
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol., 13 March 2025
Sec. Antibiotic Resistance and New Antimicrobial drugs
Volume 15 - 2025 | https://doi.org/10.3389/fcimb.2025.1540967
This article is part of the Research TopicSynergistic Approaches to Managing Gram-negative Bacterial ResistanceView all 15 articles
Background: Klebsiella pneumoniae is one of the main pathogens of nosocomial infection, among which carbapenems can be used for multidrug-resistant Klebsiella pneumoniae. However, in the past decade, the resistance rate of carbapenem-resistant Klebsiella pneumoniae has increased yearly. Tigecycline has good antibacterial activity in treating severe bacterial infections, but the reports of tigecycline resistance are increasing. This study aimed to investigate the mechanism of drug resistance and epidemiological characteristics of tigecycline-resistant Klebsiella pneumoniae (TRKP) in a large teaching hospital in southwest China, Chongqing.
Methods: We isolated 30 TRKP strains from this hospital between August 2021 and December 2023. By PCR and sequencing, we examined the presence and mutation rates of genes associated with tigecycline resistance, including acrR, oqxR, ramR, tmexC, tet(x), tet(A), tet(L), and rpsj, and performed efflux pump inhibition experiments to verify efflux pump activity. At the same time, real-time RT-PCR was used to detect the expression levels of efflux pump genes (acrB and oqxB) and ramA. To investigate the prevalence trend of TRKP in our hospital, we performed multi-site sequence typing (MLST) analysis.
Results: The mutation rates of ramR (73.3%) and tet(A) (63.3%) were significant. In efflux pump inhibition experiments, PaβN could reverse the resistance of 29 TRKP strains (96.7%) to tigecycline. Real-time RT-PCR results showed that acrB and ramA genes were up-regulated in 22 strains, while oqxB genes were overexpressed in only 4 strains. MLST analysis showed that these strains could be divided into 25 different ST subtypes, indicating that no outbreak of TRKP occurred in our hospital. In addition, two tmexCD-torpj positive strains, ST661 and ST1561, were identified for the first time.
Conclusion: The efflux pump acrB and tet(A) mutations are the primary mechanisms of resistance to tigecycline-resistant Klebsiella pneumoniae at our hospital. The ramR mutation can mediate efflux pump activity of acrB by up-regulating ramA overexpression.
With the rise of Klebsiella pneumoniae, a widely distributed pathogen, critical diseases such as endophthalmitis and bloodstream infections have become common (Lee et al., 2006, 2015; Yang et al., 2021). Carbapenems are often relied on for treating multidrug-resistant (MDR) infections; however, their overuse has resulted in the development of carbapenem-resistant Klebsiella pneumoniae (CRKP). Treatments available are increasingly limited, with only last-resort antibiotics like colistin and tigecycline remaining (Sheu et al., 2019). As the first glycylcycline antibiotic, it is a rare effective treatment for difficult-to-treat conditions, particularly those caused by CRKP (Seifert et al., 2018). Like tetracyclines, tigecycline binds reversibly to the 30S ribosomal subunit, disrupting aminoacyl-tRNA function and inhibiting bacterial translation (Pournaras et al., 2016). Tigecycline-resistant strains of K. pneumoniae have emerged rapidly since the clinical use of tigecycline, a problem likely exacerbated by the overuse of antibiotics, which has the potential to complicate treatment and pose a significant public health risk.
The current resistance mechanism of tigecycline is mainly related to the overexpression of RND efflux pumps, including AcrAB, OqxAB, MexAB-OprM, and Tmexd-toprJ (Pournaras et al., 2016; Chen et al., 2017; Lv et al., 2020; Avakh et al., 2023). The AcrAB-TolC pump is driven by the global transcriptional activator RamA and local inhibitory factor AcrR (Villa et al., 2014). The presence of a mutant in ramR, a local inhibitor of ramA, leads to elevated ramA expression and dysregulation of AcrAB expression, which ultimately leads to tigecycline resistance (Xu et al., 2021). Similarly, inactivation of OqxR enhances OqxAB transcription (Wan Nur Ismah et al., 2018). Furthermore, it has been demonstrated that mutations such as V57L in the rpsj gene can contribute to resistance even in the absence of ramR mutations (Herrera et al., 2021). The tet protein also has several known tigecycline resistance mechanisms, including tet(A), tet(L), tet(X), and tet(M) (Fiedler et al., 2016; Fan et al., 2024; Zou et al., 2024). Among them, mutations in the ribosome protection protein tet(M) can modify drug resistance by changing the binding site. Tetracycline mobile inactivating enzyme tet(X) and its variants can exist on a variety of mobile genetic elements, mediate the rapid spread of tigecycline resistance genes through horizontal transfer, and exist stably in drug-resistant strains at a very low adaptive cost, and significantly increase the level of resistance to tigecycline (Song et al., 2020; Hsieh et al., 2021).
The growing tigecycline resistance has significantly limited clinical treatment options for multidrug-resistant K. pneumoniae. Hence, it is crucial to examine TRKP isolates, particularly in regions like Southwest China, where data is lacking. This study aimed to evaluate the phenotypic characteristics, molecular prevalence and tigecycline resistance mechanism of TRKP isolates from southwest China. In brief, a drug susceptibility test was performed on clinically isolated K. pneumoniae, and the genetic relationship of TRKP isolates was studied using multi-site sequence typing (MLST) technique. At the same time, efflux pump inhibition assay was performed to verify efflux pump activity. Using PCR, DNA sequencing technology and reverse transcription PCR (RT-PCR), the determinants of drug resistance including tigecycline resistance genes, pump genes and their regulatory factors were studied.
