- 1People’s Hospital of Ningxia Hui Autonomous Region, Pharmacy Department, Yinchuan, China
- 2School of Pharmacy, Lanzhou University, Lanzhou, China
- 3Ningxia Medical University, School of Basic Medicine, Yinchuan, China
In recent years, there has been increasing interest in studying gut microbiome-derived hydrolases in relation to oral drug metabolism, particularly focusing on natural product drugs. Despite the significance of natural product drugs in the field of oral medications, there is a lack of research on the regulatory interplay between gut microbiome-derived hydrolases and these drugs. This review delves into the interaction between intestinal microbiome-derived hydrolases and natural product drugs metabolism from three key perspectives. Firstly, it examines the impact of glycoside hydrolases, amide hydrolases, carboxylesterase, bile salt hydrolases, and epoxide hydrolase on the structure of natural products. Secondly, it explores how natural product drugs influence microbiome-derived hydrolases. Lastly, it analyzes the impact of interactions between hydrolases and natural products on disease development and the challenges in developing microbial-derived enzymes. The overarching goal of this review is to lay a solid theoretical foundation for the advancement of research and development in new natural product drugs and personalized treatment.
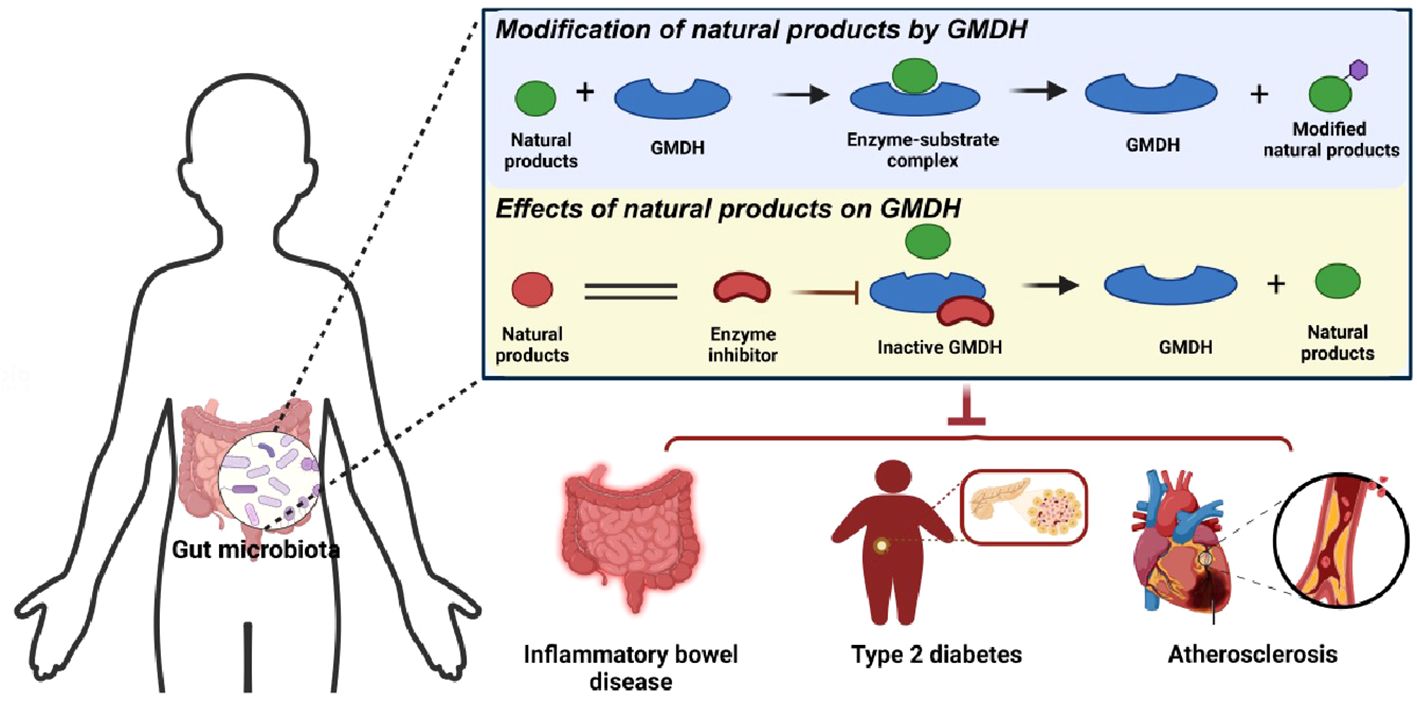
Graphical Abstract Gut microbiome derived hydrolases directly or indirectly influence the metabolic processes of natural products, thereby impacting the occurrence and development of diseases.
1 Introduction
The gut microbiota has a significant impact on the modification of oral drugs. Once oral drugs enter aims intestine, they undergo various modifications by microbiome-derived enzymes (MDE) such as hydrolases, lyases, oxidoreductases, and transferases (Xie Y. et al., 2020). These modifications alter the effectiveness and toxicity of the drugs (Zhang et al., 2018; Weersma et al., 2020). Among these, hydrolases, such as glycoside hydrolases, carboxylesterases, amide hydrolases, and bile saline hydrolases, play a significant role in drug metabolism (Tomioka et al., 2022). What’s more, It has been reported that drugs metabolized by hydrolases make up more than 30% of drugs co-metabolized by microorganisms (Zimmermann et al., 2019). Natural products are a vital component of pharmaceuticals. Over the past 40 years, 1,881 new drugs have been approved globally, with approximately 23.5% originating from natural products and their derivatives. This includes 71 natural products, accounting for 3.8%, and 356 natural product derivatives, accounting for 18.9% (Newman and Cragg, 2020). However, natural product drugs are primarily administered orally, exhibiting pharmacological activity only after microbial transformation or metabolism into more potent secondary glycosides or aglycones (Lai et al., 2010; Zhou et al., 2019). A notable example is sulforaphane, which is converted to sulforaphane isothiocyanate by hydrolase in the cecum (Lai et al., 2010; Sikorska-Zimny and Beneduce, 2021; Yuanfeng et al., 2021). And the hydrolysis of Malonyl isoflavone glucosides in the cecum produces aglycones (Yonemoto-Yano et al., 2014). In addition, it has recently been reported that dipeptidyl peptidase 4 (DPP4), a hydrolase derived from microorganisms, can collaboratively degrade in glucagon-like peptide-1 (GLP-1) in vivo, thereby influencing the metabolism of natural product drugs (Wang K. et al., 2023). This underscores the importance of investigating the metabolic influence of gut microbiome-derived hydrolases (GMDH) on natural products.
2 Regulation of natural products by gut microbiome-derived hydrolases
As a superorganism composed of 10 to 10 trillion individuals, the gut microbiome possesses a gene set that is roughly 150 times larger than the human gene set (Levan et al., 2019; Nishida et al., 2022). It is estimated that the human gut microbiome alone harbors around 1000-1500 different species, many of which remain unidentified (Qin et al., 2010). Recent advancements in genomics technology have sparked increased interest among scientific researchers in exploring the role of intestinal microorganisms and their encoded enzymes. These gut microbiota in human intestinal metabolism by providing enzymes that are not produced by the human body. These enzymes are essential for breaking down complex polysaccharides, metabolizing drugs, and carrying out various metabolic functions (Rakoff-Nahoum et al., 2016; Zhao et al., 2022; Raba and Luis, 2023). Glycoside hydrolases, amid hydrolases, carboxylic esterase, bile saline hydrolases, and epoxide hydrolases produced by microorganisms directly change the specific structures of these natural products. The following article takes these five GMDHs as examples to describe the metabolic regulation of natural products by gut microbiome-derived hydrolases.
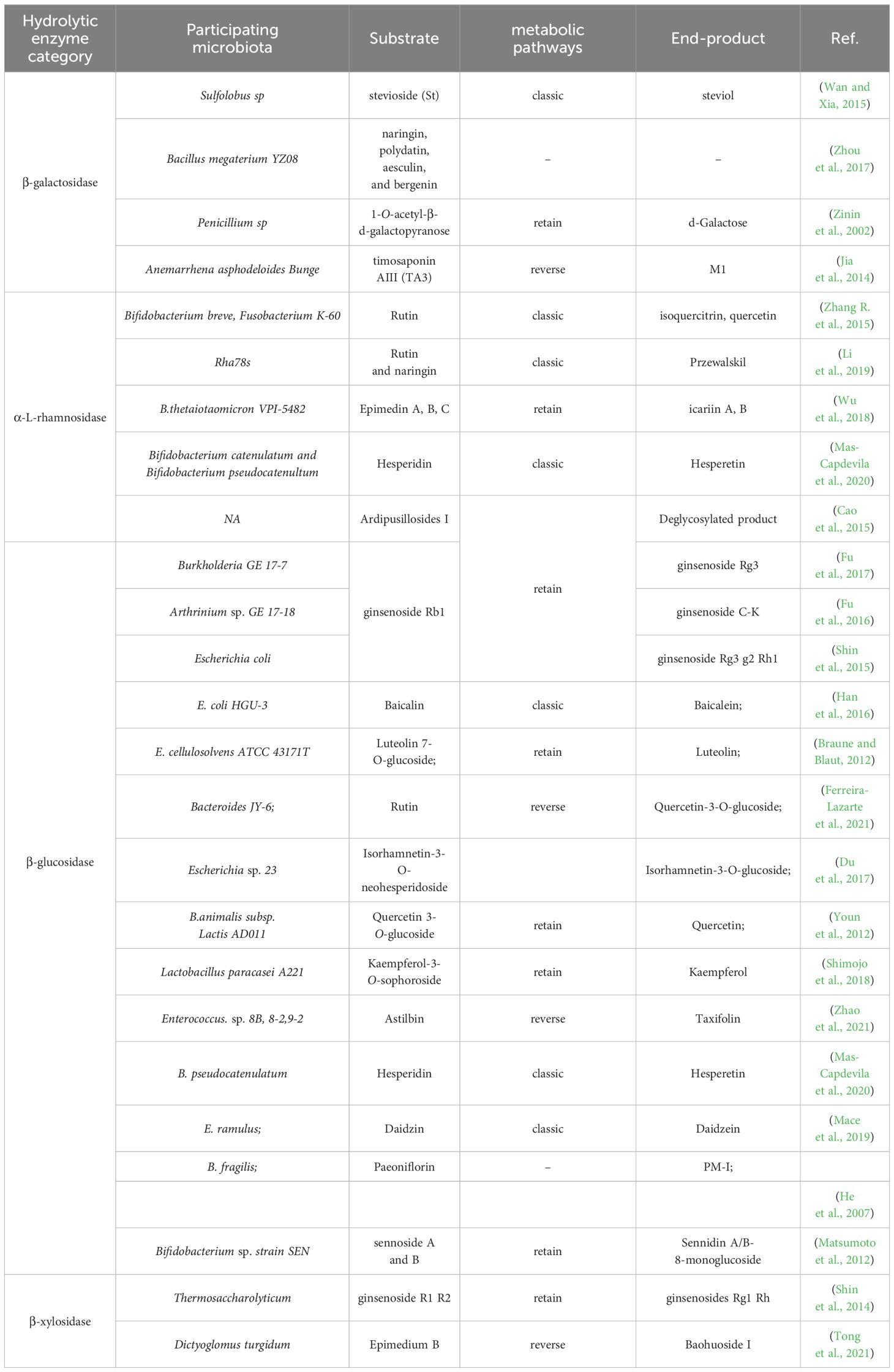
2.1 Glycoside hydrolase
Gut microbiome-derived glycoside hydrolases catalyze the hydrolysis of glycosidic bonds in glycoside natural products to produce hemiacetal and corresponding free glycoside ligands (Xu et al., 2023). These enzymes play a crucial role in altering the structure and bioavailability of glycosides, which are commonly found in natural drugs and considered ‘prodrugs’ in pharmacokinetics (Zhou et al., 2022). Interestingly, the process of gut microbiome hydrolyzing glycoside compounds is mutually beneficial. Glycoside hydrolases from gut microbiota break down glycosidic bonds in glycosides using dicarboxylic acid residues and water molecules, releasing free glycosides and glycogens. These free glycosides can serve as energy sources to support the growth of specific microorganisms in the gut (Braune and Blaut, 2016). Glycoside hydrolases commonly catalyze hydrolysis reactions using a retention mechanism, as shown in Figure 1A. However, there are cases where hydrolysis reactions take place through inversion catalysis, as shown in Figure 1B. In inverting glucosidases, the acceptor molecule attacks the anomeric carbon from the opposite direction without a second flip of the configuration, leading to the formation of products with different configurations (Coines et al., 2019). Gut microbiome-derived glycosidases, based on their substrate specificity and catalytic mechanism, can be categorized as β-D-glucosidase, β-xylosidase, β-galactosidase, α-L-mannosidase, sialidase, among others. Table 1 presents a summary of natural product drug hydrolysis by gut microbiome-derived glycosidases and their respective catalytic mechanisms, aiming to elucidate the involvement of endogenous hydrolases from the intestinal microbiome in the in vivo modification of natural product drugs.
