
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 31 May 2024
Sec. Intestinal Microbiome
Volume 14 - 2024 | https://doi.org/10.3389/fcimb.2024.1380998
Background/Aim: We employed Mendelian randomization (MR) analysis to investigate the causal relationship between the gut microbiota, acute pancreatitis, and potential inflammatory proteins.
Methods: The data for gut microbiota, acute pancreatitis, and inflammatory proteins are sourced from public databases. We conducted a bidirectional MR analysis to explore the causal relationship between gut microbiota and acute pancreatitis, and employed a two-step MR analysis to identify potential mediating inflammatory proteins. IVW is the primary analysis method, heterogeneity, pleiotropy, and sensitivity analyses were also conducted simultaneously.
Results: We identified five bacterial genera associated with the risk of acute pancreatitis, namely genus.Coprococcus3, genus.Eubacterium fissicatena group, genus.Erysipelotrichaceae UCG-003, genus.Fusicatenibacter, and genus.Ruminiclostridium6. Additionally, we have discovered three inflammatory proteins that are also associated with the occurrence of acute pancreatitis, namely interleukin-15 receptor subunit alpha (IL-15RA), monocyte chemoattractant protein-4 (CCL13), and tumor necrosis factor receptor superfamily member 9 (TNFRSF9). Following a two-step MR analysis, we ultimately identified IL-15RA as a potential intermediate factor, with a mediated effect of 0.018 (95% CI: 0.005 - 0.032).
Conclusion: Our results support the idea that genus.Coprococcus3 promotes the occurrence of acute pancreatitis through IL-15RA. Furthermore, there is a potential causal relationship between the gut microbiota, inflammatory proteins, and acute pancreatitis. These findings provide new insights for subsequent acute pancreatitis prevention.
Pancreatitis is a prevalent gastrointestinal disorder characterized by acute and chronic forms. A recent meta-analysis documented a worldwide prevalence of 33 cases per 100,000 person-years for acute pancreatitis (Xiao et al., 2016). Acute pancreatitis entails an inflammatory response, leading to self-digestion, edema, hemorrhage, and potentially necrosis of pancreatic tissues, triggered by the activation of pancreatic enzymes due to diverse etiological factors. Acute pancreatitis manifests as intense abdominal pain, nausea, vomiting, and various clinical symptoms, and in severe cases, it can lead to organ failure. Thus, it remains closely linked to high mortality, with two peaks of mortality, early and late (Garg and Singh, 2019).
There are 100 trillion microorganisms and more than 1000 different bacteria in the human intestine, which constitute the gut microbiota (Ramakrishna, 2013). Recent discoveries have shown that the gut microbiota is involved in regulating multiple host functions by producing bioactive bacterial metabolites. Therefore, it is considered a new human organ and also emerged as a key factor in the balance between health and disease (Baquero and Nombela, 2012; Lee and Hase, 2014; Schepis et al., 2021). It is now understood that dysregulation of the microbiota can lead to a range of diseases, including obesity, inflammatory bowel disease, Alzheimer’s disease, metabolic syndrome, cardiovascular disease, and even cancer (Sekirov et al., 2010; Distrutti et al., 2016; Brandi et al., 2021; Zhao et al., 2021).
The coordination of inflammatory responses involves a complex network of cells and mediators (Zhao et al., 2023), and dysregulated inflammatory proteins play a crucial role in disease progression. For example, elevated concentrations of interleukin-1α (IL-1α), interleukin-8 (IL-8), and tumor necrosis factor-α (TNF-α) are associated with an increased risk of ovarian cancer (Trabert et al., 2014). Since proteins serve as both effector molecules and drug targets in most biological processes, understanding their roles in diseases is a growing area of research.
Mendelian randomization (MR) analysis leverages genetic variation as instrumental variables (IVs) in place of exposure factors to evaluate the casual relationship between exposures and outcomes, mimicking random assignment in a study design (Wehby et al., 2008; Lee and Lim, 2019). This approach has gained popularity in recent epidemiological studies to overcome limitations inherent to observational and, to some extent, randomized controlled studies. Herein, we sought to explore the causal relationship between the gut microbiota and the onset risk of acute pancreatitis, investigating the role inflammatory proteins play in this process. Therefore, we conducted a two-step MR analysis based on genome-wide association study (GWAS) summary data.
Genetic summary statistics for acute pancreatitis were generated from a GWAS data from FinnGen (GWAS ID: finn-b-K11_ACUTPANC), and for the human gut microbiome from the published meta-analysis by the MiBioGen consortium (Kurilshikov et al., 2021). We then excluded 15 bacterial traits lacking specific nomenclature, resulting in a final set of 196 bacterial traits, encompassing 9 phyla, 16 classes, 20 orders, 32 families, and 119 genera. The data on inflammatory proteins were obtained from Jing et al (Zhao et al., 2023), who conducted a genome-wide protein quantitative trait locus (pQTL) study of 91 inflammatory proteins measured using the Olink Target platform in 14,824 European-ancestry participants. As the present study was based on public summary data, no additional ethics approval or consent to participate was required.
IVs were chosen at a significance level of p< 1.0 × 10-5. To ensure independence of the IVs from loci, we applied a linkage disequilibrium (LD) threshold of R2< 0.001 and a clumping distance of 10,000 kb in the 1000 Genomes European (EUR) data using the “TwoSampleMR” packages. Additionally, we extracted relevant information for each single nucleotide polymorphism (SNP), including effect alleles, β-value, standard deviation, and p-values. We then calculated the proportion of variation explained (R2) and F value to quantify instrument strength using the following equation:
Where “N” is the number of participants, “EAF” is the effect allele frequency, “β” is the estimated effect of the SNP (assessing its ability to uniquely predict the outcome), and “SE” is the standard error of the genetic effect (Palmer et al., 2012; Gill et al., 2019; Levin et al., 2020). A higher F value (greater than 10) indicates a lower likelihood of weak instrument bias.
