
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol., 07 July 2023
Sec. Bacteria and Host
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1232391
This article is part of the Research TopicChlamydia-Host Interaction and Its Pathogenic MechanismView all 5 articles
Chlamydia trachomatis is an obligate intracellular pathogen that actively promotes invasion of epithelial cells. A virulence-associated type III secretion system contributes to chlamydial entry and at least four effectors have been described that are deployed during this time. Two of these invasion-related effectors, the translocated membrane-associated effectors A and B (TmeA and TmeB), are encoded in a bi-cistronic operon. TmeA directly activates host N-WASP to stimulate Arp2/3-dependent actin polymerization. According to current working models, TmeA-mediated N-WASP activation contributes to invasion. TmeB has not been functionally characterized. Unlike a tmeA null strain, loss of tmeB does not impact invasion efficiency of C. trachomatis. Using strains deficient for multiple genes, we provide evidence that TmeA is dispensable for invasion in the absence of TmeB. Our data indicate that overabundance of TmeB interferes with invasion and that this activity requires active Arp2/3 complex. We further show that TmeB is capable of interfering with Arp2/3-mediated actin polymerization. In aggregate, these data point to opposing functions for TmeA and TmeB that manifest during the invasion process. These studies raise intriguing questions regarding the dynamic interplay between TmeA, TmeB, and branched actin polymerization during chlamydial entry.
Chlamydia trachomatis is a prevalent obligate intracellular bacterial pathogen that is responsible for genital and ocular diseases. Genital-specific serotypes (Serovars D-K) collectively are the most common bacterial sexually transmitted disease world-wide (Kreisel et al., 2021). Untreated C. trachomatis serovar D-K infections may lead to inflammation of the genitourinary tract tissues resulting in urethritis, proctitis, cervicitis salpingitis, pelvic inflammatory disease (PID) and perihepatitis (Gottlieb et al., 2010). C. trachomatis serovars L1-3 also infect genital tracts, but infections manifest as the more invasive disease manifestation lymphogranuloma venereum (LGV). C. trachomatis (serovars A-C) is also the leading cause of preventable blindness. Additional species pathogenic for humans include the respiratory pathogen C. pneumoniae and the zoonotic avian pathogen C. psittaci (Schachter, 1999). All Chlamydia spp exhibit a unique biphasic developmental cycle (AbdelRahman and Belland, 2005) that alternates between the extracellular-adapted, non-replicating yet infectious, elementary body (EB) and the vegetative, yet non-infectious, reticulate body (RB).
During infection, chlamydial EBs engage new host cells via initial electrostatic interactions followed by a more intimate and irreversible association (Carabeo and Hackstadt, 2001). Attachment is thought to proceed via a collection of host cell receptors that includes epithelial growth factor receptor (EGFR), fibroblast growth factor receptor (FGFR), mannose receptor (MR), protein disulfide isomerase (PDI), and mannose-6 phosphate (M6P) receptor (Elwell et al., 2016). Following receptor engagement, chlamydiae can invade host cells using multiple, likely redundant, pathways. A consensus model has emerged where entry pathways include pathogen-initiated endocytosis (Caven and Carabeo, 2019), receptor-mediated endocytosis (Gitsels et al., 2019), and capture by preformed filopodia followed by macropinocytosis (Ford et al., 2018; Jarsch et al., 2020). Efficient chlamydial entry requires a type III section system (T3SS). Anti-host, substrate proteins are pre-packaged in EBs, secreted during attachment and entry, and referred to as “early”- or “invasion-related” effectors (Mueller et al., 2014). Following internalization, the EB resides in a specialized vacuole called an inclusion and differentiates into the replicative competent RB. Development is concluded after asynchronous conversion of RBs back to EBs and subsequent exit from host cells via lysis or extrusion (Hybiske and Stephens, 2007).
Four chlamydial early effectors share the secretion chaperone Slc1 (Brinkworth et al., 2011; Pais et al., 2013) that likely governs the order of their secretion (Chen et al., 2014). These include the translocated actin recruiting phosphoprotein (Tarp), translocated early phosphor-protein (Tepp), and translocated membrane-associated effectors A and B (TmeA, TmeB) (Elwell et al., 2016), all of which are translocated into the cell within minutes of attachment (Clifton et al., 2004; Hower et al., 2009; Chen et al., 2014; Mueller and Fields, 2015). Both translocated Tarp (Jewett et al., 2008; Mehlitz et al., 2008) and Tepp (Chen et al., 2014) are phosphorylated by host cell kinases. Tarp directly nucleates actin monomers, polymerizes actin, bundles actin filaments (Jewett et al., 2006; Jiwani et al., 2013), and cooperates with branched actin polymerization (Rotty et al., 2013) mediated by the actin related protein complex (Arp2/3) (Jiwani et al., 2012). Tepp recruits phosphoinositide 3-kinases (PI3K) (Carpenter et al., 2017) and the CRK-Like proto-oncogene adapter protein (CrkL) (Chen et al., 2014) to direct host cell signaling to promote chlamydial development. TmeA associates with host membranes through a discreet membrane localization domain (Bullock et al., 2012) while the C-terminus interacts with the host scaffolding protein AHNAK (Hower et al., 2009). TmeA also directly activates host Neural-Wiskott Aldrich Syndrome Protein (N-WASP) to increase Arp2/3-directed actin polymerization (Keb et al., 2021). Residues 118-126 are required for TmeA to bind the GBD domain of N-WASP (Faris et al., 2020). TmeA therefore bypasses the need for N-WASP activation by the RhoGTPase Cdc42. N-WASP activation involves unfolding the protein, and freeing the C-terminal, Arp2/3-activating VCA domain. This domain consists of a verprolin homology sequence (V), a central (C) sequence and an acidic (A) sequence (Kelly et al., 2006). The VC domain is capable of binding monomeric actin, while the CA domain binds to and activates Arp2/3 (Padrick et al., 2011). Analysis of effector-deficient strains indicate that both Tarp (Ghosh et al., 2020) and TmeA (McKuen et al., 2017) are required for efficient C. trachomatis invasion and function synergistically (Faris et al., 2020; Keb et al., 2021) to accomplish entry.
Loss of tmeB does not impact invasion or intracellular growth (McKuen et al., 2017) and the function of TmeB has yet to be elucidated. The TmeB encoding gene is co-expressed with tmeA from a bi-cistronic operon and is also capable of associating with host cell membranes (Mueller and Fields, 2015). Given the operon structure, we hypothesized that TmeB may be functionally related to TmeA. We therefore sought to determine whether TmeB, similar to TmeA, played a role in actin dynamics during entry. In this report, we examined EB invasion kinetics of C. trachomatis strains lacking TmeB or expressing high levels of TmeB. We show that high levels of TmeB correlate with decreased rates of chlamydial entry. Moreover, we demonstrated that TmeB inhibits the activation of the host Arp2/3 complex in in vitro actin polymerization assays. Collectively, we provide evidence that TmeB functions during Chlamydia invasion by negatively regulating Arp2/3-mediated actin polymerization.
C. trachomatis serovar L2 (434/Bu) was used as the parent strain in these studies. Previously described strains include C. trachomatis Δtarp (Ghosh et al., 2020), ΔtmeB (Mueller et al., 2016), Δtepp (Keb et al., 2021), ΔtmeA (Mueller et al., 2016) (referred to herein as ΔtmeA polar), and tmeA-lx (McKuen et al., 2017) (referred to herein as non-polar ΔtmeA). C. trachomatis ΔtmeAB and ΔtmeAB-tarp were generated via FRAEM (Mueller et al., 2016) and are described below. Mobilization of respective plasmids into C. trachomatis L2 was achieved using CaCl2-mediated chemical transformation (Wang et al., 2012). Manipulations leveraging fluorescence reporting to generate engineered strains were accomplished according to established protocols (Mueller et al., 2017; Wolf et al., 2019). Chlamydiae were routinely maintained in either HeLa 229 epithelial cell monolayers (CCL-1.2; ATCC) or McCoy cell monolayers (CRL-1696; ATCC). Unless otherwise indicated, all cultures were grown in RPMI 1640 medium containing 2 mM L-glutamine (Life Technologies) supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (FBS; Sigma) at 37°C in an environment with 5% CO2 and 95% humidified air. Infections were performed using density gradient-purified EBs (Scidmore, 2005). Chlamydiae were cultivated in the presence of 600 ng/ml penicillin G (PenG; Sigma), 500 µg/ml spectinomycin (Spec; AlfaAesar), 1 µg/ml cycloheximide (Sigma), and 50 ng/ml anhydrotetracycline (aTc) where appropriate.
