
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol., 23 July 2024
Sec. Antibiotic Resistance and New Antimicrobial drugs
Volume 14 - 2024 | https://doi.org/10.3389/fcimb.2024.1445850
 Shebin Zhang1,2†;
Shebin Zhang1,2†; Jiao Long1,2†;
Jiao Long1,2†; Qiwei Li1,2†;
Qiwei Li1,2†; Mo Li1,2
Mo Li1,2 Ruiqi Yu1,2
Ruiqi Yu1,2 Yang Lu1,2
Yang Lu1,2 Xingyan Ma3
Xingyan Ma3 Yimei Cai1,2
Yimei Cai1,2 Cong Shen1,2,4
Cong Shen1,2,4 Jianming Zeng1,2,4
Jianming Zeng1,2,4 Bin Huang3*
Bin Huang3* Cha Chen1,2*
Cha Chen1,2* Jieying Pu1,2,4*
Jieying Pu1,2,4*Plasmid-mediated conjugation is a common mechanism for most bacteria to transfer antibiotic resistance genes (ARGs). The conjugative transfer of ARGs is emerging as a major threat to human beings. Although several transfer-related factors are known to regulate this process, small RNAs (sRNAs)-based regulatory roles remain to be clarified. Here, the Hfq-binding sRNA GadY in donor strain Escherichia coli (E. coli) SM10λπ was identified as a new regulator for bacterial conjugation. Two conjugation models established in our previous studies were used, which SM10λπ carrying a chromosomally integrated IncP-1α plasmid RP4 and a mobilizable plasmid pUCP24T served as donor cells, and P. aeruginosa PAO1 or E. coli EC600 as the recipients. GadY was found to promote SM10λπ-PAO1 conjugation by base-pairing with its target mRNA SdiA, an orphan LuxR-type receptor that responds to exogenous N-acylated homoserine lactones (AHLs). However, SM10λπ-EC600 conjugation was not affected due to EC600 lacking AHLs synthase. It indicates that the effects of GadY on conjugation depended on AHLs-SdiA signalling. Further study found GadY bound SdiA to negatively regulate the global RP4 repressors KorA and KorB. When under ciprofloxacin or levofloxacin treatment, GadY expression in donor strain was enhanced, and it positively regulated quinolone-induced SM10λπ-PAO1 conjugation. Thus, our study provides a novel role for sRNA GadY in regulating plasmid-mediated conjugation, which helps us better understand bacterial conjugation to counter antibiotic resistance.
Conjugation is a common mechanism for horizontal gene transfer in bacteria. It is a process by which plasmid DNA is transferred from one cell (donor) to another cell (recipient) by direct contact (Llosa et al., 2002). The dissemination of antibiotic resistance genes (ARGs) by bacterial conjugation is one of the main causes of antibiotic resistance (Cabezón et al., 2017). Most ARGs are encoded on mobile genetic elements and have been found on conjugative plasmids (McInnes et al., 2020). The plasmids carry one or more ARGs for all major classes of antibiotics currently used in clinical treatments, and they can also serve as a reservoir of ARGs to maintain antibiotic resistance even in the absence of antibiotic selection (von Wintersdorff et al., 2016; Vrancianu et al., 2020). Thus, conjugation-mediated antibiotic resistance is an accelerating crisis in bacteria, and understanding the mechanisms of bacterial conjugation is crucial for developing strategies to cope with the crisis.
The entire process of conjugation is tightly controlled by several transfer-related proteins, including TraJ, the major transcriptional activator of the transfer (tra) operon, and relaxosome components such as TraI, TraH and TraM (Pansegrau et al., 1990, 1993; Koraimann et al., 1996). Besides, the KorA and KorB proteins are present as global repressors leading to an inhibition in bacterial conjugation (Kostelidou et al., 1999). KorA inhibits seven operons including trfA on the IncP-1 plasmid RK2 (Jagura-Burdzy and Thomas, 1994). KorB acts cooperatively with KorA to function as a regulator protein in the expression of transfer genes by down-regulating the transcription of the trfA, kilA and korAB operons (Balzer et al., 1992). The mRNA expressions of these proteins are regulated at both transcriptional and post-transcriptional levels (Pansegrau et al., 1994).
Small RNAs (sRNAs) have emerged as important transcriptional and post-transcriptional regulators in bacteria in the past years (Parmeciano Di Noto et al., 2019). Bacterial sRNAs are usually between 40 and 500 nucleotides (nt) in length and most are transcribed from intergenic regions (Li et al., 2012). Under specific conditions, sRNAs expressions are generally modulated and help the bacteria rapidly adapt to environmental changes. Similar to microRNAs, their functional analogs in eukaryotes, sRNAs regulate several aspects of gene expression including transcription, translation, and mRNA stability by base-pairing with multiple target mRNAs (Dutta and Srivastava, 2018). These functions of sRNAs usually require the help of an RNA chaperon, Hfq (Murina and Nikulin, 2015; Santiago-Frangos and Woodson, 2018). Hfq is a potent regulator of many aspects of bacterial activities by influencing mRNA translation and stability via binding with sRNAs (Vogel and Luisi, 2011), and it has been reported to affect the transfer of conjugative plasmids by modulating traJ and traM transcript stability (Will and Frost, 2006). Given the close relationship between sRNAs and Hfq, the Hfq-binding sRNAs have attracted considerable attention recently. Quite a few Hfq-binding sRNAs in E. coli were identified (Gottesman, 2004; Faner and Feig, 2013; Kavita et al., 2018). Many of these sRNAs still have uncharacterized functions. In particular, whether the role of Hfq in bacterial conjugation has anything to do with these binding sRNAs is still unclear. The roles of Hfq-binding sRNAs in conjugation need further investigation.
GadY is an Hfq-binding sRNA in E. coli, and has been reported in the past as a potent regulator of acid response (Opdyke et al., 2004). The gene encoding this sRNA, gadY, is located between the gadX and gadW coding sequences on the opposite strand of the chromosome (Tramonti et al., 2008). When bacteria face mild acidic conditions, GadY is activated and binds its target mRNA GadX in a base-pairing pattern to regulate the glutamate decarboxylase encoded genes gadA and gadB, and the glutamate/GABA antiporter encoded gene gadC (Opdyke et al., 2011; Le Vo et al., 2012). It was also reported that GadY reduces acetate production and improves bacterial growth when E. coli under a mild acidic condition (Negrete and Shiloach, 2015). These current studies identify GadY’s roles in acid resistance, other biological functions of GadY however remain to be elucidated. Our previous works found the orphan LuxR-type receptor SdiA in E. coli SM10λπ regulated bacterial conjugation by responding to exogenous N-acylated homoserine lactones (AHLs) produced by P. aeruginosa PAO1 (Lu et al., 2017b), and the expression of GadY was affected by SdiA when E. coli was under acid stress (Ma et al., 2020). Whether GadY is involved in the regulation of bacterial conjugation remains to be elucidated.
To clarify the role of GadY in bacterial conjugation, gadY gene-deficient and overexpressing donor strains were generated in E. coli SM10λπ based on the conjugation models established in our previous studies (Lu et al., 2017b; Xiong et al., 2021), and PAO1 or E. coli EC600 were used as recipient strains. The E. coli SM10λπ carries a chromosomally integrated IncP-1α plasmid RP4. There is also a pUCP24T plasmid in SM10λπ, and it is mobilizable with the help of RP4 providing transfer functions. In this study, we demonstrated that GadY in donor strain promoted SM10λπ-PAO1 conjugation by base-pairing with its target mRNA SdiA. Our study provides a novel role for the Hfq-binding sRNA GadY in regulating bacterial conjugation, with implications for fighting antibiotic resistance by developing anticonjugation compounds.
The bacterial strains and plasmids used in this study are described in Supplementary Table S1. E. coli K12 SM10λπ (LMBP 3889) is available at BCCM/GeneCorner. To culture the bacteria, strains were grown in Luria-Bertani (LB) medium with shaking or on LB plates containing 1.5% agar at 37°C. In the experiments of SM10λπ-PAO1/EC600 conjugation, antibiotics were added with the following concentrations: 30 μg/ml gentamicin (GM), 50 μg/ml rifampin (RIF), 100 μg/ml ampicillin (AMP), and 16 μg/ml chloramphenicol (CM). In the experiment with exogenous AHLs replenishment, 40 µM 3-oxo-C12-HSL and 40 µM C4-HSL were used.
GadY-deficient mutant was generated in the SM10λπ strain by using a bacteriophage lambda Red recombination system, following the steps described by Datsenko and Wanner (Datsenko and Wanner, 2000). The recombinogenic plasmid pKD46 mediates lambda Red homologous recombination function. The plasmid pKD3 was used as a template and GadY-knock-F/R (Supplementary Table S2) as primers for PCR amplification. Afterwards, 100 μl cells were transformed with 200 ng PCR products using electroporation. Recombinants were selected on LB agar (CM, 16 μg/ml) and then transformed with pCP20 to eliminate the FRT-flanked cat gene. The mutants were verified by PCR with primers GadY-M1/M2 described in Supplementary Table S2. Finally, the pSTV28 plasmid which can stably replicate in E. coli was transformed into the mutant to construct the △GadY strain. Meanwhile, pSTV28 is used to construct the GadY overexpressing strain as follows: gadY gene sequence from E. coli WT genomic DNA was amplified by PCR using the cloning primers In28-gadY-F/R listed in Supplementary Table S2. The product of PCR was inserted into EcoRI/HindIII sites downstream of Plac in pSTV28. After direct sequencing (Sangon Biotech), the GadY overexpressing plasmid, and pSTV28 plasmid were transformed into SM10λπ respectively to generate the GadY+ strain, and the empty vector strain (EV).
The conjugation experiments in this study were based on the conjugation models established in our previous studies (Lu et al., 2017b; Xiong et al., 2021). E. coli SM10λπ worked as donor strains, and EC600 or PAO1 as recipient strains. GadY-deficient and overexpressing strains were constructed in SM10λπ which carries an IncP-1α plasmid RP4 (chromosomally integrated) and a mobilizable plasmid pUCP24T. The pUCP24T plasmid was generated from pUCP24 which was inserted with an oriT fragment (West et al., 1994). It is with GM resistance, and is mobilizable with the help of RP4 providing transfer functions. For mating experiments, the donor cells and recipient cells were respectively grown in LB medium overnight, 200 µl of each culture was added to 3 ml fresh LB medium for a second growth of 2 h, and then the cultures were mixed in fresh LB medium with a ratio of 1:1 (1×107 CFU/ml) at 37°C for 6 h under static conditions, total volume of the medium was 200 µl. Next, the cultures were thoroughly mixed, and then 30 µl of each mixture was spread on an LB agar plate containing GM+AMP (SM10λπ-PAO1 transconjugants) or GM+RIF (SM10λπ-EC600 transconjugants). After incubation at 37°C overnight, the numbers of transconjugant colonies were counted. In the experiment of antibiotic-induced conjugation, the overnight cultured donor cells were diluted with LB containing ciprofloxacin (CIP, 0.015625 μg/ml), levofloxacin (LEV, 0.03125 μg/ml), GM (0.3 μg/ml), azithromycin (AZM, 32 μg/ml) or CM (16 μg/ml) and were incubated at 37°C. After 6 h of incubation, the cells were washed and re-suspended in fresh LB medium without antibiotics, and used as donor strains for conjugation.
