- Department of Infectious Diseases and Clinical Microbiology, Beijing Institute of Respiratory Medicine and Beijing Chao-Yang Hospital, Capital Medical University, Beijing, China
Introduction: Vancomycin-resistant Enterococcus faecium (VRE-fm) biofilms pose a significant clinical challenge due to the limited effectiveness of traditional antibiotics. This study investigates the potential of γ-linolenic acid (GLA) as a novel antibiofilm agent.
Methods: Transcriptome analysis was performed on the V27 isolate, comparing cells in mature biofilms treated with and without GLA. The findings were further validated using qRT-PCR on six VRE-fm isolates and two E. faecalis isolates.
Results: Transcriptome analysis revealed a significant downregulation in the expression levels of genes associated with biofilm formation, including fruA, fruB, sgrA, lpxtg-cwa, tfpp, lafA, lafB, malP, fsrA, and fsrC’, while a significant upregulation was observed in the expression of fsrBD. Validation by qRT-PCR in six VRE-fm isolates confirmed the significant changes in the expression levels of all genes except for lpxtg-cwa, with statistical significance. The expression of bgsB and bgsA genes, which are the homologs of lafA and lafB genes, along with the Fsr-regulated genes gelE and sprE in E. faecalis, were also found to be downregulated by GLA. In addition, KEGG analysis identified specific metabolic pathways that were significantly downregulated by GLA.
Conclusion: GLA effectively targets multiple aspects of biofilm formation in VRE-fm, including the downregulation of key biofilm-related genes, the inhibition of quorum sensing systems, and the modulation of metabolic pathways. GLA emerges as a promising candidate for eradicating Enterococcus biofilms.
1 Introduction
Vancomycin-resistant Enterococcus faecium (VRE-fm) has become a major healthcare-associated pathogen, posing a significant threat due to its resistance to multiple antibiotics and the ability of biofilm formation (Wei et al., 2024). Extracellular polymeric substances (EPS) in biofilms provide structural integrity, protect them from environmental stresses, and facilitate communication between bacterial cells (Ch'ng et al., 2019). Biofilms promote E. faecium to form persisters and become viable but nonculturable (VBNC) (Lleò et al., 2005; Ayrapetyan et al., 2018). Biofilm formation not only increases the resistance of traditional antibiotics, but also is a critical enterococcal virulence factor, such as for infective endocarditis and urinary tract infection (Ch'ng et al., 2019). Therefore, alternative therapeutic approaches need to be developed against VRE-fm biofilm-associated infections.
Previous studies have shown that unsaturated fatty acids have antibacterial activity (Desbois, 2012; Yoon et al., 2018). However, due to the discovery and development of traditional antibiotics, research on the antibacterial properties of unsaturated fatty acids has been put on hold for a long time. With the increasingly serious problem of traditional antibiotic resistance, unsaturated fatty acids have been reconsidered as antimicrobial agents. Recent studies have shown that unsaturated fatty acids can also inhibit or eradicate biofilms formed by various microbial pathogens (Inoue et al., 2008; Kim et al., 2018; Ramanathan et al., 2018; Cui et al., 2019; Wang et al., 2022; Khan et al., 2023). Therefore, unsaturated fatty acids are very promising next-generation antibacterial agents for the treatment and prevention of biofilm-related infections.
Our previous research has found that essential fatty acids (EFAs) not only inhibit the growth of VRE-fm, but also inhibit its formation of biofilms, and even eradicate its already formed biofilms (Wei et al., 2023). In terms of eradicating biofilms, we found that γ-linolenic acid (GLA) can reduce the expression of the atlA gene, which is responsible for facilitating the release of eDNA (Xie et al., 2022; Wei et al., 2023). However, inhibition of eDNA release alone may not be enough to achieve a biofilm eradication rate of more than 60%, so we believe that there are other mechanisms that need to be further studied.
This study aims to unravel the molecular mechanisms by which GLA eradicates biofilms formed by VRE-fm using RNA sequencing (RNA-seq) technology. By providing a comprehensive snapshot of the transcriptome, RNA-seq can be used to identify genes that are differentially expressed during biofilm eradication by GLA, as well as to provide insights into the regulatory pathways involved (Willett et al., 2019; Tatta et al., 2023). In our preliminary studies of six clinical isolates of VRE-fm, GLA showed the highest biofilm eradication efficacy against isolate V27. Therefore, we selected VRE-fm isolate V27 for subsequent transcriptomic analysis. We hypothesize that GLA exerts its antibiofilm effect by modulating the expression of genes involved in various stages of biofilm formation, including adhesion, EPS production, and cell signaling. Understanding these mechanisms is crucial for developing novel strategies to combat biofilm-associated infections caused by VRE-fm.
2 Materials and methods
2.1 Strains and culture conditions
A total of six clinical VRE-fm isolates (V05, V06, V08, V09, V22, and V27) with moderate biofilm formation ability (Wei et al., 2023), and two E. faecalis isolates (efa105 and efa106) with strong biofilm formation ability were collected from the Department of Infectious Diseases and Clinical Microbiology, Beijing Chao-Yang Hospital, Capital Medical University (Beijing, China). The isolates were identified using matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) (VITEK-MS; bioMeírieux, France; IVD version 3.0). These isolates were refreshed from frozen stocks at -20°C and inoculated twice on Columbia blood agar at 35°C for 24 h before all experiments.
2.2 RNA preparation for sequencing and qRT-PCR assay
The mature biofilms of the six VRE-fm isolates (V05, V06, V08, V09, V22, and V27) were established according to the previous study (Wei et al., 2023). After the biofilms were treated with or without GLA for 24 h, the E. faecium cells were collected by centrifugation (5000 × g for 10 min). Total RNA from the E. faecium cells was extracted using the RNA extraction reagent kit (Tiangen Biotech, Beijing, China) according to the manufacturer′s instructions. The quantity and quality of the isolated RNA samples were determined using a NanoDrop One Spectrophotometer (Thermo Scientific, Waltham, MA, USA). The extracted RNA samples of V27 isolate underwent quality control and library preparation for transcriptome sequencing.
2.3 Clustering and sequencing of the V27 treated with and without GLA samples
TruSeq PE Cluster Kit v3-cBot-HS (Illumia) was used for performing the clustering of the index-coded samples on a cBot Cluster Generation System according to the manufacturer’s instructions. After cluster generation, an Illumina Novaseq platform was used for sequencing the library preparations and 150 bp paired-end reads were generated.
2.4 Data quality control of the V27 treated with and without GLA samples
Raw data of fastq format were processed through in-house perl scripts. Then, clean data were obtained by removing reads containing adapter, low-quality reads, and reads containing N base from raw data. At this step, Q20, Q30, and GC content of the clean data were calculated. All the subsequent analyses were based on high-quality clean data.
2.5 Reads mapping to the reference genome
E. faecium 1,231,408 (GenBank: GCA_000157615.1) was selected as a reference strain in this study. Its genome files were downloaded as the reference genome from the website directly. The Bowtie2-2.2.3 software was used for building an index of the reference genome and aligning clean reads to the reference genome.
2.6 Quantification of the gene expression level of the V27 treated with and without GLA samples
HTSeq v0.6.1 software was used for counting the reads numbers mapped to each gene. And then Fragments Per Kilobase of exon model per Million mapped fragments (FPKM) of each gene was calculated based on the length of the gene and reads count mapped to this gene. FPKM is currently the most common method for estimating gene expression levels.
2.7 Differential expression analysis of the V27 treated with and without GLA samples
The read counts were adjusted by the edgeR program package through one scaling normalized factor for each sequenced library before differential gene expression analysis. DEGSeq R package (1.20.0) was used for differential expression analysis of two conditions. Genes with an edgeR P value < 0.05 and |log2(Fold change)| ≥ 1 were set as the threshold for significantly differential expression.
2.8 GO and KEGG enrichment analysis of differentially expressed genes of the V27 treated with and without GLA samples
Gene Ontology (GO) enrichment analysis of differential expression genes was implemented by the GOseq R package, which was used for correcting the gene length bias. GO IDs with P value < 0.05 were considered significantly enriched by differential expression genes.
KEGG (Kyoto Encyclopedia of Genes and Genomes) is a database resource for understanding high-level functions and utilities of the biological system from molecular-level information, especially large-scale molecular datasets which are generated by high-throughput experimental technologies (http://www.genome.jp/kegg/). The KOBAS software was used for testing the statistical enrichment of differential expression genes of the V27 isolate in KEGG pathways.
