
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol. , 05 February 2024
Sec. Veterinary and Zoonotic Infection
Volume 14 - 2024 | https://doi.org/10.3389/fcimb.2024.1325346
 Saba Rafique1
Saba Rafique1 Zohra Jabeen1
Zohra Jabeen1 Treeza Pervaiz1
Treeza Pervaiz1 Farooq Rashid2,3,4
Farooq Rashid2,3,4 Sisi Luo2,3,4
Sisi Luo2,3,4 Liji Xie2,3,4
Liji Xie2,3,4 Zhixun Xie2,3,4*
Zhixun Xie2,3,4*Infectious bronchitis virus (IBV) is a positive-sense, single-stranded, enveloped RNA virus responsible for substantial economic losses to the poultry industry worldwide by causing a highly contagious respiratory disease. The virus can spread quickly through contact, contaminated equipment, aerosols, and personal-to-person contact. We highlight the prevalence and geographic distribution of all nine genotypes, as well as the relevant symptoms and economic impact, by extensively analyzing the current literature. Moreover, phylogenetic analysis was performed using Molecular Evolutionary Genetics Analysis (MEGA-6), which provided insights into the global molecular diversity and evolution of IBV strains. This review highlights that IBV genotype I (GI) is prevalent worldwide because sporadic cases have been found on many continents. Conversely, GII was identified as a European strain that subsequently dispersed throughout Europe and South America. GIII and GV are predominant in Australia, with very few reports from Asia. GIV, GVIII, and GIX originate from North America. GIV was found to circulate in Asia, and GVII was identified in Europe and China. Geographically, the GVI-1 lineage is thought to be restricted to Asia. This review highlights that IBV still often arises in commercial chicken flocks despite immunization and biosecurity measures because of the ongoing introduction of novel IBV variants and inadequate cross-protection provided by the presently available vaccines. Consequently, IB consistently jeopardizes the ability of the poultry industry to grow and prosper. Identifying these domains will aid in discerning the pathogenicity and prevalence of IBV genotypes, potentially enhancing disease prevention and management tactics.
Infectious bronchitis, which mostly affects poultry, is a viral disease. It causes substantial economic loss and morbidity in poultry industries worldwide (Cavanagh, 2007; Abozeid et al., 2017). The etiological agents are avian infectious bronchitis virus (AIBV) and avian coronavirus (Carstens and Ball, 2009), which are members of the genus Gamma coronavirus (γCoV), family Coronaviridae and order Nidovirales (Carstens, 2010; Abro et al., 2012; Durães-Carvalho et al., 2015). The Coronaviridae family is divided into four subfamilies, with four genera belonging to the Coronavirinae subfamily: alpha coronavirus (αCoV), beta coronavirus (βCoV), gammacoronavirus (γCoV) and delta coronavirus (δCoV). Avian species, including chickens, land fowl, and pheasants, are specifically infected by γCoV and δCoV (Siddell and Snijder, 2014).
The genome of IBV is composed of linear and positive single-stranded RNA strands approximately 27-28 kb long that encode a variety of structural and nonstructural proteins (NSPs). The IBV genome is organized as 5’UTR-ORF 1a/1b-S-3a-3b-E-M-4b-4c-5a-5b-N-6b-3’UTR, with an a-1 frame shift at the junction of ORF 1a/1b, resulting in the synthesis of the 1a and 1b polyproteins, which are subsequently processed to produce individual NSPs responsible for genome replication and transcription (Figure 1) (Bhuiyan et al., 2021). Furthermore, these viruses have important structural components, such as the spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins (Li et al., 2005). Notably, the spike protein, with a molecular weight of 200 kDa, is the largest glycoprotein among these proteins. Previous research has also identified two accessory genes, ORF3 and ORF5, encoding proteins 3a, 3b, 5a, and 5b (Lai and Cavanagh, 1997; Casais et al., 2005). The ORFs, 1a and 1ab are approximately 20 kb in length and constitute approximately two-thirds of the genome. NSPs are a collection of 16 proteins that play indispensable roles in viral replication, RNA synthesis, transcription, RNA proofreading, RNA capping, and host immune response modulation (Ziebuhr, 2005; Chen et al., 2009; Smith and Denison, 2012; Te Velthuis et al., 2012; Kint et al., 2016). The genes for the structural proteins and accessory proteins are ordered in the viral genome as follows.
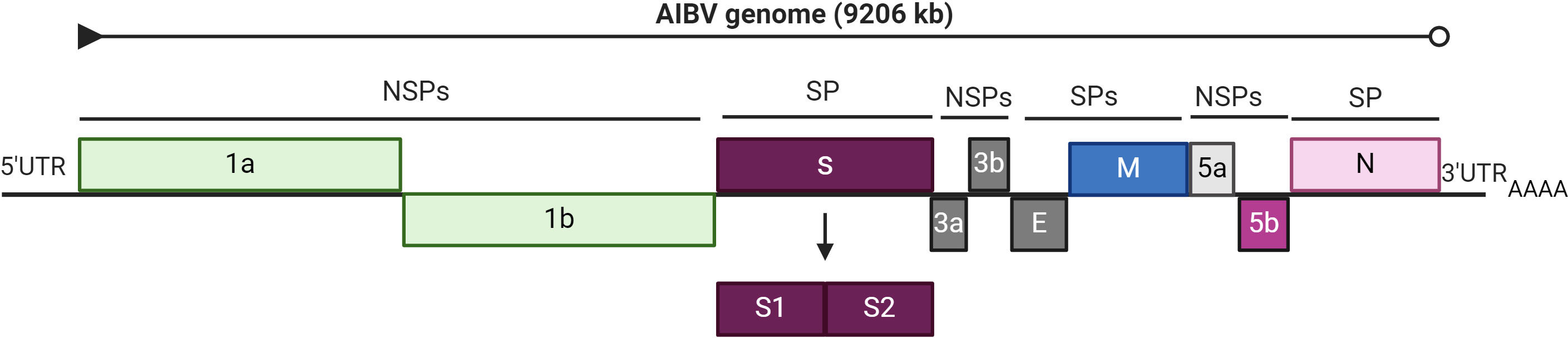
Figure 1 Genome organization of Avian infectious bronchitis virus. The total genome of AIBV is 9206 kb, showing structural proteins (SPs) and non-structural proteins (NSPs). The figure was created with BioRender.com.
Gene 2 encodes S protein and is the longest gene in this region and allows the virus to attach to host cells and mediates the fusion of viral and cellular membranes, thus contributing to viral tropism (Kant et al., 1992; Shan et al., 2018). Gene 3 is a polycistronic gene that encodes three proteins, 3a, 3b, and 3c, and is positioned downstream of gene 2. The 3c protein, generally referred to as the E protein, is required for viral particle assembly (Liu et al., 1991), but the 3a and 3b peptides have yet to be identified (Fischer et al., 1998). Gene 4 is another polycistronic gene that encodes three distinct peptides. The first ORF encodes an M protein that is a significant component of the viral envelope. The functions of the two small accessory 4b and 4c peptides are unknown. The M protein contributes to virion structure and may activate a humoral immune response in chickens, but this response is insufficient to prevent clinical signs from appearing (Ignjatovic and Galli, 1994). Gene 5 encodes two tiny accessory peptides, 5a and 5b (Youn et al., 2005; Laconi et al., 2018). Gene 6 encodes the N protein and is the last ORF of the IBV genome. This protein is a nonspecific RNA-binding protein and has numerous roles in the virus’s life cycle (Zúñiga et al., 2010; Kint et al., 2016).
IBV is traditionally thought to be a host-specific respiratory pathogen in poultry, and it replicates at the tracheal mucosa (Amarasinghe et al., 2018). However, the discovery of new IBV genotypes or serotypes has revealed a wide range of tissue tropisms, including in the bursa of Fabricius (Macdonald and McMartin, 1976; Ambali and Jones, 1990), urinary tract (Cumming, 1969; França et al., 2011), reproductive tract (Boroomand et al., 2012) and gastrointestinal tract (Cook and Ambali, 1986; Ambali and Jones, 1990; Liu et al., 2006). IBV is known to replicate in layers of the reproductive tract epithelium, resulting in decreased egg production and defective eggshell formation (Cook et al., 2012). Early infection with the IBV JX181 (GVI-1) strain is highly harmful to laying hens, causing major respiratory problems, irreversible oviduct damage, and growth retardation in the reproductive system (Bo et al., 2022). False layer syndrome, which is associated with cystic oviduct development, cystic left oviducts, signs of vent pecking, ovarian regression, and yolk coelomitis, occurs due to IBV infection in young birds aged 25-28 weeks (Ramsubeik et al., 2023). Cockerel testes can also be infected with IBV (Boltz et al., 2004).
Among all avian species, pheasants are considered the second most common natural host for IBV after poultry. (Gough et al., 1996). Antibodies against IBV have been found in quail, turkeys and free-ranging rockhopper penguins, though it is unknown whether IBV can be isolated from these animals (Karesh et al., 1999). In addition, IBV infects a variety of bird species, particularly those raised near domestic poultry, such as ducks, teals, geese, pigeons, partridges, guinea fowl, peafowl and domestic fowl (Cavanagh et al., 2005). The genome of IBV shares many similarities among various hosts. One virus found in teal and peafowl, for instance, shows 90% to 99% similarity in sequence to IBV (Liu et al., 2005). Incredibly similar nucleotide sequences were found in viruses collected from turkeys, ducks, pheasants, and whooper swans (Hughes et al., 2009). IB was first detected in the United States in 1931 (Schalk and Hawn, 1931). The causative agent was not identified, which frequently caused confusion with other viral and bacterial pathogens that can cause upper respiratory infections in chickens. Until the first report of the Connecticut isolate (Conn) (GI-1), the Massachusetts isolate (Mass) (GI-1) was the sole serotype discovered. The Conn (GI-1) causes identical symptoms but exhibits antigenic variations from the Massachusetts isolate (GI-1) (Jungherr et al., 1956). Notably, this viral pathogen has demonstrated a remarkable capacity for diversification and adaptation, resulting in the identification of numerous unique serotypes in various parts of the globe (Rafique et al., 2018b).
IBV poses no known threat to human health. Neutralizing antibodies have been found in employees who work on commercial chicken farms, though their impact is uncertain (Miller and Yates, 1968). Furthermore, there is currently little evidence to support the idea that IBV may replicate inside a human host; avians can simply transmit IBV infection to chicks mechanically (Kapikian et al., 1969). The continuing processes of genetic recombination and mutation can be attributed to the ongoing emergence of novel IBV variants. Many of these variants show noticeable antigenic differences, which are carefully displayed by cross-neutralization experiments or monoclonal antibody tests. Nonstructural 1ab and accessory proteins, including 3a, 3b, 5a, and 5b, have been reported to be potential tissue tropism regulators as well as pathogenicity determinants. Moreover, the interaction between the S1 and S2 spike subunits of the virus may control the host range and attachment site of IBV. The host immune response and infection route are additional variables that may contribute to tissue tropism (Wang and Hou, 2023).
Diverse IBV strains have been found and documented in various geographical locations. Notably, the Georgia 08 (GA08) strain Genotype 1 and lineage 27 (GI-27) of IBV are the most common and prevalent in the United States (Jackwood and Jordan, 2021). Furthermore, reports of its presence outside the United States have been documented. Similarly, GII-1 (D1466) occurs infrequently outside western Europe (Cook et al., 1999). However, the GI-13 (793B) and GI-19 (QX) variants are largely distributed in Europe, Africa, and Asia but have not been found in the United States and Australia (Tegegne et al., 2020). The S1 spike gene sequence was used to categorize IBV genotypes, resulting in the identification of nine genotypes spanning dozens of lineages.
Two conditions must be met for classification as a new IBV lineage on the basis of the S1 gene: first, well-supported statistical evidence may usually be ascertained using bootstrap or posterior probability values; second, monophyletic clusters consisting of at least three viruses collected from at least two distinct outbreaks may be present, as defined by (Valastro et al., 2016). There must be 13% or greater uncorrected pairwise distances in the nucleotide sequences for IBV lineages to be defined by the abovementioned criteria. Various mutations, including insertions, deletions, point mutations, and recombinations, exist across different strains. As a result of these events, several S1 variants have emerged and evolved (Jackwood, 2012; Hewson et al., 2014). As reported, recombination often occurs during IBV replication, resulting in the formation of chimeric viruses made up of genetic sequences from several viruses (Abro et al., 2012). Thus, the antigenic diversity of IBV is influenced by many factors.
The sporadic emergence of different genotypes of IBV across continents can be explained individually. The genetic classification of the reported genotypes and lineages was reproduced by using the S1 gene sequence of all IBV genotypes (Figure 2). The overall distribution of these genotypes from 1930 to 2022 is also represented using the Ghantt chart (Figure 3) and (Table 1) to clarify the reported patterns of the genotypic distribution of this virus. The reported patterns are explained in detail in terms of genotype.
