
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 24 November 2023
Sec. Clinical Microbiology
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1222778
 Haicong Li1
Haicong Li1 Shuo Yang2
Shuo Yang2 Dan Cao1
Dan Cao1 Qianying Wang1
Qianying Wang1 Siyu Zhang2
Siyu Zhang2 Yi Zhou1
Yi Zhou1 Di Liu2
Di Liu2 Ruifeng Yang3*
Ruifeng Yang3* Liyan Cui2*
Liyan Cui2* Zhaoqin Zhu1*
Zhaoqin Zhu1*Objectives: The aim of this study was to evaluate the performance of a new double-antigen sandwich test that is based on the light-initiated chemiluminescent assay (LiCA®) for detecting anti-hepatitis C virus antibodies (anti-HCV) in comparison to Architect®.
Methods: Analytical characteristics and diagnostic performance were tested using seroconversion panels and large pools of clinical samples. Positive results were validated by the strip immunoblot assay (RIBA) and HCV RNA.
Results: Repeatability and within-lab imprecision of LiCA® anti-HCV were 1.31%–3.27%. The C5–C95 interval was −5.44%–5.03% away from C50. LiCA® detected seroconversion in an average of 28.9 days and showed a mean of 3.7 (p = 0.0056) days earlier than Architect®. In a pool of 239 samples with known HCV genotypes 1 to 6, both assays correctly detected all subjects. In 16,305 clinical patient sera, LiCA® detected 4 false-negative (0.25‰) and 14 false-positive (0.86‰) anti-HCV cases, while Architect® recorded 6 false-negative (0.37‰) and 138 false-positive (8.46‰) subjects, respectively. Compared to Architect®, LiCA® presented a significantly better performance in specificity (99.91% vs. 99.14%, n = 16,018, p < 0.0001), positive predictive value (95.29% vs. 67.06%, n = 419, p < 0.0001), and overall accuracy (99.89% vs. 99.12%, n = 16,305, p < 0.0001), while no significant difference in sensitivity (98.61% vs. 97.91%, n = 287, p = 0.5217) and negative predictive value (99.98% vs. 99.96%, n = 15,886, p = 0.3021) was seen. An S/Co value of 3.28 was predicted to be the threshold with a positivity ≥95% for the LiCA® anti-HCV assay.
Conclusion: LiCA® anti-HCV is a precise and fully automatic chemiluminescent assay with superior sensitivity and specificity. The assay can be used as a valuable tool to supplement the diagnosis of HCV infection.
Hepatitis C virus (HCV) causes a heavy burden of liver disease globally. It is estimated that approximately 71 million people have chronic HCV infection and 20% of them might eventually develop into end-stage chronic liver diseases such as liver cirrhosis and hepatocellular carcinoma (World Health Organization, 2017a). Over the last 10 years, HCV-related liver cirrhosis and cancer have increased by 15.5% and 24.8%, respectively (Collaborators GBDCoD, 2017). Although advances in direct-acting antiviral (DAA) therapy regimens enable a high rate (>90%) of cure (Falade-Nwulia et al., 2017; Cooper, 2018), the high burden of undiagnosed infections remains a grand challenge to achieve the goals of the World Health Organization (WHO)—reducing 80% of new HCV infections and 65% of mortality by 2030 (Easterbrook and Group WHOGD, 2016; World Health Organization, 2016). To reach these objectives, at least 90% of infected individuals should be exactly identified and offered timely treatment (Wiktor, 2019).
HCV infection is generally asymptomatic and imperceptible. Laboratory testing plays an essential role for the diagnosis. As recommended by WHO and US Centers for Disease Control and Prevention (CDC), the HCV-antibody (anti-HCV) assay is the initial screening test and the polymerase chain reaction (PCR) test of HCV-RNA is used for confirmation of viremia in reflexing to a reactive anti-HCV test (Centers for Disease Control and Prevention, 2013; World Health Organization, 2017b). Sometimes, a reactive signal does not represent true-positive detection of the antibody due to an unfaithful result of the anti-HCV serological test (Alter et al., 2003). Thereby, the strip immunoblot assay (RIBA) is performed on screening-reactive tests as a supplemental assay with high specificity to confirm the serological antibody status (Alter et al., 2003). Positive results of both antibody and RNA tests indicate an active HCV infection while the antibody-only positive suggests a previously resolved infection (Ghany et al., 2020).
The screening test of anti-HCV is the initial step for the diagnosis of HCV infection. A reliable, highly sensitive and specific, easy-to-use, and affordable kit provides a foundation for access to the WHO targets by 2030, especially in developing countries (Easterbrook and Group WHOGD, 2016). This study is aimed to introduce a new double-antigen sandwich anti-HCV test that is based on the fully automatic homogeneous light-initiated chemiluminescent assay (LiCA®), and extensively validate its analytical and clinical performance in comparison to the Architect® anti-HCV assay.
We conducted this study to evaluate the analytical and clinical performance of LiCA® anti-HCV across three clinical laboratory centers in Shanghai and Beijing, China. Comprehensive assay specifications including analytical characteristics and diagnostic performance were validated as described in Figure 1. All samples were de-identified, separated into vials according to assay frequency and stored at −80°C before testing unless otherwise noted. Tests were performed with samples in freezing once only.
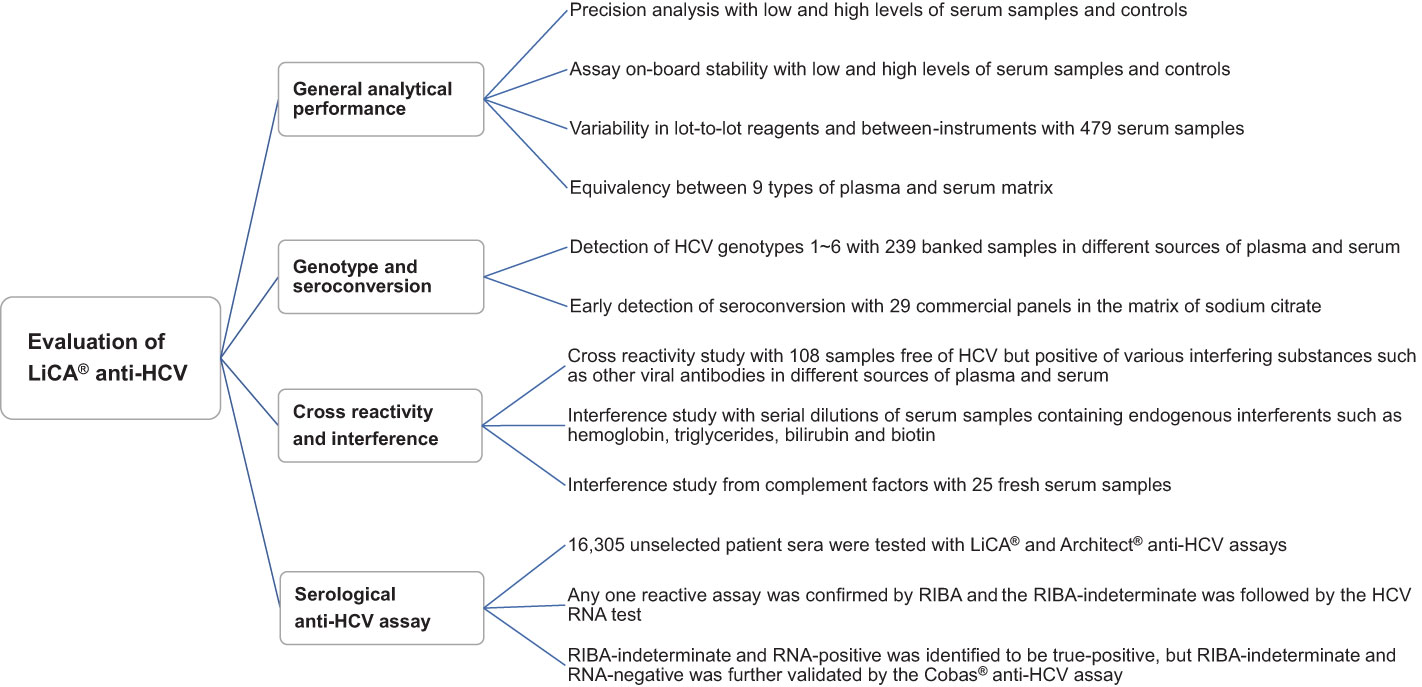
Figure 1 Multicenter performance evaluation of LiCA® anti-HCV. The multicenter evaluation study was conducted across three sites in Beijing and Shanghai, China. The experimental samples for each section were equally assigned to three sites.