2.9 Differential expression genes validation by qRT-PCR assay of the 6 VRE-fm isolates
RNA (~2 μg) from each sample was reverse transcribed for the synthesis of cDNA using SuperScript™ III First-Strand Synthesis SuperMix (Invitrogen, Carlsbad, CA, USA). The PowerUp SYBR Green Master Mix (Applied Biosystems, Life Technologies, Austin, USA) was used for quantitative reverse transcription PCR (qRT-PCR) assay in an Applied Biosystems 7500 real-time PCR system (Applied Biosystems, Waltham, MA, USA). The amplification reactions were carried out in a 20 µL volume including 10 μL of 2× PowerUp SYBR Green Master Mix, 1 μL of forward primer, 1 μL of reverse primer, 6 μL of DNase/RNase-Free water (Tiangen Biotech, Beijing, China), and 2 μL of the synthesized cDNA. The reaction conditions were 95°C for 3 min with 1 cycle, then 40 cycles (95°C for 15 s, and 60°C for 1 min) for DNA template amplification. Gene expression was normalized with the reference gene gdhA using 2−ΔΔCT method (Livak and Schmittgen, 2001). The forward and reverse primers used for the detection are shown in Supplementary Table S1.
Statistical analysis for qRT-PCR assay was performed using the IBM SPSS Statistics 25.0 software program (IBM, Armonk, NY, USA). Data were expressed as means ± standard deviation (SD) of three independent experiments. Student’s t-test was used for statistical comparison between the two groups. The standard F-test was used to test whether two populations had the same variance. A P value < 0.05 was considered statistically significant.
2.10 Evaluation of the efficacy of GLA in eradicating the biofilm of E. faecalis isolates
Biofilm eradication was assessed using two clinical isolates of E. faecalis. Mature biofilms were pre-formed in trypticase soy broth supplemented with 2% glucose (TSBG) for 24 hours. After incubation, non-adherent cells were removed by washing twice with PBS. Subsequently, TSBG containing 1 mM GLA was added to the wells. A control group was treated with fresh TSBG medium. Microtiter plates were incubated at 35°C for an additional 24 hours, and biofilm mass was quantified using the crystal violet staining method. The biofilm eradication rate (%) was calculated using the following formula: Eradication (%) = (ODcontrol - ODsample)/ODcontrol × 100% (Wang et al., 2022).
2.11 Biofilm-related gene expression in E. faecalis isolates analyzed by qRT-PCR
RNA extraction, cDNA synthesis, amplification reaction, and statistical analysis for the two E. faecalis isolates treated with and without GLA were performed using the same method as described above. The primers of efa-gdhA, bgsA, bgsB, efa-malP, gelE, and sprE genes are shown in Supplementary Table S2.
3 Results
3.1 RNA-Seq quality control and analysis
The Illumina sequencing generated a total of 15,199,804 and 15,939,512 reads for the V27 treated with GLA and V27 treated without GLA (control) samples, respectively. After eliminating low-quality reads, reads containing adapter and N base, 14,793,234 and 15,581,624 clean reads were obtained. The PHRED quality scores of Q20 of the filtered reads were 95.30% and 97.69% for the V27 treated with GLA and V27 treated without GLA samples, respectively, which confirmed the presence of high-quality sequencing reads in the transcriptome dataset. The reference genome of E. faecium 1,231,408 (GenBank: GCA_000157615.1) was selected for reference-based assembly of the transcriptome. The RNA-Seq analysis was performed by mapping filtered reads for each sample to the reference genome. The data denoted that 71.97% and 87.77% of the reads were successfully mapped to the reference genome, while 64.01% and 77.47% of reads were uniquely mapped for the V27 treated with GLA and V27 treated without GLA samples, respectively. Post read mapping the transcript quantification was performed to obtain the expression details of the transcripts for each sample.
The number of transcripts with FPKM values in the ranges of 0-1, 1-3, 3-15, 15-60, and >60 was 637 (20.15%) and 529 (16.74%), 143 (4.52%) and 47 (1.49%), 414 (13.10%) and 442 (13.98%), 481 (15.22%) and 641 (20.28%), and 1486 (47.01%) and 1502 (47.52%) for the V27 treated with GLA and V27 treated without GLA samples, respectively. The Venn diagram showed that 2519 (95.53%) genes were co-expressed (Supplementary Figure S1), and the Pearson correlation was 0.879 between the two samples.
3.2 Differential expression genes
Comparison of the transcriptome of the V27 treated with GLA to the V27 treated without GLA samples revealed that a total of 792 genes were being differentially expressed [|log2(fold change)| ≥ 1, edgeR P value < 0.05]. Among these, 263 genes were upregulated, whereas 529 genes were downregulated in the GLA treated V27 sample, which was shown in the volcano plots (Figure 1). The biofilm-associated genes, whose functions involve adhesion, synthesis and release of EPS, as well as those whose functions are unclear but can affect biofilms, were the main targets we were searching for primarily in the RNA seq results.
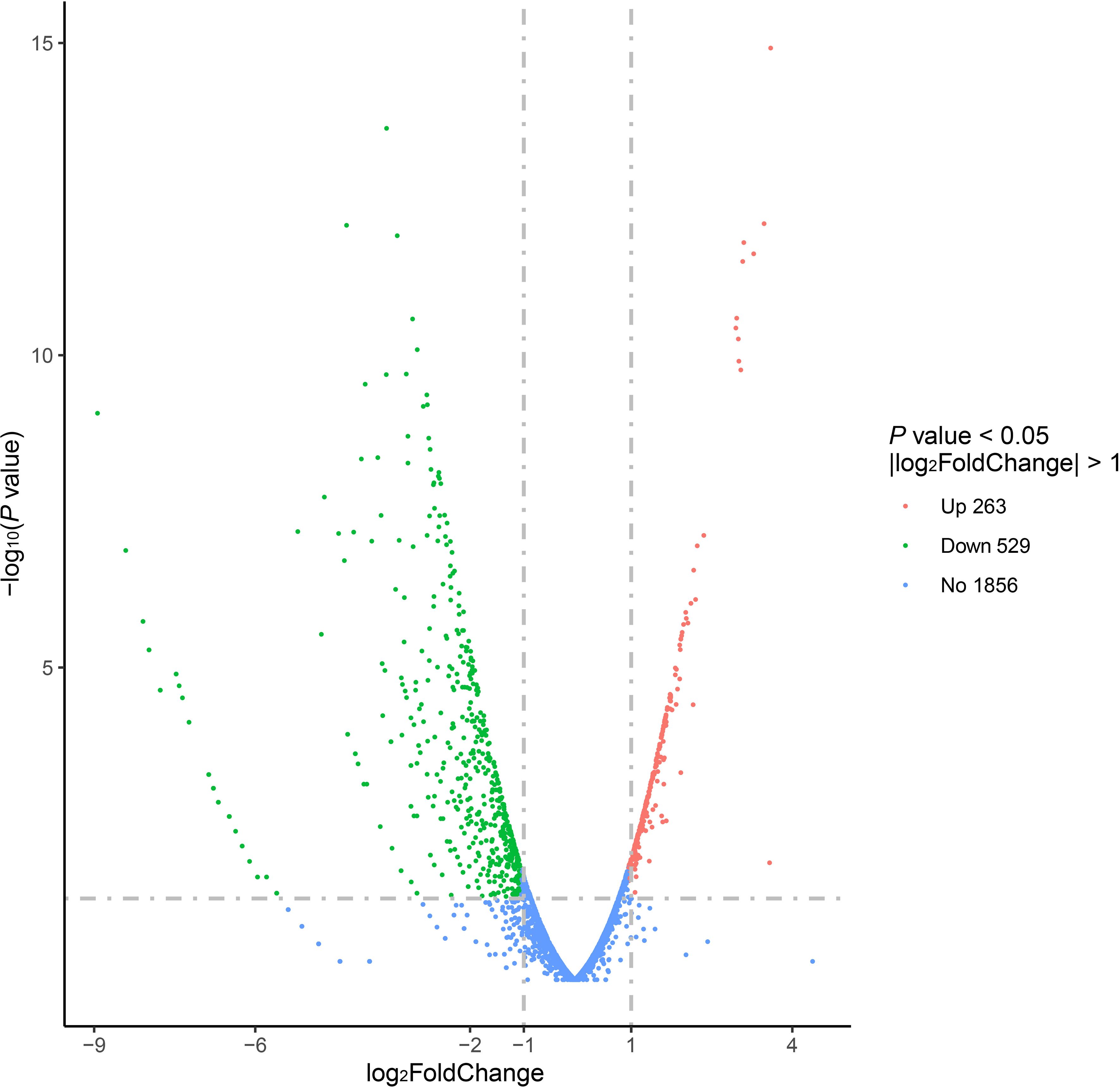
Figure 1. The volcano plots of differential expression genes for the V27 treated with GLA vs. V27 treated without GLA.
The fruA (also known as bepA) gene putatively encodes a carbohydrate phosphotransferase system (PTS) permease (Protein ID: EFF35874.1) in the E. faecium E1162 strain involved in biofilm formation (Paganelli et al., 2016). Using the online software tblastn (https://blast.ncbi.nlm.nih.gov/Blast.cgi) for similarity analysis, the protein showed the highest similarity (93.01%) with the EFUG_01291 gene in the reference genome (GenBank: GCA_000157615.1). RNA-seq showed a significant downregulation of the expression level of EFUG_01291 gene (Table 1) in the V27 treated with GLA sample. In addition, the expression level of the EFUG_01290 gene, which was 100% similar to FruB protein (Protein ID: EFF35875.1) in the E. faecium strain E1162, was also significant downregulation. The fruB gene has been reported to contribute to the fitness and virulence of Streptococcus mutans in human dental biofilms (Chakraborty et al., 2022). We have designated EFUG_01291 and EFUG_01290 genes as fruA and fruB genes, respectively.
