
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol., 11 January 2024
Sec. Antibiotic Resistance and New Antimicrobial drugs
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1324846
This article is part of the Research TopicResources for Developing Plasmid DatabasesView all 6 articles
Background: Today, the blaNDM gene is widely distributed on several plasmids from a variety of Gram-negative bacteria, primarily in transposons and gene cassettes within their multidrug-resistant (MDR) regions. This has led to the global dissemination of the blaNDM gene.
Methods: The determination of class A beta-lactamase, class B and D carbapenemases was performed according to the recommendations of the Clinical and Laboratory Standards Institute (CLSI). Antimicrobial susceptibility testing was performed using both the BioMerieux VITEK2 system and antibiotic paper diffusion methods. Plasmid transfer was then evaluated by conjugation experiments and plasmid electroporation assays. To comprehensively analyze the complete genome of K. pneumoniae strain F11 and to investigate the presence of mobile genetic elements associated with antibiotic resistance and virulence genes, Nanopore and Illumina sequencing platforms were used, and bioinformatics methods were applied to analyze the obtained data.
Results: Our findings revealed that K. pneumoniae strain F11 carried class A beta-lactamase and classes B+D carbapenemases, and exhibited resistance to commonly used antibiotics, particularly tigecycline and ceftazidime/avibactam, due to the presence of relevant resistance genes. Plasmid transfer assays demonstrated successful recovery of plasmids pA_F11 and pB_F11, with average conjugation frequencies of 2.91×10-4 and 1.56×10-4, respectively. However, plasmids pC_F11 and pD_F11 failed in both conjugation and electroporation experiments. The MDR region of plasmid pA_F11 contained rare In1765, TnAs2, and TnAs3 elements. The MDR2 region of plasmid pB_F11 functioned as a mobile genetic “island” and lacked the blaNDM-1 gene, serving as a “bridge” connecting the early composite structure of bleMBL and blaNDM-1 to the recent composite structure. Additionally, the MDR1 region of plasmid pB_F11 comprised In27, TnAs1, TnAs3, and Tn2; and plasmid pC_F11 harbored the recent composite structure of bleMBL and blaNDM-1 within Tn3000 which partially contained partial Tn125.
Conclusion: This study demonstrated that complex combinations of transposons and integron overlaps, along with the synergistic effects of different drug resistance and virulence genes, led to a lack of effective therapeutic agents for strain F11, therefore its dissemination and prevalence should be strictly controlled.
NDM-producing bacteria exhibited the ability to hydrolyze a wide range of beta- lactam antibiotics, except for monolactams (Nordmann et al., 2011), consequently, therapeutic options for infections caused by NDM-producing strains were severely limited, resulting in significantly higher mortality rates among infected patients (Logan and Weinstein, 2017; Guducuoglu et al., 2018). The blaNDM-1 gene was commonly associated with composite genes that included the aminoglycoside resistance gene aphA6 and the metallo-beta-lactamase (MBL) gene. Typically, these composite genes were located adjacent to either intact or truncated copies of ISAba125 (Toleman et al., 2012). The blaNDM-1 gene was almost always found in association with mobile genetic elements (Loh et al., 2020), contributing to its evolutionary potential and rapid dissemination (Wu et al., 2019). As a result, the blaNDM-1 gene had emerged as a major global public health concern (Walsh et al., 2011; Dortet et al., 2014). Among NDM-positive Enterobacteriaceae bacteria, K. pneumoniae accounted for over half of the cases, followed by Escherichia coli and the Enterobacter cloacae complex (Wu et al., 2019).
Furthermore, the presence of NDM had also been detected in other species, including Acinetobacter and Pseudomonas (Costa et al., 2021). Poirel et al. proposed a hypothesis suggesting that the initial integration of the blaNDM gene occurred within the chromosome of Acinetobacter originated from an unidentified environmental species. Subsequently, two copies of the insertion sequence (IS) ISAba125 captured the blaNDM gene, leading to the formation of the ISAba125 complex transposon known as Tn125 (Poirel et al., 2012). Through the transfer of the Tn125 transposon, the blaNDM gene disseminated among Enterobacteriaceae, Acinetobacter, and Pseudomonas. Consequently, both the blaNDM and its Tn125 components could be found within MDR regions of relevant bacterial plasmids or chromosomes, exhibiting various truncations and deletions (Wu et al., 2019). Notably, the truncated insertion sequence ISAba125 consistently appeared upstream of blaNDM, while the bleomycin resistance gene (bleMBL) emerged downstream of it. These genetic features were commonly observed in other typical contexts where blaNDM was present (Wu et al., 2019). Additionally, the expression of bleMBL and blaNDM was driven by the same promoter, resulting in their co-expression (Dortet et al., 2017).
In addition to transposons, the involvement of other mobile genetic elements, including IS elements and integrons, had been implicated in the mobilization of NDM (Guducuoglu et al., 2018). The rapid dissemination of these mobile genetic elements had contributed to the unprecedented and widespread prevalence of NDM, posing a significant threat to global healthcare. Consequently, there was an urgent need to focus on controlling the potentially dangerous proliferation of NDM within hospital settings. To date, a total of 31 subtypes of NDM had been identified, with NDM-1 remaining the most prevalent worldwide (https://www.bldb.eu). NDM-1 was widely distributed among various Enterobacteriaceae and other Gram-negative bacteria, while NDM-5 appeared to be more prevalent in E. coli (Wu et al., 2019). The ability of NDM-5 to efficiently dissemination among bacteria carrying incompatible plasmids was a significant worries (Bonnin et al., 2012), raising alarm about the dissemination of high-risk clones (Costa et al., 2021). Moreover, NDM-5 exhibited higher levels of antibiotic resistance compared to NDM-1 (Hornsey et al., 2011), posing a particular challenge for clinical treatment (Costa et al., 2021). NDM-producing strains had disseminated the gene to a wide range of Gram-negative bacteria via MDR plasmids, leading to serious concerns that patients infected with these bacteria might be left without effective treatment options.
In this study, the main focus was to compare and analyze the F11_plasmids, which carried the NDM-1 gene along with the rare integron In1765, in comparison with related plasmids. The study aimed to characterize the structure of these plasmids and to investigate the evolutionary processes associated with NDM.
K. pneumoniae F11 strain was isolated from the sputum of an hospitalized patient in our hospital in 2020. EC600 and E. coli DH5α were used as hosts for cloning. The almost complete 16S rRNA gene for K. pneumoniae F11 strain was amplified by PCR using the following primers AGAGTTTGATYMTGGCTCAG (forward) and TACCTTGTTACGACTT (Y, T, or C; M, A, or C) (reverse). The PCR reaction included a Taq enzyme, which was a mixture of Fermentas Taq and Pfu enzymes in a ratio of 3:1 (ThermoFisher Scientific, Burlington, VT, USA). Each 30 ml reaction contained 1.5 U of the enzyme. The PCR amplification was carried out using a temperature program consisting of 30 cycles. The program started with an initial denaturation step at 94°C for 3 minutes, followed by denaturation at 94°C for 40 seconds, annealing at 50°C for 40 seconds, extension at 72°C for 1 minute, and a final extension step at 72°C for 5 minutes. The length of the amplified fragment was approximately 1,500 bp (Frank et al., 2008). PCR products were confirmed by sequencing in both directions.
