
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol., 22 March 2024
Sec. Clinical Microbiology
Volume 14 - 2024 | https://doi.org/10.3389/fcimb.2024.1284701
This article is part of the Research TopicMicrobiology and Pathogenesis of Chlamydia, Coxiella, and RickettsiaView all 13 articles
Bacterial obligate intracellular parasites (BOIPs) represent an exclusive group of bacterial pathogens that all depend on invasion of a eukaryotic host cell to reproduce. BOIPs are characterized by extensive adaptation to their respective replication niches, regardless of whether they replicate within the host cell cytoplasm or within specialized replication vacuoles. Genome reduction is also a hallmark of BOIPs that likely reflects streamlining of metabolic processes to reduce the need for de novo biosynthesis of energetically costly metabolic intermediates. Despite shared characteristics in lifestyle, BOIPs show considerable diversity in nutrient requirements, metabolic capabilities, and general physiology. In this review, we compare metabolic and physiological processes of prominent pathogenic BOIPs with special emphasis on carbon, energy, and amino acid metabolism. Recent advances are discussed in the context of historical views and opportunities for discovery.
Bacterial obligate intracellular parasites (BOIPs) represent a unique group of bacteria that all depend on invasion of a eukaryotic host cell to reproduce (Casadevall, 2008). BOIPs are true parasites due to their negative impacts on the host cell, including opportunistic scavenging of host cell-synthesized nutrients, and active establishment and maintenance of unique replication vacuoles for some of these pathogens. We base the designation of an obligate intracellular lifestyle solely on information from the natural life cycle of the pathogen and not on the possibility of host cell-free (i.e., axenic) culture in a laboratory setting. As such, Coxiella burnetii remains a BOIP despite the possibility of culturing this organism axenically (Omsland et al., 2009). Prominent pathogenic BOIPs include species of the genera Coxiella, Chlamydia, Rickettsia, Anaplasma, Ehrlichia, and Orientia. This review will focus on the first three genera, which collectively represent variations in the obligate intracellular lifestyle and BOIP-host interaction (Figure 1). Reference will be made to BOIPs of other genera when appropriate.
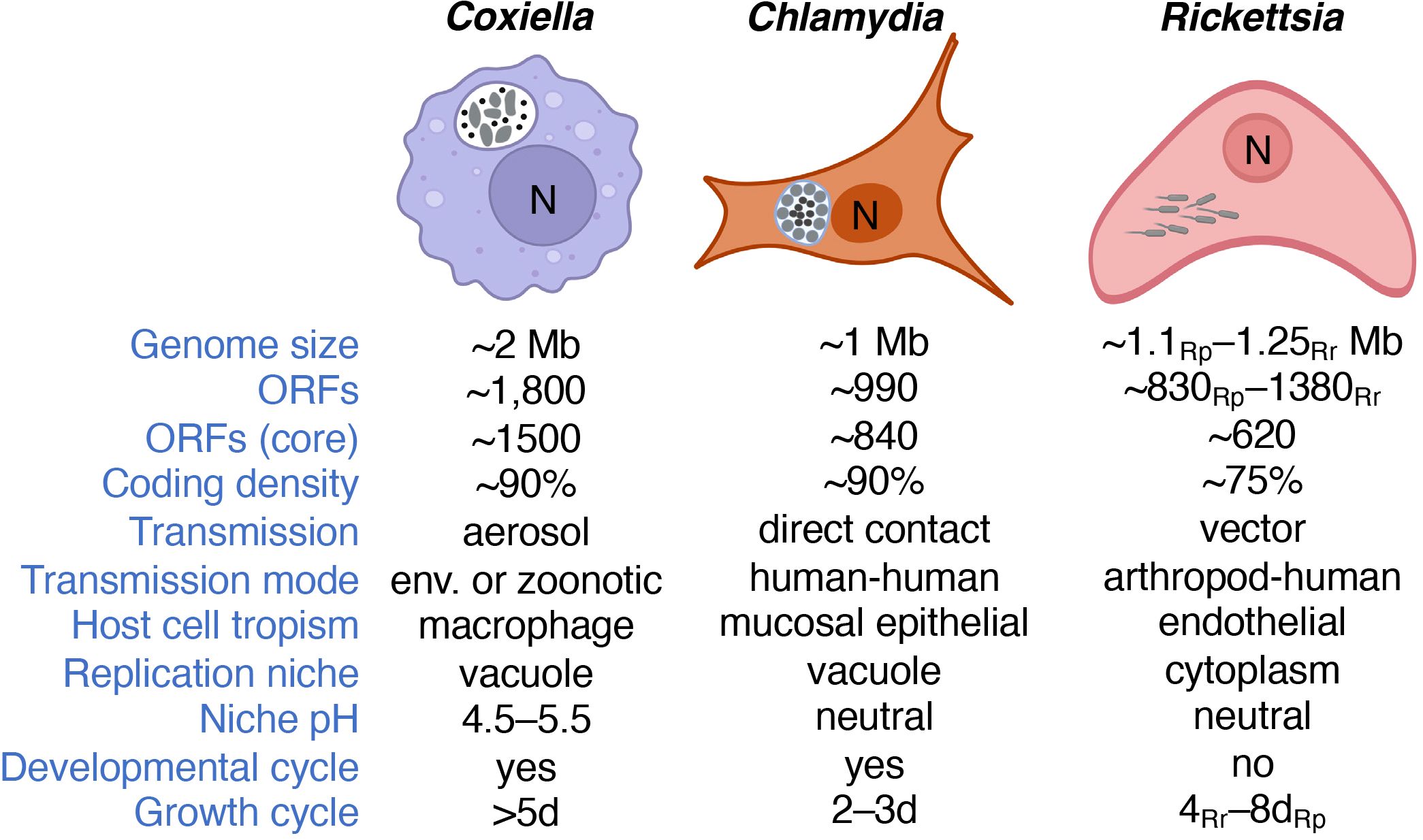
Figure 1 Major characteristics and intracellular lifestyles of Coxiella, Chlamydia and Rickettsia. Information is based on C. burnetii, C. trachomatis, and R. rickettsii unless otherwise specified. Note that differences in genome size, gene content, and/or arthropod vector exist between species of the same genus. C. burnetii is depicted as characteristic pleomorphic cells including both SCVs and LCVs. C. trachomatis is depicted with both EBs and RBs, the latter lining the inner inclusion membrane. R. rickettsii is illustrated with the characteristic actin tails that allow motility. We note that only the primary host cell type is indicated and that C. burnetii also shows tropism for placental trophoblasts. The number of ORFs refers to that of C. burnetii, C. trachomatis, R. prowazekii (Rp) and R. rickettsii (Rr), or the core genomes for the genus. R. prowazekii and R. rickettsii represent the typhus and spotted fever groups of this genus, respectively. Growth cycle refers to the approximate time required to reach stationary phase during infection of cultured Vero cells. Bacteria are shown in gray. “N” denotes the host cell nucleus and “env.” environmental (e.g., contaminated soil).
The discovery of C. burnetii appeared co-incident with an outbreak of a new febrile illness (Burnet and Freeman, 1937; Derrick, 1937), later to be called Query (Q) fever, in slaughter-house workers in Australia. Interestingly, a team of American scientists isolated C. burnetii from ticks around the same time (Davis and Cox, 1938). The pathogen has since been recognized as a major zoonotic bacterium most notably related to infection of agricultural animals, including goats (Schneeberger et al., 2014). C. burnetii is a moderate acidophile adapted to replicate within a phagolysosome-derived vacuole termed the Coxiella Containing Vacuole (CCV) (Coleman et al., 2004; Voth and Heinzen, 2007; Kohler and Roy, 2015). C. burnetii transitions between two cell forms, the replicative Large Cell Variant (LCV) and the non-replicative Small Cell Variant (SCV) that accumulates in stationary phase (Coleman et al., 2004). Both the LCV and SCV forms of C. burnetii can establish infection in cultured cells (Coleman et al., 2004; Sandoz et al., 2014).
Different species and pathotypes (aka, serovars) of the Chlamydia genus can cause a range of diseases including sexually transmitted infections, the blinding condition trachoma, as well as respiratory infections (Mishori et al., 2012; Lane and Decker, 2016; Porritt and Crother, 2016). Like Coxiella, Chlamydia species can cause disease in animals other than humans (Roulis et al., 2013; Borel et al., 2018). Also, Chlamydia species, including C. trachomatis and C. pneumoniae, replicate within a vacuole in the host cell cytoplasm termed the chlamydial inclusion (Moore and Ouellette, 2014). Development of the chlamydial inclusion depends on bacterial protein synthesis but not pathogen replication (Engström et al., 2015). Because the chlamydial inclusion does not fuse with lysosomes, the inclusion, unlike the CCV, is a non-degradative compartment with neutral pH (Heinzen et al., 1996). Members of the Chlamydia genus transition between the infectious but non-replicative Elementary Body (EB) and replicative but non-infectious Reticulate Body (RB) (Shaw et al., 2000; Belland et al., 2003b).
Pathogens in the Rickettsia genus are arthropod-borne bacteria associated with febrile illness that can cover a wide symptomatic range, including acute, chronic, and reoccurring disease (Abdad et al., 2018; Blanton, 2019). R. prowazekii and R. rickettsii, the agents of epidemic typhus and Rocky Mountain Spotted Fever, respectively, replicate within the host cell cytosol. Spotted fever group Rickettsia, including R. rickettsii, utilize actin-based motility to spread directly from an infected host cell to an adjacent non-infected cell (Teysseire et al., 1992; Heinzen et al., 1993). Members of the typhus group Rickettsia, including R. prowazekii, are non-motile or produce short actin tails (Heinzen et al., 1993). Bacteria in the Rickettsia genus can be transmitted by various arthropod vectors including lice, fleas, mites, and ticks and establish disease in various animal species (Walker and Ismail, 2008).
Neither C. burnetii nor pathogenic Chlamydia species are motile. Moreover, species of the genus Rickettsia do not transition between cell forms.
Although an understanding of the metabolic capabilities exhibited by C. burnetii, C. trachomatis and R. prowazekii saw tremendous progress during the pre-genomic era, it seems fair to say that a real sense of each pathogen’s metabolic capacity was not appreciated until their genome sequences were used as a basis for metabolic pathway reconstruction between 1998 and 2003 (Andersson et al., 1998; Stephens et al., 1998; Seshadri et al., 2003). Indeed, shortly after their discoveries, researchers questioned whether many of the pathogens known today as BOIPs were bacteria or viruses (e.g., (Ormsbee and Peacock, 1964). Early descriptions of metabolic and physiological features of these organisms were likely heavily influenced by an expectation of limited autonomous metabolic capacity. Moreover, depending on context, intracellular replication niches have been described as either nutrient rich or hostile to invasive bacteria (Moulder, 1974; Kaufmann, 2011), consistent with significant adaptability to reside intracellularly. With a relatively short history of research and severe technical challenges related to axenic culture and genetic manipulation, current understanding of BOIP metabolism and physiology remains limited. At the same time, the many knowledge gaps that exist makes this a field of research poised for discovery.
BOIPs display remarkable differences in central carbon and energy metabolism; Coxiella, Chlamydia, and Rickettsia serve as good representatives of this diversity (Figure 2). Central/core metabolism encompasses the glycolytic/gluconeogenic pathways, pentose phosphate pathway (PPP), and the tricarboxylic acid (TCA) cycle; the pathways from which core metabolites are derived (Noor et al., 2010). The general structure of the central metabolic machinery differs between Coxiella and Chlamydia in that the former has lost the oxidative branch of the PPP while the latter has lost a significant portion of the TCA cycle with the genes encoding citrate synthase, aconitase, and isocitrate dehydrogenase missing (Stephens et al., 1998). Of likely significance to the metabolic capacity and plasticity between chlamydial serovars, some strains (e.g., L2/434/Bu and L2/UCH-1/proctitis) show additional TCA cycle abnormalities at the genome level (Thomson et al., 2008; Omsland et al., 2014). While both Chlamydia (Tjaden et al., 1999) and Rickettsia (Audia and Winkler, 2006) can scavenge ATP from the host via ATP/ADP translocases, Coxiella does not rely on energy parasitism. The oxidation of glucose via glycolysis or the PPP in Coxiella (Esquerra et al., 2017) and Chlamydia (Schwöppe et al., 2002; Omsland et al., 2012) is dependent on the availability of either non-phosphorylated or phosphorylated forms of glucose, respectively. Conversely, enzymes for glycolysis/gluconeogenesis and the PPP are not encoded by species of the genus Rickettsia (Driscoll et al., 2017). In R. prowazekii, acquisition of “glucose” as a biochemical moiety for biosynthetic purposes is achieved via transport of uridine 5’-diphosphoglucose (UDP-glucose) rather than glucose itself (Winkler and Daugherty, 1986). In Chlamydia, UDP-glucose is brought into the inclusion via the host SLC35D2 transporter and used as a substrate in pathogen-driven glycogen synthesis (Gehre et al., 2016).
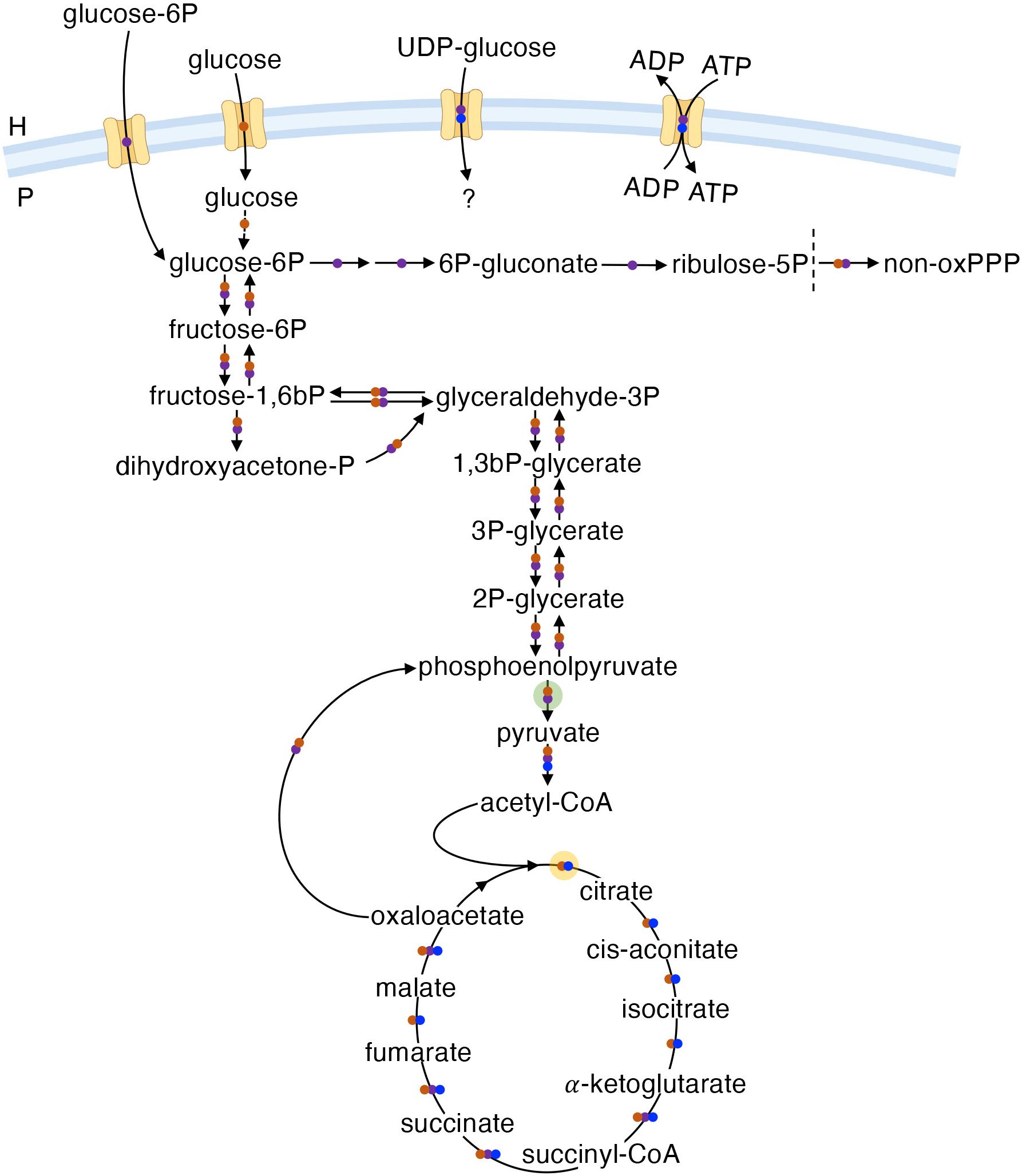
Figure 2 The central metabolic machinery. Overview of central metabolism in C. burnetii (RSA493) (●), C. trachomatis (Bu/434) (●), and R. prowazekii (Madrid E) (●). H and P denote host cell or pathogen, respectively. The broken vertical line separates the oxidative and non-oxidative branches of the PPP. The mechanism for glucose phosphorylation in C. burnetii is unknown. The question mark for UDP-glucose indicates potential use in various reactions. Transparent discs indicate bacterial enzymes with regulatory characteristics of eukaryotic proteins in Coxiella (yellow) or Chlamydia (green).
As illustrated in Figure 2, the genomes of both Coxiella and Chlamydia encode central metabolic enzymes with regulatory characteristics of their eukaryotic counterparts. For instance, citrate synthase of C. burnetii is inhibited by ATP (Heinzen and Mallavia, 1987), while pyruvate kinase from C. trachomatis is inhibited by AMP, GTP and ATP but activated by host cell-derived fructose-2,6-bisphosphate (Iliffe-Lee and McClarty, 2002). The expression of enzymes in BOIPs with regulatory characteristics akin to eukaryotic proteins may reflect adaptation to the chemical environment of the eukaryotic host cell. Moreover, these enzymes could serve as a mechanism to interconnect pathogen physiology with the physiological state of the host. Interestingly, host cell-derived pyruvate kinase (as well as aldolase A and lactate dehydrogenase) is detected at the chlamydial inclusion membrane and depletion of (host) aldolase A results in decreases in inclusion size and infectious EB progeny (Ende and Derré, 2020). The presence of host glycolytic enzymes in proximity to the chlamydial inclusion could serve to supply the pathogen and/or reactions governing the inclusion-host interaction with specific and critical glycolytic intermediates (Ende and Derré, 2020).
