- Division of Biology, Kansas State University, Manhattan, KS, United States
In insects, antibacterial immunity largely depends on the activation of downstream signaling and effector responses, leading to the synthesis and secretion of soluble effector molecules, such as antimicrobial peptides (AMPs). AMPs are acute infection response peptides secreted into the hemolymph upon bacterial stimulation. The transcription of innate immunity genes encoding for AMPs is highly dependent on several signaling cascade pathways, such as the Toll pathway. In the African malaria mosquito, Anopheles gambiae, AMPs hold a special interest as their upregulation have been shown to limit the growth of malaria parasites, bacteria, and fungi. Most of the current knowledge on the regulation of insect AMPs in microbial infection have been obtained from Drosophila. However, largely due to the lack of convenient assays, the regulation of antimicrobial activity in mosquito hemolymph is still not completely understood. In this study, we report a zone of inhibition assay to identify the contribution of AMPs and components of the Toll pathway to the antimicrobial activity of A. gambiae hemolymph. As a proof of principle, we demonstrate that Micrococcus luteus challenge induces antimicrobial activity in the adult female mosquito hemolymph, which is largely dependent on defensin 1. Moreover, by using RNAi to silence Cactus, REL1, and MyD88, we showed that Cactus kd induces antimicrobial activity in the mosquito hemolymph, whereas the antimicrobial activity in REL1 kd and MyD88 kd is reduced after challenge. Finally, while injection itself is not sufficient to induce antimicrobial activity, our results show that it primes the response to bacterial challenge. Our study provides information that increases our knowledge of the regulation of antimicrobial activity in response to microbial infections in mosquitoes. Furthermore, this assay represents an ex vivo medium throughput assay that can be used to determine the upstream regulatory elements of antimicrobial activity in A. gambiae hemolymph.
Introduction
Mosquitoes vector the causative agents of several infectious diseases worldwide (Molina-Cruz et al., 2016; Ramasamy and Surendran, 2016; Burkett-Cadena and Vittor, 2018). The Anopheles mosquito species mainly transmits Plasmodium parasites, the causative agents of malaria, one of the most widespread and devastating parasitic infections in humans (Molina-Cruz et al., 2016). In the absence of sterilizing vaccines, malaria prevention relies largely on vector control using insecticides and bed nets and, to a lesser extent, on drug treatment of most at risk populations (Lengeler, 1998; Killeen et al., 2007; Nosten and White, 2007). However, since the 1950s, insecticide resistance in mosquitoes has been reported towards different classes of insecticides, including pyrethroids, organophosphates, and carbamates (Hemingway et al., 2002; Goindin et al., 2017). Thus, new vector control strategies must be designed and implemented (Kleinschmidt et al., 2018). The mosquito immune system is a potential novel target for such alternative measures, as the immune response initiated in these insects during infection with vector-borne disease agents is a key determinant of vector competence (Huff, 1927; Collins et al., 1986; Vernick et al., 1995; Blandin et al., 2004; Osta et al., 2004; Michel et al., 2005; Habtewold et al., 2008; Edgerton et al., 2020).
To face the threat of infection due to the recurrent and diverse microbial exposure throughout their life cycle, mosquitoes deploy their innate immune system encompassing a variety of synergistic defense mechanisms (Levashina, 2004; Yassine et al., 2012; Tikhe and Dimopoulos, 2021). Binding of recognition molecules to microbe-specific molecules activates two principal arms of immune response: the humoral and cellular components. This immune response is mediated by factors that act as regulators and effectors of cellular responses—such as phagocytosis and encapsulation—and humoral responses—such as melanization and coagulation—among others (Lai et al., 2001; Lai et al., 2002; Dimopoulos, 2003; Levashina, 2004; Meister et al., 2005; Nakhleh et al., 2017). To identify and detect factors that contribute to antimicrobial immunity in A. gambiae, several assays have been developed, including the observation of phagocytosis in vivo (Levashina et al., 2001; Moita et al., 2005; Moita et al., 2006), evaluation of mosquito survival after injection of live bacteria (Blandin et al., 2002; Meister et al., 2005; Molina-Cruz et al., 2008; Gupta et al., 2009; Coggins et al., 2012; Upton et al., 2015), and quantification of bacterial proliferation after challenge (Schnitger et al., 2007; Warr et al., 2008; Schnitger et al., 2009; Yassine et al., 2012). However, these assays have a limited ability to distinguish between factors contributing to either or both arms of the immune response (Dimopoulos et al., 1997; Christophides et al., 2004; Levashina, 2004; Hillyer and Strand, 2014).
Recognition of microbial molecules also activates signaling cascades such as the Toll and Imd pathways, which, within hours after challenge, induce the synthesis of hundreds of immune-inducible molecules, including the well-characterized antimicrobial peptides (AMPs) (Levashina, 2004; Zakovic and Levashina, 2017). Knowledge on the components of the Toll pathway, as well as the interactions between these components in Drosophila, has served as a blueprint for studying it in mosquitoes. In Toll pathway signaling in A. gambiae, detection of pathogen-derived ligands by pattern recognition receptors (PRRs) triggers the proteolytic cleavage of the cytokine Spätzle, which binds to and activates the Toll receptor. This initiates signaling through the death-domain adaptor proteins MyD88, Tube, and Pelle, resulting in the phosphorylation and degradation of CACTUS, a negative regulator which binds to and inhibits the NF-κB transcription factor REL1 in the cytoplasm (Barillas-Mury et al., 1996; Cramer et al., 1999). CACTUS degradation allows REL1 to translocate into the nucleus to activate the transcription of Toll pathway-regulated genes, such as the AMPs Cecropin 1 (CEC1) and Defensin 1 (DEF1) (Frolet et al., 2006; Luna et al., 2006; Garver et al., 2009).
Upon immune induction, newly synthesized AMPs are secreted into the hemolymph where they rapidly reach micromolar concentrations, making them detectable in hemolymph after challenge (Lowenberger et al., 1999; Vizioli et al., 2001b). A variety of microorganisms have been used to induce AMP synthesis in the hemolymph of insect larvae and adults, with E. coli, M. luteus, Enterobacter cloaceae, and Pseudomonas aeruginosa being the most commonly used bacterial species (Hoffmann et al., 1981; Qu et al., 1982; Bulet et al., 1991; Sumida et al., 1992; Boulanger et al., 2002; Huberman et al., 2007a; Randolt et al., 2008). Quantification of AMP mRNA is commonly used as a measure for immune induction (Dickinson et al., 1988; Richman et al., 1996; Lemaitre et al., 1997; Lowenberger et al., 1999; Christophides et al., 2002; Dai et al., 2008; Zhang et al., 2017) and to test for the activity of the Toll and IMD pathways (Lemaitre et al., 1995; Meng et al., 1999; Meister et al., 2005; Frolet et al., 2006; Antonova et al., 2009; Meister et al., 2009; Zhong et al., 2012; Zhang et al., 2017). However, the relative contribution of signaling pathways that contribute to the expression of individual AMPs is largely unclear in mosquitoes (Meister et al., 2005; Frolet et al., 2006; Luna et al., 2006; Garver et al., 2009). Furthermore, the relationship between AMP mRNA and peptide levels can be complicated as demonstrated for defensin in A. aegypti (Bartholomay et al., 2004), and thus AMP mRNA levels may not correlate well with overall antimicrobial activity in mosquito hemolymph. AMPs can be purified instead from hemolymph or produced by recombinant DNA technologies to further study their biochemical properties and antimicrobial activity (Hultmark et al., 1983; Bulet et al., 1991; Richman et al., 1996; Otvos, 2000; Vizioli et al., 2001a; Buonocore et al., 2021). To quantify the antimicrobial activity of AMPs, in vitro antimicrobial sensitivity assays are commonly performed by measuring the zone of bacteria growth inhibition produced after immune stimulation in Antheraea pernyi (Qu et al., 1982), Trichoplusia ni (Andersons et al., 1990), Bombyx mori (Sumida et al., 1992), Stomoxys calcitrans (Boulanger et al., 2002), Apis mellifera carnica (Randolt et al., 2008), and Lucilia sericata (Huberman et al., 2007a). This method has allowed the quantification of antimicrobial activity of several AMPs such as cecropins, defensins, and gambicin in Anopheles gambiae (Vizioli et al., 2000; Vizioli et al., 2001b), attacins in Hyalophora cecropia (Hultmark et al., 1983), as well as other immune factors such as lysozymes in Gryllus bimaculatus and Heliothis virescens (Schneider, 1985; Lockey and Ourth, 1996).
In this study, we establish an assay that allows the detection of factors contributing specifically to humoral antimicrobial activity in the hemolymph of adult female mosquitoes. This assay is based on zone-of-inhibition measurements using M. luteus after bacterial challenge and/or reverse genetic manipulations. We use this assay to determine the contributions of (1) two antimicrobial peptides and (2) the Toll pathway to this antimicrobial activity. In addition, the data presented here show that this assay can also be used to detect immune priming.
Materials and methods
Mosquito strain and maintenance
The A. gambiae G3 strain (MRA-112) was reared as described previously (An et al., 2011). Briefly, mosquitoes were maintained at 27°C and 80% humidity in an environmental chamber set to 12:12-h light/dark cycle. L1 larvae were fed 2% (w/v) suspension of baker’s yeast (Fleischmann’s Active Dry Yeast, AB Mauri, St. Louis, MO, USA), and L2–L4 instars were fed on a 2:1 mixture of 2% powdered fish food (TetraMin® Tropical Flakes, Tetra, Melle, Germany) and baker’s yeast. Pupae were collected manually and placed in 173-oz plastic cages (CL Smith Co, St. Louis, MO, USA) covered with netting (JoAnn, Hudson, Ohio, USA). After emergence, adult mosquitoes were provided with 8% fructose ad libitum and heparinized horse blood (PlasVacc, Templeton, CA, USA) used as a blood meal source provided through an artificial membrane feeding system consisting of glass bells and a circulating water bath (Thermo Fisher Scientific, Waltham, MA, USA).
Bacterial and fungal strains
The following bacterial strains were used for bacterial challenge and zone of inhibition (ZOI) assays: Micrococcus luteus (lysodeikticus) (ATCC, no. 4698), live Staphylococcus aureus [strain PIG1; Liu et al., 2005)], and Escherichia coli (OP50, NCBI:txid637912). ZOI assays were also performed with Saccharomyces cerevisiae (baker’s active dried yeast from MP Biomedicals, Solon, OH, USA).
Bacterial challenges
To prepare the bacterial suspensions for mosquito challenges, lyophilized M. luteus (MilliporeSigma, Burlington, MA, USA) was resuspended in 1× phosphate-buffered saline (PBS) to an optical density of 600 nm (OD600) = 5. For live bacterial challenges, E. coli and S. aureus were cultured at 37°C in Luria-Bertani (LB) broth overnight, washed with 1× PBS, and resuspended in 1× PBS at the same optical density as mentioned above. The OD600 = 5 value corresponded to average doses of 8.5 × 108 and 9.7 × 108 colony-forming units (CFUs)/ml for live E. coli and S. aureus, respectively.
In all experiments, 2-day-old adult female mosquitoes were injected under the wing base with 50.6 nl of the bacterial challenge using a nanoinjector (Nanoject III, Drummond Scientific, Broomall, PA, USA). Sterile 1× PBS injections were used as negative controls. The experiments were performed in three independent replicates with 40 mosquitoes per treatment and per replicate.
Hemolymph collection
Unless otherwise stated, hemolymph was collected 24 h after challenge by adapting the method developed by League et al. (2017). Briefly, the lateral thorax of 40 CO2-anesthesized mosquitoes per treatment was punctured using a 30G × ½″ sterile needle. The freshly wounded mosquitoes were placed into a 0.6-ml microfuge tube whose bottom was perforated using an 18G × 1 ½× needle. This tube was then inserted into a 1.5-ml microfuge tube and centrifuged at 5,500 rpm for 5 min at 4°C. The collected hemolymph (~2 µl per treatment) was flash-frozen in liquid nitrogen immediately after collection and then stored at -80°C until use.