In accordance with a previously reported method (Chen et al., 2015), conjugation experiments were conducted using lysogeny broth (LB) as the medium. The recipient strain was strain EC600 (TaKaRa, China), while the donor was strain F11. In LB cultures, both donor and recipient bacteria experienced a brief lag phase of 0.5 to 1 hour before growth begins. Between 1.5 and 4 hours, the cultures were in the logarithmic phase and then enter the stationary phase at about 4 hours. To initiate the conjugation process, 0.5 mL of logarithmic phase cultures of both donor and recipient cells were mixed together in a total volume of 4 mL of fresh LB medium. The mixture was then incubated at a temperature of 35°C for a duration of 18 to 24 hours without shaking. Following the incubation period, transconjugants were selected by plating the conjugation mixture onto trypticase soy agar (TSA) plates supplemented using 1 ml with 10 μg/L of rifampicin and 0.02 μg/L of imipenem. These antibiotics were included in the selection medium to inhibit the growth of the donor and recipient strains, while allowing the growth of the transconjugants that acquired the resistance genes during conjugation.
In the transformation experiments, E. coli DH5α cells (TaKaRa, China) were used as the recipient cells for plasmid electroporation. The conjugation frequency was calculated as the number of transconjugants obtained per initial donor cell. To prepare electrocompetent cells for electroporation, the bacteria were grown until they reaching an optical density at 600 nm (OD600) of 0.5–0.6. Cells were then harvested by centrifugation at 4°C. Two rounds of washing and centrifugation were performed at 4°C using 1 volume of milliQ water, followed by centrifugation at 6,000 rpm (4,025×g). In the final wash step, the cells were resuspended in 1/50 volume of 10% glycerol. The cells were then aliquoted into 50 μL samples, frozen on dry ice, and stored at -70°C until further use.
For the electroporation step, aliquots of the electrocompetent cells were thawed and mixed with less than 10 ng of DNA in a 0.2 cm cuvette (Bio-Rad, California, USA). The cell-DNA mixture was subjected to an electrical pulse using specific parameters: 2.5 kV, 25 mF, and 200 Ω in a MicroPulser (Bio-Rad, California, USA). Following electroporation, the cells were added to 1 mL of LB medium and incubated at 37°C with shaking (225 rpm, 5.660×g) for 1 hour to allow for antibiotic expression. After the incubation period, the transformed cells were plated onto selective media containing antibiotics. When strain F11 plasmids were used in electroporation or transconjugation experiments, appropriate selection was performed using 1 ml with 10 μg/L of rifampicin and 0.02 μg/L of imipenem.
The detection of class A beta-lactamase, class B carbapenemase, and class D carbapenemase was performed using different methods recommended by CLSI (CLSI, 2022). These methods involved the use of paper discs containing specific inhibitors or substrates, which were placed on an agar plate inoculated with the test strain. The growth of the strain around the discs indicated the presence of the corresponding enzyme.
To specifically detect class B carbapenemases (metallo-bete-lactamases), the modified carbapenem inactivation method (mCIM) and modified carbapenem inactivation+ EDTA (eCIM) methods were used. In these methods, the test strain was incubated with a carbapenem antibiotic and growth inhibition was assessed. EDTA solution (initial concentration 0.1 mol/L, 10 μL could be added dropwise, final concentration 292 μg/tablet) was added for modified carbapenem inactivation + EDTA (eCIM). The addition of EDTA in the eCIM helped to detect metallo-beta-lactamases. An increase in the zone of inhibition by≥5 mm in the eCIM compared to the mCIM was interpreted as a positive result for metallo-beta -lactamases.
However, there was currently no definitive phenotypic detection method specifically designed for class D carbapenemases. In the absence of a specific test, the elimination method could be used. If a strain was not inhibited by a class A or class B inhibitor, it could be presumed to possess a class D carbapenemase.
Bacterial resistance was detected using the VITEK2 system from BioMérieux of France. The VITEK2 determined the minimum inhibitory concentration (MIC) values of antibiotics. Additionally, the antibiotic paper diffusion method was employed to determine the zone of inhibition in millimeters (mm) for each antibiotic. The drug susceptibility paper used in the antibiotic paper diffusion method was supplied by OXOID, UK. The results of both methods were then confirmed according to CLSI 2020 guidelines (CLSI, 2022). A total of twenty-three antibiotics and antibiotics combined with enzyme inhibitors were tested, as listed in Table 1. E. coli ATCC 25922 was used for the quality control.
The genomic sequencing of strain F11 was performed on the third-generation PacBio Sequel II platform (Pacific Biosciences, CA, USA) and the second-generation sequencing Illumina NovaSeq 6000 platform (Illumina, San Diego, USA) using a DNA library with an average size of ~15 kbp (range 10 to 20 kbp), and on a ~400 bp fragment library (range 150 bp to 600 bp). To ensure data quality, the raw data obtained from the PacBio Sequel II platform was trimmed using Canu v2.2 (https://github.com/marbl/canu) to obtain high-quality clean reads. On the other hand, the paired-end short Illumina reads were subjected to “de novo” assembly using either Unicycler v0.4.5 (https://github.com/rrwick/Unicycler) or SPAdes v3.15.3 (https://github.com/ablab/spades). After the assembly, sequence corrections were performed at the post-assembly level using Pilon v1.24 (https://github.com/broadinstitute/pilon) using the second generation sequencing reads. Overall, these multistep methods were able to generate reliable and accurate DNA sequences for strain F11.
Open reading frames (orfs) and pseudogenes were predicted using RAST2.0 (Liang et al., 2015), BLASTP/BLASTN (Boratyn et al., 2013), UniProtKB/Swiss-Prot (Boutet et al., 2016), and RefSeq databases (O’Leary et al., 2016). Resistance genes, mobile elements, and other features were annotated using online databases such as CARD 2023 (Alcock et al., 2023), ResFinder4.0 (Bortolaia et al., 2020), ISfinder (Siguier et al., 2006), INTEGRALL (Moura et al., 2009), and the Tn Number Registry (Tansirichaiya et al., 2019). The virulence genes were predicted using VFDB 2022 (Liu et al., 2022) (http://www.mgc.ac.cn/VFs/main.htm) and BacWGSTdb 2.0 (Feng et al., 2021) (http://bacdb.cn/BacWGSTdb/index.php). Clonal MLST was confirmed using MLST2.0 (https://cge.food.dtu.dk/services/MLST/) and BacWGSTdb 2.0 (Feng et al., 2021). The type of incompatible (Inc) plasmid was determined based on the comparative analysis of replication genes using BacWGSTdb 2.0 (Feng et al., 2021). MUSCLE 3.8.31 (Edgar, 2004) and BLASTN were used for multiple and pairwise sequence comparisons such as plasmid pA_F11, plasmid pB_F11, and plasmid pC_F11 with their closely related plasmids. Circos plots of plasmids were drawn with CGView (Stothard et al., 2019). Partial sequences of plasmids were compared using BLASTN to calculate identity and coverage, followed by heatmap with the R package pheatmap (https://CRAN.R-project.org/package=pheatmap). All comparative figures were generated using the R package genoPlotR v0.8.11 (http://genoplotr.r-forge.r-project.org/) and edited using Inkscape v0.48.1 (https://inkscape.org/en). These tools and databases were used to analyze and visualize various genomic features.
The sequences of F11_Chromosome, plasmid pA_F11, plasmid pB_F11, plasmid pC_F11 and plasmid pD_F11 were deposited on the GenBank database as the accession numbers of CP092901.1, CP092902.1, CP092903.1, CP092904.1 and CP092905.1, respectively. The GenBank accession numbers of the related plasmids compared with plasmid pA_F11, plasmid pB_F11 and plasmid pC_F11 were also listed in Table 2.
