
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Cell. Infect. Microbiol. , 30 August 2023
Sec. Antibiotic Resistance and New Antimicrobial drugs
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1241608
 Natasha Spottiswoode1*†
Natasha Spottiswoode1*† Samantha Hao2,3†
Samantha Hao2,3† Estella Sanchez-Guerrero3†
Estella Sanchez-Guerrero3† Angela M. Detweiler3
Angela M. Detweiler3 Honey Mekonen3
Honey Mekonen3 Norma Neff3
Norma Neff3 Henriette Macmillan1
Henriette Macmillan1 Brian S. Schwartz1
Brian S. Schwartz1 Joanne Engel1
Joanne Engel1 Joseph L. DeRisi3,4
Joseph L. DeRisi3,4 Steven A. Miller5,6
Steven A. Miller5,6 Charles R. Langelier1,3
Charles R. Langelier1,3Multidrug-resistant (MDR) Pseudomonas aeruginosa has been declared a serious threat by the United States Centers for Disease Control and Prevention. Here, we used whole genome sequencing (WGS) to investigate recurrent P. aeruginosa bloodstream infections in a severely immunocompromised patient. The infections demonstrated unusual, progressive increases in resistance to beta lactam antibiotics in the setting of active treatment with appropriate, guideline-directed agents. WGS followed by comparative genomic analysis of isolates collected over 44 days demonstrated in host evolution of a single P. aeruginosa isolate characterized by stepwise acquisition of two de-novo genetic resistance mechanisms over the course of treatment. We found a novel deletion affecting the ampC repressor ampD and neighboring gene ampE, which associated with initial cefepime treatment failure. This was followed by acquisition of a porin nonsense mutation, OprD, associated with resistance to carbapenems. This study highlights the potential for in-host evolution of P. aeruginosa during bloodstream infections in severely immunocompromised patients despite appropriate antimicrobial therapy. In addition, it demonstrates the utility of WGS for understanding unusual resistance patterns in the clinical context.
Multidrug-resistant (MDR) Pseudomonas aeruginosa is an increasing threat world-wide. To anticipate and treat MDR bacterial infections, an in-depth understanding of resistance acquisition in clinical settings is paramount. In this case, we used serial genomic analysis to study in-host evolution of a recurrent P. aeruginosa bloodstream infection in a severely immunocompromised patient. We found that infections were caused by the same strain of MDR P. aeruginosa which acquired de-novo resistance to increasingly broad spectrum, guideline-directed, anti-pseudomonal antibiotics through serial acquisition of a previously undescribed ampD deletion, and a porin mutation, respectively. This study demonstrates both the potential for in-host evolution of bacterial pathogens in immunocompromised hosts undergoing antimicrobial therapy, and highlights the utility of clinical WGS in understanding MDR organism epidemiology and in-host evolution.
Multidrug resistant (MDR) Pseudomonas aeruginosa, as defined by resistance to three or more of the major anti-pseudomonal antibiotic classes, is a global health threat of increasing importance (Magiorakos et al., 2012; CDC, 2019). P. aeruginosa can cause a multitude of infectious syndromes, including pneumonia, bloodstream infections, and wound infections. P. aeruginosa is also an infamous cause of nosocomial infections, because it can transmit effectively within the hospital environment, by contaminating surfaces such as sinks, ultrasound gel, endoscopes, or beds; it is secondary only to Acinetobacter baumanii (also designed a top threat by the CDC) in its ability to contaminate healthcare workers’ gowns and gloves (Morgan et al., 2012). Complementing this ability to transmit in-hospital, P. aeruginosa has a remarkable propensity to acquire resistance through multiple mechanisms, including alteration of porins, efflux pumps, acquisition of beta-lactamases such as ampC or extended-spectrum beta-lactamases (ESBL), carbapenemases, and aminoglycoside nucleotidyl-transferases. Recognizing this growing threat, the Infectious Disease Society recently released an updated dedicated guideline document recommending standardized treatment of treatment of MDR P. aeruginosa and other highly resistant gram-negative bacterial organisms (Tamma et al, 2023).
