- 1Center of Excellence for Vector-Borne Diseases, Department of Diagnostic Medicine/Pathobiology, College of Veterinary Medicine, Kansas State University, Manhattan, KS, United States
- 2Department of Veterinary Pathobiology, College of Veterinary Medicine, Bond Life Sciences Center, University of Missouri, Columbia, MO, United States
Ehrlichia chaffeensis is a tick-transmitted monocytic ehrlichiosis agent primarily causing the disease in people and dogs. We recently described the development and characterization of 55 random mutations in E. chaffeensis, which aided in defining the critical nature of many bacterial genes for its growth in a physiologically relevant canine infection model. In the current study, we tested 45 of the mutants for their infectivity ability to the pathogen’s tick vector; Amblyomma americanum. Four mutations resulted in the pathogen’s replication deficiency in the tick, similar to the vertebrate host. Mutations causing growth defects in both vertebrate and tick hosts included in genes coding for a predicted alpha/beta hydrolase, a putative dicarboxylate amino acid:cation symporter, a T4SS protein, and predicted membrane-bound proteins. Three mutations caused the bacterial defective growth only in the tick vector, which represented putative membrane proteins. Ten mutations causing no growth defect in the canine host similarly grew well in the tick vector. Mutations in 28 genes/genomic locations causing E. chaffeensis growth attenuation in the canine host were recognized as non-essential for its growth in the tick vector. The tick non-essential genes included genes coding for many metabolic pathway- and outer membrane-associated proteins. This study documents novel vector- and host-specific differences in E. chaffeensis for its functional gene requirements.
Introduction
The lone star tick, Amblyomma americanum, continues to be one of the most successful ticks during the past several decades and is a major contributor to the rise in documented tick-borne diseases in the USA (Rodino et al., 2020; Higuita et al., 2021). This tick has expanded its range as well as the spectrum of pathogens that it transmits (Higuita et al., 2021). As a three-host tick, all its motile life stages (larva, nymph, and adult) aggressively blood feed on a wide range of hosts, including people and dogs. This tick is the vector for Ehrlichia chaffeensis which infects several mammalian hosts, including people, dogs, coyotes, goats, and white-tailed deer (Dawson et al., 1996; Lockhart et al., 1997; Breitschwerdt et al., 1998; Dugan et al., 2000; Kocan et al., 2000; Davidson et al., 2001). This pathogen belongs to the Anaplasmataceae family which includes several obligate intracellular pathogenic bacteria of the genera Ehrlichia, Anaplasma, Rickettsia and Orientia (Dumler et al., 2001).
For most tick-borne diseases, there is a close interplay between tick vectors, vertebrate hosts and pathogenic organisms transmitted by ticks. Pathogens must navigate both the vertebrate and tick systems to efficiently adapt to the dual hosts for their continued survival despite the immune responses exerted by ticks and vertebrate hosts. In a recent study, we developed and evaluated an E. chaffeensis transposon mutant library constituting of 55 mutants to identify genes essential for its persistent growth in its natural vertebrate infection model (Wang et al., 2020). In that study, we reported that E. chaffeensis requires many functional genes. They included genes coding for three immunodominant membrane proteins; two 28 kDa outer membrane proteins (P28/OMP) and a 120 kDa surface protein. Mutations in two biotin biosynthesis pathway genes, a fatty acid biosynthesis gene, and a 3-oxoacyl-(acyl-carrier-protein) reductase gene also resulted in the E. chaffeensis rapid clearance from the vertebrate host. Mutations near genes coding for DNA repair and protein synthesis pathway proteins also caused rapid clearance of E. chaffeensis from a vertebrate host (Wang et al., 2020).
In the current study, we extended the investigations to define which genes/proteins are essential for E. chaffeensis during its replication in its tick vector. We investigated 45 of the previously characterized E. chaffeensis random mutants to define the impact of the mutations on the bacterial persistence in the tick host. We assessed the infection status of the mutants in A. americanum following needle infection and its molting from the nymphal stage to the adult stage. We discovered remarkable differences in the requirements of functional genes of E. chaffeensis for growth and persistence in A. americanum compared to the canine host. The study aided in identifying genes critical for both vertebrate and tick hosts and those exclusively needed for the bacterial replication in tick or vertebrate hosts.
Materials and methods
Ehrlichia chaffeensis in vitro cultivation
Ehrlichia chaffeensis mutant organisms (Wang et al., 2020) (45 in all) were cultivated in the canine macrophage cell line (DH82) at 37°C with 5% CO2 as described earlier (Chen et al., 1995; Cheng and Ganta, 2008). As infection progression in ticks begins with the pathogen transfer during a bloodmeal originating from a vertebrate host (such as from white tailed deer and dogs), we grew mutant organisms in DH82 for use in tick infection experiments.
Ticks and tick rearing/tick injections
Freshly engorged nymphs of A. americanum were obtained from Ecto Services Inc. (Henderson, NC). Within 48 h of dropping from the host, groups of approximately 50 engorged nymphs were washed thoroughly with 10% bleach, then rinsed three times with sterile water and dried on paper towels. The nymphs were then injected with approximately one microliter each of 5 x 108 E. chaffeensis mutant organisms using an inoculum syringe with a 27-gauge needle. The injected ticks were transferred to a humidified incubator (95% relative humidity is maintained with a saturated salt solution of potassium sulfate) maintained at room temperature with 14 h of light. These conditions allowed the molting of nymphs to the adult stage in approximately 4-5 weeks.
DNA isolation and PCR verification of E. chaffeensis mutant-injected ticks
Within four weeks of molting, genomic DNAs from 10 ticks each were individually isolated from each group of mutant-infected adult ticks. Tick genomic DNAs were isolated using a DNeasy Blood and Tissue kit as per the manufacturer’s instructions (Qiagen Sciences Inc., Germantown, MD). Purified DNA from each tick was resuspended in 200 µl of DNA elution buffer. Two microliters of DNA derived from each tick were then used for nested PCR analysis targeting to the insertion segment located within each insertion mutant organism and using the Green Taq Master Mix in a 25 µl PCR assay. The first round of amplification was carried out using the forward primer, AmtrF, 5’CTCCTAGAACGATCGCCGCATGCTAGC and the reverse primer, RRG1598, 5’TTATTTGCCGACTACCTTGGTGAT. The second round of amplification was performed using the forward primer, AmtrF2, 5’CGCGCGCACTAACAAGTGCCC and the above listed reverse primer and with 2 µl of the first round PCR product. The annealing temperature for both PCRs was 55°C for 30 sec and extension was carried out at 72°C for 2 mins for a total of 40 cycles. All PCR products were resolved in 0.9% agarose gels with the 1X tris-acetate-EDTA buffer and stained with ethidium bromide. We performed the nested PCRs two independent times to validate the results and using DNA recovered from 10 ticks per mutant group. All assays included PCR positive controls where a known template was added and negative controls containing no template DNA. Further, wild type E. chaffeensis infected and non-infected and molted ticks were included to serve as positive and negative controls, respectively.
Database searching for protein homologies and for finding protein motifs
The identification of open reading frames (ORFs) encoding proteins in E. chaffeensis was carried out using the genome entries; GenBank accession numbers CP000236 and NC_007799.1 present at the NCBI database (https://www.ncbi.nlm.nih.gov/). The secondary structure predictions and conserved domain predictions of E. chaffeensis proteins were performed using the online sequence analysis tools; SMART protein (http://smart.embl-heidelberg.de/) (Letunic et al., 2020). PRED-TMR was used for transmembrane domain predictions (http://athina.biol.uoa.gr/PRED-TMR/) (Pasquier et al., 1999). The software TMRPres2D was used to draw the prediction of transmembrane domains (Spyropoulos et al., 2004). Searches for homologous protein sequences were performed using the blastp service at the NCBI database. Multiple sequence alignments for protein sequences were done using ClustalX2 (Larkin et al., 2007).