The 16S rRNA genome sequence of strain F11 was confirmed as K. pneumoniae by BLAST and average nucleotide homology analysis. The results of drug susceptibility testing for strain F11 were shown in Table 1. The strain was highly resistant to all conventional antibiotics except erythromycin (20 mm, susceptible), especially cefoperazone/avibactam (6 mm, resistant) and tigecycline (≥8, resistant) (Table 1).
Enzyme analyses revealed that strain F11 produced class A beta-lactamase (positive ESBls) and class B metallo-beta-lactamase (class B carbapenemase); whereas class D carbapenemase was identified by analysis of plasmid sequence alignment.
In bacterial conjugative transfer and electroporation assays, transconjugants carrying the plasmid pA_F11 and plasmid pB_F11 were successfully obtained with average conjugation frequencies of 2.91×10-4 and 1.56×10-4, respectively. However, both the plasmid pC_F11 and plasmid pD_F11 could not be transferred by conjugation or electroporation.
The F11_Chromosome with a length of 5.286326 Mbp, carried four antibiotic resistance genes, including oqxB/A, blaSHV-187, and fosA (Supplementary Table S1). as well as a series of virulence genes (Supplementary Table S2). Several categories of virulence gene, such as adhesion, biofilm, regulatory, immune modulation, type VI secretion system (T6SS), siderophore uptake system, and antimicrobial activity, exhibited more than 96.5% sequence identity (Supplementary Figure S2 and Supplementary Table S2), suggesting a high degree of conservation and similarity among these gene categories in the F11_Chromosome. However, the category of virulence genes for the common pilus of E. coli showed lower sequence identities, ranging from 86.5% to 91.0% (Supplementary Figure S2 and Supplementary Table S2), indicating some level of divergence or variation in these gene category within the F11_Chromosome.
Plasmid pA_F11, classified as IncFIB/IncHI1B type (Table 2), had a length of 221.757 kbp. It contained a MDR region that spans approximately 25.8 kbp. Additionally, the plasmid carried two repB genes, four virulence genes (fimA, tssM, ipaH, and ipaH), a number of ISs, and plasmid backbone genes (Figure 1).
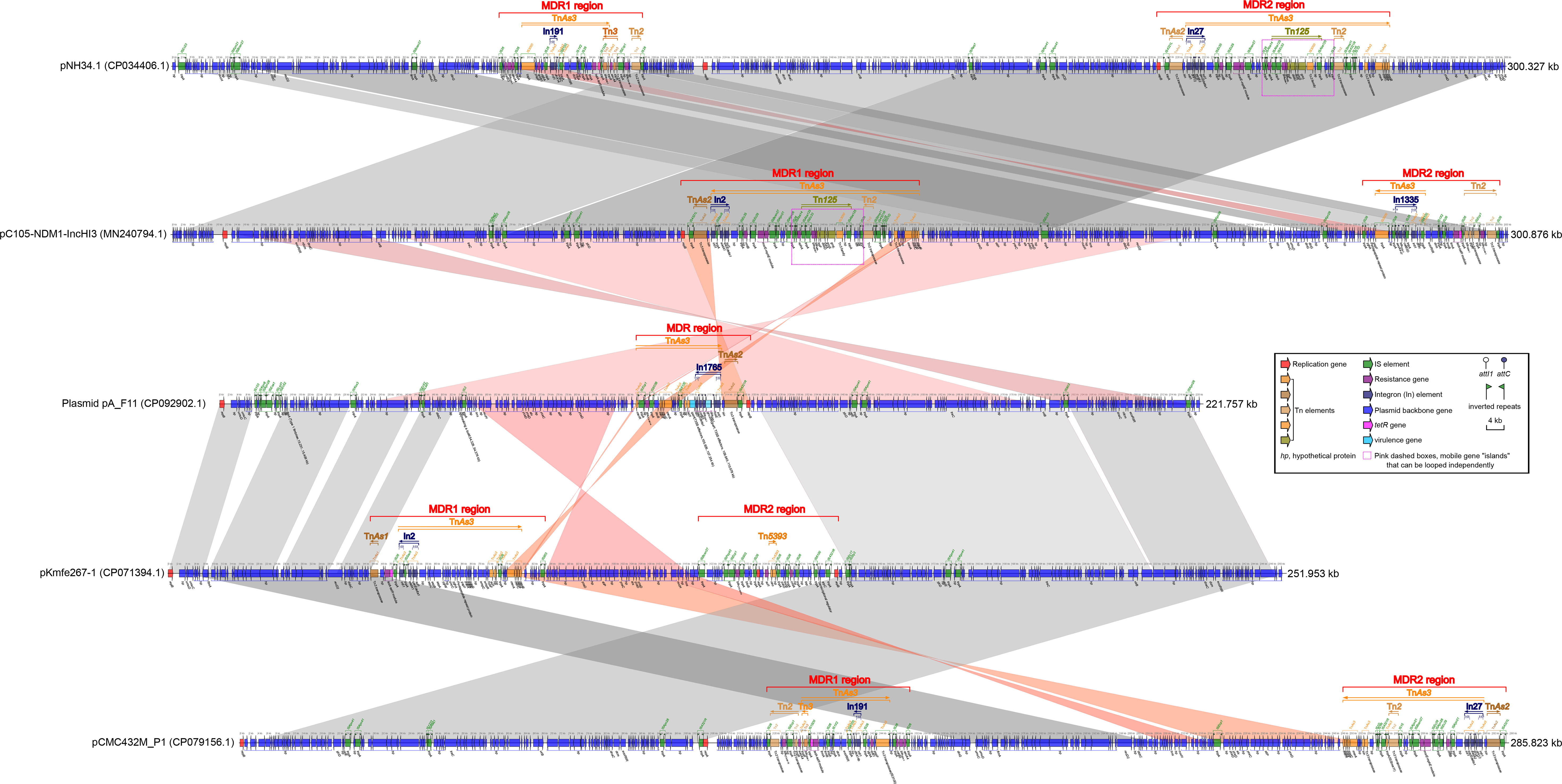
Figure 1 Comparison of plasmid pA_F11 with related plasmids. The shadow represents > 95% identity, while light gray represents the positive direction, and light pink refers to the opposite direction. The figures were created by the R pacakge genoPlotR v0.8.11 software (http://genoplotr.r-forge.r-project.org/).
The fimA gene, which belonged to the type 1 fimbriae, was located between the 15.251 kbp and 15.409 kbp sites. The tssM gene was covered by a hypothetical protein and was located between the 54.328 kbp and 54.378 kbp sites. Two copies of the ipaH gene were found within the MDR region, occupying positions from 105.926 kbp to 107.254 kbp and from 109.645 kbp to 110.978 kbp. These ipaH genes functioned as type III secretion system (T3SS) effectors (Figure 1). The MDR region of plasmid pA_F11 consisted of the transposons TnAs3, TnAs2 and the rare integron In1765 (Supplementary Table S3). Within this region, several antibiotic resistance genes were present, including blaCTX-M-14, sul1, a catB3 gene cassette, a blaOXA-1 gene cassette, and a fosE gene cassette. Notably, the blaOXA-1 gene cassette was split into two parts by the gcu206 gene cassette (ipaH) in the region of 109.645-110.978 kbp, which was a rare occurrence (Figure 1 and Supplementary Table S3).
