
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol., 24 October 2023
Sec. Molecular Bacterial Pathogenesis
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1282258
This article is part of the Research TopicSmall non-coding RNAs in Gram negative bacteriaView all 5 articles
Hfq is required by many Gram-negative bacteria to chaperone the interaction between small non-coding RNA (sRNA) and mRNA to facilitate annealing. Conversely and despite the presence of Hfq in many Gram-positive bacteria, sRNAs in Gram-positive bacteria bind the mRNA target independent of Hfq. Details provided by the Hfq structures from both Gram-negative and Gram-positive bacteria have demonstrated that despite a conserved global structure of the protein, variations of residues on the binding surfaces of Hfq results in the recognition of different RNA sequences as well as the ability of Hfq to facilitate the annealing of the sRNA to the mRNA target. Additionally, a subset of Gram-negative bacteria has an extended C-terminal Domain (CTD) that has been shown to affect the stability of the Hfq hexamer and increase the rate of release of the annealed sRNA-mRNA product. Here we review the structures of Hfq and biochemical data that have defined the interactions of the Gram-negative and Gram-positive homologues to highlight the similarities and differences in the interactions with RNA. These interactions provided a deeper understanding of the how Hfq functions to facilitate the annealing of sRNA-mRNA, the selectivity of the interactions with RNA, and the role of the CTD of Hfq in the interactions with sRNA.
Small non-coding RNAs (sRNA) have been shown to regulate a variety of different responses in bacteria by acting as a posttranscriptional regulator of translation and are often associated with adaptive stress response, including antibiotic resistance (Repoila and Darfeuille, 2009; Waters and Storz, 2009; Chakravarty and Masse, 2019; Watkins and Arya, 2019; Diallo and Provost, 2020; Avican et al., 2021; Ponath et al., 2022). The most common mechanism of mRNA regulation by sRNA is accomplished by Watson-Crick base pairing of the sRNA to the ribosomal binding site of the mRNA resulting in the silencing of the mRNA (Thisted et al., 1994; Franch et al., 1999; Brunel et al., 2002). While the silencing of the mRNA by sRNAs occurs in both Gram-negative and Gram-positive bacteria, current data suggests that there is greater use of the chaperone protein, Hfq, by Gram-negative bacteria to facilitate the interaction between the sRNA and mRNA compared to Gram-positive bacteria (Jousselin et al., 2009; Nielsen et al., 2010; Santiago-Frangos et al., 2016).
Hfq was first identified in Escherichia coli (Ec) as an essential Host factor for the replication of bacteriophage Qβ RNA (Franze de Fernandez et al., 1968; Franze de Fernandez et al., 1972), and is found in ~50% of all bacteria (Sun et al., 2002). Hfq is a pleiotropic protein and has been shown to interact with multiple types of nucleic acids including both RNA and DNA as well as various proteins (Brennan and Link, 2007; Orans et al., 2020; Cossa et al., 2022; Dendooven et al., 2023). One function of Hfq is in wild-type regulation of FtsZ abundance and cell division (Takada99).
The interactions of Hfq with RNA typically acts to suppress protein expression, but has also been shown in some instances to enhance the expression of the mRNA (Repoila and Darfeuille, 2009). Hfq has been shown to bind sRNA and mRNA independently of each other and can act to protect the mRNAs or signal the degradation of mRNA even in the absence of sRNA (Mohanty et al., 2004). However, the most common function associated with Hfq is the chaperoning of the interaction between sRNA and mRNA. This function of Hfq appears to be restricted to the homologues in Gram-negative bacteria. While the need for Hfq facilitation has been linked to the GC content of the RNA, the length of the “seed region” of the RNA, and the difference in cis-encoded and trans-encoded sRNAs (Jousselin et al., 2009), the ability of the Hfq to facilitate the interactions between mRNA and sRNA is associated with the differences in the structures of the Hfq homologues as described below.
The global structure of Hfq is a hexameric protein made up of six monomers of the Sm/Lsm family. Each subunit consists of 5 β-sheets and 1 N-terminal α−helix. All current structures of Hfq show that the global structure of the hexamer is conserved among both Gram-negative and Gram-positive bacteria and produces a “donut” shaped structure with two sides that each contain an independent RNA binding site (Schumacher et al., 2002; Sauter et al., 2003; Nikulin et al., 2005; Nielsen et al., 2007; Boggild et al., 2009; Someya et al., 2012; Kovach et al., 2014). The convex side of Hfq, with the solvent exposed α-helices of each monomer, is defined as the proximal side, and the distal side is defined as the convex surface in which most of the β-sheets solvent exposure occurs (Figure 1). A third binding site, referred to as the lateral rim, has also been identified on outer lateral surface of the Hfq core furthest from the central pore, and is made up by residues of the C-terminus of the α-helix (Robinson et al., 2014). The three binding sites allow Hfq to bind two different strands of RNA simultaneously which is fundamental to the ability of Hfq to act as a chaperone of the annealing of sRNA and mRNA (Park et al., 2021). Additionally, Hfq has a C-terminal Domain (CTD) following the Lsm core that is highly variable in length and sequence. The CTD appears to be highly flexible and typically no residues of the CTD are observed in the structures of Hfq. In fact, the CTD of Hfq was only partially observed in three of the structures of the protein (Beich-Frandsen et al., 2011; Dimastrogiovanni et al., 2014; Santiago-Frangos et al., 2019). The CTD likely has multiple functions in Hfq and has been shown to be involved in the formation of amyloids involved in the packing of DNA as well as the interactions with the cell membrane (Cossa et al., 2022; Turbant et al., 2022). However, the CTD appears to play an important, species-specific role in the chaperone activity of Hfq as described below.
