
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Cell. Infect. Microbiol. , 06 September 2023
Sec. Intestinal Microbiome
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1249069
This article is part of the Research Topic Human Microbiome and COVID-19 View all 7 articles
 Yingzhi Liu1,2
Yingzhi Liu1,2 Matthew T. V. Chan2
Matthew T. V. Chan2 Francis K. L. Chan1,3
Francis K. L. Chan1,3 William K. K. Wu2,3,4*
William K. K. Wu2,3,4* Siew C. Ng1,4,5*
Siew C. Ng1,4,5* Lin Zhang1,2,5*
Lin Zhang1,2,5*Introduction: Emerging preclinical and clinical studies suggest that altered gut microbiome composition and functions are associated with coronavirus 2019 (COVID- 19) severity and its long-term complications. We hypothesize that COVID-19 outcome is associated with gut microbiome status in population-based settings.
Methods: Gut metagenomic data of the adult population consisting of 2871 subjects from 16 countries were obtained from ExperimentHub through R, while the dynamic death data of COVID-19 patients between January 22, 2020 and December 8, 2020 in each country was acquired from Johns Hopkins Coronavirus Resource Center. An adjusted stable mortality rate (SMR) was used to represent these countries’ mortality and correlated with the mean relative abundance (mRA) of healthy adult gut microbiome species.
Results: After excluding bacterial species with low prevalence (prevalence <0.2 in the included countries), the β-diversity was significantly higher in the countries with high SMR when compared with those with median or low SMR (p <0.001). We then identified the mRA of two butyrate producers, Eubacterium rectale and Roseburia intestinalis, that were negatively correlated with SMR during the study period. And the reduction of these species was associated with severer COVID-19 manifestation.
Conclusion: Population-based microbiome signatures with the stable mortality rate of COVID-19 in different countries suggest that altered gut microbiome composition and functions are associated with mortality of COVID-19.
A previous cohort study showed that the gut microbial diversity was altered in COVID-19-infected subjects (Zhang et al., 2023). Likewise, a previous study suggested that the microbiome change in COVID-19 patients was driven by the enrichment of Ruminococcus gnavus, Ruminococcus torques, and Bacteroides dorei, and the depletion of beneficial bacterial species, including Bifidobacterium adolescentis, Faecalibacterium prausnitzii, and Eubacterium rectale (E. rectale) (Yeoh et al., 2021). However, whether COVID-19 outcome is associated with pre-existing gut microbiome status in population-based settings is unknown. Herein, gut metagenomic data of the adult population consisting of 2,871 subjects from 16 countries were obtained from ExperimentHub (STable 1, SFigure 1A), and the dynamic incidence and mortality of COVID-19 between January 22, 2020 and December 8, 2020 in each country were obtained from Johns Hopkins Coronavirus Resource Center (SFigure 1A) (Pasolli et al., 2017; Dong et al., 2020). An adjusted stable mortality rate (SMR, SFigure 1A) was used to indicate the mortality rate in these countries (SFigure 1B), and we correlated mortality data with the mean relative abundance (mRA) of healthy adult gut bacterial species. We chose the longest duration of the stable period before the introduction of the vaccination programme for all countries to calculate SMR, through which we uncovered the relationship between microbiota profiles and the SMR of COVID-19 (SFigure 1A).
Overall, although the α-diversity of the gut microbiota did not show any difference, the inverse Simpson (1-Simpson) had a marginal p-value (p = 0.054) (SFigure 1C). β-diversity was significantly higher in the countries with high SMR when compared with those with median or low SMR (p <0.001) (SFigure 1D). Importantly, after excluding bacterial species with low prevalence rates (<0.2 in the above countries), half of the top 20 bacteria (STable 2) that showed negative correlations with SMR were butyrate producers (highlighted in green).