Early studies with C. burnetii under axenic conditions at neutral pH pointed to limited ability of this organism to metabolize autonomously. Nevertheless, some enzymatic activities were detected in bacterial extracts, including that of glycolytic enzymes (Paretsky et al., 1958, 1962; Ormsbee and Peacock, 1964; McDonald and Mallavia, 1970, 1971). The discovery that C. burnetii colocalizes with lysosomal enzymes within CCVs (Burton et al., 1971; Burton et al., 1978), combined with knowledge of the acidic pH of phagolysosomes (Ohkuma and Poole, 1978), suggested C. burnetii metabolism is strictly pH-dependent. Accordingly, Hackstadt and Williams demonstrated C. burnetii reliance on moderately acidic pH for metabolic activation and catabolism of both glucose and glutamate (Hackstadt and Williams, 1981a). Shortly thereafter, glutamate was identified as the preferred energy source of C. burnetii (Hackstadt and Williams, 1981b).
Data in support of glucose utilization by C. burnetii dates to the 1960s when “hexokinase activity” was demonstrated in bacterial cytoplasmic extracts (Paretsky et al., 1962). More recently, using a C. burnetii mutant unable to undergo gluconeogenesis due to deletion of phosphoenolpyruvate carboxykinase (PEPCK, encoded by pckA, the first committed step of gluconeogenesis) in combination with the chemically defined medium D-ACM, glucose utilization by C. burnetii for biomass production has been confirmed to be nearly as efficient as growth on amino acids (Esquerra et al., 2017). Indeed, C. burnetii can acquire glucose by at least two transporters (Kuba et al., 2019). The mechanistic redundancy revealed by the ability of C. burnetii to take up glucose via more than one transporter (Kuba et al., 2019) suggests glucose is critical for the metabolic fitness of this organism. Data from genome-wide transcriptional analysis of C. burnetii during infection of mice actually indicates that glucose is the principal carbon source while fatty acids are used for energy metabolism (Kuley et al., 2015). Preferential utilization of glucose rather than amino acids by C. burnetii to drive central metabolism contradicts earlier interpretations that were based on analysis of energy charge and ATP pool stability, rather than generation of biomass. Importantly, the ability of any pathogen to use a wide variety of substrates in energy metabolism is more relevant than the identification of a “preferred” substrate, the nature of which likely depends on the type of tissue colonized within infected animals as well as the physiological state of the animal. As for C. burnetii, PEPCK is also encoded by the C. trachomatis genome. Because generation of C. trachomatis EBs and relative ATP pools are reduced in host cells cultured with gluconeogenic substrates compared to host cells cultured with excess glucose (Iliffe-Lee and McClarty, 2000), gluconeogenic capacity could be interpreted to have limited significance for C. trachomatis EB generation. Additionally, absence of a prototypical fructose 1,6-bisphosphatase (EC 3.1.3.11) could further reduce gluconeogenic capacity in C. trachomatis (Mehlitz et al., 2016). However, because pckA has been retained and is expressed maximally by replicating RBs (Skipp et al., 2016), and alternative enzymes encoded by C. trachomatis (e.g., EC 2.7.1.11 and 2.7.1.90) could serve to convert fructose 1,6-bisphosphate to fructose-6P, gluconeogenic capacity may affect replication and/or EB generation under specific, yet undetermined, conditions. Proteomic analyses have provided somewhat different pictures regarding the expression of central metabolic enzymes, including glycolytic enzymes, in EBs and RBs (Saka et al., 2011; Skipp et al., 2016). Regardless, current data point to a significant role for glycolysis in the EB form.
Emilio Weiss and colleagues described oxidation of glucose (Weiss et al., 1964) and utilization of glucose-6P or glucose in the presence of ATP (Weiss, 1965; Weiss and Wilson, 1969) by Chlamydia, also during the 1960s. In C. trachomatis, glucose metabolism is mechanistically and physiologically different from that observed in C. burnetii. First, while C. burnetii can acquire non-phosphorylated glucose (Hackstadt and Williams, 1981a; Esquerra et al., 2017; Kuba et al., 2019), C. trachomatis appears dependent on glucose-6P (Omsland et al., 2012; Gehre et al., 2016), acquired via the UhpC transporter (McClarty, 1999; Schwöppe et al., 2002). Because C. trachomatis acquires glucose-6P from the host rather than expending ATP for its phosphorylation, and because pyrophosphate (PPi) is used in formation of fructose 1,6-bisphosphate, C. trachomatis has been postulated to gain a net 4 ATP molecules rather than 2 during glycolytic activity (McClarty, 1999). The physiological impact of impaired glucose metabolism in C. trachomatis has been illustrated by incubation with KSK120, a compound that inhibits uptake and utilization of glucose-6P in this organism (Engström et al., 2014). Similar to the observed loss of infectivity upon depriving Chlamydia-infected host cells for glucose (Harper et al., 2000; Iliffe-Lee and McClarty, 2000), treatment of C. trachomatis-infected HeLa cells with KSK120 during infection reduced generation of infectious EB progeny. Thus, metabolism of glucose-6P appears to be critical for re-generation of EBs following reproduction via RB replication. While not universally relevant to Chlamydia species, loss of GlgA (glycogen synthase) activity results in impaired infectivity in C. muridarum (Gehre et al., 2016), again connecting metabolism of glucose to chlamydial virulence. Utilization of glucose-6P may be more significant for protein synthesis in EBs compared to RBs (Omsland et al., 2012), the latter of which has been shown to behave as an energy parasite by scavenging ATP, as well as other NTPs, from the host cell (Tipples and McClarty, 1993). Beyond a critical role for generation of infectious EBs, metabolism of glucose enhances infectivity of Protochlamydia amoebophila (Sixt et al., 2013). Glucose metabolism, including oxidation of glucose-6P, also enhances EB metabolism and has been suggested to underlie maintenance of EB infectivity in extracellular environments by both pathogenic and non-pathogenic Chlamydia species (Omsland et al., 2012; Sixt et al., 2013; Grieshaber et al., 2018). Utilization of glucose or glucose-6P in Coxiella and Chlamydia stand in stark contrast to current understanding of central carbon metabolism in Rickettsia. In fact, a general lack of genes related to glycolysis and gluconeogenesis in Rickettsia (Driscoll et al., 2017) is consistent with a limited role for glucose metabolism in these organisms and an absolute requirement to obtain relevant core metabolites from the host cell.
Following phosphorylation of glucose to glucose-6P, this metabolite is generally destined to be processed via one of two distinct paths in the cell, namely glycolysis or the PPP. The PPP is further divided into two branches: an oxidative branch (oxPPP), considered a major source for recovery of the reducing equivalent NADPH; and a non-oxidative branch (non-oxPPP), critical for synthesis of ribose-5P, which is ultimately required for the biosynthesis of nucleic acid precursors, or erythrose-4P, a precursor for generation of aromatic amino acids. C. burnetii lacks two enzymes of the oxPPP (glucose-6P dehydrogenase and 6-phosphogluconate dehydrogenase), suggesting negative selective pressure on this pathway in C. burnetii. Given the apparent significance of NADPH, what selective pressure would cause C. burnetii to lose the oxPPP? In a recent study, C. burnetii was transformed with the gene encoding glucose-6P dehydrogenase, zwf, to address this question (Sanchez and Omsland, 2021). While C. burnetii expressing zwf behaved similarly to the parental strain under glucose excess, the transformant showed significantly reduced ability to replicate under glucose limitation. Expression of zwf also resulted in impaired pathogen intracellular replication in J774A.1 cells but not in Vero cells, suggesting some cell types represent a glucose-limiting environment for C. burnetii. Impaired replication under glucose-limiting conditions therefore provides some explanation for why C. burnetii has lost oxPPP capacity. Identification of the NADPH-regenerating enzyme SdrA (Bitew et al., 2020) suggests C. burnetii has evolved to generate NADPH via mechanisms complimentary to the oxPPP, possibly to circumvent metabolic conflicts under low glucose availability.
Bovarnick and Schneider demonstrated over 60 years ago that both ATP generated endogenously by R. prowazekii and ATP supplemented to the medium were necessary to stimulate axenic protein synthesis in this organism (Bovarnick and Schneider, 1960). The ATP/ADP translocase utilized by Rickettsia for ATP acquisition has since been characterized in detail (Audia and Winkler, 2006). There is also evidence for the capacity of Rickettsia species to use glutamate as an energy source (Bovarnick and Miller, 1950; Rees and Weiss, 1968; Williams and Weiss, 1978). Despite the capacity to scavenge ATP from the host cell, recent data suggest that C. trachomatis also relies on ATP generated via the combined activities of the pathogen’s sodium pump (Na+-NQR) (Dibrov et al., 2004) and a Na+-permissive A1-A0-ATPase during the replicative phase of the chlamydial developmental cycle (Liang et al., 2018). The ability of C. trachomatis to generate ATP via a sodium gradient and reliance of the energy thus generated for replication adds another dimension to the energetics of chlamydial metabolism and further challenges the “energy parasite” hypothesis proposed for this organism in the 1960s–70s (Moulder, 1962, 1974), stating that the pathogen depends on host-derived ATP. Because Chlamydia-infected host cells require ATP for viability, axenic culture will likely be necessary to resolve questions about the degree to which these pathogens depend on extracellular ATP.
Aspects of central metabolic activity in Rickettsia can also take place via atypical mechanisms. In R. prowazekii, sn-glycerol-3-phosphate needed for biosynthesis of phospholipids is acquired by importing dihydroxyacetone phosphate (DHAP) with subsequent conversion to sn-glycerol-3-phosphate by GpsA, a glycerol-3P dehydrogenase (G3PDH) (Frohlich et al., 2010). Typically, G3PDH is an enzyme integrated with activities of glycolysis and gluconeogenesis as the substrate DHAP is an intermediate of these pathways. Interestingly, despite lack of genes for complete glycolytic/gluconeogenic pathways, R. prowazekii has retained sn-glycerol-3-phosphate dehydrogenase to convert DHAP to G3P to support phospholipid biosynthesis (Frohlich et al., 2010). As noted by Frohlich and colleagues, this novel mechanism can explain the evolutionary pressure to retain G3PDH in Rickettsia.
Major distinguishing metabolic features of Coxiella, Chlamydia and Rickettsia are listed in Table 1.
Genome streamlining refers to an adaptive reduction of genome size and complexity, possibly as a response to replication in a nutrient poor environment and/or to optimize the metabolic cost of biomass generation (Giovannoni et al., 2014; Bobay and Ochman, 2017). Despite uncertainty regarding underlying selective pressure(s), genome streamlining at the expense of metabolic capacity and plasticity is a hallmark of BOIPs (Andersson et al., 1998; Stephens et al., 1998; Seshadri et al., 2003; Brenner et al., 2021). From an energetic perspective, the relative benefit of genome streamlining to a pathogen can be appreciated based on the cost of biosynthesis of specific types of molecules. Intermediates of central carbon metabolism serve key roles as precursors for the biosynthesis of other molecules, including amino acids (Noor et al., 2010). Amino acids, nucleotides and fatty acids represent the most energetically costly molecules for any organism to synthesize. For C. trachomatis, tryptophan has been highlighted as the most energetically costly amino acid to produce (Carlson et al., 2006). Importantly, the cost of amino acid synthesis can be dependent on the carbon source used (Kaleta et al., 2013).
In addition to the biosynthetic cost, the pathways and thus the number of enzymes required for biosynthesis can be extensive, which results in a significant increase in genome size for organisms that have retained more extensive biosynthetic capacity. Bioinformatic analysis of the capacity to synthesize amino acids illustrates key metabolic characteristics and biologically relevant differences between C. burnetii, C. trachomatis, and R. prowazekii (Figure 3) (Ogata et al., 1999). In general agreement with bioinformatic analysis, using a chemically defined medium not supplemented with specific amino acids, C. burnetii was demonstrated unable to grow beyond a 10-fold increase in cell numbers in media lacking any one of 11 amino acids (Sandoz et al., 2016). Comparative bioinformatic analysis of the capacity for biosynthesis of amino acids in C. burnetii, C. trachomatis and R. prowazekii shows a gradual decrease in biosynthetic capacity between these organisms. The absence of key central metabolic capacity in Rickettsia, including absence of glycolytic enzymes, underlies a major challenge for de novo biosynthesis in this genus.
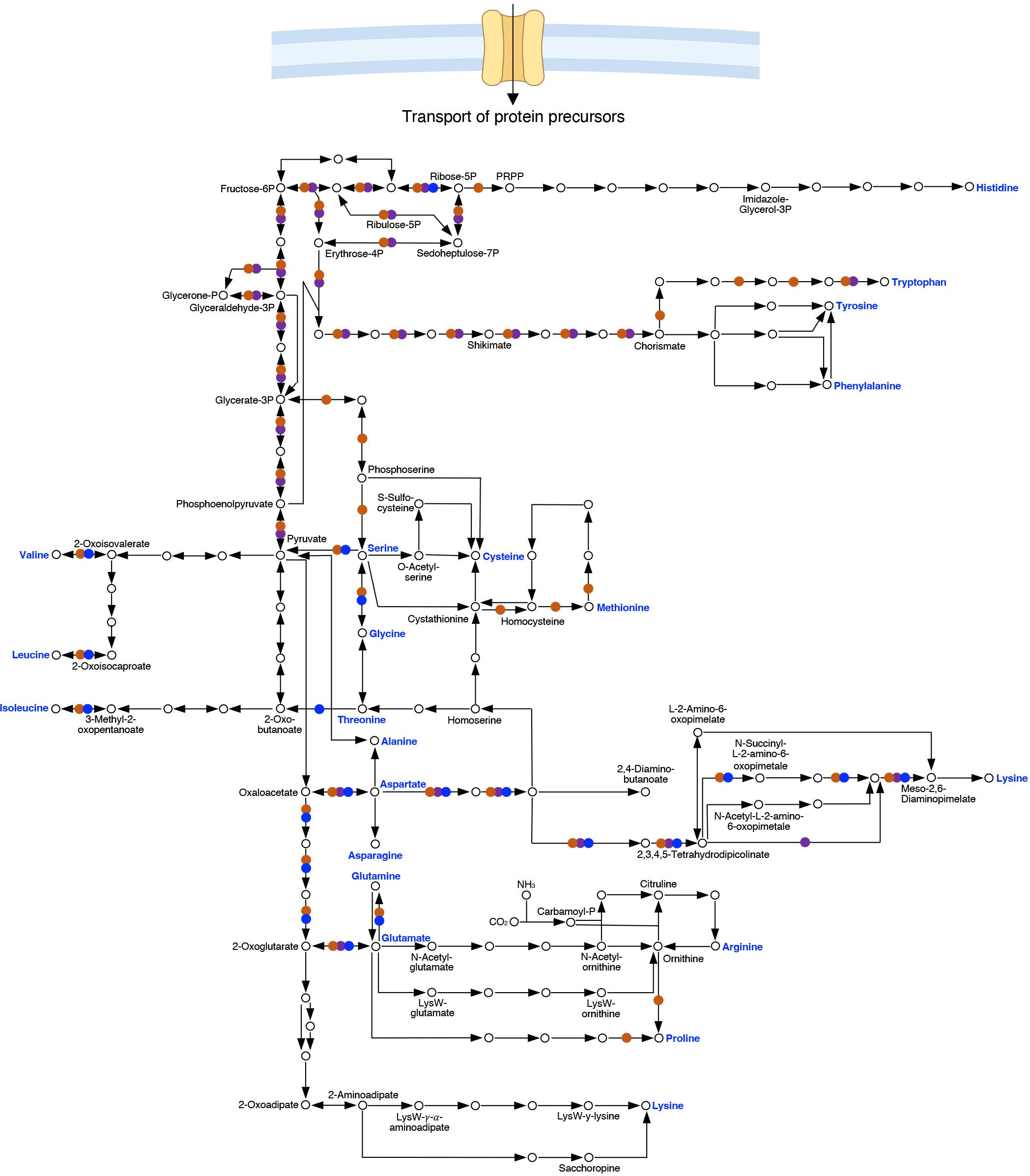
Figure 3 Comparison of pathways for biosynthesis of amino acids in Coxiella, Chlamydia and Rickettsia. Genome sequence analysis shows that the pathways for amino acid biosynthesis in C. burnetii (RSA493) (●), C. trachomatis (Bu/434) (●) and R. prowazekii (Madrid E) (●) are vastly different. Acquisition of protein precursors from the host cell, transporter specificities, and significance of such transport in virulence are largely unexplored areas of research for BOIPs.
As a phenotype, auxotrophy is not an absolute but rather entirely dependent on the availability of biosynthetic precursors and the ability of any organism to utilize such precursors. As an example, C. burnetii does not appear auxotrophic for either glutamate or glutamine in ACCM-D-based media (Sandoz et al., 2016), despite lack of glutamate synthase and thus a predicted inability to synthesize glutamate from glutamine. However, the expression of glutamate dehydrogenase (EC 1.4.1.2, reverse reaction) may fill the nutritional need for glutamate under conditions where the cell also produces α-ketoglutarate. Because protein synthesis largely takes place in the cytoplasm of eukaryotic cells, Rickettsia may experience less selective pressure to maintain pathways for amino acid synthesis than pathogens occupying replication vacuoles. Indeed, the pathways involved in amino acid synthesis reveals greater loss of biosynthetic capacity for R. prowazekii as compared to C. burnetii and C. trachomatis (Figure 3). Possibly as an adaptation to reduced capacity to synthesize protein precursors, BOIPs can show unique mechanisms to optimize utilization and recycling of amino acids. For example, in C. trachomatis, the Opp transporter has dual function as both an oligopeptide transporter serving the bacterium’s nutritional needs and at the same time functioning in peptidoglycan recycling (Singh et al., 2020).
BOIPs inhabit niches that are known to differ in terms of physicochemical characteristics. For example, while the host cell cytoplasm has a neutral pH, the C. burnetii CCV luminal pH is acidic (Grieshaber et al., 2002; Samanta et al., 2019). Additional physicochemical variables relevant to the physiology of BOIPs include osmolarity, relative concentrations of ions associated with nutrient transport, as well as availability of oxygen (O2), carbon dioxide (CO2), and temperature. A branched respiratory chain that includes cytochrome bd, a terminal oxidase associated with reduced oxygen availability (Borisov et al., 2021), motivated analysis of C. burnetii oxygen requirements (Omsland et al., 2009). Even under nutrient conditions permissive to growth, C. burnetii does not replicate optimally unless O2 tension is reduced to microaerobic levels and CO2 is available (Esquerra et al., 2017). Like C. burnetii, requirements for growth of other BOIPs is likely to be as dependent on specific physicochemical conditions as critical nutrient availability.