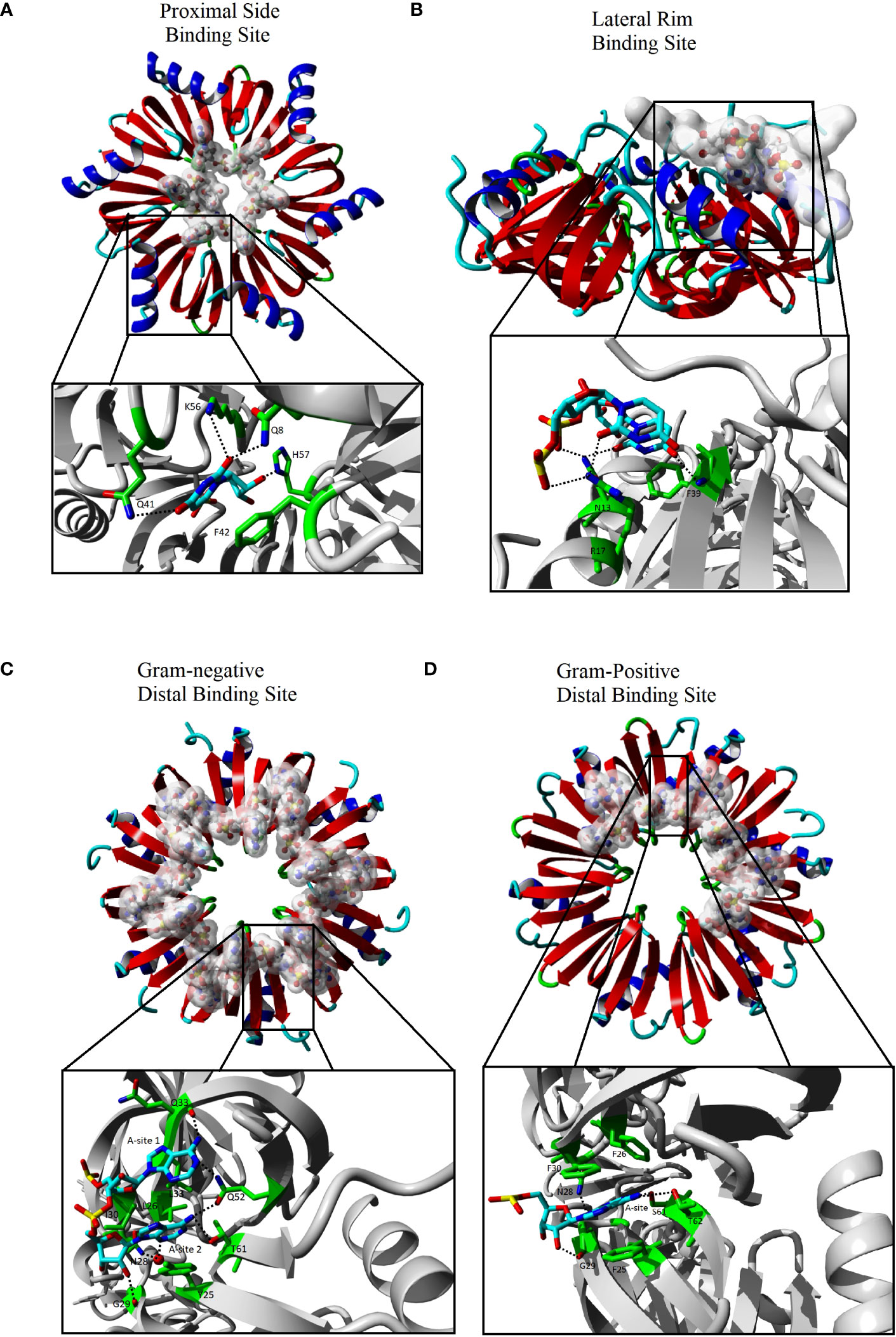
Figure 1 The interactions of Hfq with RNA. The Hfq core domain is composed of six monomers represented as red and blue ribbons. Each binding site recognizes a different sequence of RNA that is represented as sticks that are highlighted with a white cloud in the far view (C-F upper). The expanded view (C-F lower black box) shows the Hfq residues (green sticks) involved in base specific interactions with the RNA (cyan sticks). The expanded views have been rotated relative to global view for clarity. Images generated using Yasara (Krieger and Vriend, 2014; Krieger and Vriend, 2015) using Hfq species and PDBs of (A) Sa Hfq PDBID: 2ylc (B) Ec Hfq PDBID: 4v2s (C) Ec Hfq PDBID: 3gib (D) Bs Hfq PDBID: 3hsb. Some RNA residues were removed from the original models for clarity.
The proximal side of Hfq is highly conserved between homologues of Hfq and has been shown to bind single strand RNA close to the central pore of Hfq on this side (Figure 1A) (Schumacher et al., 2002; Brennan and Link, 2007; Sauer and Weichenrieder, 2011; Wang et al., 2013; Robinson et al., 2014). All structures with RNA bound to the proximal side have the single strand RNA bound to the positive electrostatic surface of Hfq in a circular pattern about the pore with one nucleotide bound to each monomer of Hfq (Figures 2A, B). The binding site on the proximal face accommodates uridine or adenine bases with a preference for U-rich sequences with a free 3’OH common in many sRNAs (Sauer and Weichenrieder, 2011). The base bound in the proximal side binding site stacks with a conserved aromatic residue (Ec, Salmonella typhimurium (Sa) and Bacillus subtilis (Bs) residue F42, Staphylococcus aureus (St) Y42). Hfq residues make a base specific contact through a conserved Q8 that recognizes an O2 of uridine or O6 of adenine, a conserved K56 residue and an amine group at position 41 (Ec and Sa Q41, Bs K41 and St N41). A conserved H57 (Ec and Sa numbering) interaction with O2’ of the ribose likely ensures a preference for RNA over DNA binding. The proximal side binding site appears to be a general binding site of all Hfq homologues and binds to single strand A/U rich RNA sequences and may participate in the rearrangement of secondary structures of the RNA (Sauer et al., 2012).
