
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 27 November 2024
Sec. Molecular Bacterial Pathogenesis
Volume 14 - 2024 | https://doi.org/10.3389/fcimb.2024.1499476
 Kassapa Ellepola1†
Kassapa Ellepola1† Lauren C. Guillot1
Lauren C. Guillot1 Bradley Comeaux1
Bradley Comeaux1 Yiran Han2
Yiran Han2 Jessica K. Kajfasz3
Jessica K. Kajfasz3 Jacob P. Bitoun4
Jacob P. Bitoun4 Grace Spatafora2
Grace Spatafora2 Jose A. Lemos3
Jose A. Lemos3 Zezhang T. Wen1,5*
Zezhang T. Wen1,5*Introduction: The sufCDSUB gene cluster, encoding the sole iron-sulfur (Fe-S) cluster assembly system in S. mutans, was recently shown to be up-regulated in response to oxidative stressors and Fe limitation.
Methods: In this study, luciferase reporter fusion assays, electrophoretic gel mobility shift assays (EMSA) and in vitro transcription assays (IVT) were used to dissect the cis- and trans-acting factors that regulate the expression of sufCDSUB.
Results and discussion: Results showed deletion of perR, for the only Fur-family transcriptional regulator in S. mutans, resulted in >5-fold increases in luciferase activity under the control of the sufCDSUB promoter (P<0.01), as compared to the parent strain, UA159 when the reporter strains were grown in medium with no supplemental iron. Site-directed mutagenesis of a PerR-box in the promoter region led to elevation of the reporter activity by >1.6-fold (P<0.01). In an EMSA, recombinant PerR (rPerR) was shown to bind to the cognate sufCDSUB promoter leading to mobility retardation. On the other hand, the reporter activity was increased by >84-fold (P<0.001) in response to the addition of cysteine at 4 mM to the culture medium. Deletion of cysR, for a LysR-type of transcriptional regulator, led to reduction of the reporter activity by >11.6-fold (P<0.001). Addition of recombinant CysR (rCysR) to an EMSA caused mobility shift of the sufCDSUB promoter probe, indicative of rCysR-promoter interaction, and rCysR was shown to enhance sufC transcription under the direction of sufCDSUB promoter in vitro. These results suggest that multiple factors are involved in the regulation of sufCDSUB expression in response to environmental cues, including cysteine and Fe availability, consistent with the important role of sufCDSUB in S. mutans physiology.
Streptococcus mutans, a keystone microorganism in the development of human dental caries, is well known for its strong ability to colonize and accumulate on the tooth surface, especially in the presence of dietary sucrose, and utilize different sugars and produce lactic acid and other weak acids that leads to enamel demineralization and development of white spots and carious lesions (Lemos et al., 2019). The bacterium possesses at least three glycosyltransferase enzymes, GtfBC&D, which use sucrose, starch and other sugars as the substrate to produce extracellular polysaccharides (better known as glucans), including the water-insoluble alpha 3,2-linked polymers, playing a central role in the cariogenicity of the bacterium (Bowen and Koo, 2011). Multiple adhesins, including WapA, GtfB and the multi-functional adhesin SpaP (aka P1, AgI/II) interact with glycoprotein GP340 in the salivary pellicles to enhance the bacterial adhesion on the tooth surface, especially in the absence of sucrose (Lemos et al., 2019). S. mutans is also instrumental in the recruitment and establishment of other cariogenic bacteria such as Candida albicans and Lactobacillus spp. in the plaque microbiota (Gregoire et al., 2011; Wen et al., 2017), and under certain conditions, causing a shift in the highly dense and diverse plaque microbiota from homeostasis to dysbiosis, leading to the development of carious lesions.
Environmental conditions, such as the pH, temperature, oxygen tension and redox potential in the oral cavity are known to constantly fluctuate and often can be harsh and detrimental to the microbes in the environment. In addition, the oral cavity is often exposed to various antimicrobial agents, such as hydrogen peroxide, fluoride, and sodium dodecyl sulfate (SDS), from oral care products and as a result of localized and systemic drug therapy and/or preventative applications. S. mutans is also known for its ability to survive and adapt under different environmental conditions, such as low pH and oxidative stressors like hydrogen peroxide. Multiple pathways are utilized by the bacterium to cope with the environmental stressors, which include the chaperones DnaK and GroEL, two-component signal transduction systems CiaHR, VicRK, and LiaSR, the caseinolytic protease complex ClpCP, and others like surface associated protein BrpA (Lemos et al., 2019).
H2O2, superoxide anion (•O2-) and hydroxyl radicals (•OH) are among the reactive oxygen species (ROS) (Imlay, 2002; Miller and Britigan, 1997), which can cause serious damages to cellular macromolecules including proteins and DNA. Although H2O2 is relatively weak in toxicity compared with that of other ROS, in the presence of iron (Fe), it can be nonenzymatically converted into highly toxic hydroxyl radicals via Fenton reactions (H2O2 + Fe+2 → •OH + -OH + Fe+3) (Enami et al., 2014). Bacterial species comprise multiple enzyme systems for the protection against ROS, such as catalase, superoxide dismutase (SOD), Dps-like protein (Dpr), alkyl hydroperoxide reductase (AhpCF), glutathione reductase, and thiol reductase (Fujishima et al., 2013). Although S. mutans lacks catalase, it contains AhpCF and Dpr that are primarily responsible for resistance to H2O2. AhpCF directly decompose H2O2, and Dpr inhibits the reaction of the Fenton pathway by capturing free Fe while SOD converts •O2- into H2O2 (Yamamoto et al., 2000; Fujishima et al., 2013).
The Fe-S clusters are known to be involved in electron transfer, sulfur mobilization, regulation and protein stabilization, catalysis and biochemical functions that are important for the biological activity of organisms including but not limited to their role as cofactors, substrate binding/activation and for iron storage (Lill, 2020, 2009). Due to the redox-active nature, Fe-S clusters are sensitive to cellular redox conditions, thus act as “molecular switches” for both transcriptional and translational regulation of gene expression (Sevilla et al., 2021). Many catalytic enzymes, including aconitase, 6-phosphogluconate dehydratase, fumarase A, NADH dehydrogenase, succinate dehydrogenase, and fumarate nitrate reductase (FNR), regulatory proteins, such as iron-sulfur cluster regulator (IscR) of Escherichia coli and SufR of Cyanobacterium spp (Vuorijoki et al., 2017), also contain Fe-S clusters directly regulating bacterial metabolism and gene expression (Fontecave et al., 2008).
Multiple Fe-S cluster biosynthesis and repair systems have been identified and described in different bacteria (Lill, 2009), including the SUF (sulfur mobilization) pathway that is encoded by the sufCDSUB operon, where sufS encodes a cysteine desulfurase. With cysteine serving as the sulfur source, cysteine desulfurase initiates the formation of Fe-S clusters by extracting the sulfur atom from cysteine generating a covalent persulfide intermediate and ferrying it to the Fe-S clusters along with alanine as a side product (Black and Dos Santos, 2015). This persulfide can then be conveyed directly to the synthesis machinery scaffold or via an intermediary carrier molecule (such as SufU or SufE) (Layer et al., 2007; Selbach et al., 2014). Unlike many other bacteria, S. mutans appears to possess only one Fe-S cluster machinery in SufCDSUB (Ellepola et al., 2021). Our recent studies by allelic exchange mutagenesis showed that SUF deficiency in S. mutans causes major defects in various cellular processes, but unlike any other bacteria that have been studied, the mutant lacking SUF is viable. Relative to the wild-type, UA159, mutants lacking sufCDSUB displayed major growth defects in an environment with low pH, oxidative and nitrosative stressors, and in the absence of certain amino acids such as leucine, isoleucine, and glutamate/glutamine whose biosynthesis require Fe-S clusters. Relative to the wild-type, mutants lacking sufCDSUB also had a significantly reduced survival rate following incubation at a low pH environment or under hydrogen peroxide challenge. Besides, the sufCDSUB mutant also tended to form aggregates and accumulated significantly less biofilms, especially during growth in medium with glucose. When analyzed using luciferase as a reporter, the expression of sufCDSUB was shown to be up-regulated in response to oxidative stresses. More than 5-fold increases were reported when the reporter strain was transferred from regular medium to medium with iron-limitation as well as medium with inclusion of an iron chelating agent.
Metalloregulatory proteins are essential for bacterial survival and pathogenesis due to their essential role in intracellular metal ion homeostasis. Among them is the ferric uptake repressor (Fur), a transcription factor which utilizes Fe2+ as a corepressor, thus involved in iron acquisition and storage, and regulates the production of some pathogenicity factors, the acid shock response, chemotaxis, and oxidative stress defenses in bacteria (Troxell and Hassan, 2013; Touati, 2000; Hantke, 2001). In Bacillus subtilis and certain other bacteria, Fur has been identified as a multifunctional protein that can also function as an iron-dependent activator (Pinochet-Barros and Helmann, 2020). The PerR (a Fur homologue) protein found in streptococci and several other bacteria with its peroxide-sensing ability is known to be associated with resistance to oxidative stresses (Brenot et al., 2005; Mongkolsuk and Helmann, 2002; Zhang et al., 2012). PerR in S. mutans, the sole Fur family protein encoded by SMU.593, regulates oxidative stress tolerance response when bound to a metal cofactor that, depending on the bacterial species, can be iron, manganese, or both (Ruxin et al., 2021; Lee and Helmann, 2006, 2007). SloR is also a metal-dependent transcription repressor, which regulates the expression of metal transporter SloABC (Spatafora et al., 2015). A recent study by Ruxin et al. has shown that SloR is regulated by repressor PerR, and together with PerR, plays essential roles in ion homeostasis and fitness of S. mutans (Ruxin et al., 2021).
Our recent studies have shown that sufCDSUB in S. mutans is regulated in response to environmental cues and regions in the sufCDSUB promoter as potential binding sites for PerR can be identified. This study is designed to further investigate how the expression of sufCDSUB is regulated by PerR and other regulatory factors. Results have shown that deletion of perR resulted in an increase of reporter activity by >5-fold, especially when grown under Fe-limitation, compared to the wild-type, but displayed no effect when growing in the presence of oxidative stressors. Unlike perR, deletion of cysR (SMU.852), encoding a LysR-type transcription regulator, led to >11-fold reduction of luciferase reporter activity. PCR-based deletions and point mutations, along with electrophoretic gel mobility shift assays (EMSA) and in vitro transcription (IVT) assays were used to further investigate how PerR and CysR regulate sufCDSUB expression. In addition, investigative proteomics was also used to elucidate the scope of the PerR-mediated regulation in S. mutans.
Bacterial strains and plasmids used in this study are listed in Table 1. S. mutans strains were maintained in brain heart infusion (BHI) medium. For biofilm formation, S. mutans was grown in modified biofilm medium (BM) with glucose (20 mM, BMG), sucrose (20 mM, BMS), or glucose (18 mM) plus sucrose (2 mM) as supplemental carbon and energy sources (BMGS) (Bitoun et al., 2012c). All solid media were prepared similarly with inclusion of Bacto agar (Difco Laboratories, Franklin Lakes, NJ) at the level of 1.5% (w/v). When needed, erythromycin (Erm, 10 µg/ml), kanamycin (Kan, 1 mg/ml), and/or spectinomycin (Spc, 1 mg/ml) were added. Unless otherwise stated, cells were grown at 37°C in an aerobic environment with 5% CO2. All E. coli strains were grown in Luria Bertani medium at 37°C aerobically, with or without inclusion of kanamycin (40 µg/ml), ampicillin (100 µg/ml), spectinomycin (100 µg/ml), and/or erythromycin (300 µg/ml).