This study employed a two-sample MR analysis to investigate the association between exposures (gut microbiota) and outcomes (the risk of acute pancreatitis). Additionally, a two-step MR analysis was conducted to assess whether inflammatory proteins mediate the potential effect of gut microbiota on pancreatitis development. Figure 1 illustrates the overall design of the study.
In this study, we employed various MR analysis methods, including inverse variance weighting (IVW), MR-Egger regression, weighted median, weighted model, and simple model. Among these, IVW provides the most precise overall effect estimate by combining a meta-analysis approach with the estimates of the effect of each genetic variant for gut microbiota on pancreatitis (Burgess et al., 2016; Li et al., 2022). Therefore, we used IVW as the primary analysis method. We conducted a series of MR analyses: first, a two-sample MR analysis for gut microbiota and acute pancreatitis. Second, a reverse MR analysis on bacteria causally associated with pancreatitis from the first analysis. Third, an MR analysis of these bacteria and 91 inflammatory proteins. Finally, we explored the relationship between inflammatory proteins and acute pancreatitis. Throughout the analyses, we employed Cochran’s Q test to assess the heterogeneity of IVs, MR-PRESSO and MR-Egger regression to check for potential horizontal pleiotropy, and the “leave-one-out” method for sensitivity analysis. P values < 0.05 were considered nominally significant. All MR analyses were performed using R version 4.2.2 (https://www.r-project.org/). MR analyses were performed using the “TwoSampleMR” package and MRPRESSO package.
Our results were visualized in a Circos plot (Figure 2). Five bacterial genera showed potential associations with acute pancreatitis. Using the IVW method, genetic predisposition to genus.Coprococcus3 and genus.Eubacterium fissicatena group may be associated with an increased risk of acute pancreatitis. Conversely, genus.Erysipelotrichaceae UCG-003, genus.Fusicatenibacter, and genus. Ruminiclostridium6 were protective against acute pancreatitis (Figure 3). Heterogeneity and pleiotropy tests yielded p-values > 0.05, indicating consistency in the results. Sensitivity analysis yielded robust and consistent results (Supplementary 1, 2).
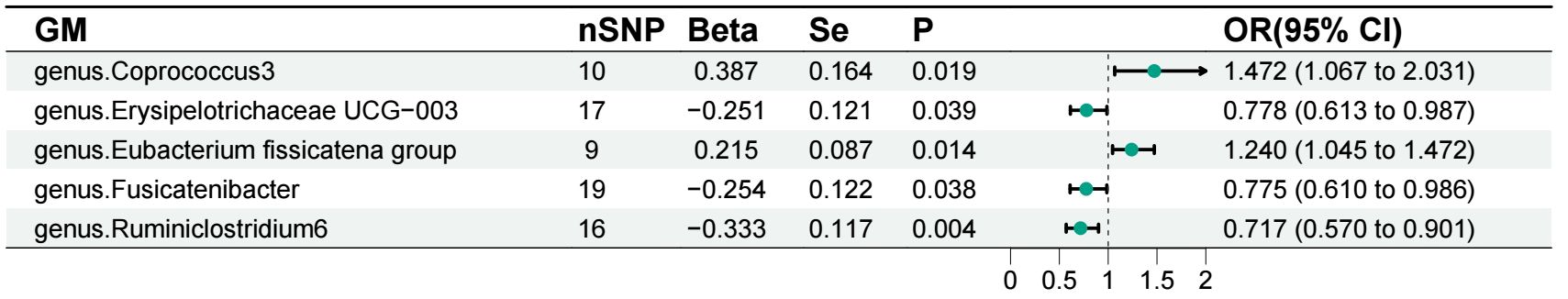
Figure 3 Forest plot of the associations between gut microbiota and acute pancreatitis. GM, gut microbiota.
Next, we evaluated the potential reverse associations of bacterial traits and acute pancreatitis using the reverse MR analyses. With the IVW method, no significant causal association was found between acute pancreatitis and any of these bacterial traits (Table 1). The results remained stable across sensitivity analyses.
Our findings based on gene prediction suggest that a positive association between increased abundance of genus.Coprococcu3 and several inflammatory proteins, including adenosine deaminase (ADA), caspase-8 (CASP-8), macrophage colony-stimulating factor-1 (CSF-1), C-X-C motif chemokine-10 (CXCL10), S100A1 protein (EN-RAGE), interleukin-15 receptor subunit alpha (IL-15RA), interleukin-18 (IL-18) and interleukin-8 (IL-8). Similarly, an increase in genus.ErysipelotrichaceaeUCG003 abundance was associated with increased levels of beta-nerve growth factor (Beta-NGF), C-X-C motif chemokine-1 (CXCL1), C-X-C motif chemokine-5 (CXCL5), C-X-C motif chemokine-6 (CXCL6), glial cell line-derived neurotrophic factor (GDNF), stem cell factor (SCF) and tumor necrosis factor ligand superfamily member 12 (TNFSF12). The genus.Ruminiclostridium6, on the other hand, showed a positive correlation with C-X-C motif chemokine 9 (CXCL9). Conversely, increased genus.Eubacterium fissicatena group abundance was associated with reduced levels of inflammatory proteins such as monocyte chemoattractant protein-1 (MCP-1) and TNFSF-12. Genus.Fusicatenibacter also had a negative impact on CXCL5, CXCL6, EN-RAGE and oncostatin-M (OSM) (Figure 4). Importantly, no significant heterogeneity or pleiotropy was observed in our data (Supplementary 1, 2).