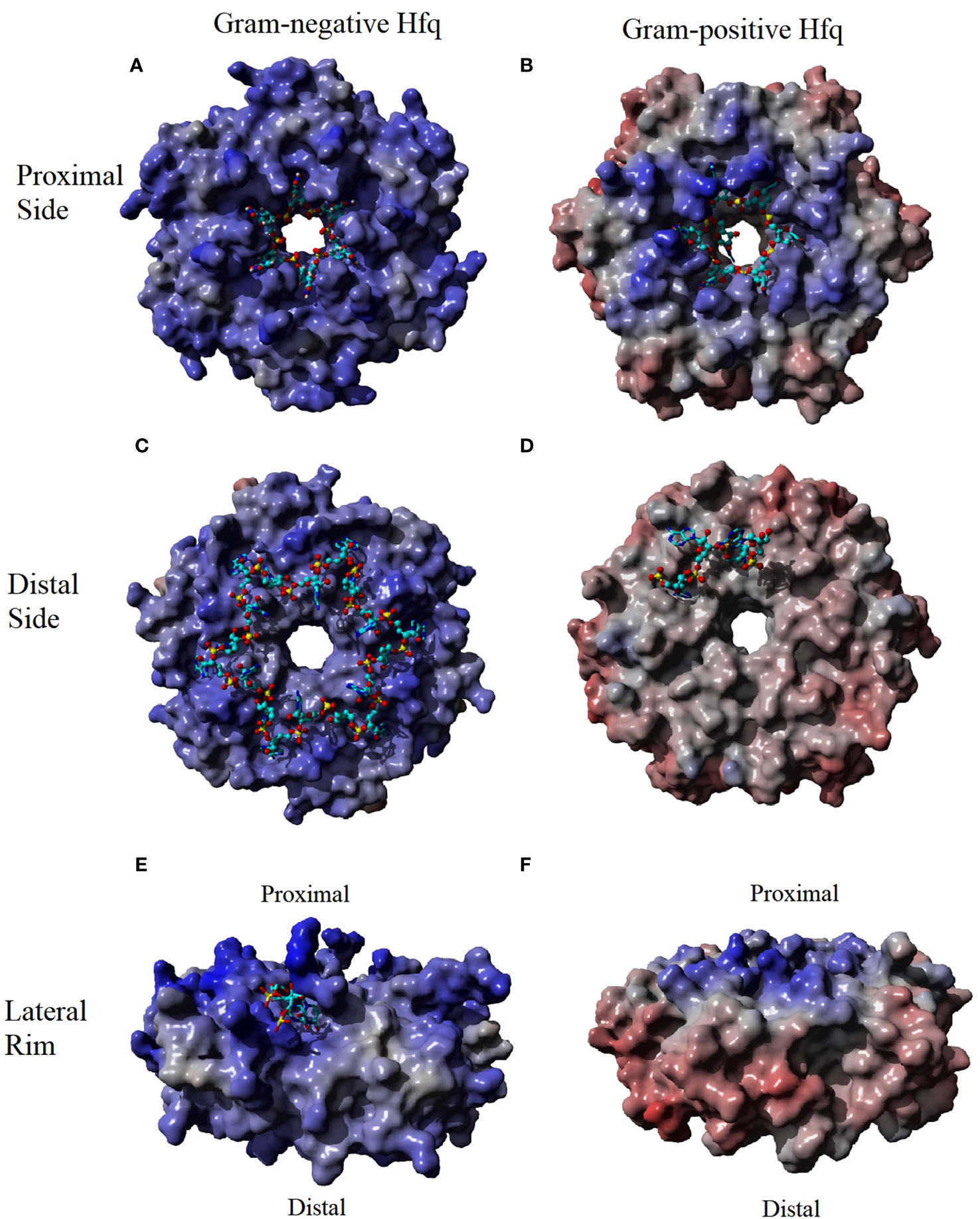
Figure 2 The Electrostatic Surface Potential (ESP) of Hfq-RNA binding surfaces. The binding surfaces of Hfq in Gram-negative bacteria have a greater positive potential (indicated as a blue surface) than the Gram-positive homologue. The RNA (cyan sticks) interacts with the positive potential of these surface. The greatest difference in the ESP occurs at the rim (compare E, F) where the potential is positive in Gram-negative and negative in Gram-positive (red surface). ESP were generated using Yasara (Krieger and Vriend, 2014; Krieger and Vriend, 2015) using an Amber96 forcefield (Kollman P et al., 1997) and a particle mesh Ewald simulation (Essman U et al., 1995) using Hfq species and PDBs of (A) Sa Hfq PDBID: 2ylc (B) Ec Hfq PDBID: 4v2s (C) Ec Hfq PDBID: 3gib (D) St Hfq PDBID: 3qsu. Some RNA residues were removed from the original models for clarity.
The various homologues of Hfq have only minor differences in the aligned primary sequences of the protein (Sonnleitner et al., 2002). However, the crystal structure of Hfq bound to RNA, along with biochemical data, have shown the changes in the sequence result in differences to distal side RNA binding sites between Gram-positive and Gram-negative bacteria (Link et al., 2009; Horstmann et al., 2012; Someya et al., 2012; Wang et al., 2013; Robinson et al., 2014). The distal binding site of Gram-negative and Gram-positive is typically responsible for recognizing the 5’-polyA sequence of mRNA, but the distal site has also been shown to act as a second binding site for some sRNAs in Gram-negative Hfq homologues as discussed below (Schu et al., 2015). The distal side of Gram-positive is less electrostatically positive compared to the Gram-negative homologue of Hfq (Figures 2C, D). In Gram-negative bacteria, the distal side contains six RNA binding sites with each site recognizing the sequence (A-A-N)n (Robinson et al., 2014). Interaction between the protein and the RNA involves two different subunits of Hfq with adenine specific contacts between the Q52 and the backbone of Q33 and a hydrophobic interaction with L32 in the same subunit of Hfq create the first A binding site of the (A-A-N)n motif of the RNA in Gram-negative homologues (Figure 1C). Residue Q52 bridges the first and second A-site making contacts to the adenine in both sites. Additional base specific contacts occur with T61 of the adjacent subunit of Hfq, and the water mediated N28 in the second A-site. Additional hydrophobic interactions are observed between the adenine in the second A-site with L26, I30 and Y25 of the adjacent subunit. The N-site has no base specificity, as no contacts between the base of the N-site and the protein have been observed. The N-site appears to bridge adjacent binding sites and may act as an entry point of the adenines (Link et al., 2009; Wang et al., 2013; Robinson et al., 2014).
