
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 17 April 2024
Sec. Bacteria and Host
Volume 14 - 2024 | https://doi.org/10.3389/fcimb.2024.1346821
This article is part of the Research Topic Role of Microbes in Ocular Surface Health and Diseases View all 8 articles
 Heba Alenezi1,2,3*
Heba Alenezi1,2,3* Grant Parnell4,5
Grant Parnell4,5 Stephen Schibeci4
Stephen Schibeci4 Jerome Ozkan2
Jerome Ozkan2 Mark Willcox2
Mark Willcox2 Andrew J. R. White2,3
Andrew J. R. White2,3 Nicole Carnt2,3,6
Nicole Carnt2,3,6Background: Microbial keratitis is one of the leading causes of blindness globally. An overactive immune response during an infection can exacerbate damage, causing corneal opacities and vision loss. This study aimed to identify the differentially expressed genes between corneal infection patients and healthy volunteers within the cornea and conjunctiva and elucidate the contributing pathways to these conditions’ pathogenesis. Moreover, it compared the corneal and conjunctival transcriptomes in corneal-infected patients to cytokine levels in tears.
Methods: Corneal and conjunctival swabs were collected from seven corneal infection patients and three healthy controls under topical anesthesia. RNA from seven corneal infection patients and three healthy volunteers were analyzed by RNA sequencing (RNA-Seq). Tear proteins were extracted from Schirmer strips via acetone precipitation from 38 cases of corneal infection and 14 healthy controls. The cytokines and chemokines IL-1β, IL-6, CXCL8 (IL-8), CX3CL1, IL-10, IL-12 (p70), IL-17A, and IL-23 were measured using an antibody bead assay.
Results: A total of 512 genes were found to be differentially expressed in infected corneas compared to healthy corneas, with 508 being upregulated and four downregulated (fold-change (FC) <−2 or > 2 and adjusted p <0.01). For the conjunctiva, 477 were upregulated, and 3 were downregulated (FC <−3 or ≥ 3 and adjusted p <0.01). There was a significant overlap in cornea and conjunctiva gene expression in patients with corneal infections. The genes were predominantly associated with immune response, regulation of angiogenesis, and apoptotic signaling pathways. The most highly upregulated gene was CXCL8 (which codes for IL-8 protein). In patients with corneal infections, the concentration of IL-8 protein in tears was relatively higher in patients compared to healthy controls but did not show statistical significance.
Conclusions: During corneal infection, many genes were upregulated, with most of them being associated with immune response, regulation of angiogenesis, and apoptotic signaling. The findings may facilitate the development of treatments for corneal infections that can dampen specific aspects of the immune response to reduce scarring and preserve sight.
Microbial keratitis is a serious infection of the cornea that classically presents with an epithelial defect with underlying infiltration by white blood cells (mostly neutrophils). During healing, microbial keratitis can give rise to corneal opacities due to scarring, which can result in blindness. Keratitis is one of the leading causes of blindness globally (Pascolini and Mariotti, 2012; Upadhyay et al., 2015; Ung et al., 2019). Pathogens can invade the cornea and cause infection when corneal defense mechanisms are decreased, such as after epithelium damage (Bourcier et al., 2003; Santacruz et al., 2015). Common identified risk factors for microbial keratitis are contact lens wear, ocular trauma, ocular surface diseases, previous ocular surgery, diabetes mellitus and immunosuppression (Bourcier et al., 2003; Keay et al., 2006; Al-Mujaini et al., 2009; Zimmerman et al., 2016; Tena et al., 2019). A wide spectrum of microorganisms can cause microbial keratitis, but bacteria represent the most common cause (Musa et al., 2010; Das et al., 2019; Green et al., 2019; Tena et al., 2019; Chen et al., 2020). Most prevalent among the bacteria are the Gram‐positive Streptococcus pneumoniae, Staphylococcus aureus and coagulase-negative staphylococci, and the Gram‐negative bacterium Pseudomonas aeruginosa (Orlans et al., 2011; Hoddenbach et al., 2014; O’Neill et al., 2014; Das et al., 2019; Green et al., 2019; Cabrera-Aguas et al., 2020).
During microbial keratitis, excessive activation of the host’s immune response contributes to tissue destruction, corneal opacities and vision loss (Willcox, 2007). Bacteria such as Pseudomonas aeruginosa produce virulence factors that can activate toll-like receptors (TLRs) on corneal epithelial cells (Willcox, 2007). TLRs recognize bacterial components such as lipopolysaccharide (LPS) or flagella from P. aeruginosa (Takeuchi et al., 1999; Barton and Medzhitov, 2003). Several TLRs (TLR2, TLR3, TLR4, TLR5, TLR8, and TLR9) are expressed by corneal epithelial cells (Zhang et al., 2003; Johnson et al., 2005; Huang et al., 2006; Mohammed et al., 2011; Chidambaram et al., 2017; Ueta et al., 2019). TLR2 is the main receptor for Gram-positive bacteria, while TLR4 regulates the LPS response (Hoshino et al., 1999; Takeuchi et al., 1999). The activation of TLRs stimulates epithelial cells to produce chemokines and cytokines (Willcox, 2007) such as interleukin-8 (IL-8), interleukin-1β (IL-1β) and interleukin-6 (IL-6) (Xue et al., 2000). These cytokines and chemokines amplify and control the response to infection by recruiting neutrophils and other white blood cells to the site of infection to eradicate the bacteria (Hazlett, 2005; Willcox, 2007). However, the persistent presence of neutrophils, and their defense systems such as enzymes and the reactive oxygen radicals can cause significant corneal damage and scarring (Chusid and Davis, 1979; Thakur et al., 2001).
Gene expression, particularly differential expression data for normal and disease tissues, may provide information that could help understand the disease’s pathogenic mechanisms. During microbial corneal infection, the host response follows particular biological pathways as the disease advances (Hazlett, 2004; Karthikeyan et al., 2013; Santacruz et al., 2015; Shrestha et al., 2021). However, there is a lack of information on the exact molecular mechanisms involved in these pathways in humans. To understand the immunopathogenesis of microbial corneal infection, this study analyzed the transcriptomes of the cornea and conjunctiva using RNA-seq of individuals with microbial infections compared to healthy controls.
The study was approved by the Western Sydney Local Health District Human Research Ethics Committee (LNR/16/WMEAD/401), and the research followed the tenets of the Declaration of Helsinki. All participants signed informed consent prior to study commencement.
Corneal infection patients undergoing a corneal scrape were recruited at the time of initial presentation at Westmead Hospital, Sydney, Australia. Healthy volunteers were recruited from the School of Optometry and Vision Science, University of New South Wales, Sydney, Australia. The cornea and conjunctiva swabs (Puritan Medical Products, ME, USA) were performed under topical anesthesia (oxybuprocaine hydrochloride 0.4% 4mg/mL minim eye drops, Bausch & Lomb, Australia). The affected eye was used for patients with corneal infection, while for healthy volunteers, the sampled eye was randomized. The individual swabs were placed in separate sterile microcentrifuge tubes containing 400 μl RNAlater® (R0901, Sigma-Aldrich) and stored at – 80°C until RNA extraction. Swabs and then tears were collected for patients following the standard diagnostic corneal scrape. Tears were collected from the same eye after swabbing, with Schirmer strips (HUB Pharmaceuticals, MI, USA) and stored at – 80°C until analysis. During the diagnostic corneal scrape, samples were collected from the corneal infection by a sterile 23-gauge needle or D25 scalpel and were plated in the clinic onto various media for culture in the diagnostic laboratory.
The present study involved the acquisition and analysis of RNA sequences from swab samples of seven patients with corneal infection and three healthy controls. In addition to the RNA cohort, tear samples were collected and analyzed from 31 further cases of corneal infection and 11 healthy controls, as detailed in Table 1.
Total RNA extraction of the corneal and conjunctival swabs was performed using NucleoSpin, RNA XS (Macherey-Nagel, Düren, Germany), following the manufacturer’s instructions. The RNA quality was checked in representative samples using the Agilent 2100 Bioanalyzer (Agilent Technologies, USA). The RNA purity and integrity of the samples were assessed according to the RNA integrity number (RIN), which can range from one (completely degraded) to 10 (fully intact) (Mueller et al., 2004; Schroeder et al., 2006). The RIN obtained was higher than seven in all samples measured and deemed suitable for RNA sequencing.
Sequencing libraries were prepared from 2 ng total RNA from each sample using the SMARTer Stranded Total RNA-Seq Kit v2 − Pico Input Mammalian (Takara Bio USA, Mountain View, CA, USA), according to the manufacturer’s protocol. The samples were processed in two batches; for the first batch, single-end (100 bp) library sequencing was performed on the Illumina HiSeq2500 Sequencing System (Illumina, San Diego, CA, USA), with three corneal infection and three control samples. Paired-end (50 bp) library sequencing was performed on the Illumina NovaSeq 6000 Sequencing System (Illumina, San Diego, CA, USA) for batch two, with four corneal infection samples.
The raw sequencing data was processed using Trimmomatic 0.39 (Bolger et al., 2014) to filter out low-quality reads and any data containing adapter/primer sequences. The filtered reads were mapped to the Homo sapiens genome (GRCh38.p13) using HISAT2 (v 2.2.1) (Kim et al., 2019). In the first batch, the number of mapped reads per sample ranged from 0.5 million to 29.8 million, while in the second batch, the range was 2.3 million to 77.3 million. To reduce uninformative variation and normalize sequencing depth between the samples, the median of ratios function provided by DESeq2 has been utilized (Anders and Huber, 2010). The PCA function of the (DESeq. 2 Bioconductor package) was used to perform principal component analysis. The expression level of genes for all the corneal infection samples and the controls was used to determine the principal components analysis (PCA) division of the corneal infection cases group and the healthy control group. Differential gene expression was calculated using the Bioconductor package, DESeq. 2 (v1.30.1) (Love et al., 2014). The differential gene expression thresholds were set with reference to the number of genes. Differential gene expression was calculated for the corneal samples based on the threshold criterion of log fold change > 1 (fold change (FC) <−2 or > 2) and adjusted P < 0.01. The differential gene expression for the conjunctiva samples was calculated based on the threshold criterion of log fold change (FC) > 1.58 (FC <−3 or ≥ 3) and adjusted P < 0.01. Due to the larger number of differentially expressed genes in the conjunctiva, a more stringent threshold for fold change was applied to obtain a manageable number of genes for downstream analysis. Gene IDs for the differentially expressed genes were linked to gene-specific information using the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/gene).
Functional annotations for the differentially expressed genes (DEGs) were performed using the Bioconductor package, clusterProfiler (v3.18.1) (Yu et al., 2012; Wu et al., 2021). The enrichGO and the enrichKEGG function from the clusterProfiler Bioconductor package were used to perform the enrichment analysis of Gene Ontology (GO) functions and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, respectively. The cut-off criteria of q value (corrected p-value) < 0.05 were used to select the significantly enriched GO terms and genes’ significantly enriched KEGG pathway.