As illustrated by the ability of C. burnetii to replicate under relatively simple nutrient conditions where amino acids and citrate (note: citrate is not required for growth) serve as the only macro-nutrients (Esquerra et al., 2017), C. burnetii may not fall into the category of a nutritionally fastidious bacterium. However, a requirement for specific nutrients and physicochemical conditions (i.e., pH, O2 and CO2) for optimal growth reflects adaptation to a unique replicative niche. It should also be noted that some genogroups of C. burnetii show poor cultivability under standard axenic culture conditions (Kersh et al., 2016), suggesting that isolates within the genus are metabolically diverse. Natural diversity in cultivability within the Coxiella genus could originate from extensive genome rearrangements (Beare et al., 2009) that affect gene expression or be a result of specific genome content. The high level of conservation between genomes of C. trachomatis serovars suggests this pathogen may show more uniform nutrient/cultivation requirements, as compared to C. burnetii. Serovar-specific ability to synthesize tryptophan de novo is a well-characterized metabolic difference between pathogenic Chlamydia species (Caldwell et al., 2003; Bommana et al., 2021). Loss of tryptophan synthase activity in ocular serovars of C. trachomatis is linked to loss of metabolic fitness due to accumulation of ammonia in indole-deficient environments (Sherchand and Aiyar, 2019).
C. muridarum has been shown to replicate in host cells maintained in the absence of oxygen, albeit at a slower rate than in host cells incubated under normoxic conditions (Sigar et al., 2020). A similar analysis showed that while C. pneumoniae replication is enhanced under microaerobic conditions, C. trachomatis is not affected (Juul et al., 2007). Analysis of C. trachomatis metabolic activity under axenic conditions showed enhanced activity over time under microaerobic conditions (2.5% O2) as compared to normoxic conditions (20% O2) for both the EB and RB cell forms (Omsland et al., 2012). It should be noted that C. trachomatis, C. pneumoniae, and C. muridarum all carry cydAB, which encodes cytochrome bd, a terminal oxidase typically used by bacteria under reduced oxygen availability (Borisov et al., 2021). Overall, these data point to clear effects of O2 tension on chlamydial metabolic activity and a positive effect of microaerobic oxygen availability. Similar to C. burnetii and C. trachomatis, cytochrome bd is also encoded by the R. prowazekii genome. In E. coli, expression of respiratory chain components is generally regulated by the FNR and ArcAB regulatory systems (Cotter et al., 1997; Levanon et al., 2005). The C. burnetii genome does not encode orthologs of these genes. It is possible that C. burnetii’s obligate intracellular lifestyle and adaptation to the specific conditions of the CCV has reduced the requirement for regulation and that C. burnetii responds by a more general upregulation of gene expression once within its replicative niche. Enhancement of C. psittaci axenic activity in the presence of CO2 (Weiss and Wilson, 1969) suggests that other species in the genus may also need CO2 for optimal activity and eventual axenic replication, as observed for C. burnetii (Esquerra et al., 2017).
Coxiella and Chlamydia are typically cultured at 37°C, generally consistent with their tissue tropisms in mammals. However, Chlamydia species show species and isolate-specific temperature preferences (Rota and Nichols, 1973; Janik et al., 2014). Additionally, under axenic conditions, C. burnetii also replicates efficiently at 27°C, albeit with an extended lag phase (Esquerra et al., 2017), potentially reflective of the organism’s ability to colonize ticks. Unlike Coxiella and Chlamydia, Rickettsia species are maintained between 28-37°C during infection of cultured cells, depending on the host cell type (e.g., mammalian vs arthropod) and pathogen species (Tello-Martin et al., 2018). For R. prowazekii, culture below 37°C does not necessarily correlate with optimal replication or the temperature of naturally infected tissue but rather a compromise between pathogen replication and maintenance of the infected host cells during infection (Pinkerton and Hass, 1932a, 1932b). Interestingly, temperature (25 vs 37°C) does not have a marked effect on gene expression in R. rickettsii (Ellison et al., 2009), although some effect of temperature has been noted for virulence-related genes (Galletti et al., 2016). Moreover, viability of R. rickettsii is better maintained at 25°C compared to 34°C during culture in Vero cells (Ellison et al., 2009), a finding potentially affected by host cell viability following infection at 25°C vs 34°C. A negative effect of elevated temperature on viability suggests that temperature could have a significant impact on the ability to culture Rickettsia species under axenic conditions. The effect of temperature on growth, virulence factor expression, and viability is likely regulated via a range of mechanisms including DNA and mRNA structure, thermo-sensitive changes in protein structure, transcription factors, and chaperone proteins (Lam et al., 2014).
Micronutrients such as transition metals and vitamins are critical for normal metabolic functions in bacteria. For BOIPs, micronutrient acquisition is in part shaped by genome streamlining. Without biosynthetic pathways to synthesize NAD+ de novo, C. trachomatis has evolved substrate promiscuity in the ATP/ADP translocase Npt1Ct to acquire NAD+ from the host (Fisher et al., 2013). Similarly, novel enzymes, including isoforms with less restrictive substrate specificity, have been identified as components of the pathway for tetrahydrofolate (an active form of vitamin B9) biosynthesis in C. trachomatis (Adams et al., 2014).
Iron is a critical micronutrient known to influence the expression of dozens of genes in free-living or facultative intracellular bacterial pathogens. Analysis in C. burnetii (Sanchez and Omsland, 2020) and C. trachomatis (Pokorzynski et al., 2019) have highlighted unique aspects or iron acquisition and metabolism in BOIPs. C. trachomatis utilizes the iron-dependent transcriptional regulator YtgR to integrate responses to iron starvation and tryptophan biosynthesis (Pokorzynski et al., 2019). In C. burnetii, apparent reliance on uptake of molecular iron via the Fe2+-specific FeoAB transporter has been linked to release of molecular iron from iron-containing molecules within the acidic microenvironment of the CCV (Sanchez and Omsland, 2020). C. burnetii cannot utilize heme as a source of iron but rather relies on de novo heme biosynthesis (Moses et al., 2017) in combination with acquisition of molecular iron (Moses et al., 2017; Sanchez and Omsland, 2020).
Responses to iron limitation also serve to illustrate a likely characteristic aspect of niche adaptation common to many BOIPs. While limiting iron access to R. rickettsii during infection of Vero cells using deferoxamine mesylate results in inhibition of growth, only 5 genes showed ≥ 3-fold differential expression in response to iron limitation (Ellison et al., 2009). Thus, R. rickettsii transcriptional responses to iron limitation appear similar to that observed in C. trachomatis whose response to iron limitation during (mid-cycle) intracellular growth following 6 h treatment with the chelator 2,2’-bipyridyl only involve 12 genes (Brinkworth et al., 2018). Complementary genome-wide transcriptional profiling to iron limitation is not available for C. burnetii, but combined bioinformatic and biochemical analysis of C. burnetii transcriptional responses to iron suggests the involvement of a limited number of genes in iron-induced stress responses (Briggs et al., 2008). As suggested for R. rickettsii (Ellison et al., 2009), BOIP responses to iron may reflect the parasites’ residence in relatively stable intracellular niches with limited requirements to respond to physiologically significant changes in iron availability. This stands in stark contrast to BOIP responses to other nutritional cues, including IFNγ-induced tryptophan starvation in C. pneumoniae where transcriptional activity is globally upregulated (Ouellette et al., 2006).
Analysis of C. burnetii growth in chemically defined media combined with metabolic pathway reconstruction suggest C. burnetii can synthesize most vitamins (Seshadri et al., 2003; Esquerra et al., 2017). Interestingly, C. burnetii has two orthologous bioC (Moses et al., 2017), allowing the initial step in biotin (vitamin B7) production. Because chemical inhibition of biotin synthesis inhibits C. burnetii replication, de novo biotin biosynthesis is likely critical in C. burnetii. Bacteria in the Rickettsia genus are unable to synthesize several cofactors and B vitamins de novo (Driscoll et al., 2017), whereas members of the Chlamydia genus are predicted to exhibit species-specific capacity for biotin synthesis (Voigt et al., 2012). Although C. trachomatis and C. psittaci can synthesize folates, strains have different capacities to scavenge folates from the host cell (Fan et al., 1992). As postulated for Rickettsia (Driscoll et al., 2017), deficiencies in cofactor and vitamin biosynthesis likely contribute to the parasitic nature of BOIPs.
BOIPs show remarkable diversity regarding their host cell interactions. Some aspects of these interactions are shaped by the interplay between pathogen metabolic capacity and continued pathoadaptation of BOIPs to their intracellular niches (Fuchs et al., 2012; Eisenreich et al., 2017). The concept of nutritional virulence (Kwaik and Bumann, 2013) frames nutrient acquisition by intracellular pathogens in terms of pathogen virulence mechanisms. For BOIPs, strategies for nutrient acquisition are interconnected with niche adaptation and genome streamlining. Advances in genetic manipulation of some BOIPs has allowed for the identification of specific genes with roles in nutrient acquisition. The molecular mechanisms for how Coxiella (Voth and Heinzen, 2007; Schaik et al., 2013; Kohler and Roy, 2015; Larson et al., 2016), Chlamydia (Elwell et al., 2016; Fischer and Rudel, 2018; Rother et al., 2019; Triboulet and Subtil, 2019), and Rickettsia (Driscoll et al., 2017; McGinn and Lamason, 2021; Voss and Rahman, 2021) interact with their respective host cells during infection has been discussed elsewhere. While this review primarily focuses on nutrient utilization and responses to nutrient limitation, recent findings regarding strategies for nutrient acquisition by BOIPs during infection warrant some discussion.
Nutritional virulence encompasses bacterial virulence strategies that target host processes to enhance pathogen access to nutrients. Because factors other than access to nutrients impact replication of BOIPs, especially those that reside in replication vacuoles, it can be challenging to distinguish the significance of underlying mechanisms. For example, in C. burnetii, several effectors are critical for normal CCV biogenesis and thus intracellular replication (Larson et al., 2013; Newton et al., 2014; Crabill et al., 2018) without necessarily affecting pathogen access to nutrients. Also, some genes, including CBU2028, are specifically linked to CCV biogenesis and not pathogen intracellular replication (Crabill et al., 2018), demonstrating that CCV size and pathogen intracellular replication are not necessarily linked. Recruitment of autophagic vesicles to the CCV may have more significance for recruitment of membranes to aid CCV expansion than pathogen replication, despite some evidence pointing to a direct effect on C. burnetii replication (Pareja et al., 2017). This may also hold true for C. trachomatis, which grows equally well in autophagy competent or incompetent mouse embryonic fibroblasts suggesting autophagy is not a critical host process for chlamydial growth (Ouellette et al., 2011). Comparison between C. trachomatis and C. pneumoniae has shown that these two species differ in their strategies to obtain protein precursors from the host cells. C. pneumoniae is primarily reliant on lysosome-derived peptides while C. trachomatis shows a preference for cytosolic amino acids (Ouellette et al., 2011). Moreover, the mechanisms by which Chlamydia obtains protein precursors from host cells can change throughout the developmental cycle. Chlamydial downregulation of host p53, a negative regulator of the PPP enzyme glucose-6P dehydrogenase, serves to enhance PPP activity in Chlamydia-infected host cells (Siegl et al., 2014). The resulting pathogen-dependent stimulation of host metabolism may promote a nutritionally favorable environment for the pathogen (Rother et al., 2019). Infection of mouse oviduct epithelial cells by C. muridarum results in upregulation of hexokinase II (George et al., 2016), consistent with pathogen-dependent stimulation of (host) production of both glucose-6P and ATP, the latter a result of glucose-6P oxidation.
The unfolded protein response (UPR) is a reaction to endoplasmic reticulum (ER) associated stress in eukaryotic cells (Celli and Tsolis, 2014). Some pathogens exploit the UPR and downstream ER-associated degradation (ERAD) of unfolded proteins as a source of amino acids. In C. burnetii-infected THP-1 macrophages, inhibition of ER stress by tauroursodeoxycholic acid reduces expansion of the CCV but does not affect pathogen replication (Brann et al., 2020), suggesting the UPR and ERAD are not critical for C. burnetii to access protein precursors. Nevertheless, normal CCV expansion and C. burnetii replication depend on activity of the UPR-related translation initiation factor eIF2α, the phosphorylation of which is reduced upon infection with bacteria unable to secrete type IVB secretion system effectors (Brann et al., 2020). Because phosphorylation of eIF2α enhances autophagy (Kouroku et al., 2006; Humeau et al., 2020), C. burnetii manipulation of UPR-related signaling may enhance pathogen recruitment of autophagosomes to the CCV and aid replication.
C. burnetii is critically dependent on moderately acidic pH for optimal transport and metabolism of specific metabolites (Hackstadt and Williams, 1981a) and replication (Esquerra et al., 2017). The discovery that C. burnetii actively manipulates the host cell to maintain CCV luminal pH to prevent activity of hydrolytic enzymes (Samanta et al., 2019) suggests C. burnetii is essentially replicating at a pH during intracellular replication that is sub-optimal for pathogen metabolism. The overall high metabolic plasticity of C. burnetii, illustrated by the ability to replicate in axenic medium composed largely of amino acids (Esquerra et al., 2017), may be important for C. burnetii to balance metabolic fitness within a CCV that is manipulated to maintain a sub-optimally high pH for pathogen metabolic activity.
Building on the discovery that EBs and RBs primarily utilize different mechanisms to obtain energy for protein synthesis (i.e., scavenging of host-derived ATP by the RB versus oxidation of glucose-6P by the EB), Grieshaber and colleagues showed that the chlamydial EB can use chemically diverse molecules, including amino acids and ATP, to maintain infectivity (Grieshaber et al., 2018). Although EBs might prefer glucose-6P to support protein synthesis (Omsland et al., 2012), the ability of EBs to respond to ATP suggests Chlamydia-dependent release of ATP from host cells (Pettengill et al., 2012; Yang et al., 2021) is a mechanism to enhance pathogen infectivity. As such, release of ATP by infected cells could have an impact on dissemination and disease progression.
Mechanisms of nutritional virulence likely differ between pathogens that replicate in the host cell cytosol and those that establish replication vacuoles (e.g., transporter requirements). Comparative analyses aimed at understanding differences in strategies employed by BOIPs for nutrient acquisition within specific intracellular niches will be important to understand mechanisms of pathoadaptation among these unique bacterial pathogens.
The virulence of bacterial pathogens is conferred by a combination of essential and subtle virulence determinants. For example, in Coxiella, secretion of effector molecules via a type IVB secretion system (Chen et al., 2010; Carey et al., 2011; Beare et al., 2012; Larson et al., 2016) is critical for establishment of the pathogen’s replication vacuole. That metabolic capacity can also affect pathogen virulence is well established; however, identifying the specific mechanism(s) for how a metabolic defect affects virulence can be challenging.
All BOIPs are associated with highly specific replication niches. The large diversity in apparent metabolic capacity (as deduced from genome sequence analysis) among BOIPs suggest that the various niches occupied by BOIPs represent vastly different nutritional environments. BOIPs of the genera Coxiella, Chlamydia, and Rickettsia are all to some degree amphotropic—capable of infecting different host organisms and host cell types—in nature. For example, C. burnetii has been shown to naturally colonize a wide range of animals [e.g., birds, cows, goats, and ticks (Canevari et al., 2018; Tokarevich et al., 2018; Tomaiuolo et al., 2020; Turcotte et al., 2021)] in addition to various tissues within infected animals (Roest et al., 2012; Gregory et al., 2019). Direct comparative analysis of the ability of different serovars of C. trachomatis to infect different host cell types indicates that the invasive C. trachomatis serovar L2, causing lymphogranuloma venereum, is moderately amphotropic compared to serovars A and D (Faris et al., 2019).
Transposon mutant libraries of C. burnetii reveal genes encoding metabolic functions whose disruption affect infection and/or intracellular replication (Martinez et al., 2014; Newton et al., 2014). Several genes have functions related to processes discussed in this review, including CO2 metabolism (carbonic anhydrase, CBU0139), respiration (cytochrome c oxidase, CBU1038-1040), glycolysis (glucose-6P isomerase, CBU0848), and metabolism of phosphorylated glucose (UTP-glucose-1-phosphate uridylyltransferase, CBU0849). Moreover, metabolic plasticity conferred by pckA has revealed specific virulence defects during C. burnetii intracellular replication in certain host cell types (Sanchez et al., 2021).
C. burnetii shows isolate-specific plasmid carriage (Long et al., 2019). Luo and colleagues recently identified a role for the QpH1 plasmid of C. burnetii during colonization of murine bone marrow-derived macrophages (Luo et al., 2021). It is not clear whether the role of the QpH1 plasmid in C. burnetii host colonization relates to virulence factor secretion or pathogen metabolic functions. Nevertheless, diversity in plasmid carriage among C. burnetii isolates may influence C. burnetii amphotropism based on potential significance of specific plasmid-associated genes in C. burnetii metabolic activities. Because genome architecture can influence gene expression, extensive differences in genome organization, including sequences containing metabolic genes, among C. burnetii isolates (Beare et al., 2009) combined with isolate-specific disease characteristics is consistent with a role of genome architecture in pathogen metabolism. We expect that research over the next decade will continue to reveal greater significance of metabolic functions in BOIP virulence and begin to provide mechanistic information related to a range of metabolic virulence determinants.
Although use of host cell-free culture is an invaluable tool to physiologically separate BOIPs from their host cells, ultimately, the physiology of BOIPs is intertwined with that of the host cell. As such, analysis of metabolic and physiological characteristics of BOIPs will require verification of biological relevance using (in vivo) animal or (ex vivo) cell culture models.
