- 1College of Veterinary Medicine, Mississippi State University, Mississippi State, MS, United States
- 2Bioinformatics & Systems Biology, Justus-Liebig-University Giessen, Giessen, Germany
- 3Thad Cochran National Warmwater Aquaculture Center, Stoneville, MS, United States
- 4Warmwater Aquaculture Research Unit, USDA-ARS, Stoneville, MS, United States
- 5Faculty of Fisheries, Recep Tayyip Erdogan University, Rize, Türkiye
Type IV pili (T4P) are versatile proteinaceous protrusions that mediate diverse bacterial processes, including adhesion, motility, and biofilm formation. Aeromonas hydrophila, a Gram-negative facultative anaerobe, causes disease in a wide range of hosts. Previously, we reported the presence of a unique Type IV class C pilus, known as tight adherence (Tad), in virulent Aeromonas hydrophila (vAh). In the present study, we sought to functionalize the role of Tad pili in the pathogenicity of A. hydrophila ML09-119. Through a comprehensive comparative genomics analysis of 170 A. hydrophila genomes, the conserved presence of the Tad operon in vAh isolates was confirmed, suggesting its potential contribution to pathogenicity. Herein, the entire Tad operon was knocked out from A. hydrophila ML09-119 to elucidate its specific role in A. hydrophila virulence. The absence of the Tad operon did not affect growth kinetics but significantly reduced virulence in catfish fingerlings, highlighting the essential role of the Tad operon during infection. Biofilm formation of A. hydrophila ML09-119 was significantly decreased in the Tad operon deletant. Absence of the Tad operon had no effect on sensitivity to other environmental stressors, including hydrogen peroxide, osmolarity, alkalinity, and temperature; however, it was more sensitive to low pH conditions. Scanning electron microscopy revealed that the Tad mutant had a rougher surface structure during log phase growth than the wildtype strain, indicating the absence of Tad impacts the outer surface of vAh during cell division, of which the biological consequences are unknown. These findings highlight the role of Tad in vAh pathogenesis and biofilm formation, signifying the importance of T4P in bacterial infections.
1 Introduction
Aquaculture helps meet the ever-growing global demand for animal protein, yet it faces a substantial threat from fish pathogens, leading to economic losses and food insecurity (Mzula et al., 2019; Irshath et al., 2023). Among these pathogens, several Aeromonas species are prominent agents of Motile Aeromonas Septicemia (MAS) in temperate and warm freshwater fish, with Aeromonas hydrophila being a significant contributor (Irshath et al., 2023).
A. hydrophila is a Gram-negative facultative anaerobe that thrives in fresh and brackish water environments. It possesses a wide host range which includes amphibians, fish, birds, reptiles, and mammals, including humans (Huys et al., 2003; Tsai et al., 2015; Laviad-Shitrit et al., 2018), causing septicemia, necrotizing fasciitis, and gastroenteritis (Parker and Shaw, 2011; Tomás, 2012; Tsai et al., 2015; Pessoa et al., 2022). Consequently, extensive research has centered on investigating the underlying virulence mechanisms of A. hydrophila. Multiple virulence factors have been identified, including iron-binding systems, flagella, adhesins, exotoxins, enterotoxins, secretion systems, S-layers, and fimbriae (Horneman et al., 2009; Tomás, 2012; Rasmussen-Ivey et al., 2016a; Davidsson et al., 2017; Barger et al., 2020). A particular clonal type of A. hydrophila affiliated with sequence type 251 (ST251) is highly virulent in catfish and carp species in aquaculture, referred hereafter as virulent A. hydrophila (vAh) (Rasmussen-Ivey et al., 2016b).
Fimbriae (pili) are long extracellular polymers that enable bacteria to interact with their external environment (Hospenthal et al., 2017). Type IV pili (T4P) are widely distributed across bacterial and archaeal phyla and allow for twitching motility, DNA uptake through natural transformation, host colonization and signaling, chemotaxis, surface sensing, and virulence protein secretion (Cehovin et al., 2013; Angelov et al., 2015; Kåhrström, 2015; Piepenbrink, 2019; Mix et al., 2021; Oliveira et al., 2022). T4P, spanning both the outer and inner membrane, are divided into two subclasses, type IV class a pili (T4aP) and type IV class b pili (T4bP), based on the length of their prepilin leader sequence (Pelicic, 2008; McCallum et al., 2019; Ellison et al., 2022).
A third subclass of T4P, Type IV class c pili (T4cP), was recently classified due to the distinct genetic organization and evolutionary history of Tight Adherence Pili (Tad) (Beeby, 2019; Denise et al., 2019). Tad pili, originally discovered in Actinobacillus actinomycetemcomitans, are crucial for forming bundled fibers that facilitate autoaggregation, adherence, colonization, and biofilm development (Kachlany et al., 2000). It has been postulated that Tad originated via horizontal acquisition from Archaea given their significant similarity to archaeal Epd-like systems (Denise et al., 2019). For example, Tad, located on single locus, lacks a retraction ATPase and instead encodes a bifunctional ATPase responsible for both extension and retraction processes (Beeby, 2019; Ellison et al., 2019; Piepenbrink, 2019).
A. hydrophila exhibits outer membrane-spanning type I pili and T4P. A. hydrophila encodes two distinct types of T4aP: Mannose-Sensitive Hemagglutinin (MSHA) and Type IV Aeromonas Pilus (Tap). MSHA bundle forming pili shares high homology to the MSHA pili of Vibrio spp., while Tap pili shares high homology to the Pil system of Vibrio and Pseudomonas spp. In addition, previous research reported the consistent presence of genes encoding Tad pili in vAh genomes (Tekedar et al., 2019). Notably, the Tad cluster was also found in a clinical A. hydrophila isolate infecting humans, suggesting potential significance in pathogenicity (Tekedar et al., 2019). However, within the context of vAh pathogenicity, the precise functions of the T4P have not been investigated previously.
The Tad system, with its diverse functions, serves as a bridge that facilitates interaction between the pathogen and its host during infection. This study aimed to delineate the function of Tad pili in vAh isolate ML09-119. We conducted an expanded comparative genomics analysis involving 170 A. hydrophila genomes sourced from the National Center for Biotechnology Information (NCBI), confirming the consistent presence of the Tad gene cluster in vAh genomes. We constructed a mutant strain with complete deletion of the Tad operon and investigated its contribution to virulence in catfish. Additionally, the influence of Tad pili on A. hydrophila physiology and response to environmental stress was determined. These results revealed the significant role of Tad pili in the virulence of vAh isolate ML09-119, highlighting its crucial function in mediating pathogenic interactions.
2 Materials and methods
2.1 Genome features and building phylogenetic tree
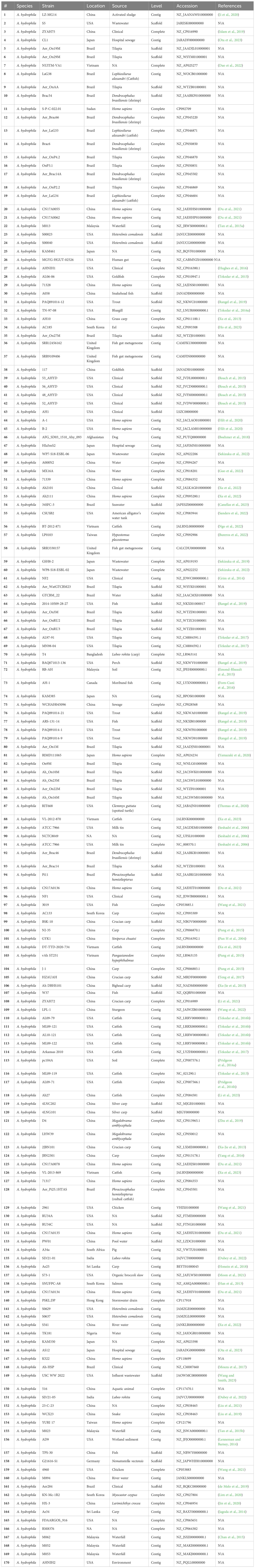
Genome sequence data for 170 A. hydrophila genomes were downloaded from the NCBI database. Genome metadata is listed in Table 1. All selected genomes shared at least 95% average nucleotide identity (ANI) among them, indicating conspecificity, and a phylogenetic tree was built from the core genome (Blom et al., 2016). Also, AAI was calculated with EDGAR 2.0 (Blom et al., 2016). Gene sets from the core genome were aligned individually using MUSCLE (Edgar, 2004) then concatenated. The concatenated alignment was used to compute a Kimura distance matrix, which was served as input for the Neighbor-Joining algorithm implemented in PHYLP (Baum, 1989). The phylogenetic tree for 170 genomes was build out of a core of 1893 genes per genome, 321810 in total. The core has 1711702 AA-residues/bp per genome, 290989340 in total.
2.2 Distribution of individual Tad operon genes in 170 A. hydrophila genomes
The virulence factor database (VFDB) (Liu et al., 2022) and MacSyFinder tool were used with default features to identify individual Tad operon genes across the 170 A. hydrophila genomes investigated (Abby et al., 2014; Abby et al., 2024). Specifically, the “unordered” dataset option was chosen because most of the evaluated genomes were incomplete drafts. Briefly, the following parameters were used: the topology of the replicon was linear/circular, maximal E-value was 1.0, maximal independent E-value was 0.001, and the minimal profile coverage was 0.5. Both mandatory genes and accessory genes were identified. The protein sequences of the 13 Tad operon genes were identified using VFDB and MacSyfinder. Based on these findings, the 13 genes were organized in an operon structure (Figure 1). The 13 Tad operon gene protein sequences were then uploaded to CLC Genomics Workbench (version 23.0.3), where BLASTp searches were conducted to compare the 13 Tad genes against the 170 A. hydrophila genomes. Only matches with E-values 1×10-20 were considered present. The distribution of the predicted 13 Tad operon genes among 170 A. hydrophila genomes was presented using the pheatmap library in R Studio (Version 2023.06.0 + 421) (Figure 2). Also, the GC content of the Tad operon was analyzed and compared to the entire genome of A. hydrophila ML09-119 using SnapGene (version 6.2.1) (Figure 1).
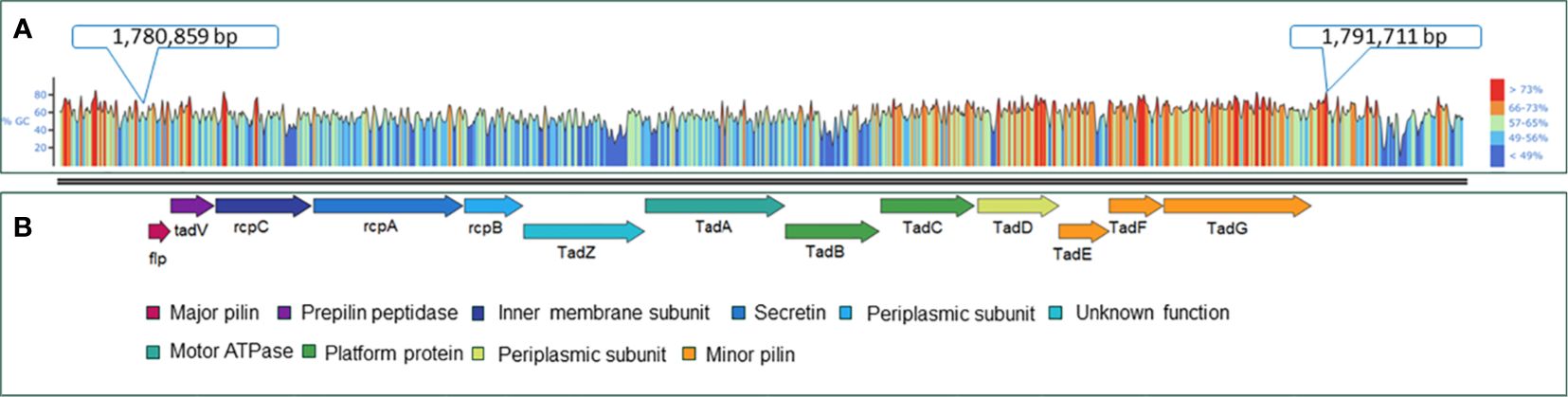
Figure 1 Genetic map of the A. hydrophila ML09-119 Tad pilus operon and its component genes. (A) GC content analysis of the Tad operon within the A. hydrophila ML09-119 genome. The genome location of the Tad operon is indicated in the figure. (B) The components of the Tad operon, along with their predicted or known functions, are indicated. The Tad genes are color-coded based on their respective functions.