Ectopic expression of TmeB was accomplished by transformation of WT or ΔtmeB chlamydiae with pCOMP-TmeB. pCOMP-TmeB was generated by mobilization of tmeB with its endogenous promoter into pComA II (Mueller et al., 2016). Amplification was accomplished using ΔtmeA genomic DNA and primers 694pro@NmPgfpF (5’-CGGTTCCTGGCCTTTTGCTGG GTACGGAAATACTATCTCCAGCTCAAAGC-3’) and 695@gfpNmPR (5’- GCCCCGCCC TGCCACTCATCGTTAGATATTCCCAACCGAAGAAGGATCTTCC-3’). For ΔtmeAB, pSUmC expressing gfp and aadA flanked by 2.2 kb of chlamydial flanking DNA was generated. Sequences 5’ of tmeA were amplified from L2 genomic DNA with ctl0063-SalI-S (5’-GCAAAAGAGCTGATCCTCCGTCACTGCAGGTACCGCTCCAGCGTTGCGTATTGTTTGTGG-3’) and ctl0063-SalI-AS (5’-CGTATAATGTATGCTATACGAAGTAGG AATGCCTCCGCCGAAGCAATAACTTTTAATTCC-3’) whereas sequences downstream of tmeB were amplified with ctl0064-SbfI-S (5’-GTATAGCATACATTATACGAAGTTA TGCATTTTCTAATAGGGAAGAGGATAAATAGCGTG-3’) and ctl0064-SbfI-AS (5’-CTTTGATCTTTCTACGGGGTCTGACGCCCGACCATTTTACTTTGGAATAAG TGTGATATC-3’). Amplicons were sequentially cloned into pSUmC using Gibson Assembly of SalI-cut or SbfI-cut plasmid, respectively. The tmeA and tmeB double deletion strain was generated by transformation of C. trachomatis L2 with pSUmC-ΔtmeAB and allelic replacement using FRAEM as described (Mueller et al., 2017). A ΔtmeAB-tarp triple mutant was subsequently generated using lateral gene transfer. McCoy cells were co-infected at an MOI of 5 with a 1:1 ratio of ΔtmeAB and Δtarp. Cultures were maintained without antibiotic selection for two 48 hr passages followed by 4 serial passages in the presence of spectinomycin and penicillin G to select for isolates carrying both the tmeAB and tarp deletions. All Chlamydia strains were clonally isolated as described (Mueller et al., 2017) by 2 sequential limiting dilution passages in 384 plates. Chromosomal deletions were confirmed using specific primers and qPCR as described for tarp (Ghosh et al., 2020), or tmeA and tmeB (Mueller et al., 2016) and whole genome sequencing (SeqCenter) was employed to verify integrity of the remaining genome.
For immunoblot analyses, proteins were concentrated from purified EBs by trichloroacetic acid precipitation. Material was separated on 4 to 15% SDS-PAGE gels (Bio-Rad) and transferred to 0.45 µm PVDF membranes (Millipore). Primary antibodies used include: MOMP (Hower et al., 2009), TmeA (Hower et al., 2009), TmeB (Mueller and Fields, 2015), Tarp (Clifton et al., 2004), Tepp (kindly provided by Dr. Raphael Valdivia, Duke University), Hsp60 (A57-B9; Santa Cruz), N-WASP (30D10; Cell Signaling), and ARPC1 (13C9; Millipore). Where appropriate, immunoblots were probed with anti-phospho-tyrosine (4G10, Millipore) to detect phosphorylated Tarp and Tepp or with anti-Flag (Sigma) to detect flag-tagged proteins. We used peroxidase-conjugated secondary antibodies (Sigma) and Amersham ECL Plus (GE Healthcare UK Limited) detection reagent to visualize proteins.
Co-purification of host proteins with TmeA-FT, TmeB-FT, or Cpn0678-FT was performed essentially as described (Keb et al., 2021). Briefly, HeLa cells were transfected with ectopic expression vectors and seeded in one 6-well plate each. Additional controls included cells expressing eGFP or Cpn0677-FT. Cultures were harvested in NP40 buffer (50 mM Tris pH7.5, 150 mM NaCl, 1 mM EDTA, 0.5% NP-40) as described (Mirrashidi Kathleen et al., 2015) after 24 hrs, and incubated on ice for 1 hr. Soluble fractions were applied to equilibrated Anti-Flag M2 Affinity gel (Sigma) and incubated overnight at 4°C. The resin was washed 3x with lysis buffer and FT proteins were eluted with 3X Flag peptide in PBS. 6x Laemmli buffer was added to eluates prior to separation on SDS-PAGE gels for immunoblotting.
HeLa229 cells were prepared in 24-well plates with 12 mm coverslips, and invasion assays were performed essentially as described (Carabeo et al., 2002). Density gradient purified EBs were used at a multiplicity of infection (MOI) of 20. Where appropriate, HeLa cells were pretreated with 200 µM CK666 (all purchased from Sigma-Aldrich) for 15 min prior to infection and maintained with 200 µM CK666 during infection and subsequent incubation. All infections were carried out on ice with rocking for 1 hr then shifted to 37°C for 45 min. Cultures were then treated with 4% paraformaldehyde, and extracellular or intracellular EBs were differentially labeled with murine LPS-specific or rabbit MOMP (Hower et al., 2009) antibodies, respectively. Secondary antibody conjugated to Alexa-594 (anti-mouse) or Alexa-488 (anti-rabbit) were used for detection. Internal and external chlamydiae were enumerated in 10 fields of view. Percent EB internalization was calculated via the formula [(total EBs –external EBs)/total red EBs] x 100 = percent (%) invasion.
Pyrene actin polymerization assays were performed as previously described (Jiwani et al., 2013). Full-length TmeB was expressed as a GST fusion in pGEX-6p-1, purified to homogeneity over GST affinity column, and eluted by cleavage of the GST-tag with PreScission protease (Sigma). Remaining components were from the commercially available pyrene actin polymerization kit (Cytoskeleton). For polymerization assays, monomeric pyrene-labeled actin was prepared by diluting lyophilized pyrene actin (Cytoskeleton) in 5mM Tris (pH 8.0) 0.2mM CaCl2 0.2mM ATP (G buffer) and collection of the supernatant of a 90 minute, 100,000 xg, 4°C spin in a Beckman Optima MAX TL Ultracentrifuge using a TLA 55 rotor (Beckman Coulter). Approximately 30 µg of pyrene-labeled actin was mixed with 1-2µg of indicated proteins (TmeB, VCA, Arp2/3, GST) in a volume of 500 µl for 5 min before the addition of 1/20th volume of polymerization buffer (500mM KCl, 20mM MgCl2, 10mM ATP). The reaction (contained in a semi-microcuvette and holder assembly) was monitored for 30 min with a LS 55 Luminescence spectrophotometer equipped with the biokinetic accessory and directed by FL Winlab software version 4.0 (Perkin-Elmer, Beaconsfield, Bucks, United Kingdom) with 2.5 nm bandwidth at 365 nm excitation wavelength and 2.5 nm bandwidth at 407 nm emission wavelength.
Unless otherwise noted, presented data are representative of triplicate experiments where quantitative data were generated from experiments containing triplicate biological replicates. Calculation of standard deviation of the mean and assessment via Student’s t-test with Welch’s correction. Statistical analyses were performed using GraphPad Prism 6 version 6.04 (Graphpad Software Inc).
We developed floxed allelic exchange mutagenesis (FLAEM) to enable marker-less gene deletion in C. trachomatis. This approach (Figure 1A) was previously used (Keb and Fields, 2020) to excise a blaM-gfp cassette replacing the tmeA locus in our original ΔtmeA null (Mueller et al., 2016) created by fluorescence-reported allelic exchange mutagenesis (FRAEM). The marker-induced polar effect on downstream tmeB was relieved, resulting in increased TmeB abundance (Keb and Fields, 2020). During the course of our TmeA functional studies, we noticed the new non-polar ΔtmeA (referred to previously as ΔtmeA-lx) strain appeared to exhibit a more robust invasion defect than the parent polar strain. We directly tested for a potential difference by performing side-by-side invasion studies (Figure 1B). Compared to WT, polar ΔtmeA reduced invasion efficiency by ca 16% whereas the non-polar strain was reduced by ca 50%. Corresponding protein abundance was examined in material concentrated from gradient-purified EBs (Figure 1C). As expected, immunoblot data confirmed loss of TmeA and lower abundance of TmeB in the polar ΔtmeA strain. Interestingly, a pronounced overabundance of TmeB was detected in non-polar ΔtmeA compared to WT EBs. TmeB overabundance was specific to the non-polar ΔtmeA strain since WT levels of TmeB were detected in strains lacking tepp or tarp (Figure S1). These data indicate an inverse correlation between invasion efficiency and TmeB abundance when TmeA is absent.