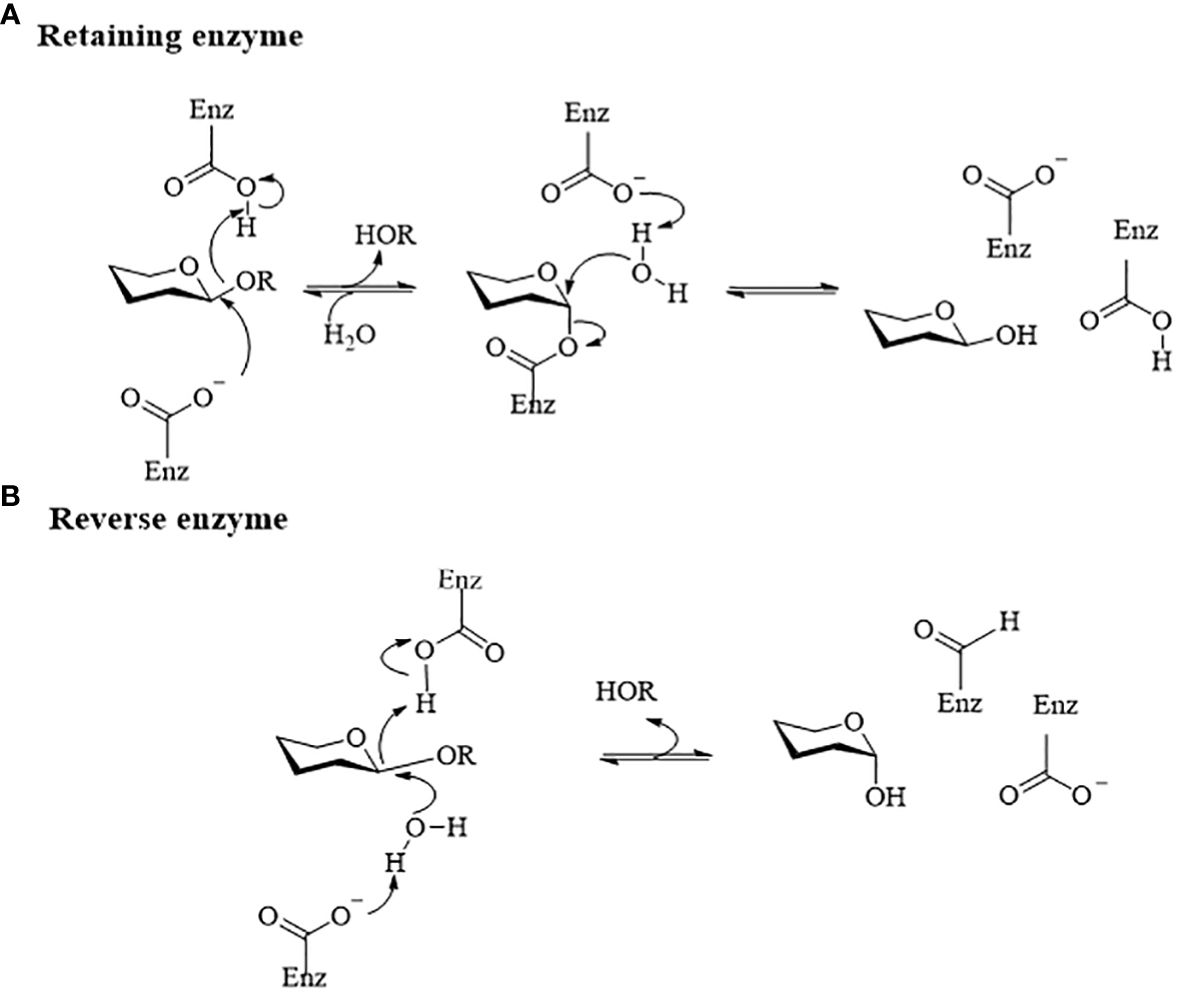
Figure 1 Diagram of the catalytic mechanism of glucoside hydrolase (Honda and Kitaoka, 2006). (A) Retention catalytic mechanism. Their catalytic mechanisms typically involve a ‘twostep’ process. In the first step, a carboxyl anion acts as a nucleophile and attacks the anomeric carbon on the glycosidic bond. Following the formation and breaking of bonds, the anomeric carbon configuration of the glycosyl molecule changes, forming an ester bond with the nucleophilic carboxyl group to produce a glycosylenzyme covalent intermediate and release a glycolipid molecule. In the second step, the active hydroxyl hydrogen of the glycosyl acceptor molecule interacts with the dissociated generalized acidbase pair carboxyl ion. This leads to the formation of an oxygencontaining carbocation-like transition state at the anomeric carbon, resulting in a second flip of the anomeric carbon configuration and the formation of a covalent bond with the acceptor hydroxyl oxygen to complete the reaction. (B) Reverting enzyme mechanism. The hydron molecule attacks the anomeric carbon from the opposite direction without a second flip of the configuration, leading to the formation of products with different configurations.
The study demonstrates that steric hindrance plays a significant role in the hydrolysis of specific flavonoids by glycosidases in the process of intestinal microbial fermentation. While there is a considerable body of research on β-D-glucosidases, there is a noticeable gap in the literature regarding the impact of other glycosidases on natural product drugs. Therefore, further investigation is warranted to delve into the alteration of natural product drugs by various types of glycosidases, in order to comprehensively grasp the mechanism by which intestinal microorganisms act on natural product drugs and lay the groundwork for personalized treatment.
2.2 Amidohydrolase
Amide bonds are a prevalent structural feature in numerous natural product drugs, with approximately 25% of them containing at least one amide bond (Jamalifard et al., 2019). Therefore, explaining the interaction between microbiome-derived amide hydrolase and natural product drugs is important for the development of new natural product drugs (Narendar Reddy et al., 2019). The catalytic mechanism of amidohydrolase is illustrated in Figure 2. However, the catalytic pathways involved in the metabolism of natural products by the body’s gut microbiota require further investigation (Wu et al., 2020). The amidohydrolase from Escherichia coli YqfB is currently the smallest monomeric amide hydrolase known, with catalytic activity towards N4-acylated cytosines found in natural products (Stanislauskienė et al., 2020). Common microbial-derived amide hydrolases are categorized into lactam hydrolases, ureases, fatty acid amide hydrolases, and broad-spectrum substrate amidases based on their respective substrates. Among these, lactam hydrolases have been extensively studied and documented (Bush, 2018). The impact of antibiotic natural product drugs on intestinal microorganisms is substantial. Following antibiotic treatment, there is a significant reduction in the species and abundance of the gastrointestinal microbiome (Maier et al., 2021). Consequently, certain intestinal microorganisms have developed amidohydrolases as a defense mechanism to degrade antibiotics and develop resistance. For instance, Pseudomonas cereus exhibits antibiotic resistance by utilizing endogenous amide hydrolase to catalyze the side chains D-phenylglycine and D-p-hydroxyphenyl glycine of β-lactam antibiotics (Pang et al., 2019; Camiade et al., 2020). Interestingly, previous perceptions of amide hydrolase produced by gut microbiota as detrimental to amide antibiotics have shifted. Recent studies indicate that introducing engineered bacteria that produce amidohydrolase in the intestine can reduce the likelihood of intestinal microorganisms developing antibiotic resistance post-treatment (Cubillos-Ruiz et al., 2022). Furthermore, urease, an amidohydrolase enzyme involved in the body’s urea cycle, seems to possess similar properties. Helicobacter pylori, for instance, sustains its colonization in the gastric mucosa by secreting urease (Fischbach and Malfertheiner, 2018). However, recent studies suggest that the proliferation of certain urease-positive bacteria like Streptococcus thermophilus and Streptococcus salivarius plays a crucial role in maintaining the balance of endogenous ammonia molecules in the body (Ni et al., 2017; Wang P. et al., 2023).
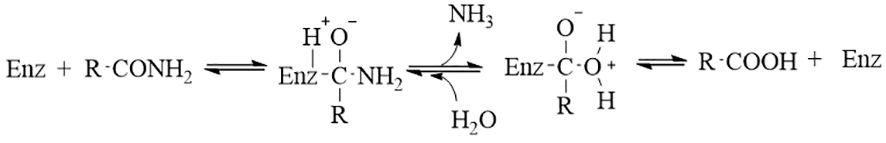
Figure 2 The catalysis mechanism of microbiome-derived amidohydrolase (Wu et al., 2020). Initially, the carbonyl amide forms an enzyme-substrate-acyl tetrahedral intermediate with the enzyme, followed by the rapid release of ammonia, resulting in the formation of an acyl-enzyme complex. Subsequently, the enzyme dissociates, leading to the production of the corresponding acid.
2.3 Carboxylesterase
Carboxylesterase (CES) are widely distributed in various tissues and organs of the body and are highly expressed in the liver, kidney, and small intestine (Hughey and Crawford, 2019). Historically, the lack of identified endogenous substrates led to the belief that the sole role of carboxylesterases was to shield cells from lipid bond-containing compounds (Hatfield et al., 2016). It is hypothesized that the hydrolysis of lipid natural products by carboxylesterase enzymes derived from intestinal microorganisms could serve as a mechanism to shield bacteria from external xenobiotics. The catalytic mechanism of CES primarily involves attacking the oxygen electrophile of the carbonyl carbon in the substrate with the help of serine residues. This forms an acylated enzyme intermediate, which is subsequently hydrolyzed to generate a new active product following the rearrangement of the carbonyl group. CES is particularly important in the microbial metabolism of natural products that contain lipid bonds in their molecular structure (Redinbo and Potter, 2005). Zhao’s study demonstrated that microbiome-derived CES catalyzes the conversion of albiflorin, an antidepressant natural product, to benzoic acid, which has a higher likelihood of crossing the blood-brain barrier and acting within the central nervous system (Zhao et al., 2018). Jin Yu and Ran Peng observed similar findings in their study on the microbial metabolism of paeoniflorin. They found that CES from Bifidobacterium can facilitate the conversion of paeoniflorin to benzoic acid (Yu et al., 2019; Peng et al., 2022). This indicates that CES from gut microorganisms may have a significant impact on the metabolism of natural antidepressants containing benzoic acid structures. Intestinal microorganism-derived CES plays a crucial role in the metabolism of lignoside by hydrolyzing it into salidroside and tyrosol (Yu H. et al., 2023). This enzyme also facilitates the conversion of certain natural product drugs with toxic effects into non-toxic products. For instance, diester diterpenoid alkaloids are transformed into less toxic monoester diterpene alkaloids through the hydrolysis of the C-8 and C-14 ester bonds (Zhang M. et al., 2015). Research conducted by Ramya revealed that CES derived from B. cereus KC985225 in P.xylosida can enhance indoxacarb degradation efficiency by up to 20% (Ramya et al., 2016). Therefore, the structural design of lipid-based natural product insecticides may need to be modified to prevent degradation by carboxylesterases in the insect gut. In summary, microbiome CES plays a crucial role in mitigating the toxicity of natural product drugs. Emphasizing their catalytic effects on natural product drugs is essential for laying the groundwork for the development of novel natural product drugs.