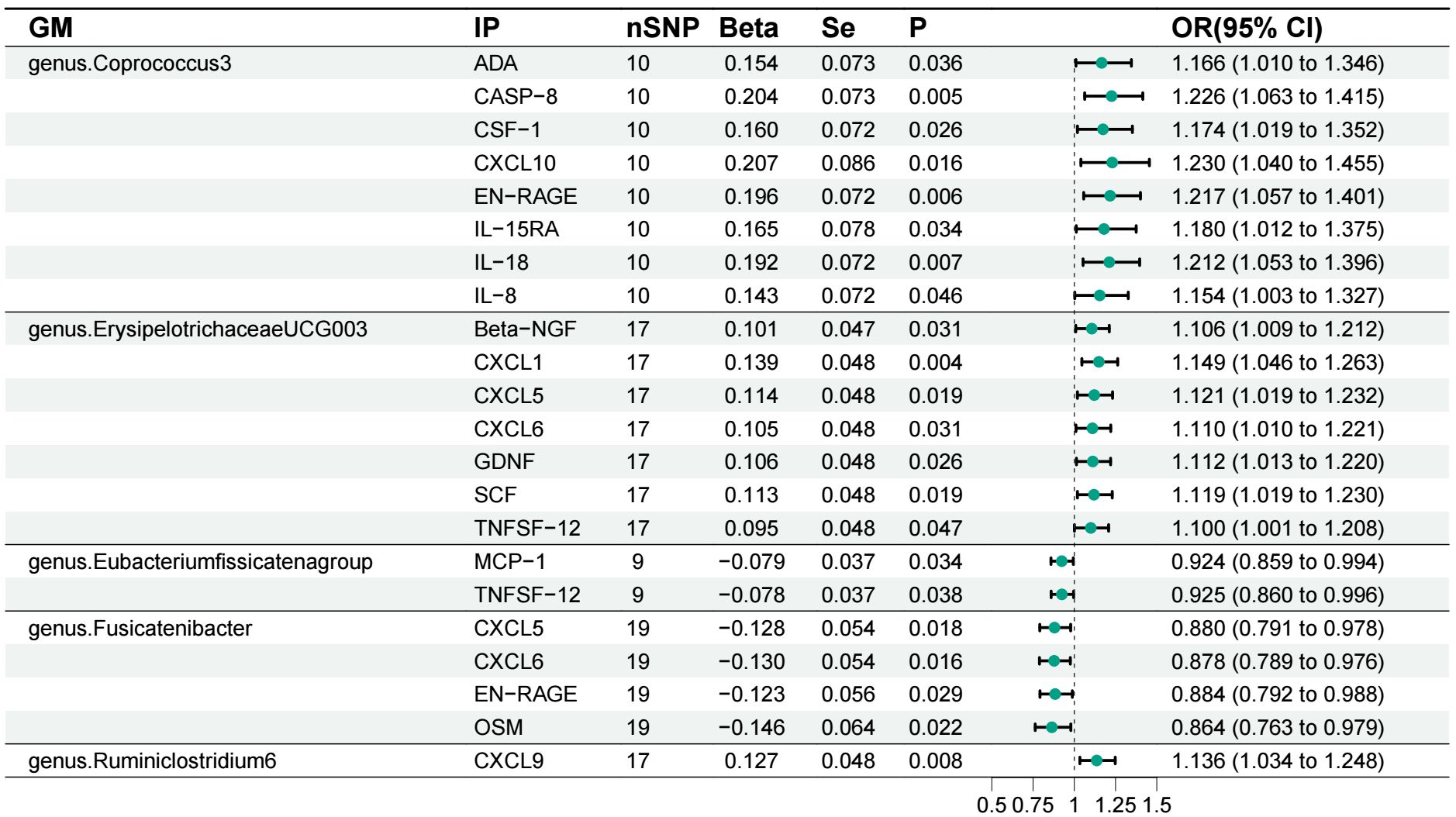
Figure 4 Forest plot of the associations between gut microbiota and inflammatory proteins. GM, gut microbiota; IP, inflammatory protein; ADA, adenosine deaminas; CASP-8, caspase-8; CSF-1, macrophage colony-stimulating factor-1; CXCL10, C-X-C motif chemokine-10; EN-RAGE, protein S100-A1; IL-15RA, Interleukin-15 receptor subunit alpha; IL-18, Interleukin-18; IL-8 Interleukin-8; Beta-NGF, beta-nerve growth factor; CXCL1, C-X-C motif chemokine-1; CXCL5, C-X-C motif chemokine-5; CXCL6, C-X-C motif chemokine-6; GDNF, Glial cell line-derived neurotrophic factor; SCF, Stem cell factor; TNFSF12, Tumor necrosis factor ligand superfamily member 12; MCP-1, monocyte chemoattractant protein-1; OSM, Oncostatin-M; CXCL9, C-X-C motif chemokine 9.
We conducted individual analyses of 91 inflammatory proteins concerning acute pancreatitis, and all the results are presented in the following figure (Figure 5). Among these, genetically predicted IL-15RA, monocyte chemoattractant protein-4 (CCL13), and tumor necrosis factor receptor superfamily member 9 (TNFRSF9) showed a significant positive correlation with the occurrence of acute pancreatitis (Figure 6).
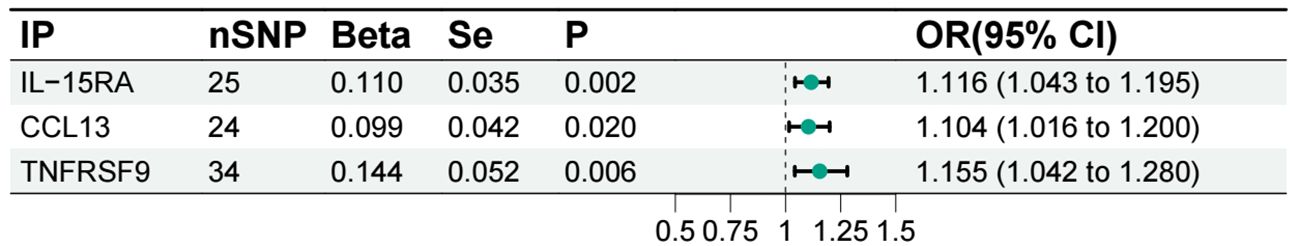
Figure 6 Forest plot of the associations between inflammatory proteins and acute pancreatitis. IP, inflammatory protein; IL-15RA, Interleukin-15 receptor subunit alpha; CCL13, Monocyte chemoattractant protein-4; TNFRSF9, Tumor necrosis factor receptor superfamily member 9.
