
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 21 August 2023
Sec. Antibiotic Resistance and New Antimicrobial drugs
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1244511
This article is part of the Research Topic Evolution, Molecular Mechanisms and the Strategies to Combat Antimicrobial Resistance (AMR): A One Health Approach View all 21 articles
 Yili Chen1†
Yili Chen1† Runshi Yang2†
Runshi Yang2† Penghao Guo1
Penghao Guo1 Pingjuan Liu1
Pingjuan Liu1 Jiankai Deng1
Jiankai Deng1 Zhongwen Wu1
Zhongwen Wu1 Qingping Wu2
Qingping Wu2 Junqi Huang1,3,4,5*
Junqi Huang1,3,4,5* Kang Liao1*
Kang Liao1*Background: The emergence of ceftazidime–avibactam (CZA) resistance among carbapenem-resistant Klebsiella pneumoniae (CRKP) is of major concern due to limited therapeutic options.
Methods: In this study, 10 CRKP strains were isolated from different samples of a patient with CRKP infection receiving CZA treatment. Whole-genome sequencing (WGS) and conjugation experiments were performed to determine the transferability of the carbapenem resistance gene.
Results: This infection began with a KPC-2-producing K. pneumoniae (CZA MIC = 2 μg/mL, imipenem MIC ≥ 16 μg/mL). After 20 days of CZA treatment, the strains switched to the amino acid substitution of T263A caused by a novel KPC-producing gene, blaKPC-145, which restored carbapenem susceptibility but showed CZA resistance (CZA MIC ≥ 256 μg/mL, imipenem MIC = 1 μg/mL). The blaKPC-145 gene was located on a 148,185-bp untransformable IncFII-type plasmid. The subsequent use of carbapenem against KPC-145-producing K. pneumoniae infection led to a reversion of KPC-2 production (CZA MIC = 2 μg/mL, imipenem MIC ≥ 16 μg/mL). WGS analysis showed that all isolates belonged to ST11-KL47, and the number of SNPs was 14. This implied that these blaKPC-positive K. pneumoniae isolates might originate from a single clone and have been colonized for a long time during the 120-day treatment period.
Conclusion: This is the first report of CZA resistance caused by blaKPC-145, which emerged during the treatment with CZA against blaKPC-2-positive K. pneumoniae-associated infection in China. These findings indicated that routine testing for antibiotic susceptibility and carbapenemase genotype is essential during CZA treatment.
With the extensive application of carbapenems, carbapenem-resistant Enterobacterales (CRE) infections have been a major threat to public healthcare, especially carbapenem-resistant Klebsiella pneumoniae (CRKP)-associated infections (Kim et al., 2017; Plazak et al., 2018). The China Antimicrobial Surveillance Network (CHINET) reported that there is an upward trend in carbapenem resistance in Klebsiella pneumoniae (Hu et al., 2019). CRE-related infections caused the deaths of 42.1% of patients with infections and 73% of severely infected patients who were in septic shock (Daikos et al., 2014; Xu et al., 2017). The production of Klebsiella pneumoniae carbapenemase (KPC), especially KPC-2, is dominant among CRKP ST11 strains in China (Yang et al., 2013; Zhang et al., 2017).
Ceftazidime–avibactam (CZA) has been approved as an effective antibiotic for CRE treatment. However, the emergency of CZA resistance rapidly appeared after its widespread application for the treatment of CRE infections (Shields et al., 2016). Some sporadic reports of CZA resistance were observed after the use of CZA for the treatment of KPC in China, such as in Shanghai (Shi et al., 2020) and Henan (Li et al., 2021a). Nevertheless, there are currently no reports of CZA developing resistance after use in southern China. In this study, we reported the dynamic evolution of CZA resistance due to a mutation between plasmid-borne blaKPC-2 and blaKPC-145 by sequencing clinical isolates from a patient with CRKP infection undergoing CZA treatment. To our knowledge, this is the first report on KPC-145 producing CRKP, which harbored a new amino acid substitution site on the basis of KPC-33.
Ten strains of K. pneumoniae were isolated from a 32-year-old man who was diagnosed with severe acute pancreatitis and multiple organ failure on admission to the First Affiliated Hospital of Sun Yat-sen University, Guangzhou. Empiric treatment with tigecycline, imipenem, and caspofungin was adopted, and surgical drainage was performed gradually from around the liver and around the pancreas. The patient developed septic shock 20 days after admission. The first K. pneumoniae strain (CZHKP-01) was obtained from a blood culture before CZA treatment. The third strain (CZHKP-03) was isolated from stool specimens. The remaining eight strains were isolated from the drainage fluid of the abdominal cavity. Species identification was performed by matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (MALDI-TOF MS) (bioMérieux, Marcy l’Etoile, France).
Antimicrobial susceptibilities for the strains were detected by Gram-negative susceptibility (GNS) cards on the Vitek-2 system (bioMérieux, Marcy l’Etoile, France). MICs of tigecycline and CZA were determined by broth microdilution method. Susceptibility testing results were interpreted under the criteria recommended by the Clinical and Laboratory Standards Institute (CLSI, 2022). The quality control strain for susceptibility testing was Escherichia coli ATCC 25922 and K. pneumoniae ATCC 700603.
The modified carbapenem inactivation method (mCIM) was carried out according to the recommendations of the CLSI (M100-S29). A carbapenemase inhibitor enhancement testing kit was purchased from Zhuhai Deere Bioengineering Co. Ltd. (Zhuhai, Guangdong, China), and the assay was performed as previously described (Tsakris et al., 2010).
Xpert Carba-R (Cepheid, Sunnyvale, CA, USA), NG-Test® CARBA 5 (NG Biotech, Guipry, France), and Goldstream Carbapenem-resistant K.N.I.V.O. Detection K-Set (Beijing Gold Mountain River Tech Development Co. Ltd., Beijing, China) were applied for rapid detection of 10 strains carbapenemase genotype according to the instructions. Targeted PCR was performed. The primers KPC1F (5′-GCTACACCTAGCTCCACCTTC-3′) and KPC1R (5′-GCATGGATTACCAACCACTGT-3′) were used to sequence the entire open reading frame (ORF) of the blaKPC gene (Smith Moland et al., 2003).