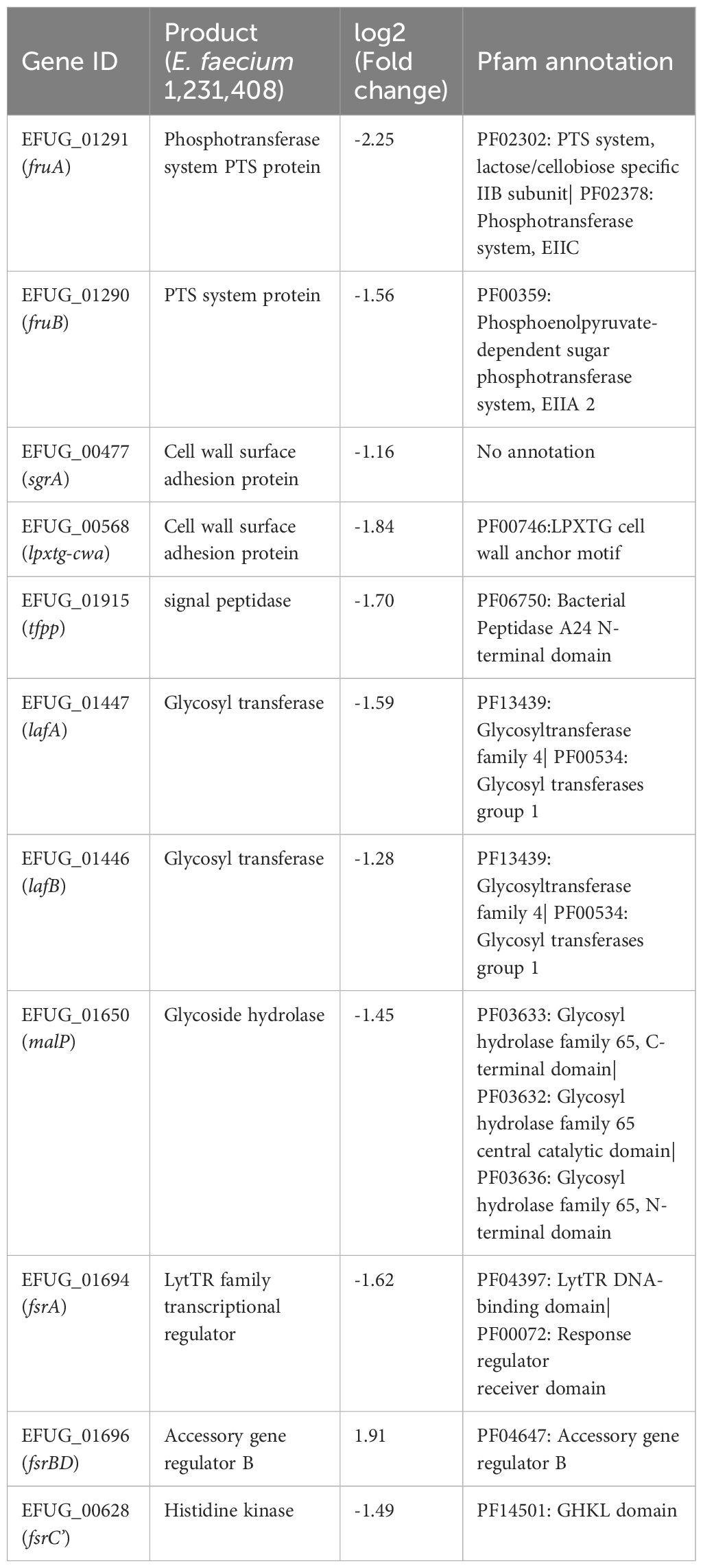
Table 1. The biofilm-associated genes with statistical differences in the V27 treated with GLA relative to the V27 treated without GLA.
The sgrA gene, encoding a nidogen-binding Leu-Pro-X-Thr-Gly, where X denotes any amino acid (LPXTG) surface adhesin protein (Protein ID: AFK59147.1) in the E. faecium DO strain implicated in biofilm formation (Hendrickx et al., 2007; Hendrickx et al., 2009a). This protein had the greatest similarity with the EFUG_00477 gene (73.77%), whose expression level was downregulation in the V27 treated with GLA sample. We have designated EFUG_00477 gene as sgrA gene.
Pfam annotation (http://pfam.xfam.org) showed that EFUG_00568 gene encodes another LPXTG-motif protein. This gene was found to be significantly downregulated in the V27 treated with GLA sample, but it has not been previously reported to be associated with biofilm formation in E. faecium. Through sequence similarity analysis, this gene exhibited the highest similarity (96.33%) to the surface protein EF3314 (Protein ID: AAO82979.1) in E. faecalis V583, which has been linked to biofilm formation (Creti et al., 2009). We have designated EFUG_00568 gene as lpxtg-cwa (LPXTG cell wall anchor) gene.
Pfam annotation showed that EFUG_01915 gene encodes peptidase A24, which is a type 4 prepilin peptidase. It catalyzes the processing of type 4 prepilin to form type 4 pilus, which in turn are involved in a variety of functions, including toxin and enzyme secretion, gene transfer, and biofilm formation (LaPointe and Taylor, 2000). The expression of EFUG_01915 gene was obviously down-regulated in the V27 treated with GLA sample in the study. We have designated EFUG_01915 gene as tfpp (type 4 prepilin peptidase) gene.
Both lafA gene and lafB gene, encoding LafA (Protein ID: WP_002287605.1) and LafB (Protein ID: WP_002296953.1) glycosyltransferases in E. faecium DO strain, are involved in lipoteichoic acid (LTA) biosynthesis (Webb et al., 2009; Mello et al., 2020). These two proteins, LafA and LafB had the greatest similarity with the EFUG_01447 gene (94.10%) and the EFUG_01446 gene (99.71%), respectively. The closest homolog of lafA and lafB genes in the E. faecalis V583 strain is the biofilm-associated glycolipid synthesis B (bgsB) and bgsA genes, which are responsible for LTA anchor formation (Theilacker et al., 2011). While no studies have reported a relationship between lafAB genes and biofilm formation, the role of bgsAB genes in promoting biofilm formation has been well-established (Theilacker et al., 2011; Haller et al., 2014; Madsen et al., 2017). The expression of both EFUG_01447 gene and EFUG_01446 gene were obviously down-regulated in the V27 treated with GLA sample in the study. We have designated EFUG_01447 and EFUG_01446 genes as lafA and lafB genes, respectively.
The malP gene, encoding a maltose phosphorylase (Protein ID: WP_002413579.1) in E. faecalis OG1RF strain, is essential for the phosphorylation of maltose to α-D-glucose and glucose-1-phosphate (Sauvageot et al., 2017). This protein catalyzes the production of monosaccharides, which are essential for exopolysaccharide synthesis (Ali et al., 2022). It had the greatest similarity with the EFUG_01650 gene (74.48%). The expression of the EFUG_01650 gene was significantly down-regulated in the V27 treated with GLA sample in the study. We have designated the EFUG_01650 gene as the malP gene.
3.3 GO and KEGG enrichment analysis
Three categories include cell component, biological process, and molecular function in GO enrichment analysis. The Gene Ratio, the count, and the corresponding P values for each GO ID comparing the V27 treated with GLA to the V27 treated without GLA samples, were presented in Figure 2A. The percentage of upregulated genes in GO enrichment analysis was shown in Supplementary Table S3. Among the top ten enriched cell components, including ribosome, ribonucleoprotein complex, cytoplasmic part, cytoplasm, organelle, non-membrane-bounded organelle, intracellular organelle, intracellular non-membrane-bounded organelle, protein-containing complex, and intracellular, at least 86.7% of genes were upregulated. Similarly, six biological processes exhibited a greater than 90% upregulation of genes, including translation, peptide metabolic process, peptide biosynthetic process, amide biosynthetic process, cellular amide metabolic process, and cellular protein metabolic process. Two additional processes showed upregulation of genes between 80-90%, and two processes showed upregulation of genes between 60-70%. Of the four molecular function categories displaying statistically significant differences, structural constituent of ribosome and structural molecule activity showed an upregulation of 93.8% of genes, while ligase activity had an upregulation of genes of 66.7%. However, active transmembrane transporter activity showed a significantly reduced upregulation of genes of only 39.3%.
