
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol., 28 March 2024
Sec. Clinical Infectious Diseases
Volume 14 - 2024 | https://doi.org/10.3389/fcimb.2024.1345706
 Ziwei Jiang1†
Ziwei Jiang1† Wei Gai2†
Wei Gai2† Xiaojing Zhang2†
Xiaojing Zhang2† Yafeng Zheng2
Yafeng Zheng2 Xuru Jin1
Xuru Jin1 Zhiqiang Han1
Zhiqiang Han1 Geriletu Ao1
Geriletu Ao1 Jiahuan He1
Jiahuan He1 Danni Shu1
Danni Shu1 Xianbing Liu1
Xianbing Liu1 Yingying Zhou1
Yingying Zhou1 Zhidan Hua1*
Zhidan Hua1*Background: Investigations assessing the value of metagenomic next-generation sequencing (mNGS) for distinguish Aspergillus infection from colonization are currently insufficient.
Methods: The performance of mNGS in distinguishing Aspergillus infection from colonization, along with the differences in patients’ characteristics, antibiotic adjustment, and lung microbiota, were analyzed.
Results: The abundance of Aspergillus significantly differed between patients with Aspergillus infection (n=36) and colonization (n=32) (P < 0.0001). Receiver operating characteristic (ROC) curve result for bronchoalveolar lavage fluid (BALF) mNGS indicated an area under the curve of 0.894 (95%CI: 0.811-0.976), with an optimal threshold value of 23 for discriminating between Aspergillus infection and colonization. The infection group exhibited a higher proportion of antibiotic adjustments in comparison to the colonization group (50% vs. 12.5%, P = 0.001), with antibiotic escalation being more dominant. Age, length of hospital stay, hemoglobin, cough and chest distress were significantly positively correlated with Aspergillus infection. The abundance of A. fumigatus and Epstein-Barr virus (EBV) significantly increased in the infection group, whereas the colonization group exhibited higher abundance of A. niger.
Conclusion: BALF mNGS is a valuable tool for differentiating between colonization and infection of Aspergillus. Variations in patients’ age, length of hospital stay, hemoglobin, cough and chest distress are observable between patients with Aspergillus infection and colonization.
Pulmonary aspergillosis is the leading pulmonary fungal infection worldwide and a significant cause of morbidity and mortality (Bongomin et al., 2017). Aspergillus is present ubiquitously in the environment and has a high propensity for colonizing the human respiratory tract (Denning and Chakrabarti, 2017; El-Baba et al., 2020; Ledoux et al., 2020). The occurrence of fungal colonization does not result in disease immediately. However, it is a prerequisite for chronic and allergic mycoses, as well as for localized airway infections in invasive mycoses (Denning et al., 2014; Kosmidis and Denning, 2015). Various methods have been employed to detect fungi, including fungal culture, galactomannan (GM) antigen testing, 1, 3-β-D-glucan (G) testing, and specific polymerase chain reaction (PCR), etc (Mengoli et al., 2009; Shin et al., 2014; Boch et al., 2016; Lamoth et al., 2021; Lamoth and Calandra, 2022). However, identifying fungal infections is a considerable challenge, particularly distinguishing between fungal colonization and infection.
Currently, the use of metagenomic next-generation sequence (mNGS) technology is increasingly prevalent in the clinical diagnosis of infectious diseases, particularly when the etiology is not confirmed by conventional microbiological testing (CMT) or when anti-infective treatment proves ineffective. Several studies have demonstrated that mNGS is highly effective in diagnosing mixed infections, and viruses’ infections (Carpenter et al., 2019; Reyes et al., 2021; Mei et al., 2023; Ogunbayo et al., 2023; Xie et al., 2023). Compared to culture and other CMT methods, mNGS has a higher positive rate, sensitivity, and specificity in diagnosing invasive fungal infections (IFA) (Song et al., 2021; Li, 2022; Wang et al., 2022). In some cases, it can even detect fungal pathogens that are difficult to diagnose using conventional methods (Liu et al., 2022; Chen et al., 2023a). Furthermore, research has investigated the diagnostic utility of mNGS in non-neutropenic patients, those with COVID-19, and immunocompromised patients with pulmonary Aspergillus infection (Bao et al., 2022; Hoenigl et al., 2023; Zhan et al., 2023). However, there is still a dearth of studies on differentiating between Aspergillus infection and colonization.
Nevertheless, recent studies using mNGS to differentiate between fungal colonization and infection have emerged. Moreover, these studies have focused on exploring the threshold of mNGS read numbers for distinguishing between infection and colonization/not infection. The study of Liu et al. revealed that bronchoalveolar lavage fluid (BALF) mNGS can distinguish between Pneumocystis jirovecii colonization and infection with an area under the curve of 0.973, and an optimal threshold value of 14 reads (Liu et al., 2021). Jia et al. discovered that the optimal cut-off value for BALF mNGS in diagnosing invasive pulmonary aspergillosis (IPA) and no-IPA was species-specific read number (SSRN) is 2.5. Additionally, they found that thresholds of 1 and 4.5 was appropriate for immunocompromised and diabetic patients with IPA, respectively (Jia et al., 2023). Studies on the clinical characteristics of patients, as well as the differences in the microbial composition of the lungs of patients with Aspergillus colonization and infection, remains inadequate.
In this study, we evaluated the effectiveness of mNGS and conventional microbiological testing (CMT) for distinguished patients with Aspergillus infection from those with colonization. We outlined the antibiotic guidance between the two groups, the variations in clinical indicators, as well as changes in the microbial composition of the lungs.
We conducted a retrospective study at the Quzhou People’s Hospital, including patients who were admitted between August 2021 and June 2023 with suspected fungal pulmonary infections and underwent BALF mNGS detection. The study was approved by the Ethics Committee of Quzhou People’s Hospital (2023-017), and the patients were anonymized for analysis. The corresponding medical records were reviewed, and the clinical data analyzed including demographic characteristics, type of underlying disease, diagnosis, clinical course, treatment, and outcome.
BALF, blood, sputum and some other types of respiratory tract samples were used for pathogen identification through CMT methods, including culture for bacteria (blood agar plates, Chocolate, and MacConkey) and fungi (Sabouraud agar plates), 1, 3-β-D-glucan (G) test (Fungi (1,3)-β-D-glucan assay kit, Ref#KTG20, BIOENDO) and galactomannan (GM) test (Platelia Aspergillus Ag Assay, Ref#62794, Bio-Rad) for fungi, serum antigen detection for Cryptococcus (Ref#CR2003, IMMY), T-spot (Ref#30230808, WANTAI Biopharm) and Gene X-pert (Ref#20173406215, Cepheid AB) for Mycobacterium, and smear microscopy for fungi (fluorescent and KOH stain) or tuberculosis (acid-fast stain). All BALF samples were cultured for bacteria and fungi identification.
BALF specimens were collected by experienced bronchoscopist under general anesthesia following standard procedures. Lesions were selected based on computed tomography images of the chest. Following general anesthesia with sufentanil and propofol (to ensure a smooth bronchoscopic procedure and prevent potential patient discomfort-related injuries), the patient was guided to the target bronchial location through the contralateral nasal cavity. Nasopharyngeal ventilation and electrocardiographic monitoring were used during the procedure. No suction was performed before entering the target location. The fiberoptic bronchoscope’s tip was tightly wedged into the opening of the target bronchial segment or subsegment. Then, 100-250 mL of 37°C sterilized saline was rapidly injected through the silicone tube via the biopsy hole. The saline was divided into 3-5 equal parts, with each part being 25-60 mL. After each perfusion, 50-100 mmHg (1mmHg=0.133kPa) negative pressure suction was used to recover the irrigation fluid. The BALF (3-5 mL) obtained from the second perfusion from each patient was collected into sterile tubes according to standard procedures and transported at 4-8 °C within a short time after collection. DNA was extracted using PathoXtract® Basic Pathogen Nucleic Acid Kit (WYXM03211S, WillingMed Corp, Beijing, China) according to the manufacturer’s protocol. DNA libraries was constructed using the Illumina® DNA Prep, (M) Tagmentation kit (20018705, Illumina). Quality control of the DNA libraries was conducted using the Agilent 2100 Bioanalyzer (Agilent Technologies) to ensure library concentration exceeded 1 ng/μL. Then, libraries with confirmed quality were sequenced by the NextSeq™ 550Dx platform with a 75 bp, single-end sequencing kit (Illumina). To exclude environmental and laboratory contaminants, an external negative control (nuclease-free water) was included in each mNGS assay run (Blauwkamp et al., 2019).
High-quality sequencing data were generated by filtered out low-quality or undetected sequences, sequences contaminated by splices, high-coverage repeats, and short-read-length sequences using Trimmomatic v0.40. The remaining sequences were aligned to four databases downloaded from NCBI, including bacteria, fungi, viruses, and parasites, for classification after removal of human host sequences with Bowtie2 v2.4.3. For identification of the pathogens, a RPTM (reads per ten million) value, which defined as detected number of pathogen specific reads per ten million, was used. The RPTM ratio metric (RPTM-r), defined as RPTMSample/RPTMNTC, set to 1 in NTC samples when the microbial taxonomic value of a given species or genus is less than 1 (Miller et al., 2019). RPTM ≥ 3 was used as an empirical threshold for virus detection. For bacteria and fungi, positive pathogens were required to meet a RPTM threshold ≥ 20 (Chen et al., 2023b). Special pathogens (including Cryptococcus and Mycobacterium) with RPTM ≥ 1 was identified as positive (Langelier et al., 2018; Chen et al., 2021). Common colonized bacteria and contaminating microorganisms, including Rothia, Corynebacterium, coagulase-negative Staphylococci, Neisseria, and Streptococcus etc., were considered as microecological bacteria in the study (Chen et al., 2020; Xie et al., 2021; Shi et al., 2022; Tan et al., 2023).