Results
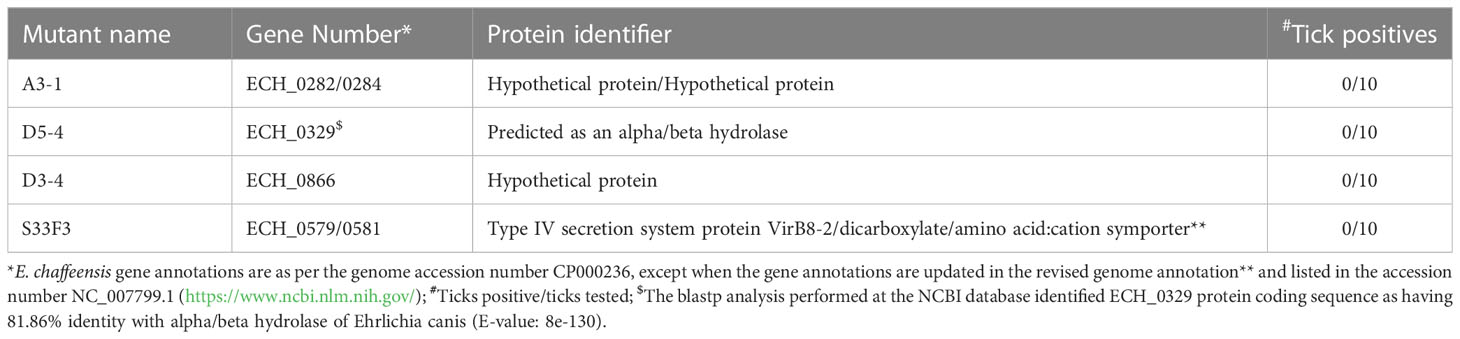
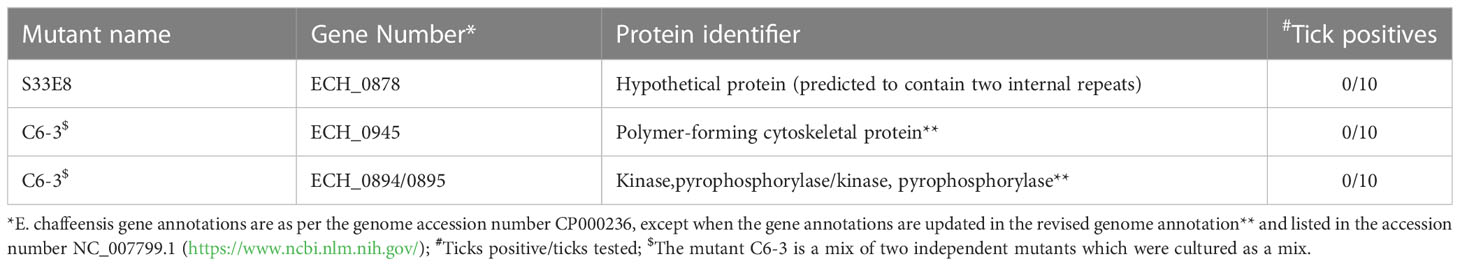
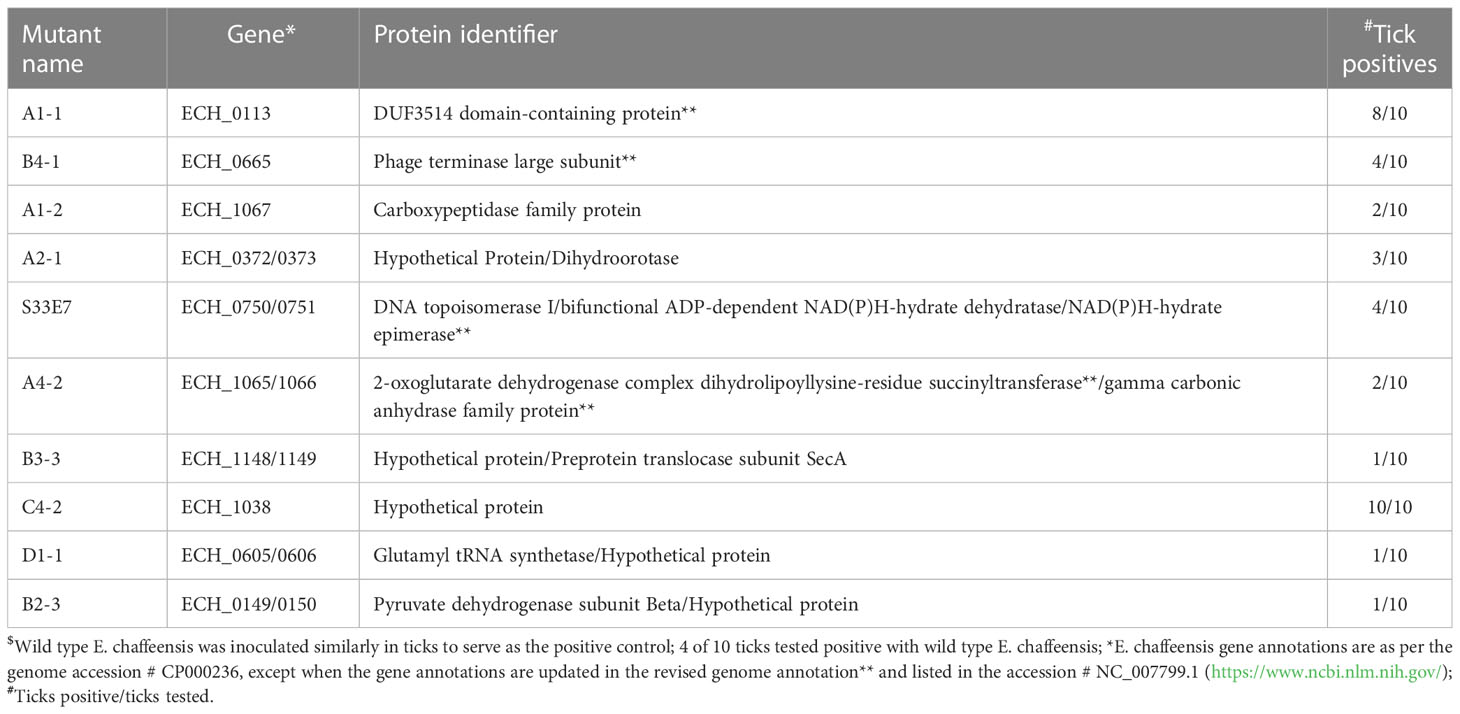
We recently reported a mutational analysis study describing 55 random mutations of E. chaffeensis. The mutants were used to define the functional gene requirements for the pathogen’s growth in a physiologically relevant canine host model (Wang et al., 2020). To determine the pathogen’s requirements for its adaptation to its tick vector (A. americanum), we screened 45 of the prior characterized E. chaffeensis mutants using our recently established tick in vivo infection assessment model (Cheng et al., 2015; Jaworski et al., 2017) (Table 1). The mutants used in the current study included 25 having insertion mutations within open reading frames (ORFs) (Table 1A), while the remaining 20 mutations were located within intergenic regions (Table 1B). Thirty-two of the mutants were previously identified as essential for the E. chaffeensis growth and persistence in the canine host (Wang et al., 2020). Contrary to this observation, only seven mutants were recognized as critical for the bacterial tick survival as they did not persist during molting from the infected nymphs to adults (Tables 2, 3), while 38 mutations had no impact on the E. chaffeensis presence in A. americanum (Tables 4, 5). Four of the mutants were also previously recognized as essential for the E. chaffeensis growth in the canine host (Table 2). Three mutations having no impact on the bacterial growth in the canine host had growth defect in the tick vector (Table 3). Mutations in the genes encoding for E. chaffeensis for the biosynthesis of biotin, protein and fatty acids, DNA repair, several outer membrane proteins, and an MDR efflux pump protein did not result in the loss of the bacterial persistence in A. americanum (Table 5). Similarly, several hypothetical proteins previously identified as essential for the E. chaffeensis persistent growth in the canine host were found to be not critical for its growth in the tick vector (Table 5).