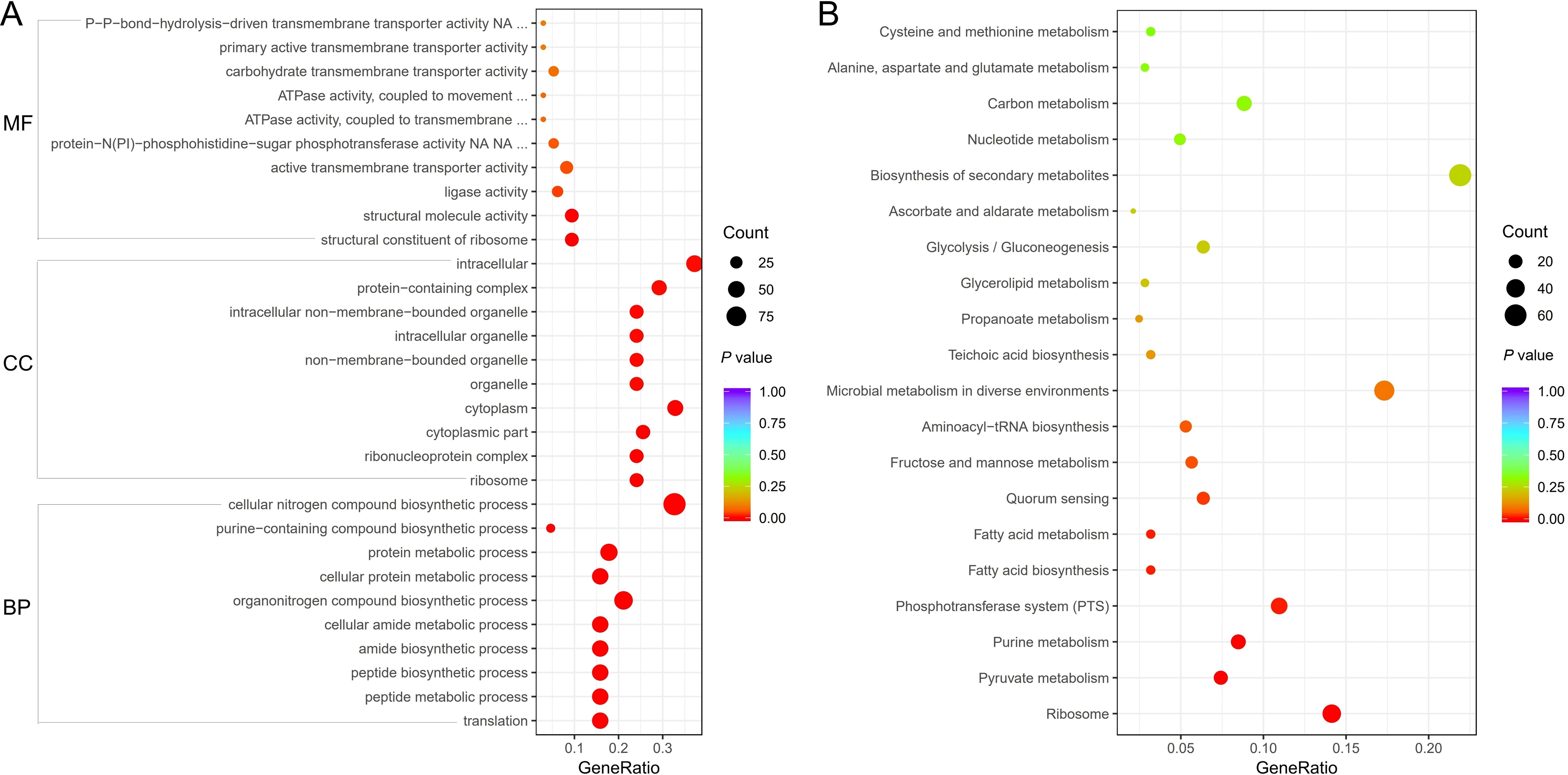
Figure 2. (A) Gene Ontology (GO) enrichment of differential expression genes between the V27 treated with GLA and V27 treated without GLA. MF, molecular function; CC, cell component; BP, biological process. (B) Kyoto Encyclopedia of Gene and Genomes (KEGG) pathway enrichment of differential expression genes between the V27 treated with GLA and V27 treated without GLA.
Enrichment analysis of KEGG pathways identified eight pathways with statistically significant enrichment (P < 0.05), fewer than those observed in GO analysis. Figure 2B showed the detailed results for these enriched pathways, including the Gene Ratio, the count, and the corresponding P values (obtained by comparing V27 samples treated with GLA to untreated V27 samples). Among these enriched pathways, only the ribosome pathway exhibited a significantly higher proportion of upregulated genes (92.5%). The remaining pathways showed lower proportions of upregulated genes, all below 50%. Specifically, pathways such as pyruvate metabolism, purine metabolism, the PTS, and fructose and mannose metabolism displayed upregulated gene proportions between 25% and 43%, whereas fatty acid biosynthesis and fatty acid metabolism showed no upregulated genes. Detailed upregulated proportions can be found in Supplementary Table S4.
The PTS and fructose and mannose metabolism pathways both contain the genes fruA and fruB. The Fsr quorum sensing system is involved in biofilm formation. In the KEGG strain E. faecium NRRL B-2354 (GenBank: CP004063.1), the regulatory genes for FsrA protein were M7W_623 and M7W_2389, for FsrB/D protein/peptide was M7W_626 gene, and for FsrC protein were M7W_624, M7W_782, and M7W_2390 genes (Figure 3). In this study, the reference strain E. faecium 1,231,408 exhibited downregulation of the EFUG_01694 and EFUG_00628 genes (Table 1), which corresponded to M7W_623 and M7W_782 genes in the KEGG strain E. faecium NRRL B-2354, respectively. Conversely, the EFUG_01696 gene, corresponding to the M7W_626 gene in the KEGG strain E. faecium NRRL B-2354, showed upregulation. We have designated EFUG_01694 and EFUG_01696 genes as fsrA and fsrB genes, respectively. Based on existing nomenclature, the M7W_624 gene, which corresponds to the EFUG_01695 gene in the reference genome, is annotated as fsrC. Therefore, we have designated M7W_782, which corresponds to EFUG_00628 in the reference genome, as fsrC’.
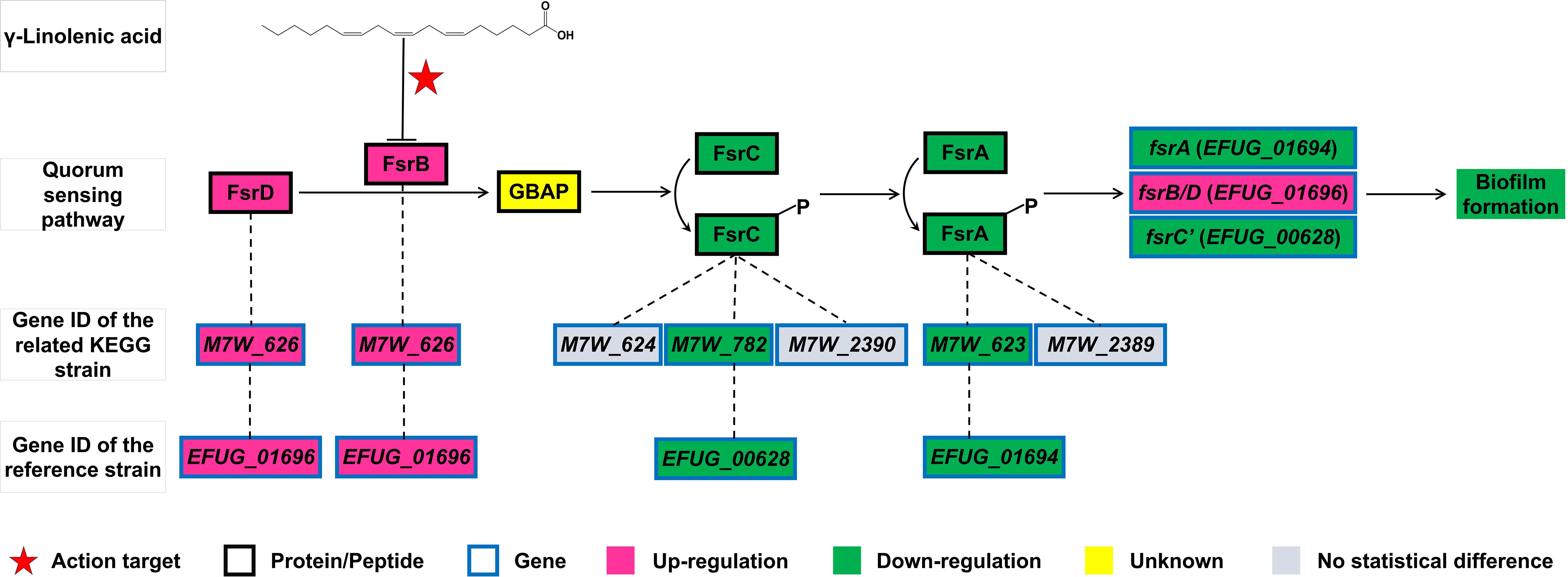
Figure 3. The mechanism of action of GLA on the Fsr quorum sensing pathway and its associated genes in this study.