Standard recombinant DNA procedures were used (Lau et al., 2002; Bitoun et al., 2013). All restriction and modifying enzymes were purchased from Invitrogen (Carlsbad, CA) or New England Biolabs (Ipswich, MA) and used as recommended by the suppliers. All primers (Supplementary Table 1) were synthesized by Integrated DNA Technologies, Inc. (Iowa City, IA). A perR deletional mutant was constructed using our well-established PCR-Ligation-Mutation protocol (Burne et al., 2011). Briefly, the 5’ and 3’ region of perR were amplified using high fidelity DNA polymerase Q5 (NEB Bioloabs) using gene-specific primers (Supplementary Table 1), and following proper restriction enzyme digestions, the flanking fragments were ligated with a non-polar spectinomycin resistant marker. Then, the ligation mix was used to transform S. mutans with inclusion of the competence stimulating peptide, and mutants lacking perR was isolated from plates with spectinomycin. Isolated mutants were verified by PCR and Sanger sequencing with gene-specific primers (Supplementary Table 1). S. mutans strains with deletion of dpr, sloR, and cysR were generated using similar strategies.
The transcription start sites were determined using the Template Switching RT Enzyme Mix (NEB #M0466) by following the 5’ RACE protocol (NEBLABS, Ipswich, MA). For total RNA, wild-type S. mutans UA159 cells were harvested at middle exponential phase (OD600 ≈ 0.3) and immediately treated with RNAProtect® (Qiagen, Inc). Total RNAs were extracted using the hot phenol method and purified using RNAeasy kit (Qiagen, Inc.), and DNase I treatment including an in-Column DNase I treatment was used to clean up the residual genomic DNA. 1 µg total RNA was used in the template switching reverse transcription, and the resulting cDNA with inclusion of a universal sequence attached to its 3’ were amplified by PCR using high fidelity DNA polymerase Q5, and the transcription initiation site was determined by Sanger sequencing.
To analyze the regulation of suf expression, a promoterless luciferase gene (luc) was used as a reporter as previously described (Kreth et al., 2005). Briefly, the cognate suf promoter region and its derivatives generated by PCR-based deletions and/or site-directed mutations were amplified using high fidelity DNA polymerase Q5 (NEB Biolabs), and following proper restriction digestions, cloned directly in front of the promoterless luc gene in integration vector pFW11-luc (Kreth et al., 2005), which also contains a Shine-Dalgarno sequence optimized for group A streptococci (Podbielski et al., 1999). Following confirmation of the cloned element by sequencing, the resulting constructs were introduced into S. mutans UA159 and its selected derivatives such as the perR deletion mutant, ΔperR, and the expression of suf were analyzed using luciferase assay by following the protocol of Podbielski (Kreth et al., 2005; Bitoun et al., 2012a, 2012). For controls, pFW11 with promoterless reporter and a luciferase reporter fused with the gyrase promoter (Pgyr) were used (Ellepola et al., 2021).
For expression and purification of recombinant PerR (rPerR), recombinant CysR (SMU.852), and recombinant Sigma factor (rSigA), the respective coding regions were amplified by PCR with gene-specific primers (Supplementary Table 1) using high fidelity DNA polymerase Q5 (NEB Biolabs), and cloned in pQE30 (Qiagen, Inc.). Following Sanger sequencing to confirm accuracy of the cloned DNA, the resulting constructs were transformed into E. coli M15, and PerR, CysR and SigA expression were induced by addition of IPTG at 0.5 mM (final conc.), and the His-tagged rPerR, rCysR and rSigA were purified using nickel resins (Fisher Scientific) under native conditions similarly as described previously (Bitoun et al., 2012c). The purified proteins were then applied to Amicon Ultra 3 Centrifugal Filters (3 kDa MWCO, Millipore) for desalting and re-constituted in 20 mM Tris, pH 7.6 plus 20% glycerol. For PerR, 10 mM EDTA was also included in the elution buffer, desalting and exchange buffers (Lee and Helmann, 2006).
For in vitro transcription assays, crude native S. mutans RNA Polymerase (RNAP) was obtained based upon the methods described by Seepersaud et al. (Seepersaud et al., 2006) and Galvão et al. (Galvao et al., 2015). Briefly, S. mutans cells of mid-exponential phase were harvested by centrifugation, resuspended and incubated in protoplast preparation buffer at 37°C for 90 minutes. Protoplasts were then pelleted and resuspended in lysis buffer, lysed by glass bead beating and, the supernatants were applied to Affi-Gel heparin resin (Bio-Rad) and eluted with a NaCl gradient of 0.1 to 1 M. Fractions containing visible subunits were pooled and applied to Macro-Prep High-Q ion exchange resin (Bio-Rad) and eluted with a gradient of 0.1–0.8 M NaCl. The desired fractions were again pooled and dialyzed.
To assess if and how transcriptional factor PerR interacts with the sufCDSUB promoter, EMSA was employed similarly as described previously (Bitoun et al., 2012a). Briefly, the intact promoter region and its derivatives with deletions and/or site-directed mutations were amplified by PCR using Q5 DNA polymerase with the primers listed in Supplementary Table 1. The PCR products, with biotinylation at the N-terminus or without (for cold-competition control), were incubated with increasing amounts of rPerR in the reaction buffer at room temperature (Bitoun et al., 2012a). The reaction mixtures were separated on a 5% polyacrylamide gel under native conditions, and DNA mobility shift was visualized via LightShift Chemiluminescent EMSA Kit by following procedures recommended by the manufacturer (Pierce, Rockford, IL) or by SYBR Gold staining (Invitrogen, CA) and a GelDoc Go Imaging system (BioRad).
In vitro transcription assay was performed by following the protocols of Dou et al (Dou et al., 2021) and Galvao et al (Galvao et al., 2015) using native S. mutans RNAP and rSigA. In some experiments, E. coli RNA polymerase holoenzyme (M0251S, NEB Biolabs) (2 units) was also used. Briefly, a DNA fragment carrying the sufCDSUB promoter plus 834 bp of the coding sequence was amplified by Q5 DNA polymerase and purified using Qiagen PCR purification kit (Qiagen, Inc.). For IVT, ~100 fM of the DNA template, 25 nM of S. mutans RNAP, and 25 nM of rSigA were incubated with and without rPerR or rCysR at different µM concentrations in buffer containing 20 mM Tris-HCl (pH 7.5), 50 mM KCl, 5 mM MgCl2, 0.1 mM DTT, 0.01% Triton X-100, 5 mM of NTP mix at 37°C for 1 hour. The resulting transcripts were analyzed using 1.4% denaturing agarose gel and visualized by SYBR Gold staining (InVitrogen, CA) similarly as described above.
Proteomics analysis was carried out similarly as described previously (Wen et al., 2006; Yue and Guidry, 2019). Briefly, cultures were grown in regular BHI until mid-exponential phase (OD600nm~0.4) and homologized in Tris-HCl buffer, 50 mM pH 7.4 containing 10% glycerol using a glass bead beater as described (Wen et al., 2006). Following protein concentration estimation by BCA assay (Pierce, ThermoFisher), 100 µg of each protein sample was digested with trypsin, and labeled using a Tandem Mass Tag (TMT) 10-plex Reagent set (Pierce, ThermoFisher). HPLC-MS/MS analysis was done in four technical replicates with a false discovery rate (FDR) of < 1% at the LSUHSC Proteomics Core by following established protocols.
At least three biological and two technical replicates each were used for all the experiments. Quantitative data were analyzed using paired Student t test. A P value of 0.05 or less is considered statistically significant.
First, rapid amplification of cDNA ends (5’RACE) was employed to determine the transcription initiation site of sufCDSUB, and it was done using total RNA isolated from mid-exponential phase cultures grown in regular BHI broth. The results showed that under these conditions, the transcription initiation site of the sufCDSUB operon was consistently mapped at nucleotide 22 from the translation initiation site ATG (Figure 1A). The promoter region was then subjected to virtual footprinting analysis (https://www.prodoric.de/vfp/), and multiple regions were identified with potential roles in sufCDSUB regulation, which include two regions, termed Fur-box1 and Fur-box2 with high similarity to the binding site of Fur (consensus: GGCAATAGTGATTACCGTC) and two regions with similarity to the PerR-binding site (consensus: TTATAATnATTATAA) (Figure 1B). To further examine the role of these respective regions in sufCDSUB expression, PCR-based mutations and deletions of the sufCDSUB promoter were made using primers listed in Supplementary Table 1, and the resulting constructs were directly cloned in front of the promoterless luciferase reporter in pFW11 (Kreth et al., 2005; Bitoun et al., 2012a). As shown in Figure 1C, mutations of the PerR-box1 from TTGGACTTTTTTTGTT to aaGatCaTTTTTTGTT (low case indicated mutated nucleotides) in PerRm resulted in elevation of the reporter activity by 1.65-fold (P<0.05), compared to the intact promoter-reporter fusion Psuf, indicative of a role as a potential binding site of a transcriptional repressor. Site-directed mutation of Fur-box 2 (AGAAGAAAAAGTAATTTTT, Box2m), where nt GAAAAAGT were changed to ctttttgt, showed no changes in the luciferase activity compared to the intact promoter-reporter fusion. Interestingly, deletion of AGAAAAAG in Fur-box 2 (dBox2, shown in red-box) led to >4.23-fold reduction of the reporter activity, while no significant effect was observed with the deletion of Fur-box1 (dBox1) (Figure 1C). The results suggest that Fur-box2 is likely a binding site for a positive regulator.
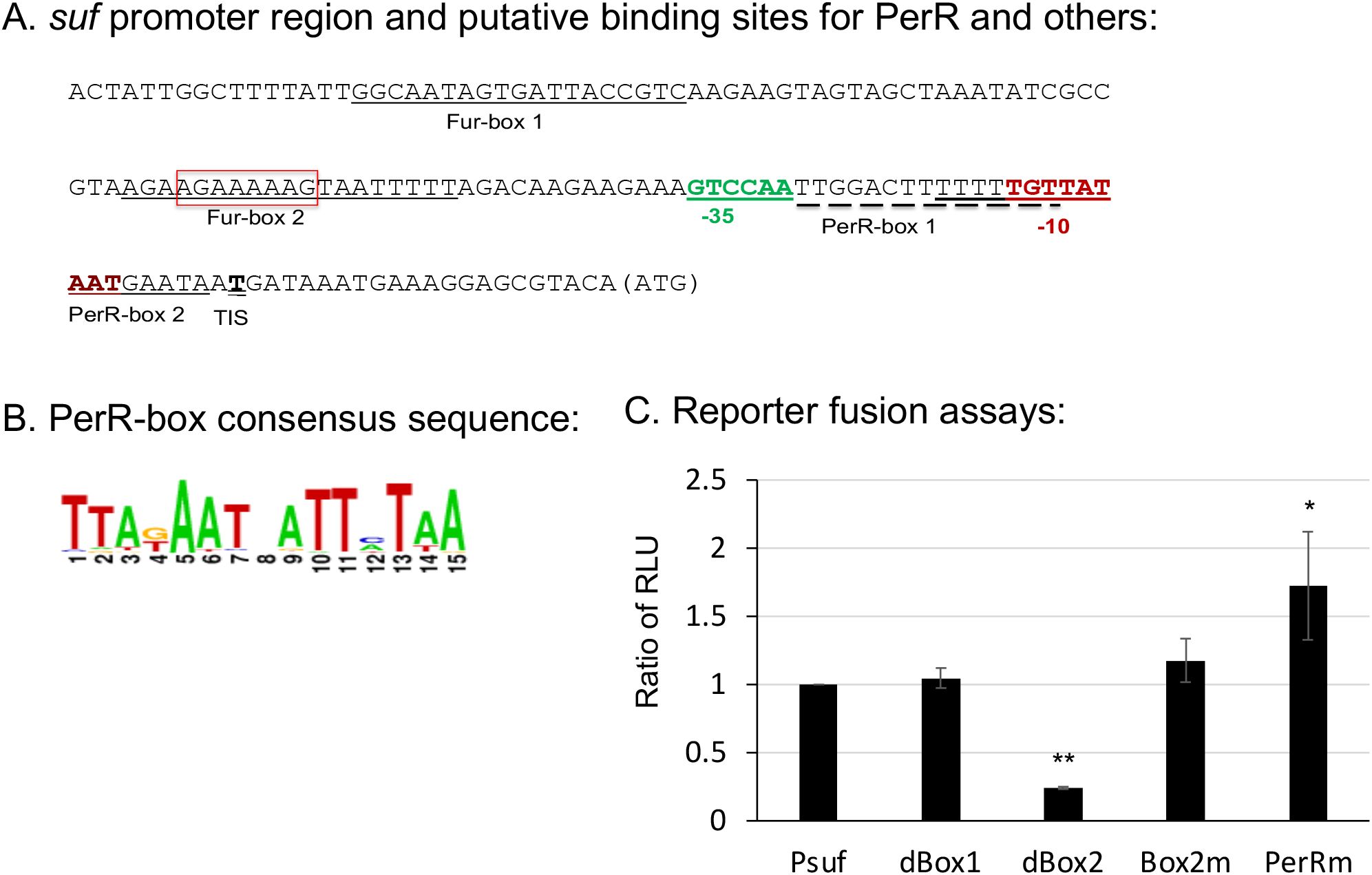
Figure 1. The sufCDSUB promoter region and the roles of different major elements in sufCDSUB expression. (A) Schematic diagram of the sufCDSUB promoter region with extended -10 region and -35 region in red and green, respectively, and the red box indicating the sequences deleted in Fur-box 2 (dBox2); (B) Consensus of PerR-boxes; (C) The effects of deletion of the conserved Fur-box 1 (dBox1), Fur-box 2 (dBox2) and site-directed mutations in Fur-box 2 (Box2m) and PerR-box 1 (PerRm) on luciferase reporter activity, as compared to the wild-type, Psuf (C). Data are expressed as ratios of the relative light units (RLU) of the respective mutant over the wild-type. *, ** P<0.05 and 0.01 vs Psuf via Student’s t test.
