- 1School of Basic Medical Sciences, Health Science Center, Ningbo University, Ningbo, Zhejiang, China
- 2Department of Blood Transfusion, Hubei No. 3 People’s Hospital of Jianghan University, Wuhan, Hubei, China
- 3Department of Rheumatology Immunology, The First People’s Hospital of Hefei, Hefei, Anhui, China
- 4Department of Laboratory, Ningbo No.2 Hospital, Ningbo, Zhejiang, China
- 5National Kunming High-level Biosafety Primate Research Center, Institute of Medical Biology, Chinese Academy of Medical Sciences and Peking Union Medical College, Kunming, Yunnan, China
Acinetobacter baumannii (A. baumannii) is a popular clinical pathogen worldwide. Biofilm-associated antibiotic-resistant A. baumannii infection poses a great threat to human health. Bacteria in biofilms are highly resistant to antibiotics and disinfectants. Furthermore, inhibition or eradication of biofilms in husbandry, the food industry and clinics are almost impossible. Phages can move across the biofilm matrix and promote antibiotic penetration. In the present study, a lytic A. baumannii phage vB_AbaM-SHI, belonging to family Straboviridae, was isolated from sauce chop factory drain outlet in Wuxi, China. The DNA genome consists of 44,180 bp which contain 93 open reading frames, and genes encoding products morphogenesis are located at the end of the genome. The amino acid sequence of vB_AbaM-SHI endolysin is different from those of previously reported A. baumannii phages in NCBI. Phage vB_AbaM-SHI endolysin has two additional β strands due to the replacement of a lysine (K) (in KU510289.1, NC_041857.1, JX976549.1 and MH853786.1) with an arginine (R) (SHI) at position 21 of A. baumannii phage endolysin. Spot test showed that phage vB_AbaM-SHI is able to lyse some antibiotic-resistant bacteria, such as A. baumannii (SL, SL1, and SG strains) and E. coli BL21 strain. Additionally, phage vB_AbaM-SHI independently killed bacteria and inhibited bacterial biofilm formation, and synergistically exerted strong antibacterial effects with antibiotics. This study provided a new perspective into the potential application value of phage vB_AbaM-SHI as an antimicrobial agent.
1 Introduction
Acinetobacter baumannii (A. baumannii) is a gram-negative opportunistic pathogen that mainly causes various infections, such as pneumonia, urinary tract infection, ventilator-acquired pneumonia and bacteremia. This bacterium was widely distributed in water, soil, food, activated sludge, and on human body (Antunes et al., 2014; Potron et al., 2015; Bello-López et al., 2020). With the widespread abuse and misuse of antibiotics, multidrug-resistant A. baumannii has been isolated from patients in hospital since the 1990s (Zheng et al., 2013; Ahmed et al., 2016). Unfortunately, more than 80% A. baumannii possess multidrug resistance (MDR) to antibiotics (such as ampicillin, amoxicillin, clavulanate, cefotaxime, ceftriaxone, aztreonam, ertapenem, trimethoprim, fosfomycin, tetracycline and colistin, etc) due to the presence of multiple genetic elements encoding antimicrobial substance (Lee et al., 2011; Pendleton et al., 2013; Wannigama et al., 2019; Srisakul et al., 2022). Moreover, A. baumannii has a strong ability to form biofilm, which exacerbates antibiotic resistance in surgical devices, catheters and medical implants (Gayoso et al., 2014). The World Health Organization classified A. baumannii as priority pathogen in 2017 because of the lack of new therapies, which highlights the urgent need for the development of novel targeted therapies (Gauba and Rahman, 2023).
Phage endolysin is a cell wall hydrolase that is synthesized by phage at the later stage of phage infection, it has the ability to selectively kill bacteria when applied from outside the cell (Fischetti, 2008; Ghose and Euler, 2020; Cui et al., 2023; Nguyen et al., 2023). The majority of endolysins with reported intrinsic antibacterial activity have a C-terminal amphipathic helix (Gutiérrez and Briers, 2021) and the phage endolysin of gram-negative bacteria is a small globular protein with a single enzyme activity domain (Briers et al., 2007). Given the broad host spectrum, phage endolysins have significant advantages in treating bacterial infections. Wang et al., reported that TSPphg lysin was a potential antimicrobial agent which could inhibit both gram-negative (Salmonella paratyphi B, Escherichia coli O157 and Klebsiella pneumoniae) and gram-positive (Bacillus subtilis and Staphylococcus aureus) bacteria growth (Wang et al., 2020a; Shein et al., 2023).
Bacterial biofilms are composed of bacteria embedded in a self-produced extracellular matrix on nonbiotic and biotic surfaces (Hall-Stoodley et al., 2004; Jakubovics et al., 2021). Bacteria in biofilms are highly resistant to antibiotics and disinfectants (Venkatesan et al., 2015; Oliveira et al., 2019), and the inhibition or eradication of biofilms in husbandry, the food industry and clinics were almost impossible (Penesyan et al., 2015; Kim et al., 2018). Although disinfectants and antibiotics play the important roles in controlling bacterial infections, the emergence of drug-resistant bacteria has changed the dominance of antibiotics (Chapman and Gunter, 2018).
As a virus, phages have the ability to lyse their bacterial hosts (Gutiérrez et al., 2016; Sharma et al., 2017). Phages can also inhibit the formation of biofilms of E. coli, Listeria monocytogenes, Salmonella, and Pseudomonas aeruginosa (de Ornellas Dutka Garcia et al., 2017; Gong and Jiang, 2017; Chegini et al., 2020; Tian et al., 2021). In particular, phages were harmless to humans and safe in the clinic and food products (Ahn et al., 2013). Therefore, phages are expected to become a good alternative or complement to antibiotics (Endersen et al., 2014; Shein et al., 2023).
In the present study, a lytic A. baumannii phage vB_AbaM-SHI was isolated from a sauce chop factory drain outlet in Wuxi, China. The genomic features of phage vB_AbaM-SHI were characterized. The capacity of phage vB_AbaM-SHI to lyse A. baumannii and inhibit the biofilm formation of A. baumannii was explored. Herein, we aimed to determine whether phage vB_AbaM-SHI represents a potential therapeutic agent for multidrug-resistant bacteria A. baumannii.
2 Materials and methods
2.1 A. baumannii strain and cultivation
A. baumannii SL was obtained from the China Microbial Culture Preservation Center (CMCPC) and grown aerobically in brain-heart infusion (BHI) broth culture or on BHI plates (Difco, MI, USA) at 37°C, and used as a host for phage isolation. A. baumannii cpx isolates (SL1, SJ9, SA5, SA3, SG, SH) were obtained from department of clinical laboratory, the first people’s hospital of Yunnan province, China. BHI soft agar containing 0.45% or 0.5% (W/V) agar was used for phage plaque confirmation. A. baumannii stock culture was stored at −80°C with 20% (V/V) glycerol.
2.2 Phage vB_AbaM-SHI isolation and purification
An A. baumannii-targeting phage was isolated from sauce chop factory drain outlet samples in Wuxi, China. Isolation of phage was performed as described in previous study (Park et al., 2012). In brief, 10 g of the sauce chop factory drain outlet sample were mixed with 20 mL of sterile Phosphate Buffered Saline (PBS) and incubated for 2 h at room temperature with shaking at 200 rpm. The sample was centrifuged at 5, 000×g for 15 min, followed by filtration with a 0.22 μm filter membrane. Ten milliliters of the filtrate were added to 40 mL of BHI broth containing a 1:100 A. baumannii SL overnight culture and incubated for 48 h. Afterwards, the culture was centrifuged at 8, 000×g for 15 min, followed by filtration with a 0.22 µm filter membrane. The filtrate was serially diluted (10-fold) and mixed with 6 mL of molten 0.5% BHI soft agar containing A. baumannii SL (2×108 cfu/mL), and then spread onto BHI plate. 24 hours later, the phage plaque was observed by naked eye and a single plaque was selected for phage purification, and the process was repeated for three times.
2.3 Evaluation of the sensitivity of the phage to physicochemical factors
The dilution (1 × 108 pfu mL-1) was performed by referring to the phage vB_AbaM-SHI titer in the storage solution with BHI broth culture. For the thermal stability experiment, one milliliter of diluted phage vB_AbaM-SHI was incubated for 1 h at the temperature of 4°C, 25°C, 37°C, 42°C, 50°C, 60°C or 90°C, respectively. As for the pH stability experiment, 0.99 mL of buffer at pH 3, 4, 5, 6, 7, 8, 9, 10 or 11 (citrate buffer, 50 mmol/L, pH 3, pH 4 or pH 5; phosphate buffer, 50 mmol/L, pH 6, pH 7 or pH 8; Tris-HCl buffer, 50 mmol/L, pH 9; sodium carbonate buffer, 50 mmol/L, pH 10 or pH 11) was mixed with phage vB_AbaM-SHI (1 × 108 pfu mL-1) in a 1.5 mL sterile centrifuge tube and incubated at 20°C for 1 h. As control, we did a complementary experiment of phage killing among the different buffers with the same pH 7. The multiplicity of infection (MOI) indicates the ratio of phage vB_AbaM-SHI to A. baumannii SL during the initial infection. Phage vB_AbaM-SHI stocks were added to the A. baumannii SL culture medium at MOIs of 100, 10, 1, 0.1, 0.01, 0.001 or 0.0001, and then cultivated at 37°C for 12 h. A. baumannii SL culture were centrifuged at 9, 000 × g for 10 min and the supernatant was filtered through a 0.22 µm filter membrane. The phage titer was determined using the double layer agar method as described previously (Park et al., 2012) and expressed as plaque-forming unit (pfu) mL-1. The experiment was repeated for three times.
2.4 Adsorption rate and one-step growth curve
To measure the adsorption rate of phage on host cell, phage vB_AbaM-SHI (1×108 pfu mL-1) and logarithmic phase A. baumannii SL were mixed at a MOI of 0.1 and incubated at 37°C. The phage titer in the supernatant was measured at 0, 2, 4, 6, 8, 10, 15, 20 and 30 min after phage infection. The one-step growth curve of phage vB_AbaM-SHI was carried out as follows. Briefly, 10 mL of exponential phase A. baumannii SL culture was centrifuged (5, 000 g, 4 min, 37°C) and the cell pellet was resuspended in 20 mL of fresh BHI to obtain an OD600 of 1. Next, 20 mL of phage vB_AbaM-SHI was added to the bacteria to reach a MOI of 1 and allowed to adsorb for 10 min at 37°C. The mixture was centrifuged at 5, 000 × g for 4 min at 37°C, and the pellet was resuspended in 10 mL of fresh BHI. Samples were taken up to 90 h, after which the supernatants were plated on BHI agar to determine the phage titer.
2.5 Transmission electron microscopy
The Phage vB_AbaM-SHI particles were analyzed using transmission electron microscopy (TEM). Dilutions of the phage vB_AbaM-SHI stock (4×108 to 4×109 pfu mL-1) were deposited on copper grids with carbon-coated formvar films for 10 min and negatively stained with 2% uranyl acetate (pH 4.0) for 2 min. The phage particles were imaged using a Philips EM 300 electron microscope operated at 80 kV at Wuxi Jiangnan University (China).
2.6 Phage vB_AbaM-SHI genomic DNA extraction
Firstly, the purified phage vB_AbaM-SHI was concentrated through a 10 kDa filter (approximately 109 to 1010 pfu mL-1). Then, the phage vB_AbaM-SHI genomic DNA was extracted using a Takara Minibest Viral RNA/DNA Extraction Kit (Cat#9766). Finally, purified phage vB_AbaM-SHI genomic DNA was treated with RNase at 37°C for 1 h. The restriction endonucleases Hind III, EcoRI, Not I and Xhol I (Takara) were used to digest the phage genomic DNA at 37°C for 1 h.
