- Department of Comparative Biomedical Sciences, College of Veterinary Medicine, Mississippi State University, Mississippi State, MS, United States
Edwardsiella ictaluri is a Gram-negative, facultative intracellular bacterium that causes enteric septicemia in catfish (ESC). The RNA chaperone Hfq (host factor for phage Qβ replication) facilitates gene regulation via small RNAs (sRNAs) in various pathogenic bacteria. Despite its significance in other bacterial species, the role of hfq in E. ictaluri remains unexplored. This study aimed to elucidate the role of hfq in E. ictaluri by creating an hfq mutant (EiΔhfq) through in-frame gene deletion and characterization. Our findings revealed that the Hfq protein is highly conserved within the genus Edwardsiella. The deletion of hfq resulted in a significantly reduced growth rate during the late exponential phase. Additionally, EiΔhfq displayed a diminished capacity for biofilm formation and exhibited increased motility. Under acidic and oxidative stress conditions, EiΔhfq demonstrated impaired growth, and we observed elevated hfq expression when subjected to in vitro and in vivo stress conditions. EiΔhfq exhibited reduced survival within catfish peritoneal macrophages, although it had no discernible effect on the adherence and invasion of epithelial cells. The infection model revealed that hfq is needed for bacterial persistence in catfish, and its absence caused significant virulence attenuation in catfish. Finally, the EiΔhfq vaccination completely protected catfish against subsequent EiWT infection. In summary, these results underscore the pivotal role of hfq in E. ictaluri, affecting its growth, motility, biofilm formation, stress response, and virulence in macrophages and within catfish host.
1 Introduction
The intracellular Gram-negative bacterium Edwardsiella ictaluri is a leading cause of enteric septicemia in catfish (ESC), posing a significant economic threat to farm-raised catfish. E. ictaluri rapidly attaches to and penetrates the host mucosal membranes and can survive and replicate inside catfish neutrophils and macrophages (Ainsworth and Chen, 1990; Booth et al., 2009). Although certain virulence factors, such as flagella, outer membrane proteins, lipopolysaccharide, type III and VI secretion systems, and extracellular proteins, have been identified (Newton and Triche, 1993; Klesius and Sealey, 1996; Lawrence et al., 2001; Bader et al., 2004; Thune et al., 2007; Kalindamar et al., 2023), the full spectrum of E. ictaluri’s virulence mechanisms remains incompletely understood.
Challenges persist despite using antimicrobials, bacterins, and live attenuated vaccines (LAV) to control ESC. Feed-additive antibiotics like Aquaflor, Romet 30, and Terramycin face reduced efficacy due to the early-onset of reduced appetite in infected fish. Furthermore, their usage may contribute to antibiotic resistance in E. ictaluri strains (Tu et al., 2008; Erickson et al., 2024). Bacterins provide limited benefits, whereas LAVs can stimulate innate and cellular immunity (Ellis, 2001; Kordon et al., 2020, 2021). Thus, E. ictaluri LAVs provide adequate protection (Cooper et al., 1996; Lawrence et al., 1997; Klesius and Shoemaker, 1999; Thune et al., 1999; Wise et al., 2015). Our research group developed two novel E. ictaluri live attenuated strains (EiΔevpB and ESC-NDKL1), providing significant protection against ESC in fry and fingerling catfish (Nho et al., 2017; Abdelhamed et al., 2018).
Small RNAs (sRNAs) have been found in both prokaryotes and eukaryotes (Meibom et al., 2009), and they are involved in the regulation of metabolism and virulence mechanisms (Oliva et al., 2015). Bacterial sRNAs respond dynamically to environmental stress, modulating transcription, translation, and RNA degradation. sRNAs employ both cis- and trans-encoded base-pairing mechanisms. Cis-encoded sRNAs exhibit complete complementarity with their target mRNA, while trans-acting sRNAs can interact with multiple mRNA targets and often necessitate the involvement of the RNA chaperone protein Hfq. While Hfq is generally considered dispensable for cis-encoded sRNA regulation, exceptions exist, underscoring the intricate involvement of Hfq in the regulatory dynamics of both cis- and trans-encoded sRNAs (Ellis et al., 2015; Brantl and Muller, 2021).
In Gram-negative bacteria, Hfq is essential for sRNAs’ activity and/or stability (Soper et al., 2010; Thomason and Storz, 2010; Wilton et al., 2015). The hfq gene is prevalent in half of all sequenced bacterial species (Sun et al., 2002). Beyond its role in virulence mechanisms, hfq governs quorum sensing, stress resistance, and various cellular functions, including osmotic stress, ethanol response, temperature shifts, and iron starvation (Fantappiè et al., 2009; Liu et al., 2011). Its impact extends to cell membranes, type III secretion system (T3SS), flagella, fimbria, biofilms, and overall bacterial fitness (Sittka et al., 2007; Chao and Vogel, 2010; Rempe et al., 2012; Caldelari et al., 2013; Cui et al., 2013; Meng et al., 2013; Kakoschke et al., 2014).
Studies on intracellular bacterial pathogens have underscored hfq’s significance in bacterial virulence (Chao and Vogel, 2010; Feliciano et al., 2016). This RNA chaperone serves as a virulence factor in diverse pathogenic bacteria, including Brucella abortus, Legionella pneumophila, Salmonella Typhimurium, and Yersinia enterocolitica (Zeng et al., 2013; Hu et al., 2014). hfq mutants display attenuated virulence in various animal models. For example, the hfq mutant of S. Typhimurium was attenuated in mice and showed reduced survival in macrophages (Sittka et al., 2007). The hfq mutant of S. Enteritidis exhibits significantly reduced virulence in chickens (Meng et al., 2013). Furthermore, in Y. enterocolitica and B. melitensis, hfq affected the metabolism, stress response, and production of virulence factors (Cui et al., 2013; Kakoschke et al., 2014). Similarly, hfq played a crucial role in the virulence of Neisseria meningitides, as shown in ex vivo and in vivo infection models (Fantappiè et al., 2009). The hfq mutant was found to be a LAV candidate against B. melitensis infection (Zhang et al., 2013). In E. tarda, the hfq mutant was attenuated in both macrophages and fish (Hu et al., 2014).
Considering the well-established role of sRNAs in regulating bacterial virulence, investigating the bacterial RNA-binding protein Hfq offers an intriguing approach to unraveling its involvement in E. ictaluri virulence. Additionally, the resulting attenuated strains could serve as valuable tools for probing the catfish immune system, potentially offering protection against E. ictaluri infections.
2 Materials and methods
2.1 Bacteria, plasmids, and media
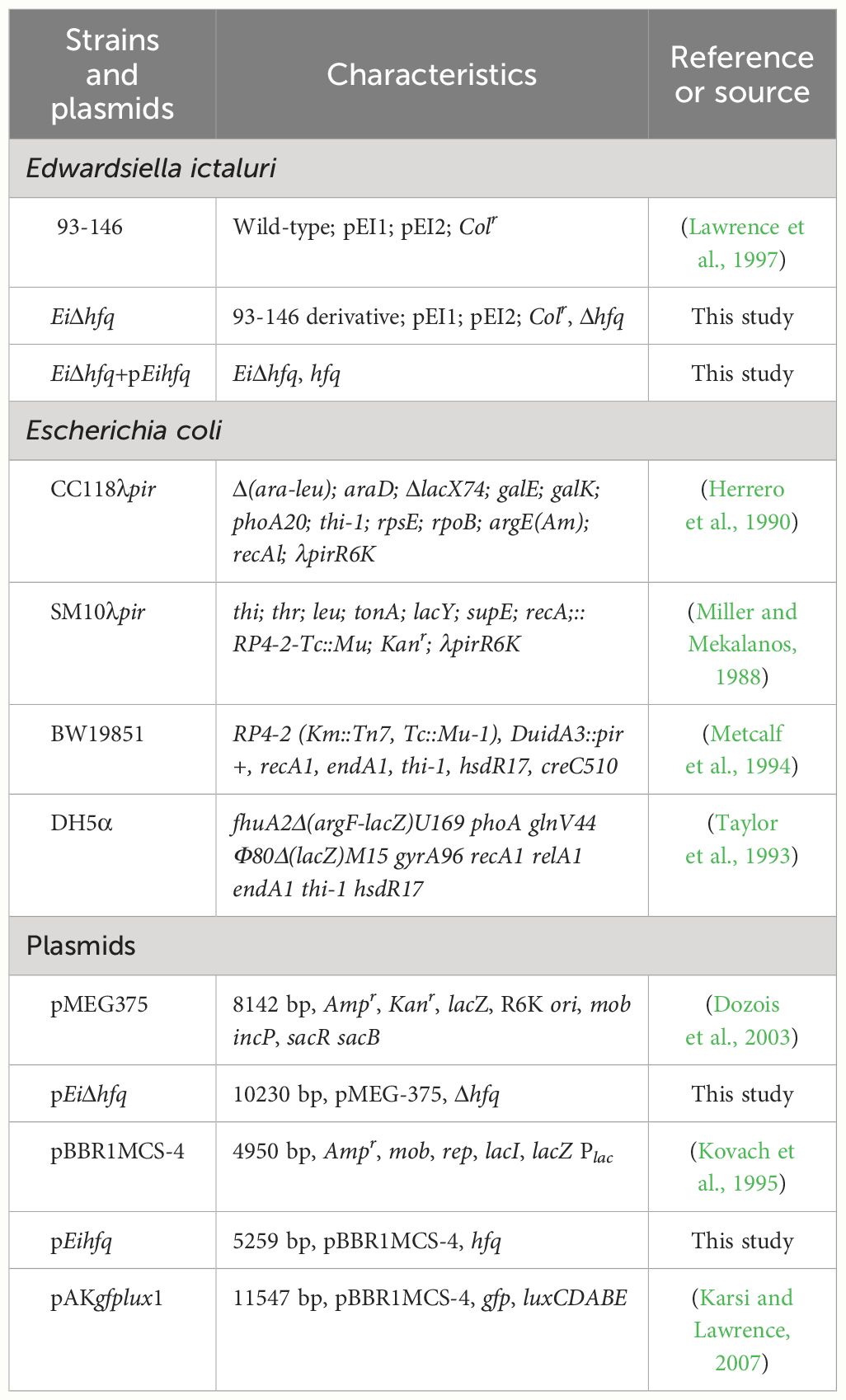
The bacterial strains and plasmids used in this study are detailed in Table 1. The wild-type E. ictaluri strain 93-146 (EiWT) was cultivated at 30°C in brain-heart infusion (BHI) broth for 16 h or on agar plates for 2 days. Escherichia coli CC118λpir and BW19851 strains were propagated in Lysogeny broth (LB) or on Lysogeny agar (LA) at 37°C for 16 h. Culture media were supplemented with ampicillin (100 mg/ml), colistin (12.5 mg/ml), sucrose (5%), and mannitol (0.35%) when required.
2.2 Comparative analysis of the Hfq protein sequences
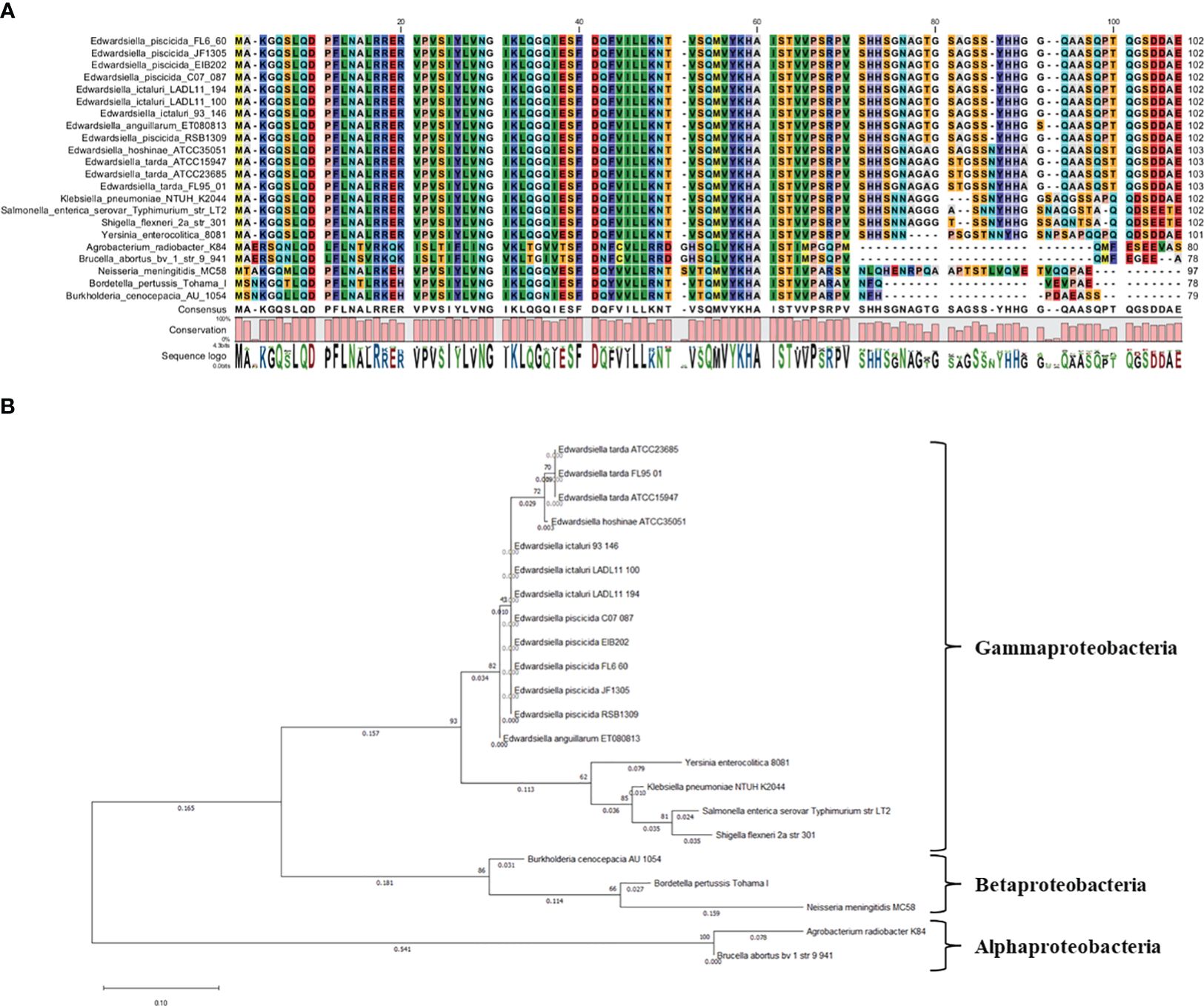
The comparative analysis involved aligning the Hfq protein sequence of E. ictaluri strain 93-146 with members of the Enterobacteriaceae family. The analysis included E. ictaluri strains LADL11-100 and LADL11-194, E. anguillarum ET080813, E. hoshinae ATCC 35051, E. piscicida strains FL6-60, C07-087, RSB1309, JF1305, and EIB202, as well as E. tarda strains ATCC 15947, ATCC 23685, and FL95-01. Members from other bacterial families, such as Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044, Salmonella enterica serovar Typhimurium str. LT2, Yersinia enterocolitica subsp. enterocolitica 8081, and Shigella flexneri 2a str. 301, were also included. Additionally, representatives from α–Proteobacteria (Brucella abortus bv.1 str. 9-941 and Agrobacterium radiobacter K84) and β–Proteobacteria (Bordetella pertussis Tohama I, Burkholderia cenocepacia AU 1054, and Neisseria meningitidis MC58) were utilized. The phylogenetic tree was constructed using MEGA v11.0 software, utilizing the maximum likelihood method with 500 bootstraps (Kumar et al., 2016). Hfq protein sequences were aligned using CLC Genomics Workbench 6.5.1 from CLC Bio.
