
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 01 August 2023
Sec. Molecular Bacterial Pathogenesis
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1203159
This article is part of the Research Topic Pathogenic mechanism and biocontrol of Xanthomonas on plants View all 6 articles
Introduction: Many Gram-negative plant- and animal-pathogenic bacteria employ type IV secretion (T4S) systems to transport proteins or DNA/protein complexes into eukaryotic or bacterial target cells. T4S systems have been divided into minimized and expanded T4S systems and resemble the VirB/VirD4 T4S system from the plant pathogen Agrobacterium tumefaciens and the Icm/Dot T4S system from the human pathogen Legionella pneumophila, respectively. The only known plant pathogen with both types of T4S systems is Xanthomonas euvesicatoria which is the causal agent of bacterial spot disease on pepper and tomato plants.
Results and discussion: In the present study, we show that virB/virD4 and icm/dot T4S genes are expressed and encode components of oligomeric complexes corresponding to known assemblies of VirB/VirD4 and Icm/Dot proteins. Both T4S systems are dispensable for the interaction of X. euvesicatoria with its host plants and do not seem to confer contact-dependent lysis of other bacteria, which was previously shown for the chromosomally encoded VirB/VirD4 T4S system from Xanthomonas axonopodis pv. citri. The corresponding chromosomal T4S gene cluster from X. euvesicatoria is incomplete, however, the second plasmid-localized vir gene cluster encodes a functional VirB/VirD4 T4S system which contributes to plasmid transfer. In agreement with this finding, we identified the predicted relaxase TraI as substrate of the T4S systems from X. euvesicatoria. TraI and additional candidate T4S substrates with homology to T4S effectors from X. axonopodis pv. citri interact with the T4S coupling protein VirD4. Interestingly, however, the predicted C-terminal VirD4-binding sites are not sufficient for T4S, suggesting the contribution of additional yet unknown mechanisms to the targeting of T4S substrates from X. euvesicatoria to both VirB/VirD4 and Icm/Dot T4S systems.
Gram-negative plant- and animal-pathogenic bacteria employ at least six types of protein secretion systems for the delivery of proteins or protein/DNA complexes into the extracellular milieu or into bacterial or eukaryotic target cells (Costa et al., 2015; Galan and Waksman, 2018). Essential for pathogenicity is often the type III secretion (T3S) system, which translocates type III effector (T3E) proteins into eukaryotic cells to manipulate metabolic pathways to the bacterial benefit (Büttner, 2012; Wagner et al., 2018; Bastedo et al., 2020; Ceulemans et al., 2021). Alternatively, some pathogens employ type IV secretion (T4S) or type VI secretion systems to translocate DNA/protein complexes or effector proteins into eukaryotic cells (Galan and Waksman, 2018). T4S systems are highly versatile nanomachines which are grouped into conjugation systems, effector translocators and contact-independent uptake or release systems (Christie, 2016; Li et al., 2019; Waksman, 2019). Conjugation systems are the largest group of T4S systems and one of the major drivers of interbacterial exchange of mobile genetic elements including antibiotic resistance and virulence genes (Christie et al., 2017; Li et al., 2019). T4S system-dependent genetic exchange allows the bacterial acquisition of novel traits, thus potentially resulting in more virulent and antibiotic-resistant strains and posing a major threat on human and animal health (Soucy et al., 2015; Shen et al., 2022).
Bacterial pathogens, which depend on T4S systems for successful infection of their host organisms, include e.g., the human pathogen Legionella pneumophila or the plant pathogen Agrobacterium tumefaciens (Kubori and Nagai, 2016; Li et al., 2019). The VirB/VirD4 T4S system from A. tumefaciens transports T-DNA (transfer DNA) and proteins into plant cells and is similar to bacterial conjugation systems (Christie, 2016; Li and Christie, 2018). VirB/VirD4-like T4S systems consist of 12 subunits including VirB1 – VirB11 and VirD4 and represent the prototype of minimized T4S systems. The designation “minimized T4S system” refers to any bacterial T4S system with a similar number of components than the VirB/VirD4 T4S system. In contrast, more complex T4S systems are grouped as “expanded” T4S systems and include e.g., the Icm/Dot (intracellular multiplication/defect in organelle trafficking) T4S system from L. pneumophila (Christie et al., 2017; Costa et al., 2021). The Icm/Dot T4S system is one of the largest known T4S systems and is assembled by more than 25 components including several counterparts of VirB/VirD4 proteins (Kitao et al., 2022; Sheedlo et al., 2022).