Between August 2021 and December 2023, we used the Vitek-2 system (Biomérieux, France) to isolate TRKP strains from southwest China. Through the MALDI-TOF mass spectrometry (Biomérieux, Craponne, France), all separate strains are recognized as K. pneumonia. The minimum inhibitory concentration (MIC) was determined with cation-regulated Mueller Hinton broth (CAMHB), and Escherichia coli ATCC 25922 was used as the control strain. Since CLSI has not yet determined the breakpoint of tigecycline, this study referred to FDA’s sensitivity guidelines for Enterobacteria (sensitivity ≤2mg/L, intermediate 4mg/L, resistance ≥ 8 mg/L) (Zheng et al., 2018). Following CLSI-2023 recommendations, the VITEK-2 system was used to carry out further susceptibility to antimicrobial investigation, and the outcomes were interpreted appropriately.
As indicated in Table 1, PCR was performed using gene-specific primers to test TRKP clinical isolates for the tigecycline resistance determinants acrR, ramR, rpsj, oqxR, tet(L), tet(A), tmexC and tet(X). In order to provide A reliable comparison basis after sequencing, so as to accurately identify and locate the mutation sites generated by our experiment, the sequences were compared to those of wild-type reference strains E. coli plasmid RP1 for tet(A) detection (GenBank accession number X00006)] and [K. pneumoniae MGH78578 (GenBank accession number CP000647) for other detection to show the mutations. A total of 35 cycles of PCR reaction conditions were as follows: predenaturation at 94°C for 5 minutes; denaturation at 94°C for 30 seconds, annealing at 56°C for 30 seconds, and extension at 72°C for 40 seconds; and finally, extension at 72°C for 5 minutes.
Utilizing the efflux pump inhibitor (EPI) Phe-Arg-β-naphthylamide (PAβN, MedChemExpress), the efflux pump activity in isolates of K. pneumoniae resistant to tigecycline was investigated. The MIC of tigecycline both with and without PAβN at a concentration of 50 mg/l was determined using the broth microdilution technique. When EPIs are present, a quadruple or higher decline in the MIC is deemed to be evidence of efflux pump efficiency.
Using qRT-PCR, the expression levels of the transcriptional regulator genes ramA and the efflux pump genes acrB and oqxB were evaluated. As directed by the manufacturer, total RNA from bacteria has been extracted using the RNAprep Pure Cell/Bacteria Kit (Tiangen, Beijing, China). The PrimeScriptTM FAST RT Reagent Kit with gDNA Eraser (TaKaRa, Kyoto, Japan) was subsequently utilized to synthesize cDNA. Each sample was analyzed in triplicate. Normalization of the target gene’s mRNA expression was done using the housekeeping gene, rpob. The relative expression level of tigecycline-sensitive bacteria ATCC 25922 was measured as a negative control. Cycle threshold (Ct) values were measured by the qRT-PCR program, and analysis of results was carried out using the 2-ΔΔCt method.
MLST has been used to analyze the genetic relationships of the strains. We download primers from PubMLST website (http://www.pasteur.fr/recherche/genopole/PF8 MLST/Kpneumoniae HTML) for seven housekeeping genes (gapA, infB, mdh, pgi, phoE, rpoB, and tonB), amplify and sequence the genes, and then analyze them using the MLST database. The analysis utilized the PubMLST site (http://www.pasteur.fr/recherche/genopole/PF8 MLST/Kpneumoniae HTML), 7 housekeeping gene synthesis listed on the website of the primer. The genes were amplified and sequenced, with subsequent analysis performed using the MLST database (Veleba et al., 2012). Sequence types (STs) were classified into clonal complexes (CCs) using the eBURST algorithm.
Statistical methods GraphPad Prism software V. 9.5.0 (GraphPad Software Inc, San Diego, CA) was used to statistically analyze the correlation between anti-tigecycline resistance and acrB, oqxB, and ramA expression levels. The calculation of gene expression differences between the groups was based on the Mann–Whitney U test and a P value less than 0.05 was considered statistically significant.
Analysis of specimen sources showed that most TRKP isolates were derived from urine (30.0%) and respiratory secretions (43.4%), of which sputum accounted for 36.7% and bronchoalveolar lavage fluid accounted for 6.7% (see Table 1). Of these 30 isolates, almost a third of the samples were taken from intensive care units (ICU). The TRKP strain affected 20 cases (66.7%) in males and 10 cases (33.3%) in women, with a median age of 68 years. (Table 1) All clinical specimens have been identified as K. pneumoniae by Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (bioMérieux, Marcy-l’Étoile, France).
In this study, the tigecycline drug sensitivity and related characteristics of TRKP clinical isolates were detected (Table 2). Based on FDA guidelines, isolates with tigecycline MICs ≥8 μg/ml were classified as resistant. Among the 30 isolates, 50% had MICs of 8 μg/ml, 30% had MICs of 16 μg/ml, and 20% had MICs of 32 μg/ml. All TRKP isolates were resistant to minocycline. These isolates were also co-resistant ciprofloxacin (25/30, 83.4%), levofloxacin (21/30,70.0%), cefoperazone (19/30, 63.4%), cefepime (19/30, 63.4%), ceftriaxone (18/30, 60.0%), aztreonam (13/30, 43.3%), Ceftazidme (12/30, 40.0%), cefoxitin (11/30, 36.7%), gentamicin (5/30, 16.7%), and amikacin (4/30, 13.4%) were also co-resistant, suggesting that the majority of the strains were MDR. However, no colistin and meropenem resistant isolates were found. We conducted efflux pump inhibition experiments to investigate the mechanism of tigecycline resistance in K. pneumoniae. After exposure to efflux pump inhibitor (EPI) phenylalanine-arginine-β-naphthylamide (PAβN), 96.6% of 30 tigecycline-resistant strains (MIC≥8 mg/L) recovered their sensitivity. Among the 30 tigecycline-resistant isolates (MIC ≥8 mg/L), one isolate exhibited a 32-fold reduction, 11 isolates had a 16-fold reduction, 15 showed an eightfold reduction, two isolates showed a fourfold reduction, and one isolate’s tigecycline MIC remained unchanged With PaβN. The effects of PaβN on mic of tigecycline are shown in Table 2.
