
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 15 March 2024
Sec. Intestinal Microbiome
Volume 14 - 2024 | https://doi.org/10.3389/fcimb.2024.1308742
This article is part of the Research Topic Advances in the Microbiome, Immunity and Cancer interplay View all 8 articles
 Kui Wang1,2†
Kui Wang1,2† Suijian Wang3†
Suijian Wang3† Xianzheng Qin1†
Xianzheng Qin1† Yifei Chen1†
Yifei Chen1† Yuhua Chen4
Yuhua Chen4 Jiawei Wang5
Jiawei Wang5 Yao Zhang1
Yao Zhang1 Qiang Guo2*
Qiang Guo2* Chunhua Zhou1*
Chunhua Zhou1* Duowu Zou1*
Duowu Zou1*Background: Growing evidence has shown that gut microbiome composition is associated with Biliary tract cancer (BTC), but the causality remains unknown. This study aimed to explore the causal relationship between gut microbiota and BTC, conduct an appraisal of the gut microbiome’s utility in facilitating the early diagnosis of BTC.
Methods: We acquired the summary data for Genome-wide Association Studies (GWAS) pertaining to BTC (418 cases and 159,201 controls) from the Biobank Japan (BBJ) database. Additionally, the GWAS summary data relevant to gut microbiota (N = 18,340) were sourced from the MiBioGen consortium. The primary methodology employed for the analysis consisted of Inverse Variance Weighting (IVW). Evaluations for sensitivity were carried out through the utilization of multiple statistical techniques, encompassing Cochrane’s Q test, the MR-Egger intercept evaluation, the global test of MR-PRESSO, and a leave-one-out methodological analysis. Ultimately, a reverse Mendelian Randomization analysis was conducted to assess the potential for reciprocal causality.
Results: The outcomes derived from IVW substantiated that the presence of Family Streptococcaceae (OR = 0.44, P = 0.034), Family Veillonellaceae (OR = 0.46, P = 0.018), and Genus Dorea (OR = 0.29, P = 0.041) exerted a protective influence against BTC. Conversely, Class Lentisphaeria (OR = 2.21, P = 0.017), Genus Lachnospiraceae FCS020 Group (OR = 2.30, P = 0.013), and Order Victivallales (OR = 2.21, P = 0.017) were associated with an adverse impact. To assess any reverse causal effect, we used BTC as the exposure and the gut microbiota as the outcome, and this analysis revealed associations between BTC and five different types of gut microbiota. The sensitivity analysis disclosed an absence of empirical indicators for either heterogeneity or pleiotropy.
Conclusion: This investigation represents the inaugural identification of indicative data supporting either beneficial or detrimental causal relationships between gut microbiota and the risk of BTC, as determined through the utilization of MR methodologies. These outcomes could hold significance for the formulation of individualized therapeutic strategies aimed at BTC prevention and survival enhancement.
Gallbladder cancer (GBC) and cholangiocarcinomas (CCAs) are cumulatively categorized under the umbrella term of biliary tract cancers (BTCs) (Benson et al., 2023). GBC is recognized as the principal malignancy impacting the biliary tract, characterized by an exceedingly poor prognosis, particularly in advanced stages. This is largely due to its aggressive invasion and the limited availability of efficacious treatment options (Roa et al., 2022). CCA is characterized as a highly lethal and heterogeneous primary liver cancer that originates from the biliary epithelium (Tomlinson et al., 2023; Ilyas et al., 2023). The incidence of BTC is on an upward trajectory globally, btc continues to constitute a significant global health concern. Projected mortality data pertaining to oncological conditions in the United States indicate that by the year 2040, hepatic and intrahepatic bile duct neoplasms are anticipated to overtake colorectal cancer, ascending to become the third leading etiology of cancer-associated fatalities (Rahib et al., 2021). However, the etiological factors contributing to CCA remain inadequately elucidated (Banales et al., 2020; Clements et al., 2020; Ouyang et al., 2021; Sung et al., 2021). The obstacles of screening and early detection persist, largely due to the infrequent manifestation of distinctive symptoms in patients (European Liver Research Association (2023); Harding et al., 2023; Kelley et al., 2023). The prompt diagnosis and categorization of BTC at its incipient stage is of critical importance to enhance the probability of therapeutic success. BTC may manifest as a consequence of the cumulative accrual of both genetic and epigenetic modifications. This pathogenesis is potentially modulated by a multitude of factors, including host immune responses, dietary habits, environmental elements, and microbial interactions (Nakamura et al., 2015; Kamisawa et al., 2017; Clements et al., 2020; Choi et al., 2022; Kendre et al., 2023; Jansson et al., 2023). The term ‘microbiome’ denotes the aggregate of genomic material stemming from microorganisms residing within a specific ecological niche (Sender et al., 2016). These microorganisms are essential in a range of host functions, including the modulation of immune responses, providing defense against pathogenic microbes, and managing metabolic regulatory processes (Honda and Littman, 2016; Brown et al., 2019).
The gastrointestinal tract and the liver share a profound anatomical and physiological interrelation, often referred to as the “gut-liver axis.” This axis governs not merely hepatic pathophysiological processes but also influences intrahepatic and systemic immune dynamics (Bubnov et al., 2019; Bubnov et al., 2017; Bubnov et al., 2015). Consequently, the gastrointestinal microbiota plays a pivotal role in regulating antineoplastic immune responses (Kudela et al., 2021). The intestinal barrier functions as the initial line of defense, a disrupted configuration of the intestinal microbiome, termed “dysbiosis,” has been linked to compromised integrity of the intestinal barrier (Peterson and Artis, 2014). The enhancement of our understanding of the microbiome’s role is substantially driven by advancements in high-throughput DNA sequencing and the refinement of computational techniques. These technological developments enable a more sophisticated examination of the microbiome’s intricacies (Wang et al., 2021).Emerging empirical data increasingly suggest that perturbations in the gut-liver axis may be instrumental in the etiopathogenesis of a myriad of hepatic disorders, including BTC (Scheufele et al., 2017; Ma et al., 2018; Schramm, 2018; Tripathi et al., 2018; Molinero et al., 2019; Wang et al., 2022). A recent study by Ma et al. confirmed that Helicobacter Species infection was associated with an increased risk of BTC (Gros et al., 2023). Zhou et al. observed a notably elevated prevalence of Helicobacter infections among individuals diagnosed with BTC, in contrast to those presenting with benign biliary conditions (Zhou et al., 2013). In the research conducted by Murphy and associates, they determined that there was a correlation between seropositivity to H. pylori proteins and an augmented likelihood of BTC onset (Murphy et al., 2014). The most efficacious approach to enhancing health outcomes appears to be through proactive prevention measures. Probiotics, supported by a robust body of evidence, demonstrate considerable potential in this preventative capacity. This presents significant prospects for the formulation of holistic strategies utilizing prebiotics for the promotion of healthful diets and the management and prophylaxis of BTC (Liu et al., 2022). Previous scholarly inquiries have indicated the potential of intestinal microbiota to act as an emergent biological indicator for the prognostic assessment and preventive measures concerning BTC (Plieskatt et al., 2013; Chng et al., 2016; Jia et al., 2020; Chen et al., 2021; Zhang Q. et al., 2021; Zhang T. et al., 2021; Mao et al., 2021; Abril et al., 2022; Binda et al., 2022; Ito et al., 2022; Okuda et al., 2022; Wheatley et al., 2022; Chai et al., 2023; Elvevi et al., 2023).
Nonetheless, the correlation between gut microbiota and BTC is susceptible to modulation by environmental variables, lifestyle choices, and additional confounding elements in observational investigations. Such conditions circumscribe the ability to draw causal inferences linking gut microbiota to BTC. The relationship between host genetic factors and the gut microbiome in the context of BTC necessitates more comprehensive exploration. Mendelian Randomization (MR), a prevalent analytical technique employed for investigating causal linkages between exposure variables and resultant outcomes, has been utilized to probe prospective causal affiliations between gut microbiota and a diverse array of medical conditions (Liu et al., 2023; Xi et al., 2023; Min et al., 2023; He et al., 2023; Li et al., 2023; Luo et al., 2023b). Consequently, we endeavored to ascertain the causal linkage between these variables at the genetic level through the application of MR techniques. Furthermore, we sought to identify potential microbial biomarkers conducive to disease prevention and amelioration in individuals diagnosed with BTC.
