
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol., 22 February 2024
Sec. Oral Microbes and Host
Volume 14 - 2024 | https://doi.org/10.3389/fcimb.2024.1357631
Streptococci are primary colonizers of the oral cavity where they are ubiquitously present and an integral part of the commensal oral biofilm microflora. The role oral streptococci play in the interaction with the host is ambivalent. On the one hand, they function as gatekeepers of homeostasis and are a prerequisite for the maintenance of oral health - they shape the oral microbiota, modulate the immune system to enable bacterial survival, and antagonize pathogenic species. On the other hand, also recognized pathogens, such as oral Streptococcus mutans and Streptococcus sobrinus, which trigger the onset of dental caries belong to the genus Streptococcus. In the context of periodontitis, oral streptococci as excellent initial biofilm formers have an accessory function, enabling late biofilm colonizers to inhabit gingival pockets and cause disease. The pathogenic potential of oral streptococci fully unfolds when their dissemination into the bloodstream occurs; streptococcal infection can cause extra-oral diseases, such as infective endocarditis and hemorrhagic stroke. In this review, the taxonomic diversity of oral streptococci, their role and prevalence in the oral cavity and their contribution to oral health and disease will be discussed, focusing on the virulence factors these species employ for interactions at the host interface.
Streptococcal species accompany us throughout our live – in the oral cavity, they are the first colonizers after birth and, onwards, they shape the establishment of a complex microbiota in health and disease (Abranches et al., 2018; Klein, 2022). Their interactions with the human host range from highly beneficial – influencing, as commensal species, proper immune system development (Lang et al., 2010) and preventing pathogen colonization - to detrimental, when dissemination in the blood stream occurs, potentially leading to no less than infective endocarditis (IE), purulent infections, brain hemorrhage, intestinal inflammation, autoimmune diseases, and bacteremia, among others (Yumoto et al., 2019). This review focuses on oral streptococci, their taxonomic diversity and roles within their ecological niche, and, first and foremost, their interaction with the host, within the oral habitat and beyond. Complementary to this review, methods and tools for the detection, identification, characterization, and spatial localization of oral streptococci have been summarized in a recent book chapter (Klein, 2022).
Streptococci are facultative anaerobic, Gram-positive bacteria. Historically, species within the genus Streptococcus were classified according to their hemolytic potential on blood agar plates as fully (β-)hemolytic, partially (α-) hemolytic and non- (γ-) hemolytic (Sherman, 1937). β-hemolytic streptococci were further subcategorized according to the carbohydrate composition of the antigens present in their cell wall (Lancefield groups) as Group A Streptococcus (GAS), such as the highly pathogenic Streptococcus pyogenes, or as Group B Streptococcus (GBS), such as Streptococcus agalactiae (Facklam, 2002; Abranches et al., 2018).
With the advent of 16S rRNA sequencing techniques, the genus Streptococcus was further categorized into eight distinct groups based on phylogenetic relationships between its members; these are the mitis, sanguinis, anginosus, salivarius, downei, mutans, pyogenic, and bovis groups (Abranches et al., 2018). While the mitis group constitutes the largest group with 20 members detected in the oral cavity, oral streptococci – also often referred to as viridans streptococci – cluster in all groups except for bovis and pyogenic (Abranches et al., 2018). Between these groups as well as species within the same group, differences in virulence and pathogenicity can be observed (Sitkiewicz, 2018). For instance, members of the mutans group, e.g., Streptococcus mutans and Streptococcus sobrinus, have been identified as species with high cariogenic potential (Richards et al., 2017). Mitis group streptococci (MGS) such as Streptococcus mitis, Streptococcus oralis, Streptococcus gordonii, Streptococcus infantis, Streptococcus sanguinis, and Streptococcus parasanguinis, on the other hand, are associated with oral health (Richards et al., 2017). Notably, especially the first three species have recently been identified as causative agents in the development of IE, after the occurrence of a blood stream infection (Chamat-Hedemand et al., 2020). Among others, S. mitis as well as S. oralis have recently been classified as members of the core oral microbiota, i.e., they belong to a group of oral bacteria that is ubiquitously present within the oral cavity (Hall et al., 2017). When it comes to their pathogenic potential, MGS as well as Streptococcus anginosus Group (SAG) members have long been considered as commensals integrated in a healthy human microbiota, however, they have more and more frequently been recognized to elicit significant health problems in humans (Mitchell, 2011; Kuryłek et al., 2022; Pilarczyk-Zurek et al., 2022).
The oral cavity is a unique and multifaceted bacterial habitat with distinct ecological niches, resulting from the presence of different surfaces for colonization and varying conditions due to considerable fluctuations in oral environmental parameters such as temperature, pH, redox potential and nutrient availability, which, in combination with behavioral aspects of the human host (e.g., dental hygiene, diet, smoking) as well as genetic predisposition and the general health status, shape the composition of the resident microbial consortia (Mark Welch et al., 2020). The non-shedding surfaces of the teeth, the dorsal and lateral surfaces of the tongue, the periodontal pockets, and the remaining epithelial surfaces of the oral mucosa offer sites for colonization by distinct microbes. Streptococci have been found to be present at all of these sites and to be the dominant genus in supra- and subgingival plaque and on soft tissues (Mager et al., 2003; Aas et al., 2005; Huse et al., 2012; Mira, 2018). As primary/pioneer colonizers, they are the first species to colonize the oral surfaces and allow for the establishment of other species such as Actinomyces, Veillonella, Fusobacterium, Prevotella, and Neisseria, initializing the process of microbial succession and establishment of a complex microbial consortium (Sampaio-Maia and Monteiro-Silva, 2014; Gomez and Nelson, 2017). Recent investigations revealed a new understanding of how the oral biofilm is formed from bacterial aggregates serving as nuclei for the development of a strong biofilm embedded in the extracellular matrix. Regarding the presence of streptococci, an interesting observation was made concerning their localization; in addition to early colonization at the biofilm basis, these were detected radially in the outermost layer of the biofilm providing a microenvironment for strict anaerobes by consuming present oxygen (Simon-Soro et al., 2022).
Formation of dental plaque on the non-shedding surfaces of the teeth is one of the best characterized multispecies biofilm community activities; dental plaque is present in healthy individuals, but it is also associated with the development of oral diseases, including caries, gingivitis and periodontal diseases (Marsh and Zaura, 2017).
Streptococci are the predominant species to initially adhere to the salivary pellicle (Diaz et al., 2006). In supragingival plaque, predominantly the dietary intake of sugars promotes the formation of extracellular polymeric substances (EPS) and acidic metabolites causing the resident microflora to shift towards aciduric and acidogenic species (Lamont et al., 2018). If persistent, this leads to the acidification of the biofilm microenvironment and, ultimately, to the demineralization of dental enamel and the development of carious lesions (Lamont et al., 2018). The role of mutans streptococci (MS), i.e., S. mutans and S. sobrinus, as cariogenic agents has long been recognized (Conrads et al., 2014; Hajishengallis et al., 2017). The cariogenic potential of MS stems from three characteristic traits – the species’ ability to synthesize large quantities of extracellular glucan from sucrose, their ability to metabolize a wide range of carbohydrates into organic acids, i.e., their acidogenicity, and their tolerance towards environmental stress conditions such as low pH, i.e., their aciduric nature (Lemos et al., 2019). Extracellular glucans form essential building blocks of the EPS as part of the extracellular biofilm matrix, which confers to the bacteria protection from shear forces and resistance to antimicrobials, influences the diffusion of oxygen, nutrients, quorum sensing signals and metabolites, and creates an acidic microenvironment in which commensal bacteria are outcompeted by cariogenic species (Bowen et al., 2018).
Carious lesions or other factors that damage the tooth’s integrity can reach the dental pulp, which is principally encased in the root canal system, exposed to the bacteria present in the oral cavity. Bacterial colonization elicits inflammation and necrosis of the dental pulp as well as inflammation of the periapical region, finally leading to bone resorption and the formation of granulomas or cysts (Graves et al., 2011). Oral streptococci, such as S. oralis, S. anginosus, S. mitis, S. sanguinis (Chávez de Paz et al., 2005; Narayanan and Vaishnavi, 2010), and S. mutans (Lima et al., 2020) are frequently among the species detected in root canal infections. In the dental pulp, bacterial pathogen associated molecular patterns (PAMPs) are recognized by macrophages, dendritic cells (DCs), odontoblasts and endothelial cells, and elicit an immune response leading to the recruitment of specialized immune cells and the initiation of bacterial clearance (Khorasani et al., 2020). The inflammatory response mechanisms, cytokine networks as well as pulpal disease pathogenesis are the focus of other reviews (Hahn and Liewehr, 2007a; Farges et al., 2015; Khorasani et al., 2020; Galler et al., 2021).
The onset of gingivitis and emergence of periodontal diseases is caused by the shift in the composition of the resident oral microbiota from commensal to dysbiotic (Hajishengallis and Lamont, 2012). In a healthy individual, bacteria in the subgingival crevice exist in a balanced state with the host immune system, under controlled, low-level inflammation keeping bacterial growth in check (Hajishengallis and Lamont, 2016). This symbiotic state can be disrupted if certain risk factors, such as disadvantageous host genetics or lifestyle, or systemic diseases apply, or if keystone pathogens are present (Loos and Van Dyke, 2020). Dysbiosis is characterized by an increase in bacterial mass and the prevalence of Gram-negative, anaerobic species, leading to chronic inflammation (Hajishengallis et al., 2020). Particularly, the emergence of inflammophilic species such as Porphyromonas gingivalis, Treponema denticola, Tannerella forsythia, Aggregatibacter actinomycetemcomitans or Filifactor alocis (Haffajee et al., 2008; Könönen et al., 2019), their modulation of the immune response and interaction with the resident polymicrobial community support an exacerbation of inflammation and bacterial overgrowth (Hajishengallis et al., 2012). Periodontal pathogens are capable of evading (Hajishengallis, 2011; Bloch et al., 2018, 2019) and subverting (Hajishengallis and Lamont, 2014; Hajishengallis and Diaz, 2020) the host immune response and thereby dysregulate immune homeostasis. A detailed description of immune mechanisms in the context of periodontal diseases can be found in several excellent reviews (Hajishengallis, 2014; Ebersole et al., 2017; Könönen et al., 2019).
Oral streptococci regulate the structure and function of the oral microbiome in a way that is beneficial to human health (Baty et al., 2022). However, Streptococcus was determined by metagenomic analyses as the predominant genus in patients with gingivitis (Park et al., 2015). As pioneer colonizers, streptococci not only enable later colonizers and pathogenic species to co-adhere, they also can be considered as accessory pathogens, as is the case for S. gordonii whose interaction with P. gingivalis and A. actinomycetemcomitans was found to substantially elevate these species’ pathogenicity (Kuboniwa et al., 2006; Daep et al., 2011; Ramsey et al., 2011). In pathogen-free mice, inoculation with P. gingivalis led to a massive increase in streptococcal cell numbers (Hajishengallis et al., 2011) and in an in vitro subgingival biofilm model, omission of the periodontal pathogen from the community resulted in invasion of gingival epithelial cells by S. oralis instead (Thurnheer et al., 2014). On the other hand, P. gingivalis failed to cause periodontal disease in germ-free mice in the absence of the commensal core microbiome –which in fact was capable of causing modest bone loss in germ-free mice on its own putting oral streptococci in the spotlight as modulators of oral immune responses (Hajishengallis et al., 2011; Abusleme et al., 2013). These findings highlight the central role oral streptococci play in the balancing act between the maintenance of host-microbiome homeostasis during health and immunomodulatory effects in disease. In the following sections, the factors employed by these bacteria to tip this balance towards a healthy or diseased state will be discussed.
Maintenance of homeostatic control of infection and inflammation is crucial to oral health. As dominant members of the resident oral microflora, streptococci exert essential functions in this process (Figure 1). In a study by Myers and coworkers, 30% of streptococci isolated from supra- and subgingival plaque samples were found to exhibit anti-inflammatory properties, specifically by downregulation of IL-8 production in epithelial cells (Myers et al., 2021). Operating as an oral commensal, S. gordonii can supress the secretion of IL-6 and IL-8 in epithelial cells, which contrasts with Fusobacterium nucleatum, an important bridging bacterium in oral biofilms that is regarded as a potential pathogen and was examined in the same study (Hasegawa et al., 2007). Other species such as S. salivarius or S. mitis contribute to a balanced immune status by attenuating pro-inflammatory immune responses towards themselves as well as towards pathogenic species (Cosseau et al., 2008; Eberhard et al., 2009; Sliepen et al., 2009a). Furthermore, S. mitis induces the release of the antimicrobial cationic peptide human β-defensin 2 (hBD-2) which is not harmful to S. mitis itself, but affects other bacteria (Eberhard et al., 2009; Mitchell, 2011) and thereby antagonizes oral colonization by pathogenic species. Expression of hBD-2 has been found to be upregulated in gingival epithelial cells in response to commensals, e.g., F. nucleatum, but not to pathogenic species, such as P. gingivalis, and is produced in the gingiva also in a non-inflamed state (Krisanaprakornkit et al., 2000). Apart from bacterial killing, hBD-2 can also inhibit bacterial biofilm formation at nanomolar concentrations, as was shown for, e.g., Pseudomonas aeruginosa and Acinetobacter baumannii (Parducho et al., 2020). In response to S. mitis challenge, monocytes produce chemotactic as well as pro-inflammatory mediators and at the same time increase the secretion of interleukin-10 (IL-10) and prostaglandin E2 (PGE2) and expression of programmed cell death protein ligand 1 (PD-L1), inhibiting neutrophil activity and T cell proliferation (Engen et al., 2018). S. mitis can, thus, trigger the recruitment of immune cells to the site of infection, while at the same time dampening the inflammatory response and thereby promote commensal bacterial tolerance and survival on site. Certain MGS and sanguinis streptococci, especially those associated with systemic infections, even display immune evasion mechanisms being less susceptible to complement opsonization and thereby escape immune surveillance (Alves et al., 2019). In S. mutans, complement resistance as well as uptake by neutrophils is controlled by the orphan response regulator CovR, a repressor of virulence factor expression (Negrini et al., 2012; Alves et al., 2016). Specifically, CovR regulates the expression of genes involved in cell wall biogenesis and surface interactions with EPS and impacts not only susceptibility to complement opsonization and survival in blood, but also the formation of cariogenic biofilms on the tooth surface (Alves et al., 2016).
