
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol., 06 February 2024
Sec. Molecular Bacterial Pathogenesis
Volume 14 - 2024 | https://doi.org/10.3389/fcimb.2024.1339131
Streptococcus equi subsp. zooepidemicus (SEZ) is an opportunistic pathogen of both humans and animals. Quorum sensing (QS) plays an important role in the regulation of bacterial group behaviors. The aim of this study was to characterize the LuxS in SEZ and evaluate its impact on biofilm formation, pathogenesis and gene expression. The wild-type SEZ and its LuxS mutant (ΔluxS) were examined for growth, biofilm formation, virulence factors, and transcriptomic profiles. Our results showed that LuxS deficiency did not affect SEZ hemolytic activity, adhesion or capsule production. For biofilm assay demonstrated that mutation in the luxS gene significantly enhances biofilm formation, produced a denser biofilm and attached to a glass surface. RAW264.7 cell infection indicated that ΔluxS promoted macrophage apoptosis and pro-inflammatory responses. In mice infection, there was no significant difference in mortality between SEZ and ΔluxS. However, the bacterial load in the spleen of mice infected with ΔluxS was significantly higher than in those infected with SEZ. And the pathological analysis further indicated that spleen damage was more severe in the ΔluxS group. Moreover, transcriptomics analysis revealed significant alterations in carbon metabolism, RNA binding and stress response genes in ΔluxS. In summary, this study provides the first evidence of AI-2/LuxS QS system in SEZ and reveals its regulatory effects on biofilm formation, pathogenicity and gene expression.
Streptococcus equi subspecies zooepidemicus (SEZ) is a Gram positive, β-hemolytic, opportunistic pathogen that commonly resides in the upper respiratory and reproductive tracts of horses (Timoney, 2004). SEZ can also infect a broad range of mammalian hosts including pigs, ruminants, cats and dogs, causing severe disease characterized by pneumonia, septicemia and meningitis (Pelkonen et al., 2013; Jie et al., 2017; Chen et al., 2020)Virulent strains of SEZ have a history of causing severe infections in pigs, including a major outbreak in China in the 1970s that resulted in over 300,000 fatalities (Wei et al., 2009). More recently, SEZ infections with high mortality have emerged in North American swine, highlighting the threat of this bacterium (Chen et al., 2020; Song et al., 2022). Additionally, SEZ also poses zoonotic risks, causing potentially fatal diseases in humans, such as sepsis, meningitis, endocarditis, and septic arthritis (Kerdsin et al., 2022).
Quorum sensing (QS) is a common phenomenon in bacteria that allows them to communicate (Xavier and Bassler, 2003). It involves bacteria producing and detecting chemical signals in order to trigger specific responses across the bacterial population (Schauder et al., 2001). QS regulates genes involved in group behaviors that require coordination among bacteria to work effectively (Abisado et al., 2018; Wang et al., 2022). These coordinated behaviors include bioluminescence, sporulation, antibiotic production, biofilm formation, and secretion of virulence factors (Whiteley et al., 2017; Abisado et al., 2018). QS relies on bacteria secreting pheromones that accumulate in the surrounding environment. When pheromones reach a high enough concentration, they are detected by receptor proteins on the surface of bacteria. These receptor proteins activate signaling pathways inside the cell through a series of enzymatic reactions, leading to phosphorylation and activation of regulatory proteins (Jimenez and Federle, 2014). This changes the expression of specific genes, allowing bacteria to mount unified, population-level responses. By using QS, bacteria can act in a coordinated manner to perform tasks that require group effort. The QS systems identified in Streptococci can be classified into four major types: the regulator gene of glucosyltransferase (Rgg) systems, Sil systems, lantibiotic systems, and AI-2/LuxS systems (Chaussee et al., 2004; Jimenez and Federle, 2014). The Rgg family represents a group of conserved transcription factors that can sense small peptide signals and accordingly modulate downstream gene expression. In SEZ, Rgg systems have been characterized to control biofilm formation and capsular polysaccharide production (Xie et al., 2019).
The autoinducer-2 (AI-2) molecule, first found to regulate bioluminescence in Vibrio harveyi, is widely synthesized by the LuxS enzyme in many Gram-negative and Gram-positive bacteria, thus being proposed as a universal signal that enables interspecies cell-cell communication (Miller and Bassler, 2001). With the increasing emergence of antibiotic resistance amongst infectious bacteria, there is a dire need for alternative therapeutic strategies to control pathogens. Many studies have emphasized the functional characteristics of LuxS quorum sensing and its potential significance as a therapeutic target (Defoirdt, 2018). Currently, there are no reports on AI-2/LuxS quorum sensing in SEZ. Therefore, this study aims to clarify the role of the quorum sensing system-related gene luxS in SEZ by comparing and assessing its characteristics in various aspects between the wild-type SEZ strain and the LuxS deletion mutant strain.
We successfully constructed a mutation strain ΔluxS, and complemented strain C-luxS. We found that the absence of LuxS does not affect the growth of SEZ under 37°C conditions (Figure 1A). Then, we used the Vibrio harveyi BB170 as a reporter strain that responds to AI-2 by inducing luminescence (Figure 1B). The ΔluxS strain exhibited the lowest luminescence at 2 hours, indicating the functionality of the AI-2/LuxS quorum sensing system in SEZ. We compared the ability of SEZ, ΔluxS, and the complemented strain C-luxS to adhere to PK-15 cells. These results show no significant differences in adhesion among the three groups (Figure 1C). Furthermore, hemolysis assays revealed no differences among the three groups (Figure 1D).
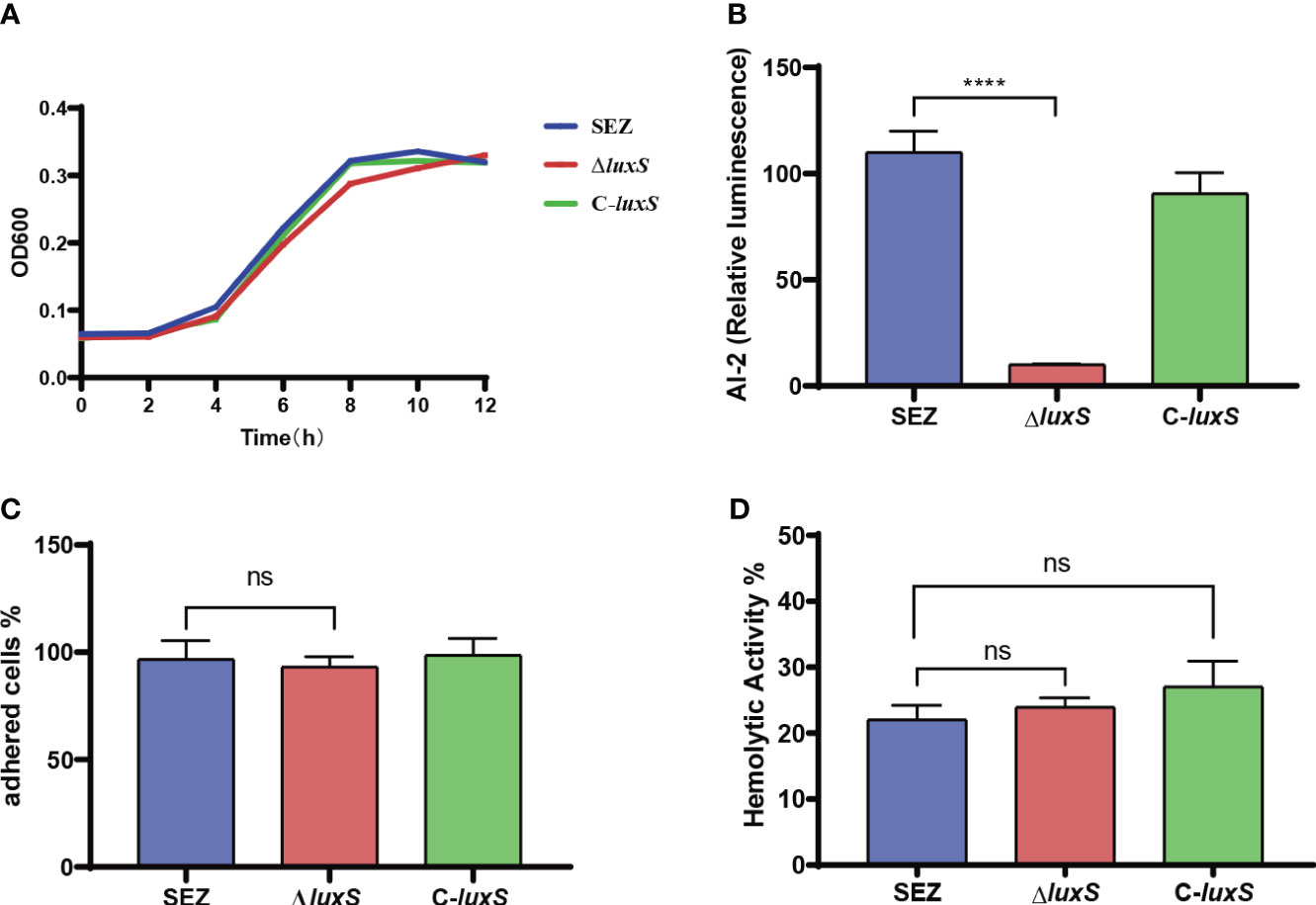
Figure 1 Characterization of the ΔluxS mutant strain. (A) Growth curves of the WT (wild type), ΔluxS (LuxS mutant), and C-luxS (complemented LuxS) strains in THB medium, monitored by measuring OD600. (B) AI-2 activity, presented as relative luminescence compared to the negative control. (C) Adherence of SEZ strains to PK-15 cells, assessed at 2 h post-inoculation. (D) Hemolytic activities of the SEZ strains, quantified using OD540 measurements in 96-well plates. Data represent mean values from three independent experiments, ns as not significant, with statistical significance denoted as ****p<0.0001.
We investigated whether LuxS has an impact on hyaluronic acid (HA) production. We first assessed hydrophobicity in SEZ strains since HA is highly hydrophilic. The results indicated that WT SEZ, ΔluxS, and C-luxS strains showed no difference in hydrophobicity and the three strains exhibited very low hydrophobicity compared to the SEZ capsule-deficient strain ΔhasB (Figure 2A). Lastly, the HA production in the three groups of strains at 10 hours showed no significant differences (Figure 2B).
