
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol., 13 June 2024
Sec. Oral Microbes and Host
Volume 14 - 2024 | https://doi.org/10.3389/fcimb.2024.1421018
Introduction: Porphyromonas gingivalis and Porphyromonas endodontalis belong to the Bacteroidota phylum. Both species inhabit the oral cavity and can be associated with periodontal diseases. To survive, they must uptake heme from the host as an iron and protoporphyrin IX source. Among the best-characterized heme acquisition systems identified in members of the Bacteroidota phylum is the P. gingivalis Hmu system, with a leading role played by the hemophore-like HmuY (HmuYPg) protein.
Methods: Theoretical analysis of selected HmuY proteins and spectrophotometric methods were employed to determine the heme-binding mode of the P. endodontalis HmuY homolog (HmuYPe) and its ability to sequester heme. Growth phenotype and gene expression analysis of P. endodontalis were employed to reveal the importance of the HmuYPe and Hmu system for this bacterium.
Results: Unlike in P. gingivalis, where HmuYPg uses two histidines for heme-iron coordination, other known HmuY homologs use two methionines in this process. P. endodontalis HmuYPe is the first characterized representative of the HmuY family that binds heme using a histidine-methionine pair. It allows HmuYPe to sequester heme directly from serum albumin and Tannerella forsythia HmuYTf, the HmuY homolog which uses two methionines for heme-iron coordination. In contrast to HmuYPg, which sequesters heme directly from methemoglobin, HmuYPe may bind heme only after the proteolytic digestion of hemoglobin.
Conclusions: We hypothesize that differences in components of the Hmu system and structure-based properties of HmuY proteins may evolved allowing different adaptations of Porphyromonas species to the changing host environment. This may add to the superior virulence potential of P. gingivalis over other members of the Bacteroidota phylum.
Bacteria belonging to the Porphyromonas genus (Gibson and Genco, 2006; Summanen et al., 2015; Guilloux et al., 2021) inhabit mainly the oral cavity, gastrointestinal tract, and urogenital tract of humans, domestic and wild animals (Paster et al., 1994; Finegold et al., 2004; Sakamoto and Ohkuma, 2013; Zamora-Cintas et al., 2018; Acuna-Amador and Barloy-Hubler, 2020; Guilloux et al., 2021; Morales-Olavarria et al., 2023). Usually, they are isolated from oral infections (e.g., gingivitis, periodontitis, endodontic infections, abscesses) but also from other body infections (e.g., abscesses, infections of amniotic fluid and umbilical cord, infected wound sites) (van Steenbergen et al., 1984; Sundqvist, 1992; Zamora-Cintas et al., 2018; Acuna-Amador and Barloy-Hubler, 2020; Morales-Olavarria et al., 2023). Some of them, like Porphyromonas gingivalis, belong to opportunistic pathogens found in healthy oral cavities in low numbers but in high numbers in patients with periodontitis (Ximenez-Fyvie et al., 2000; Gomes et al., 2005; Bik et al., 2010).
Porphyromonas endodontalis is a Gram-negative, anaerobic, asaccharolytic, black-pigmented bacterium, primarily associated with root canal infections, often identified in dental periapical abscesses of endodontic origin and orofacial odontogenic infections (Sundqvist et al., 1979; van Steenbergen et al., 1984; van Winkelhoff et al., 1985a; Sundqvist et al., 1989; Sundqvist, 1992; Gomes et al., 2005; Siqueira and Rocas, 2009; Flynn et al., 2012). The most prevalent species in persistent endodontic infections, aside from P. endodontalis, comprise other members of the Bacteroidota (formerly Bacteroidetes) phylum, mainly P. gingivalis, Tannerella forsythia, and Prevotella intermedia (Gomes et al., 2005; Siqueira and Rocas, 2009; Flynn et al., 2012; Pinto et al., 2023). P. endodontalis can also be found together with P. gingivalis, T. forsythia, and P. intermedia in diseased periodontal sites in patients with periodontitis, mainly in apical periodontitis originating from endodontic infection (Tran et al., 1997; Kumar et al., 2003; Pereira et al., 2011; Lombardo Bedran et al., 2012; Lourenco et al., 2014; Perez-Chaparro et al., 2014; Na et al., 2020; Jimenez et al., 2022; Alvarez et al., 2023). Among Porphyromonas species, the best characterized P. gingivalis is not only the main etiologic agent and keystone pathogen of periodontitis (Darveau et al., 2012; Hajishengallis and Lamont, 2012; Deng et al., 2017, 2018; Hajishengallis and Diaz, 2020) but is often associated with systemic inflammation-based diseases (Nazir, 2017; Mei et al., 2020).
Although P. endodontalis and P. gingivalis belong to the same genus, both species differ in their phenotypes. In contrast to P. gingivalis, P. endodontalis is more sensitive to oxygen, grows better in culture media supplemented with heme, hemoglobin, or PPIX, does not exhibit hemagglutination activity, does not produce trypsin-like proteolytic enzymes and gingipains (van Steenbergen et al., 1984; van Winkelhoff et al., 1985b, 1986; Zerr et al., 2000, 2001). Similar to P. gingivalis, it produces collagenases and other proteases, such as dipeptidyl-peptidases (Sorsa et al., 1992; Tran et al., 1997; Odell et al., 1999; Chang et al., 2002; Nishimata et al., 2014), and can degrade host proteins, including hemoglobin (Kilian, 1981; Carlsson et al., 1984; Jansen et al., 1994; Rosen et al., 2001). Due to the lack of genes encoding a functional heme biosynthesis pathway in the P. endodontalis genome (GenBank accession number: ACNN00000000.1), typical for almost all members of the Bacteroidota phylum, P. endodontalis must uptake this compound from the host as an iron and protoporphyrin IX (PPIX) source.
Among the best-characterized heme acquisition systems identified in members of the Bacteroidota phylum is the P. gingivalis Hmu system (Wojtowicz et al., 2009a, Wojtowicz et al., 2009b; Bielecki et al., 2018, 2020; Sieminska et al., 2021; Antonyuk et al., 2023). The Hmu system comprises six proteins (HmuYRSTUV) encoded on the P. gingivalis hmu operon. The first protein encoded on this operon, HmuYPg, is the first representative of the novel HmuY family comprising hemophore-like proteins, different from classical hemophores or other hemophore-like proteins, such as P. gingivalis HusA protein (Olczak et al., 2024). Although P. gingivalis can transport free heme directly using TonB-dependent outer-membrane receptor HmuRPg, encoded downstream of the hmuYPg gene, HmuYPg protein facilitates this process through binding of heme and its delivery to HmuRPg (Smalley and Olczak, 2017). Importantly, HmuYPg can sequester heme directly from host hemoproteins or heme-binding proteins produced by cohabitating bacteria and deliver it to HmuRPg (Smalley and Olczak, 2017; Olczak et al., 2024). The functions of the other proteins encoded on the P. gingivalis hmu operon are unknown and they most likely play a role in heme transport into the bacterial cell and/or heme metabolism (Smalley and Olczak, 2017; Rocha et al., 2019).
Our research has shown that proteins belonging to the HmuY family differ in their heme-binding properties (Olczak et al., 2024), which may influence the adaptation of bacteria to the host environment they occupy. The most significant difference among HmuY homologs is the type of amino acids engaged in heme-iron coordination, which results in different heme-binding capacities depending on the heme-iron redox state. So far characterized HmuY proteins coordinating heme iron by two methionines (HmuY homologs from P. intermedia, T. forsythia, Bacteroides vulgatus, and Bacteroides fragilis) bind heme preferentially under reducing conditions (Bielecki et al., 2018, 2020; Sieminska et al., 2021; Antonyuk et al., 2023), while P. gingivalis HmuY coordinates heme iron by two histidines which results in a high affinity of heme binding in both oxidized and reduced environments (Wojtowicz et al., 2009a, Wojtowicz et al., 2009b).
In this study, we characterized another member of the HmuY family, a hemophore-like protein produced by P. endodontalis (HmuYPe), and compared its properties with P. gingivalis HmuYPg and T. forsythia HmuYTf. This approach allowed us to understand better the function and properties of HmuY family proteins produced by different human pathogens.
P. endodontalis ATCC 35406 (Argenta, Poznań, Poland) and P. gingivalis A7436 (laboratory collection) strains were grown on Schaedler blood agar (ABA) plates (Argenta) at 37°C for 6 and 4 days, respectively, under anaerobic conditions (80% N2, 10% H2 and 10% CO2) (Whitley A35 anaerobic workstation; Bingley, UK). Then, bacteria were used to inoculate a liquid basal medium (BM) composed of 3% trypticase soy broth (Becton Dickinson, Sparks, MD, USA) and 0.5% yeast extract (Biomaxima, Lublin, Poland), supplemented with 0.05% L-cysteine (Carl Roth, Karlsruhe, Germany), 0.5 mg/l menadione (Sigma-Aldrich, St. Louis, MO, USA) (BM medium), and 7.7 µM hemin chloride (Fluka, Munich, Germany) to allow bacteria to grow in optimal, iron- and heme-rich conditions (Hm medium). Alternatively, to starve bacteria of iron and heme, no heme source was added and the medium was supplemented with 160 µM 2,2-dipyridyl (Sigma-Aldrich) to complex free iron (DIP medium). The optical density at 600 nm (OD600) at the beginning of the liquid culture was at least 0.5 and 0.2 for P. endodontalis and P. gingivalis, respectively.
To analyze growth curves, bacteria were cultured for two passages in BM medium without the addition of a heme source. BM medium (BM alone) or medium supplemented with 1.25 or 5 µM human hemoglobin (Hb), BM medium supplemented with 5 µM heme and 5 µM human serum albumin (HSA), 5 µM HmuYPg, 5 µM HmuYPe, or 5 µM HmuYTf were inoculated with P. gingivalis or P. endodontalis at starting OD600 equal to 0.2 or 0.5, respectively. Media supplemented with proteins and heme were preincubated for 16 hours at 4°C before use, to allow saturation of proteins with heme.
Escherichia coli ER2566 strain (New England Biolabs, Ipswich, MA, USA) was grown under standard aerobic conditions.
The recombinant HmuYPe, lacking the predicted signal peptide (MKTRFFLALIATSLVLGVASCRP), was overexpressed and purified using affinity chromatography. Briefly, the hmuYPe gene from P. endodontalis (GenBank accession number: POREN0001_0444) was PCR amplified using primers listed in Supplementary Table S1 and cloned into XcmI and BamHI restriction sites of a pTriEx-4 plasmid (Sigma-Aldrich) using NEBuilder HiFi DNA Assembly (New England Biolabs), resulting in the plasmid encoding HmuYPe protein with an N-terminal 6×His tag and the site recognized by Factor Xa.
To generate plasmids encoding HmuYPe variants with amino acid substitutions, the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA, USA) and primers listed in Supplementary Table S1 were used.