Our two-step MR analysis, identified IL-15RA as a crucial intermediate factor influencing the relationship between genus.Coprococcus3 and acute pancreatitis (Figure 7). We found that genus.Coprococcus3 could increase the risk of acute pancreatitis by positively influencing IL-15RA. The overall effect of genus.Coprococcus3 on acute pancreatitis was 0.387, with a direct effect of 0.369. IL-15RA has a mediating effect of 0.018 (95%CI: 0.005 - 0.032), indicating that 4.713% of the effect of genus.Coprococcus3 on pancreatitis was mediated by IL-15RA.
Our present study investigated the causal link between gut microbiota, inflammatory proteins, and the risk of acute pancreatitis using genetic prediction. We revealed that IL-15RA plays a mediating role in how genus.Coprococcus3 influences the development of acute pancreatitis. Furthermore, we identified associations between distinct microbial communities and specific inflammatory proteins associated with the disease.
IL-15RA is a high-affinity binding protein for interleukin-15 (IL-15). Its structure includes a signal peptide, sushi domain, hinge region, proline-threonine rich region, transmembrane domain and cytoplasmic domain (Schluns et al., 2005). Expressed in various cell types, IL-15RA plays a crucial role in mediating IL-15 function and T-cell biology (Giri et al., 1995). Studies have shown that the main signaling mechanism for memory T cell survival and proliferation in vivo involves the trans-delivery of IL-15 by IL-15RA on hematopoietic and non-hematopoietic cell types (Burkett et al., 2003; Burkett et al., 2004; Kenesei et al., 2021). Rowley et al. (2009) found that IL-15RA can transparent IL-15 in a cis manner to T cells, promoting the phosphorylation of signal transducer and activator of transcription-5 (STAT5), and the survival and proliferation of unstimulated CD8+ T cells. Moreover, IL-15RA expression alone promotes cell-autonomous survival and proliferation of primary unstimulated CD8+ T cells both in vitro and in vivo. Additionally, it increases proliferation and interferon-γ (IFN-γ) production in antigen-specific T cells in vitro. No prior research has investigated the relationship between IL-15RA and acute pancreatitis or genus.Coprococcus. However, elevated levels of IL-15RA play a crucial role in the pathogenesis of systemic lupus erythematosus (SLE), rheumatoid arthritis (RA), ulcerative colitis (UC), and Crohn’s disease (CD) (Baranda et al., 2005; MaChado Diaz et al., 2012; Jadidi et al., 2022). Patients with UC and CD exhibit heightened mucosal expression of IL-15RA, accompanied by increased serum levels in UC individuals (Perrier et al., 2013). Interestingly, IL-15RA is also thought to harbor anti-tumor effects. The IL-15RA rs2228059 A > C polymorphism decreased the risk of gastric cardiac adenocarcinoma and esophageal cancer in a Chinese population (Yin et al., 2014a; Yin et al., 2014b). Aerobic exercise can further promote immune mobilization and the accumulation of tumor-infiltrating IL15RA and CD8+ T cells, thereby exerting anti-tumor effects (Kurz et al., 2022). Existing research has shown that serum levels of IL-15 significantly increase in patients with severe acute pancreatitis, and it can be used for predicting complications and mortality associated with severe acute pancreatitis (Ueda et al., 2007). While the inflammatory protein data analyzed in our study lacks information on IL-15 itself, IL-15RA serves as the specific high-affinity receptor for IL-15. Upon binding, they activate JAK-STAT5 signal transduction molecules, subsequently leading to the activation of multiple signaling pathways. We speculate that these interactions between IL-15 and IL-15RA may have an impact on pancreatic tissue, although the specific mechanisms require further investigation.
Several studies have explored the relationship between specific gut bacteria and various health conditions. For instance, Alferink et al. conducted a large-scale cross-sectional study that found a weak correlation between Coprococcus and steatosis (Alferink et al., 2021). MR studies have shown that Coprococcus3 is associated with an increased risk of obstructive sleep apnea and cholelithiasis (Liu et al., 2023; Wei et al., 2023). Palm et al. observed that a member of the Erysipelotrichaceae family exhibits higher immunoglobulin A (IgA) coating compared to other gut microbes (Palm et al., 2014). Additionally, the relative abundance of Erysipelotrichi positively correlates with TNF-α (Dinh et al., 2015). In addition, Erysipelotrichaceae may also be related to metabolism. Earlier studies showed increased levels of Erysipelotrichaceae in diet-induced obese mice (Turnbaugh et al., 2008; Fleissner et al., 2010). Higher levels of Erysipelotrichaceae have also been observed in obese individuals, as well as smoking population (Zhang et al., 2009; Kaakoush, 2015; Yang et al., 2022). Several recent MR studies have yielded interesting, and sometimes seemingly contradictory, findings on specific gut bacteria. The Eubacterium fissicatena group, for example, has been shown to have a significant negative causal effect on appendicular lean mass (Zhao et al., 2023) while exhibiting a positive correlation with psoriasis (Zang et al., 2023). Animal experiments by Chonghui Yue et al. (2020) further supported this complex interplay, demonstrating a negative correlation between Eubacterium fissicatena group with various parameters, including epididymal white adipose tissue weight (eWAT weight), TNF-α, interleukin-6 (IL-6), MCP-1 in serum inflammation factors and recombinant peroxisome proliferator-activated receptor-γ (PPARγ), acetyl CoA carboxylase (ACC) in lipid metabolism-related proteins. Interestingly, some traditional Chinese medicines like Sophora japonica flowers, compound Chenpi tea, Gegen Qinlian decoction and Banxia Xiexin decoction have been linked to reducing the abundance of this bacterial group (Chen et al., 2021; Liu et al., 2022; Li et al., 2022; Wang et al., 2024). Fusicatenibacter on the other hand, plays a beneficial role. Known for producing short-chain fatty acids (Gryaznova et al., 2023) and degrading inulin polysaccharides, it also secretes interleukin-10 (IL-10), an anti-inflammatory cytokine (Takada et al., 2013; Takeshita et al., 2016). IL-10, a negative-feedback regulator cytokine, inhibits production of inflammatory cytokines like interleukin-1 (IL-1), TNF-α, and interleukin-12 (IL-12) from macrophages and suppresses T cell activation by inhibiting the expression of costimulators and MHCII on macrophages and dendritic cells (DCs) (Sziksz et al., 2015). Studies in Chinese population suggest a correlation between IL-10 and acute pancreatitis occurrence (Jia et al., 2015; Li et al., 2015; Jiang et al., 2016). Additionally, pirfenidone has been shown to augment the IL-10-driven anti-inflammatory pathway in macrophages, contributing to its effectiveness in treating acute pancreatitis (Palathingal Bava et al., 2022). While Ruminiclostridium6 is not well-studied, some MR analyses associate it with the risk of moderate to severe asthma, scoliosis, and breast inflammatory disease (Gu et al., 2023; Lai et al., 2023; Li et al., 2023). Over the past few decades, research has increasingly revealed the “gut microbiota-pancreatic axis”, highlighting the mutual influence between gut bacteria and the pancreas (Schepis et al., 2021). Under pathological conditions, bacterial translocation to the pancreas can occur. As the intestinal microbial load increases and the epithelial barrier weakens, pancreatitis severity worsens (Zheng et al., 2019; Zhang et al., 2022). Studies comparing with healthy individuals to patients with pancreatitis have shown a decrease in bacterial phyla diversity in the latter group. Specifically, Bacteroidetes, Proteobacteria, Enterococcus, and Enterobacteriaceae were found to be more abundant, while Firmicutes, Actinobacteria, Prevotella9, Baculobacter, Brucella, Lactospiraceae, and Bifidobacterium were less common (Zhang Xi et al., 2018; Zhu et al., 2018). However, our MR analysis did not identify a causal relationship between these specific bacteria and acute pancreatitis. Indeed, it should be borne in mind that MR is a genetic approach that explores potential relationships between exposures and outcomes, and differs from observational studies in its methodology.