A total of 16,305 unselected clinical patient sera were enrolled for anti-HCV screening tests in parallel on the LiCA® 500 system (Chemclin Diagnostics, Beijing, China) and the Architect® i2000SR system (Abbott Laboratories, IL, USA). LiCA® anti-HCV is a third-generation immunoassay and based on the random-access and fully automatic LiCA® system (Wang et al., 2022). The assay uses two nano-scale beads (streptavidin-coated sensitizer beads and recombinant viral antigen-coated emission beads) and biotin-labeled antigens to bridge a one-step double-antigen sandwich reaction with the biotin-streptavidin amplification format. The antigens contain targeting epitopes from core, NS3, and NS4 domains of the viral protein. After incubation, the immunocomplex can be formed if anti-HCV exists in the sample. The sensitizer and emission beads are linked through the antibody–antigen binding chain, resulting in a short distance (<200 nm) between these two beads. Therefore, the singlet oxygens that are generated from the sensitizer beads can diffuse into the emission beads and initiate a chemiluminescence. When the sample does not contain anti-HCV, there is no binding reaction. A larger distance (>200 nm) between the sensitizer and emission beads blocks the singlet oxygen shifting; thus, no chemiluminescence occurs. With the unique methodology, a LiCA® assay does not need any washing step to separate the immunocomplex from the free components, which may produce interfering signals if not washed out completely during a traditional immunoassay rather than LiCA®. Architect® anti-HCV is a two-step indirect immunoassay based on chemiluminescent microparticle technology. The particles contain antigens representing the regions of core, NS3, and NS4. Unlike the LiCA® method, the Architect® assay requires two steps of washing to elute the free components out of the reaction cuvette. Measurement with a ratio of S/Co ≥1.0 is regarded to be reactive and a negative result is considered as S/Co <1.0 for both assays.
Subjects with reactive S/Co ratios on any one assay were repeated in duplicate and identified by reflexing with the RIBA 3.0 assay (Mikrogen Diagnostics, Neuried, Germany). The RIBA results were classified as positive, negative, and indeterminate. The RIBA-indeterminate was further investigated using the real-time PCR test (Roche Diagnostics, Mannheim, Germany), which has the limit of detection (LoD) ≤15 IU/mL for HCV RNA. The RIBA-indeterminate but RNA-positive was considered to be true-positive while the RIBA-indeterminate and RNA-negative was further validated with the Roche Cobas® anti-HCV assay. Cobas® anti-HCV is a one-step double-antigen sandwich immunoassay that is based on the electrochemiluminescence method. The result is classified to be reactive as S/Co ≥1.0, non-reactive as S/Co <0.9, or borderline as S/Co within 0.9–1.0. The testing sequence is shown in Figure 2.
Precision analysis in S/Co ratios was performed using patient sera and controls following the EP15-A3 protocol of the Clinical and Laboratory Standards Institute (CLSI) (CLSI, 2014). A maximum of 15% in coefficient of variation (CV) was acceptable. The C50 test was guided by EP12-A2 using a serial of pooled sera away from both sides of the C50 target (CLSI, 2008). Acceptance was considered as the C5–C95 interval was within C50 ± 15%.
Assay on-board stability was evaluated using patient sera and controls refer to EP25-Ed2 (CLSI, 2023). Measurements were consecutively conducted over 20 days. Reloading a new reagent package and re-calibration were not allowed during study of this section. A difference of each point from the mean value of less than 3 times the standard deviation (SD) and 15% was considered to be acceptable.
Variability in lot-to-lot reagents and between-instruments was assessed refer to EP09c (CLSI, 2018) using 479 residual sera that were collected from randomly selected patients with a broad range of S/Co ratios. Comparative data were recorded in parallel. A satisfactory agreement was considered as correlation coefficient R > 0.975, slope within 0.85–1.15, intercept close to zero, and bias <15%.
To evaluate plasma equivalency to serum, we prepared 25 groups of anti-HCV-negative samples with 10 different types of matrix in serum and 9 commonly used plasma such as ethylenediaminetetraacetic acid (EDTA), heparin, and citrate. Each group was collected from the same individual free of HCV. Positive samples were prepared with the negative ones by spiking a high-positive anti-HCV specimen at 20:1 volume ratio. S/Co values were recorded for each group at the same time. A satisfactory equivalency was considered as percent difference was <15% in positive samples or S/Co difference <0.15 in negative ones.
We used 239 banked samples in different sources of plasma and serum, which has been identified with HCV genotypes by Roche real-time PCR in our labs, and 29 commercially available seroconversion panels in the matrix of sodium citrate for this study. Panels PHV913–926 were from SeraCare (n = 6, Milford, MA, USA), HCV6222–10235 were from ZeptoMetrix (n = 19, Buffalo, NY, USA), and SCP-HCV-004–012 were from BioMex (n = 4, Heidelberg, Germany). All samples were measured on LiCA® and Architect® anti-HCV in parallel.
We collected 108 samples free of HCV but positive of interfering substances, such as other viral antibodies, human anti-mouse antibody (HAMA), and auto-antibodies, to evaluate cross-reactivity from potential interferents. The specimens included different sources of plasma and serum samples. All samples were measured on LiCA® and Architect® in parallel. A significant cross-reactivity was considered as a reactive S/Co was observed in a negative sample unexpectedly.