The Omicron variant has caused an unprecedented pandemic with distinct phenotypes, and COVID-related mortality has dropped significantly with an explosion in infection rate (STable 3) (Hoffmann et al., 2022; Nyberg et al., 2022). Therefore, we applied the same analytic strategy to the Omicron pandemic to determine the replicability of our findings. Four overlapped species among the top-20s were identified, namely E. rectale, Roseburia intestinalis (R. intestinalis), Bifidobacterium angulatum, and Parabacteroides unclassified (Figure 1A, STable 2). It should be noted that a well-known beneficial butyrate producer, E. rectale, was the only species that was correlated significantly with the mortality outcome of all SARS-CoV-2 variants, i.e., the Alpha, Beta, Gamma variants, and the Omicron variant (STable 2) (Louis and Flint, 2009; Zhang et al., 2022). To validate the findings generated from the public dataset, we determined the relative abundance of the four species in the published Hong Kong COVID-19 cohorts that was conducted before the introduction of Hong Kong’s vaccination programme (Zuo et al., 2020; Yeoh et al., 2021; Zhang et al., 2022). From Figure 1B we found that the relative abundance of E. rectale and R. intestinalis was much lower in patients with severe COVID-19 compared to the control subjects or COVID-19 patients with mild or moderate symptoms.
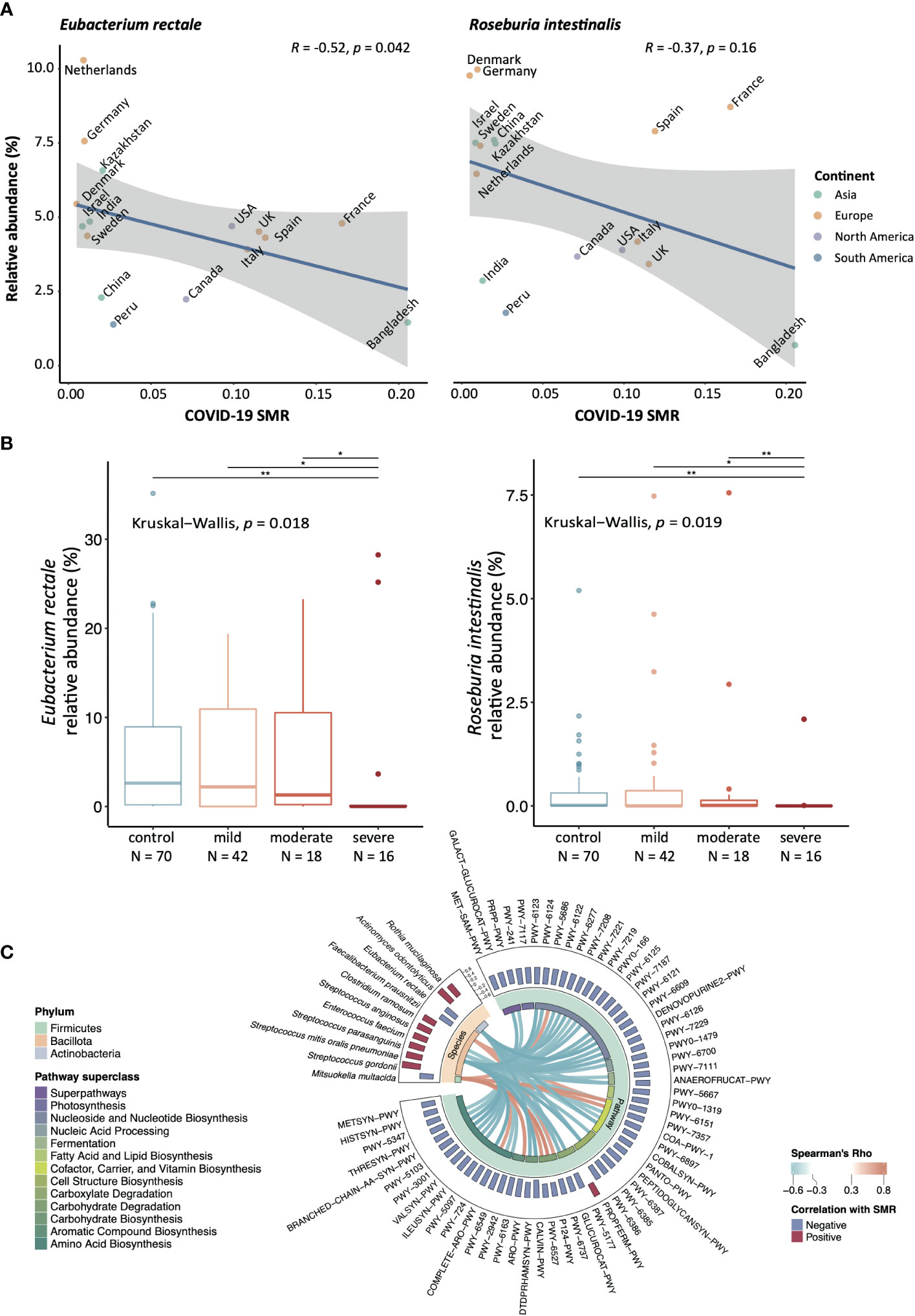
Figure 1 The bacterial species that correlate with SMR amongst different countries Significance level: *:p-value <0.05, **p-value <0.01. (A) Correlation between mRA and SMR in E. rectale and R. intestinalis, two-sided Spearman correlation test has been performed. (B) The validation cohort from Hong Kong COVID-19 cohort (Severe N = 16, moderate N = 18, mild = 42, control = 70). The relative abundance of E. rectale and R. intestinalis from the fecal metagenomic data were compared with the Kruskal−Wallis test and post-hoc tested with Wilcox rank-sum test amongst each disease/control group. The reduction of relative abundance of E. rectale in human fecal samples was reported in COVID-19 patients from the same cohort (Yeoh et al., 2021) (C) The correlations between SMR-correlated functional pathways and bacteria species were conducted using Hierarchical All-against-All association testing (HAllA), and only species/pathways with at least one significant correlation were shown.