The transferability of the carbapenem resistance phenotype was determined by conjugation experiments (Lorenzo-Diaz and Espinosa, 2009) using the streptomycin-resistant E. coli C600 strain. Transconjugants were chosen on MacConkey agar plates (Huankai Co. Ltd., Guangzhou, China) supplemented with cefotaxime (1 mg/L) and streptomycin (1,500 mg/L).
The electrotransformation experiments (Choi et al., 2006) were further performed for isolates that failed conjugation experiments. In brief, plasmid DNA was extracted using a QIAPrep Plasmid Midi Kit (QIAGEN, Hilden, Germany) and transformed into electro-competent E. coli DH5α (TaKaRa Biotechnology, Dalian, China). electrotransformants were selected on LB agar plates (Huankai Co. Ltd., Guangzhou, China) supplemented with cefotaxime (1 mg/L).
We further confirmed the transconjugants or electrotransformants by PCR for the blaKPC-2 gene and tested for antimicrobial susceptibility as described above.
Plasmid DNA was extracted from parent strains, and primers targeting the entire ORF of the blaKPC-2 gene and blaKPC-145 gene, along with NcoI and BamHI restriction sites, were designed and used for PCR. The PCR products and the plasmid vector pET28a were digested with the restriction enzymes NcoI–BamHI (TaKaRa Biotechnology, Dalian, China) and then ligated at 16°C overnight. The recombinant plasmids were transformed into E. coli BL21 (DE3); all the transformants were confirmed by PCR and sequencing analysis. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was conducted to verify the expression of the target proteins KPC-2 and KPC-145. Antimicrobial susceptibility testing for the BL21 transformants was performed as mentioned above.
Genomic DNA of the selected isolates was extracted using a DNA extraction kit (Magen, Guangzhou, China), and 150 bp paired-end reads were obtained using an Illumina NextSeq 550 system (Illumina, San Diego, CA, USA). For each whole-genome sequencing (WGS) data, at least 100-fold coverage of original reads was obtained, and the assembly draft of the sequences was generated by SPAdes (Bankevich et al., 2012). Furthermore, a representative strain (CZHKP-07) was selected for further sequencing by using the MinION platform (Oxford Nanopore Technologies, Oxford, UK).
A hybrid assembly of both short Illumina NextSeq reads and long MinION reads was built using Unicycler with the Pilon option to modify the assembled readings (Walker et al., 2014; Wick et al., 2017) and annotated with RAST (Aziz et al., 2008) and Prokka (Seemann, 2014) for functionality. The PCR-based replicon typing (PBRT) and replicon sequencing typing (RST) methods were applied to characterize IncF plasmids, as described previously (Carattoli et al., 2005; Villa et al., 2010). Prophage analysis was conducted by PHAST (http://phaster.ca/). Multilocus sequence types (MLST) and antibiotic resistance genes (ARGs) were analyzed using ABRicate (https://github.com/tseemann/abricate). Capsule serotype and virulence genes (yersiniabactin, colibactin, and other siderophore locus) were determined using Kleborate (Lam et al., 2021). Core genome alignment and calling of single nucleotide polymorphisms (SNPs) were carried out using the Snippy (https://github.com/tseemann/snippy) against reference strain CZHKP-07 and followed by purging using Gubbins (Croucher et al., 2015). The heatmap showing the SNP matrix was generated by using the “pheatmap” package in RStudio; core-genome SNPs were used to construct a maximum-likelihood phylogenetic tree by RAxML (Stamatakis, 2014) and visualized by iTOL (Letunic and Bork, 2016).
The sequence of blaKPC-145 has been deposited in GenBank under Accession No. OP626310.
In our case, after initial combination treatment with imipenem (2.0 g, IV, Q8h) and tigecycline (first dose of 100 mg, then 50 mg, IV DRIP, Q12h) for 20 days, blaKPC-2-positive K. pneumoniae strains (CZHKP-01 and CZHKP-02) were isolated from the blood culture and drainage fluid, showing resistance to imipenem and meropenem (Table 1). Subsequently, antibiotic therapy was adjusted to combination with CZA (2.5 g, IV DRIP, Q8h), polymyxin B (first dose of 1,000,000 U, then 500,000 U, IV, Q12h), tigecycline, and caspofungin, along with adequate surgical drainage (from perihepatic, lesser omental sac, right peripancreatic, and cystic duct). However, after using CZA for 20 days, the patient’s body temperature rose again. The first CZA resistance K. pneumoniae strain CZHKP-03 was isolated from fecal culture followed by CZHKP-05~CZHKP-09 (all isolated from abdominal drainage fluid). These strains restored carbapenem susceptibility but showed CZA resistance (CZA MIC ≥ 256 μg/mL, imipenem MIC = 1 μg/mL). Considering that long-term use of CZA may lead to drug-resistant strains, we stopped using CZA and switched to polymyxin B and tigecycline for anti-infection treatment.
On the 105th day of hospitalization, the anti-infection therapy was adjusted to tigecycline combined with meropenem. After over a week of treatment, the heat peak of the patient’s body temperature decreased and the drainage fluid decreased gradually. On the 113th day of hospitalization, CZHKP-10 isolated from abdominal drainage fluid culture recovered susceptibility to CZA (CZA MIC = 2 μg/mL, imipenem MIC ≥ 16 μg/mL). Through sufficient surgical drainage and reasonable anti-infection treatment, the patient’s condition ultimately improved and returned to the regular ward for further treatment on the 120th day of hospitalization. Clinical and microbiological details, timelines, and antibiotic therapies used are summarized in Figure 1.