HmuYPe protein and its site-directed mutagenesis variants were overexpressed in E. coli after induction with 0.5 mM isopropyl-ß-D-thiogalacto-pyranoside (IPTG; Carl-Roth) at 16°C for 16 hours. Proteins were purified using the soluble fraction of E. coli cell lysates and TALON Superflow resin according to the manufacturer’s instructions (Sigma-Aldrich), using 25 mM HEPES buffer, pH 7.8, supplemented with 300 mM NaCl. For the elution step, 25 mM Tris/HCl buffer, pH 7.6, supplemented with 80 mM NaCl and 150 mM imidazole (Carl-Roth) was used. To obtain un-tagged proteins, the buffer was exchanged for 25 mM Tris/HCl, pH 7.6, supplemented with 80 mM NaCl. The 6×His tag was removed using Factor Xa (New England Biolabs) and filtration through an Amicon Ultra-4 Centrifugal Ultracel-10KDa filter unit (Millipore). If necessary, another step of affinity chromatography with nickel-immobilized resin (Ni-NTA; New England Biolabs) was used to remove the 6×His tag before protein concentration (Supplementary Figure S2).
HmuYPg protein from P. gingivalis and HmuYTf protein from T. forsythia were overexpressed and purified as described previously (Slezak et al., 2020; Śmiga et al., 2023a). To determine protein concentration, empirical molar absorption coefficients for HmuYPg (ϵ280 = 36.86 mM−1 cm−1) (Wojtowicz et al., 2009b) and HmuYTf (ϵ280 = 26.32 mM−1 cm−1) (Bielecki et al., 2018) were used. For P. endodontalis HmuYPe, the empirical molar absorption coefficient was determined in this study (ϵ280 = 35.56 mM−1 cm−1), as described by others (Eakanunkul et al., 2005).
Protein samples, P. endodontalis and P. gingivalis whole cell lysates, or samples prepared from the whole bacterial cultures were analyzed by SDS-PAGE. Samples were separated on 12% polyacrylamide gels and the proteins were visualized with Coomassie Brilliant Blue G-250 (CBB G-250) or were transferred onto nitrocellulose membranes (Millipore, Billerica, MA, USA). 20 or 100 ng of HmuYPe, HmuYPg, or HmuYTf proteins in 5 µl were applied onto nitrocellulose membranes for dot blotting. Western blotting and dot blotting were performed as described before (Śmiga et al., 2015; Śmiga et al., 2023a). Briefly, membranes were incubated with rabbit anti-HmuYPg or rabbit anti-HmuYTf polyclonal antibodies (1:10,000; GenScript USA Inc.). Subsequently, goat anti-rabbit IgG antibodies conjugated with horseradish peroxidase (1:10,000; Sigma-Aldrich) were applied. Chemiluminescence staining (Perkin Elmer, Waltham, MA, USA) and ChemiDoc Imaging System (Bio-Rad Laboratories, Hercules, CA, USA) were used to visualize proteins.
Analysis of heme binding was performed as described previously (Śmiga et al., 2023b). Briefly, ~8 mg of hemin chloride (Pol-Aura, Morąg, Poland) was dissolved in 0.1 M NaOH, and its concentration was determined using empirical molar absorption coefficient (ϵ385 = 58.5 mM−1 cm−1). Proteins at 5 µM concentration were prepared in 20 mM sodium phosphate buffer, pH 7.4, containing 140 mM NaCl (PBS). Protein samples were titrated with heme, and protein-heme complexes were monitored by UV-visible absorbance (250–700 nm) spectroscopy with a double-beam Jasco V-750 spectrophotometer (Jasco GmbH, Pfungstadt, Germany). The reduced conditions were formed by the addition of sodium dithionite (Sigma-Aldrich) to the final 10 mM concentration and mineral oil overlay of the sample (Sigma-Aldrich).
Heme sequestration was examined by mixing holo- (protein-heme complex) and apo-protein (protein alone) in PBS and monitoring heme transfer by UV-visible absorbance spectroscopy or PAGE, as described before (Smalley et al., 2011; Śmiga et al., 2023b). Except for methemoglobin (metHb; Sigma-Aldrich), protein-heme complexes were prepared by mixing protein and heme at a 1:1.2 molar ratio and incubation at room temperature for 1 hour. To remove unbound heme, the solutions were passed through Zeba Spin desalting columns (Thermo Fisher, Scientific, Waltham, MA, USA).
1.25 µM metHb or 5 µM other holo-proteins were mixed with 5 µM apo-proteins. The heme sequestration process was monitored over time by recording UV-visible absorbance spectra under oxidizing and reducing conditions. Alternatively, 20 µM holo-HmuYTf protein or 10 µM other holo-proteins were mixed with 10 µM apo-proteins and incubated for 30 minutes at 37°C. To 30 µl of the sample, 10 µl of 0.4 M Tris/HCl buffer, pH 6.8, supplemented with 40% glycerol and 0.08% bromophenol blue was added, and 25 µl of the sample was immediately loaded on the 13.5% PAGE-separating gel prepared without SDS. After electrophoresis, the heme-containing complexes were first stained using 5 mM 3,3′,5,5′-tetramethylbenzidine (TMB) prepared in 100 mM Tris/HCl buffer, pH 7.5, supplemented with 140 mM NaCl and 0.05% H2O2 (TMB-H2O2 staining) up to 30 minutes. Subsequently, all proteins were stained with CBB G-250.
To determine the influence of iron and heme on P. endodontalis gene expression, bacteria were cultured in Hm medium or DIP medium for one or two 24-hour passages. Bacteria from 1 ml of culture were centrifuged and used for RNA isolation with the Total RNA Mini Kit (A&A Biotechnology, Gdańsk, Poland). Genomic DNA contamination was removed using the Clean-up RNA concentrator Kit (A&A Biotechnology). RNA was used to generate cDNA with a LunaScript RT SuperMix Kit (New England Biolabs). qPCR was performed using SensiFAST SYBR no-ROX Kit (Bioline, London, UK) and LightCycler 96 (Roche, Basel, Switzerland). PCR program comprised initial denaturation at 95°C for 120 seconds, 35 cycles of denaturation at 95°C for 5 seconds, primers annealing at 60°C for 10 seconds, and extension at 72°C for 15 seconds. After PCR, the melting curves were generated for PCR quality control. Relative change in gene expression was calculated using LightCycler 96 software (Roche) and 16S rRNA as a reference gene. All analyses were carried out in 4 biological repetitions. All primers used are listed in Supplementary Table S1.
The total proteolytic activity of whole bacterial cultures was measured using azocasein (Sigma-Aldrich) as a substrate. Briefly, to 40 μl of 1.5% azocasein solution in reaction buffer (20 mM Tris/HCl buffer, pH 7.5, supplemented with 150 mM NaCl, 5 mM CaCl2, 0.05% Tween 20, and 10 mM L-cysteine hydrochloride freshly neutralized with NaOH), 10 μl of total P. endodontalis or P. gingivalis overnight culture (grown for 24 hours) was added. Samples were incubated for 30 minutes at 37°C, and the reaction was stopped by adding 200 μl of 5% TCA (Sigma-Aldrich). Precipitated, undigested azocasein was separated by centrifugation (2000×g, 10 min). 100 μl of the supernatant was mixed with 60 μl of 0.5 M NaOH and the absorbance at 450 nm (A450) was measured using a GloMax Discover plate reader (Promega, Madison, WI, USA). The final results were presented as a change of A450 over 60 minutes caused by 1 ml of culture exhibiting OD600 equal to 1 (ΔA450/60 min/ml).
HSA (Sigma-Aldrich), metHb (Sigma-Aldrich), hemopexin (Hpx; Sigma-Aldrich), P. gingivalis HmuYPg, P. endodontalis HmuYPe, and T. forsythia HmuYTf were used to analyze their susceptibility to proteolysis performed by proteases produced by P. endodontalis. Briefly, Hm medium was supplemented with 2 µM proteins and the bacterial cultures were started with the initial OD600 equal to 0.5. Samples were collected over time and examined using SDS-PAGE and staining with CBB-G250. As a control, the fresh BM+Hm medium or P. gingivalis culture started at OD600 equal to 0.2 was used.
All experiments were performed independently at least in two biological replicates and at least in three technical repetitions each. The numerical values are represented as mean ± standard deviation (mean ± SD) or mean ± standard error (mean ± SE). All statistical analyses were done using unpaired Student’s t-test with GraphPad software (GraphPad Prism 8.0 Inc., San Diego, CA, USA).
Search for protein sequences was performed in the GenBank database using PSI-BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Protein amino acid sequences were compared with The Clustal Omega (Madeira et al., 2022) and Jalview (Waterhouse et al., 2009). Protein similarity and identity were analyzed using Sequence Manipulation Suite: Ident and Sim (Stothard, 2000). Solved (RCSB Protein Data Bank; https://www.rcsb.pdb) and predicted with AlphaFold Protein Structure Database (Jumper et al., 2021; Varadi et al., 2022) three-dimensional protein structures were visualized with UCSF Chimera (Pettersen et al., 2004). Heme binding in HmuYPe was predicted with EDock (Zhang et al., 2020). Prediction of P. endodontalis hmu operon genes was performed using an Operon mapper (https://biocomputo.ibt.unam.mx/operon_mapper/).
P. endodontalis ATCC 35406 hmu operon organization is similar to that found in P. gingivalis, including one HmuY homolog (Figure 1A). HmuYPe protein is closely related to HmuYPg with ~50% amino acid sequence identity, as well as 21–22% identity and 32% similarity to other characterized HmuY proteins produced by human oral pathogens from the Bacteroidota phylum (Figures 1B, C). Other proteins of the Hmu system in P. endodontalis show high homology to those found in P. gingivalis with amino acid sequence identity as follows: HmuR ~49%, HmuS ~ 65%, HmuT ~50%, HmuU ~68%, and HmuV ~79%. Interestingly, in P. endodontalis an additional gene (GenBank accession number: POREN0001_0445) is located upstream of the hmuYPe gene, encoding a protein containing T9SS type A sorting domain (T9SS) (Figure 1A). Using theoretical and experimental approaches, we confirmed that the POREN0001_0445 gene is a part of the hmu operon (Supplementary Figure S1A). Proteins homologous to the protein encoded by the POREN0001_0445 gene are found in other bacteria including some Porphyromonas species and P. intermedia. However, in contrast to P. endodontalis, they are not encoded as a part of operons but as orphan genes (data not shown). Despite the low amino acid sequence identity between analyzed homologs of the POREN0001_0445 protein (Supplementary Figure S1B), their predicted tertiary structure is highly similar (Supplementary Figure S1C). They form a β-barrel-like structure, which may suggest their role in transporting an unknown molecule through the outer membrane.