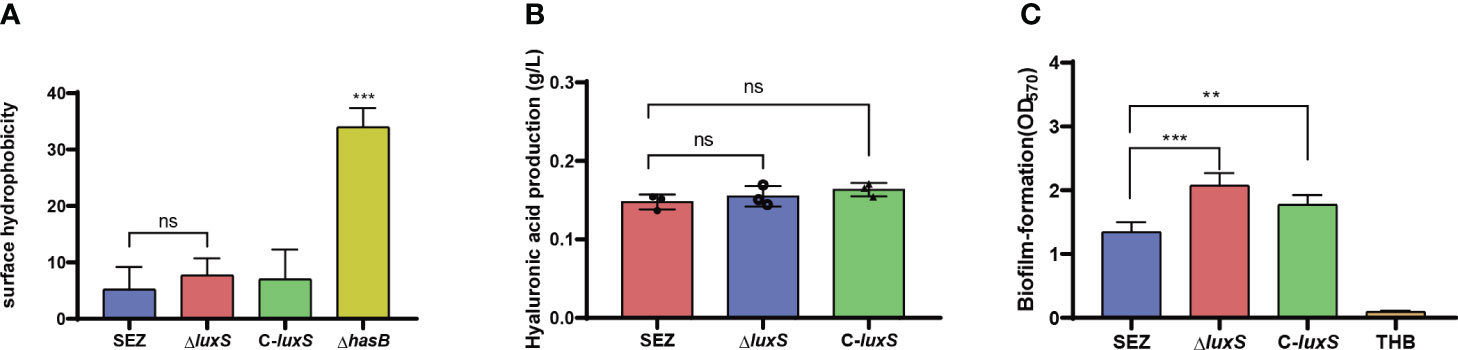
Figure 2 (A) Evaluation of surface hydrophobicity of SEZ, ΔluxS, C-luxS and capsular-deficient (ΔhasB) strains. (B) Measurement of HA production in SEZ, ΔluxS, and C-luxS strains at 10 (h) (C) Biofilm formation crystal violet assay comparing the WT-type SEZ, the ΔluxS strain and C-luxS cultured for 24 (h) The presented data are mean values derived from three independent experiments, ns as not significant, with statistical significance indicated as **p < 0.01,***p <0.001.
To investigate whether LuxS influences biofilm formation in SEZ, we initially conducted crystal violet staining using microtiter plates (Figure 2C). The ΔluxS mutant of SEZ exhibited a significantly increased biomass compared to wild-type strain. In the case of the complemented C-luxS strain, the biofilm content showed a reduction but still exhibited a significant difference compared to SEZ.
We further visualized biofilm formation through SEM and CLSM (Figure 3A). The SEM results allowed observation of the biofilm adhered to the glass surface, revealing that the biofilm formed by the ΔluxS mutant was comparatively denser than SEZ, while SEZ exhibited a more loosely formed biofilm.
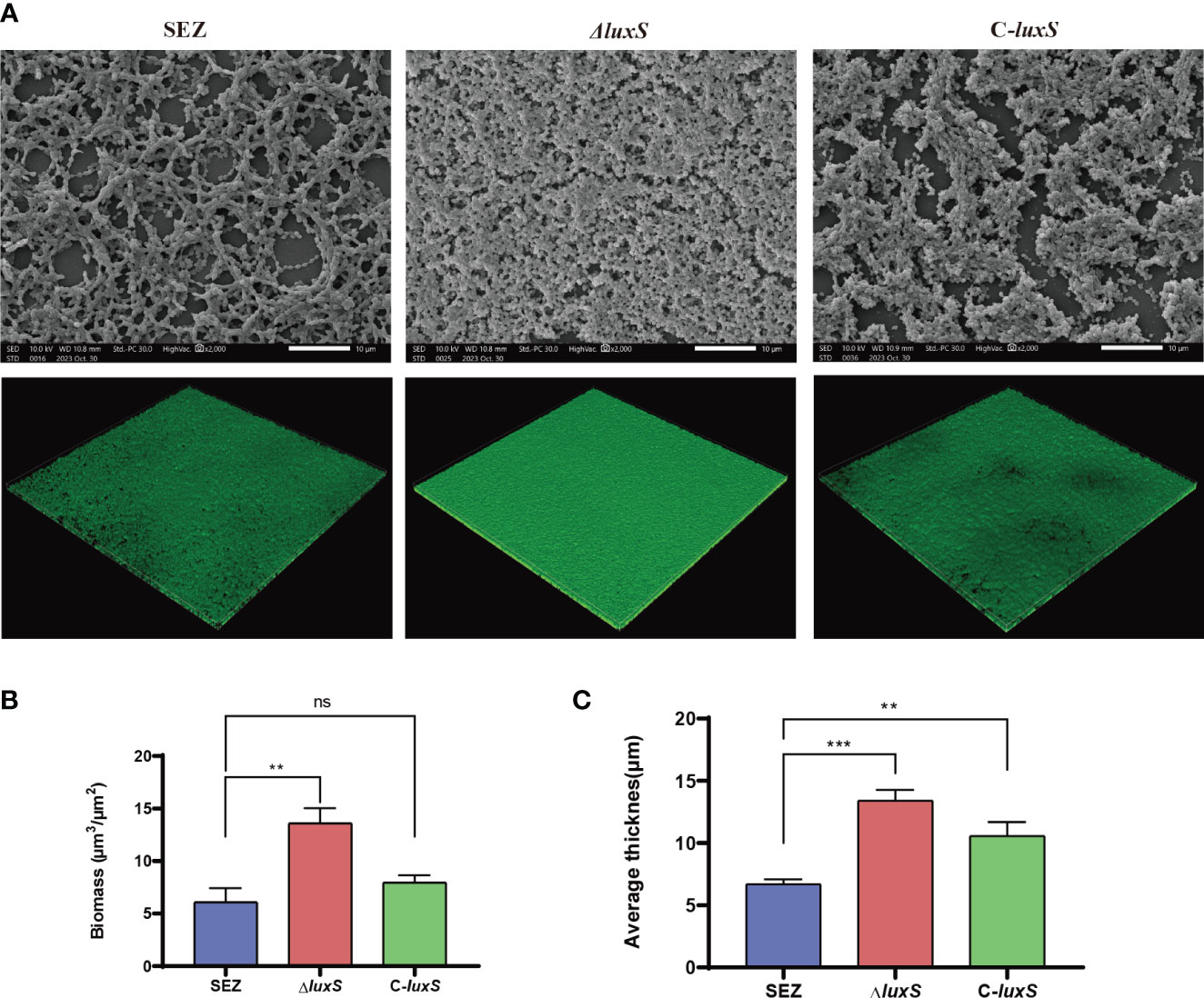
Figure 3 Scanning electron microscopy (SEM) and confocal laser scanning microscopy (CLSM) images of SEZ strains biofilm-formation after 48 h of incubation on glass coverslips. (A) SEM and CSLM image. (B, C) CSLM z-stack of biofilm thickness and biomass were calculated using COMSTAT 2(Vorregaard, 2008). The numbers we show are averages from three separate experiments, with **p < 0.01, or ***p < 0.001 showing where differences are significant, ns as not significant.
Then, we visualized SEZ, ΔluxS, and C-luxS biofilms using CLSM, converting them into z-stacks for analysis. The biofilms were characterized for structural features, thickness, and bio-volume (Figure 3B, C). The ΔluxS strain developed a thicker mature biofilm. The application of Comstat2 for biomass and average thickness determination revealed that ΔluxS was significantly higher than the wild-type SEZ strain. Collectively, these results confirm that the LuxS regulates biofilm formation of SEZ.
Previous research has indicated that the quorum sensing system plays a significant role in immune evasion (Cao et al., 2020). To further investigate this, we conducted infection experiments using mouse macrophage RAW264.7 cells. Flow cytometry analysis revealed a significant increase in apoptosis levels in the ΔluxS mutant strain compared to the SEZ-infected cells (Figures 4A, B). The ΔluxS induce higher TNF-α secretion compared to SEZ, however the complemented C-luxS strain shows no difference compared to the ΔluxS (Figure 4B). RT-PCR analysis of pro-inflammatory cytokines, including IL-1β, IL-6, IL-10, IL-18, TNF-α, and the transcription factor NF-kB, demonstrated higher transcription levels in ΔluxS (Figure 4C, D).