2.7 Complete genome sequence and bioinformatics analyses of vB_AbaM-SHI
The phage vB_AbaM-SHI genome was sequenced using the Illumina HiSeq platform (Sangon Biotech, China). The high-quality original sequencing data was assembled with de novo assembler software (Roche, Mannheim, Germany) at Sangon Biotech, China and analyzed with SPAdes software. Phage vB_AbaM-SHI genome annotation was carried out using Prokka 1.13.7. PCR amplification was performed based on a pair of primers targeting the start and end positions of the phage genome, respectively, to determine whether the phage genome is linear or circular. The complete genome sequence of phage vB_AbaM-SHI was deposited into the NCBI GenBank database (https://www.ncbi.nlm.nih.gov/nuccore) (GenBank Accession no: ON480525). The protein sequences of phage vB_AbaM-SHI were obtained using functional annotation. The Basic Local Alignment Search Tool (BLAST) was used to compare phage genome sequences from multiple databases including COG, TrEMBL, KOG, PFAM, CDD, NT, NR and SwissProt. Comparative genomic analysis among Acinetobacter phage IME-AB2 (NC_041857.1), Acinetobacter phage vB_AbaM_IME285 (MH853786.1) and Acinetobacter phage vB_AbaM-SHI were performed using the Easyfig 2.2.5 visualization tool (https://mjsull.github.io/Easyfig/). Phage vB_AbaM-SHI was identified and classified according to the International Committee on Taxonomy of Viruses (ICTV) (Adriaenssens and Brister, 2017).
2.8 Phage lysis and antibiotic sensitivity experiments of A. baumannii
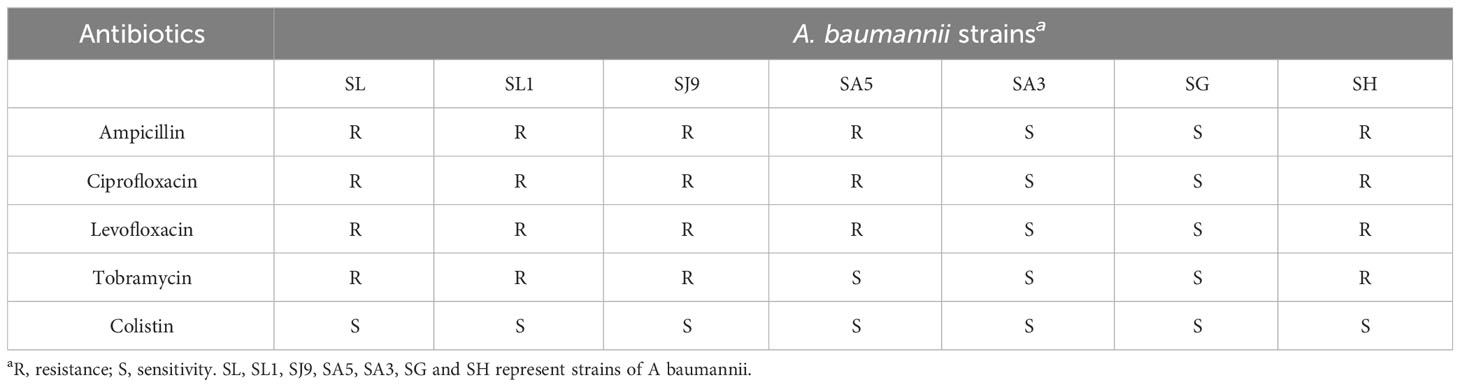
The bacterial spectrum of phage vB_AbaM-SHI was determined by the spot test method. The reference bacterial strains (S. aureus cpx, Salmonella paratyphi A NA3, Escherichia coli BL21, and A. baumannii cpx) were tested for the susceptibility to phage vB_AbaM-SHI. 200 ml of bacteria (109 cfu mL-1) were mixed with 5 ml of molten 0.5% BHI soft agar and spread onto BHI 1.8% (w/w) agar plates [colony-forming unit (cfu)]. Five minutes later, one drop of phage vB_AbaM-SHI suspension was added to the plate and incubated at 37°C for 18 h. The susceptibility of A. baumannii strain to five antibiotics (ampicillin, ciprofloxacin, levofloxacin, tobramycin and colistin) was tested using the minimal inhibitory concentration (MIC) method of serial broth microdilution.
2.9 The effects of phage vB_AbaM-SHI and kanamycin sulfate on biofilms formation
The polylysine-coated cell slides were placed to each well of 6-well plates for scanning electron analysis. The A. baumannii SL stock was inoculated into 100 mL of BHI broth culture at a ratio of 4:1000. Thereafter, 2 mL of the bacterial culture was inoculated into each well of 24-well plates. In one experiment, the test wells were added with phage vB_AbaM-SHI (MOI = 0.1), kanamycin sulfate (10 µg mL-1), or a mixture of kanamycin sulfate and phage vB_AbaM-SHI, whereas the wells receiving only BHI broth culture were used as control. In another experiment, A. baumannii SL inoculated in 24-well plate was cultivated for 12 h, followed by addition of phage vB_AbaM-SHI, kanamycin sulfate, or a mixture of kanamycin sulfate and phage vB_AbaM-SHI. Twelve hours later, the mount of A. baumannii SL in the broth culture was measured using the plate counting method. In addition, the cell slides were washed twice with PBS, fixed with pre-cold 2.5% glutaraldehyde for 2 h, and dehydrated with a gradient of ethanol solutions (15%, 30%, 40%, 50%, 70%, and 100%, respectively) for 15 min. Finally, the bacterial morphology was analyzed using a scanning electron microscope (SEM) after overnight air dry as described previously (Jiang et al., 2021b).
The A. baumannii SL stock was inoculated into BHI broth culture at a proportion of 1:250 and cultivated overnight. The diluted bacterial culture (200-fold) was added to each well of 96-well plates (200 µL/well). In one experiment, phage vB_AbaM-SHI (MOI = 0.1), kanamycin sulfate (10 µg mL-1), or a mixture of kanamycin sulfate and phage vB_AbaM-SHI was added to the test well. The control wells were added with only broth culture. The bacteria were then cultivated at 37°C for 24 h. In another experiment, A. baumannii SL was first cultivated for 12 h, and then added with phage vB_AbaM-SHI, kanamycin sulfate, or a mixture of two components. The wells given with only broth culture worked as control. The bacteria were then cultivated at 37°C for 24 h. The OD600 of each well in the plate was read using microplate reader. The supernatant in the plates was discarded and the plates were washed twice with PBS and fixed with 99% methanol for 10 min. After discarding the methanol, 2% crystal violet solution was added to each well of the plate and incubated for 10 min. The OD570 value of each was measured using microplate reader after rinsing the plate with water.
To determine the ability of phage vB_AbaM-SHI to inhibit the formation of A. baumannii SL biofilms, the cell slides were placed into each well of 12-well plates. A. baumannii SL stock was inoculated into 200 mL of BHI broth culture at a ratio of 4:1000. 1 mL of A. baumannii SL culture was inoculated into each well 12-well plates. The test wells were added with phage vB_AbaM-SHI (MOI = 1) whereas control wells were given with only broth culture. After a 36-h incubation at 37°C, the supernatants in the plates was discarded and the plates were washed twice with PBS and fixed with 99% methanol for 10 min. After discarding the methanol, 2% crystal violet solution was added to each well of the plate and incubated for 10 min. The morphology and amount of biofilms were analyzed using epifluorescence microscopy as described previously (Duan et al., 2023). Three repeated tests were performed. The experiment was repeated three times.
2.10 Tertiary structure prediction and phylogenetic analysis of A. baumannii phage endolysins
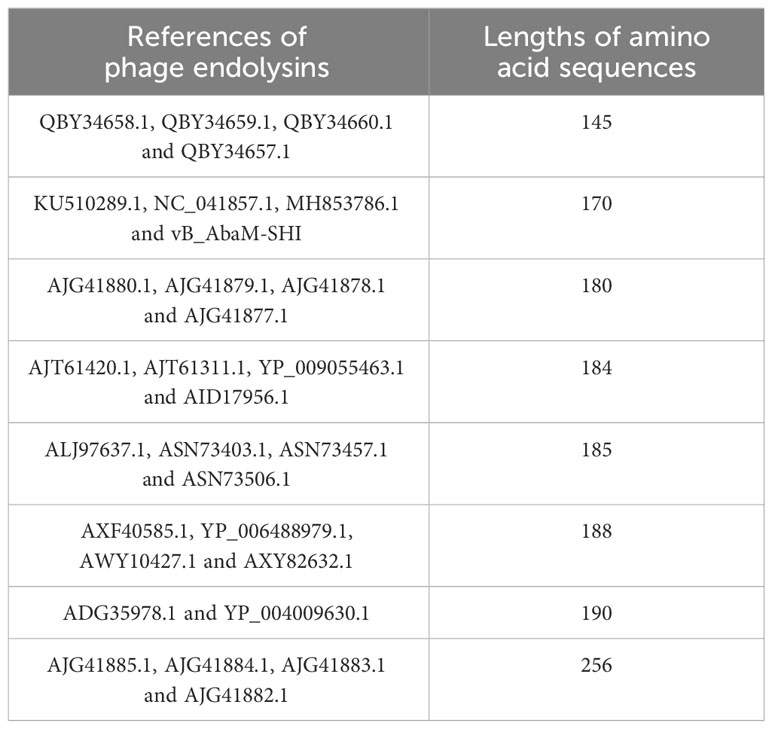
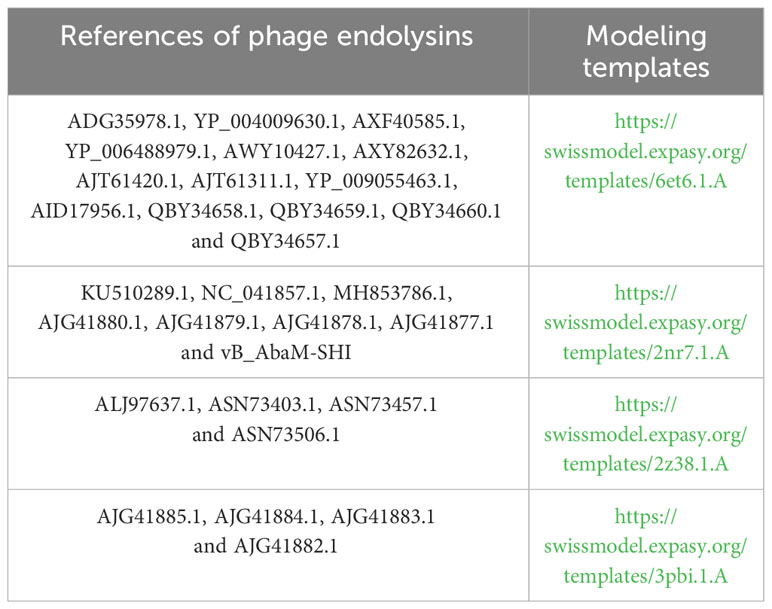
The endolysin amino acid sequences of A. baumannii phage vB_AbaM-SHI, and the other A. baumannii phages (AJG41880.1, AJG41879.1, KU510289.1, AJG41878.1, AJG41877.1, NC_041857.1, ASN73403.1, ASN73457.1, MH853786.1, AJG41885.1, AJG41884.1, ALJ97637.1, ASN73506.1, AJG41883.1, ADG35978.1, YP_004009630.1, AJG41882.1, AXF40585.1, YP_006488979.1, AXY82632.1, AJT61420.1, AWY10427.1, AJT61311.1, YP_009055463.1, QBY34658.1, QBY34659.1, AID17956.1, QBY34657.1 and QBY34660.1) were obtained from the NCBI database (https://www.ncbi.nlm.nih.gov/) and shown in Table 1. Tertiary structure homology modeling of A. baumannii phage endolysins was performed using the SWISS-MODEL online suite and the applied modeling templates were shown in Table 2. The phylogenetic analysis of A. baumannii phage endolysins was performed with the Neighbor-Joining program of Molecular Evolutionary Genetics Analysis (MEGA7.0) from multiple alignments done with MUSCLE (MEGA7.0) (Jiang et al., 2018). The number on the branch represents the bootstrap value for 1000 replicates. The amino acid specificity of endolysin was analyzed using BioEdit software.