Similar to Gram-negative bacteria, Gram-positive bacteria have six binding sites, with each binding site located at the interface of two subunits of Hfq. However, an inserted amino acid in the loop between β-strands 3 and 4 and a small number of seemingly conservative variations, particularly I30 to an aromatic (F30 in Bs, Y30 in St), in Gram-positive Hfq results in a slight shifting of the first A-site and a loss of the second A-site (Figure 1D) (Horstmann et al., 2012; Someya et al., 2012). These changes result in a distal recognition sequence of (A-L)n where the L indicates a linker between the each site and, similar to the N-site of Gram-negative Hfq, can be any nucleotide. The change in the distal binding site from an (A-A-N) to an (A-L) recognition not only alters the specificity of the sequence bound on the distal side, but also reduces the maximum number of bases bound from 18 nucleotides by Gram-negative Hfq to 12 by the Gram-positive Hfq.
The difference in the electrostatic surface potential of the lateral rim of Hfq between Gram-negative and Gram-positive homologues plays a significant role in the difference in the annealing function of Hfq homologues (Zheng et al., 2016). The surface of the rim of Gram-negative Hfq homologues is significantly more electropositive than those of Gram-positive (Figures 2E, F). Sequence alignment of the rim has shown that the Hfq of Gram-negative bacteria are enriched with arginine residues in this area compared to Gram-positive bacteria, and Ec residues R16, R17, and R19, referred to as the arginine patch, are highly conserved in Gram-negative bacteria, but less so in Gram-positive. The rate of RNA annealing increases as the number of arginine residues increases on the rim of Hfq and is independent to changes to the proximal and distal side binding sites (Sauer et al., 2012; Panja et al., 2013; Zhang et al., 2013; Cai et al., 2022). The structure of Hfq bound to a full length RhyC sRNA identified an additional RNA binding site that has specificity for single strand UU dinucleotides on the proximal face of the rim (Figure 1B) (Dimastrogiovanni et al., 2014). Additional contacts of the rim involve non-specific phosphate interactions in contact with the stem region of the RNA (Dimastrogiovanni et al., 2014). Combined with biochemical data the rim region has been implemented in the melting of the stem regions of RNA that contains the complementary seed sequences of the sRNA and the target mRNA (Panja et al., 2013). The RNA rearrangement exposes the single strand of each complementary sequence, allowing the annealing of the two strands and the formation of the sRNA-mRNA duplex (Sauer et al., 2012; Zheng et al., 2016). It has been proposed that the greater GC content of Gram-negative bacteria requires the positive arginine patch at the rim of Hfq to facilitate the rearrangement of the RNA, while the lower GC content of Gram-positive bacteria results in less stable stem regions that can rearrange independent of Hfq. This observation is supported by the lack of arginines in St homologues of Hfq and the Hfq independent annealing of sRNA to mRNA in St bacteria.
The CTD of Hfq varies greatly not just between the Gram-positive bacteria where it is largely absent or severely truncated, but also within different species of Gram-negative bacteria in which the length of the CTD varies greatly (Sonnleitner et al., 2002; Sun et al., 2002). While the length of the CTD varies, key characteristics of the CTD have been identified in most Gram-negative bacteria. The highly conserved P64 is considered the end of the core domain and defines the start of the CTD. Residue R66 (E. coli numbering) is also highly conserved but has been shown to pack against the lateral edge of the core (Santiago-Frangos et al., 2017). The middle of the CTD has the greatest variability in both length and sequence and appears to be species specific. The final six residues define the tail of the CTD and contain acidic residues with a consensus sequence of DSEETE (Arluison et al., 2004; Santiago-Frangos et al., 2017).
Two models of the Hfq facilitated sRNA-mRNA annealing in Gram-negative bacteria have been proposed based on the differences observed in the sRNA that is involved in the interactions with Hfq. Two different classes of sRNA, Class I and Class II, have been identified based on these differences in the interaction with Hfq and the recognition sequence of the sRNA (Schu et al., 2015). Both classes of sRNA interact with Hfq at two different binding surfaces. Class I of sRNA appears to be the most common and interacts with the proximal side of Hfq with the U-rich 3’ Rho-independent terminator of the sRNA. The second binding site of Class I sRNA involves the rim of Hfq at the UU binding motif. The mRNA targets of Class I sRNAs bind to the distal side binding site via an AAN motif present on the mRNA. The formation of the sRNA-Hfq-mRNA ternary complex results in the annealing of the sRNA with the mRNA seed sequence at the rim. Following the annealing the sRNA and mRNA are both degraded by RNase activity (Schu et al., 2015).
Class II sRNA binds with the same binding motif to the proximal side of Hfq as Class I but the second binding site of the sRNA is on the distal side of Hfq with an AAN motif, as opposed to the rim. The mRNA targets of Class II sRNAs are unable to bind to the distal side of Hfq even if they contain an AAN motif and must therefore bind to the rim of Hfq at the UU binding site. The formation of the sRNA-Hfq-mRNA complex again results in the annealing of the sRNA with the mRNA sequence. However, in the case of Class II sRNA, the mRNA is degraded, but the sRNA often remains bound to Hfq for multiple rounds of mRNA binding (Schu et al., 2015). As a consequence of the different modes of interactions between Class I and Class II sRNAs with Hfq, the concentration Class I sRNAs are diminished with each interaction with their target and do not accumulate in the cell. Therefore, Class I sRNAs are self-regulated. However, Class II sRNAs can remain in the cell unless “deactivated” by other cellular mechanisms. As a result, initial studies have indicated that Class I sRNAs act as response element while Class II sRNA act as silencers of cell function (Figueroa-Bossi et al., 2009; Moon and Gottesman, 2009; Overgaard et al., 2009; Thomason et al., 2012; Hoe et al., 2013).