Zone of inhibition assays
To measure the antimicrobial activity of hemolymph, ZOI assays were performed as adapted from the protocol developed by League et al. (2017). Briefly, bacterial cultures were grown overnight in LB broth at 37°C in a shaking incubator to OD600 = 10, which corresponds to average doses of 3.5 × 1010, 7.4 × 1010, and 8.1 × 1010 CFUs/ml for E. coli, S. aureus, and M. luteus, respectively. Moreover, 500 ul of the bacterial culture was used to seed 5.5 ml of 1% LB agar kept in a 55°C water bath and then poured onto a 9-cm-diameter Petri dish (Fisher Scientific, Waltham, MA, USA). Cultures and plates using S. cerevisiae were prepared as described above, substituting LB for yeast extract–peptone–dextrose. The OD600 = 10 value for this yeast species corresponded to average doses of 2.6 × 109 CFUs/ml. After the agar had solidified, equidistant wells (1.5 cm from the outer edge) were punched in the agar by using a 1-mm-tip, sterile, 9″ glass Pasteur pipette (Fisher Scientific, Waltham, MA, USA). Each well was then loaded with 1 µl of hemolymph per treatment. Once the hemolymph was absorbed by the agar, usually within a couple of minutes, the plates were incubated upside down for 16 h at 37°C, unless otherwise stated. The collected hemolymph from each treatment and replicate was tested twice using different batches of bacterial culture; hence, six ZOI trials per treatment group were performed.
Bacterial plate imaging and ZOI data analysis
The plates were imaged at 16.1 pixels per millimeter using the Azure 300 imaging system (Azure Biosystems Inc., Dublin, CA, USA), using white epi-illumination without filters. Image processing and ZOI measurement were performed in Fiji is Just Image J (Fiji, Version 2.1.0) (Schindelin et al., 2012) using the following macro: “run(“8-bit”); run(“Subtract Background…”, “rolling=100 light”); run(“Auto Threshold”, “method=Default white”); run(“Convert to Mask”); run(“Analyze Particles…”, “size=400-Infinity pixel show=Outlines display”);”. Supplementary Figure S1 provides an overview of the entire procedure.
dsRNA synthesis and RNAi in female mosquitoes
dsRNA synthesis and adult female mosquito injections were performed as described previously (An et al., 2011). The templates for dsRNA were prepared by PCR using the T7-tagged primers listed in Supplementary Table S1. In all experiments, 2-day-old adult female mosquitoes were injected with 69 nl of 3 µg/µl dsRNA for single-knockdown (kd) experiments. For kd control, the mosquitoes were injected with the same quantity of the non-related dsRNA of green fluorescent protein (GFP). To examine the genetic interactions between REL1, MyD88, and CACTUS, a double-knockdown (dkd) was performed by injecting 138 nl of a 1:1 solution of 1.5 µg/ul of each dsRNA. For dkd controls, dsGFP was added to keep the total dsRNA dose constant at 414 ng/mosquito between treatment and controls as described previously (Zhang et al., 2016).
In experiments that required immune activation after gene kd, the mosquitoes were challenged 48 h after dsRNA injection with resuspended lyophilized M. luteus, live S. aureus, and E. coli, respectively, as described in the Bacterial Challenge Section Hemolymph was collected 24 h after challenge as described in the previous section.
Real-time quantitative polymerase chain reaction
To test for gene knockdown efficiency after dsRNA treatment, eight female mosquitoes per treatment and replicate were collected 48 h after dsRNA injection for RT-qPCR. Total RNA extraction and template cDNA synthesis were performed as described previously (Rhodes et al., 2018). Purified total RNA (100 ng) was used as the template to synthesize cDNA using iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA) in a total reaction volume of 20 μl, following the manufacturer’s instructions. RT-qPCR was performed using PowerUp SYBR Green Master Mix (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. Briefly, 2 μl of 1:5 diluted cDNA was added as template for a 20-μl volume reaction, followed by amplification on the Applied Biosystems™ 7500 Real-Time PCR System using the following amplification protocol: an initial cycle of 2 min at 50°C followed by 2 min at 95°C, 40 cycles of 15 s at 95°C, and 1 min at 60°C (detection). The primers used are listed in Supplementary Table S2.
The relative expression of the genes of interest was calculated according to Pfaffl (2001), considering the primer efficiencies (Supplementary Table S2). Ribosomal protein S7 (RpS7) expression was used as reference and dsGFP treatments as calibrator conditions. Three technical replicates were measured for each sample and primer pair from three biological replicates.
Statistical analyses
Statistical analyses were performed using GraphPad Prism 6.07 Software (GraphPad Software, USA). The area of bacteria growth inhibition (mm2) data was evaluated for normality of distribution using the Kolmogorov–Smirnov normality test. Differences in the zone of inhibition were analyzed using one-way ANOVA for multiple-treatment groups, with Bonferroni’s multiple-comparisons post-test (P < 0.05).
Results
Bacterial challenge induces antimicrobial activity in mosquito hemolymph
To establish a method to quantify the antimicrobial activity in A. gambiae, we performed ZOI assays in plates seeded with M. luteus using the hemolymph of mosquitoes after lyophilized M. luteus, live E. coli, and S. aureus challenges. We selected M. luteus for the challenge as it is a well-known inducer of immunity in several insect species (Nasr and Fallon, 2003; Huberman et al., 2007b; Kajla et al., 2010). In addition, we employed E. coli and S. aureus, which had been used in previous studies of mosquito antimicrobial immunity (Dimopoulos et al., 1997; Blandin et al., 2002; Garcia-Lara et al., 2005; Dong et al., 2006; Eleftherianos et al., 2006; Rahnamaeian et al., 2015). Injection of bacteria did not alter the mosquito mortality within the first 24 h post-injection compared with PBS-injected controls, with one to two mosquitoes dying within this time frame. The antimicrobial activity of hemolymph among all five treatment groups revealed significant differences (Figures 1A, B; one-way ANOVA, df = 4, F = 137.7, P < 0.0001). The hemolymph of unchallenged (UC) mosquitoes had low antimicrobial activity, producing a ZOI of 4 to 5 mm2. This activity was not increased by PBS injection, as the size of the ZOI did not statistically significantly differ from that of the UC mosquito hemolymph. When mosquitoes were challenged with either bacterial species, the ZOI was 1.9–3.4-fold larger than that of the control hemolymph, indicating a higher antimicrobial activity after challenge. Among the different challenges, the M. luteus challenge produced the largest ZOI, with a 1.5- and 1.6-fold increase in size compared with E. coli and S. aureus, respectively. Overall, the antimicrobial activity in mosquito hemolymph was increased significantly after the bacterial challenge, with M. luteus producing a higher antimicrobial activity compared with E. coli and S. aureus (Bonferroni’s multiple comparisons test, P < 0.0001 for both comparisons).
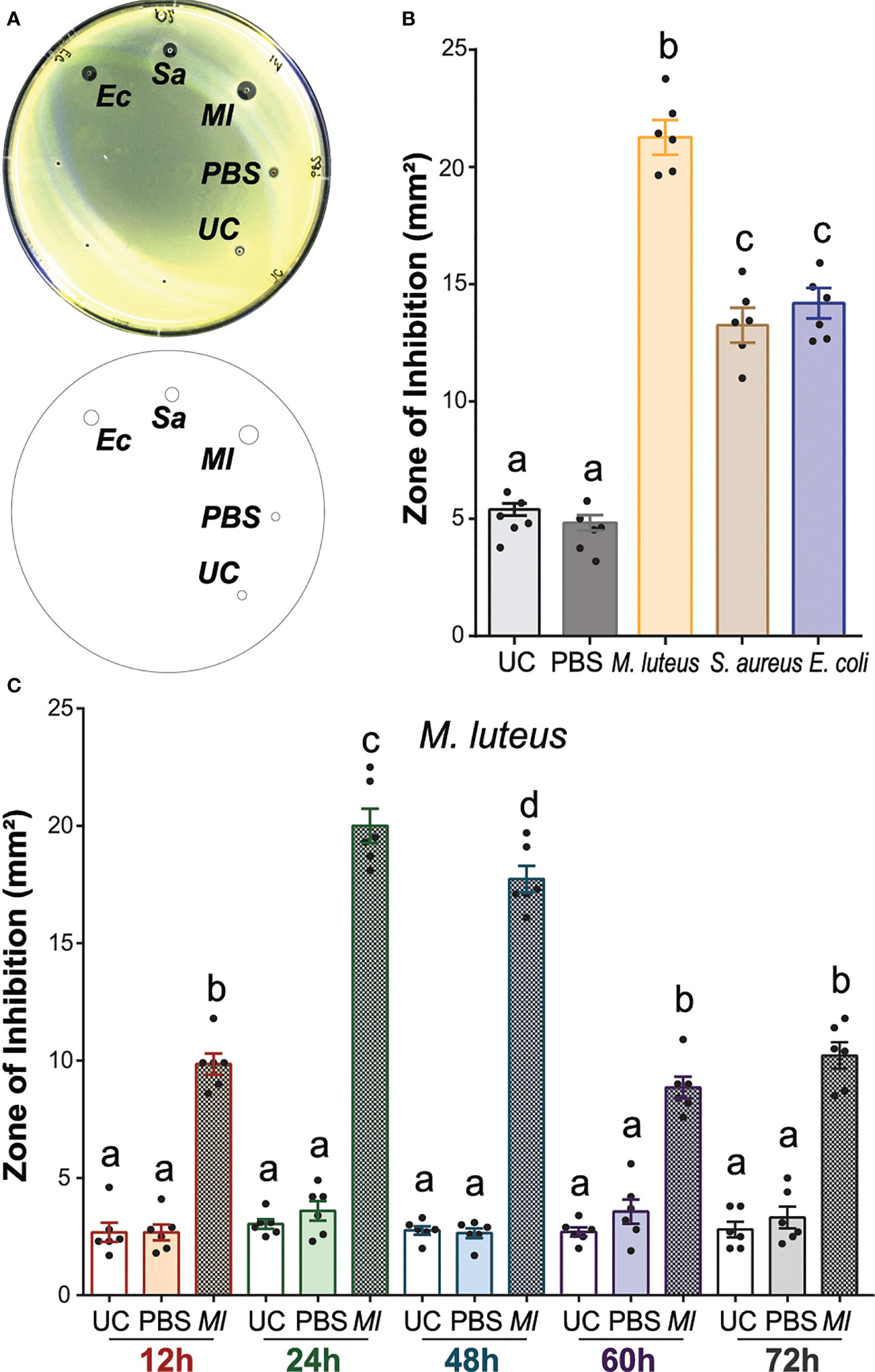
Figure 1 The antimicrobial activity of Anopheles gambiae hemolymph is induced by bacterial challenge. (A) Top: M. luteus-seeded plate showing the zone of inhibition (ZOI) produced by mosquito hemolymph collected 24 h after challenge with E coli, S. aureus, and M. luteus, respectively. The hemolymph of uninjected and phosphate-buffered-saline-injected mosquitoes acted as negative controls. Bottom: Drawing of the plate generated in Fiji is Image J to quantify the zone of inhibition (ZOI) for each treatment. (B) Zones of bacterial growth inhibition (mm2) were measured using customized macros in Fiji is Image J (C) The temporal variation of antimicrobial activity induction with M. luteus challenge was determined by collecting hemolymph at 12, 24, 48, 60, and 72 h after challenge. The ZOI was then measured as in (B). In (B) (C), data are shown as means with ±1 standard error (n = 6). One-way ANOVA, followed by Bonferroni’s multiple-comparison post-test, was performed to calculate the statistical significance (P < 0.05). Means with the same letter are not significantly different.
To determine whether the antimicrobial activity of hemolymph could inhibit the growth of other microbial species, we seeded the plates with M. luteus, E. coli, S. aureus, and S. cerevisiae, respectively, and tested the hemolymph of UC and M. luteus-challenged females. No ZOI was observed in plates seeded with E. coli, S. aureus, and S. cerevisiae, respectively (Supplementary Figure S2A). The plates seeded with M. luteus showed the same patterns of hemolymph-induced antibacterial activity as described above. To test whether the reduction in bacterial replication rates of E. coli, S. aureus, and S. cerevisiae would allow the detection of antimicrobial activity, we reduced the incubation temperature to 27°C, which is the typical temperature used in A. gambiae rearing. No ZOI was detected in either of these plates (Supplementary Figure S2B). Finally, we tested whether using the same bacterial species for both, challenging the mosquito and seeding the plates, would show inhibition of bacterial growth. For this, we collected hemolymph from mosquitoes challenged with E. coli and S. aureus, respectively, and tested those hemolymph samples in plates seeded with their corresponding bacterial species. No ZOI again was observed in either of these plates (Supplementary Figure S2C). Therefore, A. gambiae hemolymph only inhibited the growth of M. luteus, not E. coli, S. aureus, or S. cerevisiae, as measured by the ZOI assays. This was independent of the incubation temperature of the bacterial plates and the microbial challenge used to induce antimicrobial activity in hemolymph.
