
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 14 February 2025
Sec. Bacteria and Host
Volume 15 - 2025 | https://doi.org/10.3389/fcimb.2025.1522573
This article is part of the Research Topic Synergistic Approaches to Managing Gram-negative Bacterial Resistance View all 14 articles
 Kathleen Klaper1,2
Kathleen Klaper1,2 Yvonne Pfeifer1
Yvonne Pfeifer1 Lena Heinrich1
Lena Heinrich1 Marcel Prax3
Marcel Prax3 Oleg Krut3
Oleg Krut3 Isabelle Bekeredjian-Ding3†‡
Isabelle Bekeredjian-Ding3†‡ Anika Wahl1
Anika Wahl1 Martin A. Fischer1
Martin A. Fischer1 Heike Kaspar4
Heike Kaspar4 Stefan Borgmann5
Stefan Borgmann5 Roman G. Gerlach1†
Roman G. Gerlach1† Guido Werner1*
Guido Werner1*Background: Klebsiella pneumoniae is one of the most important opportunistic pathogens causing healthcare-associated and community-acquired infections worldwide. In recent years, the increase in antibiotic resistance and infections caused by hypervirulent K. pneumoniae poses great public health concerns. In this study, host-pathogen interactions of different K. pneumoniae strains of human and animal origins were analyzed in microbiological, cell-biological and immunological experiments.
Methods: In vitro infection experiments using representatives of different K. pneumoniae pathotypes and various epithelial and macrophage cell lines were executed analyzing adhesion, invasion and intracellular replication. Experimental conditions involved normoxia and hypoxia. Furthermore, survival and growth of further K. pneumoniae isolates expressing defined siderophores in blood (platelet concentrates, serum) was investigated. All experiments were done in triplicate and statistically significant differences were determined.
Results: Significant differences in adhesion and invasion capability, phagocytosis resistance and intracellular replication were measured between different K. pneumoniae pathotypes. Especially, ESBL-producing K. pneumoniae isolates demonstrated increased invasion in host cell lines and survival in macrophages. A strong cytotoxic effect on intestinal cells was observed for hypervirulent K. pneumoniae. The results from our investigations of the growth behavior of K. pneumoniae in platelets and serum showed that siderophores and/or an enlarged capsule are not essential factors for the proliferation of (hypervirulent) K. pneumoniae strains in blood components.
Conclusion: Our in vitro experiments revealed new insights into the host-pathogen interactions of K. pneumoniae strains representing different pathovars and clonal lineages in different infectious contexts and hosts. While a clear limitation of our study is the limited strain set used for both infection and as potential host, the results are a further step for a better understanding of the pathogenicity of K. pneumoniae and its properties essential for different stages of colonization and infection. When developed further, these results may offer novel approaches for future therapeutics including novel “anti-virulence strategies”.
Klebsiella pneumoniae is a Gram-negative bacterium that has emerged as a significant human pathogen. It poses a serious threat to public health by causing a wide range of infections in humans, including pneumonia, urinary tract infections, and bloodstream infections (Rahi et al., 2024). Originally recognized for its role in causing pneumonia more than 100 years ago, this microorganism has recently gained renewed attention due to its ability to adapt and evolve, resulting in enhanced virulence and evasion of host immune responses (Lam et al., 2018b; Martin and Bachman, 2018). Central to the pathogenicity of K. pneumoniae is its polysaccharide capsule, a complex outer layer that shields the bacterium from host defenses and contributes to its survival in diverse environments (Domenico et al., 1994; Sahly et al., 2000; Opoku-Temeng et al., 2019).
In the past 20 years, a continuous increase of K. pneumoniae with resistance to cephalosporines and carbapenems has been detected in healthcare settings worldwide. Extended-spectrum beta-lactamases (ESBL) are enzymes that confer resistance to a broad range of beta-lactam antibiotics, including penicillins and first- to third-generation cephalosporins (Fostervold et al., 2022; Hawkey et al., 2022). The prevalence of ESBL-producing K. pneumoniae strains from invasive infections varies considerably. For instance, rates of invasive K. pneumoniae with ESBL phenotypes ranged among European countries in 2021 from <5% to >80% with 34% on average and an overall increasing trend in recent years (Anonymous, 2023). Carbapenems are one of the few remaining treatment options for ESBL-producing and other drug-resistant Klebsiella infections. Thus, the emergence of carbapenem resistance is of major concern because the treatment options are limited only to a few remaining substances (Isler et al., 2022; Ding et al., 2023). Carbapenem resistance in K. pneumoniae is often mediated by the acquisition of carbapenemases of various types including KPC, OXA, and NDM (Argimón et al., 2021; Gorrie et al., 2022; Ding et al., 2023).
Hypervirulence in K. pneumoniae is a phenomenon characterized by strains that are able to cause severe infections in healthy individuals, in contrast to the usual susceptibility of immune-compromised patients to bacterial infections by classical healthcare pathogens (Russo and Marr, 2019; Wyres et al., 2020; Russo et al., 2024). One defining feature of hypervirulent K. pneumoniae is hypermucoviscosity, attributed to an increased production of capsular polysaccharides. This hypermucoviscous phenotype is associated with specific virulence factors such as the presence and expression of the regulators of the mucoid phenotype rmpA and/or rmpA2, which finally enhances capsule production, providing protection against phagocytosis and contributing to the overall virulence of the bacterium (Russo and Marr, 2019; Wyres et al., 2020; Mendes et al., 2023; Russo et al., 2024).
The role of siderophores in the virulence of hypervirulent K. pneumoniae strains has attracted significant attention in recent studies (Russo et al., 2021; Kumar et al., 2024; Wahl et al., 2024). Siderophores are small molecules that help bacteria acquire iron, an essential nutrient for their survival and pathogenicity. K. pneumoniae, like many other pathogens, utilizes siderophores to scavenge iron from the host environment, thereby enhancing its ability to colonize the host and cause infection. These siderophores not only contribute to the bacterial iron acquisition but also play a significant role in modulating the host immune response, ultimately increasing the pathogenicity of the bacterium (Bachman et al., 2012; Holden et al., 2016; Russo et al., 2021; Mendes et al., 2023). Several types of siderophores have been identified including colibactin clb, yersiniabactin ybt, salmochelin iro, and aerobactin iuc, among others. Each siderophore type plays a distinct role in iron acquisition and may have different implications for the pathogenicity of K. pneumoniae. Kleborate, a tool widely used for the genomic characterization of Klebsiella isolates, assesses the virulence potential of K. pneumoniae strains based on the presence of specific siderophores and assigns a virulence score (Lam et al., 2021; Russo et al., 2024); whereas the tool Kaptive delineates the capsule and polysaccharide composition (Lam et al., 2022). Traditionally, hypervirulent K. pneumoniae were less resistant to antibiotics, making them susceptible to common treatments. However, the acquisition of ESBL or carbapenemase genes by these hypervirulent strains not only enhances their ability to cause severe disease but also limits therapeutic options tremendously (Wyres et al., 2020; Mendes et al., 2023; Merla et al., 2024; Wahl et al., 2024). Specific world regions are primarily affected by this converging effect, in other regions and countries these strains occur sporadically (Liao et al., 2020; Hu et al., 2024; Merla et al., 2024; Wahl et al., 2024; Zhou et al., 2024).
Only a few studies investigated the association between increased antimicrobial resistance and virulence in K. pneumoniae (Mendes et al., 2023). Sahley et al. described that the acquisition of resistance-encoding plasmids is associated with increased fimbrial expression and increased adhesion to epithelial cells (Sahly et al., 2008). Another study investigated pathophysiological differences between antibiotic-susceptible, classical K. pneumoniae (cKp) and hypervirulent K. pneumoniae (hvKp) and demonstrated that hvKp resisted phagocyte-mediated clearance and replicated in mouse liver macrophages (Wanford et al., 2021). In the present study we follow previous observations and extend these analyses by providing new data about pathogen-host interactions of various K. pneumoniae strains of human and animal origin. Life-threatening K. pneumoniae infections include the capability of the pathogen to adhere to surfaces, to invade host cells and to translocate through cellular barriers. Therefore, we investigate adhesion, invasion and replication capacities to various human and animal cell lines as well as the phagocytosis resistance of cKp, ESBL-producing K. pneumoniae (ESBL-cKp) and hvKp isolates. The cell assays were carried out under normoxia and hypoxia conditions, the latter to better reflect the natural habitat of the bacterium. Furthermore, the resistance and growth in platelet concentrates and in human sera were investigated mimicking the infectious lifecycle of K. pneumoniae. The aim was to identify potential host associations of K. pneumoniae and to reveal novel findings about host adaptation of different pathotypes contributing to their pathogenicity and survival under challenging immunological conditions.
We used the following isolates of K. pneumoniae for most of the functional assays: 17-0683 and 17-0684, representing antibiotic-susceptible classical K. pneumoniae (cKp); two ESBL-producing isolates (16-0382 and 16-0383; ESBL-cKp) and two hypervirulent isolates (17-0609 and 18-0005; hvKp). Isolates 17-0683 and 17-0684 were isolated from blood cultures and were susceptible to all antibiotics tested, except ampicillin. Ampicillin resistance is mediated by an SHV-type beta-lactamase that is characteristic of the K. pneumoniae species (Livermore, 1995). The ESBL-producing isolates 16-0382 and 16-0383 originated from an urine culture and a throat swab, respectively. Both isolates were resistant to third-generation cephalosporins and possessed the ESBL gene blaCTX-M-15. The two selected hvKp isolates 17-0609 and 18-0005 belonged to the sequence types ST2398 and ST66, respectively (Klaper et al., 2021). Both isolates showed capsule type K2 and carried the virulence genes ybt, iro, iuc, clb and rmpA. Furthermore, these hvKp isolates were hypermucoviscous, confirmed by a positive string test (Pichler et al., 2017; Wahl et al., 2024). Characteristics of the above mentioned six isolates and further isolates used to determine serum resistance and growth in platelet concentrates are given in Table 1. The bacterial species was determined by an automated system (Vitek 2 ID card, bioMérieux, Nuertingen, Germany); and antimicrobial susceptibility testing (ampicillin, cefotaxime, ceftazidime, meropenem) was done using broth microdilution with result interpretation according to the criteria of EUCAST version v14.0 (EUCAST, 2024).
Genomic DNA was isolated by various methods for PCR and Sanger sequencing (DNeasy Blood and Tissue Kit, Qiagen, Hilden, Germany; MagAttract HMW DNA Kit, Machery & Nagel, Germany). Presence of beta-lactamase genes (blaTEM-like, blaSHV-like, blaCTX-M-Group1&9, blaNDM-like, blaOXA-48-like, blaOXA-1-like, blaOXA-9-like) and virulence genes (rmpA/A2, magA) was determined using PCR and Sanger sequencing as previously described (Wahl et al., 2024). For whole genome sequencing (WGS), isolates were grown in Brain Heart Infusion (BHI) broth (BD, Heidelberg, Germany). DNA was extracted from overnight cultures using DNeasy Blood and Tissue Kit (Qiagen). DNA was quantified using a Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific). The sequencing libraries were prepared using a Nextera XT DNA Library Prep Kit (Illumina®, San Diego, CA, USA). Sequencing was performed according to the manufacturer’s protocol on an Illumina Miseq using v3 chemistry (2 × 300 bp) according to the manufacturer’s protocol. Raw WGS data were processed as described (Wahl et al., 2024). Capsule types and virulence scores were determined using tools like Kleborate (v3.1.2) and Kaptive (Wyres et al., 2020; Lam et al., 2021; Lam et al., 2022). Kleborate is a bioinformatic tool for K. pneumoniae allowing species identification, MLST typing and resistance and virulence gene prediction from genomes. Kaptive allows K-locus (capsule) and O-locus (lipopolysaccharide) classification from genome data.
A549 human lung cells (ATCC CCL185, LGC Standards) were cultured in RPMI (Biowest, Nuaillé, France) medium supplemented with 10% FCS. HT29-MTX human colonic epithelial cells were grown in DMEM medium (high glucose, stable glutamine, sodium pyruvate) (Biowest) supplemented with 10% FCS and non-essential amino acids (Biowest). HuTu80 (Cell Line Service) human duodenum cells were kept in DMEM (Biowest) medium (high glucose, stable glutamine, sodium pyruvate) supplemented with 10% FCS. RAW246.7 murine macrophages (ATCC TIB-71™) were cultured in RPMI (sodium pyruvate) (Biowest) supplemented with 10% FCS. 100 U/mL penicillin and 100 μg/mL streptomycin (Biowest) were added to each medium. Cultures were then incubated at 37°C in a humidified atmosphere containing 5% (v/v) CO2.