An additional consequence of the difference in the interaction between Class I and Class II sRNAs is the role of the CTD of the Hfq in the regulation of the process. Early work into the role function of the CTD appeared conflicting, with some work indicating that the CTD played no role in the Hfq interaction with sRNA, while other work indicates that the CTD is required for the proper binding and annealing of sRNA to the mRNA target (Sonnleitner et al., 2002; Sonnleitner et al., 2004; Vecerek et al., 2008; Olsen et al., 2010; Updegrove and Wartell, 2011; Salim et al., 2012). While differences in experimental methods may have been partially responsible for the conflicts, later findings have shown that the conflicts likely result from the differences in the function of the CTD that depends on the type of sRNA bound to Hfq, and involves a competitive interaction between the CTD and the RNA with the positive arginine patch on the rim of Hfq (Salim et al., 2012; Santiago-Frangos et al., 2016; Santiago-Frangos et al., 2017; Sarni et al., 2022).
The facilitation of annealing of Hfq for Class I and Class II sRNA in Ec is initiated by the sRNA and mRNA binding to Hfq. Both RNAs potentially undergo a rearrangement of stem regions exposing the seed sequence at the lateral rim region of Hfq. The exposure of the single strand seed sequence of the sRNA and mRNA following the rearrangement of the stem regions allows the annealing of the sRNA-mRNA at the rim of Hfq (Hwang et al., 2011; Sauer et al., 2012; Malecka and Woodson, 2021; Cai et al., 2022). Class I sRNA binding, annealing and release appears to occur independently of the CTD. The binding and annealing of Class II sRNAs also occur independent of the CTD, but the release of the annealed sRNA-mRNA product requires or is enhanced by the CTD (Kavita et al., 2022). The double strand product has a lower affinity for the rim of Hfq and is released from Hfq as the sRNA-mRNA hybrid. Unlike Class I sRNA, however, the rate of release of Class II sRNA-mRNA relies on the presence of the acidic tail CTD to displace the RNA duplex (Santiago-Frangos et al., 2016; Santiago-Frangos et al., 2017).
The CTD also plays an important role in discriminating between Class I and Class II binding of Hfq. In the presence of the CTD, Class II sRNAs bind preferentially to Hfq over Class I. However, the removal of the CTD removes the preference for Class II sRNA (Santiago-Frangos et al., 2016). It has been shown that the CTD does not contact the RNA, does not stabilize the proximal or distal RNA-Hfq interaction, and does not speed up the annealing process. Instead, the acidic tail of Hfq competes with the RNA for the basic patch of the rim. Therefore, the binding of the Class I sRNAs would be in competition with the CTD for the basic patches of the rim while Class II sRNAs would not face competition for the binding sites on the distal face of Hfq, Thus the role of the CTD of Hfq sets up a hierarchy for sRNA binding by reducing the apparent affinity of Class I sRNAs and giving rise to preferential binding of Class II sRNAs (Schu et al., 2015; Santiago-Frangos et al., 2016; Santiago-Frangos et al., 2017; Kavita et al., 2022).
Recent studies examining the stability of the Hfq using mass spectroscopy combined with the observations described above has led to a more comprehensive model of the chaperone activity of Hfq (Sarni et al., 2022). The CTD stabilizes the association of the monomers of the hexameric toroid structure of Hfq through both intra and inter-monomer interactions between the acidic tail of the CTD and the basic patch of the rim (Sarni et al., 2022). The displacement of the CTD results in a less stable structure, and the disruption of one of the CTD interactions cascades to disrupt the network of interactions. The disruption of the interaction can be stabilized by the binding of the sRNA. However, only sRNAs that properly interact with Hfq are able to effectively stabilize the structure. Therefore, RNAs that form favorable contacts with Hfq offset the loss of stability of displacing the CTD contact with the core domain of Hfq. The coordination of the binding of the RNA and the CTD at the rim of Hfq allows the CTD to screen the RNA interactions and prevent non-specific binding of RNAs to Hfq. Additionally, because Hfq has six different interactions between the CTD and core (one per monomer) the RNA can progressively displace the CTDs from the core. The progressive type interactions would therefore allow Hfq to sample different regions of the RNA for contacts or allow the binding of different RNAs sequentially without effecting the binding of another RNA molecule (Sarni et al., 2022).
There are multiple differences in the binding surfaces of Hfq between Gram-positive bacteria compared to Gram-negative bacteria. The difference in the requirement of Hfq between the two classes of bacteria likely results in the origins of the sRNA in the different bacteria. The sRNA identified in Gram-positive bacteria are predominantly cis-encoded that are perfectly or highly complementary to the mRNA target. Gram-negative bacteria contain more trans-encoded sRNAs that have limited complementarity to the mRNA target (Peer and Margalit, 2014). Therefore, Gram-negative bacteria would benefit from Hfq that could chaperone the interaction of sRNA with its mRNA target, and it has been hypothesized that the evolution of Hfq has coincided with the evolution of trans-encoded sRNAs (Peer and Margalit, 2014). The co-evolution of sRNAs and Hfq has also been proposed to explain the greater importance of the CTD of Hfq in γ-proteobacteria. It will be interesting to see if newly discovered sRNA follow a similar trend of cis and trans-encoded sRNAs to support this hypothesis.