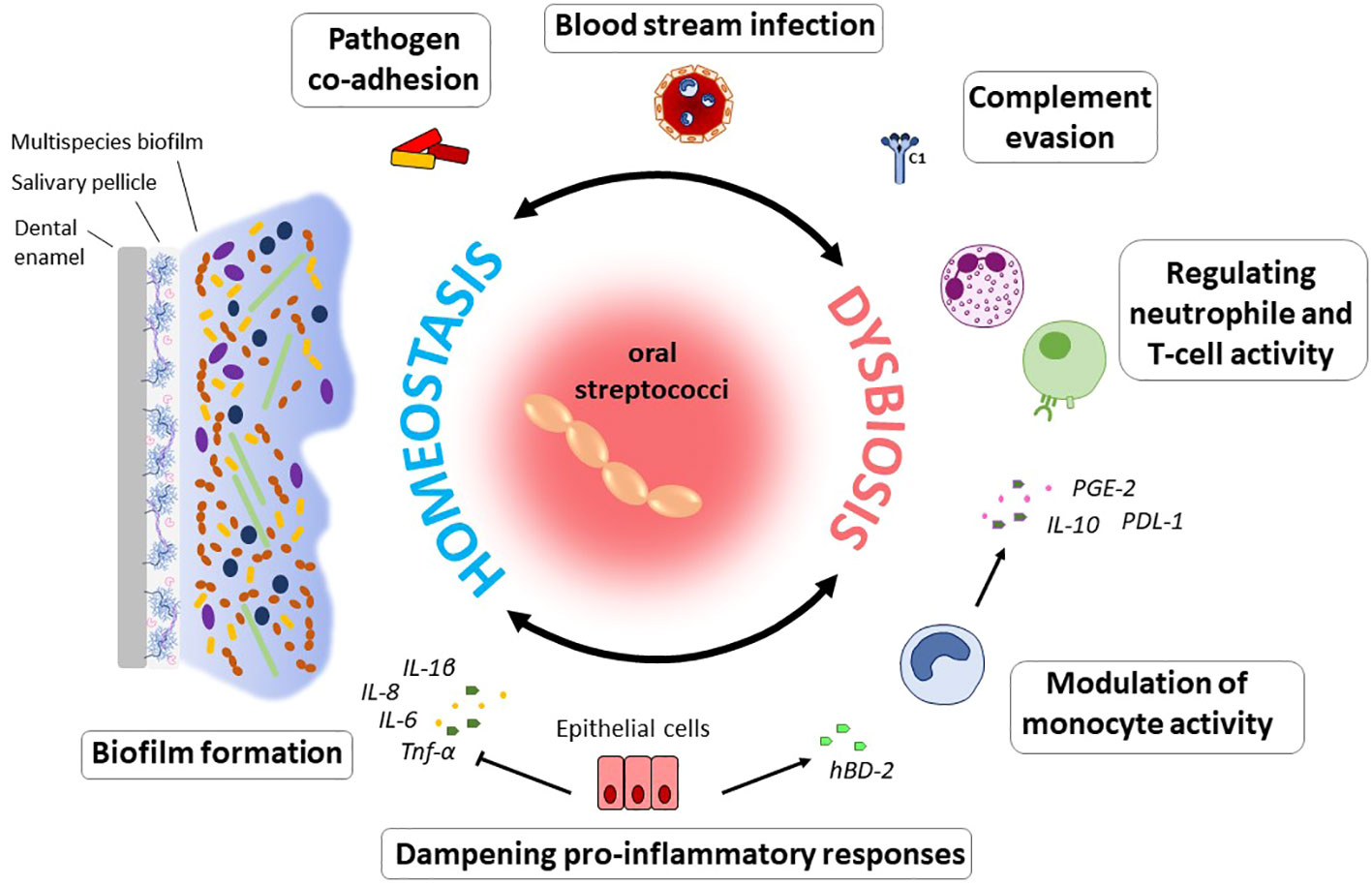
Figure 1 Immunomodulatory effects of oral commensal streptococci. The ability to form biofilms on numerous substrates can be regarded as a hallmark of oral streptococci. From within oral biofilms, streptococci interact with immune and epithelial cells, enable pathogens to co-adhere, and can disseminate into the blood stream. Modulation of the immune response to the bacterial biofilm challenge enables their persistence and survival in the host and determines the balance between immune homeostasis and dysbiosis. Specifically, through the downregulation of pro-inflammatory immune responses, oral streptococci contribute to a balanced immune status. Induction of hBD-2 released e.g., by S. mitis counteracts colonization by pathogenic species. This commensal was also found to induce a pro-inflammatory immune response in monocytes, while at the same time inhibiting neutrophil activity and T cell proliferation through the action of IL-10, PD-L1 and PGE2. In the context of systemic disease, it is beneficial for many streptococcal species that they can evade complement mediated immunity and thereby remain under the radar. Especially in periodontal disease, the role of oral streptococci to adhere to numerous substrates as primary colonizers and biofilm builders makes them a contributing factor to disease development, since they enable colonization of pathogenic species.
The salivary pellicle that forms on the tooth and mucosal surfaces constitutes the primary substrate for streptococcal colonization via specialized adhesins, which can bind to albumin, proline-rich proteins, glycoproteins, mucins, and sialic acid (Sia) (Abranches et al., 2018). Other structures that oral streptococci are able to adhere to include glucan, collagen, plasminogen, laminin, and fibrinogen (Nobbs et al., 2009). Streptococcal adhesion and colonization has been reviewed in detail by Nobbs et al. (Nobbs et al., 2009) and will be briefly discussed here.
Anchoring of streptococcal cell surface proteins typically occurs via their Leu‐Pro‐x‐Thr‐Gly (LPxTG) motif and is facilitated by sortases that catalyze the attachment of the proteins to the cell wall (Okahashi et al., 2022). The cell-wall anchored antigens I/II (AgI/II) – also named P1, SpaB, AgB, or PAc - belong to a family of adhesins that is present in most oral streptococci, and recognizes multiple host proteins thereby facilitating sucrose-independent biofilm formation, platelet aggregation, invasion of tissues, and interaction with the host immune system (Koga et al., 1990; Brady et al., 2010; Abranches et al., 2018). AgI/II proteins have been found to bind to fibronectin, salivary glycoproteins and proline-rich proteins, collagen, laminin, fibrinogen, platelets and α5ß1 integrin on epithelial and endothelial cells (Nobbs et al., 2009). S. gordonii AgI/II proteins SpaA and SpaB have also been shown to facilitate interaction with oral Actinomyces species (Jenkinson et al., 1993) and P. gingivalis, enabling invasion of dentinal tubules by the periodontopathogen (Love et al., 1997), contributing to the bacterium’s role in the development of periodontal disease (Lamont et al., 1994). Also, in fungal-bacterial communication AgI/II proteins play a pivotal role; specifically, this includes the interaction between S. gordonii SspB and the C. albicans hyphal cell wall protein Als3, which is relevant to biofilm formation (Silverman et al., 2010). Furthermore, AgI/II from S. mutans was shown to be important for the incorporation of C. albicans into a dual-species S. mutans-C. albicans biofilm and also required for increased acid production. Notably, this interaction is independent of the known streptococcal Als1 and Als3 receptors of C. albicans (Yang et al., 2018).
A group of streptococcal serine-rich repeat (Srr) glycoproteins can bind to sialoglycans present on salivary mucin MG2/MUC7, platelet glycoproteins (Takamatsu et al., 2005; Bensing et al., 2016) and polymorphonuclear neutrophils (PMNs) (Ruhl et al., 2000), and induce maturation and activation of DCs (Ko et al., 2017). Among these Srr glycoproteins are Gsp and Hsa of S. gordonii (Takahashi et al., 2002; Urano-Tashiro et al., 2016), the major fimbrial subunit Fap1 of S. parasanguinis (Wu et al., 2007) and S. oralis (Singh et al., 2017), SrpA, SrpB and SrpC of S. salivarius (Couvigny et al., 2017) and SrpA of Streptococcus cristatus (Handley et al., 2005) and S. sanguinis (Plummer et al., 2005; Deng et al., 2014). The interaction of these Sia-binding proteins with platelets may contribute to the pathogenesis of IE and potentially renders oral streptococci more virulent (Deng et al., 2014; Bensing et al., 2016).
S. gordonii additionally expresses short fibrils constituted by the CshA polypeptide on its cell surface (McNab et al., 1996). CshA as well as CshA-like fibrils present on the cell surface of other MGS such as S. oralis and S. sanguinis bind to other bacteria as well as salivary proteins (Fachon-Kalweit et al., 1985; Handley et al., 1987; Weerkamp et al., 1987; Ray et al., 1999) and immobilized fibronectin (McNab et al., 1996), thereby contributing to the establishment of the bacteria in the multispecies biofilms and colonization of various sites within the host. Other structures that oral streptococci are able to adhere to include glucan, collagen, plasminogen, laminin, and fibrinogen (Nobbs et al., 2009).
AgI/II adhesins are widely distributed among oral streptococci and constitute an important factor in the bacteria’s pathogenicity and ability to colonize oral sites (Jenkinson and Demuth, 1997) (see 5.). The AgI/II proteins consist of several structural regions, with the intertwined A- and P-regions presenting the globular, less conserved V-domain on the cell surface for ligand interaction (Manzer et al., 2020). Viridans streptococci reportedly induce the expression of proinflammatory cytokines IL-8 and IL-6 in endothelial and IL-8 in epithelial cells; in the case of S. mutans OMZ175, this is facilitated through the binding of AgI/II (as well as rhamnose-glucose polysaccharide (RGP) – see 7.3.) to glycoproteins present at the host cell surface through lectin-type interactions (Vernier et al., 1996). IL-8 stimulation occurs through binding of AgI/II to α5ß1 integrin and subsequent activation of MAPK signaling (Al-Okla et al., 1999). In primary human coronary artery endothelial cells, cellular binding of AgI/II was furthermore shown to stimulate the expression of the adhesion molecules E-selectin, ICAM-1 and VCAM-1 and thereby stimulate trans-endothelial migration of neutrophils and contribute to the exacerbation of inflammatory responses to the bacterial burden (Vernier-Georgenthum et al., 1998). Also, binding of AgI/II to monocytes occurs via a lectin-type interaction with Sia and fucose residues exposed on host cell surface-glycoproteins and results in an increased production of the proinflammatory cytokines TNF-α, Il-1β and Il-6 (Soell et al., 1994; Chatenay-Rivauday et al., 1998, 2000). Induction of pro-inflammatory cytokines in synovial cells suggests that AgI/II might even play a role in rheumatic disease, its initiation and perpetuation (Gourieux et al., 2001). Stimulation of TNF-α in THP-1 cells by the extended V-region of AgI/II was observed not only for S. mutans strains, but also for S. gordonii and the SAG members S. anginosus, Streptococcus intermedius and Streptococcus constellatus, highlighting the universal role the AgI/II protein family plays in streptococcal adhesion and virulence (Chatenay-Rivauday et al., 2000).
Another factor contributing to S. mutans’ success not only within but also outside its ecological niche is the cell surface glycoprotein Cnm which exhibits collagen- and laminin-binding capability. Cnm is present in 10-20% of all isolated strains of S. mutans (Nakano et al., 2010) and was most extensively studied in the S. mutans strain OMZ175. Cnm was found to be vital for the invasion of human coronary artery endothelial cells (HCAEC) and virulence in a Galleria mellonella infection model (Abranches et al., 2011; Aviles-Reyes et al., 2014). In the oral cavity, Cnm promotes S. mutans’ invasion of oral keratinocytes and fibroblasts and enhances binding of the bacterium to collagenated surfaces, as well as dentin and root tissues contributing to its cariogenicity (Miller et al., 2015). Furthermore, Cnm was found to influence bacterial cell permeability and therefore might play a role in the susceptibility to antimicrobial agents (Naka et al., 2022).
Together with AgI/II, Cnm plays a crucial role in the aggravation of non-alcoholic steatohepatitis (NASH) (Naka et al., 2014, 2016, 2018); it is directly involved in and a potential risk factor for hemorrhagic stroke (Nakano et al., 2011) and IE (Nomura et al., 2013) and therefore can be considered as an indispensable virulence factor in the onset and progression of systemic diseases such as IE elicited by Cnm-positive S. mutans strains. Cnm is most frequently found in strains with serotype f (see 7.3.), such as S. mutans OMZ 175, which in contrast to serotype c strains, is not most common in dental plaque, but has been specifically implicated in the pathogenesis of systemic disease (Abranches et al., 2011).