A549 cells and HT29-MTX cells were seeded in 96-well plates (Greiner bio-one Cellstar) at a density of 3×103 cells per well, respectively. The cells were allowed to grow and differentiate for 24 hours at 37°C in a humidified atmosphere containing 5% (v/v) CO2. Overnight cultures of the bacterial strains to be analyzed were adjusted in PBS to an OD600 of 0.2 (2 × 108 bacterial cells). An inoculum corresponding to a multiplicity of infection (MOI) of 100 (A549) or 25 (HT29-MTX) was prepared in DMEM and used to infect the cells. For the infections, 100 µl of medium was removed from the cells and replaced with 100 µl of the inoculum. After a 5-minute centrifugation step at 500 rpm, the bacteria adhered during a 1 h incubation at 37°C and 5% CO2. The culture medium was then removed and non-adherent cells were removed by washing twice with PBS. The cells were lysed with PBS containing 1% Elugent (Merck Millipore, Darmstadt, Germany) and 0,0625% Antifoam B (Sigma-Aldrich, Schnelldorf, Germany) by incubating on a plate rocker at 800 rpm for 20 min at 24°C. 100 µl PBS was added to the lysates and a dilution series (several 10x dilutions) was prepared in PBS, of which 50 µl were plated on MH agar plates. The inoculated agar plates were then incubated overnight at 37°C.
A549 and HuTu80 cells were seeded in 96-well plates at a density of 3×103 cells per well, respectively. To determine the invasion rate of K. pneumoniae isolates, the cell lines were infected in microtiter plates as described above using a MOI of 100 (A549) or 50 (HuTu80). Cells were washed with PBS after infection and incubated for 1 h in cell culture medium with 100 µg/ml amikacin at 37°C and 5% CO2 to kill extracellular bacteria. After 1 h, the extracellular bacteria were removed by washing with PBS and the cells were lysed, diluted (several 10x dilutions) and plated as described above.
RAW264.7 cells (ATCC TIB-71™) were seeded in 96-well plates at a density of 3x104 cells per well. To investigate the phagocytosis rate and intracellular replication of K. pneumoniae, cells in two microtiter plates were infected in parallel using the desired MOI of 5. The phagocytosis assay was conducted as described for invasion assays. In the second microtiter plate for the intracellular replication assay, the media was replaced with new RPMI with 10 µg/ml amikacin and incubated for 24 h. The cells were then lysed as described above and dilution series (several 10x dilutions) were plated to determine the replication rate using the invasion assay as a reference.
To investigate a cytotoxic effect on intestinal cells, HuTU80 cells were infected with K. pneumoniae isolates as described above. Deviating from this, after killing and removal of extracellular bacteria, live/dead staining of the eukaryotic cells was performed using the LIVE/DEAD™ Viability/Cytotoxicity Kit (Thermo Fischer Inc) according to the manufacturer’s protocol.
Ten K. pneumoniae strains (see Table 1) were examined with regard to their growth behavior in platelet concentrates. For this purpose, the K. pneumoniae strains were grown overnight in LB medium at 37°C with shaking. On the following day, subcultures were inoculated in LB medium with an OD600 of 0.1 and incubated up to an OD600 of 0.3 at 37°C and 120 rpm. A dilution series of up to 1:100,000 in 0.85% NaCl solution was then prepared from the bacterial suspensions, which corresponds to a cell count of approximately 10 CFU (colony forming unit)/ml. To determine the inoculum, the 10-5 dilution, with which the platelet concentrates were later spiked, was plated in triplicates on LB agar plates using a spiral plater (Eddy Jet, Neutec Group, Farmingdale, NY) and incubated overnight at 37°C. Buffy-coat derived pooled platelet concentrates (German Red Cross, Frankfurt, Germany) were pooled in a sterile beaker. As a sterile control, 10 ml of the concentrates were transferred to blood culture bottles and incubated for seven days in the BacT/ALERT system (bioMeriéux, Nürtingen, Germany). Furthermore, the platelet concentrates were transferred to 25 ml blood bags, spiked with 10 CFU of the K. pneumoniae isolates to be analyzed and incubated for 43 h at 22.5°C with shaking. After the first 10 h, samples were taken every 3 h, diluted and plated in triplicates with a spiral platter on LB agar plates and incubated overnight at 37°C. The last sample was taken after a total of 43 h. The bacterial colonies were counted automatically using the Sphere Flash apparatus (Neutec Group, Farmingdale, NY).
In this study, ten K. pneumoniae strains (see Table 1) were analyzed with regard to their resistance to human serum. For this purpose, the K. pneumoniae strains were cultivated overnight at 37°C in LB medium with shaking. On the following day, subcultures were inoculated in LB medium with an OD600 of 0.1 and incubated up to an OD600 of 0.3 at 37°C and 120 rpm. From the bacterial cultures, 2 ml were pelleted for 5 min at 10000 rpm. The cell pellet was washed with 0.85% NaCl solution, pelleted again for 5 min at 10,000 rpm, and the cell pellet was taken up in 1 ml of 0.85% NaCl solution, which corresponds to a cell count of approximately 2×108 CFU/ml. A dilution series up to 1:100,000 was then prepared from the bacterial suspensions in PAS-II (115.5 mM/l NaCl; 10 mM/l Na-Citrate; 30 mM/l Na-Acetate), corresponding to a cell count of approximately 2×103 CFU/ml. To determine the inoculum, the 10-5 dilution, with which the human serum was inoculated, was plated in triplicates on LB medium using a spiral platter and incubated overnight at 37°C. Furthermore, 500 µl of the 10-5 dilution was mixed with 500 µl of 50% human serum (Merck, Darmstadt, Germany), which corresponds to a cell count of 103 CFU/ml and a serum concentration of 25% (as in platelet concentrates). The preparations were incubated in a thermoshaker at 37°C and 400 rpm (Eppendorf Thermomixer® C). After 1 h and 4 h, 100 µl of the cultures were plated in triplicates with using a spiral plater on LB agar plates and incubated overnight at 37°C. The next day, the bacterial colonies were automatically counted using the Sphere Flash apparatus.
For microscopy of the cell morphology of the different K. pneumoniae pathotypes (cKp, ESBL-cKp, hvKp), various isolates were cultivated at 37°C until the stationary growth phase (isolates 16-0382, 16-0383, 17-0683, 17-0684, 17-0609, 18-0005; see Table 1). One ml of the bacterial culture was centrifuged at 14,000 rpm for 5 min. The cell pellet was resuspended in 1 ml crystal violet solution (50 µl crystal violet + 950 µl PBS) and incubated for 1 min. The culture was then centrifuged again and the cell pellet was washed in PBS. A 60% ink solution (in PBS) was added to the bacterial suspension at a ratio of 1:2. 0.2 µl of the ink stained suspension was added to a slide coated with 1.5% agarose (in PBS) and covered with a coverslip. The phase contrast of the Nikon Eclipse Ti microscope was used to analyze the cell morphology of the bacterial cells under a 100× oil immersion objective and the images were recorded using a Nikon DS-MBWc CCD camera. To determine the cell lengths, 100 bacterial cells per strain were measured in triplicates using the MicrobeJ program. MicrobeJ is a software plugin tool developed for ImageJ to detect and count bacterial cells (https://www.microbej.com/; last access 08.01.2025).
Statistical analysis was conducted using GraphPad Prism or R software. Unless otherwise specified, all analyses were performed based on three independent experiments. The results were presented as mean ± standard deviation (S.D.). Detailed information regarding the specific statistical tests employed for each analysis can be found in the corresponding figure legends.
The six clinical K. pneumoniae isolates 16-0382, 16-0383, 17-0683, 17-0684, 17-0609, 18-0005 (see Table 1) were analyzed for possible cell morphological differences using ink staining and light microscopy. Based on the microscopic images, the thick capsular layer known for hvKp was observed and appears as a clear white halo around the bacterial cells (Figure 1A). In addition, clear differences in the cell length of the hvKp in comparison to the cKp and the ESBL-cKp isolates were observed. As can be seen in Figure 1, the cells of the hvKp isolates are longer and have a filamentous shape, while the cKp and ESBL-cKp isolates have a rod shape typical of bacteria of the Enterobacterales family. In order to quantify this result, the cell lengths distribution was analyzed as visualized in microscopic images. This revealed that the median cell length of hvKp isolates is longer than that of cKp and ESBL-cKp isolates. In addition, the hvKp isolate 18-0005 had longer cells than hvKp isolate 17-609.
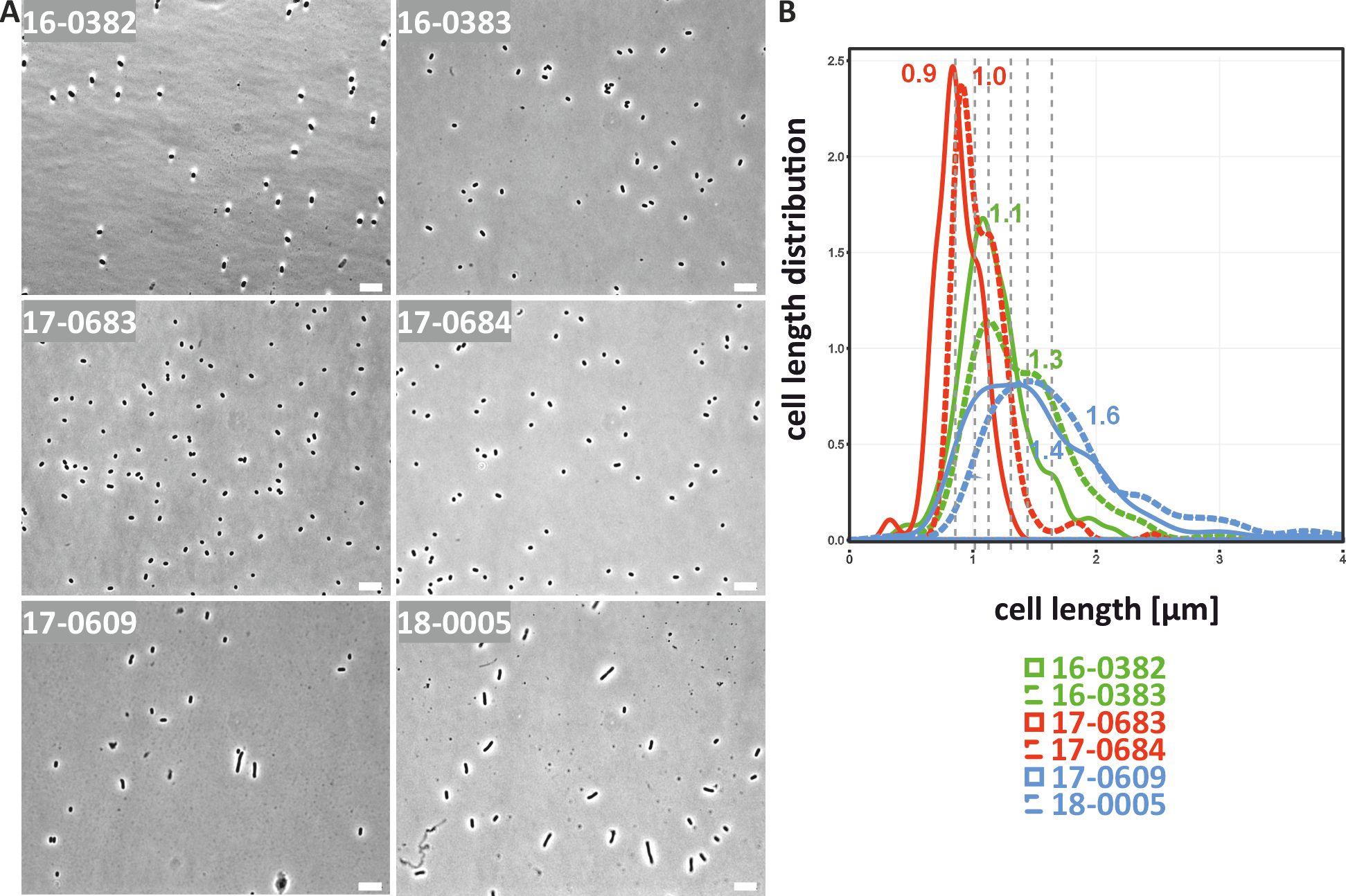
Figure 1. Cell morphological differences of cKp, ESBL-cKp and hvKp isolates under normoxia. (A) Light microscopic images of cKp (17-0683 and 17-0684), ESBL-cKp (16 0382 and 16-0383) and hvKp isolates (17-0609 and 18-0005) showed a difference in capsule thickness and cell length. (B) The distribution of cell lengths of the analyzed K. pneumoniae isolates is shown. Using MicrobeJ, triplicates of each strain with 100 cells each were measured, analyzed with R and visualized.