2.11 Statistical analyses
Statistical significance was analyzed using SPSS statistical software (ver. 11.5). The killing activity of phage vB_AbaM-SHI in the experiments were expressed as logarithmic reduction ± standard error (Zelver et al., 2001). Multiple t tests were performed to determine differences between groups with a P value of < 0.05 considered significant (*, P <0.05; **, P <0.01; ***, P <0.001).
3 Results
3.1 Biological characteristics of phage vB_AbaM-SHI
Using A. baumannii as host bacterium, we isolated a bacteriophage (named as A. baumannii phage vB_AbaM-SHI) from a sauce chop factory drain outlet in Wuxi, Jiangsu, China. Phage vB_AbaM-SHI formed the plaques with around 5 mm in diameter (Figure 1A). Phage vB_AbaM-SHI DNA was cleaved by restriction endonucleases EcoRI and Hind III, but not by Xhol I and NotI (Figure 1B). TEM images showed that phage vB_AbaM-SHI virions have a tail with a length of around 30 nm and an icosahedral head with an estimated diameter of 60 nm (Figure 1C). It was estimated from the growth curve of phage vB_AbaM-SHI in A. baumannii that the latent period of phage vB_AbaM-SHI is about 50 min, and the burst size of phage vB_AbaM-SHI is approximately 155 pfu/cell. The titers of phage vB_AbaM-SHI increased very quickly from the 4.5 h after initial phage infection, and reached the peak at the 12 h post phage infection. In addition, phage vB_AbaM-SHI has an amplification factor of approximately 5000 times (Figure 1D).
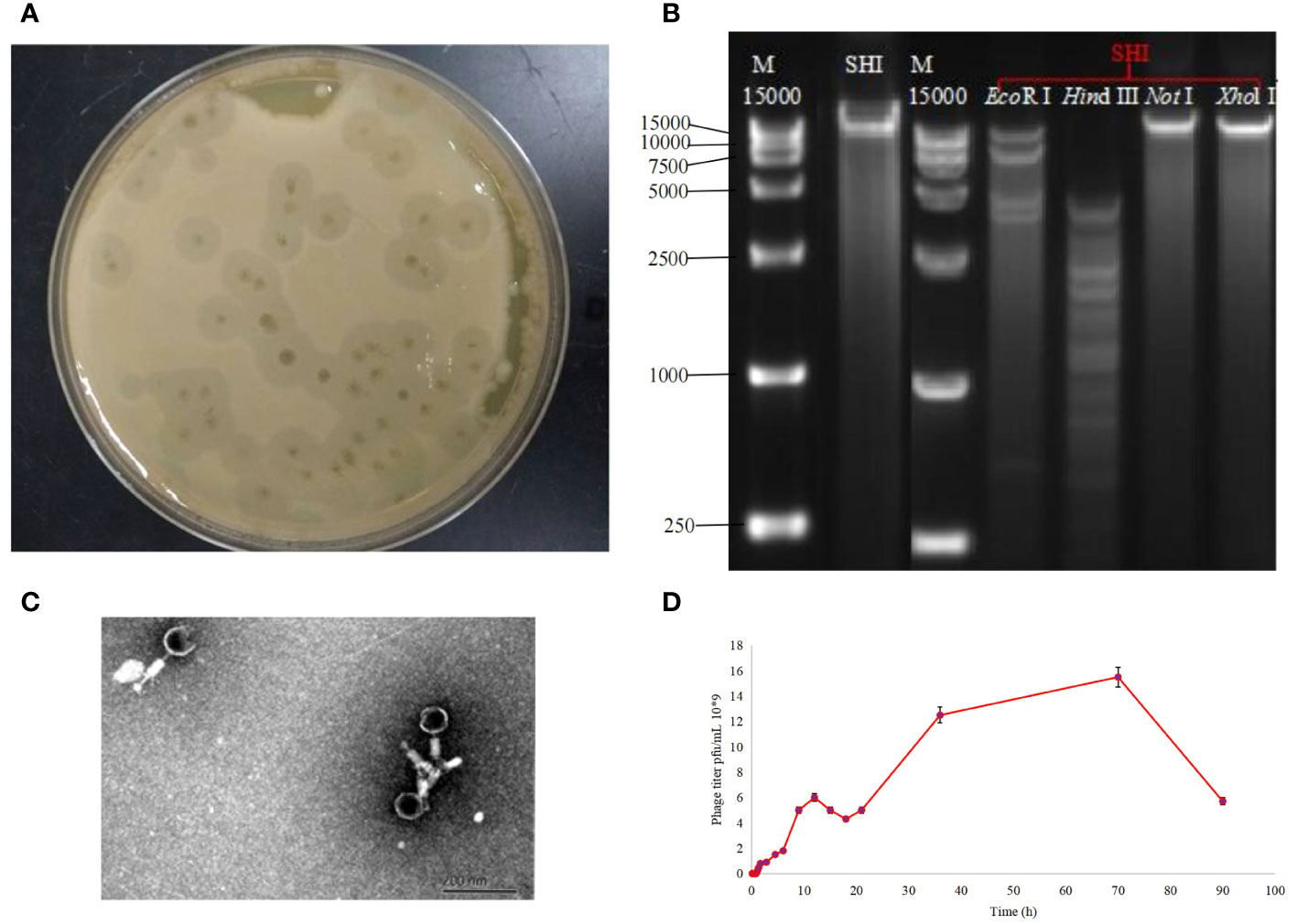
Figure 1 Biological characteristics of phage vB_AbaM-SHI. (A) Bacteriolytic plaques appearing on A. baumannii plates inoculated with phage vB_AbaM-SHI; (B) Phage vB_AbaM-SHI genomic DNA was cleaved by restriction endonucleases EcoRI and Hind III (lanes 4 and 5), but not by NotI and Xhol I (lanes 6 and 7); (C) Morphology of phage vB_AbaM-SHI under transmission electron microscopy (TEM); (D) Growth curve of phage vB_AbaM-SHI propagated in A. baumannii SL. “M”: DNA marker; “SHI”: vB_AbaM-SHI.
We next sent the extracted nucleic acid of phage vB_AbaM-SHI to a company for sequencing. The results showed that this phage genome contains 44,180 bp, with G+C contents of 38.14%. PCR amplification on primers targeting both ends of the genome did not yield products, suggesting that the phage genome should be linear. We identified 93 open reading frames (ORFs) in the complete genome of vB_AbaM-SHI (Figure 2; Supplementary Table S1). Among them, 18 ORFs encode functional proteins which can be categorized into five groups: DNA packaging and nucleic acid metabolism, cell lysis, structure proteins except tail fiber, tail-associated protein and hypothetical protein (Figure 2; Supplementary Table S1). The remaining 75 ORFs encode hypothetical proteins with unknown functions (Supplementary Table S1). In addition, no homologous sequences highly similar to published antimicrobial resistant genes (ARGs) or phage virulence factors were identified in the genome of phage vB_AbaM-SHI. Based on these facts, namely that this phage can efficiently lyse host bacteria, and there is a lack of integrase and transposon genes related to the entry of the phage into the lysogenic state in the genome of phage vB_AbaM-SHI, we carefully speculate that this phage should belong to lytic phage rather than lysogenic phage. The whole-genome alignment shows that vB_AbaM-SHI is most similar to Acinetobacter phage IME-AB2 and Acinetobacter phage vB_AbaM_IME285 in terms of genomes and module conformation (Figure 3). These three phages have more than 50% nucleotide identity, similar tRNA numbers, and similar G + C% in the genome. Therefore, according to the current criteria of the International Committee on Taxonomy of Viruses (ICTV) (Peng et al., 2014; Wang et al., 2020b), phage vB_AbaM_IME285 can be classified as family Straboviridae.
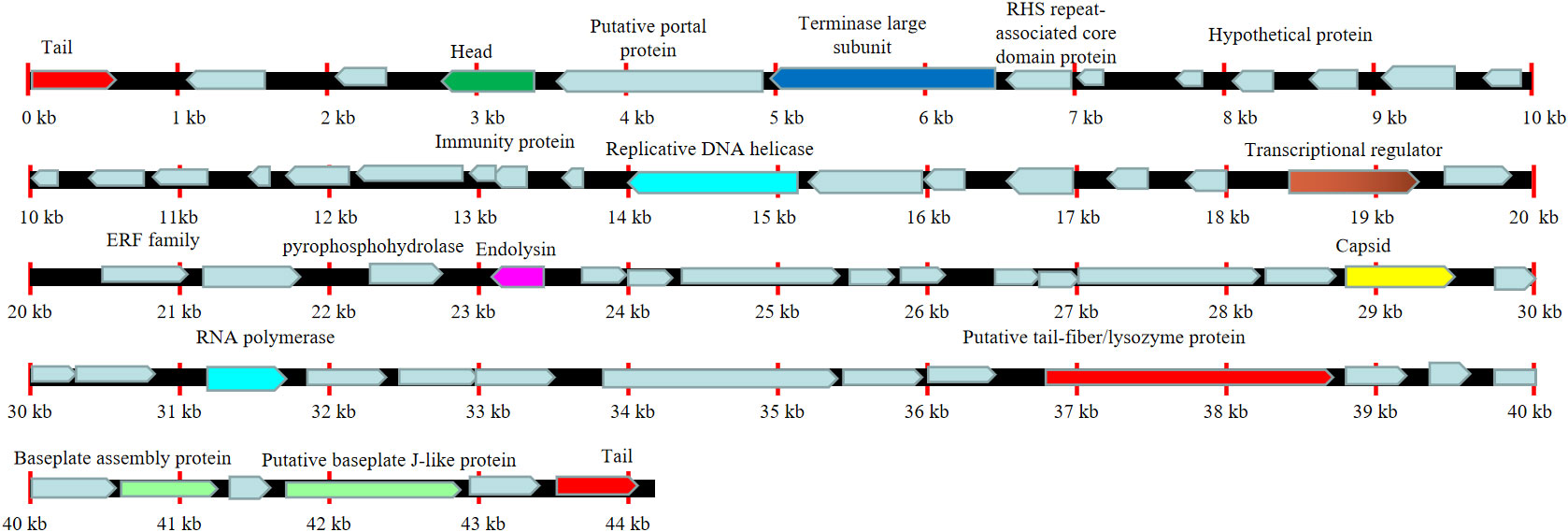
Figure 2 Line map of the A. baumannii phage vB_AbaM-SHI genome. In the vB_AbaM-SHI genome track, genes colored in red encode tail and genes colored in green encode head. The arrows represent the ORFs and point in the direction of transcription. Replicative DNA helicase and RNA polymerase were marked with cyan rectangular box, cell lysis was marked with purple rectangular box, baseplate assembly proteins were marked with light green rectangular box, and capsid was marked with yellow rectangular box.