Key to combating MDR P. aeruginosa is a detailed understanding of its epidemiology and mechanisms of resistance. Patients may acquire MDR P. aeruginosa infection endogenously by in-host evolution of P. aeruginosa under antibiotic pressure, or exogenous infection with MDR P. aeruginosa from another patient, healthcare-workers, or the healthcare environment (Johnson et al., 2009). A study of the acquisition of imipenem-resistant P. aeruginosa in intensive care units between 2001-2006 noted that approximately 50% of imipenem-resistant P. aeruginosa cases were linked to in-patient evolution and 50% were directly acquired from another patient or the hospital environment (Morgan et al., 2012).
The use of whole-genome sequencing (WGS) as part of hospital epidemiology and outbreak investigations holds considerable promise. Our hospital routinely performs WGS to evaluate suspected healthcare transmission of microbes (Crawford et al., 2020), track newly emerging pathogens (Woodworth et al., 2019), and analyze cases of unusual antimicrobial resistance patterns (Hao et al., 2020a). Here, we performed WGS of serially collected P. aeruginosa isolates to study progressive acquisition of resistance despite guideline-directed antimicrobial treatment in an immunocompromised host. We found that in-host evolution rather than exogenous acquisition of an antimicrobial resistance gene was responsible, and involved a novel deletion in ampD and ampE, and a mutation in the porin gene OprD.
Patient blood cultures were performed via inoculation into BD Bactec Plus Aerobic and Lytic Anaerobic media (Becton Dickinson). Species identification was performed using matrix-assisted laser desorption/ionization time of-flight mass spectrometry (Bruker). Minimum inhibitory concentrations (MICs) were determined using the Trek Sensititre automated broth microdilution system and the Clinical and Laboratory Standards Institute (CLSI) breakpoints.
Investigations were carried out according to a no-subject contact study protocol approved by the University of California San Francisco Institutional Review Board, which permitted analysis of deidentified leftover clinical microbiology samples from collaborating institutions and subsequent review of study participants’ electronic medical records. No decisions regarding antibiotics or other patient-specific treatment interventions were made using sequencing data.
Whole genome sequencing was performed according to a previously described protocol (Crawford et al., 2020). Briefly, DNA from each sample was sheared with fragmentase (New England Biolabs) and used to construct sequencing libraries with the NEBNext Ultra II Library Prep Kit (New England Biolabs). Adaptor ligated samples underwent amplification with dual unique indexing primers. Libraries were quantified and pooled and underwent paired end 150 base pair sequencing on an Illumina MiSeq.
Phylogenetic analyses were performed according to established methods (Hao et al., 2020b). Briefly, short reads were adapter trimmed, quality filtered with fastp v0.20.0, and analyzed using the core single-nucleotide polymorphism (SNP) detection pipeline SPID v0.4.0 (https://github.com/czbiohub/Spid.jl). Genome assembly was performed using Unicycler v0.4.8 (Wick et al., 2017), and BLAST of the largest contig identified P. aeruginosa strain F9676 (Genbank number CP012066.1) as the most closely related strain to serve a reference genome (Chen et al., 2018). INDEL analysis was performed with Snippy (Seemann, 2018). Finally, the maximum likelihood phylogeny based on the SNP alignments was built in RAxML v8.2.12 (Stamatakis, 2014).
A 55-year-old woman with a history of B-cell lymphoma, allogeneic stem cell transplant complicated by graft versus host disease, and renal failure on intermittent hemodialysis, was admitted for sepsis secondary to pan-susceptible P. aeruginosa bacteremia (Figure 1). Direct time to positivity (DTTP) blood cultures drawn through her tunneled dialysis catheter (TDC) and peripherally inserted central catheter (PICC) did not implicate the lines as the source of infection. Both lines were removed on day five, after which her cultures cleared. A transthoracic echocardiogram demonstrated no evidence of endocarditis, and a transesophageal echocardiogram (TEE) demonstrated mitral valve thickening without vegetation. New PICC and TDC lines were placed after 48 hours, and she was discharged on two weeks of cefepime for treatment of bacteremia (Figure 1).