We applied the criteria of the European Organization for Research and Treatment of Cancer and Mycoses Study Group (EORTC/MSG) for diagnose of invasive pulmonary aspergillosis (IPA) and Chronic necrotizing pulmonary aspergillosis (CNPA) (Donnelly et al., 2020). Proven IPA diagnosis criteria: histopathology, cytopathology, or direct microscopy of specimens obtained by needle aspiration or biopsy revealed hyphae, and accompanied by evidence of associated tissue damage. Alternatively, molds are cultured by aseptic procedures from specimens obtained from normally sterile, clinically, or radiologically abnormal sites consistent with an infectious disease process. Probable Aspergillus infection is definite as the presence of at least one host factor, a clinical feature and mycologic evidence. Possible cases meet the criteria of with a host factor and a clinical feature, but for which mycological evidence has not been found. The diagnostic criteria for chronic pulmonary aspergillosis (CPA) are based on the 2016 clinical guidelines from the European Society for Clinical Microbiology and Infectious Diseases (ESCMID) and the European Respiratory Society (ERS) (Denning et al., 2016).
Regarding the criteria for Aspergillus positivity in the CMT methods, positive culture results represent Aspergillus recovered by culture from a sputum, BALF or bronchial brush sample. A positive GM result is definite as a value ≥ 1.0 (Donnelly et al., 2020). Proven and probable cases were classified into Aspergillus infection group (Feys et al., 2022). Patients with positive mycological cultures or amplification of fungal DNA without signs of infection were classified as Aspergillus colonization (Donnelly et al., 2020). Therefore, cases were considered as colonization when Aspergillus was identified but without a final diagnosis of Aspergillus infection. Two experienced physicians made clinical diagnoses; when they gave different results, another senior physician made a judgement.
The chi-square test was applied to the categorical variables. A student t-test was used for continuous variables. P-value less than 0.05 was considered statistically significant. All statistical analyses were performed using GraphPad Prism (Version 9.4.1, GraphPad Software Inc) and SPSS (version 26, IBM Corp). The diagnostic performance of mNGS was evaluated using the area under the curve (AUC) of Receiver operating characteristic (ROC), where the best cut-off values were obtained. The sensitivity and specificity of the detection method were analyzed as reference (Blauwkamp et al., 2019). The alpha diversity index was calculated based on Shannon and Simpson indexes. Beta-diversity was visualized using principal coordinate analysis (PCoA), and an ANOSIM test was performed in R with the Vegan package. The stacked bar plot of the community composition was visualized in R using the ggplot2 package. Linear discriminant analysis (LDA) effect size (LEfSe) was utilized to identify significantly different species among the groups, with thresholds of log10 LDA Score ≥ 2 and P value ≤ 0.05.
A total of 68 patients were screened from a cohort of 376 individuals with suspected pulmonary infection who underwent BALF mNGS. Among them, 36 were diagnosed with Aspergillus infection and 32 were identified as cases of Aspergillus colonization. Within the group of patients with Aspergillus infection, 29 were diagnosed with invasive pulmonary aspergillosis (IPA, all proven cases), 5 with chronic pulmonary aspergillosis (CPA), and 2 with chronic necrotizing pulmonary aspergillosis (CNPA, 1 proven and 1 probable) (Figure 1).
Baseline data was compared between patients with Aspergillus infection and colonization. The patients’ ages ranged from 21 to 89 years, with a median age of 65. Of the 68 patients, 60.29% (41/68) were male. Those with Aspergillus infection were found to be significantly older than those with Aspergillus colonization (68.44 ± 11.66 vs. 61.53 ± 14.35, P = 0.032). Cough and chest distress symptoms were more frequently observed in patients with Aspergillus infection (P = 0.021, and P = 0.020). Additionally, patients with Aspergillus infection had a longer length of hospital stays (LOHS) compared with those with Aspergillus colonization (10.76 vs. 7.22 days, P = 0.011). There was no significant difference in terms of the proportion of patients who were immunosuppressed or had malignant tumors between the two groups. Pulmonary infection, pneumonia, and tuberculosis were the prevalent diseases observed in patients (Table 1). Patients with Aspergillus infection had a lower hemoglobin (Hb) level than patients with Aspergillus colonization (P = 0.027). Patchy shadow, pulmonary emphysema, pleural effusion, and nodules were common lung imaging findings that did not differ significantly between groups (Table 2).
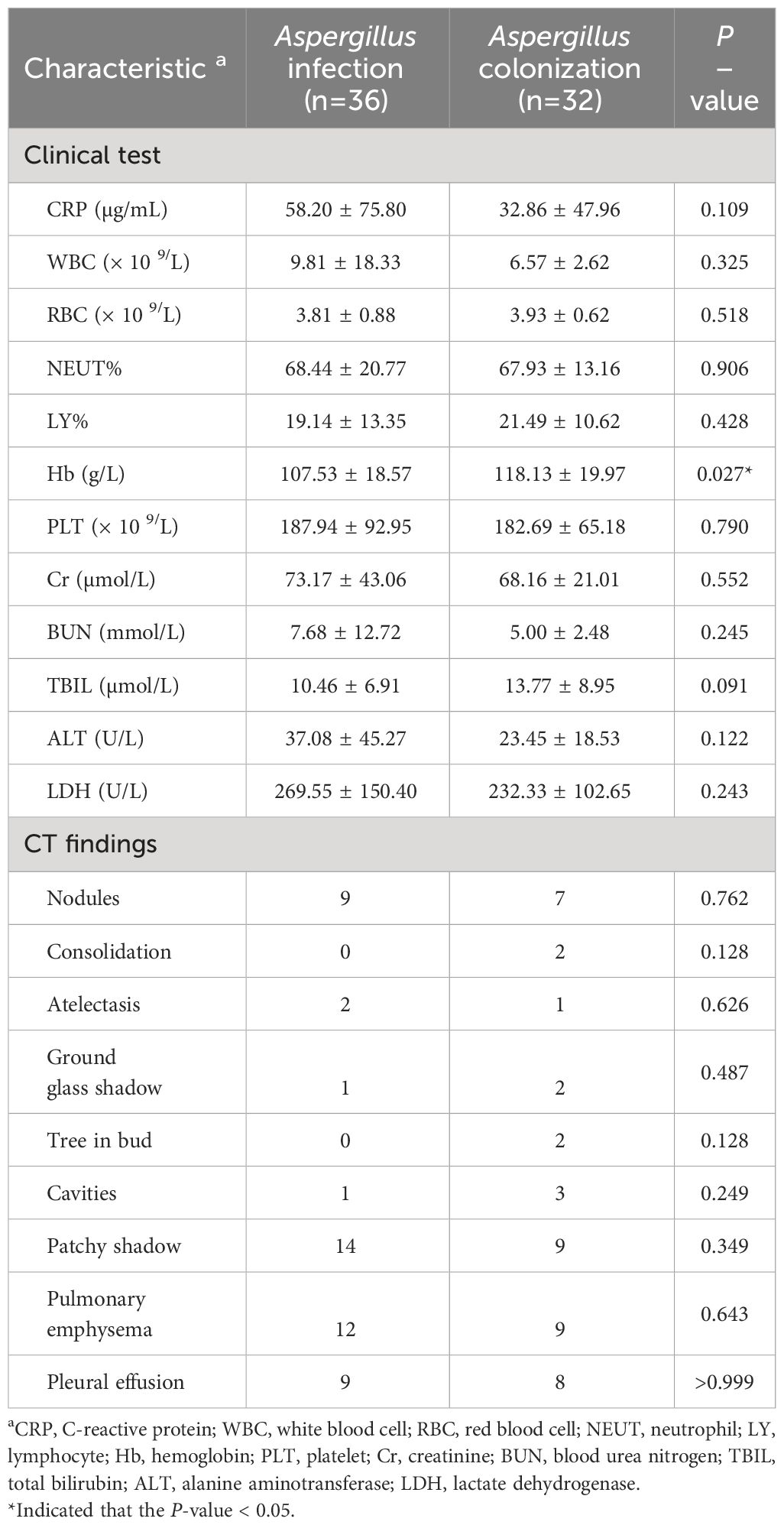
Table 2 Comparison of clinical test and chest computed tomography results of the patients with Aspergillus infection and colonization.
In the group of Aspergillus infection and colonization, mNGS identified of 5 and 13 Aspergillus species, respectively (Supplementary Table 1). Among patients with Aspergillus infection, A. fumigatus (77.78%, 28/36) was the most common species, followed by A. flavus (13.89%, 5/36) and A. oryaze (5.56%, 2/36). One patient was found to be infected with both A. flavus and A. oryzae. In patients with Aspergillus colonization, A. fumigatus (31.25%, 10/32) and A. niger (18.75%, 6/32) were the most detected species (Figure 2A).
