- 1Department of Bioengineering, Temple University, Philadelphia, PA, United States
- 2Biomedical and Data Sciences Division, CFD Research Corporation, Huntsville, AL, United States
- 3Department of Mechanical Engineering, Temple University, Philadelphia, PA, United States
- 4Department of Radiation Oncology, Lewis Katz School of Medicine, Temple University, Philadelphia, PA, United States
- 5Center for Inflammation and Lung Research, Department of Microbiology, Immunology and Inflammation, Lewis Katz School of Medicine, Temple University, Philadelphia, PA, United States
Sepsis is a global health concern accounting for more than 1 in 5 deaths worldwide. Sepsis is now defined as life-threatening organ dysfunction caused by a dysregulated host response to infection. Sepsis can develop from bacterial (gram negative or gram positive), fungal or viral (such as COVID) infections. However, therapeutics developed in animal models and traditional in vitro sepsis models have had little success in clinical trials, as these models have failed to fully replicate the underlying pathophysiology and heterogeneity of the disease. The current understanding is that the host response to sepsis is highly diverse among patients, and this heterogeneity impacts immune function and response to infection. Phenotyping immune function and classifying sepsis patients into specific endotypes is needed to develop a personalized treatment approach. Neutrophil-endothelium interactions play a critical role in sepsis progression, and increased neutrophil influx and endothelial barrier disruption have important roles in the early course of organ damage. Understanding the mechanism of neutrophil-endothelium interactions and how immune function impacts this interaction can help us better manage the disease and lead to the discovery of new diagnostic and prognosis tools for effective treatments. In this review, we will discuss the latest research exploring how in silico modeling of a synergistic combination of new organ-on-chip models incorporating human cells/tissue, omics analysis and clinical data from sepsis patients will allow us to identify relevant signaling pathways and characterize specific immune phenotypes in patients. Emerging technologies such as machine learning can then be leveraged to identify druggable therapeutic targets and relate them to immune phenotypes and underlying infectious agents. This synergistic approach can lead to the development of new therapeutics and the identification of FDA approved drugs that can be repurposed for the treatment of sepsis.
Introduction
Sepsis is a critical healthcare problem. According to a meta-analysis of literature by the World Health Organization (WHO), in 2017 approximately 48.9 million cases of sepsis were identified worldwide, accounting for 11 million sepsis-related deaths per year (World Health Organization, 2023). An estimated 20 million children younger than 5 years developed sepsis, resulting in 2.9 million global deaths in the same year. In the U.S., 1.7 million sepsis cases are reported annually, resulting in more than 270,000 deaths each year (Rhee et al., 2017) and is a leading cause of death in hospitals (Singer et al., 2016; Donnelly et al., 2017). Under the Sepsis-3 definition, sepsis is now defined as life-threatening organ dysfunction caused by a dysregulated host response to infection (Singer et al., 2016; Donnelly et al., 2017). While neutrophils are critical to host defense, neutrophil dysregulation in sepsis plays a critical role in sepsis-induced organ failure through interactions with the vascular endothelium resulting in endothelial cell (EC) damage, increased vascular barrier permeability, and enhanced neutrophil trafficking into vital organs (Guimarães-Costa et al., 2014; Lang et al., 2014; Jiao et al., 2020; Iba et al., 2022; Rosales, 2020; Sakuma et al., 2022). This neutrophil dysregulation and neutrophil-endothelial cell interactions in sepsis can lead to extravascular tissue damage and septic shock resulting in multiple organ dysfunction syndrome (MODS) and death (Kell and Pretorius, 2018).
Sepsis is a complex and multifaceted medical condition, and its manifestations can vary considerably among patients. It is not a uniform disease but rather a syndrome that results from the body’s overwhelming response to infection. The clinical presentation of sepsis can range from mild to severe, with some individuals experiencing subtle symptoms while others face life-threatening consequences, such as organ dysfunction and septic shock (Singer et al., 2016; Seymour et al., 2019). Sepsis is also known for its remarkable heterogeneity in clinical manifestations, and this diversity is often influenced by both the source of infection and the unique immune response of the affected individual. For example, sepsis originating from a respiratory infection, such as pneumonia, may present with distinctive clinical features as compared to sepsis triggered by a urinary tract infection (Rudd et al., 2020). The former may involve respiratory distress, high fever, and lung-specific complications, while the latter might primarily manifest as urinary symptoms and lower abdominal pain. These variations in sepsis presentation underscore the significance of personalized and targeted treatment approaches to address the diverse clinical trajectories of this life-threatening condition. Recent research has focused on the heterogeneity of sepsis, emphasizing the necessity of understanding the source-specific and patient-specific aspects of sepsis (Seymour et al., 2019).
To date, therapeutic approaches for the treatment of sepsis are largely supportive, but there are no specific drugs available despite promising preclinical studies in rodent models (Williams and Chambers, 2014; Jarczak et al., 2021; Niederman et al., 2021). Current treatment consists of antibiotic therapy for the underlying infection and supportive care through the administration of fluids and vasopressors (Seitz et al., 2022). Kidney dialysis and mechanical ventilation are often used to support organ failure as the disease progresses. Multiple drugs which have shown promise in preclinical models have failed in large randomized clinical trials to demonstrate a significant reduction in mortality (Osborn, 2017). This absence of clinical translation is due to a host of factors, including the incorporation of rodent models that fail at emulating the complete clinical situation that is present in humans (e.g., age, sex, demographics, comorbidities etc.) and the varied composition of systemic leukocytes in both species (Rudd et al., 2020; Iregbu et al., 2022; World Health Organization, 2017). Recent clinical trials have examined the impact of anti-cytokine receptor therapies such as anakinra (an IL-1 receptor antagonist) to mitigate the inflammatory response (Shakoory et al., 2016), and the consideration of using DNase (such as DNase-I) in addressing sepsis-associated formation of neutrophil extracellular traps (NETs), thereby highlighting the multifaceted approach required for effective sepsis treatment (Singer et al., 2016; Donnelly et al., 2017). Unfortunately, none of these treatments demonstrate targeted efficacy against sepsis. Numerous reports have highlighted the heterogeneous nature of sepsis (Scicluna et al., 2017; Fenner et al., 2021; Yang et al., 2021b; Langston et al., 2023), confirming the unlikelihood that a one-size-fits-all treatment will be appropriate for every septic patient. Thus, the lack of therapeutic options for the treatment of sepsis underscores the urgent need for further research and innovation to develop novel therapeutic interventions that can address the complexities of this life-threatening condition (Langston et al., 2023). Emerging tools such as microphysiological assays (i.e., organ-on-chip) (Kilpatrick and Kiani, 2020; Yang et al., 2021a), omics, in silico models and machine learning (Giannini et al., 2019; Stolarski et al., 2022; Honoré et al., 2023; Pan et al., 2023) are providing promising avenues for developing effective, personalized options for treating sepsis (Figure 1).
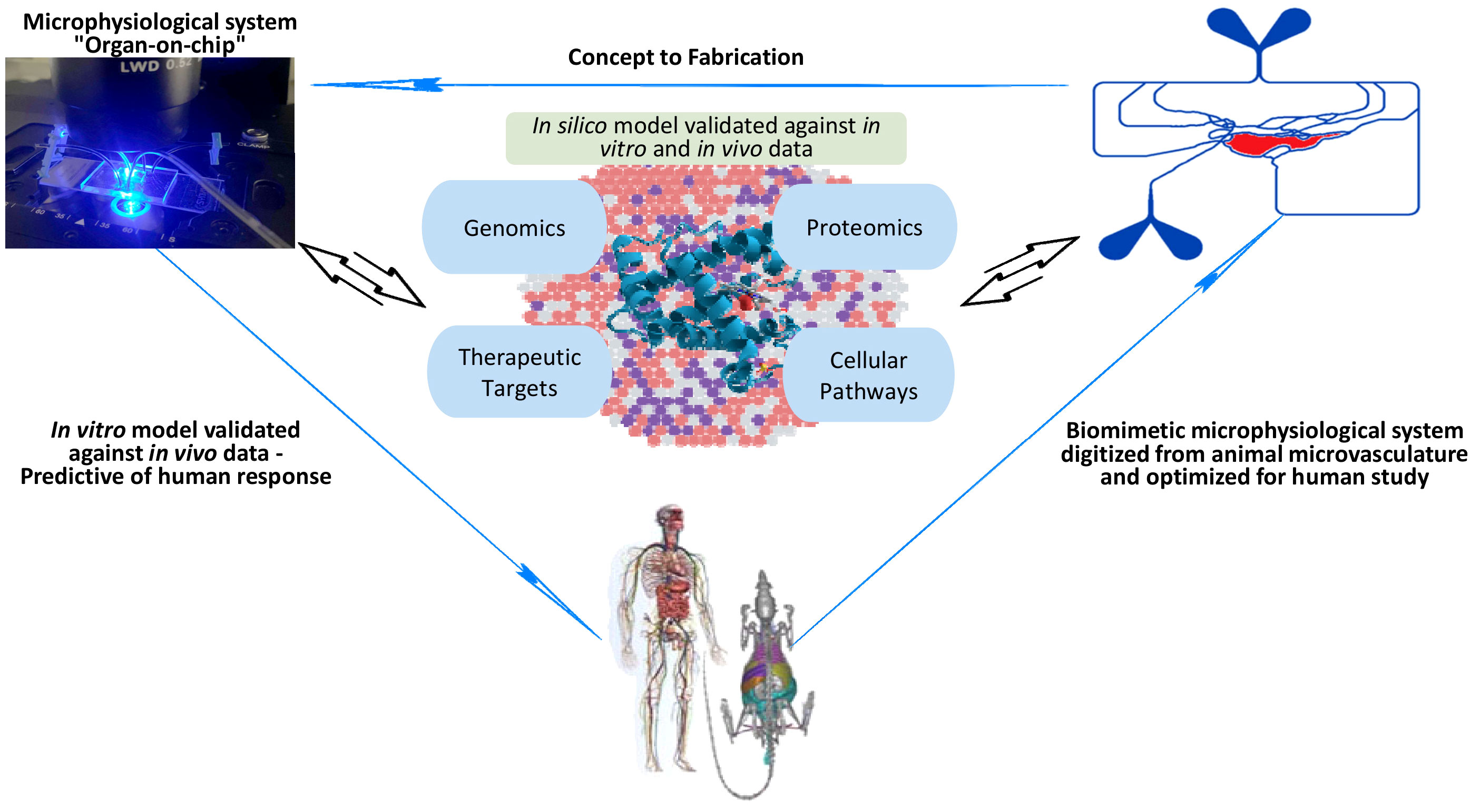
Figure 1 An illustration of how emerging tools such as biomimetic microphysiological systems (i.e., organ-on-chip), omics, in silico models and machine learning are providing promising avenues for better understanding and developing more effective, personalized options for treating sepsis.
In this review, we will examine critical aspects of neutrophil-ECs interactions during the evolution and progression of sepsis, highlighting the difficulties in development of effective therapeutic approaches. We will discuss the emergence of innovative in vitro models that mimic the complex interplay between neutrophils and ECs and the use of these models in classifying sepsis patients and testing potential therapeutics. We will focus on the importance of immune phenotyping and sepsis endotyping to gain important insight into the heterogenous nature of the disease. We will discuss the importance of omics in identifying important cellular signaling cascades involved in sepsis progression. Lastly, we will discuss promising applications of in silico models and machine learning algorithms, offering new avenues for enhancing our comprehension of sepsis.
