- 1Department of Neurosurgery, Beijing Tiantan Hospital, Capital Medical University, Beijing, China
- 2China National Clinical Research Center for Neurological Diseases, Beijing, China
- 3Center of Stroke, Beijing Institute for Brain Disorders, Beijing, China
- 4Beijing Key Laboratory of Translational Medicine for Cerebrovascular Disease, Beijing, China
- 5Beijing Translational Engineering Center for 3D Printer in Clinical Neuroscience, Beijing, China
- 6Savaid Medical School, University of Chinese Academy of Sciences, Beijing, China
- 7Department of Neurosurgery, Beijing Hospital, Beijing, China
Background and purpose: When it comes to the onset of moyamoya disease (MMD), environmental variables are crucial. Furthermore, there is confusion about the relationship between the gut microbiome, an environmental variable, and MMD. Consequently, to identify the particular bacteria that cause MMD, we examined the gut microbiome of MMD individuals and healthy controls (HC).
Methods: A prospective case-control investigation was performed from June 2021 to May 2022. The fecal samples of patients with MMD and HC were obtained. Typically, 16S rRNA sequencing was employed to examine their gut microbiota. The QIIME and R softwares were used to examine the data. The linear discriminant analysis effect size analysis was used to determine biomarkers. Multivariate analysis by linear models (MaAsLin)2 were used to find associations between microbiome data and clinical variables. Model performance was assessed using the receiver operating characteristic curve and the decision curve analysis.
Results: This investigation involved a total of 60 MMD patients and 60 HC. The MMD group’s Shannon and Chao 1 indices were substantially lower than those of the HC cohort. β-diversity was significantly different in the weighted UniFrac distances. At the phylum level, the relative abundance of Fusobacteriota/Actinobacteria was significantly higher/lower in the MMD group than that in the HC group. By MaAsLin2 analysis, the relative abundance of the 2 genera, Lachnoclostridium and Fusobacterium, increased in the MMD group, while the relative abundance of the 2 genera, Bifidobacterium and Enterobacter decreased in the MMD group. A predictive model was constructed by using these 4 genera. The area under the receiver operating characteristic curve was 0.921. The decision curve analysis indicated that the model had usefulness in clinical practice.
Conclusions: The gut microbiota was altered in individuals with MMD, and was characterized by increased abundance of Lachnoclostridium and Fusobacterium and decreased abundance of Bifidobacterium and Enterobacter. These 4 genera could be used as biomarkers and predictors in clinical practice.
Introduction
The development of intracranial carotid artery stenosis and an irregular vascular network in the brain are characteristics of moyamoya disease (MMD), an uncommon kind of cerebrovascular disorder (Suzuki and Takaku, 1969; Scott and Smith, 2009). MMD is a rare cerebrovascular condition, yet despite this, it is the most common reason for stroke in children in East Asian nations (Kuriyama et al., 2008; Kim et al., 2016). There are two primary phenotypes of MMD in these nations, according to the presentation of MMD: hemorrhagic and ischemic kind (Liu et al., 2015).
Though the epidemiology, clinical features, and treatment of MMD have been thoroughly studied (Liu et al., 2015; Acker et al., 2018), little is known about its etiology and progression. Several studies have shown that MMD may be associated with immune, inflammatory, and genetic causes (Masuda et al., 1993; Fujimura et al., 2014; Bang et al., 2016). A pathological study has revealed the existence of growing vascular smooth muscle cells accompanied with inflammatory cells in the intima of stenotic or obstructive intracranial arterial lesions (Masuda et al., 1993). Another prospective study has indicated significantly elevated circulating Treg and Th17 cells in subjects with MMD compared to healthy controls (HC) (Weng et al., 2017). A complicated procedure involving hereditary and environmental variables is likely what causes chronic inflammation (Renz et al., 2011). A significant correlation between RNF213 p.R4810K variant and MMD has also been identified (Kamada et al., 2011; Liu et al., 2011). However, the penetrance of RNF213 p.R4810K is lower than 1%, which points to a synergistic interaction with additional environmental and genetic risk factors (Koizumi et al., 2016).
The gut microbiota is a crucial environmental component that affects the host’s metabolism and immunological homeostasis, according to growing evidence. The significance of the human gut microbiome in cardiac-cerebral vascular disorders such as high blood pressure, atherosclerosis, and stroke has gotten a lot of attention recently (Jie et al., 2017; Li et al., 2017; Peh et al., 2022). Additionally, according to a number of studies, changes in gut microbiome may precipitate in development of unruptured cerebral aneurysms (Li et al., 2020; Kawabata et al., 2022). Numerous metabolites produced by the gut microbiome in these disorders are reportedly taken into the circulatory system, where they are further processed by host enzymes, causing injury to the target organs (Koeth et al., 2013; Peh et al., 2022). Mineharu et al. collected feces from 27 MMD patients and 15 normal controls. They found no difference in α-diversity or β-diversity between patients with MMD and controls. Furthermore, their data showed that increased abundance of Ruminococcus gnavus and decreased abundance of Roseburia inulinivorans were associated with higher risk of MMD (Mineharu et al., 2022). However, the sample size they included was small.
Our hypothesis was that patients with MMD and HC have different types of microbiome profiles; the intima of the intracranial artery may change pathophysiologically and develop chronic inflammation as a result of this difference. We analyzed the gut microbiome of MMD patient and HC, and isolated the bacteria associated with MMD, in order to ascertain the relationship between MMD and the microbiome.
Methods
Standard protocol verification, registrations, and participant consents
Upon reasonable request, the corresponding author will provide the information supporting the outcomes of this manuscript. The Ethics Committee of Beijing Tiantan Hospital, Capital Medical University gave its authorization to the protocol prior to the case-control research starting (approval number KY2021-072-01). Each participant signed an informed consent. The investigation is reported in compliance with the STROBE declaration. The registration number of this study is NCT04890782.
Study population
MMD was identified by digital subtraction angiography (DSA), based on Japanese recommendations released in 2012: (1) stenosis or blockage of the distal internal carotid and the proximal middle and anterior cerebral arteries and (2) unilateral or bilateral involvement (Fujimura et al., 2016). 72 adult patients (age ≥ 18 years) with MMD-type cerebrovascular disorder were prospectively enrolled between June 2021 and May 2022 from the Department of Neurosurgery, Beijing Tiantan Hospital, Capital Medical University. Among 72 adult patients, 10 patients with moyamoya syndrome and 2 with inadequate fecal sample were excluded (Scott and Smith, 2009; Kauv et al., 2019). Moyamoya syndrome refers to patients with MMD-type cerebrovascular disease accompanied by other basic diseases, such as arteriosclerosis, autoimmune disease, meningitis, and Down syndrome (Scott and Smith, 2009). In the end, 60 (83.33%) of the adult cases in the trial gave their permission. A control who had not undergo operations or taken any drugs was matched to every case with MMD. The HC were recruited from individuals who were shown not to have occlusive cerebrovascular disease by MRI and had regular medical examinations, and were matched according to their gender, age, and body mass index (BMI).
Sample collection
After a 15-minute break spent sat, the patients’ right arms were tested for systolic blood pressure (SBP) and diastolic blood pressure (DBP) using a conventional mercury manometer. Weight (kg)/height (m2) was used to determine the BMI. After the participants had not eaten or drunk anything for 12 hours, venous blood samples were obtained. Fasting blood was examined for white blood cells (WBC), glucose, albumin (ALB), uric acid, triglyceride (TG), total cholesterol, high-density lipoprotein cholesterol, low-density lipoprotein cholesterol, and homocysteine (Hcy). Patients’ RNF213 p.R4810K variation was identified. The primers were generated in the following methodology. RNF213-4810F (rs112735431): 5’-GCCCTCCATTTCTAGCACAC-3’; and RNF21-4810R: 5’-AGCTGTGGCGAAAGCTTCTA-3’.