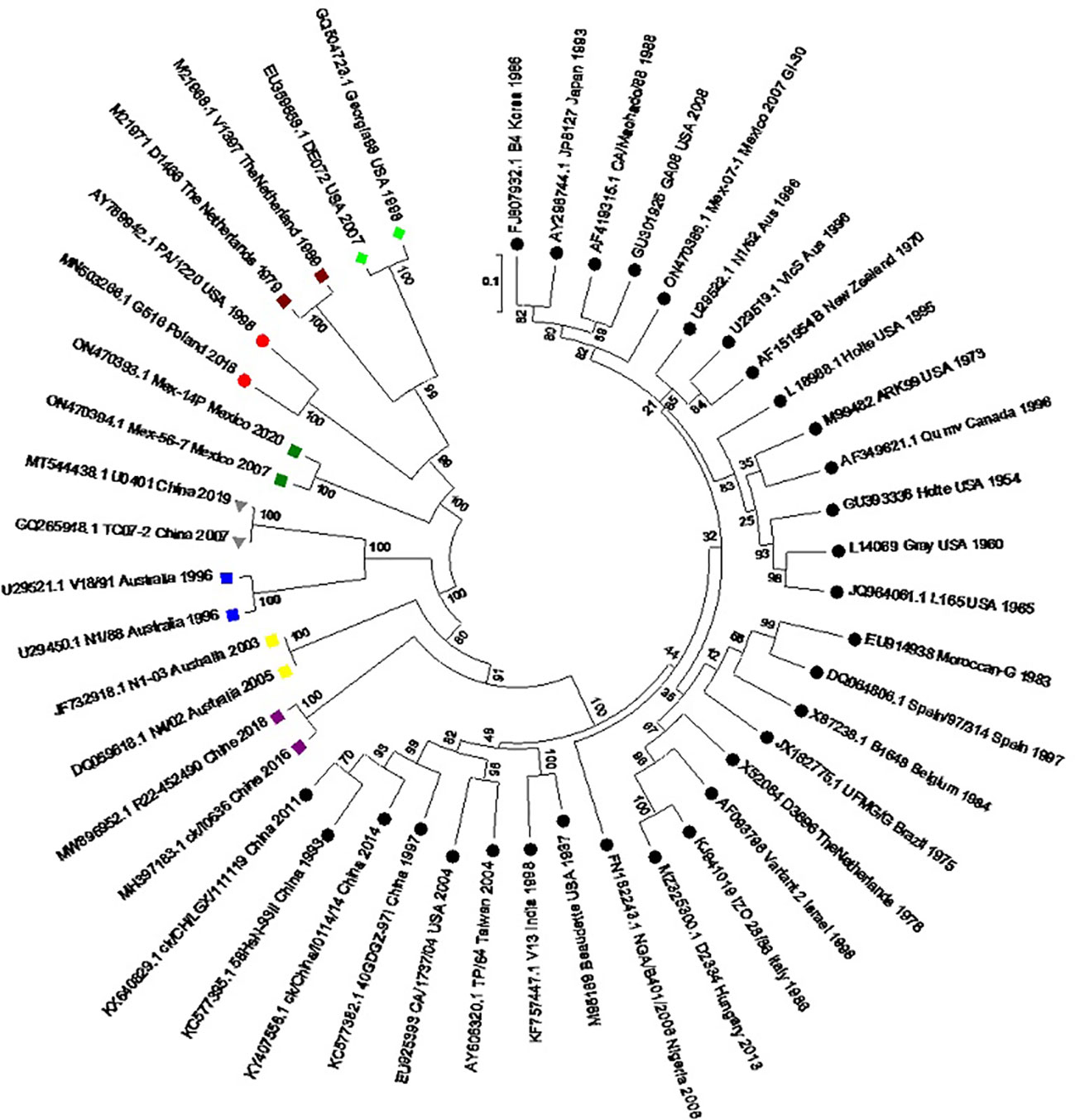
Figure 2 Molecular phylogenetic analysis of the S1 gene of the reference IBV genotypes. A circular tree was created by using the maximum likelihood tree based on the Tamura–Nei substitution model and 1000 bootstrap replicates. Different genotypes are highlighted with different shapes and colors. Black circles: GI lineages (1-31), Meroon square: GII, Blue square: GIII, Green rhombus: GIV, Yellow square: GV, Grey inverted-triangle: GVI, Purple square: GVII, Red circle: GVIII, Green square: GIX.
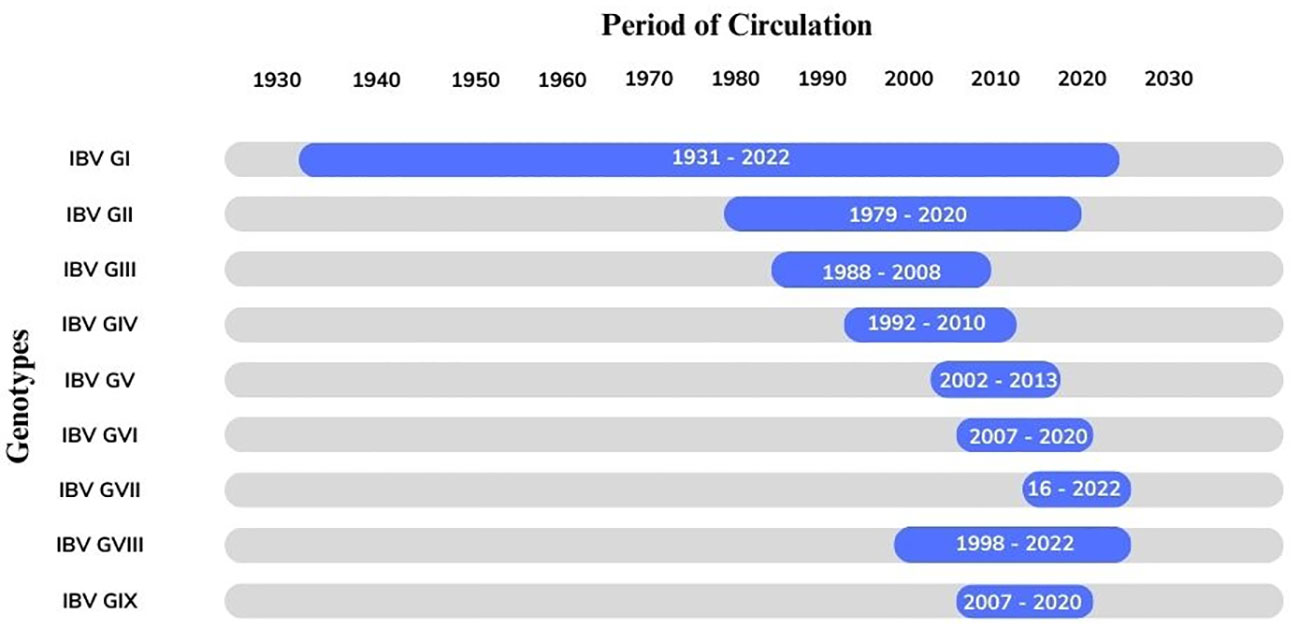
Figure 3 The Gantt chart represents the circulation of the avian infectious bronchitis virus (GI-GIX) genotype throughout the period from 1931–2022.
The first case of IBV was reported in central North American states in the spring of the early 1930s; this is an acute, deadly respiratory condition that appeared to be limited to only chicks aged 2 to 20 days. The chicks developed resistance with age, and those older than three weeks were immune. By the early 1970s, multiple distinct IBV variants had been identified, primarily through serological reports (Hitchner et al., 1966; Hitchner et al., 1966; Hopkins, 1974; Cowen and Hitchner, 1975; Johnson and Marquardt, 1976). In North America, the GI-9 and GI-27 and GIV-1 genotypes have been linked to continent-wide disease transmission and persistent viral infections (Valastro et al., 2016; Bande et al., 2017).
Previously, Arkansas (Ark) (GI-9) was the most prevalent IBV strain known in North America, but recently GA08 (GI-27) became a dominant genotype in this region due to high usage of this strain in vaccines (Fields, 1973; Jackwood and Jordan, 2021). Another significant IBV strain in the United States is DMV/1639/11 (GI-17), which was first documented in the United States in 2011 and has significantly affected broilers, breeders and layers from 2014–2015; since then, it has prevailed in certain regions of the US (Jackwood and Jordan, 2021).
The IBV 4/91 strain IBV/Ck/Can/17 038913 (GI-13) was identified from chicken flocks in eastern Canada that had reduced egg production and egg quality from 2011 to 2018. The isolate was assigned to the GI-13 genotype based on comparative genomics and phylogenetic analysis of the S1 gene, and it exhibited varying similarity with different open reading frames (ORFs) of several reference strains. Despite having wide tissue tropism in laying hens, IBV/Ck/Can/17 038913 (GI-13) did not affect egg quality or production at the infection dose used in the current investigation. However, the isolate caused considerable macrophage and T-cell migration along with tissue tropism in the kidney (Najimudeen et al., 2021). Although Ark (lineage GI-9), Holte/Iowa-97 (lineage GI-3), Mass-type (lineage GI-1), and Q1-like serotypes have been identified within commercial flocks in Mexico, a new study indicated that in 2019–2021, 793B (GI–13) and California variant CAV (GI–17)-like strains were found (Kariithi et al., 2022).
The Delmarva (DMV)/1639 variant of IBV, which is a member of GI-17, was linked to numerous nephropathogenic IBV outbreaks that occurred in the Delmarva Peninsula, United States of America (USA), in 2011. IBV strains genetically similar to the DMV/1639 (GI-17) type have been more prevalent in the poultry industry in eastern Canada since 2015. The Canadian DMV/1639 (GI-17) strain has been proven to cause noticeable pathological lesions and tissue tropism in many bodily systems, including the respiratory, reproductive and renal systems (Ali et al., 2022). Another study provided a clear illustration of the distribution of DMV strains, in which five different IBVs were isolated from Ontario, Canada. S gene analysis of these strains revealed high similarity to that of the Delmarva (DMV/1639) (GI-17) strain, and the initial detection of this virus occurred during an IBV epidemic on the Delmarva Peninsula in the USA in 2011. However, whole-genome analysis revealed its relevance to the Conn (GI-1)-type vaccine strain. These isolates were grouped according to the GI-17 genotype (Hassan et al., 2019). Furthermore, a study illustrated the importance of the GI-17 lineage in North American regions; specifically, a virus involved in the IBV epidemic that occurred in Costa Rica from May 2016 through mid-2017 was identified and categorized as a Georgia 13-like on the basis of the S1 region of the S gene. The entire genome sequence has a sequence identity with that of the strain from Georgia, USA, of 94.03%, and the least similar sequence identity with that of the strain from China is 86.03%. The Costa Rican isolate, which shows 96.89% similarity with the S1 subunit of the Georgian strain, was identified as belonging to genotype I and lineage 17 (GI-17) (Villalobos-Agüero et al., 2021).
In the 1950s, IBV spread to South America, with Brazil having the first mass (GI-1) serotype isolate (de Wit et al., 2011). IBV was recognized as a serious threat to commercial chicken populations by the mid-1980s. New mutations were discovered alongside the Conn (GI-1) and Mass (GI-1) serotypes in both layer and broiler birds (Cubillos et al., 1991).
In South America, the GI genotype has exhibited two distinct lineages, GI-11 and GI-16, which have been in extensive circulation for many years. An entirely South American lineage known as GI-11 arose in Brazil, Uruguay, and Argentina in the 1960s. Conversely, the GI-16 lineage is ubiquitous and consists of two genetic groupings that were previously thought to be independent: Q1 (GI-16) and 624/I (GI-16). The 624/I variant was initially detected in 1993 in Italy and subsequently in Slovenia, Poland, and Russia. Emerging in the late 1970s, the GI-16 lineage first appeared and is now found in the majority of South American states. Retrospective research has shown that it has been prevalent in Italy since the early 1960s, with a substantial drop in disease incidence since the 1990s. Q1 (GI-16) was also found in birds with proventriculitis in China in the 1990s and subsequently in other Asian nations, South America, Europe and the Middle East. Recent sequence analysis of the whole genomes of the Italian 624/I (GI-16) and Q1(GI-16) strains support the idea that these two genetic lineages have a common ancestor (Marandino et al., 2022). The IBV strain known as VFAR-047 was discovered in 2014 in the broiler flock and was referred to as GI-16 in Lima, Peru’s northern area. Examination of the S gene and genome sequencing allowed for the identification of this isolate (Tataje-Lavanda et al., 2019). According to a recent Peruvian study, the circulation of GI-16 has been determined along with the GI-1 (vaccine derived) lineage in the poultry industry of Peru. GI-I is prevalent in tropical areas, and GI-16 is prevalent in coastal areas (Icochea et al., 2023).
According to a report from Trinidad and Tobago, the circulating IBV strain shows more than 20% nucleotide differences from the vaccine strain M41 (GI-1) but 16.7% from the GI-14 lineage. Since the Trinidad and Tobago strains do not cluster with other lineages, they are distinct strains of a completely new lineage (Brown Jordan et al., 2020). Apart from the prevalence of the GI-11 and GI-16 genotypes in South America, the first report of the IBV GI-23 variant was recorded in Brazil in 2022. In 1998, for the first time, the GI-23 variant was found to cause disease in poultry in India. Immediately after, the virus quickly spread to other continents. Among IBV variants, GI-23 strains are significantly pathogenic to the respiratory tract of embryos and specific pathogen-free (SPF) chicks (Trevisol et al., 2023). Another study from Colombia, the country where IBV was initially discovered in 1963, reported the prevalence of GI-1 (Ramirez-Nieto et al., 2022).
In Mexico, during 2007 and 2021, 17 avian infectious bronchitis virus strains were isolated from diseased chickens. Different strains were grouped into different lineages; however, six strains in genotype 1 did not belong to any of the identified lineages. Therefore, after clustering in a well-supported clade, the cells were designated as a new lineage (GI-30) (Mendoza-González et al., 2022).
The Mass (GI-1) serotype of IBVs was considered the only disease-causing IBV until the late 1970s. Dedson & Gough announced in 1971 that other IBV variants had been found in the UK (Dawson and Gough, 1971); subsequently, employees at the Doorn Institute discovered IBVs in mass-vaccinated commercial flocks that belonged to not less than four distinct IBV serotypes linked to disease occurrence (Davelaar et al., 1984). According to a surveillance study conducted in Europe, the most prevalent IBV strains found were Massachusetts (GI-1), QX-like (GI-19), 793B (GI-13), D274 (GI-12), and D1466 (GII-1) (Worthington et al., 2008; Pohjola et al., 2014).
The GI-19 genotype was initially reported in China in the late 1990s and was formerly known as the QX (GI-19) genotype; its high prevalence has made it one of the most common IBV (Liu et al., 2005), after which it spread to other Asian, Middle Eastern and European countries in waves. The GI-19 lineage attracted increased amounts of attention when it was first discovered in the Netherlands between 2003 and 2004. This viral strain was detected in the kidneys and oviducts of infected poultry and was linked to a significant prevalence of false layer syndrome. In Italy, sequencing analysis indicated that the S1 gene shared 99% similarity with the QX (GI-19) strain. In fact, the alignment of the sequence of the hypervariable region of the S1 gene of QX (GI-19) with that of the mass (GI-1) and 4/91 (GI-13) strains was 77.1% and 81%, respectively (Terregino et al., 2008). Later, in the 2000s, the Chinese QX (GI-19) strain was detected for the first time in the United Kingdom (UK). Sequence analysis revealed 96.6% similarity to the original Chinese QX (GI-19) strain and 98.1% similarity to the common European QX-type (GI-19) IBV known as L-1148 (Gough et al., 2008).
Research carried out in Spain and Italy revealed the simultaneous spread of various IBV genotypes. The genotypes that were most prevalent in the two countries in recent years were 793B (GI-13) and QX (GI-19) (Franzo et al., 2014). However, a new IBV variant linked to the XDN variant has emerged (Ji et al., 2011). The Spanish IBV isolates and the XDN strains were found to form a homogenous clade, and both belonged to the QX (GI-19) genotype, while the Italian strains exhibit greater variation in their genomes. The large and homogeneous clade identified in Spain was thought to have descended from a single recombinant parent and then dispersed across the entire nation. In contrast, nine Italian recombinants, which were distinguished by three distinct recombination events and resulted from independent recombination events between the QX (GI-19) and 793B (GI-13) IBV variants, were considered less viable and less suited strains (Moreno et al., 2017).