For any ex vivo analysis of BOIPs, the nutrient composition of cell culture media may affect, and in some cases compromise, the utility of cell culture models to resolve phenotypes of mutants. For example, medium nutrient composition can result in accidental chemical rescue of a genetic defect. To explore how the cell culture medium can impact pathogen fitness during infection of different host cell types, a “physiological medium” (Cantor, 2019) based on the nutrient composition of interstitial fluid was designed for use with C. burnetii as the model BOIP (Sanchez et al., 2021). Use of Interstitial Fluid-modeled Medium, IFmM, proved to affect C. burnetii fitness in some but not all cell culture models. Pathogen metabolic capacity, as assessed by comparing strains capable of undergoing gluconeogenesis or not, also showed host cell type-dependent differences (Sanchez et al., 2021). Overall, this underscores the significance of the interplay between pathogen metabolic capacity and the nutritional context of the host cell.
In the study of Chlamydia species, the protein synthesis inhibitor cycloheximide is often used to promote pathogen replication by suppressing host cell protein synthesis (Alexander, 1968), overall enhancing bacterial replication by reducing host cell competition for nutrients. Notably, cycloheximide results in (host) carbon source-dependent effects on the generation of C. trachomatis EBs (Iliffe-Lee and McClarty, 2000). Because use of cycloheximide promotes chlamydial fitness during interaction with a host cell, analysis of chlamydial physiology, especially with strains that have metabolic defects, may be best done using cell culture models maintained in the absence of cycloheximide.
Any microbial pathogen must be able to respond to cues from the host to optimize its virulence potential. This may be most obvious in the context of responding to nutrient availability and physicochemical conditions such as pH and temperature. As suggested for C. burnetii (Esquerra et al., 2017), the integration of several distinct physicochemical and nutritional signals could serve as a mechanism for this organism to identify a suitable replicative niche. Similarly, Chlamydia and Rickettsia species may sense availability of ATP, a molecule largely limited to metabolically active cells, as a signal that environments suitable for growth have been encountered.
Less obvious, although the phenomenon has been known for decades (Morse and Fitzgerald, 1974; Hughes and Sperandio, 2008), is the ability of bacteria to sense and respond to mammalian (steroid) hormones, molecules unique to the host organism they infect. Unlike molecular nutrients such as glucose and amino acids that are maintained at relatively steady levels within eukaryotic host cells, hormone levels typically fluctuate, thus reflecting altered physiological states of the host organism. C. burnetii, a bacterium known to colonize placental tissue, has been shown to establish infection in cultured placental trophoblasts (Amara et al., 2010; Howard and Omsland, 2020), a major source of the steroid hormone progesterone during pregnancy. In ovariectomized mice, C. burnetii shows elevated loads in spleen and liver tissue (Leone et al., 2004), suggesting C. burnetii responses to mammalian steroid hormones has clinical significance. Data from host cell-free analysis of C. burnetii responses to progesterone suggest that this hormone has a direct inhibitory effect on C. burnetii activity (Howard and Omsland, 2020). However, in the context of animal infection and considering hormonal effects on the immune system (Bereshchenko et al., 2018; Patt et al., 2018), it is likely that steroid hormones also have indirect effects on pathogen activity and pathogenicity. Thus, while progesterone may directly inhibit C. burnetii activity, hormonal effects on host immunity may significantly confound how bacteria respond to mammalian hormones. Similar to C. burnetii, C. abortus shows tropism for placental tissue (Essig and Longbottom, 2015) and may also respond to host-derived hormones or be affected by hormone-associated signaling during colonization of placental tissue.
The physiology of BOIPs is interconnected with their metabolic capacity and plasticity. Some BOIPs, including Coxiella, Anaplasma, Ehrlichia and Chlamydia, undergo morphological transitions during their life cycles, further complicating the intricate relationships they maintain with their respective host cells. The developmental cycles of C. burnetii and C. trachomatis with leading ideas for underlying mechanisms are illustrated in Figure 4. Coleman and colleagues (Coleman et al., 2004) connected the LCV form of C. burnetii to this pathogen’s replicative phase while the SCV was shown to be a non-replicative stationary phase form. Current understanding of the physiological role of the C. burnetii SCV is limited but certain spore-like characteristics are consistent with environmental stability. In cell culture, both the C. burnetii SCV and LCV forms are infectious.
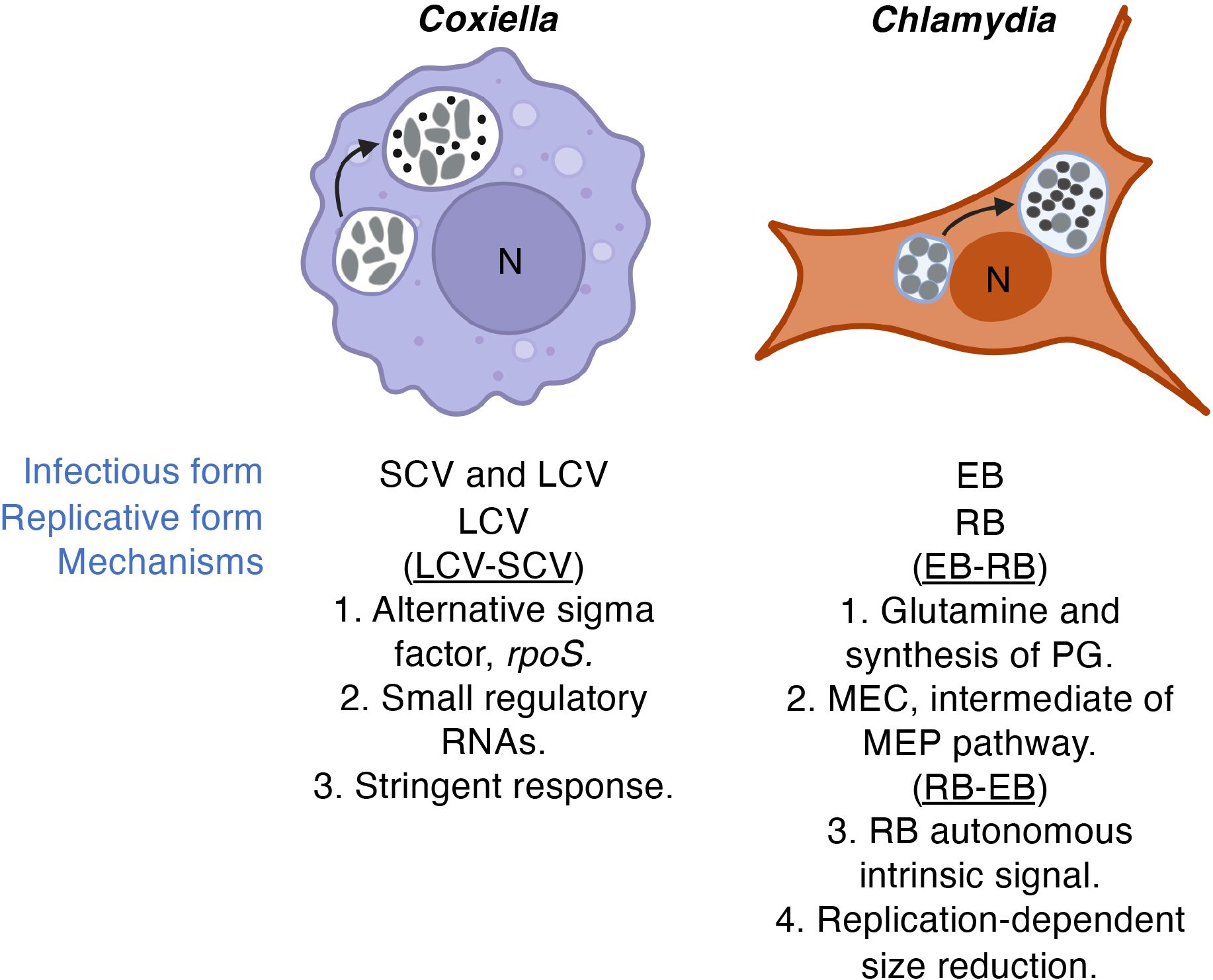
Figure 4 Developmental transitions. While recent advances based on molecular genetics are starting to shed light on the mechanism(s) of developmental transitions in Coxiella and Chlamydia, the genes and regulatory networks involved remain elusive. Leading ideas for mechanisms underlying developmental transitions are highlighted. LCVs and RBs vs SCVs and EBs are indicated by larger gray cells or smaller dark calls, respectively. Infectious form relates to infectivity of cultured cells. The direction of the developmental cycle is indicated in parentheses. "PG" and "MEC" denote peptidoglycan and 2-C-methylerythritol 2,4-cyclodiphosphate, respectively.
The physiological basis for developmental transitions in Chlamydia species appear distinct from those of C. burnetii. For example, because only the chlamydial EB form is infectious, re-generation of the chlamydial EB is critical for new rounds of infection. Despite lack of cell division by the chlamydial EB, in C. trachomatis, the EB continues to synthesize protein during infection of cultured cells (Grieshaber et al., 2018), consistent with a requirement for continued “maintenance metabolism” after RB-EB differentiation is completed.
The question of what triggers transition of the replicative LCV and RB forms of C. burnetii and C. trachomatis to their respective non-replicative SCV and EB forms remains largely unanswered. That nutrient availability can control morphological transitions in BOIPs is supported by the mechanism of developmental transitions in the intracellular bacterium Legionella pneumophila, a phylogenetic relative of C. burnetii. In L. pneumophila, the gene phtA, encoding a major facilitator superfamily transporter, influences developmental transitions in response to threonine availability (Sauer et al., 2005; Fonseca and Swanson, 2014). In part relying on axenic techniques, Rajeve et al. have produced promising data regarding a potential trigger for EB-RB development in C. trachomatis (Rajeeve et al., 2020). This group reported an interconnection between metabolism of glutamine and synthesis of peptidoglycan required for cell division in C. trachomatis (Liechti et al., 2016), establishing a critical role for glutamine in EB germination. Moreover, 2-C-methylerythritol 2,4-cyclodiphosphate (MEC), an intermediary metabolite of the methylerythritol phosphate (MEP) pathway of isoprenoid synthesis, stimulates dissociation of the chlamydial histone-like protein Hc1 from DNA thus promoting chromatin decondensation (Grieshaber et al., 2004). Because the initial step of the MEP pathway involves pyruvate and glyceraldehyde 3-phosphate (G3P), both intermediates of the central metabolic machinery, activation of central metabolism to generate pyruvate and G3P could serve as a metabolic trigger for decondensation of the EB nucleoid, a critical step in EB-RB development. To further elucidate the driving force behind C. trachomatis RB-EB transitions, Chiarelli et al. tested if signals extrinsic (i.e., extracellular) or intrinsic (i.e., intra-bacterial) to C. trachomatis are likely to be responsible for triggering RB-EB developmental transitions using mathematical models. Interestingly, they identified the triggering signal(s) to be a cell-autonomous intrinsic signal and thus not likely to be an environmental signal such as a nutrient (Chiarelli et al., 2020). Lee and colleagues established that replication-dependent size reductions of the RB controls the timing of RB-EB conversion, also in the absence of an external signal (Lee et al., 2018). It is possible that signals and mechanisms governing EB-RB versus RB-EB transitions are different in nature.
Numerous genes were implicated in developmental transitions of C. trachomatis in experiments conducted on temperature sensitive mutants (Brothwell et al., 2016). The influence of several genes in chlamydial morphological differentiation suggests redundancy in regulation. Similarly, potential involvement of small RNAs (Warrier et al., 2014) and the recently discovered significance of the alternative sigma factor RpoS in C. burnetii developmental transitions (Moormeier et al., 2019) also suggest the involvement of a larger number of genes and regulatory redundancy in the process. Regardless of what the nature of the trigger(s) for developmental transitions in (certain) BOIPs may be, understanding the makeup of the response network that allows for the orchestrated change in gene expression required to transform one cell form into another is a focus of current research.
The stringent response (SR) was identified as a regulatory mechanism that interconnects amino acid availability with RNA synthesis by providing “stringent” control of RNA synthesis under conditions of amino acid limitation (Cashel, 1969; Cashel and Gallant, 1969). The signaling nucleotide (p)ppGpp, synthesized by proteins of the RelA/SpoT homolog (RSH) family (Hauryliuk et al., 2015), and a range of co-regulatory proteins (Steinchen et al., 2020), make up the SR. In L. pneumophila, the response network involved with signaling amino acid availability via PhtA may be the SR (Sauer et al., 2005). Both phtA (Sauer et al., 2005) and an apparently intact SR network is found in C. burnetii, suggesting amino acid availability may also be directly involved in developmental transitions in this organism.
Analysis of the C. trachomatis genome suggested absence of a SR based on lack of enzymes for synthesis of (p)ppGpp (Ouellette et al., 2006). While genes for ppGpp synthesis are absent, chlamydial genomes do encode DksA, a SR-related transcriptional regulator known to work in concert with (p)ppGpp (Paul et al., 2004; Magnusson et al., 2007; Ross et al., 2016). A gene encoding GreA, a protein that in E. coli can functionally substitute for DksA (Vinella et al., 2012), has also been annotated in C. trachomatis, but the gene is larger than the E. coli ortholog and sequence homology is minimal. Overexpression of DksA in C. trachomatis has been shown to reduce generation of infectious EBs (Mandel et al., 2021). Curiously, the chlamydial dksA ortholog does not functionally complement the growth defect observed in E. coli during culture in a nutritionally minimal medium (Mandel et al., 2021), suggesting remnants of the SR machinery in C. trachomatis have evolved to acquire unique functions in this pathogen. Transcriptional analysis of Chlamydia under amino acid starvation has also revealed an apparent uncoupling of amino acid availability and transcriptional activity (Ouellette et al., 2006), again consistent with absence of “stringent” regulation of RNA synthesis. Ouellette and colleagues built on these findings and investigated the idea that chlamydial transcription during IFNγ-induced tryptophan limitation could be controlled via the density of tryptophan codons (Ouellette et al., 2016). Though it is unclear how codon content for an amino acid would regulate transcription, especially when the density of tryptophan codons does not absolutely correlate with transcript levels, the regulatory scheme points to chlamydial adaptation to amino acid starvation in the absence of a SR.
Direct comparison of transcriptional profiles of C. trachomatis cultivated in the presence or absence of IFNγ identified numerous differentially expressed genes (Belland et al., 2003a), consistent with chlamydial adaptation to an environment where an orchestrated response to tryptophan limitation is necessary for optimal fitness. In addition to the impact host-dependent regulation (i.e., synthesis vs IFNγ-dependent breakdown) of tryptophan can have on C. trachomatis, the host microbiota may also affect the Chlamydia-host interaction. Specifically, indole-producing species of the genus Prevotella, associated with bacterial vaginosis, can promote replication of C. trachomatis, thus potentially counteracting the effect of IFNγ-dependent control of tryptophan availability (Ziklo et al., 2016). Like C. trachomatis, intracellular replication of C. burnetii is also restricted by treatment of host cells with IFNγ via indolamine 2,3-dioxygenase 1-mediated breakdown of tryptophan (Ganesan and Roy, 2019). Interestingly, the inhibitory effect of IFNγ on the growth of R. prowazekii in human fibroblasts could not be rescued by addition of tryptophan to the growth medium, suggesting the involvement of alternative IFNγ-mediated inhibitory mechanisms (Turco and Winkler, 1986).
In the (facultative) intracellular pathogen L. pneumophila, the functional significance of (p)ppGpp and DksA have been demonstrated for pathogen differentiation between cell forms (Dalebroux et al., 2010). Given both phylogenetic and physiological relatedness, including morphological transitions, C. burnetii is also likely to rely on a functional SR machinery for normal physiological function. Although reliance on a SR by bacteria of the Rickettsia genus for the purpose of responding to amino acid availability has not been tested experimentally, genetic complementation of a truncated relA/spoT gene in a strain of R. rickettsii Iowa that produces lytic/clear plaques, restored the non-lytic/opaque phenotype of the plaques (Clark et al., 2011). This demonstrates the effect of relA/spoT expression in R. rickettsii Iowa and indicates the function of a response network that includes (p)ppGpp in Rickettsia. Interestingly, given the effect of (p)ppGpp levels on gene expression in free-living model organisms including E. coli (Sanchez-Vazquez et al., 2019), relA/spoT competence did not affect gene expression in R. rickettsii Iowa (late log phase) (Clark et al., 2011). The gene(s) annotated as relA/spoT in Rickettsia are shorter than the typical relA and spoT genes described in other organisms but could still encode functional (p)ppGpp synthesizing proteins because the synthase domain only accounts for a smaller part of the proteins (Andersson et al., 1998; McLeod et al., 2004; Clark et al., 2011).
The functional significance of the SR goes far beyond responses to amino acid limitation (Boutte and Crosson, 2013), the stressor first identified as a trigger for this response network (Borek, 1956; Cashel, 1969). Thus, molecular remnants of the SR in BOIPs, including Chlamydia that appear to encode a severely reduced system, may be responsive to stressors other than amino acids. Importantly, the retention of SR-related genes including dksA and greA in BOIPs could be driven by a role for these genes in processes other than those associated with the SR. For example, deletion of dksA increases accumulation of drug-induced double-stranded DNA breaks (Sivaramakrishnan et al., 2017), and both DksA and GreA/B can help resolve replication and transcription conflicts (Tehranchi et al., 2010; Satory et al., 2015). GreA has additionally been shown to serve as a chaperone (Li et al., 2012). Related yet unique functions of DksA and GreA/B is consistent with diverse selective pressures to retain a combination of SR-related genes in BOIPs. Suppression of the growth defect produced by (p)ppGpp deficiency by overexpression of dksA and greA (Vinella et al., 2012) illustrate their potential functions in the absence of a (p)ppGpp-driven SR. A comparison of SR-related genes in Coxiella, Chlamydia and Rickettsia is shown in Table 2. Because the genomes of these organisms all contain at least some components of what may be described as the canonical machinery consisting of relA, spoT, dksA and greA, unique adaptation of specific components of the network appears likely. As indicated, the predicted GreA protein of C. trachomatis is unusually large (CTL0004; 715 amino acids) compared to that of E. coli (b3181; 158 amino acids), suggesting the C. trachomatis protein is not functionally comparable to the E. coli protein.
Analysis of greA/greB/dksA triple mutants in a ppGpp0 strain of E. coli showed recovery of growth on minimal medium (not containing amino acids) upon complementation with either greA or dksA (Vinella et al., 2012). The redundancy demonstrated for the SR in E. coli, as this relates to response to amino acid starvation, hints at the molecular composition of a minimal “SR” in BOIPs, or other bacteria with reduced genomes. In addition to prototypical response systems for nutrient sensing, bacteria with a streamlined genome may well utilize expression of e.g., transporters as an “indirect” way of regulating physiological responses to nutrient availability.