Data on the MMD patients’ age, sex, and comorbidities, such as hypertension, coronary artery disease, smoking, drinking, diabetes mellitus, thyroid disorders, and initial clinical features (ischemia and hemorrhage), were gathered at the time of admission. Two neurosurgeons utilized DSA to evaluate the Suzuki stage blindly.
For HC, fecal samples were collected at home at 06:30-08:30 am. The samples were stored at -80°C within 6 hours after production. For patients, fecal samples were collected at 06:30-08:30 am within 48 hours after admission. The samples were stored at -80°C within 2 hours after production. The online Supplementary Methods provide a description of the specifics.
Bacterial DNA extraction and 16S rRNA sequencing
The CTAB technique was used to obtain microbial DNA from the stool specimens in accordance with the procedure. DNA samples that had been collected were amplified, DNA libraries had been created, and sequencing had been conducted using an Illumina NovaSeq platform. Amplified reads were analyzed. The online Supplementary Methods provide a description of the specifics.
Microbiome bioinformatics
The generated FASTQ files were imported, demultiplexed, quality filtered, and analyzed utilising QIIME pipeline (Caporaso et al., 2010). Uparse program (Uparse v7.0.1001, http://drive5.com/uparse) was employed to analyze the sequences (Bokulich et al., 2013; Edgar, 2013). Sequences with ≥ 97% similarity were attributed to the same operational taxonomic units (OTUs). The Silva Database (http://www.arb-silva.de/) was employed depending on the Mothur algorithm to annotate taxonomic data for each sample sequence (Quast et al., 2013). All samples were included since rarefaction was conducted to a depth of 21,853 base pairs (100% of the minimum sample depth) to even the sequence depth. The online Supplemental Methods come with an explanation of the details.
In order to analyze the complexity of species diversity for a sample, three indices—Chao1, Shannon, and Simpson—are used. The R software, version 4.2.0, was used to generate and show each of these indices in our samples (Oksanen et al., 2017). The variations between specimens in terms of species complexity were assessed using β-diversity assessment. β-diversity on weighted unifrac estimated using the QIIME program (Version 1.9.1). Principal Coordinate Analysis (PCoA) was used to get principal coordinates and visualize from multidimensional data. Permutational multivariate ANOVA (PERMANOVA) was carried out to examine the statistical differences in β diversity across the cohorts, which was displayed by R software (Version 4.2.0) (McMurdie and Holmes, 2013). An envfit analysis associated to the PCoA was performed to identify whether sex, smoking, drinking, or hypertension have an effect on microbial composition distribution.
To identify substantially different taxa among the cohorts, the linear discriminant analysis effect size assessment was used (Segata et al., 2011). The linear discriminant analytic score was employed to evaluate the effect size of every distinctively abundant characteristic. When comparing the relative taxa abundances between the two groups, the threshold for a linear discriminant analytic score was established as ±2.0.
Statistical analysis
The Student’s t test was employed to analyze continuous data reported as mean (standard deviation), and the Wilcoxon test was employed to analyze median with interquartile ranges. Chi-square test or Fisher’s exact analysis were used to assess categorical data, which were shown as percentages. To examine correlations between non-normal data, Spearman’s rank correlation was utilized. The Benjamin-Hochberg approach was employed to limit the false discovery rate when repeated comparisons were utilized to determine variations between 2 cohorts. By creating receiver-operating characteristic (ROC) curves and computing the area under the curve (AUC), the predictive performance of the MMD model was evaluated. For a more thorough investigation of the prediction performances, decision curve analysis (DCA) was employed. All analyses were two-sided, and statistical significance was set at p < 0.05. R program (version 4.2.0; https://www.r-project.org) and IBM SPSS Software (version 22.0; IBM Corp.) were employed for all statistical analyses.
Multivariate analysis by linear models (MaAsLin)2 is a tool to find associations between clinical metadata and bacterial abundance. We then correlated the microbiome data (generic level) with MMD via MaAsLin2 analysis adjusting for sex, age, smoking, drinking, and hypertension. All p-values were corrected for multiple testing using false discovery rate. False discovery rate adjusted p < 0.2 was considered statistically significant for taxonomic analysis.
Results
Patient’s characteristics
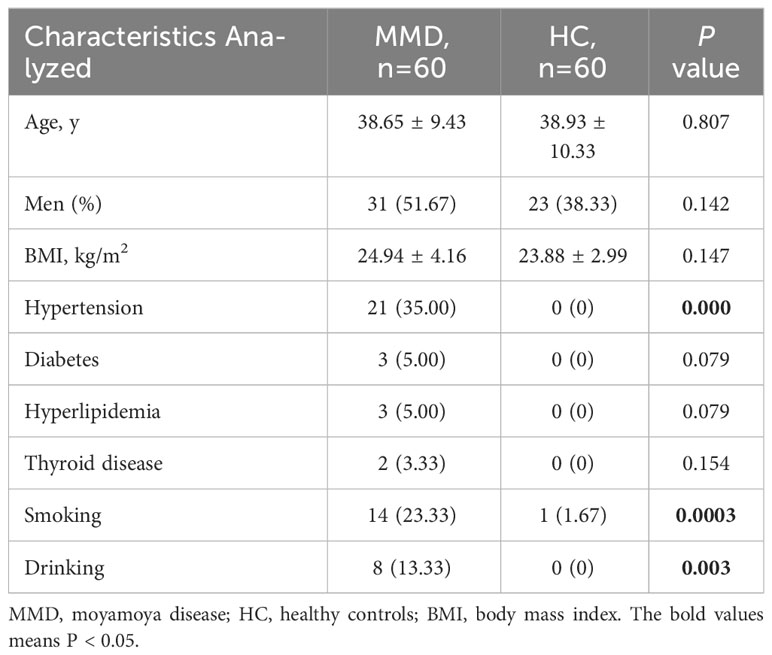
Sixty adult patients with MMD and 60 HC were consecutively recruited into our study. A total of 54 (45.00%) males and 66 (55.00%) females were enrolled, and the median age was 39.00 y (IQR, 31.00-46.50 y). Table 1 illustrates a conclusion of the features of MMD patients and HC. The characteristics, including BMI, diabetes, hyperlipidemia, and thyroid disease did not show differences between patients and HC; however, subjects with MMD had greater rates of hypertension, smoking, and drinking compared with HC (p < 0.05 for all).
Differences in gut microbiome between the MMD and HC cohorts
The 16,949 OTUs in the OUTs set of data for MMD individuals and HC were divided into 495 genera, 184 families, 108 orders, 47 classes, and 25 phyla. Figure 1A shows that although the Simpson index did not reveal a difference between the two cohorts, the Chao 1 and Shannon indices showed a substantial alteration (p < 0.001 for all). The weighted UniFrac distances between the two cohorts showed a substantial difference in β-diversity (r2 = 0.027, p = 0.001, Figure 1B).