To explore the mechanisms underlying tigecycline resistance in TRKP, we identified potential tigecycline resistance determinants through PCR and sequencing, specifically including ramR, acrR, oqxR, tet(A), tet(X), tet(L), tmexC, and rpsj (Table 2). Using published primers (Supplementary Table S1), rpsj gene was detected in all isolates. oqxR and acrR were detected in 26 isolates (86.6%), ramR was detected in 25 isolates (83.3%), and tet(A) was detected in 19 isolates (63.3%). No strains carrying tet(X) and tet(L) were detected(Table 2), Sequence alignment diagrams containing protein mutations are shown in Supplementary Figures S1-S4.
In comparison with the standard WT strain MHG78578 (GenBank number CP000647), 22 isolates (22/30,73.3%) showed nucleotide changes in ramR. Five isolates (5/22, 22.7%) had a deletion of ramR gene, and 17 isolates (17/22, 77.3%) carried frameshift or substitution mutations in ramR, most of the substitutions exist in the DNA-binding domain. Among the more common ramR mutations are A19V substitution, K63M substitution, and I141T substitution (Table 1). Seven isolates produced truncated RamR proteins, including TR17 (181 amino acids [aa]), TR7 (121 aa), TR5 (110 aa), TR28 (98 aa), TR23 (42 aa), TR24 (40 aa) and TR19 (19 aa) (Figure 1).
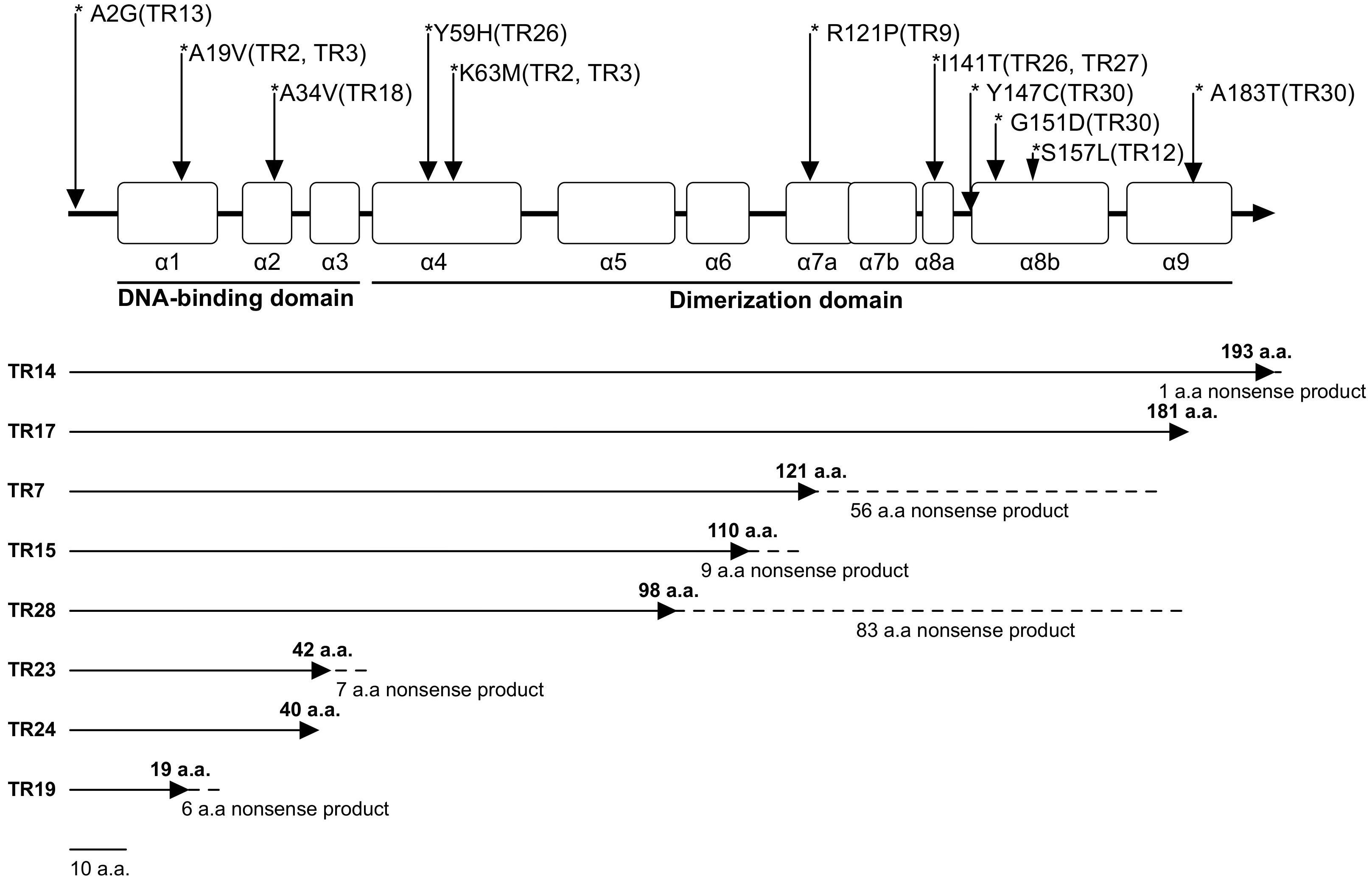
Figure 1. RamR protein mutations in TRKP clinical isolates. The open boxes indicate the nine α-helices of RamR (α1–α9). The reference sequence of K. pneumoniae MGH78578 (CP000647) was used to identify RamR mutations. Solid lines indicate efficiently transcribed RamR proteins and dashed lines indicate inefficiently transcribed RamR proteins.