To identify possible interferences from hemoglobin, triglycerides, bilirubin, and biotin, we used two patient sera containing anti-HCV of 0.71 and 4.75 S/Co, respectively. Patient samples were diluted with serial concentrations of each interferent. S/Co values were recorded on each dilution. A significant interference was considered as percent recovery change ≥15%.
Complement factors are mainly present in fresh blood samples and disappear rapidly during storage. To rule out the potential influence of complement factors on the assay, 25 fresh HCV-negative sera were spiked with a fresh serum sample in strong-positive to anti-HCV to obtain 5 low- and 20 high-positive fresh pools. All samples were stored at 2–8°C and replicate tests were performed for each dilution on days 0, 1, 2, 3, and 4. A significant influence was considered as assay difference from the baseline S/Co on day 0 was ≥15%.
All data were analyzed by the software program MedCalc 20.0.22 (MedCalc Software, Mariakerke, Belgium) and Excel 2019 (Microsoft, WA, USA). Anti-HCV screening tests were classified to be reactive as S/Co ≥1.0 or nonreactive as S/Co <1.0. Positive and negative results were confirmed upon RIBA and HCV RNA assays. A negative–positive reverse conversion was regarded to be primarily significant from the qualitative perspective. More valuable information was obtained with quantitative analysis in S/Co ratios. A statistical significance was considered as p < 0.05.
Based on the assay S/Co values, repeatability and within-lab imprecision of LiCA® anti-HCV were determined to be 1.31%–3.27% across low and high levels of quality controls (QC) and patient sera (Supplemental Table 1). From the perspective of qualitative analysis, the C5–C95 interval was identified to be −5.44%–5.03% away from the C50 target (Supplementary Figure 1). LiCA® anti-HCV presented an excellent analytical precision.
Figure 3 shows daily waves of the assay S/Co on pooled sera and controls over a consecutive 20-day monitoring and demonstrates a good on-board stability. Most measurements were narrowed in a deviation within ±2 SD from the average. Only few points jumped slightly over the line of ±2 SD. In low and high S/Co levels of serum samples, total CV was 5.21% and 3.09%, respectively. The assay difference of each point from mean was within −11.18%–11.40%. In controls, total CV was 5.22% in QC-low and 5.36% in QC-high, respectively. The assay difference of each point from mean was within −7.08%–11.32%. No negative–positive reverse conversion was observed during 4 weeks of assays.
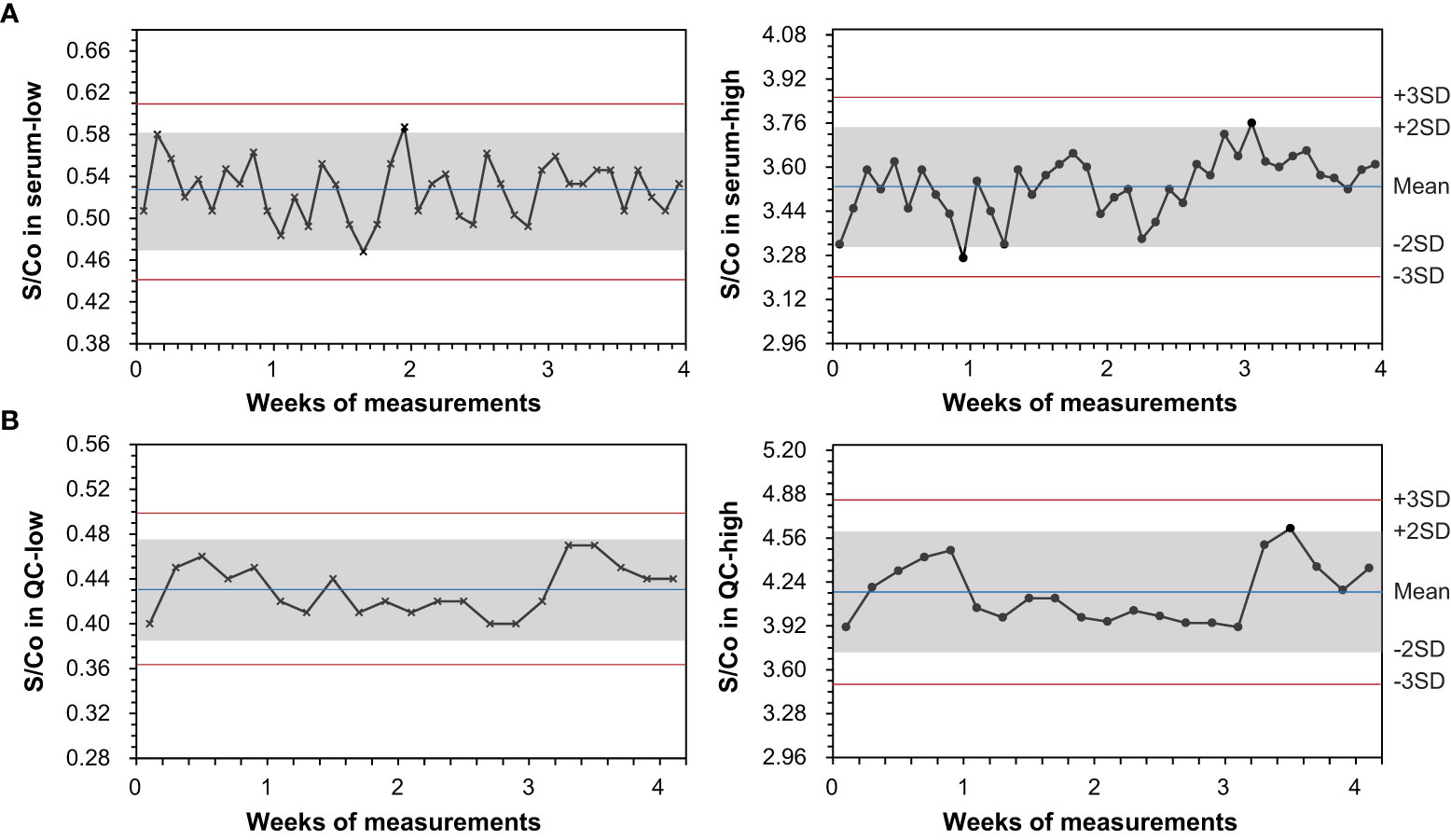
Figure 3 Evaluation of on-board stability for the LiCA® anti-HCV assay following the EP25-Ed2 protocol. Measurements were performed consecutively over 20 days, using low and high signal-to-cutoff (S/Co) ratios of pooled-serum specimens and quality controls (QC), respectively. A reactive S/Co is showing in the symbol (•) and a non-reactive S/Co is in the symbol (×).
A good concordance was validated between different reagent lots and analyzers in serum pools with either full range (0.05–380) or low levels (<10) of S/Co ratios (Supplementary Figure 2). The Spearman correlation coefficient was determined to be 0.976–0.998 and mean bias was 0.33%–4.94%. At the reactive cutoff value (S/Co =1. 0), assay difference from lot-to-lot reagents and between-instruments was estimated to be −1.32%–1.27%.