K. pneumoniae is a facultative anaerobic bacterium that grows optimally in the presence of oxygen, but can also survive in low-oxygen environments such as the intestine. For E. coli, it has already been shown that they react to oxygen reduction by filamentation of the bacterial cells and that this has an influence on the pathogenicity (Murashko and Lin-Chao, 2017). It should therefore be investigated whether cell morphological changes in K. pneumoniae can also be observed under oxygen reduction. For this purpose, the isolates were cultivated overnight at an oxygen concentration of 0.5 vol% and analyzed microscopically. As shown in Figure 2, an elongation of the bacterial cells was observed in light microscopic images and cell length measurements for representatives isolates of all three pathotypes. As previously shown under normoxia, the bacterial cells of the hvKp isolates were longer than those of the cKp and ESBL-cKp isolates. The results show an altered cell length of K. pneumoniae depending on oxygen. Due to the similarity to E. coli in the O2-dependent length distribution, we also suspect an influence of O2 on the pathogenic potential of K. pneumoniae.
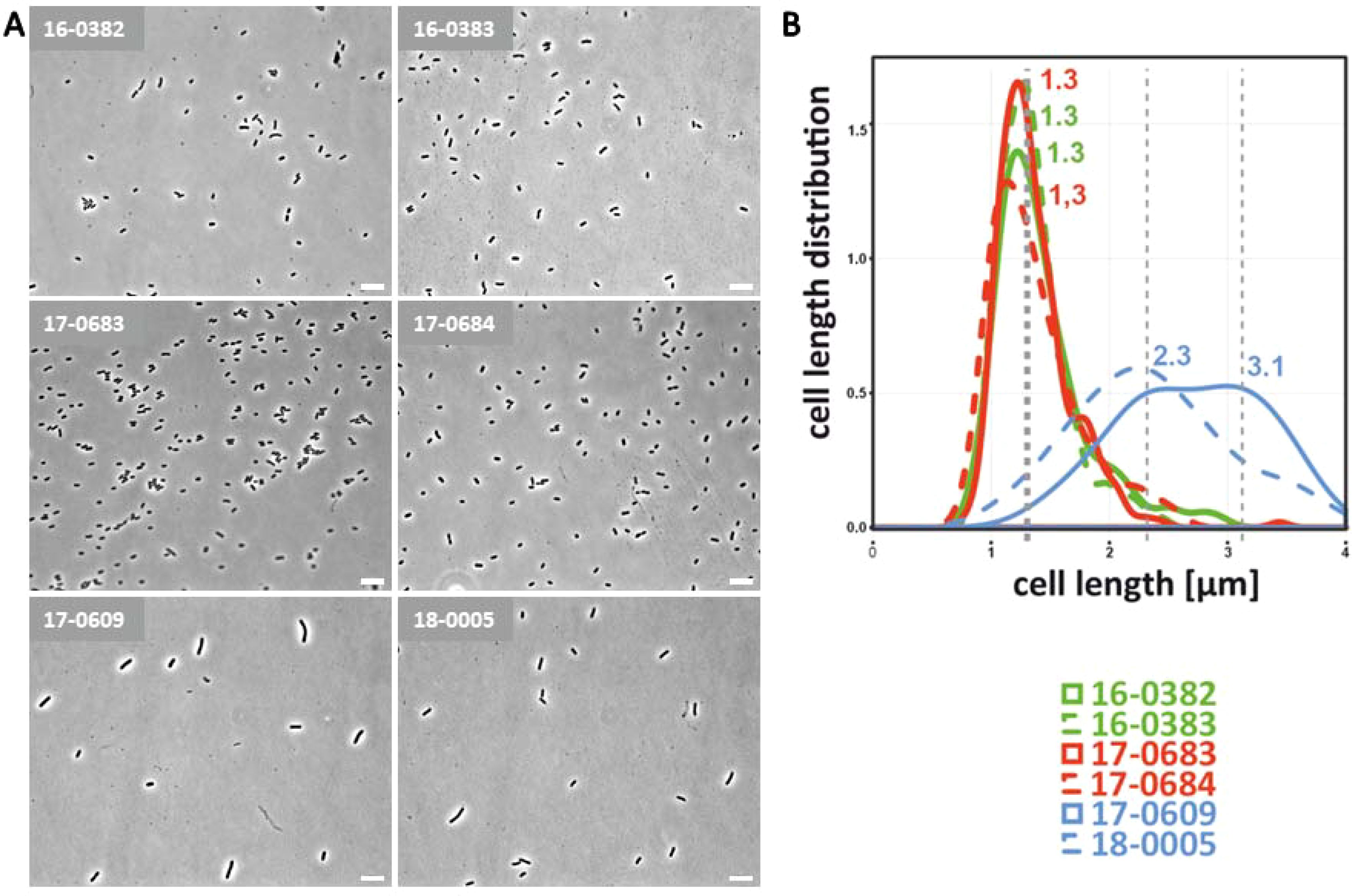
Figure 2. Cell morphological differences of cKp, ESBL-cKp and hvKp isolates under hypoxia. (A) Light microscopic images of cKp (17-0683 and 17-0684), ESBL-cKp (16 0382 and 16-0383) and hvKp isolates (17-0609 and 18-0005) showed a difference in capsule thickness and cell length. (B) Distribution of cell lengths of analyzed K. pneumoniae isolates. Using MicrobeJ, triplicates of each strain with 100 cells each were measured, analyzed with R and visualized.
Studies have shown that elongated bacterial cells can adhere better to epithelial cells in vitro due to their enlarged cell surface (Yang et al., 2016). As described above, a filamentous form was observed for the bacterial cells of hvKp isolates 17-0609 and 18-0005. In the following, it was investigated whether a difference in adhesion can be observed between the different K. pneumoniae pathotypes. Because K. pneumoniae colonizes the respiratory and intestinal tracts, the lung cell line A549 and the intestinal cell line HT29-MTX were selected as in vitro cell models. Figure 3 shows the adhesion rates as a percentage of the inoculum. The K. pneumoniae isolates analyzed adhered to a lesser extend to A549 lung cells in vitro than to HT29-MTX intestinal cells. The adhesion rate was below 5% (Figure 3A). Furthermore, no differences in adhesion capacity were observed between the different K. pneumoniae pathotypes on A549 lung cells. Although the standard deviation was large due to fluctuating adhesion rates in the individual experiments, a trend could be recognized from the data showing a better adhesion of cKp cells (independent from the ESBL status) than hvKp cells (Figure 3B). The analyzed cKp and ESBL-cKp isolates showed a higher adhesion rate (about 10% of the inoculum) to intestinal cells than to lung cells in the in vitro cell culture model. In contrast, the adhesion rate of hvKp isolates was significantly lower, the adhesion rate here was also < 5%. Thus, the results obtained in the infection experiments do not indicate a positive influence of cell morphology on the adhesion capacity.
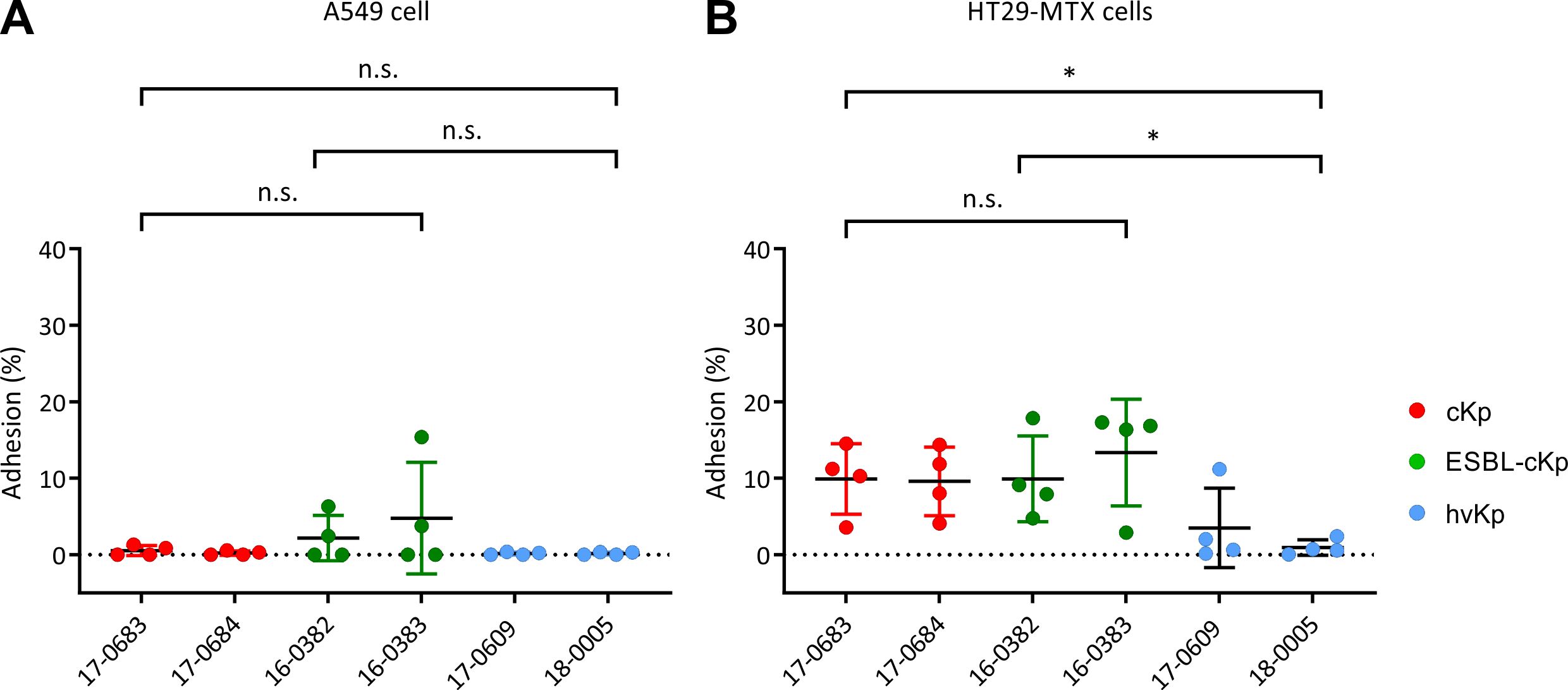
Figure 3. Quantification of the adherence of K. pneumoniae pathotypes to A549 and HT29-MTX cells. The adherence of cKp (17-0683 and 17-0684), ESBL-cKp (16-0382 and 16-0383) and hvKp isolates (17 0609 and 18-0005) to (A) A549 lung cells and (B) HT29-MTX intestinal cells is shown as a percentage of the inoculum. The data represent the results from four biologically independent triplicates. ANOVA was applied to the data to calculate significance between groups. Statistical significance is indicated as follows: n.s., not significant, *P ≤ 0.05.
We investigated whether a difference in cellular invasion can be observed between the included representative isolates of the three K. pneumoniae pathotypes. To determine the cellular invasion, the lung cell line A549 and the intestinal cell line HuTu80 were selected as models. Figure 4 shows the invasion rates as a percentage of the inoculum. The K. pneumoniae isolates analyzed showed very low invasion in vitro (invasion rate <1%) in A549 lung cells (Figure 4A) and HuTu80 intestinal cells (Figure 4B). As previously shown in the adhesion model, no significant differences were found between the different K. pneumoniae pathotypes with regard to invasion in A549 lung cells. However, differences were observed in the invasion rates determined in the cell culture model with HuTu80 intestinal cells. Here, the two ESBL-cKp isolates showed a significantly higher invasion rate than the two cKp and the two hvKp isolates. The results obtained with the infection models used did not show increased cellular invasion of the hvKp isolates compared to the cKp isolates analyzed.