2.3 In-frame deletion of the hfq gene
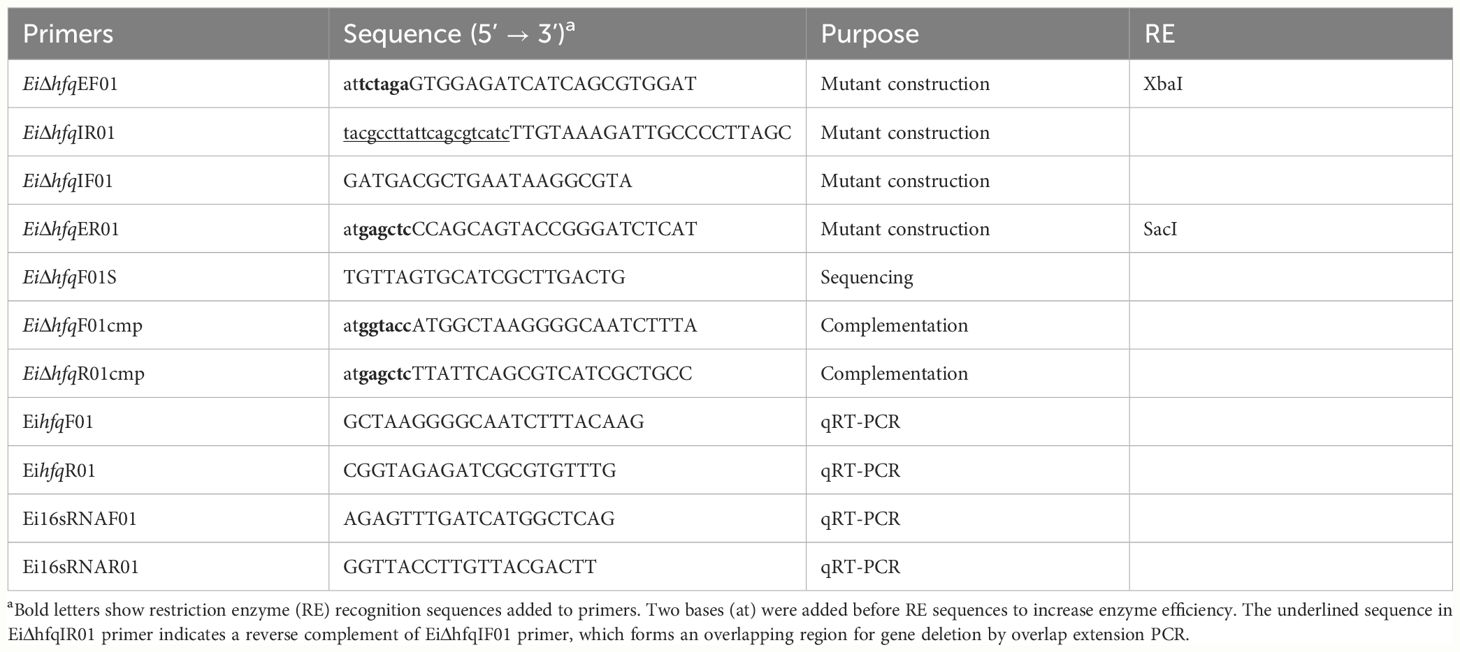
The nucleotide sequence otpf hfq (NT01EI_RS01835) was retrieved from the E. ictaluri strain 93-146 genome (GenBank accession: CP001600) (Williams et al., 2012), and the process of hfq mutant construction was illustrated in Figure 1. For the targeted in-frame deletion of the hfq gene, a set of two external and two internal primers was designed (Table 2). Genomic DNA from EiWT was extracted using the Wizard Genomic DNA Kit (Promega), and this isolated DNA served as the template for subsequent PCR amplifications.
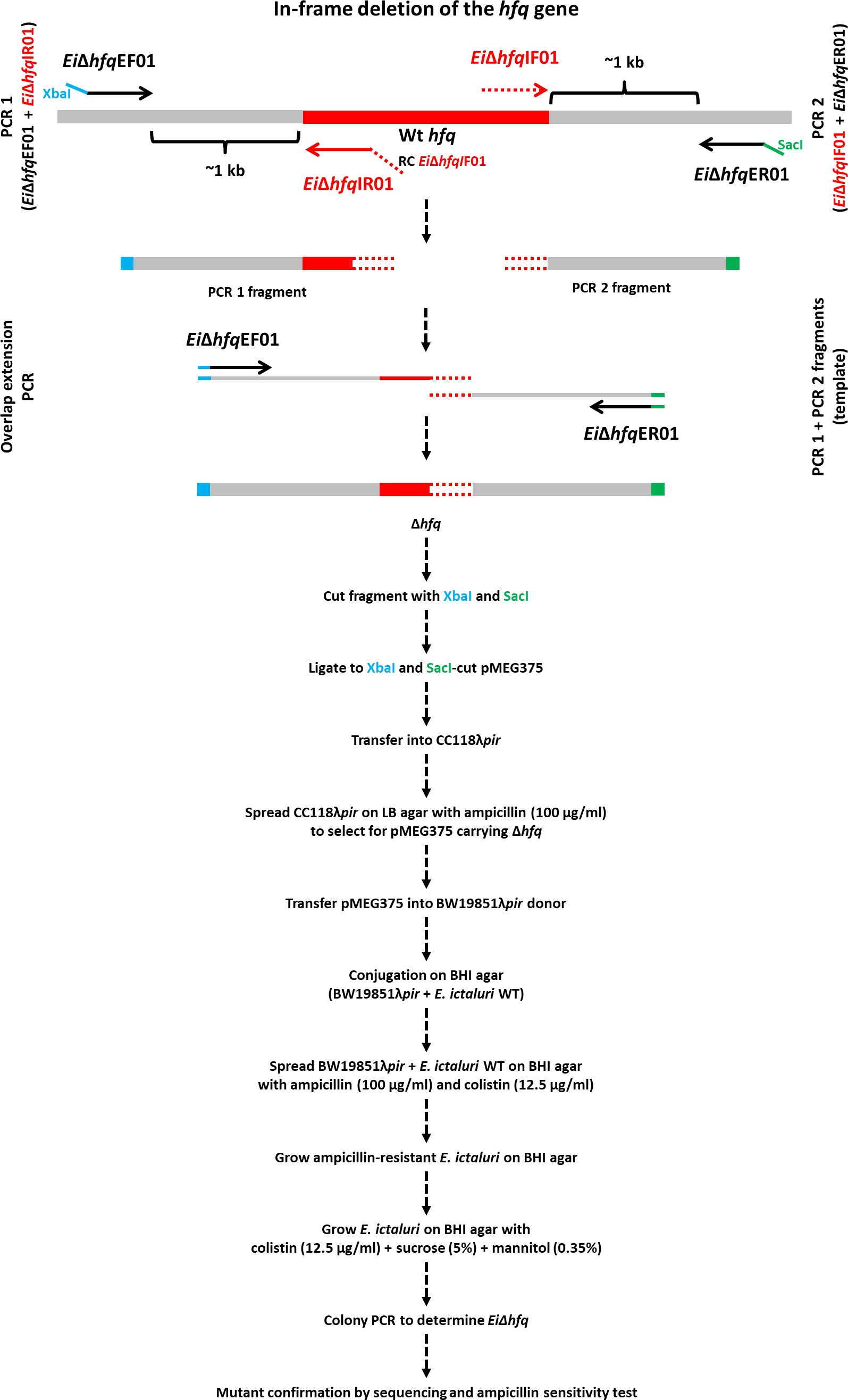
Figure 1 In-frame deletion of the hfq gene. The hfq gene is represented by a red band, and grey bands represent upstream and downstream regions. The blue and green tails at the end of EiΔhfqEF01 and EiΔhfqER01 primers indicate XbaI and SacI restriction enzymes, respectively. The red tail at the end of EiΔhfqIR01 primer represents the reverse complement of EiΔhfqIF01 primer. The overlapping region is shown in a red round dotted band.
The amplified upstream and downstream fragments underwent gel extraction using the QIAquick Gel Extraction Kit (Qiagen). These fragments were then mixed in equal proportions to serve as a template for the subsequent overlap extension PCR. The resulting in-frame deletion fragment was digested with XbaI and SacI restriction enzymes (New England Biolabs) and cleaned up.
The suicide plasmid pMEG375 was purified from an overnight E. coli culture by a QIAprep Spin Miniprep Kit (Qiagen) and cut with the same enzymes for cloning. The purified PCR product with in-frame deletion was ligated into the pMEG375 vector using T4 DNA Ligase (New England Biolabs) at 16°C overnight. E. coli CC118λpir was transformed by electroporation and plated on LB agar with ampicillin. Plasmid DNAs were prepared from the colonies, and cloning success was confirmed by plasmid size, cloned fragment size, and sequencing.
The correct plasmid, named pEiΔhfq, was transferred into E. coli BW19851λpir by chemical transformation and mobilized into EiWT by conjugation. After conjugation, plasmid integration was achieved by ampicillin selection. Then, ampicillin-resistant colonies were propagated on BHI agar to allow for the second allelic exchange. After this step, colonies were streaked on BHI plates with 5% sucrose, 0.35% mannitol, and colistin to select for loss of pMEG375. The final confirmation of plasmid loss was achieved by testing E. ictaluri colonies for ampicillin sensitivity. PCR and sequencing procedures validated the successful deletion of hfq in E. ictaluri, and the resultant mutant strain was designated as EiΔhfq.
2.4 Complementation of EiΔhfq
For complementation of EiΔhfq, a wild-type copy of the hfq gene was amplified from EiWT genomic DNA using the primers EihfqF01cmp and EihfqR01cmp, which contained KpnI and SacI restriction sites, respectively (Table 2). After digestion with both enzymes and clean-up, the amplified fragment was cloned into KpnI- and SacI-digested broad host range pBBR1MCS4 (Kovach et al., 1995). E. coli DH5α was transformed with 1 µl of the ligation mixture, and E. coli transformants containing pEihfq were identified by colony PCR, and the hfq sequence in pEihfq was confirmed by sequencing. Then, pEihfq was transformed into EiΔhfq by conjugation, and complemented colonies (EiΔhfq+pEihfq) were determined on BHI agar with ampicillin and colistin selection.
2.5 Construction of bioluminescent EiΔhfq
The pAKgfplux1 plasmid (Karsi and Lawrence, 2007) was transferred into EiΔhfq by conjugal mating to construct bioluminescent EiΔhfq. Briefly, E. coli SM10λpir carrying pAKgfplux1 was used as a donor strain to transfer the plasmid into recipient EiWT. Overnight cultures of donor and recipient were mixed at a 1:2 donor: recipient ratio. The mixture pellet was transferred onto sterile 0.45 µM filter paper on a BHI agar plate and incubated at 30°C for 24 h. Bacteria on filter paper were dissolved in BHI broth with ampicillin and colistin and then spread on BHI plates containing ampicillin and colistin. After incubation at 30°C for 24-48 h, ampicillin-resistant EiWT colonies carrying pAKgfplux1 appeared on plates, and their bioluminescence was verified by IVIS Lumina XRMS in Vivo Imaging System Series III (PerkinElmer).