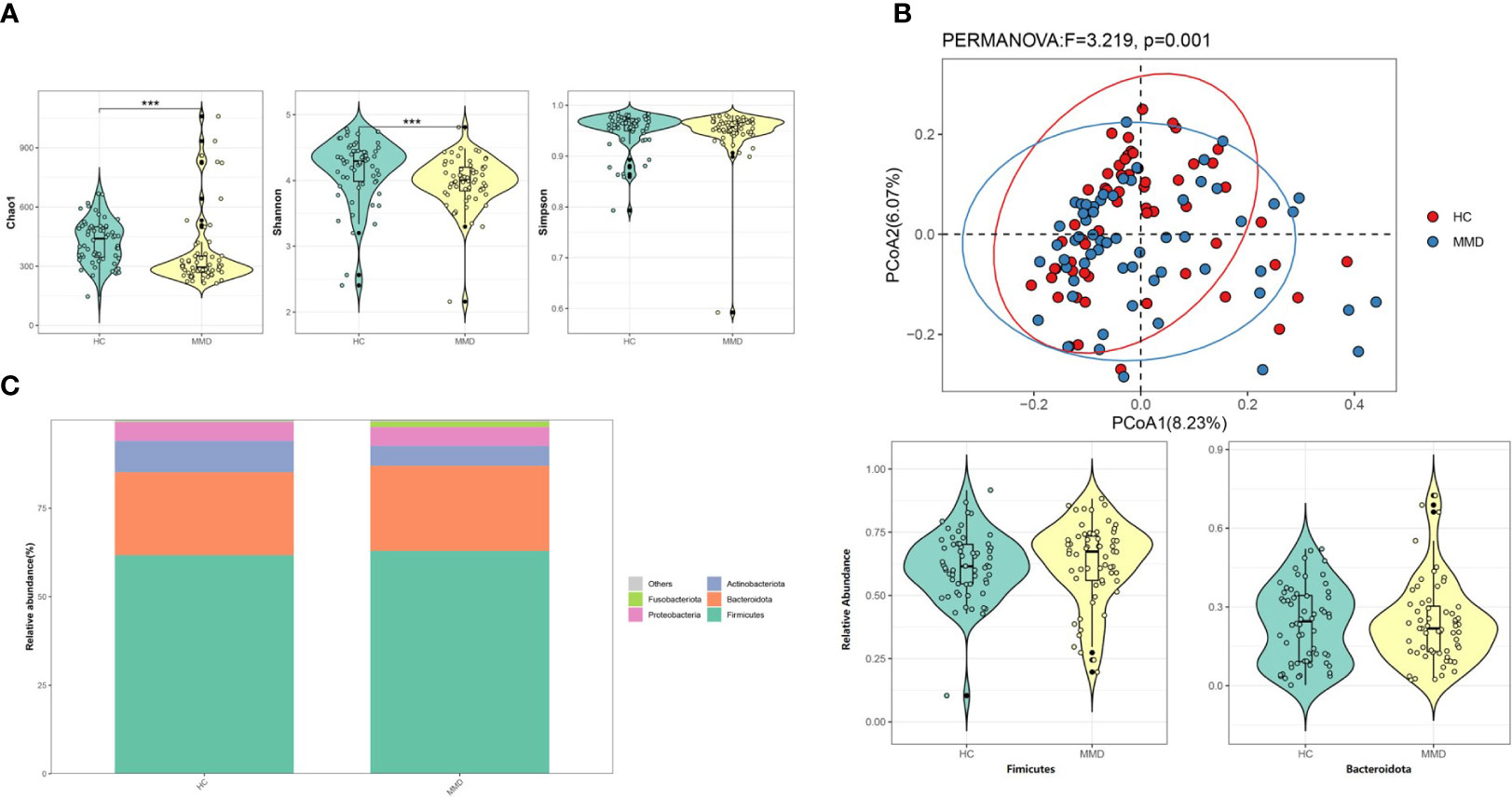
Figure 1 Comparison of microbial diversity and microbiome composition between MMD and HC groups. (A) The Chao 1 and the Shannon indices differed significantly between the two groups, while the Simpson index between the two groups did not show a difference. ***p < 0.001; (B) Principal coordinate analysis illustrating the grouping patterns of MMD and HC groups based on the weighted UniFrac distances. There was a significant difference in β-diversity (r2 = 0.027, p = 0.001); (C) Distribution of the relative abundance of bacteria at the phylum level. The relative abundance of Fimicutes and Bacteroidota did not differ significantly between the groups. MMD, moyamoya disease; HC, healthy controls.
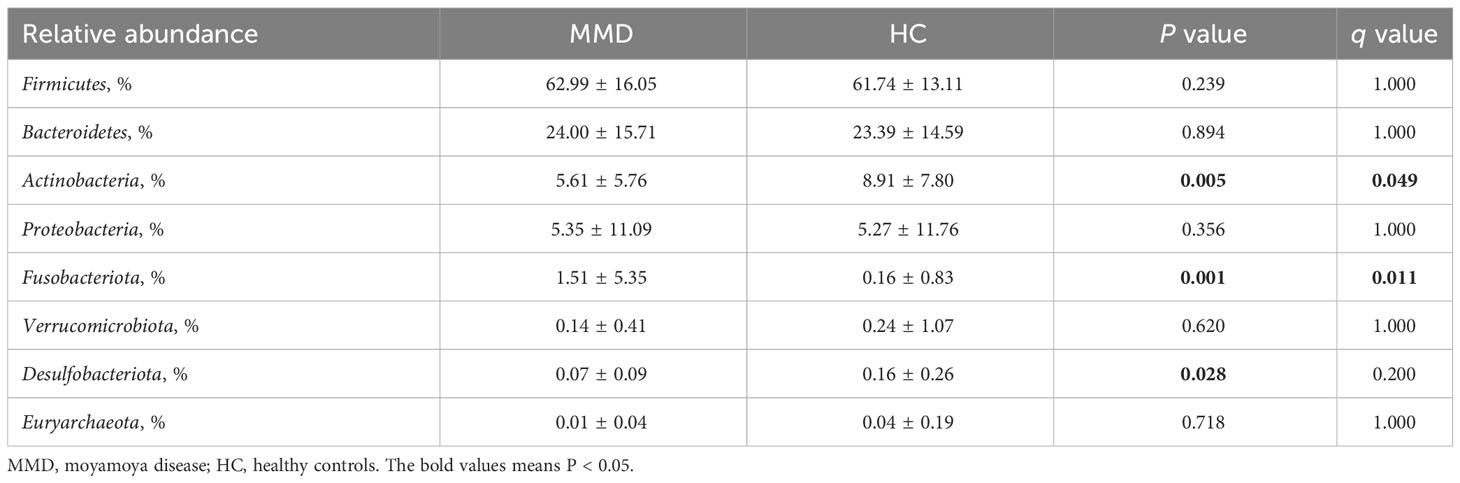
Firmicutes and Bacteroidota were the most prevalent phyla among all subjects, with 62.99 ± 16.05% and 24.00 ± 15.71 for the MMD cohort and 61.74 ± 13.11% and 23.39 ± 14.59% for the HC cohort, respectively (Table 2 and Figure 1C). Additionally, there were no variations between patients and HC in terms of the relative abundance of Firmicutes and Bacteroidota (Figure 1C). At the phylum level, the relative abundance of Fusobacteriota/Actinobacteria was substantially higher/lower in the MMD group (false discovery rate, q = 0.049/0.011; Table 2).