Comparative analysis of plasmid pA_F11 with pC105-NDM1-IncHI3 revealed minimal similarity between the MDR region of plasmid pA_F11 and the MDR1 region of pC105-NDM1-IncHI3. There was no similarity between the MDR region of plasmid pA_F11 and the MDR2 region of pC105-NDM1-IncHI3. However, the plasmid backbone region surrounding the MDR region of pA_F11 showed perfect identity with that surrounding the MDR1 region of pC105-NDM1-IncHI3 (Figure 1).
When comparing plasmid pA_F11 with pKmfe267-1, it was observed that the MDR region of plasmid pA_F11 exhibited minimal similarity to the MDR1 and MDR2 regions of pKmfe267-1. On the other hand, the backbone region of plasmid pA_F11 showed high identity with that of pKmfe267-1 (Figure 1). Comparing pC105- NDM1-IncHI3 with pNH34.1, the two plasmids exhibited a high degree of sequence identity and coverage. However, certain sequences in the front and back positions showed alterations, including shifts in two MDR regions of the plasmids. Interestingly, both the MDR1 region of pC105-NDM1-IncHI3 and the MDR2 region of pNH34.1 carried identical blaNDM-1 and Tn125 components (Figure 1). In the comparison of pKmfe267-1 with pCMC432M_P1, the backbone regions of both plasmids displayed excellent identity and coverage. However, the two MDR regions of pKmfe267-1 were not similar to those of pCMC432M_P1. Additionally, there were sequence inversions and shifts in the front and back of the backbone regions of the two plasmids (Figure 1).
Plasmid pB_F11, classified also as IncFIB/IncHI1B type (Table 2), had a length of 159.322 kbp. It contained two MDR regions, a repB gene, four virulence genes related to heavy metal resistance (arsC, arsB, arsA, and arsD virulence genes), several ISs, and plasmid backbone genes (Figure 2). The MDR1 region was composed of transposons TnAs3, TnAs2, Tn2, and In27 (Supplementary Table S3). This region carried several antibiotic resistance genes, including tetA, a dfrA12 gene cassette, an aadA2 gene cassette, sul1, qnrA1, cmlA, tetD, rmtB, and blaTEM-1 (Figure 2). The MDR2 region of plasmid pB_F11 consisted of transposons Tn125, TnAs3, and In0. Interestingly, this region did not harbor the blaNDM-1 gene, unlike other plasmids that typically carried it. However, all other Tn125 components in pB_F11 were identical to those found in plasmids known to carry blaNDM-1 (Figure 2). It suggested that plasmid pB_F11 might have lost the blaNDM-1 gene during its evolutionary process (Figure 3).
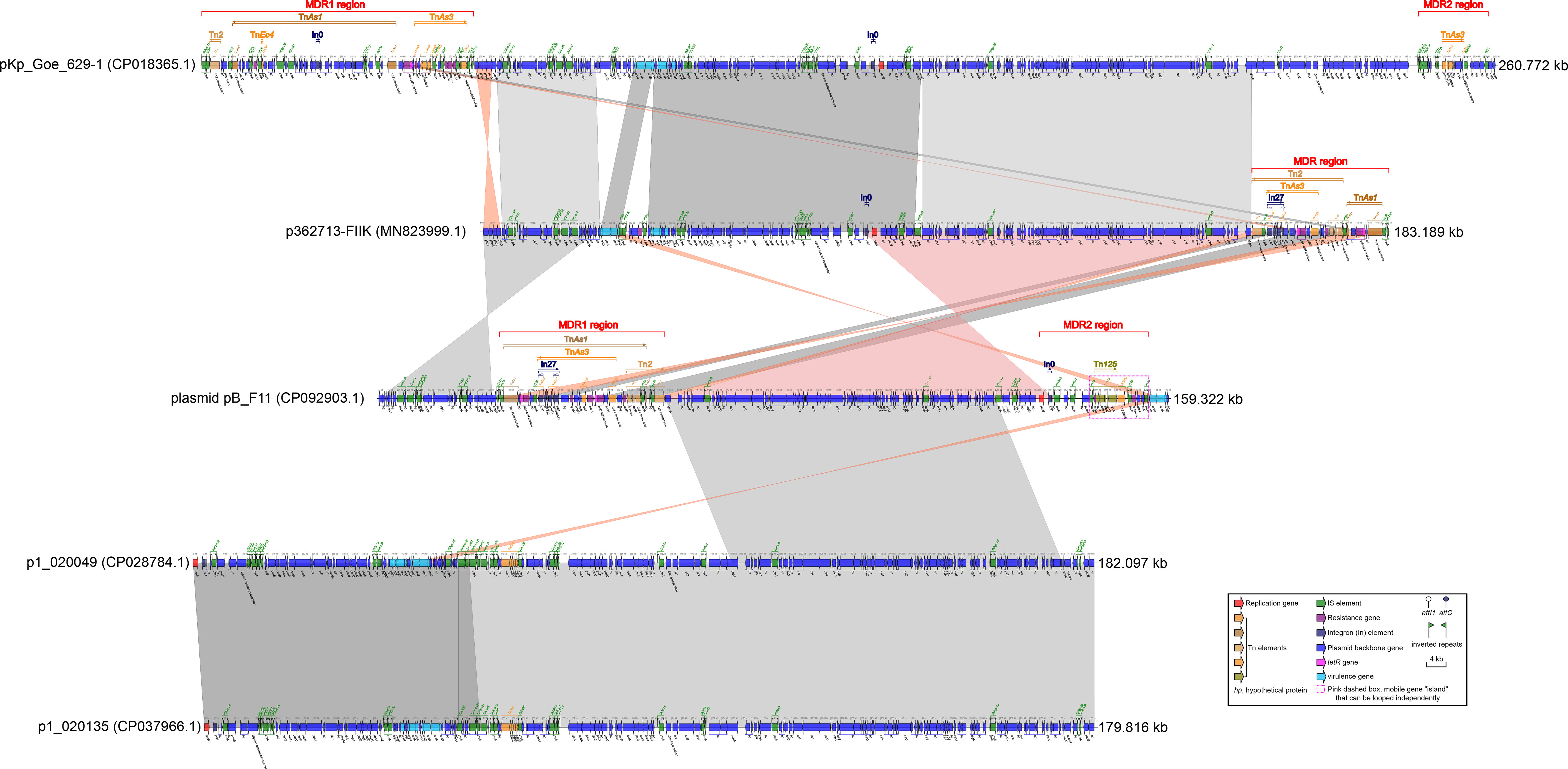
Figure 2 Comparison of plasmid pB_F11 with related plasmids. The shadow represents > 95% identity, while light gray represents the positive direction, and light pink refers to the opposite direction. The figures were also created by the R pacakge genoPlotR v0.8.11 software (http://genoplotr.r-forge.r-project.org/).