DA: Writing – review & editing. DW: Writing – original draft.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by NIH NIAID grant AI114114 to DA.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Arluison, V., Folichon, M., Marco, S., Derreumaux, P., Pellegrini, O., Seguin, J., et al. (2004). The C-terminal domain of Escherichia coli Hfq increases the stability of the hexamer. Eur. J. Biochem. 271 (7), 1258–1265. doi: 10.1111/j.1432-1033.2004.04026.x
Avican, K., Aldahdooh, J., Togninalli, M., Mahmud, A., Tang, J., Borgwardt, K. M., et al. (2021). RNA atlas of human bacterial pathogens uncovers stress dynamics linked to infection. Nat. Commun. 12 (1), 3282. doi: 10.1038/s41467-021-23588-w
Beich-Frandsen, M., Vecerek, B., Konarev, P. V., Sjoblom, B., Kloiber, K., Hammerle, H., et al. (2011). Structural insights into the dynamics and function of the C-terminus of the E. coli RNA chaperone Hfq. Nucleic Acids Res. 39 (11), 4900–4915. doi: 10.1093/nar/gkq1346
Boggild, A., Overgaard, M., Valentin-Hansen, P., Brodersen, D. E. (2009). Cyanobacteria contain a structural homologue of the Hfq protein with altered RNA-binding properties. FEBS J. 276 (14), 3904–3915. doi: 10.1111/j.1742-4658.2009.07104.x
Brennan, R. G., Link, T. M. (2007). Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 10 (2), 125–133. doi: 10.1016/j.mib.2007.03.015
Brunel, C., Marquet, R., Romby, P., Ehresmann, C. (2002). RNA loop-loop interactions as dynamic functional motifs. Biochimie 84 (9), 925–944. doi: 10.1016/s0300-9084(02)01401-3
Cai, H., Roca, J., Zhao, Y. F., Woodson, S. A. (2022). Dynamic refolding of oxyS sRNA by the hfq RNA chaperone. J. Mol. Biol. 434 (18), 167776. doi: 10.1016/j.jmb.2022.167776
Chakravarty, S., Masse, E. (2019). RNA-dependent regulation of virulence in pathogenic bacteria. Front. Cell Infect. Microbiol. 9. doi: 10.3389/fcimb.2019.00337
Cossa, A., Trepout, S., Wien, F., Groen, J., Le Brun, E., Turbant, F., et al. (2022). Cryo soft X-ray tomography to explore Escherichia coli nucleoid remodeling by Hfq master regulator. J. Struct. Biol. 214 (4), 107912. doi: 10.1016/j.jsb.2022.107912
Dendooven, T., Sonnleitner, E., Blasi, U., Luisi, B. F. (2023). Translational regulation by Hfq-Crc assemblies emerges from polymorphic ribonucleoprotein folding. EMBO J. 42 (3), e111129. doi: 10.15252/embj.2022111129
Diallo, I., Provost, P. (2020). RNA-sequencing analyses of small bacterial RNAs and their emergence as virulence factors in host-pathogen interactions. Int. J. Mol. Sci. 21 (5):1627. doi: 10.3390/ijms21051627
Dimastrogiovanni, D., Frohlich, K. S., Bandyra, K. J., Bruce, H. A., Hohensee, S., Vogel, J., et al. (2014). Recognition of the small regulatory RNA RydC by the bacterial Hfq protein. Elife 3. doi: 10.7554/eLife.05375
Essman U, P. L., Berkowitz, M. L., Darden, T., Lee, H., Pedersen, L. G. (1995). A smooth particle mesh Ewald method. J. Chem. Phys. B 103, 8577–8593. doi: 10.1063/1.470117
Figueroa-Bossi, N., Valentini, M., Malleret, L., Fiorini, F., Bossi, L. (2009). Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev. 23 (17), 2004–2015. doi: 10.1101/gad.541609
Franch, T., Petersen, M., Wagner, E. G., Jacobsen, J. P., Gerdes, K. (1999). Antisense RNA regulation in prokaryotes: rapid RNA/RNA interaction facilitated by a general U-turn loop structure. J. Mol. Biol. 294 (5), 1115–1125. doi: 10.1006/jmbi.1999.3306
Franze de Fernandez, M. T., Eoyang, L., August, J. T. (1968). Factor fraction required for the synthesis of bacteriophage Qbeta-RNA. Nature 219 (5154), 588–590. doi: 10.1038/219588a0
Franze de Fernandez, M. T., Hayward, W. S., August, J. T. (1972). Bacterial proteins required for replication of phage Q ribonucleic acid. Pruification and properties of host factor I, a ribonucleic acid-binding protein. J. Biol. Chem. 247 (3), 824–831. doi: 10.1016/S0021-9258(19)45681-0
Hoe, C. H., Raabe, C. A., Rozhdestvensky, T. S., Tang, T. H. (2013). Bacterial sRNAs: regulation in stress. Int. J. Med. Microbiol. 303 (5), 217–229. doi: 10.1016/j.ijmm.2013.04.002
Horstmann, N., Orans, J., Valentin-Hansen, P., Shelburne, S. A. (2012). Structural mechanism of Staphylococcus aureus Hfq binding to an RNA A-tract. Nucleic Acids Res. 40 (21), 11023–11035. doi: 10.1093/nar/gks809
Hwang, W., Arluison, V., Hohng, S. (2011). Dynamic competition of DsrA and rpoS fragments for the proximal binding site of Hfq as a means for efficient annealing. Nucleic Acids Res. 39 (12), 5131–5139. doi: 10.1093/nar/gkr075
Jousselin, A., Metzinger, L., Felden, B. (2009). On the facultative requirement of the bacterial RNA chaperone, Hfq. Trends Microbiol. 17 (9), 399–405. doi: 10.1016/j.tim.2009.06.003
Kavita, K., Zhang, A., Tai, C. H., Majdalani, N., Storz, G., Gottesman, S. (2022). Multiple in vivo roles for the C-terminal domain of the RNA chaperone Hfq. Nucleic Acids Res. 50 (3), 1718–1733. doi: 10.1093/nar/gkac017