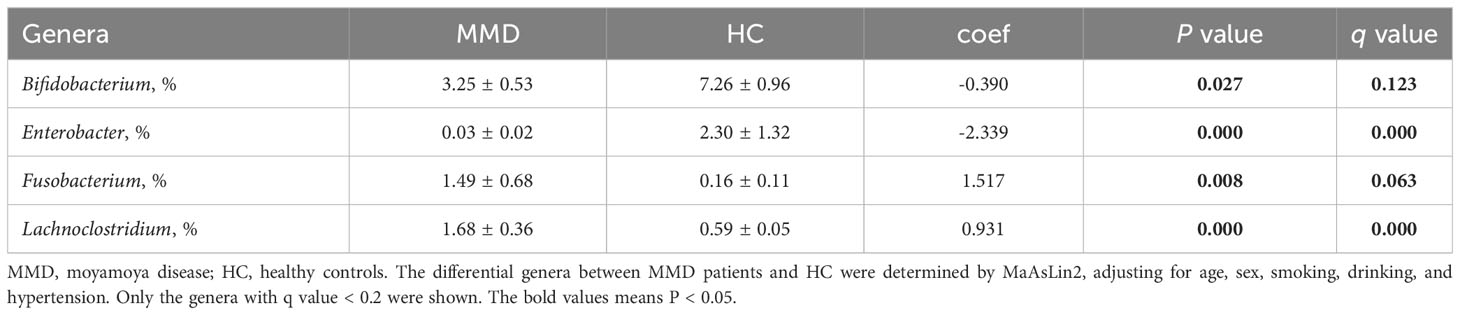
In the linear discriminant analysis effect size analysis, we identified 2 bacterial phyla, 2 classes, 4 orders, 5 families, and 6 genera that varied noticeably in their relative abundance in the MMD and HC cohorts (Figure 2A). For the examination of the abundance ratio of these 6 genera, 2 genera were enriched in the MMD cohort; 4 genera were enriched in the HC cohort (Figure 2B). The 2 genera Lachnoclostridium and Fusobacterium were more prevalent in the MMD cohort than the HC cohort (p < 0.001 for all, Figure 3). The 4 genera Bifidobacterium, Prevotella, Enterobacter, and Romboutsia were more abundant in the HC cohort as compared to those in the MMD cohort (p < 0.001 for Bifidobacterium, Prevotella, and Enterobacter; p < 0.01 for Romboutsia, Figure 3). Using MaAsLin2, we were able to identify differences in taxa abundances between MMD and HC while correcting for sex, age, smoking, drinking, and hypertension. By MaAsLin2 analysis, MMD was associated with decreased relative abundances of Enterobacter and Bifidobacterium and increased relative abundances of Lachnoclostridium and Fusobacterium (Table 3, Figure VI). We created a prediction model employing these 4 genera as a disease classifier after identifying them. The AUC value was 0.921 with 95% confidence interval of 0.873 to 0.968 between MMD and HC groups (sensitivity 91.7%; specificity 83.3%; Youden index 0.750) (Figure 4A). Due to the wide and practical ranges of the threshold probabilities, the outcomes of DCA supported and verified the utilization of these 4 genera in the anticipation of MMD (Figure 4B). There was no difference in the detection rate of Enterobacter and Fusobacterium between males and females (Figure II in the Supplementary materials). In addition, a higher proportion of sex (envfit analysis, r2 = 0.020, p = 0.328), smoking (envfit analysis, r2 = 0.018, p = 0.358), drinking (envfit analysis, r2 = 0.000, p = 0.995), and hypertension (envfit analysis, r2 = 0.035, p = 0.102), in patients with MMD did not affect the difference of microbiome between patients and controls (Figure III in the Supplementary materials).
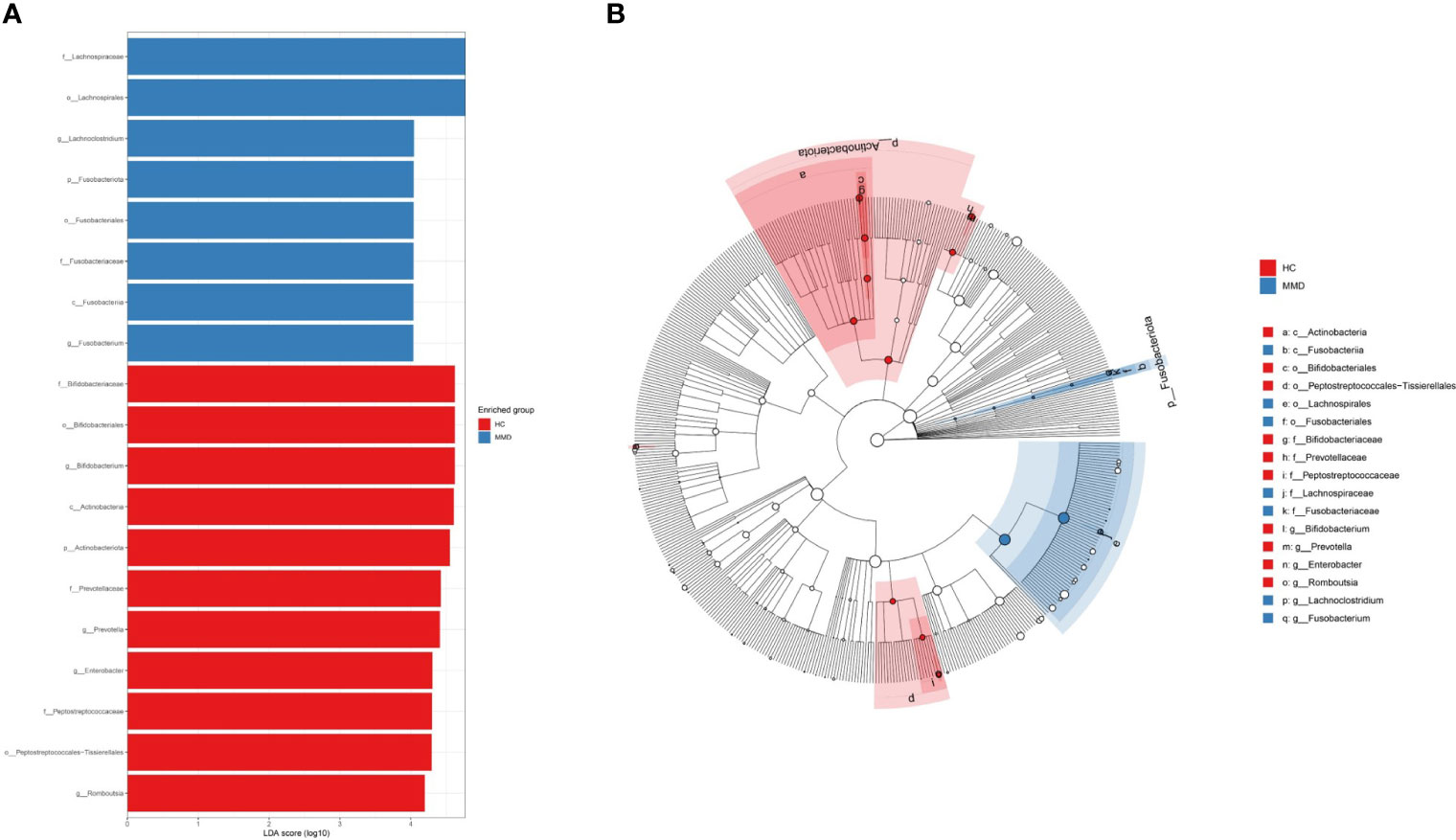
Figure 2 Discriminative taxa between MMD and HC groups. (A) Discriminative taxa between MMD and HC were determined using linear discriminant analysis effect size (LEfSe). The LEfSe analysis revealed that 2 bacterial phyla, 2 classes, 4 orders, 5 families, and 6 genera were significantly different between MMD and HC groups. The blue bar chart represents the bacteria that were more abundant in the MMD group and the red bar chart represents the bacteria that were more abundant in the HC group; (B) The cladograms report the taxa showing different abundance values according to LEfSe. The 2 genera Lachnoclostridium and Fusobacterium had higher abundance in the MMD group as compared to those in the HC group; The 4 genera Bifidobacterium, Prevotella, Enterobacter, and Romboutsia had higher abundance in the HC group as compared to those in the MMD group. MMD, moyamoya disease; HC, healthy controls.
