
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol., 04 February 2025
Sec. Fungal Pathogenesis
Volume 15 - 2025 | https://doi.org/10.3389/fcimb.2025.1497085
This article is part of the Research TopicMolecular Taxonomy of Ascomycetes and Basidiomycetes: Unveiling Fungal Diversity and EvolutionView all 10 articles
 Anusha H. Ekanayaka1,2†
Anusha H. Ekanayaka1,2† Samantha C. Karunarathna1,3†
Samantha C. Karunarathna1,3† Saowaluck Tibpromma1
Saowaluck Tibpromma1 Arun Kumar Dutta4
Arun Kumar Dutta4 Danushka Sandaruwan Tennakoon5
Danushka Sandaruwan Tennakoon5 Anuruddha Karunarathna6
Anuruddha Karunarathna6 Ekachai Chukeatirote7
Ekachai Chukeatirote7 Dong-Qin Dai1
Dong-Qin Dai1 Steven L. Stephenson8
Steven L. Stephenson8 Sajeewa S. N. Maharachchikumbura9
Sajeewa S. N. Maharachchikumbura9 Chao Liu1*
Chao Liu1* Alan J. L. Phillips10*
Alan J. L. Phillips10*The evolution of a species can be understood in the context of two major concepts—the cryptic species concept and the phenotypic noise concept. The former represents morphologically indistinguishable but genetically distinct evolutionary lineages, while the latter represents the phenotypic variations of an isogenic population. Although the concept of cryptic species currently represents a general topic, its effect on other aspects of biology, such as biodiversity, ecology, evolutionary biology, and taxonomy, is still unclear. In particular, cryptic species cause complications and prevent the development of a clear taxonomy. The phenotypic noise concept or phenotypic plasticity generally refers to the various expressions of phenotypes in different environments. Hence, the cryptic species concept refers to genetic variations, while the phenotypic noises concept is about non-genetic variations. Although both concepts are opposites, they each contribute significantly to the evolutionary process of an organism. Despite the extensive research studies and publications discussing those two concepts in separate accounts, a concise account that combines and compares both concepts are generally lacking. Nevertheless, these are essential to understand the evolutionary process clearly. This review addresses the available literature on this topic, intending to provide a general and overall discussion on both the cryptic species concept and the phenotypic noise concept and their effect on evolution, ecology, biodiversity, and taxonomy with a special focus on fungal systematics. hence, several fungal case studies representing the two concepts are presented, compared, and discussed for a better understanding.
Evolution occurs when a group of organisms undergoes gradual changes in their properties over generations. The changes in genome, physiology, or morphology make the particular group of organisms in a population different from their recent ancestors, causing those individuals in question To achieve reproductive isolation from other individuals within the population who share a particular ecological niche, maintaining the ability to interbreed (Mayr, 1982; Aldhebiani, 2018). This process is referred to as speciation. This evolutionary process or speciation event results from a complex series of circumstances that directly connect with the species concept. The species circumscription and delimitation are based on two major concepts—the cryptic species concept and the phenotypic noise concept.
The matter of delineating species has been compounded by a fundamental challenge related to what constitutes a species. This challenge arises from divergent perspectives among biologists regarding the definition of species. Over the past fifty years or so, various factions within the biological community have advocated for distinct and sometimes conflicting species concepts, as documented by (Mayden, 1997; De Queiroz, 1998; Harrison, 1998). Among them, Mayden (1997) enumerated 24 distinct species concepts with numerous additional alternative definitions. A definition here refers to a concise depiction of a concept, meaning that a single species concept may be associated with multiple definitions that vary in minor linguistic nuances. Many of these concepts and their corresponding definitions are incompatible, often leading to disparate conclusions regarding the delineation and enumeration of species. Consequently, the conundrum of species concepts, which is related to the discord of the current theoretical concept of species, is intricately bounded by the challenge of determining species’ boundaries and quantities based on empirical evidence (De Queiroz, 2007).
More commonly discussed species concepts include morphological, biological, evolutionary, and phylogenetic concepts (Taylor et al., 2000). Among them, the biological and evolutionary species concepts are widely used. Currently, there are conflicts among these terms. A biological species refers to inter-fertile populations reproductively isolated from other such groups and occupying a distinct ecological niche (Mayr, 1982). An evolutionary species refers to a single lineage of ancestor-descendant populations with a unique evolutionary history and the ability to maintain its identity from other such lineages. It also fits into its ecological niche (Simpson, 1961; Wiley, 1981; Paris et al., 1989). However, there is a congruence between the theoretical and operational species concepts. The operational species concept surpasses the theoretical one in prevalence, which causes conflicts in other related areas of evolution, ecology, biodiversity, and taxonomy (Mayden, 1997; Taylor et al., 2000). Hence, a better understanding of the causes for the congruence of the theoretical and operational species concepts is essential.
As the evolutionary species concept defines a species as a single lineage of ancestor-descendent populations, the theoretical species concept was primarily considered the evolutionary species concept (Taylor et al., 2000). However, current understandings of evolutionary consequences deviate from this primary idea about the theoretical species concept. With the slow process of evolution, specific individuals of a biological species acquire minor changes in their genotype. At a specific stage of this evolution, the newly changed individuals become non-fertile with the original population, and they thus represent a distinct evolutionary lineage (Paris et al., 1989; Struck and Cerca De Oliveira, 2019). However, both groups (original and newly changed) can still be morphologically indistinguishable (Struck and Cerca De Oliveira, 2019). This is the stage at which a cryptic species arises. It is supposed that as time passes, those two populations diverge gradually in their morphology and physiology to be suited to unique ecological conditions (Wiley, 1981; Paris et al., 1989; Struck and Cerca De Oliveira, 2019). Although most authors adopt the evolutionary species concept, they often recognize and classify species based only on distinctive phenotypic characteristics. Afterward, the classification followed also becomes morphologically based, the basis upon which the taxa can be easily identified. Generally, the evolutionary or phylogenetic species concept is superior to morphology-based and biological species concepts. Therefore, the morphological species concept is neither an evolutionary nor a biological species concept. As a result, a conflict arises between theory and practice (Paris et al., 1989; Struck and Cerca De Oliveira, 2019). Despite everything, the morphological species concept has become a more significant inference to the biological or evolutionary species concepts and the foundation for the cryptic species concept (Dobzhansky, 1982; Aldhebiani, 2018; Struck and Cerca De Oliveira, 2019). On the other hand, the morphological species concept is strongly influenced by ecological and environmental conditions. Therefore, gene expression can vary with the environment and is reflected in phenotype variations. Most importantly, even if the phenotype changes, the genotype is still the same (West-Eberhard, 2008; Levin, 2013; de Vienne, 2021).
The cryptic species concept also called the sibling species concept, describes two or several species that are indistinguishable in their phenotypes but differ in their genotypes (Zúniga-Reinoso and Benítez, 2015; Korshunova et al., 2019). Hence, in a morphological concept, they have been classified as a single nominal species (Zúniga-Reinoso and Benítez, 2015). Phenotypes of organisms within an isogenic population are not identical since they exhibit some degree of difference/variation from each other (Kaneko and Furusawa, 2008). Phenotypic noise refers to these variations in a phenotype developed by an individual genotype; it is also referred to as phenotypic plasticity (West-Eberhard, 2008). The environment and history of an organism influence the development of the variability in the cellular phenotype while the genotype is still unchanged (Raser and O’shea, 2005). As such, a single species can be given different names simply because of its phenotypic plasticity in different habitats. Both concepts are contradictory to one another and have created confusion in modern taxonomy. Both have significantly influenced other related research areas, including evolutionary biology, ecology, and biodiversity. Since morphology is a contentious marker in taxonomy, it hides cryptic species that share certain phenotypic characteristics but are otherwise distinct. As a result of the presence of cryptic species or the inability to recognize existent species, actual biodiversity levels are underestimated or overestimated (Lefébure et al., 2006; Padial et al., 2010).
Although cryptic species are more or less morphologically indistinguishable, their genotypes are significantly different (Wei et al., 2021). Therefore, molecular phylogenetic tools are the only fast, reliable, and efficient methods to discover species and face the taxonomic problems caused by cryptic species (Jörger and Schrödl, 2013; Korshunova et al., 2019). This is again applicable to the phenotypic noise concept. Molecular phylogenetic tools clearly recognize the different phenotypes influenced by the environment of the same species.
The concept of cryptic species dates back about three centuries, and the first record was provided by Derham in 1718 (in Ray and Willughby, 1718). Later authors have defined cryptic species in different ways. The first explicit definition for cryptic species was “population systems which were believed to belong to the same species until genetic evidence shows the existence of isolating mechanisms separating them” (Stebbins, 1950). Korshunova et al. (2019) recently defined cryptic species as “species which manifest low morphological but considerable genetic disparity”. Considering several definitions for cryptic species (Stebbins, 1950; Paris et al., 1989; Zúniga-Reinoso and Benítez, 2015; Korshunova et al., 2019; Struck and Cerca De Oliveira, 2019), they are considered to represent distinct evolutionary lineages and are reproductively isolated; however, morphologically indistinguishable, they have historically been misinterpreted as members of a single species.
The discussions on phenotypic noise began with the origin of the term “phenotype”. The first clear definition of phenotype was “All types of organisms, distinguishable by direct inspection or only by finer methods of measuring or description” (Johannsen, 1909). The general consideration among scientists is that the phenotype refers to the observable characteristics in an individual and reflects the genotype of an organism resulting from the expression of genes. The concept of phenotypic noise describes how the environment exerts an effect on gene expression and then how phenotypes vary based on environmental conditions (West-Eberhard, 2008; Levin, 2013). Phenotypic noise is also defined as the environmental sensitivity of a genotype (West-Eberhard, 2008).
One of the earliest examples of cryptic species is the genus Drosophila, which includes fruit flies (Dobzhansky and Epling, 1944; Dobzhansky, 1951). The two species D. pseudoobscura and D. persimilis were originally treated as one species. Later, in laboratory cultures, researchers observed these two species were reproductively isolated. Further studies identified and confirmed their differences in morphology (in wings and male genitalia), physiology, behavior, and chromosome morphology (Dobzhansky and Epling, 1944; Dobzhansky, 1951). Another example of cryptic species is provided by Adams et al. (2014), who explored the hyper-cryptic species complexes of Australian freshwater fish collections based on allozyme, mtDNA, and morphological data. They found that these species, across their broad geographic range, revealed a 1500% increase in species-level biodiversity, including 15 distinct species of phenotypically similar minnows (Phoxinus spp, a small freshwater fish) from different lakes (Adams et al., 2014). Investigations of Phoxinus communities of adjacent freshwater ecosystems of the westernmost part of the Po River basin in Italy, based on mitochondrial cytochrome oxidase I (COI) sequences from 239 specimens, revealed a complex of species of Phoxinus, consisting of P. septimaniae, P. csikii. and P. lmaireul, which are morphologically indistinct (De Santis et al., 2021). In another example, phylogenetic analyses of 37 samples from 13 taxa of the Asian forked fern genus (Dicranopteris) from five chloroplast gene regions (rbcL, atpB, rps4, matK, and trnL-trnF) showed that D. linearis is polyphyletic, which suggests that there is cryptic diversity within the species. Further comparisons erected the new species Dicranopteris austrosinensis and D. baliensis (Wei et al., 2021). Peintner et al. (2019) identified and analyzed Fomes strains from different habitats in Italy and Austria using rDNA ITS region phylogenetics. These findings proved the existence of subclades within the Fomes fomentarius clade and led to the formal recognition of the new species Fomes inzengae (Peintner et al., 2019). Researchers analyzed 96 Japanese specimens of the Hypholoma fasciculare complex, a group of common wood-decomposing fungi, using mitochondrial ribosomal RNA (mt-rRNA) sequences, nuclear ITS region, and 24 single-copy genes. The results showed that the H. fasciculare complex encompassed two species, H. fasciculare and H. subviride (Sato et al., 2020).
The phenotypic noise concept has been discussed at length in several research papers (Raser and O’shea, 2005; West-Eberhard, 2008; Levin, 2013; de Vienne, 2021). Some interesting studies on phenotypic noise include the identification of intraspecific phenotypic variation and morphological divergence of Folsomia candida by Tully and Potapov (2015) and phenotype microarray profiling of Staphylococcus aureus by von Eiff et al. (2006). Compared with the cryptic species concept, the phenotypic noise concept lacks detailed studies. However, many confusing points remain on these topics. Moreover, the combined effects of the cryptic species concept and the phenotypic noise concept on other fields, such as taxonomy, evolution, ecology, and biodiversity require further studies. Hence, the present study reviews and discusses the influence of the cryptic species concept and the phenotypic noise concept on taxonomy, evolution, ecology, and biodiversity research. Future research trends related to these concepts are summarized and discussed.
