
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 06 November 2023
Sec. Molecular Bacterial Pathogenesis
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1267748
 Liwen Yin1
Liwen Yin1 Qi Liu1
Qi Liu1 Xiaolei Pan1
Xiaolei Pan1 Chenjing Lv1
Chenjing Lv1 Yuxi Bai1
Yuxi Bai1 Fang Bai1
Fang Bai1 Zhihui Cheng1
Zhihui Cheng1 Weihui Wu1
Weihui Wu1 Un-Hwan Ha2
Un-Hwan Ha2 Yongxin Jin1*
Yongxin Jin1*Pseudomonas aeruginosa is an opportunistic human pathogen capable of causing a variety of acute and chronic infections. Its type III secretion system (T3SS) plays a critical role in pathogenesis during acute infection. ExsA is a master regulator that activates the expression of all T3SS genes. Transcription of exsA is driven by two distinct promoters, its own promoter PexsA and its operon promoter PexsC. Here, in combination with a DNA pull-down assay and mass spectrometric analysis, we found that a histone-like nucleoid-structuring (H-NS) family protein MvaT can bind to the PexsC promoter. Using EMSA and reporter assays, we further found that MvaT directly binds to the PexsC promoter to repress the expression of T3SS genes. The repression of MvaT on PexsC is independent of ExsA, with MvaT binding to the -429 to -380 bp region relative to the transcription start site of the exsC gene. The presented work further reveals the complex regulatory network of the T3SS in P. aeruginosa.
Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen that causes a variety of infections in immunocompromised individuals (Reynolds and Kollef, 2021). To successfully establish infections in the host, P. aeruginosa expresses a series of virulence determinants, including the type III secretion system (T3SS). The T3SS is a needle-like machinery deployed by P. aeruginosa to inject toxic proteins directly into host cells, including four well-known effectors ExoS, ExoT, ExoY, and ExoU (Jouault et al., 2022). It plays a critical role in the colonization and pathogenesis of P. aeruginosa during human and animal infections (Holder et al., 2001).
The regulation of the T3SS is complicated and involves varieties of factors in P. aeruginosa (Williams McMackin et al., 2019a). ExsA is a master regulator that promotes expression of the whole T3SS regulon by binding to the promoters of T3SS. ExsA also activates its own expression via the PexsC promoter which drives an operon consisting of exsC, exsE, exsB and exsA genes (Williams McMackin et al., 2019a). In addition to the operon promoter (PexsC), the expression of exsA is also stimulated by its own promoter PexsA, which is located at the intergenic region between exsB and exsA (Liang et al., 2014). Many regulatory players control the expression of the T3SS by directly or indirectly regulating exsA transcription and/or translation. Although PexsA displays a relatively weak promoter activity compared to the PexsC, several regulatory factors have been found to modulate PexsA promoter activity. Vfr, the global regulator of virulence gene expression, activates T3SS gene expression via direct binding and activation of the PexsA promoter (Marsden et al., 2016). MvaT and MvaU, histone-like nucleoid-structuring (H-NS) DNA-binding proteins, repress T3SS gene expression by direct binding and silencing of the PexsA promoter (Williams McMackin et al., 2019b). VqsM, the AraC-family transcription factor, directly binds to the promoter region of PexsA to stimulate T3SS (Liang et al., 2014). Fis, a versatile DNA binding protein, specifically binds to the intergenic region between exsB and exsA to act in transcription elongation from exsB to exsA (Deng et al., 2017). However, except for PsrA, the factors contributing to T3SS expression by controlling PexsC promoter activity remain elusive.
In this study, combining a DNA pull-down assay and mass spectrometric analysis, we identified candidate regulators that bind to and regulate the PexsC promoter. We demonstrated that MvaT also directly binds to the PexsC promoter to repress the expression of T3SS genes. The repressive function of the MvaT on PexsC is independent of ExsA, with MvaT binding to the -429 to -380 bp region relative to the transcription start site of the exsC gene. The presented work further reveals the complex regulatory network of the T3SS in P. aeruginosa.
The bacterial strains, plasmids and primers used in this study are listed in Supplementary Tables 1, 2. Bacterial cells were grown in Luria–Bertani (LB) medium (5 g/L NaCl, 5 g/L yeast extract, and 10 g/L tryptone) or on LB agar plates (LB medium containing 15 g/L agar) at 37 °C. To maintain plasmids, appropriate antibiotics were supplemented into the medium at the following concentrations: for P. aeruginosa, tetracycline at 50 μg/mL and carbenicillin at 150 μg/mL; for E. coli, tetracycline at 10 μg/mL, kanamycin at 25 µg/mL, and ampicillin at 100 μg/mL. When needed, IPTG (isopropyl β-D-1-thiogalactopyranoside) was added to the medium at a final concentration of 1 mM. For inducing T3SS of P. aeruginosa, EGTA was added to the medium at a final concentration of 5 mM.
For complementation of the mvaT gene, the mvaT gene was amplified by PCR using specific primers (Supplementary Table 2) with PAK genomic DNA as the template. The resulting PCR product was digested with BamHI-HindIII and then cloned into pMMB, generating pMMB-mvaT. PexsCmut-lacZ (EcoRI-BamHI), pMMB-exsA-His (EcoRI-HindIII) and pET28a-mvaT (NcoI-XhoI) were constructed with similar strategies. To delete the mvaT gene, two DNA fragments upstream and downstream of the mvaT gene were amplified by PCR using the specific primer pairs mvaT-UF/UR and mvaT-DF/DR (Supplementary Table 2), digested with appropriate restriction enzymes and directionally cloned into pEX18Tc, resulting in the deletion construct pEX18Tc-mvaT. To obtain the mvaT deletion mutant in the PAK and PAKΔexsA strains, pEX18Tc-mvaT was transferred into the corresponding bacterial cell via conjugation, and gene deletion was performed with a SacB-based strategy as previously described (Hoang et al., 1998).
