
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 05 July 2023
Sec. Parasite and Host
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1204707
 Sabine Bachmaier1
Sabine Bachmaier1 Matthew K. Gould1
Matthew K. Gould1 Eleni Polatoglou1
Eleni Polatoglou1 Radoslaw Omelianczyk1
Radoslaw Omelianczyk1 Ana E. Brennand1
Ana E. Brennand1 Maha A. Aloraini2
Maha A. Aloraini2 Jane C. Munday2
Jane C. Munday2 David Horn3
David Horn3 Michael Boshart1*
Michael Boshart1* Harry P. de Koning2*
Harry P. de Koning2*Cyclic AMP signalling in trypanosomes differs from most eukaryotes due to absence of known cAMP effectors and cAMP independence of PKA. We have previously identified four genes from a genome-wide RNAi screen for resistance to the cAMP phosphodiesterase (PDE) inhibitor NPD-001. The genes were named cAMP Response Protein (CARP) 1 through 4. Here, we report an additional six CARP candidate genes from the original sample, after deep sequencing of the RNA interference target pool retrieved after NPD-001 selection (RIT-seq). The resistance phenotypes were confirmed by individual RNAi knockdown. Highest level of resistance to NPD-001, approximately 17-fold, was seen for knockdown of CARP7 (Tb927.7.4510). CARP1 and CARP11 contain predicted cyclic AMP binding domains and bind cAMP as evidenced by capture and competition on immobilised cAMP. CARP orthologues are strongly enriched in kinetoplastid species, and CARP3 and CARP11 are unique to Trypanosoma. Localization data and/or domain architecture of all CARPs predict association with the T. brucei flagellum. This suggests a crucial role of cAMP in flagellar function, in line with the cell division phenotype caused by high cAMP and the known role of the flagellum for cytokinesis. The CARP collection is a resource for discovery of unusual cAMP pathways and flagellar biology.
Conservation of signalling pathways and proteins among different phyla of eukaryotes is very limited, particularly in protozoa. Cyclic nucleotides are present as second messengers in almost all organisms, and in protozoa they can regulate growth, development and metabolic adaptation. In trypanosomes an important role for cAMP is suggested by an estimated 80 adenylate cyclase encoding genes (Salmon et al., 2012b), many of which are expressed throughout the life cycle (Alexandre et al., 1996); multiple are present in the plasma membrane (Bridges et al., 2008) or mainly localised to the flagellar surface (Paindavoine et al., 1992; Saada et al., 2014). These cyclases consist of a conserved catalytic domain, a single trans-membrane domain and a variable extracellular domain (Salmon, 2018). It is thus possible that various extracellular ligands control their activity (Paindavoine et al., 1992), but none have been identified and activation by dimerization has been observed under acidic, hypotonic or other stress conditions (Nolan et al., 2000; Gould and de Koning, 2011; Salmon, 2018). In recent years, the adenylate cyclases have been implicated in cytokinesis (Salmon et al., 2012a), immune evasion in the mammalian host (Salmon et al., 2012b), and social motility in the procyclic insect stage of the parasite (Lopez et al., 2015; Oberholzer et al., 2015; Saada et al., 2015). Control of cAMP homeostasis is crucial as knockdown of the locus containing the cAMP phosphodiesterases TbrPDEB1 and TbrPDEB2 is lethal (Oberholzer et al., 2007) and inhibitors of these enzymes have potent antitrypanosomal activity (de Koning et al., 2012; de Heuvel et al., 2019). However, the effectors and cascades activated or inhibited by cAMP remain largely unknown in trypanosomes, given the absence of genes encoding the known cAMP effectors in mammalian cells. Most importantly, the homologue of Protein Kinase A (PKA) is not activated by, and does not bind, cyclic nucleotides in T. brucei (Bachmaier and Boshart, 2014; Bubis et al., 2018; Bachmaier et al., 2019). This has stimulated a screen aiming at identification of novel cAMP effectors or target proteins. We previously reported a set of four T. b. brucei genes whose RNAi repression protected against the high intracellular cAMP concentration resulting from the inhibition of TbrPDEB1/2; these genes were termed cAMP Response Protein (CARP) 1 – 4 (Gould et al., 2013). Out of these, only CARP3, a gene unique to trypanosomes, has been investigated and it was shown to be essential for social motility of procyclic trypanosomes through direct interaction with and regulation of adenylate cyclases (Bachmaier et al., 2022). CARP3 is therefore an upstream regulator of cAMP signalling and the effectors and cAMP-binding protein(s) remain to be identified.
The Trypanosoma brucei subspecies T. b. gambiense and T. b. rhodesiense are responsible for the disease known as Human African Trypanosomiasis (HAT), or sleeping sickness (Büscher et al., 2017), whereas T. b. brucei and the related trypanosomes T. congolense, T. vivax, T. evansi and T. equiperdum all contribute to the various manifestations of Animal African Trypanosomiasis (AAT), known under names such as nagana, surra and dourine (Giordani et al., 2016). Sleeping sickness has long been among the most neglected diseases, being transmitted by the tsetse fly, which makes it a problem of rural Africa, but more recently progress has been made and case numbers have dropped with more active control measures (Barrett, 2018; Franco et al., 2020) and the introduction of the first oral drug, fexinidazole, against the infection (Lindner et al., 2020). In contrast, little progress has been made in reducing the impact of AAT, which continues to have devastating effects on livestock and, consequently, on rural economies and food security in Africa and beyond, in part because T. evansi, T. vivax and T. equiperdum are not dependent on tsetse fly transmission (Jones and Dávila, 2001; Desquesnes, 2004; Desquesnes et al., 2013; Aregawi et al., 2019). No new animal trypanosomiasis drugs have been introduced for decades and the treatment options are both limited and threatened by resistance (Delespaux and de Koning, 2007; Melaku and Birasa, 2013; Giordani et al., 2016; Kasozi et al., 2022). Among the issues that have held back drug development against trypanosomiasis is the paucity of well-validated drug targets – a result of the many gaps in our knowledge of their unique biochemistry and cell biology. The limited understanding of mostly non-conserved signalling mechanisms and proteins is one reason why these pathways, preferred targets in mammalian drug development, are less exploited for the parasites.
Here we revisit the screen that identified CARP1 – 4 by deep sequencing of the RNAi library grown out after challenge with PDE inhibitor NPD-001 (Gould et al., 2013). We identify six additional genes that confer resistance to perturbation of intracellular cAMP, CARP6 – CARP11, and validate their phenotype by targeted RNAi for each individual gene. Most of the CARPs are unique to the kinetoplastidae. CARP1 and CARP11 contain potential cAMP-binding domains and cAMP binding to these proteins was confirmed by capture on cAMP-linked agarose beads. The identification of novel and trypanosomatid-specific cAMP effector candidates will elucidate the evolution of cAMP signalling and eventually provide attractive drug target candidates.
Bloodstream forms of the monomorphic Trypanosoma brucei brucei strain Lister 427 MiTat 1.2 were cultivated at 37°C and 5% CO2 in modified HMI-9 medium (Vassella et al., 1997) supplemented with 10% (v/v) heat-inactivated foetal bovine serum (FBS). For maintenance of tetracycline repressor TetR and T7 polymerase in the trypanosome cell lines MiTat 1.2 13-90 or MiTat 1.2 single marker (Wirtz et al., 1999), 2.5 µg/mL G418 and 5 µg/mL hygromycin B or only 2.5 µg/mL G418, respectively, were added to the culture medium. Cell density was monitored using a haemocytometer and was kept below 1×106 cells/mL for continuous growth of replicative long slender bloodstream forms.
Sequencing of the libraries was performed on a Thermo Scientific Ion Proton at the Glasgow Polyomics facility using the 200 base pair sequencing kit. Sequences containing a terminal RNAi-vector junction sequence (GCCTCGCGA) were mapped to the T. brucei 927 reference genome (Alsford et al., 2012), the selection allowed for a single aberration, either a base change or a single base insertion or deletion. The filtered reads were mapped to the reference genome with Bowtie2 using the local mode alignment. The aligned reads were then assigned to a region of interest, reads that either were fully contained within the region or only partially overlap the region of interest. The percentage of mapped reads for any gene is thus the number of reads aligned to that gene, divided by the total of all reads × 100. The counts for each replicate were also expressed as normalised mapped reads by dividing the total assigned to any gene by the CDS length × 100.
For in situ tagging of CARP6 and CARP10, the long primer PCR tagging strategy was used on pPOTv2 as a template (Dean et al., 2017) using primers 2CTb427.10.12390F (CGGCATTTGAGGAGATTGAGAGACGACGGCAGCAGGAAGCTGCAGCAAAGGCCGCTGCGGACGATGCTATGCCGTTAGTAactagtgtgagcaagg) and 2CTb427.10.12390R (AAAAAAAAAAGTTAGGGGACCGCGAAGGAAAAAAAGGAGTAAAGAACCAGGTCACTCCTGAATATGTACATGGTAGAGATccaatttgagagacctgtgc) for C-terminal YFP-TY (Yellow Fluorescent Protein) tagging of CARP6 and primers 2NTb427.02.5030F (TGGTGTTTTGAGTGCAATGTTTTTTTCACTTCTTTTTCTTTTCTTTACCACTTTAGCCGGAAGTTGTTGGCGGTGTCGCGgtataatgcagacctgctgc) and 2NTb427.02.5030R (ACCGTGTTACTCGACACATGATCGGTACACACAGCCTTCAAATATCTGTAAATCTGCACCACTGCTTCCTTCGTTGTCATcttgtacagctcgtccatgc) for N-terminal TY-YFP tagging of CARP10. PCR products were purified by phenol-chloroform extraction prior to transfection and transfected cells were grown in the presence of 2 µg/mL blasticidin or hygromycin, respectively.
For tetracycline-inducible knock down of putative CARPs, the following fragments were amplified from T. brucei Lister 427 MiTat 1.2 single marker genomic DNA and cloned into p2T7-177-BLE (Wickstead et al., 2002) via BamHI and XhoI restriction sites: CARP5 (Tb927.10.1740): RNAi targeting fragment: ORF nt 365-776, amplified with primers Tb427.10.1740_RNAi_Frag-F and Tb427.10.1740_RNAi_Frag-R; CARP6 (Tb927.10.12390): RNAi targeting fragment: ORF nt 256-682, amplified with primers Tb427.10.12390_RNAi_Frag-F and Tb427.10.12390_RNAi_Frag-R; CARP7 (Tb927.7.4510): RNAi targeting fragment: ORF nt 625-1177, amplified with primers Tb427.07.4510_RNAi_Frag-F and Tb427.07.4510_RNAi_Frag-R; CARP8 (Tb927.11.3910): RNAi targeting fragment: ORF nt 2076-2624, amplified with primers Tb427tmp.02.1410_RNAi_Frag-F and Tb427tmp.02.1410_RNAi_Frag-R; CARP9 (Tb927.8.4640, alternative name: CMF19, component of motile flagella 19): RNAi targeting fragment: ORF nt 601-1092, amplified with primers Tb427.08.4640_RNAi_Frag-F and Tb427.08.4640_RNAi_Frag-R; CARP10 (Tb927.11.7180): RNAi targeting fragment: ORF nt 2-439, amplified with primers Tb427tmp.02.5030_RNAi_Frag-F and Tb427tmp.02.5030_RNAi_Frag-R; CARP11 (Tb927.7.2320): RNAi targeting fragment: ORF nt 524-990, amplified with primers Tb427.07.2320_RNAi_Frag-F and Tb427.07.2320_RNAi_Frag-R. The RNAi plasmids were linearized with NotI for transfection and cells were selected by addition of 2.5 µg/mL phleomycin to the culture medium.
