- 1Department of Precision Medicine, Graduate School of Basic Medical Science, Institute for Antimicrobial Resistance Research and Therapeutics, Sungkyunkwan University School of Medicine, Suwon, Republic of Korea
- 2Department of Orthopedic Surgery, University of Pittsburgh School of Medicine, Pittsburgh, PA, United States
Pseudomonas aeruginosa is a multidrug-resistant opportunistic human pathogen that utilizes two-component systems (TCSs) to sense pathophysiological signals and coordinate virulence. P. aeruginosa contains 64 sensor histidine kinases (HKs) and 72 response regulators (RRs) that play important roles in metabolism, bacterial physiology, and virulence. However, the role of some TCSs in virulence remains uncharacterized. In this study, we evaluated the virulence potential of some uncharacterized sensor HK and RR knockouts in P. aeruginosa using a Galleria mellonella infection model. Furthermore, we demonstrated that KdpD and AauS HKs regulate virulence by affecting P. aeruginosa biofilm formation and motility. Both ΔkdpD and ΔaauS showed reduced biofilm and motility which were confirmed by restored phenotypes upon complementation. Moreover, ΔkdpD and ΔaauS exhibited increased survival of HeLa cells and G. mellonella during in vivo infection. Altered expression of the transcriptional regulators anR and lasR, along with the virulence genes lasA, pelA, cupA, pqsA, pqsB, pqsC, and pqsD in the mutant strains elucidated the mechanism by which ΔkdpD and ΔaauS affect virulence. These findings confirm that kdpD and aauS play important roles in P. aeruginosa pathogenesis by regulating biofilm formation and motility.
1 Introduction
Two-component systems (TCSs) play essential roles in the adaptation and survival of bacteria in various stressful environments, including pathophysiological conditions (Mitrophanov and Groisman, 2008; De Bentzmann and Plésiat, 2011). A TCS generally comprises a sensor histidine kinase (HK) and its cognate response regulator (RR). During pathogenesis, the HK responsible for sensing pathophysiological stimuli is phosphorylated and subsequently transfers or phosphorelays the signal to its cognate RR to activate the expression of virulence genes under its control (Stock et al., 2000). Bacterial virulence depends on virulence determinants, including biofilm formation, motility, lipopolysaccharides, capsules, alginate, pyoverdine, and pyocyanin. It also depends on the secretion of toxins, such as exotoxin A, phospholipases, and proteases, which play vital roles in disease progression caused by P. aeruginosa (Leitão, 2020; Jurado-Martín et al., 2021).
P. aeruginosa is a Gram-negative, multidrug-resistant, opportunistic bacterium responsible for severe community and nosocomial infections in immunocompromised patients (De Bentzmann and Plésiat, 2011; Wu et al., 2014; Gale et al., 2015; Moradali et al., 2017). P. aeruginosa causes approximately 7.1–7.3% of healthcare-associated infections (Magill et al., 2014; Weiner et al., 2016), and is the main cause of mortality and morbidity in patients with cystic fibrosis (CF) (Mogayzel et al., 2014). In a major multinational observational study of the point prevalence of infections in intensive care units, P. aeruginosa was reported to be responsible for 16.2% of patients’ infections and 23% of all ICU-acquired infections (Vincent et al., 2020). It is a life-threatening bacterium that was included in the priority pathogen list in 2017 (World Health Organization, 2017).
The P. aeruginosa genome contains many TCSs, including 64 sensor HKs and 72 RRs (Francis et al., 2017). These TCS-controlling networks are promising targets against antimicrobial resistance (AMR), pathogenesis, and biofilm (Francis et al., 2017; Tierney and Rather, 2019).
Biofilms are organized bacterial communities that act as buffers against external environmental stress (Yin et al., 2019). They may form colonies and persist long-term in patients by escaping the host immune system (Kostakioti et al., 2013). Moreover, extremely structured biofilms have been detected in patients with chronic lung and wound infections (Römling and Balsalobre, 2012). P. aeruginosa is a major biofilm-forming bacterium and the primary cause of 65–80% of nosocomial infections (Li et al., 2023). Motility (swarming, swimming, and twitching) is another important virulence factor in pathogenesis, as it is crucial for mobilization, colonization in multiple environments, adherence, and biofilm formation (O'Toole and Kolter, 1998). Motility is involved in biofilm formation in many model bacterial pathogens (Korber et al., 1994; Pratt and Kolter, 1998).
Previous studies have demonstrated the roles of many TCSs in virulence (Francis et al., 2017; Sultan et al., 2021). The TCS GacS/GacA and HKs RetS and LadS act as switches between planktonic and biofilm lifestyles in P. aeruginosa (Mikkelsen et al., 2011; Chambonnier et al., 2016). BfiS/BfiR, MifS/MifR, and BfmS/BfmR TCSs contribute to biofilm development in a stage-specific manner and cause irreversible attachment, microcolony formation, and maturation, respectively (Petrova and Sauer, 2009). FleS/FleR, another TCS, regulates the expression of flagellar biosynthetic genes and alters swarming motility (Dasgupta et al., 2003). Similarly, CarS/CarR is a well-recognized calcium homeostasis TCS that regulates swarming motility and calcium-induced virulence factors (Guragain et al., 2016). However, the roles of some TCSs in virulence and biofilm formation in P. aeruginosa PA14 warrant further elucidation. Therefore, it is necessary to identify uncharacterized TCSs and establish their roles in virulence and biofilm formation to better understand P. aeruginosa pathogenesis.
The functional roles of kdpD, aauS, nasT, and cheB in P. aeruginosa virulence are not well characterized (Francis et al., 2017). Therefore, we monitored their potential for infection in a G. mellonella infection model using isogenic knockout strains to investigate their role in virulence. Based on the initial test, kdpD and aauS played a more substantial role in infection than nasT and cheB. These two HKs are involved in bacterial physiology. kdpD is an essential gene for potassium (K+) transport (Freeman et al., 2013), whereas aauS is involved in acidic amino acid utilization (Sonawane and Singh, 2006). However, their roles in P. aeruginosa virulence have not been well characterized. Recently, Badal et al. (2020) developed an in vitro approach to screen the role of 112 TCS of P. aeruginosa in biofilm on endotracheal tubes. Out of 112, 56 TCS were found to be involved in biofilm. Of these, 18 new TCSs including AauS and KdpD were identified to contribute in biofilm (Badal et al., 2020). However, the current study elucidated the role of KdpD and AauS in motility; and virulence by using HeLa cell line, and G. mellonella infection model. We further demonstrated the involvement of KdpD and AauS in biofilm using functional genomics and gene expression analyses of relevant genetic cascades to elucidate the mechanism by which kdpD and aauS affect the biofilm formation, motility, and pathogenesis of P. aeruginosa PA14.
2 Materials and methods
2.1 Growth and culture conditions of bacterial strains
Non-redundant transposon mutant libraries containing ΔnasT, ΔcheB, ΔkdpD, and ΔaauS that were developed using PA14 (PA14NR) (Liberati et al., 2006) were obtained from Harvard Medical School. All strains were grown on cetrimide agar (CA) (Sigma-Aldrich, St. Louis, MO, USA) plates for 16–18 h at 37 °C. Bacterial cultures were grown in Luria-Bertani (LB) medium (Becton Dickinson, Franklin Lakes, NJ, USA) under orbital shaking culture conditions at 200 rpm. Gentamicin (15 μg/mL) was used to grow the mutants, whereas 50 µg/mL carbenicillin (Sigma-Aldrich) was used to grow the strains harboring empty pUCP18 or its derivative plasmids. The mutant complementation strains were grown in the presence of 15 μg/mL gentamicin and 50 μg/mL carbenicillin. Bacterial growth was monitored by measuring the optical density at 600 nm [OD600nm = 1.0 = 1.0 × 109 colony-forming units (CFU/mL)] using a Gen 5 BioTek microplate reader (BioTek, Winooski, VT, USA). Bacterial cells were harvested using 4000 rpm (2701 ×g) centrifugation at 4 °C. They were washed once using phosphate buffered saline (PBS, pH 7.2) in all experiments, unless otherwise stated. The strains and plasmids, as well as primers used in the study are listed in Supplementary Tables S1, S2, respectively.
2.2 Complementation of ΔkdpD and ΔaauS mutant strains
To complement the transposon mutants ΔkdpD and ΔaauS, the corresponding genes (kdpD and aauS) were PCR-amplified from P. aeruginosa PA14 genomic DNA using specific primers (Table S2). The column purified PCR-amplified DNA fragments, and the pUCP18 vector were restriction endonuclease digested with EcoRI and HindIII. The EcoRI/HindIII-digested kdpD and aauS genes and pUCP18 vector DNA were resolved on an agarose gel. This was followed by gel elution using a Cosmo Genentech gel elution kit (Seoul, Korea). The gel-eluted kdpD and aauS were cloned into EcoRI/HindIII sites of the pUCP18 vector to yield the recombinant plasmids pUCP18kdpD and pUCP18aauS, respectively. These plasmids were transformed into Escherichia coli DH5α, and clones were selected on LB agar plates containing 50 μg/mL carbenicillin. The plasmids were isolated using a Cosmo Genentech plasmid isolation kit and DNA was sequenced to verify the accuracy of the nucleotide sequences of the genes in pUCP18kdpD and pUCP18aauS. The empty control plasmid, pUCP18, was transformed into wild-type (WT), ΔkdpD and ΔaauS of P. aeruginosa PA14. The WT strain possessing pUCP18 (WT::pUCP18) was selected on LB agar plates containing 50 μg/mL carbenicillin. Furthermore, the mutant strains (ΔkdpD and ΔaauS) possessing the empty vector (ΔkdpD::pUCP18 and ΔaauS::pUCP18) were selected on LB-agar plates containing 15 μg/mL gentamicin and 50 μg/mL carbenicillin. To obtain the knockout-complemented strains (ΔkdpD::pUCP18kdpD and ΔaauS::pUCP18aauS), the recombinant plasmids pUCP18kdpD and pUCP18aauS were transformed into ΔkdpD and ΔaauS knockouts and selected on LB plates containing 50 μg/mL carbenicillin and 15 μg/mL gentamicin.
2.3 Quantification of biofilm formation using a crystal violet assay
Biofilm quantification through crystal violet (CV) staining was performed as previously described (Chen et al., 2018). Briefly, 16 h cultures of WT, mutant, and mutant-complemented strains were grown in Muller Hinton broth (MHB) (Becton Dickinson) at 37 °C and diluted to OD600 = 1 × 108 CFU/mL in fresh MHB media. An aliquot of 100 μL bacterial suspension was added to a flat-bottom 96-well polystyrene microtiter plate. After 24 or 48 h incubation at 37 °C, unattached cells were removed using two washes of PBS (pH 7.2). Next, 100 μL of 99% methanol was added to each well for 15 min, followed by methanol removal and drying of the plates at room temperature (25±1 °C) to perform biofilm fixation. Then, 100 μL of 0.04% CV was added for 20 min to stain the biofilm. Samples were washed with sterile PBS three consecutive times to remove additional colorants. Finally, 33% acetic acid was used to solubilize the biofilm-bound CV. Absorbance was measured at 630 nm using a Gen5 BioTek microplate reader (BioTek Instruments, Inc., Agilent Technologies, CA, USA). pqsA is an important biofilm producing gene so, ΔpqsA used as a negative control in the current experiment.