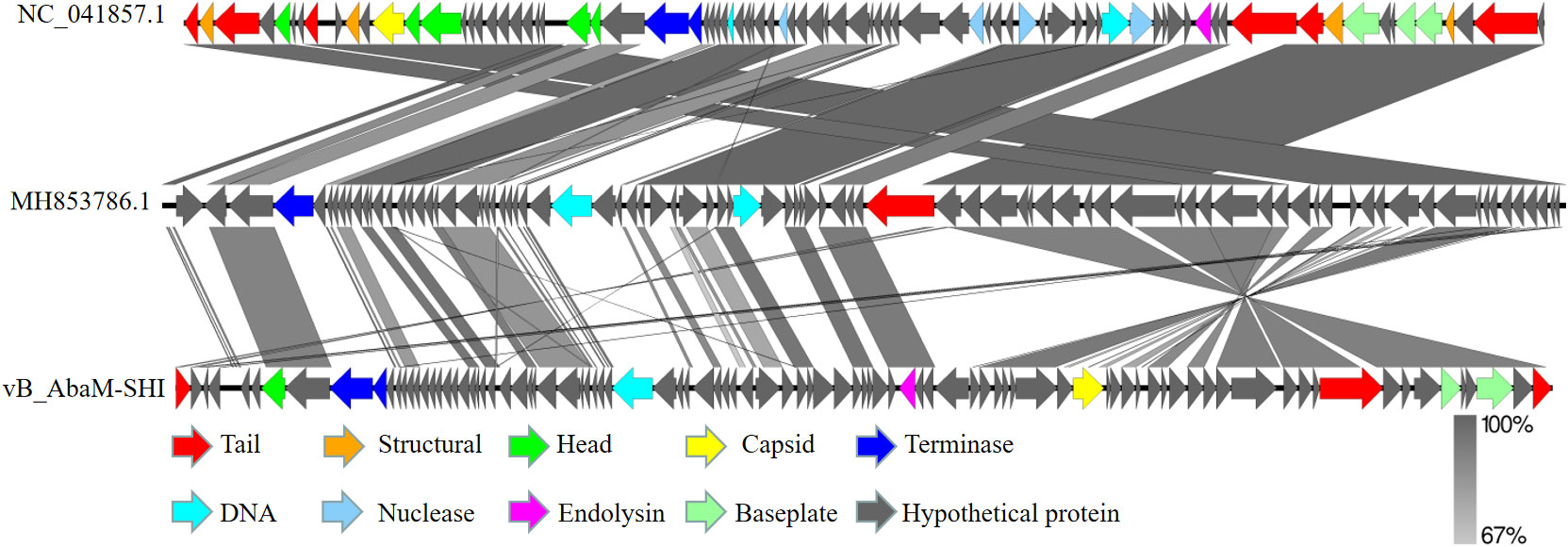
Figure 3 Easyfig output image of the genomic comparison among Acinetobacter phage IME-AB2 (top, NC_041857.1), Acinetobacter phage vB_AbaM_IME285 (middle, MH853786.1) and Acinetobacter phage vB_AbaM-SHI (bottom). Phage genomes are presented by linear visualization with coding regions shown as arrows. ORFs are color-coded according to predicted function: red, tail; orange, structural; green, head; yellow, capsid; blue, terminase; purple, endolysin; grey, hypothetical proteins. The percentage of sequence similarity is indicated by the intensity of the gray color. Vertical blocks between analyzed sequences indicate regions with at least 67% of similarity.
3.2 Optimum pH, MOI and temperature for the proliferation of phage vB_AbaM-SHI
Temperature, pH of culture medium and MOI are critical factors affecting the replication efficiency of phages, and these factors also potentially affect the bacteriolytic effect of phages. Therefore, to determine the stability of phage vB_AbaM-SHI at various temperatures and pH levels, we added phage to buffers with different pH levels or incubated them in different temperature environments, and then measured the titers of phages in the liquid. This phage exhibited a good resistance to temperatures ranging from 4°C to 37°C, and there is no significant difference in the titer of phages within this temperature range. In comparison, the titer of phages at 60°C was significantly reduced and the phages were completely inactivated at 90°C (Figure 4A). The most proper pH values to maintain phage activity ranged from 6 to 7, however, the activity of phages was significantly reduced when the phages were exposed to alkaline buffer solutions with pH 10 or pH 11 (Figure 4B). Overall, phage vB_AbaM-SHI has a high tolerance to acidic environments, as about 20% of phages remain infectious after being exposed to a pH 3 buffer for one hour, while almost all phages lost their infectivity after being exposed to a pH 11 buffer for one hour. When using bacteria to amplify phage vB_AbaM-SHI, the production of phages is inversely proportional to the MOI value during phage infection. As the MOI value increased from 0.1 to 100, the production of phages gradually decreased. When the MOI value is 0.1, the yield of phages reached the highest (Figure 4C).
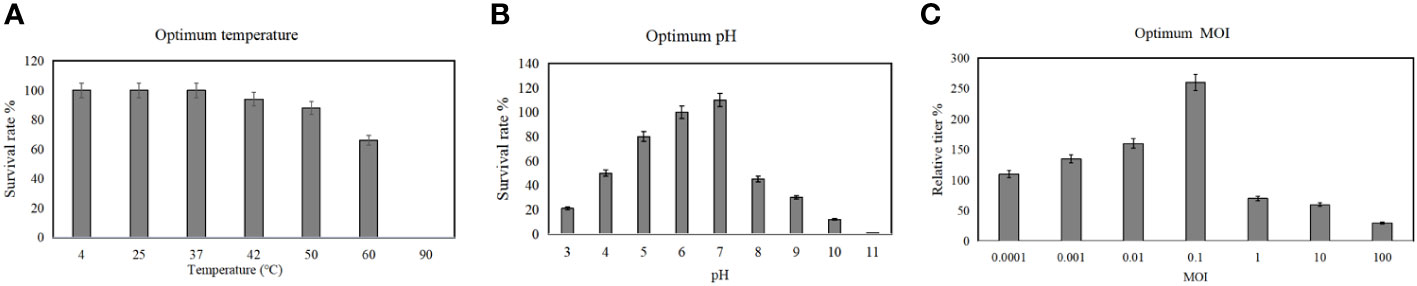
Figure 4 Optimum temperature, pH and MOI for the replication of phage vB_AbaM-SHI. (A) Stability of phage vB_AbaM-SHI at different temperatures; (B) Stability of vB_AbaM-SHI at different pH levels; (C) Optimal multiplicity of infection (MOI) determination.
3.3 Phage vB_AbaM-SHI has broad-spectrum bacteriolytic ability
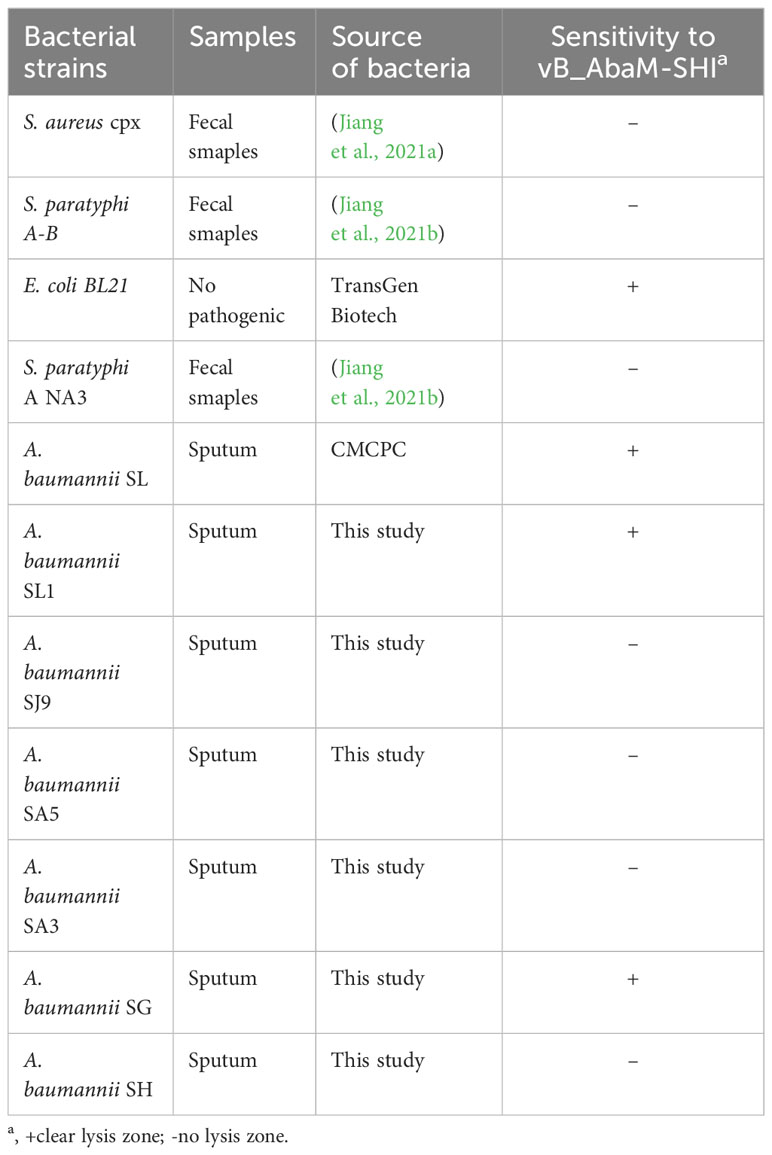
Before testing the ability of phage vB_AbaM-SHI to lyse clinically isolated A. baumannii strains, we first determine the sensitivity of A. baumannii strains to antibiotics. As a result, seven A. baumannii strains exhibited different sensitivities to 5 antibiotics tested. Among them, more than half (5/7) of the bacterial strains had resistance to 3 antibiotics, approximately half (3/7) of the bacterial strains demonstrated sensitivity to tobramycin, and all bacterial strains were sensitive to colistin (Table 3). We next tested the sensitivity of 7 A. baumannii strains and 4 other bacterial strains to phage vB_AbaM-SHI. The results showed that approximately half of (3/7) of A. baumannii strains could be lysed by the phage (the diameters of lysis zone ranging from 5 to 7 mm). In addition, E. coli BL21 strain was also lysed by this phage (with a lysis zone of 6 mm in diameter). However, both S. aureus strain and S. paratyphi strains were not sensitive to this phage lysis (without visible lysis zone) (Table 4).
3.4 Phage can independently or in combination with antibiotics inhibit the formation of bacterial biofilms
To evaluate the effect of vB_AbaM-SHI (MOI = 0.1) and kanamycin sulfate (10 µg mL-1) on the biofilm formation of bacteria, we conducted inhibition experiments of bacterial growth in 96-well plates and the OD570 or OD600 (optical density at 570 nm or 600 nm wavelength) values of each well were measured at 12 h and 24 h post bacterial inoculation. When A. baumannii SL cells were in a low-density state, all results from scanning electron micrographs (Figures 5A–C; reflecting the amount of bacterial biofilm at the bottom of the cell culture plate), OD600 values (Figures 6A; reflecting the amount of bacteria in the suspension), and OD570 values (Figures 6E, F; reflecting the amount of bacterial biofilms at the bottom of the cell culture plate) indicated that phage vB_AbaM-SHI and kanamycin sulfate has similar antibacterial effects. In contrast, when A. baumannii SL cells were locked in the high-density state, phage vB_AbaM-SHI exerted a better antibacterial effect than kanamycin sulfate (Figures 5D–F). In addition, compared to the ability to inhibit biofilm formation, phage vB_AbaM-SHI had a strong ability to control the growth of suspended cells (Figure 6). We then evaluated the ability of phage vB_AbaM-SH and kanamycin sulfate to inhibit the bacterial biofilm formation. The biofilm biomass of A. baumannii SL was measured using epifluorescence microscopy at 24 h, 36 h and 60 h post bacterial inoculation, respectively. The results showed that the amount of biofilm reached the peak at 36 h. Phage vB_AbaM-SHI significantly and persistently inhibited the formation of bacterial biofilm for up to 36 h (Figure 7). After adding phage vB_AbaM-SHI or kanamycin sulfate to A. baumannii and culturing for 24 hours, the amount of bacterial biofilm was measured. Both treatment factors can significantly inhibit the formation of bacterial biofilm, and kanamycin sulfate is superior to phage vB_AbaM-SHI (Figure 8A). Then, we first cultured the bacteria for 12 hours before adding phage vB_AbaM-SHI or kanamycin sulfate and determined the amount of bacterial biofilm. Although both can significantly reduce the amount of bacterial biofilm, phage vB_AbaM-SHI is significantly better than kanamycin sulfate (Figure 8B). In summary, phage vB_AbaM-SHI and kanamycin sulfate have distinct characteristics in inhibiting bacterial growth and controlling bacterial biofilm formation.