The DNA pull-down assay was carried out as described previously with minor modifications (Dolan et al., 2020; Modrzejewska et al., 2021). A biotinylated DNA fragment containing the PexsC promoter was amplified by PCR using primers PexsC-biot-F/R (Supplementary Table 2) with PAK genomic DNA as a template. The amplified DNA segment was purified and incubated with 100 μL of streptavidin magnetic beads (Streptavidin Mag Sepharose, Cytiva) at room temperature for 20 min. Then, the magnetic beads were washed twice with TBS buffer (50 mM Tris, 150 mM NaCl, pH 7.5) to remove the unbound DNA. In parallel, PAK cells from 200 mL cultures at the exponential phase were harvested by centrifugation at 8,000 × g for 10 min, resuspended in 3 mL TBS buffer and lysed by sonication on ice. After centrifugation at 12,000 × g for 10 min at 4°C, the supernatant was mixed with the immobilized DNA fragments on the magnetic beads and incubated at 4°C for 1 h. After that, the beads were washed three times with TBS buffer to remove nonspecifically bound proteins, and the DNA-bound proteins were released by treatment with increasing concentrations of NaCl buffer (0.2 M, 0.5 M, 1 M). PAKΔexsA/pMMB-exsA-His lysates incubated with PexsC-coupled magnetic beads served as a positive control, while PAK cell lysates incubated with DNA-uncoupled magnetic beads served as a negative control. The eluates from the ExsA-His-overproducing strain were subjected to Western blot analysis using anti-His antibodies to confirm the predicted binding of the ExsA-His protein to the PexsC promoter, validating the pull-down procedure. The eluates from the PAK strain with or without DNA-coupled beads were examined by SDS-PAGE and stained with Coomassie blue. The protein band of interest (the band observed in DNA-coupled beads, but not in the negative control) was excised from SDS-PAGE gel (from 0.2 M NaCl buffer elution) and identified by mass spectrometry analysis.
The β-galactosidase assay was carried out as described previously with minor modifications (Miller, 1972). Overnight cultures of bacteria were 50-fold diluted into fresh LB medium with 0 (for E. coli or P. aeruginosa T3SS non-inducing condition) or 5 mM EGTA (for P. aeruginosa T3SS inducing condition) and grown at 37°C with continuous agitation at 200 rpm. When the OD600 reached 1.0, 500 μL bacterial cells were collected by centrifugation and resuspended in 1.5 mL Z-buffer (60 mM Na2HPO4, 60 mM NaH2PO4, 50 mM β-mercaptoethanol, 10 mM KCl, 1 mM MgSO4). One milliliter suspension was used to measure the OD600, and 10 μL chloroform and 10 μL 0.1% SDS were added to the remaining 500 μL suspensions, followed by vortexing for 10 s. After that, 100 μL ONPG (4 mg/mL) was added to the reaction mixture and incubated at 37°C. The reaction was stopped by the addition of 500 μL of 1 M Na2CO3. The reaction time was recorded, and OD420 was measured after centrifugation at 16,000 × g for 5 min. β-galactosidase activity (Miller units) was calculated as 1000×OD420/Tmin/0.5/OD600; Tmin represents reaction time (minutes).
Overnight bacterial cultures were diluted 50-fold into 3 mL fresh LB medium containing 0 or 5 mM EGTA and then grown to an OD600 of 1.0 with continuous shaking at 200 rpm. Equivalent numbers of bacteria were harvested by centrifugation at 12,000 × g for 3 min. Samples from bacterial cells and supernatant were mixed with loading buffer, boiled for 10 min at 99 °C, and then separated on a 12% SDS-PAGE gel. After transfer onto a polyvinylidene difluoride (PVDF) membrane, the proteins were probed with the primary antibody against ExoS or RpoA (RNAP, Abcam) for 1 h at room temperature and then with the corresponding horseradish peroxidase (HRP)-conjugated secondary antibody, anti-rabbit IgG (Promega) or anti-mouse IgG (Promega), for 1 h at room temperature. All antibodies were 2000-fold diluted into PBST buffer with 5% non-fat milk. The signals were detected using an ECL Plus kit (Millipore) and visualized in a Bio-Rad molecular imager (ChemiDoc XRS+).
To express the recombinant C-terminal His-tagged MvaT, the E. coli strain BL21 (DE3) containing pET28a-mvaT was cultured at 37°C to an OD600 of 0.6. Overexpression of the recombinant MvaT-His protein was induced by the addition of 1 mM IPTG and further grown overnight at 16°C. Harvested cells were resuspended in lysis buffer (50 mM sodium phosphate, 0.3 M NaCl, pH 8.0) and lysed by sonication on ice. After centrifugation at 12,000 × g for 10 min, the supernatant was incubated with Ni-NTA resin (Qiagen) at 4°C for 1 h. Nonspecific binding proteins were washed away with lysis buffer containing 20 mM imidazole, and MvaT-His was eluted with the lysis buffer containing 200 mM imidazole. The purified protein was examined by SDS-PAGE and stained with Coomassie blue.
EMSA was carried out following a previous description with minor modifications (Williams McMackin et al., 2019b). Briefly, the PexsC DNA fragment was amplified by PCR using specific primers (Supplementary Table 2). Fifty or 20 nanogram DNA probe was incubated with increasing concentrations of purified MvaT-His protein in a 20-μL reaction (20 mM Tris, 100 mM KCl, 1 mM DTT, 10% glycerol, pH 7.5) on ice for 30 min. Each sample was loaded onto an 8% or 12% native polyacrylamide gel in 0.5 × TBE (Tris-borate-EDTA) buffer (44.5 mM Tris base, 44.5 mM boric acid, 1 mM EDTA, pH 8.0) and electrophoresed on ice at 10 mA for 1 h. The gel was then stained in 0.5 × TBE buffer containing 0.5 μg/mL ethidium bromide (EB) and imaged with a molecular imager ChemiDoc XRS+ (Bio-Rad, CA, USA).
To identify proteins that can modulate exsA transcription via the PexsC promoter and consequently T3SS expression, we used a biotin-labeled PexsC promoter fragment as bait in a DNA pull-down assay using P. aeruginosa PAK cell lysates. Proteins copurified with the PexsC fragment by streptavidin-coated beads were separated by SDS-PAGE and stained with Coomassie blue. As shown in Figure 1, a specific protein band was detected in the samples in the presence but not in the absence of the PexsC fragment. As a positive control, lysates of bacterial cells overproducing ExsA (PAKΔexsA/pMMB-exsA-His) were pulled down by the PexsC fragment and subjected to Western blot against His antibody. ExsA-His was easily detected (Supplementary Figure 1), validating our approach. Mass spectrometric analysis of the protein band collection revealed a mixture of 65 proteins (Supplementary Table 3). Among them, MvaT, the histone-like nucleoid-structuring DNA-binding protein, was detected. Interestingly, a previous study showed that MvaT regulates the T3SS via direct silencing of the PexsA promoter (Williams McMackin et al., 2019b).