Figure 1 Theoretical analysis of the P. endodontalis Hmu system. (A) Organization of P. endodontalis hmu operon and its counterpart in P. gingivalis. (B) Homology of the HmuYPe protein to the representatives of the HmuY protein family presented in the form of a heat map indicating the % identity and similarity between the amino acid sequences of the proteins. The simplified guide tree was created with Clustal Omega (https://www.ebi.ac.uk/jdispatcher/msa/clustalo). (C) The alignment of HmuY protein sequences with marked residues involved in heme-iron coordination experimentally confirmed (green) or theoretically predicted (blue). The consensus amino acid sequence is shown below the examined sequences. HmuY homologs from P. gingivalis (HmuYPg), P. gulae (HmuYPgu), P. endodontalis (HmuYPe), P. loveana (HmuYPlo), P. gingivicanis (HmuYPgc), P. circumdentaria (HmuYPcd), P. crevioricanis (HmuYPcc), T. forsythia (HmuYTf) and P. intermedia (HmuYPi-1 and HmuYPi-2). (D) Comparison of overall structures of representative HmuY proteins produced by Porphyromonas species and T. forsythia, with marked heme-iron coordinating amino acids. Structures of HmuPg and HmuYTf determined by crystallography have been deposited under PDB IDs: 6EWM and 6EU8, respectively. Other structures of HmuY homologs were predicted with AlphaFold: HmuYPgu (ID: AF-A0A099WTH2-F1), HmuYPe (ID: AF-C3JC46-F1), HmuYPlo (ID: AF-A0A2U1FHF1-F1), HmuYPgc (ID: AF-A0A0A2GAC0-F1), HmuYPcd (ID: AF-A0A1T4NY32-F1), and HmuYPcc (ID: AF-A0A0A2FIS2-F1).
Analysis of the overall three-dimensional experimentally solved (P. gingivalis HmuYPg and T. forsythia HmuYTf) or theoretically predicted (other selected Porphyromonas species) protein structures revealed that HmuY proteins identified in the Porphyromonas species are highly similar to the P. gingivalis HmuYPg (Figure 1D), with the highest similarities observed in the core region (Śmiga et al., 2023a). As in so far characterized HmuY proteins (Olczak et al., 2024), the main differences in HmuY proteins identified in Porphyromonas species are visible in the structure of heme-binding pockets and differ mainly in the size of the entrance of the heme-binding pocket (Figure 1D). The data shown in Figure 1 allowed us to verify the spatial location of predicted amino acids involved in heme binding in HmuY homologs in analyzed Porphyromonas species. To date, the characterization of HmuY proteins has included the HmuYPg protein from the P. gingivalis, which uses two histidines for heme-iron coordination, and HmuY homologs which use two methionines in this process (for example T. forsythia HmuYTf, P. intermedia HmuYPi-1 and HmuYPi-2). Our theoretical analyses showed that the majority of HmuY proteins identified in Porphyromonas species most likely coordinate heme-iron using a histidine-methionine pair or two methionines (Figures 1C, D). Only proteins closely related to the HmuYPg, namely HmuYPgu from P. gulae and HmuYPlo from P. loveana may coordinate heme iron with two histidines (Figures 1C, D).
To confirm the heme binding ability of the HmuYPe, the protein was overexpressed in E. coli and purified using chromatographic methods (Supplementary Figures S2A, S2B). Purified and concentrated protein samples exhibited a reddish color (data not shown) similar to the HmuYPg sample (Bielecki et al., 2018, 2020). The UV-visible spectroscopic analysis demonstrated that under both oxidizing and reducing conditions, the HmuYPe sample exhibited spectra similar to those of the purified HmuYPg sample (Figures 2A, B), which indicated heme binding. To confirm this, the HmuYPe protein was saturated with heme [Fe(III)heme], and heme excess was removed by desalting. A UV-Vis absorbance spectrum of the sample was characterized by maxima in both the Soret band (414 nm) and Q bands (530 nm and 559 nm) (Figure 2C). Reduction of heme in this sample [Fe(II)heme] resulted in a red shift in the Soret band (425 nm), and a shift of the Q bands to 528 nm and 558 nm, which became more intense and resolved (Figure 2D). These spectra were similar to those obtained for the HmuYPg-heme complex (Figures 2E, F).
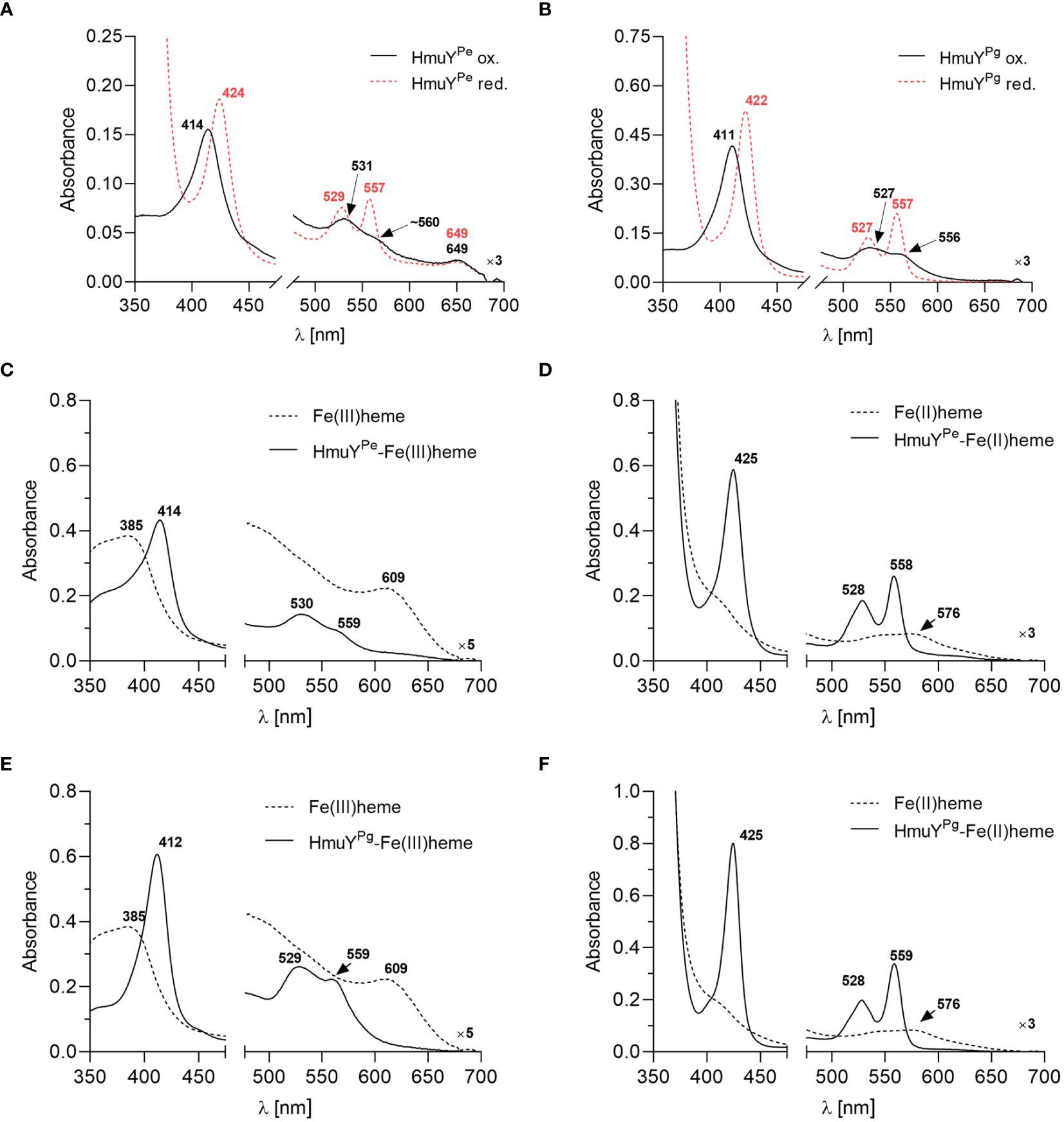
Figure 2 The heme-binding ability of HmuY proteins. The UV-visible absorbance spectrum of 50 µM purified P. endodontalis HmuYPe protein indicates heme binding (A), similar to the 50 µM purified P. gingivalis HmuYPg (B), examined under oxidizing (black lines) and reducing conditions (red lines), the latter formed by 10 mM sodium dithionite. Heme binding by HmuYPe (C, D) or HmuYPg (E, F) was confirmed by saturating the protein with heme at a 1:1.2 molar ratio and removing the excess heme by desalting. The UV-visible absorbance spectrum of the obtained sample (5 µM protein with bound heme) was compared to the spectrum of the 5 µM heme alone sample under oxidizing (C, E) and reducing conditions (D, F).
Heme binding strength was analyzed by determination of the HmuYPe-heme complex dissociation constant (Kd) (Figure 3A) by titration of apo-protein with increasing concentrations of heme (Supplementary Figure S3). Kd of HmuYPe-heme complex was lower under reducing (4.10 × 10-8 M) than oxidizing (2.45 × 10-7 M) (Figure 3A) conditions, thus being similar to HmuY homologs coordinating heme-iron with two methionine residues, e.g., T. forsythia HmuYTf, than to P. gingivalis HmuYPg (Bielecki et al., 2018).
