genetic mechanisms underlying antimicrobial resistance, whole-genome sequences of all isolates were screened for resistance genes and mutations in the quinolone resistance-determining region (QRDR). A total of 48 different resistance genes were identified across the 96 Salmonella isolates, conferring resistance or reduced susceptibility to β-lactams, aminoglycosides, quinolones, tetracyclines, chloramphenicols, sulfonamides, trimethoprims, fosfomycin, macrolides, lincosamides, and rifampicin (Figure 3). Each Salmonella isolate carried one to 21 resistances genes. They all carried cryptic aminoglycosides resistance gene aac(6’)-Iaa located on the chromosome. Additionally, one S. Sandiego and two S. Agona strains carried the chromosomal silent fosfomycin resistance gene fosA7. The amikacin- and fosfomycin- resistant S. 4,[5],12:i:- isolate carried the rmtB and fosA3 genes. Eighteen cefotaxime-resistant isolates carried either blaCTX-M (n=18) or blaCMY-2 (n=1). Three blaCTX-M variants were detected: blaCTX-M-14 (n=6), blaCTX-M-55 (n=7), and blaCTX-M-65 (n=5). One S. Agona isolate harbored both blaCTX-M-14 and blaCTX-M-55. Other β-lactamase genes were also detected, including blaTEM-1 (n=64), blaLAP-2 (n=1), blaOXA-1 (n=2), and blaOXA-10 (n=6). Thirty-seven Salmonella strains harbored one or two quinolone resistance genes, including qnrS1 (n=26), qnrB6 (n=4), qnrB19 (n=3), qnrVC1 (n=1), oqxAB (n=2), and aac(6’)-Ib-cr (n=8). Furthermore, 13 S. Enteritidis isolates contained a single mutation in gyrA (D87G or D87Y), and five S. 4,[5],12:i:- strains and three S. Typhimurium strains also had a single mutation in gyrA (D87G, D87Y, or S83F). The gyrA mutation, along with the presence of quinolone resistance genes, contributed to resistance to nalidixic acid and ciprofloxacin in two S. 4,[5],12:i:- strains and one S. Typhimurium strain. The remaining 25 nalidixic acid-resistant isolates carried a gyrA mutation (n=18) or one quinolone resistance gene (qnrB19; n=3) or two quinolone resistance genes (qnrB6 and aac(6’)-Ib-cr; n=4).
As shown in Table 1, 18 cefotaxime-resistant Salmonella isolates were collected from fecal samples, 15 of which were from patients aged less than 5 years. These isolates included nine S. 4,[5],12:i:- (ST34), three S. Enteritidis (ST11), two S. Agona (ST13), one S. Typhimurium (ST19), one S. Saintpaul (ST27), one S. Paratyphi B (ST86), and one S. Weltevreden (ST365). Notably, all three S. Enteritidis isolates carried blaCTX-M-14, and blaCTX-M-65 was identified exclusively in S. 4,[5],12:i:- strains. All ESBLs/AmpC-producing isolates exhibited resistance to ampicillin (MIC>128 mg/L) and had cefotaxime MICs ranging from 32 to >128 mg/L. Except for isolates SZ21HS63 and SZ22HS72, the remaining cefotaxime-resistant isolates were also resistant to multiple antibiotics. Thirteen of these isolates successfully transfer cefotaxime resistance to E. coli C600 via conjugation (Table 1). In addition to blaCTX-M/blaCMY, the isolates carried one to 20 other resistance genes, such as blaTEM, blaOXA, strA, strB, tet(A), qnrS1, sul2, and fosA3 (Table 1). Additionally, mutations within gyrA (D87Y or D87G) were identified in four isolates (Table 1). Interestingly, two S. 4,[5],12:i:- isolates (SZ20HS5 and SZ20HS17) and two S. Enteritidis isolates (SZ20HS10 and SZ20HS18) were obtained from the same infants sampled at different time points, respectively, sharing identical STs, resistance profiles, resistance genes, and mutations.
The lengths of blaCTX-M/blaCMY-2-carrying contigs ranged from 2,156 to 164,862 bp, and were located either on chromosome (n=1) or plasmids (n=9) (Table 1). Nine contigs were relatively short (2,156 to 12,437 bp) due to incomplete assembly and the presence of multiple insertion elements. These contigs lacked replicon genes or plasmid backbone, making it difficult to determine their exact location (Table 1; Figure 4).
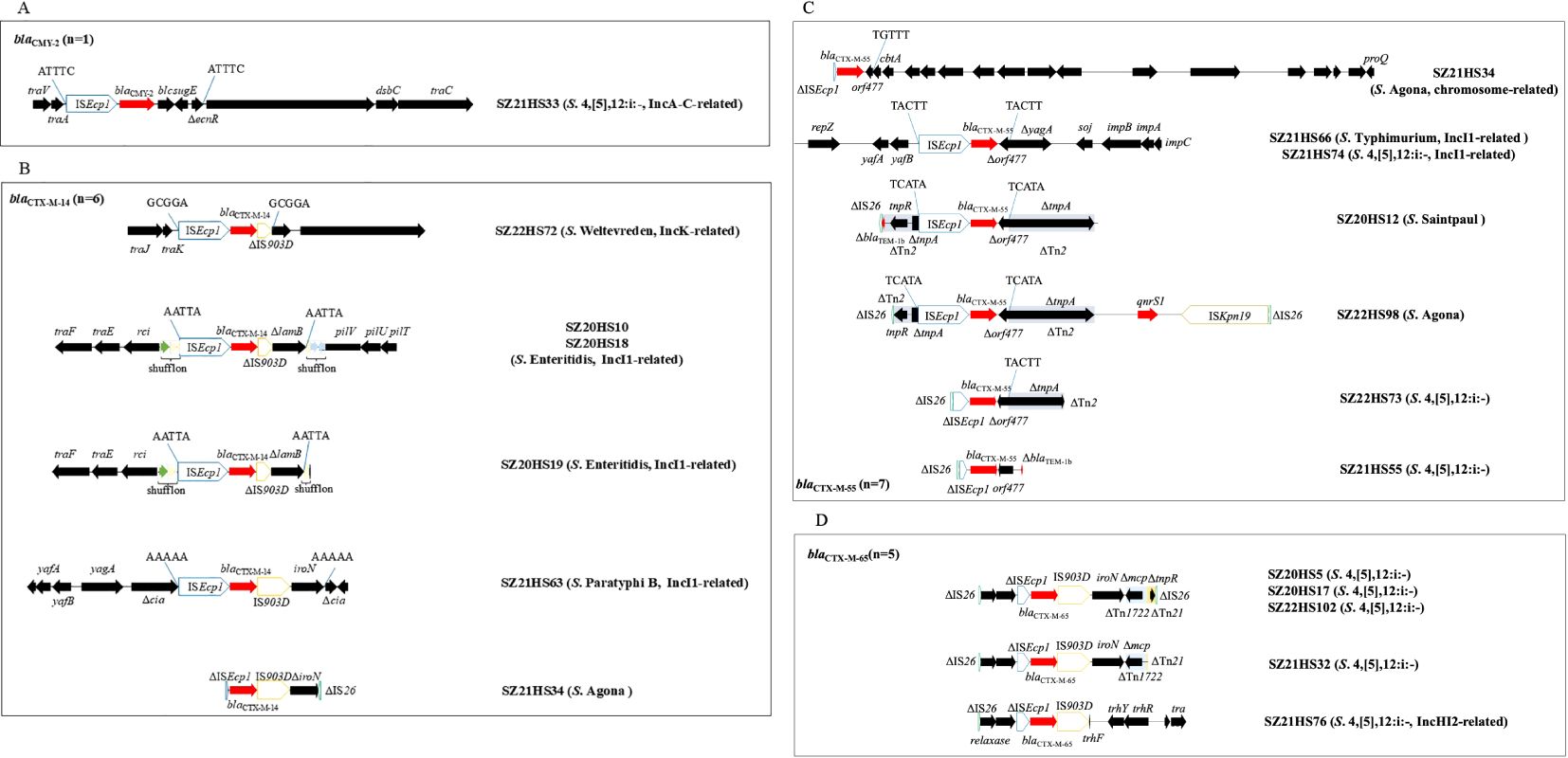
Figure 4. The genetic environments of blaCTX-M/blaCMY-2 in 18 Salmonella isolates in this study. (A) blaCMY-2; (B) blaCTX-M-14; (C) blaCTX-M-55; (D) blaCTX-M-65. The extents and directions of antibiotic resistance (red arrows) and other genes (black arrows) are indicated. ISs are shown as boxes labeled with their name. Δ indicates a truncated gene or mobile element. Arrows and sequences indicate direct repeats.