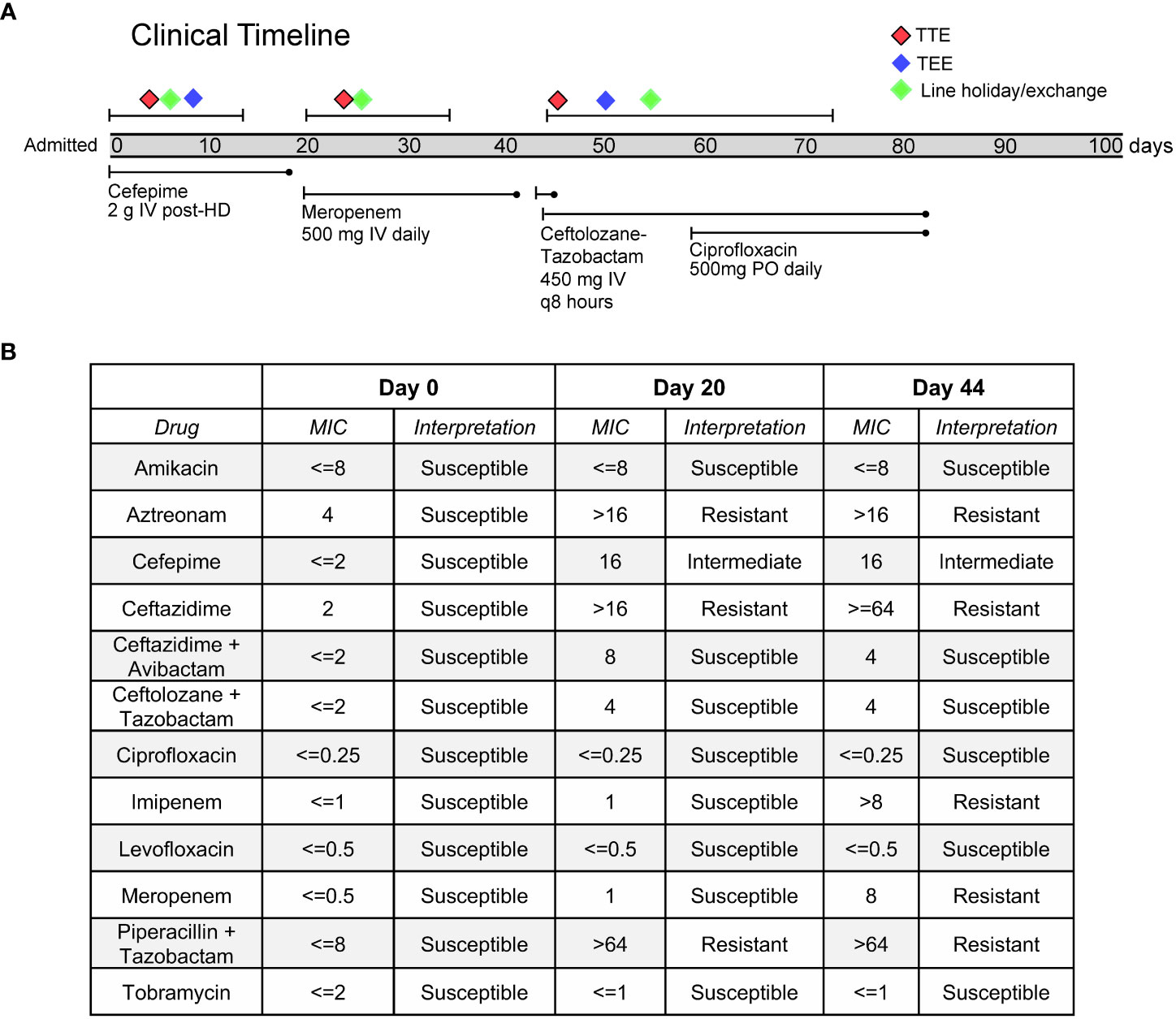
Figure 1 (A) Timeline of clinical case and evolution of antibiotic resistance in the Pseudomonas aeruginosa. (B) Phenotypic antimicrobial susceptibility testing of clinical blood culture isolates.
One day after completing her cefepime regimen, she developed fevers and re-presented to the hospital, where blood cultures drawn from her PICC, TDC, and peripheral blood again grew P. aeruginosa, but now with new intermediate resistance to cefepime, as well as resistance to aztreonam, piperacillin-tazobactam and ceftazidime. Even though DTTP blood cultures did not clearly implicate the TDC and PICC as sources of infection, both lines were again removed and later replaced, and she was prescribed a two-week course of meropenem starting from the date of culture clearance. Following clinical improvement, she was discharged home.