2.4 Confocal laser scanning microscopy
Bacteria cultures were grown in MHB at 37 °C for 16–18 h and diluted to OD600nm = 1 (1×108 CFU/mL) in fresh MHB media. Then, 500 μL bacterial suspension was added into an 8-well chambered cover glass (Lab-Tek II 1.5 Borosilicate glass, Sigma-Aldrich) and incubated at 37 °C under static culture conditions. The culture medium was changed after 24 h to remove non-attached bacterial cells. After 24 or 48 h of incubation, the biofilms were gently washed with saline [0.85% (w/v) sodium chloride (NaCl)]. Final concentrations of 30 μM propidium iodide (PI), and 5 μM SYTO9 (Sigma-Aldrich) were used for biofilm staining (Chaurasia et al., 2016). The chambered cover glass was incubated in the dark at room temperature for 20 min. The stained biofilms were washed with PBS prior to confocal microscopy. Biofilms were examined using confocal laser scanning microscopy (CLSM, Zeiss, Oberkochen, Germany) at 20× or 63× magnifications. Image stacks were collected from 20 random points on each biofilm to obtain accurate mean values for biofilm thickness measurement.
2.5 Swarming motility assay
Swarming motility was assessed as previously described, with slight modifications (Ha et al., 2014a). M8-supplemented swarming medium was used to prepare the swarm plates. Briefly, 6 g agar was added to 800 mL water (final concentration = 0.6%) and autoclaved to obtain a sterile, homogeneous agar suspension. A 5× M8 solution was prepared using 30 g Na2HPO4, 15 g KH2PO4, and 2.5 g NaCl in water by adjusting the final volume to 1 L, autoclaving, and finally adding a 5× M8 solution (200 mL) to the melted agar. Next, 20% glucose (final concentration = 0.2%), 20% casamino acids (final concentration = 0.5%), and 1 M MgSO4 (final concentration = 1 mM) were added to molten agar. The agar medium was mixed gently and cooled before being poured into Petri dishes (~25 mL/plate) and solidified at room temperature. The center of each plate was inoculated with 2.5 μL of OD600nm 1 equivalent overnight bacterial culture. Plates were incubated upright at 37 °C for 16–24 h and observed for the swarming phenotype. The sensor kinase fleS of the fleSR TCS was used as a negative control in the current experiment, as ΔfleS alters swarming motility in P. aeruginosa PA14 (Kollaran et al., 2019).
2.6 Swimming motility assay
Swimming motility was assessed using 0.3% LB agar as previously described, with slight modifications (Déziel et al., 2001). First, 25 mL media plates were solidified at room temperature for 1.5 h. The cell number was adjusted based on the OD600nm value. Then, 100 μL of OD600nm 1 bacterial culture was transferred into a 1.5 mL microcentrifuge tube. To avoid the production of swarming and twitching phenotypes, a sterile toothpick was dipped into the OD600nm 1 equivalent culture and stabilized in the middle of the agar layer without touching the top or bottom of the Petri plate. The plates were incubated upright for 20 h at 30 °C. The phenotype was observed, and the diameters of the swim zones were measured. The hook-associated gene, flgK, is essential for flagellar assembly. Additionally, its mutant showed reduced swimming motility (Ha et al., 2014b). Therefore, in the current experiment ΔflgK was used as a negative control for the no-swimming phenotypes.
2.7 Twitching motility assays
Twitching motility was tested as previously described (Déziel et al., 2001). Twitching motility was assessed on LB plates containing 1% (w/v) agar. A sterile toothpick was dipped into the OD600nm 1 equivalent bacterial culture. The toothpick touched the bottom of the Petri plate to move into the interstitial space between the basal surfaces of the agar. The plates were subsequently incubated at 37 °C for 48 h. The agar was gently removed from the Petri plate using a spatula to measure the twitch zone. The twitch zone was stained with 2 mL 0.1% (w/v) CV in water for 10 min. The CV was removed, and the plates were washed with water and allowed to air-dry at room temperature. The twitch zone diameters were measured and recorded. As the role of the TCS PilS/PilR in the regulation of type IV pilus expression has been demonstrated (Kilmury and Burrows, 2018a), we used ΔpilR as a negative control.
2.8 Quantitative reverse-transcription polymerase chain reaction analysis
The WT, mutant, and complementation strains were grown in M9 minimal medium for 10 h at 37 °C (Egli, 2015). For identification of biofilm genes bacteria was grown in 8-well chambered cover glass for 10, 24 and 48 h. Planktonic cells were discarded, and biofilm population was used for RNA isolation (Bisht et al., 2021). Total RNA was isolated using the Qiagen RNeasy kit (Hilden, Germany) according to the manufacturer’s protocol. Total RNA (1 µg) was treated with RNase-free DNase I (1 U) (amplification grade; Sigma-Aldrich) at room temperature for 15 min. The DNase I reaction was stopped using stop-buffer followed by inactivation of DNase at 70 °C for 10 min. To prepare cDNA from the isolated DNA-free mRNA, a random hexamer premix (RNA-to-cDNA EcoDryTM premix; Takara Bio, Kusatsu, Japan) was used. The qRT-PCR was performed using 55 °C annealing temperature and SYBR Green Supermix (Bio-Rad, Hercules, CA, USA). Here, rho was used as a housekeeping gene. The expression of kdpD and aauS was normalized to calculate the relative gene expression analyzed using the 2−ΔΔCT method (Schmittgen and Livak, 2008).
2.9 Invasion assay
To examine the roles of kdpD and aauS in P. aeruginosa virulence, we performed invasion assays as previously described (Kim et al., 2019). HeLa cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS) in a 5% CO2 humidified incubator at 37 °C. The cells were seeded in 6-well plates (5.0×105 cells/well) and incubated for 24 h. PBS-washed bacterial cells were placed in invasion medium (DMEM without FBS) for 2 h before infection. Then, WT and mutant strains at a multiplicity of infection (MOI) of 10 were used to infect HeLa cells for 120 min. Extracellular bacterial cells were killed through 60-min gentamicin (100 µg/mL) treatment. Residual gentamicin was removed by washing the cells twice with PBS. The infected cells were treated with 0.1% Triton-X100 to lyse the HeLa cells and retrieve intracellular bacteria, which were then serially diluted in PBS to plate 100 µL diluted cell suspension on cetrimide agar plates for CFU enumeration.
2.10 Live/dead assays
The live/dead assay was performed as previously described (Mittal et al., 2014). Briefly, DMEM with 10% FBS was used to grow HeLa cells in a 5% CO2 humidified incubator at 37 °C for 48 h. HeLa cells were seeded into 8-well chamber slides (5.0×105 cells/well) and incubated for 24 h. After two PBS washes, HeLa cells were infected with bacteria at an MOI of 10 for 120 min. Uninfected cells were used as negative (untreated) controls. After incubation, the infected HeLa cells were washed with PBS and a live/dead viability kit was used for staining. Fluorescently stained bacterial cells were observed using CLSM (Zeiss, Oberkochen, Germany) and images were captured.
2.11 G. mellonella infection model
The overnight grown cultures of P. aeruginosa strains were suspended in LB for 6 h. Bacterial cells were collected through 4000 rpm centrifugation (2701 ×g) at 4 °C and washed with PBS (pH 7.2). The number of bacterial cells was maintained by adjusting the optical density (OD600 nm = 1) in PBS. G. mellonella larvae were stored for 24 h before infection (Ménard et al., 2021). Then, the weight and length of each worm were measured. Ten waxworms of similar size (200 ± 2 mg, 2 ± 0.2 cm) were chosen for each group, including the carrier (PBS) control. Waxworms were injected with 10 bacterial cells (20 µL) in the last posterior leg using a 0.3 mL syringe (Becton Dickinson) (Desbois and Coote, 2011; Hill et al., 2014; Imdad et al., 2018). Similarly, 20 µL PBS was injected into the worms in the control group. After injection, worms were incubated at 27 °C and observed at different time points. We applied a health scoring index wherein each worm was given a score based on movement, melaninization, cocoon formation, and survival compared to the untreated control group (Champion et al., 2018). The experiment was terminated after 72 h and CFUs were counted using CA plates.
3 Results
3.1 Functional assessment of TCSs in virulence using the G. mellonella infection model
The relevance of kdpD, aauS, nasT, and cheB in P. aeruginosa virulence is yet to be well characterized (Francis et al., 2017). Therefore, we initially tested their roles in virulence using a G. mellonella infection model. The hemocytes present in G. mellonella larvae share functional similarities with mammalian phagocytic cells (Browne et al., 2013). Therefore, G. mellonella larval infection models have been used to identify virulence-related genes in various human pathogens (Jander et al., 2000; Champion et al., 2009; Lebreton et al., 2009; Peleg et al., 2009; Champion et al., 2010). rsmA, a virulence gene encoding the carbon storage and a pyocyanin production regulator, was used as a positive control to determine the potential roles of uncharacterized regulators. Mutant strains lacking rsmA, kdpD, aauS, nasT, cheB, or the WT were injected into the left posterior legs of G. mellonella larvae (n = 10) to assess their roles in the pathogenic potential of P. aeruginosa PA14. The results showed only 10% survival in WT strains 72 h post-infection, whereas the ΔrsmA showed 90% survival compared to the PBS control (100% survival) (Figure 1; Supplementary Figure S1). However, a delay in mortality was observed for ΔkdpD and ΔaauS, with 60% and 30% survival, respectively. The comparative growth profile of ΔkdpD and ΔaauS knockouts with the WT strain was monitored for physiological fitness to confirm that the difference in their infection potential was not due to their compromised growth proficiency. The growth of the WT, ΔkdpD, and ΔaauS strains was identical (Supplementary Figure S2). Furthermore, the ΔnasT and ΔcheB strains showed earlier mortality than the WT, with only 20% survival 72 h post-infection. These results demonstrate that kdpD and aauS contribute to virulence. Thus, we further investigated their relevance to the virulence potential of P. aeruginosa.
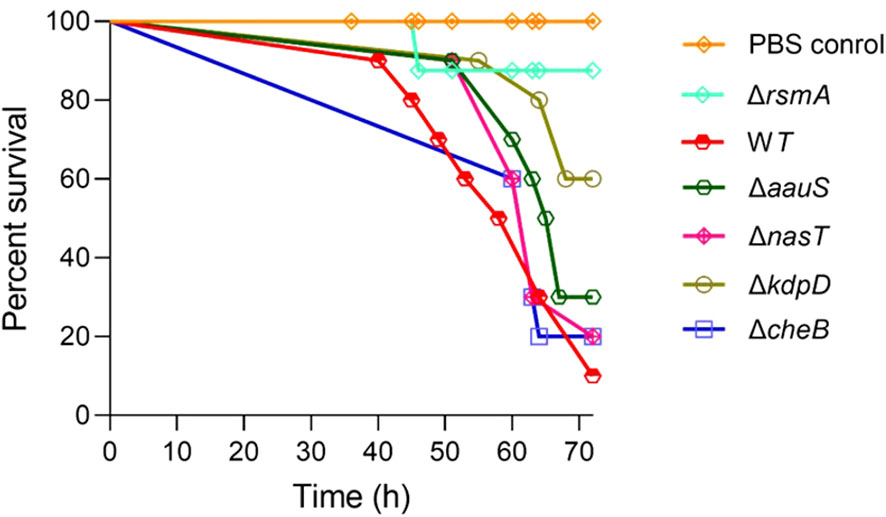
Figure 1. Screening of TCS genes relevant to virulence using a G. mellonella larvae infection model. The colony farming units (CFUs) (10 for each strain), namely the WT, ΔrsmA, ΔkdpD, ΔaauS, ΔnasT,or ΔcheB, were injected into waxworms (n =10), followed by incubation at 27 °C to examine their survival up to 72 h post-infection. A delay in mortality was observed for ΔkdpD and ΔaauS, with 60% and 30% survival, respectively.