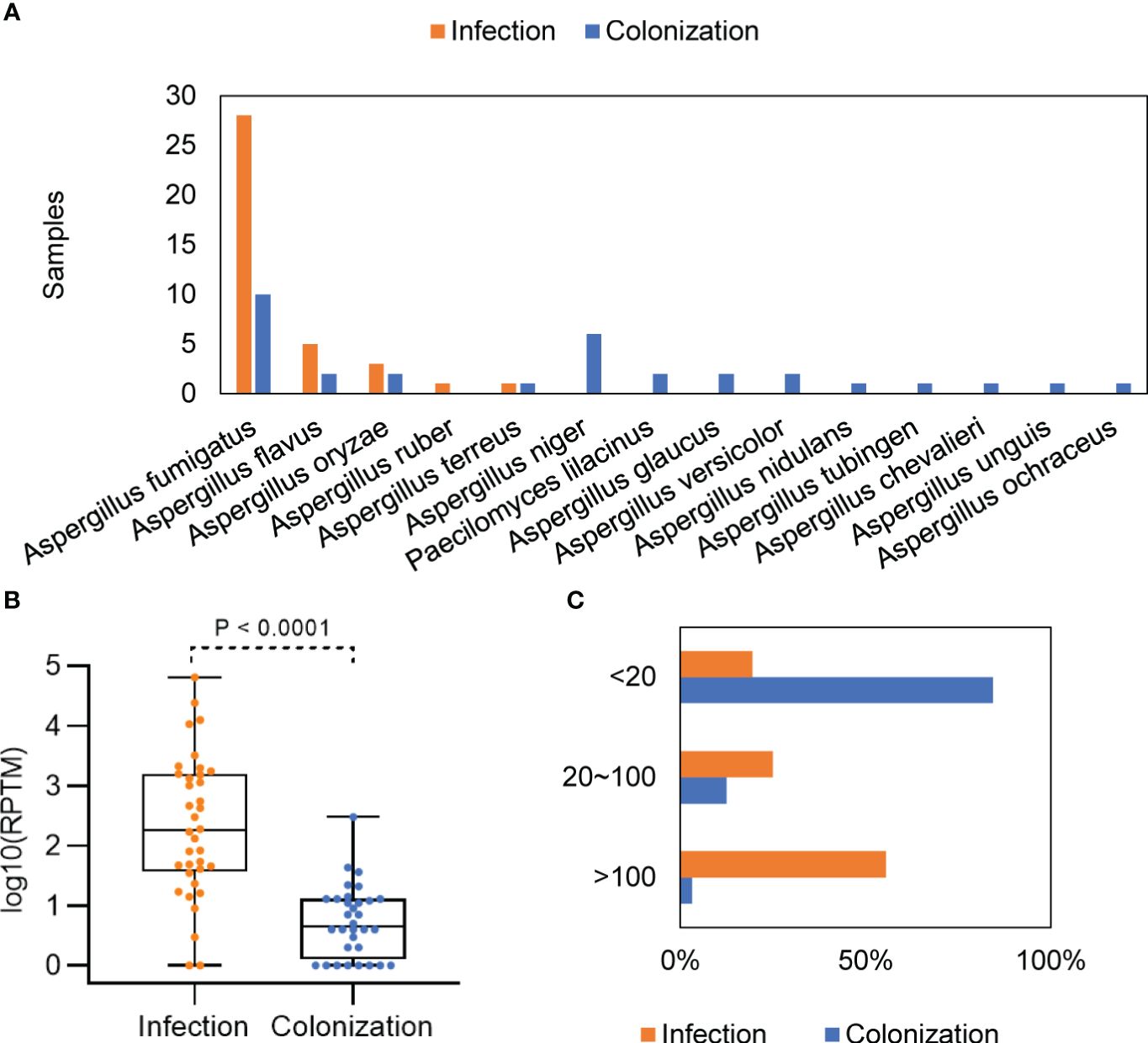
Figure 2 Distribution and abundance of Aspergillus species in patients with Aspergillus infection and colonization. (A) Comparison of Aspergillus in patients with Aspergillus infection and colonization. (B) Differences in mNGS RPTM for Aspergillus in patients with Aspergillus infection and colonization. (C) Proportion of patients with different mNGS Aspergillus reads in the infection and colonization groups.
The Aspergillus load was significantly higher in the group with Aspergillus infection compared with the group with colonization, with a median mNGS read number of 3609 ± 11358 vs. 17.88 ± 52.44 (P < 0.0001) (Figure 2B). When patients were divided into low (≤ 20), media (20 ~ 100), and high (≥ 100) groups based on the mNGS RPTM value for Aspergillus, the results showed that over 80% of patients in the infection group had an RPTM value larger than 20, while over 80% of patients in the colonization group had an RPTM value less than 20 (Figure 2C).
We generated an ROC curve using the Aspergillus RPTM of mNGS in BALF from the patients. The calculated area under curve (AUC) was 0.894 (95% CI: 0.811-0.976), with the optimal cut-off value for distinguishing Aspergillus infection from colonization was determined to be 23 (Figure 3A).
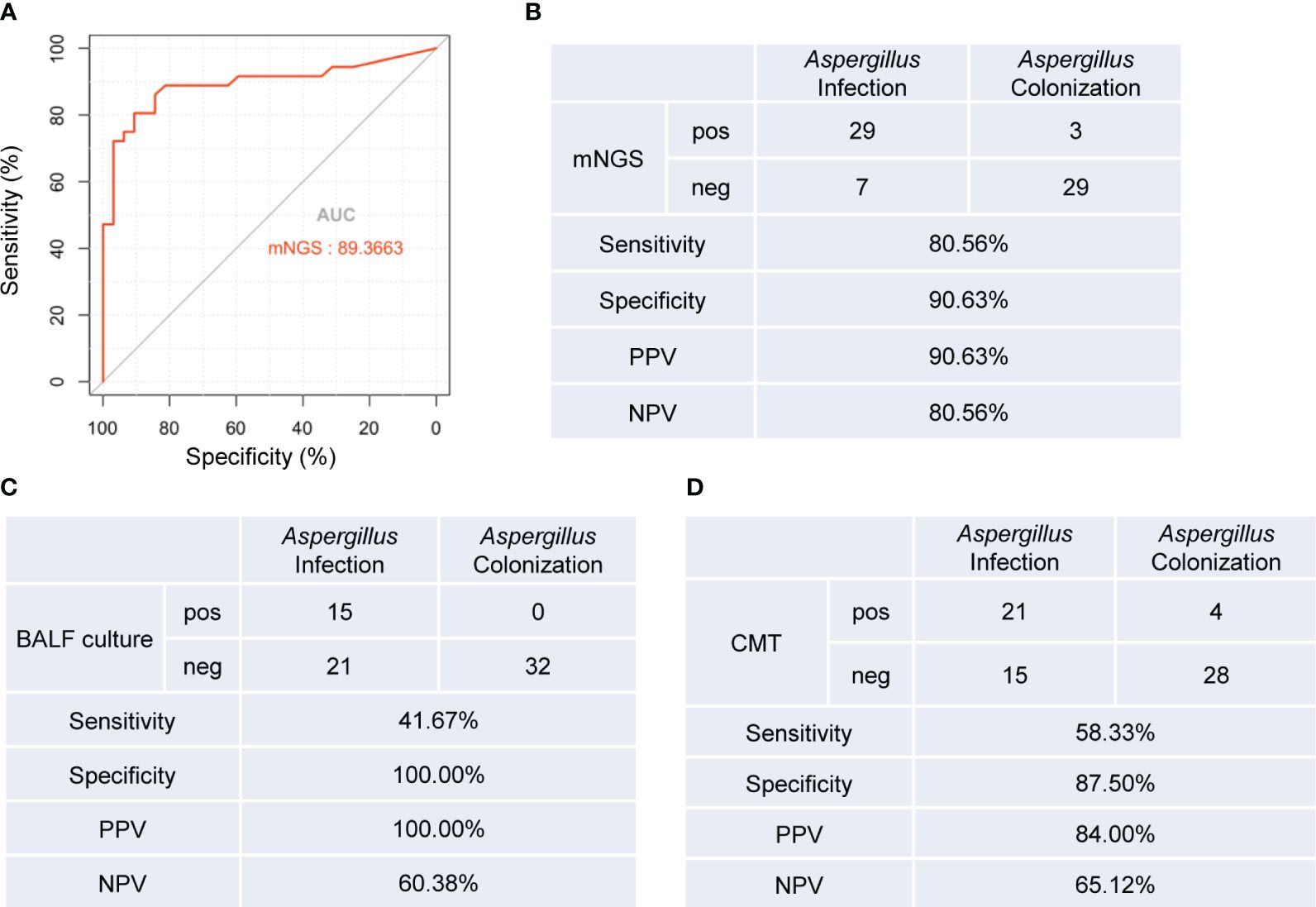
Figure 3 Diagnostic performance of mNGS, BALF culture and CMT tests for distinguishing Aspergillus infection from colonization. (A) ROC curve of mNGS for discrimination between Aspergillus infection and colonization. (B-D) Diagnostic performance of mNGS (B), BLAF culture (C), and CMT (D) methods for differentiating between Aspergillus infection and colonization. AUC, area under curve. PPV, positive predictive value; NPV, negative predictive value. pos, positive; neg, negative.