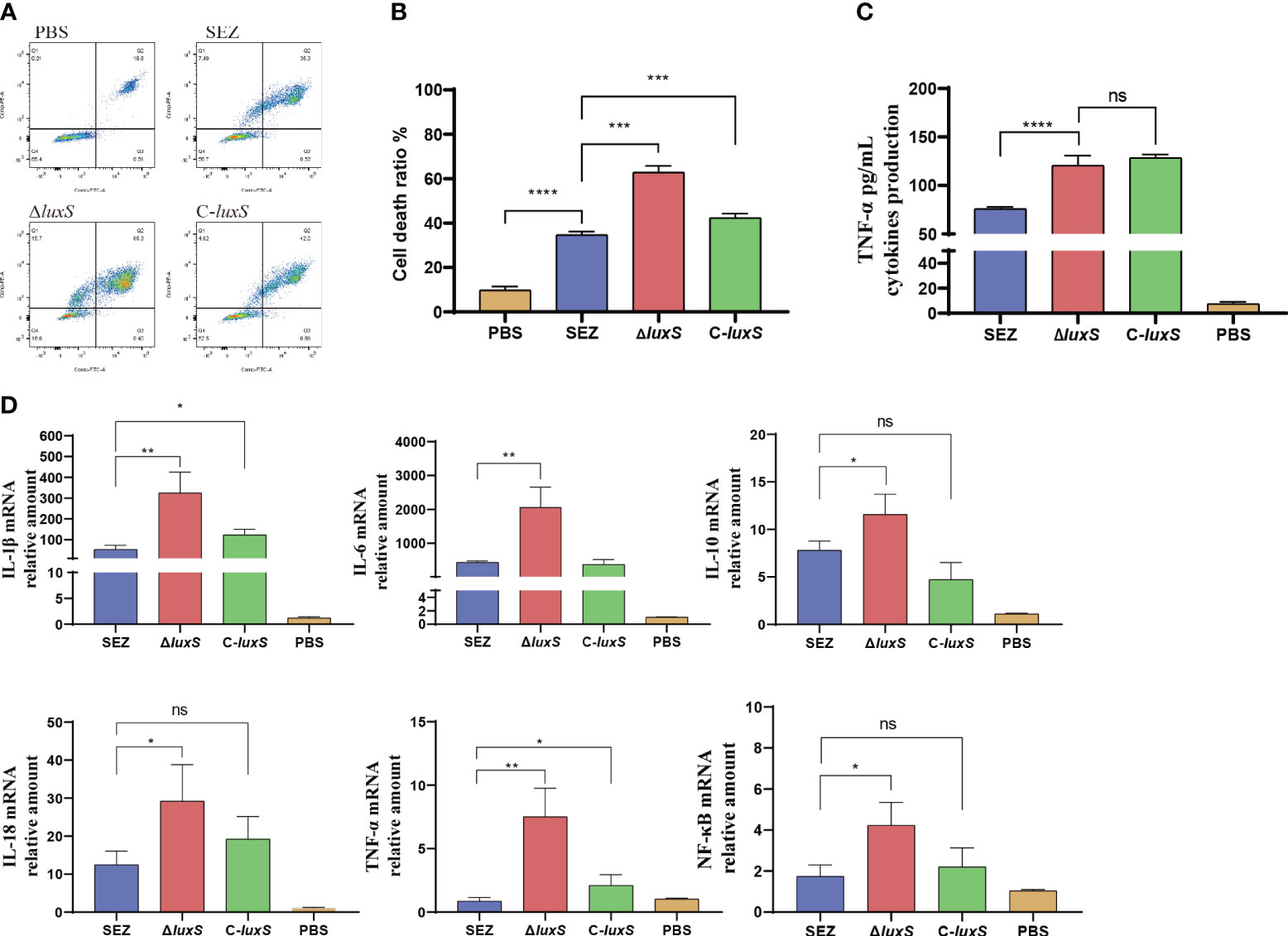
Figure 4 The deletion of luxS increases cell death and inflammatory responses in SEZ-Infected Macrophages. WT-type SEZ, ΔluxS, and C-luxS were cocultured with RAW264.7 macrophages for 12 h with MOI 1. (A, B) The apoptosis level of RAW264.7 macrophages was analyzed by flow cytometry. (C) TNF-α secretion in cell supernatant. (D) The mRNA expression levels of IL-1β, IL-6, IL-10, IL-18, TNF-α and transcription factor NF-κB. The data shown are average values from three independent experiments, with *p < 0.05, **p < 0.01, ***p < 0.001 or ****p < 0.0001 showing where differences are significant, ns as not significant.
To investigate whether the LuxS affects the virulence of SEZ, we infected three groups of mice (n=10 for each group) via the intraperitoneal route. The final mortality of mice infected with SEZ and C-luxS was 80%, while ΔluxS group was 70% (Figure 5A). Then, we evaluated the bacterial burden in different organs at 24 hours post-infection. Surprisingly, the ΔluxS strain exhibited higher bacterial loads in the spleen compared to the SEZ wild type. There were no significant differences in bacterial loads between the ΔluxS strain and the SEZ wild type in the lungs, livers and kidneys (Figure 5B). The histopathological with H&E showed notable atrophy in the splenic nodules of the ΔluxS and C-luxS groups (Figure 6). The differentiation between white and red pulp was not distinct, particularly in the ΔluxS group, which showed more severe effects. In addition, the SEZ, ΔluxS, and C-luxS groups exhibited various levels of lymphocyte deformation and necrosis. Around these necrotic areas and within the red pulp, there was a significant infiltration of inflammatory cells, mainly neutrophils, with the ΔluxS group experiencing the most severe infiltration. The data suggest that while LuxS might not significantly influence the mortality rates in mice, the tissue-specific phenomena observed highlight a complex and nuanced role for LuxS in the pathogenesis and host-pathogen of SEZ.
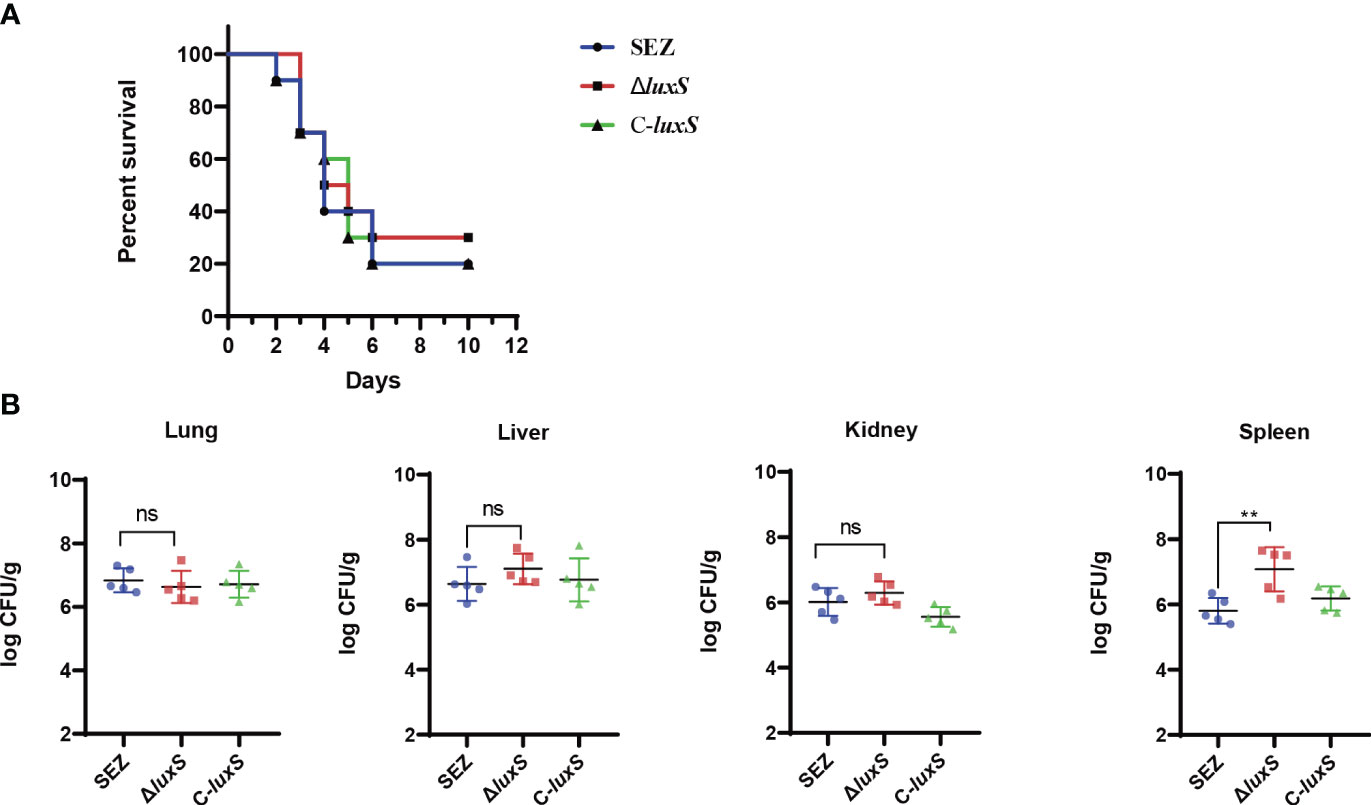
Figure 5 Survival rate and bacterial burden in mice infected WT SEZ, ΔluxS, C-luxS strains of 1×106 CFU. (A) The survival rate of mice, monitored for death 12d post-infection. n=10 (B) Bacterial burden in lungs, livers, kidneys and spleens of the infected mice (n=5 per group). 24 h post-infection. The bacterial loads were expressed as the number of CFU/g of tissue. (*p < 0.05; ns, not significant).
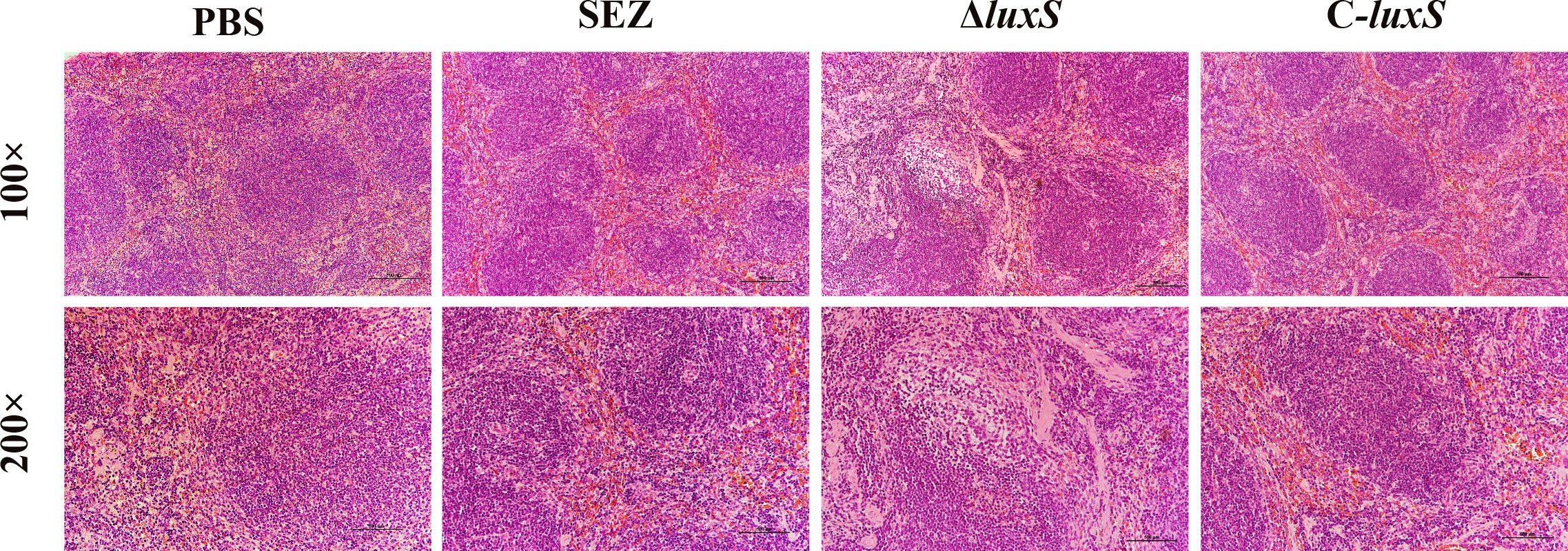
Figure 6 Histopathological changes observed in the spleens of mice (n=3 per group) Infected with SEZ, ΔluxS, and C-luxS strains, as revealed by H&E staining.
