
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol., 07 May 2024
Sec. Fungal Pathogenesis
Volume 14 - 2024 | https://doi.org/10.3389/fcimb.2024.1369301
This article is part of the Research TopicRising Stars in Fungal Pathogenesis: 2023View all 6 articles
Dual-specificity LAMMER kinases are highly evolutionarily conserved in eukaryotes and play pivotal roles in diverse physiological processes, such as growth, differentiation, and stress responses. Although the functions of LAMMER kinase in fungal pathogens in pathogenicity and stress responses have been characterized, its role in Cryptococcus neoformans, a human fungal pathogen and a model yeast of basidiomycetes, remains elusive. In this study, we identified a LKH1 homologous gene and constructed a strain with a deleted LKH1 and a complemented strain. Similar to other fungi, the lkh1Δ mutant showed intrinsic growth defects. We observed that C. neoformans Lkh1 was involved in diverse stress responses, including oxidative stress and cell wall stress. Particularly, Lkh1 regulates DNA damage responses in Rad53-dependent and -independent manners. Furthermore, the absence of LKH1 reduced basidiospore formation. Our observations indicate that Lkh1 becomes hyperphosphorylated upon treatment with rapamycin, a TOR protein inhibitor. Notably, LKH1 deletion led to defects in melanin synthesis and capsule formation. Furthermore, we found that the deletion of LKH1 led to the avirulence of C. neoformans in a systemic cryptococcosis murine model. Taken together, Lkh1 is required for the stress response, sexual differentiation, and virulence of C. neoformans.
In cell signaling cascades, protein phosphorylation and dephosphorylation serve as pivotal switches, orchestrating the activation or deactivation of signaling cascades by facilitating alterations in protein conformation, protein-protein interactions, and subcellular localization. These dynamic processes play critical roles in regulating various biological phenomena. Most protein phosphorylation occurs at serine, threonine, and tyrosine residues. Occasionally, histidine and aspartate residues are phosphorylated; however, these modifications are less prevalent and exhibit lower stability than those of serine, threonine, and tyrosine (Duclos et al., 1991). Given the importance of protein phosphorylation in cell signaling cascades, protein kinases are evolutionarily conserved across species (Manning et al., 2002).
Within the evolutionarily conserved kinases, LAMMER kinase stands out, characterized by the presence of a conserved motif, ‘EHLAMMERILG’ located in the subdomain X of the kinase catalytic domain (Yun et al., 1994). This kinase plays crucial roles in various physiological processes, such as differentiation, growth, and stress responses, from yeasts to humans. The LAMMER kinase is a dual-specific kinase that exhibits autophosphorylation activity in Drosophila, humans, and Saccharomyces cerevisiae (Lee et al., 1996). In humans, three LAMMER kinase isoforms, CLK1 (Cdc-like kinase 1), CLK2, and CLK3, are located at different chromosomal loci (Hanes et al., 1994; Talmadge et al., 1998). hCLKs play important roles in diverse cellular responses, including neuronal differentiation, cell cycle progression, and apoptosis (Myers et al., 1994; Jiang et al., 2003). Similar to humans, Arabidopsis thaliana possesses three LAMMER kinase homologs: AFC1, AFC2, and AFC3 (Savaldi-Goldstein et al., 2003). AFC1, but not AFC2, restores Ste12-dependent pseudohyphal growth in S. cerevisiae (Bender and Fink, 1994). In contrast to higher eukaryotes, which harbor several LAMMER kinase variants, the model yeasts S. cerevisiae and Schizosaccharomyces pombe have a singular LAMMER kinase. Unlike other human and plant LAMMER kinases, the deletion of KNS1, which encodes a LAMMER kinase in S. cerevisiae, does not affect the viability, growth, and sporulation efficiency (Padmanabha et al., 1991). Although the kns1Δ mutant in the S288c background does not exhibit noticeable phenotypes compared to the wild-type (WT), its regulatory mechanism has been elucidated. Kns1 undergoes autophosphorylation and its localization becomes enriched in the nucleus under TOR-suppressed conditions (Lee et al., 2012). Furthermore, Kns1 is involved in ribosome and tRNA synthesis through RNA polymerase III subunit Rpc53 phosphorylation and the regulatory subunit of the CK2 protein kinase, Ckb1 (Lee et al., 2012; Sanchez-Casalongue et al., 2015). The deletion of LKH1, which encodes a LAMMER kinase in S. pombe, results in increased susceptibility to oxidative stress by reducing the expression levels of genes encoding catalase and superoxide dismutase (Park et al., 2003). Furthermore, SpLkh1 acts as a negative filamentous growth regulator and is involved in cell wall biosynthesis (Park et al., 2003; Cho et al., 2010).
Besides the model budding and fission yeasts, the roles of LAMMER kinase and its functional regulatory mechanisms in fungal pathogens have been elucidated. In Candida albicans, CaKNS1 is involved in hyphal morphogenesis and stress responses such as cell wall stress and DNA replicative stress (Lim et al., 2020). Notably, CaKNS1 is required for the full virulence of C. albicans (Park and Park, 2011). In Aspergillus fumigatus, LkhA deletion results in growth and sexual differentiation defects. Similar to the Cakns1Δ mutant, the ΔlhkA mutant exhibits attenuated virulence in the zebrafish larvae model (Lim et al., 2021). In the plant fungal pathogen Magnaporthe oryzae, the kns1Δ mutant exhibits decreased growth and conidiation, similar to other filamentous fungi. Furthermore, the virulence of the Δkns1 mutant is significantly reduced in a rice infection model (Li et al., 2022). In Ustilago maydis, UnLhk1 is required for DNA damage, meiotic recombination, and chromosomal segregation. However, the pathogenicity of the Unlkh1Δ mutant has not yet been determined (de Sena-Tomás et al., 2015).
Cryptococcus neoformans is a human fungal pathogen belonging to the basidiomycetes and is particularly prevalent in immunocompromised patients. Without appropriate therapeutic treatment, the mortality rate increases significantly (Idnurm et al., 2005). Briefly, spores or desiccated C. neoformans from the environment are inhaled into the lungs via the respiratory tract (Ellis and Pfeiffer, 1990; Velagapudi et al., 2009). Next, the C. neoformans disseminates to other organs in the host and subsequently migrates to the central nervous system, leading to life-threatening meningitis (Gottfredsson and Perfect, 2000; Kwon-Chung et al., 2000). A recent study estimates that 152,000 cases of cryptococcal meningitis occurred in patients with HIV/AIDS, resulting in nearly 112,000 deaths in 2020 (Rajasingham et al., 2022). Furthermore, the incidence of cryptococcal infections has increased in patients with COVID-19 (Regalla et al., 2022). C. neoformans serves as an attractive fungal model for studying host-pathogen interactions (Perfect, 2014).
Although the role of LAMMER kinase Lkh1 in growth, stress response, and virulence has been clarified in ascomycetous human fungal pathogens, the pathobiological functions of the Lkh1 homolog in C. neoformans remain elusive. To explore the roles of Lkh1, we identified the LKH1 homologous gene and performed phenotypic analyses using lkh1Δ mutants. Our findings reveal that the deletion of LKH1 led to defects in growth, sexual differentiation, and stress responses. Particularly, Lkh1 was required for virulence in the mice model. Therefore, we propose that Lkh1 is vital for pathogenicity, stress response, and differentiation of the human fungal pathogen Cryptococcus.
Animal care and research were approved after deliberation by the Institutional Animal Care and Use Committee of the Experimental Animal Center at Jeonbuk National University (Approval number: JBNU 2022-092). All experiments were conducted per experimental ethics guidelines. The animal experiments were conducted at the Core Facility Center for Zoonosis Research (Jeonbuk National University, South Korea).
The C. neoformans strains used in the experiments were listed in Supplementary Table S1 and were cultured in yeast extract-peptone-dextrose (YPD) medium. The V8 medium used for the mating assay contained 5% V8 juice, 0.5 g/L of KH2PO4, and 4% bacto-agar with the pH adjusted to 5 using a KOH solution (Jung et al., 2018). For the melanin production assay, equivalent numbers of cells were spotted on a dopamine agar medium with varying glucose concentrations (0%, 0.1%, or 0.2%) (Lee et al., 2019). For the capsule assay, each strain was incubated in Littman’s medium (Littman, 1958) and capsule thickness was measured as previously described (Jang et al., 2022). To assess the growth of strains using a single amino acid as the sole nitrogen source, equivalent amounts of cells were subcultured in a minimal medium (YNB without amino acids and ammonium sulfate) containing specific nitrogen sources (2 mM proline, 2 mM isoleucine, and 2 mM lysine).
Gene disruption information was obtained from FungiDB (www.fungiDB.org). LKH1 (CNAG_00683) was disrupted by double-joint PCR using split markers and Cryptococcus transformation as described previously (Fan and Lin, 2018; Jung et al., 2018). Supplementary Table S2 describes the primers for generating the 5’- and 3’-flanking regions of LKH1 and the nourseothricin resistance (NAT) and G418 resistance (NEO) dominant selectable markers. Gel-extracted deletion cassettes, the SgRNA gene, and Cas9 were transformed using a BioRad gene pulser (Cat# 1652086). The rad53Δ lkh1Δ double mutant and mpk1Δ lkh1Δ double mutant were constructed by introducing the lkh1Δ::NEO allele into the rad53Δ mutant (YSB3785) and mpk1Δ mutant (YSB3814) through the Cryptococcus transformation. The Rad53-4×FLAG lkh1Δ strain was constructed by introducing the lkh1Δ::NAT allele into the Rad53-4×FLAG mutant (YSB3806) using the Cryptococcus transformation. To produce Lkh1-4×FLAG strains, three PCR product fragments were amplified. The 3’-exon region of LKH1 was amplified using primers J1966 and J1967 and H99 genomic DNA as the template. The 4×FLAG-HOG1ter-NEO fragment was amplified using primers B6567 and B354 and the plasmid pNEO-4×FLAG as the template (So et al., 2017). The 3’-flanking region of the LKH1 was amplified using primers J1969 and J1968 and H99 genomic DNA as the template. Next, a fusion fragment, including the 3’-exon region of LKH1 and 5’-split region of the 4×FLAG-HOG1ter-NEO fragment, was amplified using primers B1886 and J1966 and the mixed first-round PCR product as the template. A fusion fragment containing the 3’-split region of the 4×FLAG-HOG1ter-NEO fragment and the 3’-flanking region of the LKH1 gene was amplified using primers J1969 and B1887 and the combined first-round PCR products as a template. The two DJ-PCR products were mixed and introduced into the H99 strain via biolistic transformation (Jung et al., 2018). The Lkh1-4×FLAG sit4Δ strain or the Lkh1-4×FLAG sch9Δ strain were constructed by introducing the sit4Δ::NAT allele into the Lkh1-4×FLAG mutant (KW1755) using the Cryptococcus transformation or introducing sch9Δ::NAT allele into the Lkh1-4×FLAG mutant (KW1755) using the biolistic transformation. To confirm the gene disruption, Southern blot analysis was performed using a DIG-labeled gene-specific probe (Supplementary Figure S1).
To validate the observed phenotypes in the lkh1Δ mutant, we constructed the corresponding complemented strains as follows. The LKH1 gene fragment containing the promoter, open reading frame (ORF), and the terminator was divided into two fragments and amplified by PCR using the primer pairs J718/J723 and J1711/J1712 for the LKH1-L and LKH1-R parts, respectively. Each amplified gene fragment was cloned into the plasmid pJET 2.1 to produce pJET-LKH1-L (KWE174) and pJET-LKH1-R (KWE176). After confirming the DNA sequence of each part of LKH1, the BamHI restriction enzyme-digested pJET-LKH1-R insert was subcloned into pJET-LKH1-L to generate the plasmid pJET-LKH1-LR (KWE181), which contains a genomic fragment of the LKH1 gene. Next, the XhoI restriction enzyme-digested pJET-LKH1-LR insert was subcloned into plasmid pTOP-NEO containing the NEO-selection marker to produce pNEO-LKH1 (KWE188). The linearized pNEO-LKH1, digested by SnaBI, was introduced into the native LKH1 locus of the lkh1Δ mutant (KW1451) by biolistic transformation. To confirm the correct LKH1 gene insertion, we conducted diagnostic PCR using primers J1895 and J1705. To construct LKH1-KD strains, we generated LKH1 kinase-dead allele (D546A; LKH1KD) as follows. To substitute aspartic acid with alanine in LKH1, pNEO-LKH1-KD was amplified by PCR using KOD DNA polymerase (TOYOBO) with primers J2006 and J2007 and pNEO-LKH1 as a template and subsequently digested with DpnI (Enzynomics) at 37°C for 1 h to digest pNEO-LKH1. DpnI-treated PCR products were transformed into competent cells. After sequencing with no errors, the SnaBI-digested pNEO-LKH1-KD was introduced into the lkh1Δ mutant (KW1451) by biolistic transformation. The pNEO-LKH1-KD integration was confirmed by diagnostic PCR, and the LKH1-KD allele expression level in the lkh1Δ mutant was measured by qRT-PCR analysis using LKH1-specific primers, J1692 and J1908.
C. neoformans strains were cultured in 2 mL liquid YPD medium at 30°C overnight. Strains were serially diluted 10-fold (1 to 104) and spotted onto YPD medium containing diverse types of stress agents [bleomycin, methyl methanesulfonate (MMS), hydroxyurea (HU), 4-nitroquinoline 1-oxide (4-NQO), cisplatin, amphotericin B (AmpB), fluconazole (FCZ), caffeine, calcofluor white (CFW), congo-red (CR), sodium dodecyl sulfate (SDS), H2O2, diamide and menadione]. Strains that were spotted onto the YPD medium containing stress agents were incubated at 30°C for 2–5 days.
The strains were cultured in YPD medium for 16 h at 30°C and subcultured in fresh liquid YPD medium at 30°C until the optical density of the culture medium at 600 nm reached 0.6. The 50 ml of cells were harvested and pelleted by centrifugation for a basal time. The remaining cultures were treated with diverse agents. The cultures were further incubated and harvested at the indicated time points. The harvested samples were frozen in liquid nitrogen and lyophilized overnight. Total RNA was isolated using Easyblue (Intron, #17061), as previously described (Ko et al., 2009), and purified using an RNeasy Mini Kit (Qiagen, #74104) following the manufacturer’s instructions. cDNA was synthesized using the PrimeScript 1st strand cDNA synthesis kit (TAKARA, #6110A) using purified total RNAs as a template. qRT-PCR analysis was performed with the gene-specific primers listed in Supplementary Table S2 using the CFX96 real-time PCR detection system (Bio-Rad). Relative target gene expression was determined using the 2- Δ ΔCt method and ACT1 expression was used for internal control (Livak and Schmittgen, 2001). Statistical analyses were performed using a one-way analysis of variance (ANOVA) with Bonferroni’s multiple comparison test (GraphPad, USA).
C. neoformans strains were incubated overnight at 30°C. The cells were then centrifuged and washed with 1×PBS. The cell quantities were synchronized using a hemocytometer. Subsequently, synchronized cells were treated with menadione and harvested at each time point. Samples were fixed as previously described (Jung et al., 2012). Each sample was divided into two parts: one part was treated with 10 μM H2DCFDA, and the other was used as a control. All samples were incubated for 1 h in the dark at room temperature. After incubation, all samples were washed three times with 1 × PBS and divided into 96 Grenier plates and 96 flat black plates. Absorbance was measured at an optical density of 600 nm and fluorescence was measured at 488 and 522 nm.
WT (H99) and lkh1Δ strain were grown in a liquid YPD medium for 16 h at 30°C and subcultured in a fresh liquid YPD medium at 30°C until the optical density at 600 nm of the culture medium ranged between 0.7 and 0.8. The cell culture was harvested as a basal sample, and the remaining culture was treated with either caffeine (final concentration: 1 mg/mL). After treating the stress agent, 25 mL of the cell culture was harvested using an equal volume of ice-cold protein-stopping buffer (0.9% NaCl, 1 mM NaN3, 10 mM EDTA, and 50 mM NaF) at the indicated time points which are 0.5, 1, 2, and 3 h for caffeine treatment. The concentration of protein extracted from the cells was quantitatively measured using a bicinchoninic acid (BCA) kit (Thermo Fisher, #23227). A total of 50 µg of each protein was loaded onto a 10% Tris-glycine SDS-polyacrylamide gel and transferred to a nitrocellulose membrane. To detect phosphorylated Mpk1, a 1:4000 dilution of phosphor-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) rabbit monoclonal antibody (Cell Signaling Technology, #4370S) was used. To ensure equal protein loading, we used 1/4000 dilution of a Hog1-specific rabbit polyclonal antibody (Santa Cruz, SC-9079). Protein levels were measured using a ChemiDoc (Bio-Rad).
For phosphatase assay, the Rad53-4×FLAG strain and Lkh1-4×FLAG strain were inoculated in YPD medium at 30°C overnight and subcultured into 200 mL YPD medium at an OD600 of 0.2 and incubated until OD600 reached approximately 0.8. We harvested 100 mL of the culture for the basal-time sample, and the remaining sample was treated with rapamycin or MMS and further incubated for 2 h at 30°C. The collected samples were divided into two halves for phosphate treatment. One half was resuspended in lysis buffer containing a phosphatase inhibitor cocktail, and the other half was resuspended in lysis buffer without a phosphatase inhibitor cocktail. The proteins extracted from each sample were quantified using a BCA kit and adjusted to the same protein concentration. Next, protein samples were treated with PMP buffer and 800 units of phosphatase (New England Biolabs, #P0753S) for 1 h at 30°C.
The C. neoformans strains were cultured in a liquid YPD medium for 16 h at 30°C. Cell concentrations were calculated by hemocytometer, and the MATα and MATa cell concentrations were synchronized (107 cells/mL). Equal amounts of MATα and MATa cells were mixed, spotted (5 μl) onto V8 mating medium, and grown in the dark at room temperature for 6 weeks. Filamentous growth was observed and photographed once a week using a microscope equipped with a SPOT Insight digital camera (Diagnostic Instrument, Inc.).
The ergosterol content of the strains was measured as described previously (Ko et al., 2009). The strains were inoculated in YPD medium at 30°C overnight and subcultured into 150 mL YPD medium at an OD600 of 0.2 and incubated until OD600 reached about 0.8. We harvested 50 mL of the culture for the basal-time sample, and the remaining sample was treated with FCZ (10 μg/mL) and further incubated until each time point at 30°C. The ergosterol content of the sample was calculated as follows: % ergosterol = [(OD281.5/290) × F]/pellet weight – [(OD230/518) × F]/pellet weight, where F is the ethanol dilution factor and 290 and 518 are the E values (in percentage per centimeter) determined for crystalline ergosterol and 24(28) dehydroergosterol, respectively (Arthington-Skaggs et al., 1999).