S. mutans possesses a PerR (SMU.593) (Kajfasz et al., 2021), the sole Fur family of metal ion-dependent transcriptional regulator that modulates the expression of target genes by sensing and responding to oxidative stress (Bsat et al., 1998). To assess the role of PerR in sufCDSUB regulation, an allelic exchange mutant of perR was constructed with a major portion of the coding region replaced by a non-polar kanamycin resistance marker (Burne et al., 2011). As expected, deletion of perR led to de-repression of sufCDSUB expression, especially when grown in Fe-rich biofilm medium. Relative to the wild-type, the perR deletion mutant displayed an increase of >2-fold in luciferase reporter activity when the cultures were grown in regular BHI (P<0.05) (Figure 2). Such differences were increased to as much as 10-fold compared to the wild-type, when the reporter strains were transferred from BHI to semi-defined biofilm medium. Consistent with our recent studies (Ellepola et al., 2021), luciferase reporter activity of the wild-type UA159 was increased by >5.3(±1.1)-fold when the BHI-grown overnight cultures were transferred to semi-defined biofilm medium with glucose (BMG), especially when FeCl3 was omitted from the medium recipe (BMG-Fe) (Figure 2). The highest reporter activity of the wild-type was measured when the cultures were grown in BMG medium with inclusion of 2,2’-pyridyl (ACROS Organics, #366-18-7), an iron chelating agent at 50 µM. However, no such effects were measured when overnight cultures were also grown in BMG or BMG-Fe. Like the wild-type, the luciferase reporter activity of the perR mutant was also increased when it was transferred from regular BHI to BMG. Compared to the wild-type, the reporter activity of the perR mutant was increased by >10-fold of the wild-type (P<0.001). However, unlike the wild-type, no significant differences in luciferase reporter activities were measured between the perR mutant when grown in BMG and those in BMG-Fe and BMG plus 2,2’-pyridyl at 50 µM (Figure 2). These results further suggest PerR plays a major role as a repressor in sufCDSUB expression in response to Fe-availability in the culture medium, and consistent with other Fur regulators, it requires Fe as a co-factor.
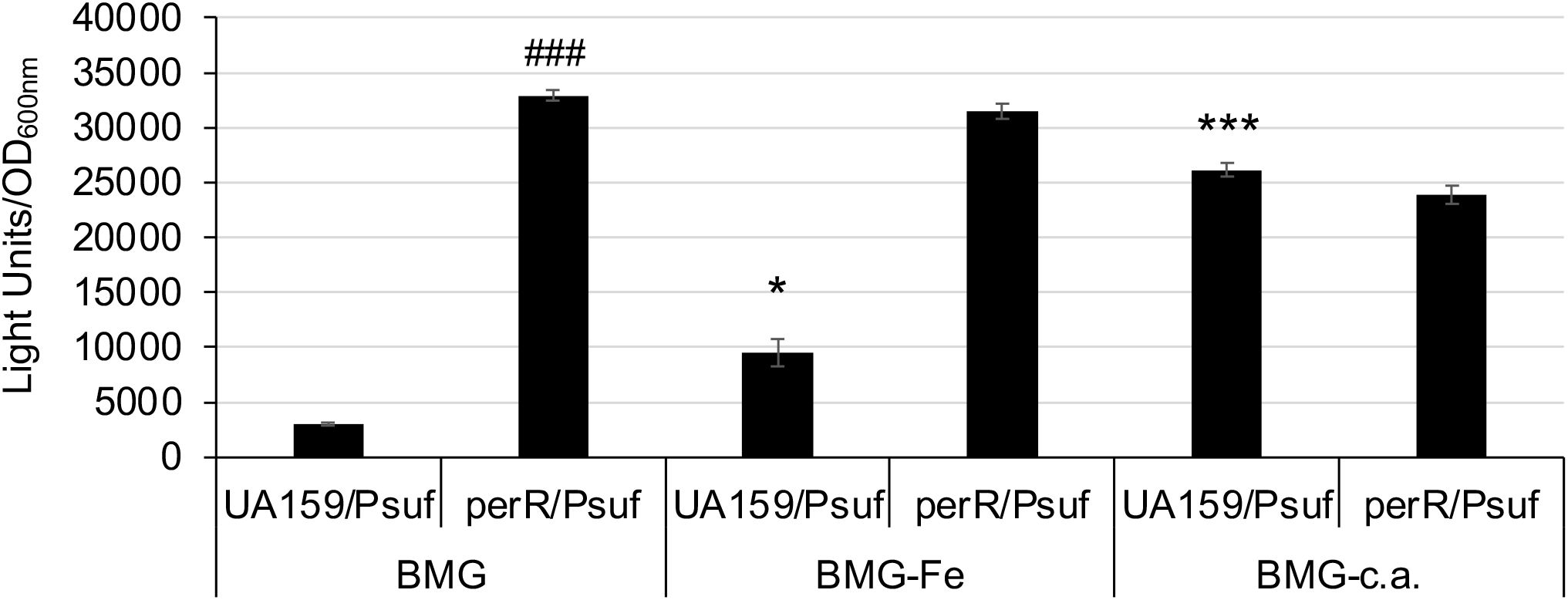
Figure 2. Luciferase reporter analysis in response to Fe-limitation. S. mutans UA159 and its perR mutant were transferred from regular BHI broth medium to semi-defined biofilm medium plus glucose with (BMG) and without inclusion of FeCl2 (BMG-Fe) and with addition of 2,2’-pyridyl, a chelating agent (BMG-c.a.) at 50 µM. *, ***, P<0.05 and 0.001 vs UA159/Psuf in BMG, respectively; ###, P<0.001 vs UA159/Psuf, via Student t-test.
To further examine if PerR interacts with the sufCDSUB promoter influencing its transcription, PerR was expressed in E. coli by isopropyl β-d-1-thiogalactopyranoside (IPTG) induction and purified using Ni2+ affinity columns, and EMSA assays were conducted using the purified rPerR. Results showed that inclusion of rPerR in the reaction mix resulted in mobility shift of the sufCDSUB promoter probe, and such activity was concentration dependent (Figure 3). The mobility shift was negated or reduced significantly when rPerR protein was first incubated with non-biotinylated promoter oligo. Reduced mobility shift was also observed when whole cell lysates of the wild-type S. mutans UA159 were used in the reaction. Interestingly, no major differences were observed in mobility shift when rPerR was mixed with oligo PerRm, which has the PerR-box1 mutated from TTGGACTTTTTTTGTT to aaGatCaTTTTTTGTT using PCR-based site-directed mutagenesis (see Figure 1). These results further suggest that PerR interacts with the sufCDSUB promoter, although the exact nucleotides that interact with PerR repressor remain unclear. In an IVT assay using native S. mutans RNA polymerase (RNAP) and recombinant Sigma factor 70 (rRpoD) with the whole promoter plus 834 bp of the coding sequence of sufC, a transcript of expected size was detected. However, inclusion of rPerR in the reaction mix only slightly reduced the amount of the transcripts, but no such effects were observed when recombinant LuxS protein or none was added as controls.
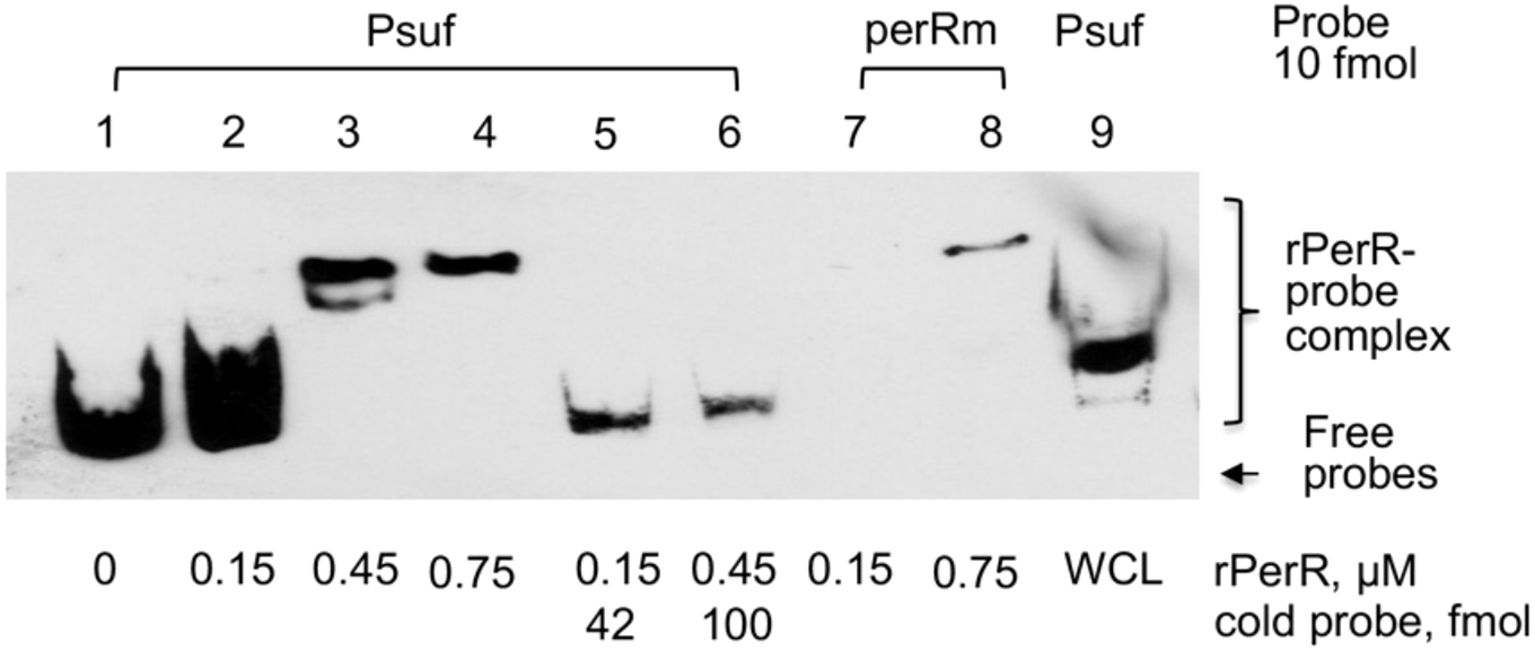
Figure 3. EMSA assays of PerR-sufCDSUB promoter interactions. Inclusion of rPerR resulted in mobility retardation of biotinylated suf promoter DNA probes (Psuf, ln 1-4), and such interaction was concentration-dependent as indicated. Addition of cold-probe led to release of biotinylated probes from the rPerR-Psuf complex (Psuf, ln 5&6). Similar effects were also observed when whole cell lysate of S. mutans UA159 (WCL) was used. However, mutations in the PerR-box 1 (perRm) showed no effect under the conditions studied.