Our MR analysis has identified a causal relationship between gut microbiota and pancreatitis, revealing the mediating role of inflammatory proteins. This approach offers new insights for future research by mitigating the influence of confounding factors and revealing a novel genetic link between these factors. From a disease prevention perspective, these findings suggest the potential for preventing pancreatitis by gut microbiota, inflammatory proteins, or relevant factors through timely adjustments. However, the limitations of the present study should be acknowledged. The majority of individuals participating in the GWAS gut microbiota coefficient meta-analysis were of European ancestry, potentially limiting the generalizability of our results to non-European populations. Furthermore, the absence of data stratified by different etiologies of pancreatitis prevented consideration of etiological influences, thereby restricting our comprehensive investigation into how various causes may impact the observed genetic associations. Additionally, future research is necessary to elucidate the exact mechanisms, targets, and pathways underlying this association. Therefore, caution is warranted when interpreting the current findings.
MR analysis revealed that genus. Coprococcus3 promotes acute pancreatitis through IL-15RA. Other intestinal bacteria, such as the genus.Eubacterium fissicatena group, are associated with an increased risk of acute pancreatitis, while, genus.Ruminiclostridium6, genus.Fusicatenibacter, and genus.Erysipelotrichaceae UCG-003 appear to be protective factors. Inflammatory proteins CCL13 and TNFRSF9 also play a promoting role in the development of acute pancreatitis. These findings may provide insights for preventing acute pancreatitis, but the specific mechanisms require further investigation.
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author/s.
PH: Formal analysis, Visualization, Writing – original draft. QL: Data curation, Formal analysis, Writing – review & editing. TZ: Formal analysis, Visualization, Writing – original draft. JY: Conceptualization, Funding acquisition, Project administration, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by Zhejiang Province’s 2024 Key R&D Plan Project (Grant No. 2024C03048); Zhejiang Provincial Traditional Chinese Medicine Science and Technology Project (Grant No. GZY-ZJ-KJ-24093); Hangzhou Science and Technology Commission (202004A14); and the Construction Fund of Medical Key Disciplines of Hangzhou (OO20190001).
This work was made possible by the generous sharing of GWAS summary statistics from the FinnGen, MiBioGen consortium, and Estonian Biobank Research Team. We offer our thanks and appreciation to the all the individual patients who provided the sample that made the data available and all the investigators who provided these data to support this study. We appreciate the Figdraw version 2.0 for providing graphical support and also thank ChatGPT version 4.0 and Researchers editorial team (www.home-for-researchers.com) for language editing services.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1380998/full#supplementary-material
Alferink, L. J. M., Radjabzadeh, D., Erler, N. S., Vojinovic, D., Medina-Gomez, C., Uitterlinden, A. G., et al. (2021). Microbiomics, metabolomics, predicted metagenomics, and hepatic steatosis in a population-based study of 1,355 adults. Hepatology 73, 968–982. doi: 10.1002/hep.31417
Baquero, F., Nombela, C. (2012). The microbiome as a human organ. Clin. Microbiol. Infect. 18 Suppl 4, 2–4. doi: 10.1111/j.1469-0691.2012.03916.x
Baranda, L., de la Fuente, H., Layseca-Espinosa, E., Portales-Pérez, D., Niño-Moreno, P., Valencia-Pacheco, G., et al. (2005). IL-15 and IL-15R in leucocytes from patients with systemic lupus erythematosus. Rheumatol. (Oxford) 44, 1507–1513. doi: 10.1093/rheumatology/kei083
Brandi, G., Turroni, S., McAllister, F., Frega, G. (2021). The human microbiomes in pancreatic cancer: Towards evidence-based manipulation strategies? Int. J. Mol. Sci. 22. doi: 10.3390/ijms22189914
Burgess, S., Dudbridge, F., Thompson, S. G. (2016). Combining information on multiple instrumental variables in Mendelian randomization: comparison of allele score and summarized data methods. Stat. Med. 35, 1880–1906. doi: 10.1002/sim.6835
Burkett, P. R., Koka, R., Chien, M., Chai, S., Chan, F., Ma, A., et al. (2003). IL-15R alpha expression on CD8+ T cells is dispensable for T cell memory. Proc. Natl. Acad. Sci. U. S. A. 100, 4724–4729. doi: 10.1073/pnas.0737048100
Burkett, P. R., Koka, R., Chien, M., Chai, S., Boone, D. L., Ma, A. (2004). Coordinate expression and trans presentation of interleukin (IL)-15Ralpha and IL-15 supports natural killer cell and memory CD8+ T cell homeostasis. J. Exp. Med. 200, 825–834. doi: 10.1084/jem.20041389
