- Department of Pharmacy, Shanghai Tenth People’s Hospital, School of Medicine, Tongji University, Shanghai, China
In the last twenty years, there has been a significant increase in invasive fungal infections, which has corresponded with the expanding population of individuals with compromised immune systems. As a result, the mortality rate linked to these infections remains unacceptably high. The currently available antifungal drugs, such as azoles, polyenes, and echinocandins, face limitations in terms of their diversity, the escalating resistance of fungi and the occurrence of significant adverse effects. Consequently, there is an urgent need to develop new antifungal medications. Vaccines and antibodies present a promising avenue for addressing fungal infections due to their targeted antifungal properties and ability to modulate the immune response. This review investigates the structure and function of cell wall proteins, secreted proteins, and functional proteins within C. albicans. Furthermore, it seeks to analyze the current advancements and challenges in macromolecular drugs to identify new targets for the effective management of candidiasis.
1 Introduction
Over the last few decades, candidiasis has gradually increased morbidity and mortality rates, primarily caused by Candida albicans (Lu et al., 2023b). Typically, C. albicans exist as non-pathogenic symbiotic fungi in healthy individuals, predominantly colonizing the surface of human mucosa, including the skin, oral and pharyngeal regions, gastrointestinal tract, and urogenital tract (Williams et al., 2013; Lu et al., 2023a). When the human immune system is compromised, C. albicans can rapidly transition from non-pathogenic to pathogenic fungi, resulting in superficial or deep candidiasis, including thrush and candidemia. Invasive candidiasis has a global annual impact on over 1,565,000 individuals, resulting in approximately 1,000,000 fatalities (Denning, 2024). Hence, candidiasis emerges as a worldwide concern, necessitating significant consideration due to its impact on human well-being.
Currently, the clinical practice utilizes only three categories of small-molecule antifungal medications: polyenes, azoles, and echinocandins (Robbins et al., 2016). However, these antifungal agents are associated with certain limitations, which impede their clinical utility for treating candidiasis (Cowen et al., 2014). Polyenes can induce significant adverse effects as a result of the structural resemblance between the intended target, ergosterol, and cholesterol, a sterol found in mammalian cell membranes (Robbins et al., 2017). Despite their potent fungicidal properties and commendable safety profile, echinocandins face limitations in terms of their narrow antifungal range, requirement for intravenous administration, and high cost, which pose challenges to their clinical application (Perlin, 2011; Pappas et al., 2016). Azoles, on the other hand, exhibit low toxicity levels and a broad spectrum of antifungal activity. However, their fungistatic effects in certain species have led to the emergence of azole-resistant isolates (Robbins et al., 2017; Nishimoto et al., 2020). Significantly, the effectiveness of these drugs in combating fungal infections is notably reduced in patients with compromised immune function when compared to individuals with an intact immune system (Logan et al., 2020). Consequently, despite advancements in antifungal therapy, patients with invasive candidiasis experience significant morbidity and mortality. Hence, the urgency for exploring and implementing alternative therapeutic strategies to manage invasive candidiasis caused by Candida species effectively cannot be overstated.
The human immune system responds to candidiasis through two distinct mechanisms: innate and adaptive immunities (Lu et al., 2023b). The innate immune system primarily consists of physical barriers, such as mucosal epithelial surfaces in the skin, mouth, upper respiratory tract, gastrointestinal tract, and urogenital tract (Nami et al., 2019). Upon fungal infection, pathogen-associated molecular patterns (PAMP) that are currently present in C. albicans are recognized by pattern recognition receptors (PRRs), such as Integrin, Lectin, and Toll-like receptors (TLRs), which trigger the production of inflammatory factors and the recruitment and activation of phagocytes to fight the infection (Vonk et al., 2006). Moreover, the fungus that has infiltrated is taken up and subjected to phagocytosis by resident macrophages, dendritic cells (DCs), and polymorphonuclear neutrophils (PMN). Additionally, macrophages and PMN release antimicrobial peptides (AMP), inflammatory cytokines, and chemokines (Netea et al., 2004; Martinez et al., 2009). Macrophages can phagocytose and destroy Candida yeast cells, reducing the fungal load as soon as an infection occurs (Jones-Carson et al., 1997). Monocytes contacted Candida early in the infection and were more effective than DCs or macrophages in destroying C. albicans (Netea et al., 2004). Monocyte-deficient mice are more susceptible to C. albicans infections, and monocytes exposed to C. albicans release tumor necrosis factor-α (TNF-α), which protects the host from invasive candidiasis (Netea et al., 1999; Ngo et al., 2014). Neutrophils can also release neutrophil extracellular traps (NETs) that capture C. albicans (Matejuk et al., 2010). Since neutrophil activation is necessary to eradicate Candida species and neutropenia is a significant risk factor for invasive fungal infections, neutrophils are vital in the host’s defense against candidiasis (Newman and Holly, 2001). Neutrophils have been shown to have a role in C. albicans death, and they are the only immune cells capable of successfully preventing the transition of C. albicans blastopores to hyphae (Lionakis and Netea, 2013; Lionakis, 2014). Natural killer (NK) cells can directly kill C. albicans by secreting cytotoxic molecules (Voigt et al., 2014). In addition, the complement system is a component of innate immunity and is critical in the host’s fight against C. albicans (Harpf et al., 2020). Along with its capacity to promote phagocytosis by opsonization, the complement system’s activation produces anaphylatoxins (C3a, C4a, and C5a) responsible for several pleiotropic effects (Brown et al., 2012). As DCs can phagocytose and present antigens, they play a significant part in the host’s defense against invasive candidiasis. This links innate and cell-mediated antifungal immunity (Newman and Holly, 2001; Romagnoli et al., 2004). Therefore, in addition to playing a crucial role in initiating early defense against fungal infections, the innate immune system also triggers various responses promoted by the adaptive immune system through DCs (Medici and Del Poeta, 2015). Cell-mediated immunity and humoral immunity are two types of adaptive immunity essential for removing invasive pathogenic fungi and creating powerful immune defenses. The host’s main defense against Candida species is cell-mediated immunity, and CD4+ T helper cells and CD8+ cytotoxic T-cells are the main players in preventing invasive candidiasis (Medici and Del Poeta, 2015). It has long been believed that the CD4+T cell immune response protects against Candida infection (Conti and Gaffen, 2010). T cells with CD4+ function participate in the antifungal immune response through Th1, Th2, and Th17 immunity (Richardson and Moyes, 2015). On the other hand, the direct cytotoxic activity and cytokine release of CD8+ T-cells are known to mediate resistance to systemic fungal infections (Ashman et al., 1999). Cytokine secretion (mainly interferon-gamma (IFN-γ) and TNF-α) has been demonstrated as one of the main effector mechanisms through which CD8+ T-cells can restrict fungal infection (Gozalbo and Gil, 2009). Antibodies primarily facilitate humoral immunity, which initiates complement activation, opsonization, and antibody-dependent cellular cytotoxicity (Casadevall et al., 2002). As a result, these antibodies neutralize toxins, hinder the adherence of pathogenic fungi to host cells, impede biofilm formation, inhibit germ tube formation, and restrict fungal load (Pathakumari et al., 2020). When the equilibrium between the host immune response and fungus is disrupted, as seen in cancer chemotherapy and immunotherapy, the heightened utilization of organ transplantation and immunosuppressive agents, along with the extensive application of broad-spectrum antibiotics, will lead to an increased pathogenic behavior of C. albicans in immunocompromised patients, resulting in the dissemination of C. albicans organs (Cassone and Cauda, 2012; Del Poeta and Casadevall, 2012).
Immunotherapeutic approaches exhibit potential as a novel strategy for treating candidiasis, owing to the significant involvement of the human immune system in managing this condition. Immunotherapies encompass therapeutic approaches aimed at targeting and impacting the immune system of the body, thereby enhancing the host’s ability to combat infections (Qadri et al., 2023). These methodologies encompass various strategies, such as augmenting the population of phagocytes, activating innate defense pathways in phagocytes and DCs, and stimulating antigen-specific immunity through means like vaccines and monoclonal antibodies. It is crucial to customize immunotherapy to suit the specific immunocompromised conditions of individuals (Segal et al., 2006). In contrast to prior literature that primarily focused on elucidating the mechanisms underlying fungal infection and the immune system’s reaction to fungi (Segal et al., 2006; Armstrong-James and Harrison, 2012; Nami et al., 2019), the present review provides a comprehensive analysis of the diverse proteins present on the surface of C. albicans, which are pivotal in the development of candidiasis (Table 1) (Figure 1). Moreover, investigations into the efficacy of antibodies and vaccines targeting these proteins have revealed promising therapeutic outcomes. This review critically examines the structural characteristics and functional roles of these proteins in candidiasis, as well as explores the associated antibodies and vaccines. This review proposes that employing immunotherapeutic approaches holds promise as an innovative treatment of candidiasis strategy.
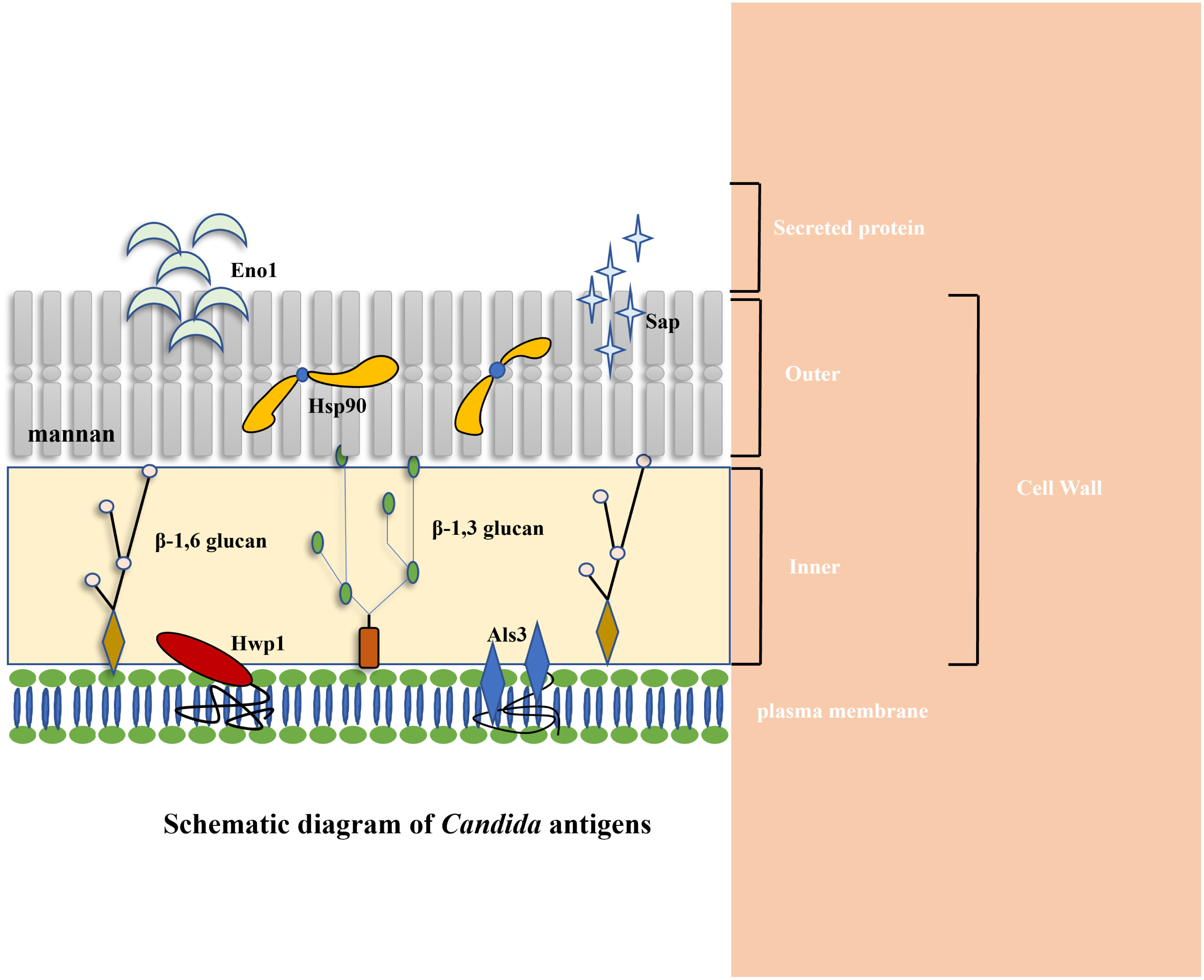
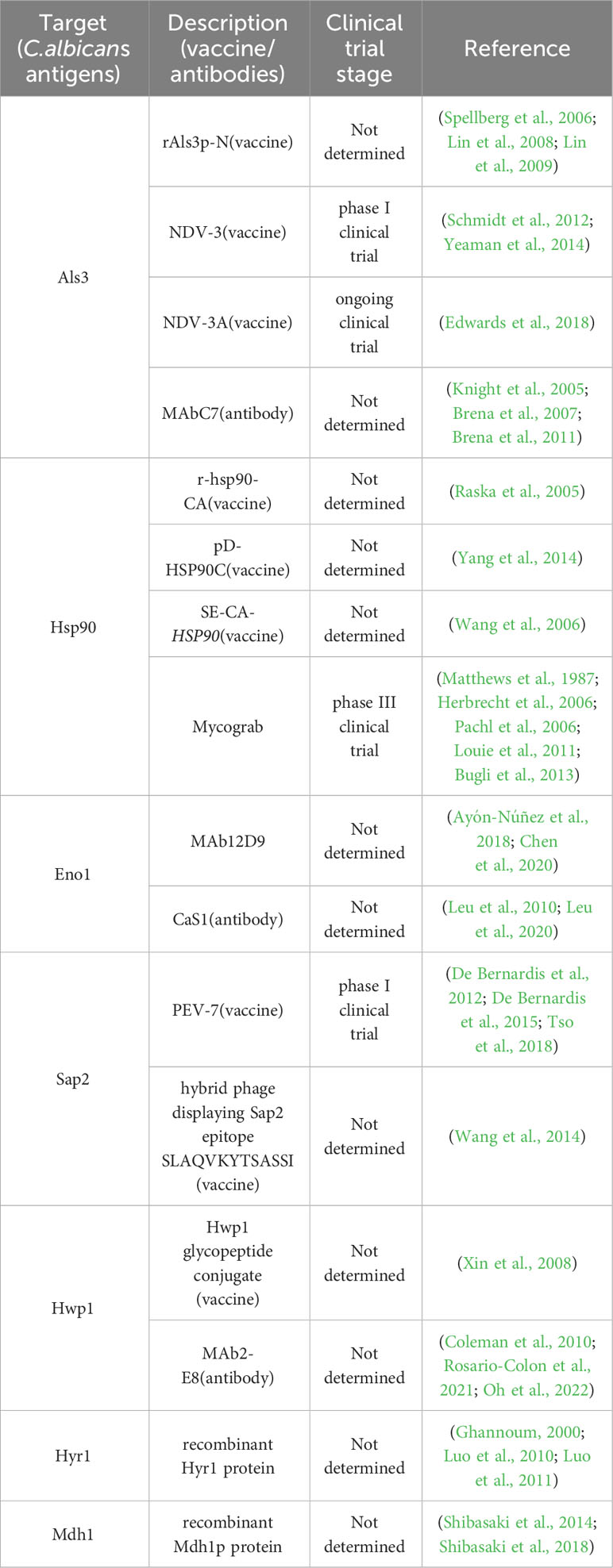
Figure 1 Schematic diagram of C. albicans antigens. Cytoplasmic location of Hsp90 and Eno1 is indicated along with various cell surface antigens (Als3 and Hwp1). Cell surface is to indicate secreted (Eno1, Sap2), and cell wall associated antigens (Hwp1, Sap2, Als3, Eno1, Hsp90) (Matthews and Burnie, 1988; Matthews and Burnie, 1989; Staab et al., 1996; Felk et al., 2002; Naglik et al., 2003; Burnie et al., 2006; Hoyer et al., 2008; Calugi et al., 2013; Silva et al., 2014; Chatterjee and Tatu, 2017; Shukla et al., 2021).