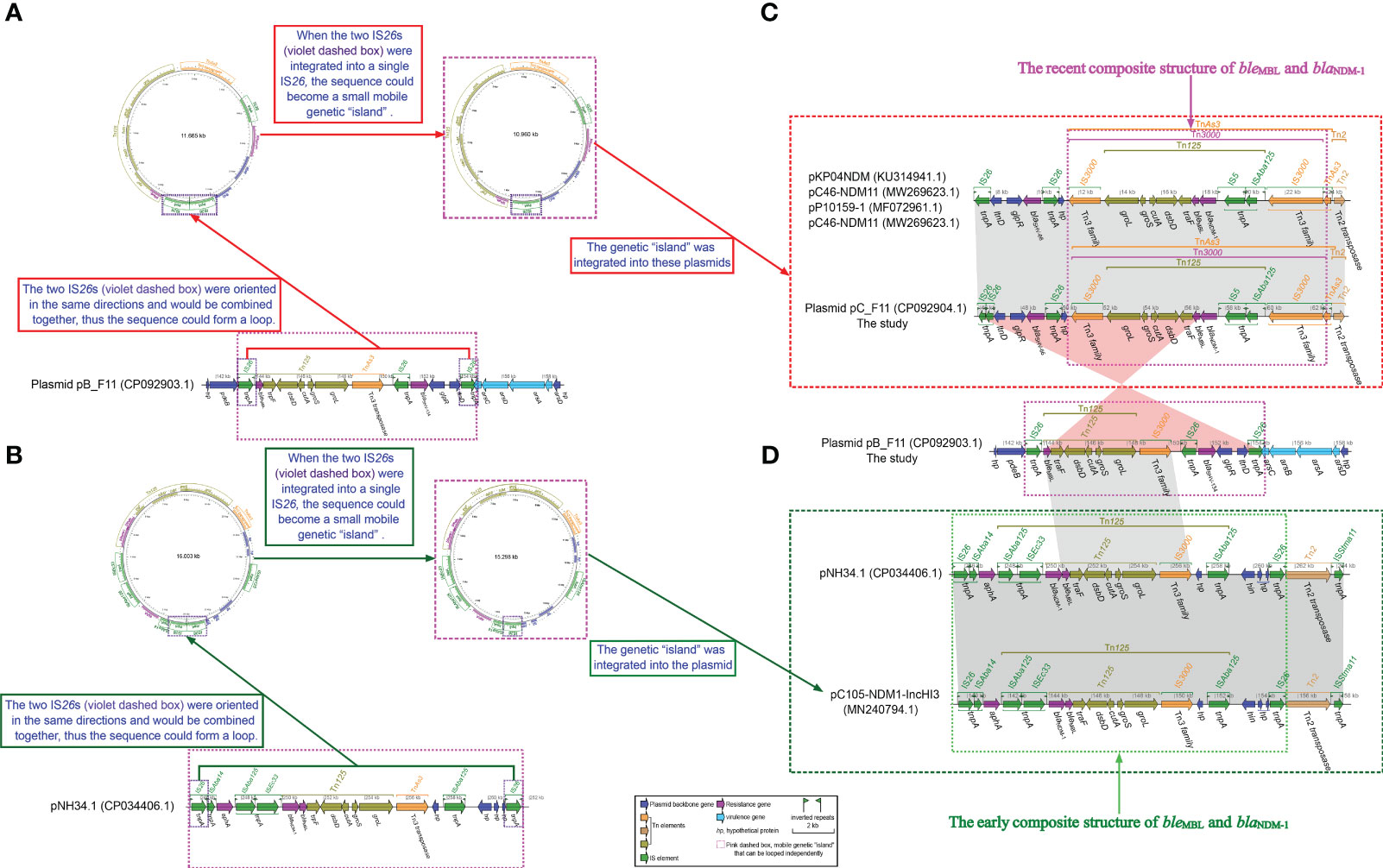
Figure 3 Schematic diagram of whether small loop intermediates could be formed by the integration of IS26s. (A) Schematic representation of a small loop intermediate that could be formed for plasmid pB_F11. (B) Schematic representation of a small loop intermediate that could be formed for pNH34.1. (C) comparisons of plasmid pC_F11, pNH34.1 and related plasmids, respectively, and (D) comparisons of plasmid pB_F11 with plasmid pC_F11 and pNH34.1. The diagram was created by the R package genoPlotR v0.8.11 software (http://genoplotr.r-forge.r-project.org/). The small loop diagrams were established using CGview v2.0.3 (https://github.com/paulstothard/cgview).
Comparing plasmid pB_F11 with p362713-FIIK, it was observed that the MDR1 region of plasmid pB_F11 showed some similarity to the MDR region of p362713-FIIK. Additionally, the backbone region of plasmid pB_F11 surrounding the MDR1 region was also similar to that of p362713-FIIK, with most sequences reversed (Figure 2). When comparing plasmid pB_F11 with p1_020049, part of the backbone region of plasmid pB_F11 was identical to that of p1_020049, except for the MDR region (Figure 2). In the comparison of p362713-FIIK with pKp_Goe_629-1, the backbone region of p362713-FIIK, which contained only one MDR region, showed good identity with that of pKp_Goe_629-1, which contained two MDR regions. However, there was no similarity between their respective MDR regions (Figure 2). Comparing p1_020049 with p1_020135, a high degree of identity was observed between the two plasmids. However, neither of them contained an MDR region (Figure 2).
Plasmid pC_F11, classified as an IncX3 type (Table 2), had a length of 92.904 kbp. It contained an MDR region, four virulence genes related to heavy metal resistance (arsC, arsB, arsA, and arsD virulence genes), several ISs, and plasmid backbone genes. However, no repB gene was found in this plasmid (Figure 4). The MDR region of plasmid pC_F11 consisted of transposons TnAs3, Tn3000, Tn125, and Tn2. This region involved antibiotic resistance genes such as blaSHV-66, bleMBL, and blaNDM-1 (Figure 4). Notably, the plasmid carried an identical blaNDM-1 gene and its associated Tn125 components, which were part of Tn3000 (Figure 4).