The CARP1 ORF was amplified by PCR from T. brucei MiTat 1.2 genomic DNA using primers Tb11.01.7890 SfiI up and Tb11.01.7890 BamHI low and cloned into plew82 (Wirtz et al., 1999) via SfiI and BamHI. Point mutations in the predicted CARP1 cyclic nucleotide binding domains were generated by site-directed mutagenesis using primers Tb11.01.7890 CNBDmut1 up and Tb11.01.7890 CNBDmut1 low (R320L, AGA to TTA), Tb11.01.7890 CNBDmut2 up and Tb11.01.7890 CNBDmut2 low (R458L, CGA to CTA), or Tb11.01.7890 CNBDmut3_R605L up and Tb11.01.7890 CNBDmut3_R605L low (R605L, AGG to CTG), respectively. The plasmids were linearized with NotI for transfection and cells were grown in presence of 2 µg/mL blasticidin.
The CARP11 ORF (Tb927.7.2320) was amplified from T. brucei MiTat 1.2 genomic DNA with primers Tb927.7.2320_SUMO_BamHI_fw (GCACTAGGATCCATGAGCATCGTCAATCAG) and Tb927.7.2320_SUMO_HindIII_rev (CCACCAAGCTTTCACTTGACACGACTGCA) and cloned into pETM11_SUMO3 via BamHI and HindIII. The protein was expressed in E. coli Rosetta (grown in TB with 1 M sorbitol, overnight induction of protein expression with 200 µM IPTG at 16°C) and the soluble fraction was used for pull-down with cAMP agarose.
The CARP1 ORF was amplified using primers Tb427tmp.01.7890.fl.f.10His (ataccatgggccaccaccaccaccaccaccaccaccaccacggcgcgggcGGTAGTTATGAATACCCAGACTAC) and Tb427tmp.01.7890.fl.b (aattcggatcctggctTTACCTTTTCGCCATGAACTCTTT) introducing an N-terminal His10 tag and cloned into pETDuet-1 via NcoI and BamHI restriction sites. The protein was expressed in E. coli Rosetta and purified under denaturing conditions using Ni-NTA columns. After separation of the concentrated protein fractions on a 10% SDS gel, rabbits were immunized with Coomassie-stained CARP1-containing gel slices according to a standard immunization protocol (Pineda, Berlin). The CARP1 antiserum was affinity-purified using His10-CARP1 according to the method of Olmsted. (Olmsted, 1981).
1×108 T. brucei cells overexpressing CARP1 were washed twice with PBS and lysed in 300 µL lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 2 mM EGTA, 0.2% NP-40, Roche Complete protease inhibitor) for 20 min at 4°C. For E. coli lysates, 12.5 mL of a logarithmically growing culture expressing His6-SUMO-CARP11 was lysed in 500 µL lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% Triton X-100, 570 µM PMSF) for 10 min at 4°C with sonication (Bioruptor, Diagenode, 10 min on ice, 30 s on/off, high power). Soluble lysates were incubated with plain agarose beads (Biolog Bremen) for 30-60 min at 4°C to remove proteins binding non-specifically to the bead matrix. Pull-downs were performed by incubation of the pre-cleared lysates (= input fractions) with 30 µL (CARP1) or 40 µL (CARP11) 2-AHA- or 8-AHA-agarose (Biolog Bremen Cat. No. A054, A028) beads slurry for 30 min – 2 h at 4°C, followed by four washes with lysis buffer. Bound proteins were eluted by boiling (5 min, 95°C) with 20 µL (CARP1) or 40 µL (CARP11) 2× Laemmli sample buffer. Equal volumes of input and bound fractions were analysed by Western blot, giving an enrichment factor in the bound fraction versus the input of 15× for CARP1 (300 µL/20 µL) and 12.5× for CARP11 (500 µL/40 µL).
NPD-001 sensitivity of the inducible CARP knock down cell lines was assessed using the Alamar blue (resazurin) cell viability assay exactly as described previously (Gould et al., 2013).
Western blot analysis was performed as previously described (Salmon et al., 2012a) with modifications for detection of CARP1. Briefly, lysates of 3-5×106 trypanosomes were separated on 10% polyacrylamide gels, transferred to a PVDF membrane via semi-dry blotting and blocked with Kem-En-Tec synthetic blocking buffer for 1 h at room temperature. The blots were incubated with primary antibodies (rabbit anti-CARP1, 1:1000; mouse anti-PFR-A/C [(Kohl et al., 1999), 1:2000; PFR, paraflagellar rod protein)] overnight at 4°C, followed by secondary antibody (IRDye680LT anti-rabbit and IRDye800CW anti-mouse, LI-COR, both 1:5000) detection for 1.5 h at room temperature. For detection of His-tagged proteins expressed in E. coli, proteins were blotted onto PVDF membranes and blocked with 5% milk for 1 h at room temperature, followed by incubation with mouse anti-His (1:1000, BioRad MCA1396GA) overnight at 4°C and secondary antibody detection with IRDye800CW anti-mouse (1:5000, LI-COR).
CARP sequences were retrieved from TriTrypDB release 58 (https://tritrypdb.org/tritrypdb/app), information on orthology was obtained from orthoMCL release 6.11 (https://orthomcl.org/orthomcl/app), and domain predictions were done with Superfamily 2 (https://supfam.mrc-lmb.cam.ac.uk/SUPERFAMILY/), Pfam version 35.0 (http://pfam.xfam.org/), and/or SMART (http://smart.embl-heidelberg.de/). AlphaFold2 structure predictions for CARP1 and CARP11 were retrieved from http://wheelerlab.net/alphafold/ and cyclic nucleotide binding domains predicted by Superfamily 2 (https://supfam.mrc-lmb.cam.ac.uk/SUPERFAMILY/) were superimposed with crystal structures of the known cAMP binding proteins E. coli CRP (PDB 4N9H) and Bos taurus PKARIa (PDB 1RGS) using Pymol (The PyMOL Molecular Graphics System, version 2.5 Schrödinger, LLC). Predicted CNBD phosphate binding cassette sequences of CARP1 and CARP11 were aligned with those of E. coli CRP, Mus musculus Epac4 and Bos taurus PKARIa using CLC Main Workbench version 21.0.5 (Qiagen). For structure similarity searches, the AlphaFold ID was obtained from TriTrypDB release 63 or directly from wheelerlab.net and the structure downloaded as a PDB file from https://alphafold.ebi.ac.uk/, which was then used to run a structure search on the Dali server (http://ekhidna2.biocenter.helsinki.fi/dali/). The top hit of the resulting output was then viewed in the RSCB protein database (PDB; https://www.rcsb.org/).
As described by Gould et al. (2013), a genome-wide RNAi library screen was carried out with bloodstream T. b. brucei under selection pressure with the phosphodiesterase inhibitor NPD-001, previously known as CpdA (de Koning et al., 2012), following established protocols (Alsford et al., 2011; Alsford et al., 2012). In the initial report, we identified four genes by sequencing PCR-fragments from amplified prominent bands on an agarose gel and designated them CARP1 – 4. The sample from this selection has now been subjected to RIT-seq deep sequencing, identifying the RNAi sequences and mapping them to the genome. This yielded a far more diverse and quantitative data set of genes that, upon RNAi knockdown, protect to any degree against the strongly elevated levels of intracellular cAMP resulting from the treatment with NPD-001. A total of 1183 genes was included in this dataset, with raw total counts between 74,563,689 and 1, out of a total of 105,662,532 bases mapped to CDS. Among the most frequent hits were CARP1 – CARP4 and an additional 7 genes were selected from the genes with highest counts, and designated CARP5 - CARP11 (Table 1). In addition, four adenylate cyclase-encoding genes were identified with high abundance, consistent with cyclase knockdown causing a reduction of cAMP levels (Salmon et al., 2012a).
Figure 1 shows a pie-chart representation of the % of total mapped reads for each of the CARPs and adenylate cyclases that together account for 98.2% of all mapped reads in the RIT-seq analysis. Clearly, the most frequent gene targeted was CARP1, with 70.6% of all mapped reads, followed by CARP4 with 13.9%.
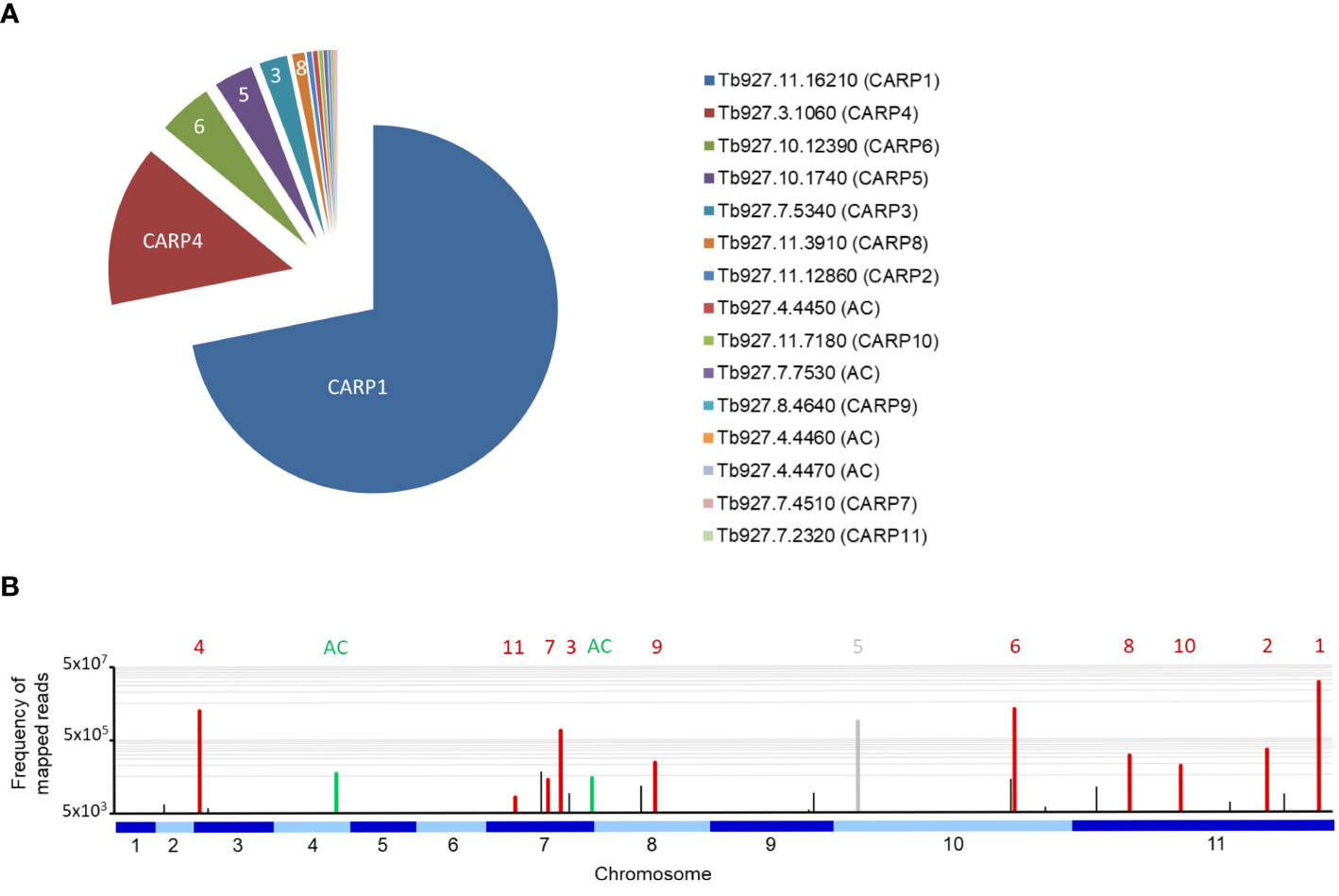
Figure 1 (A) Pie chart of the percentage of all reads mapped to each of the eleven CARP genes and to the four GRESAG4 adenylate cyclases (AC). The gene identifiers from TriTrypDB and the CARP numbering are indicated. (B) Frequency of mapped RIT-seq reads indicated on a genome map of T. brucei 927. Numbered CARPs are indicated by red bars and the corresponding numbers above them; only CARP5 is indicated in grey to indicate this was a false positive hit. Adenylate cyclases (AC) are similarly indicated in green; The green bar on chromosome 4 is an amalgamation of three bars, for genes Tb927.7.4450, Tb927.7.4460 and Tb927.7.4470. Black bars represent genes that were not followed up for validation by targeted RNAi for this study.