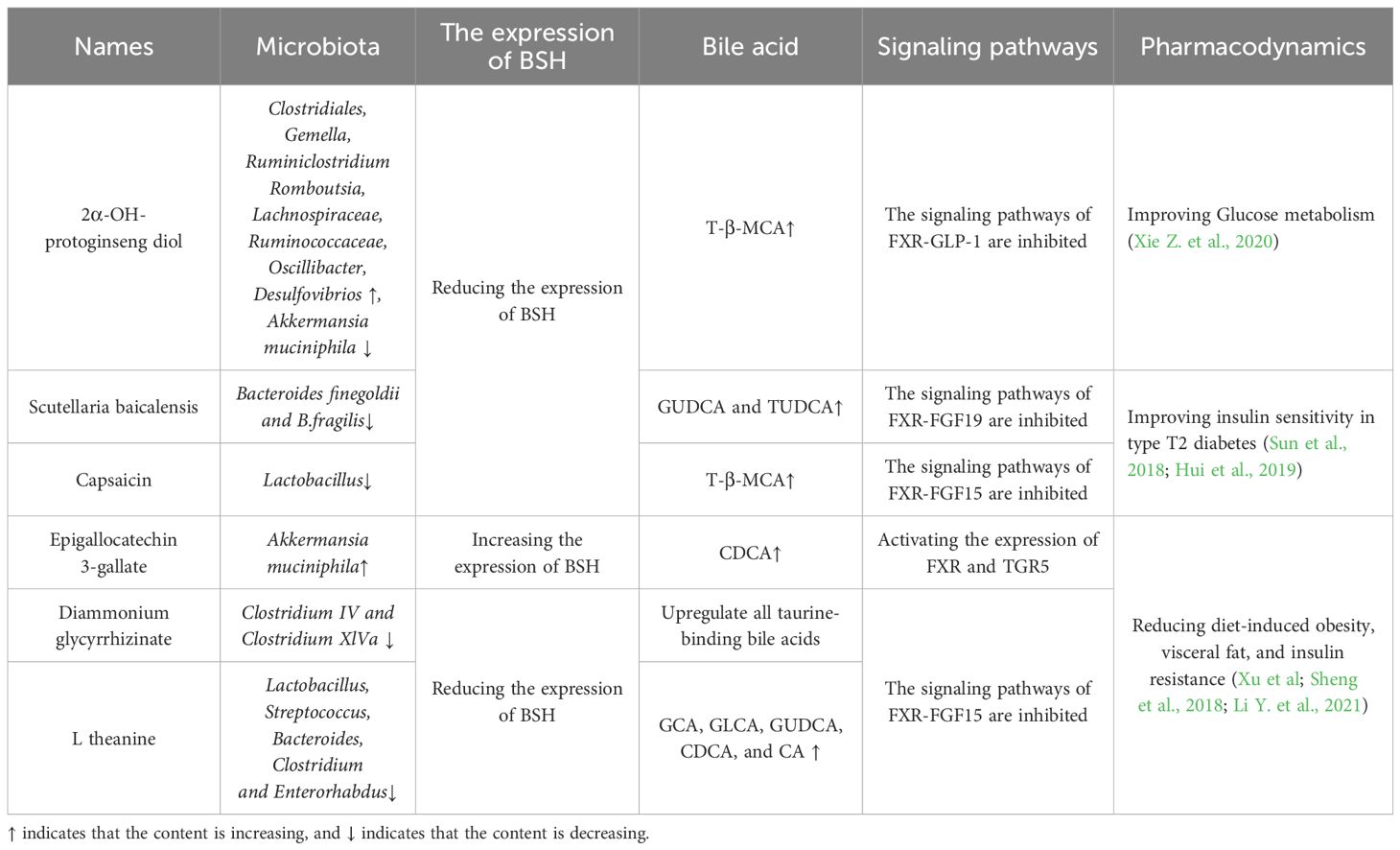
2.4 Bile salt hydrolase
Bile salt hydrolase (BSH) is a common enzyme found in the gastrointestinal tract of mammals. It is produced intracellularly by intestinal microbiota during growth and reproduction (Foley et al., 2023). The production of BSH not only assists intestinal microorganisms in dealing with high concentrations of bile salts in the intestine, but also supports the colonization and adhesion of certain beneficial bacteria (Joyce et al., 2014) The majority of gut microbiota have been found to exhibit BSH activity, including Lactobacillus plantarum, Bifidobacterium sp, Bacteroides fragilis, Bacteroides vulgatus, Clostridium perfringens, Listeria monocytogenes, Lactobacillus and Bifidobacteria (Ruiz et al., 2021, Fiorucci and Distrutti, 2015; Wang et al., 2021). As illustrated in Figure 3, BSH primarily catalyzes the hydrolysis of endogenous steroidal natural product BAs, releasing bound BAs in the intestinal bile acid pool into free bile acids and amino acid residues (Rimal et al., 2024). Numerous studies have highlighted the significance of BSH-mediated unbinding in BAs metabolism, impacting cholesterol synthesis, lipid metabolism, and glucose metabolism (Shen et al., 2021; Zhu et al., 2022). Recent research has also revealed that BSH enzymes are involved in degrading antibiotics, in addition to their role in BAs unbinding. For instance, Hiroyuki discovered that a BSH enzyme derived from Lactobacillus paracasei JCM 5343T not only shows resistance to bile salt stress but also mitigates the toxicity of β-lactam antibiotics to a certain extent (Kusada et al., 2022). In summary, In addition to BAs metabolism, there are numerous unexplored facets regarding the catalytic impact of gut microbiome-derived BSH on natural product drugs, including their response to antibiotic stress, which warrants further investigation.
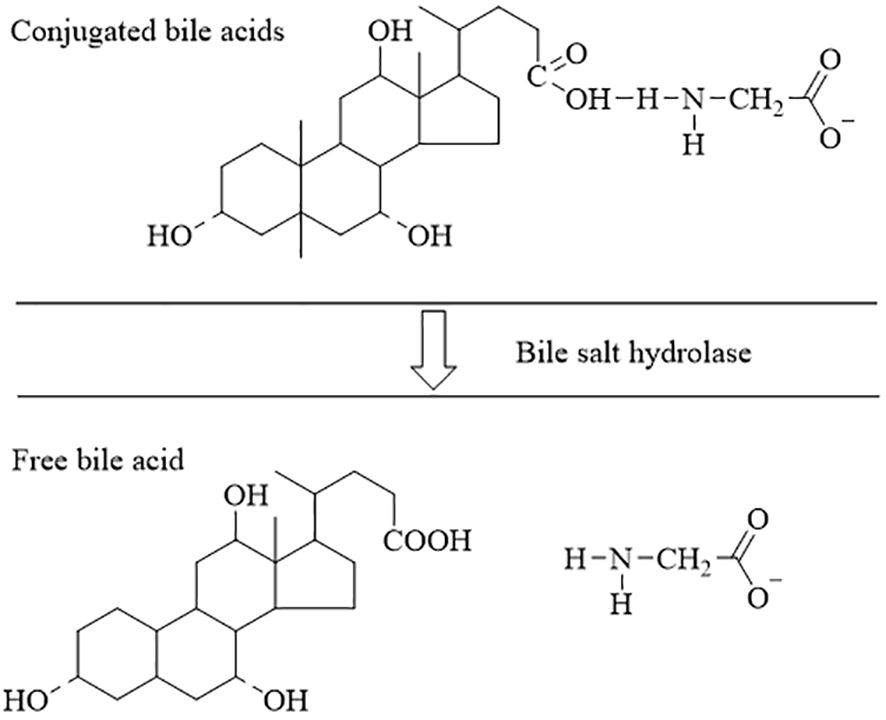
Figure 3 The process of dissociation of bile acids (Fiorucci and Distrutti, 2015; Ruiz et al., 2021; Wang et al., 2021). The conjugated bile acids are hydrolyzed to free bile acids by the action of the BSH.
2.5 Epoxide hydrolases
Epoxide hydrolases (EHs) are a significant group of catalysts that play a key role in converting epoxide compounds to decrease their reactivity. The widely acknowledged catalytic mechanisms are illustrated in Figure 4. The activity of EHs within the gut microbiota was first identified as far back as 1978 using gas chromatography-mass spectrometry (Hwang and Kelsey, 1978). However, there are still few reports on their metabolic effects on natural product drugs. Jan Madacki and colleagues discovered that Mycobacterium tuberculosis, a pathogenic microorganism responsible for intestinal and pulmonary tuberculosis in humans, can hydrolyze 9,10-cis-epoxy stearic acid into diol in vitro (Madacki et al., 2021). Previous research indicates that 12,13-diHOME, a hydrolysis product of gut microbial-EHs in newborns, could serve as a potential risk marker for childhood asthma. However, the precise substrate for this compound remains unknown (Levan et al., 2019). These findings suggest that intestinal microbial-derived EHs could play a significant role in endogenous natural product metabolism. Most studies on EHs currently focus on biocatalysts. Microbiome-derived EHs, unlike those from animals and plants, do not rely on coenzymes for catalysis, making them a promising class of biocatalysts for synthesizing complex natural products (Wang et al., 2015). For example, they have been used in the synthesis of epichlorohydrin (Liang et al., 2019), chiral phenyl ethylene glycol (Jia et al., 2011), and aryl glycidyl ether (Xu et al., 2020; Zhang et al., 2020), among others.
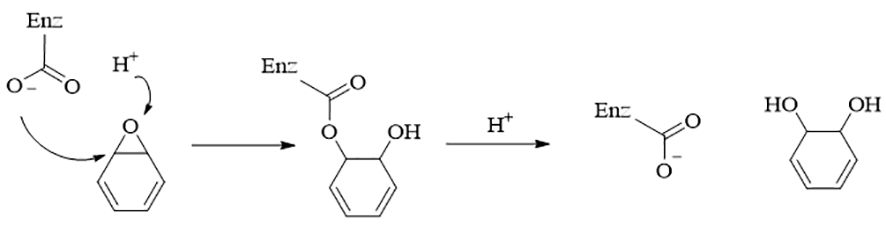
Figure 4 The catalysis mechanism of EHs. EHs form an enzyme-substrate ester-type complex with epoxide with the help of amino acid residues, which hydrolyzes to form diols (Chen et al., 2012).
3 Effects of natural product drugs on microbiologically derived hydrolases
The gut microbiome hydrolases play a crucial role in transforming the active ingredients of natural product drugs, thereby influencing the potency and bioavailability of these drugs (Zhang et al., 2018). This relationship is bidirectional, as some natural product drugs can target GMDH to modulate metabolic pathways or counteract drug effects. For instance, primary BAs have been found to inhibit microbiota-derived BSH from bacteria like Staphylococcus, Balantidium, Pneumococcus, and Enterococcus (Ridlon et al., 2016). Recent research indicates that BAs can induce DNA damage by disrupting RNA secondary structure in intestinal microbiota, impacting the levels of bile salt hydrolase. This mechanism may be crucial for maintaining bile acid pool homeostasis in the body (Watanabe et al., 2017; Bustos et al., 2018). In addition, certain natural products can also inhibit glycoside hydrolases from gut microbiota. For example, compounds like diterpenoids and linolenic acid from Rubidium strobili have demonstrated inhibition of glucosidase in E. coli (Ganzon et al., 2022; Yu et al., 2022).. Weng and colleagues conducted a comparative analysis of over 30 flavonoids to assess their ability to inhibit glycosidases derived from gut microbiota in vitro. The findings indicated that scutellarein, luteolin, baicalein, quercetin, and scutellarin exhibited significant inhibitory effects on glycoside hydrolases (Weng et al., 2017). The interaction between natural products and GMDH may exhibit dynamic relationships.