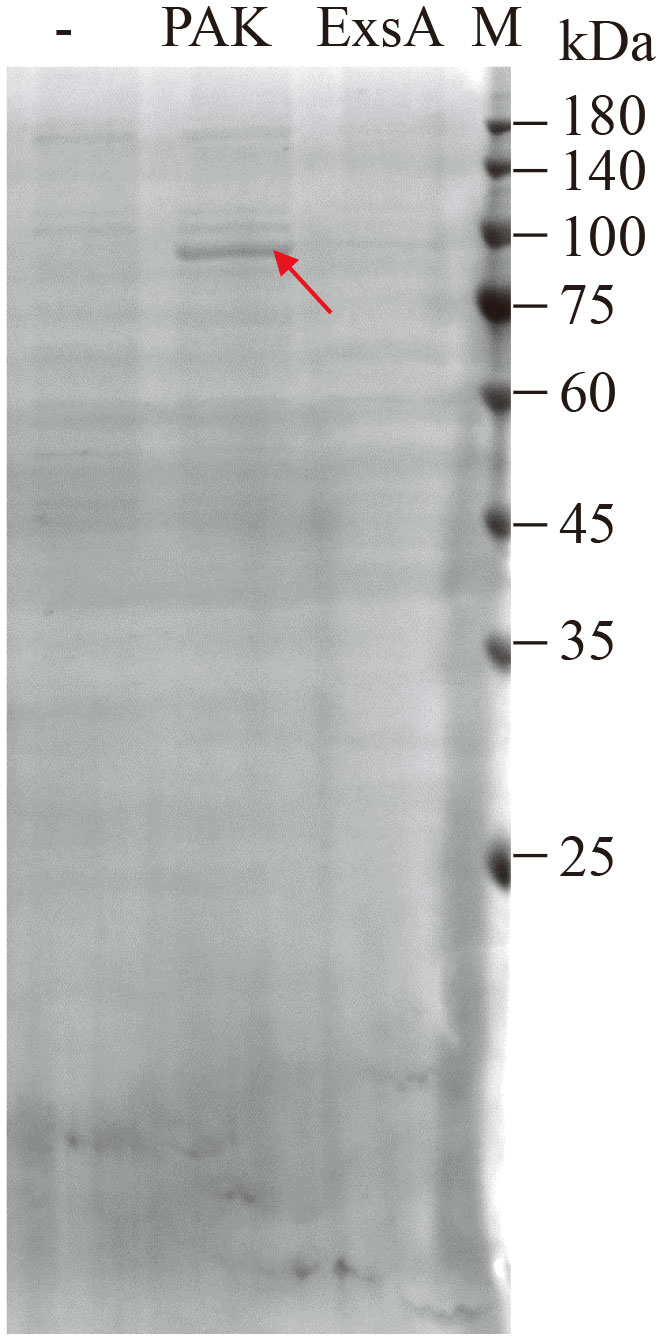
Figure 1 Identification of candidate proteins binding to the PexsC promoter. Biotin-labeled PexsC promoter fragment was incubated with cell lysates of PAK or PAKΔexsA/pMMB-exsA-His, purified with streptavidin-coated beads, separated by SDS-PAGE, and stained with Coomassie blue. The protein band different from the control sample (-, cell lysates of PAK without DNA fragment) is indicated with an arrow, excised, and analyzed by LC-MS/MS.
To determine whether MvaT, identified in the pull-down analysis, can indeed bind to the PexsC promoter, we carried out electrophoretic mobility shift assays (EMSAs) using purified MvaT-His and DNA fragments corresponding to the PexsC promoter region. The DNA fragments corresponding to PexsC, but not the negative control PalgD, were shifted upon incubation with MvaT-his (Figures 2A, B), indicating that MvaT directly binds to the promoter of PexsC in P. aeruginosa.
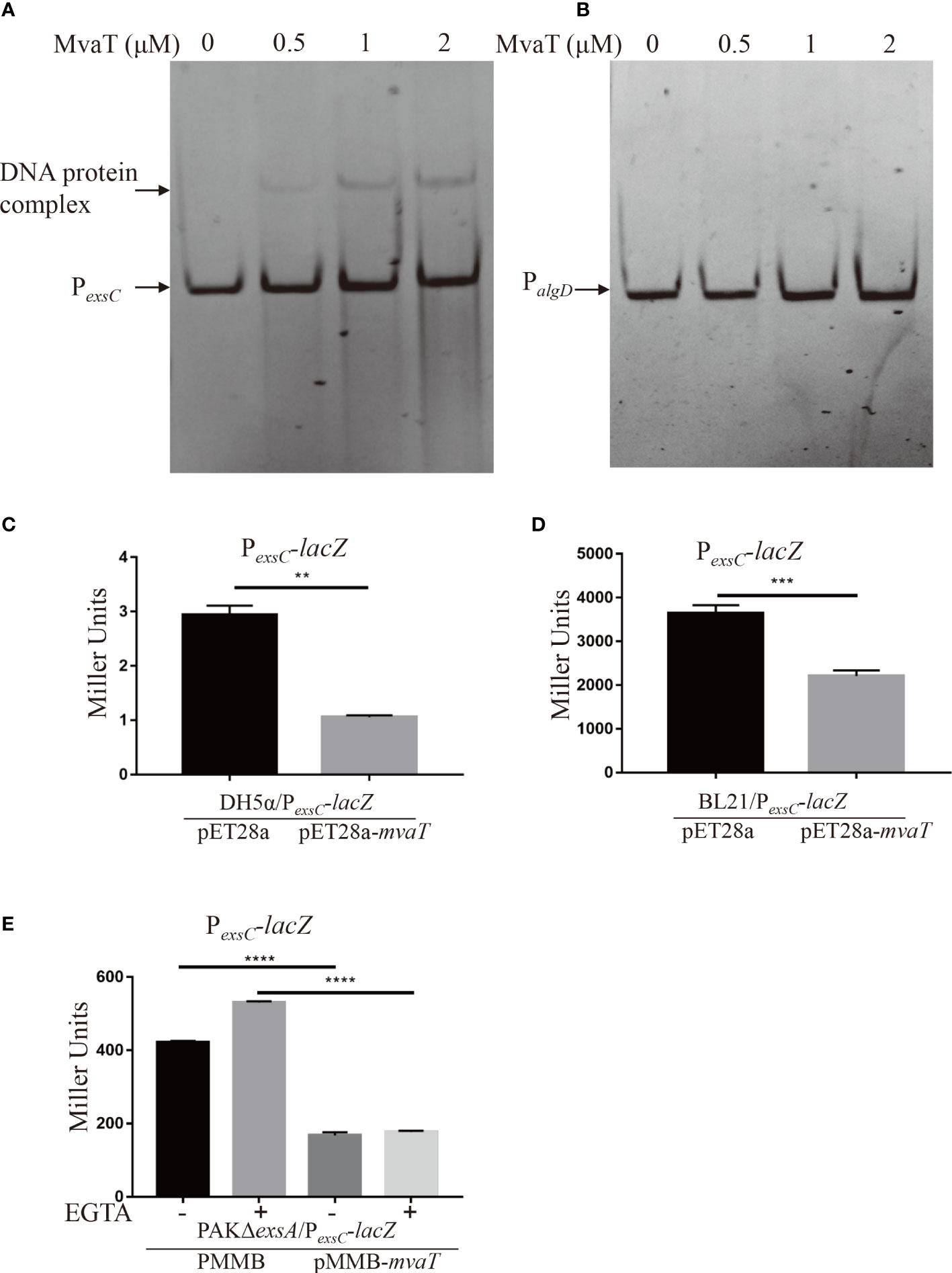
Figure 2 MvaT binds to and represses PexsC directly. (A, B) MvaT binds to the PexsC but not PalgD promoter. The DNA probe (20 ng) was incubated with 0, 0.5, 1 or 2 µM MvaT on ice for 30 min. The shifted band is indicated by an arrowhead. (C-E) To test the repression on promoter activity of PexsC by MvaT, β-galactosidase activity assays were carried out in the indicated stain backgrounds. DH5α/pET28a and DH5α/pET28a-mvaT (C), BL21/pET28a and BL21/pET28a-mvaT (D), and PAKΔexsA/pMMB and PAKΔexsA/pMMB-mvaT (E) containing the PexsC-lacZ transcriptional reporter plasmids were grown to an OD600 of 1.0 in LB (C, D) or LB with (+) or without (-) 5 mM EGTA (E) and subjected to β-galactosidase assays. Each assay was performed in triplicate, and the error bars indicate standard deviations. **P < 0.01, ***P < 0.001, ****P < 0.0001, by Student’s t test.
To further examine the influence of MvaT on the activity of the PexsC promoter, the mvaT gene was expressed under the control of PT7 in vector pET28a and introduced into DH5α and BL21 harboring the PexsC-lacZ transcriptional fusion reporter plasmid. Measurement of β-galactosidase activity showed that expression of MvaT significantly repressed PexsC activity in both DH5α and BL21 (Figures 2C, D). However, the β-galactosidase activity of PexsC-lacZ in BL21 is much higher than that in DH5α, which may be due to the absence of OmpT protease in BL21. These results suggested that MvaT acted