- Department of Biology, Purdue School of Science, Indiana University Purdue University Indianapolis, Indianapolis, IN, United States
The Gram-negative pathogen Stenotrophomonas maltophilia causes a wide range of human infections. It causes particularly serious lung infections in individuals with cystic fibrosis, leading to high mortality rates. This pathogen is resistant to most known antibiotics and harbors a plethora of virulence factors, including lytic enzymes and serine proteases, that cause acute infection in host organisms. S. maltophilia also establishes chronic infections through biofilm formation. The biofilm environment protects the bacteria from external threats and harsh conditions and is therefore vital for the long-term pathogenesis of the microbe. While studies have identified several genes that mediate S. maltophilia’s initial colonization and biofilm formation, the cascade of events initiated by these factors is poorly understood. Consequently, understanding these and other virulence factors can yield exciting new targets for novel therapeutics.
1 Introduction
Stenotrophomonas maltophilia is an opportunistic pathogen emerging globally as a multidrug-resistant organism (Brooke et al., 2017). It causes severe nosocomial infections, such as pneumonia, bacteremia, endocarditis, meningitis, mastoiditis, biliary sepsis, and catheter-related urinary tract infections (Looney et al., 2009; Chang et al., 2015). This microbe causes particularly severe infections in immunocompromised individuals, intensive care unit (ICU) patients, those taking high doses of antibiotics, burn patients, cancer patients, and patients with transplants (Nseir et al., 2006a; Demiraslan et al., 2013; Hofmann et al., 2016; Adegoke et al., 2017). A 10-year case study from 2008 to 2017 showed a ~162% increase in the rate of S. maltophilia isolates from individuals with lower respiratory tract infection (Gajdacs and Urban, 2019). It is reported that there is a higher risk of transmission of infection through contaminated healthcare associated water supplies, such as tap water, hemodialysis water, and dental unit reservoirs (Cervia et al., 2008). Notably, S. maltophilia infections exhibit a mortality rate of up to 69% (Senol et al., 2002), with higher incidences in patients with chronic kidney diseases and foley catheter usage (Jian et al., 2022). S. maltophilia also causes severe lung infection and pulmonary exacerbation (Talmaciu et al., 2000; Waters et al., 2013; Berdah et al., 2018) in the lungs of 11.9-14% of individuals with cystic fibrosis (CF) (Ewig et al., 2000; Nseir et al., 2006b; Trifonova and Strateva, 2019a; McCutcheon and Dennis, 2021). Despite the wide range of infections it can cause, there is very little known about the immune response to S. maltophilia infection. Different S. maltophilia strains show different degrees of survival inside immature dendritic cells (iDCs) (Roscetto et al., 2015). They plausibly also evade phagocytosis of these effector cells, rendering these cells incapable of antigen presentation and immune activation along with utilizing the cells as a point of bacterial dissemination for infection spread.
2 S. maltophilia infection in individuals with cystic fibrosis
The most well studied S. maltophilia infection is in the lungs of CF individuals. CF is a genetic disorder resulting from point mutation of the cystic fibrosis transmembrane conductance regulator (CFTR) gene (Knowles and Durie, 2002). Over 160,000 people worldwide are known to be suffering from CF (Guo et al., 2022). These individuals experience defective clearance of the bronchial airways and bronchopulmonary system, causing build-up of mucus in the lungs (Gardner, 2007). They eventually experience abnormal respiratory inflammation, increased mucus deposition, and periodic bacterial infections. Thus, individuals with CF have symptoms like wheezing, coughing, and shortness of breath. More than 95% of these patients die due to pulmonary disease (Gardner, 2007).
S. maltophilia is commonly isolated from the airways of CF patients (Hansen, 2012). There has been a steady increase in incidence of S. maltophilia infection associated with CF individuals over the last decade (Amin and Waters, 2016). Reports indicate that mortality or lung transplantation is 3-fold higher in CF individuals with chronic S. maltophilia infection (Gallagher et al., 2019). It is possible that the high intrinsic antibiotic resistance of S. maltophilia provides it a colonization advantage in the CF lung, as these individuals are treated with large doses of many antibiotics. The weakened immune system of these individuals can also be a risk factor for S. maltophilia infection. In any case, S. maltophilia can be isolated at high levels (105-106 CFU/mL), suggesting deterioration of the individuals’ pulmonary function (Ballestero et al., 1995; Colin and Rabin, 2011). In fact, S. maltophilia infections are associated with lower forced expiratory volume (FEV1) (47.06%) than in individuals without this bacterium (73.40%), indicating rapid deterioration of lung function (Waters et al., 2011). Studies have also shown respiratory tract co-infections of S. maltophilia with P. aeruginosa, Burkholderia cepacia, methicillin resistant Staphylococcus aureus (MRSA), methicillin sensitive S. aureus (MSSA), Aspergillus spp, Candida spp, or Achromobacter xylosoxidans (Goss et al., 2002; Colin and Rabin, 2011; Granchelli et al., 2018).
Very little research has been done on the immune responses to S. maltophilia bacterial infection in CF persons. Chronic S. maltophilia infection in CF airways presents with measurable immunological reaction (production of anti- extracellular protease and antiflagellin antibodies) (Waters et al., 2011; Hansen, 2012). Higher antibody levels have also been found in the sera of CF individuals chronically infected with S. maltophilia than CF individuals with intermittent or no infection (Brooke et al., 2017). Apart from this, it is said that the immunological response of S. maltophilia towards the pulmonary infection in CF patients is quite similar to that of P. aeruginosa (Hansen, 2012). Experimentally, mice infected with S. maltophilia via a nebulizer exhibited higher IL-6, IL-12, IL1β, IFN-γ, and TNF-α cytokine levels and lower IL-4 levels early in infection, compared to mock infected mice (Di Bonaventura et al., 2010). Later in infection, only IFN-γ was found to be significantly higher than any other cytokines in the infected mice. There were also higher keratinocyte-derived cytokine (GROα/KC), monocyte chemotactic protein 1 (MCP-1/JE), macrophage chemoattractant protein 5 (MCP-5), macrophage inflammatory protein 1α (MIP-1α), macrophage inflammatory protein 2 (MIP-2), and thymus and activation regulated chemokine (TARC) levels during early infection in the infected mice than in the control (Di Bonaventura et al., 2010). Although the studies discussed here were not with a murine model of CF, they explore the dynamic immune response that could be observed for CF based on the infection response in these mice. It will be important for future studies to elucidate this response and comprehend the complex interactions of this bacterium in a CF lung environment.
3 Antibiotic resistance
One of the most confounding features of S. maltophilia is its strong resistance toward many antibiotics, including aminoglycosides, fluroquinolones, β-lactams, cephalosporins, macrolides, carbapenems, chloramphenicol, tetracyclines, polymyxins, and sulfonamides (Brooke, 2012). Of particular concern pan-resistant strains, which are resistant to nearly all known antibiotics, have been documented (Valdezate et al., 2001). In general, though, combinational therapy of trimethoprim sulfamethoxazole (TMP-SMX) is an effective treatment (Muder et al., 1996; Muder, 2007; Nicodemo and Paez, 2007; Goldberg and Bishara, 2012). A study in Taiwan, showed that 20.5% of patients with S. maltophilia infections exhibited TMP-SMX resistance (Wang et al., 2017). Additionally, all tested fluoroquinolones were seen to effectively reduce S. maltophilia infection at one half of the MIC (Di Bonaventura et al., 2004). Similarly, in one study of patients suffering from sepsis caused by S. maltophilia, levofloxacin worked well in 70.5% of the patients (Soumya et al., 2020). However, prior levofloxacin use may induce increased S. maltophilia resistance to TMP-SMX (Wang et al., 2017).
Recently, many antibiotic resistant S. maltophilia infections have been seen in patients with certain co-morbidities. For instance, ventilator associated pneumonia patients infected with S. maltophilia showed resistance to carboxypenicillin and carbapenem (Ibn Saied et al., 2020). A case study of a 70 year old woman with hypoglycemia, chronic respiratory failure, adenocarcinoma of the lung, anemia, and other co-morbidities reported the presence of S. maltophilia in the respiratory tract; this infection showed resistance to broad spectrum antibiotics except for TMP-SMX and levofloxacin (Kanderi et al., 2020). These cases, and many others, indicate that new treatments are needed to treat this uncompromising, deadly pathogen.
3.1 Mechanism of antibiotic resistance
S. maltophilia displays antibiotic resistance through several mechanisms. S. maltophilia clinical isolate K279a carries nine Resistance Nodulation Division (RND)-type efflux pump genes that confer resistance to a number of different antibiotics (Crossman et al., 2008). For example, efflux pump operon smeABC contributes greatly to antimicrobial resistance, due to production of the outer membrane efflux lipoprotein SmeC (Li et al., 2002). Also, over-expression of smeDEF induces tetracycline, erythromycin, and fluoroquinolone resistance (Alonso and Martinez, 2000). Among the rest of the other RND efflux pumps genes, smeZ, smeJ, and smeK each contribute to aminoglycoside, fluoroquinolone, and tetracycline resistance (Gould et al., 2013). It was also seen that smeYZ conferred aminoglycoside and TMP-SMX resistance (Lin et al., 2015). Apart from this, a fusaric acid extrusion efflux pump (FuaABC), an MSF type efflux pump (EmrCABsm), and two ABC-type efflux pumps (SmrA, MacABCsm) confer antimicrobial resistance (Al-Hamad et al., 2009; Hu et al., 2012; Huang et al., 2013). Some strains of S. maltophilia encode floR, a gene found in insertion element common region (ISCR) adjacent sequences that aids in phenicol class of antibiotic resistance via expression of an exporter (Toleman et al., 2007).