The bacterial strains were cultured in LB medium at 37°C for indicated time points. After culture collection, the total RNA of bacteria was extracted by TRIzol (Invitrogen) and a spectrophotometer (NanoDrop 2000) was used to quantify the RNA. Then an RT Reagent Kit (PrimeScript, Takara) was applied to reverse transcription, in which 1 μg RNA was used and the total reaction volume was 20 μl. The quantitative PCR was performed using a SYBR Green qPCR kit (Accurate Biology) and the ViiATM 7Dx system (Applied Biosystems). The relative threshold cycle (2-ΔΔCt) method was used to normalize the quantification cycle value to the reference gene rpoD. The qRT-PCR primers were listed in Supplementary Table S2.
The online IntaRNA database (Mann et al., 2017) was used to predict the potential targets of GadY. Use the default parameters as described on the IntaRNA website (http://rna.informatik.uni-freiburg.de/IntaRNA/Input.jsp), of which the number of interactions per RNA pair was 5, and minimal number of basepairs in seed was 7. The query sRNA and target sequences were from the genome of E. coli K12 SM10λπ. Results are computed with IntaRNA version 3.3.1 linking Vienna RNA package 2.5.0.
To verify the interaction between GadY and SdiA, the pUCP30T-gfp reporter plasmid we previously generated was used (Lu et al., 2019; Pu et al., 2022). It is a plasmid containing a gentamicin resistance gene (aacC1), the origin of replication from pRO1600, oriT, and the GFP reporter region with a lac promoter that allows the insertion sequence to express transcriptional fusion with GFP. For WT SdiA plasmid, pUCP30T-sdiA-gfp, a WT sdiA mRNA sequence containing GadY putative binding site from SM10λπ genomic DNA was amplified. Next, the XbaI/NcoI sites upstream of GFP in the pUCP30T-gfp plasmid were inserted with the amplified sequence, and the plasmid was constructed. For Mut SdiA plasmid pUCP30T-mut-sdiA-gfp, or for Mut GadY overexpressing plasmid pSTV28-mut-GadY+, which carried a mutated sequence in the complementary sites for GadY, or SdiA, was generated using fusion PCR. The cloning primers listed were shown in Supplementary Table S2. After direct sequencing, the plasmid pSTV28-mut-GadY+ was transformed into SM10λπ to generate the Mut GadY+ strain, and the plasmid pUCP30T-sdiA-gfp or pUCP30T-mut-sdiA-gfp was transformed into SM10λπ EV, △GadY, or GadY+ strain respectively, or was transformed into SM10λπ EV, GadY+ or Mut GadY+ strain respectively. Incubating for 20 h overnight, the bacterial cultures were collected and OD600 values were taken, and then were washed and re-suspended in 0.9% NaCl. 100 μl of the solution was then used to determine the fluorescence intensity (F485/535) through a microplate reader (Synergy H1, BioTek), and 10 μl of the solution was applied to observe the fluorescence by a fluorescence microscope (ECLIPSE Ti2-U, Nikon).
The indicated strains were grown overnight in LB medium at 37°C, and then each 200 μl culture was added to a new 3 ml LB medium for the second growth of 2 h. Next, each culture was 1:1000 diluted and 200 μl was added to an automated 96-well microplate reader (Synergy H1, BioTek) at 37°C growing for 15 h. The OD600 values were taken at 1 h intervals with continuous shaking.
A plasmid pQF50 containing the lacZ reporter gene which codes for β-galactosidase was used in the study. The DNA sequences of the korA promoter (PkorA) ranging from -1 to -173 and the korB promoter (PkorB) ranging from -1 to -328 were amplified by PCR and inserted into the HindIII site upstream of the lacZ gene. The cloning primers were listed in Supplementary Table S2. Then the pQF50-PkorA and pQF50-PkorB plasmids were constructed, and confirmed by direct sequencing (Sangon Biotech). Next, the above two plasmids were respectively transformed into DH5α, an E. coli strain without endogenous β-galactosidase, and the pSTV28 (EV) or pSTV28-GadY+ (GadY+) plasmid was subsequently transferred. In the β-galactosidase assays, the strains were cultured at 37°C for 6 h to the mid-log phase, and the Miller method was carried out to examine the β-galactosidase activity (Giacomini et al., 1992).
At least three independent experiments were performed in each set of individual assays. Data are shown as mean ± SEM by using GraphPad Prism 9. The statistical significance between two groups was determined by the Student’s t test, and the P value was described as *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
To explore GadY’s role in bacterial conjugation, the gadY gene-deficient and overexpressing strains were constructed with SM10λπ, termed △GadY and GadY+. Fresh LB medium was used to culture the empty vector (EV), △GadY, and GadY+ strains for 6 h respectively, and the expression level of GadY was examined by qRT-PCR. GadY expression was up-regulated (~117.5-fold) in the GadY+ strain, and was undetected in the △GadY strain (Figure 1A). The bacterial growth curves showed little difference between the three strains, of which the △GadY strain grew faster under the stationary phase (Figure 1B). SM10λπ-PAO1 and SM10λπ-EC600 were two conjugation models previously generated in our lab (Lu et al., 2017b; Xiong et al., 2021), as shown in Figure 1C. We first examined the role of GadY in regulating SM10λπ-PAO1 conjugation. Compared with the EV group, overexpression of GadY significantly enhanced the conjugation between SM10λπ and PAO1, while the GadY deficiency group resulted in a reduction (Figure 1D). We subsequently used the SM10λπ-EC600 conjugation model for investigation, but found no statistical significance (Figure 1E). These results suggest GadY in SM10λπ donor strains has different roles in regulating conjugation with PAO1 or EC600 recipients.
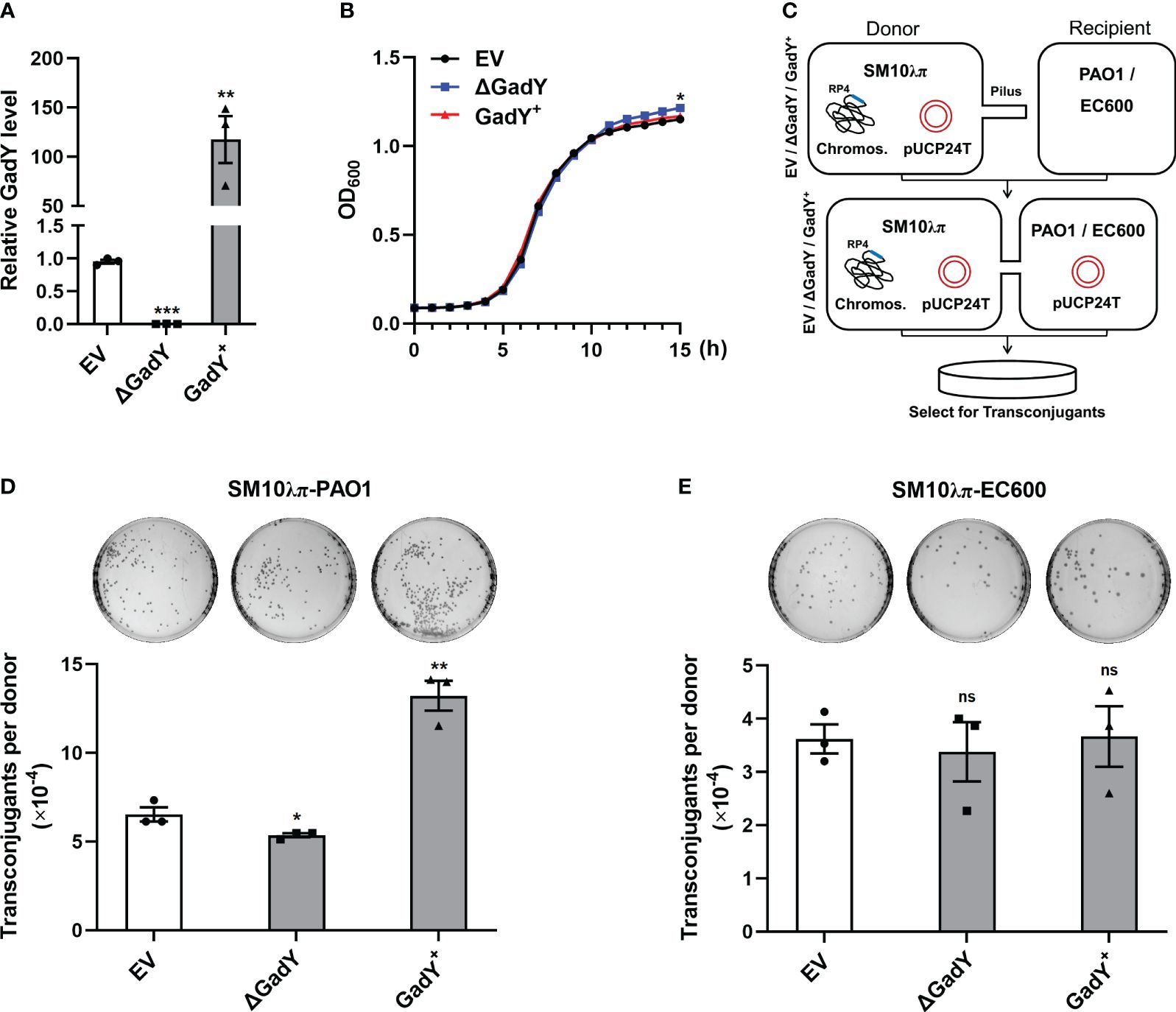
Figure 1 GadY positively regulated SM10λπ-PAO1 conjugation, but it did not affect the conjugation between SM10λπ and EC600. (A) E. coli SM10λπ carrying pSTV28 empty vector or pSTV28-gadY plasmid termed EV and GadY+ respectively, and gadY-deleted SM10λπ carrying pSTV28 plasmid termed △GadY, these three strains were grown in LB for 6 h, then the expression levels of GadY were detected by qRT-PCR. (B) Analysis of the growth curves of EV, △GadY, and GadY+ strains. (C) The conjugation models of SM10λπ-PAO1/EC600. (D, E) The donor strains SM10λπ (EV, △GadY, or GadY+) and recipient strain PAO1 (D) or EC600 (E) were mated at 37°C for 6 h (1×107 CFU/ml each), and then the numbers of transconjugant colonies were counted. Data are shown as mean ± SEM of at least three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, non-significant.