directly as a repressor of the PexsC promoter. Furthermore, we wanted to investigate whether MvaT modulates PexsC activity in P. aeruginosa. A previous study demonstrated that MvaT binds to and represses the PexsA promoter (Williams McMackin et al., 2019b). To exclude the effect of MvaT on PexsC activity derived from the altered ExsA protein, PexsC-lacZ and pMMB-mvaT were co-introduced into the PAKΔexsA strain. Consistent with the PexsC-lacZ reporter results in E. coli, expression of MvaT resulted in significant decreases in β-galactosidase activity under both T3SS inducing and noninducing conditions (Figure 2E). All these results demonstrated that MvaT represses the PexsC promoter activity directly.
To determine the influence of MvaT on T3SS gene expression, we generated a mvaT clear deletion mutant and monitored the ExoS amounts using Western blot assay. Strains were cultured under T3SS-inducing (with 5 mM EGTA) and noninducing (without EGTA) conditions for T3SS gene expression. As shown in Figure 3, the expression and secretion of ExoS were highly induced under inducing conditions in both the wild-type PAK and mvaT mutant strains. Deletion of mvaT resulted in increased expression and secretion of ExoS compared to the wild-type PAK strain, and complementation with a mvaT gene recovered the expression and secretion of ExoS in the mvaT mutant background (Figure 3A).
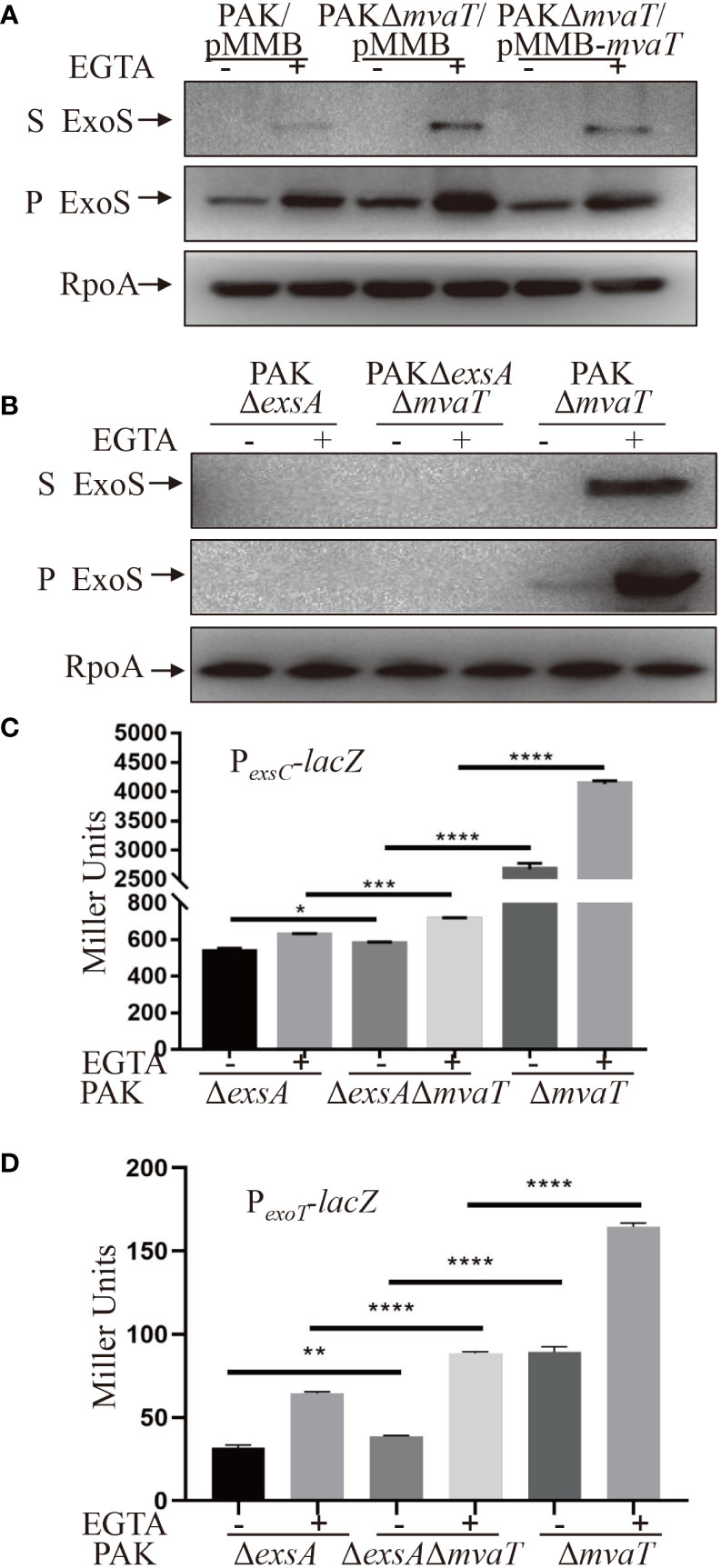
Figure 3 MvaT represses the T3SS independent of ExsA. (A, B) Expression and secretion of ExoS in the indicated strains. Bacterial cells were grown to an OD600 of 1.0 in LB with (+) or without (-) 5 mM EGTA. 1 mM IPTG was added to induce expression of mvaT in (A). Proteins in supernatants (S) and pellets (P) from equivalent bacterial cells were separated by 12% SDS-PAGE gels and probed with anti-ExoS antibody or anti-RpoA antibody. (C, D) Indicated strains containing the PexsC-lacZ or PexoT-lacZ transcriptional reporter plasmid were grown to an OD600 of 1.0 in LB with 0 (-) or 5 mM (+) EGTA and subjected to β-galactosidase assays. Each assay was performed in triplicate, and the error bars indicate standard deviations. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, by Student’s t test.