Two days after completion of her course of meropenem, the patient developed fevers, chills, and right upper quadrant pain. She re-presented to the hospital for a third time and was found to be growing P. aeruginosa from peripheral blood cultures, and the isolate was now additionally resistant to meropenem. She was started on ceftolozane-tazobactam. A repeat TEE was performed and now revealed a small mobile echodensity on the anterior leaflet of the mitral valve, thought to represent a possible vegetation. An abdominal CT scan demonstrated hepatic micro abscesses that were not amenable to drainage.
In an effort to determine whether her multiple episodes of P. aeruginosa bacteremia with progressively increased drug resistance were due to independent infection events or due to evolution of resistance within a single strain, Illumina whole-genome sequencing (WGS) was performed on DNA extracted from blood culture isolates obtained on days 0, 20 and 44, according to an established protocol (Crawford et al., 2020) (Figure 1). Phylogenetic analysis demonstrated that all isolates represented the same strain, differing by a maximum of 3 single nucleotide polymorphisms (SNPs) across a core shared genome of 6,368,008 nucleotides (Figure 2A). This confirmed that resistance had progressively evolved, and ruled-out de novo acquisition of independent infections. Prediction of multilocus sequence type (MLST) from WGS data demonstrated that the isolate was most closely related to sequence type 167, and thus differed from several well-known high-risk international strains (Jolley et al., 2018).
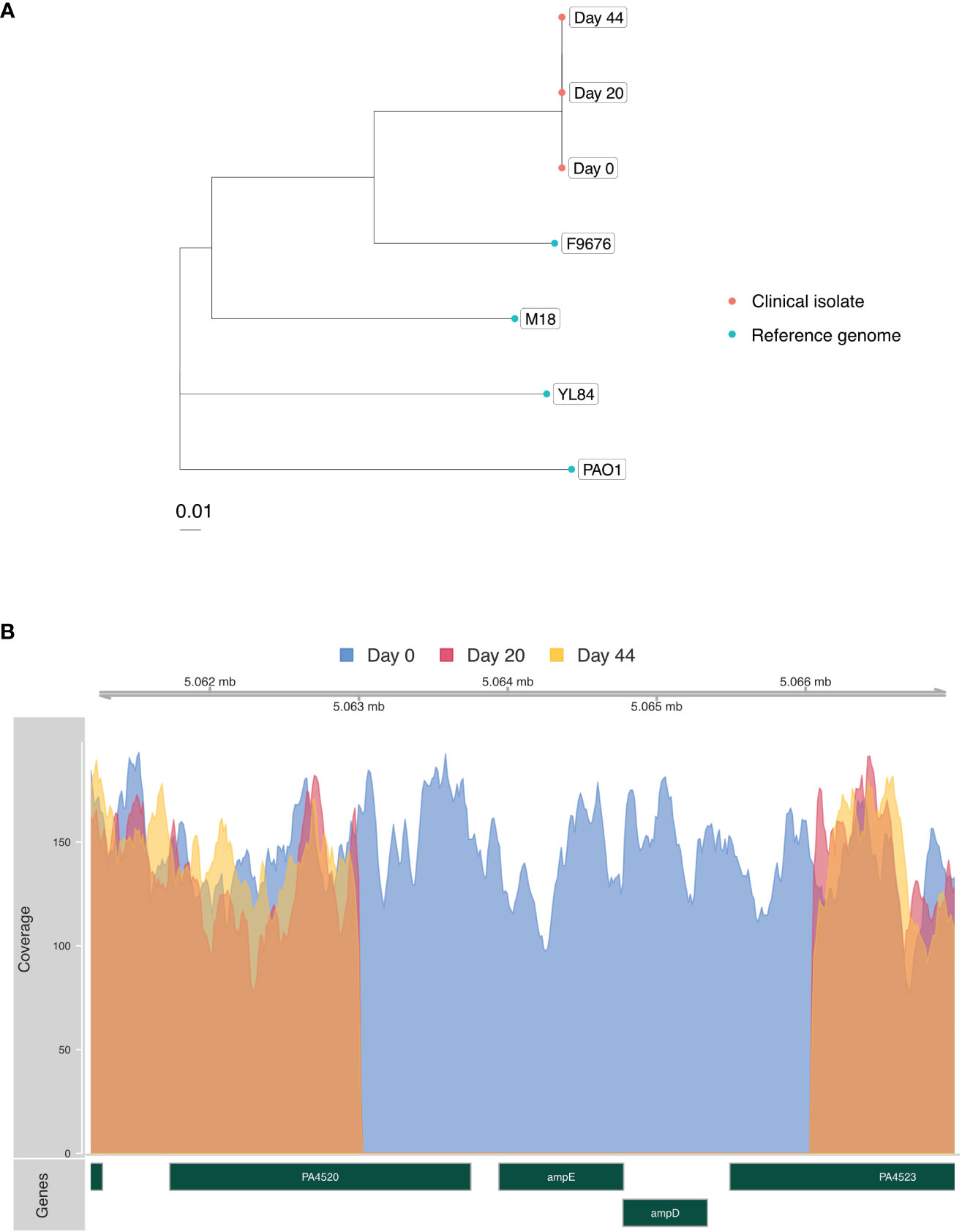
Figure 2 (A) Phylogenetic tree built using RAxML (Stamatakis, 2014) and the SNP Pipeline for Infectious Diseases (SPID) (Hao et al., 2020b) demonstrating genetic relatedness of the three P. aeruginosa isolates (red) from the patient in relation to reference genomes from NCBI Genbank (blue). Scale bar indicates substitutions per site. (B) Coverage plot of the P. aeruginosa genome assembled from clinical isolates highlighting a 3,794 base pair deletion involving the ampD and ampE genes present in the day-20 and day-44 isolates. Acquisition of this deletion, functionally predicted to de-repress the ampC beta lactamase gene, coincided with development of cefepime resistance and clinical treatment failure. rpm = P. aeruginosa reads per million reads sequenced.