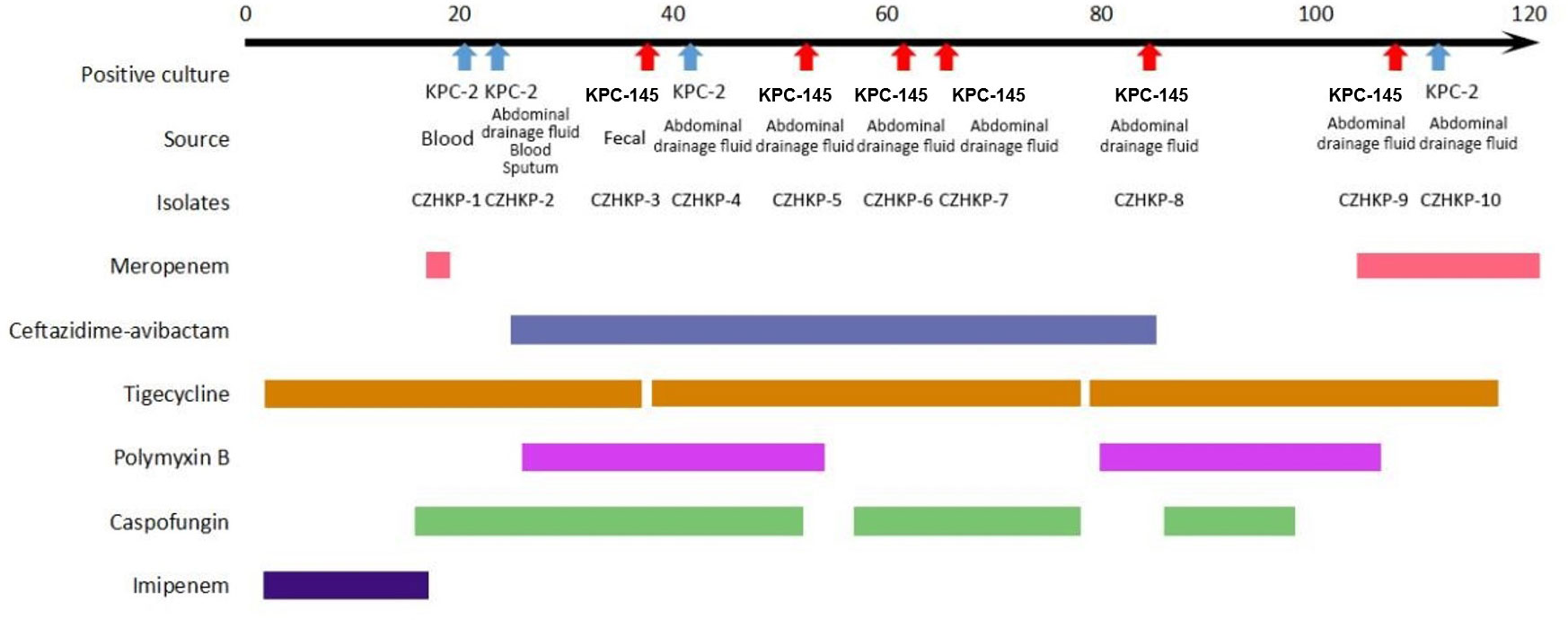
Figure 1 History of the K. pneumoniae isolations and the clinical antimicrobial treatment course of a patient with KPC-Kp infection.
The performance comparison of different carbapenem enzyme detection methods among the 10 strains is shown in Table 1. Sanger sequencing results of the blaKPC gene revealed that four CZA-susceptible isolates (CZHKP-01, CZHKP-02, CZHKP-04, CZHKP-10) carried the blaKPC-2 gene. Interestingly, the rest of the six CZA-resistant isolates (CZHKP-03, CZHKP-05, CZHKP-06, CZHKP-07, CZHKP-08, and CZHKP-09) harbored a novel blaKPC gene with a point mutation (A to G) at nucleotide position 787 compared with the blaKPC-33 gene; this mutation resulted in the amino acid substitution of threonine to alanine (T263A), assigned by GenBank as blaKPC-145.
In addition, the results of mCIM, carbapenemase inhibitor enhancement testing, and NG-Test CARBA 5 showed negative results for blaKPC-145-positive strains, which indicated that routine clinical detections have some defects on blaKPC-145-positive strains. Another immunochromatographic method, Goldstream Carbapenem-resistant K.N.I.V.O. Detection K-Set, which showed positive results in detecting blaKPC-145 strains, seems to have a better cost-to-benefit ratio. Moreover, molecular screening of GeneXpert for blaKPC-145 showed good sensitivity.
Unfortunately, despite numerous attempts conducted by conjugation assays, the transfer of blaKPC-2 and blaKPC-145 genes failed in all 10 K. pneumoniae isolates. Electrotransformations were further performed, and 10 electrotransformants were successfully obtained. All of the electrotransformants exhibited resistance to all β-lactams, aminoglycosides, and fosfomycin.
The recombinant plasmid carrying the blaKPC-145 gene in E. coli BL21, which conferred resistance to ceftazidime/avibactam (MIC = 2 mg/L) and displayed decreased carbapenem MICs (imipenem MIC = 0.125 mg/L and meropenem MIC = 0.25 mg/L) compared with blaKPC-2 in BL21. Moreover, the MICs of cefotaxime, ceftazidime, and aztreonam were 32-, 16-, and 16-fold higher than the original recipient E. coli BL21. However, compared with blaKPC-2, the MICs of blaKPC-145-carrying electrotransformants to carbapenems were 32- to 128-fold lower than those of blaKPC-2-carrying transformants.
A total of 10 KPC-producing strains were subjected to WGS. The in silico analysis showed that these 10 strains belonged to ST11-KL47 (associated with the wzi allele 209) and harbored 11 resistance genes, conferring resistance to aminoglycoside (aac3-IId, rmtB), fosfomycin (fosA3), quinolone (qnrS1), tetracycline (tetA), β-lactam (blaKPC-2 or blaKPC-145, blaCTX-M-64, blaTEM-1B, blaSHV-12), and trimethoprim/sulfamethoxazole (sulI, dfrA1). Furthermore, the representative K. pneumoniae strain (CZHKP-07) was sequenced using the MinION platform, followed by hybrid assembly. The CZHKP-07 chromosome is 5,448,939 bp in length, with a GC content of 57.44%, and harbors five plasmids with sizes of 230,198 bp (IncFIB and IncFII), 148,185 bp (IncFII), 55,161 bp, 11,970 bp (ColRNAI) and 5,596 bp (ColRNAI), named pCZHKP07-1 to pCZHKP07-5 (Supplementary Table S1). The pCZHKP07-2 was an F33:A−:B− plasmid and contained six resistance genes, including blaKPC-145. Meanwhile, the plasmid pCZHKP07-2 possessed addiction systems (pemKI, hok-sok-mok, ccdAB); these genes promote plasmid maintenance during vertical transmission.