Of the newly identified CARPs, CARP6 was the most-targeted gene and CARP11 the least targeted, accounting for 4.69% and 0.046% of mapped reads, respectively. The four listed adenylate cyclases were identified between 0.42% and 0.14% of mapped reads. A genome map of RIT-seq hits is depicted in Figure 1, which shows the designated CARPs, as well as the positions of the adenylate cyclases and other ‘hits’ that were not taken forward for verification by targeted RNAi. A listing of all 44 genes with normalised RIT-seq mapping ≥ 0.05% of mapped reads is given as Supplemental Table S2.
Previously, we confirmed that targeted RNAi knockdown of CARP1 – CARP4 did confer protection to treatment with NPD-001, i.e. a shift to a significantly higher EC50 in our standardised resazurin-based drug sensitivity test. This induced resistance was specific, as it was not afforded to treatment with control drugs pentamidine, suramin and difluoromethylornithine (Gould et al., 2013).
RNAi target sequences were chosen for each of the genes CARP5 – CARP11, in order to obtain true independent confirmation. The RNAi target fragments (Table 1) were cloned into the expression vector p2T7-177-BLE (Wickstead et al., 2002) and transfected into the T. b. brucei cell line Lister 427 MiTat 1.2 13-90 (Wirtz et al., 1999) for tetracycline (TET)-inducible expression. As knockdown measured by mRNA levels does not necessarily correlate with protein levels in T. brucei, where the regulation is mostly at the level of translation rather than transcription (Clayton, 2016), protein levels for several of the CARPs were confirmed by Western blot using CARP1-specific antibodies (Supplementary Figure S2A) or antibodies against the TY tag in situ added to CARP6 and CARP10 (Supplementary Figures S2B, C). All three CARP proteins were strongly diminished after 24 h of TET induction. The knockdown of epitope-tagged CARP1–4 was also confirmed by Western blot in our previous paper (Gould et al., 2013).
EC50 values were obtained in parallel with and without TET induction, allowing statistical comparisons. Further, the NPD-001 sensitivity tests were performed with four biological replicates (independent transfectants) and each EC50 was obtained on at least three separate occasions. Table 2 shows a summary of the CARP5 – CARP11 results, comparing the EC50 values obtained with and without TET induction and using the untransfected parental line as a further control. The RNAi knockdown resulted in significantly higher NPD-001 EC50 values for CARP6 – CARP11, but not for CARP5, which may therefore be classified as a false positive from the library screen. Indeed, further analysis showed this gene to be a frequent false hit in RIT-seq owing to the RNAi vector junction sequence GCCTCGCGA. Mis-priming from sequence GGGCCAGT within the Tb927.10.1740 (CARP5) CDS produced a 1245-bp product. For the confirmed CARPs 6–11, the level of protection against elevated cAMP by targeted RNAi was by far highest for CARP7 (17-fold), whereas the targeted knockdown of the other genes typically resulted in < 4-fold resistance gain (Table 2). The difference in response to knockdown for CARP5 and the parental control versus CARP7 is illustrated in Figure 2 with representative curves from the resazurin-based assays.
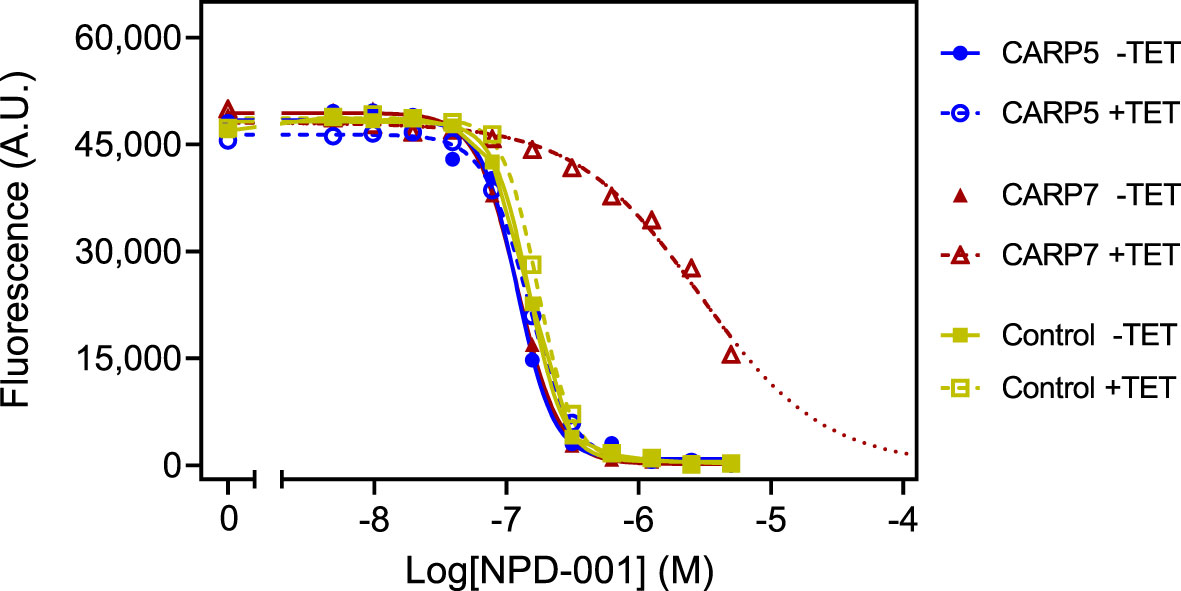
Figure 2 Representative graphs of the Alamar blue-based assay used to determine EC50 values for NPD-001. Dilution range for NPD-001 started at 5 μM and consisted of 11 doubling dilutions to 4.88 nM, and a no-drug control. The results shown are from a single experiment, representative of three independent replicates, performed with one of three clonal lines for each RNAi line (CARP5 clone 6 and CARP7 clone 4). Fluorescence read-outs in arbitrary units (A.U.), with background fluorescence subtracted, were plotted to a sigmoid curve with variable slope in Prism 9 to determine the EC50 values. Closed symbols and solid lines represent the control without tetracycline (TET), open symbols and dashed lines, with induction by TET. For the TET-induced CARP7 values the dotted line indicates extrapolation. Control cells were the parental Lister 427 ‘single marker’ strain derived from s427 by introducing the tetracycline repressor TetR and T7 polymerase (Wirtz et al., 1999).
CARP phylogeny was investigated by individual BLAST searches (blast.ncbi.nlm.nih.gov/Blast.cgi) as well as orthologue analysis based on the orthoMCL database (orthomcl.org) (Figure 3). Despite some small differences between the results of these two methods, all CARPs appear to be present in both African and American Trypanosoma species (T. brucei, T. congolense, T. vivax, T. cruzi) and most are also found in Leishmania genomes, except CARP3 and CARP11. CARP3 is also absent from the genomes of the insect gut parasites Crithidia fasciculata and Angomonas deanei, as well as the free-living trypanosomatid Bodo saltans. CARP11 is found in A. deanei and B. saltans but no orthologue could be identified in C. fasciculata.
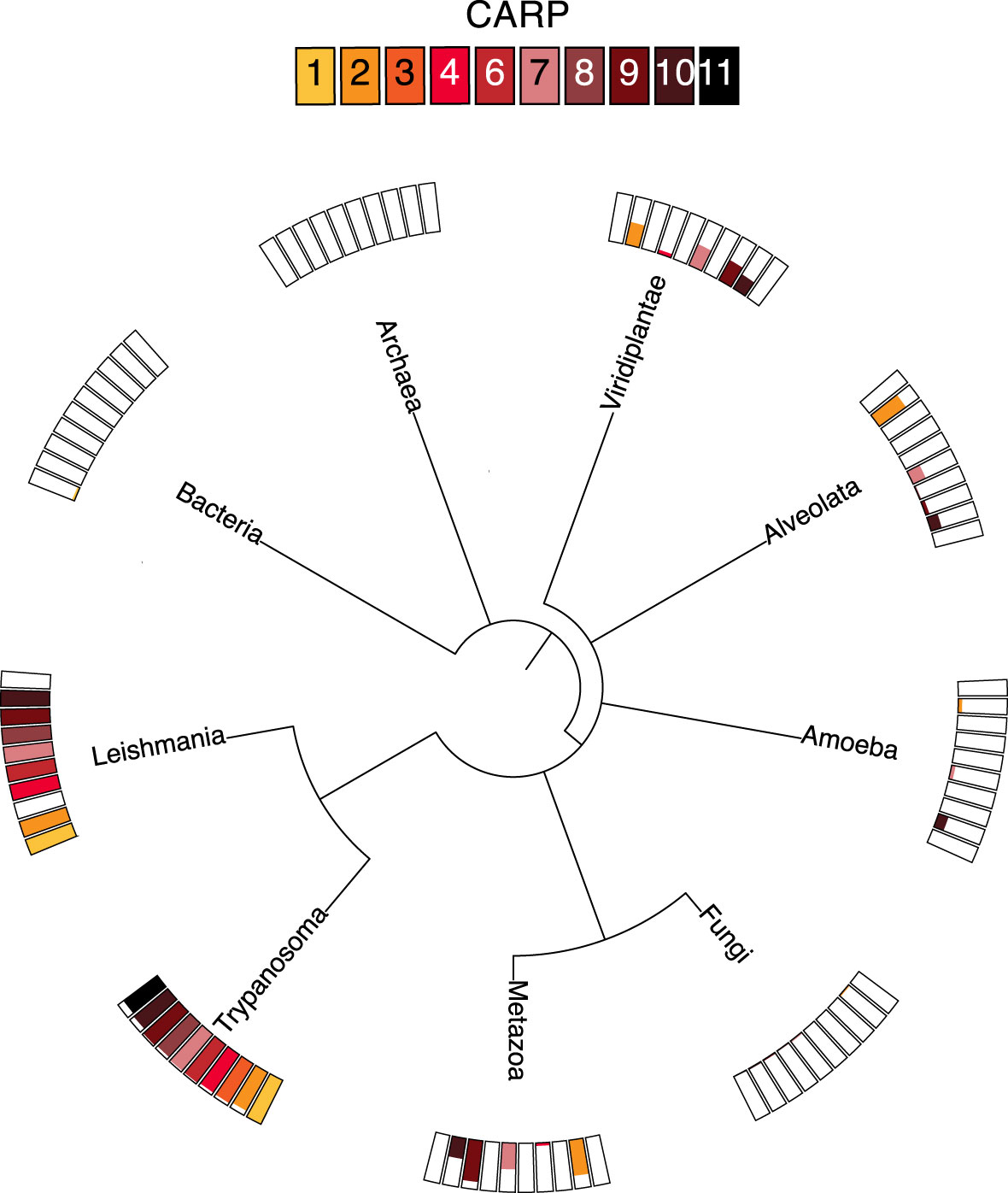
Figure 3 Distribution of ortholog groups for CARPs based on by OrthoMCL release 6.11 (https://orthomcl.org/orthomcl/app). Ortholog group IDs for CARPs were retrieved from TritrypDB release 58 (https://TritrypDB.org) and the tree was generated with phyloT version 2022.3 (https://phylot.biobyte.de/). Each CARP is shown by a differently coloured box and the size of the coloured bar represents the percentage of occurrence in the respective group of species.