The amount of intracellular ATP was measured as described previously (Garcia et al., 2008; Price et al., 2011). WT, lkh1Δ, and lkh1Δ+LKH1 strains were grown in liquid YPD medium for 16 h at 30°C and subcultured in fresh liquid YPD medium at 30°C until the optical density at 600 nm of the culture medium ranged between 0.7 and 0.8. Cultured samples were harvested and washed with PBS. Samples were resuspended in 30 mL PBS and 15 mL aliquots of each sample were centrifuged. Pellets for ATP analysis were resuspended in 1 mL of 50 mM HEPES (pH 7.7) and disrupted using a bead beater (Precellys) for 45 s. The disrupted samples were centrifuged at 5,000 rpm at 4°C for 10 min, and the supernatants were collected to determine ATP concentrations using an ATP bioluminescence assay kit (Sigma, #FLAA). Protein concentrations used to normalize the ATP concentration of each sample were measured in an aliquot of the supernatant from each sample using a BCA kit (Thermo Fisher, #23227).
Quantitative measurement of LAC (LAC1 and LAC2) genes expression was performed using a dopamine medium to induce melanin production. Strains were cultured in liquid YPD medium overnight at 30°C and subcultured in fresh liquid YPD medium and then further incubated until they reached an approximate OD600 of 0.6. Basal-level samples were harvested, and the remaining samples were washed three times using sterile water. After that, the remained cultures were resuspended in a dopamine medium and harvested after 4 h. To measure CAP10, CAP59, CAP60, and CAP64, strains were cultured in liquid YPD medium overnight at 30°C. The grown cells were subcultured in fresh liquid YPD medium at 30°C until the optical density of the culture medium ranged from 0.6 to 0.7. Next, 50 mL of the grown cells were harvested for the basal sample, and the remaining samples were washed three times using sterile water. After washing, the culture was resuspended in Littman’s medium and incubated for 4 h. Total RNA isolation and cDNA synthesis was performed as previously described (Ko et al., 2009). qRT-PCR analysis was performed using the gene-specific primers listed in Supplementary Table S2.
WT (H99) and lkh1Δ mutant were cultured in liquid YPD medium overnight at 30°C, subcultured in fresh liquid YPD medium, and then further incubated until they reached an approximate OD600 of 0.7–0.8. For basal samples, we collected 50 mL of grown cells and treated them with rapamycin (3 ng/mL) the remaining cells for 1 h. After 1 h, the cells were harvested and placed in liquid nitrogen. All samples were lyophilized and total RNA was isolated using Trizol reagent (Intron, #17061) and purified with an RNeasy mini kit (Qiagen, #74104) according to the manufacturer’s protocol. To ensure the reliability of the RNA-seq analysis, we prepared three biologically independent samples for each strain. Total RNA concentration was calculated using Quant-IT RiboGreen (Invitrogen, #R11490). To assess the integrity of the total RNA, samples were run on TapeStation RNA screen tape (Agilent, #5067-5576). Only high-quality RNA preparations (RIN greater than 7.0) were used for the RNA library construction. A library was independently prepared with 0.5 μg of total RNA for each sample by Illumina TruSeq Stranded Total RNA Library Prep Gold Kit (Illumina, Inc., San Diego, CA, USA, # 20020599). The first step in the workflow involved removing rRNA from the total RNA using the Ribo-Zero rRNA Removal Kit (Human/Mouse/Rat Gold) (Illumina, Inc., San Diego, CA, USA). Subsequently, the remaining mRNA was fragmented into small pieces using divalent cations at elevated temperatures. The cleaved RNA fragments were copied into first-strand cDNA using SuperScript II reverse transcriptase (#18064014; Invitrogen) and random primers. This was followed by the synthesis of second-strand cDNA using DNA Polymerase I, RNase H, and dUTP. These cDNA fragments then go through an end repair process, the addition of a single ‘A’ base, and then adapter ligation. The products were purified and enriched using PCR to create a final cDNA library. Libraries were quantified using KAPA Library Quantification kits for Illumina Sequencing platforms per the qPCR Quantification Protocol Guide (KAPA BIOSYSTEMS, #KK4854) and qualified using a TapeStation D1000 ScreenTape (Agilent Technologies, # 5067-5582). Indexed libraries were then subjected to Illumina NovaSeq (Illumina, Inc., San Diego, CA, USA), and paired-end (2×100 bp) sequencing was performed by Macrogen Inc. The RNA-seq data generated in this study were available from the Gene Expression Omnibus (GSE233612).
SPF/VAF-confirmed inbred 6-week-old female BALB/cAnNCrlOri mice were purchased (ORIENT BIO Inc., Republic of Korea) and used in the experiment after acclimatization to the breeding environment for one week. For infection, the strains [WT (H99), lkh1Δ (KW1451), and lkh1Δ+LKH1 (KW1633)] were inoculated in a fresh liquid YPD medium and cultured overnight in a 30°C shaking incubator. After adjusting the number to 5 × 105 yeast cells, respiratory anesthesia was induced with isoflurane, and the strains were inhaled nasally. After infection, the condition of the mice was observed daily, and the survival rate of the mice was expressed as a percentage of survival. Statistical analysis was performed using Log-rank (Mantel-Cox) test using Graphpad Prism 9.5.1.
For the BBB translocation assay, human brain microvascular endothelial cells (hCMEC/D3 cell line; Merck & CO) were cultured as previously described (Vu et al., 2009; Santiago-Tirado et al., 2017; Jin et al., 2020; Lee et al., 2020). Briefly, 5×104 hCMEC/D3 cells in EGM-2 medium (Lonza Group) were seeded on collagen (Corning, Inc)-coated 8 μm-porous membranes (BD Falcon). The day after seeding, the medium was replaced with fresh EGM™-2 medium supplemented with 2.5% human serum, and the cells were further grown for 4 days. A day before the inoculation of each strain, the medium was replaced with 0.5×diluted EGM™-2 medium (1.25% human serum), and the cells were maintained at 37°C and 5% CO2 incubator. The integrity of tight junctions between hCMEC/D3 cells was confirmed by measuring the transendothelial electrical resistance (TEER), which should be approximately 200 Ω per cm2. TEER was measured using the Epithelial Volt per Ohm Meter (EVOM2 device, World Precision Instruments). We added 5×105 cells of each strain to 500 μl of PBS, inoculated them onto the top of the porous membranes, and incubated them for 24 h at 37°C in a 5% CO2 incubator. The number of yeast cells passing through the porous membrane was determined by counting the CFU. Tight junctions between the hCMEC/D3 cells were measured using the EVOM2 device before and after inoculation with the strains.
For tissue staining, lungs were obtained from mice on day 16 after infection, weighted, and fixed in a 3.7% formaldehyde solution. After complete fixation, the tissues were dehydrated and clarified using xylene before being embedded in paraffin blocks. Tissue sections with a thickness of 5 μm were cut and stained with Grocott’s Methenamine Silver (GMS) Stain Kit (Abcam, ab287884) or Periodic Acid Schiff (PAS) Stain Kit (Mucin Stain) (Abcam, ab150680), per the manufacturer’s protocol. Slide images were captured with Primostar 3 (ZEISS, Germany) at final magnifications of 100 × and 200 ×.
The flocculation assay was performed as previously described (Cramer et al., 2006). Briefly, C. neoformans strains were cultured in liquid YPD medium for 16 h at 30°C. Subsequently, the grown cells were normalized to an OD600 of 1.5. The OD600 of each cell suspension was then measured for a duration of 4 h. Three independent biological replicates were conducted, and error bars indicate the standard error of the means (S.E.M). The adhesion assay was performed following the protocol outlined in the previous study (Lee et al., 2014). Strains were cultured in liquid YPD medium for 16 h at 30°C. Next, the 5 μl of the grown cells were spotted onto filament agar plates and incubated for 5 days at 22°C. Adherence was assessed by washing the plate under flowing distilled water for 20 sec.
For flow cytometry analysis, we prepared WT (H99) and lkh1Δ mutant strains, as previously described (Choi et al., 2024). Briefly, WT and lkh1Δ mutant strains were cultured in a liquid YPD medium for 16 h. Next, each strain was grown to an optical density at 600 nm of 0.8 and washed with phosphate-buffered saline (PBS). After cells were fixed with ethanol, cells were washed with PBS containing 1% and 0.5% bovine serum albumin (BSA). Subsequently, the cells were treated with RNase for 30 min at 37°C and stained with propidium iodide staining buffer for 2 h at room temperature in the dark. After washing, fluorescence was assessed using a BD FACS symphony A5, capturing 10,000 events per sample.
For chitin staining, the strains were cultured in YPD medium for 16 h at 30°C and subsequently subcultured in fresh liquid YPD medium until the optical density of the culture medium at 600 nm reached 0.8. A 50 mL aliquot of cells was harvested and pelleted by centrifugation to establish a basal time point. The remaining cultures were treated with 1.0 mg/mL of caffeine and further incubated before being harvested after 12 hours. Harvested samples were fixed following previously established methods (Jung et al., 2012). Fixed samples were mixed with an equal volume of calcofluor white solution (25 μg/mL) and incubated for 30 min at room temperature in darkness. Subsequently, the cells were washed twice with PBS and mixed with mounting buffer (Biomeda, cat. NO #M01) on slide glass for immobilization, followed by an additional 30 min incubation. Each sample was then photographed using fluorescence microscopy (BX51; Olympus, Tokyo, Japan), and fluorescence intensity was measured in a minimum of 50 individual cells using the ImageJ/Fiji software following previous methods (Kim et al., 2021).
To identify the LAMMER kinase ortholog in C. neoformans, BLAST searches were performed using the S. cerevisiae Kns1 protein sequence as a query. We identified a single gene (CNAG_00683) with high scores and e-values (score: 294; e-value: 3e-89). The putative LAMMER kinase harbors the conserved motif ‘EHLAMMERILG’ sequence similar to other fungal LAMMER kinases. Therefore, we named CNAG_00683 as Lkh1 in C. neoformans. Growth defect is a conserved phenotype observed in lkh1Δ mutants of other fungi (Kang et al., 2010; Park et al., 2011; Kang et al., 2013). To ascertain whether the Cryptococcus lkh1Δ mutant exhibited growth defect, we generated the lkh1Δ mutant and its complemented strain and monitored their growth at 30°C and 37°C. We found that the Cnlkh1Δ mutant displayed a growth defect compared to WT and complemented strain at both temperatures (Figure 1A and Supplementary Figure S2A). ATP synthesis is essential for fungal cell growth (Li et al., 2021; Vestergaard et al., 2022). To establish a relationship between the growth defects of the lkh1Δ mutant and intracellular ATP levels, we measured the intracellular ATP content in each strain. The ATP levels in the lkh1Δ mutant were indistinguishable from those in the WT, indicating that ATP content was not associated with the growth defects observed in the lkh1Δ mutant (Supplementary Figure S2B). Given that Lkh1 is involved in cell cycle progression by phosphorylating Rum in fission yeast (Yu et al., 2013) and lkh1Δ mutant showed slow growth in C. neoformans, we conducted a fluorescence-activated cell sorting (FACS) analysis to investigate whether CnLkh1 plays a role in cell cycle progression. We found that the WT strain showed regular G1, S, and G2 phases, whereas abnormal cell cycle patterns were observed in the lkh1Δ mutant. The lkh1Δ mutant exhibited a decreased G1 phase and increased G2 phase compared to the WT strain. Furthermore, the lkh1Δ mutant exhibited increased DNA content beyond 2N (Figures 1B, C, and Supplementary Figure S2C). Morphological analysis of the lkh1Δ mutant revealed that some of the cells were not physically separated compared to WT and its complemented strain (Supplementary Figure S2D). Therefore, these data suggest that C. neoformans Lkh1 is also involved in the cell cycle progression akin to other fungi.
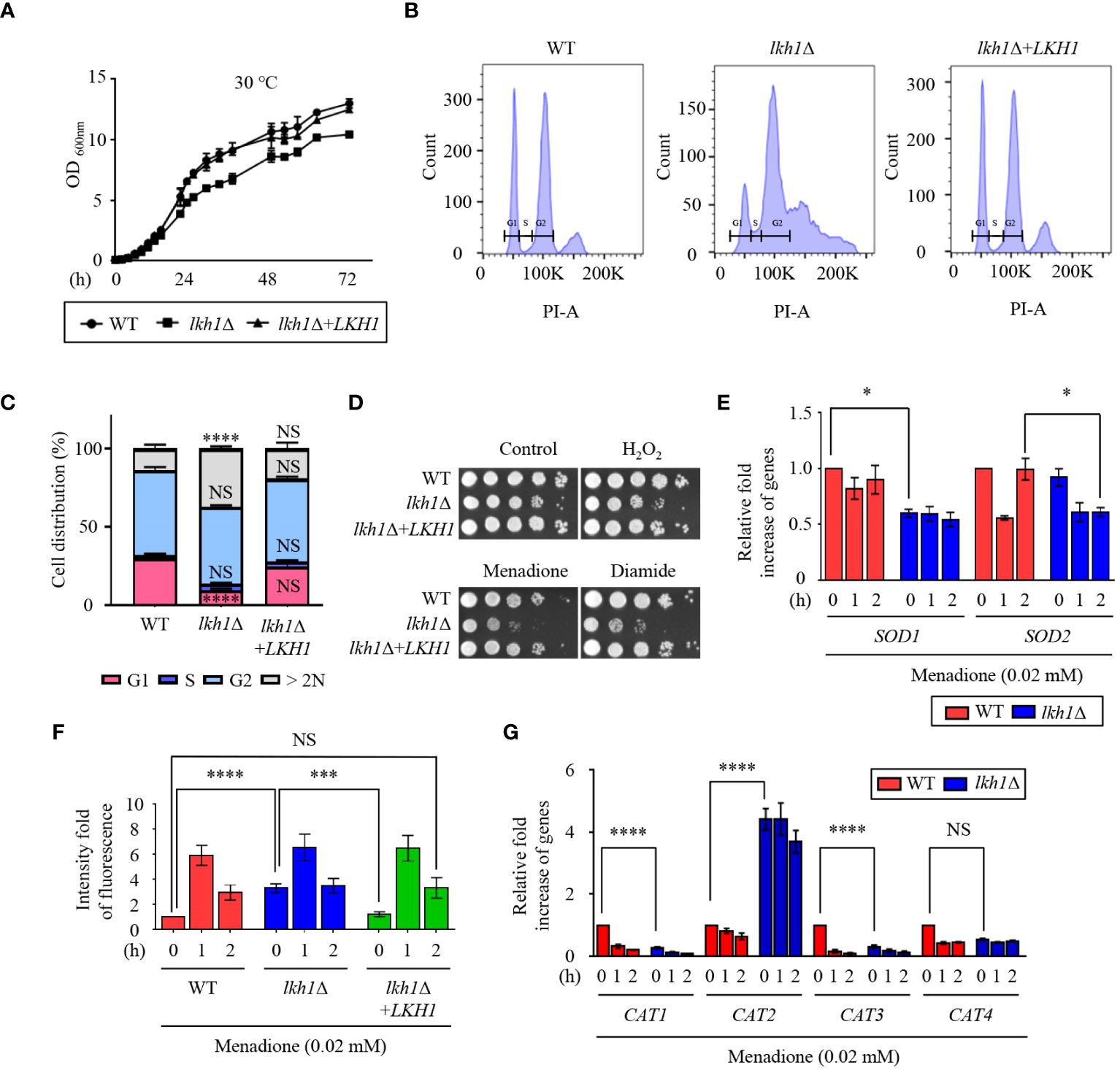
Figure 1 Lkh1 regulated growth and oxidative stress response. (A) WT (H99), lkh1Δ (KW1451), and lkh1Δ+LKH1 (KW1633) strains were grown in liquid YPD medium for 16 h at 30°C and subcultured in fresh liquid YPD medium at 30°C. Cell density was measured at an optical density of 600 nm every 2 (h) (B, C) Histogram of cell count in WT and lkh1Δ mutant by propidium iodide using flow cytometry. The provided images represent data from three biological replicates (B). Population proportion of cells were quantified. One-way ANOVA was used to dertermine statistical differences among strains (NS: not significant; * P < 0.05; *** P < 0.001; **** P < 0.0001) (D) WT (H99), lkh1Δ (KW1451), and lkh1Δ+LKH1 (KW1633) strains were cultured in liquid YPD medium overnight at 30°C, 10-fold serially diluted and spotted on YPD medium containing 0.02 mM menadione, 2 mM diamide, or 2 mM H2O2. (E, G) Expression levels of SOD genes and CAT genes were verified by qRT-PCR analysis using cDNA of WT (H99) and lkh1Δ strains with or without treatment of menadione (0.02 mM). Three independent biological experiments with duplicate technical replicates were performed. Error bars indicate the standard error of the mean (S.E.M). The statistical significance of the differences was determined by one-way ANOVA with Bonferroni’s multiple-comparison test. (NS: not significant; * P < 0.05; *** P < 0.001; **** P < 0.0001) (F) Intracellular ROS levels of WT, lkh1Δ, and lkh1Δ+LKH1 strains treated with or without MD (0.02 mM) were measured using H2DCFDA.
A previous study demonstrated that strains with deleted LKH1 showed growth defects in response to hydrogen peroxide (Park et al., 2003). To demonstrate the involvement of CnLkh1 in the oxidative stress response, we conducted a phenotypic analysis. The lkh1Δ mutant showed increased susceptibility to menadione and diamide, but not to hydrogen peroxide (Figure 1D). Menadione serves as a general superoxide generator, and Sods protect cells from the toxic effects of reactive oxygen species, which are mediated by the dismutation of superoxide radicals into hydrogen peroxide and oxygen (Miller and Britigan, 1997). To examine whether Lkh1 regulates the expression of SOD genes, we evaluated the expression of SOD genes in the WT and lkh1Δ mutant under menadione treatment. The lkh1Δ mutant exhibited diminished SOD1 expression compared to the WT, while the SOD2 expression level in the lkh1Δ mutant was lower than that in WT at 2 h post-menadione treatment (Figure 1E). Next, we sought to determine if the reduced SOD1 expression levels in the lkh1Δ mutant corresponded to changes in intracellular ROS levels. To address this, we quantified intracellular ROS levels in WT, lkh1Δ, and lkh1Δ+LKH1 strains using H2DCFDA. We observed that the intracellular ROS level in the lkh1Δ mutant was higher than that in WT at the basal level. However, ROS productions in the WT and lkh1Δ mutant were indistinguishable under menadione treatment (Figure 1F). In S. pombe, expression levels of the catalase gene, CTT1, are markedly reduced in the lkh1Δ mutant compared to those in WT (Park et al., 2003). To investigate whether Lkh1 is involved in regulating catalase genes, we assessed the expression levels of four catalase genes (CAT1, CAT2, CAT3, and CAT4) in the presence of menadione. The expression levels of CAT1 and CAT3 genes in the lkh1Δ mutant were reduced compared to those in the WT under basal conditions, whereas the CAT2 gene expression levels in the lkh1Δ mutant were significantly higher than those in the WT, regardless of the presence or absence of menadione (Figure 1G). Collectively, these results suggested that Lkh1 governs the oxidative stress response by modulating the expression of SOD and CAT genes.