A comparative transcriptomic analysis was conducted between the SEZ wild type and the ΔluxS mutant strain, providing a comprehensive insight into the role of the luxS gene in SEZ. Among a total of 242 genes displaying differential expression in the ΔluxS strain compared to SEZ, 158 genes were upregulated, while 84 genes were downregulated, with an adjusted p-value (false-discovery rate [FDR]) of ≤0.05 (Figure 7A, Supplementary Table 2).
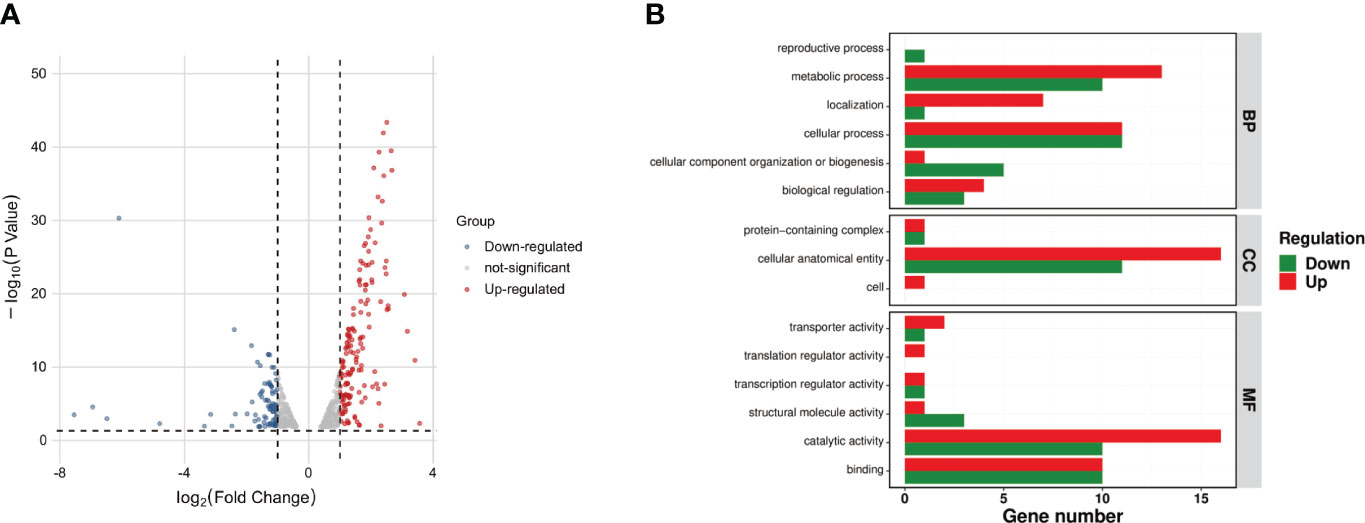
Figure 7 Comparative transcriptomic analysis between the SEZ wild-type and the LuxS mutant strain. (A)Volcano plot of differentially expressed genes between ΔluxS and WT SEZ groups. Red dots represent up-regulated genes; blue dots represent down-regulated genes; and gray dots represent non-differentially expressed genes. Among these, 158 genes were upregulated, and 84 genes were downregulated (FDR < 0.05); (B) Gene Ontology (GO) annotation analysis of differentially expressed genes. With up-regulated genes represented in red and down-regulated genes in green.
Gene ontology (GO) annotation analysis (Ashburner et al., 2000) revealed a downregulation of many genes associated with metabolic and cellular processes, while most of the downregulated genes were linked to biosynthesis, cellular localization, and structural functions in the ΔluxS mutant strain when compared to the SEZ (Figure 7B).
To provide a clearer understanding the differentially expressed genes in the ΔluxS mutant, we constructed a network (Figure 8). Cluster analysis showed that the majority of genes formed clusters related to carbohydrate metabolism, transport, iron-sulfur cluster and selenocompound metabolism, as well as the maintenance of DNA repeat elements and antiviral defense, all of which were upregulated. Downregulated genes were clustered into modules associated with ribonucleotide metabolism, peptidoglycan biosynthesis, DNA replication, and repair.
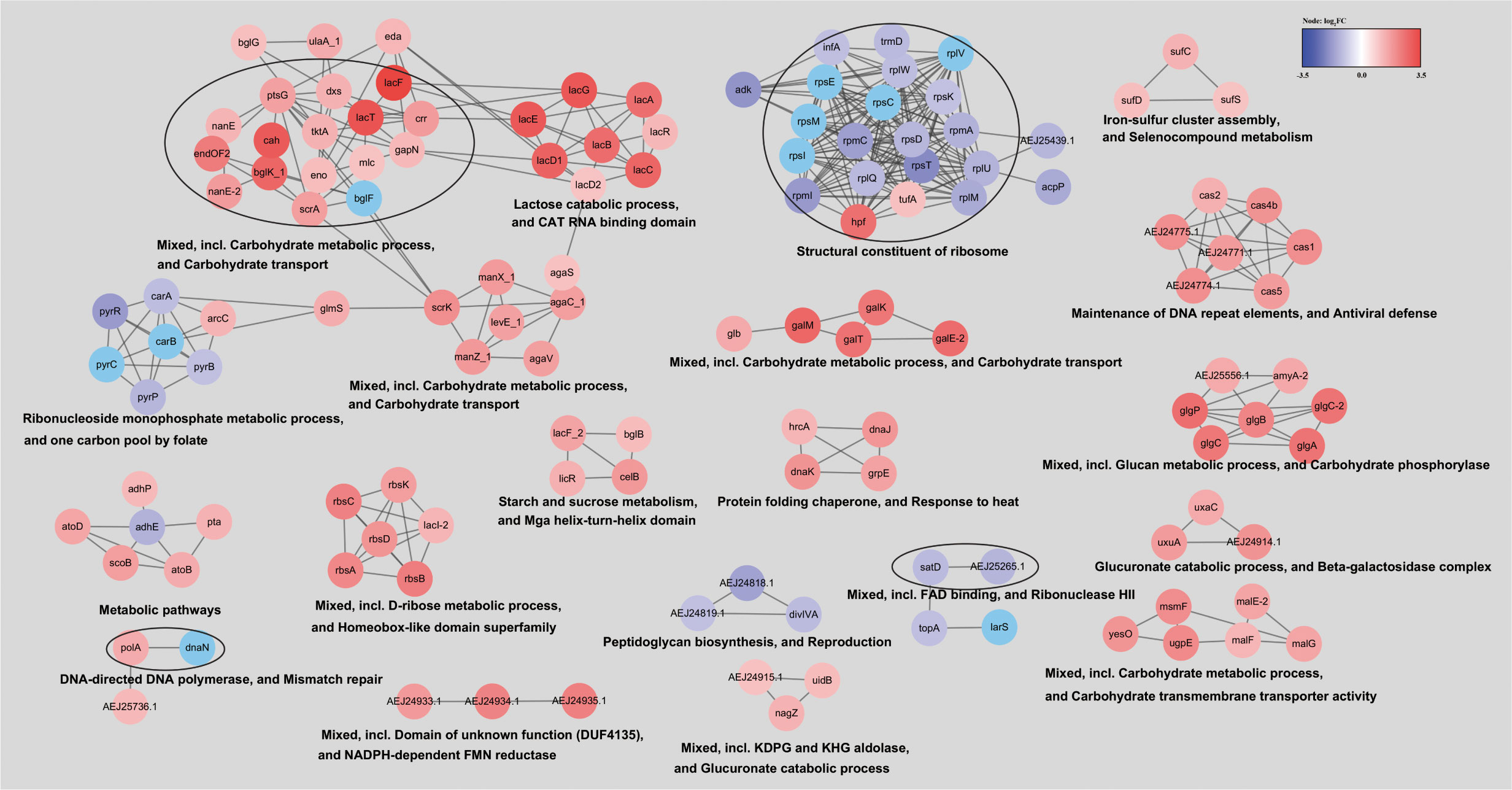
Figure 8 Network of differentially expressed genes in the ΔluxS mutant strain. To construct the network of differentially expressed genes, they were searched in a string database for SEZ ATCC 35246 organism with a 0.7 confidence (score) cutoff and a maximum of 10 additional interactors. Clustered using the MCL Cluster algorithm implemented in the Cytoscape plugin clusterMaker (Morris et al., 2011) to determine clusters and functional interactions. For each cluster, the functional enrichment was used to obtain enriched terms (available from the stringApp).
As a significant pathogen, SEZ has the potential to infect various mammals, including humans. Outbreaks in numerous animal species have been documented, and sporadic infections in humans have also been reported (Kuusi et al., 2006; Hau et al., 2022; Bosica et al., 2023; Chen et al., 2023). The exploration of immune proteins and infection mechanisms has long been a focal point of SEZ research, aiming to discover novel therapeutic approaches (Wei et al., 2013; Xie et al., 2018; Li et al., 2022; Xu et al., 2022). Many studies have shown that QS systems are responsible for group behavior, cell luminescence, antibiotic resistance, plasmid transfer, virulence factor gene expression, and biofilm formation (Waters and Bassler, 2005). Quorum sensing systems represent one of the critical targets for potential bacterial infection therapies (Bhardwaj et al., 2013; Defoirdt, 2018).