To examine the effect of MvaT on ExoS expression and secretion independent of ExsA, Western blot was carried out to compare the ExoS amounts between PAKΔexsA and PAKΔexsAΔmvaT. However, in the absence of exsA, ExoS was undetectable in both PAK and PAKΔmvaT (Figure 3B). Since the β-galactosidase activity of PexsC-lacZ was detectable in the absence of exsA in PAK (Figure 2E), we further examined and compared the PexsC promoter activities. As shown in Figure 3C, mvaT deletion resulted in increased β-galactosidase activity in PAKΔexsA/PexsC-lacZ under both T3SS-inducing and noninducing conditions. In addition, the PAKΔmvaT strain displayed a much higher β-galactosidase activity than the PAKΔexsAΔmvaT strain (Figure 3C), hinting at the master activator role of ExsA and a repressive role of MvaT on PexsA promoter (Williams McMackin et al., 2019b). Overall, these data suggest that MvaT represses PexsC transcription independent of ExsA. Furthermore, we examined the promoter activity of another T3SS effector ExoT with PexoT-lacZ transcriptional fusion reporter plasmid (Ha and Jin, 2001). Consistent with the PexsC promoter activities, absence of mvaT resulted in increased β-galactosidase activities of PexoT-lacZ in the PAKΔexsA strain under both T3SS inducing and non-inducing conditions (Figure 3D).
ExsA binds to upstream of the transcriptional start site and facilitates transcription by recruiting RNAP-σ70 to the PexsC promoter (Vakulskas et al., 2009). MvaT represses PexsC promoter activity independent of ExsA. Therefore, we wanted to determine the binding site of MvaT on the PexsC promoter. A variety of DNA fragments within PexsC (Figure 4A) were PCR amplified and used as DNA probes in the EMSA. As shown in Figure 4B, MvaT binds to the DNA fragments containing -429 to -380 bp relative to the transcription start site of the exsC gene. To examine the function of the MvaT binding site in the MvaT-mediated repression of exsC gene, we constructed an additional PexsCmut-lacZ transcriptional fusion, where a 397-bp fragment upstream of the exsC coding region without the MvaT binding site was fused to the promoterless lacZ gene. As shown in Figure 4C, removal of the MvaT binding site resulted in no difference of the β-galactosidase activity between PAKΔexsAΔmvaT and PAKΔexsA strain. This result further supports that MvaT binds to and represses PexsC independent of the master activator ExsA.
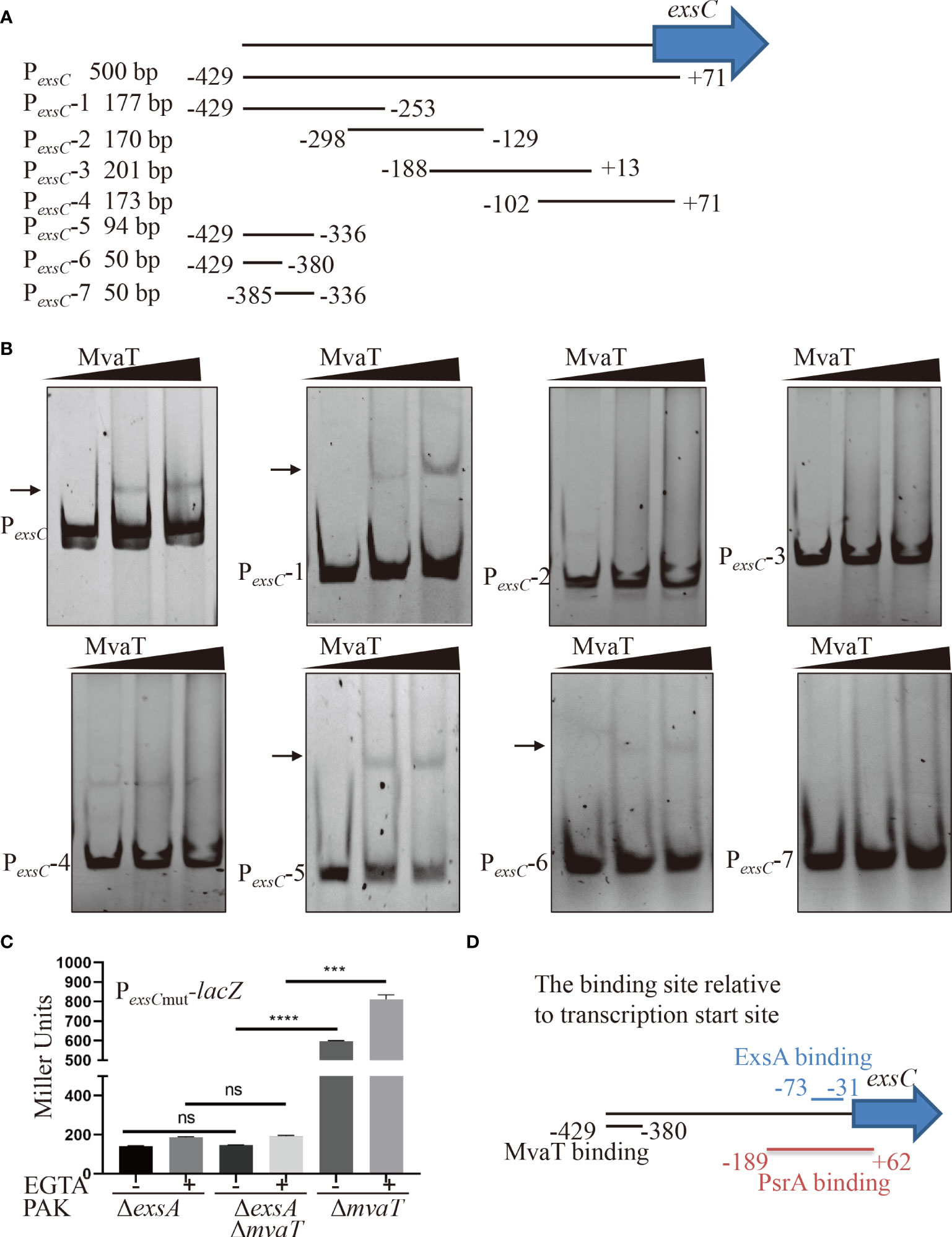
