- 1Institute of Hygiene, University Hospital Münster, Münster, Germany
- 2Department of Cardiology I – Coronary and Peripheral Vascular Disease, Heart Failure, University Hospital Münster, Münster, Germany
- 3Institute of Medical Microbiology, University Hospital Münster, Münster, Germany
Introduction: The emergence of extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae is an urgent and alarming One Health problem. This study aimed to investigate duplications of plasmid-encoded ESBL genes and their impact on antimicrobial resistance (AMR) phenotypes in clinical and screening isolates.
Methods: Multi-drug-resistant bacteria from hospitalized patients were collected during routine clinical surveillance from January 2022 to June 2023, and their antimicrobial susceptibility patterns were determined. Genotypes were extracted from long-read whole-genome sequencing data. Furthermore, plasmids and other mobile genetic elements associated with ESBL genes were characterized, and the ESBL genes were correlated to ceftazidime minimal inhibitory concentration (MIC).
Results: In total, we identified four cases of plasmid-encoded ESBL gene duplications that match four genetically similar plasmids during the 18-month surveillance period: five Escherichia coli and three Klebsiella pneumoniae isolates. As the ESBL genes were part of transposable elements, the surrounding sequence regions were duplicated as well. In-depth analysis revealed insertion sequence (IS)-mediated transposition mechanisms. Isolates with duplicated ESBL genes exhibited a higher MIC for ceftazidime in comparison to isolates with a single gene copy (3–256 vs. 1.5–32 mg/L, respectively).
Conclusion: ESBL gene duplications led to an increased phenotypic resistance against ceftazidime. Our data suggest that ESBL gene duplications by an IS-mediated transposition are a relevant mechanism for how AMR develops in the clinical setting and is part of the microevolution of plasmids.
1 Introduction
Antimicrobial resistance (AMR) is an alarming and urgent One Health problem (Ventola, 2015; Kim and Cha, 2021). AMR allows bacteria to resist and persist even though antibiotics are present. Ever since the first introduction of antimicrobials, the number of resistant bacteria has steadily increased and is becoming a major challenge for healthcare systems worldwide (Davies and Davies, 2010; O’Neill, 2014; Murray et al., 2022). The dissemination of AMR is driven by a combination of different factors such as the extensive use of antimicrobial agents (Browne et al., 2021) as well as selection pressure in almost all environments (Kizny Gordon et al., 2017; Chen et al., 2019). Especially in the hospital setting, AMR is a major concern, as they limit treatment options for serious bacterial infections significantly (Huemer et al., 2020).
Hydrolysis of the β-lactam ring of penicillin, the first discovered antibiotic agent, and their derivates by β-lactamases, is among the most important AMR mechanisms. In 1965, the first plasmid-encoded β-lactamase TEM-1 was discovered, which provided significant resistance in addition to the natural occurrence of chromosomal β-lactamase genes (Datta and Kontomichalou, 1965). Point mutations in the bla genes coding for β-lactamases generate the fast-growing family of extended-spectrum beta-lactamases (ESBLs), i.e., TEM-type and SHV-type β-lactamases (Bradford, 2001; Behzadi et al., 2020). ESBLs also provide resistance against newer antibiotic classes such as aminopenicillins and cephalosporins. The most commonly found and clinically relevant ESBL genes are blaTEM, blaSHV, and blaCTX-M (Liakopoulos et al., 2016).
ESBL-producing Enterobacteriaceae (Bradford, 2001) are of special concern in the healthcare setting, as they are listed as critical in the priority pathogens list by the WHO (Tacconelli et al., 2018). Most ESBL-producing Enterobacteriaceae in the hospital setting can be identified as Escherichia coli and Klebsiella pneumoniae. For ESBL-producing E. coli, an asymptomatic and persistent carriage in the human intestine is well-known and has been increasing in the last decades (Bezabih et al., 2021).
Most AMR genes are coded as accessory genes on mobile genetic elements (MGEs) (Partridge et al., 2018) that can be transferred among bacteria on plasmids via conjugation. This conjugation is possible not only between bacteria of the same species but also beyond species borders within a multispecies bacterial community. Therefore, plasmids are believed to be the key drivers in the development and spread of antimicrobial resistance (Carattoli, 2009). Furthermore, different types of transposons are described as MGEs used to transfer AMR genes (Frost et al., 2005). There, AMR genes are often found as accessory genes, for example, within composite transposons. These transposons are characterized by flanking insertion sequence (IS) coding a transposase, and AMR and virulence genes in between. The transposase uses the IS as recognition sites to transfer the whole transposon from one position in the DNA to another as well as from plasmids to the chromosome if a suitable site is found. Other AMR gene transpositions are carried out by unit transposons, where an IS transposase is only found on one site of the AMR gene (He et al., 2015; Varani et al., 2021).
For many years, the investigation of AMR plasmids has been labor-intensive due to the shortfalls of short-read whole-genome sequencing (WGS) technology. For instance, short-read WGS only covers parts of transposons, which leads to possible misassemblies of these sequence repetitions (Juraschek et al., 2021). Recent long-read WGS offers a significant advantage by enabling the complete coverage of plasmids or the generation of reads that span entire transposons. This capability simplifies the detection of transposons and gene duplications within the sequences.
To increase the understanding of AMR development and spread especially in ESBLs, we analyzed multi-drug-resistant Enterobacteriaceae isolated from clinical samples in more detail by long-read WGS. Although plasmids are key drivers of AMR spread, only little is known about the evolutionary and molecular mechanisms of plasmids harboring AMR genes in the clinical setting. Here, we characterize four cases of ESBL gene duplications on plasmids and demonstrate how these duplications affect the AMR phenotype.
2 Material and methods
2.1 Clinical setting and bacterial isolates
The University Hospital Münster (UHM) is a 1,300-bed tertiary care center in Münster, Germany, admitting on average 55,000 patients per year. Bacterial samples were collected from hospitalized patients at the UHM for an 18-month time period from January 2022 to June 2023 as part of the routine hospital surveillance of multi-drug-resistant bacteria (MDRBs) according to national recommendations (Bundesgesundheitsblatt, 2012). Screening specimens were cultivated on chromID ESBL Agar (biomérieux, Marcy-l’Etoile, France) and incubated at 36°C ± 1°C for 18 to 24 h. Clinical specimens were cultivated on Columbia Blood agar with 5% Sheep Blood (BD, Heidelberg, Germany; aerobic incubation at 35°C ± 2°C ambient air for up to 3 days), and MacConkey selective agar for Gram-negative bacteria (MacConkey II Agar, BD, Heidelberg, Germany; aerobic incubation at ambient air 35°C ± 2°C for up to 2 days) if applicable. During the study period, >500 MDRB Enterobacteriaceae isolates were identified.
2.2 Species identification
Species identification was performed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF/MS) (MALDI-TOF MS, Biotyper® Sirius one, Bruker, Bremen, Germany) with scores above 2.0.
2.3 Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed using VITEK®2 automated system (biomérieux, Marcy-l’Etoile, France). In addition, Etests® (biomérieux, Marcy-l’Etoile, France) for ceftazidime were performed on Müller-Hinton-Agar (Oxoid, Schwerte, Germany) to determine the minimal inhibitory concentration (MIC). All MICs were interpreted according to the 2023 European Committee on Antimicrobial Susceptibility Testing (EUCAST) clinical breakpoints version 13.1 (EUCAST, 2023).