2 Agglutinin-like sequence (Als) 3
2.1 The structure of Als3
The role of Als proteins in the adhesion of C. albicans to host cells, a necessary step for host infection, is important. Among the Als family, the fungal cell wall protein Als3 is one of the eight members, functioning as a glycosylphosphatidylinositol (GPI) anchored glycoprotein (Hoyer and Cota, 2016). The N-terminal domain of Als3 initiates with a secretory signal sequence, followed by an amyloid-forming region composed of six amino acids. Following this, it is possible for a conservative sequence abundant in threonine, comprising 103 amino acids, to coexist as a tandem copy with a variable number of repeating sequences, consisting of 36 amino acids (Lin et al., 2014). The quantity of repeat sequences differs among Als proteins, with Als3 harboring 12 such repeats. The C-terminal domain of Als3 concludes with a GPI anchoring sequence, facilitating the attachment of Als3 to the β-1,6-glucan cell wall of C. albicans (Figure 2A) (Hoyer et al., 2008).
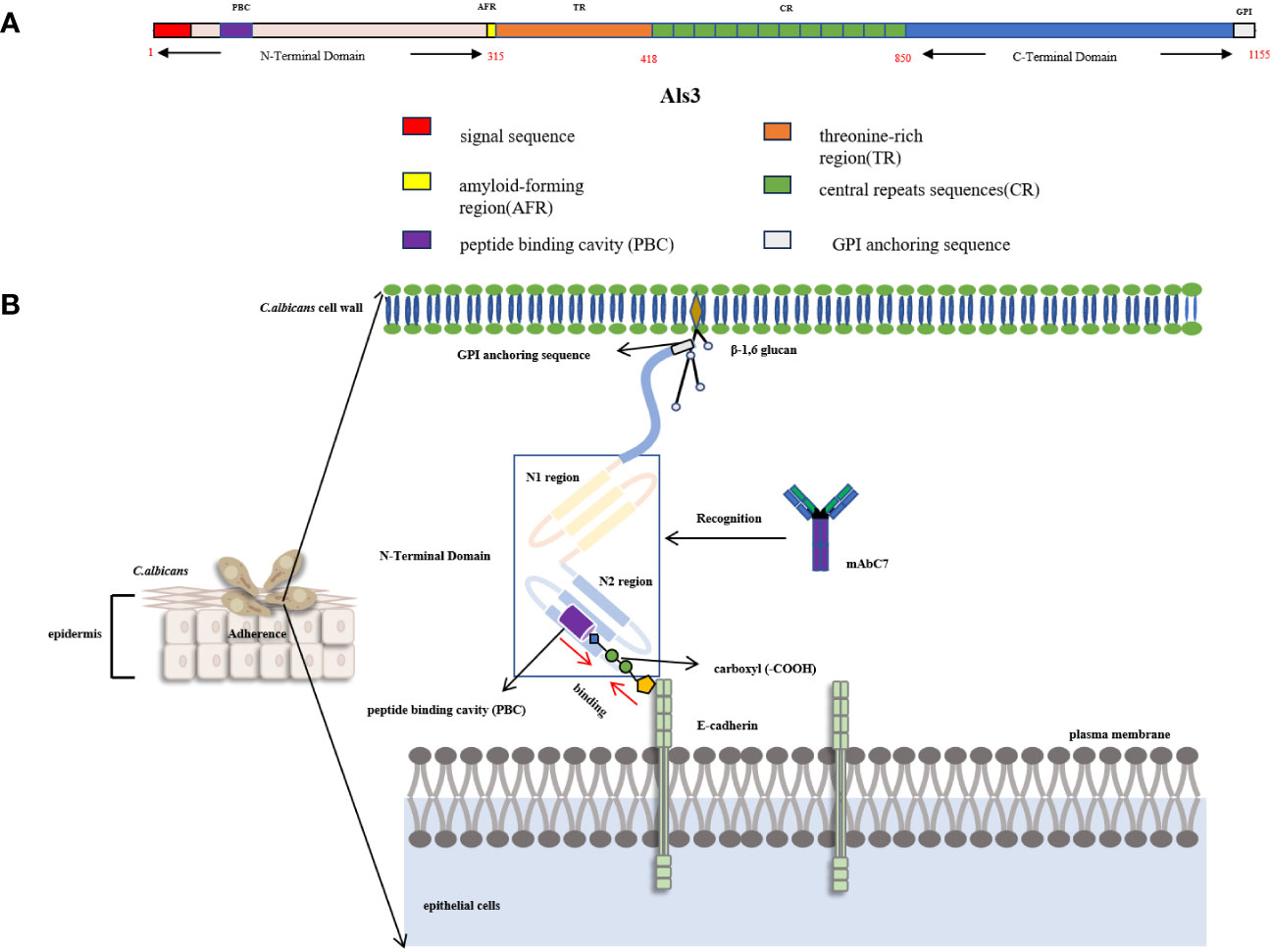
Figure 2 (A) Schematic diagram of Als3 structural domain (Hoyer et al., 2008). (B) Schematic diagram of the interaction between Als3 and E-cadherin. Als3 is comprised of two consecutive IgG-type immunoglobulin domains (N1, N2) with a peptide binding cavity (PBC) situated in the N2 region. PBC can bind to the carboxyl of E-cadherin in epithelial cells by forming hydrogen bonds. MAbC7 can recognize the N-terminal domain of Als3 and bind to it, thereby blocking the binding between C.albicans and host epithelial cells (Zhao et al., 2004; Nobbs et al., 2010; Lin et al., 2014).
Als3 is a distinctive mycelial-specific protein of C. albicans that demonstrates a wide range of substrates for adhesion, including human fibronectin, laminin, collagen, gp96, EGFR (epidermal growth factor receptor), HER2 (human epidermal growth factor receptor-2), N-cadherin, E-cadherin, fibrinogen, casein, and bovine serum albumin (Sheppard et al., 2004). The spatial structure of the N-terminal of Als3(NT-Als3) was examined through X-ray crystal diffraction analysis. Als3 is comprised of two consecutive IgG-type immunoglobulin domains (N1, N2) with a peptide binding cavity (PBC) in the N2 region. In the context of the PBC, the conserved lysine residue (K59) can interact with the ligand’s C-terminal carboxyl group (Figure 2B) (Lin et al., 2014). Upon ingress into the PBC, the ligand establishes a hydrogen bond with the neighboring parallel chain (βG2b,294TLTWWGY301). Concurrently, the water molecular network, characterized by multiple water molecules forming hydrogen bonds between the ligand proteins and PBC, facilitates the interaction between the ligand and the Als3. The variability in the arrangement of the water molecular network enables Als3 to facilitate the impact of C. albicans on endothelial and oral epithelial cells, resulting in the adherence of diverse cells and proteins, including fibrin, laminin, and salivary mucosa.
2.2 Functions of Als3 in candidiasis
Als3 serves as a structural support in promoting the attachment of C. albicans to the host cell surface. More specifically, the C-terminal region of Als3 is anchored to the cell wall by β-1,6-glucan, while its N-terminal domain extends outward to bind with the C-terminal carboxyl group of the host cell surface protein through the PBC domain. The introduction of specific point amino acid mutations (K59M, A116V, Y301F) in Als3 results in the formation of a hydrophobic patch at the terminus of the PBC, leading to a decrease in the hydrophilic interaction between the PBC and the C-terminal of the substrate protein (fibrinogen-γ). Another mutation approach (S170Y) involves the creation of a steric barrier at the PBC terminus, which serves as a “gatekeeper” to impede the entry of peptides at the PBC entrance, thereby preventing the C-terminal of the fibrinogen-γ from accessing the PBC (Lin et al., 2014). The absence of the ALS3 gene has decreased the adherence of C. albicans to endothelial and epithelial cells (Zhao et al., 2004; Nobbs et al., 2010). Further studies have demonstrated that the N-terminal domain of recombinant Als3 plays a critical role in facilitating the adhesion of Staphylococcus aureus and C. albicans in the development of a composite biofilm, highlighting its importance in the adhesion mechanism (Peters et al., 2012).
The interaction between C. albicans and epithelial cells can result in endocytosis within the epithelial cells. This process is facilitated by the PBC located in the N-terminal domain of Als3, which binds to the C-terminal carboxyl group of N-cadherin and E-cadherin in the epithelial cells. Als3 further aids in aggregating reticulin and its associated proteins (kinetin, cortisol) from the host at the binding site. This aggregation subsequently leads to the remodeling of actin cytoskeletons and plasma membranes within endothelial cells. Significantly, the deletion of the genes CDH1 and CDH2, responsible for encoding the reticulon protein, resulted in a notable decrease in the endocytosis of endothelial cells in response to C. albicans (Moreno-Ruiz et al., 2009). The study demonstrated a significant disparity in mycelial growth between the als3Δ/Δ mutant and the wild-type strain, with the former displaying a significantly diminished robustness. Additionally, there was a substantial reduction of nearly 90% in the level of endocytosis by endothelial and epithelial cells (Phan et al., 2005). These findings collectively suggest the indispensability of Als3 in facilitating C. albicans’ ability to induce endocytosis.
The role of Als3 in facilitating iron absorption by C. albicans, a crucial process for the organism’s survival, has been demonstrated (Weinberg, 1999). In general, the acquisition of iron by biological systems is carefully regulated to prevent the harmful effects of iron overload and the growth impairments caused by iron deficiency (Fourie et al., 2018). Furthermore, hosts, including humans, must restrict iron availability to pathogens. Nevertheless, C. albicans can adapt to varying levels of iron, such as the iron-rich conditions in the gastrointestinal tract and the iron deficiency encountered during systemic infection (Fourie et al., 2018). The protein Als3 plays a role in acquiring iron from the host by utilizing a reduction system that extracts iron from ferritin (Almeida et al., 2009). Als3 interacts with host ferritin, leading to the release and uptake of iron through the iron reduction pathway of C. albicans (Almeida et al., 2008; Liu and Filler, 2011). However, the specific mechanism by which this occurs is not yet fully understood. When the ALS3 gene of C. albicans is expressed in Saccharomyces cerevisiae, it enables the binding of ferritin, unlike the closely related members of the Als family, Als1 and Als5 (Almeida et al., 2008). Conversely, the absence of Als3 impairs the growth of C. albicans on ferritin plates. Moreover, the absence of transcription factors Tec1 and Bcr1, which play a role in regulating the expression of the ALS3 gene, reduces iron uptake capability and decreases the mycelial growth and invasive potential of C. albicans. Furthermore, it was observed that C. albicans hyphae, but not yeast cells, exhibit binding affinity toward ferritin, a critical factor in iron acquisition. This indicates that C. albicans can effectively utilize iron from ferritin through morphology-dependent binding facilitated by Als3, thereby highlighting the multifaceted virulence attributes of this singular protein (Lionakis, 2014). Due to its significant role in C. albicans adhesion, endocytosis, and iron uptake, Als3 presents itself as a promising candidate for immunotherapeutic interventions aimed at combating candidiasis.
2.3 Immunotherapy targeting Als3
The recombinant protein vaccine rAls3p-N, derived from S. cerevisiae, encompasses amino acids 17 to 432 in the N-terminal domain of Als3 (Lin et al., 2009). This vaccine acts as an antigen recognized by antigen-presenting cells when combined with immune adjuvants, resulting in the development of CD4+ cells by splenic and lymph node lymphocytes. Consequently, the CD4+ cells generate Th1/17 cells, which produce IL-17 and IFN-γ, exhibiting strong antifungal effects against C. albicans. The administration of this vaccine stimulates the generation of diverse helper T cells, such as Th1, Th17, and Th1/17, thereby facilitating the recruitment of neutrophils and the synthesis of pro-inflammatory factors at the site of infection (Lin et al., 2009). In mice, rAls3p-N can induce cellular immune responses, protecting against invasive C. albicans infections (Lin et al., 2008). In the pre-clinical investigation, mice were subjected to subcutaneous injections of a mixture comprising 20 μg of rAls3p-N and Complete Freund’s Adjuvant on day 0. On day 21, the mice were administered another antigen dose with Incomplete Freund’s Adjuvant. After a two-week interval, the mice were intravenously infected with C. albicans SC5314 blastospores via the tail vein, and their condition was monitored for 30 days. The findings revealed that the survival rate of mice receiving rAls3p-N injections at a dosage of 20 μg/mice increased to 50%, whereas the control group exhibited a mortality rate of 100%. In the oropharyngeal and vaginal infections model, mice that received rAls3p-N injection exhibited a notable reduction in fungal load compared to those that received adjuvant injection alone (Spellberg et al., 2006).