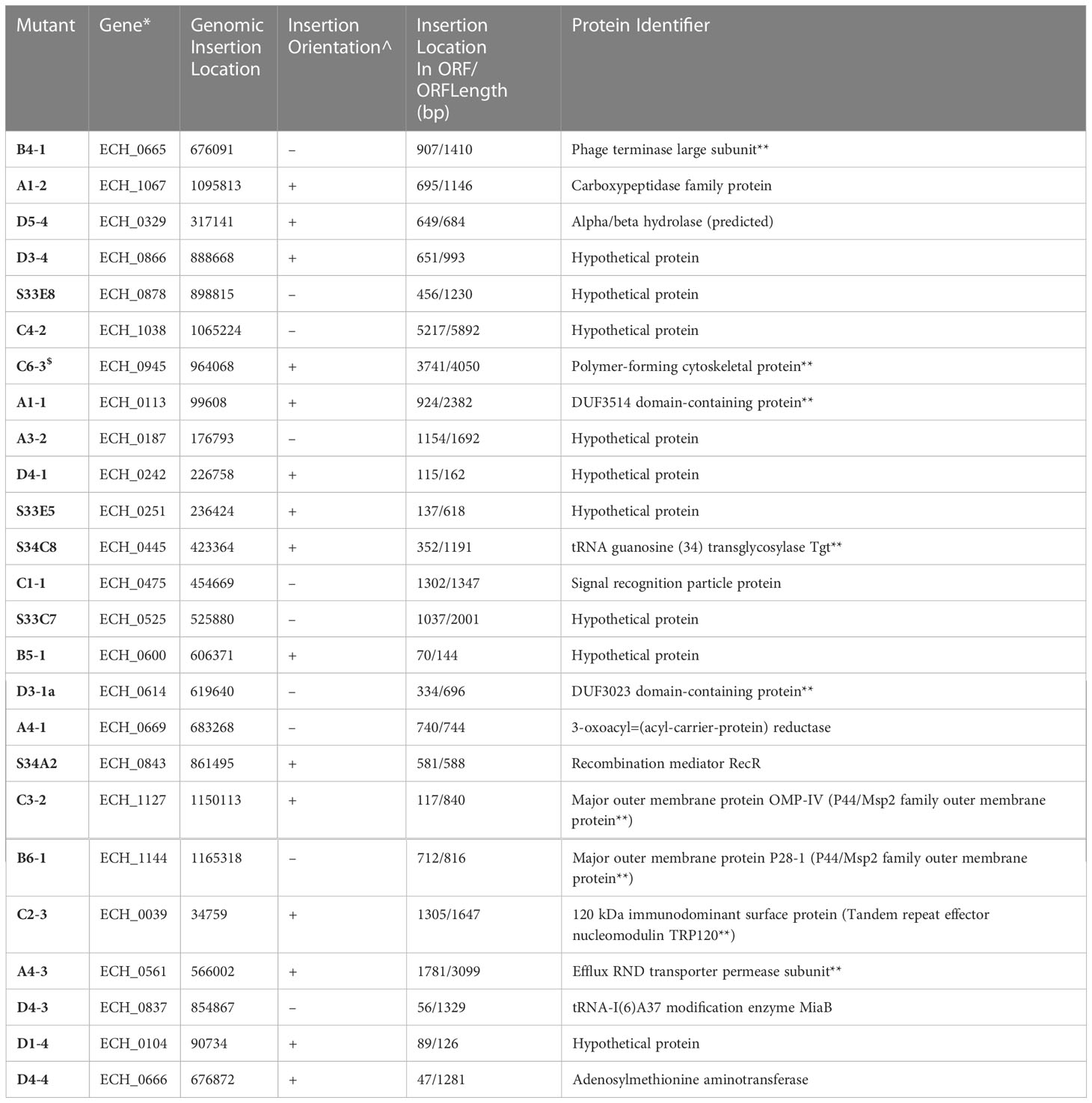
Table 1A E. chaffeensis intragenic mutants assessed for the growth and persistence in the tick host.
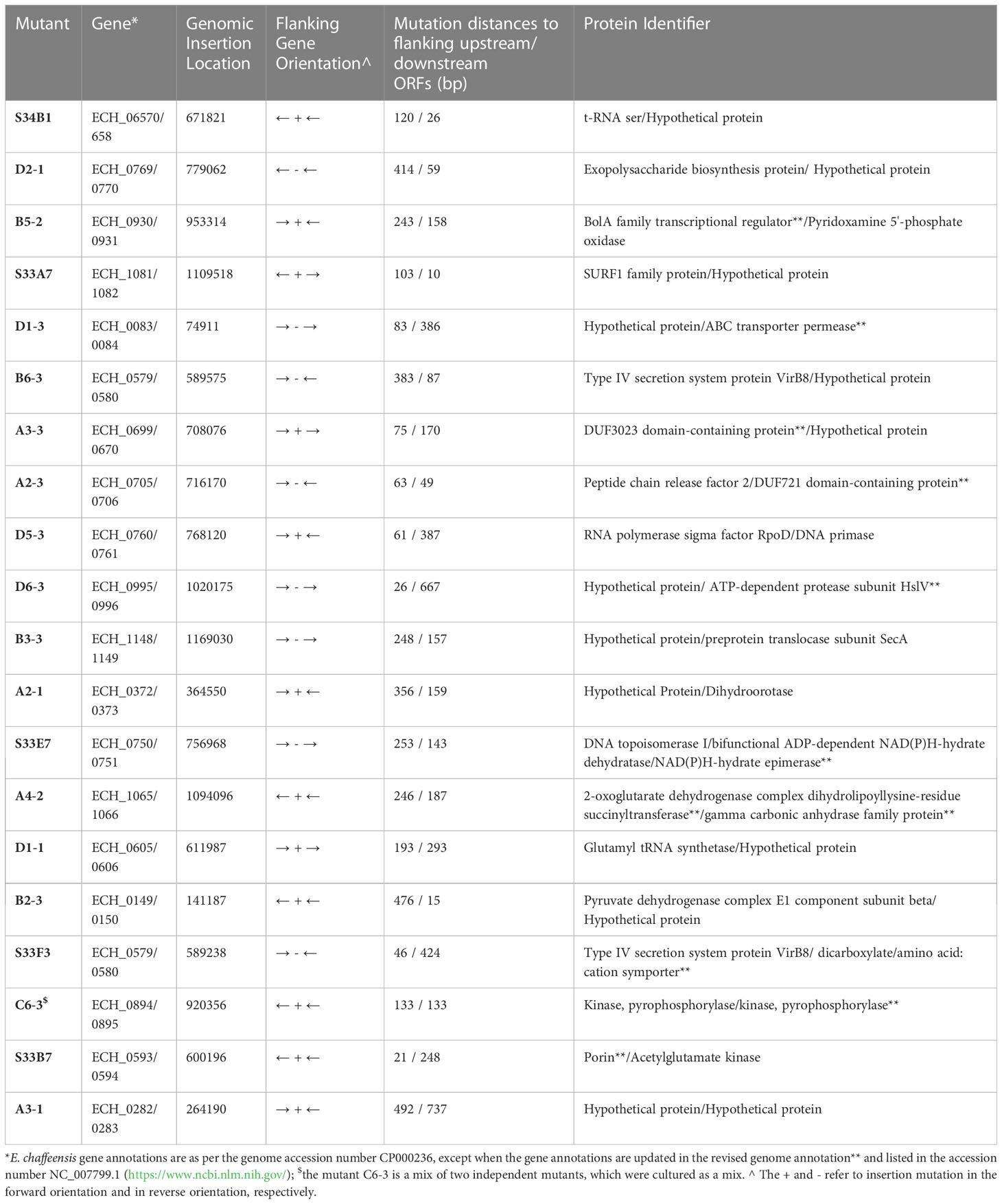
Table 1B E. chaffeensis intergenic mutants assessed for the growth and persistence in the tick host.