Over the past decade, analysis of molecular mechanisms of C. burnetii biology has been transformed with the combined access to a full complement of tools for genetic manipulation and axenic culture, allowing isolation of mutants with defects in genes needed for host cell infection and/or intracellular replication. Given recent advances in similar research tools for several other BOIPs, the next decade is likely to change the study of BOIPs. Some areas of research may be especially important and scientifically fruitful. Figure 5 illustrates the interrelatedness between the research areas described below.
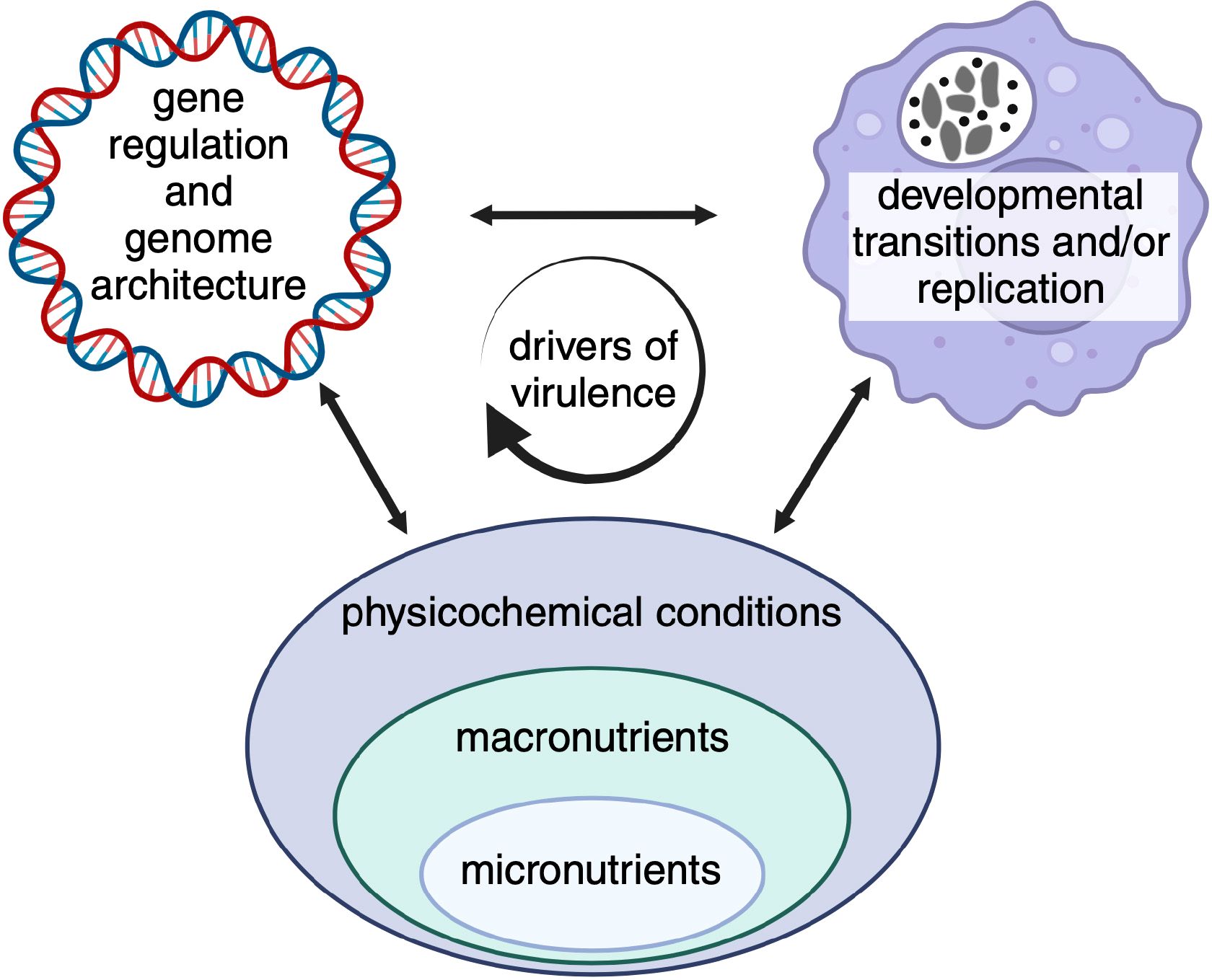
Figure 5 Opportunities for discovery. Certain areas of research appear especially relevant given recent technical and/or scientific advances. While these areas may be scientifically distinct, regulatory mechanisms are biologically intertwined and expected to influence pathogen virulence characteristics. Indicated micronutrients include, trace metals, vitamins and co-factors, macronutrients carbon sources and amino acids, and physicochemical conditions pH, oxygen and carbon dioxide.
Development of more sensitive techniques (e.g., RNA-Seq) for analysis of transcriptional responses have resulted in several studies of BOIP gene expression during host interactions. Regarding metabolic responses, metabolomic analyses of BOIPs have revealed patterns and complexities in nutrient utilization (Häuslein et al., 2017). An underexplored area and natural next step in understanding the physiology of BOIPs is combined omics analyses aimed at correlating gene and/or protein expression with the flux of metabolic intermediates during the pathogens’ life cycle. This is especially relevant for organisms that undergo developmental transitions between cells forms as data based on both gene expression and metabolic activities can identify signaling networks involved in regulating such transitions. C. burnetii would be a great model for analysis given the transition of this organism between cell forms under axenic conditions (Sandoz et al., 2014; Esquerra et al., 2017), likely also required to obtain sufficient material for optimal analysis.
Metabolic interactions between BOIPs and host cells also extends to pathogen-dependent manipulation of host transcription factors that regulate host cell metabolism. For example, C. burnetii affects the stability of the transcription factor HIF1α as well as HIF1α-regulated target genes involved in metabolism (Hayek et al., 2022). A recent analysis of host cell chromatin structure during infection with C. trachomatis (Hayward et al., 2020) provides a general view of how this pathogen impacts the host response to infection, including at the metabolic level. As discussed (Czyż et al., 2014; Rother et al., 2019), a thorough understanding of how and why BOIPs depend on a host cell for replication can be exploited to design host-directed antibacterial therapies.
Genome sequencing efforts have produced critical information about the genomes of several species and/or isolates of BOIPs. Though BOIPs exemplify a fascinating range of apparent metabolic capacity and genome stability, how these features affect pathogen physiology is largely unexplored. For example, how do differences in genome architecture between C. burnetii isolates (Beare et al., 2009) affect metabolic capabilities and how might such differences influence metabolic fitness, amphotropism, and virulence? In Chlamydia, genome sequence analysis show that metabolic capacity can be both species and isolate/strain specific (Voigt et al., 2012). Analysis of virulence following genetic complementation of metabolic capacity, as illustrated by ectopic expression of glucose-6P dehydrogenase (Sanchez and Omsland, 2021) and catalase (Mertens and Samuel, 2012) in C. burnetii, is one way to assess the selective pressures driving genome streamlining in BOIPs. Understanding how metabolic capacity correlates with niche adaptation, restriction and host range seems attractive avenues for basic research.
The list of potential genes, regulatory networks and mechanisms by which BOIPs may sense and respond to nutrient availability is expansive. The requirement for moderately acidic pH in Coxiella nutrient transport and metabolic activation (Hackstadt and Williams, 1981a), and ATP scavenging in Chlamydia are among the few mechanisms that have been experimentally tested and independently verified for how BOIPs regulate their metabolism.
Coxiella, Chlamydia and Rickettsia exhibit vastly different replication rates, a physiological characteristic implicated in virulence. As illustrated by analysis of the slow growing pathogen Mycobacterium tuberculosis (Beste et al., 2009), numerous genes can influence replication rate in bacteria. Capacity to synthesize protein and thus ribosome content has been implicated in this process. In R. prowazekii, despite only having single copies of genes encoding 16S or 23S rRNA, ribosome content has been described as an unlikely bottleneck in R. prowazekii replication (Winkler, 1995). The biological basis for replication rate and potential significance in virulence should be directly testable via currently available research tools for at least some BOIPs.
Several BOIPs undergo developmental transitions between cell forms. In part driven by the ability to genetically manipulate the organisms, both Coxiella and Chlamydia are emerging as attractive models to study the genetic and biochemical basis for such transitions. While regulation of developmental transitions in C. burnetii is likely to align with findings in L. pneumophila, including a role for the SR (Dalebroux et al., 2010), lack of a SR in C. trachomatis (Ouellette et al., 2006) and uncertainty regarding how this organism integrates responses to nutrient availability with critical physiological processes (e.g., developmental transitions) offer opportunity for significant discovery.
Despite obvious physiological significance, the effect and metabolism of micronutrients is an understudied aspect of BOIP biology. Technical challenges in separating BOIPs from their host cells to understand the effect of micronutrients directly on the pathogens is a major reason for current knowledge gaps. Of micronutrients, the role of iron has received more attention than any other, likely aided by the availability of tools, including chelators, to control pathogen access to iron. With improvements in both genetic tractability and axenic culture, including the use of chemically defined media (for C. burnetii) (Sandoz et al., 2016; Esquerra et al., 2017), analysis of micronutrients beyond iron is becoming increasingly feasible.
Inability to physically separate (most) BOIPs from their host cells essentially prevents the analysis of pathogen responses to specific physicochemical and/or nutritional conditions. Not all isolates of C. burnetii can be cultured in the axenic medium ACCM-2 (Kersh et al., 2016). Understanding the metabolic basis for why C. burnetii isolates differ in culturability has significance both for overall understanding of metabolic capabilities and potential connections to virulence. Further exploration of how glutamine may serve as a trigger for EB-RB development and continued chlamydial replication (Rajeeve et al., 2020) may be a significant discovery on the path to designing axenic culture tools for Chlamydia. Axenic culture of BOIPs is especially important to allow isolation of mutants with defects in genes required for host cell invasion and/or intracellular replication, which would be selected against in systems relying on host cells for isolation and propagation. Although genetic manipulation of BOIPs remains challenging, continued progress in this area is essential for discovery and innovation in the field (Suhan et al., 1996; Rachek et al., 1998; Felsheim et al., 2010; Burkhardt et al., 2011; Kari et al., 2011; Wang et al., 2011; Beare et al., 2012; Johnson and Fisher, 2013; Noriea et al., 2015; Larson et al., 2016; Sixt and Valdivia, 2016; McClure et al., 2017; Rahnama and Fields, 2018; Ouellette et al., 2021; Fields et al., 2022; Fu et al., 2022).
The following accession numbers were used to obtain genome sequence information for review of annotations or perform BLAST searches against relevant predicted proteins in E. coli K-12, substrain MG1655 (NC000913.3): C. burnetii RSA493 (NC002971.4), C. trachomatis L2/Bu434 (AM884176.1), C. pneumoniae J138 (NC002491.1), C. muridarum Nigg3 (NZCP009760.1), R. prowazekii Madrid E (NC000963.1), and R. rickettsii str. Sheila Smith (NC009882.1).
CM: Formal analysis, Visualization, Writing – original draft, Writing – review & editing. SS: Visualization, Writing – original draft, Writing – review & editing. CM: Visualization, Writing – original draft, Writing – review & editing. WP: Funding acquisition, Visualization, Writing – original draft, Writing – review & editing. AO: Conceptualization, Formal analysis, Funding acquisition, Resources, Supervision, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by NIH grants R21AI139992, R01AI130072, and R01AI155560 (AO), a Wellcome International Training Fellowship 220690/Z/20/Z (WP), and institutional funds from Washington State University.
While written with the intention of capturing original research and ideas from as many laboratories and individual scientists as possible, we recognize that numerous contributions have not been described at all or without the depth the work may deserve. We thank Dr. Viveka Vadyvaloo for review of the manuscript. Pathway maps were reproduced with permission from Kanehisa Laboratories, Japan. Illustrations were generated in part with BioRender.com.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
ACCM, acidified citrate cysteine medium; Arc, aerobic respiration control; BOIP, bacterial obligate intracellular parasite; CCV, Coxiella containing vacuole; EB, elementary body; ERAD, ER-associated degradation; FNR, defective in fumarate and nitrate reduction; HIF1, hypoxia-inducible factor; IFN, interferon; LCV, large cell variant; NTP, nucleotide triphosphate; PEPCK, phosphoenolpyruvate carboxykinase; PPP, pentose phosphate pathway; RB, reticulate body; SCV, small cell variant; SR, stringent response; TCA, tricarboxylic acid; UPR, unfolded protein response.
Abdad, M. Y., Abdallah, R. A., Fournier, P.-E., Stenos, J., Vasoo, S. (2018). A concise review of the epidemiology and diagnostics of rickettsioses: Rickettsia and Orientia spp. J. Clin. Microbiol. 56. doi: 10.1128/jcm.01728-17
Adams, N. E., Thiaville, J. J., Proestos, J., Juárez-Vázquez, A. L., McCoy, A. J., Barona-Gómez, F., et al. (2014). Promiscuous and adaptable enzymes fill “holes” in the tetrahydrofolate pathway in Chlamydia species. Mbio 5, e01378–e01314. doi: 10.1128/mbio.01378-14
Alexander, J. J. (1968). Separation of protein pynthesis in meningopneumonitis agent from that in L cells by differential susceptibility to cycloheximide. J. Bacteriol 95, 327–332. doi: 10.1128/jb.95.2.327-332.1968
Amara, A. B., Ghigo, E., Priol, Y. L., Lépolard, C., Salcedo, S. P., Lemichez, E., et al. (2010). Coxiella burnetii, the agent of Q fever, replicates within trophoblasts and induces a unique transcriptional response. PloS One 5, e15315. doi: 10.1371/journal.pone.0015315
Andersson, S. G. E., Zomorodipour, A., Andersson, J. O., Sicheritz-Pontén, T., Alsmark, U. C. M., Podowski, R. M., et al. (1998). The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396, 133–140. doi: 10.1038/24094
Audia, J. P., Winkler, H. H. (2006). Study of the Five Rickettsia prowazekii proteins annotated as ATP/ADP translocases (Tlc): Only Tlc1 transports ATP/ADP, while Tlc4 and Tlc5 transport other ribonucleotides. J. Bacteriol. 188, 6261–6268. doi: 10.1128/jb.00371-06
Beare, P. A., Larson, C. L., Gilk, S. D., Heinzen, R. A. (2012). Two systems for targeted gene deletion in Coxiella burnetii. Appl. Environ. Microb. 78, 4580–4589. doi: 10.1128/aem.00881-12
Beare, P. A., Unsworth, N., Andoh, M., Voth, D. E., Omsland, A., Gilk, S. D., et al. (2009). Comparative genomics reveal extensive transposon-mediated genomic plasticity and diversity among potential effector proteins within the genus Coxiella. Infect. Immun. 77, 642–656. doi: 10.1128/iai.01141-08
Belland, R. J., Nelson, D. E., Virok, D., Crane, D. D., Hogan, D., Sturdevant, D., et al. (2003a). Transcriptome analysis of chlamydial growth during IFN-γ-mediated persistence and reactivation. Proc. Natl. Acad. Sci. 100, 15971–15976. doi: 10.1073/pnas.2535394100
Belland, R. J., Zhong, G., Crane, D. D., Hogan, D., Sturdevant, D., Sharma, J., et al. (2003b). Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. 100, 8478–8483. doi: 10.1073/pnas.1331135100
Bereshchenko, O., Bruscoli, S., Riccardi, C. (2018). Glucocorticoids, sex hormones, and immunity. Front. Immunol. 9. doi: 10.3389/fimmu.2018.01332
Beste, D. J. V., Espasa, M., Bonde, B., Kierzek, A. M., Stewart, G. R., McFadden, J. (2009). The genetic requirements for fast and slow growth in mycobacteria. PloS One 4, e5349. doi: 10.1371/journal.pone.0005349
Bitew, M. A., Hofmann, J., Souza, D. P. D., Wawegama, N. K., Newton, H. J., Sansom, F. M. (2020). SdrA, an NADP(H)-regenerating enzyme, is crucial for Coxiella burnetii to resist oxidative stress and replicate intracellularly. Cell Microbiol. 22, e13154. doi: 10.1111/cmi.13154
Blanton, L. S. (2019). The rickettsioses: A practical update. Infect. Dis. Clin. N Am. 33, 213–229. doi: 10.1016/j.idc.2018.10.010