S. mutans decorates its cell surface with a rhamnose-glucose polysaccharide (RGP), a major cell wall antigen whose composition determines the strain serotype (c, e, f, and k) and which has distinct functions in cell division and morphology (Rijn and Bleiweis, 1973; Nakano and Ooshima, 2009; De et al., 2017). Also, in colonization of tooth surfaces - as well as heart muscle and kidney tissues - serotype f RGP plays a role as a putative adhesin (Stinson et al., 1980; Engels-Deutsch et al., 2003). The RGP’s contribution to adhesion was also demonstrated for THP-1, dental pulp and periodontal ligament cells, which upregulated the production of IL-6 and IL-8 in response to bacterial challenge dependent on the presence of RGP as well as AgI/II on the cell surface (Engels-Deutsch et al., 2003). RGP binds to human monocytes in a CD14-dependent manner and induces the release of TNF-α and other pro-inflammatory cytokines (Benabdelmoumene et al., 1991; Soell et al., 1995) as well as the up-regulation of RFc γ (Benabdelmoumene et al., 1991). RGP derived from S. mutans OMZ175 binds epithelial as well as endothelial cells in a dose-dependent manner and stimulates the release of IL-8 and IL-6 (Vernier et al., 1996). In rat model studies, RGP was found to render S. mutans more virulent in the induction of IE (Nagata et al., 2006) and stimulate nitric oxide synthase activity linking it to the genesis of septic shock induced by Gram-positive bacteria (Martin et al., 1997). Furthermore, the hydrophilic nature of RGP might play a role in the resistance of S. mutans to phagocytosis by PMNs and thereby contribute to the bacteria’s defiance of immune response mechanisms and survival within the host (Tsuda et al., 2000).
In Gram-positive bacteria, teichoic acids – wall teichoic acids (WTA) anchored to the cell wall peptidoglycan and lipoteichoic acids (LTA) anchored to membrane glycolipids - constitute a major part of the cell envelope and play vital roles in bacterial physiology, surface attachment, as well as interspecies and host interactions (Silhavy et al., 2010). Depending on the chemical structure, LTAs can be classified into five types (I-V) (Percy and Gründling, 2014). Type I, present e.g., in S. mutans and S. gordonii, is comprised of repeating polyglycerolphosphate units coupled with D-alanine and glucose (Lima et al., 2019); type II LTA as present in S. oralis, is composed of polyribitolphosphate repeating units (Hong et al., 2014a).
In the context of cariogenesis, LTA plays a vital role in conferring adhesive properties to bacterial cells and hydration of the cariogenic plaque in concert with streptococcal derived glucan (Rølla et al., 2009). When provided with high amounts of sucrose, S. mutans upregulates not only its glucan production, but also produces higher amounts of LTA (Rølla et al., 2009). S. mutans LTA furthermore induces apoptosis in pulpal cells from deciduous teeth indicating its involvement in the development to pulpitis (Wang et al., 2001), which has been reviewed elsewhere (Hahn and Liewehr, 2007b). In monocytic, dental pulp and periodontal ligament cells, S. mutans LTA is a less potent inducer of proinflammatory cytokines than RGP and AgI/II (Engels-Deutsch et al., 2003). In a murine macrophage cell line, LTA from both S. mutans and S. sanguinis induced the production of TNF-α and release of nitric oxide (NO) potentially contributing to septic shock elicited by these bacteria in immunocompromised patients (English et al., 1996; Hong et al., 2014a).
When analyzing S. mutans LTA-binding proteins in saliva of caries-free and caries-active subjects, Hong and coworkers found differences in protein profiles dependent on the health status, with histone H4, neutrophil defensin-1 and profilin-1 being associated with health, and cystatins and lysozyme, among others, predominantly present in saliva from caries patients (Hong et al., 2014b). In the case S. sanguinis, LTA was shown to antagonize recognition of lipopolysaccharide (LPS) on gingival fibroblasts in a CD14-dependent manner, thereby potentially dampening the cells’ immune response to Gram-negative periodontal pathogens and contributing to commensalism (Sugawara et al., 1999). LTA in contrast to LPS also stimulates expression of hepatocyte growth factor/scatter factor in gingival epithelial cells (Sugiyama et al., 1996). When stimulated with an LTA-deficient strain of S. gordonii, human dendritic cells reacted with a stronger immune response, i.e., increased phagocytic activity and production of proinflammatory markers and T-cell activation, than those treated with the corresponding parent wild-type strain, indicating an immune evasive role of LTA (Kim et al., 2023). In this context it is interesting to note, that S. gordonii cell wall lipoproteins, but not LTA have been shown to stimulate IL-8 production in periodontal ligament cells in a TLR2-dependent fashion (Kim et al., 2017; Park et al., 2020) and, in the case of lipoprotein PpiA, to suppress phagocytosis by macrophages (Cho et al., 2013).
Colonization of the oral cavity by oral streptococci depends on the production of extracellular glucans that to a large part make up the ECM of streptococcal biofilms (Nobbs et al., 2015; Bowen et al., 2018). S. mutans produces several dietary sucrose-hydrolyzing enzymes producing fructose and glucose, which are the building blocks of α-1,2- and β-1,6-linked glucans formed by the subsequent action of secreted (GtfD) and cell-wall associated (GtfB, GtfC) glucosyltransferases (Gtfs) (Nobbs et al., 2015; Bowen et al., 2018). In cariogenic biofilms, these sugars contribute to the acidification of the biofilm microenvironment and the shift to a dysbiotic microbiota, since these are catabolized along fermentative pathways leading to acid production (Bowen et al., 2018; Lamont et al., 2018). The secretion and action of Gtfs is not a specific trait of S. mutans, in which these enzymes have been most extensively studied; many oral streptococci employ Gtfs for colonization, adhesion, cohesion, and interbacterial interactions (Nobbs et al., 2009; Abranches et al., 2018). GtfB, GftC and GtfD adhere to the salivary pellicle on the tooth surface as well as to other microorganisms including bacteria and Candida albicans, essentially converting them to glucan producers contributing to the build-up of EPS and biofilm formation (Falsetta et al., 2014; Bowen, 2016). Furthermore, glucan binding proteins (Gbps) expressed by MS are prerequisites for successful biofilm formation, facilitating binding to dextrans and accumulation of MS in the biofilms (Nobbs et al., 2009). Streptococcal Gtfs can be regarded as immunogenic; a humoral immune response with anti-Gtf-specific immunoglobulin G (IgG) in serum or IgA in saliva naturally occurs in human populations (Chia et al., 1997). A Gtf-inhibitory factor (GIF) was found as an innate defense factor present in human saliva, specifically binding to the glucan binding domain of Gtfs and inhibiting Gtf function (Jespersgaard et al., 2002). GtfC and GtfD influence T-cell proliferation and modulate the immune response by monocytes, with a higher response elicited by GtfD than GtfC (Chia et al., 2001). S. mutans Gtfs furthermore robustly induce the production of IL-6 by T-cells in vitro as well as in vivo in a experimental rat model of endocarditis – pinpointing their contribution to disease development outside the oral cavity, since IL-6 levels were found to be elevated in patients with IE (Chia et al., 2001). In the same experimental model, it was also shown that S. mutans Gtfs induce the production of IL-6 in endothelial cells in infected heart valves, specifically during the acute stage of infection (Shun et al., 2005). In S. gordonii, a Gtf contributes to the bacterium’s capability to cause IE, being a prerequisite for the adhesion to endothelial as well as epithelial cells in vitro (Vacca-Smith et al., 1994).
In the resolution of bacterial infection, neutrophils play a pivotal role, clearing and killing bacterial invaders through phagocytosis, granule release, oxidative burst, and neutrophil extracellular trap (NET) formation – the release of chromatin fibers carrying antimicrobial peptides (AMPs) ready for bacterial killing and prevention of bacterial spreading (Scott and Krauss, 2012; Chapple et al., 2023). In S. sanguinis, a streptococcal wall-anchored nuclease (SWAN) confers resistance to NET killing by digesting released NET DNA (Morita et al., 2014). Another S. mutans nuclease - DeoC - enables the bacteria to escape NETs and facilitates biofilm dispersal (Liu et al., 2017). Lacking the LPxTG sorting motif for cell wall anchoring, DeoC most likely acts during late stages of biofilm growth when cell autolysis and dispersal occur and aids in bacterial escape of entrapment in NETs by chromatin degradation (Liu et al., 2017).
One characteristic trait of oral streptococci that has been getting attention for its relevance in the regulation of immune responses is the production of hydrogen peroxide as a by-product of aerobic metabolism (Zhu and Kreth, 2012). Through the production of H2O2, oral streptococci like MGS inhibit other potentially pathogenic species, among them S. mutans, and thereby gain a competitive advantage over these species (Kreth et al., 2008). By triggering the release of eDNA, H2O2 production additionally facilitates the exchange of genetic material between the bacteria (Kreth et al., 2009). In the interaction with the host, H2O2 production shows the potential to be a vital contributor to disease pathogenesis. Bacterial species such as S. sanguinis and S. oralis induce the formation of foam cells, contributing to atherosclerosis, and cause lysosome dysfunction and cell death in macrophages as well as in epithelial cells through the action of ROS, i.e., the release of H2O2 (Okahashi et al., 2011, 2013, 2014, 2016). Microarray studies of macrophages infected with S. oralis wild-type and a corresponding streptococcal pyruvate oxidase (spx)-deletion mutant deficient in H2O2 production showed that H2O2 suppresses the expression of proinflammatory mediators, especially NF-κB signaling, and cellular stress responses (Matsushima et al., 2017). Pyruvate oxidase SpxB catalyzes the conversion of inorganic phosphate and pyruvate to acetyl phosphate, carbon dioxide and H2O2 (Chen et al., 2011). Notably, SpxB is distinct from the S. mutans transcriptional regulator Spx, encoding two homologues, SpxA1 and SpxA2, involved in oxidative stress response and regulation of genes involved in cell division and cell envelope biosynthesis, respectively (Baker et al., 2020; Ganguly et al., 2020). S. oralis was furthermore found to inhibit the activation of inflammasomes through the release of H2O2 corroborating its contribution to the establishment and persistence of the oral commensal within the oral cavity and even in bloodstream infections (Erttmann and Gekara, 2019). In the context of periodontal disease, SpxB of S. oralis and S. mitis plays a vital role in enabling bacterial colonization and host homeostasis by inhibiting NF-κB signaling through the activation of nuclear factor erythroid 2-related factor 2 (Nrf2) (Tang et al., 2022). Also, through the induction of cell death in periodontal ligament cells (PDLs) by H2O2 production, S. gordonii, S. mitis, S. sanguinis and S. sobrinus play their part in the development of apical periodontitis (Park et al., 2021). In S. sanguinis, Spx was a prerequisite for bacterial survival in blood and an spx-deletion mutant failed to induce cell death and NET formation in neutrophils (Sumioka et al., 2017). Through the release of H2O2, oral streptococci can additionally dampen the proinflammatory immune response to LPS and F. nucleatum thereby influencing overall plaque development (Tang et al., 2022).
Genome mining studies have revealed an abundance of bacterial biosynthetic gene clusters (BGCs) in the oral microbiome producing small molecules and secondary metabolites that can function as signaling molecules and serve as language of interbacterial, interspecies and interkingdom communication (Donia and Fischbach, 2015; Liu et al., 2016; Aleti et al., 2019). Among these secondary metabolites are mutanobactins, mutanamide and mutanofactins - lipopeptides produced by S. mutans along a non-ribosomal peptide synthetase–polyketide synthase (NRPS–PKS) assembly line (Wu et al., 2010; Zvanych et al., 2015; Li et al., 2021). Mutanobactins act as interkingdom signaling molecules by blunting hyphae formation and inhibiting biofilm formation by the opportunistic pathogen C. albicans and as interspecies communicator by inhibiting planktonic growth of other oral bacteria (Joyner et al., 2010; Wu et al., 2010; Wang et al., 2012; Pultar et al., 2021). Given that, co-infection with S. mutans and C. albicans not only resulted in a higher bacterial-fungal burden, but also increased biofilm virulence and led to more severe carious lesion in a rat model (Falsetta et al., 2014), hyphae blunting by mutanobactin represents only a small facet of this highly complex interkingdom interaction and the implications of this process in vivo have yet to be determined. Interestingly, Zvanych et al. attested to the mutanobactins also immunomodulatory properties, since in a murine macrophage cell line pre-stimulated with LPS, mutanobactin B upregulated IL-6 and IL-12 and downregulated MCP-1, G-CSF and TNF-α production (Zvanych et al., 2015). Another small molecule produced by S. mutans with the potential to inhibit competing oral bacteria, is the tetramic acid mutanocyclin (Tang et al., 2020). In a Matrigel plug assay, mutanocyclin was found to inhibit CD45+ leukocyte infiltration thereby exerting an anti-inflammatory role (Hao et al., 2019).
For the development of microbial communities, quorum sensing (QS) is an indispensable mechanism of bacteria to regulate inter- and intra-species interactions. S. mutans utilizes multiple QS systems to regulate biofilm formation, transfer of genetic material and stress tolerance responses; one of these systems relies on the competence stimulating peptide (CSP) encoded by the comCDE gene locus (Bernabè et al., 2022). Medapati and coworkers recently explored the possibility that S. mutans CSPs might be recognized by bitter taste receptors (T2Rs) - G-protein coupled receptors (GPCRs) involved in taste chemosensation and recognition of bacterial QS molecules – thereby participating in innate immune responses to bacterial colonization (Medapati et al., 2021). The researchers demonstrated binding of CSP-1 to T2R14 expressed in gingival epithelial cells (GECs) and mediating activation of IL-6, IL-8 and TNF-α production, thus, identifying a novel mechanism of host-QS interaction and a potential target for therapeutic intervention (Medapati et al., 2021).
