Microbiological diagnostic performance of metagenomic next-generation sequencing compared with conventional culture for patients with community-acquired pneumonia
- 1Department of Intensive Care Unit, Quanzhou First Hospital Affiliated to Fujian Medical University, Quanzhou, China
- 2Shanghai Biotecan Pharmaceuticals Co., Ltd., Shanghai, China
- 3Shanghai Zhangjiang Institute of Medical Innovation, Shanghai, China
- 4Department of Clinical Laboratory, Huanghe Sanmenxia Hospital Affiliated to Henan University of Science and Technology, Sanmenxia, China
by Lin T, Tu X, Zhao J, Huang L, Dai X, Chen X, Xu Y, Li W, Wang Y, Lou J, Wu S and Zhang H (2023) Front. Cell. Infect. Microbiol. 13:1136588. doi: 10.3389/fcimb.2023.1136588
Error in Figure/Table Legend
In the published article, there was an error in the legend for Figure 3 as published. “(C) protozoa level” was included in legend of Figure 3, which should be deleted. The corrected legend appears below.
Figure 3 The comparison and overlap of infected pathogens between mNGS and laboratory culture in all 205 patients with CAP. (A) Bacteria levels; (B) fungi level; and (C) virus level.
In the published article, there was an error in the legend for Figure 4 as published. “(C) protozoa level” was included in legend of Figure 4, which should be deleted. The corrected legend appears below.
Figure 4 Infected pathogens detected by mNGS in severe and non-severe patients with CAP. (A) bacteria levels; (B) fungi level; and (C) virus level.
In the published article, there was an error in the legend for Figure 6 as published. “(C) protozoa level” was included in legend of Figure 6, which should be deleted. The corrected legend appears below.
