- 1Department of Pharmacy, Faculty of Allied Health Sciences, Daffodil International, University, Dhaka, Bangladesh
- 2Professor Joarder DNA and Chromosome Research Laboratory, Department of Genetic Engineering and Biotechnology, University of Rajshahi, Rajshahi, Bangladesh
- 3Department of Microbiology, Jagannath University, Dhaka, Bangladesh
- 4Department of Genetic Engineering and Biotechnology, University of Rajshahi, Rajshahi, Bangladesh
- 5Laboratory of Computational Research for Drug Design and Material Science, Department of Chemistry, European University of Bangladesh, Dhaka, Bangladesh
- 6Department of Pharmaceutical Sciences, School of Health and Life Sciences. North South University, Dhaka, Bangladesh
- 7Department of Pharmacy, International Islamic University Chittagong, Chittagong, Bangladesh
- 8Department of Bioinformatics and Computational Biology, Gaziantep University, Gaziantep, Türkiye
- 9Department of Food Science, Faculty of Agricultural and Food Sciences, Laval University, Quebec, QC, Canada
- 10Department of Pharmaceutics, College of Pharmacy, King Saud University, Riyadh, Saudi Arabia
- 11Department of Biology, Bahir Dar University, Bahir Dar, Ethiopia
- 12Department of Chemistry and Biochemistry, Faculty of Medicine and Pharmacy, Ibn Zohr University, Laayoune, Morocco
Tick-borne Babesiosis is a parasitic infection caused by Babesia microti that can infect both animals and humans and may spread by tick, blood transfusions, and organ transplantation. The current therapeutic options for B. microti are limited, and drug resistance is a concern. This study proposes using computational drug design approaches to find and design an effective drug against B. microti. The study investigated the potentiality of nine natural compounds against the pathogenic human B. microti parasite and identified Vasicinone and Evodiamine as the most promising drugs. The ligand structures were optimized using density functional theory, molecular docking, molecular dynamics simulations, quantum mechanics such as HOMO–LUMO, drug-likeness and theoretical absorption, distribution, metabolism, excretion, and toxicity (ADMET), and pharmacokinetics characteristics performed. The results showed that Vasicinone (−8.6 kcal/mol and −7.8 kcal/mol) and Evodiamine (−8.7 kcal/mol and −8.5 kcal/mol) had the highest binding energy and anti-parasitic activity against B. microti lactate dehydrogenase and B. microti lactate dehydrogenase apo form. The strongest binding energy was reported by Vasicinone and Evodiamine; the compounds were evaluated through molecular dynamics simulation at 100 ns, and their stability when they form complexes with the targeted receptors was determined. Finally, the pkCSM web server is employed to predict the ADMET qualities of specific molecules, which can help prevent negative effects that arise from taking the treatment. The SwissADME web server is used to assess the Lipinski rule of five and drug-likeness properties including topological polar surface area and bioavailability. The Lipinski rule is used to estimate significant drug-likeness. The theoretical pharmacokinetics analysis and drug-likeness of the selected compounds are confirmed to be accepted by the Lipinski rule and have better ADMET features. Thus, to confirm their experimental value, these mentioned molecules should be suggested to carry out in wet lab, pre-clinical, and clinical levels.
1 Introduction
Babesia are intra-erythrocytic protozoa that belong to the same species of Apicomplexa as Plasmodia. Hard-bodied ticks spread them worldwide and can infect several vertebrates, including humans (Bloch et al., 2019). Among the more than 100 Babesia species, B. microti is predominant can infect humans, and is prevalent throughout the world. It is a significant cause of endemic illness in China and the United States (US) (Kumar et al., 2021). B. microti is primarily transmitted by Ixodes scapularis, also known as the deer tick or black-legged tick (Curcio et al., 2016). Additionally, they can spread through blood transfusions and, less commonly, through organ transplants and prenatal transfer (Puri et al., 2021).
The most acute disease caused by the parasite is babesiosis, in which RBC is infected and destroyed. Fever (up to 41°C or 106°F), arthralgia, anorexia, nausea, headaches, and exhaustion are the most frequent symptoms. According to the severity, physical examination results may include hepatosplenomegaly, ocular hemorrhage, and pharyngeal erythema. Hemolytic anemia and thrombocytopenia may be consistent with typical laboratory results (Vanniere, 2008; Ortiz et al., 2020; Bastos et al., 2021). Babesia parasites in red blood cells in a blood smear can be detected under a microscope to make the diagnosis (Ortiz et al., 2020; Singh et al., 2023).
In 1888, a Hungarian pathologist and microbiologist named Victor Babes discovered that intraerythrocytic bacteria were the root of febrile hemoglobinuria in cattle (Babes, 1888; Vannier and Krause, 2012). Approximately 50 years later, the first known human case of babesiosis was discovered, and a splenectomized Croatian herdsman who had died of infection later was associated with B. divergens (Skrabalo and Deanovic, 1957; Vannier and Krause, 2012). In 1969, on Nantucket Island, off the coast of Massachusetts, the first case of B. microti infection was discovered in an immunocompetent person (Western et al., 1970; Vannier and Krause, 2012).
There has been an increase in B. microti infections worldwide in recent decades. More than 2,000 instances are reported yearly in the US, although the actual number is likely far higher. Human babesiosis has the most significant incidence in areas with high temperatures. The most dominating species, B. microti, is indigenous to southwestern China, and the northeastern and northern mid-western US (Vannier and Krause, 2012; Zhou et al., 2014; Fang et al., 2015; Krause, 2019; Kumar et al., 2021). In many parts of the world, babesiosis is now acknowledged as an emerging health concern. To date, babesiosis is categorized as a nationally notifiable disease and is acknowledged as a rising health issue in many parts of the world (Jones et al., 2008; Aguero-Rosenfeld, 2011; Vannier and Krause, 2012).
Human infections with B. microti may manifest clinically, ranging from asymptomatic infection to a severe, rapid deadly disease (Gorenflot et al., 1998). About one in five cases of babesiosis spread by blood transfusions has fatal consequences. Immunocompetent people can be infected with B. microti for up to 2 years without developing symptoms. B. microti parasitemia has been found in immunocompetent hosts for over a year who received a standard course of antibiotic therapy for more than 2 years in untreated patients (Krause et al., 1998; Moritz et al., 2016; Abraham et al., 2018). Immunocompromised patients, such as those with asplenia, HIV/AIDS, cancer, or who are on immunosuppressive medications, frequently have severe diseases that can return up to 2 years even after the treatment (Bloch et al., 2019). The severity of the illness is much higher and linked to an increased death rate in those without a spleen, as it is the first immunological barrier for babesia after transmission to the human body (Rosner et al., 1984; Krause et al., 1998; Zintl et al., 2003; Mebius and Kraal, 2005; Krause et al., 2007; Vannier and Krause, 2012; Bloch et al., 2019).
The treatment options for disease caused by B. microti include clindamycin and quinine or atovaquone and azithromycin (the preferred drug combination) (Kumar et al., 2021). However, it can produce resistance to even these two most standard combinations of drugs. The resistance to atovaquone and azithromycin is due to genetic mutation of the targeting protein of the drugs. Atovaquone’s target is the Babesia mitochondrial electron transport chain’s cytochrome B protein, while azithromycin’s target is the apicoplast protein, which prevents protein translation in these organelles (Lemieux et al., 2016; Simon et al., 2017; Bloch et al., 2019).
Due to the drug resistance and lack of a strong and permanent cure and to avoid the side effects of using broad-spectrum high-power antibiotics, the search for new drugs with minimal toxicity and high effectiveness is still ongoing. The introduction of novel therapeutic interventions, such as new anti-parasitic medications or drug combinations, enhanced anti-parasitic drug duration strategies, or immunoglobulin preparations, and also novel preventative techniques, can reduce the health burden of chronic and recrudescent babesiosis (Abraham et al., 2018; Bloch et al., 2019).
However, searching for a new drug requires a lot of resources, time, and cost to perform pre-clinical or clinical trials. Still, this does not ensure that the drugs will be available to the market. Since many adverse/side effects and toxicity are revealed during clinical or pre-clinical trials, advanced computational drug design approaches are necessary to find and design an effective drug against B. microti. This method is the best choice for fast hit identification and hit-to-lead selection against a specific pathogen or disease. Also, the time efficiency and low cost are the advantages of this effective approach to finding and designing a new pharmacologically active substance. Moreover, before going to an experimental study, computational investigation may determine the theoretical toxicity, which can also reduce the chance of drug failure during the experimental studies (Kapetanovic, 2008).