In the present study, we hypothesized that the incidence of BTC may be higher in patients with microbiota dysbiosis due to the fact that microbiota dysbiosis induces immune disruption through the gut-hepatic axis. Our objective is to attain personalized forecasting and prophylaxis of BTC by identifying specific microbiota in individual fecal samples that possess a causative association with BTC. This endeavor aims to furnish a robust evidence-based medicinal foundation for the formulation of an integrative service model encompassing BTC health prediction, diagnosis, intervention, prevention, and health augmentation.
The present MR investigation was conducted and documented in compliance with the STROBE-MR guidelines, which are designed to fortify the reporting quality of observational epidemiological studies (Skrivankova et al., 2021a; Skrivankova et al., 2021b). In order to evaluate our proposed hypothesis, we selected genetic variants pertinent to features of gut microbiota (N = 18,340) as well as BTC (comprising 418 cases and 159,201 controls) for our analysis. bidirectional MR approach was employed to scrutinize the influence of gut microbiota on the susceptibility to BTC development. MR integrates summary statistics from Genome-Wide Association Studies (GWAS), thereby attenuating the impact of confounding variables on the analysis. Figure 1 delineates the schematic representation of the research methodology. The MR investigation adhered to three critical assumptions to ensure the validity and reliability of the resultant findings: (1) Instrumental variables (IVs) were distinctly correlated with each GM taxon and exhibited no association with BTC; (2) The IVs uniquely linked with each GM taxon demonstrated no correlation with potential confounding variables; and (3) The influence exerted by the IVs on BTC was mediated exclusively through their respective associations with each GM taxon, devoid of interference from extraneous variables.
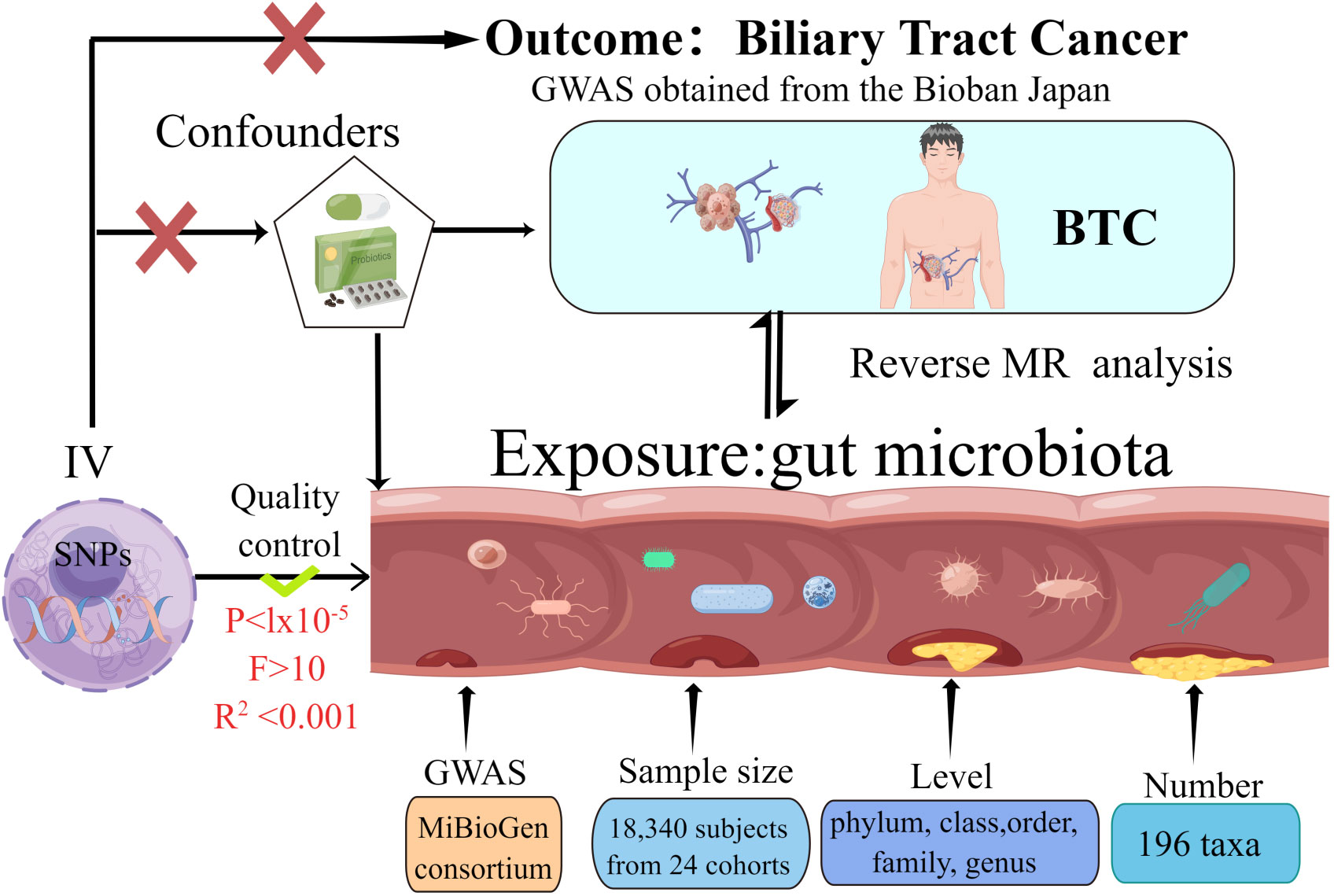
Figure 1 The study design of the present Mendelian randomization study of the associations of the gut microbiota and Biliary tract cancer risk.
We sourced Single Nucleotide Polymorphisms (SNPs) correlated with gut microbial abundance from the GWAS conducted by the MiBioGen consortium. This study encompassed 25 cohorts comprising a total of 18,340 participants and was geared toward pinpointing genetic loci that exert an impact on the relative abundance of gut microbes. This identification was achieved through the analysis of 16S rRNA sequencing profiles corresponding to the study subjects. Subsequent to their collection, summary statistics from GWAS pertaining to 196 bacterial taxa were incorporated into the MR analysis (Kurilshikov et al., 2021). We retrieved GWAS summary statistics related to BTC from the BioBank Japan (BBJ). The BBJ serves as a prospective genomic repository that has collaboratively amassed DNA and serum specimens from a consortium of 12 medical institutions across Japan. Investigations pertaining to BBJ received ethical approval from both the Institutional Review Board at RIKEN Yokohama Institute and the Ethics Committee at the Institute of Medical Science, University of Tokyo (Ishigaki et al., 2020).
A rigorous sequence of quality control measures was implemented to identify and select IVs that met the established eligibility criteria, In alignment with the prevailing standards for GM research, we selected IVs using a threshold of P < 1 × 10−5 for screening (Zhang et al., 2023). The F-statistic serves as a quantitative metric for assessing the robustness of the association between genetic variants and the exposure variable, with a higher F-statistic signifying a more potent instrumental variable. Only IVs that manifest an F-statistic exceeding 10 are retained for further analysis (Burgess et al., 2017). The calculation of linkage disequilibrium (LD) between SNPs was performed utilizing the European samples from the 1000 Genomes Project as the reference panel. SNPs manifesting the lowest P-values were retained for further study, provided they satisfied the criterion of R2 < 0.001 within a clumping window size of 10,000 kilobases. SNPs exhibiting a minor allele frequency (MAF) below the threshold of 0.01 were systematically excluded from the analysis.