Different roles of GadY in the two conjugation models were found in Figure 1. The differences may be caused by the recipient strains PAO1 and EC600. In contrast to P. aeruginosa, E. coli lacks AHLs synthase and therefore could not produce AHLs (Kumar et al., 2022). While in P. aeruginosa, quorum sensing (QS) is a vital AHLs signalling system, in which 3-oxo-C12-HSL and C4-HSL are common signals produced by LasI or RhlI respectively (Lee and Zhang, 2015). The donor SM10λπ is also an E. coli strain which does not produce AHLs, but it encodes an orphan LuxR-type receptor SdiA which responds to exogenous AHLs (Yamamoto et al., 2001). Thus, it suggests that the AHLs-SdiA signalling in the SM10λπ-PAO1 model may contribute to the different roles of GadY in the two conjugation models.
Base-pairing with its target mRNAs is an important way for sRNA to regulate various genes. To explore the molecular mechanisms of GadY affecting SM10λπ-PAO1 conjugation, the online IntaRNA database (Mann et al., 2017) was used to predict the potential targets of GadY. Surprisingly, SdiA was found to be a target mRNA, and the base-pairing of GadY occurs at the coding region (Figure 2A). The expressions of sdiA in EV, △GadY, and GadY+ strains were detected by qRT-PCR, and we found a significant up-regulation in the GadY deletion mutant and a decrease in the GadY overexpressing strain (Figure 2B). To verify the predicted data of IntaRNA, we developed a GFP (green fluorescent protein) reporter system. The pUCP30T-sdiA-gfp plasmid was first constructed by inserting the predicted mRNA sequence of sdiA upstream of the gfp gene. Next, the plasmid was transformed into SM10λπ EV, △GadY, or GadY+ strains respectively. A fluorescence microscope was used to observe and photograph the fluorescence, and a microplate reader was applied to detect the fluorescence intensity for mapping the bar graphs. As shown in Figure 2C, the intensity of GFP that carried a wild-type (WT) base-pairing site of SdiA was significantly affected by GadY, but the mutant (Mut) groups were not. To further confirm the direct interaction between GadY and SdiA, we also mutated GadY and then introduced a compensatory change in the SdiA base-pairing site to conduct mutational analyses. As expected, overexpressing mutant GadY (Mut GadY+) disrupted the inhibition role of GadY in the GFP intensity of WT SdiA, but meanwhile introducing the SdiA compensatory mutant (Mut SdiA) restored GadY’s role (Figure 2D). These results indicate that SdiA as a direct target of GadY may be involved in the role of GadY in SM10λπ-PAO1 conjugation.
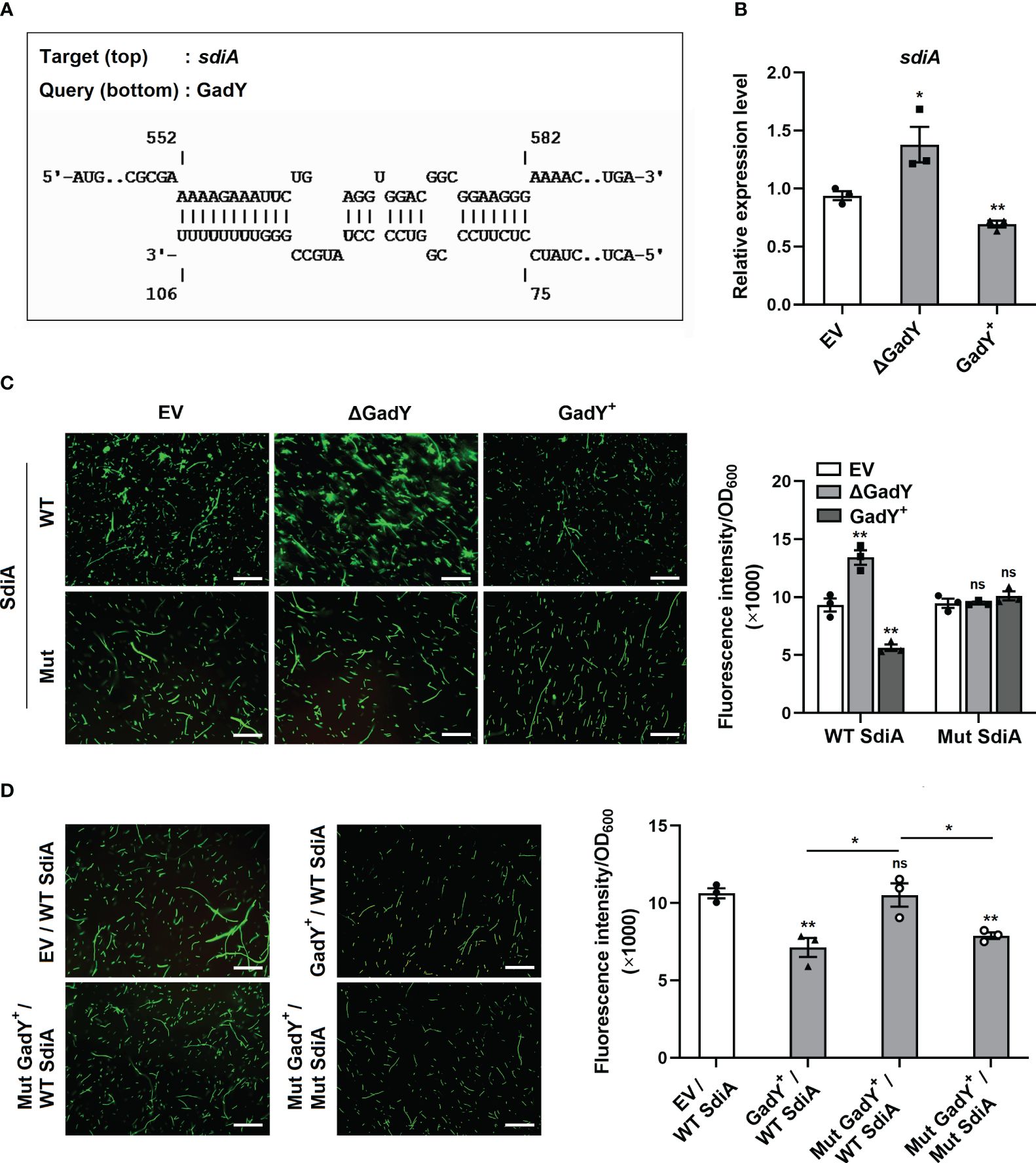
Figure 2 GadY base-paired with its target mRNA SdiA and negatively regulated its transcription. (A) Analysis of GadY’s target mRNA through IntaRNA, and found coding sequence of sdiA had a putative binding region with GadY. (B) E. coli SM10λπ EV, GadY+, and △GadY strains were grown in LB for 6 h respectively, then SdiA mRNA expression was detected by qRT-PCR. (C, D) The pUCP30T-gfp reporter plasmid was inserted with a WT or Mut sequence of sdiA mRNA, and was transformed into SM10λπ EV, △GadY or GadY+ strains (C), or EV, GadY+ or Mut GadY+ strains (D), then the fluorescence images were photographed by a microscopy, the scale bar represents 10 μm, and the fluorescence intensity normalized to OD600 was measured by a microplate reader for mapping the bar graphs. Data are shown as mean ± SEM of at least three independent experiments. *, P < 0.05; **, P < 0.01; ns, non-significant.
The AHLs receptor SdiA was found to be a target of GadY, and its mRNA expression was negatively regulated by GadY in the donor strain SM10λπ. Our previous study has demonstrated the inhibition role of AHLs-SdiA signalling in SM10λπ-PAO1 conjugation (Lu et al., 2017b). To clarify whether SdiA participates in GadY-mediated regulation of conjugation, sdiA gene-overexpressing and deficient strains in SM10λπ constructed by our previous work (Lu et al., 2017b) were used, termed sdiA+ and △sdiA. Overexpression of GadY in sdiA-deficient strain was generated, termed △sdiA/GadY+. After 6 h of culture, the expression levels of sdiA (Figure 3A) and GadY (Figure 3B) in EV, sdiA+, △sdiA, and △sdiA/GadY+ strains were confirmed by qRT-PCR. Compared with the EV group, the sdiA mRNA expression was up-regulated (~100.5-fold) in sdiA+ strain, and was undetected in △sdiA or △sdiA/GadY+ strain (Figure 3A). GadY expression was up-regulated (~24.3-fold) in △sdiA/GadY+ strain compared with the EV group, and its expression in sdiA+ or △sdiA stains showed little change (Figure 3B). There were no significant difference in the growth curves of the above four strains (Figure 3C). Next, the SM10λπ-PAO1 model was used to investigate the roles of sdiA+, △sdiA and △sdiA/GadY+ in regulating conjugation. Overexpression of sdiA significantly decreased SM10λπ-PAO1 conjugation, while an obvious increase was observed in the sdiA-deficient group (Figure 3D). The inhibitory role of SdiA in bacterial conjugation is consistent with our previous study (Lu et al., 2017). Further analysis of the result of △sdiA/GadY+ group, it showed no difference with the △sdiA group (Figure 3D). For SM10λπ-EC600 conjugation, we subsequently added exogenous AHLs in this model and measured GadY’s role. As shown in Figure 3E, exogenous addition of 3-oxo-C12-HSL and C4-HSL rescued the positive regulation of GadY in conjugation. Taken together, the data from Figures 2, 3 suggest that GadY enhances bacterial conjugation by targeting SdiA and then attenuating the AHLs-SdiA signalling.