2.4 Whole-genome sequencing
DNA from bacterial isolates was extracted using the NEB Monarch Purification Kit (New England Biolabs, Ipswich, MA, USA) and sequenced on a PacBio® Sequel IIe system (Pacific Biosciences, Menlo Park, CA, USA) as described previously (Effelsberg et al., 2021). A de novo assembly approach was used for the raw reads and analyzed using the SMRT® Link software suite v. 10 or v. 11 with default parameters.
2.5 Whole-genome data analysis, annotation, and visualization
The phylogenetic tree was generated based on WGS data by the Genome BLAST Distance Phylogeny (GBDP) approach using the Type Strain Genome Server (Meier-Kolthoff and Göker, 2019). For genotyping based on multilocus sequence typing (MLST) and core genome MLST (cgMLST) and to determine the clonal relationship, the respective schemes in Ridom SeqSphere+ software version 9.0.9 (Ridom GmbH, Münster, Germany) were used (Jünemann et al., 2013). Isolates were rated as clonal when the distance of their cgMLST allelic profiles was ≤5 alleles. Antimicrobial resistance genes were determined using the NCBI AMRFinderPlus (Feldgarden et al., 2019) implemented in SeqSphere+. If duplicated AMR ESBL genes were identified, a more detailed analysis of the respective contigs was performed. Contigs were predicted as plasmids using MOB-Suite v3.1.4 (Robertson and Nash, 2018) and further characterized by pMLST Server 2.0 (Carattoli et al., 2014). Plasmids were completely annotated using DFAST v1.6.0 (including additional scans against TIGRFAM and COG database by NCBI) (Tanizawa et al., 2018). Next, it was searched for genetically matching plasmids with the same ESBL gene but without a gene duplication in the dataset by a mash-based approach (Ondov et al., 2016) implemented in SeqSphere+. Isolates containing these plasmids were used as references and were analyzed in the same way.
Progressive Mauve (Darling et al., 2004) was used to compare and visualize the similarity of plasmid sequences and to localize the gene duplication site of the transposable regions. In addition, all mobile genetic elements were annotated using ISFinder (Siguier et al., 2006) and MobileElementFinder with the default parameters (Johansson et al., 2021). Plasmid sequences were manually curated using SnapGene® Viewer (Dotmatics; www.snapgene.com).
3 Results
3.1 Characteristics of bacterial isolates
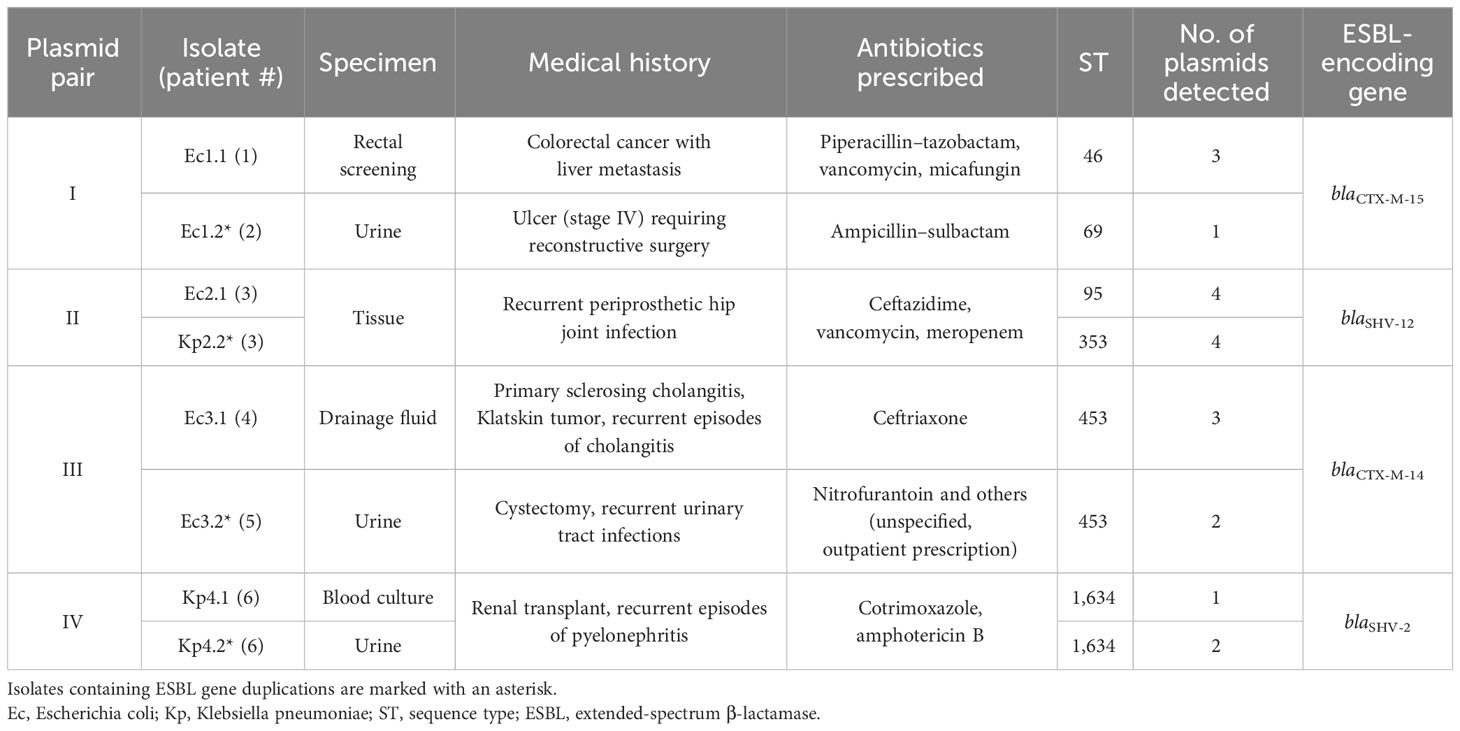
We identified four isolates (three E. coli and one K. pneumoniae) containing more than one plasmid-based ESBL gene copy within the isolates found by our routine molecular surveillance. For comparison, we matched these four isolates with more than one plasmid-based ESBL gene to four additional isolates (two E. coli and two K. pneumoniae) in the dataset containing genetically the same plasmids with only a single ESBL gene copy (Supplementary Figures 1–4). These isolate matches are named pairs I–IV and originate from different or the same patient. Details of the isolates analyzed and the patients’ characteristics are listed in Table 1. In total, bacterial isolates of six patients were included in the study: four female patients and two male patients. The median age of patients was 63 (32–94) years. According to their medical history based on the medical reports, all patients suffered from chronic or severe infections (Table 1).
The phylogenetic relation of all isolates was calculated based on the WGS data to illustrate the phylogenetic similarity between the isolates in pairs III and IV and the species difference in pair II (Figure 1A). In case the matched isolates were the same species, cgMLST typing was performed to determine whether the pairs only share a similar plasmid or in addition have a clonal relation (on the chromosomal level). The isolates of pairs I and III did not show clonality in cgMLST analysis among the E. coli isolates (Figure 1B). Pair IV was identified as a clonal lineage of K. pneumoniae isolates found in one patient (patient 6). Here, the isolates were collected 3 weeks apart in different sample materials (Table 1). The similar plasmids of pair II were shared between an E. coli isolate and a K. pneumoniae isolate that originated from the same specimen of patient 3 (Table 1). There was no clonal relationship due to the two different bacterial species.
