- 1Department of Clinical Laboratory Medicine, The First Affiliated Hospital of Shandong First Medical University & Shandong Provincial Qianfoshan Hospital, Shandong Medicine and Health Key Laboratory of Laboratory Medicine, Jinan, China
- 2School of Clinical Medicine, Jining Medical University, Jining, China
- 3Department of Bacterial Infectious Disease Control and Prevention, Shandong Center for Disease Control and Prevention, Jinan, China
- 4Center for Discovery and Innovation, Hackensack Meridian Health, Nutley, NJ, United States
- 5Department of Medical Sciences, Hackensack Meridian School of Medicine, Nutley, NJ, United States
Objective: Nontyphoidal Salmonella is a significant public health concern due to its ability to cause foodborne illnesses worldwide. This study aims to characterize the nontyphoidal Salmonella strains isolated from patients in China.
Methods: A total of 19 nontyphoidal Salmonella strains were characterized through serovar identification, antimicrobial susceptibility testing (AST), biofilm formation assessment. Genetic relatedness was determined using pulsed-field gel electrophoresis (PFGE). WGS was employed to decipher the resistance mechanism and to contextualize the S. serovar Mbandaka strains among previously sequenced isolates in China. The biofilm associated mrkA gene was examined by PCR.
Results: The predominant serovar identified was S. Enteritidis, followed by S. Mbandaka, S. Thompson, S. Livingston, S. Alachua, and S. Infantis. PFGE analysis indicated a notable genetic similarity among the S. Mbandaka isolates. Phylogenetic analysis suggested that these strains were likely derived from a single source that had persisted in China for over five years. One multidrug resistance (MDR) S. Enteritidis isolate carried a highly transferable IncB/O/K/Z plasmid with blaCTX-M-15. One S. Thompson strain, harboring the mrkABCDF operon in an IncX1 plasmid, isolated from cutaneous lesions, demonstrated robust biofilm formation. However, no mrkABCDF loci were detected in other strains.
Conclusion: Our study emphasizes the importance of persisted surveillance and prompt response to Salmonella infections to protect public health. The dissemination of blaCTX-M-15-harboring IncB/O/K/Z plasmid and the spread of virulent mrkABCDF operon among Salmonella in China and other global regions warrant close monitoring.
Introduction
Nontyphoidal Salmonella (NTS) is one of the most common agents of gastrointestinal disease globally (Amuasi and May, 2019). It is estimated that NTS cause about 129.5 million cases each year, resulting in about 100,000 to 1 million deaths per year worldwide (Wen et al., 2017; Branchu et al., 2018). Salmonella infections frequently occur in humans when they consume contaminated foods, including poultry, eggs, beef, pork, milk, seafood, and fresh fish products. Additionally, direct interactions with animals can lead to the transmission of Salmonella to humans (Zhao et al., 2003). While most infections result in self-limiting gastroenteritis, individuals such as infants, the elderly or those with compromised immune systems are susceptible to life-threatening invasive infections (Marchello et al., 2022).
To date, over 2600 Salmonella serovars have been identified, of which only a few NTS serovars are responsible for most human infections (Akinyemi et al., 2023). S. Typhimurium and S. Enteritidis accounts for the majority of NTS infections in humans (Wang et al., 2023). Alarmingly, the emergence of multi-drug-resistant (MDR) Salmonella serovars, particularly against 3rd-generation cephalosporins and fluoroquinolones, is having a great impact on the efficacy of antibiotic treatment, and an increasing prevalence of MDR strains may lead to an increase in mortality rates of Salmonella infections (Crump et al., 2015). Of note, MDR strains can carry specific virulence factors which are more virulent than their susceptible counterparts (Gebreyes et al., 2009).
In China, surveillance data spanning from 2006 to 2019 reveals an average of 62 serovars are detected annually from human origin (Wang et al., 2023). S. Typhimurium and S. Enteritidis were the most common serovar causing human infections in China. Moreover, the proportion of antimicrobial-resistant Salmonella isolates occur with increasing frequency during 2006–2019, especially beta-lactam, quinolone, tetracycline, and rifampicin resistance (Wang et al., 2023). These findings highlight the need for continuous monitoring of Salmonella infection with particular emphasis on MDR Salmonella strains.
In this study, we identified and examined a total of 19 NTS isolates from patients at a tertiary hospital in eastern China in 2022. The majority of cases were reported between July and October, with the peak incidence occurring in October. Through a combination of phenotypic and molecular analyses, we aimed to gain comprehensive insights into the characteristics of these Salmonella strains.
Materials and methods
Bacterial isolates
During the period from March 20, 2022, to December 16, 2022, a total of 19 isolates of NTS were investigated in this study (Figure 1). All strains were the first isolates collected from each patient. Ten of the isolates were obtained from feces. Other clinical specimens included blood cultures, bronchoalveolar lavage fluid, cutaneous ulcer, ascitic fluid, joint fluid. For routine use samples were inoculated on Blood Agar (TSA w/5% Sheep Blood) and selective SS Agar (Salmonella-Shigella agar) and were grown at 37°C overnight.
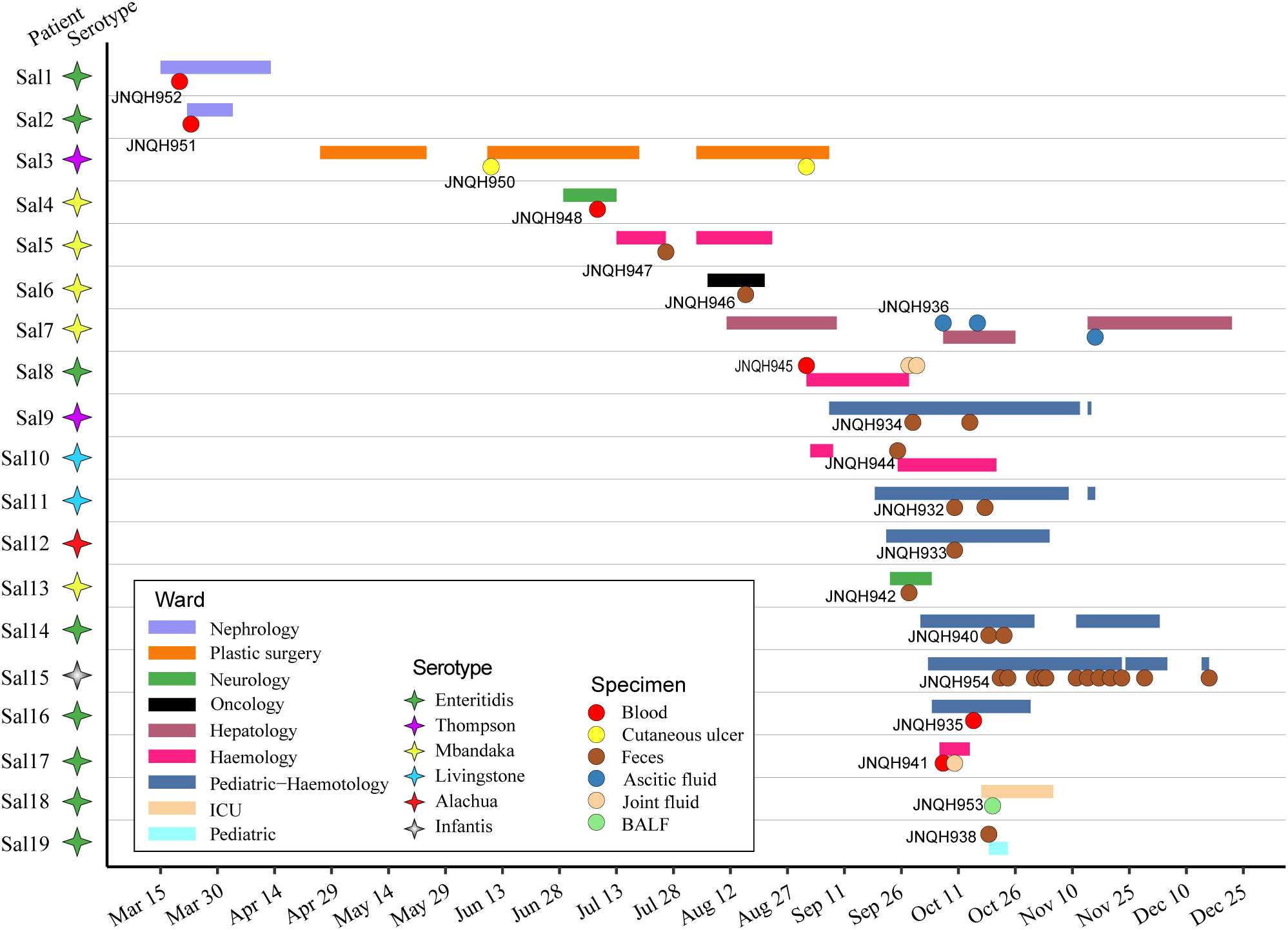
Figure 1 Time frame of stays in hospitalization for the 19 patients involved in NTS infection. Horizontal bars represent the duration of hospitalization for each patient, with distinct colors indicating the respective ward departments. Circles represent the time of isolation for each patient with colored circles representing the specimen source. Colored stars represent different serovars.