2.6 Growth kinetics of EiΔhfq
EiWT, EiΔhfq, and complemented EiΔhfq strains were streaked on BHI agar containing colistin. A single colony from BHI agar was inoculated into 5 ml of BHI broth with colistin. When optical density at 600 nm (OD600) reached 0.5, bacterial culture was diluted 50 times with 50 ml of BHI broth with colistin in a 250 ml flask, which was incubated in a rotary shaker at 200 rpm at 30°C. OD600 values were measured for 60 h by sampling bacterial cultures every 6 h. At 12, 24, and 48 h, serially diluted cultures were spread on BHI agar for colony counting. Four replicates were used for each strain.
2.7 Biofilm formation
Biofilm formation in EiΔhfq, EiWT, and complement strains was assessed using a protocol described by other authors (Cai and Arias, 2017). Briefly, 10 μl of overnight cultures were inoculated in 90 μl BHI in flat-bottom 96-well microtiter plates, and cultures were grown without disruption at 30°C for 48 h to allow biofilm formation. Following removal of planktonic cells, the wells were stained with crystal violet (150 ul, 1%) for 20 min. Then, crystal violet was removed by washing, and the remaining dye was dissolved in ethanol (96%) and quantified by measuring the OD600 values.
2.8 Motility test and scanning electron microscopy
For motility assay, bacterial strains were grown in BHI broth until the late-log phase (OD600 = 1.0), then 1 μl culture was spotted onto BHI plates containing colistin, ampicillin, 1 mM arabinose, 0.3% agar (swimming motility plate), and 0.5% agar (swarming motility plate). The plates were incubated at 30°C for 24, 48, and 72 h to observe bacterial motility zones.
For SEM analysis, aliquots of a 2 mL suspension of logarithmic-phase bacteria were incubated on sterile poly-L-lysine-coated coverslips in a sterile polystyrene 6-well plate at 25°C for 24 h to allow adhesion to occur. After the 24-h incubation, non-adherent bacteria were removed by pipetting and washed with 3 mL of sterile distilled water. The cells were fixed with 0.5% Karnovsky’s fixative in 0.1 M sodium cacodylate buffer, pH 7.2, for a minimum of 24 h. Subsequently, the cells were rinsed, postfixed in 2% osmium tetroxide for 1 h, and then dehydrated with a series of ethanol dilutions (35%, 50%, 70%, 95%, and 100%) for 15 minutes each. The adhered cells were transferred to a graded mixture of hexamethyldisilazane (HMDS) and ethanol (25%, 50%, 75%), followed by 100% HMDS for 1 h. This was followed by overnight air-drying of the samples, which were immediately coated with 45 nm of platinum in an EMS-150T ES sputter coating operation and examined using a JEOL JSM 6500 scanning electron microscope.
2.9 Survival of EiΔhfq in acidic and oxidative stress
After overnight growth, OD600 values were measured to adjust culture volumes. Subsequently, 5 µl of bacteria were inoculated into 195 µl of BHI broth with ampicillin and colistin in 96-well black plates. The medium was modified to create acidic (pH=5.5) and oxidative stress conditions (3 mM H2O2). Untreated BHI was used as control. A Cytation 5 Cell Imaging Multimode Reader (BioTek) captured bioluminescence for 3 h at 30°C. Each strain had four replicates, and the experiment was repeated three times.
2.10 Expression of hfq under in vitro and in vivo stresses
A single EiWT colony was inoculated into 5 ml of BHI broth, followed by 16-18 h incubation at 30°C with shaking at 200 rpm. For each group, 40 ml of BHI broth was inoculated and grown until OD600 reached 0.4. Then, each culture was divided into four aliquots of 10 ml, and bacteria were harvested by centrifugation at 6,000 x g for 15 min. The supernatant was removed, and cells were resuspended in 10 ml of fresh BHI supplemented with 1.5 mM H2O2 (0.05%) and BHI broth acidified with a 6 N HCl (pH=4). Cultures were incubated by shaking at 180 rpm at 30°C for 30 min. The bacteria were harvested, the supernatant was removed, and RNAlater solution was added to the pellets, which were stored for a week at -20°C until RNA isolation.
For serum treatment, EiWT was exposed to naïve channel catfish serum, with heat-inactivated catfish serum used as a control. Each treatment comprised four biological replicates. E. ictaluri cultures underwent three washes using 1.25 ml of cell wash buffer (10 mM Tris-HCl and 5 mM magnesium acetate). Subsequently, normal and heat-inactivated serum (1.25 ml) was added to the E. ictaluri pellet, mixed, and incubated for 30 min at 30°C. Following incubation, the serum-bacteria mixture was used for total RNA isolation.
For in vivo stress, eighteen-month-old specific pathogen-free channel catfish fingerlings were stocked into two tanks at a rate of 6 fish/tank. After one week of acclimation, fish were anesthetized in water containing 100 mg/L MS-222 (Sigma) and injected with bioluminescent EiWT (approximately 1x104 CFU) in 100 µl PBS. Negative control was injected with 100 µl PBS. After 30 h of bioluminescent imaging, fish were euthanized with 400 mg/L MS-222 (Sigma), and head kidney, liver, and spleen were collected in tubes with RNA later solution.
2.11 Total RNA extraction, cDNA synthesis, and qRT-PCR
The extraction of total RNA was performed using the RNeasy Protect Bacteria Mini Kit (Qiagen), following the manufacturer’s guidelines. DNase treatment was carried out using the RNase-Free DNase Set (Qiagen) to eliminate potential DNA contamination. The quality and concentration of the isolated total RNA were evaluated using NanoDrop 1000 (Thermo Fisher). Subsequently, 1 µg of total RNA was converted to cDNA utilizing the Maxima First Strand cDNA Synthesis Kit for RT-qPCR (Thermo Fisher) following the user manual.
Quantitative real-time PCR (qRT-PCR) was performed using DyNAmo SYBR Green qPCR Kit (Thermo Fisher) and an Mx3005P qPCR System (Agilent). PCR reactions contained 10 µl SYBR Green 2X mix, 0.2 µM each of forward and reverse primers, and 1 µl of 100X diluted cDNA (Table 2). The PCR was set to initial denaturation at 95°C for 3 min, 45 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s, and a final extension at 72°C for 3 min. At the end of the PCR, a melting curve program was run from 60°C to 95°C with a 0.5°C increase every 15 s. 16S RNA gene was used as an internal control (Table 2). A sample from unstressed conditions was set as a calibrator in each experiment, except heat-treated serum was used as a calibrator against normal serum. Relative expression rates were calculated by the threshold cycle changes in the sample and calibrator. The ΔΔCt method was used to calculate the expression level of related genes (Livak and Schmittgen, 2001). All expression values were normalized against 16S rRNA. ΔΔCt was calculated by ΔΔCt = ΔCt (stress condition) - ΔCt (non-stress condition), where ΔCt is the normalized signal level in a sample (ΔCt = Ct of target gene – Ct of reference gene).
2.12 Survival in peritoneal macrophages
The bacterial killing assay was performed as previously described with minor modifications (Kalindamar et al., 2023). Briefly, four days after squalene injection, peritoneal macrophages were collected from five one-year-old channel catfish (250-300 g) by injecting 10 ml cold phosphate-buffered saline (PBS) to the peritoneal cavity and harvesting macrophages by using a three-way valve. Harvested cells were washed with PBS three times, and resuspended in channel catfish macrophage medium (CCMM), including RPMI [(RPMI 1640 sans phenol red & L-glutamine, Lonza) containing 1x glutamine substitute (GlutaMAX –I CTS, Gibco)], 15 mM HEPES buffer (GIBCO), in 0.18% sodium bicarbonate solution (GIBCO), 0.05 mM 2-beta-mercaptoethanol (all from Sigma), and 5% heat-inactivated (HI) channel catfish serum.
Peritoneal macrophages and bioluminescent EiWT and EiΔhfq were suspended in a 1:1 ratio and placed in a 96-well black plate as four replicates. A negative control group without bacteria was also included in the plate. Cells and bacteria were compacted by centrifugation of the plate at 1500 rpm for 5 min at 24°C. Then, the plate was incubated with CCMM for 1 h at 30°C and centrifuged at 2000 rpm for 7-10 min. After removing the supernatant, CCMM containing 100 µg/ml gentamicin was added to the plate, which was incubated for 1 h at 30°C to kill non-phagocyted bacteria. Extracellular bacteria were removed by washing the wells 3 times with PBS. Next, the cell-bacteria mixture was incubated with CCMM containing 10 µg/ml gentamicin for 48 h with 5% CO2 at 30°C, and Cytation 5 Cell Imaging Multimode Reader (BioTek) was used to acquire bioluminescence emitted by the surviving bacteria inside the catfish peritoneal macrophages.
2.13 Cytospin and light microscopy
After incubating the cell-bacteria mixture with CCMM containing 10 µg/ml gentamicin, as described above, cells were harvested at 4 h post-challenge and washed with PBS. Subsequently, cytospin preparations were applied at 500 rpm for 1 min using a Cyto-Tek centrifuge. Following this, all samples underwent fixation and staining with Wright’s stains (Hemacolor, Merck), following previously published procedures (do Vale et al., 2002). Finally, the samples were analyzed and photographed using an Olympus BX microscope (Olympus U-TV1 X) and Infinity software.
2.14 Attachment and invasion in epithelial cells
Attachment and invasion assay was performed as previously described with minor modifications (Kalindamar et al., 2020). Channel catfish ovary (CCO) cells were suspended in DMEM medium (Sigma) with 10% fetal bovine serum and 4mM L-glutamine at the final concentration of 1x107 cells ml-1 and placed in a 24-well plate. The exact number of bioluminescent EiWT and EiΔhfq were added to the plate to achieve 1:1 multiplicity of infection. Each plate contained four biological replicates and a negative control group without bacteria. CCO and bacteria suspension were incubated 1 h at 28°C to allow bacterial attachment to cells. Following the incubation, samples were washed three times with PBS. Then, DMEM medium, including 100 µg/ml gentamicin, was used to kill non-invasive EiWT and EiΔhfq. Finally, the plate was incubated for another h at 28°C to determine bacterial invasion. Bioluminescence was captured by IVIS Lumina XRMS in Vivo Imaging System Series III and quantified from images by Living Image Software v 4.2.
2.15 Bioluminescent imaging in live catfish
Eight specific-pathogen-free (SPF) channel catfish fingerlings obtained from the MSU-CVM Hatchery (12.72 ± 5.47 cm, 24.95 ± 1.00 g) were stocked into two 50-liter tanks (four fish each) and acclimated for one week at 28°C. After lowering the tank water level to 10 liters, 100 ml cultures of EiΔhfq (treatment) and EiWT (control) were added to each tank for the immersion challenge (final dose of 5x107 CFU/ml water). After 1 h, the water flow was restored. Infected catfish were anesthetized in water containing 100 mg/L MS222 (Sigma) and immediately placed into the photon collection chamber of IVIS Lumina XRMS in Vivo Imaging System Series III for image capture. The exposure time was set to one minute to collect photon emissions from the whole fish body. After imaging, fish were placed in well-aerated water for recovery. Bioluminescent imaging was conducted at 0, 6, 12, and 24 h post-infection and subsequent daily intervals until 336 h. Living Image Software v 4.2 was used to quantify bioluminescence from fish images.
2.16 Virulence and efficacy of EiΔhfq in catfish
Virulence/vaccination and efficacy experiments were conducted as described in our earlier work (Karsi et al., 2009). Briefly, 500 specific-pathogen-free (SPF) channel catfish fingerlings obtained from the MSU-CVM Hatchery (3.81 ± 0.80 cm, 10.544 ± 0.99 g) were stocked into 20 tanks at a rate of 25 fish/tank and acclimated for one week at 28°C. Chlorine, dissolved oxygen, and temperature were monitored daily. Tanks were randomly assigned to five groups, with four tanks in each: EiΔhfq (vaccination), EiΔhfq+pEihfq (complement), EiWT (positive control), BHI (sham control), and negative control. Immersion vaccination was applied by lowering the water in each tank to 10 liters and adding 100 ml of bacterial culture (final dose of 2.1x107 CFU/ml water). After 1 h, water flow (1-1/min) was restored to each tank. Mortalities were recorded daily for 21 days, and the percent mortalities were calculated for each group. To assess the EiΔhfq as a possible vaccine candidate, all fish that survived the EiΔhfq vaccination were re-challenged with EiWT (final dose of 2.8x107 CFU/ml water) 21 days post-vaccination as described above. Fish mortalities were recorded daily for 14 days.
2.17 Statistical analysis
Statistical analysis was conducted using one-way ANOVA and two-way ANOVA procedures with Tukey’s test in SAS for Windows 9.4 (SAS Institute, Inc., Cary, NC) to determine the significance of differences between means of treatments or groups. The predetermined significance level for all tests was set at p < 0.05. Photon emissions were log10 transformed to enhance normality, and a two-independent-samples t-test was employed for symmetrical variable comparison between EiΔhfq and EiWT.
3 Results
3.1 Hfq protein is conserved in the genus Edwardsiella
We identified a single functional hfq gene in the genome of E. ictaluri strain 93-146. The average size of the Hfq protein in the Edwardsiella genus is 102-103 amino acids (aa), while the Hfq protein’s average size in α- and β-Proteobacteria ranged between 78 and 97 aa (Figure 2A). Phylogenetic analysis revealed its conservation within Enterobacteriaceae, with a closer similarity to α-Proteobacteria than to β-Proteobacteria (Figure 2B).
3.2 EiΔhfq showed decreased growth and viability in the late exponential phase
To assess the influence of hfq on E. ictaluri growth, we cultured EiΔhfq, EiWT, and complement strains in broth and agar media. Although the initial growth stages showed comparable patterns across all three strains, a distinct decline in the growth of EiΔhfq became evident in the later stages (Figure 3A). Colony counts unveiled a significant reduction in the number of EiΔhfq colonies in comparison to EiWT and complemented strains (p < 0.05) (Figure 3B).