No significant deviation was observed in nine types of commonly used plasma from serum matrix (Supplemental Table 2). In negative samples, assay S/Co values in any kind of plasma were measured to be almost equivalent to those in serum from the same individual. In positive groups, we recorded a minus but acceptable bias (−11.26 to −3.15%) in plasma from serum. No reverse conversion between negative and positive expectations was observed.
Early detection of HCV infection was evaluated with commercially available seroconversion panels in sodium citrate (Table 1). Of 29 panels studied in total, LiCA® presented 14 in earlier (mean −8.8 days, range −28 to −3 days), 3 in later (mean +5.3 days, range 3–11 days), and 12 in equal detection compared to Architect®. Overall, LiCA® detected seroconversion in an average of 28.9 (95% CI: confidence interval, 18.4–39.3) days and showed mean 3.7 (6.4–1.0) days earlier than Architect®. The Wilcoxon test demonstrated that there was a significant difference in seroconversion detection between two assays (p = 0.0056).
A total of 239 banked samples were used for detection of HCV genotypes (Supplemental Table 3). Both LiCA® and Architect® correctly detected all samples with reactive S/Co values.
No significant cross-reactivity was detected on LiCA® to 19 sources of potential interferents such as various viral antibodies, HAMA, and auto-antibodies (Supplemental Table 4). Only background signals (S/Co < 0.2) were recorded in all 108 samples free of HCV but positive of interfering substances.
A percent recovery change <15% was observed in both low and high S/Co levels of specimens (0.71 and 4.75) by spiking interferents up to 169.5 mmol/L triglycerides, 4,276.0 µmol/L bilirubin, 5.0 g/L hemoglobin, or 409.0 nmol/L biotin, respectively.
The assay difference on day 1–4 storage from the baseline S/Co on day 0 was determined to be −1.37%–8.85% and −2.65%–10.34% in 5 low-positive (mean S/Co = 5.80) and 20 high-positive (mean S/Co = 84.10) fresh serum pools, respectively. No significant influence was observed on the assay to complement factors.
Of 16,305 clinical patient serum samples, 287 were confirmed to be anti-HCV true-positive and 16,018 were true-negative (Figure 2). The diagnostic performance (95% CI) for anti-HCV detection is summarized in Table 2. Compared to Architect®, LiCA® presented a significantly better performance in specificity (99.91% vs. 99.14%, n = 16,018, p < 0.0001), positive predictive value (PPV, 95.29% vs. 67.06%, n = 419, p < 0.0001), and overall accuracy (99.89% vs. 99.12%, n = 16,305, p < 0.0001), while no significant difference in sensitivity (98.61% vs. 97.91%, n = 287, p = 0.5217) and negative predictive value (NPV, 99.98% vs. 99.96%, n = 15,886, p = 0.3021). Among 16,305 patient sera, LiCA® detected 4 false-negative (0.25‰) and 14 false-positive (0.86‰) anti-HCV cases, while Architect® recorded 6 false-negative (0.37‰) and 138 false-positive (8.46‰) subjects, respectively (Table 3). We listed all of 10 false-negative assays in terms of S/Co, positive RIBA bands, and RNA results in Table 4. LiCA® presented 100% of agreement with Cobas®. There was one case that was confirmed to be positive (857 IU/mL) by HCV RNA. LiCA® detected with a good-reactive S/Co of 12.11 but Architect® misdiagnosed with an entirely nonreactive S/Co of 0.05.
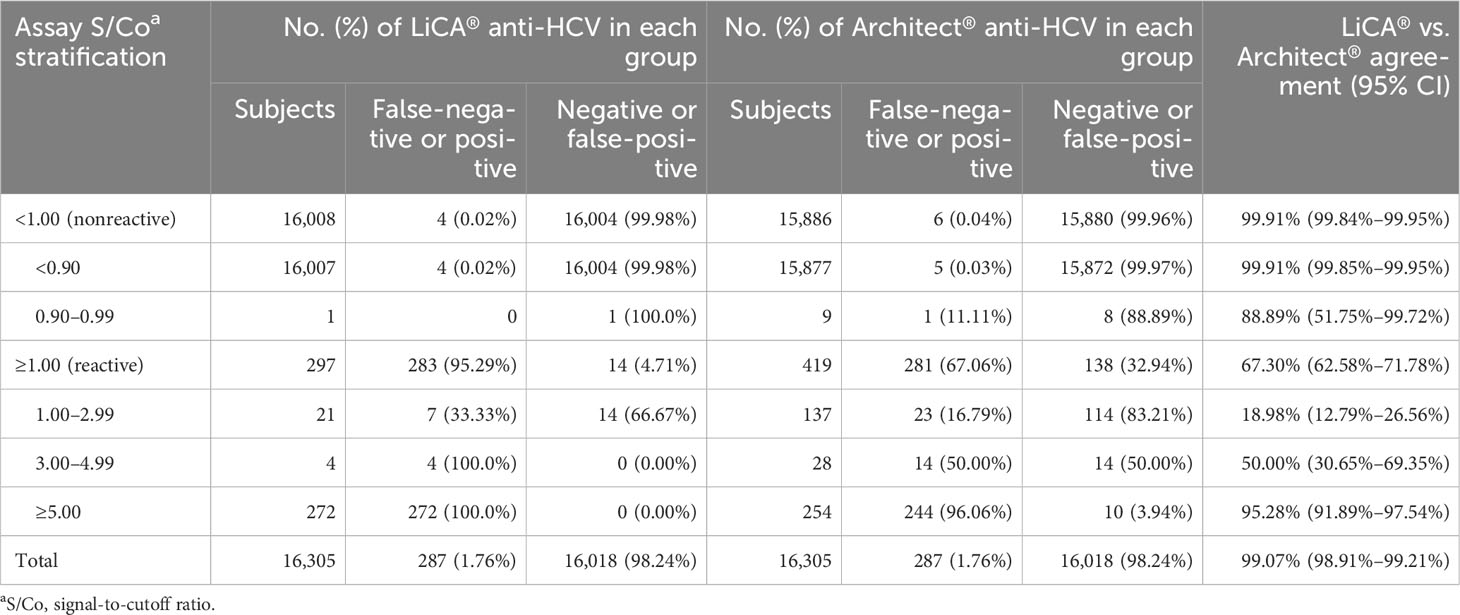
Table 3 Performance of the assays stratified by signal-to-cutoff (S/Co) ratios in clinical patient serum samples.
