- 1Department of Hospital Infection Prevention and Control, Anqing First People’s Hospital of Anhui Medical University, Anqing, Anhui, China
- 2Department of Clinical Laboratory, The Second Affiliated Hospital of Anhui Medical University, Hefei, Anhui, China
- 3Department of Hospital Infection Prevention and Control, The Second Affiliated Hospital of Anhui Medical University, Hefei, Anhui, China
- 4The Fourth Affiliated Hospital of Anhui Medical University, Hefei, Anhui, China
- 5Department of Laboratory Medicine, Shenzhen People’s Hospital, The Second Clinical Medical College, Jinan University, Shenzhen, Guangdong, China
- 6The First Affiliated Hospital, Southern University of Science and Technology, Shenzhen, Guangdong, China
Objectives: This study describes the detection and tracking of emergency neurosurgical cross-transmission infections with carbapenem-resistant Klebsiella oxytoca (CRKO).
Methods: We conducted an epidemiological investigation and a rapid screening of 66 surveillance samples using the chromogenic selective medium. Two CRKO isolates from infected patients and three from the preoperative shaving razors had similar resistance profiles identified by the clinical laboratory.
Results: The whole genome sequencing (WGS) results identified all isolates as Klebsiella michiganensis (a species in the K. oxytoca complex) with sequence type 29 (ST29) and carrying resistance genes blaKPC-2 and blaOXY-5, as well as IncF plasmids. The pairwise average nucleotide identity values of 5 isolates ranged from 99.993% to 99.999%. Moreover, these isolates displayed a maximum genetic difference of 3 among 5,229 targets in the core genome multilocus sequence typing scheme, and the razors were confirmed as the contamination source. After the implementation of controls and standardized shaving procedures, no new CRKO infections occurred.
Conclusion: Contaminated razors can be sources of neurosurgical site infections with CRKO, and standard shaving procedures need to be established. Chromogenic selective medium can help rapidly identify targeted pathogens, and WGS technologies are effective mean in tracking the transmission source in an epidemic or outbreak investigation. Our findings increase the understanding of microbial transmission in surgery to improve patient care quality.
1 Introduction
Surgical site infections (SSIs), the most prevalent healthcare-associated infections, occur in approximately 0.5% to 3% of surgery patients (Seidelman et al., 2023). Post-neurosurgical infections are a common and very harmful complication requiring re-operation, prolonging hospital stay lengths, and increasing disability and mortality rates (Li et al., 2023). Infections with antibiotic-resistant bacteria after neurosurgery have become an important treatment challenge due to the low blood-brain barrier permeation rate of most antibiotics (Li et al., 2023). Carbapenems are a potent class of broad-spectrum antibiotics with strong antibacterial activity and are considered “last-line” antibiotics (Tompkins and van Duin, 2021). Carbapenem-resistant organisms have emerged and spread worldwide (Nordmann et al., 2011). The World Health Organization (WHO) has classified these organisms as an urgent public health threat (Wyres and Holt, 2022). Carbapenem-resistant organisms are usually multidrug-resistant, extensively drug resistant, and even pandrug-resistant bacteria (Li et al., 2023). The growing prevalence and the considerable morbidity and mortality of carbapenem-resistant organisms have attracted increasing attention in neurosurgery (Li et al., 2023).
Klebsiella species often cause respiratory, urinary tract, and wound infections, and they are increasingly recognized as pathogens of emerging nosocomial infections and outbreaks, following Klebsiella pneumoniae (Guzmán-Puche et al., 2021; Neog et al., 2021). K. oxytoca has been associated with neurosurgical procedures (Tang and Chen, 1995). Recently, with the emergence of bacterial resistance to carbapenems, carbapenem-resistant K. oxytoca (CRKO) has been identified as a species associated with neurosurgical procedures (Guanghui et al., 2020); however, transmission routes of CRKO in neurosurgery have rarely been reported.
Whole genome sequencing (WGS) technologies provide rich data and capture numerous features of isolates; data helpful for identifying resistance and virulence genes, mobilizable plasmids, and evolutionary information, as well as for epidemiological tracking and pathogen surveillance (Mustafa, 2024). This study investigated neurosurgical cross-transmission infections with CRKO by applying rapid chromogenic selective medium and WGS technologies to track the source of cross-transmission and improve surgical patient care.
2 Materials and methods
2.1 Setting and epidemiological investigation
This investigation was conducted in a tertiary teaching hospital with 1,460 beds in Anqing (Anhui Province of China). Two patients with SSIs were reported in the neurosurgical intensive care unit (ICU) on October 8, 2023 after emergency neurosurgery. Two CRKO isolates with similar resistance profiles were isolated from the neurosurgical sites of these two infected patients, and an cross-transmission was suspected. We investigated the transmission and conducted an active surveillance to detect the source of infections and control the spread. The Ethical Committee of The Second Affiliated Hospital of Anhui Medical University approved the study protocol (YX2023-102).
2.2 Active surveillance cultures and microbiological methods
According to information from epidemiological investigation and interviews of medical staff in the operating room and neurosurgical ICU, we collected active surveillance samples from the potential related patients, medical staff (healthcare workers, shaver, cleaners), neurosurgery disinfectants, shared environmental and device surfaces (hanging towers, bedrails, infusion pumps, cardiovascular monitors, etc.), and equipment for preoperative shaving (such as razors and shampoo) in the operating room and neurosurgical ICU involved. We inoculated CHROMagar mSuperCARBA (CHROMagar™, France) medium, a chromogenic selective medium with 95.6% to 96.5% sensitivity to detect carbapenemase producers, with surveillance samples to rapidly screen for carbapenemase-producing organisms (Alizadeh et al., 2018). After 24 h of culture at 35°C ± 2°C, the blue colonies growing on the selective agar suggested the potential presence of CRKO. Then the suspected isolates were further identified via matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS, Germany). Antimicrobial susceptibility was tested using the VITEK2 drug sensitivity analyzer (bioMérieux, Marcy L’Étoile, France), and K. pneumoniae ATCC BAA-1705 and Escherichia coli ATCC 25922 as quality control strains. Testing results were interpreted referring to the performance standards of the Clinical and Laboratory Standards Institute (CLSI) guidelines (Wayne, 2019).