3.4 Validation by biofilm-associated genes of VRE-fm isolates by qRT-PCR
To validate transcriptome sequencing results, genes involved in biofilm expression of six VRE-fm isolates (V05, V06, V08, V09, V22, and V27) were selected as targets for qRT-PCR analysis, as shown in Figure 4. Compared to the control group, GLA down-regulated the gene expression of lpxtg-cwa (0.69-fold), tfpp (0.43-fold), sgrA (0.48-fold), lafA (0.57-fold), lafB (0.69-fold), malP (0.43-fold), fruA (0.49-fold), fruB (0.46-fold), fsrA (0.69-fold), and fsrC’ (0.46-fold), while up-regulated the gene expression of fsrB (6.34-fold). The qRT-PCR results showed a high concordance with the transcriptomic data, with all gene expression changes exhibiting statistical significance except for the decrease in lpxtg-cwa, which was not statistically different.
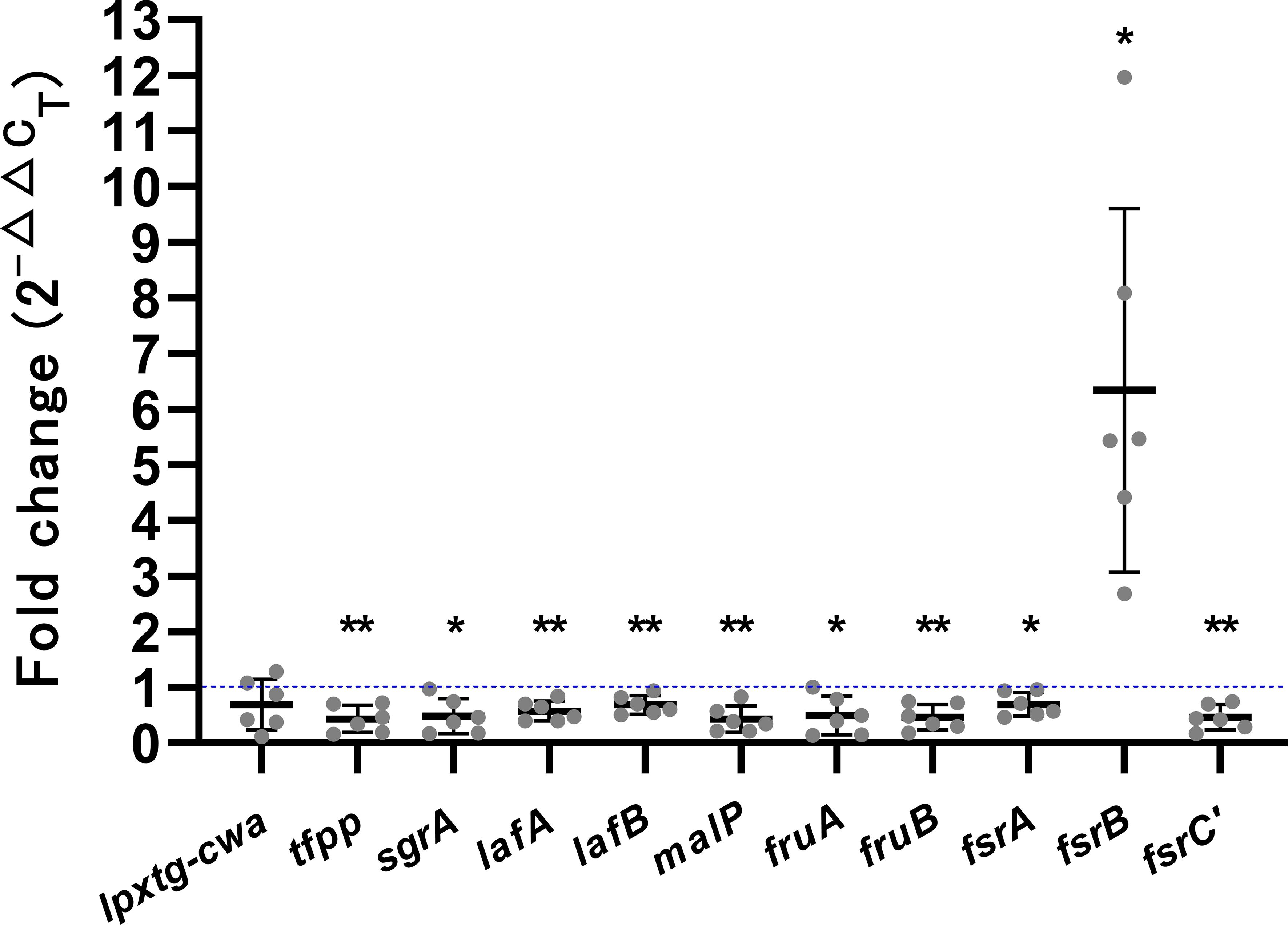
Figure 4. The expression levels of biofilm-related genes in mature biofilms of the VRE-fm isolates treated with γ-linolenic acid (GLA) comparing to the control group. *P < 0.05, and **P < 0.01.
Figure 5 further showed the regulatory effects of GLA on the expression levels of biofilm-related genes in different VRE-fm isolates. Genes including tfpp, sgrA, lafA, lafB, malP, fruA, fruB, fsrA, and fsrC were downregulated to varying degrees in all isolates. Conversely, fsrB gene was upregulated to varying degrees in all isolates. The lpxtg-cwa gene exhibited inconsistent expression patterns across the six isolates; downregulation was observed in V05, V06, V08, and V27, whereas upregulation was observed in V09 and V22.
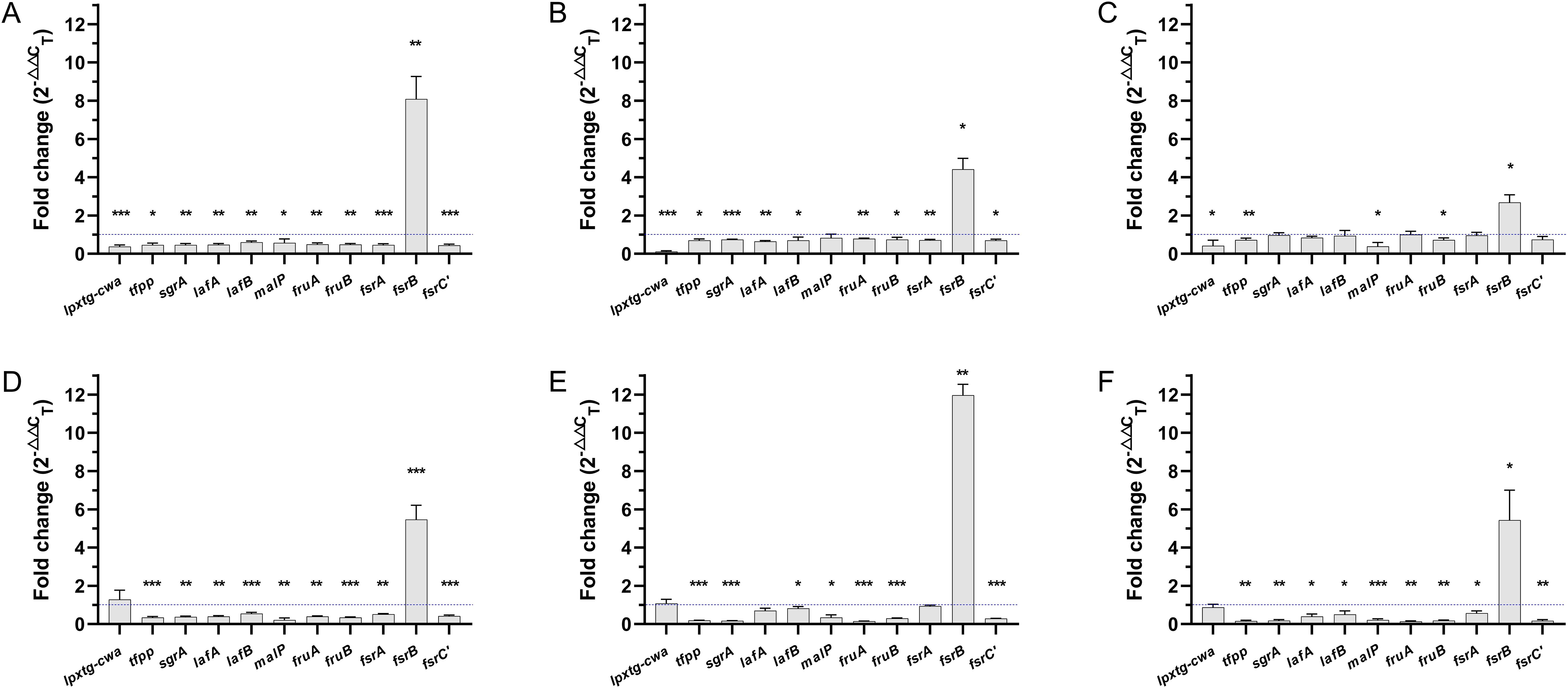
Figure 5. The expression levels of biofilm-related genes in mature biofilms of each VRE-fm isolate treated with γ-linolenic acid (GLA) comparing to the control group (A) V05; (B) V06; (C) V08; (D) V09; (E) V22; (F) V27. *P < 0.05, **P < 0.01, and ***P < 0.001.