Figure 6 Infected pathogens detected by mNGS in immunocompetent and immunocompromised patients with severe pneumonia. (A) Bacteria levels; (B) fungi level; and (C) virus level.
Error in Figure/Table
In the published article, there was an error in Figure 3 as published. Orientia tsutsugamushi was divided in Part C “Protozoa”, which should be moved to Part A “Bacteria”. The corrected Figure 3 and its caption ** The comparison and overlap of infected pathogens between mNGS and laboratory culture in all 205 patients with CAP. (A) Bacteria levels; (B) fungi level; and (C) virus level. appear below.
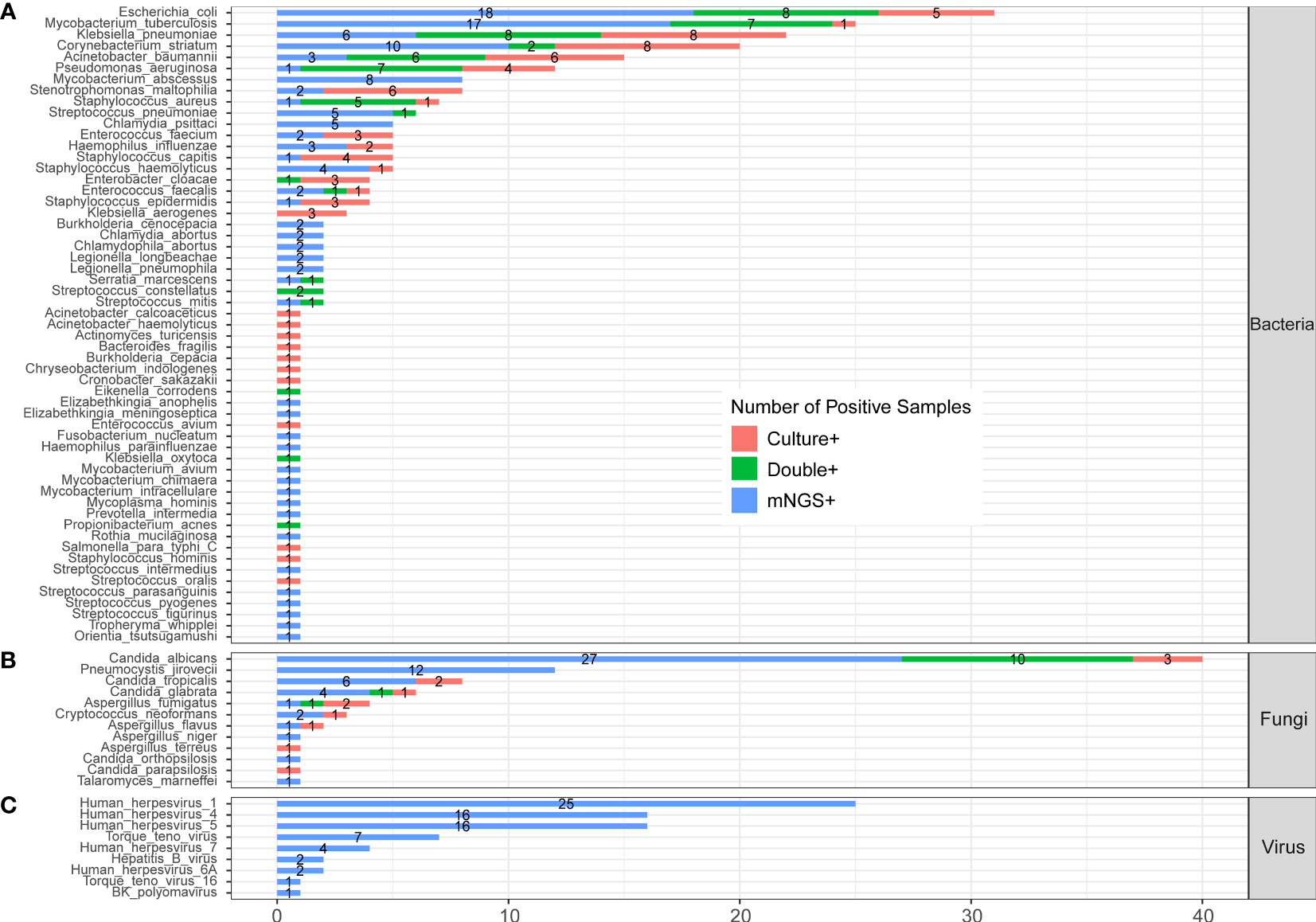
Figure 3 The comparison and overlap of infected pathogens between mNGS and laboratory culture in all 205 patients with CAP. (A) Bacteria levels; (B) fungi level; and (C) virus level.
In the published article, there was an error in Figure 4 as published. Orientia tsutsugamushi was divided in Part C “Protozoa”, which should be moved to Part A “Bacteria”. The corrected Figure 4 and its caption ** Infected pathogens detected by mNGS in severe and non-severe patients with CAP. (A) bacteria levels; (B) fungi level; and (C) virus level. appear below.
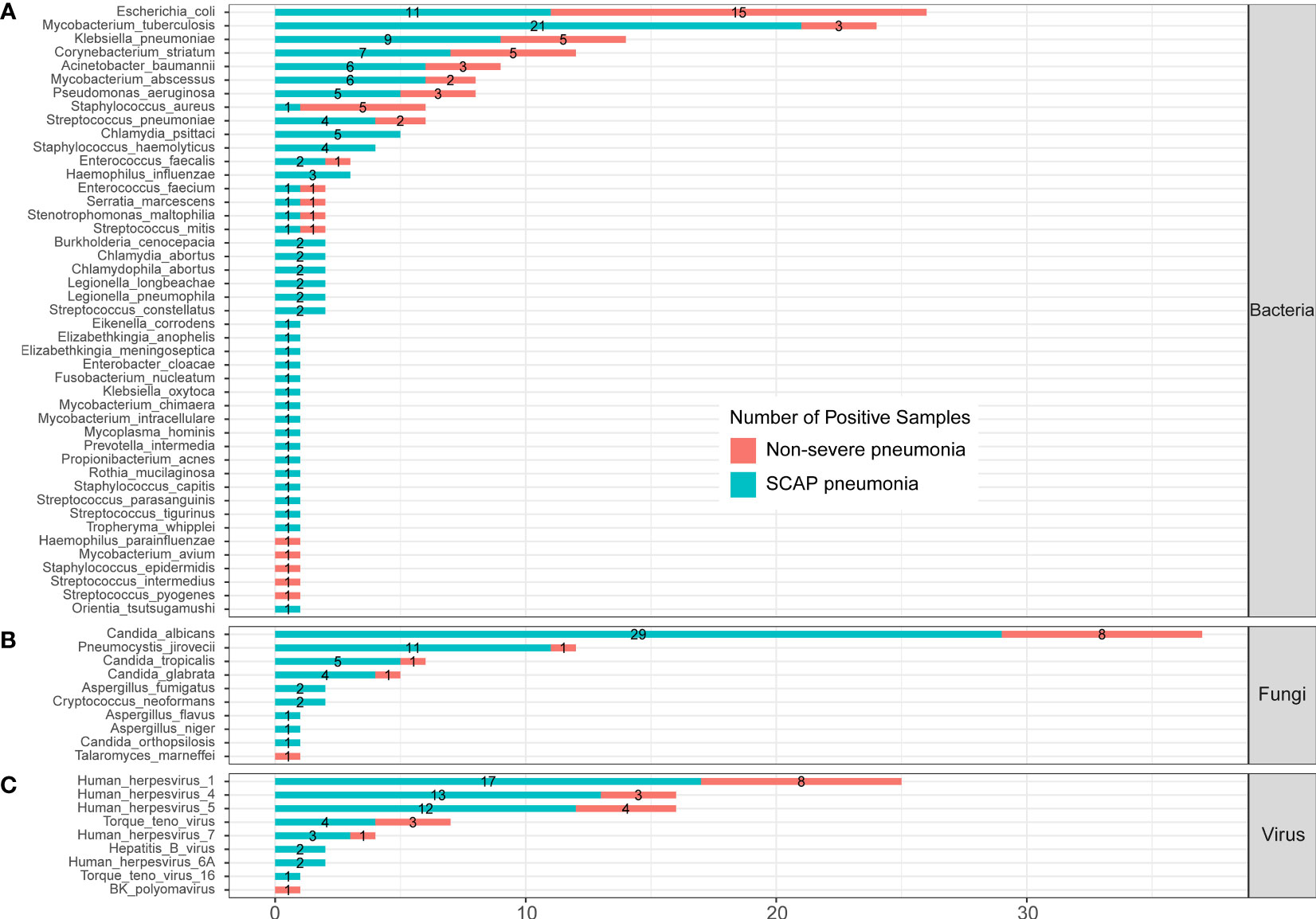
Figure 4 Infected pathogens detected by mNGS in severe and non-severe patients with CAP. (A) bacteria levels; (B) fungi level; and (C) virus level.
In the published article, there was an error in Figure 6 as published. Orientia tsutsugamushi was divided in Part C “Protozoa”, which should be moved to Part A “Bacteria”. The corrected Figure 6 and its caption ** Infected pathogens detected by mNGS in immunocompetent and immunocompromised patients with severe pneumonia. (A) Bacteria levels; (B) fungi level; and (C) virus level. appear below.