The MR investigation was integrated to assess the causative associations between 196 distinct microbial taxa and BTC. Utilizing the Bonferroni correction method, we delineated the criteria for statistical significance for the principal MR outcomes across various taxonomic levels, ranging from phylum to genus. Specifically, for a given taxonomic level that incorporates ‘n’ distinct bacterial taxa, the threshold for statistical significance, after applying Bonferroni correction, is determined as 0.05/n (Luo et al., 2023a). The inverse variance-weighted (IVW) methodology was employed as the principal analytical instrument for evaluating the influence of gut microbiota on the susceptibility to BTC (González-García et al., 2023).To enhance the robustness of our analytical approach, we incorporated four supplementary MR techniques into our investigation. These encompassed the simple mode, weighted mode, MR-Egger, and weighted median strategies. A P-value of less than 0.05 was considered to denote statistical significance in the context of MR analysis. In consideration of the limited sample size in the BTC GWAS, the statistical power for conducting a MR analysis was ascertained via computations performed on the mRnd website. (https://shiny.cnsgenomics.com/mRnd/) (Brion et al., 2013). Within the context of the IVW method, the Cochran’s Q test was employed to evaluate the extent of statistical heterogeneity among the SNPs incorporated in each individual analysis. The estimation of horizontal pleiotropy was conducted through the utilization of the MR-Egger intercept test, a P-intercept value of less than 0.05 serves as an indicative metric, suggesting the existence of horizontal pleiotropy. To mitigate the impact of potential confounding variables, we conducted an additional query using the PhenoScanner database to ascertain whether the SNPs that yielded significant MR estimates in this investigation were concomitantly linked with other risk factors for BTC. Furthermore, for those gut microbiota taxa exhibiting causal relationships, we conducted advanced pleiotropy evaluations utilizing the MR Pleiotropy RESidual Sum and Outlier (MR-PRESSO) methodology, subsequently excluding any identified outliers, Statistical evaluations were conducted utilizing the TwoSampleMR and MR-PRESSO packages within the R software environment (version 4.2.2).
To examine the potential causal relationship between BTC and various bacterial genera, we conducted a reverse Mendelian Randomization analysis, wherein BTC served as the exposure variable and the composition of the gut microbiota was the outcome variable. SNPs linked to BTC were employed as instrumental variables for this analysis.
In prior research endeavors, written informed consents were duly obtained from all involved participants. Correspondingly, these studies received the necessary approvals from relevant ethical review committees (Kurilshikov et al., 2021; Ishigaki et al., 2020).
In the present study, initial efforts were made to obtain effective IVs using rigorous quality control. These IVs were then used in an MR analysis to assess the putative causal relationship between 196 GM taxa and BTC. In every retained SNP, the F-statistic exceeded the value of 10 (as detailed in Supplementary Table), signifying an adequate level of statistical power in the correlation between the IV and their corresponding bacterial taxa. For the entirety of the MR outcomes, we undertook sensitivity assessments to scrutinize both heterogeneity, as indicated by Cochran’s Q statistic, and pleiotropic effects, as evaluated through MR-Egger regression and MR-PRESSO methodologies.
In the MR investigation conducted on gut microbiota, utilizing microbiota-associated SNPs as instruments, the initial analysis using the IVW method discerned six specific taxa that exhibited the potential for exerting causal influences on the occurrence of BTC. Utilizing the IVW analytical method, it was discerned that the Family Streptococcaceae (odds ratio (OR) 0.44, 95% confidence interval (CI) 0.21–0.94, P = 0.034); Family Veillonellaceae (OR 0.46, 95% CI 0.24–0.88, P = 0.018); Genus Dorea (OR 0.29, 95% CI 0.09–0.88, P = 0.018) exhibited a negative correlation with BTC susceptibility. Conversely, the Class Lentisphaeria (OR=2.21, 95% CI:1.15–4.25, P=0.017); Genus Lachnospiraceae FCS020 (OR=2.30, 95% CI:1.19–4.45, P=0.013) and Order Victivallales (OR=2.21, 95% CI:1.15–4.25, P=0.017) were observed to have a positive association with BTC risk (Figures 2A, B, 3A–L). The p-values derived from the Cochran Q test and the MR-Egger intercept test exceeded 0.05. This suggests compelling evidence supporting the lack of heterogeneity and the absence of pleiotropy in the study (Table 1).
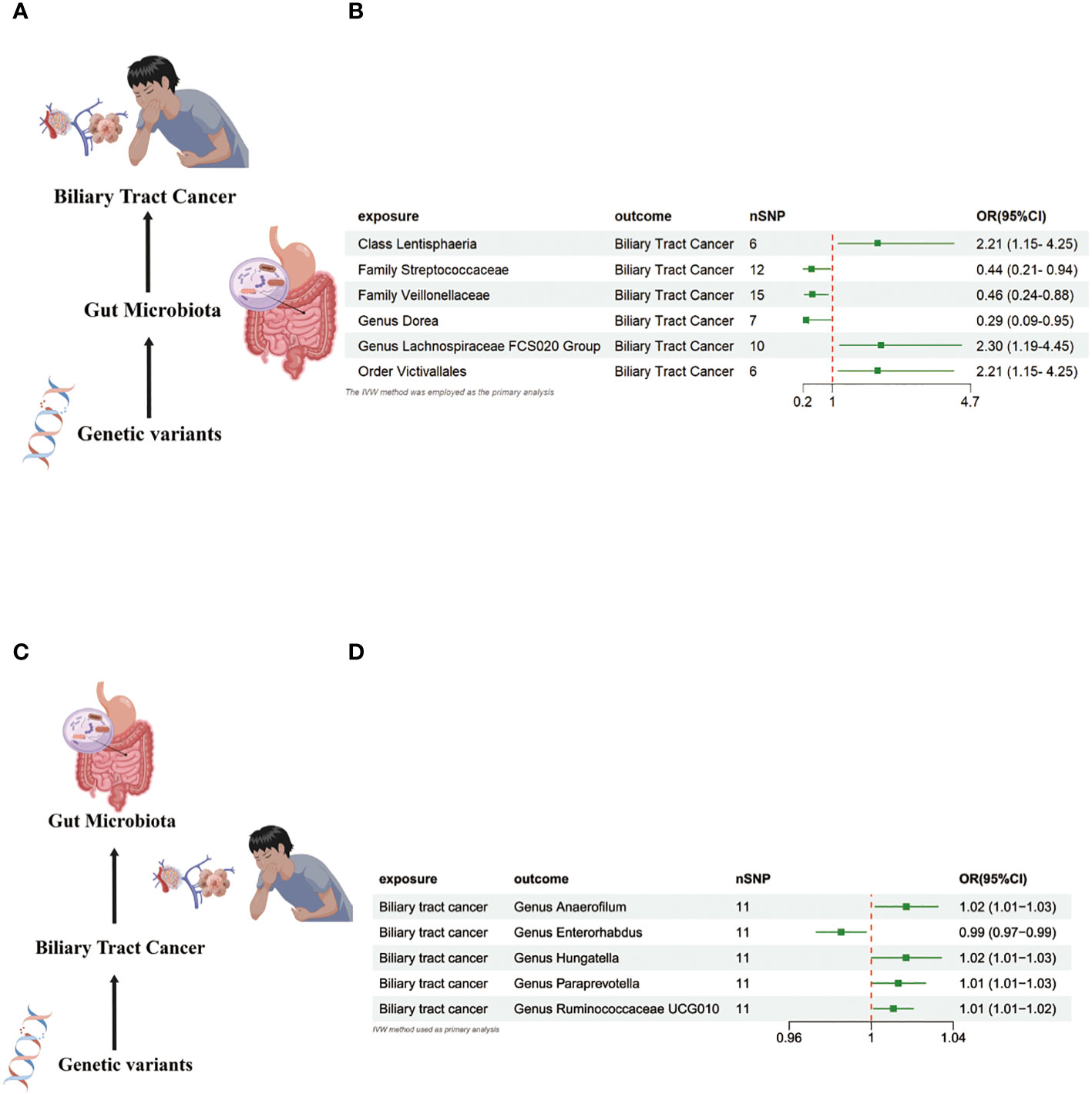
Figure 2 (A) Causal effect of gut microbiota with Biliary tract cancer Schematic representation of the MR analysis results. (B) Forest plot of the MR analysis results. (C) Causal effect of Biliary tract cancer with gut microbiota Schematic representation of the Reverse MR analysis results. (D) Forest plot of the MR analysis results. OR odds ratio, CI confidence interval, IVW inverse variance weighted method, Significant threshold was set at p-value <0.05 for the Inverse Variance Weighted method (IVW).