Patient information
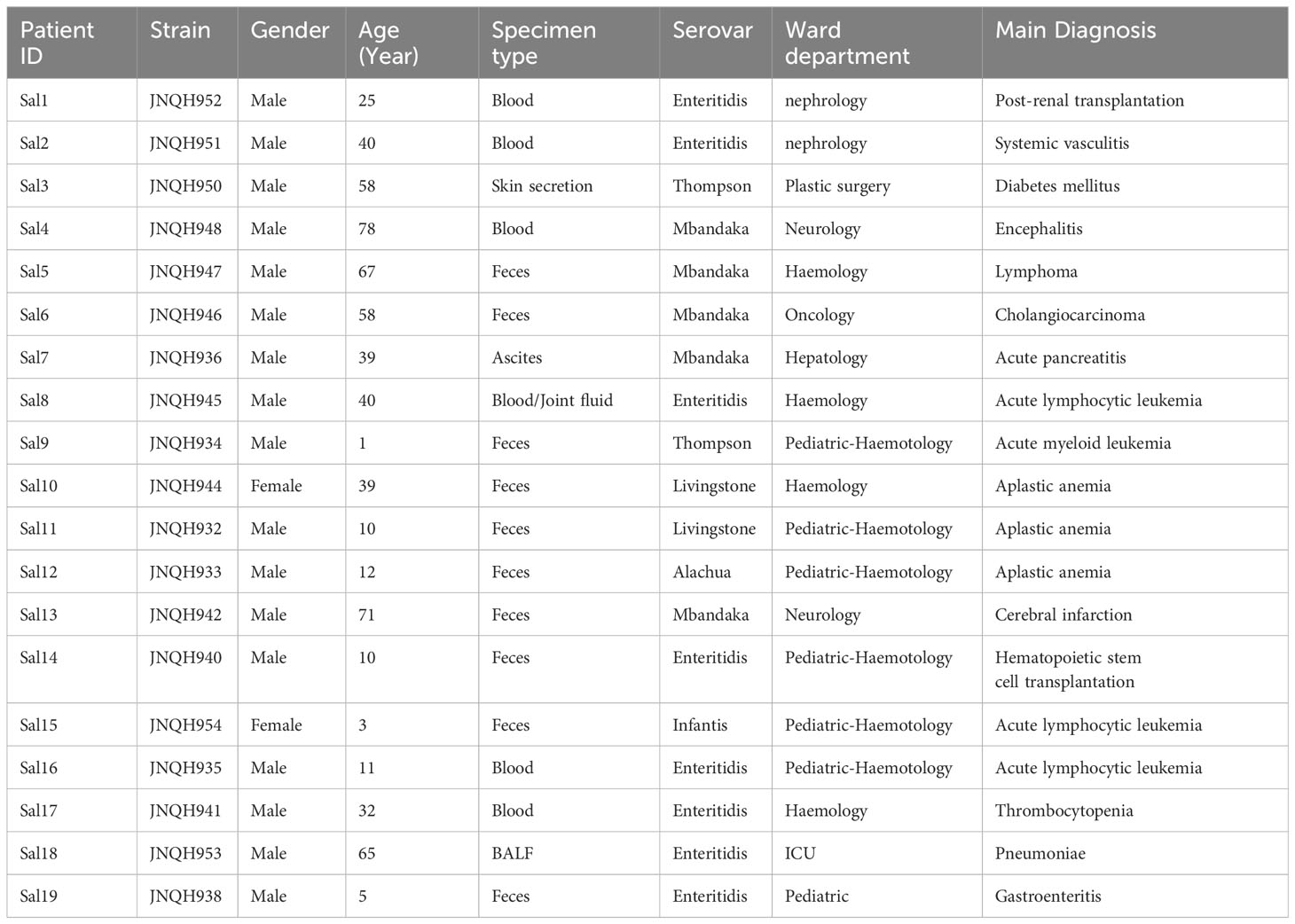
The median age of infected patients was 35.5 years (range: 1-78 years). 17 of the patients were males. The median duration of illness was 9 days (range: 5-89 days). Predominant symptoms included diarrhea (68.4%), fever (47.4%), abdominal pain (47.4%). Lower respiratory tract infection-related symptoms and signs are prominent in a patient with lung infection (patient Sal18). All the patients were administered antimicrobial agents, with 14 (73.7%) patients showing effective outcomes. Short descriptions of the investigated isolates are presented in Table 1.
Antimicrobial susceptibility testing
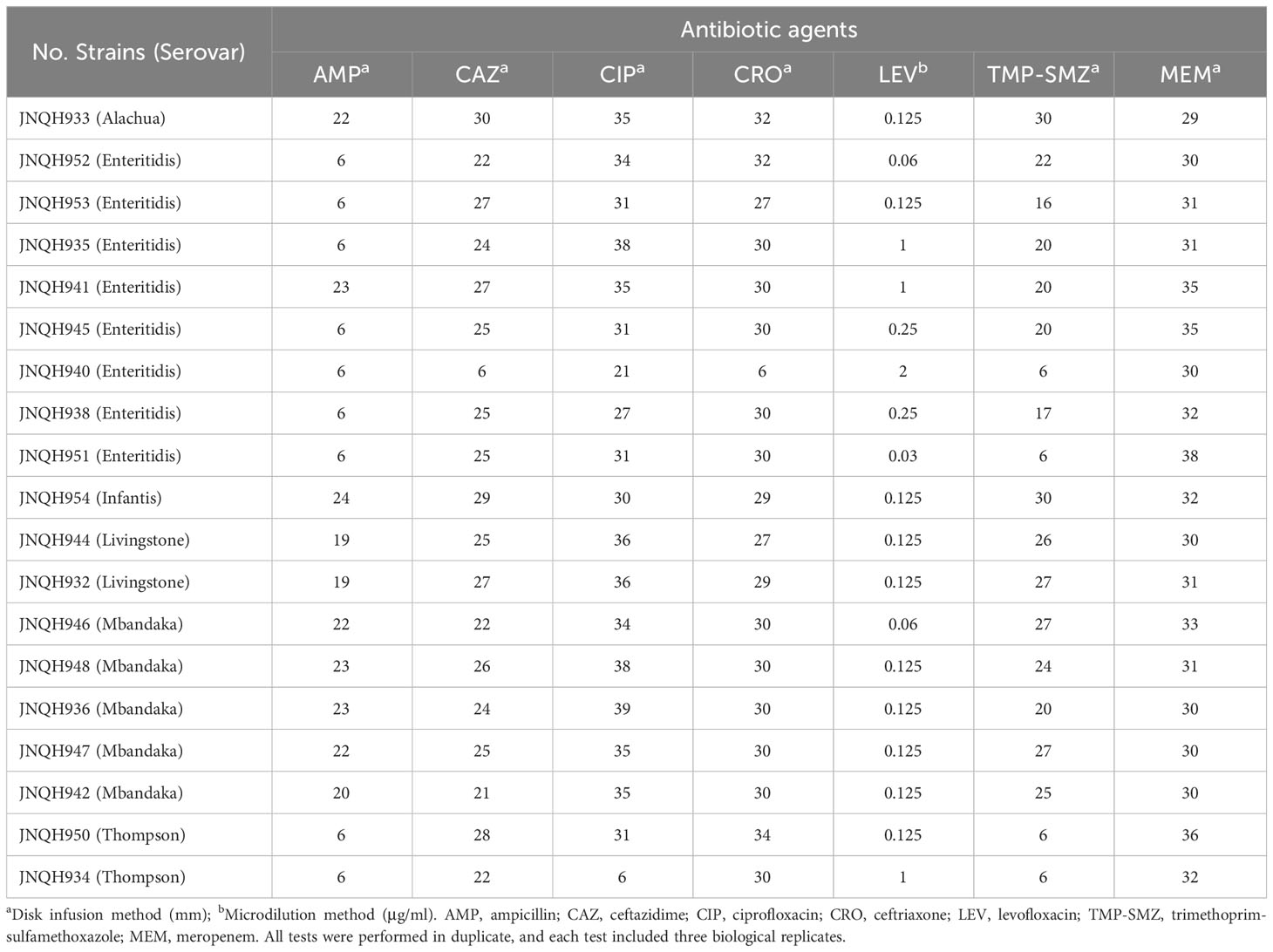
AST was performed using the disk diffusion method or microdilution method as recommended by the Clinical and Laboratory Standards Institute guidelines (CLSI, 2021). Antimicrobial discs were manufactured by Oxoid in UK. The following antimicrobial agents were tested: ampicillin (AMP, disk content 10μg), ceftazidime (CAZ, 30μg), ciprofloxacin (CIP, 5μg), ceftriaxone (CRO, 30μg), trimethoprim-sulfamethoxazole (TMP-SMZ, 1.25/23.75μg), meropenem (MEM, 10μg). Levofloxacin (LEV) was tested by microdilution method. Results were interpreted using the CLSI breakpoints (CLSI M100, 2021). MDR was defined as acquired non-susceptibility to at least one agent in three or more antimicrobial categories (Magiorakos et al., 2012). Each of the antibiotics was tested with 3 duplicates. ATCC 25922 (E. coli) were used as quality control strains for susceptibility testing.
Salmonella serotyping
The isolated Salmonella were serotyped on slides by slide agglutination as described in the manufacturer’s instructions (Statens Serum Institut, Copenhagen, Denmark). Briefly, the Salmonella strains were grown overnight on Blood Agar then colonies were transferred to the drop of antiserum. The “O” antigen type was determined based on oligosaccharides associated with lipopolysaccharide. Then the “H” antigen was determined based on flagellar proteins. The reaction was read with the naked eye by holding the slide in front of a light source against a black background. A positive reaction was seen as a visible agglutination. A negative reaction was persistence of the homogeneous milky turbidity. The serovar was assigned according to the Kauffmann-White scheme, which is a comprehensive system employed for the discrimination of serological variations within the Salmonella genus.
Pulsed-field gel electrophoresis
All isolates were analyzed for genetic relatedness by PFGE. PFGE of Xbal-digested genomic DNA samples were performed with a CHEF MAPPER XA apparatus (Bio-Rad, USA), as previously described (Ma et al., 2023). In brief, genomic DNA was prepared by embedding cells in agarose plugs, followed by XbaI digestion for 2 h at 37°C. Electrophoresis conditions consisted of one phase from 2.2 to 63.8 s at a run time of 17 h. Salmonella serovar Branderup H9812 strain was used as the reference strain. PFGE patterns were analyzed and compared using GelJ software 2.0v (Heras et al., 2015).