The LuxS quorum sensing system is one of the most extensively studied QS systems to date. The presence of this conserved quorum-sensing system remained unknown in SEZ. In this study, we have successfully confirmed the existence of an AI-2/LuxS quorum-sensing system in SEZ, which is capable of inducing luminescence in Vibrio harveyi BB170 as a reporter strain. The absence of LuxS in SEZ did not result in alterations in cell adhesion or hemolytic activity. This is primarily attributed to the fact that transcription of the hemolytic gene and the phenotypes of cell capsule remained unaltered when compared to the wild-type SEZ.
The primary component of capsular polysaccharides (CPS) in SEZ is hyaluronic acid (HA), which is produced through the hyaluronic acid synthesis (has) operon containing five genes: hasA, hasB, hasC, glmU, and pgi (Blank et al., 2008; Xu et al., 2016). Encapsulation by HA is recognized as a virulence factor for Streptococcus pyogenes, as HA can aid in protecting cells from phagocytosis and mediate the adherence of cells to host tissues(Flores et al., 2012). In SEZ, CPS is also an important virulence factor, influencing cell adhesion and phagocytosis. Our data indicate that the LuxS mutant did not alter the production of HA in SEZ.
Pathogenic bacteria exhibit an ability to form biofilms, which provide advantages for their survival against diverse stresses as well as their pathogenicity in host environments (Solano et al., 2014). The general process of biofilm formation begins with planktonic bacteria initially attaching to a surface. The bacteria then produce exopolysaccharides (EPS) that wrap around the cells, forming microcolonies. These eventually aggregate into a mature biofilm (Stoodley et al., 2002). Finally, the biofilm is dispersed to release planktonic bacteria (Kim et al., 2013). Biofilm formation is considered a virulence factor for many pathogenic bacteria. In Streptococcus suis, biofilms play a key role in causing persistent infections (Yi et al., 2016). In the present study, we conducted biofilm assays using crystal violet staining, SEM, and CLSM in a THB medium supplemented with fibrinogen. The ΔluxS mutant strain exhibited a thicker and more tightly formed biofilm, while the C-luxS strain, although showing a reduction, still displayed a discernible difference compared to the wild-type SEZ strain. In summary, these results collectively indicate that the absence of LuxS enhances the ability to form biofilms.
Bacteria employ a variety of strategies to evade host recognition and overcome the many challenges presented by the immune system, and quorum sensing systems are one such survival mechanism. For instance, the activation of the Rgg2/Rgg3 quorum sensing system in Streptococcus pyogenes can suppress NF-κB activity and reduce pro-inflammatory cytokine production in macrophages (Rahbari et al., 2021). Another study suggests that LuxS plays a role in virulence-associated traits and the immunological evasion of Streptococcus agalactiae (Cao et al., 2020). Our data indicates that ΔluxS mutation enhances some pro-inflammatory responses and promotes apoptosis in infected macrophages. Elevated pro-inflammatory cytokine expression is generally favorable for the host in clearing bacterial infections. Thought, an excessive inflammatory response can also lead to tissue damage. These results suggest that LuxS has an impact on host innate immune responses, needed further investigation into its specific mechanisms. The luxS gene has been previously recognized as a regulator of virulence, promoting it in pathogens like Streptococcus suis (Cao et al., 2011), Haemophilus parasuis (Zhang et al., 2019), Edwardsiella piscicida (Sun et al., 2020), and others (Yadav et al., 2018). However, its absence in Haemophilus influenzae exacerbates inflammatory responses and bacterial proliferation (Daines et al., 2005). In the case of Staphylococcus epidermidis, LuxS deletion paradoxically increases virulence in a rat model (Xu et al., 2006). Furthermore, in certain bacterial species such as Porphyromonas gingivalis, Bacillus anthracis and Shigella flexneri, LuxS does not appear to be essential for virulence (Day and Maurelli, 2001; Burgess et al., 2002; Bozue et al., 2012). Our mice experiments reveal the ΔluxS mutant did not significantly affect overall mortality in the mice, suggesting that it may not be an essential factor for the pathogenicity of SEZ in terms of survival outcomes. Instead, ΔluxS mutant strain had a notable influence on spleen bacterial burden. This organ-specific effect could be attributed to excessive tissue inflammation. To confirm this, we conducted further infections in mice and analyzed the pathological changes in their spleens, ΔluxS mutant showing the most significant pathologic changes. Incorporating the findings from our study, this further underscores the complex role of the LuxS quorum sensing system in modulating host-pathogen interactions.
Comparative transcriptome analysis between SEZ and the ΔluxS mutant underscores the broad impact of the LuxS on gene expression, affecting carbon source utilization, regulation, and metabolism in SEZ. Notably, in the ΔluxS strain, the LuxS transcript level was undetectable, confirming the success of the ΔluxS mutation. LuxS is not only responsible for AI-2 signaling molecule production but also plays a crucial role in central bacterial metabolism, including its involvement in the activated methyl cycle, as supported by our transcriptome results (Ramić et al., 2023).
Of particular note, we observed a significant up-regulation of the CRISPR-Cas (type I-C) system in the ΔluxS mutant (Supplementary Figure 1). It has been previously reported that CRISPR-Cas systems in SEZ likely play a crucial role in maintaining the stability of the bacterial genome (Waller and Robinson, 2013). Additionally, these CRISPR-Cas systems have been characterized in certain pathogenic bacteria, where they act as significant virulence factors, promoting biofilm formation and enhancing virulence (Nie et al., 2022). Cui et al. (2020) reported that CRISPR-cas3 of Salmonella plays a role in the regulation of QS genes, which are essential for the biofilm formation and invasion of bacteria. However, their specific effects on this bacterium have yet to be investigated and require further research. The transcript levels of grpE, hrcA, dnaK, and dnaJ were significantly up-regulated in the ΔluxS mutant, and these proteins are crucially involved in protein folding and the heat stress response in bacteria. They collectively play a role in regulating protein folding and cellular stress responses within bacterial cells. We conducted RT-PCR to assess the expression of these genes and evaluated their response to heat stress (Supplementary Figure 2). Result showed the ΔluxS mutant exhibited a notable resistance to heat shock (Supplementary Figure 3). Yi et al. (2014) employed a two-dimensional gel electrophoresis (2-DE) approach to characterize differentially expressed proteins in SEZ biofilms in comparison to their planktonic counterparts. This analysis identified 24 protein spots with varying intensities, including the upregulation of Elongation factor-Tu (EF-Tu), NADP-dependent glyceraldehyde-3-phosphate dehydrogenase (NADP-GAPDH) and glutamine–fructose-6-phosphate transaminase (glmS), which were also found in the ΔluxS mutant. It appears that we have uncovered some clues to explain the variations in biofilm formation and virulence in the ΔluxS mutant strain through the transcriptome data.
In summary, our study confirms the functionality of AI-2/LuxS quorum sensing in SEZ. LuxS does not impact capsule production, adhesion, or hemolysis. LuxS significantly enhances biofilm formation. While the loss of LuxS does not impact the lethality in mice, it modifies the bacterial load and the pathology in the spleen of infected animals, differences observed between SEZ and the ΔluxS mutant in inducing some pro-inflammatory factors and apoptosis in macrophages still reveal LuxS affects host defenses and modulates host immunity. Combined with transcriptome results, these findings provide a relatively comprehensive characterization of the role of LuxS in SEZ to date.
The bacterial strains and plasmids used in this study are detailed in (Supplementary Table 1). The SEZ ATCC35246 bacterial strain, originally isolated from an infected pig in Sichuan (Ma et al., 2011), was preserved in our laboratory for research purposes. SEZ strain were grown in Todd–Hewitt broth (THB) at 37°C. Vibrio harveyi strains BB170 were used as biosensors of the AI-2 signal and as the positive control for AI-2 bioassay, respectively. The V. harveyi strains were cultured in autoinducer bioassay medium (AB) as described (Bassler et al., 1993). The RAW264.7 macrophages and PK-15 cells were also maintained in our laboratory. These cell lines were cultured in DMEM (Gibco, USA), supplemented with 10% fetal bovine serum (FBS), and maintained in a humidified 5% CO2 incubator at 37°C.
The deletion of the luxS gene in SEZ was conducted following a previously established protocol (Takamatsu et al., 2001; Wei et al., 2012b). In sum, we initiated the process by using PCR amplification to generate upstream and downstream fragments of the luxS gene with specific primers (Supplementary Table 1). Then, these PCR products were ligated into the temperature-sensitive suicide vector, pSET-4s, using appropriate restriction enzymes. The resulting plasmid, denoted as pset4s-luxS, was introduced into SEZ through electroporation.
Bacterial cultures were under 28°C in the presence of 100 μg/mL spectinomycin (Spc). When the cultures reached the mid-logarithmic growth phase, bacteria were sub-cultured in THB supplemented with 100μg/mL Spc and further incubated at 28°C until they reached the early logarithmic phase. The culture was then shifted to 37°C for a duration of 4 hours. Subsequently, cells were plated on TSA plates and kept at 28°C. Colonies that did not exhibit resistance to Spc at 37°C were selected, indicating the loss of spectinomycin resistance conferred by the plasmid. Confirmation of the double-crossover homologous recombination mutant, designated as ΔluxS, was carried out using PCR analysis with primers luxSL-F and luxSR-R.
As for the complemented strain C-luxS, we amplified fragments that included the promoter region and the luxS gene, carried out the necessary digestion, and incorporated them into the pSET2 vector. This resulting construct was then introduced into the ΔluxS strain through electroporation to achieve complementation.
To assess the growth characteristics of SEZ, the ΔluxS mutant, and C-luxS, a single colony from each strain was cultured overnight at 37°C and adjusted to an OD600 of 0.3 using fresh THB medium. Subsequently, the cells were inoculated into fresh THB medium at a 1:1000 ratio and incubated at 37°C with shaking at 180 rpm for 24 hours. Cell densities were measured every 2 hours at OD600.
For AI-2 detection, established protocols were followed (Buck et al., 2009). The SEZ strain, ΔluxS, and the complemented C-luxS were cultured in THB medium until they reached an optical density of 0.3 at 600 nm. To obtain cell-free culture fluid (CF), the supernatant was filtered through a 0.22-μm-pore-size filter (Millipore, Bedford, MA). The reporter strain V. harveyi BB170 was diluted at a ratio of 1:5,000 in autoinducer bioassay (AB) medium, and AB medium was added to the diluted BB170 culture at a 1:10 ratio (vol/vol). This mixture was then incubated at 28°C. Aliquots of 100μL were dispensed into white, flat-bottomed, 96-well plates for AI-2 activity detection. CFs from BB170 and AB medium served as positive and negative controls, respectively. Luminescence measurements were conducted using a microplate reader.