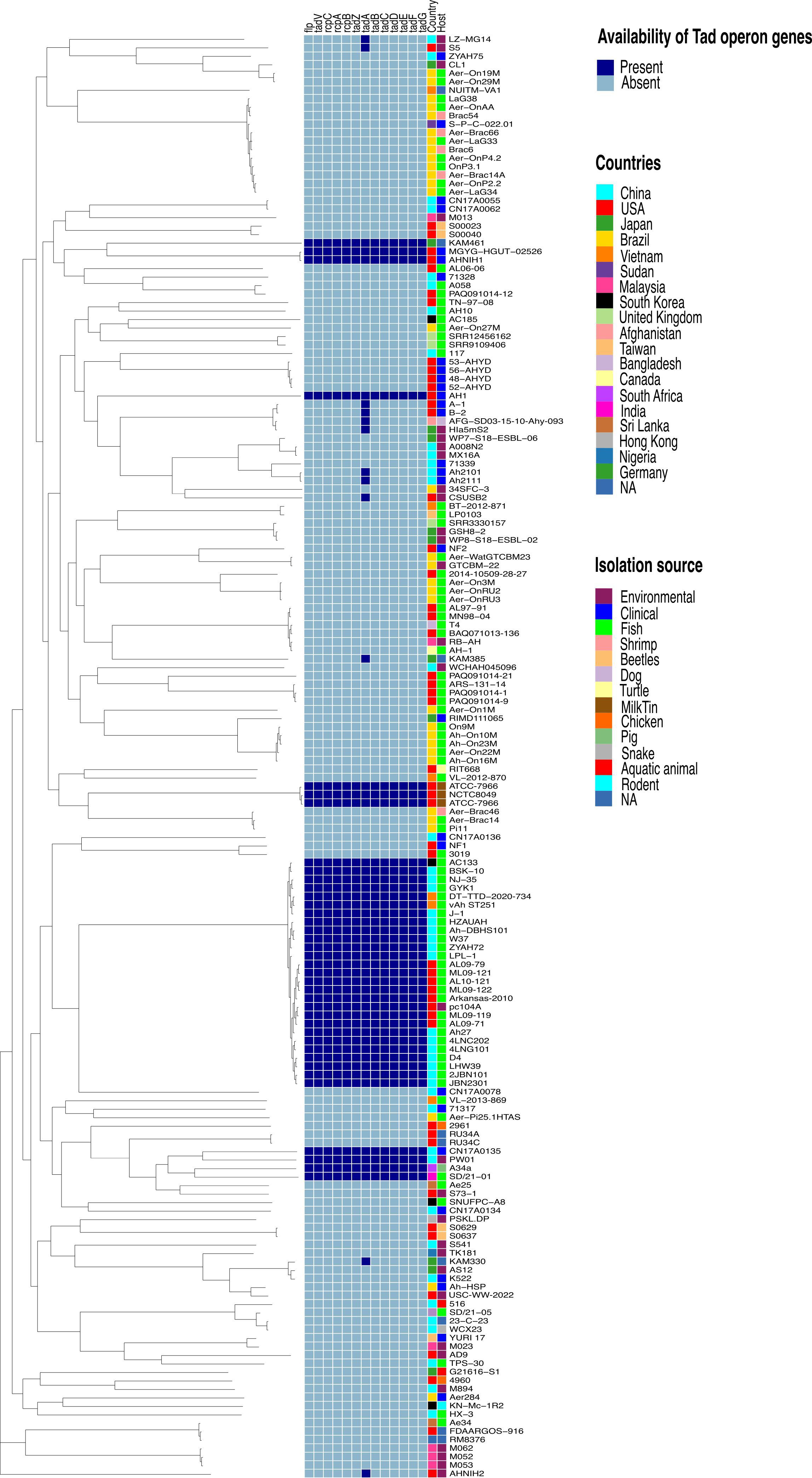
Figure 2 Phylogenetic tree based on the Aeromonas hydrophila core genome and availability of individual Tad operon genes across 170 A. hydrophila genomes. Isolation sources and geographical locations for each genome are included. Navy blue color represents the presence of Tad operon genes, whereas light blue represents absence. Branch lengths were reduced to fit the image.
2.3 Bacterial strains and plasmids
A. hydrophila strain ML09-119 (Tekedar et al., 2013) was used as a representative of the vAh clonal group impacting U.S. channel catfish aquaculture. The bacterial strains and plasmids used in this study are listed in Table 2. Escherichia coli strains CC118 λpir and BW19851 were used for cloning and conjugation purposes, respectively, and suicide plasmid pMEG-375 was used to transfer gene mutations into A. hydrophila. Brain heart infusion (BHI) agar and broth (Difco, Sparks, MD, USA) were used to grow A. hydrophila strain at 30°C. Luria–Bertani (LB) agar and broth (Difco) were used for E. coli incubation at 37°C. When needed, the following antibiotics and reagents (Sigma-Aldrich, Saint Louis, MN, USA) were used in the given concentrations: colistin (12.5 μg/mL), chloramphenicol (10-25 μg/mL), ampicillin (100 µg/mL), mannitol (0.35%), and sucrose (5%).
2.4 In-frame deletion of Tad operon in A. hydrophila ML09-119
In-frame deletion was used to create a complete Tad operon mutant in A. hydrophila ML09-119. Briefly, the Tad operon mutant was constructed by allelic exchange and homologous recombination using suicide plasmid pMEG-375 containing the counter selectable gene sacB (Dozois et al., 2003). Four primers (A, B, C, and D) were designed for the Tad operon region using Primer3 (https://primer3.ut.ee/) (Untergasser et al., 2012) (Table 3). Two specific compatible restriction enzyme sites were introduced in A and D primers for cloning, and the reverse complement of primer B was added to the 5′ end of primer C to create an overlap region to initiate the fusion of PCR fragments by overlap extension PCR (Horton et al., 1989). The upstream (AB fragment) and downstream (CD fragment) regions of the Tad operon were amplified using two sets of primers. Amplified PCR fragments AB and CD were annealed at the overlapping regions, and a single fragment was created using the A and D primers. The AD product was purified, digested, and ligated into digested pMEG-375, then transformed into E. coli CC118λpir, and the final product was grown on LB agar with ampicillin. The positive plasmids carrying the mutated Tad operon were transferred into A. hydrophila ML09-119 by conjugation using E. coli BW19851 strain. Transconjugants were selected on plates containing chloramphenicol and colistin. In this process, chloramphenicol was used to identify the integration of pMEG-375 in A. hydrophila ML09-119 chromosome, while colistin was used as counterselection against E. coli. For confirmation purposes, PCR was used to verify the vector had integrated correctly into the A. hydrophila ML09-119 chromosome. Following sucrose selection, colistin resistant and chloramphenicol sensitive colonies were selected, and the candidate mutants were screened by colony PCR using A and D primers. Mutant validation was done by sequencing AD fragments amplified from chloramphenicol sensitive mutants using Tad-ConF01 and Tad-ConR01 (Table 3). The final A. hydrophila Tad operon mutant was designated as vAhΔtad.
2.5 Verification of Tad mutant, vAhΔtad, by complete genome sequencing
The vAhΔtad was expanded in 9 mL porcine brain heart infusion broth (Becton Dickinson, Franklin Lakes, NJ, USA) in static, overnight cultures at 28°C. An aliquot (3 mL) of the expanded culture was pelleted by centrifugation (20,000 x g). High molecular weight genomic DNA was isolated from the concentrated pellet using the PureGene DNA isolation kit (QIAGEN, Hilden, Germany). Long reads were produced on a GridION platform (Oxford Nanopore Technologies, Oxford, UK) using the rapid barcoding kit (RBK004) and v9.4.1 flow cells. These sequences were filtered to an average quality 10 using NanoFilt v2.2.0 (Bolger et al., 2014) to produce 211,519,148 bases from 30,912 reads of at least 1000 bp with an average read length of 6842 bp. The genome was assembled into a single contig with Canu v1.7 (Koren et al., 2017), the consensus sequence was produced using Medaka v0.11 (Oxford Nanopore Technologies, https://github.com/nanoporetech/medaka), and the contig was circularized manually.
2.6 Virulence of the vAhΔtad mutant in catfish fingerlings
All fish experiments were conducted under a protocol approved by the Institutional Animal Care and Use Committee (IACUC) at Mississippi State University. Virulence of vAhΔtad was compared to A. hydrophila ML09-119 wild-type strain (vAh-WT) by immersion route of exposure as described previously (Abdelhamed et al., 2016). Briefly, specific-pathogen-free (SPF) channel catfish fingerlings (13.54 ± 0.54 cm, 15.32 ± 1.98 g) were stocked into twelve 40-liter flow-through tanks (14 fish/tank) and acclimated for a week. Tanks were assigned randomly to three treatment groups: vAhΔtad, vAh-WT, and BHI (sham infected). Each group included three replicate tanks. Fish were fed twice daily with a commercial catfish feed, and water temperature was maintained at 30°C (±2) throughout the experiments. On the challenge day, the water levels in each tank were decreased to 5 L, and 100 mL of overnight culture were added directly to each tank (1.9 × 109 CFU/mL water). After three hours, the water flow was restored, and fish were maintained as usual. Negative control (sham-infected group) tanks were treated similarly, but exposed to 100 mL of sterile BHI broth. During the immersion process, the water was well aerated. Fish mortalities were recorded daily for 7 days, and cumulative percent mortality calculated for each group.
2.7 Growth kinetics
Growth kinetics of vAhΔtad and vAh-WT in BHI broth were compared. Briefly, bacterial growth curves were generated by measuring the optical density at 600nm (OD600) on a multimode reader (BioTek Cytation 5) every half an hour for 72 h at 30°C. The growth assays were conducted in two independent experiments, each with 24 replicates for vAhΔtad and vAh-WT strains.
2.8 Biofilm formation
Biofilm formation of the vAhΔtad and vAh-WT strains were compared using a microtiter plate assay. Five different colonies of each strain were cultured to log-phase (OD600 = 0.6 ± 0.05) at 28°C in BHI broth and diluted to OD600 of 0.1 using freshly prepared media. A 100 µL of the diluted cultures were added into a 96-well plate and incubated at 28°C overnight. Sterile BHI media (100 µL) was added to the blank control wells. For quantitative analysis, 8 replicate wells were used for each treatment and the control group. After incubation, the bacteria were discarded by turning the plate over and shaking out the liquid, then gently submerging the plate in a tub of water. The washing step was repeated twice to remove any unbound bacteria and media before staining. To each well, 125 µL of crystal violet (CV) (0.1%) was added and the plate was incubated at room temperature for 15 min. Next, the microtiter plate was rinsed 3-4 times by submerging in sterile water as described above, followed by vigorous blotting on a stack of paper to remove any unbound bacteria and dye. The plate was air-dried and 125 µL of 95% ethanol was added to each well to solubilize the CV. The plate was incubated for 15 min and the solubilized CV was transferred to a new flat bottom microtiter plate and absorbance was measured at 595nm. The biofilm assay was conducted with 35 biological replicates for both vAhΔtad and vAh-WT strains.
2.9 Motility
The vAhΔtad and vAh-WT strains were cultured to log-phase (OD600 = 0.6 ± 0.05) at 28°C in BHI broth. Two microliters of bacterial culture were spotted on a 0.6% BHI semi-solid plate and incubated at 28°C. Replicate plates were used for statistical analysis. The diameters of bacterial growth were measured after 16 hours of incubation. Each motility assay was conducted with 7 independent biological replicates, with 3 technical replicates per biological replicate, for both the vAhΔtad and vAh-WT strains.
2.10 Temperature stress
The ability of the vAhΔtad and vAh-WT strains to withstand extreme temperatures was evaluated. Both strains were cultured to log phase (OD600 = 0.6 ± 0.05) at 28°C and then adjusted to an OD600 of 0.1 using sterile BHI broth. The bacterial suspension was subsequently incubated at 18°C, 28°C (control), and 37°C for 24h at 180 rpm. Following incubation, the cultures were washed twice with sterile phosphate-buffered saline (PBS) and spread on BHI agar plates for colony counting and statistical analysis. The temperature assay was conducted with 4 biological replicates for both vAhΔtad and vAh-WT strains.
2.11 Acid-alkali stress
The vAhΔtad and vAh-WT strains were cultured to log-phase (OD600 = 0.6 ± 0.05) at 28°C in BHI broth. Bacterial pellets were collected, washed twice with sterile PBS, and incubated in BHI at pH 5.0, pH 6.0, pH 7.0 (control), pH 9.0, and pH 10.0 for 30 minutes at 28°C. Viable bacteria were quantified by spreading on BHI agar plates after serially diluting. The two strains’ percent viability (CFUs/mL) was calculated and compared. The acid-alkali stress assay was conducted with 3 biological replicates for both vAhΔtad and vAh-WT strains.