3.2 Roles of kdpD and aauS in biofilm formation
KdpD/KdpE is an ‘adaptive regulator’ TCS involved in the virulence and intracellular survival of various pathogenic bacteria (Freeman et al., 2013). To elucidate the roles of kdpD and aauS in biofilm formation, we used a CV staining assay, followed by CLSM confirmation and image analysis to determine the quantity and thickness of the biofilm, respectively. Firstly, strains ΔkdpD and ΔaauS were tested against the WT (positive control) and ΔpqsA (negative control). After 24 and 48 h, ΔpqsA showed considerably poor biofilm formation compared to the WT. At 24 h, ΔkdpD and ΔaauS showed 69.1% and 54.9% reduction in biofilm formation, respectively (Supplementary Figure S3A). Similarly, we observed 69.7% and 72.8% inhibition of biofilm formation after 48 h in ΔkdpD and ΔaauS, respectively (Supplementary Figure S3B). Furthermore, knockouts were complemented with their corresponding genes in the pUCP18 vector to confirm the roles of kdpD and aauS in biofilm formation. Empty plasmid controls (pUCP18) were used for WT strains and knockouts. At 24 h, knockout strains with the empty plasmids ΔkdpD::pUCP18 and ΔaauS::pUCP18 showed 38.7% and 29.0% reduction in biofilm formation, respectively compared to WT::pUCP18 (Figures 2A, B). Similarly, ΔkdpD::pUCP18 and ΔaauS::pUCP18 showed 62.7% and 58.7% biofilm reduction, respectively at 48 h. The complemented strains ΔkdpD::pUCP18kdpD, and ΔaauS::pUCP18aauS showed biofilm formation similar to that of WT::pUCP18 (Figures 2C, D). These results indicate that kdpD and aauS contribute to biofilm formation. Furthermore, we investigated biofilm formation in the WT, ΔkdpD, ΔaauS, and complementation strains through CLSM using empty plasmid controls. At 24 and 48 h, ΔkdpD::pUCP18 and ΔaauS::pUCP18 showed reduced biofilm formation compared to WT::pUCP18, whereas their complemented strains ΔkdpD::pUCP18kdpD and ΔaauS::pUCP18aauS exhibited similar biofilm formation as WT::pUCP18 (Figures 2E–H). Based on these results, we concluded that kdpD and aauS play vital roles in the biofilm formation of P. aeruginosa.
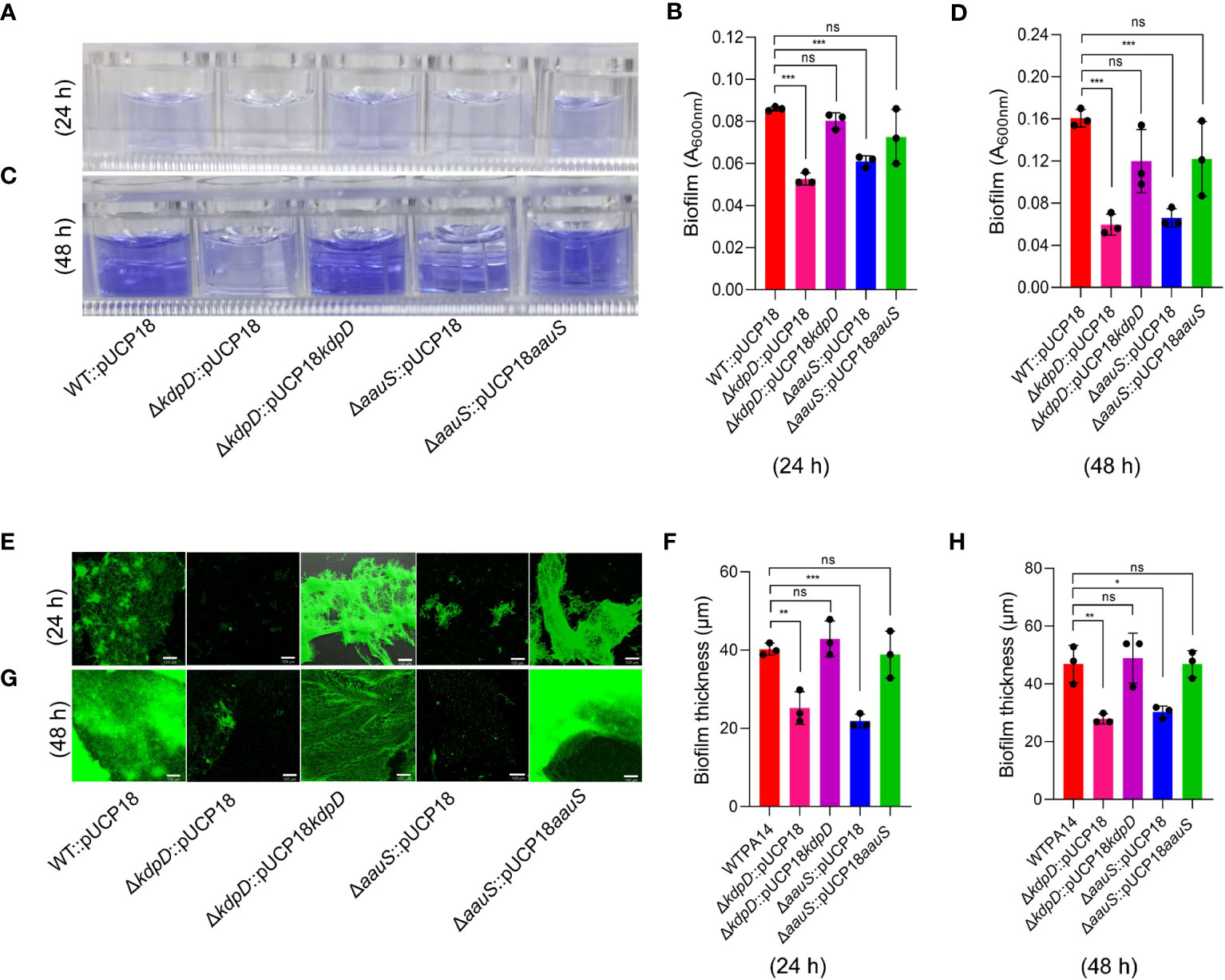
Figure 2. The roles of kdpD and aauS in biofilm formation. Qualitative and quantitative analysis of biofilm formation by WT::pUCP18, ΔkdpD::pUCP18, ΔaauS::pUCP18, ΔkdpD::pUCP18kdpD, and ΔaauS::pUCP18aauS at 24 and 48 h, performed using 96-well polystyrene plates (A–D) and 8-chambered covered glass slides (E–H). (A–D) Microtiter wells showing the CV-stained biofilm phenotype at 24 h (A) and 48 h (C), and their corresponding quantification at 24 h (B) and 48 h (D). (E–H) A SYTO9- and PI-labelled fluorescent photomicrograph showing biofilm phenotypes at 24 h (E) and 48 h (F) and their corresponding thickness, as measured through 3D images from CLSM using a 10× objective lens at 24 h (G) and 48 h (H). Mutant strains ΔkdpD::pUCP18 and ΔaauS::pUCP18 showed considerably reduced biofilm formation compared to the WT::pUCP18 strain, as determined using CV staining and fluorescence microscopic analysis. Similar to WT::pUCP18, the complemented strains (ΔkdpD::pUCP18kdpD and ΔaauS::pUCP18aauS) restored the biofilm, confirming the role of kdpD and aauS in the biofilm phenotype of P. aeruginosa. All the experiments were performed in triplicates. The significance of the data was analyzed using Student’s t-test. P < 0.05 was considered statistically significant (ns, non significant; *p < 0.05, **p < 0.01, and ***p < 0.005). The scale bar is 100 μm in confocal images.
3.3 Roles of kdpD and aauS in motility
Bacterial motility is an important virulence factor in host-pathogen interactions. Therefore, we investigated the relevance of kdpD and aauS in motility by analyzing the comparative swarming phenotypes of the WT with ΔkdpD, ΔaauS, and mutant-complemented strains. Our data revealed markedly reduced swarming (69.7%) in ΔfleS compared to the WT (Figures 3A, B). Contrastingly, ΔkdpD and ΔaauS showed 37.5% and 35.8% reduction in swarming motility, respectively compared to the WT (Figures 3A, B). These results indicate that kdpD and aauS contribute to swarming motility.
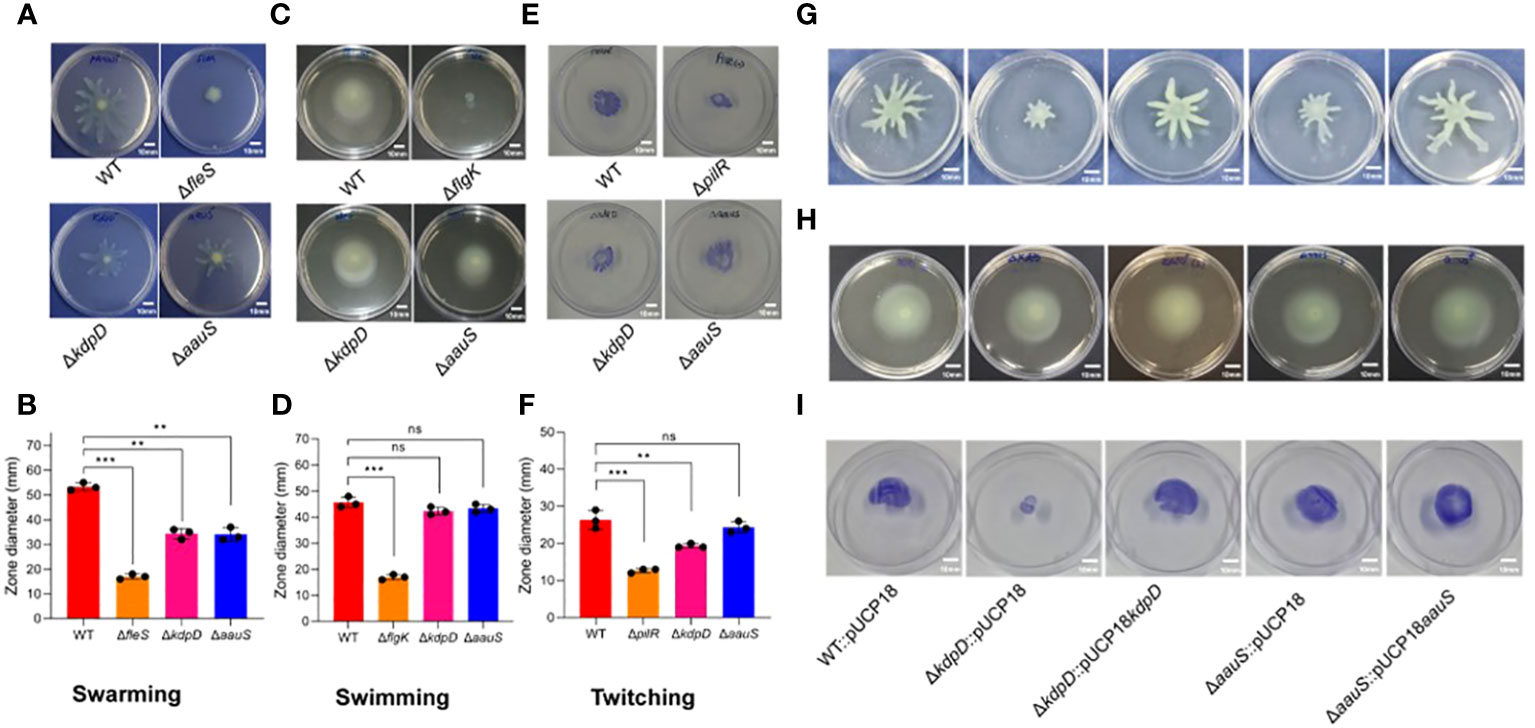
Figure 3. Assessment of the roles of kdpD and aauS in motility. Comparative qualitative and quantitative analysis of the motility phenotypes of ΔkdpD and ΔaauS mutant strains with the WT, including (A, B) swarming (A) and its quantification (B); (C, D) swimming (C) and its quantification (D); and (E, F) twitching (E) and its quantification (F). The ΔkdpD and ΔaauS mutants with empty vectors (ΔkdpD::pUCP18, ΔaauS::pUCP18) were both complemented (ΔkdpD::pUCP18kdpD and ΔaauS::pUCP18aauS). The complemented strains showed a restoration of swarming (G), swimming (H), and (I) twitching motility similar to that of the WT P. aeruginosa containing an empty vector (WT::pUCP18). All the mobility assay experiments were conducted three times. The significance of the data was analyzed using Student’s t-test. P < 0.05 was considered statistically significant (ns, non significant; **p < 0.01, and ***p < 0.005). The scale bar is 10 mm in motility plates.