2.3 Whole genome sequencing and genome-based analysis
WGS was performed for detected CRKO isolates from patients with SSIs and from surveillance samples. The concentration and quality of genomic DNA of these isolates extracted by the cetyltrimethylammonium bromide (CTAB) method were determined using a Qubit fluorometer (Invitrogen, USA) and a NanoDrop spectrophotometer (Thermo Scientific, USA). Sequencing libraries were generated using the TruSeq DNA Sample Preparation Kit (Illumina, USA) and the Template Prep Kit (Pacific Biosciences, USA). WGS was ran on an Illumina Novaseq platform at Personal Biotechnology Company (Shanghai, China). After using AdapterRemoval and SOAPec to remove adapter sequences and filter data (Lindgreen, 2012; Luo et al., 2012), the reads were assembled by the SPAdes and A5-miseq for constructing scaffolds and contigs (Bankevich et al., 2012; Coil et al., 2015). Finally, the genome sequences were obtained after correction using the Pilon software (Walker et al., 2014). Species was identified according to the PubMLST website (https://pubmlst.org/). Multilocus sequence typing (MLST) was performed via the MLST 2.0 at the Center for Genomic Epidemiology (https://cge.food.dtu.dk/services/MLST/). Resistance genes were determined using ResFinder (http://genepi.food.dtu.dk/resfinder). Plasmids were identified with PlasmidFinder (https://cge.food.dtu.dk/services/PlasmidFinder/). Virulence factors were determined in the virulence factor database (http://www.mgc.ac.cn/cgi-bin/VFs/v5/main.cgi). Pair-wise average nucleotide identity (ANI) values were calculated using the orthologous average nucleotide identity tool (OAT), using the proposed cut-off value of 95.0–96.0% for species demarcation (Lee et al., 2016). In addition, SeqSphere+ version 10.0 (Ridom, Münster, Germany) (https://www.ridom.de/news/) and the publicly available core genome MLST (cgMLST) scheme for K. oxytoca were used to calculate the minimum spanning tree (MST). A Klebsiella michiganensis isolate “Control”, which was isolated from a patient in the ICU of the same hospital on October 21, 2023, was used as the reference strain in the cgMLST analysis. Isolates with less than 9 allelic differences from 5,229 targets of the cgMLST scheme were defined as highly related (Dabernig-Heinz et al., 2024).
3 Results
3.1 Transmission investigation
Surveillance data indicated that none of the 36 patients who underwent neurosurgery in August 2023 developed SSIs. However, two of 23 patients (8.7%) undergoing neurosurgery in the neurosurgical ICU developed SSIs due to CRKO in September 2023. The two CRKO isolates with similar resistance profiles were isolated from cerebrospinal fluid and wound effusion in patients who had undergone emergency neurosurgery on September 27, 2023. Detailed characteristics of the two patients are presented in Table 1.
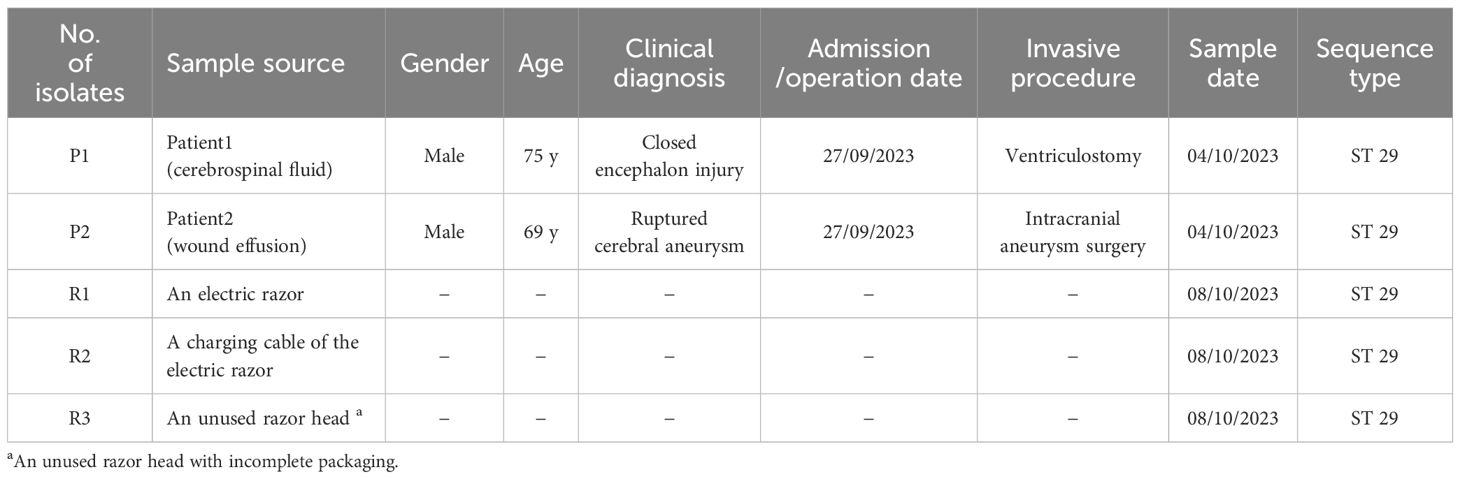
Table 1. Epidemiological and clinical characteristics of 5 Carbapenem-resistant Klebsiella oxytoca (CRKO) isolates from patients and razors.
We collected 66 samples through active surveillance and detected 10 isolates (Table 2). Among them, three CRKO isolates were obtained from a preoperative electric shaving razor, its charging cable, and an unused razor head with an incomplete packaging. The traditional preoperative shaving procedure in the hospital involved using a unique electric razor head for each patient, and disinfecting the handle of the electric razor before each use. Besides, any unused razor heads and disinfected handles were stored together. We discovered that the two patients with SSIs both had undergone preoperative shaving with these electric razors. Therefore, we suspected contaminated razors during preoperative shaving procedure as the source of transmission.