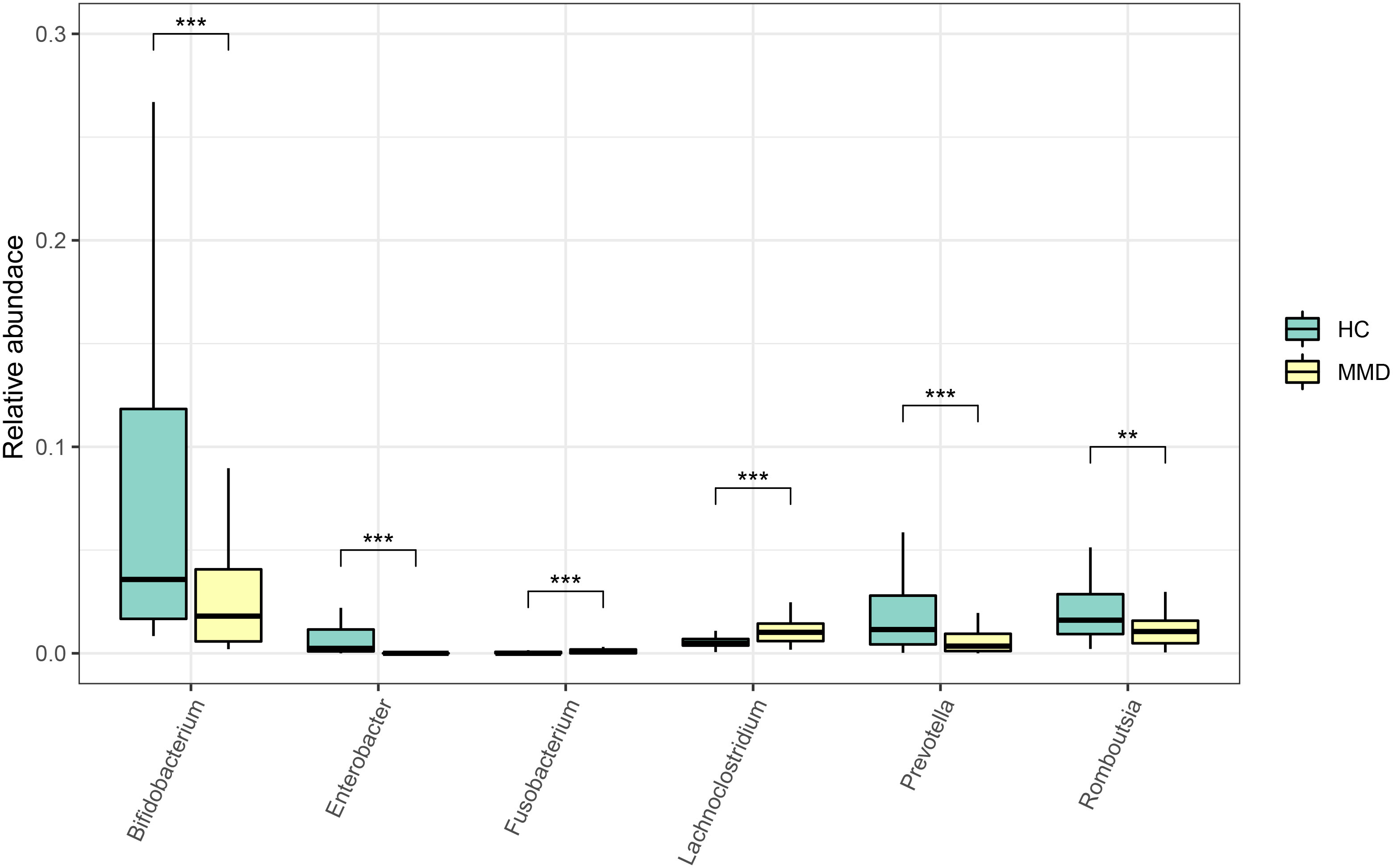
Figure 3 Comparison of the relative abundance of the differential genera between MMD and HC groups. ***p < 0.001; **0.001 < p < 0.01. MMD, moyamoya disease; HC, healthy controls.
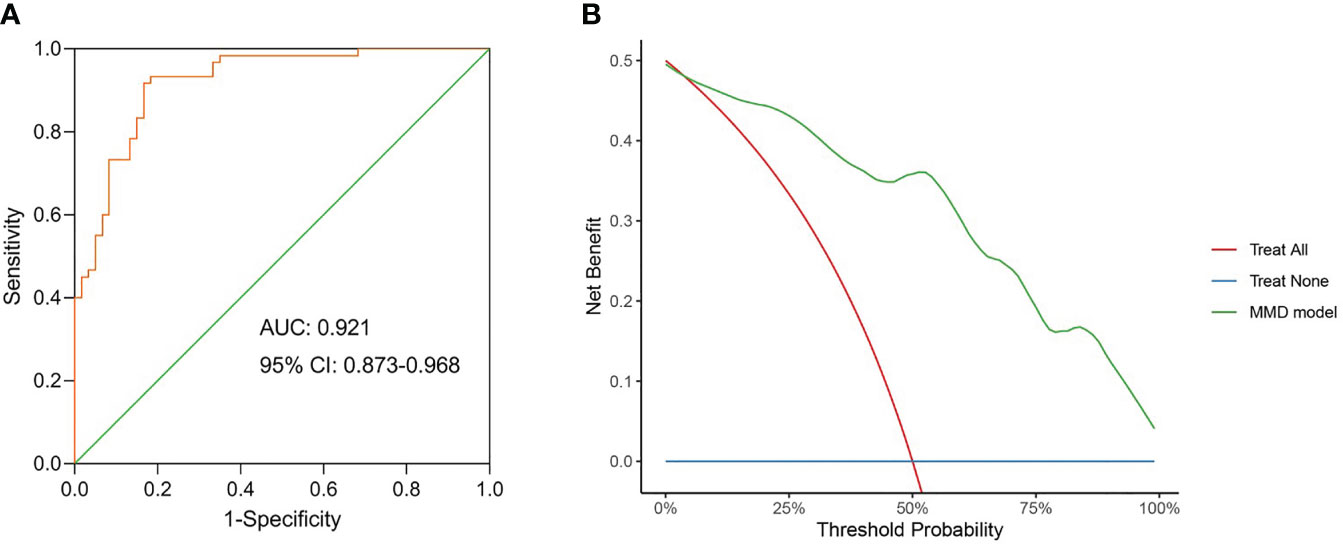
Figure 4 The model constructed using the differential genera can be used as a disease classifier to differentiate MMD patients from HC. (A) Receiver-operating characteristic (ROC) of the model constructed using the differential genera. The diagonal line in the graph marks an area under the curve (AUC) of 0.5. The AUC value was 0.921 with 95% confidence intervals (CI) of 0.873 to 0.968 (sensitivity 91.7%; specificity 83.3%; Youden index 0.750); (B) Decision curve analysis (DCA) for the model constructed using the differential genera. DCA showed wide and practical range of threshold probabilities for MMD. MMD, moyamoya disease; HC, healthy controls.
The relationship between alterations of the gut microbiota and medical indices in subjects with MMD
As shown in Figure I in the Supplementary materials, the 4 genera were significantly correlated with 4 clinical indices (p < 0.05 for all). The abundance of Enterobacter was positively correlated with serum TG levels. Furthermore, the abundance of Bifidobacterium and Lachnoclostridium were positively correlated with WBC and SBP, respectively. The abundance of Fusobacterium was negatively correlated with BMI.
Differences in gut microbiota among different MMD subtypes
The Shannon index differed substantially between the ischemic and hemorrhagic cohorts (p < 0.05, Figure 5A). However, no substantial alterations were detected in the weighted UniFrac distances between these both cohorts in β-diversity (Figure 5B). The Venn figure presenting the overlaps among these two cohorts indicated that 928 OTUs were mutual between the two cohorts (Figure 5C). In the linear discriminant analysis effect size analysis, we did not recognize that the relative abundance of any phyla, classes, orders, families, and genera differed significantly between the ischemic and hemorrhagic groups. As demonstrated in Figure IV, V in the Supplementary materials, there was also no alteration in β-diversity among MMD patients with wild-type p.R4810K variants (GG)/Suzuki stage of 0-2 and heterozygous p.R4810K variants (GA)/Suzuki stage of 3-6. Furthermore, we did not identify that the relative abundance of any phyla, classes, orders, families, and genera that differed significantly between these groups in the linear discriminant analysis effect size analysis.