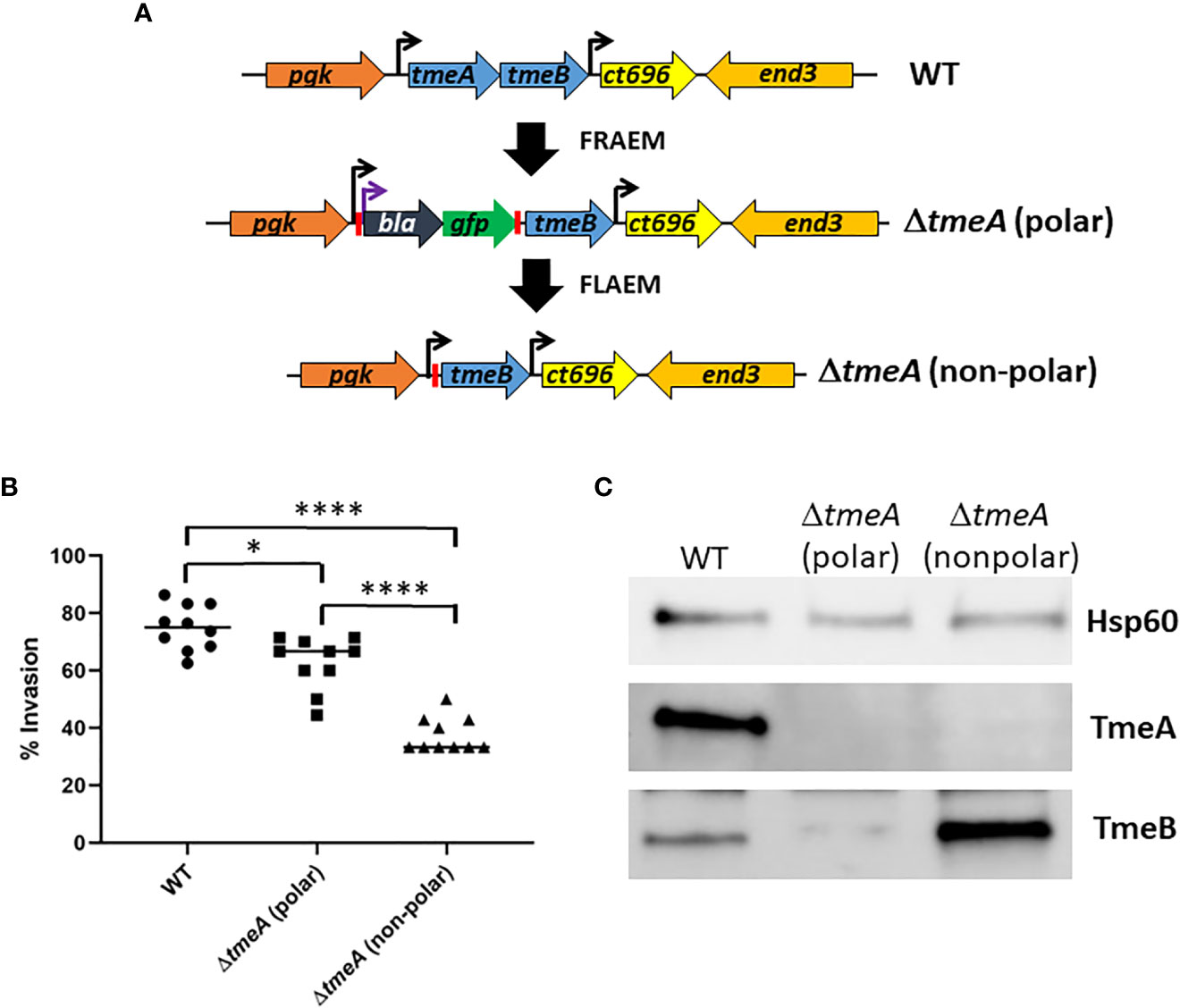
Figure 1 Abundance of TmeB correlates with severity of C. trachomatis invasion defect. (A) Schematic of C. trachomatis L2 tmeAB locus in WT, polar ΔtmeA and non-polar ΔtmeA strains. Endogenous (black) or engineered (purple) promoters are indicated by arrows. Engineered loxP sites are indicated by red bars. (B) HeLa monolayers were infected for 1 hr at 4°C with WT or mutant strains at an MOI of 10. Cultures were shifted to 37°C for 30 min and then paraformaldehyde fixed and processed for inside-out staining to assess invasion efficiency. Data from 10 fields of view are represented with means for percentage of internalized chlamydiae. Statistical significance was computed using a Student’s T-test with Welch’s correction (*P<0.005 and ****P<0.0001). (C) Immunoblot of material concentrated from equal EBs from WT, polar ΔtmeA and non-polar ΔtmeA strains. TmeA and TmeB were detected by chemilumenescence using specific antibodies and blots were probed with Hsp60-specific antibodies as a loading control.
We next generated a strain containing a complete deletion of both tmeA and tmeB to address whether the invasion defect manifested by ΔtmeA strains was due solely to the absence of TmeA. C. trachomatis L2 was transformed with pSUmC-ΔtmeAB, and FRAEM was employed to replace the entire coding region for tmeA and tmeB with a gfp-aadA cassette. Gene deletion was first confirmed in a clonal isolate via qPCR (Figure S2A) and then via immunoblot using specific antibodies to probe EB lysates (Figure 2A). Neither TmeA nor TmeB were detected whereas co-secreted effectors Tarp and Tepp remained unchanged compared to WT. Tarp is required for efficient invasion, and a triple mutant was generated using lateral gene transfer. Co-infections containing Specr ΔtmeAB and PenGr Δtarp (Ghosh et al., 2020) were cultivated in the presence of both antibiotics to select a recombinant strain carrying both mutations. Respective deletions of a clonal isolate were confirmed via qPCR (Figure S2B). Subsequent immunoblot (Figure 2B) analysis confirmed the loss of TmeA, TmeB, and Tarp in a single strain. Invasion efficiency of each strain was compared to WT (Figure 2C). While levels of ΔtmeAB invasion did not differ from WT, the additional loss of Tarp manifested a significant decrease in internalization efficiency. These data raise the possibility that TmeA, but not Tarp, is required for efficient invasion only in the presence of TmeB.
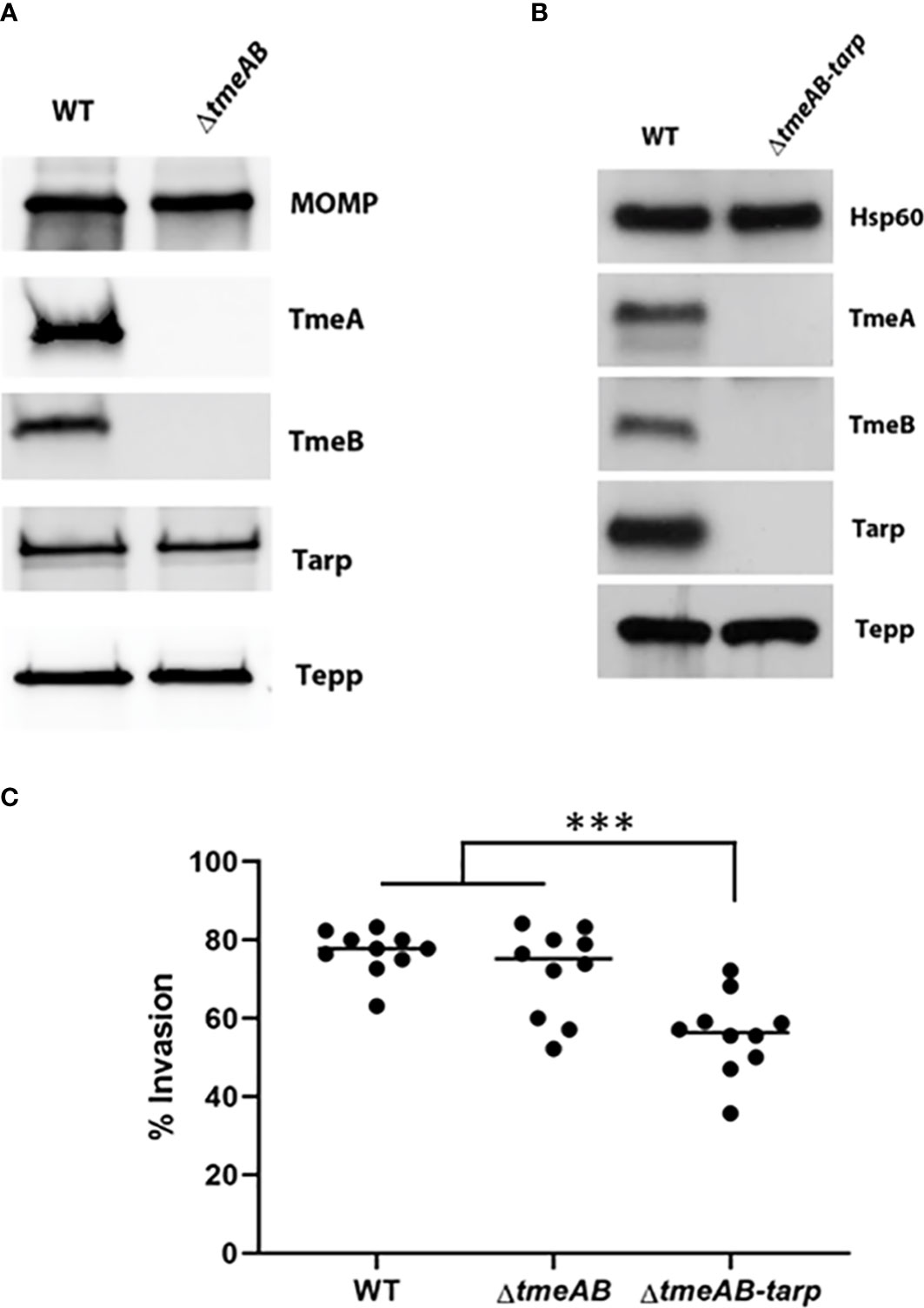
Figure 2 Invasion efficiencies of double and triple mutants. (A, B) Immunoblot analysis of material concentrated from purified WT, double mutant (ΔtmeAB), and triple mutant (ΔtmeAB-tarp) EBs. TmeA, TmeB, Tarp and Tepp were detected with specific antibodies. Antigen-specific antibodies to either MOMP or Hsp60 were used as loading controls. All proteins were visualized by chemilumenescence. (C) HeLa monolayers were infected for 1 hr at 4°C with WT or mutant strains at an MOI of 10. Cultures were paraformaldehyde fixed and processed for inside-out staining after 30 min at 37°C. Bacteria were enumerated over 10 fields of view and invasion efficiencies represented with means for percentage of internalized chlamydiae. Statistical significance was computed using One-way Anova (***P<0.0001).
Non-physiological levels of TmeB in non-polar ΔtmeA also raised the possibility that TmeB could be directly detrimental in the absence of co-expressed TmeA. C. trachomatis WT and ΔtmeB were transformed with pCOMP constitutively expressing TmeB to address this possibility. A 5-10 fold increase in TmeB abundance was evident via immunoblot analysis of EB lysates (Figure 3A). These strains were then assayed for invasion efficiency after allowing 30 min for internalization (Figure 3B). As shown previously (Keb et al., 2021), loss of tmeB did not interfere with invasion efficiency compared to WT. The presence of excess TmeB in WT and ΔtmeB, however, did correlate with decreased invasion efficiency. These data implicate non-physiological levels of TmeB as a contributing factor to the observed impairment of chlamydial entry into epithelial cells.