Antimicrobial activity in mosquito hemolymph remains elevated for 3 days post-challenge
To determine the longevity of the observed increase in antimicrobial activity in hemolymph, we next analyzed the temporal variation of the immune induction after M. luteus challenge, as this species produced the highest antimicrobial activity from the tested bacterial challenges (Figure 1C). For this, we collected hemolymph at 12, 24, 48, 60, and 72 h after M. luteus challenge and from uninjected mosquitoes (UC). Low antimicrobial activity was observed in all UC hemolymph samples, producing ZOI of 3 to 4 mm2 across all timepoints. PBS injection did not increase the antimicrobial activity at any time point. The antimicrobial activity of hemolymph was significantly increased after M. luteus challenge compared with UC and PBS-injected controls in every timepoint (one-way ANOVA, df = 14, F = 180, P < 0.0001, Bonferroni’s post-test, P < 0.0001). At the 12-h timepoint, M. luteus challenge produced a ZOI 2.7-fold larger than the UC. The antimicrobial activity after challenge peaked at 24 h, with a ZOI 5.6-fold larger than the UC and twofold larger than at the 12-h timepoint. The activity at 48 h after M. luteus injection was slightly reduced compared with the 24-h timepoint but remained high overall, with a ZOI 5.4-fold larger than the UC at the corresponding timepoint. At 60 and 72 h, the antimicrobial activity reached levels comparable with the 12-h timepoint, producing a ZOI 2.3- and 2.6-fold larger than its corresponding UC, but 55.8% and 50.1% smaller than challenge-induced hemolymph at 24 and 48 h, respectively. Overall, the antimicrobial activity in mosquito hemolymph was significantly induced after M. luteus challenge in all the timepoints that we tested. This activity peaked at 24 h and remained high at 48 h, followed by 2× decrease, after which the activity plateaued. These results together establish that changes in antimicrobial activity can be detected in the mosquito hemolymph and quantified using ZOI assays. This antimicrobial activity of hemolymph was induced with different bacterial challenges, of which M. luteus challenge induced the highest antimicrobial activity 24 h after bacterial injection.
Antimicrobial activity against Micrococcus luteus largely depends on defensin 1
The secretion of AMPs after microbial challenge increases the antimicrobial activity of insect hemolymph (Uvell and Engström, 2007). To test the contribution of AMPs on the antimicrobial activity of mosquito hemolymph, we used RNAi prior to M. luteus challenge to silence two known A. gambiae AMP genes, Defensin 1 (DEF1) and Cecropin 1 (CEC1), respectively (Figure 2). The kd efficiencies of DEF1 and CEC1 were measured by RT-qPCR 48 h after dsRNA injections (Supplementary Table S3). A comparison of the antimicrobial activity of hemolymph between treatment and control groups revealed significant differences (one-way ANOVA, df = 4, F = 202.1, P < 0.0001). As in the experiments detailed above, low antimicrobial activity was observed with the hemolymph of UC mosquitoes, producing a ZOI of 4 to 5 mm2. This activity did not increase in mosquitoes injected with dsGFP (dsRNA negative control), followed 48 h later by PBS (bacterial carrier) injection (Bonferroni’s post-test, P > 0.999). As expected, antimicrobial activity was highly increased (7.2-fold) when dsGFP-injected mosquitoes were challenged with M. luteus. dsDEF1 injection lowered the antimicrobial activity after challenge to levels comparable with the UC. Similarly, dsCEC1 injection also reduced the antimicrobial activity compared with dsGFP-injected, M. luteus-challenged mosquitoes. However, the antimicrobial activity after dsCEC1 injection remained 4.5-fold elevated compared with the UC, suggesting that DEF1 was mainly responsible of the antimicrobial activity against M. luteus in the mosquito hemolymph, while CEC1 contributed to a lesser degree.
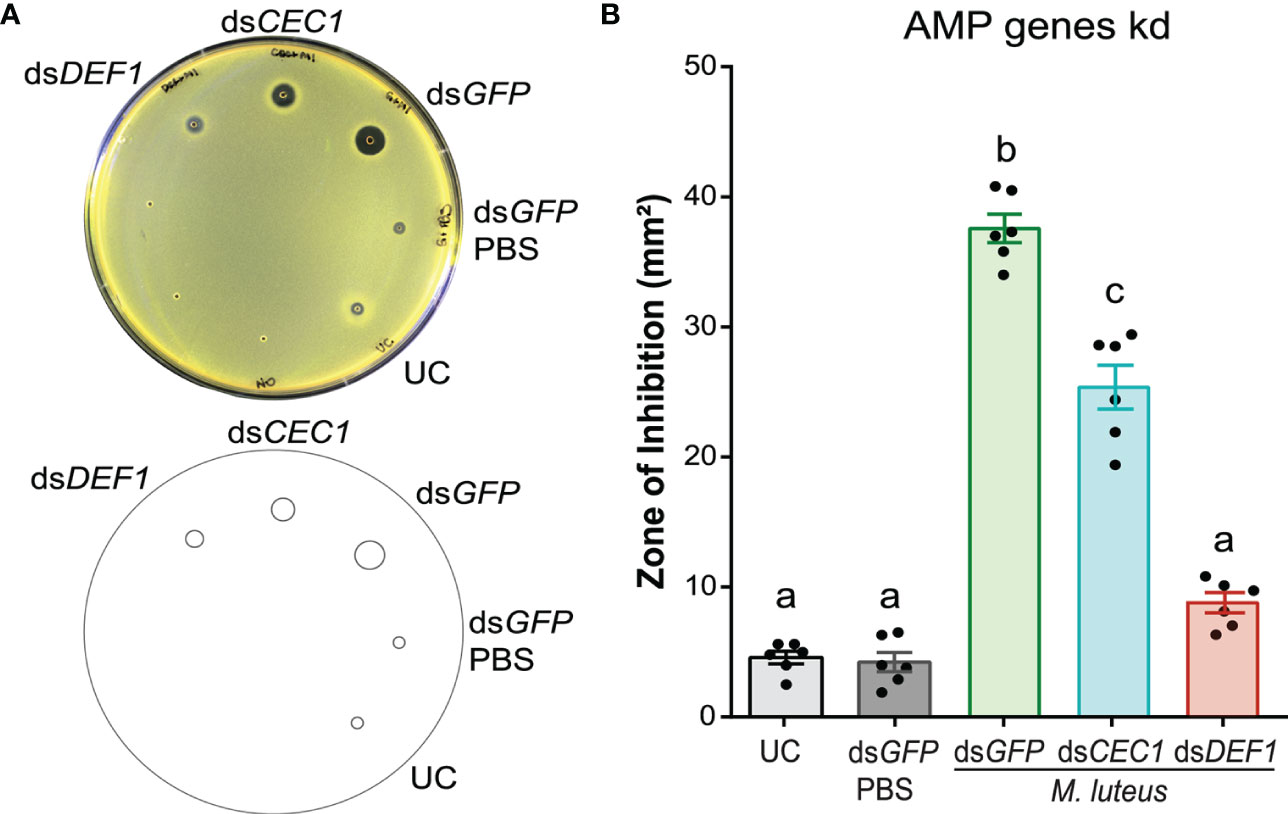
Figure 2 Defensin 1 is mainly responsible for the antimicrobial activity against M. luteus. (A) Top: M. luteus-seeded plate showing the zone of inhibition (ZOI) produced by the hemolymph of dsDEF1- and dsCEC1-injected mosquitoes. The mosquitoes were challenged 48 h after dsRNA injection with E coli, S. aureus, and M. luteus, respectively. Hemolymph was collected 24 h after challenge. Mosquitoes that were uninjected and injected with dsGFP prior to phosphate-buffered saline acted as negative controls. Bottom: Drawing of the plate generated in Fiji is Image J to quantify the ZOI for each treatment. (B) Zones of bacterial growth inhibition (mm2) were measured using customized macros in Fiji is Image J Data are shown as means with ±1 standard error (n = 6). One-way ANOVA, followed by Bonferroni’s multiple-comparison post-test, was performed to calculate the statistical significance (P < 0.05). Means with the same letter are not significantly different.
The Toll pathway contributes to antimicrobial activity in A. gambiae hemolymph
To identify the contribution of the Toll signal transduction pathway to the antimicrobial activity of A. gambiae hemolymph, we used RNAi to silence the three components of the pathway: REL1, MyD88, and Cactus (CACT) (Figure 3A). The kd efficiencies of REL1, CACT, and MyD88 were measured by RT-qPCR 48 h after dsRNA injections (Supplementary Table S3).
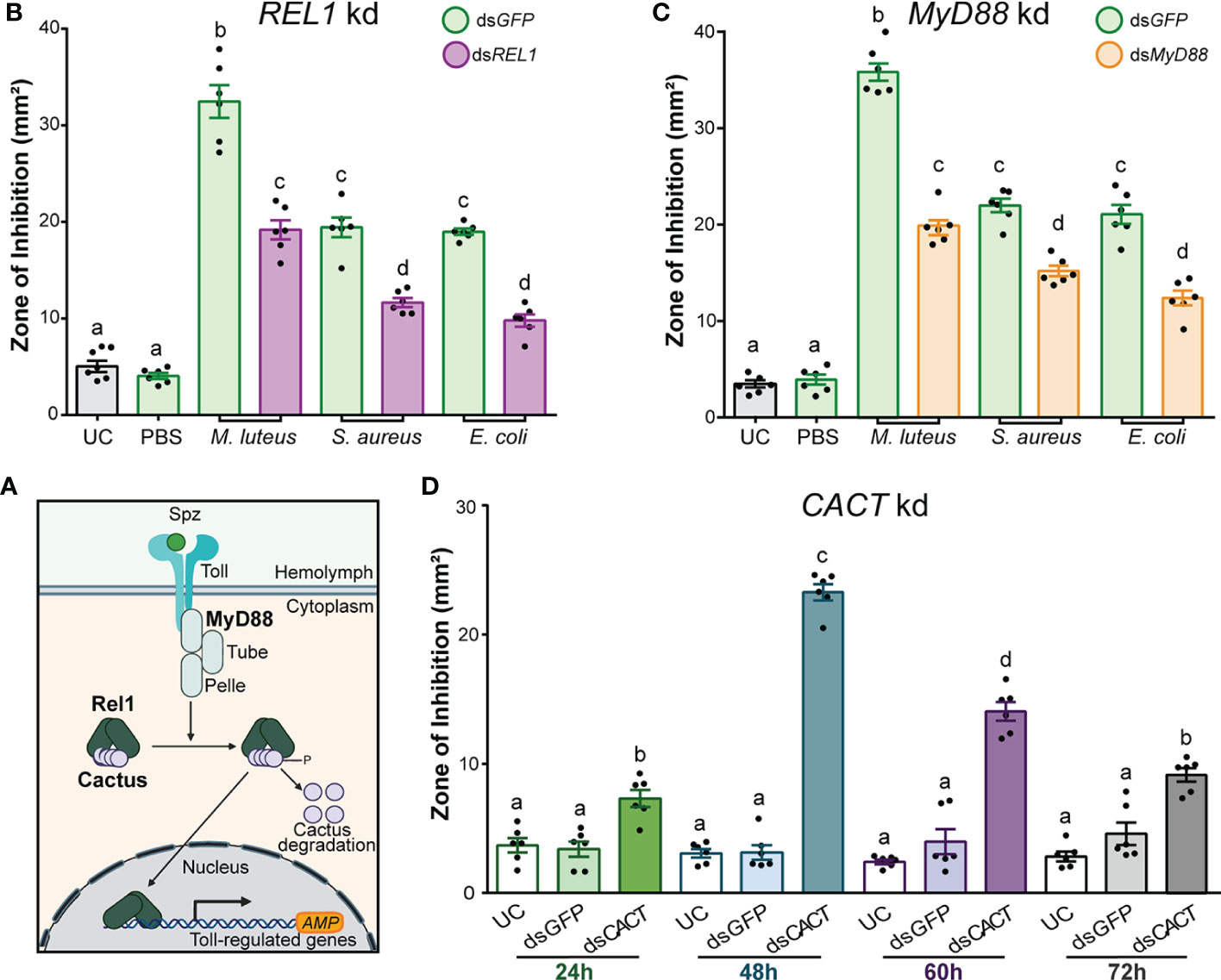
Figure 3 The antimicrobial activity of Anopheles gambiae hemolymph is regulated by the Toll pathway. (A) Overview of the Toll signal transduction pathway showing the known components that act as positive and negative regulators of the transcription of AMP genes in Anopheles gambiae. (B) Zone of inhibition produced by the hemolymph of dsREL1-injected (B) and dsMyD88-injected (C) mosquitoes. The mosquitoes were challenged 48 h after dsRNA injection with E coli, S. aureus, and M. luteus, respectively. Hemolymph was collected 24 h after challenge. Mosquitoes that were uninjected (UC) and injected with dsGFP prior to phosphate-buffered saline acted as negative controls, whereas those which had dsGFP injection prior to E coli, S. aureus, and M. luteus challenges acted as the corresponding positive controls. (D) The temporal variation of antimicrobial activity induction after CACT kd was determined by collecting hemolymph at 24, 48, 60, and 72 h after dsCACT injection. In (B), (C), and (D), data are shown as means ±1 standard error (n = 6). One-way ANOVA, followed by Bonferroni’s multiple-comparison post-test, was performed to calculate the statistical significance (P < 0.05). Means with the same letter are not significantly different.