Moreover, we identified 181 functional pathways and eleven bacteria species that were significantly correlated with SMR (STables 2, 4). To pinpoint the associated features between these bacterial species or functional pathways related to SMR, we performed all-against-all association testing to identify the adjusted correlations. Figure 1C presents the association between the differential species and pathways. Interestingly, the species negatively correlated with SMR were significantly positively correlated with the depleted pathways under the superclasses of 1) carbohydrate degradation (PWY-6737, P124-PWY, and PWY-6527); 2) cofactor, carrier, and vitamin biosynthesis (PWY-6151, PWY-7357) (Figure 1C), indicating that these functional deficiencies of the gut microbiota at the baseline might be linked to higher COVID-19 mortality.
In this study, for the first time, we identified a pre-existence mRA of E. rectale and R. intestinalis in a healthy population before the COVID-19 pandemic are associated with lower COVID-19 regional mortality in a population-based gut microbiota study. The depletion of both identified species, E. rectale and R. intestinalis have been reported not only in COVID-19 (Zuo et al., 2020; Cao et al., 2021; Yeoh et al., 2021; Zhang et al., 2022) but also in ulcerative colitis patients (Pittayanon et al., 2020; Shen et al., 2022) and is possibly linked to the reduction of host inflammatory response. The diminished abundance of E. rectale comes with the negative correlation to C-X-C motif ligand 10 (CXCL10) and tumor necrosis factor-alpha (TNF-α), two inflammation markers that indicate the origination of immune response at the early stage of COVID-19 (Yeoh et al., 2021). R. intestinalis inhibits the development of Crohn’s disease (CD) by increasing the differentiation of anti-inflammatory Tregs, which may provide the basis of new therapeutic strategies for CD (Shen et al., 2022). Such a decrease of butyrate-producing taxa, such as Blautia and Eubacterium (rectale), in influenza A virus infection was proven in the previous 16s studies (Fuentes et al., 2021; Bhar et al., 2022). Strikingly, in the pathway analysis, results displayed a strong association between the depleted species and reduced pathways, implying that the protective role of the gut microbiome in the population could be caused by their biosynthesis functions. Of them, the carbohydrate degradation pathways could be mainly contributed by butyrate-producing species (Flint et al., 2012) with the fermentation of carbohydrates.