In multispecies communities, interactions between species – through synergistic or antagonistic effects - can be a decisive factor when it comes to survival and persistence within the host. In the context of periodontal disease, for instance, P. gingivalis can subvert the host immune responses through numerous mechanisms, enabling other members of the biofilm community to escape immune surveillance and increase in cell numbers (Hajishengallis and Lamont, 2014). P. gingivalis and other late biofilm colonizers require the presence of streptococci and F. nucleatum in order to establish themselves within the biofilm community profiting from reduced oxygen tension and the provision of metabolites and nutrients by the antecedent colonizers (Kuboniwa and Lamont, 2010).
Due to their co-adhesion with P. gingivalis, MGS can be classified as accessory pathogens and contribute to the formation of pathogenic oral communities (Whitmore and Lamont, 2011). The formation of heterotypic communities between the different species has been extensively studied for S. gordonii and S. oralis with P. gingivalis. Cohesion occurs through the binding between P. gingivalis fimbriae Mfa1 and FimA to the streptococcal surface antigens SspA/B and surface-expressed glyceraldehyde-3-phosphate dehydrogenase (GAPDH), respectively (Bergmann et al., 2004; Kuboniwa and Lamont, 2010). Upon binding, a signaling cascade is triggered in P. gingivalis, leading to the suppression of Mfa1 production and the expression of a protein tyrosine phosphatase Ltp1 which regulates protease activity and thereby influences the bacterium’s pathogenic potential (Maeda et al., 2008; Kuboniwa and Lamont, 2010; Hajishengallis and Lamont, 2014).
Another species that S. gordonii is frequently associated with is F. nucleatum (Mutha et al., 2018). Co-aggregation not only results in transcriptional changes within both bacteria, but also affects their survival within macrophages and decreases the expression of pro-inflammatory cytokines and thereby influences bacterial virulence and host persistence (Liu et al., 2021). Also the commensal S. cristatus dampens the response to F. nucleatum infection by inhibiting NF-κB and IL-8 production in oral epithelial cells (Zhang et al., 2008; Zhang et al., 2011).
Co-infection studies with S. oralis and C. albicans demonstrated that in an oral thrush mouse model, C. albicans augments streptococcal colonization and that, in turn, S. oralis contributes to the exacerbation of the immune response through TLR2 signaling and neutrophil recruitment causing more severe lesions than the opportunistic fungal pathogen alone (Xu et al., 2014). In epithelial tissues, the synergy between the two bacteria caused upregulation of pro-inflammatory markers (Martorano-Fernandes et al., 2023) and the disruption of epithelial barrier integrity through activation of calpain 1, a calcium ion–dependent cysteine protease specifically cleaving E-cadherin (Xu et al., 2016). Interaction with the allegedly harmless commensal S. oralis therefore seems to be a key contributor to C. albicans virulence and tissue invasion.
When oral keratinocytes were precultured with S. cristatus, S. salivarius, S. mitis or S. sanguinis, the presence of the oral streptococci inhibited keratinocyte colonization by A. actinomycetemcomitans, with S. sanguinis exerting the strongest inhibitory effect (Teughels et al., 2007; Sliepen et al., 2009b). In a similar experiment, adhesion of periodontal pathogens, including P. gingivalis, Prevotella intermedia and A. actinomycetemcomitans, to the surface of a parallel plate flow chamber was found to be reduced in the presence of a similar set of commensal streptococci, identifying antagonistic functions that could render these bacteria suitable candidates as probiotics in the prevention of periodontal disease recurrence (Van Hoogmoed et al., 2008). Different strains of S. salivarius alone reduced the secretion of IL-6 and IL-8 by gingival fibroblasts, which is usually induced by P. gingivalis, A. actinomycetemcomitans and F. nucleatum, both in co-infection studies and when fibroblasts were pre-treated with the commensal before pathogen stimulation (MacDonald et al., 2021). Notably, the tested S. salivarius strains did not elicit enhanced pro-inflammatory cytokine secretion or microbiome alterations in healthy volunteers when administered in the form of a chewing gum and therefore might be interesting candidates for a probiotic therapy of periodontal disease (MacDonald et al., 2021).
The capacity of oral streptococci to modulate the tissue response to oral pathogens has also been discussed in the context of probiotic treatment of oral biofilm disease. Detected solely in samples from caries- and periodontitis-free patients, the rather recently discovered Streptococcus dentisani might be a potential candidate probiotic agent (Esteban-Fernández et al., 2019; Ferrer et al., 2020). In vitro studies showed that S. dentisani induced secretion of anti-inflammatory cytokine IL-10 in gingival fibroblasts, reduced the pro-inflammatory response to P. gingivalis and F. nucleatum, and inhibited growth as well as colonization of fibroblasts by these Gram-negative bacteria (Esteban-Fernández et al., 2019). This highlights the role of oral streptococci in maintaining oral health through their interaction with the host immune system and exertion of antagonistic effects towards pathogenic species.
Streptococci have a profound and life-long presence in humans, being the first colonizers in the oral cavity after birth and continuing to shape the intricate microbiota within our mouth throughout our life. Streptococcus species are commensal inhabitants, influencing the development of a properly functioning immune system and acting as barriers against the colonization of harmful bacteria – functions that are crucial for maintaining oral health and preventing disease. On the other hand, when things go awry, streptococci can become detrimental, potentially leading to serious infections and health complications, including infective endocarditis, purulent infections, brain hemorrhage, intestinal inflammation, autoimmune diseases, and bacteremia (Yumoto et al., 2019). A key characteristic in these processes is the ability of these species to adhere to numerous substrates present on host cell surfaces and within the dental pellicle and to form biofilms (Figure 1). This is facilitated through specialized surface proteins such as AgI/I, Srr proteins, Cnm and RGP formation through the action of Gtfs (Nobbs et al., 2009; Nakano et al., 2011) (Figure 2). Also, in the interaction with the host immune system these factors play a vital role by modulating the induction of proinflammatory immune responses, bacterial cell invasion and adhesion. In the context of therapeutic approaches to dental caries, the most researched virulence antigens of S. mutans are AgI/II, Gtfs and Gbps (Patel, 2020). RGP and LTA as constituents of the cell wall act as potent inducers of pro-inflammatory cytokine production and confer resistance to phagocytosis (Tsuda et al., 2000; Engels-Deutsch et al., 2003). In S. sanguis and S. mutans, SWAN confers resistance to NET killing and therefore enables these bacteria to evade immune defense mechanisms (Liu et al., 2017; Kim et al., 2023). Through the production of H2O2 commensal oral streptococci antagonize pathogens such as S. mutans or A. actinomycetemcomitans (Baty et al., 2022). While commensal streptococci such as S. mitis or S. salivarius can downregulate cytokine production in response to challenge with periodontal pathogens and thereby counteract gingival inflammation, others such as S. gordonii can be regarded as accessory pathogens and support colonization by periodontal pathogens.
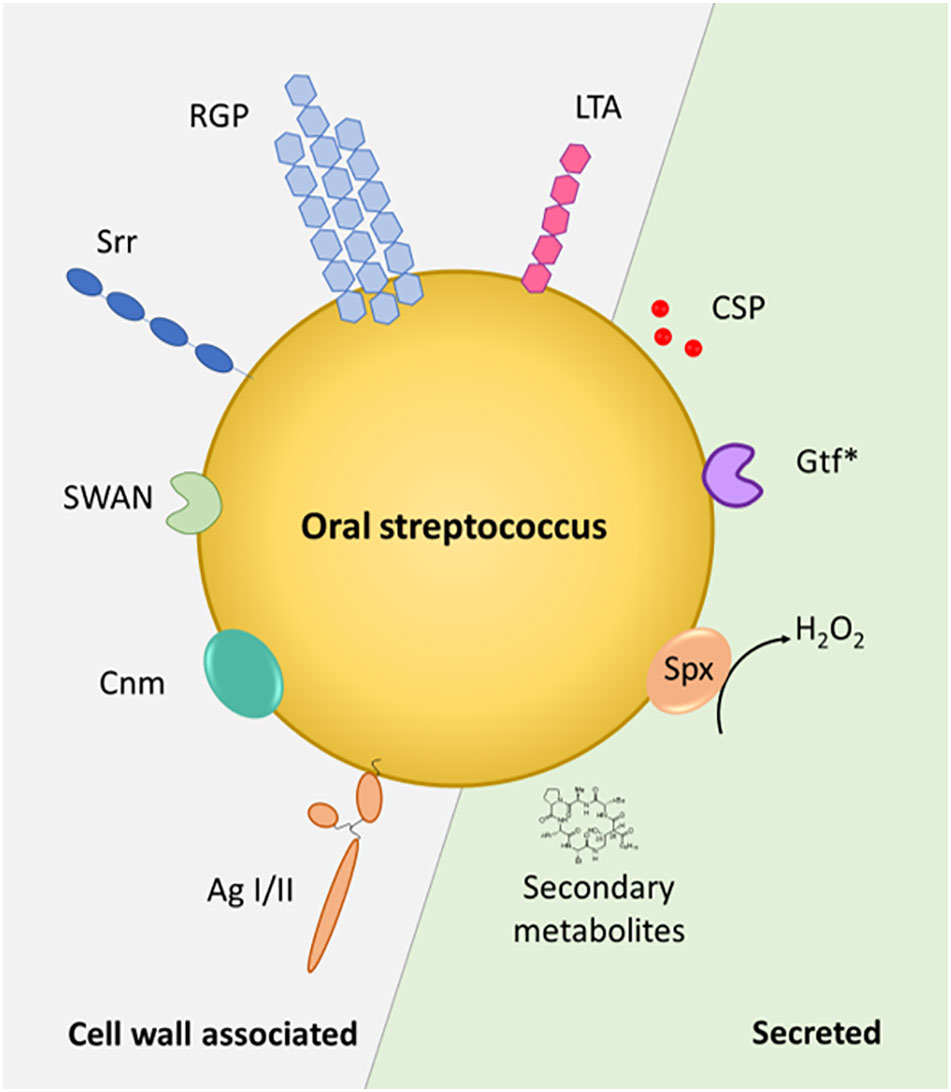
Figure 2 Streptococcal factors at the host interface. Cell wall-associated and secreted factors are depicted in the schematic representation of an oral Streptococcus sp. Gtf*, glycosyltransferases - GtfB and GtfC are cell wall associated, GtfD is secreted – Spx – streptococcal pyruvate oxidase. AgI/II – antigen I/II, Srr – serine-rich repeat protein, CSP – competence stimulating peptide, SWAN – cell-wall associated nuclease, RGP – rhamnose-glucose polymer, LTA – lipoteichoic acid, Cnm - surface glycoprotein Cnm with collagen- and laminin- binding capability.
For vaccine development and caries treatment, the identification of novel streptococcal virulence factors is a central prerequisite. Understanding virulence mechanisms and interactions within the microbial networks in oral biofilms will help to develop targeted therapeutic or preventative measures to decrease the burden of oral diseases. Promising results achieved with the use of health-associated streptococci as probiotics such as S. salivarius or S. dentisani highlight the importance of oral streptococci in oral microbial interaction networks (Ferrer et al., 2020; MacDonald et al., 2021). Streptococci are essential in the maintenance of homeostasis and controlling the composition of the oral microbiota, but their role in regulating host-microbiome interaction is still to be unraveled. The host response regulation exerted by these bacterial species could be a key factor and future studies on this topic will allow a better understanding of oral streptococci and their roles in oral health and disease.