Both VirB/VirD4- and Icm/Dot-like T4S systems consist of heterooligomeric protein complexes which span both bacterial membranes and are associated with an extracellular pilus (Li et al., 2019; Sheedlo et al., 2022). Cryo-electron tomography provided structural insights into the architecture of four different T4S systems which were isolated from intact bacteria and include a minimized T4S system from E. coli encoded on plasmid pKM101 and expanded T4S systems such as the F plasmid-encoded T4S system, the Cag T4S system from Helicobacter pylori and the Icm/Dot T4S system from L. pneumophila (Sheedlo et al., 2022). Together with cryo-electron microscopy imaging of isolated T4S system subassemblies, these studies revealed the presence of heterooligomeric protein complexes in the outer membrane (OM) and inner membrane (IM) (Costa et al., 2021; Sheedlo et al., 2022). In VirB/VirD4-like T4S systems, a central core complex in the OM and the periplasm is assembled as two ring-like layers by 14 copies of the C-terminal domain of VirB10 surrounded by VirB7 and VirB9 (Costa et al., 2021; Sheedlo et al., 2022). This OM complex is connected via a stalk structure to the IM complex which consists of 12 copies of VirB3, VirB6 and VirB8 associated with 14 copies of the N-terminal region of VirB10 (Costa et al., 2021; Sheedlo et al., 2022). Additional components of the IM complex are the three cytoplasmic ATPases VirB4, VirB11 and VirD4, which are involved in substrate recognition and transport (Costa et al., 2021; Sheedlo et al., 2022).
In expanded T4S systems such as the Icm/Dot T4S system from L. pneumophila, the central core complex is assembled by DotD, DotH, and DotG which are functional counterparts of VirB7, VirB9, and VirB10, respectively (Kitao et al., 2022; Sheedlo et al., 2022). Additional proteins specific for Icm/Dot T4S systems are involved in the formation of the membrane-spanning OM and IM complexes including the core complex components DotC and DotF as well as two cytoplasmic ATPases, DotO and DotB, which correspond to VirB4 and VirB11 (Kitao et al., 2022; Sheedlo et al., 2022). Substrate recognition by VirB/VirD4 and Icm/Dot T4S systems depends on the ATPases VirD4 and DotL, respectively, which are referred to as type IV coupling proteins (T4CPs) (Christie et al., 2017; Li et al., 2019; Costa et al., 2021; Meir et al., 2023). The cytoplasmic region of VirD4 contains an alpha-helical bundle, termed all-alpha-domain (AAD), which likely provides a binding site for T4S substrates (Christie et al., 2017; Li et al., 2019; Oka et al., 2022). In Icm/Dot T4S systems, additional adaptor proteins such as IcmS and IcmW are part of a heterooligomeric coupling complex and promote substrate recruitment by DotL (Li et al., 2019; Kitao et al., 2022; Sheedlo et al., 2022). Recognition of cargo proteins depends on different signals including a C-terminal region in T4S substrates from Xanthomonas axonopodis pv. citri, termed XVIPCD (Xanthomonas VirD4-interacting protein conserved domain), which is sequence variable but shares common features such as certain amino acid compositions or structural motifs (Li et al., 2019; Sgro et al., 2019; Costa et al., 2021).