A proteomics approach was also used to examine the protein profile of the perR mutant, and the results showed that when compared to the wild-type UA159, the expression of more than 35 proteins were altered as a result of perR deletion, including 17 up-regulated proteins and 18 down regulated (Supplementary Table 2). Among the up-regulated proteins were those with roles in oxidative stress tolerance responses including Dpr, thiol peroxidase Tpx, and putative metal transporter SMU.635, consistent with the recent RNA-seq studies by Kajfasz et al (Kajfasz et al., 2021). Interestingly, several enzymes of sugar transport and metabolism pathways were also found to be altered significantly, including down-expression of β-exo-fructanase FruA, a well-known virulence attributor, and up-expression of sucrose- and mannose-specific transporters (Supplementary Table 2). When whole cell lysates were separated by SDS-PAGE and probed in a Western blotting using polyclonal antibodies (a kind gift of Dr. RA Burne), the results further confirmed the reduction of FruA expression in the perR mutant as well as the sufCDSUB mutant (Figure 4), compared to the parent strain, UA159. However, it awaits further investigation if the expression of these genes is directly regulated by PerR.
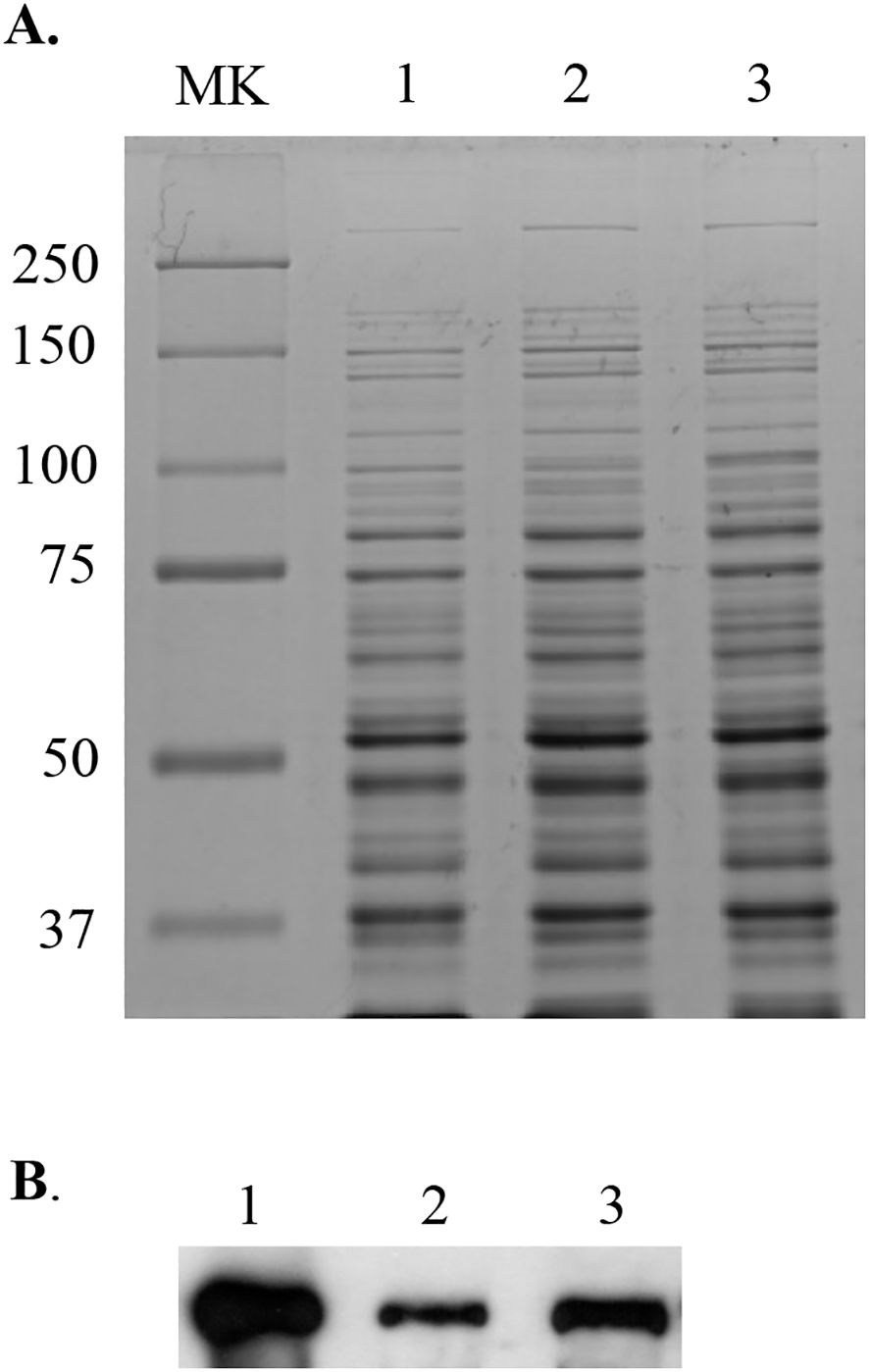
Figure 4. Western blot analysis of the perR mutant. Whole cell lysates (10 µg of total proteins) of S. mutans wild-type UA159 (lane 1) and its perR (lane 2) and sufCDSUB (lane 3) mutants were separated using 10% SDS-PAGE (A), blotted to a PVDF membrane, and then probed with polyclonal antibodies against FruA (B). MK, molecular weight marker in kilodalton.
Our previous study showed that expression of the sufCDSUB operon is regulated in response to oxidative stressors (Ellepola et al., 2021). Relative to cultures grown in regular BHI, the luciferase reporter activity was increased by >2-fold in cultures growing in the presence of methyl viologen (at 2.5 mM) (Figure 5), a molecule that induces superoxide production inside the cells. Like the wild-type, luciferase reporter activity of the perR mutant was also increased following exposure to methyl viologen at 2.5 mM, but unlike growing in regular BHI, the differences between the wild-type and the perR mutant under the condition with 2.5 mM methyl viologen were not statistically significant (Figure 5). These results suggest that factors other than PerR are likely involved in the oxidative stress mediated regulation of sufCDSUB.
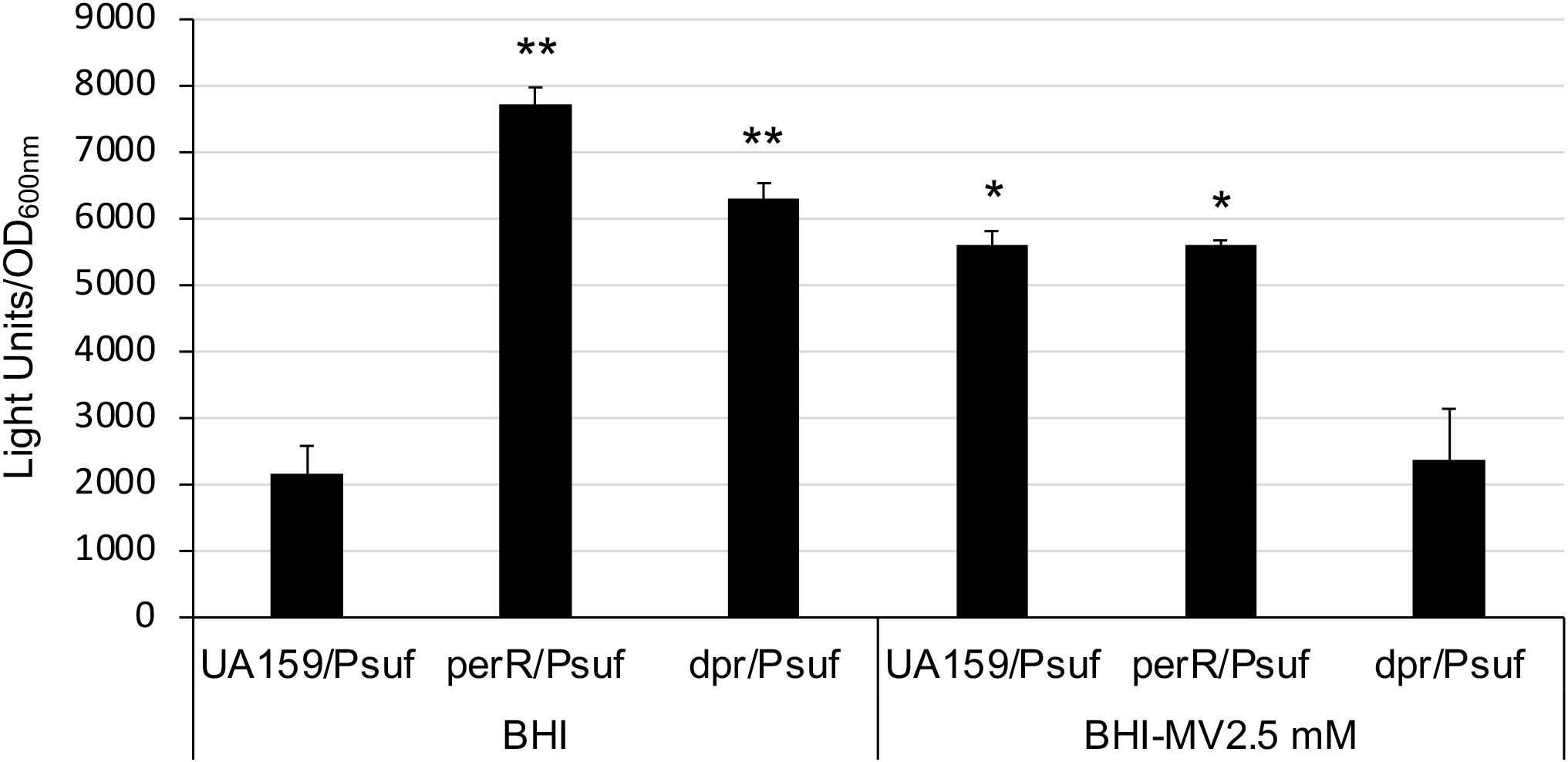
Figure 5. Luciferase reporter analysis of a dpr mutant. Overnight cultures of S. mutans UA159 and its perR and dpr mutants were washed thrice and then transferred to BHI broth with and without methyl viologen (MV) at 2.5 mM and let grow in an anaerobic box. Results showed the dpr mutant grew slowly than UA159 and the perR mutant, but similarly as UA159 and the perR mutant, could still launch oxidative stress responses and was featured with induction of luciferase reporter activity. However, unlike the parent strain and the perR mutant, no such activities were observed when it was grown in the presence of 2.5 mM MV. *, **, P<0.05 and 0.01, respectively, when compared to UA159/Psuf in BHI by Student t-test.
Dpr is known to as a major factor in resistance to reactive oxygen species. To test if Dpr mediated oxidative stress tolerance response influences sufCDSUB expression, an allelic exchange dpr mutant was constructed, and the impact of dpr deletion on sufCDSUB expression was analyzed using luciferase reporter fusion assays similarly as described above. Consistent with Ganguly et al (Ganguly et al., 2018), dpr deletion led to major reductions in growth rate and viability of the deficient mutant under aerobic conditions due to compromises in oxidative stress tolerance. Consistently, the luciferase reporter activity in the dpr mutant was increased, as compared to the wild-type, when the reporter strains were grown in an anaerobic box (Figure 5). However, no such effects were observed when methyl viologen at 2.5 mM was included in the growth medium especially under aerobic conditions, which again indicates a compromised oxidative stress tolerance response of the dpr mutant. Similar trends were also observed with the gyrase A promoter fusion as a reference. These results further suggest that oxidative stressors strongly influence the expression of sufCDSUB, and that Dpr-deficiency induces oxidative stresses of the deficient mutant, indirectly affecting sufCDSUB expression.
