- 1Department of Laboratory Medicine, West China Second University Hospital, Sichuan University, Chengdu, Sichuan, China
- 2Department of Laboratory Medicine, Meishan Women and Children’s Hospital, Alliance Hospital of West China Second University Hospital, Sichuan University, Meishan, Sichuan, China
- 3Department of Laboratory Medicine, West China Second University Hospital (Tianfu), Sichuan University/Sichuan Provincial Children’s Hospital, Meishan, Sichuan, China
- 4Key Laboratory of Birth Defects and Related Diseases of Women and Children (Sichuan University), Ministry of Education, Chengdu, Sichuan, China
The pilus is an extracellular structural part that can be detected in some Streptococcus pneumoniae (S. pneumoniae) isolates (type I pili are found in approximately 30% of strains, while type II pili are found in approximately 20%). It is anchored to the cell wall by LPXTG-like motifs on the peptidoglycan. Two kinds of pili have been discovered, namely, pilus-1 and pilus-2. The former is encoded by pilus islet 1 (PI-1) and is a polymer formed by the protein subunits RrgA, RrgB and RrgC. The latter is encoded by pilus islet 2 (PI-2) and is a polymer composed mainly of the structural protein PitB. Although pili are not necessary for the survival of S. pneumoniae, they serve as the structural basis and as virulence factors that mediate the adhesion of bacteria to host cells and play a direct role in promoting the adhesion, colonization and pathogenesis of S. pneumoniae. In addition, as candidate antigens for protein vaccines, pili have promising potential for use in vaccines with combined immunization strategies. Given the current understanding of the pili of S. pneumoniae regarding the genes, proteins, structure, biological function and epidemiological relationship with serotypes, combined with the immunoprotective efficacy of pilins as protein candidates for vaccines, we here systematically describe the research status and prospects of S. pneumoniae pili and provide new ideas for subsequent vaccine research and development.
1 Introduction
Streptococcus pneumoniae (S. pneumoniae), a common bacterium colonizing the human upper respiratory tract, is a gram-positive bacterium capable of establishing symbiotic relationships that can often lead to respiratory tract infections. After escaping the immune defense of the host, S. pneumoniae can cause local or systemic infections such as pneumonia, otitis media, meningitis, sepsis, nasosinusitis, bronchitis, abscesses, conjunctivitis, pericarditis, and arthritis, with high morbidity and mortality in children younger than 5 years and elderly people (O’Brien et al., 2009; Deloria Knoll et al., 2014). The WHO ranks pneumonia as the leading cause of death in children under 5 years of age (Bryce et al., 2005), and more than 50% of cases of severe pneumonia in children are caused by S. pneumoniae, accounting for a higher proportion of deaths than pneumonia caused by other factors (Wardlaw et al., 2006; Modell et al., 2012).
S. pneumoniae pili are long multisubunit structures that enhance the interaction between the pneumococcus and the host (Dzaraly et al., 2020). S. pneumoniae has a variety of virulence factors, including pili. As important candidate factors for S. pneumoniae protein vaccines, pili can facilitate the adherence of bacteria to the surface of host cells, recognize the extracellular matrix, and participate in biofilm formation. They also participate in invasion, which can induce an inflammatory response (Barocchi et al., 2006), and they play an important role in the processes of host tissue colonization and pathogenesis (Walker et al., 2014; Engholm et al., 2017). Additionally, anti-pilus antibodies have been observed in the serum of patients with S. pneumoniae infection (Ahmed et al., 2014). Research on S. pneumoniae pilin has yielded interesting findings, but the related reports have been relatively scattered and have lacked systematization. This review focuses on the recent findings related to the typing, structural characteristics and functions of S. pneumoniae pilins in addition to the immunoprotective efficacy of pilins as protein antigens to provide direction for follow-up research.
2 Characteristics and structure of the pilus
Similar to those of other gram-positive bacteria, the pili of S. pneumoniae are polymers formed by the covalent assembly of multiple pilin subunits, each of which contains a sortase (Srt)-specific LPXTG-like motif. Catalyzed by a specific sortase, protein subunits covalently polymerize, and the pilus anchor to the peptidoglycan on the surface of the cell wall (Naziga and Wereszczynski, 2017).
The genes encoding the pili of S. pneumoniae exist as islets. Two types of S. pneumoniae pili have been found, with the structure of the pilus islets and protein subunits differentiating the type I pilus (pilus-1) from the type II pilus (pilus-2). The former, encoded by pilus islet 1 (PI-1), is a polymer formed by the protein subunits RrgA, RrgB and RrgC. Pilus-1 was first described in 2006; it is critical in promoting virulence (Barocchi et al., 2006) and is expressed in approximately 20%-30% of S. pneumoniae strains (Moschioni et al., 2008). Pilus-2 is encoded by pilus islet-2 (PI-2) and is a polymer of the protein subunit PitB (Shaik et al., 2015). Because pilus islets have characteristics of mobile genetic elements, S. pneumoniae strains can express two types of pili, only one type of pilus or no pilus.
2.1 Characteristics of pilus-1
Pilus-1 was the first pilus found in S. pneumoniae, and it is also the most widely expressed type, with a prevalence of approximately 30% (Gholamhosseini-Moghaddam et al., 2015; Mayanskiy et al., 2017). Pilus-1 was first identified in the TIGR4 strain (Barocchi et al., 2006). PI-1 encodes pilus-1 in S. pneumoniae and includes a total of 7 genes, namely, three sortase-encoding genes, i.e., srtC-1, srtC-2, and srtC-3 (formerly known as srtB, srtC, and srtD) (Iovino et al., 2020); a positive transcriptional regulator gene, i.e., rlrA; and three surface structural protein-encoding genes, i.e., rrgA, rrgB and rrgC. Notably, rrgA, rrgB, and rrgC encode LPXTG-like motifs (rrgA: YPRTG, rrgB: IPQTG, rrgC: VPDTG) for covalent polymerization between catalytic subunits recognized by the sortase that cross-links individual pilus subunits (pilus monomers) and for anchoring pilins to the cell wall surface (Dramsi et al., 2005).
Pilus-1 (Figure 1) is present all over the surface of S. pneumoniae, and electron microscopy has shown that it has a diameter of approximately 2-6 nm (Falker et al., 2008) and a length of more than 1.5 μm. Pilus-1 is composed mainly of RrgA, RrgB and RrgC; a small amount of pilin RrgA is located at the top, and RrgC is at the bottom. RrgB is the backbone protein and plays a vital role in pilus-1 formation (Hilleringmann et al., 2009).