4 Interactions between hydrolases and natural product drugs affect disease development
The co-metabolism of drugs by the microbiome can lead to byproducts with different therapeutic effects than the original drug (Lai et al., 2010). Taking amygdalin, for instance, can effectively treat bronchitis and emphysema. However, when glycoside hydrolases from intestinal microorganisms transform amygdalin, it can produce hydrocyanic acid, leading to toxic reactions and worsening the condition (Jaswal and Palanivelu J, 2018). Therefore, investigating the combined regulation of hydrolases and natural drugs on disease occurrence and progression is essential. This synergistic regulation has been extensively documented for diseases such as ulcerative colitis (UC), nervous system disease, diabetes, and cardiovascular disease (Pavlović et al., 2012; Huang et al., 2019; Liu et al., 2021; Sun et al., 2023; Zhang ZW. et al., 2023). The following sections will discuss the impact of the interaction between microbiome hydrolases and natural products on various clinical diseases, aiming to elucidate the importance of this interaction for practical applications.
4.1 Inflammatory bowel disease
As the gut is home to tens of thousands of microorganisms, it plays an important role in the process of intestinal diseases. Inflammatory bowel disease (IBD), comprising ulcerative colitis (UC) and Crohn’s disease (CD), is a chronic inflammatory condition that primarily impacts the gastrointestinal tract. Studies indicate that various natural products may offer relief for IBD. For instance, baicalin (Zhu et al., 2016; Wu Q. et al., 2023), Sophora alopecuroides L (Jia et al., 2020), fibro mushroom polysaccharid (Nguepi Tsopmejio et al., 2023), and gallic acid (Pandurangan et al., 2015) have shown promising therapeutic benefits in animal models of chemically induced colitis. We have previously explored the regulatory relationship between GMDH and natural products. However, the role of interactions between GMDH and natural product drugs in enteritis remains to be explored. The provided Figure 5 outlines the roles of key natural products metabolized by intestinal microbiome hydrolases in enteritis, aiming to elucidate the mechanisms through which intestinal microorganisms can mitigate intestinal inflammation and lay a foundation for future research.
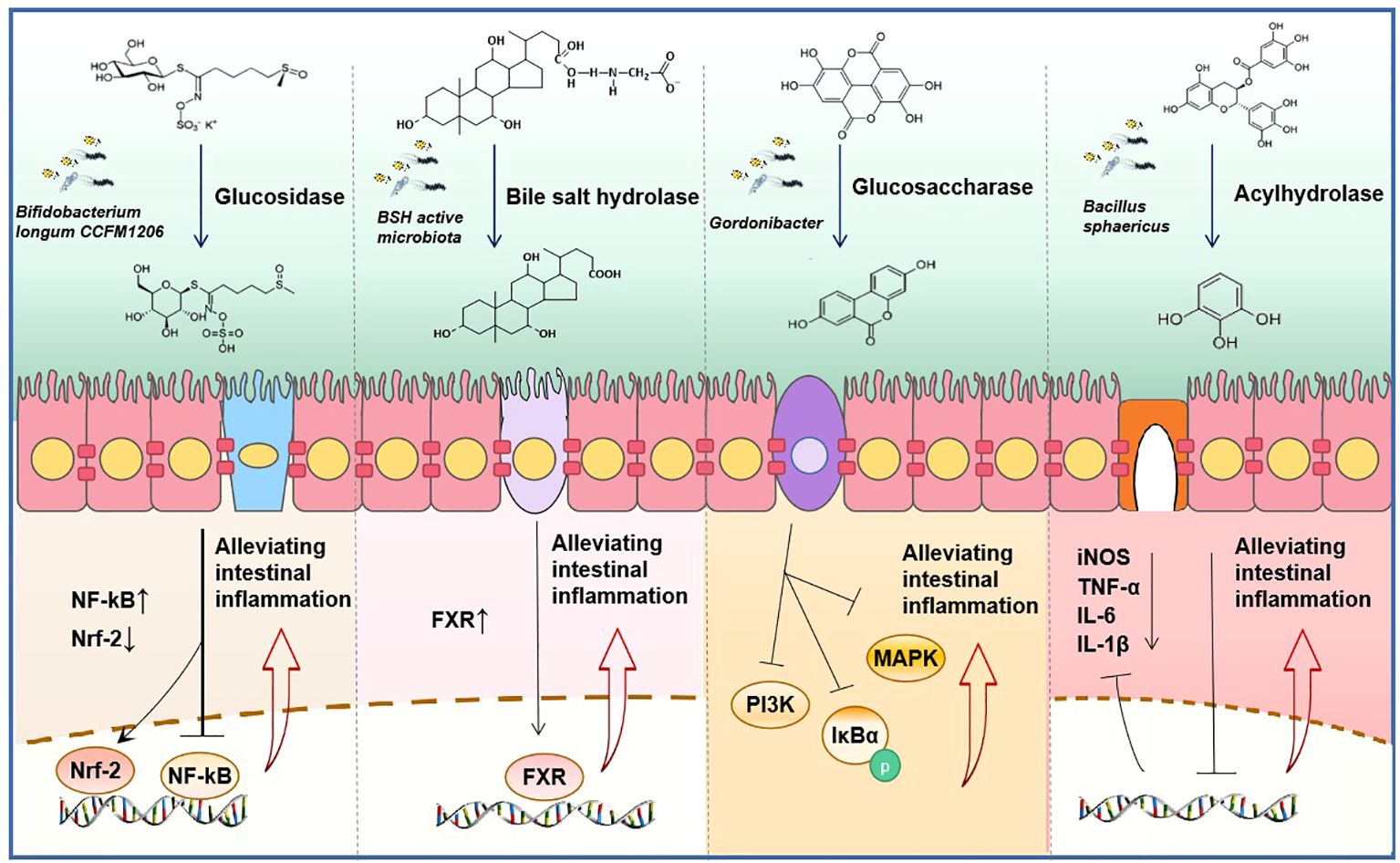
Figure 5 Synergistic anti-inflammatory mechanism of natural product drugs and get microbiome derived hydrolase (GMDH). From left to right, sulforaphane is hydrolyzed by glycosidases produced by Bifidobacterium longum CCFM1206 to its active form, reducing the expression of the inflammatory mediator nuclear factor kappa-B (NF-κB) and exerting an anti-inflammatory effect (Wu J. et al., 2023). The BSH produced by microbiota converts conjugated bile acid into free bile acid, which activates the expression of farnesoid X Receptor (FXR) and exerts an anti-inflammatory effect (Gadaleta et al., 2022). Ellagic acid is metabolized to produce urolithin A by glucosidase, which inhibits the phosphorylation of IκBα and the activation of mitogen-activated protein kinase (MAPK) and phosphatidyl inositol 3-kinase (PI3K), thus displaying anti-inflammatory properties (Selma et al., 2014; Kujawska and Jodynis-Liebert, 2020; Abdelazeem et al., 2021). Acyl hydrolase derived from Bacillus sphaericus converts catechin gallate into pyrogallol, leading to the inhibition of the secretion of inflammatory factors (Raghuwanshi et al., 2011; López de Felipe et al., 2014).
In summary, the regulatory role of gut microbiome hydrolases and natural products in the treatment and prevention of enteritis is a promising avenue for research. Exploring how to harness the benefits of gut microbiota and natural products in disease management is a growing area of interest.
4.2 Type 2 diabetes
A Mendelian randomization study conducted by Cheng demonstrated a robust association between type 2 diabetes and gastrointestinal tract diseases (Chen et al., 2023). Recent research has highlighted the importance of GMDH in the pathogenesis of type 2 diabetes. Natural products such as flavonoids, lipids, saponins, and BAs have been found to have beneficial anti-diabetic properties (Fan et al., 2023). Acarbose, a common antidiabetic medication, is subject to hydrolysis by K. grimontii TD1 bacteria in the human intestine, leading to potential resistance to acarbose (Tian et al., 2023). The previous discussion focused on the relationship between natural product drugs and gut microbes, without delving into their specific role in diabetes. Recent evidence suggests that the microbiota-derived BSH may serve as a key factor in this interaction. Natural product drugs have been found to decrease BSH levels in the intestine by inhibiting microbiota-produced BSH enzymes. This inhibition leads to alterations in downstream signaling pathways of BAs, ultimately resulting in hypoglycemic effects. Table 2 provides a list of natural products that have shown potential in reducing glucose levels through BSH modulation, offering novel insights and strategies for diabetes prevention and treatment.
In addition to BSH, the aglycone produced by some flavonoids through hydrolysis also exhibits inhibitory effects on the onset and progression of diabetes. The aglycone produced by some flavonoids through hydrolysis also exhibits inhibitory effects on the onset and progression of diabetes. For instance, ginsenoside Rb1 stimulates GLP-1 secretion (Yang et al., 2021), astragaloside IV increases the levels of intestinal butyric acid (Gong et al., 2021), and resveratrol reduces the levels of inflammatory factors in the intestine (Cai et al., 2020). In conclusion, focusing on bile salt hydrolase and glycoside hydrolase among GMDH may be crucial for the development of new anti-diabetic natural product drugs.
4.3 Atherosclerosis
Atherosclerosis is a significant type of cardiovascular disease characterized by the formation of plaques containing necrotic cores, calcified regions, accumulated modified lipids, inflamed smooth muscle cells, endothelial cells, leukocytes, and foam cells (Fan and Watanabe, 2022). Recent studies have demonstrated that hydrolysates of flavonoid microbiota can have positive impacts on atherosclerosis. For instance, 3-hydroxyphenyl, a hydrolytic metabolite of quercetin, has been shown to effectively lower arterial blood pressure and induce vascular relaxation in rats (Najmanová et al., 2016). Ginsenosides derived from steroids have demonstrated protective effects against atherosclerosis, with this protection being attributed to intestinal microbiome hydrolases (Zhang N-N. et al., 2023). The microbiome hydrolysates Rb1, Rg2, Rg3, and compound K have been found to alleviate atherosclerosis through various pathways. For instance, Rb1 was found to inhibit neointimal hyperplasia induced by balloon infusion in rats by suppressing vascular smooth muscle cell proliferation and phenotypic switching (Yang et al., 2018). Rg1 and Rg2 play a role in boosting the levels of macrophage autophagy proteins, which are vital for sustaining macrophage function in advanced atherosclerosis (Cho et al., 2013). Additionally, compound K hinders the advancement of atherosclerosis by targeting NF-κB and JNK-MAPK pathways (Coines et al., 2019). In conclusion, the hydrolysis and conversion of ginsenosides in the intestines are crucial for unleashing the therapeutic benefits of ginsenosides and easing atherosclerosis within the body.