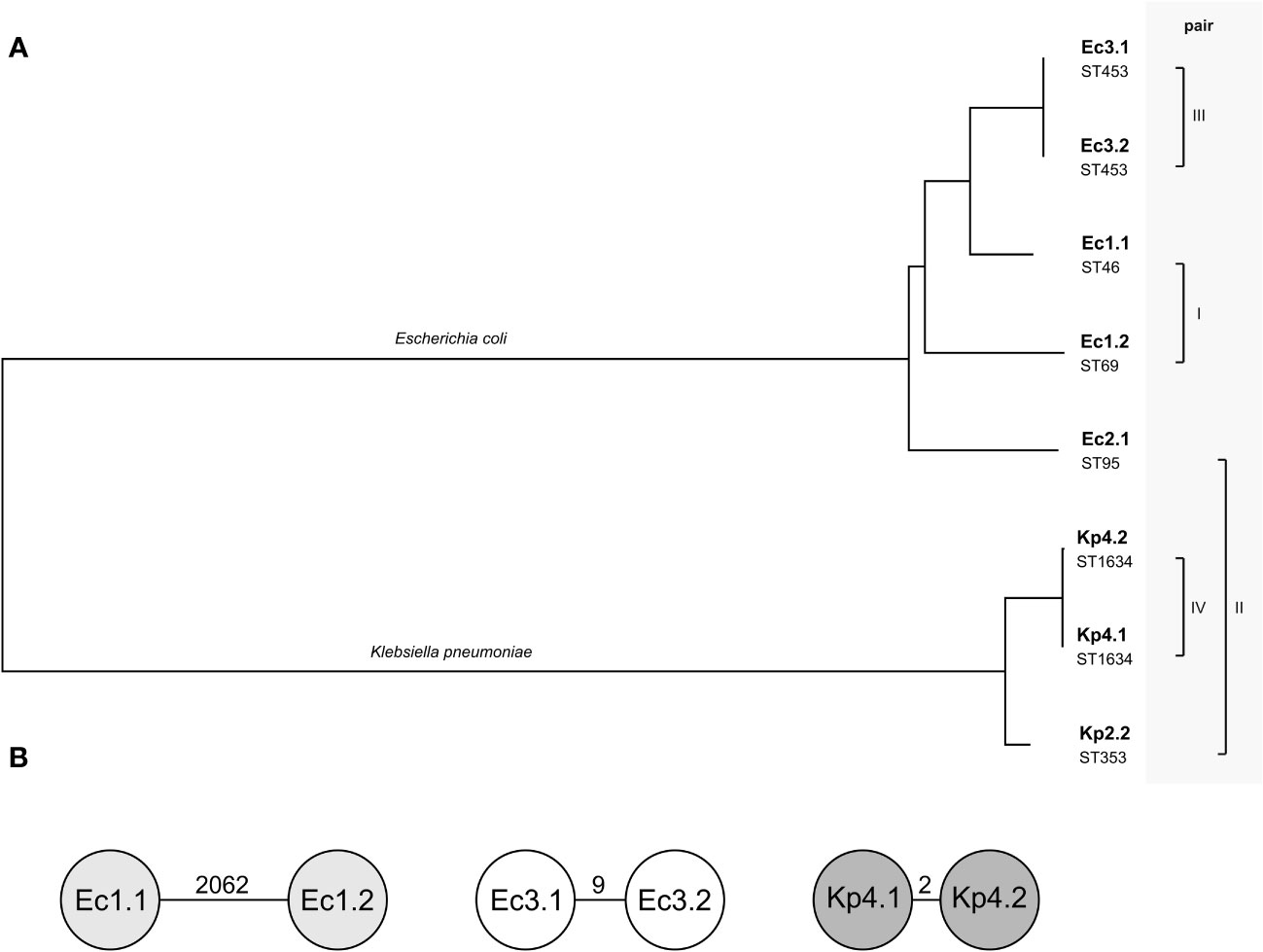
Figure 1 Relationship of the four pairs of isolates included. (A) Phylogenetic tree based on whole-genome sequences of the eight isolates calculated by Type (Strain) Genome Server. The branch lengths are scaled in terms of Genome BLAST Distance Phylogeny distance formula. The pairs describe the compared plasmids of the isolates. Below the isolate name, the sequence type (ST) of each isolate is given. (B) Detailed relation within the pairs of isolates of the same species using cgMLST. Each circle is a separate genotype based on the allelic profile of up to 2,325 genes for Escherichia coli and 2,358 genes for Klebsiella pneumoniae, with the parameter “ignoring missing genes in the pairwise comparison”. The number on the connecting lines displays the allelic distance.
3.2 In-depth analysis of plasmids harboring ESBL-encoding genes
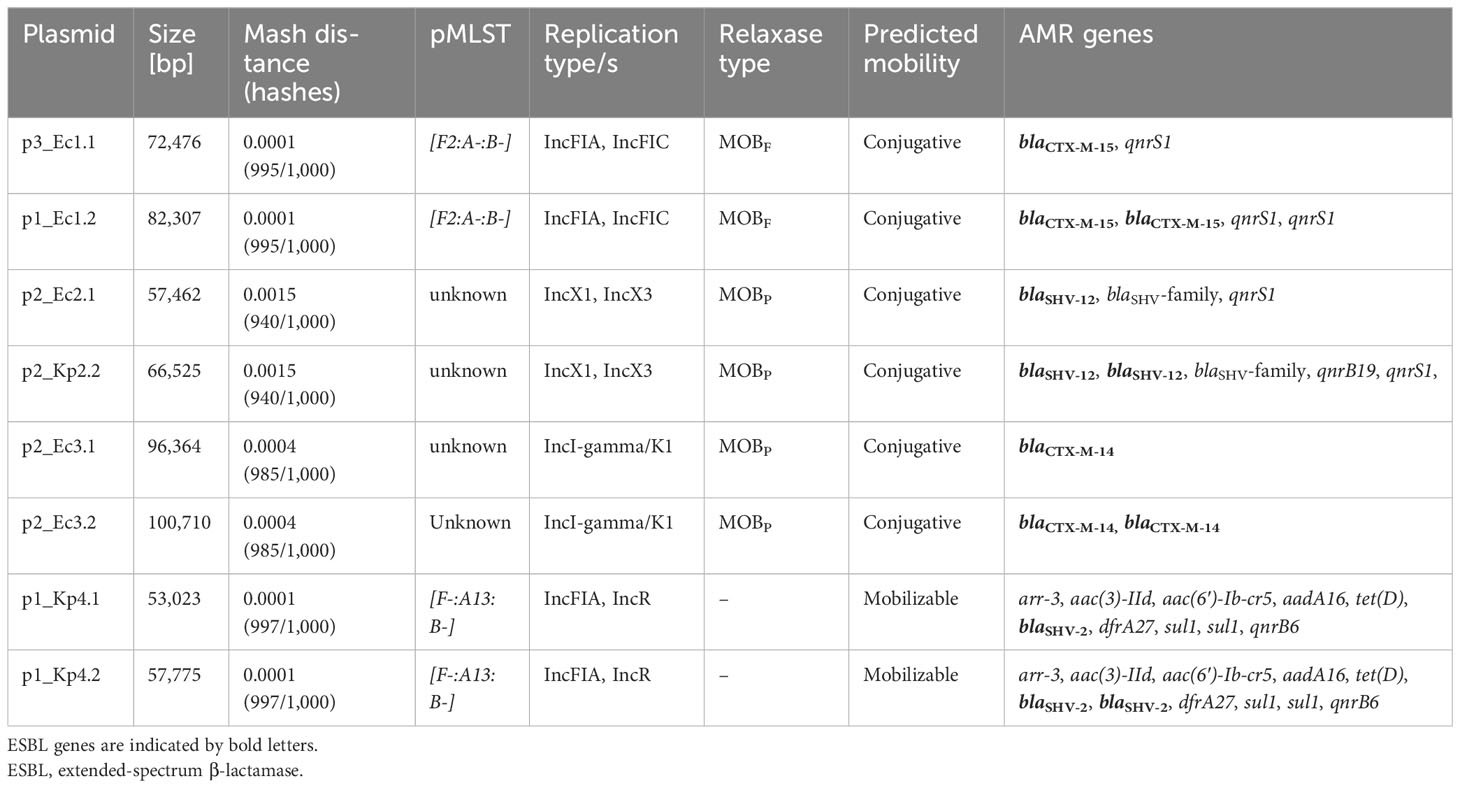
Using the MOB-suite software and AMRFinderPlus tool, we analyzed the plasmids harboring ESBL genes with respect to their sizes, replication and relaxase types, anticipated/predicted mobility, and encoded AMR genes (Table 2). Overall, the replication type, the relaxase type, and the predicted mobility of the plasmids were identical within a pair. However, due to the ESBL gene duplication, the plasmid size varied. In addition to the duplicated ESBL-encoding genes, we only found an analog AMR duplication of qnrS1 in p1_Ec1.2 mediating quinolone resistance. Of note, the similar plasmids p2_Ec2.1 and p2_Kp2.2 are most likely directly related and caused by an intra-host transmission via horizontal gene transfer (HGT) from one species to another.