The vaccine NDV-3, which consists of a tag sequence of rAls3p-N and a 6-His linkage sequence, has exhibited the capacity to induce a robust and safe immune response in individuals who are in good health. This response is characterized by notably elevated levels of anti-Als3 IgG and IgA1 antibodies (Schmidt et al., 2012). Furthermore, NDV-3 has demonstrated its ability to safeguard mice against infections caused by C. albicans and methicillin-resistant S. aureus by eliciting a potent B and T-cell response (Schmidt et al., 2012; Yeaman et al., 2014). The results of a phase I clinical trial (NCT01273922) indicated that adult participants who were administered a 30 or 300 μg dosage exhibited satisfactory tolerance, generated immune factor IFN-γ, and displayed favorable levels of IL-17A (Schmidt et al., 2012).
Furthermore, an ongoing clinical trial (NCT01926028) is presently examining the effectiveness of an alum formulation of Als3 (NDV-3A, Als3 devoid of extraneous sequences) in the treatment of individuals with a previous record of recurrent vulvovaginal candidiasis. The results of this trial demonstrate that NDV-3A is safe and capable of inducing an immune response. The trial findings have demonstrated that NDV-3A effectively treats vulvovaginal candidiasis, specifically among women under 40. The vaccine has effectively decreased the rates of disease recurrence while simultaneously upholding favorable safety and immunogenicity characteristics (Edwards et al., 2018).
The monoclonal antibody MAbC7 has been found to selectively bind to the N-terminal domain of Als3, thereby hindering the germination and mycelial formation of C. albicans. Furthermore, MAbC7 has demonstrated inhibitory properties against the attachment of C. albicans to Hep2 cells and buccal epithelial cells, resulting in inhibition rates ranging from 31.1% to 55.3% (Brena et al., 2007). Significantly, MAbC7 effectively targets the iron acquisition pathway of C. albicans, impeding the fungal cells’ ability to reduce exogenous iron. Furthermore, this antibody has been observed to enhance the expression of genes linked to iron homeostasis in C. albicans, specifically FET34, FTR1, FTR2, and SIT1 genes (Brena et al., 2011). Fet34 is an essential ferro-oxidase responsible for Fe (II) uptake. The FTR1 and FTR2 genes encode membrane iron transporters, while the SIT1 gene is involved in transporting siderophores for iron uptake. The upregulation of these genes has been shown to decrease the levels of available iron in C. albicans, leading to intracellular iron depletion in the fungus (Knight et al., 2005). Nevertheless, the exact mechanism by which MAbC7 binds to the N-terminal of Als3 remains uncertain.
3 Heat shock protein 90 (Hsp90)
3.1 The structure of Hsp90
Hsp90 is a highly conserved and indispensable molecular chaperone found in eukaryotic cells. Its structural configuration involves a homodimeric form comprising three principal constituents: an N-terminal domain that encompasses a nucleotide-binding pocket and exhibits ATP-binding activity, an ATP-binding region comprising multiple amino acid residues, and characterized by a “lid” structure that assumes a closed conformation in the presence of ATP but an open state in the ADP state, and a Middle domain (M-domain) responsible for facilitating interactions with client proteins and co-chaperones (Ali et al., 2006) (Figure 3A). The comparison of functional domains between C. albicans and human Hsp90 demonstrates that human Hsp90α share a sequence identity of 71.9%, 68.0%, and 49.2% with C. albicans Hsp90 in the N-terminal domain, M-domain, and C-terminal domain, respectively (Li et al., 2021). The M-domain of Hsp90 contains the catalytic arginine essential for ATPase activity and functions as a binding site for co-molecular chaperones, rendering it a crucial site for protein interactions (Meyer et al., 2004). Conversely, the C-terminal domain contributes to the formation of dimers in heat shock proteins (Zierer et al., 2016). Furthermore, the MEEVD motif, comprising the last five amino acids of Hsp90, serves as a binding site for interactions with ubiquitous chaperones with tetrapeptide repeat (TPR) ligands (Biebl and Buchner, 2019). Hsp90 displays restricted ATPase activity and low efficiency in ATP turnover. It is worth noting that the rate of ATP turnover for Hsp90 in yeast cells is once per minute, while in humans, it occurs 10 times per minute (Richter et al., 2001; Richter et al., 2008). Structural analysis findings indicate that Hsp90 exists in a state of dynamic equilibrium, where the binding of nucleotides can initiate a directional conformational cycle (Graf et al., 2009). Hsp90 exhibits rapid ATP binding followed by a gradual transition to the first intermediate state (I1), characterized by open N-domains and a closed ATP lid. The second intermediate state (I2) is formed through N-terminal dimerization, resulting in the repositioning and interaction of the M-domain with the N-domain. Subsequently, Hsp90 achieves a fully closed state, facilitating ATP hydrolysis. Upon hydrolysis, the N-domains dissociate, releasing ADP and inorganic phosphate (Pi), leading to the return of Hsp90 to its open conformation (Figure 3B) (Shiau et al., 2006). The protein’s nomenclature is based on its capacity to react to abrupt temperature rises in different cell types, encompassing fungal, animal, and plant cells. Inadequate levels of Hsp90 may result in heightened cell vulnerability to elevated temperatures (Borkovich et al., 1989). Nevertheless, Hsp90’s functional range surpasses its participation in heat shock adaptation, as it also assumes pivotal functions in cell signal transduction, growth, and differentiation. This is accomplished by regulating the conformation of client proteins, facilitating the folding and assembly of newly synthesized proteins, and repairing and degrading albumen damaged by external stimuli (Taipale et al., 2012).
Figure 3 (A) The conformational cycle of Hsp90 involves several distinct states. After rapid ATP binding, Hsp90 transitions slowly to the first intermediate state (I1), characterized by open N-domains but a closed ATP lid. Subsequently, the second intermediate state (I2) is formed through N-terminal dimerization, leading to the repositioning of the M-domain and its interaction with the N-domain. Once Hsp90 achieves a fully closed state, ATP hydrolysis occurs. This results in the separation of the N-domains, releasing Pi and ADP, and allowing Hsp90 to return to its open conformation (Shiau et al., 2006; Graf et al., 2009; Biebl and Buchner, 2019). (B) Schematic diagram of the recognition epitope and structure of Hsp90 (Meyer et al., 2004; Zierer et al., 2016; Biebl and Buchner, 2019). (C) The intracellular fungal Hsp90 can infiltrate and transform into extracellular Hsp90 (eHsp90,47 -kDa Fragment) within the plasma membrane, cell wall, and fungal extracellular space. eHsp90 affects the coagulation pathway as disseminated intravascular coagulation (DIC) is a recognized complication of systemic candidiasis, both eHsp90 and prothrombin carry a tripeptide VIR, which is also located in the LKVIRK epitope of eHsp90 and serves as a catalytic site in prothrombin. Mycograb can recognize and bind to epitope C, thereby blocking the effect of eHsp90 on coagulation processes (Matthews and Burnie, 1992).
3.2 Extracellular Hsp90
Moreover, the intracellular fungal Hsp90 can infiltrate and transform into extracellular Hsp90 (eHsp90) within the plasma membrane, cell wall, and fungal extracellular space (Matthews and Burnie, 1992). In the case of Aspergillus fumigatus, eHsp90 is localized within the cell wall and plays a crucial role in preserving cell wall integrity (Lamoth et al., 2012). Additionally, eHsp90 is also present on the surface of C. neoformans, and its association with the cell surface depends on the endoplasmic reticulum (ER)-Golgi classical secretory pathway (Chatterjee and Tatu, 2017). Furthermore, the study revealed that the impairment of eHsp90 functionality during and after capsulation resulted in a notable decrease in capsulation size, indicating the potential influence of eHsp90 on the virulence of C. neoformans (Chatterjee and Tatu, 2017). It is worth noting that eHsp90 is localized on the cell wall surface and the hyphal tip in C. albicans (Burnie et al., 2006). The fungal eHsp90 can also bind to diverse human serum proteins and inhibit their functionality by modulating protein folding or interaction (Matthews and Burnie, 1992). In the investigation of C. albicans, an immunodominant 47-kDa antigen originating from the C-terminal of Hsp90, which exhibited heat stability, was discovered (Matthews and Burnie, 1989). Initially, this 47-kDa antigen was acknowledged as a noteworthy focal point of the immune response in individuals afflicted with invasive candidiasis, and subsequent research has demonstrated its association with survival rates in both human and animal models of infection (Matthews and Burnie, 1988; Matthews and Burnie, 1989). The presence of antibodies to the 47-kDa antigen has been observed in the majority of patients with chronic mucocutaneous candidiasis and acquired immune deficiency syndrome (Burford-Mason et al., 1987). Furthermore, eHsp90 demonstrates exogenous biological activities within the abnormal extracellular environment. Notably, it has the potential to impact the coagulation pathway, as disseminated intravascular coagulation (DIC) is a well-established complication of systemic candidiasis. Both eHsp90 and prothrombin possess a tripeptide VIR, which is also situated within the LKVIRK epitope of eHsp90 and functions as a catalytic site in prothrombin (Matthews and Burnie, 1992) (Figure 3C). These all suggest its potential role in mediating the protective effect of humoral immunity in invasive candidiasis (Matthews et al., 1988; Matthews et al., 1991). Epitope mapping showed that patients recovering from invasive candidiasis produce antibodies against fungal Hsp90 by binding epitope C (386LKVIRK391) as the most prevalent, followed by epitope B (329LSERM333) as the second most commonly recognized and epitope H (695DEPAGE700), exclusively specific to C. albicans and located at the C-terminal, as the third commonly recognized epitope observed in approximately 50% of the subjects (Matthews, 1992).
3.3 Immunotherapy targeting Hsp90
Due to the significant contribution of eHsp90 in the progression of candidiasis, there has been substantial interest in developing vaccines and antibodies that target Hsp90. The exploration of Hsp90 antibodies can be traced back to the 1980s when Matthews et al. discovered a positive association between the levels of anti-Hsp90 antibodies and patient prognosis and mortality (Matthews et al., 1987). Mycograb, a 28-kDa recombinant human antibody fragment, encompasses the antigen binding domain of both the heavy and light antibody chains and exhibits specific binding to an epitope, 386LKVIRK391, in fungal Hsp90 (Herbrecht et al., 2006). The antifungal effects of Mycograb were synergistic when combined with amphotericin B and caspofungin. In a mouse model of systemic candidiasis with a normal immune response, Mycograb exhibited superior efficacy in killing C. albicans compared to treatment with amphotericin B alone. Furthermore, a multinational phase III clinical trial demonstrated that combining Mycograb and amphotericin B lipid-related preparations significantly reduced the mortality rate associated with Candida species in patients with invasive candidiasis, surpassing the efficacy of amphotericin B monotherapy. The combination therapy demonstrated a notable enhancement in efficacy, as evidenced by the increase in survival rate from 48% in amphotericin B monotherapy to 84% in combination therapy. This improvement in patient outcomes was further supported by the notable increase in culture-confirmed C. albicans clearance rate (Pachl et al., 2006). However, the Mycograb binding antigen epitope (386LKVIRK391) represents a relatively conserved segment within both human and fungal Hsp90. Regrettably, this characteristic also enables Mycograb to interact with the host’s Hsp90, thereby inducing a potentially severe cytokine release syndrome. As a result, the Committee for Medicinal Products for Human Use rendered an unfavorable judgment in November 2006, advising against the approval of Mycograb for marketing due to apprehensions regarding its quality and safety (Bugli et al., 2013). Considering this decision, Arnold Louie and colleagues endeavored to tackle the challenges arising from the disparities in molecular weight and conformational structure of Mycograb (Louie et al., 2011; Bugli et al., 2013). Consequently, they devised a revised version of Mycograb, known as the Mycograb C28Y variant, by substituting cysteine at position 28 with tyrosine. However, the co-administration of amphotericin B and doses up to 15 mg/kg per day of the Mycograb C28Y variant did not exhibit any enhanced efficacy compared to amphotericin B monotherapy in a murine model of invasive candidiasis under conditions of neutropenia (Louie et al., 2011).
Furthermore, some vaccines have been specifically formulated to target Hsp90. Raska et al. conducted a study in which they created a DNA vaccine expressing Hsp90 of C. albicans and a recombinant protein vaccine (r-hsp90-CA) to evaluate their effectiveness in combating candidiasis (Raska et al., 2005). The findings suggest that the levels of hsp90-specific IgG generated through DNA vaccination were approximately one-third of those produced through protein vaccination. The difference in effectiveness can be ascribed to the comparatively limited expression of the Hsp90 antigen from DNA vaccines following injection (Raska et al., 2005). Consequently, subsequent investigations have employed the concurrent administration of an immunologic adjuvant and DNA vaccine to augment the immunogenicity of the DNA vaccine. Wang et al. utilized polysaccharides derived from the fruits of Physalis alkekengi L as the immunologic adjuvant in conjunction with the DNA vaccine. The DNA vaccine pD-HSP90C consists of a recombinant plasmid containing epitope C (386LKVIRK391) of Hsp90 from C. albicans and polysaccharides. The results show that the concurrent administration of polysaccharide and DNA vaccine significantly enhances mice’s humoral and cellular (Th1, Th2) immune responses while substantially extending their survival time against systemic candidiasis (Yang et al., 2014).
Additionally, a study successfully engineered a vaccine that expressed a specific epitope H (696DEPAGE700) of C. albicans Hsp90(SE-CA-HSP90) on the surface of filamentous phage, fused to the major coat protein pVIII. The protective immune responses of the hybrid-phage vaccine were investigated in mice. In the survival rate experiment, the data showed that 60% of the mice immunized with the hybrid-phage vaccine survived after 15 days, whereas only 11% of the mice immunized with TE (Tris-EDTA) and 20% of the mice immunized with wild-type phage were observed to be alive on the 15 th day after infection. Furthermore, the presence of the SE-CA-HSP90-specific antibody was exclusively detected in the mice that received the hybrid-phage vaccine. Moreover, the delayed-type hypersensitivity (DTH) response and natural killer cell activity exhibited significant augmentation compared to the TE control group (Wang et al., 2006).