Figure 4. Quantification of the invasion of K. pneumoniae pathotypes in A549 and HuTu80 cells. The invasion of cKp (17-0683 and 17-0684), ESBL-cKp (16 0382 and 16-0383) and hvKp isolates (17-0609 and 18-0005) in (A) A549 lung cells and (B) HuTu80 intestinal cells in relation to the inoculum. The data represent the results from three biologically independent triplicates. ANOVA was applied to the data to calculate significance between groups. Statistical significance is indicated as follows: n.s., not significant, *P ≤ 0.05, ****P ≤ 0.0001.
Using an in vitro infection model with murine RAW 264.7 macrophages, the interaction of the K. pneumoniae isolates with phagocytes was determined. The aim was to characterize possible differences in phagocytosis resistance between the representative isolates of the different K. pneumoniae pathotypes. To this end, the phagocytosis rate of the six different K. pneumoniae strains was first compared. After a 1 h of infection, the number of intracellular bacteria was determined. The phagocytosis rate was calculated in relation to the inoculum. Figure 5A shows the phagocytosis rate of the six K. pneumoniae strains from three independent experiments. The hvKp isolates 17-0609 and 18-0005 showed a significantly lower phagocytosis rate in vitro compared to cKp and ESBL-cKp isolates (16-0382 and 16-0383).
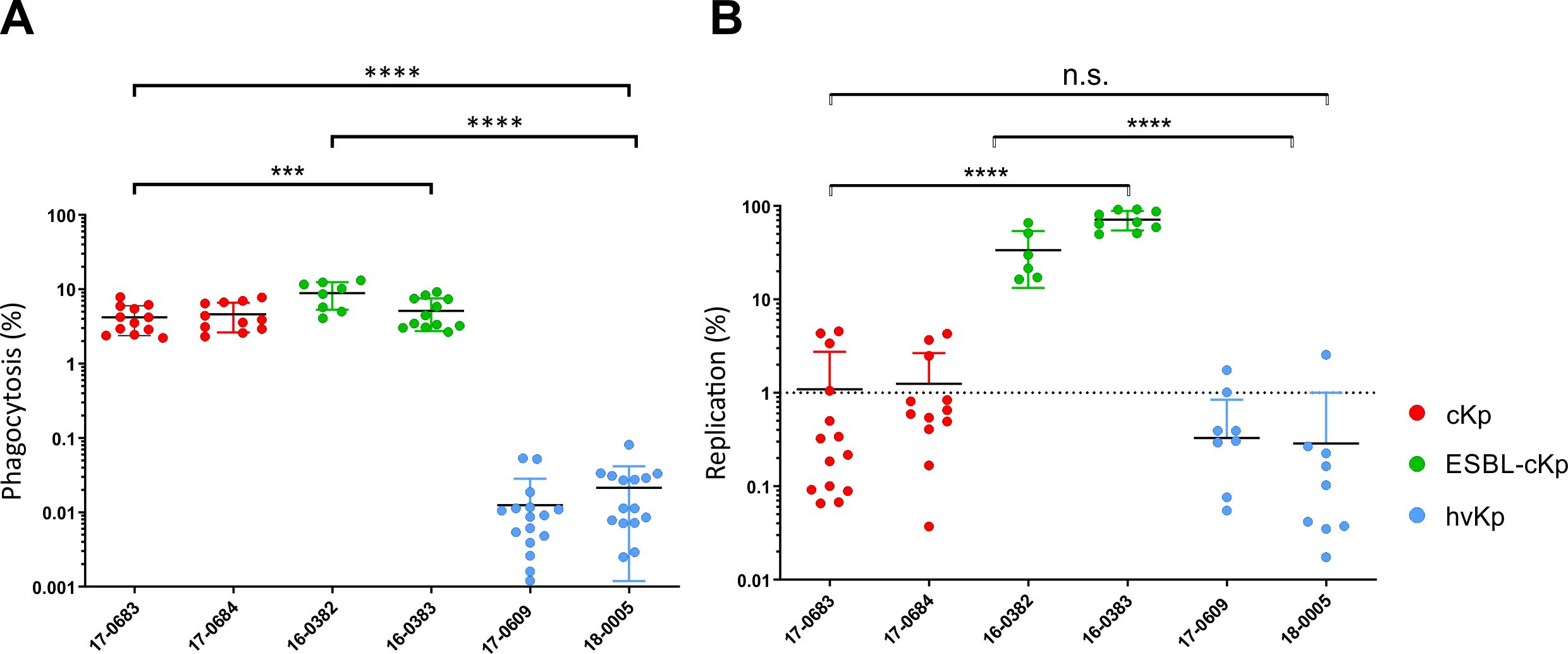
Figure 5. Quantification of phagocytosis resistance and intracellular replication of different K. pneumoniae pathotypes. (A) Phagocytosis rate of cKp (17-0683 and 17-0684), ESBL-cKp (16 0382 and 16-0383) and hvKp isolates (17-0609 and 18-0005) by murine RAW 264.7 macrophages (60,000 cells/cavity) after one hour of infection. (B) Replication rate of cKp (17-0683 and 17-0684), ESBL-cKp (16-0382 and 16-0383) and hvKp (17-0609 and 18-0005) isolates in murine RAW 264.7 macrophages (60,000 cells/cavity) after 24 hours of infection. ANOVA was applied to the data to calculate significance between groups. Statistical significance is indicated as follows: n.s., not significant, ***P ≤ 0.01 and ****P ≤ 0.0001.
To clarify whether the investigated K. pneumoniae strains survive and/or replicate intracellularly in RAW 264.7 macrophages, the number of intracellular bacterial cells was determined 24 h after infection and the replication rate was calculated in relation to the phagocytosis rate. The hvKp isolates 17-0609 and 18-0005 were killed within the macrophages as indicated by a replication rate of less than one (Figure 5B). A replication rate of about one for cKp isolates 18-0683 and 18-0684 indicated intracellular survival without replication in mouse macrophages. In contrast, both ESBL-cKp isolates showed in vitro replication rates of about 90-fold. They can persist and replicate intracellularly in macrophages.
For some enteropathogenic bacteria such as Salmonella, it has already been shown that a reduction in oxygen is associated with increased virulence of the pathogen (Marteyn et al., 2011). After filamentation of the K. pneumoniae cells under oxygen reduction has been observed (Figure 2), the aim was to investigate whether reduced oxygen availability also has an influence on the interaction with host cells. For this purpose, mouse macrophages were infected in parallel in two cell culture plates. One plate was incubated as usual under normoxia (O2 = 20.9%) and the second plate under hypoxia (O2 = 0.5%) for 24 hours. Figure 6 shows the replication rates of the analyzed six K. pneumoniae isolates. No significant difference was found in the replication rates under hypoxia. Again, only the ESBL-cKp isolates were able to replicate, while cKp isolates persisted and hvKp isolates died. No difference in intracellular replication was observed comparing hypoxic and normoxic culture conditions of the individual strains.
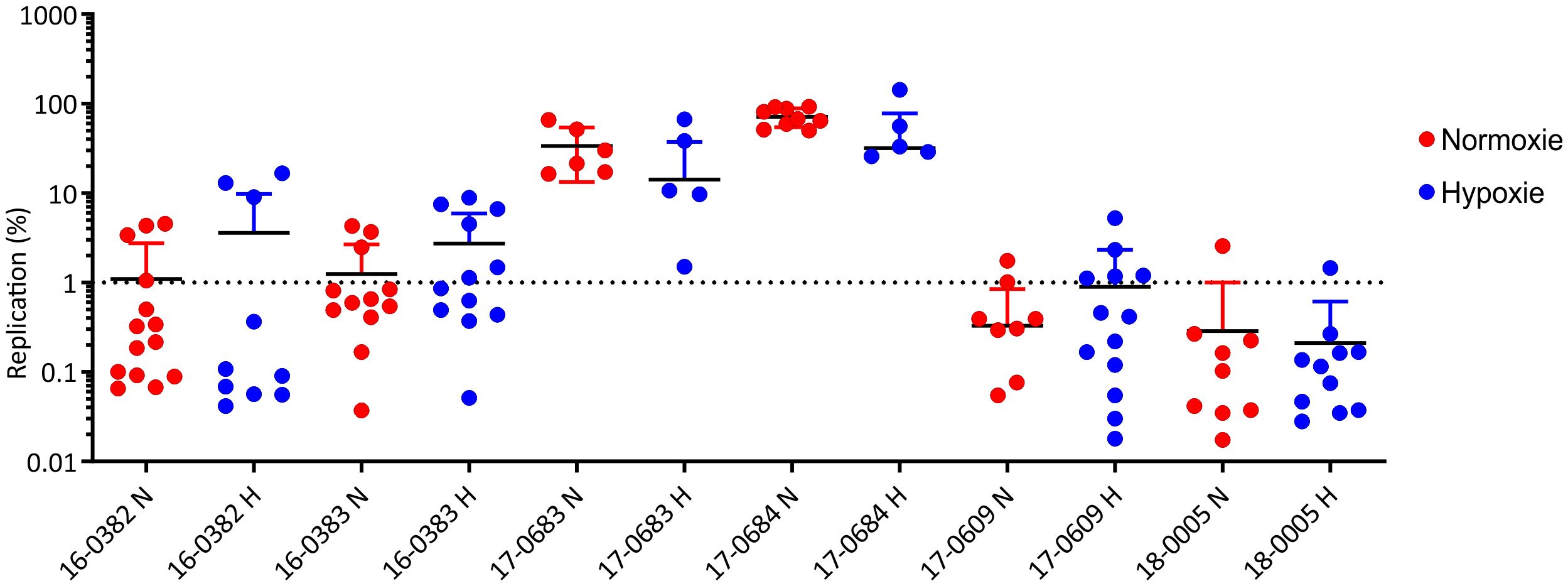
Figure 6. Comparison of the replication of K. pneumoniae isolates representing different pathotypes in RAW 264.7 mouse macrophages under normoxia (N, red) and hypoxia (H, blue). The replication rates of cKp (17-0683 and 17-0684), ESBL-cKp (16-0382 and 16-0383) and hvKp isolates (17-0609 and 18-0005) in murine RAW 264.7 macrophages (60,000 cells/cavity) under normoxia (N; O2 = 20.9%; red) and hypoxia (H; O2 = 0.5%; blue) are shown.
As described above, no cellular invasion of intestinal cells was observed for hvKp isolates. However, studies by Cano et al. had shown a cytotoxic effect of K. pneumoniae on epithelial cells (Cano et al., 2009). Therefore, we investigated whether the six K. pneumoniae isolates 16-0382, 16-0383, 17-0683, 17-0684, 17-0609 and 18-0005 have a cytotoxic effect on intestinal cells. For this purpose, HuTu80 intestinal cells were infected with the K. pneumoniae isolates as previously described. Live/dead staining with calcein AM (viable) and ethidium bromide (non-viable) was used to discriminate between viable and non-viable HuTu80 cells, which were analyzed both photometrically and microscopically. Figure 7 shows the percentages of viable and non-viable HuTu80 cells. All investigated cKp, ESBL-cKp and hvKp isolates showed a cytotoxic effect on intestinal cells in vitro. The viability of HuTu80 cells after infection was below 80% for isolates of all pathotypes and the percentage of non-viable cells was above 20%. HvKp isolates showed a stronger cytotoxic effect in vitro than cKp and ESBL-cKp isolates. Microscopic images of cells infected with hvKp isolates showed the loss of integrity of the HuTu80 monolayer, whereas a confluent cell lawn persisted after infections with cKp and ESBL-cKp isolates (Figure 7C).

Figure 7. Comparison of the cytotoxic effect of K. pneumoniae pathotypes on HuTu80 intestinal cells, showing the percentage of viable (A) and non-viable (B) HuTu80 intestinal cells after infection with cKp (17-0683 and 17-0684), ESBL-cKp (16 0382 and 16-0383) and hvKp isolates (17-0609 and 18-0005). The data represent the results of triplicates from three independent experiments. ANOVA was applied to the data to calculate significance between groups. Statistical significance is indicated as follows: *P ≤ 0.05 and **P ≤ 0.01. Transmitted light microscopy (C) and fluorescence microscopy (D) images of viable (green) and non-viable (red) HuTu80 intestinal cells after infection with K. pneumoniae. n.s., not significant.