Further, fungi are versatile and reflect their diversity in morphology, ecology, physiology, and phylogeny. Fungi rank as the second most diverse group of organisms in terms of species, followed by insects (Purvis and Hector, 2000; Antonelli, 2023). Among fungi, a remarkable range of morphological diversity exists, spanning from single-celled yeasts to substantial ‘fruiting bodies’ that can generate trillions of spores (Stajich et al., 2009). Numerous fungi’s developmental phases occur within or on intricate substrates like soil, wood, plants, or animals, rendering them challenging to observe. Consequently, developmental stages and true ecological diversity of most fungi remain undiscovered (Rodriguez et al., 2004). Both fungi that produce large reproductive structures (e.g., mushrooms and truffles) and those that make no reproductive structures apart from the meiocytes themselves (e.g., yeasts) show such complex reproductive models (Stajich et al., 2010). The most intricate formations within the fungal realm are the multicellular sexual fruiting bodies, characterized by well-defined fungal tissues and various cell types (Kües et al., 2018). Nevertheless, unicellular yeasts show huge complexities in their life cycles (Yurkov et al., 2015). They also produce numerous bioactive molecules, which are helpful for various research areas such as agriculture, industrial, and pharmaceuticals (Raja et al., 2017). The estimated number of fungal species on Earth was approximately 12 million (ranging from 11.7 to 13.2 million), a significant increase from the previous estimate of 2.2 to 3.8 million species obtained through various estimation techniques. A team of fungal experts recently assessed the fungal diversity in the world using four main academic pathways viz. scaling laws; fungus: plant ratios; actual versus previously known number of species; and DNA-based studies; according to them, there are likely to be 2–3 million species of fungi, with a best estimate of 2.5 million (Antonelli, 2023). On the other hand, molecular phylogeny has revealed the existence of numerous cryptic species within this diverse fungal kingdom (Wu et al., 2019). However, recent studies revealed that fungal species circumscription and delimitation are a considerable challenge (Raja et al., 2017; Baturo-Ciesniewska et al., 2020; Stengel et al., 2022). Specifically, fungal species delimitation is complicated by cryptic species (Crespo and Lumbsch, 2010; Shivas and Cai, 2012; Peintner et al., 2019; Sato et al., 2020; Wang et al., 2022). Phenotypic noises or plasticity also cause controversies incorrect species identification in fungi (Lehner, 2010; Behm and Kiers, 2014). Hence, this review is also expected to improve the identification of fungal species and aid in developing future fungal research applications.
To review the available literature related to cryptic species and phenotypic noises concepts, various sources such as scholarly papers, digital databases, and personal communications were used. Data on the cryptic species concept and the phenotypic noise concept were collected primarily from “Google Scholar,” “ResearchGate,” “PubMed,” and “Web of Science.” The keywords such as species concept, cryptic species, phenotypic noises, biological, phenotype, genotype, ecology, biodiversity, and evolution were used in different combinations to derive references for the review. The derived literature was reviewed and analyzed to understand the fundamentals of cryptic species and phenotypic noise concepts and an in-depth literature review was done to determine their effect on biodiversity, ecology, evolutionary biology, and taxonomy.
Several suitable examples to reflect the occurrence of cryptic species and phenotypic noises within species circumscription and delimitation, were obtained from recently published literature with the approval of the original authors (Thambugala et al., 2015; De Silva et al., 2020). Authors re-examined a few specimens to confirm their morphological characteristics viz. Marasmius imitarius specimen was loaned from the Calcutta University herbarium (CUH), Rhytidhysteron neorufulum specimens were obtained from the Mae Fah Luang University herbarium (MFLU), and Misturatosphaeria aurantiacinotata from the National Museums of Kenya/East African Herbarium (EA). The specimens of Rhytidhysteron neorufulum and Misturatosphaeria aurantiacinotata were initially examined with a Motic SMZ 168 stereomicroscope (Motic Asia, Kowloon, Hong Kong), and further observations were made with a Nikon ECLIPSE 80i compound microscope (Nikon Instruments Inc., Melville, New York, USA) with photographs recorded with a Canon 550D digital camera (Canon Inc., Ota, Tokyo, Japan). Hand-cut sections were mounted in sterile water for study and photographs. Tarosoft (R) Image Frame Work programme (Version: 1.3) was used to make measurements, and Adobe Photoshop CS3 extended version 10.0 was used to process the images used for the figures (Adobe Systems, USA). The specimen of Marasmius imitarius was studied with a Dewinter ‘crown’ trinocular microscope (Dewinter Optical Inc., New Delhi). Photographs were taken with the dedicated camera attached to the microscope.
The phylogenetic trees were reproduced using published sequence data in the NCBI database related to above mentioned examples. For each gene, sequences were aligned using MAFFT v. 7 (http://mafft.cbrc.jp/alignment/server/index.html) and manually adjusted in BioEdit v. 7.0.4 (Hall, 2004) where necessary. Datasets were concatenated with FaBox (1.41) (Villesen, 2007). Ambiguously aligned regions were excluded and gaps were treated as missing data.
As a model of evolution, the GTR+G+I substitution model was used. Maximum likelihood (ML) phylogenetic analyses were conducted using the RAxML-HPC2 Workflow on XSEDE (8.2.9) available through the CIPRES web portal (Miller et al., 2011; Stamatakis, 2014). Each ML tree bootstrap analysis was run using 1000 in-depth replicates with identical parameters. Markov chain Monte Carlo (MCMC) sampling in the CIPRES web portal (Miller et al., 2011) with MrBayes on XSEDE was used to calculate posterior probabilities (PP). Four Markov chains were run in parallel for two million generations, with tree samples taken every thousand generations. The temperature parameter of the MCMC heated chain was fixed at 0.2. Tracer v.1.5 (Rambaut and Drummond, 2009) was used to analyze the log-likelihood score distribution and determine whether more search iterations were necessary to reach convergence. Topologies below the asymptote (the top 80% of the sample) were rejected as part of the burn-in phase, and the remaining trees were used to determine posterior probabilities in the majority rule consensus tree.
The genus Marasmius is cosmopolitan in distribution and can be recognized as those taxa that possess reviving basidiomata with a sulcate or corrugated pileus, a centrally placed cartilaginous stipe, and white basidiospores (Fries, 1835). Marasmius imitarius is morphologically very similar to many other species in the same genus (Wannathes et al., 2009). In fact, the specific epithet suggests how phenotypically this species resembles others in the genus. The taxon can be identified morphologically by its light brown sulcate pileus with a darker brown to reddish brown center, its distant (10–12), cream-colored lamellae with brown edges, its clavate basidiospores with a mean range of 17.8–19.2 × 4.1–4.7 μm, the absence of pleurocystidia and caulocystidia, and the fact that it grows primarily on woody twigs (Wannathes et al., 2009). According to Wannathes et al. (2009), most of these morphological features of M. imitarius overlap with other taxa like M. bambusiniformis, M. mazatecus, M. striaepileus, and M. sierraleonis.
To confirm the observation reported by Wannathes et al. (2009), we examined one of the Indian collections of Marasmius imitarius (CUH AM078) and M. bambusiniformis (AKD 382/2014, the author’s (Arun Kumar Dutta) personal collection; Figure 1). The examination revealed that the taxon M. imitarius (CUH AM078) possesses morphological characteristics such as a small (5–17 mm diam.), obtusely conical to convex pileus that matures to convex to plano-convex with a slight central depression, often with an upturned margin showing the faces of brownish orange to reddish orange lamellae with a smooth to minutely pruinose surface; distant (L = 9−11, l = 0−1) white to cream lamellae often with brown edges; fusoid to clavate basidiospores (14.5−21.5 × 3.5–6.8 µm); absence of pleurocystidia; Siccus-type cheilocystidia with clavate to pyriform main body (7.5−14.5 × 4−9 µm) and cylindrical apical setulae (3.5−7.5 µm); a hymeniform pileipellis consisting of Siccus-type broom cells with main body measuring 10−21.5 × 6.5−10 µm, and apical setulae measuring 3.0−7.0 µm; absence of caulocystidia; and the presence of clamp-connections in all tissues.

Figure 1. Field photographs of basidiomata. (A) Marasmius imitarius (CUH AM078), (B) Marasmius bambusiniformis (author’s (Arun Kumar Dutta) personal collections, AKD 382/2014). Scale bars = 10 mm.
Further observation of M. bambusiniformis (collection no. AKD 382/2014) revealed that M. bambusiniformis consists of features such as a small (5–10 mm diam.), conic to convex or often applanate pileus, with smooth to slightly pruinose surface colored light brown to brownish yellow with brownish orange center; distant to subdistant (L = 18–21, l = 0–1) cream lamellae; fusoid to clavate basidiospores (14–19.5 × 3.5–6.0 µm); absence of pleurocystidia; Siccus-type cheilocystidia with clavate to pyriform main body (11.5–16 × 5–8 µm) and cylindrical apical setulae (3.5−8 µm); a hymeniform pileipellis consisting of Siccus-type broom cells with a main body measuring 12.5–20 × 7–8.5 µm, and apical setulae measuring 3.5−8.0 µm; absence of caulocystidia; and the presence of clamp-connections in all tissues.
Hence, our study on the detailed morphology of both taxa (like M. imitarius and M. bambusiniformis) also suggests that differentiating these two taxa based solely on morphological features is insufficient. However, phylogenetic analyses based on the nrITS DNA data of this study (Figure 2) show M. imitarius lies far away from M. bambusiniformis, and previous studies also show similar results with this study (Wannathes et al., 2009). In fact, based on phylogenetic analyses, M. imitarius is not closely related to M. bambusiniformis (Figure 2).
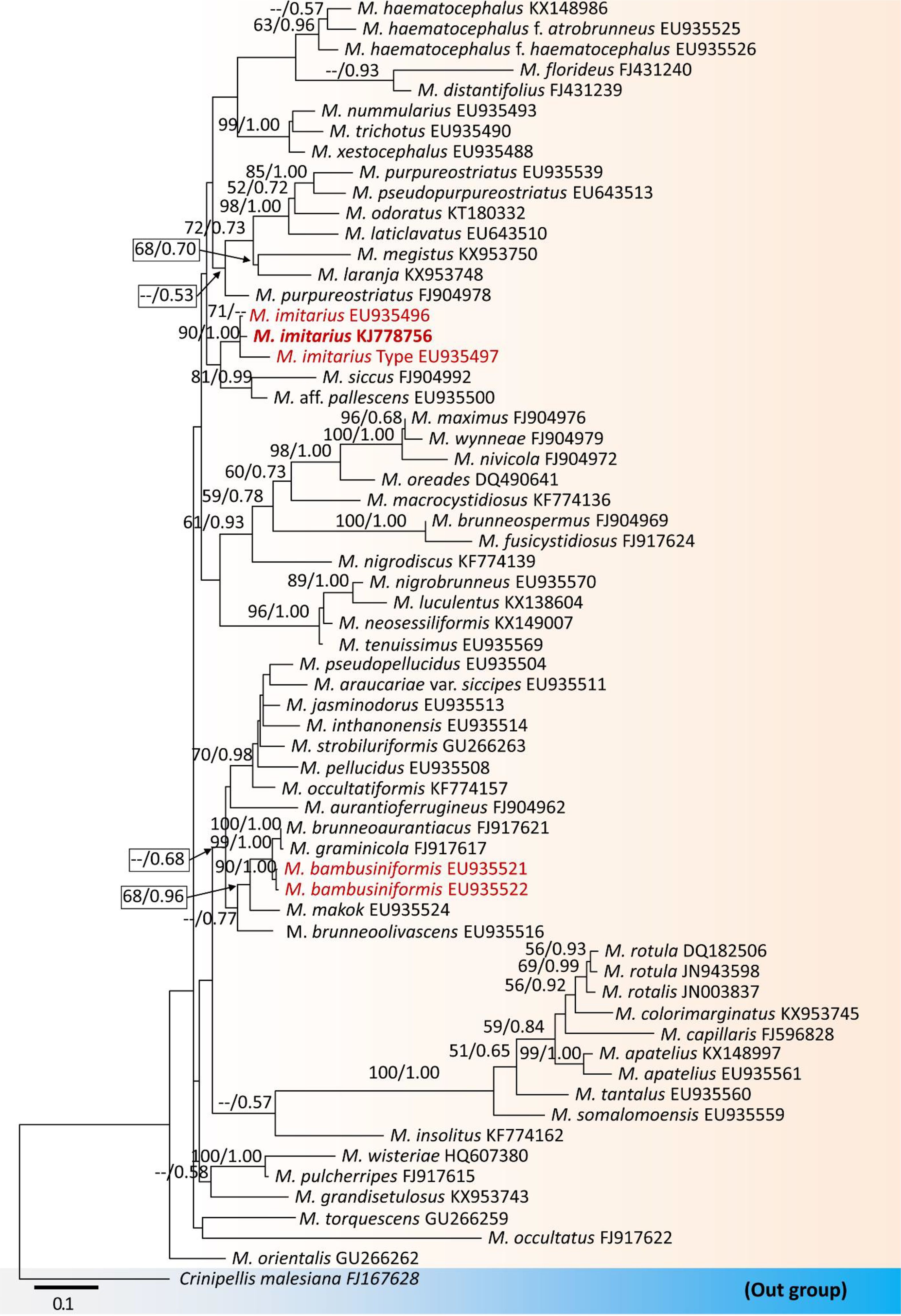
Figure 2. Maximum likelihood tree constructed using ITS rDNA sequence data. The tree is rooted with Crinipellis malesiana. Morphologically look-a-like taxa, Marasmius imitarius and M. bambusiniformis are in red. The collected specimen of M. imitarius is in bold. Bootstrap support values ≥50% from the maximum likelihood (ML) and Bayesian posterior probabilities (BYPP) values ≥0.50 are given above the nodes, respectively. The abbreviation ‘M.’ represents the genus Marasmius.
The hysteriaceous genus Rhytidisteron belongs to the family Hysteriaceae in the Dothideomycetes (Thambugala et al., 2016). These fungi have a worldwide distribution mostly as saprobes, endophytes or weak pathogens on woody plants and rarely as human pathogens (Thambugala et al., 2016; De Silva et al., 2020; Cobos-Villagrán et al., 2021). Species of Rhytidisteron are characterized by having large, conspicuous, elongated, superficial, carbonaceous to coriaceous, navicular ascomata, mostly opening by a longitudinal slit and with thick-walled ascospores (Thambugala et al., 2016). Since these fungi express varied shapes of ascomata depending on the environmental conditions, their ascomata traits are quite fascinating. For instance, their ascomata are disc-shaped (irregular opening/apothecial-shape) under wet conditions, but when dry, they fold at the margin, forming an elongated slit (hysterothecial-shape) (Thambugala et al., 2016; Cobos-Villagrán et al., 2021). In some cases, the same specimen may also contain disc-shaped and hysterothecial-shaped ascomata. For example, we observed the ascoma characteristics of several isolates of Rhytidhysteron neorufulum (MFLU 18-2644, MFLU 21-0248 and MFLU 14-0608) and found that all consist of both disc-shaped and hysterothecial-shaped ascomata (Figure 3). Thus, their apothecial or hysterothecial behavior could not be considered as a valid trait for delimitation of species of Rhytidisteron as those differences can be attributed to environmental changes.