To investigate the impact of LuxS on SEZ adhesion to cells, we conducted the following procedure (Tang et al., 2017). Cultured bacterial strains (SEZ, ΔluxS, C-luxS) were harvested, washed with sterile PBS, and then added to PK15 cells cultured in a 12-well dish at a multiplicity of infection (MOI) of 10. The cell-bacteria mixtures were co-incubated for 2 hours. Following incubation, non-adherent bacteria were removed by rinsing the cells with PBS. Subsequently, the cells were resuspended in sterile water, appropriately diluted, and plated onto tryptic soy agar (TSA) plates for colony counting.
To assess hemolysin activity in the ΔluxS mutant, we conducted a hemolytic assay following a previously described protocol (Jusuf et al., 2021). In brief, bacterial cells were washed and resuspended in PBS. Approximately 106 bacterial cells in 100 μL were added to a 96-well microtiter plate. Fresh pig blood was processed to remove serum and washed with physiological saline 2-3 times, then diluted to a 5% concentration, with 100 μL added to each well. The plate was incubated for 2 hours at 37°C. After incubation, the supernatant was collected following centrifugation, and the absorbance was measured at 420 nm. A 0.1% Triton X solution was used as a positive control to calculate the percentage of bacterial hemolysis activity.
To assess the hydrophobic properties of the bacteria, an equal volume of bacterial suspension was combined with xylene and mixed for 2 minutes. The mixture was then left to stand for 30 minutes, allowing the phases to separate, after which the OD600 of the aqueous phase was measured. The hydrophobicity percentage (H%) was calculated using the formula H% = [(H0 − H)/H0] × 100%, where H0 represents the initial absorbance before mixing with xylene, and H represents the absorbance after xylene extraction (Del Re et al., 2000). The SEZ capsule-deficient strain ΔhasB was used as the control group in this assay.
The cell-associated hyaluronic acid (HA) of SEZ was measured using a procedure inspired by existing methods, with some modifications (Velineni and Timoney, 2015). In brief, 10 ml of SEZ culture in THB, grown at 37°C, was centrifuged at 8000 g for 10 minutes. The resulting pellet was gently washed with PBS and then resuspended in 2 ml of distilled water. This suspension was transferred to a capped glass tube, to which 5 ml of chloroform was added. The mixture was then vigorously mixed for 1 hour. Following this, the sample was centrifuged at 8,500 g for 10 minutes, and the aqueous phase was carefully transferred to a new container. To this aqueous phase, two volumes of cetyltrimethylammonium bromide (CTAB) buffer (2.5 g L−1) were added, and the mixture was gently stirred. The reaction was allowed to proceed for 10 minutes at room temperature, after which the OD600 was measured (Xie et al., 2019). The concentration of HA in the sample was determined based on the OD600 value and a standard curve. This standard curve was established using a stock solution of HA prepared with standard HA (Sigma, Shanghai, China).
Biofilm formation was quantified by the crystal violet assay (Bonifait et al., 2008; Wang et al., 2011). Bacterial cultures were grown until they reached an optical density (OD600) of approximately 0.3. Then, 10 μL of the bacterial suspension was added to 96-well flat-bottomed plastic culture plates containing 90 μL of THB supplemented with 10 mg/mL of fibrinogen. The plates were incubated at 37°C for 24 hours, with each bacterial strain tested in triplicate. The culture medium was discarded, and the wells were gently washed three times with sterile PBS to remove loosely adherent cells. The remaining bacteria attached to the wells were fixed with methanol for 30 minutes and air-dried. A 1% crystal violet solution was added to each well, and the plates were stained for 10 minutes at room temperature. Excess crystal violet was removed and the wells were rinsed multiple times with distilled water. The biofilm-associated crystal violet was then solubilized with 100 μL of 95% (v/v) ethanol, and the optical density at 570 nm was measured using a microplate reader.
For scanning electron microscopy (SEM), SEZ was inoculated into a 24-well culture plate containing sterile glass slides and incubated at 37°C for 48h. The slides were then removed, washed three times with sterile PBS, and fixed in a solution of 5% glutaraldehyde, 4.4% formaldehyde, and 2.75% picric acid in 0.05% sodium cacodylate buffer for at least 1 h. The sample underwent sequential dehydration with 25%, 50%, 70%, 80%, and 95% ethanol. SEM was performed as previously described (Petruzzi et al., 2017), using a scanning electron microscope.
To study biofilms using confocal laser scanning microscopy (CLSM), each biofilm was stained with 0.3% SYTO-9 (obtained from Sigma-Aldrich, China) and incubated in darkness for 15 minutes for fluorescent labeling. The analysis of the biofilm images was conducted using Zeiss confocal software, enabling the acquisition of z-stack images. The thickness and biovolume of the biofilms were quantified using the COMSTAT2 software available at www.comstat.dk.
In the experiment with macrophages, we used RAW264.7 cells and followed a method that was a bit different from what others have done before. We put the macrophages in six-well plates. After that, we added SEZ bacteria to 12-well plates with a MOI of 1, and co-incubate for 12 h. For the negative control, we just added 100 μL of PBS.
To evaluate apoptosis, the treated cells were collected, washed twice with ice-cold PBS, and then resuspended in 1× Annexin V binding buffer at a concentration of 1×106 cells/mL. Apoptosis detection was carried out using the Annexin V-FITC Apoptosis Detection Kit (Sigma, Shanghai, China), strictly adhering to the manufacturer’s instructions. The stained cells were analyzed via flow cytometry within 1 hour of staining.
The cell supernatant was collected by centrifugation at 3,000g, and the secretion level of TNF-α was determined using an ELISA assay kit (Enzyme-linked Biotechnology, Shanghai, China).
Quantitative real-time reverse-transcription polymerase chain reaction (qRT-PCR) was used to assess the transcription levels of pro-inflammatory cytokines (IL-1β, IL-6, IL-10, IL-18, TNF-α) and NF-κB. Briefly, post-infection, the cells were washed twice with sterile PBS. Total RNA was extracted using TRIzol reagent (Sigma, Shanghai, China), followed by cDNA synthesis using the Takara reverse transcription kit. Real-time quantitative PCR was performed using SYBR Green Premix (Takara, Beijing, China) on a LightCycler 480 fast real-time cycler. Fold changes were calculated using the 2-ΔΔCt formula (Livak and Methods, 2000). The data are presented as the means ± standard deviation (SD) of triplicate reactions for each gene transcript, and the primer sequences are provided in (Supplementary Table 1).
Four-to-six-week-old BALB/c mice were obtained from Beijing Vetonlitech Experimental Animal Technology Co. Bacterial suspensions were prepared by diluting the bacteria in PBS, 1 × 106 CFU of bacteria were administered via intraperitoneal injection.
Tissue bacterial burden assays were conducted as previously described (Wei et al., 2012a). Briefly, mice were euthanized 24 hours after infection, and their spleens, livers, kidneys, and lungs were collected and weighed. Then, the tissues were homogenized and diluted in sterile saline (g/mL). The homogenized tissue samples were plated onto TSA plates for culturing, and the colonies were counted.
Briefly, the mice were infected with 1 × 106 CFU of bacteria. At 24 h post-infection, the infected mice were euthanized, and the spleen were collected and placed into 10% neutral buffered formalin. After fixation, the tissues were dehydrated and embedded in paraffin, and 4-mm sections were taken and stained with hematoxylin and eosin (H&E) for histological evaluation.
The ΔluxS mutant and wild-type strains were incubated in THB on a shaker at 37°C for 10 hours. After incubation, the cells were harvested by centrifugation at 6,000 g for 10 minutes at 4°C. Total RNA samples were extracted using the TRIzol method. RNA sample quality was assessed using Thermo NanoDrop One and Agilent 4200 Tape Station. Once qualified, the ribosomal RNA (rRNA) was removed using the Epicentre Ribo-Zero rRNA Removal Kit.
Library preparation followed a standardized protocol using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina, involving mRNA fragmentation, cDNA synthesis, double-stranded cDNA generation, purification, enzymatic degradation, end repair, adapter ligation, fragment size selection, and final PCR amplification. Libraries meeting quality control criteria underwent PE150 sequencing on the Illumina platform. Raw sequencing data were processed into FASTQ files, including read sequences and corresponding quality scores. The data sets generated for this study can be found in the NCBI Sequence Read Archive (SRA) repository under BioProject accession number PRJNA1048089.
Before conducting the differential expression analysis, we conducted quality control on the sequencing reads and trimmed the adapter sequences. The sequencing reads from each library were then used for differential expression analysis with the read counts. As a reference genome sequence, we used the complete genome sequence of Streptococcus equi subsp. zooepidemicus (ATCC_35246) and its genome annotation from the NCBI nucleotide database (accession number: NC_017582.1).
To compare gene expression between SEZ and ΔluxS. Genes with absolute log2 fold change values of ≥1 and false discovery rate (FDR) P values of ≤0.05 were considered differentially expressed. For further analysis of these differentially expressed genes, we utilized the STRING Consortium 2020, which provides functional enrichment analysis of protein-protein interaction networks through the STRING mapper tool (Szklarczyk et al., 2023).
The data in this study are expressed as means ± standard deviation (SD). For the statistical analysis, we used two-way ANOVA, conducted with Graph Pad Prism 9.0 (GraphPad Software Inc., USA). We marked significant differences with asterisks: * for p < 0.05, ** for p < 0.01, and *** for p < 0.001. Different letters represent significant differences within each strain, with p < 0.05 as the threshold for significance. All data are presented as means ± standard error of the mean (SEM), and the results are compiled from a minimum of three independent experiments.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
The animal study was approved by All operations in this study involving animal experiments were handled according to the Ethics Committee at Northwest A&F University (approval number DY2022009). The study was conducted in accordance with the local legislation and institutional requirements.
