- 1Center for Yunnan Plateau Biological Resources Protection and Utilization, College of Biological Resource and Food Engineering, Qujing Normal University, Qujing, China
- 2Department of Biology, Faculty of Science, Chiang Mai University, Chiang Mai, Thailand
- 3Department of Botany and Microbiology, College of Science, King Saud University, Riyadh, Saudi Arabia
- 4Department of Pharmacology and Toxicology, College of Pharmacy, King Saud University, Riyadh, Saudi Arabia
- 5Tropical Microbiology Research Foundation, Pannipitiya, Sri Lanka
Apiospora is an important genus in the Apiosporaceae family with a worldwide distribution. They exhibit different lifestyles including pathogenic, saprophytic, and endophytic. In this study, we aimed to explore the Apiospora associated with bamboo and collected 14 apiospora-like taxa from the forests of Yunnan Province, China. Morphological and phylogenetic analyses (combined ITS, LSU, tef1-α, and tub2 sequence data) confirmed that these collections belong to Apiospora s. str. and reports five new species (viz., Ap. dehongensis, Ap. jinghongensis, Ap. shangrilaensis, Ap. zhaotongensis, and Ap. zhenxiongensis). New sexual morphs of asexually typified Ap. globose and Ap. guangdongensis species, and a new geographical record of Ap. subglobosa are also reported. The findings of this study not only enhance the diversity of bambusicolous fungi in the region of Yunnan, but also geographical distribution of some known Apiospora species.
Introduction
Apiosporaceae (Amphisphaeriales, Sordariomycetes, Ascomycota fide Wijayawardene et al., 2022) was introduced by Hyde et al. (1998), with Apiospora Sacc. as the type genus (Saccardo, 1875). Currently, Apiosporaceae comprises three genera—Apiospora, Arthrinium Kunze, and Nigrospora Zimm (Pintos and Alvarado, 2021; Jiang et al., 2022a). The relationship among the genera in Apiosporaceae is confusing as some taxa lack sequence data and show morphological plasticity. For example, Apiospora resembles Arthrinium s. str. but is phylogenetically well-distinct (Pintos and Alvarado, 2021; Jiang et al., 2022a).
The genus Apiospora was introduced by Saccardo (1875), with Ap. montagnei Sacc. as the type species (Clements and Shear, 1931). Currently, 175 epithets are listed under the genus of Apiospora in Index Fungorum (Index Fungorum 2024; accession date: 16 June 2024). Apiospora was reported as a holomorphic genus; the sexual morph is characterized by multi-locular stromata and 1-septate (near the lower cell) ascospores (Dai et al., 2016, 2017; Bhunjun et al., 2022). The asexual morphs usually occur as coelomycetous on natural substrates or hyphomycetous on culture (e.g., MEA, OA, and PDA). The coelomycetous morph is characterized by dark brown conidia, with a longitudinal and transparent slit (Dai et al., 2016; Pintos and Alvarado, 2021; Zhao et al., 2023). The hyphomycetous morph is characterized by hyaline conidiophores, basauxic conidiogenous cells, globose to subglobose, and pale brown to brown conidia (Wang et al., 2018; Pintos and Alvarado, 2021; Tian et al., 2021; Zhao et al., 2023).
Members of Apiospora are found in different habitats such as animal tissues (including humans), air, lichens, plants, soil, and seaweeds (Liao et al., 2023). The members of Apiospora show a wide distribution and have been reported from tropical, sub-tropical, Mediterranean, and temperate regions (Pintos and Alvarado, 2021; Kwon et al., 2022). Furthermore, the species of Apiospora have been reported as endophytes, saprobes, or pathogens, and some particular species have two or three lifestyles (Samuels et al., 1981; Liao et al., 2023; Zeng et al., 2024). For instance, Ap. arundinis (Corda) Pintos & P. Alvarado has been reported as pathogens, saprobes, and endophytes (Feng et al., 2021; Liao et al., 2023). Moreover, Ap. arundinis has also been reported as a pathogen of plants, animals, and humans (Martínez-Cano et al., 1992; Mavragani et al., 2007; Bagherabadi et al., 2014; Chen et al., 2014; Jiang and Tian, 2021).
According to Monkai et al. (2022); Zhao et al. (2023), and Liu et al. (2024), asexual morphs of Apiospora are frequently observable, while sexual morphs are rare. Therefore, this study aims to explore the morphology of the sexual morph of Apiospora. Of this, a total of 14 Apiospora samples were collected from the Yunnan Province, China. Among these new collections, five new species (e.g., Ap. dehongensis, Ap. jinghongensis, Ap. shangrilaensis, Ap. zhaotongensis, and Ap. zhenxiongensis), two sexual morphs of asexually typified species (Ap. globosa and Ap. guangdongensis), and one new country record (Ap. subglobosa) are reported along with the morphological descriptions, illustrations, and updated phylogenetic trees.
Materials and methods
Collection and morphological studies
The dead and decaying bamboo culms and branches were collected from several forests in Dehong, Shangri-La, Xishuangbanna, and Zhaotong in Yunnan Province. Collected samples were kept in envelope bags and transported to the lab for further evaluation. The stromata and micro-morphological characteristics were observed and photographed using Leica S8AP0 and Olympus BX53 stereomicroscopes, respectively, which are equipped with a high-definition digital camera. The sizes of the fungal structures were measured by the Tarosoft (R) Image Frame Work program (IFW). The photo plates were processed with the Adobe Photoshop CS6 software (Adobe Systems Inc., San Jose, CA, USA).
Isolation and preservation
Pure cultures of the new collections were obtained by single spore isolation. The ascospores were picked from stromata and dispersed on sterile water droplets on a cavity slide. Spore suspensions were placed on potato dextrose agar (PDA) and stored at 27°C until germination. The germinated spores were aseptically transferred into new PDA plates and incubated. The characteristics of fungi colonies were recorded and photographed after 20 days.
Dried herbarium samples were preserved at the Mycological Herbarium of Zhongkai University of Agriculture and Engineering (MHZU) and Herbarium of Guizhou Medical University, Guiyang, China (GMB-W). Living cultures were deposited in Zhongkai University of Agriculture and Engineering (ZHKUCC) and Guizhou Medical University Culture Collection (GMBCC) Guiyang.
DNA extraction, PCR, and sequencing
The genomic DNA was extracted from fresh fungal mycelia grown on PDA [using Biospin Fungus Genomic DNA Extraction Kit (BioFlux®)]. However, for two species, single spore isolation was not successful. Thus, we used fruiting bodies to extract DNA using an E.Z.N.A. Forensic DNA Kit (BIO-TEK). The details of the primers used for PCR amplification are presented in Table 1. We followed Tian et al. (2021) for PCR amplification conditions. PCR products were sequenced at Shanghai Mayobio Biomedical Technology Co., China. All newly generated nucleotide sequence data were submitted to GenBank and the accession numbers were obtained (Table 2).
Phylogenetic analyses
All newly obtained forward and reverse sequences were assembled using Geneious 9.1.2. Those assembled sequences were searched using BLASTn (http://blast.ncbi.nlm.nih.gov/, accessed on 16 January 2024) to retrieve the sequences of closely related strains. The preliminary identification results showed that our new collections match closest with Apiospora, then all available sequences of Apiospora were downloaded from the GenBank based on previous literature (Table 2). The matrix of consensus sequences was aligned with MAFFT v. 7 (Katoh and Standley, 2013). The sequence alignments were trimmed by using trimAl.v1.2rev59 [parameters: -gt 0.7 (ITS, tub2), -gt 0.8 (LSU), -gt 0.9 (tef1-α); Capella-Gutiérrez et al., 2009] and BioEdit v. 7.0 (Hall, 2004) to remove unclear and uninformative regions. The alignments of four genes (LSU, ITS, tef1-α, and tub2) were concatenated by Matrix 1.9 (Vaidya et al., 2011). The AliView 1.26 (Larsson, 2014) was used to convert Fasta files to Phylip (for Maximum likelihood) and Nexus (for Bayesian inference) formats. Maximum likelihood analyses (ML) were performed at the CIPRES web portal using RAxML-HPC2 on XSEDE (8.2.12) (Stamatakis et al., 2008; Stamatakis, 2014) with GTRGAMMA model with 1,000 bootstrap pseudoreplicates. Bayesian inference posterior probabilities (BYPP) (Zhaxybayeva and Gogarten, 2002) were evaluated by Markov Chain Monte Carlo (MCMC) in MrBayes on XSEDE (3.2.7a) (Rannala and Yang, 1996) in the CIPRES Science Gateway web (Ronquist et al., 2012). The model of nucleotide evolution was determined by MrMTgui (Ma, 2016); GTR+I+G was the best-fit model for the ITS and tub2, SYM+I+G was the best-fit model for the LSU, and GTR+G was the best-fit model for tef1-α. Six simultaneous Markov chains were run for 1,000,000 generations and trees were sampled at every 100th generation. Phylogenetic trees were viewed in FigTree v. 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/) (Rambaut, 2012) and formatted by using Adobe Illustrator CS v. 5.
Registration of novel taxa
Newly introduced taxa were registered at the Index Fungorum (2024) and the identifiers were obtained, fulfilling the requirements as mentioned in Art. F.5.1 (International Code of Nomenclature for Algae, Fungi and Plant).
Results
Phylogenetic results
In this study, we selected 145 strains for the phylogenetic analysis and Neoarthrinium urticae (IMI 326344) as the outgroup taxon. In the phylogenetic analysis, the final alignment consisted of 2,099 characters in total, including gaps (ITS: 1–503 bp, LSU: 504–1,299 bp, tef1-α: 1,300–1,670 bp, tub2: 1,671–2,099 bp). The RAxML analysis of the combined dataset yielded the best scoring tree (Figure 1) with a final ML optimization likelihood value of −26,132.916506. The matrix had 1,073 distinct alignment patterns, with 14.23% undetermined characters or gaps. The estimated base frequencies were as follows: A = 0.231558, C = 0.249603, G = 0.260882, and T = 0.257956; substitution rates AC = 1.277568, AG = 3.415629, AT = 1.036259, CG = 0.978503, CT = 5.039154, and GT = 1.000000; and gamma distribution shape parameter α = 0.556683.