4 Eno1
4.1 The structure of Eno1
Enolase plays a crucial role in the glycolysis pathway by catalyzing the conversion of 2-phosphoglycerate to phosphoenolpyruvic acid (Kang et al., 2008). In C. albicans, the Eno1 protein comprises 440 amino acids and has a molecular weight of approximately 47 kDa. Eno1 is abundant in C. albicans, accounting for 0.7% and 2% of the total protein content during the yeast and mycelium phases (Sundstrom and Aliaga, 1994). The three-dimensional structure of Eno1 was determined through X-ray diffraction analysis of the complex formed by Eno1 and 2-phosphoglycerate. The diffraction data unveiled that four Eno1 proteins assemble into two dimers within an asymmetric unit, interconnected by four pairs of hydrogen bonds. Two Eno1 molecules form homologous dimers, exhibiting an antiparallel orientation (Li et al., 2022).
4.2 Functions of Eno1 in candidiasis
Eno1, a protein widely distributed in the interior and surface of C. albicans, possesses multifunctional properties (Gil-Bona et al., 2018). Specifically, Eno1 primarily functions in glycolysis and gluconeogenesis, exhibiting enolase activity in the cytoplasm of C. albicans. The knockout of the ENO1 gene results in the inability of C. albicans to survive on media containing glucose as the sole carbon source (Ko et al., 2013). Despite the absence of a conventional secretory signal, the surface exposure of Eno1 relies on the presence of the first 28 amino acids in the N-terminal domain and a Tlg2-dependent pathway in S. cerevisiae (Miura et al., 2012). Moreover, it has been suggested that the N-terminal domain of Eno1, consisting of 169 amino acids, may have a role in sorting processes; alternatively, Eno1 could be transported via extracellular vesicles (López-Villar et al., 2006; Karkowska-Kuleta et al., 2020). Eno1 has been found to possess transglutaminase activity on the cell wall of C. albicans and is classified as a type of cell wall integrin (Ruiz-Herrera et al., 1995). Transglutaminase functions by catalyzing N- ϵ-(γ- glutamyl lysine amide) bonds at its active center, resulting in covalent cross-linking within or between proteins (Ruiz-Herrera et al., 1995). A substrate for transglutaminase, hyphal wall protein 1 (Hwp1) is exclusively expressed during the hyphal state of C. albicans and is characterized by its high content of glutamine residues. The serine/threonine enrichment site of Hwp1 undergoes glycosylation and is subsequently secreted by a signal peptide, ultimately anchoring to the cell wall surface through GPI (Staab et al., 2004). Eno1 enzymatically interacts with Hwp1 and catalyzes covalent cross-linking between the glutamine residue of Hwp1 and the lysine residue of the N-cadherin and E-cadherin of epithelial cells, causing C. albicans to adhere to the host cell and promoting infection (Staab et al., 1999).
Extracellular Eno1 exhibits binding affinity toward various molecules, including cadherin in endothelial cells, laminin in the basal membrane, and fibronectin and vitronectin in the extracellular matrix. This binding capability contributes to the pathogenicity of fungi by disrupting the host’s homeostasis/hemostasis system through the initiation of protein hydrolysis cascades, such as fibrinolysis, complement activation, and the kallikrein-kinin system (Felk et al., 2002; Wächtler et al., 2012; Silva et al., 2014; Kozik et al., 2015; Satala et al., 2020). Additionally, Eno1 is capable of binding to human plasminogen, which is typically found on the surface of mammalian cells and within the fibrin network. This interaction is crucial in fibrin and extracellular matrix breakdown during tissue remodeling (Keragala and Medcalf, 2021). The primary binding sites for human plasminogen on Eno1 are the two C-terminal lysine residues (Derbise et al., 2004). Furthermore, employing cross-linking analysis, it has been determined that the sequence 237AGYKGKVGIAMDVASSEFYKDGK259 is a significant elongated fragment essential for the interaction between Eno1 and human plasminogen (Satala et al., 2020).
4.3 Immunotherapy targeting Eno1
The monoclonal antibody underwent screening by injecting recombinant Eno1 into mice, producing monoclonal antibodies. The findings demonstrated that 12D9 displayed selective binding properties to Eno1 in the cell wall of C. albicans, effectively impeding its interaction with plasmin. Prior research has indicated that the 251-259 position within the six-helix six sheets 6 (H6S6) loop of the EnoA structure of Taenia solium is the binding motif for plasmin (Ayón-Núñez et al., 2018). A comparative analysis involving multiple Candida species showed that the sequence composition exhibited a notably high similarity. Subsequently, it was ascertained that the 12D9 antibody specifically targeted the 254FYKDGKYDL262 motif within the Eno1 H6S6 protein, impeding its interaction with plasmin and subsequently inducing plasmin activation. The in vitro experimental results proved that 12D9 effectively alleviated the adverse impacts of C. albicans on human umbilical vein endothelial and Caco-2 cells. The administration of 12D9 in a mouse model resulted in a notable increase in the survival rate of mice and a reduction in the fungal load within their kidneys. Moreover, the combination of 12D9 with fluconazole and anidulafungin demonstrated a synergistic antifungal effect (Chen et al., 2020).
By subjecting the antibodies generated during the seventh humoral immunity process in chickens to screening and modification, researchers successfully isolated a single-chain variable antibody known as CaS1 (Leu et al., 2010). In vitro experiments revealed that the antifungal potency of scFv CaS1, at a concentration of 150 ug/mL, is comparable to that of fluconazole, effectively inhibiting the growth of C. albicans. Furthermore, by binding this antibody with various species of Candida Eno1, it was observed that only Eno1 derived from C. albicans and Candida tropicalis exhibited specific recognition by CaS1 (Leu et al., 2020). In vivo animal model experiments were conducted to investigate the effects of CaS1 on C. albicans adhesion ratio in zebrafish. The results showed that after exposure to CaS1 for 2 and 4 hours, there was a significant decrease of 21% and 35% in the adhesion ratio, respectively. Furthermore, the administration of CaS1 in mice resulted in a notable reduction in fungal burden and associated cytokine levels (Leu et al., 2020).
5 Secretory aspartate protease (Sap)
5.1 The structure of Sap
The secretion of Sap by C. albicans hyphae, an acid protease characterized by aspartic acid residues at its active site, aids in the infiltration of host cells (Mandujano-González et al., 2016). SAP genes have been observed in multiple pathogenic Candida species, such as C. albicans, C. dubliniensis, C. tropicalis, and C. parapsilosis (de Viragh et al., 1993; Gilfillan et al., 1998; Zaugg et al., 2001). Research has shown that SAP genes display unique regulatory patterns and serve specific functions at different stages of infection, likely due to the diversity of coding gene types and variations among Candida species (Schaller et al., 2001). The expressions of SAP1, SAP2, and SAP3 genes have been exclusively observed in the yeast form of C. albicans among the 10 SAP genes. Sap2 has been identified as a crucial contributor, particularly necessary for in vitro growth, specifically when bovine serum albumin is the sole carbon source (White et al., 1993). In contrast, the SAP4, SAP5, and SAP6 genes are exclusively expressed in the mycelial form and are associated with invasive candidiasis and evasion of the host immune system (Borg-von Zepelin et al., 1998). The presence of SAP7 has been observed in mouse models, but it has not been observed in any in vitro settings. Its expression is associated with the virulence of venous infections (Taylor et al., 2005). Moreover, previous research has demonstrated that SAP8 is rapidly expressed in yeast and epithelial models (Naglik et al., 2008). Additionally, the proteins encoded by the SAP9 and SAP10 genes contain GPI-anchored domains, indicating their potential role in maintaining cell wall integrity (Albrecht et al., 2006).
The synthesis of Sap initiates in the nucleus, where the newly formed mRNA is subsequently transported to the cytoplasm for translation into a proenzyme that exceeds the mature protease by 60 amino acids. In the rough endoplasmic reticulum, the N-terminal signal peptide is cleaved by the signal peptide enzyme, and the resulting zymogen is then transported to the Golgi apparatus. Within the Golgi apparatus, the zymogen undergoes additional processing facilitated by Kex2 protease, a Ca2+-dependent serine protease belonging to the subtilisin-like proprotein convertase family (Brenner and Fuller, 1992), and it has high specificity for the cleavage of C-terminal paired basic sites, most often Lys-Arg, Lys-Lys or Arg-Arg following the Lys Arg sequence (von Heijne, 1985; Togni et al., 1996). The mature form of Sap, a protein produced by C. albicans, displays a size range of 35 to 50 kDa (Monod et al., 1998). Saps possess distinctive sequence motifs, including two conserved aspartic acid residues at the active site and conserved cysteine residues that contribute to maintaining their three-dimensional structure (Naglik et al., 2003). Sap2 is the most abundant among the Saps when proteins are the sole nitrogen source (Naglik et al., 2003). Sap2 exhibits typical characteristics of Saps, featuring a monomeric structure composed of N- and C-terminal domains, which contain an active site region. The structural configuration of Sap2 is reminiscent of a bilobed structure characterized by a central fissure that can accommodate peptide substrates containing up to nine residues (Calugi et al., 2013). The N-terminal domain of Sap2 displays an Asp-Thr-Gly motif, while its C-terminal domain exhibits an Asp-Ser-Gly motif. In contrast, most mammalian aspartate proteases feature an Asp-Thr-Gly motif in both domains (Cutfield et al., 1995; Koelsch et al., 2000).
5.2 Functions of Saps in candidiasis
Saps play a pivotal role in various processes of C. albicans, including adhesion, hyphal growth, biofilm formation, and immune evasion. An Arg-Gly-Asp motif enables Sap 4, Sap5, and Sap6 to bind to an integrin receptor on the epithelial cell surface, facilitating the adhesion of C. albicans to host cells (Wu et al., 2013). An oral mouse model of candidiasis was utilized to evaluate the in vivo adhesion function of Sap6. The adhesion rate of wild-type C. albicans cells was 78 ± 1%, whereas the sap6Δ/Δ mutant exhibited a significant reduction of 60% in adhesion compared to the wild-type strain (Kumar et al., 2015).
Sap9 governs the growth of hyphae and the formation of biofilms in C. albicans utilizing a cAMP-dependent pathway that encompasses a newly identified G protein-coupled receptor, Gprl, as well as the G proteins Gpa2 and Ras2, adenylyl cyclase (AC), cyclic AMP (cAMP), and cAMP-dependent protein kinase (PKA) (Midkiff et al., 2011; Yang et al., 2020). Activation of this pathway occurs in response to environmental nutrients, such as glucose and amino acids, which are detected by the G protein-coupled receptor (GPCR) located on the cell surface. Guanine nucleotide-binding protein (G protein) activation occurs through signal detection, increasing intracellular cAMP levels during adenylate cyclase stimulation. This elevation of cAMP subsequently triggers the activation of PKA, which in turn phosphorylates downstream target proteins. In fungal pathogens, these biochemical reactions are associated with morphological alterations and the release of virulence factors (Leberer et al., 2001; Liebmann et al., 2004). Notably, studies have demonstrated the significant involvement of Sap9 in the initiation and sustenance of hyphal development (Yang et al., 2020). The precise mechanism by which Sap regulates hyphal morphogenesis remains uncertain; however, it is hypothesized that Sap9 may exert a proteolytic influence on Efg1, thereby facilitating its activation. In order to validate the impact of Sap9 on the hyphal induction pathway, this investigation documented a significant decrease of approximately 40% in Efg1 expression within strains lacking Sap9 when subjected to a hyphal induction medium. These findings suggest that Sap9 governs hyphal formation by modulating Efg1 expression via the cAMP-dependent pathway (Yang et al., 2020).
5.3 Immunotherapy targeting Sap2
The vaccine candidate Sap2, derived from C. albicans, has been the subject of extensive research due to its involvement in fungal virulence (Naglik et al., 2003). The SAP2 gene of C. albicans plays a significant role in fungal pathogenesis by degrading host proteins at epithelial sites and hydrolyzing complements (Naglik et al., 2003). De Bernardis et al. conducted a study demonstrating that Candida mutant strains lacking Sap2 displayed reduced virulence in a rat model of vaginal candidiasis compared to strains with Sap1 to Sap6 intact (De Bernardis et al., 1999). The immunization of mice using recombinant Sap2 has shown to be an effective method of protecting against invasive candidiasis. Additionally, the anti-Sap2 antibody (IgG) transfer has significantly decreased C. albicans infection (von Heijne, 1985). Classical immunization with Sap2 and alum as an adjuvant has resulted in a strong humoral response, substantially reducing C. albicans burden in the kidney during infection compared to non-immunized animals (Vilanova et al., 2004). Moreover, in a mucosal model, mice intranasally immunized with Sap2 demonstrated reduced fungal burdens following oral and vaginal challenges with C. albicans (Rahman et al., 2012). In conjunction with antibodies, several vaccines targeting Sap2 have exhibited effectiveness. An example of such a vaccine is PEV-7, which consists of a genetically engineered truncated form of Sap2 lacking 20 amino acids from the N-terminal region of the fully mature Sap2. This truncated Sap2 variant was integrated into influenza virosomes. Rats immunized with PEV-7 through the intravaginal pathway demonstrated safeguarding against vaginitis induced by C. albicans through antibodies (De Bernardis et al., 2012). In addition, the PEV-7 virosomal vaccine formulation has completed a Phase I clinical trial (NCT01067131), demonstrating that the administration of the PEV7 vaccine through intramuscular injection or intravaginal capsules induced a strong B cell-mediated immune response in vaginal and cervical samples (De Bernardis et al., 2015; Tso et al., 2018). Moreover, BALB/c mice vaccinated with either the hybrid phage displaying the Sap2 epitope 382SLAQVKYTSASSI393 or the recombinant Sap2 displayed vigorous cellular and protective humoral responses against C. albicans infection. Significantly, the findings of this study demonstrated that the hybrid phage immunization approach resulted in levels of protection that were comparable to those achieved through recombinant protein immunization. Specifically, both the group immunized with hybrid phage and the group immunized with recombinant Sap2 exhibited a significant reduction in the number of C. albicans cells at day 7 following the initial challenge. Furthermore, upon completion of the second survival challenge, mice were inoculated with 107 C. albicans cells. It became evident that the administration of hybrid phage or the recombinant Sap2 elicited an immune response and provided safeguarding against a fatal challenge in a mouse model (Wang et al., 2014).