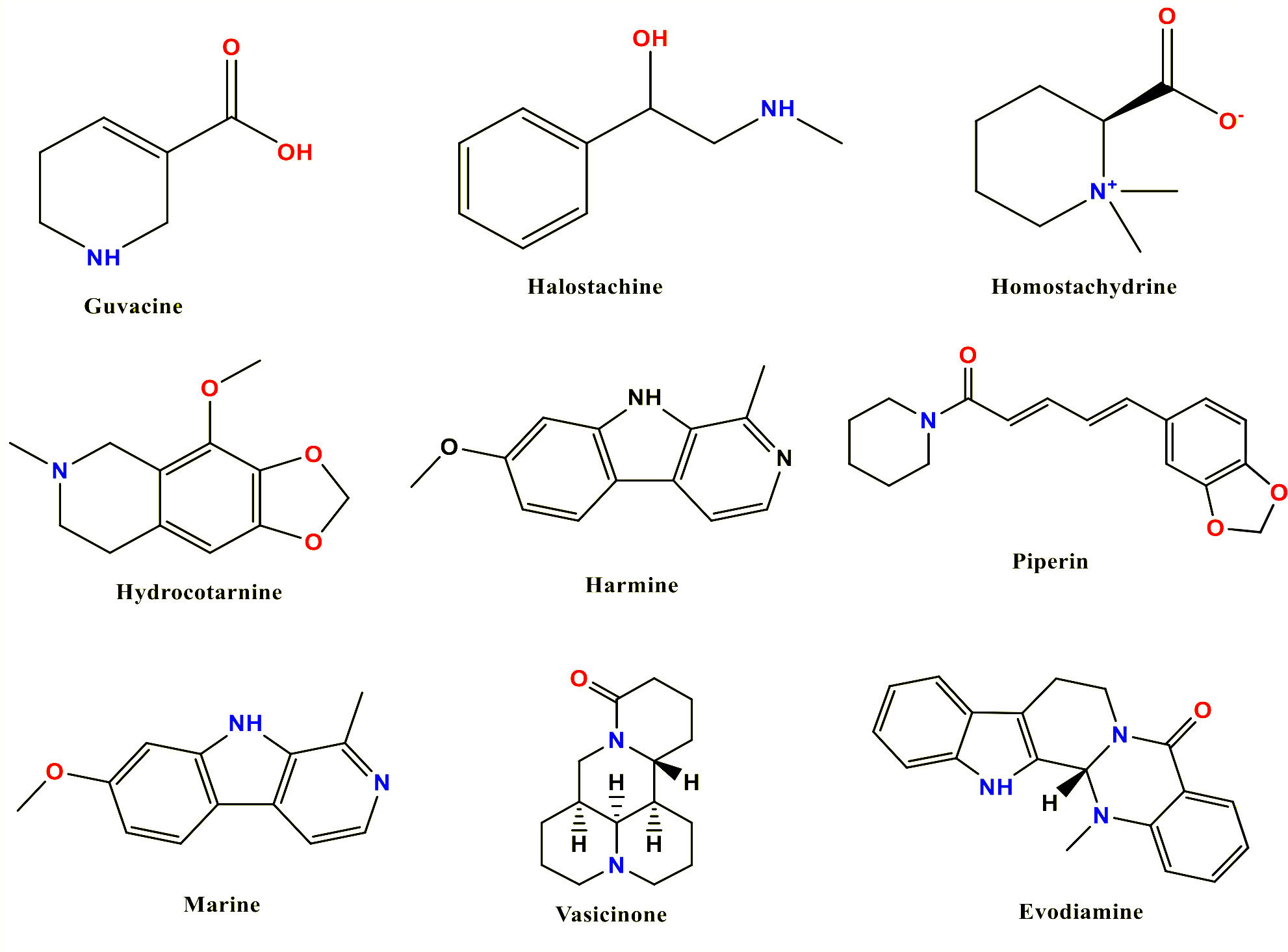
Alkaloids are a class of nitrogenous organic compounds that have a numerous range of physiological activity. The primary function of alkaloids in plants is toxin production against diseases and predators. Although plant alkaloid toxicity can be diverse, it frequently involves neurotoxicity or disruption of the cell signaling cascade. The same toxic characteristics of the plant’s defense mechanism are frequently exploited while searching for new drugs. Particular toxicities of some alkaloids may be utilized to fight certain cancerous cell types or to eradicate certain pests or microorganisms [25]. For this study, we have selected a few natural alkaloids as potential inhibitors of B. microti for the rapid development of effective and sustainable therapeutic options targeting the pathogen. In addition to numerous other activities expressed by alkaloids, they demonstrated anti-inflammatory, anticancer, analgesic, local anesthetic and pain alleviation, neuropharmacological, antibacterial, and antifungal effects, and they are also included as vitamins and diet ingredients (Kurek, 2019).
2 Materials and methods
2.1 Molecular optimization by density functional theory
Selected alkaloid’s SDF-formatted structure files were fetched from the PubChem database for optimization (Kim et al., 2019). To achieve the configuration with the lowest ground-state energy, we optimized the geometrical arrangement of the molecules and rearranged the atomic components using density functional theory (DFT) (Orio et al., 2009). This is essential since molecular docking and molecular dynamics (MD) simulation demand ligands with an optimal geometrical arrangement. For geometry optimization, all ligand structures were imported to the Gaussian 09W program and the optimization procedure was completed at DFT/B3LYP/6-31G (d’p’) levels (Rajalakshmi et al., 2015; Cao et al., 2018). After optimization, the electron acceptor and electron donor properties were determined by calculating the frontier orbital energies, including the highest occupied molecular orbital (HOMO) and the lowest unoccupied molecular orbital (LUMO), as well as the energy gap, hardness, and softness. These characteristics also provide information regarding the chemical reactivity and stability of compounds (Kumer et al., 2019). Finally, optimization molecules were saved in pdb format and again uploaded in the Material Studio program to design three-dimensional structures. Figure 1 displays the 2D chemical structure (Sharma et al., 2019).
2.2 Protein preparation and molecular docking study and visualization
To investigate and evaluate the molecular interactions between proteins and ligands at their binding sites, several bioinformatics approaches have been established. In our study, we used molecular docking, a bioinformatics tool that is extensively employed in medicinal chemistry research for the development of new therapeutic molecules that target key virulence factors for particular diseases (Vilar et al., 2008). Polar hydrogens and Kohlman charges were added before docking, and they were followed by the removal of water molecules and previously attached undesirable compounds to make it more straightforward for the ligand to be docked at its most advantageous location (Figure 2). We implemented the docking technique using PyRx free package v0.8 integrated with AutoDock Vina, assuming the entire protein surface is a receptor (Dallakyan et al., 2015). The program’s exhaustiveness parameter was set at 8 to obtain the most optimal docking position with the lowest possible margin of error. Ligands were perceived as flexible during docking. However, proteins have been perceived as stiff. Ultimately, protein-ligand docked compounds have been investigated and visualized using Pymol software and the Discovery studio program.
2.3 Determination of ADMET, Lipinski rule, and pharmacokinetics
Predicting ADMET qualities for certain lead compounds during the development of innovative drugs helps to prevent any negative effects that arise from taking the treatment (Kesharwani et al., 2020). By excluding compounds with subpar ADMET characteristics from the pipeline for medication development, this strategy can also reduce overall costs. As a result, we have estimated the critical ADMET features for our selected compounds using the pkCSM web server (https://biosig.lab.uq.edu.au/pkcsm/prediction), which works with novel graph-based signatures by employing a predictive model to compute the ADMET values for specific molecules (Pires et al., 2015). This platform is recognized for providing an efficient assessment of pharmacokinetic and toxicological characteristics to reflect the chemistry and structure of medicinal compounds. Target molecules have been loaded into the pkCSM query bar using the standard Simplified Molecular Input Line Entry System (SMILES), and the anticipated critical ADMET characteristics were recorded.
On the other hand, the commonly applied Lipinski’s rule of five is a fantastic guideline for assessing the pharmacological and biological activity of drug-like compounds (Lipinski, 2004). As a result, we assessed the Lipinski rule through the SwissADME web server (http://www.swissadme.ch/) along with estimating significant drug-like characteristics like Topological Polar Surface Area (TPSA) and bioavailability (Daina et al., 2017).