Figure 1. Phylogram retrieved from RAxML of Apiospora species using the combined dataset of LSU, ITS, tef1-α, and tub2 gene regions. The statistical values are provided at nodes as ML/PP (ML value equal to or above 60% and BI value equal to or above 0.90). The tree is rooted with Neoarthrinium urticae (IMI 326344). Ex-types and new strains are indicated by the superscript “T” and red respectively.
Furthermore, according to the phylogenetic results, two isolates (GMBCC1011 and GMBCC1012) have a close affinity to Ap. garethjonesii (KUMCC 16-0202, ex-type) with 90% ML and 0.99 BP bootstrap support. Two other isolates, GMBCC1022 and GMBCC1023, had a close affinity to Ap. guangdongensis (ZHKUCC 23-0004, ex-type). While GMBCC1019 and GMBCC1020 formed a distinct lineage with 100% ML bootstrap support and 1.00 posterior probability in BI analysis, GMBCC1017 and GMBCC1018 with GMBCC1015 and GMBCC1016 formed a sister branch with 99% ML and 1.00 BP statistical support. The isolate GMBCC1021 clustered with Ap. globosa (KUNCC 23-14210, ex-type) with 100% ML and 1.00 BP. The collection GMB-W1024 has a high similarity with Ap. subglobosa (MFLUCC 11-0397, ex-type) with 100% ML and 1.00 BP. Two isolates, GMB-W1013 and GMB-W1014, formed a distinct branch with Ap. multiloculata (MFLUCC 21-0023, ex-type) with 100% ML and 1.00 posterior probability in BI analysis (Figure 1).
Taxonomic descriptions
Apiospora Sacc., Atti Soc. Veneto-Trent. Sci. Nat., Padova, Sér. 4 4: 85 (1875); Index Fungorum Registration Identifier: IF264; Classification: Apiosporaceae, Amphisphaeriales, Sordariomycetes.
Apiospora dehongensis L.S. Han & D.Q. Dai, sp. nov. (Figure 2)
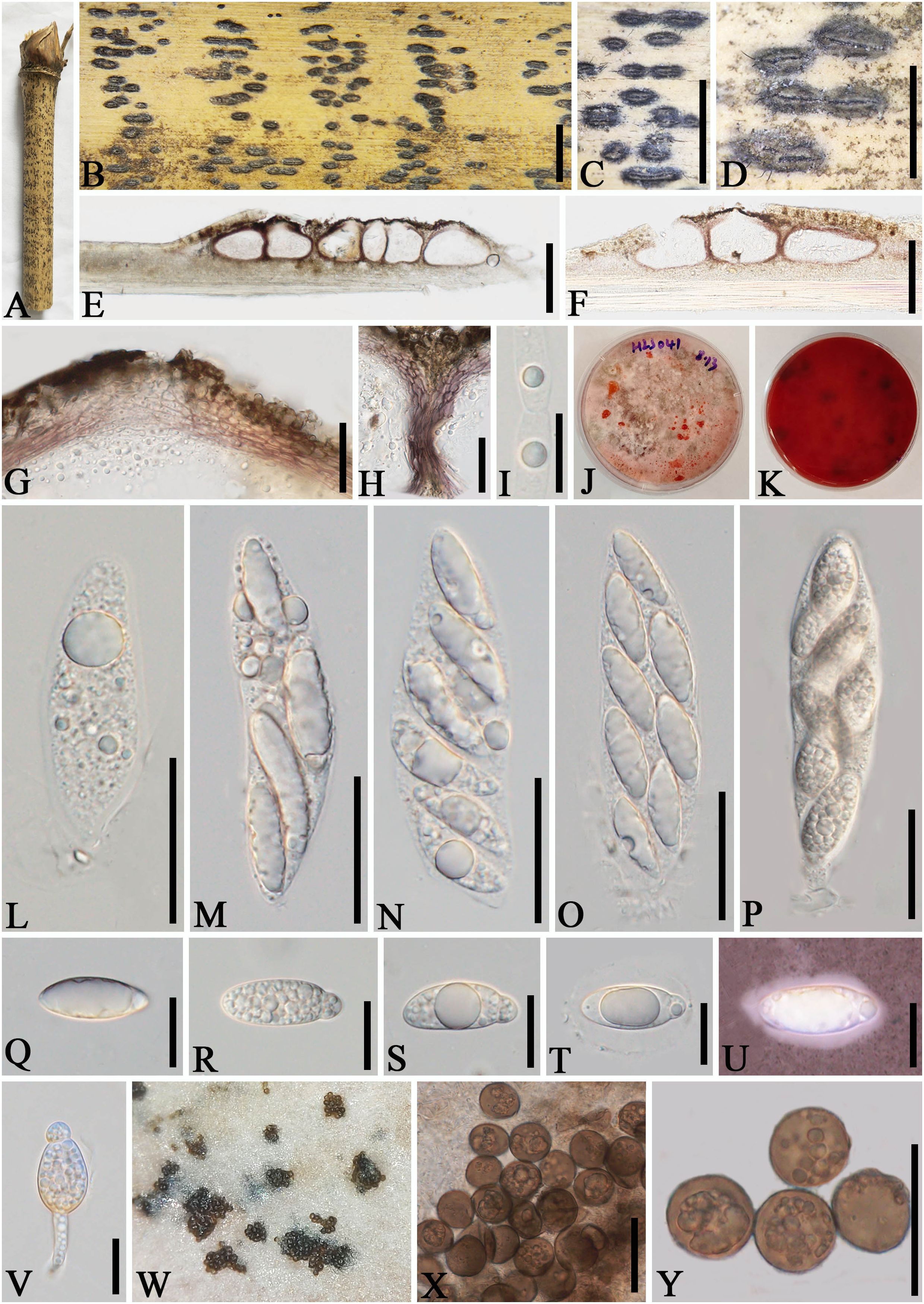
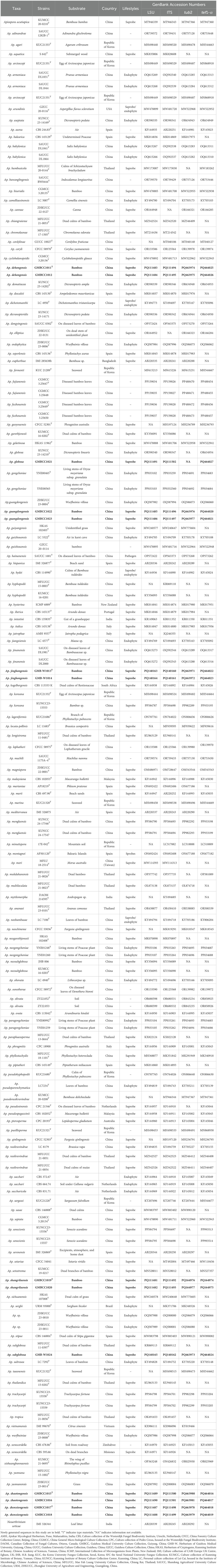
Figure 2. Apiospora dehongensis (GMB-W1011, holotype). (A) Bamboo specimen. (B–D) Stromata developing on bamboo branches. (E, F) Vertical sections of stromata. (G, H) Peridium. (I) Paraphyses. (J, K) Cultures on PDA with red pigmentation [upper (J), reverse (K)]. (L–P) Asci. (Q–T) Ascospores. (U) Ascospore stained in Indian ink showing gelatinous sheath. (V) A germinating ascospore. (W) Conidia formed in culture. (X, Y) Conidia. Scale bars: (B–D) 2 mm, (E) 200 μm, (F) 150 μm, (G, H, L–P, X, Y) 30 μm, and (I, Q–V) 15 μm.
Index Fungorum Identifier: IF902463
Etymology: Named after the location “Dehong” where the new taxon was collected.
Description: Saprobic on dead branches of bamboo. Sexual morph: Stromata 0.4–2.5 mm long, 150–400 µm wide, 125–140 µm high, dark brown, fusiform or naviculate, raised on the host surface, with a slit-like opening at the top center, immersed, scattered to gregarious, or forming groups, uniloculate to multi-loculate. Locules 100–225 μm diameter × 95–130 μm high ( = 145 × 105 µm, n = 20), gregarious, clustered, immersed in stromata, arranged in a row, obpyriform to subglobose, ostiolate at center with periphyses, membranous. Peridium 5–25 μm wide, composed of brown to purple to hyaline cells of textura angularis. Hamathecium 2.5–6.5 μm wide, filamentous, septate, unbranched, constricted at the septum. Asci 85–110 × 14–20 μm ( = 97.5 × 17.5 μm, n = 20), 6-(8)-spored, unitunicate, clavate, apedicellate, apically rounded, straight to slight curved. Ascospores 25–30 × 10–12 μm ( = 27 × 10.8 μm, n = 20), 1–3-seriate, ellipsoidal, 2-celled, with a large upper cell and a smaller lower cell, with guttules, hyaline, smooth-walled, with a gelatinous sheath. Asexual morph: Conidiophores and conidiogenous cells were not observed. Conidia forming on culture, 13–17.5 µm ( = 16 µm, n = 20), globose to subglobose, dark brown, unicellular, smooth-walled, with guttules.
Culture characteristics: Ascospores germinate on PDA within 24 h. Colonies reached 60 mm after 20 days at 27 °C. The colonies are flat, white to reddish and produce red pigment on agar medium.