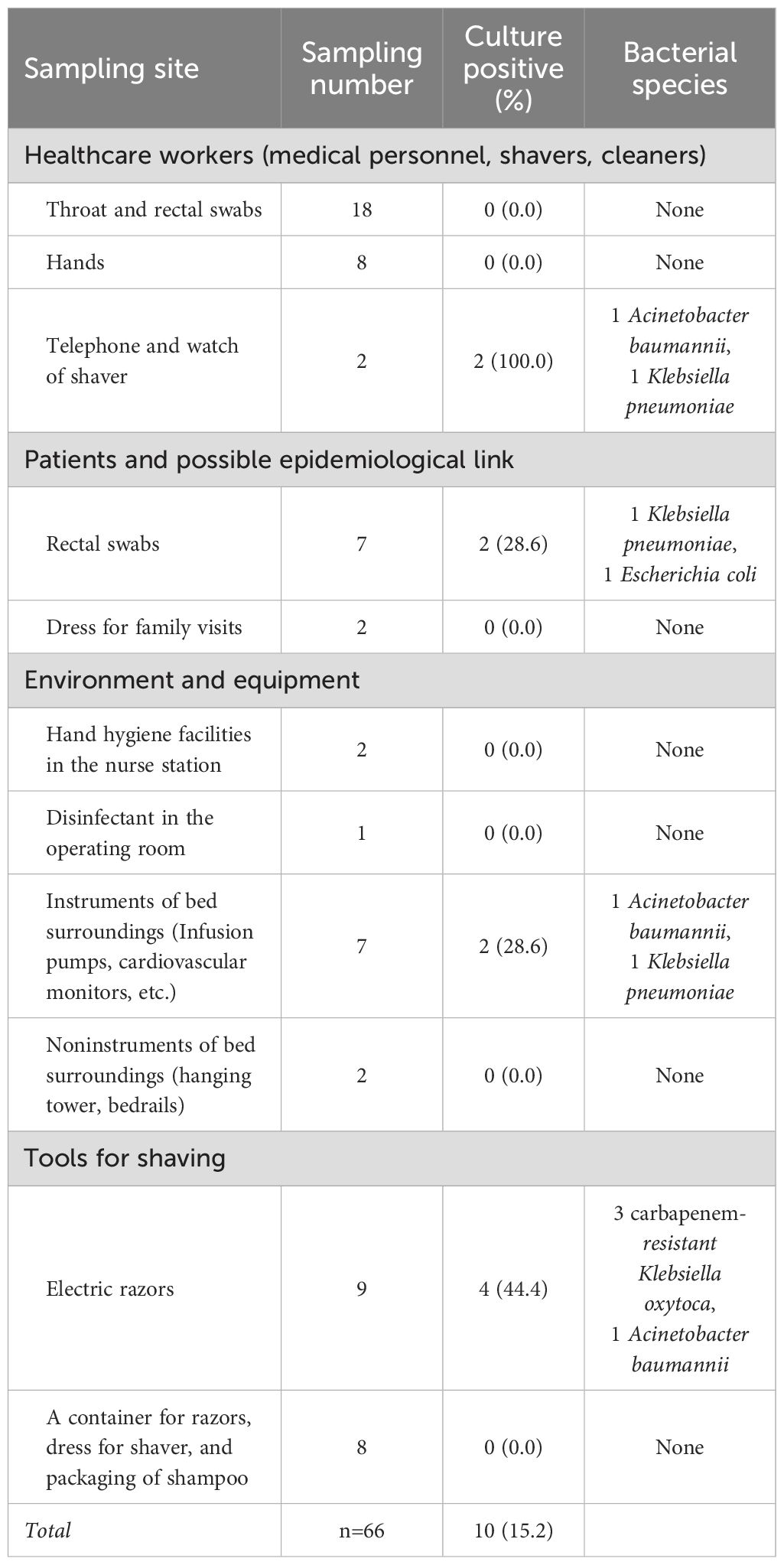
Table 2. Results of environment, healthcare worker, and patient sampling during this transmission investigation.
3.2 Implementation of control measures
Upon confirmation of CRKO infections, the two patients were immediately isolated and treated with antibiotic therapy. While investigating the transmission, additional infection control procedures were strengthened including hand hygiene, cleaning, disinfection, sterilization, surveillance cultures, and re-education of healthcare staff. Following the detection of CRKO isolates solely on the razors, when necessary, trained nurses were instructed to use disposable clippers for preoperative shaving. These cut hair close to the skin, leaving a short stubble without touching the skin. Moreover, the department of hospital infection prevention and control regularly supervised standardized preparation procedures. Finally, after the implementation of these control measures, no other patients developed SSIs with CRKO. The two infected patients were successfully treated and discharged.
3.3 Analysis of antibiotic susceptibility, resistance and virulence genes, plasmids, and genetic relatedness
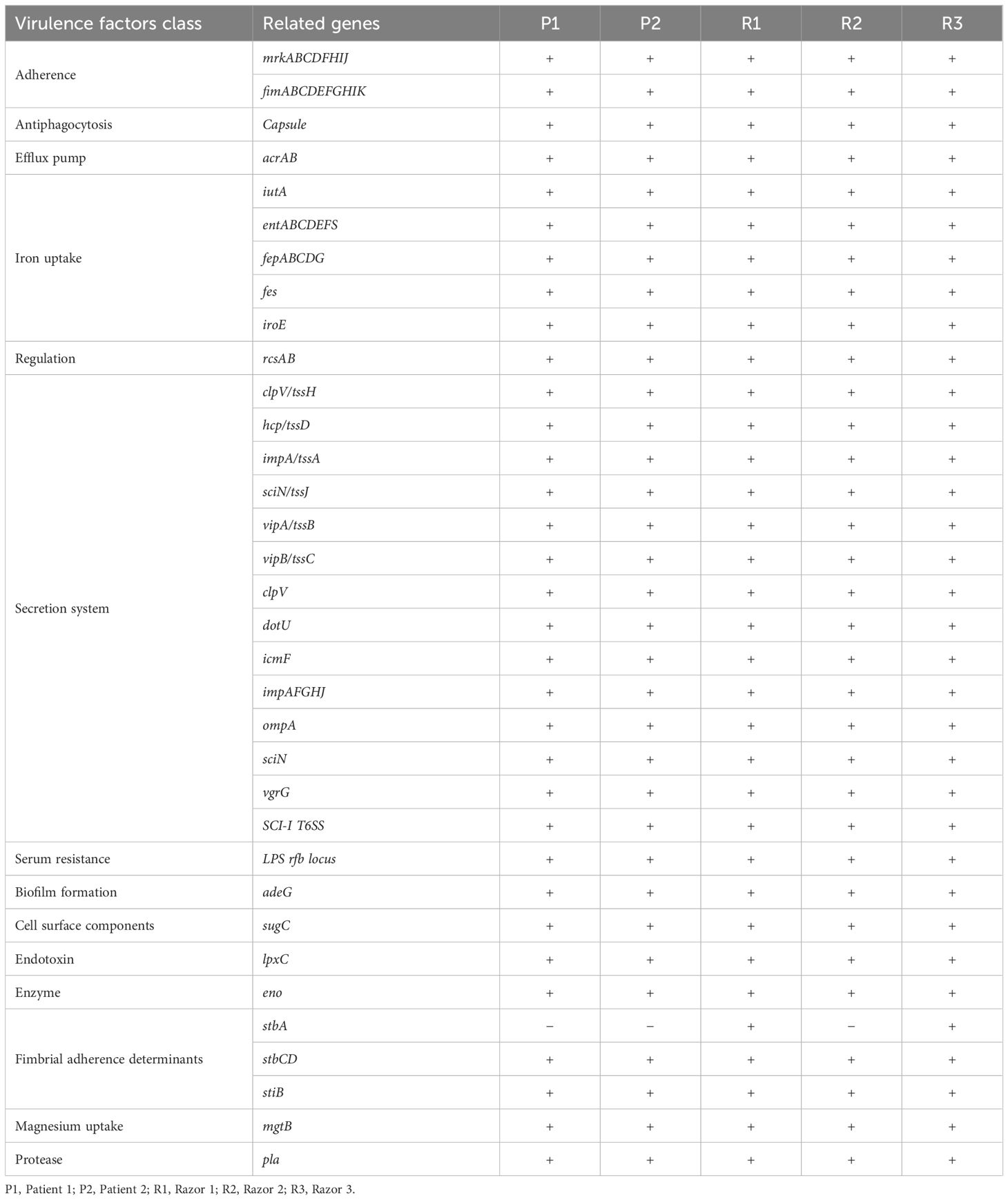