SB: Conceptualization, Formal Analysis, Writing – original draft, Writing – review & editing. FH-M: Formal Analysis, Writing – original draft, Writing – review & editing, Funding acquisition. OA: Writing – original draft, Writing – review & editing, Funding acquisition. CS: Writing – original draft, Writing – review & editing, Funding acquisition, Conceptualization, Formal Analysis, Project administration, Supervision.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Financial support came from the Austrian Science Fund FWF, projects P 32521, P 33618 and P 36398 (to CS) and P 34642 (to OA and CS), the Vienna Science and Technology Fund WWTF project LS21-007 (to CS), and the Hochschuljubiläumsstiftung der Stadt Wien, project H85594/2020 (to FH-M). The funders had no role in the design of the review article, analyses or interpretation of data, in the writing of the manuscript or in the decision to write this review.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aas, J. A., Paster, B. J., Stokes, L. N., Olsen, I., Dewhirst, F. E. (2005). Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43, 5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005
Abranches, J., Miller, J. H., Martinez, A. R., Simpson-Haidaris, P. J., Burne, R. A., Lemos, J. A. (2011). The collagen-binding protein Cnm is required for Streptococcus mutans adherence to and intracellular invasion of human coronary artery endothelial cells. Infect.Immun. 79, 2277–2284. doi: 10.1128/IAI.00767-10
Abranches, J., Zeng, L., Kajfasz, J. K., Palmer, S. R., Chakraborty, B., Wen, Z. T., et al. (2018). Biology of oral streptococci. Microbiol. Spectr. 6, GPP3-0042-2018. doi: 10.1128/microbiolspec.GPP3-0042-2018
Abusleme, L., Dupuy, A. K., Dutzan, N., Silva, N., Burleson, J. A., Strausbaugh, L. D., et al. (2013). The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 7, 1016–1025. doi: 10.1038/ismej.2012.174
Aleti, G., Baker, J. L., Tang, X., Alvarez, R., Dinis, M., Tran, N. C., et al. (2019). Identification of the bacterial biosynthetic gene clusters of the oral microbiome illuminates the unexplored social language of bacteria during health and disease. mBio 10, e00321-19. doi: 10.1128/mBio.00321-19
Al-Okla, S., Chatenay-Rivauday, C., Klein, J.-P., Wachsmann, D. (1999). Involvement of α5β1 integrins in interleukin 8 production induced by oral viridans streptococcal protein I/IIf in cultured endothelial cells. Cell. Microbiol. 1, 157–168. doi: 10.1046/j.1462-5822.1999.00016.x
Alves, L. A., de Carli, T. R., Harth-Chu, E. N., Mariano, F. S., Höfling, J. F., Stipp, R. N., et al. (2019). Oral streptococci show diversity in resistance to complement immunity. J. Med. Microbiol. 68, 600–608. doi: 10.1099/jmm.0.000955
Alves, L. A., Nomura, R., Mariano, F. S., Harth-Chu, E. N., Stipp, R. N., Nakano, K., et al. (2016). CovR regulates Streptococcus mutans susceptibility to complement immunity and survival in blood. Infect. Immun. 84, 3206–3219. doi: 10.1128/IAI.00406-16
Aviles-Reyes, A., Miller, J. H., Simpson-Haidaris, P. J., Hagen, F. K., Abranches, J., Lemos, J. A. (2014). Modification of Streptococcus mutans Cnm by PgfS contributes to adhesion, endothelial cell invasion, and virulence. J. Bacteriol. 196, 2789–2797. doi: 10.1128/JB.01783-14
Baker, J. L., Saputo, S., Faustoferri, R. C., Quivey, R. G., Jr. (2020). Streptococcus mutans SpxA2 relays the signal of cell envelope stress from LiaR to effectors that maintain cell wall and membrane homeostasis. Mol. Oral Microbiol. 35, 118–128. doi: 10.1111/omi.12282
Baty, J. J., Stoner, S. N., Scoffield, J. A. (2022). Oral commensal streptococci: Gatekeepers of the oral cavity. J. Bacteriol. 204, e0025722. doi: 10.1128/jb.00257-22
Benabdelmoumene, S., Dumont, S., Petit, C., Poindron, P., Wachsmann, D., Klein, J. P. (1991). Activation of human monocytes by Streptococcus mutans serotype f polysaccharide: immunoglobulin G Fc receptor expression and tumor necrosis factor and interleukin-1 production. Infect. Immun. 59, 3261–3266. doi: 10.1128/iai.59.9.3261-3266.1991
Bensing, B. A., Khedri, Z., Deng, L., Yu, H., Prakobphol, A., Fisher, S. J., et al. (2016). Novel aspects of sialoglycan recognition by the Siglec-like domains of streptococcal SRR glycoproteins. Glycobiology 26, 1222–1234. doi: 10.1093/glycob/cww042
Bergmann, S., Rohde, M., Hammerschmidt, S. (2004). Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus pneumoniae is a surface-displayed plasminogen-binding protein. Infect. Immun. 72, 2416–2419. doi: 10.1128/IAI.72.4.2416-2419.2004
Bernabè, G., Pauletto, A., Zamuner, A., Cassari, L., Castagliuolo, I., Brun, P., et al. (2022). Exploiting conserved quorum sensing signals in Streptococcus mutans and Streptococcus pneumoniae. Microorganisms 10, 2386. doi: 10.3390/microorganisms10122386
Bloch, S., Tomek, M. B., Friedrich, V., Messner, P., Schäffer, C. (2019). Nonulosonic acids contribute to the pathogenicity of the oral bacterium Tannerella forsythia. Interface Focus 9, 20180064. doi: 10.1098/rsfs.2018.0064
Bloch, S., Zwicker, S., Bostanci, N., Sjöling, A., Bostrom, E. A., Belibasakis, G. N., et al. (2018). Immune response profiling of primary monocytes and oral keratinocytes to different Tannerella forsythia strains and their cell surface mutants. Mol. Oral Microbiol. 33, 155–167. doi: 10.1111/omi.12208
Bowen, W. H. (2016). Dental caries - not just holes in teeth! A perspective. Mol. Oral Microbiol. 31, 228–233. doi: 10.1111/omi.12132
Bowen, W. H., Burne, R. A., Wu, H., Koo, H. (2018). Oral biofilms: pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol. 26, 229–242. doi: 10.1016/j.tim.2017.09.008
Brady, L. J., Maddocks, S. E., Larson, M. R., Forsgren, N., Persson, K., Deivanayagam, C. C., et al. (2010). The changing faces of Streptococcus antigen I/II polypeptide family adhesins. Mol. Microbiol. 77, 276–286. doi: 10.1111/j.1365-2958.2010.07212.x
Chamat-Hedemand, S., Dahl, A., Østergaard, L., Arpi, M., Fosbøl, E., Boel, J., et al. (2020). Prevalence of infective endocarditis in streptococcal bloodstream infections is dependent on streptococcal species. Circulation 142, 720–730. doi: 10.1161/CIRCULATIONAHA.120.046723
Chapple, I. L. C., Hirschfeld, J., Kantarci, A., Wilensky, A., Shapira, L. (2023). The role of the host-neutrophil biology. Periodontol. 2000, 1–47. doi: 10.1111/prd.12490
Chatenay-Rivauday, C., Yamodo, I., Sciotti, M. A., Ogier, J. A., Klein, J. P. (1998). The A and the extended V N-terminal regions of streptococcal protein I/IIf mediate the production of tumour necrosis factor alpha in the monocyte cell line THP-1. Mol. Microbiol. 29, 39–48. doi: 10.1046/j.1365-2958.1998.00881.x
Chatenay-Rivauday, C., Yamodo, I., Sciotti, M. A., Troffer-Charlier, N., Klein, J. P., Ogier, J. A. (2000). TNF-alpha release by monocytic THP-1 cells through cross-linking of the extended V-region of the oral streptococcal protein I/II. J. Leukoc. Biol. 67, 81–89. doi: 10.1002/jlb.67.1.81
Chávez de Paz, L., Svensäter, G., Dahlén, G., Bergenholtz, G. (2005). Streptococci from root canals in teeth with apical periodontitis receiving endodontic treatment. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 100, 232–241. doi: 10.1016/j.tripleo.2004.10.008
Chen, L., Ge, X., Dou, Y., Wang, X., Patel, J. R., Xu, P. (2011). Identification of hydrogen peroxide production-related genes in Streptococcus sanguinis and their functional relationship with pyruvate oxidase. Microbiology 157, 13–20. doi: 10.1099/mic.0.039669-0
Chia, J.-S., Lin, S.-W., Yang, C.-S., Chen, J.-Y. (1997). Antigenicity of a synthetic peptide from glucosyltransferases of Streptococcus mutans in humans. Infect. Immun. 65, 1126–1130. doi: 10.1128/iai.65.3.1126-1130.1997.
Chia, J.-S., You, C.-M., Hu, C.-Y., Chiang, B.-L., Chen, J.-Y. (2001). Human T-cell responses to the glucosyltransferases of Streptococcus mutans. Clin. Diagn. Lab. Immunol. 8, 441–445. doi: 10.1128/CDLI.8.2.441-445.2001
Cho, K., Arimoto, T., Igarashi, T., Yamamoto, M. (2013). Involvement of lipoprotein PpiA of Streptococcus gordonii in evasion of phagocytosis by macrophages. Mol. Oral Microbiol. 28, 379–391. doi: 10.1111/omi.12031
Conrads, G., de Soet, J. J., Song, L., Henne, K., Sztajer, H., Wagner-Döbler, I., et al. (2014). Comparing the cariogenic species Streptococcus sobrinus and Streptococcus mutans on whole genome level. J. Oral Microbiol. 6, 26189. doi: 10.3402/jom.v6.26189
Cosseau, C., Devine, D. A., Dullaghan, E., Gardy, J. L., Chikatamarla, A., Gellatly, S., et al. (2008). The commensal Streptococcus salivarius K12 downregulates the innate immune responses of human epithelial cells and promotes host-microbe homeostasis. Infect. Immun. 76, 4163–4175. doi: 10.1128/IAI.00188-08
Couvigny, B., Lapaque, N., Rigottier-Gois, L., Guillot, A., Chat, S., Meylheuc, T., et al. (2017). Three glycosylated serine-rich repeat proteins play a pivotal role in adhesion and colonization of the pioneer commensal bacterium, Streptococcus salivarius. Environ. Microbiol. 19, 3579–3594. doi: 10.1111/1462-2920.13853
Daep, C. A., Novak, E. A., Lamont, R. J., Demuth, D. R. (2011). Structural dissection and in vivo effectiveness of a peptide inhibitor of Porphyromonas gingivalis adherence to Streptococcus gordonii. Infect. Immun. 79, 67–74. doi: 10.1128/IAI.00361-10
De, A., Liao, S., Bitoun, J. P., Roth, R., Beatty, W. L., Wu, H., et al. (2017). Deficiency of RgpB causes major defects in cell division and biofilm formation, and deficiency of Lytr-Cpsa-Psr family proteins leads to accumulation of cell wall antigens in culture medium by Streptococcus mutans. Appl. Environ. Microbiol. 83, e00928-17. doi: 10.1128/aem.00928-17
Deng, L., Bensing, B. A., Thamadilok, S., Yu, H., Lau, K., Chen, X., et al. (2014). Oral streptococci utilize a Siglec-like domain of serine-rich repeat adhesins to preferentially target platelet sialoglycans in human blood. PloS Pathog. 10, e1004540. doi: 10.1371/journal.ppat.1004540
Diaz, P. I., Chalmers, N. I., Rickard, A. H., Kong, C., Milburn, C. L., Palmer, R. J., Jr., et al. (2006). Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl. Environ. Microbiol. 72, 2837–2848. doi: 10.1128/AEM.72.4.2837-2848.2006
Donia, M. S., Fischbach, M. A. (2015). Small molecules from the human microbiota. Science 349, 1254766. doi: 10.1126/science.1254766
Eberhard, J., Pietschmann, R., Falk, W., Jepsen, S., Dommisch, H. (2009). The immune response of oral epithelial cells induced by single-species and complex naturally formed biofilms. Oral Microbiol. Immunol. 24, 325–330. doi: 10.1111/j.1399-302X.2009.00518.x
Ebersole, J. L., Dawson, D., III, Emecen-Huja, P., Nagarajan, R., Howard, K., Grady, M. E., et al. (2017). The periodontal war: microbes and immunity. Periodontol. 2000 75, 52–115. doi: 10.1111/prd.12222
Engels-Deutsch, M., Pini, A., Yamashita, Y., Shibata, Y., Haikel, Y., Schöller-Guinard, M., et al. (2003). Insertional inactivation of pac and rmlB genes reduces the release of tumor necrosis factor alpha, interleukin-6, and interleukin-8 induced by Streptococcus mutans in monocytic, dental pulp, and periodontal ligament cells. Infect. Immun. 71, 5169–5177. doi: 10.1128/IAI.71.9.5169-5177.2003
Engen, S. A., Schreurs, O., Petersen, F., Blix, I. J. S., Baekkevold, E. S., Schenck, K. (2018). The regulatory role of the oral commensal Streptococcus mitis on human monocytes. Scand. J. Immunol. 87, 80–87. doi: 10.1111/sji.12636
English, B. K., Patrick, C. C., Orlicek, S. L., McCordic, R., Shenep, J. L. (1996). Lipoteichoic acid from viridans streptococci induces the production of tumor necrosis factor and nitric oxide by murine macrophages. J. Infect. Dis. 174, 1348–1351. doi: 10.1093/infdis/174.6.1348
Erttmann, S. F., Gekara, N. O. (2019). Hydrogen peroxide release by bacteria suppresses inflammasome-dependent innate immunity. Nat. Commun. 10, 3493. doi: 10.1038/s41467-019-11169-x
Esteban-Fernández, A., Ferrer, M. D., Zorraquín-Peña, I., López-López, A., Moreno-Arribas, M. V., Mira, A. (2019). In vitro beneficial effects of Streptococcus dentisani as potential oral probiotic for periodontal diseases. J. Periodontol. 90, 1346–1355. doi: 10.1002/JPER.18-0751
Fachon-Kalweit, S., Elder, B. L., Fives-Taylor, P. (1985). Antibodies that bind to fimbriae block adhesion of Streptococcus sanguis to saliva-coated hydroxyapatite. Infect. Immun. 48, 617–624. doi: 10.1128/iai.48.3.617-624.1985
Facklam, R. (2002). What happened to the streptococci: overview of taxonomic and nomenclature changes. Clin. Microbiol. Rev. 15, 613–630. doi: 10.1128/CMR.15.4.613-630.2002