Chen, J., Zhang, L. K., Gu, W. C., Zhang, X. S., Li, L., Han, T., et al. (2021). Effect of Banxia Xiexin Decoction on intestinal flora of mice with ulcerative colitis induced by dextran sodium sulfate. Zhongguo Zhong Yao Za Zhi 46, 2871–2880. doi: 10.19540/j.cnki.cjcmm.20210119.401
Dinh, D. M., Volpe, G. E., Duffalo, C., Bhalchandra, S., Tai, A. K., Kane, A. V., et al. (2015). Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J. Infect. Dis. 211, 19–27. doi: 10.1093/infdis/jiu409
Distrutti, E., Monaldi, L., Ricci, P., Fiorucci, S. (2016). Gut microbiota role in irritable bowel syndrome: New therapeutic strategies. World J. Gastroenterol. 22, 2219–2241. doi: 10.3748/wjg.v22.i7.2219
Fleissner, C. K., Huebel, N., Abd El-Bary, M. M., Loh, G., Klaus, S., Blaut, M. (2010). Absence of intestinal microbiota does not protect mice from diet-induced obesity. Br. J. Nutr. 104, 919–929. doi: 10.1017/S0007114510001303
Garg, P. K., Singh, V. P. (2019). Organ failure due to systemic injury in acute pancreatitis. Gastroenterology 156, 2008–2023. doi: 10.1053/j.gastro.2018.12.041
Gill, D., Efstathiadou, A., Cawood, K., Tzoulaki, I., Dehghan, A. (2019). Education protects against coronary heart disease and stroke independently of cognitive function: evidence from Mendelian randomization. Int. J. Epidemiol. 48, 1468–1477. doi: 10.1093/ije/dyz200
Giri, J. G., Kumaki, S., Ahdieh, M., Friend, D. J., Loomis, A., Shanebeck, K., et al. (1995). Identification and cloning of a novel IL-15 binding protein that is structurally related to the alpha chain of the IL-2 receptor. EMBO J. 14, 3654–3663. doi: 10.1002/embj.1995.14.issue-15
Gryaznova, M., Smirnova, Y., Burakova, I., Syromyatnikov, M., Chizhkov, P., Popov, E., et al. (2023). Changes in the human gut microbiome caused by the short-term impact of lactic acid bacteria consumption in healthy people. Probiotics Antimicrob. Proteins. doi: 10.1007/s12602-023-10111-4
Gu, Y., Hou, M., Chu, J., Wan, L., Yang, M., Shen, J., et al. (2023). The cause and effect of gut microbiota in development of inflammatory disorders of the breast. Eur. J. Med. Res. 28, 324. doi: 10.1186/s40001-023-01281-6
Jadidi, N., Alesaeidi, S., Arab, F., Pakzad, B., Siasi, E., Esmaeilzadeh, E. (2022). miRNA-binding site polymorphism in IL-15RA gene in rheumatoid arthritis and systemic lupus erythematosus: correlation with disease risk and clinical characteristics. Clin. Rheumatol. 41, 3487–3494. doi: 10.1007/s10067-022-06298-6
Jia, H. L., Sun, P. L., Lu, C. Q. (2015). Investigation of the association between interleukin-10 polymorphisms and risk of acute pancreatitis in a Chinese population. Genet. Mol. Res. 14, 15876–15881. doi: 10.4238/2015.December.1.39
Jiang, B. Z., Tang, L., Xue, H., Liu, D. P. (2016). Role of IL-10 gene polymorphisms in the development of acute pancreatitis. Genet. Mol. Res. 15 (2). doi: 10.4238/gmr.15027743
Kaakoush, N. O. (2015). Insights into the role of erysipelotrichaceae in the human host. Front. Cell Infect. Microbiol. 5. doi: 10.3389/fcimb.2015.00084
Kenesei, Á, Volkó, J., Szalóki, N., Mocsár, G., Jambrovics, K., Balajthy, Z., et al. (2021). IL-15 trans-presentation is an autonomous, antigen-independent process. J. Immunol. 207, 2489–2500. doi: 10.4049/jimmunol.2100277
Kurilshikov, A., Medina-Gomez, C., Bacigalupe, R., Radjabzadeh, D., Wang, J., Demirkan, A., et al. (2021). Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 53, 156–165. doi: 10.1038/s41588-020-00763-1
Kurz, E., Hirsch, C. A., Dalton, T., Shadaloey, S. A., Khodadadi-Jamayran, A., Miller, G., et al. (2022). Exercise-induced engagement of the IL-15/IL-15Rα axis promotes anti-tumor immunity in pancreatic cancer. Cancer Cell. 40, 720–737.e725. doi: 10.1016/j.ccell.2022.05.006
Lai, B., Jiang, H., Gao, Y., Zhou, X. (2023). Causal effects of gut microbiota on scoliosis: A bidirectional two-sample mendelian randomization study. Heliyon 9, e21654. doi: 10.1016/j.heliyon.2023.e21654
Lee, K., Lim, C. Y. (2019). Mendelian randomization analysis in observational epidemiology. J. Lipid Atheroscler 8, 67–77. doi: 10.12997/jla.2019.8.2.67
Lee, W. J., Hase, K. (2014). Gut microbiota-generated metabolites in animal health and disease. Nat. Chem. Biol. 10, 416–424. doi: 10.1038/nchembio.1535
Levin, M. G., Judy, R., Gill, D., Vujkovic, M., Verma, S. S., Bradford, Y., et al. (2020). Genetics of height and risk of atrial fibrillation: A Mendelian randomization study. PLoS Med. 17, e1003288. doi: 10.1371/journal.pmed.1003288
Li, R., Guo, Q., Zhao, J., Kang, W., Lu, R., Long, Z., et al. (2023). Assessing causal relationships between gut microbiota and asthma: evidence from two sample Mendelian randomization analysis. Front. Immunol. 14, 1148684. doi: 10.3389/fimmu.2023.1148684