In S. 4,[5],12:i:- isolate SZ21HS33, a 4,479-bp transposition unit (ISEcp1-blaCMY-2-blc-sugE-ΔecnR) was inserted downstream of the plasmid conjugal transfer gene traA, generating 5-bp direct repeats (DRs) (Figure 4A). This blaCMY-2-bearing contig (92,168 bp) was highly similar (>99.99%) to the corresponding region of IncA/C plasmids found in Salmonella isolates from China, such as pSa1753 (food, MT859309) and pR1041-Sal2-167k (hospital, OR095745) (Supplementary Figure S1). Additionally, seven additional resistance genes, including blaOXA-10, addA1, aac(6’)-Ib3, aac(6’)-Ib-cr, qnrVC1, dfrA14, and mph(A), were co-located within this blaCMY-2-carrying contig.
Six Salmonella isolates carried the blaCTX-M-14 gene. The blaCTX-M-14-positive contig (92,206-bp) from the S. Weltevreden isolate SZ22HS72 was similar to IncK1 plasmids from E. coli, such as pDETEC82 (patient, China, CP116171) and pF16EC0617-4 (human, South Korea, CP088378), with 85% coverage and 99.99% identity (Supplementary Figure S1). The blaCTX-M-14 gene was associated with the commonly observed 3,060-bp structure (ISEcp1-blaCTX-M-14-ΔIS903D), inserted downstream of the plasmid conjugal transfer gene trak with 5-bp DRs (5’-GCGGA-3’) (Figure 4B). The blaCTX-M-14 gene was located on an IncI1 plasmid in four isolates (SZ20HS10, SZ20HS18, SZ20HS19, and SZ21HS63). Two S. Enteritidis isolates, SZ20HS10 and SZ20HS18, shared an identical blaCTX-M-14-positive contig (92,725-bp), showing 97% coverage and 99.97% identity with IncI1 plasmid pIncI1-CTX-M-14 (MN125610), obtained from a S. Enteritidis isolate from chicken in China (Supplementary Figure S1). A similar blaCTX-M-14-bearing contig (92,219-bp) was also found in S. Enteritidis isolate SZ20HS19 (Supplementary Figure S1). In these isolates, a 4,133-bp segment (ISEcp1-blaCTX-M-14-ΔIS903D-ΔlamB) with 5-bp DRs (5’-AATTA-3’) was inserted into the shufflon region of the IncI1 plasmid (Figure 4B). In S. Paratyphi B isolate SZ21HS63, the blaCTX-M-14-carrying contig (96,781-bp) exhibited 100% similarity to IncI1 plasmid pSZB23-1 (CP107011) from a clinical S. Paratyphi B isolate in Shenzhen, China (Supplementary Figure S1). The typical transposition unit (ISEcp1-blaCTX-M-14-IS903D-iroN), flanked by 5-bp DRs (5’-AAAAA-3’), was inserted into the cia gene, which encodes a colicin-like pore-forming protein (Figure 4B). A similar structure (ΔISEcp1-blaCTX-M-14-IS903D-ΔiroN) was observed in S. Agona isolate SZ21HS34, although ISEcp1 and iroN were incomplete.
The isolate SZ21HS34 also carried another blaCTX-M, blaCTX-M-55, located on the chromosome. The blaCTX-M-55-positive contig (22,529 bp) was identical to the corresponding region in Salmonella chromosomes, such as SSDFZ54 (CP034819) and SCFS (CP051218). Furthermore, the 1,266-bp segment (ΔISEcp1-blaCTX-M-55-Δorf477) in SZ21HS34 was commonly found in plasmids and chromosomes of various species (e.g., Klebsiella pneumoniae, E. coli, and S. enterica). In SZ21HS66 (S. Typhimurium) and SZ22HS74 (S. 4,[5],12:i:-), blaCTX-M-55 was located on the IncI1 plasmid, which showed >99.99% similarity to IncI1 plasmids from S. Typhimurium strains in China, such as pST53-2 (patient, CP050747) and pS29-IncI1 (human, CP085700) (Supplementary Figure S1). The 2,971-bp transposition unit (ISEcp1-blaCTX-M-55-Δorf477) was inserted into the yagA gene, generating 5-bp DRs (5’-TACTT-3’) (Figure 4C). The identical 2,971-bp blaCTX-M-55 unit was also observed in S. Saintpaul isolate SZ20HS12, where it was inserted into the transposase gene tnpA of transposon Tn2 (blaTEM-1b-tnpR-tnpA), although the β-lactam resistance gene blaTEM-1b was truncated by an incomplete insertion sequence IS26 (Figure 4C). An identical blaCTX-M-55-carrying structure was found in S. Agona isolate SZ22HS98, followed by a 5,752-bp segment (qnrS1-ISKpn19-IS26) (Figure 4C). However, the incomplete transposon Tn2 was truncated by IS26 at the resolvase gene tnpR (Figure 4C). A 3,713-bp similar segment (ΔIS26-ΔISEcp1-blaCTX-M-55-Δorf477-ΔTn2) was observed in SZ22HS73 (S. 4,[5],12:i:-), with ISEcp1 truncated by IS26 at the 5’ end (Figure 4C). Similarly, a 2,156-bp segment was found in SZ21HS55 (S. 4,[5],12:i:-), including the commonly observed structure (ΔIS26-ΔISEcp1-blaCTX-M-55-orf477) and a truncated blaTEM-1b downstream (Figure 4C).