Next, we evaluated the diagnostic accuracy of BALF mNGS, BALF culture, and conventional microbiological testing (CMT) (Supplementary Table 1) in distinguishing Aspergillus infection from colonization. Using RPTM ≥ 23 as the threshold criterion for Aspergillus infection, mNGS demonstrated a sensitivity of 80.56%, which was significantly higher than BALF culture (41.67%, P = 0.001), and CMT methods (58.33%, P = 0.041). There was no significant difference in specificity between mNGS and BALF culture (90.63% vs. 100%, P = 0.076), nor between mNGS and CMT methods (90.63% vs. 87.5%, P = 0.689). However, the specificity of BALF culture was significantly higher than that of CMT methods (P = 0.039) (Figures 3B-D).
To explore the influence of mNGS results on antimicrobial usage, we analyzed variations in antimicrobial regimens before and after mNGS detection. The patients in the Aspergillus infection group had a higher incidence of combined bacteria and fungi infection (69%, 25/36), whereas the predominant infection in the Aspergillus colonization group was bacterial (78%, 25/32) (Supplementary Figure 1). We found that antimicrobial drug regimens were adjusted in 18 out of 36 (50%) samples from patients with Aspergillus infection, which was significantly higher than the proportion of patients with Aspergillus colonization (12.5%, P = 0.001). Among the 18 samples, 5 cases had their antibiotics changed, while 13 samples had their antibiotics escalated. The percentage of patients requiring antibiotic escalation was significantly higher in the Aspergillus infection group compared to the Aspergillus colonization group (P = 0.003) (Figure 4).
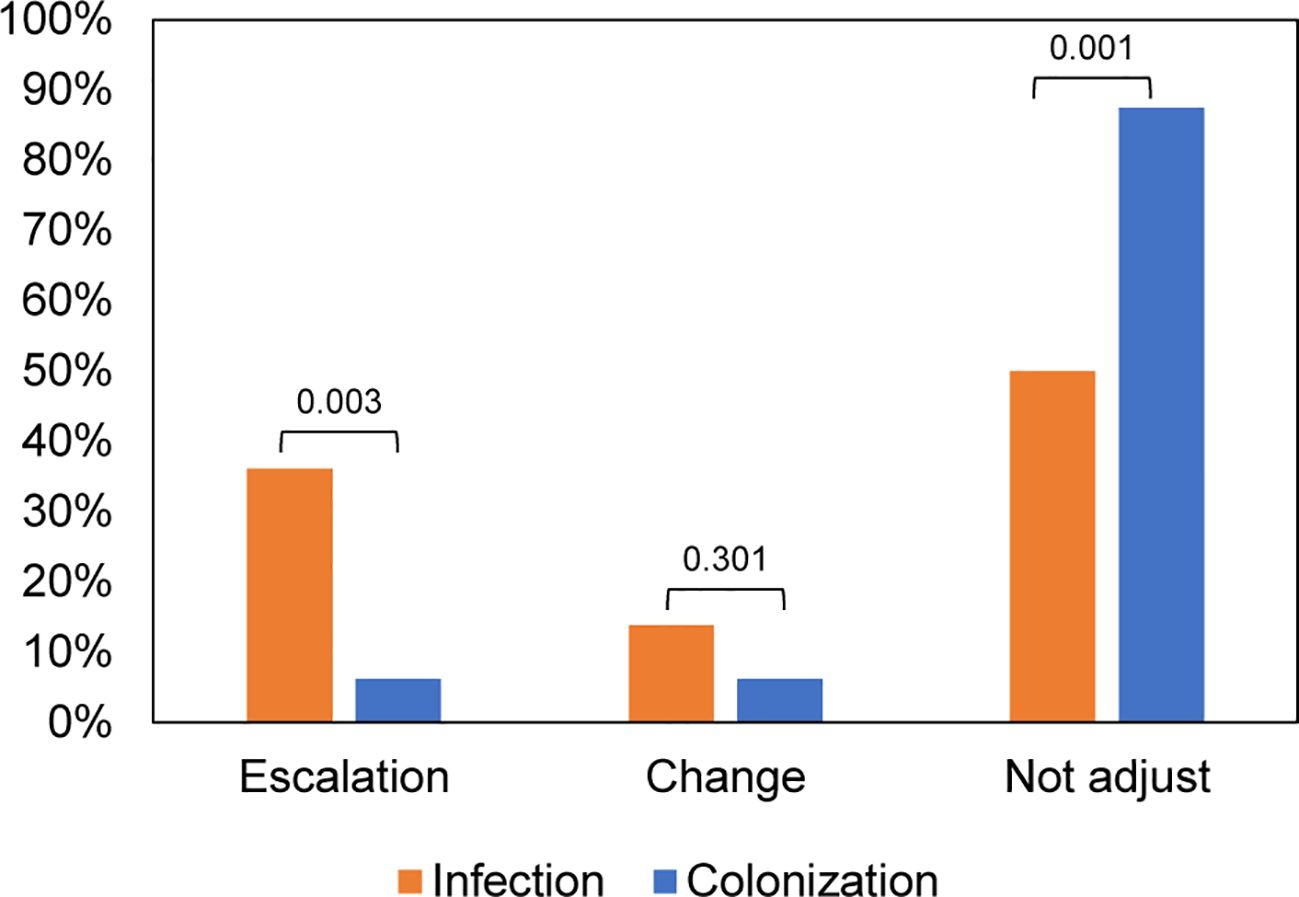
Figure 4 Impacts of mNGS on antibiotic adjustment in patients with Aspergillus infection and colonization.
We conducted Spearman correlation analyses to examine the relationship between various characteristics and Aspergillus infection. The results showed significant positive correlations between Aspergillus infection and the following variables: Aspergillus RPTM, CMT Aspergillus test results, age, LOHS, presence of cough and chest distress symptoms, and antibiotic escalation. And significant negative correlations between Aspergillus infection and Hb level were observed. Additionally, significant positive correlations were observed between Aspergillus RPTM values and the following variables: CMT Aspergillus test results, chest distress symptoms, and antibiotic adjustments and escalation. Notable, CMT Aspergillus positivity was positively correlated with diastolic blood pressure, lactate dehydrogenase (LDH), ground-glass shadow, presence of cardiovascular disease in the patient, and antibiotic escalation and change. Additionally, positive correlations were found between antibiotic adjustments and LOHS, CRP levels, and NEUT%. However, antibiotic adjustments were significantly negatively correlated with RBC and LY% (Figure 5).
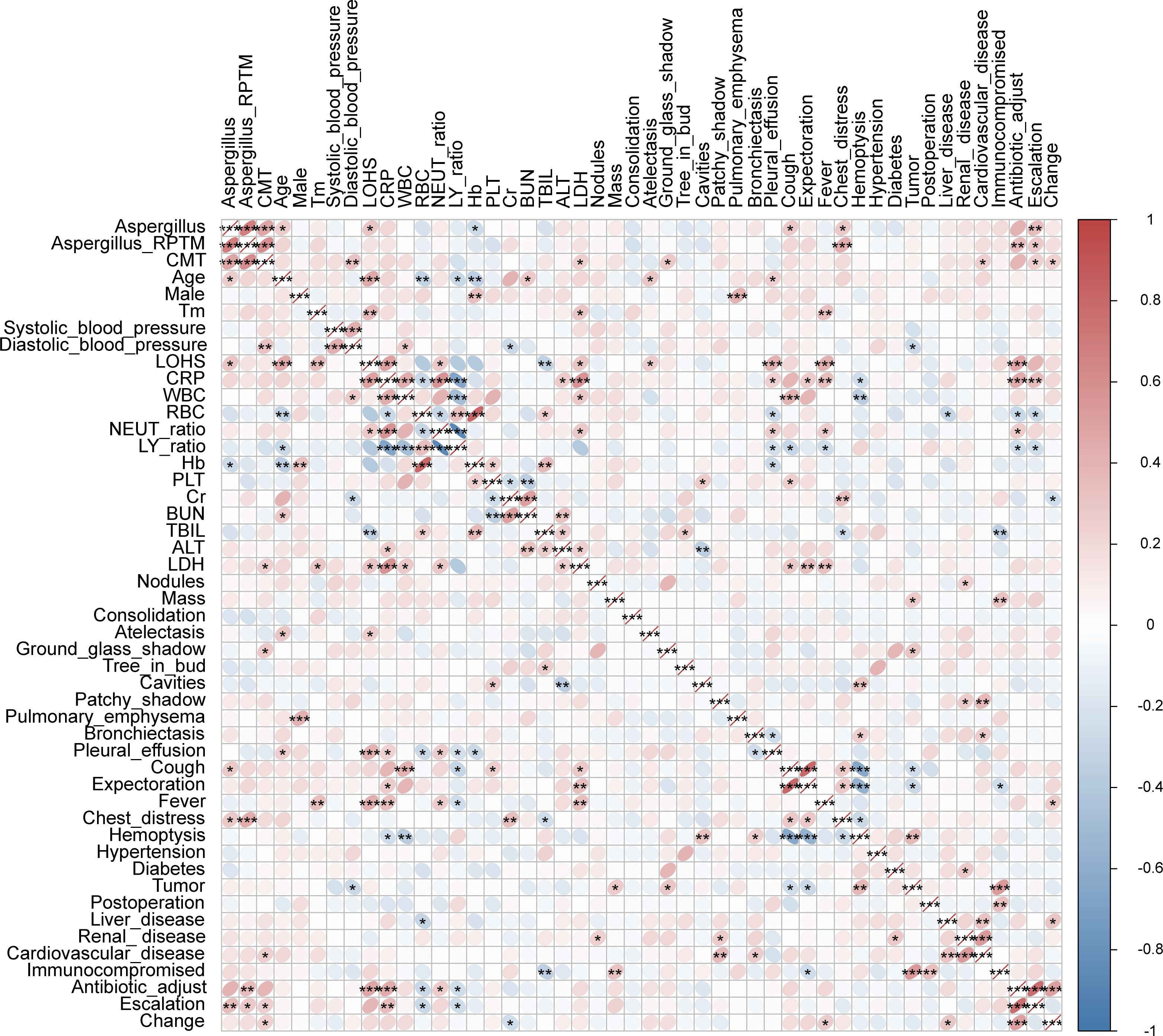
Figure 5 Spearman correlations between Aspergillus infection and characteristics of patients. Tm, temperature. “Change” indicates the adjustment of antibiotic. * P < 0.05, ** P < 0.01, *** P < 0.001.