Biofilm formation
The process of biofilm formation was assessed using crystal violet staining as described previously with minor modifications (Hao et al., 2021a). Briefly, overnight cultures of the bacteria were diluted 1:1,000 in LB medium. A total volume of 200 μl of the diluted culture was then transferred to each well of a microtiter plate. The plate was incubated at 37°C for 24 hours. After incubation, the wells were washed four times with distilled water to remove any planktonic (free-floating) bacteria. The remaining adherent bacteria were then stained with 125 μl of a 0.1% crystal violet dye solution. Following incubation for 10 minutes, the crystal violet solution was carefully removed, and the wells were washed six times with distilled water. After that, 150 μl of a 30% glacial acetic acid in water solution was added to each well. The plate was incubated for an additional 10 minutes at room temperature, allowing the acetic acid to dissolve the crystal violet-stained biofilms. The optical density (OD) of the solubilized biofilms was then measured at a wavelength of 590 nm using a microplate reader (Thermo Scientific). At least three replicates were performed for each sample. Statistical significance in biofilm assays was calculated using Student’s t-test and any differences between strains with a P value of <0.05 were considered significant.
Whole-genome sequencing
Complete sequencing was performed on four selected strains, which included a strong biofilm-forming S. Thompson strain (JNQH950), S. Mbandaka (JNQH948), as well as two resistant S. Enteritidis strains (JNQH940 and JNQH952). Genomic DNA was isolated using a WizardR Genomic DNA Purification Kit as described in the manufacturer’s instructions (Promega, Madison, WI, USA). The DNA samples were subject to next generation sequencing using the Illumina HiSeq (Illumina, San Diego, CA, USA) and Oxford Nanopore (MinION system). The hybrid assembly was performed by Unicycler v0·5.0 (Wick et al., 2017). The whole-genome sequences were annotated by Prokka (Seemann, 2014) automatically followed by manual curation.
Genomic analysis
In silico multi-locus sequence typing was performed using MLST 2.0 (Larsen et al., 2012). Seven housekeeping genes, including aroC, dnaN, hemD, hisD, purE, sucA, and thrA, were chosen for MLST analysis. The antibiotic resistome(s) were predicted by the Resistance Gene Identifier (RGI) (https://github.com/arpcard/rgi) to query the CARD database (https://card.mcmaster.ca) (Alcock et al., 2020). The virulence genes and plasmid replicons in the sequenced isolates were identified using VFDB (Liu et al., 2022) and PlasmidFinder 2.0 (Carattoli et al., 2014) respectively. In order to contextualize our isolates among the corresponding strains in China, genomes of S. Mbandaka were downloaded from EnteroBase in combination with the genomes reported in a large-scale one-health study (Wang et al., 2023). SNPs were detected using Snippy v3.2 (https://github.com/tseemann/snippy) based on recombination filtration by Gubbins v. 2.4.1 (Croucher et al., 2015) and core SNP extraction by SNP-sites v. 2.5.1 (Page et al., 2016). SNPs located in phage regions, repetitive and recombinogenic regions, were removed before phylogenetic analysis. Genomic coordinates corresponding to phage sequences and repetitive elements in the reference genome were determined using Phast (http://phaster.ca) and Mummer (Kurtz et al., 2004), respectively. All extracted SNPs in core genome regions were concatenated as pseudosequences. The approximately-maximum-likelihood phylogenetic tree was inferred from alignments of nucleotide sequences with FastTree (Price et al., 2009). The visualization and annotation of the phylogenetic tree were carried out using R ggtree package (Yu et al., 2017).
OriT Finder was used to determine the conjugation module (Li et al., 2018). ISFinder was used to identify ISs (https://isfinder.biotoul.fr/). Plasmid similarity was examined comparing against the PLSDB plasmid database (Schmartz et al., 2022). Comparison between homologous plasmids were performed using BLASTn and illustrated by Easyfig v2.2.2 (Sullivan et al., 2011). To examine the distribution and features of mrkABCDF operon in global plasmids, plasmid reference sequences were downloaded from NCBI (https://ftp.ncbi.nlm.nih.gov/refseq/release/plasmid) and compared using Mashtree (Katz et al., 2019). Serovars assigned from Kauffmann-White scheme were confirmed by SeqSero2 v1.1.1 (Zhang et al., 2019).
Conjugal transference of blaCTX-M-15 harboring plasmid
In order to examine the conjugal transference of blaCTX-M-15 harboring plasmid in JNQH940, conjugation experiments were conducted using E. coli J53AziR as recipients, following a previously described method (Hao et al., 2021b). Overnight cultures of JNQH940 and E. coli J53AziR were mixed in a 1:1 ratio and applied separately onto 0.45-μm filter paper. The filter papers were then placed on LB agar plates and incubated overnight at 37°C. Transconjugants were selected on Mueller–Hinton agar containing sodium azide (100 mg/L) and ceftazidime (16 mg/L). The presence of transconjugants was confirmed using PCR targeting the blaCTX-M-15 gene. Conjugation frequency was determined by dividing the number of transconjugants by the number of recipient cells.
Detection of mrkABCDF loci by PCR
The mrkABCDF virulence loci were identified via a colony Polymerase Chain Reaction (PCR) assay, wherein the target gene mrkA was amplified using specific primers (mrkAF: 5’-ACCAGCAAACAACAGGGCTA-3’, mrkAR: 5’-TGATTTTGTTGGTCAGCGCG-3’). The occurrence of a positive outcome in this assay was determined by the generation of a DNA fragment with a length of 106 base pairs. For the purpose of assay validation, a well-characterized K. variicola isolate JNQH473, which had previously undergone comprehensive sequencing in our prior research (Wang et al., 2021a), was utilized as a positive control.
Nucleotide sequence accession numbers
Complete sequences of the chromosomes and plasmids of strain JNQH940, 948, 950 and 952 have been deposited in the GenBank databases under accession numbers CP136141-CP136151.
Results
Serovar identification and antimicrobial susceptibility
Six different serovars were assigned according to the Kauffmann-White scheme. S. Enteritidis was the most common serovar, accounting for 42.1% of the isolates, followed by S. Mbandaka (26.3%), Thompson (10.5%), Livingston (10.5%), Alachua (5.3%), Infantis (5.3%) (Figure 1). Antimicrobial-susceptibility testing results showed that isolates of S. Mbandaka, Infantis, Livingston, and Alachua exhibited susceptibility to the tested antibiotics. In contrast, JNQH934 (S. Thompson) and JNQH940 (S. Enteritidis) demonstrated significant resistance to AMP, CIP, and TMP-SMZ, displaying multidrug resistance (Table 2).
Genetic relatedness of Salmonella isolates
PFGE analysis utilizing XbaI restriction enzyme was employed to evaluate the genetic relatedness among the isolates. The PFGE patterns of S. Enteritidis isolates were similar but distinguishable except JNQH941 and JNQH935. Two S. Thompson isolates (JNQH950, 934) had notably different PFGE patterns, indicative of genetic diversity. However, two S. Livingston isolates (JNQH932, 944) demonstrated similar PFGE patterns, suggesting a potential genetic relatedness between these isolates. Of note, the S. Mbandaka serovar isolates displayed very similar PFGE patterns, suggesting a closer genetic relationship (Figure 2). Interestingly, SNP-based phylogenetic tree uncovered a close relationship between S. Mbandaka JNQH948 and a strain (GCA_030099325.1) obtained from human feces in Zhejiang in 2017 (Figure 3), with only 8 SNP differences. These results highlight the occurrence of small-scale clonal spread of S. Mbandaka in China.
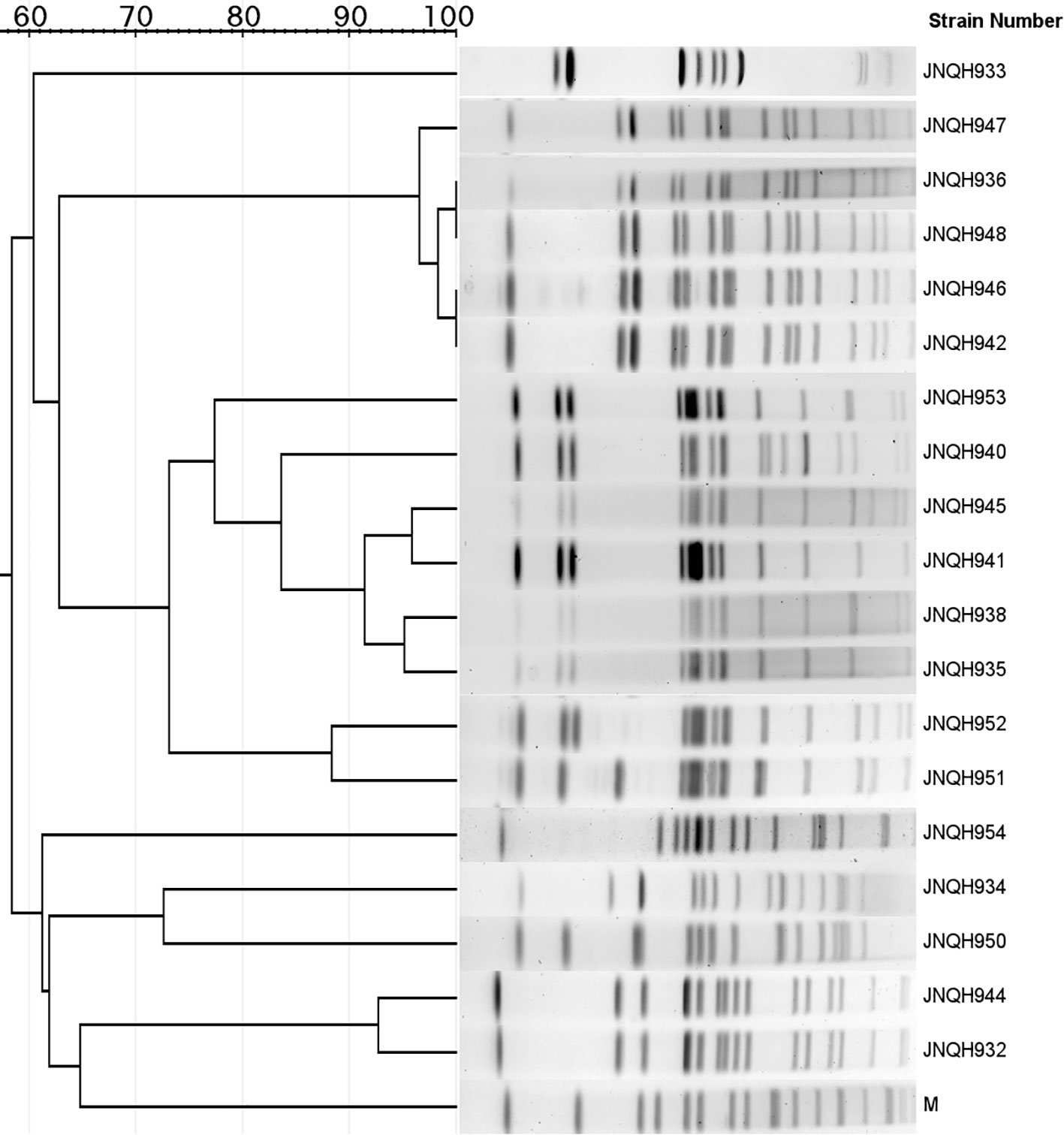
Figure 2 Dendrogram of the pulsed-field gel electrophoresis (PFGE) patterns of NTS isolates collected from this study. M, PFGE marker strain (S. Braenderup H9812).