6 Hyphal wall protein 1(Hwp1)
6.1 The structure of Hwp1
The adhesin protein Hwp1, frequently found on the surfaces of germ tubes and mycelium in Candida species, serves as a substrate for host cell transglutaminase. This enables covalent binding and cross-linking between the Candida genus and mucosal epithelial cells (Chaffin, 2008; Romeo and Criseo, 2008). The Hwp1 adhesin has also been proposed to contribute to the retention of C. albicans within biofilms in vivo (Mirhendi et al., 2011).
The HWP1 gene, responsible for encoding a protein featuring a surface-exposed N-terminal domain of Hwp1 that can bind to external ligands like antibodies, was discovered via immune screening of the germ tube cDNA library. Like Als3, Hwp1 is affixed to the cell wall through GPI through its C-terminal (Staab et al., 1996). Furthermore, the cDNA-derived amino acid sequence showcases a succession of 10 amino acid tandem repeats, constituting most of the open reading frame. The molar percentage of amino acids in the open reading frame indicates that proline accounts for 27%, glutamine accounts for 16%, and aspartic acid accounts for 12% of the composition. The C-terminal region of Hwp1 exhibits a high abundance of threonine and serine, providing numerous potential sites for O-glycosylation (Staab et al., 1996). The significant presence of proline residues and the repetitive sequence length(14 X 10 AA tandem repeats of [EVIQ]-P-[CDT]-D-[YNW]-P-[PQ]-[QI]-[QP]-[QDN]) classify Hwp1 within various proline-rich proteomes, where proline residues are recognized for their ability to maintain the extended conformation of peptide chains and facilitate non-covalent interactions between protein chains or in the case of salivary proteins, binding to toxic plant polyphenols (Williamson, 1994). The recurrence of cysteine residues potentially aids in generating extracellular disulfide bonds at consistent intervals, consequently facilitating the cross-linking of Hwp1 and other protein molecules on the cell surface (Chattaway et al., 1974). Additionally, including acidic amino acids in the repetitive sequence potentially imparts negative charges to the mycelium surface under physiological pH conditions (Holmes et al., 1992).
6.2 Functions of Hwp1 in candidiasis
The presence of Hwp1 is crucial for forming biofilms by C. albicans, both in vivo and in vitro. This protein is situated on the cellular surface and is connected to cell wall glucan through the remaining portion of its GPI anchor (Staab et al., 2004). In vitro experiments have shown that a C. albicans mutant lacking the HWP1 gene exhibits diminished biomass in biofilm production compared to the wild-type strain (Nobile et al., 2006a). Additionally, the hwp1Δ/Δ mutant produces a biofilm with a depth of approximately 100 µm that contains few hyphae. In contrast, reconstitution with a wild-type HWP1 allele permits the production of a biofilm with a depth of 200 to 300 µm, in which hyphae are readily present. The experimental findings suggest that Hwp1 plays a crucial role in the in vitro production of biofilms. Furthermore, a comparison of biofilm formation between hwp1Δ/Δ mutant and wild strains in a rat venous catheter model revealed notable distinctions that the hwp1Δ/Δ mutant had a severe biofilm defect: only sparse microcolonies were observed on the catheter, and the microcolonies were devoid of hyphae (Nobile et al., 2006b).
6.3 Immunotherapy targeting Hwp1
Recently, Hoyer and co-workers developed a monoclonal antibody against Hwp1 of C. albicans. The Gln-Pro-rich adhesive domain (CDNPPQPDQPDDN (amino acids 154 to 166 of the protein)) of Hwp1 was utilized to produce monoclonal antibody MAb 2-E8 (Oh et al., 2022). The result showed that 2-E8 can bind to Hwp1 specifically. MAb 2-E8 labeled the germ tube of the wild-type strain but not the hwp1Δ/Δ strain (Oh et al., 2022). Furthermore, assays were performed to assess the blocking potential of MAb2-E8 on adhesive interactions. As a control, Anti-Als1 MAb1-B2, which was previously shown to inhibit C. albicans adhesion to biliary epithelial cells (BECs), was included (Coleman et al., 2010). Results indicated that MAb2-E8 at a 20 µg/mL concentration effectively blocked adhesion, like Anti-Als1 (Oh et al., 2022). Furthermore, Rosario-Colon et al. found that Candida Hwp1-specific monoclonal antibodies 6H1 could protect mice against invasive C. auris infection by significantly enhancing survival and reducing fungal burdens (Rosario-Colon et al., 2021). Moreover, vaccination with glycopeptide conjugate consisting of β-mannan polysaccharide combined with Hwp1 peptide epitopes (QGETEEALIQKRSY) showed protection against experimental disseminated candidiasis in mice by favoring the production of protective and specific antibodies (Xin et al., 2008).
7 Other potential targets
In addition to the proteins mentioned above, numerous other proteins are also implicated in the pathogenesis of candidiasis. One such protein is hyphal-regulated protein 1 (Hyr1), which is crucial for hyphal growth and contributes to the virulence of C. albicans by evading phagocyte killing, a key host defense mechanism against candidiasis (Luo et al., 2010). Previous investigations have demonstrated that both active and passive immunity conferred by a recombinant N-terminal fragment of Hyr1 confer protection against lethal candidemia in mice (Luo et al., 2011). Consequently, it is postulated that vaccination with Hyr1 may exert a protective effect against candidiasis. Furthermore, it has been identified that phospholipase B serves as a virulence factor in C. albicans (Ghannoum, 2000). Moreover, the serum of individuals afflicted with invasive candidiasis exhibits the presence of antibodies that exhibit reactivity toward purified phospholipase B, thereby rendering it a compelling candidate for diagnostic and antibody preparation applications (Heilmann et al., 2011). Additionally, the indispensable role of malate dehydrogenase (Mdh1) in the tricarboxylic acid cycle, coupled with its involvement in aerobic energy production via participation in the malate-aspartate shuttle, has been established (Shibasaki et al., 2018). Mdh1 has been identified as a potential vaccine antigen for candidiasis due to its consistent presence throughout the investigated time points and lack of significant variation in relative abundance (Shibasaki et al., 2018). Investigations into the use of recombinant Mdh1 as a candidate antigen for a candidiasis vaccine have demonstrated that subcutaneous and intradermal administration in mice elicits significantly elevated antibody responses and confers significant protection against candidiasis (Shibasaki et al., 2014).
8 Conclusions and future perspectives
C. albicans, a pathogen characterized by its opportunistic nature, poses an increasing threat to human health. The proteins adhere to and invade host cells in symbiotic and pathogenic scenarios involving C. albicans. The morphological changes and accompanying physiological processes exhibited by this pathogen serve various purposes and offer promising avenues for drug investigation. Antibody-based medications targeting these proteins have exhibited positive therapeutic outcomes. In addition, antifungal antibodies present a diminished occurrence of adverse reactions and a broader array of choices when compared to small molecule drugs. Furthermore, the utilization of a synergistic combination of monoclonal antibodies and antifungal medications not only provides a more comprehensive and efficient approach to combat the escalating drug resistance observed in Candida species but also enhances specificity and holds potential for improved clinical outcomes. Additionally, the advancement of novel immunomodulatory techniques that integrate the regulation of recombinant cytokines with monoclonal antibodies presents an alternative strategy to enhance the therapeutic efficacy of the latter. Moreover, emerging technologies offer promising avenues for the treatment of life-threatening invasive fungal infections. However, despite the encouraging potential for vaccine development and antifungal therapies, additional research is necessary to comprehend their effectiveness and constraints comprehensively. It is crucial to explore approaches for establishing a persistent and uniform symbiotic model of candidiasis in mice, considering its symbiotic nature in humans. Understanding the mechanisms contributing to the effectiveness of relevant vaccines and antibodies is paramount to appropriately administering them to eligible patients and maximizing their therapeutic benefits. Additionally, it is imperative to establish a timely and dependable method for evaluating the efficacy of vaccines and antibodies while also investigating the potential of antibodies as valuable biomarkers for diagnosing Candida infection. Moreover, the primary challenge lies in securing adequate funding for clinical trials of innovative antifungal medications, particularly for vaccinations targeting opportunistic fungal pathogens that exclusively affect individuals with severe immune system deficiencies. These vaccinations may not be as commercially appealing as those designed to combat diseases that affect a significant portion of the general population. Consequently, it is anticipated that collaborative efforts involving academia, industry, and the government will be imperative in facilitating the advancement of promising and novel antifungal immunotherapeutics into clinical trials and, subsequently, the market.
Author contributions
ZF: Data curation, Formal analysis, Visualization, Writing – original draft. HL: Conceptualization, Supervision, Writing – review & editing. YJ: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study received financial support from the National Key Research and Development Program of China (No.2022YFC2303004 and No.2021YFC2300404), the National Natural Science Foundation of China (No. 82020108032), and the Innovation Program of Shanghai Municipal Education Commission (202101070007-E00094).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Albrecht, A., Felk, A., Pichova, I., Naglik, J. R., Schaller, M., de Groot, P., et al. (2006). Glycosylphosphatidylinositol-anchored proteases of Candida albicans target proteins necessary for both cellular processes and host-pathogen interactions. J. Biol. Chem. 281 (2), 688–694. doi: 10.1074/jbc.M509297200
Ali, M. M., Roe, S. M., Vaughan, C. K., Meyer, P., Panaretou, B., Piper, P. W., et al. (2006). Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature 440 (7087), 1013–1017. doi: 10.1038/nature04716
Almeida, R. S., Brunke, S., Albrecht, A., Thewes, S., Laue, M., Edwards, J. E., et al. (2008). the hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin. PloS Pathog. 4 (11), e1000217. doi: 10.1371/journal.ppat.1000217
Almeida, R. S., Wilson, D., Hube, B. (2009). Candida albicans iron acquisition within the host. FEMS Yeast Res. 9 (7), 1000–1012. doi: 10.1111/j.1567-1364.2009.00570.x
Armstrong-James, D., Harrison, T. S. (2012). Immunotherapy for fungal infections. Curr. Opin. Microbiol. 15 (4), 434–439. doi: 10.1016/j.mib.2012.06.001
Ashman, R. B., Fulurija, A., Papadimitriou, J. M. (1999). Both CD4+ and CD8+ lymphocytes reduce the severity of tissue lesions in murine systemic cadidiasis, and CD4+ cells also demonstrate strain-specific immunopathological effects. Microbiol. (Reading) 145 (Pt 7), 1631–1640. doi: 10.1099/13500872-145-7-1631
Ayón-Núñez, D. A., Fragoso, G., Espitia, C., García-Varela, M., Soberón, X., Rosas, G., et al. (2018). Identification and characterization of Taenia solium enolase as a plasminogen-binding protein. Acta Trop. 182, 69–79. doi: 10.1016/j.actatropica.2018.02.020
Biebl, M. M., Buchner, J. (2019). Structure, function, and regulation of the hsp90 machinery. Cold Spring Harb. Perspect. Biol. 11 (9). doi: 10.1101/cshperspect.a034017
Borg-von Zepelin, M., Beggah, S., Boggian, K., Sanglard, D., Monod, M. (1998). The expression of the secreted aspartyl proteinases Sap4 to Sap6 from Candida albicans in murine macrophages. Mol. Microbiol. 28 (3), 543–554. doi: 10.1046/j.1365-2958.1998.00815.x