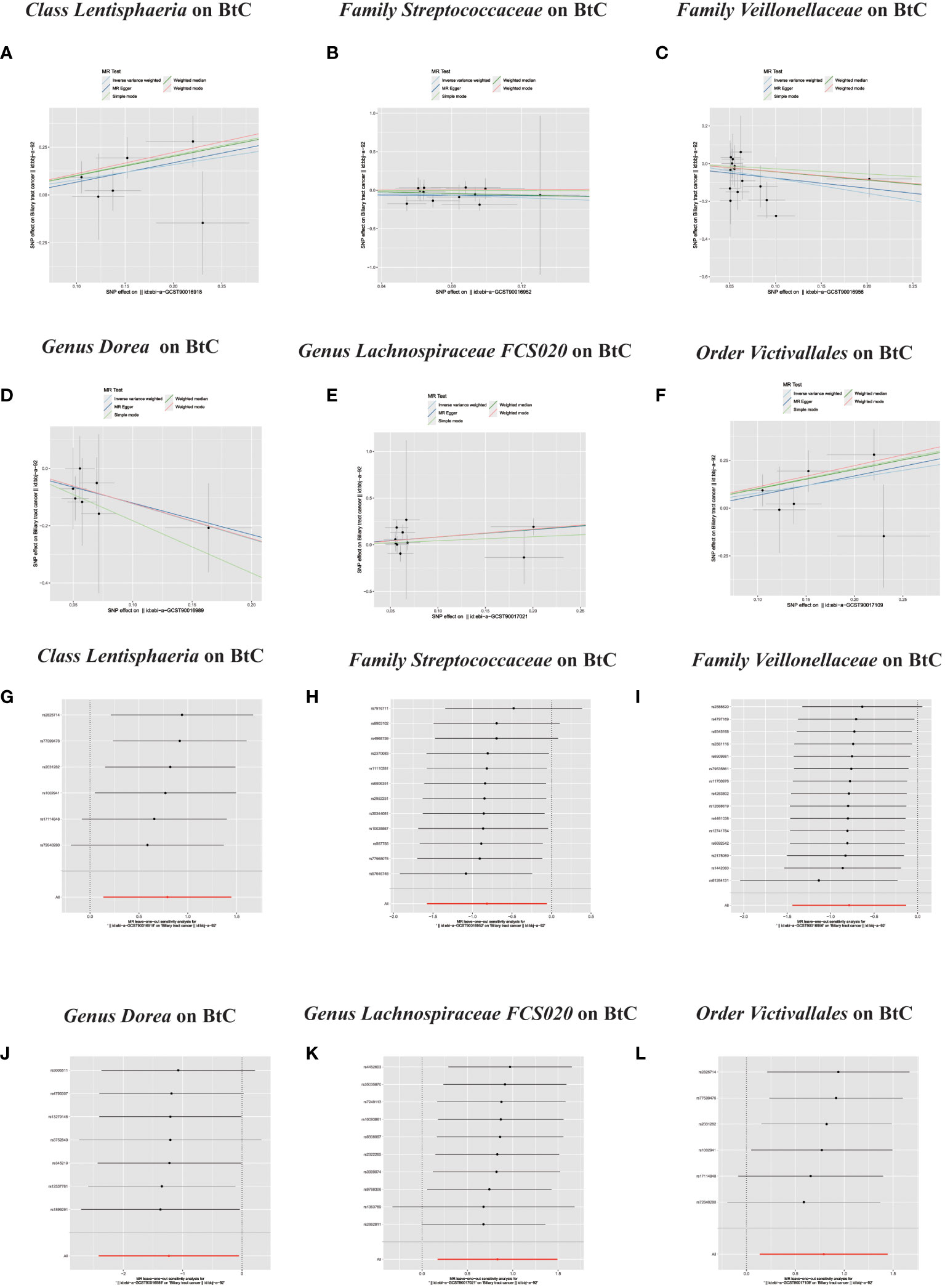
Figure 3 (A–L) Scatter plots of significant causality of the GM and BTC. Scatter plot of the effect size and 95% confidence interval of each SNP on GM and BTC risk. The horizontal axis reflects genetic effect of each SNP on GM. The vertical axis represents the genetic effect of each SNP on BTC risk. Leave-one-out analysis for the impact of individual SNPs on the association between GM and BTC risk. By leaving out exactly one SNP, it demonstrates how each individual SNP influences the overall estimate.
In the bidirectional MR study, we examined the potential causal linkage between BTC on gut microbiota. Our analysis revealed a marked causal relationship between genetically inferred BTC and an elevated abundance of the following genera: Anaerofilum (OR = 1.02, 95% CI: 1.01–1.03, P = 0.026), Hungatella (OR = 1.02, 95% CI: 1.01–1.03, P = 0.026), Paraprevotella (OR = 1.02, 95% CI: 1.01–1.03, P = 0.026), and Ruminococcaceae UCG010 (OR = 1.02, 95% CI: 1.01–1.03, P = 0.026). Conversely, a reduced abundance was observed in the Genus Enterorhabdus (OR = 0.99, 95% CI: 0.97–0.99, P = 0.018) (Figures 2C, D, 4A–J). The consistency of the outcomes was further assessed utilizing the MR Egger approach. The obtained p-values exceeding 0.05 suggest a lack of heterogeneity in our findings. Subsequent evaluation using the MR-PRESSO methodology indicated the absence of significant outliers and a lack of horizontal pleiotropy within our MR study (P > 0.05) (Table 2).