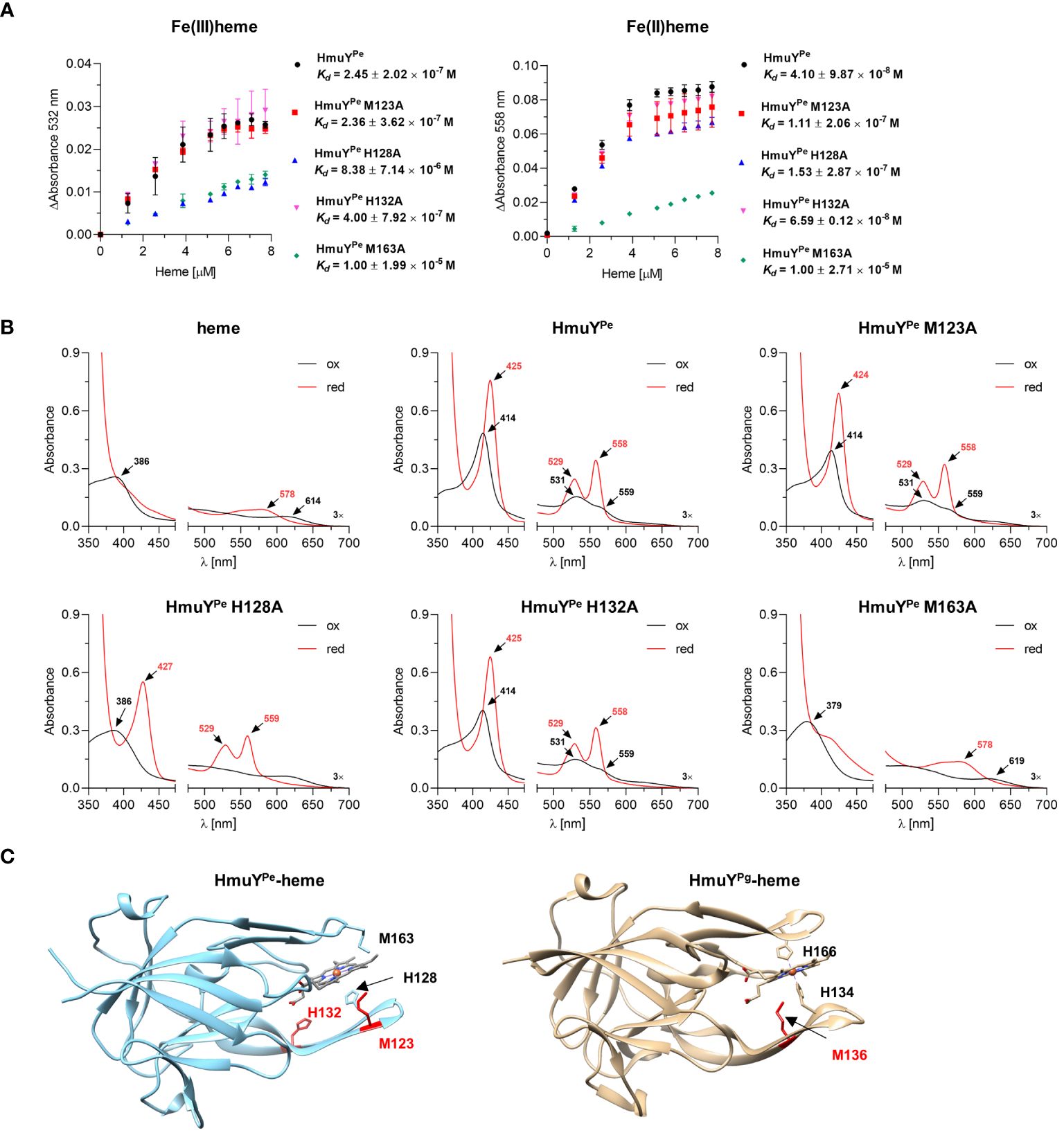
Figure 3 Analysis of P. endodontalis HmuYPe protein site-directed mutagenesis variants in complex with heme. (A) To determine the dissociation constant (Kd), HmuYPe protein variants were titrated with increasing heme concentrations. Difference absorbance spectra were used for plotting graphs of the change in the Q band absorbance maximum for oxidizing (ΔAbsorbance at 532 nm) and reducing (ΔAbsorbance at 558 nm) conditions versus heme concentration. Kd was determined using the one-site binding model. (B) Purified HmuYPe and its variants with single substitutions of methionine or histidine for alanine were saturated with heme at a 1:1 molar ratio. UV-visible absorbance spectra were recorded under oxidizing (black lines) and reducing (red lines) conditions, and compared to the spectrum of heme alone and the unmodified HmuYPe protein. The reducing conditions were formed by 10 mM sodium dithionite. (C) Comparison of overall protein structures of HmuYPe and HmuYPg in complex with heme. The experimentally solved three-dimensional structure of P. gingivalis HmuYPg in complex with heme (PDB ID: 3H8T) and modeled predicted structure of P. endodontalis HmuYPe in complex with heme are shown. The heme-iron coordinating amino acids are indicated in black, and amino acids with supporting or putatively supporting roles in heme binding are marked in red.
To experimentally confirm heme-iron coordinating amino acids, we prepared HmuYPe site-directed mutagenesis protein variants with selected histidines or methionines replaced by an alanine (M123A, H128A, H132A, and M163A), chosen based on the comparative analysis of amino acid sequences (Figure 1C). Particular single mutagenesis variants were overexpressed in E. coli and purified at similar levels as the unmodified protein (Supplementary Figure S2C). All protein variants were stable after purification, which suggests that amino acid replacements did not influence the tertiary protein structure. UV-visible absorbance spectra of HmuYPe-heme complexes showed changes mostly in the case of H128A and M163A protein variants, suggesting that these amino acids could be engaged in heme-iron coordination (Figure 3B). The determination of Kd showed that the H128A and M163A variants are characterized by a lower affinity for heme than an unmodified protein (Figure 3A). A slightly lower affinity of heme binding under reducing conditions in the case of the M123A variant (Figure 3A) may suggest local structural changes in the loop engaged in heme-iron coordination and/or supportive role in heme binding as it was shown for HmuYPg M136 (Wojtowicz et al., 2009b; Bielecki et al., 2018). This finding was confirmed by our theoretical analysis using the modeled HmuYPe structure and its comparison to the experimentally solved HmuYPg-heme structure (Figure 3C).
To compare the strength of heme binding to the HmuYPe with other HmuY proteins, we used a competitive analysis with HmuYPg and HmuYTf. For this purpose, we employed UV-visible spectroscopy and native electrophoresis (PAGE) (Figure 4). HmuYPe protein was unable to sequester heme complexed with HmuYPg (Figures 4A–C). At the same time, HmuYPg was able to capture heme from the HmuYPe-heme complex under both oxidizing and reducing conditions (Figures 4D–F). In the case of HmuYTf, we observed efficient heme sequestration of heme from the HmuYTf-heme complex by HmuYPe under oxidizing and reducing conditions, while HmuYTf was not able to capture heme from the HmuYPe complex (Figures 4G–L). In contrast to the spectroscopic method, we could visualize neither HmuYPe-heme nor HmuYTf-heme complexes with PAGE because they were hardly visible after TMB-H2O2 staining (Figures 4A, D, G, J). This effect could be caused by oxidizing conditions applied in this experiment, resulting in weak heme binding or heme release.

Figure 4 Heme sequestration capacity of P. endodontalis HmuYPe and HmuY homologs from P. gingivalis [HmuYPg; (A–F)] or T. forsythia [HmuYTf; (G–L)]. Heme transfer was examined using PAGE, staining with TMB-H2O2, and subsequent visualization of proteins by CBB G-250 staining (A, D, G, J). P. gingivalis HmuYPg was used as a control. The HmuYPe-heme complex after TMB-H2O2 staining is hardly visible due to analysis performed under aerobic conditions, causing lower heme binding to the HmuYPe under oxidizing conditions. Apo-proteins or proteins in complex with heme were incubated at equimolar concentrations under oxidizing (B, E, H, K) or reducing conditions (C, F, I, L), the latter formed by 10 mM sodium dithionite. Changes in spectra were monitored using UV-visible absorbance spectroscopy.
Further, we analyzed the ability of HmuYPe to directly sequester heme from host hemoproteins, which serve in vivo as the main heme source. Under oxidizing conditions, the affinity of HSA and HmuYPe to heme was similar because while using HSA-heme or HmuYPe-heme complex with apo-form of the counterpart, we observed an equilibrium in heme binding between the proteins (Figures 5A–F). Although PAGE results are not clear (Figures 5A, D), using the spectroscopic method we observed the spectrum shift in two experimental settings (Figures 5B, E). Under reducing conditions, HmuYPe was able to capture heme bound to HSA, whereas apo-HSA could not sequester heme from HmuYPe (Figures 5C, F). In the case of Hpx, the results are inconclusive due to the low quality of HmuYPe TMB-H2O2 staining (Figures 5G, J). In the case of these proteins, equilibrium in Hpx-heme and HmuYPe-heme complexes could also occur, which was confirmed by changes in the intensity of the analyzed UV-visible absorbance spectra over time (Figures 5H, I, K, L). However, since the absorbance maxima of both proteins in complex with heme are similar, we cannot draw firm conclusions. In contrast to HmuYPg (Smalley et al., 2011; Byrne et al., 2013; Śmiga et al., 2021), HmuYPe was unable to capture the heme associated with metHb (Supplementary Figures S4A, S4B). Moreover, heme sequestration was not observed after the chemical reduction of metHb (Supplementary Figure S4C). As a control, we used HmuYPg, which can directly sequester heme complexed with HSA (Figure 5A; Supplementary Figure S5) (Smalley et al., 2011; Sieminska et al., 2021), Hpx (Figure 5G) (Bielecki et al., 2018), and metHb (Supplementary Figure S4A) (Smalley et al., 2011).
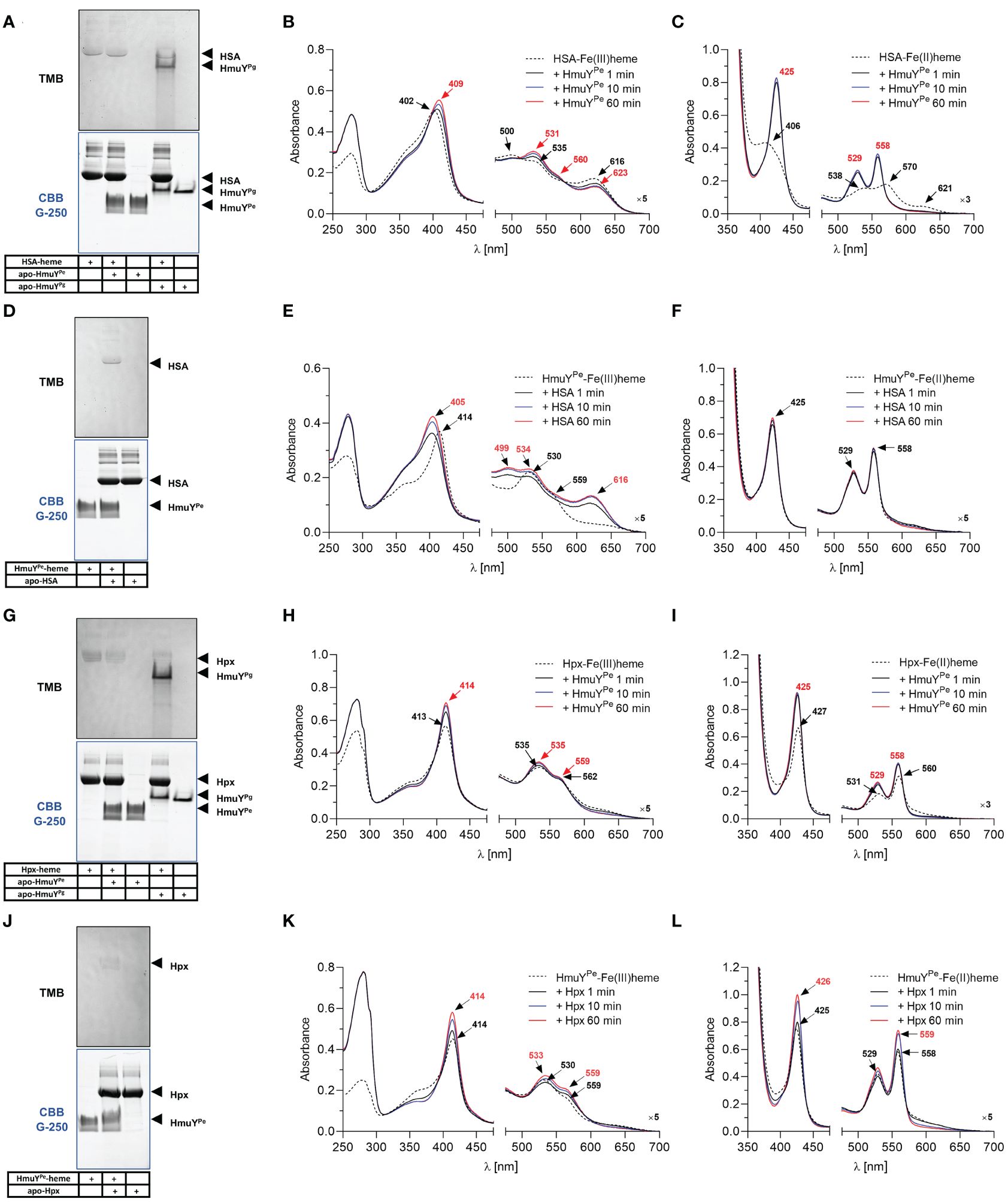
Figure 5 Heme sequestration capacity of P. endodontalis HmuYPe from human serum albumin [HSA; (A–F)] or hemopexin [Hpx; (G–L)]. Heme transfer was examined using PAGE, staining with TMB-H2O2, and subsequent visualization of proteins by CBB G-250 staining (A, D, G, J). P. gingivalis HmuYPg was used as a control. The HmuYPe-heme complex after TMB-H2O2 staining is hardly visible due to analysis performed under aerobic conditions, causing lower heme binding to the HmuYPe under oxidizing conditions. Apo-proteins or proteins in complex with heme were incubated at equimolar concentrations under oxidizing (B, E, H, K) or reducing conditions (C, F, I, L), the latter formed by 10 mM sodium dithionite. Changes in spectra were monitored using UV-visible absorbance spectroscopy.