Two CRKO isolates from infected patients and three from razors had consistent resistance patterns based on the antibiotic susceptibility testing results identified by the clinical laboratory. All the isolates were resistant to ticarcillin/clavulanic acid, piperacillin/tazobactam, aztreonam, imipenem, and meropenem (Table 3). WGS results identified these isolates as K. michiganensis (a species in the K. oxytoca complex) with sequence type 29 (ST29). All isolates carried two resistance genes (blaKPC-2 and blaOXY-5), as well as IncF plasmids (IncFIB(K) and IncFII(K)). Two isolates carried 67 virulence genes, and three isolates carried 66 virulence genes but lacked stbA (Table 4). The pairwise ANI values among the genomes of the 5 isolates ranged from 99.993% to 99.999% (Figure 1). Besides, the minimum spanning tree displayed a maximum difference of 3 among 5,229 targets in the core genome scheme between 5 isolates, whereas these 5 isolates had a significant allele difference (more than 92) to the reference strain “Control” (Figure 2), revealing the close relatedness between the isolates from patients and razors.
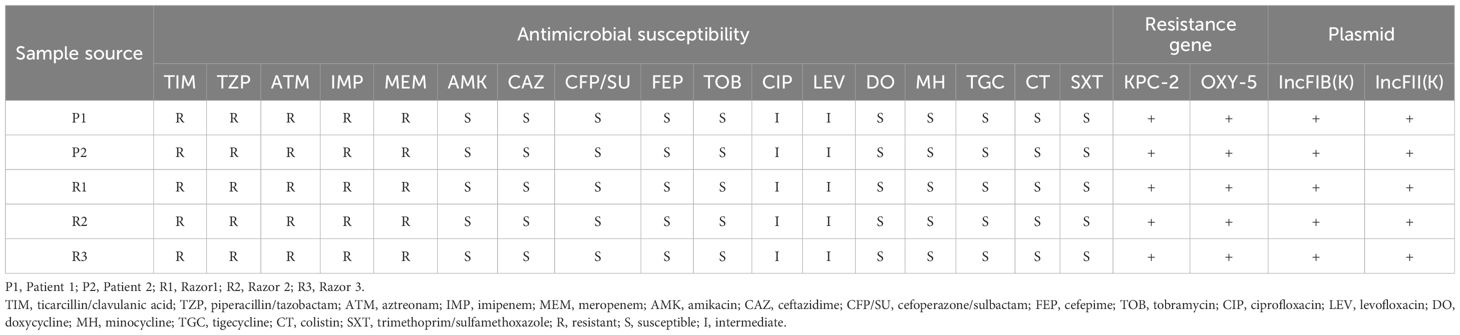
Table 3. Antimicrobial resistance phenotypes, resistance genes, and plasmids of 5 CRKO isolates from infected patients and razors.