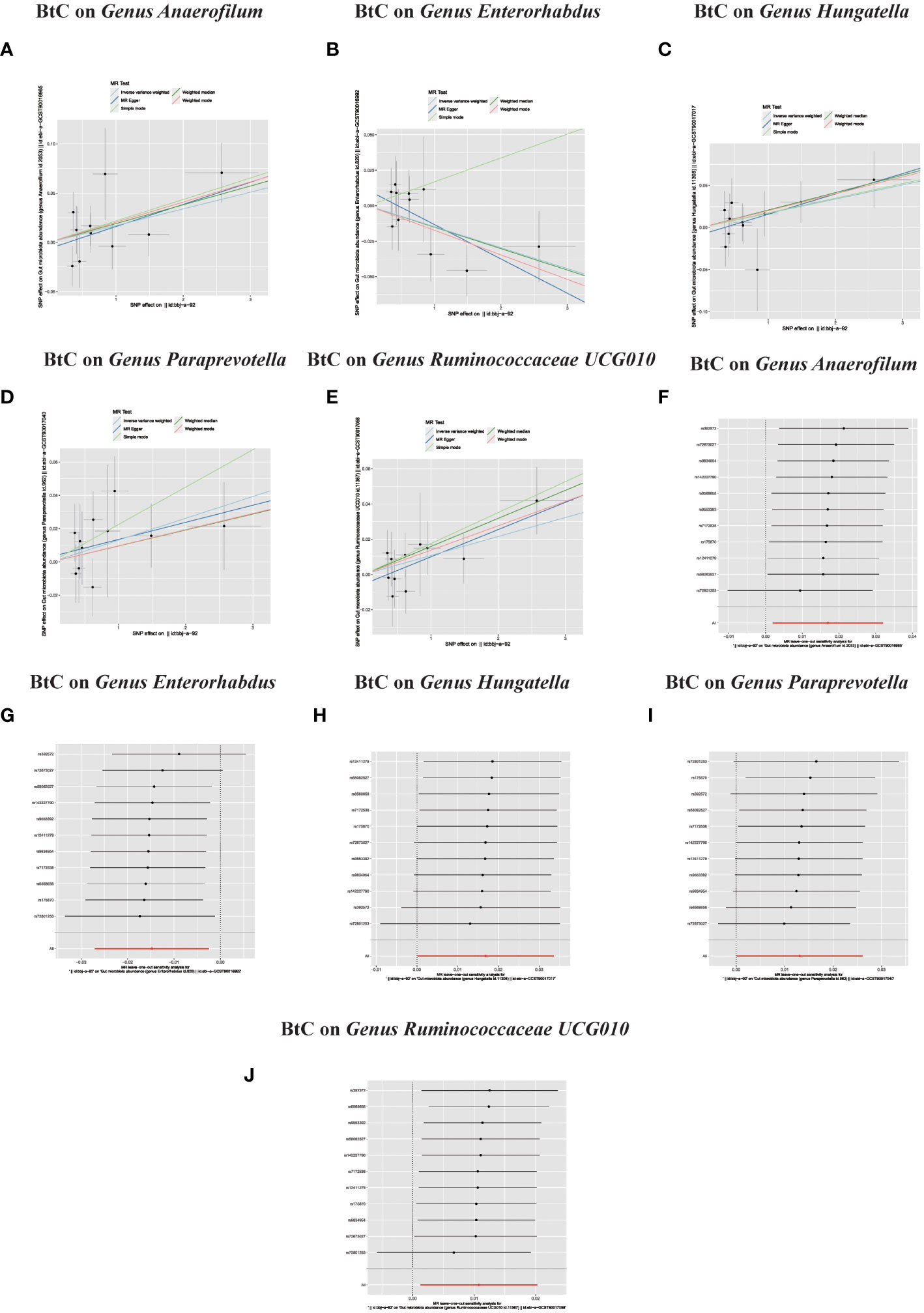
Figure 4 (A–J) In reverse MR analysis, Plots for “leave-one-out” analysis for causal effect of Biliary tract cancer on gut microbiota risk; In reverse MR analysis, The scatter plots for association between Biliary tract cancer and gut microbiota.
The study at hand validates the initial hypothesis. The adopted methodology underscores a paradigmatic transition from reactive healthcare to a framework emphasizing predictive, preventive, and personalized medicine (3PM/PPPM), targeting susceptible segments of the populace. The worldwide prevalence of BTC has consistently presented a considerable challenge to public health initiatives over an extended period (Zhang Y. et al., 2021). Biobanking is transitioning into a new epoch characterized by the prominence of big data, in the age of big data, biobanking plays a pivotal role in advancing predictive, preventive, and personalized medicine, ensuring optimal treatment for each patient at the appropriate juncture. In this research, we harnessed expansive GWAS summary-level data through MR analysis to scrutinize the putative causal linkage between the gut microbiota and BTC. The results indicate that different constituents of the gut microbiota might either augment or attenuate BTC risk. In the context of big data, biobanking facilitates the transition towards a more individualized healthcare framework. PPPM offers an innovative methodology for addressing BTC-related disorders.The current research revealed that taxa from the Family Streptococcaceae, Family Veillonellaceae, and the Genus Dorea exhibited a negative correlation with BTC susceptibility. Conversely, taxa from the Class Lentisphaeria, Order Victivallales, and the Genus Lachnospiraceae FCS020 Group demonstrated a positive association with BTC risk.
The Streptococcaceae family, particularly some strains of Streptococcus, have been investigated for their potential role in carcinogenesis (Re et al., 2023). One hypothesis is that they may induce chronic inflammation, a known risk factor for cancer. Chronic inflammation can lead to DNA damage, promoting mutations and the initiation of cancerous growths. Furthermore, certain Streptococcus species produce metabolites that may have carcinogenic properties, potentially impacting the development of BTC (Plieskatt et al., 2013; Daniel et al., 2023). Members of the Veillonellaceae family are known for their role in the fermentation of proteins and carbohydrates in the gut. The metabolites produced during this process, such as short-chain fatty acids, can have both protective and harmful effects. While some metabolites might have anti-inflammatory and anti-carcinogenic effects, others may contribute to the development of cancer. For BTC, the pathogenic mechanism may involve the alteration of bile acid metabolism and the gut-liver axis, which can lead to an imbalance in the liver’s cellular environment, potentially contributing to carcinogenesis (Mao et al., 2021; Button et al., 2023). Dorea, a less studied genus, has been linked with various gastrointestinal diseases. Its role in BTC could be associated with the modulation of the gut-liver axis and immune responses. Dorea may influence the liver’s immune environment, either promoting or inhibiting inflammation. In the context of BTC, an altered immune response in the liver could facilitate the development of cancerous cells or, conversely, provide a protective effect (Mima et al., 2017).
In the bidirectional MR study, we examined the potential causal linkage between BTC on gut microbiota. The gastrointestinal tract harbors a diverse and intricate consortium of microorganisms, collectively referred to as the gut microbiota. The portal vein conveys metabolites originating from the gut to the liver. Molecules such as bile acids, which are secreted by the liver, modulate the microbial environment. Binda et al. delineated three primary modalities through which gut microbiota potentially instigate cancerous activities: firstly, through bacterial toxins and metabolites; secondly, by altering both local and systemic immune responses of the host; and thirdly, via metabolic alterations in both the microbiota and the host (Binda et al., 2022). To a certain degree, our findings concur with earlier research (Mao et al., 2021). In clinical investigations analyzing stool samples from BTC patients, there was a notable increase in alpha-diversity compared to healthy counterparts. Specifically, the abundance of taxa such as Lactobacillus, Actinomyces, Peptostreptococcaceae, Alloscardovia, and Bifidobacteriaceae was conspicuously elevated (Jia et al., 2020). Nevertheless, the integration of such genetic information into clinical protocols presents a formidable challenge. Our research effectively established a connection between the gut microbiota and BTC by employing MR, considering genetic data as IV, and extrapolating the genetic association between the two entities. This represents the inaugural MR study elucidating the causal relationship between the gut microbiota and BTC, effectively mitigating the influence of confounders. Furthermore, the outcomes from the MR study hold significant relevance for public health. They augment prior research concerning the gut microbiota and BTC, offering a novel viewpoint on their genetic-level association. In terms of disease prevention, timely modulation of the gut microbiota can steer the prophylaxis of BTC disease. Diagnostically, it’s imperative to prioritize BTC screenings for individuals exhibiting gut microbiota disturbances. This research utilized GWAS data on intestinal flora from European subjects and BTC GWAS data from Japanese subjects. Consequently, the population sample in this investigation offers a representative cross-section. It is imperative to acknowledge the constraints inherent to our research. First, the genetic information pertaining to BTC was sourced from GWAS. However, the relatively small sample size of cancer cases could potentially skew the results of the GWAS, given the imbalanced ratios between cases and controls; Second, our projections may also be susceptible to inherent limitations associated with Mendelian Randomization analysis, including potential selection bias.
From a broader perspective, these bacteria and their interactions with the host’s immune system, as well as their metabolic by-products, are of significant interest in understanding BTC. The gut microbiota can modulate systemic inflammation, immune surveillance, and the metabolome, all of which are crucial factors in carcinogenesis. It is essential to note that while these associations are promising, they are complex and require further research. Future studies should focus on elucidating the precise mechanisms by which these bacteria influence BTC, potentially leading to new therapeutic strategies. The rapidly evolving field of microbiome research continually provides new insights into the intricate relationship between the gut flora and various cancers, including BTC.