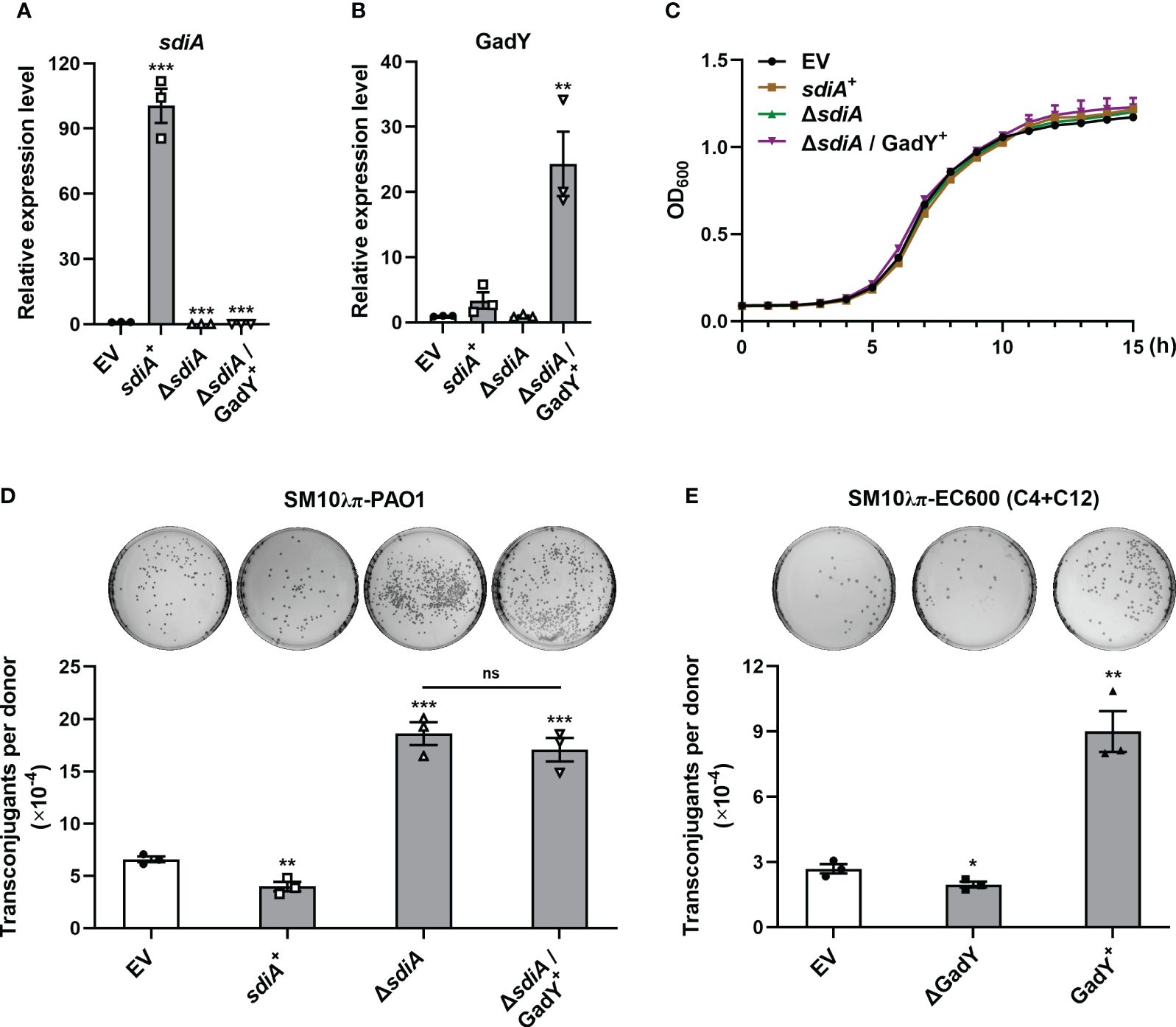
Figure 3 AHLs-SdiA signalling was involved in GadY’s effect on bacterial conjugation. (A, B) E. coli SM10λπ carrying pSTV28 empty vector or pSTV28-sdiA plasmid termed EV and sdiA + respectively, and sdiA-deleted SM10λπ carrying pSTV28 or pSTV28-gadY plasmid termed △sdiA and △sdiA/GadY+ respectively, these four strains were grown in LB for 6 h, then sdiA (A) and GadY (B) expressions were examined by qRT-PCR. (C) Analysis of the growth curves of EV, sdiA+, △sdiA, and △sdiA/GadY+ strains. (D) The donor strains SM10λπ (EV, sdiA+, △sdiA, or △sdiA/GadY+) and recipient strain PAO1 were mated at 37°C for 6 h (1×107 CFU/ml each), and then counted the numbers of transconjugant colonies. (E) The donor strains SM10λπ (EV, △GadY, or GadY+) were cultured in the presence of 40 µM 3-oxo-C12-HSL and C4-HSL, followed by EC600 conjugation, the mixtures were incubated at 37°C for 6 h (1×107 CFU/ml each), and then counted the numbers of transconjugant colonies. Data are shown as mean ± SEM of at least three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, non-significant.
The conjugation process is initiated by the assembly of a relaxosome at the transfer origin (oriT), which requires the interaction of transfer-related proteins such as TraJ and TraI (Pansegrau et al., 1990, 1993; Koraimann et al., 1996). Thus, we first examined the role of GadY in regulating TraJ and TraI mRNA expressions in the donor strain SM10λπ. The expression levels of traJ (Supplementary Figure S1A) and traI (Supplementary Figure S1B) were decreased in the GadY deletion mutant. However, both traJ and traI expressions were not significantly affected in the GadY overexpressing strain (Supplementary Figures S1A, B). Encoded in the IncP plasmid’s central control region, the KorA and KorB proteins are global repressors leading to a potent decrease in conjugative transfer (Balzer et al., 1992; Jagura-Burdzy and Thomas, 1994; Kostelidou et al., 1999). We subsequently measured KorA and KorB mRNA expressions in the GadY-deficient or overexpressing donor strains. As shown in Figures 4A, B, the results showed an obvious up-regulation of korA and korB expressions in the GadY deletion mutant, and a decrease in the GadY overexpressing strain. It suggests that GadY promotes bacterial conjugation by down-regulation of KorA and KorB. In Figures 2 and 3, we have demonstrated that GadY enhances conjugation by targeting SdiA. To further clarify whether SdiA involves in the effects of GadY on KorA and KorB, sdiA gene-deficient and overexpressing strains were used. As shown in Figures 4C, D, both korA and korB expressions were significantly up-regulated in sdiA+ strain, but decreased in the △sdiA strain and △sdiA/GadY+ strain. The expressions of korA and korB in △sdiA/GadY+ strain had no difference with the △sdiA group (Figures 4C, D). These results indicate that the negative regulation of KorA and KorB by GadY depends on base-pairing with its target mRNA SdiA.
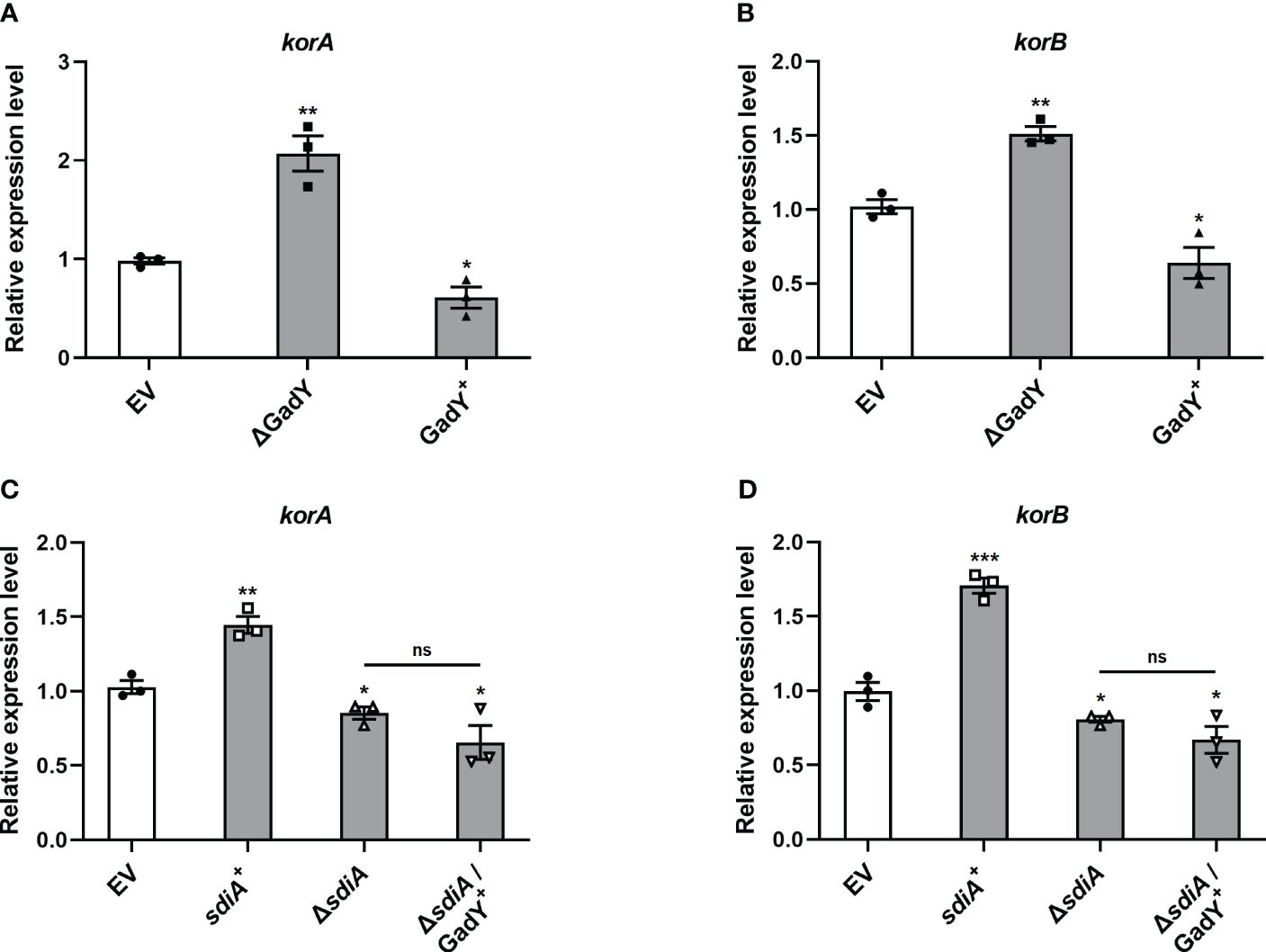
Figure 4 The mRNA expressions of global RP4 repressors KorA and KorB were regulated by GadY and SdiA. (A, B) E. coli SM10λπ EV, GadY+, and △GadY strains were grown in LB for 6 h respectively, then korA (A) and korB (B) expressions were examined by qRT-PCR. (C, D) E. coli SM10λπ EV, sdiA+, △sdiA, and △sdiA/GadY+ strains were grown in LB for 6 h respectively, then korA (C) and korB (D) expressions were detected by qRT-PCR. Data are shown as mean ± SEM of at least three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, non-significant.
The qRT-PCR results in Figure 4 preliminarily showed GadY down-regulated korA and korB expressions at the transcriptional level. To further confirm this, we constructed a transcriptional lacZ fusion to the korA or korB promoter. The DNA sequence of korA or korB promoter, namely PkorA or PkorB, was inserted respectively upstream of the β-galactosidase gene lacZ in the pQF50 reporter plasmid (Figures 5A, B). Compared with the EV group, the β-galactosidase activity from the PkorA-lacZ fusion was inhibited by GadY (Figure 5A). More significantly, GadY greatly impaired the activity from the PkorB-lacZ fusion (Figure 5B). These findings establish that the transcriptional activities of korA and korB promoter regions are repressed by GadY.