Neutrophil activation during sepsis
Neutrophils are key cells of the human innate immune system and critical elements of host defense against pathogens. Neutrophils identify infectious agents through pattern recognition receptors (PRRs), such as Toll-like receptors, on the cell surface that recognize a wide variety of PAMPs and DAMPS that are released by invading pathogens and damaged tissue (Iba and Ogura, 2018). Upon activation, neutrophils rapidly traffic to sites of infection or inflammation (Ley et al., 2007). Neutrophils clear pathogens through phagocytosis, degranulation and the release of proteases and the production of reactive oxygen species (ROS), which can kill pathogens directly by causing oxidative damage (Perobelli et al., 2015; Glémain et al., 2022; Rosales, 2018). Neutrophils possess numerous types of granules within their cytoplasm that are sequentially released upon stimulation and degranulation. Neutrophils contain primary (azurophilic), secondary (specific) and tertiary (gelatinase) granules, as well as secretory vesicles encompassing a repertoire of more than 300 proteins involved in cell adhesion, migration and bactericidal activities (Fox et al., 2010). A number of potent proteases are located in these granules including myeloperoxidase (MPO), neutrophil elastase and cathepsin G (Borregaard, 2010). The release of these proteases is tightly regulated to minimize host tissue damage, but during sepsis and immune dysregulation, the release of these granule contents can damage ECs and host tissue. Neutrophils can also immobilize and kill pathogens through the release of NETs (Perobelli et al., 2015; Herrero-Cervera et al., 2022; Rosales et al., 2023). NETs are composed of extruded strands of nuclear material (such as DNA) which form a web-like structure that is composed of decondensed chromatin fibers decorated with antimicrobial enzymes released from granules, such as neutrophil elastase, MPO and cathepsin, as well as nuclear proteins (Perobelli et al., 2015; Döring et al., 2017; Jiao et al., 2020). NETs can be a double-edged sword; they are critical for microbicidal activity, but uncontrolled NETs release during sepsis can also exacerbate inflammation, damage ECs, and cause tissue injury (Chang, 2019; Glémain et al., 2022).
Thus, neutrophil activation plays a pivotal role during sepsis, with a complex interplay between various cellular and molecular mechanisms. The activation of neutrophils leads to a cascade of events, including adhesion, migration, degranulation, and the formation of NETs. However, dysregulated neutrophil activation contributes to EC damage and organ dysfunction.
Endothelial cell activation during sepsis
Under normal conditions, ECs act as an important regulator of hemodynamic balance throughout the body. ECs possess a remarkable ability to sense and react to various extracellular stimuli originating from the microenvironment (Yang et al., 2021b). These stimuli encompass both biomechanical factors (e.g., shear forces, pressure, and cyclical strain) as well as biochemical signals (e.g., growth factors, hormones, cytokines, chemokines, nitric oxide, oxygen and reactive oxygen species) (Rosales, 2018). The ability to sense hemodynamic stimuli allows ECs to quickly respond to microenvironmental changes in the blood vessel environment, such as sensing the pro-inflammatory cytokines released by neutrophils during sepsis (Iba et al., 2022). Notably, in response to these stimuli, ECs exhibit distinct phenotypes characterized by spatial and temporal tissue-specific heterogeneity (Aird, 2012).
The diverse morphological characteristics of endothelium are directly related to their functional heterogeneity (Yang et al., 2021b). For example, within arteries and arterioles, ECs align and elongate in parallel to the direction of blood flow, playing a crucial role in regulating vascular tone (Yang et al., 2021b). In contrast, ECs in veins and venules typically assume a polygonal shape rather than an elongated shape, lacking specific orientation (Yang et al., 2021b). Moreover, venous ECs tend to have higher permeability compared to arterial ECs (Sriram et al., 2015).Notably, post-capillary venules serve as primary sites for leukocyte extravasation during inflammation and sepsis, facilitating the movement of immune cells from the bloodstream into the surrounding tissues (Chi et al., 2003). These contrasting characteristics of ECs are closely linked to variances in hemodynamic environments and the functional properties of the vessels they line (Young and Simmons, 2010).
Endothelial cells (ECs) also exhibit remarkable heterogeneity across different organs, and their distinct phenotypic profiles can play varying roles in sepsis. For instance, lung microvascular ECs are characterized by high levels of tissue factor expression, which contributes to the initiation of coagulation pathways during sepsis (Wang et al., 2008). In contrast, hepatic ECs possess unique functional features, including the clearance of endotoxins and bacterial products where the EC structure and alignment are discontinuous, allowing the exchange of large solutes between plasma and intestinal environment (Shetty et al., 2018). On the other hand, kidney EC exhibits a fenestrated alignment that features intracellular pores with a diaphragm that penetrate the endothelium layer, which warrants the rapid exchange of water (Yang et al., 2021b). This specific EC alignment in kidney plays a crucial role in sepsis by potentially enabling enhanced vascular permeability and leukocyte trafficking, thereby contributing to the pathophysiological processes and kidney dysfunction during sepsis progression. These organ-specific variation in EC function during sepsis is critical for understanding the differential responses observed in discrete organs. At the molecular level, these organ-specific disparities can be attributed to differences in the expression of adhesion molecules, cytokine receptors, and signaling pathways. For instance, the lung ECs express higher levels of vascular cell adhesion molecule 1 (VCAM-1) and intercellular adhesion molecule 1 (ICAM-1), which are involved in leukocyte adhesion and recruitment (Ley et al., 2007). Liver ECs, on the other hand, express receptors for bacterial toxins, enabling them to actively participate in detoxification processes (Dejager et al., 2011). Vascular permeability is regulated by tight and adherens junctions between ECs, therefore, the disassembly of occludin, claudin-5 and VE-cadherin from tight junctions can be used as an indicator of increased vascular permeability (Kondo et al., 2009). Aslan et al. reported that baseline expression levels of occludin, claudin-5, and VE-cadherin markedly differed between kidney and lung, with claudin-5 and VE-cadherin being highly expressed in the lung, and occludin being highly expressed in the kidney during sepsis in mice and humans, indicating an organ-specific molecular manifestation of ECs (Aslan et al., 2017). This intricate web of organ-specific heterogeneity in EC function during sepsis not only enhances our understanding of the diverse organ responses but also underscores the pivotal role of these cells in shaping the outcomes of this complex and life-threatening condition.
The endothelial glycocalyx (eGC) is a protective layer on the cell surface of vascular ECs (Fatmi et al., 2022) that plays a critical role during inflammation. This glycoprotein layer has an intricate architecture consisting of different components integrating either plasma- or endothelium-derived soluble molecules, including syndecan-1 (SDC-1), Heparan sulphaste (HS), Hyaluronan (HA), chondroitin sulphate (CS), and other cell adhesion molecules (CAMs) such as VCAM-1 and ICAM-1 (Puchwein-Schwepcke et al., 2021; Fatmi et al., 2022). These important surface molecules play critical roles during the leukocyte adhesion cascade in sepsis (Ley et al., 2007; Iba and Ogura, 2018; Iba et al., 2022). Activated neutrophils can damage the protective EC glycocalyx leading to increased vascular barrier permeability and neutrophil trafficking into vital organs (Chang, 2019; Iba et al., 2022).
ECs are tightly connected through junctional adhesion molecules (JAMs) including JAM-A, -B, -C and endothelial cell-selective adhesion molecules (ESAM) which are crucial for maintaining the endothelial junctional complexes that regulate vascular transmigration and permeability (Woodfin et al., 2011; Amalakuhan et al., 2016). During sepsis, these junctional functions are disrupted and become dysregulated, leading to increased vascular permeability and, fluid leakage into the surrounding tissue and edema, causing further organ damage (Figure 2) (Nourshargh and Alon, 2014; Uchimido et al., 2019; Duong and Vestweber, 2020). Other junctional components include platelet endothelial cell adhesion molecule (PECAM)-1 and nectin; the disruption of these components also contribute to the uncontrolled neutrophil influx (Duong and Vestweber, 2020).
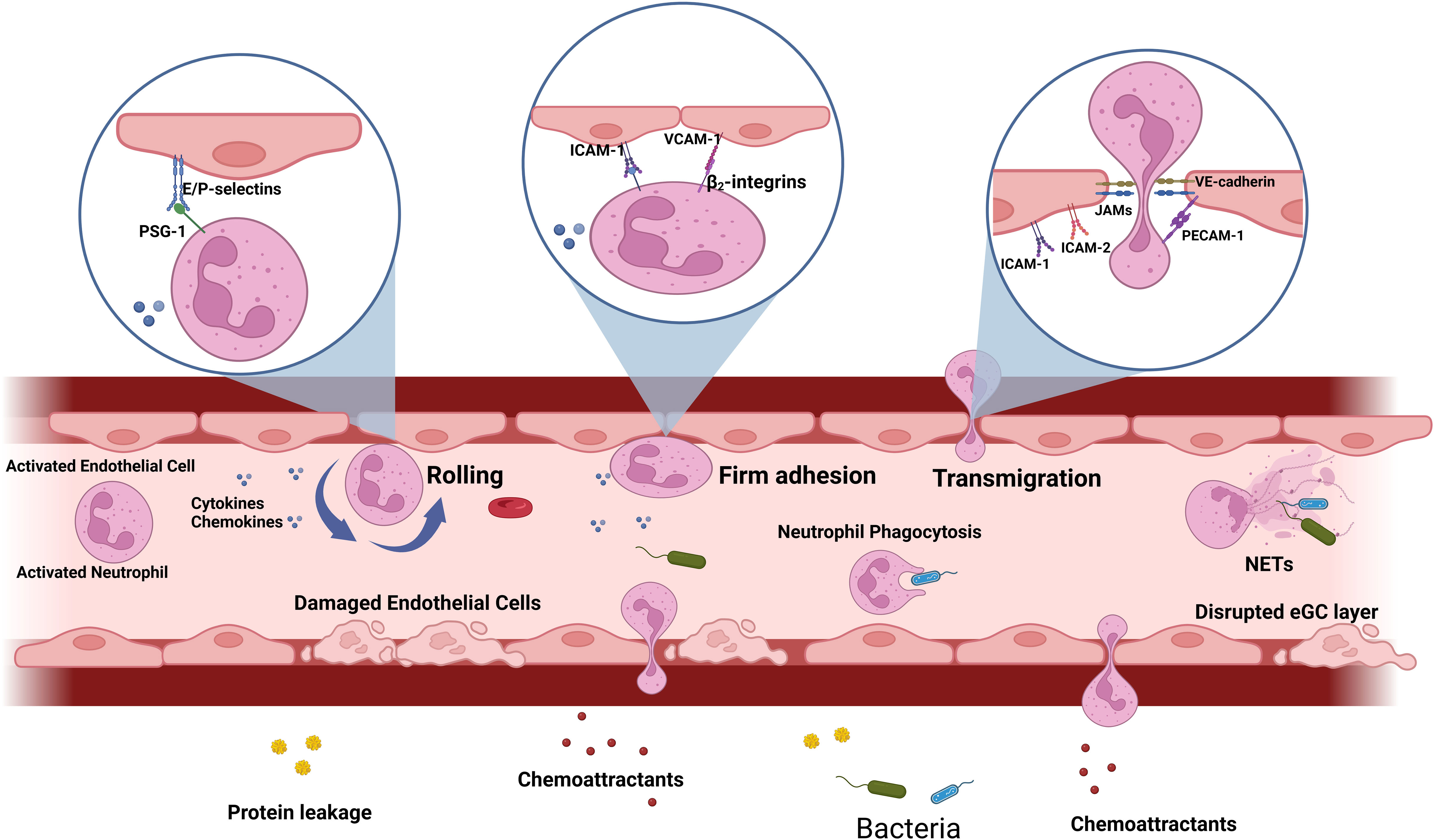
Figure 2 Leukocyte adhesion cascade during sepsis: During sepsis, both PAMPs and DAMPs trigger the activation of neutrophils and endothelial cells, prompting the production of cytokines and chemoattractants. Consequently, neutrophils display surface molecules that interact with adhesion molecules expressed by activated endothelium. The initial rolling step involves interactions between E/P-selectin and their ligands, such as PSGL-1, which slows down the neutrophil. Subsequently, firm adhesion is mediated by endothelial adhesion molecules like ICAM-1, ICAM-2, and VCAM-1, interacting with neutrophil ligands, such as β2 integrins. In response to chemoattractants, adhered neutrophils migrate through endothelial junctions, facilitated by PECAM-1, VE-cadherin and JAMs. Concurrently, activated neutrophils release cytokines, reactive oxygen species (ROS), and proteases, or undergo the formation of neutrophil extracellular traps (NETs). During sepsis, the endothelial glycocalyx (eGC) is degraded, endothelial cell tight junctions are damaged, and there is an increase in endothelial cell apoptosis, ultimately leading to compromised barrier function and increased permeability.