3.5 GLA affects biofilm-related genes in Enterococcus faecalis
Because the effects of certain genes on biofilm formation in E. faecium were predicted from homologous genes in E. faecalis, we validated these predictions by investigating the effect of GLA on these homologous genes in E. faecalis. Figure 6A showed that two clinical isolates of E. faecalis (efa105 and efa106) both exhibited strong biofilm formation ability. After treatment with GLA, the biofilm biomass of both isolates decreased by 75.2% and 75.6%, respectively. Figures 6B, C showed the effect of GLA on the gene expression levels of bgsA, bgsB, efa-malP, gelE, and sprE in isolates efa105 and efa106, with the latter two genes being positively regulated by the Fsr quorum sensing system. All gene expression levels in isolate efa105 were downregulated, while in isolate efa106, all gene expression levels except for efa-malP were also downregulated.
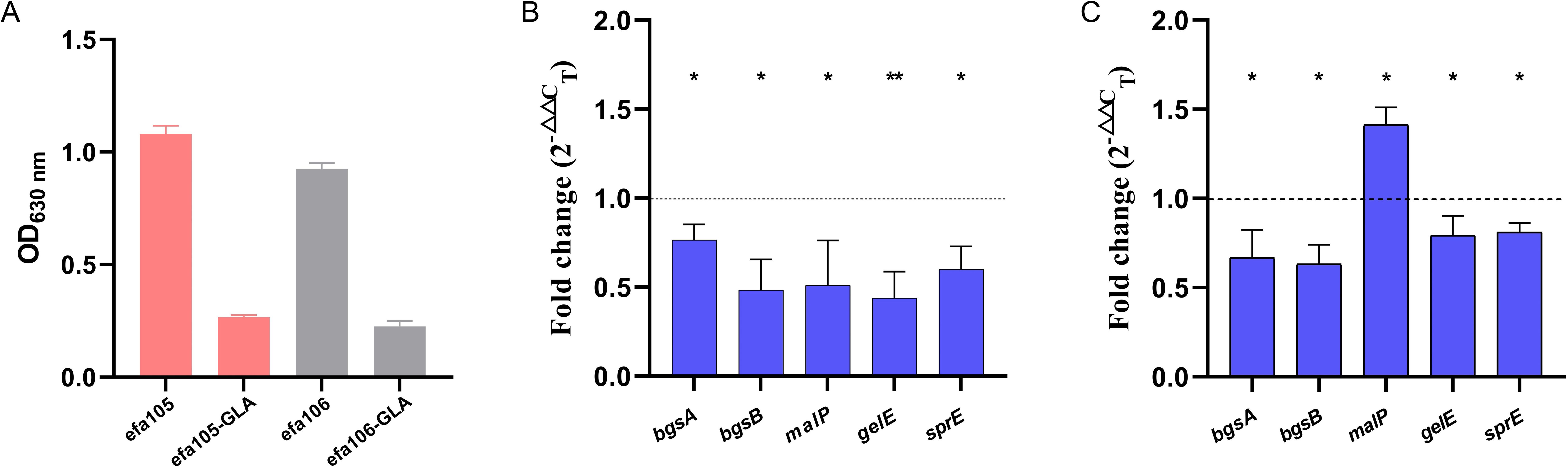
Figure 6. (A) The biofilm biomass of two clinical isolates of E. faecalis (efa105 and efa106) before and after treated with γ-linolenic acid (GLA); The expression levels of biofilm-related genes in mature biofilms of (B) efa105 and (C) efa106 treated with γ-linolenic acid (GLA) comparing to the control group. *P < 0.05, and **P < 0.01.
4 Discussion
Biofilm formation significantly contributes to the pathogenesis of Enterococcus, especially in infective endocarditis and urinary tract infections, where it promotes bacterial persistence and resistance to antibiotics (Wei et al., 2024). An intriguing study revealed that a unique metabolite, putatively a short-chain fatty acid (SCFA) produced by Bacteroides spp. (dominant members of the gut microbiota), demonstrated a strong association with VRE-fm decolonization (Deleu et al., 2021; MaChado et al., 2021). Similarly, our previous study demonstrated that essential fatty acids can inhibit and eradicate E. faecium biofilms. Preliminary mechanistic investigations revealed a significant downregulation of atlA gene expression (Wei et al., 2023). However, the downregulation of a single gene might not be sufficient to explain the substantial eradication of preformed biofilms. Therefore, in this study, we employed transcriptome analysis to further elucidate the mechanisms.
Transcriptome analysis revealed a significant downregulation in the expression levels of genes associated with biofilm formation, including fruA, fruB, sgrA, lpxtg-cwa, tfpp, lafA, lafB, malP, fsrA, and fsrC’, while a significant upregulation was observed in the expression of fsrBD. (Table 1). Validation by qRT-PCR in six VRE-fm isolates confirmed the significant changes in the expression levels of all genes except for lpxtg-cwa, with statistical significance (Figure 4).
Fernanda et al. reported that deletion of the fruA (bepA) gene in E. faecium strain E1162 impaired biofilm formation and metabolism of β-methyl-D-glucoside. Since β-glucoside metabolism is linked to the metabolism of glycosaminoglycans, which are exposed on injured heart valves where bacteria attach and form vegetations, the authors propose that the PTS permease FruA (BepA) influences biofilm formation through this pathway (Paganelli et al., 2016). The fruAB genes constitute an operon, and the fruB gene is coordinately regulated with the fruA gene (Burne et al., 1999; Chakraborty et al., 2022). In S. mutans, the fruB gene encodes an endolevanase FruB, that enhances levan metabolism, and provides the bacteria with energy and carbon sources. It plays a crucial role in biofilm formation and pathogenesis (Chakraborty et al., 2022). The current study found that GLA simultaneously downregulated the expression of both fruA and fruB genes. However, the involvement of fruB in biofilm formation in E. faecium remains unclear and requires further investigation. Additionally, it is yet to be confirmed whether GLA downregulates fruAB expression by inhibiting their promoter.
Biofilm formation in Enterococcus species is initiated by adhesion, a critical step that sets the stage for subsequent biofilm development (Ch'ng et al., 2019). SgrA belongs to the LPxTG-type surface proteins, a class of proteins characterized by a C-terminal LPxTG-like motif (Hendrickx et al., 2009a). This motif is recognized and cleaved by a transpeptidase (sortase), which covalently anchors the surface protein to the cell wall peptidoglycan (Hendrickx et al., 2009b). SgrA has been shown to bind to the extracellular matrix molecules nidogens and fibrinogen. Deletion of the sgrA gene significantly impairs biofilm formation. This study found that GLA significantly decreased the expression of sgrA gene (Hendrickx et al., 2009a), suggesting a potential mechanism for the antibiofilm activity of GLA. Although a decrease in the expression of another LPxTG-type surface protein-encoding gene (lpxtg-cwa) was observed in some individual isolates, this reduction was not statistically significant for all VRE-fm isolates.
Another important structure involved in Enterococcus adhesion is pilus (Hendrickx et al., 2009b). Type IV pilus formation relies on the cleavage of type IV prepilins by type IV prepilin peptidase (TFPP). Following cleavage, type IV prepilins not only contribute to type IV pilus assembly but also participate in a variety of biological processes, including toxin and enzyme secretion, gene transfer, and biofilm formation (LaPointe and Taylor, 2000). In this study, GLA significantly downregulated the expression of the tfpp gene. Therefore, GLA can also inhibit biofilm formation by interfering with type IV pilus formation.
Both lafA and lafB genes, encoding glycosyltransferase, are involved in the biosynthesis of lipoteichoic acid (LTA) (Mello et al., 2020). The glycolipid anchor of LTA is able to insert into eukaryotic membranes. In a study of E. faecalis, deletion of the bgsB and bgsA genes, which are homologous to lafA and lafB genes, significantly impaired biofilm formation in a rat model of infective endocarditis (Haller et al., 2014). In this study, GLA significantly downregulated the expression of lafA and lafB genes. Based on this homology, we further investigated two clinical isolates of E. faecalis and found that GLA could also downregulate the expression of bgsA and bgsB genes. However, the potential role of lafA and lafB genes in influencing biofilm formation in E. faecium remains to be elucidated and warrants further investigation. Suelen et al. reported that deletion of the lafB gene resulted in increased susceptibility to daptomycin (Mello et al., 2020). Therefore, GLA may enhance daptomycin susceptibility in E. faecium.