5 Limitations of research on gut microbiome-derived enzymes
The development of intestinal microbial-derived enzymes faces three main challenges. Firstly, pinpointing a specific intestinal microorganism during the metabolism of natural product drugs is challenging. Researchers commonly employ methods like fecal bacterial transplantation, antibiotic treatment, and 16S rRNA analysis to study the intestinal microbial populations involved in drug metabolism (Li Q. et al., 2021; Zeng et al., 2023). However, these methods have limitations due to inter-individual variability and variances in the gastrointestinal tract between human and animal models (Yu Y. et al., 2023). Moreover, besides bacteria, the intestine also harbors fungi, viruses, and phages. The current analysis methods primarily focus on bacteria, neglecting the roles of other microorganisms in drug metabolism (Shao et al., 2023). The advancement of high-throughput single-cell genome sequencing technology holds promise for enhancing the resolution of intestinal microorganisms (Zheng et al., 2022). The second challenge involves incubating differential strains in vitro. Bacteria can only be studied and identified through in vitro culturing, yet isolating specific groups from the extensive intestinal microbiome proves challenging. The microbiome-derived metabolism screening platform, created by Bahar Javdan, enables high-throughput screening of bacterial groups related to natural product metabolism. However, there are limitations to the strains that can be cultured in vitro (Javdan et al., 2020). Utilizing culturomics methods may enable the isolation of a greater diversity of bacteria in the future, potentially offering a solution to this technical challenge (Chang et al., 2019). A third challenge is the identification of key gut microbe-derived enzymes, as discrepancies persist between enzymes found in human gut microbes and data from gut microbial genomes. Approximately 40% of protein sequences lack functional annotation, making it difficult to link many enzyme sequences to specific functions (Almeida et al., 2021). Moreover, the conservation of similar gene sequences often leads to misannotation and over-prediction of enzyme molecular functions in public databases, hindering the identification of derivative enzymes in the gut microbiome (Schnoes et al., 2013; Jia et al., 2017). The advancement of bioinformatics and computer-assisted techniques can be valuable tools in overcoming this challenge (Sharma et al., 2017; Guthrie et al., 2019; Malwe et al., 2023). In summary, the advancement of single-cell genome sequencing technology, culture-omics technology, and bioinformatics analysis platforms holds promise for enhancing our comprehension of microbial drug metabolism in the future.
6 Conclusion
Although natural products are often valuable as lead compounds, they are seldom directly applicable in clinical settings. The bioactivation and potential health benefits of many natural products, such as flavonoids, alkaloids, and lignin, heavily rely on gut microbes acting as substrate processing plants (Singh et al., 2020).To enhance their clinical utility, it is imperative to modify the structure of natural products. Oral natural product drugs come into direct contact with the intestinal microbiota. Structural alterations of drugs by hydrolases derived from the intestinal microbiota are a crucial strategy to enhance drug efficacy, refine chemical structure, and mitigate adverse reactions (Zhang et al., 2018; Weersma et al., 2020; Xie Y. et al., 2020). Individual variations in the intestinal microbiome may account for differences in the bioavailability of certain natural product drugs among various populations, offering insights for personalized treatment approaches. The impact of natural products on microbiome hydrolase suggests a need for increased vigilance regarding potential drug interactions when combining natural product drugs. Certain drugs may interact with clinical drugs that undergo enterohepatic circulation via microbiome hydrolase-mediated mechanisms, highlighting the importance of studying combinations of natural product drugs and traditional Chinese medicine prescriptions. For instance, antibiotics like cefalexin, tetracycline, and erythromycin have been shown to notably alter the oral pharmacokinetic properties of baicalin (Kang et al., 2014). In studies, it has been found that low concentrations of clove water extract can competitively inhibit the activity of the β-D-glucuronide enzyme from Escherichia coli by producing the hydrolyzed product urolitin M5 (Bai et al., 2021). Additionally, amentoflavone was found to strongly inhibit the hydrolysis of 6, 8-dichloro-7-hydroxy-9,9-dimethylacridin by β-D-glucuronide (Tian et al., 2021). It is crucial to consider gut microbial hydrolases when investigating the interactions between oral natural products and drugs. All in all, the interplay of GMDH with natural product drugs significantly impacts host health, disease progression, and therapeutic outcomes, offering valuable insights into disease mechanisms and potential drug targets.
Author contributions
JH: Writing – original draft. XL: Writing – review & editing. JZ: Writing – review & editing. RW: Writing – review & editing. XC: Supervision, Writing – original draft. GL: Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (No. 82173738), Natural Science Foundation of Gansu Province (20JR10RA010), the Key Research and Development Plan of Ningxia Hui Autonomous Region (2020BEG03060), and the scientific research project of Ningxia Medical University (XT2020013).
Acknowledgments
The authors thank Xiaofeng Liu, Rong Wang, and Junmin Zhang for their constructive comments, as well as Xinyuan Cao and Ge Liu for their assistance with the figures. The authors also thank the scientific drawing website BioRender, where the graphical abstract was created.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdelazeem, K. N. M., Kalo, M. Z., Beer-Hammer, S., Lang, F. (2021). The gut microbiota metabolite urolithin a inhibits NF-κb activation in LPS stimulated BMDMs. Sci. Rep. 11, 7117. doi: 10.1038/s41598-021-86514-6
Almeida, A., Nayfach, S., Boland, M., Strozzi, F., Beracochea, M., Shi, Z. J., et al. (2021). A unified catalog of 204,938 reference genomes from the human gut microbiome. Nat. Biotechnol. 39, 105–114. doi: 10.1038/s41587-020-0603-3
Bai, Y., Chen, L., Cao, Y. F., Hou, X. D., Jia, S. N., Zhou, Q., et al. (2021). Beta-glucuronidase inhibition by constituents of mulberry bark. Planta Med. 87, 631–641. doi: 10.1055/a-1402-6431
Braune, A., Blaut, M. (2012). Intestinal bacterium eubacterium cellulosolvens deglycosylates flavonoid c- and o-glucosides. Appl. Environ. Microbiol. 78 (22), 8151–8153. doi: 10.1128/AEM.02115-12
Braune, A., Blaut, M. (2016). Bacterial species involved in the conversion of dietary flavonoids in the human gut. Gut Microbes 7 (3), 216–234. doi: 10.1080/19490976.2016.1158395
Bush, K. (2018). Past and present perspectives on β-lactamases. Antimicrobial. Agents Chemother. 62 (10). doi: 10.1128/AAC.01076-18
Bustos, A. Y., Font de Valdez, G., Fadda, S., Taranto, M. P. (2018). New insights into bacterial bile resistance mechanisms: The role of bile salt hydrolase and its impact on human health. Food Res. Int. 112, 250–262. doi: 10.1016/j.foodres.2018.06.035
Cai, T.-T., Ye, X.-L., Li, R.-R., Chen, H., Wang, Y.-Y., Yong, H.-J., et al. (2020). Resveratrol modulates the gut microbiota and inflammation to protect against diabetic nephropathy in mice. Front. Pharmacol. 11. doi: 10.3389/fphar.2020.01249
Camiade, M., Bodilis, J., Chaftar, N., Riah-Anglet, W., Gardères, J., Buquet, S., et al. (2020). Antibiotic resistance patterns of pseudomonas spp. Isolated from faecal wastes in the environment and contaminated surface water. FEMS Microbiol. Ecol. 96 (2). doi: 10.1093/femsec/fiaa008
Cao, W. Y., Wang, Y. N., Wang, P. Y., Lei, W., Feng, B., Wang, X. J. (2015). Ardipusilloside-I metabolites from human intestinal bacteria and their antitumor activity. Molecules. 20, 20569–20581. doi: 10.3390/molecules201119719
Chang, Y., Hou, F., Pan, Z., Huang, Z., Han, N., Bin, L., et al. (2019). Optimization of culturomics strategy in human fecal samples. Front. Microbiol. 10, 2891. doi: 10.3389/fmicb.2019.02891
Chen, Y., Chen, C., Wu, X. (2012). Dicarbonyl reduction by single enzyme for the preparation of chiral diols. Chem. Soc. Rev. 41 (5), 1742–1753. doi: 10.1039/c1cs15230k
Chen, J., Yuan, S., Fu, T., Ruan, X., Qiao, J., Wang, X., et al. (2023). Gastrointestinal consequences of type 2 diabetes mellitus and impaired glycemic homeostasis: A mendelian randomization study. Diabetes Care. doi: 10.2337/dc22-1385
Cho, Y. S., Kim, C. H., Ha, T. S., Lee, S. J., Ahn, H. Y. (2013). Ginsenoside rg2 inhibits lipopolysaccharide-induced adhesion molecule expression in human umbilical vein endothelial cell. Korean J. Physiol. Pharmacol. 17, 133–137. doi: 10.4196/kjpp.2013.17.2.133
Coines, J., Raich, L., Rovira, C. (2019). Modeling catalytic reaction mechanisms in glycoside hydrolases. Curr. Opin. Chem. Biol. 53, 183–191. doi: 10.1016/j.cbpa.2019.09.007
Cubillos-Ruiz, A., Alcantar, M. A., Donghia, N. M., Cárdenas, P., Avila-Pacheco, J., Collins, J. J. (2022). An engineered live biotherapeutic for the prevention of antibiotic-induced dysbiosis. Nat. Biomed. Eng. 6, 910–921. doi: 10.1038/s41551-022-00871-9
Du, L. Y., Zhao, M., Tao, J. H., Qian, D. W., Jiang, S., Shang, E. X., et al. (2017). The metabolic profiling of isorhamnetin-3-o-neohesperidoside produced by human intestinal flora employing UPLC-Q-TOF/MS. J. Chromatogr. Sci. 55, 243–250. doi: 10.1093/chromsci/bmw176
Fan, J., Watanabe, T. (2022). Atherosclerosis: known and unknown. Pathol. Int. 72 (3), 151–160. doi: 10.1111/pin.13202
Fan, X., Zhang, Y., Song, Y., Zhao, Y., Xu, Y., Guo, F., et al. (2023). Compound danshen dripping pills moderate intestinal flora and the TLR4/MyD88/NF-κB signaling pathway in alleviating cognitive dysfunction in type 2 diabetic kk-ay mice. Phytomed.: Int. J. Phytother. Phytopharmacol. 111, 154656. doi: 10.1016/j.phymed.2023.154656
Ferreira-Lazarte, A., Plaza-Vinuesa, L., de Las Rivas, B., Villamiel, M., Muñoz, R., Moreno, F. J., et al. (2021). Production of α-rhamnosidases from Lactobacillus plantarum WCFS1 and their role in deglycosylation of dietary flavonoids naringin and rutin. Int. J. Biol. Macromol. 193, 1093–1102. doi: 10.1016/j.ijbiomac.2021.11.053
Fiorucci, S., Distrutti, E. (2015). Bile acid-activated receptors, intestinal microbiota, and the treatment of metabolic disorders. Trends Mol. Med. 21 (11), 702–714. doi: 10.1016/j.molmed.2015.09.001
Fischbach, W., Malfertheiner, P. (2018). Helicobacter pylori infection. Deutsches Arzteblatt Int. 115 (25), 429–436. doi: 10.3238/arztebl.2018.0429
Foley, M. H., Walker, M. E., Stewart, A. K., O'Flaherty, S., Gentry, E. C., Patel, S., et al. (2023). Bile salt hydrolases shape the bile acid landscape and restrict clostridioides difficile growth in the murine gut. Nat. Microbiol. 8 (4), 611–628. doi: 10.1038/s41564-023-01337-7
Fu, Y., Yin, Z. H., Wu, L. P., Yin, C. R. (2016). Biotransformation of ginsenoside Rb1 to ginsenoside C-K by endophytic fungus Arthrinium sp. GE 17-18 isolated from Panax ginseng. Lett. Appl. Microbiol. 63 (6), 196–201. doi: 10.1111/lam.12606
Fu, Y., Yin, Z. H., Yin, C. Y. (2017). Biotransformation of ginsenoside Rb1 to ginsenoside Rg3 by endophytic bacterium Burkholderia sp. GE 17-7 isolated from Panax ginseng. J. Appl. Microbiol. 122, 1579–1585. doi: 10.1111/jam.2017.122.issue-6
Gadaleta, R. M., Cariello, M., Crudele, L., Moschetta, A. (2022). Bile salt hydrolase-competent probiotics in the management of IBD: Unlocking the “bile acid code”. Nutrients 14 (15). doi: 10.3390/nu14153212
Ganzon, J. G., Liaw, C. C., Lin, Y. C., Lin, Z. H., Wang, C. H., Chen, C. Y., et al. (2022). New ent-kaurene and germacrene derivatives from Mesona procumbens Hemseley and their biological activity. Nat Prod Res, 1–9. doi: 10.1080/14786419.2022.2034811
Gong, P., Xiao, X., Wang, S., Shi, F., Liu, N., Chen, X., et al. (2021). Hypoglycemic effect of astragaloside IV via modulating gut microbiota and regulating AMPK/SIRT1 and PI3K/AKT pathway. J. Ethnopharmacol. 281, 114558. doi: 10.1016/j.jep.2021.114558
Guthrie, L., Wolfson, S., Kelly, L. (2019). The human gut chemical landscape predicts microbe-mediated biotransformation of foods and drugs. eLife 8. doi: 10.7554/eLife.42866.022
Han, D. H., Lee, Y., Ahn, J. H. (2016). Biological synthesis of baicalein derivatives using Escherichia coli. J. Microbiol. Biotechnol. 26 (11), 1918–1923. doi: 10.4014/jmb.1605.05050
Hatfield, M. J., Umans, R. A., Hyatt, J. L., Edwards, C. C., Wierdl, M., Tsurkan, L., et al. (2016). Carboxylesterases: General detoxifying enzymes. Chem Biol. Interact. 259, 327–331. doi: 10.1016/j.cbi.2016.02.011
He, J. X., Goto, E., Akao, T., Tani, T. (2007). Interaction between Shaoyao-Gancao-Tang and a laxative with respect to alteration of paeoniflorin metabolism by intestinal bacteria in rats. Phytomedicine 14 (7-8), 452–459. doi: 10.1016/j.phymed.2006.09.014
Honda, Y., Kitaoka, M. (2006). The first glycosynthase derived from an inverting glycoside hydrolase. J. Biol. Chem. 281 (3), 1426–1431. doi: 10.1074/jbc.M511202200
Huang, F., Zheng, X., Ma, X., Jiang, R., Zhou, W., Zhou, S., et al. (2019). Theabrownin from Pu-erh tea attenuates hypercholesterolemia via modulation of gut microbiota and bile acid metabolism. Nat. Commun. 10 (1), 4971. doi: 10.1038/s41467-019-12896-x
Hughey, C. C., Crawford, P. A. (2019). Pyruvate carboxylase wields a double-edged metabolic sword. Cell Metab. 29 (6), 1236–1238. doi: 10.1016/j.cmet.2019.05.013
Hui, S., Liu, Y., Chen, M., Wang, X., Lang, H., Zhou, M., et al. (2019). Capsaicin improves glucose tolerance and insulin sensitivity through modulation of the gut microbiota-bile acid-FXR axis in type 2 diabetic db/db mice. Mol. Nutr. Food Res. 63 (23). doi: 10.1002/mnfr.201900608
Hwang, K. K., Kelsey, M. I. (1978). Evidence of epoxide hydrase activity in human intestinal microflora. Cancer Biochem. Biophys. 3 (1), 31–35.