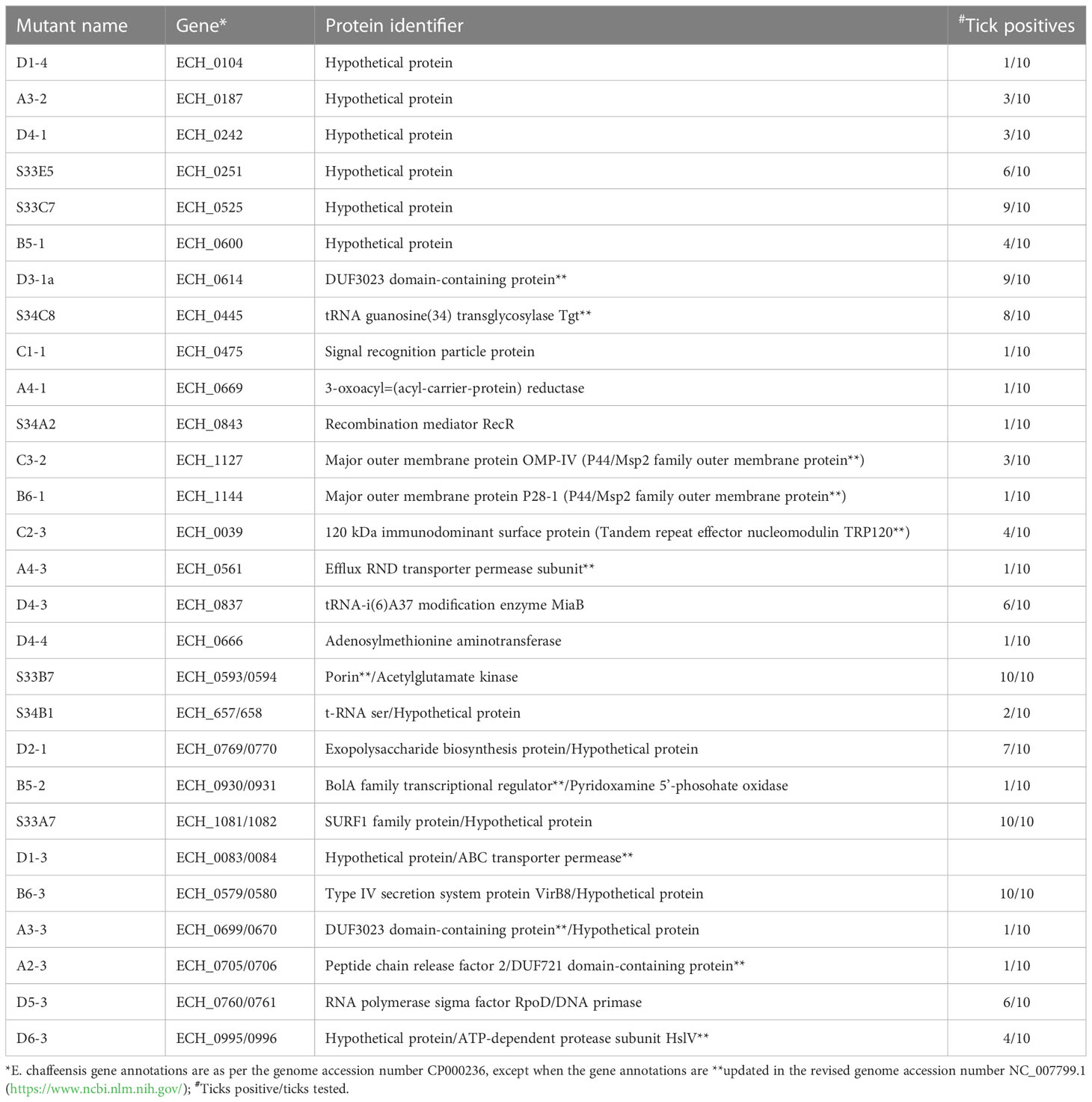
Table 5 E. chaffeensis mutants found as non-essential for the tick host which were previously found as essential for a vertebrate host (Wang et al., 2020).
Mutation within the ORF of ECH_0329 coding for a predicted alpha/beta hydrolase (Figure 1) caused growth defect of the organism in both the tick vector and in the canine host (Table 2). The remaining three mutations causing rapid clearance from the tick and canine hosts (Table 2) were within or near the genes identified as coding for hypothetical proteins (ECH_0866, ECH_0282 and 0284), a T4SS protein VirB8-2 (ECH_0579), and dicarboxylate amino acid:cation symporter protein (ECH_0581). Analysis using the PRED-TMR (a transmembrane protein prediction tool) (Pasquier et al., 1999; Spyropoulos et al., 2004), predicted the ORFs of ECH_0282, ECH_0284 and ECH_0866 as having one transmembrane domain each located at N-terminal regions (Figure 2). The second intergenic mutation is located downstream to the coding region of VirB8-2 (ECH_0579) and upstream to dicarboxylate amino acid:cation symporter protein (ECH_0581). PRED-TMR similarly predicted VirB8-2 as having one transmembrane domain at N-terminal. Dicarboxylate amino acid:cation symporter proteins is predicted to contain 8 transmembrane domains distributed throughout the protein sequence (Pasquier et al., 1999; Spyropoulos et al., 2004) (Figure 2).

Figure 1 Alignment of the predicted alpha/beta hydrolase of E. chaffeensis (GenBank # WP_006011217) encoded by gene ECH_0329, E. canis (GenBank # WP_011304785) and Serratia fonticola (GenBank # WP_202728975). Identical amino acids are shaded. Amino acid numbers of each protein are shown on the right.
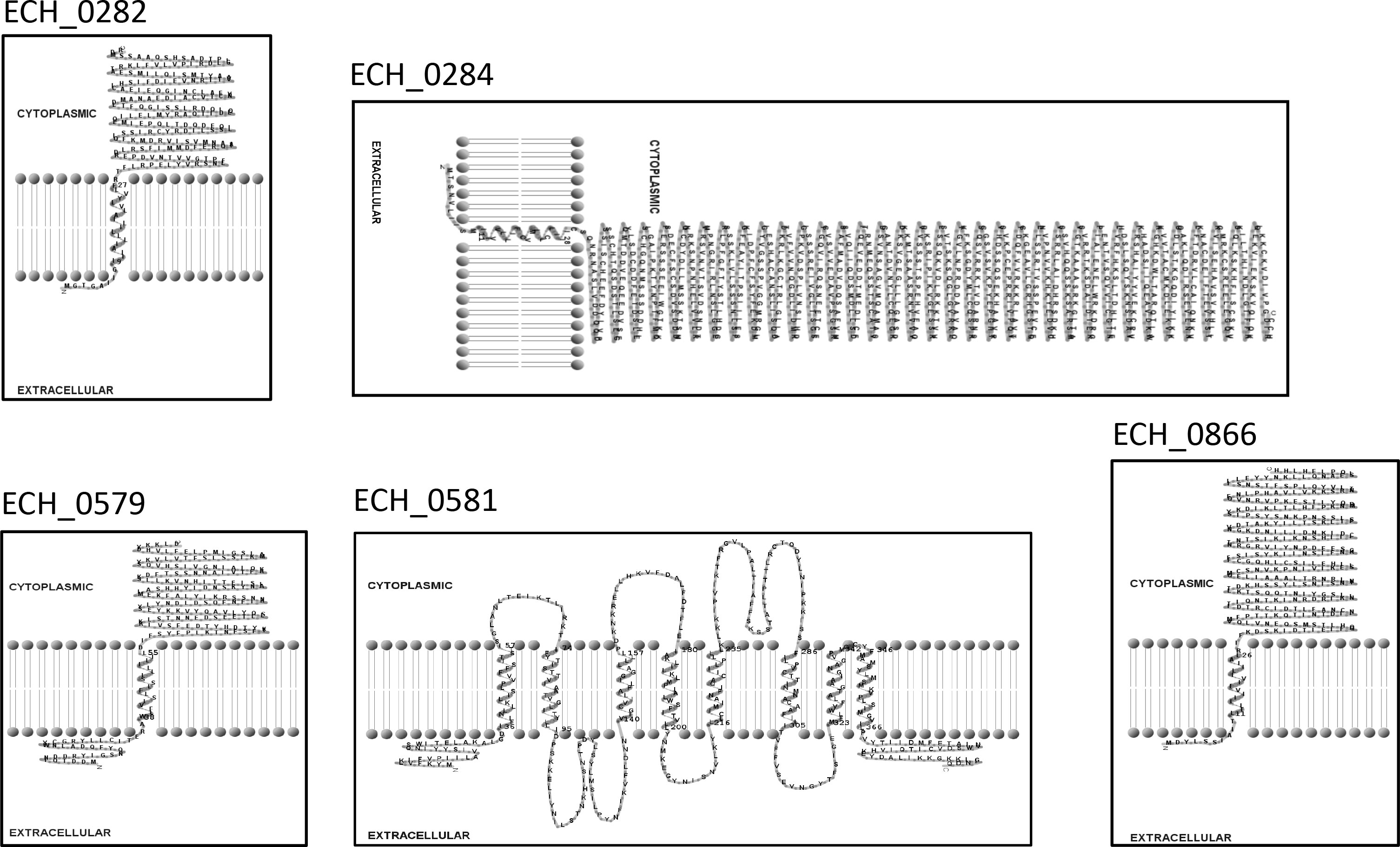
Figure 2 Transmembrane segments for five proteins of E. chaffeensis encoded by ECH_0282, ECH_0284, ECH_0579, ECH_0581 and ECH_0866 which were predicted by PRED-TMR (Pasquier et al., 1999). The software TMR Pres2D (Spyropoulos et al., 2004) was used to draw the predicted transmembrane domain.