In summary, ECs play a pivotal role in maintaining vascular health and homeostasis, and their injury can trigger a cascade of events leading to tissue and organ dysfunction. EC injury can occur due to various insults, such as inflammation, oxidative stress or mechanical damage. At the molecular level, the loss of EC integrity disrupts the endothelial barrier, exposing underlying tissues to a pro-inflammatory environment. Moreover, injured ECs release vasoactive molecules, such as endothelin-1, leading to vasoconstriction and impaired blood flow (Aird, 2012). Disruption of the endothelial barrier can also result in increased vascular permeability, allowing the leakage of proteins and immune cells into the interstitium, further exacerbating inflammation (Chistiakov et al., 2015). Over time, these processes can lead to chronic inflammation, tissue damage, and ultimately, organ dysfunction. Understanding the molecular mechanisms underlying EC injury and dysfunction is crucial for developing targeted therapeutic interventions to mitigate the consequences of endothelial damage (Libby et al., 2011).
Neutrophil-endothelium interaction during sepsis
Septic patients often die from MODS, in part caused by the dysregulated neutrophil influx into tissues leading to organ damage (Yang et al., 2021a). During infection, pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) are released and activate the innate immune system. This activation results in the release of cytokines, chemokines and other proinflammatory mediators which can activate neutrophils and endothelial cells (ECs) leading to neutrophil-EC interaction.
The leukocyte adhesion cascade is a process by which leukocytes, such as neutrophils, roll, firmly adhere and then migrate through activated ECs (Ley et al., 2007; Futosi et al., 2013; Tecchio et al., 2014; Rosales, 2018; Rosales, 2020). This process is mediated on ECs by pro-adhesive and other effectors molecules through the Nuclear factor-kappa B (NF-κB) signaling pathway (Kojok et al., 2018). Pattern recognition receptors (PRRs) are widely expressed on neutrophils and ECs (Iba and Ogura, 2018). When these receptors interact with PAMPs and DAMPs, cytokines [e.g., Tumor necrosis factor-alpha (TNF-α) and Interleukin-1 Beta (IL-1β)] and chemokines [e.g., Interleukin-8 (IL-8) and Monocyte chemoattractant protein 1 (MCP-1)] are synthesized and released into the bloodstream (Zhu et al., 2017; Kojok et al., 2018; Wang et al., 2018; Georgescu et al., 2020). These pro-inflammatory cytokines disrupt the integrity of the endothelial glycocalyx (eGC) surface layer, exposing adhesion molecules, such as VCAM-1, ICAM-1 and E-selectin and result in increased number of neutrophils rolling, firmly adhering and transmigrating through the ECs (Schmidt et al., 2012; Yang et al., 2021b). As sepsis advances, VCAM-1 and ICAM-1, which are expressed in large quantities on the activated EC surface of septic patients, are also cleaved and released into the circulation (Amalakuhan et al., 2016). Figure 2 illustrates the key steps of the leukocyte adhesion cascade process during the onset of sepsis and highlights the unique characteristics of neutrophils and ECs that allow them to interact effectively during sepsis.
Better understanding of neutrophil-endothelium interaction can help identify potential therapeutic targets to prevent the development of organ dysfunction in sepsis.
Sepsis manifestations and existing models for sepsis research and their limitations
Animal models and in vitro models employing cell culture and organoids, have proven invaluable for unraveling some of the intricacies of sepsis pathophysiology. The classical approach in exploring the mechanisms of inflammatory diseases involves the use of murine models. However, the lack of alignment between murine models and human septic manifestations is a growing concern (Soroush et al., 2020). Additionally, the significant phenotypic variation observed among various types of endothelial cells has raised questions about the translatability of findings from these models to human diseases (Seok et al., 2013; Sznajder et al., 2013). Consequently, a notable limitation of murine models is the potential for a given therapeutic intervention to yield distinct outcomes in mice compared to humans (Seok et al., 2013; Sznajder et al., 2013). These concerns gain support from recent research employing bulk and single-cell transcriptomics to delineate innate immune responses. This work has demonstrated substantial interspecies variations in the expression of cytokines, chemokines, and their corresponding receptors (Hagai et al., 2018). While rodent models remain a valuable tool in sepsis research, an international panel of experts has emphasized the pressing need for models that more realistically recapitulate human disease, acknowledging the significant limitations of murine models (Zingarelli et al., 2019). Hence, there exists a considerable demand for the development of “in vitro reconstitution of disease-related cell types or tissues” employing human cells to investigate human inflammatory diseases more effectively (Kilpatrick and Kiani, 2020; Soroush et al., 2020; Yang et al., 2021a). Reconstruction of more in vivo human relevant models could bridge the gap between traditional murine models and the intricacies of human inflammatory disease, potentially advancing our understanding and therapeutic strategies in this field.
Traditional cell culture models have been used to study molecular and cellular responses to septic insults. These models typically involve specific cell types, such as macrophages or endothelial cells, and exposing them to PAMPs, such as endotoxin, or proinflammatory cytokines to mimic septic conditions. These in vitro studies allow for the investigation of host responses, such as the activation of PRRs which activate NF-κB and other transcription factors to produce proinflammatory mediators such as cytokines and chemokines. (Ploppa et al., 2012; Mai et al., 2013; Wicherska-pawłowska et al., 2021). Despite their utility and ease of use, cell culture models have significant limitations. They often lack the intricate three-dimensional tissue architecture seen in vivo, which is crucial for studying cell-cell interactions. Moreover, these models may not fully replicate the complexity of the immune response in sepsis, as they often lack multiple cell types important for cell-cell communication (Chistiakov et al., 2015). More importantly, static cell culture models do not take in to account the impact of physiologically relevant flow conditions which is a critical parameter in cell activation and signaling and needed to model in vivo conditions. Organoids represent another promising approach for in vitro studies of sepsis. These miniature 3D organ-like structures are developed by culturing organ-specific cells, such as intestinal tissue (Siwczak et al., 2021), lung tissue (Bosáková et al., 2022), or brain tissue (Lim et al., 2022) under conditions that promote self-organization and differentiation. Recent advancements have allowed researchers to create lung, liver, and intestinal organoids to mimic in vivo inflammation more realistically. These models provide valuable insights into the tissue-specific molecular responses during inflammatory conditions (Lim et al., 2022), including the dysregulation of tight junction proteins, altered metabolic pathways and the release of proinflammatory cytokines (Lee et al., 2014). However, organoids, too, have their limitations. They may not fully capture the systemic effects of sepsis, given their focus on individual organs. Additionally, their utility is currently restricted by challenges in scaling up for high-throughput screening and maintaining long-term viability. Last but not least, the vascular network of organoids is hard to reconstruct in vitro, therefore, these organoid models often lack realistic blood flow representation as well.
In summary, organoids offer insights into tissue-specific responses while murine animal models could provide a more translational understanding to human in vivo conditions. These models enhance our understanding of sepsis pathophysiology, but researchers must be mindful of their limitations, such as the lack of tissue complexity in cell cultures, the limited systemic perspective of organoids, and the significant limitations of animal models. Addressing these challenges is essential for optimizing the translational potential of these models in sepsis research.
Emerging in vitro models – the promise of the organ-on-chip technology
As discussed previously, excessive neutrophil infiltration into tissues causes organ damage, often resulting in organ failure and death during sepsis. Employing screening models that mimic neutrophil-endothelium interactions under inflammatory conditions have become important tools for developing precision therapeutics.
Development of “organ-on-chip” assays, also known as microphysiological systems or 3-D biomimetic microfluidic assays, offer a number of advantages including the ability to observe and measure in real time neutrophil-endothelial cell interactions such as chemotaxis, upregulation of adhesion molecules and changes in response to various therapeutic agents (Ren et al., 2022; Man et al., 2023). The ability to use primary human cells in microphysiological systems increases their clinical relevance. For example, Muldur et al. utilized a microfluidic system to examine the distinct effects of different types of Complement 5 (C5) cleavage inhibitors on human neutrophils activities including anti-microbial functions, chemotaxis and swarming (Muldur et al., 2021). Zhang et al. developed a microfluidic system to measure affinity separation to capture the concentration of Cluster of differentiation (CD64)+ cells and provided evidence that neutrophil CD64 expression is upregulated in sepsis patients (Zhang et al., 2018).
The formation of NETs is a critical step in antimicrobial defense through the immobilization of pathogens (Jiao et al., 2020). Microfluidic assays have been used to investigate NETs. For example, Sakuma et al. developed a microfluidic assay to capture and measure NETs in a drop of human blood using immunostaining and fluorescence microscopy (Sakuma et al., 2022). In this system, using as little as 10 uL of blood, without neutrophil or plasma separation, the assay consistently measures the sum of intact and degraded NETs (Sakuma et al., 2022). “Open-well” barrier based microfluidic designs could potentially replicate the compartmentalization found in vivo (Byrne et al., 2017). In these assays, primary human cells can be seeded directly onto the membrane that physically separates the different compartments while permitting soluble factor communication between the cell populations (Byrne et al., 2017). The conventional transwell assay design has been used to study the blood-brain barrier (Stone et al., 2019), alveolar-capillary interface (Janga et al., 2018) and glomerular barrier (Moriyama et al., 2017). However, these transwell model designs lack fluid flow, an important component in the in vivo microenvironments. In order to address this constraint, Mehran et al. integrated laminar flow into the open-well model concept and developed an open-well modular system (m-uSiM) utilizing a silicon membrane (thickness=100nm, porosity=15%), establishing a human endothelial cell layer in the device that could be visualized under a microscope (Mansouri et al., 2022).
Aside from the visualization of the cell-cell interaction, another important advantage of these assays is the ability to study neutrophil behavior and functions in real-time. For example, studies have shown that when using biomimetic microfluidic assays, neutrophil migration (Boribong et al., 2019) and NETosis (Petchakup et al., 2023) can be observed. Furthermore, biomimetic microfluidic assays can be used to observe the process of neutrophils eliminating pathogens such as fungal clusters (Alex et al., 2020) and bacteria (Ellett et al., 2019) in real-time.
Many of the microfluidic devices used for studies of the inflammatory response do not completely reproduce the geometry and structure of microvascular networks observed in vivo. To better model the neutrophil-endothelium interaction during sepsis using primary human cells in a more realistic environment, our group has developed a biomimetic microfluidic assay that reproduces complete microvascular networks on a chip to study the effect of inflammation and sepsis on endothelial permeability and neutrophil-endothelial cell interaction (Yang et al., 2021a). This system can be used to observe the entire leukocyte adhesion cascade including rolling, firm adhesion, spreading and migration of neutrophils into the tissue compartment (Figure 3), mediated by the presence of chemoattractants such as fMLP in the tissue compartment. Neutrophil adhesion and migration could then be measured under physiologically relevant shear stress levels in response to, for example, pro-inflammatory cytokines (Soroush et al., 2016). We have used this microfluidic assay to show that a novel Protein Kinase C-delta (PKCδ) inhibitor not only reduces neutrophil adhesion to and migration across ECs but also decreases the expression of ICAM-1 and other adhesion molecules on ECs during the onset of inflammation (Soroush et al., 2016; Kilpatrick and Kiani, 2020; Soroush et al., 2020). The findings from our microfluidic system have been validated against in vivo observations in sepsis animal models (Soroush et al., 2016). In a rodent model of sepsis, administration of the PKCδ inhibitor significantly decreased sepsis-induced neutrophil influx into the lungs and kidneys (Liverani et al., 2020). Furthermore, PKCδ inhibition was lung protective and decreased sepsis-induced VCAM-1 and ICAM-1 endothelial expression (Mondrinos et al., 2014).