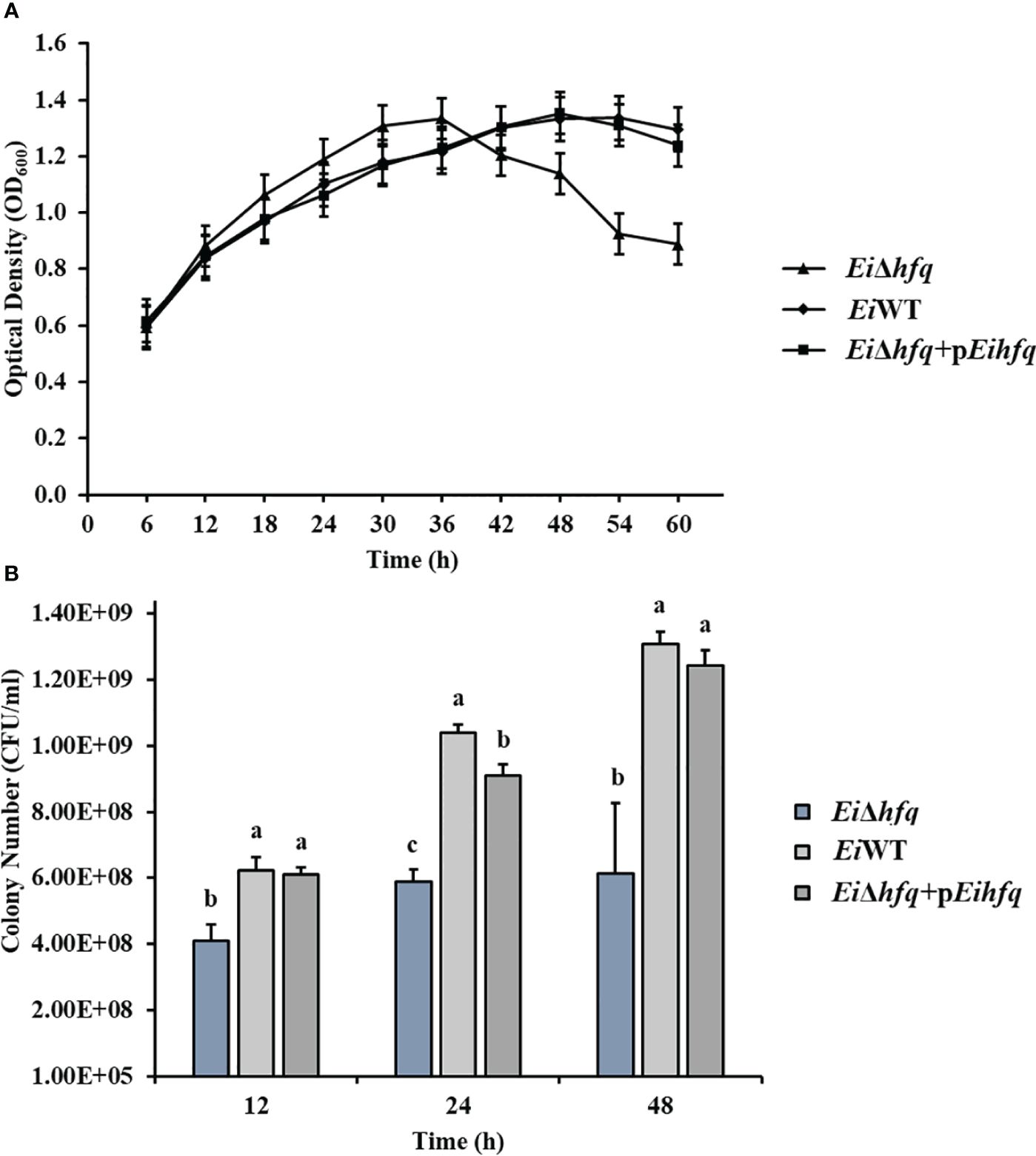
Figure 3 The growth kinetics of EiΔhfq, EiWT, and complemented (EiΔhfq+pEihfq) strains were evaluated under different conditions. (A) Optical density at 600 nm (OD600) was measured over 60 h at 30°C using a shaker incubator in Brain Heart Infusion (BHI) broth. The OD600 values provide insights into the bacterial growth trends over time. (B) Colony forming units per milliliter (CFU/ml) were determined during bacterial growth at 12, 24, and 48 h The cultures were incubated in a 30°C shaker incubator in BHI broth. Serial dilutions were performed, and colonies were counted after plating on BHI agar plates. This method allows for quantifying viable bacterial cells at different time points. Small letters indicate statistical differences between treatments (p < 0.05).
3.3 Deletion of hfq affects biofilm formation
EiΔhfq formed significantly less biofilm compared to EiWT and complement strains, while no significant differences were observed between EiWT and complement strains at 48 h (p < 0.05) (Figure 4).
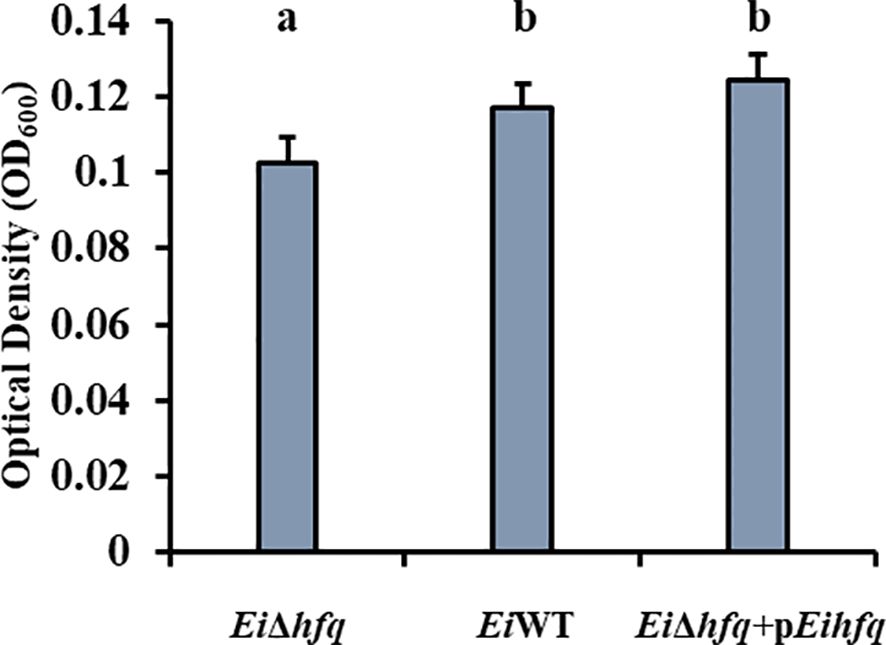
Figure 4 Biofilm formation of EiΔhfq, EiWT, and complement strains at 48 h. Small letters indicate statistical differences between treatments (p < 0.05).
3.4 Deletion of hfq affects motility with little effect on flagella
EiΔhfq exhibited swimming motility comparable to EiWT at 24 h. However, the motility of EiΔhfq was significantly higher than that of EiWT at 48 h and 72 h (Figure 5A). SEM imaging of EiΔhfq and EiWT revealed the presence of flagella in both strains. There were numerous thin and few dense lateral flagellar filaments in EiWT and EiΔhfq, respectively (Figure 5B).
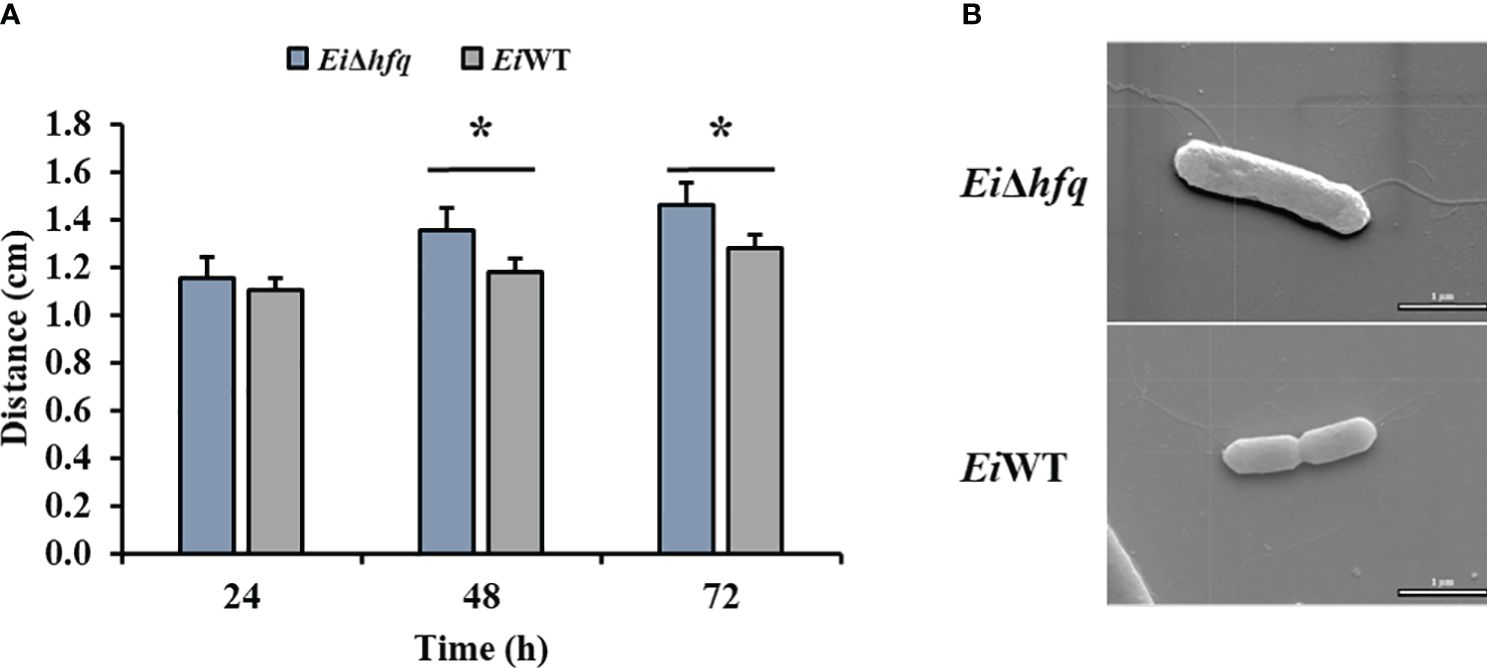
Figure 5 Motility and SEM. (A) The swimming zones of EiΔhfq and EiWT were determined at 24, 48, and 72 h by measuring diameters (cm). Statistical significance is shown by an asterisk (p < 0.05). (B) SEM photomicrograph of representaive EiΔhfq and EiWT strains with flagellar filaments (scale bar is 1 µm).
3.5 hfq is essential for survival under acidic and oxidative stresses
Exposure of EiΔhfq to acidic and oxidative stresses resulted in a three- to four-fold reduction in its bioluminescence compared to EiWT (Figure 6), indicating the mutant’s diminished capability to cope with these stresses.
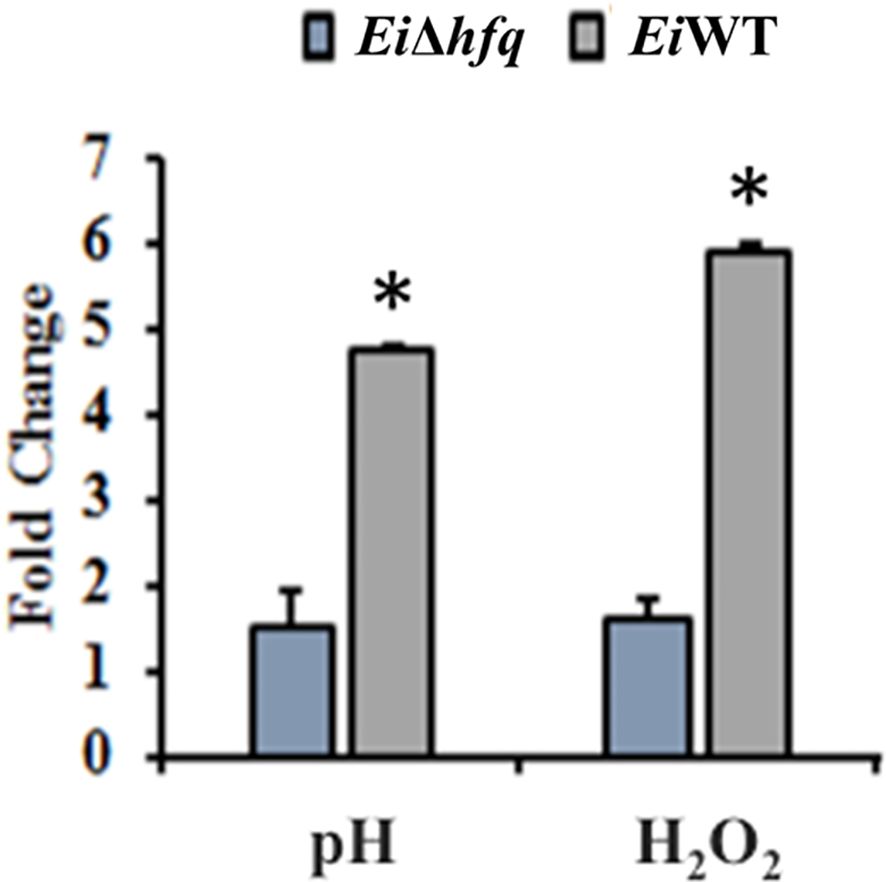
Figure 6 Fold change of bioluminescence in acidic (pH 5.5) and oxidative (3 mM H2O2) stresses. Untreated BHI was used as a control to calculate fold changes in each treatment. The (*) symbol indicates a significant difference between treatments (p < 0.01). The data are representative of three independent experiments.