The growth behavior of 10 K. pneumoniae isolates, which harbor different siderophore genes (Table 1), was compared with that of the K. pneumoniae reference strain PEI-B-P-08 [143] in platelet concentrates. For this purpose, a 50 ml platelet concentrate was spiked with 10 CFU of one of the K. pneumoniae isolates to be tested and incubated under conditions specified by haemotherapy guidelines (Spindler-Raffel et al., 2017; Prax et al., 2019). Figure 8 shows the growth kinetics of the analyzed K. pneumoniae isolates over a period of 43 hours. For reference strain PEI-B-P-08 (cKp), hvKp reference strain NCTC 14052 and test strains 18-0030 (cKp) and 17-0609 (hvKp), an increase in bacterial count was observed after 10 h, with the bacterial count of isolates NCTC 14052 and PEI-B-P-08 being one order of magnitude higher than that of isolates 18-0030 and 17-0609. Furthermore, the growth of isolates NCTC 14052 and PEI-B-P-08 was exponential, while the growth of isolates 18-0030 and 17-0609 showed a linear increase. After 37-43 h, the growth of isolates NCTC 14052, PEI-B-P-08, 18-0030 and 17-0609 was in the stationary phase with bacterial counts of about 107. The isolates 19-0213 (hvKp) and ATCC 700721 (cKp) exhibited a long lag phase of over 22 h. Only after 38 h a significant increase in bacterial counts was detected for these isolates. Four further isolates analyzed (ATCC13883 (cKp), 17-0736 (hvKp), 18-0414 (ESBL-cKp) and 19-0036 (hvKp) were unable to grow in platelet concentrates. This growth behavior could be due to different serum resistances (Doorduijn et al., 2016).
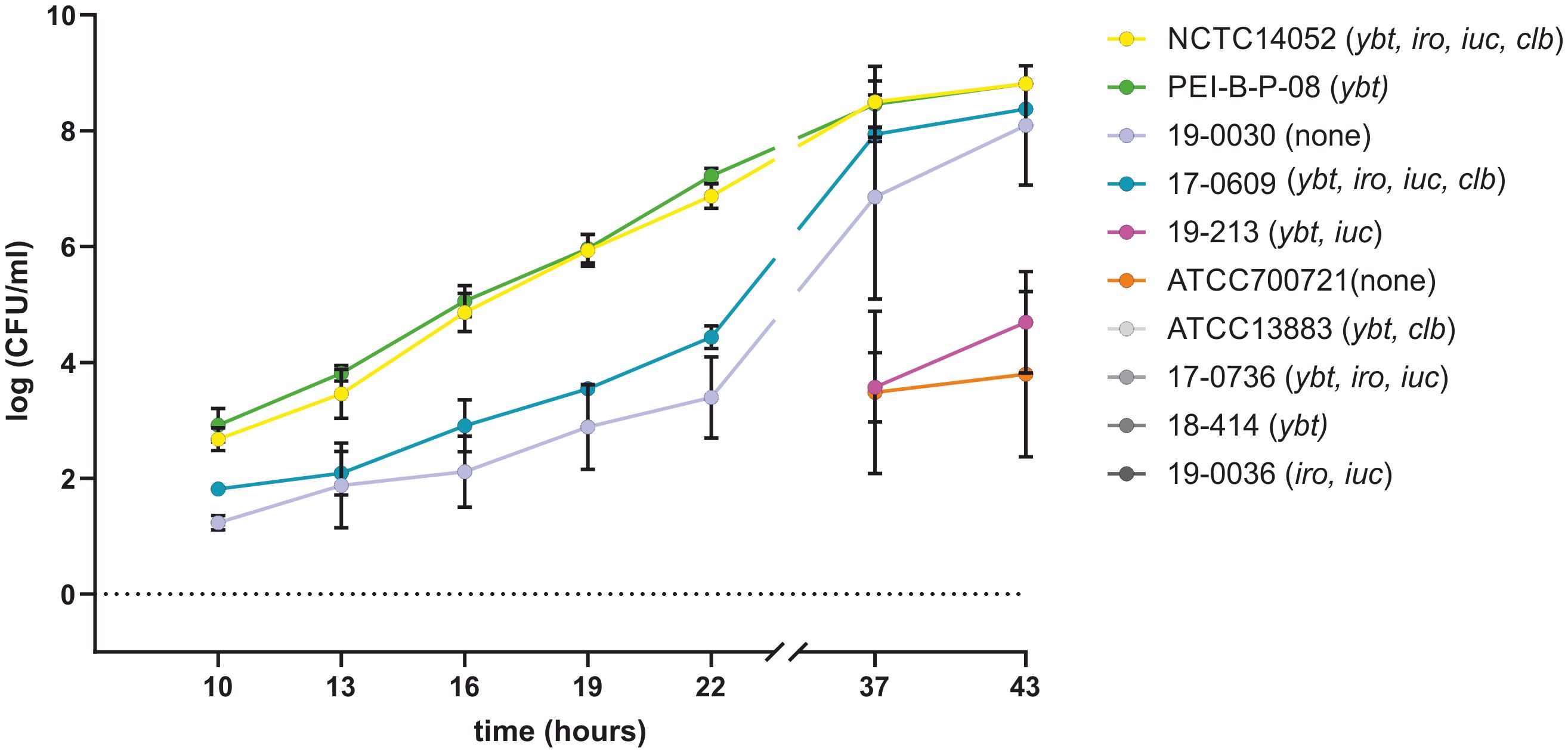
Figure 8. Comparison of the growth behavior of K. pneumoniae isolates in platelet concentrates. The growth curves of K. pneumoniae isolates ATCC 13883, ATCC 700721, NCTC 14052, PEI-B-P-08, 17-0609, 17-0736, 18-0030, 18-0414, 19-0036 and 19-0213 at 22.5°C in platelet concentrates are shown. No growth could be detected for the four isolates labelled with grey colors. The data represent the results of triplicates from three independent experiments.
In order to investigate possible serum resistance of the ten K. pneumoniae isolates, human serum was inoculated with 103 CFU of the isolates to be analyzed. Figure 9 shows the results of the bacterial counts determined after 1 h and 4 h. The isolates PEI-B-P-08 (cKp), NCTC 14052 (hvKp), 17-0609 (hvKp) 18-0030 (cKp), 19-0213 (hvKp) and ATCC 700721 (cKp), were resistant to human serum. While the bacterial counts of isolates ATCC 700721 remained stable, increased bacterial counts were observed for isolates NCTC 14052, PEI-B-P-08, 18-0030, 17-0609, and 19-0213. The isolates ATCC 13883 (cKp), 17-0736 (hvKp), 18-0414 (ESBL-cKp) and 19-0036 (hvKp) are not resistant to human serum, here bacterial counts decreased over 4 h.
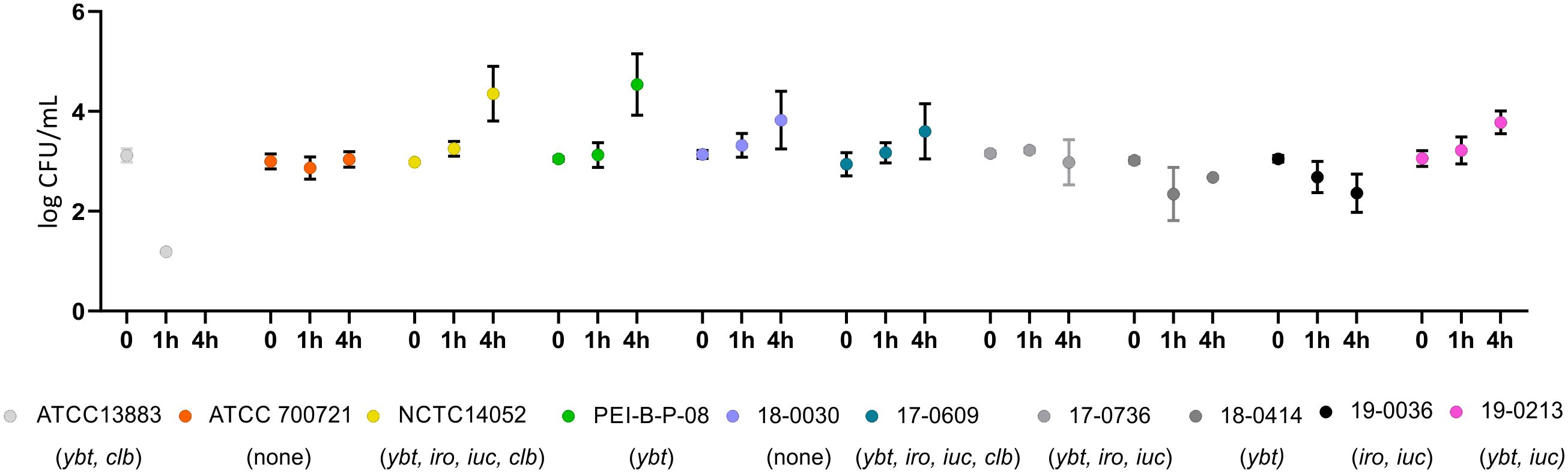
Figure 9. Comparison of the serum resistance of ten K. pneumoniae isolates. Serum resistance of K. pneumoniae isolates ATCC 13883, ATCC 700721, NCTC 14052, PEI-B-P-08, 17-0609, 17-0736, 18-0030, 18-0414, 19-0036 and 19-0213 at 37°C in 25% human serum. No growth could be detected for isolates labelled in different grey colors. The data represent the results of triplicates from three independent experiments.
Previous studies have focused on genome-wide association studies of globally distributed K. pneumoniae lineages (“high-risk clones”) and epidemiological studies to identify risk factors that favor nosocomial K. pneumoniae infections (Cano et al., 2009; Meatherall et al., 2009; Martin and Bachman, 2018; Cienfuegos-Gallet et al., 2019). However, in the face of increasing antibiotic resistance and the global spread of hypervirulent strains, a better understanding of the pathogenesis of K. pneumoniae is essential for the development of preventive measures and more targeted treatments including “anti-virulence strategies”. Therefore, the present study investigated properties of different K. pneumoniae pathotypes in order to be able to make statements about host adaptations that contribute to the successful spread of this bacterium in the different habitats.
First, differences in the cell morphology of hvKp isolates compared to cKp and ESBL-cKp isolates were determined and documented by means of light microscopy (Figures 1, 2). The analyzed hvKp isolates showed a filamentous phenotype. In the literature, filamentation of bacterial cells is often described as the result of impaired cell division (Klein et al., 2015; Cayron et al., 2023). However, some bacteria are able to react to stress conditions such as oxidative stress, nutrient deficiency or the defense mechanisms of the host immune system by a temporary change in cell morphology (Justice et al., 2008). For E. coli, it has been observed that the enlargement of the cell surface associated with a filamentous phenotype has a positive effect on adhesion to surfaces and protects the bacteria from phagocytosis by immune cells (Klein et al., 2015). In our study, cell filamentation of hvKp was much more pronounced when compared to cKp and ESBL-cKp, but cell lengths’ variation among individual hvKp isolates was also remarkable suggesting a potential greater polymorphic population than among cKp and ESBL-cKp. However, adhesion of hvKp to two different cell lines was weaker compared to cKp (Figures 1–3). Thus, the positive effect of cell filamentation might be compensated by the protective effect of the larger capsule (Figure 3). Under hypoxia, an increase in cell length was also observed for cKp and ESBL-cKp isolates, but to a much lesser extent than for hvKp isolates. It is known that filamentous bacteria have an advantage in adherence to epithelial cells. As hypoxia conditions prevail in the gut, reduced oxygen levels could be a reason for K. pneumoniae to alter cell morphology in order to adhere better to the intestinal epithelium.
The cell culture model was used to investigate whether representative isolates of the three K. pneumoniae pathotypes show differences in the pathogen-host interaction. Since respiratory and gastrointestinal colonization of the bacterium is considered to be the main reservoir for K. pneumoniae infections, the cell infection experiments were carried out with the lung cell line A549 and the intestinal cell lines HT29-MTX and HuTu80 (Sun et al., 2019). In previous cell infection experiments, the adhesion and cellular invasion of encapsulated and non-encapsulated K. pneumoniae isolates were compared (Sahly et al., 2000; Cortes et al., 2002; Struve and Krogfelt, 2003; Opoku-Temeng et al., 2019). A direct correlation between the adhesion to and invasion of lung epithelial cells and the ability of K. pneumoniae isolates to express capsular polysaccharide has been observed (Fumagalli et al., 1997; Sahly et al., 2000; Struve and Krogfelt, 2003; de Astorza et al., 2004). No differences of the included representative strains representing the three K. pneumoniae pathotypes cKp, ESBL-cKp and hvKp in adherence to and invasion of A549 lung cells were found (Figures 3A, 4A). In general, adhesion/invasion rates are lower with the lung cell lines compared to intestinal cells. These results suggest that adhesion and invasion of lung cells is not a classical pathophysiological mechanism of K. pneumoniae. One possible way in which the bacterium is able to overcome the cell barrier in the respiratory tract is by disrupting the cell integrity of the epithelium through injury in the respiratory tract or, as described by Cano et al., via a cytotoxic effect on A549 lung cells by encapsulated K. pneumoniae isolates (Cano et al., 2009).