Li, Y., Li, N., Liu, J., Wang, T., Dong, R., Ge, D., et al. (2022). Gegen qinlian decoction alleviates experimental colitis and concurrent lung inflammation by inhibiting the recruitment of inflammatory myeloid cells and restoring microbial balance. J. Inflamm. Res. 15, 1273–1291. doi: 10.2147/JIR.S352706
Li, D., Li, J., Wang, L., Zhang, Q. (2015). Association between IL-1β, IL-8, and IL-10 polymorphisms and risk of acute pancreatitis. Genet. Mol. Res. 14, 6635–6641. doi: 10.4238/2015.June.18.6
Li, P., Wang, H., Guo, L., Gou, X., Chen, G., Lin, D., et al. (2022). Association between gut microbiota and preeclampsia-eclampsia: a two-sample Mendelian randomization study. BMC Med. 20, 443. doi: 10.1186/s12916-022-02657-x
Liu, Y., Huang, W., Ji, S., Wang, J., Luo, J., Lu, B. (2022). Sophora japonica flowers and their main phytochemical, rutin, regulate chemically induced murine colitis in association with targeting the NF-κB signaling pathway and gut microbiota. Food Chem. 393, 133395. doi: 10.1016/j.foodchem.2022.133395
Liu, X., Qi, X., Han, R., Mao, T., Tian, Z. (2023). Gut microbiota causally affects cholelithiasis: a two-sample Mendelian randomization study. Front. Cell Infect. Microbiol. 13. doi: 10.3389/fcimb.2023.1253447
MaChado Diaz, A. C., Chico Capote, A., Arrieta Aguero, C. A., Rodríguez Alvarez, Y., García Del Barco Herrera, D., Estévez Del Toro, M., et al. (2012). Proinflammatory soluble interleukin-15 receptor alpha is increased in rheumatoid arthritis. Arthritis 2012, 943156. doi: 10.1155/2012/943156
Palathingal Bava, E., George, J., Tarique, M., Iyer, S., Sahay, P., Gomez Aguilar, B., et al. (2022). Pirfenidone increases IL-10 and improves acute pancreatitis in multiple clinically relevant murine models. JCI Insight 7 (2), e141108. doi: 10.1172/jci.insight.141108
Palm, N. W., de Zoete, M. R., Cullen, T. W., Barry, N. A., Stefanowski, J., Hao, L., et al. (2014). Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 158, 1000–1010. doi: 10.1016/j.cell.2014.08.006
Palmer, T. M., Lawlor, D. A., Harbord, R. M., Sheehan, N. A., Tobias, J. H., Timpson, N. J., et al. (2012). Using multiple genetic variants as instrumental variables for modifiable risk factors. Stat. Methods Med. Res. 21, 223–242. doi: 10.1177/0962280210394459
Perrier, C., Arijs, I., Staelens, D., Breynaert, C., Cleynen, I., Covens, K., et al. (2013). Interleukin-15 receptor α expression in inflammatory bowel disease patients before and after normalization of inflammation with infliximab. Immunology. 138, 47–56. doi: 10.1111/imm.12014
Ramakrishna, B. S. (2013). Role of the gut microbiota in human nutrition and metabolism. J. Gastroenterol. Hepatol. 28 Suppl 4, 9–17. doi: 10.1111/jgh.12294
Rowley, J., Monie, A., Hung, C. F., Wu, T. C. (2009). Expression of IL-15RA or an IL-15/IL-15RA fusion on CD8+ T cells modifies adoptively transferred T-cell function in cis. Eur. J. Immunol. 39, 491–506. doi: 10.1002/eji.200838594
Schepis, T., De Lucia, S. S., Nista, E. C., Manilla, V., Pignataro, G., Ojetti, V., et al. (2021). Microbiota in pancreatic diseases: A review of the literature. J. Clin. Med. 10 (24), 5920. doi: 10.3390/jcm10245920
Schluns, K. S., Stoklasek, T., Lefrançois, L. (2005). The roles of interleukin-15 receptor alpha: trans-presentation, receptor component, or both? Int. J. Biochem. Cell Biol. 37, 1567–1571. doi: 10.1016/j.biocel.2005.02.017
Sekirov, I., Russell, S. L., Antunes, L. C., Finlay, B. B. (2010). Gut microbiota in health and disease. Physiol. Rev. 90, 859–904. doi: 10.1152/physrev.00045.2009
Sziksz, E., Pap, D., Lippai, R., Béres, N. J., Fekete, A., Szabó, AJ., et al. (2015). Fibrosis related inflammatory mediators: Role of the IL-10 cytokine family. Mediators Inflamm. 2015, 764641. doi: 10.1155/2015/764641
Takada, T., Kurakawa, T., Tsuji, H., Nomoto, K. (2013). Fusicatenibacter saccharivorans gen. nov., sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 63, 3691–3696. doi: 10.1099/ijs.0.045823-0
Takeshita, K., Mizuno, S., Mikami, Y., Sujino, T., Saigusa, K., Matsuoka, K., et al. (2016). A single species of clostridium subcluster XIVa decreased in ulcerative colitis patients. Inflamm. Bowel Dis. 22, 2802–2810. doi: 10.1097/MIB.0000000000000972
Trabert, B., Pinto, L., Hartge, P., Kemp, T., Black, A., Sherman, M. E., et al. (2014). Pre-diagnostic serum levels of inflammation markers and risk of ovarian cancer in the prostate, lung, colorectal and ovarian cancer (PLCO) screening trial. Gynecol. Oncol. 135, 297–304. doi: 10.1016/j.ygyno.2014.08.025
Turnbaugh, P. J., Bäckhed, F., Fulton, L., Gordon, J. I. (2008). Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 3, 213–223. doi: 10.1016/j.chom.2008.02.015