Figure 3. Ascomata variation in Rhytidhysteron neorufulum. (A, B) Ascomata of MFLU 18-2644. (C) Ascomata of MFLU 21-0248. (D–F) Ascomata of MFLU 14-0608. (G) Vertical section of MFLU 14-0608. (H, I) Vertical section of MFLU 18-2644. Scale bars: (A–C, E) = 500 μm, (D) = 2000 μm, (F) = 1000 μm, (G–I) = 300 μm.
We reexamined the ascospore characteristics of three isolates of R. neorufulum (MFLU 18-2644, MFLU 21-0248 and MFLU 14-0608) and found that they are highly diverse in their shape, color, and septation (Figure 4). Ascospores can range from ellipsoidal to fusiform, ellipsoid to oblong or narrowly fusiform, and with pointed or obtuse ends (Thambugala et al., 2016; De Silva et al., 2020). Indeed, a single specimen can possess all these shapes (MFLU 18-2644). Ascospores of R. neorufulum have a variety of colors, including hyaline, pale brown, golden brown, and dark brown. The ascospores can be mono-septate, symmetrical, or asymmetrical and constricted at the central septum, while others are 2–3-septate, symmetrical, or asymmetrical and slightly constricted at the central septa. In addition, some ascospores have enlarged second cells from the apex. Rhytidhysteron neorufulum has a high phenotypic plasticity of ascospores and ascomata characteristics, and those characteristics may be considered insufficient for delimitation of species of Rhytidhysteron. Therefore, species-level delimitation in Rhytidhysteron is best accomplished using a multi-gene phylogeny (Figure 5) in combination with morphological characteristics.

Figure 4. Variations in Rhytidhysteron neorufulum ascospores. (A, B) Asci of MFLU 18-2644. (C, D) Asci of MFLU 14-0608. (E–J) Ascospores of MFLU 14-0608. (K–R) Ascospores of MFLU 18-2644. (S–AC) Ascospores of MFLU 21-0248. Scale bars: (A–D) = 50 μm, (E–J) = 15 μm, (K–AC) = 12 μm.
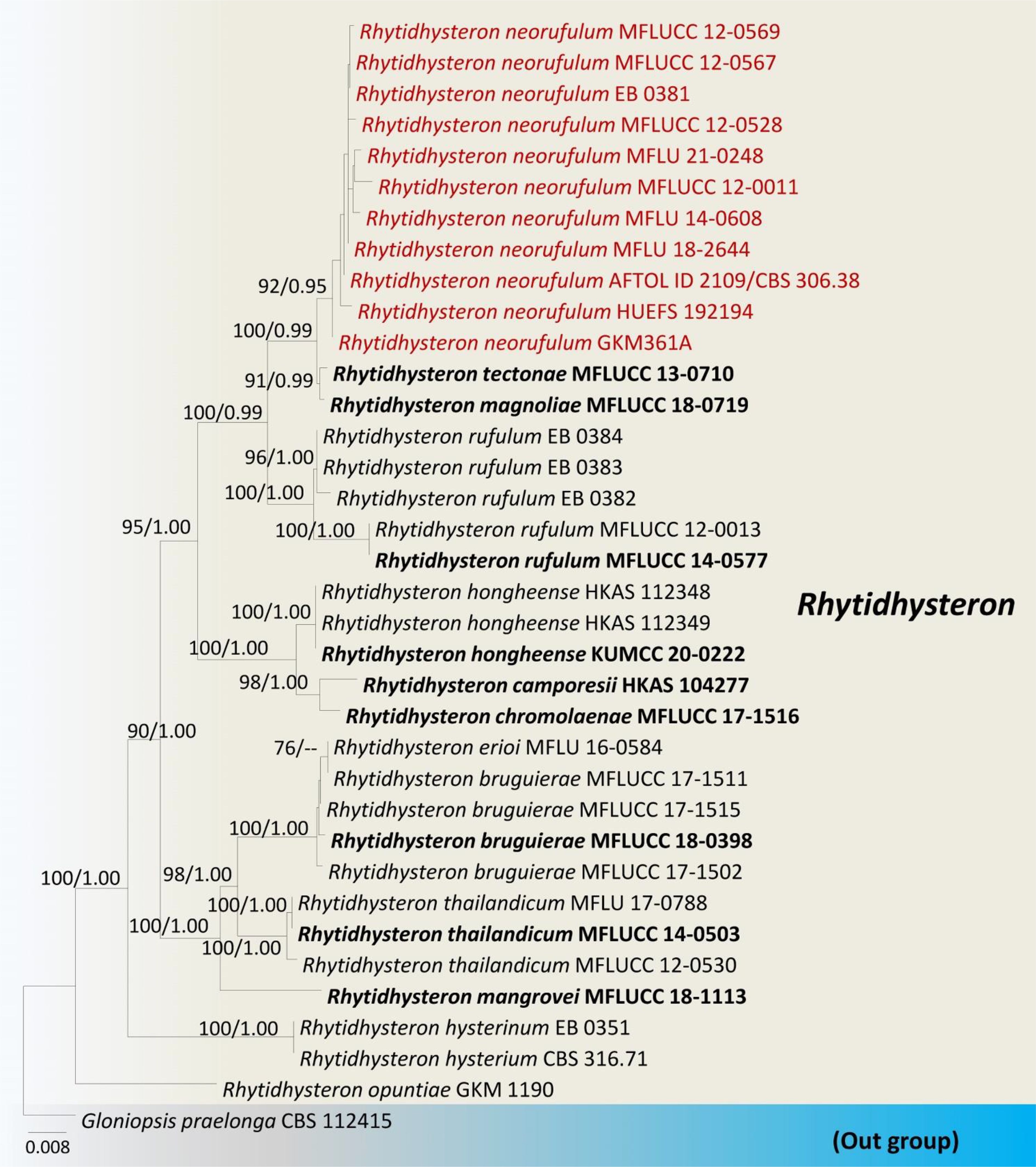
Figure 5. Phylogram generated from maximum likelihood analysis based on combined LSU, SSU, ITS and tef1 sequence data. The tree is rooted with Gloniopsis praelonga (CBS112415). The ex-type strains are in bold, and the Rhytidhysteron neorufulum isolates are in red. Bootstrap support values ≥75% from the maximum likelihood (ML) and Bayesian posterior probabilities (BYPP) values ≥0.95 are given above the nodes, respectively.
Misturatosphaeria is a member of the family Teichosporaceae in the Dothideomycetes. This genus was introduced by Mugambi and Huhndorf (2009) based on mixed ascospore morphological characteristics of this group. It is undeniably true that ascospores of this genus exhibit high phenotypic plasticity even within the same species (GKM 1238). For instance, the ascospore characteristics of Misturatosphaeria aurantiacinotata vary from hyaline to light brown or dark brown, are 1–3-septate, deeply or slightly constricted at the central septum and some have a mucilaginous sheath while some lack this feature (Figure 6). As such, their ascospores display highly diverse morphological characteristics (Mugambi and Huhndorf, 2009; Thambugala et al., 2015; Tennakoon et al., 2021). Moreover, it is very important to select fruiting bodies of the same level of maturity for morphological examinations, especially with respect to ascospores. Otherwise, there is a possibility of misidentifying a specimen as an entirely different species. In the case of M. aurantiacinotata, when only immature ascomata are present on a substrate, it is possible to observe hyaline, 1-septate ascospores that are deeply constricted at the central septum have a distinct mucilaginous sheath. But in mature ascomata, one can observe light brown to dark brown, 3-septate ascospores that lack a mucilaginous sheath (Figure 6). Thus, multi-gene phylogeny takes on an important role in the classification of species of Misturatosphaeria aurantiacinotata (Figure 7).
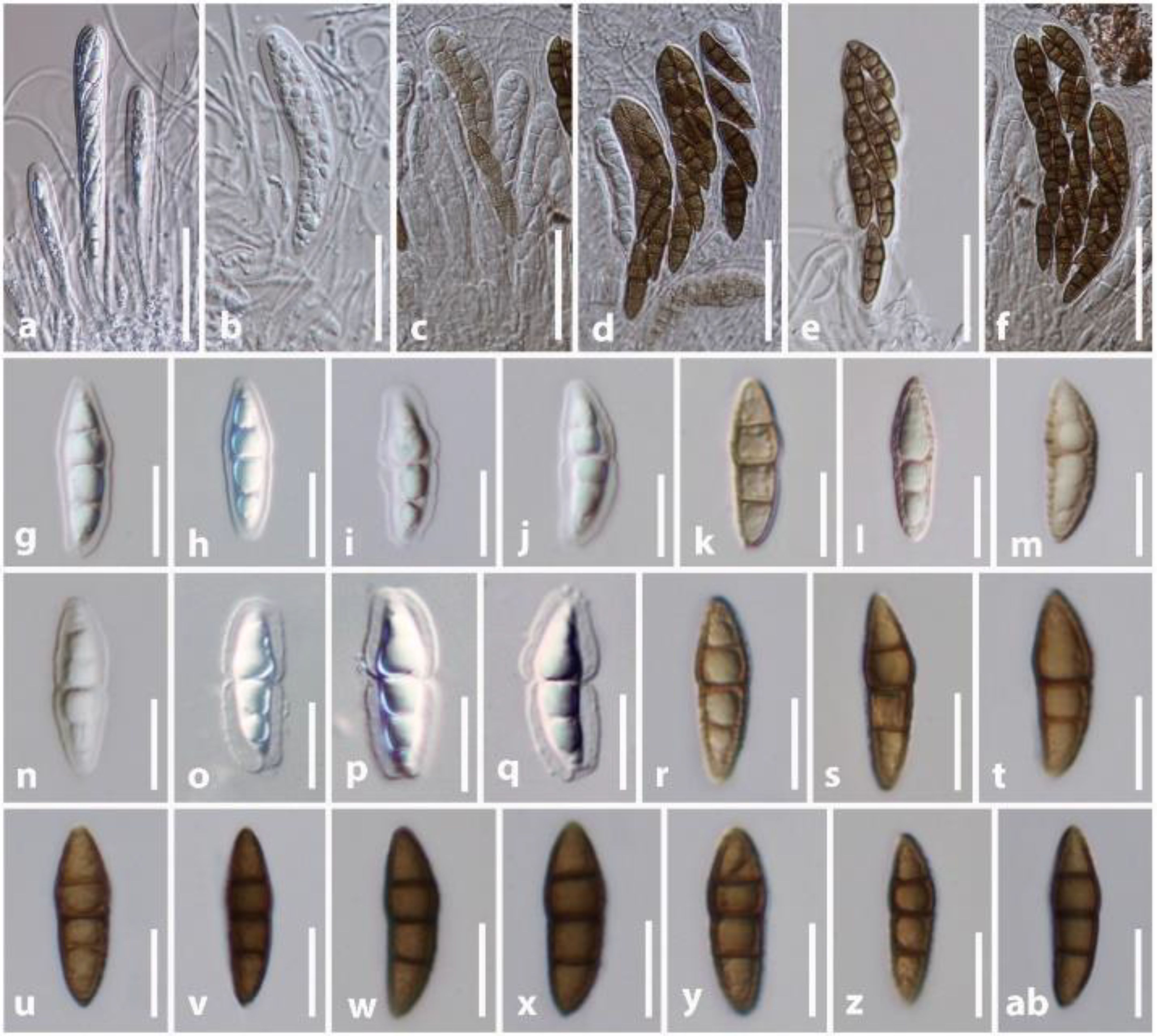
Figure 6. Ascospores variation in Misturatosphaeria aurantiacinotata (GKM 1238). (A–F) Asci. (G–AB) Ascospores. Scale bars: (A–F) = 50 μm, (G–AB) = 10 μm.