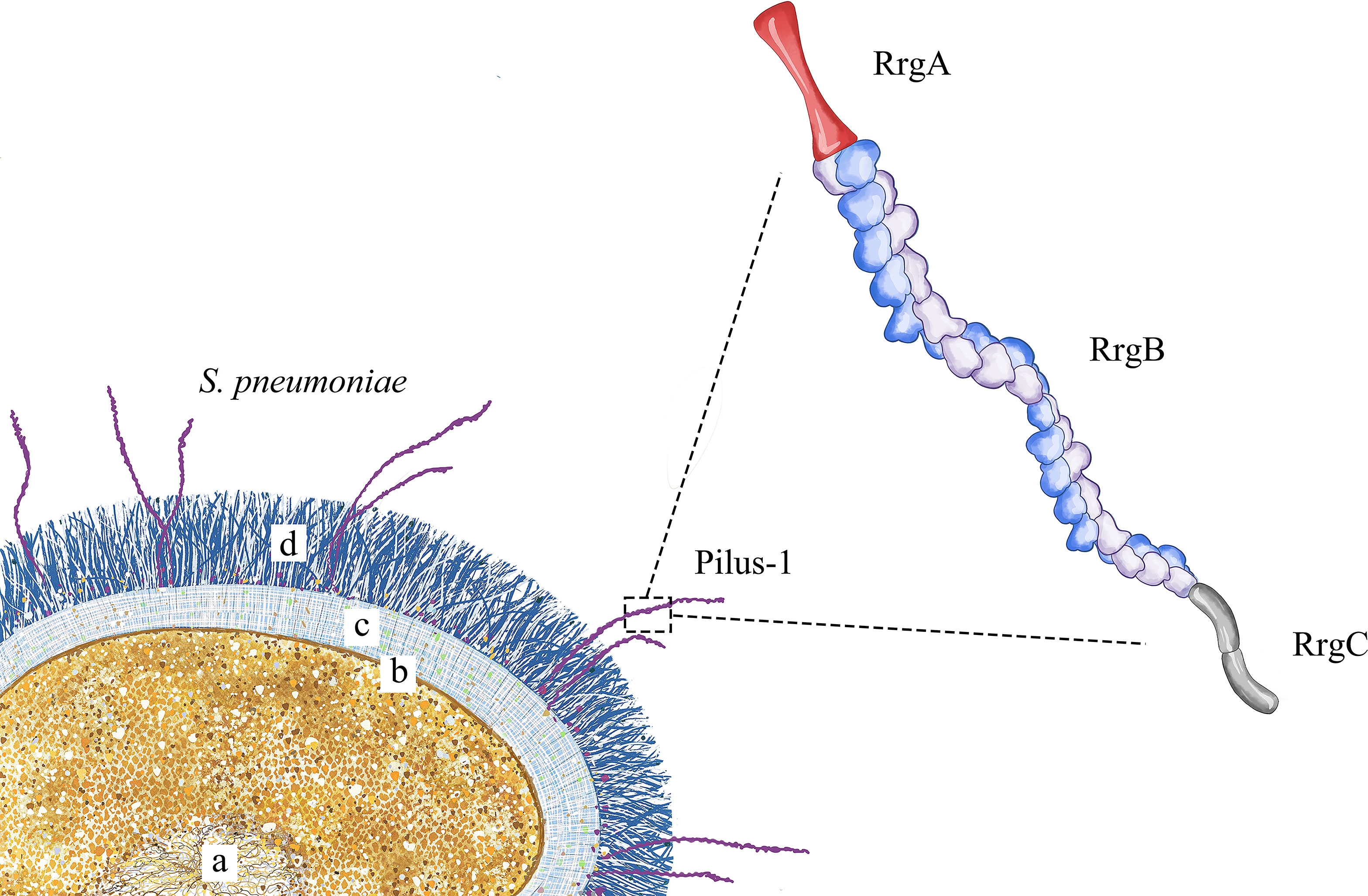
Figure 1 Pilus-1 schematic diagram. S. pneumoniae is shown in the lower left, where (a) is the nucleoid, (b) is the cell membrane, (c) is the cell wall, and (d) is the capsule. The enlarged structure on the right is pilus-1, including RrgA at the top, the backbone protein RrgB in the middle and RrgC at the bottom.
2.2 Structure of pilus-1
RrgB (PDB ID: 3RPK) is composed of four domains (Figure 2A), with a nose-like bulge in spatial configuration and polarity. The protein subunits are connected head to tail to form protofilaments, and two protofilaments are intertwined to form the main structure of S. pneumoniae pilus-1 (Hilleringmann et al., 2008). Due to the sequence variability of PI-1, there are currently three isoforms (RrgB clade I, RrgB clade II, RrgB clade III), and the protein homology among the different isoforms ranges from 48% to 60%. The homology among the proteins is >99% (Moschioni et al., 2008), and the sequences of the same isoforms are highly conserved. The crystal structure of the C-terminal major part of RrgB (residues 184–627) has been solved at 2.8 Å resolution, revealing three independently folded protein domains (D2–D4). The N-terminal independently folded domain D1 rarely interacts with the remaining RrgB structure, suggesting that D1 may be more flexible than D2–D4 (Paterson and Baker, 2011). One study tested single recombinant RrgB domains D1–D4 in active and passive immunization, and the results showed that the functional domain D1 was the most effective, providing a level of protection comparable to that of the full-length protein (Gentile et al., 2011).
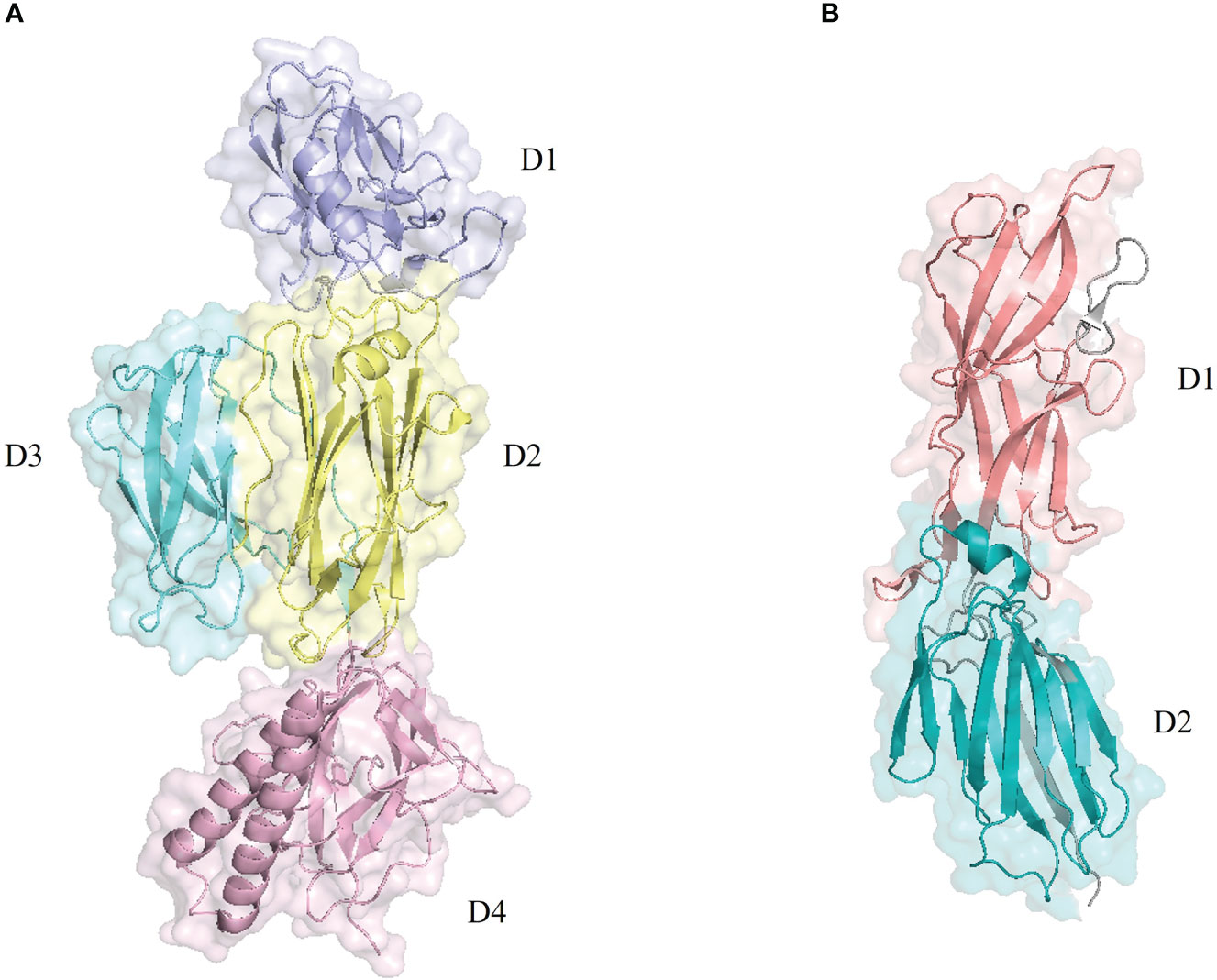
Figure 2 Pilus backbone protein structure diagram. The structures were retrieved from the RCSB Protein Data Bank and visualized with PyMOL. (A) The backbone protein RrgB (PDB: 3RPK) of pilus-1 includes four structural domains: domain 1 (D1) in purple, domain 2 (D2) in yellow, domain 3 (D3) in light blue, and domain 4 (D4) in light pink. (B) The backbone protein pitB (PDB: 7F7Y) of pilus-2 includes two structural domains: domain 1 (D1) in pink and domain 2 (D2) in blue.
