- 1Office of Research Administration, Chiang Mai University, Chiang Mai, Thailand
- 2Center of Excellence in Microbial Diversity and Sustainable Utilization, Chiang Mai University, Chiang Mai, Thailand
- 3Guangdong Provincial Key Laboratory for Plant Epigenetics, Shenzhen Key Laboratory of Microbial Genetic Engineering, College of Life Science and Oceanography, Shenzhen University, Shenzhen, China
- 4Department of Education and Training Management, Tashkent International University of Education, Tashkent, Uzbekistan
- 5Central Asian Center for Development Studies, New Uzbekistan University, Tashkent, Uzbekistan
- 6Mycology Laboratory, Institute of Botany, Academy of Sciences of Republic of Uzbekistan, Tashkent, Uzbekistan
- 7Department of Ecology and Botany, Faculty of Natural Sciences, Andijan State University, Andijan, Uzbekistan
Most species of Dothiora are known from the dead parts of various host plants as saprobic fungi in terrestrial habitats occurring in tropical and temperate regions. In the present study, samples of Dothiora were collected from dead twigs and branches of Capparis spinosa, Rhaponticum repens, and an unknown angiosperm plant from the Tashkent and Jizzakh regions of Uzbekistan. Multi-gene phylogenetic analyses based on a combined ITS, LSU, SSU, TEF1, and TUB2 sequence data revealed their taxonomic positions within the Dothideaceae. Three new species of Dothiora, namely, Dothiora capparis, Dothiora rhapontici, and Dothiora uzbekistanica were proposed by molecular and morphological data. Likewise, the phylogenetic relationship and morphology of Dothiora are discussed. In addition, we provide a list of accepted Dothiora species, including host information, distribution, morphology descriptions, and availability of sequence data, to enhance the current knowledge of the diversity within Dothiora.
Introduction
Dothideales Lindau is an order in the class Dothideomycetes that comprises four families, including Dothideaceae, Neocelosporiaceae, Saccotheciaceae, and Zalariaceae (Hongsanan et al., 2020; Wijayawardene et al., 2022). Members of this order are saprobic and occasionally pathogenic to plants in terrestrial habitats and humans, including house dust (Thambugala et al., 2014; Crous et al., 2018a; Hongsanan et al., 2020). In addition, some species have been used as potential biocontrol for pest management (Zajc et al., 2019; Cignola et al., 2023). To date, 26 genera were accepted in Dothideales, while six genera were proposed in Dothideales genera incertae sedis (Wijayawardene et al., 2022). Dothideaceae Chevall. was introduced to accommodate the genus Dothidea, with D. gibberulosa as the type species (Chevallier, 1826; Fuckel, 1869). Currently, 14 genera are accepted in Dothideaceae, viz., Delphinella (Sacc.) Kuntze, Dictyodothis Theiss. & Syd., Dothidea Fr., Dothiora Fr., Endoconidioma Tsuneda, Endodothiora Petr., Kabatina R. Schneid. & Arx, Neocylindroseptoria Thambug. & K.D. Hyde, Neodothiora Crous, G.C. Adams & Winton, Phaeocryptopus Naumov, Plowrightia Sacc., Stylodothis Arx & E. Müll., Sydowia Bres., and Uleodothis Theiss. & Syd (Wijayawardene et al., 2022).
Dothiora was established by Fries (1894), typified by D. pyrenophora Berk. ex Sacc. The sexual morph of Dothiora is characterized by immersed to erumpent ascostromata, lacking pseudoparaphyses, eight or more spored, bitunicate asci, hyaline to yellow or pale brown, and one septate or muriform ascospores (Thambugala et al., 2014; Crous and Groenewald, 2017). Dothiora has dothichiza-like asexual morph via culture studies, which is characterized by pycnidial conidiomata, phialidic conidiogenous cells, hyaline, aseptate conidia, forming a hormonema-like synasexual morph (Thambugala et al., 2014; Crous and Groenewald, 2016; Crous and Groenewald, 2017). Earlier, Dothiora was treated in different families, such as Dothideaceae and Dothioraceae, by several authors (Barr, 1972; Barr, 1979; Barr, 1987; Froidevaux, 1972; Luttrell, 1973; von Arx and Müller, 1975; Hawksworth et al., 1995; Lumbsch and Huhndorf, 2010); however, Dothioraceae was synonymized under Dothideaceae based on its phylogenetic placement within Dothideales (Thambugala et al., 2014). Although 89 epithets are listed in Index Fungorum (accessed on 13 March 2024), most species have not been well studied since their introduction, and many species have been transferred to other genera such as Dothiorella, Myriangium, Protoscypha, and Saccothecium. Furthermore, only 28 Dothiora species have been sequenced and confirmed their phylogenetic placements (Hyde et al., 2020; Boonmee et al., 2021). Dothiora species are distributed worldwide on woody plants in terrestrial habitats as saprobes and pathogens causing leaf spots or possibly weak pathogens on stressed plant tissues (Crous and Groenewald, 2016; Crous and Groenewald, 2017; Hyde et al., 2020). Not only Dothiora infuscans Rodr.-Andr., Stchigel, Guarro & Cano is reported on the blackened wall of an industrial warehouse (Crous et al., 2018a), but also Dothiora sp. is recorded as an endophytic fungus that produces compounds with cytotoxic activity against cancer cell lines (Pérez-Bonilla et al., 2017).
Most of the Dothiora species are reported from Europe and North America (Hyde et al., 2020). Targeting underexplored regions such as Central Asia, including Uzbekistan, might be helpful for the discovery of new fungi (Gafforov, 2017; Cheek et al., 2020). Recent studies have led to the discovery of several new genera and species of ascomycetous microfungi in Uzbekistan (Gafforov and Rakhimov, 2017; Pem et al., 2018; Pem et al., 2019a; Pem et al., 2019b; Gafforov et al., 2019; Abdurazakov et al., 2021; Appadoo et al., 2021; Htet et al., 2021; Lestari et al., 2021; Aluthmuhandiram et al., 2022; Dong et al., 2023). However, Dothiora is still poorly known in Asia including Central Asian regions. The aim of the present study was to clarify the taxonomic position of Dothiora and to identify new taxa through multi-gene phylogeny and morphological examination. Fresh specimes collected from Uzbekistan were examined and their DNA sequence data were obtained for use in multi-gene phylogenetic analyses. Moreover, an updated list of Dothiora species worldwide is also provided.
Materials and methods
Sample collection and specimen examination
Specimens were collected from Capparis spinosa, Rhaponticum repens, and an unknown angiosperm plant from the Tashkent and Jizzakh regions of Uzbekistan. The collected specimens were brought to the laboratory in small plastic bags. Ascomata were sectioned by hand, examined, and captured under a Nikon SMZ800N stereomicroscope. The slides were prepared by mounting the materials in double-distilled water (ddH2O), lactophenol, and Indian ink stain. The micro-morphological characters were examined and captured using a Nikon DS-Ri2 camera connected with a Nikon ECLIPSE Ni (Tokyo, Japan) compound microscope. The measurement of structures was done by the Tarosoft® Image Framework program (v.0.9.0.7). Adobe Photoshop Version: 22.4.2 (Adobe Systems U.S.A.) was used to make the photographic plates. The specimens were deposited in the Herbarium of the Department of Biology (CMUB), Faculty of Science, Chiang Mai University, Thailand, and the Tashkent Mycological Herbarium (TASM) of the Institute of Botany, Uzbekistan Academy of Sciences, Uzbekistan.
DNA extraction, PCR amplification, and sequencing
To obtain pure cultures, single ascospore isolation was carried out following the methods of Senwanna et al. (2019) and Senanayake et al. (2020). However, no germinated ascospores were found on Petri dishes containing 2% malt extract agar (MEA; Gibco, Life Technologies Corporation, USA), 2% water agar (WA), and potato dextrose agar (PDA; BD Difco™, Becton, Dickinson and Company, USA) after incubation at 25°C to 30°C in the dark for 24–96 h. Fungal fruiting bodies, thus, were picked up and placed in a 1.5-mL sterilized tube. Genomic DNA was directly extracted, using E.Z.N.A.® Genomic DNA Isolation Kits (OMEGA Bio-Tek, Georgia) following the manufacturer’s protocol. Polymerase chain reaction (PCR) amplification was carried out using the primer pairs as follows: ITS5 and ITS4 (White et al., 1990) to amplify the partial gene regions of internal transcribed spacers (ITS); LR0R and LR5 (Vilgalys and Hester, 1990) to amplify the 28S large subunit (LSU); NS1 and NS4 (White et al., 1990) to amplify the 18S small subunit (SSU); and EF1-728F (Carbone and Kohn, 1999) and EF2 (O’Donnell et al., 1998) to amplify the protein coding region for the translation elongation factor 1-alpha gene (TEF1). The PCR mixture contained 6 µL of double-distilled water (ddH2O),10 μL of 2 × Quick TaqTM HS DyeMix (TOYOBO, Japan), 2 µL of genomic DNA, and 1 μL of each forward and reverse primer. The PCR thermal cycle programs for ITS, LSU, and SSU amplification were as follows: initial denaturing step of 95°C for 5 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 52°C for 45 s, elongation at 72°C for 1 min, and final extension at 72°C for 10 min. The PCR thermal cycle programs for TEF1 amplification were as follows: initial denaturing step of 94°C for 5 min, followed by 40 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 30 s, elongation at 72°C for 1 min, and final extension at 72°C for 10 min. PCR products were examined on 1% agarose electrophoresis gels under UV light. PCR products were purified using the PCR clean-up Gel extraction NucleoSpin® Gel and PCR Clean-up Kit (Macherey-Nagel, Germany) following the manufacturer’s protocol. PCR fragments were performed and sequenced at 1st BASE Company (Kembangan, Malaysia).