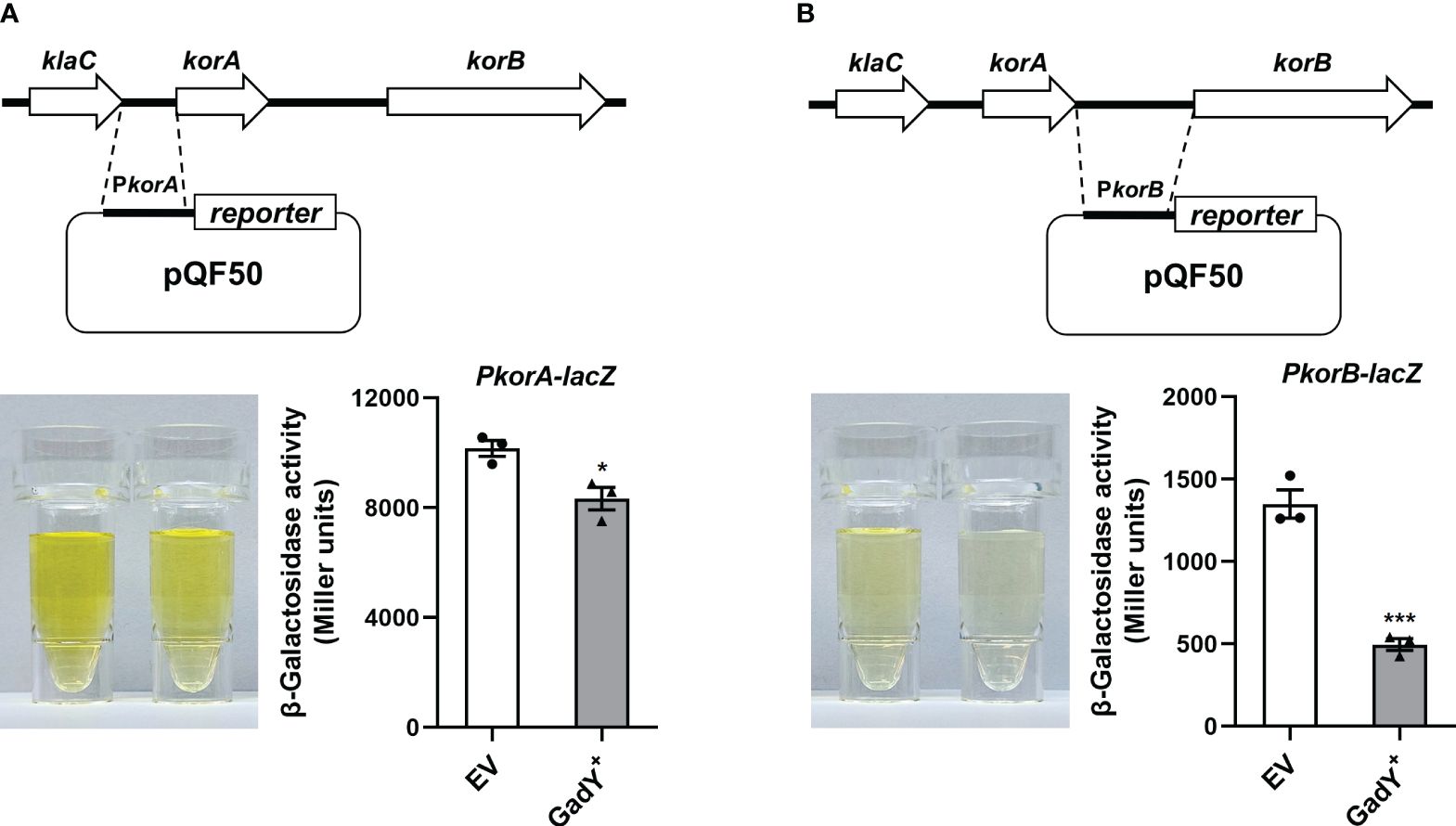
Figure 5 The promoter activities of korA and korB were affected by GadY. (A, B) Diagram of the korA-korB operon and the investigation of the promoter activities of korA (A) and korB (B). The E. coli DH5α strains carrying the pQF50-PkorA (A) or pQF50-PkorB (B) reporter plasmid combined with pSTV28 (EV) or pSTV28-GadY+ (GadY+) were cultured at 37 °C for 6 h to the mid-log phase, and the β-galactosidase activity was measured. Data are shown as mean ± SEM of at least three independent experiments. *, P < 0.05; ***, P < 0.001.
To further investigate the roles of GadY in antibiotics-induced conjugation, several types of antibiotics including ciprofloxacin (CIP), levofloxacin (LEV), gentamicin (GM), azithromycin (AZM) and chloramphenicol (CM) were separately added to the SM10λπ-PAO1 conjugation model. These antibiotics positively regulate bacterial conjugation, as demonstrated in our previous studies (Shun-Mei et al., 2018; Lu et al., 2017a). In the EV group, the conjugations under the above antibiotics treatments were enhanced, while deletion of GadY significantly decreased the numbers of transconjugants under CIP, LEV or GM treatment (Figures 6A, B; Supplementary Figure S2A). In order to demonstrate the roles of GadY in SM10λπ-PAO1 conjugation under antibiotics treatments more clearly, relative conjugation levels by calculating the transconjugant ratio of antibiotic to vehicle were analyzed. As shown in Figures 6C, D, the increase of LEV on SM10λπ-PAO1 conjugation was much inhibited in △GadY group than in EV group, and there was also a slight decline under CIP treatment. However, the relative conjugation levels were not affected by GadY deletion under GM, AZM or CM treatment (Supplementary Figures S2B-D). The role of antibiotics in the expression of GadY in SM10λπ was finally examined, and we found the presence of either CIP or LEV significantly increased GadY expression (Figure 6E), but no changes were observed in GM, AZM or CM group (Supplementary Figure S2E). These results indicate that quinolone antibiotics mediate the up-regulated expression of GadY, which positively regulates SM10λπ-PAO1 conjugation.
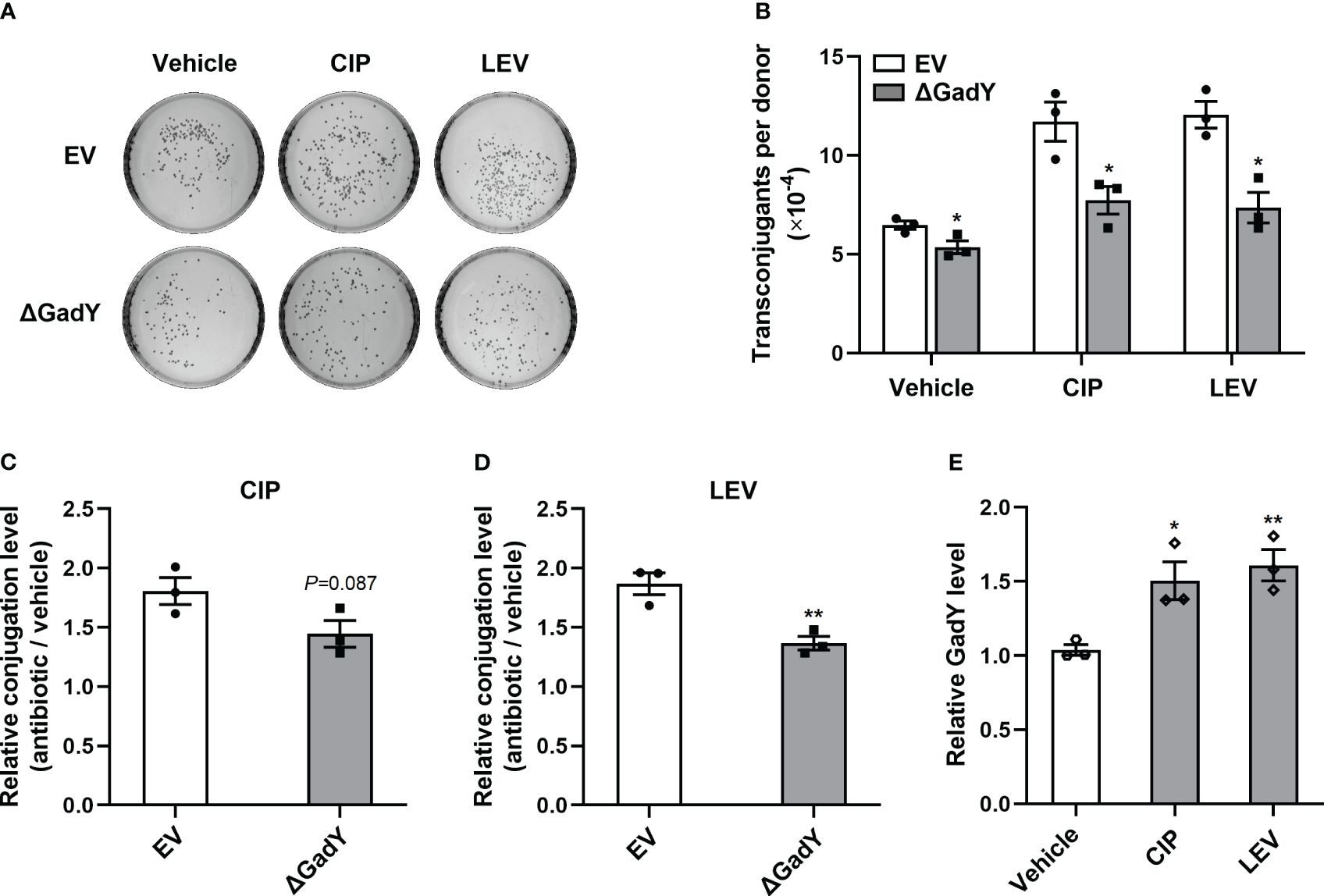
Figure 6 Quinolone antibiotics induced up-regulated expression of GadY to promote SM10λπ-PAO1 conjugation. (A, B) E. coli SM10λπ (EV or △GadY) were treated with sub-MIC of CIP, LEV or untreated (vehicle) for 6 h at 37°C, then were mated with PAO1 (1×107 CFU/ml each) for 6 h, and transconjugant colonies numbers were counted. (C, D) The relative conjugation levels by calculating the transconjugant ratio of antibiotic to vehicle were analyzed, and CIP (C), LEV (D). (E) SM10λπ was treated with sub-MIC of CIP, LEV or untreated (vehicle) for 6 h at 37°C, and the expression levels of GadY was examined by qRT-PCR. Data are shown as mean ± SEM of at least three independent experiments. *, P < 0.05; **, P < 0.01.
In this study, the Hfq-binding sRNA GadY in donor strain E. coli SM10λπ is found to be involved in regulating bacterial conjugation (Figure 7). GadY promoted SM10λπ-PAO1 conjugation by base-pairing with its target mRNA SdiA. In E. coli, the orphan LuxR-type receptor SdiA responds to exogenous AHLs from other bacterial species like P. aeruginosa (Yamamoto et al., 2001). Given this, we further clarified that the effects of GadY on SM10λπ-PAO1 conjugation depended on AHLs-SdiA signalling. Additional studies found GadY decreased the expression levels of KorA and KorB mRNA by targeting SdiA. The expression of GadY in donor strain was up-regulated by CIP or LEV treatment, and it promoted quinolone-induced conjugation. Therefore, the Hfq-binding sRNA GadY from donor strain E. coli is a novel regulator involved in bacterial conjugation.