Previous studies have reported a critical role for U. maydis Lkh1 in maintaining genome stability and facilitating DNA repair processes (de Sena-Tomás et al., 2015). To demonstrate the requirement of Lkh1 in DNA damage response, survival tests were conducted under DNA damage stress conditions. Consistent with observation in the U. maydis lkh1Δ mutant, the lkh1Δ mutant exhibited a significant growth defect in response to various types of DNA damage stress (Figure 2A). In C. neoformans, the Rad53-Bdr1 pathway plays a central role in the response to DNA damage stress by regulating the expression of DNA repair genes (Jung et al., 2016, 2019, 2021). To demonstrate that Lkh1 controls the DNA damage response via the Rad53-Bdr1 pathway, we measured the expression levels of BDR1 and its downstream genes. Notably, the lkh1Δ mutant displayed inherently higher expression levels of BDR1, RAD51, RAD54, and RDH54 compared to the WT, whereas induction levels upon MMS treatment were similar between WT and lkh1Δ strains (Figure 2B). This result led us to compare Rad53 phosphorylation levels in WT and lkh1Δ strains because Rad53 phosphorylation induces the expression of DNA repair genes. To compare Rad53 phosphorylation, we constructed lkh1Δ mutant strains in the Rad53-4×FLAG background and monitored Rad53 phosphorylation in both WT and lkh1Δ mutant. We observed that Rad53 was more phosphorylated in lkh1Δ mutant compared to WT under a basal condition (Figure 2C). These findings indicate that DNA damage responses in the lkh1Δ mutant are more impaired than those in WT at the basal condition.
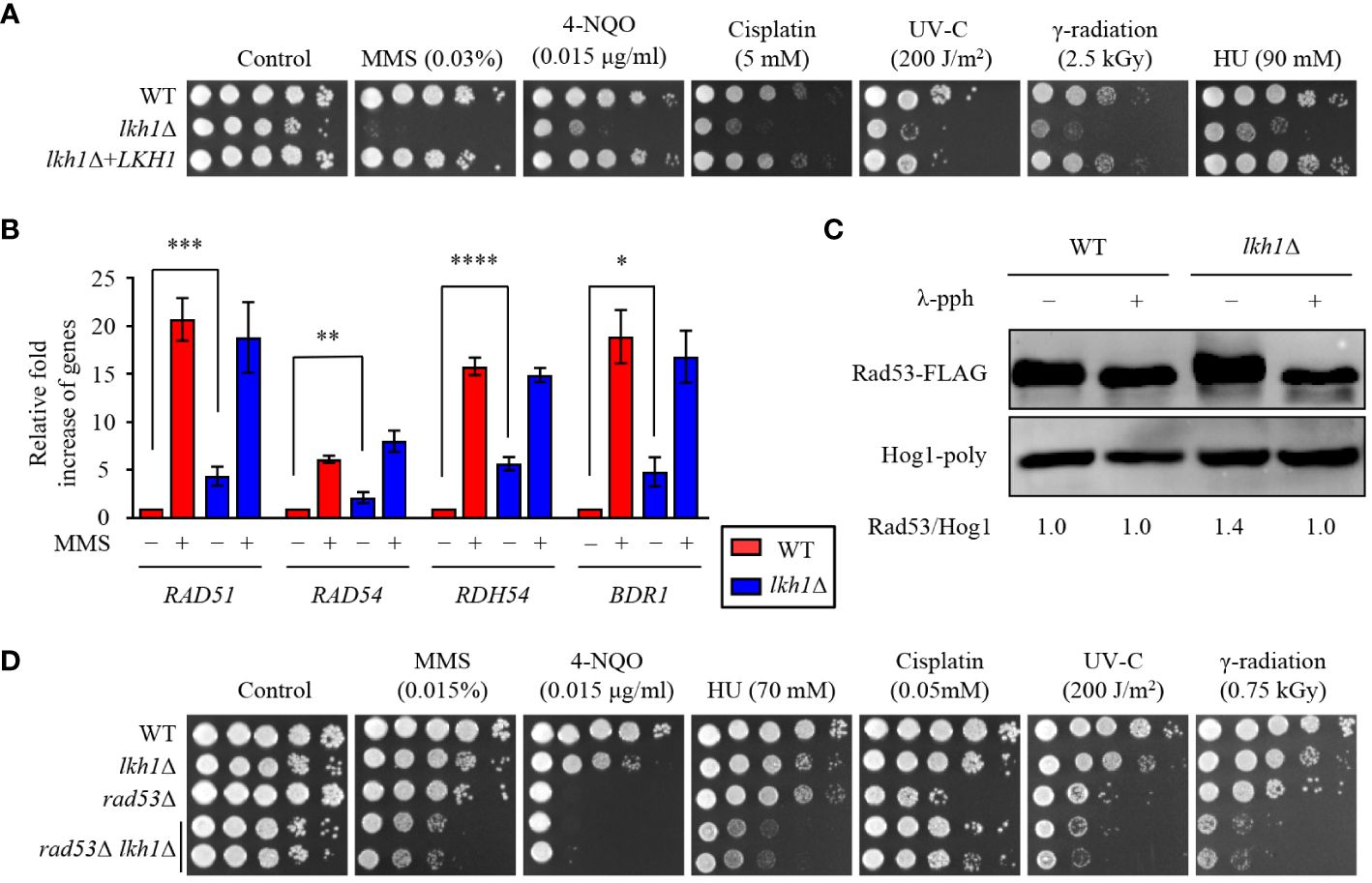
Figure 2 Lkh1 regulated DNA damage response. (A) WT (H99), lkh1Δ (KW1451), and lkh1Δ+LKH1 (KW1633) strains were cultured in a liquid YPD medium overnight at 30°C, 10-fold serially diluted and spotted on YPD medium containing the indicated concentration of DNA damage insults. For UV-C and γ-radiation tests, the serially diluted cells were spotted onto YPD medium and exposed to the indicated dose of UV-C (200 J/m2) or γ-radiation (2.5 kGy). Next, cells were further incubated at 30 °C and photographed daily for 2-4 days. (B) Expression levels of the BDR1 gene and Rad53-Bdr1 pathway downstream genes were verified by qRT-PCR analysis using cDNA of WT and lkh1Δ mutant with or without treating MMS (0.02%). Three independent biological experiments with duplicate technical replicates were performed. Error bars indicate S.E.M. Statistical significances of the differences were determined by one-way ANOVA with Bonferroni’s multiple-comparison test. (* P < 0.05; ** P < 0.01; *** P < 0.001; **** P < 0.0001) (C) Phosphorylation of Rad53 was monitored by analysis of the reduced electrophoretic migration using western blotting with anti-FLAG antibody. The Rad53-4×FLAG strain (YSB3806) and lkh1Δ Rad53-4×FLAG (KW1806) were grown to the mid-logarithmic phase and the cell extract was incubated at 30°C for 1 h with or without λ-phosphatase (PPase). (D) WT (H99), lkh1Δ (KW1451), rad53Δ (YSB3785), and rad53Δ lkh1Δ (KW1610 and KW1612) strains were cultured in a liquid YPD medium overnight at 30°C, 10-fold serially diluted and spotted on YPD medium containing the indicated concentration of DNA damage insults. For UV-C and γ-radiation tests, the serially diluted cells were spotted onto a YPD medium and exposed to the indicated dose of UV-C or γ-radiation. Next, cells were further incubated at 30°C and photographed daily for 2-4 days.
Because Lkh1 controlled DNA damage response in both Rad53-dependent and independent manner, we hypothesized that rad53Δ lkh1Δ double mutants show more pronounced growth defect under DNA damage stress compared to the individual lkh1Δ and rad53Δ mutant. To validate this hypothesis, we constructed two additional independent rad53Δ lkh1Δ double mutants and performed phenotypic analyses using them. Expectedly, the rad53Δ lkh1Δ double mutants were more susceptibility to DNA damage insults compared to the individual lkh1Δ and rad53Δ mutant (Figure 2D). Therefore, Lkh1 plays an important role in the DNA damage response in both Rad53-dependent and -independent manners.
In C. albicans, deletion of KNS1 results in growth defects in response to cell stress agents and alterations in cell wall composition (Park and Park, 2011; Choi et al., 2014). Similar to the kns1Δ mutant in C. albicans, the lkh1Δ mutant showed growth defects in response to cell wall stress agents, such as caffeine, calcofluor white, and congo-red (Figure 3A). In C. neoformans, the Mpk1 MAPK pathway plays a central role in regulating cell wall integrity through phosphorylation in response to cell wall stress agents (Kraus et al., 2003). To ascertain the potential involvement of Lkh1 in Mpk1 phosphorylation during cell wall stress, we monitored the phosphorylation status of Mpk1 in lkh1Δ mutant under cell wall stress conditions. We observed that Mpk1 phosphorylation levels in lkh1Δ mutant were slightly increased than those in WT in the presence of caffeine (Figure 3B).
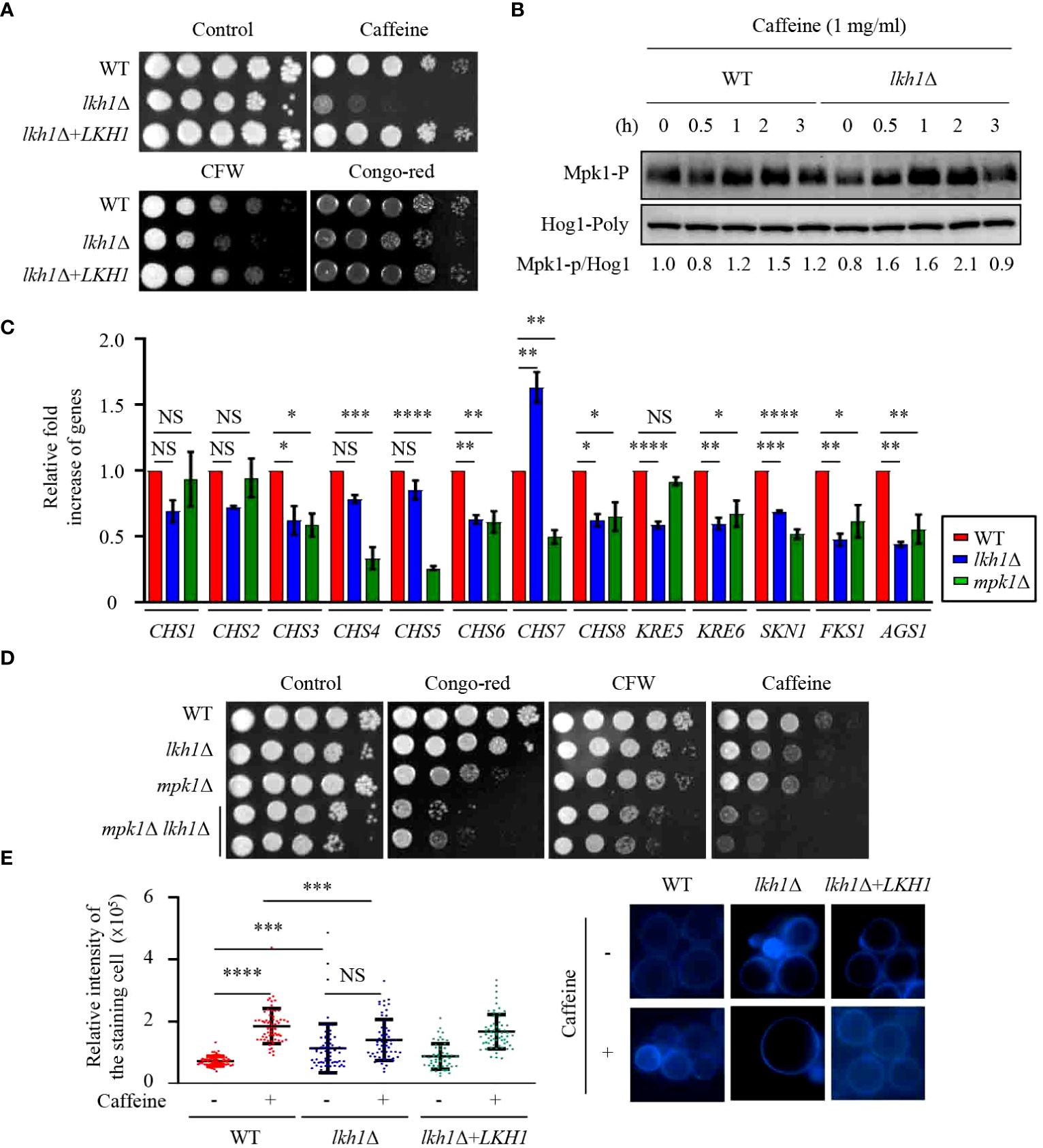
Figure 3 Lkh1 was involved in the cell wall damage stress. (A) WT (H99), lkh1Δ (KW1451), and lkh1Δ+LKH1 (KW1633) strains were cultured in a liquid YPD medium overnight at 30°C, 10-fold serially diluted and spotted on a YPD medium containing 60 mg/mL CFW, 1 mg/mL caffeine, or 1% congo-red. (B) Mpk1 phosphorylation was monitored using western blotting with an anti-phospho-Erk1 antibody. The WT (H99) and lkh1Δ strains were grown to the mid-logarithmic phase and then treated with caffeine (1 mg/mL) for 3 h. Next, total protein was extracted from each strain for immunoblot analysis. (C) Expression levels of chitin synthesis-related genes and glucan synthesis-related genes were verified by qRT-PCR analysis using cDNA of WT (H99) and lkh1Δ strains. Three independent biological experiments with duplicate technical replicates were performed. Error bars indicate S.E.M. Statistical significances of the differences were determined by one-way ANOVA with Bonferroni’s multiple-comparison test. (* P < 0.05; ** P < 0.01; **** P < 0.0001; NS: not significant) (D) WT (H99), lkh1Δ (KW1451), mpk1Δ (YSB3814), and mpk1Δ lkh1Δ (KW1858 and KW1860) strains were cultured in liquid YPD medium overnight at 30°C, 10-fold serially diluted and spotted on a YPD medium containing 1 mg/mL CFW, 0.3 mg/mL caffeine, or 0.04% congo-red. (E) Chitin staining with CFW. The quantitative fluorescence intensity of 50 individual cells of each strain were measured suing ImageJ/Fiji software. The error bar indicates the standard deviations (SD). The statistical significance of the differences was determined by one-way ANOVA using Bonferroni’s multiple-comparison test. (*** P < 0.001; **** P < 0.0001; NS: not significant).
The cell wall of C. neoformans is composed of α- and β-glucan as well as chitin. C. neoformans harbors eight putative chitin synthase encoding genes, seven β-glucan synthesis-related genes, and one α-glucan synthase encoding gene (Thompson et al., 1999; Reese and Doering, 2003; Banks et al., 2005; Reese et al., 2007; Wang et al., 2018). Given that the lkh1Δ mutant exhibited growth defects in response to cell wall stress agents, we wondered whether Lkh1 controls the expression of genes involved in cell wall biosynthesis. To test this hypothesis, we measured the expression of chitin synthase-associated genes. The expressions of CHS3, CHS6, and CHS8 in the lkh1Δ mutant were reduced compared to WT, while the expression of CHS7 in the lkh1Δ mutant was elevated relative to the WT (Figure 3C). β-1,6-glucan is the predominant polysaccharide in the C. neoformans cell wall compared to β-1,3-glucan (Zaragoza et al., 2009; Gilbert et al., 2010). To confirm Lkh1 involvement in β-glucan synthesis in C. neoformans, we measured the expression levels of related genes. The expressions of KRE6 (β-1,6-glucan synthase-related gene), FKS1 (encoding β-1,3-glucan synthase), and ASG1 (α-1,3-glucan synthase) were reduced in both mpk1Δ mutant and lkh1Δ mutant compared to WT. The expression of the KRE5 (β-1,6-glucan synthase-related gene) in the lkh1Δ mutant was reduced compared to WT but remained unaffected in the mpk1Δ mutant (Figure 3C). Next, to reinforce the notion that Lkh1 is involved in cell wall integrity in an Mpk1-independent manner, we constructed mpk1Δ lkh1Δ double mutants and subjected them to a cell wall stress assay. Expectedly, mpk1Δ lkh1Δ double mutants showed significantly higher susceptibility to cell wall damage agents compared to the individual lkh1Δ and mpk1Δ mutants (Figure 3D). Given that expression levels of genes related to chitin synthesis were diminished in lkh1Δ mutant compared to those in WT, we quantitatively measured chitin levels in WT and lkh1Δ mutant in the presence or absence of caffeine. Under basal conditions, the chitin content in the lkh1Δ mutant was higher than those in WT and its complemented strain. However, in the presence of caffeine, the chitin content in WT and lkh1Δ+LKH1 strain increased, whereas that in the lkh1Δ mutant was not enhanced to the same extent as in WT (Figure 3E). Collectively, these findings suggest that Lkh1 controls the cell wall stress response in an Mpk1-dependent and -independent manner.
The fungal cell membrane is a key target for antifungal drugs, primarily because of its specific fungal components such as ergosterol. Azole drugs inhibit ergosterol biosynthesis by binding to Erg11 and inducing the accumulation of 14-methylated sterols, compromising cell membrane integrity (Joseph-Horne et al., 1995). Amphotericin B interacts with the lipid bilayer membrane, induces pore formation, and increases cell permeability (Joseph-Horne et al., 1995). Given the observed susceptibility of the lkh1Δ mutant to cell membrane destabilizer (Figure 4A), an antifungal drug test was conducted. The lkh1Δ mutant showed susceptibility to amphotericin B whereas it exhibited resistance to fluconazole (Figure 4B). This contrasting response to azole and polyene drugs suggests a potential role of Lkh1 in the regulation of ergosterol biosynthesis. To test this hypothesis, we measured the expression levels of ERG1, ERG11, and HMG1, which participate in the rate-limiting step of ergosterol biosynthesis (Veen et al., 2003). We observed that expression levels of ERG1 and ERG11 in lkh1Δ mutant were similar to those in WT. However, the induction of HMG1 in the lkh1Δ mutant was slightly lower than compared to the WT (Figure 4C). Although Lkh1 did not appear to be critical for the regulation of ergosterol biosynthesis genes, the ergosterol content was determined to further elucidate the regulatory role of Lkh1. Similar to expression levels of ergosterol biosynthesis genes, the ergosterol contents in the lkh1Δ were indistinguishable from those in WT (Figure 4D). In addition to ergosterol biosynthesis, several studies have demonstrated that ABC transporters are also critical for the acquisition of azole resistance in C. neoformans (Sanguinetti et al., 2006; Chang et al., 2018). To investigate whether Lkh1 regulates ABC transporters, we measured expression levels of these genes under fluconazole treatment. Contrary to our expectation, expression levels of ABC transporter genes such as AFR1 and MDR1 in lkh1Δ mutant were lower than those in WT under fluconazole treatment. However, the expression levels of AFR2 and AFR3 were similar in both lkh1Δ and WT strain in the presence of fluconazole (Figure 4E). Collectively, these findings suggest that Lkh1 controls azole resistance in an ergosterol biosynthesis-and ABC transporters-independent manners.