Three mutants causing failed growth of E. chaffeensis (Table 3) only in the tick vector represented one each insertion mutation within hypothetical protein genes; ECH_0878 and ECH_0945 and the 3rd mutation located between genes ECH_0894 and ECH_0895 encoding for partial ORFs for a kinase/pyrophosphorylase (GenBank # NC_007799.1). The SMART protein and conserved domain search analysis (Letunic et al., 2020) found ECH_0878 ORF as having two highly homologous repeats with 64% sequence identity (Figure 3). Similarly, ECH_0945 codes for a polymer-forming cytoskeletal protein having two internal repeat sequences with 38% identity. This gene also has a similarity to bactofilin (Figure 4).
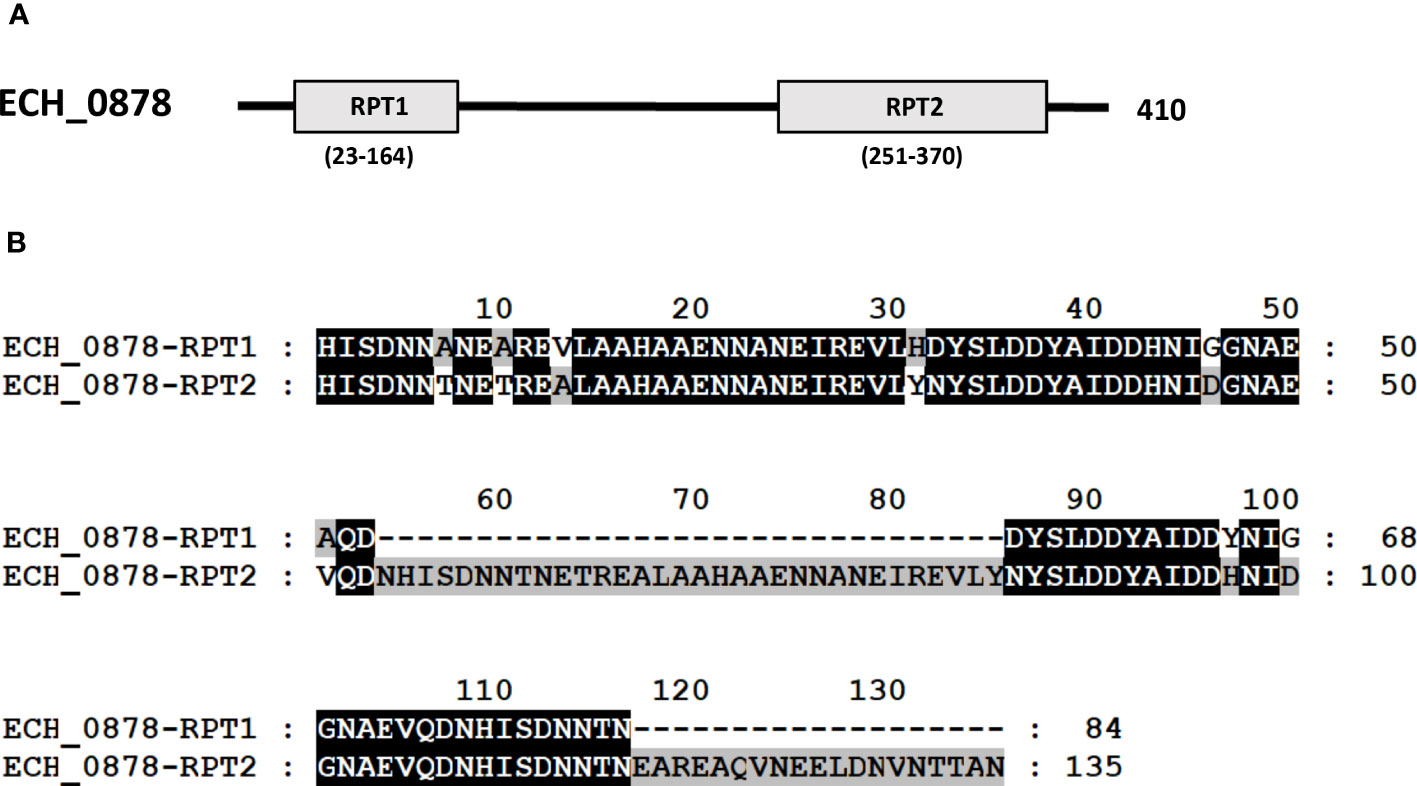
Figure 3 E chaffeensis ECH_0878 ORF was analyzed using the SMART protein and conserved domain search analysis (Letunic et al., 2020). (A) A scheme described that the protein encoded by gene ECH_0878 is identified to contain two major repeats, RPT1 and RPT2. (B) Alignment of two repeats of the protein encoded by gene ECH_0878.
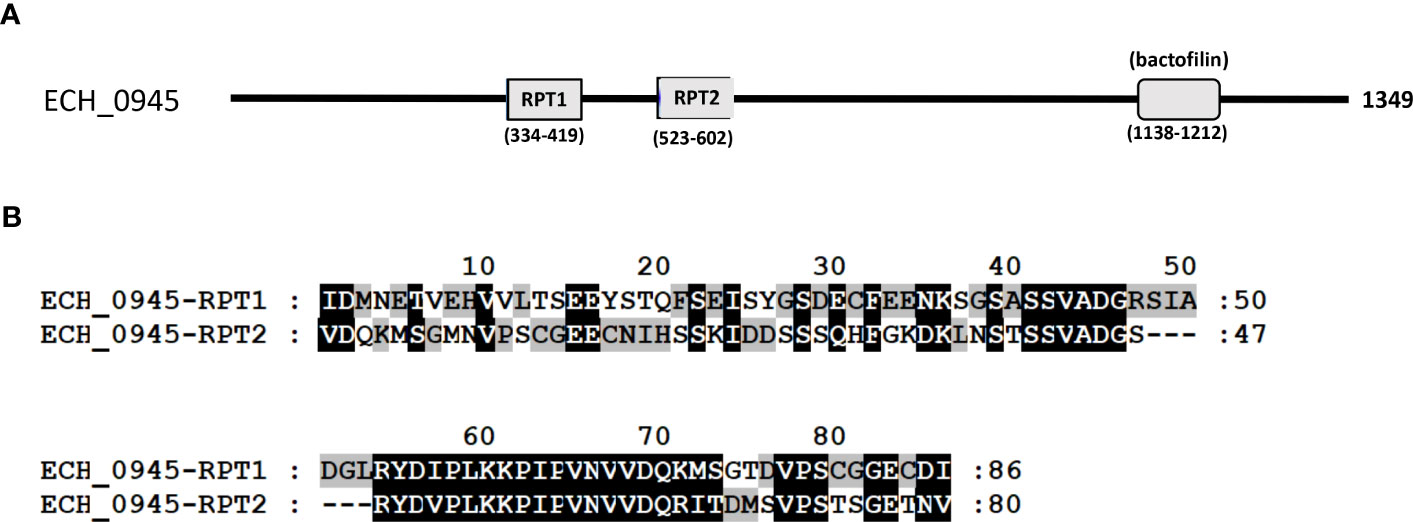
Figure 4 The polymer-forming cytoskeletal protein of E chaffeensis encoded by ECH_0945 was analyzed by SMART protein and conserved domain search (Letunic et al., 2020). (A) A scheme described that the protein encoded by gene ECH_0945 is identified to contain two major repeats (RPT1 and RPT2) and a conserved bactofilin. (B) Alignment of two repeats of the polymer-forming cytoskeletal protein encoded by gene ECH_0945.
Discussion
During the last four decades, several tick-transmitted rickettsial pathogens have been recognized as causing diseases in people and several vertebrate hosts (Rodino et al., 2020). E. chaffeensis is first identified in 1987 as a human pathogen transmitted from A. americanum tick and followed by the discovery of additional rickettsial pathogen infections caused by A. phagocytophilum, E. ewingii and E. muris eauclairensis (Dawson et al., 1991; Chen et al., 1994; Buller et al., 1999; Pritt et al., 2017). Despite several recent advances, much remains to be understood regarding how these broad host range rickettsial pathogens have evolved in adapting to vertebrate and tick hosts. Progress in molecular genetic studies focused on disrupting genes and their use to define the importance of rickettsial pathogens is very limited (Cheng et al., 2013; Crosby et al., 2015; McClure et al., 2017; Wang et al., 2020). Such studies are important in discovering proteins critical for host-pathogen interactions and in devising effective methods of controlling infections (Liu et al., 2007; Chen et al., 2012; Cheng et al., 2013; Crosby et al., 2014; Crosby et al., 2015; Nair et al., 2015; Lamason et al., 2018; Bekebrede et al., 2020; Wang et al., 2020; Hove et al., 2022). Despite low mutation efficiency in Ehrlichia and Anaplasma species, we recently reported the development of a small library of mutants for E. chaffeensis and described its value in identifying many genes critical for the bacterial persistent growth in a physiologically relevant vertebrate host infection model (Wang et al., 2020). Our prior studies demonstrated that function disruption mutations in many pathogen genes can be detrimental to E. chaffeensis persistence in the canine host, while not having an impact on the in vitro growth conditions (Cheng et al., 2013; McGill et al., 2016; Wang et al., 2020). The prior studies also demonstrated that the pathogen is exposed to unique challenges during its growth in vivo and that it requires many functional genes to facilitate its adaptation and persistence in a vertebrate host.