Borkovich, K. A., Farrelly, F. W., Finkelstein, D. B., Taulien, J., Lindquist, S. (1989). hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol. Cell Biol. 9 (9), 3919–3930. doi: 10.1128/mcb.9.9.3919-3930.1989
Brena, S., Cabezas-Olcoz, J., Moragues, M. D., Fernández de Larrinoa, I., Domínguez, A., Quindós, G., et al. (2011). Fungicidal monoclonal antibody C7 interferes with iron acquisition in Candida albicans. Antimicrob. Agents Chemother. 55 (7), 3156–3163. doi: 10.1128/aac.00892-10
Brena, S., Omaetxebarría, M. J., Elguezabal, N., Cabezas, J., Moragues, M. D., Pontón, J. (2007). Fungicidal monoclonal antibody C7 binds to Candida albicans Als3. Infect. Immun. 75 (7), 3680–3682. doi: 10.1128/iai.01840-06
Brenner, C., Fuller, R. S. (1992). Structural and enzymatic characterization of a purified prohormone-processing enzyme: secreted, soluble Kex2 protease. Proc. Natl. Acad. Sci. U.S.A. 89 (3), 922–926. doi: 10.1073/pnas.89.3.922
Brown, G. D., Denning, D. W., Gow, N. A., Levitz, S. M., Netea, M. G., White, T. C. (2012). Hidden killers: human fungal infections. Sci. Transl. Med. 4 (165), 165rv13. doi: 10.1126/scitranslmed.3004404
Bugli, F., Cacaci, M., Martini, C., Torelli, R., Posteraro, B., Sanguinetti, M., et al. (2013). Human monoclonal antibody-based therapy in the treatment of invasive candidiasis. Clin. Dev. Immunol. 2013, 403121. doi: 10.1155/2013/403121
Burford-Mason, A. P., Matthews, R. C., Williams, J. R. (1987). Transient abrogation of immunosuppression in a patient with chronic mucocutaneous candidiasis following vaccination with Candida albicans. J. Infect. 14 (2), 147–157. doi: 10.1016/s0163-4453(87)91977-3
Burnie, J. P., Carter, T. L., Hodgetts, S. J., Matthews, R. C. (2006). Fungal heat-shock proteins in human disease. FEMS Microbiol. Rev. 30 (1), 53–88. doi: 10.1111/j.1574-6976.2005.00001.x
Calugi, C., Guarna, A., Trabocchi, A. (2013). Insight into the structural similarity between HIV protease and secreted aspartic protease-2 and binding mode analysis of HIV-Candida albicans inhibitors. J. Enzyme Inhib Med. Chem. 28 (5), 936–943. doi: 10.3109/14756366.2012.696245
Casadevall, A., Feldmesser, M., Pirofski, L. A. (2002). Induced humoral immunity and vaccination against major human fungal pathogens. Curr. Opin. Microbiol. 5 (4), 386–391. doi: 10.1016/s1369-5274(02)00337-5
Cassone, A., Cauda, R. (2012). Candida and candidiasis in HIV-infected patients: where commensalism, opportunistic behavior and frank pathogenicity lose their borders. Aids 26 (12), 1457–1472. doi: 10.1097/QAD.0b013e3283536ba8
Chaffin, W. L. (2008). Candida albicans cell wall proteins. Microbiol. Mol. Biol. Rev. 72 (3), 495–544. doi: 10.1128/mmbr.00032-07
Chattaway, F. W., Shenolikar, S., Barlow, A. J. (1974). The release of acid phosphatase and polysaccharide- and protein-containing components from the surface of the dimorphic forms of Candida albicans by treatment with dithiothreitol. J. Gen. Microbiol. 83 (2), 423–425. doi: 10.1099/00221287-83-2-423
Chatterjee, S., Tatu, U. (2017). Heat shock protein 90 localizes to the surface and augments virulence factors of Cryptococcus neoformans. PloS Negl. Trop. Dis. 11 (8), e0005836. doi: 10.1371/journal.pntd.0005836
Chen, S. M., Zou, Z., Guo, S. Y., Hou, W. T., Qiu, X. R., Zhang, Y., et al. (2020). Preventing Candida albicans from subverting host plasminogen for invasive infection treatment. Emerg. Microbes Infect. 9 (1), 2417–2432. doi: 10.1080/22221751.2020.1840927
Coleman, D. A., Oh, S. H., Zhao, X., Hoyer, L. L. (2010). Heterogeneous distribution of Candida albicans cell-surface antigens demonstrated with an Als1-specific monoclonal antibody. Microbiol. (Reading) 156 (Pt 12), 3645–3659. doi: 10.1099/mic.0.043851-0
Conti, H. R., Gaffen, S. L. (2010). Host responses to Candida albicans: Th17 cells and mucosal candidiasis. Microbes Infect. 12 (7), 518–527. doi: 10.1016/j.micinf.2010.03.013
Cowen, L. E., Sanglard, D., Howard, S. J., Rogers, P. D., Perlin, D. S. (2014). Mechanisms of antifungal drug resistance. Cold Spring Harb. Perspect. Med. 5 (7), a019752. doi: 10.1101/cshperspect.a019752
Cutfield, S. M., Dodson, E. J., Anderson, B. F., Moody, P. C., Marshall, C. J., Sullivan, P. A., et al. (1995). The crystal structure of a major secreted aspartic proteinase from Candida albicans in complexes with two inhibitors. Structure 3 (11), 1261–1271. doi: 10.1016/s0969-2126(01)00261-1
De Bernardis, F., Amacker, M., Arancia, S., Sandini, S., Gremion, C., Zurbriggen, R., et al. (2012). A virosomal vaccine against candidal vaginitis: immunogenicity, efficacy and safety profile in animal models. Vaccine 30 (30), 4490–4498. doi: 10.1016/j.vaccine.2012.04.069
De Bernardis, F., Arancia, S., Morelli, L., Hube, B., Sanglard, D., Schäfer, W., et al. (1999). Evidence that members of the secretory aspartyl proteinase gene family, in particular SAP2, are virulence factors for Candida vaginitis. J. Infect. Dis. 179 (1), 201–208. doi: 10.1086/314546
De Bernardis, F., Arancia, S., Sandini, S., Graziani, S., Norelli, S. (2015). Studies of immune responses in candida vaginitis. Pathogens 4 (4), 697–707. doi: 10.3390/pathogens4040697
Del Poeta, M., Casadevall, A. (2012). Ten challenges on Cryptococcus and cryptococcosis. Mycopathologia 173 (5-6), 303–310. doi: 10.1007/s11046-011-9473-z
Denning, D. W. (2024). Global incidence and mortality of severe fungal disease. Lancet Infect. Dis. doi: 10.1016/S1473-3099(23)00692-8
Derbise, A., Song, Y. P., Parikh, S., Fischetti, V. A., Pancholi, V. (2004). Role of the C-terminal lysine residues of streptococcal surface enolase in Glu- and Lys-plasminogen-binding activities of group A streptococci. Infect. Immun. 72 (1), 94–105. doi: 10.1128/iai.72.1.94-105.2004
de Viragh, P. A., Sanglard, D., Togni, G., Falchetto, R., Monod, M. (1993). Cloning and sequencing of two Candida parapsilosis genes encoding acid proteases. J. Gen. Microbiol. 139 (2), 335–342. doi: 10.1099/00221287-139-2-335
Edwards, J. E., Jr., Schwartz, M. M., Schmidt, C. S., Sobel, J. D., Nyirjesy, P., Schodel, F., et al. (2018). A fungal immunotherapeutic vaccine (NDV-3A) for treatment of recurrent vulvovaginal candidiasis-A phase 2 randomized, double-blind, placebo-controlled trial. Clin. Infect. Dis. 66 (12), 1928–1936. doi: 10.1093/cid/ciy185
Felk, A., Kretschmar, M., Albrecht, A., Schaller, M., Beinhauer, S., Nichterlein, T., et al. (2002). Candida albicans hyphal formation and the expression of the Efg1-regulated proteinases Sap4 to Sap6 are required for the invasion of parenchymal organs. Infect. Immun. 70 (7), 3689–3700. doi: 10.1128/iai.70.7.3689-3700.2002
Fourie, R., Kuloyo, O. O., Mochochoko, B. M., Albertyn, J., Pohl, C. H. (2018). Iron at the Centre of Candida albicans Interactions. Front. Cell Infect. Microbiol. 8. doi: 10.3389/fcimb.2018.00185
Ghannoum, M. A. (2000). Potential role of phospholipases in virulence and fungal pathogenesis. Clin. Microbiol. Rev. 13 (1), 122–143. doi: 10.1128/cmr.13.1.122
Gil-Bona, A., Amador-García, A., Gil, C., Monteoliva, L. (2018). The external face of Candida albicans: A proteomic view of the cell surface and the extracellular environment. J. Proteomics 180, 70–79. doi: 10.1016/j.jprot.2017.12.002
Gilfillan, G. D., Derek, J. S., Parkinson, T., Coleman, D. C., Gow, N. A. R. (1998). Candida dubliniensis: phylogeny and putative virulence factors. Microbiol. (Reading) 144 (Pt 4), 829–838. doi: 10.1099/00221287-144-4-829
Gozalbo, D., Gil, M. L. (2009). IFN-gamma in Candida albicans infections. Front. Biosci. (Landmark Ed) 14 (5), 1970–1978. doi: 10.2741/3356
Graf, C., Stankiewicz, M., Kramer, G., Mayer, M. P. (2009). Spatially and kinetically resolved changes in the conformational dynamics of the Hsp90 chaperone machine. EMBO J. 28 (5), 602–613. doi: 10.1038/emboj.2008.306
Harpf, V., Rambach, G., Würzner, R., Lass-Flörl, C., Speth, C. (2020). Candida and complement: new aspects in an old battle. Front. Immunol. 11. doi: 10.3389/fimmu.2020.01471
Heilmann, C. J., Sorgo, A. G., Siliakus, A. R., Dekker, H. L., Brul, S., de Koster, C. G., et al. (2011). Hyphal induction in the human fungal pathogen Candida albicans reveals a characteristic wall protein profile. Microbiol. (Reading) 157 (Pt 8), 2297–2307. doi: 10.1099/mic.0.049395-0
Herbrecht, R., Fohrer, C., Nivoix, Y. (2006). Mycograb for the treatment of invasive candidiasis. Clin. Infect. Dis. 43 (8), 1083–1084. doi: 10.1086/507547
Holmes, A. R., Cannon, R. D., Shepherd, M. G. (1992). Mechanisms of aggregation accompanying morphogenesis in Candida albicans. Oral. Microbiol. Immunol. 7 (1), 32–37. doi: 10.1111/j.1399-302x.1992.tb00017.x
Hoyer, L. L., Cota, E. (2016). Candida albicans agglutinin-like sequence (Als) family vignettes: A review of als protein structure and function. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00280
Hoyer, L. L., Green, C. B., Oh, S. H., Zhao, X. (2008). Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family–a sticky pursuit. Med. Mycol 46 (1), 1–15. doi: 10.1080/13693780701435317
Jones-Carson, J., Vazquez-Torres, F. A., Balish, E. (1997). B cell-independent selection of memory T cells after mucosal immunization with Candida albicans. J. Immunol. 158 (9), 4328–4335. doi: 10.4049/jimmunol.158.9.4328
Kang, H. J., Jung, S. K., Kim, S. J., Chung, S. J. (2008). Structure of human alpha-enolase (hENO1), a multifunctional glycolytic enzyme. Acta Crystallogr. D Biol. Crystallogr. 64 (Pt 6), 651–657. doi: 10.1107/s0907444908008561
Karkowska-Kuleta, J., Kulig, K., Karnas, E., Zuba-Surma, E., Woznicka, O., Pyza, E., et al. (2020). Characteristics of Extracellular Vesicles Released by the Pathogenic Yeast-Like Fungi Candida glabrata, Candida parapsilosis and Candida tropicalis. Cells 9 (7). doi: 10.3390/cells9071722
Keragala, C. B., Medcalf, R. L. (2021). Plasminogen: an enigmatic zymogen. Blood 137 (21), 2881–2889. doi: 10.1182/blood.2020008951
Knight, S. A., Vilaire, G., Lesuisse, E., Dancis, A. (2005). Iron acquisition from transferrin by Candida albicans depends on the reductive pathway. Infect. Immun. 73 (9), 5482–5492. doi: 10.1128/iai.73.9.5482-5492.2005
Ko, H. C., Hsiao, T. Y., Chen, C. T., Yang, Y. L. (2013). Candida albicans ENO1 null mutants exhibit altered drug susceptibility, hyphal formation, and virulence. J. Microbiol. 51 (3), 345–351. doi: 10.1007/s12275-013-2577-z
Koelsch, G., Tang, J., Loy, J. A., Monod, M., Jackson, K., Foundling, S. I., et al. (2000). Enzymic characteristics of secreted aspartic proteases of Candida albicans. Biochim. Biophys. Acta 1480 (1-2), 117–131. doi: 10.1016/s0167-4838(00)00068-6
Kozik, A., Karkowska-Kuleta, J., Zajac, D., Bochenska, O., Kedracka-Krok, S., Jankowska, U., et al. (2015). Fibronectin-, vitronectin- and laminin-binding proteins at the cell walls of Candida parapsilosis and Candida tropicalis pathogenic yeasts. BMC Microbiol. 15, 197. doi: 10.1186/s12866-015-0531-4
Kumar, R., Saraswat, D., Tati, S., Edgerton, M. (2015). Novel aggregation properties of candida albicans secreted aspartyl proteinase sap6 mediate virulence in oral candidiasis. Infect. Immun. 83 (7), 2614–2626. doi: 10.1128/iai.00282-15
Lamoth, F., Juvvadi, P. R., Fortwendel, J. R., Steinbach, W. J. (2012). Heat shock protein 90 is required for conidiation and cell wall integrity in Aspergillus fumigatus. Eukaryot Cell 11 (11), 1324–1332. doi: 10.1128/ec.00032-12
Leberer, E., Harcus, D., Dignard, D., Johnson, L., Ushinsky, S., Thomas, D. Y., et al. (2001). Ras links cellular morphogenesis to virulence by regulation of the MAP kinase and cAMP signalling pathways in the pathogenic fungus Candida albicans. Mol. Microbiol. 42 (3), 673–687. doi: 10.1046/j.1365-2958.2001.02672.x
Leu, S. J., Lee, Y. C., Lee, C. H., Liao, P. Y., Chiang, C. W., Yang, C. M., et al. (2020). Generation and Characterization of Single Chain Variable Fragment against Alpha-Enolase of Candida albicans. Int. J. Mol. Sci. 21 (8). doi: 10.3390/ijms21082903
Leu, S. J., Lee, Y. C., Shih, N. Y., Huang, I. J., Liu, K. J., Lu, H. F., et al. (2010). Generation and characterization of anti-alpha-enolase single-chain antibodies in chicken. Vet. Immunol. Immunopathol. 137 (3-4), 251–260. doi: 10.1016/j.vetimm.2010.06.001
Li, L., Lu, H., Zhang, X., Whiteway, M., Wu, H., Tan, S., et al. (2022). Baicalein Acts against Candida albicans by Targeting Eno1 and Inhibiting Glycolysis. Microbiol. Spectr. 10 (4), e0208522. doi: 10.1128/spectrum.02085-22