2.4 Molecular dynamics simulation
Yet Another Scientific Artificial Reality Application (YASARA) dynamics software was used to perform MD simulation using Assisted Model Building with Energy Refinement (AMBER) 14 force field (Wang and Wolf, 2004; Krieger and Vriend, 2014). The docked complexes were the earliest cleanup and optimized, and optimization of the hydrogen bond network was also performed. GAFF (general AMBER force field) and assigning AM1BCC charges were used to generate the topology files of ligands. A cubic simulation cell was created by the TIP3P solvation model having a periodic boundary condition (Harrach and Drossel, 2014). To neutralize the simulated system, the physiological conditions were set at 0.9% NaCl, 310 K, and pH 7.4 (Krieger and Vriend, 2015). To determine the long-range electrostatic interaction with an 8.0-Å cutoff radius, the particle mesh Ewald (PME) method was used (Essman et al., 1995). A time step of 2.0 fs was chosen for the simulation. By using simulated annealing techniques, the primary energy minimization was carried out using the steepest gradient algorithms (5,000 cycles). From the simulated trajectories, the RMSD, Rg, SASA, hydrogen bond, and Molecular Mechanics/Position-Boltzmann Surface Area (MMPBSA) binding free energy were evaluated. The simulation was conducted for 100 ns, and the simulation trajectories were captured after every 100 ps (Cheung and Sundram, 2017; Baildya et al., 2021; Patil et al., 2021).
3 Results and discussion
3.1 Lipinski rule and pharmacokinetics
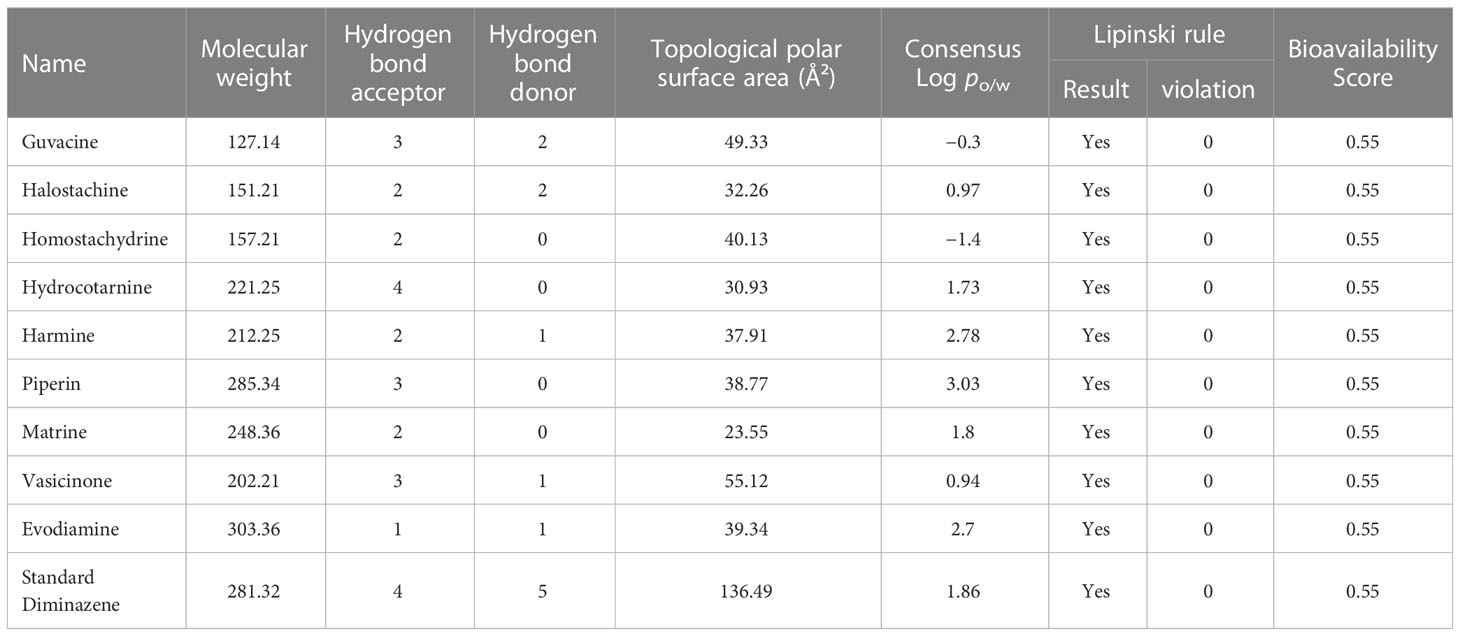
The structure files of selected ligands and standard Diminazene were uploaded to the SwissADME web server to generate their respective canonical SMILES so that these SMILES can be used to obtain respective pharmacokinetics characteristics and Lipinski’s rule statement.
According to the results generated by SwissADME, all of the selected nine alkaloids and Diminazene follow Lipinski’s rule with no violations (Table 1). The rule states that the molecular weight for any drug-like molecules must not cross 500 Daltons. We can observe that all of our selected alkaloids have molecular weights of <500 Daltons. Moreover, none of the compounds has more than five hydrogen bond donors; in fact, only Diminazene presented five hydrogen bond donors, whereas the remaining compounds have a maximum of 2 hydrogen bond donors (Guvacine and Halostachine). In addition, the number of hydrogen bond acceptors for all the compounds did not cross 10, which is acceptable according to the Lipinski rule. Ultimately, Consensus Log Po/w values for potential lead molecules were also assessed and were found to be in an acceptable range. The TPSA is an effective method to estimate polar surface area since it eliminates the requirement to determine the necessary biological conformation or conformations or determine the drug’s three-dimensional arrangement (Prasanna and Doerksen, 2009). However, a TPSA score less than 140 Ų is ideal for drug-like molecules; here, all our selected alkaloids followed this threshold (Prasanna and Doerksen, 2009). Finally, the bioavailability scores were also calculated, revealing that the selected nine alkaloid compounds and Diminazene had acceptable (0.55) bioavailability scores.
3.2 Molecular docking analysis against targeted receptor of Babesia microti
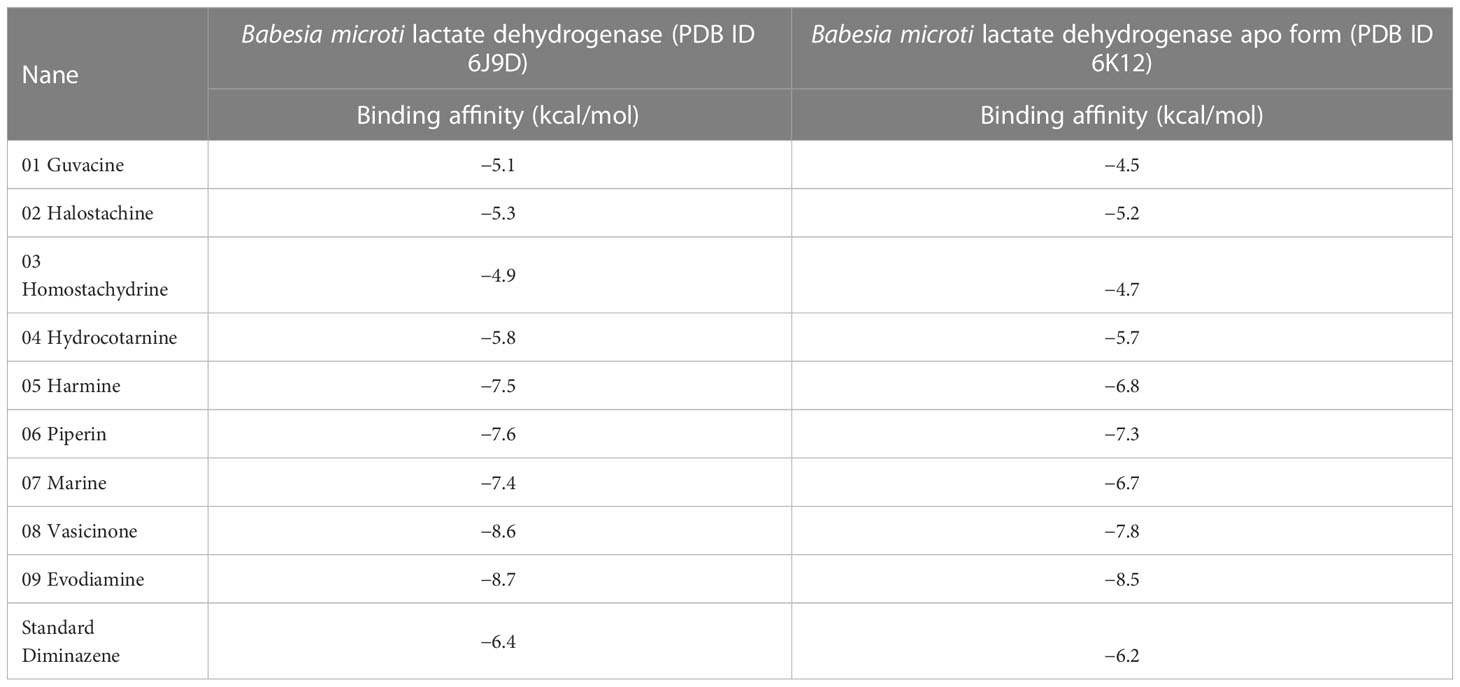
Pharmacological research and development must comprehend the mechanisms by which small-molecule ligands identify and engage with proteins. Therefore, molecular docking is most frequently used to achieve this objective as it offers investigations of important molecular processes, such as ligand binding mechanisms and the related intermolecular interactions that maintain the ligand–receptor complex (Ferreira et al., 2015). Babesia microti lactate dehydrogenase and B. microti lactate dehydrogenase apo form were selected as target macromolecules for molecular docking against the selected nine (Guvacine, Halostachine, Homostachydrine, Hydrocotarnine, Harmine, Piperin, Matrine, Vasicinone, and Evodiamine) natural compounds and standard drug (Diminazene). The primary objective of performing this step was to identify small molecules with satisfactory binding affinity to the selected receptors so that they could be considered for MD simulations.