The swimming phenotype is also important for flagellar motility, which plays an important role in infection by various pathogenic bacteria (Horstmann et al., 2017; Corral et al., 2020). Therefore, the involvement of kdpD and aauS was investigated to assess their roles in swimming motility. Here, ΔflgK showed a reduction of 62.9% in swimming compared to WT, whereas ΔkdpD and ΔaauS did not (Figures 3C, D). These findings suggest that these two genes are not involved in swimming motility in P. aeruginosa. Furthermore, we compared the twitching phenotypes of ΔkdpD and ΔaauS knockouts with those of the WT strain. We observed 62.9% and 26.5% reductions in twitching activity in ΔpilR and ΔkdpD compared to the WT. However, no reduction was observed in the twitching phenotype of the ΔaauS mutant strain (Figures 3E, F).
We confirmed these results by investigating the motility of WT::pUCP18, ΔkdpD::pUCP18, ΔaauS::pUCP18, ΔkdpD::pUCP18kdpD, and ΔaauS::pUCP18aauS. In this study, ΔkdpD::pUCP18 and ΔaauS::pUCP18 showed reduced swarming compared to the empty vector control (WT::pUCP18). In contrast, their corresponding mutant-complemented strains ΔkdpD::pUCP18kdpD and ΔaauS::pUCP18aauS exhibited a swarming phenotype similar to that of WT::pUCP18 (Figure 3G). These results indicate that kdpD and aauS contributory roles into the swarming mobility of P. aeruginosa. Complementation of the sensor kinases aauS and kdpD further confirmed that these genes have no role in swimming motility (Figure 3H). aauS displayed no role in twitching, whereas kdpD played a considerable role in twitching mobility. Similarly, ΔkdpD::pUCP18 showed reduced twitching, and ΔkdpD::pUCP18kdpD restored the phenotype to WT::pUCP18 (Figure 3I). These results indicate that kdpD contributes to swarming and twitching motility, whereas aauS only affects the swarming motility of P. aeruginosa.
3.4 The roles of kdpD and aauS in the inhibition of genes involved in biofilm and motility
Well-known genes were initially considered to assess the relative gene expression in ΔkdpD and ΔaauS knockouts with the empty plasmids to elucidate the role of kdpD and aauS in biofilm formation, swarming, and twitching mobility. Many genes play important roles in P. aeruginosa biofilm formation. For example, lasA plays a key role in biofilm development (Thi et al., 2020) while the pelA markedly contributes to the pellicle matrix formation of biofilm in P. aeruginosa (Friedman and Kolter, 2004; Sakuragi and Kolter, 2007; Colvin et al., 2012; Wei and Ma, 2013). Additionally, cupA is a crucial fimbria gene that contributes to biofilm formation via adhesion (Vallet et al., 2001; Vallet-Gely et al., 2007). Therefore, lasA, pelA, and cupA expression was investigated in the WT with empty plasmids and knockout with the empty plasmid strains. We found reduction in lasA expression at 10 h by 62.5% and 39.1% in ΔkdpD::pUCP18 and ΔaauS::pUCP18, respectively (Figure 4A). Complemented strains ΔkdpD::pUCP18kdpD and ΔaauS::pUCP18aauS restored expression levels to the level of of WT::pUCP18 (Figure 4A). Furthermore, lasA expression was also found to be reduced at 24 and 48 h time points by 25.9% and 19.1% in ΔkdpD::pUCP18; and 22.0% and 26.0% in case of ΔaauS::pUCP18, respectively (Figures 4B, C). These results demonstrate that kdpD and aauS contribute to initial biofilm formation by controlling lasA expression. We found no significant reduction in pelA and cupA in both the ΔkdpD::pUCP18 and ΔaauS::pUCP18 strains at 10 and 24 h (Supplementary Figures S4A–D). However, at 48 h pelA showed slightly reduced expression of 13.0% and 31.0% in the ΔkdpD::pUCP18 and ΔaauS::pUCP18 strains, respectively (Figure 4D). Furthermore, cupA also showed reduced expression of 21.9% and 15.3% at 48 h in the ΔkdpD::pUCP18 and ΔaauS::pUCP18 strains, respectively (Figure 4E). Complemented strains ΔkdpD::pUCP18kdpD and ΔaauS::pUCP18aauS restored expression levels to the level of WT::pUCP18 (Figure 4E). Overall, these results indicate that kdpD and aauS contribute to biofilm formation by modulating the expression of lasA, pelA and cupA genes.
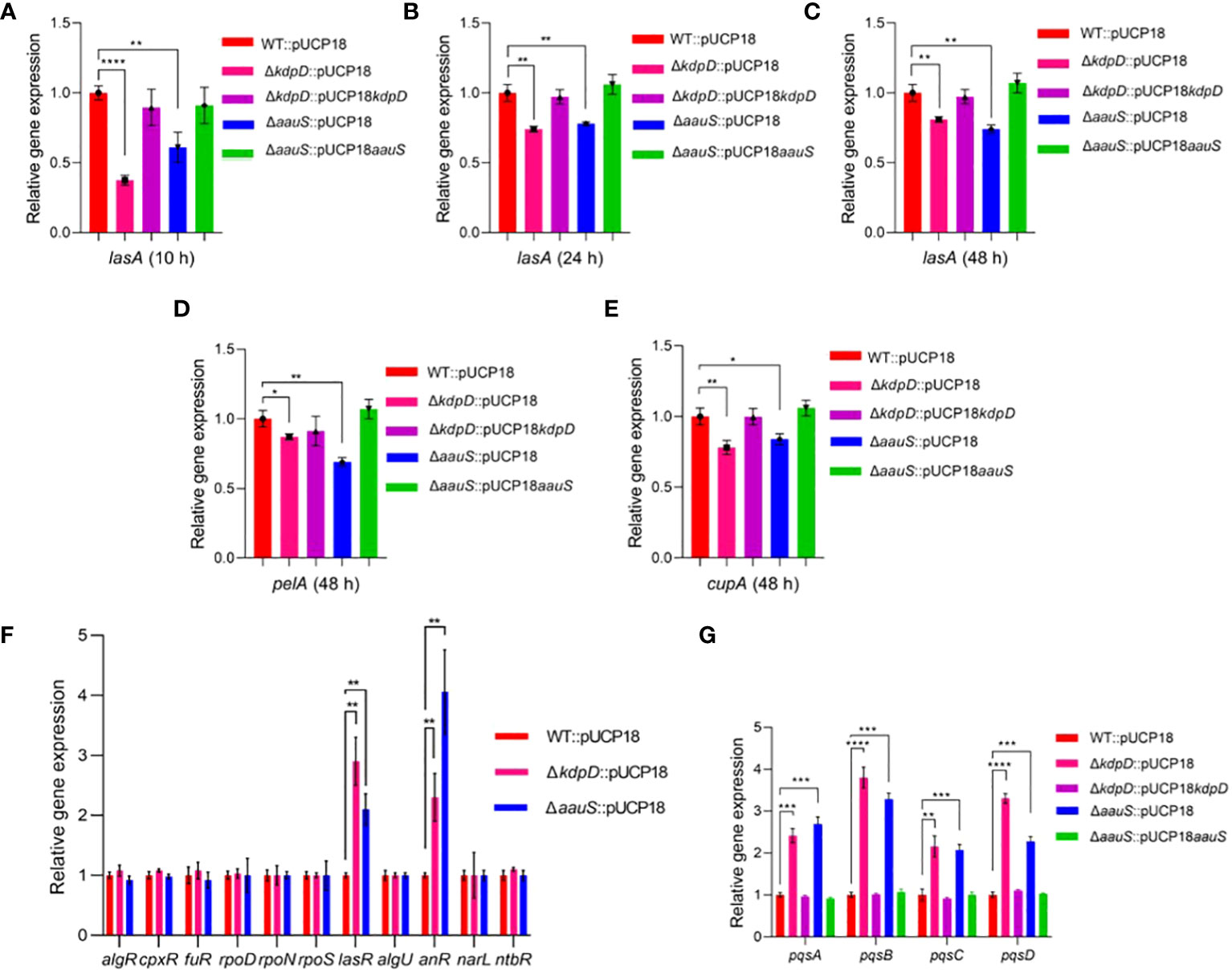
Figure 4. Quantitative-RT PCR analysis of the transcription factors (TFs) and their downstream genes in kdpD and aauS knockouts affecting biofilm formation, motility, and pathogenic potentials. (A–C) Relative expression of biofilm-regulated lasA in kdpD::pUCP18 and ΔaauS::pUCP18 mutants compared to WT::pUCP18 and their complemented ΔkdpD::pUCP18kdpD and ΔaauS::pUCP18aauS strains at 10, 24 and 48 h; (D) Relative expression of pelA in kdpD::pUCP18 and ΔaauS::pUCP18 mutants compared to WT::pUCP18 and their complemented ΔkdpD::pUCP18kdpD and ΔaauS::pUCP18aauS strains at 48 h; (E) Relative expression of cupA in kdpD::pUCP18 and ΔaauS::pUCP18 mutants compared to WT::pUCP18 and their complemented ΔkdpD::pUCP18kdpD and ΔaauS::pUCP18aauS strains at 48 h; (F) Relative gene expression of various transcription regulators (algR, cpxR, fuR, rpoD, rpoN, rpoS, lasR, algU, anR, narL, and ntbR in the kdpD::pUCP18 and ΔaauS::pUCP18 mutants) compared to the WT::pUCP18 strain; and (G) Relative gene expression of the PQS operon (pqsA, pqsB pqsC and pqsD) in ΔkdpD::pUCP18 and ΔaauS::pUCP18 mutants compared to the WT::pUCP18 strain and their complimented strains ΔkdpD::pUCP18kdpD and ΔaauS::pUCP18aauS. All the experiments were performed in triplicates. The significance of the data was analyzed using Student’s t-test. P < 0.05 was considered statistically significant (*p < 0.05, **p < 0.01, and ***p < 0.005, ****p < 0.0001).