Phylogenetic analyses
The generated sequence data were assembled using SeqMan 5.00, and the consensus sequences were subjected to BLASTn searches of the NCBI nucleotide database (http://blast.ncbi.nlm.nih.gov/; accessed on 2 November 2023) to determine their most probable closely related taxa. The representative taxa used in the analyses were selected from GenBank based on the BLASTn searches and recently published data (Boonmee et al., 2021; Gao et al., 2021) (Table 1). Each gene alignment was carried out with MAFFT version 7 (Katoh et al., 2019; http://mafft.cbrc.jp/alignment/server/; accessed on 6 November 2023) and was improved manually where necessary. The phylogenetic tree was carried out using the maximum likelihood (ML). The single gene datasets were then combined using BioEdit v.7.0.9.1 (Hall, 1999). The final alignments of the combined ITS, LSU, SSU, TEF1, and beta-tubulin (TUB2) datasets were analyzed, and the phylogenetic trees were inferred based on ML and Bayesian inference (BI) analyses.
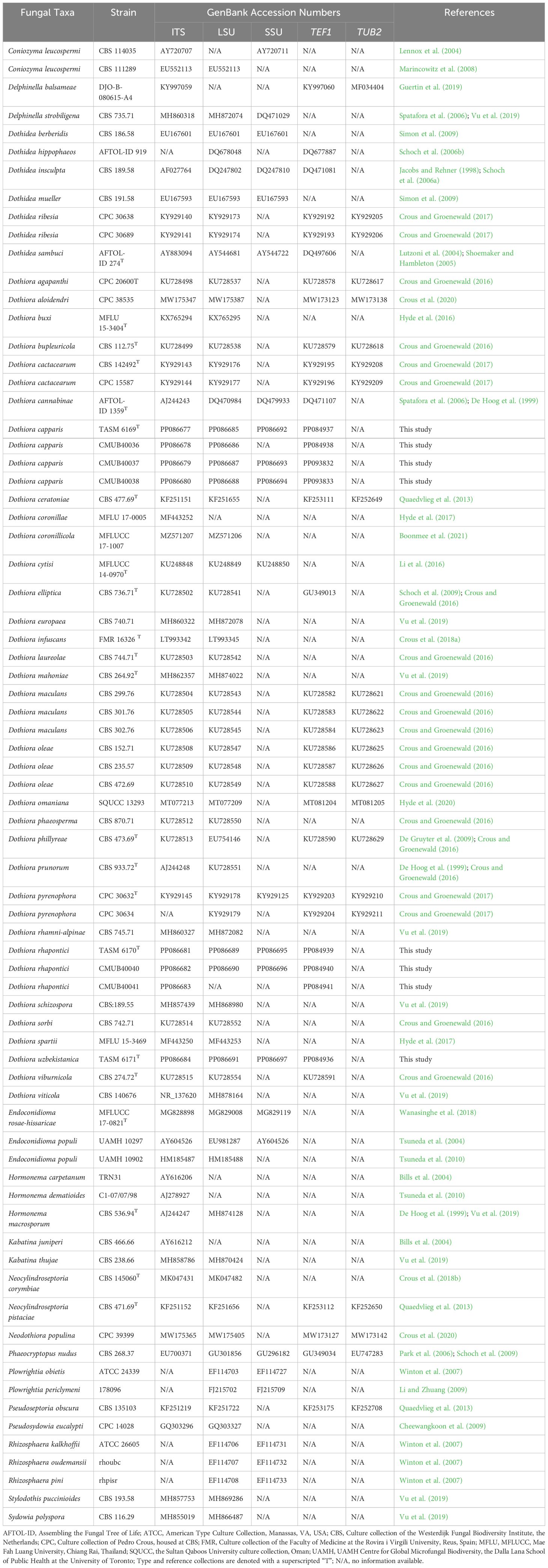
Table 1 Taxa names, strain numbers, and GenBank accession numbers of sequences used in the phylogenetic analyses of this study.
The ML tree was accomplished using the RAxML-HPC2 on XSEDE (v. 8.2.12) (Stamatakis, 2014) under the GTRGAMMA substitution model of nucleotide substitution with 1,000 bootstrap (BS) iterations. For BI analyses, the best-fit model of the sequence evolution of each locus was estimated using the Akaike information criterion (AIC) in MrModeltest v. 2.3 (Nylander, 2008) implemented in PAUP v. 4.0b10 (Swofford, 2002). The GTR+G+I substitution model was the best-fit model for all loci. The BI tree was executed with MrBayes v. 3.2.6 (Ronquist et al., 2012) to evaluate posterior probabilities (PP) (Rannala and Yang, 1996; Zhaxybayeva and Gogarten, 2002) by Markov Chain Monte Carlo sampling (BMCMC). Four simultaneous Markov chains were run for 5,000,000 generations, with the trees sampled every 100th generation resulting in 50,000 trees. The run was stopped when the standard deviation of split frequencies reached below 0.01. The first 12,500 trees were discarded as the burn-in phase of the analyses, while the remaining 37,500 trees were calculated for PP in the majority rule consensus tree. The resulting phylogenetic trees were drawn using FigTree v1.4.0 (Rambaut, 2016) and edited using Adobe Illustrator Version 25.2.3 and Adobe Photoshop Version 22.4.2 (Adobe Systems., U.S.A.). ML bootstrap values ≥50% and Bayesian PP ≥0.95 were placed above each node (Figure 1). The new nucleotide sequence data are deposited in GenBank (Table 1). The final alignment and tree were deposited in TreeBASE (http://www.treebase.org/) under the accession number S31239 and URL http://purl.org/phylo/treebase/phylows/study/TB2:S31239?x-access-code=12b27b6543ffd8301eed3a89eb09aed8&format=html.