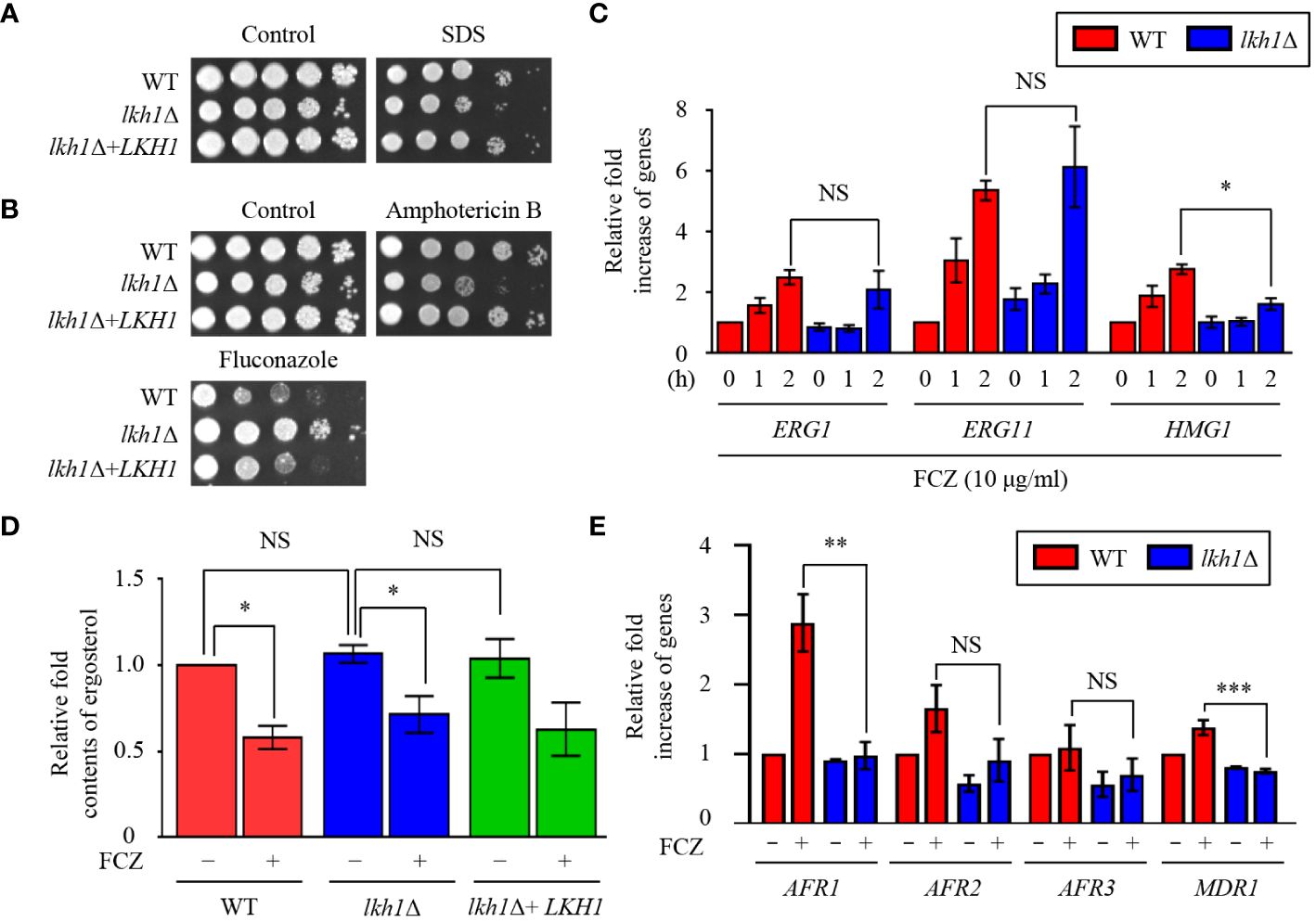
Figure 4 Lkh1 was involved in antifungal drug resistance. (A, B) WT (H99), lkh1Δ (KW1451), and lkh1Δ+LKH1 (KW1633) strains were cultured in a liquid YPD medium overnight at 30°C, 10-fold serially diluted and spotted on YPD medium containing 0.02% SDS, 1.5 μg/mL amphotericin B, 14 μg/mL fluconazole. (C, E) Expression levels of ergosterol synthesis-related genes and ABC transporter genes were verified by qRT-PCR analysis using cDNA of WT (H99) and lkh1Δ strains with or without treatment of fluconazole (16 μg/mL). Three independent biological experiments with duplicate technical replicates were performed. Error bars indicate S.E.M. Statistical significances of the differences were determined by one-way ANOVA with Bonferroni’s multiple-comparison test. (* P < 0.05; ** P < 0.01; *** P < 0.001; NS: not significant) (D) The relative ergosterol contents of WT, lkh1Δ, and lkh1Δ+LKH1 strains were assessed with or without fluconazole treatment.
Deletion of LKH1 results in reduced meiotic recombination in U. maydis and LkhA plays a pivotal role in the sexual development of Aspergillus nidulans (de Sena-Tomás et al., 2015, Kang et al., 2013). To demonstrate whether Lkh1 in C. neoformans is indispensable for sexual differentiation, we generated an LKH1 deletion strain in a MATa background and performed a mating assay. In unilateral crosses with the wild-type strains (H99 or KN99a strains) and lkh1Δ mutants, there was a noticeable delay in basidiospores production. However, basidiospores failed to form in bilateral crosses between the MATα lkh1Δ mutant and MATa lkh1Δ mutant (Figure 5A). These results indicate that Lkh1 is required for basidiospore formation in C. neoformans. Previous studies have reported that Lkh1 is involved in adhesion and flocculation in other fungi (Kim et al., 2001; Lim et al., 2021). Therefore, we performed flocculation and adhesion assays using WT, lkh1Δ, and lkh1Δ+LKH1 complemented strains. We found that the lkh1Δ mutant exhibited quicker settling out of suspension than WT and lkh1Δ+LKH1 strains (Figure 5B). In C. neoformans, mannoproteins (MPs) play a role in adhesion to host cells (Teixeira et al., 2014; Lee et al., 2023). Supporting this result, the expression level of adhesion-related gene CDA2 in lkh1Δ mutant was reduced compared to that in WT (Figure 5C). This result indicates that Lkh1 plays a negative role in flocculation. Contrary to flocculation, the lkh1Δ mutant exhibited reduced agar adherence compared to the WT strain (Figure 5D).
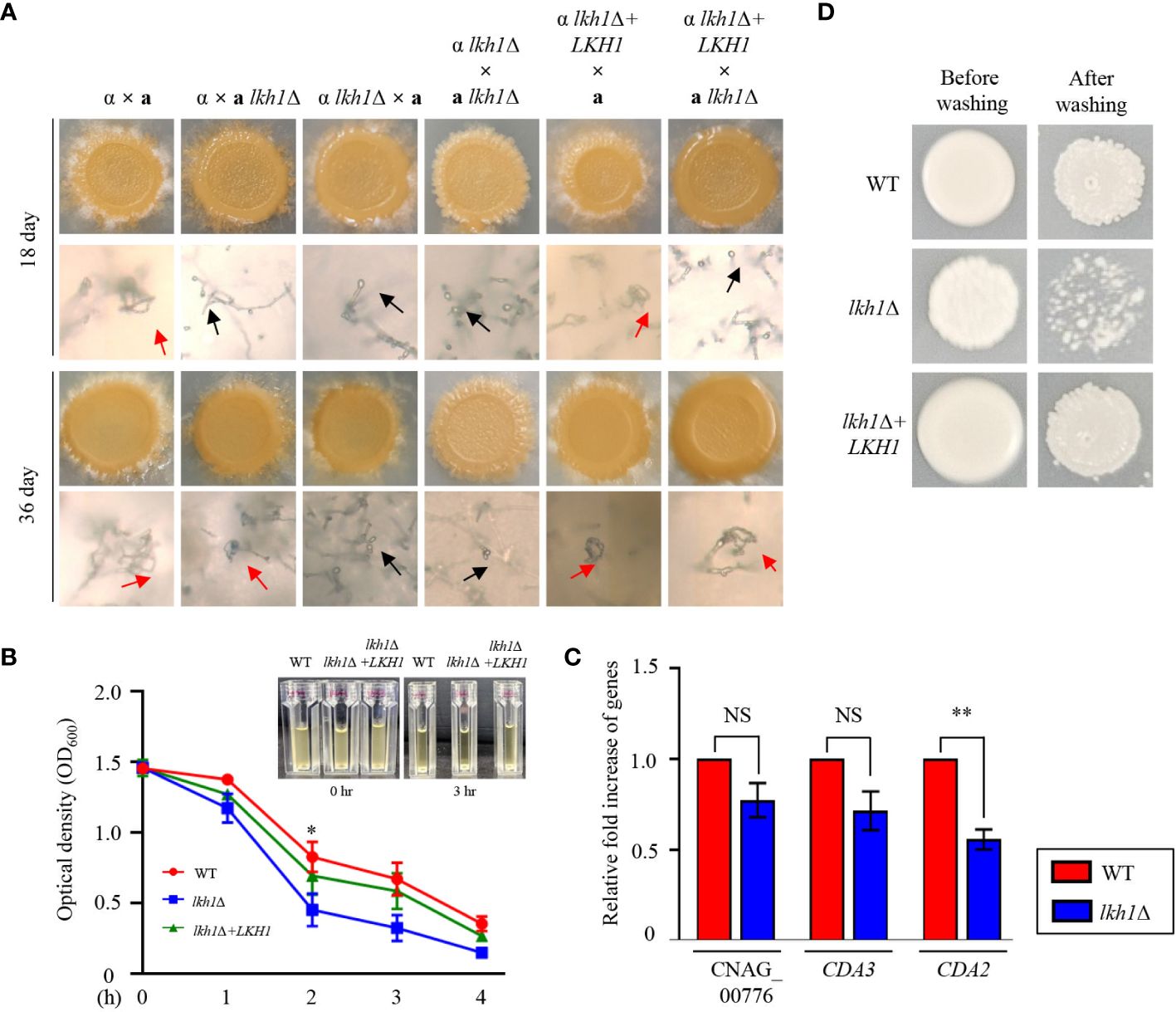
Figure 5 Lkh1 was required for basidiospores formation in C neoformans. (A) Serotype A MATα and MATa strains were co-cultured on a V8 medium (pH 5.0) for 5 weeks at room temperature in the dark: WT α × WT a (H99 and KN99), WT α × a lkh1Δ (H99 and KW1529), α lkh1Δ × a (KW1451 and KN99), α lkh1Δ × a lkh1Δ (KW1451 and KW1529), α lkh1Δ+LKH1 × a (KW1633 and KN99), and α lkh1Δ+LKH1 × a lkh1Δ (KW1633 and KW1529). The images were photographed after 18 days and 36 days. The black arrows and red arrows indicate basidium and basidiospores, respectively. (B) WT, lkh1Δ, and lkh1Δ+LKH1 strains were cultured in a liquid YPD medium, and each strain was normalized to an OD600 of 1.5. The OD600 was measured to determine flocculation at the indicated time points. The error bars indicate the standard error of the means (S.E.M). (C) The expression levels of adhesion-related genes in lkh1Δ mutant. Three independent biological experiments with duplicate technical replicates were performed. Error bars indicate the S.E.M. The statistical significance of the differences was determined by one-way ANOVA with Bonferroni’s multiple-comparison test. (NS: not significant; * P < 0.05; ** P < 0.01) (D) The same concentration of each strain was spotted on filament agar. After 5 days of incubation at room temperature, cells were washed with flowing water, followed by a subsequent incubation at 30°C for 2 days.
In S. cerevisiae, the TOR signaling pathway regulates ribosome and tRNA synthesis through the negative regulation of LAMMER kinase (Lee et al., 2012). To reveal the relationship between Lkh1 and Tor1, we conducted a spotting assay using rapamycin, a Tor1 target. We observed lkh1Δ mutant showed resistance to rapamycin compared to WT (Figure 6A), suggesting that Lkh1 negatively regulates rapamycin resistance. To elucidate whether C. neoformans Lhk1 is phosphorylated by Tor1 kinase, we constructed strains containing LKH1-4×FLAG and confirmed the functionality of the Lkh1-4×FLAG fusion protein (Supplementary Figure S3). Next, we monitored the shift of Lkh1-4×FLAG in the presence of rapamycin. We found that Lkh1 was a slight upshift following rapamycin treatment (Figure 6B). To further demonstrate that the upshift in Lkh1 was caused by phosphorylation rather than by increased Lkh1 production, we compared the protein mobility of Lkh1 in the presence or absence of λ-phosphatase or phosphatase inhibitors. Notably, the mobility shift of Lkh1 was abolished in the presence of λ-phosphatase under untreated conditions, indicating that C. neoformans Lkh1 is intrinsically phosphorylated under basal conditions. In both the rapamycin and the λ-phosphatase treatments, the abolishment of the Lkh1 mobility shift was even more apparent (Figure 6B).
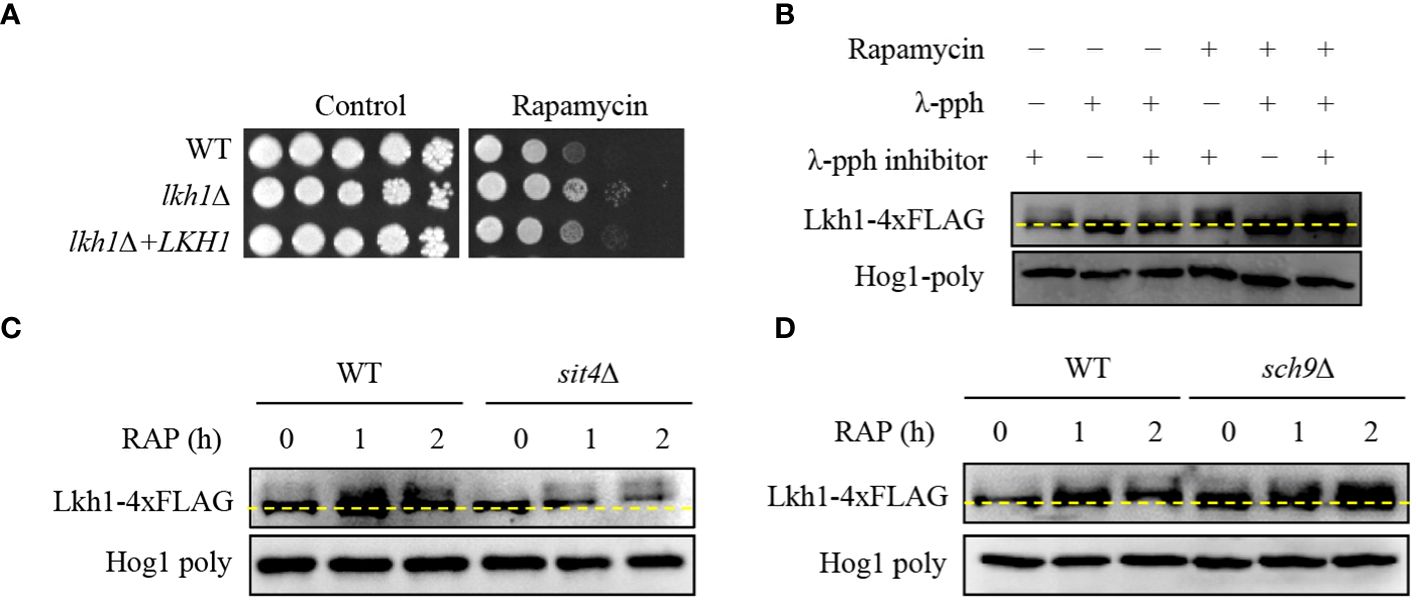
Figure 6 Phosphorylation of Lkh1 in a TOR1-dependent manner. (A) WT (H99), lkh1Δ (KW1451), and lkh1Δ+LKH1 (KW1633) strains were cultured in a liquid YPD medium overnight at 30°C, 10-fold serially diluted and spotted on a YPD medium containing rapamycin (9 μg/mL). (B–D) Lkh1 phosphorylation was monitored using western blotting with an anti-FLAG antibody. The Lkh1-4×FLAG strain (KW1755), sit4Δ Lkh1-4×FLAG strain (KW2058), and sch9Δ Lkh1-4×FLAG strain (KW2091) were grown to the mid-logarithmic phase and then treated with rapamycin (3 ng/mL) for 2 h. The cell extract was incubated at 30°C for 1 h with or without λ-phosphatase (PPase) and PPase inhibitor.
The fact that Lkh1 was phosphorylated under TOR kinase inhibition indicates that TOR-dependent kinases and phosphatases directly affect Lkh1 phosphorylation. In S. cerevisiae, the TOR kinase negatively regulates Sit4 phosphatase by facilitating complex formation between Tap42 and Sit4 (Di Como and Arndt, 1996). To demonstrate whether Sit4 is required for Lkh1 phosphorylation, we constructed a sit4Δ mutant in the background of the Lkh1-4×FLAG strain and monitored the Lkh1 protein shift. The data revealed that the Lkh1 protein in the sit4Δ mutant exhibited an increased mobility pattern compared to that in WT under both the absence and presence of rapamycin (Figure 6C). These data indicate that Sit4 indirectly influences Lkh1 phosphorylation. In S. cerevisiae, the TOR kinase directly phosphorylates the ACK kinase Sch9 (Urban et al., 2007). AKT, an ortholog of Sch9 in yeast, phosphorylates CLK1, CLK2, and murine LAMMER kinases (Song et al., 2023). To reveal whether Sch9 is required for Lkh1 phosphorylation, we constructed an sch9Δ mutant in the background of the Lkh1-4×FLAG strain. We observed that the Lkh1 protein in the sch9Δ mutant showed a significant upshift pattern compared to that of WT under both the absence and presence of rapamycin conditions (Figure 6D). This suggests that Sch9 is indirectly implicated in Lkh1 phosphorylation in an opposite manner to that of Sit4. Therefore, the Tor1-dependent pathway is responsible for Lkh1 phosphorylation.