Many genes and transcription regulators involved in flagellar biosynthesis, assembly and regulation are relevant to motility (Dasgupta et al., 2003). For example, fleQ is an important transcription regulator in P. aeruginosa, contributing to transcription of many flagellar genes (Arora et al., 1997). Similarly, FleS/FleR is a crucial TCS that modulates flagellar motility and adhesion (Ritchings et al., 1995). FleN is an anti-activator of FleQ, which plays a vital role in retaining a single flagellum (Dasgupta et al., 2000). In addition, sigma factor σ54 encoded by rpoN contributes to the regulation of flagellar expression (Totten et al., 1990). Furthermore, flgM is an important flagellar gene whose interactions with the sigma factor fliA contribute to flagellar biogenesis (Frisk et al., 2002). We analyzed the expression of several flagellar genes and transcription regulators in ΔkdpD and ΔaauS strains to investigate kdpD and aauS contribution to motility. There were no significant differences in the expression levels of the major motility-regulated genes fleS, fleR, fleQ, flgK, fliC, fliD, flgD, and flgE (Supplementary Figure S5). Therefore, a top-down approach was applied to comprehensively elucidate the mechanistic roles of kdpD and aauS in biofilm, motility, and pathogenesis. The expression of known transcription factors (TF) in ΔkdpD::pUCP18 and ΔaauS::pUCP18 compared to WT::pUCP18 was examined using q-RT-PCR. Accordingly, the transcriptional regulators (algR, cpxR, furR, rpoD, rpoN, rpoS, lasR, algU, anR, narL, and nbtR) that are directly or indirectly involved in biofilm formation, motility, and virulence gene expression have been assessed (Pusic et al., 2021). In this study, the expression levels of lasR was enhanced by 190% and 110% in ΔkdpD::pUCP18 and ΔaauS::pUCP18, respectively, compared to WT::pUCP18 (Figure 4F). Secondly, the anR expression was increased by 130% and 306% in ΔkdpD::pUCP18 and ΔaauS::pUCP18, respectively; compared to WT::pUCP18 (Figure 4F). anR and lasR contribute to biofilm formation and motility by controlling the Pseudomonas quinolone signal (PQS) quorum sensing (QS) system (Pusic et al., 2021). The las-QS system is essential for PQS-stimulated biofilm formation in P. aeruginosa PA14 (Christiaen et al., 2014). These results partially demonstrate that kdpD and aauS contribute to biofilm formation by controlling lasA expression through the lasR TF. PQS suppresses swarming motility in P. aeruginosa PA14 (Guo et al., 2014; García-Reyes et al., 2021). Therefore, we confirmed the expression levels of pqs in ΔkdpD::pUCP18 and ΔaauS::pUCP18. Our results showed increased pqsA, pqsB, pqsC, and pqsD expression (141%, 208%, 115% and 230%) in ΔkdpD::pUCP18, and (169%, 228%, 107% and 127%) in ΔaauS::pUCP18 strains (Figure 4G). Their expression was restored to that observed in WT::pUCP18 in the complemented (ΔkdpD::pUCP18kdpD and ΔaauS::pUCP18aauS) strains. Overall, these results indicate that kdpD and aauS contribute to swarming motility by altering the expression of the PQS quorum sensing system controlled by anR and lasR transcriptional regulators.
3.5 ΔkdpD and ΔaauS attenuate HeLa cell invasion and increased host cell survival
P. aeruginosa mainly invades mammalian epithelial cells via the type III secretion system (T3S) (Hernández-Padilla et al., 2017; Kroken et al., 2018). Therefore, we investigated the invasive activity of mutant strains with empty plasmids to evaluate the roles of kdpD and aauS in cell invasion. HeLa cells were infected with bacteria at an MOI of 10 for 120 min, and the number of intracellular bacterial cells was measured through CFU enumeration. Strains ΔkdpD::pUCP18 and ΔaauS::pUCP18 showed reduced invasion activity, with intracellular CFUs difference of 2.0×103 and 2.3×103 CFU/mL, respectively compared to WT::pUCP18. In contrast, the intracellular CFU of the WT::pUCP18 strain was 3.7×103 CFU/mL (Figure 5A). Moreover, the complemented strains ΔkdpD::pUCP18kdpD and ΔaauS::pUCP18aauS restored the intracellular CFU count to that of WT::pUCP18.
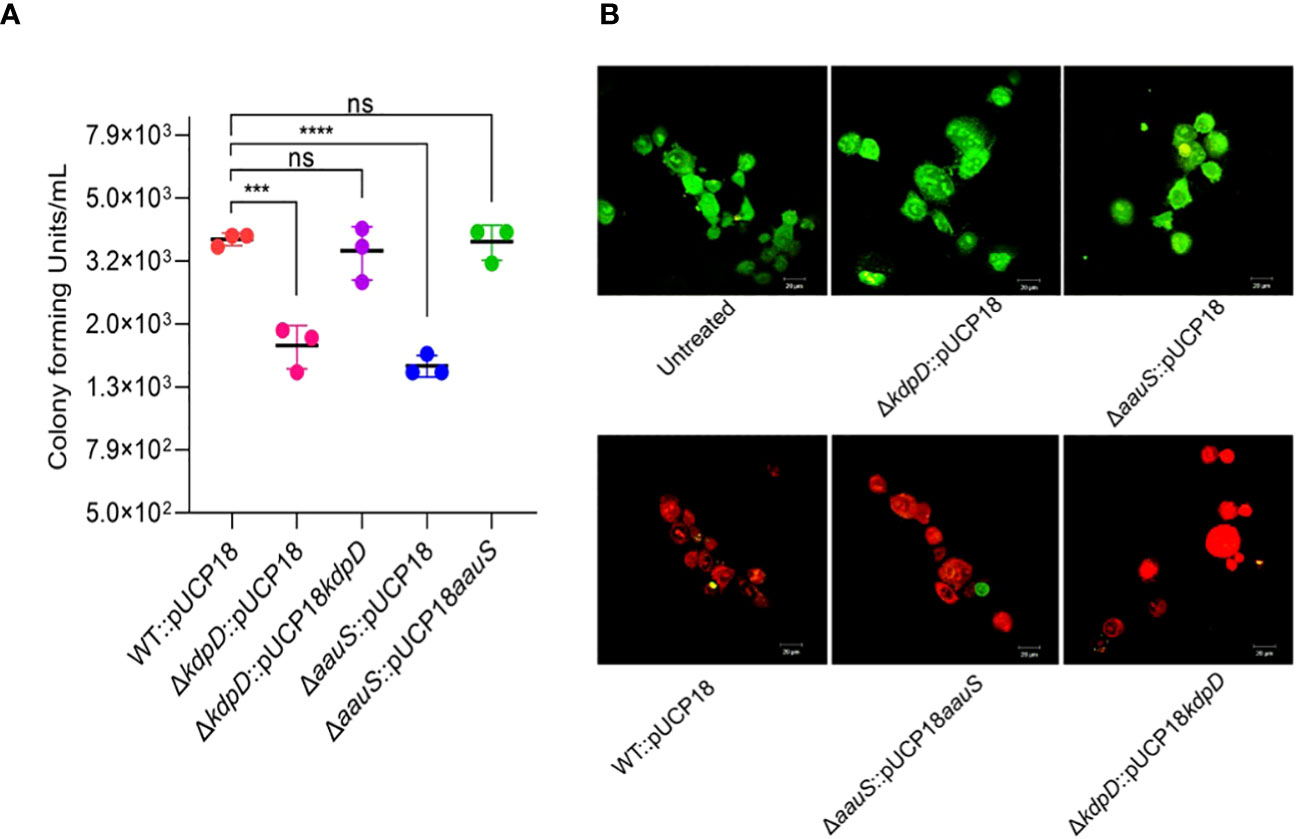
Figure 5. Role of kdpD and aauS in host cell survival in mammalian cell culture conditions. (A) Assessment of role of kdpD and aauS the internalization potential of using GPA. HeLa cells were infected with bacteria at an MOI of 10 for 120 min in a 5% CO2 environment at 37 °C. Extracellular bacteria were killed through gentamycin treatment, and internalized bacterial cells were assessed through CFU determination. (B) Assessment of the in vitro pathogenic potential of various strains using live/dead staining of the host-pathogen complex after infection, wherein HeLa cells were infected with WT::pUCP18, ΔkdpD::pUCP18, ΔaauS::pUCP18, ΔkdpD::pUCP18kdpD, and ΔaauS::pUCP18aauS at an MOI of 10. Untreated cells were used as negative controls. After 2 h infection, the cells were stained with PI (red fluorescence) and fluorescein diacetate (green fluorescence) and visualized using CLSM. Green represents live cells, whereas red represents dead cells. Knockout strains of kdpD and aauS with empty vectors (ΔkdpD::pUCP18, ΔaauS::pUCP18) were the least toxic to host cells compared to the wild-type with empty vectors (WT::pUCP18) and complemented strains (ΔkdpD::pUCP18kdpD and ΔaauS::pUCP18aauS). The experiments were performed in triplicates. The significance of the data was analyzed using Student’s t-test. P < 0.05 was considered statistically significant (ns, non significant; ***p < 0.005, ****p < 0.0001). The scale bar is 20 μm in live dead images.
The host-pathogen complex was stained with live/dead (green/red) reagents to further evaluate the roles of kdpD and aauS in HeLa cell infection. Uninfected HeLa cells mostly retained green fluorophores (Untreated, Figure 5B). A high number of dead host cells was observed when WT::pUCP18 was added (Figure 5B). Moreover, a markedly increased number of live cells was observed when host cells were infected with ΔkdpD::pUCP18 or ΔaauS::pUCP18 (Figure 5B). Contrastingly, a considerable number of dead host cells was observed when the complemented strains ΔkdpD::pUCP18kdpD and ΔaauS::pUCP18aauS were used to infect HeLa cells. These results demonstrate the involvement of kdpD and aauS in host cell invasion and infection.
3.6 kdpD and aauS play critical roles in G. mellonella infection
G. mellonella larvae were challenged with WT::pUCP18, ΔkdpD::pUCP18, ΔaauS::pUCP18, ΔkdpD::pUCP18kdpD, and ΔaauS::pUCP18aauS at 10 CFU/larvae to further analyze the roles of kdpD and aauS in virulence. The WT::pUCP18 bacterial load was designed to allow 10% survival 72 h post-infection. Then, survival percentages of the larvae and their health indices were recorded. Our results indicated delayed mortality, with increased survival in ΔkdpD::pUCP18 (60%) and ΔaauS::pUCP18 (40%) compared to WT::pUCP18, after 72 h. This is consistent with the results obtained in the initial screening (Figure 1). We further investigated the infection properties of complemented ΔkdpD::pUCP18kdpD and ΔaauS::pUCP18aauS strains. The results showed a survival comparable to that of WT::pUCP18 cells (Figure 6A). We also recorded the health index of each strain based on movement, cocoon formation, melaninization, and survival. Our results showed greater movement, full cocoon formation with less melaninization, and increased survival in larvae infected with ΔkdpD::pUCP18 and ΔaauS::pUCP18 than in those infected with WT::pUCP18. However, larvae infected with ΔkdpD::pUCP18kdpD and ΔaauS::pUCP18aauSshowed less movement, greater melaninization, less cocoon formation, and decreased survival 72 h post-infection, demonstrating results similar to those of WT::pUCP18 (Figures 6B, C). Next, we determined the bacterial burden in the worms by measuring the number of bacteria in the infected larvae. Larvae infected with ΔkdpD::pUCP18 and ΔaauS::pUCP18 showed 1.7×105 and 3.4×105CFU/mL, respectively. These were lower than the 9.3×105 CFU/mL observed in the WT::pUCP18 strain (Figure 6D). However, strains ΔkdpD::pUCP18kdpD and ΔaauS::pUCP18aauS showed no significant reduction in CFU compared to WT::pUCP18 (Figure 6D). These results were consistent with the survival and health index results, indicating that kdpD and aauS play important roles in G. mellonella infection.