For an in-depth analysis of the plasmids, we annotated the whole plasmids with DFAST and used ISFinder and MobileElementFinder (Supplementary Figures 1–4). All duplicated ESBL genes were part of a longer transposable DNA sequence, which was duplicated in full length (Figure 2; Supplementary Table 1).
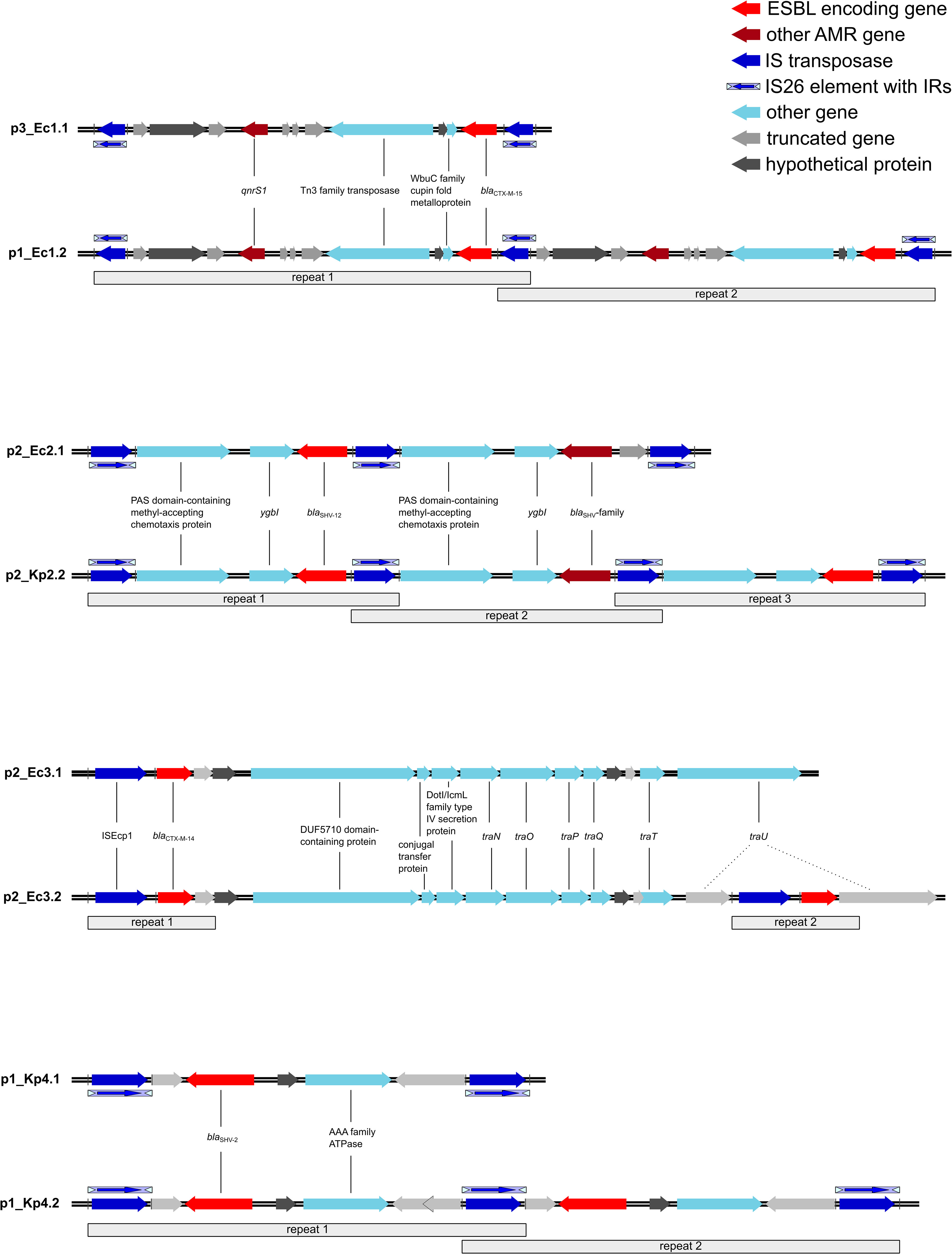
Figure 2 Transposons harboring the ESBL genes and the duplicated regions. All repeated regions harbor the same genes unless indicated otherwise. The blaSHV-family gene is a mutated variant of blaSHV-12. Ec, Escherichia coli; Kp, Klebsiella pneumoniae; ESBL, extended-spectrum β-lactamase; AMR, antimicrobial resistance; IS, insertion sequence; IR, inverted repeats; ESBL, extended-spectrum β-lactamase.
In pair I, the duplication occurred in p1_Ec1.2 as a tandem duplication with flanking IS26 elements (Figure 2), notably with three IS26 elements in comparison to p3_Ec1.1 instead of two flanking IS26 elements for each duplication. The transposon was classified as a composite transposon and harbors the ESBL gene blaCTX-M-15 as well as the quinolone resistance gene qnrS1. In addition to AMR genes, a WbuC family cupin fold metalloprotein and Tn3 transposase were encoded.
In p2_Ec2.1, the reference plasmid of pair II, we already detected an IS26 tandem duplication of a transposon harboring the ESBL gene blaSHV-12. However, there was a difference in the sequence length of one of the transposons caused by an incomplete IS26 element in the second repeat (Figure 2) and a deletion in the ESBL gene (blaSHV-12/blaSHV-family). A BLAST analysis revealed a deletion of three nucleotides in the ESBL gene coding originally for leucine 53 in blaSHV-12. In addition, the transposon contains genes for a PAS domain-containing methyl-accepting chemotaxis protein and for the DNA-binding transcriptional repressor YgbI. While the reference plasmid already shows a duplication, we found a tandem triplication of the transposon in p2_Kp2.2. The coded genes remain the same as in the reference p2_Ec2.1. Nevertheless, one of the three blaSHV genes shows a nucleotide deletion as well. The deletion corresponding to threonine 18 in blaSHV-12 results in a frame shift and an early stop codon, thereby shortening the potentially encoded β-lactamase.