Kollman, P., Dixon, R., Cornell, W., Fox, T., Chipot, C., Pohorille, A. (1997). The development/application of a ‘minimalist’ organic/biochemical molecular mechanic force field using a combination of ab initio calculations and experimental data, in Computer Simulation of Biomolecular Systems: Theoretical and Experimental Applications, eds. van Gunsteren, W.F., Weiner, P.K., Wilkinson, A.J.. (Dordrecht:Springer Netherlands), 83–96. doi: 10.1007/978-94-017-1120-3_2
Kovach, A. R., Hoff, K. E., Canty, J. T., Orans, J., Brennan, R. G. (2014). Recognition of U-rich RNA by Hfq from the Gram-positive pathogen Listeria monocytogenes. RNA 20 (10), 1548–1559. doi: 10.1261/rna.044032.113
Krieger, E., Vriend, G. (2014). YASARA View - molecular graphics for all devices - from smartphones to workstations. Bioinformatics 30 (20), 2981–2982. doi: 10.1093/bioinformatics/btu426
Krieger, E., Vriend, G. (2015). New ways to boost molecular dynamics simulations. J. Comput. Chem. 36 (13), 996–1007. doi: 10.1002/jcc.23899
Link, T. M., Valentin-Hansen, P., Brennan, R. G. (2009). Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl. Acad. Sci. U. S. A. 106 (46), 19292–19297. doi: 10.1073/pnas.0908744106
Malecka, E. M., Woodson, S. A. (2021). Stepwise sRNA targeting of structured bacterial mRNAs leads to abortive annealing. Mol. Cell 81 (9), 1988–1999.e1984. doi: 10.1016/j.molcel.2021.02.019
Mohanty, B. K., Maples, V. F., Kushner, S. R. (2004). The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol. Microbiol. 54 (4), 905–920. doi: 10.1111/j.1365-2958.2004.04337.x
Moon, K., Gottesman, S. (2009). A PhoQ/P-regulated small RNA regulates sensitivity of Escherichia coli to antimicrobial peptides. Mol. Microbiol. 74 (6), 1314–1330. doi: 10.1111/j.1365-2958.2009.06944.x
Nielsen, J. S., Boggild, A., Andersen, C. B., Nielsen, G., Boysen, A., Brodersen, D. E., et al. (2007). An Hfq-like protein in archaea: crystal structure and functional characterization of the Sm protein from Methanococcus jannaschii. RNA 13 (12), 2213–2223. doi: 10.1261/rna.689007
Nielsen, J. S., Lei, L. K., Ebersbach, T., Olsen, A. S., Klitgaard, J. K., Valentin-Hansen, P., et al. (2010). Defining a role for Hfq in Gram-positive bacteria: evidence for Hfq-dependent antisense regulation in Listeria monocytogenes. Nucleic Acids Res. 38 (3), 907–919. doi: 10.1093/nar/gkp1081
Nikulin, A., Stolboushkina, E., Perederina, A., Vassilieva, I., Blaesi, U., Moll, I., et al. (2005). Structure of Pseudomonas aeruginosa Hfq protein. Acta Crystallogr. D Biol. Crystallogr. 61 (Pt 2), 141–146. doi: 10.1107/S0907444904030008
Olsen, A. S., Moller-Jensen, J., Brennan, R. G., Valentin-Hansen, P. (2010). C-terminally truncated derivatives of Escherichia coli Hfq are proficient in riboregulation. J. Mol. Biol. 404 (2), 173–182. doi: 10.1016/j.jmb.2010.09.038
Orans, J., Kovach, A. R., Hoff, K. E., Horstmann, N. M., Brennan, R. G. (2020). Crystal structure of an Escherichia coli Hfq Core (residues 2-69)-DNA complex reveals multifunctional nucleic acid binding sites. Nucleic Acids Res. 48 (7), 3987–3997. doi: 10.1093/nar/gkaa149
Overgaard, M., Johansen, J., Moller-Jensen, J., Valentin-Hansen, P. (2009). Switching off small RNA regulation with trap-mRNA. Mol. Microbiol. 73 (5), 790–800. doi: 10.1111/j.1365-2958.2009.06807.x
Panja, S., Schu, D. J., Woodson, S. A. (2013). Conserved arginines on the rim of Hfq catalyze base pair formation and exchange. Nucleic Acids Res. 41 (15), 7536–7546. doi: 10.1093/nar/gkt521
Park, S., Prevost, K., Heideman, E. M., Carrier, M. C., Azam, M. S., Reyer, M. A., et al. (2021). Dynamic interactions between the RNA chaperone Hfq, small regulatory RNAs, and mRNAs in live bacterial cells. Elife 10. doi: 10.7554/eLife.64207
Peer, A., Margalit, H. (2014). Evolutionary patterns of Escherichia coli small RNAs and their regulatory interactions. RNA 20 (7), 994–1003. doi: 10.1261/rna.043133.113
Ponath, F., Hor, J., Vogel, J. (2022). An overview of gene regulation in bacteria by small RNAs derived from mRNA 3' ends. FEMS Microbiol. Rev. 46 (5). doi: 10.1093/femsre/fuac017
Repoila, F., Darfeuille, F. (2009). Small regulatory non-coding RNAs in bacteria: physiology and mechanistic aspects. Biol. Cell 101 (2), 117–131. doi: 10.1042/BC20070137
Robinson, K. E., Orans, J., Kovach, A. R., Link, T. M., Brennan, R. G. (2014). Mapping Hfq-RNA interaction surfaces using tryptophan fluorescence quenching. Nucleic Acids Res. 42 (4), 2736–2749. doi: 10.1093/nar/gkt1171
Salim, N. N., Faner, M. A., Philip, J. A., Feig, A. L. (2012). Requirement of upstream Hfq-binding (ARN)x elements in glmS and the Hfq C-terminal region for GlmS upregulation by sRNAs GlmZ and GlmY. Nucleic Acids Res. 40 (16), 8021–8032. doi: 10.1093/nar/gks392
Santiago-Frangos, A., Frohlich, K. S., Jeliazkov, J. R., Malecka, E. M., Marino, G., Gray, J. J., et al. (2019). Caulobacter crescentus Hfq structure reveals a conserved mechanism of RNA annealing regulation. Proc. Natl. Acad. Sci. U. S. A. 116 (22), 10978–10987. doi: 10.1073/pnas.1814428116
Santiago-Frangos, A., Jeliazkov, J. R., Gray, J. J., Woodson, S. A. (2017). Acidic C-terminal domains autoregulate the RNA chaperone Hfq. Elife 6. doi: 10.7554/eLife.27049