Our inaugural systematic Mendelian randomization assessment furnishes empirical support suggesting a potential causal linkage between various gut microbiota taxa and BTC. This finding could offer salient biomarkers that are advantageous for the early, non-invasive detection of BTC. Furthermore, it might delineate prospective targets for therapeutic strategies in addressing the ailment. While extensive research is still needed to elucidate the connections between the gut microbiome and BTC, burgeoning mechanistic understandings are paving the way for innovative interventions, including potential strategies for microbiota modulation. Furthermore, these insights are shaping public health recommendations concerning dietary and lifestyle habits to preemptively combat these fatal malignancies. Additional scholarly investigations are imperative to both authenticate particular gut microbes linked to BTC and to substantiate the underlying mechanism and causal relationship through animal models and clinical studies. Ultimately, this progression will facilitate the shift from reactive medical interventions to a PPPM approach in the oversight of BTC.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The studies involving humans were approved by All the data utilized in this investigation are publicly accessible and fall within the public domain. All participants granted informed consent, and the study protocols received approval from their respective local ethical committees. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article. Consensus among all authors was achieved regarding the manuscript.
KW: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing, Supervision. SW: Writing – original draft, Writing – review & editing, Data curation, Methodology, Supervision, Conceptualization, Formal analysis, Validation, Investigation, Visualization, Software. XQ: Conceptualization, Investigation, Methodology, Software, Writing – original draft, Writing – review & editing, Project administration, Supervision, Validation, Data curation. YFC: Conceptualization, Investigation, Methodology, Software, Writing – original draft, Writing – review & editing, Project administration, Validation, Supervision. YHC: Writing – review & editing, Data curation, Methodology, Supervision, Conceptualization, Validation, Investigation, Software. JW: Conceptualization, Data curation, Formal analysis, Investigation, Project administration, Software, Writing – original draft. YZ: Investigation, Methodology, Project administration, Software, Supervision, Validation, Writing – review & editing. QG: Conceptualization, Data curation, Investigation, Methodology, Project administration, Supervision, Writing – review & editing. CZ: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Writing – review & editing. DZ: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by grants from the National Nature Science Foundation of China [grant number: 82270667].
We are very grateful to MiBioGen Consortium and Biobank Japan (BBJ) database for their selfless public sharing of GWAS summary data, which provides us with great convenience to carry out this research. We are grateful for expertise assistance in Figure from Figdraw (www.figdraw.com).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1308742/full#supplementary-material
Supplementary Table 1 | Instrumental variables (selected SNPs) used in MR analysis of the association between gut microbiota and Biliary tract cancer.
Supplementary Table 2 | Instrumental variables (selected SNPs) used in bidirectional MR analysis of the association between Biliary tract cancer and gut microbiota.
BTC, Biliary tract cancer; MR, Mendelian Randomization; GWAS, Genome-wide association study; IVW, Inverse variance weighting; PPPM, Predictive, Preventive, and Personalized Medicine; GBC, Gallbladder cancer; CCAs, cholangiocarcinomas; SNP, Single-nucleotide polymorphism.
Abril, A. G., Villa, T. G., Sánchez-Pérez, Á., Notario, V., Carrera, M. (2022). The role of the gallbladder, the intestinal barrier and the gut microbiota in the development of food allergies and other disorders. Int. J. Mol. Sci. 23. doi: 10.3390/ijms232214333
Banales, J. M., Marin, J. J. G., Lamarca, A., Rodrigues, P. M., Khan, S. A., Roberts, L. R., et al. (2020). Cholangiocarcinoma 2020: the next horizon in mechanisms and management. Nat. Rev. Gastroenterol. Hepatol. 17, 557–588. doi: 10.1038/s41575-020-0310-z
Benson, A. B., D’Angelica, M. I., Abrams, T., Abbott, D. E., Ahmed, A., Anaya, D. A., et al. (2023). Nccn guidelines® Insights: biliary tract cancers, version 2.2023. J. Natl. Compr. Canc Netw. 21, 694–704. doi: 10.6004/jnccn.2023.0035
Binda, C., Gibiino, G., Coluccio, C., Sbrancia, M., Dajti, E., Sinagra, E., et al. (2022). Biliary diseases from the microbiome perspective: how microorganisms could change the approach to benign and malignant diseases. Microorganisms 10. doi: 10.3390/microorganisms10020312
Brion, M.-J. A., Shakhbazov, K., Visscher, P. M. (2013). Calculating statistical power in mendelian randomization studies. Int. J. Epidemiol. 42, 1497–1501. doi: 10.1093/ije/dyt179
Brown, E. M., Kenny, D. J., Xavier, R. J. (2019). Gut microbiota regulation of T cells during inflammation and autoimmunity. Annu. Rev. Immunol. 37, 599–624. doi: 10.1146/annurev-immunol-042718-041841
Bubnov, R., Babenko, L., Lazarenko, L., Kryvtsova, M., Shcherbakov, O., Zholobak, N., et al. (2019). Can tailored nanoceria act as a prebiotic? Report on improved lipid profile and gut microbiota in obese mice. EPMA J. 10, 317–335. doi: 10.1007/s13167-019-00190-1
Bubnov, R. V., Babenko, L. P., Lazarenko, L. M., Mokrozub, V. V., Demchenko, O. A., Nechypurenko, O. V., et al. (2017). Comparative study of probiotic effects of lactobacillus and bifidobacteria strains on cholesterol levels, liver morphology and the gut microbiota in obese mice. EPMA J. 8, 357–376. doi: 10.1007/s13167-017-0117-3
Bubnov, R. V., Spivak, M. Y., Lazarenko, L. M., Bomba, A., Boyko, N. V. (2015). Probiotics and immunity: provisional role for personalized diets and disease prevention. EPMA J. 6, 14. doi: 10.1186/s13167-015-0036-0
Burgess, S., Small, D. S., Thompson, S. G. (2017). A review of instrumental variable estimators for mendelian randomization. Stat. Methods Med. Res. 26, 2333–2355. doi: 10.1177/0962280215597579
Button, J. E., Cosetta, C. M., Reens, A. L., Brooker, S. L., Rowan-Nash, A. D., Lavin, R. C., et al. (2023). Precision modulation of dysbiotic adult microbiomes with a human-milk-derived synbiotic reshapes gut microbial composition and metabolites. Cell Host Microbe 31, 1523–38.e10. doi: 10.1016/j.chom.2023.08.004
Chai, X., Wang, J., Li, H., Gao, C., Li, S., Wei, C., et al. (2023). Intratumor microbiome features reveal antitumor potentials of intrahepatic cholangiocarcinoma. Gut Microbes 15, 2156255. doi: 10.1080/19490976.2022.2156255
Chen, Y.-W., Su, S.-L., Li, C.-W., Tsai, C.-S., Lo, C.-L., Syue, L.-S., et al. (2021). Pancreaticobiliary cancers and aeromonas isolates carrying type III Secretion system genes ascf-ascg are associated with increased mortality: an analysis of 164 aeromonas infection episodes in southern Taiwan. Front. Cell Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.749269