Interrogation of the WGS data was then carried out to identify a molecular explanation for the observed resistance. Intriguingly, a 3,794 base pair deletion involving ampD and ampE was identified in isolates from days 20 and 44 (Figure 2B) that coincided with acquisition of new beta lactam resistance and cefepime clinical treatment failure. AmpD represses the expression of the ampC beta lactamase gene, and in vitro deletions in ampD have been associated with augmented expression of ampC and increased beta lactam resistance (Zamorano et al., 2010). Only one prior case of P. aeruginosa resistance attributed to an ampD mutation has been reported, and this involved an ampD missense mutation implicated in the rapid acquisition of resistance to multiple beta-lactam antibiotics (Barbosa et al., 2020).
In addition to the novel deletion, the blood culture isolate from day 44 was also found to contain a C748T nonsense mutation in OprD, paralleling the acquisition of carbapenem resistance during the patient’s secondary treatment regimen with meropenem. OprD encodes a 443-amino acid porin that forms a monomeric 18-stranded β barrel with 9 loops; it interacts with carbapenems and permits their entrance to the cell (Li et al., 2012; Jumper et al., 2021; Varadi et al., 2022). The mutation we identified, C748T, would be predicted to generate a truncated 249-amino acid protein that would most likely be non-functional. Prior work has associated other OprD mutations with carbapenem resistance (Marvig et al., 2013; Wheatley et al., 2021). Increased resistance to ceftazidime also characterized the day 44 isolate, which could potentially be due to OprD loss of function, which has been associated with increased resistance to ceftazidime in a large study of clinical isolates (Castanheira et al., 2014).
The patient was ultimately treated for endocarditis with four weeks of ceftolozane-tazobactam plus ciprofloxacin, according to clinical guidelines (Baddour et al., 2015). Following completion of therapy, the patient has not experienced any further recurrent episodes of bacteremia due to MDR P. aeruginosa.