3.6 The hfq gene is highly expressed under in vitro and in vivo stress conditions
The relative expression of hfq in EiWT increased significantly under in vitro stresses (pH 4 and 1.5 mM H2O2) (Figure 7A). EiWT demonstrated replication in fish over time (Figures 7B, C), and growth in the head kidney, spleen, and liver environments induced hfq expression, particularly in the spleen (Figure 7D).
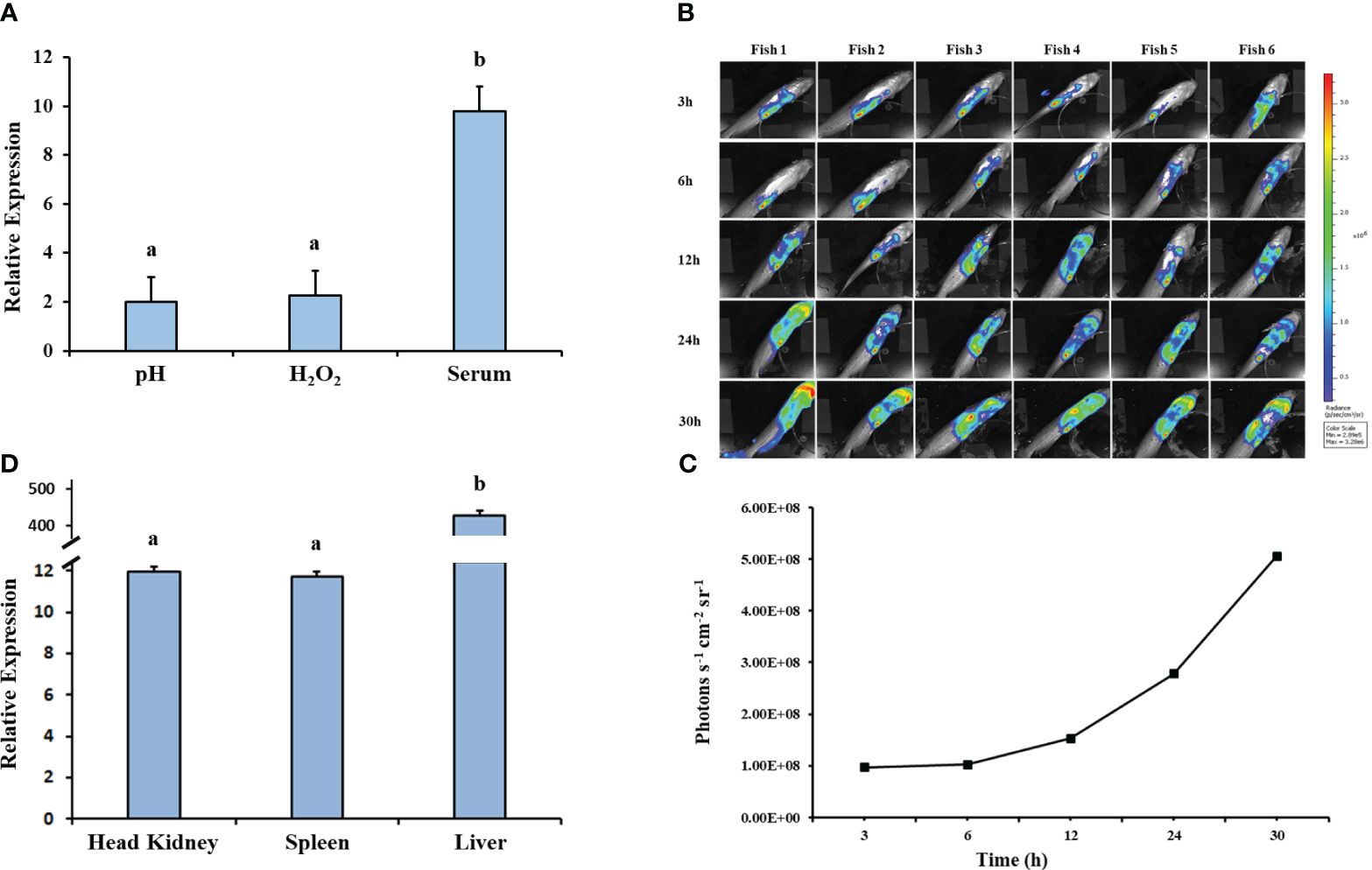
Figure 7 Gene expression analysis of hfq. (A) Relative gene expression values of hfq in E. ictaluri exposed to in vitro stress factors: low pH (4), H2O2 (1.5mM), and channel catfish serum. The gene expression was relative to the BHI growth of E. ictaluri. Small letters indicate statistical differences between treatments (p < 0.05). (B) Bioluminescent imaging of E. ictaluri in six live catfish after intraperitoneal injection. The imaging times are marked on the left, and the color scale on the right shows photon emission intensity from low (blue) to high (red). (C) Numerical values of photon emissions at each time point. Head kidney, spleen, and liver tissues were collected after imaging catfish at 30 h, and (D) hfq expression relative to the BHI-grown E. ictaluri was determined. The expression values were normalized by 16S rRNA. Small letters indicate statistical differences between treatments (p < 0.05).
3.7 Deletion of hfq resulted in increased bacterial killing in peritoneal macrophages
Catfish peritoneal macrophages were able to uptake EiΔhfq and EiWT (Figure 8A). Following the uptake, the bioluminescence of EiΔhfq decreased; hence, the bacterial killing increased over time, while EiWT’s bioluminescence, hence, the viability was up until 6 h, then it decreased (Figure 8B). At 6 h, the macrophage killing of EiΔhfq was significantly higher than that of EiWT (p < 0.05), while there were no significant differences at other time points.
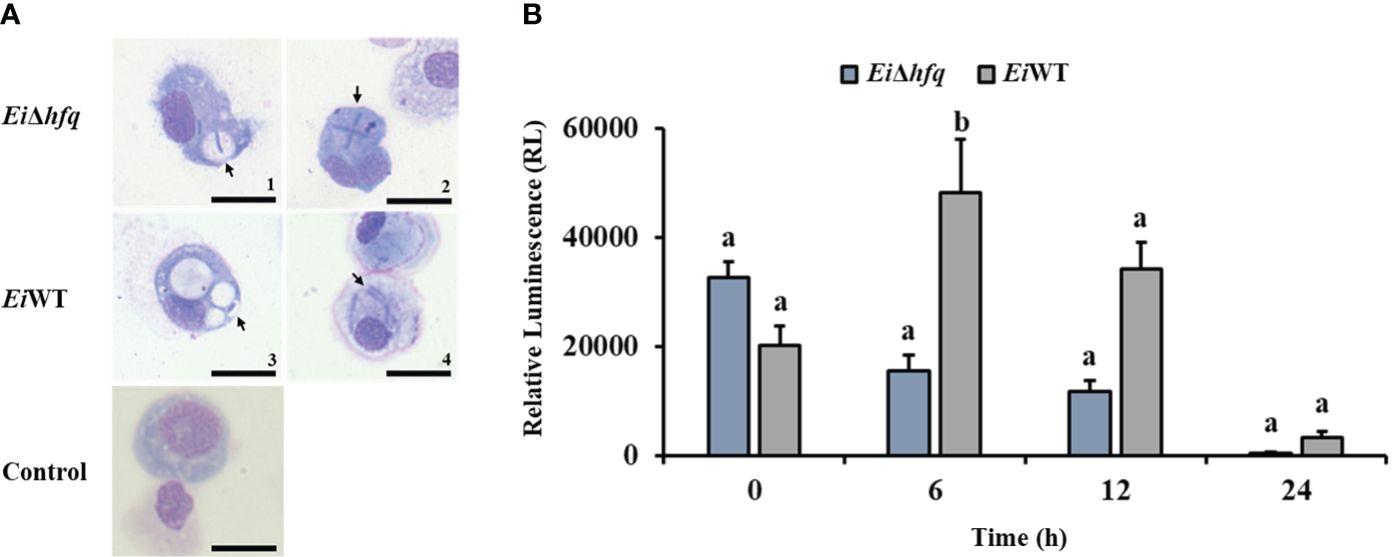
Figure 8 Replication of EiΔhfq and EiWT inside catfish peritoneal macrophages. (A) The active uptake of EiΔhfq and EiWT in catfish peritoneal macrophages. Each picture was taken 4 h post-treatment at 100 x magnification (scale bar is 20 µm). (B) The mean of photon exposure was obtained from each EiΔhfq and EiWT. The data represented the mean of each treatment ± SD. Small letters indicate statistical differences between treatments (p < 0.05).
3.8 Loss of hfq did not affect bacterial attachment and invasion of catfish epithelial cells
Bioluminescence imaging of EiΔhfq and EiWT (Figure 9A) indicated that attachment (Figure 9B) and invasion (Figure 9C) characteristics of both strains were similar in CCO cells (p > 0.05).
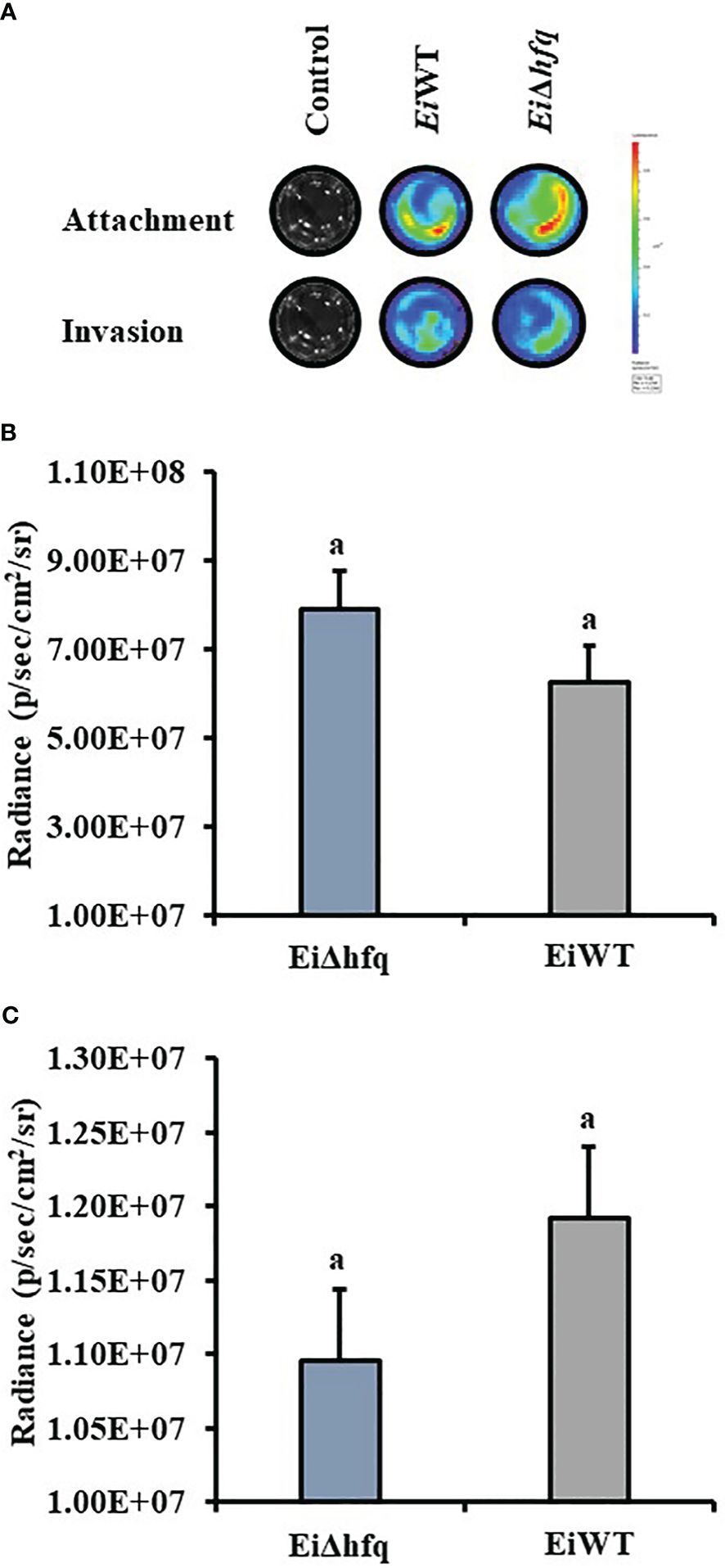
Figure 9 Attachment and invasion of EiΔhfq and EiWT to CCO. (A) The bioluminescent imaging of CCO cells treated with bioluminescent EiΔhfq, EiWT, and control (non-treated). (B) Attachment assay. The mean of photon exposure was obtained from each well in a 24-well plate incubated with EiΔhfq, EiWT, and control after an h of incubation. The bar graph indicates the mean of photons obtained from four biological replicas. Small letters indicate statistical differences between treatments (p < 0.05). (C) Invasion assay. The mean of photon exposure was obtained from the same 24-well plate incubated with EiΔhfq, EiWT, and control, including gentamycin, for an h after attachment. The bar graph indicates the mean of photons obtained from four biological replicas. Small letters indicate statistical differences between treatments (p < 0.05).