Most CARPs are scarcely found outside the trypanosomatids, although there are some exceptions. CARP2 and CARP7 are found in some species of Viridiplantae, including the green algae Chlamydomonas, in some Alveolata, specifically the Apicomplexa, and in some metazoa including Mammalia; CARP9 is also present in Chlamydomonas and is quite widespread in metazoa, but is only found in very few Alveolata. Conversely, CARP4 is present in a few species of Viridiplantae and Metazoa but not in other non-trypanosomatids. Only CARP10 had a few orthologues in Amoebozoa and Alveolata and some in Viridiplantae and Metazoa. None of the CARPs had detectable orthologues in prokaryotes, fungi or in Arabidopsis. Overall, most CARPs are (almost) exclusive to trypanosomatids, with CARP3 limited to the genus Trypanosoma.
The domain structure and subcellular localization of the 10 confirmed CARPs (i.e., excluding CARP5) is summarised in Figure 4. Domains could be identified in profile database searches for all except CARP8, although some of these are Domains of Unknown Function (DUF), specifically DUF4464 as the main conserved domain in CARP2 and DUF4201 as the only domain detected in CARP7. Modelled structures were obtained from AlphaFold and used for structural similarity searches in the RCSB Protein Data Base (PDB); the top returns for each validated CARP are listed in Supplemental Table S1.
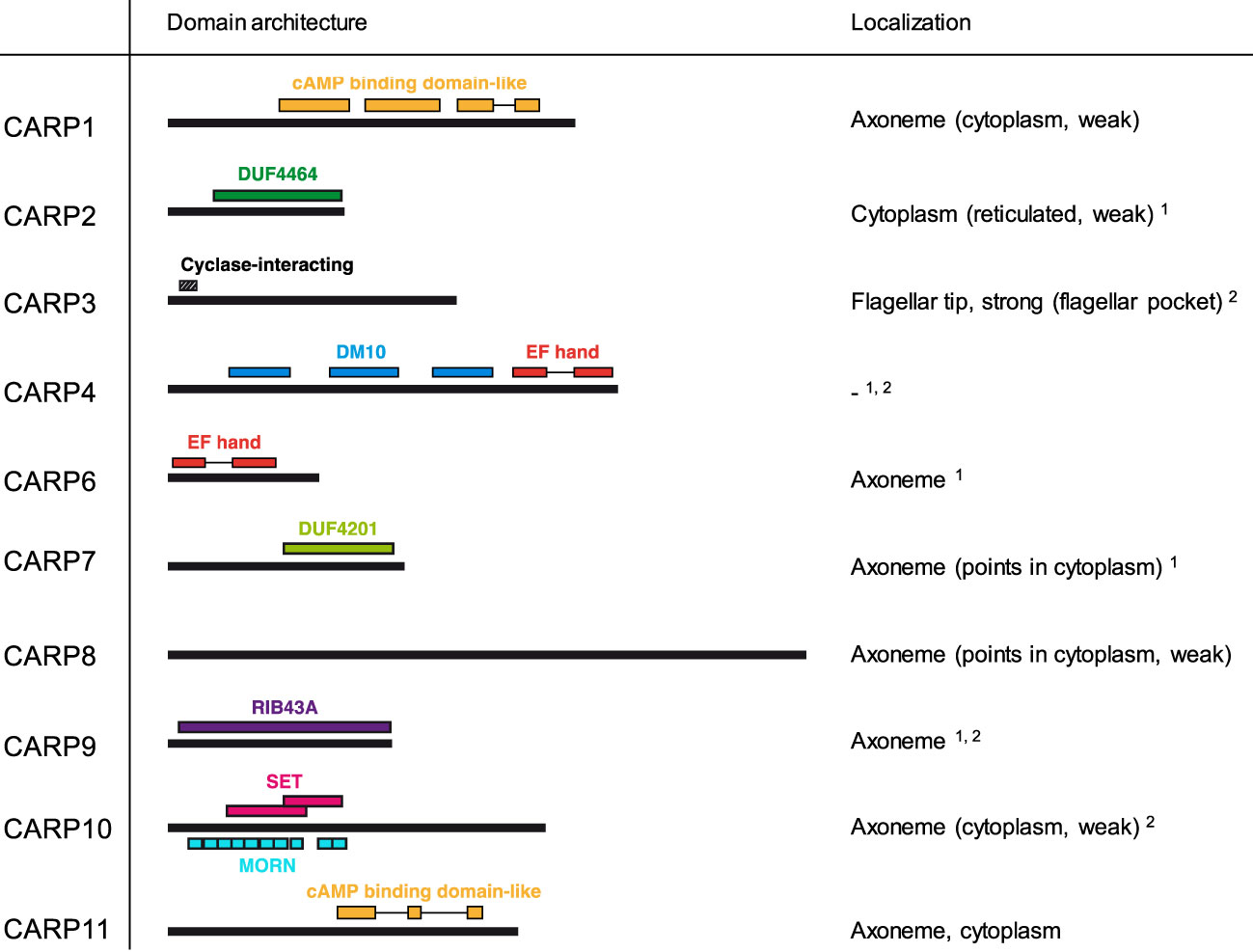
Figure 4 Domain predictions for CARPs by the domain profile databases Superfamily 2.0 (https://supfam.org/), Pfam version 35.0 (http://pfam.xfam.org/), and/or SMART (http://smart.embl-heidelberg.de/) are shown as schematic representation. CARP proteins are shown as black bars (to scale) and predicted domains are displayed by coloured boxes with IDs as indicated in the figure. For CARP3, the region required for flagellar tip localization and cyclase interaction (designated ‘Cyclase-interacting’) according to Bachmaier et al. (2022) is indicated. Subcellular localization of CARP proteins is given exactly as annotated in the TrypTag.org database (Dean et al., 2017). Additional evidence for axonemal1 or flagellar2 localization is taken from Broadhead et al. (2006) and Subota et al. (2014), respectively. DUF, domain of unknown function; MORN, Membrane Occupation and Recognition Nexus.
Strikingly, the TrypTag genome-wide tagging project (Dean et al., 2017; Billington et al., 2023) revealed an axonemal localization for 8/9 CARPs successfully tagged in procyclic T. b. brucei, which closely aligned with Gene Ontology (GO) assignments in TriTrypDB (Supplemental Table S2). The exception was CARP2, for which no GO assignments were available, and which showed a cytoplasmic distribution as N-terminal mNeonGreen fusion protein in TrypTag. However, CARP2 and CARPs 3–7, 9 and 10 are all present in at least one flagellar proteome of procyclic trypanosomes (Broadhead et al., 2006; Subota et al., 2014). Also, many CARP2 orthologues from other organisms are annotated as cilia- and flagella-associated proteins, and a bioinformatic study by Baron et al. (2007), identified CARP2 as a conserved component of motile cilia and flagella, supporting a flagellar localization. Structural similarity searches using AlphaFold and the Dali server highlighted as top hit a Legionella pneumophilia protein, LvgA, which is part of the complex that recognises effector proteins to be exported by their type IVB secretion system. Kim et al. (2020) describe this protein as an adaptor, recruiting multiple other effector proteins to the complex.
Baron et al. (2007) also identified CARP4, the only CARP without localization information on TrypTag.org (accessed 02/04/2023), as a flagellar protein. CARP4 is orthologous to Chlamydomonas reinhardtii Rib72 (Chlre5_6|10141; BLASTP E-value = 3.1E-28), which appears to be involved in linking microtubules of the outer doublets of the axoneme (Ikeda et al., 2003). Like CARP4, CrRIB72 contains three DM10 domains and a C-terminal EF-hand domain, which binds Ca2+ and is implicated in regulatory and signalling responses (Nelson et al., 2002). Moreover, CARP4 is orthologous to the human EF Hand Containing protein EFHC1 (BLASTP E-value = 5E-55) that is linked to Juvenile Myoclonic epilepsy (King, 2006) and is believed to be part of the structure of cilia (King, 2006; Suzuki et al., 2020). Structurally, CARP4 aligned with other EF Hand domain-containing proteins, specifically the Ca2+ sensor Case16 (Leder et al., 2010) and an EF-hand-containing domain of a spectraplakin that cross-links microtubule and actin and binds two Ca2+ (Lane et al., 2017). It is thus likely that CARP4 is a Ca2+-binding protein involved in direct interactions between flagellar microtubules and, possibly, other effector proteins.
CARP9 is annotated as Component of Motile Flagella 19 (CMF19) in TriTrypDB. It contains a RIB43A domain, a structure that was first identified in Chlamydomonas, and is associated with protofilament ribbons and basal bodies of ciliary and flagellar microtubules (Norrander et al., 2000). As the microtubular ribbons in Chlamydomonas mostly consist of RIB43A and RIB72 in addition to tubulin (King, 2006), it is likely that CARP4 and CARP9 play a similar role in T. brucei, with the EF-hand domain providing Ca2+-mediated regulation. Curiously, CARP9 was co-purified with the T. brucei Mitochondrial Calcium Uniporter (TbMCU; (Huang and Docampo, 2020)), although it is not immediately clear how a putative axonemal protein and an integral membrane protein of the inner mitochondrial membrane might associate in vivo. The CARP9 structure consists of two long alpha-helical rods, connected by a third, inter-strand helix (alphafold.ebi.ac.uk/entry/Q57UY7) and structural similarity searches were not very informative regarding a specific function. CARP6 also contains an EF-hand domain (Figure 4), reinforcing the link to Ca2+ signalling somewhere in the response to high cAMP; it is structurally related to calmodulin (Ishida et al., 2000).
CARP3’s predicted structure shows a highly structured N-terminus essential for localization of the protein at the distal tip of the flagellum in procyclic trypanosomes (Bachmaier et al., 2022) where it interacts with several adenylate cyclases and regulates social motility and tsetse fly transmission (Bachmaier et al., 2022; Shaw et al., 2022). Interestingly, the closest structural match for CARP3 was Bacillus subtilis RapH, a phosphatase response regulator (Parashar et al., 2011), which is consistent with a regulatory role in the cAMP response.
CARP10 contains a series of ten predicted Membrane Occupation and Recognition Nexus (MORN) domains as well as two copies of the Histone H3 K4-specific methyltransferase SET7/9 N-terminal domain superfamily (101-220 and 201-300). Although the MORN domain has been identified in a total of 113,928 proteins in many species (SMART database; http://smart.embl.de/smart/; 25/07/2022), the function of this domain is still poorly understood (Zhou et al., 2022). However, the structure of the domain in CARP10 is closely related to H. sapiens MORN4, which Li et al. (2019) describe as containing an extended single-layered β-sheet structure that uses a U-shaped groove to bind the long α-helix tail of myosin 3A. CARP10, being localised to the axoneme based on GO terms and TrypTag and sharing the U-shaped β-sheet (‘long open tube’) structure of HsMORN4, could therefore be implicated in coupling regulatory proteins to the flagellar motor function.