P. gingivalis can use a variety of heme sources thanks to the direct sequestration of heme from hemoproteins by HmuYPg or its synergistic cooperation with highly active proteases – gingipains (Smalley and Olczak, 2017; Śmiga et al., 2023b; Olczak et al., 2024). Although P. endodontalis does not encode gingipain homologs, it produces proteases, albeit less active compared to P. gingivalis proteases (Figure 6A). Nevertheless, they can efficiently degrade metHb in the P. endodontalis culture (Figure 6B). None of the additionally analyzed proteins were degraded by P. endodontalis proteases, including human hemoproteins and representatives of HmuY homologs produced by other human pathogens (Figure 6B). These results are consistent with P. endodontalis growing in the planktonic form with different heme sources, the best achieved in the presence of free heme or metHb (Supplementary Figure S6; Figure 6C).
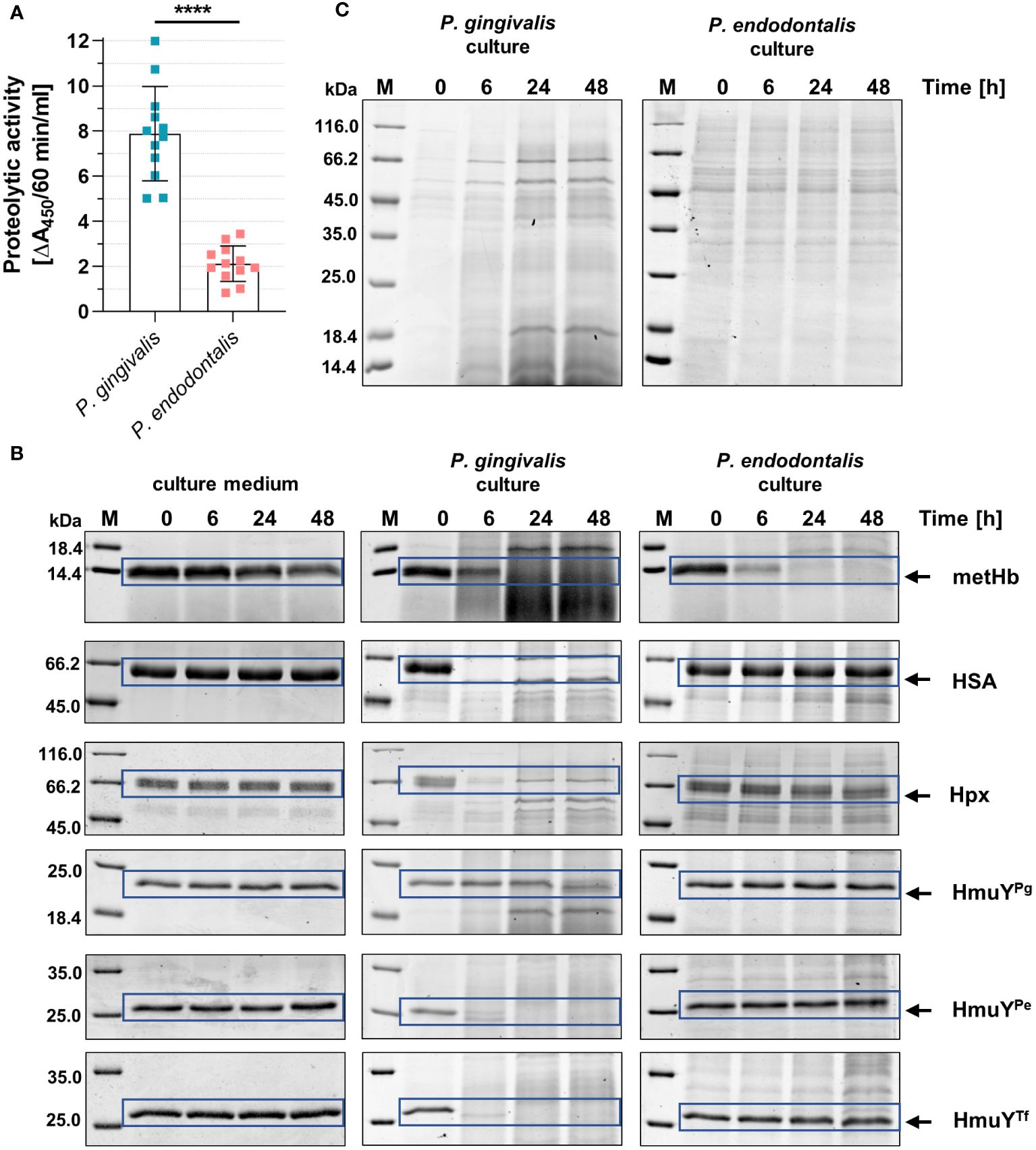
Figure 6 Comparison of the proteolytic activity of P. endodontalis and P. gingivalis. (A) Total proteolytic activity in whole bacterial cultures (iron- and heme-rich medium; Hm medium) was determined using azocasein as a substrate. The activity is shown as an increase in absorbance at 450 nm (A450) caused by the release of a colored product within 60 minutes by 1 ml of bacterial culture with an optical density at 600 nm of 1. (B) Susceptibility of human hemoproteins and selected HmuY proteins to degradation by proteases produced by P. endodontalis or P. gingivalis. Purified proteins were added to the bacterial cultures (Hm medium) at a final 2 µM concentration, samples were incubated, collected at the indicated time points, and analyzed by SDS-PAGE and CBB G-250 staining. As a control, Hm medium alone was used instead of bacterial cultures. (C) Protein pattern of P. gingivalis or P. endodontalis proteins in iron- and heme-rich culture medium (Hm medium) without adding host hemoproteins or HmuY proteins. metHb, methemoglobin; HSA, serum albumin; Hpx, hemopexin; HmuYPg, P. gingivalis HmuY; HmuYPe, P. endodontalis HmuY homolog; HmuYTf, T. forsythia HmuY homolog; M, protein molecular mass markers. ****P<0.0001.
Similar to other HmuY proteins that were characterized previously (Bielecki et al., 2018, 2020; Sieminska et al., 2021; Antonyuk et al., 2023), transcript encoding HmuYPe was produced at higher levels when bacteria were grown in iron- and heme-depleted conditions, formed as shown in Figure 7A. However, the increase of the hmuYPe gene expression was only up to 6 times, whereas for hmuYPg the expression increased up to several hundred times (Bielecki et al., 2018, 2020), resulting in a significant increase in produced HmuYPg protein (Figure 7B) (Olczak et al., 2010). This dissimilarity may result from weaker growth of P. endodontalis under laboratory conditions, especially under iron and heme starvation (Supplementary Figure S6).
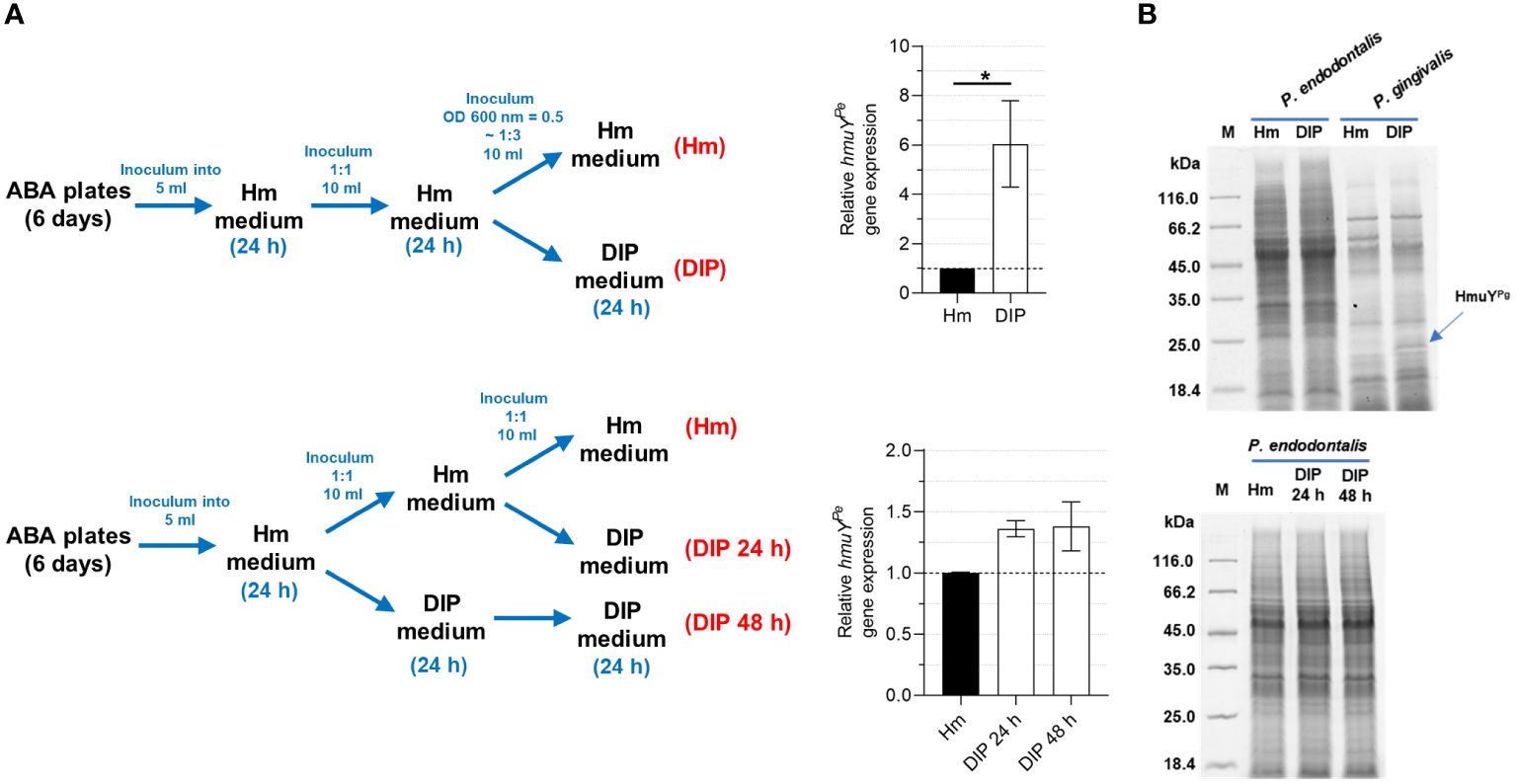
Figure 7 Influence of iron and heme starvation on P. endodontalis HmuYPe expression. (A) Determination of relative hmuYPe gene expression examined by RT-qPCR with 16S rRNA as a reference gene. Relative expression fold change represents mRNA levels in bacteria starved of iron and heme (DIP medium) versus bacteria grown in rich iron and heme conditions (Hm medium), the latter set as 1.0. (B) The influence of iron and heme starvation on the overall production of proteins in P. endodontalis was examined by SDS-PAGE and CBB G-250 staining. P. gingivalis was used as a control and a band indicating HmuYPg is shown with a blue arrow. M, protein molecular mass markers. *P<0.05.