MDR P. aeruginosa infections can arise in the hospital from direct human to human transmission, environmental acquisition, or from endogenous evolution in the setting of antibiotic selective pressure. While the latter appears to be the least common, it may account for as many as 19% of hospital-onset carbapenem-resistant P. aeruginosa infections (Johnson et al., 2009). The progressive acquisition of increasing resistance in the setting of appropriate anti-pseudomonal antibiotics, however, has not been widely reported. Reflecting this, guidelines recommend a non-carbapenem anti-pseudomonal agent as a first-line approach for susceptible isolates (Tamma et al., 2023).
In this case, initial imaging did not demonstrate discrete foci of infection amenable to source control, or an intravascular infection. It is possible that clinically occult endocarditis or an abdominal infection could have been present at the time of the patient’s initial hospitalization, but was too subtle to be confidently detected by echocardiography or CT scan. It is perhaps more likely that the patient’s underlying immunocompromised state impaired effective clearance of the pathogen, facilitating de-novo evolution of resistance. This phenomenon has been recently well described in the setting of SARS-CoV-2 respiratory infections in severely immunocompromised individuals (Scherer et al., 2022), but is not well described in the setting of bacteremia.
WGS is increasingly becoming a tool of choice for understanding the epidemiology and drug resistance mechanisms of MDR P. aeruginosa (Sundermann et al., 2021). While WGS is an established tool for outbreak investigation and pathogen surveillance (Stanton et al., 2022), its use to inform care for individual patient cases has been more limited. Several case studies have used WGS to identify the mechanisms underlying P. aeruginosa drug resistance in single patients (Alamri et al., 2020; Goldenberger et al., 2020; Hao et al., 2021). Few studies, however, have longitudinally assessed in host evolution using WGS, and those to date have focused exclusively on pulmonary infections (Datar et al., 2021; Wheatley et al., 2021; Rojas et al., 2022). An important recent study described the progressive acquisition, and loss, of carbapenem resistance during a prolonged P. aeruginosa pulmonary infection associated with the acquisition of mutations in oprD and other genes (Wheatley et al., 2021).
This study extends prior in vivo longitudinal genomic analyses of this pathogen to the setting of bacterial bloodstream infections. Furthermore, it highlights the potential clinical utility of WGS for understanding progressive and unexplained drug resistance during treatment, and for disambiguating hospital-acquired infections from cases of in-host evolution. Clinical deployment of WGS is not yet widely available, but may soon offer a sufficiently rapid turn-around time to directly impact clinical care.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA888413.
The studies involving humans were approved by University of California San Francisco Institutional Review Board. The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from a by- product of routine care or industry. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
NS, SH, and EG: Equal contributions. NS lead writing, clinical data review and integration. SH lead genomic and computational analyses. EG lead microbiology and whole genome sequencing. AD, HMe, NN, and HMa provided computational analysis and pipeline support. BS and JE advised clinical data review and microbiological interpretation. JLD, SM, and CL oversaw all aspects of study and writing. All authors contributed to the article and approved the submitted version.
This work was supported by the National Heart, Lung, and Blood Institute [grant number K23HL138461-01A1 to CL] and the Chan Zuckerberg Biohub, San Francisco, CA.
Author SM was employed by the company Illumina, Inc, and by the company Delve Bio, Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alamri, A. M., Alfifi, S., Aljehani, Y., Alnimr, A. (2020). Whole genome sequencing of ceftolozane-tazobactam and ceftazidime-avibactam resistant pseudomonas aeruginosa isolated from a blood stream infection reveals VEB and chromosomal metallo-beta lactamases as genetic determinants: A case report. Infect. Drug Resist. 13, 4215–4222. doi: 10.2147/IDR.S285293
Baddour, L. M., Wilson, W. R., Bayer, A. S., Fowler, V. G., Jr., Tleyjeh, I. M., Rybak, M. J., et al. (2015). Infective endocarditis in adults: diagnosis, antimicrobial therapy, and management of complications: A scientific statement for healthcare professionals from the american heart association. Circulation 132, 1435–1486. doi: 10.1161/CIR.0000000000000296
Barbosa, C., Gregg, K. S., Woods, R. J. (2020). Variants in ampD and dacB lead to in vivo resistance evolution of Pseudomonas aeruginosa within the central nervous system. J. Antimicrob. Chemother. 75, 3405–3408. doi: 10.1093/jac/dkaa324
Castanheira, M., Mills, J. C., Farrell, D. J., Jones, R. N. (2014). Mutation-driven beta-lactam resistance mechanisms among contemporary ceftazidime-nonsusceptible Pseudomonas aeruginosa isolates from U.S. hospitals. Antimicrob. Agents Chemother. 58, 6844–6850. doi: 10.1128/AAC.03681-14
CDC (2019). Antibiotic resistance threats in the unted states (Atlanta GA: U.S. Department of Health and Human Services). Available at: www.cdc.gov/DrugResistance/Biggest-Threats.html.