Chng, K. R., Chan, S. H., Ng, A. H. Q., Li, C., Jusakul, A., Bertrand, D., et al. (2016). Tissue microbiome profiling identifies an enrichment of specific enteric bacteria in opisthorchis viverrini associated cholangiocarcinoma. EBioMedicine 8, 195–202. doi: 10.1016/j.ebiom.2016.04.034
Choi, W. J., Williams, P. J., Claasen, M. P. A. W., Ivanics, T., Englesakis, M., Gallinger, S., et al. (2022). Systematic review and meta-analysis of prognostic factors for early recurrence in intrahepatic cholangiocarcinoma after curative-intent resection. Ann. Surg. Oncol. 29, 4337–4353. doi: 10.1245/s10434-022-11463-x
Clements, O., Eliahoo, J., Kim, J. U., Taylor-Robinson, S. D., Khan, S. A. (2020). Risk factors for intrahepatic and extrahepatic cholangiocarcinoma: A systematic review and meta-analysis. J. Hepatol. 72. doi: 10.1016/j.jhep.2019.09.007
Daniel, N., Genua, F., Jenab, M., Mayén, A. L., Chrysovalantou Chatziioannou, A., Keski-Rahkonen, P., et al. (2023). The role of the gut microbiome in the development of hepatobiliary cancers. Hepatology. doi: 10.1097/hep.0000000000000406
Elvevi, A., Laffusa, A., Gallo, C., Invernizzi, P., Massironi, S. (2023). Any role for microbiota in cholangiocarcinoma? A comprehensive review. Cells 12. doi: 10.3390/cells12030370
European Liver Research Association (2023). Easl-ilca clinical practice guidelines on the management of intrahepatic cholangiocarcinoma. J. Hepatol. 79, 181–208. doi: 10.1016/j.jhep.2023.03.010
González-García, B. P., Marí, S., Cilleros-Portet, A., Hernangomez-Laderas, A., Fernandez-Jimenez, N., García-Santisteban, I., et al. (2023). Two-sample mendelian randomization detects bidirectional causality between gut microbiota and celiac disease in individuals with high genetic risk. Front. Immunol. 14. doi: 10.3389/fimmu.2023.1082862
Gros, B., Gómez Pérez, A., Pleguezuelo, M., Serrano Ruiz, F. J., de la Mata, M., Rodríguez-Perálvarez, M. (2023). Helicobacter species and hepato-biliary tract Malignancies: A systematic review and meta-analysis. Cancers (Basel) 15. doi: 10.3390/cancers15030595
Harding, J. J., Fan, J., Oh, D.-Y., Choi, H. J., Kim, J. W., Chang, H.-M., et al. (2023). Zanidatamab for her2-amplified, unresectable, locally advanced or metastatic biliary tract cancer (Herizon-btc-01): A multicentre, single-arm, phase 2b study. Lancet Oncol. 24, 772–782. doi: 10.1016/S1470-2045(23)00242-5
He, G., Cao, Y., Ma, H., Guo, S., Xu, W., Wang, D., et al. (2023). Causal effects between gut microbiome and myalgic encephalomyelitis/chronic fatigue syndrome: A two-sample mendelian randomization study. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1190894
Honda, K., Littman, D. R. (2016). The microbiota in adaptive immune homeostasis and disease. Nature 535, 75–84. doi: 10.1038/nature18848
Ilyas, S. I., Affo, S., Goyal, L., Lamarca, A., Sapisochin, G., Yang, J. D., et al. (2023). Cholangiocarcinoma - novel biological insights and therapeutic strategies. Nat. Rev. Clin. Oncol. 20, 470–486. doi: 10.1038/s41571-023-00770-1
Ishigaki, K., Akiyama, M., Kanai, M., Takahashi, A., Kawakami, E., Sugishita, H., et al. (2020). Large-scale genome-wide association study in a Japanese population identifies novel susceptibility loci across different diseases. Nat. Genet. 52, 669–679. doi: 10.1038/s41588-020-0640-3
Ito, Z., Koido, S., Kato, K., Odamaki, T., Horiuchi, S., Akasu, T., et al. (2022). Dysbiosis of the fecal and biliary microbiota in biliary tract cancer. Cancers (Basel) 14. doi: 10.3390/cancers14215379
Jansson, H., Villard, C., Nooijen, L. E., Ghorbani, P., Erdmann, J. I., Sparrelid, E. (2023). Prognostic influence of multiple hepatic lesions in resectable intrahepatic cholangiocarcinoma: A systematic review and meta-analysis. Eur. J. Surg. Oncol. 49, 688–699. doi: 10.1016/j.ejso.2023.01.006
Jia, X., Lu, S., Zeng, Z., Liu, Q., Dong, Z., Chen, Y., et al. (2020). Characterization of gut microbiota, bile acid metabolism, and cytokines in intrahepatic cholangiocarcinoma. Hepatology 71, 893–906. doi: 10.1002/hep.30852
Kamisawa, T., Kaneko, K., Itoi, T., Ando, H. (2017). Pancreaticobiliary maljunction and congenital biliary dilatation. Lancet Gastroenterol. Hepatol. 2, 610–618. doi: 10.1016/S2468-1253(17)30002-X
Kelley, R. K., Ueno, M., Yoo, C., Finn, R. S., Furuse, J., Ren, Z., et al. (2023). Pembrolizumab in combination with gemcitabine and cisplatin compared with gemcitabine and cisplatin alone for patients with advanced biliary tract cancer (Keynote-966): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 401, 1853–1865. doi: 10.1016/S0140-6736(23)00727-4
Kendre, G., Murugesan, K., Brummer, T., Segatto, O., Saborowski, A., Vogel, A. (2023). Charting co-mutation patterns associated with actionable drivers in intrahepatic cholangiocarcinoma. J. Hepatol. 78, 614–626. doi: 10.1016/j.jhep.2022.11.030
Kudela, E., Liskova, A., Samec, M., Koklesova, L., Holubekova, V., Rokos, T., et al. (2021). The interplay between the vaginal microbiome and innate immunity in the focus of predictive, preventive, and personalized medical approach to combat hpv-induced cervical cancer. EPMA J. 12, 199–220. doi: 10.1007/s13167-021-00244-3
Kurilshikov, A., Medina-Gomez, C., Bacigalupe, R., Radjabzadeh, D., Wang, J., Demirkan, A., et al. (2021). Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 53, 156–165. doi: 10.1038/s41588-020-00763-1
Li, W., Ren, A., Qin, Q., Zhao, L., Peng, Q., Ma, R., et al. (2023). Causal associations between human gut microbiota and cholelithiasis: A mendelian randomization study. Front. Cell Infect. Microbiol. 13. doi: 10.3389/fcimb.2023.1169119
Liu, K., Cai, Y., Song, K., Yuan, R., Zou, J. (2023). Clarifying the effect of gut microbiota on allergic conjunctivitis risk is instrumental for predictive, preventive, and personalized medicine: A mendelian randomization analysis. EPMA J. 14, 235–248. doi: 10.1007/s13167-023-00321-9
Liu, J.-X., Liu, M., Yu, G.-Z., Zhao, Q.-Q., Wang, J.-L., Sun, Y.-H., et al. (2022). Clonorchis sinensis infection induces hepatobiliary injury via disturbing sphingolipid metabolism and activating sphingosine 1-phosphate receptor 2. Front. Cell Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.1011378
Luo, M., Cai, J., Luo, S., Hong, X., Xu, L., Lin, H., et al. (2023a). Causal effects of gut microbiota on the risk of chronic kidney disease: A mendelian randomization study. Front. Cell Infect. Microbiol. 13. doi: 10.3389/fcimb.2023.1142140
Luo, M., Sun, M., Wang, T., Zhang, S., Song, X., Liu, X., et al. (2023b). Gut microbiota and type 1 diabetes: A two-sample bidirectional mendelian randomization study. Front. Cell Infect. Microbiol. 13. doi: 10.3389/fcimb.2023.1163898
Ma, C., Han, M., Heinrich, B., Fu, Q., Zhang, Q., Sandhu, M., et al. (2018). Gut microbiome-mediated bile acid metabolism regulates liver cancer via nkt cells. Science 360. doi: 10.1126/science.aan5931
Mao, J., Wang, D., Long, J., Yang, X., Lin, J., Song, Y., et al. (2021). Gut microbiome is associated with the clinical response to anti-pd-1 based immunotherapy in hepatobiliary cancers. J. Immunother. Cancer 9. doi: 10.1136/jitc-2021-003334