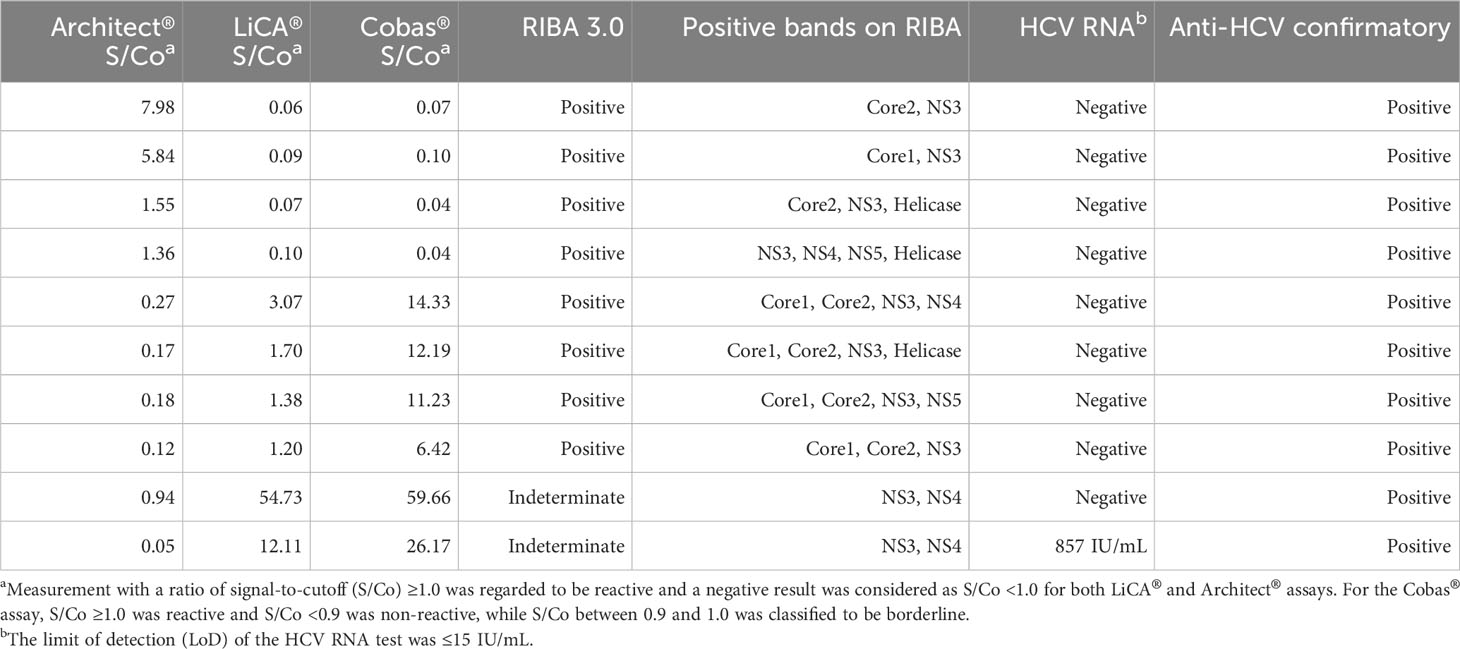
Table 4 Overview of false-negative assays on LiCA® and Architect® anti-HCV in a cohort of 16,305 patient sera (n = 10).
With stratified S/Co ratios in a range of 1.00–2.99, 3.00–4.99, and ≥5.00, and overall tested-reactive results (≥1.00), the proportion of confirmatory positive results was determined to be 33.33% (n = 21), 100% (n = 4), and 100% (n = 272), and 95.29% (n = 297) on LiCA® in contrast to 16.79% (n = 137), 50.00% (n = 28), and 96.06% (n = 254), and 67.06% (n = 419) on Architect®, respectively (Table 3). The distribution of S/Co ratios in more detailed stratification clearly revealed that Architect® had much more false-positive tests especially in a range of 1.0–5.0 S/Co (Figure 4). LiCA® reported the results with much wider distribution of S/Co ratios (0–475.87 vs. 0–24.94) and less count in the “borderline” range of 0.9–5.0 S/Co (26 vs. 174) than Architect® did.
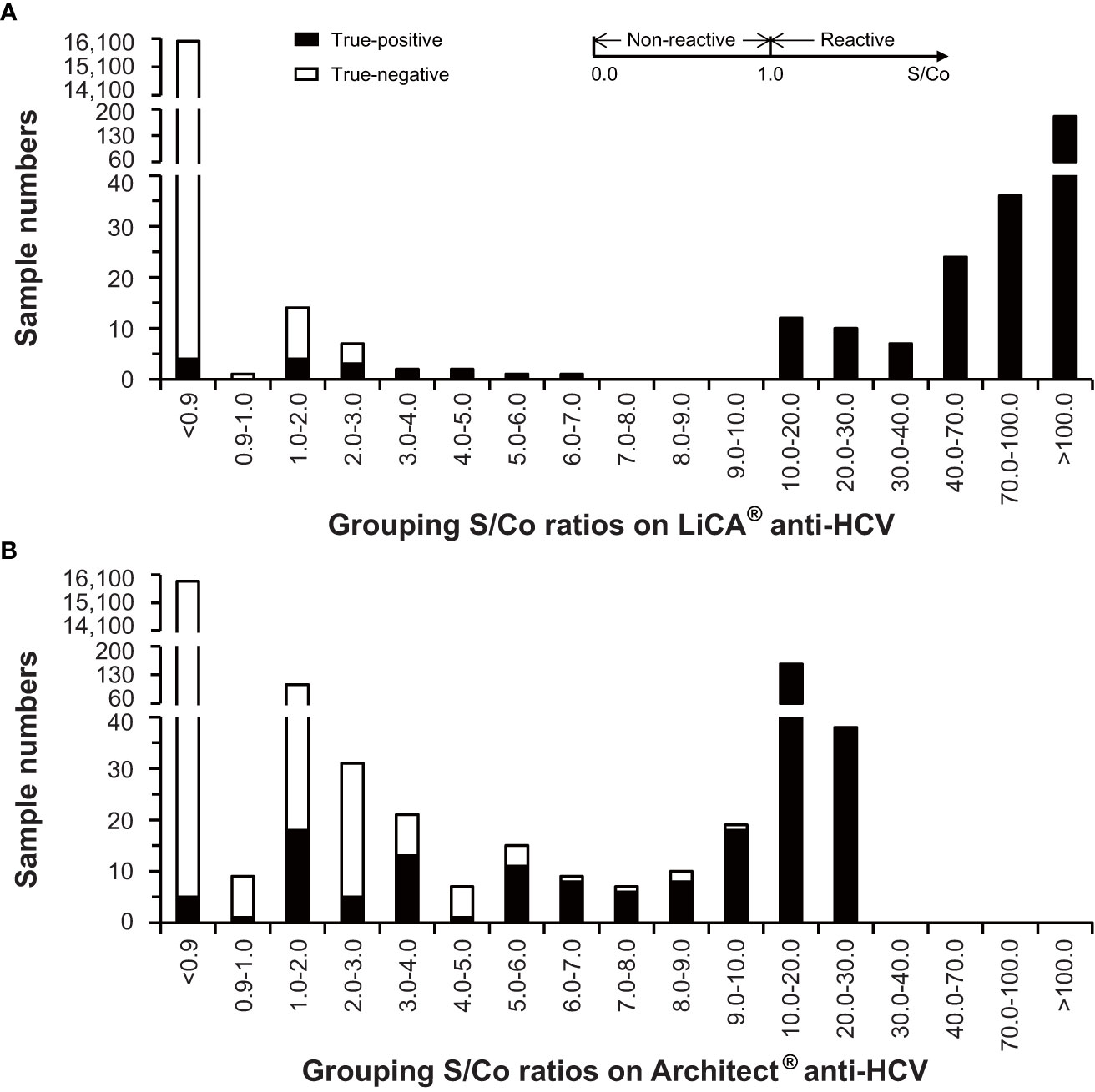
Figure 4 Distribution of signal-to-cutoff (S/Co) ratios on LiCA® and Architect® anti-HCV assays in clinical patient sera (n=16,305).