In pair III, we observed another duplication pattern. The ESBL target gene blaCTX-M-14 in p2_Ec3.2 and the reference plasmid p2_Ec3.1 was carried in an ISEcp1-mediated unit transposon. ISEcp1 could be aligned with 100% and 99.94% sequence identities. In addition to the β-lactamase gene, only potential gene residues were transferred within the transposon. The comparison revealed that the point of insertion of the second repeat of the unit transposon in p2_Ec3.2 is the traU gene (Figure 2).
The plasmid p1_Kp4.2 belongs to pair IV. A tandem duplication of IS26-mediated composite transposons could be detected. The one nucleotide difference in the first repeat in p1_Kp4.2 was caused by an insertion in the already truncated oligosaccharide MFS transporter gene as indicated in Figure 2. The transposon contains the ESBL target gene blaSHV-2 and a gene for an AAA family ATPase.
Taken together, IS26-mediated duplications were found to be tandem duplications, where one IS26 element is shared between two transposons. This mechanism was identified in three out of four described cases. An alternative mechanism was observed in pair II. Here, a unit transposon was duplicated in an ISEcp1-mediated mode.
3.3 Correlation of phenotypic cephalosporin resistance and ESBL gene copies
To investigate the correlation between the phenotypic antimicrobial susceptibility and the presence of AMR resistance genes, we manually curated the VITEK®2 data and used the NCBI AMRFinderPlus to identify matching resistance genes. Furthermore, we used Etests for ceftazidime as a reference substance to determine the cephalosporin resistance more specifically and compare it with the number of ESBL gene copies in the AMR plasmids. In total, AMRFinderPlus annotated 37 different AMR genes within the eight WGS datasets with four to 13 different genes per isolate; including duplications, six to 25 AMR-encoding genes were detected. The lowest numbers of resistance genes were found in Ec1.2 with the duplication of the ESBL gene blaCTX-M-15 and the qnrS1, and cya_S352T and glpT_E448K. The broadest resistance spectrum showed the three K. pneumoniae isolates, with the highest number of AMR genes in Kp4.2. A complete overview of the resistome of the eight analyzed bacterial isolates, categorized into chromosome and plasmid-encoded genes, can be found in the Supplement (Supplementary Table 2).
The phenotypic resistance pattern of the bacterial isolates was analyzed by comparing up to 12 different antibiotic substances (Supplementary Table 3). All isolates were susceptible to carbapenems, and no carbapenemases were detected in the genomic data. In contrast, all isolates were resistant to ampicillin in line with the expression of at least one β-lactamase also found in the genotypic annotation (Supplementary Table 2). To determine the MIC of ceftazidime, we performed Etests®. The MICs within each pair were found to be more than two times higher for the isolates with the ESBL gene duplications compared to the references (Figure 3). The elevated resistance to ceftazidime correlates in all pairs with the higher number of the ESBL target genes.
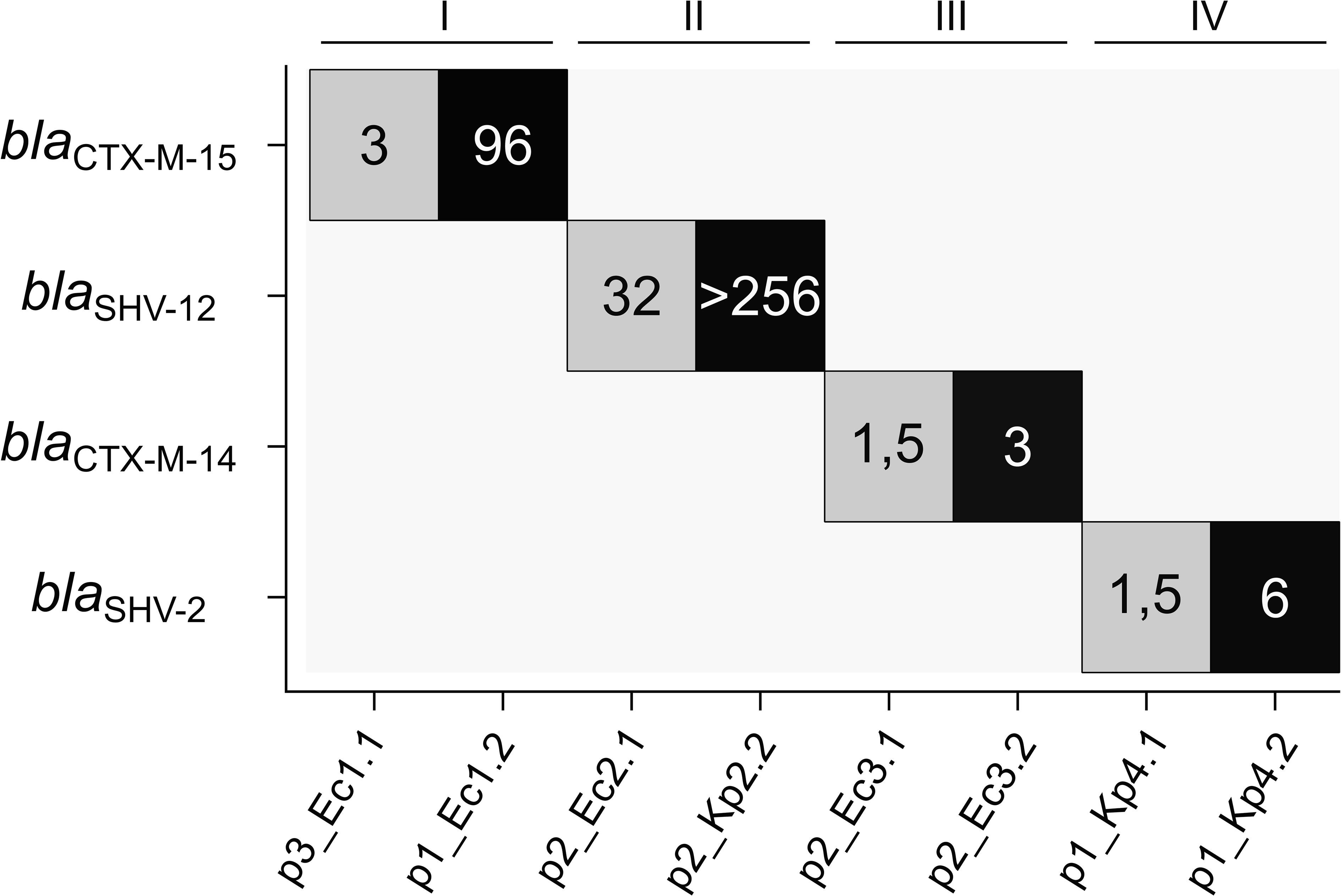
Figure 3 Number of ESBL gene copies in the analyzed plasmids in relation to ceftazidime MIC. ESBL gene copy numbers are color-coded (gray, single copy; black, two copies), and the numbers display the MICs for ceftazidime [mg/L]. ESBL, extended-spectrum β-lactamase; MIC, minimal inhibitory concentration.