EPS production plays a crucial role in biofilm maturation. Our previous study has demonstrated that GLA inhibits eDNA release by downregulating the expression of atlA gene (Wei et al., 2023). In this study, we further observed that GLA downregulates the expression of the malP gene. In E. faecalis, the homologous gene efa-malP is essential for the phosphorylation of intracellular maltose to α-D-glucose and glucose-1-phosphate. Downregulation of malP results in a decrease in the availability of monosaccharides needed for exopolysaccharide synthesis (Ali et al., 2022). Based on this homology, we also investigated the expression of the efa-malP gene in E. faecalis. We observed contrasting effects of GLA on efa-malP expression in two E. faecalis isolates, with a decrease in one isolate and an increase in the other. Therefore, further studies with a larger sample size are needed to confirm the effect of GLA on efa-malP gene expression in E. faecalis.
Quorum sensing is a type of cell-to-cell communication used by bacteria. It allows bacteria to coordinate their behavior based on the density of their population (Mukherjee and Bassler, 2019). The Fsr quorum sensing system plays a role in biofilm formation in Enterococcus (Tsikrikonis et al., 2012; Ali et al., 2017). The FsrD propeptide is processed by FsrB to produce gelatinase biosynthesis-activating pheromone (GBAP) (Figure 3). Accumulated GBAP is sensed by the sensor protein, FsrC, leading to its phosphorylation and activation of the response regulator, FsrA. Activated FsrA promotes the expression of the fsr locus, gelE, and sprE genes. Upregulation of gelE expression, which encodes a gelatinase, promotes biofilm formation in E. faecalis (Ali et al., 2017). The precise mechanisms by which the Fsr system promotes biofilm formation in E. faecium remain unclear. However, a study has shown that deletion of the fsrB gene inhibits biofilm formation (Tsikrikonis et al., 2012). In this study, we demonstrated that GLA decreased the expression of the fsrA and fsrC’ genes but significantly increased the expression of the fsrB gene. This result contradicts the expectation that reduced fsrA expression would inhibit the expression of the fsr locus. A previous study has suggested that the function of FsrB can be inhibited by ambuic acid, thereby reducing the production of the signaling molecule GBAP (Nakayama et al., 2009). Therefore, we hypothesize that GLA, similar to Ambuic Acid, can also inhibit the function of FsrB, thereby suppressing biofilm formation. The observed increase in fsrB gene expression may be attributed to negative feedback regulation resulting from FsrB inhibition. Further investigation is required to elucidate the specific mechanisms involved in this interaction. While the specific genes regulated by the Fsr system in E. faecium remain unclear, this system is known to regulate the expression of gelE and sprE genes in E. faecalis. Therefore, we investigated the effects of GLA on the expression of gelE and sprE in E. faecalis. The results demonstrated that GLA reduced the expression of gelE and sprE genes in two E. faecalis clinical isolates. These findings provide further evidence that GLA inhibits the Fsr system, highlighting its potential as a novel anti-biofilm agent.
In addition to downregulating biofilm-related genes, GLA also alters metabolic pathways, further supporting its biofilm eradicating effects. GO analysis revealed a significantly higher proportion of upregulated genes across all GO IDs except for the active transmembrane transporter activity, suggesting a more active metabolic state in GLA-treated cells compared to biofilm-embedded cells. This finding aligns with our understanding that cells within biofilms exhibit a slower metabolic rate (Vestby et al., 2020). Furthermore, KEGG analysis identified specific metabolic pathways that were significantly downregulated by GLA, including purine, phosphotransferase system, and fatty acid metabolism. These molecules may play a crucial role in cell survival, repair, and biofilm formation (Zhang et al., 2024).
In summary, we investigated the effects of GLA on VRE-fm using transcriptomic analysis and qRT-PCR. Our results revealed that GLA significantly reduced the expression of numerous biofilm-associated genes in VRE-fm. Furthermore, GLA inhibited the Fsr quorum sensing system by suppressing the function of FsrB. The expression of bgsB and bgsA genes, which are the homologs of lafA and lafB genes, along with the Fsr-regulated genes gelE and sprE in E. faecalis, were also found to be downregulated by GLA. GO analysis revealed a more active metabolic state in GLA-treated VRE-fm cells compared to biofilm-embedded cells, suggesting a shift in metabolic activity. KEGG analysis identified specific metabolic pathways were significantly downregulated by GLA. Overall, these findings demonstrate that GLA effectively targets multiple aspects of biofilm formation in VRE-fm, including the downregulation of key biofilm-related genes, the inhibition of quorum sensing systems, and the modulation of metabolic pathways. GLA emerges as a promising candidate for eradicating Enterococcus biofilms.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/sra/PRJNA1178430.
Ethics statement
The study was carried out according to the Declaration of Helsinki and was approved by the Ethics Committee of Beijing Chao-Yang Hospital, Capital Medical University (2023-3-10-3). Patient consent was waived due to the retrospective nature of the study.
Author contributions
MW: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft. PW: Methodology, Writing – original draft. TL: Investigation, Writing – original draft. JL: Validation, Writing – original draft. YW: Formal analysis, Writing – original draft. LG: Supervision, Writing – review & editing. SW: Funding acquisition, Visualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the National Natural Science Foundation of China (Grant No. 82302551); Beijing Natural Science Foundation (Grant No. 7234369); and the Reform and Development Program of Beijing Institute of Respiratory Medicine (Grant No. Ggyfz202419 and Ggyfz202425).
Acknowledgments
The authors would like to thank all members of the Department of Infectious Diseases and Clinical Microbiology staff at Beijing Chao-Yang Hospital (Beijing, China) for contributing to this work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1525581/full#supplementary-material
References
Ali, L., Goraya, M. U., Arafat, Y., Ajmal, M., Chen, J. L., Yu, D. (2017). Molecular mechanism of quorum-sensing in Enterococcus faecalis: Its role in virulence and therapeutic approaches. Int. J. Mol. Sci. . 18, 960. doi: 10.3390/ijms18050960
Ali, I. A. A., Lévesque, C. M., Neelakantan, P. (2022). Fsr quorum sensing system modulates the temporal development of Enterococcus faecalis biofilm matrix. Mol. Oral. Microbiol. . 37, 22–30. doi: 10.1111/omi.12357
Ayrapetyan, M., Williams, T., Oliver, J. D. (2018). Relationship between the viable but nonculturable state and antibiotic persister cells. J. Bacteriol . 200, e00249–e00218. doi: 10.1128/jb.00249-18
Burne, R. A., Wen, Z. T., Chen, Y. Y., Penders, J. E. (1999). Regulation of expression of the fructan hydrolase gene of Streptococcus mutans GS-5 by induction and carbon catabolite repression. J. Bacteriol . 181, 2863–2871. doi: 10.1128/jb.181.9.2863-2871.1999
Ch'ng, J. H., Chong, K. K. L., Lam, L. N., Wong, J. J., Kline, K. A. (2019). Biofilm-associated infection by enterococci. Nat. Rev. Microbiol. . 17, 82–94. doi: 10.1038/s41579-018-0107-z
Chakraborty, B., Zeng, L., Burne, R. A. (2022). The fruB gene of Streptococcus mutans encodes an endo-levanase that enhances growth on levan and influences global gene expression. Microbiol. Spectr. . 10, e0052222. doi: 10.1128/spectrum.00522-22
Creti, R., Fabretti, F., Koch, S., Huebner, J., Garsin, D. A., Baldassarri, L., et al. (2009). Surface protein EF3314 contributes to virulence properties of Enterococcus faecalis. Int. J. Artif. Organs . 32, 611–620. doi: 10.1177/039139880903200910
Cui, C., Song, S., Yang, C., Sun, X., Huang, Y., Li, K., et al. (2019). Disruption of quorum sensing and virulence in Burkholderia cenocepacia by a structural analogue of the cis-2-dodecenoic acid signal. Appl. Environ. Microbiol. . 85, e00105–e00119. doi: 10.1128/aem.00105-19