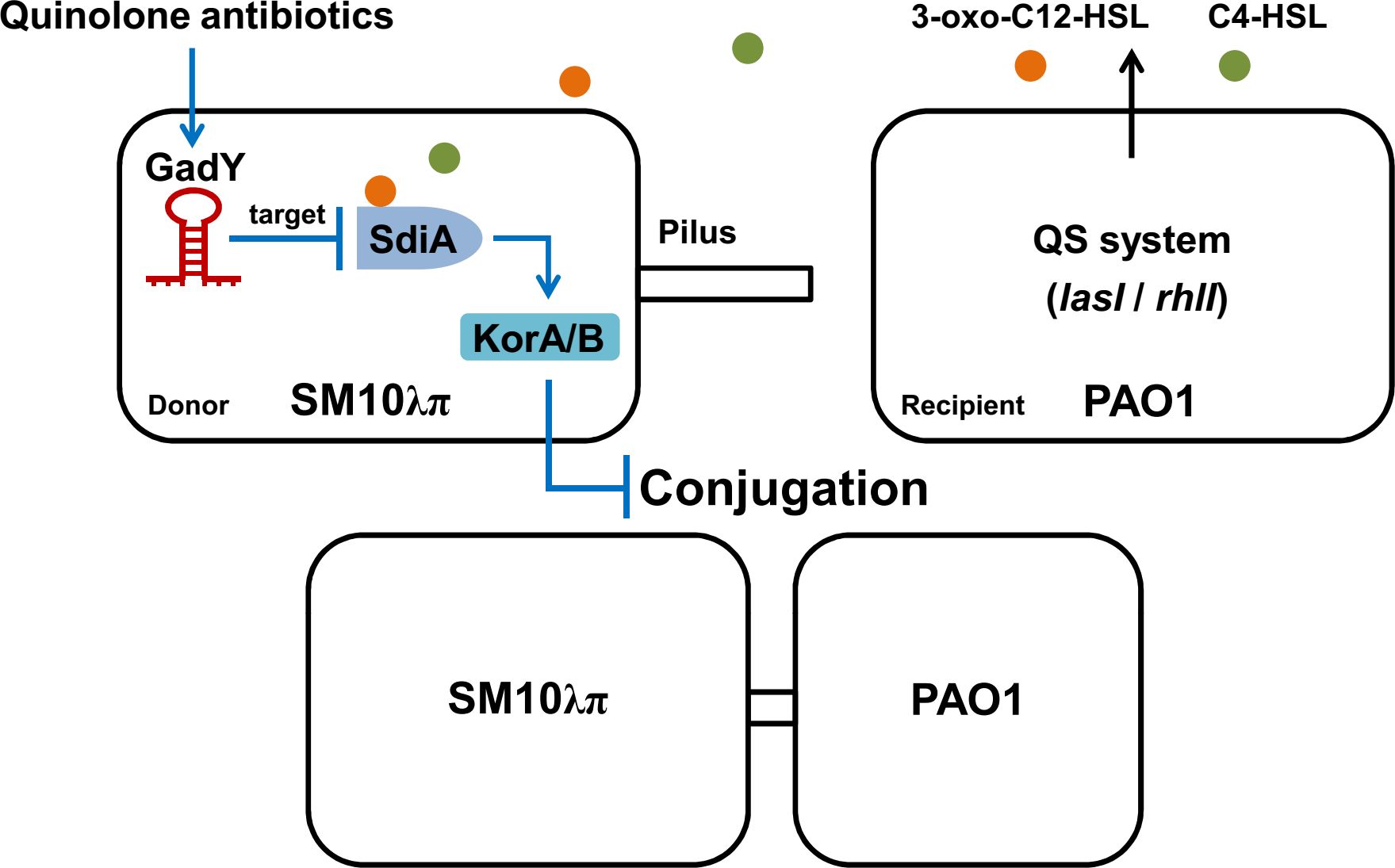
Figure 7 A working model of how GadY promotes SM10λπ-PAO1 conjugation by targeting SdiA under quinolone antibiotics treatments. GadY expression in donor strain SM10λπ was up-regulated by ciprofloxacin or levofloxacin treatment. Then GadY promoted SM10λπ-PAO1 conjugation by base-pairing with its target mRNA SdiA, an orphan LuxR-type receptor that responded to exogenous AHLs produced by the recipient strain PAO1. The transcription of the global RP4 repressors KorA and KorB was subsequently inhibited by GadY.
GadY is an Hfq-binding sRNA and has been reported in the past as a potent regulator of acid response in E. coli (Opdyke et al., 2004). Other biological functions of GadY are currently still unknown. Given that its chaperone Hfq plays important roles in bacterial conjugation by regulating the transcript stability of transfer-related genes traM and traJ (Will and Frost, 2006), GadY’s roles in bacterial conjugation arouse our interest. Based on the SM10λπ-PAO1/EC600 conjugation models established in our previous studies (Lu et al., 2017b; Xiong et al., 2021), gadY gene-deficient and overexpressing donor strains (SM10λπ) were thus constructed, and PAO1 or EC600 were used as recipient strains. Intriguingly, GadY positively enhanced the conjugation between SM10λπ and PAO1, but had no significance in SM10λπ-EC600 conjugation (Figure 1). We speculate that the different roles of GadY in regulating SM10λπ’s conjugation with PAO1 or EC600 may be related to the AHLs produced by recipient strains. PAO1 has autoinducer synthase from QS system to synthesize various AHLs including 3-oxo-C12-HSL, C4-HSL, quinolones and IQS signals (Papenfort and Bassler, 2016; García-Reyes et al., 2020). In contrast to PAO1, EC600 could not produce AHLs because it lacks AHLs synthase.
A previous study in our group has demonstrated that 3-oxo-C12-HSL and C4-HSL from PAO1 activate the SdiA receptor of SM10λπ, which results in the attenuation of SM10λπ-PAO1 conjugation (Lu et al., 2017b). SdiA is an orphan receptor in bacterial species such as Escherichia, Klebsiella, Salmonella, and Shigella, which responds to a wide range of exogenous AHLs (Smith et al., 2011). These organisms are nonAHL-producing bacteria because they lack AHLs synthase (Mayer et al., 2023). Therefore, the different roles of GadY in SM10λπ-PAO1/EC600 conjugation may be closely related to the AHLs-SdiA signalling. Base-pairing with its target mRNAs is an important way for sRNA to regulate various genes (Desgranges et al., 2019; Jørgensen et al., 2020). Using a GFP reporter system including WT and Mut experiments, SdiA was surprisingly demonstrated as a direct target of GadY (Figure 2). Adding AHL in the SM10λπ-EC600 model rescued GadY’s roles in conjugation (Figure 3). It indicates that GadY-mediated regulation of bacterial conjugation depends on the AHLs-SdiA signalling between the donor and recipient cells.
The interaction between sRNA and its target mRNA can affect translation initiation, transcription termination or mRNA stability. The binding sites of sRNA can occur at different regions of the target mRNAs, such as the 5’ untranslated region (5’-UTR), 3’ untranslated region (3’-UTR), and the coding region (Waters and Storz, 2009). Most of the intergenic sRNAs frequently bind to the 5’-UTR of the target mRNAs where the ribosome accommodation usually appears (Storz et al., 2011). The coding sequence is also a usual region for some sRNA to interact with its target mRNA. The Hfq-binding sRNA DsrA in E. coli base-pairs far downstream in the coding sequence of rbsD and induces mRNA decay (Lalaouna et al., 2015). MicC as an Hfq-associated sRNA in Salmonella binds to the coding sequence of ompD to accelerate RNase E-dependent mRNA decay (Pfeiffer et al., 2009). The base-pairing region of GadY was around 552~582 nt from the start codon of sdiA (Figure 2). Given the coding sequence of sdiA is 723 nt, GadY interacted downstream of the coding region. Likewise, GadY is also an Hfq-binding sRNA as DsrA and MicC, and the sites for GadY interacting with SdiA are comprised of a poly-U tail, an AU-rich single-stranded region which is often required for Hfq binding on sRNA (Link et al., 2009). It suggests that base-paring with the coding sequence and then affecting the stability and translation of mRNA may be an alternative way for Hfq-binding sRNA to regulate its target.
The donor strain E. coli SM10λπ used in our conjugation model carries a chromosomally integrated IncP-1α plasmid RP4. The transfer of conjugative DNA, a pUCP24T plasmid, mediated by RP4 starts at oriT. In this process, an oriT-recognizing protein TraJ is encoded by the traJ gene and is essential for the initiation of transfer DNA replication. The TraJ protein binds oriT and begins the formation of a relaxosome, and then the TraI protein, a key component of the relaxosome, combines the TraJ oriT complex to promote the transfer process (Fürste et al., 1989; Pansegrau et al., 1994). However, overexpression of GadY did not significantly affect the expressions of traJ and traI, although GadY deletion had some changes (Supplementary Figure S1). It suggests GadY’s roles in conjugation are not achieved by regulating TraJ and TraI.
In addition to transfer-related proteins, the global transcriptional repressors KorA and KorB are major factors in controlling the regulatory circuit of RP4 plasmid (Young et al., 1987). KorA has a dimerization module shared by the RP4 protein TrbA, another global repressor which inhibits the transcription of trfA (Jagura-Burdzy et al., 1992; Jagura-Burdzy and Thomas, 1994). KorA also causes derepression of the promoter for trbA (Jagura-Burdzy and Thomas, 1994). In conjunction with KorA, KorB works as a regulator protein in modulating transfer genes expression by suppressing the transcription of the trfA, kilA and korAB operons (Schmidhauser et al., 1989; Bingle et al., 2005). We found that the korA and korB mRNA expressions were affected by GadY and SdiA (Figure 4). The transcriptional activities of korA and korB promoter regions were also analyzed. It was reported that korA and korB are co-transcribed from the upstream promoter of korA, and korB is primarily expressed in an operon with korA (Bechhofer and Figurski, 1983; Smith and Thomas, 1984; Bechhofer et al., 1986). However, when a heterologous promoter upstream of korB is present, PkorA is nonessential to korB transcription (Bechhofer et al., 1986; Young et al., 1987). Our results from the β-galactosidase assays showed that PkorB has weaker promoter activity than PkorA, and GadY inhibited the activities of both of them (Figure 5). The results from Figures 4, 5 suggest that the inhibitory effects on KorA and KorB enable GadY to promote bacterial conjugation, and this process depends on the base-pairing between GadY and its target mRNA SdiA.