RrgA is the main pilus adhesin (Nelson et al., 2007). It is distributed primarily in the distal end of pilus-1, is arranged in clusters, and is the apical protein of pilus-1, mediating the adhesion function of the pilus and interacting with collagen, fibronectin and laminin. Similar to findings in other gram-positive bacteria, RrgA is not required for the initial formation of S. pneumoniae pilus-1. The RrgA protein contains four domains, including an integrin I collagen recognition domain consisting of two intervening “arms” folded into a positively charged cradle structure and three stem-forming domains (Shaik et al., 2014). Because the homology of clade I and clade III RrgAs encoded by PI-1 is >99%, only two isoforms of RrgA exist; the variation between the two isoforms is mainly concentrated in the protein head, while the slender stem is conserved (Moschioni et al., 2010b). RrgA has 893 residues, and its crystal structure reveals a 195 Å long elongated protein with four domains that compactly fold with a limited size. The binding of the main part of RrgA to RrgB occurs via an interaction catalyzed by the specific sortase at the C-terminal end of its D4 domain, and the four domains of RrgA rarely come in contact with each other, indicating potential flexibility (Izore et al., 2010).
RrgC is distributed mainly at the end of S. pneumoniae pilus-1 near the bacterial cell surface. It contains three domains connected by two isopeptide bonds, and the structure is curved and rod-like. During the polymerization of subunits, RrgC does not depend on the sortase encoded by PI-1; rather, this process is catalyzed by the housekeeping sortase SrtA, which anchors S. pneumoniae pilus-1 on the bacterial surface (Shaik et al., 2014). Unlike RrgA and RrgB, the RrgC protein subunit is highly homologous (>98%) among S. pneumoniae strains.
2.3 Characteristics of pilus-2
PI-2 encoding S. pneumoniae pilus-2 consists of five genes: the signal peptide-like protein gene sipA; pitA and pitB, which encode two surface structural proteins, and the sortase genes srtG1 and srtG2. The entire islet is located between the protease T gene (PepT) and the ferrochelatase gene (HemH) (Bagnoli et al., 2008; Shaik et al., 2015). In recent years, the prevalence of S. pneumoniae pilus-2 has increased to approximately 20%, and it is mainly associated with serotypes 1, 2, 7F, 19A and 19F (Zahner et al., 2010; Hjalmarsdottir et al., 2015). Pilus-2 is composed predominantly of repeating units of the structural protein PitB, whose covalent polymerization is catalyzed by the homologous sortase SrtG1 and SipA. PitB contains two domains, D1 and D2, which are linked by SrtG1 through recognition of the VTPTG motif. PitA and SrtG2 are not necessary for pilus-2 (Bagnoli et al., 2008).
2.4 Structure of pilus-2
In contrast to pilus-1 of S. pneumoniae, whose complex structure has been well studied, pilus-2 is thus far poorly understood. The structure of PitB (PDB ID: 7F7Y), the backbone pilin, has been resolved to 2.8 Å. It is 110 Å long and 40 Å wide and consists of two domains, D1 and D2 (Figure 2B), whose covalent binding is catalyzed by the sortase SrtG1 (Yadav and Krishnan, 2022). The two domains of PitB present an irregular structure; the N-terminal D1 domain consists of 12 chains, and the C-terminal D2 domain consists of 11 chains (Shaik et al., 2015). The interaction between D1 and D2 is similar to that observed in RrgB (Paterson and Baker, 2011; El Mortaji et al., 2012). However, the structural protein PitA and the sortase SrtG2 do not play important roles in the formation of pilus-2.
3 Functions of S. pneumoniae pili and their correlations with serotypes
Although pili are not essential structures for the survival of S. pneumoniae, they play important roles in the initial stage of S. pneumoniae infection, forming a structural basis and acting as virulence factors that mediate bacterial adhesion to host cells. Studies have shown that pili play direct roles in promoting the adhesion, colonization and pathogenesis of S. pneumoniae (Barocchi et al., 2006; Kreikemeyer et al., 2011).
3.1 Biological functions of pili
Local invasive infection is the basis for bacteremia. In the context of respiratory infection with S. pneumoniae, in one study on the adhesion function of S. pneumoniae pilus-1, Nelson et al. (Nelson et al., 2007) used human alveolar epithelial A549 cells as host cells and found that although rrgA-knockout bacteria formed pili, their adhesion ability was significantly reduced. In contrast, the rrgB- and rrgC-knockout bacteria did not form pili, but their adhesion ability was similar to that of the wild-type bacteria with pili. These findings suggest that the adhesion function of S. pneumoniae pilus-1 is determined by RrgA rather than the pilus backbone protein. In a preliminary study on the immune mechanism of S. pneumoniae pilus-1, Nelson et al. found that compared with pilus-deficient strains, pilus-carrying strains not only had advantages in colonization, pathogenicity and septicemia induction but also effectively induced the release of tumor necrosis factor alpha (TNF-α) and interleukin 6 (IL-6) and enhanced the host’s inflammatory response. Subsequently, it was found that this proinflammatory effect of pilus-1 is mediated by the surface-exposed domain 3 of RrgA, particularly the 49-amino-acid sequence, which acts as a TLR2 receptor agonist (Basset et al., 2013). In a study on the S. pneumoniae pilus-1 protein subunit RrgA and the mechanism of host immune damage, Orrskog et al. (Orrskog et al., 2012) used rrgA knockout and nonknockout strains through intranasal and intraperitoneal challenge experiments and found that infection with strains expressing RrgA led to earlier development of sepsis and more rapid disease progression in wild-type mice than infection with complement receptor 3 (CR3) antibody and infection in CR3-deficient mice, suggesting that RrgA can influence macrophage function and systemic infection status by interacting with CR3. The uptake of S. pneumoniae by murine and human macrophages is enhanced for strains that express RrgA, and RrgA-CR3-mediated phagocytosis promotes the spread of localized to systemic infections. Basset et al. (Basset et al., 2013) showed that RrgA can enhance virulence and the inflammatory response by activating TLR2.
Infections other than respiratory system infections caused by S. pneumoniae mostly develop from bacteremia caused by respiratory tract invasion by pathogenic bacteria. S. pneumoniae is the main cause of bacterial meningitis (Iovino et al., 2016a). With regard to meningeal infection caused by S. pneumoniae, Iovino et al. (Iovino et al., 2016b) conducted an intravenous challenge experiment and found that the bacterial loads of the pilus-carrying strains were higher in the brain; when rrgA was knocked out, the load of the pilus-carrying strains in the brain decreased. Additionally, strains that expressed RrgA more readily dispersed to form a single coccus, and based on findings by high-resolution immunofluorescence microscopy, these strains were more likely to adhere to the vascular endothelium of the blood−brain barrier, promoting the passage of bacteria through the blood−brain barrier. This finding suggests that the adhesin RrgA of pilus-1, as a virulence factor associated with S. pneumoniae meningitis, plays an important role in promoting the adhesion of individual S. pneumoniae to the vascular endothelium of the blood−brain barrier. Further research on adhesion ligands (Iovino et al., 2017) has shown that RrgA can bind to polymeric immunoglobulin receptor (pIgR) and platelet endothelial cell adhesion molecule (PECAM-1) on the surface of blood−brain barrier epithelial cells and that downregulation of these two receptors has the potential to prevent S. pneumoniae meningitis. Blocking the receptors pIgR and PECAM-1 or RrgA in the pilus significantly reduces brain invasion by S. pneumoniae and increases mouse survival (Iovino et al., 2018). These findings offer promising prospects for new treatments.