In the intestinal cell model, however, differences in adherence and invasion between the isolates representing the three different K. pneumoniae pathotypes were observed (Figures 3B, 4B). While the cKp and ESBL-cKp isolates are equally capable of adhering to intestinal cells, no adhesion was observed for the hvKp isolates analyzed. These results are congruent with the observations that a thick capsular layer reduces the adhesion capacity of K. pneumoniae (Sahly et al., 2000). Furthermore, the hypothesis that an enlarged cell surface increases adhesion due to a filamentous phenotype was not confirmed under normoxic conditions. Hus et al. were able to show in a cell culture model that K. pneumoniae can overcome the barrier function of the intestinal epithelium via a transcellular pathway (Hsu et al., 2015). This could only be confirmed for ESBL-cKp in the present study. The cKp and hvKp isolates showed no cellular invasion in vitro in the intestinal cell line used. These results are comparable to previous data by Sahly et al. who observed an increased cellular invasion of ESBL-producing K. pneumoniae isolates (Sahly et al., 2008), an effect that might potentially derive from additional features encoded on ESBL-containing plasmids than from the pure presence of the ESBL gene itself.
The clinical pictures of hvKp infections include invasive and necrotizing infections, which suggests a cytoxic effect of hvKp isolates (Evangelista et al., 2018). Our study showed that the investigated hypervirulent isolates had a significantly higher cytotoxic effect on intestinal epithelial cells than cKp or ESBL-cKp isolates. Holes in the monolayer were observed after infection demonstrating the loss of epithelial integrity (Figure 7C). It can be assumed that the proportion of non-viable cells was even higher, as the detached (and subsequently washed away) cells could not be detected. This indicates that hvKp do not invade intestinal cells but (Figure 4), due to their cytotoxic effect, damage the intestinal epithelium and overcome the intestinal barrier in this way. Recent studies in particular describe virulence factors that support this hypothesis. For example, a genotoxic effect on eukaryotic cells was observed in vitro for colibactin-producing K. pneumoniae isolates (Lai et al., 2014; Lu et al., 2017). It is assumed that colibactin leads to apoptosis of intestinal cells and thus enables the intestinal translocation of hvKp (Secher et al., 2015; Wyres et al., 2020).
The capsule of K. pneumoniae is the most important virulence factor (Huynh et al., 2017; Opoku-Temeng et al., 2019). It protects the bacteria from antibiotics, the complement system of the immune defense and phagocytosis (Kabha et al., 1995). It is known from the literature that encapsulated K. pneumoniae are resistant to opsonization in vivo and to uptake by phagocytes (Domenico et al., 1994; Alvarez et al., 2000). The studies also compared encapsulated K. pneumoniae isolates and capsule mutants with each other. However, only the general role of the capsule on host interaction was investigated. No conclusions can be drawn about differences between the K. pneumoniae pathotypes. The present study investigated the extent to which the abilities of phagocytosis resistance, intracellular persistence and replication differ between the included isolates representing the various K. pneumoniae pathotypes. Using a mouse macrophage infection model, significant differences were observed, both cKp and ESBL-cKp were equally phagocytosed by the immune cells, while the hvKp isolates were not (Figure 5).
Our results also showed no intracellular persistence or replication for hvKp isolates (Figure 6). There are several possible reasons for this. Firstly, increased capsule synthesis is characteristic of hvKp isolates, which makes them less accessible to macrophages. Furthermore, as described above, filamentation of the hvKp bacterial cells was observed. For some bacteria it is known that they change from the bacillary to the filamentous cell form and are therefore less accessible for phagocytotic cells (Prashar et al., 2013). However, the comparatively low number of phagocytosed hvKp must be considered as a limitation, which led to a higher variability of the individual experiments despite a large number of replicates.
The ability of ESBL-cKp to increase intracellular persistence compared to cKp as described by Cano et al. was confirmed in the present study (Cano et al., 2015) (Figures 5, 6). Interestingly, the ESBL-Kp isolates analyzed were also able to replicate in RAW macrophages. While hvKp were able to evade macrophage access, ESBL-cKp have found a strategy to replicate in macrophages similar to classical intracellular pathogens such as Legionella pneumophila and Salmonella enterica (Price and Vance, 2014).
There is currently a lack of a standardized method for virulence prediction of K. pneumoniae and thus a derivation of markers for laboratory diagnostics. To date, virulence analyses have concentrated on genome comparisons and genome-wide association studies (Spadar et al., 2022; Cheung et al., 2023). A conventional assessment of the virulence potential and the differentiation of cKp and hvKp isolates is currently under discussion and is based, among other things, on the detection of specific iron transport systems (Holt et al., 2015; Lam et al., 2018b; Lam et al., 2018a). Molecular epidemiological studies have shown that the prevalence of iron transport systems is higher in hvKp isolates than in cKp isolates (Russo et al., 2015; Russo and Marr, 2019).
In this study, the growth of hvKp isolates with different siderophore systems (Table 1) was compared with that of the reference strain PEI-BP-08 in platelet concentrates. The PEI-BP-08 reference isolate (ST48-K62), which only possesses ybt, and the hvKp reference strain NCTC 14052 (ST23-K1) harboring all relevant siderophore genes (ybt, iro, iuc, clb) and capsule regulator genes (rmpA/rmpA2), in addition, were among the fast-growing isolates. cKp isolate 18-0030 (ST310-K1) and hvKp isolate 17-0609 (ST3895-K2) were also able to proliferate in platelet concentrates, but their proliferation was delayed. Interestingly, for the hvKp isolate 19-0213 (ST23-KL107) and ESBL-cKp isolate ATCC 700721 (ST38-K15), proliferation was only detected after an incubation period of 37 hours. Bacteria with such a long lag phase represent a major challenge in transfusion medicine, as such bacterial contamination is difficult to detect and harbors the risk of transfusion-related bacterial infections. Despite the presence of siderophores, no growth could be observed for four of the analyzed isolates of various classifications and with presence of several siderophore genes.
In contrast to the present knowledge and working hypotheses (Holt et al., 2015; Russo and Marr, 2019), our study results indicate that siderophores are not essential factors for the proliferation of K. pneumoniae in blood components. It was shown that virulence prediction by the presence or absence of siderophores alone is not necessarily associated with the ability to proliferate in blood components, which calls into question the reliability of the proposed classification of hvKp isolates based on the presence of specific iron transport systems only (Holt et al., 2015; Lam et al., 2021). Instead, the results of the present work provide evidence that the capsule plays the decisive role in the pathogenesis of K. pneumoniae. Current studies are also increasingly focusing on the investigation of the capsule of K. pneumoniae and its regulation and expression as well as synthesis mechanisms (Chu et al., 2023; Muner et al., 2024; Sun et al., 2024; Tang et al., 2024). For hvKp isolates in particular, more and more genetic factors are being described that lead to attenuated capsule synthesis compared to other pathotypes and offer a survival advantage in the host organism (Walker et al., 2020). In particular, the presence of the transcriptional regulators RmpA and RmpA2 is associated with a hypermucoviscous phenotype and increased virulence (Lai et al., 2003; Cheng et al., 2010; Hsu et al., 2015; Lin et al., 2020; Chu et al., 2023).
Just recently, Russo et al. published a study where they investigated 49 K. pneumoniae strains possessing varying combinations of siderophore and other virulence genes associated with a hypervirulence phenotype and with different antibiotic resistance properties (Russo et al., 2024). Altogether 16 isolates were categorized as hvKp and 33 as cKp based on their behavior in a murine infection model. Biomarker presence, siderophore production, mucoviscosity, virulence plasmid homogeneities, and Kleborate virulence scores were measured and evaluated to accurately differentiate pathotypes. The presence of all the investigated biomarkers iucA, iroB, peg-344, rmpA, and rmpA2 was most accurate (94%) to predict hypervirulence; whereas the presence of ≥4 of these biomarkers was most sensitive (100%). Even when using such an extensive and well-defined strain collection and informative models and logistic regressions, predictions of hypervirulence in K. pneumoniae isolates remains challenging.
In a recent paper, Masson et al., describe that K. pneumoniae O1 antigen prevents complement mediated killing when these isolates were compared to a larger collection of K. pneumoniae isolates with other O antigens (Masson et al., 2024). We did not see any link between pathotypes, presence of O antigens and growth and survival in serum samples or platelet concentrates (Figures 8, 9). Admittedly, our strain collection was less variable in terms of O-antigen structure. However, K. pneumoniae isolates with identical O1-antigens behaved differently in the two mentioned experimental settings suggesting the important role of components other than the O locus involved in growth and survival in blood or similar compartments.
Our study has several limitations. First, the classification into the three groups cKp, ESBL-cKp and hvKp is oversimplified because it ignores major differences in genome content, in addition to the group designations, between the isolates within a given group and across different categories. These differences may potentially also influence cell-biological behavior in the given experiments. Second, we might have validated our experimental findings by including more than two isolates per given group. The limitation in isolate number per group is due to the fact that each experiment had been executed three times at least due to statistical validations, and that we included quite a high number of different adhesion, invasion and replication experiments and various cell line types. We have extensively tested and analyzed a larger set of cell lines and experimental settings beforehand, which is not described in greater details in this manuscript. Third, we are well aware that using isogenic strains would be beneficial for the experimental settings. For instance, using two strain types just differing by a blaESBL-containing plasmid (e.g., as a cKp vs. ESBL-cKp) would better specify some findings and let assume to point towards physiological and immunological behavior to a few genomic differences only. However, well-defined and characterized K. pneumoniae recipients were not available when we planned and started our experiments some years ago. Additionally, genetic strategies to cure plasmids safely from K. pneumoniae strains were not established. Forth, it would have been advantageous, if we have used a complete and entire strain set through all the experiments included. The selection and limitation to a smaller or slightly different isolate selection was due to organizational and infrastructural requirements and demands of QM/QC at the different research institutions involved.
In summary, the data from the in vitro infection experiments showed that K. pneumoniae isolates representing various pathotypes (cKp, ESBL-cKp, hvKp) differ in the host-pathogen interactions with regard to their invasion, resistance to phagocytosis and replication potential in macrophages. Thus, evidence was found that the successful spread of strains representing distinct K. pneumoniae lineages is not solely due to a survival advantage under selective pressure by antibiotics, which contributes to a better understanding of pathogenic mechanisms of K. pneumoniae. A stronger cytotoxic effect on intestinal cells was observed for hvKp, which presumably favors their translocation. Furthermore, it could be shown that the pure presence of siderophore genes have no positive influence on the proliferation of K. pneumoniae in blood.
The data presented in the study are deposited in the ENA repository, project accession number PRJEB85663.
KK: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. YP: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Validation, Writing – original draft, Writing – review & editing. LH: Formal analysis, Investigation, Methodology, Writing – review & editing. MP: Formal analysis, Investigation, Methodology, Supervision, Validation, Writing – review & editing. OK: Formal analysis, Investigation, Methodology, Supervision, Validation, Writing – review & editing. IB: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Validation, Writing – review & editing. AW: Investigation, Methodology, Validation, Visualization, Writing – review & editing. MF: Data curation, Investigation, Methodology, Validation, Writing – review & editing. HK: Conceptualization, Data curation, Resources, Writing – review & editing. SB: Data curation, Formal analysis, Investigation, Writing – review & editing. RG: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Supervision, Validation, Writing – review & editing. GW: Supervision, Writing – original draft, Writing – review & editing, Conceptualization, Funding acquisition, Project administration, Resources.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The study received support by a research grant from the Robert Koch Institute as part of the GOHI initiative (https://www.gohi.online/GOHI/EN/Home/Homepage_node.html, accessed on 08.01.2025) to GW and KK.