Probit regression estimated that the predicting S/Co value (95% CI) with a positivity ≥95% was 3.28 (2.74–4.12) on LiCA® (Supplementary Figure 3). Using the predicting value as a cutoff, the data set of this study presented 100% of PPV and 99.93% of NPV.
LiCA® showed a weak correlation with Architect® based on the assays with S/Co ≥1.0 (Supplementary Figure 4A, R = 0.296, p < 0.0001). An improved correlation was observed with logarithmic transformation of S/Co ratios (Supplementary Figure 4B, R = 0.650, p < 0.0001). Overall agreement (95% CI) between both methods was 99.07% (98.91%–99.21%, n = 16,305) with Cohen’s kappa 0.78 (0.75–0.82). Positive and negative agreements were 67.30% (62.58%–71.78%, n = 419) and 99.91% (99.84%–99.95%, n = 15,886), respectively (Table 3).
A total of 152 (0.93%, n = 16,305) discordant measurements were observed between these two assays (Supplemental Table 5). Most discrepancies (133/152, 87.50%) resulted from the false-positive assays on Architect®, especially from those with a lower reactive S/Co between 1.0 and 5.0 (123/133, 92.48%). For these discordant assays between LiCA® and Architect®, LiCA® presented a high agreement with Cobas® (143/152, 94.08%). There were 47 (0.28%, n = 16,305) discordances with RIBA-indeterminate but RNA-negative results. The Cobas® anti-HCV assay was used for further validation and confirmed that 46/47 were false-positive and 1/47 was false-negative from Architect®. A perfect consistency (47/47, 100%) was observed between LiCA® and Cobas® assays. The detailed records were summarized in Supplemental Table 6.
Typically, HCV RNA can be detected in blood within 1–3 weeks while the antibody seropositivity occurs from several weeks to months after viral exposure (Cloherty et al., 2016; Ghany et al., 2020). Among patients with acute HCV infection, approximately 50%–80% individuals become chronic cases while 20%–50% of the subjects achieve spontaneous clearance (Kamal, 2008). In general, a positive HCV RNA test confirms an early infection during the “window” stage of seroconversion and the viremia status of a current infection (acute and chronic) while the anti-HCV-positive indicates a prior resolved infection or a current virus carrier. During the early stage of infection, people may develop different levels of antibodies due to varied immunogenicity of HCV antigens and different reactivity of individuals. The detection capability for a given anti-HCV assay is highly associated with the antigen configuration of the reagent kit because each HCV antigen may have unequal contribution to antibody detection (Warkad et al., 2019; Jiang et al., 2021). Both WHO and US CDC guidelines on HCV tests recommend that the diagnosis of infection is initially detected with a serological screening of anti-HCV and followed by a reflexive PCR test of HCV RNA (Centers for Disease Control and Prevention, 2013; World Health Organization, 2017b). A probable false-negative anti-HCV test due to the window period (World Health Organization, 2017b) or delayed seroconversion (Thomson et al., 2009) may lead to a misdiagnosis while too many false-positive screening detections would cause high burden of expensive RNA assays and unnecessary medical visits (Alter et al., 2003; Centers for Disease Control and Prevention, 2013). Thereby, a reliable anti-HCV screening kit with high sensitivity and specificity is essential (Easterbrook and Group WHOGD, 2016).
In this study, LiCA® demonstrated superior sensitivity to Architect® for the early detection of HCV infection. Of the 29 seroconversion panels studied in total, LiCA® detected 14 in earlier (mean −8.8 days) and only 3 in later (mean +5.3 days) detection than Architect® did. LiCA® presented a significantly shorter window phase (overall −3.7 days, p = 0.0056) in comparison to Architect®. LiCA® also correctly detected all subjects in a pool of 239 samples with known HCV genotype 1–6. Using a large pool of clinical patient sera (n = 16,305), LiCA® showed a slightly better performance in diagnostic sensitivity (98.61% vs. 97.91%) compared to Architect®. Notably, LiCA® and Architect® respectively detected four and five false-negative cases that were confirmed to be anti-HCV-positive but RNA-negative. Undetectable HCV RNA may indicate a prior resolved infection in which the virus has been eradicated from the circulation (El Ekiaby et al., 2015). Inconsistency of detection performance in the same cohort may be explained by the specific antigen configuration of different anti-HCV assays (Warkad et al., 2019; Jiang et al., 2021). An additional one that was misdiagnosed on Architect® with an entirely nonreactive S/Co (0.05) was well-reactive (12.11) on LiCA® and confirmed to be positive (857 IU/mL) by HCV RNA. The indeterminate RIBA test showed positive bands to antigens representing NS3 and NS4 regions. This case might be associated with the early stage of an acute infection. It has been demonstrated that high rates of the viral transmission occurred at all levels of viremia, whether anti-HCV was positive or negative (Operskalski et al., 2003). Factors for misdiagnosis on Architect® could be less sensitivity in detecting antibodies to NS3 and NS4 proteins (Myrmel et al., 2005) and lack of capture to anti-HCV IgM on an indirect immunoassay (Wu et al., 2008).
Among of 16,305 clinical samples enrolled, the comparative Architect® assay recorded 138 false-positive results. In low levels of S/Co ratios (1.0–5.0) on Architect®, over three-fourths of assays were exclusive of true positivity. Compared to Architect®, LiCA® detected only 14 false-positive cases in the same cohort and presented a significantly better specificity (99.91% vs. 99.14%, n = 16,018, p < 0.0001) and PPV (95.29% vs. 67.06%, n = 419, p < 0.0001). Cross-reactivity and interfering experiments also demonstrated that LiCA® anti-HCV did not show any response to 19 sources of potential interferents studied. The exceptionally high specificity plus high sensitivity enables LiCA® to deliver a more accurate diagnosis, in which negative and positive cases can be better discriminated with a wider distribution of S/Co but with a much lower count in the “borderline” (S/Co 0.9–5.0) range (Figure 4), thus facilitating a timely treatment. Using the threshold of 3.28 S/Co that was determined herein to predict positivity ≥95% may further improve reflexive testing sequence for HCV infection (Lai et al., 2011; Choi et al., 2018).