Finally, we aimed to find whether, despite 50% amino acid sequence identity (Figure 1) and almost identical tertiary structures of HmuYPe and HmuYPg (Figure 3C), HmuYPe could potentially serve as a specific marker for P. endodontalis. We analyzed the purified HmuYPe protein, both in the form of native and denatured protein and showed cross-reactivity neither with IgG antibodies raised against HmuYPg nor HmuYTf (Supplementary Figure S7).
Interactions between microorganisms and the host play an important role in the etiopathogenesis of many human diseases. Although the main ecological niches of P. gingivalis and P. endodontalis (subgingival pockets and root canals, respectively) are different, both species can be found in the same polymicrobial consortia, including periodontal pockets, often with other members of the Bacteroidota phylum (Kumar et al., 2003; Pereira et al., 2011; Lombardo Bedran et al., 2012; Lourenco et al., 2014). Proteomic analysis revealed that the type and number of proteins associated with virulence were more similar between P. endodontalis and the more virulent P. gingivalis W83 strain as compared to the less virulent P. gingivalis ATCC 33277 strain (Li et al., 2016). One of the main virulence factors expressed by the members of the Bacteroidota phylum is the Hmu heme acquisition system. Recently, we demonstrated that the expression of heme acquisition systems, mainly the Hmu system, is different between more and less virulent P. gingivalis strains, expressed and used more effectively in more virulent strains (Śmiga et al., 2024). Since one of its components, namely the HmuY protein is considered to be one of the main virulence factors of opportunistic pathogens, in this study, we performed a comparative analysis of the P. endodontalis HmuYPe protein and HmuY proteins produced by P. gingivalis (HmuYPg) and T. forsythia (HmuYTf).
The main heme sources for P. endodontalis, similar to P. gingivalis, are serum heme-sequestering proteins, heme complexed to bacterial proteins, and periodically, hemoglobin released from erythrocytes (Grenier and Mayrand, 1986; Sundqvist, 1992; Zerr et al., 1998, 2000, 2001; Smalley and Olczak, 2017). In humans, oral pathogens are provided with heme mainly by HSA, the main component of the gingival crevicular fluid. This is also the case of HmuYPe which captures heme directly from the HSA-heme complex, with higher ability under reducing conditions. This finding is consistent with literature data reporting Kd of HSA-heme of 10-7-10-8 M, which is similar to that determined for the HmuYPe, as well as by the fact that heme can be more easily released from the HSA-heme complex under reducing conditions (Cao et al., 2012). This property is also exploited by HmuY homologs in which two methionines are used to coordinate heme-iron (Bielecki et al., 2018, 2020; Sieminska et al., 2021; Antonyuk et al., 2023).
Although hemoglobin is unavailable in higher concentrations in the main niche occupied by P. endodontalis, namely root canals, this heme source can be available for this bacterium during treatment procedures, or when the bacterium is found in deep periodontitis sites, where in advanced stages of disease bleeding may occur. As shown in this study, even though the total proteolytic activity of P. endodontalis cultures was significantly lower compared to P. gingivalis cultures, the degradation capacity of metHb was comparable in both bacterial cultures. In contrast to P. gingivalis, which prefers to convert oxyhemoglobin possessing Fe(II)heme to metHb possessing Fe(III)heme, the process facilitating hemoglobin degradation by gingipains (Smalley et al., 2007, 2008) and direct heme sequestration by P. gingivalis HmuYPg (Smalley et al., 2011), HmuYPe was unable to sequester heme bound to metHb. It has been demonstrated that P. endodontalis can reduce metHb under anaerobic conditions (Zerr et al., 2001). However, we showed that this ability does not allow HmuYPe for heme sequestration from chemically reduced metHb in the experiment carried out under limited oxygen access. Therefore, based on better P. endodontalis growth in a culture medium supplemented with metHb, we assume that the high ability of metHb degradation by this bacterium may be used to efficiently release heme from metHb which can be subsequently bound by HmuYPe, instead of direct heme capture by HmuYPe.
P. gingivalis expresses several features engaged in its higher adaptation to the changing host environment and inhibitory activity against other bacteria, including P. endodontalis (van Winkelhoff et al., 1987). Many Porphyromonas species are obligate anaerobes and can survive temporarily in the presence of oxygen, P. gingivalis being the best example (Smalley et al., 1998; Mark Welch et al., 2016; Slezak et al., 2020). But this is not the case for P. endodontalis which is highly sensitive to oxygen (van Winkelhoff et al., 1985b, 1986). Data obtained in our study showed that one of the P. gingivalis adaptive features can be assigned not only to better tolerance to oxygen and higher proteolytic activity but also to different properties of heme binding by HmuYPg as compared to HmuYPe. The strength of heme binding by HmuY proteins depends on the redox state of the external environment (Olczak et al., 2024) and based on our results is as follows: HmuYPg>HmuYPe>HmuYTf. HmuYPg, coordinating heme iron with two histidines, binds heme efficiently under both oxidizing and reducing conditions. When two methionine residues are involved in this process in HmuY homologs, including T. forsythia HmuYTf, they bind heme preferentially under reducing conditions (Olczak et al., 2024). P. endodontalis HmuYPe, coordinating heme-iron with the histidine-methionine pair, is a missing link between the HmuY proteins mentioned above. Although P. endodontalis HmuYPe is the first characterized protein among HmuY family members with a histidine-methionine pair engaged in heme-iron coordination, our theoretical analysis showed that histidine-methionine pair may be commonly used by HmuY homologs from other Porphyromonas species. This feature allows HmuYPe and possibly other HmuY homologs for better heme sequestration from other hemophore-like proteins that use two methionines to coordinate heme-iron, for example from T. forsythia HmuYTf, allowing P. endodontalis and other Porphyromonas species to compete for heme source. However, HmuYPe is unable to capture heme bound to HmuYPg, which confirms the predominance of P. gingivalis in heme acquisition in polymicrobial consortia.
Identification and characterization of bacterial antigens expressed by the members of the Bacteroidota phylum allows not only for improved knowledge of pathogens’ phenotypes and their pathogenicity but also for the development of diagnostic and therapeutic strategies. One of the targets in such methods can be the host’s immune response toward components of bacterial heme acquisition systems, including HmuY proteins. The amino acid residues that form the core of the protein structure in HmuY family members are most conservatively preserved, whereas the greatest variability of epitopes exists on the surface of HmuY proteins (Śmiga et al., 2023a). Previously, in patients with periodontitis, we reported higher serum levels of antibodies directed against total P. gingivalis antigens and HmuYPg protein, as well as HmuY homolog from P. intermedia (HmuYPi-2) (Trindade et al., 2012; Śmiga et al., 2023a). Others demonstrated that in patients with apical periodontitis, P. gingivalis and P. endodontalis correlated with higher serum levels of IgG antibodies directed toward both bacterial species (Flynn et al., 2012). Importantly, we showed that there is no cross-reactivity between P. gingivalis HmuYPg and T. forsythia HmuYTf (Śmiga et al., 2015; Śmiga et al., 2023a) and that IgG antibodies raised toward these proteins did not recognize HmuYPe (this study). This suggests that HmuY proteins produced by oral pathogens differ in their epitopes sufficiently to be considered candidates for the development of diagnostic methods or biological markers to monitor polymicrobial diseases.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
MŚ Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Project administration, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. TO: Conceptualization, Formal Analysis, Funding acquisition, Validation, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by grant number 2023/49/B/NZ6/00129 (to TO) from the National Science Center (NCN, Narodowe Centrum Nauki, Kraków, Poland).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1421018/full#supplementary-material
Acuna-Amador, L., Barloy-Hubler, F. (2020). Porphyromonas spp. have an extensive host range in ill and healthy individuals and an unexpected environmental distribution: A systematic review and meta-analysis. Anaerobe. 66, 102280. doi: 10.1016/j.anaerobe.2020.102280
Alvarez, G., Arredondo, A., Isabal, S., Teughels, W., Laleman, I., Contreras, J. M., et al. (2023). Association of nine pathobionts with periodontitis in four South American and European countries. J. Oral. Microbiol. 15, 2188630. doi: 10.1080/20002297.2023.2188630
Antonyuk, S. V., Sieminska, K., Smiga, M., Strange, R. W., Wagner, M., Barnett, K. J., et al. (2023). Bacteroides fragilis expresses three proteins similar to Porphyromonas gingivalis HmuY: hemophore-like proteins differentially evolved to participate in heme acquisition in oral and gut microbiomes. FASEB J. 37, e22981. doi: 10.1096/fj.202300366R
Bielecki, M., Antonyuk, S., Strange, R. W., Sieminska, K., Smalley, J. W., Mackiewicz, P., et al. (2020). Prevotella intermedia produces two proteins homologous to Porphyromonas gingivalis HmuY but with different heme coordination mode. Biochem. J. 477, 381–405. doi: 10.1042/BCJ20190607