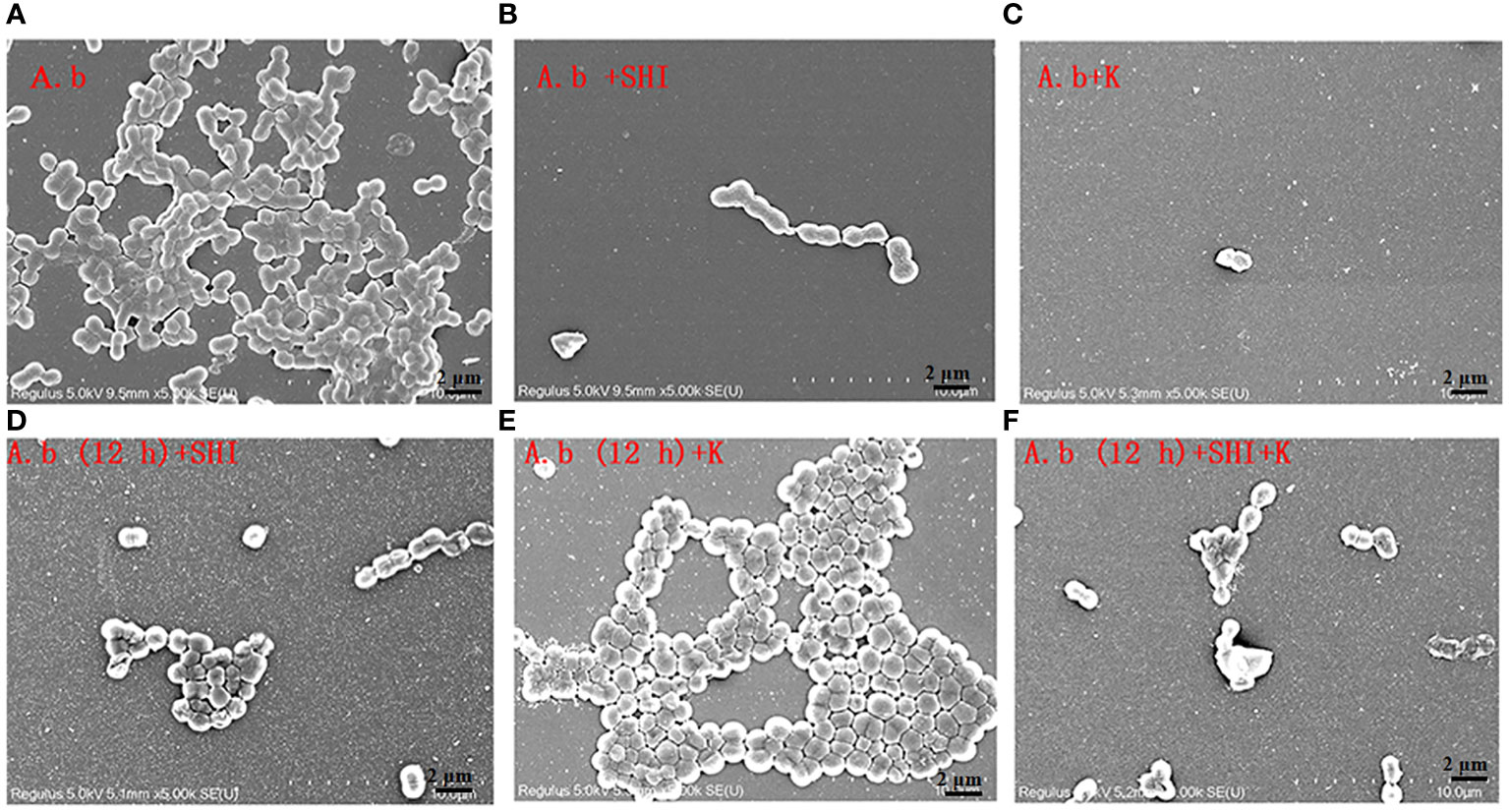
Figure 5 Scanning electron microscopy (SEM) image of A. baumannii SL colonization in biofilms formed on a round coverslip before and after phage vB_AbaM-SHI and kanamycin sulfate treatment. (A) A. baumannii SL was diluted in the overnight culture (1:250) and cultured for 24 hours before the addition of (B) phage vB_AbaM-SHI (MOI = 0.1) or (C) kanamycin sulfate (10 µg mL-1); (D) A. baumannii SL was diluted in the overnight culture (1:250) and cultured for 12 hours, and then phage vB_AbaM-SHI (MOI = 0.1) was added and cultured for another 12 hours; (E) A. baumannii SL was diluted in the overnight culture (1:250) and cultured for 12 hours, and then kanamycin sulfate (10 µg mL-1) was added and cultured for another 12 hours; (F) A. baumannii SL was diluted in the overnight culture (1:250) and cultured for 12 hours, and then both phage vB_AbaM-SHI (MOI = 0.1) and kanamycin sulfate (10 µg mL-1) were added and cultured for another 12 hours. The colonization of bacteria on coverslip was analyzed using scanning electron microscopy (5, 000× magnification). “A.b”: A. baumannii; “SHI”: vB_AbaM-SHI; “K”: kanamycin sulfate.
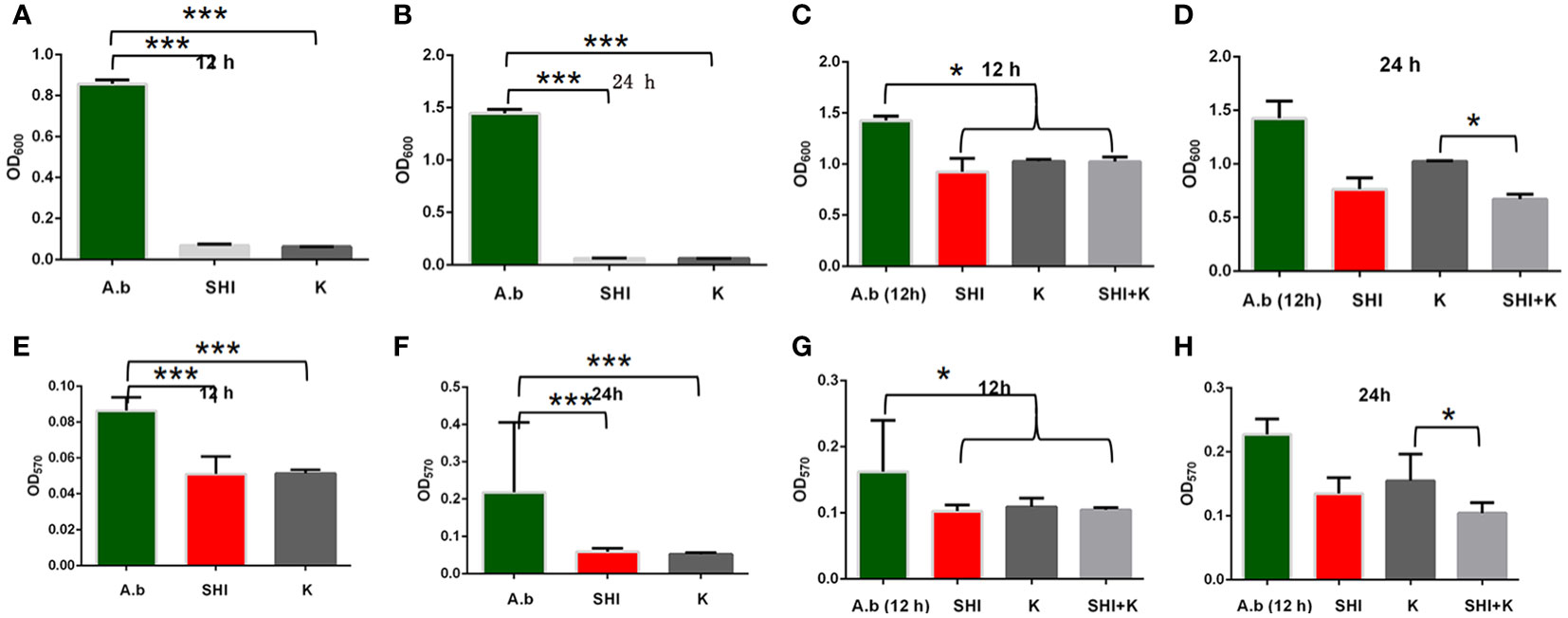
Figure 6 Effects of phage vB_AbaM-SHI and kanamycin sulfate on planktonic and bacterial biofilm formation. (A, B) Effects of phage vB_AbaM-SHI and kanamycin sulfate (10 µg mL-1) on the growth of A. baumannii SL after pre-cultivation for 12 h (A) and 24 h (B). (C, D) Effects of phage vB_AbaM-SHI and kanamycin sulfate (10 µg mL-1) on A. baumannii SL (inoculated at a concentration of 4‰) growth after first being cultured for 12 h, followed by phage A. baumannii SL and kanamycin sulfate (10 µg mL-1) addition and culturing for 12 h (C) and 24 h (D). (E, F) Effects of phage vB_AbaM-SHI and kanamycin sulfate (10 µg mL-1) on A. baumannii SL (inoculated at a concentration of 4‰) biofilm formation after pre-cultivation for 12 h (E) and 24 h (F). (G, H) Effects of phage vB_AbaM-SHI and kanamycin sulfate (10 µg mL-1) on A. baumannii SL (inoculated at a concentration of 4‰) biofilm formation after an initial culture for 12 h, followed by the addition of phage vB_AbaM-SHI and kanamycin sulfate (10 µg mL-1) and culture for 12 (G) and 24 h (H). *P <0.05, ***P <0.001. “A.b”: A. baumannii; “SHI”: vB_AbaM-SHI; “K”: kanamycin sulfate.
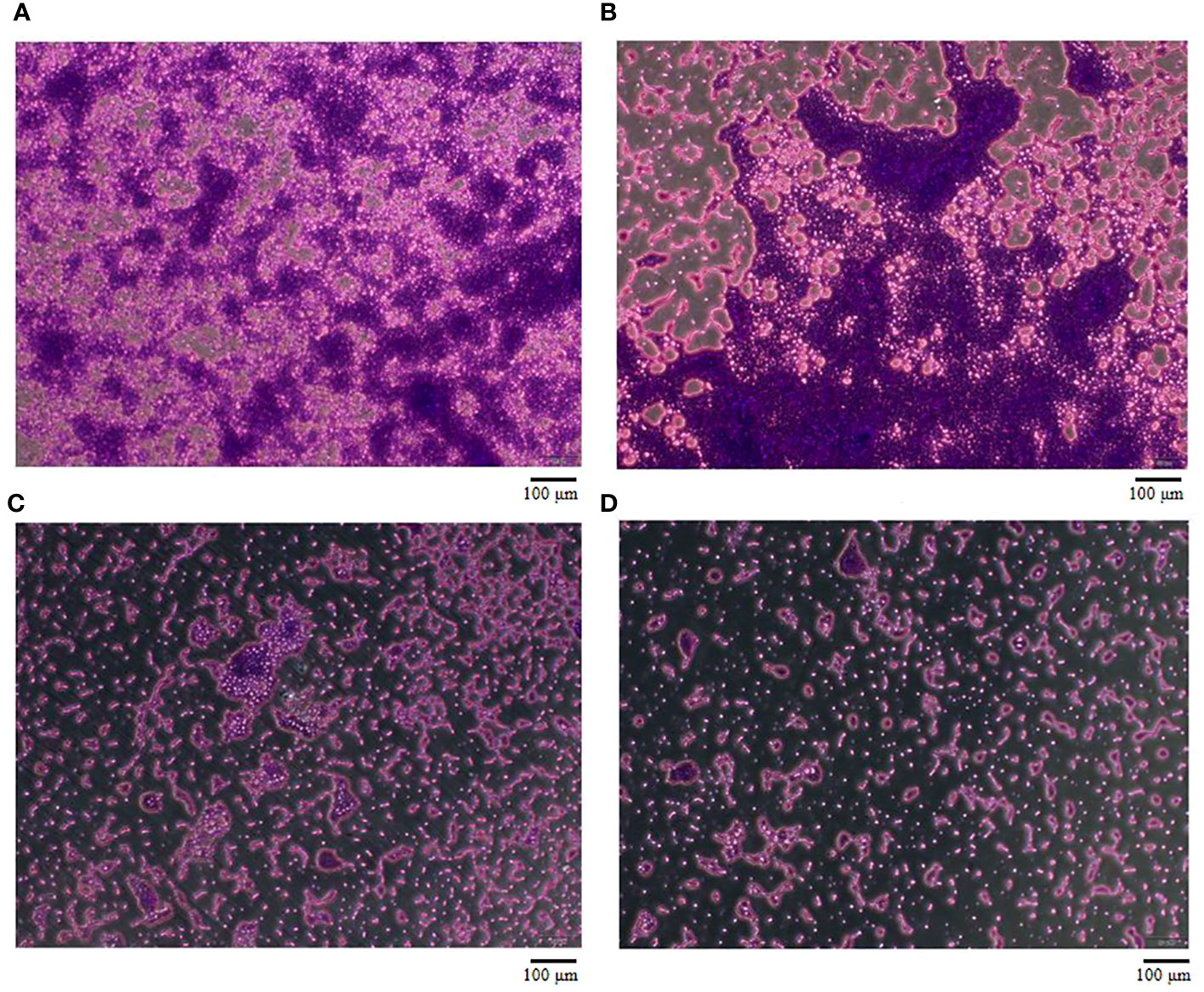
Figure 7 The effect of Phage vB_AbaM-SHI on the biofilm formation of A. baumannii SL. (A-C) Biofilm formed by A. baumannii SL at 24 h, 36 h and 60 h, respectively. (D) Phage vB_AbaM-SHI inhibited the formation of A. baumannii SL biofilm (MOI = 1, 36 h).