The GI-23 lineage IBV variant known as Israel strain 2 was initially identified in Israel in 1998 (Meir et al., 2004). The first incidence of IBV VAR2 from the GI-23 genotype was found in Poland in December 2015. The whole-genome analysis of the Polish GI-23 lineage revealed that the mosaic pattern of the viral genome from other IBV strains descended from the QX-like (GI-19), 793B-like (GI-13), and mass-like (GI-1) serotypes. The exact origin of the virus that infects the Polish chicken population is still unknown (Lisowska et al., 2017).
However, a German broiler farm also reported the IBV Israel strain 2. This was the first time that the GI-23 lineage of Middle Eastern origin was isolated in Germany, adding to the increasing concern about the spread of IBV variants across Europe (Fischer et al., 2020). The birds exhibited respiratory and nephropathogenic symptoms. Phylogenetic analysis suggested that the current isolates are members of the GI-23 lineage, indicating that they are most genetically related to Polish variants and the IBV VAR206 (GI-23) vaccine. The possibility of recombination events between Polish GI-23 and German field or vaccine strains is evident given the potential of GI-23 strains to undergo recombination (Zanaty et al., 2016a).
IBV emerged in southern Africa in the early 1980s, resulting in severe disease conditions such as swollen head syndrome (Morley and Thomson, 1984). It was first identified in Mediterranean basin countries. Although IBV is frequently found in both symptomatic and asymptomatic poultry, its effect on the African continent is unknown (Bali et al., 2022).
Although several lineages of IBV have been recovered in Africa, only GI-26 is considered indigenous to the continent and is predominantly composed of strains from North and West Africa (Valastro et al., 2016). Other Eurasian-originating African IBV strains include those from the lineages GI-1 (linked with sporadic IBV outbreaks in many countries), GI-12 (Nigeria), GI-13 (South Africa, Morocco, Ethiopia, Sudan, and Algeria), GI-14 (Nigeria & Cameroon), GI-16 (Nigeria & Côte d’Ivoire), GI-19 (Ghana, Nigeria, South Africa, Zimbabwe, and Algeria), and GI-23 (Nigeria & Egypt), as well as various lineages from Algeria, Tunisia, Libya, and Ethiopia (Valastro et al., 2016; Bande et al., 2017; Moharam et al., 2020).
IBV has not been reported from Kenya or the East or Central African regions up until this point, except Cameroon in Central Africa (Bali et al., 2022). It is possible that the Kenyan strains A374/17 and A376/17 are gastroenteric. S1 gene analysis of this strain revealed its resemblance to GI-23 viruses rather than GI-16 viruses. The four lineages GI-1, GI-13, GI-14, and GI-19 are used to categorize other strains of African IBV. In contrast, the Kenyan isolate A376/17 resembles turkey coronaviruses (TCoVs) from Asia and France more closely than those from North America. However, unlike all other TCoVs that have been studied, TCoVs exhibit distinct differences (Kariithi et al., 2023).
In Ethiopia, sequencing analysis confirmed the presence of 793B genotypes (GI-13). Due to the high frequency of 793B (GI-13) live vaccine administration, these Ethiopian IBV isolates exhibited substantial genetic variation, indicating that they are most likely to be field strains. Only backyard poultry showed positive results, indicating that this type of setting is more conducive to IBV circulation (Tegegne et al., 2020).
There are several different IBV genotypes in Egypt, comprising GI-13, GI-23, GI-1, and GI-16, each with unique genetic and pathogenic characteristics. Based on phylogenetic analysis, the circulating IBV strains in Egypt can be divided into two major groups. The first group consists of strains from the GI-1 lineage that have developed through genetic drift from live attenuated traditional vaccines (Zanaty et al., 2016b). The second group consisted of strains from the GI-23 lineage, including circulating Egyptian variants. According to recombination analysis, a minimum of three distinct IBV strains, 4/91 (GI-13), H120 (GI-1), and QX-type (GI-19), tend to undergo recombination events (Abozeid et al., 2017).
In North African nations such as Tunisia, Egypt, and Morocco, the combination of endemic and classic IBV variants prevails, contributing to nearly all the information on IBV strains in Africa (de Wit et al., 2011). A QX-like (GI-19) IBV strain was recently discovered in Zimbabwe. The IBV strain QX L-1148 (GI-19) from China shows the maximum nucleotide sequence similarity (98.6%) to the S1 gene. Notably, this is the first reported instance of QX-like (GI-19) IBV circulation in Africa (Toffan et al., 2011).
This GI-19 lineage was introduced in 2009, likely as a result of a single introduction event from Germany (Andreopoulou et al., 2019). In Sudan, two IBV strains were identified from commercial broiler farms that exhibited respiratory symptoms and increased mortality. The Chinese IBV strain, the Italian IBV strain, and the 4/91 (GI-13) vaccine strain had the highest nucleotide sequence similarity for the whole genome (93%). The IBV/Ck/Sudan/AR251-15/2014 strain was genetically distinct from formerly defined IBV strains in Africa and worldwide on the basis of S1 gene analysis and was clustered with viruses of the GI-19 lineage (Naguib et al., 2016). Greece has a high incidence of IBV field strains, with 20 or more of the GI-19 lineage (Andreopoulou et al., 2019).
In a study in Africa, the complete genome sequences of five IBV strains from the sub-Saharan region were analyzed. Based on phylogenetic analysis, three (GI-14, GI-16, and GI-19) lineages prevailed in that region. However, it has also been observed that a strain isolated from the Ivory Coast, D2334/11/2/13/CI, belongs to a distinct lineage within the GI-31 genotype (Bali et al., 2022).
A novel IBV genotype known as “IBADAN” has recently been reported. Analysis of the S1 gene sequence indicated that NGA/A116E7/2006 had 24-25% amino acid and nucleotide variation from the nearest strain of a different cluster, whereas ITA/90254/2005 shows 14% nucleotide and 16% amino acid variation from its closest strain. When the whole genomes of NGA/A116E7/2006 and ITA/90254/2005 were analyzed, 10% nucleotide variation was found, with the IBADAN strain exhibiting a high genetic distance from 9.7% to 16.4% when compared to other known IBV sequences. The S1 protein amino acid sequence revealed particular positions shared by NGA/A116E7/2006 and ITA/90254/2005, differentiating them from other strains (Ducatez et al., 2009).
Several lineages, such as GI-7, -15, -18, -22, and -24, and GVI-1, are exclusive to Asia (Valastro et al., 2016). In Japan, IBV strains have been genotyped via incomplete nucleotide sequence information that included the HVR-1 and HVR-2 regions (Mase et al., 2004; Mase et al., 2021a). The strains were divided into seven genotypes based on this classification: JP-I (GI-18), JP-II (GI-7), JP-III (GI-19), JP-IV (GVI-1), Mass (GI-1), 4/91 (GI-13), and Gray (GI-3). The JP/KH/64 strain (GI-18) was identified from chickens exhibiting respiratory symptoms in 1964 in Japan, and it was the first JP-I genotype to be identified in Japan (Mase et al., 2021a).
The complete genome sequence of the primary genotype JP-I (GI-18) strains was retrieved to further understand the distribution of IBV variations in Japan. Phylogenetic analysis of the full coding sequence of the S1 gene revealed that JP/KH/64 is part of the GI-18 lineage and groups in a single category with the lineage prototype strain JP8127, which was discovered in Japan. Because the JP-I (GI-18) genotype has been reported in China, Taiwan, and Japan (12, 13), studying the evolution of IBV in East Asian countries may be aided by this knowledge of the JP/KH/64 strain (Mase et al., 2021b). Another study in Japan determined and studied the entire S1 gene of 61 IBV strains. Except for three strains (JP/Nagasaki/2013, JP/Kochi/2013, and JP/Nagasaki/2016), all strains grouped into the seven genotypes indicated above and were thought to be produced from recombination within the IBV G1-13 and GI-19 S1 genes (Mase et al., 2022). IBV was first characterized in South Korea in 1986 after respiratory signs were detected in poultry. It was initially classified as Korean group I (K-I) and subsequently assigned to the GI-15 lineage (Song et al., 1998). A nephropathogenic form of IBV, currently known as the Korean group II (K-II) (GI-19) subgroup KM91-like variant (Lee et al., 2004), devastated the South Korean chicken industry in 1990. The QX (GI-19) genotype, originally discovered in 1993 in China, is now widespread worldwide and was introduced to South Korea in 2002-2003. Presently, this particular strain is referred to as the QX-like (GI-19) K-II subgroup variant (Lee et al., 2008). Novel variants of the KM91-like (GI-19) and QX-like (GI-19) strains, termed ‘K40/09-like’, appeared in 2009 and have since become the prevalent strains in South Korea (Lim et al., 2011). According to Valastro et al. (2016), by using the IBV S1 gene categorization method, all K-II (GI-19) subgroups (KM91-like, K40/09-like, and QX-like) and QX genotypes belong to the GI-19 lineage. The S1 gene sequences of 60 South Korean IBV isolates were analyzed, and it was discovered that in addition to the GI-15 and GI-19 IBV lineages, five other subgroups of GI-19 cocirculated and expanded in South Korea (Lee et al., 2021). Since its discovery in 1996, Lineage GI-19 (QX-type) has been the most prevalent IBV strain in China 1996 (Feng et al., 2017; Xu et al., 2018) Furthermore, GI-7 (TW-type) and GI-13 (4/91-type) lineages have been recognized to be significant IBV strains in China, with rare infections of GI-1 (Mass-type), GI-9 (Ark-type), and GI-28 (LDT3-type) also reported (Zhao et al., 2016; Lian et al., 2021). The latest studies found 19 IBV variants in clinical specimens collected in China amid January 2021 and June 2022; these included 12 variants of GI-19, three variants of GI-7, and one each of GI-1, GI-9, GI-13, and GI-28. The most common IBV lineages in China according to these studies are GI-19 and GI-7. The progression and dissemination of IBV GI-7 were also detected, and it was proposed that the region of Taiwan might be the source of the IBV lineage GI-7 and that South China has a significant role in IBV transmission (Chen et al., 2023).
Recently, the respiratory type Mass 41 (GI-1) was the most prevalent type of IBV in India (Kumanan et al., 2004). Elankumaran et al. (1999), reported the presence of the IBV variant 793/B by serological testing but did not detect the virus. Bayry et al. (2005), found a single strain of nephropathogenic IBV (PDRC/Pune/Ind/1/00) in India. Later, Gaba et al. (2010), and Sumi et al. (2012), identified and genotyped viruses, confirming the presence of 793/B, an IBV strain. Current investigations revealed that the Mass 41 (GI-1) genotype was present in field isolates (Patel et al., 2015; Parveen et al., 2017; Jakhesara et al., 2018). From 2003 to 2011, Raja et al. (2020), examined 20 IBV field strains from India, three of which were identified as novel IBV genotypes. These authors confirmed that these strains are similar to the GI-24 genetic lineage (Valastro et al., 2016) but not to formerly reported strains (Bande et al., 2017).
The IS-1494 Mahed (variant-2; GI-23) nephropathogenic IBV strain was originally discovered in Iran in December 2015. The strains were grouped with QX (GI-19) and 4/91 (GI-13) according to nucleoprotein gene analysis. However, phylogenetic analysis revealed it to be a chimeric strain (Mousavi et al., 2018).
In Pakistan, there have been few reports of IBV circulation. Ahmed et al. (2007) documented the dissemination of several European IBV variants in addition to the Massachusetts type. Many IBV vaccines were not effective locally, particularly those with D-1466 (GII-1), 4/91 (GI-13), D-274 (GI-12), or Mass-41 (GI-1), indicating the existence of unidentified IBV variants in the field. Rafique et al. (2018b), detected the existence of IBV in a one-year study that monitored the spread of IBV in Pakistan. Their results indicated the presence of mass (GI-1)- and 4/91 (GI-13)-type IBV in 43% and 51%, respectively, of the isolates, but only 5% of the various untyped IBV variants were present. Current analysis also revealed one unique Pak-973 isolate (KX013102_Ck/Rwp/NARC-973/2015_Pakistan). Compared to the 4/91 (GI-13) and mass (GI-1) vaccine strains, the isolate had distinct mutations. Another study by Rafique et al. (2018a), molecularly characterized the field strain KU145467_ NARC/786_Pakistan_2013 (also known as Pak-786) in the same year. The Pak-786 isolate is a member of the GI-13 lineage and includes both vaccine and field strains formerly ascribed to the 793B group (Rafique, 2018). A recent study revealed that IBV-17 of the GI-24 lineage differs from the vaccine strain GI-23, which has been widely used in Pakistan. These variations can result in changes that allow GI-24 lineage viruses to avoid vaccines designed for the GI-23 lineage. It appears that GI-24 has taken over as the main lineage of Pakistani field IBV isolates throughout the past few years (2017-2020) (Saleem et al., 2024).
A ‘HFT-IBV’ variant was discovered in a layer chicken flock in Israel that was routinely vaccinated against 4/91 (GI-13) and H120 (GI-1) variants. The disease was caused by the Israeli variant IS/1494/06, which belongs to the GI-23 lineage. The HFT-IBV isolate shares 97.7% nucleotide sequence similarity with the IS-Var2-like isolates but less than 90% nucleotide sequence similarity with the M41-related (GI-1) (H120, Ma5, M41) and 4/91 (GI-13) (793/B, Moroccan G/83, and CR88) strains. IS-Var2-like genotypes have been assigned to the GI-23 lineage, whereas M41 was assigned to the GI-1 lineage; 4/91-like viruses were assigned to the GI-13 lineage (Öngör et al., 2021). The IS var-2 IBV field isolates recovered from commercial broiler flocks in Turkey from 2014 to 2019 were discovered to be on the same branch as the GI-23 genotype, which has 99% similarity and is one of the most common wild-type clusters in the Middle East (Müştak et al., 2022).
In 2014, three infectious bronchitis virus (IBV) strains were isolated and identified from hens suspected of being infected with IBV in Guangxi Province, China: CoV/ck/China/I0111/14, CoV/ck/China/I0114/14, and CoV/ck/China/I0118/14. S1 sequencing and phylogenetic analysis revealed that the three IBV isolates are genetically distinct from other known IBV types, indicating the presence of a novel genotype (GI-29) (Jiang et al., 2017).