In addition to efflux pumps, several antibiotic modifying enzymes make this bacterium resistant to many antimicrobials. L1 and L2 β-lactamases produced by S. maltophilia hydrolyse β-lactams (Avison et al., 2001). The L1 enzyme is a metallo-β-lactamase conferring resistance to all β-lactams, including penicillins, cephalosporins, and carbapenems (Saino et al., 1982; Paton et al., 1994; Walsh et al., 1994; Crowder et al., 1998), whereas L2 is a cephalosporinase (Saino et al., 1984; Walsh et al., 1997). S. maltophilia also carries an mphBM gene, which displays high homology to a macrolide phosphotransferase in S. aureus that makes S. aureus resistant to erythromycin (Alonso et al., 2000). It is possible that this factor does the same in S. maltophilia. Presence of sul1, sul2, and the ISCR element have been found in some strains of S. maltophilia rendering them resistant to TMP-SMX and other sulfonamides (Barbolla et al., 2004; Toleman et al., 2007; Chung et al., 2015). dfr genes expressing the enzymes dihydrofolate reductase has been also helpful in making strains resistant to trimethoprim. Thus both sul and dfr genes are seen in the TMP-SMX resistance strains (Hu et al., 2011; Hu et al., 2016). Other resistance genes have also been found in ISCR adjacent sequences such as dhfrA10, dhfrA9, dhfrA20 (aiding in trimethoprim resistance), tetR (aiding in resistance to the tetracycline class), and strA (aiding in streptomycin resistance) (Toleman et al., 2007). Interestingly, research has shown that some innately resistant environmental bacteria, such as S. maltophilia, can metabolize the carbon from antimicrobial agents to aid in growth (Martinez, 2008) (Dantas et al., 2008). Additionally, a truncated phosphoglucosamine mutase, encoded by glmM, indirectly counters the effects of antibiotics that target bacterial cell wall peptidoglycan (Avison et al., 2001), and the qnr gene protects bacteria from quinolone antibiotics targeting DNA gyrase and topoisomerase (Zhang et al., 2023). However, prevalence of most of these resistance factors is unknown.
4 Metal resistance
S. maltophilia can also grow in the presence of most heavy metals such as copper, zinc, cobalt, nickel, mercury, silver, antimony, tellurite, selenite, lead, and molybdenum among other metals (Pages et al., 2008; Deredjian et al., 2016; Yu et al., 2018; Zhang et al., 2023). For instance, S. maltophilia strain D457R, a multiresistant derivative of clinical isolate D457, displays cadmium resistance due to the cadmium efflux pump encoded by cadA (Pages et al., 2008). cadC, a regulator of cadA gene expression aids in making the bacteria cadmium resistant (Alonso et al., 2000; Pages et al., 2008). S. maltophilia was seen to express a SulP family protein, potentially involved in sulfate permease activities responsible for uptake of oxyanions such as selenate and selenite (Yu et al., 2018). A phosphate uptake protein with a PitA-like sequence had also been found in S. maltophilia conferring arsenate resistance. Genes putatively encoding homologs of metal transporters MgtA, MgtE, CorA, MntH, and ZupT have been identified, and these potential transporters can cause accumulation of Mg²+, Mn²+, Ni²+, Zn²+, Cd²+, Co²+, and Cu²+ (Yu et al., 2018). Additionally, genome annotations have revealed multicopper oxidases and copper transporting, binding, and storage proteins conferring bacterial resistance to copper. Furthermore, genome analysis has revealed MdtABC efflux pumps of zinc; CzcA, CzcB, and CzcC proteins causing efflux of zinc, cadmium, and cobalt; and ArsC, arsenite reductase, mercury reductase, and other arsenite and mercury transporter proteins conferring arsenic and mercury resistance (Yu et al., 2018). This metal resistance, combined with the extreme antibiotic resistance that S. maltophilia displays, makes this pathogen extremely difficult to kill (Edet et al., 2023).
5 Virulence factors
Though traditionally considered a low virulence pathogen, S. maltophilia contains genes with homology to numerous classical virulence factors, though many have not been experimentally validated (Trifonova and Strateva, 2019a). Here, we highlight the vast arsenal that S. maltophilia contains with which it conducts pathogenesis.
5.1 Lytic enzymes
S. maltophilia can secrete a variety of different extracellular enzymes, including phospholipase, DNAase, RNAse, esterases, gelatinase, lipases, proteinase, proteases, heparinase, hyaluronidase, and hemolysin (Crossman et al., 2008; Thomas et al., 2014) (Figure 1). Serine proteases StmPr1, StmPr2, and StmPr3, have particularly been shown to make S. maltophilia highly cytotoxic. Among other effects, these proteases sever the host cell extracellular matrix (ECM) proteins like human type I collagen, fibrinogen, and fibronectin (DuMont and Cianciotto, 2017). These proteases also show caseinolytic, gelatinase, and hydrolytic activities, and they are associated with loss of mammalian cell structural viability, changes in actin cytoskeleton, loss of integrin/ECM connections, cell detachment, and degradation of IL-8. These effects can trigger the process of apoptosis (anoikis) by activating a cascade of caspases starting from caspase 3, 6, and 7 (DuMont and Cianciotto, 2017).
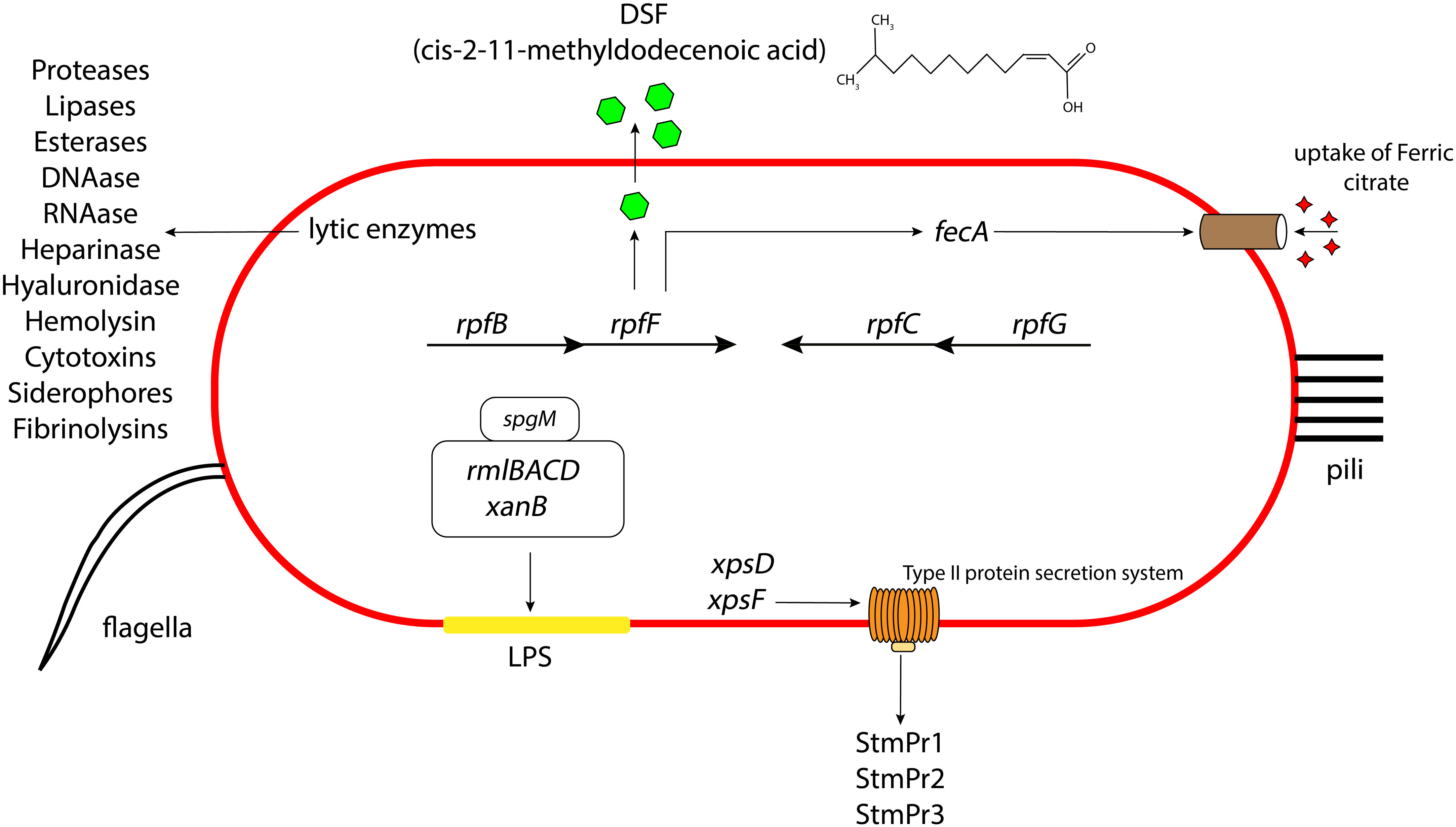
Figure 1 Schematic representation of the different virulence and attachment factors in S. maltophilia. The role of LPS, lytic enzymes, quorum sensing, secretion systems, iron uptake mechanisms contributing to the overall bacterial pathogenesis, along with the presence of pili and flagella aiding in their initial attachment and colonization. Not drawn to scale.
5.2 Secretion systems
S. maltophilia also produces type I, type II, type IV, type V, type VI secretion systems. Many of the above lytic enzymes are secreted through one of these apparatuses. For instance, the StmPr proteases, secreted through the xps-encoded type II secretion (T2S) system, causes cell rounding, actin rearrangement, degradation of extracellular matrix proteins, degradation of IL-8, cell detachment, and ultimately cell death of alveolar epithelial cells (Karaba et al., 2013; DuMont et al., 2015; DuMont and Cianciotto, 2017) (Figure 1). S. maltophilia also express a type IV secretion system (T4SS) whose effector molecules render the pathogen advantage in interbacterial competition with P. aeruginosa (Nas et al., 2021) and also lyses infected mammalian macrophages (Nas et al., 2019). It is suggested that the VirB/D4 apparatus of T4SS has pro- and anti-killing effector molecules which have disparate actions on normal and infected mammalian cells and heterologous bacteria inhabiting the human host (Nas et al., 2019). Another study suggests that there are two effector molecules of the T4SS in S. maltophilia, namely TcfA and TcfB, which on expression kill the target bacterial cell in a mixed bacterial environment (Nas et al., 2021). Based on assays, sequence analysis of the effector protein and structural prediction, it is hypothesized that in a coculture environment, TcfA, a lipase enzyme, on injection to the outer or inner membrane of the targeted competitor cell causes cell death. Sequence similarity and software prediction also shows that TcfB is anticipated to belong to a family of endolysins that exhibit lethal properties linked to an activity resembling lysozyme. It is hypothesized to interact and degrade the peptidoglycan for effective killing of the targeted cell. S. maltophilia environmental and clinical isolates also possess a type VI secretion system (T6SS) (Crisan et al., 2023). Genome annotation suggest that this system expresses proteins with putative phospholipase, endopeptidase, and lysozyme like activity. There is also evidence that pulmonary S. maltophilia strain STEN00241 can utilize a T6SS to compete with P. aeruginosa, and outcompete B. cenocepacia and E. coli in a co-culture environment (Crisan et al., 2023). Finally, genome annotations indicate the presence of type I secretion systems in strains K279a and R551-3 (Rocco et al., 2009) and type V secretion system (autotransporter) in strain K279a (Crossman et al., 2008). Intriguingly, this Gram-negative pathogen does not make a type III secretion system.