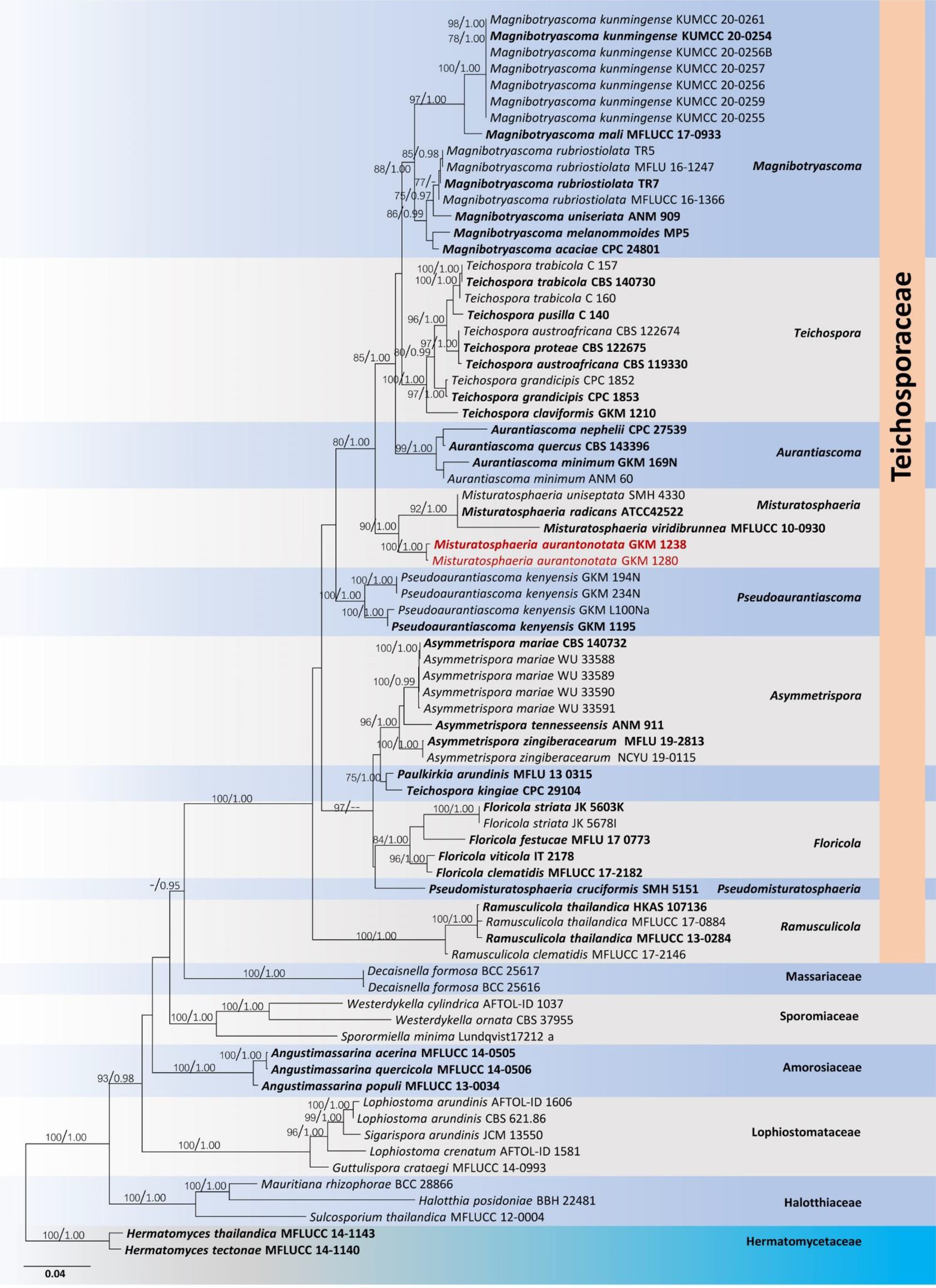
Figure 7. Phylogram generated from a maximum likelihood analysis based on combined LSU, SSU, ITS, tef1 and rpb2 sequence data. The tree is rooted with Hermatomyces tectonae (MFLUCC 14-1140) and H. thailandica (MFLUCC 14-1143). The ex-type strains are in bold, and the Misturatosphaeria aurantiacinotata isolates are in red. Bootstrap support values ≥75% from the maximum likelihood (ML) and Bayesian posterior probabilities (BYPP) values ≥0.95 are given above the nodes, respectively.
A comprehensive taxonomic framework serves as a cornerstone in many biological research endeavors. It is imperative to thoroughly scrutinize and articulate the specific delineation of species within a taxonomic group, as well as to define its boundaries, to establish a robust taxonomic structure. The presence of cryptic species and phenotypic noise often contributes to the lack of taxonomic clarity and leads to confusion within the field (Bochner, 2008; Delić et al., 2017). In some cases, the same species has been listed under different taxonomic names; in other cases, different species have been classified under the same name.
Distinct species may be hidden within species complexes because the characteristics expressed by the phenotype are not sufficient or strong enough to classify them as different species even though they are genotypically different and reproductively isolated. Hence, it causes difficulties in setting up species circumscriptions and delimitations. Furthermore, the same species having several taxonomic names results in an inaccurate taxonomic system, leading to errors in taxonomic applications in other research areas.
There are practical difficulties in detailed observations of microorganisms and even microstructures of both macro- and microorganisms; thus, detecting cryptic species by their micromorphology could be more difficult. However, in the case of pseudo-cryptic species, a detailed re-examination of phenotypic data provides better solutions for correctly identifying cryptic species (Lajus et al., 2015).
In some cases, detailed anatomical characteristics are not supportive enough to get a clear taxonomy. Instead, molecular tools provide better comparisons and more effectively reveal cryptic species (Karanovic and Cooper, 2012; Karanovic et al., 2016). The detailed microanatomical re-description for Pontohedyle verrucosa was insufficient to reveal reliable characteristics for diagnosing the two major clades identified within the genus. Thus, the molecular analyses based on four genetic markers (mitochondrial cytochrome c oxidase subunit I, 16S rRNA, nuclear 28S, and 18S rRNA) provided sufficient characterization to discover and formally describe nine cryptic new species (Jörger and Schrödl, 2013).
Moreover, fast-evolving molecular markers are even more effective in revealing cryptic species (Karanovic et al., 2016). For example, the 5´-end of the mitochondrial COI gene has been recommended as the barcoding gene for all species of animals (Hebert et al., 2004). The main benefit of this mtCOI gene is that it has little genetic variation within a species but considerable variation between species (Karanovic et al., 2016). Furthermore, metabarcoding has recently emerged as a prominent tool for species identification. This approach proves particularly valuable when conventional morphological identification of species encounters difficulties, as it hinges on the distinctive genetic information inherent to individual species (Semmouri et al., 2021). Metabarcoding can help distinguish cryptic species by targeting specific barcoding DNA regions (e.g., COI for animals, rbcL for plants, and ITS for fungi). In some cases, phenotypic noise, which includes variation in appearance or phenotype within a single species, can make traditional identification methods less reliable. Metabarcoding can overcome this challenge by focusing on the underlying genetic information, which tends to be more consistent. This helps reduce the impact of phenotypic noise and cryptic species on species identification (De Luca et al., 2021; McInnes et al., 2021; Semmouri et al., 2021). However, when molecular markers are inadequate in clarifying taxonomic uncertainties, whole-genome sequencing emerges as a promising approach to establishing a consistent taxonomy (Thompson et al., 2021; Cai et al., 2023). In bacterial taxonomy, common genetic markers encompass the 16S small subunit ribosomal RNA, a hypervariable region within the genome. Nevertheless, the utility of 16S rRNA genes in Microcystis taxonomy has demonstrated their limited informativeness, primarily attributed to the high sequence similarities (>97%) (Harke et al., 2016; Dick et al., 2021). Moreover, while the selected genetic markers may not accurately represent the entire genomic evolutionary landscape, whole-genome sequencing offers a potential solution for establishing a stable taxonomy (Rodriguez-R et al., 2018; Thompson et al., 2021).
In this context, revisiting old taxonomy with modern tools is important to resolve taxonomic confusion. The recent research by Hongsanan et al. (2020) on refining families of the Dothideomycetes expanded information on families in the Dothideomycetidae using new data and molecular tools. Furthermore, the paper provided a refined, updated document on orders and families incertae sedis of the Dothideomycetes. When considering phenotypic noise, morphological changes are more readily visible in macroorganisms than microorganisms. Therefore, the morphology of macroorganisms can tell a different story than their genotype. However, physiological characteristics are easily detectable in microorganisms, especially bacteria and viruses. As a result, in many group phenotypes (collectively morphology and physiology) express variations independently from genotypes, which can result in a conflicting taxonomy (Dussex et al., 2018). However, deviations also can be seen in this regard, especially in yeast (Yurkov et al., 2015).
Taxa with large geographic distributions are comprised of cryptic species and phenotypic noise, thus leading to errors in a taxonomic system. For example, many marine taxa have an exceedingly wide geographical distribution (Knowlton, 1993, further discussion in the “Cryptic species–phenotypic noise and biodiversity section”). Hence, marine specimens from widely separated areas are possibly misidentified as a single species but later recognized as a cryptic species complex. Similarly, a huge geographical distribution can lead to phenotypic variations within a single species, resulting in several different species names being applied. Until those different names for the same species are synonymized, probably following molecular phylogenetic analyses, they confuse taxonomic systems.
Integrated taxonomy (taxonomy based on characters from different sources (e.g., from morphology, molecules, ecology or distribution) plays a major in minimizing the effect of cryptic species and phenotypic noise (Padial and De la Riva, 2010; Jörger and Schrödl, 2013). Furthermore, using discriminate characters, based on their quality and suitability in species delineation, instead of adding more and more data is also important in developing a clear taxonomy. Hence, the taxonomic experts of a particular group are there to guide the respective set(s) of characters that will provide the best backbone for the diagnosis of a specific group (Jörger and Schrödl, 2013).
Although cryptic species are phenotypically similar and occur in most groups, those with a well-studied morphology and that contain specimens of a large size show this phenomenon to a lesser extent (Karanovic et al., 2016). Molecular studies or integrated taxonomy of morphology and genetics can resolve this situation and incorporate cryptic species into their component species (Karanovic et al., 2016). For example, four possible cryptic species were found after barcoding 260 North American bird species (Hebert et al., 2004). In contrast, numerous cryptic species were found in subterranean faunas based on molecular analyses with deep morphological examinations (Guzik et al., 2011). In the case of phenotypic noise, molecular phylogenetic tools are essential for the identification of species phylogeny and assigning these to their correct taxonomic ranks.
Cryptic species contribute to taxonomic incompleteness, impacting the elucidation of evolutionary events and significantly influencing the delineation and classification of a given taxon. Moreover, these cryptic species obscure the evolutionary trajectories of traits within a specific lineage, thereby complicating species delineation efforts (Karanovic et al., 2016).
Cryptic species are in different evolutionary states. Some cryptic species are in the transition stage from populations to species. They are evolutionarily young, and their morphological descendants have not yet diverged (Delić et al., 2017). Other cryptic species are reproductively isolated from each other, with strong biological barriers between them. These cryptic species are evolutionarily old, and their morphologies provide delimiting characters when examined in detail (Delić et al., 2017). Both of these cryptic species and their evolutionary states are important to develop well resolved evolutionary relationships.
Stochasticity in gene expression causes phenotypic noise (Raser and O’shea, 2005). Within a cell or an individual organism, gene expression can be affected by either extrinsic or intrinsic factors, resulting in gene expression variations (Kaneko and Furusawa, 2008). Intrinsic factors are cell signaling molecules and minor genetic alterations. Examples for those genetic alterations include such things as haploinsufficiency and epistasis (Deutschbauer et al., 2005; Eckardt, 2008; Wang et al., 2014). These genetic alterations are inheritable and cause significant variations in the phenotype. The extrinsic factors are environmental factors. These factors are not significant enough to cause a particular species to separate into different species as described in the species concept (Raser and O’shea, 2005; Eckardt, 2008; Wang et al., 2014); hence it results fluctuations in phenotypes, which are phenotypic noises. These phenotypic noises are at sub-species level of a taxonomically described species. They are actually environmentally sensitive subpopulations of the same genotype. Hence, it is assumed that the actual genotype is being shielded by the external environment, which results in a conflict between genotype and phenotype (de Vienne, 2021). Consequently, correct identification of the distinct phenotypic and genotypic state of a particular species is essential for the understanding of its evolutionary relationships.
Moreover, this is the initial stage of the evolution of isogenic populations with phenotypic noise. This is a deviation from the general evolution process based on the inheritance of beneficial mutations of an organism in a population. Environmentally sensitive subpopulations characterized by phenotypic noise can adapt to a variety of environmental conditions without any genetic base, however with changes in their pattern of gene expression influenced by the environment. This spontaneous adaptation by noise can later be developed to a new environmentally well adapted isogenic population, when the deviation of gene expression level is significant enough to separate them from the main population (Kaneko and Furusawa, 2008).
The physiological characters of a species are even more related to ecology. As such, phenotypic noise species are highly influenced by their environment. Although cryptic species are phenotypically undifferentiated, their ecology provides more possibilities for their discovery. As an example, Peintner et al. (2019) examined the use of ecological factors in resolving cryptic species in the fungal genus Fomes, and they concluded that volatile organic compounds serve a better outcome in species delimitation and discovering cryptic species in the future (Peintner et al., 2019). In addition, Bickford et al. (2007) mentioned that cryptic species are more common in insects and reptiles, and they tend to occur more in the tropics than temperate regions.
Cryptic species are common and can be found in any kingdom of the Tree of Life (McGuire, 2018). However, cryptic plant species are comparatively unclear and rare because genetic and molecular tools used to identify cryptic species in other organisms do not really fit with plants (Shneyer and Kotseruba, 2015). Furthermore, more records of cryptic plant species can be determined with more detailed examination of plants and with the improvement of new tools such as nuclear ribosomal DNA sequencing (Okuyama and Kato, 2009; Shneyer and Kotseruba, 2015). Okuyama and Kato (2009) examined the Asian endemic perennial lineage of Mitella (Asimitellaria; Saxifragaceae) based on nuclear ribosomal DNA sequences and discovered ten distinct biological species, including two new species. A multidisciplinary study, which combined cytogenetic analyses and phylogenetic analyses on two plastid and five nuclear genes, revealed the taxonomic separation of three distinct species in the Brachypodium distachyon complex, a model grass for cereals (Catalán et al., 2012). Moreover, cryptic species in plants can be differentiated by the secondary metabolites they produce. These secondary metabolites associated with pest and disease susceptibility, stress tolerance, interactions with soil microbes, attraction of pollinators, and palatability to herbivores or simply the ecological niche of a particular plant species (Shneyer and Kotseruba, 2015).
Gene expression variations associated with non-genetic factors are closely connected with ecology. Hence, phenotypic noise species are adaptive subpopulations to a particular habitat. The long-term ecological influences on those subpopulations or specific phenotypic noise also increase the evolution rate of the population. Moreover, those specific phenotypic noise species bear acquired characters, which can be referred to as adaptations to their environments. In multicellular organisms, acquired characters would be in germplasm (genotype) or somatic cells (phenotype) (de Vienne, 2021). Somatic cell DNA mutations can impact individuals but not their progeny (Hurle, 2022). However, recent agricultural advances in tissue culture techniques and somatic cell genetics allow the regeneration of plants from cells in culture. In somatic cell genetics, even the changes in somatic cells can be transferred to the next generation (NCBI, 2022).
Agricultural research and ecology-based applications are more affected by cryptic species. One of the best examples is the development of biocontrol agents. The success of a biocontrol agent is based on a solid understanding of the biocontrol agent and the target organism, the details of their life cycle, preferred habitats, and foods (Walter, 2003; McGuire, 2018). The presence of cryptic species within either the pest population or the biocontrol agent population can fail the control method for no apparent reason (Walter, 2003; McGuire, 2018). Distinct species react differently to control methods. Correct identification and a better understanding of host and pathogen species are very important in developing control strategies (Walter, 2003; McGuire, 2018). This applies not only to biological control but also to any of the pathogen control methods affected by cryptic species.