S. mutans possesses two Spx homologues, SpxA1 and SpxA2, which serve as the activator of genes involved in oxidative stress response (Galvao et al., 2015; Kajfasz et al., 2015). By RNA-seq analysis, Kajfasz et al (Kajfasz et al., 2017) found that members of the sufCDSUB operon were up-regulated in the wild-type following exposure to sub-lethal amount of hydrogen peroxide, but they were significantly reduced in spxA1 mutant and more so in a spxA1/spxA2 double mutant, compared to the wild-type, UA159. To further investigate how the SpxA regulators influence the expression of the SUF machinery, the suf promoter-reporter fusion construct was transformed into the allelic exchange spxA1 and spxA2 mutants, and the impact of spxA deletion on sufCDSUB expression was analyzed using luciferase reporter assays with the gyrase gyrA promoter PgyrA-reporter fusion serving as a reference. As compared to the wild-type, the luciferase reporter activities under the direction of Psuf promoter were reduced by >2.8-fold (P<0.01) (Figure 6) in the spxA2 mutant, which in part can also be attributed to the fact that the spxA2 mutants grew more slowly as compared to the wild-type, UA159 (Galvao et al., 2015). No significant differences were observed between the spxA1 mutant and the wild-type, UA159. The reference PgyrA-reporter fusion showed no significant change in expression under the conditions tested.
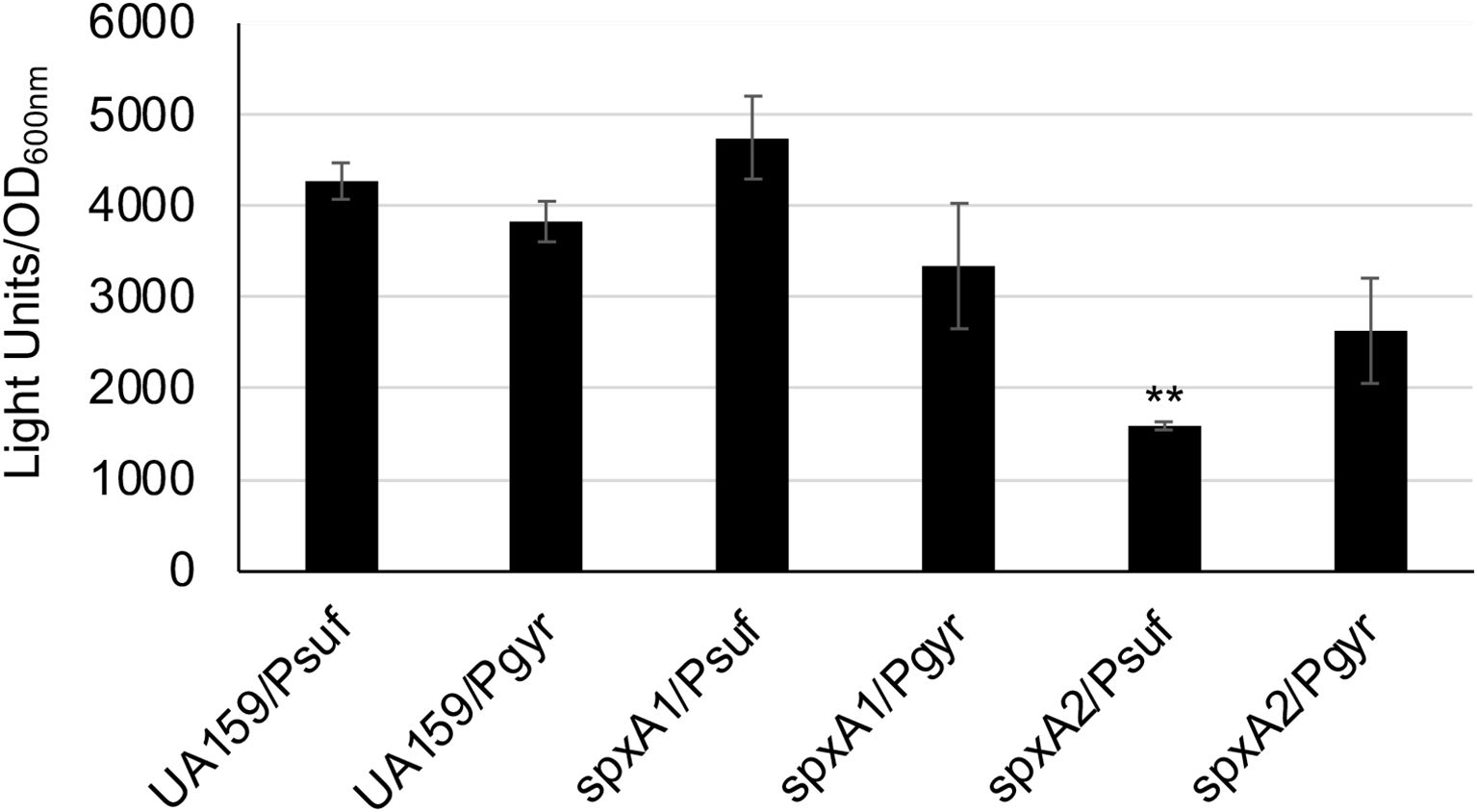
Figure 6. Luciferase reporter fusion analysis of the spxA mutants. S. mutans wild-type (UA159) and its spxA1 (spxA1) & spxA2 (spxA2) deficient mutants carrying the suf promoter-reporter fusion (Psuf) or the gyrA promoter-reporter fusion (Pgyr) as a reference were grown in BHI broth to mid-exponential phase (OD600nm ~0.4), when luciferase activities were analyzed. **, P<0.01 vs UA159 under the same condition via Student t-test.
SloR is known as a metal-dependent repressor of sloABC when Fe is plentiful in the growth environment (Rolerson et al., 2006). SloR also facilitates its own expression as a positive autoregulator (Monette et al., 2018). To investigate potential roles of SloR in sufCDSUB expression, an allelic exchange sloR (SMU.186) mutant was constructed and further analyzed, along with a perR/sloR double mutant, using luciferase reporter assays as described above. Relative to the parent strain, UA159 and the perR mutant, the sloR mutant displayed some aberrant growth phenotypes including extended lag phase (>24 hours) and aggregations when growing in regular BMG medium. However, both the sloR single and sloR/perR double mutant grew similarly well as the wild-type in BMG medium with omission of iron in the recipe (BMG-Fe) or BMG with inclusion of chelating agent 2,2’-pyridyl at 50 µM. When analyzed with the full promoter-reporter fusion construct (Psuf), luciferase reporter activity in the sloR mutant was reduced by >2-fold (P<0.01) when the strains were grown in BMG medium, especially in BMG with exclusion of Fe (BMG-Fe), as compared to the parent strain under the same conditions (Figure 7). Similar results were measured with the sloR/perR double mutant when grown in BMG, although no significant differences were measured when the double mutant was grown in BMG-Fe or BMG plus chelating agent (Figure 7).
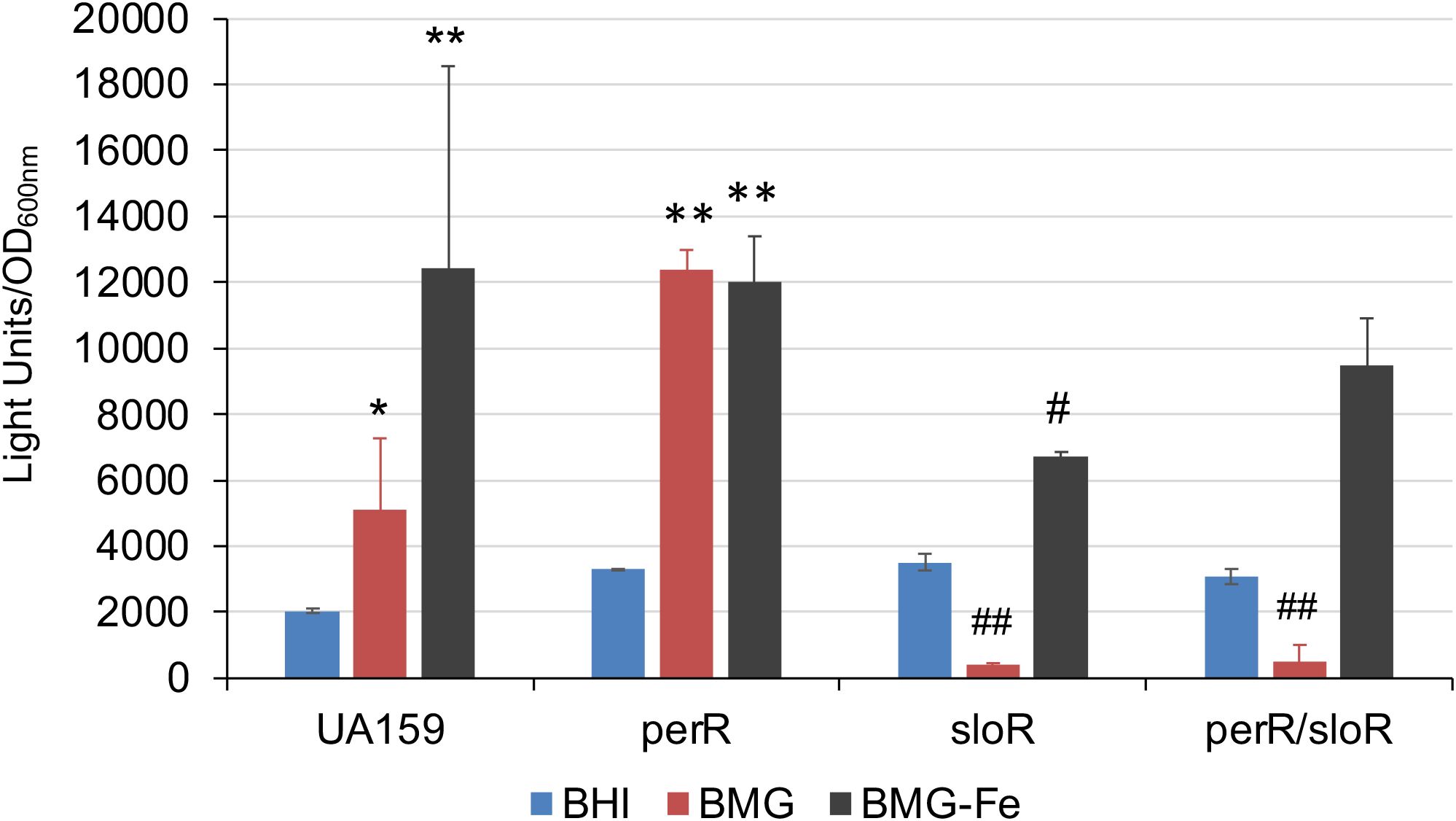
Figure 7. Luciferase reporter analysis of a sloR mutant and a sloR/perR mutant. S. mutans wild-type UA159 and its sloR and sloR/perR mutants were grown in BHI, BM plus glucose with (BMG) and without inclusion of FeCl2 (BMG-Fe). *, **, P<0.05 and 0.01, respectively, when compared to growth in BHI; #, ##, P<0.05 and 0.01 vs UA159 in BMG-Fe and BMG, respectively, while analyzed by Student t test.
To examine if SloR directly interacts with the promoter, regulating sufCDSUB expression, recombinant SloR (rSloR) was used in an EMSA assay with rSloR-sloA promoter interaction as a positive control (Spatafora et al., 2015). As expected, the inclusion of rSloR led to mobility shift of sloA probe. However, no apparent effect was observed when rSloR was mixed with the sufCDSUB probe on its gel mobility under the conditions studied (Supplementary Figure 1). Similar results were observed when S. mutans whole cell lysates were used in the EMSA assay. These results suggest that SloR is not directly involved in regulation of sufCDSUB expression, although it is also possible that other unknown factor(s) are involved in the SloR-mediated regulation, and in the absence of its co-factor, SloR will not bind to its promoter DNA in EMSA.