Considering our prior published evidence documenting that E. chaffeensis protein expression is highly variable during its replication in vertebrate and tick host background (Singu et al., 2005; Singu et al., 2006; Seo et al., 2008), in the current study we tested the hypothesis that the pathogen has unique functional gene requirements. Specifically, we investigated which E. chaffeensis genes were critical for the pathogen’s growth in its tick host; A. americanum. We observed remarkable differences in the E. chaffeensis genes required for its growth in the tick compared to the vertebrate host. We utilized our previously well-established needle infection method to screen 45 E. chaffeensis mutants for their abilities to infect A. americanum nymphs and their transstadial transmission during molting to the adult stage (Cheng et al., 2015; Jaworski et al., 2017). We observed that only 7 mutations in E. chaffeensis caused growth impact for the pathogen in A. americanum, unlike the documentation of many mutations as essential for the bacterial persistence in the canine host (Wang et al., 2020). Notably, 28 mutations caused growth defects of E. chaffeensis in the canine host grew normally in A. americanum suggesting that E. chaffeensis adaptation strategies differ for the tick compared to the vertebrate host. E. chaffeensis genes essential for its continued growth in a vertebrate host included genes encoding for three immunodominant membrane proteins; the 120 kDa protein and two p28 Omps (OMP 1V and p28-1) and those involved in the biosynthesis of biotin, protein and fatty acids, DNA repair and MDR efflux pump. Mutations in all these genes, however, did not prevent the bacterial replication within its tick host. While different membrane protein expressions are anticipated, it is not clear how the deficiency resulting from mutations in several metabolic genes of E. chaffeensis is compensated during its replication and transstadial transmission in the tick. Possible explanations are that the deficiency of the metabolic pathway proteins is compensated from other sources within the tick. Hard ticks cohost numerous pathogenic and non-pathogenic bacteria, including several rickettsial endosymbionts (Wikel, 2018; Rodino et al., 2020; Hirunkanokpun et al., 2022; Qi et al., 2022). Recently, Wang et al. described the detailed overview of how ticks and other blood sucking ectoparasites depend on symbiotic microorganisms for completing essential aspects of their biology, such as development and reproduction (Wang et al., 2023). Endosymbionts and other microorganisms co-existing in ticks may likely also benefit from each other. The deficiencies resulting from mutations leading to the deprivation of essential metabolites and proteins in E. chaffeensis are possibly compensated by other bacteria which coexist in A. americanum. This hypothesis, however, remains to be tested.
The bacterial survival with deficiency for the immunodominant outer membrane proteins may reflect that the three previously identified membrane proteins are only critical for the E. chaffeensis growth in the vertebrate host. These data are consistent with the previous report demonstrating that the two p28-Omp proteins (OMP 1V and p28-1) expressed in a vertebrate host are not among the E. chaffeensis proteins found in the tick host (Unver et al., 2002). Of the three mutants that were exclusively absent in the tick vector, one of the mutations is located near a gene (ECH_0895) which encodes for a kinase/pyrophosphorylase. This protein is among the outer membrane expressed proteins of E. chaffeensis when it is cultured in tick cells (ISE6) (Seo et al., 2008). Another mutation within ECH_0945 gene codes for polymer-forming cytoskeletal protein having high homology to bactofilins. Bactofilins are known to have diverse functions, including cell stalk formation to chromosome segregation and motility in bacteria (Deng et al., 2019). Together, the unique differences noted in the E. chaffeensis outer membrane-expressed proteins suggest that the pathogen has distinct needs for the cell surface expressed proteins when replicating in the tick host compared to a vertebrate host.
A mutation in the ECH_0329 gene coding for an alpha/beta hydrolase caused rapid clearance from both vertebrate and tick hosts. This protein is expressed on the E. chaffeensis morula membrane during the bacterial replication in a macrophage cell line (Kondethimmanahalli and Ganta, 2022) and another alpha/beta hydrolase (ECH_0326 gene product) is present on the bacterial outer membrane (Seo et al., 2008). Alpha/beta hydrolases are a large family of proteins with many known functions and are present in several pathogenic bacteria. For example, Mycobacterium tuberculosis has 105 alpha/beta hydrolases, and they are involved in lipid metabolism, evasion and modulation of immune responses, detoxification, and metabolic adaptations including growth (Yang et al., 2014; Johnson, 2017). These enzymes in M. tuberculosis are known to contribute to the bacterial virulence (Johnson, 2017). The clearance of E. chaffeensis alpha/beta hydrolase mutant suggests that this multifunctional enzyme and likely its homologs may be important for the bacterial survival in both vertebrate and tick hosts.
Additional mutations causing the pathogen clearance from A. americanum included those present in intergenic locations. They included mutations near genes coding for proteins having transmembrane domains, such as the protein encoded from ECH_0282 gene, T4SS protein VirB8-2 (ECH_0579), dicarboxylate amino acid:cation symporter (ECH_0581), and kinase/pyrophosphorylase (ECH_0895). The dicarboxylate amino acid:cation symporter is critical for the uptake of C(4)-dicarboxylates, such as succinate or l-malate, in bacteria (Youn et al., 2009). Similarly, T4SS-associated proteins are critical for supporting the bacterial evasion of host responses mediated by the release of T4SS effectors into infected host cells (Rikihisa, 2021; Lin et al., 2023). While it is unclear how the intergenic mutations described may have impacted the functions of genes 5’ and 3’ to insertion sites, our previously published detailed investigation demonstrates that such mutations, as well as some intragenic mutations, cause polar effects impacting gene expressions from genes upstream and/or down to the mutated regions (Cheng et al., 2015). For example, an intergenic mutation downstream to ECH_0230 prevented the transcription from the gene and caused in vivo growth defect. A mutation between ECH_0479 and ECH_0480 caused the transcriptionally silent ECH_0480 gene to be transcriptionally active; intergenic mutations between ECH_0202 and ECH_0203 and between ECH_0284 and ECH_0285 caused RNA expression enhancement from ECH_0203 and ECH_0285, respectively. Similarly, mutations within ECH_0379 and ECH_0660 resulted in preventing transcription from these genes as well as the genes located 5’ to them; ECH_0378 and ECH_0659, respectively. These two mutations also caused the bacterial growth defect in vertebrate hosts. Thus, it is not surprising that the intergenic mutations have detrimental outcomes for the bacterium’s in vivo growth. Intergenic space sequences typically include promoter regions and other regulating domains where small non-coding RNAs and DNA binding proteins bind and contribute to transcriptional regulation (Wassarman, 2002; Raghavan et al., 2011; Schroeder et al., 2015). The functional roles of all the predicted essential membrane expressed proteins remains to be determined for E. chaffeensis replication in both vertebrate and tick hosts.
Considering the exposure of outer membrane domains to host cells, the detection of several bacterial membrane-bound proteins of E. chaffeensis as essential for the pathogen’s continued survival in vertebrate and tick hosts will be valuable for furthering our understanding of bacterial pathogenesis, host-pathogen interactions and in devising effective methods of control and prevention. In particular, very little information is currently available regarding what bacterial factors define the survival of E. chaffeensis and other related tick-borne rickettsiales in dual host environments and in causing pathogenesis. This mutational analysis study is the first to identify many E. chaffeensis genes uniquely required for its adaptation to tick and vertebrate hosts and that the data will be valuable for advancing our understanding of the bacterial host-pathogen interactions.
Data availability statement
The datasets presented in this article are not readily available. Requests to access the datasets should be directed to cm9tYW5nYW50YUBtaXNzb3VyaS5lZHU=.