The ecological niche of the organism is very important in discovering cryptic species within pathogenic taxa. More studies on pathogen-host-environment interactions are required to increase the understanding of the ecology of cryptic species (Jung et al., 2017). Also, there is a higher possibility of misleading species identification by phenotypic noise. As the plant host plays a major role in speciation, the physiological characters of the pathogenic organism have also turned out to be species-specific (Peintner et al., 2019). Hence, the same species can have different phenotypes within different hosts. Conversely, host interactions can use as a delimiting character to resolve cryptic species. An example for the host and cryptic species relationship showed up in an analysis of anther smuts (Genus Antherospora) on Muscari spp. The molecular phylogeny revealed three distinct lineages that were correlated with host plants with slight morphological differences. These lineages were assigned to three cryptic species: Antherospora hortensis sp. nov. on Muscari armeniacum, A. muscari-botryoidis comb. nov. (syn. Ustilago muscari-botryoidis) on M. botryoides, and A. vaillantii s. str. on M. comosum and M. tenuiflorum (Piątek et al., 2013). Furthermore, new cryptic species of Teratosphaeria, which is a serious pathogen of Eucalyptus were discovered from the phylogenetic analyses of gene regions of Internal transcribed spacer (ITS), β-tubulin (tub2), and translation elongation factor (tef1) (Andjic et al., 2016). Crespo and Lumbsch (2010) also examined cryptic species on lichens.
Furthermore, species complexes such as Microcystis are ecologically much bounded. The taxa within complexes vary across the environmental and seasonal conditions (Frangeul et al., 2008; Cai et al., 2023). Hence, the phenotypic characters become useless in species delimitation, and molecular tools provide promising results. However, the high genome variability of Microcystis due to high genome plasticity and horizontal gene transfer causes the use of genetic markers to be inadequate for species delimitation. In such cases, advanced molecular tools such as pangenomics reveal cryptic diversity within those complexes (Cai et al., 2023).
The “species concept” or “species level” concept plays a fundamental role in biodiversity (Lefébure et al., 2006). Estimating species numbers is an important criterion for both fundamental and applied biodiversity perspectives. Determining the boundaries of species is essential for species number and diversity estimations (Sites and Marshall, 2003). Although many species boundaries exist, their delimitation has no clear and operational criterion (Lefébure et al., 2006). The presence of cryptic species or failures in detecting existing species results in an underestimation of true levels of biodiversity (Lefébure et al., 2006; Padial et al., 2010). Conversely, the same species can be identified by different names as it has many phenotypic noise species, which falsely increases the number of species.
Moreover, biodiversity conservation has also been disturbed by cryptic species. This is because there are species hidden within cryptic species complexes, and these are often rare, have low populations, are seriously threatened, and thus are highly endangered and require urgent conservation efforts. However, conservation programs for undiscovered and unavailable species cannot be carried out. In addition, the formal naming of cryptic species is essential to enable them to be added to conservation policies and faunal listings (Delić et al., 2017). Recent research on the South European cryptic complex of the subterranean amphipod Niphargus stygius sensu lato used uni- and multilocus delineation methods. Molecular analyses showed that the newly discovered species came from several different subgroups within the genus, and that these subgroups coexist while showing no signs of lineage sharing. Those newly discovered cryptic species have increased the number of subterranean endemics in Slovenia by 12 and in Croatia by four previously underestimated species. Moreover, the new taxonomic additions renewed the national Red Lists, as it previously included mostly species with large ranges but omitted critically endangered single-site endemics (Delić et al., 2017).
True cryptic species within a species complex cause huge errors in biodiversity analyses, as discussed above. However, the morphological analyses have considerable limitations in discriminating species and need to be more robust for describing biodiversity at the species level. Therefore, nonmorphological techniques such as genetic analyses and investigations of behavioral, physiological, and other traits must be employed (Lajus et al., 2015). Furthermore, as some studies have included thorough morphological and molecular examinations to detect cryptic species, they do not lead to species descriptions (Pante et al., 2015). This results in an inflation of alpha diversity estimation (Chessman et al., 2007; Morrison et al., 2009; Karanovic et al., 2016). In this context, the phylogenetic species concept is essential in recognizing a far greater number of much less inclusive units. Moreover, the phylogenetic species concept closely examines the same group of organisms, resulting in different species identities, species ranges, and number of individuals (Agapow et al., 2004). Literature reviewing and analyzing organisms that were categorized under phylogenetic and nonphylogenetic concepts show a significant difference, in analyses based on a phylogenetic species concept showing 48% more species than nonphylogenetic concept analyses (Agapow et al., 2004).
The traditional molecular techniques faced challenges when dealing with extensive sample analyses, which often hindered conducting large-scale biodiversity assessments. As a result, researchers outside the taxonomy field have adopted metabarcoding tools for specimen identification. Metabarcoding also holds promise for tasks such as the detection of new species, the revelation of cryptic species, the identification of non-native or invasive species, and the assessment of taxonomically meaningful variations within species that have broad or geographically scattered distributions (De Luca et al., 2021; McInnes et al., 2021; Semmouri et al., 2021). Marine protists have been perceived as having low diversity and a widespread distribution. However, recent research has revealed that many protist species are, in fact, intricate assemblages of cryptic species, often confined to specific biogeographic regions. Nevertheless, the detection of these cryptic species is frequently hindered by limitations in sampling coverage and the application of methods, such as phylogenetic trees, which are less suited for identifying relatively recent divergence and ongoing gene flow (De Luca et al., 2021). In such scenarios, metabarcoding emerges as an ideal solution. De Luca et al. (2021), successfully unraveled the complexities within the Chaetoceros curvisetus (Bacillariophyta) species complex by employing two complementary metabarcoding datasets. Species are diverse and common, especially in marine habitats (Knowlton, 1993). In situ observation of marine taxa is difficult. Furthermore, many marine invertebrate taxa require in situ detailed observations of live specimens by experts or well preserved and fixed specimens to be properly identified, conditions that are hard to fulfill, especially when it comes to large-scale marine inventories. Moreover, since it is possible to spread larvae/adults in a homogenous sea by ocean currents, marine organisms have a wider geographical distribution than terrestrial ones (Knowlton, 1993; Sundberg et al., 2009). This huge distribution of marine organisms results in errors in identifications and later gives rise to cryptic species complexes. Misidentification of the same species based on its phenotypic noise is even possible. Some taxa are affected by these factors more than others, such as small, soft-bodied, marine invertebrates with few morphological characters useful for taxonomy (Knowlton, 1993; Sundberg et al., 2009). Furthermore, certain organisms remain concealed within vast ecosystems because of their unculturable nature and significantly smaller population sizes (He et al., 2015; Ryberg, 2015). However, studies on fungi inhabiting unique environments have revealed that numerous fungi previously considered ‘unculturable’ can be cultivated on specific substrates and under particular conditions. Additionally, high-throughput amplicon sequencing, shotgun metagenomics, single-cell genomics, and Molecular Operational Taxonomic Units (MOTUs) serve as a valuable means to unveil these hidden organisms and offer a promising tool for delineating species boundaries and quantifying biodiversity (He et al., 2015; Ryberg, 2015; Wu et al., 2019).
This paper discusses the two major concepts, cryptic species and phenotypic noise, which affect species concepts. The species concept is the basis of taxonomy, evolution, ecology, and biodiversity. As the kingdom of fungi has made many contributions to biotechnology and applied sciences, this review is expected to improve the identification of fungal species and aid in developing future fungal research applications.
Several branches of species concept are currently used, viz. morphological species concept, biological species concept, evolutionary species concept, and phylogenetic species concept (Taylor et al., 2000). A better understanding of each concept is essential for investigations on cryptic species and phenotypic noise concepts. Cronquist (1978) adopted the morphological species concept and refers to a community with distinctive morphological characters sufficient to entitle them to a specific name (Aldhebiani, 2018).
The biological species concept applies to a group with a distinct ecological niche that is reproductively isolated (Mayr, 1982), while an evolutionary species refers to a single lineage of ancestor-descendant populations with a unique evolutionary history (Simpson, 1961; Wiley, 1981; Paris et al., 1989). The phylogenetic species concept is outside of cladistic analysis and focuses on possible evolutionary processes contributing to species formation, including biotic and abiotic (even random) factors (Wheeler, 1999). A new concept being proposed is the modern species concept, which is focused on the relationship (phenetic or phylogenetic) between individuals (Aldhebiani, 2018).
However, there is no clear congruence between intraspecific variations and species boundaries since some taxonomic studies and many identifications are still based on morphological characters. This simply results in cryptic species, which are phenotypically similar but genotypically distinct, as well as phenotypic noise from species that are phenotypically distinct but genotypically identical. Hence, the cryptic species represent genetic variations of a population. Non-genetic variations are phenotypic variations that arise from different expressions of a genotype. Phenotypic variations are persuaded by varying environmental cues and categorized into two types: phenotypic plasticity and developmental noise (Saito et al., 2023). Phenotypic plasticity is a common act of a biological population or among individuals of a single species as a response to their recent environment by changing their morphology, physiology, or behavior. This is the foundation of how organisms learn, interact with their environment, and adapt to their environment during their lifetime (Matthey-Doret et al., 2020). Therefore, phenotypic plasticity refers to expressing traits that depend on the environment. However, the traits that change unpredictably in different environments are usually said to be ‘noisy’ rather than plastic (Gomulkiewicz and Stinchcombe, 2022). Hence, in this article, we are addressing the concept of phenotypic plasticity using the term phenotypic noises.
Nowadays, DNA sequence information has become the only truly reliable factor for species identification. However, morphological data still play a crucial role in taxonomy, biodiversity, and ecological studies as these data are the primary key to identifying a species, and sometimes there are practical difficulties in obtaining molecular data. Moreover, the concept of phenotypic noise can also result in the occurrence of different individuals or groups of individuals within an isogenic population (Engl, 2019). Hence, using a combination of molecular phylogenetic approaches and morphological and physiological data make a fine-scale taxonomy possible (Jörger and Schrödl, 2013). Moreover, once cryptic species are discovered as a result of the availability of molecular data, there must then be a detailed examination and well documented description of newly discovered taxa using visible external or internal differences to avoid misidentifications in the future.
Most traditional taxonomic studies for some fungal groups, especially those that are unable or difficult to grow on artificial growth media, such as lichenized fungi and discomycetes, are still based on morphological characteristics. Hence, this results in classifying different species under the same taxonomic name. Later, these can be recognized as cryptic species, which causes a major conflict in natural classification systems. Furthermore, setting up evolutionary relationships is difficult with cryptic species. Generally, for most organisms, genetic changes tend to happen slowly than morphological changes (Hanken and Carl, 1996; Carroll, 2005; Hall and Strickberger, 2008). However, when genetic changes happen more rapidly than morphological changes, we see only the phylogenetic differences whereas the morphological changes are not yet discernible. The concept of phenotypic noise results in morphologically and physiologically diverse individuals within isogenic populations (Engl, 2019). Furthermore, Fungi have a unique mode of growth that allows them to adapt quickly to changing environments. Unlike other multicellular organisms (animals and plants), fungi evolved complex multicellularity through filamentous intermediate stages (Nagy et al., 2017). Hence they can exhibit a wide range of phenotypic or functional plasticity in response to environmental cues (Nagy et al., 2017). These changes can occur rapidly and do not always require genetic changes (Naranjo-Ortiz and Gabaldón, 2019). Moreover, since genetic changes can contribute to morphological changes in fungi, it is not accurate to say that genetic changes happen more rapidly than morphological changes in this group of organisms. The relationship between genetic and morphological changes in fungi is complex and varies depending on the specific context (Naranjo-Ortiz and Gabaldón, 2019). This would also result in confusion not only in taxonomy and evolution but also in biodiversity and ecology (Korshunova et al., 2019).
Here, we provide three fungal examples to understand the cryptic species concept and phenotypic noises concept clearly. Around 2–11 million estimated species and around 3 million predicted species are in kingdom fungi, and only around 150,000, a tiny fraction of the total estimation, have been formally described (Phukhamsakda et al., 2022). Compared to plants and animals, fungi have simple body plans that are often morphologically and ecologically concealed and continuously challenging for precise identifications and species delimitations (Lücking et al., 2020). Plant pathogenic fungi cover one of the most significant fractions of total estimated fungi and play an important ecological and economic role; hence, they are directly involved with the development of effective food security, crop protection, and pest risk assessments (Cai et al., 2011; Li et al., 2020). For example, rust and smut fungi collectively cover 10% of all known fungi. Therefore, DNA sequence data have recently been extensively employed in developing stable taxonomy for plant pathogenic fungi (Cai et al., 2011). The next crucial ecological group of fungi is saprobes for the earth’s nutrient cycling. The examples provided here are saprobes, which evolve fast with environmental stimuli (Naranjo-Ortiz and Gabaldón, 2019). This causes fast-evolving species concepts and delimitations. Hence, the fungi kingdom needs extensive analyses and comprehensive species identification methods. Recently, a conceptual framework was analyzed and suggested for identifying fungi (Lücking et al., 2020). Further, the approach encourages the integrative (polyphasic) taxonomy for species delimitation, which combines phylogeny, phenotype, and reproductive biology. This facilitates evaluating a wide range of diagnostic characteristics, either phenotypic, molecular, or both (Lücking et al., 2020). However, polyphasic approaches are being limited by the availability of data and analyzing capacity of particular groups, while it is well suited for some groups. The effectiveness of polyphasic approaches for identifying yeast as a case study was widely investigated (Yurkov et al., 2015; Boekhout et al., 2021). The advantage of polyphasic approaches over traditional morphological character-based approaches is that species ultimately depend on species recognition approaches and the number of characters in traditional methods. Considering the dichotomous key-like approach, species recognition is based on a few characteristics, such as categorical (spore types, color) or numerical range variables (spore size). In polyphasic approaches, taxonomy based on many characteristics [e.g., assimilation tests for yeasts (Yurkov et al., 2015; Boekhout et al., 2021)] and results in an in-depth understanding of the phenotype and/or niche concepts which aid to recognize cryptic species and phenotypic noises and minimize the confliction in stable systematics.