2.12 Osmotic stress
Under optimal growth conditions at 28°C, the vAhΔtad and vAh-WT strains were cultured to log-phase with an optical density of 0.6 ± 0.05 at 600nm. Bacterial pellets were collected, washed with PBS, and resuspended in fresh BHI broth containing 0.5 M sodium chloride (NaCl), which was then adjusted to OD600 of 0.1. Subsequently, 200 μL of each bacterial suspension was dispensed into separate wells of a 96-well microplate. To the control wells, 200 μL of sterile BHI broth was added. Growth was monitored every 1 h for 16 h at 28°C in an automatic microplate reader using absorbance at 600nm. The osmotic stress assay was conducted with four biological replicates for both vAhΔtad and vAh-WT strains.
2.13 Oxidative stress
To evaluate the viability of the strains against environmental oxidants, both vAhΔtad and vAh-WT strains were cultured to log-phase (OD600 = 0.6 ± 0.05) at 28°C in BHI broth. Bacterial pellets were collected and washed twice with sterile PBS solution before treatment with 0.1M hydrogen peroxide (H2O2), using PBS as control. Bacteria were incubated for 20 min at 28°C, and the viable bacteria were quantified by spreading them onto BHI agar. The percentage viability of the two strains in the treatment group (H2O2) compared to the control group (PBS) was calculated and compared between the mutant and wild-type strain. The oxidative stress assay was conducted with four biological replicates for both vAhΔtad and vAh-WT strains.
2.14 Scanning electron microscopy
The effects of Tad pili on the membrane structure of A. hydrophila was evaluated using scanning electron microscopy. Both the vAhΔtad and vAh-WT strains were grown at 30°C. Pellets were collected from log phase and overnight cultures, and bacteria were fixed in 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1M sodium cacodylate buffer for 2 hours at room temperature prior to fixation in 1% osmium tetroxide for one hour. Fixed pellets were then dehydrated in a graded ethanol series (10%, 20%, 30%, 50%, 70%, 80%, 85%, 95%, 100%), dried via a critical point dryer, mounted on aluminum stubs, and sputter coated with 30nm platinum. Images were captured using a JEOL 6500F field emission SEM operating at 10KeV at the Mississippi State University Institute for Imaging and Analytical Technologies.
2.15 Statistical analysis
Mean cumulative percent mortality data were arcsine transformed, and analysis of variance (ANOVA) was applied using PROC GLM in SAS for Windows v9.4 (SAS Institute, Inc., Cary, NC). For the characterization assays, the colony forming units/mL between the vAhΔtad and vAh-WT strains were compared and analyzed using one-way analysis of variance (ANOVA). An alpha level of 0.05 was used in all analyses.
3 Results
3.1 Distribution of Tad operon in A. hydrophila isolates
This study comprehensively analyzed 170 A. hydrophila genome sequences derived from diverse geographical locations and hosts (Table 1), several of which originated from catfish disease cases in Alabama and Mississippi (Tekedar et al., 2013; Tekedar et al., 2015; Tekedar et al., 2016a; Tekedar et al., 2016b; Tekedar et al., 2017). Isolate identity was confirmed by ANI, and all selected genomes displayed ANI values of 95% or higher, indicating they are all the same species (Goris et al., 2007). Subsequently, a phylogenetic tree was constructed for the 170 A. hydrophila strains based on a core set of 1893 genes per genome, totaling 321,810 genes. The core genome contained 1,711,702 amino acid residues per base pair or 290,989,340 in total. Notably, the vAh isolates formed a distinct cluster with 100% branch conservation, and this separation was verified by ANI and average amino acid identity (AAI) values (data not shown here).
Furthermore, analysis of the A. hydrophila ML09-119 genome revealed a 10,852 bp operon containing 13 genes (flp, tadV, rcpC, rcpA, rcpB, tadZ, tadA, tadB, tadC, tadD, tadE, tadF, tadG) within the range of AHML_RS08095 to AHML_RS08155, collectively encoding the Tad operon (Figure 1). The GC content of the Tad operon was 64.82%, while the average GC ratio of the entire genome was 60.82% (Figure 1). In silico analysis of Tad operon distribution across the 170 A. hydrophila genomes confirmed the presence of the Tad operon in all U.S. and Chinese (24 genomes) vAh isolates (Figure 2). Additionally, human clinical isolates, particularly from the U.S. (strains AH1, MGYG-HGUT-02526, and AHNIH1) and China (strain CN17A0135) encoded Tad operon (Figure 2). Interestingly, most non-vAh isolates did not encode the Tad operon, with the exception of isolates PW01 from swimming pool water, A34a from swine, SD/21-01 from rohu carp (Labeo rohita), KAM461 (origin unknown), and NCTC8049 and ATCC 7966 from milk tins (Figure 2). Sequencing of the Tad mutant, vAhΔtad, confirmed deletion of the Tad operon and the absence of additional random mutations outside the targeted locus.
3.2 Tad operon deletion impairs virulence and biofilm formation of A. hydrophila ML09-119
To investigate the role of Tad pili in A. hydrophila ML09-119 virulence, infectivity trials were conducted in catfish fingerlings. The vAhΔtad strain exhibited a significantly lower mortality rate (14.65%) compared to the vAh-WT strain (74.36%) 72 hour post-infection (Figure 3A). Next, the impact of deletion of the Tad operon on growth kinetics of vAh isolate ML09-119 in BHI broth was assessed, revealing no measurable differences in growth (Figure 3B). Furthermore, the role of the Tad pili on biofilm formation was evaluated. The vAhΔtad strain displayed significantly reduced (p ≤ 0.05) biofilm formation compared to the vAh-WT strain (Figure 3C), indicating that Tad pili are crucial for biofilm formation. No significant difference in motility was detected between the mutant and wild type parent (Figure 3D).
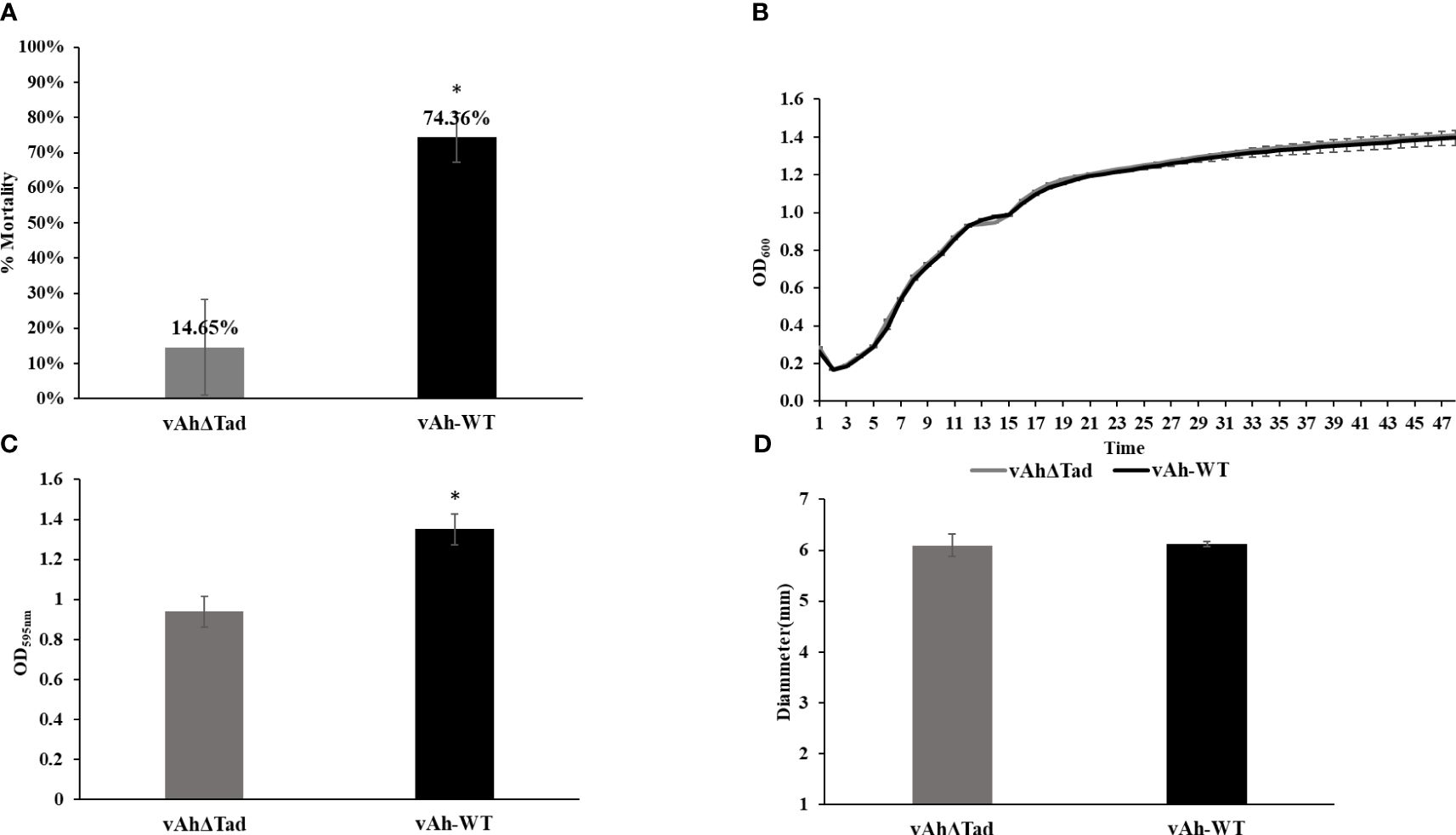
Figure 3 Evaluating the role of Tad operon in A. hydrophila ML09-119. The vAhΔtad and vAh-WT strains were compared and analyzed using one-way analysis of variance (ANOVA). An alpha level of 0.05 was used for all analyses, with asterisks (*) indicating statistical significance. (A) Percent mortalities of fish immersion challenge. The fish were exposed to 2 treatments, A. hydrophila and vAhΔtad. Fish mortalities were recorded daily for 7 days, and percent mortalities was calculated for each treatment. (B) Growth Curve. Bacterial curves were generated by measuring the optical density at 600nm (OD600) on a multimode reader every half an hour for 72h at 30°C. (C) Biofilm assay. The strains were cultured to log phase and adjusted to OD600 of 0.1. The cultures were added to 96-well plate and incubated at 28°C for 24h. The cultures were washed and air-dried post staining with crystal violet (CV) for 15min at room temperature. The CV was solubilized with 95% ethanol for 15min, and absorbance was measured at 595nm. (D) Motility assay. The strains were cultured to log-phase. Two microliters of the culture were spotted on 0.6% BHI semi-solid plates and incubated at 28°C for 24h. The motility diameter for both strains were measured in millimeters.
3.3 Role of Tad pili in stress resistance
The influence of Tad in the tolerance of A. hydrophila to environmental stressors, including temperature, pH, NaCl, and H2O2 was evaluated. The vAhΔtad mutant strain exhibited growth to a higher density at 28°C than vAh-WT strain, but there were no observed differences in growth at 18°C and 37°C (Figure 4A). Under acid-alkali stress, both strains demonstrated similar growth at neutral pH 7.0, with a baseline percent viability of 100% (Figure 4B). However, as the pH deviated from neutrality, distinct trends emerged. Under acidic conditions (pH 5.0) the vAhΔtad strain demonstrated significantly reduced viability compared to the vAh-WT strain (p ≤ 0.05) (Figure 4B). Under osmotic stress conditions, no significant difference was observed in the growth kinetics between the vAhΔtad mutant and the vAh-WT strain. However, the vAhΔtad strain had a shorter log-phase than the vAh-WT strain, suggesting a potential adaptation that renders it slightly quicker to adapt to osmotic stress (Figure 4C). Following exposure to hydrogen peroxide (H2O2), both the vAhΔtad mutant and the vAh-WT strain exhibited decreased viability, with no observed differences between the mutant and the wild-type, indicating that Tad does not confer tolerance to oxidative stress in A. hydrophila (Figure 4D). Collectively, these results indicate an impact of Tad pili on acid tolerance in vAh strain ML09-119, but not temperature or osmotic stress.