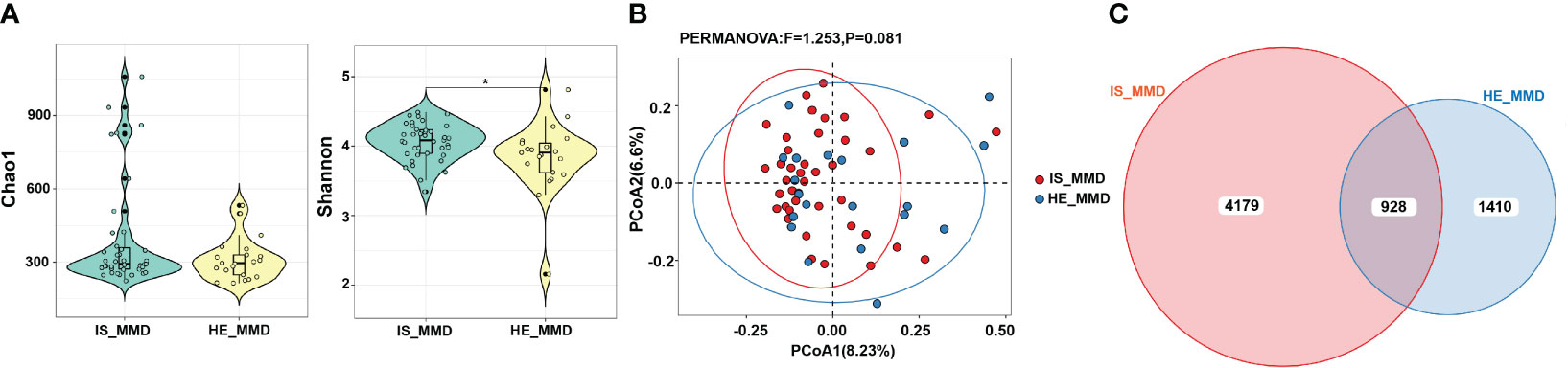
Figure 5 Comparison of microbial diversity between ischemic (IS) and hemorrhagic (HE) MMD. (A) The Shannon index differed between the two groups; (B) There was no significant difference in β-diversity (p > 0.05); (C) The Venn diagram displaying the overlaps between these two groups indicated that 928 OTUs were shared among the two groups. *0.01 < p < 0.05. MMD, moyamoya disease; IS_MMD, ischemic moyamoya disease; HE_MMD, hemorrhagic moyamoya disease.
Discussion
In this study, at the phylum level, the relative abundance of Fusobacteriota/Actinobacteria was considerably higher/lower in the MMD cohort than that in the HC cohort. In the linear discriminant analysis effect size analysis, 2 bacterial phyla, 2 classes, 4 orders, 5 families, and 6 genera were detected. We discovered that the abundance of the 2 genera, Lachnoclostridium and Fusobacterium, rose in the MMD group, while the abundance of the 2 genera, Bifidobacterium and Enterobacter diminished in the MMD group. Additionally, we created a disease classification model employing these 4 markers (genera), and it was shown to be beneficial for medical prediction.
The Chao 1, Shannon, and Simpson indices are frequently employed to estimate the diversity within a single environment or sample (α-diversity). Rare species are given greater weight by the Chao 1 and Shannon indices than by the Simpson index, which favors common species (Qian et al., 2020). The Simpson index did not vary substantially between the two cohort in this study, but the Chao 1 and Shannon indices did, suggesting that the rare species may have a stronger impact on the differences than the common species. β-diversity index varied among the two cohorts, indicating that the gut microbiota of the two groups was distinct.
The results of a diversity study showed that the microbiome structure of both cohorts differed. Univariate community assessment further emphasized the variations in the both cohorts’ microbiome. Fusobacterium is an opportunistic commensal anaerobe in the oral cavity or colon (Brennan and Garrett, 2019). In recent years, many investigations have exhibited that Fusobacterium is involved in the growth and progression of colorectal cancer by regulating immune inflammatory factors (Kostic et al., 2013; Rubinstein et al., 2013). In addition, Fusobacterium can promote the production of pro-inflammatory factors and reactive oxygen species, which may play a key role in chronic inflammation (Kang et al., 2019). In this investigation, the abundance of Fusobacterium was higher in the MMD group than in the HC group. Therefore, it is reasonable to speculate that chronic inflammation mediated by Fusobacterium may have a potential relationship with the development of MMD. By digesting type IV collagen, matrix metalloproteinase (MMP)-9 destroys endothelium basal lamina and tight junction proteins, and its over activation causes endothelial instability (Murphy and Nagase, 2008). The serum MMP-9 may act as a biomarker for bleeding anticipation in MMD, according to a prospective study that used plasma samples from 84 subjects with MMD (Lu et al., 2021). It has been shown that Fusobacterium can induce the production of MMP-9 (Suzuki et al., 2022). So, it is also possible that Fusobacterium mediates the pathogenesis of MMD by inducing the production of MMP-9. More in-depth studies are needed to confirm the above ideas.
The extremely polyphyletic class Clostridia includes the recently founded genus Lachnoclostridium (Yutin and Galperin, 2013). Additionally, compared to normal controls, colorectal cancer patients had a greater relative abundance of Lachnoclostridium (Liang et al., 2020). Wang et al. firstly suggested a dose-dependent relationship between plasma levels of trimethylamine-N-oxide (TMAO) and risk of cardiovascular disorders in individuals with heart problems in 2011 (Wang et al., 2011). Furthermore, the levels of TMAO were positively correlated with the prevalence of hypertension or hyperhomocysteinemia (Nie et al., 2018; Ge et al., 2020b). Trimethylamine (TMA), a byproduct of intestinal microbial metabolism, is converted to TMAO in the liver via flavin-containing monooxygenase 3 (Bennett et al., 2013). A recent study demonstrated that Lachnoclostridium can enhance the synthesis of TMA and so promote atherosclerosis progression (Cai et al., 2022). Previous research has shown that elevated Hcy was connected to a greater risk of MMD (Ge et al., 2020a). In this investigation, the abundance of Lachnoclostridium was higher in the MMD cohort than in the HC cohort. Therefore, we hypothesized that Lachnoclostridium may influence the levels of Hcy and thus involve with the onset of MMD by promoting the production of TMAO. What’s more interesting is that among individuals with MMD, we discovered a positive association between the relative abundance of Lachnoclostridium and SBP. This can also be explained by the effect of Lachnoclostridium on TMAO synthesis. The relative abundance of Lachnoclostridium might be predictive of blood pressure in patients with MMD.
Bifidobacterium is an important intestinal beneficial microorganism. Previous research has already indicated that Bifidobacterium was negatively correlated with plasma TMAO and TMA (Chen et al., 2016). In this experiment, the MMD cohort had a lesser abundance of Bifidobacterium than the HC cohort. As mentioned earlier, the emergence of this trend was logical. Enterobacter may exert a probiotic effect in the gastrointestinal tract of humans (Robinson, 2014). The relative abundance of Enterobacter in MMD patients were also reduced in this study. Bifidobacterium and Enterobacter may have antagonistic effect on the occurrence or development of MMD.
Mineharu et al. collected feces from 27 MMD patients and 15 normal controls. They found no difference in α-diversity or β-diversity between patients with MMD and controls (Mineharu et al., 2022). We speculated that the reason their findings were not consistent with ours is because of their small sample size. Furthermore, they found that increased abundance of Ruminococcus gnavus and Peptostreptococcaceae and decreased abundance of Roseburia inulinivorans in gut microbiota in MMD patients. However, our data yielded the opposite conclusion. The present study demonstrated an increase in the abundance of Peptostreptococcaceae in healthy controls. In addition, although we found a higher abundance of Actinobacteria in the HC group, we did not find a difference in the abundance of Roseburia between the two groups. Although species level was not involved in this study, there were significant differences between our study and the previous study. We speculated that gut microbiota in population may also change depending on geographic location. Therefore, international multicenter replication studies are needed to confirm the results of the present study.