Chen, S., Zhou, Y., Chen, Y., Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Crawford, E., Kamm, J., Miller, S., Li, L. M., Caldera, S., Lyden, A., et al. (2020). Investigating transfusion-related sepsis using culture-independent metagenomic sequencing. Clin. Infect. Dis. 71, 1179–1185. doi: 10.1093/cid/ciz960
Datar, R., Coello Pelegrin, A., Orenga, S., Chalansonnet, V., MIrande, C., Dombrecht, J., et al. (2021). Phenotypic and genomic variability of serial peri-lung transplantation pseudomonas aeruginosa isolates from cystic fibrosis patients. Front. Microbiol. 12, 604555. doi: 10.3389/fmicb.2021.604555
Goldenberger, D., Vuichard-Gysin, D., Herzog, K., Hinic, V., Seth-Smith, H., Egli, A. (2020). First case of IMP-15-producing Pseudomonas aeruginosa in Switzerland: Challenging laboratory diagnosis using whole genome sequencing (WGS). IDCases 22, e00933. doi: 10.1016/j.idcr.2020.e00933
Hao, S., Abdelghany, M., Lyden, A., Sit, R., Tan, M., Tato, C. M., et al. (2020a). Genomic profiling of evolving daptomycin resistance in a patient with recurrent staphylococcus argenteus sepsis. Antimicrob. Agents Chemother. 64. doi: 10.1128/AAC.00961-20
Hao, M., Ma, W., Dong, X., Li, X., Cheng, F., Wang, Y. (2021). Comparative genome analysis of multidrug-resistant Pseudomonas aeruginosa JNQH-PA57, a clinically isolated mucoid strain with comprehensive carbapenem resistance mechanisms. BMC Microbiol. 21, 133. doi: 10.1186/s12866-021-02203-4
Hao, S., Veuthey, T., Caldera, S., Serpa, P. H., Haller, B., Tan, M., et al. (2020b). Draft genome sequence of an extensively drug-resistant salmonella enterica serovar typhi strain from a returned traveler from Pakistan. Microbiol. Resour Announc 9. doi: 10.1128/MRA.00427-20
Johnson, J. K., Smith, G., Lee, M. S., Venezia, R. A., Stine, O. C., Nataro, J. P., et al. (2009). The role of patient-to-patient transmission in the acquisition of imipenem-resistant Pseudomonas aeruginosa colonization in the intensive care unit. J. Infect. Dis. 200, 900–905. doi: 10.1086/605408
Jolley, K. A., Bray, J. E., Maiden, M. C. J. (2018). Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 3, 124. doi: 10.12688/wellcomeopenres.14826.1
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Li, H., Luo, Y. F., Williams, B. J., Blackwell, T. S., Xie, C. M. (2012). Structure and function of OprD protein in Pseudomonas aeruginosa: from antibiotic resistance to novel therapies. Int. J. Med. Microbiol. 302, 63–68. doi: 10.1016/j.ijmm.2011.10.001
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Marvig, R. L., Johansen, H. K., Molin, S., Jelsbak, L. (2013). Genome analysis of a transmissible lineage of pseudomonas aeruginosa reveals pathoadaptive mutations and distinct evolutionary paths of hypermutators. PloS Genet. 9, e1003741. doi: 10.1371/journal.pgen.1003741
Morgan, D. J., Rogawski, E., Thom, K. A., Johnson, J. K., Perencevich, E. N., Shardell, M., et al. (2012). Transfer of multidrug-resistant bacteria to healthcare workers' gloves and gowns after patient contact increases with environmental contamination. Crit. Care Med. 40, 1045–1051. doi: 10.1097/CCM.0b013e31823bc7c8
Rojas, L. J., Yasmin, M., Benjamino, J., Marshall, S. M., Deronde, K. J., Krishnan, N. P., et al. (2022). Genomic heterogeneity underlies multidrug resistance in Pseudomonas aeruginosa: A population-level analysis beyond susceptibility testing. PloS One 17, e0265129. doi: 10.1371/journal.pone.0265129
Scherer, E. M., Babiker, A., Adelman, M. W., Allman, B., Key, A., Kleinhenz, J. M., et al. (2022). SARS-coV-2 evolution and immune escape in immunocompromised patients. N Engl. J. Med. 386, 2436–2438. doi: 10.1056/NEJMc2202861
Seemann, T. (2018) Snippy. Available at: https://github.com/tseemann/snippy (Accessed 2023).