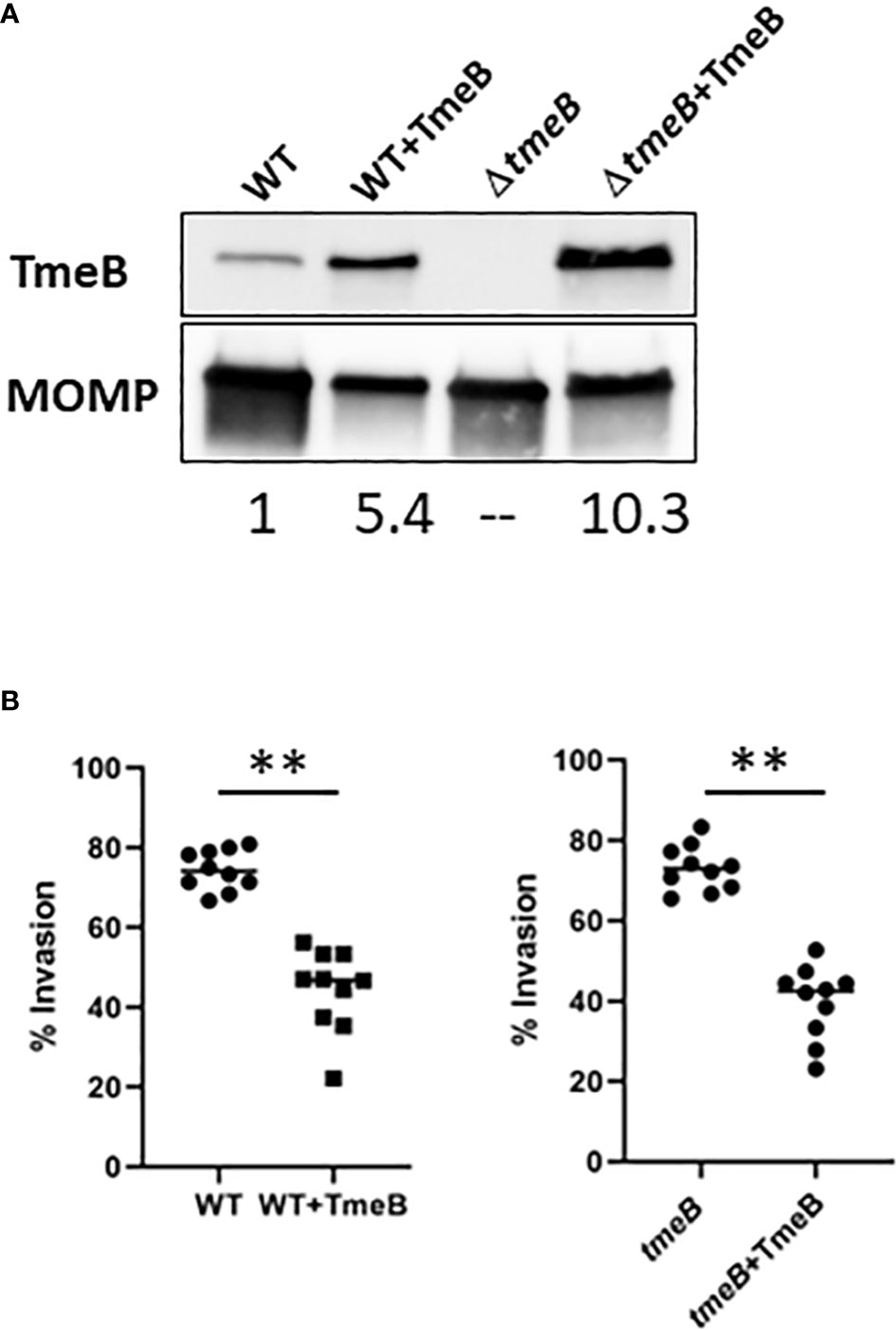
Figure 3 The impact of TmeB overexpression during invasion. (A) Immunoblot analysis of material concentrated from purified EBs from WT and ΔtmeB alone or expressing pCOMP-TmeB (+TmeB). Blots were probed with TmeB-specific antibodies or MOMP antibodies as a loading control and visualized via chemilumenescence. TmeB band intensities were determined using ImageJ and normalized based on MOMP signal with WT being nominally set to 1. (B) HeLa monolayers were infected for 1 hr at 4°C with respective strains at an MOI of 10. Cultures were paraformaldehyde fixed and processed for inside-out staining after 30 min at 37°C. Bacteria were enumerated over 10 fields of view and invasion efficiencies represented with means for percentage of internalized chlamydiae. Statistical significance was computed using a Student’s T-test with Welch’s correction (**P<0.001).
Possibilities for TmeB-specific interference with invasion include indirect effects such as competition for the secretion chaperone Slc1 within chlamydiae or more directly by manifesting a dysregulated activity within host cells. To test the former, HeLa cultures were infected for 1 hr with WT or ΔtmeB strains alone or ectopically expressing TmeB. Material from whole-cell lysates was probed with phospho-tyrosine-specific antibodies to detect translocated Tarp and Tepp (Figure S3). Both were readily detected in all strains, indicating that negative impact of excess TmeB likely does not manifest by interfering with type III secretion. Given that operon-encoded genes are often functionally related, we next reasoned that TmeB could have an activity related to TmeA. HeLa cells were transiently transfected with plasmids encoding Flag-tagged TmeA or TmeB. HeLa cells were also nucleofected with plasmids expressing eGFP as a mock control. Plasmids encoding Flag-tagged C. pneumoniae Cpn0677 or Cpn0678, which are encoded by tmeA-syntenic genes found in C. pneumoniae, were used as specificity controls. Flag-tagged proteins were immunoprecipitated under mild lysis conditions using Flag-beads, and material was probed in immunoblots for N-WASP, which is a molecular target of TmeA (Figure 4A). We also probed for the Arp2/3 complex using antibodies specific for the ARPC1 subunit. As expected, all Flag-tagged proteins were recovered and neither N-WASP nor ARPC1 co-precipitated with Cpn0678. A cross-reactive band was detected in the GFP control with Flag antibodies, but neither N-WASP nor ARPC1 were detected. N-WASP was detected in the presence of TmeA and C. pneumonia Cpn0677. We also detected ARPC1 which presumably co-precipitates with the activated N-WASP complex. N-WASP did not co-precipitate with TmeB, yet ARPC1 was detected, raising the possibility that TmeB targets the Arp2/3 complex. We tested this hypothesis by assessing comparative invasion in the presence or absence of the pharmacologic Arp2/3 inhibitor CK666 (Figure 4B). We chose a CK666 dose sufficient to inhibit WT invasion ca. 50%. HeLa cells were infected with C. trachomatis WT, ΔtmeB or ΔtmeB+TmeB with or without 200 µM CK666 and processed for internalization counts at 30 min. Consistent with previous studies, cultivation with CK666 significantly decreased WT invasion. This was also detected for ΔtmeB. Interestingly, CK666 failed to additionally inhibit ΔtmeB+TmeB, indicating that the negative impact was not synergistic. Instead, the negative effect of over-expressed TmeB appears to be manifested through Arp2/3. In aggregate, these data suggest that TmeB targets the host Arp2/3 complex during invasion in an inhibitory fashion.