A linear sequence comparison of the plasmids pCZHKP07-2, pHN7A8 (Accession No. JN232517), and p1068-KPC (Accession No. MF168402) was performed. The genetic structure suggested that pCZHKP07-2 possessed a similar backbone structure as F33:A−:B− plasmid pHN7A8, but a large segment of the conjugative transfer-associated genes (tra and trb) was absent (Figure 2A), which might be the reason for conjugation failure. Therefore, we speculate that the insertion of an extra copy of IS26 at target site TAACTGAA on the traI gene in the pHN7N8 plasmid, followed by homologous recombination between IS26 elements, may have led to the loss of the conjugative transfer regions. Meanwhile, two variable regions and a prophage region were embedded in the pCZHKP07-2 (Figure 2A). The first variable region (VR1) was 15,338 bp in length and was flanked at both ends by fragments of IS1 found in pCZHKP07-2, which consists of four different resistance genes, blaCTX-M-65, fosA3, blaTEM-1B, and rmtB, and abundant complete or truncated transposons. This structure was similar to the pHN7A8 except for the fosA3 resistance module with an opposite orientation, which was flanked by two intact copies of IS26 at both ends (Figure 2B). The second variable region (VR2) possessed blaKPC-145 and blaSHV modules. The blaKPC-145 and blaSHV regions in pCZHKP07-2 were disrupted into two separate parts, which may be caused by homologous recombination after IS26 insertion. The genetic structure of blaKPC-145 was composed of Tn1722-based unit transposon including ISKpn27-blaKPC-145-ISKpn6-korC-klcA-ΔrepB, and the blaSHV module was flanked by two intact or truncated IS26, resulting the differed from p1068-KPC with the inversion of this segment (Figure 2C). Moreover, the prophage region (30,305 bp) of pCZHKP07-2, flanked by 8 bp ATCCTCCT direct repeats (DRs), was inserted into the plasmid pIR12183_unnamed1 ATPase gene and bound at both ends by IS26. A maximum likelihood phylogenetic tree and SNP matrix were constructed based on core-genome SNPs (cgSNPs) from the 10 genomes in this study. We identified four KPC-2-positive K. pneumoniae and six KPC-145-positive K. pneumoniae that were separated into two clades and shared 14 SNPs in total (Figure 3).
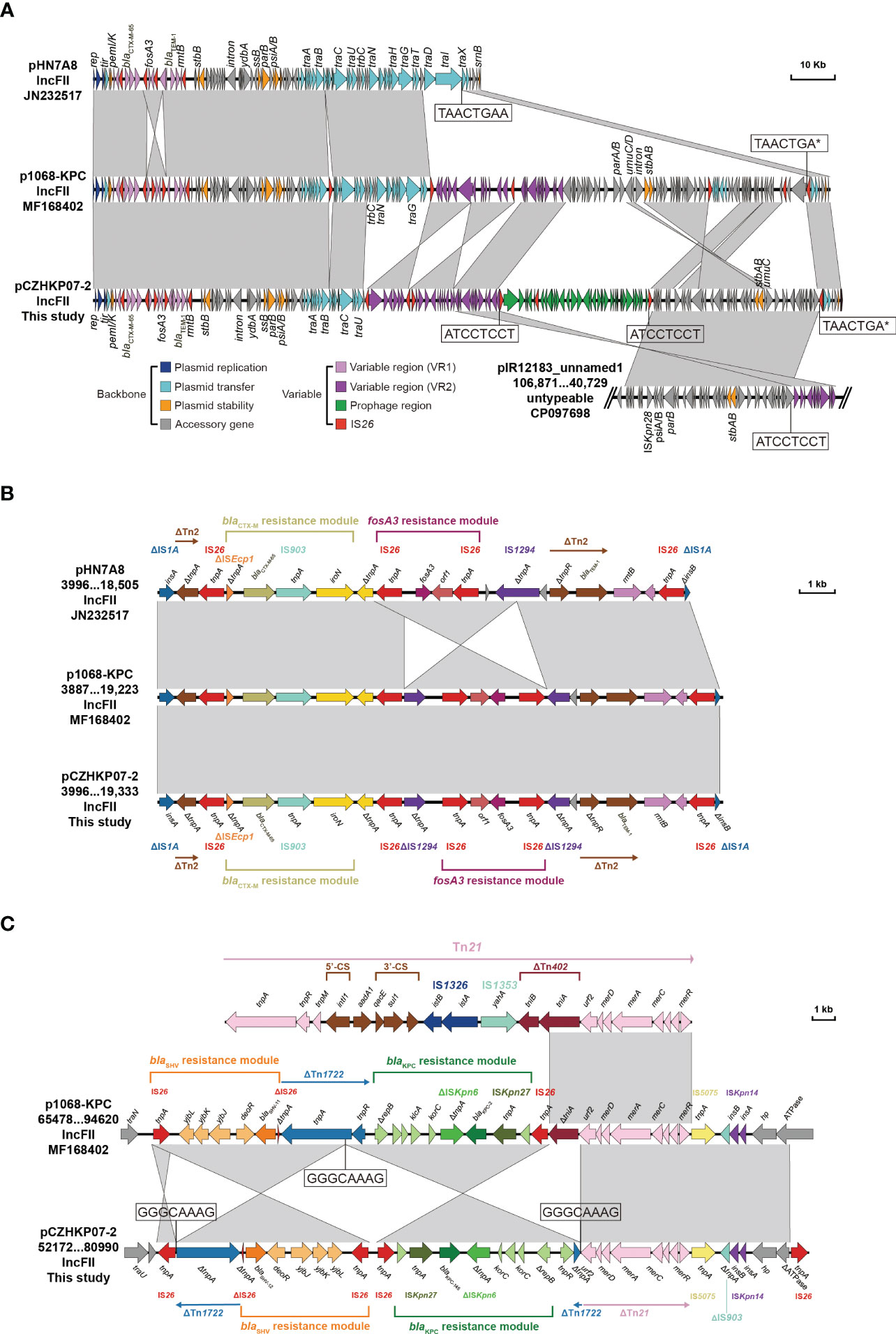
Figure 2 Linear comparison of the KPC-145-positive plasmid pCZHKP07-2 identified in this study with plasmid pHN7A8, p1068-KPC, and pIR12183_unnamed1 (A) and two variable regions (B, C). The arrows indicate the positions and transcriptional directions of the ORFs, while homologous regions are represented in gray.