The study compared the overall composition and diversity of the lung microbial signature in patients with Aspergillus infection and colonization. Although no significant difference was observed, patients with Aspergillus infection showed a higher α diversity according to both the Shannon and Simpson indices, indicating a trend towards increased richness and evenness of microbial composition (Figure 6A). Principal co-ordinates analysis (PCoA) results indicated that the samples from both groups were intermixed. However, the colonization group displayed a wider spread of data compared to the infection group (Figure 6B). Moreover, a significant difference in the microbial community structure between the two groups was observed (Figure 6C).
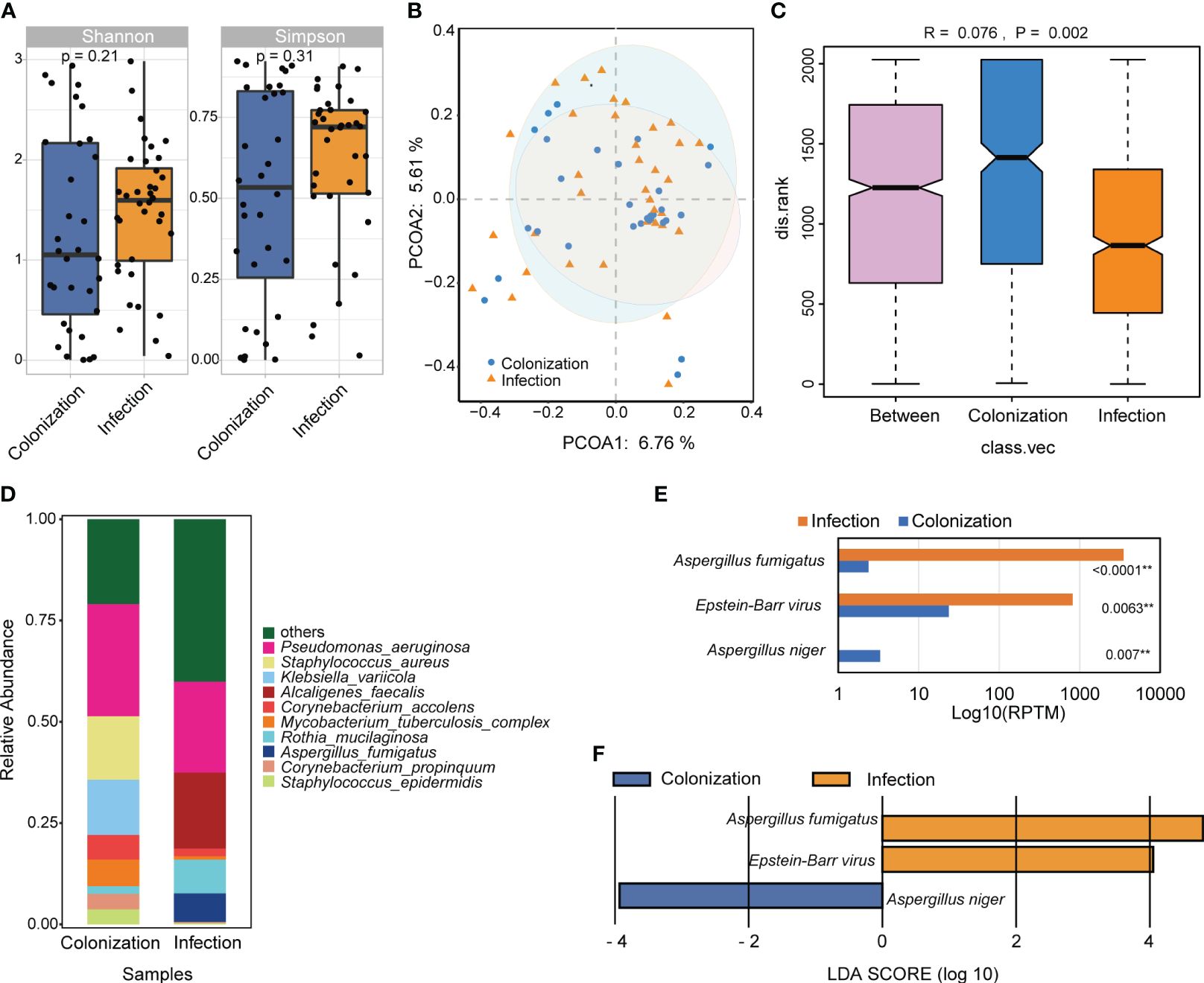
Figure 6 The difference of microbial composition for patients with Aspergillus infection and colonization. (A) Alpha diversity was showed by Shannon and Simpson index. (B) PCoA analysis of the microbial composition. (C) ANOSIM for the analysis of microbial community structure. (D) Barplot showed the top 10 species with the highest abundance between two groups. (E) Significant different analysis of the species between two groups with Kruskal-Wallis test. (F) Lefse analysis for enriched species for the two groups. ** P < 0.01.
The relative abundance of the top 10 species were examined, which included Pseudomonas aeruginosa, Staphylococcus aureus, Klebsiella variicola, Alcaligenes faecalis, Corynebacterium accolens, Mycobacterium tuberculosis complex, Rothia mucilaginosa, Aspergillus fumigatus, Corynebacterium propinquum, and Staphylococcus epidermidis. Among these species, only A. fumigatus showed significant differences between the two groups (Figures 6D, E). Additionally, three species with LDA scores ≥ 2 and a significance level of P < 0.05 were identified. A. fumigatus and Epstein-Barr virus (EBV) were significantly more abundant in patients with Aspergillus infection, whereas A. niger was more enriched in cases with Aspergillus colonization (Figure 6F).
The prevalence of pulmonary aspergillosis has significantly increased in recent years and is associated with the high mortality rates (Bongomin et al., 2017). Aspergillus colonization may indicate a transient presence within the airway, as long-term non-threatening carriage is often identified in patients with restrictive structural or functional lung defects. However, it can serve as an early warning sign before the development of apparent Aspergillus infection (Soubani et al., 2004; Khasawneh et al., 2006). Timely and precise diagnosis of Aspergillus infection and colonization is crucial for effective treatment, but poses significant clinical challenges (Thompson and Young, 2021). Although Aspergillus infection and colonization have clear definitions (Donnelly et al., 2020), different diagnostic methods vary in their performance for identifying Aspergillus (Mengoli et al., 2009; Shin et al., 2014; Boch et al., 2016; Lamoth et al., 2021; Lamoth and Calandra, 2022). However, studies aimed at distinguishing between Aspergillus infection and colonization are still inadequate. This study evaluated the efficacy of mNGS in differentiating Aspergillus infection from colonization. Furthermore, it outlined the distributional characteristics of Aspergillus, the distinctions in clinical characteristics and antibiotic adjustment among patients with Aspergillus infection and colonization, as well as the variations in lung microbiota.
Aspergillus is a ubiquitous fungus found worldwide. Among the over 200 species in the genus Aspergillus, only a small percentage of them are known to infect humans (Nabili et al., 2016; Khojasteh et al., 2023). Notably, A. fumigatus, A. flavus, A. niger, and A. terreus are the most common species causing disease in humans (Sugui et al., 2014; Lamoth and Calandra, 2022). Consistent with previous studies, our study identified 14 Aspergillus species among the patients, with only 5 species leading to Aspergillus infection. Among these, A. fumigatus, A. flavus, and A. oryaze were the most prevalent in patients with Aspergillus infection. A. niger, except for A. fumigatus, was the most common species in patients with Aspergillus colonization (Figure 2A). Although A. niger only played a role only in colonization in our study, it has the potential to cause pneumonia. In a study of Japan, A. niger was also identified as the dominant species in patients with Aspergillus colonization, and it was most commonly isolated in patients with allergic bronchopulmonary aspergillosis (Tashiro et al., 2011). A case report described a chronically steroid-treated COPD patient presenting with hemoptysis and pleuritic chest pain, where the presence of calcium oxalate crystals in histopathological specimens was a typical sign of A. niger infection (Person et al., 2010). A. flavus was found to be more prevalent in certain regions of Africa or Asia (Lamoth and Calandra, 2022). Several cases of pulmonary infection caused by a combination of A. flavus and other pathogens have been identified. In one case, a 60-year-old man exhibited patchy high-density shadows and a few pleural effusions in the transplanted lung, indicating a mixed infection of Klebsiella pneumoniae, Ureaplasma urealyticum, and A. flavus (Liu et al., 2023). Another case involved a 62-year-old male with co-infection of A. flavus and Klebsiella pneumoniae. This patient exhibited leukopenia, thrombocytopenia, acute kidney failure, and severe myelosuppression (Xu et al., 2022).