The cytokines and chemokines IL-1β, IL-6, IL-8, CX3CL1, IL-10, IL-12 (p70), IL-17A and IL-23 were measured in the tear samples of 38 corneal infection cases and 14 healthy controls. Tear proteins were extracted from Schirmer strips using acetone precipitation for maximum yield (Powell et al., 2010) The measurement was carried out in duplicate using Millipore’s MILLIPLEX® MAP Human High Sensitivity T Cell Magnetic Bead Panel kit (Millipore Corporation, Billerica, MA, USA) and a Luminex 100/200 system (Luminex Corporation, Austin, TX). The kit reagent was used to dilute the samples considering the sample volume and normalized in preparation for analysis. Standard curves for the Luminex assay were produced utilizing known duplicate dilutions. Samples that contained concentrations lower than the detection limit were designated as not detectable, while those exceeding the lower detection limit were adjusted according to the dilution factor. The data were analyzed using GraphPad Prism 10 software (GraphPad Software, La Jolla, CA). As a crucial step in data analysis, the Robust Regression and Outlier Removal (ROUT) methodology has effectively identified outliers with a Q value of 0.1% (Motulsky and Brown, 2006). It should be noted that Q denotes the maximum acceptable false discovery rate. Unpaired samples were compared using the Mann-Whitney test, a non-parametric method. The results have been adjusted for the multiple comparisons utilizing the Bonferroni method. A two-tailed p-value of less than 0.05 determined significance and the results were reported as median & interquartile ranges (IQR).
Of seven corneal infection samples, one case was identified as polymicrobial with the presence of Staphylococcus aureus, coagulase-negative Staphylococcus, and Varicella-Zoster virus (VZV) cultured in the diagnostic microbiology laboratory of the hospital from the corneal scrapes. One infection was caused by Streptococcus pneumonia. Cultured scrapes from three cases were culture negative. Data on the microbes could not be retrieved for the remaining two cases.
The RNA-Seq data for all the samples were subjected to principal components analysis (Supplementary Figure 1) and showed cornea and conjunctiva infection samples clustered together and the control samples clustered together. There was no discernible differentiation between the samples obtained from the cornea and those collected from the conjunctiva. This consistency was found to be the case across different batches. In addition, differential expression analysis identified several differentially expressed genes (DEGs) in the cornea and conjunctiva infection samples compared to healthy volunteers.
In the cornea and conjunctiva of patients with corneal infection compared to controls, 945 differentially expressed genes (DEGs) were identified at a significance level of adjusted P < 0.01. Among these DEGs, 465 were differentially expressed in the cornea only, 433 were differentially expressed in the conjunctiva only, and 47 were differentially expressed in both tissues (Figure 1) (Supplementary Table S1).
The 47 differentially expressed genes common to the cornea and conjunctiva, which were all upregulated genes, were analyzed to investigate their biological function. The enrichGO function from the clusterProfiler Bioconductor package was used to identify significantly enriched Gene Ontology (GO)terms. The cut-off criteria used for this analysis was a q-value of < 0.05. Gene Ontology is divided into three categories: Biological Process (BP), Cellular Component (CC), and Molecular Function (MF). The most significantly enriched GO terms in the Biological Process category were those for neutrophil activation, leukocyte migration, acute inflammatory response, response to molecule of bacterial origin, phagocytosis, humoral immune response, and cytokine secretion (Figure 2A).
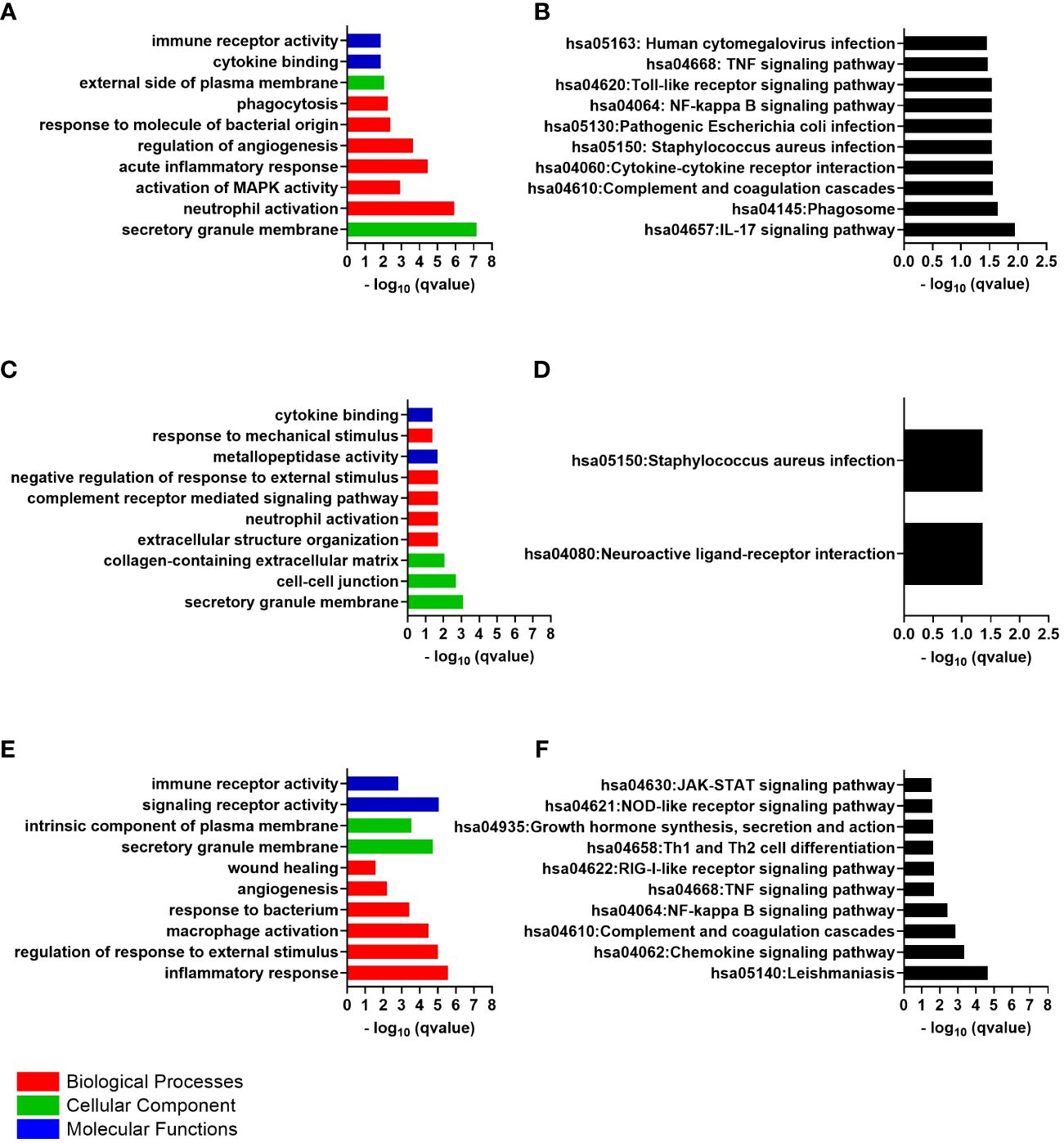
Figure 2 The top 10 gene ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of differentially expressed genes, ranked by their -log p values. (A) Common GO Enrichment analysis; (B) Common KEGG pathway enrichment analysis; (C) Cornea GO Enrichment analysis; (D) Cornea KEGG pathway enrichment analysis; (E) Conjunctiva GO Enrichment analysis; (F) Conjunctiva KEGG pathway enrichment analysis.
The particular genes that were found to be enriched included C-X-C motif chemokine ligand 8 (CXCL8) and its receptor C-X-C motif chemokine receptor 2 (CXCR2), interleukin 1 beta (IL1B), triggering receptor expressed on myeloid cells 1 (TREM1 also called CD354), complement C5a receptor 1 (C5AR1 also called CD88), colony-stimulating factor 3 receptor (CSF3R also called CD114), solute carrier family 11 member 1 (SLC11A1), adhesion G protein-coupled receptor E3 (ADGRE3), Fos proto-oncogene, AP-1 transcription factor subunit (FOS also called or p55), serglycin (SRGN) and NADPH oxidase complex subunit neutrophil cytosolic factor 2 (NCF2). In addition, cell adhesion molecules such as integrin subunit alpha X (ITGAX also called CD11C) and selectin L (SELL also called CD62L) were also enriched in these immune response terms.
Numerous terms were also significantly enriched in the “biological process”, including the regulation of angiogenesis and apoptotic signaling pathway. Within these terms, genes such as thrombospondin 1 (THBS1), prostaglandin-endoperoxide synthase 2 (PTGS2), plasminogen activator, urokinase receptor (PLAUR also called CD87), and G0/G1 switch 2 (G0S2) were enriched. Other enriched terms in the biological process included activation of MAPK activity, activation of protein kinase activity, positive regulation of protein serine/threonine kinase activity, response to cAMP, and positive regulation of reactive oxygen species metabolic process. These terms were found to be enriched with genes such as C5AR1, THBS1, SLC11A1, IL1B, neurotrophic receptor tyrosine kinase 3 (NTRK3), prokineticin 2 (PROK2), FOS, PTGS2, solute carrier family 8 member A1(SLC8A1) and aquaporin 9 (AQP9).
The most significantly enriched GO terms in the “cellular component” were the secretory granule membrane and the external side of plasma membrane. Within these terms, genes such as Fc gamma receptor IIa (FCGR2A also called CD32) and Fc gamma receptor IIIa (FCGR3A also called or CD16) were enriched. The significantly enriched GO terms were cytokine binding and immune receptor activity in the molecular function. The cytokine receptor genes CXCR2 and interleukin 1 receptor type 2 (IL1R2 also called CD121b), were enriched within these terms. Supplementary Table S2 contains the details of the GO terms.
An analysis was conducted on the common DEGs list in the cornea and conjunctiva using the KEGG pathway enrichment method and the most significant KEGG pathways were identified (Figure 2B) (Table 2) (Supplementary Table S3). These were IL-17 signaling pathway, Hematopoietic cell lineage, Neutrophil extracellular trap formation, Phagosome, Complement and coagulation cascades, Cytokine-cytokine receptor interaction, Staphylococcus aureus infection, Pathogenic Escherichia coli infection, NF-kappa B signaling pathway, Toll-like receptor signaling pathway, TNF signaling pathway and Human cytomegalovirus infection.