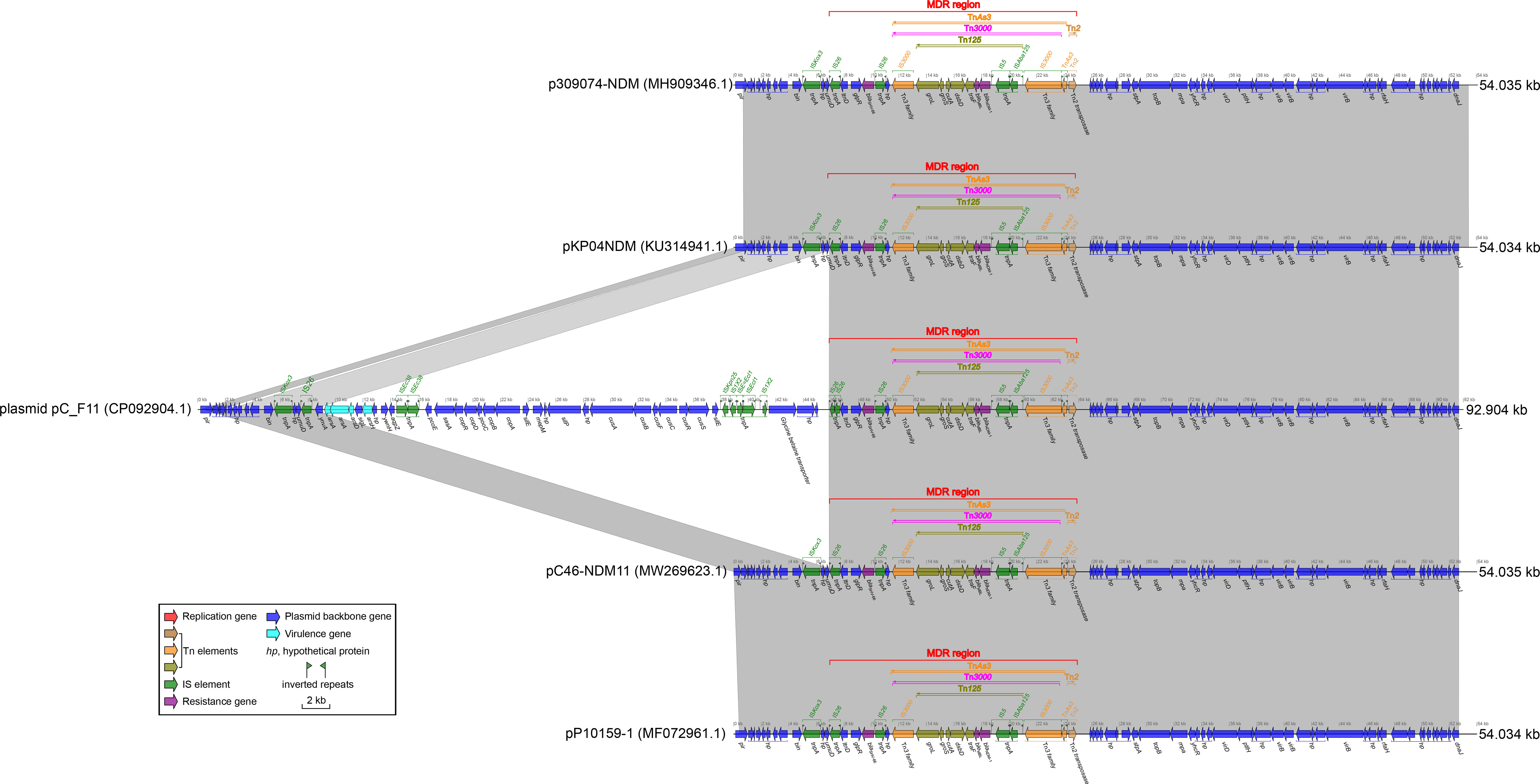
Figure 4 Comparison of plasmid pC_F11 with related plasmids. The shadow represents > 95% identity, while light gray represents the positive direction, and light pink refers to the opposite direction. The Figures were also created by the R pacakge genoPlotR v0.8.11 software (http://genoplotr.r-forge.r-project.org/).
When comparing plasmid pC_F11 with plasmids pKP04NDM, pC46-NDM11, p309074-NDM, and pP10159-1, a high degree of identity was observed. However, there was an interesting difference in plasmid pC_F11, where an approximately 37.69 kbp-long sequence was inserted between the two IS26 sites at positions 7.537 to 8.241 kbp and 45.931 to 46.260 kbp (Figure 4). Furthermore, the partial sequence of plasmid pB_F11 (indicated by pink dashed boxes in Figure 3C) was almost identical to the sequences of plasmids: plasmid pC_F11, p362713-FIIK, p1_020049, pKp_Goe_629-1, and p1_020135, all of which carried the blaNDM-1 gene. The only difference was that plasmid pB_F11 lacked the blaNDM-1 gene (Figure 3C).
Plasmid pD_F11 was characterized as a plasmid fragment with a length of 2.927 kbp. It consisted of only four hypothetical proteins and did not contain a repA gene or any antibiotic resistance genes (Supplementary Figure S2).
An detailed analysis of the plasmid pB_F11 sequence revealed that from 143.212 kbp to 154.876 kbp, it carried a genetic assay consisting of IS26 (forward)–(bleMBL–trpF–dsbB–cutA–groS–groL) (Tn125, forward)–IS3000(forward)–IS26 (reverse)–blaSHV-134 (forward)–glpR (reverse)–ltnD (forward)–IS26 (forward), but did not contain the blaNDM gene (Figure 3A). Upon integration of the two forward IS26 elements into a single IS26, it formed independent genetic “islands” (Figure 3A). This identical structure was also found to be integrated in plasmids such as pKP04NDM, pC46-NDM11, pP10159-1, pC46-NDM11, and plasmid pC_F11 (Figure 3C). By performing a BLASTN analysis using the sequence of the genetic “islands”, it was discovered that this integratable structure was present in 50 plasmids. Interestingly, all 50 of these plasmids carried the blaNDM gene (Supplementary Figure S1; Supplementary Tables S4).
In the case of the pNH34.1 sequence, from 245.506 kbp to 261.607 kbp, it contained a genetic assay as follows, IS26 (forward)–ISAba14 (forward)–aphA (forward)–ISAba125 (forward)–ISEc33 (forward)–(blaNDM-1–bleMBL–trpF–dsbB–cutA–groS–groL) (Tn125, forward)–IS3000 (forward)-hp (reverse)–ISAba125 (forward)–hin (reverse)–hp (reverse)–hp (forward)–hp (forward)–hp (forward)–IS26 (forward)(Figure 3B). The integration of two forward IS26 elements into a single IS26, as observed in the pNH34.1 sequence, also led to the formation of independent genetic islands” (Figure 4B). This structure also appeared in plasmids pC105-NDM1- IncHI3 (Figure 3C). Sequence BLASTN analysis revealed that only pNH34.1 and pC105-NDM1-IncHI3 had this specific structure. None of the other plasmids examined showed an equivalent assay.
The blaNDM gene was commonly found on plasmids containing different backbone components (Acman et al., 2022). IncHI-like plasmids, which were typically larger than 200 kbp in size, had a wide distribution in the host population (Rozwandowicz et al., 2018) and played a crucial role in the dissemination of heavy metal resistance genes and antibiotic resistance genes (Cain and Hall, 2012). For example, IncHI5-like plasmids were often associated with different carbapenemase genes (Zhu et al., 2020), and two IncHI5-like plasmids were previously reported to carry both carbapenem resistance genes and tigecycline resistance modules (Qin et al., 2021). To date, at least 20 different types of incompatible plasmids had been associated with the blaNDM gene. These plasmids included IncFIB, IncFII, IncA/C (IncC), IncX3, IncH, IncL/M, as well as untyped plasmids (Acman et al., 2022).
Prior to 1985, the blaNDM gene was typically found within the Tn125 transposon, containing the IS26 insertion sequence. However, the global prevalence of IS26 did not occur until after 2015 (Acman et al., 2022). Tn125 played a important role in the early plasmid-mediated dissemination of the blaNDM gene (Figure 3D), but in recent years, it had been replaced by mobile elements such as Tn3000 (Acman et al., 2022). The plasmids pC105- NDM-IncHI3 and pNH34.1, which showed high similarity to the non-MDR region of plasmid pA_F11, shared an identical relatively complete Tn125 structure in the MDR1 region of pC105-NDM1-IncHI3 and the MDR2 region of pNH34.1. This structure represented an early composite structure of bleMBL and blaNDM-1 between the two ends of ISAba15 (Figure 3D). Plasmid pC_F11 exhibited a high degree of identity in both the MDR and non-MDR regions with pKP04NDM, pC46-NDM11, p309074-NDM, and pP10159-1, except for the insertion of a non-MDR sequence of approximately 37.69 kbp in length. In the MDR region of these plasmids, the composite structure of bleMBL and blaNDM-1, located between the two IS3000 elements, contained a portion of the Tn125 structure. This composite structure had been classified as the recently prevalent Tn3000 structure (Acman et al., 2022) (Figure 3C). Several mobile elements were recognized to play a critical role in the dissemination of these genetic elements, including ISAba125, IS3000, IS26, IS5, ISCR1, Tn3, Tn125, and Tn3000 (Acman et al., 2022). Tn3000 was flanked by two copies of IS3000 and had a length of approximately 11.800 kbp (Campos et al., 2015). The first copy of IS3000 truncated the upstream portion of the ISAba125 element, followed by the blaNDM-1 gene. Downstream of the blaNDM-1 gene, there was the bleMBL gene, followed by the trpF, dsbD, and cutA genes. The groL and groS genes were linked to the second copy of IS3000, forming the complete Tn3000 structure. This structure was also observed in plasmids such as pC_F11, pKP04NDM, pC46-NDM11, p309074-NDM, and pP10159-1 (Figures 3C, 4).