In recent years, the benefits of using mNGS for fungal detection have become increasingly apparent. A meta-analysis has shown that mNGS has a combined sensitivity and specificity of 78% (95% confidence interval [CI]: 67-87%; I2 = 92%) and 77% (95% CI: 64-94%; I2 = 74%), respectively, for fungal diagnosis. Subgroup analysis revealed that the sensitivity of mNGS was significantly affected by severe or immunocompromised patients with pulmonary infection (P < 0.001) (Chen et al., 2022). This suggests that mNGS testing, even before CMT results are available, is a highly effective option. Early detection of pathogenic microorganisms in patients with severe infections is crucial for providing appropriate clinical intervention and targeted antibiotics as soon as possible. There are many reasons for this heterogeneity of mNGS for fungi identification, one of which may be that the thick cell wall of Aspergillus makes it difficult to break the wall and release nucleic acid, which may cause false negative mNGS results (Bittinger et al., 2014). However, the diagnostic performance of mNGS for Aspergillus has improved with the optimization of DNA extraction methods (Gu et al., 2021). Additionally, the positive threshold criteria for mNGS may also be an important influencing factor (Chen et al., 2022). Thus, it is crucial to determine the positive threshold of mNGS in clinical applications, but there is currently no unified international standard. Our study has laid the foundation for the establishment of this standard to some extent.
Our study found that BALF mNGS demonstrated superior accuracy in diagnosing Aspergillus infection and distinguishing it from colonization when compared to BALF culture and CMT methods (P < 0.05). The optimal cut-off value of RPTM for mNGS was 23. At this threshold, mNGS achieves a sensitivity of 80.56% and a specificity of 90.63% for the final diagnosis (Figure 3). While there have been numerous studies investigating the diagnostic ability of mNGS for Aspergillus infection, there remains a lack of research regarding the discrimination between Aspergillus colonization and infection (Jia et al., 2023). In addition, the value of Bio-Rad Platelia Aspergillus IgG (Platelia Aspergillus IgG) enzyme-linked immunosorbent assay (ELISA) method and the Aspergillus precipitin test for distinguish pulmonary aspergillosis from colonization were assessed. Platelia Aspergillus IgG had a higher positive rate than Aspergillus precipitin test for Aspergillus detection, and showed a sensitivity of 74.5% and a specificity of 94.8% for distinguishing Aspergillus infection from colonization (Shinfuku et al., 2023). Their diagnostic accuracy was slightly lower than that of mNGS. Nevertheless, the current research serves as a valuable reference for analyzing patients with Aspergillus colonization and infection. While mNGS serves as a precise pathogenetic test and holds potential in clinical diagnosis, the final diagnosis of the disease depends on clinical experts who integrate the patient’s symptoms, clinical test results, and pathogenetic findings to make a comprehensive decision.
We examined the clinical characteristics of patients with Aspergillus infection and colonization and found that patients with Aspergillus infection were older and had longer lengths of hospital stay (Table 1). This trend was also found in COVID-19 patients with or without fungal coinfection (Negm et al., 2023). Common non-specific symptoms observed in patients with Aspergillus infection included persistent fever, dry cough, hemoptysis, and chest pain (Guo et al., 2023). In this study, cough, expectoration, and chest distress were found to be more prevalent, and the proportion of patients experiencing cough and chest distress symptoms was higher in the Aspergillus infection group. Although immunosuppression is a crucial risk factor for Aspergillus infections (Patterson et al., 2016), there was no significant difference in immunosuppression status between the two groups (Table 1). Among routine blood indicators, only hemoglobin levels showed significant differences, with a marked increase in the Aspergillus colonization group compared to the infection group. CT findings indicated that Aspergillus infection and colonization were characterized by patchy shadows, pulmonary emphysema, pleural effusion, and nodules (Table 2). Correlation analysis confirmed the significance of these characteristics in relation to Aspergillus infection and colonization. In addition, a significant positive correlation was observed between Aspergillus infection and mNGS determined antibiotic adjustment (Figure 5). We observed no significant differences in species abundance and diversity between the groups with Aspergillus infection and colonization, although these indexes were higher in the infection group (Figure 6). A. fumigatus and EBV appeared more frequently in patients with Aspergillus infection. EBV is a member of the gamma herpesviruses and is one of the most common human viruses. During childhood, all people are infected with multiple herpesviruses, and after clearing an acute infection, herpesviruses enter a latent state (Barton et al., 2007). EBV is commonly associated with infectious mononucleosis, lymphoma, and cancer in adolescents and adults (Epstein et al., 1964; Ok et al., 2015). However, cases of pneumonitis due to EBV infection in adults are rare (McManus et al., 2009; Niazi et al., 2020; Liao et al., 2023). In our study, only two immunocompetent patients were diagnosed with EBV infection, one with A. fumigatus and one with A. oryzae. In addition, EBV was also detected in another 12 patients with Aspergillus infection and 4 patients with Aspergillus colonization in this study. A previous study observed a significantly different microbial structure between IPA and non-IPA patients, with cytomegalovirus and A. fumigatus appeared more frequently in patients with IPA (Ao et al., 2023). Further exploration is necessary to understand the relationship and potential interaction mechanism between the two pathogens. Several factors may account for this disparity of the different results of microbiome analysis, including the advanced age of our patients, their relatively low mortality rate, and the absence of any restrictions on the type of diseases they presented.
In this investigation, we conducted a comprehensive study to analyze the clinical characteristics and lung microbiota changes in individuals with Aspergillus infection and colonization. Moreover, we evaluated the efficacy of BALF mNGS in distinguishing between Aspergillus infection and colonization. While our study was meticulously designed and analyzed, we acknowledge its limitations. Bacterial and fungal cultures of BALF were performed in all patients, but not all patients underwent G/GM tests, resulting in a lack of corresponding comparative diagnostic performance results. Furthermore, this study was conducted in a single-center and the sample collection was limited to a two years period, which may introduce bias in the outcomes.
In this study, the performance of mNGS in distinguishing Aspergillus infection from colonization, along with the differences in patients’ characteristics, antibiotic adjustment, and lung microbiota, were analyzed. We found that BALF mNGS has a high diagnostic efficacy for distinguishing Aspergillus infection and colonization, which was superior to BLAF culture and the CMT methods used in this study. Furthermore, mNGS plays a more important role in the guidance of medication in patients with Aspergillus infection. Age, LOHS, cough, chest distress, and Hb were significant different indicators between patients with Aspergillus infection from colonization.
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author/s.
The studies involving humans were approved by the Ethics Committee of Quzhou People’s Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. The ethics committee/institutional review board waived the requirement of written informed consent for participation from the participants or the participants’ legal guardians/next of kin because Written informed consent was waived by the ethics committee due to the retrospective nature of study design. We analyzed the data anonymously.
ZJ: Conceptualization, Data curation, Formal analysis, Visualization, Writing – original draft, Writing – review & editing. WG: Conceptualization, Formal analysis, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. XZ: Conceptualization, Data curation, Formal analysis, Visualization, Writing – original draft, Writing – review & editing. YaZ: Conceptualization, Formal analysis, Supervision, Visualization, Writing – original draft, Writing – review & editing. XJ: Data curation, Resources, Supervision, Writing – review & editing. ZH: Data curation, Resources, Supervision, Writing – review & editing. GA: Data curation, Resources, Supervision, Writing – review & editing. JH: Data curation, Supervision, Writing – review & editing. DS: Data curation, Resources, Supervision, Writing – review & editing. XL: Data curation, Resources, Supervision, Writing – review & editing. YiZ: Data curation, Resources, Supervision, Writing – review & editing. ZH: Funding acquisition, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was funded by 2024 Science and Technology Program for Medicine and Health in Zhejiang Province (Grant No. 2024KY1761) and Quzhou Science and Technology Program (Grant No. 2023K112).