5.3 Diffusible signaling factor
Pathogenesis in S. maltophilia is governed by a quorum sensing molecule called diffusible signaling factor (DSF), or cis-11-methyl-2-dodecenoic acid. DSF synthesis is encoded by rpfF (An and Tang, 2018) (Figure 1). rpfF is transcribed as part of an operon with rpfB, encoding a long chain fatty acyl coA ligase (on which the secretion of DSF is partially dependent) (Yero et al., 2020). A convergently transcribed operon consisting of rpfC and rpfG encodes a 2-component system of sensor kinase and response regulator, respectively, which are involved in DSF transcriptional regulation. DSF has been shown to regulate S. maltophilia biofilm formation, synthesis of extracellular polymeric substance (EPS), and secretion of protease (Huang et al., 2006; Ryan et al., 2015; Yero et al., 2020). It also contributes to bacterial motility. Furthermore, DSF was seen to influence the production of β-lactamase, making the bacterium resistant to β-lactams. This finding was the first evidence for the role of DSF signaling in antibiotic resistance (Alcaraz et al., 2019). DSF can also regulate the expression of an outer membrane receptor FecA, used for the uptake of ferric citrate in S. maltophilia (Matera et al., 2004). Some S. maltophilia strains contain an extra DSF-related gene, rpfS, which contributes to epiphytic disease (An et al., 2014). Intriguingly, DSF also mediates interspecies communication and can thus mediate coculture of S. maltophilia with other species (Ryan et al., 2015).
5.4 Lipopolysaccharide
The LPS layer of Gram-negative bacteria, including S. maltophilia, can greatly influence the antimicrobial activities of the cell. The LPS in S. maltophilia is primarily made of O-specific polymer formed of D-glucose, L-rhamnose, and D-fucose (Winn et al., 1993). The endotoxicity of the LPS layer is associated with neutrophil activation or IL-8 production inside the host which gives rise to a systemic immune response. Various combinations of antibiotics interacting with the LPS could increase (ceftazidime) or decrease (β-lactams and aminoglycosides) this immune response (Matera et al., 2004). In S. maltophilia, the spgM gene, encoding for phosphoglucomutase and phosphomannomutase enzyme activities (like the phosphoglucomutase producing gene algC in P. aeruginosa), contributes to the production of a thicker LPS layer (McKay et al., 2003) (Figure 1). A strain with mutation of the spgM gene exhibited a two to four fold increased susceptibility to aminoglycosides, quinolones, vancomycin, polymyxin B and E, and many other antibiotics. The spgM mutant was also unable to colonize rat lungs, suggesting that the gene is an important factor in the virulence of the bacterium. Other genes affecting S. maltophilia LPS production include xanB, rmlA, and rmlC (Huang et al., 2006). Mutation of rmlAC and xanB results in lowered O-antigen production in the bacterial LPS, leading to a defective outer membrane layer and impaired flagellar production, motility, adherence, and biofilm formation (Huang et al., 2006).
5.5 Attachment factors
5.5.1 Pili
Microscopy studies have demonstrated the presence of peritrichous semiflexible pili on the surface of S. maltophilia (de Oliveira-Garcia et al., 2003) (Figure 1). These pili have been named S. maltophilia fimbriae-1, or Smf-1. Electron microscopy images reveal that Smf-1 has a size of 5-7nm in width and forms primarily at 37°C (Flores-Trevino et al., 2019; Kalidasan and Neela, 2020). The Smf-1 N-terminal amino acid sequence is similar to that of the CupA fimbriae in P. aeruginosa and similar chaperone/usher pili in other Gram-negative bacteria (de Oliveira-Garcia et al., 2003). Smf-1 is capable of agglutinating red blood cells from several animals, including humans (de Oliveira-Garcia et al., 2003). It also binds to several abiotic and biotic surfaces, such as medical devices and human epithelial cells (de Oliveira-Garcia et al., 2003). Additionally, this pilus is highly immunogenic. Mice administered purified Smf-1 showed a high innate immune response with increase in pro-inflammatory cytokines (e.g. IL-1β, and TNF-α). These mice also exhibited elevated IL-8 levels in the bladder, suggesting neutrophil chemotaxis, followed by high nitric oxide production (Zgair and Al-Adressi, 2013).
Through genome annotations, it has been reported that S. maltophilia expresses a type I pilus that is assembled via the chaperone usher pathway (Rocco et al., 2009). These types of pili are comprised of a shaft made of a primary fimbrial protein and two other ancillary proteins. This S. maltophilia pilus has been linked to bacterial adhesion and initial stages of biofilm formation (Ryan et al., 2009).
Type IV pili also play a role in S. maltophilia virulence (Trifonova and Strateva, 2019b). Type IV pili in S. maltophilia CF isolates contribute to the bacterial biofilm development (Pompilio et al., 2011). They also putatively mediate adherence to surfaces, auto-aggregation, and twitching motility of the bacterium (Ryan et al., 2009). Importantly, the S. maltophilia type IV pilus is also a receptor for several bacteriophages such as DLP1 and DLP2 (McCutcheon et al., 2018). Additionally, AXL3 bacteriophage has been shown to interact with the PilA subunit present in the type IV pilus rod, and the subsequent viral penetration is facilitated as the physical retraction of the type IV pilus allows the attached phage to reach the cell surface (McCutcheon et al., 2020). If we can understand and control this interaction, it is possible that this mechanism can be exploited in developing specialized tools and treatments tailored to fight this multidrug resistant pathogen.
5.5.2 Flagella
S. maltophilia produces a polar flagellum, which is considered a significant attachment factor aiding colonization of host cells (Figure 1). The main structural protein of S. maltophilia flagella is a 38kDa protein called FliC (de Oliveira-Garcia et al., 2002). Studies have shown that, in several clinical isolates of S. maltophilia, presence of flagella correlates with binding to mouse tracheal mucus (Zgair and Chhibber, 2011). The flagellin protein acts as an adhesin, and anti-flagellin antibody significantly lowers bacterial adherence to mucus. Additionally, a homolog of the Xanthomonas campestris flil gene (encoding a flagellin associated ATPase) demonstrated an important role in flagellar production and bacterial colonization of S. maltophilia strains isolated from individuals with cystic fibrosis (Di Bonaventura et al., 2007). Another study showed that mutation of the ompA gene (encoding a porin protein) severely compromised the integrity of the S. maltophilia outer membrane. This action caused a defect in flagellar assembly which resulted in failure of bacterial swimming motility (Liao et al., 2021). In S. maltophilia, BsmR (biofilm and swimming motility regulator) regulates the expression of 349 genes, including fsnR, which in turn switches on the expression of 2 operons required for flagella production (Kang et al., 2015; Liu et al., 2017). With the activation of these operons, flagella production is increased, which increases the bacterial motility.
5.5.3 Phosphoglycerate mutase impacts bacterial attachment
A recent genetic screen of S. maltophilia strain K279a identified gpmA, encoding glycolytic enzyme phosphoglycerate mutase, as affecting biofilm formation (Ramos-Hegazy et al., 2020). Subsequent isogenic deletion of gpmA resulted in defects in adhesion to biotic and abiotic surfaces. Initial attachment of the ΔgpmA strain was greatly reduced at early time points compared to the wild type strain on polystyrene plates. Nevertheless, there was no difference in biofilm formation at later time points, which suggests that this gene is required for early attachment and development of biofilms on abiotic surfaces. It was also observed that the ΔgpmA strain exhibited a 100-fold reduction in binding to CF bronchial epithelial cells (CFBE) compared to the wild type strain at 1 hour. As with polystyrene plate biofilms, though, there was no significant difference between the wild type and ΔgpmA strains at later time points. It is unclear why a metabolic gene affects attachment, though this finding suggests an intriguing link between metabolism and virulence in S. maltophilia.
5.6 Other virulence factors
Genome annotation show that S. maltophilia strain K279a also produces a YadA-like protein, of the BuHA family of proteins (Nseir et al., 2006a). First identified in Burkholderia mallei (Tiyawisutsri et al., 2007), members of this protein family have been shown to have filamentous hemagglutinin, invasin, and hemolysin properties, and they are considered as important virulence factors in the pathogenesis and rapid spread of bacteria (Colombi et al., 2006; Holden et al., 2009). Indeed reports mention the presence of filamentous hemagglutinins via genome annotation in various environmental and clinical strains of S. maltophilia that conduct virulence through cell to cell aggregation (Ryan et al., 2009). S. maltophilia strains were also found to produce outer membrane vesicles that are cytotoxic to alveolar epithelial cells and induce a strong inflammatory response in both in vitro and in vivo environments (Kim et al., 2016). Additionally, the bacterium can protect itself from host defenses through expression of enzymes such as alkyl hydroperoxidase, superoxide dismutase, catalase, and melanin pigments that can disrupt or counteract host defense products (Thomas et al., 2014; Jair et al., 2019; Li et al., 2020). Southern blot hybridization and PCR analysis have shown S. maltophilia to carry a phage gene sequence mildly homologous to a sequence in Vibrio cholerae producing the zonula occludens toxin (zot) (Hagemann et al., 2006). This toxin impairs the intercellular junctions in host cells allowing easy access for infection (Fasano et al., 1991).
6 S. maltophilia biofilm formation
Most microorganisms in nature attach to and grow on surfaces (Armbruster and Parsek, 2018). This attachment facilitates the aggregation of the bacteria into microcolonies, which increase in size as bacteria multiply and other cells attach to the cluster. The constituent microbial cells produce a polymer matrix that surrounds the cluster, creating a slimy layer. This process is called biofilm formation (Hernández-Jiménez et al., 2013; Deepigaa, 2017). Bacteria generally remain in sessile communities within the biofilm, as the biofilm matures (Crouzet et al., 2014). Eventually, as the biofilm ages, cells disperse and spread to other locations. Because of the chemical and physical conditions within biofilms, the bacterial community is protected from adverse environmental conditions, such as extreme temperatures and extreme pH, high salt concentrations, UV radiation, depletion of nutrients, host immune responses, antimicrobial agents, peptides (LL37, β-defensins, dermcidin, for example), and even antibiotics (Hoiby et al., 2010; Yasir et al., 2018; Yin et al., 2019). Biofilms can be found in nature, in medical settings, and in industrial settings (Mah and O’Toole, 2001; Hall-Stoodley et al., 2012), and during infection, biofilms contribute to pathogenesis due to their highly resistant nature (Yin et al., 2019). Some estimates indicate that in humans, about 60-80% of microbial infections involve a biofilm component (Costerton et al., 1999). Thus, it is important to understand the pathways that trigger this physiological process so we can better fight the causative organisms.