Bielecki, M., Antonyuk, S., Strange, R. W., Smalley, J. W., Mackiewicz, P., Smiga, M., et al. (2018). Tannerella forsythia Tfo belongs to Porphyromonas gingivalis HmuY-like family of proteins but differs in heme-binding properties. Biosci. Rep. 38, BSR20181325. doi: 10.1042/BSR20181325
Bik, E. M., Long, C. D., Armitage, G. C., Loomer, P., Emerson, J., Mongodin, E. F., et al. (2010). Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 4, 962–974. doi: 10.1038/ismej.2010.30
Byrne, D. P., Potempa, J., Olczak, T., Smalley, J. W. (2013). Evidence of mutualism between two periodontal pathogens: co-operative haem acquisition by the HmuY hemophore of Porphyromonas gingivalis and the cysteine protease interpain A (InpA) of Prevotella intermedia. Mol. Oral. Microbiol. 28, 219–229. doi: 10.1111/omi.12018
Cao, Y., Nicoletti, F. P., De Sanctis, G., Bocedi, A., Ciaccio, C., Gullota, F., et al. (2012). Evidence for pH-dependent multiple conformers in iron(II) heme-human serum albumin: spectroscopic and kinetic investigation of carbon monoxide binding. J. Biol. Inorg Chem. 17, 133–147. doi: 10.1007/s00775-011-0837-0
Carlsson, J., Hofling, J. F., Sundqvist, G. K. (1984). Degradation of albumin, hemopexin, haptoglobin and transferrin, by black-pigmented Bacteroides species. J. Med. Microbiol. 18, 39–46. doi: 10.1099/00222615-18-1-39
Chang, Y. C., Lai, C. C., Yang, S. F., Chan, Y., Hsieh, Y. S. (2002). Stimulation of matrix metalloproteinases by black-pigmented Bacteroides in human pulp and periodontal ligament cell cultures. J. Endod. 28, 90–93. doi: 10.1097/00004770-200202000-00010
Darveau, R. P., Hajishengallis, G., Curtis, M. A. (2012). Porphyromonas gingivalis as a potential community activist for disease. J. Dent. Res. 91, 816–820. doi: 10.1177/0022034512453589
Deng, Z. L., Szafranski, S. P., Jarek, M., Bhuju, S., Wagner-Dobler, I. (2017). Dysbiosis in chronic periodontitis: Key microbial players and interactions with the human host. Sci. Rep. 7, 3703. doi: 10.1038/s41598–017-03804–8
Deng, Z. L., Sztajer, H., Jarek, M., Bhuju, S., Wagner-Dobler, I. (2018). Worlds apart – transcriptome profiles of key oral microbes in the periodontal pocket compared to single laboratory culture reflect synergistic interactions. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00124
Eakanunkul, S., Lukat-Rodgers, G. S., Sumithran, S., Ghosh, A., Rodgers, K. R., Dawson, J. H., et al. (2005). Characterization of the periplasmic heme-binding protein ShuT from the heme uptake system of Shigella dysenteriae. Biochemistry. 44, 13179–13191. doi: 10.1021/bi050422r
Finegold, S. M., Vaisanen, M. L., Rautio, M., Eerola, E., Summanen, P., Molitoris, D., et al. (2004). Porphyromonas uenonis sp. nov., a pathogen for humans distinct from P. asaccharolytica and P. endodontalis. J. Clin. Microbiol. 42, 5298–5301. doi: 10.1128/JCM.42.11.5298-5301.2004
Flynn, T. R., Paster, B. J., Stokes, L. N., Susarla, S. M., Shanti, R. M. (2012). Molecular methods for diagnosis of odontogenic infections. J. Oral. Maxillofac. Surg. 70, 1854–1859. doi: 10.1016/j.joms.2011.09.009
Gibson, F. C., 3rd, Genco, C. A. (2006). “The genus porphyromonas,” in The prokaryotes: A handbook on the biology of bacteria, 3rd edition, vol. 7 . Eds. Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K. H., Stackebrandt, E. (Springer, New York), 428–454. doi: 10.1007/0–387-30747–8_15
Gomes, B. P., Jacinto, R. C., Pinheiro, E. T., Sousa, E. L., Zaia, A. A., Ferraz, C. C. R., et al. (2005). Porphyromonas gingivalis, Porphyromonas endodontalis, Prevotella intermedia and Prevotella nigrescens in endodontic lesions detected by culture and by PCR. Oral. Microbiol. Immunol. 20, 211–215. doi: 10.1111/j.1399-302X.2005.00214.x
Grenier, D., Mayrand, D. (1986). Nutritional relationships between oral bacteria. Infect. Immun. 53, 616–620. doi: 10.1128/iai.53.3.616-620.1986
Guilloux, C. A., Lamoureux, C., Beauruelle, C., Hery-Arnaud, G. (2021). Porphyromonas: A neglected potential key genus in human microbiomes. Anaerobe. 68, 102230. doi: 10.1016/j.anaerobe.2020.102230
Hajishengallis, G., Diaz, P. I. (2020). Porphyromonas gingivalis: Immune subversion activities and role in periodontal dysbiosis. Curr. Oral. Health Rep. 7, 12–21. doi: 10.1007/s40496-020-00249-3
Hajishengallis, G., Lamont, R. J. (2012). Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol. Oral. Microbiol. 27, 409–419. doi: 10.1111/j.2041-1014.2012.00663.x
Jansen, H. J., van der Hoeven, J. S., van den Kieboom, C. W., Goertz, J. H., Camp, P. J., Bakkeren, J. A. (1994). Degradation of immunoglobulin G by periodontal bacteria. Oral. Microbiol. Immunol. 9, 345–351. doi: 10.1111/j.1399-302X.1994.tb00284.x
Jimenez, C., Garrido, M., Pussinen, P., Bordagaray, M. J., Fernandez, A., Vega, C., et al. (2022). Systemic burden and cardiovascular risk to Porphyromonas species in apical periodontitis. Clin. Oral. Invest. 26, 993–1001. doi: 10.1007/s00784-021-04083-4
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature. 596, 583–589. doi: 10.1038/s41586-021-03819-2
Kilian, M. (1981). Degradation of immunoglobulins A1, A2 and G by suspected principal periodontal pathogens. Infect. Immun. 34, 757–765. doi: 10.1128/iai.34.3.757-765.1981
Kumar, P. S., Griffen, A. L., Barton, J. A., Paster, B. J., Moeschberger, M. L., Leys, E. J. (2003). New bacterial species associated with chronic periodontitis. J. Dent. Res. 82, 338–344. doi: 10.1177/154405910308200503
Li, H., Ji, H., Wu, S. S., Hou, B. X. (2016). Analysis of virulence factors of Porphyromonas endodontalis based on comparative proteomics technique. Zhonghua Kou Qiang Yi Xue Za Zhi. 51, 746–752. doi: 10.3760/cma.j.issn.1002–0098.2016.12.009
Lombardo Bedran, T. B., Marcantonio, R. A. C., Spin Neto, R., Alves Mayer, M. P., Grenier, D., Spolidorio, L. C., et al. (2012). Porphyromonas endodontalis in chronic periodontitis: a clinical and microbiological cross-sectional study. J. Oral. Microbiol. 4, 10123. doi: 10.3402/jom.v4i0.10123
Lourenco, T. G. B., Heller, D., Silva-Boghossian, C. M., Cotton, S. L., Paster, B. J., Colombo, A. P. V. (2014). Microbial signature profiles of periodontally healthy and diseased patients. J. Clin. Periodontol. 41, 1027–1036. doi: 10.1111/jcpe.12302
Madeira, F., Pearce, M., Tivey, A. R., Basutkar, P., Lee, J., Edbali, O., et al. (2022). Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 50, W276–W279. doi: 10.1093/nar/gkac240
Mark Welch, J. L., Rossetti, B. J., Rieken, C. W., Dewhirst, F. E., Borisy, G. G. (2016). Biogeography of a human oral microbiome at the micron scale. Proc. Natl. Acad. Sci. U.S.A. 113, E791–E800. doi: 10.1073/pnas.1522149113
Mei, F., Xie, M., Huang, X., Long, Y., Lu, X., Wang, X., et al. (2020). Porphyromonas gingivalis and its sytemic impact. Current status. Pathogens. 9, 944. doi: 10.3390/pathogens9110944
Morales-Olavarria, M., Nunez-Belmar, J., Gonzalez, D., Vicencio, E., Rivas-Pardo, J. A., Cortez, C., et al. (2023). Phylogenomic analysis of the Porphyromonas gingivalis - Porphyromonas gulae duo: approaches to the origin of periodontitis. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1226166
Na, H. S., Kim, S. Y., Han, H., Kim, H. J., Lee, J. Y., Lee, J. H., et al. (2020). Identification of potential oral microbial biomarkers for the diagnosis of periodontitis. J. Clin. Med. 9, 1549. doi: 10.3390/jcm9051549
Nazir, M. A. (2017). Prevalence of periodontal disease, its association with systemic diseases and prevention. Int. J. Health Sci. (Qassim). 11, 72–80.
Nishimata, H., Ohara-Nemoto, Y., Baba, T. T., Hoshino, T., Fujiwara, T., Shimoyama, Y., et al. (2014). Identification of dipeptidyl-peptidase (DPP)5 and DPP7 in Porphyromonas endodontalis, distinct from those in. Porphyromonas gingivalis PloS One 9, e114221. doi: 10.1371/journal.pone.0114221
Odell, L. J., Baumgartner, J. C., Xia, T., David, L. L. (1999). Survey for collagenase gene prtC in Porphyromonas gingivalis and Porphyromonas endodontalis isolated from endodontic infections. J. Endod. 25, 555–558. doi: 10.1016/S0099-2399(99)80379-3
Olczak, T., Smiga, M., Antonyuk, S. V., Smalley, J. W. (2024). Hemophore-like proteins of the HmuY family in the oral and gut microbiome: unraveling the mystery of their evolution. Microbiol. Mol. Biol. Rev. 88, e0013123. doi: 10.1128/mmbr.00131-23
Olczak, T., Wojtowicz, H., Ciuraszkiewicz, J., Olczak, M. (2010). Species specificity, surface exposure, protein expression, immunogenicity, and participation in biofilm formation of Porphyromonas gingivalis HmuY. BMC Microbiol. 10, 134. doi: 10.1186/1471-2180-10-134
Paster, B. J., Dewhirst, F. E., Olsen, I., Fraser, G. J. (1994). Phylogeny of Bacteroides, Prevotella, and Porphyromonas spp. and related bacteria. J. Bacteriol. 176, 725–732. doi: 10.1128/jb.176.3.725-732.1994
Pereira, C. V., Stipp, R. N., Fonseca, D. C., Pereira, L. J., Hofling, J. F. (2011). Detection and clonal analysis of anaerobic bacteria associated to endodontic-periodontal lesions. J. Periodontol. 82, 1767–1775. doi: 10.1902/jop.2011.110063
Perez-Chaparro, P., Goncalves, C., Figueiredo, L. C., Faveri, M., Lobao, E., Tamashiro, N., et al. (2014). Newly identified pathogens associated with periodontitis. J. Dent. Res. 93, 846–858. doi: 10.1177/0022034514542468
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 5, 1605–1612. doi: 10.1002/jcc.20084
Pinto, K. P., Barbosa, A. F. A., Silva, E. J. N. L., Santos, A. P. P., Sassone, L. M. (2023). What is the microbial profile in persistent endodontic infections? A scoping review. J. Endod. 49, 786–798.e7. doi: 10.1016/j.joen.2023.05.010