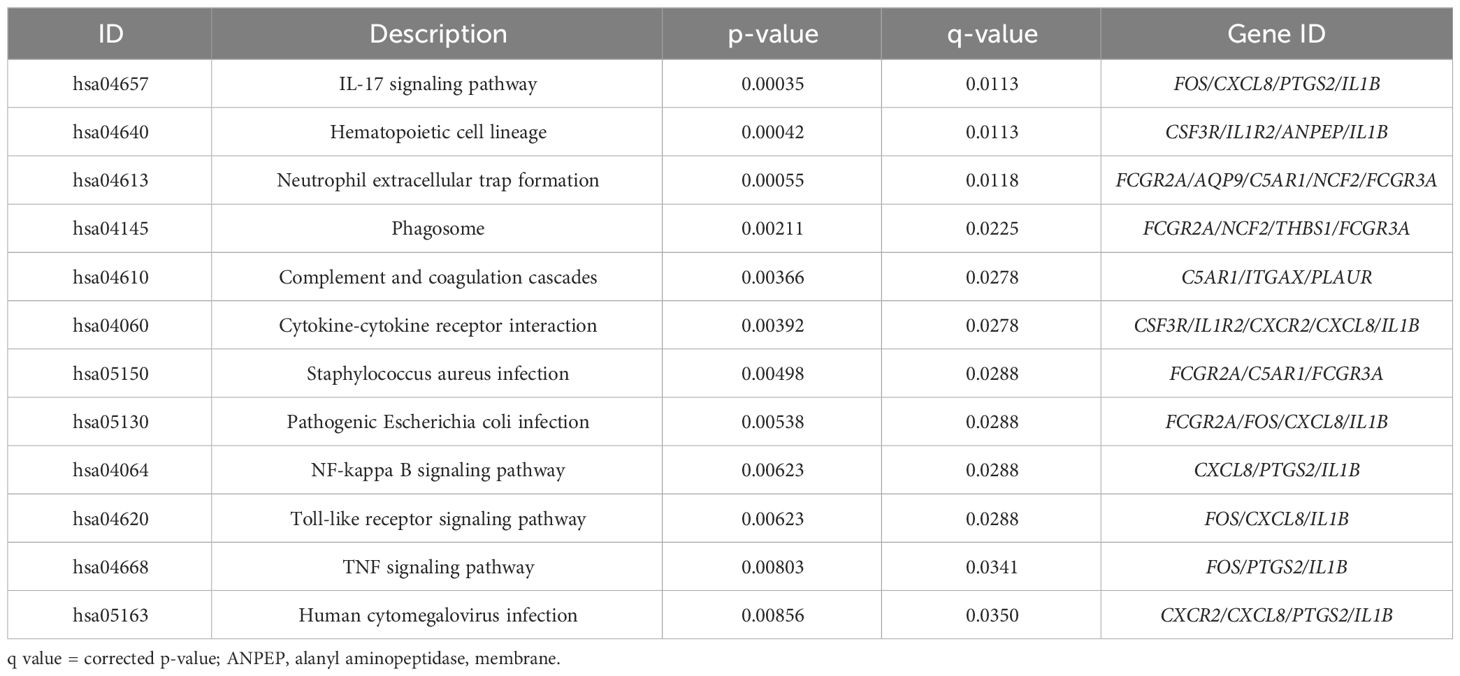
Table 2 The KEGG pathways are ranked in order of likely biological relevance. q value = corrected p-value.
A total of 512 differentially expressed genes were identified after comparing corneal infection samples and healthy volunteers’ samples based on the log fold change threshold criterion > 1 and adjusted P < 0.01. Of these, 508 genes were upregulated, and four were downregulated in the corneal infection samples (Supplementary Table S4).
The three most strongly upregulated genes in the cornea in response to infection were CXCL8, solute carrier family 2 member 3 (SLC2A3) and TREM1. While the downregulated genes were ribosomal protein S27a (RPS27A), ribosomal protein L13a (RPL13A), BAG cochaperone 1(BAG1) and pancreatic progenitor cell differentiation and proliferation factor (PPDPF).
The GO Enrichment analysis indicated that the significantly enriched “biological process” terms were relevant to neutrophil activation, complement receptor-mediated signaling pathway, extracellular structure organization, negative regulation of response to external stimulus and response to mechanical stimulus (Figure 2C). Examples of genes enriched within the Biological Process were CXCL8, CXCR2, ITGAX, IL1B, TNF receptor superfamily member 1B (TNFRSF1B), CD300a molecule (CD300A), formyl peptide receptor 1 and 2 (FPR1, FPR2), adhesion G protein-coupled receptor E3 (ADGRE3), alpha-2-macroglobulin (A2M), fibronectin leucine rich transmembrane protein 2 (FLRT2), ficolin 1 (FCN1), semaphorin 6A (SEMA6A), NLR family pyrin domain containing 3 (NLRP3), and the pattern recognition receptor (PRR) the toll-like receptor 4 (TLR4). Many genes that encode an extracellular matrix protein were upregulated and enriched within these terms, including tenascin C (TNC), laminin subunit alpha 1,2 and 3 (LAMA1, LAMA2 and LAMA3), matrix metallopeptidase 10 and 25 (MMP10 and MMP25). In addition, genes that related to the regulation of cell differentiation and proliferation were also upregulated, such as FOS and FosB proto-oncogene, AP-1 transcription factor subunit (FOSB).
The secretory granule membrane, cell-cell junction, and collagen-containing extracellular matrix were the most significantly enriched GO terms in the “cellular component”, with upregulated genes such as cell adhesion molecule 1 (CADM1), IL2 inducible T cell kinase (ITK), TIMP metallopeptidase inhibitor 3(TIMP3) and FRAS1-related extracellular matrix 1 (FREM1) enriched within these terms. Additionally, genes that contribute to epithelial cell-cell junctions and wound healing, such as desmoglein 3 (DSG3), tenascin C (TNC), collagen type XVII alpha 1 chain (COL17A1) and collagen type VI alpha 6 chain (COL6A6), were upregulated and enriched in the CC terms.
The GO terms significantly enriched in the molecular function category include metallopeptidase activity and cytokine binding. ADAM metallopeptidase domain 23 (ADAM23), chloride channel accessory 2 (CLCA2), MMP10 and MMP25.Additionally, the genes are responsible for encoding the cytokines, and cytokines/chemokines receptors were enriched within the MF terms, such as interleukin 36 receptor antagonist (IL36RN), interleukin 5 receptor subunit alpha (IL5RA), IL1R2 and CXCR2. Supplementary Table S5 contains the details of the GO terms.
The KEGG pathway analysis revealed that the cornea’s differentially expressed genes were significantly enriched in only two pathways: the neuroactive ligand-receptor interaction pathway and the Staphylococcus aureus infection pathway (Figure 2D). Some examples of these genes include C5AR1, FPR1, and FPR2. The Staphylococcus aureus infection pathway showed more upregulated genes, including FCGR2A, keratin 14 (KRT14), and keratin 17 (KRT17).
In comparing conjunctiva samples from corneal infection patients with controls, 480 genes showed differential expression between the two groups. Among these genes, 477 were upregulated, and three were downregulated in the conjunctiva of patients with corneal infection (Supplementary Table S6). This determination was based on a threshold criterion of log fold change > 1.58 and adjusted P < 0.01.
The three most highly upregulated genes in the conjunctiva in response to infection were integrin subunit alpha X (ITGAX), myeloid cell nuclear differentiation antigen (MNDA) and pleckstrin (PLEK) triggering. While the three downregulated genes were ribosomal protein L6 (RPL6), HOP homeobox (HOPX) and ribosomal protein L32 (RPL32).
According to the GO Enrichment analysis, the top “biological process” terms that were significantly enriched relate to the inflammatory response, regulation of response to external stimulus, cytokine production, macrophage activation, and response to bacterium (Figure 2E). These immune response terms included genes like the pattern recognition receptor, the toll-like receptor 1(TLR1), and TLR2. In addition, chemokine genes and chemokine receptor genes were also upregulated: C-X-C motif chemokine ligand 1 (CXCL1), CXCL2, CXCL8, CXCR2 and C-C motif chemokine receptor 1(CCR1). Cytokine genes and cytokine receptors were also present in the immune terms, including IL1B and its receptor IL1R2, interleukin 1 receptor accessory protein (IL1RAP), interferon-gamma receptor 1(IFNGR1), interleukin 4 and 6 receptors (IL4R and IL6R), interleukin 1 receptor associated kinase 3 (IRAK3), additionally to the genes involved in the signaling cascades from these receptors, such as spleen-associated tyrosine kinase (SYK). The analysis also revealed the presence of cytokines production activator such as adhesion G protein-coupled receptor E2 (ADGRE2) and the cytokine signaling negative regulators, the suppressor of cytokine signaling 3 (SOCS3), all of which were enriched within the immune response terms. Genes associated with cell migration and genes promoting cell adhesion, such as FGR proto-oncogene, Src family tyrosine kinase (FGR) and the integrins ITGAM and ITGAX, were also enriched. The genes that related to the regulation of inflammatory response were also involved in the immune response terms like phosphoinositide-3-kinase adaptor protein 1 (PIK3AP1), MEFV innate immunity regulator, pyrin (MEFV), TNF receptor superfamily member 1A (TNFRSF1A) and TNF superfamily member 14 (TNFSF14).
The angiogenesis and wound healing terms were also identified as biological process enriched terms. Those genes that showed considerable differences in expression levels within this pathway included notch receptor 1 (NOTCH1) and integrin subunit alpha 5 (ITGA5). Regarding cellular components, the secretory granule membrane and intrinsic component of plasma membrane were the most significantly enriched GO terms. CD53 molecule (CD53), CD93 molecule (CD93), CD46 molecule (CD46), CD59 molecule (CD59), CD163 molecule (CD163) and transforming growth factor beta receptor 1 (TGFBR1) were found to be upregulated and enriched in the CC terms. Further analysis of differentially expressed genes determined that their primary molecular function is related to signaling receptor activity and immune receptor activity. Supplementary Table S7 contains the details of the GO terms.
An analysis was carried out on the DEGs list in the conjunctiva using the KEGG pathway enrichment method. As a result, the most significant KEGG pathways were identified, including Leishmaniasis, Chemokine signaling pathway, Complement and coagulation cascades, NF-kappa B signaling pathway, TNF signaling pathway, RIG-I-like receptor signaling pathway, Viral protein interaction with cytokine and cytokine receptor, Th17 cell differentiation, Th1 and Th2 cell differentiation, Growth hormone synthesis, secretion and action, NOD-like receptor signaling pathway and JAK-STAT signaling pathway (Figure 2F). Supplementary Table S8 contains the details of the KEGG pathway identified genes.
Out of total 38 corneal infection tear samples analyzed, microbial diagnostic results could be identified for 33, 12 (36%) of which were culture positive. Seven were gram-positive bacteria and five gram-negative bacteria with six cases of polymicrobial (6/12, 50%) keratitis, usually with Herpes Simplex or Varicella Zoster Virus (Supplementary Table S9).
The cytokine concentrations (median and interquartile ranges) were determined for the case and control tear samples, including mild and severe cases versus controls, and severe versus mild cases. Severe keratitis was defined as the final best-corrected visual acuity (BCVA) of ≤ 20/40, repeat scrape or culture positive. Cases of microbial keratitis that did not meet these criteria were classified as mild. Medians and interquartile ranges were tabulated due to the skewed data distribution Table 3. The concentration of IL-8 was slightly but not significantly elevated in corneal infection samples compared to the control group (P > 0.05). The concentration of IL-6 was not significantly higher in corneal infection samples than in the controls after applying the Bonferroni correction for multiple comparisons (P = 0.0067) (corrected P = 0.0536). Patients with severe corneal infection (n = 10) exhibited slightly but significantly higher tear concentrations of IL-6 compared to the controls (P = 0.0303) (corrected P = 0.2424) (Figure 3). As previously mentioned, severe keratitis is defined as having a repeat scrape or positive culture and a final best-corrected visual acuity (BCVA) of ≤ 20/40. There were also no statistically significant differences in the concentration of IL-6 in tears of patients with mild corneal infection (n = 7) compared to the controls and between the severe and mild groups (P = 0.0556) (corrected P = 0.4448) and (P = 0.0878) (corrected P = 0.7024), respectively. IL-1β levels were observed in the patients’ samples; however, they were below the minimum detectable limit in the control samples. Therefore, it was concluded that performing the Mann-Whitney test was not feasible. The study results indicated that CX3CL1 was detected in all participants with no significant variations between the groups. Conversely, in a small number of cases the cytokines IL-10, IL-12 (p70), IL-17A, and IL-23 could be detected, but these cytokines were below the minimum limit of detection in most of the case and control samples. Therefore, it was not feasible to perform the Mann-Whitney test, except for IL-23, which was detected in two control samples (P > 0.9999). The proportion of detectable samples for each cytokine is outlined in Table 4.