Bobay, L.-M., Ochman, H. (2017). The evolution of bacterial genome architecture. Front. Genet. 8. doi: 10.3389/fgene.2017.00072
Bommana, S., Somboonna, N., Richards, G., Tarazkar, M., Dean, D. (2021). Tryptophan operon diversity reveals evolutionary trends among geographically disparate Chlamydia trachomatis ocular and urogenital strains affecting tryptophan repressor and synthase function. Mbio 12, e00605–e00621. doi: 10.1128/mbio.00605-21
Borek, J. R. A. R. E. (1956). Studies on a mutant of Escherichia coli with unbalanced ribonucleic acid synthesis. J. Bacteriology 71, 318–323. doi: 10.1128/jb.71.3.318-323.1956
Borel, N., Polkinghorne, A., Pospischil, A. (2018). A review on chlamydial diseases in animals: still a challenge for pathologists? Vet. Pathol. 55, 374–390. doi: 10.1177/0300985817751218
Borisov, V. B., Siletsky, S. A., Paiardini, A., Hoogewijs, D., Forte, E., Giuffrè, A., et al. (2021). Bacterial oxidases of the cytochrome bd family: redox enzymes of unique structure, function, and utility as drug targets. Antioxid Redox Sign 34, 1280–1318. doi: 10.1089/ars.2020.8039
Boutte, C. C., Crosson, S. (2013). Bacterial lifestyle shapes stringent response activation. Trends Microbiol. 21, 174–180. doi: 10.1016/j.tim.2013.01.002
Bovarnick, M. R., Miller, J. C. (1950). Oxidation and transamination of glutamate by typhus rickettsiae. J. Biol. Chem. 184, 661–676. doi: 10.1016/S0021-9258(19)51000-6
Bovarnick, M. R., Schneider, L. (1960). The incorporation of glycine-1-C14 by typhus rickettsiae. J. Biol. Chem. 235, 1727–1731. doi: 10.1016/s0021-9258(19)76871-9
Brann, K. R., Fullerton, M. S., Voth, D. E. (2020). Coxiella burnetii requires host eukaryotic initiation factor 2α activity for efficient intracellular replication. Infect. Immun. 88. doi: 10.1128/iai.00096-20
Brenner, A. E., Muñoz-Leal, S., Sachan, M., Labruna, M. B., Raghavan, R. (2021). Coxiella burnetii and related tick endosymbionts evolved from pathogenic ancestors. Genome Biol. Evol. 13. doi: 10.1093/gbe/evab108
Briggs, H. L., Pul, N., Seshadri, R., Wilson, M. J., Tersteeg, C., Russell-Lodrigue, K. E., et al. (2008). Limited role for iron regulation in Coxiella burnetii pathogenesis▿ †. Infect. Immun. 76, 2189–2201. doi: 10.1128/iai.01609-07
Brinkworth, A. J., Wildung, M. R., Carabeo, R. A. (2018). Genome wide transcriptional responses of iron-starved Chlamydia trachomatis reveal prioritization of metabolic precursor synthesis over protein translation. Msystems 3, e00184–e00117. doi: 10.1128/msystems.00184-17
Brothwell, J. A., Muramatsu, M. K., Toh, E., Rockey, D. D., Putman, T. E., Barta, M. L., et al. (2016). Interrogating genes that mediate Chlamydia trachomatis survival in cell culture using conditional mutants and recombination. J. Bacteriol 198, 2131–2139. doi: 10.1128/jb.00161-16
Burkhardt, N. Y., Baldridge, G. D., Williamson, P. C., Billingsley, P. M., Heu, C. C., Felsheim, R. F., et al. (2011). Development of shuttle vectors for transformation of diverse Rickettsia species. PloS One 6, e29511. doi: 10.1371/journal.pone.0029511
Burnet, F. M., Freeman, M. (1937). Experimental studies on the virus of “Q” fever. Med. J. Aust. 2, 299–305. doi: 10.5694/j.1326-5377.1937.tb43744.x
Burton, P. R., Kordová, N., Paretsky, D. (1971). Electron microscopic studies of the rickettsia Coxiella burnetii: entry, lysosomal response, and fate of rickettsial DNA in L-cells. Canadian Journal of Microbiology 17, 143–150. doi: 10.1139/m71-025
Burton, P. R., Stueckemann, J., Welsh, R. M., Paretsky, D. (1978). Some ultrastructural effects of persistent infections by the rickettsia Coxiella burnetii in mouse L cells and green monkey kidney (Vero) cells. Infection Immun. 21, 556–566. doi: 10.1128/iai.21.2.556-566.1978
Caldwell, H. D., Wood, H., Crane, D., Bailey, R., Jones, R. B., Mabey, D., et al. (2003). Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. J. Clin. Invest. 111, 1757–1769. doi: 10.1172/jci17993
Canevari, J. T., Firestone, S. M., Vincent, G., Campbell, A., Tan, T., Muleme, M., et al. (2018). The prevalence of Coxiella burnetii shedding in dairy goats at the time of parturition in an endemically infected enterprise and associated milk yield losses. BMC Vet. Res. 14, 353. doi: 10.1186/s12917-018-1667-x
Cantor, J. R. (2019). The rise of physiologic media. Trends Cell Biol. 29, 854–861. doi: 10.1016/j.tcb.2019.08.009
Carey, K. L., Newton, H. J., Lührmann, A., Roy, C. R. (2011). The Coxiella burnetii Dot/Icm system delivers a unique repertoire of type IV effectors into host cells and is required for intracellular replication. PloS Pathog. 7, e1002056. doi: 10.1371/journal.ppat.1002056
Carlson, J. H., Wood, H., Roshick, C., Caldwell, H. D., McClarty, G. (2006). In vivo and in vitro studies of Chlamydia trachomatis TrpR : DNA interactions. Mol. Microbiol. 59, 1678–1691. doi: 10.1111/j.1365-2958.2006.05045.x
Casadevall, A. (2008). Evolution of intracellular pathogens. Annu. Rev. Microbiol. 62, 19–33. doi: 10.1146/annurev.micro.61.080706.093305
Cashel, M. (1969). The control of ribonucleic acid synthesis in Escherichia coli IV. Relevance of unusual phosphorylated compounds from amino acid-starved stringent strains. J. Biol. Chem. 244, 3133–3141. doi: 10.1016/S0021-9258(18)93106-6
Cashel, M., Gallant, J. (1969). Two compounds implicated in the function of the RC gene of Escherichia coli. Nature 221, 838–841. doi: 10.1038/221838a0
Celli, J., Tsolis, R. M. (2014). Bacteria, the endoplasmic reticulum and the unfolded protein response: friends or foes? Nat. Rev. Microbiol. 13, 71–82. doi: 10.1038/nrmicro3393
Chen, C., Banga, S., Mertens, K., Weber, M. M., Gorbaslieva, I., Tan, Y., et al. (2010). Large-scale identification and translocation of type IV secretion substrates by Coxiella burnetii. Proc. Natl. Acad. Sci. 107, 21755–21760. doi: 10.1073/pnas.1010485107
Chiarelli, T. J., Grieshaber, N. A., Omsland, A., Remien, C. H., Grieshaber, S. S. (2020). Single-inclusion kinetics of Chlamydia trachomatis development. Msystems 5, e00689–e00620. doi: 10.1128/msystems.00689-20
Clark, T. R., Ellison, D. W., Kleba, B., Hackstadt, T. (2011). Complementation of Rickettsia rickettsii RelA/SpoT restores a nonlytic plaque phenotype. Infect. Immun. 79, 1631–1637. doi: 10.1128/iai.00048-11
Coleman, S. A., Fischer, E. R., Howe, D., Mead, D. J., Heinzen, R. A. (2004). Temporal analysis of Coxiella burnetii morphological differentiation. J. Bacteriol 186, 7344–7352. doi: 10.1128/jb.186.21.7344-7352.2004
Cotter, P. A., Melville, S. B., Albrecht, J. A., Gunsalus, R. P. (1997). Aerobic regulation of cytochrome d oxidase (cydAB) operon expression in Escherichia coli: roles of Fnr and ArcA in repression and activation. Mol. Microbiol. 25, 605–615. doi: 10.1046/j.1365-2958.1997.5031860.x
Crabill, E., Schofield, W. B., Newton, H. J., Goodman, A. L., Roy, C. R. (2018). Dot/Icm-translocated proteins important for biogenesis of the Coxiella burnetii-containing vacuole identified by screening of an effector mutant sublibrary. Infect. Immun. 86, e00758–e00717. doi: 10.1128/iai.00758-17
Czyż, D. M., Potluri, L.-P., Jain-Gupta, N., Riley, S. P., Martinez, J. J., Steck, T. L., et al. (2014). Host-directed antimicrobial drugs with broad-spectrum efficacy against intracellular bacterial pathogens. Mbio 5, e01534–e01514. doi: 10.1128/mbio.01534-14
Dalebroux, Z. D., Yagi, B. F., Sahr, T., Buchrieser, C., Swanson, M. S. (2010). Distinct roles of ppGpp and DksA in Legionella pneumophila differentiation. Mol. Microbiol. 76, 200–219. doi: 10.1111/j.1365-2958.2010.07094.x
Davis, G. E., Cox, H. R. (1938). A filter-passing infectious agent isolated from ticks. Public Heal Reports 1896-1970 53, Chapter I isolation from Dermacentor andersoni, reactions in animals, and filtration experiments. Sage Publication, Inc. doi: 10.2307/4582746
Derrick, E. H. (1937). “Q” fever, a new fever entity: clinical features, diagnosis and laboratory investigation. Med. J. Aust. 2, 281–299. doi: 10.5694/j.1326-5377.1937.tb43743.x
Dibrov, P., Dibrov, E., Pierce, G. N., Galperin, M. Y. (2004). Salt in the wound: a possible role of Na+ gradient in chlamydial infection. J. Mol. Microb. Biotech. 8, 1–6. doi: 10.1159/000082075
Driscoll, T. P., Verhoeve, V. I., Guillotte, M. L., Lehman, S. S., Rennoll, S. A., Beier-Sexton, M., et al. (2017). Wholly Rickettsia! Reconstructed metabolic profile of the quintessential bacterial parasite of eukaryotic cells. Mbio 8, e00859–e00817. doi: 10.1128/mbio.00859-17
Eisenreich, W., Rudel, T., Heesemann, J., Goebel, W. (2017). To eat and to be eaten: mutual metabolic adaptations of immune cells and intracellular bacterial pathogens upon infection. Front. Cell Infect. Mi 7. doi: 10.3389/fcimb.2017.00316
Ellison, D. W., Clark, T. R., Sturdevant, D. E., Virtaneva, K., Hackstadt, T. (2009). Limited transcriptional responses of Rickettsia rickettsii exposed to environmental stimuli. PloS One 4, e5612. doi: 10.1371/journal.pone.0005612
Elwell, C., Mirrashidi, K., Engel, J. (2016). Chlamydia cell biology and pathogenesis. Nat. Rev. Microbiol. 14, 385–400. doi: 10.1038/nrmicro.2016.30
Ende, R. J., Derré, I. (2020). Host and bacterial glycolysis during Chlamydia trachomatis Infection. Infect. Immun. 88. doi: 10.1128/iai.00545-20
Engström, P., Bergström, M., Alfaro, A. C., Krishnan, K. S., Bahnan, W., Almqvist, F., et al. (2015). Expansion of the Chlamydia trachomatis inclusion does not require bacterial replication. Int. J. Med. Microbiol. Ijmm 305, 378–382. doi: 10.1016/j.ijmm.2015.02.007
Engström, P., Krishnan, K. S., Ngyuen, B. D., Chorell, E., Normark, J., Silver, J., et al. (2014). A 2-pyridone-amide inhibitor targets the glucose metabolism pathway of Chlamydia trachomatis. Mbio 6, e02304–e02314. doi: 10.1128/mbio.02304-14
Esquerra, E. V., Yang, H., Sanchez, S. E., Omsland, A. (2017). Physicochemical and nutritional requirements for axenic replication suggest physiological basis for Coxiella burnetii niche restriction. Front. Cell Infect. Mi 7. doi: 10.3389/fcimb.2017.00190
Essig, A., Longbottom, D. (2015). Chlamydia abortus: new aspects of infectious abortion in sheep and potential risk for pregnant women. Curr. Clin. Microbiol. Rep. 2, 22–34. doi: 10.1007/s40588-015-0014-2
Fan, H., Brunham, R. C., McClarty, G. (1992). Acquisition and synthesis of folates by obligate intracellular bacteria of the genus Chlamydia. J. Clin. Invest. 90, 1803–1811. doi: 10.1172/jci116055
Faris, R., Andersen, S. E., McCullough, A., Gourronc, F., Klingelhutz, A. J., Weber, M. M. (2019). Chlamydia trachomatis serovars drive differential production of proinflammatory cytokines and chemokines depending on the type of cell infected. Front. Cell Infect. Mi 9. doi: 10.3389/fcimb.2019.00399
Felsheim, R. F., Chávez, A. S. O., Palmer, G. H., Crosby, L., Barbet, A. F., Kurtti, T. J., et al. (2010). Transformation of Anaplasma marginale. Vet. Parasitol. 167, 167–174. doi: 10.1016/j.vetpar.2009.09.018
Fields, K. A., Bodero, M. D., Scanlon, K. R., Jewett, T. J., Wolf, K. (2022). A minimal replicon enables efficacious, species-specific gene deletion in Chlamydia and extension of gene knockout studies to the animal model of infection using Chlamydia muridarum. Infect. Immun. 90, e00453–e00422. doi: 10.1128/iai.00453-22
Fischer, A., Rudel, T. (2018). Safe haven under constant attack-The Chlamydia-containing vacuole. Cell Microbiol. 20, e12940. doi: 10.1111/cmi.12940
Fisher, D. J., Fernández, R. E., Maurelli, A. T. (2013). Chlamydia trachomatis transports NAD via the Npt1 ATP/ADP translocase. J. Bacteriol 195, 3381–3386. doi: 10.1128/jb.00433-13
Fonseca, M. V., Swanson, M. S. (2014). Nutrient salvaging and metabolism by the intracellular pathogen Legionella pneumophila. Front. Cell Infect. Mi 4. doi: 10.3389/fcimb.2014.00012
Frohlich, K. M., Roberts, R. A. W., Housley, N. A., Audia, J. P. (2010). Rickettsia prowazekii uses an sn-glycerol-3-phosphate dehydrogenase and a novel dihydroxyacetone phosphate transport system to supply triose phosphate for phospholipid biosynthesis. J. Bacteriol 192, 4281–4288. doi: 10.1128/jb.00443-10
Fu, M., Liu, Y., Wang, G., Wang, P., Zhang, J., Chen, C., et al. (2022). A protein–protein interaction map reveals that the Coxiella burnetii effector CirB inhibits host proteasome activity. PloS Pathog. 18, e1010660. doi: 10.1371/journal.ppat.1010660
Fuchs, T. M., Eisenreich, W., Heesemann, J., Goebel, W. (2012). Metabolic adaptation of human pathogenic and related nonpathogenic bacteria to extra- and intracellular habitats. FEMS Microbiol. Rev. 36, 435–462. doi: 10.1111/j.1574-6976.2011.00301.x
Galletti, M. F. B. M., Fujita, A., Rosa, R. D., Martins, L. A., Soares, H. S., Labruna, M. B., et al. (2016). Virulence genes of Rickettsia rickettsii are differentially modulated by either temperature upshift or blood-feeding in tick midgut and salivary glands. Parasite Vector 9, 331. doi: 10.1186/s13071-016-1581-7
Ganesan, S., Roy, C. R. (2019). Host cell depletion of tryptophan by IFNγ-induced Indoleamine 2,3-dioxygenase 1 (IDO1) inhibits lysosomal replication of Coxiella burnetii. PloS Pathog. 15, e1007955. doi: 10.1371/journal.ppat.1007955
Gehre, L., Gorgette, O., Perrinet, S., Prevost, M.-C., Ducatez, M., Giebel, A. M., et al. (2016). Sequestration of host metabolism by an intracellular pathogen. Elife 5, e12552. doi: 10.7554/elife.12552
George, Z., Omosun, Y., Azenabor, A. A., Partin, J., Joseph, K., Ellerson, D., et al. (2016). The roles of unfolded protein response pathways in Chlamydia pathogenesis. J. Infect. Dis. 215, jiw569. doi: 10.1093/infdis/jiw569
Giovannoni, S. J., Thrash, J. C., Temperton, B. (2014). Implications of streamlining theory for microbial ecology. Isme J. 8, 1553–1565. doi: 10.1038/ismej.2014.60