In S. cerevisiae, the Asp (D440) residue is required for the kinase activity of Kns1, and kinase-dead alleles decrease the Kns1 auto-phosphorylation (Lee et al., 2012). To assess the significance of the kinase activity of CnLkh1 for its functionality, we constructed the Lkh1 kinase-dead allele (LKH1D546A) and introduced it into the lkh1Δ mutant (Figure 7A). We confirmed the LKH1D546A allele expression level in the lkh1Δ mutant was similar to that in the WT (Figure 7B). Next, we observed that the resistance of LKH1D546A strains in response to various stress inducers except for fluconazole was similar to that of the lkh1Δ mutant (Figure 7C). These findings suggest that most Lkh1 functions are controlled by the kinase activity of Lkh1, except for regulating resistance to fluconazole.
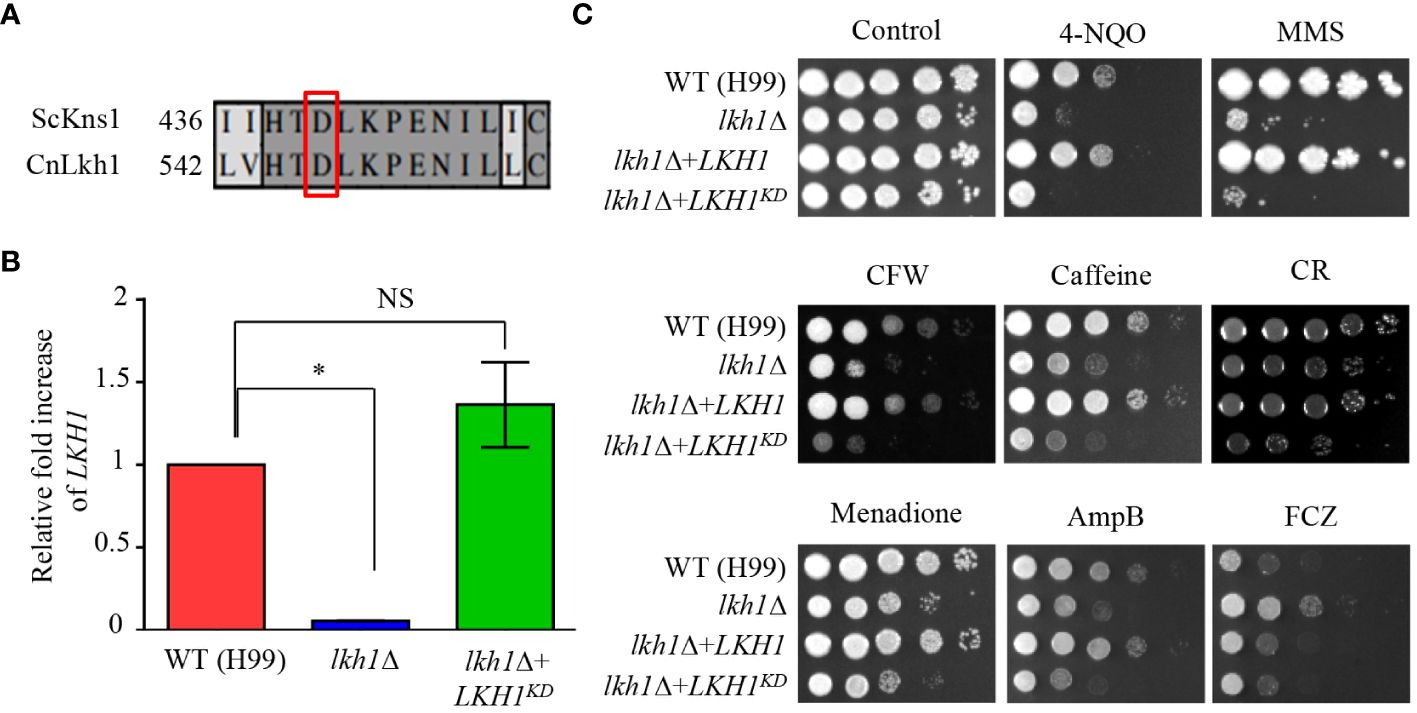
Figure 7 Kinase activity of Lkh1 was required for Lkh1 function. (A) Sequence alignment of the catalytic region for the kinase activity of S. cerevisiae Kns1 and C neoformans Lkh1. (B) The LKH1 expression level in the LKH1KD strain. The qRT-PCR analysis was performed with cDNA synthesized from WT (H99), lkh1Δ (KW1451), and LKH1KD (KW1958) strains grown to the mid-log phase. Error bars indicate standard deviation. The statistical significance of difference was determined by one-way analysis of variance with Bonferroni’s multiple comparison tests. (* P < 0.05 and NS: non-significant) (C) WT (H99), lkh1Δ (KW1451), lkh1Δ+LKH1 (KW1633), and LKH1KD (KW1958) strains were cultured in liquid YPD medium overnight at 30°C, 10-fold serially diluted and spotted on YPD medium containing stress-inducing agents. For the UV-C resistance test, the serially-diluted cells were spotting onto the YPD medium, and the indicated dose of UV-C was exposed. The plates were further incubated at 30°C and daily photographed for 2-4 days.
In this study, we observed that Tor1 acts as an upstream regulator of Lkh1. Our previous study reported that Tor1 regulates the expression of genes involved in transmembrane transport, ribosome synthesis, and metabolic processes (So et al., 2019). To elucidate the signaling circuitry downstream of CnLkh1 in the TOR1-Lkh1 pathway, we conducted RNA-seq analysis comparing the transcriptome profiles of the WT strain and the lkh1Δ mutants with or without rapamycin treatment. Under the basal condition, the expression levels of 409 genes in the lkh1Δ mutant were either increased (276 genes) or decreased (133 genes) compared to WT in a statistically significant manner (P < 0.05, | log2 (fold change) | >= 2) (Supplementary Figure S4A and Supplementary Table S3). Under rapamycin treatment, the expression levels of 452 genes in lkh1Δ mutant were either increased (290 genes) or decreased (162 genes) compared to WT in a statistically significant manner (P < 0.05, | log2 (fold change) | >= 2) (Supplementary Figure S4B and Supplementary Table S3). Based on the KEGG pathway analysis, we observed that CnLkh1 was involved in amino acid metabolism, DNA replication, and base excision repair in the absence of rapamycin treatment. Under rapamycin treatment, CnLkh1 was observed to be involved in ribosomal biogenesis (Figure 8A). To determine whether Lkh1 regulates the expression of genes involved in ribosome synthesis and small nuclear RNA processing [CNAG_02382 (BRX1), CNAG_04365 (MAK21), CNAG_02437 (RRP5), CNAG_04072 (NIP7), CNAG_06318 (YTM1), and CNAG_06535 (UTP30)], qRT-PCR analysis was performed. Notably, only BRX1 demonstrated lower expression in the lkh1Δ mutant compared to that in WT. Conversely, the MAK21, RRP5, NIP7, YTM1, and UTP30 expression levels demonstrated no significant differences between WT and lkh1Δ mutants (Figure 8B and Supplementary Figure S4C). The transcriptome analysis showed that expression levels of amino acid transporter or amino acid metabolism genes [CNAG_02049 (PUT1), CNAG_07902 (AAP2), CNAG_05345 (AAP7), and CNAG_00597 (AAP4)] were reduced in the lkh1Δ mutant. We observed that the PUT1, AAP2, AAP4, and AAP7 expression levels were similar between WT and lkh1Δ mutant under basal conditions. However, the induction levels in the lkh1Δ mutant were lower than those in WT (Figure 8C). Put1 is involved in proline production (Lee et al., 2011). Aap4 and Aap2 are required for transporting lysine and isoleucine, respectively, as the sole nitrogen source (Martho et al., 2016). To determine whether Lkh1 is required for cell growth when a single amino acid serves as the sole nitrogen source, we monitored the growth of each strain in a minimal medium supplemented with various nitrogen sources, including lysine, isoleucine, and proline. We observed that the growth of the lkh1Δ mutant in YNB including proline or lysine, but not isoleucine, was partially restored compared to that of the lkh1Δ mutant in the YNB medium alone (Figure 8D). However, the growth of the lkh1Δ mutant in YNB medium supplemented with all amino acids (proline, isoleucine, and lysine) was similar to those in YNB supplemented with either proline (Figure 8D). Collectively, Lkh1 is required for proline- and lysine-dependent growth of C. neoformans.
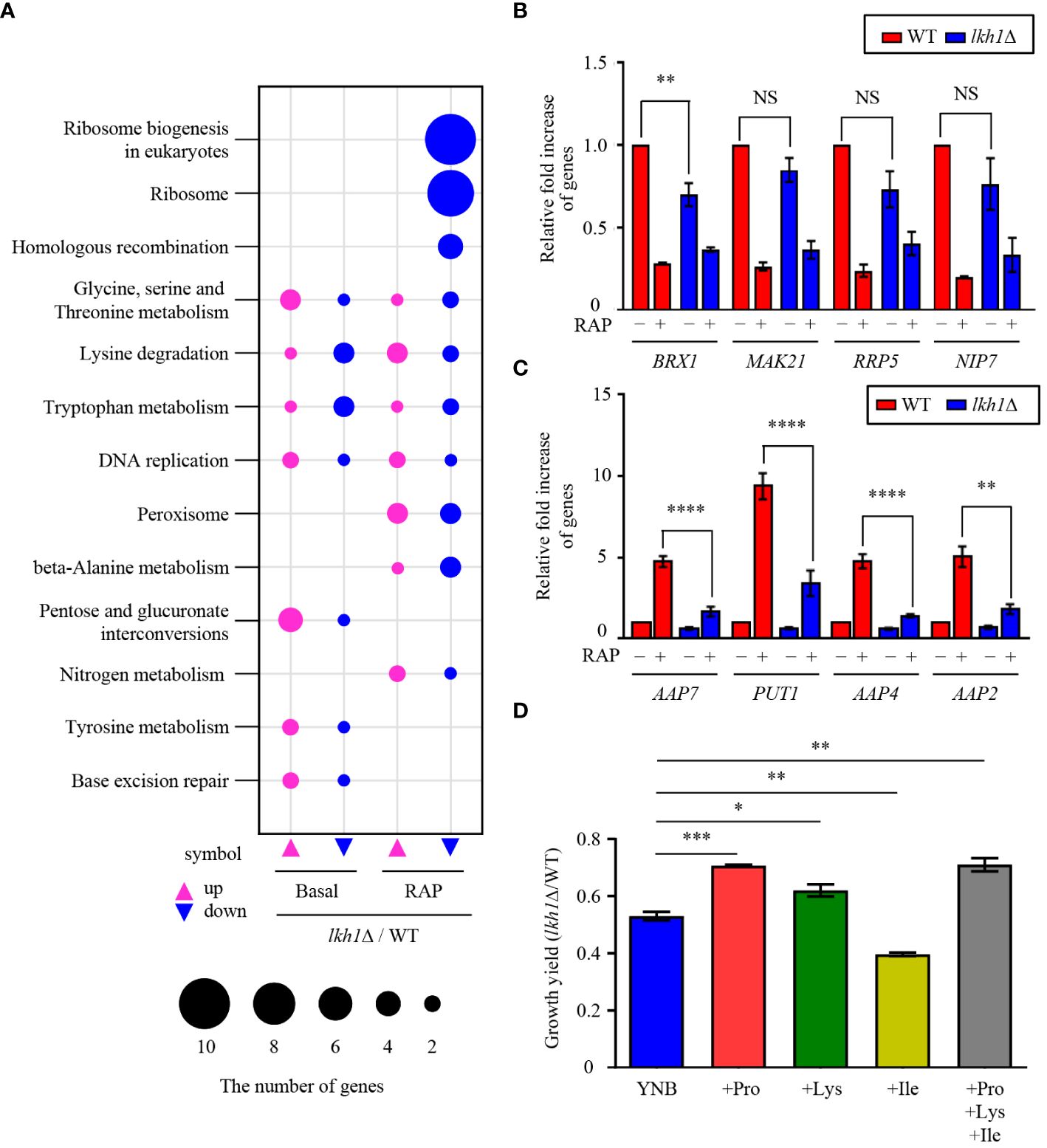
Figure 8 Transcriptome analysis of lkh1Δ mutant in response to rapamycin. (A) Significant differences in KEGG pathway analysis were observed between basal and rapamycin-treatment conditions (3 ng/mL) in lkh1Δ mutants. (B, C) Expression levels of genes encoding genes related to ribosome synthesis, small nuclear RNA, and amino acid transporter were verified by qRT-PCR analysis using cDNA of WT and lkh1Δ strains with or without rapamycin treatment (3 ng/mL). Three independent biological experiments with duplicate technical replicates were performed. Error bars indicate S.E.M. Statistical significances of the differences were determined by one-way ANOVA with Bonferroni’s multiple-comparison test (NS: not significant; ** P < 0.01; **** P < 0.0001). (D) Growth yield was calculated by measuring the growth curve of each strain. Each strain was grown in a minimal medium containing certain nitrogen sources (2 mM proline, 2 mM isoleucine, 2 mM lysine, and all together) Error bars indicate S.E.M. Statistical significances of the differences were determined by one-way ANOVA with Bonferroni’s multiple-comparison test. (* P < 0.05; ** P < 0.01; *** P < 0.001).
C. neoformans possesses several virulence factors for survival and proliferation within the host. One such factor is melanin, a dark brown polyphenolic pigment with antiphagocytic and antioxidant properties (Eisenman and Casadevall, 2012). To investigate the role of Lkh1 in melanin production, we assessed the melanin production levels of the lhk1Δ mutant on the L-DOPA medium. We observed that the lkh1Δ mutant produced less melanin compared to WT and its complemented strain (Figure 9A). In high glucose concentration conditions, which is a melanin-repressed condition (Jin et al., 2020), the difference in melanin production between lkh1Δ and WT strains was obvious. Despite a long incubation period (7 days), melanin production in lkh1Δ mutant did not recover to WT level (Data not shown). Given that the lkh1Δ mutant exhibited growth defect, we wondered whether the reduced melanin production in the lkh1Δ mutant originated from its growth defect or the influence on melanin-producing genes encoding laccases (LAC1 and LAC2). To test this hypothesis, we measured the LAC1 and LAC2 expression levels under melanin-induced conditions. We observed that the induction levels of LAC1 in the lkh1Δ mutant were significantly lower compared to the WT, and the LAC2 expression level in the lkh1Δ mutant was slightly lower than that in WT (Figure 9B).
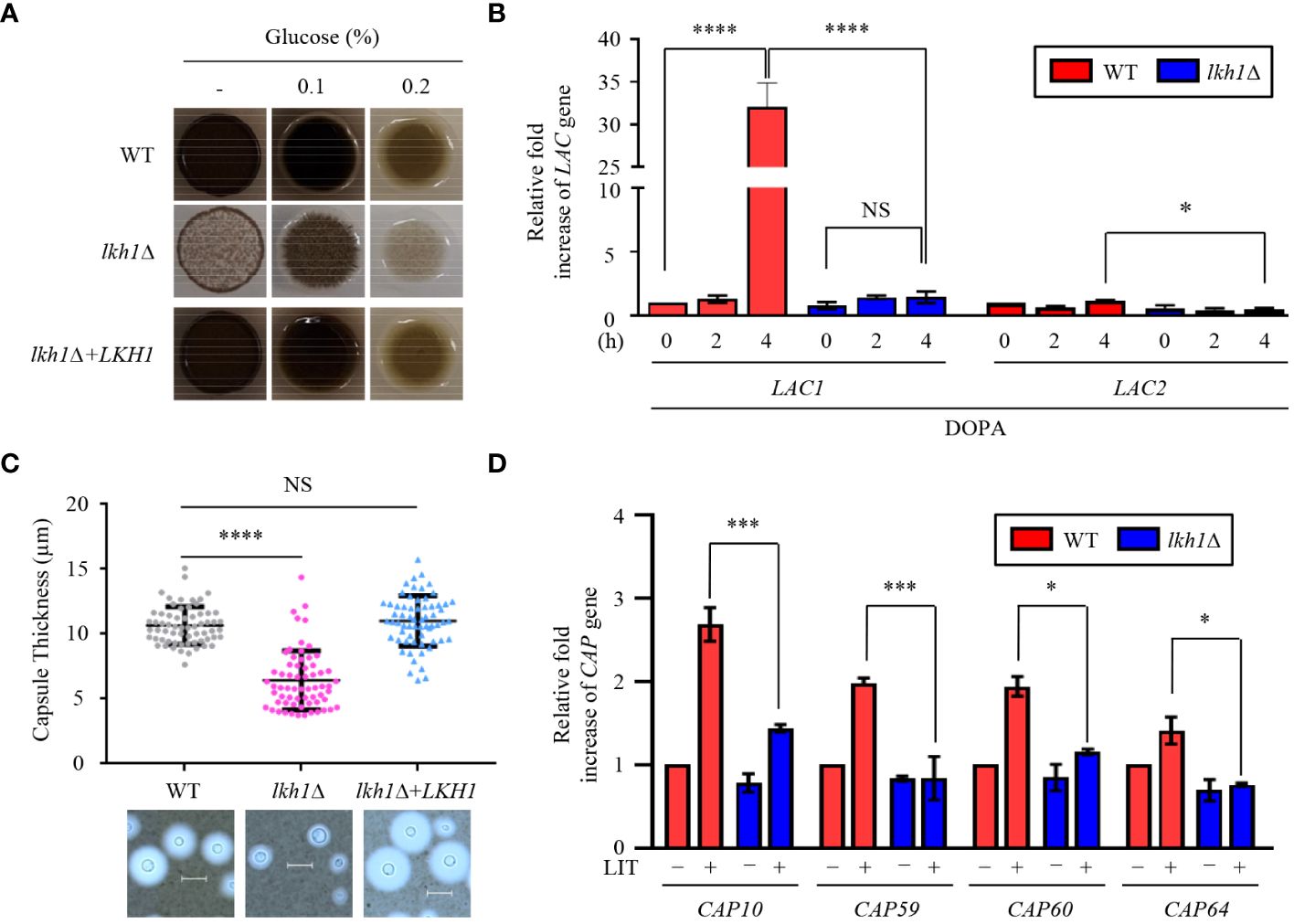
Figure 9 Lkh1 was involved in capsule and melanin production of C. neoformans. (A) WT (H99), lkh1Δ (KW1451), and lkh1Δ+LKH1 (KW1633) strains were spotted on L-DOPA medium (0.1%, 0.2% glucose, and without glucose) at 30°C for 3 days. (B, D) Expression levels of laccase genes (LAC1 and LAC2) and capsule-related genes (CAP10, CAP59, CAP60, and CAP64) were verified by qRT-PCR analysis using cDNA of WT and lkh1Δ mutant. Three independent biological experiments with duplicate technical replicates were performed. Error bars indicate S.E.M. Statistical significances of the differences were determined by one-way ANOVA with Bonferroni’s multiple-comparison test. (* P < 0.05; *** P < 0.001; **** P < 0.0001; NS: not significant) (C) The WT, lkh1Δ, and lkh1Δ+LKH1 strains were cultured on a Littman medium for capsule production at 30°C for 3 days. The scale bar indicates 10 μm.