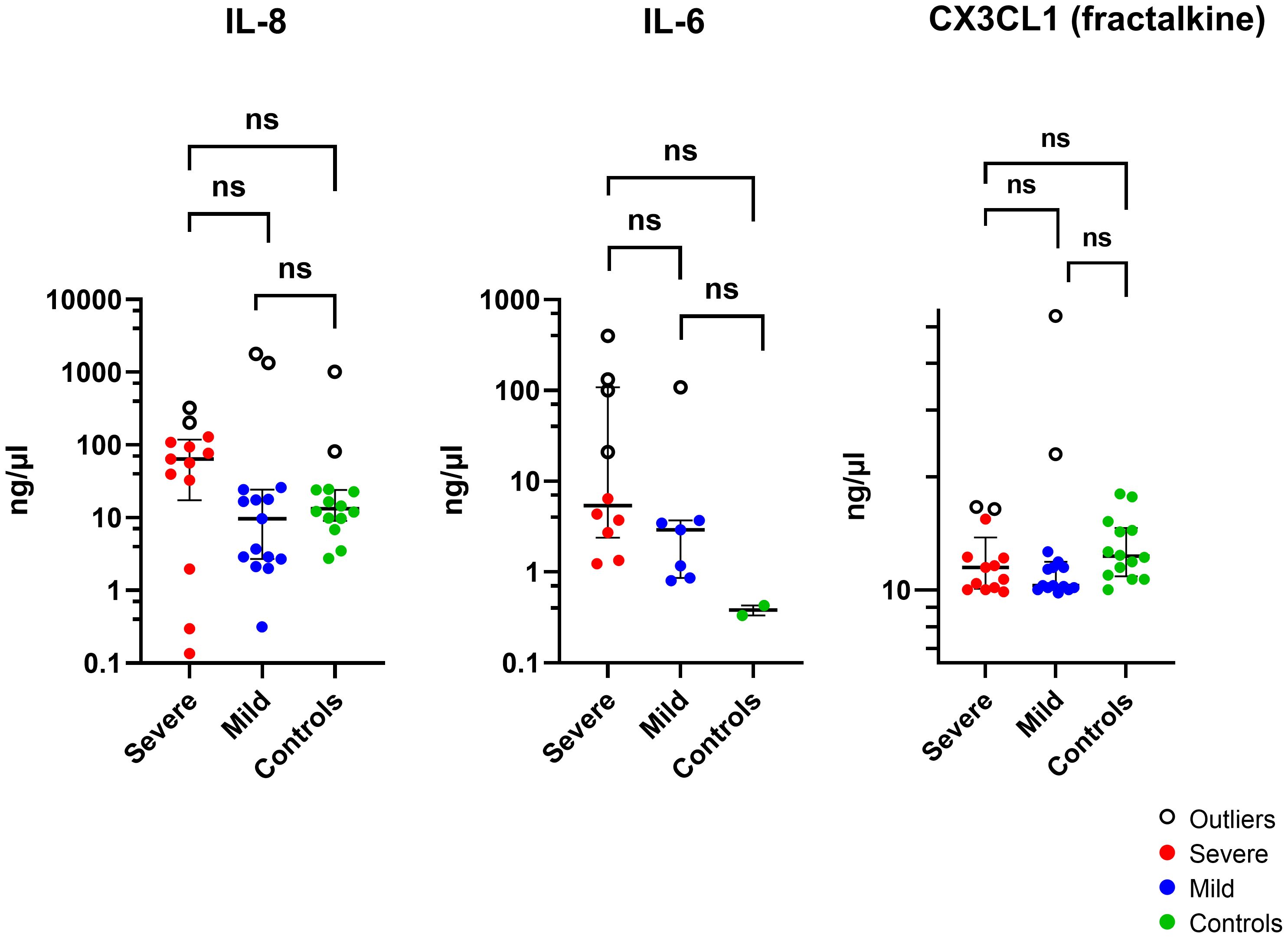
Figure 3 The Log scale scatter plot displays the median ± IQR of tear cytokine levels among individuals with severe and mild corneal infections and controls. Statistical significance between unpaired samples was assessed using a two-tailed Mann-Whitney non-parametric test. NS, not significant (corrected P >0.05).
The RNA cohort had higher levels of CX3CL1 (P = 0.02; corrected P = 0.16) and IL-1β (P = 0.02 corrected P = 0.16) compared to the tear film only cohort, but there was no difference in IL-6 (P = 0.08) and IL-8 (P = 0.17) levels, performed with Mann-Whitney test.
After excluding outliers shown in (Figure 3) (Supplementary Table S10) using the ROUT method, the analysis revealed that there was no statistically significant variation in the concentration of IL-8 protein in tears of patients with corneal infection as compared to the control group (P = 0.7184). A higher concentration of IL-6 was observed in the samples of corneal infection patients compared to the control samples (P = 0.0117; corrected P = 0.0936), but the difference was not statistically significant (P > 0.05). There was also no significant difference observed between the severe and mild groups and the control group (P = 0.0714). Moreover, the concentration of CX3CL1 was higher in corneal infection patients’ samples than in the control group (P = 0.0017; corrected P = 0.0136). Individuals with mild corneal infection had significantly higher CX3CL1 concentrations than the control group (P = 0.0049; corrected P = 0.0392). Nevertheless, there wasn’t any statistically significant difference in the concentration of CX3CL1 in the tears of patients with severe corneal infection compared to healthy volunteers or between the severe and mild groups (P = 0.0505) and (P = 0.5201), respectively.
The study has identified genes, biological processes and pathways that may play a pivotal role in the pathogenesis of corneal infection. Differential expression of genes was observed in both the cornea and conjunctiva of corneal infection patients compared to controls. The upregulated genes that overlapped in both the cornea and conjunctiva had primary associations to immune response, regulation of angiogenesis, and apoptotic signaling pathways. These pathways align with the known biological pathways of keratitis progression (Zhou et al., 2010; Narimatsu et al., 2019; Shrestha et al., 2021; Lin et al., 2022).
CXCL8 (encodes the IL-8 protein) had the most significant upregulation among the differentially expressed genes (DEGs) in the cornea of patients with corneal infection and was upregulated in the conjunctiva. An increased concentration of the IL-8 protein was found in the tears of patients compared to controls, but the difference observed was not statistically significant. CXCL8 is expressed by various ocular surface cell types, including corneal and conjunctival epithelial cells, corneal keratocytes, PMNs, and macrophages, in response to the infection (Elner et al., 1991; Oakes et al., 1993; Scapini et al., 2000; Venza et al., 2007; Waugh and Wilson, 2008; Shtein et al., 2012). Animal studies have shown that homologues of IL-8, such as macrophage inflammatory protein-2 (also known as CXCL2) and keratinocyte-derived chemokine (also known as CXCL1), are also highly expressed in the corneas of rats and mice infected with Pseudomonas aeruginosa or Staphylococcus aureus (Cole et al., 2000; Xue et al., 2003; Hume et al., 2005). As well, the receptor for these CXC chemokines, CXCR2, which was significantly upregulated in the cornea and conjunctiva of the corneal infection patients, plays a significant role in the pathogenesis of bacterial keratitis (Khan et al., 2007; Cole et al., 2014). These studies in animals have shown that CXC chemokines are produced by corneal epithelial cells and infiltrating neutrophils (Cole et al., 2000; Cole et al., 2005) (the conjunctiva of animals was not studied). Furthermore, studies blocking the action of CXCL2 demonstrated its involvement in the neovascularisation of the cornea and recruitment of neutrophils (Xue et al., 2002; Xue et al., 2003).
Cell culture of human corneal epithelial cells confirmed that CXCL8 was also able to be upregulated when the cells were exposed to P. aeruginosa (Xue et al., 2000). Human corneal stromal cells exposed to lipopolysaccharide (LPS) display a time-dependent expression of IL-8 (Shtein et al., 2012). An in vitro study on human adenovirus keratitis found that the IL-8 produced in the process binds to the basement membrane of epithelial tissues, creating tissue reservoirs (Rajaiya et al., 2015).
Polymorphonuclear neutrophils (PMNs) attracted to persistent reservoirs produce MMPs, which result in collagen degradation (Ghasemi et al., 2011). IL-8 plays a crucial role in optimizing this process by increasing MMP production and activation in PMNs (Li et al., 2003a; Ellis et al., 2018; Raziyeva et al., 2021). A previous study has shown that IL-8 plays a role in upregulating the mRNA expression of MMP-2 and MMP-9 in human endothelial cells (Li et al., 2003b).This might have been involved in the upregulation of MMP10 and MMP25 in the cornea and MMP25 in the conjunctiva in the current study.
The ocular surface, particularly the cornea, is known to have a certain level of immune privilege (de Paiva et al., 2022) due to the absence of blood vessels. On the other hand, the conjunctiva possesses immunity and competence (de Paiva et al., 2022). The conjunctiva and to a lesser extent, the cornea are responsible for determining whether to initiate protective mechanisms or tolerate insults (de Paiva et al., 2022). Critical components of conjunctival mucosal tolerance in the ocular surface include CD11c+ antigen-presenting cells (APCs) (Dang et al., 2014; de Paiva et al., 2022). In this study, among the differentially expressed genes in the conjunctiva, ITGAX (CD11c) was the most upregulated. The upregulation of this gene was also observed in the cornea but to a lesser degree. Significant upregulation of ITGAX in patients with late-stage microbial keratitis has been reported compared to normal corneal tissue (Chidambaram et al., 2017). These elevated expression levels may be attributed to the expression of CD11c on myeloid dendritic cells or macrophages (Merad et al., 2013; Mrugacz et al., 2021). This gene was a member in many GO terms in the current study, including inflammatory response, neutrophil activation and degranulation, extracellular structure organization, signaling receptor activity, and angiogenesis. A study has revealed that CD11c expression is notably elevated in individuals with atopic keratoconjunctivitis, a condition characterized by persistent inflammation of the conjunctiva (Matsuda et al., 2019). The macrophage response and phenotype were analyzed in a murine model of corneal inflammation. It was observed that macrophages accumulated on both the corneal endothelium and stroma (Chinnery et al., 2014). According to a study on mice, the central and peripheral corneas showed a higher density of dendritic cells during the initial stages of Herpes simplex virus type 1 keratitis (Jamali et al., 2020).
CD11c has been identified as a significant contributor to the cornea’s immune response and inflammatory processes during P. aeruginosa infection. Previous murine studies have demonstrated the crucial role of CD11c in preventing adhesion and penetration of P. aeruginosa in healthy cornea (Metruccio et al., 2017). CD11c has also been shown to play a significant role in activating immune and inflammatory responses during P. aeruginosa infections, thus contributing to the defense against the pathogen (Metruccio et al., 2017). Recent research on murine subjects has demonstrated that CD11c plays a significant role in regulating the neutrophil maturation process and its associated effector functions (Hou et al., 2023). In a mouse study on P. aeruginosa corneal infections, initiating apoptosis of neutrophils at an earlier stage resulted in better disease outcomes (Zhou et al., 2008). Considering this, targeting the CD11c to control the neutrophil maturation process during corneal infections could be a promising strategy for enhancing disease outcomes. While the present study has identified significant transcriptional changes in CD11c in the conjunctiva and cornea of corneal infection patients, it is necessary to conduct further investigation to determine whether these gene expression changes correspond to protein expression differences.