Li, W., Shrivastava, M., Lu, H., Jiang, Y. (2021). Calcium-calcineurin signaling pathway in Candida albicans: A potential drug target. Microbiol. Res. 249, 126786. doi: 10.1016/j.micres.2021.126786
Liebmann, B., Müller, M., Braun, A., Brakhage, A. A. (2004). The cyclic AMP-dependent protein kinase a network regulates development and virulence in Aspergillus fumigatus. Infect. Immun. 72 (9), 5193–5203. doi: 10.1128/iai.72.9.5193-5203.2004
Lin, L., Ibrahim, A. S., Avanesian, V., Edwards, J. E., Jr., Fu, Y., Baquir, B., et al. (2008). Considerable differences in vaccine immunogenicities and efficacies related to the diluent used for aluminum hydroxide adjuvant. Clin. Vaccine Immunol. 15 (3), 582–584. doi: 10.1128/cvi.00427-07
Lin, L., Ibrahim, A. S., Xu, X., Farber, J. M., Avanesian, V., Baquir, B., et al. (2009). Th1-Th17 cells mediate protective adaptive immunity against Staphylococcus aureus and Candida albicans infection in mice. PloS Pathog. 5 (12), e1000703. doi: 10.1371/journal.ppat.1000703
Lin, J., Oh, S. H., Jones, R., Garnett, J. A., Salgado, P. S., Rusnakova, S., et al. (2014). The peptide-binding cavity is essential for Als3-mediated adhesion of Candida albicans to human cells. J. Biol. Chem. 289 (26), 18401–18412. doi: 10.1074/jbc.M114.547877
Lionakis, M. S. (2014). New insights into innate immune control of systemic candidiasis. Med. Mycol 52 (6), 555–564. doi: 10.1093/mmy/myu029
Lionakis, M. S., Netea, M. G. (2013). Candida and host determinants of susceptibility to invasive candidiasis. PloS Pathog. 9 (1), e1003079. doi: 10.1371/journal.ppat.1003079
Liu, Y., Filler, S. G. (2011). Candida albicans Als3, a multifunctional adhesin and invasin. Eukaryot Cell 10 (2), 168–173. doi: 10.1128/ec.00279-10
Logan, C., Martin-Loeches, I., Bicanic, T. (2020). Invasive candidiasis in critical care: challenges and future directions. Intensive Care Med. 46 (11), 2001–2014. doi: 10.1007/s00134-020-06240-x
López-Villar, E., Monteoliva, L., Larsen, M. R., Sachon, E., Shabaz, M., Pardo, M., et al. (2006). Genetic and proteomic evidences support the localization of yeast enolase in the cell surface. Proteomics 6 Suppl 1, S107–S118. doi: 10.1002/pmic.200500479
Louie, A., Stein, D. S., Zack, J. Z., Liu, W., Conde, H., Fregeau, C., et al. (2011). Dose range evaluation of Mycograb C28Y variant, a human recombinant antibody fragment to heat shock protein 90, in combination with amphotericin B-desoxycholate for treatment of murine systemic candidiasis. Antimicrob. Agents Chemother. 55 (7), 3295–3304. doi: 10.1128/aac.01324-10
Lu, H., Hong, T., Jiang, Y., Whiteway, M., Zhang, S. (2023a). Candidiasis: From cutaneous to systemic, new perspectives of potential targets and therapeutic strategies. Adv. Drug Delivery Rev. 199, 114960. doi: 10.1016/j.addr.2023.114960
Lu, H., Li, W., Whiteway, M., Wang, H., Zhu, S., Ji, Z., et al. (2023b). A small molecule inhibitor of erg251 makes fluconazole fungicidal by inhibiting the synthesis of the 14α-methylsterols. mBio 14 (1), e0263922. doi: 10.1128/mbio.02639-22
Luo, G., Ibrahim, A. S., French, S. W., Edwards, J. E., Jr., Fu, Y. (2011). Active and passive immunization with rHyr1p-N protects mice against hematogenously disseminated candidiasis. PloS One 6 (10), e25909. doi: 10.1371/journal.pone.0025909
Luo, G., Ibrahim, A. S., Spellberg, B., Nobile, C. J., Mitchell, A. P., Fu, Y. (2010). Candida albicans Hyr1p confers resistance to neutrophil killing and is a potential vaccine target. J. Infect. Dis. 201 (11), 1718–1728. doi: 10.1086/652407
Mandujano-González, V., Villa-Tanaca, L., Anducho-Reyes, M. A., Mercado-Flores, Y. (2016). Secreted fungal aspartic proteases: A review. Rev. Iberoam Micol 33 (2), 76–82. doi: 10.1016/j.riam.2015.10.003
Martinez, F. O., Helming, L., Gordon, S. (2009). Alternative activation of macrophages: an immunologic functional perspective. Annu. Rev. Immunol. 27, 451–483. doi: 10.1146/annurev.immunol.021908.132532
Matejuk, A., Leng, Q., Begum, M. D., Woodle, M. C., Scaria, P., Chou, S. T., et al. (2010). Peptide-based antifungal therapies against emerging infections. Drugs Future 35 (3), 197. doi: 10.1358/dof.2010.035.03.1452077
Matthews, R. C. (1992). The 14th C. L. Oakley Lecture. Candida albicans HSP 90: link between protective and auto immunity. J. Med. Microbiol. 36 (6), 367–370. doi: 10.1099/00222615-36-6-367
Matthews, R., Burnie, J. (1988). Diagnosis of systemic candidiasis by an enzyme-linked dot immunobinding assay for a circulating immunodominant 47-kilodalton antigen. J. Clin. Microbiol. 26 (3), 459–463. doi: 10.1128/jcm.26.3.459-463.1988
Matthews, R., Burnie, J. (1989). Cloning of a DNA sequence encoding a major fragment of the 47 kilodalton stress protein homologue of Candida albicans. FEMS Microbiol. Lett. 51 (1), 25–30. doi: 10.1016/0378-1097(89)90071-2
Matthews, R., Burnie, J. (1992). The role of hsp90 in fungal infection. Immunol. Today 13 (9), 345–348. doi: 10.1016/0167-5699(92)90169-8
Matthews, R. C., Burnie, J. P., Howat, D., Rowland, T., Walton, F. (1991). Autoantibody to heat-shock protein 90 can mediate protection against systemic candidosis. Immunology 74 (1), 20–24.
Matthews, R., Burnie, J., Smith, D., Clark, I., Midgley, J., Conolly, M., et al. (1988). Candida and AIDS: evidence for protective antibody. Lancet 2 (8605), 263–266. doi: 10.1016/s0140-6736(88)92547-0
Matthews, R. C., Burnie, J. P., Tabaqchali, S. (1987). Isolation of immunodominant antigens from sera of patients with systemic candidiasis and characterization of serological response to Candida albicans. J. Clin. Microbiol. 25 (2), 230–237. doi: 10.1128/jcm.25.2.230-237.1987
Medici, N. P., Del Poeta, M. (2015). New insights on the development of fungal vaccines: from immunity to recent challenges. Mem Inst Oswaldo Cruz 110 (8), 966–973. doi: 10.1590/0074-02760150335
Meyer, P., Prodromou, C., Liao, C., Hu, B., Mark Roe, S., Vaughan, C. K., et al. (2004). Structural basis for recruitment of the ATPase activator Aha1 to the Hsp90 chaperone machinery. EMBO J. 23 (3), 511–519. doi: 10.1038/sj.emboj.7600060
Midkiff, J., Borochoff-Porte, N., White, D., Johnson, D. I. (2011). Small molecule inhibitors of the Candida albicans budded-to-hyphal transition act through multiple signaling pathways. PloS One 6 (9), e25395. doi: 10.1371/journal.pone.0025395
Mirhendi, H., Bruun, B., Schønheyder, H. C., Christensen, J. J., Fuursted, K., Gahrn-Hansen, B., et al. (2011). Differentiation of Candida glabrata, C. nivariensis and C. bracarensis based on fragment length polymorphism of ITS1 and ITS2 and restriction fragment length polymorphism of ITS and D1/D2 regions in rDNA. Eur. J. Clin. Microbiol. Infect. Dis. 30 (11), 1409–1416. doi: 10.1007/s10096-011-1235-9
Miura, N., Kirino, A., Endo, S., Morisaka, H., Kuroda, K., Takagi, M., et al. (2012). Tracing putative trafficking of the glycolytic enzyme enolase via SNARE-driven unconventional secretion. Eukaryot Cell 11 (8), 1075–1082. doi: 10.1128/ec.00075-12
Monod, M., Hube, B., Hess, D., Sanglard, D. (1998). Differential regulation of SAP8 and SAP9, which encode two new members of the secreted aspartic proteinase family in Candida albicans. Microbiol. (Reading) 144 (Pt 10), 2731–2737. doi: 10.1099/00221287-144-10-2731
Moreno-Ruiz, E., Galán-Díez, M., Zhu, W., Fernández-Ruiz, E., d’Enfert, C., Filler, S. G., et al. (2009). Candida albicans internalization by host cells is mediated by a clathrin-dependent mechanism. Cell Microbiol. 11 (8), 1179–1189. doi: 10.1111/j.1462-5822.2009.01319.x
Naglik, J. R., Challacombe, S. J., Hube, B. (2003). Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol. Mol. Biol. Rev. 67 (3), 400–428. doi: 10.1128/mmbr.67.3.400-428.2003
Naglik, J. R., Moyes, D., Makwana, J., Kanzaria, P., Tsichlaki, E., Weindl, G., et al. (2008). Quantitative expression of the Candida albicans secreted aspartyl proteinase gene family in human oral and vaginal candidiasis. Microbiol. (Reading) 154 (Pt 11), 3266–3280. doi: 10.1099/mic.0.2008/022293-0
Nami, S., Aghebati-Maleki, A., Morovati, H., Aghebati-Maleki, L. (2019). Current antifungal drugs and immunotherapeutic approaches as promising strategies to treatment of fungal diseases. BioMed. Pharmacother. 110, 857–868. doi: 10.1016/j.biopha.2018.12.009
Netea, M. G., Gijzen, K., Coolen, N., Verschueren, I., Figdor, C., van der Meer, J. W., et al. (2004). Human dendritic cells are less potent at killing Candida albicans than both monocytes and macrophages. Microbes Infect. 6 (11), 985–989. doi: 10.1016/j.micinf.2004.05.013
Netea, M. G., van Tits, L. J., Curfs, J. H., Amiot, F., Meis, J. F., van der Meer, J. W., et al. (1999). Increased susceptibility of TNF-alpha lymphotoxin-alpha double knockout mice to systemic candidiasis through impaired recruitment of neutrophils and phagocytosis of Candida albicans. J. Immunol. 163 (3), 1498–1505. doi: 10.4049/jimmunol.163.3.1498
Newman, S. L., Holly, A. (2001). Candida albicans is phagocytosed, killed, and processed for antigen presentation by human dendritic cells. Infect. Immun. 69 (11), 6813–6822. doi: 10.1128/iai.69.11.6813-6822.2001
Ngo, L. Y., Kasahara, S., Kumasaka, D. K., Knoblaugh, S. E., Jhingran, A., Hohl, T. M. (2014). Inflammatory monocytes mediate early and organ-specific innate defense during systemic candidiasis. J. Infect. Dis. 209 (1), 109–119. doi: 10.1093/infdis/jit413
Nishimoto, A. T., Sharma, C., Rogers, P. D. (2020). Molecular and genetic basis of azole antifungal resistance in the opportunistic pathogenic fungus Candida albicans. J. Antimicrob. Chemother. 75 (2), 257–270. doi: 10.1093/jac/dkz400
Nobbs, A. H., Vickerman, M. M., Jenkinson, H. F. (2010). Heterologous expression of Candida albicans cell wall-associated adhesins in Saccharomyces cerevisiae Reveals differential specificities in adherence and biofilm formation and in binding oral Streptococcus gordonii. Eukaryot Cell 9 (10), 1622–1634. doi: 10.1128/ec.00103-10
Nobile, C. J., Andes, D. R., Nett, J. E., Smith, F. J., Yue, F., Phan, Q. T., et al. (2006a). Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PloS Pathog. 2 (7), e63. doi: 10.1371/journal.ppat.0020063
Nobile, C. J., Nett, J. E., Andes, D. R., Mitchell, A. P. (2006b). Function of Candida albicans adhesin Hwp1 in biofilm formation. Eukaryot Cell 5 (10), 1604–1610. doi: 10.1128/ec.00194-06
Oh, S. H., Martin-Yken, H., Coleman, D. A., Dague, E., Hoyer, L. L. (2022). Development and use of a monoclonal antibody specific for the candida albicans cell-surface protein hwp1. Front. Cell Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.907453
Pachl, J., Svoboda, P., Jacobs, F., Vandewoude, K., van der Hoven, B., Spronk, P., et al. (2006). A randomized, blinded, multicenter trial of lipid-associated amphotericin B alone versus in combination with an antibody-based inhibitor of heat shock protein 90 in patients with invasive candidiasis. Clin. Infect. Dis. 42 (10), 1404–1413. doi: 10.1086/503428
Pappas, P. G., Kauffman, C. A., Andes, D. R., Clancy, C. J., Marr, K. A., Ostrosky-Zeichner, L., et al. (2016). Clinical practice guideline for the management of candidiasis: 2016 update by the infectious diseases society of america. Clin. Infect. Dis. 62 (4), e1–50. doi: 10.1093/cid/civ933
Pathakumari, B., Liang, G., Liu, W. (2020). Immune defence to invasive fungal infections: A comprehensive review. BioMed. Pharmacother. 130, 110550. doi: 10.1016/j.biopha.2020.110550
Perlin, D. S. (2011). Current perspectives on echinocandin class drugs. Future Microbiol. 6 (4), 441–457. doi: 10.2217/fmb.11.19
Peters, B. M., Ovchinnikova, E. S., Krom, B. P., Schlecht, L. M., Zhou, H., Hoyer, L. L., et al. (2012). Staphylococcus aureus adherence to Candida albicans hyphae is mediated by the hyphal adhesin Als3p. Microbiol. (Reading) 158 (Pt 12), 2975–2986. doi: 10.1099/mic.0.062109-0
Phan, Q. T., Fratti, R. A., Prasadarao, N. V., Edwards, J. E., Jr., Filler, S. G. (2005). N-cadherin mediates endocytosis of Candida albicans by endothelial cells. J. Biol. Chem. 280 (11), 10455–10461. doi: 10.1074/jbc.M412592200
Qadri, H., Shah, A. H., Alkhanani, M., Almilaibary, A., Mir, M. A. (2023). Immunotherapies against human bacterial and fungal infectious diseases: A review. Front. Med. (Lausanne) 10. doi: 10.3389/fmed.2023.1135541