HX: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Supervision, Visualization, Writing – original draft, Writing – review & editing. RZ: Data curation, Formal analysis, Software, Visualization, Writing – review & editing. RG: Investigation, Methodology, Writing – review & editing. YZ: Conceptualization, Formal analysis, Writing – review & editing. JZ: Project administration, Writing – review & editing. HL: Formal analysis, Writing – review & editing. QF: Resources, Validation, Writing – review & editing. XW: Funding acquisition, Project administration, Resources, Validation, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the China Wool sheep Industry and Technology System project (No. CARS-39-14).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1339131/full#supplementary-material
Abisado, R. G., Benomar, S., Klaus, J. R., Dandekar, A. A., Chandler, J. R. (2018). Bacterial quorum sensing and microbial community interactions. MBio 9 (3), 10–1128. doi: 10.1128/mbio.02331-17
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25 (1), 25–29. doi: 10.1038/75556
Bassler, B. L., Wright, M., Showalter, R. E., Silverman, M. R. (1993). Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9 (4), 773–786. doi: 10.1111/j.1365-2958.1993.tb01737.x
Bhardwaj, A. K., Vinothkumar, K., Rajpara, N. (2013). Bacterial quorum sensing inhibitors: attractive alternatives for control of infectious pathogens showing multiple drug resistance. Recent Pat. Antiinfect Drug Discovery 8 (1), 68–83. doi: 10.2174/157489113805290809
Blank, L. M., Hugenholtz, P., Nielsen, L. K. (2008). Evolution of the hyaluronic acid synthesis (has) operon in Streptococcus zooepidemicus and other pathogenic streptococci. J. Mol. Evol. 67 (1), 13–22. doi: 10.1007/s00239-008-9117-1
Bonifait, L., Grignon, L., Grenier, D. (2008). Fibrinogen induces biofilm formation by Streptococcus suis and enhances its antibiotic resistance. Appl. Environ. Microbiol. 74 (15), 4969–4972. doi: 10.1128/AEM.00558-08
Bosica, S., Chiaverini, A., De Angelis, M. E., Petrini, A., Averaimo, D., Martino, M., et al. (2023). Severe Streptococcus equi Subspecies zooepidemicus Outbreak from Unpasteurized Dairy Product Consumption, Italy. Emerg. Infect. Dis. 29 (5), 1020–1024. doi: 10.3201/eid2905.221338
Bozue, J., Powell, B. S., Cote, C. K., Moody, K. L., Gelhaus, H. C., Vietri, N. J., et al. (2012). Disrupting the luxS quorum sensing gene does not significantly affect Bacillus anthracis virulence in mice or Guinea pigs. Virulence 3 (6), 504–509. doi: 10.4161/viru.21934
Buck, B. L., Azcarate-Peril, M. A., Klaenhammer, T. R. (2009). Role of autoinducer-2 on the adhesion ability of Lactobacillus acidophilus. J. Appl. Microbiol. 107 (1), 269–279. doi: 10.1111/j.1365-2672.2009.04204.x
Burgess, N. A., Kirke, D. F., Williams, P., Winzer, K., Hardie, K. R., Meyers, N. L., et al. (2002). LuxS-dependent quorum sensing in Porphyromonas gingivalis modulates protease and haemagglutinin activities but is not essential for virulence. Microbiol. (Reading) 148 (Pt 3), 763–772. doi: 10.1099/00221287-148-3-763
Cao, M., Feng, Y., Wang, C., Zheng, F., Li, M., Liao, H., et al. (2011). Functional definition of LuxS, an autoinducer-2 (AI-2) synthase and its role in full virulence of Streptococcus suis serotype 2. J. Microbiol. 49 (6), 1000–1011. doi: 10.1007/s12275-011-1523-1
Cao, Q., Ma, K., Nie, M., Dong, Y., Lu, C., Liu, Y. (2020). Role of luxS in immune evasion and pathogenicity of piscine Streptococcus agalactiae is not dependent on autoinducer-2. Fish Shellfish Immunol. 99, 274–283. doi: 10.1016/j.fsi.2020.02.016
Chaussee, M. A., Callegari, E. A., Chaussee, M. S. (2004). Rgg regulates growth phase-dependent expression of proteins associated with secondary metabolism and stress in Streptococcus pyogenes. J. Bacteriol 186 (21), 7091–7099. doi: 10.1128/JB.186.21.7091-7099.2004
Chen, X., Mou, K., Lu, W., Schumacher, L., Resende-De-Macedo, N., Sitthicharoenchai, P., et al. (2023). Genomic characterization of Streptococcus equi subspecies zooepidemicus from a 2021 outbreak in Indiana with increased sow mortality. mSphere 8 (6), e0040423. doi: 10.1128/msphere.00404-23
Chen, X., Resende-De-Macedo, N., Sitthicharoenchai, P., Sahin, O., Burrough, E., Clavijo, M., et al. (2020). Genetic characterization of Streptococcus equi subspecies zooepidemicus associated with high swine mortality in the United States. Transbound Emerg. Dis. 67 (6), 2797–2808. doi: 10.1111/tbed.13645
Cui, L., Wang, X., Huang, D., Zhao, Y., Feng, J., Lu, Q., et al. (2020). CRISPR-cas3 of Salmonella upregulates bacterial biofilm formation and virulence to host cells by targeting quorum-sensing systems. Pathogens 9 (1), 53. doi: 10.3390/pathogens9010053
Daines, D. A., Bothwell, M., Furrer, J., Unrath, W., Nelson, K., Jarisch, J., et al. (2005). Haemophilus influenzae luxS mutants form a biofilm and have increased virulence. Microb. Pathog. 39 (3), 87–96. doi: 10.1016/j.micpath.2005.06.003
Day, W. A., Jr., Maurelli, A. T. (2001). Shigella flexneri LuxS quorum-sensing system modulates virB expression but is not essential for virulence. Infect. Immun. 69 (1), 15–23. doi: 10.1128/IAI.69.1.15-23.2001
Defoirdt, T. (2018). Quorum-sensing systems as targets for antivirulence therapy. Trends Microbiol. 26 (4), 313–328. doi: 10.1016/j.tim.2017.10.005
Del Re, B., Sgorbati, B., Miglioli, M., Palenzona, D. (2000). Adhesion, autoaggregation and hydrophobicity of 13 strains of Bifidobacterium longum. Lett. Appl. Microbiol. 31 (6), 438–442. doi: 10.1046/j.1365-2672.2000.00845.x
Flores, A. R., Jewell, B. E., Fittipaldi, N., Beres, S. B., Musser, J. M. (2012). Human disease isolates of serotype m4 and m22 group a streptococcus lack genes required for hyaluronic acid capsule biosynthesis. MBio 3 (6), e00413–e00412. doi: 10.1128/mBio.00413-12
Hau, S. J., Lantz, K., Stuart, K. L., Sitthicharoenchai, P., Macedo, N., Derscheid, R. J., et al. (2022). Replication of Streptococcus equi subspecies zooepidemicus infection in swine. Vet. Microbiol. 264, 109271. doi: 10.1016/j.vetmic.2021.109271
Jie, P., Zhe, M., Chengwei, H., Huixing, L., Hui, Z., Chengping, L., et al. (2017). Virulent and Vaccine Strains of Streptococcus equi ssp. zooepidemicus Have Different Influences on Phagocytosis and Cytokine Secretion of Macrophages. J. Proteome Res. 16 (1), 77–86. doi: 10.1021/acs.jproteome.6b00571
Jimenez, J. C., Federle, M. J. (2014). Quorum sensing in group A Streptococcus. Front. Cell Infect. Microbiol. 4, 127. doi: 10.3389/fcimb.2014.00127
Jusuf, S., Dong, P. T., Hui, J., Ulloa, E. R., Liu, G. Y., Cheng, J. X. (2021). Granadaene photobleaching reduces the virulence and increases antimicrobial susceptibility of Streptococcus agalactiae. Photochem. Photobiol. 97 (4), 816–825. doi: 10.1111/php.13389
Kerdsin, A., Chopjitt, P., Hatrongjit, R., Boueroy, P., Gottschalk, M. (2022). Zoonotic infection and clonal dissemination of Streptococcus equi subspecies zooepidemicus sequence type 194 isolated from humans in Thailand. Transbound Emerg. Dis. 69 (4), e554–e565. doi: 10.1111/tbed.14331
Kim, S. M., Park, J. H., Lee, H. S., Kim, W. B., Ryu, J. M., Han, H. J., et al. (2013). LuxR homologue SmcR is essential for Vibrio vulnificus pathogenesis and biofilm detachment, and its expression is induced by host cells. Infect. Immun. 81 (10), 3721–3730. doi: 10.1128/IAI.00561-13
Kuusi, M., Lahti, E., Virolainen, A., Hatakka, M., Vuento, R., Rantala, L., et al. (2006). An outbreak of Streptococcus equi subspecies zooepidemicus associated with consumption of fresh goat cheese. BMC Infect. Dis. 6, 36. doi: 10.1186/1471-2334-6-36
Li, G., Zong, X., Cheng, Y., Xu, J., Deng, J., Huang, Y., et al. (2022). miR-223-3p contributes to suppressing NLRP3 inflammasome activation in Streptococcus equi ssp. zooepidemicus infection. Vet. Microbiol. 269, 109430. doi: 10.1016/j.vetmic.2022.109430
Livak, K., Methods, T. S. J. (2000). Analysis of relative gene expression data using real-time quantitative PCR and the 2-△△Ct method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Ma, Z., Geng, J., Zhang, H., Yu, H., Yi, L., Lei, M., et al. (2011). Complete genome sequence of Streptococcus equi subsp. zooepidemicus strain ATCC 35246. J. Bacteriol 193 (19), 5583–5584. doi: 10.1128/jb.05700-11
Miller, M. B., Bassler, B. L. (2001). Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199. doi: 10.1146/annurev.micro.55.1.165