The tet(A) variant is a major cause of tigecycline resistance: type 1 (n=17) and type 3 (n=2). Type 3 tet(A) showed a single amino acid difference from type 1 and a 28-bp nucleotide deletion.
The amino acid substitution involved in two acrR mutants (S215P, Y114F, V165I in TR6 and E200V in TR18) has not been previously reported. Of the three oqxR mutant isolates (E24R in TR1, C100Y in TR19, and 226-227 base deletion in TR29), E24R mutation in TR1 isolates did not lead to overexpression of oqxB. Recently, mutations in the rpsj gene encoding the ribosomal protein S10 have been reported to be associated with tigecycline resistance in Klebsiella pneumoniae. Although rpsj gene was present in all 30 TRKP isolates in our study, no rpsj gene mutation was found. The tmexC gene was detected in 2 TRKP isolates. The ST1561 TmexCD-ToprJ-positive TRKP strain did not use tigecycline during treatment, and eventually the patient recovered and was discharged successfully. Resistance gene screening revealed the presence of type 1 tet(A) mutants in this strain, along with A2G mutations in ramR, showing expression of rpsj and oqxr genes, but not acrR. In contrast, the ST661 strain was treated with tigecycline, but the patient sadly died. The strain expresses type 1 tet(A) mutant, rpsj, ramR and acrR, but does not express oqxr. Additionally, no mobilized tigecycline resistance genes, such as tet(X), were detected.
We evaluated expression levels of efflux pump AcrAB-TolC, OqxAB, and transcriptional regulatory gene ramA. The qRT-PCR analysis showed that 22 of the 30 Tigecycline-resistant strains had overexpression of acrB gene (5.49- to 48.49- fold) and ramA gene (5.20-83.93 fold). However, in the OqxAB efflux pump pathway, only 4 strains overexpressed the oqxB gene (9.02- to 60.07- fold) (Figures 2A-C). TRKP collected was divided into three groups according to MIC value (MIC=8ug/ml, MIC=16ug/ml, MIC=32ug/ml). Compared with the group with MIC of 8ug/ml, acrB was statistically significant when MIC was 32 groups(p 0.0302). There were significant differences in ramA when MIC was 16 (p 0.0248) (Figure 2D).
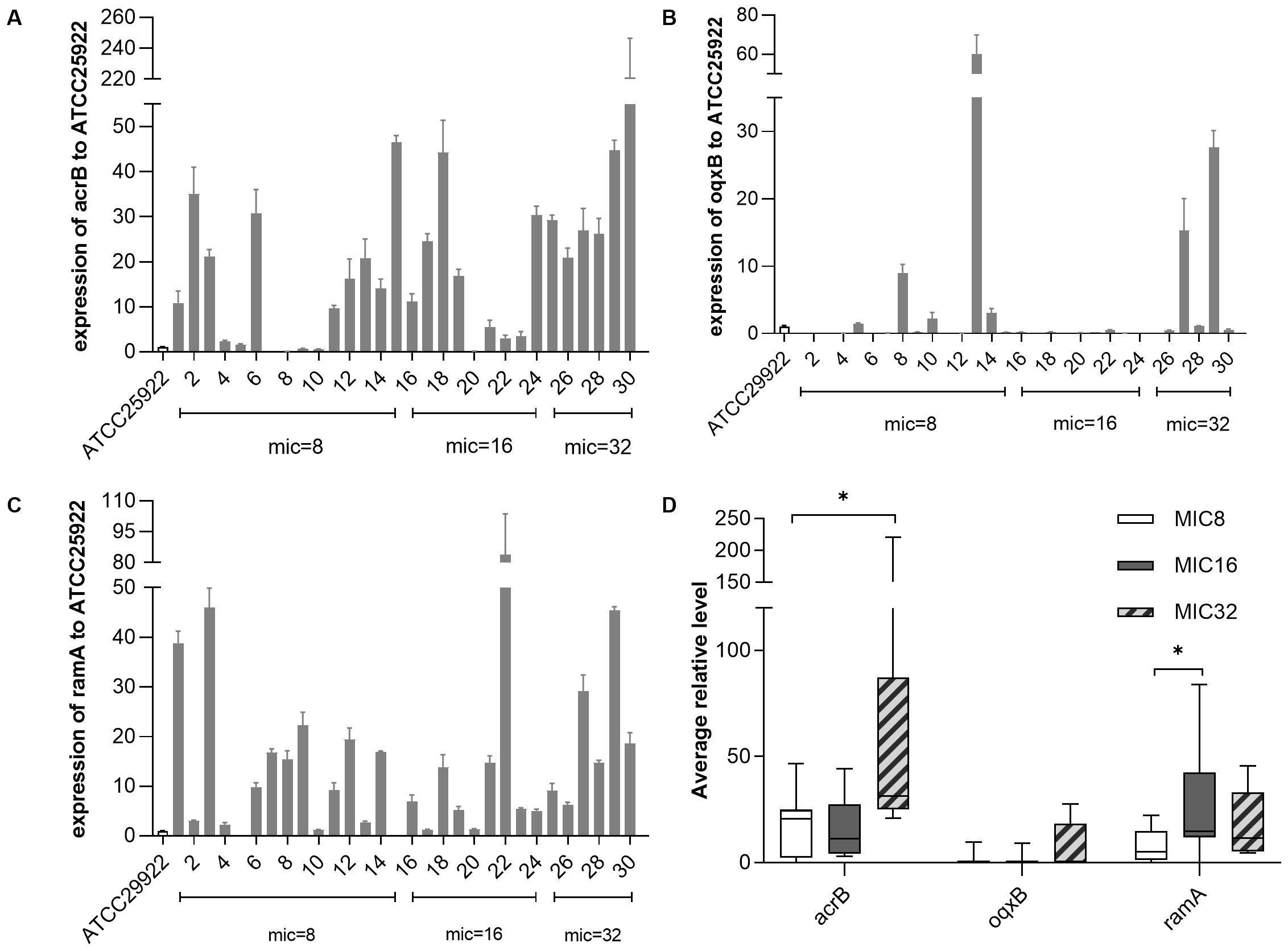
Figure 2. The expression levels of resistance-nodulation-cell division (RND) efflux pump gene and ramA gene in Klebsiella pneumonia. The expression levels of resistance-nodulation-cell division (RND) efflux pump gene and ramA gene in K. pneumoniae were determined by qPCR and the target gene expression levels were divided into three groups according to the MIC value of tigecycline (=8 mg/L, = 16mg/L, = 32mg/L) for comparison, with tigecycline-susceptible Escherichia coli ATCC 25922 as control (expression = 1). (A) Relative expression (RE) level of acrB in TRKP strains; (B) RE level of oqxB in TRKP strains; (C) RE level of ramR in TRKP strains; (D)Average RE of acrB, oqxB, and ramA in TRKP isolates treated with different tigecycline MIC values. The bars represent the average value and the error bars represent the standard error of the mean value. Data were analyzed by Mann–Whitney U test (*P <0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
Twenty-five different STs were observed in 30 TRKP isolates. ST307 was the dominant strain, accounting for 4 strains (11.8%), followed by ST15 type, accounting for 2 strains (6.7%). No novel ST types were detected in this study.