Five S. 4,[5],12:i:- isolates carried blaCTX-M-65. As shown in Figure 4D, three isolates (SZ20HS5, SZ20HS17 and SZ22HS102) shared an identical blaCTX-M-65 region, including the typical transposition unit (ΔISEcp1-blaCTX-M-65-IS903-iroN), followed by three incomplete mobile elements (ΔTn1722, ΔTn21, and ΔIS26). A 1,205-bp region, containing two hypothetical proteins and an incomplete IS26 (76-bp), was found upstream of the blaCTX-M-65 unit. A similar region with 277-bp deletions of ΔTn21 and ΔIS26 was identified in SZ21HS32. Similarly, a 3,645-bp blaCTX-M-65 region was observed in SZ21HS76, including the upstream 1,205-bp segment and the blaCTX-M-65 unit (ΔISEcp1-blaCTX-M-65-IS903). This region was followed by IncHI2 plasmid conjugal transfer genes, such as ΔtrhF, trhY, trhR, and traH. The blaCTX-M-65-positive contig (164,862-bp) from isolate SZ21HS76 displayed >99.9% similarity to multiple IncHI2 plasmids, such as pS304_1 (S. Typhimurium, human, CP061127) and pHH194M-228K (E. coli, migratory bird, CP101516) (Supplementary Figure S1).
So far, among the more than 2,600 identified Salmonella serovars, many are known to cause human salmonellosis (EFSA and ECDC, 2023; Monte and Sellera, 2020). In this study, among the 96 Salmonella isolates representing 17 different serovars, the most prevalent was S. Typhimurium monophasic variant (S. 4,[5],12:i:-), accounting for 37.50%, followed by S. Enteritidis (15.63%) and S. Typhimurium (14.58%). This is consistent with previous study indicating that S. 4,[5],12:i:-, S. Enteritidis, and S. Typhimurium were the most common serovars among outpatients in Shaoxing, China (Chen et al., 2022). Similarly, an analysis of over 35,000 S. enterica strains from human and non-human sources across 23 Chinese provinces or municipal cities from 2006 to 2019 reported S. Typhimurium (including its monophasic variant) as the most common serovar, followed by S. Enteritidis (Wang et al., 2022a). In Europe, the three most commonly reported serovars since 2014 are S. Enteritidis, S. Typhimurium, and its monophasic variant S. 4,[5],12:i:-, which together account for more than 70% of human cases (EFSA and ECDC, 2023). S. Typhimurium has been identified as one of the most common serovars responsible for foodborne gastroenteritis worldwide (CDC, 2019; EFSA and ECDC, 2023; Nambiar et al., 2024; Van Puyvelde et al., 2023). Its monophasic variant, S. 4,[5],12:i:-, which lacks the ability to express the second-phase flagellar antigen, has become increasingly prevalent and is now a dominant serotype causing human salmonellosis (Wang et al., 2023a). In the United States, infections caused by S. 4,[5],12:i:- increased between 2009 and 2018, and this serotype has become the fifth most commonly reported serotype causing human illness (Plumb et al., 2023). Recently, an outbreak of multidrug-resistant S. 4,[5],12:i:- ST34 infection linked to chocolate products has been reported globally (Lund et al., 2022).
The widespread use of antimicrobial agents has led to an increase in MDR Salmonella. In this study, Salmonella isolates carried between one and 21 resistance genes, with 70.83% classified as MDR, severely limiting therapeutic options for clinical infections. Resistance to several important antibiotics was observed in our study. For instance, Fosfomycin exhibits strong antimicrobial activity against both Gram-negative and Gram-positive bacteria (Falagas et al., 2016). However, the widespread dissemination of fosfomycin resistance genes has led to an increase in reports of fosfomycin-resistant Salmonella (Aghamali et al., 2019; Fang et al., 2020). In Gram-negative bacteria, several plasmid-mediated fosfomycin resistance genes have been identified, with fosA3 being the most prevalent in Enterobacteriaceae, including Salmonella (Aghamali et al., 2019; Fang et al., 2020). In this study, one S. 4,[5],12:i:- isolate carrying fosA3 was found to be resistant to fosfomycin. Although fosA7 was detected in three Salmonella isolates in our study, it is present as a silent gene located on the chromosomes of certain Salmonella serotypes, and does not contribute to fosfomycin resistance (Monte et al., 2023; Wang et al., 2024). The fosfomycin-resistant S. 4,[5],12:i:- isolate carried the rmtB gene, conferring resistance to gentamicin and amikacin. The 16S rRNA methylase genes mediate high-level resistance to aminoglycosides, with armA and rmtB being the most prevalent in various Gram-negative bacteria, including Escherichia coli, Salmonella, K. pneumoniae, and Acinetobacter baumannii (Doi et al., 2016). Resistance to colistin, the last-resort antibiotic for Gram-negative pathogens, was observed in four S. Enteritidis isolates in our study. However, neither the colistin resistance gene mcr nor mutations in PmrA/B-PhoP/Q were identified, suggesting that their resistance was probably intrinsic, due to the O-antigen epitope of group D Salmonella governing the levels of colistin susceptibility (Ricci et al., 2020).
Fluoroquinolones and third-generation cephalosporins, such as ciprofloxacin and cefotaxime, are first-line treatments for Salmonella infections (Ruiz et al., 2004). Fluoroquinolone resistance in Gram-negative bacteria is primarily associated with mutations in the chromosomal QRDRs of the gyrA and parC genes, as well as plasmid-mediated quinolone resistance (PMQR) genes (Hooper and Jacoby, 2015). In this study, mutations in the gyrA gene were identified in 21 Salmonella isolates, mainly at positions D87G/D87Y, with S83F also being detected. However, mutations in gyrB, parC, or parE were not identified in this study. GyrA position 87 is a common mutation site in Salmonella, and mutations at gyrA position 83 and parC position 80 are also frequently observed, whereas the frequency of gyrB and parE mutations is relatively low (Wang et al., 2022a). Additionally, 37 Salmonella strains harbored at least one PMQR gene in our study, with qnrS1 being the most prevalent. Worryingly, a high prevalence (10.45%) of 1,962 Salmonella isolates in China carried the PMQR genes, with qnrS1 being the most common (Wang et al., 2022a). As previously described (Bai et al., 2016; Wang et al., 2023b), the combination of chromosomal gyrA mutations and PMQR genes contribute to resistance to nalidixic acid and ciprofloxacin in Salmonella.