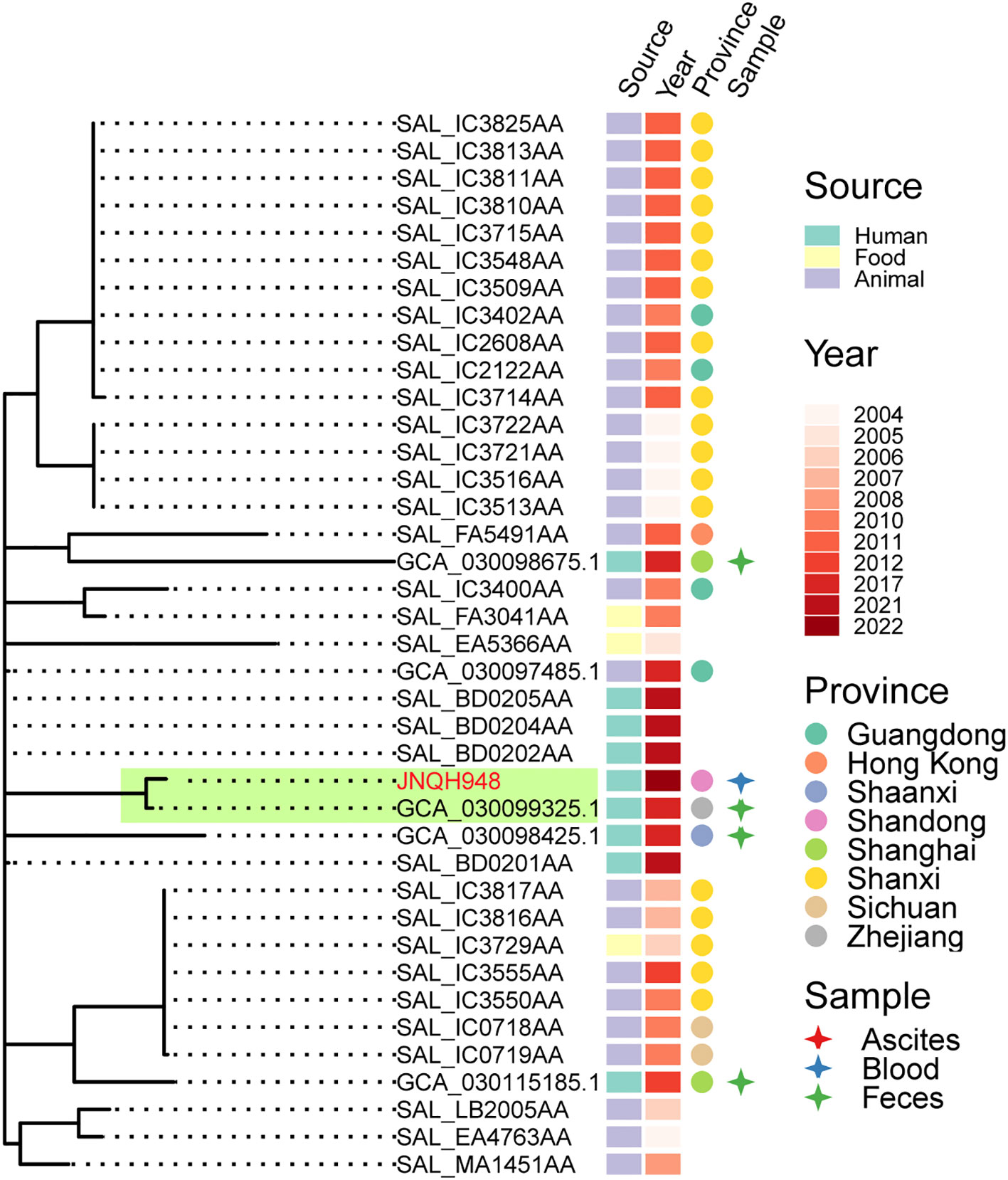
Figure 3 Phylogenetic relationships within S. Mbandaka strain JNQH948 and publicly available sequences from EnteroBase and a Chinese study. Phylogeny (left) showing the relationships. The strain source, collection year, location and specimen types were shown for each isolate. Subclade containing the JNQH948 and the closely related S. Mbandaka strain were indicated in light green shade.
Genomic and phenotypic characterization of Salmonella strains
Whole-genome sequences were obtained for four strains (JNQH948, 950, 952, 940). S. Enteritidis strains JNQH940 and JNQH952 harbored three and two plasmids respectively. S. Thompson strain JNQH950 carried two plasmids whereas no plasmids were identified in S. Mbandaka strain JNQH948. The serovars of all isolates predicted to be consistent with Salmonella serotyping by slide agglutination. MLST revealed the S. Mbandaka isolate (JNQH948) belonged to ST413. S. Enteritidis (JNQH940, 952) and S. Thompson (JNQH950) were classified as ST11 and ST26 respectively. The resistance genotypes predicted by RGI revealed ampicillin resistance were mediated by blaTEM-1. For the MDR JNQH940, resistance to quinolone (CIP) was conferred by qnrS1. The resistance of TMP-SMZ was associated with the presence of trimethoprim resistant sul1/sul2 genes. JNQH940 was found to carry the blaCTX-M-15 gene. Notably, the virulence cluster of mrkABCDF was detected in S. Thompson strain JNQH950 but absent in other strains.
blaCTX-M-15 was carried by a highly transferrable IncB/O/K/Z plasmid in S. Enteritidis strain JNQH940
The blaCTX-M-15-harboring plasmid in strain JNQH940 was determined to be an IncB/O/K/Z plasmid type, with a length of 127,061bp base pairs. The plasmid contains genes associated with plasmid conjugation, as well as a region comprising genes to encode a type IV pilus system. The conjugation elements included the origin of transfer site (oriT), type IV coupling protein gene (T4CP), and genes encoding the relaxase and some type IV secretion components (T4SS) (Figure 4). Conjugation assays showed the IncB/O/K/Z plasmid was successfully transferred into E. coli J53 from JNQH940 strain. The conjugation frequency was 10-3 per recipient cell, which was further confirmed by PCR targeting the blaCTX-M-15 gene and IncB/O/K/Z replicon. In addition to blaCTX-M-15, the plasmid also harbored 9 additional antimicrobial resistance genes encoding resistance to fluroquinolone (qnrS1), macrolide (mphA, mrx), sulfonamide (sul1/sul2), antiseptics (qacE) and aminoglycoside [aadA5, APH(3’)-Ib, APH(6)-Id].