Author contributions
DJ, YW, AN, HL, and RG contributed to experimental design and execution, drafting, and finalizing the manuscript. RG conceived the project and secured funding. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the PHS grant #s AI070908 and AI152418 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, USA. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Author RG is on the editorial board as a specialty Chief Editor for the Veterinary and Zoonotic Infection section of the Frontiers of Cellular and Infection Microbiology.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bekebrede, H., Lin, M., Teymournejad, O., Rikihisa, Y. (2020). Discovery of in vivo virulence genes of obligatory intracellular bacteria by random mutagenesis. Front. Cell. Infect Microbiol. 10, 2–2. doi: 10.3389/fcimb.2020.00002
Breitschwerdt, E. B., Hegarty, B. C., Hancock, S. I. (1998). Sequential evaluation of dogs naturally infected with ehrlichia canis, ehrlichia chaffeensis, ehrlichia equi, ehrlichia ewingii, or bartonella vinsonii. J. Clin. Microbiol. 36, 2645–2651. doi: 10.1128/JCM.36.9.2645-2651.1998
Buller, R. S., Arens, M., Hmiel, S. P., Paddock, C. D., Sumner, J. W., Rikhisa, Y., et al. (1999). Ehrlichia ewingii, a newly recognized agent of human ehrlichiosis. N. Engl. J. Med. 341, 148–155. doi: 10.1056/NEJM199907153410303
Chen, S. M., Dumler, J. S., Bakken, J. S., Walker, D. H. (1994). Identification of a granulocytotropic ehrlichia species as the etiologic agent of human disease. J. Clin. Microbiol. 32, 589–595. doi: 10.1128/jcm.32.3.589-595.1994
Chen, S. M., Popov, V. L., Feng, H. M., Wen, J., Walker, D. H. (1995). Cultivation of ehrlichia chaffeensis in mouse embryo, vero, BGM, and L929 cells and study of ehrlichia-induced cytopathic effect and plaque formation. Infect. Immun. 63, 647–655. doi: 10.1128/iai.63.2.647-655.1995
Chen, G., Severo, M. S., Sakhon, O. S., Choy, A., Herron, M. J., Felsheim, R. F., et al. (2012). Anaplasma phagocytophilum dihydrolipoamide dehydrogenase 1 affects host-derived immunopathology during microbial colonization. Infect. Immun. 80, 3194–3205. doi: 10.1128/IAI.00532-12
Cheng, C., Ganta, R. R. (2008). Laboratory maintenance of Ehrlichia chaffeensis and ehrlichia canis and recovery of organisms for molecular biology and proteomics studies. Curr. Protoc. Microbiol. 3. doi: 10.1002/9780471729259.mc03a01s9
Cheng, C., Nair, A. D. S., Indukuri, V. V., Gong, S., Felsheim, R. F., Jaworski, D., et al. (2013). Targeted and random mutagenesis of Ehrlichia chaffeensis for the identification of genes required for In vivo infection. PLoS Pathog. 9, e1003171. doi: 10.1371/journal.ppat.1003171
Cheng, C., Nair, A. D. S., Jaworski, D. C., Ganta, R. R. (2015). Mutations in Ehrlichia chaffeensis causing polar effects in gene expression and differential host specificities. PLoS One 10, e0132657. doi: 10.1371/journal.pone.0132657
Crosby, F. L., Brayton, K. A., Magunda, F., Munderloh, U. G., Kelley, K. L., Barbet, A. F. (2015). Reduced infectivity in cattle for an outer membrane protein mutant of anaplasma marginale. Appl. Environ. Microbiol. 81, 2206–2214. doi: 10.1128/AEM.03241-14
Crosby, F. L., Wamsley, H. L., Pate, M. G., Lundgren, A. M., Noh, S. M., Munderloh, U. G., et al. (2014). Knockout of an outer membrane protein operon of anaplasma marginale by transposon mutagenesis. BMC Genomics 15, 278–278. doi: 10.1186/1471-2164-15-278
Davidson, W. R., Lockhart, J. M., Stallknecht, D. E., Howerth, E. W., Dawson, J. E., Rechav, Y. (2001). Persistent ehrlichia chaffeensis infection in white-tailed deer. J. Wildl Dis. 37, 538–546. doi: 10.7589/0090-3558-37.3.538
Dawson, J. E., Anderson, B. E., Fishbein, D. B., Sanchez, J. L., Goldsmith, C. S., Wilson, K. H., et al. (1991). Isolation and characterization of an ehrlichia sp. from a patient diagnosed with human ehrlichiosis. J. Clin. Microbiol. 29, 2741–2745. doi: 10.1128/jcm.29.12.2741-2745.1991
Dawson, J. E., Biggie, K. L., Warner, C. K., Cookson, K., Jenkins, S., Levine, J. F., et al. (1996). Polymerase chain reaction evidence of Ehrlichia chaffeensis, an etiologic agent of human monocytic ehrlichiosis, in dogs, from southeast Virginia. Am. J. Vet. Res. 57, 1175–1179.
Deng, X., Gonzalez Llamazares, A., Wagstaff, J. M., Hale, V. L., Cannone, G., Mclaughlin, S. H., et al. (2019). The structure of bactofilin filaments reveals their mode of membrane binding and lack of polarity. Nat. Microbiol. 4, 2357–2368. doi: 10.1038/s41564-019-0544-0
Dugan, V. G., Little, S. E., Stallknecht, D. E., Beall, A. D. (2000). Natural infection of domestic goats with Ehrlichia chaffeensis. J. Clin. Microbiol. 38, 448–449. doi: 10.1128/JCM.38.1.448-449.2000
Dumler, J. S., Barbet, A. F., Bekker, C. P., Dasch, G. A., Palmer, G. H., Ray, S. C., et al. (2001). Reorganization of genera in the families rickettsiaceae and anaplasmataceae in the order rickettsiales: unification of some species of ehrlichia with anaplasma, cowdria with ehrlichia and ehrlichia with neorickettsia, descriptions of six new species combinations and designation of ehrlichia equi and 'HGE agent' as subjective synonyms of ehrlichia phagocytophila. Int. J. systematic evolutionary Microbiol. 51, 2145–2165. doi: 10.1099/00207713-51-6-2145
Higuita, N. I. A., Franco-Paredes, C., Henao-Martínez, A. F. (2021). The expanding spectrum of disease caused by the lone star tick, amblyomma americanum. Infez Med. 29, 378–385. doi: 10.53854/liim-2903-8
Hirunkanokpun, S., Ahantarig, A., Baimai, V., Pramual, P., Rakthong, P., Trinachartvanit, W. (2022). Spotted fever group rickettsia, anaplasma and coxiella-like endosymbiont in haemaphysalis ticks from mammals in Thailand. Vet. Res. Commun. 46, 1209–1219. doi: 10.1007/s11259-022-09980-x
Hove, P., Madesh, S., Nair, A., Jaworski, D., Liu, H., Ferm, J., et al. (2022). Targeted mutagenesis in anaplasma marginale to define virulence and vaccine development against bovine anaplasmosis. PLoS Pathog. 18, e1010540. doi: 10.1371/journal.ppat.1010540
Jaworski, D. C., Cheng, C., Nair, A. D. S., Ganta, R. R. (2017). Amblyomma americanum ticks infected with in vitro cultured wild-type and mutants of Ehrlichia chaffeensis are competent to produce infection in naïve deer and dogs. Ticks tick-borne Dis. 8, 60–64. doi: 10.1016/j.ttbdis.2016.09.017
Johnson, G. (2017). The α/β hydrolase fold proteins of mycobacterium tuberculosis, with reference to their contribution to virulence. Curr. Protein Pept. Sci. 18, 190–210. doi: 10.2174/1389203717666160729093515
Kocan, A. A., Levesque, G. C., Whitworth, L. C., Murphy, G. L., Ewing, S. A., Barker, R. W. (2000). Naturally occurring Ehrlichia chaffeensis infection in coyotes from Oklahoma. Emerg. Infect. Dis. 6, 477–480. doi: 10.3201/eid0605.000505
Kondethimmanahalli, C., Ganta, R. R. (2022). Proteome analysis of ehrlichia chaffeensis containing phagosome membranes revealed the presence of numerous bacterial and host proteins. Front. Cell. Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.1070356