IBV lineages GI-1, GI-13 and GI-19 are prevalent. According to a study confirmed the presence of various IBV lineages in Thai chicken flocks, along with a novel recombinant IBV variant that originated from the GI-19 and GI-13 lineage viruses (Munyahongse et al., 2020). In another study, variant IBV isolates were categorized into four groups in a study conducted on IBV infection in chickens in Thailand between 2014 and 2016: QX-like IBV (GI-19), Massachusetts (GI-1), 4/91 (GI-13), and a novel variant. QX-like was the most prevalent IBV genotype among these groups in Thailand (Munyahongse et al., 2020). In a recent study, vaccine strains were compared with local common Malaysian IBV strains with two isolates of ACoVs from guinea fowl and jungle fowl. The two isolates from the sample were categorized as genotype I and placed in the GI-13 lineage alongside three other frequently occurring local vaccine strains, namely, CR88, 793B, and 4/91. Molecular characterization revealed homology to the common IBV vaccine strain 4/91 (GI-13) among ACoV isolates from diagnostic cases of jungle fowl (isolate 2015) and guinea fowl (isolate 2016; Besar et al., 2023). In the Vietnamese provinces of Hanoi, Thainguyen, and Haiphong, three strains of IBV, known as VNUA3, VNUA8, and VNUA11, were isolated from sick or infected chickens. The Vietnam isolates belonged to three genotypes: Q1-like (GI-16) (VNUA3), QX-like (GI-19) (VNUA8), and TC07-2-like (GVI-1) (VNUA11). This finding indicates that at least three different IBV genotypes are circulating in North Vietnamese poultry (Le et al., 2019). A study conducted at four farms in Myanmar revealed through phylogenetic analysis of the S1 gene that the IBVs are closely related to the C-78 (GI-18) IBV vaccine strain, while the IBV found at farm 2 was found to be closely related to the GN strain (both of which are categorized as JP-1 (GI-18) types). The isolates from farms 4 and 5 are similar to K446–01 (mass type) (GI-1) and TM86 (JP-2 type (GI-7)), respectively. All of the identified IBV types are extensively distributed, and commercial vaccines targeting their weakened strains are accessible. The JP-1 (GI-18), JP-2 (GI-7), and Mass (GI-1) types of IBV were found to be present in Myanmar poultry farms during this survey (Fujisawa et al., 2019).
In this geographically isolated country, IBV evolved distinctly from the rest of the world (Ignjatovic et al., 2006). Since the early 1960s, several distinct IBV variants have been identified and characterized (Cumming, 1963), and several lineages (GIII-1, GI-6, GV-1, GI-5, and GI-10) have been found to be exclusive to Australia and New Zealand (Valastro et al., 2016).
The traditional IBV variants from Australia have been divided into two distinct lineages, GI-5 and GI-6, which can be identified by the VicS and N1/62 vaccine strains. Surprisingly, both the N1/62 and VicS strains were discovered in 1962, albeit in different parts of Victoria and New South Wales (Cumming, 1963). Although their S1 gene sequences are 83% identical, their distinction into distinct lineages indicates either a unique ancestral origin or major divergence from a parental strain at the time of isolation (Sapats et al., 1996; Mardani et al., 2008; Quinteros et al., 2015).
IBV was not prevalent in New Zealand until the 1970s, when the first case of IB emerged (Lohr, 1976). Notably, strains from New Zealand are included in the GI-5 and GI-6 lineages. New Zealand’s Strain A clustered in the GI-6 lineage (Sapats et al., 1996; Mardani et al., 2008; Quinteros et al., 2015). Six indigenous New Zealand viruses, three of which were identified in the 1970s and the remainder in the 2000s, are part of the GI-10 lineage (McFarlane and Verma, 2008). This IBV strain was identified in the region for the first time in 1967 (Pohl, 1967). According to virus neutralization testing, 4 serotypes (A, B, C, and D) of the virus were identified in New Zealand in 1976; these serotypes differ from those found in other countries (Lohr, 1976). Four new IBV variants, K43, T6, K32, and K87, were discovered later in 2008 in clinically infected flocks. The sequence homology between these strains and the previously mentioned B, C, and D strains is more than 99% (McFarlane and Verma, 2008). Phylogenetic analysis of New Zealand strains confirmed the relevance of these strains to Australian (Vic S) (GI-6) strains rather than European or North American strains (Ignjatovic et al., 2006).
In the late 1970s, the GII-1 lineage of IBV (also known as D1466 (GII-1) or D212 variation) was identified initially in the Netherlands as an etiological factor associated with egg loss (Davelaar et al., 1984; de Wit et al., 2011). Due to differences between its S1 coding region and those of other European IBV strains, the variant was grouped with the Dutch V1397 strain under the GII genotype (Valastro et al., 2016). Many studies indicate that compared to other IBV strains, this variant has considerably different antigenic and molecular characteristics (Lin and Chen, 2017).
For many years, the D1466 (GII-1) strain was only sporadically recognized, but the findings of a genetic investigation carried out between 2005 and 2006 revealed that this variant was causing additional problems in Western European countries (Worthington et al., 2008). Only a few infections are detected by the D1466 (GII-1) mutation in the UK and France; nevertheless, the disease dynamics are increasing in other European countries. In countries such as Germany, the Netherlands, and Belgium, the prevalence of the D1466-like (GII-1) virus was on average between 3% and 5% in 2005 and increased to 10%, 7%, and 16%, respectively, in 2006. In Poland, the conventional nested RT−PCR technique was used from November 2011 to December 2013. The first D1466 (GII-1) IBV was detected during this time, resulting in 26 positive samples or a prevalence of the variant in 11.7% of the surveyed chicken flocks. Our findings revealed that the prevalence of GII-1 IBV is gradually decreasing in Poland. Certain D1466-positive chicken flocks were declared healthy by healthcare personnel, indicating that the virulence and pathogenicity of the GII-1 strains are not severe (Domanska-Blicharz et al., 2017).
GII-2 (D181) is a novel IBV strain that emerged from an unexpected report in 2017 and became the second most isolated IBV strain among the breeders and layers in the Netherlands in 2018. This strain was also found in Belgian and German samples. According to the entire S1 gene and maximum likelihood analysis, D181 is more closely related to GII-1, commonly known as D1466 (GII-1), than to any other IBV strain, the latter of which shares 90% of the sequence. The remaining 10% are mutations that are distributed throughout the whole S1 gene, and a recombination study provided no evidence that the S1 gene resulted from recombination between D1466 (GII-1) and other IBV strains (Molenaar et al., 2020).
The IBV genotype GII lineage was identified in Georgia, USA, in 2000, and its whole genome was submitted to GenBank under accession number AF274435.1 (Icochea et al., 2023).
In an investigation, RT−PCR was employed to identify the IBV D1466 strain of genotype GII-1 (Khaltabadi Farahani et al., 2020). This strategy was validated by studying the spike protein-encoding area in the proprietary S gene of the GII-1 pedigree (pseudo-D1466 strain), which corresponds to IBV(Domanska-Blicharz et al., 2017).
The GIII-1 lineage originated in 1988 and was given the name Australian subgroup II (Sapats et al., 1996). The IBV isolates Q3/88 (GIII-2) and N1/88 (GIII-1) were identified from these outbreaks (Ignjatovic and McWaters, 1991; Sapats et al., 1996); they are genetically and antigenically different from all formerly identified classical variants, assigned as new or subgroup 2 variants, and were subsequently categorized as the GIII genotype (GIII-1 and GIII-2 lineages, respectively) (Valastro et al., 2016).
Phylogenetic analysis of the 27 identified IBV strains revealed that amino acid residues of the S1 glycoprotein align with the H120 (GI-1) vaccine variant. The isolated viruses were divided into three genotypes based on their genetic origins (genotype I, II and III). Among these isolates, Li et al. (2010) identified these variants at the genotype III level; these included 3 isolates from 2004, 4 from 2006, 1 from 2007, and 6 from 2008 (Li et al., 2010).
GIV lineage 1 (GIV-1) is the only North American lineage with a unique genotype. This category included (n = 24) vaccinated and field strains identified during 1992 and 2003. The Delaware variant (DE or DE072) (GIV-1) was one of these variants and first discovered in commercial broiler flocks infected with severe respiratory infections in 1992. This difference was attributed to the distinct genotype and novel serotype of this strain compared to the other strains (Gelb et al., 1997; Mondal et al., 2001). The IBV strains that were once known as GA98 (GIV-1) were found to be similar to the DE variants despite having a different serotype and are still considered a part of the same lineage (Lee et al., 2001). It has been proposed that the GA98 (GIV-1) variant emerged as a result of immunological selection triggered by the DE072 (GIV-1) attenuated live vaccine that was administered throughout the country during 1993 (Lee and Jackwood, 2001). Additionally, in 2000, this lineage included viruses that were found in layer flocks and led to decreased egg production (Mondal et al., 2001).
Two previously known Taiwanese IBV isolates evolved into a single genotype (GIV), indicating that the development phenomenon in Taiwan was isolated (Li et al., 2010).
The GV-1 lineage, known as Australian subgroup III, was reported in 2002; approximately 14 years later, GIII was identified (Ignjatovic et al., 2006). Respiratory and endemic Australian diseases (4 and 7 variants, respectively) have been characterized in GV lineages. The variants Q1/13 and V1/07 were isolated from broilers with respiratory symptoms in Victoria and Queensland in 2007 and 2013, respectively. These results are identical to those for N1/03 at the genetic level, indicating increased geographic dissemination of genotype GV strains (Hewson et al., 2009).
A GVI-1 strain was initially discovered in Guangxi Province, China, in 2007. The TC07-2 strain was shown to be significantly evolutionarily distant from six other main genotypes (Li et al., 2010). GVI was subsequently isolated in South Korea and Japan, among other Asian nations (Mase et al., 2010; Jang et al., 2018). The respiratory tract tropism of GVI-1 strains may be attributed to extensive recombination of gene 3 with the S gene (Ren et al., 2019). Nonetheless, not all GVI-1 variants descended from a single common ancestor.
A variant of IBV GVI is a newly found strain that is not particularly infectious to poultry, but coinfection with an epidemic variant may occur and harm China’s poultry industry. In China, IBV genotype VI (GVI-1) was identified in two separate studies from 2019 to 2020 (Wang et al., 2022). A comparison of the whole genomes of two IBV variant strains in the present study with those of other genotype variants indicated only minor similarities in the 5a, 5b, M, and N genes, with few previously identified GVI-1 variants, but greater similarities with the GI-19, GI-22, and GVII-1 genotype variants. The S gene of GVI IBV was substantially dissimilar to the S genes of the QX (GI-19) and YN (GI-22) strains (Sun et al., 2021).
Furthermore, genotype VI was produced by three classical American strains and one Japanese strain (GI-18), and the isolated TC07-2 (GVI-1) and published DE/072/92 (GIV-1) strains had the greatest evolutionary distances to all six major genotypes, but their significance was unclear (Li et al., 2010). GVI-I was isolated from Japan, including Ibaraki/168-1/2009, JP/Chiba/2010, and JP/Kagoshima-3/2014 and shown to cause clinical symptoms such as a reduction in egg production, nephritis and respiratory problems, respectively (Mase et al., 2022).
Recently, the circulation of the GVI-1 lineage, which was geographically assumed to be contained in Asia, was detected in research in Colombia (Ramirez-Nieto et al., 2022).
In addition, another novel genotype emerged from China in the late 2020s and grouped as GVII from the I0636/1 isolate and the GX-NN130021 reference strain because it did not resemble other established lineages. In a study, an in-depth comparison and phylogenetic analysis of 74 complete sequences on the basis of the S gene were carried out that involved 73 representatives from each lineage and genotype along with the I0636/1 strain. Additionally, within the S gene of GI-18, at least two recombination sites are replaced with an unidentified sequence that most likely originated from another IBV strain. As a result, a new serotype with limited respiratory tract tropism in poultry emerged (Ma et al., 2019).
A novel IBV variant was discovered in Poland. This variant differs from previously discovered viruses and is closely related to the North American isolate PA/1220/98. The variant was identified as a distinct isolate on the basis of the S1 coding region and shows homology to other recognized GVII IBV genotypes. This lineage was categorized as distinct within the novel GVIII genotype using the standard criteria for designating a novel IBV genotype or lineage. The nucleotide identity of this strain with any known IBV genotype ranges from 52.7 to 58.1%, with maximum identity (81.4%) with the North American variant. This novel strain was subsequently identified in three other flocks of chickens with poor egg production. Notably, the virus has not yet been found in broilers (Domanska-Blicharz et al., 2020).
The newly identified genotype GVIII was found in two Mexican samples closely related to unique Mexican strains (UNAM-97/AF288467) (Valastro et al., 2016). However, GVIII-1 exhibits very little intra-lineage variation, with only a 2% difference in nucleotides and a 3% difference in amino acids. The evidence supporting their classification as a new genotype includes their distinct isolation in the flock and clustering within the phylogenetic tree; a significant divergence of 28% in nucleotides and 45% in amino acids from the nearest GIV-1; and 30 different amino acid alterations. The nucleotide and amino acid sequences of IBV genotypes vary by more than 29% (Mendoza-González et al., 2022).
Another genotype that emerged in North America in North America is called GIX-1, and distinct clustering of the two Mexican isolates was observed on the phylogenetic tree. The samples were collected 13 years ago from different parts of the country. With 35% nucleotide variation and almost 50% amino acid variation from the closest genotype, GVII-1, this novel genotype has diverged significantly. The same lineage also exhibits a 6% divergence in nucleotides and a 10% in amino acids. It contains 24 distinct residues and has two distinct amino acid insertions (Mendoza-González et al., 2022).
Diverse genotypes are thoroughly documented alongside their respective pathogenicity indexes and tissue tropism in the (Table 2) provided below. The pathogenicity indexes reveal diversity even within identical genotypes, highlighting the complex nature of strain-specific variations. Furthermore, a careful examination reveals that tissue tropism contradicts a one-to-one link with any specific lineage, highlighting the complexities of host-pathogen interactions. This in-depth investigation sheds light on the complex interactions between genotypes, pathogenicity, and tissue tropism, setting the framework for a more sophisticated understanding of microbial dynamics.