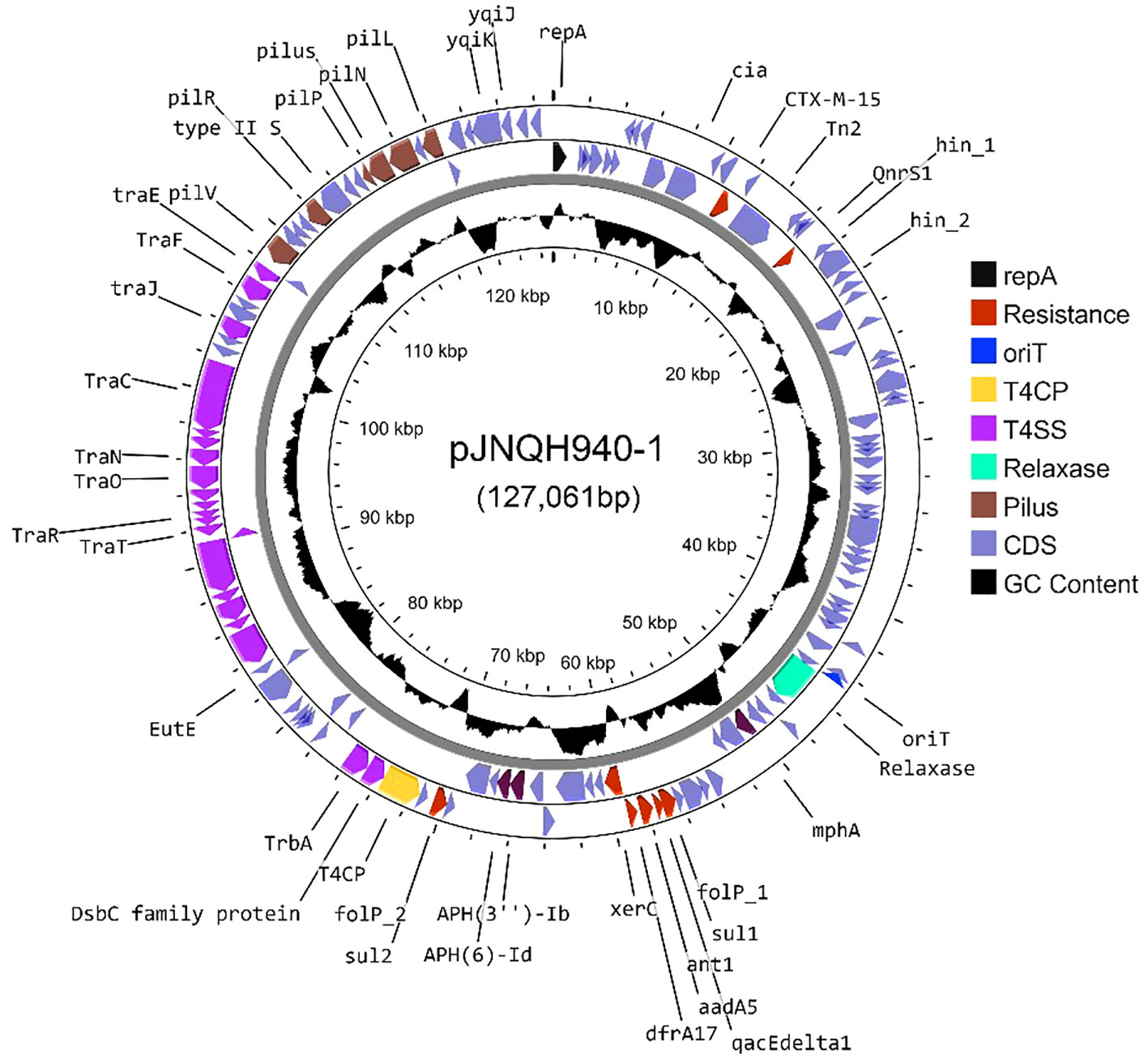
Figure 4 A circle map of the blaCTX-M-15-bearing IncB/O/K/Z type plasmid pJNQH940-1 in strain JNQH940. Open reading frames (ORFs) of pJNQH940-1 are shown as the outermost ring, with plasmid replicons, transconjugation elements (T4CP, T4SS, oriT, relaxase), type IV pilus system and antimicrobial resistance genes highlighted.
JNQH950 exhibited significantly higher biofilm-forming capabilities mediated by mrkABCDF operon in an IncX1 plasmid
We investigated the biofilm-forming capabilities of various Salmonella strains. The OD590 for JNQH950 was determined to be 1.187, while the other strains exhibited an OD590 measurement below 0.29. ATCC14028, which served as a reference, was measured to be 0.284. These results underscore the unique biofilm-forming potential of JNQH950 when compared to other tested strains (Figure 5). Further WGS analysis revealed the backbone structure of mrkABCDF cluster harboring plasmid belonged to an IncX1 plasmid, with a length of 40,394 base pairs. The mrkABCDF cassettes were contained within two IS1A elements of the IS1 family, constituting a transposon of Tn6011. These mrkABCDF cassettes encode the main structural subunit and assembly machinery of type 3 fimbriae, which are associated with biofilm formation. In addition, the MGEs associated with plasmid replication, stability, transfer integration was predicted. BLAST query against GenBank database showed that plasmid was closely related to plasmid pCP8-3-IncX1 (NZ_CP053740.1, with 90% coverage and 99.65% identity), pOLA52 (NC_010378.1, with 95% coverage and 99.47% identity), and some others plasmids hosted by Enterobacterales (Figure 6).
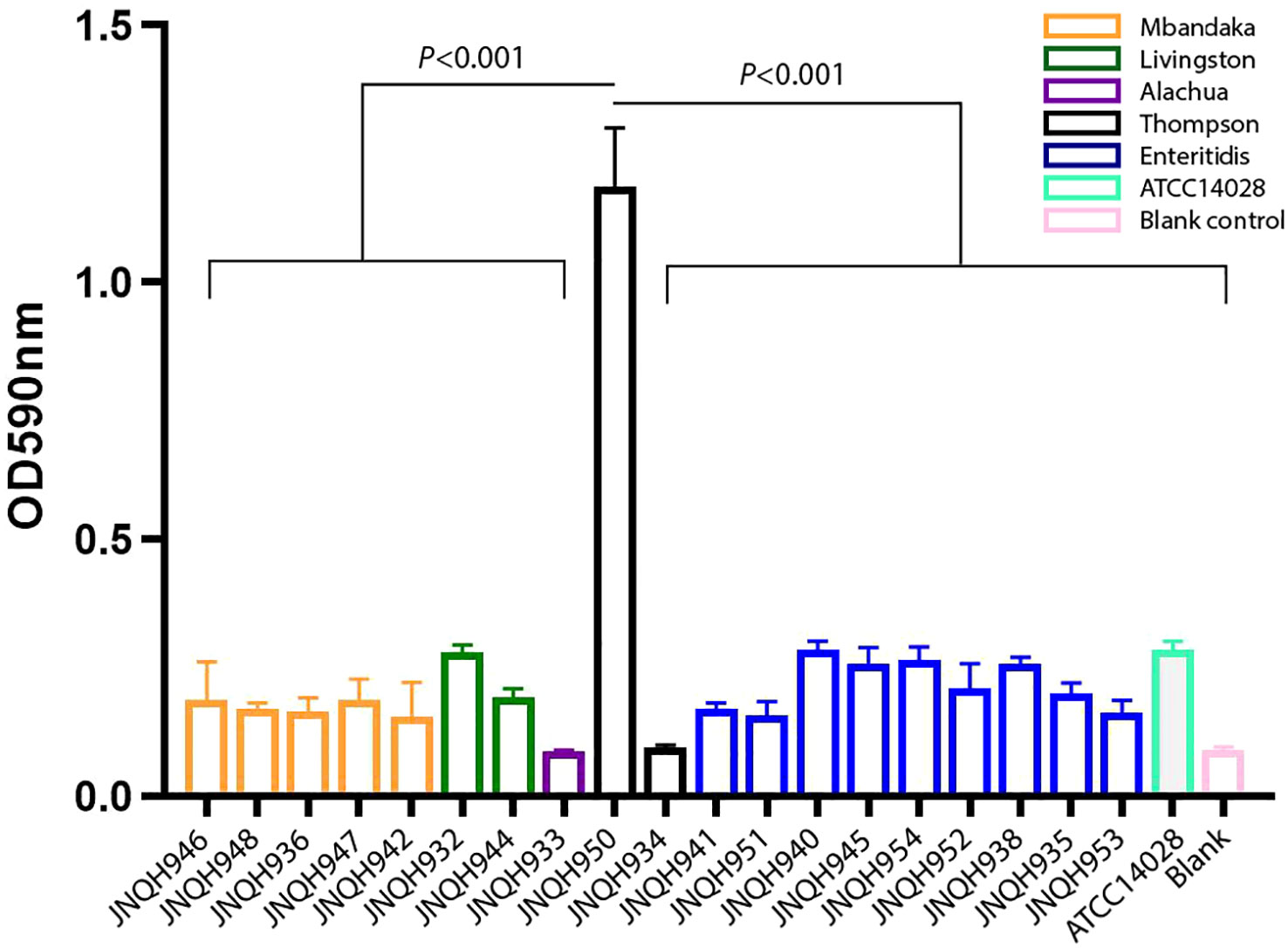
Figure 5 Biofilm formation of NTS strains. Data are represented as mean ± standard deviation from at least three independent experiments.
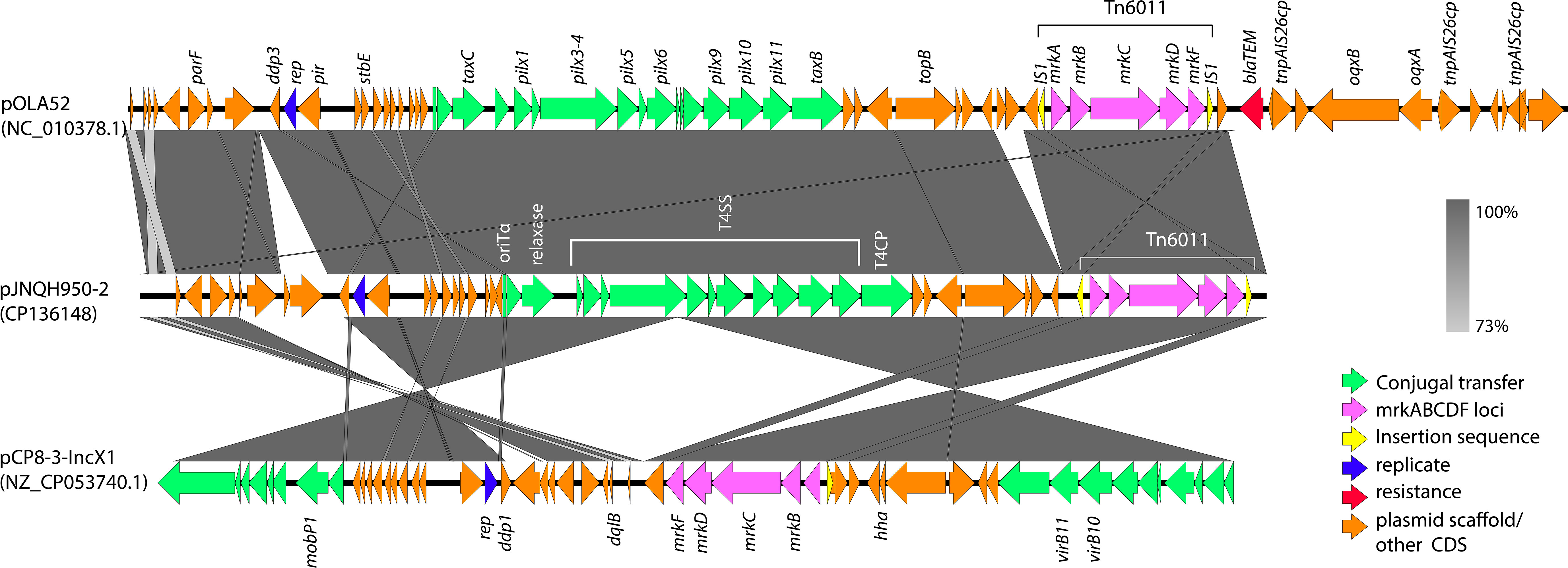
Figure 6 Genetic comparison of mrkABCDF harboring plasmid pJNQH950-2 and two closely related to plasmid pCP8-3-IncX1 (NZ_CP053740.1), pOLA52 (NC_010378.1). mrkABCDF transposon (Tn6011) are shown as the outermost ring. The genes are exhibited in arrows with different colors to note on their functional classes. Shades indicate shared regions of homology. The plasmid accession numbers are listed below the plasmid names.