Santiago-Frangos, A., Kavita, K., Schu, D. J., Gottesman, S., Woodson, S. A. (2016). C-terminal domain of the RNA chaperone Hfq drives sRNA competition and release of. Proc. Natl. Acad. Sci. U. S. A. 113 (41), E6089–E6096. doi: 10.1073/pnas.1613053113
Sarni, S. H., Roca, J., Du, C., Jia, M., Li, H., Damjanovic, A., et al. (2022). Intrinsically disordered interaction network in an RNA chaperone revealed by native mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 119 (47), e2208780119. doi: 10.1073/pnas.2208780119
Sauer, E., Schmidt, S., Weichenrieder, O. (2012). Small RNA binding to the lateral surface of Hfq hexamers and structural rearrangements upon mRNA target recognition. Proc. Natl. Acad. Sci. U. S. A. 109 (24), 9396–9401. doi: 10.1073/pnas.1202521109
Sauer, E., Weichenrieder, O. (2011). Structural basis for RNA 3'-end recognition by Hfq. Proc. Natl. Acad. Sci. U. S. A. 108 (32), 13065–13070. doi: 10.1073/pnas.1103420108
Sauter, C., Basquin, J., Suck, D. (2003). Sm-like proteins in Eubacteria: the crystal structure of the Hfq protein from Escherichia coli. Nucleic Acids Res. 31 (14), 4091–4098. doi: 10.1093/nar/gkg480
Schu, D. J., Zhang, A., Gottesman, S., Storz, G. (2015). Alternative Hfq-sRNA interaction modes dictate alternative mRNA recognition. EMBO J. 34 (20), 2557–2573. doi: 10.15252/embj.201591569
Schumacher, M. A., Pearson, R. F., Moller, T., Valentin-Hansen, P., Brennan, R. G. (2002). Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 21 (13), 3546–3556. doi: 10.1093/emboj/cdf322
Someya, T., Baba, S., Fujimoto, M., Kawai, G., Kumasaka, T., Nakamura, K. (2012). Crystal structure of Hfq from Bacillus subtilis in complex with SELEX-derived RNA aptamer: insight into RNA-binding properties of bacterial Hfq. Nucleic Acids Res. 40 (4), 1856–1867. doi: 10.1093/nar/gkr892
Sonnleitner, E., Moll, I., Blasi, U. (2002). Functional replacement of the Escherichia coli hfq gene by the homologue of Pseudomonas aeruginosa. Microbiol. (Reading) 148, 883–891. doi: 10.1099/00221287-148-3-883
Sonnleitner, E., Napetschnig, J., Afonyushkin, T., Ecker, K., Vecerek, B., Moll, I., et al. (2004). Functional effects of variants of the RNA chaperone Hfq. Biochem. Biophys. Res. Commun. 323 (3), 1017–1023. doi: 10.1016/j.bbrc.2004.08.190
Sun, X., Zhulin, I., Wartell, R. M. (2002). Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res. 30 (17), 3662–3671. doi: 10.1093/nar/gkf508
Thisted, T., Sorensen, N. S., Wagner, E. G., Gerdes, K. (1994). Mechanism of post-segregational killing: Sok antisense RNA interacts with Hok mRNA via its 5'-end single-stranded leader and competes with the 3'-end of Hok mRNA for binding to the mok translational initiation region. EMBO J. 13 (8), 1960–1968. doi: 10.1002/j.1460-2075.1994.tb06465.x
Thomason, M. K., Fontaine, F., De Lay, N., Storz, G. (2012). A small RNA that regulates motility and biofilm formation in response to changes in nutrient availability in Escherichia coli. Mol. Microbiol. 84 (1), 17–35. doi: 10.1111/j.1365-2958.2012.07965.x
Turbant, F., Waeytens, J., Campidelli, C., Bombled, M., Martinez, D., Grelard, A., et al. (2022). Unraveling membrane perturbations caused by the bacterial riboregulator hfq. Int. J. Mol. Sci. 23 (15):8739. doi: 10.3390/ijms23158739
Updegrove, T. B., Wartell, R. M. (2011). The influence of Escherichia coli Hfq mutations on RNA binding and sRNA*mRNA duplex formation in rpoS riboregulation. Biochim. Biophys. Acta 1809 (10), 532–540. doi: 10.1016/j.bbagrm.2011.08.006
Vecerek, B., Rajkowitsch, L., Sonnleitner, E., Schroeder, R., Blasi, U. (2008). The C-terminal domain of Escherichia coli Hfq is required for regulation. Nucleic Acids Res. 36 (1), 133–143. doi: 10.1093/nar/gkm985
Wang, W., Wang, L., Wu, J., Gong, Q., Shi, Y. (2013). Hfq-bridged ternary complex is important for translation activation of rpoS by DsrA. Nucleic Acids Res. 41 (11), 5938–5948. doi: 10.1093/nar/gkt276
Waters, L. S., Storz, G. (2009). Regulatory RNAs in bacteria. Cell 136 (4), 615–628. doi: 10.1016/j.cell.2009.01.043
Watkins, D., Arya, D. P. (2019). Regulatory roles of small rnas in prokaryotes: Parallels and contrast with eukaryotic mirna. Non-coding RNA Investig. 3 (10.21037):29. doi: 10.21037/ncri.2019.10.02
Zhang, A., Schu, D. J., Tjaden, B. C., Storz, G., Gottesman, S. (2013). Mutations in interaction surfaces differentially impact E. coli Hfq association with small RNAs and their mRNA targets. J. Mol. Biol. 425 (19), 3678–3697. doi: 10.1016/j.jmb.2013.01.006
Keywords: RNA, bacteria, E. coli, Hfq, protein
Citation: Watkins D and Arya D (2023) Models of Hfq interactions with small non-coding RNA in Gram-negative and Gram-positive bacteria. Front. Cell. Infect. Microbiol. 13:1282258. doi: 10.3389/fcimb.2023.1282258
Received: 23 August 2023; Accepted: 05 October 2023;
Published: 24 October 2023.
Edited by:
Bindu Subhadra, Long Island University, United StatesReviewed by:
Suganya Sivagurunathan, Broad Institute, United StatesCopyright © 2023 Watkins and Arya. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dev Arya, ZHBhcnlhQG1haWwuY2xlbXNvbi5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.