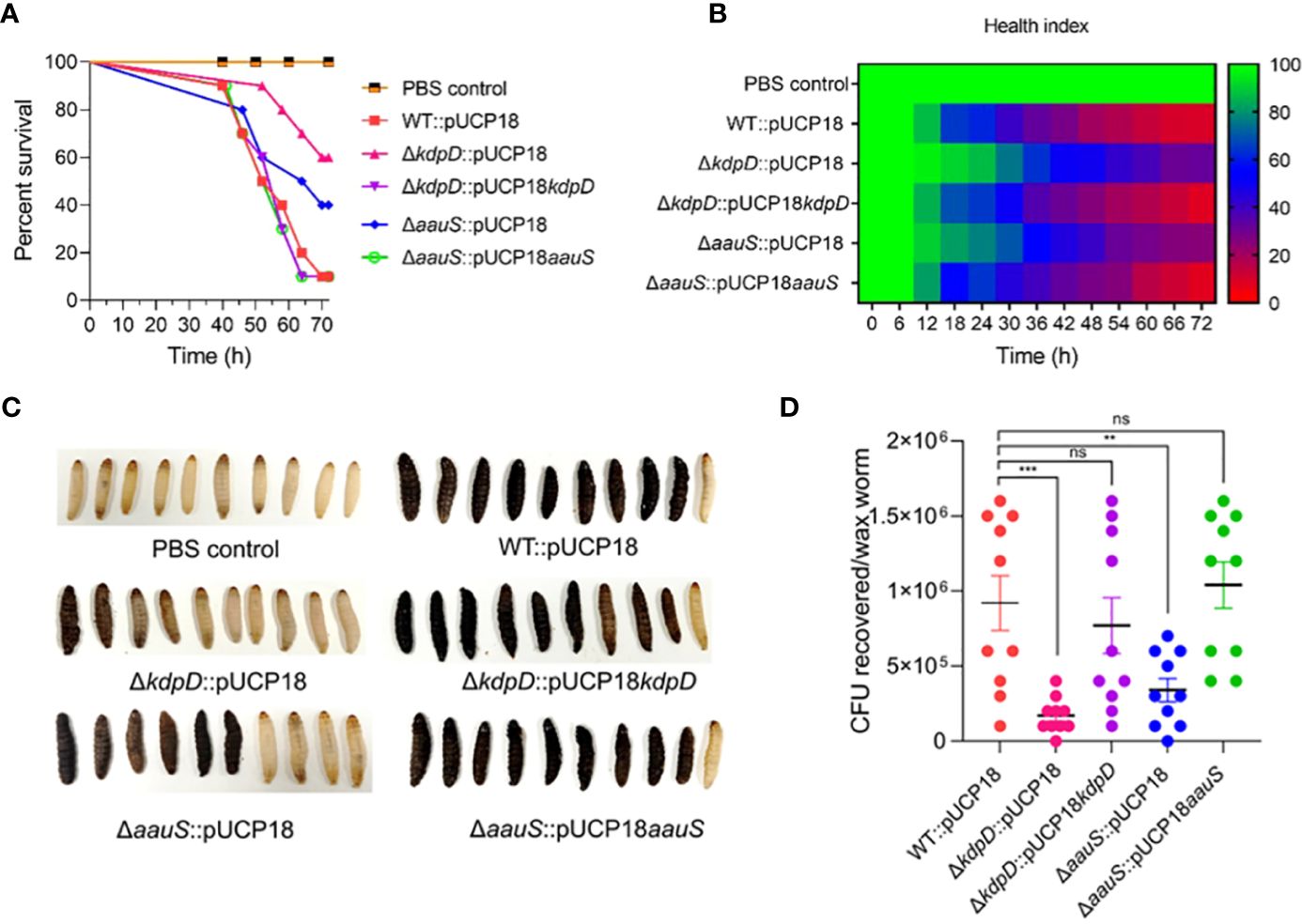
Figure 6. Confirming the roles of kdpD and aauS in the virulence potential of P. aeruginosa using the G. mellonella infection model. (A) Survival of G. mellonella larvae infected with 10 CFU of WT::pUCP18, ΔkdpD::pUCP18, ΔaauS::pUCP18, ΔkdpD::pUCP18kdpD, and ΔaauS::pUCP18aauS strains. The number of larvae in each group was 10 (n =10). The larvae were injected with 20 µL bacterial solution, and their survival was examined for 72 (h) The control group received 20 µL PBS. (B) The health index of larvae is plotted as an average of each group at multiple time points based on the health scoring index (movement, melaninization, cocoon formation, and survival). (C) An image showing the differential melaninization and death of waxworms infected with different bacterial strains; and (D) Assessment of CFUs recovered on selective media CA after 72 h of infection. The experiments were performed in triplicates. The significance of the data was analyzed using Student’s t-test. P < 0.05 was considered statistically significant (ns, non significant; **p < 0.01, and ***p < 0.005).
4 Discussion
P. aeruginosa is a gram-negative environmental MDR pathogen responsible for acute and chronic diseases, owing to its extensive biofilm formation. Protease, elastase, and phospholipase (Vasil, 1986) determine P. aeruginosa biofilm production, motility, pigment secretion, and pathogenic potential. These virulence determinants play important roles in bacterial colonization and host tissue invasion, which may lead to life-threatening infections. Chronic infections caused by biofilm formation include chronic pneumonia in patients with CF, colonization of ventilators and urinary catheters, and infected burn wounds (Bjarnsholt, 2013). Moreover, pseudomonad biofilm is a major cause of antimicrobial therapy failure because the biofilm enhances antimicrobial resistance by 2–3 log values (Yin et al., 2022).
P. aeruginosa uses TCSs to sense signals and coordinate bacterial virulence. The sensor HKs and RRs considerably contribute to P. aeruginosa metabolism and pathophysiology. However, only a few of these TCSs have been studied to investigate their role in virulence. Therefore, we screened a few P. aeruginosa TCSs whose functions in virulence are not clearly understood. We assessed the virulence potential of their mutant strains and identified two HKs that are involved in P. aeruginosa virulence. Furthermore, we investigated and confirmed the roles of these TCSs in the biofilm formation and motility of P. aeruginosa.
KdpD senses environmental signals to activate the expression of the K+ transport system in E. coli (Epstein, 2016). Previous studies have emphasized its role in the virulence of other bacteria (Zhao et al., 2010; Xue et al., 2020). KdpD/KdpE is responsible for the virulence and motility of avian pathogenic E. coli (Xue et al., 2020). This TCS regulates the transcription of the flagellar-associated genes fliR and fliP. The deletion of KdpD/KdpE modulates the expression of several genes, including cap (synthesis of capsule), hla (alpha-toxin), spa (surface protein), geh (lipase gene), and hlgB (gamma-hemolysin), in S. aureus (Zhao et al., 2010). The AauS/AauR TCS is involved in acidic amino acid utilization in Pseudomonas putida (Sonawane and Singh, 2006). In P. putida, ΔaauS and ΔaauR cannot consume glutamate, aspartate, or glutamine as the sole nitrogen and carbon sources (Sonawane and Singh, 2006). Additionally, KdpD/KdpE regulation responds to virulence-related conditions, including phagocytosis, exposure to microbicides, host hormones, and quorum sensing (QS) signals (Freeman et al., 2013; Ali et al., 2017). However, the role of aauS in the virulence of other bacteria, including P. aeruginosa has not been intensively investigated until now. Therefore, our results provide clear evidence of kdpD and aauS-mediated P. aeruginosa virulence.
Several TCSs contribute to biofilm formation in P. aeruginosa by sensing signals, eventually causing the bacteria to enter the sessile phase (Mikkelsen et al., 2011). GacS/GacA acts along with RetS and LadS to regulate the switch between planktonic and biofilm lifestyles (Mikkelsen et al., 2011; Chambonnier et al., 2016). Biofilm formation occurs at different stages, with BfiS/BfiR, MifS/MifR, and BfmS/BfmR playing essential roles in irreversible attachment, microcolony formation, and maturation of biofilm (Petrova and Sauer, 2009). Furthermore, microarray analysis and promoter fusion confirmed the role of FleSR in biofilm formation (Dasgupta et al., 2003). Our quantitative biofilm results and confocal microscopy data showed kdpD and aauS involvement in the regulation of biofilm formation (Figure 2). Therefore, in addition to known biofilm-associated TCSs, we propose that KdpD/KdpE and AauR/AauS are responsible for biofilm formation. In P. aeruginosa, QS systems play vital roles in biofilm formation (Solano et al., 2014; Thi et al., 2020). Therefore, the HK KdpD sensor could activate its cognate RR to express the genes involved in biofilm formation. Our gene expression results showed reduced expression of pelA, cupA, increased lasR TF expression and reduced expression of the downstream gene lasA in ΔkdpD::pUCP18 and ΔaauS::pUCP18, confirming that kdpD and aauS involvement in biofilm formation occurs via the pelA, cupA and las-QS system (Figures 4A–E). The role of KdpD and AauS in P. aeruginosa biofilm is consistent with study of Badal et al. (2020) wherein KdpD and AauS was identified to involve in biofilm formation of P. aeruginosa on endotracheal tubes (Badal et al., 2020).
The pathogenesis of P. aeruginosa depends on motility, which modulates its mobilization and colonization during biofilm formation (O'Toole and Kolter, 1998), as demonstrated in studies investigating P. fluorescens and E. coli (Korber et al., 1994; Pratt and Kolter, 1998). Many TCSs and genes are associated with swarming, swimming, and twitching motility (Sultan et al., 2021). For example, FleS/FleR regulates the expression of flagellar biosynthetic genes and alters swarming motility. Similarly, CarS/CarR senses and modulates Ca2+ homeostasis. This crucial TCS further contributes to swarming motility via carP (Guragain et al., 2016). PilS/PilR is another important TCS responsible for twitching and flagellum-dependent swimming motility (Kilmury and Burrows, 2018a). As we did not observe any changes in pilS, pilR, carS, or carR in the kdpD or aauS knockout strains, their contribution to motility seemed to be independent of PilS/PilR or CarS/CarR involvement. Similarly, we did not observe any mechanistic role of kdpD and aauS in motility by FleS/FleR, as there were no significant differences in the expression of flagellar genes. The metabolic regulator anR and quorum-sensing regulator lasR control the PQS system to regulate biofilm formation and motility (Pusic et al., 2021). PQS suppresses swarming motility in P. aeruginosa PA14 (Guo et al., 2014; García-Reyes et al., 2021). In this study, anR and lasR expression was increased in kdpD::pUCP18 and ΔaauS::pUCP18 knockouts, confirming the roles of kdpD and aauS in swarming motility, which is conferred by these transcription regulators. The expression of PQS genes increases when anR and lasR expression is upregulated (Pusic et al., 2021). In our study, pqsA, pqsB, pqsC, and pqsD expression was also increased in ΔkdpD::pUCP18 and ΔaauS::pUCP18 knockouts (Figure 4G). This expression was restored to the level of WT::pUCP18 in the complement strains ΔkdpD::pUCP18kdpD and ΔaauS::pUCP18aauS. Overall, these results confirmed that kdpD plays a role in swarming motility by altering the expression of PQS quorum sensing system controlled by anR and lasR transcription regulators.
Several swarming-deficient mutants of P. aeruginosa showed poor biofilm formation, suggesting a strong relationship between swarming and biofilm formation (Overhage et al., 2007). Consistent with this finding, kdpD and aauS were associated with the swarming phenotype and biofilm formation in our study. However, twitching activity was not altered by aauS but was affected by kdpD. Furthermore, kdpD and aauS exhibited no roles in swimming motility. These results suggest that KdpD and AauS sensor HKs contribute to P. aeruginosa virulence through swarming motility.
In this study, we identified the roles of kdpD and aauS in the biofilm formation and motility of P. aeruginosa via several key genes involved in these virulence phenotypes. These roles were validated through cell invasion (Figure 5) and a G. mellonella infection model (Figure 6). The effects of kdpD and aauS were minimal on phenotype reduction in their knockout strains compared with the genes used as controls, such as pqsA for biofilm formation and fleS, flgK, and pilR for motility. However, our results clearly demonstrate the contributions of kdpD and aauS to the virulence phenotype. This was confirmed by the reduced infection activity of the knockout strain in the infection model and reduced expression of genes that control virulence. Therefore, we propose that kdpD and aauS are virulence-controlling TCSs. This study represents a milestone in the development of methods for identifying and validating key genes involved in bacterial virulence. Furthermore, our findings provide a molecular basis for understanding the virulence of P. aeruginosa and contribute to the design of inhibitors that target TCSs to overcome biofilm-mediated AMR.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material. Further inquiries can be directed to the corresponding authors.
Ethics statement
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because We do not use animals for experiments.