The analysis of T4S systems in Gram-negative bacteria suggests that Icm/Dot T4S systems are predominantly present in human and animal pathogens where they act as important pathogenicity factors in the delivery of effector proteins into eukaryotic host cells (Nagai and Kubori, 2011; Li et al., 2019; Liu and Shin, 2019; Costa et al., 2021). In contrast, plant-pathogenic bacteria such as A. tumefaciens depend on VirB/VirD4 T4S systems to interact with their hosts (Li et al., 2019). In Xanthomonas spp., VirB/VirD4 T4S systems, also designated Xanthomonadales-like or X-T4S systems, were initially identified as conjugation machines for plasmid transfer (El Yacoubi et al., 2007; Kaur et al., 2019; Sgro et al., 2019). In X. axonopodis pv. citri and the related human-pathogenic bacterium Stenotrophomonas maltophilia, however, VirB/VirD4 T4S systems deliver toxins into bacterial competitor cells (Souza et al., 2015; Bayer-Santos et al., 2019; Sgro et al., 2019). Candidate substrates were identified by yeast two-hybrid analyses as interactors of the T4CP VirD4 which serves as substrate acceptor site (Sgro et al., 2019). The identified Xanthomonas VirD4-interacting proteins (XVIPs) include Xanthomonadales-like T4S system effectors (X-Tfes) and share a C-terminal domain of approximately 120 residues with conserved amino acid motifs, designated XVIP conserved domain (XVIPCD) (Alegria et al., 2005; Souza et al., 2015). The XVIPCD presumably provides the binding site for VirD4 and is required for T4S (Souza et al., 2015; Oka et al., 2022).
In the present study, we analyzed the function of T4S systems in Xanthomonas euvesicatoria, which is the causal agent of bacterial spot disease in pepper and tomato. X. euvesciatoria is the only known plant pathogen with a putative Icm/Dot T4S system and a VirB/VirD4 T4S system. Corresponding genes are located on the chromosome and on plasmids pXCV183 and pXCV38 (Thieme et al., 2005). A possible contribution of T4S systems from X. euvesicatoria to the host-pathogen interaction and to conjugation or effector delivery has not yet been investigated. It was previously shown that pathogenicity of X. euvesicatoria depends on a T3S system which translocates type III effectors (T3Es) into plant cells and thus allows the manipulation of cellular processes to the advantage of the pathogen (Büttner and Bonas, 2010; Alvarez-Martinez et al., 2021). In addition to T3Es, substrates of the Xps type II secretion (T2S) system contribute to virulence and likely degrade components of the plant cell wall to facilitate the delivery of T3Es (Szczesny et al., 2010; Solé et al., 2015). In the present study, we show that virulence of X. euvesicatoria is independent of T4S genes which are expressed in vitro and in planta. The plasmid-encoded VirB/VirD4 T4S system likely contributes to plasmid transfer between different X. euvesicatoria strains, suggesting a role as conjugation machine. Candidate T4S substrates interact with VirD4 and include the putative relaxase TraI and XVIPCD-containing homologs of X-Tfes from X. axonopodis pv. citri. Interestingly, our data suggests that T4S substrates are targeted to both T4S systems and that the XVIPCD is not sufficient for T4S.
Bacterial strains and plasmids used in this study are listed in Table S1. Escherichia coli strains were grown at 37°C in lysogeny broth (LB) medium, and X. euvesicatoria at 30°C in nutrient-yeast extract-glycerol (NYG) medium. Antibiotics were added to the media at the following final concentrations: ampicillin, 100 μg/ml; kanamycin, 25 μg/ml; rifampicin, 100 μg/ml; spectinomycin, 100 μg/ml; streptomycin, 2.5 µg/ml and gentamicin, 15 μg/ml. Plasmids were introduced into E. coli by transformation and into X. euvesicatoria by electroporation or triparental conjugation, using pRK2013 as helper plasmid.
For infection assays, X. euvesicatoria strains were resuspended in 1 mM MgCl2 at an optical density (OD600nm) of 0.1 which corresponds to 1×108 colony-forming units (CFU) ml−1. Bacterial suspensions were infiltrated into leaves of the pepper cultivar Early Cal Wonder (ECW) using a needleless syringe. Infected plants were incubated in growth chambers for 16 hours of light at 28°C and 8 hours of darkness at 22°C. Disease symptoms were photographed seven days post inoculation (dpi). In planta growth curves were performed in three ECW plants as described (Bonas et al., 1991). To monitor translocation of dTALE-2, X. euvesicatoria strains were resuspended in 1 mM MgCl2 at a density of 4×108 CFU ml−1 and infiltrated into leaves of gfp-transgenic Nicotiana benthamiana plants (Werner et al., 2011). Infected N. benthamiana plants were incubated for 16 hours of light at 20°C and 8 hours of darkness at 18°C. GFP fluorescence was documented four dpi using a chemiluminescence/fluorescence imager (Vilber Fusion FX Edge). Infection experiments were performed at least three times with different transconjugants; representative results are shown.