Authors WG, XZ, and YaZ are employed by WillingMed Technology Beijing Co., Ltd, Beijing, China.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1345706/full#supplementary-material
Ao, Z., Xu, H., Li, M., Liu, H., Deng, M., Liu, Y. (2023). Clinical characteristics, diagnosis, outcomes and lung microbiome analysis of invasive pulmonary aspergillosis in the community-acquired pneumonia patients. BMJ Open Respir. Res. 10, e001358. doi: 10.1136/bmjresp-2022-001358
Bao, S., Song, H., Chen, Y., Zhong, C., Tang, H. (2022). Metagenomic next-generation sequencing for the diagnosis of pulmonary aspergillosis in non-neutropenic patients: a retrospective study. Front. Cell Infect. Microbiol. 12, 925982. doi: 10.3389/fcimb.2022.925982
Barton, E. S., White, D. W., Cathelyn, J. S., Brett-Mcclellan, K. A., Engle, M., Diamond, M. S., et al. (2007). Herpesvirus latency confers symbiotic protection from bacterial infection. Nature 447, 326–329. doi: 10.1038/nature05762
Bittinger, K., Charlson, E. S., Loy, E., Shirley, D. J., Haas, A. R., Laughlin, A., et al. (2014). Improved characterization of medically relevant fungi in the human respiratory tract using next-generation sequencing. Genome Biol. 15, 487. doi: 10.1186/s13059-014-0487-y
Blauwkamp, T. A., Thair, S., Rosen, M. J., Blair, L., Lindner, M. S., Vilfan, I. D., et al. (2019). Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat. Microbiol. 4, 663–674. doi: 10.1038/s41564-018-0349-6
Boch, T., Spiess, B., Cornely, O. A., Vehreschild, J. J., Rath, P. M., Steinmann, J., et al. (2016). Diagnosis of invasive fungal infections in hematological patients by combined use of galactomannan, 1,3-beta-D-glucan, Aspergillus PCR, multifungal DNA-microarray, and Aspergillus azole resistance PCRs in blood and bronchoalveolar lavage samples: results of a prospective multicenter study. Clin. Microbiol. Infect. 22, 862–868. doi: 10.1016/j.cmi.2016.06.021
Bongomin, F., Gago, S., Oladele, R. O., Denning, D. W. (2017). Global and multi-national prevalence of fungal diseases-estimate precision. J. Fungi (Basel) 3, 57. doi: 10.3390/jof3040057
Carpenter, M. L., Tan, S. K., Watson, T., Bacher, R., Nagesh, V., Watts, A., et al. (2019). Metagenomic next-generation sequencing for identification and quantitation of transplant-related DNA viruses. J. Clin. Microbiol. 57, e0113–19. doi: 10.1128/JCM.01113-19
Chen, S., Kang, Y., Li, D., Li, Z. (2022). Diagnostic performance of metagenomic next-generation sequencing for the detection of pathogens in bronchoalveolar lavage fluid in patients with pulmonary infections: Systematic review and meta-analysis. Int. J. Infect. Dis. 122, 867–873. doi: 10.1016/j.ijid.2022.07.054
Chen, G., Ma, N., Zhu, D., Zhou, H., Liang, Q., Meng, J., et al. (2023a). Accurate diagnosis of bronchopulmonary Talaromyces marneffei infection in an anti-IFN-γ autoantibodies positive patient assisted by endobronchial ultrasound-guided TBNA and mNGS: a case report. Front. Cell Infect. Microbiol. 13, 1186335. doi: 10.3389/fcimb.2023.1186335
Chen, H., Yin, Y., Gao, H., Guo, Y., Dong, Z., Wang, X., et al. (2020). Clinical utility of in-house metagenomic next-generation sequencing for the diagnosis of lower respiratory tract infections and analysis of the host immune response. Clin. Infect. Dis. 71, S416–S426. doi: 10.1093/cid/ciaa1516
Chen, H., Zhang, Y., Zheng, J., Shi, L., He, Y., Niu, Y., et al. (2021). Application of mNGS in the etiological diagnosis of thoracic and abdominal infection in patients with end-stage liver disease. Front. Cell Infect. Microbiol. 11, 741220. doi: 10.3389/fcimb.2021.741220
Chen, H., Zheng, Y., Zhang, X., Liu, S., Yin, Y., Guo, Y., et al. (2023b). Clinical evaluation of cell-free and cellular metagenomic next-generation sequencing of infected body fluids. J. Adv. Res. 119–129. doi: 10.1016/j.jare.2023.02.018
Denning, D. W., Cadranel, J., Beigelman-Aubry, C., Ader, F., Chakrabarti, A., Blot, S., et al. (2016). Chronic pulmonary aspergillosis: rationale and clinical guidelines for diagnosis and management. Eur. Respir. J. 47, 45–68. doi: 10.1183/13993003.00583-2015
Denning, D. W., Chakrabarti, A. (2017). Pulmonary and sinus fungal diseases in non-immunocompromised patients. Lancet Infect. Dis. 17, e357–e366. doi: 10.1016/S1473-3099(17)30309-2
Denning, D. W., Pashley, C., Hartl, D., Wardlaw, A., Godet, C., Del Giacco, S., et al. (2014). Fungal allergy in asthma-state of the art and research needs. Clin. Transl. Allergy 4, 14. doi: 10.1186/2045-7022-4-14
Donnelly, J. P., Chen, S. C., Kauffman, C. A., Steinbach, W. J., Baddley, J. W., Verweij, P. E., et al. (2020). Revision and update of the consensus definitions of invasive fungal disease from the European organization for research and treatment of cancer and the mycoses study group education and research consortium. Clin. Infect. Dis. 71, 1367–1376. doi: 10.1093/cid/ciz1008
El-Baba, F., Gao, Y., Soubani, A. O. (2020). Pulmonary aspergillosis: what the generalist needs to know. Am. J. Med. 133, 668–674. doi: 10.1016/j.amjmed.2020.02.025
Epstein, M. A., Achong, B. G., Barr, Y. M. (1964). Virus particles in cultured lymphoblasts from burkitt’s lymphoma. Lancet 1, 702–703. doi: 10.1016/S0140-6736(64)91524-7
Feys, S., Goncalves, S. M., Khan, M., Choi, S., Boeckx, B., Chatelain, D., et al. (2022). Lung epithelial and myeloid innate immunity in influenza-associated or COVID-19-associated pulmonary aspergillosis: an observational study. Lancet Respir. Med. 10, 1147–1159. doi: 10.1016/S2213-2600(22)00259-4
Gu, W., Deng, X., Lee, M., Sucu, Y. D., Arevalo, S., Stryke, D., et al. (2021). Rapid pathogen detection by metagenomic next-generation sequencing of infected body fluids. Nat. Med. 27, 115–124. doi: 10.1038/s41591-020-1105-z
Guo, L., Wu, X., Wu, X. (2023). Aspergillus infection in chronic obstructive pulmonary diseases. Clin. Respir. J. 17, 129–138. doi: 10.1111/crj.13585
Hoenigl, M., Egger, M., Price, J., Krause, R., Prattes, J., White, P. L. (2023). Metagenomic next-generation sequencing of plasma for diagnosis of COVID-19-associated pulmonary aspergillosis. J. Clin. Microbiol. 61, e0185922. doi: 10.1128/jcm.01859-22
Jia, H., Liu, H., Tu, M., Wang, Y., Wang, X., Li, J., et al. (2023). Diagnostic efficacy of metagenomic next generation sequencing in bronchoalveolar lavage fluid for proven invasive pulmonary aspergillosis. Front. Cell Infect. Microbiol. 13, 1223576. doi: 10.3389/fcimb.2023.1223576
Khasawneh, F., Mohamad, T., Moughrabieh, M. K., Lai, Z., Ager, J., Soubani, A. O. (2006). Isolation of Aspergillus in critically ill patients: a potential marker of poor outcome. J. Crit. Care 21, 322–327. doi: 10.1016/j.jcrc.2006.03.006
Khojasteh, S., Abastabar, M., Haghani, I., Valadan, R., Ghazanfari, S., Abbasi, K., et al. (2023). Five-year surveillance study of clinical and environmental Triazole-Resistant Aspergillus fumigatus isolates in Iran. Mycoses 66, 98–105. doi: 10.1111/myc.13535
Kosmidis, C., Denning, D. W. (2015). The clinical spectrum of pulmonary aspergillosis. Thorax 70, 270–277. doi: 10.1136/thoraxjnl-2014-206291
Lamoth, F., Akan, H., Andes, D., Cruciani, M., Marchetti, O., Ostrosky-Zeichner, L., et al. (2021). Assessment of the role of 1,3-beta-d-glucan testing for the diagnosis of invasive fungal infections in adults. Clin. Infect. Dis. 72, S102–S108. doi: 10.1093/cid/ciaa1943
Lamoth, F., Calandra, T. (2022). Pulmonary aspergillosis: diagnosis and treatment. Eur. Respir. Rev. 31, 220114. doi: 10.1183/16000617.0114-2022
Langelier, C., Kalantar, K. L., Moazed, F., Wilson, M. R., Crawford, E. D., Deiss, T., et al. (2018). Integrating host response and unbiased microbe detection for lower respiratory tract infection diagnosis in critically ill adults. Proc. Natl. Acad. Sci. U.S.A. 115, E12353–E12362. doi: 10.1073/pnas.1809700115
Ledoux, M. P., Guffroy, B., Nivoix, Y., Simand, C., Herbrecht, R. (2020). Invasive pulmonary aspergillosis. Semin. Respir. Crit. Care Med. 41, 80–98. doi: 10.1055/s-0039-3401990
Li, H. (2022). Editorial: mNGS for fungal pulmonary infection diagnostics. Front. Cell Infect. Microbiol. 12, 864163. doi: 10.3389/fcimb.2022.864163
Liao, H., Zhu, M., Cheng, Z. (2023). Epstein-Barr virus (EBV) induced pneumonitis in a patient with breast cancer receiving neoadjuvant chemotherapy: A case report. Respir. Med. Case Rep. 45, 101849. doi: 10.1016/j.rmcr.2023.101849
Liu, L., Wu, H., Liang, J., Wu, W., Peng, Y., Zhou, H., et al. (2022). Rhizomucor pusillus infection in a patient with acute myeloid leukemia after hematopoietic stem cell transplantation: clinical impact of metagenomics next-generation sequencing. Infect. Drug Resist. 15, 4285–4290. doi: 10.2147/IDR.S376045
Liu, H., Xu, H., Liu, H., Zhao, Z., Zhang, X. (2023). Metagenomic next-generation sequencing in the diagnose of pulmonary infection with airway complications in a lung transplant recipient. Heliyon 9, e19611. doi: 10.1016/j.heliyon.2023.e19611
Liu, L., Yuan, M., Shi, Y., Su, X. (2021). Clinical Performance of BAL Metagenomic Next-Generation Sequence and Serum (1,3)-beta-D-Glucan for Differential Diagnosis of Pneumocystis jirovecii Pneumonia and Pneumocystis jirovecii Colonization. Front. Cell Infect. Microbiol. 11, 784236. doi: 10.3389/fcimb.2021.784236
McManus, T. E., Coyle, P. V., Lawson, J., Js, E., Jc., K. (2009). Epstein - barr virus pneumonitis. Ulster Med. J. 78, 137–138.