Jamalifard, S., Mokhtari, J., Mirjafary, Z. (2019). One-pot synthesis of amides via the oxidative amidation of aldehydes and amines catalyzed by a copper-MOF. RSC Adv. 9 (39), 22749–22754. doi: 10.1039/C9RA04216D
Jaswal, V., Palanivelu, J., Ramalingam, C. (2018). Effects of the gut microbiota on amygdalin and its use as an anti-cancer therapy: Substantial review on the key components involved in altering dose efficacy and toxicity. Biochem. Biophys. Rep. 14, 125–132. doi: 10.1016/j.bbrep.2018.04.008
Javdan, B., Lopez, J. G., Chankhamjon, P., Lee, Y. J., Hull, R., Wu, Q., et al. (2020). Personalized mapping of drug metabolism by the human gut microbiome. Cell 181, 1661–1679.e1622. doi: 10.1016/j.cell.2020.05.001
Jia, B., Jia, X., Kim, K. H., Jeon, C. O. (2017). Integrative view of 2-oxoglutarate/Fe(II)-dependent oxygenase diversity and functions in bacteria. Biochim. Biophys. Acta Gen. Subj. 1861, 323–334. doi: 10.1016/j.bbagen.2016.12.001
Jia, Y., Wu, B., Fan, M., Wang, J., Huang, J., Huang, C. (2014). High-performance liquid chromatography-electrospray ionization tandem mass spectrometry for metabolism study of timosaponin AIII. J. Chromatogr. Sci. 52 (5), 418–422. doi: 10.1093/chromsci/bmt052
Jia, X., Xu, Y., Li, Z. (2011). Regio- and stereoselective concurrent oxidations with whole cell biocatalyst: Simple and green syntheses of enantiopure 1,2-Diols via oxidative kinetic resolution. ACS Catal. 1 (6), 591–596. doi: 10.1021/cs200099k
Jia, Y. Q., Yuan, Z. W., Zhang, X. S., Dong, J. Q., Liu, X. N., Peng, X. T., et al. (2020). Total alkaloids of sophora alopecuroides L. Ameliorated murine colitis by regulating bile acid metabolism and gut microbiota. J. Ethnopharmacol. 255, 112775. doi: 10.1016/j.jep.2020.112775
Joyce, S. A., MacSharry, J., Casey, P. G., Kinsella, M., Murphy, E. F., Shanahan, F., et al. (2014). Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc. Natl. Acad. Sci. U. S. A. 111 (20), 7421–7426. doi: 10.1073/pnas.1323599111
Kang, M. J., Ko, G. S., Oh, D. G., Kim, J. S., Noh, K., Kang, W., et al. (2014). Role of metabolism by intestinal microbiota in pharmacokinetics of oral baicalin. Arch. Pharmacal. Res. 37 (3), 371–378. doi: 10.1007/s12272-013-0179-2
Kujawska, M., Jodynis-Liebert, J. (2020). Potential of the ellagic acid-derived gut microbiota metabolite - Urolithin a in gastrointestinal protection. World J. Gastroenterol. 26 (23), 3170–3181. doi: 10.3748/wjg.v26.i23.3170
Kusada, H., Arita, M., Tohno, M., Tamaki, H. (2022). Bile salt hydrolase degrades β-lactam antibiotics and confers antibiotic resistance on Lactobacillus paragasseri. Front. Microbiol. 13, 858263. doi: 10.3389/fmicb.2022.858263
Lai, R. H., Miller, M. J., Jeffery, E. (2010). Glucoraphanin hydrolysis by microbiota in the rat cecum results in sulforaphane absorption. Food Funct. 1 (2), 161–166. doi: 10.1039/c0fo00110d
Levan, S. R., Stamnes, K. A., Lin, D. L., Panzer, A. R., Fukui, E., McCauley, K., et al. (2019). Elevated faecal 12,13-Dihome concentration in neonates at high risk for asthma is produced by gut bacteria and impedes immune tolerance. Nat. Microbiol. 4 (11), 1851–1861. doi: 10.1038/s41564-019-0498-2
Li, Q., Cui, Y., Xu, B., Wang, Y., Lv, F., Li, Z., et al. (2021). Main active components of Jiawei Gegen Qinlian decoction protects against ulcerative colitis under different dietary environments in a gut microbiota-dependent manner. Pharmacol. Res. 170, 105694. doi: 10.1016/j.phrs.2021.105694
Li, Y., Hou, H., Wang, X., Dai, X., Zhang, W., Tang, Q., et al. (2021). Diammonium glycyrrhizinate ameliorates obesity through modulation of gut microbiota-conjugated BAs-FXR signaling. Front. Pharmacol. 12. doi: 10.3389/fphar.2021.796590
Li, B. C., Zhang, T., Li, Y. Q., Ding, G. B. (2019). Target discovery of novel α-l-rhamnosidases from human fecal metagenome and application for biotransformation of natural flavonoid glycosides. Appl. Biochem. Biotechnol. 189 (4), 1245–1261. doi: 10.1007/s12010-019-03063-5
Liang, Y., Jiao, S., Wang, M., Yu, H., Shen, Z. (2019). Overexpression of epoxide hydrolase in Rhodococcus ruber with high robustness for the synthesis of chiral epichlorohydrin. Process Biochem. 79, 49–56. doi: 10.1016/j.procbio.2018.12.023
Liu, S., He, F., Zheng, T., Wan, S., Chen, J., Yang, F., et al. (2021). Ligustrum robustum alleviates atherosclerosis by decreasing serum tmao, modulating gut microbiota, and decreasing bile acid and cholesterol absorption in mice. Mol. Nutr. Food Res. 65, e2100014. doi: 10.1002/mnfr.202100014
López de Felipe, F., de Las Rivas, B., Muñoz, R. (2014). Bioactive compounds produced by gut microbial tannase: Implications for colorectal cancer development. Front. Microbiol. 5, 684. doi: 10.3389/fmicb.2014.00684
Mace, T. A., Ware, M. B., King, S. A., Loftus, S., Farren, M. R., McMichael, E., et al. (2019). Soy isoflavones and their metabolites modulate cytokine-induced natural killer cell function. Sci. Rep. 9 (1), 5068. doi: 10.1038/s41598-019-41687-z
Madacki, J., Kopál, M., Jackson, M., Korduláková, J. (2021). Mycobacterial epoxide hydrolase EphD is inhibited by urea and thiourea derivatives. Int. J. Mol. Sci. 22 (6). doi: 10.3390/ijms22062884
Maier, L., Goemans, C. V., Wirbel, J., Kuhn, M., Eberl, C., Pruteanu, M., et al. (2021). Unravelling the collateral damage of antibiotics on gut bacteria. Nature 599 (7883), 120–124. doi: 10.1038/s41586-021-03986-2
Malwe, A. S., Srivastava, G. N., Sharma, V. K. (2023). GutBug: A tool for prediction of human gut bacteria mediated biotransformation of biotic and xenobiotic molecules using machine learning. J. Mol. Biol. 435 (14), 168056. doi: 10.1016/j.jmb.2023.168056
Mas-Capdevila, A., Teichenne, J., Domenech-Coca, C., Caimari, A., Del Bas, J. M., Escoté, X., et al. (2020). Effect of hesperidin on cardiovascular disease risk factors: The role of intestinal microbiota on hesperidin bioavailability. Nutrients 12 (5). doi: 10.3390/nu12051488
Matsumoto, M., Ishige, A., Yazawa, Y., Kondo, M., Muramatsu, K., Watanabe, K. (2012). Promotion of intestinal peristalsis by Bifidobacterium spp. Capable of hydrolysing sennosides in mice. PLoS One 7 (2), e31700. doi: 10.1371/journal.pone.0031700
Najmanová, I., Pourová, J., Vopršalová, M., Pilařová, V., Semecký, V., Nováková, L., et al. (2016). Flavonoid metabolite 3-(3-hydroxyphenyl)propionic acid formed by human microflora decreases arterial blood pressure in rats. Mol. Nutr. Food Res. 60, 981–991. doi: 10.1002/mnfr.201500761
Narendar Reddy, T., Beatriz, A., Jayathirtha Rao, V., de Lima, D. P. (2019). Carbonyl compounds’ journey to amide bond formation. Chem. Asian J. 14, 344–388. doi: 10.1002/asia.201801560
Newman, D. J., Cragg, G. M. (2020). Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 83 (3), 770–803. doi: 10.1021/acs.jnatprod.9b01285
Nguepi Tsopmejio, I. S., Yuan, J., Diao, Z., Fan, W., Wei, J., Zhao, C., et al. (2023). Auricularia polytricha and flammulina velutipes reduce liver injury in DSS-induced inflammatory bowel disease by improving inflammation, oxidative stress, and apoptosis through the regulation of TLR4/NF-κB signaling pathways. J. Nutr. Biochem. 111, 109190. doi: 10.1016/j.jnutbio.2022.109190
Ni, J., Shen, T. D., Chen, E. Z., Bittinger, K., Bailey, A., Roggiani, M., et al. (2017). A role for bacterial urease in gut dysbiosis and Crohn’s disease. Sci. Trans. Med. 9 (416).