The majority of TRKP strains in our hospital come from older adults and those in the ICU with catheters. Severe pneumonia caused by TRKP often results in extended hospital stays (3-370 days) and poor outcomes, with a 36.7% mortality rate. K. pneumoniae comes from a variety of specimens and can cause infections of the lungs, urinary tract, tissues, bile, and ducts. Antimicrobial susceptibility tests show that TRKP isolates exhibit high resistance to most antibiotics. The increased MIC of tigecycline may be linked to the use of other antibiotics, which are also expelled via the AcrAB-TolC pump (Roy et al., 2013). In this study, 80% of patients had no prior exposure to tigecycline, confirming that resistance can occur without direct exposure. Efflux pump inhibitors (EPIs) down-regulate their expression by interfering with the combination of efflux pump-related proteins, blocking the energy supply effect, and inhibiting the substrate’s passage through efflux pump channels [16]. To detect the presence of overexpression of efflux pumps in TRKP strains, we used efflux pump inhibitor (EPI) PAβN to evaluate efflux pump activity. In our study, 96.6% of TRKP strains showed a fourfold or greater reduction when PAβN was present. The MIC of tigecycline remained unchanged when PAβN was present in TR23. We believe that the following factors may be involved: First, changes in membrane permeability, such as mutations of ompK35 and ompK36 genes, may lead to decreased permeability of the outer membrane to tigecycline; Second, in the presence of PAβN, other efflux pumps may still expel tigecycline. Finally, the metabolic status of bacteria may also affect their susceptibility to drugs. We plan to further explore these resistance mechanisms through whole genome sequencing. However, most of the existing efflux pump inhibitors have obvious toxicity, and how to develop clinical drugs with high specificity, low toxicity, and high safety remains to be further explored.
Tet (A) belongs to the MFS efflux pump family with mutations that allow tigecycline to accumulate within bacterial cells leading to resistance (Linkevicius et al., 2016). In 2017, Chiu et al. first discovered that type 1 tet(A) raised the MIC of tigecycline by a factor of 8. It was also demonstrated that in the case of tet(A) mutation, loss of RamR protein has a synergistic effect on tigecycline resistance in K. pneumoniae (Chiu et al., 2017). Five years later, Peng et al. demonstrated through cloning experiments that type 3 tet (A) could raise the MIC of tigecycline fourfold (Peng et al., 2022a). In our experiments, we found that almost all isolates carried tet (A) mutations or ramR mutations. No ramR was detected in five of the isolates, suggesting that their ramR gene may have been truncated or deleted, similar to the fully ramR deletion mutant of Klebsiella pneumoniae strain KPBj1 M3 Lev (Bialek-Davenet et al., 2013). Furthermore, the deletion and insertion of various fragments among the seven isolates contributed to the premature appearance of the stop codon. This may result in the loss of the α8-α9 region, disrupt dimerization, and ultimately lead to a loss of function (Yamasaki et al., 2013). Fortunately, A large study has shown that although ramR mutants can enhance bacterial resistance, they also induce an enhanced immune response by modulating the structure of lipid A, thereby reducing the pathogen’s ability to kill in organs and blood (Yu et al., 2024).