3.9 hfq is vital for bacterial persistence in catfish
Bioluminescent imaging indicated that catfish fingerlings exposed to EiWT died completely within four days. In contrast, three out of four catfish fingerlings exposed to EiΔhfq effectively cleared it and survived (Figure 10A). The photon counts showed that both strains reached the highest numbers in catfish in 4 days (Figure 10B).
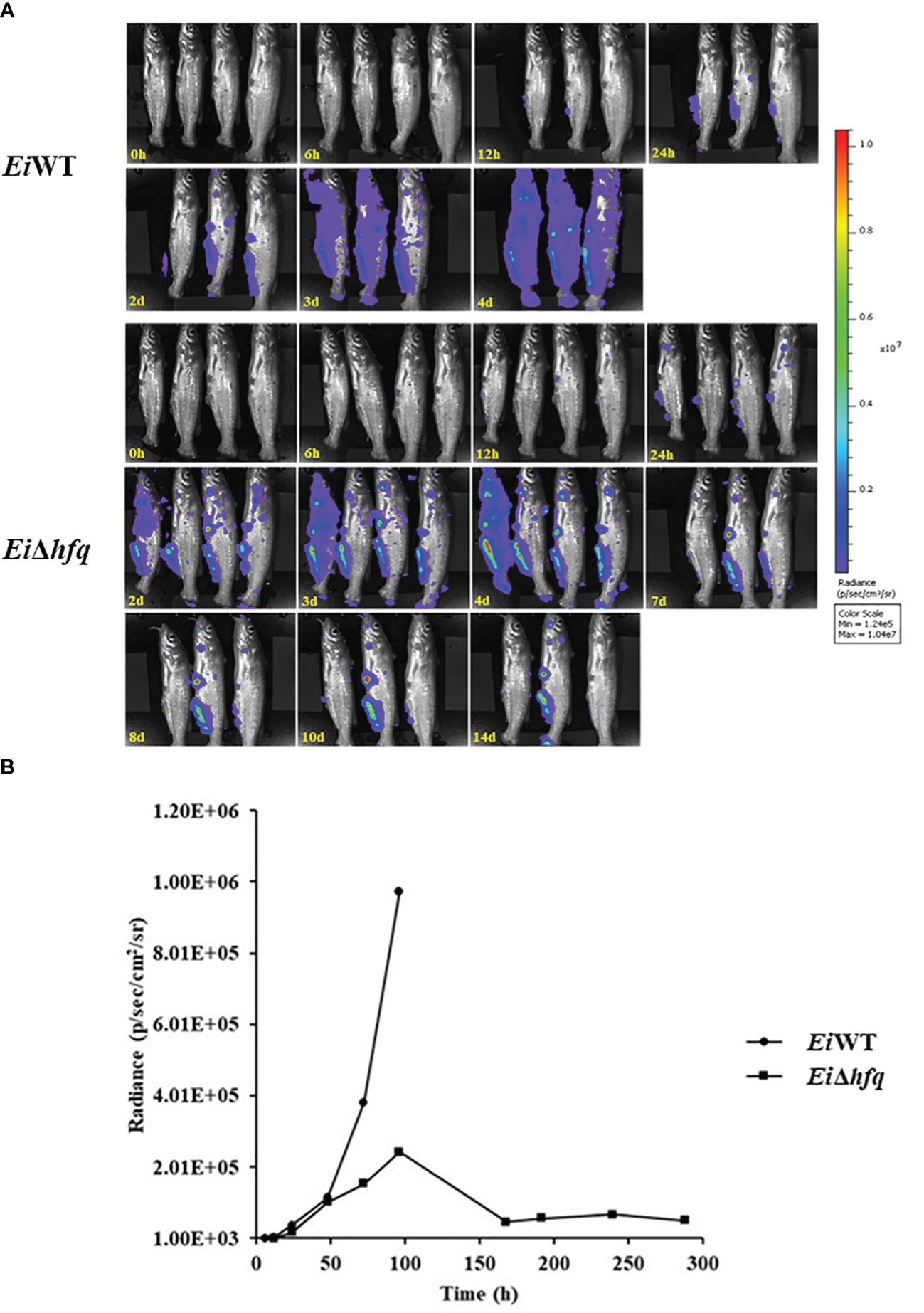
Figure 10 Bioluminescent imaging of catfish fingerlings immersion exposure challenged with EiΔhfq and EiWT. (A) The bioluminescent image was taken with four fish exposed to EiΔhfq and EiWT in 0, 6, 12, 24 h, and subsequent daily intervals until 14 days. (B) The total photon emissions from four fish exposed to EiΔhfq and EiWT.
3.10 EiΔhfq protects catfish against EiWT
Assessment of virulence showed that the percent survival of catfish challenged by EiΔhfq (83.76% survival) and complement (63.92% survival) strains were significantly higher than those of EiWT (18.95% survival) (p < 0.05) (Figure 11A). The catfish fingerlings immunized with EiΔhfq and challenged with EiWT showed complete protection (100% survival) compared to non-vaccinated sham control (14% survival) (p < 0.05) (Figure 11B).
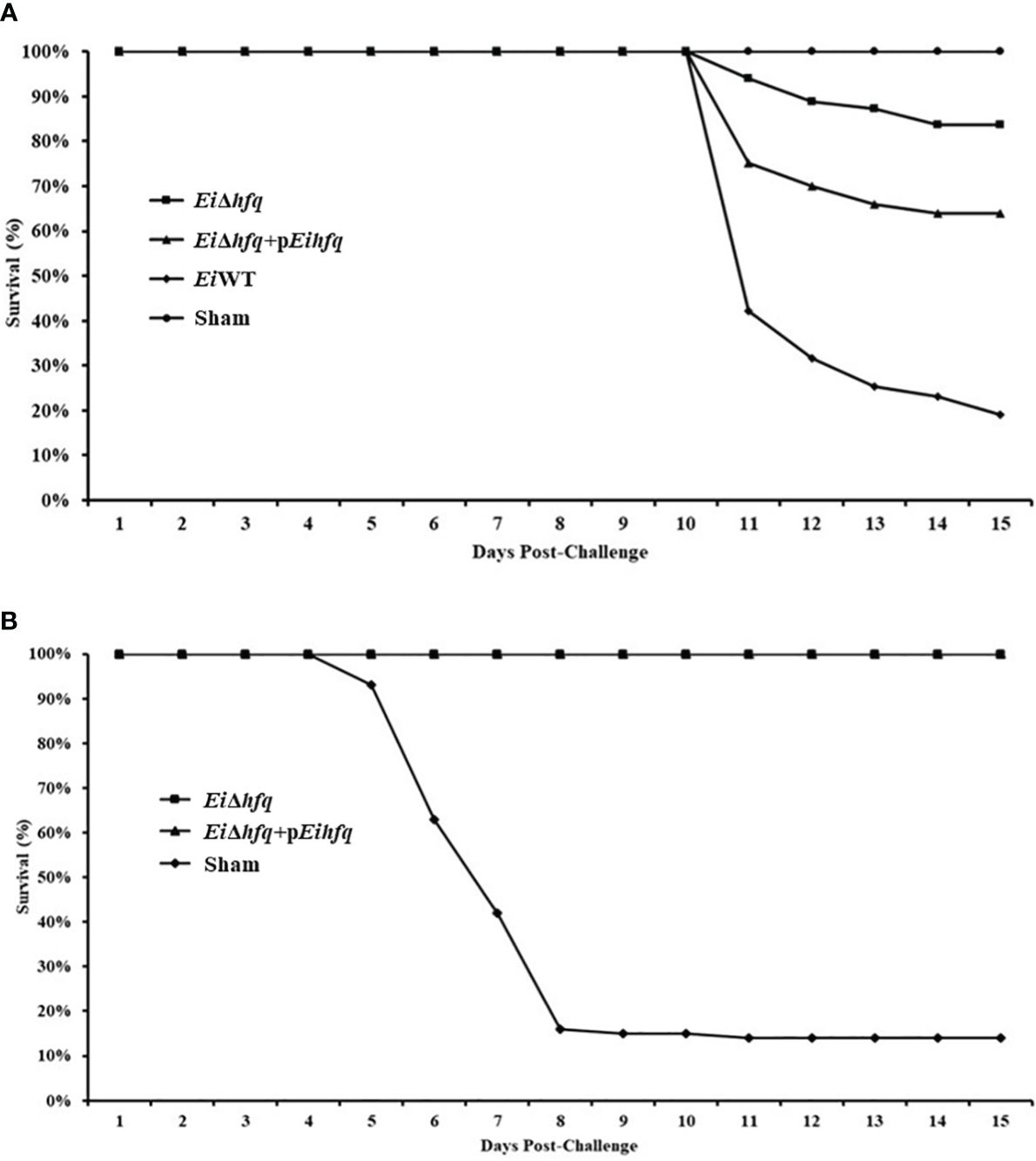
Figure 11 Vaccination and efficacy studies using EiΔhfq, EiWT, and complemented (EiΔhfq+pEihfq) strains in catfish fingerlings. (A) Percent survival of catfish fingerlings during immersion vaccination. (B) Percent survival of immunized catfish fingerlings after re-challenge with EiWT.
4 Discussion
The objective of this study was to construct an hfq in-frame deletion mutant strain (EiΔhfq) and characterize the role of hfq in E. ictaluri’s growth, biofilm formation, motility, in vitro and in vivo stress response, replication in macrophages, cell attachment and invasion, and virulence. Further, we determined the vaccine potential of the EiΔhfq strain.
The identification of a single functional hfq gene in the genome of E. ictaluri underscores its importance in this bacterium. The average size of the Hfq protein in the Edwardsiella genus differs from that observed in α- and β-Proteobacteria, suggesting potential functional variations in Hfq across bacterial taxa.
Loss of hfq in E. ictaluri led to slightly faster growth than EiWT during the exponential growth phase. Later, rapid decreases in growth and viability in the stationary growth phase were observed. These results underscore the essential role of hfq in regulating bacterial metabolism based on nutrient availability. Other potential factors could contribute to this phenomenon, including accumulation of toxic metabolites, impaired ability to respond to stress, and cell lysis. hfq is vital in cell growth in the complex and minimal medium, where pleiotropic phenotypes affect growth rate and cell morphology. Deletion of hfq has a modest growth effect on Francisella novicida and Shewanella oneidensis (Chambers and Bender, 2011; Brennan et al., 2013). Interestingly, the hfq mutation had no growth effect and stress tolerance in a rich medium in Haemophilus influenzae (Hempel et al., 2013). Besides, hfq had a different effect on the growth rate in Yersinia species. Although loss of hfq caused a slower growth rate in Y. pestis and Y. enterocolitica, loss of hfq did not affect growth in Y. pseudotuberculosis (Geng et al., 2009; Schiano et al., 2010; Kakoschke et al., 2014). In E. tarda, the hfq mutant showed a slower growth rate than the wild-type but reached the same level of growth at the stationary phase (Hu et al., 2014).
Significant reduction in biofilm formation in EiΔhfq at 48 h suggests that the absence of hfq has a notable impact on the ability of E. ictaluri to form robust biofilms. hfq is required for biofilm production in the flea’s proventriculus (Rempe et al., 2012), suggesting that hfq enables Y. pestis transmission from flea to mammalian host (Hinnebusch and Erickson, 2008). hfq is also involved in biofilm produced by E. coli and V. cholerae. These studies suggested that hfq’s contribution to biofilm formation might be conserved in the bacterial world (Kulesus et al., 2008; Bardill et al., 2011; Schiano and Lathem, 2012).
Our SEM analysis confirmed the presence of few flagella in the EiΔhfq strain. Our findings indicate that the increased motility observed in the EiΔhfq strain could be due to the dysregulation of flagellar gene expression or global regulatory networks rather than changes in flagella structure. It has been shown that hfq was involved in motility in diverse bacterial species (Sonnleitner et al., 2003; Ding et al., 2004; Sittka et al., 2007). For example, a significant impairment in motility was shown in Bacillus subtilis and S. typhimurium (Sittka et al., 2007; Jagtap et al., 2016). Similarly, a non-flagellar increase in the motility of the hfq mutant strain was observed in Y. pseudotuberculosis (Schiano et al., 2010). Non-flagellar motility might be utilized during the free-living phase in the environment or could inhibit or slow down swarming when the bacteria enter their hosts (Schiano and Lathem, 2012).
The diminished growth of EiΔhfq in response to heightened acidity and oxidative stress suggests a crucial role for hfq in the regulatory networks associated with acidic and oxidative stress response mechanisms. E. ictaluri may encounter other stresses in a host environment, such as osmotic stress and nutrient deprivation, which could differentially impact the bacterium’s physiology, gene expression, and virulence. Future studies could expand the scope of stress conditions. hfq is critically important in the virulence of several bacterial pathogens. hfq mutants have challenges when grown under stressors such as H2O2, salt, and antimicrobial peptides (Lenz et al., 2004; Sittka et al., 2007; Fantappiè et al., 2009). The stress sensitivity we observed in the E. ictaluri hfq mutant is consistent with the altered stress responses observed for other bacteria (Robertson and Roop, 1999; Christiansen et al., 2004; Fantappiè et al., 2009). hfq contributes to the resistance to oxidative stress in Vibrio alginolyticus (Deng et al., 2016). These support that hfq is important for growth and survival in harsh environments.
The expression analysis of E. ictaluri hfq showed that hfq responds to in vivo stresses more than in vitro stresses. The elevated hfq gene expression in the host, especially in the spleen, indicates a potential role for hfq in facilitating E. ictaluri’s adaptation and survival within specific host tissues. The hfq gene regulates many genes involved in metabolism, virulence, stress responses, and adaptation in B. melitensis (Cui et al., 2013). Interestingly, hfq appeared to govern the expression of genes indirectly by affecting sigma factor (RpoS and RpoE) dependent genes and modulating the physiological fitness and virulence of K. pneumonia (Chiang et al., 2011). Another study showed that hfq controls virulence through the positive regulation of T3SS, and importantly, hfq is a key factor regulating acid stress tolerance and virulence in S. flexneri (Yang et al., 2015).