Figure 4 Identification of the binding region of MvaT in the promoter of PexsC. (A) Diagram of the DNA probe in the PexsC. The number indicated the position relative to the transcription start site of exsC gene. (B) Different DNA fragments within PexsC were incubated with 0, 2 or 4 µM MvaT on ice for 30 min. The shifted band is indicated by an arrowhead. (C) Indicated strains containing the PexsCmut-lacZ transcriptional reporter plasmid were grown to an OD600 of 1.0 in LB with 0 (-) or 5 mM (+) EGTA and subjected to β-galactosidase assays. Each assay was performed in triplicate, and the error bars indicate standard deviations. ns, not significant, ***P < 0.001, ****P < 0.0001, by Student’s t test. (D) The binding model of MvaT and previously identified ExsA and PsrA (Hovey and Frank, 1995; Shen et al., 2006).
MvaT is a member of the H-NS family of proteins, which are encoded by many Gram-negative bacteria, including Pseudomonas (Tendeng et al., 2003). In P. aeruginosa, MvaT serves as a global regulator and regulates the expression of more than 150 genes (Vallet et al., 2004; Castang et al., 2008), including virulence factor-encoding genes (Vallet et al., 2004). Similar to other H-NS-like proteins in enteric bacteria, MvaT acts as a transcriptional silencer of foreign DNA by preferentially binding to AT-rich elements of chromosomes in P. aeruginosa (Castang et al., 2008). MvaT negatively controls the expression of fimbrial cup gene clusters and biofilm formation (Vallet et al., 2004; Vallet-Gely et al., 2005). MvaT modulates chloramphenicol resistance in P. aeruginosa by controlling the expression of the multidrug efflux pump MexEF-OprN (Westfall et al., 2006). Recently, MvaT was shown to control T3SS gene expression via direct silencing of the PexsA promoter and indirectly via the RsmY/Z-RsmA pathway (Williams McMackin et al., 2019b). In this study, we found that MvaT also directly binds to the PexsC promoter to repress the expression of T3SS genes. Combining these findings, a model has been proposed for the MvaT-mediated regulation of the T3SS in P. aeruginosa (Figure 5). In addition to its function as a repressor, MvaT was also reported to positively control the expression of the exotoxin A regulatory gene ptxS by directly binding to its upstream region in P. aeruginosa (Westfall et al., 2004).
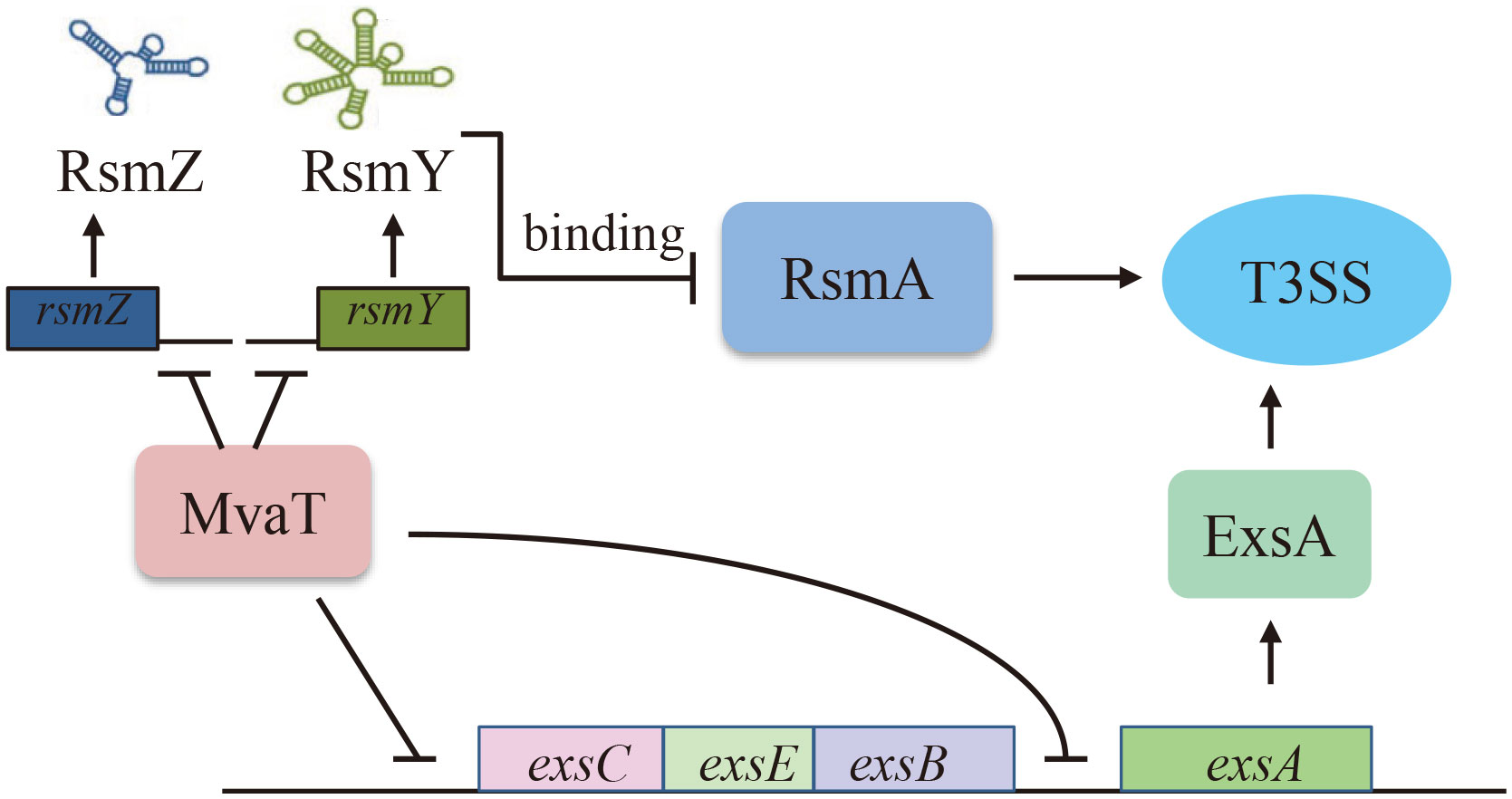
Figure 5 Proposed model of MvaT-mediated T3SS regulation in P. aeruginosa. MvaT regulates T3SS via direct binding and silencing the PexsC and PexsA promoters, as well as RsmY/Z-RsmA pathway indirectly (Williams McMackin et al., 2019b).