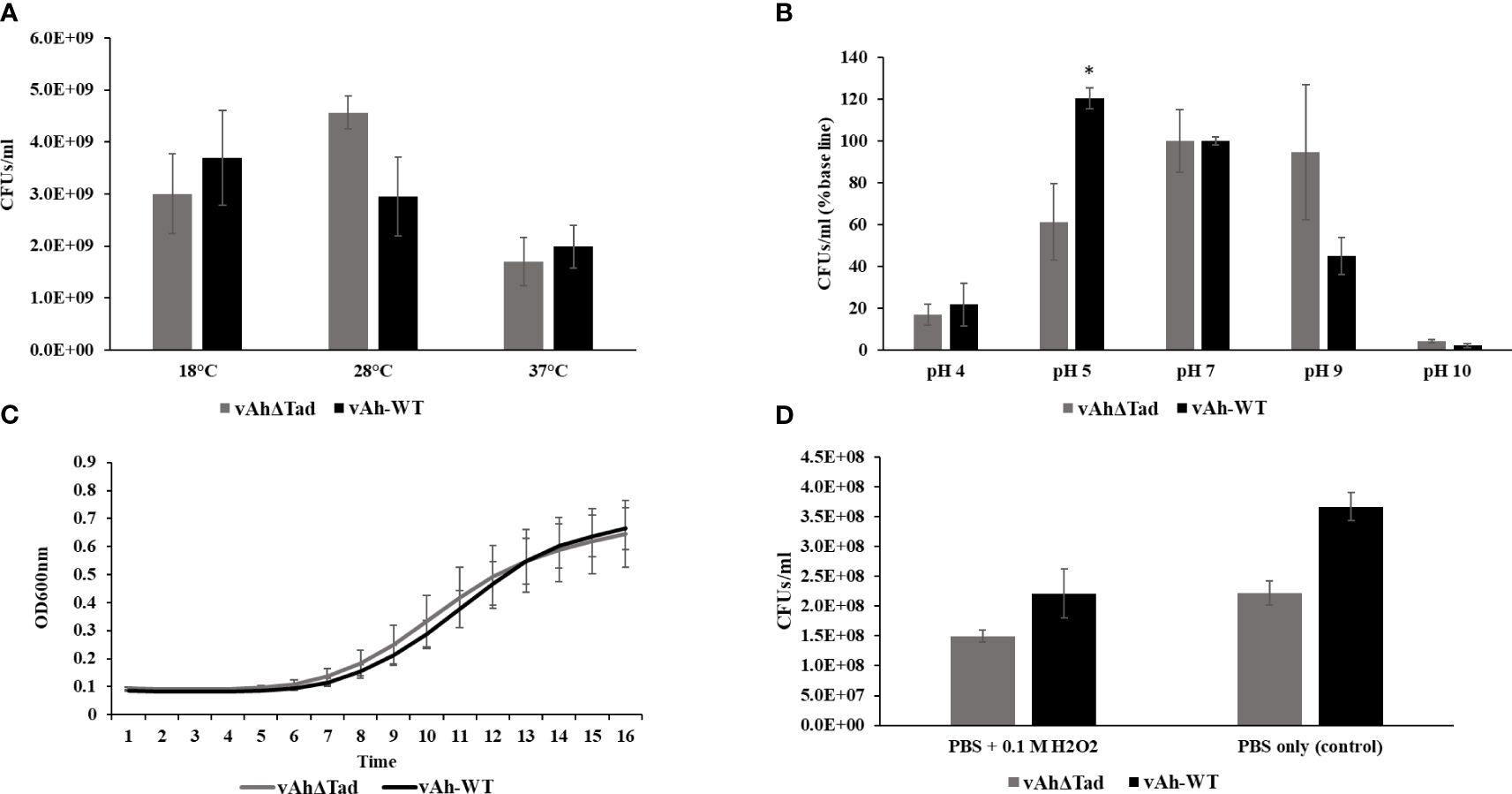
Figure 4 Characterization of vAhΔtad under different stress conditions. For the characterization assays, vAhΔtad and vAh-WT strains were compared and analyzed using one-way analysis of variance (ANOVA). An alpha level of 0.05 was used for all analyses, with asterisks (*) indicating statistical significance. (A) Temperature stress assay. The strains were cultured to log phase and OD600 was adjusted to 0.1 with BHI medium. The bacterial suspension was incubated at 18°C, 28°C (as a control) and 37°C for 24h. The cultures were double diluted with sterile PBS and plated onto BHI agar, for colony counting and statistical analysis. (B) Acid-Alkali tolerance. The strains were cultured at log-phase and the pellet was collected, washed twice with sterile PBS, and subjected to treatment in BHI at pH 4.0, pH 5.0, pH 7.0 (as a control), pH 9.0, and pH 10.0 for 30 minutes at 28°C. Viable cells were counted and compared for both strains by plating on BHI agar after serial dilutions. (C) Osmotic stress assay. The strains were cultured to log phase; pellets were collected and resuspended in BHI with 0.5M NaCl to adjust the OD600 of 0.1. The OD600 was monitored every 1h for 16h at 28°C. (D) Oxidative stress assay. The strains were cultured to log phase; pellets were collected and treated for 20min in PBS with 0.1M H2O2 and PBS without H2O2 as a control. Viable cells were counted on BHI plates after incubating for 24h.
3.4 Tad pili affect bacterial surface structure in A. hydrophila ML09-119
Scanning electron microscopy was used to evaluate the effects of Tad pili on the surface structure of A. hydrophila ML09-119. While overnight cultures showed no apparent differences (data not shown), we observed changes in the bacterial surface morphology during the log-phase. In the log phase, vAh-WT strain displayed a distinctly smooth material covering its surface, a feature absent in the vAhΔtad, which exhibited a surface that appeared rough (Figures 5B, D). In addition, the vAh-WT strain possessed prominent protrusions on its surface, while vAhΔtad lacked these distinctive surface features (Figures 5A, C).
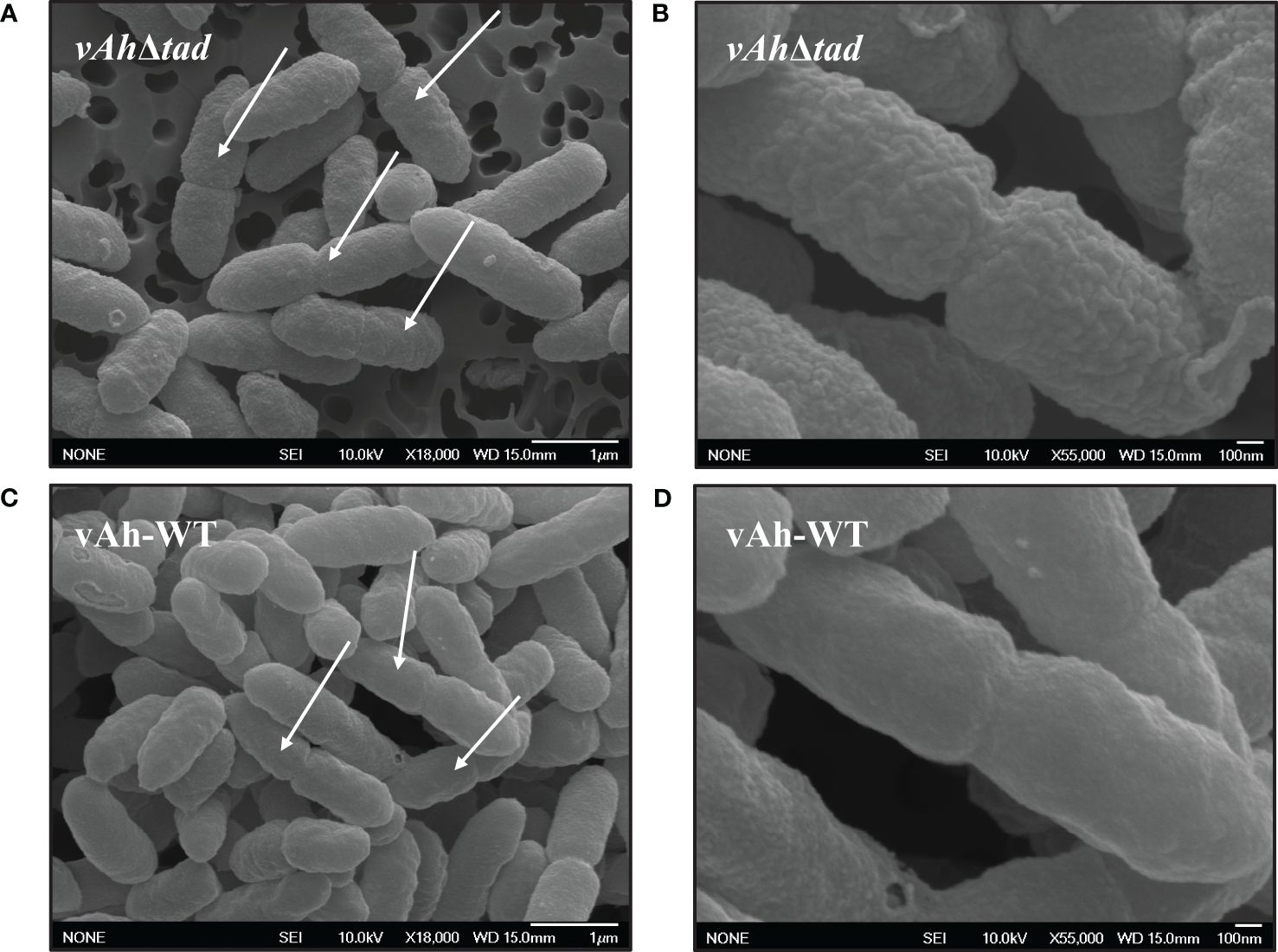
Figure 5 Ultrastructural observation of vAhΔtad and vAh-WT. Bacterial strains were grown to log-phase in BHI at 30°C and specimens were examined with JEOL 6500F field emission SEM operating at 10 KeV. The white arrows indicate dividing cells. (A) Cell structure of vAhΔtad at 18,000x and (B) at 55,000x under SEM. (C) Cell structure of vAh-WT at 18,000x and (D) at 55,000x under SEM.
4 Discussion
Amidst the dynamics of host-pathogen interactions, pathogens are driven to persistently evolve, adapt to varied ecological niches, and evade the host’s immune defenses. These adaptations often occur by acquiring unique genes, toxins, and systems through horizontal gene transfer, gene fissions, or gene duplications, which leaves a distinct imprint on the pathogen’s genetic makeup. Comparative genomics allows identification of these unique genetic adaptations, which can be specific for a bacterial species or for a genetic lineage within a species. A. hydrophila is a genetically diverse species that has many unique lineages with varied environmental adaptations, virulence, and host specificities.
Based on previous work, vAh isolates consistently encode a unique T4cP, known as tight adherence (Tad), which is largely absent from non-vAh isolates (Tekedar et al., 2019). Based on this key finding, it was hypothesized that acquisition of the Tad operon is an adaptive advantage, potentially contributing to the pathogenicity of vAh isolates. To test this hypothesis, a comprehensive comparative genome analysis was conducted with 170 A. hydrophila genomes, sourced from diverse hosts and distinct geographical locations. All evaluated genomes shared an ANI ≥95%, indicating conspecificity (Kim et al., 2014). A core-genome based phylogenetic tree revealed that vAh isolates from different geographical origins form a discrete subgroup (Hossain et al., 2014; Tekedar et al., 2019). These analyses also support the notion that U.S. and Chinese vAh isolates originated from the same monophyletic clade. Furthermore, in silico analysis confirmed previous findings that vAh isolates consistently carry the Tad operon.
Homologs of Tad loci are widely distributed across plant, human, and animal pathogens such as Sinorhizobium meliloti (Zatakia et al., 2014), Yersinia pestis (Tomich et al., 2007), Bifidobacterium breve (O'Connell Motherway et al., 2011; Ellison et al., 2019; O'Connell Motherway et al., 2019; Alessandri et al., 2021), Vibrio vulnificus (Pu et al., 2018a; Duong-Nu et al., 2019; Zhang et al., 2021), Myxococcus xanthus (Seef et al., 2021), Mycobacterium tuberculosis (Hosseini et al., 2014; Ramsugit and Pillay, 2015; Mann et al., 2016; Alteri et al., 2022), Caulobacter crescentus (Skerker and Shapiro, 2000; Mignolet et al., 2018; Hershey et al., 2019; Sangermani et al., 2019; Mignolet et al., 2021), and Cutibacterium acnes (Davidsson et al., 2017). The presence of the Tad system in such diverse bacterial species hints at an adaptive advantage, substantiated by its diverse roles. Tad pili have been implicated in biofilm formation, which provides a protected environment that enables increased resistance to antibiotics, biocides, and immune defenses. Therefore, biofilms facilitate environmental persistence and chronic bacterial infections (Pu et al., 2018a; Duong-Nu et al., 2019). Additionally, Tad pili are associated with enhanced resistance to serum bactericidal activity, a critical factor in bacterial survival within the host (Duong-Nu et al., 2019). What distinguishes Tad from other Type IV Pilus (T4P) systems is its versatility, encompassing participation in predator-prey interactions (Seef et al., 2021), invasion of host tissue (Duong-Nu et al., 2019), establishment of symbiotic relationships (Nykyri et al., 2013; Zatakia et al., 2014), modulation of cellular signaling within the host (Bernard et al., 2009; O'Connell Motherway et al., 2019), persistent infections in prostate tissue (Davidsson et al., 2017), and natural competence (Angelov et al., 2015; Cai et al., 2021). Furthermore, investigations into other bacterial species have highlighted the importance of the Tad pili in host invasion, survival, and resistance to complement activation (Duong-Nu et al., 2019).