In this study, 18.75% of Salmonella isolates exhibited resistance to the third-generation cephalosporin cefotaxime, due to the presence of blaCTX-M, with one strain carrying blaCMY-2. Since the discovery of blaCTX-M, its prevalence has increased globally, making it one of the most common ESBL gene worldwide (Bevan et al., 2017). In China, blaCTX-M is also the predominant ESBL gene in Salmonella strains from human, animal, and food sources (Bao et al., 2024; Wang et al., 2022a). High detection rates of blaCTX-M have been observed in several Salmonella serotypes, such as S. Typhimurium, S. Kentucky, and S. Indiana (Sun et al., 2022; Wang et al., 2020, 2023b). Currently, more than 260 different subtypes of blaCTX-M have been identified globally (https://www.ncbi.nlm.nih.gov/pathogens/refgene/), with blaCTX-M-14 and blaCTX-M-15 being the most prevalent subtypes globally, and blaCTX-M-55 being predominant mainly in Asia such as China (Bevan et al., 2017). In this study, three blaCTX-M subtypes were identified: blaCTX-M-55 (38.89%), blaCTX-M-14 (33.33%), and blaCTX-M-65 (27.78%). In a previous study, 2,283 Salmonella strains from human feces and animal-derived food samples (chicken, pork, and seafood) collected across five provinces in China were analyzed, 200 of them were positive for blaCTX-M with blaCTX-M-65, blaCTX-M-123 and blaCTX-M-14 being the most prevalent (Wang et al., 2020). Similarly, Wang et al. (2022a) performed whole-genome sequencing on 1,962 Salmonella isolates (from both human and non-human sources) across 22 provinces and municipalities in China, and identified 13 distinct blaCTX-M subtypes, including blaCTX-M-55, blaCTX-M-14, and blaCTX-M-65. Interestingly, despite the global dominance of blaCTX-M-15, it was not detected in our study, reflecting its lower prevalence in Salmonella from China (Bevan et al., 2017; Jiang et al., 2022; Wang et al., 2022a). Although co-resistance to both cephalosporins and fluoroquinolones in Salmonella has been increasingly reported (Bai et al., 2016; Liu et al., 2023; Nambiar et al., 2024; Wang et al., 2023b), only one S. 4,[5],12:i:- isolate in our study exhibited co-resistance to cefotaxime and ciprofloxacin.
The global dissemination of blaCTX-M is partly driven by some successful clones. For example, E. coli ST131, which produces CTX-M-15, is a high-risk international clone, particularly in hospitals (Becerra-Aparicio et al., 2023; Bevan et al., 2017; Nicolas-Chanoine et al., 2014). Similarly, S. 4,[5],12:i:- ST34 and S. Kentucky ST198 have disseminated globally, facilitating the spread of resistance genes such as blaCTX-M-65, blaCTX-M-55, and blaCTX-M-14 (Hawkey et al., 2019; Protonotariou et al., 2022; Wang et al., 2023b). Notably, two S. 4,[5],12:i:- isolates and two S. Enteritidis isolates in this study, obtained from the same infants at different times, shared identical STs, resistance profiles, resistance genes, mutations, and blaCTX-M-carrying contigs. This suggests long-term colonization of ESBL-producing S. 4,[5],12:i:- or S. Enteritidis clones in individual patients for periods exceeding three or six weeks, likely contributing to the persistence and transmission of antimicrobial resistance within the local population. Similar long-term persistence of bacterial clones has been previously reported, such as the S. Kentucky ST198 clone in a patient with inflammatory bowel disease (Jiang et al., 2024), and CTX-M-producing E. coli in healthy food handlers for durations ranging from three months to two years (Nakane et al., 2016). Many bacterial species can establish persistent infections in their hosts, even after antibiotic treatment, due to factors such as host immunocompromise, bacterial immune evasion, and/or inadequate eradication of the pathogen by antibiotics (Fisher et al., 2017). Given that antimicrobial drugs are sometime ineffective in eliminating long-term colonization, it is crucial to explore alternative strategies, such as phage therapy, for bacterial decolonization in humans to prevent infections and reduce the spread of MDR organisms (Fang et al., 2024).
Pandemic plasmids play a significant role in the global transmission of blaCTX-M in Enterobacteriaceae, with plasmids such as IncI1, IncK1, IncHI2, IncF, and IncN acting as key vectors for horizontal gene transfer (Bevan et al., 2017; Rozwandowicz et al., 2018). The IncI1 plasmid, one of the most common types in Enterobacteriaceae from humans, animals and the environment, is an important carrier of blaCTX-M genes (Carattoli et al., 2021; Rozwandowicz et al., 2018). IncI1 plasmids are linked to the global spread of multiple blaCTX-M variants, such as blaCTX-M-14 and blaCTX-M-55 in this study, as well as blaCTX-M-1, blaCTX-M-3, blaCTX-M-15, and blaCTX-M-101 (Irrgang et al., 2018; Protonotariou et al., 2022; Qin and Zhang, 2023; Saidani et al., 2019; Yu et al., 2024). Other plasmids, such as IncK1, are also crucial for the dissemination of blaCTX-M in Enterobacteriaces from diverse sources. For example, IncK1 plasmids are common vectors for horizontal transfer of blaCTX-M-14 in E. coli isolates from both humans and animals in Europe (Rozwandowicz et al., 2017; Stokes et al., 2012; Valverde et al., 2009) and in E. coli isolates from healthy volunteers in Yangzhou, China (Wang et al., 2022b). Similarly, IncHI2 plasmids, which are prevalent in Salmonella isolates and often associated with MDR (Chen et al., 2016; McMillan et al., 2020), are important carriers of blaCTX-M-14/-55/-65 in E. coli and Salmonella isolates from humans, animals, and food products in China (Jiang et al., 2022; Li et al., 2022; Wang et al., 2018; 2022b; Tian et al., 2022).
The IncA/C plasmid has also emerged as a major vector for resistance genes, particularly in the spread of cephalosporinase genes like blaCMY-2 (Rozwandowicz et al., 2018). IncA/C plasmids carrying blaCMY-2 have been frequently detected in E. coli and Salmonella isolates from both animals and humans (He et al., 2021; Liu et al., 2023; Zhang et al., 2024; Zheng et al., 2022). In this study, one S. 4,[5],12:i:- isolate carried blaCMY-2 on an IncA/C plasmid highly similar to other IncA/C plasmids from Salmonella. In addition to blaCMY-2, IncA/C plasmids have been described to mediate the spread of ESBL genes (e.g., blaCTX-M) and carbapenemase genes (e.g., blaNDM-1) in E. coli, Salmonella, and K. pneumoniae isolates from various sources (Papa-Ezdra et al., 2021; Tello et al., 2022; Villa et al., 2015; Wailan et al., 2016; Wasyl et al., 2015).