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stanton, R. A., Campbell, D., Mcallister, G. A., Breaker, E., Adamczyk, M., Daniels, J. B., et al. (2022). Whole-Genome Sequencing Reveals Diversity of Carbapenem-Resistant Pseudomonas aeruginosa Collected through CDC's Emerging Infections Program, United State-2018. Antimicrob. Agents Chemother. 66, e0049622. doi: 10.1128/aac.00496-22
Sundermann, A. J., Chen, J., Miller, J. K., Saul, M. I., Shutt, K. A., Griffith, M. P., et al. (2021). Outbreak of pseudomonas aeruginosa infections from a contaminated gastroscope detected by whole genome sequencing surveillance. Clin. Infect. Dis. 73, e638–e642. doi: 10.1093/cid/ciaa1887
Tamma, P. D., Aitken, S. L., Bonomo, R. A., Mathers, A. J., van Duin, D., Clancy, C. J. (2023) Infectious diseases society of america antimicrobial-resistant treatment guidance: gram-negative bacterial infections.Version 3.0. Available at: https://www.idsociety.org/practice-guideline/amr-guidance/ (Accessed July 17 2023).
Varadi, M., Anyango, S., Deshpande, M., Nair, S., Natassia, C., Yordanova, G., et al. (2022). AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444. doi: 10.1093/nar/gkab1061
Wheatley, R., Diaz Caballero, J., Kapel, N., De Winter, F. H. R., Jangir, P., Quinn, A., et al. (2021). Rapid evolution and host immunity drive the rise and fall of carbapenem resistance during an acute Pseudomonas aeruginosa infection. Nat. Commun. 12, 2460. doi: 10.1038/s41467-021-22814-9
Wick, R. R., Judd, L. M., Gorrie, C. L., Holt, K. E. (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PloS Comput. Biol. 13, e1005595. doi: 10.1371/journal.pcbi.1005595
Woodworth, M. H., Dynerman, D., Crawford, E. D., Doernberg, S. B., Ramirez-Avila, L., Serpa, P. H., et al. (2019). Sentinel case of candida auris in the western United States following prolonged occult colonization in a returned traveler from India. Microb. Drug Resist. 25, 677–680. doi: 10.1089/mdr.2018.0408
Keywords: antimicrobial resistance, whole-genome sequencing, gram-negative bacteria, gram negative, gram negative (G -) bacteria, hospital epidemiology
Citation: Spottiswoode N, Hao S, Sanchez-Guerrero E, Detweiler AM, Mekonen H, Neff N, Macmillan H, Schwartz BS, Engel J, DeRisi JL, Miller SA and Langelier CR (2023) In host evolution of beta lactam resistance during active treatment for Pseudomonas aeruginosa bacteremia. Front. Cell. Infect. Microbiol. 13:1241608. doi: 10.3389/fcimb.2023.1241608
Received: 16 June 2023; Accepted: 14 August 2023;
Published: 30 August 2023.
Edited by:
Zichen Yang, Xinqiao Hospital, ChinaReviewed by:
Daniela Visaggio, Roma Tre University, ItalyCopyright © 2023 Spottiswoode, Hao, Sanchez-Guerrero, Detweiler, Mekonen, Neff, Macmillan, Schwartz, Engel, DeRisi, Miller and Langelier. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Natasha Spottiswoode, bmF0YXNoYS5zcG90dGlzd29vZGVAdWNzZi5lZHU=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.