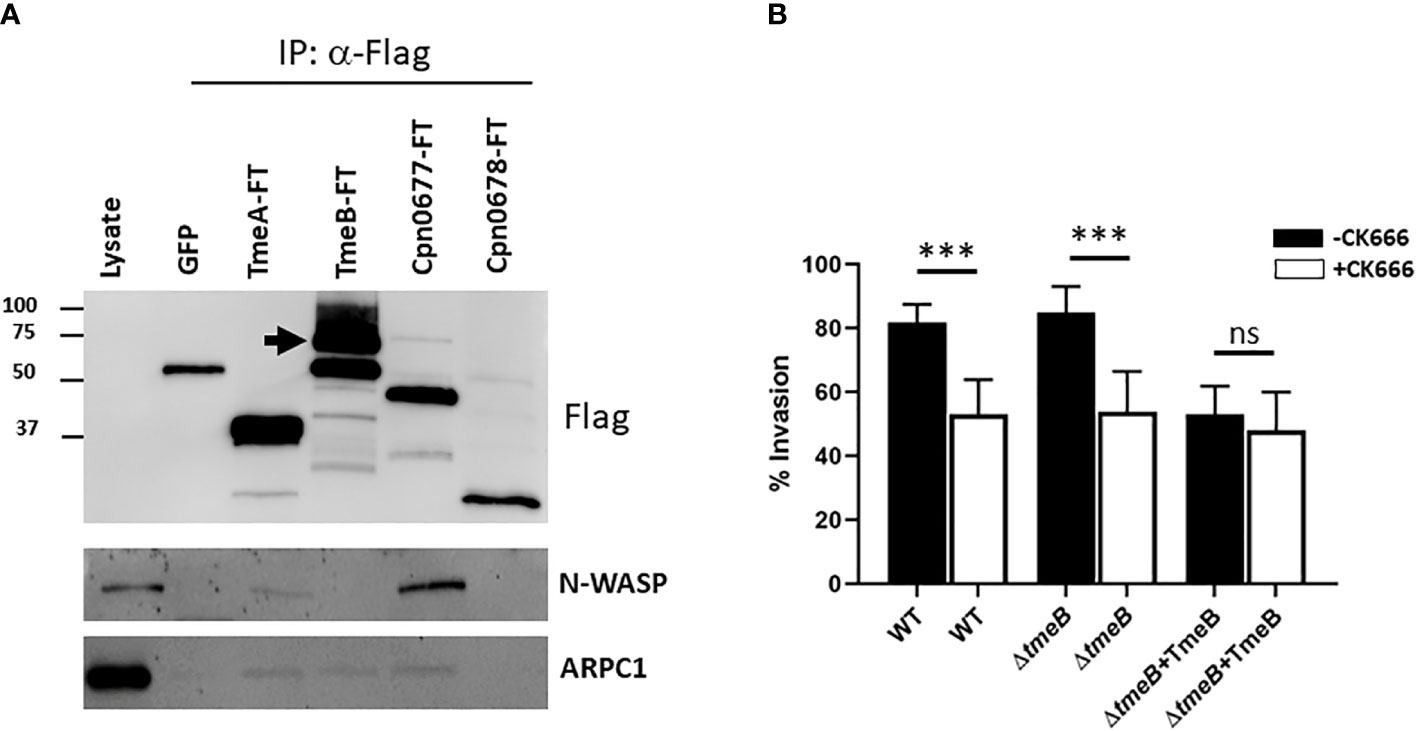
Figure 4 TmeB targets the Arp2/3 complex. (A) Flag-tagged proteins were immunoprecipitated from whole-cell lysates of HeLa cells transiently expressing GFP, TmeA-FT, TmeB-FT, Cpn0677-FT, or Cpn0678-FT for 24 hrs. Eluted material was probed in immunoblots for tagged chlamydial proteins using anti-Flag antibodies. Host proteins were detected using antigen-specific antibodies, and HeLa whole-cell lysates were included as a positive control for these antibodies. Arrow indicated position of full-length TmeB-FT. (B) HeLa monolayers were infected for 1 hr at 4°C with WT, ΔtmeB or ΔtmeB+TmeB strains at an MOI of 10. Infections were carried in the absence (black bars) or presence (white bars) of 200 µM CK666. Cultures were shifted to 37°C and maintained for 30 min, with or without drug, then paraformaldehyde fixed and processed for inside-out staining to assess invasion efficiency. Data are represented as mean values for percentage of internalized chlamydiae and are shown with standard deviations. Statistical significance was computed using a Student’s T test with Welch’s correction (***P<0.0001). ns, not significant.
The pyrene-based actin fluorescence assay is a well-established approach to functionally investigate factors impacting the state of actin polymerization. Indeed, we have leveraged this approach to demonstrate Tarp-dependent actin polymerization (Jewett et al., 2006) and activation of N-WASP-dependent polymerization by TmeA (Keb et al., 2021). The verprolin homology, central, acidic (VCA) domain of N-WASP was used to activate Arp2/3 and test the impact of TmeB on actin polymerization. Purified preparations (Figure 5A) of protein components were incubated alone or in combination with pyrene-conjugated actin. As expected, the combination of Arp2/3 and VCA increased polymerization rates, as indicated by fluorescence intensity, compared to actin alone (Figure 5B). Addition of control GST did not interfere with this polymerization. In the presence of pure TmeB, the fluorescence intensity profile was similar to that of actin alone. TmeB-mediated inhibition of actin polymerization was dose-dependent as increasing levels of TmeB resulted in progressively dampened actin polymerization rates (Figure 5C). This decrease in actin polymerization depended on Arp2/3 since, in the absence of Arp2/3 and VCA, TmeB alone had no impact on polymerization rates (Figure S4). In aggregate, these data indicate that one function of TmeB is to antagonize branched actin polymerization by interfering with Arp2/3 activity.
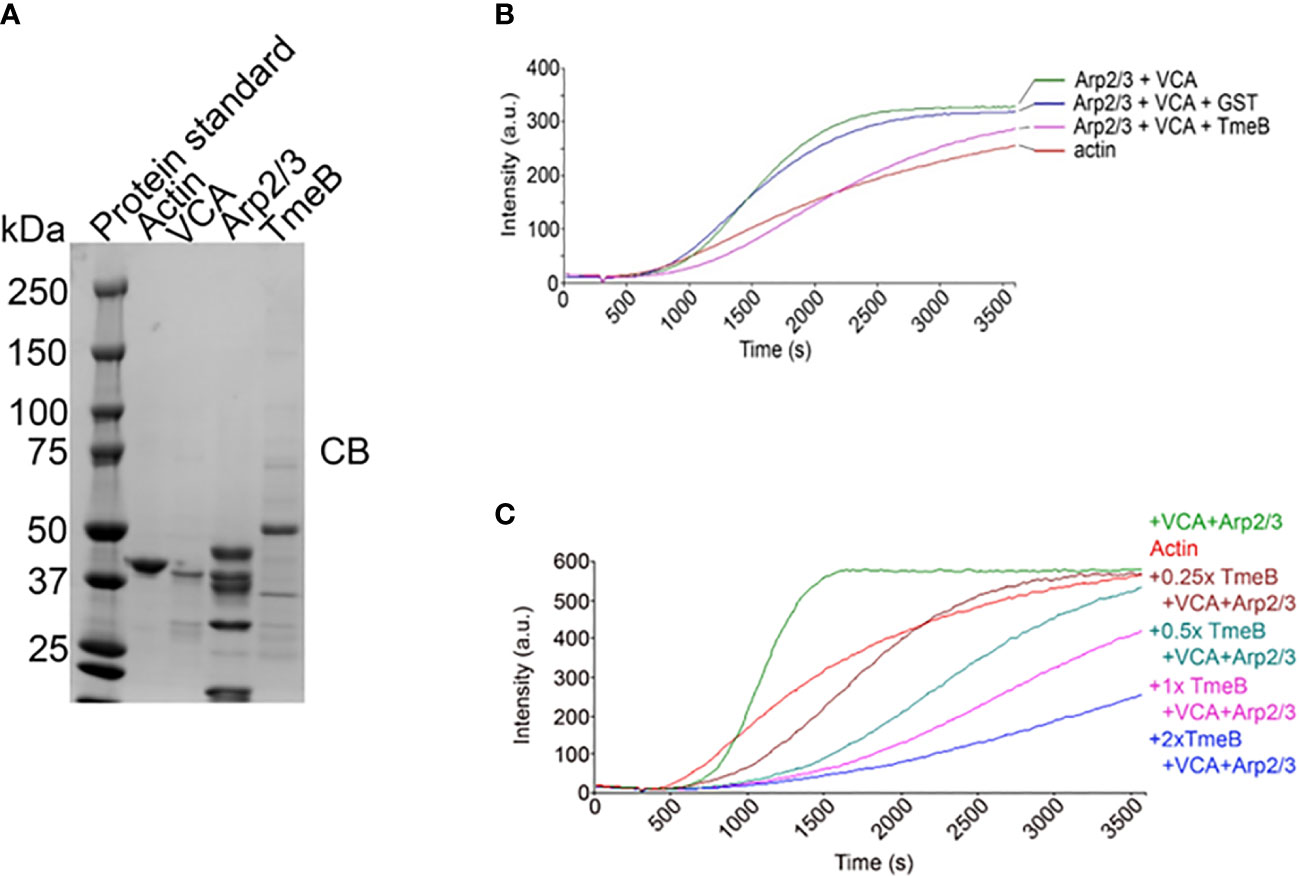
Figure 5 Impact of TmeB on Arp2/3-mediated actin polymerization. (A) Coomassie stained preparations of the purified Actin, Arp2/3 complex, the Verprolin homology, Central, Acidic domain of N-WASP, and TmeB employed in the pyrene actin polymerization assay. Increase in actin polymerization after the addition of polymerization buffer at 300 s was measured as the arbitrary fluorescence intensity (arbitrary units [a.u.] over time [s]) with excitation and emission wavelengths of 365 and 407 nm respectively. (B) TmeB, Arp2/3 and VCA were added individually or in combination to monomeric pyrene-labeled actin and actin alone served as a negative control. (C) Like the assay shown in (A) but with 2-fold increases in the amount of pure TmeB included in mixtures. Maximal actin polymerization occurred in the presence of Arp2/3 and VCA and this was inhibited by the presence of TmeB.
The VCA domain of N-WASP exhibits constitutive activity, and we previously used a version of N-WASP (residues 151-501) to show that TmeA activates N-WASP to promote Arp2/3-dependent actin polymerization (Keb et al., 2021). We therefore examined whether TmeB could interfere with actin polymerization stimulated by the TmeA-N-WASP complex by combining these proteins in the pyrene assay (Figure 6A). As expected, addition of TmeA-N-WASP with Arp2/3 enhanced fluorescence kinetics and intensity. Inclusion of TmeB interfered with this activation and slowed the overall polymerization rate. We next tested whether TmeB acts synergistically with CK666 (Figure 6B). Addition of low levels (17 µM) of CK666 to VCA + Arp2/3 resulted in inhibition of actin polymerization as expected. Inclusion of TmeB shifted the fluorescence profile further downward indicating a synergistic effect between CK666 and TmeB. This additive effect was not detected at a saturating concentration (100 µM) of CK666 (Figure S4B). Collectively, these data suggest that CK666 and TmeB-mediated inhibition of Arp2/3 may manifest via a similar mechanism.