To generate X. euvesicatoria T4S deletion mutants, DNA fragments flanking individual (virB4, virD4, virG, virA and icmE) or multiple (virB11virB1, virB6virB8virB9virB10 and dotCB) T4S genes were amplified by PCR and cloned into the Golden Gate-compatible suicide vector pOGG2 using the type IIs restriction enzyme BsaI and ligase (Engler et al., 2008). The resulting constructs were transferred into strain 85-10 by triparental conjugation. Transconjugants were selected as described previously and double crossovers resulted in T4S deletion mutant strains (Huguet et al., 1998). Multiple deletions of T4S genes or gene regions resulted from the successive introduction of corresponding deletion constructs into X. euvesicatoria strains.
To insert the gentamicin resistance gene into the genome of X. euvesicatoria strain 85-10, the gene was amplified by PCR from plasmid pBRM and cloned into the Golden Gate-compatible suicide vector pLAND downstream of the lac promoter using BsaI and ligase. pLAND allows the insertion of genes into the hpaFG region adjacent to the T3S gene cluster (Lorenz et al., 2012). The corresponding construct pLAND-gentR was introduced into strain 85-10 by conjugation and homologous recombination resulted in strain 85-10::gentR as described previously (Huguet et al., 1998; Lorenz et al., 2012).
To insert a kanamycin resistance gene and gfp (green fluorescent protein) into X. euvesicatoria, DNA fragments flanking a non-coding region at nucleotide position 2831 on plasmid pXCV183 were amplified by PCR from strain 85-10. Similarly, the promoter of the gentamicin resistance gene on plasmid pBRM, the kanamycin resistance gene from plasmid pKT25 and the gfp gene including the lac promoter were amplified by PCR using plasmid pBRM-sfgfp as template. Flanking BsaI sites with matching overhangs were introduced by the primer sequences. The corresponding amplicons were first subcloned in vector pICH41021 using the blunt-end cutter SmaI and ligase in a cut-ligation. All five amplicons were subsequently assembled in the Golden Gate-compatible suicide vector pOGG2 using BsaI and ligase. The resulting construct pOGG2-kanRgfp(pXCV183) was transferred into strain 85-10 by triparental conjugation. Double crossovers resulted in X. euvesicatoria strain 85-10 containing the kanamycin resistance gene and gfp on plasmid pXCV183.
For the generation of XCV0332, XCV1120 and XCV3751 expression constructs, genes were amplified by PCR from X. euvesicatoria strain 85-10 and individually cloned into the Golden Gate-compatible expression vector pBRNM using BsaI in a single restriction/ligation reaction. Vector pBRNM allows the expression of genes under control of a lac promoter in frame with an N-terminal 3 × c-Myc epitope-encoding sequence. To generate the traI expression construct, the promoter of XCV4361, the 3 × c-Myc epitope-encoding sequence and traI were amplified by PCR. Primer sequences introduced BsaI restriction sites with matching overhangs. When compared with the lac promoter, the promoter upstream of XCV4361 results in lower expression levels of downstream genes as was previously shown for the predicted xylanase gene XCV4360 (Solé et al., 2015). All three DNA fragments were assembled by Golden Gate cloning in vector pBRM-P which contains an integrated stop codon downstream of the insert.
For in cis expression of virB4 in X. euvesicatoria, virB4 was amplified by PCR and cloned into the Golden Gate-compatible suicide vector pLAND downstream of the lac promoter and in frame with a C-terminal 3 x c-Myc epitope-encoding sequence using BsaI and ligase. To insert virB4-c-myc into the genome of X. euvesicatoria, pLAND-virB4 was transferred into strain 85-10ΔvirB4. Double homologous recombination events led to the insertion of virB4-c-myc into the hpaFG region. For the in cis expression of icmE, icmE and the promoter of XCV0160 were amplified by PCR from strain 85-10 and assembled in the Golden Gate-compatible suicide vector pLAND-P in frame with a C-terminal 3 × c-Myc epitope-encoding sequence using BsaI and ligase. The promoter of XCV0160 was previously shown to mediate gene expression in X. euvesicatoria and presumably results in lower expression levels than the constitutive lac promoter (Scheibner and Büttner, unpublished data). The resulting construct pLAND-P-icmE was transferred into strain 85-10ΔicmE and double homologous recombination events led to the insertion of icmE-c-myc into the hpaFG region.