Of note, our mass spectrometric analysis also revealed PA3981 as a candidate PexsC binding protein (Supplementary Table 3). PA3981, also named YbeZ, comprises an ATP binding domain and a nucleoside triphosphate hydrolase domain. Our previous study demonstrated that YbeZ controls the T3SS through RetS in P. aeruginosa (Xia et al., 2021). Whether YbeZ also regulates T3SS through direct binding to and modulating the PexsC promoter remains elusive and warrants further study.
Transcription of the exsA gene is driven by two distinct promoters, PexsA and PexsC. It has been demonstrated that the PexsA promoter is modulated by several regulatory factors, including activators of Vfr and VqsM and silencers of MvaT and MvaU (Williams McMackin et al., 2019a). Although not binding to and regulating PexsA promoter activity, ExsA primarily binds to and activates PexsC promoter activity, leading to a positive feedback loop (Yahr and Frank, 1994). In addition, the PexsC promoter was also directly bound and stimulated by PsrA, a TetR family transcriptional regulator in P. aeruginosa (Shen et al., 2006). Previous studies showed that ExsA binds to the -52 bp region relative to the transcriptional start site of the exsC gene (Hovey and Frank, 1995), while the PsrA binding site is located between -207 bp and +44 bp relative to the translational start site (-189 bp to 62 bp relative to the transcription start site) of the exsC gene (Shen et al., 2006). In this study, we found that MvaT can also bind to but repress PexsC promoter activity. According to the EMSA results, the binding site of MvaT on the PexsC promoter is located between -429 and -380 bp relative to the transcription start site of the exsC gene (Figure 4D). Since the binding regions of MvaT do not include the -35 or -10 sequences within the PexsC promoter, the repressive mechanism might not be caused by the occlusion of RNAP by MvaT from the PexsC promoter. Therefore, the possible repressive mechanism of MvaT on PexsC is through preventing RNAP from escaping the PexsC promoter.
Phylogenetic analysis revealed that the T3SS is evolutionarily acquired by horizontal gene transmission (Nguyen et al., 2000). H-NS family DNA-binding proteins play important roles in driving evolution by permitting and regulating horizontally acquired genes (Navarre, 2016). From our experimental results and those of others (Williams McMackin et al., 2019b), both the PexsA and PexsC promoters of exsA were directly silenced by the MvaT, hinting at the complication of T3SS evolution and regulation.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
LY: Investigation, Methodology, Writing – original draft. QL: Investigation, Methodology, Writing – review & editing. XP: Methodology, Formal Analysis, Writing – review & editing. CL: Formal Analysis, Methodology, Writing – review & editing. YB: Formal Analysis, Methodology, Writing – review & editing. FB: Formal Analysis, Funding acquisition, Writing – review & editing. ZC: Formal Analysis, Funding acquisition, Supervision, Writing – review & editing. WW: Formal Analysis, Funding acquisition, Writing – review & editing. UH: Formal Analysis, Writing – review & editing. YJ: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Key Research and Development Project of China (2021YFE0201300 and 2021YFE0101700), the Science and Technology Committee of Tianjin (22JCYBJC00790 and 22JCYBJC00560), the National Science Foundation of China (31970179, 31970680, 32170177, 82061148018, and 32170199), and the Fundamental Research Funds for the Central Universities, Nankai University (63231048 and 63231047). The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1267748/full#supplementary-material
Castang, S., McManus, H. R., Turner, K. H., Dove, S. L. (2008). H-NS family members function coordinately in an opportunistic pathogen. Proc. Natl. Acad. Sci. U.S.A. 105 (48), 18947–18952. doi: 10.1073/pnas.0808215105
Deng, X., Li, M., Pan, X., Zheng, R., Liu, C., Chen, F., et al. (2017). Fis Regulates Type III Secretion System by Influencing the Transcription of exsA in Pseudomonas aeruginosa Strain PA14. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00669
Dolan, S. K., Pereira, G., Silva-Rocha, R., Welch, M. (2020). Transcriptional regulation of central carbon metabolism in Pseudomonas aeruginosa. Microb. Biotechnol. 13 (1), 285–289. doi: 10.1111/1751-7915.13423
Ha, U., Jin, S. (2001). Growth phase-dependent invasion of Pseudomonas aeruginosa and its survival within HeLa cells. Infect. Immun. 69 (7), 4398–4406. doi: 10.1128/iai.69.7.4398-4406.2001
Hoang, T. T., Karkhoff-Schweizer, R. R., Kutchma, A. J., Schweizer, H. P. (1998). A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212 (1), 77–86. doi: 10.1016/s0378-1119(98)00130-9
Holder, I. A., Neely, A. N., Frank, D. W. (2001). Type III secretion/intoxication system important in virulence of Pseudomonas aeruginosa infections in burns. Burns 27 (2), 129–130. doi: 10.1016/s0305-4179(00)00142-x
Hovey, A. K., Frank, D. W. (1995). Analyses of the DNA-binding and transcriptional activation properties of ExsA, the transcriptional activator of the Pseudomonas aeruginosa exoenzyme S regulon. J. Bacteriol 177 (15), 4427–4436. doi: 10.1128/jb.177.15.4427-4436.1995
Jouault, A., Saliba, A. M., Touqui, L. (2022). Modulation of the immune response by the Pseudomonas aeruginosa type-III secretion system. Front. Cell Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.1064010
Liang, H., Deng, X., Li, X., Ye, Y., Wu, M. (2014). Molecular mechanisms of master regulator VqsM mediating quorum-sensing and antibiotic resistance in Pseudomonas aeruginosa. Nucleic Acids Res. 42 (16), 10307–10320. doi: 10.1093/nar/gku586
Marsden, A. E., Intile, P. J., Schulmeyer, K. H., Simmons-Patterson, E. R., Urbanowski, M. L., Wolfgang, M. C., et al. (2016). Vfr Directly Activates exsA Transcription To Regulate Expression of the Pseudomonas aeruginosa Type III Secretion System. J. Bacteriol 198 (9), 1442–1450. doi: 10.1128/jb.00049-16
Miller, J. H. (1972). “Assay of β-galactosidase,” in Experiments in Molecular Genetics (Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press).