Morris, J. H., Apeltsin, L., Newman, A. M., Baumbach, J., Wittkop, T., Su, G., et al. (2011). clusterMaker: a multi-algorithm clustering plugin for Cytoscape. BMC Bioinf. 12, 436. doi: 10.1186/1471-2105-12-436
Nie, M., Dong, Y., Cao, Q., Zhao, D., Ji, S., Huang, H., et al. (2022). CRISPR contributes to adhesion, invasion, and biofilm formation in Streptococcus agalactiae by repressing capsular polysaccharide production. Microbiol. Spectr. 10 (4), e0211321. doi: 10.1128/spectrum.02113-21
Pelkonen, S., Lindahl, S. B., Suomala, P., Karhukorpi, J., Vuorinen, S., Koivula, I., et al. (2013). Transmission of Streptococcus equi subspecies zooepidemicus infection from horses to humans. Emerg. Infect. Dis. 19 (7), 1041–1048. doi: 10.3201/eid1907.121365
Petruzzi, B., Briggs, R. E., Tatum, F. M., Swords, W. E., De Castro, C., Molinaro, A., et al. (2017). Capsular polysaccharide interferes with biofilm formation by Pasteurella multocida serogroup A. MBio 8 (6), 10–1128. doi: 10.1128/mBio.01843-17
Rahbari, K. M., Chang, J. C., Federle, M. J. (2021). A streptococcus quorum sensing system enables suppression of innate immunity. MBio 12 (3), 10–1128. doi: 10.1128/mBio.03400-20
Ramić, D., Jug, B., Šimunović, K., Žnidarič, M. T., Kunej, U., Toplak, N., et al. (2023). The Role of luxS in Campylobacter jejuni Beyond Intercellular Signaling. Microbiol. Spectr. 11 (2), e0257222. doi: 10.1128/spectrum.02572-22
Schauder, S., Shokat, K., Surette, M. G., Bassler, B. L. (2001). The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41 (2), 463–476. doi: 10.1046/j.1365-2958.2001.02532.x
Solano, C., Echeverz, M., Lasa, I. (2014). Biofilm dispersion and quorum sensing. Curr. Opin. Microbiol. 18, 96–104. doi: 10.1016/j.mib.2014.02.008
Song, H., Yuan, C., Zhang, Y., Pan, F., Fan, H., Ma, Z. (2022). Protection Efficacy of Monoclonal Antibodies Targeting Different Regions of Specific SzM Protein from Swine-Isolated Streptococcus equi ssp. zooepidemicus Strains. Microbiol. Spectr. 10 (6), e0174222. doi: 10.1128/spectrum.01742-22
Stoodley, P., Sauer, K., Davies, D. G., Costerton, J. W. (2002). Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56, 187–209. doi: 10.1146/annurev.micro.56.012302.160705
Sun, Y., Li, Y., Luo, Q., Huang, J., Chen, J., Zhang, R., et al. (2020). LuxS/AI-2 quorum sensing system in Edwardsiella piscicida promotes biofilm formation and pathogenicity. Infect. Immun. 88 (5), 10–1128. doi: 10.1128/IAI.00907-19
Szklarczyk, D., Kirsch, R., Koutrouli, M., Nastou, K., Mehryary, F., Hachilif, R., et al. (2023). The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51 (D1), D638–d646. doi: 10.1093/nar/gkac1000
Takamatsu, D., Osaki, M., Sekizaki, T. (2001). Thermosensitive suicide vectors for gene replacement in Streptococcus suis. Plasmid 46 (2), 140–148. doi: 10.1006/plas.2001.1532
Tang, B., Liang, H., Zhao, P., Wei, Z. (2017). miR-194b-3p partially inhibits Streptococcus equi subsp. zooepidemicus adherence to PK15 cells. Vet. Microbiol. 210, 38–42. doi: 10.1016/j.vetmic.2017.08.023
Timoney, J. F. (2004). The pathogenic equine streptococci. Vet. Res. 35 (4), 397–409. doi: 10.1051/vetres:2004025
Velineni, S., Timoney, J. F. (2015). Capsular hyaluronic acid of equine isolates of Streptococcus zooepidemicus is upregulated at temperatures below 35 °C. Equine Vet. J. 47 (3), 333–338. doi: 10.1111/evj.12272
Vorregaard, M. (2008). Comstat2-a modern 3D image analysis environment for biofilms (Lyngby, Denmark: Technical University of Denmark, DTU, DK-2800 Kgs).
Waller, A. S., Robinson, C. (2013). Streptococcus zooepidemicus and Streptococcus equi evolution: the role of CRISPRs. Biochem. Soc. Trans. 41 (6), 1437–1443. doi: 10.1042/BST20130165
Wang, Y., Bian, Z., Wang, Y. (2022). Biofilm formation and inhibition mediated by bacterial quorum sensing. Appl. Microbiol. Biotechnol. 106 (19-20), 6365–6381. doi: 10.1007/s00253-022-12150-3
Wang, Y., Zhang, W., Wu, Z., Zhu, X., Lu, C. (2011). Functional analysis of luxS in Streptococcus suis reveals a key role in biofilm formation and virulence. Vet. Microbiol. 152 (1-2), 151–160. doi: 10.1016/j.vetmic.2011.04.029
Waters, C. M., Bassler, B. L. (2005). Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21, 319–346. doi: 10.1146/annurev.cellbio.21.012704.131001
Wei, Z., Fu, Q., Chen, Y., Cong, P., Xiao, S., Mo, D., et al. (2012a). The capsule of Streptococcus equi ssp. zooepidemicus is a target for attenuation in vaccine development. Vaccine 30 (31), 4670–4675. doi: 10.1016/j.vaccine.2012.04.092
Wei, Z., Fu, Q., Chen, Y., Li, M., Cong, P., Mo, D., et al. (2013). Streptococcus equi ssp. zooepidemicus C5a peptidase, a putative invasin, induces protective immune response in mice. Res. Vet. Sci. 95 (2), 444–450. doi: 10.1016/j.rvsc.2013.03.026
Wei, Z., Fu, Q., Liu, X., Chen, Y. (2012b). Attenuated Streptococcus equi ssp. zooepidemicus as a bacterial vector for expression of porcine circovirus type 2 capsid protein. FEMS Microbiol. Lett. 332 (1), 20–26. doi: 10.1111/j.1574-6968.2012.02569.x
Wei, Z., Li, R., Zhang, A., He, H., Hua, Y., Xia, J., et al. (2009). Characterization of Streptococcus suis isolates from the diseased pigs in China between 2003 and 2007. Vet. Microbiol. 137 (1-2), 196–201. doi: 10.1016/j.vetmic.2008.12.015
Whiteley, M., Diggle, S. P., Greenberg, E. P. (2017). Progress in and promise of bacterial quorum sensing research. Nature 551 (7680), 313–320. doi: 10.1038/nature24624
Xavier, K. B., Bassler, B. L. (2003). LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6 (2), 191–197. doi: 10.1016/S1369-5274(03)00028-6
Xie, Z., Meng, K., Yang, X., Liu, J., Yu, J., Zheng, C., et al. (2019). Identification of a quorum sensing system regulating capsule polysaccharide production and biofilm formation in Streptococcus zooepidemicus. Front. Cell Infect. Microbiol. 9, 121. doi: 10.3389/fcimb.2019.00121
Xie, H., Wei, Z., Ma, C., Li, S., Liu, X., Fu, Q. (2018). Characterization of SeseC_01411 as a surface protective antigen of Streptococcus equi ssp. zooepidemicus. Res. Vet. Sci. 118, 517–521. doi: 10.1016/j.rvsc.2018.05.007
Xu, G., Guo, Z., Liu, Y., Yang, Y., Lin, Y., Li, C., et al. (2022). Gasdermin D protects against Streptococcus equi subsp. zooepidemicus infection through macrophage pyroptosis. Front. Immunol. 13, 1005925. doi: 10.3389/fimmu.2022.1005925
Xu, L., Li, H., Vuong, C., Vadyvaloo, V., Wang, J., Yao, Y., et al. (2006). Role of the luxS quorum-sensing system in biofilm formation and virulence of Staphylococcus epidermidis. Infect. Immun. 74 (1), 488–496. doi: 10.1128/IAI.74.1.488-496.2006
Xu, B., Pei, X., Su, Y., Ma, Z., Fan, H. (2016). Capsule of Streptococcus equi subsp. zooepidemicus hampers the adherence and invasion of epithelial and endothelial cells and is attenuated during internalization. FEMS Microbiol. Lett. 363 (16), fnw164. doi: 10.1093/femsle/fnw164
Yadav, M. K., Vidal, J. E., Go, Y. Y., Kim, S. H., Chae, S. W., Song, J. J. (2018). The LuxS/AI-2 quorum-sensing system of Streptococcus pneumoniae is required to cause disease, and to regulate virulence- and metabolism-related genes in a rat model of middle ear infection. Front. Cell Infect. Microbiol. 8, 138. doi: 10.3389/fcimb.2018.00138
Yi, L., Wang, Y., Ma, Z., Zhang, H., Li, Y., Zheng, J. X., et al. (2014). Biofilm formation of Streptococcus equi ssp. zooepidemicus and comparative proteomic analysis of biofilm and planktonic cells. Curr. Microbiol. 69 (3), 227–233. doi: 10.1007/s00284-014-0574-z
Yi, L., et al. (2016). Identification and characterization of a Streptococcus equi ssp. zooepidemicus immunogenic GroEL protein involved in biofilm formation. Veterinary Res. 47.1, 1–9.
Keywords: Streptococcus equi subsp. zooepidemicus, LuxS, biofilm, capsular polysaccharide, virulence, infection
Citation: Xie H, Zhang R, Guo R, Zhang Y, Zhang J, Li H, Fu Q and Wang X (2024) Characterization of AI-2/LuxS quorum sensing system in biofilm formation, pathogenesis of Streptococcus equi subsp. zooepidemicus. Front. Cell. Infect. Microbiol. 14:1339131. doi: 10.3389/fcimb.2024.1339131
Received: 15 November 2023; Accepted: 24 January 2024;
Published: 06 February 2024.
Edited by:
Florent Sebbane, Institut National de la Santé et de la Recherche Médicale (INSERM), FranceReviewed by:
Sébastien Bontemps-Gallo, Institut Pasteur de Lille, FranceCopyright © 2024 Xie, Zhang, Guo, Zhang, Zhang, Li, Fu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinglong Wang, d3hsb25nQG53c3VhZi5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.