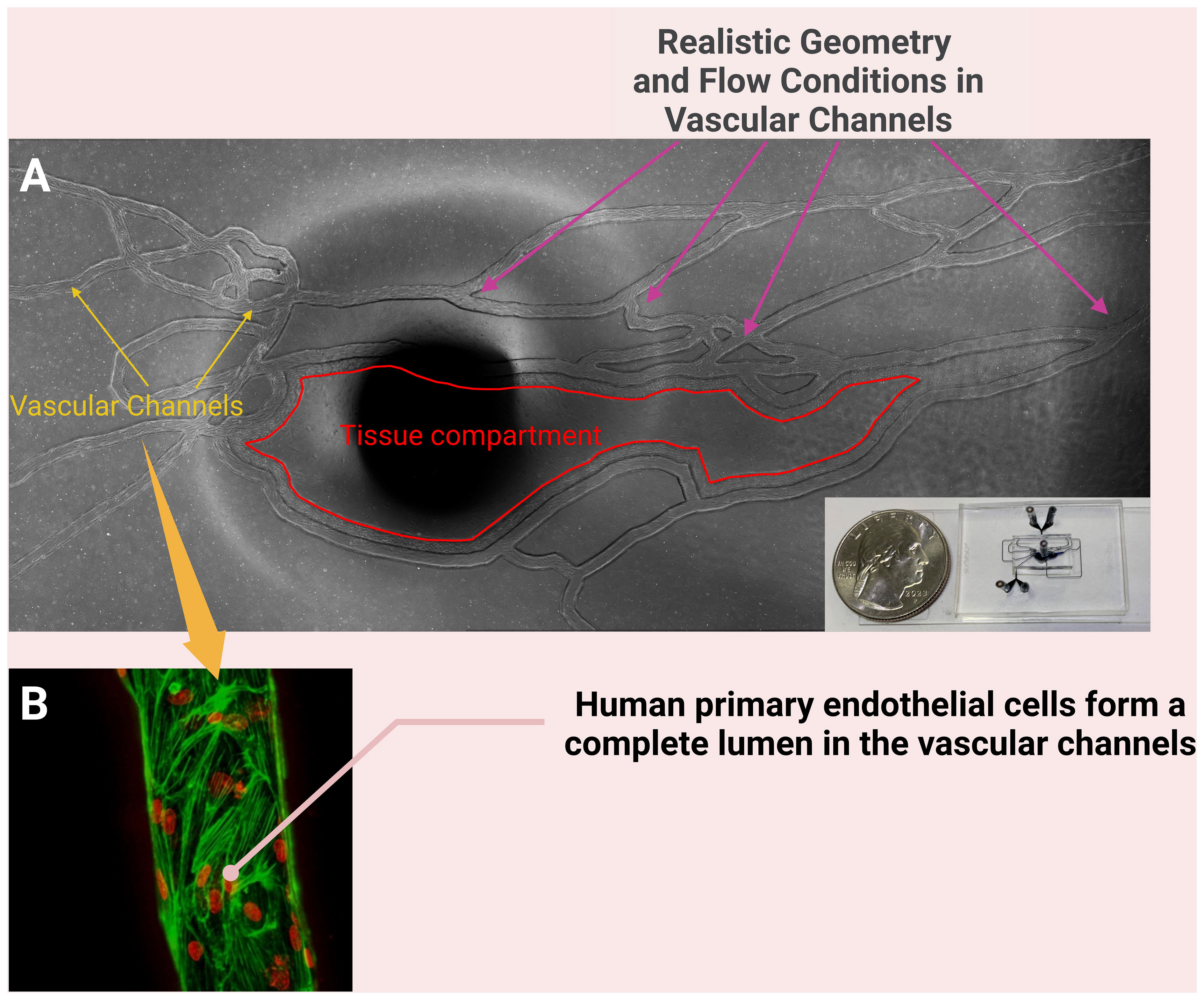
Figure 3 A biomimetic microfluidic assay that reproduces complete microvascular networks on a chip to study the effect of inflammation and sepsis on endothelial permeability and of neutrophil-endothelial cell interaction (A); the dark spot in the middle of (A) is the port for connecting tubing to the tissue compartment. The vascular channels of this system are cultured with primary human endothelial cells (B) and the neutrophil-endothelial cell interaction can be studied under physiologically relevant shear stress levels in the presence of chemoattractants such as fMLP in the tissue compartment. [(B) reproduced with permission (Soroush et al., 2020)].
ECs are heterogeneous, in part due to organ type, and it is therefore critical to develop organ-on-chip assays that realistically represent the conditions in each organ. Our group, for example, has employed a novel blood-brain-barrier on-a-chip (B3C) microfluidic assay to study the role of PKCδ in mediating human brain microvascular endothelial cell (HBMVEC) permeability, junctional protein expression and leukocyte adhesion and migration (Tang et al., 2018). Our findings indicated that TNF-α activates PKCδ and its translocation in HBMVEC (Tang et al., 2018). Inhibition of PKCδ significantly reduced TNF-α mediated hyperpermeability and increased TEER in activated HBMVEC (Tang et al., 2018). Using this B3C microfluidic assay, we demonstrated a more critical role of PKCδ in regulating neutrophil transmigration as compared to neutrophil adhesion, providing important mechanistic insight into the mechanism of action of PKCδ. Our in vitro results agree with our in vivo studies in a rodent model of sepsis where PKCδ inhibition reduced BBB permeability as measured by Evans Blue uptake into the brain (Tang et al., 2018).
It is important to recognize that microphysiological systems, despite their utility in studying EC function, have inherent limitations. While they are often referred to as “organ-on-chip” systems, it is crucial not to interpret this term literally (Yang et al., 2021a). These systems typically involve the co-culture of a limited number of cell types within a predominantly mechanically rigid synthetic or gel scaffold. Consequently, they often lack the presence of other critical in vivo components, such as additional cell types, the extracellular matrix or other components. Experiments using microphysiological systems should therefore be carefully designed, and there is a need for cautious interpretation when extrapolating findings from these models to the more complex dynamics of in vivo systems (Yang et al., 2021b).
Clinical research - sepsis heterogenicity and endotyping of patients
Devising a single standardized treatment for the heterogeneous sepsis patient population has proven problematic, emphasizing the recognition of the need to classify sepsis patients into distinct endotype classes that define specific host response subgroups (Leligdowicz and Matthay, 2019). Therefore, phenotyping sepsis patients in the clinical environment could identify patients for individualized treatment options.
Traditional methods to classify septic patients into distinct endotypes utilizes statistical models such as (k-means) clustering; most of these models suffer from lack of accuracy and/or validation (Li et al., 2022). For example, Li et al. collected 17 published clinical studies that focused on phenotyping adult septic patients (Li et al., 2022). However, only 6 out of 17 studies in the report were given a low risk of bias rating by the authors; the rest of the trials were deemed medium risk (8/17) and high risk of bias (3/17) due to lack of either internal validation or external validation or both (Li et al., 2022). This report implies that the traditional clustering models might not always yield reliable outcome to phenotype sepsis patients. In pediatric sepsis clinical research, Lin et al. suggested classifying new or progressive multiple organ dysfunction syndrome (NPMODS) during the 7-day observation as a distinct phenotype for better clinical outcomes (Lin et al., 2017). The authors report that the hospital mortality in patients with NPMODS was 51% compared with patients with new multiple organ dysfunction syndrome (28%) and those with single-organ dysfunction without multiple organ dysfunction syndrome (10%) (p < 0.001) (Lin et al., 2017). This study provides important clinical insight, but multi-institutional prospective clinical trials are needed for validation. There are many reports suggesting the use of biomarkers to phenotype patients For example, plasma lipoprotein level (Barker et al., 2021; Guirgis et al., 2021), coagulation markers (Barker et al., 2021; Kudo et al., 2021), hypoxia-inducible factor-1 (HIF-1) in macrophages (Peyssonnaux et al., 2007), source of contracting sepsis (Rosales et al., 2023) and vital signs such as hypotension or elevated lactate (Aldewereld et al., 2022) were used as criteria for phenotyping clusters. Most of these studies were done retrospectively using unsupervised clustering approaches. Although they provide important information for future research, prospective clinical trials are needed for validation. Advanced and more clinically relevant phenotyping mechanisms need to be developed for septic patients to receive customized treatment for better clinical outcomes.
Sepsis is an aggressive, heterogeneous disease that requires novel tools to characterize critical stages of the disease in each patient; omics analysis can help discover and characterize sepsis-related biomarkers (Langston et al., 2022). Specifically, omics can help in the identification of a) prognostic biomarkers, b) diagnostic biomarkers and c) biomarkers for personalized patient response to therapeutic intervention (Průcha et al., 2018; Langston et al., 2022). Since one biomarker is unlikely to represent a patient’s comprehensive sepsis condition, a synergistic panel of biomarkers that could satisfy the aforementioned criteria is needed (Průcha et al., 2018). A recent review analyzed 7 biomarkers that were most studied in sepsis: CRP, sTREM-1, LBP, Presepsin, CD64, PCT and IL-6. Results indicated that IL-6, PCT, LBP, sTREM-1 and presepsin (98) had the highest diagnostic accuracy (Teggert et al., 2020). However, limitations of current biomarkers include: prognostic and diagnostic accuracy, variability in concentration during the different stages of sepsis, lack of studies comparing biomarkers and no consensus on diagnostic cut-off values for analysis (Pierrakos et al., 2020; Teggert et al., 2020).
Several studies have classified sepsis patients into endotypes based on modeling methodologies and genome and transcriptome data (Wong et al., 2009; Davenport et al., 2016; Scicluna et al., 2017; Sweeney et al., 2018; Seymour et al., 2019). These studies have correlated the features of each endotype with clinical outcomes, demonstrating the heterogeneity of sepsis in patients. Since sepsis affects a myriad of biological compartments and cell types, omics can interrogate patient-specific genome, transcriptome and proteome expression in cells and tissues and, in combination with clinical and functional studies, decipher how biological pathways are dysregulated (Hasin et al., 2017; Langston et al., 2022). These findings will allow individual, molecular sub-typing of patients and distinguish the mechanisms in each endotype (Itenov et al., 2018; Olivier et al., 2019; Langston et al., 2022). A seminal genome-wide study identified three subclasses, via hierarchical clustering, in pediatric septic shock patients (Wong et al., 2009). Subclass A patients exhibited an immunocompromised presentation (due to repressed gene expression (e.g., Linker of activated T cells (LAT) and T-cell receptor-associated transmembrane adapter 1 (TRAT) in immunity) and thus had the highest mortality compared to subclasses B and C (Wong et al., 2009). Repressed genes were correlated with glucocorticoid and B-cell pathway signaling which further confirms that this group of patients did not have strong immunity (Wong et al., 2009). In a second study, the Molecular Diagnosis and Risk Stratification of Sepsis (MARS) project was designed to evaluate an 8 gene signature in classifying ICU sepsis patients into distinct endotypes (Scicluna et al., 2017). The endotypes generated from this study included MARS 1 (a group of patients showing decreased acquired and innate immunity gene expression), MARS 2 (patients exhibiting increased chemokine expression), MARS 3 (patients presenting increased acquired immunity expression) and MARS 4 (patients displaying increased NF-кB and interferon expression) (Scicluna et al., 2017). When developing a biomarker signature based on omics for distinguishing the heterogeneity of sepsis between patients, it should be simple and sensitive enough for patient classification to be feasible for use at the bedside (Langston et al., 2022). Furthermore, comparing endotype studies is complicated due to the differences in genome/transcriptome expression at various times (Langston et al., 2022). For example, most of the cited studies collected data in the first 48 hours from the time of hospital admission; however, it has been found that half of patients can change endotypes within the first 100 hours of admission (Leligdowicz and Matthay, 2019). Thus, omic expression needs to be carefully tracked as the disease progresses.
Even though the methodologies used to stratify patients into unique endotypes were different, there are similarities across endotypes which may be helpful for future therapeutic research (Langston et al., 2022). Low mortality endotypes [MARS 3 (Scicluna et al., 2017), subclasses B and C (Wong et al., 2009)] had increased immune signaling compared to high mortality endotypes [MARS 1 (Scicluna et al., 2017), subclass A (Wong et al., 2009)] which had repressed immune function (Langston et al., 2022). Overall, endotyping could be used to discover subphenotypes of a disease for tailoring of therapeutics, but it should not be used as a definitive, prognosis tool (Wong et al., 2017). Furthermore, additional, endotype studies are needed to examine their signatures across populations beyond their original intent and to observe if combinations of different endotypes can unravel novel, biological entities between populations leading to clinical presentation (Langston et al., 2022). However, before these advanced studies can be executed, there needs to be standardization by which endotypes are defined and standardized in various studies (Langston et al., 2022). Most importantly, endotype studies must be validated in prospective studies across institutions with sepsis patients who are at different stages of sepsis progression before they can be clinically relevant (Langston et al., 2022).
In silico models – phenotyping using omic methodologies, machine learning and artificial intelligence
Because of the heterogenous nature of sepsis at omic, functional, clinical and temporal levels which results in phenotype differences (e.g., hypoinflammatory vs. hyperinflammatory phenotypes), data incorporated into in silico models are often collected and analyzed from diverse, reputable sources in order to comprehensively capture a patient’s condition. Most importantly, the integration of these sources of data into a model can expedite the drug discovery process and generate novel hypotheses that can be tested experimentally. In silico models offer a promising approach to integrate omics findings with functional measures from innovative tools like microphysiological systems and clinical parameters, enabling the generation of testable hypotheses and expediting the drug discovery process. These models also can unravel complex mechanisms of action that may not be readily apparent through traditional statistical analysis and/or reductive studies (Langston et al., 2022). Multiple modeling methods have been applied to sepsis that are quantitative (e.g., data-driven, multivariate regression, PCA) or qualitative (e.g., network) (Vodovotz and Billiar, 2013). Most models have focused on dysregulation of the immune response (Shi et al., 2015; Stan et al., 2018) but have not identified novel drugs or druggable targets that can be repurposed for sepsis. In silico modeling studies that not only incorporate large sets of data from different repositories but also focus on investigating new drugs or repurposing drugs for sepsis are urgently needed to address this life-threatening condition.