Diminazene exhibited moderate binding affinity towards selected receptor macromolecules. On the contrary, our five (Harmine, Piperin, Matrine, Vasicinone, and Evodiamine) selected alkaloids revealed better binding affinity than Diminazene. Evodiamine expressed the most satisfactory docking outcome among all selected compounds, with a binding affinity of −8.7 kcal/mol for B. microti lactate dehydrogenase and −8.5 kcal/mol for B. microti lactate dehydrogenase apo form. Meanwhile, the docking outcome for Vasicinone was also noticeable as it had a binding affinity of −8.6 kcal/mol for B. microti lactate dehydrogenase and −7.8 kcal/mol for B. microti lactate dehydrogenase apo form. These two small molecules were selected for further investigation for expressing consistent results for both receptors even though Harmine, Piperin, and Matrine also conveyed promising results (Table 2).
3.3 Molecular docking pose and interaction analysis
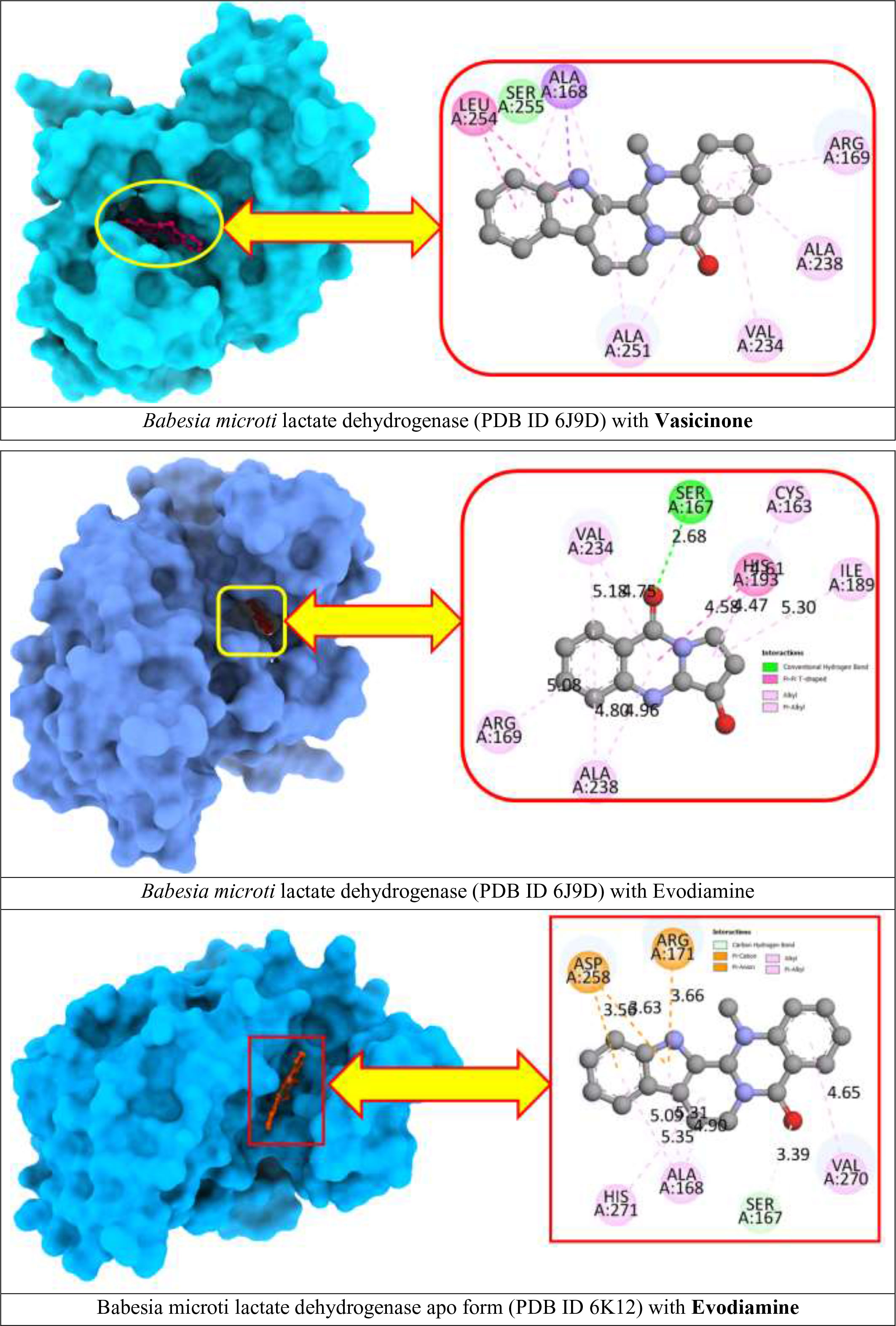
To better understand the molecular interaction between selected compounds and receptor macromolecules, we decided to visualize ligand–protein docked complexes in BIOVIA Discovery Studio Visualizer. From the previous step, Vasicinone and Evodiamine expressed the strongest binding affinity for both receptor proteins since they showed the better binding energy scores. Hence, ligand–protein complexes (Figure 3) for these two compounds are thoroughly assessed in this step.
B. microti lactate dehydrogenase docked with Evodiamine showed four different types of interactions that include the following: Conventional Hydrogen Bond with SER A:167 at 2.68 Å distance; Pi-Pi T-shaped bond with HIS A:193 at 4.58 Å distance; Alkyl and Pi-Alkyl bond with CYS A:163, ARG A:169, ILE A:189, VAL A:234, and ALA A:238. On the other hand, Evodiamine docked with B. microti lactate dehydrogenase apo form macromolecule showed different interaction profiles with five different types of interactions. The ligand formed a Carbon–Hydrogen bond with SER A:167 at 3.39 Å distance; a Pi–Cation bond and Pi–Anion bond with ARG A:171 and ASP A:258 at reasonable distances; and Alkyl and Pi–Alkyl bond with ALA A:168, VAL A:270, and HIS A:271. Moreover, visualization for B. microti lactate dehydrogenase docked with Vasicinone also showed strong interactions with ALA A:168, ARG A:169, VAL A:234, ALA A:238, ALA A:251, LEU A:254, and LEU A:255.
3.4 Molecular dynamics simulation
To determine the stability of hit complexes, an MD study was conducted at the 100-ns simulation period. Two best-hit compounds (Vasicinone and Evodiamine) along with apoprotein and standard Diminazene were used for performing 100-ns dynamic simulation to confirm whether the ligands were sustaining at the protein’s active site. The root mean square deviation (RMSD), the radius of gyration (Rg), solvent accessible surface area (SASA), hydrogen bonds, and MMPBSA binding free energy of the hit complexes were examined.
The RMSD value is used to determine the standard measure of structural distance between coordinates. In our findings, all complexes exhibited satisfactory RMSD values with an average of 2.43, 3.80, 1.69, and 3.03 Å for C1 (Evodiamine complex), C2 (Vasicinone complex), C3 (Docked protein), and C4 (Standard Diminazene), respectively. Initially, all complexes showed increasing movement of RMSD value until 15 ns, and thereafter they maintain their stability during the simulation period with different RMSD profiles (Figure 4A).

Figure 4 The molecular dynamics simulation of C1, C2, C3, and C4 complexes, where (A–D) designate RMSD, Rg, SASA, and hydrogen bond respectively.
During the 10 ns simulation period, the Rg profile of C2 and C4 complexes followed an increasing trend whereas C1 exhibited decreasing movement. Then, all three complexes C1, C2, and C4 showed stability and almost similar profiles until 65 ns. However, C1 and C2 met nearly the same point whereas C4 showed increasing movement at the end of the simulation period. Conversely, the C3 complex showed a higher and different Rg profile than the other three complexes although it maintains stability throughout the simulation period (Figure 4B).
From the very beginning to the end of the simulation period, all four complexes displayed amazing stability in terms of SASA profile; however, the C3 complex showed comparatively higher SASA value than other complexes (Figure 4C). Similarly, the hydrogen bond of all four complexes exhibited extraordinary stability and almost similar profile except the C3 complex during the 100-ns simulation period due to the folding and change in the confirmation of the complexes (Figure 4).