Rocha, E. R., Bergonia, H. A., Gerdes, S., Smith, C. J. (2019). Bacteroides fragilis requires the ferrous-iron transporter FeoAB and the CobN-like proteins BtuS1 and BtuS2 for assimilation of iron released from heme. Microbiologyopen. 8, e00669. doi: 10.1002/mbo3.669
Rosen, G., Shoshani, M., Naor, R., Sela, M. N. (2001). The purification and characterization of an 88-kDa Porphyromonas endodontalis 35406 protease. Oral. Microbiol. Immunol. 16, 326–331. doi: 10.1034/j.1399-302X.2001.160602.x
Sakamoto, M., Ohkuma, M. (2013). Porphyromonas crevioricanis is an earlier heterotypic synonym of Porphyromonas cansulci and has priority. Int. J. Syst. Evol. Microbiol. 63, 454–457. doi: 10.1099/ijs.0.042531-0
Sieminska, K., Cierpisz, P., Smiga, M., Olczak, T. (2021). Porphyromonas gingivalis HmuY and Bacteroides vulgatus Bvu – a novel competitive heme acquisition strategy. Int. J. Mol. Sci. 22, 2237. doi: 10.3390/ijms22052237
Siqueira, J. F., Jr, Rocas, I. N. (2009). Diversity of endodontic microbiota revisited. J. Dent. Res. 88, 969–981. doi: 10.1177/0022034509346549
Slezak, P., Smiga, M., Smalley, J. W., Sieminska, K., Olczak, T. (2020). Porphyromonas gingivalis HmuY and Streptococcus gordonii GAPDH—novel heme acquisition strategy in the oral microbiome. Int. J. Mol. Sci. 21, 4150. doi: 10.3390/ijms21114150
Smalley, J. W., Birss, A. J., Szmigielski, B., Potempa, J. (2007). Sequential action of R and K-specific gingipains of Porphyromonas gingivalis in the generation of the haem containing pigment from oxyhaemoglobin. Arch. Biochem. Biophys. 465, 44–49. doi: 10.1016/j.abb.2007.05.011
Smalley, J. W., Birss, A. J., Szmigielski, B., Potempa, J. (2008). Mechanism of methaemoglobin breakdown by the lysine-specific gingipain of the periodontal pathogen Porphyromonas gingivalis. Biol. Chem. 389, 1235–1238. doi: 10.1515/BC.2008.140
Smalley, J. W., Byrne, D. P., Birss, A. J., Wojtowicz, H., Sroka, A., Potempa, J., et al. (2011). HmuY haemophore and gingipain proteases constitute a unique syntrophic system of haem acquisition by Porphyromonas gingivalis. PloS One 6, e17182. doi: 10.1371/annotation/8658727f-cc10-47ba-9c7e-8726f1f94b91
Smalley, J. W., Olczak, T. (2017). Heme acquisition mechanisms of Porphyromonas gingivalis - strategies used in a polymicrobial community in a heme-limited host environment. Mol. Oral. Microbiol. 32, 1–23. doi: 10.1111/omi.12149
Smalley, J. W., Silver, J., Marsh, P. J., Birss, A. J. (1998). The periodontopathogen Porphyromonas gingivalis binds iron protoporphyrin IX in the mu-oxo dimeric form: an oxidative buffer and possible pathogenic mechanism. Biochem. J. 331, 681–685. doi: 10.1042/bj3310681
Śmiga, M., Bielecki, M., Olczak, M., Smalley, J. W., Olczak, T. (2015). Anti-HmuY antibodies specifically recognize Porphyromonas gingivalis HmuY protein but not homologous proteins in other periodontopathogens. PloS One 10, e0117508. doi: 10.1371/journal.pone.0117508
Śmiga, M., Sieminska, K., Trindade, S. C., Gomes-Filho, I. S., Nobre dos Santos, E. K., Olczak, T. (2023a). Hemophore-like proteins produced by periodontopathogens are recognized by the host immune system and react differentially with IgG antibodies. J. Oral. Microbiol. 15, 2214455. doi: 10.1080/20002297.2023.2214455
Śmiga, M., Slezak, P., Olczak, T. (2024). Comparative analysis of Porphyromonas gingivalis A7436 and ATCC 33277 strains reveals differences in the expression of heme acquisition systems. Microbiol. Spectr. 12, e0286523. doi: 10.1128/spectrum.02865-23
Śmiga, M., Slezak, P., Wagner, M., Olczak, T. (2023b). Interplay between Porphyromonas gingivalis hemophore-like protein HmuY and Kgp/RgpA gingipains plays a superior role in heme supply. Microbiol. Spectr. 11, e0459322. doi: 10.1128/spectrum.04593-22
Śmiga, M., Smalley, J. W., Slezak, P., Brown, J. L., Sieminska, K., Jenkins, R. E., et al. (2021). Glycation of host proteins increases pathogenic potential of Porphyromonas gingivalis. Int. J. Mol. Sci. 22, 12084. doi: 10.3390/ijms222112084
Sorsa, T., Ingman, T., Suomalainen, K., Haapasalo, M., Konttinen, Y. T., Lindy, O., et al. (1992). Identification of proteases from periodontopathogenic bacteria as activators of latent human neutrophil and fibroblast-type interstitial collagenases. Infect. Immun. 60, 4491–4495. doi: 10.1128/iai.60.11.4491-4495.1992
Stothard, P. (2000). The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques. 28, 1102, 1104. doi: 10.2144/00286ir01
Summanen, P., Finegold, S. M., Summanen, P., Finegold, S. M. (2015). Bergey’s manual of systematics of archaea and bacteria (Hoboken, NJ, USA: John Wiley & Sons, Ltd), 1–14. doi: 10.1002/9781118960608.gbm00246
Sundqvist, G. (1992). Associations between microbial species in dental root canal infections. Oral. Microbiol. Immunol. 7, 257–262. doi: 10.1111/j.1399-302X.1992.tb00584.x
Sundqvist, G., Eckerbom, M. I., Larsson, A. P., Sjogren, U. (1979). Capacity of anaerobic bacteria from necrotic dental pulps to induce purulent infections. Infect. Immun. 25, 685–693. doi: 10.1128/iai.25.2.685-693.1979
Sundqvist, G., Johansson, E., Sjogren, U. (1989). Prevalence of black-pigmented Bacteroides species in root canal infections. J. Endod. 15, 13–19. doi: 10.1016/S0099-2399(89)80092-5
Tran, T., Flynn, M. J., Chen, C., Slots, J. (1997). Porphyromonas endodontalis in subgingival plaque. Clin. Infect. Dis. 25, S222–S223. doi: 10.1086/516232
Trindade, S. C., Olczak, T., Gomes-Filho, I. S., Moura-Costa, L. F., Cerqueira, E. M. M., Galdino-Neto, M., et al. (2012). Induction of interleukin (IL)-1β, IL-10, IL-8 and immunoglobulin G by Porphyromonas gingivalis in humans. J. Periodontal Res. 47, 27–32. doi: 10.1111/j.1600-0765.2011.01401.x
van Steenbergen, T. J. M., van Winkelhoff, A. J., Mayrand, D., Grenier, D., de Graaff, J. (1984). Bacteroides endodontalis sp. nov., as an asaccharolytic black-pigmented Bacteroides species from infected dental root canals. Int. J. Syst. Bacteriol. 34, 118–120. doi: 10.1099/00207713-34-2-118
van Winkelhoff, A. J., Carlee, C. W., de Graaff, J. (1985a). B. endodontalis and other black-pigmented Bacteroides species in odontogenic abscesses. Infect. Immun. 49, 494–497. doi: 10.1128/iai.49.3.494-497.1985
van Winkelhoff, A. J., Kippuw, N., de Graaff, J. (1985b). Further characterization of Bacteroides endodontalis, an asaccharolytic black-pigmented Bacteroides species from the oral S cavity. J. Clin. Microbiol. 22, 75–79. doi: 10.1128/jcm.22.1.75-79.1985
van Winkelhoff, A. J., Kippuw, N., de Graaff, J. (1987). Cross-inhibition between black-pigmented Bacteroides species. J. Dent. Res. 66, 1663–1667. doi: 10.1177/00220345870660111201
van Winkelhoff, A. J., van Steenbergen, T. J. M., de Graaff, J. (1986). Oxygen tolerance of oral and non-oral black-pigmented Bacteroides species. FEMS Microbiol. Lett. 33, 215–218. doi: 10.1111/fml.1986.33.issue-2-3
Varadi, M., Anyango, S., Deshpande, M., Nair, S., Natassia, C., Yordanova, G., et al. (2022). AlphaFold Protein Structure Database: massively expanding the structural coverage of protein sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444. doi: 10.1093/nar/gkab1061
Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M., Barton, G. J. (2009). Jalview Version 2 - a multiple sequence alignment editor and analysis workbench. Bioinformatics. 25, 1189–1191. doi: 10.1093/bioinformatics/btp033
Wojtowicz, H., Guevara, T., Tallant, C., Olczak, M., Sroka, A., Potempa, J., et al. (2009a). Unique structure and stability of HmuY, a novel heme-binding protein of Porphyromonas gingivalis. PloS Pathog. 5, e1000419. doi: 10.1371/journal.ppat.1000419
Wojtowicz, H., Wojaczynski, J., Olczak, M., Kroliczewski, J., Latos-Grazynski, L., Olczak, T. (2009b). Heme environment in HmuY, the heme-binding protein of Porphyromonas gingivalis. Biochem. Biophys. Res. Commun. 383, 178–182. doi: 10.1016/j.bbrc.2009.03.148
Ximenez-Fyvie, L. A., Haffajee, A. D., Socransky, S. S. (2000). Comparison of the microbiota of supra- and subgingival plaque in health and periodontitis. J. Clin. Periodontol. 27, 649–657. doi: 10.1034/j.1600–051x.2000.027009648.x
Zamora-Cintas, M., Marin, M., Quiroga, L., Martinez, A., Fernández-Chico, M. A., Bouza, E., et al. (2018). Identification of Porphyromonas isolates from clinical origin using MALDI-TOF mass spectrometry. Anaerobe. 54, 197–200. doi: 10.1016/j.anaerobe.2018.06.017
Zerr, M. A., Cox, C. D., Johnson, W. T., Drake, D. R. (1998). Effect of red blood cells on the growth of Porphyromonas endodontalis and microbial community development. Oral. Microbiol. Immunol. 13, 106–112. doi: 10.1111/j.1399-302X.1998.tb00720.x
Zerr, M. A., Cox, C. D., Johnson, W. T., Drake, D. R. (2000). Growth stimulation of Porphyromonas endodontalis by hemoglobin and protoporphyrin IX. Oral. Microbiol. Immunol. 15, 365–370. doi: 10.1034/j.1399-302x.2000.150605.x
Zerr, M., Drake, D., Johnson, W., Cox, C. D. (2001). Porphyromonas endodontalis binds, reduces and grows on human hemoglobin. Oral. Microbiol. Immunol. 16, 229–234. doi: 10.1034/j.1399-302X.2001.160406.x
Keywords: Porphyromonas endodontalis, Porphyromonas gingivalis, Tannerella forsythia, hemophore-like protein, HmuY, heme
Citation: Śmiga M and Olczak T (2024) Porphyromonas endodontalis HmuY differentially participates in heme acquisition compared to the Porphyromonas gingivalis and Tannerella forsythia hemophore-like proteins. Front. Cell. Infect. Microbiol. 14:1421018. doi: 10.3389/fcimb.2024.1421018
Received: 21 April 2024; Accepted: 03 June 2024;
Published: 13 June 2024.
Edited by:
Masae Kuboniwa, Osaka University, JapanReviewed by:
Keiji Nagano, Health Sciences University of Hokkaido, JapanCopyright © 2024 Śmiga and Olczak. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michał Śmiga, bWljaGFsLnNtaWdhQHV3ci5lZHUucGw=
†ORCID: Michał Śmiga, orcid.org/0000–0002-9236–4236
Teresa Olczak, orcid.org/0000–0002-6140–8144
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.