For the generation of promoter-reporter constructs, upstream regions of virB2, virB5, virD4, virG, dotA, dotD, icmL, hrpB1 as well as 129 bp downstream of the annotated start codon of icmL were amplified by PCR. Each amplicon was ligated with the sfgfp reporter gene in the Golden Gate-compatible vector pBRM-P+T, which contains a terminator upstream of the cloning site. The resulting constructs were transferred into X. euvesicatoria strains 85-10 and 85-10ΔvirG. vir and icm/dot promoter regions were additionally cloned upstream of the dTALE-2 (designer transcription activator-like effector 2) gene in a Golden Gate-compatible expression vector using the modular cloning (MoClo) system (Weber et al., 2011). The MoClo system is based on different cloning vectors (designated level -2, -1, 0, 1 and 2 vectors) which allow the stepwise assembly of gene and promoter modules into multigene constructs by the alternate use of BsaI and BpiI restriction enzymes (Weber et al., 2011). vir and icm/dot promoter regions were first cloned into vector pAGM9121 using BpiI and ligase, thus resulting in level 0 constructs, and were subsequently transferred to vector pICH47732 using BsaI and ligase to create level 1 promoter constructs. For the assembly of dTALE-2, modules encoding the N-terminal region (pICH70781), the central repeats (pICH73079, pICH73081, pICH73093) and the C-terminal region of dTALE-2 (pICH72151) were assembled in the level 0 construct pICH73103. The resulting insert was subsequently cloned into vector pICH50251 using BsaI and ligase to create the level 1 dTALE-2 construct pICH79631. For the generation of dTALE-2 expression constructs containing vir or icm/dot promoters, modules with promoter fragments, the dTALE-2-encoding sequence (pICH79631), and a transcription terminator (pICH50122) were assembled in the level 2 vector pICH77739 using BpiI and ligase.
For bacterial adenylate cyclase-based two-hybrid (BACTH) assays, T4S genes were cloned into the Golden Gate-compatible vectors pUT18GG, pUT18CGG, pKT25GG and pKNT25GG using BsaI and ligase. Additionally, the putative T4S substrate genes XCV0332, traI, XCV3751 and XCV1120 were amplified by PCR from X. euvesicatoria strain 85-10, subcloned into pICH41021 as blunt-end fragments using SmaI and ligase, and the resulting inserts were cloned into the BsaI sites of vectors pUT18GG, pUT18CGG, pKT25GG and pKNT25GG by Golden Gate cloning. Primers and plasmids used in this study are listed in Tables S1, S2.
For in vitro T4S assays, X. euvesicatoria strains were grown over night in NYG medium with antibiotics, resuspended in fresh NYG medium containing 50 μg ml−1 bovine serum albumin (BSA) at a cell density of 2 × 108 CFU/ml, and incubated on a rotary shaker at 30°C for 4 h. Bacterial cells and secreted proteins were separated by filtration as described previously (Rossier et al., 1999). Proteins in 2 ml of the culture supernatants were precipitated with trichloroacetic acid and resuspended in 20 μl of Laemmli buffer. Total cell extracts and culture supernatants were analyzed by SDS-PAGE and immunoblotting, using antibodies directed against the c-Myc epitope (Sigma Aldrich) and the β-subunit of the RNA polymerase (Invitrogen), respectively. Horseradish peroxidase-labelled anti-mouse and anti-rabbit antibodies were used as secondary antibodies. Binding of antibodies was visualized using a chemiluminescence imager (Vilber Fusion FX Edge). Results were reproduced at least two times.