Material examined: CHINA, Yunnan Province, Dehong, Ruili, Jiexiang town (23°97′17″ N, 97°73′03″ E, 926 m), on dead branches of bamboo, 23 July 2023, L.S. Han & D.Q. Dai, HLS41 (GMB-W1011, holotype), ex-type GMBCC1011; ibid. (MHZU 24-0623, isotype), ex-isotype ZHKUCC 24-1160; ibid. HLS90 (GMB-W1012, isotype), ex-isotype GMBCC1012.
Notes: Phylogenetic analyses showed that newly generated strains GMBCC1011 and GMBCC1012 formed a sister branch to Ap. garethjonesii (D.Q. Dai & H.B. Jiang) Pintos & P. Alvarado (KUMCC 16-0202, ex-type) with 90% ML and 0.99 BI support (Figure 1). However, tef1-α and tub2 data of Ap. garethjonesii (KUMCC 16-0202, ex-type) are unavailable in GenBank. Morphologically, Ap. dehongensis differs from Ap. garethjonesii in having smaller asci (85–110 × 14–20 μm vs. 125–154 × 35–42 μm) and ascospores (25–30 × 10–12 μm vs. 30–42 × 11–16 μm) (Dai et al., 2016). Moreover, our cultures produced red pigment on PDA (Figures 2J, K), which was not observed in Ap. garethjonesii (Dai et al., 2016). Hence, based on morphological and culture characteristics and DNA sequence analyses, we introduce our new collection as a new species, viz., Ap. dehongensis.
Apiospora jinghongensis L.S. Han & D.Q. Dai, sp. nov. (Figure 3)
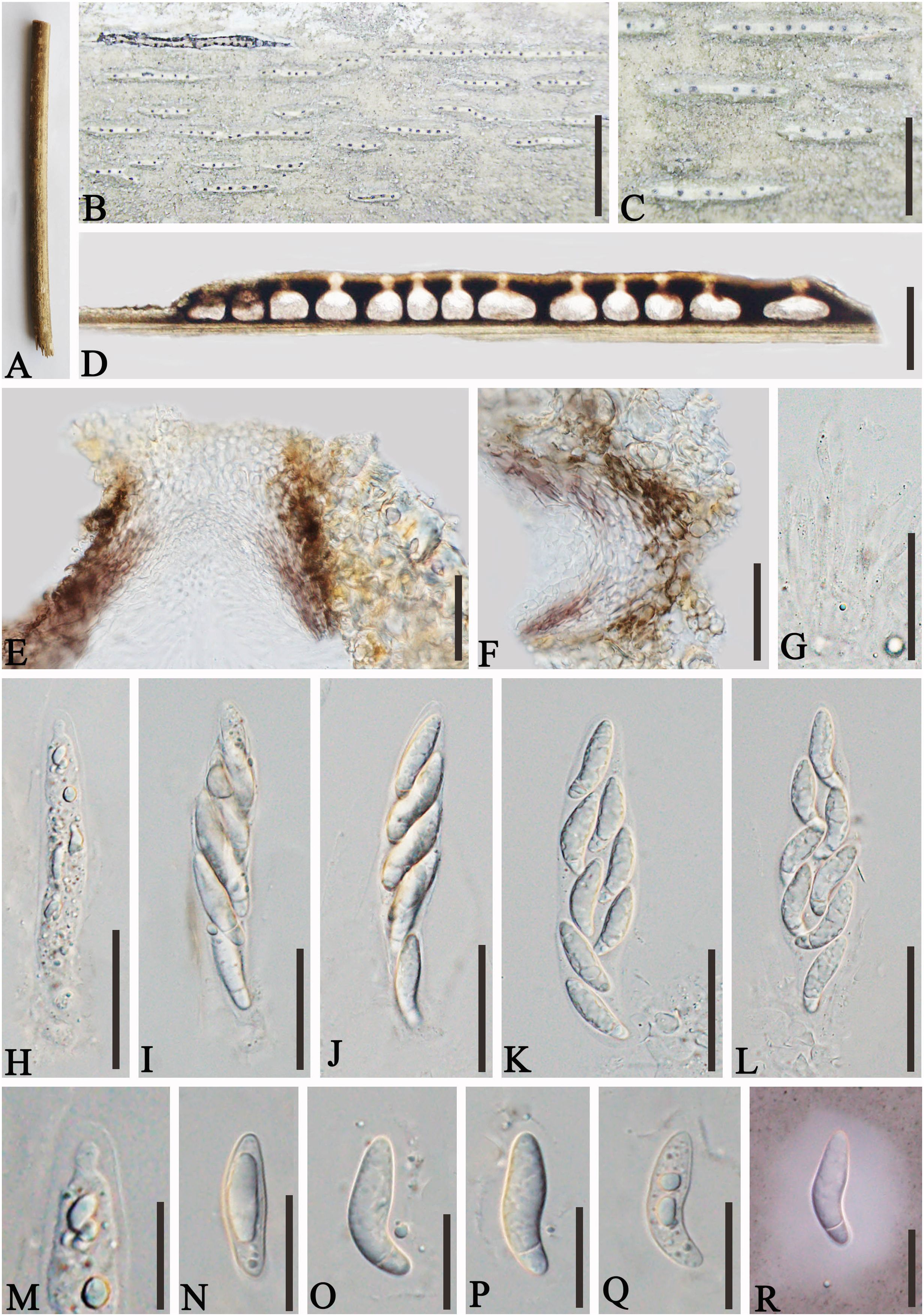
Figure 3. Apiospora jinghongensis (GMB-W1013, holotype). (A) Bamboo specimen. (B, C) Stromata developing on bamboo branches. (D) Vertical sections of stromata. (E, F) Peridium. (G) Paraphyses. (H–L) Asci. (M) Ascus with the rounded and smooth apex. (N–R) Ascospores [ascospore stained in Indian ink showing gelatinous sheath (R)]. Scale bars: (B) 2 mm, (C) 1 mm, (D) 300 μm, (E–L) 30 μm, and (M–R) 15 μm.
Index Fungorum Identifier: IF902464
Etymology: Named after the location “Jinghong” where the new taxon was collected.
Description: Saprobic on dead branches of bamboo. Sexual morph: Stromata 0.6–5.5 mm long, 250–400 μm wide, 175–200 high, filiform, with parallel black spots, raised when mature, still under the host surface, scattered to gregarious, 2–20-loculate. Locules 110–240 μm diameter × 150–185 μm high ( = 160 × 170 µm, n = 20), clustered, gregarious, immersed in stromata, forming groups, arranged in a row, ampulliform to obpyriform, usually with flattened base, brown to dark brown, membranous, with a periphysate ostiole in the center. Peridium 15–40 μm thick, composed of several layers, brown to hyaline cells of textura angularis. Hamathecium 1.5–4 μm wide, filamentous, hyaline, septate, unbranched paraphyses. Asci 85–105 × 13–20 μm ( = 91.5 × 16 μm, n = 20), 8-spored, unitunicate, clavate to cylindrical, apically rounded, slightly curved, short pedicel. Ascospores 22–28 × 6.5–7.5 μm ( = 23 × 6.7 μm, n = 20), biseriate, ellipsoidal, 1-septate, upper cell larger, and lower cell smaller, hyaline, smooth-walled, rounded at both ends, curved at the bottom, surrounded a gelatinous sheath. Asexual morph: Undetermined.
Material examined: CHINA, Yunnan Province, Xishuangbanna, Jinghong city, Manzhang, Mengla (21°91′97″ N, 101°20′42″ E, 617 m), on dead branches of bamboo, 16 August 2020, L.S. Han & D.Q. Dai, DDQ1033 (GMB-W1013, holotype); ibid. (MHZU 24-0624, isotype); ibid. DDQ1033-1 (GMB-W1014, isotype).
Notes: The phylogenetic tree shows that our new collections GMB-W1013 and GMB-W10134 formed a distinct sister branch to Ap. multiloculata Zhang et al. (MFLUCC 21-0023) with 100% ML and 1.00 BI support (Figure 1). Additionally, the nucleotide pairwise of new collections and Ap. multiloculata in ITS, LSU, and tub2 have 6.9% (37/535 bp), 0.8% (7/793 bp), and 9.7% (41/421 bp) differences, respectively. Morphologically, our new collection resembles Ap. multiloculata in having filiform stromata with central ostiole, 1-septate ascospores curved at the lower cell, but differs by having wider locules (110–240 μm vs. 88–160 μm) (Bhunjun et al., 2022). Although the new taxon morphologically resembles Ap. multiloculata, based on multigene phylogenetic analyses, we introduced Ap. jinghongensis.
Apiospora shangrilaensis L.S. Han & D.Q. Dai, sp. nov. (Figure 4)
Figure 4. Apiospora shangrilaensis (GMB-W1019, holotype). (A) Bamboo specimen. (B) Stromata developing on bamboo branches. (C, D) Vertical sections of stromata. (E) Peridium. (F) Paraphyses. (G–J) Asci. (K–P) Ascospores [a ascospore stained in Indian ink showing gelatinous sheath (P)]. (Q) The germinating ascospore. (R, S) Cultures on PDA [upper (R), reverse (S)]. Scale bars: (C) 500 μm, (D, E, G–Q) 50 μm, (F) 15 μm.
Index Fungorum Identifier: IF902465
Etymology: Named after the location “Shangri-La” where the new taxon was collected.
Description: Saprobic on dead culms of bamboo. Sexual morph: Stromata 1–3.2 mm long, 200–400 μm wide, 185–210 μm high, elongated fusiform, raised with long, black axis broken at the apex, immersed, multi-loculate. Locules 200–250 μm diameter × 120–190 μm high ( = 228 × 157.5 µm, n = 20), gregarious, clustered, immersed in stromata, arranged in a row, ampulliform to subglobose with poor development base, pseudothecial, brown to dark brown. Peridium 10–25 µm wide, composed of three to five layers of hyaline to brown, cells of textura angularis. Hamathecium 2.5–4.5 µm wide, long, septate, slightly taping at the top, unbranched paraphyses. Asci 130–150 × 30–40 µm ( = 140 × 36.6 µm, n = 20), 8-spored, unitunicate, clavata, apically rounded, straight to slightly curved, with short pedicel. Ascospores 40–55 × 14–17 µm ( = 46.4 × 15.4 µm, n = 20), 1–2-seriate, ellipsoidal, aseptate when immature, 1-septate when mature, with a larger upper cell, and a smaller lower cell, occasionally with guttules, hyaline, smooth-walled, surrounded by a gelatinous sheath. Asexual morph: Undetermined.