Phylogeny-based clade-specific evolutionary histories are mostly used to verify species identifications, a pivotal step in taxonomy. However, no single tool for identifying fungi and internal transcribed spacer (ITS) is still considered the first diagnosis, particularly in metabarcoding studies. Secondary DNA barcodes are long-established for resolving taxonomy and for precise identifications where they do not provide sufficient distinctness (Lücking et al., 2020). When considering the molecular markers for resolving cryptic species, it is important to know that identifying “what we call cryptic” depends on the markers used. Therefore, it is more important to ensure congruence between the different markers. Simply adding more and more increasingly variable gene loci can identify more lineages in a population, but the lineages may not necessarily represent species. This problem of applying species names to lineages within a species is becoming more common in the literature with respect to the diversity of fungi lineages being described as species. These pitfalls can be resolved using single-copy orthologs genes and coalescence-based methods (Shen et al., 2016; Maharachchikumbura et al., 2021). Single-copy orthologs are genes that are present only once in the genome of each species, and have descended from a single ancestral gene present in their last common ancestor (Aguileta et al., 2008; Ren et al., 2016; Shen et al., 2016; Maharachchikumbura et al., 2021). Applications of whole genome sequencing are becoming increasingly common in fungal studies, as complete genomes provide in-depth knowledge and comparisons on the evolvability of the genomes and, consequently, reveal the species’ true ancestry. Furthermore, using complete genome information is more beneficial than drafting genomes, as it provides a better understanding of species and their genome and genetic dynamics (Boekhout et al., 2021). The species recognition in some groups of fungi is tightly connected to species concepts of their hosts and vectors. This is especially important for parasitic fungi, depending on their host species. Some plant-pathogenic fungi have a broad host range, while the fungal family Erysiphaceae causes considerable damage to many crop plants, including Grapevine, Eucalyptus, Rubber, cucumber, tomato, onion, pepper and potato (Ekanayaka et al., 2019), some are highly limited in the range of plant species [e.g. Fungal family Cyttariaceae is host specific on Nothofagus spp (Ekanayaka et al., 2019)] or even cultivars that they cause diseases (strict host-parasite association) (Li et al., 2020). Several recent taxonomic studies of fungi include the host of fungi collected into their descriptions, and the host used as a character in species recognition (Hongsanan et al., 2020; Hyde et al., 2020), which would be helpful in accurate species identification. Furthermore, Cryptic sibling species can be specialized in different ecological conditions and often become host-specific. For instance, the fungus Microbotryum violaceum causes another smut disease in Caryophyllaceae plants, and the degree of specialization and gene flow between strains on different hosts are becoming controversial. Molecular phylogenetic analyses using single-copy nuclear genes help resolve taxonomic confusion in M. violaceum from 23 host species and different geographic origins (Le Gac et al., 2007). Hence, molecular phylogenetic analyses play a crucial role in identifying cryptic species. Moreover, host specificity indexes, such as STD, which measures the average taxonomic distinctness among the host species used by a parasite, weighted for the parasite’s prevalence in the different hosts (Poulin and Mouillot, 2005) and specific analyses such as Global and individual ParaFit tests to construct a plant–pathogen evolutionary association network (Zeng et al., 2019; Chen et al., 2022) can be applied to analyze to investigate host and fungal relationship and their coevolution. This also provides a broad spectrum of data on those fungal groups, specifically in niche concepts.
Correct fungal species identification is vital as the kingdom of fungi plays a crucial role in applied sciences. For instance, disease diagnosis and medical prescriptions are based on correct species identifications in the pharmaceutical industry and medical sciences. Cryptic Aspergillus species cause cryptic aspergillosis and are becoming more prevalent in humans and can cause significant morbidity and mortality in immunocompromised individuals. Moreover, cryptic Aspegillus species are causing invasive diseases and are usually more resistant to common antifungal therapies. Therefore, the correct identification and characterization of these fungi have both epidemiological and clinical implications when evaluating the impact of such species in aspergillosis (Fernandez-Pittol et al., 2022; Ninan et al., 2022). Furthermore, accurate species identification can reveal important information regarding possible biochemical properties of a particular species, which can be explicitly applied to drug discoveries (Raja et al., 2017).
Cryptic species represent a portion of the missing fraction of biodiversity (Vodă et al., 2015). Consequently, resolving cryptic species is important as it reveals the true level of biodiversity and thus contributes to conservation efforts. Some cryptic species may be seriously threatened but unavailable for conservation programs as they remain undescribed (Delić et al., 2017). The evolutionary state of the cryptic species (See cryptic species and evolution) is important with respect to conservation programs. However, the biological properties and their relationship to the recent ancestors relevant for the conservation of cryptic species (i.e., sexual reproduction, spore germination, etc.) are often not known (Delić et al., 2017). Inappropriate management of endangered species can result in these being more threatened than previously thought (Niemiller et al., 2013; Brodersen and Seehausen, 2014; Delić et al., 2017).
During the analyses of highly diverse species, mislabeling and insufficient investigations lead to identifying the same species in different names because of phenotypic plasticity. Later, this can be corrected in taxonomic revisions, and the misidentifications are synonymized under the correct names. However, having many synonyms for a single species still needs to be more accurate. In this regard, conducting taxonomic revisions [e.g (Cruz-Morales et al., 2019; El-Banhawy et al., 2021)] and taxonomic outlines [e.g (Wijayawardene et al., 2020)] that provide recent taxonomic updates on taxa at one place is very important and helpful for precise species identifications.
Resolving cryptic species complexes and identifying phenotypic plasticity of a species are highly demanding but critically important in taxonomy and taxonomic applications in other areas such as evolution, ecology, and biodiversity. It provides a better understanding of species, their distribution patterns in the Tree of Life, and the underlying story of their evolution. We hope this review will provide an overall understanding of cryptic species, phenotypic noise, and their effects. Also, this should encourage more detailed analyses of morphological characters with the combination of molecular phylogenetic analyses, which can provide fine scale taxonomy.
“Cryptic species” refers to a situation in which two or more species are morphologically indistinguishable in their original descriptions and have been erroneously classified under the same scientific name. “Phenotypic noise” refers to different phenotypic states of the same isogenic population. Both are contributing to evolution but in two opposite ways.
Cryptic species and phenotypic noise create confusion in taxonomic structure and leave huge gaps in describing the flow of the evolutionary process. Molecular tools are the only way to detect them easily. However, a detailed examination of macro-, micro-, external, and internal morphological characters also provides a better endorsement of taxonomic status. Both concepts hide the real biodiversity and inhibit a clear understanding of species evolution and their ecological significance.
Although members of a particular species may appear to be similar, several to many distinct species can be present. In other words, many unrecognized species may be hidden as cryptic species. Many important taxa may need to be protected but are already extinct within cryptic species complexes. Conversely, species limitations must be well identified as the same species could be differently identified based on its phenotypic plasticity for different environmental conditions. Cryptic species and phenotypic variations of the same species also affect studies of ecology and agricultural research, especially in relation to plant pathology and developing biocontrol agents, where these concepts can pose significant obstacles.
Cryptic species result from the species evolution process. Discovering cryptic species and identifying the phenotypic noise of particular species helps develop a fine-scale taxonomic system and improve its applications in areas such as evolution, ecology, and biodiversity.
AE: Conceptualization, Formal analysis, Methodology, Writing – original draft, Writing – review & editing. SK: Conceptualization, Formal analysis, Investigation, Methodology, Supervision, Writing – original draft, Writing – review & editing. ST: Formal analysis, Methodology, Software, Visualization, Writing – review & editing. AD: Formal analysis, Methodology, Software, Writing – review & editing. DT: Formal analysis, Methodology, Writing – review & editing. AK: Formal analysis, Methodology, Writing – review & editing. EC: Formal analysis, Methodology, Writing – review & editing. DD: Formal analysis, Methodology, Writing – review & editing. SS: Formal analysis, Methodology, Validation, Writing – review & editing. SM: Formal analysis, Investigation, Methodology, Writing – review & editing. CL: Formal analysis, Funding acquisition, Methodology, Resources, Writing – review & editing. AP: Formal analysis, Investigation, Methodology, Validation, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. National Natural Science Foundation of China Grant No. 32060710, 32260004, 31760013, and 31950410558.
AP acknowledges the support from UID/04046/2025 Centre grant from FCT, Portugal (to BioISI). SK, ST, and D-QD thank the High-Level Talent Recruitment Plan of Yunnan Provinces (“Young Talents and High-End Foreign Experts” Programs) for their support. CL thanks the National Natural Science Foundation of China Grant No. 32060710 for the support. The authors would like to thank the Program of Doctoral Innovation Research Team from Qujing Normal University, and Yunnan Provincial Department of Science and Technology “Zhihui Yunnan” plan (202403AM140023) for the support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adams, M., Raadik, T. A., Burridge, C. P., Georges, A. (2014). Global biodiversity assessment and hyper-cryptic species complexes: more than one species of elephant in the room? Systematic Biol. 63, 518–533. doi: 10.1093/sysbio/syu017
Agapow, P.-M., Bininda-Emonds, O. R., Crandall, K. A., Gittleman, J. L., Mace, G. M., Marshall, J. C., et al. (2004). The impact of species concept on biodiversity studies. Q. Rev. Biol. 79, 161–179. doi: 10.1086/383542
Aguileta, G., Marthey, S., Chiapello, H., Lebrun, M.-H., Rodolphe, F., Fournier, E., et al. (2008). Assessing the performance of single-copy genes for recovering robust phylogenies. Systematic Biol. 57, 613–627. doi: 10.1080/10635150802306527
Aldhebiani, A. Y. (2018). Species concept and speciation. Saudi J. Biol. Sci. 25, 437–440. doi: 10.1016/j.sjbs.2017.04.013
Andjic, V., Maxwell, A., Hardy, G. E., Burgess, T. I. (2016). New cryptic species of Teratosphaeria on Eucalyptus in Australia. IMA Fungus 7, 253–263. doi: 10.5598/imafungus.2016.07.02.05
Antonelli, A. F. (2023). State of the World’s Plants and Fungi 2023 (Kew, UK: Royal Botanic Gardens, Kew).
Baturo-Ciesniewska, A., Pusz, W., Patejuk, K. (2020). Problems, limitations, and challenges in species identifcation of Ascomycota members on the basis of ITS regions. Acta Mycologica 55. doi: 10.5586/am.5512
Behm, J. E., Kiers, E. T. (2014). A phenotypic plasticity framework for assessing intraspecific variation in arbuscular mycorrhizal fungal traits. J. Ecol. 102, 315–327. doi: 10.1111/jec.2014.102.issue-2
Bickford, D., Lohman, D. J., Sodhi, N. S., Ng, P. K., Meier, R., Winker, K., et al. (2007). Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 22, 148–155. doi: 10.1016/j.tree.2006.11.004
Bochner, B. R. (2008). Global phenotypic characterization of bacteria. FEMS Microbiol. Rev. 33, 191–205. doi: 10.1111/j.1574-6976.2008.00149.x
Boekhout, T., Aime, M. C., Begerow, D., Gabaldón, T., Heitman, J., Kemler, M., et al. (2021). The evolving species concepts used for yeasts: from phenotypes and genomes to speciation networks. Fungal Diversity 109, 27–55. doi: 10.1007/s13225-021-00475-9
Brodersen, J., Seehausen, O. (2014). Why evolutionary biologists should get seriously involved in ecological monitoring and applied biodiversity assessment programs. Evolutionary Appl. 7, 968–983. doi: 10.1111/eva.2014.7.issue-9
Cai, H., McLimans, C. J., Beyer, J. E., Krumholz, L. R., Hambright, K. D. (2023). Microcystis pangenome reveals cryptic diversity within and across morphospecies. Sci. Adv. 9, eadd3783. doi: 10.1126/sciadv.add3783
Cai, L., Giraud, T., Zhang, N., Begerow, D., Cai, G., Shivas, R. G. (2011). The evolution of species concepts and species recognition criteria in plant pathogenic fungi. Fungal Diversity 50, 121–133. doi: 10.1007/s13225-011-0127-8
Carroll, S. B. (2005). Endless forms most beautiful: The new science of evo devo and the making of the animal kingdom (WW Norton and Co: WW Norton & Company).
Catalán, P., Müller, J., Hasterok, R., Jenkins, G., Mur, L. A., Langdon, T., et al. (2012). Evolution and taxonomic split of the model grass Brachypodium distachyon. Ann. Bot. 109, 385–405. doi: 10.1093/aob/mcr294
Chen, L., Yang, A.-L., Li, Y.-X., Zhang, H.-B. (2022). Virulence and host range of fungi associated with the invasive plant Ageratina adenophora. Front. Microbiol. 13, 857796. doi: 10.3389/fmicb.2022.857796
Chessman, B., Williams, S., Besley, C. (2007). Bioassessment of streams with macroinvertebrates: effect of sampled habitat and taxonomic resolution. J. North Am. Benthological Soc. 26, 546–565. doi: 10.1899/06-074.1
Cobos-Villagrán, A., Valenzuela, R., Hernández-Rodríguez, C., Calvillo-Medina, R. P., Villa-Tanaca, L., Mateo-Cid, L. E., et al. (2021). Three new species of Rhytidhysteron (Dothideomycetes, Ascomycota) from Mexico. MycoKeys 83, 123. doi: 10.3897/mycokeys.83.68582
Crespo, A., Lumbsch, H. T. (2010). Cryptic species in lichen-forming fungi. IMA Fungus 1, 167–170. doi: 10.5598/imafungus.2010.01.02.09
Cronquist, A. (1978). Once again, what is a species? Biosystematics in agriculture (Biosystematics in agriculture), Vol. 2. 3–20.