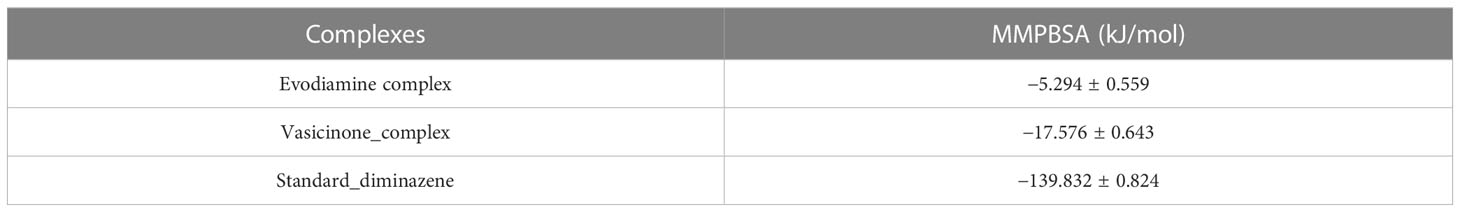
MMPBSA was used to calculate the binding free energy of interactions between protein–ligand complexes. Figure 5 shows the MMPBSA binding free energy of three complexes at the 100-ns simulation period. The average MMPBSA binding free energies of the Evodiamine complex, Vasicinone complex, and standard Diminazene complexes were −5.294 ± 0.559, −17.576 ± 0.643, and −139.832 ± 0.824 kJ/mol, respectively (Table 3). The negative MMPBSA energy confirms that the ligands were sustained at the protein’s active site.

Figure 5 The MMPBSA binding free energy of the Evodiamine complex, Vasicinone complex, and Standard Diminazene complexes. The overall MD simulation study suggests that these two hit complexes could be used as a lead compound.
3.5 Dynamic behavior and confirmational change of protein–ligand complex
Correlative motions play an essential role in recognizing and interacting with bio-macromolecular systems, which can be attained through the covariance equation of molecular perturbation generated by the MD simulation trajectory. Using the dynamic cross-correlation matrix (DCCM), the relative motions of various simulation systems were analyzed. The conformational alterations of the Evodiamine complex, Vasicinone complex, docked protein, and standard Diminazene were further analyzed using DCCM analysis. The DCCM fluctuations are illustrated in Figure 6, which depicts time-correlated information among the protein residues.

Figure 6 Ca-residue cross-correlation profiles for the Evodiamine complex (A), Vasicinone complex (B), docked protein (C), and the standard Diminazene (D).
The greater the color intensity, the more positively or negatively correlated motions between structures are highlighted. The color yellow represents a significant positive correlation, while the color blue represents a negative correlation.
The dynamic cross-correlation map analysis was utilized to visualize the residual correlative motion in protein structures for four entities, namely, the Evodiamine complex (A), the Vasicinone complex (B), the docked protein (C), and the standard Diminazene (D). Strong correlative motions (1 to 1) are depicted on the right side of each panel using a four-dimensional caricature model. In every instance, correlations were depicted as a spectrum of colors from red to blue. The yellow color is indicative of a positive correlation between two residues, implying that both residues exhibit synchronous movement. In contrast, residues with opposite motion are designated as having anti-correlative motion and are colored blue.
The docked protein increased both correlated and anti-correlated motions in PKD1; however, it generated random motions in the majority of the crucial motifs. Positive and negative correlations were found to be stronger inbound proteins, particularly in the interloop residues and beta-sheet. The anti-correlated motion was detected in the residue range of 227 to 243 for the Vasicinone complex protein. Conversely, the Evodiamine complex protein exhibited a reduction in anti-correlated motion when compared to standard Diminazene.
3.6 ADMET data analysis
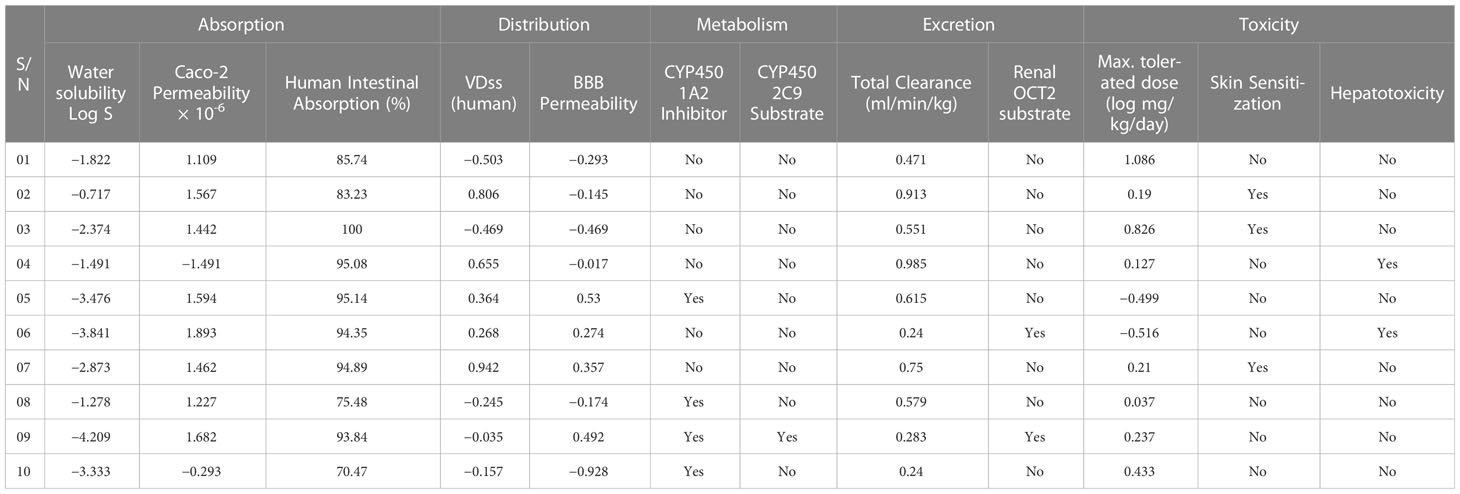
Predicting ADMET properties plays a critical role in ensuring the overall efficacy and safety of therapeutic molecules as more than 50% of drugs fail to reach clinical trials for not having proper ADMET attributes. pkCSM using its novel graph-based method predicted important ADMET properties for the selected therapeutic molecules (Table 4).
Drugs molecules’ absorption ability plays a critical role in their successful application. Therefore, in the absorption panel, we have paid close attention to important absorption properties such as “water solubility”, “Caco-2 permeability”, and “Human intestine absorption”. Therapeutic molecules with proper water solubility competence have high absorption in the system while the water solubility (calculated in Log S) ability can be determined from the following range: Insoluble < −10 poorly < −6, moderately < −4 soluble < −2 very < 0 < highly (Lipinski et al., 1997; Lipinski and Methods, 2000). Diminazene (compound no. 10) and compounds 03, 05, 06, and 07 were predicted as soluble in water. In addition to this, compounds 01, 02, 04, and 08 showed better water solubility than the previous group and were declared as “very soluble”. Only compound 09 was predicted as moderately soluble for having a water solubility score below −4.00 Log S. Absorption properties for orally administrated drugs are assessed by the substance’s ability to cross the Caco-2 cell line (established by Human colorectal adenocarcinoma epithelial cells) (Kumar et al., 2010). High Caco-2 permeability (value >0.9) denotes a high absorption capacity of drugs. According to the server’s predictive model, Diminazene expressed low Caco-2 permeability along with compound 04 whereas all the remaining compounds had high Caco-2 permeability. However, human intestine absorption was also predicted where Diminazene had the lowest absorption rate (70.476%). Interestingly, all our selected natural alkaloids had excellent human intestine absorption percentages (much higher than the standard drug); compound 03 had the most promising outcome (100%).
Drug distribution is the process of unmetabolized drug movement via the body’s blood and tissues. This process bears massive significance for being an indicator of the efficacy or toxicity of a drug. For the distribution panel, we have considered two properties: Volume of distribution (VDss) and Blood–Brain Barrier (BBB) permeability. A higher VDss value indicates renal failure and dehydration for the drug being distributed in the tissue rather than plasma. According to the server’s guideline, the VDss value < −0.15 indicates low, and >0.45 VDss value is considered high. Conforming to the prediction, Diminazene and compounds 01, 03, and 08 have low VDss scores while compounds 02, 04, and 07 have high VDss scores. In addition to this, compounds 05, 06, and 09 can have moderate VDss scores as their score fell between the ranges of high and low VDss scores. Our brain is protected with BBB from any non-native compounds; that is why predicting any drug’s ability to penetrate BBB could be an essential property to be considered (Daneman and Prat, 2015). Poor dispersion is indicated by a BBB permeability value of < −1, whereas a score of >0.3 denotes strong BBB permeability. Compounds 01, 02, 03, and 08 and Diminazene expressed poor BBB permeability while compounds 05, 07, and 09 were declared as high BBB permeable. The human liver contains the detoxification enzyme family, which includes cytochrome P450, which is crucial for efficient drug metabolism(Ogu and Maxa, 2000). Cytochrome P450 can be inhibited by several drugs. Therefore, it is crucial to carefully consider how well the chemicals can block this enzyme. For our experiment, the inhibition values of two cytochrome P450 isoforms—CYP450 1A2 and CYP450 2C9—were calculated. Compounds 05, 08, and 09, and Diminazene inhibited CYP450 1A2 activity while the remaining substances did not inhibit this enzyme. Besides, none of the compounds were reported to inhibit CYP450 2C9 enzyme.