4 Discussion
The emergence of ESBL-producing Enterobacteriaceae is reported worldwide and poses a substantial threat to public health (Murray et al., 2022). There is consensus that plasmids are key drivers for the dissemination of AMR genes such as ESBL genes (Carattoli, 2013). However, little is known about the underlying mechanisms that increase MICs of isolates harboring AMR-encoding genes in the clinical context. Therefore, the aim of our study was to investigate long-read WGS data of clinical bacterial samples with a focus on AMR plasmids and gain insights into the spread of AMR genes in the hospital setting.
We found that ESBL gene duplications occur by transposition events within plasmids and increase the phenotypic resistance against cephalosporins due to the higher gene copy number. Four different cases of gene duplications in plasmids harboring ESBL genes were identified after analyzing the surveillance dataset. Interestingly, all described cases share the IS-mediated transposition with ESBL gene duplication as an in vivo evolution event of AMR plasmids and indicate a genotype–phenotype correlation between ESBL gene copy number and increased ceftazidime MIC. Our study highlights the potential of long-read WGS data to better understand AMR and the underlying genetic adaptation mechanisms.
4.1 Role of plasmids in AMR spread
All presented cases show gene duplications within the plasmid. The association between plasmids, AMR genes, and a high number of plasmid-borne AMR genes is well known (Vrancianu et al., 2020). Despite the metabolic burden required to maintain a plasmid, most studies could demonstrate that the benefit of the encoded resistance genes is much higher for bacteria or is compensated by additional mutations (Loftie-Eaton et al., 2017; San Millan and MacLean, 2017). This seems plausible for the described cases, as all samples were collected from patients who frequently receive antibiotic treatment. Hence, all isolates have experienced selection pressure.
Interestingly, our study observed highly similar plasmids in different bacterial isolates that showed no genetic relation based on cgMLST (pairs I and III). This could be an example illustrating how plasmids are shared within a community of bacteria with different genetic backgrounds. This HGT is an important mechanism to exchange genetic information known for decades (Thomas and Nielsen, 2005) and well-studied in the adaptive evolution of bacteria (Arnold et al., 2022; Dimitriu, 2022). For the E. coli Ec2.1 and the K. pneumoniae Kp2.2 (pair II), we assume the HGT of the plasmid harboring the ESBL gene because the species borders have to be crossed. However, it is not possible to determine clearly the chronological sequence of the HGT and the duplication event. On note, even if we discuss predominantly the duplication event, the relation between the compared isolates could also be a loss of a transposable region except in pair IV, where the timeline is clear. Kp4.2 was isolated 3 weeks after Kp4.1 from the same patient.
4.2 IS-mediated transposition and other mobile genetic elements associated with ESBL genes
IS-mediated transposition is often observed with accessory genes including AMR genes. All duplication events investigated used this mode of action but with either an IS26 or an ISEcp1 transposition. IS26 transposition events that could arrange transposons containing AMR genes with only three remaining IS26 elements were described before including the mechanistic details (He et al., 2015; Harmer and Hall, 2019; Harmer and Hall, 2021). Our data indicate that tandem duplication of the same transposon is also a quite common transposition event as observed in three out of four cases, i.e., in pairs I, III, and IV. In accordance with this observation, a few cases of IS26-mediated ESBL gene duplications were described before (Garza-Ramos et al., 2009; Tamamura-Andoh et al., 2021; van Almsick et al., 2022), and also the association of IS26 and blaCTX-M genes was already described in Enterobacteriaceae in a systematic study (Shropshire et al., 2022). Interestingly, neither documented IS26 transposon with ESBL genes nor a high sequence similarity could be found in the databases of TnCentral (Ross et al., 2021) or Transposon Registry (Tansirichaiya et al., 2019) yet. In the fourth analyzed case (pair II), the transposition was ISEcp1 mediated. The detected unit transposon contains a blaCTX-M-14 gene, which also was found in the E. coli isolates of Shropshire et al (Shropshire et al., 2022).
4.3 Gene duplications in evolution and correlation to the phenotypic susceptibility
Gene duplication is a common mechanism in evolutionary biology to develop new traits and adaptations (Innan and Kondrashov, 2010; Vosseberg et al., 2021). A new variant to improve resistance by mutation might be observed in pair II annotated as blaSHV genes in p2_Ec2.1 and p2_Kp2.2 (Figure 2). Both investigated plasmids in this pair have different deletions of nucleotides within one gene copy of the ancestor blaSHV-12 gene. However, it is unclear whether these mutated genes code for a functional and optimized β-lactamase. Schuster et al. found similar results; they identified a novel blaCTX-M gene in this context (Schuster et al., 2022). In that study, the encoded β-lactamase only differed by one amino acid from the ancestor gene that was found in a similar gene array as the new gene. In accordance with this, the evolution of blaSHV genes from blaTEM with the high sequence similarities underlines this as a plausible mechanism for the generation of novel and better-adapted β-lactamases (Liakopoulos et al., 2016).
In addition, we hypothesize that the increased resistance level to ceftazidime correlates with the number of ESBL gene copies encoded on the plasmid. Only very few reported cases of ESBL gene duplications found this correlation between genotypic and phenotypic resistance so far (Tamamura-Andoh et al., 2021; van Almsick et al., 2022). However, in accordance, this observation has been described before for other AMR genes (Coppi et al., 2020; Han et al., 2021). Nevertheless, increased antibiotic resistance has also been linked to point mutations in resistance genes (Finci et al., 2022). To gain more insights into the genetic resistance patterns of Enterobacteriaceae, more genetic analyses based on studies with higher sample sizes are required.
4.4 Study limitations
One limitation of our study is the relatively small number of isolates included in the study. During the 18-month period, four cases of ESBL gene duplications were identified. While this result indicates that ESBL gene duplication is a possible mechanism for developing AMR in the clinical context, higher sample sizes are needed to estimate how frequently this phenomenon happens. Moreover, the comparison of different species and different clonal lineages with variable resistomes has an influence on MIC determination (Girlich et al., 2021). To consider these aspects, we listed additional β-lactamases encoded on other plasmids or integrated into the chromosome for all isolates (Supplementary Table 1). Furthermore, the plasmid copy numbers and other regulatory mechanisms were not covered and are out of the scope of this study (Nelson et al., 2017).
5 Conclusion
We identified IS-mediated AMR gene duplications as a mechanism to overcome antibiotic selection pressure in the hospital setting and to generate new genetic variants of the ancestor resistance gene. Our results also highlight the genetic potential of Enterobacteriaceae for adaptation and evolution of AMR.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA1030289.
Ethics statement
Ethical approval was not required for the study involving humans in accordance with the local legislation and institutional requirements. Written informed consent to participate in this study was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and the institutional requirements.
Author contributions
AS: Investigation, Data curation, Formal analysis, Software, Visualization, Writing – original draft. NS: Software, Methodology, Validation, Writing – review & editing. FS: Methodology, Validation, Writing – review & editing, Formal analysis, Resources. SB: Validation, Writing – review & editing, Conceptualization. AM: Conceptualization, Data curation, Project administration, Resources, Software, Supervision, Validation, Writing – review & editing. VS: Conceptualization, Data curation, Formal analysis, Methodology, Project administration, Supervision, Validation, Writing – review & editing. VA: Conceptualization, Funding acquisition, Investigation, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. AS and VA are supported by a grant (SEED 019/23) from the Interdisciplinary Center of Clinical Research (IZKF), University of Münster.