Insertion sequence (IS) facilitate the horizontal transfer of blaCTX-M genes between plasmids and chromosomes, with ISEcp1 and IS26 being key elements in this process (Castanheira et al., 2021; Partridge et al., 2018). ISEcp1, a member of the IS1380 family, transposes by recognizing the right inverted repeat sequence or its similar sequence and can carry adjacent structures including drug resistance genes, often generating 5-bp DRs (Partridge et al., 2018). In this study, ISEcp1 mediated the transfer of blaCMY-2 and blaCTX-M genes. ISEcp1 typically locates upstream of blaCTX-M, providing a promoter for its expression (Partridge et al., 2018; Poirel et al., 2003). Interestingly, the blaCTX-M-55 gene was found on the chromosome of a S. Agona isolate, associated with an incomplete ISEcp1 insertion upstream. Although blaCTX-M is frequently plasmid-borne, there is increasing evidence of its integration into chromosomal DNA, mediated by mobile elements, in E. coli, Salmonella, and K. pneumoniae isolates (Huang et al., 2017; Jiang et al., 2023; Shen et al., 2016; Yoon et al., 2020). For instance, ISEcp1-mediated blaCTX-M-14 integration into the type VI secretion system on the chromosome of S. Kentucky ST198.2-1 clade has been reported in Europe and China (Hawkey et al., 2019; Wang et al., 2023b). Once integrated into the chromosome, resistance genes such as blaCTX-M can be vertically transmitted to daughter cells, becoming intrinsic resistance determinants. Furthermore, various mobile elements are involved in the acquisition and spread of blaCTX-M and blaCMY genes, such as the class I integron-ISCR1 complex (blaCTX-M-2 and blaCTX-M-9) and IS1294 (blaCMY-2) (Castanheira et al., 2021; Tagg et al., 2014).
However, our study has several limitations. Our study provides limited information and insights into Salmonellosis surveillance, since the analysis was based on only 96 Salmonella isolates collected from a single hospital in China. The small sample size and restricted geographic scope may not adequately represent the overall epidemiology of Salmonellosis or the prevalence of antimicrobial resistance in this region. Furthermore, although 17 serotypes were identified, the limited number of isolates for each serotype may have resulted in the underrepresentation of other important serotypes. The small sample size per serotype also hinders comparisons of resistance patterns across different serotypes. Therefore, continuous and expanded surveillance of Salmonellosis involving multiple hospitals and a larger number of samples, is essential to provide a comprehensive understanding of its epidemiology and antimicrobial resistance patterns.
In conclusion, S. 4,[5],12:i:-, S. Enteritidis, and S. Typhimurium are the predominant serotypes in this clinical setting, exhibiting a concerning prevalence of MDR isolates. The dissemination of blaCTX-M/blaCMY-2 among clinical Salmonella isolates is primarily mediated through horizontal gene transfer facilitated by global successful pandemic plasmids (e.g., IncI1) and associated mobile elements (ISEcp1). The findings highlight the urgent need for implementation of targeted antimicrobial stewardship programs, reinforcement of infection control measures, and the development of alternative therapeutic strategies, such as phage therapy, to manage persistent infections and curb the spread of MDR strains. Long-term colonization and plasmid-mediated resistance spread reveal gaps in our understanding of transmission dynamics, underscoring the necessity of incorporating molecular surveillance into routine clinical practice to enhance prevention and control measures.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The studies involving humans were approved by Jiangsu Key Laboratory of Zoonosis, Yangzhou University. The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from a by- product of routine care or industry. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
JW: Conceptualization, Formal Analysis, Funding acquisition, Investigation, Project administration, Supervision, Writing – original draft. Z-HD: Investigation, Writing – original draft. X-YZ: Formal Analysis, Investigation, Resources, Writing – review & editing. Q-CM: Investigation, Writing – review & editing. Z-YW: Formal Analysis, Writing – review & editing. DCL: Investigation, Resources, Writing – review & editing. Y-FH: Investigation, Resources, Writing – review & editing. CZ: Investigation, Resources, Writing – review & editing. XJ: Funding acquisition, Writing – review & editing. DL: Investigation, Resources, Supervision, Writing – review & editing. QL: Conceptualization, Formal Analysis, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported in part by the National Key Research and Development Program of China (no. 2022YFC2604200), the National Natural Science Foundation of China (no. 31902319), and the fifth phase of the “333 project” scientific research project in Jiangsu Province (no. BRA2020002).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1544757/full#supplementary-material
Aghamali, M., Sedighi, M., Zahedi Bialvaei, A., Mohammadzadeh, N., Abbasian, S., Ghafouri, Z., et al. (2019). Fosfomycin: mechanisms and the increasing prevalence of resistance. J. Med. Microbiol. 68, 11–25. doi: 10.1099/jmm.0.000874
Bai, L., Zhao, J., Gan, X., Wang, J., Zhang, X., Cui, S., et al. (2016). Emergence and diversity of Salmonella enterica serovar Indiana isolates with concurrent resistance to ciprofloxacin and cefotaxime from patients and food-producing animals in China. Antimicrob. Agents Chemother. 60, 3365–3371. doi: 10.1128/AAC.02849-15
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bao, D., Chen, L., Shen, W., Xu, X., Zhu, L., Wang, Y., et al. (2024). Genomic epidemiology of ceftriaxone-resistant non-typhoidal Salmonella enterica strain in China. BMC Genomics 25, 974. doi: 10.1186/s12864-024-10890-2
Becerra-Aparicio, F., Gómez-Zorrilla, S., Hernández-García, M., Gijón, D., Siverio, A., Berbel, D., et al. (2023). Significant increase of CTX-M-15-ST131 and emergence of CTX-M-27-ST131 Escherichia coli high-risk clones causing healthcare-associated bacteraemia of urinary origin in Spain (ITUBRAS-2 project). J. Antimicrob. Chemother. 78, 2291–2296. doi: 10.1093/jac/dkad234
Bevan, E. R., Jones, A. M., Hawkey, P. M. (2017). Global epidemiology of CTX-M β-lactamases: temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 72, 2145–2155. doi: 10.1093/jac/dkx146
Carattoli, A., Villa, L., Fortini, D., García-Fernández, A. (2021). Contemporary IncI1 plasmids involved in the transmission and spread of antimicrobial resistance in Enterobacteriaceae. Plasmid 118, 102392. doi: 10.1016/j.plasmid.2018.12.001
Castanheira, M., Simner, P. J., Bradford, P. A. (2021). Extended-spectrum β-lactamases: an update on their characteristics, epidemiology and detection. JAC Antimicrob. Resist. 3, dlab092. doi: 10.1093/jacamr/dlab092
CDC (2019). Antibiotic Resistance Threats in the United State (Atlanta, GA: U.S. Department of Health and Human Services). doi: 10.15620/cdc:82532
Chen, J., Ed-Dra, A., Zhou, H., Wu, B., Zhang, Y., Yue, M. (2022). Antimicrobial resistance and genomic investigation of non-typhoidal Salmonella isolated from outpatients in Shaoxing city, China. Front. Public Health 10. doi: 10.3389/fpubh.2022.988317
Chen, W., Fang, T., Zhou, X., Zhang, D., Shi, X., Shi, C. (2016). IncHI2 plasmids are predominant in antibiotic-resistant Salmonella isolates. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01566
CLSI (2012). Clinical and Laboratory Standards Institute (CLSI). Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. CLSI document M07-A9. 9th ed (Wayne, PA: CLSI).