Plasmid pA_F11 showed high sequence identity with related plasmids in the non-MDR regions, but low sequence identity in the MDR region. It was constructed mainly from a composite of TnAs3, TnAs2, and In1765. The sequence structure of plasmid pA_F11 with only one MDR region, especially In1765, was rare and had not been identified in the literature or GenBank. Plasmid pB_F11 consisted of two MDR regions, MDR1 and MDR2. The MDR regions of plasmid pB_F11 and plasmid p362713-FIIK shared identical In27 and part of the TnAs3 components. The analysis revealed that the MDR2 region of plasmid pB_F11 independently formed a circular genetic “island” characterized as a mobile element. This genetic “island” contained genetic arrays including IS26–(bleMBL–trpF–dsbB–cutA–groS–groL)–IS3000–IS26–blaSHV-134–glpR–ltnD–IS26 (Figure 3), which was part of the composite structure of bleMBL and blaNDM-1, except for the absence of blaNDM-1. Further BLASTN analysis using these sequences of genetic “island” as templates, it showed highly consistent results, except that all 50 identified sequences carried the blaNDM-1 gene (Supplementary Figure S1; Supplementary Table S3). These sequences represented the popular composite structure observed in recent years (Acman et al., 2022). The results indicated that the MDR2 region of plasmid pB_F11 evolved as a unique structure due to the loss of the blaNDM-1 gene during the plasmid evolution process. Similarly, the MDR2 region of pNH34.1 and the MDR1 region of pC105-NDM1-IncHI3 independently developed circular genetic “islands” characterized as mobile elements (Figure 3). These genetic “islands” contained genetic arrays including IS26–ISAba14–aphA–ISAba125–ISEc33–(blaNDM-1–bleMBL–trpF–dsbB–cutA–groS–groL)–IS3000–hp–ISAba125–hin–hp–hp–hp–hp–IS26, which belonged to the early composite structure of bleMBL and blaNDM-1 (Acman et al., 2022) (Figure 3D). BLASTN comparison using the sequences of genetic “islands” as templates revealed that only pNH34.1 and pC105-NDM1-IncHI3 had the identical structure, both representing the early composite structure of bleMBL and blaNDM-1 (Acman et al., 2022) (Figure 3D). The MDR2 region of plasmid pB_F11 served as an independent mobile element while acting as a “bridge” connecting the early composite structure of bleMBL and blaNDM-1 (Figure 3D) to the recent composite structure of bleMBL and blaNDM-1 (Acman et al., 2022) (Figure 3C).
The strain F11 demonstrated a high level of antibiotic resistance, including resistance to conventional antibiotics such as cefoperazone/avibactam and tigecycline (Table 1). This resistance was attributed to the complex combination of transposons and gene cassettes carrying different antibiotic resistance and virulence genes on the F11_chromosome and several F11_plasmids. The strain was resistant to almost all antibiotics tested, limiting treatment options. Although erythromycin was found to be sensitive to strain F11 in vitro, clinical treatment with this antibiotic was ineffective. This might be due to the involvement of multiple virulence genes in the strain, which could contribute to its pathogenicity and ability to evade antibiotic treatment. As a result, no effective drugs were available to effectively treat infections caused by strain F11.
The combination of complex overlapping transposons and gene cassettes, along with the synergistic effects of multiple antibiotic resistance and heavy metal resistance genes (virulence genes), contributed to the lack of an effective drug to treat infections caused by strain F11. It was essential to prevent the emergence and dissemination of strain F11 through strict control measures and the promotion of responsible antibiotic use.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: The sequences of F11_Chromosome, plasmid pA_F11, plasmid pB_F11, plasmid pC_F11 and plasmid pD_F11 were deposited on the GenBank database as the accession numbers of CP092901.1, CP092902.1, CP092903.1, CP092904.1 and CP092905.1, respectively.
This study was approved by the Ethics Committee of Taizhou University, Zhejiang, China, and written informed consent was obtained from each of the participants in accordance with the Declaration of Helsinki. The rights of the research subjects were protected throughout, and we confirm that this study was conducted in our school. The use of human specimens and all related experimental protocols were approved by the Committee on Human Research of Taizhou University, and the protocols were carried out in accordance with approved guidelines.
LM: Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Supervision, Visualization, Writing – review & editing, Funding acquisition. YQ: Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Supervision, Validation, Visualization, Writing – review & editing. WW: Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Supervision, Validation, Visualization, Writing – review & editing. DW: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Zhejiang Health Department of China (2023KY1319).
We thank Yolanda Elisabet Gonzalez Flores, Ph.D, from Université de Limoge, for analyzing the integrons.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1324846/full#supplementary-material
Supplementary Figure 1 | Heatmap analysis of sequence coverage and identity using BLASTN with sequence of genetic “island” from plasmid pB_F11 as a template. The figure was created with the R package pheatmap (https://CRAN.R-project.org/package=pheatmap) base on Supplementary Table S4.
Supplementary Figure 2 | Distribution of the location for virulence genes and antibiotic resistance genes on F11_chromosome.
Supplementary Figure 3 | Circle diagram of plasmid pD_F11.
Acman, M., Wang, R., van Dorp, L., Shaw, L. P., Wang, Q., Luhmann, N., et al. (2022). Role of mobile genetic elements in the global dissemination of the carbapenem resistance gene blaNDM. Nat. Commun. 13 (1), 1131. doi: 10.1038/s41467-022-28819-2
Alcock, B. P., Huynh, W., Chalil, R., Smith, K. W., Raphenya, A. R., Wlodarski, M. A., et al. (2023). SCARD 2023: expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 51 (D1), D690–D699. doi: 10.1093/nar/gkac920
Bonnin, R. A., Poirel, L., Carattoli, A., Nordmann, P. (2012). Characterization of an IncFII plasmid encoding NDM-1 from Escherichia coli ST131. Escherichia coli ST131. PloS One 7 (4), e34752. doi: 10.1371/journal.pone.0034752
Boratyn, G. M., Camacho, C., Cooper, P. S., Coulouris, G., Fong, A., Ma, N., et al. (2013). BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 41 (W), W29–W33. doi: 10.1093/nar/gkt282
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75 (12), 3491–3500. doi: 10.1093/jac/dkaa345
Boutet, E., Lieberherr, D., Tognolli, M., Schneider, M., Bansal, P., Bridge, A. J., et al. (2016). UniProtKB/Swiss-Prot, the manually annotated section of the UniProt KnowledgeBase: how to use the entry view. Methods Mol. Biol. 1374, 23–54. doi: 10.1007/978-1-4939-3167-5_2
Cain, A. K., Hall, R. M. (2012). Evolution of IncHI2 plasmids via acquisition of transposons carrying antibiotic resistance determinants. J. Antimicrob. Chemother. 67 (5), 1121–1127. doi: 10.1093/jac/dks004
Campos, J. C., da Silva, M. J., dos Santos, P. R., Barros, E. M., Pereira Mde, O., Seco, B. M., et al. (2015). Characterization of Tn3000, a transposon responsible for blaNDM-1 dissemination among enterobacteriaceae in Brazil, Nepal, Morocco, and India. Antimicrob. Agents Chemother. 59 (12), 7387–7395. doi: 10.1128/AAC.01458-15
Chen, Z., Li, H., Feng, J., Li, Y., Chen, X., Guo, X., et al. (2015). NDM-1 encoded by a pNDM- BJ01-like plasmid p3SP-NDM in clinical Enterobacter aerogenes. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00294
CLSI (2022). Performance Standards for Antimicrobial Susceptibility Testing. 32nd Edition (Wayne, PA: CLSI). CLSI supplement M100.