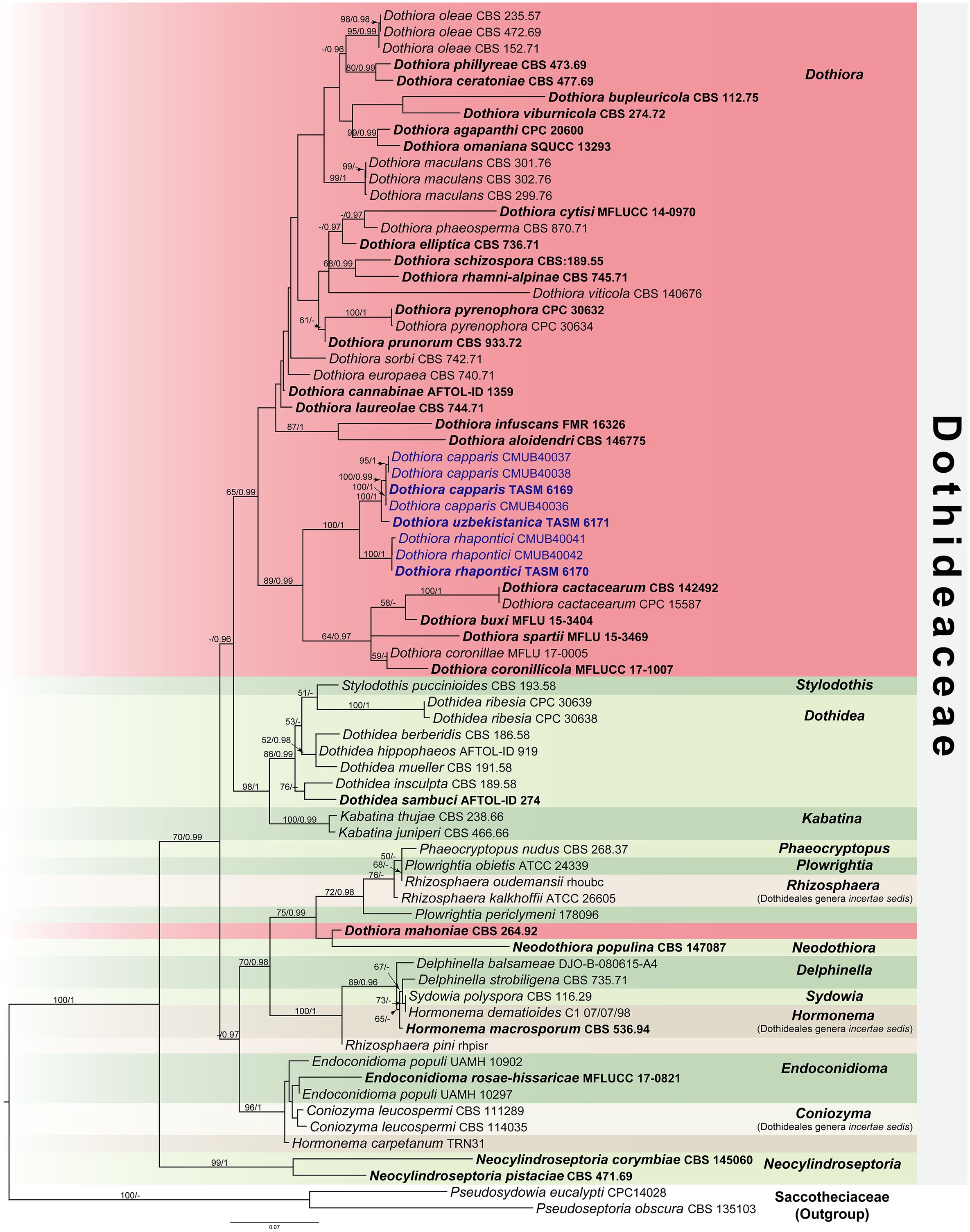
Figure 1 RAxML tree generated by maximum likelihood analysis of combined LSU, ITS, SSU, TEF1, and TUB2 sequence data representing Dothideaceae. Bootstrap support values for maximum likelihood (ML, left) ≥50% and Bayesian posterior probabilities (PP, right) ≥0.95 are indicated above the nodes. Hyphens (-) represent support values <50% ML/0.95 PP. The tree is rooted to Pseudoseptoria obscura (CBS 135103) and Pseudosydowia eucalypti (CPC14028). The ex-type strains are in bold, and the newly generated sequences in this study are in blue.
Results
Phylogenetic analyses
The combined dataset of LSU, ITS, SSU, TEF1, and TUB2 sequence data comprises 74 taxa, which represent strains from Dothideaceae and two outgroup taxa in Saccotheciaaceae, Pseudoseptoria obscura Quaedvl., Verkley & Crous (CBS 135103) and Pseudosydowia eucalypti (Verwoerd & du Plessis) Thambug. & K.D. Hyde (CPC14028) (Table 1). The combined sequence alignment consisting of 4,055 characters was analyzed by ML and BI. A best scoring RAxML tree with a final likelihood value of −19,627.809539 is presented in Figure 1. The matrix of the combined dataset had 1,384 distinct alignment patterns and 53.70% of undetermined characters or gaps. Estimated base frequencies were A = 0.253918, C = 0.235773, G = 0.266686, T = 0.243623; substitution rates were AC = 1.637334, AG = 2.504185, AT = 1.806129, CG = 1.203612, CT = 7.247662, GT = 1.000000; and gamma distribution shape parameter α = 0.504007. Bayesian posterior probabilities (PP) from MCMC were evaluated with the final average standard deviation of split frequencies = 0.009894. The Bayesian analysis resulted in a tree with similar topology and clades as the ML tree. Phylogenetic analyses of a combined LSU, ITS, SSU, TEF1, and TUB2 sequence data (Figure 1) show that three novel species of Dothiora in this study form a clade within the Dothideaceae with high support (100% ML and 1 PP) and sister to the clade containing Dothiora buxi Jayasiri, Camporesi & K.D. Hyde, D. cactacearum Crous, D. coronillae Dissan., Camporesi & K.D. Hyde, D. coronilicola Dissan., Camporesi & K.D. Hyde, and D. spartii Dissan., Camporesi & K.D. Hyde.
Taxonomic descriptions
Dothiora capparis Senwanna, N. Suwannar., & Gafforov, sp. nov. (Figure 2).

Figure 2 Dothiora capparis (TASM 6169, holotype). (A) Appearance of ascomata on host. (B, C) Fruiting bodies under a stereo microscope [superficial mycelia; blue arrows in panel (B)]. (D) Section through the ascoma. (E) Squash mount. (F) Peridium. (G–O) Asci. (P–T) Ascospores (mounted in ddH2O). (U, V) Ascospore with mucilaginous sheath (mounted in Indian ink). Scale bars: (B) 1000 µm, (C) 200 µm, (D, E) 50 µm, (F–O) 20 µm, and (P–V) 10 µm.
MycoBank number: MB851612
Etymology: Name reflects the host genus Capparis from which it was isolated.
Saprobic on dead twigs and branches of Capparis spinosa L. Sexual morph: Mycelium partly immersed on the substrate, simple to branched, septate, smooth-walled, pale brown to brown hyphae. Ascomata 90–115 µm diam. × 55–100 µm high, semi-immersed to erumpent through the epidermis, solitary or clustered, scattered, globose, dark brown to black, with single locules. Peridium (13–)17–28(–33) µm wide, composed of cells of textura angularis, an outer layer dark brown to black, thick-walled, an inner layer hyaline, thin-walled. Hamathecium lacking pseudoparaphyses. Asci (47.5–)54–82(–88) × (16–)21–28(–34) µm (x̅ = 66 × 24 µm, n = 40), 8-spored, bitunicate, fissitunicate, cylindro-clavate, pedicellate, apically rounded, with a small ocular chamber. Ascospores (15–)19–28 × (6–)9–12 µm (x̅ = 24 × 10 µm, n = 50), overlapping 1–2-seriate, fusoid to ovoid, one end narrower than other, hyaline, aseptate, smooth-walled with granular contents, surrounded by a distinct mucilaginous sheath, 4–10 µm wide at sides. Asexual morph: undetermined.
Material examined: UZBEKISTAN, Jizzakh Region, Forish District, Yangiqishloq village, dead twigs and branches of Capparis spinosa L. (Capparaceae), 05 May 2021, Y. Gafforov, M. Yarasheva, YG-F-2-1 (TASM 6169, holotype; CMUB40035, paratype); ibid, Capparis spinosa, 05 May 2021, Y. Gafforov, M. Yarasheva, YG-F-2-2 (CMUB40036); ibid, Nurota District, Nurota, dead twigs and branches of Capparis spinosa, 07 May 2021, Y. Gafforov, YG-F-5-1 (CMUB40037); ibid., dead twigs of Capparis spinosa, 07 May 2021, Y. Gafforov, YG-F-5-2 (CMUB40038).
Notes: In a BLASTn search of NCBI GenBank, the closest match of the LSU sequence of D. capparis (TASM 6169, holotype) was D. spartii (strain MFLU 15-3469; MF443250) with 99.52% similarity; the closest match of the ITS sequence with 98.36% similarity was D. coronillae (MFLU 17-0005; NR157481); the closest matches of the SSU sequence with 100% similarity were D. pyrenophora (CPC 30632; KY929125), D. prunorum (C. Dennis & Buhagiar) Crous (CBS 933.72; EU707926), and D. cannabinae Froid. (CBS 737.71; NG062696), respectively; while the closest matches of the TEF1 sequence with 97.77% similarity were D. oleae (DC.) Crous (SAG 68856-SF; KY613610, CBS 472.69; KU728588, CBS 235.57; KU728587, and CBS 152.71; KU728586) and D. ceratoniae (Quaedvl., Verkley & Crous) Crous (CBS 441.75; KU728581). Based on the multi-gene phylogenetic analyses, D. capparis (TASM6169, CMUB40036, CMUB40037, and CMUB40038) form a distinct lineage with 100% ML and 0.99 PP statistical support (Figure 1) and is closely related to D. uzbekistanica. Albeit the phylogenetic relationships of those four strains clustering in two different clades, a comparison of ITS and TEF1 nucleotides shows that strains TASM 6169 and CMUB40036 differ from strains CMUB40037 and CMUB40038 in 1/589 bp (0.17%) and 2/479 bp (0.42%), respectively. Moreover, D. capparis differs from D. uzbekistanica in having ascospores with a distinct mucilaginous sheath, while the latter lacks this character. A nucleotide comparison of the TEF1 gene indicated that all strains of D. capparis differ from D. uzbekistanica by 20/414 bp (4.83%). According to Jeewon and Hyde (2016), a nucleotide comparison of reliable genes must reveal a difference of more than 1.5% to confirm the existence of a different species. Therefore, D. capparis and D. uzbekistanica are different species.
Dothiora rhapontici Senwanna, N. Suwannar., & Gafforov, sp. nov. (Figure 3).