Silencing REL1 (the NF-κB transcription factor purportedly downstream of TOLL) prior to M. luteus, E. coli, and S. aureus challenge significantly reduced the hemolymph antimicrobial activity compared with dsGFP-injected control mosquitoes (Figure 3B; one-way ANOVA, df = 7, F = 187, P < 0.0001; Bonferroni’s post-test, P < 0.0001 for all challenge comparisons). The bacterial challenge after dsGFP injection produced a several-fold increase in ZOI compared with UC hemolymph. Fold induction was highest after the challenge with M. luteus (5.5-fold), followed by S. aureus (2.9-fold) and E. coli (2.8-fold). Injection of dsREL1 reduced the fold induction of antimicrobial activity by 40%–50%, compared with the dsGFP-injected mosquitoes challenged with the same bacterial species. Therefore, the antimicrobial activity in the A. gambiae hemolymph was significantly reduced in REL1-silenced mosquitoes after the bacterial challenge independent of the microbial species that we tested. While not the focus of this current study, dsRNA injection targeting REL2, the transcription factor downstream of the IMD pathway in mosquitoes, reduced the antimicrobial activity in A. gambiae hemolymph similarly to REL1 kd (Supplementary Figure S3). The kd efficiency of REL2 was measured 48 h after dsRNA injections (Supplementary Table S3).
A similar effect was observed after silencing MyD88, one of the death domain adaptor proteins in the Toll intracellular signaling cascade (Figures 3A, C). Silencing MyD88 prior to M. luteus, E. coli, and S. aureus challenge produced a significantly lower antimicrobial activity than in the dsGFP-injected control mosquitoes (Figure 3C; one-way ANOVA, df = 7, F = 216.2, P < 0.0001; Bonferroni’s post-test, P < 0.0001 for all challenge comparisons). The M. luteus challenge after dsGFP injection produced a ZOI 9.3-fold larger than the UC, whereas the ZOI after dsMyD88 injection was only 4.7-fold larger. Thus, MyD88 kd reduced the ZOI by 44.5% compared with the dsGFP control. Similarly, injection of dsMyD88 reduced the fold induction of antimicrobial activity after the E. coli (43.6%) and S. aureus (46.3%) challenge compared with the dsGFP-injected mosquitoes challenged with the same bacterial species. Therefore, silencing MyD88 negatively impacted the antimicrobial activity in the hemolymph after the bacterial challenge, phenocopying REL1-silencing.
In its unphosphorylated state, the inhibitor of (I)κB CACT is bound to REL1, preventing it from entering the nucleus. Thus, removal of CACT leads to a constitutive activation of the Toll pathway (Frolet et al., 2006; Riehle et al., 2008; Garver et al., 2009) (Figure 3A). To determine the constitutive immune activation and to explore the temporal variation of antimicrobial activity after injection of dsCACT, we collected hemolymph at 24, 48, 60, and 72 h after dsRNA injections. The antimicrobial activity of hemolymph was significantly increased in dsCACT-injected mosquitoes compared with UC and dsGFP-injected controls at every timepoint (Figure 3D; one-way ANOVA, df = 11, F = 102.4, P < 0.0001, Bonferroni’s post-test, P < 0.0001). At the 24-h timepoint, hemolymph from dsCACT-injected mosquitoes produced a twofold larger ZOI than the hemolymph UC. The antimicrobial activity after dsCACT injection further increased at 48 h, with a ZOI 6.6-fold larger than the UC and 2.2-fold larger than at the 24-h timepoint. The activity at 60 h after dsCACT injection was reduced by 39.8% compared with the 48-h timepoint but remained high overall, with a ZOI 4.8-fold larger than the UC at the same timepoint. At 72 h, the antimicrobial activity was reduced to levels comparable with the 24-h timepoint, producing a ZOI 2.3-fold larger than the hemolymph of UC but 60.8% and 34.9% smaller than the hemolymph of dsCACT-injected mosquitoes at 48 and 60 h, respectively. Overall, these data confirmed that constitutive immune activation through dsCACT injection can be detected through ZOI assays. The antimicrobial activity peaked at 48 h after dsCACT injection, after which the activity plateaued but remained elevated for at least an additional 24 h.
To confirm that the elevated antimicrobial activity observed after dsCACT injection can be explained by canonical Toll signaling (Figure 3A), we tested the effect of co-silencing REL1 and CACTUS. The kd efficiency of CACT/REL1 was measured 48 h after dsRNA injections (Supplementary Table S3). The antimicrobial activity of hemolymph was significantly reduced in dsREL1/dsCACT-injected mosquitoes compared with the dsCACT-injected controls (Figure 4A; one-way ANOVA, df = 4, F = 382.2, P < 0.0001, Bonferroni’s post-test, P < 0.0001). Hemolymph antimicrobial activity did not increase by doubling the amount of injected dsGFP nor by injecting dsRELl1/dsGFP compared with the UC (Bonferroni’s post-test, P > 0.999). Similar to the previously observed increases in ZOI in the single-knockdown assays (Figure 3A), the antimicrobial activity was increased 6.1-fold in hemolymph from dsCACT/dsGFP-injected mosquitoes compared with the UC. Co-injection of dsCACT and dsREL1 reduced the ZOI by 45% when compared with dsCACT/dsGFP-injected mosquitoes, demonstrating that REL1 is required for the antimicrobial activity induced by dsCACT injection.
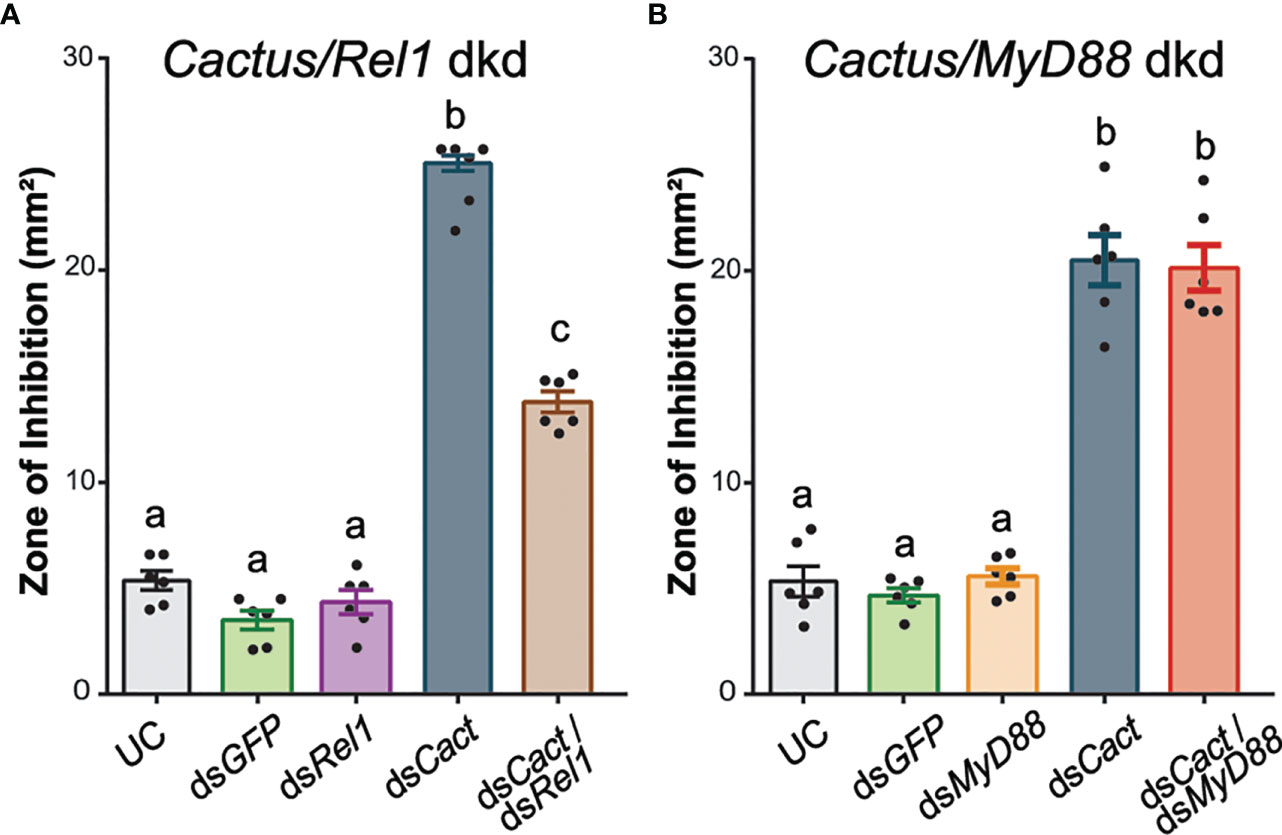
Figure 4 DsREL1 injection partially reverses the antimicrobial activity induced by CACT kd. (A) Zone of inhibition (ZOI) produced by the hemolymph of mosquitoes injected with dsREL1/dsGFP, dsCACT/dsGFP, and dsCACT/dsREL1. Uninjected and dsGFP-injected mosquitoes acted as negative controls. (B) ZOI produced by the hemolymph of mosquitoes injected with dsMyD88/dsGFP, dsCACT/dsGFP, and dsCACT/dsMyD88. Uninjected (UC) and dsGFP-injected mosquitoes acted as negative controls. Data are shown as means ±1 standard error (n = 6). One-way ANOVA, followed by Bonferroni’s multiple-comparison post-test, was performed to calculate the statistical significance (P < 0.05). Means with the same letter are not significantly different.
In contrast, the antimicrobial activity in hemolymph collected from mosquitoes co-injected with dsMyD88 and dsCACT was comparable with that of hemolymph from dsCACT/dsGFP-injected mosquitoes (Figure 4B; one-way ANOVA, df = 4, F = 103.0, P < 0.0001, Bonferroni’s post-test, P > 0.999). The kd efficiency of CACT/MyD88 was measured 48 h after dsRNA injections (Supplementary Table S3). The ZOI produced by hemolymph collected from dsMyD88/dsGFP-injected mosquitoes was not statistically significantly different from that of UC mosquitoes (Bonferroni’s post-test, P > 0.999). The antimicrobial activity after dsCACT/dsGFP injections was increased compared with that of UC hemolymph and not statistically significantly different from that of dsCACT/dsMyD88-injected mosquitoes. Thus, dsMyD88 injection did not affect the antimicrobial activity induced by dsCACT injection.
These data altogether demonstrate that ZOI assays provide a convenient means to evaluate the contribution of the Toll pathway to the antimicrobial activity in A. gambiae hemolymph. Our data demonstrate that REL1 and MyD88 are required for the antimicrobial activity against M. luteus and that this activity is inhibited by CACT. In addition, the dynamic range of the assay is sufficient to perform epistatic analyses and confirm that MyD88 is upstream of CACT in the Toll pathway in A. gambiae.
Injection induces a priming response
Across all the experiments that required immune activation via bacterial challenge, we consistently observed a larger ZOI when dsGFP was injected prior to challenge, suggesting a priming response. To determine whether the injury itself or the presence of dsRNA was responsible for this effect, we injected mosquitoes with either dsGFP resuspended in sterile deionized water (H2O) or H2O only, followed by M. luteus challenge (Figure 5). The antimicrobial activity of the hemolymph of UC mosquitoes was low and did not increase in the hemolymph of H2O- or dsGFP-injected mosquitoes that were injected 48 h later with PBS (one-way ANOVA, df = 5, F = 235.3, P < 0.0001, Bonferroni’s post-test, P > 0.999). As demonstrated previously (Figures 1B, C), M. luteus challenge significantly increased the antimicrobial activity (threefold) in hemolymph compared with the UC (Bonferroni’s post-test, P < 0.0001). Interestingly, the antimicrobial activity in the hemolymph of H2O- and dsGFP-injected mosquitoes subsequently challenged with M. luteus was significantly higher compared with that of mosquitoes only challenged with M. luteus (41.7% and 44.5%, Bonferroni’s post-tests, P < 0.0001 for both). The antimicrobial activity was comparable between the hemolymph of H2O- and dsGFP-injected mosquitoes subsequently challenged with M. luteus (Bonferroni’s post-test, P > 0.999). These results demonstrate that antimicrobial activity induction in mosquito hemolymph is primed by injection. This effect, however, is not exacerbated by the injection of dsRNA molecules.