The 4 genera found in our investigation were employed to create a predictive model. We utilized ROC and DCA to assess the predictive power of the model. The AUC value of the model was. The results of DCA also indicated that the model has good predictive power. It’s still not apparent if adding the clinical characteristics will help the prediction model get any better. Nevertheless, for the diagnosis of MMD, DSA is still necessary. The results of this study may provide some clues to the etiology of MMD. Moreover, we did not observe differences in gut microbiota between subgroups of MMD. Therefore, we hypothesized that similar alterations in gut microbiota may lead to different phenotypes of MMD. RNF213 p.R4810K variant did not appear to affect the gut microbiota of MMD patients. The underlying mechanisms of these hypotheses need to be confirmed by further studies.
This study has a number of restrictions. First, metabolomics and heredity as well as other risk variables were not studied. Second, these findings could not be extended to children or other races, as only Chinese adult patients with MMD were included. Third, to ascertain the cause-and-effect connection between the gut microbiome and the onset of MMD, more research, including longitudinal human studies, is required. Fourth, unequal distribution of clinical variables (smoking, drinking, use of medication, etc) between cases and controls may affect the distribution of gut microbiome to some extent. Finally, this is a prospective study using clinical information from a single institute. The results’ applicability is constrained, necessitating additional validation in a different cohort or location.
Conclusions
In conclusion, patients with MMD had changed gut microbiota, which were distinguished by elevated abundance of Lachnoclostridium and Fusobacterium and reduced abundance of Bifidobacterium and Enterobacter. Additionally, these 4 genera utilized in the model’s construction were able to accurately identify whether a person had MMD or not, indicating that these genera may be applied in clinical settings as predictors and biomarkers.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The studies involving humans were approved by The Ethics Committee of Beijing Tiantan Hospital, Capital Medical University. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
Conceptualization and methodology: PG and DZ. Data curation and writing original draft: XiY. Visualization and investigation: YuZ and WL. Supervision: QZ, XuY, and JZ. Software and validation: XL, RW, and YaZ. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Key Technology Research and Development Programme of the Ministry of Science and Technology of China (grants 2021YFC2500502).
Acknowledgments
We thank all the participants involved in this study for their important contributions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1252681/full#supplementary-material
Abbreviations
MMD, moyamoya disease; HC, healthy controls; SBP, systolic blood pressure; DBP, diastolic blood pressure; BMI, body mass index; WBC, white blood cells; ALB, albumin; TG, triglyceride; Hcy, homocysteine; DSA, digital subtraction angiography; OTUs, operational taxonomic units; ROC, receiver-operating characteristic; AUC, area under the curve; DCA, decision curve analysis; MMP-9, matrix metalloproteinase-9; TMAO, trimethylamine-N-oxide; TMA, trimethylamine.
References
Acker, G., Fekonja, L., Vajkoczy, P. (2018). Surgical management of moyamoya disease. Stroke. 49, 476–482. doi: 10.1161/strokeaha.117.018563
Bang, O. Y., Fujimura, M., Kim, S. K. (2016). The pathophysiology of moyamoya disease: An update. J. Stroke. 18, 12–20. doi: 10.5853/jos.2015.01760
Bennett, B. J., de Aguiar Vallim, T. Q., Wang, Z., Shih, D. M., Meng, Y., Gregory, J., et al. (2013). Trimethylamine-N-oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation. Cell Metab. 17, 49–60. doi: 10.1016/j.cmet.2012.12.011
Bokulich, N. A., Subramanian, S., Faith, J. J., Gevers, D., Gordon, J. I., Knight, R., et al. (2013). Quality-filtering vastly improves diversity estimates from illumina amplicon sequencing. Nat. Methods 10, 57–59. doi: 10.1038/nmeth.2276
Brennan, C. A., Garrett, W. S. (2019). Fusobacterium nucleatum - symbiont, opportunist and oncobacterium. Nat. Rev. Microbiol. 17, 156–166. doi: 10.1038/s41579-018-0129-6
Cai, Y. Y., Huang, F. Q., Lao, X., Lu, Y., Gao, X., Alolga, R. N., et al. (2022). Integrated metagenomics identifies a crucial role for trimethylamine-producing Lachnoclostridium in promoting atherosclerosis. NPJ Biofilms Microbiomes. 8, 11. doi: 10.1038/s41522-022-00273-4
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). Qiime allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chen, M. L., Yi, L., Zhang, Y., Zhou, X., Ran, L., Yang, J., et al. (2016). Resveratrol attenuates trimethylamine-N-oxide (TMAO)-induced atherosclerosis by regulating TMAO synthesis and bile acid metabolism via remodeling of the gut microbiota. mBio. 7, e02210–e02215. doi: 10.1128/mBio.02210-15
Edgar, R. C. (2013). Uparse: Highly accurate otu sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Fujimura, M., Bang, O. Y., Kim, J. S. (2016). Moyamoya disease. Front. Neurol. Neurosci. 40, 204–220. doi: 10.1159/000448314
Fujimura, M., Sonobe, S., Nishijima, Y., Niizuma, K., Sakata, H., Kure, S., et al. (2014). Genetics and biomarkers of moyamoya disease: Significance of RNF213 as a susceptibility gene. J. Stroke. 16, 65–72. doi: 10.5853/jos.2014.16.2.65
Ge, P., Zhang, Q., Ye, X., Liu, X., Deng, X., Wang, J., et al. (2020a). Modifiable risk factors associated with moyamoya disease: A case-control study. Stroke. 51, 2472–2479. doi: 10.1161/strokeaha.120.030027
Ge, X., Zheng, L., Zhuang, R., Yu, P., Xu, Z., Liu, G., et al. (2020b). The gut microbial metabolite trimethylamine N-oxide and hypertension risk: A systematic review and dose-response meta-analysis. Adv. Nutr. 11, 66–76. doi: 10.1093/advances/nmz064
Jie, Z., Xia, H., Zhong, S. L., Feng, Q., Li, S., Liang, S., et al. (2017). The gut microbiome in atherosclerotic cardiovascular disease. Nat. Commun. 8, 845. doi: 10.1038/s41467-017-00900-1
Kamada, F., Aoki, Y., Narisawa, A., Abe, Y., Komatsuzaki, S., Kikuchi, A., et al. (2011). A genome-wide association study identifies RNF213 as the first moyamoya disease gene. J. Hum. Genet. 56, 34–40. doi: 10.1038/jhg.2010.132
Kang, W., Jia, Z., Tang, D., Zhang, Z., Gao, H., He, K., et al. (2019). Fusobacterium nucleatum facilitates apoptosis, ROS generation, and inflammatory cytokine production by activating AKT/MAPK and NF-κB signaling pathways in human gingival fibroblasts. Oxid. Med. Cell Longev. 2019, 1681972. doi: 10.1155/2019/1681972
Kauv, P., Gaudré, N., Hodel, J., Tuilier, T., Habibi, A., Oppenheim, C., et al. (2019). Characteristics of moyamoya syndrome in sickle-cell disease by magnetic resonance angiography: An adult-cohort study. Front. Neurol. 10. doi: 10.3389/fneur.2019.00015