CLSI (2022). Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. CLSI Supplement M100. 32th Edn (Wayne, PA: CLSI).
Crump, J. A., Sjölund-Karlsson, M., Gordon, M. A., Parry, C. M. (2015). Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive Salmonella infections. Clin. Microbiol. Rev. 28, 901–937. doi: 10.1128/CMR.00002-15
Doi, Y., Wachino, J. I., Arakawa, Y. (2016). Aminoglycoside resistance: the emergence of acquired 16S ribosomal RNA methyltransferases. Infect. Dis. Clin. North Am. 30, 523–537. doi: 10.1016/j.idc.2016.02.011
EFSA, ECDC (2023). The European union one health 2022 zoonoses report. EFSA J. 21, e8442. doi: 10.2903/j.efsa.2023.8442
Falagas, M. E., Vouloumanou, E. K., Samonis, G., Vardakas, K. Z. (2016). Fosfomycin. Clin. Microbiol. Rev. 29, 321–347. doi: 10.1128/CMR.00068-15
Fang, L. X., Jiang, Q., Deng, G. H., He, B., Sun, R. Y., Zhang, J. F., et al. (2020). Diverse and flexible transmission of fosA3 associated with heterogeneous multidrug resistance regions in Salmonella enterica serovar Typhimurium and Indiana isolates. Antimicrob. Agents Chemother. 64, e02001-19. doi: 10.1128/AAC.02001-19
Fang, Q., Yin, X., He, Y., Feng, Y., Zhang, L., Luo, H., et al. (2024). Safety and efficacy of phage application in bacterial decolonisation: a systematic review. Lancet Microbe 5, e489–e499. doi: 10.1016/S2666-5247(24)00002-8
Fisher, R. A., Gollan, B., Helaine, S. (2017). Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 15, 453–464. doi: 10.1038/nrmicro.2017.42
Hawkey, J., Le Hello, S., Doublet, B., Granier, S. A., Hendriksen, R. S., Fricke, W. F., et al. (2019). Global phylogenomics of multidrug-resistant Salmonella enterica serotype Kentucky ST198. Microb. Genom. 5, e000269. doi: 10.1099/mgen.0.000269
He, D. D., Cui, M. M., Zhang, T. L., Hu, G. Z., Liu, J. H., Pan, Y. S. (2021). Characterization of blaCMY-2-carrying IncC and rmtB-carrying IncI1/ST136 plasmids in an avian Escherichia coli ST224 strain. Plasmid 114, 102555. doi: 10.1016/j.plasmid.2021.102555
Hooper, D. C., Jacoby, G. A. (2015). Mechanisms of drug resistance: quinolone resistance. Ann. N. Y. Acad. Sci. 1354, 12–31. doi: 10.1111/nyas.12830
Huang, W., Wang, G., Sebra, R., Zhuge, J., Yin, C., Aguero-Rosenfeld, M. E., et al. (2017). Emergence and evolution of multidrug-resistant Klebsiella pneumoniae with both blaKPC and blaCTX-M integrated in the chromosome. Antimicrob. Agents Chemother. 61, e00076-17. doi: 10.1128/AAC.00076-17
Irrgang, A., Hammerl, J. A., Falgenhauer, L., Guiral, E., Schmoger, S., Imirzalioglu, C., et al. (2018). Diversity of CTX-M-1-producing E. coli from German food samples and genetic diversity of the blaCTX-M-1 region on IncI1 ST3 plasmids. Vet. Microbiol. 221, 98–104. doi: 10.1016/j.vetmic.2018.06.003
Jiang, Q., Ke, B. X., Wu, D. S., Wang, D., Fang, L. X., Sun, R. Y., et al. (2022). Epidemiology of blaCTX-M-positive Salmonella Typhimurium from diarrhoeal outpatients in Guangdong, China 2010-2017. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.865254
Jiang, Y., Wang, Z. Y., Li, Q. C., Lu, M. J., Wu, H., Mei, C. Y., et al. (2023). Characterization of extensively drug-resistant Salmonella enterica serovar Kentucky sequence type 198 isolates from chicken meat products in Xuancheng, China. Microbiol. Spectr. 11, e0321922. doi: 10.1128/spectrum.03219-22
Jiang, Y., Yang, H., Wang, Z. Y., Lin, D. C., Jiao, X., Hu, Y., et al. (2024). Persistent colonization of ciprofloxacin-resistant and extended-spectrum β-lactamase (ESBL)-producing Salmonella enterica serovar Kentucky ST198 in a patient with inflammatory bowel disease. Infect. Drug Resist. 17, 1459–1466. doi: 10.2147/IDR.S447971
Li, L., Olsen, R. H., Xiao, J., Meng, H., Peng, S., Shi, L. (2022). Genetic context of blaCTX-M-55 and qnrS1 genes in a foodborne Salmonella enterica serotype Saintpaul isolate from China. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.899062
Liu, J., Wei, S., Ma, J., Zeng, Z., Lü, D., Yang, G., et al. (2007). Detection and characterization of CTX-M and CMY-2 β-lactamases among Escherichia coli isolates from farm animals in Guangdong province of China. Int. J. Antimicrob. Agents 29, 576–581. doi: 10.1016/j.ijantimicag.2006.12.015
Liu, M., Zhu, K., Li, X., Han, Y., Yang, C., Liu, H., et al. (2023). Genetic characterization of a Salmonella enterica serovar Typhimurium isolated from an infant with concurrent resistance to ceftriaxone, ciprofloxacin and azithromycin. J. Glob. Antimicrob. Resist. 35, 252–256. doi: 10.1016/j.jgar.2023.09.016
Lund, S., Tahir, M., Vohra, L. I., Hamdana, A. H., Ahmad, S. (2022). Outbreak of monophasic Salmonella Typhimurium sequence type 34 linked to chocolate products. Ann. Med. Surg. 82, 104597. doi: 10.1016/j.amsu.2022.104597
McMillan, E. A., Jackson, C. R., Frye, J. G. (2020). Transferable plasmids of Salmonella enterica associated with Antibiotic resistance genes. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.562181
Monte, D. F. M., Doi, Y., Lincopan, N. (2023). High prevalence and global distribution of fosfomycin resistance genes in Salmonella serovars. Lancet Microbe 4, e968. doi: 10.1016/S2666-5247(23)00261-6