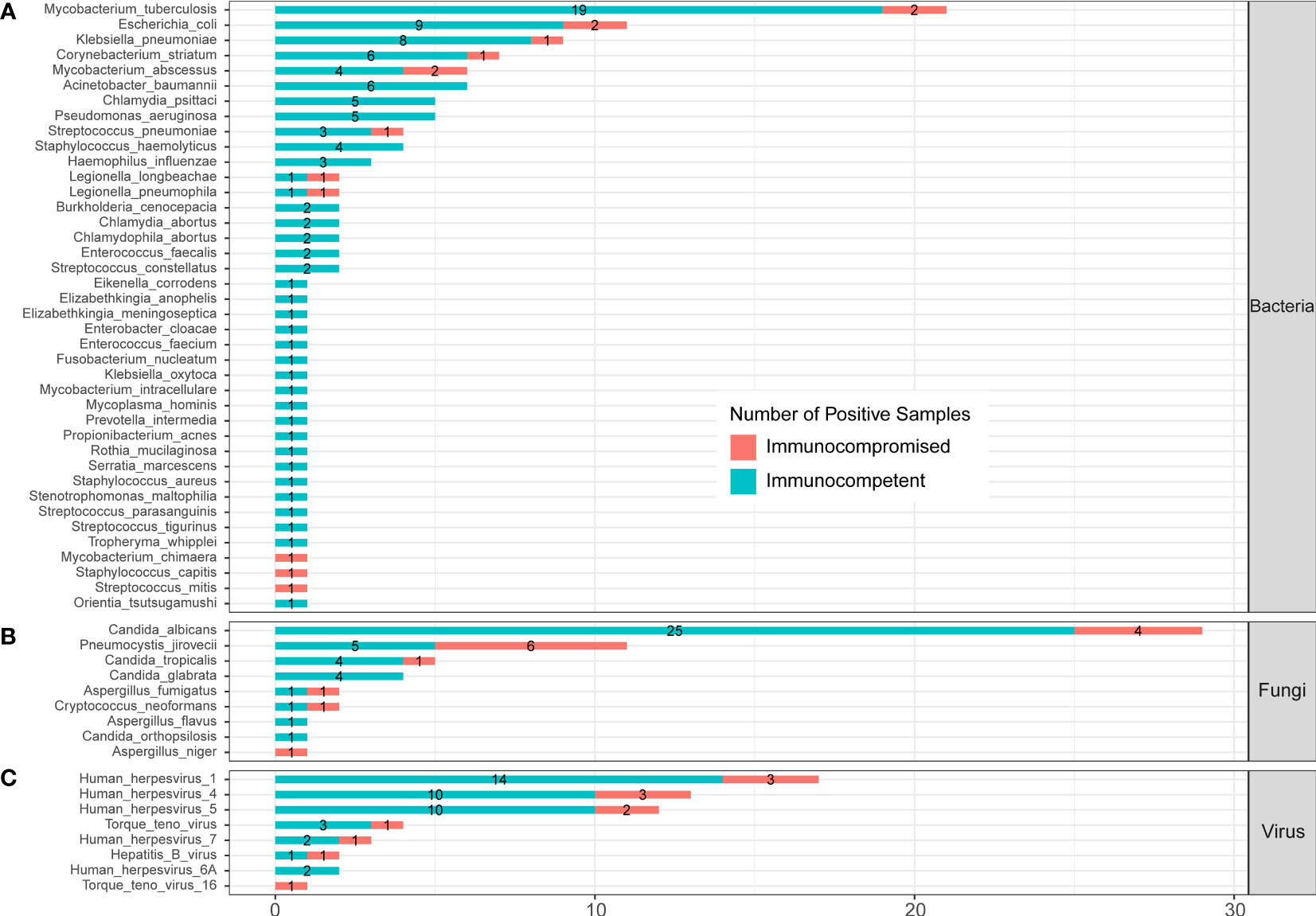
Figure 6 Infected pathogens detected by mNGS in immunocompetent and immunocompromised patients with severe pneumonia. (A) Bacteria levels; (B) fungi level; and (C) virus level.
Incorrect Supplementary Material
In the published article, there was an error in Supplementary Figure 2. Orientia tsutsugamushi was divided in Part C “Protozoa”, which should be moved to Part A “Bacteria”.
Supplementary Figure 2. The comparison and overlap of infected pathogens between metagenomic next-generation sequencing (mNGS) and laboratory culture in 186 patients with CAP whose sample type was consistent between two methods. (A) Bacteria levels; (B) Fungi level; (C) Virus level.
In the published article, there was an error in Supplementary Table 2. Orientia tsutsugamushi was divided in Part of “Protozoa”, which should be moved to Part of “Bacteria”.
Supplementary Table 2. Comparison of pathogens detected by mNGS between non-severe CAP and SCAP groups.
In the published article, there was an error in Supplementary Table 3. Orientia tsutsugamushi was divided in Part of “Protozoa”, which should be moved to Part of “Bacteria”.
Supplementary Table 3. Comparison of pathogens detected by mNGS between immunocompetent and immunocompromised patients with SCAP.
Text Correction
In the published article, there was an error. Orientia tsutsugamushi was wrongly be categorized as Protozoa, which should be a bacterium. So, the number of detected bacteria and protozoan should be corrected.