IBV has been linked to a number of clinical symptoms in its host, the domestic chicken. The virus appears to reach host cells via viropexis after initially replicating in the upper respiratory tract (Patterson and Bingham, 1976). Tissue tropism varies between strains, although the reason for this variation is unknown. Variations in IBV tissue tropism contribute to differences in clinical symptoms in infected birds. In general, these distinctions allow viruses to be classified as proventriculus, respiratory, reproductive, or nephropathogenic based on the major clinical presentations (Quinteros et al., 2022). On the basis of pathogenicity, IBV characterized as low moderate, and high pathogenic. These conditions can also have turned vice versa based on secondary and opportunistic microorganism infection. Nephropathogenic IBV strains induce nephritis in hens and are the most pathogenic IBVs, having mortality more than respiratory and reproductive strains (Chen et al., 1996). Proventriculitis causes ruffled feathers and respiratory symptoms such as tracheal rales, nasal discharge, sneezing, and coughing in birds. Ulcers and hemorrhages were found in the proventriculus papillae later on, and the condition is serious (Yu et al., 2001).
To control infectious diseases on modern poultry farms, biosecurity measures, and a productive management system are essential. For the avian influenza virus, this concept depends solely on having appropriate knowledge concerning the variables that affect viral transmission (Boender et al., 2007; Singh et al., 2018). Consequently, there is a dire need to thoroughly analyze the epidemiological factors responsible for the transmission of IBV, particularly the determinants of spread (Franzo et al., 2020; Najimudeen et al., 2020).
Vaccination is still the most effective way to manage IBV infection, despite its limitations, which include serious adverse effects of vaccination in young birds, the need for periodic vaccine replacement due to viral mutation, and the likelihood of viral recombination (Franzo et al., 2016; Ji et al., 2020). Effective and meticulously administered vaccines can reduce the viral load, rate of infection, and occurrence of clinical symptoms (Franzo et al., 2016). Several factors influence the strength and period of response to vaccination include chick age, vaccine immunogenicity, field strain virulence, vaccine administration strategy, extent of maternal immunity, and the time frame between vaccination and challenge. Vaccinated chickens may remain immune for several months, and in the case of broilers, this immunity may persist throughout their lifetime (Bijlenga et al., 2004). The majority of commercial chicken flocks are currently immunized against IBV. The IBV immunization protocol may change based on the type of vaccine used and the particular circumstances of the poultry farm. To maintain immunity, chicks are vaccinated throughout their life beginning at one day old. Booster agents can be administered at 7–10 days, 3–4 weeks, and then every 5–6 weeks afterward. The idea of a protectotype has become more widely accepted for regulating IBV due to its variants circulating worldwide.
Numerous vaccination strategies have been designed to manufacture IBV vaccines that are effective. However, the complex immune protection process prevents the extensive use of new vaccine approaches, which are still in the laboratory research stage (Kapczynski et al., 2003; Yang et al., 2009; Bande et al., 2015). The process of developing in ovo vaccination is also at the research stage. The vaccine will be based on the type of IBV strain and will not kill the embryos (Tarpey et al., 2006). The market offers a variety of IBV vaccines, which might differ in nature and vaccination strains based on local isolates and recombination in variants isolated from various countries with distinct laws and regulations. Heterologous IBV vaccinations effectively provide immunization against the 793B-type (GI-13) variant, which was previously proven to persist with live attenuated IBV vaccines and to be effective against the QX (GI-19) and Italy 02 (GI-21) strains (Cook et al., 1999). In a study in Korea, it was demonstrated that the K2 vaccine may be more potent for preventing and controlling novel IBV recombinants and variants that are spreading (Shirvani and Samal, 2020). To manage IBV in China, H120 (GI-1) vaccination is frequently used in conjunction with the indigenous FNO-55 (GI-13, 4/91-like), QXL87 (GI-19, QX-like), or LDT3-A (GI-28, YN-like) variants.
For serotypes, including Arkansas (GI-9), Massachusetts (GI-1), and Conn (GI-1), improved live vaccines and killed oil-based emulsions are available in North America. The Georgia 98 (GIV-1) and California (GI-17) strain vaccines were obtained from the USA. The vaccinations, designated “Holland variants” D274 (GI-12) and D1466 (GII-1), are generally produced in Europe. Conversely, vaccines based on the H120 (GI-1) strain are being used throughout Europe.
The degree of immunity may rely on regional sources to produce varying levels of immunity and an unusual ability to cross-protect against a few heterologous IBV strains Florida (GI-1) and JMK (GI-3) in the U.S. Overall, combining the IBV 4/91 (GI-13) and Ma5 (GI-1) variant vaccines may provide excellent protection against heterologous IBV strains. Although QX-type (GI-19) live vaccines have been developed in Europe, their use is restricted (Cook et al., 2012). A new generation of IBV vaccines against the regionally dominant D274 (GI-12) variant has been produced for future layer and breeding stocks.
One study demonstrated that the type of tissue, inoculation route and vaccine strain all affect the pattern of IBV replication. The distribution and elimination of the vaccine viruses for Massachusetts (GI-1) and 793B (GI-13) were slower when the viruses were administered by drinking water (DW) than when they were administered via the oculonasal (ON) route. Both vaccines were able to induce similar levels of mucosal immunity when administered via the ON route. Regardless of the vaccination technique, the Mass IBV vaccine induces cellular immunity at comparable levels. The 793B vaccine produced noticeably greater levels of humoral immunity when administered via the ON or DW route (Al-Rasheed et al., 2021). In a study, birds were given bivalent live attenuated IB vaccines containing the Mass and Conn serotypes at intervals of two, five, nine, and fourteen weeks after they were first primed with a monovalent live attenuated IB vaccine (mass serotype) at one day old. There was no apparent difference in the ability of the two vaccination regimens to protect laying hens against mass IBV challenge. These findings suggest that the vaccine strain may have a greater level of protection when faced with homologous IBV strains (Ali et al., 2023). The probability of postvaccination challenges is infrequent, but postvaccination challenges may lead to reversion to virulence in immunocompromised or unvaccinated chickens, which eventually causes significant mortality and the intermittent spread of IBV (Hiscox et al., 2001; Bhuiyan et al., 2021). The inability of birds to generate an adequate immune response after vaccination is the cause of vaccine failure (Bosha and Nongo, 2012). It is noteworthy that vaccination techniques are subject to change over time in response to the introduction of novel IBV genotypes and improvements in vaccine technology. Furthermore, local elements influence vaccination programs differently across different regions. These include disease prevalence, farm size, biosecurity measures so on.
This review emphasizes the critical relevance of understanding genotypic variations to implement effective control measures. The identification of region-specific genotypes provides poultry stakeholders with customized vaccination, biosecurity, and management measures. With the ever-changing IBV landscape, the incorporation of genotypic information into control systems has emerged as a critical tool. This knowledge synthesis not only improves our understanding of IBV epidemiology but also allows for the creation of customized therapies to reduce the impact of this economically significant poultry virus. This review, in essence, serves as a foundation for expanding the understanding and control of IBV, hence encouraging sustainable and resilient poultry production systems worldwide.
SR: Writing – original draft, Writing – review & editing. ZJ: Writing – review & editing. TP: Writing – review & editing. FR: Writing – review & editing. SL: Investigation, Writing – review & editing. LX: Formal analysis, Writing – review & editing. ZX: Conceptualization, Funding acquisition, Resources, Supervision, Validation, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The Guangxi Science Base and Talents Special Program (AD17195083) and the Guangxi BaGui Scholars Program Foundation (2019A50) were used.
Authors SR, ZJ, and TP were employed by Sadiq Poultry Pvt. Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abozeid, H. H., Paldurai, A., Khattar, S. K., Afifi, M. A., El-Kady, M. F., El-Deeb, A. H., et al. (2017). Complete genome sequences of two avian infectious bronchitis viruses isolated in Egypt: Evidence for genetic drift and genetic recombination in the circulating viruses. Infect. Genet. Evol. 53, 7–14. doi: 10.1016/j.meegid.2017.05.006
Abro, S. H., Renström, L. H. M., Ullman, K., Belák, S., Baule, C. (2012). Characterization and analysis of the full-length genome of a strain of the European QX-like genotype of infectious bronchitis virus. Arch. Virol. 157, 1211–1215. doi: 10.1007/s00705-012-1284-0
Ahmed, Z., Naeem, K., Hameed, A. (2007). Detection and seroprevalence of infectious bronchitis virus strains in commercial poultry in Pakistan. Poult. Sci. 86, 1329–1335. doi: 10.1093/ps/86.7.1329
Ali, A., Hassan, M. S. H., Najimudeen, S. M., Farooq, M., Shany, S., El-Safty, M. M., et al. (2023). Efficacy of two vaccination strategies against infectious bronchitis in laying hens. Vaccines 11, 338. doi: 10.3390/vaccines11020338
Ali, A., Ojkic, D., Elshafiee, E. A., Shany, S., EL-Safty, M. M., Shalaby, A. A., et al. (2022). Genotyping and in silico analysis of delmarva (DMV/1639) infectious bronchitis virus (IBV) spike 1 (S1) glycoprotein. Genes (Basel) 13, 1617. doi: 10.3390/genes13091617
Al-Rasheed, M., Ball, C., Ganapathy, K. (2021). Route of infectious bronchitis virus vaccination determines the type and magnitude of immune responses in table egg laying hens. Vet. Res. 52, 1–15. doi: 10.1186/s13567-021-01008-7
Amarasinghe, A., De Silva Senapathi, U., Abdul-Cader, M. S., Popowich, S., Marshall, F., Cork, S. C., et al. (2018). Comparative features of infections of two Massachusetts (Mass) infectious bronchitis virus (IBV) variants isolated from Western Canadian layer flocks. BMC Vet. Res. 14, 1–12. doi: 10.1186/s12917-018-1720-9
Ambali, A. G., Jones, R. C. (1990). Early pathogenesis in chicks of infection with an enterotropic strain of infectious bronchitis virus. Avian Dis., 809–817. doi: 10.2307/1591367
Andreopoulou, M., Franzo, G., Tucciarone, C. M., Prentza, Z., Koutoulis, K. C., CecChinato, M., et al. (2019). Molecular epidemiology of infectious bronchitis virus and avian metapneumovirus in Greece. Poult. Sci. 98, 5374–5384. doi: 10.3382/ps/pez360
Bali, K., Kaszab, E., Marton, S., Hamdiou, S. H., Bentaleb, R. K., Kiss, I., et al. (2022). Novel lineage of infectious bronchitis virus from sub-Saharan Africa identified by random amplification and next-generation sequencing of viral genome. Life 12, 475. doi: 10.3390/life12040475
Bande, F., Arshad, S. S., Hair Bejo, M., Moeini, H., Omar, A. R. (2015). Progress and challenges toward the development of vaccines against avian infectious bronchitis. J. Immunol. Res. 2015. doi: 10.1155/2015/424860
Bande, F., Arshad, S. S., Omar, A. R., Hair-Bejo, M., Mahmuda, A., Nair, V. (2017). Global distributions and strain diversity of avian infectious bronchitis virus: A review. Anim. Heal. Res. Rev. 18, 70–83. doi: 10.1017/S1466252317000044
Bayry, J., Goudar, M. S., Nighot, P. K., Kshirsagar, S. G., Ladman, B. S., Gelb, J., et al. (2005). Emergence of a nephropathogenic avian infectious bronchitis virus with a novel genotype in India. J. Clin. Microbiol. 43, 916–918. doi: 10.1128/jcm.43.4.2039.2005
Besar, S. A., Arshad, S. S., Ramanoon, S. Z., Omar, A. R. (2023). Isolation and characterization of avian coronavirus from diagnostic cases of selected bird species in Malaysia. Pertanika J. Trop. Agric. Sci. 46, 503–516. doi: 10.47836/pjtas.46.2.08
Bhuiyan, M. S. A., Amin, Z., Bakar, A. M. S. A., Saallah, S., Yusuf, N. H. M., Shaarani, S. M., et al. (2021). Factor influences for diagnosis and vaccination of avian infectious bronchitis virus (Gammacoronavirus) in chickens. Vet. Sci. 8, 47. doi: 10.3390/vetsci8030047
Bijlenga, G., Cook, J. K. A., Gelb, J., De Wit, J. J. (2004). Development and use of the H strain of avian infectious bronchitis virus from the Netherlands as a vaccine: A review. Avian Pathol. 33, 550–557. doi: 10.1080/03079450400013154
Bo, Z., Chen, S., Zhang, C., Guo, M., Cao, Y., Zhang, X., et al. (2022). Pathogenicity evaluation of GVI-1 lineage infectious bronchitis virus and its long-term effects on reproductive system development in SPF hens. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.1049287
Boender, G. J., Hagenaars, T. J., Bouma, A., Nodelijk, G., Elbers, A. R. W., De Jong, M. C. M., et al. (2007). Risk maps for the spread of highly pathogenic avian influenza in poultry. PloS Comput. Biol. 3, e71. doi: 10.1371/journal.pcbi.0030071
Boltz, D. A., Nakai, M., Bahr, J. M. (2004). Avian infectious bronchitis virus: A possible cause of reduced fertility in the rooster. Avian Dis. 48, 909–915. doi: 10.1637/7192-040808R1
Boroomand, Z., Asasi, K., Mohammadi, A. (2012). Pathogenesis and tissue distribution of avian infectious bronchitis virus isolate IRFIBV32 (793/B Serotype) in experimentally infected broiler chickens. Sci. World J. 2012. doi: 10.1100/2012/402537
Bosha, J., Nongo, N. (2012). Common breaches in poultry vaccine handling and administration in makurdi metropolis: A recurrent phenomenon in the tropics. Vom. J. Vet. Sci. 9, 11–16.