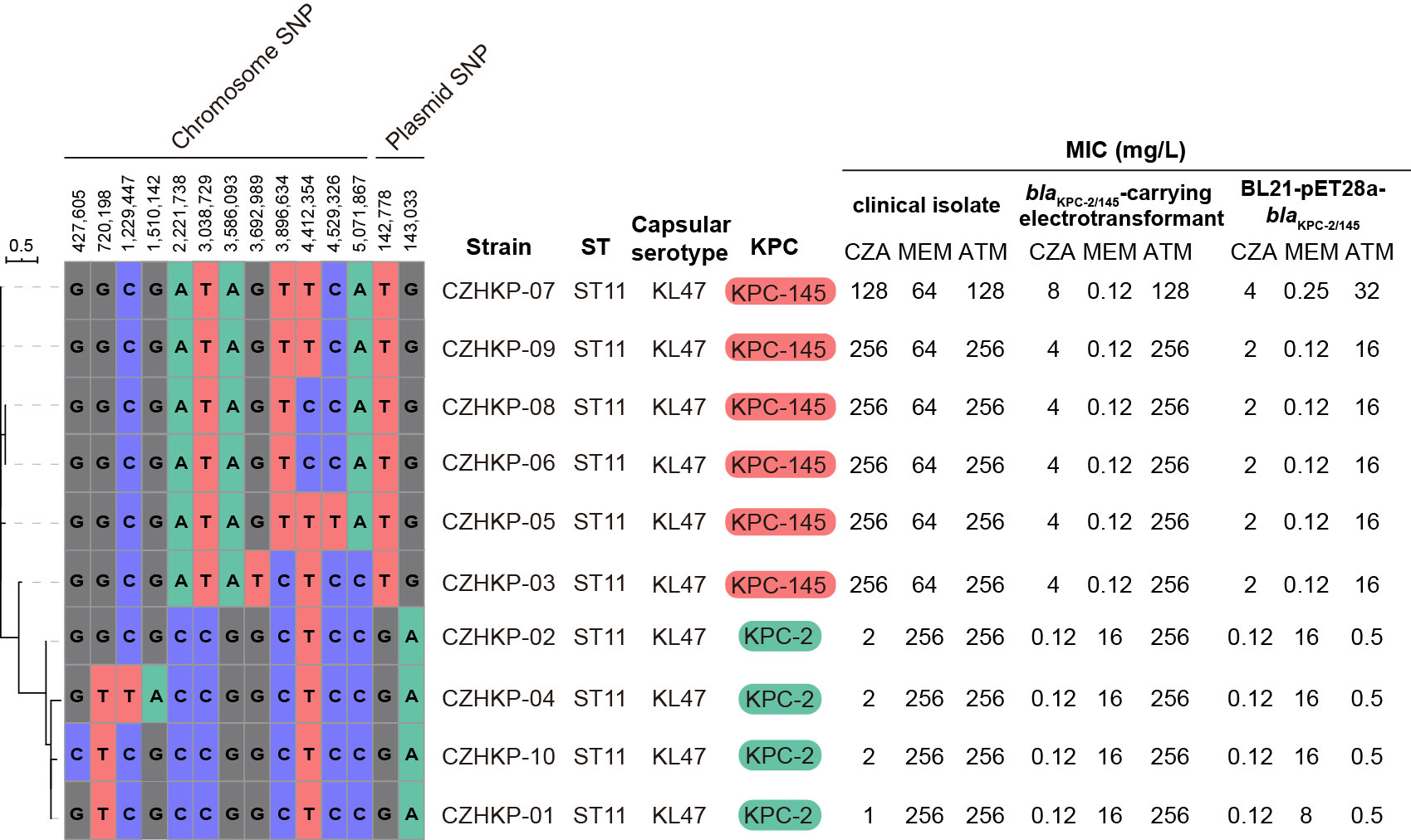
Figure 3 The maximum likelihood phylogenetic tree generated from the core-genome sequences of the 10 KPC-positive strains identified in this study. Each isolate is listed with its ST, capsular serotype, blaKPC allele, and antimicrobial susceptibility results.
KPC-2-producing strains, especially those that belong to ST11, are the majority population of K. pneumoniae resistant to carbapenems and become one of the most urgent public health issues in China (Li et al., 2021b). Avibactam is a β-lactamase inhibitor with a wide range of therapeutic effects against serine β-lactamases, including KPC. Combined with ceftazidime, CZA has become an important therapeutic option for treating KPC-producing K. pneumoniae infections (Tumbarello et al., 2021). Unfortunately, the development of CZA resistance has increased since it was clinically approved. In the International Network for Optimal Resistance Monitor (INFORM) surveillance program (2015–2017), the susceptibility rate of CZA against CRE was 73.0% (Spiliopoulou et al., 2020). A recent study from the CHINET revealed that CZA exhibited potent activity against Enterobacterales (93.6% susceptible), while only 65.2% of CRE remained susceptible to CZA (Guo et al., 2022).