To test if cysteine also plays a direct role in SUF expression, the modified semi-defined biofilm medium with the omission of cysteine in the amino acid mix was used to grow the luciferase reporter strains, and the reporter activities were measured with and without addition of cysteine one hour prior to the luciferase assay. The results showed that reporter activity without cysteine was at its base line, while addition of cysteine at 4 mM was found to induce the reporter activity by 84.6 (± 7.4)-fold, compared to the ones receiving solvent control (P<0.001) (Figure 8). Addition of serine at 4 mM also led to increases of reporter activity, but to a much less degree with an average of 3.63(± 0.43)-fold (P<0.05). SMU.852, also cysR (Sperandio et al., 2010), encodes a LysR-family transcriptional regulator, which we have recently identified in a study of the regulation of brpA expression. To analyze if CysR plays a role in sufCDSUB expression, an allelic exchange mutant, TW660, was constructed, and relative to its parent strain, UA159, the cysR mutant displayed a reduced growth rate and optical density overnight when grown in regular BHI broth (Supplementary Figure 2). When transformed with the Psuf promoter luciferase reporter fusion, the reporter activity in the cysR mutant was shown to be reduced by >11.06-fold, compared to the parent strain (P<0.001) (Figure 8). However, the reporter activity of the cysR mutant was still inducible with the addition of cysteine, when grown in the modified BM medium similarly as the wild-type, albeit to a reduced degree, which in part could be attributed to the reduced growth rate of the cysR mutant (Supplementary Figure 2). These results suggest that expression of sufCDSUB requires cysteine as an inducer, and CysR is a positive regulator of sufCDSUB expression.
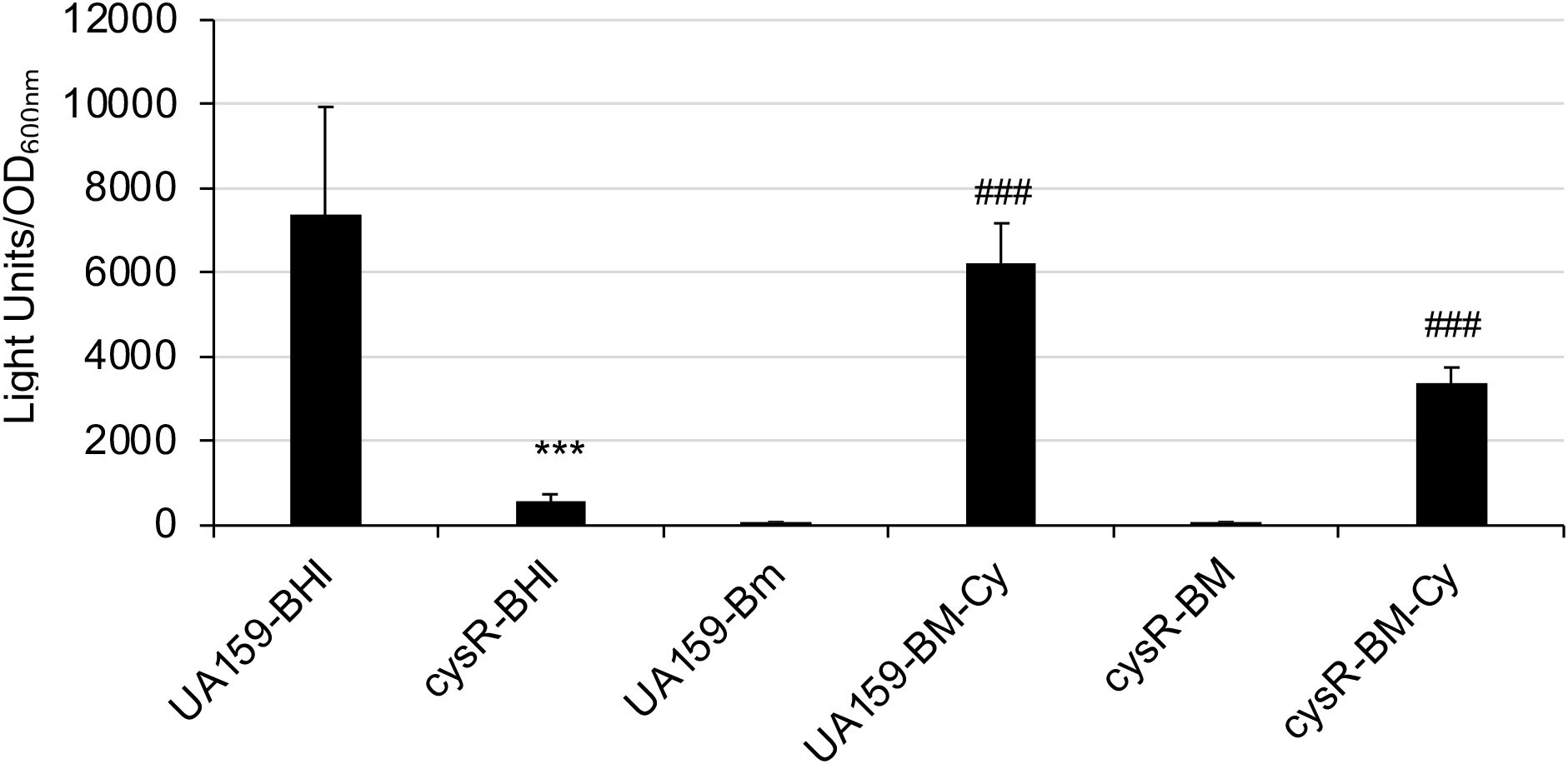
Figure 8. Luciferase reporter assays of a cysR mutant. S. mutans wild-type (UA159) and its cysR mutant (cysR) carrying the suf promoter-reporter were grown to mid-exponential phase in plain BHI (BHI) or modified BM (BM) medium with and without inclusion of cysteine (Cy, 4 mM). Results showed luciferase reporter expression under the direction of the sufCDSUB promoter was dramatically reduced in response to cysR deletion; and it was at its minimum when grown in BM medium without cysteine. ***, P<0.001, cysR vs UA159 in BHI; ###, P<0.001, BM-with cysteine vs BM without cysteine, when analyzed by Student t-test.
When analyzed, two regions with some similarities to the consensus of the LysR-type of regulators (https://regprecise.lbl.gov/sites.jsp?regulog_id=4369) were identified in the promoter region with one overlapping the -10 and the perR-box2 region and the other overlapping the Fur-Box1 site (Supplementary Figure 3). To test if CysR interacts with the sufCDSUB promoter, CysR was expressed and purified using affinity chromatography, and as shown in Figure 9, inclusion of the recombinant CysR protein (rCysR) was shown to cause mobility shift of the promoter probes, indicative of rCysR-promoter interaction. Consistent with the luciferase reporter fusion assays, rCysR was also shown in an IVT assay to enhance the transcription of sufC under the direction of sufCDSUB promoter (Figure 9). These results further support the role of CysR as a positive regulator of sufCDSUB in S. mutans.
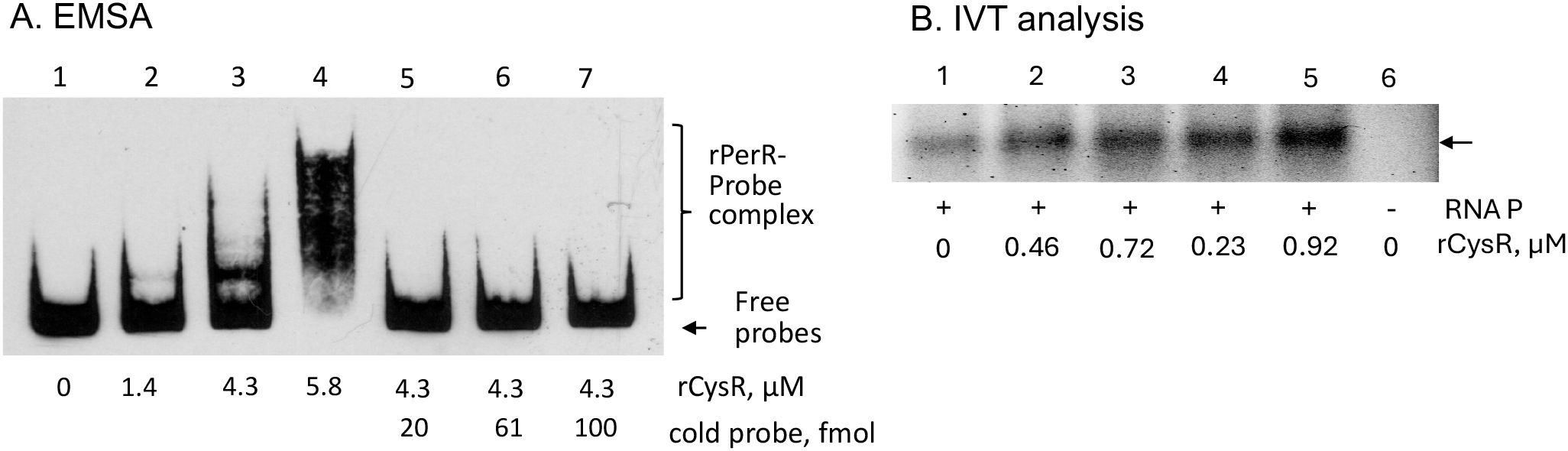
Figure 9. EMSA (A) and IVT (B) analysis of CysR. Inclusion of rCysR resulted in mobility retardation of biotinylated suf promoter DNA probes (Psuf, lanes 2-4), and such interaction was concentration-dependent as indicated. Addition of cold-probe led to release of biotinylated probes from the rPerR-promoter complex (lanes 5-7). (B) For IVT assay, inclusion of rCysR led to increase of sufC transcription (indicated by an arrow) under the direction of suf promoter. No RNA polymerase served as a negative control.
Results presented here further demonstrated that consistent with our previous findings, the sufCDSUB operon in S. mutans is highly regulated in response to environmental cues and that multiple cis- and trans-acting factors have been shown to play a significant role in the regulation of sufCDSUB expression. These include transcription repressor PerR and Fe2+, as a co-effector, and positive regulator CysR. In addition, SpxA2, a global regulator in oxidative stress responses, is also required for sufCDSUB expression.
In Synechocystis spp. and several others, the expression of suf gene cluster is governed by sufR, which lies in front of the suf operon and acts as a repressor of the suf expression and an auto regulator of its own expression (Vuorijoki et al., 2017; Willemse et al., 2018; Cheng et al., 2020). Although S. mutans genome does not possess an apparent sufR, it contains a perR gene that encodes PerR, a peroxide stress regulator of the Fur family, widespread in many Gram-negative and low-GC Gram-positive bacteria such as Bacillus spp. and S. pyogenes (Bsat et al., 1998; Kajfasz et al., 2021). PerR is also iron-dependent, but unlike SufR, it does not directly interact with Fe-S clusters. Active PerR exists in homodimers, and each possesses two metal binding centers, with one for Zn2+required for conformation and dimerization and the other for Fe2+ or Mn2+ for promoter DNA interaction and regulation of target gene expression (Traore et al., 2006). However, recent studies have shown that in the presence of excessive iron (FeCl2) and upon exposure to hydrogen peroxide, the cysteine and histidine residues in the regulatory metal center can get oxidized quickly, which triggers a conformational change (Marinho et al., 2014; Lee and Helmann, 2006). Consequently, the regulator protein dissociates from the promoter, and the target gene gets transcribed. Like in group A streptococci, PerR in S. mutans has recently been shown by RNA-seq to be involved in regulation of a large amount of known genes including many involved in oxidative stress responses (Kajfasz et al., 2021; Ruxin et al., 2021). However, to the best of our knowledge, the current study is the first instance a direct link of PerR in regulation of sufCDSUB expression and Fe-S cluster homeostasis has been established.
Consistent with the role of PerR as a repressor in sufCDSUB expression, deletion of perR by allelic exchange led to significant increases of luciferase reporter activity, a de-repression of luciferase reporter expression. This is further supported by the result of site-directed mutations of a PerR-box adjacent to the extended -10 site-box, where EMSA showed that recombinant PerR interacts with the sufCDSUB promoter resulting in a mobility shift, and the result of IVT assay that the PerR-mediated repression is Fe2+-dependent. However, as mutations of selected nucleotides did not seem to significantly alter the PerR-promoter interaction, the exact nucleotides that interact with PerR await further investigation. It is also worth noting that PerR is known to be easily inactivated when exposed to iron and oxygen, weakening its capacity to bind to its target promoter, and such oxidation can be prevented by Zn2+ or Mn2+ (Lee and Helmann, 2006). Consistently, the inclusion of Zn2+ in the reaction mixture was shown to significantly enhance the rPerR binding and the consequent mobility shift in an EMSA.