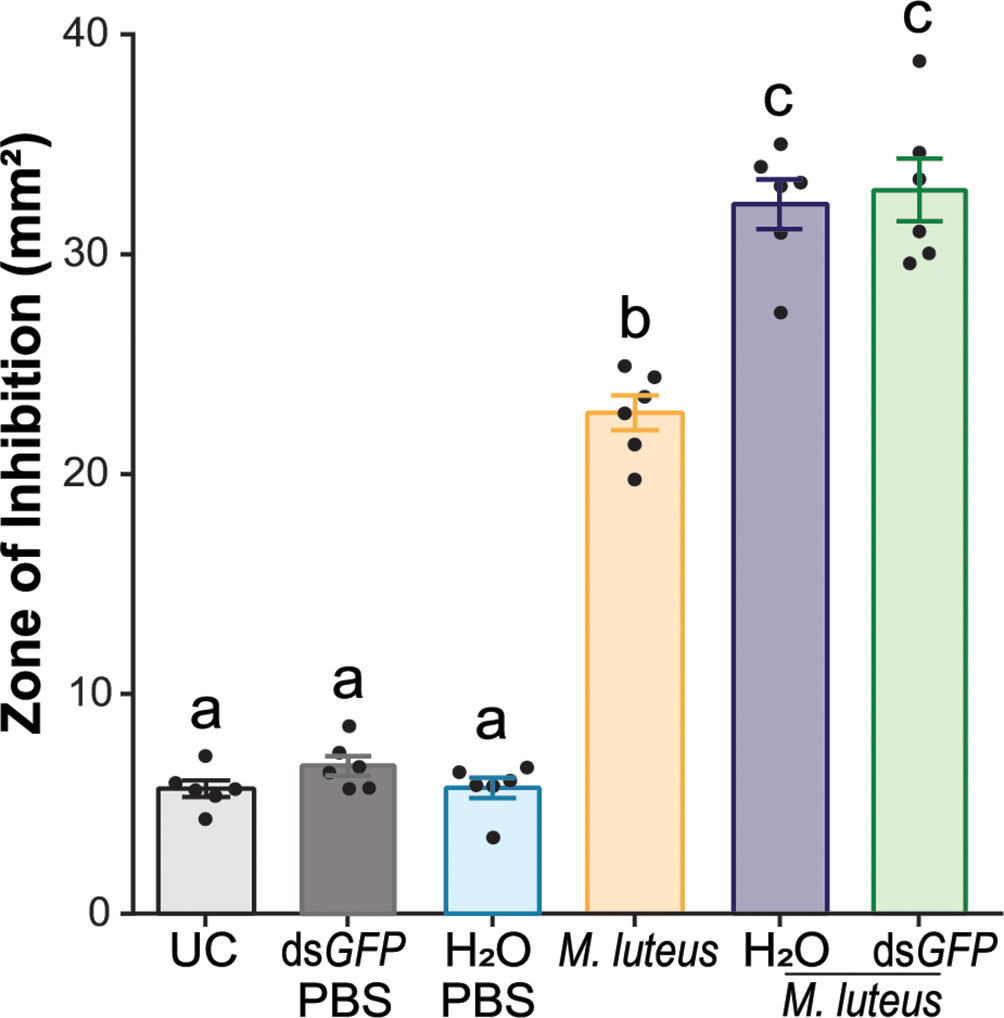
Figure 5 Intrathoracic injection enhances the efficacy of bacterial growth inhibition after bacterial challenge. The graph shows the zone of inhibition (ZOI) produced by the hemolymph of mosquitoes injected with sterile deionized water (H2O) or dsGFP, followed by phosphate-buffered saline injection, M. luteus challenge only, and H2O and dsGFP followed by M. luteus challenge. The mosquitoes were challenged 48 h after dsRNA or H2O injection. The hemolymph of uninjected mosquitoes acted as negative controls. Data are shown as means ±1 standard error (n = 6). One-way ANOVA, followed by Bonferroni’s multiple-comparison post-test, was performed to calculate the statistical significance (P < 0.05). Means with the same letter are not significantly different.
Discussion
Using ZOI assays to quantify humoral antimicrobial activity in A. gambiae
In this study, we propose a new application of ZOI assays based on reverse genetics approach, which aims to identify the contribution of specific immune factors to the antimicrobial activity of A. gambiae hemolymph. Previous assays to identify factors that contribute to antimicrobial immunity in A. gambiae have largely relied on in vivo assays, including mosquito survival after injection of live bacteria (Blandin et al., 2002; Gupta et al., 2009; Coggins et al., 2012), observations of phagocytosis in vivo (Moita et al., 2005), and measurements of bacterial proliferation after challenge (Schnitger et al., 2007; Yassine et al., 2012). However, survival and bacterial proliferation are dependent on both cellular and humoral immune responses (Dimopoulos et al., 1997; Christophides et al., 2004; Levashina, 2004; Hillyer and Strand, 2014), and thus these assays are not able to distinguish between factors contributing to phagocytosis, humoral immunity, or both. ZOI assays specifically measure the humoral arm of the antimicrobial immune response and thus provide a much-needed tool for the analysis of immunity in A. gambiae.
The humoral antimicrobial activity that we detected in the ZOI assays was not specific to bacterial species that were used in the challenges. Indeed all challenges that we employed—lyophilized M. luteus, live E. coli, and S. aureus—induced antimicrobial activity against M. luteus, further supporting the literature that different microbial challenges induce an overlapping repertoire of humoral innate immune responses that broadly defend against an array of bacterial species (Coggins et al., 2012; Rhodes et al., 2018).
Despite our efforts, we could, however, not establish similar ZOI assays against other microbial species, including E. coli, S. aureus, and S. cerevisiae. This parallels the findings by League et al. (2017), where the hemolymph of A. gambiae larvae also was only able to limit the growth of M. luteus and not E. coli. Similarly, the hemolymph of adult Tenebrio molitor did not limit the growth of E. coli but only that of Arthrobacter globiformis, and M. luteus was not tested in this study (Haine et al., 2008). The authors speculated that the replication rate of the microbe tested may be responsible for these observations, suggesting that fast-replicating species in this in vitro assay outpace the antimicrobial activity of hemolymph, thus precluding the measurements of ZOI (Haine et al., 2008; League et al., 2017). However, when we reduced the incubation temperature to slow down bacterial replication, we still could not observe a ZOI. Another potential explanation for the lack of ZOIs against E. coli in A. gambiae hemolymph may be the more limited sensitivity of the ZOI assay when using microbial species other than M. luteus. Such limited sensitivity would disproportionately affect ZOI assays using insect species with a small individual hemolymph volume. Indeed several studies that use hemolymph from larger insect species, including Manduca sexta, Hyalophora cecropia, Antheraea mylitta, Reticulitermes flavipes, and Diatraea saccharalis, have indeed detected ZOIs in plates seeded with E. coli, Staphylococcus aureus, and Klebsiella pneumoniae (Hultmark et al., 1983; Booth et al., 2015; Rocha et al., 2016; Zeng et al., 2016; Chowdhury et al., 2021). Given the size of mosquitoes and the volume of hemolymph that we collected and used for the assay, it is possible that using higher volumes of hemolymph would show bacterial growth inhibition in plates. Supplementary Table S4 summarizes the uses and limitations of ZOI assays compared with other assays commonly used to assess humoral antimicrobial activity in mosquitoes. In the future, the sensitivity of ZOI assays using A. gambiae hemolymph may be further increased by adding commercial chicken egg white lysozyme into the E. coli-seeded agarose, as demonstrated by Chalk et al. (1994).
AMPs are responsible for the antimicrobial activity in mosquito hemolymph
Our data show that performing gene knockdown by dsRNA injection prior to the ZOI assays can be used to identify the major factors underlying the antimicrobial activity in A. gambiae. We tested the contribution of AMP genes to antimicrobial activity and showed that, in the hemolymph of adult female A. gambiae, DEF1 is largely responsible for the antimicrobial activity against M. luteus. This ex vivo observation of the antimicrobial activity of DEF1 using ZOI assays is consistent with in vitro results of the antimicrobial activity spectrum of recombinant DEF1. Most of the Gram-positive bacterial species tested were sensitive in a range of concentrations from 0.1 to 0.75 μM of recombinant DEF1. At 1 µM, DEF1 exhibited a strong bactericidal effect on M. luteus, killing almost all bacteria within 60 s (Vizioli et al., 2001b). In addition, our data corroborate the DEF1 kd findings by Blandin et al. (2002) that showed that this AMP is required for the mosquito antimicrobial defense against Gram-positive bacteria in female A. gambiae. In this study, DEF1 depletion rendered the mosquitoes more susceptible to M. luteus (20% lethality) and highly susceptible to S. aureus (80% lethality) infections.
In addition to DEF1, we also found that CEC1 contributed to antimicrobial activity in A. gambiae hemolymph, albeit to a lesser extent than DEF1. CEC1 is produced in bacteria-inoculated adults, where mature CEC1 can be found in two forms—C-terminally amidated and glycine-extended (non-amidated) forms—showing minimal differences in antimicrobial spectrum (Vizioli et al., 2000). Both forms of CEC1 show a strong bactericidal effect on M. luteus, with 0.5–1 and 1–2.5 uM minimal inhibitory concentrations (MIC) for the amidated and glycine-extended forms, respectively (Vizioli et al., 2000). The in vitro bactericidal effects of DEF1 and CEC1 on M. luteus are similar and thus do not explain the smaller ZOIs that we observed after CEC1 kd. The underlying cause of this difference is currently unclear. It is possible that this difference can be attributed to differences in kd levels, which could be addressed by producing CRISPR-knockout strains. An alternative and currently equal parsimonious explanation is that the DEF1 and CEC1 peptide levels differ in the hemolymph of adult female A. gambiae after M. luteus challenge.
Overall, our results provide further evidence that not only AMPs are required for antimicrobial activity in A. gambiae hemolymph but also the induction of antimicrobial activity against M. luteus after bacterial challenge is largely explained by DEF1.
Humoral antimicrobial activity in A. gambiae hemolymph is regulated by the Toll pathway
Our results confirm previous reports that the Toll pathway is the major regulator of antimicrobial activity in the mosquito hemolymph through several lines of evidence (Christophides et al., 2002). First, humoral antimicrobial activity was reduced in the hemolymph of bacterially challenged adult female insects injected with dsREL1. This confirms the results in previous studies in A. gambiae that identified AMPs, including DEF1, as REL1-regulated immune genes (Frolet et al., 2006; Luna et al., 2006). Additionally, our data show that the antimicrobial activity in the mosquito hemolymph increased after silencing CACT. This increase is explained by the gene overexpression of several AMPs, including CEC1, DEF1, Gambicin 1 (GAM1), and CEC3 induced by the silencing of CACT in A. gambiae (Garver et al., 2009). This increase in antimicrobial activity is partially reversed in dsCACT/dsREL1-injected mosquitoes, further supporting that the increased activation of humoral antimicrobial activity by CACT depletion is dependent on the downstream NF-kB transcription factor REL1 as shown previously at the transcriptional level (Garver et al., 2009). Our data parallel previous findings on antimicrobial activity against the fungal entomopathogen Beauveria bassiana as well as on antiparasitic activity against the rodent malaria parasite Plasmodium berghei, both of which can be boosted by CACT kd in a REL1-dependent manner (Frolet et al., 2006; Rhodes et al., 2018).
Furthermore, to our knowledge, we provide the first evidence of the death-domain adapter protein MyD88 to be required for humoral antimicrobial immunity in A. gambiae. In D. melanogaster, MyD88 is a component of the Toll pathway, acting upstream of CACT (Horng and Medzhitov, 2001; Tauszig-Delamasure et al., 2002). MyD88 in A. gambiae was first identified as a 1:1 ortholog during the initial genome annotation of the A. gambiae PEST strain (Christophides et al., 2002). An injection of dsMyD88 reduced the antimicrobial activity of A. gambiae hemolymph after bacterial challenge, phenocopying the effect of dsREL1 injection. However, injection of dsMyD88 did not impact the dsCACT phenotype in dsCACT/dsMyD88-injected mosquitoes, supporting a placement of MyD88 in the Toll pathway upstream of CACT.