Point mutations in the local suppressor oqxR have been shown to cause the OqxAB efflux pump to become overactive, giving the isolates increased virulence and multidrug-resistant characteristics (Bialek-Davenet et al., 2015). Previous studies have shown that amino acid substitution mutations such as V102G and V130A confer resistance to tigecycline (Bialek-Davenet et al., 2015; Chiu et al., 2017). This paper identifies for the first time two additional amino acid substitutions (E24R and C100Y) and frameshift deletions involving amino acids 226 to 227, which may negatively impact function. Therefore, the effect of these mutations on drug resistance requires further investigation. The recent discovery of the novel tetracycline-inactivating enzyme tet(X) homologs and the efflux pump gene clusters Tmexd-toprj has been linked to high levels of tigecycline resistance. These resistance mechanisms can be horizontally transferred via mobile elements like plasmids, spreading to humans, animals, and the environment, and posing a significant public health threat (Guo et al., 2024; Pan et al., 2024). A report identified 237 bacterial strains worldwide carrying the tmexCD-toprJ gene, with 92.83% originating from China (Dong et al., 2022). These strains represent 50 unique sequence types. Our study marks the first identification of two novel tmexCD-toprJ-positive strains, ST661 and ST1561, offering fresh insights into microbial research and underscoring the significance of bacterial diversity alongside its vast research potential. Tmexcd-positive strains were previously found mainly on chromosomes, but the discovery of plasmids in the past five years suggests a possible transmission mechanism. Despite attempts to perform conjugation experiments to determine whether tmexC is located on the plasmid and its ability to transfer, they were unsuccessful, suggesting that they may indeed be located on chromosomes. To further test this hypothesis, whole genome sequencing is planned to confirm the specific location and function of tmexCD-toprJ. In addition, patients infected with ST1561 were observed to survive, while those infected with ST661 died, a difference that may reflect significant differences in the pathogenicity and virulence of the strains. rpsj mutations linked to tigecycline resistance are generally found in the amino acids located between positions 53 and 60 of the S10 ribosomal protein (Beabout et al., 2015). Fortunately, we did not find any rpsj mutations related to tigecycline in the strains, nor did we detect any cases of tet(X) carrying.
Both AcrAB and OqxAB efflux pumps are common resistance mechanisms in enterobacteria (Liu et al., 2018). Research shows that OqxAB overexpression can lead to resistance to several antimicrobials, including chloramphenicol, quinolones, furantoin, and tigecycline (Li et al., 2019). In our study, acrB expression increased with varying MIC levels, while oqxB was overexpressed in only 4 bacterial strains. This aligns with findings by Perez et al., showing that while OqxAB is widely distributed in K. pneumoniae, it is not always overexpressed (Perez et al., 2013). It is important to note that the ramR gene was absent from the highly resistant strain TR4 (MIC 8µg/ml), and efflux pump inhibition tests demonstrated that the resistance mechanism was linked to the efflux pump even though acrB expression was only 2.32 times higher. Furthermore, no known resistance determinants were discovered, indicating that resistance may emerge through other pathways such as KpgABC or MacAB-TolC efflux pumps. Mutation of ramR, as an inhibitor of ramA, leads to up-regulation of ramA expression, which in turn increases the expression of acrAB or oqxAB efflux pump. In this study, three isolates (TR10, TR16, TR25) showed elevated ramA expression without any ramR mutations. This may be linked to mutations in the RamR recognition sites (PI and PII promoters) or Lon protease mutations (Rosenblum et al., 2011; Ricci et al., 2014), requiring further investigation to clarify this finding. Although it has been established that efflux mechanisms generally only lead to low levels of resistance to tigecycline, we speculate that multiple mechanisms may be at work at the same time, with possible synergistic effects. Therefore, more in-depth research is urgently needed to explore the interrelationships and effects of these mechanisms.
Additionally, 30 K. pneumoniae isolates were classified into 25 distinct ST types, reflecting the significant genetic diversity of TRKP. Previous research has indicated that most TRKP strains are resistant to multiple antibiotics and carry virulence factors, heightening the risk of resistance and virulence gene transfer. Monitoring the spread of these clones is essential to prevent their emergence as clinical pathogens (Peng et al., 2022a).
This study’s limitation is its single-center scope, which may limit representativeness. Additionally, only certain resistance genes were screened, leaving other mechanisms unexplored. Future efforts will expand sample size through multi-center studies and broaden genetic screening to better understand resistance mechanisms and enhance the study’s clinical relevance. In conclusion, acrB overexpression and tet(A) mutations are key contributors to tigecycline resistance in K. pneumoniae in southwest China.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
The study protocol was approved by the Ethics Committee of The First Affiliated Hospital of Chongqing Medical University (2024-373-01).