Drug repurposing or repositioning is an emerging area in drug discovery. The focus of the field is to identify novel uses for approved therapeutics that are outside the scope of the original disease (e.g., breast cancer) or indication (e.g., hematology) of interest (Pushpakom et al., 2018). The key advantage of this approach is the low risk of failure since the drug has already been approved for a specific disease/indication and has passed the extensive safety assessments. Furthermore, drug repurposing saves time and costs associated with moving a drug from the bench to the bedside, since on average, de novo drug development is a 10–17-year process with a cost of ~$2 billion. Moreover, the probability of a drug entering the market is below 10%; thus, alternative approaches are needed (Rumienczyk et al., 2022). Drug repurposing in sepsis has not yet reached its full potential, but efforts are underway; this is especially important since all ~150 drugs recently designed to treat sepsis have been successful in rodent models but have failed in clinical trials (Drake, 2013). This absence of clinical translation is due to a host of factors, including the incorporation of rodent models that fail at emulating the complete clinical situation that is present in humans (e.g., age, sex, demographics, comorbidities etc.) and the varied composition of systemic leukocytes in both species (humans have higher numbers of circulating neutrophils compared to mice) (Drake, 2013; Efron et al., 2015). These factors could significantly alter the trajectory of the disease (Langston et al., 2023). A recent review discussed the repurposing of oncology drugs to treat sepsis and identified several potential compounds (e.g., Topotecan, Olaparib, Trametinib) that are currently being evaluated in sepsis models (Rumienczyk et al., 2022). Another review identified several drugs (e.g., Methylthiouracil, Simvastatin, Mangiferin) used in endocrinology and oncology that are being repurposed as possible treatment approaches for sepsis (Prakash et al., 2020). However, there have not been many studies that use in silico modeling and drug repurposing to investigate neutrophil-EC dysregulation in sepsis.
We are using a synergistic combination of omics, in silico modeling and drug repurposing to investigate neutrophil-endothelial interaction in sepsis/inflammatory conditions. Recently, we implemented proteomic analysis of endothelial cells (from lung, liver and kidney) to investigate differences in protein expression levels between organ-specific ECs under inflammatory conditions across time. As time progressed, the number of differentially expressed proteins shared between ECs increased (Rossi et al., 2022). These findings suggest that sepsis progressively affects protein expression in different organ-specific ECs over time. Currently, we are identifying signaling pathways that have a mechanism of action associated with FDA approved drugs for repurposing. Using a combination of functional responses integrated with proteomic and genomic analysis, we will be able to evaluate the feasibility of repurposing these drugs for treating sepsis.
Machine learning (ML), a branch of Artificial intelligence (AI), can be used to improve sepsis diagnosis, prognosis and clinical/drug monitoring of the disease (Langston et al., 2023). For example, Goto et al. utilized a ML approach to identify sepsis patients with distinct phenotypes that could benefit from recombinant human thrombomodulin (rhTM) therapy (Goto et al., 2022). With this methodology, the authors developed a web-based application to identify rhTM target phenotype. Although promising, this model has not been validated in prospective studies (Vincent et al., 2019; Goto et al., 2022). ML has also been used to create a proteomic panel that distinguishes acute respiratory distress syndrome patients from sepsis patients and to predict early sepsis onset and prognosis (Yehya et al., 2022; Moor et al., 2021). However, there have not been any ML studies that specifically investigate neutrophil-endothelial interactions in sepsis and identify repurposed therapeutics which are urgently needed.
To advance the field, it is essential to explore the integration of genomic, transcriptomic, proteomic and metabolomic approaches, since multi-omics holds the potential for improved prognostication and the selection of more suitable treatments (von Groote and Meersch-Dini, 2022). However, the effectiveness of these approaches in improving clinical outcomes needs to be demonstrated through further rigorous clinical studies for validation (Arcaroli et al., 2005). Exciting advancements in omics technology offer promise in enhancing our understanding of disease pathophysiology at an individual patient level, thus facilitating the progression of precision medicine in sepsis and aiding in the development and validation of biomarkers (Davenport et al., 2016). Moreover, adopting more comprehensive tissue interrogation techniques, such as single-nuclei RNA sequencing and single-cell proteomics (Bennett et al., 2023), has the potential to augment biomarker-driven approaches by providing deeper insights into injury mechanisms and the localization of damage (Davenport et al., 2016). These strategies could provide the granularity required to implement a true precision medicine treatment approach for sepsis-related complications, such as acute kidney injury (AKI) (Kiryluk et al., 2018). Given the large volume of data generated from omics studies and the need to synergistically combine and validate this data with those obtained from microphysiological systems and clinical data, in silico models and ML will likely play a critical role in integrating these multiple types of data to allow for a comprehensive understanding of sepsis.
Future directions
In this review, we suggest that leveraging a multidisciplinary approach that incorporates innovative technologies such as microphysiological systems, translational clinical research, and in silico methods incorporating omics, machine learning and artificial intelligence (AI) in sepsis research will enhance our understanding of sepsis and bring about the promise of precisions medicine (Figure 4). Microphysiological systems offer a promising avenue for studying sepsis in vitro by mimicking the physiological conditions of the human body and providing a more accurate representation of the complex interactions between different cell types, tissues and the immune system. Future research studies should aim to optimize and standardize these platforms so that findings from different studies can be compared. This is particularly important as the recent FDA Modernization Act 2.0 now allows for such alternatives to animal testing to bring a drug to human trials (Wadman, 2023). Integration of biomimetic microfluidic assays with advanced imaging techniques and real-time monitoring systems can provide valuable insights into dynamic cellular and molecular changes during sepsis progression.
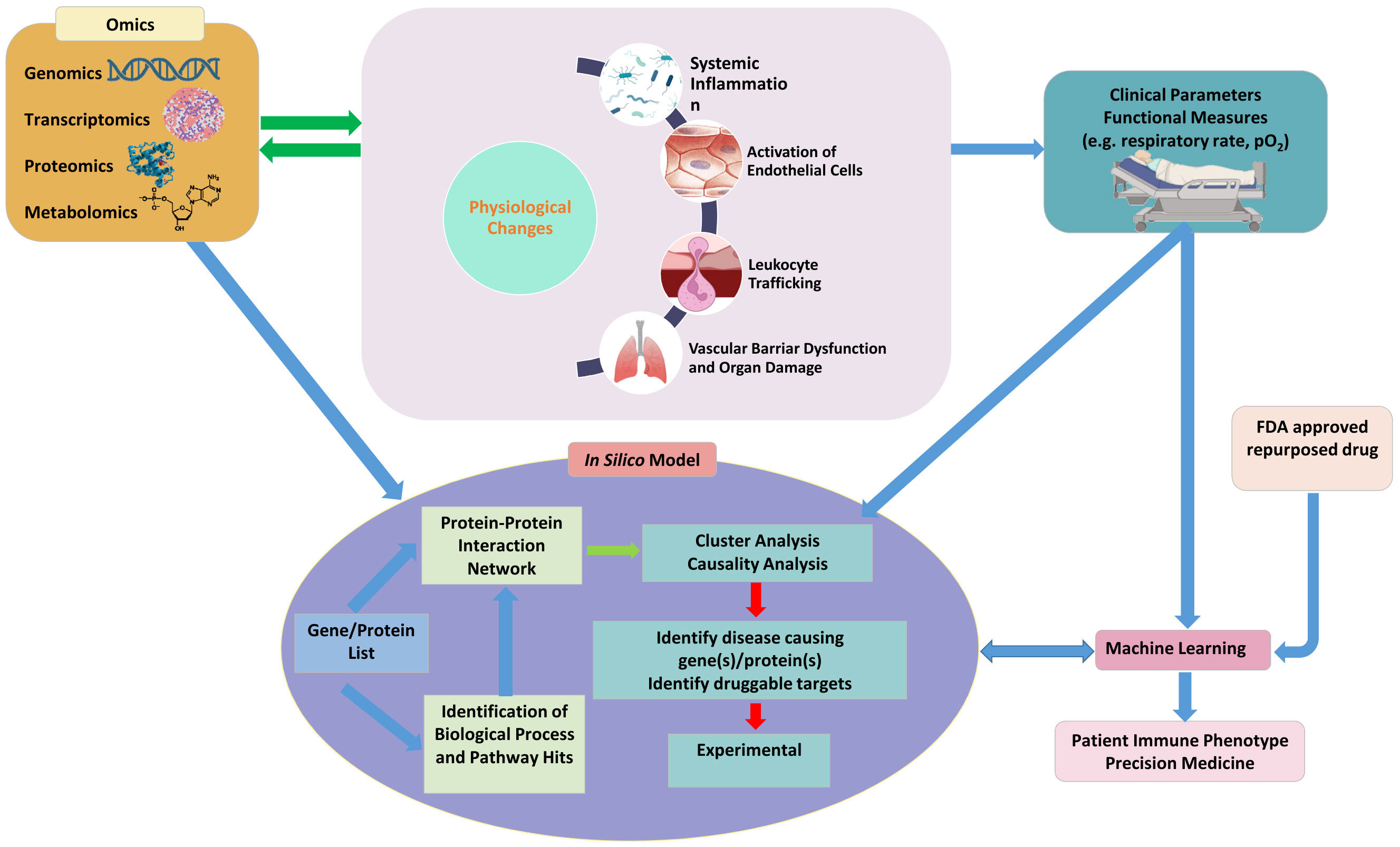
Figure 4 A synergistic platform incorporating emerging technologies such as microphysiological systems to study physiological function, clinical parameters, omics and in silico models combined with machine learning and artificial intelligence will not only provide a better understanding of the pathophysiology of sepsis but also outline a roadmap for precision medicine for treating sepsis.
Complementing in vitro studies with in vivo clinical research remains crucial for translating laboratory findings into clinical applications. Clinical trials that involve large cohorts of sepsis patients can help validate the efficacy and safety of potential treatments. These studies should focus on identifying patient subgroups with distinct sepsis endotypes, allowing for targeted interventions and personalized treatment strategies. In addition, investigating the long-term outcomes of sepsis survivors and identifying potential biomarkers associated with prognosis and response to therapy will be essential.
Furthermore, synergistic incorporation and interpretation of large volumes of diverse data from multiple sources requires the use of emerging techniques such as in silico models and machine learning algorithms which can analyze and interpret vast amounts of data and assist in the prediction of disease progression, patient outcomes and treatment responses. These models can provide a holistic view of the dynamics of sepsis leading to a deeper understanding of its pathophysiology, facilitating the development of novel therapeutics which can ultimately improve patient care.
Author contributions
DL: Visualization, Writing – original draft, Writing – review & editing. JL: Writing – original draft, Writing – review & editing. BP: Writing – original draft. MK: Conceptualization, Supervision, Validation, Writing – review & editing, Funding acquisition, Visualization. LK: Conceptualization, Funding acquisition, Supervision, Validation, Visualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. JL is an NIH NRSA F31 Predoctoral Fellow (1-F31AI164870-01). This work was supported by the Defence Threat Reduction Agency (HDTRA11910012).