The polysaccharide capsule, located outside the fungal cell wall and enabling the fungus to evade immune recognition and destruction (Chen et al., 2022), is another major virulence factor of C. neoformans. Next, we compared capsule production between WT and lkh1Δ mutants. Similar to the melanin production, lkh1Δ mutant showed reduced capsule production compared to WT and complemented strains (Figure 9C). We wondered whether the decrease in capsule production in the lkh1Δ mutant resulted from its growth defect or differences in gene expression related to capsule production. We observed that induction levels of capsule production-related genes including CAP10, CAP59, CAP60, and CAP64 in the lkh1Δ mutant were significantly lower compared to those in WT under capsule induction conditions (Figure 9D). To determine whether the kinase activity of Lkh1 influences the formation of virulence factors, we performed melanin and capsule production assays. Strikingly, the LKH1D546A strains produced less melanin and capsule production similar to the lkh1Δ mutant (Supplementary Figure S5). Taken together, these findings indicate that Lkh1 is involved in the formation of both capsule and melanin in C. neoformans.
Our experiment results, which showed that the lkh1Δ mutant displayed a growth defect at 37°C, along with reduced melanin and capsule productions, indicate that it plays an essential role in virulence. To demonstrate this hypothesis, we assessed the virulence of the lkh1Δ mutant in a murine model of cryptococcosis. We found that the lkh1Δ mutant was completely avirulent. Although the mice infected with lkh1Δ+LKH1 showed delayed symptoms compared to mice infected with the WT, all the mice were symptomatic within 30 days post-infection (Figure 10A). To investigate the effect of Lkh1 on pulmonary growth, we observed the lung tissues of infected mice at 16 days post-infection. Lung tissues were much heavier in WT and lkh1Δ+LKH1 strain-infected mice compared to lkh1Δ mutant-infected mice (Figure 10B). In particular, no evidence of fungal infection was observed around the bronchioles of lkh1Δ mutant-infected lung (Figure 10C, upper panel for fungal organisms; Figure 10C, lower panel for immune cell clusters). To further demonstrate that Lkh1 is required for BBB-traversal activity, we performed an in vitro BBB transmigration assay. Expectedly, BBB transmigration of the lkh1Δ mutant was much lower than that of the WT or lkh1Δ+LKH1 strains (Figure 10D). These results indicate that Lkh1 plays a crucial role in the virulence of C. neoformans by governing its BBB crossing capacity.
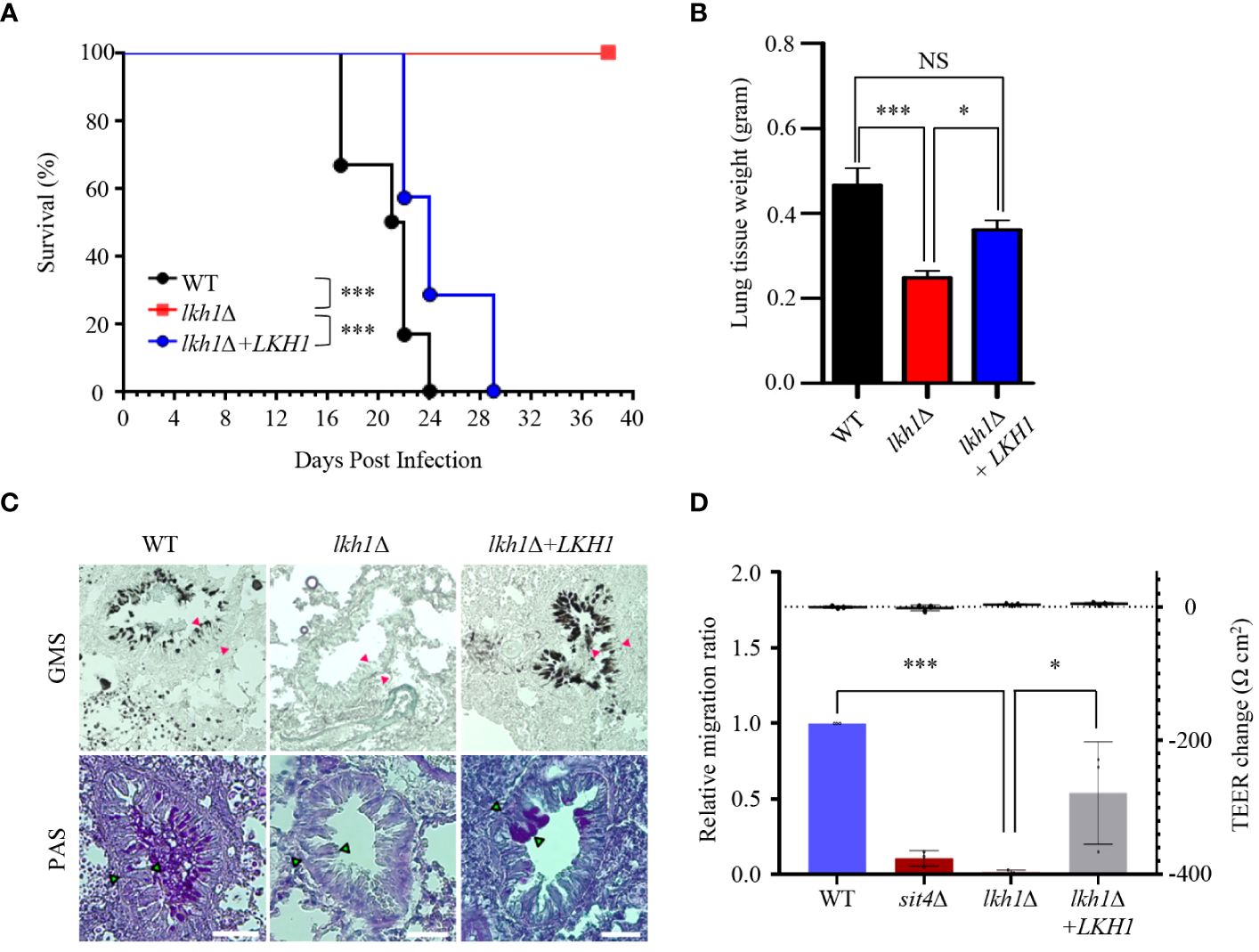
Figure 10 Lkh1 was essential for the virulence of C neoformans. (A) C neoformans WT (H99), lkh1Δ (KW1451), and lkh1Δ+LKH1 (KW1633) strains were nasally inoculated into the 7-week-old female Balb/c mice and monitored. Survival curve of infected mice (n=7). (B) Mice were sacrificed for histochemistry at 16 dpi (n=5). (B) Comparison of lung tissue weight measurements. (C) Grocott’s methenamine silver (GMS) and Periodic Acid-Schiff (PAS) staining of lung tissues of mice sacrificed 16 days after infection. Red arrows indicate the walls of small bronchioles. Scale bar means 0.04 mm. (D) BBB translocation assays using hCMEC/D3-coated transwell. Plates with an inoculum of 105 cells of WT, lkh1Δ, lkh1 Δ+LKH1, and sit4Δ strains were incubated at 37°C in a CO2 incubator for 24 h, and the number of yeast cells passing through the hCMEC/D3-coated transwell was measured by CFU. The sit4Δ mutant was used as a negative control. The left and right Y axes indicate the migrated CFU and the trans-endothelial electrical resistance (TEER) value, respectively. Three independent biological experiments with duplicate technical replicates were performed. Error bars indicate S.E.M. Statistical significances of the differences were determined by one-way ANOVA with Bonferroni’s multiple-comparison test. (* P < 0.05; *** P < 0.001).
In this study, we demonstrated the pleiotropic roles of CnLkh1 in diverse stress responses, sexual differentiation, and antifungal drug resistance. Furthermore, we characterized its downstream regulatory networks governed by CnLkh1 using transcriptome analysis and confirmed that Lkh1 acts as one of the downstream kinases in the TOR signaling pathway (Figure 11).
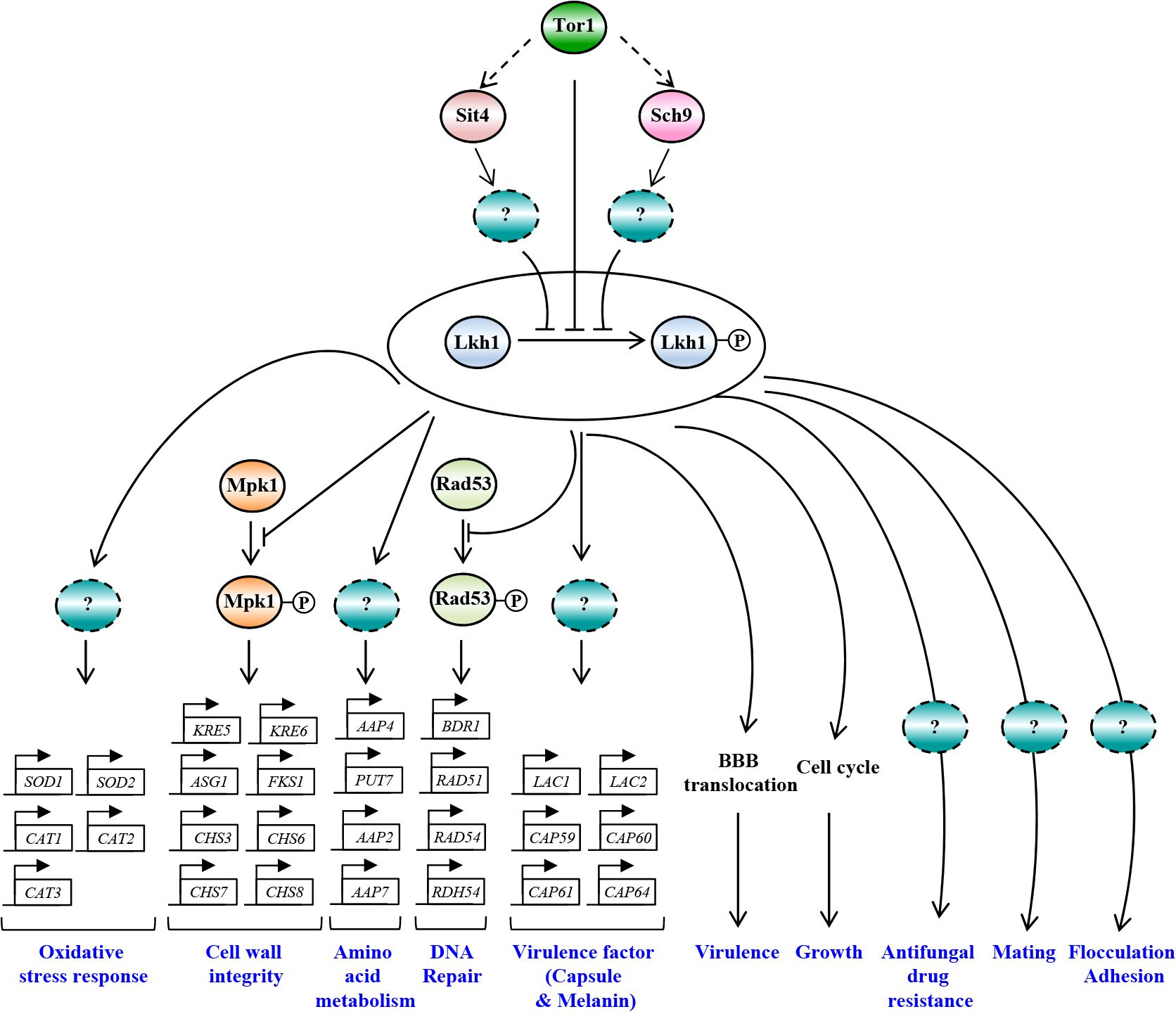
Figure 11 The proposed model of Lkh1-dependent pathway in C. neoformans. In response to extraneous stress stimuli, Lkh1 orchestrates a myriad of stress responses including oxidative stress response, preservation of cell wall integrity, DNA damage response, and modulation of amino acid metabolism via the regulation of downstream target gene expression. Additionally, Lkh1 contributes to antifungal drug resistance, mating, flocculation, adhesion, and intrinsic growth. Notably, Lkh1 plays a pivotal role in virulence by modulating the translocalization of the blood-brain barrier (BBB).
Although Cryptococcus LKH1 deletion caused defects in growth and stress responses akin to other fungal species lacking the LKH1 homologous gene, its regulatory mechanisms appeared to be distinct among fungi. In U. maydis, Lkh1 regulates the DNA damage response in a homologous recombination (HR) pathway-independent manner (de Sena-Tomás et al., 2015). M. oryzae Kns1 controls the expression of genes involved in DNA damage checkpoints and cell cycle regulation (Li et al., 2022). Our study reported that CnLkh1 regulates DNA damage response in both Rad53-dependent and -independent manners. Second, in S. cerevisiae, both catalytic and non-catalytic activities of LAMMER kinase are required for the stress response (Lee et al., 2012). MoKns1 physically interacts with Atg18 via a non-kinase domain (Li et al., 2022). Furthermore, the strain with a mutation at Q488 in the kinase domain of Lkh1 showed greater susceptibility to DNA damage stress than the strain lacking the entire LKH1 gene in U. maydis (de Sena-Tomás et al., 2015). However, the catalytic activity of Lkh1 in C. neoformans is mainly responsible for most stress responses and virulence factor formation, except for fluconazole resistance. Third, our study revealed that Lkh1 phosphorylation was dependent on the TOR signaling pathway. A unique finding was that Cryptococcus Lkh1 was already phosphorylated in the absence of external stress. In this context, numerous genes were intrinsically up-regulated or down-regulated in the lkh1Δ mutant compared to WT at basal condition. These data indicate that Lkh1 in C. neoformans intrinsically contributes to diverse cellular responses regardless of external stress.
Fungal cell walls serve multifaceted roles in host interaction and are indispensable for maintaining cellular integrity. Integral constituents within the cell wall are recognized as major fungal pathogen-associated molecular patterns (PAMPs). The perturbation in cell composition resulting from the deletion of LKH1 influences PAMPs, thereby diminishing virulence in A. fumigatus (Lim et al., 2021). In C. albicans, genes responsible for encoding GPI-anchored proteins exhibit substantial upregulation in the kns1Δ mutant compared to WT (Lim et al., 2020). Our investigation also elucidated that perturbation in LKH1 changes in chitin content. This observation is substantiated by the impact of Lkh1 on flocculation and adhesion activities in C. neoformans. Consequently, modifications in the cell wall components of the lkh1Δ mutant may contribute to the attenuation of virulence in C. neoformans.
In this study, we made a notable observation that ergosterol contents in the lkh1Δ mutant were similar to that in WT, although the lkh1Δ mutant exhibited opposite resistance to amphotericin B and fluconazole, respectively. This result contrasts with strains showing the opposite resistance to polyene and azole drugs, which typically contain more or less ergosterol contents in yeast (Young et al., 2003; Ko et al., 2009). However, susceptibility or resistance to azole/polyene drugs does not depend on the ergosterol content alone. Supporting this, no significant correlation was observed between azole/polyene susceptibility patterns and ergosterol content in clinical yeast strains such as C. albicans and C. neoformans (Gomez-Lopez et al., 2011). Given that antifungal drugs penetrate the cell wall before interacting with the cell membrane, changes in cell wall constituents may contribute to susceptibility to polyene or azole drugs. Remarkably, reduced chitin content in the cell wall has been shown to decrease the susceptibility to amphotericin B in Kluyveromyces and Candida strains (Bahmed et al., 2003). Similarly, increased chitin levels were observed in fluconazole-resistant Candida auris clinical isolates (Shahi et al., 2022). There, alteration in cell wall constituents of the lkh1Δ mutant might induce polyene or azole drug susceptibility.
Our finding that Lkh1 is essential for virulence reinforces the pivotal role of the TOR signaling pathway in the pathogenicity of C. neoformans. Our previous studies revealed that TOR-dependent cellular processes such as ribosome genesis, carbon utilization, and amino acid metabolism are required for BBB infection and survival in the brain (Lee et al., 2020). Supporting this notion, Sch9 kinase and Sit4 phosphatase have been identified as necessary components for BBB infection (Jin et al., 2020). Recently, the TOR pathway has been implicated in adaptation to the host environment through Ypk1-dependent phospholipid asymmetry remodeling (Ristow et al., 2023). Besides kinases and phosphatases, Zfp1, which is homologous to Sfp1 and is involved in nutrient sensing and the cell cycle as a downstream transcription factor of TOR signaling in S. cerevisiae, is required for virulence in C. neoformans. Especially, the zfp1Δ mutant shows a reduced fungal burden in the brain and spleen (Fan et al., 2019). A previous study reported that TF mutants (hlh5Δ, bzp1Δ, hob1Δ, ada2Δ, crz1Δ, skn7Δ, and atf1Δ mutants) exhibit susceptibility or resistance in response to rapamycin and some of them display reduced virulence (Jung et al., 2015; So et al., 2019). Therefore, the TOR pathway appears essential for virulence in C. neoformans.