Evidence suggests that besides CD11c, TREM1 is also a critical receptor in regulating immune responses during the early stages of bacterial infection (Bleharski et al., 2003; Colonna, 2003; Lagler et al., 2009; Hommes et al., 2014; Colonna, 2023). The current study has unveiled a significant upregulation in TREM1 in both the cornea and conjunctiva of the corneal infection patient. A study on the P. aeruginosa murine keratitis model showed that TREM1 has an inflammatory amplification effect during infection (Wu et al., 2011). TREM1 expression is detectable in different types of immune cells, such as PMNs, monocytes, and macrophages (Bouchon et al., 2000; Schenk et al., 2007; Ferat-Osorio et al., 2008), which is likely the driver of the increased expression in the present study. TREM1 stimulates the release of IL-8 and induces the upregulation of adhesion molecules, including CD11c (Bouchon et al., 2000). These findings suggest that TREM1 may be involved in the upregulation of these genes in the current study. In cases of unilateral bacterial keratitis, there is a notable elevation in the concentration of TREM1 in both eyes (Yamaguchi et al., 2014). Blocking TREM1 in a mouse model of P. aeruginosa keratitis resulted in a reduction of corneal disease severity (Wu et al., 2011). Conversely, activating TREM1 signaling exacerbated the disease, leading to an earlier corneal perforation (Wu et al., 2011). Further research is required to assess the viability of TREM1 as a diagnostic indicator for determining the extent of corneal infections and predicting unfavorable outcomes.
The findings of the current study indicate that most downregulated genes in both the cornea and conjunctiva were those encoding ribosomal proteins. It is noteworthy that among the identified genes, RPS27A plays a crucial role in the biogenesis of ribosomes and cellular growth (Nosrati et al., 2015; Zhou et al., 2015). RPS27A has been shown to exert a regulatory effect on cell cycle progression, promote proliferation, and have an inhibition effect on apoptosis of leukemia cells (Wang et al., 2014). In addition to ribosomal protein genes, this study found that the BAG1 was downregulated in the cornea. BAG1 is known to possess anti-apoptotic properties and may have a significant role in preventing cell death through autophagy activation (Portt et al., 2011), which is the cellular mechanism of recycling (Kamil and Mohan, 2021). The reduced expression of ribosomal protein genes and BAG1 in the cornea suggests that corneal cells could potentially be at a higher vulnerability to apoptotic triggers (Portt et al., 2011). In the current study, PPDPF is identified as one of the down-regulated genes in the cornea. Previous research has shown that PPDPF is expressed in the corneal limbus of healthy individuals (Li et al., 2022). This may indicate the important role of this gene in regulating corneal cell differentiation and proliferation. In a previous study conducted on rats, it was found that the expression level of this gene increased with the onset of cataracts in an ex vivo model (Nagaya et al., 2022). Cataracts are an eye condition that causes cloudiness and opaqueness of the lens (Nagaya et al., 2022). Whereas, in conjunctiva, the study found that the expression of HOPX was significantly reduced, in addition to the ribosomal protein genes. As part of the investigation into the differentiation of limbal stem cells in homeostasis, a significant increase in the expression of the HOPX was observed in wing cells (Lin et al., 2023). This indicates that HOPX plays a crucial role in regulating the differentiation and regenerative processes of epithelial tissues, as seen in other similar tissues (Yang et al., 2010; Ota et al., 2018). Silencing HOPX in human keratinocytes (Yang et al., 2010) and murine alveolar epithelial cell lines (Ota et al., 2018) increases cellular proliferation (Ota et al., 2018) and differentiation markers (Yang et al., 2010). Taken together, these findings suggest that corneal infection results in an increase in apoptosis and a decrease in differentiation compared to a non-infected cornea. The apoptosis of the keratocytes can serve as a protective mechanism against the posterior spread of infectious pathogens in corneal injuries (Wilson and Esposito, 2009). These apoptosis signals attract inflammatory cells to the area, which are crucial for healing (Yoon et al., 2020; Kamil and Mohan, 2021). Further research is required to establish the exact role of BAG1, PPDPF, HOPX, and ribosomal protein genes in the cornea and conjunctiva, both in normal and pathological states.
A further objective of this current research was to analyze cytokines and chemokines levels in the tears of patients with corneal infections. The study compared those with mild infection to those with severe infection and healthy controls. The results revealed that although the difference in IL-8 concentration did not show statistical significance, it is noteworthy that the levels were relatively higher in patients than in the control group. This finding was consistent with RNA-seq results. However, mRNA-protein correlation can depend upon factors such as post-transcriptional regulation, including protein degradation and protein buffering which may contribute to this imbalance (Buccitelli and Selbach, 2020). Furthermore, IL-6 was significantly detected in the tears of patients with severe corneal infections compared to healthy volunteers, but it was not statistically significant after the Bonferroni correction. Previous research has shown that increased levels of IL-8 and IL-6 have been observed in patients suffering from bacterial keratitis caused by gram-negative or gram-positive bacteria compared to healthy controls (Yu et al., 2012; Wu et al., 2021). Moreover, the levels of IL-8 and IL-6 reported in these studies were higher than those observed in the present study. The presence of CX3CL1 was observed in all study participants. However, no statistically significant variances were observed between the groups and did not appear in this study’s list of genes that were differentially expressed. However, IL-10, IL-12 (p70), IL-17A, and IL-23 were detected in only a small number of cases and were found to be below the minimum limit of detection in most samples. These cytokines were not found in the list of genes that exhibited differential expressions. The findings of the present study align with previous research on bacterial keratitis, indicating that IL-17A levels were not significantly increased in the affected eye compared to healthy controls (Powell et al., 2010). However, the study found that IL-17A levels were significantly elevated in the unaffected contralateral eyes (Powell et al., 2010). These results suggest that IL-17A may play a role in the pathogenesis of bacterial keratitis, and further research is required to understand its effects fully. The level of IL-10 and IL-12 (p70) in the tear of bacterial keratitis patients was higher in previous studies (Yu et al., 2012), than in this study. Differences observed between the current study’s cytokines levels and previous research might be due to the sample size. Therefore, a more comprehensive investigation of these proteins with a larger sample size was required.
Although there are few studies on human microbial keratitis gene expression, this study’s results show some similarity to previous ocular expression studies. Earlier studies on expressed genes in human corneal tissue and scrapings showed that bacterial keratitis leads to an increase in the expression of TLR4 and TLR2 genes (Karthikeyan et al., 2013; Chidambaram et al., 2017; Tian et al., 2020). The findings of this study are consistent with previous research, revealing significant regulation of TLR4. In the conjunctiva, TLR1 and TLR2 are also regulated. The current study identified 47 upregulated genes overlapping between the cornea and conjunctiva. Notably, 21 of these genes were previously listed in the transcriptional profile of the late-stage human bacterial keratitis microarray study (Chidambaram et al., 2017). Interestingly, these genes included CXCL8, ITGAX, TREM1, IL1B, SLC11A1, C5AR1, PTGS2, G0S2, AQP9, IL1R2, and CD163. It is worth noting that another 21 genes were also upregulated, specifically in the cornea in the current study and the previous study (Chidambaram et al., 2017). These genes included NLRP3, FOSB, FPR1, FPR2, and MMP10. There were notable dissimilarities in the findings of both studies. It’s worth noting that while the previous study (Chidambaram et al., 2017) found that among the chemokines, CXCL2 showed the highest fold change, the current study observed up-regulation of genes for CXCR2, THBS1, FOS and MMP25, which were not included in the gene list of the previous study (Chidambaram et al., 2017). THBS1 and FOS play essential roles in the process of tissue regeneration and wound healing (Verrier et al., 1986; Blanco-Mezquita et al., 2013). The precise role of MMP25 in wound healing is not yet fully understood (Caley et al., 2015). Nonetheless, its expression in leukocytes suggests that MMP25 may significantly influence IL-8 secretion and respiratory bursts (Cui et al., 2017). The observed distinction may be attributable to the use of microbial keratitis corneal tissues for the microarray study, whereas the control incorporated normal cadaver corneas. Furthermore, the limitations of microarray technology, such as cross-hybridization issues and probe selection biases, could also contribute to the disparity (Liu et al., 2007; Roy et al., 2011). RNA sequencing can provide an unbiased identification of all transcripts in a sample, offering a more comprehensive gene expression analysis (Liu et al., 2007; Roy et al., 2011). Moreover, the observed differences between the two studies could also be due to a heightened emphasis on late-stage keratitis within a median of 15 days following the onset of symptoms in the previous study. These differences highlight the need for continued research to understand the complex mechanisms of microbial keratitis pathogenesis.
A recent study analyzed the molecular changes in the human cornea during bacterial/fungal keratitis using RNA sequences (Lapp et al., 2024). It is worth noting that 31/47 genes upregulated in the cornea and conjunctiva in the current study were also found to be significantly upregulated in this recent study using similar RNA sequence technology (Lapp et al., 2024). These genes included CXCL8, ITGAX, TREM1, CD163, C5AR1, CXCR2, MMP25 and THBS1. Two of the 47 upregulated genes in the current study were significantly downregulated in the recent RNA sequences study (Lapp et al., 2024). However, the function of these genes is unlikely to be important in this context. These differences in differentially expressed genes are likely affected by the limited sample size in both studies, differences in the sample processing techniques and the nature of the samples. For example, the current study collected swab samples from the corneal lesion of the patients at the time of diagnosis. Meanwhile, in the previous research, corneal tissue was obtained from people who had undergone keratoplasty, where the tissue was processed by fixation in formalin and subsequent embedding in paraffin (Lapp et al., 2024). Even with these differences, the upregulated genes of both studies show significant similarities. This suggests a common immune response mechanism to microbial keratitis and provides a strong basis for further research into the mechanisms of microbial keratitis pathogenesis.
There are several limitations of the current study. Due to the limited samples available, the statistical power of the experiment was restricted. To ensure a more comprehensive analysis, additional samples from both corneal infection patients and controls are required. It is unknown specifically what type of cells are collected with the swabs, nor the exact number of cells. However, the major cell type that was most probably collected is superficial epithelial cells (Anagnostopoulou-Fotinopoulou and Rammou-Kinia, 1993; Kobayashi et al., 1997; Yağmur et al., 1997). Furthermore, only two of seven corneal infection samples were culture-positive for bacteria. Several research studies have conducted comparative analyses of keratitis cases, including culture-positive and culture-negative cases (Sharma et al., 2007; Bhadange et al., 2015; Atta et al., 2023). According to these studies, several possible reasons exist for the absence of microbial growth in culture-negative cases. These reasons include using antimicrobials before presentation, inadequate specimen collection due to the smaller size of the defects, and the unavailability of appropriate types of culture (Sharma et al., 2007; Bhadange et al., 2015; Atta et al., 2023). The positivity rate of 40% in the current is within normal limits of corneal cultures (Ng et al., 2015; Oliveira-Ferreira et al., 2019). While positive corneal culture may have different presenting and prognostic factors, a larger sample size would be required to determine differences between culture positive and negative cases gene expression. A strength of the current study was the ability to detect gene expression and to part replicate the results in protein concentration for some of the targets. To better comprehend the mechanisms underlying the observed changes in gene expression, it is crucial to conduct targeted gene expression analysis of immune response to culture positive and culture negative, gram-positive and gram-negative bacteria, as well as patient severity outcomes.