As a common mechanism of horizontal transfer of ARGs, plasmid-mediated conjugation contributes to the rapid spread of antibiotic resistance among bacteria. Therefore, we finally investigated the roles of GadY in conjugation induced by different antibiotics, including CIP, LEV, GM, AZM and CM. A previous work in our group found sub-minimal inhibitory concentrations (sub-MIC) of CIP or LEV treating the donor strain SM10λπ significantly increased conjugation frequency (Shun-Mei et al., 2018). We also found PAO1 treated with GM, AZM or CM promoted SM10λπ-PAO1 conjugation (Lu et al., 2017a). Thus, these five antibiotics were used in the current study. The results showed that GadY deletion decreased SM10λπ-PAO1 conjugation under sub-MIC CIP or LEV treatment, but had little influence on GM, AZM or CM group (Figure 6; Supplementary Figure S2). It may be related to the different ways in which the several types of antibiotics regulate bacterial conjugation.
CIP and LEV are synthetic quinolone antibiotics with a broad spectrum of antimicrobial activity for treating various types of infections in humans (Bolon, 2009). However, the increasing emergence of antibiotic resistance compromised their use. The acquisition of quinolone resistance has multiple causes. Plasmid-mediated conjugation is one of the main resistance mechanisms, besides genomic mutations of the bacteria (Strahilevitz et al., 2009). Our previous work showed treating the donor stains SM10λπ with sub-MIC (1/8-1/32 MIC) of CIP or LEV promote plasmid transfer into the recipient strains PAO1 through conjugation (Shun-Mei et al., 2018). An earlier study also found low concentrations of CIP increase the transfer frequencies of resistance plasmids (Ohlsen et al., 2003). GM, AZM and CM have been identified as QS-inhibiting antibiotics which reduce P. aeruginosa AHLs production (Skindersoe et al., 2008; Lu et al., 2017a; Zeng et al., 2016). This suggests GM, AZM and CM positively regulate SM10λπ-PAO1 conjugation by suppressing the AHLs-SdiA signalling between the donor and recipient cells. Thus, CIP and LEV promote bacterial conjugation by influencing the donor strain SM10λπ, while GM, AZM and CM function by affecting the recipient strain PAO1. Intriguingly, our further study found sub-MIC CIP or LEV treatment significantly increased GadY expression in donor strains, while GM, AZM or CM treatment did not (Figure 6; Supplementary Figure S2). It suggests that the up-regulated expression of GadY in donor strains by quinolone antibiotics increases SM10λπ-PAO1 conjugation.
In conclusion, a novel role for the Hfq-binding sRNA GadY in regulating conjugation between E. coli and other bacteria is demonstrated in this study. Positively induced by quinolone antibiotics, GadY involves in regulating SM10λπ-PAO1 conjugation by targeting the orphan LuxR-type receptor SdiA, and then decreases the expressions of global repressors KorA and KorB. Thus, our findings disclose a novel function of the Hfq-binding sRNA GadY and shed new light on developing a promising approach to counter antibiotic resistance by inhibition of conjugation.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
SZ: Data curation, Formal Analysis, Methodology, Project administration, Writing – original draft. JL: Data curation, Formal Analysis, Project administration, Writing – original draft. QL: Data curation, Formal Analysis, Project administration, Writing – original draft. ML: Data curation, Formal Analysis, Writing – review & editing. RY: Data curation, Formal Analysis, Writing – review & editing. YL: Resources, Writing – review & editing. XM: Formal Analysis, Writing – review & editing. YC: Formal Analysis, Writing – review & editing. CS: Resources, Writing – review & editing. JZ: Resources, Writing – review & editing. BH: Conceptualization, Supervision, Writing – review & editing. CC: Conceptualization, Supervision, Writing – review & editing. JP: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (Grant No. 82202590; 82072345; 82372308), the Natural Science Foundation of Guangdong Province (Grant No. 2024A1515011306; 2023A1515011011), the Guangzhou Science Technology and Innovation Commission (Grant No. 2024A03J0111; 2023A03J0225; 202201011470), and the Guangdong Provincial Hospital of Traditional Chinese Medicine (Grant No. YN2022QN19).
We are very grateful to Prof. B.L. Wanner (Department of Biological Sciences, Purdue University, West Lafayette, USA) for generously providing the lambda red recombination system, and Prof. H. P. Schweizer (Department of Microbiology and Infectious Diseases, University of Calgary Health Sciences Center, Alberta, Canada) for providing the plasmid pUCP24.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1445850/full#supplementary-material
Balzer, D., Ziegelin, G., Pansegrau, W., Kruft, V., Lanka, E. (1992). KorB protein of promiscuous plasmid RP4 recognizes inverted sequence repetitions in regions essential for conjugative plasmid transfer. Nucleic Acids Res. 20, 1851–1858. doi: 10.1093/nar/20.8.1851
Bechhofer, D. H., Figurski, D. H. (1983). Map location and nucleotide sequence of korA, a key regulatory gene of promiscuous plasmid RK2. Nucleic Acids Res. 11, 7453–7469. doi: 10.1093/nar/11.21.7453
Bechhofer, D. H., Kornacki, J. A., Firshein, W., Figurski, D. H. (1986). Gene control in broad host range plasmid RK2: expression, polypeptide product, and multiple regulatory functions of korB. Proc. Natl. Acad. Sci. U.S.A. 83, 394–398. doi: 10.1073/pnas.83.2.394
Bingle, L. E., Macartney, D. P., Fantozzi, A., Manzoor, S. E., Thomas, C. M. (2005). Flexibility in repression and cooperativity by KorB of broad host range IncP-1 plasmid RK2. J. Mol. Biol. 349, 302–316. doi: 10.1016/j.jmb.2005.03.062
Bolon, M. K. (2009). The newer fluoroquinolones. Infect. Dis. Clin. North Am. 23, 1027–1051. doi: 10.1016/j.idc.2009.06.003
Cabezón, E., de la Cruz, F., Arechaga, I. (2017). Conjugation inhibitors and their potential use to prevent dissemination of antibiotic resistance genes in bacteria. Front. Microbiol. 30, 8:2329. doi: 10.3389/fmicb.2017.02329
Datsenko, K. A., Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
Desgranges, E., Marzi, S., Moreau, K., Romby, P., Caldelari, I. (2019). Noncoding RNA. Microbiol. Spectr. 7. doi: 10.1128/microbiolspec.GPP3-0038-2018
Dutta, T., Srivastava, S. (2018). Small RNA-mediated regulation in bacteria: A growing palette of diverse mechanisms. Gene 656, 60–72. doi: 10.1016/j.gene.2018.02.068
Faner, M. A., Feig, A. L. (2013). Identifying and characterizing Hfq-RNA interactions. Methods 63, 144–159. doi: 10.1016/j.ymeth.2013.04.023
Fürste, J. P., Pansegrau, W., Ziegelin, G., Kröger, M., Lanka, E. (1989). Conjugative transfer of promiscuous IncP plasmids: interaction of plasmid-encoded products with the transfer origin. Proc. Natl. Acad. Sci. U.S.A. 86, 1771–1775. doi: 10.1073/pnas.86.6.1771
García-Reyes, S., Soberón-Chávez, G., Cocotl-Yanez, M. (2020). The third quorum-sensing system of Pseudomonas aeruginosa: Pseudomonas quinolone signal and the enigmatic PqsE protein. J. Med. Microbiol. 69, 25–34. doi: 10.1099/jmm.0.001116
Giacomini, A., Corich, V., Ollero, F. J., Squartini, A., Nuti, M. P. (1992). Experimental conditions may affect reproducibility of the beta-galactosidase assay. FEMS Microbiol. Lett. 100, 87–90. doi: 10.1111/j.1574-6968.1992.tb05687.x
Gottesman, S. (2004). The small RNA regulators of Escherichia coli: roles and mechanisms*. Annu. Rev. Microbiol. 58, 303–328. doi: 10.1146/annurev.micro.58.030603.123841
Jagura-Burdzy, G., Khanim, F., Smith, C. A., Thomas, C. M. (1992). Crosstalk between plasmid vegetative replication and conjugative transfer: repression of the trfA operon by trbA of broad host range plasmid RK2. Nucleic Acids Res. 20, 3939–3944. doi: 10.1093/nar/20.15.3939
Jagura-Burdzy, G., Thomas, C. M. (1994). KorA protein of promiscuous plasmid RK2 controls a transcriptional switch between divergent operons for plasmid replication and conjugative transfer. Proc. Proc. Natl. Acad. Sci. U.S.A. 91, 10571–10575. doi: 10.1073/pnas.91.22.10571
Jørgensen, M. G., Pettersen, J. S., Kallipolitis, B. H. (2020). sRNA-mediated control in bacteria: An increasing diversity of regulatory mechanisms. Biochim. Biophys. Acta Gene Regul. Mech. 1863, 194504. doi: 10.1016/j.bbagrm.2020.194504
Kavita, K., de Mets, F., Gottesman, S. (2018). New aspects of RNA-based regulation by Hfq and its partner sRNAs. Curr. Opin. Microbiol. 42, 53–61. doi: 10.1016/j.mib.2017.10.014
Koraimann, G., Teferle, K., Markolin, G., Woger, W., Högenauer, G. (1996). The FinOP repressor system of plasmid R1: analysis of the antisense RNA control of traJ expression and conjugative DNA transfer. Mol. Microbiol. 21, 811–821. doi: 10.1046/j.1365-2958.1996.361401.x
Kostelidou, K., Jones, A. C., Thomas, C. M. (1999). Conserved C-terminal region of global repressor KorA of broad-host-range plasmid RK2 is required for co-operativity between KorA and a second RK2 global regulator, KorB. J. Mol. Biol. 289, 211–221. doi: 10.1006/jmbi.1999.2761
Kumar, L., Patel, S. K. S., Kharga, K., Kumar, R., Kumar, P., Pandohee, J., et al. (2022). Molecular mechanisms and applications of N-Acyl homoserine lactone-mediated quorum sensing in bacteria. Molecules 27, 7584. doi: 10.3390/molecules27217584
Lalaouna, D., Morissette, A., Carrier, M. C., Massé, E. (2015). DsrA regulatory RNA represses both hns and rbsD mRNAs through distinct mechanisms in Escherichia coli. Mol. Microbiol. 98, 357–369. doi: 10.1111/mmi.13129