Culture characteristics: Ascospores germinating on PDA within 24 h. Colonies reached 30 mm diameter in 20 days under dark and at 27°C conditions. Colonies flat, circular, cottony, irregular edge, white from above, yellow in the center and outward gradually becoming pale yellow to white from below.
Materials examined: CHINA, Yunnan Province, Shangri-La City, Bigu mountain (27°36′56.9″N, 99°42′6.4″E, 3,460 m), on dead culms of bamboo, 21 July 2020, L.S. Han & D.Q. Dai, DDQ00801 (GMB-W1019, holotype), ex-type, GMBCC1019, ibid. (MHZU 24-0625, isotype), ex-isotype ZHKUCC 24-1161, ibid. DDQ00920 (GMB-W1020), living culture GMBCC1020.
Notes: In the phylogenetic analyses, our new isolates, GMBCC1019 and GMBCC1020, formed a distinct branch (Figure 1). Morphologically, the new species is resembling Ap. hydei (Crous) Pintos & P. Alvarado (CBS 114990, ex-type) in having immersed, multi-loculate ascostromata, unitunicate asci, 1–septate, smooth-walled ascospores with a gelatinous sheath. However, our new collections can be distinguished from Ap. hydei (CBS 114990, ex-type) by having longer and wider asci (130–150 × 30–40 µm vs. 110–130 × 17–24 µm) and wider ascospores (40–55 × 14–17 µm vs. 35–45 × 8.5–11 µm) (Dai et al., 2016; Zeng et al., 2022). Hence, we introduced Ap. shangrilaensis as a new member of Apiospora based on morphological characteristics and phylogeny.
Apiospora zhaotongensis L.S. Han & D.Q. Dai, sp. nov. (Figure 5)
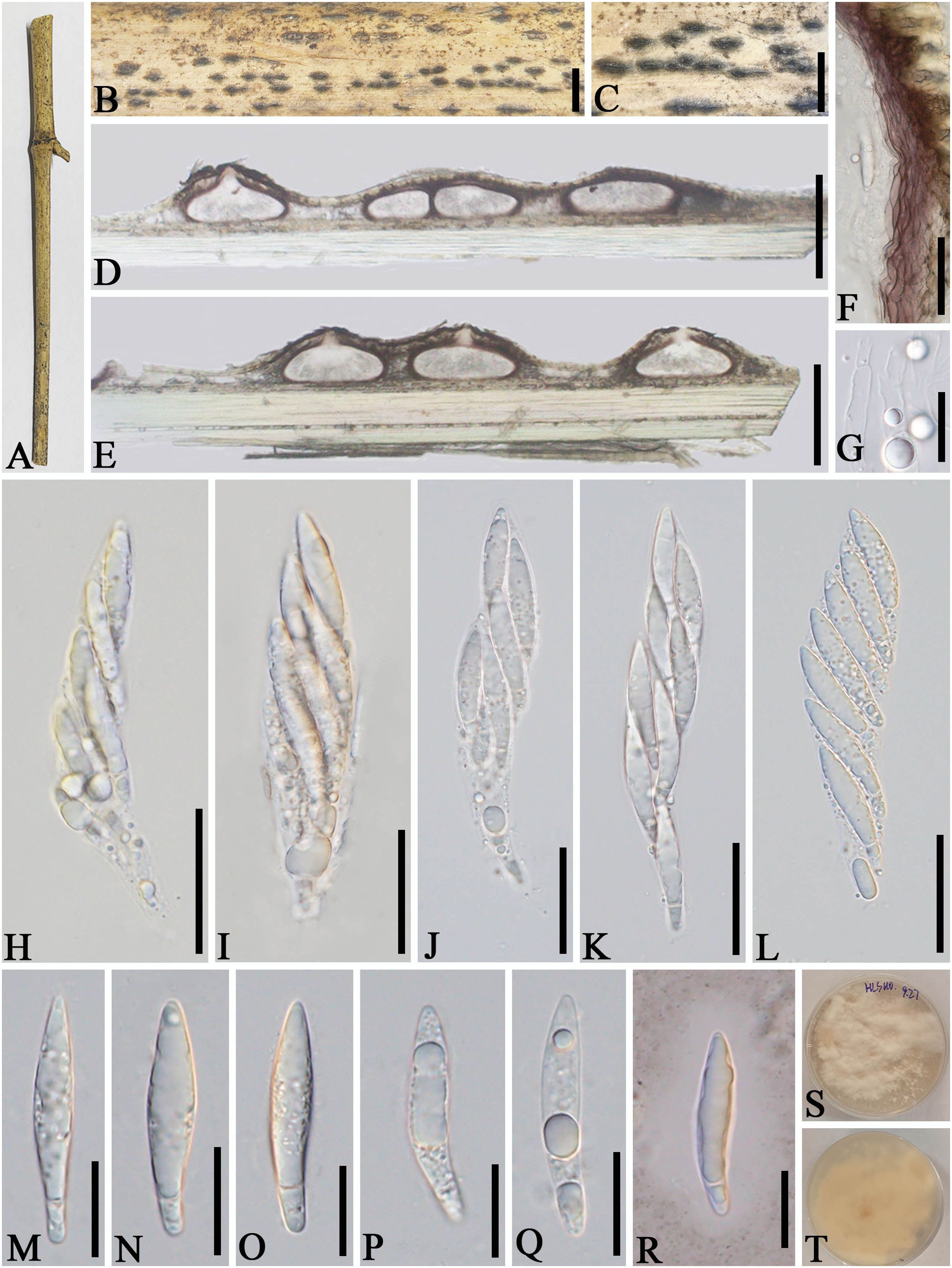
Figure 5. Apiospora zhaotongensis (GMB-W1015, holotype). (A) Bamboo specimen. (B, C) Stromata developing on bamboo branches. (D, E) Vertical sections of stromata. (F) Peridium. (G) Paraphyses. (H–L) Asci. (M–R) Ascospores. (R) The ascospore stained in Indian ink showing gelatinous sheath. (S, T) Cultures on PDA [upper (S), reverse (T)]. Scale bars: (B) 1.5 mm, (C) 1 mm, (D, E) 300 μm, (F, H–L) 30 μm, and (G, M–R) 15 μm.
Index Fungorum Identifier: IF902466
Etymology: Named after the location “Zhaotong” where the new taxon was discovered.
Description: Saprobic on dead branches of bamboo. Sexual morph: Stromata 55–270 μm long, 250–450 μm wide, 140–180 μm high, naviculate or filiform, raised but still under on the host tissue with a slit-like opening at the top, scattered to gregarious, uniloculate to multi-loculate, black. Locules 150–290 diameter × 100–170 μm high ( = 250 × 130 µm, n = 20), immersed in stromata, scattered or clustered, dark brown to black, obpyriform to subglobose, with a central ostiole, papillate. Peridium 25–45 μm thick, composed of several layers, dark brown to hyaline cells of textura angularis. Hamathecium 2.5–4.5 μm wide, filamentous distinctly septate, constricted at the septum, unbranched paraphyses, with guttules. Asci 85–110 × 14–20 μm ( = 97.5 × 17.5 μm, n = 20), 8-spored, unitunicate, cylindrical, straight to slightly curved, apically rounded, apedicellate. Ascospores 30–40 × 6–7.5 μm ( = 36 × 6.5 μm, n = 20), overlapping, 2-seriate, 1-sepetate, conical at both ends, with a larger upper cell and a smaller lower cell, some with guttules, hyaline, smooth-walled, mostly straight, sometimes slightly curved, surrounded by a gelatinous sheath. Asexual morph: Undetermined.
Culture characteristics: Ascospores germinate on PDA within 24 h. Colonies reached 60 mm after 20 days at 27°C. The colonies are white, fluffy, cottony, with irregular edge.
Material examined: CHINA, Yunnan Province, Zhaotong City, Zhenxiong Town (27°62′52″N, 104°81′98″E), on dead branches of bamboo, 4 August 2023, L.S. Han & D.Q. Dai, HLS110 (GMB-W1015, holotype), ex-type GMBCC1015; ibid. (MHZU 24-0626, isotype), ex-isotype, ZHKUCC 24-1162; ibid. HLS110-1 (GMB-W1016, isotype), GMBCC1016 (ex-isotype).
Notes: Two newly generated strains, GMBCC1015 and GMBCC1016, are phylogenetically close to Ap. zhenxiongensis (GMBCC1017 ex-type, GMBCC1018) (Figure 1). Morphologically, the new taxon can be distinguished from Ap. zhenxiongensis in having ascospores conical at both ends, whereas Ap. zhenxiongensis ascospores are rounded at both ends. Moreover, Ap. zhaotongensis has straighter ascospores at the bottom than Ap. zhenxiongensis. Therefore, we introduce a novel species, Ap. zhaotongensis, to accommodate our new collection based on morphology and phylogeny.