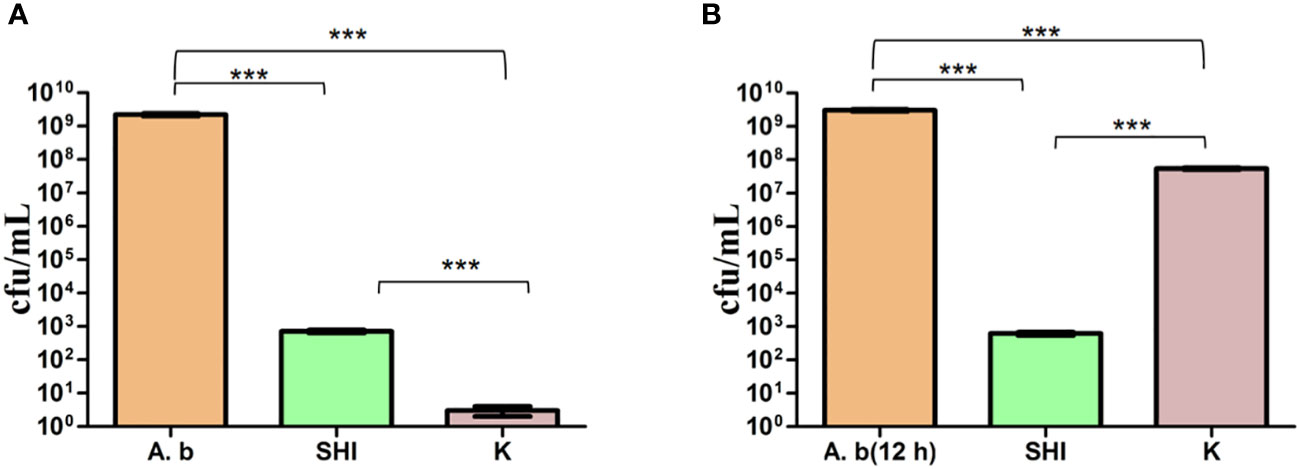
Figure 8 Phage vB_AbaM-SHI and kanamycin sulfate inhibit the growth of A. baumannii SL. (A) Effects of phage vB_AbaM-SHI and kanamycin sulfate (10 µg mL-1) on the growth of A. baumannii SL (diluted in the overnight culture [1:250]) cultured for 24 h. (B) Effects of phage vB_AbaM-SHI and kanamycin sulfate (10 µg mL-1) on A. baumannii SL (diluted in the overnight culture [1:250]) after an initial culture for 12 hours followed by the addition of phage vB_AbaM-SHI and kanamycin sulfate (10 µg mL-1) and culture for 12 h. ***P <0.001. “A.b”: A. baumannii; “SHI”: vB_AbaM-SHI; “K”: kanamycin sulfate.
3.5 Structure and molecular specificity of vB_AbaM-SHI endolysin
Phage endolysin is a cell wall hydrolase that is encoded by a phage gene and synthesized at the late stage of phage infection (Fischetti, 2008). In general, endolysin needs to work in synergy with holin or spanin protein to lyse gram-positive or gram-negative bacteria, respectively (Singh et al., 2022). By comparing the genome sequence of phage vB_AbaM-SHI with database, we found that there is a lack of genes encoding holin or spanin in phage vB_AbaM-SHI, suggesting endolysin in phage vB_AbaM-SHI could be able to exert bacteriolytic effect alone. In fact, although it is difficult for endolysin alone to penetrate the outer membrane of gram-negative bacteria and destroy the peptidoglycans in bacterial cell wall, recent studies reported that some phage endolysins can independently lyse gram-negative bacteria. For instance, the recombinant endolysin LysSS exhibited bacteriolytic activity against A. baumannii, E. coli, K. pneumoniae, Salmonella sp. and P. aeruginosa without pre-treatment with an outer membrane permeabilizer (Kim et al., 2020). Similarly, another A. baumannii phage endolysin LysAB54 with high antibacterial activity against multiple multidrug-resistant A. baumannii and other gram-negative bacteria, including E. coli, P. aeruginosa and K. pneumoniae, in the absence of outer membrane permeabilizer (Khan et al., 2021). Moreover, a recent study reported a novel lysis mechanism that Salmonella endolysin M4Lys exerted its bacteriolytic activity, which was not dependent on either holin or the Sec pathway (Bai et al., 2020). Considering that endolysin may be an independent factor for phage vB_AbaM-SHI to exert bacteriolytic effect, we next compared and analyzed the phylogenetic relationship between our endolysin and similar endolysins. In general, the endolysins of A. baumannii phages have significant differences in size, amino acid sequence, and homology. The lengths of KU510289.1, NC_041857.1, AJG41879.1, AJG41878.1, ASN73457.1 AJG41884.1, YP_004009630.1 AWY10427.1 AID17956.1, QBY34659.1 and QBY34657.1 range from 145 to 256 amino acids (Supplementary Figure S1; Table 1). The tertiary structures predicted using SWISS-MODEL (https://swissmodel.expasy.org/) showed that these endolysins share common modeling template (Table 2). A. baumannii phage endolysins with similar structures are located in very close branch of evolution, for example, KU510289.1, NC_ 041857.1, MH853786.1, and vB_AbaM-SHI are in the same evolutionary branch (Supplementary Figure S1). The structures of A. baumannii phage endolysins demonstrated significant differences in β strands and α helices (Supplementary Figure S1).
By comparing the amino acid sequences of endolysins, it can be found that KU510289.1, NC_041857.1, JX976549.1, vB_AbaM-SHI and MH853786.1 have amino acid sequence differences mainly at positions 2, 9, 21, 99, 125, 126 and 129 (Figure 9). Intriguingly, we identified that phage vB_AbaM-SHI endolysin contains two additional β strands, attributing to the amino acid change at position 21 [lysine (K) in KU510289.1, NC_041857.1, JX976549.1 and MH853786.1 was replaced with arginine (R) in vB_AbaM-SHI].
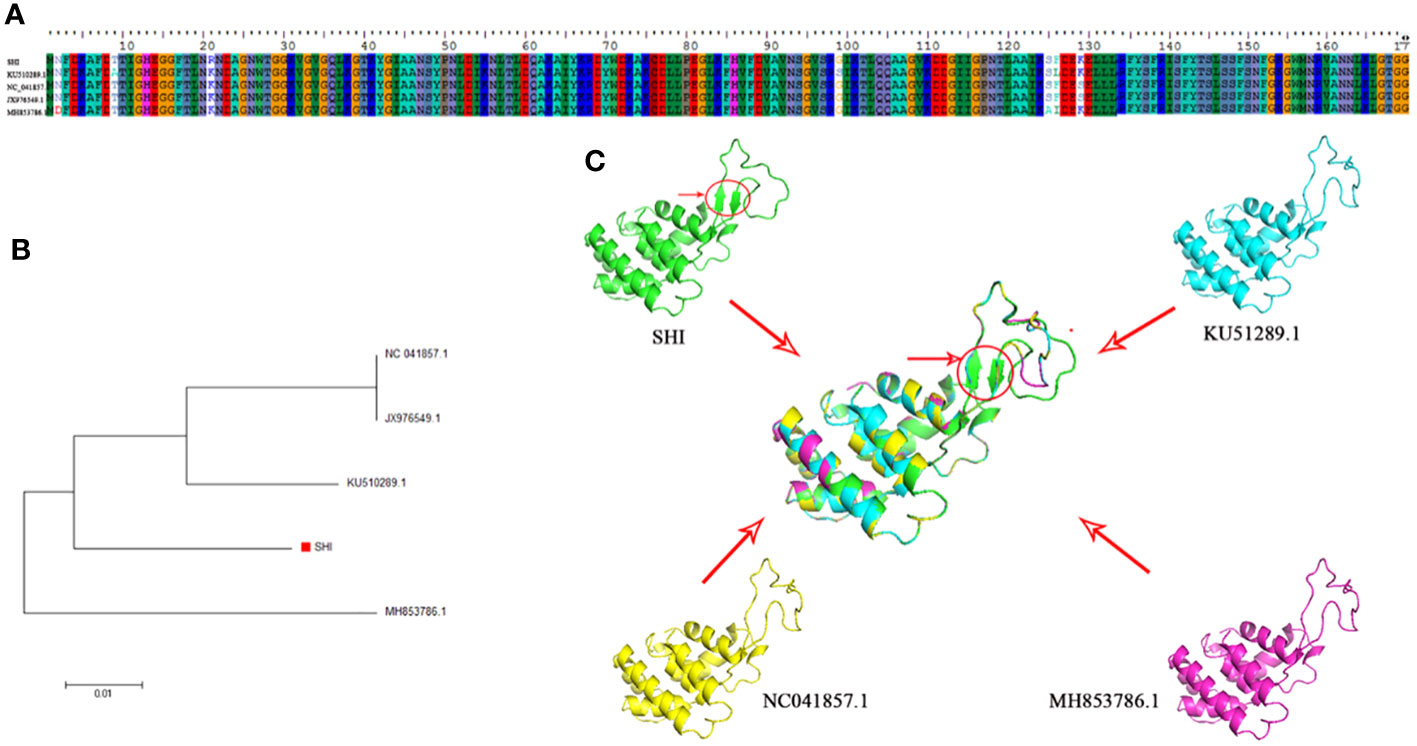
Figure 9 Amino acid sequence alignment, phylogenetic analysis and predicted tertiary structure of A. baumannii phage endolysins (A) Amino acid sequence alignment of endolysin proteins among vB_AbaM-SHI, KU510289.1, NC_041857.1, and MH853786.1. (B) Phylogenetic tree of A. baumannii phage endolysins. (The red square denotes the A. baumannii phage vB_AbaM-SHI endolysin). (C) Predicted tertiary structure of the A. baumannii phage endolysin protein. Superposition of vB_AbaM-SHI (green), KU510289.1 (cyano), NC_041857.1 (yellow), and MH853786.1 (purple) structures (https://swissmodel.expasy.org/templates/2nr7.1), which was reproduced/adapted from Structural Genomics, the crystal structure of putative secretion activator protein from Porphyromonas gingivalis W83, by Tan, K., Bigelow, L., Gu, M., Joachimiak, A., Midwest Center for Structural Genomics (MCSG), https://swissmodel.expasy.org/templates/2nr7.1, licensed CC-BY-SA-4.0. “SHI”: vB_AbaM-SHI.