Conflict of interest
Author BP was employed by the company CFD Research Corporation.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
References
Aird, W. C. (2012). Endothelial cell heterogeneity. Cold Spring Harbor Perspect. Med. 2 (1), a006429. doi: 10.1101/cshperspect.a006429
Aldewereld, Z. T., Zhang, L. A., Urbano, A., Parker, R. S., Swigon, D., Banerjee, I., et al. (2022). Identification of clinical phenotypes in septic patients presenting with hypotension or elevated lactate. Front. Med. 9. doi: 10.3389/fmed.2022.794423
Alex, H., Scherer, A., Kreuzburg, S., Abers, M. S., Zerbe, C. S., Dinauer, M. C., et al. (2020). Neutrophil swarming delays the growth of clusters of pathogenic fungi. Nat. Commun. 11, 1. doi: 10.1038/s41467-020-15834-4
Amalakuhan, B., Habib, S. A., Mangat, M., Reyes, L. F., Rodriguez, A. H., Hinojosa, C. A., et al. (2016). Endothelial adhesion molecules and multiple organ failure in patients with severe sepsis. Cytokine 88, 267–273. doi: 10.1016/j.cyto.2016.08.028
Arcaroli, J., Fessler, M. B., Abraham, E. (2005). Genetic polymorphisms and sepsis. Shock 24 (4), 300–312. doi: 10.1097/01.shk.0000180621.52058.e1
Aslan, A., Van Meurs, M., Moser, J., Popa, E. R., Jongman, R. M., Zwiers, P. J., et al. (2017). Organ-specific differences in endothelial permeability-regulating molecular responses in mouse and human sepsis. Shock 48 (1), 69–77. doi: 10.1097/SHK.0000000000000841
Barker, G., Leeuwenburgh, C., Brusko, T., Moldawer, L., Reddy, S. T., Guirgis, F. W. (2021). Review lipid and lipoprotein dysregulation in sepsis: Clinical and mechanistic insights into chronic critical illness. J. Clin. Med. 10 (8), 1693. doi: 10.3390/jcm10081693
Bennett, H. M., Stephenson, W., Rose, C. M., Darmanis, S. (2023). Single-cell proteomics enabled by next-generation sequencing or mass spectrometry. Nat. Methods 20 (3), 363–374. doi: 10.1038/s41592-023-01791-5
Boribong, B. P., Lenzi, M. J., Li, L., Jones, C. N. (2019). Super-low dose lipopolysaccharide dysregulates neutrophil migratory decision-making. Front. Immunol. 10. doi: 10.3389/fimmu.2019.00359
Borregaard, N. (2010). Neutrophils, from marrow to microbes. Immunity 33 (5), 657–670. doi: 10.1016/j.immuni.2010.11.011
Bosáková, V., De Zuani, M., Sládková, L., Garlíková, Z., Jose, S. S., Zelante, T., et al. (2022). Lung organoids—The ultimate tool to dissect pulmonary diseases? Front. Cell Dev. Biol 10:899368. doi: 10.3389/fcell.2022.899368
Byrne, M. B., Leslie, M. T., Patel, H. S., Gaskins, H. R., Kenis, P. J. A. (2017). Design considerations for open-well microfluidic platforms for hypoxic cell studies. Biomicrofluidics 11 (5), 054116. doi: 10.1063/1.4998579
Chang, J. C. (2019). Sepsis and septic shock: Endothelial molecular pathogenesis associated with vascular microthrombotic disease. Thromb. J. 17 (1), 10. doi: 10.1186/s12959-019-0198-4
Chi, J.-T., Chang, H. Y., Haraldsen, G., Jahnsen, F. L., Troyanskaya, O. G., Chang, D. S., et al. (2003). Endothelial cell diversity revealed by global expression profiling. Proc Nat Acad Sci USA 20, 197–210. doi: 10.1038/s41569-022-00770-1
Chistiakov, D. A., Orekhov, A. N., Bobryshev, Y. V. (2015). Endothelial barrier and its abnormalities in cardiovascular disease. Front. Physiol. 6. doi: 10.3389/fphys.2015.00365
Davenport, E. E., Burnham, K. L., Radhakrishnan, J., Humburg, P., Hutton, P., Mills, T. C., et al. (2016). Genomic landscape of the individual host response and outcomes in sepsis: A prospective cohort study. Lancet Respir. Med. 4 (4), 259–271. doi: 10.1016/S2213-2600(16)00046-1
Dejager, L., Pinheiro, I., Dejonckheere, E., Libert, C. (2011). Cecal ligation and puncture: The gold standard model for polymicrobial sepsis? Trends Microbiol. 19 (4), 198–208. doi: 10.1016/j.tim.2011.01.001
Donnelly, J. P., Safford, M. M., Shapiro, N. I., Baddley, J. W., Wang, H. E. (2017). Application of the Third International Consensus Definitions for Sepsis (Sepsis-3) Classification: a retrospective population-based cohort study. Lancet Infect. Dis. 17 (6), 661–670. doi: 10.1016/S1473-3099(17)30117-2
Döring, Y., Soehnlein, O., Weber, C. (2017). Neutrophil extracellular traps in atherosclerosis and atherothrombosis. Circ. Res. 120 (4), 736–743. doi: 10.1161/CIRCRESAHA.116.309692
Drake, A. C. (2013). Of mice and men: What rodent models don’t tell us. Cell. Mol. Immunol. 10 (4), 284–285. doi: 10.1038/cmi.2013.21
Duong, C. N., Vestweber, D. (2020). Mechanisms ensuring endothelial junction integrity beyond VE-cadherin. Front. Physiol. 11. doi: 10.3389/fphys.2020.00519
Efron, P. A., Mohr, A. M., Moore, F. A., Moldawer, L. L. (2015). The future of murine sepsis and trauma research models. J. Leukoc. Biol. 98 (6), 945–952. doi: 10.1189/jlb.5mr0315-127r
Ellett, F., Jalali, F., Marand, A. L., Jorgensen, J., Mutlu, B. R., Lee, J., et al. (2019). Microfluidic arenas for war games between neutrophils and microbes. Lab. Chip 19 (7), 1205–1216. doi: 10.1039/c8lc01263f
Fatmi, A., Saadi, W., Beltrán-García, J., García-Giménez, J. L., Pallardó, F. V. (2022). The endothelial glycocalyx and neonatal sepsis. Int. J. Mol. Sci. 24 (1), 364. doi: 10.3390/ijms24010364
Fenner, B. P., Darden, D. B., Kelly, L. S., Rincon, J., Brakenridge, S. C., Larson, S. D., et al. (2021). Immunological endotyping of chronic critical illness after severe sepsis. Front. Med. 7. doi: 10.3389/fmed.2020.616694
Fox, S., Leitch, A. E., Duffin, R., Haslett, C., Rossi, A. G. (2010). Neutrophil apoptosis: Relevance to the innate immune response and inflammatory disease. J. Innate Immun. 2 (3), 216–227. doi: 10.1159/000284367
Futosi, K., Fodor, S., Mócsai, A. (2013). Neutrophil cell surface receptors and their intracellular signal transduction pathways. Int. Immunopharmacol. 17 (3), 638–650. doi: 10.1016/j.intimp.2013.06.034
Georgescu, A. M., Banescu, C., Azamfirei, R., Hutanu, A., Moldovan, V., Badea, I., et al. (2020). Evaluation of TNF-α genetic polymorphisms as predictors for sepsis susceptibility and progression. BMC Infect. Dis. 20 (1), 221. doi: 10.1186/s12879-020-4910-6
Giannini, H. M., Ginestra, J. C., Chivers, C., Draugelis, M., Hanish, A., Schweickert, W. D., et al. (2019). A machine learning algorithm to predict severe sepsis and septic shock: development, implementation, and impact on clinical practice. Crit. Care Med. 47 (11), 1485–1492. doi: 10.1097/CCM.0000000000003891
Glémain, A., Néel, M., Néel, A., André-Grégoire, G., Gavard, J., Martinet, B., et al. (2022). Neutrophil-derived extracellular vesicles induce endothelial inflammation and damage through the transfer of miRNAs. J. Autoimmun. 129, 102826. doi: 10.1016/j.jaut.2022.102826
Goto, T., Kudo, D., Uchimido, R., Hayakawa, M., Yamakawa, K., Abe, T., et al. (2022). Web-based application for predicting the potential target phenotype for recombinant human thrombomodulin therapy in patients with sepsis: analysis of three multicentre registries. Crit. Care 26 (1), 145. doi: 10.1186/s13054-022-04020-1
Guimarães-Costa, A. B., DeSouza-Vieira, T. S., Paletta-Silva, R., Freitas-Mesquita, A. L., Meyer-Fernandes, J. R., Saraiva, E. M. (2014). 3’-nucleotidase/nuclease activity allows Leishmania parasites to escape killing by neutrophil extracellular traps. Infect. Immun. 82 (4), 1732–1740. doi: 10.1128/IAI.01232-13
Guirgis, F. W., Black, L. P., Henson, M., Labilloy, G., Smotherman, C., Hopson, C., et al. (2021). A hypolipoprotein sepsis phenotype indicates reduced lipoprotein antioxidant capacity, increased endothelial dysfunction and organ failure, and worse clinical outcomes. Crit. Care 25 (1), 341. doi: 10.1186/s13054-021-03757-5
Hagai, T., Chen, X., Miragaia, R. J., Rostom, R., Gomes, T., Kunowska, N., et al. (2018). Gene expression variability across cells and species shapes innate immunity. Nature 563 (7730), 197–202. doi: 10.1038/s41586-018-0657-2
Hasin, Y., Seldin, M., Lusis, A. (2017). Multi-omics approaches to disease. Genome Biol. 18 (1), 83. doi: 10.1186/s13059-017-1215-1
Herrero-Cervera, A., Soehnlein, O., Kenne, E. (2022). Neutrophils in chronic inflammatory diseases. Cell. Mol. Immunol. 19 (2), 177–191. doi: 10.1038/s41423-021-00832-3
Honoré, A., Forsberg, D., Adolphson, K., Chatterjee, S., Jost, K., Herlenius, E. (2023). Vital sign-based detection of sepsis in neonates using machine learning. Acta Paediatrica 112 (4), 686–696. doi: 10.1111/apa.16660
Iba, T., Levi, M., Levy, J. H. (2022). Intracellular communication and immunothrombosis in sepsis. J. Thromb. Haemost. 20 (11), 2475–2484. doi: 10.1111/jth.15852
Iba, T., Ogura, H. (2018). Role of extracellular vesicles in the development of sepsis-induced coagulopathy. J. Intensive Care 6 (1), 68. doi: 10.1186/s40560-018-0340-6
Iregbu, K., Dramowski, A., Milton, R., Nsutebu, E., Howie, S. R. C., Chakraborty, M., et al. (2022). Global health systems’ data science approach for precision diagnosis of sepsis in early life. Lancet Infect. Dis. 22 (5), e143–e152. doi: 10.1016/S1473-3099(21)00645-9
Itenov, T. S., Murray, D. D., Jensen, J. U. S. (2018). Sepsis: Personalized medicine utilizing ‘omic’ technologies-a paradigm shift? Healthcare 6 (3), 111. doi: 10.3390/healthcare6030111
Jarczak, D., Kluge, S., Nierhaus, A. (2021). Sepsis-pathophysiology and therapeutic concepts. Front. Med. 8. doi: 10.3389/fmed.2021.628302
Janga, H., Cassidy, L., Wang, F., Spengler, D., Oestern-Fitschen, S., Krause, M. F., et al. (2018). Site-specific and endothelial-mediated dysfunction of the alveolar-capillary barrier in response to lipopolysaccharides. J. Cell. Mol. Med. 22 (2), 982–998. doi: 10.1111/jcmm.13421
Jiao, Y., Li, W., Wang, W., Tong, X., Xia, R., Fan, J., et al. (2020). Platelet-derived exosomes promote neutrophil extracellular trap formation during septic shock. Crit. Care 24 (1), 380. doi: 10.1186/s13054-020-03082-3
Kell, D. B., Pretorius, E. (2018). To what extent are the terminal stages of sepsis, septic shock, systemic inflammatory response syndrome, and multiple organ dysfunction syndrome actually driven by a prion/amyloid form of fibrin? Semin. Thromb. Hemost. 44 (3), 224–238. doi: 10.1055/s-0037-1604108
Kilpatrick, L. E., Kiani, M. F. (2020). Experimental approaches to evaluate leukocyte-endothelial cell interactions in sepsis and inflammation. Shock 53 (5), 585–595. doi: 10.1097/SHK.0000000000001407