Tracking key virulence loci encoding mrkABCDF operon in global plasmids revealed their prevalence in Enterobacteriaceae
A total of 195 plasmids harboring mrkABCDF operon were identified in the reference plasmids in NCBI database, most of which are carried by Enterobacteriaceae. K. pneumoniae is the dominant host strain, followed by E. coli, E. hormaechei, K. quasipneumoniae. These plasmids belong to several incompatibility groups, predominant by IncFIB, IncFII, IncFIA, IncX, IncR. Plasmid sequence analysis by oriTfinder revealed the presence of four crucial conjugation elements (oriT, T4SS, T4CP, relaxase gene) in 31.2% (61/195) of the plasmids. The majority of these plasmids were found in host strains isolated in the environmental sources. The top 5 countries were UK, China, USA, India and Austria. Notably, plasmids originating from homo sapiens were most commonly detected in urine samples. It is noteworthy that 60 of the analyzed plasmids coharbored antibiotic resistance genes, including 6 carbapenem resistance genes and 17 mcr genes. mrkABCDF operon harboring plasmids has been present in E. coli strains dating back to the 1950s. Eight plasmids comprised of a cluster which had a close relationship with our plasmids, all of which belonged to IncX1 plasmid type and were carried by E. coli strains (Figure 7).
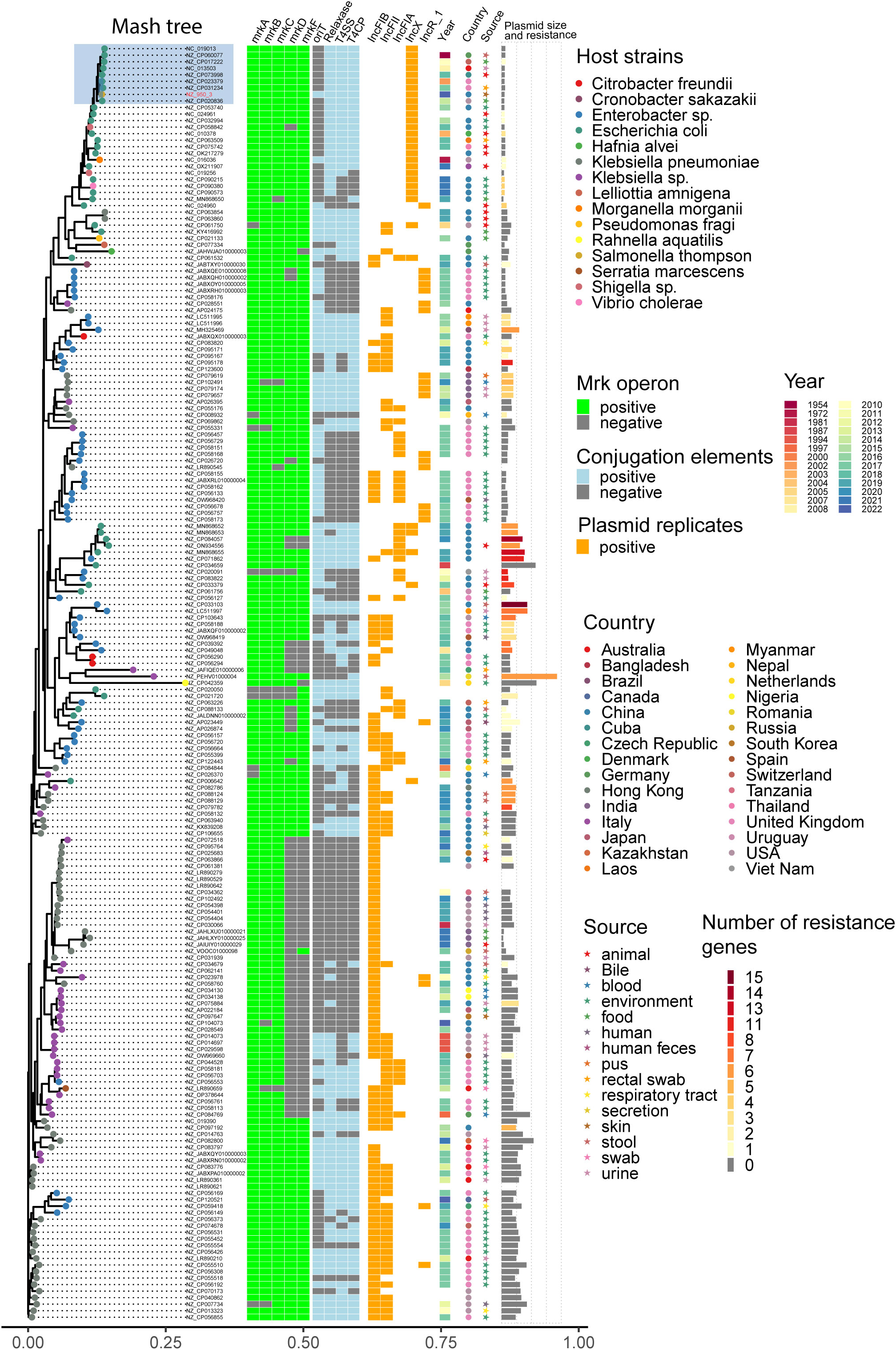
Figure 7 Distance tree of plasmids harboring mrkABCDF operon in reference NCBI database. Phylogeny (left) showing the relationships between 195 mrkABCDF operons harboring plasmids. The presence of mrkABCDF virulence loci, plasmid conjugation elements (oriT, T4SS, T4CP, relaxase), top five plasmid types, host strain properties (collection year, country, source), as well as the plasmid size and number of resistance genes were illustrated for each isolate. Grey tiles indicate gene’s absence. The isolates’ tip points are colored by the host species.
Discussion
During the year 2022, our hospital diagnosed a total of 19 patients with Salmonella infections, as determined through microbiologic cultures. The most commonly identified serovar was S. Enteritidis, closely followed by S. Mbandaka and some other sporadic serovars including S. Thompson, Livingston, Alachua and Infantis. The majority of cases were reported between the months of July and October, with the highest peak occurring in October. Genetic relatedness analysis suggested that S. Mbandaka isolates were part of a single clonal expansion whereas S. Enteritidis strains were composed of diverse clonal linages. A significant observation was that S. Alachua was isolated from the feces of a patient suffering from a gastro-infection, marking the first observation of this particular serovar in China.
Non-typhoidal Salmonella infections typically manifest as mild and self-limiting acute enterocolitis in the majority of individuals (Crump et al., 2015). Parenteral infections, resulting from the dissemination of the bacterium beyond the gastrointestinal tract, are infrequent complications of gastroenteritis (Graham et al., 2000). However, reports of nontyphoidal Salmonella causing intravascular, bone, and joint infections are widespread on a global scale. In Africa, NTS strains cause invasive disease with bacteraemia more often in children, with 4100 deaths per year (Crump et al., 2015; Akinyemi et al., 2023). Such invasive infections are particularly associated with various forms of immunocompromise, with individuals at the extremes of age or those with compromised immune systems (Crump et al., 2011; Parry et al., 2013; Crump et al., 2015; Akinyemi et al., 2021). In our study, nine of our patients experienced extra-intestinal infections, underscoring the potential severity of the encountered Salmonella strains. Our cases demonstrated a heightened vulnerability to these infections, possibly due to the presence of multiple comorbidities, including acute leukemia, diabetes, autoimmune diseases, and organ transplantation. Given the impaired immunity in these patients, swift and comprehensive clinical intervention is essential. This is because Salmonella can penetrate the compromised intestinal barrier and disseminate to multiple organs, potentially leading to sepsis and severe complications. Therefore, proactive and diligent medical management is crucial for these individuals to minimize adverse outcomes.
Cutaneous infections caused by Salmonella are exceedingly rare occurrences. Only a few reports have documented cutaneous manifestations in patients with S. enterica infection (Marzano et al., 2003; Desikan et al., 2009; Wiegand et al., 2009; Alfouzan et al., 2017). In our study, we observed that S. Thompson strain JNQH950 isolated from the skin secretion displayed a remarkable capacity for robust biofilm formation. Notably, this strain persisted for three months in the toe secretion of a 58-year-old male patient with type 2 diabetes mellitus. Despite antibiotic treatment and undergoing three operations of toe amputation and skin grafting, the condition worsened, leading to the necessity of foot amputation due to progressive tissue necrosis. Phenotypic analysis showed the S. Thompson strain JNQH950 could form strong biofilm compared with other strains. It is suspected that bacteria are difficult to eradicate when protected within biofilms, thus acquire the enhanced ability to persist and spread in the human body or the environment (Lamas et al., 2018; Gual-De-Torrella et al., 2022).