To investigate middle ear infections caused by S. pneumoniae, Figueira et al. (Figueira et al., 2014) created a middle ear infection model by nasal challenge and analyzed the middle ear fluid. The results showed that although S. pneumoniae pilus-1 was nonessential for S. pneumoniae infection of the middle ear, the bacterial loads were higher for pilus-carrying strains than for pilus-deficient strains, suggesting that S. pneumoniae pilus-1 promotes middle ear infections. The results of the above studies demonstrate that S. pneumoniae pilus-1 plays a vital role in bacterial adhesion, invasion, host immune damage, and metastasis of infection foci and that the RrgA protein subunit is the main adhesion factor and immune mediator.
All the above studies have shown that although the skeletal protein of S. pneumoniae pilus-1 is RrgB, the main functional protein is RrgA. S. pneumoniae pilus-1 has adhesion functions and virulence effects, exacerbates inflammatory damage in the host and is vital in the local and systemic immune response.
In contrast to S. pneumoniae pilus-1, Bagnoli et al. (Bagnoli et al., 2008) found, through an adhesion experiment with A549 cells, that pilus-2 aids in adhesion through the backbone protein PitB and that the adhesion ability is weaker than that of RrgA. Collagen, fibronectin and laminin are the common adhesion ligands of pilus-1 and pilus-2.
3.2 Relationships between pilus and vaccine serotypes
The relationships between the epidemiology of S. pneumoniae pili and the serotypes covered by vaccines have received extensive attention. Kawaguchiya et al. found that 19F, 23F and 19A were vaccine-covered serotypes among the isolates carrying PI-1, while 6E, 15B and 35B were nonvaccine types, and that pneumococcal conjugate vaccine (PCV) covered most of the pilus-1-carrying strains (Kawaguchiya et al., 2017). Pilus-1 is not widely distributed among the 100 known S. pneumoniae serotypes (Ganaie et al., 2020); analyses of global isolates have shown that the overall frequency of pilus-1 is 30%, suggesting that the islet is associated with pneumonia. There is a certain correlation among the genotypes of S. pneumoniae isolates (Aguiar et al., 2008; Moschioni et al., 2008). Aguiar et al. (Aguiar et al., 2008) found an association between 83% of rlrA-positive isolates and vaccine serotypes 4, 6B, 9V, and 14 (all covered by PCV7) in their study of the pilus regulation gene rlrA; in other studies, this gene has been found to be absent in nonvaccine serotypes (1, 7F, 8 and 12B). Among the 305 S. pneumoniae isolates collected worldwide, pilus-2 has been found in 16.4%. Most pilus-2 strains are associated with serotypes 1, 2, 7F, 19A, and 19F (all vaccine-covered serotypes) (Bagnoli et al., 2008). Serotypes 7F and 19A are the serotypes that most frequently carry PI-2, followed by 1 and 19F (Zahner et al., 2010). A study from Iceland showed that among 398 isolates, 88.1% (118 isolates) and 81.6% (31 isolates) of PI-1-positive and PI-2-positive isolates, respectively, belonged to vaccine serotypes. PI-1-positive isolates were most common in serotypes 19F and 6B, and PI-2-positive isolates were most common in serotype 19F.
Overall, S. pneumoniae pili are generally associated with serotypes covered by PCVs. However, in the long term, the ongoing phenomenon of serotype replacement with vaccine introduction (Lo et al., 2022) means that pilus isolates could also be replaced. One study reported a prevalence of 23.7% for pilus-1 isolates in 2001, one year after the introduction of PCV7 in the United States, and 14.9% in 2004. However, by 2007, the prevalence of nonvaccine pilus isolates had increased to 26.1% (Regev-Yochay et al., 2010). The analysis showed that the decline in the prevalence of pilus-1 among S. pneumoniae isolates from 2001 to 2004 was due primarily to the replacement of vaccine-type (VT) strains with nonvaccine-type (NVT) strains. However, between 2004 and 2007, the increase in prevalence was due mainly to the increased frequency of pili in NVT strains. The results reported in this review indicate that the application of PCVs will have an impact on the emergence of new cloned strains and provide new ideas for future S. pneumoniae pathogenic surveillance.
4 Potential of S. pneumoniae pilins as vaccine candidates
4.1 Limitations of traditional vaccines
At present, the traditional pneumococcal polysaccharide vaccine (PPV) and PCV are widely used in many countries and regions. Due to the immaturity of the immune systems of young children, the polysaccharide antigens used in PPV cannot produce ideal protective efficacy in infants under 2 years of age (Kadioglu et al., 2008). This limitation prompted the creation of the pneumococcal 7-valent conjugate vaccine (PCV7), which covers serotypes 4, 6B, 9V, 14, 18C, 19F, and 23F. In addition, PCV10 and PCV13 have also been included in immunization programs in more than 120 countries (Ubukata et al., 2015). Although PCV can elicit an effective immune response, it has limited serotype coverage and is expensive to manufacture. It has been reported (Regev-Yochay et al., 2010; Regev-Yochay et al., 2016) that after planned immunization with PCV7, the prevalence of S. pneumoniae in the population does not change significantly, but the proportion of nonvaccine strains among S. pneumoniae epidemic strains gradually increases. This result suggests that after the large-scale use of serotype-dependent vaccines, serotype replacement occurs in the S. pneumoniae epidemic strain, which reduces the protective efficacy of the vaccines.
4.2 Application prospects of pilin
Many studies have shown that a variety of S. pneumoniae virulence factors can be used as candidate antigens for novel protein vaccines (Kim et al., 2017), providing broad-spectrum protection against S. pneumoniae infection. These include, for example, choline-binding proteins (CBPs) (Bittaye and Cash, 2015), pneumoniae surface proteins A and C (PspA/PspC) (Tostes et al., 2017), heat shock proteins (Hsp40/DnaJ) (Cui et al., 2011), pneumococcal surface adhesin (PsaA) (Converso et al., 2017) and pneumolysin (Ply) (Brooks et al., 2015). However, there is currently no commercialized protein vaccine that can protect against S. pneumoniae infection. Continued discovery of new protein antigenic sites may not only effectively promote the development of protein vaccines with better protective efficacy but also facilitate the identification of drug targets through the study of immune protection mechanisms.
Studies (Moschioni et al., 2010a; Regev-Yochay et al., 2010) have found that serotypes and S. pneumoniae pili are correlated. In the initial stage after traditional S. pneumoniae vaccine immunization, the expression of pilus-1 in epidemic strains decreased sharply in a short period due to the rapid reductions in vaccine-type strains; however, the expression has increased yearly, even exceeding the level before the popularization of traditional vaccines, especially in vaccine-type strains and drug-resistant strains. This finding indicates that the S. pneumoniae pilus islet has mobile genetic elements and that pilus-1 plays an important role in the pathogenicity and resistance of epidemic strains. Pilus-1 can thus be used as a candidate antigen target for the development of protein vaccines and further in-depth research. In addition, there is a strong link between pili and antimicrobial resistance, and the above findings could lay a foundation for using pili as potential antigens for future protein vaccines.