BACTH assays were performed using the Euromedex BACTH system kit and Golden Gate-compatible expression vectors (Table S1). To monitor protein synthesis, expression constructs encoding T25 and T18 fusion proteins were transformed into JM109 E. coli cells, and gene expression was induced with IPTG (isopropyl-β-D-thiogalactopyranoside; 2 mM final concentration) at an OD600 of 0.6 - 0.8. After induction, cultures were incubated on a rotary shaker for 2 h at 37°C. Bacterial cells were collected by centrifugation, resuspended in Laemmli buffer, and analyzed by immunoblotting, using a FLAG epitope-specific antibody.
Protein-protein interaction studies were performed as described previously (Karimova et al., 2005; Otten and Büttner, 2021). Briefly, expression constructs encoding T18- and T25-fusion proteins were cotransformed into chemically competent BTH101 E. coli cells and transformants were plated on LB plates containing kanamycin and gentamicin. At least three colonies per transformant were cultivated over night at 30°C on a rotary shaker in liquid LB medium with appropriate antibiotics. Two microliters of each culture were spotted on LB plates containing kanamycin, gentamicin, X-gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranosid; 40 μg/ml) and 2 mM IPTG. Colonies were photographed over a period of three to 5 days. Every co-transformation was performed at least three times; representative results are shown.
Growth of E. coli during cocultivation with X. euvesicatoria was analyzed as described previously (Bayer-Santos et al., 2019). X. euvesicatoria and E. coli strains were grown overnight in NYG or LB medium, respectively, with appropriate antibiotics. Cells were harvested by centrifugation, resuspended at OD600nm of 0.3 in fresh medium and grown to exponential phase for 4 h on a rotary shaker. The OD600nm of all cultures was measured and adjusted to 1. Serial dilutions (1:2) of E. coli cultures were prepared in 96 well plates and equal volumes of E. coli and X. euvesicatoria cultures were mixed in each well. Five microliters of every suspension were spotted on LB agar plates containing 100 μM IPTG and 40 μg/mL X-gal. Plates were incubated for 24 h at 30˚C and photographed. The experiment was performed three times with similar results.
For quantitative growth competition assays, X. euvesicatoria and E. coli were cocultivated according to a previously described protocol (Souza et al., 2015). For this, X. euvesicatoria strains were grown over night on NYG agar plates containing rifampicin and E. coli strain Top 10 was grown over night in 5 ml LB liquid medium in the presence of streptinomycin. Cells were resuspended to an OD600nm of 0.2 in liquid NYG or LB medium and cultures were incubated for five hours on a rotary shaker at 30°C for X. euvesicatoria or at 37°C for E. coli. After incubation, the OD600nm of all cultures was measured and X. euvesicatoria was mixed with E. coli in triplicates in a final volume of 1 ml at a ratio of 100: 1 (X. euvesicatoria: E. coli) corresponding to a final OD600nm for X. euvesicatoria of 0.05 and for E. coli of 0.0005. To measure bacterial growth at 0 hours, we prepared serial dilutions of 100 µl of the mixtures which were plated on LB plates containing streptinomycin for selection of Top 10 and on NYG plates containing rifampicin for selection of X. euvesicatoria. For the analysis of bacterial growth after cocultivation of X. euvesciatoria and E. coli, 100 µl of the mixtures were spotted on LB agar plates and cultivated at 30°C. After 38 hours, bacteria were resuspended in 1 ml of NYG medium and all mixtures were adjusted to the same OD600nm. Serial dilutions were subsequently plated on LB plates containing streptinomycin for selection of Top 10 and on NYG plates containing rifampicin for selection of X. euvesicatoria and the final X. euvesicatoria – E. coli ratios were calculated after counting the colonies.