Gregory, A. E., Schaik, E. J., Russell-Lodrigue, K. E., Fratzke, A. P., Samuel, J. E. (2019). Coxiella burnetii intratracheal aerosol infection model in mice, Guinea pigs, and nonhuman primates. Infect. Immun. 87. doi: 10.1128/iai.00178-19
Grieshaber, N. A., Fischer, E. R., Mead, D. J., Dooley, C. A., Hackstadt, T. (2004). Chlamydial histone-DNA interactions are disrupted by a metabolite in the methylerythritol phosphate pathway of isoprenoid biosynthesis. P Natl. Acad. Sci. U.S.A. 101, 7451–7456. doi: 10.1073/pnas.0400754101
Grieshaber, S., Grieshaber, N., Yang, H., Baxter, B., Hackstadt, T., Omsland, A. (2018). Impact of active metabolism on Chlamydia trachomatis elementary body transcript profile and infectivity. J. Bacteriol 200, e00065–e00018. doi: 10.1128/jb.00065-18
Grieshaber, S., Swanson, J. A., Hackstadt, T. (2002). Determination of the physical environment within the Chlamydia trachomatis inclusion using ion-selective ratiometric probes. Cell Microbiol. 4, 273–283. doi: 10.1046/j.1462-5822.2002.00191.x
Hackstadt, T., Williams, J. C. (1981a). Biochemical stratagem for obligate parasitism of eukaryotic cells by Coxiella burnetii. Proc. Natl. Acad. Sci. 78, 3240–3244. doi: 10.1073/pnas.78.5.3240
Hackstadt, T., Williams, J. C. (1981b). Stability of the adenosine 5′-triphosphate pool in Coxiella burnetii: influence of pH and substrate. J. Bacteriology 148, 419 425. doi: 10.1128/jb.148.2.419-425.1981
Harper, A., Pogson, C. I., Jones, M. L., Pearce, J. H. (2000). Chlamydial development is adversely affected by minor changes in amino acid supply, blood plasma amino acid levels, and glucose deprivation. Infect. Immun. 68, 1457 1464. doi: 10.1128/iai.68.3.1457-1464.2000
Hauryliuk, V., Atkinson, G. C., Murakami, K. S., Tenson, T., Gerdes, K. (2015). Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 13, 298–309. doi: 10.1038/nrmicro3448
Häuslein, I., Cantet, F., Reschke, S., Chen, F., Bonazzi, M., Eisenreich, W. (2017). Multiple substrate usage of Coxiella burnetii to feed a bipartite metabolic network. Front. Cell Infect. Mi 7. doi: 10.3389/fcimb.2017.00285
Hayek, I., Szperlinski, M., Lührmann, A. (2022). Coxiella burnetii affects HIF1α accumulation and HIF1α target gene expression. Front. Cell. Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.867689
Hayward, R. J., Marsh, J. W., Humphrys, M. S., Huston, W. M., Myers, G. S. (2020). Chromatin accessibility dynamics of Chlamydia-infected epithelial cells. Epigenet. Chromatin 13, 45. doi: 10.1186/s13072-020-00368-2
Heinzen, R. A., Hayes, S. F., Peacock, M. G., Hackstadt, T. (1993). Directional actin polymerization associated with spotted fever group Rickettsia infection of Vero cells. Infect. Immun. 61, 1926–1935. doi: 10.1128/iai.61.5.1926-1935.1993
Heinzen, R. A., Mallavia, L. P. (1987). Cloning and functional expression of the Coxiella burnetii citrate synthase gene in Escherichia coli. Infect. Immun. 55, 848–855. doi: 10.1128/iai.55.4.848-855.1987
Heinzen, R. A., Scidmore, M. A., Rockey, D. D., Hackstadt, T. (1996). Differential interaction with endocytic and exocytic pathways distinguish parasitophorous vacuoles of Coxiella burnetii and Chlamydia trachomatis. Infection Immun. 64, 796 809. doi: 10.1128/iai.64.3.796-809.1996
Howard, Z. P., Omsland, A. (2020). Selective inhibition of Coxiella burnetii replication by the steroid hormone progesterone. Infect. Immun. 88. doi: 10.1128/iai.00894-19
Hughes, D. T., Sperandio, V. (2008). Inter-kingdom signalling: communication between bacteria and their hosts. Nat. Rev. Microbiol. 6, 111–120. doi: 10.1038/nrmicro1836
Humeau, J., Leduc, M., Cerrato, G., Loos, F., Kepp, O., Kroemer, G. (2020). Phosphorylation of eukaryotic initiation factor-2α (eIF2α) in autophagy. Cell Death Dis. 11, 433. doi: 10.1038/s41419-020-2642-6
Iliffe-Lee, E. R., McClarty, G. (2000). Regulation of carbon metabolism in Chlamydia trachomatis. Mol. Microbiol. 38, 20–30. doi: 10.1046/j.1365-2958.2000.02102.x
Iliffe-Lee, E. R., McClarty, G. (2002). Pyruvate kinase from Chlamydia trachomatis is activated by fructose-2,6-bisphosphate: Pyruvate kinase from C. trachomatis. Mol. Microbiol. 44, 819–828. doi: 10.1046/j.1365-2958.2002.02924.x
Janik, K., Bode, J., Dutow, P., Laudeley, R., Geffers, R., Sommer, K., et al. (2014). Temperature and host cell-dependent changes in virulence of Chlamydia pneumoniae CWL029 in an optimized mouse infection model. Pathog. Dis. 73, 1–8. doi: 10.1093/femspd/ftu001
Johnson, C. M., Fisher, D. J. (2013). Site-specific, insertional inactivation of incA in Chlamydia trachomatis using a group II intron. PloS One 8, e83989. doi: 10.1371/journal.pone.0083989
Juul, N., Jensen, H., Hvid, M., Christiansen, G., Birkelund, S. (2007). Characterization of in vitro chlamydial cultures in low-oxygen atmospheres. J. Bacteriol 189, 6723–6726. doi: 10.1128/jb.00279-07
Kaleta, C., Schäuble, S., Rinas, U., Schuster, S. (2013). Metabolic costs of amino acid and protein production in Escherichia coli. Biotechnol. J. 8, 1105–1114. doi: 10.1002/biot.201200267
Kang, P. J., Craig, E. A. (1990). Identification and characterization of a new Escherichia coli gene that is a dosage-dependent suppressor of a dnaK deletion mutation. J. Bacteriol 172, 2055–2064. doi: 10.1128/jb.172.4.2055-2064.1990
Kari, L., Goheen, M. M., Randall, L. B., Taylor, L. D., Carlson, J. H., Whitmire, W. M., et al. (2011). Generation of targeted Chlamydia trachomatis null mutants. Proc. Natl. Acad. Sci. 108, 7189–7193. doi: 10.1073/pnas.1102229108
Kaufmann, S. H. E. (2011). Intracellular pathogens: living in an extreme environment: Intracellular pathogens. Immunol. Rev. 240, 5–10. doi: 10.1111/j.1600-065x.2010.01001.x
Keasling, J. D., Bertsch, L., Kornberg, A. (1993). Guanosine pentaphosphate phosphohydrolase of Escherichia coli is a long-chain exopolyphosphatase. Proc. Natl. Acad. Sci. 90, 7029–7033. doi: 10.1073/pnas.90.15.7029
Kersh, G., Priestley, R., Hornstra, H., Self, J., Fitzpatrick, K., Biggerstaff, B., et al. (2016). Genotyping and axenic growth of Coxiella burnetii isolates found in the United States environment. Vector-borne Zoonot 16, vbz.2016.1972. doi: 10.1089/vbz.2016.1972
Kohler, L. J., Roy, C. R. (2015). Biogenesis of the lysosome-derived vacuole containing Coxiella burnetii. Microbes Infect. 17, 766–771. doi: 10.1016/j.micinf.2015.08.006
Kouroku, Y., Fujita, E., Tanida, I., Ueno, T., Isoai, A., Kumagai, H., et al. (2006). ER stress (PERK/eIF2α phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ 14, 230–239. doi: 10.1038/sj.cdd.4401984
Kuba, M., Neha, N., Souza, D. P. D., Dayalan, S., Newson, J. P. M., Tull, D., et al. (2019). Coxiella burnetii utilizes both glutamate and glucose during infection with glucose uptake mediated by multiple transporters. Biochem. J. 476, BCJ20190504. doi: 10.1042/bcj20190504
Kuley, R., Bossers-deVries, R., Smith, H. E., Smits, M. A., Roest, H. I. J., Bossers, A. (2015). Major differential gene regulation in Coxiella burnetii between in vivo and in vitro cultivation models. BMC Genomics 16, 953. doi: 10.1186/s12864-015-2143-7
Kwaik, Y. A., Bumann, D. (2013). Microbial quest for food in vivo: ‘Nutritional virulence’ as an emerging paradigm. Cell Microbiol. 15, 882–890. doi: 10.1111/cmi.12138
Lam, O., Wheeler, J., Tang, C. M. (2014). Thermal control of virulence factors in bacteria: a hot topic. Virulence 5, 852–862. doi: 10.4161/21505594.2014.970949
Lane, A. B., Decker, C. F. (2016). Chlamydia trachomatis infections. Dis 62, 269–273. doi: 10.1016/j.disamonth.2016.03.010
Larson, C. L., Beare, P. A., Howe, D., Heinzen, R. A. (2013). Coxiella burnetii effector protein subverts clathrin-mediated vesicular trafficking for pathogen vacuole biogenesis. Proc. Natl. Acad. Sci. 110, E4770 4779. doi: 10.1073/pnas.1309195110
Larson, C. L., Martinez, E., Beare, P. A., Jeffrey, B., Heinzen, R. A., Bonazzi, M. (2016). Right on Q: genetics begin to unravel Coxiella burnetii host cell interactions. Future Microbiol. 11, 919–939. doi: 10.2217/fmb-2016-0044
Lee, J. K., Enciso, G. A., Boassa, D., Chander, C. N., Lou, T. H., Pairawan, S. S., et al. (2018). Replication-dependent size reduction precedes differentiation in Chlamydia trachomatis. Nat. Commun. 9, 45. doi: 10.1038/s41467-017-02432-0
Léger, L., Byrne, D., Guiraud, P., Germain, E., Maisonneuve, E. (2021). NirD curtails the stringent response by inhibiting RelA activity in Escherichia coli. Elife 10, e64092. doi: 10.7554/elife.64092
Leone, M., Honstettre, A., Lepidi, H., Capo, C., Bayard, F., Raoult, D., et al. (2004). Effect of sex on Coxiella burnetii infection: protective role of 17β-estradiol. J. Infect. Dis. 189, 339–345. doi: 10.1086/380798
Levanon, S. S., San, K.-Y., Bennett, G. N. (2005). Effect of oxygen on the Escherichia coli ArcA and FNR regulation systems and metabolic responses. Biotechnol. Bioeng 89, 556–564. doi: 10.1002/bit.20381
Li, K., Jiang, T., Yu, B., Wang, L., Gao, C., Ma, C., et al. (2012). Transcription elongation factor GreA has functional chaperone activity. PloS One 7, e47521. doi: 10.1371/journal.pone.0047521
Liang, P., Rosas-Lemus, M., Patel, D., Fang, X., Tuz, K., Juárez, O. (2018). Dynamic energy dependency of Chlamydia trachomatis on host cell metabolism during intracellular growth: role of sodium-based energetics in chlamydial ATP generation. J. Biol. Chem. 293, 510–522. doi: 10.1074/jbc.m117.797209
Liechti, G., Kuru, E., Packiam, M., Hsu, Y.-P., Tekkam, S., Hall, E., et al. (2016). Pathogenic Chlamydia lack a classical sacculus but synthesize a narrow, mid-cell peptidoglycan ring, regulated by MreB, for cell division. PloS Pathog. 12, e1005590. doi: 10.1371/journal.ppat.1005590
Long, C. M., Beare, P. A., Cockrell, D. C., Larson, C. L., Heinzen, R. A. (2019). Comparative virulence of diverse Coxiella burnetii strains. Virulence 10, 133–150. doi: 10.1080/21505594.2019.1575715
Luo, S., Lu, S., Fan, H., Chen, Z., Sun, Z., Hu, Y., et al. (2021). The Coxiella burnetii QpH1 plasmid is a virulence factor for colonizing bone marrow-derived murine macrophages. J. Bacteriol 203. doi: 10.1128/jb.00588-20
Magnusson, L. U., Gummesson, B., Joksimović, P., Farewell, A., Nyström, T. (2007). Identical, independent, and opposing roles of ppGpp and DksA in Escherichia coli. J. Bacteriol 189, 5193–5202. doi: 10.1128/jb.00330-07
Mandel, C., Yang, H., Buchko, G. W., Abendroth, J., Grieshaber, N., Chiarelli, T., et al. (2021). Expression and structure of the Chlamydia trachomatis DksA ortholog. Pathog. Dis. 80, ftac007. doi: 10.1093/femspd/ftac007
Martinez, E., Cantet, F., Fava, L., Norville, I., Bonazzi, M. (2014). Identification of OmpA, a Coxiella burnetii protein involved in host cell invasion, by multi-phenotypic high-content screening. PloS Pathog. 10, e1004013. doi: 10.1371/journal.ppat.1004013
McClarty, G. (1999). “Chlamydial metabolism as inferred from the complete genome sequence,” in Chlamydia intracellular biology, pathogenesis, and immunity. (Washington D.C.: American Society for Microbiology). Ed. Stephens, R. S., 69–100.
McClure, E. E., Chávez, A. S. O., Shaw, D. K., Carlyon, J. A., Ganta, R. R., Noh, S. M., et al. (2017). Engineering of obligate intracellular bacteria: progress, challenges and paradigms. Nat. Rev. Microbiol. 15, 544–558. doi: 10.1038/nrmicro.2017.59
McDonald, T. L., Mallavia, L. (1970). Biochemistry of Coxiella burnetti: 6-phosphogluconic acid dehydrogenase. J. Bacteriol 102, 1–5. doi: 10.1128/jb.102.1.1-5.1970
McDonald, T. L., Mallavia, L. (1971). Biochemistry of Coxiella burnetii: Embden-Meyerhof pathway. J. Bacteriol 107, 864–869. doi: 10.1128/jb.107.3.864-869.1971
McGinn, J., Lamason, R. L. (2021). The enigmatic biology of rickettsiae: recent advances, open questions and outlook. Pathog. Dis. 79, ftab019. doi: 10.1093/femspd/ftab019
McLeod, M. P., Qin, X., Karpathy, S. E., Gioia, J., Highlander, S. K., Fox, G. E., et al. (2004). Complete genome sequence of Rickettsia typhi and comparison with sequences of other rickettsiae. J. Bacteriol 186, 5842–5855. doi: 10.1128/jb.186.17.5842-5855.2004
Mehlitz, A., Eylert, E., Huber, C., Lindner, B., Vollmuth, N., Karunakaran, K., et al. (2016). Metabolic adaptation of Chlamydia trachomatis to mammalian host cells. Mol. Microbiol. 103, 1004–1019. doi: 10.1111/mmi.13603
Mertens, K., Samuel, J. E. (2012). Coxiella burnetii: recent advances and new perspectives in research of the Q fever bacterium. Adv. Exp. Med. Biol. 984, 39–63. doi: 10.1007/978-94-007-4315-1_3
Mishori, R., McClaskey, E. L., WinklerPrins, V. J. (2012). Chlamydia trachomatis infections: screening, diagnosis, and management. Am. Fam Physician 86, 1127–1132.
Moore, E. R., Ouellette, S. P. (2014). Reconceptualizing the chlamydial inclusion as a pathogen-specified parasitic organelle: an expanded role for Inc proteins. Front. Cell Infect. Mi 4. doi: 10.3389/fcimb.2014.00157
Moormeier, D. E., Sandoz, K. M., Beare, P. A., Sturdevant, D. E., Nair, V., Cockrell, D. C., et al. (2019). Coxiella burnetii RpoS regulates genes involved in morphological differentiation and intracellular growth. J. Bacteriol 201. doi: 10.1128/jb.00009-19
Morse, S. A., Fitzgerald, T. J. (1974). Effect of progesterone on Neisseria gonorrhoeae. Infect. Immun. 10, 1370–1377. doi: 10.1128/iai.10.6.1370-1377.1974
Moses, A. S., Millar, J. A., Bonazzi, M., Beare, P. A., Raghavan, R. (2017). Horizontally acquired biosynthesis genes boost Coxiella burnetii’s physiology. Front. Cell Infect. Mi 7. doi: 10.3389/fcimb.2017.00174
Moulder, J. W. (1962). The biochemistry of intracellular parasitism (Chicago: University of Chicago Press), 122–124.