Kawabata, S., Takagaki, M., Nakamura, H., Oki, H., Motooka, D., Nakamura, S., et al. (2022). Dysbiosis of gut microbiome is associated with rupture of cerebral aneurysms. Stroke. 53, 895–903. doi: 10.1161/strokeaha.121.034792
Kim, T., Oh, C. W., Bang, J. S., Kim, J. E., Cho, W. S. (2016). Moyamoya disease: Treatment and outcomes. J. Stroke. 18, 21–30. doi: 10.5853/jos.2015.01739
Koeth, R. A., Wang, Z., Levison, B. S., Buffa, J. A., Org, E., Sheehy, B. T., et al. (2013). Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19, 576–585. doi: 10.1038/nm.3145
Koizumi, A., Kobayashi, H., Hitomi, T., Harada, K. H., Habu, T., Youssefian, S. (2016). A new horizon of moyamoya disease and associated health risks explored through RNF213. Environ. Health Prev. Med. 21, 55–70. doi: 10.1007/s12199-015-0498-7
Kostic, A. D., Chun, E., Robertson, L., Glickman, J. N., Gallini, C. A., Michaud, M., et al. (2013). Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14, 207–215. doi: 10.1016/j.chom.2013.07.007
Kuriyama, S., Kusaka, Y., Fujimura, M., Wakai, K., Tamakoshi, A., Hashimoto, S., et al. (2008). Prevalence and clinicoepidemiological features of moyamoya disease in Japan: Findings from a nationwide epidemiological survey. Stroke. 39, 42–47. doi: 10.1161/strokeaha.107.490714
Li, H., Xu, H., Li, Y., Jiang, Y., Hu, Y., Liu, T., et al. (2020). Alterations of gut microbiota contribute to the progression of unruptured intracranial aneurysms. Nat. Commun. 11, 3218. doi: 10.1038/s41467-020-16990-3
Li, J., Zhao, F., Wang, Y., Chen, J., Tao, J., Tian, G., et al. (2017). Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome. 5, 14. doi: 10.1186/s40168-016-0222-x
Liang, J. Q., Li, T., Nakatsu, G., Chen, Y. X., Yau, T. O., Chu, E., et al. (2020). A novel faecal Lachnoclostridium marker for the non-invasive diagnosis of colorectal adenoma and cancer. Gut. 69, 1248–1257. doi: 10.1136/gutjnl-2019-318532
Liu, W., Morito, D., Takashima, S., Mineharu, Y., Kobayashi, H., Hitomi, T., et al. (2011). Identification of RNF213 as a susceptibility gene for moyamoya disease and its possible role in vascular development. PloS One 6, e22542. doi: 10.1371/journal.pone.0022542
Liu, X. J., Zhang, D., Wang, S., Zhao, Y. L., Teo, M., Wang, R., et al. (2015). Clinical features and long-term outcomes of moyamoya disease: A single-center experience with 528 cases in China. J. Neurosurg. 122, 392–399. doi: 10.3171/2014.10.Jns132369
Lu, J., Wang, J., Lin, Z., Shi, G., Wang, R., Zhao, Y., et al. (2021). MMP-9 as a biomarker for predicting hemorrhagic strokes in moyamoya disease. Front. Neurol. 12. doi: 10.3389/fneur.2021.721118
Masuda, J., Ogata, J., Yutani, C. (1993). Smooth muscle cell proliferation and localization of macrophages and T cells in the occlusive intracranial major arteries in moyamoya disease. Stroke. 24, 1960–1967. doi: 10.1161/01.str.24.12.1960
McMurdie, P. J., Holmes, S. (2013). Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PloS One 8, e61217. doi: 10.1371/journal.pone.0061217
Mineharu, Y., Nakamura, Y., Sato, N., Kamata, T., Oichi, Y., Fujitani, T., et al. (2022). Increased abundance of Ruminococcus gnavus in gut microbiota is associated with moyamoya disease and non-moyamoya intracranial large artery disease. Sci. Rep. 12, 20244. doi: 10.1038/s41598-022-24496-9
Murphy, G., Nagase, H. (2008). Progress in matrix metalloproteinase research. Mol. Aspects Med. 29, 290–308. doi: 10.1016/j.mam.2008.05.002
Nie, J., Xie, L., Zhao, B. X., Li, Y., Qiu, B., Zhu, F., et al. (2018). Serum Trimethylamine N-oxide concentration is positively associated with first stroke in hypertensive patients. Stroke. 49, 2021–2028. doi: 10.1161/strokeaha.118.021997
Oksanen, J., Blanchet, F., Kindt, R., Legendre, P., Minchin, P., O’hara, R., et al. (2017). Ordination methods, diversity analysis and other functions for community and vegetation ecologists. Vegan: Community Ecol. Package., 05–26. Available at: https://cran.r-project.org/web/packages/vegan/index.html.
Peh, A., O'Donnell, J. A., Broughton, B. R. S., Marques, F. Z. (2022). Gut microbiota and their metabolites in stroke: A double-edged sword. Stroke. 53, 1788–1801. doi: 10.1161/strokeaha.121.036800
Qian, X., Liu, Y. X., Ye, X., Zheng, W., Lv, S., Mo, M., et al. (2020). Gut microbiota in children with juvenile idiopathic arthritis: Characteristics, biomarker identification, and usefulness in clinical prediction. BMC Genomics 21, 286. doi: 10.1186/s12864-020-6703-0
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Renz, H., von Mutius, E., Brandtzaeg, P., Cookson, W. O., Autenrieth, I. B., Haller, D. (2011). Gene-environment interactions in chronic inflammatory disease. Nat. Immunol. 12, 273–277. doi: 10.1038/ni0411-273
Robinson, R. K. (2014). Encyclopedia of food microbiology (American Academic Press: Academic press).
Rubinstein, M. R., Wang, X., Liu, W., Hao, Y., Cai, G., Han, Y. W. (2013). Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 14, 195–206. doi: 10.1016/j.chom.2013.07.012
Scott, R. M., Smith, E. R. (2009). Moyamoya disease and moyamoya syndrome. N Engl. J. Med. 360, 1226–1237. doi: 10.1056/NEJMra0804622
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60. doi: 10.1186/gb-2011-12-6-r60
Suzuki, R., Kamio, N., Sugimoto, K., Maruoka, S., Gon, Y., Kaneko, T., et al. (2022). Periodontopathic bacterium Fusobacterium nucleatum affects matrix metalloproteinase-9 expression in human alveolar epithelial cells and mouse lung. In Vivo. 36, 649–656. doi: 10.21873/invivo.12749
Suzuki, J., Takaku, A. (1969). Cerebrovascular "moyamoya" disease. Disease showing abnormal net-like vessels in base of brain. Arch. Neurol. 20, 288–299. doi: 10.1001/archneur.1969.00480090076012
Wang, Z., Klipfell, E., Bennett, B. J., Koeth, R., Levison, B. S., Dugar, B., et al. (2011). Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 472, 57–63. doi: 10.1038/nature09922
Weng, L., Cao, X., Han, L., Zhao, H., Qiu, S., Yan, Y., et al. (2017). Association of increased Treg and Th17 with pathogenesis of moyamoya disease. Sci. Rep. 7, 3071. doi: 10.1038/s41598-017-03278-8
Keywords: gut microbiota, characteristics, biomarker, moyamoya disease, adults
Citation: Yu X, Ge P, Zhai Y, Liu W, Zhang Q, Ye X, Liu X, Wang R, Zhang Y, Zhao J and Zhang D (2023) Gut microbiota in adults with moyamoya disease: characteristics and biomarker identification. Front. Cell. Infect. Microbiol. 13:1252681. doi: 10.3389/fcimb.2023.1252681
Received: 04 July 2023; Accepted: 26 September 2023;
Published: 17 October 2023.
Edited by:
Veeranoot Nissapatorn, Walailak University, ThailandReviewed by:
Yohei Mineharu, Kyoto University, JapanWenfeng Feng, Southern Medical University, China
Gang Wang, Southern Medical University, China
Copyright © 2023 Yu, Ge, Zhai, Liu, Zhang, Ye, Liu, Wang, Zhang, Zhao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peicong Ge, Z2VwZWljb25nQDE2My5jb20=; Dong Zhang, emhhbmdkb25nMDY2MEBhbGl5dW4uY29t
 Xiaofan Yu
Xiaofan Yu Peicong Ge
Peicong Ge Yuanren Zhai
Yuanren Zhai Wei Liu
Wei Liu Qian Zhang
Qian Zhang Xun Ye1,2,3,4,5
Xun Ye1,2,3,4,5 Xingju Liu
Xingju Liu Rong Wang
Rong Wang Yan Zhang
Yan Zhang Jizong Zhao
Jizong Zhao Dong Zhang
Dong Zhang