Cruz-Morales, P., Orellana, C. A., Moutafis, G., Moonen, G., Rincon, G., Nielsen, L. K., et al. (2019). Revisiting the evolution and taxonomy of Clostridia, a phylogenomic update. Genome Biol. Evol. 11, 2035–2044. doi: 10.1093/gbe/evz096
Delić, T., Trontelj, P., Rendoš, M., Fišer, C. (2017). The importance of naming cryptic species and the conservation of endemic subterranean amphipods. Sci. Rep. 7, 1–12. doi: 10.1038/s41598-017-02938-z
De Luca, D., Piredda, R., Sarno, D., Kooistra, W. H. (2021). Resolving cryptic species complexes in marine protists: phylogenetic haplotype networks meet global DNA metabarcoding datasets. ISME J. 15, 1931–1942. doi: 10.1038/s41396-021-00895-0
De Queiroz, K. (1998). “The general lineage concept of species, species criteria, and the process of speciation,” in Endless forms: species and speciation.
De Queiroz, K. (2007). Species concepts and species delimitation. Systematic Biol. 56, 879–886. doi: 10.1080/10635150701701083
De Santis, V., Delmastro, G. B., Vanetti, I., Britton, J. R., Zaccara, S. (2021). Species composition of introduced and natural minnow populations of the Phoxinus cryptic complex in the westernmost part of the Po River Basin (north Italy). Biol. Invasions 23, 657–668. doi: 10.1007/s10530-020-02406-2
De Silva, N., Tennakoon, D., Thambugala, K., Karunarathna, S., Lumyong, S., Hyde, K. (2020). Morphology and multigene phylogeny reveal a new species and a new record of Rhytidhysteron (Dothideomycetes, Ascomycota) from China. Asian J. Mycology 3, 295–306. doi: 10.5943/ajom/3/1/4
Deutschbauer, A. M., Jaramillo, D. F., Proctor, M., Kumm, J., Hillenmeyer, M. E., Davis, R. W., et al. (2005). Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics 169, 1915–1925. doi: 10.1534/genetics.104.036871
de Vienne, D. (2021). What is a phenotype? History and new developments of the concept. Genetica, 1–6. doi: 10.1007/s10709-021-00134-6
Dick, G. J., Duhaime, M. B., Evans, J. T., Errera, R. M., Godwin, C. M., Kharbush, J. J., et al. (2021). The genetic and ecophysiological diversity of Microcystis. Environ. Microbiol. 23, 7278–7313. doi: 10.1111/1462-2920.15615
Dobzhansky, T. (1951). Experiments on sexual isolation in Drosophila: X. Reproductive isolation between Drosophila pseudoobscura and Drosophila persimilis under natural and under laboratory conditions. Proc. Natl. Acad. Sci. United States America 37, 792. doi: 10.1073/pnas.37.12.792
Dobzhansky, T., Epling, C. (1944). Contributions to the genetics, taxonomy, and ecology of Drosophila pseudoobscura and its relatives (Carnegie institution of Washington).
Dussex, N., Taylor, H. R., Irestedt, M., Robertson, B. C. (2018). When genetic and phenotypic data do not agree: the conservation implications of ignoring inconvenient taxonomic evidence. New Z. J. Ecol. 42, 284–290. doi: 10.20417/nzjecol.42.13
Eckardt, N. A. (2008). Epistasis and genetic regulation of variation in the Arabidopsis metabolome. doi: 10.1105/tpc.108.061051
Ekanayaka, A., Hyde, K., Gentekaki, E., McKenzie, E., Zhao, Q., Bulgakov, T., et al. (2019). Preliminary classification of leotiomycetes. Mycosphere 10, 310–489. doi: 10.5943/mycosphere/10/1/7
El-Banhawy, A., Nour, I. H., Acedo, C., ElKordy, A., Faried, A., Al-Juhani, W., et al. (2021). Taxonomic revisiting and phylogenetic placement of two endangered plant species: Silene leucophylla Boiss. and Silene schimperiana Boiss.(Caryophyllaceae). Plants 10, 740. doi: 10.3390/plants10040740
Engl, C. (2019). Noise in bacterial gene expression. Biochem. Soc. Trans. 47, 209–217. doi: 10.1042/BST20180500
Fernandez-Pittol, M., Alejo-Cancho, I., Rubio-García, E., Cardozo, C., Puerta-Alcalde, P., Moreno-García, E., et al. (2022). Aspergillosis by cryptic Aspergillus species: A case series and review of the literature. Rev. Iberoamericana Micología 39, 44–49. doi: 10.1016/j.riam.2022.04.002
Frangeul, L., Quillardet, P., Castets, A.-M., Humbert, J.-F., Matthijs, H. C., Cortez, D., et al. (2008). Highly plastic genome of Microcystis aeruginosa PCC 7806, a ubiquitous toxic freshwater cyanobacterium. BMC Genomics 9, 1–20. doi: 10.1186/1471-2164-9-274
Fries, E. (1835). Corpus florarum provincialium Sueciae: I. Floram scanicam (Typis Palmblad, Sebell & Company).
Gomulkiewicz, R., Stinchcombe, J. R. (2022). Phenotypic plasticity made simple, but not too simple. Am. J. Bot. 109, 1519. doi: 10.1002/ajb2.v109.10
Guzik, M. T., Austin, A. D., Cooper, S. J., Harvey, M. S., Humphreys, W. F., Bradford, T., et al. (2011). Is the Australian subterranean fauna uniquely diverse? Invertebrate Systematics 24, 407–418. doi: 10.1071/IS10038
Hall, T. (2004). BioEdit version 7.0. 0. Available online at: www.mbio.ncsu.edu/BioEdit/bioedit.html.
Hanken, J., Carl, T. F. (1996). The shape of life: Genes, development, and the evolution of animal form. Ed. Rudolf, A. (Raff University of Chicago Press), 520ISBN 0226702650.
Harke, M. J., Steffen, M. M., Gobler, C. J., Otten, T. G., Wilhelm, S. W., Wood, S. A., et al. (2016). A review of the global ecology, genomics, and biogeography of the toxic cyanobacterium, Microcystis spp. Harmful Algae 54, 4–20. doi: 10.1016/j.hal.2015.12.007
He, Y., Caporaso, J. G., Jiang, X.-T., Sheng, H.-F., Huse, S. M., Rideout, J. R., et al. (2015). Stability of operational taxonomic units: an important but neglected property for analyzing microbial diversity. Microbiome 3, 1–10. doi: 10.1186/s40168-015-0081-x
Hebert, P. D. N., Stoeckle, M. Y., Zemlak, T. S., Francis, C. M., Godfray, C. (2004). Identification of birds through DNA barcodes. PloS Biol. 2, e312. doi: 10.1371/journal.pbio.0020312
Hongsanan, S., Hyde, K. D., Phookamsak, R., Wanasinghe, D. N., McKenzie, E. H., Sarma, V. V., et al. (2020). Refined families of Dothideomycetes: orders and families incertae sedis in Dothideomycetes. Fungal Diversity 105, 17–318. doi: 10.1007/s13225-020-00462-6
Hurle, B. (2022). Somatic Cells (National Human Genome Research Institute). Available online at: https://www.genome.gov/genetics-glossary/Somatic-Cells (Accessed September 21, 2022).
Hyde, K. D., Dong, Y., Phookamsak, R., Jeewon, R., Bhat, D. J., Jones, E. G., et al. (2020). Fungal diversity notes 1151–1276: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 100, 5–277. doi: 10.1007/s13225-020-00439-5
Jörger, K. M., Schrödl, M. (2013). How to describe a cryptic species? Practical challenges of molecular taxonomy. Front. Zoology 10, 1–27. doi: 10.1186/1742-9994-10-59
Jung, T., Jung, M. H., Cacciola, S. O., Cech, T., Bakonyi, J., Seress, D., et al. (2017). Multiple new cryptic pathogenic Phytophthora species from Fagaceae forests in Austria, Italy and Portugal. IMA Fungus 8, 219–244. doi: 10.5598/imafungus.2017.08.02.02
Kaneko, K., Furusawa, C. (2008). Relevance of phenotypic noise to adaptation and evolution. IET Syst. Biol. 2, 234–246. doi: 10.1049/iet-syb:20070078
Karanovic, T., Cooper, S. J. (2012). Explosive radiation of the genus Schizopera on a small subterranean island in Western Australia (Copepoda: Harpacticoida): unravelling the cases of cryptic speciation, size differentiation and multiple invasions. Invertebrate Systematics 26, 115–192. doi: 10.1071/IS11027
Karanovic, T., Djurakic, M., Eberhard, S. M. (2016). Cryptic species or inadequate taxonomy? Implementation of 2D geometric morphometrics based on integumental organs as landmarks for delimitation and description of copepod taxa. Systematic Biol. 65, 304–327. doi: 10.1093/sysbio/syv088
Knowlton, N. (1993). Sibling species in the sea. Annu. Rev. Ecol. Systematics 24, 189–216. doi: 10.1146/annurev.es.24.110193.001201
Korshunova, T., Picton, B., Furfaro, G., Mariottini, P., Pontes, M., Prkić, J., et al. (2019). Multilevel fine-scale diversity challenges the ‘cryptic species’ concept. Sci. Rep. 9, 1–23. doi: 10.1038/s41598-019-42297-5
Kües, U., Khonsuntia, W., Subba, S. (2018). Complex fungi. Fungal Biol. Rev. 32, 205–218. doi: 10.1016/j.fbr.2018.08.001
Lajus, D., Sukhikh, N., Alekseev, V. (2015). Cryptic or pseudocryptic: can morphological methods inform copepod taxonomy? An analysis of publications and a case study of the Eurytemora affinis species complex. Ecol. Evol. 5, 2374–2385. doi: 10.1002/ece3.2015.5.issue-12
Lefébure, T., Douady, C., Gouy, M., Gibert, J. (2006). Relationship between morphological taxonomy and molecular divergence within Crustacea: proposal of a molecular threshold to help species delimitation. Mol. Phylogenet. Evol. 40, 435–447. doi: 10.1016/j.ympev.2006.03.014
Le Gac, M., Hood, M. E., Fournier, E., Giraud, T. (2007). Phylogenetic evidence of host-specific cryptic species in the anther smut fungus. Evolution 61, 15–26. doi: 10.1111/j.1558-5646.2007.00002.x
Lehner, B. (2010). Conflict between noise and plasticity in yeast. PloS Genet. 6, e1001185. doi: 10.1371/journal.pgen.1001185
Li, J., Cornelissen, B., Rep, M. (2020). Host-specificity factors in plant pathogenic fungi. Fungal Genet. Biol. 144, 103447. doi: 10.1016/j.fgb.2020.103447
Lücking, R., Aime, M. C., Robbertse, B., Miller, A. N., Ariyawansa, H. A., Aoki, T., et al. (2020). Unambiguous identification of fungi: where do we stand and how accurate and precise is fungal DNA barcoding? IMA Fungus 11, 1–32. doi: 10.1186/s43008-020-00033-z
Maharachchikumbura, S. S., Chen, Y., Ariyawansa, H. A., Hyde, K. D., Haelewaters, D., Perera, R. H., et al. (2021). Integrative approaches for species delimitation in Ascomycota. Fungal Diversity 109, 155–179. doi: 10.1007/s13225-021-00486-6
Matthey-Doret, R., Draghi, J. A., Whitlock, M. C. (2020). Plasticity via feedback reduces the cost of developmental instability. Evol. Lett. 4, 570–580. doi: 10.1002/evl3.202
Mayden, R. L. (1997). A hierarchy of species concepts: the denouement in the saga of the species problem.
McGuire, A. (2018). Cryptic Species, Agriculture, and Confirmation Bias (Center for Sustaining Agriculture and Natural Resources (CSANR), Washington State University). Available online at: https://csanr.wsu.edu/cryptic-species/:~:text=For%20example%2C%20cryptic%20species%20in,et%20al.%2C%202012 (Accessed May 28, 2022).
McInnes, J. C., Bird, J. P., Deagle, B. E., Polanowski, A. M., Shaw, J. D. (2021). Using DNA metabarcoding to detect burrowing seabirds in a remote landscape. Conserv. Sci. Pract. 3, e439. doi: 10.1111/csp2.v3.7
Miller, M. A., Pfeiffer, W., Schwartz, T. (2011). The CIPRES science gateway: a community resource for phylogenetic analyses. 1–8.
Morrison, W., III, Lohr, J., Duchen, P., Wilches, R., Trujillo, D., Mair, M., et al. (2009). The impact of taxonomic change on conservation: does it kill, can it save, or is it just irrelevant? Biol. Conserv. 142, 3201–3206. doi: 10.1016/j.biocon.2009.07.019
Mugambi, G., Huhndorf, S. (2009). Molecular phylogenetics of pleosporales: Melanommataceae and Lophiostomataceae re-circumscribed (Pleosporomycetidae, Dothideomycetes, Ascomycota). Stud. Mycology 64, 103–121. doi: 10.3114/sim.2009.64.05
Nagy, L. G., Tóth, R., Kiss, E., Slot, J., Gácser, A., Kovacs, G. M. (2017). “Six key traits of fungi: their evolutionary origins and genetic bases,” in The fungal kingdom, 35–56.
Naranjo-Ortiz, M. A., Gabaldón, T. (2019). Fungal evolution: major ecological adaptations and evolutionary transitions. Biol. Rev. 94, 1443–1476. doi: 10.1111/brv.2019.94.issue-4
NCBI (2022). Somatic Cell Genetics - Genetic Engineering of Plants - NCBI Bookshelf (National Center for Biotechnological Information). Available online at: https://www.ncbi.nlm.nih.gov/books/NBK216404/ (Accessed September 21, 2022).