In addition to the upstream regulators of LAMMER kinases, the genome-wide downstream genes regulated by LAMMER kinases in fungal pathogens remain elusive. To the best of our knowledge, this is the first transcriptome analysis of a LAMMER kinase mutant in a human fungal pathogen. The transcriptome analyses in this study provide insights into LKH1 deletion-induced phenotypic changes in response to diverse stress responses and sexual differentiation. First, among Lkh1-dependent genes, expression levels of nucleotide metabolism-related genes were changed. YNK1, which encodes a nucleoside diphosphate kinase, catalyzes the transfer of phosphates from nucleoside triphosphates and is induced during DNA replication stress (Yang et al., 2009). Likewise, HAM1, which encodes nucleoside triphosphate pyrophosphohydrolase, is involved in non-canonical purine and pyrimidine metabolism, and its protein level increases in response to DNA replication stress (Takayama et al., 2007). Our previous studies reveal that Cryptococcus YNK1 appears to be essential for growth, while the ham1Δ mutant showed WT-levels of resistance in response to stress responses (Jin et al., 2020). Supporting this, nucleotide metabolism is crucial for DNA damage stress as well as DNA replication stress (Jung et al., 2022). These data indicate that Cryptococcus Lkh1 may function in response to DNA damage stress by regulating nucleotide metabolism. Secondly, nitrogen metabolism has been characterized by its role in virulence, stress response, autophagy, sexual differentiation, and capsule and melanin production in C. neoformans (Kmetzsch et al., 2011; Lee et al., 2011; Frazzitta et al., 2013; Jiang et al., 2020). Although the TOR signaling pathway controls nitrogen metabolism in S. cerevisiae (Crespo et al., 2002), the regulatory mechanism of TOR-mediated nitrogen metabolism has not been elucidated in C. neoformans. Our transcriptome data provided evidence that Lkh1 controls nitrogen metabolism by regulating the expression of amino acid transporter genes. Supporting this, the phenomenon that lkh1Δ mutant showed reduced mating efficiency, capsule production, and melanin biosynthesis, is similar to the phenotype observed in the strains deleted with genes related to amino acid metabolism (Kmetzsch et al., 2011; Lee et al., 2011). However, in this state, we need to identify TOR-dependent transcription factors that regulate nitrogen metabolism and further elucidate the relationship between these transcription factors and Lkh1 in C. neoformans.
Although LAMMER kinases in fungi are evolutionarily conserved and play pleiotropic roles in stress responses and differentiation, information about crosstalks between LAMMER kinase and other signaling pathways is limited. However, our study revealed that Lkh1 is partially interconnected with the Mpk1 pathway for cell wall stress response and the Rad53-Bdr1 pathway for the DNA damage response. In yeast, crosstalk between TOR and cAMP/PKA pathway has been partially elucidated. For instance, activation signals of Rim15, a protein kinase involved in cell proliferation in response to nutrient, are integrated from the TOR and PKA pathways (Pedruzzi et al., 2003). Furthermore, the constitutive activation of the RAS/cAMP pathway inhibits rapamycin-induced responses (Schmelzle et al., 2004). Defects in the capsule and melanin productions in lkh1Δ mutant were also observed, akin to when the PKA/cAMP pathway is impaired. Therefore, LAMMER kinase, Lkh1 may have crosstalk with the cAMP/PKA pathway as the downstream factor of the TOR pathway in C. neoformans and further research will be necessary to address this relationship in the future.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/geo/, GSE233612.
The animal study was approved by Korea Zoonosis Research Institute. The study was conducted in accordance with the local legislation and institutional requirements.
SK: Validation, Supervision, Writing – review & editing, Writing – original draft, Methodology, Investigation, Formal analysis, Data curation. YC: Validation, Writing – review & editing, Writing – original draft, Methodology, Investigation, Data curation. E-SK: Writing – review & editing, Writing – original draft, Methodology, Investigation, Formal analysis. K-TL: Writing – review & editing, Writing – original draft, Supervision, Investigation. Y-SB: Writing – review & editing, Writing – original draft, Supervision, Investigation. K-WJ: Formal analysis, Writing – review & editing, Writing – original draft, Validation, Supervision, Investigation, Funding acquisition, Data curation, Conceptualization.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MIST) (No. 2020R1C1C1005468 and 2022R1C1C2003274) and the KAERI Institutional R&D Program (523610-24).
We thanks Macrogen for helping the RNAseq data submission to NCBI and Yu-Byeong Jang for technical support of microscopy measurement.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1369301/full#supplementary-material
Supplementary Figure 1 | Construction of the lkh1Δ, rad53Δ lkh1Δ, mpk1Δ lkh1Δ, Rad53-4×FLAG lkh1Δ, LKH1-4×FLAG, LKH1-4×FLAG sit4Δ, and LKH1-4×FLAG sch9Δ mutants. (A, B, H, J, L) Diagram for LKH1 gene disruption in serotype A (MATα) H99 strain, rad53Δ, mpk1Δ, Rad53-4×FLAG mutant, MATa KN99, LKH1-4×FLAG, LKH1-4×FLAG sit4Δ, and LKH1-4×FLAG sch9Δ mutants. (C, D, E, F, G, I, K, and M) The correct gene disruptions were confirmed by Southern blot analysis using genomic DNAs digested with the indicated restriction enzyme.
Supplementary Figure 2 | Lkh1 was involved in the cell cycle progression. (A) WT (H99), lkh1Δ (KW1451), and lkh1Δ+LKH1 (KW1633) strains were grown in liquid YPD medium for 16 h at 30°C and subcultured in fresh liquid YPD medium at 37°C. Cell density was measured at an optical density of 600 nm every 2 h. (B) Intracellular measurement of ATP level in lkh1Δ mutant. WT (H99), lkh1Δ (KW1451), and lkh1Δ+LKH1 (KW1633) strains were grown in liquid YPD medium for 16 h at 30°C and washed using PBS. The ATP contents of strains were calculated by measuring luminescence. ATP concentrations were normalized using soluble-protein concentrations for each sample. Three independent biological experiments with duplicate technical replicates were performed. Error bars indicate S.E.M. Statistical significances of the differences were determined by one-way ANOVA with Bonferroni’s multiple-comparison test. (NS: not significant) (C) Cell cycle analysis of lkh1Δ mutant via flow cytometry. Cells were stained with propidium iodine (PI). The three biological replicates were performed. (D) Morphological test of lkh1Δ mutant. Cells were fixed with ethanol and observed using differential interference contrast (DIC) (Nikon Eclipse, Japan).
Supplementary Figure 3 | Spotting assay to confirm the functionality of Lkh1-4×FLAG protein. WT (H99), lkh1Δ (KW1451), lkh1Δ+LKH1 (KW1633), and Lkh1-4×FLAG strain (KW1755) strains were cultured in liquid YPD medium overnight at 30°C, 10-fold serially diluted and spotted on YPD medium containing 2 μg/mL bleomycin, 0.8 mg/mL caffeine, 0.8 mg/mL CFW, 1.2% CR, 0.02% MMS, 0.15 μg/mL 4-NQO, 90 mM HU, 5 mM cisplatin, 1 μg/mL Amphotericin B, or 18 μg/mL fluconazole. For the UV-C resistance test, the serially-diluted cells were spotting onto the YPD medium, and the indicated dose of UV-C was exposed. The plates were further incubated at 30°C and photographed daily for 2-4 days.
Supplementary Figure 4 | Volcano plot and expression of genes using data from transcriptome analysis. (A) Volcano plots were shown between WT and lkh1Δ mutant (KW1451) at basal time. Yellow dots mean genes in a statistically significant manner (log2 Fold Change [lkh1Δ/WT] >= 2 and P < 0.05) and blue dots mean genes in a statistically significant manner (log2 Fold Change [lkh1Δ/WT] <= -2 and P < 0.05). (B) Volcano plots were shown between WT and lkh1Δ mutant (KW1451)) under rapamycin (3 ng/mL) treatment. Yellow dots mean genes in a statistically significant manner (log2 Fold Change [lkh1Δ/WT] >= 2 and P < 0.05) and blue dots mean genes in a statistically significant manner (log2 Fold Change [lkh1Δ/WT] <= -2 and P < 0.05) (C) Expression levels of genes encoding genes related ribosome synthesis were verified by qRT-PCR analysis using cDNA of WT and lkh1Δ strains with or without rapamycin treatment (3 ng/mL). Three independent biological experiments with duplicate technical replicates were performed. Error bars indicate S.E.M. Statistical significances of the differences were determined by one-way ANOVA with Bonferroni’s multiple-comparison test (NS: not significant).
Supplementary Figure 5 | Catalytic activity of Lkh1 was involved in the capsule and melanin production of C. neoformans. (A) The WT (H99), lkh1Δ (KW1451), lkh1Δ+LKH1 (KW1633), and LKH1D546A (KW1958) strains were spotted on L-DOPA medium with 0.2% glucose and without glucose at 30°C and 37°C for 3 days. (B) The WT (H99), lkh1Δ (KW1451), lkh1Δ+LKH1 (KW1633), and LKH1D546A (KW1958) strains were cultured on a Littman medium for capsule production at 30°C for 3 days. The scale bar indicates 10 μm.
Arthington-Skaggs, B. A., Jradi, H., Desai, T., Morrison, C. J. (1999). Quantitation of ergosterol content: novel method for determination of fluconazole susceptibility of Candida albicans. J. Clin. Microbiol. 37, 3332–3337. doi: 10.1128/JCM.37.10.3332-3337.1999
Bahmed, K., Bonaly, R., Coulon, J. (2003). Relation between cell wall chitin content and susceptibility to amphotericin B in Kluyveromyces, Candida and Schizosaccharomyces species. Res. Microbiol. 154, 215–222. doi: 10.1016/S0923-2508(03)00049-4
Banks, I. R., Specht, C. A., Donlin, M. J., Gerik, K. J., Levitz, S. M., Lodge, J. K. (2005). A chitin synthase and its regulator protein are critical for chitosan production and growth of the fungal pathogen Cryptococcus neoformans. Eukaryot. Cell 4, 1902–1912. doi: 10.1128/EC.4.11.1902-1912.2005
Bender, J., Fink, G. R. (1994). Afc1, A LAMMER kinase from Arabidopsis thaliana, activates ste12-dependent processes in yeast. Proc. Natl. Acad. Sci. U. S. A., 12105–12109, 91. doi: 10.1073/pnas.91.25.12105
Chang, M., Sionov, E., Khanal Lamichhane, A., Kwon-Chung, K. J., Chang, Y. C. (2018). Roles of three Cryptococcus neoformans and Cryptococcus gattii efflux pump-coding genes in response to drug treatment. Antimicrob. Agents Chemother. 62, 10.1128/Aac. 01751–17. doi: 10.1128/AAC.01751-17
Chen, Y., Shi, Z. W., Strickland, A. B., Shi, M. (2022). Cryptococcus neoformans infection in the central nervous system: the battle between host and pathogen. J. Fungi 8, 1069. doi: 10.3390/jof8101069
Cho, S. J., Kim, Y. H., Park, H. M., Shin, K. S. (2010). Possible roles of LAMMER kinase Lkh1 in fission yeast by comparative proteome analysis. Mycobiology 38, 108–112. doi: 10.4489/MYCO.2010.38.2.108
Choi, Y. K., Kang, E.-H., Park, H.-M. (2014). Role of lammer kinase in cell wall biogenesis during vegetative growth of Aspergillus nidulans. Mycobiology 42, 422–426. doi: 10.5941/MYCO.2014.42.4.422
Choi, Y., Yu, S.-R., Lee, Y., Na, A.-Y., Lee, S., Heitman, J., et al. (2024). Casein kinase 2 complex: A central regulator of multiple pathobiological signaling pathways in Cryptococcus neoformans. Mbio 15, E03275–E03223. doi: 10.1128/mbio.03275-23
Cramer, K. L., Gerrald, Q. D., Nichols, C. B., Price, M. S., Alspaugh, J. A. (2006). Transcription factor nrg1 mediates capsule formation, stress response, and pathogenesis in Cryptococcus neoformans. Eukaryot. Cell 5, 1147–1156. doi: 10.1128/EC.00145-06
Crespo, J. L., Powers, T., Fowler, B., Hall, M. N. (2002). The TOR-controlled transcription activators Gln3, Rtg1, and Rtg are regulated in response to intracellular levels of glutamine. Proc. Natl. Acad. Sci. U. S. A. 99, 6784–6789. doi: 10.1073/pnas.102687599
de Sena-Tomás, C., Sutherland, J. H., Milisavljevic, M., Nikolic, D. B., Përez-Martïn, J., Kojic, M., et al. (2015). Lammer kinase contributes to genome stability in Ustilago maydis. DNA Repair 33, 70–77. doi: 10.1016/j.dnarep.2015.05.011
Di Como, C. J., Arndt, K. T. (1996). Nutrients, via the TOR proteins, stimulate the association of Tap42 with type 2a phosphatases. Genes. Dev. 10, 1904–1916. doi: 10.1101/gad.10.15.1904
Duclos, B., Marcandier, S., Cozzone, A. J. (1991). Chemical properties and separation of phosphoamino acids by thin-layer chromatography and/or electrophoresis. Methods Enzymol. 201, 10–21. doi: 10.1016/0076-6879(91)01004-L
Eisenman, H. C., Casadevall, A. (2012). Synthesis and assembly of fungal melanin. Appl. Microbiol. Biotechnol. 93, 931–940. doi: 10.1007/s00253-011-3777-2
Ellis, D. H., Pfeiffer, T. J. (1990). Ecology, life cycle, and infectious propagule of Cryptococcus neoformans. Lancet 336, 923–925. doi: 10.1016/0140-6736(90)92283-N
Fan, C.-L., Han, L.-T., Jiang, S.-T., Chang, A.-N., Zhou, Z.-Y., Liu, T.-B. (2019). The Cys2His2 zinc finger protein Zfp1 regulates sexual reproduction and virulence in Cryptococcus neoformans. Fungal. Genet. Biol. 124, 59–72. doi: 10.1016/j.fgb.2019.01.002
Fan, Y., Lin, X. (2018). Multiple applications of A transient Cryptococcus coupled with electroporation (Trace) system in the Cryptococcus neoformans species complex. Genetics 208, 1357–1372. doi: 10.1534/genetics.117.300656
Frazzitta, A. E., Vora, H., Price, M. S., Tenor, J. L., Betancourt-Quiroz, M., Toffaletti, D. L., et al. (2013). Nitrogen source-dependent capsule induction in human-pathogenic Cryptococcus species. Eukaryot. Cell 12, 1439–1450. doi: 10.1128/EC.00169-13
Garcia, J., Shea, J., Alvarez-Vasquez, F., Qureshi, A., Luberto, C., Voit, E. O., et al. (2008). Mathematical modeling of pathogenicity of Cryptococcus neoformans. Mol. Syst. Biol. 4, 183. doi: 10.1038/msb.2008.17
Gilbert, N. M., Donlin, M. J., Gerik, K. J., Specht, C. A., Djordjevic, J. T., Wilson, C. F., et al. (2010). Kre genes are required for β-1, 6-glucan synthesis, maintenance of capsule architecture and cell wall protein anchoring in Cryptococcus neoformans. Mol. Microbiol. 76, 517–534. doi: 10.1111/j.1365-2958.2010.07119.x
Gomez-Lopez, A., Buitrago, M. J., Rodriguez-Tudela, J. L., Cuenca-Estrella, M. (2011). In vitro antifungal susceptibility pattern and ergosterol content in clinical yeast strains. Rev. Iberoam. Micol. 28, 100–103. doi: 10.1016/j.riam.2010.12.003
Gottfredsson, M., Perfect, J. R. (2000). Fungal meningitis. Semin. Neurol., 20, 307–322. doi: 10.1055/s-2000-9394
Hanes, J., Von Der Kammer, H., Klaudiny, J., Scheit, K. H. (1994). Characterization by cDNA cloning of two new human protein kinases. Evidence by sequence comparison of A new family of mammalian protein kinases. J. Mol. Biol. 244, 665–672. doi: 10.1006/jmbi.1994.1763
Idnurm, A., Bahn, Y. S., Nielsen, K., Lin, X., Fraser, J. A., Heitman, J. (2005). Deciphering the model pathogenic fungus Cryptococcus neoformans. Nat. Rev. Microbiol. 3, 753–764. doi: 10.1038/nrmicro1245
Jang, E-H, Kim, J-S, Yu, S-R, Bahn, Y-S (2022). Unraveling Capsule Biosynthesis and Signaling Networks in Cryptococcus neoformans. Microbiol. Spectr. 10, e02866–22. doi: 10.1128/spectrum.02866-22
Jiang, N., Benard, C. Y., Kebir, H., Shoubridge, E. A., Hekimi, S. (2003). Human clk2 links cell cycle progression, apoptosis, and telomere length regulation. J. Biol. Chem. 278, 21678–21684. doi: 10.1074/jbc.M300286200
Jiang, S. T., Chang, A. N., Han, L. T., Guo, J. S., Li, Y. H., Liu, T. B. (2020). Autophagy regulates fungal virulence and sexual reproduction in Cryptococcus neoformans. Front. Cell. Dev. Biol. 8, 374. doi: 10.3389/fcell.2020.00374
Jin, J.-H., Lee, K.-T., Hong, J., Lee, D., Jang, E.-H., Kim, J.-Y., et al. (2020). Genome-wide functional analysis of phosphatases in the pathogenic fungus Cryptococcus neoformans. Nat. Commun. 11, 4212. doi: 10.1038/s41467-020-18028-0
Joseph-Horne, T., Hollomon, D., Loeffler, R., Kelly, S. L. (1995). Cross-resistance to polyene and azole drugs in Cryptococcus neoformans. Antimicrob. Agents. Chemother. 39, 1526–1529. doi: 10.1128/AAC.39.7.1526
Jung, K.-W., Jung, J.-H., Park, H.-Y. (2021). Functional roles of homologous recombination and non-homologous end joining in DNA damage response and microevolution in Cryptococcus neoformans. J. Fungi 7, 566. doi: 10.3390/jof7070566