Monte, D. F. M., Sellera, F. P. (2020). Salmonella. Emerg. Infect. Dis. 26, 2955. doi: 10.3201/eid2612.et2612
Nakane, K., Kawamura, K., Goto, K., Arakawa, Y. (2016). Long-term colonization by blaCTX-M-harboring Escherichia coli in healthy Japanese people engaged in food handling. Appl. Environ. Microbiol. 82, 1818–1827. doi: 10.1128/AEM.02929-15
Nambiar, R. B., Elbediwi, M., Ed-Dra, A., Wu, B., Yue, M. (2024). Epidemiology and antimicrobial resistance of Salmonella serovars Typhimurium and 4,[5],12:i- recovered from hospitalized patients in China. Microbiol. Res. 282, 127631. doi: 10.1016/j.micres.2024.127631
Nicolas-Chanoine, M. H., Bertrand, X., Madec, J. Y. (2014). Escherichia coli ST131, an intriguing clonal group. Clin. Microbiol. Rev. 27, 543–574. doi: 10.1128/CMR.00125-13
Papa-Ezdra, R., Cordeiro, N. F., Di, P. V., Chiarelli, A., Pallecchi, L., Garcia-Fulgueiras, V., et al. (2021). Description of novel resistance islands harbouring blaCTX-M-2 in IncC type 2 plasmids. J. Glob. Antimicrob. Resist. 26, 37–41. doi: 10.1016/j.jgar.2021.03.031
Partridge, S. R., Kwong, S. M., Firth, N., Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31, e00088-17. doi: 10.1128/CMR.00088-17
Plumb, I. D., Brown, A. C., Stokes, E. K., Chen, J. C., Carleton, H., Tolar, B., et al. (2023). Increased multidrug-resistant Salmonella enterica I serotype 4,[5],12:i:- infections associated with pork, United States 2009-2018. Emerg. Infect. Dis. 29, 314–322. doi: 10.3201/eid2902.220950
Poirel, L., Decousser, J. W., Nordmann, P. (2003). Insertion sequence ISEcp1B is involved in expression and mobilization of a blaCTX-M β-lactamase gene. Antimicrob. Agents Chemother. 47, 2938–2945. doi: 10.1128/AAC.47.9.2938-2945.2003
Protonotariou, E., Meletis, G., Papadopoulos, T., Kagkalou, G., Tychala, A., Chattaway, M. A., et al. (2022). Phenotypic and molecular characterization of blaCTX-M-3 and blaCTX-M-55-producing monophasic Salmonella enterica serovar Typhimurium in Greece. J. Glob. Antimicrob. Resist. 30, 75–80. doi: 10.1016/j.jgar.2022.05.017
Qin, X., Zhang, Z. (2023). Emergence of a hybrid IncI1-Iα plasmid-encoded blaCTX-M-101 conferring resistance to cephalosporins in Salmonella enterica serovar Enteritidis. Microorganisms 11, 1275. doi: 10.3390/microorganisms11051275
Ricci, V., Zhang, D., Teale, C., Piddock, L. J. V. (2020). The O-antigen epitope governs susceptibility to colistin in Salmonella enterica. mBio 11, e02831-19. doi: 10.1128/mBio.02831-19
Rozwandowicz, M., Brouwer, M. S. M., Fischer, J., Wagenaar, J. A., Gonzalez-Zorn, B., Guerra, B., et al. (2018). Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 73, 1121–1137. doi: 10.1093/jac/dkx488
Rozwandowicz, M., Brouwer, M. S., Zomer, A. L., Bossers, A., Harders, F., Mevius, D. J., et al. (2017). Plasmids of distinct IncK lineages show compatible phenotypes. Antimicrob. Agents Chemother. 61, e01954-16. doi: 10.1128/AAC.01954-16
Ruiz, M., Rodríguez, J. C., Escribano, I., Royo, G. (2004). Available options in the management of non-typhi Salmonella. Expert Opin. Pharmacother. 5, 1737–1743. doi: 10.1517/14656566.5.8.1737
Saidani, M., Messadi, L., Mefteh, J., Chaouechi, A., Soudani, A., Selmi, R., et al. (2019). Various Inc-type plasmids and lineages of Escherichia coli and Klebsiella pneumoniae spreading blaCTX-M-15,blaCTX-M-1 and mcr-1 genes in camels in Tunisia. J. Glob. Antimicrob. Resist. 19, 280–283. doi: 10.1016/j.jgar.2019.05.007
Shen, P., Yi, M., Fu, Y., Ruan, Z., Du, X., Yu, Y., et al. (2016). Detection of an Escherichia coli sequence type 167 strain with two tandem copies of blaNDM-1 in the chromosome. J. Clin. Microbiol. 55, 199–205. doi: 10.1128/JCM.01581-16
Stokes, M. O., Cottell, J. L., Piddock, L. J., Wu, G., Wootton, M., Mevius, D. J., et al. (2012). Detection and characterization of pCT-like plasmid vectors for blaCTX-M-14 in Escherichia coli isolates from humans, Turkeys and cattle in England and Wales. J. Antimicrob. Chemother. 67, 1639–1644. doi: 10.1093/jac/dks126
Sun, R. Y., Guo, W. Y., Zhang, J. X., Wang, M. G., Wang, L. L., Lian, X. L., et al. (2022). Phylogenomic analysis of Salmonella Indiana ST17, an emerging MDR clonal group in China. J. Antimicrob. Chemother. 77, 2937–2945. doi: 10.1093/jac/dkac243
Tagg, K. A., Iredell, J. R., Partridge, S. R. (2014). Complete sequencing of IncI1 sequence type 2 plasmid pJIE512b indicates mobilization of blaCMY-2 from an IncA/C plasmid. Antimicrob. Agents Chemother. 58, 4949–4952. doi: 10.1128/AAC.02773-14
Tello, M., Oporto, B., Lavín, J. L., Ocejo, M., Hurtado, A. (2022). Characterization of a carbapenem-resistant Escherichia coli from dairy cattle harbouring blaNDM-1 in an IncC plasmid. J. Antimicrob. Chemother. 77, 843–845. doi: 10.1093/jac/dkab455
Tian, T., Dai, S., Liu, D., Wang, Y., Qiao, W., Yang, M., et al. (2022). Occurrence and transfer characteristics of blaCTX-M genes among Escherichia coli in anaerobic digestion systems treating swine waste. Sci. Total Environ. 834, 155321. doi: 10.1016/j.scitotenv.2022.155321
Valverde, A., Cantón, R., Garcillán-Barcia, M. P., Novais, A., Galán, J. C., Alvarado, A., et al. (2009). Spread of blaCTX-M-14 is driven mainly by IncK plasmids disseminated among Escherichia coli phylogroups A, B1, and D in Spain. Antimicrob. Agents Chemother. 53, 5204–5212. doi: 10.1128/AAC.01706-08