In particular, Tad pili serve a unique function in the early stages of infection by acting as an adhesive bridge, facilitating the attachment of pathogens to host cells. For instance, Tad pili are indispensable for Bifidobacterium breve to colonize the human gut. In particular, the tadE gene contributes to host signaling that promotes colonic host epithelial proliferation (O'Connell Motherway et al., 2019). Similarly, in S. meliloti, the Tad locus is necessary for competitive bacterial-plant symbiotic interactions (Zatakia et al., 2014).
In vAh ML09-119, a set of 13 Tad genes organized within an operon structure were identified (Figure 1). The function of Tad in vAh was investigated through the generation of a knockout mutant that encompassed the entire operon. Compared to the wild-type strain, there was significantly reduced mortality in catfish fingerlings exposed to vAhΔtad indicating the Tad operon plays a critical role in the virulence of A. hydrophila ML09-119 (Figure 3A). This finding is consistent with the role of the Tad operon in diverse host-pathogen systems.
In contrast to these findings, studies on Aeromonas salmonicida (Boyd et al., 2008) and the human pathogen M. tuberculosis, which primarily focused on disruptions in a subset of the Tad operon genes, revealed the Tad operon was not essential for virulence in those host-pathogen interactions (Mann et al., 2016). Comparably, the current study involved a complete deletion of the Tad operon, resulting in significant attenuation. These contrasting outcomes could be driven by species- or host-specific nuances during host-pathogen interactions. For example, deleting Tad genes in a human pathogen, M. tuberculosis, did not impact biofilm formation or virulence in a mouse model (Davidsson et al., 2017). In contrast, the Tad operon in M. tuberculosis was upregulated upon interaction with human epithelial cells (Alteri et al., 2022), suggesting that differences in host environment may affect the function of Tad pili. Interestingly, Cutibacterium acnes type II isolates carrying plasmids with four Tad genes from human prostate cancer specimens were speculated to trigger infection-induced prostate cancer (Davidsson et al., 2017). Herein, a complete Tad operon was identified from some clinical A. hydrophila isolates from humans in the USA and China (Figure 2), suggesting Tad may facilitate A. hydrophila invasion of human cells. However, despite these observations, the precise role of Tad pili in A. hydrophila isolated from human cases remains a subject for future investigation.
A prominent hypothesis in studies focused on pathogens including A. actinomycetemcomitans, Haemophilus ducreyi, Pasteurella multocida, V. vulnificus, C. crescentus, M. tuberculosis and S. meliloti, suggests the Tad system has been horizontally transferred (Planet et al., 2003; Zatakia et al., 2014; Ramsugit and Pillay, 2015; Pu et al., 2018b; Beeby, 2019; Ellison et al., 2019). Evolutionary analyses suggest this genetic transfer coincided with the development of a ‘EppA-dependent’ (Epd) pilus homologue, acquiring a secretin, tracing their origins to an ancient genetic exchange with the Archaea (Denise et al., 2019; Sonani et al., 2023). It is thought the Tad operon may provide a competitive edge by facilitating niche adaptation, allowing the pathogen to persist in diverse environments.
Tad systems show considerable diversity among bacterial species, revealing distinct arrangements and functional roles. For instance, M. tuberculosis maintains only five Tad genes, diverging from the typical functional Tad loci which consists of 12-14 genes organized in a single operon. Other organisms like Burkholderia pseudomallei, Mesorhizobium loti, and V. vulnificus harbor three Tad loci (Hossain et al., 2014). In V. vulnificus, the deletion of all three Tad loci was necessary to mitigate pathogenicity (O'Connell Motherway et al., 2011). S. meliloti, on the other hand, encodes two Tad loci, situated in the chromosome and a megaplasmid (Kim et al., 2014). In comparison, C. crescentus has seven genes homologous to the Flp pilus genes; in this species, Flp pili serve as surface sensors altering secondary molecules controlling cell cycle events (Ellison et al., 2017; Sangermani et al., 2019). In Mycobacterium luteus, two Tad clusters located 1.2 Mbp apart are crucial for natural competence (Angelov et al., 2015).
In A. hydrophila ML09-119, the Tad operon has higher GC content (64.82%) than the average for the rest of the genome (60.82%) (Figure 1), suggesting acquisition from horizontal transfer. Interestingly, vAh isolates consistently encode the Tad operon while exhibiting the absence of any type III secretion system (T3SS) (data not shown here). This suggests the mechanism of pathogenesis for vAh, particularly how vAh manipulates function of host cells, is quite different from pathogens that utilize T3SS to inject effector proteins directly into host cells, and Tad may be an important adaptation in the absence of T3SS.
In addition to its role in pathogenicity, the effects of the Tad operon knockout on critical factors such as growth kinetics, motility, and biofilm formation in A. hydrophila was also assessed. Although deletion of the Tad operon had no effect on growth rate and motility, it significantly reduced biofilm formation. Tad pili have been reported as a crucial adhesion factors for initial surface attachment, a vital step in microcolony formation. They facilitate cell-cell interactions and aggregation, promoting the development and maturation of structured biofilms. Biofilms shield pathogens from environmental stressors and host defenses, enhancing survival against antimicrobial peptides and phagocytosis. Biofilms are a characteristic feature of chronic infections in numerous pathogens, including Y. pestis (causing bubonic plague) (Cui et al., 2020), Vibrio cholerae (responsible for gastrointestinal infections) (Huang et al., 2023), M. tuberculosis (Alteri et al., 2022), and Bordetella pertussis (causing whooping cough) (Tomich et al., 2007; Serra et al., 2011). Notably, in V. vulnificus, Tad pili mediate biofilms, which can facilitate resistance to serum killing (Duong-Nu et al., 2019). Notably, A. hydrophila isolated from biofilms results in chronic necrosis within 24 hours when injected into channel catfish muscle. Comparably, planktonic A. hydrophila failed to produce similar lesions, even up to seven days post-injection (Stewart and Franklin, 2008; Barger et al., 2021).
The significance of T4P extends beyond their conventional roles in attachment and motility, including the capacity to perceive cues within the environment and orchestrate responses. In the current study, Tad pili did not contribute to A. hydrophila resistance against most stress factors investigated (Figure 4). However, a significant contribution to survival at pH 5 was observed. Interestingly, in the human pathogen M. tuberculosis, acidic pH of the phagosomal vacuole was shown to trigger assembly of the Tad pilin (Alteri, 2005). In A. hydrophila, Tad pili may function as a sensor regulator for low pH, inducing expression of other adaptive mechanisms, or it may directly mediate resistance to low pH.
Structural observations under SEM revealed distinct characteristics in actively dividing A. hydrophila. vAh-WT exhibited a smooth layer absent in vAhΔtad bacteria, which displayed rough, irregular protrusions with a coarse texture (Figure 5). Interestingly, these distinct surface modifications were only observed in the log-phase of growth and were absent during the stationary phase. Because Tad pili are a surface structure, it is not surprising that surface alterations were detected by SEM, but the significance of the rough surface during log phase is not currently known. In C. crescentus, Tad pili regulate secondary signaling pathways through mechanical sensing of surface cues, leading to the production of highly efficient progeny equipped for sustained attachment (Sangermani et al., 2019; Snyder et al., 2020). It is possible the bacterial surface alterations in vAhΔtad impacted host cell adhesion and/or induction of virulence mechanisms through signaling pathways resulting in reduced virulence, but further research is needed.
Collectively, these investigations into the function of Tad pili in vAh isolate A. hydrophila ML09-119 provides valuable insights into its role in virulence and environmental adaptation. Comparative genomic analysis confirmed the consistent presence of the Tad operon in the vAh subclade of A. hydrophila, suggesting Tad is essential for vAh virulence in catfish, biofilm formation and resistance to acid stress. This work highlights the multifaceted role of Tad pili in A. hydrophila pathogenicity and physiology. The presence of the Tad operon in some human A. hydrophila isolates suggests it may contribute to pathogenesis in humans as well, but further investigation is needed. Regardless, these findings demonstrate a clear role of Tad pili in contributing to virulence of vAh in catfish, which is one of the most important diseases affecting catfish aquaculture in the southeastern United States. Further investigation is warranted to delineate the role of Tad pili in vAh pathogenesis and explore its potential for treatment and prevention of motile Aeromonas septicemia.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, Further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was approved by Institutional Animal Care and Use Committee of the Mississippi State University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
HT: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. FP: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. JB: Data curation, Resources, Software, Visualization, Writing – review & editing. MG: Data curation, Resources, Writing – review & editing. GW: Data curation, Resources, Writing – review & editing. SK: Conceptualization, Data curation, Methodology, Writing – review & editing. HA: Data curation, Methodology, Writing – review & editing. VD: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. LH: Formal analysis, Methodology, Resources, Writing – review & editing. ML: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Mississippi State University College of Veterinary Medicine and the USDA Agricultural Research Service project 310048-182020-021000-371060.
Acknowledgments
The authors thank the Laboratory Animal Resources and Care (LARAC) at the College of Veterinary Medicine for providing the SPF channel catfish and Dr. Orion S. Rivers for his assistance with the SEM imaging.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1425624/full#supplementary-material
References
Abby, S. S., Denise, R., Rocha, E. P. C. (2024). Identification of protein secretion systems in bacterial genomes using macSyFinder version 2. Methods Mol. Biol. 2715, 1–25. doi: 10.1007/978-1-0716-3445-5_1
Abby, S. S., Néron, B., Ménager, H., Touchon, M., Rocha, E. P. C. (2014). MacSyFinder: A program to mine genomes for molecular systems with an application to CRISPR-cas systems. PLoS One 9, e110726. doi: 10.1371/journal.pone.0110726
Abdelhamed, H., Nho, S. W., Turaga, G., Banes, M. M., Karsi, A., Lawrence, M. L. (2016). Protective efficacy of four recombinant fimbrial proteins of virulent Aeromonas hydrophila strain ML09-119 in channel catfish. Vet. Microbiol. 197, 8–14. doi: 10.1016/j.vetmic.2016.10.026
Alessandri, G., Van Sinderen, D., Ventura, M. (2021). The genus Bifidobacterium: from genomics to functionality of an important component of the mammalian gut microbiota. Comput. Struct. Biotechnol. J. 19, 1472–1487. doi: 10.1016/j.csbj.2021.03.006
Alteri, C. J. (2005). Novel pili of Mycobacterium tuberculosis. Ph.D. Thesis. (University of Arizona;).