Modrzejewska, M., Kawalek, A., Bartosik, A. A. (2021). The lysR-type transcriptional regulator bsrA (PA2121) controls vital metabolic pathways in pseudomonas aeruginosa. mSystems 6 (4), e0001521. doi: 10.1128/mSystems.00015-21
Navarre, W. W. (2016). The impact of gene silencing on horizontal gene transfer and bacterial evolution. Adv. Microb. Physiol. 69, 157–186. doi: 10.1016/bs.ampbs.2016.07.004
Nguyen, L., Paulsen, I. T., Tchieu, J., Hueck, C. J., Saier, M. H., Jr. (2000). Phylogenetic analyses of the constituents of Type III protein secretion systems. J. Mol. Microbiol. Biotechnol. 2 (2), 125–144.
Reynolds, D., Kollef, M. (2021). The epidemiology and pathogenesis and treatment of pseudomonas aeruginosa infections: an update. Drugs 81 (18), 2117–2131. doi: 10.1007/s40265-021-01635-6
Shen, D. K., Filopon, D., Kuhn, L., Polack, B., Toussaint, B. (2006). PsrA is a positive transcriptional regulator of the type III secretion system in Pseudomonas aeruginosa. Infect. Immun. 74 (2), 1121–1129. doi: 10.1128/iai.74.2.1121-1129.2006
Tendeng, C., Soutourina, O. A., Danchin, A., Bertin, P. N. (2003). MvaT proteins in Pseudomonas spp.: a novel class of H-NS-like proteins. Microbiol. (Reading) 149 (Pt 11), 3047–3050. doi: 10.1099/mic.0.C0125-0
Vallet, I., Diggle, S. P., Stacey, R. E., Cámara, M., Ventre, I., Lory, S., et al. (2004). Biofilm formation in Pseudomonas aeruginosa: fimbrial cup gene clusters are controlled by the transcriptional regulator MvaT. J. Bacteriol 186 (9), 2880–2890. doi: 10.1128/jb.186.9.2880-2890.2004
Vallet-Gely, I., Donovan, K. E., Fang, R., Joung, J. K., Dove, S. L. (2005). Repression of phase-variable cup gene expression by H-NS-like proteins in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 102 (31), 11082–11087. doi: 10.1073/pnas.0502663102
Vakulskas, C. A., Brady, K. M., Yahr, T. L. (2009). Mechanism of transcriptional activation by Pseudomonas aeruginosa ExsA. J. Bacteriol 191 (21), 6654–6664. doi: 10.1128/jb.00902-09
Westfall, L. W., Carty, N. L., Layland, N., Kuan, P., Colmer-Hamood, J. A., Hamood, A. N. (2006). mvaT mutation modifies the expression of the Pseudomonas aeruginosa multidrug efflux operon mexEF-oprN. FEMS Microbiol. Lett. 255 (2), 247–254. doi: 10.1111/j.1574-6968.2005.00075.x
Westfall, L. W., Luna, A. M., Francisco, M. S., Diggle, S. P., Worrall, K. E., Williams, P., et al. (2004). The Pseudomonas aeruginosa global regulator MvaT specifically binds to the ptxS upstream region and enhances ptxS expression. Microbiol. (Reading) 150 (Pt 11), 3797–3806. doi: 10.1099/mic.0.27270-0
Williams McMackin, E. A., Djapgne, L., Corley, J. M., Yahr, T. L. (2019a). Fitting Pieces into the Puzzle of Pseudomonas aeruginosa Type III Secretion System Gene Expression. J. Bacteriol 201 (13). doi: 10.1128/jb.00209-19
Williams McMackin, E. A., Marsden, A. E., Yahr, T. L. (2019b). H-NS family members mvaT and mvaU regulate the pseudomonas aeruginosa type III secretion system. J. Bacteriol 201 (14). doi: 10.1128/jb.00054-19
Xia, Y., Xu, C., Wang, D., Weng, Y., Jin, Y., Bai, F., et al. (2021). YbeY controls the type III and type VI secretion systems and biofilm formation through RetS in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 87 (5). doi: 10.1128/aem.02171-20
Keywords: P. aeruginosa, T3SS, MvaT, transcriptional regulation, PexsC, PexsA
Citation: Yin L, Liu Q, Pan X, Lv C, Bai Y, Bai F, Cheng Z, Wu W, Ha U-H and Jin Y (2023) MvaT binds to the PexsC promoter to repress the type III secretion system in Pseudomonas aeruginosa. Front. Cell. Infect. Microbiol. 13:1267748. doi: 10.3389/fcimb.2023.1267748
Received: 27 July 2023; Accepted: 19 October 2023;
Published: 06 November 2023.
Edited by:
George P. Munson, University of Miami, United StatesReviewed by:
Ritesh Ranjan Pal, Indian Association for the Cultivation of Science (IACS), IndiaCopyright © 2023 Yin, Liu, Pan, Lv, Bai, Bai, Cheng, Wu, Ha and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongxin Jin, eXhqaW5AbmFua2FpLmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.