Acknowledgments
We thank the sequencing team of the Institute of Hygiene at the University Hospital Münster and the laboratory team of the Institute of Microbiology at University Hospital Münster for excellent technical assistance.
Conflict of interest
NS is a part-time employee of Ridom GmbH.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1343858/full#supplementary-material
References
Arnold, B. J., Huang, I.-T., Hanage, W. P. (2022). Horizontal gene transfer and adaptive evolution in bacteria. Nat. Rev. Microbiol. 20, 206–218. doi: 10.1038/s41579-021-00650-4
Behzadi, P., García-Perdomo, H. A., Karpiński, T. M., Issakhanian, L. (2020). Metallo-ß-lactamases: a review. Mol. Biol. Rep. 47, 6281–6294. doi: 10.1007/s11033-020-05651-9
Bezabih, Y. M., Sabiiti, W., Alamneh, E., Bezabih, A., Peterson, G. M., Bezabhe, W. M., et al. (2021). The global prevalence and trend of human intestinal carriage of ESBL-producing Escherichia coli in the community. J. Antimicrob. Chemother. 76, 22–29. doi: 10.1093/jac/dkaa399
Bradford, P. A. (2001). Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14, 933–951. doi: 10.1128/CMR.14.4.933-951.2001
Browne, A. J., Chipeta, M. G., Haines-Woodhouse, G., Kumaran, E. P. A., Hamadani, B. H. K., Zaraa, S., et al. (2021). Global antibiotic consumption and usage in humans 2000-18: a spatial modelling study. Lancet Planet Health 5, e893–e904. doi: 10.1016/S2542-5196(21)00280-1
Bundesgesundheitsblatt (2012). Hygienemaßnahmen bei Infektionen oder Besiedlung mit multiresistenten gramnegativen Stäbchen. Empfehlung der Kommission für Krankenhaushygiene und Infektionsprävention (KRINKO) beim Robert Koch-Institut (RKI). Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz 55, 1311–1354. doi: 10.1007/s00103-012-1549-5
Carattoli, A. (2009). Resistance plasmid families in Enterobacteriaceae. Antimicrobial. Agents Chemother. 53, 2227–2238. doi: 10.1128/AAC.01707-08
Carattoli, A. (2013). Plasmids and the spread of resistance. Int. J. Med. Microbiol. 303, 298–304. doi: 10.1016/j.ijmm.2013.02.001
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using plasmidFinder and plasmid multilocus sequence typing. Antimicrobial. Agents Chemother. 58, 3895–3903. doi: 10.1128/aac.02412-14
Chen, K., Dong, N., Chan, E. W.-C., Chen, S. (2019). Transmission of ciprofloxacin resistance in Salmonella mediated by a novel type of conjugative helper plasmids. Emerg. Microbes Infect. 8, 857–865. doi: 10.1080/22221751.2019.1626197
Coppi, M., Di Pilato, V., Monaco, F., Giani, T., Conaldi, P. G., Rossolini, G. M. (2020). Ceftazidime-Avibactam Resistance Associated with Increased blaKPC-3 Gene Copy Number Mediated by pKpQIL Plasmid Derivatives in Sequence Type 258 Klebsiella pneumoniae. Antimicrobial. Agents Chemother. 64. doi: 10.1128/AAC.01816-19
Darling, A. C. E., Mau, B., Blattner, F. R., Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
Datta, N., Kontomichalou, P. (1965). Penicillinase synthesis controlled by infectious R factors in Enterobacteriaceae. Nature 208, 239–241. doi: 10.1038/208239a0
Davies, J., Davies, D. (2010). Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74, 417–433. doi: 10.1128/MMBR.00016-10
Dimitriu, T. (2022). Evolution of horizontal transmission in antimicrobial resistance plasmids. Microbiol. (Reading) 168. doi: 10.1099/mic.0.001214
Effelsberg, N., Kobusch, I., Linnemann, S., Hofmann, F., Schollenbruch, H., Mellmann, A., et al. (2021). Prevalence and zoonotic transmission of colistin-resistant and carbapenemase-producing Enterobacterales on German pig farms. One Health 13, 100354. doi: 10.1016/j.onehlt.2021.100354
EUCAST (2023) Breakpoint tables for interpretation of MICs and zone diameters. Version 13.1. Available online at: http://www.eucast.org.
Feldgarden, M., Brover, V., Haft, D. H., Prasad, A. B., Slotta, D. J., Tolstoy, I., et al. (2019). Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrobial. Agents Chemother. 63. doi: 10.1128/aac.00483-19
Finci, I., Albertini, A., Merker, M., Andres, S., Bablishvili, N., Barilar, I., et al. (2022). Investigating resistance in clinical Mycobacterium tuberculosis complex isolates with genomic and phenotypic antimicrobial susceptibility testing: a multicentre observational study. Lancet Microbe 3, e672–e682. doi: 10.1016/S2666-5247(22)00116-1
Frost, L. S., Leplae, R., Summers, A. O., Toussaint, A. (2005). Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3, 722–732. doi: 10.1038/nrmicro1235
Garza-Ramos, U., Davila, G., Gonzalez, V., Alpuche-Aranda, C., López-Collada, V. R., Alcantar-Curiel, D., et al. (2009). The blaSHV-5 gene is encoded in a compound transposon duplicated in tandem in Enterobacter cloacae. Clin. Microbiol. Infect. 15, 878–880. doi: 10.1111/j.1469-0691.2009.02790.x
Girlich, D., Bonnin, R. A., Proust, A., Naas, T., Dortet, L. (2021). Undetectable production of the VIM-1 carbapenemase in an atlantibacter hermannii clinical isolate. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.741972
Han, X., Shi, Q., Mao, Y., Quan, J., Zhang, P., Lan, P., et al. (2021). Emergence of ceftazidime/avibactam and tigecycline resistance in carbapenem-resistant klebsiella pneumoniae due to in-host microevolution. Front. Cell Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.757470
Harmer, C. J., Hall, R. M. (2019). An analysis of the IS6/IS26 family of insertion sequences: IS it a single family? Microbial. Genomics 5. doi: 10.1099/mgen.0.000291
Harmer, C. J., Hall, R. M. (2021). IS26 cannot move alone. J. Antimicrob. Chemother. 76, 1428–1432. doi: 10.1093/jac/dkab055
He, S., Hickman, A. B., Varani, A. M., Siguier, P., Chandler, M., Dekker, J. P., et al. (2015). Insertion sequence IS26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. mBio 6. doi: 10.1128/mBio.00762-15
Huemer, M., Mairpady Shambat, S., Brugger, S. D., Zinkernagel, A. S. (2020). Antibiotic resistance and persistence-Implications for human health and treatment perspectives. EMBO Rep. 21, e51034. doi: 10.15252/embr.202051034
Innan, H., Kondrashov, F. (2010). The evolution of gene duplications: classifying and distinguishing between models. Nat. Rev. Genet. 11, 97–108. doi: 10.1038/nrg2689
Johansson, M. H. K., Bortolaia, V., Tansirichaiya, S., Aarestrup, F. M., Roberts, A. P., Petersen, T. N. (2021). Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 76, 101–109. doi: 10.1093/jac/dkaa390
Jünemann, S., Sedlazeck, F. J., Prior, K., Albersmeier, A., John, U., Kalinowski, J., et al. (2013). Updating benchtop sequencing performance comparison. Nat. Biotechnol. 31, 294–296. doi: 10.1038/nbt.2522