We appreciate excellent technical assistance by Sybille Mueller-Bertling and Kirstin Ganske.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alvarez, D., Merino, S., Tomás, J. M., Benedí, V. J., Albertí, S. (2000). Capsular polysaccharide is a major complement resistance factor in lipopolysaccharide O side chain-deficient Klebsiella pneumoniae clinical isolates. Infect. Immun. 68, 953–955. doi: 10.1128/IAI.68.2.953-955.2000
Anonymous (2023). Antimicrobial resistance in the EU/EEA (EARS-Net) - Annual Epidemiological Report 2022 (Stockholm, Sweden: European Centre for Disease Prevention and Control).
Argimón, S., David, S., Underwood, A., Abrudan, M., Wheeler, N. E., Kekre, M., et al. (2021). Rapid genomic characterization and global surveillance of Klebsiella pneumoniae using Pathogenwatch. Clin. Infect. Dis. 73, S325–SS35. doi: 10.1093/cid/ciab784
Bachman, M. A., Lenio, S., Schmidt, L., Oyler, J. E., Weiser, J. N. (2012). Interaction of lipocalin 2, transferrin, and siderophores determines the replicative niche of Klebsiella pneumoniae during pneumonia. Mbio 3, e00224–e00211. doi: 10.1128/mBio.00224-11
Becker, L., Fuchs, S., Pfeifer, Y., Semmler, T., Eckmanns, T., Korr, G., et al. (2018). Whole genome sequence analysis of CTX-M-15 producing Klebsiella iIsolates allowed dissecting a polyclonal outbreak scenario. Front. Microbiol. 9, 322. doi: 10.3389/fmicb.2018.00322
Cano, V., March, C., Insua, J. L., Aguiló, N., Llobet, E., Moranta, D., et al. (2015). Klebsiella pneumoniae survives within macrophages by avoiding delivery to lysosomes. Cell Microbiol. 17, 1537–1560. doi: 10.1111/cmi.12466
Cano, V., Moranta, D., Llobet-Brossa, E., Bengoechea, J. A., Garmendia, J. (2009). Klebsiella pneumoniae triggers a cytotoxic effect on airway epithelial cells. BMC Microbiol. 9, 156. doi: 10.1186/1471-2180-9-156
Cayron, J., Dedieu-Berne, A., Lesterlin, C. (2023). Bacterial filaments recover by successive and accelerated asymmetric divisions that allow rapid post-stress cell proliferation. Mol. Microbiol. 119, 237–251. doi: 10.1111/mmi.v119.2
Cheng, H. Y., Chen, Y. S., Wu, C. Y., Chang, H. Y., Lai, Y. C., Peng, H. L. (2010). RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43. J. Bacteriol. 192, 3144–3158. doi: 10.1128/JB.00031-10
Cheung, B. H., Alisoltani, A., Kochan, T. J., Lebrun-Corbin, M., Nozick, S. H., Axline, C. M. R., et al. (2023). Genome-wide screens reveal shared and strain-specific genes that facilitate enteric colonization by Klebsiella pneumoniae. Mbio 14, e0212823. doi: 10.1128/mbio.02128-23
Chu, W. H. W., Tan, Y. H., Tan, S. Y., Chen, Y., Yong, M., Lye, D. C., et al. (2023). Acquisition of regulator on virulence plasmid of hypervirulent Klebsiella allows bacterial lifestyle switch in response to iron. Mbio 14, e0129723. doi: 10.1128/mbio.01297-23
Cienfuegos-Gallet, A. V., Ocampo de Los Rios, A. M., Sierra Viana, P., Ramirez Brinez, F., Restrepo Castro, C., Roncancio Villamil, G., et al. (2019). Risk factors and survival of patients infected with carbapenem-resistant Klebsiella pneumoniae in a KPC endemic setting: a case-control and cohort study. BMC Infect. Dis. 19, 830. doi: 10.1186/s12879-019-4461-x
Cortes, G., Alvarez, D., Saus, C., Alberti, S. (2002). Role of lung epithelial cells in defense against Klebsiella pneumoniae pneumonia. Infect. Immun. 70, 1075–1080. doi: 10.1128/IAI.70.3.1075-1080.2002
de Astorza, B., Cortes, G., Crespi, C., Saus, C., Rojo, J. M., Alberti, S. (2004). C3 promotes clearance of Klebsiella pneumoniae by A549 epithelial cells. Infect. Immun. 72, 1767–1774. doi: 10.1128/IAI.72.3.1767-1774.2004
Ding, L., Shen, S., Chen, J., Tian, Z., Shi, Q., Han, R., et al. (2023). Klebsiella pneumoniae carbapenemase variants: the new threat to global public health. Clin. Microbiol. Rev. 36, e0000823. doi: 10.1128/cmr.00008-23
Domenico, P., Salo, R. J., Cross, A. S., Cunha, B. A. (1994). Polysaccharide capsule-mediated resistance to opsonophagocytosis in Klebsiella pneumoniae. Infect. Immun. 62, 4495–4499. doi: 10.1128/iai.62.10.4495-4499.1994
Doorduijn, D. J., Rooijakkers, S. H., van Schaik, W., Bardoel, B. W. (2016). Complement resistance mechanisms of Klebsiella pneumoniae. Immunobiology 221, 1102–1109. doi: 10.1016/j.imbio.2016.06.014
Evangelista, V., Goncalves, C. V., Almeida, R., Henriques, C., Baptista, A. M., da Graca, J. P., et al. (2018). Klebsiella pneumoniae invasive syndrome. Eur. J. Case Rep. Intern. Med. 5, 000800. doi: 10.12890/2018_000800
Fostervold, A., Hetland, M. A. K., Bakksjo, R., Bernhoff, E., Holt, K. E., Samuelsen, O., et al. (2022). A nationwide genomic study of clinical Klebsiella pneumoniae in Norway 2001-15: introduction and spread of ESBLs facilitated by clonal groups CG15 and CG307. J. Antimicrob. Chemoth. 77, 665–674. doi: 10.1093/jac/dkab463
Fumagalli, O., Tall, B. D., Schipper, C., Oelschlaeger, T. A. (1997). N-glycosylated proteins are involved in efficient internalization of Klebsiella pneumoniae by cultured human epithelial cells. Infect. Immun. 65, 4445–4451. doi: 10.1128/iai.65.11.4445-4451.1997
Gorrie, C. L., Mirceta, M., Wick, R. R., Judd, L. M., Lam, M. M. C., Gomi, R., et al. (2022). Genomic dissection of Klebsiella pneumoniae infections in hospital patients reveals insights into an opportunistic pathogen. Nat. Commun. 13, 3017. doi: 10.1038/s41467-022-30717-6
Hawkey, J., Wyres, K. L., Judd, L. M., Harshegyi, T., Blakeway, L., Wick, R. R., et al. (2022). ESBL plasmids in Klebsiella pneumoniae: diversity, transmission and contribution to infection burden in the hospital setting. Genome Med. 14, 97. doi: 10.1186/s13073-022-01103-0
Holden, V. I., Breen, P., Houle, S., Dozois, C. M., Bachman, M. A. (2016). Klebsiella pneumoniae siderophores induce inflammation, bacterial dissemination, and HIF-1α stabilization during pneumonia. Mbio 7, e01397–e01316. doi: 10.1128/mBio.01397-16
Holt, K. E., Wertheim, H., Zadoks, R. N., Baker, S., Whitehouse, C. A., Dance, D., et al. (2015). Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. U.S.A. 112, E3574–E3581. doi: 10.1073/pnas.1501049112
Hsu, C. R., Pan, Y. J., Liu, J. Y., Chen, C. T., Lin, T. L., Wang, J. T. (2015). Klebsiella pneumoniae translocates across the intestinal epithelium via Rho GTPase- and phosphatidylinositol 3-kinase/Akt-dependent cell invasion. Infect. Immun. 83, 769–779. doi: 10.1128/IAI.02345-14
Hu, F. P., Pan, Y. Q., Li, H., Han, R. R., Liu, X., Ma, R. J., et al. (2024). Carbapenem-resistant Klebsiella pneumoniae capsular types, antibiotic resistance and virulence factors in China: a longitudinal, multi-centre study. Nat. Microbiol. 9, 814–829. doi: 10.1038/s41564-024-01612-1
Huynh, D. T. N., Kim, A. Y., Kim, Y. R. (2017). Identification of pathogenic factors in Klebsiella pneumoniae using impedimetric sensor equipped with biomimetic surfaces. Sensors (Basel). 17, 1406. doi: 10.3390/s17061406
Isler, B., Aslan, A. T., Akova, M., Harris, P., Paterson, D. L. (2022). Treatment strategies for OXA-48-like and NDM producing Klebsiella pneumoniae infections. Expert Rev. Anti Infect. Ther. 20, 1389–1400. doi: 10.1080/14787210.2022.2128764
Justice, S. S., Hunstad, D. A., Cegelski, L., Hultgren, S. J. (2008). Morphological plasticity as a bacterial survival strategy. Nat. Rev. Microbiol. 6, 162–168. doi: 10.1038/nrmicro1820
Kabha, K., Nissimov, L., Athamna, A., Keisari, Y., Parolis, H., Parolis, L. A. S., et al. (1995). Relationships among capsular structure, phagocytosis, and mouse virulence in Klebsiella pneumoniae. Infect. Immun. 63, 847–852. doi: 10.1128/iai.63.3.847-852.1995
Klaper, K., Wendt, S., Lubbert, C., Lippmann, N., Pfeifer, Y., Werner, G. (2021). Hypervirulent Klebsiella pneumoniae of lineage ST66-K2 caused tonsillopharyngitis in German patient. Microorganisms 9, 133. doi: 10.3390/microorganisms9010133
Klein, K., Palarasah, Y., Kolmos, H. J., Moller-Jensen, J., Andersen, T. E. (2015). Quantification of filamentation by uropathogenic Klebsiella pneumoniae during experimental bladder cell infection by using semi-automated image analysis. J. Microbiol. Meth. 109, 110–116. doi: 10.1016/j.mimet.2014.12.017
Kumar, A., Chakravorty, S., Yang, T. H., Russo, T. A., Newton, S. M., Klebba, P. E. (2024). Siderophore-mediated iron acquisition by Klebsiella pneumoniae. J. Bacteriol. 206, e0002424. doi: 10.1128/jb.00024-24
Lai, Y. C., Lin, A. C., Chiang, M. K., Dai, Y. H., Hsu, C. C., Lu, M. C., et al. (2014). Genotoxic Klebsiella pneumoniae in Taiwan. PLoS One 9, e96292. doi: 10.1371/journal.pone.0096292
Lai, Y. C., Peng, H. L., Chang, H. Y. (2003). RmpA2, an activator of capsule biosynthesis in Klebsiella pneumoniae CG43, regulates K2 cps gene expression at the transcriptional level. J. Bacteriol. 185, 788–800. doi: 10.1128/JB.185.3.788-800.2003
Lam, M. M. C., Wick, R. R., Judd, L. M., Holt, K. E., Wyres, K. L. (2022). Kaptive 2.0: updated capsule and lipopolysaccharide locus typing for the Klebsiella pneumoniae species complex. Microb. Genomics 8(3), 000800. doi: 10.1099/mgen.0.000800
Lam, M. M. C., Wick, R. R., Watts, S. C., Cerdeira, L. T., Wyres, K. L., Holt, K. E. (2021). A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 12, 4188. doi: 10.1038/s41467-021-24448-3
Lam, M. M. C., Wick, R. R., Wyres, K. L., Gorrie, C. L., Judd, L. M., Jenney, A. W. J., et al. (2018a). Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microb. Genom. 4, e000196. doi: 10.1099/mgen.0.000196
Lam, M. M. C., Wyres, K. L., Judd, L. M., Wick, R. R., Jenney, A., Brisse, S., et al. (2018b). Tracking key virulence loci encoding aerobactin and salmochelin siderophore synthesis in Klebsiella pneumoniae. Genome Med. 10, 77. doi: 10.1186/s13073-018-0587-5
Liao, W., Liu, Y., Zhang, W. (2020). Virulence evolution, molecular mechanisms of resistance and prevalence of ST11 carbapenem-resistant Klebsiella pneumoniae in China: A review over the last 10 years. J. Glob Antimicrob. Resist. 23, 174–180. doi: 10.1016/j.jgar.2020.09.004