A correction has been made to 3 Results, 3.4 Pathogens’ profile of all CAP patients according to detection methods, Paragraph 1. This sentence previously stated:
“The detected pathogens were divided into four kingdoms, namely, bacteria, fungi, protozoa, and viruses. A total of 56 bacteria (Figure 3A), 12 fungi (Figure 3B), 1 protozoan (Figure 3C), and 9 viruses (Figure 3D) were detected by mNGS and the CMT.”
The corrected sentence appears below:
“The detected pathogens were divided into three kingdoms, namely, bacteria, fungi, and viruses. A total of 57 bacteria (Figure 3A), 12 fungi (Figure 3B), and 9 viruses (Figure 3C) were detected by mNGS and the CMT.”
In the published article, there was an error. Orientia tsutsugamushi was wrongly be categorized as Protozoa, which should be a bacterium. So, the number of detected bacteria and protozoan should be corrected.
A correction has been made to 3 Results, 3.4.1 Profile of bacteria, Paragraph 1. This sentence previously stated:
“A total of 22 bacteria were only detected by mNGS, including Mycobacterium abscessus (M. abscessus, n=8), Chlamydia psittaci (C. psittaci, n=5), Burkholderia cenocepacia (n=2), and Chlamydia abortus (n=2).”
The corrected sentence appears below:
“A total of 23 bacteria were only detected by mNGS, including Mycobacterium abscessus (M. abscessus, n=8), Chlamydia psittaci (C. psittaci, n=5), Burkholderia cenocepacia (n=2), and Chlamydia abortus (n=2).”
In the published article, there was an error. Orientia tsutsugamushi was wrongly be categorized as Protozoa, which should be a bacterium. So, result of protozoa should be deleted.