Kiryluk, K., Bomback, A. S., Cheng, Y. L., Xu, K., Camara, P. G., Rabadan, R., et al. (2018). Precision medicine for acute kidney injury (AKI): redefining AKI by agnostic kidney tissue interrogation and genetics. Semin. Nephrol. 38 (1), 40–51. doi: 10.1016/j.semnephrol.2017.09.006
Kojok, K., El Akoum, S., Mohsen, M., Mourad, W., Merhi, Y. (2018). CD40L priming of platelets via NF-κB activation is CD40-and TAK1-dependent. J. Am. Heart Assoc. 7 (23), e03677. doi: 10.1161/JAHA.118.009636
Kondo, N., Ogawa, M., Wada, H., Nishikawa, S. I. (2009). Thrombin induces rapid disassembly of claudin-5 from the tight junction of endothelial cells. Exp. Cell Res. 315 (17), 2879–2887. doi: 10.1016/j.yexcr.2009.07.031
Kudo, D., Goto, T., Uchimido, R., Hayakawa, M., Yamakawa, K., Abe, T., et al. (2021). Coagulation phenotypes in sepsis and effects of recombinant human thrombomodulin: an analysis of three multicentre observational studies. Crit. Care 25 (1), 114. doi: 10.1186/s13054-021-03541-5
Lang, F., Abed, M., Lang, E., Föller, M. (2014). Oxidative stress and suicidal erythrocyte death. Antioxid. Redox Signaling 21 (1), 138–153. doi: 10.1089/ars.2013.5747
Langston, J. C., Rossi, M. T., Yang, Q., Ohley, W., Perez, E., Kilpatrick, L. E., et al. (2022). Omics of endothelial cell dysfunction in sepsis. Vasc. Biol. 4 (1), R15–R34. doi: 10.1530/vb-22-0003
Langston, J. C., Yang, Q., Kiani, M. F., Kilpatrick, L. E. (2023). Leukocyte phenotyping in sepsis using omics, functional analysis, and in silico modelling. Shock 59 (2), 224–231. doi: 10.1097/SHK.0000000000002047
Lee, J. H., Bhang, D. H., Beede, A., Huang, T. L., Stripp, B. R., Bloch, K. D., et al. (2014). Lung stem cell differentiation in mice directed by endothelial cells via a BMP4-NFATc1-thrombospondin-1 axis. Cell 156 (3), 440–455. doi: 10.1016/j.cell.2013.12.039
Leligdowicz, A., Matthay, M. A. (2019). Heterogeneity in sepsis: New biological evidence with clinical applications. Crit. Care 23 (1), 80. doi: 10.1186/s13054-019-2372-2
Ley, K., Laudanna, C., Cybulsky, M. I., Nourshargh, S. (2007). Getting to the site of inflammation: The leukocyte adhesion cascade updated. Nat. Rev. Immunol. 7 (9), 678–689. doi: 10.1038/nri2156
Li, H., Markal, A., Balch, J. A., Loftus, T. J., Efron, P. A., Ozrazgat-Baslanti, T., et al. (2022). Methods for phenotyping adult patients in sepsis and septic shock: A scoping review. Crit. Care Explor. 4 (4), e0672. doi: 10.1097/cce.0000000000000672
Libby, P., Ridker, P. M., Hansson, G. K. (2011). Progress and challenges in translating the biology of atherosclerosis. Nature 473 (7347), 317–325. doi: 10.1038/nature10146
Lim, J. Y., Lee, J. E., Kim, H. K., Park, Y. J., Jeon, J. H., Park, S. A., et al. (2022). Human palatine tonsils are linked to alzheimer’s disease through function of reservoir of amyloid beta protein associated with bacterial infection. Cells 11 (15), 2285. doi: 10.3390/cells11152285
Lin, J. C., Spinella, P. C., Fitzgerald, J. C., Tucci, M., Bush, J. L., Nadkarni, V. M., et al. (2017). New or progressive multiple organ dysfunction syndrome in pediatric severe sepsis: A sepsis phenotype with higher morbidity and mortality. Pediatr. Crit. Care Med. 18 (1), 8–16. doi: 10.1097/PCC.0000000000000978
Liverani, E., Tursi, S. A., Cornwell, W. D., Mondrinos, M. J., Sun, S., Buttaro, B. A., et al. (2020). Protein kinase C-delta inhibition is organ-protective, enhances pathogen clearance, and improves survival in sepsis. FASEB J. 34 (2), 2497–2510. doi: 10.1096/fj.201900897R
Mai, J., Virtue, A., Shen, J., Wang, H., Yang, X. F. (2013). An evolving new paradigm: Endothelial cells - Conditional innate immune cells. J. Hematol. Oncol. 6 (1), 61. doi: 10.1186/1756-8722-6-61
Man, Y., Kucukal, E., Liu, S., An, R., Goreke, U., Wulftange, W. J., et al. (2023). A microfluidic device for assessment of E-selectin-mediated neutrophil recruitment to inflamed endothelium and prediction of therapeutic response in sickle cell disease. Biosens. Bioelectron. 222, 114921. doi: 10.1016/j.bios.2022.114921
Mansouri, M., Ahmed, A., Ahmad, S. D., McCloskey, M. C., Joshi, I. M., Gaborski, T. R., et al. (2022). The modular µSiM reconfigured: integration of microfluidic capabilities to study in vitro barrier tissue models under flow. Adv. Healthc. Mater. 11 (21), e2200802. doi: 10.1002/adhm.202200802
Mondrinos, M. J., Zhang, T., Sun, S., Kennedy, P. A., King, D. J., Wolfson, M. R., et al. (2014). Pulmonary endothelial protein kinase C-Delta (PKCδ) regulates neutrophil migration in acute lung inflammation. Am. J. Pathol. 184 (1), 200–213. doi: 10.1016/j.ajpath.2013.09.010
Moor, M., Rieck, B., Horn, M., Jutzeler, C. R., Borgwardt, K. (2021). Early prediction of sepsis in the ICU using machine learning: A systematic review. Front. Med. 8. doi: 10.3389/fmed.2021.607952
Moriyama, T., Sasaki, K., Karasawa, K., Uchida, K., Nitta, K. (2017). Intracellular transcytosis of albumin in glomerular endothelial cells after endocytosis through caveolae. J. Cell. Physiol. 232 (12), 3565–3573. doi: 10.1002/jcp.25817
Muldur, S., Vadysirisack, D. D., Ragunathan, S., Tang, Y., Ricardo, A., Sayegh, C. E., et al. (2021). Human neutrophils respond to complement activation and inhibition in microfluidic devices. Front. Immunol. 12. doi: 10.3389/fimmu.2021.777932
Niederman, M. S., Baron, R. M., Bouadma, L., Calandra, T., Daneman, N., DeWaele, J., et al. (2021). Initial antimicrobial management of sepsis. Crit. Care 25 (1), 307. doi: 10.1186/s13054-021-03736-w
Nourshargh, S., Alon, R. (2014). Leukocyte migration into inflamed tissues. Immunity 41 (5), 694–707. doi: 10.1016/j.immuni.2014.10.008
Olivier, M., Asmis, R., Hawkins, G. A., Howard, T. D., Cox, L. A. (2019). The need for multi-omics biomarker signatures in precision medicine. Int. J. Mol. Sci. 20 (19), 4781. doi: 10.3390/ijms20194781
Osborn, T. M. (2017). Severe sepsis and septic shock trials (ProCESS, ARISE, proMISe): what is optimal resuscitation? Crit. Care Clinics 33 (2), 323–344. doi: 10.1016/j.ccc.2016.12.004
Pan, X., Xie, J., Zhang, L., Wang, X., Zhang, S., Zhuang, Y., et al. (2023). Evaluate prognostic accuracy of SOFA component score for mortality among adults with sepsis by machine learning method. BMC Infect. Dis. 23 (1), 76. doi: 10.1186/s12879-023-08045-x
Perobelli, S. M., Galvani, R. G., Gonçalves-Silva, T., Xavier, C. R., Nóbrega, A., Bonomo, A. (2015). Plasticity of neutrophils reveals modulatory capacity. Braz. J. Med. Biol. Res. 48 (8), 665–675. doi: 10.1590/1414-431X20154524
Petchakup, C., Wong, S. O., Dalan, R., Hou, H. W. (2023). Label-free virtual staining of neutrophil extracellular traps (NETs) in microfluidics. Lab. Chip 23:3936–44. doi: 10.1039/d3lc00398a
Peyssonnaux, C., Cejudo-Martin, P., Doedens, A., Zinkernagel, A. S., Johnson, R. S., Nizet, V. (2007). Cutting edge: essential role of hypoxia inducible factor-1α in development of lipopolysaccharide-induced sepsis. J. Immunol. 178 (12), 7516–7519. doi: 10.4049/jimmunol.178.12.7516
Pierrakos, C., Velissaris, D., Bisdorff, M., Marshall, J. C., Vincent, J. L. (2020). Biomarkers of sepsis: Time for a reappraisal. Crit. Care 24 (1), 287. doi: 10.1186/s13054-020-02993-5
Ploppa, A., Kampmann, M., Johannes, T., Haeberle, H. A., Nohé, B. (2012). Effects of different leukocyte subpopulations and flow conditions on leukocyte accumulation during reperfusion. J. Vasc. Res. 49 (2), 169–180. doi: 10.1159/000335147
Prakash, A. V., Park, J. W., Seong, J. W., Kang, T. J. (2020). Repositioned drugs for inflammatory diseases such as sepsis, asthma, and atopic dermatitis. Biomol. Ther. 28 (3), 222–229. doi: 10.4062/biomolther.2020.001
Průcha, M., Zazula, R., Russwurm, S. (2018). Sepsis diagnostics in the era of “Omics“ Technologies. Prague Med. Rep. 119 (1), 9–29. doi: 10.14712/23362936.2018.2
Puchwein-Schwepcke, A., Genzel-Boroviczény, O., Nussbaum, C. (2021). The endothelial glycocalyx: physiology and pathology in neonates, infants and children. Front. Cell Dev. Biol. 9. doi: 10.3389/fcell.2021.733557
Pushpakom, S., Iorio, F., Eyers, P. A., Escott, K. J., Hopper, S., Wells, A., et al. (2018). Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discovery 18 (1), 41–58. doi: 10.1038/nrd.2018.168
Ren, J., Wang, N., Guo, P., Fan, Y., Lin, F., Wu, J. (2022). Recent advances in microfluidics-based cell migration research. Lab. Chip 22 (18), 3361–3376. doi: 10.1039/d2lc00397j
Rhee, C., Dantes, R., Epstein, L., Murphy, D. J., Seymour, C. W., Iwashyna, T. J., et al. (2017). Incidence and trends of sepsis in US hospitals using clinical vs claims data 2009-2014. JAMA J. Am. Med. Assoc. 318 (13), 1241–1249. doi: 10.1001/jama.2017.13836
Rosales, C. (2018). Neutrophil: A cell with many roles in inflammation or several cell types? Front. Physiol. 9. doi: 10.3389/fphys.2018.00113
Rosales, C. (2020). Neutrophils at the crossroads of innate and adaptive immunity. J. Leukoc. Biol. 108 (1), 377–396. doi: 10.1002/JLB.4MIR0220-574RR
Rosales, J., Ireland, M., Gonzalez-Gallo, K., Wisler, J., Jalilvand, A. (2023). Characterization of mortality by sepsis source in patients admitted to the surgical intensive care unit. J. Surg. Res. 283, 1117–1123. doi: 10.1016/j.jss.2022.10.096
Rossi, M. T., Langston, J. C., Singh, N., Merali, C., Yang, Q., Merali, S., et al. (2022). Molecular framework of mouse endothelial cell dysfunction during inflammation: A proteomics approach. Int. J. Mol. Sci. 23 (15), 8399. doi: 10.3390/ijms23158399
Rudd, K. E., Johnson, S. C., Agesa, K. M., Shackelford, K. A., Tsoi, D., Kievlan, D. R., et al. (2020). Global, regional, and national sepsis incidence and mortality 1990–2017: analysis for the global burden of disease study. Lancet 395 (10219), 200–211. doi: 10.1016/S0140-6736(19)32989-7
Rumienczyk, I., Kulecka, M., Statkiewicz, M., Ostrowski, J., Mikula, M. (2022). Oncology drug repurposing for sepsis treatment. Biomedicines 10 (4), 921. doi: 10.3390/biomedicines10040921
Sakuma, M., Wang, X., Ellett, F., Edd, J. F., Babatunde, K. A., Viens, A., et al. (2022). Microfluidic capture of chromatin fibres measures neutrophil extracellular traps (NETs) released in a drop of human blood. Lab. Chip 22 (5), 936–944. doi: 10.1039/d1lc01123e