4 Discussion
Lytic phages were usually isolated from sewage (Carey-Smith et al., 2006). In the present study, phage vB_AbaM-SHI was obtained from sauce chop factory drain outlet samples in Wuxi, China and classified as the family Straboviridae. Phage vB_AbaM-SHI has a genome with 44,180 bp and G+C contents of 38.14%. In comparison, the genomes of A. baumannii phage vB-GEC_Ab-M-G7 and phage BΦ62, two members of family Straboviridae, are 90 kb and 44,844 bp, respectively (Jeon et al., 2016; Kusradze et al., 2016). In addition, 93 ORFs were identified in the genome of vB_AbaM-SHI, whereas phage 5W has 61 putative ORFs in the genome (Peng et al., 2021). The phylogenetic analysis showed that phage vB_AbaM-SHI endolysin has high amino acid sequence homology with endolysins described in previous studies. No antibiotic resistance and virulence associated genes were identified in vB_AbaM-SHI genome.
With the increase in antibiotic misused, the emergence of multidrug-resistant and extremely resistant bacteria has become a serious threat to human health. Alternatives to antibiotics are urgently needed. Hopefully, phages represent an alternative solution to the issue of drug resistance. Although vB_AbaM-SHI barely lysed bacteria from other genera, vB_AbaM-SHI exhibited broad-spectrum bacteriolytic activity against multidrug-resistant A. baumannii that were resistant to one or more commonly used antibiotics. Lytic phages usually show a narrow lytic spectrum. Different bacterial strains have distinct differences in the sensitivity to the phage, which restrict the potential application of phage. For example, Wintachai et al., reported that phage vABWU2101 has the ability to kill 70% of the tested MDR A. baumannii strains (Wintachai et al., 2022). In contrast, Peng et al., demonstrated that phage 5W only lyse 21% A. baumannii isolates (Peng et al., 2021). Herein, phage vB_AbaM-SHI could lyse 42.8% of A. baumannii strains which were isolated from clinical patients, suggesting that vB_AbaM-SHI has the potential to be used for the treatment of corresponding clinical bacterial infections.
Biofilm is usually defined as a surface-attached microbial community, which mainly protects bacteria from the external dangerous environment (Qi et al., 2016). The ability of bacteria to form biofilms on different surfaces increases the risk of contamination, particularly in poultry products, clinics and food industries (Liu et al., 2019; Lianou et al., 2020). Although the exploitation of treatments that interrupt multidrug efflux pumps and quorum sensing interference strategies are important for inhibiting biofilm formation (Subhadra et al., 2018), developing effective strategies to eliminate biofilms remains a challenge and ideal agents for bacteria biofilm control are not available (Joshi et al., 2021; Li et al., 2021). The biofilm formation of A. baumannii was affected by the phenotype, genotype, physicochemical characteristics of bacteria (Eze et al., 2018; Yang et al., 2019). In the present study, kanamycin sulfate displayed good inhibiting ability of bacterial biofilm formation in low-density A. baumannii SL culture. But, phage vB_AbaM-SHI exerted better anti-biofilm formation and sterilization effects than kanamycin sulfate in a high-density A. baumannii SL culture. Additionally, the combination of phage vB_AbaM-SHI and kanamycin sulfate displayed strong anti-biofilm formation and antibacterial effects than phage vB_AbaM-SHI or kanamycin sulfate alone in a low-density A. baumannii SL culture. However, compared to killing suspended A. baumannii SL strain, phage vB_AbaM-SHI has weaker ability to lyse bacteria in biofilms, suggesting this phage has weak penetrating ability. Therefore, reagents or strategies that help phages penetrate biofilms are with great significance. Taken together, phage vB_AbaM-SHI is expected to become a promising agent to control multidrug-resistant A. baumannii. In fact, Grygorcewicz et al., recently revealed that a A. baumannii phages cocktail in combination with antibiotics had strong lytic ability to destroy biofilm in human urine (Grygorcewicz et al., 2021).
In summary, we identified a broad-spectrum phage vB_AbaM-SHI that exhibited a potent lytic effect on A. baumannii strains isolated in clinic. vB_AbaM-SHI can independently kill bacteria and inhibit bacterial biofilm formation, and can also synergistically exert strong antibacterial effects with antibitics, indicating vB_AbaM-SHI is a promising agent for the bio-control of multi-drug resistant A. baumannii strains and preventing the biofilm formation on medical devices.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: ON480525.1.
Author contributions
LJ: Conceptualization, Writing – original draft, Writing – review & editing. QX: Writing – original draft. YW: Formal analysis, Writing – review & editing. XZ: Resources, Conceptualization, Formal analysis, Validation, Writing – original draft. ZC: Methodology, Writing – original draft. QS: Software, Writing – original draft. JW: Conceptualization, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by the National Natural Science Foundation of China (82160403, 42206093, 32070162), the Yunnan Science and Technology Commission (202101AY070001-247) of the Yunnan provincial Science and Technology Department and Kunming Medical University, Young and middle-aged academic and technical leaders talent project, Grant No. 202305AC160023, the Respiratory Clinical Medical Center of Yunnan, Grant No.2020LCZXKF-HX01, the Medical Scientific Research Foundation of Zhejiang Province, Grant No.2022KY1124, the Natural Science Foundation of Ningbo (2021J118, 2021J113), and the K.C. Wong Magna Fund at Ningbo University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1351993/full#supplementary-material
Supplementary Figure 1 | The characteristics of A. baumannii phage endolysin. a, Amino acid substitutions of A. baumannii phage endolysin proteins among vB_AbaM-SHI (A), AJG41880.1 (B), ALJ97637.1 (C), AJG41885.1 (D), ADG35978.1 (E), AXF40585.1 (F), AJT61420.1 (G) and QBY34658.1 (H). b, Phylogenetic tree of A. baumannii phage endolysin proteins among vB_AbaM-SHI (A), AJG41880.1 (B), ALJ97637.1 (C), AJG41885.1 (D), ADG35978.1 (E), AXF40585.1 (F), AJT61420.1 (G) and QBY34658.1 (H). (C), Predicted tertiary structures of A. baumannii phage endolysin proteins among vB_AbaM-SHI (A), AJG41880.1 (B), ALJ97637.1 (C), AJG41885.1 (D), ADG35978.1 (E), AXF40585.1 (F), AJT61420.1 (G) and QBY34658.1 (H).
References
Adriaenssens, E., Brister, J. R. (2017). How to name and classify your phage: an informal guide. Viruses 9, 70. doi: 10.3390/v9040070
Ahmed, S. S., Alp, E., Ulu-Kilic, A., Dinc, G., Aktas, Z., Ada, B., et al. (2016). Spread of carbapenem-resistant international clones of Acinetobacter baumannii in Turkey and Azerbaijan: a collaborative study. Eur. J. Clin. Microbiol. Infect. Dis. 35, 1463–1468. doi: 10.1007/s10096-016-2685-x
Ahn, J., Kim, S., Jung, L. S., Biswas, D. (2013). In vitro assessment of the susceptibility of planktonic and attached cells of foodborne pathogens to bacteriophage p22-mediated salmonella lysates. J. Food Prot. 76, 2057–2062. doi: 10.4315/0362-028X.JFP-13-183
Antunes, L. C., Visca, P., Towner, K. J. (2014). Acinetobacter baumannii: evolution of a global pathogen. Pathog. Dis. 71, 292–301. doi: 10.1111/2049-632X.12125
Bai, J., Lee, S., Ryu, S. (2020). Identification and in vitro characterization of a novel phage endolysin that targets gram-negative bacteria. Microorganisms. 8, 447. doi: 10.3390/microorganisms8030447
Bello-López, E., Rocha-Gracia, R. D. C., Castro-Jaimes, S., Cevallos, M. A., Vargas-Cruz, M., Verdugo-Yocupicio, R., et al. (2020). Antibiotic resistance mechanisms in Acinetobacter spp. strains isolated from patients in a paediatric hospital in Mexico. J. Glob. Antimicrob. Resist. 23, 120–129. doi: 10.1016/j.jgar.2020.08.014
Briers, Y., Lavigne, R., Volckaert, G., Hertveldt, K. (2007). A standardized approach for accurate quantification of murein hydrolase activity in high-throughput assays. J. Biochem. Biophys. Methods. 70, 531–533. doi: 10.1016/j.jbbm.2006.10.009
Carey-Smith, G. V., Billington, C., Cornelius, A. J., Hudson, J. A., Heinemann, J. A. (2006). Isolation and characterization of bacteriophages infecting Salmonella spp. FEMS Microbiol. Lett. 258, 182–186. doi: 10.1111/fml.2006.258.issue-2
Chapman, B., Gunter, C. (2018). Local food systems food safety concerns. Microbiol. Spectr. 6, 1–9. doi: 10.1128/microbiolspec.PFS-0020-2017
Chegini, Z., Khoshbayan, A., Taati Moghadam, M., Farahani, I., Jazireian, P., Shariati, A. (2020). Bacteriophage therapy against Pseudomonas aeruginosa biofilms: a review. Ann. Clin. Microbiol. Antimicrob. 19, 45. doi: 10.1186/s12941-020-00389-5
Cui, L., Veeranarayanan, S., Thitiananpakorn, K., Wannigama, D. L. (2023). Bacteriophage bioengineering: A transformative approach for targeted drug discovery and beyond. Pathogens. 12, 1179. doi: 10.3390/pathogens12091179
de Ornellas Dutka Garcia, K. C., de Oliveira Corrêa, I. M., Pereira, L. Q., Silva, T. M., de Souza Ribeiro Mioni, M., de Moraes Izidoro, A. C., et al. (2017). Bacteriophage use to control Salmonella biofilm on surfaces present in chicken slaughterhouses. Poult. Sci. 96, 3392–3398. doi: 10.3382/ps/pex124
Duan, X., Jiang, L., Guo, M., Li, C. (2023). Isolation, characterization and application of a lytic phage vB_VspM_VS1 against Vibrio splendidus biofilm. PloS One. 18, e0289895. doi: 10.1371/journal.pone.0289895
Endersen, L., O'Mahony, J., Hill, C., Ross, R. P., McAuliffe, O., Coffey, A. (2014). Phage therapy in the food industry. Annu. Rev. Food Sci. Technol. 5, 327–349. doi: 10.1146/annurev-food-030713-092415
Eze, E. C., Chenia, H. Y., El Zowalaty, M. E. (2018). Acinetobacter baumannii biofilms: effects of physicochemical factors, virulence, antibiotic resistance determinants, gene regulation, and future antimicrobial treatments. Infect. Drug. Resist. 11, 2277–2299. doi: 10.2147/IDR.S169894
Fischetti, V. A. (2008). Bacteriophage lysins as effective antibacterials. Curr Opin Microbiol. 11, 393–400. doi: 10.1016/j.mib.2008.09.012
Gauba, A., Rahman, K. M. (2023). Evaluation of antibiotic resistance mechanisms in gram-negative bacteria. Antibiotics (Basel). 12, 1590. doi: 10.3390/antibiotics12111590
Gayoso, C. M., Mateos, J., Méndez, J. A., Fernández-Puente, P., Rumbo, C., Tomás, M., et al. (2014). Molecular mechanisms involved in the response to desiccation stress and persistence in Acinetobacter baumannii. J. Proteome. Res. 13, 460–476. doi: 10.1021/pr400603f
Ghose, C., Euler, C. W. (2020). Gram-negative bacterial lysins. Antibiotics (Basel). 9, 74. doi: 10.3390/antibiotics9020074