Nishida, A., Nishino, K., Ohno, M., Sakai, K., Owaki, Y., Noda, Y., et al. (2022). Update on gut microbiota in gastrointestinal diseases. World J. Clin. Cases 10 (22), 7653–7664. doi: 10.12998/wjcc.v10.i22.7653
Pandurangan, A. K., Mohebali, N., Esa, N. M., Looi, C. Y., Ismail, S., Saadatdoust, Z. (2015). Gallic acid suppresses inflammation in dextran sodium sulfate-induced colitis in mice: Possible mechanisms. Int. Immunopharmacol. 28 (2), 1034–1043. doi: 10.1016/j.intimp.2015.08.019
Pang, Z., Raudonis, R., Glick, B. R., Lin, T. J., Cheng, Z. (2019). Antibiotic resistance in pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 37, 177–192. doi: 10.1016/j.biotechadv.2018.11.013
Pavlović, N., Stankov, K., Mikov, M. (2012). Probiotics–interactions with bile acids and impact on cholesterol metabolism. Appl. Biochem. Biotechnol. 168, 1880–1895. doi: 10.1007/s12010-012-9904-4
Peng, R., Han, P., Fu, J., Zhang, Z.-W., Ma, S.-R., Pan, L.-B., et al. (2022). Esterases from bifidobacteria exhibit the conversion of albiflorin in gut microbiota. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.880118
Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K. S., Manichanh, C., et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65. doi: 10.1038/nature08821
Raba, G., Luis, A. S. (2023). Mucin utilization by gut microbiota: Recent advances on characterization of key enzymes. Essays Biochem. 67 (3), 345–353. doi: 10.1042/EBC20220121
Raghuwanshi, S., Dutt, K., Gupta, P., Saxena, R. K. (2011). Bacillus sphaericus: the highest bacterial tannase producer with potential for gallic acid synthesis. J. Biosci. Bioeng. 111, 635–640. doi: 10.1016/j.jbiosc.2011.02.008
Rakoff-Nahoum, S., Foster, K. R., Comstock, L. E. (2016). The evolution of cooperation within the gut microbiota. Nature 533 (7602), 255–259. doi: 10.1038/nature17626
Ramya, S. L., Venkatesan, T., Murthy, K. S., Jalali, S. K., Verghese, A. (2016). Detection of carboxylesterase and esterase activity in culturable gut bacterial flora isolated from diamondback moth, Plutella xylostella (Linnaeus), from India and its possible role in indoxacarb degradation. Braz. J. Microbiol. 47 (2), 327–336. doi: 10.1016/j.bjm.2016.01.012
Redinbo, M. R., Potter, P. M. (2005). Mammalian carboxylesterases: From drug targets to protein therapeutics. Drug Discov. Today 10 (5), 313–325. doi: 10.1016/S1359-6446(05)03383-0
Ridlon, J. M., Harris, S. C., Bhowmik, S., Kang, D. J., Hylemon, P. B. (2016). Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 7, 22–39. doi: 10.1080/19490976.2015.1127483
Rimal, B., Collins, S. L., Tanes, C. E., Rocha, E. R., Granda, M. A., Solanki, S., et al. (2024). Bile salt hydrolase catalyses formation of amine-conjugated bile acids. Nature 626 (8000), 859–863. doi: 10.1038/s41586-023-06990-w
Ruiz, L., Sanchez, B., Margolles, A. (2021). Determination of bile salt hydrolase activity in bifidobacteria. Methods Mol. Biol. (Clifton N.J.) 2278, 149–155. doi: 10.1007/978-1-0716-1274-3_13
Schnoes, A. M., Ream, D. C., Thorman, A. W., Babbitt, P.C., Friedberg, I. (2013). Biases in the experimental annotations of protein function and their effect on our understanding of protein function space. PLoS Comput. Biol. 9, e1003063. doi: 10.1371/journal.pcbi.1003063
Selma, M. V., Beltrán, D., García-Villalba, R., Espín, J. C., Tomás-Barberán, F. A. (2014). Description of urolithin production capacity from ellagic acid of two human intestinal Gordonibacter species. Food Funct. 5, 1779–1784. doi: 10.1039/C4FO00092G
Shao, T., Hsu, R., Hacein-Bey, C., Zhang, W., Gao, L., Kurth, M. J., et al. (2023). The evolving landscape of fecal microbial transplantation. Clin. Rev. Allergy Immunol. 65, 101–120. doi: 10.1007/s12016-023-08958-0
Sharma, A. K., Jaiswal, S. K., Chaudhary, N., Sharma, V. K. (2017). A novel approach for the prediction of species-specific biotransformation of xenobiotic/drug molecules by the human gut microbiota. Sci. Rep. 7, 9751. doi: 10.1038/s41598-017-10203-6
Shen, H., Ding, L., Baig, M., Tian, J., Wang, Y., Huang, W. (2021). Improving glucose and lipids metabolism: Drug development based on bile acid related targets. Cell Stress 5, 1–18. doi: 10.15698/cst2021.01.239
Sheng, L., Jena, P. K., Liu, H.-X., Hu, Y., Nagar, N., Bronner, D. N., et al (2018). Obesity treatment by epigallocatechin-3-gallate-regulated bile acid signaling and its enriched Akkermansia muciniphila. FASEB J. 32, 6371–6384. doi: 10.1096/fj.201800370R
Shimojo, Y., Ozawa, Y., Toda, T., Igami, K., Shimizu, T.. (2018). Probiotic Lactobacillus paracasei A221 improves the functionality and bioavailability of kaempferol-glucoside in kale by its glucosidase activity. Sci. Rep. 8, 9239. doi: 10.1038/s41598-018-27532-9
Shin, K. C., Lee, H. J., Oh, D. K. (2015). Substrate specificity of β-glucosidase from Gordonia terrae for ginsenosides and its application in the production of ginsenosides Rg3, Rg2, and Rh1 from ginseng root extract. J. Biosci. Bioeng. 119, 497–504. doi: 10.1016/j.jbiosc.2014.10.004
Shin, K. C., Seo, M. J., Oh, D. K. (2014). Characterization of β-xylosidase from Thermoanaerobacterium thermosaccharolyticum and its application to the production of ginsenosides Rg1 and Rh 1 from notoginsenosides R 1 and R 2. Biotechnol. Lett. 36, 2275–2281. doi: 10.1007/s10529-014-1604-4
Sikorska-Zimny, K., Beneduce, L. (2021). The metabolism of glucosinolates by gut microbiota. Nutrients 13 (8). doi: 10.3390/nu13082750
Singh, A., Poling, H. M., Spence, J. R., Wells, J. M., Helmrath, M. A. (2020). Gastrointestinal organoids: A next-generation tool for modeling human development. Am. J. Physiol. Gastrointestinal liver Physiol. 319, G375–Gg81. doi: 10.1152/ajpgi.00199.2020
Stanislauskienė, R., Laurynėnas, A., Rutkienė, R., Aučynaitė, A., Tauraitė, D., Meškienė, R., et al. (2020). Yqfb protein from Escherichia coli: An atypical amidohydrolase active towards N(4)-acylcytosine derivatives. Sci. Rep. 10, 788. doi: 10.1038/s41598-020-57664-w
Sun, X., Chen, Z., Yu, L., Zeng, W., Sun, B., Fan, H., et al. (2023). Bacteroides dorei BDX-01 alleviates DSS-induced experimental colitis in mice by regulating intestinal bile salt hydrolase activity and the FXR-NLRP3 signaling pathway. Front. Pharmacol. 14, 1205323. doi: 10.3389/fphar.2023.1205323
Sun, L., Xie, C., Wang, G., Wu, Y., Wu, Q., Wang, X., et al. (2018). Gut microbiota and intestinal FXR mediate the clinical benefits of metformin. Nat. Med. 24, 1919–1929. doi: 10.1038/s41591-018-0222-4
Tian, J., Li, C., Dong, Z., Yang, Y., Xing, J., Yu, P., et al. (2023). Inactivation of the antidiabetic drug acarbose by human intestinal microbial-mediated degradation. Nat. Metab. 5, 896–909. doi: 10.1038/s42255-023-00796-w
Tian, X. G., Yan, J. K., Sun, C. P., Li, J. X., Ning, J., Wang, C., et al. (2021). Amentoflavone from selaginella tamariscina as a potent inhibitor of gut bacterial β-glucuronidase: Inhibition kinetics and molecular dynamics stimulation. Chem. Biol. Interact. 340, 109453. doi: 10.1016/j.cbi.2021.109453
Tomioka, S., Seki, N., Sugiura, Y., Akiyama, M., Uchiyama, J., Yamaguchi, G., et al. (2022). Cooperative action of gut-microbiota-accessible carbohydrates improves host metabolic function. Cell Rep. 40, 111087. doi: 10.1016/j.celrep.2022.111087
Tong, X., Qi, Z., Zheng, D., Pei, J., Li, Q., Zhao, L. (2021). High-level expression of a novel multifunctional GH3 family β-xylosidase/α-arabinosidase/β-glucosidase from Dictyoglomus turgidum in Escherichia coli. Bioorgan. Chem. 111, 104906. doi: 10.1016/j.bioorg.2021.104906
Wan, H. D., Xia, Y. M. (2015). Enzymatic transformation of stevioside using a β-galactosidase from Sulfolobus sp. Food Funct. 6 (10), 3291–3295. doi: 10.1039/C5FO00631G
Wang, B., Guo, F., Ren, J., Ai, G., Aigle, B., Fan, K., et al. (2015). Identification of Alp1U and Lom6 as epoxy hydrolases and implications for kinamycin and lomaiviticin biosynthesis. Nat. Commun. 6, 7674. doi: 10.1038/ncomms8674
Wang, P., Wu, P. F., Wang, H. J., Liao, F., Wang, F., Chen, J. G., et al. (2023). Gut microbiome-derived ammonia modulates stress vulnerability in the host. Nat. Metab. 5, 1986–2001. doi: 10.1038/s42255-023-00909-5
Wang, G., Yu, H., Feng, X., Tang, H., Xiong, Z., Xia, Y., et al. (2021). Specific bile salt hydrolase genes in Lactobacillus plantarum AR113 and relationship with bile salt resistance. Lwt-Food Sci. Technol. 145. doi: 10.1016/j.lwt.2021.111208
Wang, K., Zhang, Z., Hang, J., Liu, J., Guo, F., Ding, Y., et al. (2023). Microbial-host-isozyme analyses reveal microbial dpp4 as a potential antidiabetic target. Science 381 (6657), eadd5787. doi: 10.1126/science.add5787
Watanabe, M., Fukiya, S., Yokota, A. (2017). Comprehensive evaluation of the bactericidal activities of free bile acids in the large intestine of humans and rodents. J. Lipid Res. 58 (6), 1143–1152. doi: 10.1194/jlr.M075143
Weersma, R. K., Zhernakova, A., Fu, J. (2020). Interaction between drugs and the gut microbiome. Gut 69 (8), 1510–1519. doi: 10.1136/gutjnl-2019-320204
Weng, Z. M., Wang, P., GE, G. B., Dai, Z. R., Wu, D. C., Zou, L. W., et al. (2017). Structure-activity relationships of flavonoids as natural inhibitors against E. coli β-glucuronidase. Food Chem. Toxicol. 109, 975–983. doi: 10.1016/j.fct.2017.03.042