Author contributions
MS: Writing – original draft, Writing – review & editing, Data curation, Investigation, Methodology. RA: Writing – review & editing, Conceptualization, Supervision. AC: Conceptualization, Supervision, Writing – review & editing, Investigation, Writing – original draft. KK: Conceptualization, Writing – original draft, Writing – review & editing, Funding acquisition.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Research Foundation (NRF) of Korea grant to KKK (2017M3A9E4078553 and 2021M3A9I2080487). The funders had no role in the study design, data collection, analysis, interpretation, or the decision to submit this work for publication.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1270667/full#supplementary-material
References
Ali, M. K., Li, X., Tang, Q., Liu, X., Chen, F., Xiao, J., et al. (2017). Regulation of inducible potassium transporter KdpFABC by the KdpD/KdpE two-component system in Mycobacterium smegmatis. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00570
Arora, S.K., Ritchings, B.W., Almira, E.C., Lory, S., Ramphal, R. (1997). A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J. Bacterial. 179 (17), 5574–5581. doi: 10.1128/jb.179.17.5574-5581.1997
Badal, D., Jayarani, A. V., Kollaran, M. A., Kumar, A., Singh, V. (2020). Pseudomonas aeruginosa biofilm formation on endotracheal tubes requires multiple two-component systems. J. Med. Microbiol. 69 (6), 906–919. doi: 10.1099/jmm.0.001199
Bisht, K., Moore, J. L., Caprioli, R. M., Skaar, E. P., Wakeman, C. A. (2021). Impact of temperature-dependent phage expression on Pseudomonas aeruginosa biofilm formation. NPJ Biofilms Microbiomes 7 (1), 22. doi: 10.1038/s41522-021-00194-8
Bjarnsholt, T. (2013). The role of bacterial biofilms in chronic infections. APMIS 121, 1–58. doi: 10.1111/apm.12099
Browne, N., Heelan, M., Kavanagh, K. (2013). An analysis of the structural and functional similarities of insect hemocytes and mammalian phagocytes. Virulence 4 (7), 597–603. doi: 10.4161/viru.25906
Chambonnier, G., Roux, L., Redelberger, D., Fadel, F., Filloux, A., Sivaneson, M., et al. (2016). The hybrid histidine kinase LadS forms a multicomponent signal transduction system with the GacS/GacA two-component system in Pseudomonas aeruginosa. PloS Genet. 12 (5), e1006032. doi: 10.1371/journal.pgen.1006032
Champion, O. L., Cooper, I. A., James, S. L., Ford, D., Karlyshev, A., Wren, B. W., et al. (2009). Galleria mellonella as an alternative infection model for Yersinia pseudotuberculosis. Microbiology 155 (5), 1516–1522. doi: 10.1099/mic.0.026823-0
Champion, O. L., Karlyshev, A. V., Senior, N. J., Woodward, M., La Ragione, R., Howard, S. L., et al. (2010). Insect infection model for Campylobacter jejuni reveals that O-methyl phosphoramidate has insecticidal activity. J. Infect. Dis. 201 (5), 776–782. doi: 10.1086/650494
Champion, O. L., Titball, R. W., Bates, S. (2018). Standardization of G. mellonella larvae to provide reliable and reproducible results in the study of fungal pathogens. J. Fungi. 4 (3), 108. doi: 10.3390/jof4030108
Chaurasia, A. K., Thorat, N. D., Tandon, A., Kim, J.-H., Park, S. H., Kim, K. K. (2016). Coupling of radiofrequency with magnetic nanoparticles treatment as an alternative physical antibacterial strategy against multiple drug resistant bacteria. Sci. Rep. 6 (1), 33662. doi: 10.1038/srep33662
Chen, H., Wubbolts, R. W., Haagsman, H. P., Veldhuizen, E. J. (2018). Inhibition and eradication of Pseudomonas aeruginosa biofilms by host defence peptides. Sci. Rep. 8 (1), 1–10. doi: 10.1038/s41598-018-28842-8
Christiaen, S. E., Matthijs, N., Zhang, X.-H., Nelis, H. J., Bossier, P., Coenye, T. (2014). Bacteria that inhibit quorum sensing decrease biofilm formation and virulence in Pseudomonas aeruginosa PAO1. Pathog. Dis. 70 (3), 271–279. doi: 10.1111/2049-632X.12124
Colvin, K. M., Irie, Y., Tart, C. S., Urbano, R., Whitney, J. C., Ryder, C., et al. (2012). The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ. Microbiol. 14 (8), 1913–1928. doi: 10.1111/j.1462-2920.2011.02657.x
Corral, J., Sebastià, P., Coll, N. S., Barbé, J., Aranda, J., Valls, M. (2020). Twitching and swimming motility play a role in Ralstonia solanacearum pathogenicity. mSphere 5 (2), e00740–e00719. doi: 10.1128/msphere.00740-19
Dasgupta, N., Arora, S. K., Ramphal, R. (2000). fleN, a gene that regulates flagellar number in Pseudomonas aeruginosa. J. Bacteriol. 182 (2), 357–364. doi: 10.1128/jb.182.2.357-364.2000
Dasgupta, N., Wolfgang, M. C., Goodman, A. L., Arora, S. K., Jyot, J., Lory, S., et al. (2003). A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50 (3), 809–824. doi: 10.1046/j.1365-2958.2003.03740.x
De Bentzmann, S., Plésiat, P. (2011). The Pseudomonas aeruginosa opportunistic pathogen and human infections. Environ. Microbiol. 13 (7), 1655–1665. doi: 10.1111/j.1462-2920.2011.02469.x
Desbois, A. P., Coote, P. J. (2011). Wax moth larva (Galleria mellonella): an in vivo model for assessing the efficacy of antistaphylococcal agents. J. Antimicrob. Chemother. 66 (8), 1785–1790. doi: 10.1093/jac/dkr198
Déziel, E., Comeau, Y., Villemur, R. (2001). Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J. Bacteriol. 183 (4), 1195–1204. doi: 10.1128/jb.183.4.1195-1204.2001
Egli, T. (2015). Microbial growth and physiology: a call for better craftsmanship. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00287
Epstein, W. (2016). The KdpD sensor kinase of Escherichia coli responds to several distinct signals to turn on expression of the Kdp transport system. J. Bacteriol. 198 (2), 212–220. doi: 10.1128/jb.00602-15
Francis, V. I., Stevenson, E. C., Porter, S. L. (2017). Two-component systems required for virulence in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 364 (11), fnx104. doi: 10.1093/femsle/fnx104
Freeman, Z. N., Dorus, S., Waterfield, N. R. (2013). The KdpD/KdpE two-component system: integrating K+ homeostasis and virulence. PloS Pathog. 9 (3), e1003201. doi: 10.1371/journal.ppat.1003201
Friedman, L., Kolter, R. (2004). Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51 (3), 675–690. doi: 10.1046/j.1365-2958.2003.03877.x
Frisk, A., Jyot, J., Arora, S., Ramphal, R. (2002). Identification and functional characterization of flgM, a gene encoding the anti-sigma 28 factor in Pseudomonas aeruginosa. J. Bacteriol. 184 (6), 1514–1521. doi: 10.1128/jb.184.6.1514-1521.2002
Gale, M. J., Maritato, M. S., Chen, Y.-L., Abdulateef, S. S., Ruiz, J. E. (2015). Pseudomonas aeruginosa causing inflammatory mass of the nasopharynx in an immunocompromised HIV infected patient: A mimic of Malignancy. IDCases 2 (2), 40–43. doi: 10.1016/j.idcr.2015.01.004
García-Reyes, S., Cocotl-Yañez, M., Soto-Aceves, M. P., González-Valdez, A., Servín-González, L., Soberón-Chávez, G. (2021). PqsR-independent quorum-sensing response of Pseudomonas aeruginosa ATCC 9027 outlier-strain reveals new insights on the PqsE effect on RhlR activity. Mol. Microbiol. 116 (4), 1113–1123. doi: 10.1111/mmi.14797
Guo, Q., Kong, W., Jin, S., Chen, L., Xu, Y., Duan, K. (2014). PqsR-dependent and PqsR-independent regulation of motility and biofilm formation by PQS in Pseudomonas aeruginosa PAO1. J. Basic Microbiol. 54 (7), 633–643. doi: 10.1002/jobm.201300091
Guragain, M., King, M. M., Williamson, K. S., Pérez-Osorio, A. C., Akiyama, T., Khanam, S., et al. (2016). The Pseudomonas aeruginosa PAO1 two-component regulator CarSR regulates calcium homeostasis and calcium-induced virulence factor production through its regulatory targets CarO and CarP. J. Bacteriol. 198 (6), 951–963. doi: 10.1128/jb.00963-15
Ha, D.-G., Kuchma, S. L., O’Toole, G. A. (2014a). “Plate-based assay for swarming motility in Pseudomonas aeruginosa,” in Pseudomonas methods and protocols. (Humana, New York, NY: Springer) 1149, 67–72. doi: 10.1007/978-1-4939-0473-0_8
Ha, D.-G., Kuchma, S. L., O’Toole, G. A. (2014b). “Plate-based assay for swimming motility in Pseudomonas aeruginosa,” in Pseudomonas methods and protocols. (Humana, New York, NY: Springer) 1149, 59–65. doi: 10.1007/978-1-4939-0473-0_7
Hernández-Padilla, L., Vázquez-Rivera, D., Sánchez-Briones, L. A., Díaz-Pérez, A. L., Moreno-Rodríguez, J., Moreno-Eutimio, M. A., et al. (2017). The antiproliferative effect of cyclodipeptides from Pseudomonas aeruginosa PAO1 on HeLa cells involves inhibition of phosphorylation of Akt and S6k kinases. Molecules 22 (6), 1024. doi: 10.3390/molecules22061024
Hill, L., Veli, N., Coote, P. J. (2014). Evaluation of Galleria mellonella larvae for measuring the efficacy and pharmacokinetics of antibiotic therapies against Pseudomonas aeruginosa infection. Int. J. @ Antimicrob. Agents 43 (3), 254–261. doi: 10.1016/j.ijantimicag.2013.11.001
Horstmann, J. A., Zschieschang, E., Truschel, T., de Diego, J., Lunelli, M., Rohde, M., et al. (2017). Flagellin phase-dependent swimming on epithelial cell surfaces contributes to productive Salmonella gut colonisation. Cell. Microbiol. 19 (8), e12739. doi: 10.1111/cmi.12739
Imdad, S., Batool, N., Pradhan, S., Chaurasia, A. K., Kim, K. K. (2018). Identification of 2′, 4′-dihydroxychalcone as an antivirulence agent targeting HlyU, a master virulence regulator in Vibrio vulnificus. Molecules 23 (6), 1492. doi: 10.3390/ijms22063128
Jander, G., Rahme, L. G., Ausubel, F. M. (2000). Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J. Bacteriol. 182 (13), 3843–3845. doi: 10.1128/jb.182.13.3843-3845.2000
Jurado-Martín, I., Sainz-Mejías, M., McClean, S. (2021). Pseudomonas aeruginosa: An audacious pathogen with an adaptable arsenal of virulence factors. Int. J. Mol. Sci. 22 (6), 3128. doi: 10.3390/ijms22063128
Kilmury, S. L., Burrows, L. L. (2018a). The Pseudomonas aeruginosa PilSR two-component system regulates both twitching and swimming motilities. mBio 9 (4), e01310–e01318. doi: 10.1128/mbio.01310-18
Kim, J.-H., Chaurasia, A. K., Batool, N., Ko, K. S., Kim, K. K. (2019). Alternative enzyme protection assay to overcome the drawbacks of the gentamicin protection assay for measuring entry and intracellular survival of staphylococci. Infect. Immun. 87 (5), e00119–e00119. doi: 10.1128/iai.00119-19