Plasmid transfer between X. euvesicatoria strains was analyzed according to a protocol by Stall et al. (1986). For this, selected X. euvesicatoria strains were grown over night on NYG agar plates containing appropriate antibiotics. Cells were resuspended at an OD600nm of 0.3 in liquid NYG medium without antibiotics. After six hours of incubation at 30°C on a rotary shaker, donor and recipient strains were mixed in 1 ml of NYG medium at a final OD600nm of 0.4 for each strain. Bacterial suspensions were subsequently dropped on a cellulose nitrate filter (NC20, membrane filters, 0.2 µm, diameter 50 mm, Whatman) which was transferred onto an NYG agar plate containing rifampicin. Bacteria were covered with 1 ml 1% water agar and incubated over night at 30°C. Bacterial conjugation mixtures were then transferred from the cellulose nitrate filters to 1 ml of NYG medium using a spatula and adjusted to an OD600nm of 0.05. Serial dilutions were plated on NYG plates (1.5% agar) with appropriate antibiotics. The plates were incubated for two days at 30°C and the transconjugants were counted. The number of transconjugants obtained with the wild-type strain as donor was set to 100%.
For the isolation of native plasmids, X. euvesicatoria strains were resuspended at an OD600nm of 0.05 in 50 ml NYG medium containing rifampicin and grown over night at 30°C. Plasmids were isolated using the GeneJET plasmid midiprep kit (Thermo Scientific) according to the manufacturer’s instructions. Twenty microliters of each plasmid preparation were loaded on an 0.7% agarose gel containing 5% ethidium bromide. The electrophoresis was performed in 1 x TBE (Tris-borate-EDTA [ethylene diamine tetraacetic acid]) buffer at 120 mA for 25 min and DNA was visualized using an UV imager. The agarose gel electrophoresis was performed three times with similar results.
Previous genome sequence analyses identified predicted virB/virD4 and icm/dot T4S genes on the chromosome and on plasmids pXCV38 and pXCV183 of X. euvesicatoria strain 85-10 (Thieme et al., 2005). The chromosomal T4S gene cluster contains virB7, virB8, virB9, virD4 and four copies of virB6 (Figure 1A). virB7, virB8, virB9 and virD4 genes encode predicted core components of the T4S system which are homologous (76 – 100% amino acid identity) to corresponding proteins from X. axonopodis pv. citri (Figure 1A; Table S3). VirB6 proteins are less conserved (40 – 42% amino acid identity) (Figure 1A). In contrast to the chromosomally encoded VirB/VirD4 T4S system from X. axonopodis pv. citri, which was previously shown to deliver toxins into bacterial cells (Souza et al., 2015), the chromosomal vir gene cluster from X. euvesicatoria is incomplete because it lacks virB1 to virB5 as well as virB10 and virB11 (Figure 1A). Chromosomal vir genes of X. euvesicatoria, however, include virA and virG (Figure 1A), which encode putative regulators with homology to the corresponding sensor and response regulator of a VirA/VirG two-component system from A. tumefaciens (Gelvin, 2003; Wise and Binns, 2015). VirA from A. tumefaciens is autophosphorylated and subsequently phosphorylates VirG which binds to vir box elements (consensus [5’-TG(A/T)AA(C/T)-3’]) in promoters of target genes (Winans et al., 1987; Jin et al., 1990; Tamamoto et al., 1990; Winans, 1990). Notably, predicted vir boxes are located upstream of several vir and icm/dot genes from X. euvesicatoria (Table 1).