In the present study, the first CZA resistance strain was isolated after treatment with CZA for 20 days, consistent with the previous reports that CZA resistance commonly appears after 10 to 19 days and sometimes after 33 days (Shi et al., 2020). Our study also showed that the blaKPC-145 gene-mediated CZA resistance was accompanied by a substantial decrease in carbapenem MICs. Moreover, when the therapy was subsequently adjusted to meropenem, it took only 4 days for the strains to change from blaKPC-145 back to blaKPC-2. It was observed that the in vivo evolution of wild-type KPC-2 changed into KPC-145 and then reversed to its original wild-type KPC-2. Alternatively, the combination of CZA and carbapenems may be an ideal option, which has been proven to be effective in some clinical cases (Bianco et al., 2020; Shi et al., 2020; Li et al., 2021a). In addition, meropenem–vaboractam has been receiving increasing attention as a potential antibiotic for clinics, and there are no K. pneumoniae-harboring mutant blaKPC resistant to meropenem–vaborbactam (Wilson et al., 2019).
So far, there are three principal CZA resistance mechanisms (Galani et al., 2021), including the production of metallo-β-lactamases, increased expression of KPC carbapenemases with porin mutations, and amino acid changes of KPC carbapenemases. Among them, the primary CZA resistance mechanism is a single mutation within the blaKPC gene allele, which is commonly observed in several pivotal regions, especially the omega loop (amino acid positions 164–179), the 240-loop (Galani et al., 2021) (aa 238–243) and 270-loop (aa 263–277) (Venditti et al., 2021). The novel variant, KPC-145, with two amino acid substitutions (D179Y and T263A), was identified in our study. These two amino acid substitutions were observed in KPC-3, named KPC-70 (Carattoli et al., 2021). Our results revealed that KPC-145 mediated the CZA resistance and restored susceptibility to carbapenems, similar to KPC-70 in the previous report. The D179Y mutation within the omega loop may be the main mechanism for resistance change for the KPC-145 enzyme, which causes enhanced affinity to ceftazidime and restricts avibactam binding (Winkler et al., 2015). In addition, it is noteworthy that the number of KPC variants has been increasing rapidly in recent years; this situation poses a significant threat to public health.
Therefore, rapid and accurate detection of KPC-subtype-producing strains plays an important role in clinical drug selection and patient prognosis. However, in our study, the results of mCIM, carbapenemase inhibitor enhancement testing, and NG-Test® CARBA 5 showed negative results for blaKPC-145-positive strains, which indicated deficiencies in routine clinical testing for blaKPC-145-positive strains. Both K.N.I.V.O. Detection K-Set and GeneXpert showed satisfactory diagnostic performance in detecting blaKPC-145, which was consistent with other studies on the detection ability of KPC-2 variants (Ding et al., 2021).
The dissemination of resistance genes is also associated with sequence types and plasmid replicon types. Since the emergence of KPC-producing K. pneumoniae in 2001 (Yigit et al., 2001), ST11 has become the most prevalent CRKP clone in Asia, especially in China. In this study, we found that all isolates belonged to ST11-KL47, and the number of SNPs was 14. This implies that these blaKPC-positive K. pneumoniae isolates might originate from a single clone and long-term colonization in this patient during the 120-day therapy. Meanwhile, CZA resistance encoded by blaKPC variants has been increasingly reported in ST11 K. pneumoniae in China, including blaKPC-33 (Shi et al., 2020), blaKPC-51, blaKPC-52 (Sun et al., 2020), blaKPC-74 (Li et al., 2021b), and blaKPC-93 (Wu et al., 2022). Otherwise, ST11 is a single-locus variant of the globally popular high-risk clone ST258 through the acquisition of the tonB allele (Breurec et al., 2013). These two sequence types together with ST512 and a few other SNP variants comprising the clonal group CG258, responsible for the global dissemination of CRKP. This underscores the importance of coordinated international action to surveillance and control the high-risk clone type. IncF is a narrow host plasmid that is widely found in Enterobacteriaceae. Numerous studies have shown that the IncF subtype IncFII plasmid carries a variety of ARGs and plays a significant role in the transmission of specific resistance genes, including the blaKPC gene (Shi et al., 2018). In our study, the untransferable IncFII plasmid carried by K. pneumoniae encodes the resistance genes blaKPC-145, blaTEM-1B, blaSHV-12, blaCTX-M-64, fosA3, and rmtB, which are associated with CZA, cephalosporins, fosfomycin, and aminoglycosides, suggesting that IncFII plasmid contribute to the transmission of resistant strains and ARGs. The mobile genetic elements were widely distributed in the bacterial genomes and played a crucial role in the dissemination of antibiotic resistance (Partridge et al., 2018). The diverse structure of variable regions may be caused by a series of molecular module rearrangements, acquisitions, or loss events mediated by insertion sequences such as IS26 by transposition or homologous recombination. It is also worth pointing out that IS26 not only mediated the inversion of the resistance modules but also caused the absence of large segments of the conjugative transfer region.
In this study, we reported the first case of KPC-145-producing ST11-KL47 K. pneumoniae strains conferring resistance to CZA after exposure to CZA for 20 days. This novel variant mediated CZA resistance and restored susceptibility to carbapenems, indicating that the detection and identification of mutant blaKPC isolates may be a key to antimicrobial management.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
YC, RY, QW, JH, and KL conceived and designed the experiments. YC, JD, and ZW collected clinical data. YC, RY, and PG performed laboratory work. RY performed whole-genome sequencing and bioinformatics analysis. YC, RY, and PL analyzed clinical and microbiological data. YC, RY, and PC prepared the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the Guangdong Natural Science Foundation - General Program. Yili Chen is a grant recipient of the project (2023A1515011252).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1244511/full#supplementary-material
Aziz, R. K., Bartels, D., Best, A. A., Dejongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9, 75. doi: 10.1186/1471-2164-9-75
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bianco, G., Boattini, M., Iannaccone, M., Cavallo, R., Costa, C. (2020). Bloodstream infection by two subpopulations of Klebsiella pneumoniae ST1685 carrying KPC-33 or KPC-14 following ceftazidime/avibactam treatment: considerations regarding acquired heteroresistance and choice of carbapenemase detection assay. J. Antimicrob. Chemother. 75, 3075–3076. doi: 10.1093/jac/dkaa283
Breurec, S., Guessennd, N., Timinouni, M., Le, T. A., Cao, V., Ngandjio, A., et al. (2013). Klebsiella pneumoniae resistant to third-generation cephalosporins in five African and two Vietnamese major towns: multiclonal population structure with two major international clonal groups, CG15 and CG258. Clin. Microbiol. Infect. 19, 349–355. doi: 10.1111/j.1469-0691.2012.03805.x
Carattoli, A., Arcari, G., Bibbolino, G., Sacco, F., Tomolillo, D., Di Lella, F. M., et al. (2021). Evolutionary Trajectories toward Ceftazidime-Avibactam Resistance in Klebsiella pneumoniae Clinical Isolates. Antimicrob. Agents Chemother. 65, e0057421. doi: 10.1128/AAC.00574-21
Carattoli, A., Bertini, A., Villa, L., Falbo, V., Hopkins, K. L., Threlfall, E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63, 219–228. doi: 10.1016/j.mimet.2005.03.018
Clinical and Laboratory Standards Institute. (2022). Performance standards for antimicrobial susceptibility testing, 32nd ed. Clinical and Laboratory Standards Institute, Wayne, PA.