4.3 Status of pilin research
Early developed commercial S. pneumoniae vaccines used capsular polysaccharide (CPS) as the antigen (PPV23), aiming to reduce S. pneumoniae infection by inhibiting the colonization of specific serotype strains (Duke and Avci, 2023). To improve the defect of poor immunogenicity, conjugation technology was introduced to covalently combine CPS with immunogenic carrier proteins to develop glycoconjugate vaccines (Avci, 2013; Avci et al., 2019). Multiple CPS-protein conjugate vaccines including V116 (Platt et al., 2023), VAX24 (Fairman et al., 2021) and PNEUMOSIL (Duke and Avci, 2023) have been approved to enter clinical trials. Although the majority of invasive pneumococcal disease (IPD) strains are encapsulated organisms with typed serotypes, nontypeable isolates (including nonencapsulated isolates) can also cause IPD (Scott et al., 2012). Nonencapsulated strains are easily transformable due to the lack of capsules (Pearce et al., 2002) and adhere well to the respiratory epithelial cells, which has advantages in nasopharyngeal colonization, further exacerbating the limitations of CPS-based vaccine applications. As an alternative or supplementary strategy to conjugate vaccines (PCVs), pneumococcal protein vaccines aim to overcome serotype dependence and promisingly provide broad-spectrum protection at a relatively low cost, providing an option for children in developing countries.
Preliminary research on S. pneumoniae pilin vaccines has focused mainly on S. pneumoniae pilus-1, which has filamentous structures and can be effectively identified by the host immune system. Gianfaldoni et al. (Gianfaldoni et al., 2007) used recombinant pilus-1 protein for the first time to evaluate the protective efficacy of different proteins against S. pneumoniae in mice and found that RrgA, RrgB and RrgC stimulate the production of specific antibodies. Among them, RrgA and RrgB have immunoprotective efficacy that can significantly prolong the survival time of mice challenged by a lethal S. pneumoniae dose; notably, the protective efficacy of RrgC immunization was poor. Further studies by Moschioni et al. (Moschioni et al., 2010b) and Harfouche et al. (Harfouche et al., 2012) have found that there is cross-immunity between the two isoforms of RrgA and that immunization with RrgA clade II antigens can protect mice from lethal-dose challenge with the RrgA clade I strain. This result indicates that RrgA is not only immunogenic but also well conserved, making it an ideal candidate protein for pilus vaccines. However, there are no cross-protective effects among the three isoforms of RrgB. To address this problem, researchers have used the rrgB of S. pneumoniae TIGR4 (RrgB clade I), S. pneumoniae 6BSPEC (RrgB clade II), and S. pneumoniae 35BSME15 (RrgB clade III) to construct a fusion protein, RrgB321, containing three RrgB isoforms. The subunits of the different isoforms are connected by Gly-Ser-Gly-Gly-Gly-Gly-Gly so that the fusion protein can fold normally. The newly constructed fusion protein antigen RrgB321 exhibits good protective efficacy against S. pneumoniae in mice after active and passive immunization. In addition, the level of complement-dependent bacterial phagocytosis in pilus strains is comparable to that observed with PCV7, indicating that the RrgB321 component vaccine leads to successful immune protection against S. pneumoniae with pilus-1, covering more than 30% of the total S. pneumoniae strains. Unlike RrgB321, a single fusion protein component, PnuBioVax developed by Entwisle et al. is a whole-cell vaccine produced from genetically modified S. pneumoniae TIGR4 (Entwisle et al., 2017). Its enriched surface-exposed antigens include but are not limited to pilins RrgB and RrgA, pneumococcal surface adhesin A (PsaA) and non-toxic pneumolysin (Ply). Antibody responses to the TIGR4-type pilins RrgB and RrgA in subjects were shown to increase by more than 2-fold.
An RrgB fusion protein vaccine developed by Novartis is in preclinical trials (Harfouche et al., 2012). Intranasal mucosal immunization can reduce bacterial load in the early middle ear fluid of mice with experimental S. pneumoniae-related otitis media. However, due to limited antigenic targets, the pilin vaccine provides immune protection only against pilus-carrying strains, not against pilus-deficient strains. In addition, in the presence of pilin antibodies, the bacteria can selectively not express pili to evade immune attack. To overcome the limitations of pilins and enhance the immune coverage of vaccines, multivalent vaccines can be prepared in combination with bacterially conserved proteins. For example, a triple-protein vaccine consisting of pneumococcal choline-binding protein A (PcpA), pneumococcal histidine trimer protein D (PhtD), and detoxified pneumolysin (dPly) and a triple-protein vaccine composed of Haemophilus influenzae protein D, dPly, and PhtD have entered clinical phase II trials (Feldman and Anderson, 2014). As important components of protein-based vaccines, dPly, PcpA and PhtD participate in the compatibility of various protein vaccines (Brooks et al., 2015; Odutola et al., 2017; Odutola et al., 2019) and have entered clinical trials. Pili, with strong immunogenicity, are important for pathogenicity and induction of protective immunity against S. pneumoniae (Mora et al., 2005; Ahmed et al., 2014). Combining pilins with mature protein components is promising to exploit the dual advantages of easy recognition by the immune system and high conservation of thalline proteins. Furthermore, the study by Narciso et al. used the highly conserved lipoproteins MalX and PrsA in membrane particles (MPs) as antigens and provided us with new ideas (Narciso et al., 2022). MPs could be used as a platform for the preparation of protein-based vaccines against pneumococcal infections, with MalX and PrsA as the main antigens responsible for cross-protection. Membrane vesicles can promote immune responses and contribute to the transportation of virulence factors, and are promising for use as vaccine adjuvants and carriers (Jan, 2017; Malekan et al., 2020).
In addition to the licensed carrier proteins tetanus toxoid (TT), diphtheria toxoid (DT), CRM197, Haemophilus protein D (PD), etc., researchers have begun to pay attention to the impact of different types of carriers on the protective efficacy of vaccines, including outer membrane vesicles (OMVs) and generalized modules for membrane antigens (GMMA), virus-like particles (VLP) and nanomaterials, etc. (Gong et al., 2022; van der Put et al., 2023). Different immunization doses, immunization routes and adjuvants can also strongly affect the effects of pilin vaccines. Adjusting the protein compatibility ratio between different antigens of a combined vaccine, trying various immunization routes, and selecting appropriate immune adjuvants can help to optimize the protective efficacy provided by a pilin vaccine (Principi and Esposito, 2018).
5 Discussion
S. pneumoniae pili mediate bacterial adhesion, tissue colonization, and barrier invasion; induce inflammatory damage in the body; and play a vital role in the infection process. Research on the structures and functions of pili has gradually expanded, but the protective efficacy of pili as protein vaccine candidates and the mechanisms of their effects need to be further studied. After the popularization of PPV and PCV, expectations are high for S. pneumoniae vaccines in the face of changing epidemic strains and the rebound of pilus expression. Protein vaccines have become a hot topic in the field of S. pneumoniae vaccine research due to their low cost, strong conservation, and lack of serotype-specific restrictions. Although current research on protein vaccines involves multiple levels, there is no ideal protein vaccine on the market yet. It is expected that universal protein vaccines with broad-spectrum immune protection will be developed. Pili not only play important roles in the pathogenic process of S. pneumoniae but also meet the selection criteria for vaccine candidate proteins in terms of their biological functions, immunogenicity, extracellular expression, and induction of antibody production. They thus have great potential for use in vaccine research and development. Current research on recombinant S. pneumoniae pilin vaccines is still relatively limited. Continued discovery of new protein antigen targets and evaluation of protein vaccine combination modes remain promising research directions for universal S. pneumoniae protein vaccines.
Author contributions
CM: Visualization, Writing – original draft, Software. YC: Writing – original draft, Supervision. ZY: Writing – original draft, Visualization. YJ: Supervision, Writing – review & editing, Funding acquisition.