Brown Jordan, A., Fusaro, A., Blake, L., Milani, A., Zamperin, G., Brown, G., et al. (2020). Characterization of novel, pathogenic field strains of infectious bronchitis virus (IBV) in poultry in Trinidad and Tobago. Transbound Emerg. Dis. 67, 2775–2788. doi: 10.1111/tbed.13637
Carstens, E. B. (2010). Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses, (2009). Arch. Virol. 155, 133–146. doi: 10.1007/s00705-009-0547-x
Carstens, E. B., Ball, L. A. (2009). Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses, (2008). Arch. Virol. 154, 1181–1188. doi: 10.1007/s00705-009-0400-2
Casais, R., Davies, M., Cavanagh, D., Britton, P. (2005). Gene 5 of the avian coronavirus infectious bronchitis virus is not essential for replication. J. Virol. 79, 8065–8078. doi: 10.1128/jvi.79.13.8065-8078.2005
Cavanagh, D. (2007). Coronavirus avian infectious bronchitis virus. Vet. Res. 38, 281–297. doi: 10.1051/vetres:2006055
Cavanagh, D., Picault, J. P., Gough, R. E., Hess, M., Mawditt, K., Britton, P. (2005). Variation in the spike protein of the 793/B type of infectious bronchitis virus, in the field and during alternate passage in chickens and embryonated eggs. Avian Pathol. 34, 20–25. doi: 10.1080/03079450400025414
Chen, B. Y., Hosi, S., Nunoya, T., Itakura, C., et al. (1996). Histopathology and immunohistochemistry of renal lesions due to infectious bronchitis virus in chicks. Avian Pathol. 25, 269–283. doi: 10.1080/03079459608419141
Chen, Y., Cai, H., Pan, J., Xiang, N., Tien, P., Ahola, T., et al. (2009). Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. U. S. A. 106, 3484–3489. doi: 10.1073/pnas.0808790106
Chen, L., Jiang, W., Wu, W., Zhang, S., Cai, J., Lv, T., et al. (2023). Insights into the epidemiology, phylodynamics, and evolutionary changes of lineage GI-7 infectious bronchitis virus. Transbound Emerg. Dis. 2023. doi: 10.1155/2023/9520616
Chen, Y., Jiang, L., Zhao, W., Liu, L., Zhao, Y., Shao, Y., et al. (2017). Identification and molecular characterization of a novel serotype infectious bronchitis virus (GI-28) in China. Vet. Microbiol. 198, 108–115. doi: 10.1016/j.vetmic.2016.12.017
Cook, J. K. A., Ambali, A. G. (1986). The isolation and characterisation of six avian infectious bronchitis viruses isolated in Morocco. Avian Pathol. 15, 93–105. doi: 10.1080/03079458608436269
Cook, J. K. A., Jackwood, M., Jones, R. C. (2012). The long view: 40 years of infectious bronchitis research. Avian Pathol. 41, 239–250. doi: 10.1080/03079457.2012.680432
Cook, J. K. A., Orbell, S. J., Woods, M. A., Huggins, M. B. (1999). Breadth of protection of the respiratory tract provided by different live-attenuated infectious bronchitis vaccines against challenge with infectious bronchitis viruses of heterologous serotypes. Avian Pathol. 28, 477–485. doi: 10.1080/03079459994506
Cowen, B. S., Hitchner, S. B. (1975). Serotyping of avian infectious bronchitis viruses by the virus-neutralization test. Avian Dis., 583–595. doi: 10.2307/1589084
Cubillos, A., Ulloa, J., Cubillos, V., Cook, J. K. A. (1991). Characterisation of strains of infectious bronchitis virus isolated in Chile. Avian Pathol. 20, 85–99. doi: 10.1080/03079459108418744
Cumming, R. B. (1963). Infectious avian nephrosis (Uraemia) in Australia. Aust. Vet. J. 39, 145–147. doi: 10.1111/j.1751-0813.1963.tb04255.x
Cumming, R. B. (1969). The control of OF avian infectious bronchitis/nephrosis in Australia. Aust. Vet. J. 45, 200–203. doi: 10.1111/j.1751-0813.1969.tb01930.x
Davelaar, F. G., Kouwenhoven, B., Burger, A. G. (1984). Occurrence and significance of infectious bronchitis virus variant strains in egg and broiler production in the Netherlands. Vet. Q. 6, 114–120. doi: 10.1080/01652176.1984.9693924
Dawson, P. S., Gough, R. E. (1971). Antigenic variation in strains of avian infectious bronchitis virus. Arch. Gesamte Virusforsch. 34, 32–39. doi: 10.1007/BF01250243
de Wit, J. J. S., Cook, J. K. A., van der Heijden, H. M. J. F. (2011). Infectious bronchitis virus variants: A review of the history, current situation and control measures. Avian Pathol. 40, 223–235. doi: 10.1080/03079457.2011.566260
Domanska-Blicharz, K., Lisowska, A., Pikuła, A., Sajewicz-Krukowska, J. (2017). Specific detection of GII-1 lineage of infectious bronchitis virus. Lett. Appl. Microbiol. 65, 141–146. doi: 10.1111/lam.12753
Domanska-Blicharz, K., Sajewicz-Krukowska, J., Lisowska, A. (2020). New PA/1220/98-like variant of infectious bronchitis virus in Poland. Avian Pathol. 49, 380–388. doi: 10.1080/03079457.2020.1754332
Ducatez, M. F., Martin, A. M., Owoade, A. A., Olatoye, I. O., Alkali, B. R., Maikano, I., et al. (2009). Characterization of a new genotype and serotype of infectious bronchitis virus in Western Africa. J. Gen. Virol. 90, 2679–2685. doi: 10.1099/vir.0.012476-0
Durães-Carvalho, R., Caserta, L. C., Barnabé, A. C. S., Martini, M. C., Simas, P. V. M., Santos, M. M. B., et al. (2015). Phylogenetic and phylogeographic mapping of the avian coronavirus spike protein-encoding gene in wild and synanthropic birds. Virus Res. 201, 101–112. doi: 10.1016/j.virusres.2015.03.002
Elankumaran, S., Balachandran, C., Chandran, N. D., Roy, P., Albert, A., Manickam, R. (1999). Serological evidence for a 793/B related avian infectious bronchitis virus in India. Vet. Rec. 144, 299–300. doi: 10.1128/JCM.43.4.1982-1984.2005
Feng, K., Wang, F., Xue, Y., Zhou, Q., Chen, F., Bi, Y., et al. (2017). Epidemiology and characterization of avian infectious bronchitis virus strains circulating in southern China during the period from 2013-2015. Sci. Rep. 7, 6576. doi: 10.1038/s41598-017-06987-2
Fields, D. B. (1973). Arkansas 99, a new infectious bronchitis serotype. Avian Dis. 17, 659–661. doi: 10.2307/1589167
Fischer, S., Klosterhalfen, D., Wilms-Schulze Kump, F., Casteel, M. (2020). Research Note: First evidence of infectious bronchitis virus Middle-East GI-23 lineage (Var2-like) in Germany. Poult. Sci. 99, 797–800. doi: 10.1016/j.psj.2019.10.031
Fischer, F., Stegen, C. F., Masters, P. S., Samsonoff, W. A. (1998). Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J. Virol. 72, 7885–7894. doi: 10.1128/jvi.72.10.7885-7894.1998
França, M., Woolcock, P. R., Yu, M., Jackwood, M. W., Shivaprasad, H. L. (2011). Nephritis associated with infectious bronchitis virus Cal99 variant in game chickens. Avian Dis. 55, 422–428. doi: 10.1637/9417-060510-Reg.1
Franzo, G., Naylor, C. J., Lupini, C., Drigo, M., Catelli, E., Listorti, V., et al. (2014). Continued use of IBV 793B vaccine needs reassessment after its withdrawal led to the genotype’s disappearance. Vaccine 32, 6765–6767. doi: 10.1016/j.vaccine.2014.10.006
Franzo, G., Tucciarone, C. M., Blanco, A., Nofrarías, M., Biarnés, M., Cortey, M., et al. (2016). Effect of different vaccination strategies on IBV QX population dynamics and clinical outbreaks. Vaccine 34, 5670–5676. doi: 10.1016/j.vaccine.2016.09.014
Franzo, G., Tucciarone, C. M., Moreno, A., Legnardi, M., Massi, P., Tosi, G., et al. (2020). Phylodynamic analysis and evaluation of the balance between anthropic and environmental factors affecting IBV spreading among Italian poultry farms. Sci. Rep. 10, 7289. doi: 10.1038/s41598-020-64477-4
Fujisawa, S., Murata, S., Takehara, M., Katakura, K., Hmoon, M. M., Win, S. Y., et al. (2019). Molecular detection and genetic characterization of Mycoplasma gallisepticum, Mycoplama synoviae, and infectious bronchitis virus in poultry in Myanmar. BMC Vet. Res. 15, 1–7. doi: 10.1186/s12917-019-2018-2
Gaba, A., Dave, H., Pal, J. K., Prajapati, K. S. (2010). Isolation, identification and molecular characterization of IBV variant from out break of visceral gout in commercial broilers. Vet. World 3, 375. doi: 10.5455/vetworld.2010.375-377
Gelb, J., Keeler, C. L., Nix, W. A., Rosenberger, J. K., Cloud, S. S. (1997). Antigenic and S-1 genomic characterization of the Delaware variant serotype of infectious bronchitis virus. Avian Dis., 661–669. doi: 10.2307/1592158
Gough, R. E., Cox, W. J., Gutierrez, E., MacKenzie, G., Wood, A. M., Dagless, M. D. (1996). Isolation of “variant” strains of infectious bronchitis virus from vaccinated chickens in Great Britain. Vet. Rec. 139, 552.
Gough, R. E., Cox, W. J., Welchman, D. D. B., Worthington, K. J., Jones, R. C. (2008). Chinese QX strain of infectious bronchitis virus isolated in the UK [2]. Vet. Rec. 162, 99. doi: 10.1136/vr.162.3.99
Hassan, M. S. H., Ojkic, D., Coffn, C. S., Cork, S. C., Van Der Meer, F., Abdul-Careem, M. F. (2019). Delmarva (dmv/1639) infectious bronchitis virus (ibv) variants isolated in eastern Canada show evidence of recombination. Viruses 11, 1054. doi: 10.3390/v11111054
Hewson, K. A., Noormohammadi, A. H., Devlin, J. M., Browning, G. F., Schultz, B. K., Ignjatovic, J. (2014). Evaluation of a novel strain of infectious bronchitis virus emerged as a result of spike gene recombination between two highly diverged parent strains. Avian Pathol. 43, 249–257. doi: 10.1080/03079457.2014.914624
Hewson, K., Noormohammadi, A. H., Devlin, J. M., Mardani, K., Ignjatovic, J. (2009). Rapid detection and non-subjective characterisation of infectious bronchitis virus isolates using high-resolution melt curve analysis and a mathematical model. Arch. Virol. 154, 649–660. doi: 10.1007/s00705-009-0357-1
Hiscox, J. A., Wurm, T., Wilson, L., Britton, P., Cavanagh, D., Brooks, G. (2001). The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J. Virol. 75, 506–512. doi: 10.1128/jvi.75.1.506-512.2001
Hitchner, S. B., Winterfield, R. W., Appleton, G. S. (1966). Infectious bronchitis virus types: incidence in the United States. Avian Dis. 10, 98–102. doi: 10.2307/1588213
Hopkins, S. R. (1974). Serological comparisons of strains of infectious bronchitis virus using plaque purified isolants. Avian Dis., 231–239. doi: 10.2307/1589131
Hughes, L. A., Savage, C., Naylor, C., Bennett, M., Chantrey, J., Jones, R. (2009). Genetically diverse coronaviruses in wild bird populations of northern England. Emerg. Infect. Dis. 15, 1091. doi: 10.3201/eid1507.090067
Icochea, E., González, R., Castro-Sanguinetti, G., Maturrano, L., Alzamora, L., Sesti, L., et al. (2023). Genetic analysis of infectious bronchitis virus S1 gene reveals novel amino acid changes in the GI-16 lineage in Peru. Microorganisms 11, 691. doi: 10.3390/microorganisms11030691
Ignjatovic, J., Galli, L. (1994). The S1 glycoprotein but not the N or M proteins of avian infectious bronchitis virus induces protection in vaccinated chickens. Arch. Virol. 138, 117–134. doi: 10.1007/BF01310043
Ignjatovic, J., Gould, G., Sapats, S. (2006). Isolation of a variant infectious bronchitis virus in Australia that further illustrates diversity among emerging strains. Arch. Virol. 151, 1567–1585. doi: 10.1007/s00705-006-0726-y
Ignjatovic, J., McWaters, P. G. (1991). Monoclonal antibodies to three structural proteins of avian infectious bronchitis virus: Characterization of epitopes and antigenic differentiation of Australian strains. J. Gen. Virol. 72, 2915–2922. doi: 10.1099/0022-1317-72-12-2915
Jackwood, M. W. (2012). Review of infectious bronchitis virus around the world. Avian Dis. 56, 634–641. doi: 10.1637/10227-043012-Review.1
Jackwood, M. W., Jordan, B. J. (2021). Molecular evolution of infectious bronchitis virus and the emergence of variant viruses circulating in the United States. Avian Dis. 65, 631–636. doi: 10.1637/aviandiseases-D-21-00104
Jakhesara, S. J., Nath, B., Pal, J. K., Joshi, C. G., Kumar, S.. (2018). Emergence of a genotype I variant of avian infectious bronchitis virus from Northern part of India. Acta Trop. 183, 57–60. doi: 10.1016/j.actatropica.2018.04.004
Jang, I., Lee, H. J., Bae, Y. C., Park, S. C., Lee, H. S., Choi, K. S. (2018). Genetic and pathologic characterization of a novel recombinant TC07-2-type avian infectious bronchitis virus. Avian Dis. 62, 109–113. doi: 10.1637/11764-103017-ResNote.1
Ji, W., Wang, W., Zhao, X., Zai, J., Li, X. (2020). Cross-species transmission of the newly identified coronavirus 2019-nCoV. J. Med. Virol. 92, 433–440. doi: 10.1002/jmv.25682
Ji, J., Xie, J., Chen, F., Shu, D., Zuo, K., Xue, C., et al. (2011). Phylogenetic distribution and predominant genotype of the avian infectious bronchitis virus in China during 2008-2009. Virol. J. 8, 1–9. doi: 10.1186/1743-422X-8-184
Jiang, L., Zhao, W., Han, Z., Chen, Y., Zhao, Y., Sun, J., et al. (2017). Genome characterization, antigenicity and pathogenicity of a novel infectious bronchitis virus type isolated from south China. Infect. Genet. Evol. 54, 437–446. doi: 10.1016/j.meegid.2017.08.006
Johnson, R. B., Marquardt, W. W. (1976). Strains of infectious bronchitis virus on the Delmarva peninsula and in Arkansas. Avian Dis., 382–386. doi: 10.2307/1589277
Jungherr, E. L., Chomiak, T. W., Luginbuhl, R. E. (1956). “Immunologic differences in strains of infectious bronchitis virus,” in Proc. 60th Ann. Meet. U. S. Livest. Sanit. Assoc, Chicago. Ill. (Chicago).