Apiospora zhenxiongensis L.S. Han & D.Q. Dai, sp. nov. (Figure 6)
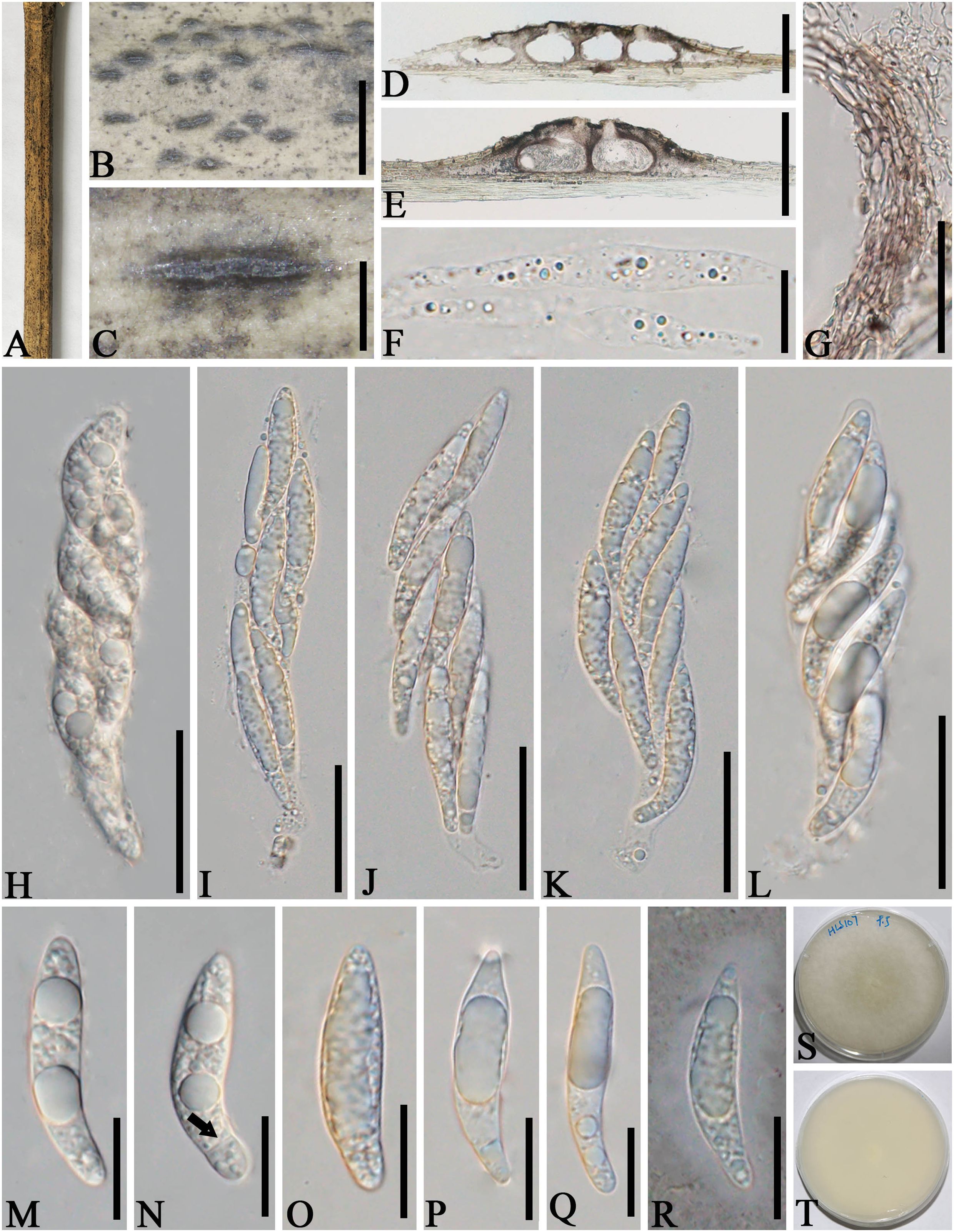
Figure 6. Apiospora zhenxiongensis (GMB-W1017, holotype). (A) Bamboo specimen. (B, C) Stromata developing on bamboo branches. (D, E) Vertical sections of stromata. (F) Paraphyses. (G) Peridium. (H–L) Asci. (M–R) Ascospores [a ascospore stained in Indian ink showing gelatinous sheath (R)]. (S, T) Cultures on PDA [upper (S), reverse (T)]. Scale bars: (B) 1.5 mm, (C) 500 μm, (D, E) 300 μm, (F, M–R) 15 μm, and (G, H–L) 30 μm.
Index Fungorum Identifier: IF902467
Etymology: Named after the location “Zhenxiong” where the new taxon was discovered.
Description: Saprobic on dead branches of bamboo. Sexual morph: Stromata 0.45–1.6 mm long, 200–450 μm wide, 140–160 μm high, raised, with a slit-like opening at the top, dark brown to black, scattered to gregarious, naviculate or filiform, multi-loculate. Locules 130–230 μm diameter × 90–150 μm high ( = 183 × 128 µm, n = 20), immersed in stromata, arranged in a row, obpyriform to ampulliform, dark brown to black. Ostiole 30–60 µm wide, 35–65 µm high, with a black papillate. Peridium 5–25 μm thick, composed of several layers of brown cells of textura angularis. Hamathecium 3–8 μm wide, hyaline, septate, unbranched paraphyses. Asci 80–110 × 15–25 μm ( = 95 × 19 μm, n = 20), 8-spored, unitunicate, cylindrical, apically rounded, with short pedicel. Ascospores 30–40 × 6–8.5 μm ( = 34 × 7.5 μm, n = 20), overlapping, biseriate, ellipsoidal, rounded at both ends, 1-sepetate, cell above septa larger than those below, with guttules, hyaline, smooth-walled, distinctly curved at lower cell when mature, with a gelatinous sheath. Asexual morph: Undetermined.
Culture characteristics: Ascospores germinate on PDA within 24 h. Colonies reached 60 mm after 20 days at 27°C. The colonies are flat, white from above and below, dense, circular, cottony, with regular edge.
Material examined: CHINA, Yunnan Province, Zhaotong City, Zhenxiong (27°63′28″ N, 104°81′85″ E, 1,559 m), on dead branches of bamboo, 4 August 2023, L.S. Han & D.Q. Dai, HLS107 (GMB-W1017, holotype), ex-type GMBCC1017; ibid. (MHZU 24-0627, isotype), ex-isotype, ZHKUCC 24-1163; ibid. HLS136 (GMB-W1018), living culture GMBCC1018.
Notes: In our phylogenetic analyses, GMBCC1017 and GMBCC1018 formed a sister branch to Ap. zhaotongensis (GMBCC1015, ex-type and GMBCC1016) (99% ML, 1.00 BP, Figure 1). Morphologically, the new collections can be distinguished from Ap. zhaotongensis in having ascospores with rounded ends at both ends, while Ap. zhaotongensis ascospores have conical ends. Moreover, the ascospores of the new isolate are more curved at the lower cell than Ap. zhaotongensis. Hence, we introduced Ap. zhenxiongensis to accommodate our new collections based on morphological comparisons coupled with molecular data.
Apiospora globosa J.Y. Zhang & Y.Z. Lu, Journal of Fungi 9 (no. 1,096) (2023) (Figure 7)

Figure 7. Apiospora globosa (GMB-W1021) (A) Bamboo specimen. (B, C) Stromata developing on bamboo branches. (D) Vertical sections of stromata. (E) Peridium. (F–J) Asci. (K–O) Ascospores surrounded by a gelatinous sheath. (P) A germinating ascospore. (Q, R) Cultures on PDA [upper (Q), reverse (R)]. Scale bars: (D) 300 μm, (E–J) 50 μm, and (K–P) 20 μm.
Index Fungorum Identifier: IF 901402
Description: Saprobic on dead culms of bamboo. Sexual morph: Stromata 0.45–3.3 mm long, 200–300 µm wide, 260–300 µm high, brown to black, fusiform, with stromata breaking through raised cracks at the black center, immersed, gregarious, multi-loculate. Locules 180–255 μm diameter × 100–240 μm high ( = 222 × 164.5 µm, n = 20), gregarious, clustered, immersed in stromata, arranged in a row, obpyriform to ampulliform, ostiole with periphyses, membranous, brown to dark brown. Peridium 10–50 µm thick, composed of several layers of brown to hyaline, cells of textura angularis. Asci 100–135 × 21–25 µm ( = 115.8 × 22.4 µm, n = 20), 4-(8)-spored, unitunicate, broadly cylindrical to clavate, with a short pedicel, straight to slightly curved, apically rounded. Ascospores 32–40 × 10–12.5 µm ( = 34.6 × 11.3 µm, n = 20), 1–2-seriate, elliptical, 1–septate, with a larger upper cell, and a small lower cell, hyaline, with many guttules, smooth-walled, curved, constricted at the septum, surrounded by an entire gelatinous sheath. Asexual morph: Endophytic in the stems of Dicranopteris linearis, see Zhang et al. (2023a).
Culture characteristics: Ascospores germinate on PDA within 24 h. Colonies reached 55 mm after 20 days at 27°C. The colonies are white to pale reddish from above, pale reddish from below, circular, cottony, flat, spreading, with irregular edge.
Material examined: CHINA, Yunnan Province, Zhaotong City, Zhenxiong town (27°63′36″N, 104°81′84″E, 1,577 m), on dead culms of bamboo, 4 August 2023, L.S. Han & D.Q. Dai, HLS126 (GMB-W1021, first report of the sexual morph), living culture, GMBCC1021.
Notes: Apiospora globosa J.Y. Zhang & Y.Z. Lu was originally described by Zhang et al. (2023a) based on the asexual morph from a healthy stem of Dicranopteris linearis (KUNCC 23-14210, ex-type) collected from Guizhou Province, China. Our phylogenetic results (Figure 1) indicated that the strain GMBCC1021 is identical to the ex-type of Ap. globosa (KUNCC 23-14210, ex-type) with 100% MLBP and 1.00 BYPP statistic support. Moreover, the base pair arrangement of our collections with KUNCC 23-14210 is identical. Hence, in this study, we report the sexual morph of Ap. globosa for the first time.