S. maltophilia is a known biofilm producer. In one study, 98.7% of 150 clinical S. maltophilia isolates produced biofilm (Azimi et al., 2020). Among this percentage, 46% had strong biofilm producing capabilities. In a few molecular studies of S. maltophilia biofilm, smf-1, rmlA, rpfF, and spgM genes were found to influence S. maltophilia biofilm initiation and development (Azimi et al., 2020); these genes are widespread among S. maltophilia strains (Azimi et al., 2020).
Additionally, studies have shown that iron regulation in S. maltophilia influences biofilm formation (García et al., 2015). For instance, the Fur protein regulator that maintains iron homeostasis in the cell also regulates certain virulence factors and mediates oxidative stress response (Kalidasan et al., 2018a). Isogenic deletion of the fur gene results in decreased biofilm in iron depleted conditions, compared to iron-rich environments. Moreover, upregulation of two iron repressed outer membrane proteins (IROMPs) in such conditions also affects biofilm. These proteins are putatively required for scavenging iron from the environment to induce higher biofilm growth.
While S. maltophilia strains producing strong biofilms on abiotic surfaces show significant resistance to many antibiotics, like ticarcillin clavulanate, ceftazidime, ciprofloxacin, and doxycycline (Azimi et al., 2020), some antibiotics have been found to be quite effective in treating biofilms of this microbe. For instance, TMP-SMX was found to be active in reducing bacterial viability of S. maltophilia in biofilm (Di Bonaventura et al., 2004). The fluoroquinolone moxifloxacin was also useful in stopping bacterial adherence, by inducing cell lysis, resulting in detachment of bacterial cell surface glycocalyx from polystyrene surfaces (Di Bonaventura et al., 2004). Another study showed that levofloxacin could inhibit biofilm formation of clinical S. maltophilia strains in the presence of erythromycin (Sun et al., 2016). Erythromycin can also act synergistically with cefoperazone/sulbactam, and piperacillin in increasing susceptibility among constituents of biofilms (Sun et al., 2016). This indicates a potential use of combination therapy of certain antibiotics with macrolides in combating S. maltophilia biofilm formation.
Interestingly, one study found an inverse relationship between antibiotic resistance and the biofilm formation of S. maltophilia (Pompilio et al., 2020). It was seen that pathogenic strains of S. maltophilia susceptible to ceftazidime, levofloxacin, colistin, and ticarcillin clavulanate showed greater biofilm formation capabilities than pathogenic strains resistant to the same antibiotic. However, there was no significant difference in the level of biofilm for non-pathogenic S. maltophilia strains for any antibiotic (Pompilio et al., 2020). Studies also showed mutation of smeYZ genes, conferring aminoglycoside and TMP-SMX resistance to the bacteria, produced lesser biofilm than the wild type clinical isolate S. maltophilia KJ (Lin et al., 2015).
7 S. maltophilia and metabolism
The name “Stenotrophomonas” means “narrow eating unit”, which refers to the comparative lack of substrates utilized by this microorganism for growth. “Maltophilia” means “maltose lover” (Denton and Kerr, 1998). Historically, it has been thought that this pathogen mainly utilizes hexose sugars, and growth studies somewhat support that assumption. Beyond sugar, though, S. maltophilia displays some intriguing metabolic characteristics. In fact, recent studies have made connections between S. maltophilia metabolism and virulence behaviors, including biofilm formation (Figure 2).
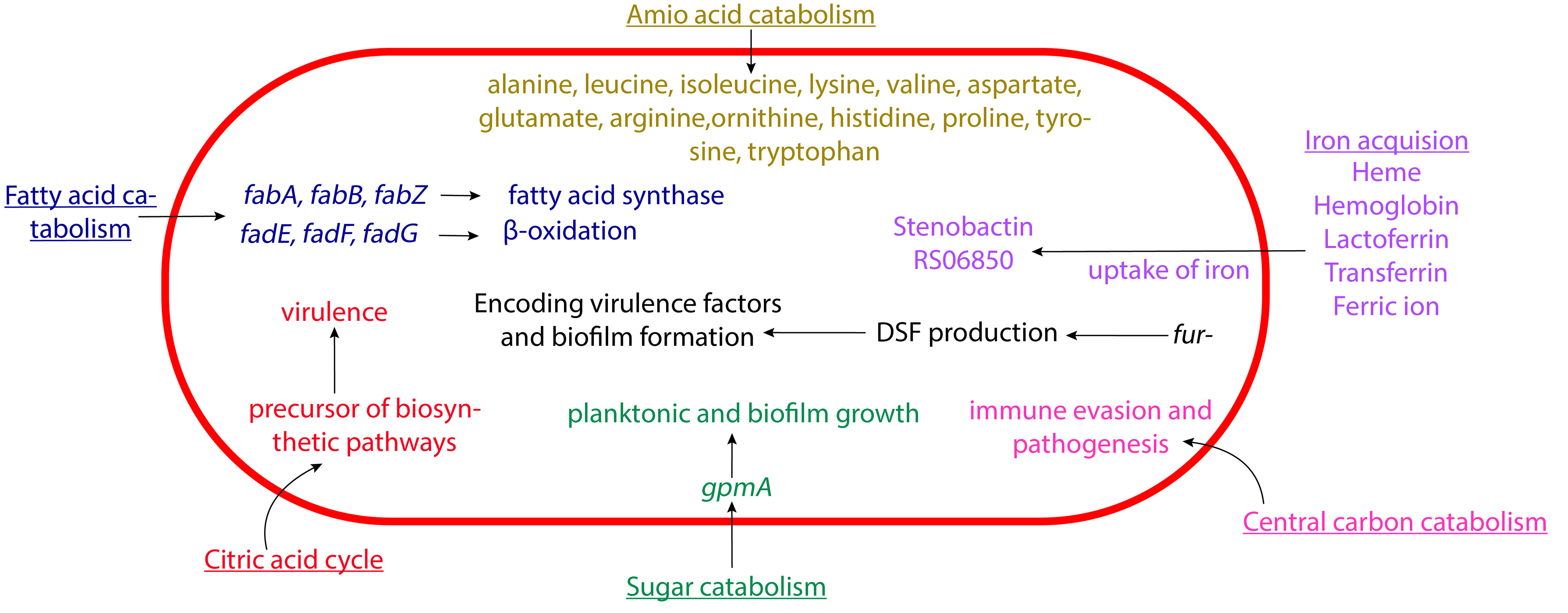
Figure 2 Schematic representation of the metabolism of S. maltophilia affecting its pathogenesis. The fatty acid catabolism, citric acid cycle, sugar catabolism, amino acid catabolism, central carbon catabolism, and iron acquisition come together in contributing to the overall bacterial virulence. Not drawn to scale.
Iron acquisition has an important role in S. maltophilia metabolism (Kalidasan et al., 2018b). This includes the use of siderophores like enterobactin and stenobactin, alongside multiple other pathways that can utilize various iron sources (Ryan et al., 2009; Adamek et al., 2014; Suvorova and Rodionov, 2016; Nas and Cianciotto, 2017; Kalidasan et al., 2018b; Azman et al., 2019; Brooke, 2021). The microbes’s central carbon catabolism is crucial for biosynthesis and energy production (Munoz-Elias and McKinney, 2006). S. maltophilia contains genes for several glycolytic pathway, including Embden-Meyerhof-Parnas, Entner-Doudoroff, and pentose phosphate pathways (Stanier et al., 1966; Munoz-Elias and McKinney, 2006; Crossman et al., 2008). As mentioned above, glycolytic gene gpmA appears to affect S. maltophilia attachment. gpmA encodes phosphoglycerate mutase, which catalyzes the interconversion of 2-phospho-D-glycerate and 3-phospho-D-glycerate. It was further discovered that without gpmA, S. maltophilia planktonic growth and biofilm formation are severely decreased when grown on different substrates (Isom et al., 2022; Ramos-Hegazy et al., 2020). Intriguingly, recent work suggested that under low nutrient conditions this pathogen preferentially utilizes amino acid over glucose as carbon source, potentially shuttling carbons through GpmA (Isom et al., 2022). Beyond iron acquisition pathways and glycolysis, S. maltophilia contains genes for the citric acid cycle (Kanehisa et al., 2023), β-oxidation and fatty acid synthesis (Haydon and Guest, 1991; Heath and Rock, 1996; Rock and Cronan, 1996; Textor et al., 1997; Cronan and Subrahmanyam, 1998; DiRusso and Nyström, 1998; Wang and Cronan, 2004; Fujita et al., 2007; Shuman, 2010; Kanehisa et al., 2023), and utilization of various amino acids like alanine, leucine, isoleucine, lysine, valine, aspartate, glutamate, arginine, ornithine, histidine, proline, tyrosine, and tryptophan (Omenn, 2010). Investigating the possible connection of these metabolic pathways to pathogenesis will be an intriguing and potentially fruitful avenue of research in the future.
8 Conclusion
S. maltophilia is a fascinating microorganism that presents us with extensive challenges and opportunities. Because of the lack of knowledge of its virulence mechanisms and basic biology, there is a huge potential for further research to understand the evolution, adaptability, and the versatility of this pathogen. It is evident that we know quite a bit about the antibiotic resistance of S. maltophilia, the types of virulence factors it can produce, and its role in infection of the bronchopulmonary system. Additionally, much work has been done regarding S. maltophilia multi-species biofilm, roles of iron, DSF factor, different lytic enzymes, and treatment options for CF individuals colonized with this microbe, but very little work has been done to understand the mechanism of biofilm formation, the regulators in biofilm growth, the specific immune responses to the formation of S. maltophilia biofilm, and the cascade of virulence factor events during an infection. An integrated disease model should be established to develop newer understanding into such various topics. These efforts may aid in understanding infection patterns and pathogenesis and yield insights about novel molecular and cellular drug targets. Moreover, greater efforts should also be placed on surveillance of the worldwide drug resistance situation and the role opportunistic pathogens like S. maltophilia play in genetic transfer of resistance determinants. Apart from this, climate change has been identified as a significant factor in altering the transmission patterns of S. maltophilia, rendering it an extremely hazardous pathogen (Lafferty, 2009; Omenn, 2010; Shuman, 2010; Brooke, 2012). According to current research, the global average temperature will increase 1.8°C to 5.8°C over the next 100 years and that may alter the hydrolytic cycle of the environment, increase pollution, and create poorer sanitation (Shuman, 2010). It is also expected that this will change the geographical distribution of waterborne S. maltophilia, alter its microbial evolution pattern, and give the bacterium a susceptible environment to increase its spread (Brooke, 2012; Brooke, 2021). Further investigation could ultimately lead us to update our knowledge about this emerging global opportunistic pathogen, S. maltophilia, and to identify novel therapies to inhibit its growth and spread.