Falsetta, M. L., Klein, M. I., Colonne, P. M., Scott-Anne, K., Gregoire, S., Pai, C. H., et al. (2014). Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect. Immun. 82, 1968–1981. doi: 10.1128/IAI.00087-14
Farges, J.-C., Alliot-Licht, B., Renard, E., Ducret, M., Gaudin, A., Smith, A. J., et al. (2015). Dental pulp defence and repair mechanisms in dental caries. Mediators Inflamm. 2015, 230251. doi: 10.1155/2015/230251
Ferrer, M. D., López-López, A., Nicolescu, T., Perez-Vilaplana, S., Boix-Amorós, A., Dzidic, M., et al. (2020). Topic application of the probiotic Streptococcus dentisani improves clinical and microbiological parameters associated with oral health. Front. Cell. Infect. Microbiol. 10. doi: 10.3389/fcimb.2020.00465
Galler, K. M., Weber, M., Korkmaz, Y., Widbiller, M., Feuerer, M. (2021). Inflammatory response mechanisms of the dentine-pulp complex and the periapical tissues. Int. J. Mol. Sci. 22, 1480. doi: 10.3390/ijms22031480
Ganguly, T., Kajfasz, J. K., Abranches, J., Lemos, J. A. (2020). Regulatory circuits controlling Spx levels in Streptococcus mutans. Mol. Microbiol. 114, 109–126. doi: 10.1111/mmi.14499
Gomez, A., Nelson, K. E. (2017). The oral microbiome of children: development, disease, and implications beyond oral health. Microb. Ecol. 73, 492–503. doi: 10.1007/s00248-016-0854-1
Gourieux, B., Al-Okla, S., Schöller-Guinard, M., Klein, J., Sibilia, J., Wachsmann, D. (2001). Pro-inflammatory cytokine production by synoviocytes following exposure to protein I/II, a modulin from oral streptococci. FEMS Immunol. Med. Microbiol. 30, 13–19. doi: 10.1111/fim.2001.30.issue-1
Graves, D. T., Oates, T., Garlet, G. P. (2011). Review of osteoimmunology and the host response in endodontic and periodontal lesions. J. Oral Microbiol. 3, 5304. doi: 10.3402/jom.v3i0.5304
Haffajee, A., Socransky, S., Patel, M., Song, X. (2008). Microbial complexes in supragingival plaque. Oral Microbiol. Immunol. 23, 196–205. doi: 10.1111/j.1399-302X.2007.00411.x
Hahn, C.-L., Liewehr, F. R. (2007a). Innate immune responses of the dental pulp to caries. J. Endodont. 33, 643–651. doi: 10.1016/j.joen.2007.01.001
Hahn, C.-L., Liewehr, F. R. (2007b). Relationships between caries bacteria, host responses, and clinical signs and symptoms of pulpitis. J. Endodont. 33, 213–219. doi: 10.1016/j.joen.2006.11.008
Hajishengallis, G. (2011). Immune evasion strategies of Porphyromonas gingivalis. J. Oral Biosci. 53, 233–240. doi: 10.1016/S1349-0079(11)80006-X
Hajishengallis, G. (2014). Immunomicrobial pathogenesis of periodontitis: keystones, pathobionts, and host response. Trends Immunol. 35, 3–11. doi: 10.1016/j.it.2013.09.001
Hajishengallis, G., Chavakis, T., Lambris, J. D. (2020). Current understanding of periodontal disease pathogenesis and targets for host-modulation therapy. Periodontol. 2000 84, 14–34. doi: 10.1111/prd.12331
Hajishengallis, G., Darveau, R. P., Curtis, M. A. (2012). The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 10, 717–725. doi: 10.1038/nrmicro2873
Hajishengallis, G., Diaz, P. I. (2020). Porphyromonas gingivalis: Immune subversion activities and role in periodontal dysbiosis. Curr. Oral Health Rep. 7, 12–21. doi: 10.1007/s40496-020-00249-3
Hajishengallis, G., Lamont, R. J. (2012). Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol. Oral Microbiol. 27, 409–419. doi: 10.1111/j.2041-1014.2012.00663.x
Hajishengallis, G., Lamont, R. J. (2014). Breaking bad: manipulation of the host response by Porphyromonas gingivalis. Eur. J. Immunol. 44, 328–338. doi: 10.1002/eji.201344202
Hajishengallis, G., Lamont, R. J. (2016). Dancing with the stars: how choreographed bacterial interactions dictate nososymbiocity and give rise to keystone pathogens, accessory pathogens, and pathobionts. Trends Microbiol. 24, 477–489. doi: 10.1016/j.tim.2016.02.010
Hajishengallis, G., Liang, S., Payne, M. A., Hashim, A., Jotwani, R., Eskan, M. A., et al. (2011). Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe 10, 497–506. doi: 10.1016/j.chom.2011.10.006
Hajishengallis, E., Parsaei, Y., Klein, M. I., Koo, H. (2017). Advances in the microbial etiology and pathogenesis of early childhood caries. Mo.l Oral Microbiol. 32, 24–34. doi: 10.1111/omi.12152
Hall, M. W., Singh, N., Ng, K. F., Lam, D. K., Goldberg, M. B., Tenenbaum, H. C., et al. (2017). Inter-personal diversity and temporal dynamics of dental, tongue, and salivary microbiota in the healthy oral cavity. NPJ Biofilms Microbiomes 3, 2. doi: 10.1038/s41522-016-0011-0
Handley, P. S., Correia, F. F., Russell, K., Rosan, B., DiRienzo, J. M. (2005). Association of a novel high molecular weight, serine-rich protein (SrpA) with fibril-mediated adhesion of the oral biofilm bacterium Streptococcus cristatus. Oral Microbiol. Immunol. 20, 131–140. doi: 10.1111/j.1399-302X.2004.00190.x
Handley, P. S., Harty, D. W., Wyatt, J. E., Brown, C. R., Doran, J. P., Gibbs, A. C. (1987). A comparison of the adhesion, coaggregation and cell-surface hydrophobicity properties of fibrillar and fimbriate strains of Streptococcus salivarius. J. Gen. Microbiol. 133, 3207–3217. doi: 10.1099/00221287-133-11-3207
Hao, T., Xie, Z., Wang, M., Liu, L., Zhang, Y., Wang, W., et al. (2019). An anaerobic bacterium host system for heterologous expression of natural product biosynthetic gene clusters. Nat. Commun. 10, 3665. doi: 10.1038/s41467-019-11673-0
Hasegawa, Y., Mans, J. J., Mao, S., Lopez, M. C., Baker, H. V., Handfield, M., et al. (2007). Gingival epithelial cell transcriptional responses to commensal and opportunistic oral microbial species. Infect. Immun. 75, 2540–2547. doi: 10.1128/IAI.01957-06
Hong, S. W., Baik, J. E., Kang, S. S., Yun, C. H., Seo, D. G., Han, S. H. (2014a). Lipoteichoic acid of Streptococcus mutans interacts with Toll-like receptor 2 through the lipid moiety for induction of inflammatory mediators in murine macrophages. Mol. Immunol. 57, 284–291. doi: 10.1016/j.molimm.2013.10.004
Hong, S. W., Seo, D.-G., Baik, J. E., Cho, K., Yun, C.-H., Han, S. H. (2014b). Differential profiles of salivary proteins with affinity to Streptococcus mutans lipoteichoic acid in caries-free and caries-positive human subjects. Mol. Oral Microbiol. 29, 208–218. doi: 10.1111/omi.12057
Huse, S. M., Ye, Y., Zhou, Y., Fodor, A. A. (2012). A core human microbiome as viewed through 16S rRNA sequence clusters. PloS One 7, e34242. doi: 10.1371/journal.pone.0034242
Jenkinson, H. F., Demuth, D. R. (1997). Structure, function and immunogenicity of streptococcal antigen I/II polypeptides. Mol. Microbiol. 23, 183–190. doi: 10.1046/j.1365-2958.1997.2021577.x
Jenkinson, H. F., Terry, S. D., McNab, R., Tannock, G. W. (1993). Inactivation of the gene encoding surface protein SspA in Streptococcus gordonii DL1 affects cell interactions with human salivary agglutinin and oral actinomyces. Infect. Immun. 61, 3199–3208. doi: 10.1128/iai.61.8.3199-3208.1993
Jespersgaard, C., Hajishengallis, G., Russell, M. W., Michalek, S. M. (2002). Identification and characterization of a nonimmunoglobulin factor in human saliva that inhibits Streptococcus mutans glucosyltransferase. Infect. Immun. 70, 1136–1142. doi: 10.1128/IAI.70.3.1136-1142.2002
Joyner, P. M., Liu, J., Zhang, Z., Merritt, J., Qi, F., Cichewicz, R. H. (2010). Mutanobactin A from the human oral pathogen Streptococcus mutans is a cross-kingdom regulator of the yeast-mycelium transition. Org. Biomol. Chem. 8, 5486–5489. doi: 10.1039/c0ob00579g
Khorasani, M. M., Hassanshahi, G., Brodzikowska, A., Khorramdelazad, H. (2020). Role (s) of cytokines in pulpitis: latest evidence and therapeutic approaches. Cytokine 126, 154896. doi: 10.1016/j.cyto.2019.154896
Kim, A. R., Ahn, K. B., Kim, H. Y., Seo, H. S., Kum, K. Y., Yun, C. H., et al. (2017). Streptococcus gordonii lipoproteins induce IL-8 in human periodontal ligament cells. Mol. Immunol. 91, 218–224. doi: 10.1016/j.molimm.2017.09.009
Kim, S. K., Im, J., Ko, E. B., Lee, D., Seo, H. S., Yun, C. H., et al. (2023). Lipoteichoic acid of Streptococcus gordonii as a negative regulator of human dendritic cell activation. Front. Immunol. 14. doi: 10.3389/fimmu.2023.1056949
Klein, M. I. (2022). “"Oral streptococci,",” in Molecular Typing in Bacterial Infections, Volume I. Ed. de Filippis, I. (Springer International Publishing, Cham), 125–137.
Ko, E. B., Kim, S. K., Seo, H. S., Yun, C. H., Han, S. H. (2017). Serine-rich repeat adhesins contribute to Streptococcus gordonii-induced maturation of human dendritic cells. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00523
Koga, T., Okahashi, N., Takahashi, I., Kanamoto, T., Asakawa, H., Iwaki, M. (1990). Surface hydrophobicity, adherence, and aggregation of cell surface protein antigen mutants of Streptococcus mutans serotype c. Infect. Immun. 58, 289–296. doi: 10.1128/iai.58.2.289-296.1990
Könönen, E., Gursoy, M., Gursoy, U. K. (2019). Periodontitis: A multifaceted disease of tooth-supporting tissues. J. Clin. Med. 8, 1135. doi: 10.3390/jcm8081135
Kreth, J., Vu, H., Zhang, Y., Herzberg, M. C. (2009). Characterization of hydrogen peroxide-induced DNA release by Streptococcus sanguinis and Streptococcus gordonii. J. Bacteriol. 191, 6281–6291. doi: 10.1128/JB.00906-09
Kreth, J., Zhang, Y., Herzberg, M. C. (2008). Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii Interference with Streptococcus mutans. J. Bacteriol. 190, 4632–4640. doi: 10.1128/JB.00276-08
Krisanaprakornkit, S., Kimball, J. R., Weinberg, A., Darveau, R. P., Bainbridge, B. W., Dale, B. A. (2000). Inducible expression of human beta-defensin 2 by Fusobacterium nucleatum in oral epithelial cells: multiple signaling pathways and role of commensal bacteria in innate immunity and the epithelial barrier. Infect. Immun. 68, 2907–2915. doi: 10.1128/IAI.68.5.2907-2915.2000
Kuboniwa, M., Lamont, R. J. (2010). Subgingival biofilm formation. Periodontol. 2000 52, 38–52. doi: 10.1111/(ISSN)1600-0757
Kuboniwa, M., Tribble, G. D., James, C. E., Kilic, A. O., Tao, L., Herzberg, M. C., et al. (2006). Streptococcus gordonii utilizes several distinct gene functions to recruit Porphyromonas gingivalis into a mixed community. Mol. Microbiol. 60, 121–139. doi: 10.1111/j.1365-2958.2006.05099.x
Kuryłek, A., Stasiak, M., Kern-Zdanowicz, I. (2022). Virulence factors of Streptococcus anginosus – a molecular perspective. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.1025136
Lamont, R. J., Gil, S., Demuth, D. R., Malamud, D., Rosan, B. (1994). Molecules of Streptococcus gordonii that bind to Porphyromonas gingivalis. Microbiology 140, 867–872. doi: 10.1099/00221287-140-4-867
Lamont, R. J., Koo, H., Hajishengallis, G. (2018). The oral microbiota: dynamic communities and host interactions. Nat. Rev. Microbiol. 16, 745–759. doi: 10.1038/s41579-018-0089-x
Lang, M. L., Zhu, L., Kreth, J. (2010). Keeping the bad bacteria in check: interactions of the host immune system with oral cavity biofilms. Endodont. Topics 22, 17–32. doi: 10.1111/j.1601-1546.2012.00278.x
Lemos, J. A., Palmer, S. R., Zeng, L., Wen, Z. T., Kajfasz, J. K., Freires, I. A., et al. (2019). The biology of Streptococcus mutans. Microbiol. Spectr. 7, GPP3-0051-2018. doi: 10.1128/microbiolspec.GPP3-0051-2018
Li, Z. R., Sun, J., Du, Y., Pan, A., Zeng, L., Maboudian, R., et al. (2021). Mutanofactin promotes adhesion and biofilm formation of cariogenic Streptococcus mutans. Nat. Chem. Biol. 17 (5), 576–584. doi: 10.1038/s41589-021-00745-2
Lima, A. R., Ganguly, T., Walker, A. R., Acosta, N., Francisco, P. A., Pileggi, R., et al. (2020). Phenotypic and genotypic characterization of Streptococcus mutans strains isolated from endodontic infections. J. Endodont. 46, 1876–1883. doi: 10.1016/j.joen.2020.09.002
Lima, B. P., Kho, K., Nairn, B. L., Davies, J. R., Svensäter, G., Chen, R., et al. (2019). Streptococcus gordonii type I lipoteichoic acid contributes to surface protein biogenesis. mSphere 4, e00814-19. doi: 10.1128/mSphere.00814-19
Liu, L., Hao, T., Xie, Z., Horsman, G. P., Chen, Y. (2016). Genome mining unveils widespread natural product biosynthetic capacity in human oral microbe Streptococcus mutans. Sci. Rep. 6, 37479. doi: 10.1038/srep37479
Liu, J., Sun, L., Liu, W., Guo, L., Liu, Z., Wei, X., et al. (2017). A nuclease from Streptococcus mutans facilitates biofilm dispersal and escape from killing by neutrophil extracellular traps. Front. Cell. Infect. Microbiol. 7. doi: 10.3389/fcimb.2017.00097