Choi, K. H., Kumar, A., Schweizer, H. P. (2006). A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64, 391–397. doi: 10.1016/j.mimet.2005.06.001
Croucher, N. J., Page, A. J., Connor, T. R., Delaney, A. J., Keane, J. A., Bentley, S. D., et al. (2015). Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 43, e15. doi: 10.1093/nar/gku1196
Daikos, G. L., Tsaousi, S., Tzouvelekis, L. S., Anyfantis, I., Psichogiou, M., Argyropoulou, A., et al. (2014). Carbapenemase-producing Klebsiella pneumoniae bloodstream infections: lowering mortality by antibiotic combination schemes and the role of carbapenems. Antimicrob. Agents Chemother. 58, 2322–2328. doi: 10.1128/AAC.02166-13
Ding, L., Shi, Q., Han, R., Yin, D., Wu, S., Yang, Y., et al. (2021). Comparison of four carbapenemase detection methods for blaKPC-2 variants. Microbiol. Spectr. 9, e0095421. doi: 10.1128/Spectrum.00954-21
Galani, I., Karaiskos, I., Giamarellou, H. (2021). Multidrug-resistant Klebsiella pneumoniae: mechanisms of resistance including updated data for novel beta-lactam-beta-lactamase inhibitor combinations. Expert Rev. Anti Infect. Ther. 19, 1457–1468. doi: 10.1080/14787210.2021.1924674
Guo, Y., Han, R., Jiang, B., Ding, L., Yang, F., Zheng, B., et al. (2022). In Vitro Activity of New beta-Lactam-beta-Lactamase Inhibitor Combinations and Comparators against Clinical Isolates of Gram-Negative Bacilli: Results from the China Antimicrobial Surveillance Network (CHINET) in 2019. Microbiol. Spectr. 10, e0185422. doi: 10.1128/spectrum.01854-22
Hu, F., Guo, Y., Yang, Y., Zheng, Y., Wu, S., Jiang, X., et al. (2019). Resistance reported from China antimicrobial surveillance network (CHINET) in 2018. Eur. J. Clin. Microbiol. Infect. Dis. 38, 2275–2281. doi: 10.1007/s10096-019-03673-1
Kim, D., Ahn, J. Y., Lee, C. H., Jang, S. J., Lee, H., Yong, D., et al. (2017). Increasing resistance to extended-spectrum cephalosporins, fluoroquinolone, and carbapenem in gram-negative bacilli and the emergence of carbapenem non-susceptibility in klebsiella pneumoniae: analysis of Korean antimicrobial resistance monitoring system (KARMS) data from 2013 to 2015. Ann. Lab. Med. 37, 231–239. doi: 10.3343/alm.2017.37.3.231
Lam, M. M. C., Wick, R. R., Watts, S. C., Cerdeira, L. T., Wyres, K. L., Holt, K. E. (2021). A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 12, 4188. doi: 10.1038/s41467-021-24448-3
Letunic, I., Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. doi: 10.1093/nar/gkw290
Li, D., Li, K., Dong, H., Ren, D., Gong, D., Jiang, F., et al. (2021a). Ceftazidime-Avibactam Resistance in Klebsiella pneumoniae Sequence Type 11 Due to a Mutation in Plasmid-Borne bla kpc-2 to bla kpc-33, in Henan, China. Infect. Drug Resist. 14, 1725–1731. doi: 10.2147/IDR.S306095
Li, X., Quan, J., Ke, H., Wu, W., Feng, Y., Yu, Y., et al. (2021b). Emergence of a KPC variant conferring resistance to ceftazidime-avibactam in a widespread ST11 carbapenem-resistant klebsiella pneumoniae clone in China. Front. Microbiol. 12, 724272. doi: 10.3389/fmicb.2021.724272
Lorenzo-Diaz, F., Espinosa, M. (2009). Lagging-strand DNA replication origins are required for conjugal transfer of the promiscuous plasmid pMV158. J. Bacteriol 191, 720–727. doi: 10.1128/JB.01257-08
Partridge, S. R., Kwong, S. M., Firth, N., Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31(4):e00088-17. doi: 10.1128/CMR.00088-17
Plazak, M. E., Tamma, P. D., Heil, E. L. (2018). The antibiotic arms race: current and emerging therapy for Klebsiella pneumoniae carbapenemase (KPC) - producing bacteria. Expert Opin. Pharmacother. 19, 2019–2031. doi: 10.1080/14656566.2018.1538354
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shi, L., Feng, J., Zhan, Z., Zhao, Y., Zhou, H., Mao, H., et al. (2018). Comparative analysis of blaKPC-2- and rmtB-carrying IncFII-family pKPC-LK30/pHN7A8 hybrid plasmids from Klebsiella pneumoniae CG258 strains disseminated among multiple Chinese hospitals. Infect. Drug Resist. 11, 1783–1793. doi: 10.2147/IDR.S171953
Shi, Q., Yin, D., Han, R., Guo, Y., Zheng, Y., Wu, S., et al. (2020). Emergence and Recovery of Ceftazidime-avibactam Resistance in blaKPC-33-Harboring Klebsiella pneumoniae Sequence Type 11 Isolates in China. Clin. Infect. Dis. 71, S436–S439. doi: 10.1093/cid/ciaa1521
Shields, R. K., Potoski, B. A., Haidar, G., Hao, B., Doi, Y., Chen, L., et al. (2016). Clinical outcomes, drug toxicity, and emergence of ceftazidime-avibactam resistance among patients treated for carbapenem-resistant enterobacteriaceae infections. Clin. Infect. Dis. 63, 1615–1618. doi: 10.1093/cid/ciw636