Any drug-like molecules should have an acceptable excretion profile, which primarily depends on two major properties: total clearance and Renal OCT2 substrate identification. Based on the combined data of hepatic and renal clearance, total clearance produces a score and displays a distinct excretion profile for each medicine (Dowd et al., 2010). The maximum total clearance score was expressed by compound 04 and the minimum score was calculated from compound 06 and Diminazene. Meanwhile, the kidney’s ability to remove drugs, as well as additional medications from the body, depends on the renal transporter OCT2. Compounds 06 and 09 were declared as potential OCT2 substrates while the remaining compounds represented negative results.
Finally, toxicity prediction revealed that compounds 02, 03, and 07 can induce skin sensitization, and compounds 04 and 06 might provoke hepatotoxicity; therefore, precautions should be taken before applications of these selected compounds. For interpreting results for maximum tolerated dose (MTD), a value less than or equal to 0.477 indicates low MTD and a value greater than 0.477 denotes high MTD. In keeping with the given range, compounds 01 and 03 represented high MTD and the rest of the compounds had low MTD.
3.7 Frontier molecular orbital and molecular properties analysis
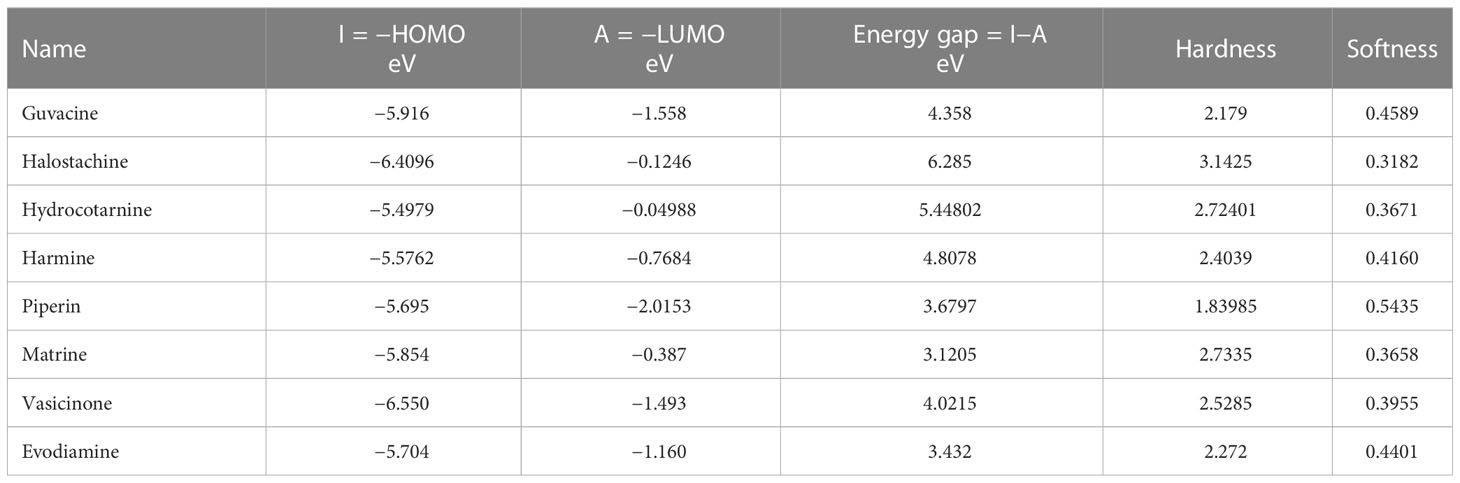
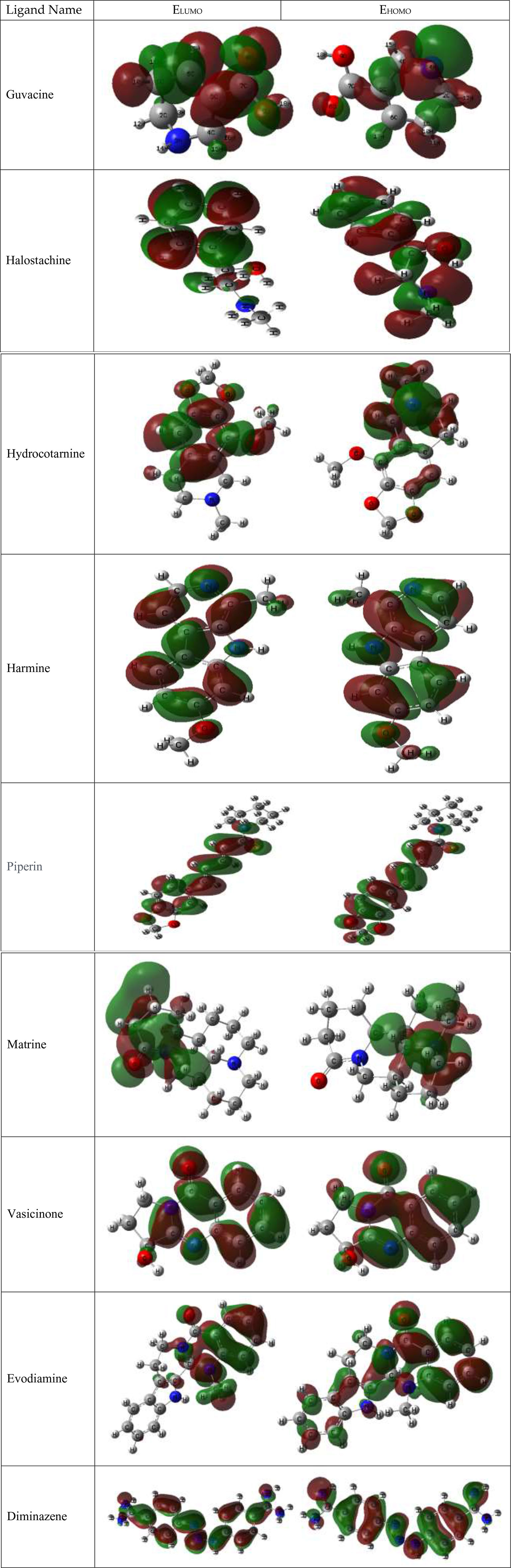
Table 5 presents the HOMO–LUMO energies, hardness, softness, and energy gap of all compounds. The statistical profiles were computed utilizing the DFT function. The energy difference between the HOMO and the LUMO is a commonly employed metric for evaluating the chemical reactivity of molecules. If the energy gap between the HOMO and LUMO levels of a molecule is large, it indicates that the molecule is chemically unstable and highly unreactive. The principal cause of this phenomenon is the obstruction of the electronic transition, which is attributed to a substantial energy disparity between the ground state and the excited state. Typically, a molecule exhibiting a small HOMO–LUMO gap is indicative of a high degree of stability. The values of chemical hardness, softness, and chemical potential are contingent upon the energy of the HOMO and the LUMO. The results presented in Table 5 indicate that the HOMO–LUMO gaps of the chemicals being studied are situated within the range of 3.1205 eV to 6.285 eV. The compounds Matrine and Evodiamine exhibited the lowest HOMO–LUMO energy gap (3.1205–3.432 eV) among the listed compounds. On the other hand, the ligand Hydrocotarnine demonstrated the highest hardness value and the widest energy gap. Typically, compounds with higher hardness values exhibit greater resistance to alterations in electron configuration at the molecular level and the obtained hardness data suggest that a prolonged period may have been necessary for the specimens to undergo disintegration after they arrived at the physiological system. Also, the ligand Piperin was found to possess compounds with a maximum softness value of 0.5435, indicating a higher rate of dissolution for this drug. The HOMO–LUMO data and Frontier molecular orbital diagram are shown in Table 5 and Figure 7.