Apiospora guangdongensis C.F. Liao & Doilom, Journal of Fungi 9 (no. 1,087): 12 (2023) (Figure 8)

Figure 8. Apiospora guangdongensis (GMB-W1022) (A) Bamboo specimen. (B, C) Stromata developing on bamboo branches. (D) Vertical sections of stromata. (E) Peridium. (F) Paraphyses. (G–K) Asci. (L–Q) Ascospore [a ascospore stained in Indian ink showing gelatinous sheath (Q)]. (R) A germinating ascospore. (S, T) Cultures on PDA [upper (S), reverse (T)]. Scale bars: (D) 150 μm, (E, G–K) 30 μm, (F) 10 μm, and (L–R) 15 μm.
Index Fungorum Identifier: IF 225951
Description: Saprobic on dead culms of bamboo. Sexual morph: Stromata 0.4–2.8 mm long, 250–350 mm wide, 130–190 μm high, raised on the host surface, with blackspots on slit-like opening, immersed, scattered to gregarious, 1–5-loculate, fusiform, brown. Locules perithecial, 210–380 μm diameter × 110–180 μm high ( = 269 × 145 µm, n = 20), gregarious, clustered, immersed in stromata, arranged in a row, obpyriform to ampulliform to subglobose. Ostiole central, with periphyses. Peridium 5–25 μm wide, composed of dark brown to purple to hyaline cells of textura angularis. Hamathecium 2.5–4.5 μm wide, hyaline, septate, constricted at the septum, unbranched, not anastomosed paraphyses. Asci 90–120 × 16–21 μm ( = 102 × 18 μm, n = 20), 8-spored, unitunicate, cylindrical, apically rounded, with a short pedicel, slightly curved. Ascospores 26–35 × 6.5–10 μm ( = 31.5 × 8 μm, n = 20), biseriate, ellipsoidal, 2-celled, with a larger upper cell and a smaller lower cell, with guttules, hyaline, smooth-walled, rounded at both ends, with a gelatinous sheath.
Culture characteristics: Ascospores germinate on PDA within 24 h. Colonies reached 60 mm after 20 days at 27°C. The colonies are floccose, white, circular, cottony, with regular edge, no pigment.
Materials examined: CHINA, Yunnan Province, Zhaotong City, Zhenxiong town (27°63′28″ N, 104°81′88″ E, 1,557 m), on dead culms of bamboo, 4 August 2023, L.S. Han & D.Q. Dai, HLS51 (GMB-W1022, first report of the sexual morph), living culture GMBCC1022, GMB-W1023; ibid. HLS133 (GMB-W1023), living culture GMBCC1023.
Notes: Asexually typified, endophytic species, Apiospora guangdongensis C.F. Liao & Doilom (ZHKUCC 23-0004, ex-type) (from the leaves of Wurfbainia villosa) was originally described by Liao et al. (2023) from Guangdong Province, China. Our multi-gene phylogenetic tree (Figure 1) showed that our new isolates GMBCC1022 and GMBCC1023 grouped with Ap. guangdongensis (ZHKUCC 23-0004, ex-type). Moreover, the base pair arrangement of our collections with ZHKUCC 23-0004 was identical. Therefore, we reported the sexual morph of Ap. guangdongensis for the first time in this study.
Apiospora subglobosa (D.Q. Dai & K.D. Hyde) Pintos & P. Alvarado, Fungal Systematics and Evolution 7: 207 (2021) (Figure 9)

Figure 9. Apiospora subglobosa (GMB-W1024) (A) Bamboo specimen. (B–D) Stromata developing on bamboo branches. (E) Vertical sections of stromata. (F) Peridium. (G–K) Asci. (L) Paraphyses. (M–R) Ascospores surrounded by a gelatinous sheath. Scale bars: (B–D) 2 mm, (E) 300 μm, (F–I, K) 30 μm, and (L–R) 15 μm.
Index Fungorum Identifier: IF 837715
See Senanayake et al. (2015) for the description.
Material examined: CHINA, Yunnan Province, Dehong, Mang City, Xuangang town (24°45′41″N, 98°43′83″E, 919 m), on dead culms of bamboo, 22 July 2023, L.S. Han & D.Q. Dai, HLS84 (GMB-W1024, new geographical record in China).
Known distributions: Thailand (Senanayake et al., 2015) and China (this study).
Known hosts: Bamboo (Senanayake et al., 2015, this study).
Notes: Senanayake et al. (2015) introduced Arthrinium subglobosum D.Q. Dai & K.D. Hyde, but later, Pintos and Alvarado (2021) transferred it to Apiospora s. str. as Apiospora subglobosa (D.Q. Dai & K.D. Hyde) Pintos & P. Alvarado. In our phylogenetic tree, our new collection GMB-W1024 clustered with Ap. subglobosa (MFLUCC 11-0397, ex-type) with 100% ML and 1.00 BI support (Figure 1). Morphologically, our new collection is similar to Ap. subglobosa in muti-loculate stromata with black slit-like opening, straight or curved, apical cell large, with smaller basal cell ascospores. However, the asci of GMB-W1024 are narrower than in MFLU 15-0384 (20–25 µm vs. 27–36 µm) (holotype). The ascospores of GMB-W1024 are longer than those of MFLU 15-0384 (25–33 µm vs. 24–28 µm) but narrower (7–10 μm vs. 8.5–12.5 μm), possibly due to the environmental change leading to slight differences in size. Nevertheless, we confirmed that our new collection (GMB-W1024) is Ap. subglobosa based on phylogenetic analyses (Figure 1).
4 Discussion
Fungal diversity in southwestern China is very high and a large number of species are introduced annually (Wijayawardene et al., 2021a; Lu et al., 2024; Du et al., 2023; Zhang et al., 2023b, 2024; Chen et al., 2024; Liu et al., 2023a; Tian et al., 2024; Xu et al., 2024; Zhang et al., 2024). Fungi associated with bamboo is one of the popular research topics among the mycologists in this region and several new species have been published in recent studies (e.g., Bambusicola hongheensis fide Phookamsak et al., 2024, Parabambusicola yunnanensis fide Han et al., 2023, Paramphibambusa bambusicola fide Han et al., 2024). However, a large number of taxa are yet to be discovered in this region and from bamboo plants, although it has been a well-studied host (Wijayawardene et al., 2021b, 2022).
In this study, we introduced five new species, viz., Apiospora dehongensis, Ap. jinghongensis, Ap. shangrilaensis, Ap. zhaotongensis, and Ap. zhenxiongensis, and two new sexual morph reports, viz., Ap. globosa and Ap. guangdongensis. Furthermore, one new geographical record of Ap. subglobosa was also reported based on morphological and multi-locus phylogenetic analyses. All the species were found as saprobic taxa on decaying bamboo branches and culms. Three specimens were collected from western Yunnan Province, China (Dehong), two specimens were obtained from the southwestern part (Xishuangbanna), seven specimens were collected from the northeastern section (Zhaotong), and two specimens were gathered from the northwestern part (Shangri-La), which displayed the highly hidden species richness of Apiospora in the different regions of Yunnan. Jiang et al. (2022b) and Wang et al. (2018) also emphasized the high species richness of bambusicolous ascomycetes in southwest China, with Apiospora as one of the genera with high species diversity. Thus, ongoing research on the genus Apiospora is essential.
According to the recently published studies, 90 species of Apiospora have been reported to have only an asexual morph, 19 species have been reported to have only a sexual morph, and 22 species have both morphs based on molecular data, including this study (Pintos and Alvarado, 2021; Li et al., 2023; Zhang et al., 2023a; Zhao et al., 2023; Ai et al., 2024; Dissanayake et al., 2024; Liu et al., 2024; Tian et al., 2024; Yan and Zhang, 2024; Zhao et al., 2024). Moreover, regarding the reports of Apiospora discovered on bamboo (based on molecular data), 17 species have been identified solely by their asexual morph, while 15 species have sexual morph only. However, 12 species have been reported with both morphs (Pintos and Alvarado, 2021; Liao et al., 2023; Zhao et al., 2023, 2024; Zhang et al., 2023a; Ai et al., 2024; Dissanayake et al., 2024). Therefore, it is necessary to continue studying bambusicolous Apiospora to explore more asexual or sexual morphs of known or unknown species. Furthermore, because some Apiospora species were reported to have only asexual or sexual forms, it is crucial to collect more specimens in nature to clarify the status of these species in the genus Apiospora.
Note that most of the Apiospora species reported from bamboo are saprobes, while only Ap. dongyingensis Liu et al. and Ap. hainanensis Liu et al. have been reported as pathogens (Liao et al., 2023; Liu et al., 2023b). Jiang et al. (2020) emphasized the importance of researching on bambusicolous pathogenic fungi, because pathogenic fungi have the potential to hinder the advancement of the bamboo industry and even lead to ecological problems. So far, more than 190 bambusicolous pathogenic fungi have been discovered (Kuai, 1996). The continuous study of bamboo pathogenic fungi is related to the conservation and utilization of bamboo resources, which is of great significance to the promotion of sustainable development. Thus, more search works on bambusicolous pathogenic fungi are needed.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, ITS: PQ140160, PQ140161, PQ140162, PQ111492, PQ111493, PQ111494, PQ111495, PQ111496, PQ111497, PQ111498, PQ111499, PQ111500, PQ111501, PQ111502; LSU: PQ140163, PQ140164, PQ140165, PQ111481,PQ111482, PQ111483, PQ111484, PQ111485, PQ111486PQ111487 PQ111488, PQ111489, PQ111490, PQ111491; tub2: PQ463971, PQ463972, PQ463973, PQ463974, PQ463975, PQ463976, PQ463977, PQ463978 PQ463979, PQ463980, PQ463981, PQ164976, PQ164977; tef1-α: PQ464016, PQ464017, PQ464018, PQ464019, PQ464020, PQ464021, PQ464022, PQ464023, PQ464024, PQ464025, PQ464026, PQ464027, PQ164974, PQ164975.