Van Puyvelde, S., de Block, T., Sridhar, S., Bawn, M., Kingsley, R. A., Ingelbeen, B., et al. (2023). A genomic appraisal of invasive Salmonella Typhimurium and associated antibiotic resistance in sub-Saharan Africa. Nat. Commun. 14, 6392. doi: 10.1038/s41467-023-41152-6
Villa, L., Guerra, B., Schmoger, S., Fischer, J., Helmuth, R., Zong, Z., et al. (2015). IncA/C plasmid carrying blaNDM-1, blaCMY-16, and fosA3 in a Salmonella enterica serovar Corvallis strain isolated from a migratory wild bird in Germany. Antimicrob. Agents Chemother. 59, 6597–6600. doi: 10.1128/AAC.00944-15
Wailan, A. M., Sidjabat, H. E., Yam, W. K., Alikhan, N. F., Petty, N. K., Sartor, A. L., et al. (2016). Mechanisms involved in acquisition of blaNDM genes by IncA/C2 and IncFIIY plasmids. Antimicrob. Agents Chemother. 60, 4082–4088. doi: 10.1128/AAC.00368-16
Wang, Z., Gu, D., Hong, Y., Hu, Y., Gu, J., Tang, Y., et al. (2023a). Microevolution of Salmonella 4,[5],12:i:- derived from Salmonella enterica serovar Typhimurium through complicated transpositions. Cell Rep. 42, 113227. doi: 10.1016/j.celrep.2023.113227
Wang, J., Huang, X. Y., Xia, Y. B., Guo, Z. W., Ma, Z. B., Yi, M. Y., et al. (2018). Clonal Spread of Escherichia coli ST93 carrying mcr-1-harboring IncN1-IncHI2/ST3 plasmid among companion animals, China. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.02989
Wang, Z. Y., Jiang, Y., Shao, Y. Q., Lu, H. F., Lu, M. J., Jiao, X., et al. (2022b). Nasal carriage of CTX-M-55-producing Escherichia coli ST8369 in a healthy cohort in the city of Yangzhou, China. Front. Cell. Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.970940
Wang, Z., Jiang, Y., Xu, H., Jiao, X., Wang, J., Li, Q. (2023b). Poultry production as the main reservoir of ciprofloxacin- and tigecycline-resistant extended-spectrum β-lactamase (ESBL)-producing Salmonella enterica serovar Kentucky ST198.2-2 causing human infections in China. Appl. Environ. Microbiol. 89, e0094423. doi: 10.1128/aem.00944-23
Wang, J., Li, Q., Jiang, Y., Wang, Z., Jiao, X. (2024). fosA7: a silent fosfomycin resistance gene in Salmonella? Lancet Microbe 5, e211. doi: 10.1016/S2666-5247(23)00342-7
Wang, Y., Liu, Y., Lyu, N., Li, Z., Ma, S., Cao, D., et al. (2022a). The temporal dynamics of antimicrobial-resistant Salmonella enterica and predominant serovars in China. Natl. Sci. Rev. 10, nwac269. doi: 10.1093/nsr/nwac269
Wang, W., Zhao, L., Hu, Y., Dottorini, T., Fanning, S., Xu, J., et al. (2020). Epidemiological study on prevalence, serovar diversity, multidrug resistance, and CTX-M-type extended-spectrum β-lactamases of Salmonella spp. from patients with diarrhea, food of animal origin, and pets in several provinces of China. Antimicrob. Agents Chemother. 64, e00092-20. doi: 10.1128/AAC.00092-20
Wasyl, D., Kern-Zdanowicz, I., Domańska-Blicharz, K., Zając, M., Hoszowski, A. (2015). High-level fluoroquinolone resistant Salmonella enterica serovar Kentucky ST198 epidemic clone with IncA/C conjugative plasmid carrying blaCTX-M-25 gene. Vet. Microbiol. 175, 85–91. doi: 10.1016/j.vetmic.2014.10.014
Yoon, E. J., Gwon, B., Liu, C., Kim, D., Won, D., Park, S. G., et al. (2020). Beneficial chromosomal integration of the genes for CTX-M extended-spectrum β-lactamase in Klebsiella pneumoniae for stable propagation. mSystems 5, e00459-20. doi: 10.1128/mSystems.00459-20
Yoshida, C. E., Kruczkiewicz, P., Laing, C. R., Lingohr, E. J., Gannon, V. P., Nash, J. H., et al. (2016). The Salmonella In Silico Typing Resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PloS One 11, e0147101. doi: 10.1371/journal.pone.0147101
Yu, K., Huang, Z., Xiao, Y., Gao, H., Bai, X., Wang, D. (2024). Global spread characteristics of CTX-M-type extended-spectrum β-lactamases: A genomic epidemiology analysis. Drug Resist. Updat. 73, 101036. doi: 10.1016/j.drup.2023.101036
Zhang, Z., Kuang, D., Xu, X., Zhan, Z., Ren, H., Shi, C. (2024). Dissemination of IncC plasmids in Salmonella enterica serovar Thompson recovered from seafood and human diarrheic patients in China. Int. J. Food Microbiol. 417, 110708. doi: 10.1016/j.ijfoodmicro.2024.110708
Keywords: non-typhoidal Salmonella, S. 4, [5], 12:i:-, S. Enteritidis, S. Typhimurium, blaCTX-M, Typhimurium monophasic variant S. 4
Citation: Wang J, Dong Z-H, Zhou X-Y, Ma Q-C, Wang Z-Y, Lin D, Huang Y-F, Zhang C, Jiao X, Li D and Li Q (2025) Stool carriage of CTX-M/CMY-producing Salmonella enterica in a Chinese tertiary hospital in Shenzhen, China. Front. Cell. Infect. Microbiol. 15:1544757. doi: 10.3389/fcimb.2025.1544757
Received: 13 December 2024; Accepted: 26 February 2025;
Published: 13 March 2025.
Edited by:
Juan Carlos Rodriguez Diaz, Hospital General Universitario de Alicante, SpainReviewed by:
Daniel F. M. Monte, North Carolina State University, United StatesCopyright © 2025 Wang, Dong, Zhou, Ma, Wang, Lin, Huang, Zhang, Jiao, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Deng Li, 294495121@qq.com; Qiuchun Li, qcli@yzu.edu.cn
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.