4 Conclusion
An advanced computational investigation has been conducted by nine natural compounds against the pathogenic human B. microti parasite. In this view, the binding scores and stability of the protein–ligand complexes of selected compounds with targeted B. microti lactate dehydrogenase (PDB ID 6J9D) and B. microti lactate dehydrogenase apo form (PDB ID 6K12) have been applied, and their binding energy, bioactivity, MD simulation, and pharmacokinetic properties have been determined. In the initial stages, all the molecules were optimized by applying the DFT method. Among all the natural biomolecules, the highest value binding energy was reported for Vasicinone (−8.6 kcal/mol) and Evodiamine (−8.7 kcal/mol) against B. microti lactate dehydrogenase (PDB ID 6J9D) similar to these two ligands are also most potential against B. microti lactate dehydrogenase apo form (PDB ID 6K12). Furthermore, the anti-parasitic activity analysis exhibited that all the selected compounds had outstanding efficiency in comparison to the standard Diminazene. Molecular docking result indicated that Vasicinone and Evodiamine might play an essential role in the inhibition of B. microti lactate dehydrogenase and B. microti lactate dehydrogenase apo form and may act as potential anti-parasitic agents to fight the B. microti parasite. Then, the MD simulations were applied to the two best-docked compounds along with standard Diminazene for 100 ns to confirm the stability of drug–protein complexes. The stability of drug–protein complexes was predicted by analyzing RMSD, Rg, SASA, hydrogen bonds, and MMPBSA. This two-hypothetical value confirmed the atomic mobility, structural stability, and residue flexibility during the formation of drug–protein complex structures. Given the MD simulation, Vasicinone and Evodiamine have been suggested to form more stable complexes with the receptor than the standard Diminazene. We were also determined the theoretical pharmacokinetics and drug-likeness. It is confirmed that all the molecules are accepted by the Lipinski rule and have better ADMET features.
4.1 Limitations of the study
It is a theoretical investigation; to validate this investigation, and develop newer and safer drugs from natural sources, these phytochemical compounds must be carried out from computational (in vitro and in vivo), pre-clinical, and clinical trials, to find out their practical value.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
Conceptualization: SA, MEH, and SM. Methodology: SA, MEH, AK, SM, SJS, SS, and AK. writing original draft, and analysis: SA, MEH, SJS, and SM. Formal analysis: SA, AK, MEH, SS, IB, and SJ. Data curation and analysis: AK, SS, IB, and SJ. Writing and review: SA MEH, SJS and SM. Editing: HA-N, AK, and MB. Supervision: AK, SAN, and YJ, Resources, project administration, and data validation: AK, AM HA-N, and SA. All authors contributed to the article and approved the submitted version.
Acknowledgments
The authors extend their appreciation to the Deputyship for Research & innovation, “Ministry of Education” in Saudi Arabia for funding this research (IFKSUOR3-155-1).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abraham, A., Thekkiniath, J., Kilian, N., Lawres, L., Gao, R., Debus, K., et al. (2018). Establishment of a continuous In vitro culture of babesia duncani in human erythrocytes reveals unusually high tolerance to recommended therapies. J. Of. Biol. Chem. 293, 19974–19981. doi: 10.1074/jbc.AC118.005771
Aguero-Rosenfeld, M. (2011). “Committee on Lyme disease and other tick-borne diseases: the state of the science, institute of medicine,” in Critical needs and gaps in understanding prevention, amelioration, and resolution of Lyme and other tick-borne diseases [Electronic resource]: the short-term and long-term outcomes: workshop report [Internet] (Washington, DC: National Academies Press).
Baildya, N., Khan, A. A., Ghosh, N. N., Dutta, T., Chattopadhyay, A. P. (2021). Screening of potential drug from azadirachta indica (Neem) extracts for SARS-Cov-2: an insight from molecular docking and MD-simulation studies. J. Of. Mol. Structure. 1227, 129390. doi: 10.1016/j.molstruc.2020.129390
Bastos, R. G., Thekkiniath, J., Ben Mamoun, C., Fuller, L., Molestina, R. E., Florin-Christensen, M., et al. (2021). Babesia microti immunoreactive rhoptry-associated protein-1 paralogs are ancestral members of the piroplasmid-confined rap-1 family. Pathogens 10, 1384. doi: 10.3390/pathogens10111384
Bloch, E. M., Kumar, S., Krause, P. J. (2019). Persistence of babesia microti infection in humans. Pathogens 8, 102. doi: 10.3390/pathogens8030102
Cao, W., Ding, Z., Hang, X., Xu, K., Song, J., Huang, J., et al. (2018). Theoretical study of a series of 4, 4′-Azo-1 h-1, 2, 4-Triazol-5-One based nitrogen-rich salts as potential energetic compounds. RSC. Adv. 8, 23805–23816. doi: 10.1039/C7RA13424J
Cheung, G., Sundram, F. (2017). Understanding the progression from physical illness to suicidal behavior: a case study based on a newly developed conceptual model. Clin. Gerontol. 40, 124–129. doi: 10.1080/07317115.2016.1217962
Curcio, S. R., Tria, L. P., Gucwa, A. L. (2016). Seroprevalence of babesia microti in individuals with Lyme disease. Vector-Borne. And. Zoonotic. Dis. 16, 737–743. doi: 10.1089/vbz.2016.2020
Daina, A., Michielin, O., Zoete, V. J. S. R. (2017). Swissadme: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 7, 42717. doi: 10.1038/srep42717
Dallakyan, S., Olson, A J J C B M, Protocols (2015). Small-molecule library screening by docking with pyrx. 243–250.
Daneman, R., Prat, A J C S H P I B (2015). The blood–brain barrier. Cold Spring Harb. Perspect. Biol. 7, A020412. doi: 10.1101/cshperspect.a020412
Dowd, F. J., Yagiela, J. A., Johnson, B., Mariotti, A., Neidle, E. A. (2010). Pharmacology and therapeutics for dentistry-E-Book (Elsevier Health Sciences).
Essman, U., Perera, L., Berkowitz, M. L., Darden, T., Lee, H., Pedersen, L. (1995). A smooth particle mesh ewald potential. J. Chem. Phys. 103, 8577–8592. doi: 10.1063/1.470117
Fang, L.-Q., Liu, K., Li, X.-L., Liang, S., Yang, Y., Yao, H.-W., et al. (2015). Emerging tick-borne infections in mainland China: an increasing public health threat. Lancet Infect. Dis. 15, 1467–1479. doi: 10.1016/S1473-3099(15)00177-2
Ferreira, L. G., Dos Santos, R. N., Oliva, G., Andricopulo, A. D. J. M. (2015). Molecular docking and structure-based drug design strategies. Molecules 20, 13384–13421. doi: 10.3390/molecules200713384
Gorenflot, A., Moubri, K., Precigout, E., Carcy, B., Schetters, T. (1998). Human babesiosis. Ann. Of. Trop. Med. Parasitol. 92, 489–501. doi: 10.1080/00034983.1998.11813307
Harrach, M. F., Drossel, B. (2014). Structure and dynamics of Tip3p, Tip4p, and Tip5p water near smooth and atomistic walls of different hydroaffinity. J. Of. Chem. Phys. 140, 174501. doi: 10.1063/1.4872239
Jones, K. E., Patel, N. G., Levy, M. A., Storeygard, A., Balk, D., Gittleman, J. L., et al. (2008). Global trends in emerging infectious diseases. Nature 451, 990–993. doi: 10.1038/nature06536
Kapetanovic, I. (2008). Computer-aided drug discovery and development (Caddd): in silico-Chemico-Biological approach. Chemico-Biol. Interact. 171, 165–176. doi: 10.1016/j.cbi.2006.12.006
Kesharwani, R. K., Vishwakarma, V. K., Keservani, R. K., Singh, P., Katiyar, N., Tripathi, S J C -A D D (2020). Role of admet tools in current scenario: application and limitations. 71–87.
Kim, S., Chen, J., Cheng, T., Gindulyte, A., He, J., He, S., et al. (2019). Pubchem 2019 update: improved access to chemical data. Nucleic Acids Res. 47, D1102–D1109. doi: 10.1093/nar/gky1033
Krause, P. J. (2019). Human babesiosis. Int. J. For. Parasitol. 49, 165–174. doi: 10.1016/j.ijpara.2018.11.007
Krause, P. J., Daily, J., Telford, S. R., Vannier, E., Lantos, P., Spielman, A. (2007). Shared features in the pathobiology of babesiosis and malaria. Trends In. Parasitol. 23, 605–610. doi: 10.1016/j.pt.2007.09.005
Krause, P. J., Spielman, A., Telford, S. R., Sikand, V. K., Mckay, K., Christianson, D., et al. (1998). Persistent parasitemia after acute babesiosis. New Engl. J. Of. Med. 339, 160–165. doi: 10.1056/NEJM199807163390304
Krieger, E., Vriend, G. (2014). Yasara view–molecular graphics for all devices–from smartphones to workstations. Bioinformatics 30, 2981–2982. doi: 10.1093/bioinformatics/btu426
Krieger, E., Vriend, G. (2015). New ways to boost molecular dynamics simulations. J. Of. Comput. Chem. 36, 996–1007. doi: 10.1002/jcc.23899
Kumar, K. K., Karnati, S., Reddy, M. B., Chandramouli, R J J O B, Pharmacy, C. (2010). Caco-2 cell lines in drug discovery-an updated perspective. JBCP 1, 63.