These data together provide strong evidence that the Toll pathway directly regulates humoral anti-bacterial activity in the hemolymph of adult A. gambiae. Importantly, the data also show that the dynamic range of the A. gambiae hemolymph ZOI assays is sufficient to perform epistasis-like analyses by dsRNA injections. The latter will be invaluable to decipher regulatory components of the Toll pathway upstream of its receptor.
Bacteria challenge induces the highest antimicrobial activity 24 h after bacterial injection
While not the focus of our study, measurement of antimicrobial activity after bacterial challenge and dsCACT injection revealed the temporal nature of the humoral immune responses to bacterial challenge in adult female A. gambiae. In our study, hemolymph antimicrobial activity, upon bacterial challenge, reached maximal levels between 24 and 48 h and peaked at 48 h after dsCACT-injection. Several studies previously described the rapid induction of RNA levels of DEF1, CEC1, and GAM1 in adult A. gambiae (Dimopoulos et al., 1997; Vizioli et al., 2000; Vizioli et al., 2001a; Vizioli et al., 2001b). In addition, previous reports from other insects such as Zophobas atratus and Bombus terrestris show that the time at which hemolymph antimicrobial activity peaks after a challenge varies between 24 and 48 h (Bulet et al., 1991; Korner and Schmid-Hempel, 2004).
Furthermore, we observed a persistence of elevated antimicrobial activity for at least up to 72 h. Antimicrobial activity lasting at least 5 days has been observed in the butterfly Pieris brassicae and the wax moth Galleria mellonella (Jarosz, 1993) and up to 44 days after challenge in the dragonfly Aeschna cyanea (Bulet et al., 1992). In A. gambiae, E. coli challenge stably induces the expression of DEF1 mRNA over a period of at least 8–12 days (Gorman and Paskewitz, 2000; Blandin et al., 2002), whereas the gene expression of DEFA, the 1:1 ortholog of DEF1 in the mosquito Aedes aegypti, remains abundant for at least 21 days (Lowenberger et al., 1999). However, whether this elevated gene expression is translated to sustained elevated peptide levels is currently unknown, especially because data on A. aegypti show that the expression of DEFA is regulated post-transcriptionally and that elevated mRNA levels do not translate to elevated peptide levels (Bartholomay et al., 2004).
Future studies will have to determine whether the persistence of antimicrobial activity in A. gambiae after M. luteus challenge is due to continued elevated expression levels and/or the persistence of certain antimicrobial peptides.
Injection is a stressor that triggers antimicrobial activity
The lasting effect of antimicrobial activity in the hemolymph suggests that, in secondary challenges, initial AMP levels may be elevated compared with those in hemolymph from naïve mosquitoes. Our results show that increased antimicrobial activity upon a secondary challenge is a consequence of previously activated defenses. This phenomenon of immune memory, reliant on the innate immune system, is referred to as immune priming or trained immunity (Boman et al., 1972; Moret and Schmid-Hempel, 2001; Apidianakis et al., 2005; Schmid-Hempel, 2005; Pham et al., 2007; Sun et al., 2009; Netea et al., 2011; Christofi and Apidianakis, 2013).
Immune priming in A. gambiae has been described in response to injury, bacterial challenge, and malaria parasite infection (Rodrigues et al., 2010; Powers et al., 2020). Injury or bacterial challenge leads, in secondary bacterial infections, to a decreased bacterial load and increased survival in adult female A. gambiae (Powers et al., 2020). Similarly, malaria parasite infection loads were decreased if mosquitoes were previously infected with the same parasite species (Rodrigues et al., 2010). While the molecular mechanisms underlying the priming effect of bacteria or injury is unknown, priming by parasite infection is mediated by hemocyte differentiation (Ramirez et al., 2014).
Our data suggest that priming by injury is, at least in part, based on humoral factors, as the ZOI assays are cell-free. While we cannot exclude that injury is accompanied by the introduction of a small number of environmental bacteria from the mosquito surface or environmental bacteria present in dH2O or dsGFP, the priming effect that we observed is not explained simply by the continued elevation of AMP levels, as injury through either injection of water or dsRNA did not increase the antimicrobial activity upon the initial injury itself or subsequent secondary injury. Increased antimicrobial activity was instead only observed after secondary bacterial challenge following the initial injury. Future studies will have to determine the molecular underpinnings of this injury-based priming and will have to rely on reverse genetics methodology that does not require injury.
In conclusion, the novel use of ZOI assays that we propose in this study represents an ex vivo low-biosafety-level, medium-throughput assay that can be used to provide information on the regulation of antimicrobial activity in response to microbial infections in mosquitoes. Modifications to this assay, such as the use of a device that will allow us to produce uniform and equally spaced wells at the same time, will further streamline the generation of plates. Alternatively, using a MIC assay based on broth dilution methods may allow us to measure the differences in antimicrobial activity with bacterial species besides M. luteus. Importantly, beyond differences in antimicrobial activity after silencing the genes involved in the intracellular cascade of Toll pathway, this assay can be extended to determine the extracellular regulatory elements of antimicrobial activity in A. gambiae hemolymph.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
BM and KM conceived the original idea and designed the experiments. BM performed the experiments and analyzed the data. KM supervised the project. BM and KM drafted the manuscript and contributed to the final version of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by funding from the National Institutes of Health (grant number R01AI140760) and USDA National Institute of Food and Agriculture Hatch (project 1021223). This is contribution no. 22-131-J from the Kansas Agricultural Experiment Station. The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the funding agencies.
Acknowledgments
BM and KM thank all members of the Michel laboratory for their help with insect rearing, laboratory maintenance, and fruitful scientific discussions. In addition, the authors thank Dr. Michi Tobler, Division of Biology, Kansas State University, for the use of their photography setup to take color images of ZOI plates.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.891577/full#supplementary-material
References
An, C., Budd, A., Kanost, M. R., Michel, K. (2011). Characterization of a regulatory unit that controls melanization and affects longevity of mosquitoes. Cell Mol. Life Sci. 68, 1929–1939. doi: 10.1007/s00018-010-0543-z
Andersons, D., Gunne, H., Hellers, M., Johansson, H., Steiner, H. (1990). Immune responses in Trichoplusia ni challenged with bacteria or baculoviruses. Insect Biochem. 20, 537–543. doi: 10.1016/0020-1790(90)90037-U
Antonova, Y., Alvarez, K. S., Kim, Y. J., Kokoza, V., Raikhel, A. S. (2009). The role of NF-κB factor REL2 in the. Aedes aegypti Immune response Insect Biochem. Mol. Biol. 39, 303–314. doi: 10.1016/j.ibmb.2009.01.007
Apidianakis, Y., Mindrinos, M. N., Xiao, W., Lau, G. W., Baldini, R. L., Davis, R. W., et al. (2005). Profiling early infection responses: Pseudomonas aeruginosa eludes host defenses by suppressing antimicrobial peptide gene expression. PNAS 102, 2573–2578. doi: 10.1073/pnas.0409588102
Barillas-Mury, C., Charlesworth, A., Gross, I., Richman, A., Hoffmann, J. A., Kafatos, F. C. (1996). Immune factor Gambif1, a new rel family member from the human malaria vector, Anopheles gambiae. EMBO J. 15, 4691–4701. doi: 10.1002/j.1460-2075.1996.tb00846.x
Bartholomay, L. C., Fuchs, J. F., Cheng, L.-L., Beck, E. T., Vizioli, J., Lowenberger, C., et al. (2004). Reassessing the role of defensin in the innate immune response of the mosquito, Aedes aegypti. Insect Mol. Biol. 13, 125–132. doi: 10.1111/j.0962-1075.2004.00467.x
Blandin, S., Moita, L. F., Köcher, T., Wilm, M., Kafatos, F. C., Levashina, E. A. (2002). Reverse genetics in the mosquito Anopheles gambiae: targeted disruption of the Defensin gene. EMBO Rep. 3, 852–856. doi: 10.1093/embo-reports/kvf180
Blandin, S., Shiao, S.-H., Moita, L. F., Janse, C. J., Waters, A. P., Kafatos, F. C., et al. (2004). Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell 116, 661–670. doi: 10.1016/s0092-8674(04)00173-4
Boman, H. G., Nilsson, I., Rasmuson, B. (1972). Inducible antibacterial defence system in Drosophila. Nature 237, 232–235. doi: 10.1038/237232a0
Booth, K., Cambron, L., Fisher, N., Greenlee, K. J. (2015). Immune defense varies within an instar in the tobacco hornworm, Manduca sexta. Physiol. Biochem. Zool. 88, 226–236. doi: 10.1086/680054
Boulanger, N., Munks, R. J. L., Hamilton, J. V., Vovelle, F., Brun, R., Lehane, M. J., et al. (2002). Epithelial innate immunity: A novel antimicrobial peptide with antiparasitic activity in the blood-sucking insect Stomoxys calcitrans. J. Biol. Chem. 277, 49921–49926. doi: 10.1074/jbc.M206296200
Bulet, P., Cociancich, S., Dimarcq, J. L., Lambert, J., Reichhart, J. M., Hoffmann, D., et al. (1991). Insect immunity. isolation from a coleopteran insect of a novel inducible antibacterial peptide and of new members of the insect defensin family. J. Biol. Chem. 266, 24520–24525. doi: 10.1016/S0021-9258(18)54260-5
Bulet, P., Cociancich, S., Reuland, M., Sauber, F., Bischoff, R., Hegy, G., et al. (1992). A novel insect defensin mediates the inducible antibacterial activity in larvae of the dragonfly Aeschna cyanea (Paleoptera, odonata). Eur. J. Biochem. 209, 977–984. doi: 10.1111/j.1432-1033.1992.tb17371.x
Buonocore, F., Fausto, A. M., Della Pelle, G., Roncevic, T., Gerdol, M., Picchietti, S. (2021). Attacins: A promising class of insect antimicrobial peptides. Antibiotics 10, 212. doi: 10.3390/antibiotics10020212
Burkett-Cadena, N. D., Vittor, A. Y. (2018). Deforestation and vector-borne disease: forest conversion favors important mosquito vectors of human pathogens. Basic Appl. Ecol. 26, 101–110. doi: 10.1016/j.baae.2017.09.012
Chalk, R., Townson, H., Natori, S., Desmond, H., Ham, P. J. (1994). Purification of an insect defensin from the mosquito, Aedes aegypti. Insect Biochem. Mol. Biol. 24, 403–410. doi: 10.1016/0965-1748(94)90033-7
Chowdhury, T., Mandal, S. M., Dutta, S., Ghosh, A. K. (2021). Identification of a novel proline-rich antimicrobial protein from the hemolymph of Antheraea mylitta. Arch. Insect Biochem. 106, e21771. doi: 10.1002/arch.21771
Christofi, T., Apidianakis, Y. (2013). Drosophila immune priming against Pseudomonas aeruginosa is short-lasting and depends on cellular and humoral immunity. F1000Res 2, 76. doi: 10.12688/f1000research.2-76.v1
Christophides, G. K., Vlachou, D., Kafatos, F. C. (2004). Comparative and functional genomics of the innate immune system in the malaria vector Anopheles gambiae. Immunol. Rev. 198, 127–148. doi: 10.1111/j.0105-2896.2004.0127.x
Christophides, G. K., Zdobnov, E., Barillas-Mury, C., Birney, E., Blandin, S., Blass, C., et al. (2002). Immunity-related genes and gene families in Anopheles gambiae. Science 298, 159–165. doi: 10.1126/science.1077136
Coggins, S. A., Estévez-Lao, T. Y., Hillyer, J. F. (2012). Increased survivorship following bacterial infection by the mosquito Aedes aegypti as compared to Anopheles gambiae correlates with increased transcriptional induction of antimicrobial peptides. Dev. Comp. Immunol. 37, 390–401. doi: 10.1016/j.dci.2012.01.005
Collins, F. H., Sakai, R. K., Vernick, K. D., Paskewitz, S., Seeley, D. C., Miller, L. H., et al. (1986). Genetic selection of a plasmodium-refractory strain of the malaria vector Anopheles gambiae. Science 234, 607–610. doi: 10.1126/science.3532325
Cramer, P., Varrot, A., Barillas-Mury, C., Kafatos, F. C., Müller, C. W. (1999). Structure of the specificity domain of the Dorsal homologue Gambif1 bound to DNA. Structure 7, 841–852. doi: 10.1016/s0969-2126(99)80107-5
Dai, H., Rayaprolu, S., Gong, Y., Huang, R., Prakash, O., Jiang, H. (2008). Solution structure, antibacterial activity, and expression profile of Manduca sexta moricin. J. Pept. Sci. 14, 855–863. doi: 10.1002/psc.1016
Dickinson, L., Russell, V., Dunn, P. E. (1988). A family of bacteria-regulated, cecropin d-like peptides from manduca sexta. J. Biol. Chem. 263, 19424–19429. doi: 10.1016/S0021-9258(19)77650-9
Dimopoulos, G. (2003). Insect immunity and its implication in mosquito-malaria interactions. Cell Microb. 5, 3–14. doi: 10.1046/j.1462-5822.2003.00252.x