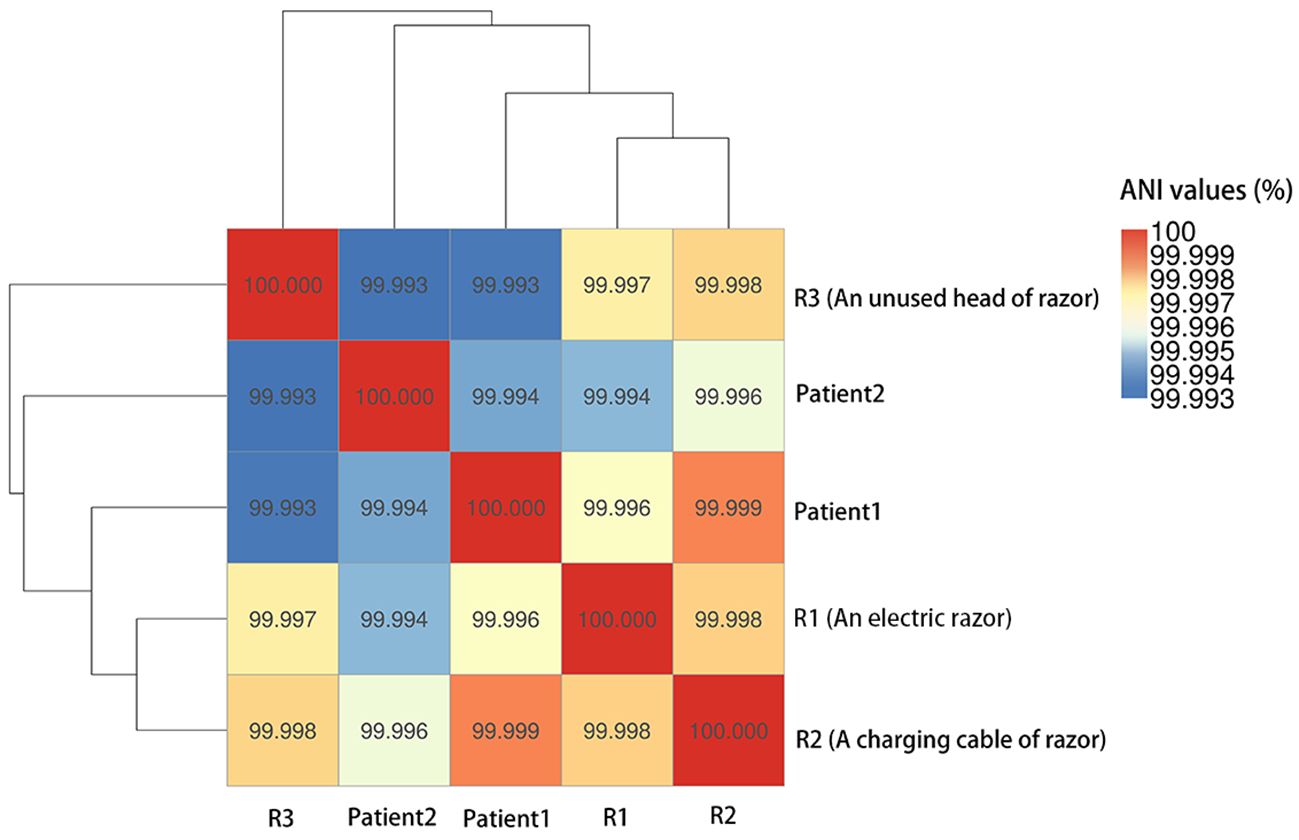
Figure 1. Pairwise average nucleotide identities (ANI) values among the genomes of the 5 CRKO isolates from infected patients and razors.
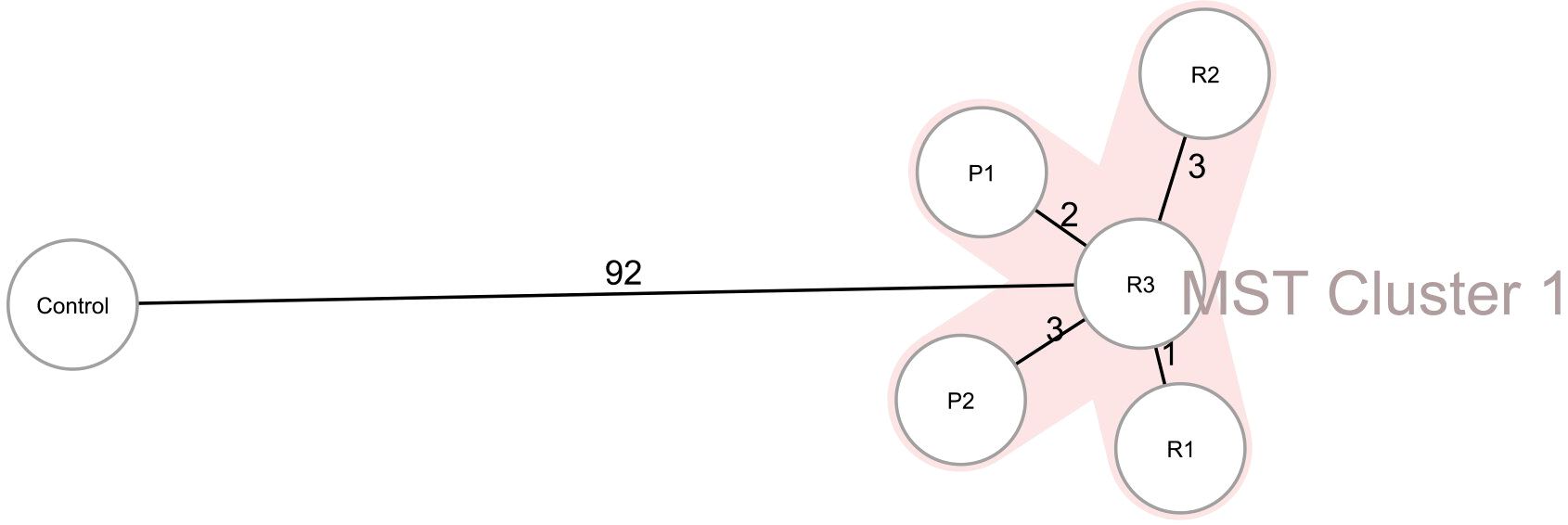
Figure 2. Ridom SeqSphere+ minimum spanning tree (MST) for 5 CRKO isolates from infected patients and razors based on 5,229 target genes (2536 cgMLST and 2693 accessory) with missing values are an own category. MST Cluster distance threshold: 9. P1, Patient 1; P2, Patient 2; R1, Razor 1; R2, Razor 2; R3, Razor 3.
4 Discussion
Despite great advances in neurosurgical technology, SSIs after neurosurgery remain a common problem (Velnar et al., 2023). The incidence of SSIs post-craniotomy ranges between 5.1% and 6.2% (He et al., 2024; Lee et al., 2024). Post-craniotomy SSIs can significantly impact patients recovery and lead to increased morbidity, mortality, hospital stay lengths, pain, and costs, and require re-operation (Velnar et al., 2023). Cerebrospinal fluid leaks, infratentorial surgery, emergency surgery, re-operation, and other factors have been associated with post-craniotomy SSIs (Magni et al., 2023). SSIs can be cause by bacteria from the patients themselves or from medical staff, equipment, or the environment entering surgical incision sites (Tanner and Melen, 2021). One previous study has reported a carbapenemase-producing K. pneumonia outbreak caused by a barber’s contaminated shaving razor (Dai et al., 2014). Despite K. oxytoca being the 2nd most clinically prevalent Klebsiella species, there is limited information about it, especially about the carbapenem-resistant species and its infectivity and epidemiology in neurosurgery (Neog et al., 2021). According to data from the worldwide bacterial collection of SENTRY program, the resistance rate of K. oxytoca to carbapenems is 1.8% with an alarming increasing rate (Yang et al., 2022). K. oxytoca is known to cause healthcare associated infections outbreaks (Yang et al., 2022). In this study, the CRKO isolates from infected patients and the razors shared the same resistance profiles, and had pairwise ANI values higher than 99.993% and a maximum difference of 3 among 5229 targets in the core genome scheme, indicating strong relatedness. A cgMLST analysis may enhance the MLST analysis by assigning allele numbers correlated to the exact genetic sequence of thousands of coding regions throughout the genome, providing a very high resolution and facilitating transmission and outbreak investigations (Dabernig-Heinz et al., 2024). Our findings confirm that contaminated razors can be the sources of CRKO transmission. Thus, inadequate preoperative shaving procedures can cause neurosurgical site infections. Moreover, K. oxytoca seems well adapted to healthcare environments (Yang et al., 2022), and implementation of sanitation protocols, equipment handling, aseptic technique, and antibiotic stewardship strategies should be emphasized, especially in preparation of surgical procedures.