Alteri, C. J., Rios-Sarabia, N., de la Cruz, M. A., González-y-Merchand, J. A., Soria-Bustos, J., Maldonado-Bernal, C., et al. (2022). The Flp type IV pilus operon of Mycobacterium tuberculosis is expressed upon interaction with macrophages and alveolar epithelial cells. Front. Cell Infect. Microbiol. 12, 916247. doi: 10.3389/fcimb.2022.916247
Angelov, A., Bergen, P., Nadler, F., Hornburg, P., Lichev, A., Ãœbelacker, M., et al. (2015). Novel Flp pilus biogenesis-dependent natural transformation. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00084
Barger, P. C., Liles, M. R., Beck, B. H., Newton, J. C. (2021). Differential production and secretion of potentially toxigenic extracellular proteins from hypervirulent Aeromonas hydrophila under biofilm and planktonic culture. BMC Microbiol. 21, 8. doi: 10.1186/s12866-020-02065-2
Barger, P. C., Liles, M. R., Newton, J. C. (2020). Type II secretion is essential for virulence of the emerging fish pathogen, hypervirulent aeromonas hydrophila. Front. Vet. Sci. 7, 574113. doi: 10.3389/fvets.2020.574113
Baum, B. R. (1989). PHYLIP: phylogeny inference package. Version 3.2. Joel felsenstein. Q. Rev. Biol. 64, 539–541. doi: 10.1086/416571
Beeby, M. (2019). Evolution of a family of molecular Rube Goldberg contraptions. PLoS Biol. 17, e3000405. doi: 10.1371/journal.pbio.3000405
Bernard, C. S., Bordi, C., Termine, E., Filloux, A., De Bentzmann, S. (2009). Organization and PprB-Dependent Control of the Pseudomonas aeruginosa tad Locus, Involved in Flp Pilus Biology. J. Bacteriol. 191, 1961–1973. doi: 10.1128/JB.01330-08
Blom, J., Kreis, J., Spanig, S., Juhre, T., Bertelli, C., Ernst, C., et al. (2016). EDGAR 2.0: an enhanced software platform for comparative gene content analyses. Nucleic Acids Res. 44, W22–W28. doi: 10.1093/nar/gkw255
Boehmer, T., Vogler, A. J., Thomas, A., Sauer, S., Hergenroether, M., Straubinger, R. K., et al. (2018). Phenotypic characterization and whole genome analysis of extended-spectrum beta-lactamase-producing bacteria isolated from dogs in Germany. PLoS One 13, e0206252. doi: 10.1371/journal.pone.0206252
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boyd, J. M., Dacanay, A., Knickle, L. C., Touhami, A., Brown, L. L., Jericho, M. H., et al. (2008). Contribution of Type IV Pili to the Virulence of Aeromonas salmonicida subsp. salmonicida in Atlantic Salmon (Salmo salar L.). Infect. Immun. 76, 1445–1455. doi: 10.1128/IAI.01019-07
Bureros, K. J., Chiu, Y. C., Liou, C. Y., Ma, C. Y., Wang, L. C. (2022). Complete genome sequence of a suckermouth catfish outbreak isolate, aeromonas hydrophila strain LP0103. Microbiol. Resour Announc. 11, e0040822. doi: 10.1128/mra.00408-22
Cai, L., Jain, M., Sena-Vélez, M., Jones, K. M., Fleites, L. A., Heck, M., et al. (2021). Tad pilus-mediated twitching motility is essential for DNA uptake and survival of Liberibacters. PLoS One 16, e0258583. doi: 10.1371/journal.pone.0258583
Canellas, A. L. B., de Oliveira, B. F. R., Laport, M. S. (2023). Hiding in plain sight: Characterization of aeromonas species isolated from a recreational estuary reveals the carriage and putative dissemination of resistance genes. Antibiotics. 12, 84. doi: 10.3390/antibiotics12010084
Cehovin, A., Simpson, P. J., McDowell, M. A., Brown, D. R., Noschese, R., Pallett, M., et al. (2013). Specific DNA recognition mediated by a type IV pilin. Proc. Natl. Acad. Sci. U.S.A. 110, 3065–3070. doi: 10.1073/pnas.1218832110
Chan, K. G., Tan, W. S., Chang, C. Y., Yin, W. F., Mumahad Yunos, N. Y. (2015). Genome sequence analysis reveals evidence of quorum-sensing genes present in aeromonas hydrophila strain M062, isolated from freshwater. Genome Announc. 3. doi: 10.1128/genomeA.00100-15
Cui, Y., Schmid, B. V., Cao, H., Dai, X., Du, Z., Ryan Easterday, W., et al. (2020). Evolutionary selection of biofilm-mediated extended phenotypes in Yersinia pestis in response to a fluctuating environment. Nat. Commun. 11, 281. doi: 10.1038/s41467-019-14099-w
Dao, T. D., Yano, H., Takemura, T., Hirabayashi, A., Trang, L. T., Tran, H. H., et al. (2022). Strand-biased circularizing integrative elements spread tmexCD -toprJ gene clusters encoding RND-type multidrug efflux pumps by repeated transpositions. bioRxiv. 2022, 09.22.508988. doi: 10.1101/2022.09.22.508988
Davidsson, S., Carlsson, J., Mölling, P., Gashi, N., Andrén, O., Andersson, S.-O., et al. (2017). Prevalence of flp pili-encoding plasmids in cutibacterium acnes isolates obtained from prostatic tissue. Front. Microbiol. 8, 2241. doi: 10.3389/fmicb.2017.02241
de Melo, B. S. T., Mendes-Marques, C. L., Campos, T., Almeida, A., Leal, N. C., Xavier, D. E. (2019). High-resolution genome-wide analysis is essential for the identification of ambiguous Aeromonas strains. FEMS Microbiol. Letters. 366. doi: 10.1093/femsle/fnz245
Denise, R., Abby, S. S., Rocha, E. P. C. (2019). Diversification of the type IV filament superfamily into machines for adhesion, protein secretion, DNA uptake, and motility. PLoS Biol. 17, e3000390. doi: 10.1371/journal.pbio.3000390
Dozois, C. M., Daigle, F., Curtiss, R., 3rd. (2003). Identification of pathogen-specific and conserved genes expressed in vivo by an avian pathogenic Escherichia coli strain. Proc. Natl. Acad. Sci. United States America. 100, 247–252. doi: 10.1073/pnas.232686799
Du, X., Wang, M., Zhou, H., Li, Z., Xu, J., Li, Z., et al. (2021). Comparison of the multiple platforms to identify various aeromonas species. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.625961
Dubey, S., Ager-Wick, E., Kumar, J., Karunasagar, I., Karunasagar, I., Peng, B., et al. (2022). Aeromonas species isolated from aquatic organisms, insects, chicken, and humans in India show similar antimicrobial resistance profiles. Front. Microbiol. 13, 1008870. doi: 10.3389/fmicb.2022.1008870
Duong-Nu, T.-M., Jeong, K., Hong, S. H., Puth, S., Kim, S. Y., Tan, W., et al. (2019). A stealth adhesion factor contributes to Vibrio vulnificus pathogenicity: Flp pili play roles in host invasion, survival in the blood stream and resistance to complement activation. PLoS Pathog. 15, e1007767. doi: 10.1371/journal.ppat.1007767
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Ellison, C. K., Kan, J., Chlebek, J. L., Hummels, K. R., Panis, G., Viollier, P. H., et al. (2019). A bifunctional ATPase drives tad pilus extension and retraction. Sci. Advances. 5, eaay2591. doi: 10.1126/sciadv.aay2591
Ellison, C. K., Kan, J., Dillard, R. S., Kysela, D. T., Ducret, A., Berne, C., et al. (2017). Obstruction of pilus retraction stimulates bacterial surface sensing. Science. 358, 535–538. doi: 10.1126/science.aan5706
Ellison, C. K., Whitfield, G. B., Brun, Y. V. (2022). Type IV Pili: dynamic bacterial nanomachines. FEMS Microbiol. Rev. 46, fuab053. doi: 10.1093/femsre/fuab053
Emond-Rheault, J. G., Vincent, A. T., Trudel, M. V., Brochu, F., Boyle, B., Tanaka, K. H., et al. (2015). Variants of a genomic island in Aeromonas salmonicida subsp. salmonicida link isolates with their geographical origins. Vet. Microbiol. 175, 68–76. doi: 10.1016/j.vetmic.2014.11.014
Forn-Cuní, G., Tomás, J. M., Merino, S. (2016). Whole-genome sequence of aeromonas hydrophila strain AH-1 (Serotype O11). Genome Announc. 4. doi: 10.1128/genomeA.00920-16
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., Tiedje, J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
Griffin, M. J., Goodwin, A. E., Merry, G. E., Liles, M. R., Williams, M. A., Ware, C., et al. (2013). Rapid quantitative detection of Aeromonas hydrophila strains associated with disease outbreaks in catfish aquaculture. J. Vet. Diagn. Invest. 25, 473–481. doi: 10.1177/1040638713494210
Grim, C. J., Kozlova, E. V., Ponnusamy, D., Fitts, E. C., Sha, J., Kirtley, M. L., et al. (2014). Functional genomic characterization of virulence factors from necrotizing fasciitis-causing strains of Aeromonas hydrophila. Appl. Environ. Microbiol. 80, 4162–4183. doi: 10.1128/AEM.00486-14
Guo, Y., Zeng, C., Ma, C., Cai, H., Jiang, X., Zhai, S., et al. (2022). Comparative genomics analysis of the multidrug-resistant Aeromonas hydrophila MX16A providing insights into antibiotic resistance genes. Front. Cell Infect. Microbiol. 12, 1042350. doi: 10.3389/fcimb.2022.1042350
Han, J. E., Kim, J. H., Choresca, C., Shin, S. P., Jun, J. W., Park, S. C. (2013). Draft Genome Sequence of a Clinical Isolate, Aeromonas hydrophila SNUFPC-A8, from a Moribund Cherry Salmon (Oncorhynchus masou masou). Genome Announc. 1. doi: 10.1128/genomeA.00133-12
Herrero, M., de Lorenzo, V., Timmis, K. N. (1990). Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. bacteriology. 172, 6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990
Hershey, D. M., Porfírio, S., Black, I., Jaehrig, B., Heiss, C., Azadi, P., et al. (2019). Composition of the holdfast polysaccharide from caulobacter crescentus. J. Bacteriol. 201. doi: 10.1128/JB.00276-19
Hilt, E. E., Fitzwater, S. P., Ward, K., de St. Maurice, A., Chandrasekaran, S., Garner, O. B., et al. (2020). Carbapenem Resistant Aeromonas hydrophila Carrying blacphA7 Isolated From Two Solid Organ Transplant Patients. Front. Cell Infect. Microbiol. 10. doi: 10.3389/fcimb.2020.563482
Ho, D. T., Lee, Y., Kim, N., Roh, H., Kim, A., Kim, D. H. (2023). Complete genomic sequences of two aeromonas hydrophila isolates derived from diseased fish in South Korea. Microbiol. Resour Announc. 12, e0078622. doi: 10.1128/mra.00786-22
Honein, K., Jagoda, S., Arulkanthan, A., Ushio, H., Asakawa, S. (2018). Draft Genome Sequence of Aeromonas hydrophila Strain Ae25, Isolated from a Septicemic Moribund Koi Carp (Cyprinus carpio) in Sri Lanka. Genome Announc. 6. doi: 10.1128/genomeA.01523-17
Horneman, A. J., Chopra, A. K., Graf, J., Horneman, A. J., Johnson, J. A. (2009). Virulence factor–activity relationships (VFAR) with specific emphasis on Aeromonas species (spp. ). J. Water Health 07, S29. doi: 10.2166/wh.2009.053