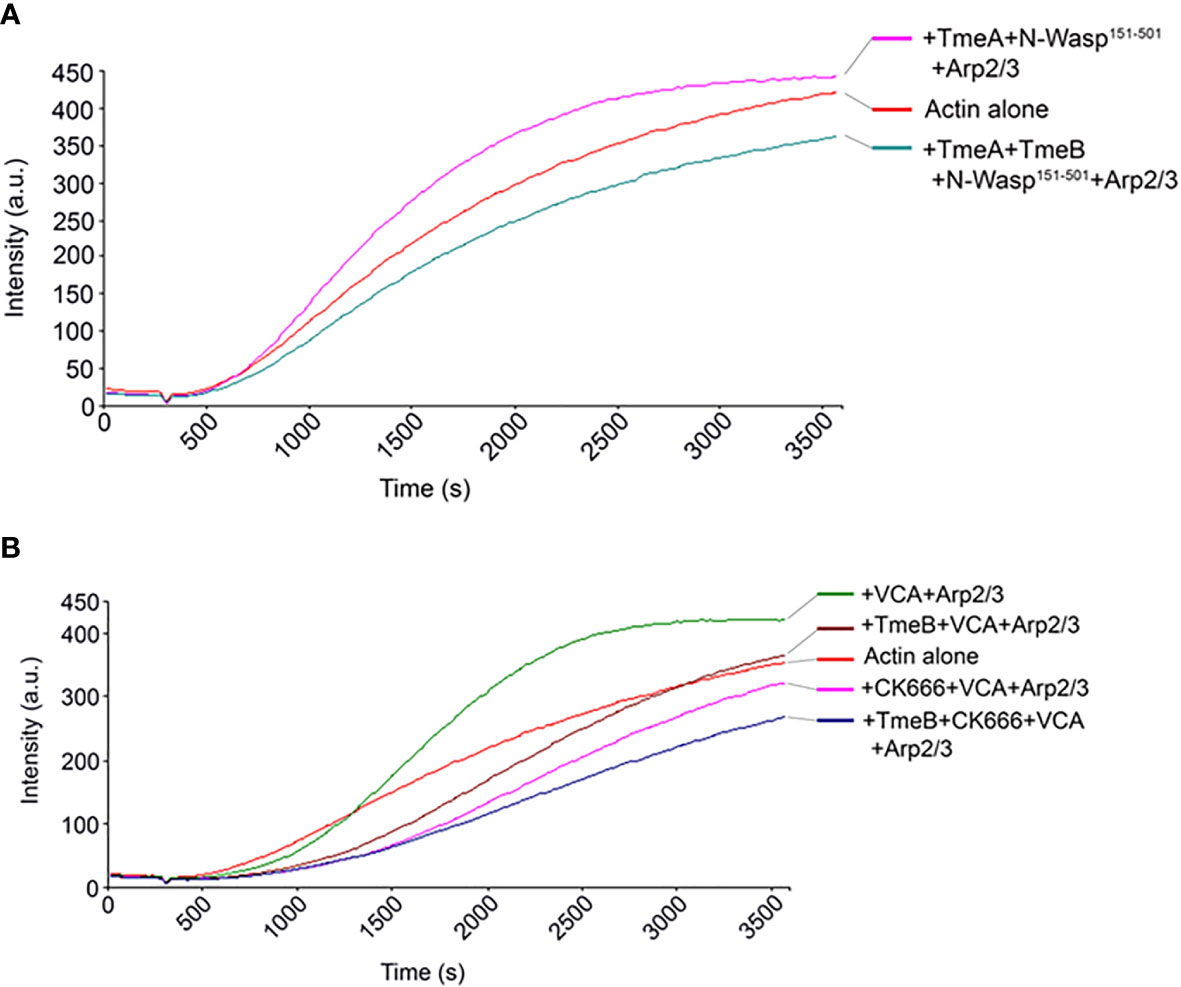
Figure 6 TmeB inhibition in the presence of TmeA-N-WASP or CK666. Actin polymerization was tested measured as the arbitrary fluorescence intensity (arbitrary units [a.u.] over time [s]) with excitation and emission wavelengths of 365 and 407 nm respectively. Polymerization buffer was added at 300 s. Monomeric pyrene-labeled actin and actin alone served as a negative control. (A) TmeA and N-WASP151-501 were added individually or in combination with TmeB. (B) TmeB, Arp2/3 and VCA were added individually or in combination with 17 µM of the Arp2/3 inhibitor CK666.
TmeA and TmeB are two C. trachomatis early effectors that are encoded in a bi-cistronic operon, share the secretion chaperone Slc1, and are translocated into human cells during invasion via T3S (Hower et al., 2009). Both effectors also peripherally associate with host membranes through N-terminally localized helices (Bullock et al., 2012; Mueller and Fields, 2015). While we previously demonstrated that invasion efficiency of a polar ΔtmeA mutant differs moderately from WT (McKuen et al., 2017), invasion efficiency by the non-polar ΔtmeA mutant is more robustly reduced (Keb et al., 2021). We provide evidence herein that this difference in invasion efficiency is likely manifested by strain-specific differences in TmeB abundance. The elevated level of TmeB in the non-polar ΔtmeA strain could be due to increased message levels (Keb et al., 2018) or translation efficiency. During EB entry of a host cell, TmeA is required to recruit host N-WASP to sites of invasion (Faris et al., 2020; Keb et al., 2021). TmeA directly induces activation of N-WASP to promote Arp2/3-directed actin polymerization (Keb et al., 2021) and this activity is postulated to contribute to remodeling of the actin cytoskeleton necessary to drive chlamydial entry (Faris et al., 2020; Keb et al., 2021). Loss of TmeB does not manifest in an entry defect (Keb et al., 2021), and functional characterization of this effector has been lacking. Using genetic and biochemical approaches, we have now demonstrated that TmeB exerts an inhibitory role in Arp2/3-mediated actin polymerization. Taken together, these data suggest a regulatory axis between TmeA and TmeB, wherein TmeB is capable of negatively regulating the ability of TmeA to incite Arp2/3-dependent actin polymerization.
Epithelial cell ARPC1, but not N-WASP, co-precipitated with ectopically expressed TmeB, raising the possibility of a direct interaction with the Arp2/3 complex. Although TmeB interfered with TmeA-N-WASP-mediated Arp2/3 activation in the in vitro actin polymerization assay, our additional data indicate that TmeB directly inhibits Arp2/3-mediated actin polymerization. Moreover, our efforts have failed to indicate any direct interaction of TmeA and TmeB (Fields, unpublished). CK-666 is a small molecule inhibitor of the Arp2/3 complex that binds in the pocket between Arp2 and Arp3 blocking the two subunits from forming a short-pitched dimer necessary to bind actin (Hetrick et al., 2013). Our data showed that CK-666 did not synergistically inhibit invasion alongside over expression of TmeB. These data implicate the TmeB inhibitory activity manifesting during chlamydial infection. Interestingly, TmeB’s ability to inhibit the Arp2/3 complex appears to be similar to that of the host cell regulating protein Arpin. TmeB and Arpin both share an unstructured, acidic domain implicated in the negative regulation of the Arp2/3 complex (Dang et al., 2013). The TmeB acidic domain contains 19 acidic residues in the first 80 amino acids with 10 acidic residues between amino acids 21-40 of the TmeB protein. The acidic domain of Arpin was found to associate with the Arp3 subunit leading to Arp2/3 inhibition (Fregoso et al., 2022). It is intriguing to speculate that TmeB, like Arpin binds, to and disrupts a specific subunit of the Arp2/3 complex. We observed that CK666 and TmeB inhibitory effects showed synergy and could indicate that TmeB also impacts formation of the short-pitched Arp2-Arp3 dimer. Future studies will be required to determine the specificity of the interaction between the Arp2/3 complex and TmeB, and how TmeB is able to inhibit Arp2/3-mediated actin polymerization.
C. trachomatis utilizes several early effectors to manipulate the host actin cytoskeleton to promote invasion of epithelial cells (Caven and Carabeo, 2019). In addition to TmeA, chlamydial Tarp can alter actin polymerization dynamics directly via interactions with monomeric or filamentous actin (Jewett et al., 2006; Jiwani et al., 2012) or indirectly by nucleating host factors culminating in Rac-1 dependent Arp2/3 activation (Caven and Carabeo, 2019). Our observation that a clean tmeA-tmeB double mutant does not manifest an invasion defect implies a functional link between TmeA and TmeB activities. These data indicate that TmeA is only essential for invasion in the presence of TmeB. N-WASP is likely required since trans-complementation with TmeA deficient for N-WASP binding failed to restore WT levels of entry (Faris et al., 2020). Tarp-dependent entry is seemingly not linked since the tmeA-tmeB-tarp triple mutant showed decreased invasion efficiency. This would be consistent with the notion that TmeA and Tarp contribute independently to invasion (Faris et al., 2020; Keb et al., 2021) and that direct actin polymerization and bundling Tarp activity is the primary requirement during chlamydial entry (Ghosh et al., 2020). Analysis of the early chlamydial effectors ability to associate with the T3S chaperone Slc1 suggests a hierarchical and temporal release of early effectors during chlamydial invasion in which Tarp and TmeA are translocated first with the subsequent recycling of chaperone Slc1 being free to translocate a second wave of effectors such as Tepp and TmeB (Chen et al., 2014). The consequence of the preferred Tarp and TmeA chaperone binding and delivery may function to fine tune actin dynamics during EB invasion by initially promoting actin filament formation followed by the immediate inhibition of Arp2/3 complex by TmeB.