Ueda, T., Takeyama, Y., Yasuda, T., Shinzeki, M., Sawa, H., Nakajima, T., et al. (2007). Serum interleukin-15 level is a useful predictor of the complications and mortality in severe acute pancreatitis. Surgery. 142, 319–326. doi: 10.1016/j.surg.2007.05.002
Wang, J., Hao, J., Miao, D., Xiao, P., Jiang, X., L., E.H., et al. (2024). Compound chenpi tea consumption reduces obesity-related metabolic disorders by modulating gut microbiota and serum metabolites in mice. J. Sci. Food Agric. 104, 431–442. doi: 10.1002/jsfa.12940
Wehby, G. L., Ohsfeldt, R. L., Murray, J. C. (2008). 'Mendelian randomization' equals instrumental variable analysis with genetic instruments. Stat. Med. 27, 2745–2749. doi: 10.1002/sim.3255
Wei, Y., Huang, L., Liu, C., Qi, M. (2023). Causal relationship between Gut Microbiota and Obstructive sleep apnea. Arch. Gerontol Geriatr. 113, 105052. doi: 10.1016/j.archger.2023.105052
Xiao, A. Y., Tan, M. L., Wu, L. M., Asrani, V. M., Windsor, J. A., Yadav, D., et al. (2016). Global incidence and mortality of pancreatic diseases: a systematic review, meta-analysis, and meta-regression of population-based cohort studies. Lancet Gastroenterol. Hepatol. 1, 45–55. doi: 10.1016/S2468-1253(16)30004-8
Yang, H. T., Xiu, W. J., Liu, J. K., Yang, Y., Zhang, Y. J., Zheng, Y. Y., et al. (2022). Characteristics of the intestinal microorganisms in middle-aged and elderly patients: Effects of smoking. ACS Omega 7, 1628–1638. doi: 10.1021/acsomega.1c02120
Yin, J., Liu, C., Wang, X., Wang, L., Shi, Y., Tang, W., et al. (2014a). Interleukin 15 receptor alpha rs2228059 A > C polymorphism decreased risk of gastric cardiac adenocarcinoma in a Chinese population. Tumour Biol. 35, 6593–6600. doi: 10.1007/s13277-014-1872-6
Yin, J., Wang, L., Shi, Y., Shao, A., Tang, W., Wang, X., et al. (2014b). IL-15 receptor alpha rs2228059 A>C polymorphism was associated with a decreased risk of esophageal cancer in a Chinese population. Mol. Biol. Rep. 41, 1951–1957. doi: 10.1007/s11033-014-3042-8
Yue, C., Li, M., Li, J., Han, X., Zhu, H., Yu, G., et al. (2020). Medium-, long- and medium-chain-type structured lipids ameliorate high-fat diet-induced atherosclerosis by regulating inflammation, adipogenesis, and gut microbiota in ApoE(-/-) mice. Food Funct. 11, 5142–5155. doi: 10.1039/D0FO01006E
Zang, C., Liu, J., Mao, M., Zhu, W., Chen, W., Wei, B. (2023). Causal associations between gut microbiota and psoriasis: A mendelian randomization study. Dermatol. Ther. (Heidelb). 13, 2331–2343. doi: 10.1007/s13555-023-01007-w
Zhang, H., DiBaise, J. K., Zuccolo, A., Kudrna, D., Braidotti, M., Yu, Y., et al. (2009). Human gut microbiota in obesity and after gastric bypass. Proc. Natl. Acad. Sci. U. S. A. 106, 2365–2370. doi: 10.1073/pnas.0812600106
Zhang, Z., Tanaka, I., Pan, Z., Ernst, PB., Kiyono, H., Kurashima, Y. (2022). Intestinal homeostasis and inflammation: Gut microbiota at the crossroads of pancreas-intestinal barrier axis. Eur. J. Immunol. 52, 1035–1046. doi: 10.1002/eji.202149532
Zhang Xi, M., Zhang Zheng, Y., Zhang Chen, H., Wu, J., Wang You, X., Zhang Guo, X.. (2018). Intestinal microbial community differs between acute pancreatitis patients and healthy volunteers. Biomed. Environ. Sci. 31, 81–86. doi: 10.3967/bes2018.010
Zhao, Y., Jaber, V., Lukiw, W. J. (2021). Gastrointestinal tract microbiome-derived pro-inflammatory neurotoxins in alzheimer's disease. J. Aging Sci. 9 (Suppl 5), 002.
Zhao, J., Liang, R., Song, Q., Song, S., Yue, J., Wu, C. (2023). Investigating association between gut microbiota and sarcopenia-related traits: a Mendelian randomization study. Precis Clin. Med. 6, pbad010. doi: 10.1093/pcmedi/pbad010
Zhao, J. H., Stacey, D., Eriksson, N., Macdonald-Dunlop, E., Hedman, Å K., Kalnapenkis, A., et al. (2023). Genetics of circulating inflammatory proteins identifies drivers of immune-mediated disease risk and therapeutic targets. Nat. Immunol. 24, 1540–1551. doi: 10.1038/s41590-023-01588-w
Zheng, J., Lou, L., Fan, J., Huang, C., Mei, Q., Wu, J., et al. (2019). Commensal Escherichia coli Aggravates Acute Necrotizing Pancreatitis through Targeting of Intestinal Epithelial Cells. Appl. Environ. Microbiol. 85 (12), e00059–19. doi: 10.1128/AEM.00059-19
Keywords: acute pancreatitis, gut microbiota, inflammatory proteins, Mendelian randomization, single nucleotide polymorphism
Citation: Huang P, Liu Q, Zhang T and Yang J (2024) Gut microbiota influence acute pancreatitis through inflammatory proteins: a Mendelian randomization analysis. Front. Cell. Infect. Microbiol. 14:1380998. doi: 10.3389/fcimb.2024.1380998
Received: 02 February 2024; Accepted: 13 May 2024;
Published: 31 May 2024.
Edited by:
Abbas Yadegar, Shahid Beheshti University of Medical Sciences, IranCopyright © 2024 Huang, Liu, Zhang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianfeng Yang, eWpmMzMwM0B6anUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.