A correction has been made to 3 Results, 3.4.2 Profile of fungi and protozoa, sub-title and Paragraph 1. This paragraph previously stated:
“3.4.2 Profile of fungi and protozoa
In the fungi level (Figure 3B), Candida albicans (C. albicans) detected in 40 patients was the most frequent fungus, and 27 of cases were only detected by mNGS. Pneumocystis jirovecii (P. jirovecii) was the second common fungus detected in 12 cases by mNGS only. One case was positive of Orientia tsutsugamushi, which was detected by mNGS only (Figure 3C).”
The corrected paragraph appears below:
“3.4.2 Profile of fungi
In the fungi level (Figure 3B), Candida albicans (C. albicans) detected in 40 patients was the most frequent fungus, and 27 of cases were only detected by mNGS. Pneumocystis jirovecii (P. jirovecii) was the second common fungus detected in 12 cases by mNGS only.”
In the published article, there was an error. Orientia tsutsugamushi was wrongly be categorized as Protozoa, which should be a bacterium. So, the number of detected bacteria should be corrected.
A correction has been made to 3 Results, 3.5 Comparison of pathogens detected by metagenomic next-generation sequencing between severe and non-severe community-acquired pneumonia patients, Paragraph 1. These sentences previously stated:
“A total of 44 bacteria were identified by mNGS in 205 CAP patients. There were 14 bacteria found in both severe and non-severe CAP patients, 5 bacteria were found only in infected non-severe patients, and 25 bacteria were only detected in SCAP patients (Figure 4A).”
The corrected sentence appears below:
“A total of 45 bacteria were identified by mNGS in 205 CAP patients. There were 14 bacteria found in both severe and non-severe CAP patients, 5 bacteria were found only in infected non-severe patients, and 26 bacteria were only detected in SCAP patients (Figure 4A).”
In the published article, there was an error. Orientia tsutsugamushi was wrongly be categorized as Protozoa, which should be a bacterium. So, the number of detected bacteria should be corrected.
A correction has been made to 3 Results, 3.6 Comparison of pathogens between immunocompromised and immunocompetent patients with severe community-acquired pneumonia, Paragraph 2. These sentences previously stated:
“A total of 39 bacteria were identified by mNGS from 144 SCAP patients. Among them, 28 bacteria were detected in immunocompetent cases only, 3 bacteria were detected in immunocompromised cases only, and 8 bacteria were found in both immunocompetent and immunocompromised groups.”
The corrected sentence appears below:
“A total of 40 bacteria were identified by mNGS from 144 SCAP patients. Among them, 29 bacteria were detected in immunocompetent cases only, 3 bacteria were detected in immunocompromised cases only, and 8 bacteria were found in both immunocompetent and immunocompromised groups.”
In the published article, there was an error. Orientia tsutsugamushi was wrongly be categorized as Protozoa, which should be a bacterium. So, the word “Protozoa” should be deleted in Conclusions.
A correction has been made to Conclusions. These sentences previously stated:
“mNGS is superior in detecting MTB, NTM, viruses, P. jirovecii, chlamydia, and protozoa.”
The corrected sentence appears below:
“mNGS is superior in detecting MTB, NTM, viruses, P. jirovecii, and chlamydia.”
The authors apologize for these errors and state that this do not change the scientific conclusions of the article in any way. The original article has been updated.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: metagenomic next-generation sequencing, culture, community-acquired pneumonia, conventional microbiological test, pathogen detection
Citation: Lin T, Tu X, Zhao J, Huang L, Dai X, Chen X, Xu Y, Li W, Wang Y, Lou J, Wu S and Zhang H (2023) Corrigendum: Microbiological diagnostic performance of metagenomic next-generation sequencing compared with conventional culture for patients with community-acquired pneumonia. Front. Cell. Infect. Microbiol. 13:1205802. doi: 10.3389/fcimb.2023.1205802
Received: 14 April 2023; Accepted: 07 July 2023;
Published: 14 August 2023.
Edited and Reviewed by:
Christoph Gabler, Freie Universität Berlin, GermanyCopyright © 2023 Lin, Tu, Zhao, Huang, Dai, Chen, Xu, Li, Wang, Lou, Wu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingwei Lou, amluZ3dlaWxvdUBiaW90ZWNhbi5jb20=; Shouxin Wu, c3d1QGJpb3RlY2FuLmNvbQ==; Hongling Zhang, emhhbmdob25nbGluZ3lzakAxNjMuY29t
†These authors have contributed equally to this work and share first authorship
 Tianlai Lin1†
Tianlai Lin1† Jiangman Zhao
Jiangman Zhao