Consistent with our previous finding that sufCDSUB expression is regulated in response to iron availability in the growth environment, cultures transferred from regular biofilm medium to medium with omission of FeCl3 or with inclusion of chelating agent 2,2’-pyridyl displayed >5-fold increases of luciferase reporter activity, but not in cells that were transferred from medium without inclusion of FeCl3, indicative of iron as a co-repressor. The in vitro transcription assay leading to complete inhibition of suf transcription in the presence of recombinant PerR and Fe2+ corroborates the reporter activity findings. Iron is the second most abundant metal on earth, of which most exist in iron ions, Fe2+ and Fe3+. It is apparent that the amount of iron in the water and the basal medium components are sufficient for the bacterium to grow. It is the state of iron depletion that the bacterium senses from regular iron-rich condition to iron-limiting condition that triggers the up-regulation of the sufCDSUB expression. The incapability of the perR mutant to launch such a response to iron limitation is also consistent with the nature of its iron-dependence. The potential redox sensing ability that allows PerR-Fe2+ complexes to bind with the DNA and regulate gene expression of sufCDSUB needs further investigation.
Recent studies in group A streptococci and others have shown that PerR is a major oxidative stress-sensing repressor of H2O2 resistance (King et al., 2000; Ricci et al., 2002). Among other proteins and factors, PerR-deficiency leads to up-regulation of Dps-like peroxide resistance protein Dpr, a major H2O2 resistance factor. Dpr has been identified as a major factor in the resistance of S. mutans to ROS during growth in the presence of oxygen (Yamamoto et al., 2004). Intracellular free iron and hydrogen peroxide can oxidize a wide range of substrates and cause detrimental alterations of macromolecules and their related biological processes. The reaction, referred to as the Fenton reaction, is complex and capable of generating both hydroxyl radicals and higher oxidation states of the iron creating oxidative stress in the cell. Consistent with Fujishima et al. and Kajfasz et al (Fujishima et al., 2013; Kajfasz et al., 2021), deletion of perR in S. mutans led to up- and down-regulation of >35 proteins, including several sugar transporters and fructose polymer utilization. Of the up-regulated genes are those for major components of the oxidative stress tolerance pathways, including Dpr. Further study of dpr by allelic exchange mutagenesis further revealed that lacking of Dpr in S. mutans led to major defects in oxidative stress tolerance, significantly reduced its growth rate, and elevated autolysis of the dpr mutant, consistent with Kajfasz et al (Kajfasz et al., 2021). When compared to the wild-type, luciferase reporter activity in the Dpr-deficient mutant was elevated by >3-fold when grown in regular BHI, but no major differences were measured when grown in BHI broth plus methyl viologen at 2.5 mM (Supplementary Figure 3). The results suggest that unlike PerR, Dpr is a major factor that plays a direct role in oxidative stressor-mediated regulation of sufCDSUB expression in S. mutans.
Metalloregulator SloR is mostly known as a repressor to regulate the expression of target genes in response to manganese availability, including transporter SloABC for Mn2+/Fe2+in S. mutans. In S. mutans, it also positively regulates its own expression (Monette et al., 2018). However, unlike the repressor SloR, the binding site(s) for positive regulator SloR remains unclear and awaits further investigation. It is apparent that SloR-deficiency resulted in significant reduction of luciferase reporter activity, indicative of a potential role as a positive regulator in sufCDSUB expression, although EMSA showed no SloR-promoter DNA interaction under the condition studied. Considering the sloR deficient mutant also displayed growth defects when it was growing in Fe-rich medium, such growth defects could be in part attributed to the altered expression. However, it is also possible that other unknown factors are involved in SloR-mediated regulation of sufCDSUB expression. It is possible that SloR functions as a positive regulator, especially under iron depletion conditions, when PerR is mostly inactive, which also explains why in the presence of chelating agent the differences were not as significant, especially for the sloR/perR double mutant. Of note, the expression of SloR in S. mutans is also regulated by repressor PerR (Ruxin et al., 2021). Without SloR, the mutant can still take up the Fe in the culture medium, but its ability to assemble it into the Fe-S cluster is significantly reduced. Consequently, accumulation of free irons leads to the production of reactive oxygen species that causes cytotoxicity and growth defects including an extended lag phase. However, the exact role of SloR, as well as its binding site, in sufCDSUB regulation awaits further investigation. The results presented here also suggest that in S. mutans, repressor PerR and probably SloR, as a positive regulator, work in concert to regulate sufCDSUB expression most likely in response to the Fe-availability in the oral cavity, although how they coordinate in fine tuning the sufCDSUB expression remains unclear.
S. mutans possesses two global oxidative stress regulators, SpxA1 and SpA2, which have recently been shown by RNA-seq analysis to be required for expression of major oxidative stress genes including members of the sufCDSUB cluster (Galvao et al., 2015; Kajfasz et al., 2017). This study used luciferase reporter fusion assays and consistently, the results showed that the mutant lacking SpxA2, but not SpxA1, displayed major reductions in sufCDSUB expression. SpxA proteins are known to interact with the RNA polymerase in regulation of its target genes.
In Fe-S cluster assembly, cysteine is utilized by the desulfurase SufS to generate the sulfur for Fe-S cluster assembly (Lill, 2009; Sperandio et al., 2010). The result that addition of cysteine to the growth environment triggers dramatic increases of luciferase reporter activity suggests that the expression of sufCDSUB is regulated in response to cysteine as an inducer. A LysR family transcriptional regulator, CysR was previously found to play a role as an activator in regulating the expression of genes involved in cysteine biosynthesis and utilization in response to the availability of cysteine in the environment (Sperandio et al., 2010). In addition, it also indirectly regulates the expression of the genes involved in biosynthesis of methionine. An important amino acid, cysteine is also involved in various cellular processes, including protein synthesis and redox regulation. Methionine is an essential amino acid and serves as a methyl donor in various methylation reactions, playing a crucial role in epigenetic regulation and other cellular processes. It is therefore not surprising that the cysR mutant displays some major growth defects in growth rate and optical density. It is worth noting that no significant differences were observed in the wild-type for the growth rate and culture density between the cultures grown with and without inclusion of cysteine under the conditions studied. Like the wild-type, cysteine was able to induce the expression of the sufCDSUB gene cluster albeit to a less degree, which can be in part attributed to the reduced growth rate of the cysR mutant. The result also suggests that cysteine likely serves as a co-factor of CysR-mediated regulation of sufCDSUB expression.
In summary, the results presented here demonstrate that multiple factors work in concert to regulate sufCDSUB in S. mutans in response to iron and cysteine availability, other environment cues such as oxidative stressors, and other signals related to sulfur metabolism. These results also suggest that a keystone cariogenic bacterium, S. mutans inhabiting a unique niche in the oral cavity, may have evolved specific adaptations in its sole Fe-S cluster assembly machinery to cope with the challenges posed by its environment, such as fluctuations in pH, oxygen tension, and nutrient availability.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
KE: Formal Analysis, Investigation, Writing – original draft. LG: Investigation, Writing – review & editing. BC: Investigation, Writing – review & editing. YH: Writing – review & editing, Investigation. JK: Investigation, Writing – review & editing. JB: Conceptualization, Writing – review & editing. GS: Conceptualization, Writing – review & editing, Formal Analysis. JL: Conceptualization, Writing – review & editing. ZW: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported in part by NIH grant DE19452 to ZW.
We are grateful to Dr. Hemendra Pal Singh Dhaked, Ms. Jyoti N. Prajapati, and Mr. Grayson D. Lambert for their assistance on certain aspects of the experiments. We also want to thank Dr. Robert A. Burne for his generous gift of the polyclonal antibodies against FruA.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1499476/full#supplementary-material
Bitoun, J. P., Liao, S., Mckey, B. A., Yao, X., Fan, Y., Abranches, J., et al. (2013). Psr is involved in regulation of glucan production, and double deficiency of BrpA and Psr is lethal in Streptococcus mutans. Microbiol. (Reading) 159, 493–506. doi: 10.1099/mic.0.063032-0
Bitoun, J. P., Liao, S., Yao, X., Ahn, S. J., Isoda, R., Nguyen, A. H., et al. (2012a). BrpA is involved in regulation of cell envelope stress responses in Streptococcus mutans. Appl. Environ. Microbiol. 78, 2914–2922. doi: 10.1128/AEM.07823-11
Bitoun, J. P., Liao, S., Yao, X., Xie, G. G., Wen, Z. T. (2012c). The redox-sensing regulator rex modulates central carbon metabolism, stress tolerance response and biofilm formation by streptococcus mutans. PloS One 7, e44766. doi: 10.1371/journal.pone.0044766
Black, K. A., Dos Santos, P. C. (2015). Shared-intermediates in the biosynthesis of thio-cofactors: Mechanism and functions of cysteine desulfurases and sulfur acceptors. Biochim. Biophys. Acta 1853, 1470–1480. doi: 10.1016/j.bbamcr.2014.10.018
Bowen, W. H., Koo, H. (2011). Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res. 45, 69–86. doi: 10.1159/000324598
Brenot, A., King, K. Y., Caparon, M. G. (2005). The PerR regulon in peroxide resistance and virulence of Streptococcus pyogenes. Mol. Microbiol. 55, 221–234. doi: 10.1111/j.1365-2958.2004.04370.x
Bsat, N., Herbig, A., Casillas-Martinez, L., Setlow, P., Helmann, J. D. (1998). Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol. Microbiol. 29, 189–198. doi: 10.1046/j.1365-2958.1998.00921.x
Burne, R. A., Abranches, J., Ahn, S. J., Lemos, J. A., Wen, Z. T., Zeng, L. (2011). “Functional Genomics of Streptococcus mutans,” in Oral Microbial Communities: Genomic Inquires and Interspecies Communication. Ed. Kolenbrander, P. E. (ASM Press).