Liu, T., Yang, R., Zhou, J., Lu, X., Yuan, Z., Wei, X., et al. (2021). Interactions between Streptococcus gordonii and Fusobacterium nucleatum altered bacterial transcriptional profiling and attenuated the immune responses of macrophages. Front. Cell. Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.783323
Loos, B. G., Van Dyke, T. E. (2020). The role of inflammation and genetics in periodontal disease. Periodontol. 2000 83, 26–39. doi: 10.1111/prd.12297
Love, R. M., McMillan, M. D., Jenkinson, H. F. (1997). Invasion of dentinal tubules by oral streptococci is associated with collagen recognition mediated by the antigen I/II family of polypeptides. Infect. Immun. 65, 5157–5164. doi: 10.1128/iai.65.12.5157-5164.1997
MacDonald, K. W., Chanyi, R. M., Macklaim, J. M., Cadieux, P. A., Reid, G., Burton, J. P. (2021). Streptococcus salivarius inhibits immune activation by periodontal disease pathogens. BMC Oral Health 21, 245. doi: 10.1186/s12903-021-01606-z
Maeda, K., Tribble, G. D., Tucker, C. M., Anaya, C., Shizukuishi, S., Lewis, J. P., et al. (2008). A Porphyromonas gingivalis tyrosine phosphatase is a multifunctional regulator of virulence attributes. Mol. Microbiol. 69, 1153–1164. doi: 10.1111/j.1365-2958.2008.06338.x
Mager, D. L., Ximenez-Fyvie, L. A., Haffajee, A. D., Socransky, S. S. (2003). Distribution of selected bacterial species on intraoral surfaces. J. Clin. Periodontol. 30, 644–654. doi: 10.1034/j.1600-051X.2003.00376.x
Manzer, H. S., Nobbs, A. H., Doran, K. S. (2020). The multifaceted nature of streptococcal antigen I/II proteins in colonization and disease pathogenesis. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.602305
Mark Welch, J. L., Ramírez-Puebla, S. T., Borisy, G. G. (2020). Oral microbiome geography: micron-scale habitat and niche. Cell Host Microbe 28, 160–168. doi: 10.1016/j.chom.2020.07.009
Marsh, P. D., Zaura, E. (2017). Dental biofilm: ecological interactions in health and disease. J. Clin. Periodontol. 44, S12–S22. doi: 10.1111/jcpe.12679
Martin, V., Kleschyov, A. L., Klein, J. P., Beretz, A. (1997). Induction of nitric oxide production by polyosides from the cell walls of Streptococcus mutans OMZ 175, a gram-positive bacterium, in the rat aorta. Infect. Immun. 65, 2074–2079. doi: 10.1128/iai.65.6.2074-2079.1997
Martorano-Fernandes, L., Brito, A. C. M., Araújo, E., Almeida, L., Wei, X.-Q., Williams, D. W., et al. (2023). Epithelial responses and Candida albicans pathogenicity are enhanced in the presence of oral streptococci. Braz. Dent. J. 34, 73–81. doi: 10.1590/0103-6440202305420
Matsushima, H., Kumagai, Y., Vandenbon, A., Kataoka, H., Kadena, M., Fukamachi, H., et al. (2017). Microarray analysis of macrophage response to infection with Streptococcus oralis reveals the immunosuppressive effect of hydrogen peroxide. Biochem. Biophys. Res. Commun. 485, 461–467. doi: 10.1016/j.bbrc.2017.02.048
McNab, R., Holmes, A. R., Clarke, J. M., Tannock, G. W., Jenkinson, H. F. (1996). Cell surface polypeptide CshA mediates binding of Streptococcus gordonii to other oral bacteria and to immobilized fibronectin. Infect. Immun. 64, 4204–4210. doi: 10.1128/iai.64.10.4204-4210.1996
Medapati, M. R., Singh, N., Bhagirath, A. Y., Duan, K., Triggs-Raine, B., Batista, E. L., Jr., et al. (2021). Bitter taste receptor T2R14 detects quorum sensing molecules from cariogenic Streptococcus mutans and mediates innate immune responses in gingival epithelial cells. FASEB J. 35, e21375. doi: 10.1096/fj.202000208R
Miller, J. H., Avilés-Reyes, A., Scott-Anne, K., Gregoire, S., Watson, G. E., Sampson, E., et al. (2015). The collagen binding protein Cnm contributes to oral colonization and cariogenicity of Streptococcus mutans OMZ175. Infect. Immun. 83, 2001–2010. doi: 10.1128/IAI.03022-14
Mira, A. (2018). Oral microbiome studies: potential diagnostic and therapeutic implications. Adv. Dent. Res. 29, 71–77. doi: 10.1177/0022034517737024
Mitchell, J. (2011). Streptococcus mitis: walking the line between commensalism and pathogenesis. Mol. Oral Microbiol. 26, 89–98. doi: 10.1111/j.2041-1014.2010.00601.x
Morita, C., Sumioka, R., Nakata, M., Okahashi, N., Wada, S., Yamashiro, T., et al. (2014). Cell wall-anchored nuclease of Streptococcus sanguinis contributes to escape from neutrophil extracellular trap-mediated bacteriocidal activity. PloS One 9, e103125. doi: 10.1371/journal.pone.0103125
Mutha, N. V. R., Mohammed, W. K., Krasnogor, N., Tan, G. Y. A., Choo, S. W., Jakubovics, N. S. (2018). Transcriptional responses of Streptococcus gordonii and Fusobacterium nucleatum to coaggregation. Mol. Oral Microbiol. 33, 450–464. doi: 10.1111/omi.12248
Myers, S., Do, T., Meade, J. L., Tugnait, A., Vernon, J. J., Pistolic, J., et al. (2021). Immunomodulatory streptococci that inhibit CXCL8 secretion and NFkappaB activation are common members of the oral microbiota. J. Med. Microbiol. 70, 1329. doi: 10.1099/jmm.0.001329
Nagata, E., Okayama, H., Ito, H.-O., Yamashita, Y., Inoue, M., Oho, T. (2006). Serotype-specific polysaccharide of Streptococcus mutans contributes to infectivity in endocarditis. Oral Microbiol. Immunol. 21, 420–423. doi: 10.1111/j.1399-302X.2006.00317.x
Naka, S., Hatakeyama, R., Takashima, Y., Matsumoto-Nakano, M., Nomura, R., Nakano, K. (2016). Contributions of Streptococcus mutans Cnm and PA antigens to aggravation of non-alcoholic steatohepatitis in mice. Sci. Rep. 6, 36886. doi: 10.1038/srep36886
Naka, S., Matsuoka, D., Goto, K., Misaki, T., Nagasawa, Y., Ito, S., et al. (2022). Cnm of Streptococcus mutans is important for cell surface structure and membrane permeability. Front. Cell. Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.994014
Naka, S., Nomura, R., Takashima, Y., Okawa, R., Ooshima, T., Nakano, K. (2014). A specific Streptococcus mutans strain aggravates non-alcoholic fatty liver disease. Oral Dis. 20, 700–706. doi: 10.1111/odi.12191
Naka, S., Wato, K., Hatakeyama, R., Okawa, R., Nomura, R., Nakano, K. (2018). Longitudinal comparison of Streptococcus mutans-induced aggravation of non-alcoholic steatohepatitis in mice. J. Oral Microbiol. 10, 1428005. doi: 10.1080/20002297.2018.1428005
Nakano, K., Hokamura, K., Taniguchi, N., Wada, K., Kudo, C., Nomura, R., et al. (2011). The collagen-binding protein of Streptococcus mutans is involved in haemorrhagic stroke. Nat. Commun. 2, 485. doi: 10.1038/ncomms1491
Nakano, K., Nomura, R., Taniguchi, N., Lapirattanakul, J., Kojima, A., Naka, S., et al. (2010). Molecular characterization of Streptococcus mutans strains containing the cnm gene encoding a collagen-binding adhesin. Arch. Oral Biol. 55, 34–39. doi: 10.1016/j.archoralbio.2009.11.008
Nakano, K., Ooshima, T. (2009). Serotype classification of Streptococcus mutans and its detection outside the oral cavity. Future Microbiol. 4, 891–902. doi: 10.2217/fmb.09.64
Narayanan, L. L., Vaishnavi, C. (2010). Endodontic microbiology. J. Conserv. Dent. 13, 233–239. doi: 10.4103/0972-0707.73386
Negrini, T. C., Duque, C., Vizoto, N. L., Stipp, R. N., Mariano, F. S., Höfling, J. F., et al. (2012). Influence of VicRK and CovR on the interactions of Streptococcus mutans with phagocytes. Oral Dis. 18, 485–493. doi: 10.1111/j.1601-0825.2011.01896.x
Nobbs, A. H., Jenkinson, H. F., Everett, D. B. (2015). Generic determinants of Streptococcus colonization and infection. Infect. Gen. Evol. 33, 361–370. doi: 10.1016/j.meegid.2014.09.018
Nobbs, A. H., Lamont, R. J., Jenkinson, H. F. (2009). Streptococcus adherence and colonization. Microbiol. Mol. Biol. Rev. 73, 407–450. doi: 10.1128/MMBR.00014-09
Nomura, R., Naka, S., Nemoto, H., Inagaki, S., Taniguchi, K., Ooshima, T., et al. (2013). Potential involvement of collagen-binding proteins of Streptococcus mutans in infective endocarditis. Oral Dis. 19, 387–393. doi: 10.1111/odi.12016
Okahashi, N., Nakata, M., Kuwata, H., Kawabata, S. (2016). Streptococcus oralis induces lysosomal impairment of macrophages via bacterial hydrogen peroxide. Infect. Immun. 84, 2042–2050. doi: 10.1128/IAI.00134-16
Okahashi, N., Nakata, M., Kuwata, H., Kawabata, S. (2022). Oral mitis group streptococci: A silent majority in our oral cavity. Microbiol. Immunol. 66, 539–551. doi: 10.1111/1348-0421.13028
Okahashi, N., Nakata, M., Sumitomo, T., Terao, Y., Kawabata, S. (2013). Hydrogen peroxide produced by oral streptococci induces macrophage cell death. PloS One 8, e62563. doi: 10.1371/journal.pone.0062563
Okahashi, N., Okinaga, T., Sakurai, A., Terao, Y., Nakata, M., Nakashima, K., et al. (2011). Streptococcus sanguinis induces foam cell formation and cell death of macrophages in association with production of reactive oxygen species. FEMS Microbiol. Lett. 323, 164–170. doi: 10.1111/fml.2011.323.issue-2
Okahashi, N., Sumitomo, T., Nakata, M., Sakurai, A., Kuwata, H., Kawabata, S. (2014). Hydrogen peroxide contributes to the epithelial cell death induced by the oral mitis group of streptococci. PloS One 9, e88136. doi: 10.1371/journal.pone.0088136
Parducho, K. R., Beadell, B., Ybarra, T. K., Bush, M., Escalera, E., Trejos, A. T., et al. (2020). The antimicrobial peptide human beta-defensin 2 inhibits biofilm production of Pseudomonas aeruginosa without compromising metabolic activity. Front. Immunol. 11. doi: 10.3389/fimmu.2020.00805
Park, O. J., Kim, A. R., So, Y. J., Im, J., Ji, H. J., Ahn, K. B., et al. (2021). Induction of apoptotic cell death by oral streptococci in human periodontal ligament cells. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.738047
Park, O. J., Kwon, Y., Park, C., So, Y. J., Park, T. H., Jeong, S., et al. (2020). Streptococcus gordonii: Pathogenesis and host response to its cell wall components. Microorganisms 8, 1852. doi: 10.3390/microorganisms8121852
Park, O.-J., Yi, H., Jeon, J., Kang, S.-S., Koo, K.-T., Kum, K.-Y., et al. (2015). Pyrosequencing analysis of subgingival microbiota in distinct periodontal conditions. J. Dent. Res. 94, 921–927. doi: 10.1177/0022034515583531
Patel, M. (2020). Dental caries vaccine: are we there yet? Lett. Appl. Microbiol. 70, 2–12. doi: 10.1111/lam.13218
Percy, M. G., Gründling, A. (2014). Lipoteichoic acid synthesis and function in Gram-positive bacteria. Annu. Rev. Microbiol. 68, 81–100. doi: 10.1146/annurev-micro-091213-112949
Pilarczyk-Zurek, M., Sitkiewicz, I., Koziel, J. (2022). The clinical view on Streptococcus anginosus group – Opportunistic pathogens coming out of hiding. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.956677
Plummer, C., Wu, H., Kerrigan, S. W., Meade, G., Cox, D., Ian Douglas, C. W. (2005). A serine-rich glycoprotein of Streptococcus sanguis mediates adhesion to platelets via GPIb. Br. J. Haematol. 129, 101–109. doi: 10.1111/j.1365-2141.2005.05421.x
Pultar, F., Hansen, M. E., Wolfrum, S., Böselt, L., Fróis-Martins, R., Bloch, S., et al. (2021). Mutanobactin D from the human microbiome: Total synthesis, configurational assignment, and biological evaluation. J. Am. Chem. Soc 143, 10389–10402. doi: 10.1021/jacs.1c04825
Ramsey, M. M., Rumbaugh, K. P., Whiteley, M. (2011). Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PloS Pathog. 7, e1002012. doi: 10.1371/journal.ppat.1002012
Ray, C. A., Gfell, L. E., Buller, T. L., Gregory, R. L. (1999). Interactions of Streptococcus mutans fimbria-associated surface proteins with salivary components. Clin. Diagn. Lab. Immunol. 6, 400–404. doi: 10.1128/CDLI.6.3.400-404.1999
Richards, V. P., Alvarez, A. J., Luce, A. R., Bedenbaugh, M., Mitchell, M. L., Burne, R. A., et al. (2017). Microbiomes of site-specific dental plaques from children with different caries status. Infect. Immun. 85, e00106–e00117. doi: 10.1128/IAI.00106-17
Rijn, I. V. D., Bleiweis, A. S. (1973). Antigens of Streptococcus mutans I. Characterization of a serotype-specific determinant from Streptococcus mutans. Infect. Immun. 7, 795–804. doi: 10.1128/iai.7.5.795-804.1973
Rølla, G., Oppermann, R. V., Bowen, W. H., Ciardi, J. E., Knox, K. W. (2009). High amounts of lipoteichoic acid in sucrose-induced plaque in vivo. Caries Res. 14, 235–238. doi: 10.1159/000260459
Ruhl, S., Cisar, J. O., Sandberg, A. L. (2000). Identification of polymorphonuclear leukocyte and HL-60 cell receptors for adhesins of Streptococcus gordonii and Actinomyces naeslundii. Infect. Immun. 68, 6346–6354. doi: 10.1128/IAI.68.11.6346-6354.2000
Sampaio-Maia, B., Monteiro-Silva, F. (2014). Acquisition and maturation of oral microbiome throughout childhood: An update. Dent. Res. J. 11, 291–301.