Dimopoulos, G., Richman, A., Müller, H.-M., Kafatos, F. C. (1997). Molecular immune responses of the mosquito Anopheles gambiae to bacteria and malaria parasites. Proc. Natl. Acad. Sci. 94, 11508–11513. doi: 10.1073/pnas.94.21.11508
Dong, Y., Aguilar, R., Xi, Z., Warr, E., Mongin, E., Dimopoulos, G. (2006). Anopheles gambiae immune responses to human and rodent Plasmodium parasite species. PloS Pathog. 2, e52. doi: 10.1371/journal.ppat.0020052
Edgerton, E. B., McCrea, A. R., Berry, C. T., Kwok, J. Y., Thompson, L. K., Watson, B., et al. (2020). Activation of mosquito immunity blocks the development of transmission-stage filarial nematodes. Proc. Natl. Acad. Sci. 117, 3711–3717. doi: 10.1073/pnas.1909369117
Eleftherianos, I., Marokhazi, J., Millichap, P. J., Hodgkinson, A. J., Sriboonlert, A., ffrench-Constant, R. H., et al. (2006). Prior infection of Manduca sexta with non-pathogenic Escherichia coli elicits immunity to pathogenic Photorhabdus luminescens: Roles of immune-related proteins shown by RNA interference. Insect Biochem. Mol. Biol. 36, 517–525. doi: 10.1016/j.ibmb.2006.04.001
Frolet, C., Thoma, M., Blandin, S., Hoffmann, J. A., Levashina, E. A. (2006). Boosting NF-κB-dependent basal immunity of anopheles gambiae aborts development of Plasmodium berghei. Immunity 25, 677–685. doi: 10.1016/j.immuni.2006.08.019
Garcia-Lara, J., Needham, A. J., Foster, S. J. (2005). Invertebrates as animal models for Staphylococcus aureus pathogenesis: a window into host–pathogen interaction. FEMS Microbiol. Immunol. 43, 311–323. doi: 10.1016/j.femsim.2004.11.003
Garver, L. S., Dong, Y., Dimopoulos, G. (2009). Caspar Controls resistance to Plasmodium falciparum in diverse anopheline species. PloS Pathog. 5, e1000335. doi: 10.1371/journal.ppat.1000335
Goindin, D., Delannay, C., Gelasse, A., Ramdini, C., Gaude, T., Faucon, F., et al. (2017). Levels of insecticide resistance to deltamethrin, malathion, and temephos, and associated mechanisms in Aedes aegypti mosquitoes from the Guadeloupe and saint martin islands (French West indies). Infect. Dis. Poverty. 6, 38. doi: 10.1186/s40249-017-0254-x
Gorman, M. J., Paskewitz, S. M. (2000). Persistence of infection in mosquitoes injected with bacteria. J. Invertebr. Pathol. 75, 296–297. doi: 10.1006/jipa.2000.4930
Gupta, L., Molina-Cruz, A., Kumar, S., Rodrigues, J., Dixit, R., Zamora, R. E., et al. (2009). The STAT pathway mediates late-phase immunity against Plasmodium in the mosquito Anopheles gambiae. Cell Host Microbe 5, 498–507. doi: 10.1016/j.chom.2009.04.003
Habtewold, T., Povelones, M., Blagborough, A. M., Christophides, G. K. (2008). Transmission blocking immunity in the malaria non-vector mosquito Anopheles quadriannulatus species a. PloS Pathog. 4, e1000070. doi: 10.1371/journal.ppat.1000070
Haine, E. R., Pollitt, L. C., Moret, Y., Siva-Jothy, M. T., Rolff, J. (2008). Temporal patterns in immune responses to a range of microbial insults (Tenebrio molitor). J. Insect Physiol. 54, 1090–1097. doi: 10.1016/j.jinsphys.2008.04.013
Hemingway, J., Field, L., Vontas, J. (2002). An overview of insecticide resistance. Science 298, 96–97. doi: 10.1126/science.1078052
Hillyer, J. F., Strand, M. R. (2014). Mosquito hemocyte-mediated immune responses. Curr. Opin. Insect Sci. 3, 14–21. doi: 10.1016/j.cois.2014.07.002
Hoffmann, D., Hultmark, D., Boman, H. G. (1981). Insect immunity: Galleria mellonella and other lepidoptera have cecropia-P9-like factors active against gram negative bacteria. Insect Biochem. 11, 537–548. doi: 10.1016/0020-1790(81)90022-6
Horng, T., Medzhitov, R. (2001). Drosophila MyD88 is an adapter in the toll signaling pathway. Proc. Natl. Acad. Sci. 98, 12654–12658. doi: 10.1073/pnas.231471798
Huberman, L., Gollop, N., Mumcuoglu, K. Y., Block, C., Galun, R. (2007a). Antibacterial properties of whole body extracts and haemolymph of Lucilia sericata maggots. J. Wound Care. 16:123–127 doi: 10.12968/jowc.2007.16.3.27011
Huberman, L., Gollop, N., Mumcuoglu, K. Y., Breuer, E., Bhusare, S. R., Shai, Y., et al. (2007b). Antibacterial substances of low molecular weight isolated from the blowfly, Lucilia sericata. Med. Vet. Entomol. 21, 127–131. doi: 10.1111/j.1365-2915.2007.00668.x
Huff, C. (1927). Studies on the infectivity of plasmodia of birds for mosquitoes, with special reference to the problem of immunity in the mosquito. Am. J. Epidemiol. 7, 706–734. doi: 10.1093/oxfordjournals.aje.a120440
Hultmark, D., Engström, A., Andersson, K., Steiner, H., Bennich, H., Boman, H. G. (1983). Attacins, a family of antibacterial proteins from Hyalophora cecropia. EMBO J. 2, 571–576. doi: 10.1002/j.1460-2075.1983.tb01465.x
Jarosz, J. (1993). Induction kinetics of immune antibacterial proteins in pupae of Galleria mellonella and Pieris brassicae. Comp. Biochem. Physiol. B. 106, 415–421. doi: 10.1016/0305-0491(93)90322-v
Kajla, M. K., Andreeva, O., Gilbreath, T. M., Paskewitz, S. M. (2010). Characterization of expression, activity and role in antibacterial immunity of Anopheles gambiae lysozyme c-1. Comp. Biochem. Physiol. B. 155, 201–209. doi: 10.1016/j.cbpb.2009.11.012
Killeen, G. F., Smith, T. A., Ferguson, H. M., Mshinda, H., Abdulla, S., Lengeler, C., et al. (2007). Preventing childhood malaria in Africa by protecting adults from mosquitoes with insecticide-treated nets. PloS Med. 4, e229. doi: 10.1371/journal.pmed.0040229
Kleinschmidt, I., Bradley, J., Knox, T. B., Mnzava, A. P., Kafy, H. T., Mbogo, C., et al. (2018). Implications of insecticide resistance for malaria vector control with long-lasting insecticidal nets: a WHO-coordinated, prospective, international, observational cohort study. Lancet Infect. Dis. 18, 640–649. doi: 10.1016/S1473-3099(18)30172-5
Korner, P., Schmid-Hempel, P. (2004). In vivo dynamics of an immune response in the bumble bee Bombus terrestris. J. Invertebr. Pathol. 87, 59–66. doi: 10.1016/j.jip.2004.07.004
Lai, S.-C., Chen, C.-C., Hou, R. F. (2001). Electron microscopic observations on wound-healing in larvae of the mosquito Armigeres subalbatus (Diptera: Culicidae). J. Med. Entomol. 38, 836–843. doi: 10.1603/0022-2585-38.6.836
Lai, S.-C., Chen, C.-C., Hou, R. F. (2002). Immunolocalization of prophenoloxidase in the process of wound healing in the mosquito Armigeres subalbatus (Diptera: Culicidae). J. Med. Entomol. 39, 266–274. doi: 10.1603/0022-2585-39.2.266
League, G. P., Estevez-Lao, T. Y., Yan, Y., Garcia-Lopez, V. A., Hillyer, J. F. (2017). Anopheles gambiae larvae mount stronger immune responses against bacterial infection than adults: evidence of adaptive decoupling in mosquitoes. Parasite Vector 10, 367. doi: 10.1186/s13071-017-2302-6
Lemaitre, B., Kromer-Metzger, E., Michaut, L., Nicolas, E., Meister, M., Georgel, P., et al. (1995). A recessive mutation, immune deficiency (imd), defines two distinct control pathways in the Drosophila host defense. PNAS 92, 9465–9469. doi: 10.1073/pnas.92.21.9465
Lemaitre, B., Reichhart, J.-M., Hoffmann, J. A. (1997). Drosophila host defense: Differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. PNAS 94, 14614–14619. doi: 10.1073/pnas.94.26.14614
Lengeler, C. (1998). Insecticide treated bednets and curtains for malaria control. Cochrane Database Syst. Rev. 2, CD000363. doi: 10.1002/14651858.CD000363.pub2
Levashina, E. A. (2004). Immune responses in Anopheles gambiae. Insect Biochem. Mol. Biol. 34, 673–678. doi: 10.1016/j.ibmb.2004.03.020
Levashina, E. A., Moita, L. F., Blandin, S., Vriend, G., Lagueux, M., Kafatos, F. C. (2001). Conserved role of a complement-like protein in phagocytosis revealed by dsRNA knockout in cultured cells of the mosquito, Anopheles gambiae. Cell 104, 709–718. doi: 10.1016/S0092-8674(01)00267-7
Liu, G. Y., Essex, A., Buchanan, J. T., Datta, V., Hoffman, H. M., Bastian, J. F., et al. (2005). Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J. Exp. Med. 202, 209–215. doi: 10.1084/jem.20050846
Lockey, T. D., Ourth, D. D. (1996). Purification and characterization of lysozyme from hemolymph of Heliothis virescens larvae. Biochem. Biophys. Res. Commun. 220, 502–508. doi: 10.1006/bbrc.1996.0434
Lowenberger, C. A., Smartt, C. T., Bulet, P., Ferdig, M. T., Severson, D. W., Hoffmann, J. A., et al. (1999). Insect immunity: molecular cloning, expression, and characterization of cDNAs and genomic DNA encoding three isoforms of insect defensin in Aedes aegypti. Insect Mol. Biol. 8, 107–118. doi: 10.1046/j.1365-2583.1999.810107.x
Luna, C., Hoa, N. T., Lin, H., Zhang, L., Nguyen, H. L. A., Kanzok, S. M., et al. (2006). Expression of immune responsive genes in cell lines from two different anopheline species. Insect Mol. Biol. 15, 721–729. doi: 10.1111/j.1365-2583.2006.00661.x
Meister, S., Agianian, B., Turlure, F., Relógio, A., Morlais, I., Kafatos, F. C., et al. (2009). Anopheles gambiae PGRPLC-mediated defense against bacteria modulates infections with malaria parasites. PloS Pathog. 5, e1000542. doi: 10.1371/journal.ppat.1000542
Meister, S., Kanzok, S. M., Zheng, X., Luna, C., Li, T.-R., Hoa, N. T., et al. (2005). Immune signaling pathways regulating bacterial and malaria parasite infection of the mosquito Anopheles gambiae. Proc. Natl. Acad. Sci. 102, 11420–11425. doi: 10.1073/pnas.0504950102
Meng, X., Khanuja, B. S., Ip, Y. T. (1999). Toll receptor-mediated Drosophila immune response requires dif, an NF-κB factor. Genes Dev. 13, 792–797. doi: 10.1101/gad.13.7.792
Michel, K., Budd, A., Pinto, S., Gibson, T. J., Kafatos, F. C. (2005). Anopheles gambiae SRPN2 facilitates midgut invasion by the malaria parasite Plasmodium berghei. EMBO Rep. 6, 891–897. doi: 10.1038/sj.embor.7400478
Moita, L. F., Vriend, G., Mahairaki, V., Louis, C., Kafatos, F. C. (2006). Integrins of Anopheles gambiae and a putative role of a new β integrin, BINT2, in phagocytosis of E. coli. Insect Biochem. Mol. Biol. 36, 282–290. doi: 10.1016/j.ibmb.2006.01.004
Moita, L. F. L. F., Wang-Sattler, R., Michel, K., Zimmermann, T., Blandin, S., Levashina, E. A., et al. (2005). In vivo identification of novel regulators and conserved pathways of phagocytosis in A. gambiae. Immunity 23, 65–73. doi: 10.1016/j.immuni.2005.05.006
Molina-Cruz, A., DeJong, R. J., Charles, B., Gupta, L., Kumar, S., Jaramillo-Gutierrez, G., et al. (2008). Reactive oxygen species modulate Anopheles gambiae immunity against bacteria and plasmodium. J. Biol. Chem. 283, 3217–3223. doi: 10.1074/jbc.M705873200
Molina-Cruz, A., Zilversmit, M. M., Neafsey, D. E., Hartl, D. L., Barillas-Mury, C. (2016). Mosquito vectors and the globalization of Plasmodium falciparum malaria. Annu. Rev. Genet. 50, 447–465. doi: 10.1146/annurev-genet-120215-035211
Moret, Y., Schmid-Hempel, P. (2001). Immune defense in bumble-bee offspring. Nature 414, 506–506. doi: 10.1038/35107138
Nakhleh, J., El Moussawi, L., Osta, M. A. (2017). The melanization response in insect immunity. Adv. Insect Phys. 52, 83–109. doi: 10.1016/bs.aiip.2016.11.002
Nasr, N. M., Fallon, A. M. (2003). Detection of lysozyme-like enzymatic activity secreted by an immune-responsive mosquito cell line. J. Invertebr. Pathol. 82, 162–166. doi: 10.1016/S0022-2011(03)00030-2
Netea, M. G., Quintin, J., van der Meer, J. W. M. (2011). Trained immunity: a memory for innate host defense. Cell Host Microbe 9, 355–361. doi: 10.1016/j.chom.2011.04.006
Nosten, F., White, N. J. (2007). Artemisinin-based combination treatment of falciparum malaria. Am. J. Trop. Med. 77, 181–192. doi: 10.4269/ajtmh.2007.77.181
Osta, M. A., Christophides, G. K., Kafatos, ,. F. C. (2004). Effects of mosquito genes on Plasmodium development. Science 303, 2030–2032. doi: 10.1126/science.1091789
Otvos, L. (2000). Antibacterial peptides isolated from insects. J. Pept. Sci. 6, 497–511. doi: 10.1002/1099-1387(200010)6:10<497
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, 45e–445. doi: 10.1093/nar/29.9.e45
Pham, L. N., Dionne, M. S., Shirasu-Hiza, M., Schneider, D. S. (2007). A specific primed immune response in Drosophila is dependent on phagocytes. PloS Pathog. 3, e26. doi: 10.1371/journal.ppat.0030026
Powers, J. C., Turangan, R., Joosse, B. A., Hillyer, J. F. (2020). Adult mosquitoes infected with bacteria early in life have stronger antimicrobial responses and more hemocytes after reinfection later in life. Insects 11, 331. doi: 10.3390/insects11060331
Qu, X., Steiner, H., Engström, Å., Bennich, H., Boman, H. G. (1982). Insect immunity: Isolation and structure of cecropins b and d from pupae of the Chinese oak silk moth, Antheraea pernyi. Eur. J. Biochem. 127, 219–224. doi: 10.1111/j.1432-1033.1982.tb06858.x
Rahnamaeian, M., Cytryńska, M., Zdybicka-Barabas, A., Dobslaff, K., Wiesner, J., Twyman, R. M., et al. (2015). Insect antimicrobial peptides show potentiating functional interactions against gram-negative bacteria. Proc. Natl. Acad. Sci. 282, 20150293. doi: 10.1098/rspb.2015.0293
Ramasamy, R., Surendran, S. N. (2016). Mosquito vectors developing in atypical anthropogenic habitats: global overview of recent observations, mechanisms and impact on disease transmission. J. Vector Borne Dis. 53, 91.