This study investigated the changes in the immune transcriptomic profile in the cornea and conjunctiva, as well as the tear cytokines of patients with corneal infection. The study findings revealed a significant overlap in the differential gene expression observed in the cornea and conjunctiva and identified a set of genes and pathways that play a role in corneal infection pathogenesis. It was also observed that the corneal infection genes were highly associated with immune response, regulation of angiogenesis, and regulation of apoptotic signaling pathways. Although this study observed moderate alignment between gene and tear protein expression, the integration of multi-omics datasets in future studies is likely to provide a better understanding of the complex genome to phenotype relationship and hold insights for better therapeutics. This study’s outcomes can offer valuable insights for further research. In future, these may contribute to the development of targeted therapeutic interventions to reduce tissue damage and control infection severity by targeting the underlying genetic and immunological factors associated with corneal infection.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1050754. Repository, accession number PRJNA1050754.
The studies involving humans were approved by the Western Sydney Local Health District Human Research Ethics Committee (LNR/16/WMEAD/401), and the research followed the tenets of the Declaration of Helsinki. All participants signed informed consent prior to study commencement. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
HA: Data curation, Formal Analysis, Investigation, Software, Writing – original draft, Writing – review & editing. GP: Methodology, Supervision, Writing – review & editing. SS: Methodology, Writing – review & editing. JO: Supervision, Writing – review & editing. MW: Supervision, Writing – review & editing. AW: Supervision, Writing – review & editing. NC: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the 2015 American Optometric Foundation Allergan Foundation Research Grant.
Scientia Support funding to NC contributed to the cost of consumables, sequencing and data analysis. This study is supported via funding from Prince Sattam bin Abdulaziz University project number (PSAU/2023/R/1445).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1346821/full#supplementary-material
FC, Fold-Change; TLRs, Toll-like Receptors; LPS, Lipopolysaccharide; SD, Standard Deviation; RIN, RNA Integrity Number; PCA, Principal Components Analysis; NCBI, National Center for Biotechnology Information; DEGs, Differentially Expressed Genes; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; IQR, Interquartile Ranges; BP, Biological Process; CC, Cellular Component; MF, Molecular Function; PMNs, Polymorphonuclear Neutrophils; APCs, Antigen-Presenting Cells.
Al-Mujaini, A., Al-Kharusi, N., Thakral, A., Wali, U. K. (2009). Bacterial keratitis: perspective on epidemiology, clinico-pathogenesis, diagnosis and treatment. Sultan Qaboos Univ Med. J. 9, 184–195.
Anagnostopoulou-Fotinopoulou, I., Rammou-Kinia, R. (1993). Cytobrush sampling in conjunctival cytology. Diagn. Cytopathol. 9, 113–115. doi: 10.1002/dc.2840090123
Anders, S., Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11, R106. doi: 10.1186/gb-2010-11-10-r106
Atta, S., Singh, R. B., Samanthapudi, K., Perera, C., Omar, M., Nayyar, S., et al. (2023). Clinical characterization and outcomes of culture- and polymerase chain reaction-negative cases of infectious keratitis. Diagnostics (Basel) 13 (15), 2528. doi: 10.3390/diagnostics13152528
Barton, G. M., Medzhitov, R. (2003). Toll-like receptor signaling pathways. Science 300, 1524–1525. doi: 10.1126/science.1085536
Bhadange, Y., Das, S., Kasav, M. K., Sahu, S. K., Sharma, S. (2015). Comparison of culture-negative and culture-positive microbial keratitis: cause of culture negativity, clinical features and final outcome. Br. J. ophthalmology. 99, 1498–1502. doi: 10.1136/bjophthalmol-2014-306414
Blanco-Mezquita, J. T., Hutcheon, A. E., Zieske, J. D. (2013). Role of thrombospondin-1 in repair of penetrating corneal wounds. Invest. Ophthalmol. Vis. Sci. 54, 6262–6268. doi: 10.1167/iovs.13-11710
Bleharski, J. R., Kiessler, V., Buonsanti, C., Sieling, P. A., Stenger, S., Colonna, M., et al. (2003). A role for triggering receptor expressed on myeloid cells-1 in host defense during the early-induced and adaptive phases of the immune response. J. Immunol. 170, 3812–3818. doi: 10.4049/jimmunol.170.7.3812
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bouchon, A., Dietrich, J., Colonna, M. (2000). Cutting edge: inflammatory responses can be triggered by TREM-1, a novel receptor expressed on neutrophils and monocytes. J. Immunol. 164, 4991–4995. doi: 10.4049/jimmunol.164.10.4991
Bourcier, T., Thomas, F., Borderie, V., Chaumeil, C., Laroche, L. (2003). Bacterial keratitis: predisposing factors, clinical and microbiological review of 300 cases. Br. J. Ophthalmology. 87, 834–838. doi: 10.1136/bjo.87.7.834
Buccitelli, C., Selbach, M. (2020). mRNAs, proteins and the emerging principles of gene expression control. Nat. Rev. Genet. 21, 630–644. doi: 10.1038/s41576-020-0258-4
Cabrera-Aguas, M., Khoo, P., George, C. R. R., Lahra, M. M., Watson, S. L. (2020). Antimicrobial resistance trends in bacterial keratitis over 5 years in Sydney, Australia. Clin. Exp. Ophthalmol. 48, 183–191. doi: 10.1111/ceo.13672
Caley, M. P., Martins, V. L., O'Toole, E. A. (2015). Metalloproteinases and wound healing. Adv. Wound Care (New Rochelle). 4, 225–234. doi: 10.1089/wound.2014.0581
Chen, C. A., Hsu, S. L., Hsiao, C. H., Ma, D. H., Sun, C. C., Yu, H. J., et al. (2020). Comparison of fungal and bacterial keratitis between tropical and subtropical Taiwan: a prospective cohort study. Ann. Clin. Microbiol. Antimicrob. 19, 11. doi: 10.1186/s12941-020-00353-3
Chidambaram, J. D., Kannambath, S., Srikanthi, P., Shah, M., Lalitha, P., Elakkiya, S., et al. (2017). Persistence of innate immune pathways in late stage human bacterial and fungal keratitis: results from a comparative transcriptome analysis. Front. Cell Infect. Microbiol. 7, 193. doi: 10.3389/fcimb.2017.00193
Chinnery, H. R., Leong, C. M., Chen, W., Forrester, J. V., McMenamin, P. G. (2014). TLR9 and TLR7/8 activation induces formation of keratic precipitates and giant macrophages in the mouse cornea. J. Leukocyte Biol. 97, 103–110. doi: 10.1189/jlb.3AB0414-216R
Chusid, M. J., Davis, S. D. (1979). Experimental bacterial keratitis in neutropenic Guinea pigs: polymorphonuclear leukocytes in corneal host defense. Infect. Immun. 24, 948–952. doi: 10.1128/iai.24.3.948-952.1979
Cole, N., Bao, S., Thakur, A., Willcox, M., Husband, A. J. (2000). KC production in the cornea in response to Pseudomonas aeruginosa challenge. Immunol. Cell Biol. 78, 1–4. doi: 10.1046/j.1440-1711.2000.00860.x
Cole, N., Hume, E. B., Khan, S., Garthwaite, L., Conibear, T. C., Willcox, M. D. (2014). The role of CXC chemokine receptor 2 in Staphylococcus aureus keratitis. Exp. Eye Res. 127, 184–189. doi: 10.1016/j.exer.2014.07.016
Cole, N., Hume, E., Khan, S., Madigan, M., Husband, A. J., Garthwaite, L., et al. (2005). Contribution of the cornea to cytokine levels in the whole eye induced during the early phase of Pseudomonas aeruginosa challenge. Immunol. Cell Biol. 83, 301–306. doi: 10.1111/j.1440-1711.2005.01324.x
Colonna, M. (2003). TREMs in the immune system and beyond. Nat. Rev. Immunol. 3, 445–453. doi: 10.1038/nri1106
Colonna, M. (2023). The biology of TREM receptors. Nat. Rev. Immunol. 23, 580–594. doi: 10.1038/s41577-023-00837-1
Cui, N., Hu, M., Khalil, R. A. (2017). Biochemical and biological attributes of matrix metalloproteinases. Prog. Mol. Biol. Transl. Sci. 147, 1–73. doi: 10.1016/bs.pmbts.2017.02.005
Dang, Z., Kuffová, L., Liu, L., Forrester, J. V. (2014). Soluble antigen traffics rapidly and selectively from the corneal surface to the eye draining lymph node and activates T cells when codelivered with CpG oligonucleotides. J. Leukocyte Biol. 95, 431–440. doi: 10.1189/jlb.0612294
Das, S., Samantaray, R., Mallick, A., Sahu, S. K., Sharma, S. (2019). Types of organisms and in-vitro susceptibility of bacterial isolates from patients with microbial keratitis: A trend analysis of 8 years. Indian J. Ophthalmology. 67, 49–53. doi: 10.4103/ijo.IJO_500_18
de Paiva, C. S., St. Leger, A. J., Caspi, R. R. (2022). Mucosal immunology of the ocular surface. Mucosal Immunol. 15, 1143–1157. doi: 10.1038/s41385-022-00551-6
Ellis, S., Lin, E. J., Tartar, D. (2018). Immunology of wound healing. Curr. Dermatol. Rep. 7, 350–358. doi: 10.1007/s13671-018-0234-9
Elner, V. M., Strieter, R. M., Pavilack, M. A., Elner, S. G., Remick, D. G., Danforth, J. M., et al. (1991). Human corneal interleukin-8. IL-1 and TNF-induced gene expression and secretion. Am. J. Pathol. 139, 977–988.