Jung, K.-W., Kwon, S., Jung, J.-H., Bahn, Y.-S. (2022). Essential roles of ribonucleotide reductases under DNA damage and replication stresses in Cryptococcus neoformans. Microbiol. Spectr. 10, E01044–E01022. doi: 10.1128/spectrum.01044-22
Jung, K.-W., Lee, Y., Huh, E. Y., Lee, S. C., Lim, S., Bahn, Y.-S. (2019). Rad53-and Chk1-dependent DNA damage response pathways cooperatively promote fungal pathogenesis and modulate antifungal drug susceptibility. Mbio 10, E01726–E01718. doi: 10.1128/mBio.01726-18
Jung, K. W., Lee, K. T., So, Y. S., Bahn, Y. S. (2018). Genetic manipulation of Cryptococcus neoformans. Curr. Protoc. Microbiol. 50, E59. doi: 10.1002/cpmc.59
Jung, K.-W., Strain, A. K., Nielsen, K., Jung, K.-H., Bahn, Y.-S. (2012). Two cation transporters ena1 and nha1 cooperatively modulate ion homeostasis, antifungal drug resistance, and virulence of Cryptococcus neoformans via the hog pathway. Fungal. Genet. Biol. 49, 332–345. doi: 10.1016/j.fgb.2012.02.001
Jung, K.-W., Yang, D.-H., Kim, M.-K., Seo, H. S., Lim, S., Bahn, Y.-S. (2016). Unraveling fungal radiation resistance regulatory networks through the genome-wide transcriptome and genetic analyses of Cryptococcus neoformans. Mbio 7, E01483–E01416. doi: 10.1128/mBio.01483-16
Jung, K.-W., Yang, D.-H., Maeng, S., Lee, K.-T., So, Y.-S., Hong, J., et al. (2015). Systematic functional profiling of transcription factor networks in Cryptococcus neoformans. Nat. Commun. 6, 1–14. doi: 10.1038/ncomms7757
Kang, E.-H., Kim, J.-A., Oh, H.-W., Park, H.-M. (2013). Lammer kinase LkhA plays multiple roles in the vegetative growth and asexual and sexual development of Aspergillus nidulans. PloS One 8, E58762. doi: 10.1371/journal.pone.0058762
Kang, W.-H., Park, Y.-H., Park, H.-M. (2010). The lammer kinase homolog, Lkh1, regulates tup transcriptional repressors through phosphorylation in Schizosaccharomyces pombe. J. Biol. Chem. 285, 13797–13806. doi: 10.1074/jbc.M110.113555
Kim, K.-H., Cho, Y.-M., Kang, W.-H., Kim, J.-H., Byun, K.-H., Park, Y.-D., et al. (2001). Negative regulation of filamentous growth and flocculation by Lkh1, A fission yeast lammer kinase homolog. Biochem. Biophys. Res. Commun. 289, 1237–1242. doi: 10.1006/bbrc.2001.6128
Kim, J.-S., Lee, K.-T., Lee, M. H., Cheong, E., Bahn, Y.-S. (2021). Adenylyl cyclase and protein kinase A play redundant and distinct roles in growth, differentiation, antifungal drug resistance, and pathogenicity of Candida auris. Mbio 12. doi: 10.1128/mBio.02729-21
Kmetzsch, L., Staats, C. C., Simon, E., Fonseca, F. L., Oliveira, D. L., Joffe, L. S., et al. (2011). The gata-type transcriptional activator gat1 regulates nitrogen uptake and metabolism in the human pathogen Cryptococcus neoformans. Fungal Genet. Biol. 48, 192–199. doi: 10.1016/j.fgb.2010.07.011
Ko, Y.-J., Yu, Y. M., Kim, G.-B., Lee, G.-W., Maeng, P. J., Kim, S., et al. (2009). Remodeling of global transcription patterns of Cryptococcus neoformans genes mediated by the stress-activated HOG signaling pathways. Eukaryot. Cell 8, 1197–1217. doi: 10.1128/EC.00120-09
Kraus, P. R., Fox, D. S., Cox, G. M., Heitman, J. (2003). The Cryptococcus neoformans MAP kinase Mpk1 regulates cell integrity in response to antifungal drugs and loss of calcineurin function. Mol. Microbiol. 48, 1377–1387. doi: 10.1046/j.1365-2958.2003.03508.x
Kwon-Chung, K., Sorrell, T., Dromer, F., Fung, E., Levitz, S. (2000). Cryptococcosis: clinical and biological aspects. Med. Mycol. 38, 205–213. doi: 10.1080/mmy.38.s1.205.213
Lee, K.-T., Byun, H.-J., Jung, K.-W., Hong, J., Cheong, E., Bahn, Y.-S. (2014). Distinct and redundant roles of protein tyrosine phosphatases Ptp1 and Ptp2 in governing the differentiation and pathogenicity of Cryptococcus neoformans. Eukaryot. Cell. 13, 796–812. doi: 10.1128/EC.00069-14
Lee, I. R., Chow, E. W., Morrow, C. A., Djordjevic, J. T., Fraser, J. A. (2011). Nitrogen metabolite repression of metabolism and virulence in the human fungal pathogen Cryptococcus neoformans. Genetics 188, 309–323. doi: 10.1534/genetics.111.128538
Lee, K., Du, C., Horn, M., Rabinow, L. (1996). Activity and autophosphorylation of lammer protein kinases. J. Biol. Chem. 271, 27299–27303. doi: 10.1074/jbc.271.44.27299
Lee, K.-T., Hong, J., Lee, D.-G., Lee, M., Cha, S., Lim, Y.-G., et al. (2020). Fungal kinases and transcription factors regulating brain infection in Cryptococcus neoformans. Nat. Commun. 11, 1521. doi: 10.1038/s41467-020-15329-2
Lee, D., Jang, E.-H., Lee, M., Kim, S.-W., Lee, Y., Lee, K.-T., et al. (2019). Unraveling melanin biosynthesis and signaling networks in Cryptococcus neoformans. Mbio 10. doi: 10.1128/mBio.02267-19
Lee, J., Moir, R. D., Mcintosh, K. B., Willis, I. M. (2012). TOR signaling regulates ribosome and tRNA synthesis via LAMMER/Clk and GSK-3 family kinases. Mol. Cell 45, 836–843. doi: 10.1016/j.molcel.2012.01.018
Lee, S.-B., Mota, C., Thak, E. J., Kim, J., Son, Y. J., Oh, D.-B., et al. (2023). Effects of altered N-glycan structures of Cryptococcus neoformans mannoproteins, MP98 (Cda2) and MP84 (Cda3), on interaction with host cells. Sci. Rep. 13, 1175. doi: 10.1038/s41598-023-27422-9
Li, S., Zhao, Y., Zhang, Y., Zhang, Y., Zhang, Z., Tang, C., et al. (2021). The Δ subunit of F1fo-atp synthase is required for pathogenicity of Candida albicans. Nat. Commun. 12, 6041. doi: 10.1038/s41467-021-26313-9
Li, L., Zhu, X.-M., Wu, J.-Q., Cao, N., Bao, J.-D., Liu, X.-H., et al. (2022). The lammer kinase MoKns1 regulates growth, conidiation and pathogenicity in Magnaporthe oryzae. Int. J. Mol. Sci. 23, 8104. doi: 10.3390/ijms23158104
Lim, J.-Y., Kim, Y. J., Woo, S. A., Jeong, J. W., Lee, Y.-R., Kim, C.-H., et al. (2021). The lammer kinase, LkhA, affects Aspergillus fumigatus pathogenicity by modulating reproduction and biosynthesis of cell wall pamps. Front. Cell. Infect. Microbiol. 1001. doi: 10.3389/fcimb.2021.756206
Lim, J.-Y., Park, Y.-H., Pyon, Y.-H., Yang, J.-M., Yoon, J.-Y., Park, S. J., et al. (2020). The lammer kinase is involved in morphogenesis and response to cell wall-and DNA-damaging stresses in Candida albicans. Med. Mycol. 58, 240–247. doi: 10.1093/mmy/myz049
Littman, M. (1958). Capsule synthesis by Cryptococcus neoformans. Trans. Ny Acad. Sci. 20, 623–648. doi: 10.1111/j.2164-0947.1958.tb00625.x
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative pcr and the 2– Δδct method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Manning, G., Plowman, G. D., Hunter, T., Sudarsanam, S. (2002). Evolution of protein kinase signaling from yeast to man. Trends. Biochem. Sci. 27, 514–520. doi: 10.1016/S0968-0004(02)02179-5
Martho, K. F., De Melo, A. T., Takahashi, J. P., Guerra, J. M., Santos, D. C., Purisco, S. U., et al. (2016). Amino acid permeases and virulence in Cryptococcus neoformans. PloS One 11, E0163919. doi: 10.1371/journal.pone.0163919
Miller, R. A., Britigan, B. E. (1997). Role of oxidants in microbial pathophysiology. Clin. Microbiol. Rev. 10, 1–18. doi: 10.1128/CMR.10.1.1
Myers, M. P., Murphy, M. B., Landreth, G. (1994). The dual-specificity Clk kinase induces neuronal differentiation of PC12 cells. Mol. Cell. Biol. 14, 6954–6961. doi: 10.1128/mcb.14.10.6954-6961.1994
Padmanabha, R., Gehrung, S., Snyder, M. (1991). The kns1 gene of Saccharomyces cerevisiae encodes A nonessential protein kinase homologue that is distantly related to members of the cdc28/cdc2 gene family. Mol. Gen. Genet. 229, 1–9. doi: 10.1007/BF00264206
Park, Y.-D., Kang, W.-H., Yang, W.-S., Shin, K.-S., Bae, K. S., Park, H.-M. (2003). Lammer kinase homolog, Lkh1, is involved in oxidative-stress response of fission yeast. Biochem. Biophys. Res. Commun. 311, 1078–1083. doi: 10.1016/j.bbrc.2003.10.110
Park, Y.-H., Park, H.-M. (2011). Disruption of the dual specificity kinase gene causes the reduction of virulence in Candida albicans. Kor. J. Mycol. 39, 85–87. doi: 10.4489/KJM.2011.39.1.085
Park, Y.-H., Yang, J.-M., Yang, S.-Y., Kim, S.-M., Cho, Y.-M., Park, H.-M. (2011). Function of dual specificity kinase, ScKns1, in adhesive and filamentous growth of Saccharomyces cerevisiae. Korean J. Microbiol. 47, 110–116.
Pedruzzi, I., Dubouloz, F., Cameroni, E., Wanke, V., Roosen, J., Winderickx, J., et al. (2003). TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Mol. Cell. Biol. 12, 1607–1613. doi: 10.1016/S1097-2765(03)00485-4
Perfect, J. R. (2014). Cryptococcosis: A model for the understanding of infectious diseases. J. Clin. Invest. 124, 1893–1895. doi: 10.1172/JCI75241
Price, M. S., Betancourt-Quiroz, M., Price, J. L., Toffaletti, D. L., Vora, H., Hu, G., et al. (2011). Cryptococcus neoformans requires A functional glycolytic pathway for disease but not persistence in the host. Mbio 2, E00103–E00111. doi: 10.1128/mBio.00103-11
Rajasingham, R., Govender, N. P., Jordan, A., Loyse, A., Shroufi, A., Denning, D. W., et al. (2022). The global burden of hiv-associated cryptococcal infection in adults in 2020: A modelling analysis. Lancet Infect. Dis. 22, 1748–1755. doi: 10.1016/S1473-3099(22)00499-6
Reese, A. J., Doering, T. L. (2003). Cell wall α-1, 3-glucan is required to anchor the Cryptococcus neoformans capsule. Mol. Microbiol. 50, 1401–1409. doi: 10.1046/j.1365-2958.2003.03780.x
Reese, A. J., Yoneda, A., Breger, J. A., Beauvais, A., Liu, H., Griffith, C. L., et al. (2007). Loss of cell wall alpha (1-3) glucan affects Cryptococcus neoformans from ultrastructure to virulence. Mol. Microbiol. 63, 1385–1398. doi: 10.1111/j.1365-2958.2006.05551.x
Regalla, D., Vannatta, M., Alam, M., Malek, A. E. (2022). Covid-19-associated Cryptococcus infection (Caci): A review of literature and clinical pearls. Infection 50, 1007–1012. doi: 10.1007/s15010-022-01805-y
Ristow, L. C., Jezewski, A. J., Chadwick, B. J., Stamnes, M. A., Lin, X., Krysan, D. J. (2023). Cryptococcus neoformans adapts to the host environment through TOR-mediated remodeling of phospholipid asymmetry. Nat. Commun. 14, 6587. doi: 10.1038/s41467-023-42318-y
Sanchez-Casalongue, M. E., Lee, J., Diamond, A., Shuldiner, S., Moir, R. D., Willis, I. M. (2015). Differential phosphorylation of A regulatory subunit of protein kinase CK2 by target of rapamycin complex 1 signaling and the Cdc-like kinase Kns1. J. Biol. Chem. 290, 7221–7233. doi: 10.1074/jbc.M114.626523
Sanguinetti, M., Posteraro, B., La Sorda, M., Torelli, R., Fiori, B., Santangelo, R., et al. (2006). Role of afr1, an abc transporter-encoding gene, in the in vivo response to fluconazole and virulence of Cryptococcus neoformans. Infect. Immun. 74, 1352–1359. doi: 10.1128/IAI.74.2.1352-1359.2006
Santiago-Tirado, F. H., Onken, M. D., Cooper, J. A., Klein, R. S., Doering, T. L. (2017). Trojan horse transit contributes to blood-brain barrier crossing of A eukaryotic pathogen. Mbio 8, E02183–E02116. doi: 10.1128/mBio.02183-16
Savaldi-Goldstein, S., Aviv, D., Davydov, O., Fluhr, R. (2003). Alternative splicing modulation by A lammer kinase impinges on developmental and transcriptome expression. Plant Cell 15, 926–938. doi: 10.1105/tpc.011056
Schmelzle, T., Beck, T., Martin, D. E., Hall, M. N. (2004). Activation of the RAS/cyclic AMP pathway suppresses A TOR deficiency in yeast. Mol. Cell. Biol. 24, 338–351. doi: 10.1128/MCB.24.1.338-351.2004
Shahi, G., Kumar, M., Skwarecki, A. S., Edmondson, M., Banerjee, A., Usher, J., et al. (2022). Fluconazole resistant Candida auris clinical isolates have increased levels of cell wall chitin and increased susceptibility to A glucosamine-6-phosphate synthase inhibitor. Cell. Surf. 8, 100076. doi: 10.1016/j.tcsw.2022.100076
So, Y.-S., Lee, D.-G., Idnurm, A., Ianiri, G., Bahn, Y.-S. (2019). The TOR pathway plays pleiotropic roles in growth and stress responses of the fungal pathogen Cryptococcus neoformans. Genetics 212, 1241–1258. doi: 10.1534/genetics.119.302191
So, Y.-S., Yang, D.-H., Jung, K.-W., Huh, W.-K., Bahn, Y.-S. (2017). Molecular characterization of adenylyl cyclase complex proteins using versatile protein-tagging plasmid systems in Cryptococcus neoformans. J. Microbiol. Biotechnol. 27, 357–364. doi: 10.4014/jmb.1609.09036
Song, M., Pang, L., Zhang, M., Qu, Y., Laster, K. V., Dong, Z. (2023). Cdc2-like kinases: structure, biological function, and therapeutic targets for diseases. Signal. Transduction Targeting Ther. 8, 148. doi: 10.1038/s41392-023-01409-4
Takayama, S., Fujii, M., Kurosawa, A., Adachi, N., Ayusawa, D. (2007). Overexpression of HAM1 gene detoxifies 5-bromodeoxyuridine in the yeast Saccharomyces cerevisiae. Curr. Genet. 52, 203–211. doi: 10.1007/s00294-007-0152-z
Talmadge, C. B., Finkernagel, S., Sumegi, J., Sciorra, L., Rabinow, L. (1998). Chromosomal mapping of three human lammer protein-kinase-encoding genes. Hum. Genet. 103, 523–524. doi: 10.1007/s004390050861
Teixeira, P. A., Penha, L. L., Mendonéa-Previato, L., Previato, J. O. (2014). Mannoprotein mp84 mediates the adhesion of Cryptococcus neoformans to epithelial lung cells. Front. Cell. Infect. Microbiol. 4, 106. doi: 10.3389/fcimb.2014.00106
Thompson, J. R., Douglas, C. M., Li, W., Jue, C. K., Pramanik, B., Yuan, X., et al. (1999). A glucan synthase fks1 homolog in Cryptococcus neoformans is single copy and encodes an essential function. J. Bacteriol. 181, 444–453. doi: 10.1128/JB.181.2.444-453.1999
Urban, J., Soulard, A., Huber, A., Lippman, S., Mukhopadhyay, D., Deloche, O., et al. (2007). Sch9 is A major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 26, 663–674. doi: 10.1016/j.molcel.2007.04.020
Veen, M., Stahl, U., Lang, C. (2003). Combined overexpression of genes of the ergosterol biosynthetic pathway leads to accumulation of sterols in Saccharomyces cerevisiae. FEMS Yeast Res. 4, 87–95. doi: 10.1016/S1567-1356(03)00126-0
Velagapudi, R., Hsueh, Y. P., Geunes-Boyer, S., Wright, J. R., Heitman, J. (2009). Spores as infectious propagules of Cryptococcus neoformans. Infect. Immun. 77, 4345–4355. doi: 10.1128/IAI.00542-09
Vestergaard, M., Bald, D., Ingmer, H. (2022). Targeting the atp synthase in bacterial and fungal pathogens: beyond Mycobacterium tuberculosis. J. Glob. Antimicrob. Resist. 29, 29–41. doi: 10.1016/j.jgar.2022.01.026
Vu, K., Weksler, B., Romero, I., Couraud, P.-O., Gelli, A. (2009). Immortalized human brain endothelial cell line hcmec/D3 as A model of the blood-brain barrier facilitates in vitro studies of central nervous system infection by Cryptococcus neoformans. Eukaryot. Cell. 8, 1803–1807. doi: 10.1128/EC.00240-09
Wang, Z. A., Li, L. X., Doering, T. L. (2018). Unraveling synthesis of the cryptococcal cell wall and capsule. Glycobiology 28, 719–730. doi: 10.1093/glycob/cwy030
Yang, M., Jarrett, S. G., Craven, R., Kaetzel, D. M. (2009). Ynk1, the yeast homolog of human metastasis suppressor NM23, is required for repair of UV radiation-and etoposide-induced DNA damage. Mutat. Res. 660, 74–78. doi: 10.1016/j.mrfmmm.2008.09.015
Young, L. Y., Hull, C. M., Heitman, J. (2003). Disruption of ergosterol biosynthesis confers resistance to amphotericin B in Candida lusitaniae. Antimicrob. Agents. Chemother. 47, 2717–2724. doi: 10.1128/AAC.47.9.2717-2724.2003
Yu, E.-Y., Lee, J.-H., Kang, W.-H., Park, Y.-H., Kim, L., Park, H.-M. (2013). Fission yeast lammer kinase Lkh1 regulates the cell cycle by phosphorylating the Cdk-inhibitor Rum1. Biochem. Biophys. Res. Commun. 432, 80–85. doi: 10.1016/j.bbrc.2013.01.082
Yun, B., Farkas, R., Lee, K., Rabinow, L. (1994). The DOA locus encodes A member of A new protein kinase family and is essential for eye and embryonic development in. Drosophila Melanogaster. Genes Dev. 8, 1160–1173. doi: 10.1101/gad.8.10.1160
Keywords: LAMMER kinase, Cryptococcus, stress response, virulence, antifungal drug resistance
Citation: Kwon S, Choi Y, Kim E-S, Lee K-T, Bahn Y-S and Jung K-W (2024) Pleiotropic roles of LAMMER kinase, Lkh1 in stress responses and virulence of Cryptococcus neoformans. Front. Cell. Infect. Microbiol. 14:1369301. doi: 10.3389/fcimb.2024.1369301
Received: 12 January 2024; Accepted: 27 March 2024;
Published: 07 May 2024.
Edited by:
Giuseppe Ianiri, University of Molise, ItalyReviewed by:
Larissa Fernandes Matos, University of Brasilia, BrazilCopyright © 2024 Kwon, Choi, Kim, Lee, Bahn and Jung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kwang-Woo Jung, a3dqdW5nQGthZXJpLnJlLmty
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.