There were several limitations of this study. First, the mechanism between gut microorganisms and immune functions was not further explored in the study, and it would be beneficial to understand how microbiota derive metabolites or immune activation against infection for future applications in medication or prevention. In addition, due to the limitation of the online metagenomic database, the study did not consider antibiotic usage at an individual or regional level, which could mask the metagenomic profiling and mortality rate in a country. The alteration of the bacterial community structure after antibiotic treatment has been reported (Hill et al., 2010). On the other hand, long-term antibiotic exposure may be a risk factor for all-cause mortality (Heianza et al., 2020; Verdecchia et al., 2020). Another disadvantage is that the study applied an imbalanced sample size from different countries to represent the populational species’ relative abundance. Unlike epidemiology-based cohorts, due to the heterogeneity of each country and individual, metagenomic-based cohorts, restricted by their expense and feasibility, could only serve as a nation miniature. As a compromise, we conducted the validation cohort in the Hong Kong population to confirm our findings. Yet, a more extensive study with larger sample sizes is required to validate the predictive capability of these two butyrate producers. Third, this study considered COVID-19 mortality as the only outcome, which may neglect that the low abundance of E. rectale may also imply the association with other disease conditions (Vermeiren et al., 2012; Zhu et al., 2018; Su et al., 2022). Therefore, although the metadata from the public datasets was limited, all the included subjects were labelled as a healthy condition to eliminate the potential bias. To our best knowledge, there are lack of comparative metagenomic-based gut microbiome-related studies related to other emerging infectious diseases at the time of our study endpoint. However, the reduction of beneficial microbial products, especially butyrate, in influenza A virus infection and sepsis has been described (Adelman et al., 2020; Sencio et al., 2020).
Presenting the negative association between the populational gut abundance of two butyrate producers and the COVID-19 regional mortality, our study offered hope that microbiota modulation could be one of the keys to reducing COVID-19-related mortality. In particular, to develop butyrate-producing probiotics with high-fibber diet may assist the enrichment of such beneficial species (Kasahara et al., 2018). Nonetheless, future assessment of these potential next-generation probiotics in animal models of COVID-19 and clinical trials in humans is warranted.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethical approval was not required for the study involving humans in accordance with the local legislation and institutional requirements. Written informed consent to participate in this study was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and the institutional requirements.
LZ, WW, MC, FC and SN conceived and designed the study. WW, LZ and YL developed the detailed methodology. LZ and YL were responsible for data collection and data analysis, interpreted the results, and drafted the manuscript. WW, SN, MC and FC reviewed the manuscript with critical suggestions. All authors critically reviewed and approved the final version. All authors had full access to all the data in the study and accepted the responsibility to submit it for publication. LZ and SN had final responsibility for the decision to submit for publication. All authors contributed to the article and approved the submitted version.
The Health and Medical Research Fund (HMRF: 6905543) and General Research Fund (GRF: 2141153).
LZ and SN were partially supported by InnoHK, The Government of Hong Kong, Special Administrative Region of the People’s Republic of China. SN is also supported by the Croucher Senior Medical Research Fellowship. We would like to thank all healthcare workers working in isolation wards of the Prince of Wales Hospital and the United Christian Hospital, Hong Kong SAR, China. We thank all health care workers working in the isolation wards of Prince of Wales Hospital, Hong Kong, China. We thank Jessica Ching, Chun Pan Cheung, Miu Ling Chin, Apple C.M. Yeung, Wendy C.S. Ho, and other staff/students for their technical contribution to this study including sample collection, inventory, and processing. We also want to acknowledge Dr. X Wang and Ms. X Hu for the advice on the methodology parts and Dr. F Zhang’s technical support on metagenomic and clinical data.
FC is Board Member of CUHK Medical Centre. He is a co-founder, non-executive Board Chairman and non-executive scientific advisor, Chief Medical Officer and shareholder of GenieBiome Ltd. shareholder of GenieBiome Ltd. He receives patent royalties through his affiliated institutions. He has received fees as an advisor and honoraria as a speaker for Eisai Co. Ltd., AstraZeneca, Pfizer Inc., Takeda Pharmaceutical Co., and Takeda China Holdings Co. Ltd. SN has served as an advisory board member for Pfizer, Ferring, Janssen, and Abbvie and received honoraria as a speaker for Ferring, Tillotts, Menarini, Janssen, Abbvie, and Takeda. SN has received research grants through her affiliated institutions from Olympus, Ferring, and Abbvie. SN is a founder member, non-executive director, non-executive scientific advisor, and shareholder of GenieBiome Ltd. SN is a scientific co-founder and shareholder of GenieBiome Ltd. SN receives patent royalties through her affiliated institutions. FC, SN and LZ are named inventors of patent applications held by the CUHK and MagIC that cover the therapeutic and diagnostic use of microbiome.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1249069/full#supplementary-material
COVID-19, Coronavirus Disease 2019; CXCL10, C-X-C motif ligand 10; CD, Crohn’s disease; mRA, Mean relative abundance; SMR, Stable mortality rate; TNF-α, Tumor necrosis factor-alpha; PWY, Pathway.