The nature of high sensitivity and specificity on LiCA® anti-HCV can be attributed to the assay methodology. LiCA® anti-HCV is based on the double-antigen sandwich format. Two levels of antigen specifically clamp the antibody to reduce nonspecific binding minimally. In the indirect method that is used on Architect®, however, the labeled second anti-IgG antibodies can recognize not only the targeting antibodies but all human IgG molecules, thus detecting signals in combination of nonspecific bindings and giving false-positive results (Wu et al., 2008). Previous studies confirmed a high rate of false-positive detection on Architect® anti-HCV especially in low S/Co (Ha et al., 2019; Yang et al., 2019; Lucey et al., 2022). A two-screening-test strategy was proposed to relieve the high burden of further confirmatory tests because a false-positive result was rarely seen on two different serological assays (Vermeersch et al., 2008; Easterbrook and Group WHOGD, 2016). However, ruling out the discordant reactive anti-HCV by repeating tests may yield more false-negative diagnoses (Parry et al., 2017). Hence a reliable screening test with high sensitivity and specificity is critical. Interestingly, a high agreement (94.08%) between LiCA® and Cobas® was observed in the cohort of 152 discrepancies between LiCA® and Architect®. Sharing a similar methodology in a double-antigen sandwich format on both LiCA® and Cobas® might explain this. Generally, sample dilution is used to mitigate interference from nonspecific IgG captures in the indirect format. In contrast, the sandwich method allows assay in an undiluted specimen, consequently offering better detection despite the low levels of antibodies. Another advantage ascribed to the sandwich format is the enhanced detectability to anti-HCV IgM that is from a newly infected individual but cannot be recognized in the indirect assay (Wu et al., 2008). Improved sensitivity on LiCA® can also be obtained from the bigger specific surface area on the nano-scale particles and from signal amplification on the biotin-streptavidin mechanism (Wu et al., 2008; Yu et al., 2023). Recent findings have validated the superior assay capability of the LiCA® method in detecting thyrotropin, cardiac troponin, and severe acute respiratory syndrome coronavirus type-2 (SARS-CoV-2) antigen (Wang et al., 2022; Yang et al., 2022; Yu et al., 2023).
Along with high sensitivity and specificity, LiCA® anti-HCV was characteristic of excellent analytical precision and stability in day-to-day, lot-to-lot, different instruments, and sample matrix. Moreover, the assay was performed on a no-wash homogeneous immunoassay platform with a fully automatic walkaway model and gained advantages in flexibility, productivity, and cost-effectiveness to adapt varied testing demand towards ending HCV worldwide.
The limitation of this study is incomplete RNA testing data and missing medical history and patient follow-up. Although discriminating the true antibody-positive from the false-positive by RIBA is informative of infection status (Rafik et al., 2016), a further investigation of the correlation of HCV-antibody titer to active viremia is necessary.
In conclusion, LiCA® anti-HCV is a precise and fully automatic chemiluminescent assay with superior sensitivity and specificity. The assay can be used as a valuable tool to supplement the diagnosis of HCV infection.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Research involving human subjects complied with all relevantnational regulation and institutional policies and is in accordancewith the tenets of the Helsinki Declaration (as revised in 2013), and has been approved by the Ethics Committee of Shanghai PublicHealth Clinical Center (2018-E091-01).
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by the National Natural Science Foundation of China (Nos. 82101847 and 81671636). The funding organization(s) played no role in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the report for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1222778/full#supplementary-material
Alter, M. J., Kuhnert, W. L., Finelli, L., Centers for Disease Control and Prevention (2003). Guidelines for laboratory testing and result reporting of antibody to hepatitis C virus. MMWR Recomm Rep. 52 (RR-3), 1–13.
Centers for Disease Control and Prevention (2013). Testing for HCV infection: an update of guidance for clinicians and laboratorians. MMWR Morb Mortal Wkly Rep. 62 (18), 362–365.
Choi, M. S., Lee, K., Hong, Y. J., Song, E. Y., Kim, D. S., Song, J. (2018). The role of the signal-to-cutoff ratio in automated anti-HCV chemiluminescent immunoassays by referring to the nucleic acid amplification test and the recombinant immunoblot assay. Ann. Lab. Med. 38 (5), 466–472. doi: 10.3343/alm.2018.38.5.466
Cloherty, G., Talal, A., Coller, K., Steinhart, C., Hackett, J., Jr., Dawson, G., et al. (2016). Role of serologic and molecular diagnostic assays in identification and management of hepatitis C virus infection. J. Clin. Microbiol. 54 (2), 265–273. doi: 10.1128/JCM.02407-15
CLSI (2008). “User protocol for evaluation of qualitative test performance; approved guideline,” in CLSI document EP12-A2, 2nd edition (Wayne, PA: Clinical and Laboratory Standards Institute).
CLSI (2014). “User verification of precision and estimation of bias; approved guidline,” in CLSI document EP15-A3, 3rd edition (Wayne, PA: Clinical and Laboratory Standards Institute).
CLSI (2018). “Measurement procedure comparison and bias estimation using patient samples,” in CLSI guideline EP09c, 3rd edition (Wayne, PA: Clinical and Laboratory Standards Institute).
CLSI (2023). “Evaluation of stability of in vitro medical laboratory test reagents,” in CLSI guideline EP25, 2nd edition (Wayne, PA: Clinical and Laboratory Standards Institute).
Collaborators GBDCoD (2017). Global, regional, and national age-sex specific mortality for 264 causes of death, 1980-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 390 (10100), 1151–1210.