Finally, cAMP binding motifs were identified in the sequences of two CARP proteins, CARP1 and CARP11, i.e. the genes to which the highest and the lowest percentage of reads, respectively, were mapped in the RIT-seq screen. In CARP1, three cAMP binding domains are detected at a.a. positions 233–346, 370–491 and 519–577, 612–651, whereas in CARP11 a single cyclic nucleotide binding domain is detected, with insertions, at positions 293–358, 415–437 and 519–545.
The putative phosphate binding cassette (PBC) sequences of the predicted CARP1 and CARP11 cAMP binding domains were aligned with well-characterised but diverse cAMP binding domains from different species: E. coli CRP (EcCRP, Uniprot P0ACJ8), Mus musculus EPAC4 (MmEpac4, Uniprot Q9EQZ6) and Bos taurus PKARIα (BtPKARIα, Uniprot P00514) in Figure 5. The structural searches also turned up diverse cyclic nucleotide binding domains for both CARPs, including the regulatory subunit of Toxoplasma gondii PKA as the top hit for CARP11 (El Bakkouri et al., unpublished; www.rcsb.org/structure/5J3U). A PBC consensus sequence that was compiled for PKA regulatory subunits, F-G-E-[LIV]-A-L-[LIMV]-x(3)-[PV]-R-[ANQV]-A (Canaves & Taylor, 2002; Kannan et al., 2007), is also displayed for comparison. Highly conserved residues of the PBC include an invariant arginine (R211 in BtPKARIα) known to interact with the phosphate group of cAMP and a glutamate (E202 in BtPKARIα) that interacts with the 2’OH group of the ribose (Berman et al., 2005). While CNBD-A of CARP1 and CARP11 match very well with the PKA consensus, CNBD-B and -C of CARP1 have important deviations and the validity of the alignment and cAMP binding prediction are questionable (Figures 5A, C). Structural alignment of the CNBDs in the AlphaFold2-predicted CARP1 structure (retrieved from http://wheelerlab.net/alphafold/; Wheeler, 2021) with published crystal structures of diverse cAMP binding proteins (EcCRP, PDB 4N9H; BtPKARIα, PDB 1RGS) showed a very good structural preservation including overlay of R320 in CARP1 CNBD-A with the conserved arginines of the reference protein structures (Figure 5B, RMSD 1.168 with EcCRP CNBD; RMSD 0.782 with BtPKARIα CNBD-A). The same observation is made for R520 in the CARP11 CNBD (Figure 5D, RMSD 1.039 with EcCRP CNBD; RMSD 0.602 with BtPKARIα CNBD-A). In contrast, the conserved arginine in the reference structures aligned with Lys459 and Tyr629 in CNBD-B and CNBD-C of CARP1, respectively (Supplemental Figures S1A, B).
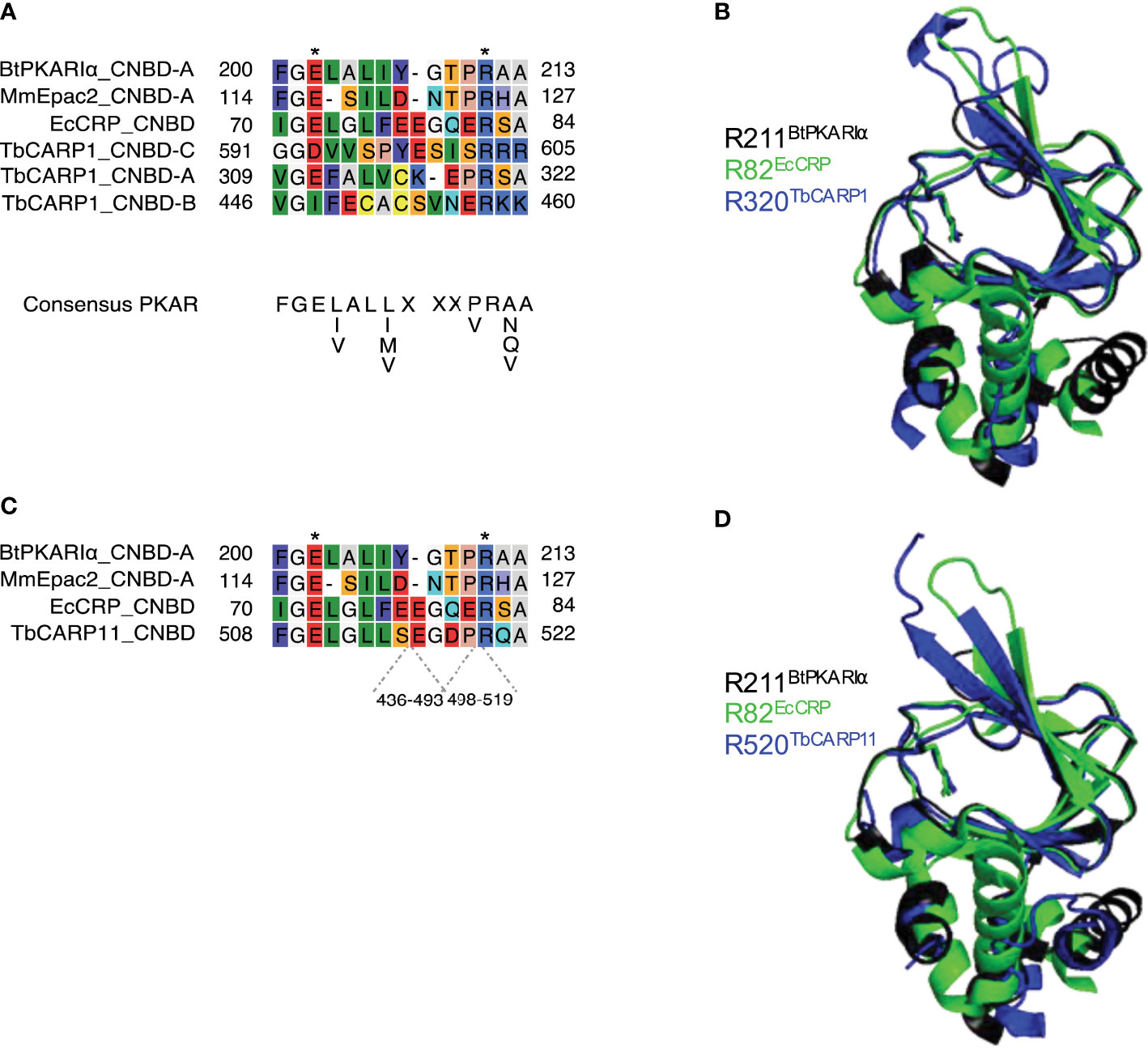
Figure 5 Sequence comparisons (A, C) of the predicted phosphate binding cassettes (PBCs) of T. brucei CARP1 (A, CNBD-A, -B, -C) or CARP11 (C) with Escherichia coli CRP (EcCRP), Mus musculus EPAC2 (MmEpac2) and Bos taurus PKARIα (BtPKARIα). The amino acid numbers of the predicted sequence insertions within the CARP11 PBC are indicated below the alignment. Asterisks indicate conserved Glu and Arg residues known to be essential for cAMP binding (Berman et al., 2005). (B, D) Structural alignments of CARP1 CNBD-A (B, dark blue) or CARP11 CNBD (D, dark blue) with BtPKARIa CNBD-A (PDB 1RGS, black) and EcCRP CNBD (PDB 4N9H, green) with the conserved arginines R320 of CARP1 and R520 of CARP11, respectively, overlaying the conserved arginines R211 of BtPKARIa CNBD-A and R82 of EcCRP CNBD. The structures were retrieved from http://wheelerlab.net/alphafold/ and superimposed with Pymol version 2.5.4.
In order to test whether CARP1 and CARP11 do bind cAMP, we performed a pull-down experiment with cAMP immobilised on agarose beads via a flexible aminohexyl linker on position 8 (8-(6-aminohexylamino)-adenosine-3’, 5’-cyclic monophosphate; 8-AHA-cAMP-agarose). Lysates of T. brucei overexpressing CARP1 were first incubated with agarose beads without attached cAMP in order to remove proteins non-specifically binding to the bead matrix. Subsequent incubation with 8-AHA-cAMP-agarose allowed the pulldown of a fraction that was highly enriched in cAMP-binding proteins. This procedure was performed in the presence of increasing cAMP concentrations (0 – 50 µM) intended to outcompete the binding to 8-AHA-cAMP. Subsequent elution of the protein fraction from the agarose beads, followed by Western blot with a rabbit anti-CARP1 serum gave a quantifiable amount of CARP1 that was plotted against the cAMP concentration to derive an approximation of cAMP binding affinity. The antiserum had been validated using a panel of T. brucei cell lines, showing an increased signal in the CARP1-overexpressing strain and a strong decrease upon RNAi knockdown (Supplemental Figure S2A). Figure 6 shows the average of two such determinations plotted with a sigmoidal curve with variable slope (standard inhibition; r2 = 0.977) yielding an EC50 value of 32.4 nM (95% CI 14.6 – 76.1 nM). Approximately 0.6–7.5% of the input CARP1 was pulled down in the competition series from 0 to 50 µM. Most likely this represents the active, properly folded fraction of the highly insoluble CARP1. A further pull-down with cAMP-agarose beads was performed using a set of differently linked cAMP analogues. Beads with various linkers connected to the purine ring at positions 2, 6, or 8 all showed similar capacity to pull down CARP1, but two different linkers attached to the 2’ position of the ribose failed to pull down any CARP1 (Supplemental Figure S3A), consistent with cAMP interaction within the CNB pocket primarily via the ribose and cyclic phosphate.

Figure 6 Cyclic AMP affinity purification of CARPs with predicted cAMP binding domains. (A) CARP1 was pulled down by 8-AHA-cAMP agarose from T. brucei cells overexpressing CARP1 (2 replicates). (B) CARP11 was expressed in E. coli with an N-terminal His6-SUMO tag and pulled down by 2-AHA- (top) or 8-AHA-cAMP agarose (bottom). The Western blots show the input fractions and the pulled down material in presence of increasing concentrations of cAMP (0 nM – 50 µM) for competition. The load of the bound fractions relative to the input is 15× for CARP1 and 12.5× for CARP11. Binding curves from the fluorescence quantifications are shown below; the amount of CARP pulled down without competition was set to 100%.
The same procedure was performed with CARP11 expressed with a His6-SUMO tag in E. coli (Figure 6), using 8-AHA-cAMP and 2-AHA-cAMP agarose beads. For Western blots, a mouse anti-His serum was used for detection of the tagged CARP11. The competition titrations with cAMP in Figure 6 show similar binding to 8-AHA-cAMP and 2-AHA-cAMP agarose beads with almost identical EC50 estimates: 309 nM for 2-AHA-cAMP and 276 nM for 8-AHA-cAMP. Approximately 0.6–24.5% of the input CARP11 was pulled down in the competition series from 0 to 50 µM (Supplemental Figure S3B). We conclude CARP1 and CARP11 are cAMP-binding proteins and that CARP1 has a ~10× higher affinity for the cyclic nucleotide than CARP11.