Lamason, R. L., Kafai, N. M., Welch, M. D. (2018). A streamlined method for transposon mutagenesis of rickettsia parkeri yields numerous mutations that impact infection. PLoS One 13, e0197012. doi: 10.1371/journal.pone.0197012
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. A., Mcwilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Letunic, I., Khedkar, S., Bork, P. (2020). SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460. doi: 10.1093/nar/gkaa937
Lin, M., Koley, A., Zhang, W., Pei, D., Rikihisa, Y. (2023). Inhibition of ehrlichia chaffeensis infection by cell-permeable macrocyclic peptides that bind type IV secretion effector etf-1. PNAS Nexus. 2, 1-15 doi: 10.1093/pnasnexus/pgad017
Liu, Z. M., Tucker, A. M., Driskell, L. O., Wood, D. O. (2007). Mariner-based transposon mutagenesis of rickettsia prowazekii. Appl. Environ. Microbiol. 73, 6644–6649. doi: 10.1128/AEM.01727-07
Lockhart, J. M., Davidson, W. R., Stallknecht, D. E., Dawson, J. E., Howerth, E. W. (1997). Isolation of ehrlichia chaffeensis from wild white-tailed deer (Odocoileus virginianus) confirms their role as natural reservoir hosts. J. Clin. Microbiol. 35, 1681–1686. doi: 10.1128/jcm.35.7.1681-1686.1997
McClure, E. E., Chávez, A. S. O., Shaw, D. K., Carlyon, J. A., Ganta, R. R., Noh, S. M., et al. (2017). Engineering of obligate intracellular bacteria: progress, challenges and paradigms. Nat. Rev. Microbiol. 15, 544. doi: 10.1038/nrmicro.2017.59
McGill, J. L., Nair, A. D. S., Cheng, C., Rusk, R. A., Jaworski, D. C., Ganta, R. R. (2016). Vaccination with an attenuated mutant of Ehrlichia chaffeensis induces pathogen-specific CD4+ T cell immunity and protection from tick-transmitted wild-type challenge in the canine host. PLoS One 11, e0148229. doi: 10.1371/journal.pone.0148229
Nair, A. D. S., Cheng, C., Jaworski, D. C., Ganta, S., Sanderson, M. W., Ganta, R. R. (2015). Attenuated mutants of Ehrlichia chaffeensis induce protection against wild-type infection challenge in the reservoir host and in an incidental host. Infection Immun. 83, 2827–2835. doi: 10.1128/IAI.00487-15
Pasquier, C., Promponas, V. J., Palaios, G. A., Hamodrakas, J. S., Hamodrakas, S. J. (1999). A novel method for predicting transmembrane segments in proteins based on a statistical analysis of the SwissProt database: the PRED-TMR algorithm. Protein Eng. 12, 381–385. doi: 10.1093/protein/12.5.381
Pritt, B. S., Allerdice, M. E. J., Sloan, L. M., Paddock, C. D., Munderloh, U. G., Rikihisa, Y., et al. (2017). Proposal to reclassify Ehrlichia muris as Ehrlichia muris subsp. muris subsp. nov. and description of Ehrlichia muris subsp. eauclairensis subsp. nov., a newly recognized tick-borne pathogen of humans. Int. J. Systematic Evolutionary Microbiol. 67, 2121–2126. doi: 10.1099/ijsem.0.001896
Qi, Y., Ai, L., Zhu, C., Ye, F., Lv, R., Wang, J., et al. (2022). Wild hedgehogs and their parasitic ticks coinfected with multiple tick-borne pathogens in jiangsu province, Eastern China. Microbiol. Spectr. 10, e0213822. doi: 10.1128/spectrum.02138-22
Raghavan, R., Groisman, E. A., Ochman, H. (2011). Genome-wide detection of novel regulatory RNAs in e. coli. Genome Res. 21, 1487–1497. doi: 10.1101/gr.119370.110
Rikihisa, Y. (2021). The "Biological weapons" of ehrlichia chaffeensis: novel molecules and mechanisms to subjugate host cells. Front. Cell Infect. Microbiol. 11, 830180. doi: 10.3389/fcimb.2021.830180
Rodino, K. G., Theel, E. S., Pritt, B. S. (2020). Tick-borne diseases in the united states. Clin. Chem. 66, 537–548. doi: 10.1093/clinchem/hvaa040
Schroeder, C. L. C., Narra, H. P., Rojas, M., Sahni, A., Patel, J., Khanipov, K., et al. (2015). Bacterial small RNAs in the genus rickettsia. BMC Genomics 16, 1075. doi: 10.1186/s12864-015-2293-7
Seo, G.-M., Cheng, C., Tomich, J., Ganta, R. R. (2008). Total, membrane, and immunogenic proteomes of macrophage- and tick cell-derived Ehrlichia chaffeensis evaluated by liquid chromatography-tandem mass spectrometry and MALDI-TOF methods. Infect. Immun. 76, 4823–4832. doi: 10.1128/IAI.00484-08
Singu, V., Liu, H., Cheng, C., Ganta, R. R. (2005). Ehrlichia chaffeensis expresses macrophage- and tick cell-specific 28-kilodalton outer membrane proteins. Infection Immun. 73, 79–87. doi: 10.1128/IAI.73.1.79-87.2005
Singu, V., Peddireddi, L., Sirigireddy, K. R., Cheng, C., Munderloh, U., Ganta, R. R. (2006). Unique macrophage and tick cell-specific protein expression from the p28/p30-outer membrane protein multigene locus in Ehrlichia chaffeensis and Ehrlichia canis. Cell. Microbiol. 8, 1475–1487. doi: 10.1111/j.1462-5822.2006.00727.x
Spyropoulos, I. C., Liakopoulos, T. D., Bagos, P. G., Hamodrakas, S. J. (2004). TMRPres2D: high quality visual representation of transmembrane protein models. Bioinformatics 20, 3258–3260. doi: 10.1093/bioinformatics/bth358
Unver, A., Rikihisa, Y., Stich, R. W., Ohashi, N., Felek, S. (2002). The omp-1 major outer membrane multigene family of Ehrlichia chaffeensis is differentially expressed in canine and tick hosts. Infection Immun. 70, 4701–4704. doi: 10.1128/IAI.70.8.4701-4704.2002
Wang, J., Gao, L., Aksoy, S. (2023). Microbiota in disease-transmitting vectors. Nat. Rev. Microbiol. doi: 10.1038/s41579-023-00901-6
Wang, Y., Nair, A. D. S., Alhassan, A., Jaworski, D. C., Liu, H., Trinkl, K., et al. (2020). Multiple Ehrlichia chaffeensis genes critical for its persistent infection in a vertebrate host are identified by random mutagenesis coupled with in vivo infection assessment. Infection Immun. 88, e00316-20. doi: 10.1128/IAI.00316-20
Wassarman, K. M. (2002). Small RNAs in bacteria: diverse regulators of gene expression in response to environmental changes. Cell 109, 141–144. doi: 10.1016/S0092-8674(02)00717-1
Wikel, S. K. (2018). Ticks and tick-borne infections: complex ecology, agents, and host interactions. Veterinary Sci. 5, 60. doi: 10.3390/vetsci5020060
Yang, Y., Kulka, K., Montelaro, R. C., Reinhart, T. A., Sissons, J., Aderem, A., et al. (2014). A hydrolase of trehalose dimycolate induces nutrient influx and stress sensitivity to balance intracellular growth of mycobacterium tuberculosis. Cell Host Microbe 15, 153–163. doi: 10.1016/j.chom.2014.01.008
Keywords: Ehrlichia chaffeensis, rickettsial genetics, tick-borne pathogens, tick- and host-specific Ehrlichia genes, mutational studies
Citation: Jaworski DC, Wang Y, Nair A, Liu H and Ganta RR (2023) Multiple Ehrlichia chaffeensis genes critical for persistent infection in a vertebrate host are identified as nonessential for its growth in the tick vector; Amblyomma americanum. Front. Cell. Infect. Microbiol. 13:1220025. doi: 10.3389/fcimb.2023.1220025
Received: 09 May 2023; Accepted: 13 June 2023;
Published: 27 June 2023.
Edited by:
Adela Oliva Chavez, Texas A and M University, United StatesReviewed by:
Mingqun Lin, The Ohio State University, United StatesXin-Ru Wang, Upstate Medical University, United States
Copyright © 2023 Jaworski, Wang, Nair, Liu and Ganta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Roman R. Ganta, cm9tYW5nYW50YUBtaXNzb3VyaS5lZHU=
 Deborah C. Jaworski1
Deborah C. Jaworski1 Huitao Liu
Huitao Liu Roman R. Ganta
Roman R. Ganta