Funding
The authors declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the West China Second University Hospital (No. KL066) and the Sichuan Science and Technology Program (No. 2022YFS0239).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aguiar, S. I., Serrano, I., Pinto, F. R., Melo-Cristino, J., Ramirez, M. (2008). The presence of the pilus locus is a clonal property among pneumococcal invasive isolates. BMC Microbiol. 8, 41. doi: 10.1186/1471-2180-8-41
Ahmed, M. S., Derbyshire, S., Flanagan, B., Loh, C., McCormick, M., Barocchi, M., et al. (2014). Immune responses to pneumococcal pilus RrgA and RrgB antigens and their relationship with pneumococcal carriage in humans. J. Infect. 68, 562–571. doi: 10.1016/j.jinf.2014.01.013
Avci, F. Y. (2013). Novel strategies for development of next-generation glycoconjugate vaccines. Curr. Top. Med. Chem. 13, 2535–2540. doi: 10.2174/15680266113136660180
Avci, F., Berti, F., Dull, P., Hennessey, J., Pavliak, V., Prasad, A. K., et al. (2019). Glycoconjugates: what it would take to master these well-known yet little-understood immunogens for vaccine development. mSphere 4, e00520-19. doi: 10.1128/mSphere.00520-19
Bagnoli, F., Moschioni, M., Donati, C., Dimitrovska, V., Ferlenghi, I., Facciotti, C., et al. (2008). A second pilus type in Streptococcus pneumoniae is prevalent in emerging serotypes and mediates adhesion to host cells. J. Bacteriol. 190, 5480–5492. doi: 10.1128/JB.00384-08
Barocchi, M. A., Ries, J., Zogaj, X., Hemsley, C., Albiger, B., Kanth, A., et al. (2006). A pneumococcal pilus influences virulence and host inflammatory responses. Proc. Natl. Acad. Sci. U. S. A. 103, 2857–2862. doi: 10.1073/pnas.0511017103
Basset, A., Zhang, F., Benes, C., Sayeed, S., Herd, M., Thompson, C., et al. (2013). Toll-like receptor (TLR) 2 mediates inflammatory responses to oligomerized RrgA pneumococcal pilus type 1 protein. J. Biol. Chem. 288, 2665–2675. doi: 10.1074/jbc.M112.398875
Bittaye, M., Cash, P. (2015). Streptococcus pneumoniae proteomics: determinants of pathogenesis and vaccine development. Expert Rev. Proteomics 12, 607–621. doi: 10.1586/14789450.2015.1108844
Brooks, W. A., Chang, L. J., Sheng, X., Hopfer, R., Team P. P. R. S (2015). Safety and immunogenicity of a trivalent recombinant PcpA, PhtD, and PlyD1 pneumococcal protein vaccine in adults, toddlers, and infants: A phase I randomized controlled study. Vaccine 33, 4610–4617. doi: 10.1016/j.vaccine.2015.06.078
Bryce, J., Boschi-Pinto, C., Shibuya, K., Black, R. E., Group, W. H. O. C. H. E. R (2005). WHO estimates of the causes of death in children. Lancet 365, 1147–1152. doi: 10.1016/S0140-6736(05)71877-8
Converso, T. R., Goulart, C., Darrieux, M., Leite, L. C. C. (2017). A protein chimera including PspA in fusion with PotD is protective against invasive pneumococcal infection and reduces nasopharyngeal colonization in mice. Vaccine 35, 5140–5147. doi: 10.1016/j.vaccine.2017.08.010
Cui, Y., Zhang, X., Gong, Y., Niu, S., Yin, N., Yao, R., et al. (2011). Immunization with DnaJ (hsp40) could elicit protection against nasopharyngeal colonization and invasive infection caused by different strains of Streptococcus pneumoniae. Vaccine 29, 1736–1744. doi: 10.1016/j.vaccine.2010.12.126
Deloria Knoll, M., Park, D. E., Johnson, T. S., Chandir, S., Nonyane, B. A., Conklin, L., et al. (2014). Systematic review of the effect of pneumococcal conjugate vaccine dosing schedules on immunogenicity. Pediatr. Infect. Dis. J. 33 Suppl 2, S119–S129. doi: 10.1097/INF.0000000000000079
Dramsi, S., Trieu-Cuot, P., Bierne, H. (2005). Sorting sortases: a nomenclature proposal for the various sortases of Gram-positive bacteria. Res. Microbiol. 156, 289–297. doi: 10.1016/j.resmic.2004.10.011
Duke, J. A., Avci, F. Y. (2023). Emerging vaccine strategies against the incessant pneumococcal disease. NPJ Vaccines 8, 122. doi: 10.1038/s41541-023-00715-w
Dzaraly, N. D., Muthanna, A., Mohd Desa, M. N., Taib, N. M., Masri, S. N., Rahman, N. I. A., et al. (2020). Pilus islets and the clonal spread of piliated Streptococcus pneumoniae: A review. Int. J. Med. Microbiol. 310, 151449. doi: 10.1016/j.ijmm.2020.151449
El Mortaji, L., Contreras-Martel, C., Moschioni, M., Ferlenghi, I., Manzano, C., Vernet, T., et al. (2012). The full-length Streptococcus pneumoniae major pilin RrgB crystallizes in a fibre-like structure, which presents the D1 isopeptide bond and provides details on the mechanism of pilus polymerization. Biochem. J. 441, 833–841. doi: 10.1042/BJ20111397
Engholm, D. H., Kilian, M., Goodsell, D. S., Andersen, E. S., Kjaergaard, R. S. (2017). A visual review of the human pathogen Streptococcus pneumoniae. FEMS Microbiol. Rev. 41, 854–879. doi: 10.1093/femsre/fux037
Entwisle, C., Hill, S., Pang, Y., Joachim, M., McIlgorm, A., Colaco, C., et al. (2017). Safety and immunogenicity of a novel multiple antigen pneumococcal vaccine in adults: A Phase 1 randomised clinical trial. Vaccine 35, 7181–7186. doi: 10.1016/j.vaccine.2017.10.076
Fairman, J., Agarwal, P., Barbanel, S., Behrens, C., Berges, A., Burky, J., et al. (2021). Non-clinical immunological comparison of a Next-Generation 24-valent pneumococcal conjugate vaccine (VAX-24) using site-specific carrier protein conjugation to the current standard of care (PCV13 and PPV23). Vaccine 39, 3197–3206. doi: 10.1016/j.vaccine.2021.03.070
Falker, S., Nelson, A. L., Morfeldt, E., Jonas, K., Hultenby, K., Ries, J., et al. (2008). Sortase-mediated assembly and surface topology of adhesive pneumococcal pili. Mol. Microbiol. 70, 595–607. doi: 10.1111/j.1365-2958.2008.06396.x
Feldman, C., Anderson, R. (2014). Review: current and new generation pneumococcal vaccines. J. Infect. 69, 309–325. doi: 10.1016/j.jinf.2014.06.006
Figueira, M., Moschioni, M., De Angelis, G., Barocchi, M., Sabharwal, V., Masignani, V., et al. (2014). Variation of pneumococcal Pilus-1 expression results in vaccine escape during Experimental Otitis Media [EOM]. PloS One 9, e83798. doi: 10.1371/journal.pone.0083798
Ganaie, F., Saad, J. S., McGee, L., van Tonder, A. J., Bentley, S. D., Lo, S. W., et al. (2020). A new pneumococcal capsule type, 10D, is the 100th serotype and has a large cps fragment from an oral streptococcus. mBio 11, e00937–e00920. doi: 10.1128/mBio.00937-20
Gentile, M. A., Melchiorre, S., Emolo, C., Moschioni, M., Gianfaldoni, C., Pancotto, L., et al. (2011). Structural and functional characterization of the Streptococcus pneumoniae RrgB pilus backbone D1 domain. J. Biol. Chem. 286, 14588–14597. doi: 10.1074/jbc.M110.202739
Gholamhosseini-Moghaddam, T., Rad, M., Mousavi, S. F., Ghazvini, K. (2015). Detection of lytA, pspC, and rrgA genes in Streptococcus pneumoniae isolated from healthy children. Iran J. Microbiol. 7, 156–160.