Cheng, Y., Lyu, M., Yang, R., Wen, Y., Song, Y., Li, J., et al. (2020). SufR, a [4Fe-4S] Cluster-containing transcription factor, represses the sufRBDCSU operon in Streptomyces avermitilis iron-sulfur cluster assembly. Appl. Environ. Microbiol. (18), e01523–20. doi: 10.1128/AEM.01523-20
Dou, Y., Rutanhira, H., Schormann, N., Deivanayagam, C., Fletcher, H. M. (2021). PG1659 functions as anti-sigma factor to extracytoplasmic function sigma factor RpoE in Porphyromonas gingivalis W83. Mol. Oral. Microbiol. 36, 80–91. doi: 10.1111/omi.12329
Ellepola, K., Huang, X., Riley, R. P., Bitoun, J. P., Wen, Z. T. (2021). Streptococcus mutans Lacking sufCDSUB Is Viable, but Displays Major Defects in Growth, Stress Tolerance Responses and Biofilm Formation. Front. Microbiol. 12, 671533. doi: 10.3389/fmicb.2021.671533
Enami, S., Sakamoto, Y., Colussi, A. J. (2014). Fenton chemistry at aqueous interfaces. Proc. Natl. Acad. Sci. U.S.A. 111, 623–628. doi: 10.1073/pnas.1314885111
Fontecave, M., Py, B., Ollagnier De Choudens, S., Barras, F. (2008). From iron and cysteine to iron-sulfur clusters: the biogenesis protein machineries. EcoSal Plus 3(1). doi: 10.1128/ecosal.3.6.3.14
Fujishima, K., Kawada-Matsuo, M., Oogai, Y., Tokuda, M., Torii, M., Komatsuzawa, H. (2013). dpr and sod in Streptococcus mutans are involved in coexistence with S. sanguinis, and PerR is associated with resistance to H2O2. Appl. Environ. Microbiol. 79, 1436–1443. doi: 10.1128/AEM.03306-12
Galvao, L. C., Miller, J. H., Kajfasz, J. K., Scott-Anne, K., Freires, I. A., Franco, G. C., et al. (2015). Transcriptional and phenotypic characterization of novel spx-regulated genes in streptococcus mutans. PloS One 10, e0124969. doi: 10.1371/journal.pone.0124969
Ganguly, T., Kajfasz, J. K., Miller, J. H., Rabinowitz, E., Galvao, L. C. C., Rosalen, P. L., et al. (2018). Disruption of a Novel Iron Transport System Reverses Oxidative Stress Phenotypes of a dpr Mutant Strain of Streptococcus mutans. J. Bacteriol. 200 (14), e00062–18. doi: 10.1128/JB.00062-18
Gregoire, S., Xiao, J., Silva, B. B., Gonzalez, I., Agidi, P. S., Klein, M. I., et al. (2011). Role of glucosyltransferase B in interactions of Candida albicans with Streptococcus mutans and with an experimental pellicle on hydroxyapatite surfaces. Appl. Environ. Microbiol. 77, 6357–6367. doi: 10.1128/AEM.05203-11
Hantke, K. (2001). Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4, 172–177. doi: 10.1016/S1369-5274(00)00184-3
Imlay, J. A. (2002). How oxygen damages microbes: oxygen tolerance and obligate anaerobiosis. Adv. Microb. Physiol. 46, 111–153. doi: 10.1016/S0065-2911(02)46003-1
Kajfasz, J. K., Ganguly, T., Hardin, E. L., Abranches, J., Lemos, J. A. (2017). Transcriptome responses of Streptococcus mutans to peroxide stress: identification of novel antioxidant pathways regulated by Spx. Sci. Rep. 7, 16018. doi: 10.1038/s41598-017-16367-5
Kajfasz, J. K., Rivera-Ramos, I., Scott-Anne, K., Gregoire, S., Abranches, J., Lemos, J. A. (2015). Transcription of Oxidative Stress Genes Is Directly Activated by SpxA1 and, to a Lesser Extent, by SpxA2 in Streptococcus mutans. J. Bacteriol 197, 2160–2170. doi: 10.1128/JB.00118-15
Kajfasz, J. K., Zuber, P., Ganguly, T., Abranches, J., Lemos, J. A. (2021). Increased oxidative stress tolerance of a spontaneously occurring perR gene mutation in Streptococcus mutans UA159. J. Bacteriol. 203 (8). doi: 10.1128/JB.00535-20
King, K. Y., Horenstein, J. A., Caparon, M. G. (2000). Aerotolerance and peroxide resistance in peroxidase and PerR mutants of Streptococcus pyogenes. J. Bacteriol 182, 5290–5299. doi: 10.1128/JB.182.19.5290-5299.2000
Kreth, J., Merritt, J., Shi, W., Qi, F. (2005). Co-ordinated bacteriocin production and competence development: a possible mechanism for taking up DNA from neighbouring species. Mol. Microbiol. 57, 392–404. doi: 10.1111/j.1365-2958.2005.04695.x
Lau, P. C., Sung, C. K., Lee, J. H., Morrison, D. A., Cvitkovitch, D. G. (2002). PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49, 193–205. doi: 10.1016/S0167-7012(01)00369-4
Layer, G., Gaddam, S. A., Ayala-Castro, C. N., Ollagnier-De Choudens, S., Lascoux, D., Fontecave, M., et al. (2007). SufE transfers sulfur from SufS to SufB for iron-sulfur cluster assembly. J. Biol. Chem. 282, 13342–13350. doi: 10.1074/jbc.M608555200
Lee, J.-W., Helmann, J. D. (2006). The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature 440, 363–367. doi: 10.1038/nature04537
Lee, J.-W., Helmann, J. D. (2007). Functional specialization within the Fur family of metalloregulators. BioMetals 20, 485–499. doi: 10.1007/s10534-006-9070-7
Lemos, J. A., Palmer, S. R., Zeng, L., Wen, Z. T., Kajfasz, J. K., Freires, I. A., et al. (2019). The biology of streptococcus mutans. Microbiol. Spectr. 7 (1). doi: 10.1128/microbiolspec.GPP3-0051-2018
Lill, R. (2009). Function and biogenesis of iron-sulphur proteins. Nature 460, 831–838. doi: 10.1038/nature08301
Lill, R. (2020). From the discovery to molecular understanding of cellular iron-sulfur protein biogenesis. Biol. Chem. 401, 855–876. doi: 10.1515/hsz-2020-0117
Marinho, H. S., Real, C., Cyrne, L., Soares, H., Antunes, F. (2014). Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol. 2, 535–562. doi: 10.1016/j.redox.2014.02.006
Miller, R. A., Britigan, B. E. (1997). Role of oxidants in microbial pathophysiology. Clin. Microbiol. Rev. 10, 1–18. doi: 10.1128/CMR.10.1.1
Monette, P., Brach, R., Cowan, A., Winters, R., Weisman, J., Seybert, F., et al. (2018). Autoregulation of the streptococcus mutans sloR metalloregulator is constitutive and driven by an independent promoter. J. Bacteriol. 200 (14), e00214–18. doi: 10.1128/JB.00214-18
Mongkolsuk, S., Helmann, J. D. (2002). Regulation of inducible peroxide stress responses. Mol. Microbiol. 45, 9–15. doi: 10.1046/j.1365-2958.2002.03015.x
Pinochet-Barros, A., Helmann, J. D. (2020). Bacillus subtilis Fur Is a Transcriptional Activator for the PerR-Repressed pfeT Gene, Encoding an Iron Efflux Pump. J. Bacteriol. 202 (8), e00697–19. doi: 10.1128/JB.00697-19
Podbielski, A., Woischnik, M., Leonard, B. A., Schmidt, K. H. (1999). Characterization of nra, a global negative regulator gene in group A streptococci. Mol. Microbiol. 31, 1051–1064. doi: 10.1046/j.1365-2958.1999.01241.x
Ricci, S., Janulczyk, R., Björck, L. (2002). The regulator PerR is involved in oxidative stress response and iron homeostasis and is necessary for full virulence of Streptococcus pyogenes. Infect. Immun. 70, 4968–4976. doi: 10.1128/IAI.70.9.4968-4976.2002
Rolerson, E., Swick, A., Newlon, L., Palmer, C., Pan, Y., Keeshan, B., et al. (2006). The SloR/Dlg metalloregulator modulates Streptococcus mutans virulence gene expression. J. Bacteriol 188, 5033–5044. doi: 10.1128/JB.00155-06
Ruxin, T. R., Schwartzman, J. A., Davidowitz, C. R., Peters, Z., Holtz, A., Haney, R. A., et al. (2021). Regulatory involvement of the perR and sloR metalloregulators in the streptococcus mutans oxidative stress response. J. Bacteriol. 203 (11), e00678–20. doi: 10.1128/jb.00678-20
Seepersaud, R., Needham, R. H., Kim, C. S., Jones, A. L. (2006). Abundance of the delta subunit of RNA polymerase is linked to the virulence of Streptococcus agalactiae. J. Bacteriol 188, 2096–2105. doi: 10.1128/JB.188.6.2096-2105.2006
Selbach, B. P., Chung, A. H., Scott, A. D., George, S. J., Cramer, S. P., Dos Santos, P. C. (2014). Fe-S cluster biogenesis in Gram-positive bacteria: SufU is a zinc-dependent sulfur transfer protein. Biochemistry 53, 152–160. doi: 10.1021/bi4011978
Sevilla, E., Bes, M. T., Peleato, M. L., Fillat, M. F. (2021). Fur-like proteins: Beyond the ferric uptake regulator (Fur) paralog. Arch. Biochem. Biophys. 701, 108770. doi: 10.1016/j.abb.2021.108770
Spatafora, G., Corbett, J., Cornacchione, L., Daly, W., Galan, D., Wysota, M., et al. (2015). Interactions of the metalloregulatory protein sloR from streptococcus mutans with its metal ion effectors and DNA binding site. J. Bacteriol 197, 3601–3615. doi: 10.1128/JB.00612-15
Sperandio, B., Gautier, C., Pons, N., Ehrlich, D. S., Renault, P., Guedon, E. (2010). Three paralogous LysR-type transcriptional regulators control sulfur amino acid supply in Streptococcus mutans. J. Bacteriol 192, 3464–3473. doi: 10.1128/JB.00119-10
Touati, D. (2000). Iron and oxidative stress in bacteria. Arch. Biochem. Biophys. 373, 1–6. doi: 10.1006/abbi.1999.1518
Traore, D. A., El Ghazouani, A., Ilango, S., Dupuy, J., Jacquamet, L., Ferrer, J. L., et al. (2006). Crystal structure of the apo-PerR-Zn protein from Bacillus subtilis. Mol. Microbiol. 61, 1211–1219. doi: 10.1111/j.1365-2958.2006.05313.x
Troxell, B., Hassan, H. M. (2013). Transcriptional regulation by Ferric Uptake Regulator (Fur) in pathogenic bacteria. Front. Cell Infect. Microbiol. 3, 59. doi: 10.3389/fcimb.2013.00059
Vuorijoki, L., Tiwari, A., Kallio, P., Aro, E. M. (2017). Inactivation of iron-sulfur cluster biogenesis regulator SufR in Synechocystis sp. PCC 6803 induces unique iron-dependent protein-level responses. Biochim. Biophys. Acta Gen. Subj 1861, 1085–1098. doi: 10.1016/j.bbagen.2017.02.020
Wen, Z. T., Baker, H. V., Burne, R. A. (2006). Influence of BrpA on critical virulence attributes of Streptococcus mutans. J. Bacteriol. 188, 2983–2992. doi: 10.1128/JB.188.8.2983-2992.2006
Wen, Z. T., Liao, S., Bitoun, J. P., De, A., Jorgensen, A., Feng, S., et al. (2017). Streptococcus mutans displays altered stress responses while enhancing biofilm formation by Lactobacillus casei in mixed-species consortium. Front. Cell Infect. Microbiol. 7, 524. doi: 10.3389/fcimb.2017.00524
Willemse, D., Weber, B., Masino, L., Warren, R. M., Adinolfi, S., Pastore, A., et al. (2018). Rv1460, a SufR homologue, is a repressor of the suf operon in Mycobacterium tuberculosis. PloS One 13, e0200145. doi: 10.1371/journal.pone.0200145
Yamamoto, Y., Fukui, K., Koujin, N., Ohya, H., Kimura, K., Kamio, Y. (2004). Regulation of the intracellular free iron pool by Dpr provides oxygen tolerance to Streptococcus Mutans. J. Bacteriol 186, 5997–6002. doi: 10.1128/JB.186.18.5997-6002.2004
Yamamoto, Y., Higuchi, M., Poole, L. B., Kamio, Y. (2000). Role of the dpr product in oxygen tolerance in Streptococcus mutans. J. Bacteriol. 182, 3740–3747. doi: 10.1128/JB.182.13.3740-3747.2000
Yue, X., Guidry, J. J. (2019). Differential protein expression profiles of bronchoalveolar lavage fluid following lipopolysaccharide-induced direct and indirect lung injury in mice. Int. J. Mol. Sci. 20 (14), 3401. doi: 10.3390/ijms20143401
Keywords: Streptococcus mutans, sufCDSUB, gene expression, PerR, CysR, SloR, SpxA, Dpr
Citation: Ellepola K, Guillot LC, Comeaux B, Han Y, Kajfasz JK, Bitoun JP, Spatafora G, Lemos JA and Wen ZT (2024) Multiple factors regulate the expression of sufCDSUB in Streptococcus mutans. Front. Cell. Infect. Microbiol. 14:1499476. doi: 10.3389/fcimb.2024.1499476
Received: 20 September 2024; Accepted: 05 November 2024;
Published: 27 November 2024.
Edited by:
Ozlem Yilmaz, Medical University of South Carolina, United StatesReviewed by:
Avishek Mitra, Oklahoma State University, United StatesCopyright © 2024 Ellepola, Guillot, Comeaux, Han, Kajfasz, Bitoun, Spatafora, Lemos and Wen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zezhang T. Wen, endlbkBsc3Voc2MuZWR1
†Present address: Kassapa Ellepola, Department of Oral and Craniofacial Biology, College of Dentistry, University of Illinois Chicago, Chicago, IL, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.