Although the benefits of hair removal for preventing SSIs remain controversial in the literature, preoperative shaving at the intended surgical incision site is a traditional practice that reduces the interference of hair at the surgical site and incision suture, promoting cleanliness and facilitating dressing applications (Kose et al., 2016; Tanner and Melen, 2021; Liu et al., 2022). Preoperative hair removal may result in skin trauma, which in turn contributes to SSIs (Tanner and Melen, 2021). A meta-analysis has indicated that, when compared to the SSIs risk in patients without hair removal, the risk increases in patients who undergo hair removal using a razor (RR: 1.82) (Tanner and Melen, 2021). WHO Guidelines recommend that hair should not be routinely removed in patients undergoing surgical procedures, and that it should be removed with a clipper if absolutely necessary (World Health Organization).
Numerous advanced techniques have been applied for the prevention and control of infections and for promoting patient care. Timely evaluation of transmission sources is critical for implementing intervention strategies to contain the spread. In this study, we successfully applied rapid chromogenic screening and WGS technologies to investigate and control the transmission, and these practicabilities were well confirmed. We used chromogenic-based selective medium to screen for carbapenemase-producing organisms from active surveillance samples, and rapidly detected three suspected CRKO isolates on the razors (within 24 hours) to help timely implementation of precise control measures. The range of available chromogenic culture medium has experienced a rapid expansion. Chromogenic medium are based on synthetic chromogenic enzyme substrates to target specific species (Perry, 2017). Compared with traditional culture medium, chromogenic medium often save costs by reducing labor time and reagents use, while contributing to rapid pathogens identifications (Perry, 2017).
WGS technologies have contributed to the diagnosis, treatment, surveillance, and epidemiological investigations of bacterial infections (Mustafa, 2024). Utilizing WGS and genome-based analysis, these K. oxytoca isolates were identified as the species of K. michiganensis with ST29. K. oxytoca is described as a complex of nine species including K. michiganensis (Yang et al., 2022). K. michiganensis was first recovered from a toothbrush holder in 2012 (Saha et al., 2013). However, the clinical significance including the prevalence, disease spectrum, and severity of each species in the K. oxytoca complex remains largely unknown (Yang et al., 2022). By applying a genome-based analysis, we achieved the precise species identification, providing comprehensive information on the species in the K. oxytoca complex as well as on patient treatment, surveillance, and infection control interventions (Yang et al., 2022). We found that the isolates carried blaKPC-2 and blaOXY-5 resistance genes and IncF plasmids. In addition to the intrinsic blaOXY of K. oxytoca, a number of genes encoding β-lactamases have been reported, and blaKPC-2 is the most common carbapenemase gene and is often carried on IncF plasmids (Yang et al., 2022). The horizontal transmission of blaKPC-2-carrying plasmids across different genera of bacteria has been demonstrated (Schweizer et al., 2019). We also identified virulence factors such as mrk, fim, iutA, and iroE genes in these isolates. The mrk gene cluster can encode mannose-resistant Klebsiella-like hemagglutinins, allowing Klebsiella spp. to attach to surfaces and form biofilms (Yang et al., 2022). Particularly, the iutA gene seems to be important for hypervirulence (Lee et al., 2017). Based on our findings, we believe that K. oxytoca can become a potential major threat to public health due to their ability to acquire resistance to antimicrobials and carry a large number of virulence genes (Yang et al., 2022). Supplemented with epidemiological data, we further validated cgMLST as a useful tool for investigating the source and route of pathogen transmission. cgMLST had been applied to guide interventions, determine durations, and evaluate effects of interventions strategies for other pathogens (Kjær Hansen et al., 2021; Knudsen et al., 2022). Also based on our results, WGS technologies are effective to precisely track transmission sources and provide abundant information on the complete genome of pathogens in an epidemic or outbreak investigation.
Increase in the numbers of multidrug-resistant bacteria, particularly of carbapenem-resistant organisms, have proved fatal to patients undergoing neurosurgery (Li et al., 2023). The surgical team and any professionals directly providing surgical care have an important responsibility to prevent SSIs (World Health Organization; Tvedt et al., 2017). The safety of surgical patients is of the utmost importance, and a list of evidence-based best practices including hand hygiene, preoperative skin antisepsis, antimicrobial irrigation, and others are routinely employed to promote a safe outcomes for surgical patients (Bashaw and Keister, 2019). Evidence-based information is needed to clarify the risk factors of SSIs and improve the quality of care (World Health Organization, 2018). With this study based on WGS, we demonstrated that contaminated razors can lead to SSIs with carbapenem-resistant organisms; therefore, a standard preoperative shaving procedure should be established and executed. Moreover, updated techniques such as chromogenic selective medium and WGS technology can be applied for prevention and control of SSIs and promoting surgical patient care.
We are aware of our study’s limitations. First, although several virulence genes such as the hypervirulence iutA were detected via WGS, the virulence phenotypes were not tested. Second, although we confirmed that razors can transmit pathogens, including carbapenem-resistant organisms, and lead to SSIs, the survival time of these pathogens in the environment such as in razors needs to be further investigated. Finally, the cross-transmission involved a small number of isolates with the same sequence type. Therefore, further research is needed to account for the genetic diversity of CRKO complex bacteria.
To conclude, a cross-transmission of CRKO infections caused by contaminated preoperative shaving razors was confirmed during emergency neurosurgery, and revealed that contaminated shaving razors can transmit CRKO. Whenever hair removal is necessary, it should be removed in the proper area, and clippers may be a more proper alternative to razors (Knudsen et al., 2022; Seidelman et al., 2023). Chromogenic-based selective medium is a sensitive and convenient means to rapidly detect suspected isolates of carbapenem-resistant organisms in an epidemic or outbreak investigation (Alizadeh et al., 2018). We recommend applying WGS technology to track the microbial movement from the original to the infection site. Findings of this study will contribute to the understanding of microbial transmission in surgery and improving of infection prevention and control and patient care.