Juraschek, K., Borowiak, M., Tausch, S. H., Malorny, B., Käsbohrer, A., Otani, S., et al. (2021). Outcome of different sequencing and assembly approaches on the detection of plasmids and localization of antimicrobial resistance genes in commensal escherichia coli. Microorganisms 9, 598. doi: 10.3390/microorganisms9030598
Kim, D.-W., Cha, C.-J. (2021). Antibiotic resistome from the One-Health perspective: understanding and controlling antimicrobial resistance transmission. Exp. Mol. Med. 53, 301–309. doi: 10.1038/s12276-021-00569-z
Kizny Gordon, A. E., Mathers, A. J., Cheong, E. Y. L., Gottlieb, T., Kotay, S., Walker, A. S., et al. (2017). The hospital water environment as a reservoir for carbapenem-resistant organisms causing hospital-acquired infections—A systematic review of the literature. Clin. Infect. Dis. 64, 1435–1444. doi: 10.1093/cid/cix132
Liakopoulos, A., Mevius, D., Ceccarelli, D. (2016). A review of SHV extended-spectrum β-lactamases: neglected yet ubiquitous. Front. Microbiol. 7, 1374. doi: 10.3389/fmicb.2016.01374
Loftie-Eaton, W., Bashford, K., Quinn, H., Dong, K., Millstein, J., Hunter, S., et al. (2017). Compensatory mutations improve general permissiveness to antibiotic resistance plasmids. Nat. Ecol. Evol. 1, 1354–1363. doi: 10.1038/s41559-017-0243-2
Meier-Kolthoff, J. P., Göker, M. (2019). TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 10. doi: 10.1038/s41467-019-10210-3
Murray, C. J. L., Ikuta, K. S., Sharara, F., Swetschinski, L., Robles Aguilar, G., Gray, A., et al. (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655. doi: 10.1016/S0140-6736(21)02724-0
Nelson, K., Hemarajata, P., Sun, D., Rubio-Aparicio, D., Tsivkovski, R., Yang, S., et al. (2017). Resistance to ceftazidime-avibactam is due to transposition of KPC in a porin-deficient strain of klebsiella pneumoniae with increased efflux activity. Antimicrobial. Agents Chemother. 61. doi: 10.1128/AAC.00989-17
Ondov, B. D., Treangen, T. J., Melsted, P., Mallonee, A. B., Bergman, N. H., Koren, S., et al. (2016). Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 17. doi: 10.1186/s13059-016-0997-x
O’Neill, J. (2014). Antimicrobial resistance: tackling a crisis for the health and wealth of nations 2014. Rev. Antimicrobial. Resist.
Partridge, S. R., Kwong, S. M., Firth, N., Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31. doi: 10.1128/CMR.00088-17
Robertson, J., Nash, J. H. E. (2018). MOB-suite: software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microbial. Genomics 4. doi: 10.1099/mgen.0.000206
Ross, K., Varani, A. M., Snesrud, E., Huang, H., Alvarenga, D. O., Zhang, J., et al. (2021). TnCentral: a prokaryotic transposable element database and web portal for transposon analysis. mBio 12, e0206021. doi: 10.1128/mBio.02060-21
San Millan, A., MacLean, R. C. (2017). Fitness costs of plasmids: a limit to plasmid transmission. Microbiol. Spectr. 5. doi: 10.1128/microbiolspec.mtbp-0016-2017
Schuster, C. F., Weber, R. E., Weig, M., Werner, G., Pfeifer, Y. (2022). Ultra-deep long-read sequencing detects IS-mediated gene duplications as a potential trigger to generate arrays of resistance genes and a mechanism to induce novel gene variants such as blaCTX-M-243. J. Antimicrob. Chemother. 77, 381–390. doi: 10.1093/jac/dkab407
Shropshire, W. C., Konovalova, A., McDaneld, P., Gohel, M., Strope, B., Sahasrabhojane, P., et al. (2022). Systematic analysis of mobile genetic elements mediating β-lactamase gene amplification in noncarbapenemase-producing carbapenem-resistant enterobacterales bloodstream infections. mSystems 7, e0047622. doi: 10.1128/msystems.00476-22
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Tacconelli, E., Carrara, E., Savoldi, A., Harbarth, S., Mendelson, M., Monnet, D. L., et al. (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327. doi: 10.1016/S1473-3099(17)30753-3
Tamamura-Andoh, Y., Niwa, H., Kinoshita, Y., Uchida-Fujii, E., Arai, N., Watanabe-Yanai, A., et al. (2021). Duplication of blaCTX-M-1 and a class 1 integron on the chromosome enhances antimicrobial resistance in Escherichia coli isolated from racehorses in Japan. J. Global Antimicrobial. Resist. 27, 225–227. doi: 10.1016/j.jgar.2021.10.004
Tanizawa, Y., Fujisawa, T., Nakamura, Y. (2018). DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 34, 1037–1039. doi: 10.1093/bioinformatics/btx713
Tansirichaiya, S., Rahman, M. A., Roberts, A. P. (2019). The transposon registry. Mobile DNA 10, 40. doi: 10.1186/s13100-019-0182-3
Thomas, C. M., Nielsen, K. M. (2005). Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3, 711–721. doi: 10.1038/nrmicro1234
van Almsick, V., Schuler, F., Mellmann, A., Schwierzeck, V. (2022). The Use of Long-Read Sequencing Technologies in Infection Control: Horizontal Transfer of a blaCTX-M-27 Containing lncFII Plasmid in a Patient Screening Sample. Microorganisms 10, 491. doi: 10.3390/microorganisms10030491
Varani, A., He, S., Siguier, P., Ross, K., Chandler, M. (2021). The IS6 family, a clinically important group of insertion sequences including IS26. Mobile DNA 12, 11. doi: 10.1186/s13100-021-00239-x
Ventola, C. L. (2015). The antibiotic resistance crisis: part 1: causes and threats. P T 40, 277–283.
Vosseberg, J., van Hooff, J. J. E., Marcet-Houben, M., van Vlimmeren, A., van Wijk, L. M., Gabaldón, T., et al. (2021). Timing the origin of eukaryotic cellular complexity with ancient duplications. Nat. Ecol. Evol. 5, 92–100. doi: 10.1038/s41559-020-01320-z
Keywords: antimicrobial resistance, ESBL, plasmid, transposons, gene duplication, antibiotic susceptibility
Citation: Sobkowiak A, Scherff N, Schuler F, Bletz S, Mellmann A, Schwierzeck V and van Almsick V (2024) Plasmid-encoded gene duplications of extended-spectrum β-lactamases in clinical bacterial isolates. Front. Cell. Infect. Microbiol. 14:1343858. doi: 10.3389/fcimb.2024.1343858
Received: 24 November 2023; Accepted: 02 February 2024;
Published: 26 February 2024.
Edited by:
Marion Skalweit, Case Western Reserve University, United StatesReviewed by:
Anand Bahadur Karki, Sam Houston State University, United StatesIbrahim Bitar, Charles University, Czechia
Copyright © 2024 Sobkowiak, Scherff, Schuler, Bletz, Mellmann, Schwierzeck and van Almsick. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vincent van Almsick, VmluY2VudEZyZWRlcmljLnZhbkFsbXNpY2tAdWttdWVuc3Rlci5kZQ==
 Annika Sobkowiak
Annika Sobkowiak Natalie Scherff1
Natalie Scherff1 Alexander Mellmann
Alexander Mellmann Vera Schwierzeck
Vera Schwierzeck Vincent van Almsick
Vincent van Almsick