Figure 3 Dothiora rhapontici (TASM 6170, holotype). (A) Appearance of ascomata on host. (B–D) Fruiting bodies under a stereo microscope. (E) Section through the ascoma. (F) Peridium. (G–J) Asci (mounted in ddH2O). (K) Asci (mounted in lactophenol). (L–T) Ascospores (mounted in ddH2O). (U) Ascospores with mucilaginous sheath (mounted in Indian ink). Scale bars: (B) 1,000 µm, (C, D) 100 µm, (E) 50 µm, (F–K) 20 µm, and (L–U) 10 µm.
MycoBank number: MB851613
Etymology: The name reflects the host genus Rhaponticum from where it was isolated.
Saprobic on dead twigs of Rhaponticum repens L. Sexual morph: Ascomata 80–155 µm diam. × 60–120 µm high, semi-immersed to erumpent through the epidermis, solitary or clustered, scattered, globose to subglobose, dark brown to black, with single locules. Peridium 12–21(–23) µm wide, composed of cells of textura angularis, an outer layer dark brown to black, thick-walled, an inner layer hyaline, thin-walled. Hamathecium lacking pseudoparaphyses. Asci 63–80(–113) × (23–)29–39 µm (x̅ = 73 × 34 µm, n = 20), polysporous (24 or more spores), bitunicate, cylindro-clavate, pedicellate, apically rounded. Ascospores (14–)16–26(–28) × 6–7(–8) µm (x̅ = 19 × 7 µm, n = 40), multi-seriate, fusoid to ovoid, one end narrower than other, hyaline, aseptate, with a central concave depression, smooth-walled with granular contents, surrounded with a mucilaginous sheath, 2–4.5 µm wide at sides.
Material examined: UZBEKISTAN, Tashkent Region, Ugam-Chatkal National Park, Chimyon, Western Tien-Shan Mountains, dead twigs of Rhaponticum repens (L.) Hidalgo (Asteraceae), 21 July 2019, Y. Gafforov, M. Yarasheva, YG-S-22-4 (TASM 6170, holotype; CMUB40039, paratype); Jizzakh Region, Zaamin District, Zaamin National State Park, dead branches of Capparis spinosa L. (Capparaceae), 27 August 2020, Y. Gafforov, A. Abdurazakov, YG-ZMB-40-1 (CMUB40040); ibid., dead branches of Capparis spinosa, 27 August 2020, Y. Gafforov, A. Abdurazakov, YG-ZMB-40-2 (CMUB 40041).
Notes: In a BLASTn search of NCBI GenBank, the closest match of the LSU sequence of D. rhapontici (TASM6170, holotype) was D. cannabinae (AFTOL-ID 1359; MF443250) with 99.66% similarity; the closest match of the ITS sequence with 98.16% similarity was D. coronillae (MFLU 17-0005; NR157481); the closest matches of the SSU sequence with 100% similarity were D. pyrenophora (CPC 30632; KY929125), D. prunorum (CBS 933.72; EU707926), and D. cannabinae (CBS 737.71; NG062696), respectively; while the closest matches of the TEF1 sequence with 96.09% similarity was D. phillyreae (CBS 473.69; KU728590). The phylogenetic analysis reveals that D. rhapontici formed a distinct sister clade with D. capparis and D. uzbekistanica, which is statistically supported (100% ML and 1 PP) (Figure 1). Dothiora rhapontici shares similar morphological features of ascospores with related species; however, D. rhapontici can be distinguished from those latter species by having polysporous asci and having longer and wider ascospores.
Dothiora uzbekistanica Senwanna, N. Suwannar., & Gafforov, sp. nov. (Figure 4).
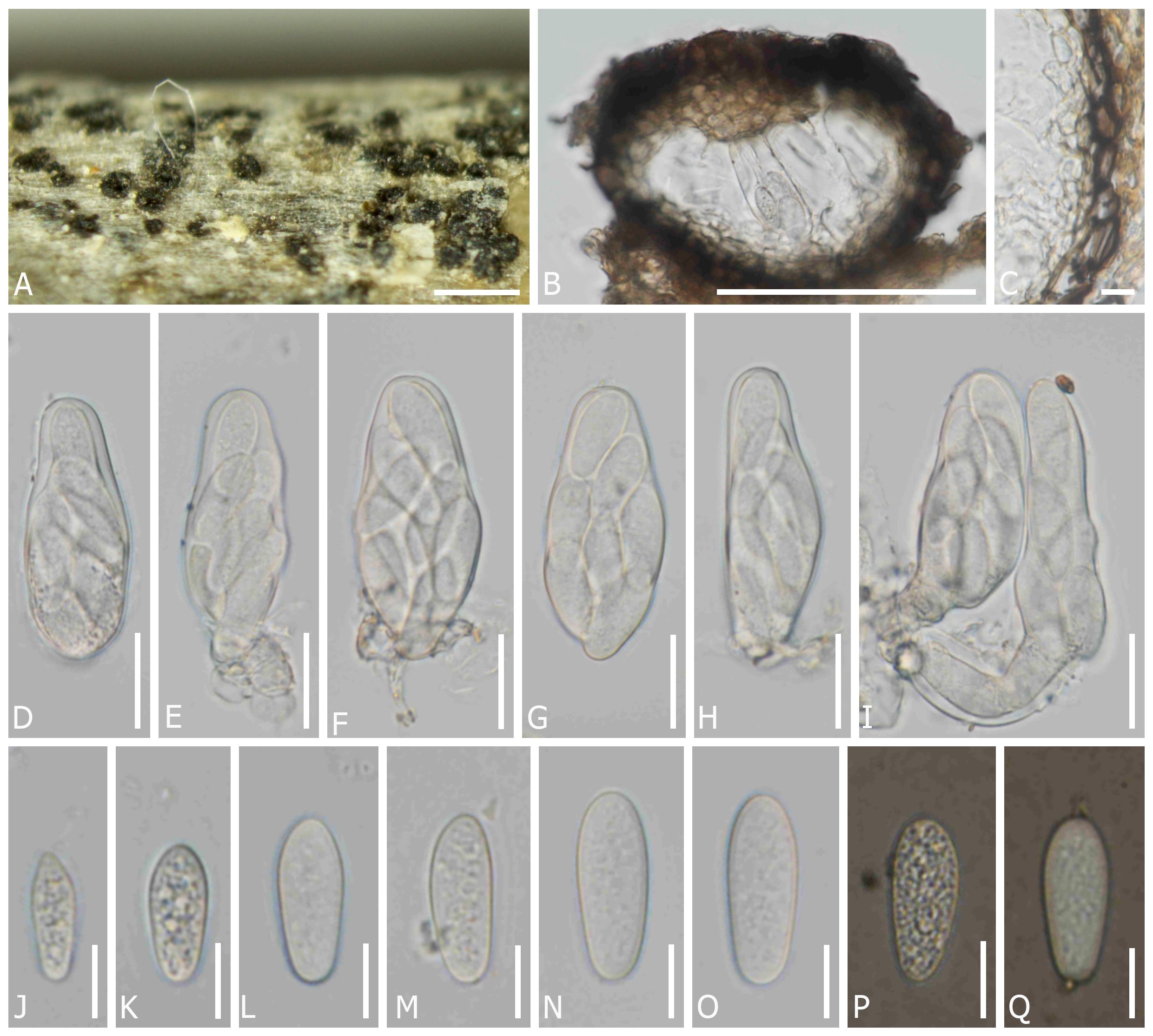
Figure 4 Dothiora uzbekistanica (TASM 6171, holotype). (A) Fruiting bodies under a stereo microscope. (B) Section through the ascoma. (C) Peridium. (D–I) Asci. (J–O) Ascospores (mounted in ddH2O). (P, Q) Ascospores (mounted in Indian ink). Scale bars: (A) 200 µm, (B) 100 µm, (C, J–Q) 10 µm, and (D–I) 20 µm.
MycoBank number: MB851614
Etymology: The name refers to the country Uzbekistan, where it was collected.
Saprobic on dead twigs of unknown angiosperm perennial plants. Sexual morph: Ascomata 95–155 µm diam. × 80–135 µm high, semi-immersed to erumpent through the epidermis, solitary or clustered, globose to subglobose, black, with single locules. Peridium (10–)15–25(–30) µm wide, composed of cells of textura angularis, an outer layer dark brown to black, thick-walled, an inner layer hyaline, thin-walled. Hamathecium lacking pseudoparaphyses. Asci (50–)54–66(–70) × (19–)21–27 µm (x̅ = 58 × 22 µm, n = 20), 8-spored, bitunicate, fissitunicate, cylindro-clavate, pedicellate, apically rounded. Ascospore (16–)19–26 × (5–)9–11 µm (x̅ = 22 × 9.5 µm, n = 45), overlapping one to two-seriate, fusoid to ovoid, one end narrower than other, hyaline, aseptate, smooth-walled with granular contents, lacking a mucilaginous sheath. Asexual morph: undetermined.