Data availability statement
The raw sequence data in this study can be found in the online repository. The name of the repository and accession number can be found below: NCBI Sequence Read Archive, PRJNA1171638.
Ethics statement
The studies involving humans were approved by Ethical Committee of The Second Affiliated Hospital of Anhui Medical University (YX2023-102). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
Y-LW: Conceptualization, Formal Analysis, Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review & editing. Y-LJ: Conceptualization, Funding acquisition, Investigation, Methodology, Writing – original draft. Y-YL: Conceptualization, Methodology, Writing – review & editing. L-LL: Data curation, Investigation, Writing – review & editing. Z-PL: Data curation, Investigation, Writing – review & editing. DL: Data curation, Investigation, Writing – review & editing. J-HT: Data curation, Formal Analysis, Investigation, Writing – review & editing. X-QH: Formal Analysis, Methodology, Writing – review & editing. W-HZ: Formal Analysis, Methodology, Writing – review & editing. W-WC: Formal Analysis, Funding acquisition, Methodology, Writing – review & editing. XZ: Funding acquisition, Methodology, Writing – review & editing. WH: Conceptualization, Formal Analysis, Project administration, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Natural Science Foundation of the Republic of China (No. 82202572), the Anqing municipal medical category of science and technology project (No. 2023Z2019), and the Natural Science Research Project Funding of Higher Education Institutions of Anhui Province (No. KJ2021A0321, No. KJ2021A0332; No. 2023AH053175).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alizadeh, N., Rezaee, M. A., Kafil, H. S., Barhaghi, M. H. S., Memar, M. Y., Milani, M., et al. (2018). Detection of carbapenem-resistant Enterobacteriaceae by chromogenic screening media. J. Microbiol. Methods 153, 40–44. doi: 10.1016/j.mimet.2018.09.001
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bashaw, M. A., Keister, K. J. (2019). Perioperative strategies for surgical site infection prevention. AORN J. 109, 68–78. doi: 10.1002/aorn.2019.109.issue-1
Coil, D., Jospin, G., Darling, A. E. (2015). A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 31, 587–589. doi: 10.1093/bioinformatics/btu661
Dabernig-Heinz, J., Wagner, G. E., Prior, K., Lipp, M., Kienesberger, S., Ruppitsch, W., et al. (2024). Core genome multilocus sequence typing (cgMLST) applicable to the monophyletic Klebsiella oxytoca species complex. J. Clin. Microbiol. 62, e0172523. doi: 10.1128/jcm.01725-23
Dai, Y., Zhang, C., Ma, X., Chang, W., Hu, S., Jia, H., et al. (2014). Outbreak of carbapenemase-producing Klebsiella pneumoniae neurosurgical site infections associated with a contaminated shaving razor used for preoperative scalp shaving. Am. J. Infect. Control. 42, 805–806. doi: 10.1016/j.ajic.2014.03.023
Guanghui, Z., Jing, L., Guojun, Z., Hong, L. (2020). Epidemiology and risk factors of neurosurgical bacterial meningitis/encephalitis induced by carbapenem resistant Enterobacteriaceae. J. Infect. Chemother. 26, 101–106. doi: 10.1016/j.jiac.2019.07.023
Guzmán-Puche, J., Jenayeh, R., Pérez-Vázquez, M., Manuel-Causse Asma, F., Jalel, B., Oteo-Iglesias, J., et al. (2021). Characterization of OXA-48-producing Klebsiella oxytoca isolates from a hospital outbreak in Tunisia. J. Glob Antimicrob. Resist. 24, 306–310. doi: 10.1016/j.jgar.2021.01.008
He, K., Li, Y. Y., Liu, H. L. (2024). Risk and protective factors associated with wound infection after neurosurgical procedures: A meta-analysis. Int. Wound J. 21, e14699. doi: 10.1111/iwj.14699
Kjær Hansen, S., Andersen, L., Detlefsen, M., Holm, A., Roer, L., Antoniadis, P., et al. (2021). Using core genome multilocus sequence typing (cgMLST) for vancomycin-resistant Enterococcus faecium isolates to guide infection control interventions and end an outbreak. J. Glob Antimicrob. Resist. 24, 418–423. doi: 10.1016/j.jgar.2021.02.007
Knudsen, M. J. S., Rubin, I. M. C., Gisselø, K., Mollerup, S., Petersen, A. M., Pinholt, M., et al. (2022). The use of core genome multilocus sequence typing to determine the duration of vancomycin-resistant Enterococcus faecium outbreaks. APMIS. 130, 323–329. doi: 10.1111/apm.v130.6
Kose, G., Tastan, S., Kutlay, M., Bedir, O. (2016). The effects of different types of hair shaving on the body image and surgical site infection in elective cranial surgery. J. Clin. Nurs. 25, 1876–1885. doi: 10.1111/jocn.2016.25.issue-13pt14
Lee, K. S., Borbas, B., Plaha, P., Ashkan, K., Jenkinson, M. D., Price, S. J. (2024). Incidence and risk factors of surgical site infection after cranial surgery for patients with brain tumors: A systematic review and meta-analysis. World Neurosurg. 185, e800–e819. doi: 10.1016/j.wneu.2024.02.133
Lee, C. R., Lee, J. H., Park, K. S., Jeon, J. H., Kim, Y. B., Cha, C. J., et al. (2017). Antimicrobial resistance of hypervirulent klebsiella pneumoniae: epidemiology, hypervirulence-associated determinants, and resistance mechanisms. Front. Cell Infect. Microbiol. 7, 483. doi: 10.3389/fcimb.2017.00483
Lee, I., Ouk Kim, Y., Park, S. C., Chun, J. (2016). OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103. doi: 10.1099/ijsem.0.000760