Kumar, A., O’bryan, J., Krause, P. J. (2021). The global emergence of human babesiosis. Pathogens 10, 1447. doi: 10.3390/pathogens10111447
Kumer, A., Sarker, M. N., Sunanda, P. (2019). The theoretical investigation of homo, lumo, thermophysical properties and qsar study of some aromatic carboxylic acids using hyperchem programming. Int. J. Of. Chem. And. Technol. 3, 26–37. doi: 10.32571/ijct.478179
Lemieux, J. E., Tran, A. D., Freimark, L., Schaffner, S. F., Goethert, H., Andersen, K. G., et al. (2016). A global map of genetic diversity in babesia microti reveals strong population structure and identifies variants associated with clinical relapse. Nat. Microbiol. 1, 1–7. doi: 10.1038/nmicrobiol.2016.79
Lipinski, C. A. J. D. D. T. T. (2004). Lead-and drug-like compounds: the rule-Of-Five revolution. Drug Discov. Today Technol. 1, 337–341. doi: 10.1016/j.ddtec.2004.11.007
Lipinski, C. A., Lombardo, F., Dominy, B. W., Feeney, P J J A D D R (1997). Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 23, 3–25. doi: 10.1016/S0169-409X(96)00423-1
Lipinski, C A J J O P, Methods, T. (2000). Drug-like properties and the causes of poor solubility and poor permeability. J Pharmacol Toxicol Methods 44, 235–249. doi: 10.1016/S1056-8719(00)00107-6
Mebius, R. E., Kraal, G. (2005). Structure and function of the spleen. Nat. Rev. Immunol. 5, 606–616. doi: 10.1038/nri1669
Moritz, E. D., Winton, C. S., Tonnetti, L., Townsend, R. L., Berardi, V. P., Hewins, M.-E., et al. (2016). Screening for babesia microti in the us blood supply. New Engl. J. Of. Med. 375, 2236–2245. doi: 10.1056/NEJMoa1600897
Ogu, C. C., Maxa, J. L. “Drug interactions due to cytochrome P450,” in Baylor University medical center proceedings (2000) (Taylor & Francis) 13(4), 421–423.
Orio, M., Pantazis, D. A., Neese, F. J. P. R. (2009). Density functional theory. SPRINGER LINK 102, 443–453. doi: 10.1007/s11120-009-9404-8
Ortiz, J. F., Millhouse, P. W., Cox, ÁM., Campoverde, L., Kaur, A., Wirth, M., et al. (2020). Babesiosis: appreciating the pathophysiology and diverse sequela of the infection. Cureus 12. doi: 10.7759/cureus.11085
Patil, S. M., Maruthi, K., Bajpe, S. N., Vyshali, V., Sushmitha, S., Akhila, C., et al. (2021). Comparative molecular docking and simulation analysis of molnupiravir and remdesivir with sars-Cov-2 rna dependent rna polymerase (Rdrp). Bioinformation 17, 932. doi: 10.6026/97320630017932
Pires, D. E., Blundell, T. L., Ascher, D. B. J. J. O. M. C. (2015). Pkcsm: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 58, 4066–4072. doi: 10.1021/acs.jmedchem.5b00104
Prasanna, S., Doerksen, R. J. C. M. C. (2009). Topological polar surface area: a useful descriptor in 2d-qsar. Curr. Med. Chem. 16, 21–41. doi: 10.2174/092986709787002817
Puri, A., Bajpai, S., Meredith, S., Aravind, L., Krause, P. J., Kumar, S. (2021). Babesia microti: pathogen genomics, genetic variability, immunodominant antigens, and pathogenesis. Front. In. Microbiol. 12, 2416. doi: 10.3389/fmicb.2021.697669
Rajalakshmi, K., Gunasekaran, S., Kumaresan, S. (2015). Density functional theory, comparative vibrational spectroscopic studies, highest occupied molecular orbital and lowest unoccupied molecular orbital analysis of linezolid. Indian J. Of. Phys. 89, 525–538. doi: 10.1007/s12648-014-0618-z
Rosner, F., Zarrabi, M. H., Benach, J. L., Habicht, G. S. (1984). Babesiosis in splenectomized adults: review of 22 reported cases. Am. J. Of. Med. 76, 696–701. doi: 10.1016/0002-9343(84)90298-5
Sharma, S., Kumar, P., Chandra, R J M D S O N U B M S, Lammps, G. (2019). Applications of biovia materials studio, lammps, and gromacs in various fields of science and engineering. 329–341.
Simon, M. S., Westblade, L. F., Dziedziech, A., Visone, J. E., Furman, R. R., Jenkins, S. G., et al. (2017). Clinical and molecular evidence of atovaquone and azithromycin resistance in relapsed babesia microti infection associated with rituximab and chronic lymphocytic leukemia. Clin. Infect. Dis. 65, 1222–1225. doi: 10.1093/cid/cix477
Singh, P., Lonardi, S., Liang, Q., Vydyam, P., Khabirova, E., Fang, T., et al. (2023). Babesia duncani multi-omics identifies virulence factors and drug targets. Nat. Microbiol., 1–15. doi: 10.1038/s41564-023-01360-8
Skrabalo, Z., Deanovic, Z. (1957). Piroplasmosis in man report on a case documenta de medicina geographica et tropica. CAB Direct 9, 11–16.
Vannier, E., Krause, P. J. (2012). Human babesiosis. New Engl. J. Of. Med. 366, 2397–2407. doi: 10.1056/NEJMra1202018
Vilar, S., Cozza, G., Moro, S J C T I M C (2008). Medicinal chemistry and the molecular operating environment (Moe): application of qsar and molecular docking to drug discovery. Curr. Top. Med 8, 1555–1572. doi: 10.2174/156802608786786624
Wang, W., Wolf, R. (2004). J ; Caldwell, jw; kollman, pa; case. Da. J. Of. Comput. Chem. 25, 92. doi: 10.1002/jcc.20035
Western, K. A., Benson, G. D., Gleason, N. N., Healy, G. R., Schultz, M. G. (1970). Babesiosis in a Massachusetts resident. New Engl. J. Of. Med. 283, 854–856. doi: 10.1056/NEJM197010152831607
Zhou, X., Xia, S., Huang, J.-L., Tambo, E., Zhuge, H.-X., Zhou, X.-N. (2014). Human babesiosis, an emerging tick-borne disease in the people’s republic of China. Parasites Vectors 7, 1–10. doi: 10.1186/s13071-014-0509-3
Keywords: molecular docking, DFT, molecular dynamic simulation, ADMET, B. microti, and drug design
Citation: Akash S, Hosen ME, Mahmood S, Supti SJ, Kumer A, Sultana S, Jannat S, Bayıl I, Nafidi H-A, Jardan YAB, Mekonnen AB and Bourhia M (2023) Anti-parasitic drug discovery against Babesia microti by natural compounds: an extensive computational drug design approach. Front. Cell. Infect. Microbiol. 13:1222913. doi: 10.3389/fcimb.2023.1222913
Received: 15 May 2023; Accepted: 20 June 2023;
Published: 16 August 2023.
Edited by:
Heba Alzan, Washington State University, United StatesReviewed by:
Hussein Al-Dahmoshi, University of Babylon, IraqMuhammad Arshad, Northern Alberta Institute of Technology (NAIT), Canada
Ali H. Al-Marzoqi, University of Babylon, Iraq
Copyright © 2023 Akash, Hosen, Mahmood, Supti, Kumer, Sultana, Jannat, Bayıl, Nafidi, Jardan, Mekonnen and Bourhia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amare Bitew Mekonnen, YW1hcmViaXRldzhAZ21haWwuY29t; Mohammed Bourhia, Ym91cmhpYW1vaGFtbWVkQGdtYWlsLmNvbQ==
 Shopnil Akash
Shopnil Akash Md. Eram Hosen
Md. Eram Hosen Sajjat Mahmood
Sajjat Mahmood Sumaiya Jahan Supti4
Sumaiya Jahan Supti4 Ajoy Kumer
Ajoy Kumer Shamima Sultana
Shamima Sultana Imren Bayıl
Imren Bayıl Yousef A. Bin Jardan
Yousef A. Bin Jardan Amare Bitew Mekonnen
Amare Bitew Mekonnen Mohammed Bourhia
Mohammed Bourhia