By genome-wide RNAi screening for cAMP resistance in bloodstream stage trypanosomes, we have identified a set of proteins with a predicted function in cAMP metabolism or cAMP signalling. The selection was based on the essentiality of cAMP metabolism (Oberholzer et al., 2007; Shakur et al., 2011; Salmon et al., 2012a) and a strong cell division phenotype resulting from genetic or pharmacological perturbations of cellular cAMP concentrations (de Koning et al., 2012; Salmon et al., 2012a; Gould et al., 2013). Strikingly, for all identified cAMP response proteins (CARPs) we found publicly available experimental evidence (summarized in Figure 4) for association with the flagellar axoneme or the flagellum, suggesting an important impact of cAMP signalling on flagellar biology. An intact and functional flagellum was previously shown to be essential for cell division of bloodstream stage trypanosomes, as RNAi repression of many genes with flagellar localization or function result in a lethal cell division phenotype (Broadhead et al., 2006; Marques et al., 2022). This may explain why the screen returned specifically the observed set of proteins. Initiation of flagellar replication is the necessary first step in trypanosome cell division (Vaughan, 2010; Sun et al., 2013), and impaired flagellar function results in a block in cytokinesis. This is exactly the lethal phenotype seen with non-physiologically elevated cAMP (de Koning et al., 2012; Gould et al., 2013). Consequently, cyclic nucleotide phosphodiesterases (PDEs) of trypanosomes are considered good drug targets (Blaazer et al., 2018; de Heuvel et al., 2019).
A priori, we expected to get from our RNAi screening the following protein categories (1) proteins involved in cAMP production like adenylate cyclases (ACs) or proteins affecting the activity or abundance of ACs, (2) effectors binding cAMP and (3) a diverse group of targets downstream of the effectors or modulating the activity of those effectors. As expected, the hit list included several AC genes, but at relatively low RIT-seq representation. The T. brucei genome contains more than 70 receptor-type ACs, the catalytic domains of which share homology with mammalian class III cyclases (Pays & Nolan, 1998; Salmon et al., 2012b; Salmon, 2018; Durante et al., 2020). Many of the T. brucei ACs have been shown to be developmentally regulated (Saada et al., 2014; Naguleswaran et al., 2018), while others are expressed throughout the life cycle (Alexandre et al., 1996). The fact that we observed only relatively few ACs, and at relatively low RIT-seq representation (Figure 1), may be explained by the limited impact on the cAMP concentration upon RNAi of any individual AC family members (Salmon et al., 2012a). A higher RIT-seq representation was found for CARP3, a membrane-associated protein that interacts with several ACs, including the ESAG4 cyclase that is abundant in bloodstream forms, and positively regulates their abundance (Bachmaier et al., 2022). It is therefore likely that CARP3 repression confers cAMP resistance in bloodstream forms by concomitant downregulation of a significant fraction of ACs, particularly the dominant ESAG4, and thereby lower cAMP production.
At the effector level, we have identified two novel cAMP-binding proteins from the RNAi screen, CARP1 (Gould et al., 2013) and CARP11. The T. cruzi CARP1 homolog (TcCLB.508523.80) also seems to bind cAMP (Jäger et al., 2014). CARP1 has three predicted CNB domains; the phosphate binding cassettes (PBCs) of the first (CNBD-A) and of the CNB domain of CARP11 match a consensus sequence for the PBCs of PKA of diverse species very well (Figure 5A, C). In contrast, CNBD-B and CNBD-C of CARP1 deviate in critical residues and therefore, we can neither predict nor exclude their contribution to cAMP binding in our immobilised cAMP assay (see also Figure S1). Interestingly, CARP11 has two longer sequence insertions within the CNBD that apparently do not compromise the cAMP binding function. AlphaFold2 modelling of the domain and overlay with reference CNB crystal structures (Figure 5D) show indeed a well conserved CNB structure in CARP11, suggesting that the insertions have accessory functions compatible with cAMP binding. The binding affinity, estimated by EC50 of cAMP competition, is ~30 nM for CARP1, a value in the range of the free regulatory subunit of PKA from other organisms (Moll et al., 2007). The EC50 for CARP11 is ~300 nM and closer to effectors like mammalian EPAC (Dao et al., 2006) and to physiologically relevant cAMP concentrations in cAMP microdomains (Anton et al., 2022).
The remaining seven CARP genes identified in the genome-wide RNAi screen cannot be assigned yet to a specific biochemical function. Some of them are most likely targets downstream of the cAMP signal, supported by the identified domains and available data on potential orthologs in higher eukaryotes (e.g. CARPs 2, 4, 9). For others (CARPs 6, 7, 8, 10), a function at the level of cAMP signal production, as found for CARP3, is equally possible. EF-hand domains (Nelson et al., 2002) in CARP4 and 6 suggest crosstalk between cAMP signalling and regulatory processes involving Ca2+.
Most importantly, the experimentally determined subcellular localization in trypanosomes retrieved from the database provided by the TrypTag project (Dean et al., 2017; Billington et al., 2023) assigned 7/10 CARPs to the flagellar axoneme and 2/10 (CARPs 2 and 4) were assigned to the flagellum based on proteome data (Broadhead et al., 2006; Subota et al., 2014). CARP3 is associated with the flagellar membrane and the axonemal cap via FLAM8 (Bachmaier et al., 2022). Thus, all CARPs are predicted to have a role in flagellar biology. By inference, cAMP signalling is important for regulation of flagellar functions. The impact is not limited to trypanosomes: 5/7 of the CARPs present in the Leishmania genome (Figure 3) have been found enriched in the flagellar insoluble fraction by proteome analysis (CARPs 2, 4, 6, 9, 10 (Beneke et al., 2019)). The CARPs thus provide a valuable resource to functionally dissect important flagellar biology of kinetoplastid parasites. Indeed the longer list of 44 genes with normalised RIT-seq mapped reads ≥ 0.05 (Supplemental Table S2) was highly enriched with axonemal (16/44) and other flagellar (7/44) localizations (TrypTag.org) and was significantly enriched for the GO component terms ‘axoneme’ (6-fold; P = 2.7×10-8), ‘ciliary plasm’ (2.3-fold; P = 7.7×10-5) and ‘cilium’ (1.85-fold; P = 5.7×10-4). One potential entry of interest is hypothetical protein Tb927.7.4100, which encodes a 500-a.a. protein that like CARP7 features a DUF4201 domain and is located to the flagellum by proteomic analysis (Subota et al, 2014) and to the axoneme by TrypTag. This dataset could thus provide a starting point for further investigation into flagellar biology as well as novel cAMP-regulated pathways essential for parasite growth, survival and transmission. These pathways are potential targets for drug development against kinetoplastid diseases.
One example of an in-depth analysis is provided by CARP3. This protein localizes to the flagellar membrane in bloodstream forms but is restricted to a specialised microdomain at the tip of the flagellum in the procyclic stage (Bachmaier et al., 2022). In that stage, cAMP signalling via tip-localized adenylate cyclases (ACs) has been shown to regulate social motility “SoMo” on agarose plates (Lopez et al., 2015; Oberholzer et al., 2015), a phenotype that is now interpreted as chemotaxis along a pH gradient towards a more basic environment (Shaw et al., 2022). CARP3 interacts with multiple ACs in a flagellar tip complex and is essential for “SoMo” and for colonization of tsetse fly tissues and thus transmission (Bachmaier et al., 2022; Shaw et al., 2022; Shaw & Roditi, 2023). CARP3 differentially regulates the abundance of ACs, and a model predicts that it controls the relative abundance of functionally different cyclases for integration of various external signals (Bachmaier et al., 2022). The flagellar tip and membrane localization of CARP3 was also seen in T. cruzi (Won et al., 2023). The specific role in signalling from the environment to the cell, e.g. pH chemotaxis, is consistent with the membrane-associated localization of CARP3. In contrast, the other CARPs are localized to the axoneme and therefore more likely play roles in cell autonomous pathways, regulating flagellar motility. The fact that flagellar motility per se is unaffected in CARP3-depleted SoMo-negative cells (Bachmaier et al., 2022) also argues for distinct pathways. In mammalian cells, cAMP is involved in regulating ciliar motility within the axoneme (Wirschell et al., 2011) as well as distinct cilial sensing pathways (Nachury & Mick, 2019).
The novel cAMP-binding protein CARP1 appears in the genome of Euglenozoa (Figure 3), also supported by detection of CARP1, 4, 6, 8, 9, 10 in the flagellar proteome of Euglena gracilis (Hammond et al., 2021). In contrast, CARP11 is only detected in Trypanosoma. We notice a striking correlation between the exclusive presence of these cAMP effectors in Euglenozoa and the absence of a cAMP-dependent protein kinase A. PKA in T. brucei and T. cruzi is structurally conserved but repurposed for binding and regulation by nucleoside analogs (Bachmaier et al., 2019). Future dissection of the novel flagellar cAMP signalling mechanisms may indicate why repurposing of PKA for a different second messenger or activation mechanism might have conferred a selective evolutionary advantage to organisms with novel cAMP effectors.
The datasets presented in this study can be found in online repositories. The RIT-seq data was submitted to the European Nucleotide Archive (https://www.ebi.ac.uk/ena/browser/home), with submission number PRJEB60531.
MB, DH and HK designed and supervised research; SB, MG, EP, RO, AB, MA, JM performed research; SB, MG, HK, MB and DH analyzed data; HK, MB and SB wrote the paper. All authors contributed to the article and approved the submitted version.
Work in Munich was funded by Marie Curie PIEF-GA-2012-626034 “TrypCARPinteractors” (MG) and BioNa young scientists award of the Faculty of Biology LMU (SB). MA was funded by a studentship from the Ministry of Higher Education of Saudi Arabia. JM and HK were supported by an FP7 grant from the European Commission (project reference 602666) and a grant from the Medical Research Council (G0701258). DH is supported by a Wellcome Trust Investigator Award [217105/Z/19/Z].
We thank Frank Schwede (Biolog, Bremen) for cAMP agarose beads and advice and the Tryptag.org team for a public resource that was invaluable for interpretation of the CARPs’ phenotype. The authors are grateful to Graham Hamilton of the Glasgow Polyomics facility and Jon Wilkes of the Wellcome Centre for Molecular Parasitology of the University of Glasgow for expert assistance with the RIT-sequencing and the mapping of the reads to the genome, respectively. The contributions of Jacqueline Cordes and Lisa Dietsche at the Boshart lab are also acknowledged.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1204707/full#supplementary-material
Alexandre, S., Paindavoine, P., Hanocq-Quertier, J., Paturiaux-Hanocq, F., Tebabi, P., Pays, E. (1996). ‘Families of adenylate cyclase genes in Trypanosoma brucei.’. Mol. Biochem. Parasitol. 77, 173–182. doi: 10.1016/0166-6851(96)02591-1
Alsford, S., Eckert, S., Baker, N., Glover, L., Sanchez-Flores, A., Leung, K. F., et al. (2012). ‘High-throughput decoding of antitrypanosomal drug efficacy and resistance’. Nature 482, 232–236. doi: 10.1038/nature10771
Alsford, S., Turner, D. J., Obado, S. O., Sanchez-Flores, A., Glover, L., Berriman, M., et al. (2011). ‘High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome.’. Genome Res. 21, 915–924. doi: 10.1101/gr.115089.110
Anton, S. E., Kayser, C., Maiellaro, I., Nemec, K., Möller, J., Koschinski, A., et al. (2022). ‘Receptor-associated independent cAMP nanodomains mediate spatiotemporal specificity of GPCR signaling.’. Cell 185, 1130–1142. doi: 10.1016/j.cell.2022.02.011
Aregawi, W. G., Agga, G. E., Abdi, R. D., Büscher, P. (2019). ‘Systematic review and meta-analysis on the global distribution, host range, and prevalence of Trypanosoma evansi.’. Parasitol. Vectors 12, 67. doi: 10.1186/s13071-019-3311-4
Bachmaier, S., Boshart, M. (2014). “Kinetoplastid AGC kinases,” in Protein phosphorylation in parasites. Eds. Doerig, C., Späth, G., Wiese, M. (Weilheim, Germany: Wiley-VHC Verlag GmbH & Co. KGaA), 99–122.