Kant, A., Koch, G., Van Roozelaar, D. J., Kusters, J. G., Poelwijk, F. A. J., van der Zeijst, B. A. M. (1992). Location of antigenic sites defined by neutralizing monoclonal antibodies on the S1 avian infectious bronchitis virus glycopolypeptide. J. Gen. Virol. 73, 591–596. doi: 10.1099/0022-1317-73-3-591
Kapczynski, D. R., Hilt, D. A., Shapiro, D., Sellers, H. S., Jackwood, M. W. (2003). Protection of chickens from infectious bronchitis by in ovo and intramuscular vaccination with a DNA vaccine expressing the S1 glycoprotein. Avian Dis. 47, 272–285. doi: 10.1637/0005-2086(2003)047[0272:POCFIB]2.0.CO;2
Kapikian, A. Z., James, H. D., Kelly, S. J., Dees, J. H., Turner, H. C., McIntosh, K., et al. (1969). Isolation from man of “avian infectious bronchitis virus-like” viruses (coronaviruses) similar to 229e virus, with some epidemiological observations. J. Infect. Dis. 119, 282–290. doi: 10.1093/infdis/119.3.282
Karesh, W. B., Uhart, M. M., Frere, E., Gandini, P., Braselton, W. E., Puche, H., et al. (1999). Health evaluation of free-ranging rockhopper penguins (Eudyptes chrysocomes) in Argentina. J. Zoo Wildl. Med., 25–31.
Kariithi, H. M., Volkening, J. D., Goraichuk, I. V., Ateya, L. O., Williams-Coplin, D., Olivier, T. L., et al. (2023). Unique variants of avian coronaviruses from indigenous chickens in Kenya. Viruses 15, 264. doi: 10.3390/v15020264
Kariithi, H. M., Volkening, J. D., Leyson, C. M., Afonso, C. L., Christy, N., Decanini, E. L., et al. (2022). Genome sequence variations of infectious bronchitis virus serotypes from commercial chickens in Mexico. Front. Vet. Sci. 9. doi: 10.3389/fvets.2022.931272
Khaltabadi Farahani, R., Ghalyanchilangeroudi, A., Fallah Mehrabadi, M. H., Ghafouri, S. A., Maghsoudloo, H. (2020). Molecular monitoring of D1466 genotype of avian infectious bronchitis virus in Iran: A retrospective study in 2013-2017. Arch. Razi Inst. 75, 163. doi: 10.22092/ARI.2018.121512.1214
Kint, J., Langereis, M. A., Maier, H. J., Britton, P., van Kuppeveld, F. J., Koumans, J., et al. (2016). Infectious bronchitis coronavirus limits interferon production by inducing a host shutoff that requires accessory protein 5b. J. Virol. 90, 7519–7528. doi: 10.1128/jvi.00627-16
Kumanan, K., Thamizh Selvam, N., Dhinakar Raj, G., Nachimuthu, K. (2004). Molecular epizootiology of infectious bronchitis virus isolates in Tamil Nadu indicating the possible involvement of a vaccine strain. Indian Vet. J. 81, 1307–1312.
Laconi, A., van Beurden, S. J., Berends, A. J., Krämer-Kühl, A., Jansen, C. A., Spekreijse, D., et al. (2018). Deletion of accessory genes 3a, 3b, 5a or 5b from avian coronavirus infectious bronchitis virus induces an attenuated phenotype both in vitro and in vivo. J. Gen. Virol. 99, 1381–1390. doi: 10.1099/jgv.0.001130
Lai, M. M., Cavanagh, D. (1997). The molecular biology of coronaviruses. Adv. Virus Res. 66, 193–292. doi: 10.1016/s0065-3527(08)60286-9
Le, T. B., Lee, H.-J., Le, V. P., Choi, K.-S. (2019). Multiple genotypes of avian infectious bronchitis virus circulating in Vietnam. Korean J. Poult. Sci. 46, 127–136. doi: 10.5536/kjps.2019.46.2.127
Lee, C. W., Hilt, D. A., Jackwood, M. W. (2001). Identification and analysis of the Georgia 98 serotype, a new serotype of infectious bronchitis virus. Avian Dis., 164–172. doi: 10.2307/1593024
Lee, C. W., Jackwood, M. W. (2001). Origin and evolution of Georgia 98 (GA98), a new serotype of avian infectious bronchitis virus. Virus Res. 80, 33–39. doi: 10.1016/S0168-1702(01)00345-8
Lee, E. K., Jeon, W. J., Lee, Y. J., Jeong, O. M., Choi, J. G., Kwon, J. H., et al. (2008). Genetic diversity of avian infectious bronchitis virus isolates in Korea between 2003 and 2006. Avian Dis. 52, 332–337. doi: 10.1637/8117-092707-ResNote.1
Lee, H. C., Jeong, S., Cho, A. Y., Kim, K. J., Kim, J. Y., Park, D. H., et al. (2021). Genomic analysis of avian infectious bronchitis viruses recently isolated in South Korea reveals multiple introductions of GI-19 lineage (Qx genotype). Viruses 13, 1045. doi: 10.3390/v13061045
Lee, S. K., Sung, H. W., Kwon, H. M. (2004). S1 glycoprotein gene analysis of infectious bronchitis viruses isolated in Korea. Arch. Virol. 149, 481–494. doi: 10.1007/s00705-003-0225-3
Li, W., Shi, Z., Yu, M., Ren, W., Smith, C., Epstein, J. H., et al. (2005). Bats are natural reservoirs of SARS-like coronaviruses. Sci. (80-.) 310, 676–679. doi: 10.1126/science.1118391
Li, L., Xue, C., Chen, F., Qin, J., Xie, Q., Bi, Y., et al. (2010). Isolation and genetic analysis revealed no predominant new strains of avian infectious bronchitis virus circulating in South China during 2004-2008. Vet. Microbiol. 143, 145–154. doi: 10.1016/j.vetmic.2009.11.022
Lian, J., Wang, Z., Xu, Z., Chen, T., Shao, G., Zhang, X., et al. (2021). Distribution and molecular characterization of avian infectious bronchitis virus in southern China. Poult. Sci. 100, 101169. doi: 10.1016/j.psj.2021.101169
Lim, T. H., Lee, H. J., Lee, D. H., Lee, Y. N., Park, J. K., Youn, H. N., et al. (2011). An emerging recombinant cluster of nephropathogenic strains of avian infectious bronchitis virus in Korea. Infect. Genet. Evol. 11, 678–685. doi: 10.1016/j.meegid.2011.01.007
Lin, S. Y., Chen, H. W. (2017). Infectious bronchitis virus variants: Molecular analysis and pathogenicity investigation. Int. J. Mol. Sci. 18, 2030. doi: 10.3390/ijms18102030
Lisowska, A., Sajewicz-Krukowska, J., Fusaro, A., Pikula, A., Domanska-Blicharz, K. (2017). First characterization of a Middle-East GI-23 lineage (Var2-like) of infectious bronchitis virus in Europe. Virus Res. 242, 43–48. doi: 10.1016/j.virusres.2017.09.010
Liu, D. X., Cavanagh, D., Green, P., Inglis, S. C. (1991). A polycistronic mRNA specified by the coronavirus infectious bronchitis virus. Virology 184, 531–544. doi: 10.1016/0042-6822(91)90423-9
Liu, S., Chen, J., Chen, J., Kong, X., Shao, Y., Han, Z., et al. (2005). Isolation of avian infectious bronchitis coronavirus from domestic peafowl (Pavo cristatus) and teal (Anas). J. Gen. Virol. 86, 719–725. doi: 10.1099/vir.0.80546-0
Liu, S., Chen, J., Han, Z., Zhang, Q., Shao, Y., Kong, X., et al. (2006). Infectious bronchitis virus: S1 gene characteristics of vaccines used in China and efficacy of vaccination against heterologous strains from China. Avian Pathol. 35, 394–399. doi: 10.1080/03079450600920984
Lohr, J. E. (1976). Serologic differences between strains of infectious bronchitis virus from New Zealand, Australia, and the United States. Avian Dis., 478–482. doi: 10.2307/1589380
Ma, T., Xu, L., Ren, M., Shen, J., Han, Z., Sun, J., et al. (2019). Novel genotype of infectious bronchitis virus isolated in China. Vet. Microbiol. 230, 178–186. doi: 10.1016/j.vetmic.2019.01.020
Macdonald, J. W., McMartin, D. A. (1976). Observations on the effects of the H52 and H120 vaccine strains of infectious bronchitis virus in the domestic fowl. Avian Pathol. 5, 157–173. doi: 10.1080/03079457608418182
Marandino, A., Vagnozzi, A., Tomás, G., Techera, C., Gerez, R., Hernández, M., et al. (2022). Origin of new lineages by recombination and mutation in avian infectious bronchitis virus from South America. Viruses 14, 2095. doi: 10.3390/v14102095
Mardani, K., Noormohammadi, A. H., Hooper, P., Ignjatovic, J., Browning, G. F. (2008). Infectious bronchitis viruses with a novel genomic organization. J. Virol. 82, 2013–2024. doi: 10.1128/jvi.01694-07
Mase, M., Gotou, M., Inoue, D., Watanabe, S., Iseki, H. (2021a). Genotyping of infectious bronchitis viruses isolated in Japan during 2008–2019. J. Vet. Med. Sci. 83 (3), 522–526. doi: 10.1292/jvms.20-0620
Mase, M., Hiramatsu, K., Watanabe, S., Iseki, H. (2021b). Complete genome sequence of infectious bronchitis virus strain JP/KH/64, isolated in Japan. Microbiol. Resour. Announc. 10, 10–1128. doi: 10.1128/mra.00665-21
Mase, M., Hiramatsu, K., Watanabe, S., Iseki, H. (2022). Genetic analysis of the complete S1 gene in Japanese infectious bronchitis virus strains. Viruses. 83(3):522-526. doi: 10.3390/v14040716
Mase, M., Kawanishi, N., Ootani, Y., Murayama, K., Karino, A., Inoue, T., et al. (2010). A novel genotype of avian infectious bronchitis virus isolated in Japan in 2009. J. Vet. Med. Sci. 72, 1265–1268. doi: 10.1292/jvms.10-0080
Mase, M., Tsukamoto, K., Imai, K., Yamaguchi, S. (2004). Phylogenetic analysis of avian infectious bronchitis virus strains isolated in Japan. Arch. Virol. 149, 2069–2078. doi: 10.1007/s00705-004-0369-9
McFarlane, R., Verma, R. (2008). Sequence analysis of the gene coding for the S1 glycoprotein of infectious bronchitis virus (IBV) strains from New Zealand. Virus Genes 37, 351–357. doi: 10.1007/s11262-008-0273-6
Meir, R., Rosenblut, E., Perl, S., Kass, N., Ayali, G., Hemsani, E., et al. (2004). Identification of a novel nephropathogenic infectious bronchitis virus in Israel. Avian Dis. 48, 635–641. doi: 10.1637/7107
Mendoza-González, L., Marandino, A., Panzera, Y., Tomás, G., Williman, J., Techera, C., et al. (2022). Research Note: High genetic diversity of infectious bronchitis virus from Mexico. Poult. Sci. 101, 102076. doi: 10.1016/j.psj.2022.102076
Miller, L. T., Yates, V. J. (1968). Neutralization of infectious bronchitis virus human sera. Am. J. Epidemiol. 88, 406–409. doi: 10.1093/oxfordjournals.aje.a120901
Moharam, I., Sultan, H., Hassan, K., Ibrahim, M., Shany, S., Shehata, A. A., et al. (2020). Emerging infectious bronchitis virus (IBV) in Egypt: Evidence for an evolutionary advantage of a new S1 variant with a unique gene 3ab constellation. Infect. Genet. Evol. 85, 104433. doi: 10.1016/j.meegid.2020.104433
Molenaar, R. J., Dijkman, R., de Wit, J. J. (2020). Characterization of infectious bronchitis virus D181, a new serotype (GII-2). Avian Pathol. 49, 243–250. doi: 10.1080/03079457.2020.1713987
Mondal, S. P., Lucio-Martínez, B., Naqi, S. A. (2001). Isolation and characterization of a novel antigenic subtype of infectious bronchitis virus serotype DE072. Avian Dis., 1054–1059. doi: 10.2307/1592888
Moreno, A., Franzo, G., Massi, P., Tosi, G., Blanco, A., Antilles, N., et al. (2017). A novel variant of the infectious bronchitis virus resulting from recombination events in Italy and Spain. Avian Pathol. 46, 28–35. doi: 10.1080/03079457.2016.1200011
Morley, A. J., Thomson, D. K. (1984). Swollen-head syndrome in broiler chickens. Avian Dis. 26, 139–154. doi: 10.2307/1590147
Mousavi, F. S., Ghalyanchilangeroudi, A., Hosseini, H., Nayeri Fasaei, B., Ghafouri, S. A., Abdollahi, H., et al. (2018). Complete genome analysis of Iranian IS-1494 like avian infectious bronchitis virus. VirusDisease 29, 390–394. doi: 10.1007/s13337-018-0462-4
Munyahongse, S., Pohuang, T., Nonthabenjawan, N., Sasipreeyajan, J., Thontiravong, A. (2020). Genetic characterization of infectious bronchitis viruses in Thailand 2014–2016: identification of a novel recombinant variant. Poult. Sci. 99, 1888–1895. doi: 10.1016/j.psj.2019.11.044
Müştak, B., Müştak, H. K., Bilgen, N. (2022). S1 gene based phylogeny of Israel variant-2 infectious bronchitis virus isolated in Turkey in a five year period. Pol. J. Vet. Sci., 45–50. doi: 10.24425/pjvs.2022.140839
Naguib, M. M., Höper, D., Arafa, A. S., Setta, A. M., Abed, M., Monne, I., et al. (2016). Full genome sequence analysis of a newly emerged QX-like infectious bronchitis virus from Sudan reveals distinct spots of recombination. Infect. Genet. Evol. 46, 42–49. doi: 10.1016/j.meegid.2016.10.017
Najimudeen, S. M., Hassan, M. S. H., Cork, S. C., Abdul-Careem, M. F. (2020). Infectious bronchitis coronavirus infection in chickens: Multiple system disease with immune suppression. Pathogens 9, 779. doi: 10.3390/pathogens9100779
Najimudeen, S. M., Hassan, M. S. H., Goldsmith, D., Ojkic, D., Cork, S. C., Boulianne, M., et al. (2021). Molecular characterization of 4/91 infectious bronchitis virus leading to studies of pathogenesis and host responses in laying hens. Pathogens 10, 624. doi: 10.3390/pathogens10050624
Öngör, H., Tımurkaan, N., Çöven, F., Karabulut, B., Eröksüz, H., Çetınkaya, B., et al. (2021). Detection of Israel variant 2 (IS/1494/06) genotype of infectious bronchitis virus in a layer chicken flock. Ankara Univ. Vet. Fak. Derg. 68, 167-172. doi: 10.33988/auvfd.756970
Parveen, R., Farooq, I., Ahangar, S., Nazki, S., Dar, Z., Dar, T., et al. (2017). Genotyping and phylogenetic analysis of infectious bronchitis virus isolated from broiler chickens in Kashmir. VirusDisease 28, 434–438. doi: 10.1007/s13337-017-0416-2
Patel, B. H., Bhimani, M. P., Bhanderi, B. B., Jhala, M. K.. (2015). Isolation and molecular characterization of nephropathic infectious bronchitis virus isolates of Gujarat state, India. VirusDisease 26, 42–47. doi: 10.1007/s13337-015-0248-x
Patterson, S., Bingham, R. W.. (1976). Electron microscope observations on the entry of avian infectious bronchitis virus into susceptible cells. Arch. Virol. 52, 191–200. doi: 10.1007/BF01348016
Pereira, N. A., Alessi, A. C., Montassier, H. J., Pereira, R. J. G., Taniwaki, S. A., Botosso, V. F., et al. (2019). Gonadal pathogenicity of an infectious bronchitis virus strain from the Massachusetts genotype. Braz. J. Microbiol. 50, 313–320. doi: 10.1007/s42770-018-0007-4
Pohjola, L. K., Ek-Kommonen, S. C., Tammiranta, N. E., Kaukonen, E. S., Rossow, L. M., Huovilainen, T. A. (2014). Emergence of avian infectious bronchitis in a non-vaccinating country. Avian Pathol. 43, 244–248. doi: 10.1080/03079457.2014.913770
Pohl, R. M. (1967). Infectious bronchitis in chickens. N. Z. Vet. J. 151. doi: 10.1080/00480169.1967.33717
Quinteros, J. A., Ignjatovic, J., Chousalkar, K. K., Noormohammadi, A. H., Browning, G. F. (2021). Infectious bronchitis virus in Australia: a model of coronavirus evolution–a review. Avian Pathol. 50, 295–310. doi: 10.1080/03079457.2021.1939858
Quinteros, J. A., Markham, P. F., Lee, S. W., Hewson, K. A., Hartley, C. A., Legione, A. R., et al. (2015). Analysis of the complete genomic sequences of two virus subpopulations of the Australian infectious bronchitis virus vaccine VicS. Avian Pathol. 44, 182–191. doi: 10.1080/03079457.2015.1022857
Quinteros, J. A., Noormohammadi, A. H., Lee, S. W., Browning, G. F., Diaz-Méndez, A. (2022). Genomics and pathogenesis of the avian coronavirus infectious bronchitis virus. Aust. Vet. J. 100, 496–512. doi: 10.1111/avj.13197
Rafique, S. (2018). Seroprevalence and molecular characterization of Infectious bronchitis virus variants from poultry in Pakistan. (Islamabad, Pakistan: Department of Microbiology, Quaid-i- Azam University).