Material examined: UZBEKISTAN, Jizzakh Region, Forish District, Yangiqishloq village, dead twigs of unknown angiosperm plants, 05 May 2021, Y. Gafforov, M. Yarasheva, YG-F-4-2 (TASM 6171, holotype; CMUB40042, paratype).
Notes: In a BLASTn search of NCBI GenBank, the closest matches of the LSU sequence of D. uzbekistanica (TASM 6171, holotype) is D. spartii (strain MFLU 15-3469; MF443250) with 99.52% similarity, the closest matches of the ITS sequence with 98.16% similarity, was D. coronillae (MFLU 17-0005; NR157481), the closest matches of the SSU sequence with 100% similarity was D. pyrenophora (CPC 30632; KY929125), D. prunorum (CBS 933.72; EU707926) and D. cannabinae (CBS 737.71; NG062696), respectively, while the closest matches of the TEF1 sequence with 97.21% similarity was D. oleae (SAG 68856-SF; KY613610, CBS 472.69; KU728588, CBS 235.57; KU728587, and CBS 152.71; KU728586) and D. ceratoniae (CBS 441.75; KU728581). In the phylogenetic analysis, D. uzbekistanica forms a distinct lineage basal to D. capparis with 100% ML and 1 PP bootstrap support (Figure 1). Moreover, the different characteristics of the microscopic features and the nucleotide comparison data of D. uzbekistanica differ from D. capparis have been mentioned above.
Discussion
Historically, Dothiora has relied on morphological studies, and only a few sequences of species are available in GenBank. In this study, three novel species of Dothiora are introduced in the family Dothideaceae from the Central Asian region based on their morphological distinctiveness and phylogenetic analyses. Although a living culture from an isolated ascospore could not be obtained, the fungal DNA was extracted directly from the ascomata. The connection between sexual and asexual morphs is likewise unknown. The individual phylogenetic analyses of ITS or LSU separated Dothiora species from other genera in Dothideaceae, but their placement was otherwise unresolved (Crous and Groenewald, 2017; Crous et al., 2018a; Crous et al., 2018b). Therefore, the combination of LSU, ITS, and SSU sequence data was previously used to clarify the relationships among the species in Dothideaeceae, although there is no strong statistical support (Thambugala et al., 2014; Hyde et al., 2017; Crous et al., 2020, Crous et al., 2022; Hongsanan et al., 2020; Boonmee et al., 2021). Gao et al. (2021) recommended using a combination of the nuclear ribosomal region (ITS, LSU, and SSU) and the protein-coding gene regions (TEF1 and TUB2) to clarify the relationships of Dothideaceae. Our attempts to obtain TUB2 sequence data for our new strains were unsuccessful; however, the data from the combined sequence analyses of the ITS, LSU, SSU, and TEF1 loci are not well-resolved for most Dothiora species. Thus, a phylogenetic analysis based on a combination of five loci was generated for a better phylogenetic relationship within the family and genus. Our multigene phylogeny (Figure 1) revealed that the generic placement within Dothideaceae, comprising 14 genera and three Dothideales genera incertae sedis, viz., Coniozyma, Hormonema, and Rhizosphaera, was similar to those of Hongsanan et al. (2020) and Gao et al. (2021). Most Dothiora taxa clustered together in its own clade with 65% ML, 1 PP statistical support, excepting D. mahoniae (A.W. Ramaley) Crous (strain CBS 264.92) (Figure 1).
A checklist of 69 accepted Dothiora species, including details of each species based on recorded from Index Fungorum (2024), MycoBank (2024), and published articles, is provided in Table 2. Dothiora have a cosmopolitan distribution and are mainly saprobic, found in decaying wood and plant litter in terrestrial environments. Members of Dothiora have been recorded on 36 host plant families, viz., Aceraceae, Adoxaceae, Amaryllidaceae, Apiaceae, Apocynaceae, Araliaceae, Asphodelaceae, Asteraceae, Berberidaceae, Buxaceae, Cactaceae, Capparaceae, Caprifoliaceae, Celastraceae, Convolvulaceae, Cupressaceae, Elaeagnaceae, Ericaceae, Fabaceae, Grossulariaceae, Lythraceae, Magnoliaceae, Oleaceae, Pinaceae, Plantaginaceae, Podocarpaceae, Rhamnaceae, Rosaceae, Rubiaceae, Salicaceae, Staphyleaceae, Symplococeae, Tamaricaceae, Taxaceae, Thymelaeaceae, and Vitaceae. This study is the first record of Dothiora on Asteraceae and Capparaceae. The genus has mainly been reported in the USA (22 species), Italy (10 species), Canada (8 species), and Switzerland (7 species). The identification of Dothiora species was initially based on its sexual morph. A total of 37 species of Dothiora are known only for their sexual morph, 15 species are known only for their asexual morph, and another eight species have no information available. There are only eight Dothiora species that have asexual-sexual morph connections, viz., D. buxi, D. cytisi (Wanas., Camporesi, E.B.G. Jones & K.D. Hyde) Crous, D. lonicerae Fuckel, D. pyrenophora, D. schizospora Luttr., D. sorbi (Wahlenb.) Fuckel, D. sphaeroides (Pers.) Fr., and D. taxicola (Peck) M.E. Barr. To date, 31 species have been reported based on molecular data, and only in four species have been proven the connectivity of sexual and asexual morphs through sequence data and culture studies.

Table 2 Morphology, host information, locality, sequence data, and related references Dothiora reported worldwide based on the record of Species Fungorum and MycoBank database 2024.
Although the sexual morph of Dothiora species has been delimited according to morphological criteria by having one or more septate or muriform ascospores (Thambugala et al., 2014; Crous and Groenewald, 2017), many species with aseptate ascospores were also classified as Dothiora based on morphological characteristics and phylogenetic analyses (Hyde et al., 2016; Hyde et al., 2017; Boonmee et al., 2021). Most Dothiora species with hyaline to pale brown, one or more septate or muriform ascospores, form a separate clade without statistical support (Figure 1), while three new taxa, D. capparis, D. rhapontici, and D. uzbekistanica, cluster with D. buxi, D. coronillae, D. coronillicola, and D. spartii, which also have hyaline, aseptate, and fusoid to ovoid ascospores with 89% ML and 0.99 PP statistical support in different clades, agreeing with previous studies (Boonmee et al., 2021). Dothiora rhapontici and D. buxi differ from related species in having polysporous asci, while other species have octosporous asci. These species can be distinguished from each other based on their asci and ascospores sizes; moreover, D. capparis can be distinguished from D. coronillae, D. coronillicola, D. spartii, and D. uzbekistanica by the presence of a mucilaginous sheath surrounding the ascospores. Dothiora cactacearum also clustered among these species; however, its morphological characteristics cannot be compared with others, as it is known only by its asexual morph. Dothiora buxi is phylogenetically close to D. cactacearum, and both species have similar asexual morphs to the generic, as described by Thambugala et al. (2014) and Crous and Groenewald (2017). The morphology of ascospores of Dothiora buxi, D. capparis, D. coronillae, D. coronillicola, D. rhapontici, D. spartii, and D. uzbekistanica, was clearly distinct from that of most Dothiora; however, only this evidence did not have enough support to accommodate a distinct lineage in Dothideaceae. Furthermore, an asexual morph of these species has not been cultured or reported to verify its morphological features, except for D. buxi. Due to the fact that most of the available sequences of Dothiora are only LSU and/or ITS, their taxonomic position remains uncertain. The taxonomic classification of Dothiora species is still incomplete; further investigations of freshly collected specimens in different regions and sequence data are needed to better understand their natural classification.
Crous et al. (2018b) transferred Kabatina mahoniae A.W. Ramaley to Dothiora as D. mahoniae. According to the multigene analyses herein (Figure 1), Dothiora mahoniae (strain CBS 264.92) clustered with Neodothiora populina Crous, G.C. Adams & Winton (strain CBS 147087) with no statistical support for this relationship and separated from Dothiora species, which is consistent with Crous et al. (2020). Based on a comparison of morphology, Dothiora mahoniae fits well with the generic concepts of Kabatina rather than Dothiora and Neodothiora (Ramaley, 1992; Thambugala et al., 2014; Crous and Groenewald, 2017; Crous et al., 2020). In addition, only LSU and ITS are available for D. mahoniae in GenBank. Thus, the species is retained until more evidence of fresh collections with DNA sequence data is available to resolve its phylogenetic placement within the family.