We found that the absence of hfq does not significantly impact adherence and invasion of CCO cells. However, the lack of functional hfq exhibited a continual decrease in viability inside catfish macrophages over time. These results suggest that hfq may not play an essential role in the initial steps of infection but implies a potential role in modulating E. ictaluri survival within host immune cells. Disruption of the hfq gene caused reduced adhesion to host epithelial cells, impaired survivability within macrophages, and less virulence in mice infection models of S. Typhimurium and S. Enteritidis (Sittka et al., 2007; Meng et al., 2013). Similarly, lack of hfq caused less adherence, invasion, and survivability inside macrophages in Proteus mirabilis, Cronobacter sakazakii, and Acinetobacter baumannii (Wang et al., 2014; Kim et al., 2015; Kuo et al., 2017). Unlikely, a mutation in hfq did not affect adherence and invasion of in vitro epithelial and macrophage cell lines in E. coli (Kulesus et al., 2008). Specific adhesion molecules, such as pili, fimbriae, or surface proteins, often mediate initial bacterial attachment to host cells. Potential constitutive or hfq-independent expression of these molecules could minimize the impact of hfq mutation on cell attachment.
The contrasting outcomes observed in catfish fingerlings exposed to the EiΔhfq and EiWT strains underscore the importance of the hfq in the virulence of E. ictaluri. Moreover, the protective efficacy of EiΔhfq was evident in catfish fingerlings immunized and subsequently challenged with EiWT 21 days post-vaccination. Several studies about hfq showed that it broadly affected the virulence of pathogenic bacteria. The hfq mutant failed to cause a systemic infection in a mouse model of K. pneumonia (Chiang et al., 2011). Deletion of hfq in Salmonella Typhimurium caused attenuated in cell culture and animal models, and oral immunization with the Salmonella hfq mutant protected mice against subsequent oral challenge with virulent Salmonella Typhimurium (Allam et al., 2011). Mutation in hfq caused increased expression of T3SS and resulted in attenuation in Shigella flexneri, and it caused a protective immunity against Shigella strains; therefore, the vaccine potential of hfq mutant was established in two guinea pig models (Mitobe et al., 2017). hfq is a global coordinator of surface virulence determinants and essential for the virulence of Y. enterocolitica, in mice (Kakoschke et al., 2016). Our virulence/vaccination study shows that EiΔhfq exhibits a five-fold decrease in virulence (16.26% vs 81.05% mortality with EiWT). Survivors of the vaccination are fully protected against EiWT infection, suggesting promising outcomes for vaccine development by adjusting the vaccination dose in future vaccine safety studies.
5 Conclusion
This study elucidates the essential role of hfq in E. ictaluri. The highly conserved Hfq protein significantly impacts bacterial growth, motility, biofilm formation, and stress response. Deletion of hfq resulted in attenuated virulence, emphasizing its importance for bacterial persistence in catfish. Furthermore, vaccination with EiΔhfq conferred protection against pathogenic EiWT infection. These findings underscore the multifaceted significance of hfq in E. ictaluri physiology, highlighting its potential as a target for understanding and managing bacterial infections.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author/s.
Ethics statement
This study followed a protocol approved by the Institutional Animal Care and Use Committee of the Mississippi State University (Protocol Number: 15-043).
Author contributions
AA: Formal analysis, Methodology, Visualization, Writing – original draft, Writing – review & editing. SK: Formal analysis, Methodology, Visualization, Writing – original draft, Writing – review & editing. AdK: Formal analysis, Methodology, Visualization, Writing – review & editing. HA: Formal analysis, Methodology, Visualization, Writing – review & editing. II: Formal analysis, Methodology, Visualization, Writing – review & editing. HT: Formal analysis, Methodology, Visualization, Writing – review & editing. AtK: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the USDA NIFA project number MISV-371790. Bioluminescence imaging was supported by USDA-ARS Biophotonics Initiative #58-6402-3-018. SK was supported by a fellowship from the Ministry of National Education of Türkiye.
Acknowledgments
We thank Dr. Larry A. Hanson for providing channel catfish ovary cell lines and Dr. Ayfer Akgul for technical help.
Conflict of interest
The authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdelhamed, H., Lawrence, M. L., Karsi, A. (2018). Development and characterization of a novel live attenuated vaccine against enteric septicemia of catfish. Front. Microbiol. 9, 1819. doi: 10.3389/fmicb.2018.01819
Ainsworth, A. J., Chen, D. X. (1990). Differences in the phagocytosis of four bacteria by channel catfish neutrophils. Dev. Comp. Immunol. 14, 201–209. doi: 10.1016/0145-305X(90)90091-R
Allam, U. S., Krishna, M. G., Lahiri, A., Joy, O., Chakravortty, D. (2011). Salmonella enterica serovar Typhimurium lacking hfq gene confers protective immunity against murine typhoid. PloS One 6, e16667. doi: 10.1371/journal.pone.0016667
Bader, J. A., Shoemaker, C. A., Klesius, P. H. (2004). Immune response induced by N-lauroylsarcosine extracted outer-membrane proteins of an isolate of Edwardsiella ictaluri in channel catfish. Fish. Shellfish. Immunol. 16, 415–428. doi: 10.1016/j.fsi.2003.07.003
Bardill, J. P., Zhao, X., Hammer, B. K. (2011). The Vibrio cholerae quorum sensing response is mediated by Hfq-dependent sRNA/mRNA base pairing interactions. Mol. Microbiol. 80, 1381–1394. doi: 10.1111/j.1365-2958.2011.07655.x
Booth, N. J., Beekman, J. B., Thune, R. L. (2009). Edwardsiella ictaluri encodes an acid-activated urease that is required for intracellular replication in channel catfish (Ictalurus punctatus) macrophages. Appl. Environ. Microbiol. 75, 6712–6720. doi: 10.1128/AEM.01670-09
Brantl, S., Muller, P. (2021). Cis- and trans-encoded small regulatory RNAs in bacillus subtilis. Microorganisms 9 (9), 1865. doi: 10.3390/microorganisms9091865
Brennan, C. M., Keane, M. L., Hunt, T. M., Goulet, M. T., Mazzucca, N. Q., Sexton, Z., et al. (2013). Shewanella oneidensis Hfq promotes exponential phase growth, stationary phase culture density, and cell survival. BMC Microbiol. 13, 33. doi: 10.1186/1471-2180-13-33
Cai, W., Arias, C. R. (2017). Biofilm formation on aquaculture substrates by selected bacterial fish pathogens. J. Aquat. Anim. Health 29, 95–104. doi: 10.1080/08997659.2017.1290711
Caldelari, I., Chao, Y., Romby, P., Vogel, J. (2013). RNA-mediated regulation in pathogenic bacteria. Cold Spring Harb. Perspect. Med. 3, a010298. doi: 10.1101/cshperspect.a010298
Chambers, J. R., Bender, K. S. (2011). The RNA chaperone hfq is important for growth and stress tolerance in francisella novicida. PloS One 6, e19797. doi: 10.1371/journal.pone.0019797
Chao, Y., Vogel, J. (2010). The role of Hfq in bacterial pathogens. Curr. Opin. Microbiol. 13, 24–33. doi: 10.1016/j.mib.2010.01.001
Chiang, M. K., Lu, M. C., Liu, L. C., Lin, C. T., Lai, Y. C. (2011). Impact of Hfq on global gene expression and virulence in Klebsiella pneumoniae. PloS One 6, e22248. doi: 10.1371/journal.pone.0022248
Christiansen, J. K., Larsen, M. H., Ingmer, H., Sogaard-Andersen, L., Kallipolitis, B. H. (2004). The RNA-binding protein Hfq of Listeria monocytogenes: role in stress tolerance and virulence. J. Bacteriol. 186, 3355–3362. doi: 10.1128/JB.186.11.3355-3362.2004
Cooper, R. K., Shotts, E. B., Nolan, L. K. (1996). Use of a mini-transposon to study chondroitinase activity associated with edwardsiella ictaluri. J. Aquat. Anim. Health 8, 319–324. doi: 10.1577/1548-8667(1996)008<0319:UOAMTT>2.3.CO;2
Cui, M., Wang, T., Xu, J., Ke, Y., Du, X., Yuan, X., et al. (2013). Impact of hfq on global gene expression and intracellular survival in brucella melitensis. PloS One 8, e71933. doi: 10.1371/journal.pone.0071933
Deng, Y., Chen, C., Zhao, Z., Zhao, J., Jacq, A., Huang, X., et al. (2016). The RNA chaperone hfq is involved in colony morphology, nutrient utilization and oxidative and envelope stress response in vibrio alginolyticus. PloS One 11, e0163689–e0163689. doi: 10.1371/journal.pone.0163689
Ding, Y., Davis, B. M., Waldor, M. K. (2004). Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol. Microbiol. 53, 345–354. doi: 10.1111/j.1365-2958.2004.04142.x
do Vale, A., Afonso, A., Silva, M. T. (2002). The professional phagocytes of sea bass (Dicentrarchus labrax L.): cytochemical characterisation of neutrophils and macrophages in the normal and inflamed peritoneal cavity. Fish. Shellfish. Immunol. 13, 183–198. doi: 10.1006/fsim.2001.0394
Dozois, C. M., Daigle, F., Curtiss, R., 3rd. (2003). Identification of pathogen-specific and conserved genes expressed in vivo by an avian pathogenic Escherichia coli strain. Proc. Natl. Acad. Sci. U. S. A. 100, 247–252. doi: 10.1073/pnas.232686799
Ellis, M. J., Trussler, R. S., Haniford, D. B. (2015). A cis-encoded sRNA, Hfq and mRNA secondary structure act independently to suppress IS200 transposition. Nucleic Acids Res. 43, 6511–6527. doi: 10.1093/nar/gkv584
Ellis, R. W. (2001). Technologies for the design, discovery, formulation and administration of vaccines. Vaccine 19, 2681–2687. doi: 10.1016/S0264-410X(00)00504-1
Erickson, V. I., Dung, T. T., Khoi, L. M., Hounmanou, Y. M. G., Phu, T. M., Dalsgaard, A. (2024). Genomic Insights into Edwardsiella ictaluri: Molecular Epidemiology and Antimicrobial Resistance in Striped Catfish (Pangasianodon hypophthalmus) Aquaculture in Vietnam. Microorganisms 12, 1182. doi: 10.3390/microorganisms12061182
Fantappiè, L., Metruccio, M. M. E., Seib, K. L., Oriente, F., Cartocci, E., Ferlicca, F., et al. (2009). The RNA chaperone hfq is involved in stress response and virulence in neisseria meningitidis and is a pleiotropic regulator of protein expression. Infect. Immunity. 77, 1842–1853. doi: 10.1128/IAI.01216-08
Feliciano, J. R., Grilo, A. M., Guerreiro, S. I., Sousa, S. A., Leitao, J. H. (2016). Hfq: a multifaceted RNA chaperone involved in virulence. Future Microbiol. 11, 137–151. doi: 10.2217/fmb.15.128
Geng, J., Song, Y., Yang, L., Feng, Y., Qiu, Y., Li, G., et al. (2009). Involvement of the post-transcriptional regulator Hfq in Yersinia pestis virulence. PloS One 4, e6213. doi: 10.1371/journal.pone.0006213
Hempel, R. J., Morton, D. J., Seale, T. W., Whitby, P. W., Stull, T. L. (2013). The role of the RNA chaperone Hfq in Haemophilus influenzae pathogenesis. BMC Microbiol. 13, 134–134. doi: 10.1186/1471-2180-13-134
Herrero, M., de Lorenzo, V., Timmis, K. N. (1990). Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172, 6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990
Hinnebusch, B. J., Erickson, D. L. (2008). Yersinia pestis biofilm in the flea vector and its role in the transmission of plague. Curr. Topics. Microbiol. Immunol. 322, 229–248. doi: 10.1007/978-3-540-75418-3_11
Hu, Y. H., Li, Y. X., Sun, L. (2014). Edwardsiella tarda Hfq: impact on host infection and global protein expression. Vet. Res. 45, 23. doi: 10.1186/1297-9716-45-23
Jagtap, C. B., Kumar, P., Rao, K. K. (2016). Bacillus subtilis Hfq: A role in chemotaxis and motility. J. Biosci. 41, 347–358. doi: 10.1007/s12038-016-9618-9
Kakoschke, T., Kakoschke, S., Magistro, G., Schubert, S., Borath, M., Heesemann, J., et al. (2014). The RNA chaperone Hfq impacts growth, metabolism and production of virulence factors in Yersinia enterocolitica. PloS One 9, e86113. doi: 10.1371/journal.pone.0086113