Deleu, S., Machiels, K., Raes, J., Verbeke, K., Vermeire, S. (2021). Short chain fatty acids and its producing organisms: An overlooked therapy for IBD? EBioMedicine . 66, 103293. doi: 10.1016/j.ebiom.2021.103293
Desbois, A. P. (2012). Potential applications of antimicrobial fatty acids in medicine, agriculture and other industries. Recent Pat. Antiinfect Drug Discovery . 7, 111–122. doi: 10.2174/157489112801619728
Haller, C., Berthold, M., Wobser, D., Kropec, A., Lauriola, M., Schlensak, C., et al. (2014). Cell-wall glycolipid mutations and their effects on virulence of E. faecalis in a rat model of infective endocarditis. PLoS One . 9, e91863. doi: 10.1371/journal.pone.0091863
Hendrickx, A. P., van Luit-Asbroek, M., Schapendonk, C. M., van Wamel, W. J., Braat, J. C., Wijnands, L. M., et al. (2009a). SgrA, a nidogen-binding LPXTG surface adhesin implicated in biofilm formation, and EcbA, a collagen binding MSCRAMM, are two novel adhesins of hospital-acquired Enterococcus faecium. Infect. Immun. . 77, 5097–5106. doi: 10.1128/iai.00275-09
Hendrickx, A. P., van Wamel, W. J., Posthuma, G., Bonten, M. J., Willems, R. J. (2007). Five genes encoding surface-exposed LPXTG proteins are enriched in hospital-adapted Enterococcus faecium clonal complex 17 isolates. J. Bacteriol . 189, 8321–8332. doi: 10.1128/jb.00664-07
Hendrickx, A. P., Willems, R. J., Bonten, M. J., van Schaik, W. (2009b). LPxTG surface proteins of enterococci. Trends Microbiol. . 17, 423–430. doi: 10.1016/j.tim.2009.06.004
Inoue, T., Shingaki, R., Fukui, K. (2008). Inhibition of swarming motility of Pseudomonas aeruginosa by branched-chain fatty acids. FEMS Microbiol. Lett. . 281, 81–86. doi: 10.1111/j.1574-6968.2008.01089.x
Khan, N. A., Barthes, N., McCormack, G., O'Gara, J. P., Thomas, O. P., Boyd, A. (2023). Sponge-derived fatty acids inhibit biofilm formation of MRSA and MSSA by down-regulating biofilm-related genes specific to each pathogen. J. Appl. Microbiol. 134, lxad152. doi: 10.1093/jambio/lxad152
Kim, Y. G., Lee, J. H., Raorane, C. J., Oh, S. T., Park, J. G., Lee, J. (2018). Herring oil and omega fatty acids inhibit Staphylococcus aureus biofilm formation and virulence. Front. Microbiol. . 9. doi: 10.3389/fmicb.2018.01241
LaPointe, C. F., Taylor, R. K. (2000). The type 4 prepilin peptidases comprise a novel family of aspartic acid proteases. J. Biol. Chem. . 275, 1502–1510. doi: 10.1074/jbc.275.2.1502
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods . 25, 402–408. doi: 10.1006/meth.2001.1262
Lleò, M. M., Bonato, B., Benedetti, D., Canepari, P. (2005). Survival of enterococcal species in aquatic environments. FEMS Microbiol. Ecol. . 54, 189–196. doi: 10.1016/j.femsec.2005.03.016
MaChado, M. G., Sencio, V., Trottein, F. (2021). Short-chain fatty acids as a potential treatment for infections: a closer look at the lungs. Infect. Immun. . 89, e0018821. doi: 10.1128/iai.00188-21
Madsen, K. T., Skov, M. N., Gill, S., Kemp, M. (2017). Virulence factors associated with Enterococcus faecalis infective endocarditis: A mini review. Open Microbiol. J. 11, 1–11. doi: 10.2174/1874285801711010001
Mello, S. S., Van Tyne, D., Lebreton, F., Silva, S. Q., Nogueira, M. C. L., Gilmore, M. S., et al. (2020). A mutation in the glycosyltransferase gene lafB causes daptomycin hypersusceptibility in Enterococcus faecium. J. Antimicrob. Chemother. . 75, 36–45. doi: 10.1093/jac/dkz403
Mukherjee, S., Bassler, B. L. (2019). Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol. . 17, 371–382. doi: 10.1038/s41579-019-0186-5
Nakayama, J., Uemura, Y., Nishiguchi, K., Yoshimura, N., Igarashi, Y., Sonomoto, K. (2009). Ambuic acid inhibits the biosynthesis of cyclic peptide quormones in gram-positive bacteria. Antimicrob. Agents Chemother. . 53, 580–586. doi: 10.1128/aac.00995-08
Paganelli, F. L., Huebner, J., Singh, K. V., Zhang, X., van Schaik, W., Wobser, D., et al. (2016). Genome-wide screening identifies phosphotransferase system permease BepA to be involved in Enterococcus faecium endocarditis and biofilm formation. J. Infect. Dis. . 214, 189–195. doi: 10.1093/infdis/jiw108
Ramanathan, S., Ravindran, D., Arunachalam, K., Arumugam, V. R. (2018). Inhibition of quorum sensing-dependent biofilm and virulence genes expression in environmental pathogen Serratia marcescens by petroselinic acid. Antonie Van Leeuwenhoek . 111, 501–515. doi: 10.1007/s10482-017-0971-y
Sauvageot, N., Mokhtari, A., Joyet, P., Budin-Verneuil, A., Blancato, V. S., Repizo, G. D., et al. (2017). Enterococcus faecalis uses a phosphotransferase system permease and a host colonization-related ABC transporter for maltodextrin uptake. J. Bacteriol . 199, e00878–e00816. doi: 10.1128/jb.00878-16
Tatta, E. R., Paul, S., Kumavath, R. (2023). Transcriptome analysis revealed the synergism of novel rhodethrin inhibition on biofilm architecture, antibiotic resistance and quorum sensing in Enterococcus faecalis. Gene . 871, 147436. doi: 10.1016/j.gene.2023.147436
Theilacker, C., Sava, I., Sanchez-Carballo, P., Bao, Y., Kropec, A., Grohmann, E., et al. (2011). Deletion of the glycosyltransferase bgsB of Enterococcus faecalis leads to a complete loss of glycolipids from the cell membrane and to impaired biofilm formation. BMC Microbiol. . 11, 67. doi: 10.1186/1471-2180-11-67
Tsikrikonis, G., Maniatis, A. N., Labrou, M., Ntokou, E., Michail, G., Daponte, A., et al. (2012). Differences in biofilm formation and virulence factors between clinical and fecal enterococcal isolates of human and animal origin. Microb. Pathog. . 52, 336–343. doi: 10.1016/j.micpath.2012.03.003
Vestby, L. K., Grønseth, T., Simm, R., Nesse, L. L. (2020). Bacterial biofilm and its role in the pathogenesis of disease. Antibiotics (Basel) . 9, 59. doi: 10.3390/antibiotics9020059
Wang, S., Wang, P., Liu, J., Yang, C., Wang, Q., Su, M., et al. (2022). Antibiofilm activity of essential fatty acids against Candida albicans from vulvovaginal candidiasis and bloodstream infections. Infect. Drug Resist. . 15, 4181–4193. doi: 10.2147/idr.s373991
Webb, A. J., Karatsa-Dodgson, M., Gründling, A. (2009). Two-enzyme systems for glycolipid and polyglycerolphosphate lipoteichoic acid synthesis in Listeria monocytogenes. Mol. Microbiol. . 74, 299–314. doi: 10.1111/j.1365-2958.2009.06829.x
Wei, Y., Palacios Araya, D., Palmer, K. L. (2024). Enterococcus faecium: evolution, adaptation, pathogenesis and emerging therapeutics. Nat. Rev. Microbiol. . 22, 705–721. doi: 10.1038/s41579-024-01058-6
Wei, M., Wang, P., Li, T., Wang, Q., Su, M., Gu, L., et al. (2023). Antimicrobial and antibiofilm effects of essential fatty acids against clinically isolated vancomycin-resistant Enterococcus faecium. Front. Cell Infect. Microbiol. . 13. doi: 10.3389/fcimb.2023.1266674
Willett, J. L. E., Ji, M. M., Dunny, G. M. (2019). Exploiting biofilm phenotypes for functional characterization of hypothetical genes in Enterococcus faecalis. NPJ Biofilms Microbiomes . 5, 23. doi: 10.1038/s41522-019-0099-0
Xie, Y., Wang, L., Yang, Y., Zha, L., Zhang, J., Rong, K., et al. (2022). Antibacterial and anti-biofilm activity of diarylureas against Enterococcus faecium by suppressing the gene expression of peptidoglycan hydrolases and adherence. Front. Microbiol. . 13. doi: 10.3389/fmicb.2022.1071255
Yoon, B. K., Jackman, J. A., Valle-González, E. R., Cho, N. J. (2018). Antibacterial free fatty acids and monoglycerides: Biological activities, experimental testing, and therapeutic applications. Int. J. Mol. Sci. . 19, 1114. doi: 10.3390/ijms19041114
Keywords: vancomycin-resistant Enterococcus faecium, biofilm, γ-linolenic acid, eradication, molecular mechanism
Citation: Wei M, Wang P, Li T, Liu J, Wang Y, Gu L and Wang S (2025) Transcriptome analysis reveals the molecular mechanism of γ-linolenic acid eradicating the biofilm of vancomycin-resistant Enterococcus faecium. Front. Cell. Infect. Microbiol. 15:1525581. doi: 10.3389/fcimb.2025.1525581
Received: 11 November 2024; Accepted: 06 January 2025;
Published: 31 January 2025.
Edited by:
Manuel Espinosa-Urgel, Spanish National Research Council (CSIC), SpainReviewed by:
Ram Nageena Singh, South Dakota School of Mines and Technology, United StatesPang Li Mei, A*STAR Infectious Disease Labs, Singapore
Copyright © 2025 Wei, Wang, Li, Liu, Wang, Gu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuai Wang, d2FuZ3BrcUAxNjMuY29t; Li Gu, ZGlkY20yMDA2QG1haWwuY2NtdS5lZHUuY24=
 Ming Wei
Ming Wei Peng Wang
Peng Wang Tianmeng Li
Tianmeng Li Shuai Wang
Shuai Wang