Why TmeB may balance TmeA activity depends on the true role of TmeA during invasion. Faris, et al. showed that the non-polar ΔtmeA strain failed to manifest actin-based pedestal formation, leading them to postulate that TmeA-mediated N-WASP activation leads to protrusions associated with EBs (Faris et al., 2020). Given the newly appreciated role of excess TmeB in this strain, this effect could also be spuriously manifested by excess TmeB. N-WASP is transiently recruited to sites of invasion and associated with macropinocytosis and the Bin-Amphiphysin-Rvs (BAR) domain protein Sorting Nexin 9 (SNX9) found in F-actin based pedestals present at sites of chlamydial entry (Ford et al., 2018). While SNX9 is involved in biogenesis of filipodial structures, chlamydiae appear to associate with preformed structures instead of driving de novo production (Jarsch et al., 2020). Interestingly, our control protein Cpn0677 also co-immunoprecipitated with N-WASP. C. pneumoniae Cpn0677 is not homologous to TmeA but is encoded within a syntenic locus (Hower et al., 2009). Cpn0677 (renamed secretion effector of membrane and actin dynamics, SemD) was recently proposed as a sorting platform effector where SemD induced membrane curvature to recruit SNX9 and the subsequent interaction with N-WASP drives engulfment and scission of the nascent inclusion (Spona et al., 2023). Genetic manipulation of C. pneumoniae is currently lagging so the model has not been confirmed genetically. If C. trachomatis TmeA functions similarly, TmeB could contribute to cessation of actin polymerization around the nascent inclusion. This notion is reinforced by the fact that C. pneumoniae Cpn 0676 is homologous to C. trachomatis TmeB (Hower et al., 2009) and could function similarly. Regardless, future studies will require direct chromosomal manipulation to create in-frame TmeA domain mutants to separate TmeA and TmeB functionally.
Beyond TmeA, there are additional possibilities regarding the overarching functional consequences of Arp2/3 inhibition by TmeB. Other bacterial effectors have been shown to inhibit the Arp2/3 complex. Legionella pneumophila kinase LegK2 is a type IV secretion system effector protein capable of inhibiting Arp2/3 through phosphorylation of the ARPC1 and Arp3 subunits (Michard et al., 2015). TmeB has yet to be implicated as a kinase, though this is an interesting area of study that remains to be investigated. The Legionella pneumophila type IV secretion system phosphotyrosine-phosphatase effector protein WipA has also been shown to disrupt Arp2/3-directed actin polymerization, through a different mechanism than LegK2. WipA reduces the tyrosine phosphorylation of N-WASP and Arp3 during infection resulting in a reduction of actin polymerization (He et al., 2019). Both LegK2 and WipA hijacking of the N-Wasp/Arp2/3 pathway is speculated to promote intracellular survival and protection of the Legionella containing vacuoles from the endocytic pathway (Michard et al., 2015; He et al., 2019). TmeB could therefore contribute to avoidance of the nascent inclusion with fusion with vesicles of the endocytic pathway. Redundant mechanisms would be required since the ΔtmeB strain grows similarly to WT (McKuen et al., 2017). More broadly, dysregulated actin polymerization can be detrimental via activation of cell-autonomous immunity (Mostowy and Shenoy, 2015; Garcia et al., 2020). We postulate that at least one role of TmeB is to return host cells to a basal physiological state similar to, but mechanistically distinct from, SptP of Salmonella Typhimurium (Fu and Galan, 1999). Activation of the Rho-GTPases Rac1 and Cdc42 triggered by invasion-promoting effectors such as SopE can activate innate immune signaling through the NOD pattern recognition receptors (Keestra et al., 2013). SptP reverses Rho-GTPase activation (Fu and Galan, 1999) to antagonize immune detection and NF-κB (Keestra et al., 2013). In aggregate, we favor a similar working model in which the delivery of TmeB into the host cell restores cytoskeletal homeostasis by resetting structural and mechanical properties of the cytoskeleton to pre-infection levels to favor chlamydial survival, growth, and immune evasion within the nascent inclusion.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
KS and GK contributed equally to this work. All authors contributed to the article and approved the submitted version.
This work was supported by Public Health Service grants from the National Institutes of Health, NIAID (grants R01AI065530 and R01AI175220), to KF and (R01AI139242) to TJ.
The authors would like to thank C. Hawk, G. Eicher, and Drs. P. Nguyen and M. Jewett for critical reading of the manuscript and R. W. Hayman for excellent technical assistance.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1232391/full#supplementary-material
AbdelRahman, Y., Belland, R. (2005). The chlamydial developmental cycle. FEMS Microbiol. Rev. 29, 949–959. doi: 10.1016/j.femsre.2005.03.002
Brinkworth, A. J., Malcolm, D. S., Pedrosa, A. T., Roguska, K., Shahbazian, S., Graham, J. E., et al. (2011). Chlamydia trachomatis Slc1 is a type III secretion chaperone that enhances the translocation of its invasion effector substrate TARP. Mol. Microbiol. 82, 131–144. doi: 10.1111/j.1365-2958.2011.07802.x
Bullock, H. D., Hower, S., Fields, K. A. (2012). Domain analyses reveal that c. trachomatis CT694 belongs to the membrane-localized family of type III effector proteins. J. Biol. Chem. 287, 28078–28086. doi: 10.1074/jbc.M112.386904
Carabeo, R., Grieshaber, S., Fischer, E., Hackstadt, T. (2002). Chlamydia trachomatis induces remodeling of the actin cytoskeleton during attachment and entry into HeLa cells. Infect. Immun. 70, 3793–3803. doi: 10.1128/IAI.70.7.3793-3803.2002
Carabeo, R. A., Hackstadt, T. (2001). Isolation and characterization of a mutant Chinese hamster ovary cell line that is resistant to Chlamydia trachomatis infection at a novel step in the attachment process. Infect. Immun. 69, 5899–5904. doi: 10.1128/IAI.69.9.5899-5904.2001
Carpenter, V., Chen, Y.-S., Dolat, L., Valdivia, R. H. (2017). The effector TepP mediates recruitment and activation of phosphoinositide 3-kinase on early Chlamydia trachomatis vacuoles. mSphere 2. doi: 10.1128/mSphere.00207-17
Caven, L., Carabeo, R. A. (2019). Pathogenic puppetry: manipulation of the host actin cytoskeleton by Chlamydia trachomatis. Int. J. Mol. Sci. 21. doi: 10.3390/ijms21010090
Chen, Y.-S., Bastidas, R. J., Saka, H. A., Carpenter, V. K., Richards, K. L., Plano, G. V., et al. (2014). The Chlamydia trachomatis type III secretion chaperone Slc1 engages multiple early effectors, including TepP, a tyrosine-phosphorylated protein required for the recruitment of CrkI-II to nascent inclusions and innate immune signaling. PloS Pathog. 10, e1003954. doi: 10.1371/journal.ppat.1003954
Clifton, D., Fields, K., Grieshaber, N., Dooley, C., Fischer, E., Mead, D., et al. (2004). A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. PNAS 101, 10166–10171. doi: 10.1073/pnas.0402829101
Dang, I., Gorelik, R., Sousa-Blin, C., Derivery, E., Guérin, C., Linkner, J., et al. (2013). Inhibitory signalling to the Arp2/3 complex steers cell migration. Nature 503, 281–284. doi: 10.1038/nature12611
Elwell, C., Mirrashidi, K., Engel, J. (2016). Chlamydia cell biology and pathogenesis. Nat. Rev. Micro. 14, 385–400. doi: 10.1038/nrmicro.2016.30
Faris, R., McCullough, A., Andersen, S. E., Moninger, T. O., Weber, M. M. (2020). The Chlamydia trachomatis secreted effector TmeA hijacks the n-WASP-ARP2/3 actin remodeling axis to facilitate cellular invasion. PloS Pathog. 16, e1008878. doi: 10.1371/journal.ppat.1008878
Ford, C., Nans, A., Boucrot, E., Hayward, R. D. (2018). Chlamydia exploits filopodial capture and a macropinocytosis-like pathway for host cell entry. PloS Pathog. 14, e1007051. doi: 10.1371/journal.ppat.1007051
Fregoso, F. E., van Eeuwen, T., Simanov, G., Rebowski, G., Boczkowska, M., Zimmet, A., et al. (2022). Molecular mechanism of Arp2/3 complex inhibition by arpin. Nat. Commun. 13, 628. doi: 10.1038/s41467-022-28112-2
Fu, Y., Galan, J. E. (1999). A Salmonella protein antagonizes rac-1 and Cdc42 to mediate host-cell recovery after bacterial invasion. Nature 401, 293–297. doi: 10.1038/45829
Garcia, A. M., Paredes, S. C., Schnoor, M. (2020). The role of the Arp2/3 inhibitory protein arpin in the regulation of endothelial barrier functions. Faseb J. 34. 1–1. doi: 10.1096/fasebj.2020.34.s1.02453