Wu, J., Cui, S., Tang, X., Zhang, Q., Jin, Y., Zhao, J., et al. (2023). Bifidobacterium longum ccfm1206 promotes the biotransformation of glucoraphanin to sulforaphane that contributes to amelioration of dextran-sulfate-sodium-induced colitis in mice. J. Agric. Food Chem. 71, 1100–1112. doi: 10.1021/acs.jafc.2c07090
Wu, Z., Liu, C., Zhang, Z., Zheng, R., Zheng, Y. (2020). Amidase as a versatile tool in amide-bond cleavage: From molecular features to biotechnological applications. Biotechnol. Adv. 43, 107574. doi: 10.1016/j.biotechadv.2020.107574
Wu, T., Pei, J., GE, L., Wang, Z., Ding, G., Xiao, W., et al. (2018). Characterization of a α-l-rhamnosidase from bacteroides thetaiotaomicron with high catalytic efficiency of epimedin C. Bioorgan. Chem. 81, 461–467. doi: 10.1016/j.bioorg.2018.08.004
Wu, Q., Wu, X., Wang, M., Liu, K., Li, Y., Ruan, X., et al. (2023). Therapeutic mechanism of baicalin in experimental colitis analyzed using network pharmacology and metabolomics. Drug Des. Devel. Ther. 17, 1007–1024. doi: 10.2147/DDDT.S399290
Xie, Y., Hu, F., Xiang, D., Lu, H., Li, W., Zhao, A., et al. (2020). The metabolic effect of gut microbiota on drugs. Drug Metab. Rev. 52, 139–156. doi: 10.1080/03602532.2020.1718691
Xie, Z., Jiang, H., Liu, W., Zhang, X., Chen, D., Sun, S., et al. (2020). The triterpenoid sapogenin (2α-OH-protopanoxadiol) ameliorates metabolic syndrome via the intestinal FXR/GLP-1 axis through gut microbiota remodelling. Cell Death Dis. 11, 770. doi: 10.1038/s41419-020-02974-0
Xu, X.-F., Hu, D., Hu, B.-C., Li, C., Liu, Y.-Y., Wu, M.-C. (2020). Near-perfect kinetic resolution of o-methylphenyl glycidyl ether by RpEH, a novel epoxide hydrolase from Rhodotorula paludigena JNU001 with high stereoselectivity. Appl. Microbiol. Biotechnol. 104, 6199–6210. doi: 10.1007/s00253-020-10694-w
Xu, W., Kong, Y., Zhang, T., Gong, Z., Xiao, W. L-theanine regulates lipid metabolism by modulating gut microbiota and bile acid metabolism. J. Sci. Food Agric. doi: 10.1002/jsfa.12222
Xu, H., Yu, H., Fu, J., Zhang, Z. W., Hu, J. C., Lu, J. Y., et al. (2023). Metabolites analysis of plantamajoside based on gut microbiota-drug interaction. Phytomedicine 116, 154841. doi: 10.1016/j.phymed.2023.154841
Yang, X., Dong, B., An, L., Zhang, Q., Chen, Y., Wang, H., et al. (2021). Ginsenoside Rb1 ameliorates glycemic disorder in mice with high fat diet-induced obesity via regulating gut microbiota and amino acid metabolism. Front. Pharmacol. 12. doi: 10.3389/fphar.2021.756491
Yang, P., Ling, L., Sun, W., Yang, J., Zhang, L., Chang, G., et al. (2018). Ginsenoside Rg1 inhibits apoptosis by increasing autophagy via the AMPK/mTOR signaling in serum deprivation macrophages. Acta Biochim. Biophys. Sin. 50, 144–155. doi: 10.1093/abbs/gmx136
Yonemoto-Yano, H., Maebuchi, M., Fukui, K., Tsuzaki, S., Takamatsu, K., Uehara, M. (2014). Malonyl isoflavone glucosides are chiefly hydrolyzed and absorbed in the colon. J. Agric. Food Chem. 62, 2264–2270. doi: 10.1021/jf404378r
Youn, S. Y., Park, M. S., Ji, G. E. (2012). Identification of the beta-glucosidase gene from Bifidobacterium animalis subsp. Lactis and its expression in B. bifidum BGN4. J. Microbiol. Biotechnol. 22, 1714–1723. doi: 10.4014/jmb
Yu, H.-F., Cheng, Y., Wu, C., Ran, K., Wei, B., Xu, Y.-k., et al. (2022). Diverse diterpenoids with α-glucosidase and β-glucuronidase inhibitory activities from Euphorbia milii. Phytochemistry 196, 113106. doi: 10.1016/j.phytochem.2022.113106
Yu, Y., Wang, W., Zhang, F. (2023). The next generation fecal microbiota transplantation: To transplant bacteria or virome. Adv Sci (Weinh) 10, e2301097. doi: 10.1002/advs.202301097
Yu, H., Xu, H., Yang, X., Zhang, Z., Hu, J., Lu, J., et al. (2023). Gut microbiota-based pharmacokinetic-pharmacodynamic study and molecular mechanism of specnuezhenide in the treatment of colorectal cancer targeting carboxylesterase. J. Pharm. Anal. 13, 1024–1040. doi: 10.1016/j.jpha.2023.06.012
Yu, J.-B., Zhao, Z.-X., Peng, R., Pan, L.-B., Fug, J., Ma, S.-R., et al. (2019). Gut microbiota-based pharmacokinetics and the antidepressant mechanism of paeoniflorin. Front. Pharmacol. 10. doi: 10.3389/fphar.2019.00268
Yuanfeng, W., Chengzhi, L., Ligen, Z., Juan, S., Xinjie, S., Yao, Z., et al. (2021). Approaches for enhancing the stability and formation of sulforaphane. Food Chem. 345, 128771. doi: 10.1016/j.foodchem.2020.128771
Zeng, Y., Wu, R., Wang, F., Li, S., Li, L., Li, Y., et al. (2023). Liberation of daidzein by gut microbial β-galactosidase suppresses acetaminophen-induced hepatotoxicity in mice. Cell Host Microbe 31, 766–80.e7. doi: 10.1016/j.chom.2023.04.002
Zhang, Z. W., Han, P., Fu, J., Yu, H., Xu, H., Hu, J. C., et al. (2023). Gut microbiota-based metabolites of Xiaoyao Pills (a typical Traditional Chinese medicine) ameliorate depression by inhibiting fatty acid amide hydrolase levels in brain. J. Ethnopharmacol. 313, 116555. doi: 10.1016/j.jep.2023.116555
Zhang, N.-N., Jiang, Z.-M., Li, S.-Z., Yang, X., Liu, E.H. (2023). Evolving interplay between natural products and gut microbiota. Eur. J. Pharmacol., 175557. doi: 10.1016/j.ejphar.2023.175557
Zhang, C., Liu, Y., Li, C., Xu, Y., Su, Y., Li, J., et al. (2020). Significant improvement in catalytic activity and enantioselectivity of a Phaseolus vulgaris epoxide hydrolase, PVEH3, towards ortho-cresyl glycidyl ether based on the semi-rational design. Sci. Rep. 10 (1). doi: 10.1038/s41598-020-58693-1
Zhang, M., Peng, C. S., Li, X. B. (2015). In vivo and in vitro metabolites from the main diester and monoester diterpenoid alkaloids in a Traditional Chinese herb, the aconitum species. Evid Based Complement Alternat Med. 2015, 252434. doi: 10.1155/2015/252434
Zhang, J. H., Zhang, J. M., Wang, R. (2018). Gut microbiota modulates drug pharmacokinetics. Drug Metab. Rev. 50, 357–368. doi: 10.1080/03602532.2018.1497647
Zhang, R., Zhang, B. L., Xie, T., Li, G. C., Tuo, Y., Xiang, Y. T., et al. (2015). Biotransformation of rutin to isoquercitrin using recombinant α-L-rhamnosidase from Bifidobacterium breve. Biotechnol. Lett. 37, 1257–1264. doi: 10.1007/s10529-015-1792-6
Zhao, Z.-X., Fu, J., Ma, S.-R., Peng, R., Yu, J.-B., Cong, L., et al. (2018). Gut-brain axis metabolic pathway regulates antidepressant efficacy of albiflorin. Theranostics 8, 5945–5959. doi: 10.7150/thno.28068
Zhao, L., Qi, Z., Yi, L., Li, J., Cui, Y., Ur Rehman, F., et al. (2021). The interaction between gut microbiota and flavonoid extract from Smilax glabra Roxb. And its potent alleviation fatty liver. Food Funct. 12, 7836–7850. doi: 10.1039/D1FO00727K
Zhao, Y., Zhong, X., Yan, J., Sun, C., Zhao, X., Wang, X. (2022). Potential roles of gut microbes in biotransformation of natural products: An overview. Front. Microbiol. 13, 956378. doi: 10.3389/fmicb.2022.956378
Zheng, W., Zhao, S., Yin, Y., Zhang, H., Needham, D. M., Evans, E. D., et al. (2022). High-throughput, single-microbe genomics with strain resolution, applied to a human gut microbiome. Science 376 (6597), eabm1483. doi: 10.1126/science.abm1483
Zhou, L., Li, Z. K., Li, C. Y., Liang, Y. Q., Yang, F. (2022). Anticancer properties and pharmaceutical applications of ginsenoside compound K: A review. Chem. Biol. Drug Des. 99, 286–300. doi: 10.1111/cbdd.13983
Zhou, Y., Liu, K., Zhang, J., Chu, J., He, B. (2017). Mg(2+)-induced stabilization of β-galactosidase from Bacillus megaterium and its application in the galactosylation of natural products. Biotechnol. Lett. 39, 1175–1181. doi: 10.1007/s10529-017-2344-z
Zhou, P., Xie, W., He, S., Sun, Y., Meng, X., Sun, G., et al. (2019). Ginsenoside Rb1 as an anti-diabetic agent and its underlying mechanism analysis. Cells 8. doi: 10.3390/cells8030204
Zhu, W., Jin, Z., Yu, J., Liang, J., Yang, Q., Li, F., et al. (2016). Baicalin ameliorates experimental inflammatory bowel disease through polarization of macrophages to an M2 phenotype. Int. Immunopharmacol. 35, 119–126. doi: 10.1016/j.intimp.2016.03.030
Zhu, H., Zhao, F., Zhang, W., Xia, W., Chen, Y., Liu, Y., et al. (2022). Cholesterol-lowering effect of bile salt hydrolase from a Lactobacillus johnsonii strain mediated by FXR pathway regulation. Food Funct. 13, 725–736. doi: 10.1039/D1FO03143K
Zimmermann, M., Zimmermann-Kogadeeva, M., Wegmann, R., Wegmann, R., Goodman, A. L. (2019). Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 570, 462–467. doi: 10.1038/s41586-019-1291-3
Zinin, A. I., Eneyskaya, E. V., Shabalin, K. A., Kulminskaya, A. A., Shishlyannikov, S. M., Neustroev, K. N. (2002). 1-O-acetyl-beta-D-galactopyranose: A novel substrate for the transglycosylation reaction catalyzed by the beta-galactosidase from Penicillium sp. Carbohydr. Res. 337, 635–642. doi: 10.1016/S0008-6215(02)00027-7
Keywords: natural product, microorganisms, gut microbiome-derived hydrolases, secondary metabolism, mechanism of enzymatic deconstruction
Citation: He J, Liu X, Zhang J, Wang R, Cao X and Liu G (2024) Gut microbiome-derived hydrolases—an underrated target of natural product metabolism. Front. Cell. Infect. Microbiol. 14:1392249. doi: 10.3389/fcimb.2024.1392249
Received: 28 February 2024; Accepted: 16 May 2024;
Published: 10 June 2024.
Edited by:
Aaron T. Wright, Baylor University, United StatesCopyright © 2024 He, Liu, Zhang, Wang, Cao and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ge Liu, biohazard_1220@163.com
 Jiaxin He
Jiaxin He