Ramirez, J. L., Garver, L. S., Brayner, F. A., Alves, L. C., Rodrigues, J., Molina-Cruz, A., et al. (2014). The role of hemocytes in Anopheles gambiae antiplasmodial immunity. J. Innate Immun. 6, 119–128. doi: 10.1159/000353765
Randolt, K., Gimple, O., Geissendörfer, J., Reinders, J., Prusko, C., Mueller, M. J., et al. (2008). Immune-related proteins induced in the hemolymph after aseptic and septic injury differ in honey bee worker larvae and adults. Arch. Insect Biochem. 69, 155–167. doi: 10.1002/arch.20269
Rhodes, V. L., Thomas, M. B., Michel, K. (2018). The interplay between dose and immune system activation determines fungal infection outcome in the African malaria mosquito, Anopheles gambiae. Dev. Comp. Immunol. 85, 125–133. doi: 10.1016/j.dci.2018.04.008
Richman, A. M., Bulet, P., Hetru, C., Barillas-Mury, C., Hoffmann, J. A., Kafatos, F. C. (1996). Inducible immune factors of the vector mosquito Anopheles gambiae: biochemical purification of a defensin antibacterial peptide and molecular cloning of preprodefensin cDNA. Insect Mol. Biol. 5, 203–210. doi: 10.1111/j.1365-2583.1996.tb00055.x
Riehle, M. M., Xu, J., Lazzaro, B. P., Rottschaefer, S. M., Coulibaly, B., Sacko, M., et al. (2008). Anopheles gambiae APL1 is a family of variable LRR proteins required for Rel1-mediated protection from the malaria parasite, Plasmodium berghei. PloS One 3, e3672. doi: 10.1371/journal.pone.0003672
Rocha, I. F., Maller, A., de Cássia Garcia Simão, R., Kadowaki, M. K., Angeli Alves, L. F., Huergo, L. F., et al. (2016). Proteomic profile of hemolymph and detection of induced antimicrobial peptides in response to microbial challenge in Diatraea saccharalis (Lepidoptera: Crambidae). Biochem. Biophys. Res. Commun. 473, 511–516. doi: 10.1016/j.bbrc.2016.03.091
Rodrigues, J., Brayner, F. A., Alves, L. C., Dixit, R., Barillas-Mury, C. (2010). Hemocyte differentiation mediates innate immune memory in Anopheles gambiae mosquitoes. Science 329, 1353–1355. doi: 10.1126/science.1190689
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi: 10.1038/nmeth.2019
Schmid-Hempel, P. (2005). Natural insect host-parasite systems show immune priming and specificity: puzzles to be solved. Bioessays 27, 1026–1034. doi: 10.1002/bies.20282
Schneider, P. M. (1985). Purification and properties of three lysozymes from hemolymph of the cricket, Gryllus bimaculatus (De geer). Insect Biochem. 15, 463–470. doi: 10.1016/0020-1790(85)90058-7
Schnitger, A. K. D., Kafatos, F. C., Osta, M. A. (2007). The melanization reaction is not required for survival of Anopheles gambiae mosquitoes after bacterial infections. J. Biol. Chem. 282, 21884–21888. doi: 10.1074/jbc.M701635200
Schnitger, A. K. D., Yassine, H., Kafatos, F. C., Osta, M. A. (2009). Two c-type lectins cooperate to defend Anopheles gambiae against gram-negative bacteria. J. Biol. Chem. 284, 17616–17624. doi: 10.1074/jbc.M808298200
Sumida, M., Ichimori, H., Johchi, S., Takaoka, A., Yuhki, T., Mori, H., et al. (1992). Antibacterial activity inducible in the haemolymph of the silkworm, Bombyx mori, by injection of formalin-treated Escherichia coli K-12 during the fifth larval instar and pharate adult development. Comp. Biochem. Physiol. B. 101, 165–171. doi: 10.1016/0305-0491(92)90173-O
Sun, J. C., Beilke, J. N., Lanier, L. L. (2009). Adaptive immune features of natural killer cells. Nature 457, 557–561. doi: 10.1038/nature07665
Tauszig-Delamasure, S., Bilak, H., Capovilla, M., Hoffmann, J. A., Imler, J.-L. (2002). Drosophila MyD88 is required for the response to fungal and gram-positive bacterial infections. Nat. Immunol. 3, 91–97. doi: 10.1038/ni747
Tikhe, C. V., Dimopoulos, G. (2021). Mosquito antiviral immune pathways. Dev. Comp. Immunol. 116, 103964. doi: 10.1016/j.dci.2020.103964
Upton, L. M., Povelones, M., Christophides, G. K. (2015). Anopheles gambiae blood feeding initiates an anticipatory defense response to Plasmodium berghei. J. Innate Immun. 7, 74–86. doi: 10.1159/000365331
Uvell, H., Engström, Y. (2007). A multilayered defense against infection: combinatorial control of insect immune genes. Trends Genet. 23, 342–349. doi: 10.1016/j.tig.2007.05.003
Vernick, K. D., Fujioka, H., Seeley, D. C., Tandler, B., Aikawa, M., Miller, L. H. (1995). Plasmodium gallinaceum: a refractory mechanism of ookinete killing in the mosquito, Anopheles gambiae. Exp. Parasitol. 80, 583–595. doi: 10.1006/expr.1995.1074
Vizioli, J., Bulet, P., Charlet, M., Lowenberger, C., Blass, C., Müller, H.-M., et al. (2000). Cloning and analysis of a cecropin gene from the malaria vector mosquito, Anopheles gambiae. Insect Mol. Biol. 9, 75–84. doi: 10.1046/j.1365-2583.2000.00164.x
Vizioli, J., Bulet, P., Hoffmann, J. A., Kafatos, F. C., Müller, H.-M., Dimopoulos, G. (2001a). Gambicin: A novel immune responsive antimicrobial peptide from the malaria vector Anopheles gambiae. Proc. Natl. Acad. Sci. 98, 12630–12635. doi: 10.1073/pnas.221466798
Vizioli, J., Richman, A. M., Uttenweiler-Joseph, S., Blass, C., Bulet, P. (2001b). The defensin peptide of the malaria vector mosquito Anopheles gambiae: antimicrobial activities and expression in adult mosquitoes. Insect Biochem. Mol. Biol. 31, 241–248. doi: 10.1016/S0965-1748(00)00143-0
Warr, E., Das, S., Dong, Y., Dimopoulos, G. (2008). The gram-negative bacteria-binding protein gene family: Its role in the innate immune system of anopheles gambiae and in anti-Plasmodium defense. Insect Mol. Biol. 17, 39–51. doi: 10.1111/j.1365-2583.2008.00778.x
Yassine, H., Kamareddine, L., Osta, M. A. (2012). The mosquito melanization response is implicated in defense against the entomopathogenic fungus Beauveria bassiana. PloS Pathog. 8, e1003029. doi: 10.1371/journal.ppat.1003029
Zakovic , S., Levashina, E. A. (2017). NF-κB-Like signaling pathway REL2 in immune defenses of the malaria vector Anopheles gambiae . Front. Cell. Infect. Microbiol. 7, 1–7. doi: 10.3389/fcimb.2017.00258
Zeng, Y., Hu, X. P., Suh, S.-J. (2016). Characterization of antibacterial activities of Eastern subterranean termite, Reticulitermes flavipes, against human pathogens. PloS One 11, e0162249. doi: 10.1371/journal.pone.0162249
Zhang, X., An, C., Sprigg, K., Michel, K. (2016). CLIPB8 is part of the prophenoloxidase activation system in Anopheles gambiae mosquitoes. Insect Biochem. Mol. Biol. 71, 106–115. doi: 10.1016/j.ibmb.2016.02.008
Zhang, R., Zhu, Y., Pang, X., Xiao, X., Zhang, R., Cheng, G. (2017). Regulation of antimicrobial peptides in Aedes aegypti Aag2 cells. Front. Cell Infect. Microbiol. 7. doi: 10.3389/fcimb.2017.00022
Keywords: innate immunity, mosquito, toll pathway, antimicrobial peptide (AMP), defensin 1, ZOI (zone of inhibition)
Citation: Morejon B and Michel K (2023) A zone-of-inhibition assay to screen for humoral antimicrobial activity in mosquito hemolymph. Front. Cell. Infect. Microbiol. 13:891577. doi: 10.3389/fcimb.2023.891577
Received: 07 March 2022; Accepted: 10 January 2023;
Published: 26 January 2023.
Edited by:
Layla Kamareddine, Qatar University, QatarReviewed by:
Yiorgos Apidianakis, University of Cyprus, Nicosia, CyprusYuemei Dong, Johns Hopkins Malaria Research Institute, Johns Hopkins University, United States
Copyright © 2023 Morejon and Michel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kristin Michel, a21pY2hlbEBrc3UuZWR1
 Bianca Morejon
Bianca Morejon Kristin Michel
Kristin Michel