Costa, A., Figueroa-Espinosa, R., Gaudenzi, F., Lincopan, N., Fuga, B., Ghiglione, B., et al. (2021). Co-occurrence of NDM-5 and rmtb in a clinical isolate of Escherichia coli belonging to CC354 in Latin America. Front. Cell Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.654852
Dortet, L., Girlich, D., Virlouvet, A., Poirel, L., Nordmann, P., Iorga, B. I., et al. (2017). Characterization of BRPMBL, the bleomicin resistance protein associated with the carbapenemase NDM. Antimicrob. Agents Chemother. 61 (3), e02413–e02416. doi: 10.1128/AAC.02413-16
Dortet, L., Poirel, L., Nordmann, P. (2014). Worldwide dissemination of the NDM- type carbapenemases in Gram-negative bacteria. BioMed. Res. Int. 2014, 249856. doi: 10.1155/2014/249856
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 (5), 1792–1797. doi: 10.1093/nar/gkh340
Feng, Y., Zou, S., Chen, H., Yu, Y., Ruan, Z. (2021). BacWGSTdb 2.0: a one-stop repository for bacterial whole-genome sequence typing and source tracking. Nucleic Acids Res. 49 (D1), D644–D650. doi: 10.1093/nar/gkaa821
Frank, J. A., Reich, C. I., Sharma, S., Weisbaumm, J. S., Wilson, B. A., Olsen, G. J. (2008). Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 74 (8), 2461–2470. doi: 10.1128/AEM.02272-07
Guducuoglu, H., Gursoy, N. C., Yakupogullari, Y., Parlak, M., Karasin, G., Sunnetcioglu, M., et al. (2018). Hospital outbreak of a colistin-resistant, NDM-1- and OXA-48- producing Klebsiella pneumoniae: high mortality from pandrug resistance. Microb. Drug Resist. 24 (7), 966–972. doi: 10.1089/mdr.2017.0173
Hornsey, M., Phee, L., Wareham, D. W. (2011). A novel variant, NDM-5, of the New Delhi metallo-β-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob. Agents Chemother. 55 (12), 5952–5954. doi: 10.1128/AAC.05108-11
Liang, Q., Yin, Z., Zhao, Y., Liang, L., Feng, J., Zhan, Z., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5, 8365. doi: 10.1016/j.ijantimicag.2017.01.021
Liu, B., Zheng, D., Zhou, S., Chen, L., Yang, J. (2022). VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. 50 (D1), D912–D917. doi: 10.1093/nar/gkab1107
Logan, L. K., Weinstein, R. A. (2017). The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J. Infect. Dis. 215 (suppl_1), S28–S36. doi: 10.1093/infdis/jiw282
Loh, B., Chen, J., Manohar, P., Yu, Y., Hua, X., Leptihn, S. (2020). A biological inventory of prophages in A. baumannii genomes reveal distinct distributions in classes, length, and genomic positions. Front. Microbiol. 11, 579802. doi: 10.3389/fmicb.2020.579802
Moura, A., Soares, M., Pereira, C., Leitão, N., Henriques, I., Correia, A. (2009). INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25 (8), 1096–1098. doi: 10.1093/bioinformatics/btp105
Nordmann, P., Poirel., L., Walsh, T. R., Livermore, D. M. (2011). The emerging NDM carbapenemases. Trends Microbiol. 19 (12), 588–595. doi: 10.1016/j.tim.2011.09.005
O’Leary, N. A., Wright, M. W., Brister, J. R., Ciufo, S., Haddad, D., McVeigh, R., et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44 (D1), D733–DD745. doi: 10.1093/nar/gkv1189
Poirel, L., Bonnin, R. A., Boulanger, A., Schrenzel, J., Kaase, M., Nordmann, P. (2012). Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii. Antimicrob. Agents Chemother. 56 (2), 1087–1089. doi: 10.1128/aac.05620-11
Qin, S., Peng, J., Deng, R., Peng, K., Yan, T., Chen, F., et al. (2021). Identification of two plasmids co-harboring carbapenemase genes and tmexCD1-Toprj1 in clinical Klebsiella pneumoniae ST2667. Antimicrob. Agents Chemother. 65 (6), e00625–e00621. doi: 10.1128/aac.00625-21
Rozwandowicz, M., Brouwer., M. S. M., Fischer, J., Wagenaar, J. A., Gonzalez Zorn, B., Guerra, B., et al. (2018). Plasmids carrying antimicrobial resistance genes in enterobacteriaceae. J. Antimicrob. Chemother. 73 (5), 1121–1137. doi: 10.1093/jac/dkx488
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34 (D), D32–D6. doi: 10.1093/nar/gkj014
Stothard, P., Grant, J. R., Van Domselaar, G. (2019). Visualizing and comparing circular genomes using the CGView family of tools. Brief Bioinform. 20 (4), 1576–1582. doi: 10.1093/bib/bbx081
Tansirichaiya, S., Rahman, M. A., Roberts, A. P. (2019). The transposon registry. Mob. DNA 10, 40. doi: 10.1186/s13100-019-0182-3
Toleman, M. A., Spencer, J., Jones, L., Walsh, T. R. (2012). blaNDM-1 is a chimera likely constructed in Acinetobacter baumannii. Antimicrob. Agents Chemother. 56 (5), 2773–2776. doi: 10.1128/AAC.06297-11
Walsh, T. R., Weeks, J., Livermore, D. M., Toleman, M. A. (2011). Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect. Dis. 11 (5), 355–362. doi: 10.1016/S1473-3099(11)70059-7
Wu, W., Feng, Y., Tang, G., Qiao, F., McNally, A., Zong, Z. (2019). NDM metallobeta -lactamases and their bacterial producers in health care settings. Clin. Microbiol. Rev. 32 (2), e00115–e00118. doi: 10.1128/CMR.00115-18
Keywords: Klebsiella pneumoniae, plasmid pA_F11, plasmid pB_F11, plasmid pC_F11, drug resistance, In1765, the composite structure of bleMBL and blaNDM-1
Citation: Ma L, Qu Y, Wang W and Wang D (2024) Characterization of Klebsiella pneumoniae carrying the blaNDM-1 gene in IncX3 plasmids and the rare In1765 in an IncFIB-IncHI1B plasmid. Front. Cell. Infect. Microbiol. 13:1324846. doi: 10.3389/fcimb.2023.1324846
Received: 20 October 2023; Accepted: 15 December 2023;
Published: 11 January 2024.
Edited by:
Sanket Kaushik, Amity University Jaipur, IndiaReviewed by:
Manju Narwal, The Pennsylvania State University (PSU), United StatesCopyright © 2024 Ma, Qu, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongguo Wang, d2RndHpzQDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.