Kollaran, A. M., Joge, S., Kotian, H. S., Badal, D., Prakash, D., Mishra, A., et al. (2019). Context-specific requirement of forty-four two-component loci in Pseudomonas aeruginosa swarming. iScience 13, 305–317. doi: 10.1016/j.isci.2019.02.028
Korber, D. R., Lawrence, J. R., Caldwell, D. E. (1994). Effect of motility on surface colonization and reproductive success of Pseudomonas fluorescens in dual-dilution continuous culture and batch culture systems. Appl. Eviron. Microbiol. 60 (5), 1421–1429. doi: 10.1128/aem.60.5.1421-1429.194
Kostakioti, M., Hadjifrangiskou, M., Hultgren, S. J. (2013). Bacterial biofilms: development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb. Perspect. Med. 3 (4), a010306. doi: 10.1101/cshperspect.a010306
Kroken, A. R., Chen, C. K., Evans, D. J., Yahr, T. L., Fleiszig, S. M. (2018). The impact of ExoS on Pseudomonas aeruginosa internalization by epithelial cells is independent of fleQ and correlates with bistability of type three secretion system gene expression. Mbio 9 (3), e00668–e00618. doi: 10.1128/mbio.00668-18
Lebreton, F., Riboulet-Bisson, E., Serror, P., Sanguinetti, M., Posteraro, B., Torelli, R., et al. (2009). ace, which encodes an adhesin in Enterococcus faecalis, is regulated by Ers and is involved in virulence. Infect. Immun. 77 (7), 2832–2839. doi: 10.1128/iai.01218-08
Leitão, J. H. (2020). Microbial virulence factors. Int. J. Mol. Sci. 21 (15), 5320. doi: 10.3390/ijms21155320
Li, X., Gu, N., Huang, T. Y., Zhong, F., Peng, G. (2023). Pseudomonas aeruginosa: A typical biofilm forming pathogen and an emerging but underestimated pathogen in food processing. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.1114199
Liberati, N. T., Urbach, J. M., Miyata, S., Lee, D. G., Drenkard, E., Wu, G., et al. (2006). An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. 103 (8), 2833–2838. doi: 10.1073/pnas.0511100103
Magill, S. S., Edwards, J. R., Bamberg, W., Beldavs, Z. G., Dumyati, G., Kainer, M. A., et al. (2014). Multistate point-prevalence survey of health care–associated infections. N. Engl. J. @ Med. 370 (13), 1198–1208. doi: 10.1056/NEJMoa1306801
Ménard, G., Rouillon, A., Cattoir, V., Donnio, P.-Y. (2021). Galleria mellonella as a suitable model of bacterial infection: past, present and future. Front. Cell. Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.782733
Mikkelsen, H., Sivaneson, M., Filloux, A. (2011). Key two-component regulatory systems that control biofilm formation in Pseudomonas aeruginosa. Environ. Microbiol. 13 (7), 1666–1681. doi: 10.1111/j.1462-2920.2011.02495.x
Mitrophanov, A. Y., Groisman, E. A. (2008). Signal integration in bacterial two-component regulatory systems. Genes Dev. 22 (19), 2601–2611. doi: 10.1101/gad.1700308
Mittal, R., Grati, M., Gerring, R., Blackwelder, P., Yan, D., Li, J.-D., et al. (2014). In vitro interaction of Pseudomonas aeruginosa with human middle ear epithelial cells. PloS One 9 (3), e91885. doi: 10.1371/journal.pone.0091885
Mogayzel, P. J., Jr., Naureckas, E. T., Robinson, K. A., Brady, C., Guill, M., Lahiri, T., et al. (2014). Cystic Fibrosis Foundation pulmonary guideline. Pharmacologic approaches to prevention and eradication of initial Pseudomonas aeruginosa infection. Ann. Am. Thorac. Soc 11 (10), 1640–1650. doi: 10.1513/AnnalsATS.201404-166OC
Moradali, M. F., Ghods, S., Rehm, B. H. (2017). Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front. Cell. Infect. Microbiol. 7. doi: 10.3389/fcimb.2017.00039
O'Toole, G. A., Kolter, R. (1998). Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30 (2), 295–304. doi: 10.1046/j.1365-2958.1998.01062.x
Overhage, J., Lewenza, S., Marr, A. K., Hancock, R. E. (2007). Identification of genes involved in swarming motility using a Pseudomonas aeruginosa PAO1 mini-Tn 5-lux mutant library. J. Bacteriol. 189 (5), 2164–2169. doi: 10.1128/jb.01623-06
Peleg, A. Y., Monga, D., Pillai, S., Mylonakis, E., Moellering, R. C., Jr., Eliopoulos, G. M. (2009). Reduced susceptibility to vancomycin influences pathogenicity in Staphylococcus aureus infection. J. Infect. Dis. 199 (4), 532–536. doi: 10.1086/596511
Petrova, O. E., Sauer, K. (2009). A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PloS Pathog. 5 (11), e1000668. doi: 10.1371/journal.ppat.1000668
Pratt, L. A., Kolter, R. (1998). Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30 (2), 285–293. doi: 10.1046/j.1365-2958.1998.01061.x
Pusic, P., Sonnleitner, E., Bläsi, U. (2021). Specific and global RNA regulators in Pseudomonas aeruginosa. Int. J. Mol. Sci. 22 (16), 8632. doi: 10.3390/ijms22168632
Ritchings, B. W., Almira, E. C., Lory, S., Ramphal, R. (1995). Cloning and phenotypic characterization of fleS and fleR, new response regulators of Pseudomonas aeruginosa which regulate motility and adhesion to mucin. Infect. Immun. 63 (12), 4868–4876. doi: 10.1128/iai.63.12.4868-4876.1995
Römling, U., Balsalobre, C. (2012). Biofilm infections, their resilience to therapy and innovative treatment strategies. J. Intern. Med. 272 (6), 541–561. doi: 10.1111/joim.12004
Sakuragi, Y., Kolter, R. (2007). Quorum-sensing regulation of the biofilm matrix genes (pel) of Pseudomonas aeruginosa. J. Bacteriol. 189 (14), 5383–5386. doi: 10.1128/jb.00137-07
Schmittgen, T. D., Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3 (6), 1101–1108. doi: 10.1038/nprot.2008.73
Solano, C., Echeverz, M., Lasa, I. (2014). Biofilm dispersion and quorum sensing. Curr. Opin. Microbiol. 18, 96–104. doi: 10.1016/j.mib.2014.02.008
Sonawane, A. M., Singh, B. (2006). The AauR-AauS two-component system regulates uptake and metabolism of acidic amino acids in Pseudomonas putida. Appl. Environ. Microbiol. 72 (10), 6569–6577. doi: 10.1128/AEM.00830-06
Stock, A. M., Robinson, V. L., Goudreau, P. N. (2000). Two-component signal transduction. Annu. Rev. Biochem. 69 (1), 183–215. doi: 10.1146/annurev.biochem.69.1.183
Sultan, M., Arya, R., Kim, K. K. (2021). Roles of two-component systems in Pseudomonas aeruginosa virulence. Int. J. Mol. Sci. 22 (22), 12152. doi: 10.3390/ijms222212152
Thi, M. T. T., Wibowo, D., Rehm, B. H. (2020). Pseudomonas aeruginosa biofilms. Int. J. Mol. Sci. 21 (22), 8671. doi: 10.3390/ijms21228671
Tierney, A. R., Rather, P. N. (2019). Roles of two-component regulatory systems in antibiotic resistance. Future Microbiol. 14 (6), 533–552. doi: 10.2217/fmb-2019-0002
Totten, P. A., Lara, J. C., Lory, S. (1990). The rpoN gene product of Pseudomonas aeruginosa is required for expression of diverse genes, including the flagellin gene. J. Bacteriol. 172 (1), 389–396. doi: 10.1128/jb.172.1.389-396.1990
Vallet, I., Olson, J. W., Lory, S., Lazdunski, A., Filloux, A. (2001). The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc. Natl. Acad. Sci. 98 (12), 6911–6916. doi: 10.1073/pnas.111551898
Vallet-Gely, I., Sharp, J. S., Dove, S. L. (2007). Local and global regulators linking anaerobiosis to cupA fimbrial gene expression in Pseudomonas aeruginosa. J. Bacteriol. 189 (23), 8667–8676. doi: 10.1128/jb.01344-07
Vasil, M. L. (1986). Pseudomonas aeruginosa: biology, mechanisms of virulence, epidemiology. J. Pediatr. 108 (5), 800–805. doi: 10.1016/S0022-3476(86)80748-X
Vincent, J.-L., Sakr, Y., Singer, M., Martin-Loeches, I., MaChado, F. R., Marshall, J. C., et al. (2020). Prevalence and outcomes of infection among patients in intensive care units in 2017. Jama 323 (15), 1478–1487. doi: 10.1001/2Fjama.2020.2717
Wei, Q., Ma, L. Z. (2013). Biofilm matrix and its regulation in Pseudomonas aeruginosa'. Int. J. Mol. Sci. 14 (10), 20983–21005. doi: 10.3390/ijms141020983
Weiner, L. M., Webb, A. K., Limbago, B., Dudeck, M. A., Patel, J., Kallen, A. J., et al. (2016). Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention 2011–2014. Infect. Control Hosp. Epidemiol. 37 (11), 1288–1301. doi: 10.1017/ice.2016.174
World Health Organization. (2017). Prioritization of pathogens to guide discovery, research and development of new antibiotics for drug-resistant bacterial infections, including tuberculosis (WHO, Geneva: World Health Organization). Available at: https://www.who.int/publications/i/item/WHO-EMP-IAU-2017.12.
Wu, W., Jin, Y., Bai, F., Jin, S. (2014). Pseudomonas aeruginosa. Mol. Med. Microbiol. 2–3, 753–767. doi: 10.1016/B978-0-12-397169-2.00041-X
Xue, M., Raheem, M. A., Gu, Y., Lu, H., Song, X., Tu, J., et al. (2020). The KdpD/KdpE two-component system contributes to the motility and virulence of avian pathogenic Escherichia coli. Res. Vet. Sci. 131, 24–30. doi: 10.1016/j.rvsc.2020.03.024
Yin, R., Cheng, J., Wang, J., Li, P., Lin, J. (2022). Treatment of Pseudomonas aeruginosa infectious biofilms: challenges and strategies. Front. Microbiol. 13, 3325. doi: 10.3389/fmicb.2022.955286
Yin, W., Wang, Y., Liu, L., He, J. (2019). Biofilms: the microbial “protective clothing” in extreme environments. Int. J. Mol. Sci. 20 (14), 3423. doi: 10.3390/ijms20143423
Keywords: Pseudomonas aeruginosa, two-component system (TCS), sensor histidine kinase, KdpD, AauS, quorum sensing (QS), virulence
Citation: Sultan M, Arya R, Chaurasia AK and Kim KK (2023) Sensor histidine kinases kdpD and aauS regulate biofilm and virulence in Pseudomonas aeruginosa PA14. Front. Cell. Infect. Microbiol. 13:1270667. doi: 10.3389/fcimb.2023.1270667
Received: 01 August 2023; Accepted: 25 September 2023;
Published: 10 October 2023.
Edited by:
Ahmed Sherif Attia, New Giza University, EgyptReviewed by:
Karishma Bisht, Princeton University, United StatesReham Wasfi, Faculty of Pharmacy, MSA University, Egypt
Copyright © 2023 Sultan, Arya, Chaurasia and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kyeong Kyu Kim, a3llb25na3l1QHNra3UuZWR1; Rekha Arya, cmVraHVhcnlhQGdtYWlsLmNvbQ==; Akhilesh Kumar Chaurasia, Y2hhdXJhc2lhLmFrQGdtYWlsLmNvbQ==
 Maria Sultan
Maria Sultan Rekha Arya
Rekha Arya Akhilesh Kumar Chaurasia
Akhilesh Kumar Chaurasia Kyeong Kyu Kim
Kyeong Kyu Kim