In this study, three new species of hyaline-spored Dothiora (D. capparis, D. rhapontici, and D. uzbekistanica) are described and illustrated. It is interesting to note that Dothiora have morphological variability in their spores. Thus, it is inadequate to determine Dothiora spp. based solely on morphological data. It can be seen that phylogenetic analyses are necessary to confirm morphology-based identifications and detect species new to science. Many Dothiora on the list (Table 2) have not been verified yet based on the molecular data; however, these species have characteristics that match the generic description. Additionally, the classification of several species remains unclear due to the variability in some morphological characters, a lack of molecular information regarding protein-coding genes, and no sexual–asexual links. Hence, these species are not excluded from Dothiora until increased taxon samplings and sequence data are available. Further sampling is necessary to improve our knowledge of the diversity, ecology, and impacts of hyaline-spored Dothiora species on flowering plants in arid and semi-arid habitats.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, ITS: PP086677, PP086678, PP086679, PP086680, PP086681, PP086682, PP086683 and PP086684; LSU: PP086685, PP086686, PP086687, PP086688, PP086689, PP086690 and PP086691; SSU: PP086692, PP086693, PP086694, PP086695, PP086696 and PP086697; TEF1: PP084936, PP084937, PP084938, PP084939, PP084940, PP084941, PP093832 and PP093833.
Author contributions
CS: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. SH: Conceptualization, Formal analysis, Project administration, Supervision, Visualization, Writing – review & editing. SK: Formal analysis, Investigation, Visualization, Writing – review & editing. JK: Data curation, Investigation, Software, Supervision, Visualization, Writing – original draft, Writing – review & editing. MY: Investigation, Resources, Visualization, Writing – review & editing. YG: Conceptualization, Data curation, Investigation, Project administration, Resources, Supervision, Visualization, Writing – review & editing. AA: Investigation, Resources, Visualization, Writing – review & editing. NS: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Chiang Mai University, Thailand, State Scientific and Technical Program of the Institute of Botany of Uzbekistan Academy of Sciences (2021–2025) and Agency for Innovative Development of the Republic of Uzbekistan (Project no. AL 2021090820) research support. Chanokned Senwanna expresses appreciation to the CMU Proactive Researcher, Chiang Mai University (Grant number 911/2566).
Acknowledgments
Shaun Pennycook is thanked for the nomenclatural review. Milan C. Samarakoon is thanked for his valuable suggestions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author SH declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdurazakov, A. A., Bulgakov, T. S., Kholmuradova, T. N., Gafforov, Y. S. (2021). Powdery mildew fungi (Erysiphaceae) of the Fergana Valley (within Uzbekistan): A first annotated checklist. Nov. Sist. Nizshikh Rastenii 55, 55–78. doi: 10.31111/nsnr/2021.55.1.55
Ahn, Y. M., Shearer, C. A. (1998). Reexamination of taxa in Leptosphaeria originally described on host species in Ranunculaceae, Papaveraceae, and Magnoliaceae. Canad. J. Bot. 76, 258–280. doi: 10.1139/cjb-76-2-258
Aluthmuhandiram, J. V. S., Wanasinghe, D. N., Chethana, K. W. T., Gafforov, Y., Saichana, N., Li, X. H., et al. (2022). Lophiostomataceae (Dothideomycetes): Introducing Lophiostoma khanzadakirgizbaeva sp. nov. and Paucispora xishanensis sp. nov. Phytotaxa 559, 247–262. doi: 10.11646/phytotaxa.559.3.3
Appadoo, M. A., Wanasinghe, D. N., Gafforov, Y., Chetana, K. T., Abdurazakov, A., Hyde, K. D., et al. (2021). Morphological and phylogenetic insights reveal Cucurbitaria berberidicola (Cucurbitariaceae, Pleosporales) as a new species from Uzbekistan. Phytotaxa 518, 1–13. doi: 10.11646/phytotaxa.518.1.1
Barr, M. E. (1972). Preliminary studies on the Dothideales in temperate North America. Contr. Univ. Michigan Herb. 9, 523–638.
Barr, M. E. (1981). The genus Curreya: an example of taxonomic confusion in the Ascomycetes. Mycologia 73, 599–609. doi: 10.2307/3759486
Barr, M. E. (1987). Prodomus to class Loculoascomycetes (Amherst, Massachusetts, University of Massachusetts, USA).
Bills, G. F., Collado, J., Ruibal, C., Peláez, F., Platas, G. (2004). Hormonema carpetanum sp. nov., a new lineage of dothideaceous black yeasts from Spain. Stud. Mycol. 50, 149–157.
Boonmee, S., Wanasinghe, D. N., Calabon, M. S., Huanraluek, N., Chandrasiri, S. K. U., Jones, G. E. B., et al. (2021). Fungal diversity notes 1387–1511: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 111, 1–335. doi: 10.1007/s13225-021-00489-3
Carbone, I., Kohn, L. M. (1999). A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91, 553–556. doi: 10.1080/00275514.1999.12061051
Cheek, M., Lughadha, E. N., Kirk, P. M., Lindon, H. L., Carretero, J. M., Looney, B. P., et al. (2020). New scientific discoveries: Plants and fungi. Plants People Planet 2, 371–388. doi: 10.1002/ppp3.10148
Cheewangkoon, R., Groenewald, J. Z., Summerell, B. A., Hyde, K. D., To-Anun, C., Crous, P. W. (2009). Myrtaceae, a cache of fungal biodiversity. Persoonia 23, 55–85. doi: 10.3767/003158509X474752
Cignola, R., Boato, A., Sadallah, A., Firrao, G., Di Francesco, A. (2023). Molecular characterization of Aureobasidium spp. strains isolated during the cold season. A preliminary efficacy evaluation as novel potential biocontrol agents against postharvest pathogens. Eur. J. Plant Pathol. 167, 221–233. doi: 10.1007/s10658-023-02696-x
Crous, P. W., Begoude, B. A. D., Boers, J., Braun, U., Declercq, B., Dijksterhuis, J., et al. (2022). New and interesting fungi. 5. Fungal Syst. Evol. 10, 19–90. doi: 10.3114/fuse.2022.10.02
Crous, P. W., Cowan, D. A., Maggs-Kölling, G., Yilmaz, N., Larsson, E., Angelini, C., et al. (2020). Fungal Planet description sheets: 1112–1181. Persoonia 45, 251–409. doi: 10.3767/persoonia.2020.45.10
Crous, P. W., Groenewald, J. Z. (2016). They seldom occur alone. Fungal Biol. 120, 1392–1415. doi: 10.1016/j.funbio.2016.05.009
Crous, P. W., Groenewald, J. Z. (2017). The genera of fungi – G 4: Camarosporium and Dothiora. IMA Fungus 8, 131–152. doi: 10.5598/imafungus.2017.08.01.10
Crous, P. W., Luangsa-ard, J. J., Wingfield, M. J., Carnegie, A. J., Hernández-Restrepo, M., Lombard, L., et al. (2018b). Fungal Planet description sheets: 785–867. Persoonia 41, 238–417. doi: 10.3767/persoonia.2018.41.12
Crous, P. W., Wingfield, M. J., Burgess, T. I., Hardy, G. E., St., J., Gené, J., et al. (2018a). Fungal Planet description sheets: 716–784. Persoonia 40, 240–393. doi: 10.3767/persoonia.2018.40.00
Crous, P. W., Wingfield, M. J., Guarro, J., Hernández-Restrepo, M., Sutton, D. A., Acharya, K., et al. (2015). Fungal Planet description sheets: 320-370. Persoonia. 34, 167–266. doi: 10.3767/003158515X688433
De Gruyter, J., Aveskamp, M. M., Woudenberg, J. H., Verkley, G. J., Groenewald, J. Z., Crous, P. W. (2009). Molecular phylogeny of Phoma and allied anamorph genera: towards a reclassification of the Phoma complex. Mycol. Res. 113, 408–419. doi: 10.1016/j.mycres.2009.01.002
De Hoog, G. D., Zalar, P., Urzi, C., De Leo, F., Yurlova, N. A., Sterflinger, K. (1999). Relationships of dothideaceous black yeasts and meristematic fungi based on 5.8S and ITS2 rDNA sequence comparison. Stud. Mycol. 43, 31–37.
Dennis, C., Buhagiar, R. W. M. (1973). Comparative study of Aureobasidium pullulans, A. prunorum sp. nov. and Trichosporon pullulans. Trans. Br. Mycol. Soc 60, 567–575. doi: 10.1016/S0007-1536(73)80041-5
Dennis, R. W. G., Reid, D. A., Spooner, B. M. (1977). The fungi of the azores. Kew Bull. 32, 85–136. doi: 10.2307/4117263
Dong, W., Hyde, K. D., Jeewon, R., Liao, C. F., Zhao, H. J., Kularathnage, N. D., et al. (2023). Mycosphere notes 449–468: saprobic and endophytic fungi in China, Thailand, and Uzbekistan. Mycosphere 14, 2208–2262. doi: 10.5943/mycosphere/14/1/26
Duan, J. X., Wu, W. P., Liu, X. Z. (2007). Reinstatement of Coleonaema for Coleophoma oleae and notes on Coleophoma. Fungal Divers. 26, 187–204.