Lee, J., Zhang, L. (2015). The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 6, 26–41. doi: 10.1007/s13238-014-0100-x
Le Vo, T. D., Kim, T. W., Hong, S. H. (2012). Effects of glutamate decarboxylase and gamma-aminobutyric acid (GABA) transporter on the bioconversion of GABA in engineered Escherichia coli. Bioprocess Biosyst. Eng. 35, 645–650. doi: 10.1007/s00449-011-0634-8
Li, W., Ying, X., Lu, Q., Chen, L. (2012). Predicting sRNAs and their targets in bacteria. Genomics Proteomics Bioinf. 10, 276–284. doi: 10.1016/j.gpb.2012.09.004
Link, T. M., Valentin-Hansen, P., Brennan, R. G. (2009). Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl. Acad. Sci. U.S.A. 106, 19292–19297. doi: 10.1073/pnas.0908744106
Llosa, M., Gomis-Rüth, F. X., Coll, M., de la Cruz Fd, F. (2002). Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 45, 1–8. doi: 10.1046/j.1365-2958.2002.03014.x
Lu, Y., Li, H., Pu, J., Xiao, Q., Zhao, C., Cai, Y., et al. (2019). Identification of a novel RhlI/R-PrrH-LasI/Phzc/PhzD signalling cascade and its implication in P. aeruginosa virulence. Emerg. Microbes Infect. 8, 1658–1667. doi: 10.1080/22221751.2019.1687262
Lu, Y., Zeng, J., Wang, L., Lan, K., E, S., Wang, L., et al. (2017a). Antibiotics promote Escherichia coli-Pseudomonas aeruginosa conjugation through inhibiting quorum sensing. Antimicrob. Agents Chemother. 61, e01284–e01217. doi: 10.1128/AAC.01284-17
Lu, Y., Zeng, J., Wu, B., E, S., Wang, L., Cai, R., et al. (2017b). Quorum sensing N-acyl homoserine lactones-SdiA suppresses Escherichia coli-Pseudomonas aeruginosa conjugation through inhibiting traI expression. Front. Cell Infect. Microbiol. 7, 7. doi: 10.3389/fcimb.2017.00007
Ma, X., Zhang, S., Xu, Z., Li, H., Xiao, Q., Qiu, F., et al. (2020). SdiA improves the acid tolerance of E. coli by regulating gadW and gadY expression. Front. Microbiol. 11, 1078. doi: 10.3389/fmicb.2020.01078
Mann, M., Wright, P. R., Backofen, R. (2017). IntaRNA 2.0: enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res. 45, W435–W439. doi: 10.1093/nar/gkx279
Mayer, C., Borges, A., Flament-Simon, S. C., Simões, M. (2023). Quorum sensing architecture network in Escherichia coli virulence and pathogenesis. FEMS Microbiol. Rev. 47, fuad031. doi: 10.1093/femsre/fuad031
McInnes, R. S., McCallum, G. E., Lamberte, L. E., van Schaik, W. (2020). Horizontal transfer of antibiotic resistance genes in the human gut microbiome. Curr. Opin. Microbiol. 53, 35–43. doi: 10.1016/j.mib.2020.02.002
Murina, V. N., Nikulin, A. D. (2015). Bacterial small regulatory RNAs and Hfq protein. Biochem. (Mosc) 80, 1647–1654. doi: 10.1134/S0006297915130027
Negrete, A., Shiloach, J. (2015). Constitutive expression of the sRNA GadY decreases acetate production and improves E. coli growth. Microb. Cell Fact. 14, 148. doi: 10.1186/s12934-015-0334-1
Ohlsen, K., Ternes, T., Werner, G., Wallner, U., Löffler, D., Ziebuhr, W., et al. (2003). Impact of antibiotics on conjugational resistance gene transfer in Staphylococcus aureus in sewage. Environ. Microbiol. 5, 711–716. doi: 10.1046/j.1462-2920.2003.00459.x
Opdyke, J. A., Fozo, E. M., Hemm, M. R., Storz, G. (2011). RNase III participates in GadY-dependent cleavage of the gadX-gadW mRNA. J. Mol. Biol. 406, 29–43. doi: 10.1016/j.jmb.2010.12.009
Opdyke, J. A., Kang, J. G., Storz, G. (2004). GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 186, 6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004
Pansegrau, W., Balzer, D., Kruft, V., Lurz, R., Lanka, E. (1990). In vitro assembly of relaxosomes at the transfer origin of plasmid RP4. Proc. Natl. Acad. Sci. U.S.A. 87, 6555–6559. doi: 10.1073/pnas.87.17.6555
Pansegrau, W., Lanka, E., Barth, P. T., Figurski, D. H., Guiney, D. G., Haas, D., et al. (1994). Complete nucleotide sequence of Birmingham IncP alpha plasmids. Compilation and comparative analysis. J. Mol. Biol. 239, 623–663. doi: 10.1006/jmbi.1994.1404
Pansegrau, W., Schröder, W., Lanka, E. (1993). Relaxase (TraI) of IncP alpha plasmid RP4 catalyzes a site-specific cleaving-joining reaction of single-stranded DNA. Proc. Natl. Acad. Sci. U.S.A. 90, 2925–2929. doi: 10.1073/pnas.90.7.2925
Papenfort, K., Bassler, B. L. (2016). Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 14, 576–588. doi: 10.1038/nrmicro.2016.89
Parmeciano Di Noto, G., Molina, M. C., Quiroga, C. (2019). Insights into non-coding RNAs as novel antimicrobial drugs. Front. Genet. 10, 57. doi: 10.3389/fgene.2019.00057
Pfeiffer, V., Papenfort, K., Lucchini, S., Hinton, J. C., Vogel, J. (2009). Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat. Struct. Mol. Biol. 16, 840–846. doi: 10.1038/nsmb.1631
Pu, J., Zhang, S., He, X., Zeng, J., Shen, C., Luo, Y., et al. (2022). The small RNA AmiL regulates quorum sensing-mediated virulence in Pseudomonas aeruginosa PAO1. Microbiol. Spectr. 10, e0221121. doi: 10.1128/spectrum.02211-21
Santiago-Frangos, A., Woodson, S. A. (2018). Hfq chaperone brings speed dating to bacterial sRNA. Wiley Interdiscip. Rev. RNA 9, e1475. doi: 10.1002/wrna.1475
Schmidhauser, T. J., Bechhofer, D. H., Figurski, D. H., Helinski, D. R. (1989). Host-specific effects of the korA-korB operon and oriT region on the maintenance of miniplasmid derivatives of broad host-range plasmid RK2. Plasmid 21, 99–112. doi: 10.1016/0147-619X(89)90053-X
Shun-Mei, E., Zeng, J. M., Yuan, H., Lu, Y., Cai, R. X., Chen, C. (2018). Sub-inhibitory concentrations of fluoroquinolones increase conjugation frequency. Microb. Pathog. 114, 57–62. doi: 10.1016/j.micpath.2017.11.036
Skindersoe, M. E., Alhede, M., Phipps, R., Yang, L., Jensen, P. O., Rasmussen, T. B., et al. (2008). Effects of antibiotics on quorum sensing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 52, 3648–3663. doi: 10.1128/AAC.01230-07
Smith, J. L., Fratamico, P. M., Yan, X. (2011). Eavesdropping by bacteria: the role of SdiA in Escherichia coli and Salmonella enterica serovar Typhimurium quorum sensing. Foodborne Pathog. Dis. 8, 169–178. doi: 10.1089/fpd.2010.0651
Smith, C. A., Thomas, C. M. (1984). Molecular gentic analysis of the trfB and korB region of broad host range plasmid RK2. J. Gen. Microbiol. 130, 1651–1663. doi: 10.1099/00221287-130-7-1651
Storz, G., Vogel, J., Wassarman, K. M. (2011). Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell 43, 880–891. doi: 10.1016/j.molcel.2011.08.022
Strahilevitz, J., Jacoby, G. A., Hooper, D. C., Robicsek, A. (2009). Plasmid-mediated quinolone resistance: a multifaceted threat. Clin. Microbiol. Rev. 22, 664–689. doi: 10.1128/CMR.00016-09
Tramonti, A., De Canio, M., De Biase, D. (2008). GadX/GadW-dependent regulation of the Escherichia coli acid fitness island: transcriptional control at the gadY-gadW divergent promoters and identification of four novel 42 bp GadX/GadW-specific binding sites. Mol. Microbiol. 70, 965–982. doi: 10.1111/j.1365-2958.2008.06458.x
Vogel, J., Luisi, B. F. (2011). Hfq and its constellation of RNA. Nat. Rev. Microbiol. 9, 578–589. doi: 10.1038/nrmicro2615
von Wintersdorff, C. J., Penders, J., van Niekerk, J. M., Mills, N. D., Majumder, S., van Alphen, L. B., et al. (2016). Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 7, 173. doi: 10.3389/fmicb.2016.00173
Vrancianu, C. O., Popa, L. I., Bleotu, C., Chifiriuc, M. C. (2020). Targeting plasmids to limit acquisition and transmission of antimicrobial resistance. Front. Microbiol. 11, 761. doi: 10.3389/fmicb.2020.00761
Waters, L. S., Storz, G. (2009). Regulatory RNAs in bacteria. Cell 136, 615–628. doi: 10.1016/j.cell.2009.01.043
West, S. E., Schweizer, H. P., Dall, C., Sample, A. K., Runyen-Janecky, L. J. (1994). Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148, 81–86. doi: 10.1016/0378-1119(94)90237-2
Will, W. R., Frost, L. S. (2006). Hfq is a regulator of F-plasmid TraJ and TraM synthesis in Escherichia coli. J. Bacteriol. 188, 124–131. doi: 10.1128/JB.188.1.124-131.2006
Xiong, R., Liu, Y., Pu, J., Liu, J., Zheng, D., Zeng, J., et al. (2021). Indole inhibits IncP-1 conjugation system mainly through promoting korA and korB expression. Front. Microbiol. 12, 628133. doi: 10.3389/fmicb.2021.628133
Yamamoto, K., Yata, K., Fujita, N., Ishihama, A. (2001). Novel mode of transcription regulation by SdiA, an Escherichia coli homologue of the quorum-sensing regulator. Mol. Microbiol. 41, 1187–1198. doi: 10.1046/j.1365-2958.2001.02585.x
Young, C., Burlage, R. S., Figurski, D. H. (1987). Control of the kilA gene of the broad-host-range plasmid RK2: involvement of korA, korB, and a new gene, korE. J. Bacteriol. 169, 1315–1320. doi: 10.1128/jb.169.3.1315-1320.1987
Keywords: GadY, small RNA, plasmid conjugation, Escherichia coli, SdiA
Citation: Zhang S, Long J, Li Q, Li M, Yu R, Lu Y, Ma X, Cai Y, Shen C, Zeng J, Huang B, Chen C and Pu J (2024) Small RNA GadY in Escherichia coli enhances conjugation system of IncP-1 by targeting SdiA. Front. Cell. Infect. Microbiol. 14:1445850. doi: 10.3389/fcimb.2024.1445850
Received: 08 June 2024; Accepted: 08 July 2024;
Published: 23 July 2024.
Edited by:
Xiancai Rao, Army Medical University, ChinaCopyright © 2024 Zhang, Long, Li, Li, Yu, Lu, Ma, Cai, Shen, Zeng, Huang, Chen and Pu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin Huang, aHVhbmdiM0BtYWlsLnN5c3UuZWR1LmNu; Cha Chen, Y2hlbmNoYTkwNkAxNjMuY29t; Jieying Pu, cHVqeTExMDVAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.