Scott, D. A., Krauss, J. (2012). Neutrophils in periodontal inflammation. Front. Oral Biol. 15, 56–83. doi: 10.1159/000329672
Shun, C. T., Lu, S. Y., Yeh, C. Y., Chiang, C. P., Chia, J. S., Chen, J. Y. (2005). Glucosyltransferases of viridans streptococci are modulins of interleukin-6 induction in infective endocarditis. Infect. Immun. 73, 3261–3270. doi: 10.1128/IAI.73.6.3261-3270.2005
Silhavy, T. J., Kahne, D., Walker, S. (2010). The bacterial cell envelope. Cold Spring Harbor Perspect. Biol. 2, a000414–a000414. doi: 10.1101/cshperspect.a000414
Silverman, R. J., Nobbs, A. H., Vickerman, M. M., Barbour, M. E., Jenkinson, H. F. (2010). Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect. Immun. 78, 4644–4652. doi: 10.1128/IAI.00685-10
Simon-Soro, A., Ren, Z., Krom, B. P., Hoogenkamp, M. A., Cabello-Yeves, P. J., Daniel, S. G., et al. (2022). Polymicrobial aggregates in human saliva build the oral biofilm. mBio 13, e0013122. doi: 10.1128/mbio.00131-22
Singh, A. K., Woodiga, S. A., Grau, M. A., King, S. J. (2017). Streptococcus oralis neuraminidase modulates aherence to multiple crbohydrates on platelets. Infect. Immun. 85, e00774–e00716. doi: 10.1128/IAI.00774-16
Sitkiewicz, I. (2018). How to become a killer, or is it all accidental? Virulence strategies in oral streptococci. Mol. Oral Microbiol. 33, 1–12. doi: 10.1111/omi.12192
Sliepen, I., Van Damme, J., Van Essche, M., Loozen, G., Quirynen, M., Teughels, W. (2009a). Microbial interactions influence inflammatory host cell responses. J. Dent. Res. 88, 1026–1030. doi: 10.1177/0022034509347296
Sliepen, I., Van Essche, M., Loozen, G., Van Eldere, J., Quirynen, M., Teughels, W. (2009b). Interference with Aggregatibacter actinomycetemcomitans: colonization of epithelial cells under hydrodynamic conditions. Oral Microbiol. Immunol. 24, 390–395. doi: 10.1111/j.1399-302X.2009.00531.x
Soell, M., Holveck, F., Schöller, M., Wachsmann, R. D., Klein, J. P. (1994). Binding of Streptococcus mutans SR protein to human monocytes: production of tumor necrosis factor, interleukin 1, and interleukin 6. Infect. Immun. 62, 1805–1812. doi: 10.1128/iai.62.5.1805-1812.1994
Soell, M., Lett, E., Holveck, F., Schöller, M., Wachsmann, D., Klein, J. P. (1995). Activation of human monocytes by streptococcal rhamnose glucose polymers is mediated by CD14 antigen, and mannan binding protein inhibits TNF-alpha release. J. Immunol. 154, 851–860. doi: 10.4049/jimmunol.154.2.851
Stinson, M. W., Nisengard, R. J., Bergey, E. J. (1980). Binding of streptococcal antigens to muscle tissue in vitro. Infect. Immun. 27, 604–613. doi: 10.1128/iai.27.2.604-613.1980
Sugawara, S., Arakaki, R., Rikiishi, H., Takada, H. (1999). Lipoteichoic acid acts as an antagonist and an agonist of lipopolysaccharide on human gingival fibroblasts and monocytes in a CD14-dependent manner. Infect. Immun. 67, 1623–1632. doi: 10.1128/IAI.67.4.1623-1632.1999
Sugiyama, A., Arakaki, R., Ohnishi, T., Arakaki, N., Daikuhara, Y., Takada, H. (1996). Lipoteichoic acid and interleukin 1 stimulate synergistically production of hepatocyte growth factor (scatter factor) in human gingival fibroblasts in culture. Infect. Immun. 64, 1426–1431. doi: 10.1128/iai.64.4.1426-1431.1996
Sumioka, R., Nakata, M., Okahashi, N., Li, Y., Wada, S., Yamaguchi, M., et al. (2017). Streptococcus sanguinis induces neutrophil cell death by production of hydrogen peroxide. PloS One 12, e0172223. doi: 10.1371/journal.pone.0172223
Takahashi, Y., Konishi, K., Cisar, J. O., Yoshikawa, M. (2002). Identification and characterization of hsa, the gene encoding the sialic acid-binding adhesin of Streptococcus gordonii DL1. Infect. Immun. 70, 1209–1218. doi: 10.1128/IAI.70.3.1209-1218.2002
Takamatsu, D., Bensing, B. A., Cheng, H., Jarvis, G. A., Siboo, I. R., López, J. A., et al. (2005). Binding of the Streptococcus gordonii surface glycoproteins GspB and Hsa to specific carbohydrate structures on platelet membrane glycoprotein Ibalpha. Mol. Microbiol. 58, 380–392. doi: 10.1111/j.1365-2958.2005.04830.x
Tang, X., Kudo, Y., Baker, J. L., LaBonte, S., Jordan, P. A., McKinnie, S. M. K., et al. (2020). Cariogenic Streptococcus mutans produces tetramic acid strain-specific antibiotics that impair commensal colonization. ACS Infect. Dis. 6, 563–571. doi: 10.1021/acsinfecdis.9b00365
Tang, Y. L., Sim, T. S., Tan, K. S. (2022). Oral streptococci subvert the host innate immune response through hydrogen peroxide. Sci. Rep. 12, 656. doi: 10.1038/s41598-021-04562-4
Teughels, W., Kinder Haake, S., Sliepen, I., Pauwels, M., Van Eldere, J., Cassiman, J. J., et al. (2007). Bacteria interfere with A. actinomycetemcomitans colonization. J. Dent. Res. 86, 611–617. doi: 10.1177/154405910708600706
Thurnheer, T., Belibasakis, G. N., Bostanci, N. (2014). Colonisation of gingival epithelia by subgingival biofilms in vitro: Role of "red complex" bacteria. Arch. Oral Biol. 59, 977–986. doi: 10.1016/j.archoralbio.2014.05.023
Tsuda, H., Yamashita, Y., Toyoshima, K., Yamaguchi, N., Oho, T., Nakano, Y., et al. (2000). Role of serotype-specific polysaccharide in the resistance of Streptococcus mutans to phagocytosis by human polymorphonuclear leukocytes. Infect. Immun. 68, 644–650. doi: 10.1128/IAI.68.2.644-650.2000
Urano-Tashiro, Y., Takahashi, Y., Oguchi, R., Konishi, K. (2016). Two arginine residues of Streptococcus gordonii sialic acid-binding adhesin hsa are essential for interaction to host cell receptors. PloS One 11, e0154098. doi: 10.1371/journal.pone.0154098
Vacca-Smith, A. M., Jones, C. A., Levine, M. J., Stinson, M. W. (1994). Glucosyltransferase mediates adhesion of Streptococcus gordonii to human endothelial cells in vitro. Infect. Immun. 62, 2187–2194. doi: 10.1128/iai.62.6.2187-2194.1994
Van Hoogmoed, C. G., Geertsema-Doornbusch, G. I., Teughels, W., Quirynen, M., Busscher, H. J., van der Mei, H. C. (2008). Reduction of periodontal pathogens adhesion by antagonistic strains. Oral Microbiol. Immunol. 23, 43–48. doi: 10.1111/j.1399-302X.2007.00388.x
Vernier, A., Diab, M., Soell, M., Haan-Archipoff, G., Beretz, A., Wachsmann, D., et al. (1996). Cytokine production by human epithelial and endothelial cells following exposure to oral viridans streptococci involves lectin interactions between bacteria and cell surface receptors. Infect. Immun. 64, 3016–3022. doi: 10.1128/iai.64.8.3016-3022.1996
Vernier-Georgenthum, A., al-Okla, S., Gourieux, B., Klein, J. P., Wachsmann, D. (1998). Protein I/II of oral viridans streptococci increases expression of adhesion molecules on endothelial cells and promotes transendothelial migration of neutrophils in vitro. Cell. Immunol. 187, 145–150. doi: 10.1006/cimm.1998.1327
Wang, X., Du, L., You, J., King, J. B., Cichewicz, R. H. (2012). Fungal biofilm inhibitors from a human oral microbiome-derived bacterium. Org. Biomol. Chem. 10, 2044–2050. doi: 10.1039/c2ob06856g
Wang, P. L., Shirasu, S., Daito, M., Ohura, K. (2001). Streptococcus mutans lipoteichoic acid-induced apoptosis in cultured dental pulp cells from human deciduous teeth. Biochem. Biophys. Res. Commun. 281, 957–961. doi: 10.1006/bbrc.2001.4451
Weerkamp, A. H., van der Mei, H. C., Slot, J. (1987). Relationship of cell surface morphology and composition of Streptococcus salivarius K+ to adherence and hydrophobicity. Infect. Immun. 55, 438–445. doi: 10.1128/iai.55.2.438-445.1987
Whitmore, S. E., Lamont, R. J. (2011). The pathogenic persona of community-associated oral streptococci. Mol. Microbiol. 81, 305–314. doi: 10.1111/j.1365-2958.2011.07707.x
Wu, H., Bu, S., Newell, P., Chen, Q., Fives-Taylor, P. (2007). Two gene determinants are differentially involved in the biogenesis of Fap1 precursors in Streptococcus parasanguis. J. Bacteriol. 189, 1390–1398. doi: 10.1128/JB.00836-06
Wu, C., Cichewicz, R., Li, Y., Liu, J., Roe, B., Ferretti, J., et al. (2010). Genomic island TnSmu2 of Streptococcus mutans harbors a nonribosomal peptide synthetase-polyketide synthase gene cluster responsible for the biosynthesis of pigments involved in oxygen and H2O2 tolerance. Appl. Environ. Microbiol. 76, 5815–5826. doi: 10.1128/AEM.03079-09
Xu, H., Sobue, T., Bertolini, M., Thompson, A., Dongari-Bagtzoglou, A. (2016). Streptococcus oralis and Candida albicans synergistically activate μ-Calpain to degrade E-cadherin from oral epithelial junctions. J. Infect. Dis. 214, 925–934. doi: 10.1093/infdis/jiw201
Xu, H., Sobue, T., Thompson, A., Xie, Z., Poon, K., Ricker, A., et al. (2014). Streptococcal co-infection augments Candida pathogenicity by amplifying the mucosal inflammatory response. Cell. Microbiol. 16, 214–231. doi: 10.1111/cmi.12216
Yang, C., Scoffield, J., Wu, R., Deivanayagam, C., Zou, J., Wu, H. (2018). Antigen I/II mediates interactions between Streptococcus mutans and Candida albicans. Mol. Oral Microbiol. 33, 283–291. doi: 10.1111/omi.12223
Yumoto, H., Hirota, K., Hirao, K., Ninomiya, M., Murakami, K., Fujii, H., et al. (2019). The pathogenic factors from oral streptococci for systemic diseases. Int. J. Mol. Sci. 20, 4571. doi: 10.3390/ijms20184571
Zhang, G., Chen, R., Rudney, J. D. (2008). Streptococcus cristatus attenuates Fusobacterium nucleatum-induced interleukin-8 expression in oral epithelial cells. J. Periodont. Res. 43, 408–416. doi: 10.1111/j.1600-0765.2007.01057.x
Zhang, G., Chen, R., Rudney, J. D. (2011). Streptococcus cristatus modulates the Fusobacterium nucleatum-induced epithelial interleukin-8 response through the nuclear factor-kappa B pathway. J. Periodont. Res. 46, 558–567. doi: 10.1111/j.1600-0765.2011.01373.x
Zhu, L., Kreth, J. (2012). The role of hydrogen peroxide in environmental adaptation of oral microbial communities. Oxid. Med. Cell. Longev. 2012, 717843. doi: 10.1155/2012/717843
Keywords: biofilm, host interaction, oral diseases, Streptococcus sp., virulence factors
Citation: Bloch S, Hager-Mair FF, Andrukhov O and Schäffer C (2024) Oral streptococci: modulators of health and disease. Front. Cell. Infect. Microbiol. 14:1357631. doi: 10.3389/fcimb.2024.1357631
Received: 18 December 2023; Accepted: 05 February 2024;
Published: 22 February 2024.
Edited by:
Garrit Koller, King’s College London, United KingdomReviewed by:
Michał Śmiga, University of Wrocław, PolandCopyright © 2024 Bloch, Hager-Mair, Andrukhov and Schäffer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christina Schäffer, Y2hyaXN0aW5hLnNjaGFlZmZlckBib2t1LmFjLmF0; Susanne Bloch, c3VzYW5uZS5ibG9jaEBtZWR1bml3aWVuLmFjLmF0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.