Salmonella sp. has the capability to develop biofilms, and bacteria within these biofilms exhibit increased resistance to drugs, chemicals, physical and mechanical stresses, as well as evasion of host immune responses (Lamas et al., 2018). The primary constituents of Salmonella biofilms are curli fimbriae and cellulose (Lamas et al., 2018). Interestingly, genetic analysis revealed an intact mrkABCDF operon in strain JNQH950, which had been demonstrated associated with type 3 fimbriae expression, surface attachment, and biofilm formation in K. pneumoniae (Norman et al., 2008; Ong et al., 2009). The mrkABCDF operon harboring plasmid had highly conserved plasmid synteny and structure with a previously reported pOLA52 plasmid in E.coli, which also is an IncX1 plasmid and showed a high conjugative capability (Norman et al., 2008). In addition, the gene cluster had acquired mobility flanking by IS1 (Tn6011) compared to its original gene cluster (Norman et al., 2008). Further genetic analysis showed the mrkABCDF gene cluster has now spread among various enterobacterial species, but was not reported in S. enterica strains (Figure 7). In this study, although the conjugation was not performed due to the lack of selective marker, we can suspect the mrkABCDF loci could be mobilized through plasmid conjugation or genetic loci transposon. Taken together, to the best of our knowledge, this was the first report of mrkABCDF operon within S. enterica responsible for human infection. The emergence of such Salmonella strain emphasizes the urgent need for meticulous monitoring and surveillance to address potential public health implications. Nevertheless, the molecular mechanism responsible for the contribution of mrkABCDF to biofilm formation and enhanced antibiotic treatment resistance in S. enterica is yet to be elucidated.
The increasing prevalence of antibiotic resistance, particularly towards fluoroquinolones and third-generation cephalosporins, presents formidable challenges in clinical management (Fierer, 2022). Infections caused by antimicrobial-resistant Salmonella, especially those resistant to quinolones, pose a significant threat to human populations, leading to higher morbidity and mortality rates (Varma et al., 2005; Xia et al., 2009). Of particular concern is the emergence of nontyphoidal Salmonella strains that exhibit resistance to extended-spectrum cephalosporins, such as ceftazidime and ceftriaxone, which raises substantial public health concerns (Crump et al., 2015; Fakorede et al., 2023). These third-generation cephalosporins play a crucial role in managing invasive Salmonella infections, especially in vulnerable populations like children, for whom fluoroquinolones may not be the preferred treatment. Our study identified two MDR isolates that were resistant to AMP, CIP and TMP-SMZ, one of which exhibited resistance to third-generation cephalosporins. The presence of blaCTX-M-15 gene, conferring cephalosporin resistance, was located on a transferable IncB/O/K/Z plasmid. The plasmid carried all essential elements necessary for conjugation and, importantly, also harbored the type IV pilus system, known to significantly enhance conjugation both in vivo and in vitro (Neil et al., 2020). Consequently, the combination of fluoroquinolone and cephalosporin resistance, carried on a highly transferable plasmid in these Salmonella isolates, raises serious concerns about the potential dissemination of these resistance traits. Vigilant monitoring and the implementation of comprehensive strategies are urgently needed to address this pressing public health issue.
Our data also showed that S. Enteritidis were the most common serovars detected in our hospital, which is consistent with the findings in other studies in China (Wang et al., 2015; Wang et al., 2023). In addition, the proportion of S. Mbandaka was reported to be 0.25% of the total NTS collection, most of which were from non-human origin (Wang et al., 2023). Genetic analysis revealed that the S. Enteritidis strains in this study displayed significant genetic diversity, in line with findings from an epidemiological study in China (Wang et al., 2015). On the other hand, our analysis of S. Mbandaka isolates revealed a different scenario. These isolates formed a single clonal lineage, indicating a common origin. A recent study in China had reported S. Mbandaka could be clonally transmitted between broiler farm and slaughterhouse (Wang et al., 2021b). Further comparison with a large-scale collection of S. Mbandaka in China identified one closely related strain, which was isolated from human feces in Zhejiang in 2017. Given previous studies suggesting S. Mbandaka’s adaptation to survival in the farm environment (Hayward et al., 2016), our findings suggest that this clone in this study likely originated from a single source that persisted over 5 years in China.
In summary, our research provides the phenotypic and molecular overview of current serovar prevalence of NTS strains isolated from human origins in China. S. Enteritidis strains displayed significant genetic diversity while S. Mbandaka strains were suspected to derive from a single clonal expansion. The further spread of the blaCTX-M-15 harboring IncB/O/K/Z plasmids and spread of virulent mrkABCDF operon into Salmonella in China and other global regions should be closely monitored.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
The studies involving humans were approved by The First Affiliated Hospital of Shandong First Medical University. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributions
WM: Conceptualization, Methodology, Supervision, Writing – original draft. XC: Data curation, Methodology, Writing – original draft. XD: Writing – original draft. XL: Methodology, Writing – original draft. KL: Visualization, Writing – original draft. YW: Software, Writing – original draft. XS: Project administration, Writing – original draft. LC: Validation, Visualization, Writing – review & editing. MH: Conceptualization, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Natural Science Foundation of Shandong Province, China [grant number ZR2021MH078]; Clinical & Medical Science and Technology Innovation Program of Jinan, Shandong Province [grant number 202134040]; and Cultivate Fund from The First Affiliated Hospital of Shandong First Medical University & Shandong Provincial Qianfoshan Hospital [grant numbers QYPY2022NSFC0802].
Acknowledgments
We like to thank Bingqing Li for the gift of S. Typhimurium ATCC14028 strain.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Akinyemi, K. O., Ajoseh, S. O., Fakorede, C. O. (2021). A systemic review of literatures on human Salmonella enterica serovars in Nigeria, (1999-2018). J. Infect. Dev. Ctries. 15, 1222–1235. doi: 10.3855/jidc.12186
Akinyemi, K. O., Fakorede, C. O., Linde, J., Methner, U., Wareth, G., Tomaso, H., et al. (2023). Whole genome sequencing of Salmonella enterica serovars isolated from humans, animals, and the environment in Lagos, Nigeria. BMC Microbiol. 23, 164. doi: 10.1186/s12866-023-02901-1
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Alfouzan, W., Bulach, D., Izumiya, H., Albassam, K., Sheikh, S., Alrubai’aan, N., et al. (2017). Carbuncle due to Salmonella Enteritidis: a novel presentation. Gut Pathog. 9, 51. doi: 10.1186/s13099-017-0200-2
Amuasi, J. H., May, J. (2019). Non-typhoidal salmonella: invasive, lethal, and on the loose. Lancet Infect. Dis. 19, 1267–1269. doi: 10.1016/S1473-3099(19)30521-3
Branchu, P., Bawn, M., Kingsley, R. A. (2018). Genome variation and molecular epidemiology of Salmonella enterica serovar typhimurium pathovariants. Infect. Immun. 86, e00079–18. doi: 10.1128/IAI.00079-18
Carattoli, A., Zankari, E., Garcia-Fernandez, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
CLSI (2021). Performance Standards for Antimicrobial Susceptibility Testing, M100. 31st Edition (Wayne, PA: Clinical and Laboratory Standards Institute).