Mei, J., Hu, H., Zhu, S., Ding, H., Huang, Z., Li, W., et al. (2023). Diagnostic role of mNGS in polymicrobial periprosthetic joint infection. J. Clin. Med. 12, 1838. doi: 10.3390/jcm12051838
Mengoli, C., Cruciani, M., Barnes, R. A., Loeffler, J., Donnelly, J. P. (2009). Use of PCR for diagnosis of invasive aspergillosis: systematic review and meta-analysis. Lancet Infect. Dis. 9, 89–96. doi: 10.1016/S1473-3099(09)70019-2
Miller, S., Naccache, S. N., Samayoa, E., Messacar, K., Arevalo, S., Federman, S., et al. (2019). Laboratory validation of a clinical metagenomic sequencing assay for pathogen detection in cerebrospinal fluid. Genome Res. 29, 831–842. doi: 10.1101/gr.238170.118
Nabili, M., Shokohi, T., Moazeni, M., Khodavaisy, S., Aliyali, M., Badiee, P., et al. (2016). High prevalence of clinical and environmental triazole-resistant Aspergillus fumigatus in Iran: is it a challenging issue? J. Med. Microbiol. 65, 468–475. doi: 10.1099/jmm.0.000255
Negm, E. M., Mohamed, M. S., Rabie, R. A., Fouad, W. S., Beniamen, A., Mosallem, A., et al. (2023). Fungal infection profile in critically ill COVID-19 patients: a prospective study at a large teaching hospital in a middle-income country. BMC Infect. Dis. 23, 246. doi: 10.1186/s12879-023-08226-8
Niazi, M. R., Iqbal, Q. Z., Mishiyev, D., Narula, N., Abdul Sattar, S. B., Zia, Z., et al. (2020). Epstein-Barr virus (EBV) induced pneumonitis in an immunocompetent adult: A case report. Respir. Med. Case Rep. 31, 101262. doi: 10.1016/j.rmcr.2020.101262
Ogunbayo, A. E., Mogotsi, M. T., Sondlane, H., Sabiu, S., Nyaga, M. M. (2023). Metagenomics characterization of respiratory viral RNA pathogens in children under five years with severe acute respiratory infection in the Free State, South Africa. J. Med. Virol. 95, e28753. doi: 10.1002/jmv.28753
Ok, C. Y., Li, L., Young, K. H. (2015). EBV-driven B-cell lymphoproliferative disorders: from biology, classification and differential diagnosis to clinical management. Exp. Mol. Med. 47, e132–e132. doi: 10.1038/emm.2014.82
Patterson, T. F., Thompson, G. R., 3rd, Denning, D. W., Fishman, J. A., Hadley, S., Herbrecht, R., et al. (2016). Practice guidelines for the diagnosis and management of aspergillosis: 2016 update by the infectious diseases society of america. Clin. Infect. Dis. 63, e1–e60. doi: 10.1093/cid/ciw326
Person, A. K., Chudgar, S. M., Norton, B. L., Tong, B. C., Stout, J. E. (2010). Aspergillus Niger: an unusual cause of invasive pulmonary aspergillosis. J. Med. Microbiol. 59, 834–838. doi: 10.1099/jmm.0.018309-0
Reyes, A., Carbo, E. C., Harinxma Thoe Slooten, J. S. V., Kraakman, M. E. M., Sidorov, I. A., Claas, E. C. J., et al. (2021). Viral metagenomic sequencing in a cohort of international travelers returning with febrile illness. J. Clin. Virol. 143, 104940. doi: 10.1016/j.jcv.2021.104940
Shi, Y., Peng, J. M., Qin, H. Y., Du, B. (2022). Metagenomic next-generation sequencing: A promising tool for diagnosis and treatment of suspected pneumonia in rheumatic patients with acute respiratory failure: Retrospective cohort study. Front. Cell Infect. Microbiol. 12, 941930. doi: 10.3389/fcimb.2022.941930
Shin, B., Koh, W. J., Jeong, B. H., Yoo, H., Park, H. Y., Suh, G. Y., et al. (2014). Serum galactomannan antigen test for the diagnosis of chronic pulmonary aspergillosis. J. Infect. 68, 494–499. doi: 10.1016/j.jinf.2014.01.005
Shinfuku, K., Suzuki, J., Takeda, K., Kawashima, M., Morio, Y., Sasaki, Y., et al. (2023). Validity of platelia aspergillus igG and aspergillus precipitin test to distinguish pulmonary aspergillosis from colonization. Microbiol. Spectr. 11, e0343522. doi: 10.1128/spectrum.03435-22
Song, N., Li, X., Liu, W. (2021). Metagenomic next-generation sequencing (mNGS) for diagnosis of invasive fungal infectious diseases: a narrative review. J. Lab. Precis. Med. 6, 29–29. doi: 10.21037/jlpm
Soubani, A. O., Khanchandani, G., Ahmed, H. P. (2004). Clinical significance of lower respiratory tract Aspergillus culture in elderly hospitalized patients. Eur. J. Clin. Microbiol. Infect. Dis. 23, 491–494. doi: 10.1007/s10096-004-1137-1
Sugui, J. A., Kwon-Chung, K. J., Juvvadi, P. R., Latge, J. P., Steinbach, W. J. (2014). Aspergillus fumigatus and related species. Cold Spring Harb. Perspect. Med. 5, a019786. doi: 10.1101/cshperspect.a019786
Tan, Y., Chen, Z., Zeng, Z., Wu, S., Liu, J., Zou, S., et al. (2023). Microbiomes detected by bronchoalveolar lavage fluid metagenomic next-generation sequencing among HIV-infected and uninfected patients with pulmonary infection. Microbiol. Spectr. 11, e0000523. doi: 10.1128/spectrum.00005-23
Tashiro, T., Izumikawa, K., Tashiro, M., Takazono, T., Morinaga, Y., Yamamoto, K., et al. (2011). Diagnostic significance of Aspergillus species isolated from respiratory samples in an adult pneumology ward. Med. Mycol 49, 581–587. doi: 10.3109/13693786.2010.548084
Thompson, G. R., 3rd, Young, J. H. (2021). Aspergillus infections. N Engl. J. Med. 385, 1496–1509. doi: 10.1056/NEJMra2027424
Wang, C., You, Z., Fu, J., Chen, S., Bai, D., Zhao, H., et al. (2022). Application of metagenomic next-generation sequencing in the diagnosis of pulmonary invasive fungal disease. Front. Cell Infect. Microbiol. 12, 949505. doi: 10.3389/fcimb.2022.949505
Xie, Y., Dai, B., Zhou, X., Liu, H., Wu, W., Yu, F., et al. (2023). Diagnostic value of metagenomic next-generation sequencing for multi-pathogenic pneumonia in HIV-infected patients. Infect. Drug Resist. 16, 607–618. doi: 10.2147/IDR.S394265
Xie, G., Zhao, B., Wang, X., Bao, L., Xu, Y., Ren, X., et al. (2021). Exploring the clinical utility of metagenomic next-generation sequencing in the diagnosis of pulmonary infection. Infect. Dis. Ther. 10, 1419–1435. doi: 10.1007/s40121-021-00476-w
Xu, Y., Wang, Y., Wu, J., Zhao, X., Huang, G., Fang, J. (2022). Coinfection with Hypervirulent Klebsiella pneumoniae and Aspergillus flavus in a Critically Ill Patient with Aspergillus Overlap Syndrome: A Case Report. Infect. Drug Resist. 15, 7823–7830. doi: 10.2147/IDR.S394004
Keywords: pulmonary aspergillosis, Aspergillus, colonization, metagenomic next-generation sequencing, optimal threshold value
Citation: Jiang Z, Gai W, Zhang X, Zheng Y, Jin X, Han Z, Ao G, He J, Shu D, Liu X, Zhou Y and Hua Z (2024) Clinical performance of metagenomic next-generation sequencing for diagnosis of pulmonary Aspergillus infection and colonization. Front. Cell. Infect. Microbiol. 14:1345706. doi: 10.3389/fcimb.2024.1345706
Received: 28 November 2023; Accepted: 18 March 2024;
Published: 28 March 2024.
Edited by:
Hossein Zarrinfar, Mashhad University of Medical Sciences, IranReviewed by:
Çağrı Ergin, Pamukkale University, TürkiyeCopyright © 2024 Jiang, Gai, Zhang, Zheng, Jin, Han, Ao, He, Shu, Liu, Zhou and Hua. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhidan Hua, aHVhemhpZGFuQHdtdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.