Gianfaldoni, C., Censini, S., Hilleringmann, M., Moschioni, M., Facciotti, C., Pansegrau, W., et al. (2007). Streptococcus pneumoniae pilus subunits protect mice against lethal challenge. Infect. Immun. 75, 1059–1062. doi: 10.1128/IAI.01400-06
Gong, X., Gao, Y., Shu, J., Zhang, C., Zhao, K. (2022). Chitosan-based nanomaterial as immune adjuvant and delivery carrier for vaccines. Vaccines (Basel) 10, 1906. doi: 10.3390/vaccines10111906
Harfouche, C., Filippini, S., Gianfaldoni, C., Ruggiero, P., Moschioni, M., Maccari, S., et al. (2012). RrgB321, a fusion protein of the three variants of the pneumococcal pilus backbone RrgB, is protective in vivo and elicits opsonic antibodies. Infect. Immun. 80, 451–460. doi: 10.1128/IAI.05780-11
Hilleringmann, M., Giusti, F., Baudner, B. C., Masignani, V., Covacci, A., Rappuoli, R., et al. (2008). Pneumococcal pili are composed of protofilaments exposing adhesive clusters of Rrg A. PloS Pathog. 4, e1000026. doi: 10.1371/journal.ppat.1000026
Hilleringmann, M., Ringler, P., Muller, S. A., De Angelis, G., Rappuoli, R., Ferlenghi, I., et al. (2009). Molecular architecture of Streptococcus pneumoniae TIGR4 pili. EMBO J. 28, 3921–3930. doi: 10.1038/emboj.2009.360
Hjalmarsdottir, M. A., Petursdottir, B., Erlendsdottir, H., Haraldsson, G., Kristinsson, K. G. (2015). Prevalence of pilus genes in pneumococci isolated from healthy preschool children in Iceland: association with vaccine serotypes and antibiotic resistance. J. Antimicrob. Chemother. 70, 2203–2208. doi: 10.1093/jac/dkv096
Iovino, F., Engelen-Lee, J. Y., Brouwer, M., van de Beek, D., van der Ende, A., Valls Seron, M., et al. (2017). pIgR and PECAM-1 bind to pneumococcal adhesins RrgA and PspC mediating bacterial brain invasion. J. Exp. Med. 214, 1619–1630. doi: 10.1084/jem.20161668
Iovino, F., Hammarlof, D. L., Garriss, G., Brovall, S., Nannapaneni, P., Henriques-Normark, B. (2016a). Pneumococcal meningitis is promoted by single cocci expressing pilus adhesin RrgA. J. Clin. Invest 126, 2821–2826. doi: 10.1172/JCI84705
Iovino, F., Nannapaneni, P., Henriques-Normark, B., Normark, S. (2020). The impact of the ancillary pilus-1 protein RrgA of Streptococcus pneumoniae on colonization and disease. Mol. Microbiol. 113, 650–658. doi: 10.1111/mmi.14451
Iovino, F., Seinen, J., Henriques-Normark, B., van Dijl, J. M. (2016b). How does streptococcus pneumoniae invade the brain? Trends Microbiol. 24, 307–315. doi: 10.1016/j.tim.2015.12.012
Iovino, F., Thorsdottir, S., Henriques-Normark, B. (2018). Receptor blockade: A novel approach to protect the brain from pneumococcal invasion. J. Infect. Dis. 218, 476–484. doi: 10.1093/infdis/jiy193
Izore, T., Contreras-Martel, C., El Mortaji, L., Manzano, C., Terrasse, R., Vernet, T., et al. (2010). Structural basis of host cell recognition by the pilus adhesin from Streptococcus pneumoniae. Structure 18, 106–115. doi: 10.1016/j.str.2009.10.019
Jan, A. T. (2017). Outer membrane vesicles (OMVs) of gram-negative bacteria: A perspective update. Front. Microbiol. 8, 1053. doi: 10.3389/fmicb.2017.01053
Kadioglu, A., Weiser, J. N., Paton, J. C., Andrew, P. W. (2008). The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat. Rev. Microbiol. 6, 288–301. doi: 10.1038/nrmicro1871
Kawaguchiya, M., Urushibara, N., Aung, M. S., Shinagawa, M., Takahashi, S., Kobayashi, N. (2017). Serotype distribution, antimicrobial resistance and prevalence of pilus islets in pneumococci following the use of conjugate vaccines. J. Med. Microbiol. 66, 643–650. doi: 10.1099/jmm.0.000479
Kim, G. L., Seon, S. H., Rhee, D. K. (2017). Pneumonia and Streptococcus pneumoniae vaccine. Arch. Pharm. Res. 40, 885–893. doi: 10.1007/s12272-017-0933-y
Kreikemeyer, B., Gamez, G., Margarit, I., Giard, J. C., Hammerschmidt, S., Hartke, A., et al. (2011). Genomic organization, structure, regulation and pathogenic role of pilus constituents in major pathogenic Streptococci and Enterococci. Int. J. Med. Microbiol. 301, 240–251. doi: 10.1016/j.ijmm.2010.09.003
Lo, S. W., Mellor, K., Cohen, R., Alonso, A. R., Belman, S., Kumar, N., et al. (2022). Emergence of a multidrug-resistant and virulent Streptococcus pneumoniae lineage mediates serotype replacement after PCV13: an international whole-genome sequencing study. Lancet Microbe 3, e735–e743. doi: 10.1016/S2666-5247(22)00158-6
Malekan, M., Siadat, S. D., Aghasadeghi, M., Shahrokhi, N., Afrough, P., Behrouzi, A., et al. (2020). Evaluation of protective immunity responses against pneumococcal PhtD and its C-terminal in combination with outer-membrane vesicles as adjuvants. J. Med. Microbiol. 69, 465–477. doi: 10.1099/jmm.0.001103
Mayanskiy, N., Savinova, T., Alyabieva, N., Ponomarenko, O., Brzhozovskaya, E., Lazareva, A., et al. (2017). Antimicrobial resistance, penicillin-binding protein sequences, and pilus islet carriage in relation to clonal evolution of Streptococcus pneumoniae serotype 19A in Russi 2002-2013. Epidemiol. Infect. 145, 1708–1719. doi: 10.1017/S0950268817000541
Modell, B., Berry, R. J., Boyle, C. A., Christianson, A., Darlison, M., Dolk, H., et al. (2012). Global regional and national causes of child mortality. Lancet 380, 1556–1557. doi: 10.1016/S0140-6736(12)61878-9
Mora, M., Bensi, G., Capo, S., Falugi, F., Zingaretti, C., Manetti, A. G., et al. (2005). Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc. Natl. Acad. Sci. U. S. A. 102, 15641–15646. doi: 10.1073/pnas.0507808102
Moschioni, M., De Angelis, G., Melchiorre, S., Masignani, V., Leibovitz, E., Barocchi, M. A., et al. (2010a). Prevalence of pilus-encoding islets among acute otitis media Streptococcus pneumoniae isolates from Israel. Clin. Microbiol. Infect. 16, 1501–1504. doi: 10.1111/j.1469-0691.2009.03105.x
Moschioni, M., Donati, C., Muzzi, A., Masignani, V., Censini, S., Hanage, W. P., et al. (2008). Streptococcus pneumoniae contains 3 rlrA pilus variants that are clonally related. J. Infect. Dis. 197, 888–896. doi: 10.1086/528375
Moschioni, M., Emolo, C., Biagini, M., Maccari, S., Pansegrau, W., Donati, C., et al. (2010b). The two variants of the Streptococcus pneumoniae pilus 1 RrgA adhesin retain the same function and elicit cross-protection in vivo. Infect. Immun. 78, 5033–5042. doi: 10.1128/IAI.00601-10