Kakoschke, T. K., Kakoschke, S. C., Zeuzem, C., Bouabe, H., Adler, K., Heesemann, J., et al. (2016). The RNA chaperone hfq is essential for virulence and modulates the expression of four adhesins in yersinia enterocolitica. Sci. Rep. 6, 29275. doi: 10.1038/srep29275
Kalindamar, S., Abdelhamed, H., Kordon, A. O., Tekedar, H. C., Pinchuk, L., Karsi, A. (2023). Characterization of Type VI secretion system in Edwardsiella ictaluri. PloS One 18, e0296132. doi: 10.1371/journal.pone.0296132
Kalindamar, S., Kordon, A. O., Abdelhamed, H., Tan, W., Pinchuk, L. M., Karsi, A. (2020). Edwardsiella ictaluri evpP is required for colonisation of channel catfish ovary cells and necrosis in anterior kidney macrophages. Cell Microbiol. 22, e13135. doi: 10.1111/cmi.13135
Karsi, A., Gulsoy, N., Corb, E., Dumpala, P. R., Lawrence, M. L. (2009). High-throughput bioluminescence-based mutant screening strategy for identification of bacterial virulence genes. Appl. Environ. Microbiol. 75, 2166–2175. doi: 10.1128/AEM.02449-08
Karsi, A., Lawrence, M. L. (2007). Broad host range fluorescence and bioluminescence expression vectors for Gram-negative bacteria. Plasmid 57, 286–295. doi: 10.1016/j.plasmid.2006.11.002
Kim, S., Hwang, H., Kim, K. P., Yoon, H., Kang, D. H., Ryu, S. (2015). Hfq plays important roles in virulence and stress adaptation in Cronobacter sakazakii ATCC 29544. Infect. Immun. 83, 2089–2098. doi: 10.1128/IAI.03161-14
Klesius, P. H., Sealey, W. M. (1996). Chemotactic and chemokinetic responses of channel catfish macrophages to exoantigen from edwardsiella ictaluri. J. Aquat. Anim. Health 8, 314–318. doi: 10.1577/1548-8667(1996)008<0314:CACROC>2.3.CO;2
Klesius, P. H., Shoemaker, C. A. (1999). Development and use of modified live Edwardsiella ictaluri vaccine against enteric septicemia of catfish. Adv. Vet. Med. 41, 523–537. doi: 10.1016/S0065-3519(99)80039-1
Kordon, A. O., Abdelhamed, H., Karsi, A., Pinchuk, L. M. (2021). Adaptive immune responses in channel catfish exposed to Edwardsiella ictaluri live attenuated vaccine and wild type strains through the specific gene expression profiles. Dev. Comp. Immunol. 116, 103950. doi: 10.1016/j.dci.2020.103950
Kordon, A. O., Kalindamar, S., Majors, K., Abdelhamed, H., Tan, W., Karsi, A., et al. (2020). Live attenuated Edwardsiella ictaluri vaccines enhance the protective innate immune responses of channel catfish B cells. Dev. Comp. Immunol. 109, 103711. doi: 10.1016/j.dci.2020.103711
Kovach, M. E., Elzer, P. H., Hill, D. S., Robertson, G. T., Farris, M. A., Roop, R. M., 2nd, et al. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176. doi: 10.1016/0378-1119(95)00584-1
Kulesus, R. R., Diaz-Perez, K., Slechta, E. S., Eto, D. S., Mulvey, M. A. (2008). Impact of the RNA chaperone Hfq on the fitness and virulence potential of uropathogenic Escherichia coli. Infect. Immun. 76, 3019–3026. doi: 10.1128/IAI.00022-08
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kuo, H. Y., Chao, H. H., Liao, P. C., Hsu, L., Chang, K. C., Tung, C. H., et al. (2017). Functional characterization of acinetobacter baumannii lacking the RNA chaperone hfq. Front. Microbiol. 8, 2068. doi: 10.3389/fmicb.2017.02068
Lawrence, M. L., Banes, M. M., Williams, M. L. (2001). Phenotype and virulence of a transposon-derived lipopolysaccharide O side-chain mutant strain of edwardsiella ictaluri. J. Aquat. Anim. Health 13, 291–299. doi: 10.1577/1548-8667(2001)013<0291:PAVOAT>2.0.CO;2
Lawrence, M. L., Cooper, R. K., Thune, R. L. (1997). Attenuation, persistence, and vaccine potential of an Edwardsiella ictaluri purA mutant. Infect. Immun. 65, 4642–4651. doi: 10.1128/iai.65.11.4642-4651.1997
Lenz, D. H., Mok, K. C., Lilley, B. N., Kulkarni, R. V., Wingreen, N. S., Bassler, B. L. (2004). The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118, 69–82. doi: 10.1016/j.cell.2004.06.009
Liu, H., Wang, Q., Liu, Q., Cao, X., Shi, C., Zhang, Y. (2011). Roles of Hfq in the stress adaptation and virulence in fish pathogen Vibrio alginolyticus and its potential application as a target for live attenuated vaccine. Appl. Microbiol. Biotechnol. 91, 353–364. doi: 10.1007/s00253-011-3286-3
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Meibom, K. L., Forslund, A. L., Kuoppa, K., Alkhuder, K., Dubail, I., Dupuis, M., et al. (2009). Hfq, a novel pleiotropic regulator of virulence-associated genes in Francisella tularensis. Infect. Immun. 77, 1866–1880. doi: 10.1128/IAI.01496-08
Meng, X., Meng, X., Zhu, C., Wang, H., Wang, J., Nie, J., et al. (2013). The RNA chaperone Hfq regulates expression of fimbrial-related genes and virulence of Salmonella enterica serovar Enteritidis. FEMS Microbiol. Lett. 346, 90–96. doi: 10.1111/fml.2013.346.issue-2
Metcalf, W. W., Jiang, W., Wanner, B. L. (1994). Use of the rep technique for allele replacement to construct new Escherichia coli hosts for maintenance of R6K gamma origin plasmids at different copy numbers. Gene 138, 1–7. doi: 10.1016/0378-1119(94)90776-5
Miller, V. L., Mekalanos, J. J. (1988). A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170, 2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988
Mitobe, J., Sinha, R., Mitra, S., Nag, D., Saito, N., Shimuta, K., et al. (2017). An attenuated Shigella mutant lacking the RNA-binding protein Hfq provides cross-protection against Shigella strains of broad serotype. PloS Negl. Trop. Dis. 11, e0005728. doi: 10.1371/journal.pntd.0005728
Newton, J. C., Triche, P. L. (1993). Isolation and characterization of flagella from edwardsiella ictaluri. J. Aquat. Anim. Health 5, 16–22. doi: 10.1577/1548-8667(1993)005<0016:IACOFF>2.3.CO;2
Nho, S. W., Abdelhamed, H., Karsi, A., Lawrence, M. L. (2017). Improving safety of a live attenuated Edwardsiella ictaluri vaccine against enteric septicemia of catfish and evaluation of efficacy. Vet. Microbiol. 210, 83–90. doi: 10.1016/j.vetmic.2017.09.004
Oliva, G., Sahr, T., Buchrieser, C. (2015). Small RNAs, 5′ UTR elements and RNA-binding proteins in intracellular bacteria: impact on metabolism and virulence. FEMS Microbiol. Rev. 39, 331–349. doi: 10.1093/femsre/fuv022
Rempe, K. A., Hinz, A. K., Vadyvaloo, V. (2012). Hfq regulates biofilm gut blockage that facilitates flea-borne transmission of yersinia pestis. J. Bacteriol. 194, 2036–2040. doi: 10.1128/JB.06568-11
Robertson, G. T., Roop, R. M., Jr. (1999). The Brucella abortus host factor I (HF-I) protein contributes to stress resistance during stationary phase and is a major determinant of virulence in mice. Mol. Microbiol. 34, 690–700. doi: 10.1046/j.1365-2958.1999.01629.x
Schiano, C. A., Bellows, L. E., Lathem, W. W. (2010). The small RNA chaperone Hfq is required for the virulence of Yersinia pseudotuberculosis. Infect. Immun. 78, 2034–2044. doi: 10.1128/IAI.01046-09
Schiano, C. A., Lathem, W. W. (2012). Post-transcriptional regulation of gene expression in Yersinia species. Front. Cell Infect. Microbiol. 2, 129. doi: 10.3389/fcimb.2012.00129
Sittka, A., Pfeiffer, V., Tedin, K., Vogel, J. (2007). The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 63, 193–217. doi: 10.1111/j.1365-2958.2006.05489.x
Sonnleitner, E., Hagens, S., Rosenau, F., Wilhelm, S., Habel, A., Jäger, K.-E., et al. (2003). Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Microbial. Pathogene. 35, 217–228. doi: 10.1016/S0882-4010(03)00149-9
Soper, T., Mandin, P., Majdalani, N., Gottesman, S., Woodson, S. A. (2010). Positive regulation by small RNAs and the role of Hfq. Proc. Natl. Acad. Sci. 107, 9602–9607. doi: 10.1073/pnas.1004435107
Sun, X., Zhulin, I., Wartell, R. M. (2002). Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res. 30, 3662–3671. doi: 10.1093/nar/gkf508
Taylor, R. G., Walker, D. C., McInnes, R. R. (1993). E. coli host strains significantly affect the quality of small scale plasmid DNA preparations used for sequencing. Nucleic Acids Res. 21, 1677–1678. doi: 10.1093/nar/21.7.1677
Thomason, M. K., Storz, G. (2010). Bacterial antisense RNAs: How many are there and what are they doing? Annu. Rev. Genet. 44, 167–188. doi: 10.1146/annurev-genet-102209-163523
Thune, R. L., Fernandez, D. H., Battista, J. R. (1999). An aroA Mutant of Edwardsiella ictaluri Is Safe and Efficacious as a Live, Attenuated Vaccine. J. Aquat. Anim. Health 11, 358–372. doi: 10.1577/1548-8667(1999)011<0358:AAMOEI>2.0.CO;2
Thune, R. L., Fernandez, D. H., Benoit, J. L., Kelly-Smith, M., Rogge, M. L., Booth, N. J., et al. (2007). Signature-tagged mutagenesis of Edwardsiella ictaluri identifies virulence-related genes, including a salmonella pathogenicity island 2 class of type III secretion systems. Appl. Environ. Microbiol. 73, 7934–7946. doi: 10.1128/AEM.01115-07
Tu, T. D., Haesebrouck, F., Nguyen, A. T., Sorgeloos, P., Baele, M., Decostere, A. (2008). Antimicrobial susceptibility pattern of Edwardsiella ictaluri isolates from natural outbreaks of bacillary necrosis of Pangasianodon hypophthalmus in Vietnam. Microb. Drug Resist. 14, 311–316. doi: 10.1089/mdr.2008.0848
Wang, M.-C., Chien, H.-F., Tsai, Y.-L., Liu, M.-C., Liaw, S.-J. (2014). The RNA chaperone hfq is involved in stress tolerance and virulence in uropathogenic proteus mirabilis. PloS One 9, e85626. doi: 10.1371/journal.pone.0085626
Williams, M. L., Gillaspy, A. F., Dyer, D. W., Thune, R. L., Waldbieser, G. C., Schuster, S. C., et al. (2012). Genome sequence of Edwardsiella ictaluri 93-146, , a strain associated with a natural channel catfish outbreak of enteric septicemia of catfish. J. Bacteriol. 194, 740–741.
Wilton, J., Acebo, P., Herranz, C., Gómez, A., Amblar, M. (2015). Small regulatory RNAs in Streptococcus pneumoniae: discovery and biological functions. Front. Genet. 6, 126. doi: 10.3389/fgene.2015.00126
Wise, D. J., Greenway, T. E., Byars, T. S., Griffin, M. J., Khoo, L. H. (2015). Oral Vaccination of Channel Catfish against Enteric Septicemia of Catfish Using a Live Attenuated Edwardsiella ictaluri Isolate. J. Aquat. Anim. Health 27, 135–143. doi: 10.1080/08997659.2015.1032440
Yang, G., Wang, L., Wang, Y., Li, P., Zhu, J., Qiu, S., et al. (2015). hfq regulates acid tolerance and virulence by responding to acid stress in Shigella flexneri. Res. Microbiol. 166, 476–485. doi: 10.1016/j.resmic.2015.06.007
Zeng, Q., McNally, R. R., Sundin, G. W. (2013). Global small RNA chaperone Hfq and regulatory small RNAs are important virulence regulators in Erwinia amylovora. J. Bacteriol. 195, 1706–1717. doi: 10.1128/JB.02056-12
Keywords: Edwardsiella ictaluri, ESC, Hfq, SRNAs, stress response, virulence
Citation: Akgul A, Kalindamar S, Kordon AO, Abdelhamed H, Ibrahim I, Tekedar HC and Karsi A (2024) The RNA chaperone Hfq has a multifaceted role in Edwardsiella ictaluri. Front. Cell. Infect. Microbiol. 14:1394008. doi: 10.3389/fcimb.2024.1394008
Received: 29 February 2024; Accepted: 01 July 2024;
Published: 19 July 2024.
Edited by:
Jose Ramos-Vivas, Universidad Europea del Atlántico, SpainReviewed by:
Rajagopal Kammara, Central Food Technological Research Institute (CSIR), IndiaYibin Yang, Chinese Academy of Fishery Sciences (CAFS), China
Copyright © 2024 Akgul, Kalindamar, Kordon, Abdelhamed, Ibrahim, Tekedar and Karsi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Attila Karsi, a2Fyc2lAY3ZtLm1zc3RhdGUuZWR1
†Present address: Ali Akgul, Merck & Co., Inc., West Point, PA, United States Safak Kalindamar, Department of Molecular Biology and Genetics, Faculty of Arts and Sciences, Ordu University, Ordu, Türkiye Iman Ibrahim, Department of Pathology, Faculty of Veterinary Medicine, Mansoura University, Mansoura, Egypt
‡These authors have contributed equally to this work and share first authorship
 Ali Akgul
Ali Akgul Safak Kalindamar
Safak Kalindamar Adef O. Kordon
Adef O. Kordon Hossam Abdelhamed
Hossam Abdelhamed Iman Ibrahim
Iman Ibrahim Hasan C. Tekedar
Hasan C. Tekedar Attila Karsi
Attila Karsi