Adelman, M. W., Woodworth, M. H., Langelier, C., Busch, L. M., Kempker, J. A., Kraft, C. S., et al. (2020). The gut microbiome’s role in the development, maintenance, and outcomes of sepsis. Crit. Care 24, 278. doi: 10.1186/s13054-020-02989-1
Bhar, A., Gierse, L. C., Meene, A., Wang, H., Karte, C., Schwaiger, T., et al. (2022). Application of a maximal-clique based community detection algorithm to gut microbiome data reveals driver microbes during influenza A virus infection. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.979320
Cao, J., Wang, C., Zhang, Y., Lei, G., Xu, K., Zhao, N., et al. (2021). Integrated gut virome and bacteriome dynamics in COVID-19 patients. Gut Microbes 13, 1–21. doi: 10.1080/19490976.2021.1887722
Dong, E., Du, H., Gardner, L. (2020). An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 20, 533–534. doi: 10.1016/S1473-3099(20)30120-1
Flint, H. J., Scott, K. P., Duncan, S. H., Louis, P., Forano, E. (2012). Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3, 289–306. doi: 10.4161/gmic.19897
Fuentes, S., den Hartog, G., Nanlohy, N. M., Wijnands, L., Ferreira, J. A., Nicolaie, M. A., et al. (2021). Associations of faecal microbiota with influenza-like illness in participants aged 60 years or older: an observational study. Lancet Healthy Longev. 2, e13–e23. doi: 10.1016/S2666-7568(20)30034-9
Heianza, Y., Ma, W., Li, X., Cao, Y., Chan, A. T., Rimm, E. B., et al. (2020). Duration and life-stage of antibiotic use and risks of all-cause and cause-specific mortality: prospective cohort study. Circ. Res. 126, 364–373. doi: 10.1161/CIRCRESAHA.119.315279
Hill, D. A., Hoffmann, C., Abt, M. C., Du, Y., Kobuley, D., Kirn, T. J., et al. (2010). Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 3, 148–158. doi: 10.1038/mi.2009.132
Hoffmann, M., Kruger, N., Schulz, S., Cossmann, A., Rocha, C., Kempf, A., et al. (2022). The Omicron variant is highly resistant against antibody-mediated neutralization: Implications for control of the COVID-19 pandemic. Cell 185, 447–456 e411. doi: 10.1016/j.cell.2021.12.032
Kasahara, K., Krautkramer, K. A., Org, E., Romano, K. A., Kerby, R. L., Vivas, E. I., et al. (2018). Interactions between Roseburia intestinalis and diet modulate atherogenesis in a murine model. Nat. Microbiol. 3, 1461–1471. doi: 10.1038/s41564-018-0272-x
Louis, P., Flint, H. J. (2009). Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 294, 1–8. doi: 10.1111/j.1574-6968.2009.01514.x
Nyberg, T., Ferguson, N. M., Nash, S. G., Webster, H. H., Flaxman, S., Andrews, N., et al. (2022). Comparative analysis of the risks of hospitalisation and death associated with SARS-CoV-2 omicron (B.1.1.529) and delta (B.1.617.2) variants in England: a cohort study. Lancet 399, 1303–1312. doi: 10.1016/S0140-6736(22)00462-7
Pasolli, E., Schiffer, L., Manghi, P., Renson, A., Obenchain, V., Truong, D. T., et al. (2017). Accessible, curated metagenomic data through ExperimentHub. Nat. Methods 14, 1023–1024. doi: 10.1038/nmeth.4468
Pittayanon, R., Lau, J. T., Leontiadis, G. I., Tse, F., Yuan, Y., Surette, M., et al. (2020). Differences in gut microbiota in patients with vs without inflammatory bowel diseases: a systematic review. Gastroenterology 158, 930–946 e931. doi: 10.1053/j.gastro.2019.11.294