Ferat-Osorio, E., Esquivel-Callejas, N., Wong-Baeza, I., Aduna-Vicente, R., Arriaga-Pizano, L., Sánchez-Fernández, P., et al. (2008). The increased expression of TREM-1 on monocytes is associated with infectious and noninfectious inflammatory processes. J. Surg. Res. 150, 110–117. doi: 10.1016/j.jss.2007.12.805
Ghasemi, H., Ghazanfari, T., Yaraee, R., Faghihzadeh, S., Hassan, Z. M. (2011). Roles of IL-8 in ocular inflammations: A review. Ocul Immunol. Inflamm. 19, 401–412. doi: 10.3109/09273948.2011.618902
Green, M., Carnt, N., Apel, A., Stapleton, F. (2019). Queensland microbial keratitis database: 2005–2015. Br. J. Ophthalmology. 103, 1481–1486. doi: 10.1136/bjophthalmol-2018-312881
Hazlett, L. D. (2004). Corneal response to Pseudomonas aeruginosa infection. Prog. Retin Eye Res. 23, 1–30. doi: 10.1016/j.preteyeres.2003.10.002
Hazlett, L. D. (2005). Role of innate and adaptive immunity in the pathogenesis of keratitis. Ocul Immunol. Inflamm. 13, 133–138. doi: 10.1080/09273940490912362
Hoddenbach, J. G., Boekhoorn, S. S., Wubbels, R., Vreugdenhil, W., Van Rooij, J., Geerards, A. J. M. (2014). Clinical presentation and morbidity of contact lens–associated microbial keratitis: a retrospective study. Graefe's Arch. Clin. Exp. Ophthalmology. 252, 299–306. doi: 10.1007/s00417-013-2514-1
Hommes, T. J., Hoogendijk, A. J., Dessing, M. C., Van't Veer, C., Florquin, S., Colonna, M., et al. (2014). Triggering receptor expressed on myeloid cells-1 (TREM-1) improves host defence in pneumococcal pneumonia. J. Pathol. 233, 357–367. doi: 10.1002/path.4361
Hoshino, K., Takeuchi, O., Kawai, T., Sanjo, H., Ogawa, T., Takeda, Y., et al. (1999). Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J. Immunol. 162, 3749–3752. doi: 10.4049/jimmunol.162.7.3749
Hou, L., Voit, R. A., Shibamura-Fujiogi, M., Koutsogiannaki, S., Li, Y., Chen, Y., et al. (2023). CD11c regulates neutrophil maturation. Blood Adv. 7, 1312–1325. doi: 10.1182/bloodadvances.2022007719
Huang, X., Du, W., McClellan, S. A., Barrett, R. P., Hazlett, L. D. (2006). TLR4 is required for host resistance in Pseudomonas aeruginosa keratitis. Invest. Ophthalmol. Vis. Sci. 47, 4910–4916. doi: 10.1167/iovs.06-0537
Hume, E. B., Cole, N., Khan, S., Garthwaite, L. L., Aliwarga, Y., Schubert, T. L., et al. (2005). A Staphylococcus aureus mouse keratitis topical infection model: cytokine balance in different strains of mice. Immunol. Cell Biol. 83, 294–300. doi: 10.1111/j.1440-1711.2005.01326.x
Jamali, A., Hu, K., Sendra, V. G., Blanco, T., Lopez, M. J., Ortiz, G., et al. (2020). Characterization of resident corneal plasmacytoid dendritic cells and their pivotal role in herpes simplex keratitis. Cell Rep. 32, 108099. doi: 10.1016/j.celrep.2020.108099
Johnson, A. C., Heinzel, F. P., Diaconu, E., Sun, Y., Hise, A. G., Golenbock, D., et al. (2005). Activation of toll-like receptor (TLR)2, TLR4, and TLR9 in the mammalian cornea induces myD88-dependent corneal inflammation. Invest. Ophthalmol. Vis. Sci. 46, 589–595. doi: 10.1167/iovs.04-1077
Kamil, S., Mohan, R. R. (2021). Corneal stromal wound healing: Major regulators and therapeutic targets. Ocul Surf. 19, 290–306. doi: 10.1016/j.jtos.2020.10.006
Karthikeyan, R. S., Priya, J. L., Leal, S. M., Jr., Toska, J., Rietsch, A., Prajna, V., et al. (2013). Host Response and Bacterial Virulence Factor Expression in Pseudomonas aeruginosa and Streptococcus pneumoniae Corneal Ulcers. PloS One 8, e64867. doi: 10.1371/journal.pone.0064867
Keay, L., Edwards, K., Naduvilath, T., Taylor, H. R., Snibson, G. R., Forde, K., et al. (2006). Microbial keratitis: predisposing factors and morbidity. Ophthalmology 113, 109–116. doi: 10.1016/j.ophtha.2005.08.013
Khan, S., Cole, N., Hume, E. B., Garthwaite, L., Conibear, T. C., Miles, D. H., et al. (2007). The role of CXC chemokine receptor 2 in Pseudomonas aeruginosa corneal infection. J. Leukoc. Biol. 81, 315–318. doi: 10.1189/jlb.0506344
Kim, D., Paggi, J. M., Park, C., Bennett, C., Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Kobayashi, T. K., Ueda, M., Nishino, T., Ishida, Y., Takamura, E., Tsubota, K. (1997). Cytologic evaluation of conjunctival epithelium using Cytobrush-S: value of slide preparation by ThinPrep technique. Cytopathology 8, 381–387. doi: 10.1111/j.1365-2303.1997.tb00568.x
Lagler, H., Sharif, O., Haslinger, I., Matt, U., Stich, K., Furtner, T., et al. (2009). TREM-1 activation alters the dynamics of pulmonary IRAK-M expression in vivo and improves host defense during pneumococcal pneumonia. J. Immunol. 183, 2027–2036. doi: 10.4049/jimmunol.0803862
Lapp, T., Kammrath Betancor, P., Schlunck, G., Auw-Hädrich, C., Maier, P., Lange, C., et al. (2024). Transcriptional profiling specifies the pathogen-specific human host response to infectious keratitis. Front. Cell Infect. Microbiol. 13. doi: 10.3389/fcimb.2023.1285676
Li, A., Dubey, S., Varney, M. L., Dave, B. J., Singh, R. K. (2003a). IL-8 directly enhanced endothelial cell survival, proliferation, and matrix metalloproteinases production and regulated angiogenesis1. J. Immunol. 170, 3369–3376. doi: 10.4049/jimmunol.170.6.3369
Li, A., Dubey, S., Varney, M. L., Dave, B. J., Singh, R. K. (2003b). IL-8 directly enhanced endothelial cell survival, proliferation, and matrix metalloproteinases production and regulated angiogenesis. J. Immunol. 170, 3369–3376. doi: 10.4049/jimmunol.170.6.3369
Li, Y., Jeong, J., Song, W. (2022). Molecular characteristics and distribution of adult human corneal immune cell types. Front. Immunol. 13, 798346. doi: 10.3389/fimmu.2022.798346
Lin, W., Chen, M., Cissé, Y., Chen, X., Bai, L. (2022). Roles and mechanisms of regulated necrosis in corneal diseases: progress and perspectives. J. Ophthalmol. 2022, 2695212. doi: 10.1155/2022/2695212
Lin, J. B., Shen, X., Pfeifer, C. W., Shiau, F., Santeford, A., Ruzycki, P. A., et al. (2023). Dry eye disease in mice activates adaptive corneal epithelial regeneration distinct from constitutive renewal in homeostasis. Proc. Natl. Acad. Sci. U S A. 120, e2204134120. doi: 10.1073/pnas.2204134120
Liu, F., Jenssen, T. K., Trimarchi, J., Punzo, C., Cepko, C. L., Ohno-MaChado, L., et al. (2007). Comparison of hybridization-based and sequencing-based gene expression technologies on biological replicates. BMC Genomics 8, 153. doi: 10.1186/1471-2164-8-153
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Matsuda, A., Asada, Y., Suita, N., Iwamoto, S., Hirakata, T., Yokoi, N., et al. (2019). Transcriptome profiling of refractory atopic keratoconjunctivitis by RNA sequencing. J. Allergy Clin. Immunol. 143, 1610–4.e6. doi: 10.1016/j.jaci.2018.11.007
Merad, M., Sathe, P., Helft, J., Miller, J., Mortha, A. (2013). The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu. Rev. Immunol. 31, 563–604. doi: 10.1146/annurev-immunol-020711-074950
Metruccio, M. M. E., Tam, C., Evans, D. J., Xie, A. L., Stern, M. E., Fleiszig, S. M. J. (2017). Contributions of MyD88-dependent receptors and CD11c-positive cells to corneal epithelial barrier function against Pseudomonas aeruginosa. Sci. Rep. 7, 13829. doi: 10.1038/s41598-017-14243-w
Mohammed, I., Abedin, A., Tsintzas, K., Abedin, S. A., Otri, A. M., Hopkinson, A., et al. (2011). Increased expression of hepcidin and toll-like receptors 8 and 10 in viral keratitis. Cornea 30, 899–904. doi: 10.1097/ICO.0b013e31820126e5
Motulsky, H. J., Brown, R. E. (2006). Detecting outliers when fitting data with nonlinear regression – a new method based on robust nonlinear regression and the false discovery rate. BMC Bioinf. 7, 123. doi: 10.1186/1471-2105-7-123
Mrugacz, M., Bryl, A., Falkowski, M., Zorena, K. (2021). Integrins: an important link between angiogenesis, inflammation and eye diseases. Cells 10 (7), 1703. doi: 10.3390/cells10071703
Mueller, O., Lightfoot, S., Schroeder, A. (2004). RNA integrity number (RIN)–standardization of RNA quality control. Agilent Appl. note publication. 1, 1–8.
Musa, F., Tailor, R., Gao, A., Hutley, E., Rauz, S., Scott, R. A. H. (2010). Contact lens-related microbial keratitis in deployed British military personnel. Br. J. Ophthalmology. 94, 988–993. doi: 10.1136/bjo.2009.161430
Nagaya, M., Yamaoka, R., Kanada, F., Sawa, T., Takashima, M., Takamura, Y., et al. (2022). Histone acetyltransferase inhibition reverses opacity in rat galactose-induced cataract. PloS One 17, e0273868. doi: 10.1371/journal.pone.0273868
Narimatsu, A., Hattori, T., Koike, N., Tajima, K., Nakagawa, H., Yamakawa, N., et al. (2019). Corneal lymphangiogenesis ameliorates corneal inflammation and edema in late stage of bacterial keratitis. Sci. Rep. 9, 2984. doi: 10.1038/s41598-019-39876-x
Ng, A. L.-K., To, K. K.-W., Choi, C. C.-L., Yuen, L. H., Yim, S.-M., Chan, K. S.-K., et al. (2015). Predisposing factors, microbial characteristics, and clinical outcome of microbial keratitis in a tertiary centre in Hong Kong: A 10-year experience. J. ophthalmology. 2015, 769436–769439. doi: 10.1155/2015/769436
Nosrati, N., Kapoor, N. R., Kumar, V. (2015). DNA damage stress induces the expression of Ribosomal Protein S27a gene in a p53-dependent manner. Gene 559, 44–51. doi: 10.1016/j.gene.2015.01.014
Oakes, J. E., Monteiro, C. A., Cubitt, C. L., Lausch, R. N. (1993). Induction of interleukin-8 gene expression is associated with herpes simplex virus infection of human corneal keratocytes but not human corneal epithelial cells. J. Virol. 67, 4777–4784. doi: 10.1128/jvi.67.8.4777-4784.1993
Oliveira-Ferreira, C., Leuzinger-Dias, M., Tavares-Ferreira, J., Torrão, L., Falcão-Reis, F. (2019). Microbiological profile of infectious keratitis in a portuguese tertiary centre. J. ophthalmology. 2019, 1–6. doi: 10.1155/2019/6328058
O’Neill, E. C., Yeoh, J., Fabinyi, D. C. A., Cassidy, D., Vajpayee, R. B., Allen, P., et al. (2014). Risk factors, microbial profiles and prognosis of microbial keratitis-associated endophthalmitis in high-risk eyes. Graefe's Arch. Clin. Exp. Ophthalmology. 252, 1457–1462. doi: 10.1007/s00417-014-2732-1