Author contributions
L-SH: Writing – original draft, Writing – review & editing, Data curation, Methodology, Software. CL: Writing – review & editing, Data curation, Writing – original draft. D-QD: Funding acquisition, Methodology, Writing – review & editing, Visualization. IP: Methodology, Writing – review & editing, Software, Visualization. AE: Funding acquisition, Methodology, Writing – review & editing. SA-R: Writing – review & editing, Funding acquisition. QL: Writing – review & editing, Funding acquisition, Methodology. NW: Methodology, Writing – review & editing, Data curation, Software.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Yunnan Revitalization Talents Support Plan (Young Talents Program and High-End Foreign Experts Program), the National Natural Science Foundation of China (No. NSFC 32460002 and No. NSFC 32060710), and the Key Laboratory of Yunnan Provincial Department of Education of the Deep-Time Evolution on Biodiversity from the Origin of the Pearl River for supporting this study. The authors further extend their appreciation to the Meemann Chang Academician Workstation in Yunnan Province (202225AF150002), Yunnan Province Young and Middle-aged Academic and Technical Leaders Reserve Talents Program (202305AC350252), and the General Programs of the Provincial Department of Science and Technology (202101BA070001-076). L-SH would like to thank the Faculty of Science and Graduate School, Chiang Mai University, for supporting the tuition fee for MSc. In addition, the authors extend their appreciation to the Researchers supporting Project Number (RSP2024R120) King Saud University, Riyadh, Saudi Arabia.
Conflict of interest
The authors declare the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ai, C. C., Dong, Z. X., Yun, J. X., Zhang, Z. X., Xia, J. W., Zhang, X. G. (2024). Phylogeny, taxonomy and morphological characteristics of Apiospora (Amphisphaeriales, Apiosporaceae). Microorganisms. 12, 1372. doi: 10.3390/microorganisms12071372
Bagherabadi, S., Zafari, D., Anvar, F. G. (2014). First report of leaf spot caused by Arthrinium arundinis on rosemary in Iran. J. Plant Pathol. 96, 4–126. doi: 10.4454/JPP.V96I4.017
Bhunjun, C. S., Niskanen, T., Suwannarach, N., Wannathes, N., Chen, Y. J., McKenzie, E. H., et al. (2022). The numbers of fungi: are the most speciose genera truly diverse? Fungal Diversity. 114, 387–462. doi: 10.1007/s13225-022-00501-4
Capella-Gutiérrez, S., Silla-Martínez, J. M., Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Carbone, I., Kohn, L. M. (1999). A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 91, 553–556. doi: 10.1080/00275514.1999.12061051
Chen, K., Wu, X. Q., Huang, M. X., Han, Y. Y. (2014). First report of brown culm streak of Phyllostachys praecox caused by Arthrinium arundinis in Nanjing, China. Plant Dis. 98, 1274. doi: 10.1094/PDIS-02-14-0165-PDN
Chen, X. M., Tang, X., Ma, J., Liu, N. G., Tibpromma, S., Karunarathna, S. C., et al. (2024). Identification of two new species and a new host record of Distoseptispora (Distoseptisporaceae, Distoseptisporales, Sordariomycetes) from terrestrial and freshwater habitats in Southern China. MycoKeys. 102, 83. doi: 10.3897/mycokeys.102.115452
Clements, F. E., Shear, C. L. (1931). The genera of fungi (New York, USA: H.W. Wilson company publishing), 1–496.
Dai, D. Q., Jiang, H. B., Tang, L. Z., Bhat, D. J. (2016). Two new species of Arthrinium (Apiosporaceae, Xylariales) associated with bamboo from Yunnan, China. Mycosphere. 7, 1332–1345. doi: 10.5943/mycosphere/7/9/7
Dai, D. Q., Phookamsak, R., Wijayawardene, N. N., Li, W. J., Bhat, D. J., Xu, J. C., et al. (2017). Bambusicolous fungi. Fungal Diversity. 82, 1–105. doi: 10.1007/s13225-016-0367-8
Dissanayake, L. S., Samarakoon, M. C., Maharachchikumbura, S. S. N., Hyde, K. D., Tang, X., Mortimer, P. E., et al (2024). Exploring the taxonomy and phylogeny of Sordariomycetes taxa emphasizing Xylariomycetidae in Southwestern China. Mycosphere. 15, 1675–1793. doi: 10.5943/mycosphere/15/1/15
Du, T. Y., Dai, D. Q., Mapook, A., Lu, L., Stephenson, S. L., Suwannarach, N., et al. (2023). Additions to rhytidhysteron (Hysteriales, dothideomycetes) in China. J. Fungi. 9, 148. doi: 10.3390/jof9020148
Feng, Y., Liu, J. K., Lin, C. G., Chen, Y. Y., Xiang, M. M., Liu, Z. Y. (2021). Additions to the genus arthrinium (Apiosporaceae) from bamboos in China. Front. Microbiol. 12, e661281. doi: 10.3389/fmicb.2021.661281
Glass, N. L., Donaldson, G. C. (1995). Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 61, 1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995
Hall, T. (2004). BioEdit (Carlsbad, CA, 92008, USA: Ibis Therapeutics). Available online at: http://www.mbio.ncsu.edu/BioEdit/bioedit.html/ (Accessed 26 Jan 2024).
Han, L. S., Dai, D. Q., Du, T. Y., Wijayawardene, N. N., Promputtha, I., Bhat, D. J., et al. (2023). Taxonomy and phylogenetic studies revealed Parabambusicola yunnanensis sp. nov. (Parabambusicolaceae, Pleosporales) on bamboo from Yunnan, China. Phytotaxa. 589, 245–258. doi: 10.11646/phytotaxa.589.3.3
Han, L. S., Wijayawardene, N. N., Liu, C., Han, L. H., Promputtha, I., Li, Q., et al. (2024). Paramphibambusa bambusicola gen. et. sp. nov., Arecophila xishuangbannaensis and A. zhaotongensis spp. nov. in Cainiaceae from Yunnan, China. MycoKeys. 104, 113. doi: 10.3897/mycokeys.104.117872
Hyde, K. D., Fröhlich, J., Taylor, J. E. (1998). Fungi from palms XXXVI - Refl ections on unitunicate ascomycetes with apiospores. Sydowia. 50, 21–80.
Index Fungorum (2024). Available online at: http://www.indexfungorum.org (Accessed 17 Jan 2024).
Jiang, H. B., Phookamsak, R., Hongsanan, S., Bhat, D. J., Mortimer, P. E., Suwannarach, N., et al. (2022b). A review of bambusicolous Ascomycota in China with an emphasis on species richness in southwest China. Stud. Fungi. 7, 1–33. doi: 10.48130/SIF-2022-0020
Jiang, N., Tian, C. M. (2021). The holomorph of Arthrinium setariae sp. nov. (Apiosporaceae, Xylariales) from China. Phytotaxa. 483, 149–159. doi: 10.11646/phytotaxa.483.2.7
Jiang, N., Liang, Y. M., Tian, C. M. (2020). A novel bambusicolous fungus from China, Arthrinium chinense (Xylariales). Sydowia. 72, 77–83. doi: 10.12905/0380.sydowia72-2020-0077
Jiang, N., Voglmayr, H., Ma, C. Y., Xue, H., Piao, C. G., Li, Y. (2022a). A new Arthrinium-like genus of Amphisphaeriales in China. MycoKeys. 92, 27–43. doi: 10.3897/mycokeys.92.86521
Katoh, K., Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kuai, S. Y. (1996). A check-list of pathogenic bambusicolous fungi of mainland China and Taiwan. J. For. Sci. Technology. 4, 64–71.
Kwon, S. L., Cho, M., Lee, Y. M., Lee, H., Kim, C., Kim, G. H., et al. (2022). Diversity of the bambusicolous fungus apiospora in Korea: discovery of new apiospora species. Mycobiology. 50, 302–316. doi: 10.1080/12298093.2022.2133808
Larsson, A. (2014). AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics. 30, 3276–3278. doi: 10.1093/bioinformatics/btu531
Li, S., Peng, C., Yuan, R., Tian, C. (2023). Morphological and phylogenetic analyses reveal three new species of Apiospora in China. MycoKeys. 99, 297. doi: 10.3897/mycokeys.99.108384
Liao, C. F., Senanayake, I. C., Dong, W., Thilini Chethana, K. W., Tangtrakulwanich, K., Zhang, Y., et al. (2023). Taxonomic and phylogenetic updates on Apiospora: introducing four new species from Wurfbainia villosa and grasses in China. J. Fungi. 9, 1087. doi: 10.3390/jof9111087
Liu, R. Y., Li, D. H., Zhang, Z. X., Liu, S. B., Liu, X. Y., Wang, Y. X., et al. (2023b). Morphological and phylogenetic analyses reveal two new species and a new record of Apiospora (Amphisphaeriales, Apiosporaceae) in China. MycoKeys. 95, 27. doi: 10.3897/mycokeys.95.96400
Liu, X. F., Tibpromma, S., Hughes, A. C., Chethana, K. W. T., Wijayawardene, N. N., Dai, D. Q., et al. (2023a). Culturable mycota on bats in central and southern Yunnan Province, China. Mycosphere. 14, 497–662. doi: 10.5943/mycosphere/14/1/7
Liu, X., Zhang, Z., Wang, S., Zhang, X. (2024). Three new species of Apiospora (Amphisphaeriales, Apiosporaceae) on Indocalamus longiauritus, Adinandra glischroloma and Machilus nanmu from Hainan and Fujian, China. J. Fungi. 10, 74. doi: 10.3390/jof10010074
Lu, W. H., Suwannarach, N., Lumyong, S., Elgorban, A. M., Dai, D. Q., Dutta, A. K., et al. (2024). Molecular phylogeny and morphology reveal two new species of Conocybe (Bolbitiaceae, Agaricales) from southwest China. New Z. J. Botany., 1–18. doi: 10.1080/0028825X.2024.2327117
Ma, X. (2016). Study on complete mitochondrial genome of Cypridopsis vidua and molecular phylogeny of Ostracoda. East China Normal University, Shanghai, China.