Author contributions
RB: Conceptualization, Writing – original draft, Writing – review & editing. NA: Conceptualization, Writing – original draft, Writing – review & editing. GA: Conceptualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by BRIDGE funding to GA from IUPUI.
Acknowledgments
We thank E. Klein, L. Ramos-Hegazy, S. Chakravarty, C. Isom, A. Beard, O. Harrigan, J. Hall for their helpful discussions and suggestions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adamek, M., Linke, B., Schwartz, T. (2014). Virulence genes in clinical and environmental Stenotrophomas maltophilia isolates: a genome sequencing and gene expression approach. Microb. Pathog. 67-68, 20–30. doi: 10.1016/j.micpath.2014.02.001
Adegoke, A. A., Stenstrom, T. A., Okoh, A. I. (2017). Stenotrophomonas maltophilia as an emerging ubiquitous pathogen: looking beyond contemporary antibiotic therapy. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02276
Alcaraz, E., Garcia, C., Friedman, L., de Rossi, B. P. (2019). The rpf/DSF signalling system of Stenotrophomonas maltophilia positively regulates biofilm formation, production of virulence-associated factors and beta-lactamase induction. FEMS Microbiol. Lett. 366 (6), fnz069. doi: 10.1093/femsle/fnz069
Al-Hamad, A., Upton, M., Burnie, J. (2009). Molecular cloning and characterization of SmrA, a novel ABC multidrug efflux pump from Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 64, 731–734. doi: 10.1093/jac/dkp271
Alonso, A., Martinez, J. L. (2000). Cloning and characterization of SmeDEF, a novel multidrug efflux pump from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 44, 3079–3086. doi: 10.1128/AAC.44.11.3079-3086.2000
Alonso, A., Sanchez, P., Martinez, J. L. (2000). Stenotrophomonas maltophilia D457R contains a cluster of genes from gram-positive bacteria involved in antibiotic and heavy metal resistance. Antimicrob. Agents Chemother. 44, 1778–1782. doi: 10.1128/AAC.44.7.1778-1782.2000
Amin, R., Waters, V. (2016). Antibiotic treatment for Stenotrophomonas maltophilia in people with cystic fibrosis. Cochrane Database Syst. Rev. 7, CD009249. doi: 10.1002/14651858.CD009249.pub4
An, S. Q., Tang, J. L. (2018). Diffusible signal factor signaling regulates multiple functions in the opportunistic pathogen Stenotrophomonas maltophilia. BMC Res. Notes 11, 569. doi: 10.1186/s13104-018-3690-1
An, S. Q., Allan, J. H., McCarthy, Y., Febrer, M., Dow, J. M., Ryan, R. P. (2014). The PAS domain-containing histidine kinase RpfS is a second sensor for the diffusible signal factor of X anthomonas campestris. Mol. Microbiol. 92, 586–597. doi: 10.1111/mmi.12577
Armbruster, C. R., Parsek, M. R. (2018). New insight into the early stages of biofilm formation. Proc. Natl. Acad. Sci. 115, 4317–4319. doi: 10.1073/pnas.1804084115
Avison, M. B., Higgins, C. S., von Heldreich, C. J., Bennett, P. M., Walsh, T. R. (2001). Plasmid location and molecular heterogeneity of the L1 and L2 beta-lactamase genes of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 45, 413–419. doi: 10.1128/AAC.45.2.413-419.2001
Azimi, A., Aslanimehr, M., Yaseri, M., Shadkam, M., Douraghi, M. (2020). Distribution of smf-1, rmlA, spgM and rpfF genes among Stenotrophomonas maltophilia isolates in relation to biofilm-forming capacity. J. Global Antimicrobial Resistance 23, 321–326. doi: 10.1016/j.jgar.2020.10.011
Azman, A., Vasodavan, K., Joseph, N., Kumar, S., Hamat, R. A., Nordin, S. A., et al. (2019). Physiological and proteomic analysis of Stenotrophomonas maltophilia grown under the iron-limited condition. Future Microbiol. 14, 1417–1428. doi: 10.2217/fmb-2019-0174
Ballestero, S., Virseda, I., Escobar, H., Suarez, L., Baquero, F. (1995). Stenotrophomonas maltophilia in cystic fibrosis patients. Eur. J. Clin. Microbiol. Infect. Dis. 14, 728–729. doi: 10.1007/BF01690887
Barbolla, R., Catalano, M., Orman, B. E., Famiglietti, A., Vay, C., Smayevsky, J., et al. (2004). Class 1 integrons increase trimethoprim-sulfamethoxazole MICs against epidemiologically unrelated Stenotrophomonas maltophilia isolates. Antimicrob. Agents Chemother. 48, 666–669. doi: 10.1128/AAC.48.2.666-669.2004
Berdah, L., Taytard, J., Leyronnas, S., Clement, A., Boelle, P. Y., Corvol, H. (2018). Stenotrophomonas maltophilia: a marker of lung disease severity. Pediatr. pulmonology 53, 426–430. doi: 10.1002/ppul.23943
Brooke, J. S. (2012). Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin. Microbiol. Rev. 25, 2–41. doi: 10.1128/CMR.00019-11
Brooke, J. S. (2021). Advances in the microbiology of stenotrophomonas maltophilia. Clin. Microbiol. Rev. 34, e0003019. doi: 10.1128/CMR.00030-19
Brooke, J. S., Di Bonaventura, G., Berg, G., Martinez, J. L. (2017). Editorial: A Multidisciplinary Look at Stenotrophomonas maltophilia: An Emerging Multi-Drug-Resistant Global Opportunistic Pathogen. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.01511
Cervia, J. S., Ortolano, G. A., Canonica, F. P. (2008). Hospital tap water as a source of Stenotrophomonas maltophilia infection. Clin. Infect. Dis. 46, 1485–1487. doi: 10.1086/587180
Chang, Y. T., Lin, C. Y., Chen, Y. H., Hsueh, P. R. (2015). Update on infections caused by Stenotrophomonas maltophilia with particular attention to resistance mechanisms and therapeutic options. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00893
Chung, H. S., Kim, K., Hong, S. S., Hong, S. G., Lee, K., Chong, Y. (2015). The sul1 gene in Stenotrophomonas maltophilia with high-level resistance to trimethoprim/sulfamethoxazole. Ann. Lab. Med. 35, 246–249. doi: 10.3343/alm.2015.35.2.246
Colin, A. A., Rabin, H. R. (2011). Stenotrophomonas maltophilia in cystic fibrosis: guilty or innocent? Am. J. Respir. Crit. Care Med. 183, 564–566. doi: 10.1164/rccm.201010-1668ED
Colombi, D., Oliveira, M. L., Campos, I. B., Monedero, V., Pérez-Martinez, G., Ho, P. L. (2006). Haemagglutination Induced by Bordetella pertussis Filamentous Haemagglutinin Adhesin (FHA) Is Inhibited by Antibodies Produced Against FHA 430–873 Fragment Expressed in Lactobacillus casei. Curr. Microbiol. 53, 462–466. doi: 10.1007/s00284-005-0388-0
Costerton, J. W., Stewart, P. S., Greenberg, E. P. (1999). Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322. doi: 10.1126/science.284.5418.1318
Crisan, C. V., Van Tyne, D., Goldberg, J. B. (2023). The type VI secretion system of the emerging pathogen Stenotrophomonas maltophilia complex has antibacterial properties. Msphere 8 (6), e00584–e00523. doi: 10.1128/msphere.00584-23
Cronan, J. E., Jr., Subrahmanyam, S. (1998). FadR, transcriptional co-ordination of metabolic expediency. Mol. Microbiol. 29 (4), 937–943. doi: 10.1046/j.1365-2958.1998.00917.x
Crossman, L. C., Gould, V. C., Dow, J. M., Vernikos, G. S., Okazaki, A., Sebaihia, M., et al. (2008). The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol. 9, R74. doi: 10.1186/gb-2008-9-4-r74
Crouzet, M., Le Senechal, C., Brözel, V., Costaglioli, P., Barthe, C., Bonneu, M., et al. (2014). Exploring early steps in biofilm formation: set-up of an experimental system for molecular studies. BMC Microbiol. 14, 253. doi: 10.1186/s12866-014-0253-z
Crowder, M. W., Walsh, T. R., Banovic, L., Pettit, M., Spencer, J. (1998). Overexpression, purification, and characterization of the cloned metallo-beta-lactamase L1 from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 42, 921–926. doi: 10.1128/AAC.42.4.921
Dantas, G., Sommer, M. O., Oluwasegun, R. D., Church, G. M. (2008). Bacteria subsisting on antibiotics. Science 320, 100–103. doi: 10.1126/science.1155157
Deepigaa, M. (2017). Antibacterial resistance of bacteria in biofilms. Res. J. Pharm. Technol. 10, 4019–4023. doi: 10.5958/0974-360X.2017.00728.4
Demiraslan, H., Sevim, M., Pala, Ç., Durmaz, S., Berk, V., Kaynar, L., et al. (2013). Risk factors influencing mortality related to Stenotrophomonas maltophilia infection in hematology-oncology patients. Int. J. Hematol. 97, 414–420. doi: 10.1007/s12185-013-1296-x
Denton, M., Kerr, K. G. (1998). Microbiological and clinical aspects of infection associated with Stenotrophomonas maltophilia. Clin. Microbiol. Rev. 11, 57–80. doi: 10.1128/CMR.11.1.57
de Oliveira-Garcia, D., Dall’Agnol, M., Rosales, M., Azzuz, A. C.G.S., Martinez, M. B., Girón, J. A. (2002). Characterization of flagella produced by clinical strains of Stenotrophomonas maltophilia. Emerg. Infect. Dis. 8, 918–923. doi: 10.3201/eid0809.010535
de Oliveira-Garcia, D., Dall’Agnol, M., Rosales, M., Azzuz, A. C.G.S., Alcántara, N., Martinez, M. B., et al. (2003). Fimbriae and adherence of Stenotrophomonas maltophilia to epithelial cells and to abiotic surfaces. Cell Microbiol. 5, 625–636. doi: 10.1046/j.1462-5822.2003.00306.x