Horton, R. M., Hunt, H. D., Ho, S. N., Pullen, J. K., Pease, L. R. (1989). Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 77, 61–68. doi: 10.1016/0378-1119(89)90359-4
Hospenthal, M. K., Costa, T. R. D., Waksman, G. (2017). A comprehensive guide to pilus biogenesis in Gram-negative bacteria. Nat. Rev. Microbiol. 15, 365–379. doi: 10.1038/nrmicro.2017.40
Hossain, M. J., Sun, D., McGarey, D. J., Wrenn, S., Alexander, L. M., Martino, M. E., et al. (2014). An asian origin of virulent aeromonas hydrophila responsible for disease epidemics in United States-farmed catfish. mBio. 5, e00848–e00814. doi: 10.1128/mBio.00848-14
Hosseini, H., Fooladi, A. A. I., Arjomandzadegan, M., Emami, N., Bornasi, H. (2014). Genetics study and transmission electron microscopy of pili in susceptible and resistant clinical isolates of Mycobacterium tuberculosis. Asian Pacific J. Trop. Med. 7, S199–S203. doi: 10.1016/S1995-7645(14)60232-7
Huang, X., Nero, T., Weerasekera, R., Matej, K. H., Hinbest, A., Jiang, Z., et al. (2023). Vibrio cholerae biofilms use modular adhesins with glycan-targeting and nonspecific surface binding domains for colonization. Nat. Commun. 14, 2104. doi: 10.1038/s41467-023-37660-0
Hughes, H. Y., Conlan, S. P., Lau, A. F., Dekker, J. P., Michelin, A. V., Youn, J. H., et al. (2016). Detection and whole-genome sequencing of carbapenemase-producing aeromonas hydrophila isolates from routine perirectal surveillance culture. J. Clin. Microbiol. 54, 1167–1170. doi: 10.1128/JCM.03229-15
Huys, G., Pearson, M., Kämpfer, P., Denys, R., Cnockaert, M., Inglis, V., et al. (2003). Aeromonas hydrophila subsp. ranae subsp. nov., isolated from septicaemic farmed frogs in Thailand. Int. J. Systematic Evolutionary Microbiol. 53, 885–891. doi: 10.1099/ijs.0.02357-0
Irshath, A. A., Rajan, A. P., Vimal, S., Prabhakaran, V.-S., Ganesan, R. (2023). Bacterial pathogenesis in various fish diseases: Recent advances and specific challenges in vaccine development. Vaccines. 11, 470. doi: 10.3390/vaccines11020470
Islam, M. S., Raz, A., Liu, Y., Elbassiony, K. R. A., Dong, X., Zhou, P., et al. (2019). Complete genome sequence of aeromonas phage ZPAH7 with halo zones, isolated in China. Microbiol. Resour Announc 8. doi: 10.1128/MRA.01678-18
Jagoda, S. S., Tan, E., Arulkanthan, A., Kinoshita, S., Watabe, S., Asakawa, S. (2014). Draft Genome Sequence of Aeromonas hydrophila Strain Ae34, Isolated from a Septicemic and Moribund Koi Carp (Cyprinus carpio koi), a Freshwater Aquarium Fish. Genome Announc. 2. doi: 10.1128/genomeA.00572-14
Ji, J., Kulshreshtha, S., Kakade, A., Majeed, S., Li, X., Liu, P. (2020). Bioaugmentation of membrane bioreactor with Aeromonas hydrophila LZ-MG14 for enhanced malachite green and hexavalent chromium removal in textile wastewater. Int. Biodeterioration Biodegradation. 150, 104939. doi: 10.1016/j.ibiod.2020.104939
Jin, L., Chen, Y., Yang, W., Qiao, Z., Zhang, X. (2020). Complete genome sequence of fish-pathogenic Aeromonas hydrophila HX-3 and a comparative analysis: insights into virulence factors and quorum sensing. Sci. Rep. 10, 15479. doi: 10.1038/s41598-020-72484-8
Kachlany, S. C., Planet, P. J., Bhattacharjee, M. K., Kollia, E., DeSalle, R., Fine, D. H., et al. (2000). Nonspecific adherence by actinobacillus actinomycetemcomitans requires genes widespread in bacteria and archaea. J. Bacteriol. 182, 6169–6176. doi: 10.1128/JB.182.21.6169-6176.2000
Kåhrström, C. T. (2015). Type IV pili function as mechanosensors. Nat. Rev. Microbiol. 13, 399. doi: 10.1038/nrmicro3517
Kim, M., Oh, H.-S., Park, S.-C., Chun, J. (2014). Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Systematic Evolutionary Microbiol. 64, 346–351. doi: 10.1099/ijs.0.059774-0
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., Phillippy, A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736. doi: 10.1101/gr.215087.116
Laviad-Shitrit, S., Izhaki, I., Arakawa, E., Halpern, M. (2018). Wild waterfowl as potential vectors of Vibrio cholerae and Aeromonas species. Trop. Med. Int. Health 23, 758–764. doi: 10.1111/tmi.13069
Lenneman, E. M., Barney, B. M. (2014). Draft Genome Sequences of the Alga-Degrading Bacteria Aeromonas hydrophila Strain AD9 and Pseudomonas pseudoalcaligenes Strain AD6. Genome Announc. 2. doi: 10.1128/genomeA.00709-14
Li, J., Ma, S., Li, Z., Yu, W., Zhou, P., Ye, X., et al. (2021). Construction and characterization of an aeromonas hydrophila multi-gene deletion strain and evaluation of its potential as a live-attenuated vaccine in grass carp. Vaccines (Basel) 9. doi: 10.3390/vaccines9050451
Li, X., Shang, B., Zhang, X., Zhang, H., Li, Z., Shen, X., et al. (2023). Complete genome sequence data of multidrug-resistant Aeromonas hydrophila Ah27 isolated from intussusception channel catfish (Ictalurus punctatus). Gene Rep. 33, 101807. doi: 10.1016/j.genrep.2023.101807
Lim, S. R., Lee, D.-H., Kim, Y.-C., Park, S. Y., Kwon, H., Han, J. E., et al. (2020). Complete genome sequence of multidrug-resistant Aeromonas hydrophila strain KN-Mc-1R2 isolated from the wild nutria (Myocastor coypus ) in Korea. Korean J. Microbiol. 56, 340–342. doi: 10.7845/kjm.2020.0078
Liu, J., Xiao, G., Zhou, W., Yang, J., Sun, Z. (2021). Hypothetical Protein gene1038 Contributes to Colistin Resistance in Aeromonas hydrophila. Antimicrob. Agents Chemother. 65, e0150321. doi: 10.1128/AAC.01503-21
Liu, J., Xie, L., Zhao, D., Yang, T., Hu, Y., Sun, Z., et al. (2019). A fatal diarrhoea outbreak in farm-raised Deinagkistrodon acutus in China is newly linked to potentially zoonotic Aeromonas hydrophila. Transboundary Emerging Dis. 66, 287–298. doi: 10.1111/tbed.13020
Liu, B., Zheng, D., Zhou, S., Chen, L., Yang, J. (2022). VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. 50, D912–D9D7. doi: 10.1093/nar/gkab1107
Mann, K. M., Pride, A. C., Flentie, K., Kimmey, J. M., Weiss, L. A., Stallings, C. L. (2016). Analysis of the contribution of MTP and the predicted Flp pilus genes to Mycobacterium tuberculosis pathogenesis. Microbiology. 162, 1784–1796. doi: 10.1099/mic.0.000368
McCallum, M., Burrows, L. L., Howell, P. L. (2019). The dynamic structures of the type IV pilus. Microbiol. Spectr. 7, 7.2.02. doi: 10.1128/microbiolspec.PSIB-0006-2018
Metcalf, W. W., Jiang, W., Wanner, B. L. (1994). Use of the rep technique for allele replacement to construct new Escherichia coli hosts for maintenance of R6K gamma origin plasmids at different copy numbers. Gene 138, 1–7. doi: 10.1016/0378-1119(94)90776-5
Mignolet, J., Mathelié-Guinlet, M., Viljoen, A., Dufrêne, Y. F. (2021). AFM force-clamp spectroscopy captures the nanomechanics of the Tad pilus retraction. Nanoscale Horiz. 6, 489–496. doi: 10.1039/D1NH00158B
Mignolet, J., Panis, G., Viollier, P. H. (2018). More than a Tad: spatiotemporal control of Caulobacter pili. Curr. Opin. Microbiol. 42, 79–86. doi: 10.1016/j.mib.2017.10.017
Mix, A.-K., Goob, G., Sontowski, E., Hauck, C. R. (2021). Microscale communication between bacterial pathogens and the host epithelium. Genes Immun. 22, 247–254. doi: 10.1038/s41435-021-00149-1
Moon, S. H., Udaondo, Z., Li, X., Yang, X., Jun, S.-R., Huang, E. (2021). Isolation and characterisation of carbapenemase-producing and polymyxin B-resistant Enterobacter bugandensis from a vegetable. J. Global Antimicrobial Resistance. 26, 264–265. doi: 10.1016/j.jgar.2021.07.013
Moura, Q., Fernandes, M. R., Cerdeira, L., Santos, A. C. M., de Souza, T. A., Ienne, S., et al. (2017). Draft genome sequence of a multidrug-resistant Aeromonas hydrophila ST508 strain carrying rmtD and blaCTX-M-131 isolated from a bloodstream infection. J. Global Antimicrobial Resistance. 10, 289–290. doi: 10.1016/j.jgar.2017.07.007
Mzula, A., Wambura, P. N., Mdegela, R. H., Shirima, G. M. (2019). Current state of modern biotechnological-based aeromonas hydrophila vaccines for aquaculture: A systematic review. BioMed. Res. Int. 2019, 1–11. doi: 10.1155/2019/3768948
Ngo, T. P. H., Vu, H. T. T., Le, T. T. T., Bui, H. C. N., Liles, M. R., Rodkhum, C. (2022). Comparative genomic analysis of hypervirulent Aeromonas hydrophila strains from striped catfish (Pangasianodon hypophthalmus) in Vietnam. Aquaculture. 558, 738364. doi: 10.1016/j.aquaculture.2022.738364
Nykyri, J., Mattinen, L., Niemi, O., Adhikari, S., Kõiv, V., Somervuo, P., et al. (2013). Role and Regulation of the Flp/Tad Pilus in the Virulence of Pectobacterium atrosepticum SCRI1043 and Pectobacterium wasabiae SCC3193. PLoS One 8, e73718. doi: 10.1371/journal.pone.0073718
O'Connell Motherway, M., Houston, A., O’Callaghan, G., Reunanen, J., O’Brien, F., O’Driscoll, T., et al. (2019). A Bifidobacterial pilus-associated protein promotes colonic epithelial proliferation: Bifidobacterial proliferation factor. Mol. Microbiol. 111, 287–301. doi: 10.1111/mmi.14155
O'Connell Motherway, M., Zomer, A., Leahy, S. C., Reunanen, J., Bottacini, F., Claesson, M. J., et al. (2011). Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc. Natl. Acad. Sci. U.S.A. 108, 11217–11222. doi: 10.1073/pnas.1105380108
Oliveira, N. M., Wheeler, J. H. R., Deroy, C., Booth, S. C., Walsh, E. J., Durham, W. M., et al. (2022). Suicidal chemotaxis in bacteria. Nat. Commun. 13, 7608. doi: 10.1038/s41467-022-35311-4
Ota, Y., Prah, I., Mahazu, S., Gu, Y., Nukui, Y., Koike, R., et al. (2023). Novel insights into genetic characteristics of blaGES-encoding plasmids from hospital sewage. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1209195
Pang, M., Jiang, J., Xie, X., Wu, Y., Dong, Y., Kwok, A. H., et al. (2015). Novel insights into the pathogenicity of epidemic Aeromonas hydrophila ST251 clones from comparative genomics. Sci. Rep. 5, 9833. doi: 10.1038/srep09833
Pan H, W. S., Dong, C., Shi, C., Ye, M., Lin, T., Huang, Z. (2004). Identification, virulence, hemolytic activity of GYK1, a strain of pathogenic Aeromonas hydrophila isolated from mandarinfish. J. Shanghai Fish Univ. 13.