Ghosh, S., Ruelke, E. A., Ferrell, J. C., Bodero, M. D., Fields, K. A., Jewett, T. J. (2020). Fluorescence-reported allelic exchange mutagenesis-mediated gene deletion indicates a requirement for Chlamydia trachomatis tarp during In vivo infectivity and reveals a specific role for the c terminus during cellular invasion. Infect. Immun. 88, e00841–e00819. doi: 10.1128/IAI.00841-19
Gitsels, A., Sanders, N., Vanrompay, D. (2019). Chlamydial infection from outside to inside. Front. Microbiol. 10, 2329. doi: 10.3389/fmicb.2019.02329
Gottlieb, S. L., Martin, D. H., Xu, F., Byrne, G. I., Brunham, R. C. (2010). Summary: the natural history and immunobiology of Chlamydia trachomatis genital infection and implications for Chlamydia control. J. Infect. Dis. 201, S190–S204. doi: 10.1086/652401
He, L., Lin, Y., Ge, Z. H., He, S. Y., Zhao, B. B., Shen, D., et al. (2019). The Legionella pneumophila effector WipA disrupts host f-actin polymerisation by hijacking phosphotyrosine signalling. Cell Microbiol. 21, e13014. doi: 10.1111/cmi.13014
Hetrick, B., Han, M. S., Helgeson, L. A., Nolen, B. J. (2013). Small molecules CK-666 and CK-869 inhibit actin-related protein 2/3 complex by blocking an activating conformational change. Chem. Biol. 20, 701–712. doi: 10.1016/j.chembiol.2013.03.019
Hower, S., Wolf, K., Fields, K. A. (2009). Evidence that CT694 is a novel Chlamydia trachomatis T3S substrate capable of functioning during invasion or early cycle development. Mol. Microbiol. 72, 1423–1437. doi: 10.1111/j.1365-2958.2009.06732.x
Hybiske, K., Stephens, R. S. (2007). Mechanisms of host cell exit by the intracellular bacterium Chlamydia. PNAS 104, 11430–11435. doi: 10.1073/pnas.0703218104
Jarsch, I. K., Gadsby, J. R., Nuccitelli, A., Mason, J., Shimo, H., Pilloux, L., et al. (2020). A direct role for SNX9 in the biogenesis of filopodia. J. Cell Biol. 219, e201909178. doi: 10.1083/jcb.201909178
Jewett, T., Dooley, C., Mead, D., Hackstadt, T. (2008). Chlamydia trachomatis tarp is phosphorylated by src family tyrosine kinases. Biochem. Biophys. Res. Commun. 371, 339–344. doi: 10.1016/j.bbrc.2008.04.089
Jewett, T., Fischer, E., Mead, D., Hackstadt, T. (2006). Chlamydial TARP is a bacterial nucleator of actin. PNAS 103, 15599–15604. doi: 10.1073/pnas.0603044103
Jiwani, S., Alvarado, S., Ohr, R. J., Romero, A., Nguyen, B., Jewett, T. J. (2013). Chlamydia trachomatis tarp harbors distinct G and f actin binding domains that bundle actin filaments. J. Bacteriol. 195, 708–716. doi: 10.1128/JB.01768-12
Jiwani, S., Ohr, R. J., Fischer, E. R., Hackstadt, T., Alvarado, S., Romero, A., et al. (2012). Chlamydia trachomatis tarp cooperates with the Arp2/3 complex to increase the rate of actin polymerization. Biochem. Biophys. Res. Commun. 420, 816–821. doi: 10.1016/j.bbrc.2012.03.080
Keb, G., Ferrell, J. C., Scanlon, D., Jewett, T. J., Fields, K. A. (2021). Chlamydia trachomatis TmeA directly activates n-WASP to promote actin polymerization and functions synergistically with TarP during invasion. mBio 12, e02861–e02820. doi: 10.1128/mBio.02861-20
Keb, G., Fields, K. A. (2020). Markerless gene deletion by floxed cassette allelic exchange mutagenesis in Chlamydia trachomatis. J. Visualized Experiments JoVE 30. doi: 10.3791/60848
Keb, G., Hayman, R., Fields, K. A. (2018). Floxed-cassette allelic exchange mutagenesis enables marker-less gene deletion in Chlamydia trachomatis and can reverse cassette-induced polar effects. J. Bacteriol. 200, 00418, e00479. doi: 10.1128/JB.00479-18
Keestra, A. M., Winter, M. G., Auburger, J. J., Frässle, S. P., Xavier, M. N., Winter, S. E., et al. (2013). Manipulation of small rho GTPases is a pathogen-induced process detected by NOD1. Nature 496, 233–237. doi: 10.1038/nature12025
Kelly, A. E., Kranitz, H., Dötsch, V., Mullins, R. D. (2006). Actin binding to the central domain of WASP/Scar proteins plays a critical role in the activation of the Arp2/3 complex. J. Biol. Chem. 281, 10589–10597. doi: 10.1074/jbc.M507470200
Kreisel, K. M., Weston, E. J., St Cyr, S. B., Spicknall, I. H. (2021). Estimates of the prevalence and incidence of Chlamydia and Gonorrhea among US men and women, 2018. Sex Transm. Dis. 48, 222–231. doi: 10.1097/OLQ.0000000000001382
McKuen, M. J., Mueller, K. E., Bae, Y. S., Fields, K. A. (2017). Fluorescence-reported allelic exchange mutagenesis reveals a role for Chlamydia trachomatis TmeA in invasion that is independent of host AHNAK. Infect. Immun. 85, e00640–e00617. doi: 10.1128/IAI.00640-17
Mehlitz, A., Banhart, S., Hess, S., Selback, M., Meyer, T. F. (2008). Complex kinase requirements for Chlamydia trachomatis tarp phosphorylation. FEMS Microbiol. Lett. 289, 233–240. doi: 10.1111/j.1574-6968.2008.01390.x
Michard, C., Sperandio, D., Baïlo, N., Pizarro-Cerdá, J., LeClaire, L., Chadeau-Argaud, E., et al. (2015). The Legionella kinase LegK2 targets the ARP2/3 complex to inhibit actin nucleation on phagosomes and allow bacterial evasion of the late endocytic pathway. mBio 6, e00354–e00315. doi: 10.1128/mBio.00354-15
Mirrashidi Kathleen, M., Elwell Cherilyn, A., Verschueren, E., Johnson Jeffrey, R., Frando, A., Von Dollen, J., et al. (2015). Global mapping of the inc-human interactome reveals that retromer restricts Chlamydia infection. Cell Host Microbe 18, 109–121. doi: 10.1016/j.chom.2015.06.004
Mostowy, S., Shenoy, A. R. (2015). The cytoskeleton in cell-autonomous immunity: structural determinants of host defence. Nat. Rev. Immunol. 15, 559–573. doi: 10.1038/nri3877
Mueller, K. E., Fields, K. A. (2015). Application of β-lactamase reporter fusions as an indicator of protein secretion during infections with the obligate intracellular pathogen Chlamydia trachomatis. PloS One 10, e0135295. doi: 10.1371/journal.pone.0135295
Mueller, K. E., Plano, G. V., Fields, K. A. (2014). New frontiers in type III secretion biology: the Chlamydia perspective. Infection Immun. 82, 2–9. doi: 10.1128/IAI.00917-13
Mueller, K. E., Wolf, K., Fields, K. A. (2016). Gene deletion by fluorescence-reported allelic exchange mutagenesis (FRAEM) in Chlamydia trachomatis. mBio 7, e01817–e01815. doi: 10.1128/mBio.01817-15
Mueller, K. E., Wolf, K., Fields, K. A. (2017). Chlamydia trachomatis transformation and allelic exchange mutagenesis. Curr. Protocol Microbiol. 45, 11A.13.11–11A.13.15. doi: 10.1002/cpmc.31
Padrick, S. B., Doolittle, L. K., Brautigam, C. A., King, D. S., Rosen, M. K. (2011). Arp2/3 complex is bound and activated by two WASP proteins. Proc. Natl. Acad. Sci. U.S.A. 108, E472–E479. doi: 10.1073/pnas.1100236108
Pais, S. V., Milho, C., Almeida, F., Mota, L. J. (2013). Identification of novel type III secretion chaperone-substrate complexes of Chlamydia trachomatis. PLoS One 8, e56292. doi: 10.1371/journal.pone.0056292
Rotty, J. D., Wu, C., Bear, J. E. (2013). New insights into the regulation and cellular functions of the ARP2/3 complex. Nat. Rev. Mol. Cell Biol. 14, 7–12. doi: 10.1038/nrm3492
Schachter, J. (1999). “Infection and disease epidemiology,” in Chlamydia; intracellular biology, pathogenesis, and immunity, vol. p . Ed. Stephens, R. S. (Washington, D.C: ASM Press), 139–169.
Scidmore, M. A. (2005). Cultivation and laboratory maintenance of Chlamydia trachomatis. Curr. Protocol Microbiol. 11A.1. doi: 10.1002/9780471729259.mc11a01s00
Spona, D., Hanisch, P. T., Hegemann, J. H., Mölleken, K. (2023). A single chlamydial protein reshapes the plasma membrane and serves as recruiting platform for central endocytic effector proteins. Commun. Biol. 6, 520. doi: 10.1038/s42003-023-04913-z
Wang, Y., Kahane, S., Cutcliffe, L. T., Skilton, R. J., Lambden, P. R., Clarke, I. N. (2012). Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PloS Pathog. 7, e1002258. 11A.1. doi: 10.1371/journal.ppat.1002258
Keywords: type III secretion, cytoskeleton, invasion, actin, pathogenesis
Citation: Scanlon KR, Keb G, Wolf K, Jewett TJ and Fields KA (2023) Chlamydia trachomatis TmeB antagonizes actin polymerization via direct interference with Arp2/3 activity. Front. Cell. Infect. Microbiol. 13:1232391. doi: 10.3389/fcimb.2023.1232391
Received: 31 May 2023; Accepted: 26 June 2023;
Published: 07 July 2023.
Edited by:
Chunfu Yang, Southern University of Science and Technology, ChinaReviewed by:
Stéphanie M. Pontier, Cégep de l’Outaouais, CanadaCopyright © 2023 Scanlon, Keb, Wolf, Jewett and Fields. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kenneth A. Fields, S2VuLmZpZWxkc0B1a3kuZWR1
†Present address: Gabrielle Keb, Charles River Laboratories, Mattawan, MI, United States
‡These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.