Cooper, C. L. (2018). Now is the time to quickly eliminate barriers along the hepatitis C cascade of care. J. Infect. Dis. 217 (12), 1858–1860. doi: 10.1093/infdis/jiy117
Easterbrook, P. J., Group WHOGD (2016). Who to test and how to test for chronic hepatitis C infection - 2016 WHO testing guidance for low- and middle-income countries. J. Hepatol. 65 (1 Suppl), S46–S66. doi: 10.1016/j.jhep.2016.08.002
El Ekiaby, M., Moftah, F., Goubran, H., van Drimmelen, H., LaPerche, S., Kleinman, S., et al. (2015). Viremia levels in hepatitis C infection among Egyptian blood donors and implications for transmission risk with different screening scenarios. Transfusion. 55 (6), 1186–1194. doi: 10.1111/trf.13061
Falade-Nwulia, O., Suarez-Cuervo, C., Nelson, D. R., Fried, M. W., Segal, J. B., Sulkowski, M. S. (2017). Oral direct-acting agent therapy for hepatitis C virus infection: A systematic review. Ann. Intern. Med. 166 (9), 637–648. doi: 10.7326/M16-2575
Ghany, M. G., Morgan, T. R., Panel A-IHCG (2020). Hepatitis C guidance 2019 update: American Association for the Study of Liver Diseases-Infectious Diseases Society of America recommendations for testing, managing, and treating hepatitis C virus infection. Hepatology. 71 (2), 686–721. doi: 10.1002/hep.31060
Ha, J., Park, Y., Kim, H. S. (2019). Signal-to-cutoff ratios of current anti-HCV assays and a suggestion of new algorithm of supplementary testing. Clin. Chim. Acta 498, 11–15. doi: 10.1016/j.cca.2019.08.002
Jiang, X., Chang, L., Yan, Y., Ji, H., Sun, H., Guo, F., et al. (2021). A study based on four immunoassays: Hepatitis C virus antibody against different antigens may have unequal contributions to detection. Virol. J. 18 (1), 137. doi: 10.1186/s12985-021-01608-x
Kamal, S. M. (2008). Acute hepatitis C: a systematic review. Am. J. Gastroenterol. 103 (5), 1283–1297. doi: 10.1111/j.1572-0241.2008.01825.x
Lai, K. K., Jin, M., Yuan, S., Larson, M. F., Dominitz, J. A., Bankson, D. D. (2011). Improved reflexive testing algorithm for hepatitis C infection using signal-to-cutoff ratios of a hepatitis C virus antibody assay. Clin. Chem. 57 (7), 1050–1056. doi: 10.1373/clinchem.2010.158691
Lucey, O., Acana, S., Olupot-Olupot, P., Muhindo, R., Ayikobua, R., Uyoga, S., et al. (2022). High false discovery rate of the Architect anti-HCV screening test in blood donors in Uganda and evaluation of an algorithm for confirmatory testing. Vox Sang. 117 (12), 1360–1367. doi: 10.1111/vox.13364
Myrmel, H., Navaratnam, V., Asjo, B. (2005). Detection of antibodies to hepatitis C virus: false-negative results in an automated chemiluminescent microparticle immunoassay (ARCHITECT Anti-HCV) compared to a microparticle enzyme immunoassay (AxSYM HCV Version 3.0). J. Clin. Virol. 34 (3), 211–215. doi: 10.1016/j.jcv.2005.05.013
Operskalski, E. A., Mosley, J. W., Tobler, L. H., Fiebig, E. W., Nowicki, M. J., Mimms, L. T., et al. (2003). HCV viral load in anti-HCV-reactive donors and infectivity for their recipients. Transfusion. 43 (10), 1433–1441. doi: 10.1046/j.1537-2995.2003.00475.x
Parry, J. V., Easterbrook, P., Sands, A. R. (2017). One or two serological assay testing strategy for diagnosis of HBV and HCV infection? The use of predictive modelling. BMC Infect. Dis. 17 (Suppl 1), 705–716. doi: 10.1186/s12879-017-2774-1
Rafik, M., Bakr, S., Soliman, D., Mohammed, N., Ragab, D., ElHady, W. A., et al. (2016). Characterization of differential antibody production against hepatitis C virus in different HCV infection status. Virol. J. 13, 116–124. doi: 10.1186/s12985-016-0572-9
Thomson, E. C., Nastouli, E., Main, J., Karayiannis, P., Eliahoo, J., Muir, D., et al. (2009). Delayed anti-HCV antibody response in HIV-positive men acutely infected with HCV. AIDS. 23 (1), 89–93. doi: 10.1097/QAD.0b013e32831940a3
Vermeersch, P., Van Ranst, M., Lagrou, K. (2008). Validation of a strategy for HCV antibody testing with two enzyme immunoassays in a routine clinical laboratory. J. Clin. Virol. 42 (4), 394–398. doi: 10.1016/j.jcv.2008.02.015
Wang, M., Li, J., Huang, Y., Chen, T., Dong, S., Zhang, R., et al. (2022). Analytical validation of the LiCA high-sensitivity human thyroid stimulating hormone assay. Clin. Biochem. 101, 42–49. doi: 10.1016/j.clinbiochem.2021.11.018
Warkad, S. D., Song, K. S., Pal, D., Nimse, S. B. (2019). Developments in the HCV screening technologies based on the detection of antigens and antibodies. Sensors (Basel). 19 (19). doi: 10.3390/s19194257
Wiktor, S. (2019). How feasible is the global elimination of HCV infection. Lancet. 393 (10178), 1265–1267. doi: 10.1016/S0140-6736(18)32750-8
World Health Organization (2016). Global health sector strategy on viral hepatitis 2016-2021: Towards ending viral hepatitis. Available at: https://www.who.int/publications/i/item/WHO-HIV-2016.06.
World Health Organization (2017a). Global hepatitis report. Available at: https://www.who.int/publications/i/item/9789241565455.
World Health Organization (2017b). Guidelines on hepatitis B and C testing. Available at: https://www.who.int/publications/i/item/9789241549981.
Wu, F. B., Ouyan, H. Q., Tang, X. Y., Zhou, Z. X. (2008). Double-antigen sandwich time-resolved immunofluorometric assay for the detection of anti-hepatitis C virus total antibodies with improved specificity and sensitivity. J. Med. Microbiol. 57 (Pt 8), 947–953. doi: 10.1099/jmm.0.47835-0
Yang, S., Yang, R., Zhang, S., Liu, D., Zhou, J., Wang, T., et al. (2019). Clinical diagnostic performance of light-initiated chemiluminescent assay compared with the Architect chemiluminescence immunoassay for detection of HCV antibody. J. Clin. Lab. Anal. 33 (7), e22928. doi: 10.1002/jcla.22928
Yang, S., Zhang, Q., Yang, B., Li, Z., Sun, W., Cui, L. (2022). Analytical and clinical performance evaluation of a new high-sensitivity cardiac troponin I assay. Clin. Chem. Lab. Med. 60 (8), 1299–1307. doi: 10.1515/cclm-2021-1136
Keywords: hepatitis C virus, LiCA® anti-HCV, Architect® anti-HCV, double-antigen sandwich, performance evaluation
Citation: Li H, Yang S, Cao D, Wang Q, Zhang S, Zhou Y, Liu D, Yang R, Cui L and Zhu Z (2023) A new double-antigen sandwich test based on the light-initiated chemiluminescent assay for detecting anti-hepatitis C virus antibodies with high sensitivity and specificity. Front. Cell. Infect. Microbiol. 13:1222778. doi: 10.3389/fcimb.2023.1222778
Received: 16 May 2023; Accepted: 20 October 2023;
Published: 24 November 2023.
Edited by:
Maria-Cristina Navas, Universidad de Antioquia, ColombiaReviewed by:
Jean-Pierre Allain, University of Cambridge, United KingdomCopyright © 2023 Li, Yang, Cao, Wang, Zhang, Zhou, Liu, Yang, Cui and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhaoqin Zhu, emhhb3FpbnpodUAxNjMuY29t; Liyan Cui, Y2xpeWFuQDE2My5jb20=; Ruifeng Yang, eWFuZ3J1aWZlbmdAcGt1cGguZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.