Bachmaier, S., Giacomelli, G., Calvo-Alvarez, E., Rezende Vieira, L., Van Den Abbeele, J., Aristodemou, A., et al. (2022). ‘A multi-adenylate cyclase regulator at the flagellar tip controls African trypanosome transmission.’. Nat. Commun. 13, 5445. doi: 10.1038/s41467-022-33108-z
Bachmaier, S., Volpato Santos, Y., Kramer, S., Githure, G. B., Klöckner, T., Pepperl, J., et al. (2019). ‘Nucleoside analogue activators of cyclic AMP-independent protein kinase a of Trypanosoma.’. Nat. Commun. 10, 1421. doi: 10.1038/s41467-019-09338-z
Baron, D. M., Ralston, K. S., Kabututu, Z. P., Hill, K. L. (2007). ‘Functional genomics in Trypanosoma brucei identifies evolutionarily conserved components of motile flagella.’. J. Cell Sci. 120, 478–491. doi: 10.1242/jcs.03352
Barrett, M. P. (2018). ‘The elimination of human African trypanosomiasis is in sight: report from the third WHO stakeholders meeting on elimination of gambiense human African trypanosomiasis.’. PloS Negl. Trop. Dis. 12, e0006925. doi: 10.1371/journal.pntd.0006925
Beneke, T., Demay, F., Hookway, E., Ashman, N., Jeffery, H., Smith, J., et al. (2019). ‘Genetic dissection of a Leishmania flagellar proteome demonstrates requirement for directional motility in sand fly infections.’. PloS Path. 15, e1007828. doi: 10.1371/journal.ppat.1007828
Berman, H. M., Ten Eyck, L. F., Goodsell, D. S., Haste, N. M., Kornev, A., Taylor, S. S. (2005). ‘The cAMP binding domain: an ancient signaling module.’. Proc. Natl. Acad. Sci. U.S.A. 102, 45–50. doi: 10.1073/pnas.0408579102
Billington, K., Halliday, C., Madden, R., Dyer, P., Carrington, M., Vaughan, S., et al. (2023). ‘Genome-wide subcellular protein localisation in the flagellate parasite Trypanosoma brucei.’. Nat. Microbiol. 8, 533–547. doi: 10.1038/s41564-022-01295-6
Blaazer, A. R., Singh, A. K., de Heuvel, E., Edink, E., Orrling, K. M., Veerman, J. J. N., et al. (2018). ‘Targeting a subpocket in Trypanosoma brucei phosphodiesterase B1 (TbrPDEB1) enables the structure-based discovery of selective inhibitors with trypanocidal activity.’. J. Med. Chem. 61, 3870–3888. doi: 10.1021/acs.jmedchem.7b01670
Bridges, D. J., Pitt, A. R., Hanrahan, O., Brennan, K., Voorheis, H. P., Herzyk, P., et al. (2008). ‘Characterisation of the plasma membrane subproteome of bloodstream form Trypanosoma brucei.’. Proteomics 8, 83–99. doi: 10.1002/pmic.200700607
Broadhead, R., Dawe, H. R., Farr, H., Griffiths, S., Hart, S. R., Portman, N., et al. (2006). ‘Flagellar motility is required for the viability of the bloodstream trypanosome.’. Nature 440, 224–227. doi: 10.1038/nature04541
Bubis, J., Martinez, J. C., Calabokis, M., Ferreira, J., Sanz-Rodriguez, C. E., Navas, V., et al. (2018). ‘The gene product of a Trypanosoma equiperdum ortholog of the cAMP-dependent protein kinase regulatory subunit is a monomeric protein that is not capable of binding cyclic nucleotides.’. Biochimie 146, 166–180. doi: 10.1016/j.biochi.2017.12.010
Büscher, P., Cecchi, G., Jamonneau, V., Priotto, G. (2017). ‘Human African trypanosomiasis.’. Lancet 390, 2397–2409. doi: 10.1016/S0140-6736(17)31510-6
Canaves, J. M., Taylor, S. S. (2002). ‘Classification and phylogenetic analysis of the cAMP-dependent protein kinase regulatory subunit family.’. J. Mol. Evol. 54, 17–29. doi: 10.1007/s00239-001-0013-1
Clayton, C. E. (2016). ‘Gene expression in kinetoplastids.’. Curr. Opin. Microbiol. 32, 46–51. doi: 10.1016/j.mib.2016.04.018
Dao, K. K., Teigen, K., Kopperud, R., Hodneland, E., Schwede, F., Christensen, A. E., et al. (2006). ‘Epac1 and cAMP-dependent protein kinase holoenzyme have similar cAMP affinity, but their cAMP domains have distinct structural features and cyclic nucleotide recognition.’. J. Biol. Chem. 281, 21500–21511. doi: 10.1074/jbc.M603116200
Dean, S., Sunter, J. D., Wheeler, R. J. (2017). ‘TrypTag.org: a trypanosome genome-wide protein localisation resource.’. Trends Parasitol. 33, 80–82. doi: 10.1016/j.pt.2016.10.009
de Heuvel, E., Singh, A. K., Edink, E., van der Meer, T., van der Woude, M., Sadek, P., et al. (2019). ‘Alkynamide phthalazinones as a new class of TbrPDEB1 inhibitors.’. Bioorg. Med. Chem. 27, 3998–4012. doi: 10.1016/j.bmc.2019.06.027
de Koning, H. P., Gould, M. K., Sterk, G. J., Tenor, H., Kunz, S., Luginbuehl, E., et al. (2012). ‘Pharmacological validation of Trypanosoma brucei phosphodiesterases as novel drug targets.’. J. Infect. Dis. 206, 229–237. doi: 10.1093/infdis/jir857
Delespaux, V., de Koning, H. P. (2007). ‘Drugs and drug resistance in African trypanosomiasis.’. Drug Resist. Updat. 10, 30–50. doi: 10.1016/j.drup.2007.02.004
Desquesnes, M. (2004). ‘Livestock trypanosomoses and their vectors in Latin america’ (Paris, France: OIE), 183.
Desquesnes, M., Dargantes, A., Lai, D. H., Lun, Z. R., Holzmuller, P., Jittapalapong, S. (2013). ‘Trypanosoma evansi and surra: a review and perspectives on transmission, epidemiology and control, impact, and zoonotic aspects.’. Biomed. Res. Int. 2013, 321237. doi: 10.1155/2013/321237
Durante, I. M., Butenko, A., Rašková, V., Charyyeva, A., Svobodová, M., Yurchenko, V., et al. (2020). ‘Large-scale phylogenetic analysis of trypanosomatid adenylate cyclases reveals associations with extracellular lifestyle and host-pathogen interplay.’. Genome Biol. Evol. 12, 2403–2416. doi: 10.1093/gbe/evaa226
Franco, J. R., Cecchi, G., Priotto, G., Paone, M., Diarra, A., Grout, L., et al. (2020). ‘Monitoring the elimination of human African trypanosomiasis at continental and country level: update to 2018.’. PloS Negl. Trop. Dis. 14, e0008261. doi: 10.1371/journal.pntd.0008261
Giordani, F., Morrison, L. J., Rowan, T. G., de Koning, H. P., Barrett, M. P. (2016). ‘The animal trypanosomiases and their chemotherapy: a review.’. Parasitology 143, 1862–1889. doi: 10.1017/S0031182016001268
Gould, M. K., Bachmaier, S., Ali, J. A., Alsford, S., Tagoe, D. N., Munday, J. C., et al. (2013). ‘Cyclic AMP effectors in African trypanosomes revealed by genome-scale RNA interference library screening for resistance to the phosphodiesterase inhibitor CpdA.’. Antimicrob. Agents Chemother. 57, 4882–4893. doi: 10.1128/AAC.00508-13
Gould, M. K., de Koning, H. P. (2011). ‘Cyclic-nucleotide signalling in protozoa.’. FEMS Microbiol. Rev. 35, 515–541. doi: 10.1111/j.1574-6976.2010.00262.x
Hammond, M., Zoltner, M., Garrigan, J., Butterfield, E., Varga, V., Lukeš, J., et al. (2021). ‘The distinctive flagellar proteome of Euglena gracilis illuminates the complexities of protistan flagella adaptation.’. New Phytol. 232, 1323–1336. doi: 10.1111/nph.17638
Huang, G., Docampo, R. (2020). ‘The mitochondrial calcium uniporter interacts with subunit c of the ATP synthase of trypanosomes and humans.’. mBio 11, e00268–e00220. doi: 10.1128/mBio.00268-20
Ikeda, K., Brown, J. A., Yagi, T., Norrander, J. M., Hirono, M., Eccleston, E., et al. (2003). ‘Rib72, a conserved protein associated with the ribbon compartment of flagellar A-microtubules and potentially involved in the linkage between outer doublet microtubules.’. J. Biol. Chem. 278, 7725–7734. doi: 10.1074/jbc.M210751200
Ishida, H., Takahashi, K., Nakashima, K. I., Kumaki, Y., Nakata, M., Hikichi, K., et al. (2000). ‘Solution structures of the N-terminal domain of yeast calmodulin: Ca2+-dependent conformational change and its functional implication.’. Biochemistry 39, 13660–13668. doi: 10.1021/bi000582x
Jäger, A. V., De Gaudenzi, J. G., Mild, J. G., Mc Cormack, B., Pantano, S., Altschuler, D. L., et al. (2014). ‘Identification of novel cyclic nucleotide binding proteins in Trypanosoma cruzi.’. Mol. Biochem. Parasitol. 198, 104–112. doi: 10.1016/j.molbiopara.2015.02.002
Jones, T. W., Dávila, A. M. (2001). ‘Trypanosoma vivax – out of africa.’. Trends Parasitol. 17, 99–101. doi: 10.1016/S1471-4922(00)01777-3
Kannan, N., Wu, J., Anand, G. S., Yooseph, S., Neuwald, A. F., Venter, J. C., et al. (2007). ‘Evolution of allostery in the cyclic nucleotide binding module.’. Genome Biol. 8, R264. doi: 10.1186/gb-2007-8-12-r264
Kasozi, K. I., MacLeod, E. T., Welburn, S. C. (2022). ‘African animal trypanocide resistance: a systematic review and meta-analysis.’ Front. Vet. Sci. 9, 950248. doi: 10.3389/fvets.2022.950248
Kim, H., Kubori, T., Yamazaki, K., Kwak, M. J., Park, S. Y., Nagai, H., et al. (2020). ‘Structural basis for effector protein recognition by the Dot/Icm type IVB coupling protein complex.’. Nat. Commun. 11, 2623. doi: 10.1038/s41467-020-16397-0
King, S. M. (2006). ‘Axonemal protofilament ribbons, DM10 domains, and the link to juvenile myoclonic epilepsy.’. Cell Motil. Cytoskeleton 63, 245–253. doi: 10.1002/cm.20129
Kohl, L., Sherwin, T., Gull, K. (1999). ‘Assembly of the paraflagellar rod and the flagellum attachment zone complex during the Trypanosoma brucei cell cycle.’. J. Eukaryot. Microbiol. 46, 105–109. doi: 10.1111/j.1550-7408.1999.tb04592.x
Lane, T. R., Fuchs, E., Slep, K. C. (2017). ‘Structure of the ACF7 EF-hand-GAR module and delineation of microtubule binding determinants.’. Structure 25, 1130–1138. doi: 10.1016/j.str.2017.05.006
Li, J., Liu, H., Raval, M. H., Wan, J., Yengo, C. M., Liu, W., et al. (2019). Structure of the MORN4/Myo3a tail complex reveals MORN repeats as protein binding modules;. Eukaryot. Cell 27, 1366–1374.