YL: Writing – original draft, Writing – review & editing, Formal Analysis. ST: Writing – review & editing. QH: Supervision, Writing – review & editing. PX: Supervision, Writing – review & editing. TS: Supervision, Writing – review & editing. YS: Supervision, Writing – review & editing. YX: Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1540967/full#supplementary-material
Avakh, A., Grant, G. D., Cheesman, M. J., Kalkundri, T., Hall, S. (2023). The art of war with pseudomonas aeruginosa: targeting mex efflux pumps directly to strategically enhance antipseudomonal drug efficacy. Antibiotics (Basel) 12, 1304. doi: 10.3390/antibiotics12081304
Beabout, K., Hammerstrom, T. G., Perez, A. M., Magalhães, B. F., Prater, A. G., Clements, T. P., et al. (2015). The ribosomal S10 protein is a general target for decreased tigecycline susceptibility. Antimicrob. Agents Chemother. 59, 5561–5566. doi: 10.1128/aac.00547-15
Bialek-Davenet, S., Lavigne, J. P., Guyot, K., Mayer, N., Tournebize, R., Brisse, S., et al. (2015). Differential contribution of AcrAB and OqxAB efflux pumps to multidrug resistance and virulence in Klebsiella pneumoniae. J. Antimicrob. Chemother. 70, 81–88. doi: 10.1093/jac/dku340
Bialek-Davenet, S., Leflon-Guibout, V., Tran Minh, O., Marcon, E., Moreau, R., Nicolas-Chanoine, M. H. (2013). Complete deletion of the ramR gene in an in vitro-selected mutant of Klebsiella pneumoniae overexpressing the AcrAB efflux pump. Antimicrob. Agents Chemother. 57, 672–673. doi: 10.1128/aac.01410-12
Chen, Y., Hu, D., Zhang, Q., Liao, X. P., Liu, Y. H., Sun, J. (2017). Efflux Pump Overexpression Contributes to Tigecycline Heteroresistance in Salmonella enterica serovar Typhimurium. Front. Cell Infect. Microbiol. 7. doi: 10.3389/fcimb.2017.00037
Chiu, S. K., Huang, L. Y., Chen, H., Tsai, Y. K., Liou, C. H., Lin, J. C., et al. (2017). Roles of ramR and tet(A) Mutations in Conferring Tigecycline Resistance in Carbapenem-Resistant Klebsiella pneumoniae Clinical Isolates. Antimicrob. Agents Chemother. 61, e00391-17. doi: 10.1128/aac.00391-17
Dong, N., Zeng, Y., Wang, Y., Liu, C., Lu, J., Cai, C., et al. (2022). Distribution and spread of the mobilised RND efflux pump gene cluster tmexCD-toprJ in clinical Gram-negative bacteria: a molecular epidemiological study. Lancet Microbe 3, e846–e856. doi: 10.1016/s2666-5247(22)00221-x
Fan, X. Y., Jiang, Y., Wu, H., Liu, J., Gu, Q. Y., Wang, Z. Y., et al. (2024). Distribution and spread of tigecycline resistance gene tet(X4) in Escherichia coli from different sources. Front. Cell Infect. Microbiol. 14. doi: 10.3389/fcimb.2024.1399732
Fiedler, S., Bender, J. K., Klare, I., Halbedel, S., Grohmann, E., Szewzyk, U., et al. (2016). Tigecycline resistance in clinical isolates of Enterococcus faecium is mediated by an upregulation of plasmid-encoded tetracycline determinants tet(L) and tet(M). J. Antimicrob. Chemother. 71, 871–881. doi: 10.1093/jac/dkv420
Guo, H., Li, L., Zhang, Y., Zhang, Y., Song, C., Wu, Y., et al. (2024). Global genomic epidemiology and transmission dynamics of plasmid-borne tmexCD-toprJ-carrying Klebsiella pneumoniae in a one health context. Sci. Total Environ. 953, 176065. doi: 10.1016/j.scitotenv.2024.176065
Herrera, M., Gregorio, S. D., Haim, M. S., Posse, G., Mollerach, M., Di Conza, J. (2021). Genetic changes associated with tigecycline resistance in Staphylococcus aureus in vitro-selected mutants belonging to different lineages. Int. J. Antimicrob. Agents 57, 106304. doi: 10.1016/j.ijantimicag.2021.106304
Hsieh, Y. C., Wu, J. W., Chen, Y. Y., Quyen, T. L. T., Liao, W. C., Li, S. W., et al. (2021). An Outbreak of tet(X6)-Carrying Tigecycline-Resistant Acinetobacter baumannii Isolates with a New Capsular Type at a Hospital in Taiwan. Antibiotics (Basel) 10, 1239. doi: 10.3390/antibiotics10101239
Lee, H. C., Chuang, Y. C., Yu, W. L., Lee, N. Y., Chang, C. M., Ko, N. Y., et al. (2006). Clinical implications of hypermucoviscosity phenotype in Klebsiella pneumoniae isolates: association with invasive syndrome in patients with community-acquired bacteraemia. J. Intern. Med. 259, 606–614. doi: 10.1111/j.1365-2796.2006.01641.x
Lee, W. H., Choi, H. I., Hong, S. W., Kim, K. S., Gho, Y. S., Jeon, S. G. (2015). Vaccination with Klebsiella pneumoniae-derived extracellular vesicles protects against bacteria-induced lethality via both humoral and cellular immunity. Exp. Mol. Med. 47, e183. doi: 10.1038/emm.2015.59
Li, J., Zhang, H., Ning, J., Sajid, A., Cheng, G., Yuan, Z., et al. (2019). The nature and epidemiology of OqxAB, a multidrug efflux pump. Antimicrob. Resist. Infect. Control 8, 44. doi: 10.1186/s13756-019-0489-3
Linkevicius, M., Sandegren, L., Andersson, D. I. (2016). Potential of tetracycline resistance proteins to evolve tigecycline resistance. Antimicrob. Agents Chemother. 60, 789–796. doi: 10.1128/aac.02465-15
Liu, B., Wu, H., Zhai, Y., He, Z., Sun, H., Cai, T., et al. (2018). Prevalence and molecular characterization of oqxAB in clinical Escherichia coli isolates from companion animals and humans in Henan Province, China. Antimicrob. Resist. Infect. Control 7, 18. doi: 10.1186/s13756-018-0310-8
Lv, L., Wan, M., Wang, C., Gao, X., Yang, Q., Partridge, S. R., et al. (2020). Emergence of a plasmid-encoded resistance-nodulation-division efflux pump conferring resistance to multiple drugs, including tigecycline, in klebsiella pneumoniae. mBio 11, e02930-19. doi: 10.1128/mBio.02930-19
Pan, Y., Zeng, J., Zhang, L., Hu, J., Hao, H., Zeng, Z., et al. (2024). The fate of antibiotics and antibiotic resistance genes in Large-Scale chicken farm Environments: Preliminary view of the performance of National veterinary Antimicrobial use reduction Action in Guangdong, China. Environ. Int. 191, 108974. doi: 10.1016/j.envint.2024.108974