Li, C., Zhou, P., Liu, Y., Zhang, L. (2023). Treatment of ventriculitis and meningitis after neurosurgery caused by carbapenem-resistant enterobacteriaceae (CRE): A challenging topic. Infect. Drug Resist. 16, 3807–3818. doi: 10.2147/IDR.S416948
Lindgreen, S. (2012). AdapterRemoval: easy cleaning of next-generation sequencing reads. BMC Res. Notes. 2, 337. doi: 10.1186/1756-0500-5-337
Liu, W. J., Duan, Y. C., Chen, M. J., Tu, L., Yu, A. P., Jiang, X. L. (2022). Effectiveness of preoperative shaving and postoperative shampooing on the infection rate in neurosurgery patients: A meta-analysis. Int. J. Nurs. Stud. 131, 104240. doi: 10.1016/j.ijnurstu.2022.104240
Luo, R., Liu, B., Xie, Y., Li, Z., Huang, W., Yuan, J., et al. (2012). SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 1, 18. doi: 10.1186/2047-217X-1-18
Magni, F., Al-Omari, A., Vardanyan, R., Rad, A. A., Honeyman, S., Boukas, A. (2023). An update on a persisting challenge: a systematic review and meta-analysis of the risk factors for surgical site infection post craniotomy. Am. J. Infect. Control. 52, 650–658. doi: 10.1016/j.ajic.2023.11.005
Mustafa, A. S. (2024). Whole genome sequencing: applications in clinical microbiology. Med. Princ Pract. 33, 185–197. doi: 10.1159/000538002
Neog, N., Phukan, U., Puzari, M., Sharma, M., Chetia, P. (2021). Klebsiella oxytoca and emerging nosocomial infections. Curr. Microbiol. 78, 1115–1123. doi: 10.1007/s00284-021-02402-2
Nordmann, P., Naas, T., Poirel, L. (2011). Global spread of carbapenemase-producing enterobacteriaceae. Emerg. Infect. Dis. 17, 1791–1798. doi: 10.3201/eid1710.110655
Perry, J. D. (2017). A decade of development of chromogenic culture media for clinical microbiology in an era of molecular diagnostics. Clin. Microbiol. Rev. 30, 449–479. doi: 10.1128/CMR.00097-16
Saha, R., Farrance, C. E., Verghese, B., Hong, S., Donofrio, R. S. (2013). Klebsiella michiganensis sp. nov., a new bacterium isolated from a tooth brush holder. Curr. Microbiol. 66, 72–78. doi: 10.1007/s00284-012-0245-x
Schweizer, C., Bischoff, P., Bender, J., Kola, A., Gastmeier, P., Hummel, M., et al. (2019). Plasmid-mediated transmission of KPC-2 carbapenemase in enterobacteriaceae in critically ill patients. Front. Microbiol. 10, 276. doi: 10.3389/fmicb.2019.00276
Seidelman, J. L., Mantyh, C. R., Anderson, D. J. (2023). Surgical site infection prevention: A review. JAMA. 329, 244–252. doi: 10.1001/jama.2022.24075
Tang, L. M., Chen, S. T. (1995). Klebsiella oxytoca meningitis: frequent association with neurosurgical procedures. Infection. 23, 163–167. doi: 10.1007/BF01793857
Tanner, J., Melen, K. (2021). Preoperative hair removal to reduce surgical site infection. Cochrane Database Syst. Rev. 8, CD004122. doi: 10.1002/14651858.CD004122.pub5
Tompkins, K., van Duin, D. (2021). Treatment for carbapenem-resistant Enterobacterales infections: recent advances and future directions. Eur. J. Clin. Microbiol. Infect. Dis. 40, 2053–2068. doi: 10.1007/s10096-021-04296-1
Tvedt, C., Sjetne, I. S., Helgeland, J., Løwer, H. L., Bukholm, G. (2017). Nurses' reports of staffing adequacy and surgical site infections: A cross-sectional multi-centre study. Int. J. Nurs. Stud. 75, 58–64. doi: 10.1016/j.ijnurstu.2017.07.008
Velnar, T., Kocivnik, N., Bosnjak, R. (2023). Clinical infections in neurosurgical oncology: An overview. World J. Clin. Cases. 11, 3418–3433. doi: 10.12998/wjcc.v11.i15.3418
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PloS One 9, e112963. doi: 10.1371/journal.pone.0112963
Wayne, P. (2019). Performance standards for antimicrobial susceptibility testing; 29th informational supplement. CLSI document M100-S29 (Maryland, USA: Clinical and Laboratory Standards Institute 2019).
World Health Organization Global Guidelines for the Prevention of Surgical Site Infection. 2nd ed. Available online at: https://www.who.int/publications/i/item/9789241550475 (Accessed 11 March 2024).
World Health Organization (2018). Global Guidelines for the Prevention of Surgical Site Infection. 2nd ed. (Geneva: World Health Organization).
Wyres, K., Holt, K. (2022). Regional differences in carbapenem-resistant Klebsiella pneumoniae. Lancet Infect. Dis. 22, 309–310. doi: 10.1016/S1473-3099(21)00425-4
Keywords: surgery, infections, Klebsiella oxytoca, carbapenems, whole genome sequencing
Citation: Jiang Y-L, Lyu Y-Y, Liu L-L, Li Z-P, Liu D, Tai J-H, Hu X-Q, Zhang W-H, Chu W-W, Zhao X, Huang W and Wu Y-L (2024) Carbapenem-resistant Klebsiella oxytoca transmission linked to preoperative shaving in emergency neurosurgery, tracked by rapid detection via chromogenic medium and whole genome sequencing. Front. Cell. Infect. Microbiol. 14:1464411. doi: 10.3389/fcimb.2024.1464411
Received: 14 July 2024; Accepted: 25 September 2024;
Published: 17 October 2024.
Edited by:
Stefano Stracquadanio, University of Catania, ItalyReviewed by:
William R. Schwan, University of Wisconsin–La Crosse, United StatesHumberto Barrios Camacho, National Institute of Public Health, Mexico
Copyright © 2024 Jiang, Lyu, Liu, Li, Liu, Tai, Hu, Zhang, Chu, Zhao, Huang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yi-Le Wu, d3V5aWxlYmFua0AxNjMuY29t; Wei Huang, d2h1YW5nX3N6QDE2My5jb20=
†These authors have contributed equally to this work
 Yun-Lan Jiang1†
Yun-Lan Jiang1† Yi-Yu Lyu
Yi-Yu Lyu Li-Li Liu
Li-Li Liu Jie-Hao Tai
Jie-Hao Tai Wen-Wen Chu
Wen-Wen Chu Wei Huang
Wei Huang Yi-Le Wu
Yi-Le Wu