Deredjian, A., Alliot, N., Blanchard, L., Brothier, E., Anane, M., Cambier, P., et al. (2016). Occurrence of Stenotrophomonas maltophilia in agricultural soils and antibiotic resistance properties. Res. Microbiol. 167, 313–324. doi: 10.1016/j.resmic.2016.01.001
Di Bonaventura, G., Prosseda, G., Del Chierico, F., Cannavacciuolo, S., Cipriani, P., Petrucca, A., et al. (2007). Molecular characterization of virulence determinants of Stenotrophomonas maltophilia strains isolated from patients affected by cystic fibrosis. Int. J. Immunopathol. Pharmacol. 20, 529–537. doi: 10.1177/039463200702000311
Di Bonaventura, G., Pompilio, A., Zappacosta, R., Petrucci, F., Fiscarelli, E., Rossi, C., et al. (2010). Role of excessive inflammatory response to Stenotrophomonas maltophilia lung infection in DBA/2 mice and implications for cystic fibrosis. Infect. Immun. 78, 2466–2476. doi: 10.1128/IAI.01391-09
Di Bonaventura, G., Spedicato, I., D’Antonio, D., Robuffo, I., Piccolomini, R. (2004). Biofilm formation by Stenotrophomonas maltophilia: modulation by quinolones, trimethoprim-sulfamethoxazole, and ceftazidime. Antimicrob. Agents Chemother. 48, 151–160. doi: 10.1128/AAC.48.1.151-160.2004
DiRusso, C. C., Nyström, T. (1998). The fats of Escherichia coli during infancy and old age: regulation by global regulators, alarmones and lipid intermediates. Mol. Microbiol. 27 (1), 1–8. doi: 10.1046/j.1365-2958.1998.00645.x
DuMont, A. L., Cianciotto, N. P. (2017). Stenotrophomonas maltophilia serine protease stmPr1 induces matrilysis, anoikis, and protease-activated receptor 2 activation in human lung epithelial cells. Infect. Immun. 85 (12), iai.00544-17. doi: 10.1128/IAI.00544-17
DuMont, A. L., Karaba, S. M., Cianciotto, N. P. (2015). Type II secretion-dependent degradative and cytotoxic activities mediated by stenotrophomonas maltophilia serine proteases stmPr1 and StmPr2. Infect. Immun. 83, 3825–3837. doi: 10.1128/IAI.00672-15
Edet, U. O., Bassey, I. U., Joseph, A. P. (2023). Heavy metal co-resistance with antibiotics amongst bacteria isolates from an open dumpsite soil. Heliyon 9, e13457. doi: 10.1016/j.heliyon.2023.e13457
Ewig, S., Soler, N., Gonzalez, J., Celis, R., El-Ebiary, M., Torres, A. (2000). Evaluation of antimicrobial treatment in mechanically ventilated patients with severe chronic obstructive pulmonary disease exacerbations. Crit. Care Med. 28, 692–697. doi: 10.1097/00003246-200003000-00015
Fasano, A., Baudry, B., Pumplin, D. W., Wasserman, S. S., Tall, B. D., Ketley, J. M., et al. (1991). Vibrio cholerae produces a second enterotoxin, which affects intestinal tight junctions. Proc. Natl. Acad. Sci. 88, 5242–5246. doi: 10.1073/pnas.88.12.5242
Flores-Trevino, S., Bocanegra-Ibarias, P., Camacho-Ortiz, A., Morfín-Otero, R., Salazar-Sesatty, H. A., Garza-González, E. (2019). Stenotrophomonas maltophilia biofilm: its role in infectious diseases. Expert Rev. Anti Infect. Ther. 17, 877–893. doi: 10.1080/14787210.2019.1685875
Gajdacs, M., Urban, E. (2019). Prevalence and antibiotic resistance of stenotrophomonas maltophilia in respiratory tract samples: A 10-year epidemiological snapshot. Health Serv. Res. Manag Epidemiol. 6, 2333392819870774. doi: 10.1177/2333392819870774
Fujita, Y., Matsuoka, H., Hirooka, K. (2007). Regulation of fatty acid metabolism in bacteria. Mol. Microbiol. 66 (4), 829–839. doi: 10.1111/j.1365-2958.2007.05947.x
Gallagher, T., Phan, J., Oliver, A., Chase, A. B., England, W. E., Wandro, S., et al. (2019). Cystic Fibrosis-Associated Stenotrophomonas maltophilia Strain-Specific Adaptations and Responses to pH. J. Bacteriol 201 (7), jb.00478-18. doi: 10.1128/JB.00478-18
García, C. A., Alcaraz, E. S., Franco, M. A., Passerini de Rossi, B. N. (2015). Iron is a signal for Stenotrophomonas maltophilia biofilm formation, oxidative stress response, OMPs expression, and virulence. Front. Microbiol. 6, 926. doi: 10.3389/fmicb.2015.00926
Gardner, J. (2007). What you need to know about cystic fibrosis. Nursing2021 37, 52–55. doi: 10.1097/01.NURSE.0000279437.30155.1e
Goldberg, E., Bishara, J. (2012). Contemporary unconventional clinical use of co-trimoxazole. Clin. Microbiol. Infect. 18, 8–17. doi: 10.1111/j.1469-0691.2011.03613.x
Goss, C. H., Otto, K., Aitken, M. L., Rubenfeld, G. D. (2002). Detecting Stenotrophomonas maltophilia does not reduce survival of patients with cystic fibrosis. Am. J. Respir. Crit. Care Med. 166, 356–361. doi: 10.1164/rccm.2109078
Gould, V. C., Okazaki, A., Avison, M. B. (2013). Coordinate hyperproduction of SmeZ and SmeJK efflux pumps extends drug resistance in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 57, 655–657. doi: 10.1128/AAC.01020-12
Granchelli, A. M., Adler, F. R., Keogh, R. H., Kartsonaki, C., Cox, D. R., Liou, T. G. (2018). Microbial interactions in the cystic fibrosis airway. J. Clin. Microbiol. 56 (8), jcm.00354-18. doi: 10.1128/JCM.00354-18
Guo, J., Garratt, A., Hill, A. (2022). Worldwide rates of diagnosis and effective treatment for cystic fibrosis. J. Cyst Fibros 21, 456–462. doi: 10.1016/j.jcf.2022.01.009
Hagemann, M., Hasse, D., Berg, G. (2006). Detection of a phage genome carrying a zonula occludens like toxin gene (zot) in clinical isolates of Stenotrophomonas maltophilia. Arch. Microbiol. 185, 449–458. doi: 10.1007/s00203-006-0115-7
Hall-Stoodley, L., Stoodley, P., Kathju, S., Høiby, N., Moser, C., Costerton, J. W., et al. (2012). Towards diagnostic guidelines for biofilm-associated infections. FEMS Immunol. Med. Microbiol. 65, 127–145. doi: 10.1111/j.1574-695X.2012.00968.x
Hansen, C. R. (2012). Stenotrophomonas maltophilia: to be or not to be a cystic fibrosis pathogen. Curr. Opin. Pulm Med. 18, 628–631. doi: 10.1097/MCP.0b013e328358d4f8
Haydon, D. J., Guest, J. R. (1991). A new family of bacterial regulatory proteins. FEMS Microbiol. Lett. 63 (2-3), 291–295. doi: 10.1016/0378-1097(91)90101-f
Heath, R. J., Rock, C. O. (1996). Roles of the FabA and FabZ beta-hydroxyacyl-acyl carrier protein dehydratases in Escherichia coli fatty acid biosynthesis. J. Biol. Chem. 271 (44), 27795–27801. doi: 10.1074/jbc.271.44.27795
Hernández-Jiménez, E., Del Campo, R., Toledano, V., Vallejo-Cremades, M. T., Muñoz, A., Largo, C., et al. (2013). Biofilm vs. planktonic bacterial mode of growth: which do human macrophages prefer? Biochem. Biophys. Res. Commun. 441, 947–952. doi: 10.1016/j.bbrc.2013.11.012
Hofmann, P., Hombach, M., Seifert, B., Schuurmans, M. M., Bürgi, U., Isenring, B., et al. (2016). Isolation of Stenotrophomonas maltophilia in asymptomatic lung transplant recipients: effects of treatment on eradication and outcome. Clin. Transplant. 30, 857–863. doi: 10.1111/ctr.12770
Hoiby, N., Bjarnsholt, T., Givskov, M., Molin, S., Ciofu, O. (2010). Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 35, 322–332. doi: 10.1016/j.ijantimicag.2009.12.011
Holden, M. T., Seth-Smith, H. M., Crossman, L. C., Sebaihia, M., Bentley, S. D., Cerdeño-Tárraga, A. M., et al. (2009). The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J. Bacteriol 191, 261–277. doi: 10.1128/JB.01230-08
Hu, L.-F., Chang, X., Ye, Y., Wang, Z. X., Shao, Y. B., Shi, W., et al. (2011). Stenotrophomonas maltophilia resistance to trimethoprim/sulfamethoxazole mediated by acquisition of sul and dfrA genes in a plasmid-mediated class 1 integron. Int. J. antimicrobial Agents 37, 230–234. doi: 10.1016/j.ijantimicag.2010.10.025
Hu, L. F., Chen, G. S., Kong, Q. X., Gao, L. P., Chen, X., Ye, Y., et al. (2016). Increase in the prevalence of resistance determinants to trimethoprim/sulfamethoxazole in clinical stenotrophomonas maltophilia isolates in China. PloS One 11, e0157693. doi: 10.1371/journal.pone.0157693
Hu, R. M., Liao, S. T., Huang, C. C., Huang, Y. W., Yang, T. C. (2012). An inducible fusaric acid tripartite efflux pump contributes to the fusaric acid resistance in Stenotrophomonas maltophilia. PloS One 7, e51053. doi: 10.1371/journal.pone.0051053
Huang, T. P., Somers, E. B., Wong, A. C. (2006). Differential biofilm formation and motility associated with lipopolysaccharide/exopolysaccharide-coupled biosynthetic genes in Stenotrophomonas maltophilia. J. Bacteriol 188, 3116–3120. doi: 10.1128/JB.188.8.3116-3120.2006
Huang, Y. W., Hu, R. M., Chu, F. Y., Lin, H. R., Yang, T. C. (2013). Characterization of a major facilitator superfamily (MFS) tripartite efflux pump EmrCABsm from Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 68, 2498–2505. doi: 10.1093/jac/dkt250
Ibn Saied, W., Merceron, S., Schwebel, C., Le Monnier, A., Oziel, J., Garrouste-Orgeas, M., et al. (2020). Ventilator-associated pneumonia due to Stenotrophomonas maltophilia: risk factors and outcome. J. Infection 80, 279–285. doi: 10.1016/j.jinf.2019.10.021