Narciso, A. R., Iovino, F., Thorsdottir, S., Mellroth, P., Codemo, M., Spoerry, C., et al. (2022). Membrane particles evoke a serotype-independent cross-protection against pneumococcal infection that is dependent on the conserved lipoproteins MalX and PrsA. Proc. Natl. Acad. Sci. U. S. A. 119, e2122386119. doi: 10.1073/pnas.2122386119
Naziga, E. B., Wereszczynski, J. (2017). Molecular mechanisms of the binding and specificity of streptococcus pneumoniae sortase C enzymes for pilin subunits. Sci. Rep. 7, 13119. doi: 10.1038/s41598-017-13135-3
Nelson, A. L., Ries, J., Bagnoli, F., Dahlberg, S., Falker, S., Rounioja, S., et al. (2007). RrgA is a pilus-associated adhesin in Streptococcus pneumoniae. Mol. Microbiol. 66, 329–340. doi: 10.1111/j.1365-2958.2007.05908.x
O’Brien, K. L., Wolfson, L. J., Watt, J. P., Henkle, E., Deloria-Knoll, M., McCall, N., et al. (2009). Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374, 893–902. doi: 10.1016/S0140-6736(09)61204-6
Odutola, A., Ota, M. O. C., Antonio, M., Ogundare, E. O., Saidu, Y., Foster-Nyarko, E., et al. (2017). Efficacy of a novel, protein-based pneumococcal vaccine against nasopharyngeal carriage of Streptococcus pneumoniae in infants: A phase 2, randomized, controlled, observer-blind study. Vaccine 35, 2531–2542. doi: 10.1016/j.vaccine.2017.03.071
Odutola, A., Ota, M. O. C., Antonio, M., Ogundare, E. O., Saidu, Y., Owiafe, P. K., et al. (2019). Immunogenicity of pneumococcal conjugate vaccine formulations containing pneumococcal proteins, and immunogenicity and reactogenicity of co-administered routine vaccines - A phase II, randomised, observer-blind study in Gambian infants. Vaccine 37, 2586–2599. doi: 10.1016/j.vaccine.2019.03.033
Orrskog, S., Rounioja, S., Spadafina, T., Gallotta, M., Norman, M., Hentrich, K., et al. (2012). Pilus adhesin RrgA interacts with complement receptor 3, thereby affecting macrophage function and systemic pneumococcal disease. mBio 4, e00535-00512. doi: 10.1128/mBio.00535-12
Paterson, N. G., Baker, E. N. (2011). Structure of the full-length major pilin from Streptococcus pneumoniae: implications for isopeptide bond formation in gram-positive bacterial pili. PloS One 6, e22095. doi: 10.1371/journal.pone.0022095
Pearce, B. J., Iannelli, F., Pozzi, G. (2002). Construction of new unencapsulated (rough) strains of Streptococcus pneumoniae. Res. Microbiol. 153, 243–247. doi: 10.1016/s0923-2508(02)01312-8
Platt, H., Omole, T., Cardona, J., Fraser, N. J., Mularski, R. A., Andrews, C., et al. (2023). Safety, tolerability, and immunogenicity of a 21-valent pneumococcal conjugate vaccine, V116, in healthy adults: phase 1/2, randomised, double-blind, active comparator-controlled, multicentre, US-based trial. Lancet Infect. Dis. 23, 233–246. doi: 10.1016/S1473-3099(22)00526-6
Principi, N., Esposito, S. (2018). Development of pneumococcal vaccines over the last 10 years. Expert Opin. Biol. Ther. 18, 7–17. doi: 10.1080/14712598.2018.1384462
Regev-Yochay, G., Hanage, W. P., Trzcinski, K., Rifas-Shiman, S. L., Lee, G., Bessolo, A., et al. (2010). Re-emergence of the type 1 pilus among Streptococcus pneumoniae isolates in Massachusetts, USA. Vaccine 28, 4842–4846. doi: 10.1016/j.vaccine.2010.04.042
Regev-Yochay, G., Jaber, H., Hamdan, A., Daana, M., Nammouz, H., Thalji, A., et al. (2016). Vaccine escape of piliated Streptococcus pneumoniae strains. Vaccine 34, 2787–2792. doi: 10.1016/j.vaccine.2016.04.064
Scott, J. R., Hinds, J., Gould, K. A., Millar, E. V., Reid, R., Santosham, M., et al. (2012). Nontypeable pneumococcal isolates among navajo and white mountain apache communities: are these really a cause of invasive disease? J. Infect. Dis. 206, 73–80. doi: 10.1093/infdis/jis307
Shaik, M. M., Lombardi, C., Maragno Trindade, D., Fenel, D., Schoehn, G., Di Guilmi, A. M., et al. (2015). A structural snapshot of type II pilus formation in Streptococcus pneumoniae. J. Biol. Chem. 290, 22581–22592. doi: 10.1074/jbc.M115.647834
Shaik, M. M., Maccagni, A., Tourcier, G., Di Guilmi, A. M., Dessen, A. (2014). Structural basis of pilus anchoring by the ancillary pilin RrgC of Streptococcus pneumoniae. J. Biol. Chem. 289, 16988–16997. doi: 10.1074/jbc.M114.555854
Tostes, R. O., Rodrigues, T. C., da Silva, J. B., Schanoski, A. S., Oliveira, M. L., Miyaji, E. N. (2017). Protection Elicited by Nasal Immunization with Recombinant Pneumococcal Surface Protein A (rPspA) Adjuvanted with Whole-Cell Pertussis Vaccine (wP) against Co-Colonization of Mice with Streptococcus pneumoniae. PloS One 12, e0170157. doi: 10.1371/journal.pone.0170157
Ubukata, K., Chiba, N., Hanada, S., Morozumi, M., Wajima, T., Shouji, M., et al. (2015). Serotype changes and drug resistance in invasive pneumococcal diseases in adults after vaccinations in children, Japan 2010-2013. Emerg. Infect. Dis. 21, 1956–1965. doi: 10.3201/eid2111.142029
van der Put, R. M. F., Metz, B., Pieters, R. J. (2023). Carriers and antigens: new developments in glycoconjugate vaccines. Vaccines (Basel) 11, 219. doi: 10.3390/vaccines11020219
Walker, M. J., Barnett, T. C., McArthur, J. D., Cole, J. N., Gillen, C. M., Henningham, A., et al. (2014). Disease manifestations and pathogenic mechanisms of Group A Streptococcus. Clin. Microbiol. Rev. 27, 264–301. doi: 10.1128/CMR.00101-13
Wardlaw, T., Salama, P., Johansson, E. W., Mason, E. (2006). Pneumonia: the leading killer of children. Lancet 368, 1048–1050. doi: 10.1016/S0140-6736(06)69334-3
Yadav, R. K., Krishnan, V. (2022). New structural insights into the PI-2 pilus from Streptococcus oralis, an early dental plaque colonizer. FEBS J. 289, 6342–6366. doi: 10.1111/febs.16527
Keywords: Streptococcus pneumoniae, pilus, structure, pilin, protein vaccine
Citation: Miao C, Cui Y, Yan Z and Jiang Y (2023) Pilus of Streptococcus pneumoniae: structure, function and vaccine potential. Front. Cell. Infect. Microbiol. 13:1270848. doi: 10.3389/fcimb.2023.1270848
Received: 01 August 2023; Accepted: 04 September 2023;
Published: 20 September 2023.
Edited by:
Ying Xu, First Affiliated Hospital of Chengdu Medical College, ChinaReviewed by:
Marie-stephanie Aschtgen, Karolinska Institutet (KI), SwedenPing-Hua Qu, Guangzhou University of Chinese Medicine, China
Copyright © 2023 Miao, Cui, Yan and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ziyi Yan, yan_z_y2@qq.com; Yongmei Jiang, jiangyongmei-1@163.com
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work and share last authorship
 Chenglin Miao
Chenglin Miao