Parker, J. L., Shaw, J. G. (2011). Aeromonas spp. clinical microbiology and disease. J. Infection. 62, 109–118. doi: 10.1016/j.jinf.2010.12.003
Pelicic, V. (2008). Type IV pili: e pluribus unum? Mol. Microbiol. 68, 827–837. doi: 10.1111/j.1365-2958.2008.06197.x
Pessoa, R. B. G., Oliveira, W. F. D., Correia, M. T. D. S., Fontes, A., Coelho, L. C. B. B. (2022). Aeromonas and human health disorders: Clinical approaches. Front. Microbiol. 13, 868890. doi: 10.3389/fmicb.2022.868890
Piepenbrink, K. H. (2019). DNA uptake by type IV filaments. Front. Mol. Biosci. 6, 1. doi: 10.3389/fmolb.2019.00001
Planet, P. J., Kachlany, S. C., Fine, D. H., DeSalle, R., Figurski, D. H. (2003). The widespread colonization island of actinobacillus actinomycetemcomitans. Nat. Genet. 34, 193–198. doi: 10.1038/ng1154
Pridgeon, J. W., Zhang, D., Zhang, L. (2014a). Complete Genome Sequence of a Moderately Virulent Aeromonas hydrophila Strain, pc104A, Isolated from Soil of a Catfish Pond in West Alabama. Genome Announc. 2. doi: 10.1128/genomeA.00554-14
Pridgeon, J. W., Zhang, D., Zhang, L. (2014b). Complete genome sequence of the highly virulent aeromonas hydrophila AL09-71 isolated from diseased channel catfish in west alabama. Genome Announc. 2. doi: 10.1128/genomeA.00450-14
Pu, M., Duriez, P., Arazi, M., Rowe-Magnus, D. A. (2018a). A conserved tad pilus promotes Vibrio vulnificus oyster colonization: A tad pilus promotes V. vulnificus oyster colonization. Environ. Microbiol. 20, 828–841. doi: 10.1111/1462-2920.14025
Pu, M., Duriez, P., Arazi, M., Rowe-Magnus, D. A. (2018b). A conserved tad pilus promotes Vibrio vulnificus oyster colonization. Environ. Microbiol. 20, 828–841. doi: 10.1111/1462-2920.14025
Ramsugit, S., Pillay, M. (2015). Pili of Mycobacterium tuberculosis: current knowledge and future prospects. Arch. Microbiol. 197, 737–744. doi: 10.1007/s00203-015-1117-0
Rangel, L. T., Marden, J., Colston, S., Setubal, J. C., Graf, J., Gogarten, J. P. (2019). Identification and characterization of putative Aeromonas spp. T3SS effectors. PLoS One 14, e0214035. doi: 10.1371/journal.pone.0214035
Rasmussen-Ivey, C. R., Figueras, M. J., McGarey, D., Liles, M. R. (2016a). Virulence factors of aeromonas hydrophila: in the wake of reclassification. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01337
Rasmussen-Ivey, C. R., Hossain, M. J., Odom, S. E., Terhune, J. S., Hemstreet, W. G., Shoemaker, C. A., et al. (2016b). Classification of a hypervirulent aeromonas hydrophila pathotype responsible for epidemic outbreaks in warm-water fishes. Front. Microbiol. 7, 1615. doi: 10.3389/fmicb.2016.01615
Roach, D. J., Burton, J. N., Lee, C., Stackhouse, B., Butler-Wu, S. M., Cookson, B. T., et al. (2015). A year of infection in the intensive care unit: Prospective whole genome sequencing of bacterial clinical isolates reveals cryptic transmissions and novel microbiota. PLoS Genet. 11, e1005413. doi: 10.1371/journal.pgen.1005413
Sanders, C. I., Ne Ville, C. J., Orwin, P. M. (2022). Complete genome sequences of four isolated bacteria from an undergraduate microbiology course using a hybrid assembly approach. Microbiol. Resour Announc. 11, e0102221. doi: 10.1128/mra.01022-21
Sangermani, M., Hug, I., Sauter, N., Pfohl, T., Jenal, U. (2019). Tad pili play a dynamic role in caulobacter crescentus surface colonization. mBio. 10, e01237–e01219. doi: 10.1128/mBio.01237-19
Seef, S., Herrou, J., De Boissier, P., My, L., Brasseur, G., Robert, D., et al. (2021). A Tad-like apparatus is required for contact-dependent prey killing in predatory social bacteria. eLife. 10, e72409. doi: 10.7554/eLife.72409
Sekizuka, T., Inamine, Y., Segawa, T., Hashino, M., Yatsu, K., Kuroda, M. (2019). Potential KPC-2 carbapenemase reservoir of environmental Aeromonas hydrophila and Aeromonas caviae isolates from the effluent of an urban wastewater treatment plant in Japan. Environ. Microbiol. Rep. 11, 589–597. doi: 10.1111/1758-2229.12772
Sekizuka, T., Tanaka, R., Hashino, M., Yatsu, K., Kuroda, M. (2022). Comprehensive genome and plasmidome analysis of antimicrobial resistant bacteria in wastewater treatment plant effluent of tokyo. Antibiotics (Basel). 11. doi: 10.3390/antibiotics11101283
Serra, D. O., Conover, M. S., Arnal, L., Sloan, G. P., Rodriguez, M. E., Yantorno, O. M., et al. (2011). FHA-mediated cell-substrate and cell-cell adhesions are critical for Bordetella pertussis biofilm formation on abiotic surfaces and in the mouse nose and the trachea. PLoS One 6, e28811. doi: 10.1371/journal.pone.0028811
Seshadri, R., Joseph, S. W., Chopra, A. K., Sha, J., Shaw, J., Graf, J., et al. (2006). Genome sequence of Aeromonas hydrophila ATCC 7966T: jack of all trades. J. Bacteriol. 188, 8272–8282. doi: 10.1128/JB.00621-06
Skerker, J. M., Shapiro, L. (2000). Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J. 19, 3223–3234. doi: 10.1093/emboj/19.13.3223
Snyder, R. A., Ellison, C. K., Severin, G. B., Whitfield, G. B., Waters, C. M., Brun, Y. V. (2020). Surface sensing stimulates cellular differentiation in Caulobacter crescentus. Proc. Natl. Acad. Sci. U.S.A. 117, 17984–17991. doi: 10.1073/pnas.1920291117
Sonani, R. R., Sanchez, J. C., Baumgardt, J. K., Kundra, S., Wright, E. R., Craig, L., et al. (2023). Tad and toxin-coregulated pilus structures reveal unexpected diversity in bacterial type IV pili. Proc. Natl. Acad. Sci. U.S.A. 120, e2316668120. doi: 10.1073/pnas.2316668120
Stewart, P. S., Franklin, M. J. (2008). Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 6, 199–210. doi: 10.1038/nrmicro1838
Tan, W. S., Yin, W. F., Chan, K. G. (2015a). Insights into the Quorum-Sensing Activity in Aeromonas hydrophila Strain M013 as Revealed by Whole-Genome Sequencing. Genome Announc. 3. doi: 10.1128/genomeA.01372-14
Tan, W. S., Yin, W. F., Chang, C. Y., Chan, K. G. (2015b). Whole-genome sequencing analysis of quorum-sensing aeromonas hydrophila strain M023 from freshwater. Genome Announc. 3. doi: 10.1128/genomeA.01548-14
Tekedar, H. C., Abdelhamed, H., Kumru, S., Blom, J., Karsi, A., Lawrence, M. L. (2019). Comparative Genomics of Aeromonas hydrophila Secretion Systems and Mutational Analysis of hcp1 and vgrG1 Genes From T6SS. Front. Microbiol. 9, 3216. doi: 10.3389/fmicb.2018.03216
Tekedar, H. C., Karsi, A., Akgul, A., Kalindamar, S., Waldbieser, G. C., Sonstegard, T., et al. (2015). Complete genome sequence of fish pathogen aeromonas hydrophila AL06-06. Genome Announc 3. doi: 10.1128/genomeA.00368-15
Tekedar, H. C., Kumru, S., Kalindamar, S., Karsi, A., Waldbieser, G. C., Sonstegard, T., et al. (2017). Draft genome sequences of three aeromonas hydrophila isolates from catfish and tilapia. Genome Announc 5. doi: 10.1128/genomeA.01509-16
Tekedar, H. C., Kumru, S., Karsi, A., Waldbieser, G. C., Sonstegard, T., Schroeder, S. G., et al. (2016a). Draft genome sequence of aeromonas hydrophila TN97-08. Genome Announc 4. doi: 10.1128/genomeA.00436-16
Tekedar, H. C., Kumru, S., Karsi, A., Waldbieser, G. C., Sonstegard, T., Schroeder, S. G., et al. (2016b). Draft genome sequences of four virulent aeromonas hydrophila strains from catfish aquaculture. Genome Announc 4. doi: 10.1128/genomeA.00860-16
Tekedar, H. C., Waldbieser, G. C., Karsi, A., Liles, M. R., Griffin, M. J., Vamenta, S., et al. (2013). Complete genome sequence of a channel catfish epidemic isolate, aeromonas hydrophila strain ML09-119. Genome Announc 1. doi: 10.1128/genomeA.00755-13
Teng, L., Deng, L., Dong, X., Wei, S., Li, J., Li, N., et al. (2017). Genome sequence of hypervirulent aeromonas hydrophila strain HZAUAH. Genome Announc 5. doi: 10.1128/genomeA.00012-17
Thomas, S. G., Glover, M. A., Parthasarathy, A., Wong, N. H., Shipman, P. A., Hudson, A. O. (2020). Expression of a Shiga-Like Toxin during Plastic Colonization by Two Multidrug-Resistant Bacteria, Aeromonas hydrophila RIT668 and Citrobacter freundii RIT669, Isolated from Endangered Turtles (Clemmys guttata). Microorganisms. 8, 1172. doi: 10.3390/microorganisms8081172
Tomás, J. M. (2012). The main aeromonas pathogenic factors. ISRN Microbiol. 2012, 1–22. doi: 10.5402/2012/256261
Tomich, M., Planet, P. J., Figurski, D. H. (2007). The tad locus: postcards from the widespread colonization island. Nat. Rev. Microbiol. 5, 363–375. doi: 10.1038/nrmicro1636
Tsai, Y.-H., Shen, S.-H., Yang, T.-Y., Chen, P.-H., Huang, K.-C., Lee, M. S. (2015). Monomicrobial Necrotizing Fasciitis Caused by Aeromonas hydrophila and Klebsiella pneumoniae. Med. Princ Pract. 24, 416–423. doi: 10.1159/000431094
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3—new capabilities and interfaces. Nucleic Acids Res. 40, e115–e11e. doi: 10.1093/nar/gks596
Wang, Y., Hou, N., Rasooly, R., Gu, Y., He, X. (2021). Prevalence and genetic analysis of chromosomal mcr-3/7 in aeromonas from U.S. Animal-derived samples. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.667406
Wang, P., Smith, A. L. (2023). Emergence of mobile colistin resistance genes within los angeles county wastewater. Environ. Sci. Technol. Letters. 10, 316–321. doi: 10.1021/acs.estlett.3c00080
Wang, Y., Wei, Y., Shang, N., Li, P. (2022). Synergistic inhibition of plantaricin E/F and lactic acid against aeromonas hydrophila LPL-1 reveals the novel potential of class IIb bacteriocin. Front. Microbiol. 13, 774184. doi: 10.3389/fmicb.2022.774184
Xu, L., Fan, J., Fu, H., Yang, Y., Luo, Q., Wan, F. (2022). The variants of polymyxin susceptibility in different species of genus Aeromonas. Front. Microbiol. 13, 1030564. doi: 10.3389/fmicb.2022.1030564
Xu, T., Rasmussen-Ivey, C. R., Moen, F. S., Fernandez-Bravo, A., Lamy, B., Beaz-Hidalgo, R., et al. (2023). A Global Survey of Hypervirulent Aeromonas hydrophila (vAh) Identified vAh Strains in the Lower Mekong River Basin and Diverse Opportunistic Pathogens from Farmed Fish and Other Environmental Sources. Microbiol. Spectr. 11, e0370522. doi: 10.1128/spectrum.03705-22
Xu, Z., Shen, W., Zhang, R., Cai, J. (2022). Clonal Dissemination of Aeromonas hydrophila With Binary Carriage of blaKPC-2-Bearing Plasmids in a Chinese Hospital. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.918561
Xu, L., Wang, H., Yang, X., Lu, L. (2013). Integrated pharmacokinetics/pharmacodynamics parameters-based dosing guidelines of enrofloxacin in grass carp Ctenopharyngodon idella to minimize selection of drug resistance. BMC Vet. Res. 9, 126. doi: 10.1186/1746-6148-9-126
Xu-Jie, Z., Yang, W.-M., Tong-Tong, L. I., Ai-Hua, L. I. (2013). The genetic diversity and virulence characteristics of Aeromonas hydrophila isolated from fishponds with disease outbreaks in Hubei province. Acta Hydrobiologica Sinica. 37, 458–466.
Yamazaki, K., Kashimoto, T., Niwano, A., Yamasaki, M., Nomura, M., Akeda, Y., et al. (2020). Expansion of necrosis depending on hybrid motor-driven motility of aeromonas hydrophila in a murine wound infection model. Microorganisms. 9. doi: 10.3390/microorganisms9010010
Yang, W., Li, N., Li, M., Zhang, D., An, G. (2016). Complete genome sequence of fish pathogen aeromonas hydrophila JBN2301. Genome Announc. 4. doi: 10.1128/genomeA.01615-15
Zatakia, H. M., Nelson, C. E., Syed, U. J., Scharf, B. E. (2014). ExpR coordinates the expression of symbiotically important, bundle-forming flp pili with quorum sensing in sinorhizobium meliloti. Appl. Environ. Microbiol. 80, 2429–2439. doi: 10.1128/AEM.04088-13
Zhang, J., Huang, Y., Xu, H., Ying, S., Pan, H., Yu, W. (2021). Genomic and phenotypic characteristics for vibrio vulnificus infections. IDR. Volume 14, 3721–3726. doi: 10.2147/IDR.S331468
Keywords: Aeromonas hydrophila, virulence, type IV pili, Tad (tight adherence), host-pathogen interaction
Citation: Tekedar HC, Patel F, Blom J, Griffin MJ, Waldbieser GC, Kumru S, Abdelhamed H, Dharan V, Hanson LA and Lawrence ML (2024) Tad pili contribute to the virulence and biofilm formation of virulent Aeromonas hydrophila. Front. Cell. Infect. Microbiol. 14:1425624. doi: 10.3389/fcimb.2024.1425624
Received: 30 April 2024; Accepted: 01 July 2024;
Published: 31 July 2024.
Edited by:
Jose Ramos-Vivas, Universidad Europea del Atlántico, SpainReviewed by:
Parisa Noorian, University of Technology Sydney, AustraliaMatej Medvecky, University of Warwick, United Kingdom
Copyright © 2024 Tekedar, Patel, Blom, Griffin, Waldbieser, Kumru, Abdelhamed, Dharan, Hanson and Lawrence. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hasan C. Tekedar, aGN0MzdAbXNzdGF0ZS5lZHU=
 Hasan C. Tekedar
Hasan C. Tekedar Fenny Patel1
Fenny Patel1 Jochen Blom
Jochen Blom Matt J. Griffin
Matt J. Griffin Geoffrey C. Waldbieser
Geoffrey C. Waldbieser Salih Kumru
Salih Kumru Hossam Abdelhamed
Hossam Abdelhamed Mark L. Lawrence
Mark L. Lawrence