Leder, L., Stark, W., Freuler, F., Marsh, M., Meyerhofer, M., Stettler, T., et al. (2010). ‘The structure of Ca2+ sensor Case16 reveals the mechanism of reaction to low Ca2+ concentrations.’. Sensors 10, 8143–8160. doi: 10.3390/s100908143
Lindner, A. K., Lejon, V., Chappuis, F., Seixas, J., Kazumba, L., Barrett, M. P., et al. (2020). ‘New WHO guidelines for treatment of gambiense human African trypanosomiasis including fexinidazole: substantial changes for clinical practice.’. Lancet Infect. Dis. 20, e38–e46. doi: 10.1016/S1473-3099(19)30612-7
Lopez, M. A., Saada, E. A., Hill, K. L. (2015). ‘Insect stage-specific adenylate cyclases regulate social motility in African trypanosomes.’. Eukaryot. Cell 14, 104–112. doi: 10.1128/EC.00217-14
Marques, C. A., Ridgway, M., Tinti, M., Cassidy, A., Horn, D. (2022). ‘Genome-scale RNA interference profiling of Trypanosoma brucei cell cycle progression defects.’. Nat. Commun. 13, 5326. doi: 10.1038/s41467-022-33109-y
Melaku, A., Birasa, B. (2013). ‘Drugs and drug resistance in African animal trypanosomosis: a review.’. Eur. J. Appl. Sci. 5, 84–91. doi: 10.5829/idosi.ejas.2013.5.3.75164
Moll, D., Schweinsberg, S., Hammann, C., Herberg, F. W. (2007). ‘Comparative thermodynamic analysis of cyclic nucleotide binding to protein kinase A. Biol. Chem. 388, 163–172. doi: 10.1515/BC.2007.018
Nachury, M. V., Mick, D. U. (2019). ‘Establishing and regulating the composition of cilia for signal transduction.’. Nat. Rev. Mol. Cell Biol. 20, 389–405. doi: 10.1038/s41580-019-0116-4
Naguleswaran, A., Doiron, N., Roditi, I. (2018). ‘RNA-seq analysis validates the use of culture-derived Trypanosoma brucei and provides new markers for mammalian and insect life-cycle stages.’. BMC Genomics 19, 227. doi: 10.1186/s12864-018-4600-6
Nelson, M. R., Thulin, E., Fagan, P. A., Forsén, S., Chazin, W. J. (2002). ‘The EF-hand domain: a globally cooperative structural unit.’. Protein Sci. 11, 198–205. doi: 10.1110/ps.33302
Nolan, D. P., Rolin, S., Rodriguez, J. R., Van Den Abbeele, J., Pays, E. (2000). ‘Slender and stumpy bloodstream forms of Trypanosoma brucei display a differential response to extracellular acidic and proteolytic stress.’. Eur. J. Biochem. 267, 18–27. doi: 10.1046/j.1432-1327.2000.00935.x
Norrander, J. M., deCathelineau, A. M., Brown, J. A., Porter, M. E., Linck, R. W. (2000). ‘The Rib43a protein is associated with forming the specialized protofilament ribbons of flagellar microtubules in Chlamydomonas.’. Mol. Biol. Cell 11, 201–215. doi: 10.1091/mbc.11.1.201
Oberholzer, M., Marti, G., Baresic, M., Kunz, S., Hemphill, A., Seebeck, T. (2007). ‘The Trypanosoma brucei cAMP phosphodiesterases TbrPDEB1 and TbrPDEB2: flagellar enzymes that are essential for parasite virulence.’. FASEB J. 21, 720–731. doi: 10.1096/fj.06-6818com
Oberholzer, M., Saada, E. A., Hill, K. L. (2015). ‘Cyclic AMP regulates social behavior in African trypanosomes.’. mBio 6, e01954–14. doi: 10.1128/mBio.01954-14
Olmsted, J. B. (1981). ‘Affinity purification of antibodies from diazotized paper blots of heterogeneous protein samples.’. J. Biol. Chem. 256, 11955–11957. doi: 10.1016/S0021-9258(18)43211-5
Paindavoine, P., Rolin, S., Van Assel, S., Geuskens, M., Jauniaux, J. C., Dinsart, C., et al. (1992). ‘A gene from the variant surface glycoprotein expression site encodes one of several transmembrane adenylate cyclases located on the flagellum of Trypanosoma brucei.’. Mol. Cell Biol. 12, 1218–1225. doi: 10.1128/mcb.12.3.1218-1225.1992
Parashar, V., Mirouze, N., Dubnau, D. A., Neiditch, M. B. (2011). ‘Structural basis of response regulator dephosphorylation by rap phosphatases.’. PloS Biol. 9, e1000589. doi: 10.1371/journal.pbio.1000589
Pays, E., Nolan, D. P. (1998). ‘Expression and function of surface proteins in Trypanosoma brucei.’. Mol. Biochem. Parasitol. 91, 3–36. doi: 10.1016/S0166-6851(97)00183-7
Saada, E. A., DeMarco, S. F., Shimogawa, M. M., Hill, K. L. (2015). ‘"With a little help from my friends" – social motility in Trypanosoma brucei.’. PloS Path. 11, e1005272. doi: 10.1371/journal.ppat.1005272
Saada, E. A., Kabututu, Z. P., Lopez, M., Shimogawa, M. M., Langousis, G., Oberholzer, M., et al. (2014). ‘Insect stage-specific receptor adenylate cyclases are localized to distinct subdomains of the Trypanosoma brucei flagellar membrane.’. Eukaryot. Cell 13, 1064–1076. doi: 10.1128/EC.00019-14
Salmon, D. (2018). Adenylate cyclases of Trypanosoma brucei, environmental sensors and controllers of host innate immune response.’. Pathogens 7, 48. doi: 10.3390/pathogens7020048
Salmon, D., Bachmaier, S., Krumbholz, C., Kador, M., Gossmann, J. A., Uzureau, P., et al. (2012a). ‘Cytokinesis of Trypanosoma brucei bloodstream forms depends on expression of adenylyl cyclases of the ESAG4 or ESAG4-like subfamily.’. Mol. Microbiol. 84, 225–242. doi: 10.1111/j.1365-2958.2012.08013.x
Salmon, D., Vanwalleghem, G., Morias, Y., Denoeud, J., Krumbholz, C., Lhomme, F., et al. (2012b). ‘Adenylate cyclases of Trypanosoma brucei inhibit the innate immune response of the host.’. Science 337, 463–466. doi: 10.1126/science.1222753
Shakur, Y., de Koning, H. P., Ke, H., Kambayashi, J., Seebeck, T. (2011). ‘Therapeutic potential of phosphodiesterase inhibitors in parasitic diseases.’. Handb. Exp. Pharmacol. 2011, 487–510. doi: 10.1007/978-3-642-17969-3_20
Shaw, S., Knüsel, S., Abbühl, D., Naguleswaran, A., Etzensperger, R., Benninger, M., et al. (2022). ‘Cyclic AMP signalling and glucose metabolism mediate pH taxis by African trypanosomes.’. Nat. Commun. 13, 603. doi: 10.1038/s41467-022-28293-w
Shaw, S., Roditi, I. (2023). The sweet and sour sides of trypanosome social motility. Trends Parasitol. 4, 242–250. doi: 10.1016/j.pt.2023.01.001
Subota, I., Julkowska, D., Vincensini, L., Reeg, N., Buisson, J., Blisnick, T., et al. (2014). ‘Proteomic analysis of intact flagella of procyclic Trypanosoma brucei cells identifies novel flagellar proteins with unique sub-localization and dynamics.’. Mol. Cell Proteomics 13, 1769–1786. doi: 10.1074/mcp.M113.033357
Sun, S. Y., Wang, C., Yuan, Y. A., He, C. Y. (2013). ‘An intracellular membrane junction consisting of flagellum adhesion glycoproteins links flagellum biogenesis to cell morphogenesis in Trypanosoma brucei.’. J. Cell Sci. 126, 520–531. doi: 10.1242/jcs.113621
Suzuki, T., Inoue, I., Yamakawa, K. (2020). ‘Epilepsy protein Efhc1/myoclonin1 is expressed in cells with motile cilia but not in neurons or mitotic apparatuses in brain.’. Sci. Rep. 10, 22076. doi: 10.1038/s41598-020-79202-4
Vassella, E., Reuner, B., Yutzy, B., Boshart, M. (1997). ‘Differentiation of African trypanosomes is controlled by a density sensing mechanism which signals cell cycle arrest via the cAMP pathway.’. J. Cell Sci. 110, 2661–2671. doi: 10.1242/jcs.110.21.2661
Vaughan, S. (2010). ‘Assembly of the flagellum and its role in cell morphogenesis in Trypanosoma brucei.’. Curr. Opin. Microbiol. 13, 453–458. doi: 10.1016/j.mib.2010.05.006
Wheeler, R. J. (2021). A resource for improved predictions of Trypanosoma and Leishmania protein three-dimensional structure. PloS One 16, e0259871. doi: 10.1371/journal.pone.0259871
Wickstead, B., Ersfeld, K., Gull, K. (2002). ‘Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei.’. Mol. Biochem. Parasitol. 125, 211–216. doi: 10.1016/S0166-6851(02)00238-4
Wirschell, M., Yamamoto, R., Alford, L., Gokhale, A., Gaillard, A., Sale, W. S. (2011). ‘Regulation of ciliary motility: conserved protein kinases and phosphatases are targeted and anchored in the ciliary axoneme.’. Arch. Biochem. Biophys. 510, 93–100. doi: 10.1016/j.abb.2011.04.003
Wirtz, E., Leal, S., Ochatt, C., Cross, G. A. (1999). ‘A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei.’. Mol. Biochem. Parasitol. 99, 89–101. doi: 10.1016/S0166-6851(99)00002-X
Won, M. M., Baublis, A., Burleigh, B. A. (2023). ‘Proximity-dependent biotinylation and identification of flagellar proteins in Trypanosoma cruzi.’. mSphere, e00088-23. doi: 10.1128/msphere.00088-23
Keywords: Trypanosoma brucei, cAMP, CARP, flagellum, adenylate cyclase, RNAi screening, NPD-001
Citation: Bachmaier S, Gould MK, Polatoglou E, Omelianczyk R, Brennand AE, Aloraini MA, Munday JC, Horn D, Boshart M and de Koning HP (2023) Novel kinetoplastid-specific cAMP binding proteins identified by RNAi screening for cAMP resistance in Trypanosoma brucei. Front. Cell. Infect. Microbiol. 13:1204707. doi: 10.3389/fcimb.2023.1204707
Received: 12 April 2023; Accepted: 14 June 2023;
Published: 05 July 2023.
Edited by:
Igor Cestari, McGill University, CanadaReviewed by:
Suzanne McDermott, Seattle Children’s Research Institute, United StatesCopyright © 2023 Bachmaier, Gould, Polatoglou, Omelianczyk, Brennand, Aloraini, Munday, Horn, Boshart and de Koning. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael Boshart, Ym9zaGFydEBsbXUuZGU=; Harry P. de Koning, SGFycnkuRGUtS29uaW5nQGdsYXNnb3cuYWMudWs=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.