Froidevaux, L. (1972). Contribution à l'étude des Dothioracées (Ascomycètes) (Doctoral dissertation, ETH Zurich). Nova Hedwigia 23, 679–734. doi: 10.3929/ethz-a-000091271
Fuckel, L. (1870). Symbolae mycologicae. Beiträge zur Kenntniss der Rheinischen Pilze. Jahrbücher des Nassauischen Vereins für Naturkunde: Wiesbaden (Germany). 23–24, 1–459.
Fuckel, L. (1871). Symbolae mycologicae. Beiträge zur Kenntniss der rheinischen Pilze. Erster Nachtrag, Vol. 25–26. 287–346.
Fuckel, L. (1875). Symbolae mycologicae. Beiträge zur Kenntniss der rheinischen Pilze, Vol. 29–30. 1–39.
Gafforov, Y. S. (2017). A preliminary checklist of ascomycetous microfungi from Southern Uzbekistan. Mycosphere 8, 660–696. doi: 10.5943/mycosphere/8/4/12
Gafforov, Y., Rakhimov, D. (2017). Diplodia and Dothiorella species (Botryosphaeriaceae, ascomycota) from Uzbekistan. J. Bot. Res. Inst. 11, 455–467. doi: 10.17348/jbrit.v11.i2.1083
Gafforov, Y., Phookamsak, R., Jiang, H. B., Wanasinghed, D. N., Juliev, M. (2019). Ophiobolus hydei sp. nov. (Phaeosphaeriaceae, Ascomycota) from Cirsium and Phlomoides in Uzbekistan. Botany 97, 671–680. doi: 10.1139/cjb-2019-0118
Gao, Y., Monkai, J., Gentekaki, E., Ren, G. C., Wanasinghe, D. N., Xu, J. C., et al. (2021). Dothidea kunmingensis, a novel asexual species of Dothideaceae on Jasminum nudiflorum (winter jasmine) from Southwestern China. Phytotaxa 529, 43–56. doi: 10.11646/phytotaxa.529.1.3
Guertin, J. F., Zitouni, M., Tanguay, P., Hogue, R., Beaulieu, C. (2019). Detection of Delphinella shoot blight in plantations of balsam fir (Abies balsamea) Christmas trees in Quebec, Canada. Can. J. Plant Pathol. 41, 87–97. doi: 10.1080/07060661.2018.1547791
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hawksworth, D. L., Kirk, P. M., Sutton, B. C., Pegler, D. N. (1995). Ainsworth & Bisby’s Dictionary of the Fungi. 8 Edn (Wallingford, UK: CAB International).
Hongsanan, S., Hyde, K. D., Phookamsak, R., Wanasinghe, D. N., McKenzie, E. H. C., Sarma, V. V., et al. (2020). Refined families of dothideomycetes: dothideomycetidae and pleosporomycetidae. Mycosphere 11, 1553–2107. doi: 10.5943/mycosphere/11/1/13
Htet, Z. H., Mapook, A., Gafforov, Y., Chethana, K. T., Lumyong, S., Hyde, K. D. (2021). Molecular phylogeny and diversity of Laburnicola (Didymosphaeriaceae): a new species from Uzbekistan. Phytotaxa 527, 177–190. doi: 10.11646/phytotaxa.527.3.2
Hyde, K. D., Hongsanan, S., Jeewon, R., Bhat, D. J., McKenzie, E. H. C., Jones, E. B. G., et al. (2016). Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 80, 1–270. doi: 10.1007/s13225-016-0373-x
Hyde, K. D., Jeewon, R., Chen, Y. J., Bhunjun, C. S., Calabon, M. S., Jiang, H. B., et al. (2020). The numbers of fungi: is the descriptive curve flattening? Fungal Divers. 103, 219–271. doi: 10.1007/s13225-020-00458-2
Hyde, K. D., Norphanphoun, C., Abreu, V. P., Bazzicalupo, A., Chethana, K. W. T., Clericuzio, M., et al. (2017). Fungal diversity notes 603–708: taxonomic and phylogenetic notes on genera and species. Fungal Divers. 87, 1–235. doi: 10.1007/s13225-017-0391-3
Index Fungorum. (2024). Available at: http://www.indexfungorum.org (Accessed 13 March 2024).
Jacobs, K. A., Rehner, S. A. (1998). Comparison of cultural and morphological characters and ITS sequences in anamorphs of Botryosphaeria and related taxa. Mycologia 90, 601–610. doi: 10.1080/00275514.1998.12026949
Jeewon, R., Hyde, K. D. (2016). Establishing species boundaries and new taxa among fungi: Recommendations to resolve taxonomic ambiguities. Mycosphere 7, 1669–1677. doi: 10.5943/mycosphere/7/11/4
Katoh, K., Rozewicki, J., Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Lennox, C. L., Serdani, M., Groenewald, J. Z., Crous, P. W. (2004). Prosopidicola mexicana gen. et sp. nov., causing a new pod disease of Prosopis species. Stud. Mycol. 50, 187–194.
Lestari, A. S., Wanasinghe, D. N., Gafforov, Y., Tennakoon, N. S., Chethana, K. W. T., Aburazakov, A., et al. (2021). Taxonomy and phylogenetic appraisal of Leptosphaeria chatkalica sp. nov. (Leptosphaeriaceae, Pleosporales) from Uzbekistan. Phytotaxa 520, 155–168. doi: 10.11646/phytotaxa.520.2.3
Li, G. J., Hyde, K. D., Zhao, R. N., Hongsanan, S., Abdel-Aziz, F. A., AbdelWahab, M. A., et al. (2016). Fungal Diversity notes 253–366: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 78, 1–237. doi: 10.1007/s13225-016-0366-9
Li, W. Y., Zhuang, W. Y. (2009). Preliminary study on relationships of Dothideales and its allies. Mycosystem 28, 161–170.
Lumbsch, H. T., Huhndorf, S. M. (2010). Myconet volume 14. Part one. Outline of Ascomycota—2009. Part two. Notes on Ascomycete systematics. Nos. 4751–5113. Fieldiana Life Earth Sci. 1, 1–64. doi: 10.3158/1557.1
Luttrell, E. S. (1960). The morphology of an undescribed species of Dothiora. Mycologia 52, 64–79. doi: 10.1080/00275514.1960.12024880
Luttrell, E. S. (1973). “Loculoascomycetes,” in The fungi, an advanced treatise, a taxonomic review with keys: ascomycetes and fungi imperfecti. Eds. Ainsworth, G. C., Sparrow, F. K., Sussman, A. S. (Academic, New York).
Lutzoni, F., Kauff, F., Cox, C. J., McLaughlin, D., Celio, G., Dentinger, B., et al. (2004). Assembling the fungal tree of life: progress, classification, and evolution of subcellular traits. Am. J. Bot. 91, 1446–1480. doi: 10.3732/ajb.91.10.1446
Marincowitz, S., Crous, P. W., Groenewald, J. Z., Wingfield, M. J. (2008). Microfungi occurring on Proteaceae in the fynbos. CBS biodiversity series, no. 7 (Utrecht, the Netherlands: CBS Fungal Biodiversity Centre).
MycoBank (2024). Available online at: https://www.mycobank.org (Accessed 13 March 2024).
Nylander, J. (2008). MrModeltest2 version 2.3 (program for selecting DNA substitution models using PAUP*). Uppsala, Sweden: Evolutionary Biology Centre.
O’Donnell, K., Kistler, H. C., Cigelnik, E., Ploetz, R. C. (1998). Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. 95, 2044–2049. doi: 10.1073/pnas.95.5.2044
Park, J. Y., Kim, K. M., Jung, H. S. (2006). Rhizosphaera densiflorae sp. nov. is described as a new species based on morphological and molecular data. Abstract retrieved Abstracts Meeting Mycological Soc. Japan 38, 38–38. doi: 10.11556/msj7abst.50.0.38.0
Pem, D., Gafforov, Y., Jeewon, R., Hongsanan, S., Promputtha, I., Doilom, M., et al. (2018). Multigene phylogeny coupled with morphological characterization reveal two new species of Holmiella and taxonomic insights within Patellariaceae. Cryptogam. Mycol. 39, 193–209. doi: 10.7872/crym/v39.iss2.2018.193
Pem, D., Jeewon, J., Bulgakov, T., Gafforov, Y., Hongsanan, S., Phookamsak, R., et al. (2019a). Taxonomy and molecular phylogeny of Thyrostroma ephedricola sp. nov. (Dothidotthiaceae) and proposal for Thyrostroma jaczewskii comb. nov. Phytotaxa 416, 243–256. doi: 10.11646/phytotaxa.416.4.3
Pem, D., Jeewon, R., Gafforov, Y., Hongsanan, S., Phukhamsakda, C., Promputtha, I., et al. (2019b). Melanocamarosporioides ugamica gen. et sp. nov., a novel member of the family Melanommataceae from Uzbekistan. Mycol. Prog. 18, 471–481. doi: 10.1007/s11557-018-1448-8
Pérez-Bonilla, M., González-Ménendez, V., Pérez-Victoria, I., de Pedro, N., Martín, J., Molero-Mesa, J., et al. (2017). Hormonemate derivatives from Dothiora sp., an endophytic fungus. J. Nat. Prod. 80, 845–853. doi: 10.1021/acs.jnatprod.6b00680
Petrak, F. (1937). Verzeichnis der neuen Arten, Varietäten, Formen, Namen und wichtigsten Synonyme. Just’s Botanischer Jahresbericht. 56, 291–697.