Isom, C. M., Fort, B., Anderson, G. G. (2022). Evaluating metabolic pathways and biofilm formation in stenotrophomonas maltophilia. J. Bacteriol 204, e0039821. doi: 10.1128/JB.00398-21
Jair, H. W., Lu, H. F., Huang, Y. W., Pan, S. Y., Lin, I. L., Huang, H. H., et al. (2019). Roles of the two-mnSOD system of stenotrophomonas maltophilia in the alleviation of superoxide stress. Int. J. Mol. Sci. 20 (7), ijms20071770. doi: 10.3390/ijms20071770
Jian, J., Xie, Z., Chen, L. (2022). Risk factors for mortality in hospitalized patients with stenotrophomonas maltophilia bacteremia. Infect. Drug Resist. 15, 3881–3886. doi: 10.2147/IDR.S371129
Kalidasan, V., Azman, A., Joseph, N., Kumar, S., Awang Hamat, R., et al. (2018a). Putative iron acquisition systems in stenotrophomonas maltophilia. Molecules 23 (8), molecules23082048. doi: 10.3390/molecules23082048
Kalidasan, V., Joseph, N., Kumar, S., Awang Hamat, R., Neela, V. K. (2018b). Iron and virulence in stenotrophomonas maltophilia: all we know so far. Front. Cell Infect. Microbiol. 8 (401), fcimb.2018.00401. doi: 10.3389/fcimb.2018.00401
Kalidasan, V., Neela, V. K. (2020). Twitching motility of Stenotrophomonas maltophilia under iron limitation: In-silico, phenotypic and proteomic approaches. Virulence 11, 104–112. doi: 10.1080/21505594.2020.1713649
Kanderi, T., Shrimanker, I., Mansoora, Q., Shah, K., Yumen, A., Komanduri, S. (2020). Stenotrophomonas maltophilia: an emerging pathogen of the respiratory tract. Am. J. Case Rep. 21, e921466. doi: 10.12659/AJCR.921466
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M., Ishiguro-Watanabe, M. (2023). KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51 (D1), D587–D592. doi: 10.1093/nar/gkac963
Kang, X.-M., Wang, F.-F., Zhang, H., Zhang, Q., Qian, W. (2015). Genome-wide identification of genes necessary for biofilm formation by nosocomial pathogen Stenotrophomonas maltophilia reveals that orphan response regulator FsnR is a critical modulator. Appl. Environ. Microbiol. 81, 1200–1209. doi: 10.1128/AEM.03408-14
Karaba, S. M., White, R. C., Cianciotto, N. P. (2013). Stenotrophomonas maltophilia encodes a type II protein secretion system that promotes detrimental effects on lung epithelial cells. Infect. Immun. 81, 3210–3219. doi: 10.1128/IAI.00546-13
Kim, Y. J., Jeon, H., Na, S. H., Kwon, H. I., Selasi, G. N., Nicholas, A., et al. (2016). Stenotrophomonas maltophilia outer membrane vesicles elicit a potent inflammatory response in vitro and in vivo. Pathog. Dis. 74 (8), ftw104. doi: 10.1093/femspd/ftw104
Lafferty, K. D. (2009). The ecology of climate change and infectious diseases. Ecology 90, 888–900. doi: 10.1890/08-0079.1
Li, L.-H., Shih, Y. L., Huang, J. Y., Wu, C. J., Huang, Y. W., Huang, H. H., et al. (2020). Protection from hydrogen peroxide stress relies mainly on AhpCF and KatA2 in Stenotrophomonas maltophilia. J. Biomed. Sci. 27, 1–9. doi: 10.1186/s12929-020-00631-4
Li, X. Z., Zhang, L., Poole, K. (2002). SmeC, an outer membrane multidrug efflux protein of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 46, 333–343. doi: 10.1128/AAC.46.2.333-343.2002
Liao, C.-H., Chang, C. L., Huang, H. H., Lin, Y. T., Li, L.H., Yang, T. C. (2021). Interplay between OmpA and RpoN regulates flagellar synthesis in Stenotrophomonas maltophilia. Microorganisms 9, 1216. doi: 10.3390/microorganisms9061216
Lin, Y. T., Huang, Y. W., Chen, S. J., Chang, C. W., Yang, T. C. (2015). The SmeYZ efflux pump of Stenotrophomonas maltophilia contributes to drug resistance, virulence-related characteristics, and virulence in mice. Antimicrob. Agents Chemother. 59, 4067–4073. doi: 10.1128/AAC.00372-15
Liu, W., Tian, X. Q., Wei, J. W., Ding, L. L., Qian, W., Liu, Z., et al. (2017). BsmR degrades c-di-GMP to modulate biofilm formation of nosocomial pathogen Stenotrophomonas maltophilia. Sci. Rep. 7, 4665. doi: 10.1038/s41598-017-04763-w
Looney, W. J., Narita, M., Muhlemann, K. (2009). Stenotrophomonas maltophilia: an emerging opportunist human pathogen. Lancet Infect. Dis. 9, 312–323. doi: 10.1016/S1473-3099(09)70083-0
Mah, T. F., O’Toole, G. A. (2001). Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9, 34–39. doi: 10.1016/s0966-842x(00)01913-2
Martinez, J. L. (2008). Antibiotics and antibiotic resistance genes in natural environments. Science 321, 365–367. doi: 10.1126/science.1159483
Matera, G., Barreca, G. S., Puccio, R., Quirino, A., Liberto, M. C., De Rosa, M., et al. (2004). Stenotrophomonas maltophilia lipopolysaccharide (LPS) and antibiotics:” in vitro” effects on inflammatory mediators. Le Infezioni Medicina 12, 227–238.
McCutcheon, J. G., Dennis, J. J. (2021). The Potential of Phage Therapy against the Emerging Opportunistic Pathogen Stenotrophomonas maltophilia. Viruses 13 (6), v13061057. doi: 10.3390/v13061057
McCutcheon, J. G., Lin, A., Dennis, J. J. (2020). Isolation and Characterization of the Novel Bacteriophage AXL3 against Stenotrophomonas maltophilia. Int. J. Mol. Sci. 21 (17), ijms21176338. doi: 10.3390/ijms21176338
McCutcheon, J. G., Peters, D. L., Dennis, J. J. (2018). Identification and characterization of type IV pili as the cellular receptor of broad host range Stenotrophomonas maltophilia bacteriophages DLP1 and DLP2. Viruses 10, 338. doi: 10.3390/v10060338
McKay, G. A., Woods, D. E., MacDonald, K. L., Poole, K. (2003). Role of phosphoglucomutase of Stenotrophomonas maltophilia in lipopolysaccharide biosynthesis, virulence, and antibiotic resistance. Infection Immun. 71, 3068–3075. doi: 10.1128/IAI.71.6.3068-3075.2003
Muder, R. R. (2007). Optimizing therapy for Stenotrophomonas maltophilia. Semin. Respir. Crit. Care Med. 28, 672–677. doi: 10.1055/s-2007-996414
Muder, R. R., Harris, A. P., Muller, S., Edmond, M., Chow, J. W., Papadakis, K., et al. (1996). Bacteremia due to Stenotrophomonas (Xanthomonas) maltophilia: a prospective, multicenter study of 91 episodes. Clin. Infect. Dis. 22, 508–512. doi: 10.1093/clinids/22.3.508
Munoz-Elias, E. J., McKinney, J. D. (2006). Carbon metabolism of intracellular bacteria. Cell Microbiol. 8, 10–22. doi: 10.1111/j.1462-5822.2005.00648.x
Nas, M. Y., Cianciotto, N. P. (2017). Stenotrophomonas maltophilia produces an EntC-dependent catecholate siderophore that is distinct from enterobactin. Microbiology 163, 1590. doi: 10.1099/mic.0.000545
Nas, M. Y., Gabell, J., Cianciotto, N. P. (2021). Effectors of the Stenotrophomonas maltophilia Type IV Secretion System Mediate Killing of Clinical Isolates of Pseudomonas aeruginosa. mBio 12, e0150221. doi: 10.1128/mBio.01502-21
Nas, M. Y., White, R. C., DuMont, A. L., Lopez, A. E., Cianciotto, N. P. (2019). Stenotrophomonas maltophilia encodes a VirB/VirD4 type IV secretion system that modulates apoptosis in human cells and promotes competition against heterologous bacteria, including Pseudomonas aeruginosa. Infection Immun. 87 (9), iai.00457-19. doi: 10.1128/iai.00457-00419
Nicodemo, A. C., Paez, J. I. (2007). Antimicrobial therapy for Stenotrophomonas maltophilia infections. Eur. J. Clin. Microbiol. Infect. Dis. 26, 229–237. doi: 10.1007/s10096-007-0279-3
Nseir, S., Di Pompeo, C., Brisson, H., Dewavrin, F., Tissier, S., Diarra, M., et al. (2006a). Intensive care unit-acquired Stenotrophomonas maltophilia: incidence, risk factors, and outcome. Crit. Care 10, R143. doi: 10.1186/cc5063
Nseir, S., Di Pompeo, C., Cavestri, B., Jozefowicz, E., Nyunga, M., Soubrier, S., et al. (2006b). Multiple-drug-resistant bacteria in patients with severe acute exacerbation of chronic obstructive pulmonary disease: Prevalence, risk factors, and outcome. Crit. Care Med. 34, 2959–2966. doi: 10.1097/01.CCM.0000245666.28867.C6
Omenn, G. S. (2010). Evolution and public health. Proc. Natl. Acad. Sci. 107, 1702–1709. doi: 10.1073/pnas.0906198106
Pages, D., Rose, J., Conrod, S., Cuine, S., Carrier, P., Heulin, T., et al. (2008). Heavy metal tolerance in Stenotrophomonas maltophilia. PloS One 3, e1539. doi: 10.1371/journal.pone.0001539
Paton, R., Miles, R. S., Amyes, S. G. (1994). Biochemical properties of inducible beta-lactamases produced from Xanthomonas maltophilia. Antimicrob. Agents Chemother. 38, 2143–2149. doi: 10.1128/AAC.38.9.2143
Pompilio, A., Pomponio, S., Crocetta, V., Gherardi, G., Verginelli, F., Fiscarelli, E., et al. (2011). Phenotypic and genotypic characterization of Stenotrophomonas maltophilia isolates from patients with cystic fibrosis: genome diversity, biofilm formation, and virulence. BMC Microbiol. 11, 159. doi: 10.1186/1471-2180-11-159
Pompilio, A., Ranalli, M., Piccirilli, A., Perilli, M., Vukovic, D., Savic, B., et al. (2020). Biofilm formation among Stenotrophomonas maltophilia isolates has clinical relevance: the ANSELM prospective multicenter study. Microorganisms 9, 49. doi: 10.3390/microorganisms9010049