Schmidt, E. P., Yang, Y., Janssen, W. J., Gandjeva, A., Perez, M. J., Barthel, L., et al. (2012). The pulmonary endothelial glycocalyx regulates neutrophil adhesion and lung injury during experimental sepsis. Nat. Med. 18 (8), 1217–1223. doi: 10.1038/nm.2843
Scicluna, B. P., van Vught, L. A., Zwinderman, A. H., Wiewel, M. A., Davenport, E. E., Burnham, K. L., et al. (2017). Classification of patients with sepsis according to blood genomic endotype: a prospective cohort study. Lancet Respir. Med. 5 (10), 816–826. doi: 10.1016/S2213-2600(17)30294-1
Seitz, K. P., Qian, E. T., Semler, M. W. (2022). Intravenous fluid therapy in sepsis. Nutr. Clin. Pract. 37 (5), 990–1003. doi: 10.1002/ncp.10892
Seok, J., Warren, H. S., Alex, G. C., Michael, N. M., Henry, V. B., Xu, W., et al. (2013). Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. U States America 110 (9), 3507–3512. doi: 10.1073/pnas.1222878110
Seymour, C. W., Kennedy, J. N., Wang, S., Chang, C. C. H., Elliott, C. F., Xu, Z., et al. (2019). Derivation, validation, and potential treatment implications of novel clinical phenotypes for sepsis. JAMA J. Am. Med. Assoc. 321 (20), 2003–2017. doi: 10.1001/jama.2019.5791
Shakoory, B., Carcillo, J. A., Chatham, W. W., Amdur, R. L., Zhao, H., Dinarello, C. A., et al. (2016). Interleukin-1 receptor blockade is associated with reduced mortality in sepsis patients with features of macrophage activation syndrome: reanalysis of a prior phase III trial∗. Crit. Care Med. 44 (2), 275–281. doi: 10.1097/CCM.0000000000001402
Shetty, S., Lalor, P. F., Adams, D. H. (2018). Liver sinusoidal endothelial cells - gatekeepers of hepatic immunity. Nat Rev Gastroenterol Hepatol. 15(9), 555–567. doi: 10.1038/s41575-018-0020-y
Shi, Z., Wu, C. H. J., Ben-Arieh, D., Simpson, S. Q. (2015). Mathematical model of innate and adaptive immunity of sepsis: A modeling and simulation study of infectious disease. BioMed. Res. Int. 2015, 504259. doi: 10.1155/2015/504259
Singer, M., Deutschman, C. S., Seymour, C., Shankar-Hari, M., Annane, D., Bauer, M., et al. (2016). The third international consensus definitions for sepsis and septic shock (sepsis-3). JAMA J. Am. Med. Assoc. 315 (8), 801–810. doi: 10.1001/jama.2016.0287
Siwczak, F., Loffet, E., Kaminska, M., Koceva, H., Mahe, M. M., Mosig, A. S. (2021). Intestinal stem cell-on-chip to study human host-microbiota interaction. Front. Immunol. 12. doi: 10.3389/fimmu.2021.798552
Soroush, F., Tang, Y., Mustafa, O., Sun, S., Yang, Q., Kilpatrick, L. E., et al. (2020). Neutrophil-endothelial interactions of murine cells is not a good predictor of their interactions in human cells. FASEB J. 34 (2), 2691–2702. doi: 10.1096/fj.201900048R
Soroush, F., Zhang, T., King, D. J., Tang, Y., Deosarkar, S., Prabhakarpandian, B., et al. (2016). A novel microfluidic assay reveals a key role for protein kinase C δ in regulating human neutrophil–endothelium interaction. J. Leukoc. Biol. 100 (5), 1027–1035. doi: 10.1189/jlb.3ma0216-087r
Sriram, G., Tan, J. Y., Islam, I., Rufaihah, A. J., Cao, T. (2015). Efficient differentiation of human embryonic stem cells to arterial and venous endothelial cells under feeder- and serum-free conditions. Stem Cell Res. Ther. 6 (1), 261. doi: 10.1186/s13287-015-0260-5
Stan, R. C., Soriano, F. G., De Camargo, M. M. (2018). A mathematical model relates intracellular TLR4 oscillations to sepsis progression. BMC Res. Notes 11 (1), 462. doi: 10.1186/s13104-018-3561-9
Stolarski, A. E., Kim, J., Nudel, J., Gunn, S., Remick, D. G. (2022). Defining sepsis phenotypes - two murine models of sepsis and machine learning. Shock 57 (6), 268–273. doi: 10.1097/SHK.0000000000001935
Stone, N. L., England, T. J., O’Sullivan, S. E. (2019). A novel transwell blood brain barrier model using primary human cells. Front. Cell. Neurosci. 13. doi: 10.3389/fncel.2019.00230
Sweeney, T. E., Azad, T. D., Donato, M., Haynes, W. A., Perumal, T. M., Henao, R., et al. (2018). Unsupervised analysis of transcriptomics in bacterial sepsis across multiple datasets reveals three robust clusters. Crit. Care Med. 46 (6), 915–925. doi: 10.1097/CCM.0000000000003084
Sznajder, J. I., Brochard, L., Scott Budinger, G. R., Erzurum, S. C., Israel, E., Schumacker, P. (2013). Humanizing the mouse: in defense of murine models of critical illness. Am. J. Respir. Crit. Care Med. 187 (9), 897–898. doi: 10.1164/rccm.201303-0409ED
Tang, Y., Soroush, F., Sun, S., Liverani, E., Langston, J. C., Yang, Q., et al. (2018). Protein kinase C-delta inhibition protects blood-brain barrier from sepsis-induced vascular damage. J. Neuroinflamm. 15 (1), 309. doi: 10.1186/s12974-018-1342-y
Tecchio, C., Micheletti, A., Cassatella, M. A. (2014). Neutrophil-derived cytokines: Facts beyond expression. Front. Immunol. 5. doi: 10.3389/fimmu.2014.00508
Teggert, A., Datta, H., Ali, Z. (2020). Biomarkers for point-of-care diagnosis of sepsis. Micromachines 11 (3), 286. doi: 10.3390/mi11030286
Uchimido, R., Schmidt, E. P., Shapiro, N. I. (2019). The glycocalyx: A novel diagnostic and therapeutic target in sepsis. Crit. Care 23 (1), 16. doi: 10.1186/s13054-018-2292-6
Vincent, J. L., Francois, B., Zabolotskikh, I., Daga, M. K., Lascarrou, J. B., Kirov, M. Y., et al. (2019). Effect of a recombinant human soluble thrombomodulin on mortality in patients with sepsis-associated coagulopathy: the SCARLET randomized clinical trial. JAMA J. Am. Med. Assoc. 321 (20), 1993–2002. doi: 10.1001/jama.2019.5358
Vodovotz, Y., Billiar, T. R. (2013). In silico modeling: Methods and applications to trauma and sepsis. Crit. Care Med. 41 (8), 2008–2014. doi: 10.1097/CCM.0b013e31829a6eb4
von Groote, T., Meersch-Dini, M. (2022). Biomarkers for the Prediction and Judgement of Sepsis and Sepsis Complications: A Step towards precision medicine? J. Clin. Med. 11 (19), 5782. doi: 10.3390/jcm11195782
Wadman, M. (2023). FDA no longer has to require animal testing for new drugs. Science 379 (6628), 127–128. doi: 10.1126/science.adg6276
Wang, L., Bastarache, J. A., Ware, L. B. (2008). The coagulation cascade in sepsis. Curr. Pharm. Design 14 (19), 1860–1869. doi: 10.2174/138161208784980581
Wang, Y., Liu, Q., Liu, T., Zheng, Q., Xu, X., Liu, X., et al. (2018). Early plasma monocyte chemoattractant protein 1 predicts the development of sepsis in trauma patients. Medicine 97 (14), e0356. doi: 10.1097/MD.0000000000010356
Wicherska-pawłowska, K., Wróbel, T., Rybka, J. (2021). Toll-like receptors (Tlrs), nod-like receptors (nlrs) and rig-i-like receptors (rlrs) in innate immunity. tlrs, nlrs and rlrs ligands as immunotherapeutic agents for hematopoietic diseases. Int. J. Mol. Sci. 22 (24), 13397. doi: 10.3390/ijms222413397
Williams, A. E., Chambers, R. C. (2014). The mercurial nature of neutrophils: still an enigma in ARDS? Am. J. Physiol. Lung Cell Mol. Physiol. 306 (3), L217–L230. doi: 10.1152/ajplung.00311.2013
Wong, H. R., Cvijanovich, N., Lin, R., Allen, G. L., Thomas, N. J., Willson, D. F., et al. (2009). Identification of pediatric septic shock subclasses based on genome-wide expression profiling. BMC Med. 7, 34. doi: 10.1186/1741-7015-7-34
Wong, H. R., Sweeney, T. E., Hart, K. W., Khatri, P., Lindsell, C. J. (2017). Pediatric sepsis endotypes among adults with sepsis. Crit. Care Med. 45, 12, e1289–e1291. doi: 10.1097/CCM.0000000000002733
Woodfin, A., Voisin, M. B., Beyrau, M., Colom, B., Caille, D., Diapouli, F. M., et al. (2011). The junctional adhesion molecule JAM-C regulates polarized transendothelial migration of neutrophils in vivo. Nat. Immunol. 12 (8), 761–769. doi: 10.1038/ni.2062
World Health Organization (2023) Sepsis. Available at: https://www.who.int/news-room/fact-sheets/detail/sepsis (Accessed November 15, 2023).
Yang, Q., Langston, J. C., Tang, Y., Prabhakarpandian, B., Kilpatrick, L. E., Kiani, M. F. (2021a). A microphysiological system to study leukocyte-endothelial cell interaction during inflammation. J. Visual. Exp. 178. doi: 10.3791/63312
Yang, Q., Wijerathne, H., Langston, J. C., Kiani, M. F., Kilpatrick, L. E. (2021b). Emerging approaches to understanding microvascular endothelial heterogeneity: A roadmap for developing anti-inflammatory therapeutics. Int. J. Mol. Sci. 22 (15), 7770. doi: 10.3390/ijms22157770
Yehya, N., Fazelinia, H., Taylor, D. M., Lawrence, G. G., Spruce, L. A., Thompson, J. M., et al. (2022). Differentiating children with sepsis with and without acute respiratory distress syndrome using proteomics. Am. J. Physiol. Lung Cell Mol. Physiol. 322 (3), L365–L372. doi: 10.1152/ajplung.00164.2021
Young, E. W. K., Simmons, C. A. (2010). Macro- and microscale fluid flow systems for endothelial cell biology. Lab. Chip 10 (2), 143–160. doi: 10.1039/b913390a
Zhang, Y., Li, W., Zhou, Y., Johnson, A., Venable, A., Hassan, A., et al. (2018). Detection of sepsis in patient blood samples using CD64 expression in a microfluidic cell separation device. Analyst 143 (1), 241–249. doi: 10.1039/c7an01471f
Zhu, T., Liao, X., Feng, T., Wu, Q., Zhang, J., Cao, X., et al. (2017). Plasma monocyte chemoattractant protein 1 as a predictive marker for sepsis prognosis: A prospective cohort study. Tohoku J. Exp. Med. 241 (2), 139–147. doi: 10.1620/tjem.241.139
Keywords: biomimetic microfluidic assay, endothelial cells, inflammation, in silico modeling, neutrophils, omics, organ-on-chip, sepsis
Citation: Liu D, Langston JC, Prabhakarpandian B, Kiani MF and Kilpatrick LE (2024) The critical role of neutrophil-endothelial cell interactions in sepsis: new synergistic approaches employing organ-on-chip, omics, immune cell phenotyping and in silico modeling to identify new therapeutics. Front. Cell. Infect. Microbiol. 13:1274842. doi: 10.3389/fcimb.2023.1274842
Received: 09 August 2023; Accepted: 18 December 2023;
Published: 08 January 2024.
Edited by:
Peibin Zeng, Sichuan University, ChinaReviewed by:
Jennifer Catherine Brazil, University of Michigan, United StatesChristian Con Yost, The University of Utah, United States
Yang Zhao, Institute of Zoology (CAS), China
Copyright © 2024 Liu, Langston, Prabhakarpandian, Kiani and Kilpatrick. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Laurie E. Kilpatrick, bGF1cmllLmtpbHBhdHJpY2tAdGVtcGxlLmVkdQ==
 Dan Liu
Dan Liu Jordan C. Langston1
Jordan C. Langston1 Laurie E. Kilpatrick
Laurie E. Kilpatrick