Mima, K., Nakagawa, S., Sawayama, H., Ishimoto, T., Imai, K., Iwatsuki, M., et al. (2017). The microbiome and hepatobiliary-pancreatic cancers. Cancer Lett. 402, 9–15. doi: 10.1016/j.canlet.2017.05.001
Min, Q., Geng, H., Gao, Q., Xu, M. (2023). The association between gut microbiome and pcos: evidence from meta-analysis and two-sample mendelian randomization. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1203902
Molinero, N., Ruiz, L., Milani, C., Gutiérrez-Díaz, I., Sánchez, B., Mangifesta, M., et al. (2019). The human gallbladder microbiome is related to the physiological state and the biliary metabolic profile. Microbiome 7, 100. doi: 10.1186/s40168-019-0712-8
Murphy, G., Michel, A., Taylor, P. R., Albanes, D., Weinstein, S. J., Virtamo, J., et al. (2014). Association of seropositivity to helicobacter species and biliary tract cancer in the atbc study. Hepatology 60, 1963–1971. doi: 10.1002/hep.27193
Nakamura, H., Arai, Y., Totoki, Y., Shirota, T., Elzawahry, A., Kato, M., et al. (2015). Genomic spectra of biliary tract cancer. Nat. Genet. 47, 1003–1010. doi: 10.1038/ng.3375
Okuda, S., Hirose, Y., Takihara, H., Okuda, A., Ling, Y., Tajima, Y., et al. (2022). Unveiling microbiome profiles in human inner body fluids and tumor tissues with pancreatic or biliary tract cancer. Sci. Rep. 12, 8766. doi: 10.1038/s41598-022-12658-8
Ouyang, G., Liu, Q., Wu, Y., Liu, Z., Lu, W., Li, S., et al. (2021). The global, regional, and national burden of gallbladder and biliary tract cancer and its attributable risk factors in 195 countries and territories, 1990 to 2017: A systematic analysis for the global burden of disease study 2017. Cancer 127, 2238–2250. doi: 10.1002/cncr.33476
Peterson, L. W., Artis, D. (2014). Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 14, 141–153. doi: 10.1038/nri3608
Plieskatt, J. L., Deenonpoe, R., Mulvenna, J. P., Krause, L., Sripa, B., Bethony, J. M., et al. (2013). Infection with the carcinogenic liver fluke opisthorchis viverrini modifies intestinal and biliary microbiome. FASEB J. 27, 4572–4584. doi: 10.1096/fj.13-232751
Rahib, L., Wehner, M. R., Matrisian, L. M., Nead, K. T. (2021). Estimated projection of us cancer incidence and death to 2040. JAMA Netw. Open 4, e214708. doi: 10.1001/jamanetworkopen.2021.4708
Re, O. L., López-López, V., Balaguer-Román, A., Martínez-Sánchez, M. A., Eshmuminov, D., Llamoza-Torres, C. J., et al. (2023). New challenges in cholangiocarcinoma candidates for elective surgery: harnessing the microbiome dysbiosis. Langenbeck’s Arch. Surg. 408, 134. doi: 10.1007/s00423-023-02867-8
Roa, J. C., García, P., Kapoor, V. K., Maithel, S. K., Javle, M., Koshiol, J. (2022). Gallbladder cancer. Nat. Rev. Dis. Primers 8, 69. doi: 10.1038/s41572-022-00398-y
Scheufele, F., Aichinger, L., Jäger, C., Demir, I. E., Schorn, S., Sargut, M., et al. (2017). Effect of preoperative biliary drainage on bacterial flora in bile of patients with periampullary cancer. Br. J. Surg. 104, e182–e1e8. doi: 10.1002/bjs.10450
Schramm, C. (2018). Bile acids, the microbiome, immunity, and liver tumors. N Engl. J. Med. 379, 888–890. doi: 10.1056/NEJMcibr1807106
Sender, R., Fuchs, S., Milo, R. (2016). Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell 164, 337–340. doi: 10.1016/j.cell.2016.01.013
Skrivankova, V. W., Richmond, R. C., Woolf, B. A. R., Davies, N. M., Swanson, S. A., VanderWeele, T. J., et al. (2021a). Strengthening the reporting of observational studies in epidemiology using mendelian randomisation (Strobe-mr): explanation and elaboration. BMJ 375, n2233. doi: 10.1136/bmj.n2233
Skrivankova, V. W., Richmond, R. C., Woolf, B. A. R., Yarmolinsky, J., Davies, N. M., Swanson, S. A., et al. (2021b). Strengthening the reporting of observational studies in epidemiology using mendelian randomization: the strobe-mr statement. JAMA 326, 1614–1621. doi: 10.1001/jama.2021.18236
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249. doi: 10.3322/caac.21660
Tomlinson, J. L., Valle, J. W., Ilyas, S. I. (2023). Immunobiology of cholangiocarcinoma. J. Hepatol. 79, 867–875. doi: 10.1016/j.jhep.2023.05.010
Tripathi, A., Debelius, J., Brenner, D. A., Karin, M., Loomba, R., Schnabl, B., et al. (2018). The gut-liver axis and the intersection with the microbiome. Nat. Rev. Gastroenterol. Hepatol. 15, 397–411. doi: 10.1038/s41575-018-0011-z
Wang, T., Rong, X., Zhao, C. (2022). Circadian rhythms coordinated with gut microbiota partially account for individual differences in hepatitis B-related cirrhosis. Front. Cell Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.936815
Wang, R., Tang, R., Li, B., Ma, X., Schnabl, B., Tilg, H. (2021). Gut microbiome, liver immunology, and liver diseases. Cell Mol. Immunol. 18. doi: 10.1038/s41423-020-00592-6
Wheatley, R. C., Kilgour, E., Jacobs, T., Lamarca, A., Hubner, R. A., Valle, J. W., et al. (2022). Potential influence of the microbiome environment in patients with biliary tract cancer and implications for therapy. Br. J. Cancer 126, 693–705. doi: 10.1038/s41416-021-01583-8
Xi, Y., Zhang, C., Feng, Y., Zhao, S., Zhang, Y., Duan, G., et al. (2023). Genetically predicted the causal relationship between gut microbiota and infertility: bidirectional mendelian randomization analysis in the framework of predictive, preventive, and personalized medicine. EPMA J. 14, 405–416. doi: 10.1007/s13167-023-00332-6
Zhang, Q., Ma, C., Duan, Y., Heinrich, B., Rosato, U., Diggs, L. P., et al. (2021). Gut microbiome directs hepatocytes to recruit mdscs and promote cholangiocarcinoma. Cancer Discov. 11, 1248–1267. doi: 10.1158/2159-8290.CD-20-0304
Zhang, Y., Yang, C., Huang, M. (2021). Global, regional, and national burden of biliary tract cancer from 1990 to 2017 based on the 2017 global burden of disease study. Cancer Epidemiol. 73, 101949. doi: 10.1016/j.canep.2021.101949
Zhang, T., Zhang, S., Jin, C., Lin, Z., Deng, T., Xie, X., et al. (2021). A predictive model based on the gut microbiota improves the diagnostic effect in patients with cholangiocarcinoma. Front. Cell Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.751795
Zhang, R., Zhao, W., Zhao, R., Zhao, Y., Zhang, Y., Liang, X. (2023). Causal relationship in gut microbiota and upper urinary urolithiasis using mendelian randomization. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1170793
Keywords: genome-wide association study, comprehensive bidirectional mendelian randomization, gut microbiota, biliary tract cancer, probiotics
Citation: Wang K, Wang S, Qin X, Chen Y, Chen Y, Wang J, Zhang Y, Guo Q, Zhou C and Zou D (2024) The causal relationship between gut microbiota and biliary tract cancer: comprehensive bidirectional Mendelian randomization analysis. Front. Cell. Infect. Microbiol. 14:1308742. doi: 10.3389/fcimb.2024.1308742
Received: 07 October 2023; Accepted: 29 February 2024;
Published: 15 March 2024.
Edited by:
Qihang Yuan, Dalian Medical University, ChinaCopyright © 2024 Wang, Wang, Qin, Chen, Chen, Wang, Zhang, Guo, Zhou and Zou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiang Guo, Z3FrajAwM0BzaW5hLmNvbQ==; Chunhua Zhou, NzIwMDAxMjIyOEBzaHNtdS5lZHUuY24=; Duowu Zou, emR3cmp4aDY2QHNqdHUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.