Moulder, J. W. (1974). Intracellular parasitism: life in an extreme environment. J. Infect. Dis. 130, 300–306. doi: 10.1093/infdis/130.3.300
Newton, H. J., Kohler, L. J., McDonough, J. A., Temoche-Diaz, M., Crabill, E., Hartland, E. L., et al. (2014). A screen of Coxiella burnetii mutants reveals important roles for Dot/Icm effectors and host autophagy in vacuole biogenesis. PloS Pathog. 10, e1004286. doi: 10.1371/journal.ppat.1004286
Noor, E., Eden, E., Milo, R., Alon, U. (2010). Central carbon metabolism as a minimal biochemical walk between precursors for biomass and energy. Mol. Cell 39, 809–820. doi: 10.1016/j.molcel.2010.08.031
Noriea, N. F., Clark, T. R., Hackstadt, T. (2015). Targeted knockout of the Rickettsia rickettsii OmpA surface antigen does not diminish virulence in a mammalian model system. Mbio 6, e00323–e00315. doi: 10.1128/mbio.00323-15
Ogata, H., Goto, S., Sato, K., Fujibuchi, W., Bono, H., Kanehisa, M. (1999). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 27, 29–34. doi: 10.1093/nar/27.1.29
Ohkuma, S., Poole, B. (1978). Fluorescence probe measurement of the intra-lysosomal pH in living cells and the perturbation of pH by various agents. Proc. Natl. Acad. Sci. 75, 3327–3331. doi: 10.1073/pnas.75.7.3327
Omsland, A., Cockrell, D. C., Howe, D., Fischer, E. R., Virtaneva, K., Sturdevant, D. E., et al. (2009). Host cell-free growth of the Q fever bacterium Coxiella burnetii. Proc. Natl. Acad. Sci. 106, 4430–4434. doi: 10.1073/pnas.0812074106
Omsland, A., Sager, J., Nair, V., Sturdevant, D. E., Hackstadt, T. (2012). Developmental stage-specific metabolic and transcriptional activity of Chlamydia trachomatis in an axenic medium. Proc. Natl. Acad. Sci. 109, 19781 19785. doi: 10.1073/pnas.1212831109
Omsland, A., Sixt, B. S., Horn, M., Hackstadt, T. (2014). Chlamydial metabolism revisited: interspecies metabolic variability and developmental stage-specific physiologic activities. FEMS Microbiol. Rev. 38, 779–801. doi: 10.1111/1574-6976.12059
Ormsbee, R. A., Peacock, M. G. (1964). Metabolic activity in Coxiella burnetii. J. Bacteriol 88, 1205–1210. doi: 10.1128/jb.88.5.1205-1210.1964
Ouellette, S. P., Blay, E. A., Hatch, N. D., Fisher-Marvin, L. A. (2021). CRISPR interference to inducibly repress gene expression in Chlamydia trachomatis. Infect. Immun. 89, e00108–e00121. doi: 10.1128/iai.00108-21
Ouellette, S. P., Dorsey, F. C., Moshiach, S., Cleveland, J. L., Carabeo, R. A. (2011). Chlamydia species-dependent differences in the growth requirement for lysosomes. PloS One 6, e16783. doi: 10.1371/journal.pone.0016783
Ouellette, S. P., Hatch, T. P., AbdelRahman, Y. M., Rose, L. A., Belland, R. J., Byrne, G. I. (2006). Global transcriptional upregulation in the absence of increased translation in Chlamydia during IFNγ-mediated host cell tryptophan starvation. Mol. Microbiol. 62, 1387–1401. doi: 10.1111/j.1365-2958.2006.05465.x
Ouellette, S. P., Rueden, K. J., Rucks, E. A. (2016). Tryptophan codon-dependent transcription in Chlamydia pneumoniae during gamma interferon-mediated tryptophan limitation. Infect. Immun. 84, 2703–2713. doi: 10.1128/iai.00377-16
Pareja, M. E. M., Bongiovanni, A., Lafont, F., Colombo, M. I. (2017). Alterations of the Coxiella burnetii replicative vacuole membrane integrity and interplay with the autophagy pathway. Front. Cell Infect. Mi 7. doi: 10.3389/fcimb.2017.00112
Paretsky, D., Consigli, R. A., Downs, C. M. (1962). Studies on the physiology of rickettsiae iii. Glucose phosphorylation and hexokinase activity in Coxiella burnetii. J. Bacteriology 83, 538 543. doi: 10.1128/jb.83.3.538-543.1962
Paretsky, D., Downs, C. M., Consigli, R. A., Joyce, B. K. (1958). Studies on the physiology of rickettsiae I. Some enzyme systems of Coxiella burnetii. J. Infect. Dis. 103, 6–11. doi: 10.1093/infdis/103.1.6
Patt, M. W., Conte, L., Blaha, M., Plotkin, B. J. (2018) (Downers Grove, IL: Midwestern University), 60515. doi: 10.3934/molsci.2018.1.117
Paul, B. J., Barker, M. M., Ross, W., Schneider, D. A., Webb, C., Foster, J. W., et al. (2004). DksA a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 118, 311–322. doi: 10.1016/j.cell.2004.07.009
Persky, N. S., Ferullo, D. J., Cooper, D. L., Moore, H. R., Lovett, S. T. (2009). The ObgE/CgtA GTPase influences the stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol. 73, 253–266. doi: 10.1111/j.1365-2958.2009.06767.x
Pettengill, M. A., Marques-da-Silva, C., Avila, M. L., Oliveira, S., d’Arc dos, S., Lam, V. W., et al. (2012). Reversible inhibition of Chlamydia trachomatis infection in epithelial cells due to stimulation of P2X 4 receptors. Infect. Immun. 80, 4232–4238. doi: 10.1128/iai.00441-12
Pinkerton, H., Hass, G. M. (1932a). Typhus fever. J. Exp. Med. 56, 131–143. doi: 10.1084/jem.56.1.131
Pinkerton, H., Hass, G. M. (1932b). Typhus fever. J. Exp. Med. 56, 145–150. doi: 10.1084/jem.56.1.145
Pokorzynski, N. D., Brinkworth, A. J., Carabeo, R. (2019). A bipartite iron-dependent transcriptional regulation of the tryptophan salvage pathway in Chlamydia trachomatis. Elife 8, e42295. doi: 10.7554/elife.42295
Porritt, R. A., Crother, T. R. (2016). Chlamydia pneumoniae infection and inflammatory diseases. Forum Immunopathol. Dis. Ther. 7, 237–254. doi: 10.1615/forumimmundisther.2017020161
Rachek, L. I., Tucker, A. M., Winkler, H. H., Wood, D. O. (1998). Transformation of Rickettsia prowazekii to rifampin resistance. J. Bacteriol. 180, 2118–2124. doi: 10.1128/jb.180.8.2118-2124.1998
Rahnama, M., Fields, K. A. (2018). Transformation of Chlamydia: current approaches and impact on our understanding of chlamydial infection biology. Microbes Infect. 20, 445–450. doi: 10.1016/j.micinf.2018.01.002
Rajeeve, K., Vollmuth, N., Janaki-Raman, S., Wulff, T. F., Baluapuri, A., Dejure, F. R., et al. (2020). Reprogramming of host glutamine metabolism during Chlamydia trachomatis infection and its key role in peptidoglycan synthesis. Nat. Microbiol. 5, 1390–1402. doi: 10.1038/s41564-020-0762-5
Rees, H. B., Weiss, E. (1968). Glutamate catabolism of Rickettsia rickettsii and factors affecting retention of metabolic activity. J. Bacteriol. 95, 389–396. doi: 10.1128/jb.95.2.389-396.1968
Roest, H.-J., Gelderen, B., Dinkla, A., Frangoulidis, D., van Zijderveld, F., Rebel, J., et al. (2012). Q Fever in pregnant goats: pathogenesis and excretion of Coxiella burnetii. PloS One 7, e48949. doi: 10.1371/journal.pone.0048949
Ross, W., Sanchez-Vazquez, P., Chen, A. Y., Lee, J.-H., Burgos, H. L., Gourse, R. L. (2016). ppGpp binding to a site at the RNAP-DksA interface accounts for its dramatic effects on transcription initiation during the stringent response. Mol. Cell 62, 811–823. doi: 10.1016/j.molcel.2016.04.029
Rota, T. R., Nichols, R. L. (1973). Chlamydia trachomatis in cell culture. I. Comparison of efficiencies of infection in several chemically defined media, at various pH and temperature values, and after exposure to diethylaminoethyl-dextran. Appl. Microbiol. 26, 560–565. doi: 10.1128/am.26.4.560-565.1973
Rother, M., Costa, A. R. T., Zietlow, R., Meyer, T. F., Rudel, T. (2019). Modulation of host cell metabolism by Chlamydia trachomatis. Microbiol. Spectr. 7. doi: 10.1128/microbiolspec.bai-0012-2019
Roulis, E., Polkinghorne, A., Timms, P. (2013). Chlamydia pneumoniae: modern insights into an ancient pathogen. Trends Microbiol. 21, 120–128. doi: 10.1016/j.tim.2012.10.009
Saka, H. A., Thompson, J. W., Chen, Y. S., Kumar, Y., Dubois, L. G., Moseley, M. A., et al. (2011). Quantitative proteomics reveals metabolic and pathogenic properties of Chlamydia trachomatis developmental forms. Mol. Microbiol. 82, 1185 1203. doi: 10.1111/j.1365-2958.2011.07877.x
Samanta, D., Clemente, T. M., Schuler, B. E., Gilk, S. D. (2019). Coxiella burnetii type 4B secretion system-dependent manipulation of endolysosomal maturation is required for bacterial growth. PloS Pathog. 15, e1007855. doi: 10.1371/journal.ppat.1007855
Sanchez, S. E., Goodman, A. G., Omsland, A. (2021). Metabolic plasticity aids amphotropism of Coxiella burnetii. Infect. Immun. 82 (12), IAI0013521. doi: 10.1128/iai.00135-21
Sanchez, S. E., Omsland, A. (2020). Critical role for molecular iron in Coxiella burnetii replication and viability. Msphere 5, e00458–e00420. doi: 10.1128/msphere.00458-20
Sanchez, S. E., Omsland, A. (2021). Conditional impairment of Coxiella burnetii by glucose-6P dehydrogenase activity. Pathog. Dis. 79, ftab034. doi: 10.1093/femspd/ftab034
Sanchez-Vazquez, P., Dewey, C. N., Kitten, N., Ross, W., Gourse, R. L. (2019). Genome-wide effects on Escherichia coli transcription from ppGpp binding to its two sites on RNA polymerase. P Natl. Acad. Sci. U.S.A. 116, 8310–8319. doi: 10.1073/pnas.1819682116
Sandoz, K. M., Beare, P. A., Cockrell, D. C., Heinzen, R. A. (2016). Complementation of arginine auxotrophy for genetic transformation of Coxiella burnetii by use of a defined axenic medium. Appl. Environ. Microb. 82, 3042–3051. doi: 10.1128/aem.00261-16
Sandoz, K. M., Sturdevant, D. E., Hansen, B., Heinzen, R. A. (2014). Developmental transitions of Coxiella burnetii grown in axenic media. J. Microbiol. Meth 96, 104–110. doi: 10.1016/j.mimet.2013.11.010
Satory, D., Gordon, A. J. E., Wang, M., Halliday, J. A., Golding, I., Herman, C. (2015). DksA involvement in transcription fidelity buffers stochastic epigenetic change. Nucleic Acids Res. 43, 10190–10199. doi: 10.1093/nar/gkv839
Sauer, J.-D., Bachman, M. A., Swanson, M. S. (2005). The phagosomal transporter A couples threonine acquisition to differentiation and replication of Legionella pneumophila in macrophages. P Natl. Acad. Sci. U.S.A. 102, 9924–9929. doi: 10.1073/pnas.0502767102
Schaik, E. J., Chen, C., Mertens, K., Weber, M. M., Samuel, J. E. (2013). Molecular pathogenesis of the obligate intracellular bacterium Coxiella burnetii. Nat. Rev. Microbiol. 11, 561–573. doi: 10.1038/nrmicro3049
Schneeberger, P. M., Wintenberger, C., van der Hoek, W., Stahl, J. P. (2014). Q fever in the Netherlands – 2007–2010: What we learned from the largest outbreak ever. Médecine Et Maladies Infect. 44, 339–353. doi: 10.1016/j.medmal.2014.02.006
Schwöppe, C., Winkler, H. H., Neuhaus, H. E. (2002). Properties of the glucose-6-phosphate transporter from Chlamydia pneumoniae (HPTcp) and the glucose-6-phosphate sensor from Escherichia coli (UhpC). J. Bacteriol 184, 2108–2115. doi: 10.1128/jb.184.8.2108-2115.2002
Seshadri, R., Paulsen, I. T., Eisen, J. A., Read, T. D., Nelson, K. E., Nelson, W. C., et al. (2003). Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proc. Natl. Acad. Sci. 100, 5455–5460. doi: 10.1073/pnas.0931379100
Shaw, E. I., Dooley, C. A., Fischer, E. R., Scidmore, M. A., Fields, K. A., Hackstadt, T. (2000). Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Mol. Microbiol. 37, 913–925. doi: 10.1046/j.1365-2958.2000.02057.x
Sherchand, S. P., Aiyar, A. (2019). Ammonia generation by tryptophan synthase drives a key genetic difference between genital and ocular Chlamydia trachomatis isolates. P Natl. Acad. Sci. U.S.A. 116, 12468–12477. doi: 10.1073/pnas.1821652116
Siegl, C., Prusty, B. K., Karunakaran, K., Wischhusen, J., Rudel, T. (2014). Tumor suppressor p53 alters host cell metabolism to limit Chlamydia trachomatis infection. Cell Rep. 9, 918–929. doi: 10.1016/j.celrep.2014.10.004
Sigar, I. M., Kaminski, A., Ito, B., Christoffersen-Cebi, J., Vidovich, A., Macarulay, C., et al. (2020). Comparison of in vitro Chlamydia muridarum infection under aerobic and anaerobic conditions. Curr. Microbiol. 77, 1580–1589. doi: 10.1007/s00284-020-01966-9
Singh, R., Liechti, G., Slade, J. A., Maurelli, A. T. (2020). Chlamydia trachomatis oligopeptide transporter performs dual functions of oligopeptide transport and peptidoglycan recycling. Infect. Immun. 88. doi: 10.1128/iai.00086-20
Sivaramakrishnan, P., Sepúlveda, L. A., Halliday, J. A., Liu, J., Núñez, M. A. B., Golding, I., et al. (2017). The transcription fidelity factor GreA impedes DNA break repair. Nature 550, 214–218. doi: 10.1038/nature23907
Sixt, B. S., Siegl, A., Müller, C., Watzka, M., Wultsch, A., Tziotis, D., et al. (2013). Metabolic features of Protochlamydia amoebophila elementary bodies – A link between activity and infectivity in chlamydiae. PloS Pathog. 9, e1003553. doi: 10.1371/journal.ppat.1003553
Sixt, B. S., Valdivia, R. H. (2016). Molecular genetic analysis of Chlamydia species. Annu. Rev. Microbiol. 70, 179–198. doi: 10.1146/annurev-micro-102215-095539
Skipp, P. J. S., Hughes, C., McKenna, T., Edwards, R., Langridge, J., Thomson, N. R., et al. (2016). Quantitative proteomics of the infectious and replicative forms of Chlamydia trachomatis. PloS One 11, e0149011. doi: 10.1371/journal.pone.0149011
Somerville, C. R., Ahmed, A. (1979). Mutants of Escherichia coli defective in the degradation of guanosine 5′-triphosphate, 3′-diphosphate (pppGpp). Mol. Gen. Genet. Mgg 169, 315–323. doi: 10.1007/bf00382277
Steinchen, W., Zegarra, V., Bange, G. (2020). (p)ppGpp: Magic modulators of bacterial physiology and metabolism. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.02072
Stephens, R. S., Kalman, S., Lammel, C., Fan, J., Marathe, R., Aravind, L., et al. (1998). Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282, 754–759. doi: 10.1126/science.282.5389.754
Suhan, M. L., Chen, S.-Y., Thompson, H. A. (1996). Transformation of Coxiella burnetii to ampicillin resistance. Journal of Bacteriology 178, 2701 2708. doi: 10.1128/jb.178.9.2701-2708.1996
Tehranchi, A. K., Blankschien, M. D., Zhang, Y., Halliday, J. A., Srivatsan, A., Peng, J., et al. (2010). The transcription factor DksA prevents conflicts between DNA replication and transcription machinery. Cell 141, 595–605. doi: 10.1016/j.cell.2010.03.036
Tello-Martin, R., Dzul-Rosado, K., Zavala-Castro, J., Lugo-Caballero, C. (2018). Approaches for the successful isolation and cell culture of American Rickettsia species. J. Vector Dis. 55, 258. doi: 10.4103/0972-9062.256560
Teysseire, N., Chiche-Portiche, C., Raoult, D. (1992). Intracellular movements of Rickettsia conorii and R. typhi based on actin polymerization. Res. Microbiol. 143, 821–829. doi: 10.1016/0923-2508(92)90069-z
Thomson, N. R., Holden, M. T. G., Carder, C., Lennard, N., Lockey, S. J., Marsh, P., et al. (2008). Chlamydia trachomatis: Genome sequence analysis of lymphogranuloma venereum isolates. Genome Res. 18, 161–171. doi: 10.1101/gr.7020108
Tipples, G., McClarty, G. (1993). The obligate intracellular bacterium Chlamydia trachomatis is auxotrophic for three of the four ribonucleoside triphosphates. Mol. Microbiol. 8, 1105–1114. doi: 10.1111/j.1365-2958.1993.tb01655.x
Tjaden, J., Winkler, H. H., Schwöppe, C., Laan, M. V. D., Möhlmann, T., Neuhaus, H. E. (1999). Two nucleotide transport proteins in Chlamydia trachomatis, one for net nucleoside triphosphate uptake and the other for transport of energy. J. Bacteriology 181, 1196–1202. doi: 10.1128/JB.181.4.1196-1202.1999
Tokarevich, N. K., Panferova, Y. A., Freylikhman, O. A., Blinova, O. V., Medvedev, S. G., Mironov, S. V., et al. (2018). Coxiella burnetii in ticks and wild birds. Ticks Tick-borne Dis. 10, 377–385. doi: 10.1016/j.ttbdis.2018.11.020
Tomaiuolo, S., Boarbi, S., Fancello, T., Michel, P., Desqueper, D., Grégoire, F., et al. (2020). Phylogeography of human and animal Coxiella burnetii strains: genetic fingerprinting of Q Fever in Belgium. Front. Cell Infect. Mi 10. doi: 10.3389/fcimb.2020.625576
Triboulet, S., Subtil, A. (2019). Make it a sweet home: responses of Chlamydia trachomatis to the challenges of an intravacuolar lifestyle. Microbiol. Spectr. 7. doi: 10.1128/microbiolspec.bai-0005-2019
Turco, J., Winkler, H. H. (1986). Gamma-interferon-induced inhibition of the growth of Rickettsia prowazekii in fibroblasts cannot be explained by the degradation of tryptophan or other amino acids. Infect. Immun. 53, 38–46. doi: 10.1128/iai.53.1.38-46.1986
Turcotte, M.-È., Buczinski, S., Leboeuf, A., Harel, J., Bélanger, D., Tremblay, D., et al. (2021). Epidemiological study of Coxiella burnetii in dairy cattle and small ruminants in Québec, Canada. Prev. Vet. Med. 191, 105365. doi: 10.1016/j.prevetmed.2021.105365
Vinella, D., Potrykus, K., Murphy, H., Cashel, M. (2012). Effects on growth by changes of the balance between GreA, GreB, and DksA suggest mutual competition and functional redundancy in Escherichia coli. J. Bacteriol 194, 261–273. doi: 10.1128/jb.06238-11
Voigt, A., Schöfl, G., Saluz, H. P. (2012). The Chlamydia psittaci genome: A comparative analysis of intracellular pathogens. PloS One 7, e35097. doi: 10.1371/journal.pone.0035097
Voss, O. H., Rahman, M. S. (2021). Rickettsia-host interaction: strategies of intra-cytosolic host colonization. Pathog. Dis. 79, ftab015. doi: 10.1093/femspd/ftab015
Voth, D. E., Heinzen, R. A. (2007). Lounging in a lysosome: the intracellular lifestyle of Coxiella burnetii. Cell Microbiol. 9, 829–840. doi: 10.1111/j.1462-5822.2007.00901.x
Walker, D. H., Ismail, N. (2008). Emerging and re-emerging rickettsioses: endothelial cell infection and early disease events. Nat. Rev. Microbiol. 6, 375–386. doi: 10.1038/nrmicro1866
Wang, Y., Kahane, S., Cutcliffe, L. T., Skilton, R. J., Lambden, P. R., Clarke, I. N. (2011). Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PloS Pathog. 7, e1002258. doi: 10.1371/journal.ppat.1002258
Warrier, I., Hicks, L. D., Battisti, J. M., Raghavan, R., Minnick, M. F. (2014). Identification of novel small RNAs and characterization of the 6S RNA of Coxiella burnetii. PloS One 9, e100147. doi: 10.1371/journal.pone.0100147
Weiss, E. (1965). Adenosine triphosphate and other requirements for the utilization of glucose by agents of the psittacosis-trachoma group. J. Bacteriol 90, 243–253. doi: 10.1128/jb.90.1.243-253.1965
Weiss, E., Myers, W. F., Dressler, H. R., Chun-Hoon, H. (1964). Glucose metabolism by agents of the psittacosis-trachoma group. Virology 22, 551–562. doi: 10.1016/0042-6822(64)90076-5
Weiss, E., Wilson, N. N. (1969). Role of exogenous adenosine triphosphate in catabolic and synthetic activities of Chlamydia psittaci. J. Bacteriol 97, 719–724. doi: 10.1128/jb.97.2.719-724.1969
Williams, J. C., Weiss, E. (1978). Energy metabolism of Rickettsia typhi: pools of adenine nucleotides and energy charge in the presence and absence of glutamate. J. Bacteriol. 134, 884–892. doi: 10.1128/jb.134.3.884-892.1978
Winkler, H. (1995). Rickettsia prowazekii, ribosomes and slow growth. Trends Microbiol. 3, 196–198. doi: 10.1016/s0966-842x(00)88920-9
Winkler, H. H., Daugherty, R. M. (1986). Acquisition of glucose by Rickettsia prowazekii through the nucleotide intermediate uridine 5’-diphosphoglucose. J. Bacteriol 167, 805–808. doi: 10.1128/jb.167.3.805-808.1986
Yang, C., Lei, L., Collins, J. W. M., Briones, M., Ma, L., Sturdevant, G. L., et al. (2021). Chlamydia evasion of neutrophil host defense results in NLRP3 dependent myeloid-mediated sterile inflammation through the purinergic P2X7 receptor. Nat. Commun. 12, 5454. doi: 10.1038/s41467-021-25749-3
Keywords: obligate, nutrient, genome streamlining, physiology, metabolism, auxotrophy
Citation: Mandel CG, Sanchez SE, Monahan CC, Phuklia W and Omsland A (2024) Metabolism and physiology of pathogenic bacterial obligate intracellular parasites. Front. Cell. Infect. Microbiol. 14:1284701. doi: 10.3389/fcimb.2024.1284701
Received: 28 August 2023; Accepted: 01 February 2024;
Published: 22 March 2024.
Edited by:
Matteo Bonazzi, Centre National de la Recherche Scientifique (CNRS), FranceReviewed by:
Janakiram Seshu, University of Texas at San Antonio, United StatesCopyright © 2024 Mandel, Sanchez, Monahan, Phuklia and Omsland. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anders Omsland, YW5kZXJzLm9tc2xhbmRAd3N1LmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.