Sencio, V., Barthelemy, A., Tavares, L. P., Machado, M. G., Soulard, D., Cuinat, C., et al. (2020). Gut Dysbiosis during Influenza Contributes to Pulmonary Pneumococcal Superinfection through Altered Short-Chain Fatty Acid Production. Cell Rep. 30, 2934–2947 e2936. doi: 10.1016/j.celrep.2020.02.013
Shen, Z., Luo, W., Tan, B., Nie, K., Deng, M., Wu, S., et al. (2022). Roseburia intestinalis stimulates TLR5-dependent intestinal immunity against Crohn’s disease. EBioMedicine 85, 104285. doi: 10.1016/j.ebiom.2022.104285
Su, Q., Liu, Q., Lau, R. I., Zhang, J., Xu, Z., Yeoh, Y. K., et al. (2022). Faecal microbiome-based machine learning for multi-class disease diagnosis. Nat. Commun. 13, 6818. doi: 10.1038/s41467-022-34405-3
Verdecchia, P., Angeli, F., Cavallini, C., Reboldi, G. (2020). Use of antibiotics and mortality in women: does duration of exposure matter? Circ. Res. 126, 374–376. doi: 10.1161/CIRCRESAHA.119.316406
Vermeiren, J., Van den Abbeele, P., Laukens, D., Vigsnaes, L. K., De Vos, M., Boon, N., et al. (2012). Decreased colonization of fecal Clostridium coccoides/Eubacterium rectale species from ulcerative colitis patients in an in vitro dynamic gut model with mucin environment. FEMS Microbiol. Ecol. 79, 685–696. doi: 10.1111/j.1574-6941.2011.01252.x
Yeoh, Y. K., Zuo, T., Lui, G. C., Zhang, F., Liu, Q., Li, A. Y., et al. (2021). Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut 70, 698–706. doi: 10.1136/gutjnl-2020-323020
Zhang, F., Wan, Y., Zuo, T., Yeoh, Y. K., Liu, Q., Zhang, L., et al. (2022). Prolonged impairment of short-chain fatty acid and L-isoleucine biosynthesis in gut microbiome in patients with COVID-19. Gastroenterology 162, 548–561 e544. doi: 10.1053/j.gastro.2021.10.013
Zhang, F., Lau, R. I., Liu, Q., Su, Q., Chan, F. K. L., Ng, S. C. (2023). Gut microbiota in COVID-19: key microbial changes, potential mechanisms and clinical applications. Nat. Rev. Gastroenterol. Hepatol. 20, 323–337. doi: 10.1038/s41575-022-00698-4
Zhu, Q., Gao, R., Zhang, Y., Pan, D., Zhu, Y., Zhang, X., et al. (2018). Dysbiosis signatures of gut microbiota in coronary artery disease. Physiol. Genomics 50, 893–903. doi: 10.1152/physiolgenomics.00070.2018
Keywords: gut microbiome, Eubacterium rectale, COVID-19, mortality, population based
Citation: Liu Y, Chan MTV, Chan FKL, Wu WKK, Ng SC and Zhang L (2023) Lower gut abundance of Eubacterium rectale is linked to COVID-19 mortality. Front. Cell. Infect. Microbiol. 13:1249069. doi: 10.3389/fcimb.2023.1249069
Received: 28 June 2023; Accepted: 18 August 2023;
Published: 06 September 2023.
Edited by:
Kundlik Gadhave, Johns Hopkins University, United StatesReviewed by:
Ramhari Kumbhar, Johns Hopkins Medicine, United StatesCopyright © 2023 Liu, Chan, Chan, Wu, Ng and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: William K. K. Wu, d3VrYWtlaUBjdWhrLmVkdS5oaw==; Siew C. Ng, c2lld2NoaWVubmdAY3Voay5lZHUuaGs=; Lin Zhang, bGluemhhbmdAY3Voay5lZHUuaGs=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.