Orlans, H. O., Hornby, S. J., Bowler, I. C. J. W. (2011). In vitro antibiotic susceptibility patterns of bacterial keratitis isolates in Oxford, UK: a 10-year review. Eye (Lond). 25, 489–493. doi: 10.1038/eye.2010.231
Ota, C., Ng-Blichfeldt, J. P., Korfei, M., Alsafadi, H. N., Lehmann, M., Skronska-Wasek, W., et al. (2018). Dynamic expression of HOPX in alveolar epithelial cells reflects injury and repair during the progression of pulmonary fibrosis. Sci. Rep. 8, 12983. doi: 10.1038/s41598-018-31214-x
Pascolini, D., Mariotti, S. P. (2012). Global estimates of visual impairment: 2010. Br. J. Ophthalmology. 96, 614–618. doi: 10.1136/bjophthalmol-2011-300539
Portt, L., Norman, G., Clapp, C., Greenwood, M., Greenwood, M. T. (2011). Anti-apoptosis and cell survival: A review. Biochim. Biophys. Acta (BBA) - Mol. Cell Res. 1813, 238–259. doi: 10.1016/j.bbamcr.2010.10.010
Powell, D. R., Thangavelu, M., Chandler, H. L., Nichols, K. K., Nichols, J. J. (2010). Evaluation of extractants and precipitants in tear film proteomic analyses. Optom Vis. Sci. 87, 854–860. doi: 10.1097/OPX.0b013e3181f6fb71
Rajaiya, J., Zhou, X., Barequet, I., Gilmore, M. S., Chodosh, J. (2015). Novel model of innate immunity in corneal infection. In Vitro Cell. Dev. Biol. - Animal. 51, 827–834. doi: 10.1007/s11626-015-9910-2
Raziyeva, K., Kim, Y., Zharkinbekov, Z., Kassymbek, K., Jimi, S., Saparov, A. (2021). Immunology of acute and chronic wound healing. Biomolecules 11 (5), 700. doi: 10.3390/biom11050700
Roy, N. C., Altermann, E., Park, Z. A., McNabb, W. C. (2011). A comparison of analog and Next-Generation transcriptomic tools for mammalian studies. Briefings Funct. Genomics 10, 135–150. doi: 10.1093/bfgp/elr005
Santacruz, C., Linares, M., Garfias, Y., Loustaunau, L. M., Pavon, L., Perez-Tapia, S. M., et al. (2015). Expression of IL-8, IL-6 and IL-1β in tears as a main characteristic of the immune response in human microbial keratitis. Int. J. Mol. Sci. 16, 4850–4864. doi: 10.3390/ijms16034850
Scapini, P., Lapinet-Vera, J. A., Gasperini, S., Calzetti, F., Bazzoni, F., Cassatella, M. A. (2000). The neutrophil as a cellular source of chemokines. Immunol. Rev. 177, 195–203. doi: 10.1034/j.1600-065X.2000.17706.x
Schenk, M., Bouchon, A., Seibold, F., Mueller, C. (2007). TREM-1–expressing intestinal macrophages crucially amplify chronic inflammation in experimental colitis and inflammatory bowel diseases. J. Clin. Invest. 117, 3097–3106. doi: 10.1172/JCI30602
Schroeder, A., Mueller, O., Stocker, S., Salowsky, R., Leiber, M., Gassmann, M., et al. (2006). The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 7, 3. doi: 10.1186/1471-2199-7-3
Sharma, S., Taneja, M., Gupta, R., Upponi, A., Gopinathan, U., Nutheti, R., et al. (2007). Comparison of clinical and microbiological profiles in smear-positive and smear-negative cases of suspected microbial keratitis. Indian J. ophthalmology. 55, 21–25. doi: 10.4103/0301-4738.29490
Shrestha, G. S., Vijay, A. K., Stapleton, F., Henriquez, F. L., Carnt, N. (2021). Understanding clinical and immunological features associated with Pseudomonas and Staphylococcus keratitis. Cont Lens Anterior Eye. 44, 3–13. doi: 10.1016/j.clae.2020.11.014
Shtein, R. M., Elner, S. G., Bian, Z.-M., Elner, V. M. (2012). IL-8 and MCP gene expression and production by LPS-stimulated human corneal stromal cells. Int. J. Inflam. 2012, 714704. doi: 10.1155/2012/714704
Takeuchi, O., Hoshino, K., Kawai, T., Sanjo, H., Takada, H., Ogawa, T., et al. (1999). Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity 11, 443–451. doi: 10.1016/S1074-7613(00)80119-3
Tena, D., Rodríguez, N., Toribio, L., González-Praetorius, A. (2019). Infectious keratitis: microbiological review of 297 cases. Jpn J. Infect. Dis. 72, 121–123. doi: 10.7883/yoken.JJID.2018.269
Thakur, A., Kyd, J., Xue, M., Willcox, M. D., Cripps, A. (2001). Effector mechanisms of protection against Pseudomonas aeruginosa keratitis in immunized rats. Infect. Immun. 69, 3295–3304. doi: 10.1128/IAI.69.5.3295-3304.2001
Tian, R., Zou, H., Wang, L., Liu, L., Song, M., Zhang, H. (2020). Analysis of differentially expressed genes in bacterial and fungal keratitis. Indian J. Ophthalmol. 68, 39–46. doi: 10.4103/ijo.IJO_65_19
Ueta, M., Hamuro, J., Ohsako, S., Kinoshita, S. (2019). Distinctly regulated functions and mobilization of CD11c-positive cells elicited by TLR3- and IPS-1 signaling in the cornea. Immunol. Lett. 206, 49–53. doi: 10.1016/j.imlet.2018.12.004
Ung, L., Acharya, N. R., Agarwal, T., Alfonso, E. C., Bagga, B., Bispo, P. J., et al. (2019). Infectious corneal ulceration: a proposal for neglected tropical disease status. Bull. World Health Organ. 97, 854–856. doi: 10.2471/BLT.19.232660
Upadhyay, M. P., Srinivasan, M., Whitcher, J. P. (2015). Diagnosing and managing microbial keratitis. Community Eye Health 28, 3–6.
Venza, I., Cucinotta, M., Caristi, S., Mancuso, G., Teti, D. (2007). Transcriptional regulation of IL-8 by Staphylococcus aureus in human conjunctival cells involves activation of AP-1. Invest. Ophthalmol. Vis. Sci. 48, 270–276. doi: 10.1167/iovs.06-0081
Verrier, B., Müller, D., Bravo, R., Müller, R. (1986). Wounding a fibroblast monolayer results in the rapid induction of the c-fos proto-oncogene. EMBO J. 5, 913–917. doi: 10.1002/embj.1986.5.issue-5
Wang, H., Yu, J., Zhang, L., Xiong, Y., Chen, S., Xing, H., et al. (2014). RPS27a promotes proliferation, regulates cell cycle progression and inhibits apoptosis of leukemia cells. Biochem. Biophys. Res. Commun. 446, 1204–1210. doi: 10.1016/j.bbrc.2014.03.086
Waugh, D. J. J., Wilson, C. (2008). The interleukin-8 pathway in cancer. Clin. Cancer Res. 14, 6735–6741. doi: 10.1158/1078-0432.CCR-07-4843
Willcox, M. D. (2007). Pseudomonas aeruginosa infection and inflammation during contact lens wear: a review. Optometry Vision Science. 84, 273–278. doi: 10.1097/OPX.0b013e3180439c3e
Wilson, S. E., Esposito, A. (2009). Focus on molecules: interleukin-1: a master regulator of the corneal response to injury. Exp. Eye Res. 89, 124–125. doi: 10.1016/j.exer.2009.02.011
Wu, T., Hu, E., Xu, S., Chen, M., Guo, P., Dai, Z., et al. (2021). clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2 (3), 100141. doi: 10.1016/j.xinn.2021.100141
Wu, M., Peng, A., Sun, M., Deng, Q., Hazlett, L. D., Yuan, J., et al. (2011). TREM-1 amplifies corneal inflammation after Pseudomonas aeruginosa infection by modulating Toll-like receptor signaling and Th1/Th2-type immune responses. Infect. Immun. 79, 2709–2716. doi: 10.1128/IAI.00144-11
Xue, M., Thakur, A., Lutze-Mann, L., Willcox, M. (2000). Pro-inflammatory cytokine/chemokine gene expression in human corneal epithelial cells colonized by Pseudomonas aeruginosa. Clin. Exp. Ophthalmology. 28, 197–200. doi: 10.1046/j.1442-9071.2000.00286.x
Xue, M. L., Thakur, A., Willcox, M. (2002). Macrophage inflammatory protein-2 and vascular endothelial growth factor regulate corneal neovascularization induced by infection with Pseudomonas aeruginosa in mice. Immunol. Cell Biol. 80, 323–327. doi: 10.1046/j.1440-1711.2002.01094.x
Xue, M. L., Thakur, A., Willcox, M. D. P., Zhu, H., Lloyd, A. R., Wakefield, D. (2003). Role and regulation of CXC-chemokines in acute experimental keratitis. Exp. Eye Res. 76, 221–231. doi: 10.1016/S0014-4835(02)00270-1
Yağmur, M., Ersöz, C., Ersöz, T. R., Varinli, S. (1997). Brush technique in ocular surface cytology. Diagn. Cytopathology. 17, 88–91. doi: 10.1002/(ISSN)1097-0339
Yamaguchi, T., Calvacanti, B. M., Cruzat, A., Qazi, Y., Ishikawa, S., Osuka, A., et al. (2014). Correlation between human tear cytokine levels and cellular corneal changes in patients with bacterial keratitis by in vivo confocal microscopy. Invest. Ophthalmol. Vis. Sci. 55, 7457–7466. doi: 10.1167/iovs.14-15411
Yang, J. M., Sim, S. M., Kim, H. Y., Park, G. T. (2010). Expression of the homeobox gene, HOPX, is modulated by cell differentiation in human keratinocytes and is involved in the expression of differentiation markers. Eur. J. Cell Biol. 89, 537–546. doi: 10.1016/j.ejcb.2010.01.005
Yoon, H., Choi, S.-I., Kim, E. K. (2020). Uptake of cell debris and enhanced expression of inflammatory factors in response to dead cells in corneal fibroblast cells. Exp. Eye Res. 194, 108017. doi: 10.1016/j.exer.2020.108017
Yu, G., Wang, L.-G., Han, Y., He, Q.-Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS: A J. Integr. Biol. 16, 284–287. doi: 10.1089/omi.2011.0118
Zhang, J., Xu, K., Ambati, B., Yu, F.-S. X. (2003). Toll-like receptor 5-mediated corneal epithelial inflammatory responses to pseudomonas aeruginosa flagellin. Invest. Ophthalmol. Vis. Sci. 44, 4247–4254. doi: 10.1167/iovs.03-0219
Zhou, Z., Barrett, R. P., McClellan, S. A., Zhang, Y., Szliter, E. A., van Rooijen, N., et al. (2008). Substance P Delays Apoptosis, Enhancing Keratitis after Pseudomonas aeruginosa Infection. Invest. Ophthalmol. Vis. Sci. 49, 4458–4467. doi: 10.1167/iovs.08-1906
Zhou, X., Liao, W.-J., Liao, J.-M., Liao, P., Lu, H. (2015). Ribosomal proteins: functions beyond the ribosome. J. Mol. Cell Biol. 7, 92–104. doi: 10.1093/jmcb/mjv014
Zhou, Z., Wu, M., Barrett, R. P., McClellan, S. A., Zhang, Y., Hazlett, L. D. (2010). Role of the Fas pathway in Pseudomonas aeruginosa keratitis. Invest. Ophthalmol. Vis. Sci. 51, 2537–2547. doi: 10.1167/iovs.09-4152
Keywords: corneal infection, conjunctiva, ocular surface, keratitis, bacteria, transcriptome, gene expression
Citation: Alenezi H, Parnell G, Schibeci S, Ozkan J, Willcox M, White AJR and Carnt N (2024) Ocular surface immune transcriptome and tear cytokines in corneal infection patients. Front. Cell. Infect. Microbiol. 14:1346821. doi: 10.3389/fcimb.2024.1346821
Received: 30 November 2023; Accepted: 19 March 2024;
Published: 17 April 2024.
Edited by:
Poonam Mudgil, Charles Sturt University, AustraliaReviewed by:
Berit Sletbakk Brusletto, Oslo University Hospital, NorwayCopyright © 2024 Alenezi, Parnell, Schibeci, Ozkan, Willcox, White and Carnt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Heba Alenezi, aGguYWxlbmV6aUBwc2F1LmVkdS5zYQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.