Gong, C., Jiang, X. (2017). Application of bacteriophages to reduce Salmonella attachment and biofilms on hard surfaces. Poult. Sci. 96, 1838–1848. doi: 10.3382/ps/pew463
Grygorcewicz, B., Wojciuk, B., Roszak, M., Łubowska, N., Błażejczak, P., Jursa-Kulesza, J., et al. (2021). Environmental phage-based cocktail and antibiotic combination effects on acinetobacter baumannii biofilm in a human urine model. Microb. Drug Resist. 27, 25–35. doi: 10.1089/mdr.2020.0083
Gutiérrez, D., Briers, Y. (2021). Lysins breaking down the walls of Gram-negative bacteria, no longer a no-go. Curr. Opin. Biotechnol. 68, 15–22. doi: 10.1016/j.copbio.2020.08.014
Gutiérrez, D., Rodríguez-Rubio, L., Martínez, B., Rodríguez, A., García, P. (2016). Bacteriophages as weapons against bacterial biofilms in the food industry. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00825
Hall-Stoodley, L., Costerton, J. W., Stoodley, P. (2004). Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2, 95–108. doi: 10.1038/nrmicro821
Jakubovics, N. S., Goodman, S. D., Mashburn-Warren, L., Stafford, G. P., Cieplik, F. (2021). The dental plaque biofilm matrix. Periodontol. 2000. 86, 32–56. doi: 10.1111/prd.12361
Jeon, J., Ryu, C. M., Lee, J. Y., Park, J. H., Yong, D., Lee, K. (2016). In vivo application of bacteriophage as a potential therapeutic agent to control OXA-66-like carbapenemase-producing acinetobacter baumannii strains belonging to sequence type 357. Appl. Environ. Microbiol. 82, 4200–4208. doi: 10.1128/AEM.00526-16
Jiang, L., Ma, D., Ye, C., Li, L., Li, X., Yang, J., et al. (2018). Molecular characterization of dengue virus serotype 2 cosmospolitan genotype from 2015 dengue outbreak in yunnan, China. Front. Cell. Infect. Microbiol. 8. doi: 10.3389/fcimb.2018.00219
Jiang, Y., Xu, Q., Jiang, L., Zheng, R. (2021a). Isolation and Characterization of a Lytic Staphylococcus aureus Phage WV against Staphylococcus aureus Biofilm. Intervirology 64, 169–177. doi: 10.1159/000515282
Jiang, L., Zheng, R., Sun, Q., Li, C. (2021b). Isolation, characterization, and application of Salmonella paratyphi phage KM16 against Salmonella paratyphi biofilm. Biofouling 37, 276–288. doi: 10.1080/08927014.2021.1900130
Joshi, R. V., Gunawan, C., Mann, R. (2021). We are one: multispecies metabolism of a biofilm consortium and their treatment strategies. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.635432
Khan, F. M., Gondil, V. S., Li, C., Jiang, M., Li, J., Yu, J., et al. (2021). A novel acinetobacter baumannii bacteriophage endolysin lysAB54 with high antibacterial activity against multiple gram-negative microbes. Front. Cell. Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.637313
Kim, H. S., Choi, S. J., Yoon, K. S. (2018). Efficacy Evaluation of Control Measures on the Reduction of Staphylococcus aureus in Salad and Bacillus cereus in Fried Rice Served at Restaurants. Foodborne. Pathog. Dis. 15, 198–209. doi: 10.1089/fpd.2017.2334
Kim, S., Lee, D. W., Jin, J. S., Kim, J. (2020). Antimicrobial activity of LysSS, a novel phage endolysin, against Acinetobacter baumannii and Pseudomonas aeruginosa. J. Glob. Antimicrob. Resist. 22, 32–39. doi: 10.1016/j.jgar.2020.01.005
Kusradze, I., Karumidze, N., Rigvava, S., Dvalidze, T., Katsitadze, M., Amiranashvili, I., et al. (2016). Characterization and Testing the Efficiency of Acinetobacter baumannii Phage vB-GEC_Ab-M-G7 as an Antibacterial Agent. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01590
Lee, K., Yong, D., Jeong, S. H., Chong, Y. (2011). Multidrug-resistant Acinetobacter spp.: increasingly problematic nosocomial pathogens. Yonsei Med. J. 52, 879–891. doi: 10.3349/ymj.2011.52.6.879
Li, Q., Liu, L., Guo, A., Zhang, X., Liu, W., Ruan, Y. (2021). Formation of multispecies biofilms and their resistance to disinfectants in food processing environments: A review. J. Food Prot. 84, 2071–2083. doi: 10.4315/JFP-21-071
Lianou, A., Nychas, G. E., Koutsoumanis, K. P. (2020). Strain variability in biofilm formation: A food safety and quality perspective. Food Res. Int. 137, 109424. doi: 10.1016/j.foodres.2020.109424
Liu, L., Ye, C., Soteyome, T., Zhao, X., Xia, J., Xu, W., et al. (2019). Inhibitory effects of two types of food additives on biofilm formation by foodborne pathogens. Microbiologyopen. 8, e00853. doi: 10.1002/mbo3.853
Nguyen, H. M., Watanabe, S., Sharmin, S., Kawaguchi, T., Tan, X. E., Wannigama, D. L., et al. (2023). RNA and single-stranded DNA phages: unveiling the promise from the underexplored world of viruses. Int. J. Mol. Sci. 24, 17029. doi: 10.3390/ijms242317029
Oliveira, G. S., Lopes, D. R. G., Andre, C., Silva, C. C., Baglinière, F., Vanetti, M. C. D. (2019). Multispecies biofilm formation by the contaminating microbiota in raw milk. Biofouling 35, 819–831. doi: 10.1080/08927014.2019.1666267
Park, M., Lee, J. H., Shin, H., Kim, M., Choi, J., Kang, D. H., et al. (2012). Characterization and comparative genomic analysis of a novel bacteriophage, SFP10, simultaneously inhibiting both Salmonella enterica and Escherichia coli O157:H7. Appl. Environ. Microbiol. 78, 58–69. doi: 10.1128/AEM.06231-11
Pendleton, J. N., Gorman, S. P., Gilmore, B. F. (2013). Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti. Infect. Ther. 11, 297–308. doi: 10.1586/eri.13.12
Penesyan, A., Gillings, M., Paulsen, I. T. (2015). Antibiotic discovery: combatting bacterial resistance in cells and in biofilm communities. Molecule 20, 5286–5298. doi: 10.3390/molecules20045286
Peng, F., Mi, Z., Huang, Y., Yuan, X., Niu, W., Wang, Y., et al. (2014). Characterization, sequencing and comparative genomic analysis of vB_AbaM-IME-AB2, a novel lytic bacteriophage that infects multidrug-resistant Acinetobacter baumannii clinical isolates. BMC Microbiol. 14, 181. doi: 10.1186/1471-2180-14-181
Peng, W., Zeng, F., Wu, Z., Jin, Z., Li, W., Zhu, M., et al. (2021). Isolation and genomic analysis of temperate phage 5W targeting multidrug-resistant Acinetobacter baumannii. Arch. Microbiol. 204, 58. doi: 10.1007/s00203-021-02618-7
Potron, A., Poirel, L., Nordmann, P. (2015). Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: Mechanisms and epidemiology. Int. J. Antimicrob. Agents. 45, 568–585. doi: 10.1016/j.ijantimicag.2015.03.001
Qi, L., Li, H., Zhang, C., Liang, B., Li, J., Wang, L., et al. (2016). Relationship between antibiotic resistance, biofilm formation, and biofilm-specific resistance in Acinetobacter baumannii. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00483
Sharma, S., Chatterjee, S., Datta, S., Prasad, R., Dubey, D., Prasad, R. K., et al. (2017). Bacteriophages and its applications: an overview. Folia Microbiol. (Praha). 62, 17–55. doi: 10.1007/s12223-016-0471-x
Shein, A. M. S., Wannigama, D. L., Hurst, C., Monk, P. N., Amarasiri, M., Badavath, V. N., et al. (2023). Novel intranasal phage-CaEDTA-ceftazidime/avibactam triple combination therapy demonstrates remarkable efficacy in treating Pseudomonas aeruginosa lung infection. Biomedicine Pharmacotherapy. 168, 115793. doi: 10.1016/j.biopha.2023.115793
Singh, A., Padmesh, S., Dwivedi, M., Kostova, I. (2022). How good are bacteriophages as an alternative therapy to mitigate biofilms of nosocomial infections. Infection Drug resistance. 15, 503–532. doi: 10.2147/IDR.S348700
Srisakul, S., Wannigama, D. L., Higgins, P. G., Hurst, C., Abe, S., Hongsing, P., et al. (2022). Overcoming addition of phosphoethanolamine to lipid A mediated colistin resistance in Acinetobacter baumannii clinical isolates with colistin-sulbactam combination therapy. Sci. Rep. 12, 11390. doi: 10.1038/s41598-022-15386-1
Subhadra, B., Kim, D. H., Woo, K., Surendran, S., Choi, C. H. (2018). Control of biofilm formation in healthcare: recent advances exploiting quorum-sensing interference strategies and multidrug efflux pump inhibitors. Materials (Basel). 11, 1676. doi: 10.3390/ma11091676
Tian, F., Li, J., Nazir, A., Tong, Y. (2021). Bacteriophage - A promising alternative measure for bacterial biofilm control. Infect. Drug Resist. 14, 205–217. doi: 10.2147/IDR.S290093
Venkatesan, N., Perumal, G., Doble, M. (2015). Bacterial resistance in biofilm-associated bacteria. Future Microbiol. 10, 1743–1750. doi: 10.2217/fmb.15.69
Wang, F., Ji, X., Li, Q., Zhang, G., Peng, J., Hai, J., et al. (2020a). TSPphg lysin from the extremophilic thermus bacteriophage TSP4 as a potential antimicrobial agent against both gram-negative and gram-positive pathogenic bacteria. Viruses. 12, 192. doi: 10.3390/v12020192
Wang, C., Li, P., Zhu, Y., Huang, Y., Gao, M., Yuan, X., et al. (2020b). Identification of a novel acinetobacter baumannii phage-derived depolymerase and its therapeutic application in mice. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01407
Wannigama, D. L., Hurst, C., Pearson, L., Saethang, T., Singkham-In, U., Luk-In, S., et al. (2019). Simple fluorometric-based assay of antibiotic effectiveness for Acinetobacter baumannii biofilms. Sci. Rep. 9, 6300. doi: 10.1038/s41598-019-42353-0
Wintachai, P., Surachat, K., Singkhamanan, K. (2022). Isolation and characterization of a novel autographiviridae phage and its combined effect with tigecycline in controlling multidrug-resistant acinetobacter baumannii-associated skin and soft tissue infections. Viruses. 14, 194. doi: 10.3390/v14020194
Yang, C. H., Su, P. W., Moi, S. H., Chuang, L. Y. (2019). Biofilm formation in acinetobacter baumannii: genotype-phenotype correlation. Molecules. 24, 1849. doi: 10.3390/molecules2410184910.3390/molecules24101849
Zelver, N., Hamilton, M., Goeres, D., Heersink, J. (2001). Development of a standardized antibiofilm test. Methods Enzymol. 337, 363–376. doi: 10.1016/S0076-6879(01)37025-8
Keywords: Acinetobacter baumannii, phage, bacterial biofilm, antibiotic, inhibition
Citation: Jiang L, Xu Q, Wu Y, Zhou X, Chen Z, Sun Q and Wen J (2024) Characterization of a Straboviridae phage vB_AbaM-SHI and its inhibition effect on biofilms of Acinetobacter baumannii. Front. Cell. Infect. Microbiol. 14:1351993. doi: 10.3389/fcimb.2024.1351993
Received: 15 December 2023; Accepted: 22 February 2024;
Published: 08 March 2024.
Edited by:
Zichen Yang, Xinqiao Hospital, ChinaReviewed by:
Agnieszka Kwiatek, University of Warsaw, PolandDhammika Leshan Wannigama, Yamagata Prefectural Central Hospital, Japan
Copyright © 2024 Jiang, Xu, Wu, Zhou, Chen, Sun and Wen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinsheng Wen, d2Vuamluc2hlbmdAbmJ1LmVkdS5jbg==
†These authors have contributed equally to this work
 Liming Jiang
Liming Jiang Qian Xu2†
Qian Xu2† Qiangming Sun
Qiangming Sun Jinsheng Wen
Jinsheng Wen