Rafique, S., Naeem, K., Siddique, N., Abbas, M. A., Shah, A. A., Ali, A., et al. (2018a). Determination of genetic variability in avian infectious bronchitis virus (AIBV) isolated from Pakistan. Pak. J. Zool. 50, 695–701. doi: 10.17582/JOURNAL.PJZ/2018.50.2.695.701
Rafique, S., Siddique, N., Abbas, M. A., Shah, A. A., Sharif, A., Naeem, K. (2018b). Isolation and molecular characterization of Infectious Bronchitis Virus (IBV) variants circulating in commercial poultry in Pakistan. Pak. Vet. J. 38, 365–370. doi: 10.29261/pakvetj/2018.071
Raja, A., Dhinakar Raj, G., Kumanan, K. (2020). Emergence of variant avian infectious bronchitis virus in India. Iran. J. Vet. Res. 21, 33.
Ramirez-Nieto, G., Mir, D., Almansa-Villa, D., Cordoba-Argotti, G., Beltran-Leon, M., Rodriguez-Osorio, N., et al. (2022). New insights into avian infectious bronchitis virus in Colombia from whole-genome analysis. Viruses 14, 2562. doi: 10.3390/v14112562
Ramsubeik, S., Stoute, S., Gallardo, R. A., Crossley, B., Rejmanek, D., Jude, R., et al. (2023). Infectious bronchitis virus california variant CA1737 isolated from a commercial layer flock with cystic oviducts and poor external egg quality. Avian Dis. 67, 212–218. doi: 10.1637/aviandiseases-D-23-00014
Ren, M., Sheng, J., Ma, T., Xu, L., Han, Z., Li, H., et al. (2019). Molecular and biological characteristics of the infectious bronchitis virus TC07-2/GVI-1 lineage isolated in China. Infect. Genet. Evol. 75, 103942. doi: 10.1016/j.meegid.2019.103942
Saleem, W., Vereecke, N., Zaman, M. G., Afzal, F., Reman, I., Khan, S.U.-H., et al. (2024). Genotyping and phylogeography of infectious bronchitis virus isolates from Pakistan show unique linkage to GI-24 lineage. Poult. Sci. 103, 103236. doi: 10.1016/j.psj.2023.103236
Sapats, S. I., Ashton, F., Wright, P. J., Ignjatovic, J. (1996). Sequence analysis of the S1 glycoprotein of infectious bronchitis viruses: Identification of a novel genotypic group in Australia. J. Gen. Virol. 77, 413–418. doi: 10.1099/0022-1317-77-3-413
Schalk, A. F., Hawn, M. C. (1931). An apparently new respiratory disease of baby chicks. J. Am. Vet. Med. Assoc. 78, 413–423.
Shan, D., Fang, S., Han, Z., Ai, H., Zhao, W., Chen, Y., et al. (2018). Effects of hypervariable regions in spike protein on pathogenicity, tropism, and serotypes of infectious bronchitis virus. Virus Res. 250, 104–113. doi: 10.1016/j.virusres.2018.04.013
Shirvani, E., Samal, S. K. (2020). Comparative protective efficacies of novel avian paramyxovirus-vectored vaccines against virulent infectious bronchitis virus in chickens. Viruses 12, 697. doi: 10.3390/v12070697
Siddell, S., Snijder, E. J. (2014). “An introduction to nidoviruses,” in Nidoviruses. (ASM press), 1–13. doi: 10.1128/9781555815790.ch1
Singh, M., Toribio, J. A., Scott, A. B., Groves, P., Barnes, B., Glass, K., et al. (2018). Assessing the probability of introduction and spread of avian influenza (AI) virus in commercial Australian poultry operations using an expert opinion elicitation. PloS One 13, e0193730. doi: 10.1371/journal.pone.0193730
Smith, E. C., Denison, M. R. (2012). Implications of altered replication fidelity on the evolution and pathogenesis of coronaviruses. Curr. Opin. Virol. 2, 519–524. doi: 10.1016/j.coviro.2012.07.005
Song, C. S., Lee, Y. J., Kim, J. H., Sung, H. W., Lee, C. W., Izumiya, Y., et al. (1998). Epidemiological classification of infectious bronchitis virus isolated in Korea between 1986 and 1997. Avian Pathol. 27, 409–416. doi: 10.1080/03079459808419360
Sumi, V., Singh, S. D., Dhama, K., Gowthaman, V., Barathidasan, R., Sukumar, K. (2012). Isolation and molecular characterization of infectious bronchitis virus from recent outbreaks in broiler flocks reveals emergence of novel strain in India. Trop. Anim. Health Prod. 44, 1791–1795. doi: 10.1007/s11250-012-0140-2
Sun, L., Tang, X., Qi, J., Zhang, C., Zhao, J., Zhang, G., et al. (2021). Two newly isolated GVI lineage infectious bronchitis viruses in China show unique molecular and pathogenicity characteristics. Infect. Genet. Evol. 94, 105006. doi: 10.1016/j.meegid.2021.105006
Tarpey, I., Orbell, S. J., Britton, P., Casais, R., Hodgson, T., Lin, F., et al. (2006). Safety and efficacy of an infectious bronchitis virus used for chicken embryo vaccination. Vaccine 24, 6830–6838. doi: 10.1016/j.vaccine.2006.06.040
Tataje-Lavanda, L., Izquierdo-Lara, R., Ormeño-Vásquez, P., Huamán-Gutiérrez, K., Zimic-Peralta, M., Fernández-Díaz, M. (2019). Near-complete genome sequence of infectious bronchitis virus strain VFAR-047 (GI-16 lineage), isolated in Peru. Microbiol. Resour. Announc. 8, 10–1128. doi: 10.1128/mra.01555-18
Tegegne, D., Deneke, Y., Sori, T., Abdurahaman, M., Kebede, N., CecChinato, M., et al. (2020). Molecular epidemiology and genotyping of infectious bronchitis virus and avian metapneumovirus in backyard and commercial chickens in Jimma Zone, Southwestern Ethiopia. Vet. Sci. 7, 187. doi: 10.3390/vetsci7040187
Terregino, C., Toffan, A., Serena Beato, M., De Nardi, R., Vascellari, M., Meini, A., et al. (2008). Pathogenicity of a QX strain of infectious bronchitis virus in specific pathogen free and commercial broiler chickens, and evaluation of protection induced by a vaccination programme based on the Ma5 and 4/91 serotypes. Avian Pathol. 37, 487–493. doi: 10.1080/03079450802356938
Te Velthuis, A. J. W., Van Den Worm, S. H. E., Snijder, E. J. (2012). The SARS-coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 40, 1737–1747. doi: 10.1093/nar/gkr893
Toffan, A., Monne, I., Terregino, C., Cattoli, G., Hodobo, C. T., Gadaga, B., et al. (2011). QX-like infectious bronchitis virus in Africa. Vet. Rec. 169, 589. doi: 10.1136/vr.d7636
Trevisol, I. M., Caron, L., Mores, M. A. Z., Voss-Rech, D., da Silva Zani, G., Back, A., et al. (2023). Pathogenicity of GI-23 avian infectious bronchitis virus strain isolated in Brazil. Viruses 15, 1200. doi: 10.3390/v15051200
Valastro, V., Holmes, E. C., Britton, P., Fusaro, A., Jackwood, M. W., Cattoli, G., et al. (2016). S1 gene-based phylogeny of infectious bronchitis virus: An attempt to harmonize virus classification. Infect. Genet. Evol. 39, 349–364. doi: 10.1016/j.meegid.2016.02.015
Villalobos-Agüero, R. A., Ramírez-Carvajal, L., Zamora-Sanabria, R., León, B., Karkashian-Córdoba, J. (2021). Molecular characterization of an avian GA13-like infectious bronchitis virus full-length genome from Costa Rica. VirusDisease 32, 347–353. doi: 10.1007/s13337-021-00667-6
Wang, C., Hou, B. (2023). A pathogenic and recombinant infectious bronchitis virus variant (CK/CH/GX/202109) with multiorgan tropism. Vet. Res. 54, 54. doi: 10.1186/s13567-023-01182-w
Wang, C. Y., Luo, Z. B., Shao, G. Q., Hou, B. (2022). Genetic and pathogenic characteristics of a novel infectious bronchitis virus strain in genogroup VI (CK/CH/FJ/202005). Vet. Microbiol. 266, 109352. doi: 10.1016/j.vetmic.2022.109352
Worthington, K. J., Currie, R. J. W., Jones, R. C. (2008). A reverse transcriptase-polymerase chain reaction survey of infectious bronchitis virus genotypes in Western Europe from 2002 to 2006. Avian Pathol. 37, 247–257. doi: 10.1080/03079450801986529
Xu, L., Han, Z., Jiang, L., Sun, J., Zhao, Y., Liu, S. (2018). Genetic diversity of avian infectious bronchitis virus in China in recent years. Infect. Genet. Evol. 66, 82–94. doi: 10.1016/j.meegid.2018.09.018
Yan, S., Chen, Y., Zhao, J., Xu, G., Zhao, Y., Zhang, G. Z. (2016). Pathogenicity of a TW-like strain of infectious bronchitis virus and evaluation of the protection induced against it by a QX-like strain. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01653
Yang, T., Wang, H. N., Wang, X., Tang, J. N., Lu, D., Zhang, Y. F., et al. (2009). The protective immune response against infectious bronchitis virus induced by multi-epitope based peptide vaccines. Biosci. Biotechnol. Biochem. 73, 1500–1504. doi: 10.1271/bbb.80864
Youn, S., Leibowitz, J. L., Collisson, E. W. (2005). In vitro assembled, recombinant infectious bronchitis viruses demonstrate that the 5a open reading frame is not essential for replication. Virology 332, 206–215. doi: 10.1016/j.virol.2004.10.045
Yu, L., Jiang, Y., Low, S., Wang, Z., Nam, S. J., Liu, W., et al. (2001). Characterization of three infectious bronchitis virus isolates from China associated with proventriculus in vaccinated chickens. Avian Dis., 416–424. doi: 10.2307/1592981
Zanaty, A., Arafa, A.-S., Hagag, N., El-Kady, M. (2016a). Genotyping and pathotyping of diversified strains of infectious bronchitis viruses circulating in Egypt. World J. Virol. 5, 125. doi: 10.5501/wjv.v5.i3.125
Zanaty, A., Naguib, M. M., El-Husseiny, M. H., Mady, W., Hagag, N., Arafa, A. S. (2016b). The sequence of the full spike S1 glycoprotein of infectious bronchitis virus circulating in Egypt reveals evidence of intra-genotypic recombination. Arch. Virol. 161, 3583–3587. doi: 10.1007/s00705-016-3042-1
Zhao, Y., Zhang, H., Zhao, J., Zhong, Q., Jin, J. H., Zhang, G. Z. (2016). Evolution of infectious bronchitis virus in China over the past two decades. J. Gen. Virol. 97, 1566. doi: 10.1099/jgv.0.000464
Zhong, Q., Hu, Y. X., Jin, J. H., Zhao, Y., Zhao, J., Zhang, G. Z. (2016). Pathogenicity of virulent infectious bronchitis virus isolate YN on hen ovary and oviduct. Vet. Microbiol. 193, 100–105. doi: 10.1016/j.vetmic.2016.08.017
Keywords: AIBV, distribution, continents, prevalence, genotype
Citation: Rafique S, Jabeen Z, Pervaiz T, Rashid F, Luo S, Xie L and Xie Z (2024) Avian infectious bronchitis virus (AIBV) review by continent. Front. Cell. Infect. Microbiol. 14:1325346. doi: 10.3389/fcimb.2024.1325346
Received: 21 October 2023; Accepted: 15 January 2024;
Published: 05 February 2024.
Edited by:
Anan Jongkaewwattana, National Center for Genetic Engineering and Biotechnology (BIOTEC), ThailandReviewed by:
Aunyaratana Thontiravong, Chulalongkorn University, ThailandCopyright © 2024 Rafique, Jabeen, Pervaiz, Rashid, Luo, Xie and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhixun Xie, eGllemhpeHVuQDEyNi5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.