Ramos-Hegazy, L., Chakravarty, S., Anderson, G. G. (2020). Phosphoglycerate mutase affects Stenotrophomonas maltophilia attachment to biotic and abiotic surfaces. Microbes Infect. 22, 60–64. doi: 10.1016/j.micinf.2019.08.001
Rocco, F., De Gregorio, E., Colonna, B., Di Nocera, P. P. (2009). Stenotrophomonas maltophilia genomes: a start-up comparison. Int. J. Med. Microbiol. 299, 535–546. doi: 10.1016/j.ijmm.2009.05.004
Rock, C. O., Cronan, J. E. (1996). Escherichia coli as a model for the regulation of dissociable (type II) fatty acid biosynthesis. Biochim. Biophys. Acta. 1302 (1), 1–16. doi: 10.1016/0005-2760(96)00056-2
Roscetto, E., Vitiello, L., Muoio, R., Soriano, A. A., Iula, V. D., Vollaro, A., et al. (2015). In vitro interaction of Stenotrophomonas maltophilia with human monocyte-derived dendritic cells. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00723
Ryan, R. P., An, S. Q., Allan, J. H., McCarthy, Y., Dow, J. M. (2015). The DSF family of cell-cell signals: an expanding class of bacterial virulence regulators. PloS Pathog. 11, e1004986. doi: 10.1371/journal.ppat.1004986
Ryan, R. P., Monchy, S., Cardinale, M., Taghavi, S., Crossman, L., Avison, M. B., et al. (2009). The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat. Rev. Microbiol. 7, 514–525. doi: 10.1038/nrmicro2163
Saino, Y., Inoue, M., Mitsuhashi, S. (1984). Purification and properties of an inducible cephalosporinase from Pseudomonas maltophilia GN12873. Antimicrob. Agents Chemother. 25, 362–365. doi: 10.1128/AAC.25.3.362
Saino, Y., Kobayashi, F., Inoue, M., Mitsuhashi, S. (1982). Purification and properties of inducible penicillin beta-lactamase isolated from Pseudomonas maltophilia. Antimicrob. Agents Chemother. 22, 564–570. doi: 10.1128/AAC.22.4.564
Senol, E., DesJardin, J., Stark, P. C., Barefoot, L., Snydman, D. R. (2002). Attributable mortality of Stenotrophomonas maltophilia bacteremia. Clin. Infect. Dis. 34, 1653–1656. doi: 10.1086/340707
Shuman, E. K. (2010). Global climate change and infectious diseases. N Engl. J. Med. 362, 1061–1063. doi: 10.1056/NEJMp0912931
Soumya, J., Kabbin, J. S., Ambica, R. (2020). Stenotrophomonas maltophilia: An emerging pathogen in sepsis. MRIMS J. Health Sci. 8, 64. doi: 10.4103/mjhs.mjhs_15_20
Stanier, R. Y., Palleroni, N. J., Doudoroff, M. (1966). The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol. 43, 159–271. doi: 10.1099/00221287-43-2-159
Sun, E., Liang, G., Wang, L., Wei, W., Lei, M., Song, S., et al. (2016). Antimicrobial susceptibility of hospital acquired Stenotrophomonas maltophilia isolate biofilms. Braz. J. Infect. Dis. 20, 365–373. doi: 10.1016/j.bjid.2016.04.002
Suvorova, I. A., Rodionov, D. A. (2016). Comparative genomics of pyridoxal 5’-phosphate-dependent transcription factor regulons in Bacteria. Microb. Genom 2, e000047. doi: 10.1099/mgen.0.000047
Talmaciu, I., Varlotta, L., Mortensen, J., Schidlow, D. V. (2000). Risk factors for emergence of Stenotrophomonas maltophilia in cystic fibrosis. Pediatr. pulmonology 30, 10–15. doi: 10.1002/1099-0496(200007)30:1<10::AID-PPUL3>3.0.CO;2-Q
Textor, S., Wendisch, V. F., De Graaf, A. A., Müller, U., Linder, M. I., Linder, D., et al. (1997). Propionate oxidation in Escherichia coli: evidence for operation of a methylcitrate cycle in bacteria. Arch. Microbiol. 168 (5), 428–436. doi: 10.1007/s002030050518
Thomas, R., Hamat, R. A., Neela, V. (2014). Extracellular enzyme profiling of Stenotrophomonas maltophilia clinical isolates. Virulence 5, 326–330. doi: 10.4161/viru.27724
Tiyawisutsri, R., Holden, M. T., Tumapa, S., Rengpipat, S., Clarke, S. R., Foster, S. J., et al. (2007). Burkholderia Hep_Hag autotransporter (BuHA) proteins elicit a strong antibody response during experimental glanders but not human melioidosis. BMC Microbiol. 7, 19. doi: 10.1186/1471-2180-7-19
Toleman, M. A., Bennett, P. M., Bennett, D. M., Jones, R. N., Walsh, T. R. (2007). Global emergence of trimethoprim/sulfamethoxazole resistance in Stenotrophomonas maltophilia mediated by acquisition of sul genes. Emerg. Infect. Dis. 13, 559–565. doi: 10.3201/eid1304.061378
Trifonova, A., Strateva, T. (2019a). Stenotrophomonas maltophilia - a low-grade pathogen with numerous virulence factors. Infect. Dis. (Lond) 51, 168–178. doi: 10.1080/23744235.2018.1531145
Trifonova, A., Strateva, T. (2019b). Stenotrophomonas maltophilia–a low-grade pathogen with numerous virulence factors. Infect. Dis. 51, 168–178. doi: 10.1080/23744235.2018.1531145
Valdezate, S., Vindel, A., Loza, E., Baquero, F., Canton, R. (2001). Antimicrobial susceptibilities of unique Stenotrophomonas maltophilia clinical strains. Antimicrob. Agents Chemother. 45, 1581–1584. doi: 10.1128/AAC.45.5.1581-1584.2001
Walsh, T. R., Hall, L., Assinder, S. J., Nichols, W. W., Cartwright, S. J., MacGowan, A. P., et al. (1994). Sequence analysis of the L1 metallo-beta-lactamase from Xanthomonas maltophilia. Biochim. Biophys. Acta 1218, 199–201. doi: 10.1016/0167-4781(94)90011-6
Walsh, T. R., MacGowan, A. P., Bennett, P. M. (1997). Sequence analysis and enzyme kinetics of the L2 serine beta-lactamase from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 41, 1460–1464. doi: 10.1128/AAC.41.7.1460
Wang, C. H., Lin, J. C., Chang, F. Y., Yu, C. M., Lin, W. S., Yeh, K. M., et al. (2017). Risk factors for hospital acquisition of trimethoprim-sulfamethoxazole resistant Stenotrophomonas maltophilia in adults: A matched case-control study. J. Microbiol. Immunol. Infect. 50, 646–652. doi: 10.1016/j.jmii.2016.12.007
Wang, H., Cronan, J. E. (2004). Functional replacement of the FabA and FabB proteins of Escherichia coli fatty acid synthesis by Enterococcus faecalis FabZ and FabF homologues. J. Biol. Chem. 279 (33), 34489–95. doi: 10.1074/jbc.M403874200
Waters, V., Yau, Y., Prasad, S., Lu, A., Atenafu, E., Crandall, I., et al. (2011). Stenotrophomonas maltophilia in cystic fibrosis: serologic response and effect on lung disease. Am. J. Respir. Crit. Care Med. 183, 635–640. doi: 10.1164/rccm.201009-1392OC
Waters, V., Atenafu, E. G., Lu, A., Yau, Y., Tullis, E., Ratjen, F. (2013). Chronic Stenotrophomonas maltophilia infection and mortality or lung transplantation in cystic fibrosis patients. J. Cystic Fibrosis 12, 482–486. doi: 10.1016/j.jcf.2012.12.006
Winn, A. M., Galbraith, L., Temple, G. S., Wilkinson, S. G. (1993). Structure of the O19 antigen of Xanthomonas maltophilia. Carbohydr. Res. 247, 249–254. doi: 10.1016/0008-6215(93)84257-7
Yasir, M., Willcox, M. D. P., Dutta, D. (2018). Action of antimicrobial peptides against bacterial biofilms. Materials (Basel) 11 (12), ma11122468. doi: 10.3390/ma11122468
Yero, D., Huedo, P., Conchillo-Solé, O., Martínez-Servat, S., Mamat, U., Coves, X., et al. (2020). Genetic variants of the DSF quorum sensing system in Stenotrophomonas maltophilia influence virulence and resistance phenotypes among genotypically diverse clinical isolates. Front. Microbiol. 11, 1160. doi: 10.3389/fmicb.2020.01160
Yin, W., Wang, Y., Liu, L., He, J. (2019). Biofilms: the microbial “protective clothing” in extreme environments. Int. J. Mol. Sci. 20, 3423. doi: 10.3390/ijms20143423
Yu, W., Chen, X., Sheng, Y., Hong, Q. (2018). Genomic analysis for heavy metal resistance in S. maltophilia. BioRxiv, 404954. doi: 10.1101/404954
Zgair, A. K., Al-Adressi, A. M. (2013). Stenotrophomonas maltophilia fimbrin stimulates mouse bladder innate immune response. Eur. J. Clin. Microbiol. Infect. Dis. 32, 139–146. doi: 10.1007/s10096-012-1729-0
Zgair, A. K., Chhibber, S. (2011). Adhesion of Stenotrophomonas maltophilia to mouse tracheal mucus is mediated through flagella. J. Med. Microbiol. 60, 1032–1037. doi: 10.1099/jmm.0.026377-0
Keywords: Stenotrophomonas maltophilia, antibiotic, biofilm, virulence, chronic infection, opportunistic
Citation: Bhaumik R, Aungkur NZ and Anderson GG (2024) A guide to Stenotrophomonas maltophilia virulence capabilities, as we currently understand them. Front. Cell. Infect. Microbiol. 13:1322853. doi: 10.3389/fcimb.2023.1322853
Received: 16 October 2023; Accepted: 20 December 2023;
Published: 11 January 2024.
Edited by:
Manuel L. Lemos, University of Santiago de Compostela, SpainReviewed by:
Nicholas Cianciotto, Northwestern University, United StatesInês Bezerra Gomes, University of Porto, Portugal
Copyright © 2024 Bhaumik, Aungkur and Anderson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gregory G. Anderson, Z2EyQGl1cHVpLmVkdQ==
 Radhika Bhaumik
Radhika Bhaumik Nabiha Zumana Aungkur
Nabiha Zumana Aungkur Gregory G. Anderson
Gregory G. Anderson