Quaedvlieg, W. G., Verkley, G. J., Shin, H. D., Barreto, R. W., Alfenas, A. C., Swart, W. J., et al. (2013). Sizing up septoria. Stud. Mycol. 75, 307–390. doi: 10.3114/sim0017
Ramaley, A. W. (1992). Tectacervulus mahoniae, Kabatina mahoniae, and Selenophoma mahoniae, three new fungi on Mahonia repens. Mycotaxon 43, 437–452.
Rambaut, A. (2016). FigTree, version 1.4.3. Institute of Evolutionary Biology, University of Edinburgh, UK. Available at: http://tree. bio. ed. ac. uk/software/figtree (Accessed 12 December 2023).
Rannala, B., Yang, Z. (1996). Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J. Mol. Evol. 43, 304–311. doi: 10.1007/BF02338839
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Saccardo, P. A., Sydow, P. (1902). Sylloge Fungorum 16 Vol. i-viii. Ed. Saccardo, P. A. (Italy), 1–1291.
Schoch, C. L., Crous, P. W., Groenewald, J. Z., Boehm, E. W. A., Burgess, T. I., de Gruyter, J., et al. (2009). A class-wide phylogenetic assessment of Dothideomycetes. Stud. Mycol. 64, 1–15. doi: 10.3114/sim.2009.64.01
Schoch, C. L., Kohlmeyer, J., Volkmann-Kohlmeyer, B., Tsui, C. K., Spatafora, J. W. (2006a). The halotolerant fungus Glomerobolus gelineus is a member of the Ostropales. Mycol. Res. 110, 257–263. doi: 10.1016/j.mycres.2005.10.001
Schoch, C. L., Shoemaker, R. A., Seifert, K. A., Hambleton, S., Spatafora, J. W., Crous, P. W. (2006b). A multigene phylogeny of the Dothideomycetes using four nuclear loci. Mycologia 98, 1041–1052. doi: 10.3852/mycologia.98.6.1041
Senanayake, I. C., Rathnayaka, A. R., Marasinghe, D. S., Calabon, M. S., Gentekaki, E., Lee, H. B., et al. (2020). Morphological approaches in studying fungi: collection, examination, isolation, sporulation and preservation. Mycosphere 11, 2678–2754. doi: 10.5943/mycosphere/11/1/20
Senwanna, C., Wanasinghe, D. N., Bulgakov, T. S., Wang, Y., Bhat, D. J., Tang, A. M. C., et al. (2019). Towards a natural classification of Dothidotthia and Thyrostroma in Dothidotthiaceae (Pleosporineae, Pleosporales). Mycosphere 10, 701–738. doi: 10.5943/mycosphere/10/1/15
Shear, C. L., Davidson, R. W. (1940). A new species of Dothiora on aspen and willow. Mycologia 32, 105–111. doi: 10.1080/00275514.1940.12017397
Shoemaker, R. A., Babcock, C. E. (1987). Wettsteinina. Canad. J. Bot. 65, 373–405. doi: 10.1139/b87-048
Shoemaker, R. A., Hambleton, S. (2005). Dothidea sambuci and Diaporthe spiculose. Canad. J. Bot. 83, 484–490. doi: 10.1139/b05-023
Simon, U. K., Groenewald, J. Z., Crous, P. W. (2009). Cymadothea trifolii, an obligate biotrophic leaf parasite of Trifolium, belongs to Mycosphaerellaceae as shown by nuclear ribosomal DNA analyses. Persoonia 22, 49–55. doi: 10.3767/003158509X425350
Spatafora, J. W., Sung, G. H., Johnson, D., Hesse, C., O’Rourke, B., Serdani, M., et al. (2006). A five-gene phylogeny of Pezizomycotina. Mycologia 98, 1018–1028. doi: 10.1080/15572536.2006.11832630
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Swofford, D. L. (2002). PAUP*: phylogenetic analysis using parsimony (*and other methods). Version 4 (Sinauer Associates, Sunderland, Massachusetts, USA).
Thambugala, K. M., Ariyawansa, H. A., Li, Y. M., Boonmee, S., Hongsanan, S., Tian, Q., et al. (2014). Dothideales. Fungal Divers. 68, 105–158. doi: 10.1007/s13225-014-0303-8
Tibpromma, S., Hyde, K. D., Jeewon, R., Maharachchikumbura, S. S. N., Liu, J. K., Bhat, D. J., et al. (2017). Fungal diversity notes 491–602: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 83, 1–261. doi: 10.1007/s13225-017-0378-0
Tsuneda, A., Hambleton, S., Currah, R. S. (2004). Morphology and phylogenetic placement of Endoconidioma, a new endoconidial genus from trembling aspen. Mycologia 96, 1128–1135. doi: 10.1080/15572536.2005.11832910
Tsuneda, A., Hambleton, S., Currah, R. S. (2010). Endoconidioma populi from aspen and alder: phylogeny, and variations in cleistopycnidial morphology and their ecological implications. Botany 88, 675–684. doi: 10.1139/B10-043
Vilgalys, R., Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
von Arx, J. A., Müller, E. (1975). A re-evaluation of the bitunicate ascomycetes with keys to families and genera. Stud. Mycol 9, 1–159.
Vu, D., Groenewald, M., De Vries, M., Gehrmann, T., Stielow, B., Eberhardt, U., et al. (2019). Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 92, 135–154. doi: 10.1016/j.simyco.2018.05.001
Wanasinghe, D. N., Phukhamsakda, C., Hyde, K. D., Jeewon, R., Lee, H. B., Jones, E. B. G., et al. (2018). Fungal diversity notes 709–839: taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Divers. 89, 1–236. doi: 10.1007/s13225-018-0395-7
White, T., Bruns, T., Lee, S., Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR protocols: a guide to methods and applications. Eds. Innis, M., Gelfand, D., Shinsky, J., White, T. (Academic Press, New York). 315–322 pp.
Wijayawardene, N. N., Hyde, K. D., Dai, D. Q., Sánchez-García, M., Goto, B. T., Saxena, R. K., et al. (2022). Outline of Fungi and fungus-like taxa – 2021. Mycosphere 13, 53–453. doi: 10.5943/mycosphere/13/1/2
Winton, L. M., Stone, J. K., Hansen, E. M., Shoemaker, R. A. (2007). The systematic position of Phaeocryptopus gaeumannii. Mycologia 99, 240–252. doi: 10.1080/15572536.2007.11832584
Zajc, J., Gostinčar, C., Černoša, A., Gunde-Cimerman, N. (2019). Stress-tolerant yeasts: opportunistic pathogenicity versus biocontrol potential. Genes 10, 42. doi: 10.3390/genes10010042
Keywords: Asia, Dothideomycetes, Dothideales, fungal taxonomy, new species, saprobic fungi
Citation: Senwanna C, Hongsanan S, Khuna S, Kumla J, Yarasheva M, Gafforov Y, Abdurazakov A and Suwannarach N (2024) Insights into the molecular phylogeny and morphology of three novel Dothiora species, along with a worldwide checklist of Dothiora. Front. Cell. Infect. Microbiol. 14:1367673. doi: 10.3389/fcimb.2024.1367673
Received: 09 January 2024; Accepted: 20 March 2024;
Published: 19 April 2024.
Edited by:
Asha Janadaree Dissanayake, University of Electronic Science and Technology of China, ChinaReviewed by:
Subashini Chathumini Jayasiri, Mae Fah Luang University, ThailandKasun Thambugala, University of Sri Jayewardenepura, Sri Lanka
Copyright © 2024 Senwanna, Hongsanan, Khuna, Kumla, Yarasheva, Gafforov, Abdurazakov and Suwannarach. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nakarin Suwannarach, c3V3YW5fNDYxQGhvdG1haWwuY29t
 Chanokned Senwanna
Chanokned Senwanna Sinang Hongsanan
Sinang Hongsanan Surapong Khuna
Surapong Khuna Jaturong Kumla
Jaturong Kumla Manzura Yarasheva
Manzura Yarasheva Yusufjon Gafforov
Yusufjon Gafforov Aziz Abdurazakov7
Aziz Abdurazakov7 Nakarin Suwannarach
Nakarin Suwannarach