Rahman, D., Mistry, M., Thavaraj, S., Naglik, J. R., Challacombe, S. J. (2012). Murine model of concurrent oral and vaginal Candida albicans colonisation. Methods Mol. Biol. 845, 527–535. doi: 10.1007/978-1-61779-539-8_38
Raska, M., Beláková, J., Wudattu, N. K., Kafková, L., Růzicková, K., Sebestová, M., et al. (2005). Comparison of protective effect of protein and DNA vaccines hsp90 in murine model of systemic candidiasis. Folia Microbiol. (Praha) 50 (1), 77–82. doi: 10.1007/bf02931297
Richardson, J. P., Moyes, D. L. (2015). Adaptive immune responses to Candida albicans infection. Virulence 6 (4), 327–337. doi: 10.1080/21505594.2015.1004977
Richter, K., Muschler, P., Hainzl, O., Buchner, J. (2001). Coordinated ATP hydrolysis by the Hsp90 dimer. J. Biol. Chem. 276 (36), 33689–33696. doi: 10.1074/jbc.M103832200
Richter, K., Soroka, J., Skalniak, L., Leskovar, A., Hessling, M., Reinstein, J., et al. (2008). Conserved conformational changes in the ATPase cycle of human Hsp90. J. Biol. Chem. 283 (26), 17757–17765. doi: 10.1074/jbc.M800540200
Robbins, N., Caplan, T., Cowen, L. E. (2017). Molecular evolution of antifungal drug resistance. Annu. Rev. Microbiol. 71, 753–775. doi: 10.1146/annurev-micro-030117-020345
Robbins, N., Wright, G. D., Cowen, L. E. (2016). Antifungal drugs: the current armamentarium and development of new agents. Microbiol. Spectr. 4 (5). doi: 10.1128/microbiolspec.FUNK-0002-2016
Romagnoli, G., Nisini, R., Chiani, P., Mariotti, S., Teloni, R., Cassone, A., et al. (2004). The interaction of human dendritic cells with yeast and germ-tube forms of Candida albicans leads to efficient fungal processing, dendritic cell maturation, and acquisition of a Th1 response-promoting function. J. Leukoc. Biol. 75 (1), 117–126. doi: 10.1189/jlb.0503226
Romeo, O., Criseo, G. (2008). First molecular method for discriminating between Candida africana, Candida albicans, and Candida dubliniensis by using hwp1 gene. Diagn. Microbiol. Infect. Dis. 62 (2), 230–233. doi: 10.1016/j.diagmicrobio.2008.05.014
Rosario-Colon, J., Eberle, K., Adams, A., Courville, E., Xin, H. (2021). Candida Cell-Surface-Specific Monoclonal Antibodies Protect Mice against Candida auris Invasive Infection. Int. J. Mol. Sci. 22 (11). doi: 10.3390/ijms22116162
Ruiz-Herrera, J., Iranzo, M., Elorza, M. V., Sentandreu, R., Mormeneo, S. (1995). Involvement of transglutaminase in the formation of covalent cross-links in the cell wall of Candida albicans. Arch. Microbiol. 164 (3), 186–193. doi: 10.1007/bf02529970
Satala, D., Satala, G., Karkowska-Kuleta, J., Bukowski, M., Kluza, A., Rapala-Kozik, M., et al. (2020). Structural insights into the interactions of candidal enolase with human vitronectin, fibronectin and plasminogen. Int. J. Mol. Sci. 21 (21). doi: 10.3390/ijms21217843
Schaller, M., Januschke, E., Schackert, C., Woerle, B., Korting, H. C. (2001). Different isoforms of secreted aspartyl proteinases (Sap) are expressed by Candida albicans during oral and cutaneous candidosis in vivo. J. Med. Microbiol. 50 (8), 743–747. doi: 10.1099/0022-1317-50-8-743
Schmidt, C. S., White, C. J., Ibrahim, A. S., Filler, S. G., Fu, Y., Yeaman, M. R., et al. (2012). NDV-3, a recombinant alum-adjuvanted vaccine for Candida and Staphylococcus aureus, is safe and immunogenic in healthy adults. Vaccine 30 (52), 7594–7600. doi: 10.1016/j.vaccine.2012.10.038
Segal, B. H., Kwon-Chung, J., Walsh, T. J., Klein, B. S., Battiwalla, M., Almyroudis, N. G., et al. (2006). Immunotherapy for fungal infections. Clin. Infect. Dis. 42 (4), 507–515. doi: 10.1086/499811
Sheppard, D. C., Yeaman, M. R., Welch, W. H., Phan, Q. T., Fu, Y., Ibrahim, A. S., et al. (2004). Functional and structural diversity in the Als protein family of Candida albicans. J. Biol. Chem. 279 (29), 30480–30489. doi: 10.1074/jbc.M401929200
Shiau, A. K., Harris, S. F., Southworth, D. R., Agard, D. A. (2006). Structural Analysis of E. coli hsp90 reveals dramatic nucleotide-dependent conformational rearrangements. Cell 127 (2), 329–340. doi: 10.1016/j.cell.2006.09.027
Shibasaki, S., Aoki, W., Nomura, T., Karasaki, M., Sewaki, T., Ueda, M. (2014). Evaluation of Mdh1 protein as an antigenic candidate for a vaccine against candidiasis. Biocontrol Sci. 19 (1), 51–55. doi: 10.4265/bio.19.51
Shibasaki, S., Karasaki, M., Aoki, W., Ueda, M. (2018). Molecular and physiological study of candida albicans by quantitative proteome analysis. Proteomes 6 (3). doi: 10.3390/proteomes6030034
Shukla, M., Chandley, P., Rohatgi, S. (2021). The role of B-cells and antibodies against candida vaccine antigens in invasive candidiasis. Vaccines (Basel) 9 (10). doi: 10.3390/vaccines9101159
Silva, R. C., Padovan, A. C., Pimenta, D. C., Ferreira, R. C., da Silva, C. V., Briones, M. R. (2014). Extracellular enolase of Candida albicans is involved in colonization of mammalian intestinal epithelium. Front. Cell Infect. Microbiol. 4. doi: 10.3389/fcimb.2014.00066
Spellberg, B. J., Ibrahim, A. S., Avanesian, V., Fu, Y., Myers, C., Phan, Q. T., et al. (2006). Efficacy of the anti-Candida rAls3p-N or rAls1p-N vaccines against disseminated and mucosal candidiasis. J. Infect. Dis. 194 (2), 256–260. doi: 10.1086/504691
Staab, J. F., Bahn, Y. S., Tai, C. H., Cook, P. F., Sundstrom, P. (2004). Expression of transglutaminase substrate activity on Candida albicans germ tubes through a coiled, disulfide-bonded N-terminal domain of Hwp1 requires C-terminal glycosylphosphatidylinositol modification. J. Biol. Chem. 279 (39), 40737–40747. doi: 10.1074/jbc.M406005200
Staab, J. F., Bradway, S. D., Fidel, P. L., Sundstrom, P. (1999). Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 283 (5407), 1535–1538. doi: 10.1126/science.283.5407.1535
Staab, J. F., Ferrer, C. A., Sundstrom, P. (1996). Developmental expression of a tandemly repeated, proline-and glutamine-rich amino acid motif on hyphal surfaces on Candida albicans. J. Biol. Chem. 271 (11), 6298–6305. doi: 10.1074/jbc.271.11.6298
Sundstrom, P., Aliaga, G. R. (1994). A subset of proteins found in culture supernatants of Candida albicans includes the abundant, immunodominant, glycolytic enzyme enolase. J. Infect. Dis. 169 (2), 452–456. doi: 10.1093/infdis/169.2.452
Taipale, M., Krykbaeva, I., Koeva, M., Kayatekin, C., Westover, K. D., Karras, G. I., et al. (2012). Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell 150 (5), 987–1001. doi: 10.1016/j.cell.2012.06.047
Taylor, B. N., Hannemann, H., Sehnal, M., Biesemeier, A., Schweizer, A., Röllinghoff, M., et al. (2005). Induction of SAP7 correlates with virulence in an intravenous infection model of candidiasis but not in a vaginal infection model in mice. Infect. Immun. 73 (10), 7061–7063. doi: 10.1128/iai.73.10.7061-7063.2005
Togni, G., Sanglard, D., Quadroni, M., Foundling, S. I., Monod, M. (1996). Acid proteinase secreted by Candida tropicalis: functional analysis of preproregion cleavages in C. tropicalis and Saccharomyces cerevisiae. Microbiol. (Reading) 142 (Pt 3), 493–503. doi: 10.1099/13500872-142-3-493
Tso, G. H. W., Reales-Calderon, J. A., Pavelka, N. (2018). The elusive anti-candida vaccine: lessons from the past and opportunities for the future. Front. Immunol. 9. doi: 10.3389/fimmu.2018.00897
Vilanova, M., Teixeira, L., Caramalho, I., Torrado, E., Marques, A., Madureira, P., et al. (2004). Protection against systemic candidiasis in mice immunized with secreted aspartic proteinase 2. Immunology 111 (3), 334–342. doi: 10.1111/j.1365-2567.2004.01819.x
Voigt, J., Hünniger, K., Bouzani, M., Jacobsen, I. D., Barz, D., Hube, B., et al. (2014). Human natural killer cells acting as phagocytes against Candida albicans and mounting an inflammatory response that modulates neutrophil antifungal activity. J. Infect. Dis. 209 (4), 616–626. doi: 10.1093/infdis/jit574
von Heijne, G. (1985). Signal sequences. The limits of variation. J. Mol. Biol. 184 (1), 99–105. doi: 10.1016/0022-2836(85)90046-4
Vonk, A. G., Netea, M. G., van der Meer, J. W., Kullberg, B. J. (2006). Host defence against disseminated Candida albicans infection and implications for antifungal immunotherapy. Expert Opin. Biol. Ther. 6 (9), 891–903. doi: 10.1517/14712598.6.9.891
Wächtler, B., Citiulo, F., Jablonowski, N., Förster, S., Dalle, F., Schaller, M., et al. (2012). Candida albicans-epithelial interactions: dissecting the roles of active penetration, induced endocytosis and host factors on the infection process. PloS One 7 (5), e36952. doi: 10.1371/journal.pone.0036952
Wang, Y., Su, Q., Dong, S., Shi, H., Gao, X., Wang, L. (2014). Hybrid phage displaying SLAQVKYTSASSI induces protection against Candida albicans challenge in BALB/c mice. Hum. Vaccin Immunother. 10 (4), 1057–1063. doi: 10.4161/hv.27714
Wang, G., Sun, M., Fang, J., Yang, Q., Tong, H., Wang, L. (2006). Protective immune responses against systemic candidiasis mediated by phage-displayed specific epitope of Candida albicans heat shock protein 90 in C57BL/6J mice. Vaccine 24 (35-36), 6065–6073. doi: 10.1016/j.vaccine.2006.05.022
Weinberg, E. D. (1999). Iron loading and disease surveillance. Emerg. Infect. Dis. 5 (3), 346–352. doi: 10.3201/eid0503.990305
White, T. C., Miyasaki, S. H., Agabian, N. (1993). Three distinct secreted aspartyl proteinases in Candida albicans. J. Bacteriol 175 (19), 6126–6133. doi: 10.1128/jb.175.19.6126-6133.1993
Williams, D. W., Jordan, R. P., Wei, X. Q., Alves, C. T., Wise, M. P., Wilson, M. J., et al. (2013). Interactions of Candida albicans with host epithelial surfaces. J. Oral. Microbiol. 5. doi: 10.3402/jom.v5i0.22434
Williamson, M. P. (1994). The structure and function of proline-rich regions in proteins. Biochem. J. 297 (Pt 2), 249–260. doi: 10.1042/bj2970249
Wu, H., Downs, D., Ghosh, K., Ghosh, A. K., Staib, P., Monod, M., et al. (2013). Candida albicans secreted aspartic proteases 4-6 induce apoptosis of epithelial cells by a novel Trojan horse mechanism. FASEB J. 27 (6), 2132–2144. doi: 10.1096/fj.12-214353
Xin, H., Dziadek, S., Bundle, D. R., Cutler, J. E. (2008). Synthetic glycopeptide vaccines combining beta-mannan and peptide epitopes induce protection against candidiasis. Proc. Natl. Acad. Sci. U.S.A. 105 (36), 13526–13531. doi: 10.1073/pnas.0803195105
Yang, H., Han, S., Zhao, D., Wang, G. (2014). Adjuvant effect of polysaccharide from fruits of Physalis alkekengi L. @ in DNA vaccine against systemic candidiasis. Carbohydr Polym 109, 77–84. doi: 10.1016/j.carbpol.2014.03.054
Yang, H., Tsang, P. C. S., Pow, E. H. N., Lam, O. L. T., Tsang, P. W. (2020). Potential role of Candida albicans secreted aspartic protease 9 in serum induced-hyphal formation and interaction with oral epithelial cells. Microb. Pathog. 139, 103896. doi: 10.1016/j.micpath.2019.103896
Yeaman, M. R., Filler, S. G., Chaili, S., Barr, K., Wang, H., Kupferwasser, D., et al. (2014). Mechanisms of NDV-3 vaccine efficacy in MRSA skin versus invasive infection. Proc. Natl. Acad. Sci. U.S.A. 111 (51), E5555–E5563. doi: 10.1073/pnas.1415610111
Zaugg, C., Borg-Von Zepelin, M., Reichard, U., Sanglard, D., Monod, M. (2001). Secreted aspartic proteinase family of Candida tropicalis. Infect. Immun. 69 (1), 405–412. doi: 10.1128/iai.69.1.405-412.2001
Zhao, X., Oh, S. H., Cheng, G., Green, C. B., Nuessen, J. A., Yeater, K., et al. (2004). ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesin; functional comparisons between Als3p and Als1p. Microbiol. (Reading) 150 (Pt 7), 2415–2428. doi: 10.1099/mic.0.26943-0
Keywords: candidiasis, immunotherapy, antibody, vaccine, Candida albicans
Citation: Feng Z, Lu H and Jiang Y (2024) Promising immunotherapeutic targets for treating candidiasis. Front. Cell. Infect. Microbiol. 14:1339501. doi: 10.3389/fcimb.2024.1339501
Received: 16 November 2023; Accepted: 29 January 2024;
Published: 09 February 2024.
Edited by:
Michal Adam Olszewski, University of Michigan, United StatesReviewed by:
Eman A. Gouda M. Youssef, Lundquist Institute for Biomedical Innovation, United StatesWenjie Fang, Shanghai Changzheng Hospital, China
Kristie Goughenour, University of Michigan, United States
Copyright © 2024 Feng, Lu and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Lu, bHVodWkyMDE5QHRvbmdqaS5lZHUuY24=; Yuanying Jiang, amlhbmd5eUB0b25namkuZWR1LmNu
 Zhe Feng
Zhe Feng Hui Lu
Hui Lu Yuanying Jiang
Yuanying Jiang