Lin, Z. W., Zheng, J. X., Bai, B., Xu, G. J., Lin, F. J., Chen, Z., et al. (2020). Characteristics of hypervirulent Klebsiella pneumoniae: Does low expression of rmpA contribute to the absence of hypervirulence? Front. Microbiol. 11, 436. doi: 10.3389/fmicb.2020.00436
Livermore, D. M. (1995). beta-Lactamases in laboratory and clinical resistance. Clin. Microbiol. Rev. 8, 557–584. doi: 10.1128/CMR.8.4.557
Lu, M. C., Chen, Y. T., Chiang, M. K., Wang, Y. C., Hsiao, P. Y., Huang, Y. J., et al. (2017). Colibactin contributes to the hypervirulence of pks(+) K1 CC23 Klebsiella pneumoniae in mouse meningitis infections. Front. Cell Infect. Microbiol. 7, 103. doi: 10.3389/fcimb.2017.00103
Marteyn, B., Scorza, F. B., Sansonetti, P. J., Tang, C. (2011). Breathing life into pathogens: the influence of oxygen on bacterial virulence and host responses in the gastrointestinal tract. Cell Microbiol. 13, 171–176. doi: 10.1111/j.1462-5822.2010.01549.x
Martin, R. M., Bachman, M. A. (2018). Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front. Cell Infect. Microbiol. 8, 4. doi: 10.3389/fcimb.2018.00004
Masson, F. M., Karadottir, S., van der Lans, S. P. A., Doorduijn, D. J., de Haas, C. J. C., Rooijakkers, S. H. M., et al. (2024). Klebsiella LPS O1-antigen prevents complement-mediated killing by inhibiting C9 polymerization. Sci. Rep. 14, 20701. doi: 10.1038/s41598-024-71487-z
Meatherall, B. L., Gregson, D., Ross, T., Pitout, J. D., Laupland, K. B. (2009). Incidence, risk factors, and outcomes of Klebsiella pneumoniae bacteremia. Am. J. Med. 122, 866–873. doi: 10.1016/j.amjmed.2009.03.034
Mendes, G., Santos, M. L., Ramalho, J. F., Duarte, A., Caneiras, C. (2023). Virulence factors in carbapenem-resistant hypervirulent Klebsiella pneumoniae. Front. Microbiol. 14, 1325077. doi: 10.3389/fmicb.2023.1325077
Merla, C., Kuka, A., Mileto, I., Petazzoni, G., Gaiarsa, S., De Vitis, D., et al. (2024). One-year surveillance for hypervirulent Klebsiella pneumoniae detected carbapenem-resistant superbugs. Microbiol. Spectr. 12, e0329223. doi: 10.1128/spectrum.03292-23
Muner, J. J., de Oliveira, P. A. A., Baboghlian, J., Moura, S. C., de Andrade, A. G., de Oliveira, M. M., et al. (2024). The transcriptional regulator Fur modulates the expression of uge, a gene essential for the core lipopolysaccharide biosynthesis in Klebsiella pneumoniae. BMC Microbiol. 24, 279. doi: 10.1186/s12866-024-03418-x
Murashko, O. N., Lin-Chao, S. (2017). Klebsiella pneumoniae responds to environmental changes using enolasic degradosomes and stabilized DicF sRNA to alter cellular morphology. P Natl. Acad. Sci. U.S.A. 114, E8025–E8E34. doi: 10.1073/pnas.1703731114
Opoku-Temeng, C., Kobayashi, S. D., DeLeo, F. R. (2019). Klebsiella pneumoniae capsule polysaccharide as a target for therapeutics and vaccines. Comput. Struct. Biotec. 17, 1360–1366. doi: 10.1016/j.csbj.2019.09.011
Pichler, C., Buchsel, M., Rossen, J. W., Vavra, M., Reuter, S., Kern, W. V., et al. (2017). First report of invasive liver abscess syndrome with endophthalmitis caused by a K2 serotype ST2398 hypervirulent Klebsiella pneumoniae in Germany, 2016. New Microbes New Infect. 17, 77–80. doi: 10.1016/j.nmni.2017.02.006
Prashar, A., Bhatia, S., Gigliozzi, D., Martin, T., Duncan, C., Guyard, C., et al. (2013). Filamentous morphology of bacteria delays the timing of phagosome morphogenesis in macrophages. J. Cell Biol. 203, 1081–1097. doi: 10.1083/jcb.201304095
Prax, M., Bekeredjian-Ding, I., Krut, O. (2019). Microbiological screening of platelet concentrates in Europe. Transfus Med. Hemoth. 46, 76–86. doi: 10.1159/000499349
Price, J. V., Vance, R. E. (2014). The macrophage paradox. Immunity 41, 685–693. doi: 10.1016/j.immuni.2014.10.015
Rahi, P., Rodriguez, C., Brisse, S. (2024). “Klebsiella spp,” in Bergey’s Manual of Systematics of Archea and Bacteria. Ed. Whitman, W. B. (John Wiley & Sons). Update based on the original article by Patrick A.D. Grimont and Francine Grimont in Bergey’s Manual of Systematics of Archaea and Bacteria, published by John Wiley & Sons.
Russo, T. A., Alvarado, C. L., Davies, C. J., Drayer, Z. J., Carlino-MacDonald, U., Hutson, A., et al. (2024). Differentiation of hypervirulent and classical Klebsiella pneumoniae with acquired drug resistance. Mbio 15, e0286723. doi: 10.1128/mbio.02867-23
Russo, T. A., MacDonald, U., Hassan, S., Camanzo, E., LeBreton, F., Corey, B., et al. (2021). An assessment of siderophore production, mucoviscosity, and mouse infection models for defining the virulence spectrum of hypervirulent Klebsiella pneumoniae. Msphere 6, e00045–e00021. doi: 10.1128/mSphere.00045-21
Russo, T. A., Marr, C. M. (2019). Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 32, e00001–e00019. doi: 10.1128/CMR.00001-19
Russo, T. A., Olson, R., MacDonald, U., Beanan, J., Davidson, B. A. (2015). Aerobactin, but not yersiniabactin, salmochelin, or enterobactin, enables the growth/survival of hypervirulent (hypermucoviscous) Klebsiella pneumoniaeex vivo and in vivo. Infect. Immun. 83, 3325–3333. doi: 10.1128/IAI.00430-15
Sahly, H., Navon-Venezia, S., Roesler, L., Hay, A., Carmeli, Y., Podschun, R., et al. (2008). Extended-spectrum β-lactamase production is associated with an increase in cell invasion and expression of fimbrial adhesins in Klebsiella pneumoniae. Antimicrob. Agents Ch 52, 3029–3034. doi: 10.1128/AAC.00010-08
Sahly, H., Podschun, R., Oelschlaeger, T. A., Greiwe, M., Parolis, H., Hasty, D., et al. (2000). Capsule impedes adhesion to and invasion of epithelial cells by Klebsiella pneumoniae. Infect. Immun. 68, 6744–6749. doi: 10.1128/IAI.68.12.6744-6749.2000
Secher, T., Payros, D., Brehin, C., Boury, M., Watrin, C., Gillet, M., et al. (2015). Oral tolerance failure upon neonatal gut colonization with Klebsiella pneumoniae producing the genotoxin colibactin. Infect. Immun. 83, 2420–2429. doi: 10.1128/IAI.00064-15
Spadar, A., Perdigao, J., Campino, S., Clark, T. G. (2022). Genomic analysis of hypervirulent Klebsiella pneumoniae reveals potential genetic markers for differentiation from classical strains. Sci. Rep. 12, 13671. doi: 10.1038/s41598-022-17995-2
Spindler-Raffel, E., Benjamin, R. J., McDonald, C. P., Ramirez-Arcos, S., Aplin, K., Bekeredjian-Ding, I., et al. (2017). Enlargement of the WHO international repository for platelet transfusion-relevant bacteria reference strains. Vox Sang. 112, 713–722. doi: 10.1073/pnas.1703731114
Struve, C., Krogfelt, K. A. (2003). Role of capsule in Klebsiella pneumoniae virulence: lack of correlation between in vitro and in vivo studies. FEMS Microbiol. Lett. 218, 149–154. doi: 10.1111/j.1574-6968.2003.tb11511.x
Sun, Q. L., Gu, D., Wang, Q., Hu, Y., Shu, L., Hu, J., et al. (2019). Dynamic colonization of Klebsiella pneumoniae isolates in gastrointestinal tract of intensive care patients. Front. Microbiol. 10, 230. doi: 10.3389/fmicb.2019.00230
Sun, W., Rong, C., Chen, L., Li, J., An, Z., Yue, J., et al. (2024). Microaerobic-mediated suppression of Klebsiella pneumoniae mucoviscosity is restored by rmpD overexpression. J. Appl. Microbiol. 135, lxae192. doi: 10.1093/jambio/lxae192
Tang, M., Zhao, D., Zhang, Y., Qian, C., Chen, H., Chen, L., et al. (2024). Impact of LuxS on virulence and pathogenicity in Klebsiella pneumoniae exhibiting varied mucoid phenotypes. Infect. Immun. 92, e0001224. doi: 10.1128/iai.00012-24
EUCAST 2024. Breakpoint tables for interpretation of MICs and zone diameters. Version 11.0, 2021. Available online at: http://www.eucast.org. (Accessed February 7, 2025)
Wahl, A., Fischer, M. A., Klaper, K., Müller, A., Borgmann, S., Friesen, J., et al. (2024). Presence of hypervirulence-associated determinants in Klebsiella pneumoniae from hospitalised patients in Germany. Int. J. Med. Microbiol. 314, 151601. doi: 10.1016/j.ijmm.2024.151601
Walker, K. A., Treat, L. P., Sepulveda, V. E., Miller, V. L. (2020). The small protein RmpD drives hypermucoviscosity in Klebsiella pneumoniae. Mbio 11, e01750–e01720. doi: 10.1128/mBio.01750-20
Wanford, J. J., Hames, R. G., Carreno, D., Jasiunaite, Z., Chung, W. Y., Arena, F., et al. (2021). Interaction of Klebsiella pneumoniae with tissue macrophages in a mouse infection model and ex-vivo pig organ perfusions: an exploratory investigation. Lancet Microbe 2, E695–E703. doi: 10.1016/S2666-5247(21)00195-6
Wyres, K. L., Nguyen, T. N. T., Lam, M. M. C., Judd, L. M., Chau, N. V., Dance, D. A. B., et al. (2020). Genomic surveillance for hypervirulence and multi-drug resistance in invasive Klebsiella pneumoniae from South and Southeast Asia. Genome Med. 12, 11. doi: 10.1186/s13073-019-0706-y
Yang, D. C., Blair, K. M., Salama, N. R. (2016). Staying in shape: the impact of cell shape on bacterial survival in diverse environments. Microbiol. Mol. Biol. Rev. 80, 187–203. doi: 10.1128/MMBR.00031-15
Keywords: adhesion, invasion, replication, Klebsiella pneumoniae, serum resistance, platelet concentrate, cell morphology, normoxia
Citation: Klaper K, Pfeifer Y, Heinrich L, Prax M, Krut O, Bekeredjian-Ding I, Wahl A, Fischer MA, Kaspar H, Borgmann S, Gerlach RG and Werner G (2025) Enhanced invasion and survival of antibiotic- resistant Klebsiella pneumoniae pathotypes in host cells and strain-specific replication in blood. Front. Cell. Infect. Microbiol. 15:1522573. doi: 10.3389/fcimb.2025.1522573
Received: 04 November 2024; Accepted: 23 January 2025;
Published: 14 February 2025.
Edited by:
Andreas Erich Zautner, University Hospital Magdeburg, GermanyReviewed by:
Hagen Frickmann, Bundeswehr Hospital Hamburg, GermanyCopyright © 2025 Klaper, Pfeifer, Heinrich, Prax, Krut, Bekeredjian-Ding, Wahl, Fischer, Kaspar, Borgmann, Gerlach and Werner. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guido Werner, V2VybmVyR0Bya2kuZGU=
†Present addresses: Isabelle Bekeredjian-Ding, Institute of Medical Microbiology and Hospital Hygiene, University Hospital Marburg, Marburg, Germany
Roman G. Gerlach, Institute of Clinical Microbiology, Immunology and Hygiene, University Hospital of Erlangen and Friedrich-Alexander-University (FAU) Erlangen-Nuremberg, Germany
‡ORCID: Beredjian-Ding, orcid.org/0000-0001-6646-5888
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.