Niemiller, M. L., Graening, G. O., Fenolio, D. B., Godwin, J. C., Cooley, J. R., Pearson, W. D., et al. (2013). Doomed before they are described? The need for conservation assessments of cryptic species complexes using an amblyopsid cavefish (Amblyopsidae: Typhlichthys) as a case study. Biodiversity Conserv. 22, 1799–1820. doi: 10.1007/s10531-013-0514-4
Ninan, M. M., Kurien, R., Fouzia, N. A., Sahni, R. D., Michael, J. S. (2022). Cryptic aspergillosis: a rare entity and a diagnostic challenge. Access Microbiol. 4, 000344. doi: 10.1099/acmi.0.000344
Okuyama, Y., Kato, M. (2009). Unveiling cryptic species diversity of flowering plants: successful biological species identification of Asian Mitellausing nuclear ribosomal DNA sequences. BMC Evolutionary Biol. 9, 1–16. doi: 10.1186/1471-2148-9-105
Padial, J. M., De la Riva, I. (2010). A response to recent proposals for integrative taxonomy. Biol. J. Linn. Soc. 101, 747–756. doi: 10.1111/j.1095-8312.2010.01528.x
Padial, J. M., Miralles, A., De la Riva, I., Vences, M. (2010). The integrative future of taxonomy. Front. Zoology 7, 1–14. doi: 10.1186/1742-9994-7-16
Pante, E., Schoelinck, C., Puillandre, N. (2015). From integrative taxonomy to species description: one step beyond. Systematic Biol. 64, 152–160. doi: 10.1093/sysbio/syu083
Paris, C. A., Wagner, F. S., Wagner, W. H. (1989). Cryptic species, species delimitation, and taxonomic practice in the homosporous ferns. Am. Fern J. 79, 46–54. doi: 10.2307/1547159
Peintner, U., Kuhnert-Finkernagel, R., Wille, V., Biasioli, F., Shiryaev, A., Perini, C. (2019). How to resolve cryptic species of polypores: an example in Fomes. IMA Fungus 10, 1–21. doi: 10.1186/s43008-019-0016-4
Phukhamsakda, C., Nilsson, R. H., Bhunjun, C. S., de Farias, A. R. G., Sun, Y.-R., Wijesinghe, S. N., et al. (2022). The numbers of fungi: Contributions from traditional taxonomic studies and challenges of metabarcoding. Fungal Diversity 114, 327–386. doi: 10.1007/s13225-022-00502-3
Piątek, M., Lutz, M., Chater, A. O. (2013). Cryptic diversity in the Antherospora vaillantii complex on Muscari species. IMA Fungus 4, 5–19. doi: 10.5598/imafungus.2013.04.01.02
Poulin, R., Mouillot, D. (2005). Combining phylogenetic and ecological information into a new index of host specificity. J. Parasitol. 91, 511–514. doi: 10.1645/GE-398R
Purvis, A., Hector, A. (2000). Getting the measure of biodiversity. Nature 405, 212–219. doi: 10.1038/35012221
Raja, H. A., Miller, A. N., Pearce, C. J., Oberlies, N. H. (2017). Fungal identification using molecular tools: a primer for the natural products research community. J. Natural Products 80, 756–770. doi: 10.1021/acs.jnatprod.6b01085
Rambaut, A., Drummond, A. (2009). Tracer v. 1.5. Available online at:http.ac.uk/Tracer (Accessed January 1, 2013).
Raser, J. M., O’shea, E. K. (2005). Noise in gene expression: origins, consequences, and control. Science 309, 2010–2013. doi: 10.1126/science.1105891
Ray, J., Willughby, F. (1718). Philosophical Letters Between the Late Learned Mr. Ray and Several of His Ingenious Correspondents, Natives and Foreigners: To which are Added Those of Francis Willughby Esq (W. and J. Innys).
Ren, R., Sun, Y., Zhao, Y., Geiser, D., Ma, H., Zhou, X. (2016). Phylogenetic resolution of deep eukaryotic and fungal relationships using highly conserved low-copy nuclear genes. Genome Biol. Evol. 8, 2683–2701. doi: 10.1093/gbe/evw196
Rodriguez, R. J., Cullen, D., Kurtzman, C. P., Khachatourians, G. G., Hegedus, D. D. (2004). “Molecular methods for discriminating taxa, monitoring species, and assessing fungal diversity,” in Biodiversity of fungi: inventory and monitoring methods, 77–102. doi: 10.1016/B978-012509551-8/50009-X
Rodriguez-R, L. M., Castro, J. C., Kyrpides, N. C., Cole, J. R., Tiedje, J. M., Konstantinidis, K. T. (2018). How much do rRNA gene surveys underestimate extant bacterial diversity? Appl. Environ. Microbiol. 84, e00014–e00018.
Ryberg, M. (2015). Molecular operational taxonomic units as approximations of species in the light of evolutionary models and empirical data from Fungi. doi: 10.1111/mec.2015.24.issue-23
Saito, K., Tsuboi, M., Takahashi, Y. (2023). Developmental noise and phenotypic plasticity are correlated in Drosophila simulans. bioRxiv. doi: 10.1093/evlett/qrad069
Sato, H., Ohta, R., Murakami, N. (2020). Molecular prospecting for cryptic species of the Hypholoma fasciculare complex: toward the effective and practical delimitation of cryptic macrofungal species. Sci. Rep. 10, 1–13. doi: 10.1038/s41598-020-70166-z
Semmouri, I., De Schamphelaere, K. A., Willemse, S., Vandegehuchte, M. B., Janssen, C. R., Asselman, J. (2021). Metabarcoding reveals hidden species and improves identification of marine zooplankton communities in the North Sea. ICES J. Mar. Sci. 78, 3411–3427. doi: 10.1093/icesjms/fsaa256
Shen, X.-X., Zhou, X., Kominek, J., Kurtzman, C. P., Hittinger, C. T., Rokas, A. (2016). Reconstructing the backbone of the Saccharomycotina yeast phylogeny using genome-scale data. G3: Genes Genomes Genet. 6, 3927–3939. doi: 10.1534/g3.116.034744
Shivas, R. G., Cai, L. (2012). Cryptic fungal species unmasked. Microbiol. Aust. 33, 36–37. doi: 10.1071/MA12036
Shneyer, V., Kotseruba, V. (2015). Cryptic species in plants and their detection by genetic differentiation between populations. Russian J. Genetics: Appl. Res. 5, 528–541. doi: 10.1134/S2079059715050111
Sites, J. W., Jr., Marshall, J. C. (2003). Delimiting species: a Renaissance issue in systematic biology. Trends Ecol. Evol. 18, 462–470. doi: 10.1016/S0169-5347(03)00184-8
Stajich, J. E., Berbee, M. L., Blackwell, M., Hibbett, D. S., James, T. Y., Spatafora, J. W., et al. (2009). The fungi. Curr. Biol. 19, R840–R845. doi: 10.1016/j.cub.2009.07.004
Stajich, J. E., Wilke, S. K., Ahrén, D., Au, C. H., Birren, B. W., Borodovsky, M., et al. (2010). Insights into evolution of multicellular fungi from the assembled chromosomes of the mushroom Coprinopsis cinerea (Coprinus cinereus). Proc. Natl. Acad. Sci. 107, 11889–11894. doi: 10.1073/pnas.1003391107
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stengel, A., Stanke, K. M., Quattrone, A. C., Herr, J. R. (2022). Improving taxonomic delimitation of fungal species in the age of genomics and Phenomics. Front. Microbiol. 13, 847067. doi: 10.3389/fmicb.2022.847067
Struck, T. H., Cerca De Oliveira, J. (2019). “Cryptic species and their evolutionary significance,” in Encyclopedia of Life Sciences, 1–9.
Sundberg, P., Vodoti, E. T., Zhou, H., Strand, M. (2009). Polymorphism hides cryptic species in Oerstedia dorsalis (Nemertea, Hoplonemertea). Biol. J. Linn. Soc. 98, 556–567. doi: 10.1111/j.1095-8312.2009.01310.x
Taylor, J. W., Jacobson, D. J., Kroken, S., Kasuga, T., Geiser, D. M., Hibbett, D. S., et al. (2000). Phylogenetic species recognition and species concepts in fungi. Fungal Genet. Biol. 31, 21–32. doi: 10.1006/fgbi.2000.1228
Tennakoon, D. S., Jeewon, R., Thambugala, K. M., Gentekaki, E., Wanasinghe, D. N., Promputtha, I., et al. (2021). Biphasic taxonomic approaches for generic relatedness and phylogenetic relationships of Teichosporaceae. Fungal Diversity 110, 199–241. doi: 10.1007/s13225-021-00492-8
Thambugala, K. M., Hyde, K. D., Eungwanichayapant, P. D., Romero, A. I., Liu, Z.-Y. (2016). Additions to the genus rhytidhysteron in hysteriaceae. Cryptogamie Mycologie 37, 99–116. doi: 10.7872/crym/v37.iss1.2016.99
Thambugala, K. M., Hyde, K. D., Tanaka, K., Tian, Q., Wanasinghe, D. N., Ariyawansa, H. A., et al. (2015). Towards a natural classification and backbone tree for Lophiostomataceae, Floricolaceae, and Amorosiaceae fam. nov. Fungal Diversity 74, 199–266. doi: 10.1007/s13225-015-0348-3
Thompson, C. C., Vidal, L., Salazar, V., Swings, J., Thompson, F. L. (2021). “Microbial genomic taxonomy,” in Trends in the systematics of bacteria and fungi, 168–178.
Tully, T., Potapov, M. (2015). Intraspecific Phenotypic Variation and Morphological Divergence of Strains of Folsomia candida (Willem)(Collembola: Isotomidae), the” Standard” Test Springtaill. PloS One 10, e0136047. doi: 10.1371/journal.pone.0136047
Villesen, P. (2007). FaBox: an online toolbox for fasta sequences. Mol. Ecol. Notes 7, 965–968. doi: 10.1111/j.1471-8286.2007.01821.x
Vodă, R., Dapporto, L., Dincă, V., Vila, R. (2015). Cryptic matters: overlooked species generate most butterfly beta-diversity. Ecography 38, 405–409. doi: 10.1111/ecog.00762
von Eiff, C., McNamara, P., Becker, K., Bates, D., Lei, X.-H., Ziman, M., et al. (2006). Phenotype microarray profiling of Staphylococcus aureus menD and hemB mutants with the small-colony-variant phenotype. J. Bacteriology 188, 687–693. doi: 10.1128/JB.188.2.687-693.2006
Wang, R., Tsui, C. K., You, C. (2022). Cryptic species diversity and phylogenetic relationship in the rust genus Chrysomyxa from China. J. Fungi 8, 83. doi: 10.3390/jof8010083
Wang, G., Yang, E., Brinkmeyer-Langford, C. L., Cai, J. J. (2014). Additive, epistatic, and environmental effects through the lens of expression variability QTL in a twin cohort. Genetics 196, 413–425. doi: 10.1534/genetics.113.157503
Wannathes, N., Desjardin, D., Lumyong, S. (2009). Four new species of Marasmius section Globulares from northern Thailand. Fungal Diversity 36, 155–163.
Wei, Z., Xia, Z., Shu, J., Shang, H., Maxwell, S. J., Chen, L., et al. (2021). Phylogeny and taxonomy on cryptic species of forked ferns of asia. Front. Plant Sci., 2786. doi: 10.3389/fpls.2021.748562
West-Eberhard, M. (2008). “Phenotypic Plasticity,” in Encyclopedia of Ecology. Eds. Jørgensen, S. E., Fath, B. D..
Wijayawardene, N. N., Hyde, K. D., Al-Ani, L. K. T., Tedersoo, L., Haelewaters, D., Rajeshkumar, K. C., et al. (2020). Outline of Fungi and fungus-like taxa. Mycosphere Online: J. Fungal Biol. 11, 1060–1456. doi: 10.5943/mycosphere/11/1/8
Wu, B., Hussain, M., Zhang, W., Stadler, M., Liu, X., Xiang, M. (2019). Current insights into fungal species diversity and perspective on naming the environmental DNA sequences of fungi. Mycology 10, 127–140. doi: 10.1080/21501203.2019.1614106
Yurkov, A., Inácio, J., Chernov, I. Y., Fonseca, A. (2015). Yeast biogeography and the effects of species recognition approaches: the case study of widespread basidiomycetous species from birch forests in Russia. Curr. Microbiol. 70, 587–601. doi: 10.1007/s00284-014-0755-9
Zeng, X.-Y., Wu, H.-X., Hongsanan, S., Jeewon, R., Wen, T.-C., Maharachchikumbura, S. S., et al. (2019). Taxonomy and the evolutionary history of Micropeltidaceae. Fungal Diversity 97, 393–436. doi: 10.1007/s13225-019-00431-8
Keywords: biodiversity, ecology, evolution, phylogeny, taxonomy
Citation: Ekanayaka AH, Karunarathna SC, Tibpromma S, Dutta AK, Tennakoon DS, Karunarathna A, Chukeatirote E, Dai D-Q, Stephenson SL, Maharachchikumbura SSN, Liu C and Phillips AJL (2025) Species evolution: cryptic species and phenotypic noise with a particular focus on fungal systematics. Front. Cell. Infect. Microbiol. 15:1497085. doi: 10.3389/fcimb.2025.1497085
Received: 16 September 2024; Accepted: 07 January 2025;
Published: 04 February 2025.
Edited by:
Sinang Hongsanan, Shenzhen University, ChinaReviewed by:
Rajesh Jeewon, University of Mauritius, MauritiusCopyright © 2025 Ekanayaka, Karunarathna, Tibpromma, Dutta, Tennakoon, Karunarathna, Chukeatirote, Dai, Stephenson, Maharachchikumbura, Liu and Phillips. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alan J. L. Phillips, YWxhbi5qbC5waGlsbGlwc0BnbWFpbC5jb20=; Chao Liu, TGl1Y2hhb184MEAxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.