Peng, K., Wang, Q., Li, Y., Wang, M., Kurekci, C., Li, R., et al. (2022a). Molecular mechanisms and genomic basis of tigecycline-resistant Enterobacterales from swine slaughterhouses. Microbiological Res. 264, 127151. doi: 10.1016/j.micres.2022.127151
Perez, F., Rudin, S. D., Marshall, S. H., Coakley, P., Chen, L., Kreiswirth, B. N., et al. (2013). OqxAB, a quinolone and olaquindox efflux pump, is widely distributed among multidrug-resistant Klebsiella pneumoniae isolates of human origin. Antimicrob. Agents Chemother. 57, 4602–4603. doi: 10.1128/aac.00725-13
Pournaras, S., Koumaki, V., Spanakis, N., Gennimata, V., Tsakris, A. (2016). Current perspectives on tigecycline resistance in Enterobacteriaceae: susceptibility testing issues and mechanisms of resistance. Int. J. Antimicrob. Agents 48, 11–18. doi: 10.1016/j.ijantimicag.2016.04.017
Ricci, V., Blair, J. M., Piddock, L. J. (2014). RamA, which controls expression of the MDR efflux pump AcrAB-TolC, is regulated by the Lon protease. J. Antimicrob. Chemother. 69, 643–650. doi: 10.1093/jac/dkt432
Rosenblum, R., Khan, E., Gonzalez, G., Hasan, R., Schneiders, T. (2011). Genetic regulation of the ramA locus and its expression in clinical isolates of Klebsiella pneumoniae. Int. J. Antimicrob. Agents 38, 39–45. doi: 10.1016/j.ijantimicag.2011.02.012
Roy, S., Datta, S., Viswanathan, R., Singh, A. K., Basu, S. (2013). Tigecycline susceptibility in Klebsiella pneumoniae and Escherichia coli causing neonatal septicaemia, (2007-10) and role of an efflux pump in tigecycline non-susceptibility. J. Antimicrob. Chemother. 68, 1036–1042. doi: 10.1093/jac/dks535
Seifert, H., Blondeau, J., Dowzicky, M. J. (2018). In vitro activity of tigecycline and comparators, (2014-2016) among key WHO 'priority pathogens' and longitudinal assessment, (2004-2016) of antimicrobial resistance: a report from the T.E.S.T. study. Int. J. Antimicrob. Agents 52, 474–484. doi: 10.1016/j.ijantimicag.2018.07.003
Sheu, C. C., Chang, Y. T., Lin, S. Y., Chen, Y. H., Hsueh, P. R. (2019). Infections caused by carbapenem-resistant enterobacteriaceae: an update on therapeutic options. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00080
Song, H., Liu, D., Li, R., Fu, Y., Zhai, W., Liu, X., et al. (2020). Polymorphism Existence of Mobile Tigecycline Resistance Gene tet(X4) in Escherichia coli. Antimicrob. Agents Chemother. 64, e01825-19. doi: 10.1128/aac.01825-19
Veleba, M., Higgins, P. G., Gonzalez, G., Seifert, H., Schneiders, T. (2012). Characterization of RarA, a novel AraC family multidrug resistance regulator in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 56, 4450–4458. doi: 10.1128/aac.00456-12
Villa, L., Feudi, C., Fortini, D., García-Fernández, A., Carattoli, A. (2014). Genomics of KPC-producing Klebsiella pneumoniae sequence type 512 clone highlights the role of RamR and ribosomal S10 protein mutations in conferring tigecycline resistance. Antimicrob. Agents Chemother. 58, 1707–1712. doi: 10.1128/aac.01803-13
Wan Nur Ismah, W. A. K., Takebayashi, Y., Findlay, J., Heesom, K. J., Avison, M. B. (2018). Impact of OqxR loss of function on the envelope proteome of Klebsiella pneumoniae and susceptibility to antimicrobials. J. Antimicrob. Chemother. 73, 2990–2996. doi: 10.1093/jac/dky293
Xu, Q., Sheng, Z., Hao, M., Jiang, J., Ye, M., Chen, Y., et al. (2021). RamA upregulates multidrug resistance efflux pumps AcrAB and OqxAB in Klebsiella pneumoniae. Int. J. Antimicrob. Agents 57, 106251. doi: 10.1016/j.ijantimicag.2020.106251
Yamasaki, S., Nikaido, E., Nakashima, R., Sakurai, K., Fujiwara, D., Fujii, I., et al. (2013). The crystal structure of multidrug-resistance regulator RamR with multiple drugs. Nat. Commun. 4, 2078. doi: 10.1038/ncomms3078
Yang, Y., Yang, Y., Chen, G., Lin, M., Chen, Y., He, R., et al. (2021). Molecular characterization of carbapenem-resistant and virulent plasmids in Klebsiella pneumoniae from patients with bloodstream infections in China. Emerg. Microbes Infect. 10, 700–709. doi: 10.1080/22221751.2021.1906163
Yu, W.-T., Jia, P., Chu, X., Li, S., Jia, X., Zhu, Y., et al. (2024). Dual role of ramR mutation in enhancing immune activation and elevating eravacycline resistance in Klebsiella pneumoniae. iMetaOmics. 1, e39. doi: 10.1002/imo2.v1.2
Zheng, J. X., Lin, Z. W., Sun, X., Lin, W. H., Chen, Z., Wu, Y., et al. (2018). Overexpression of OqxAB and MacAB efflux pumps contributes to eravacycline resistance and heteroresistance in clinical isolates of Klebsiella pneumoniae. Emerg. Microbes Infect. 7, 139. doi: 10.1038/s41426-018-0141-y
Keywords: resistance mechanism, Klebsiella pneumoniae, tigecycline-resistance, tet(A), RND efflux pump
Citation: Li Y, Tang S, Han Q, Xia P, Si T, Song Y and Xia Y (2025) The investigation of molecular epidemiological characteristics and resistance mechanism of tigecycline resistant Klebsiella pneumoniae from a large teaching hospital in southwest China, Chongqing. Front. Cell. Infect. Microbiol. 15:1540967. doi: 10.3389/fcimb.2025.1540967
Received: 06 December 2024; Accepted: 21 February 2025;
Published: 13 March 2025.
Edited by:
Percy Schröttner, Technische Universität Dresden, GermanyReviewed by:
Ariadnna Cruz-Córdova, Federico Gómez Children’s Hospital, MexicoCopyright © 2025 Li, Tang, Han, Xia, Si, Song and Xia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yun Xia, eGlheXVuMTJjbkBhbGl5dW4uY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.