Croucher, N. J., Page, A. J., Connor, T. R., Delaney, A. J., Keane, J. A., Bentley, S. D., et al. (2015). Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 43, e15. doi: 10.1093/nar/gku1196
Crump, J. A., Medalla, F. M., Joyce, K. W., Krueger, A. L., Hoekstra, R. M., Whichard, J. M., et al. (2011). Antimicrobial resistance among invasive nontyphoidal Salmonella enterica isolates in the United States: National Antimicrobial Resistance Monitoring System 1996 to 2007. Antimicrob. Agents Chemother. 55, 1148–1154. doi: 10.1128/AAC.01333-10
Crump, J. A., Sjolund-Karlsson, M., Gordon, M. A., Parry, C. M. (2015). Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive Salmonella infections. Clin. Microbiol. Rev. 28, 901–937. doi: 10.1128/CMR.00002-15
Desikan, P., Kumar, Y., Pande, H. K., Jain, A., Panwalkar, N., Verma, M., et al. (2009). Isolated ulcerative skin lesion caused by Salmonella Weltevreden. J. Infect. Dev. Ctries. 3, 569–571. doi: 10.3855/jidc.477
Fakorede, C. O., Amisu, K. O., Saki, M., Akinyemi, K. O. (2023). Co-existence of extended-spectrum beta-lactamases bla(CTX-M-9) and bla(CTX-M-15) genes in Salmonella species isolated from febrile and diarrhoeagenic patients in Lagos, Nigeria: a cross-sectional study. Eur. J. Med. Res. 28, 3. doi: 10.1186/s40001-022-00960-0
Fierer, J. (2022). Invasive non-typhoidal Salmonella (iNTS) infections. Clin. Infect. Dis. 75, 732–738. doi: 10.1093/cid/ciac035
Gebreyes, W. A., Thakur, S., Dorr, P., Tadesse, D. A., Post, K., Wolf, L. (2009). Occurrence of spvA virulence gene and clinical significance for multidrug-resistant Salmonella strains. J. Clin. Microbiol. 47, 777–780. doi: 10.1128/JCM.01660-08
Graham, S. M., Molyneux, E. M., Walsh, A. L., Cheesbrough, J. S., Molyneux, M. E., Hart, C. A. (2000). Nontyphoidal Salmonella infections of children in tropical Africa. Pediatr. Infect. Dis. J. 19, 1189–1196. doi: 10.1097/00006454-200012000-00016
Gual-De-Torrella, A., Delgado-Valverde, M., Perez-Palacios, P., Oteo-Iglesias, J., Rojo-Molinero, E., Macia, M. D., et al. (2022). Prevalence of the fimbrial operon mrkABCD, mrkA expression, biofilm formation and effect of biocides on biofilm formation in carbapenemase-producing Klebsiella pneumoniae isolates belonging or not belonging to high-risk clones. Int. J. Antimicrob. Agents 60, 106663. doi: 10.1016/j.ijantimicag.2022.106663
Hao, M., Ma, W., Dong, X., Li, X., Cheng, F., Wang, Y. (2021a). Comparative genome analysis of multidrug-resistant Pseudomonas aeruginosa JNQH-PA57, a clinically isolated mucoid strain with comprehensive carbapenem resistance mechanisms. BMC Microbiol. 21, 133. doi: 10.1186/s12866-021-02203-4
Hao, M., Schuyler, J., Zhang, H., Shashkina, E., Du, H., Fouts, D. E., et al. (2021b). Apramycin resistance in epidemic carbapenem-resistant Klebsiella pneumoniae ST258 strains. J. Antimicrob. Chemother. 76, 2017–2023. doi: 10.1093/jac/dkab131
Hayward, M. R., Petrovska, L., Jansen, V. A., Woodward, M. J. (2016). Population structure and associated phenotypes of Salmonella enterica serovars Derby and Mbandaka overlap with host range. BMC Microbiol. 16, 15. doi: 10.1186/s12866-016-0628-4
Heras, J., Dominguez, C., Mata, E., Pascual, V., Lozano, C., Torres, C., et al. (2015). GelJ–a tool for analyzing DNA fingerprint gel images. BMC Bioinf. 16, 270. doi: 10.1186/s12859-015-0703-0
Katz, L. S., Griswold, T., Morrison, S. S., Caravas, J. A., Zhang, S., Den Bakker, H. C., et al. (2019). Mashtree: a rapid comparison of whole genome sequence files. J. Open Source Softw. 4, 1762. doi: 10.21105/joss.01762
Kurtz, S., Phillippy, A., Delcher, A. L., Smoot, M., Shumway, M., Antonescu, C., et al. (2004). Versatile and open software for comparing large genomes. Genome Biol. 5, R12. doi: 10.1186/gb-2004-5-2-r12
Lamas, A., Regal, P., Vazquez, B., Miranda, J. M., Cepeda, A., Franco, C. M. (2018). Salmonella and Campylobacter biofilm formation: a comparative assessment from farm to fork. J. Sci. Food Agric. 98, 4014–4032. doi: 10.1002/jsfa.8945
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Li, X., Xie, Y., Liu, M., Tai, C., Sun, J., Deng, Z., et al. (2018). oriTfinder: a web-based tool for the identification of origin of transfers in DNA sequences of bacterial mobile genetic elements. Nucleic Acids Res. 46, W229–W234. doi: 10.1093/nar/gky352
Liu, B., Zheng, D., Zhou, S., Chen, L., Yang, J. (2022). VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. 50, D912–D917. doi: 10.1093/nar/gkab1107
Ma, W., Zhu, B., Wang, W., Wang, Q., Cui, X., Wang, Y., et al. (2023). Genetic and enzymatic characterization of two novel bla(NDM-36, -37) variants in Escherichia coli strains. Eur. J. Clin. Microbiol. Infect. Dis. 42, 471–480. doi: 10.1007/s10096-023-04576-y
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Marchello, C. S., Birkhold, M., Crump, J. A., Vacc-I, N.T.S.C.C. (2022). Complications and mortality of non-typhoidal salmonella invasive disease: a global systematic review and meta-analysis. Lancet Infect. Dis. 22, 692–705. doi: 10.1016/S1473-3099(21)00615-0
Marzano, A. V., Mercogliano, M., Borghi, A., Facchetti, M., Caputo, R. (2003). Cutaneous infection caused by Salmonella typhi. J. Eur. Acad. Dermatol. Venereol. 17, 575–577. doi: 10.1046/j.1468-3083.2003.00797.x
Neil, K., Allard, N., Grenier, F., Burrus, V., Rodrigue, S. (2020). Highly efficient gene transfer in the mouse gut microbiota is enabled by the Incl(2) conjugative plasmid TP114. Commun. Biol. 3, 523. doi: 10.1038/s42003-020-01253-0
Norman, A., Hansen, L. H., She, Q., Sorensen, S. J. (2008). Nucleotide sequence of pOLA52: a conjugative IncX1 plasmid from Escherichia coli which enables biofilm formation and multidrug efflux. Plasmid 60, 59–74. doi: 10.1016/j.plasmid.2008.03.003
Ong, C. L., Beatson, S. A., Mcewan, A. G., Schembri, M. A. (2009). Conjugative plasmid transfer and adhesion dynamics in an Escherichia coli biofilm. Appl. Environ. Microbiol. 75, 6783–6791. doi: 10.1128/AEM.00974-09
Page, A. J., Taylor, B., Delaney, A. J., Soares, J., Seemann, T., Keane, J. A., et al. (2016). SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb. Genom. 2, e000056. doi: 10.1099/mgen.0.000056
Parry, C. M., Thomas, S., Aspinall, E. J., Cooke, R. P., Rogerson, S. J., Harries, A. D., et al. (2013). A retrospective study of secondary bacteraemia in hospitalised adults with community acquired non-typhoidal Salmonella gastroenteritis. BMC Infect. Dis. 13, 107. doi: 10.1186/1471-2334-13-107
Price, M. N., Dehal, P. S., Arkin, A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Schmartz, G. P., Hartung, A., Hirsch, P., Kern, F., Fehlmann, T., Muller, R., et al. (2022). PLSDB: advancing a comprehensive database of bacterial plasmids. Nucleic Acids Res. 50, D273–D278. doi: 10.1093/nar/gkab1111
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Sullivan, M. J., Petty, N. K., Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Varma, J. K., Molbak, K., Barrett, T. J., Beebe, J. L., Jones, T. F., Rabatsky-Ehr, T., et al. (2005). Antimicrobial-resistant nontyphoidal Salmonella is associated with excess bloodstream infections and hospitalizations. J. Infect. Dis. 191, 554–561. doi: 10.1086/427263
Wang, Y. T., Lei, C. W., Liu, S. Y., Chen, X., Gao, Y. F., Zhang, Y., et al. (2021b). Tracking Salmonella enterica by whole genome sequencing of isolates recovered from broiler chickens in a poultry production system. Int. J. Food Microbiol. 350, 109246. doi: 10.1016/j.ijfoodmicro.2021.109246
Wang, Y., Liu, Y., Lyu, N., Li, Z., Ma, S., Cao, D., et al. (2023). The temporal dynamics of antimicrobial-resistant Salmonella enterica and predominant serovars in China. Natl. Sci. Rev. 10, nwac269. doi: 10.1093/nsr/nwac269
Wang, Y., Yang, B., Wu, Y., Zhang, Z., Meng, X., Xi, M., et al. (2015). Molecular characterization of Salmonella enterica serovar Enteritidis on retail raw poultry in six provinces and two National cities in China. Food Microbiol. 46, 74–80. doi: 10.1016/j.fm.2014.07.012
Wang, Y., Zhu, B., Liu, M., Dong, X., Ma, J., Li, X., et al. (2021a). Characterization of IncHI1B Plasmids Encoding Efflux Pump TmexCD2-ToprJ2 in Carbapenem-Resistant Klebsiella variicola, Klebsiella quasipneumoniae, and Klebsiella michiganensis Strains. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.759208
Wen, S. C., Best, E., Nourse, C. (2017). Non-typhoidal Salmonella infections in children: Review of literature and recommendations for management. J. Paediatr. Child Health 53, 936–941. doi: 10.1111/jpc.13585
Wick, R. R., Judd, L. M., Gorrie, C. L., Holt, K. E. (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PloS Comput. Biol. 13, e1005595. doi: 10.1371/journal.pcbi.1005595
Wiegand, G., Rauch, R., Hermann, M., Apitz, C., Hofbeck, M., Heininger, U. (2009). [Salmonella enteritidis infection presenting with septic shock, renal failure and cutaneous manifestations]. Klin. Padiatr. 221, 41–43. doi: 10.1055/s-2007-984366
Xia, S., Hendriksen, R. S., Xie, Z., Huang, L., Zhang, J., Guo, W., et al. (2009). Molecular characterization and antimicrobial susceptibility of Salmonella isolates from infections in humans in Henan Province, China. J. Clin. Microbiol. 47, 401–409. doi: 10.1128/JCM.01099-08
Yu, G., Smith, D. K., Zhu, H., Guan, Y., Lam, T. T. Y. (2017). ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36. doi: 10.1111/2041-210X.12628
Zhang, S., Den Bakker, H. C., Li, S., Chen, J., Dinsmore, B. A., Lane, C., et al. (2019). SeqSero2: rapid and improved Salmonella serotype determination using whole-genome sequencing data. Appl. Environ. Microbiol. 85, e01746–19. doi: 10.1128/AEM.01746-19
Keywords: nontyphoidal Salmonella, antibiotic resistance, mrkABCDF operon, phylogenetic analysis, biofilm
Citation: Ma W, Cui X, Dong X, Li X, Liu K, Wang Y, Shi X, Chen L and Hao M (2024) Characterization of nontyphoidal Salmonella strains from a tertiary hospital in China: serotype diversity, multidrug resistance, and genetic insights. Front. Cell. Infect. Microbiol. 13:1327092. doi: 10.3389/fcimb.2023.1327092
Received: 24 October 2023; Accepted: 20 December 2023;
Published: 09 January 2024.
Edited by:
Manuel Gerardo Ballesteros Monrreal, University of Sonora, MexicoReviewed by:
María Cristina González Vázquez, Benemérita Universidad Autónoma de Puebla, MexicoAsive Myataza, National Health Laboratory Service (NHLS), South Africa
Copyright © 2024 Ma, Cui, Dong, Li, Liu, Wang, Shi, Chen and Hao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingju Hao, aGFvbWluZ2p1QDE2My5jb20=
†These authors contribute equally to this work
 Wanshan Ma1†
Wanshan Ma1† Yujiao Wang
Yujiao Wang Xiaohong Shi
Xiaohong Shi Liang Chen
Liang Chen Mingju Hao
Mingju Hao