Martínez-Cano, C., Grey, W. E., Sands, D. C. (1992). First report of Arthrinium arundinis causing kernel blight on barley. Plant Dis. 76, e1077. doi: 10.1094/PD-76-1077B
Mavragani, D. C., Abdellatif, L., McConkey, B., Hamel, C., Vujanovic, V. (2007). First report of damping-off of durum wheat caused by Arthrinium sacchari in the semi-arid Saskatchewan fields. Plant Disease. 91, e469. doi: 10.1094/PDIS-91-4-0469A
Monkai, J., Phookamsak, R., Tennakoon, D. S., Bhat, D. J., Xu, S., Li, Q., et al. (2022). Insight into the taxonomic resolution of Apiospora: introducing novel species and records from bamboo in China and Thailand. Diversity. 14, 918. doi: 10.3390/d14110918
O'Donnell, K., Kistler, H. C., Cigelnik, E., Ploetz, R. C. (1998). Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. U.S.A. 95, 2044–2049. doi: 10.1073/pnas.95.5.2044
Phookamsak, R., Hongsanan, S., Bhat, D. J., Wanasinghe, D. N., Promputtha, I., Suwannarach, N., et al. (2024). Exploring ascomycete diversity in Yunnan II: Introducing three novel species in the suborder Massarineae (Dothideomycetes, Pleosporales) from fern and grasses. MycoKeys 104, 9–50. doi: 10.3897/mycokeys.104.112149
Pintos, Á., Alvarado, P. (2021). Phylogenetic delimitation of apiospora and arthrinium. Fungal Systematics Evol. 7, 197–221. doi: 10.3114/fuse.2021.07.10
Rambaut, A. (2012). FigTree v1. 4.0. a Graphical viewer of phylogenetictrees. Available online at: http://tree.bio.ed.ac.uk/software/figtree/ (Accessed 3 January 2023).
Rannala, B., Yang, Z. (1996). Probability distribution of molecular evolutionary trees, a new method of phylogenetic inference. J. Mol. Evolution. 43, 304–311. doi: 10.1007/BF02338839
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Saccardo, P. (1875). Conspectus generum pyrenomycetum italicorum additis speciebus fungorum Venetorum novis vel criticis, systemate carpologico dispositorum. Atti Soc Veneziana-Trent.-Istriana Sci. Nat. 4, 77–100.
Samuels, G., McKenzie, E., Buchanan, D. E. (1981). Ascomycetes of New Zealand 3. Two new species of Apiospora and their Arthrinium anamorphs on bamboo. N. Z. J. Bot. 19, 137–149. doi: 10.1080/0028825X.1981.10425113
Senanayake, I. C., Maharachchikumbura, S. S., Hyde, K. D., Bhat, J. D., Jones, E. G., McKenzie, E. H., et al. (2015). Towards unraveling relationships in Xylariomycetidae (Sordariomycetes). Fungal Diversity. 73, 73–144. doi: 10.1007/s13225-015-0340-y
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stamatakis, A., Hoover, P., Rougemont, J. (2008). A rapid bootstrap algorithm for the ML web servers. Syst. Biol. 57, 758–771. doi: 10.1080/10635150802429642
Tian, X. G., Bao, D. F., Karunarathna, S. C., Jayawardena, R. S., Hyde, K. D., Bhat, D. J., et al. (2024). Taxonomy and phylogeny of ascomycetes associated with selected economicallyimportant monocotyledons in China and Thailand. Mycosphere. 15, 1–274. doi: 10.5943/mycosphere/15/1/1
Tian, X. G., Karunarathna, S. C., Mapook, A., Promputtha, I., Xu, J., Bao, D. F., et al. (2021). One new species and two new host records of Apiospora from bamboo and maize in Northern Thailand with thirteen new combinations. Life. 11, 1071. doi: 10.3390/life11101071
Vaidya, G., Lohman, D. J., Meier, R. (2011). Sequence Matrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 27, 171–180. doi: 10.1111/j.1096-0031.2010.00329.x
Vilgalys, R., Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
Wang, M., Tan, X. M., Liu, F., Cai, L. (2018). Eight new Arthrinium species from China. MycoKeys. 1, 1–24. doi: 10.3897/mycokeys.34.24221
White, T., Bruns, T., Lee, S., Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR protocols: a guide to methods and applications. Eds. Innis, M., Gelfand, D., Shinsky, J., White, T. (Academic Press, New York), 315–322 pp.
Wijayawardene, N. N., Dissanayake, L. S., LI, Q. R., Dai, D. Q., Xiao, Y., Wen, T. C., et al. (2021a). Yunnan–Guizhou Plateau: a mycological hotspot. Phytotaxa. 523, 1–31. doi: 10.11646/phytotaxa.523.1.1
Wijayawardene, N. N., Hyde, K. D., Dai, D. Q., Sánchez-García, M., Goto, B. T., Saxena, R. K., et al. (2022). Outline of fungi and fungus-like taxa – 2021. Mycosphere. 13, 53–453. doi: 10.5943/mycosphere/13/1/2
Wijayawardene, N. N., Phillips, A. J. L., Tibpromma, S., Dai, D. Q., Selbmann, L., Monteiro, J. S., et al. (2021b). Looking for the undiscovered asexual taxa; case studies from lesser studied life modes and habitats. Mycosphere. 12, 1186–1229. doi: 10.5943/mycosphere/12/1/17
Xu, R. F., Karunarathna, S. C., Phukhamsakda, C., Dai, D. Q., Elgorban, A. M., Suwannarach, N., et al. (2024). Four new species of Dothideomycetes (Ascomycota) from Pará Rubber (Heveabrasiliensis) in Yunnan Province, China. MycoKeys. 103, 71. doi: 10.3897/mycokeys.103.117580
Yan, X. N., Zhang, C. L. (2024). Three new endophytic Apiospora species (Apiosporaceae, Amphisphaeriales) from China. MycoKeys. 105, 295. doi: 10.1094/PDIS-06-20-1159-PDN
Zeng, Y., Luo, M., Wei, T., Zhang, H., Jia, W., Jiang, Y. (2024). First report of Apiospora hysterina causing leaf spot on faba bean (Vicia faba). Crop Prot. 184, 106778. doi: 10.1016/j.cropro.2024.106778
Zeng, Q., Lv, Y. C., Xu, X. L., Deng, Y., Wang, F. H., Liu, S. Y., et al. (2022). Morpho-molecular characterization of microfungi associated with Phyllostachys (Poaceae) in Sichuan, China. J. Fungi. 8, 702. doi: 10.3390/jof8070702
Zhang, G. Q., Dai, D. Q., Wijayawardene, N. N., Promputtha, I., Bhat, D. J., Dawoud, T. M., et al. (2023b). Taxonomy and phylogeny of Hypoxylon zhaotongensis sp. nov. (Hypoxylaceae, Xylariales), a bambusicolous fungus from Yunnan, China. Phytotaxa. 598, 111–123. doi: 10.11646/phytotaxa.598.2.1
Zhang, J. Y., Chen, M. L., Boonmee, S., Wang, Y. X., Lu, Y. Z. (2023a). Four new endophytic Apiospora species isolated from three Dicranopteris species in Guizhou, China. J. Fungi. 9, 1096. doi: 10.3390/jof9111096
Zhang, X., Karunarathna, S. C., Tibpromma, S., Du, T. Y., Elgorban, A. M., Lumyong, S., et al. (2024). Morphology and phylogenetic analyses reveal Neopalmiascoma gen. nov. (Bambusicolaceae, Pleosporales) on Macadamia integrifolia in Yunnan Province, China. Phytotaxa 633, 230–240. doi: 10.11646/phytotaxa.633.3.3
Zhao, H. J., Dong, W., Shu, Y., Mapook, A., Manawasinghe, I., Doilom, M., et al. (2023). Bambusicolous fungi in Guangdong, China: establishing Apiospora magnispora sp. nov. (Apiosporaceae, Amphisphaeriales) based on morphological and molecular evidence. Curr. Res. Environ. Appl. Mycol. 13, 1–15. doi: 10.5943/cream/13/1/1
Zhao, Z. Z., Mu, T. C., Keyhani, N. O., Pu, H. L., Lin, Y. S., Lv, Z. Y., et al. (2024). Diversity and new species of ascomycota from bamboo in China. J. Fungi. 10, 454. doi: 10.3390/jof10070454
Keywords: Apiosporaceae, phylogeny, saprobes, taxonomy, bambusicolous fungi
Citation: Han L-S, Liu C, Dai D-Q, Promputtha I, Elgorban AM, Al-Rejaie S, Li Q and Wijayawardene NN (2024) Five new species, two new sexual morph reports, and one new geographical record of Apiospora (Amphisphaeriales, Sordariomycetes) isolated from bamboo in Yunnan, China. Front. Cell. Infect. Microbiol. 14:1476066. doi: 10.3389/fcimb.2024.1476066
Received: 05 August 2024; Accepted: 30 October 2024;
Published: 11 December 2024.
Edited by:
Sinang Hongsanan, Shenzhen University, ChinaReviewed by:
Chihiro Kadooka, Sojo University, JapanYanpeng Chen, Helmholtz Association of German Research Centers (HZ), Germany
Copyright © 2024 Han, Liu, Dai, Promputtha, Elgorban, Al-Rejaie, Li and Wijayawardene. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiang Li, TFFAbWFpbC5xam51LmVkdS5jbg==; Dong-Qin Dai, Y2ljaWRhaWRvbmdxaW5AZ21haWwuY29t
†These authors have contributed equally to this work
 Li-Su Han
Li-Su Han Chao Liu
Chao Liu Dong-Qin Dai
Dong-Qin Dai Itthayakorn Promputtha2
Itthayakorn Promputtha2 Abdallah M. Elgorban
Abdallah M. Elgorban Nalin N. Wijayawardene
Nalin N. Wijayawardene