Smith Moland, E., Hanson, N. D., Herrera, V. L., Black, J. A., Lockhart, T. J., Hossain, A., et al. (2003). Plasmid-mediated, carbapenem-hydrolysing beta-lactamase, KPC-2, in Klebsiella pneumoniae isolates. J. Antimicrob. Chemother. 51, 711–714. doi: 10.1093/jac/dkg124
Spiliopoulou, I., Kazmierczak, K., Stone, G. G. (2020). In vitro activity of ceftazidime/avibactam against isolates of carbapenem-non-susceptible Enterobacteriaceae collected during the INFORM global surveillance programme, (2015-17). J. Antimicrob. Chemother. 75, 384–391. doi: 10.1093/jac/dkz456
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Sun, L., Chen, W., Li, H., Li, L., Zou, X., Zhao, J., et al. (2020). Phenotypic and genotypic analysis of KPC-51 and KPC-52, two novel KPC-2 variants conferring resistance to ceftazidime/avibactam in the KPC-producing Klebsiella pneumoniae ST11 clone background. J. Antimicrobial Chemother. 75, 3072–3074. doi: 10.1093/jac/dkaa241
Tsakris, A., Poulou, A., Pournaras, S., Voulgari, E., Vrioni, G., Themeli-Digalaki, K., et al. (2010). A simple phenotypic method for the differentiation of metallo-beta-lactamases and class A KPC carbapenemases in Enterobacteriaceae clinical isolates. J. Antimicrob. Chemother. 65, 1664–1671. doi: 10.1093/jac/dkq210
Tumbarello, M., Raffaelli, F., Giannella, M., Mantengoli, E., Mularoni, A., Venditti, M., et al. (2021). Ceftazidime-Avibactam Use for Klebsiella pneumoniae Carbapenemase-Producing K. pneumoniae Infections: A Retrospective Observational Multicenter Study. Clin. Infect. Dis. 73, 1664–1676. doi: 10.1093/cid/ciab176
Venditti, C., Butera, O., Meledandri, M., Balice, M. P., Cocciolillo, G. C., Fontana, C., et al. (2021). Molecular analysis of clinical isolates of ceftazidime-avibactam-resistant Klebsiella pneumoniae. Clin. Microbiol. Infect. 27, 1040.e1041–1040 e1046. doi: 10.1016/j.cmi.2021.03.001
Villa, L., Garcia-Fernandez, A., Fortini, D., Carattoli, A. (2010). Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 65, 2518–2529. doi: 10.1093/jac/dkq347
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9, e112963. doi: 10.1371/journal.pone.0112963
Wick, R. R., Judd, L. M., Gorrie, C. L., Holt, K. E. (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13, e1005595. doi: 10.1371/journal.pcbi.1005595
Wilson, W. R., Kline, E. G., Jones, C. E., Morder, K. T., Mettus, R. T., Doi, Y., et al. (2019). Effects of KPC variant and porin genotype on the in vitro activity of meropenem-vaborbactam against carbapenem-resistant enterobacteriaceae. Antimicrob. Agents Chemother. 63(3):e02048-18. doi: 10.1128/AAC.02048-18
Winkler, M. L., Papp-Wallace, K. M., Bonomo, R. A. (2015). Activity of ceftazidime/avibactam against isogenic strains of Escherichia coli containing KPC and SHV beta-lactamases with single amino acid substitutions in the Omega-loop. J. Antimicrob. Chemother. 70, 2279–2286. doi: 10.1093/jac/dkv094
Wu, Y., Yang, X., Liu, C., Zhang, Y., Cheung, Y. C., Wai Chi Chan, E., et al. (2022). Identification of a KPC variant conferring resistance to ceftazidime-avibactam from ST11 carbapenem-resistant Klebsiella pneumoniae strains. Microbiol. Spectr. 10, e0265521. doi: 10.1128/spectrum.02655-21
Xu, L., Sun, X., Ma, X. (2017). Systematic review and meta-analysis of mortality of patients infected with carbapenem-resistant Klebsiella pneumoniae. Ann. Clin. Microbiol. Antimicrob. 16, 18. doi: 10.1186/s12941-017-0191-3
Yang, J., Ye, L., Guo, L., Zhao, Q., Chen, R., Luo, Y., et al. (2013). A nosocomial outbreak of KPC-2-producing Klebsiella pneumoniae in a Chinese hospital: dissemination of ST11 and emergence of ST37, ST392 and ST395. Clin. Microbiol. Infect. 19, E509–E515. doi: 10.1111/1469-0691.12275
Yigit, H., Queenan, A. M., Anderson, G. J., Domenech-Sanchez, A., Biddle, J. W., Steward, C. D., et al. (2001). Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45, 1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001
Keywords: carbapenem resistance, Klebsiella pneumoniae, ceftazidime-avibactam resistance, KPC-2, KPC-145
Citation: Chen Y, Yang R, Guo P, Liu P, Deng J, Wu Z, Wu Q, Huang J and Liao K (2023) Dynamic evolution of ceftazidime–avibactam resistance due to interchanges between blaKPC-2 and blaKPC-145 during treatment of Klebsiella pneumoniae infection. Front. Cell. Infect. Microbiol. 13:1244511. doi: 10.3389/fcimb.2023.1244511
Received: 22 June 2023; Accepted: 31 July 2023;
Published: 21 August 2023.
Edited by:
Ronni Mol Joji, Arabian Gulf University, BahrainCopyright © 2023 Chen, Yang, Guo, Liu, Deng, Wu, Wu, Huang and Liao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junqi Huang, aHVhbmdqcUBtYWlsLnN5c3UuZWR1LmNu; Kang Liao, bGlhb2thbmc4NDYxQDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.