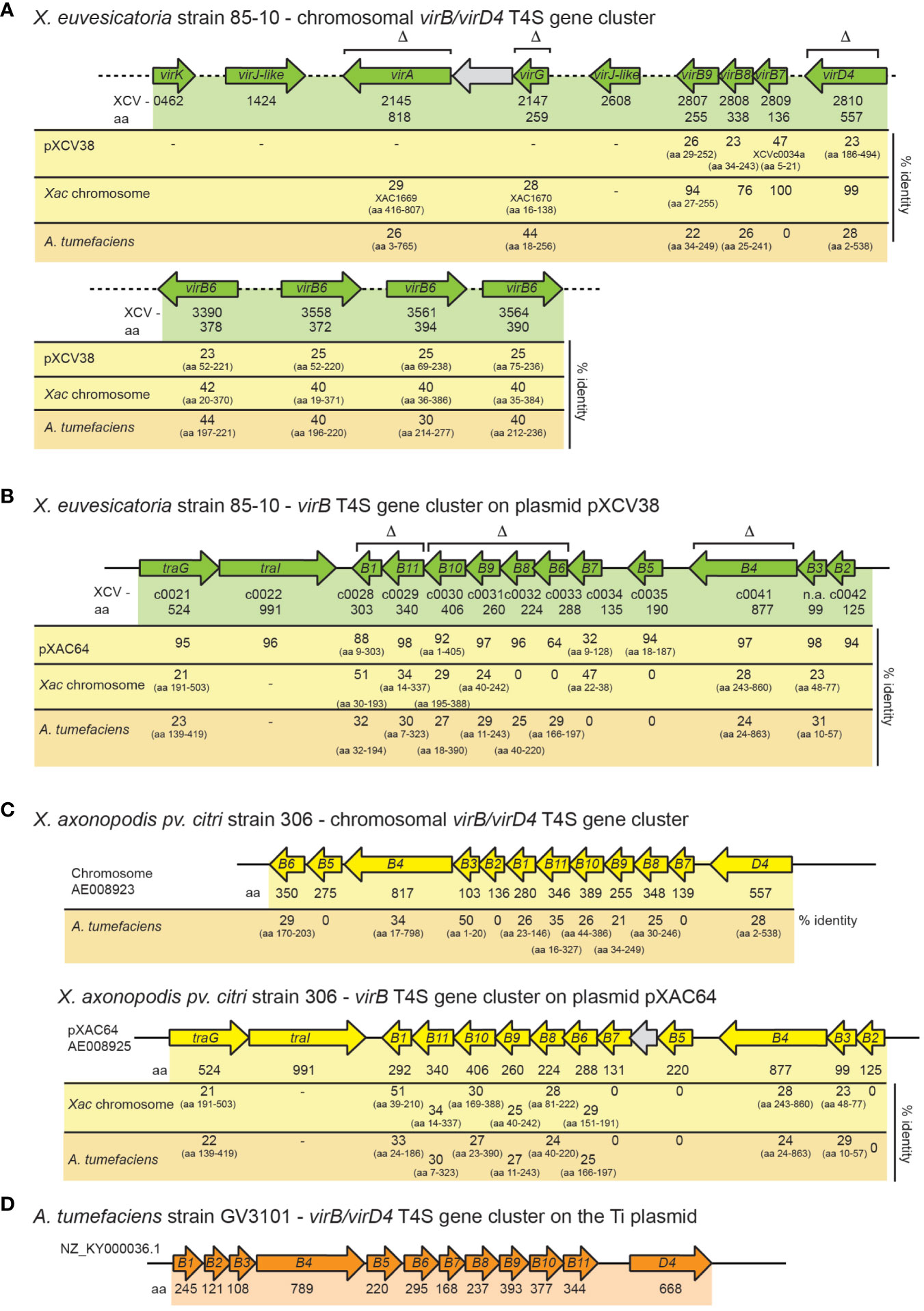
Figure 1 Genetic organization of vir gene clusters in X. euvesicatoria, X. axonopodis pv. citri and A. tumefaciens. (A) Organization of chromosomal vir genes from X. euvesicatoria strain 85-10. Genes are represented by arrows. Gene numbers, the length of the corresponding gene products in amino acids (aa) and the percentage of amino acid identity to corresponding proteins encoded on plasmid pXCV38 from X. euvesicatoria strain 85-10, on the chromosome of X. axonopodis pv. citri (Xac) strain 306 and of A. tumefaciens is indicated. The symbol Δ refers to genes which were deleted in the present study. Accession numbers and predicted functions of all proteins are summarized in Table S3. (B) Schematic representation of the vir gene cluster on plasmid pXCV38 from X. euvesicatoria strain 85-10. virB3 and virB7 were identified by bioinformatic analyses based on sequence homologies of the corresponding gene products with known VirB3 and VirB7 proteins. Gene numbers, the size of corresponding gene products and the amino acid identity to homologous proteins is indicated as described in (A). (C) Organization of the chromosomal and plasmid-localized vir gene clusters from X. axonopodis pv. citri strain 306. Gene numbers, the size of corresponding gene products and the amino acid identity to homologous proteins is indicated as described in (A). (D) Schematic representation of the vir gene cluster from A. tumefaciens. Numbers refer to the size of the corresponding gene products in amino acids.
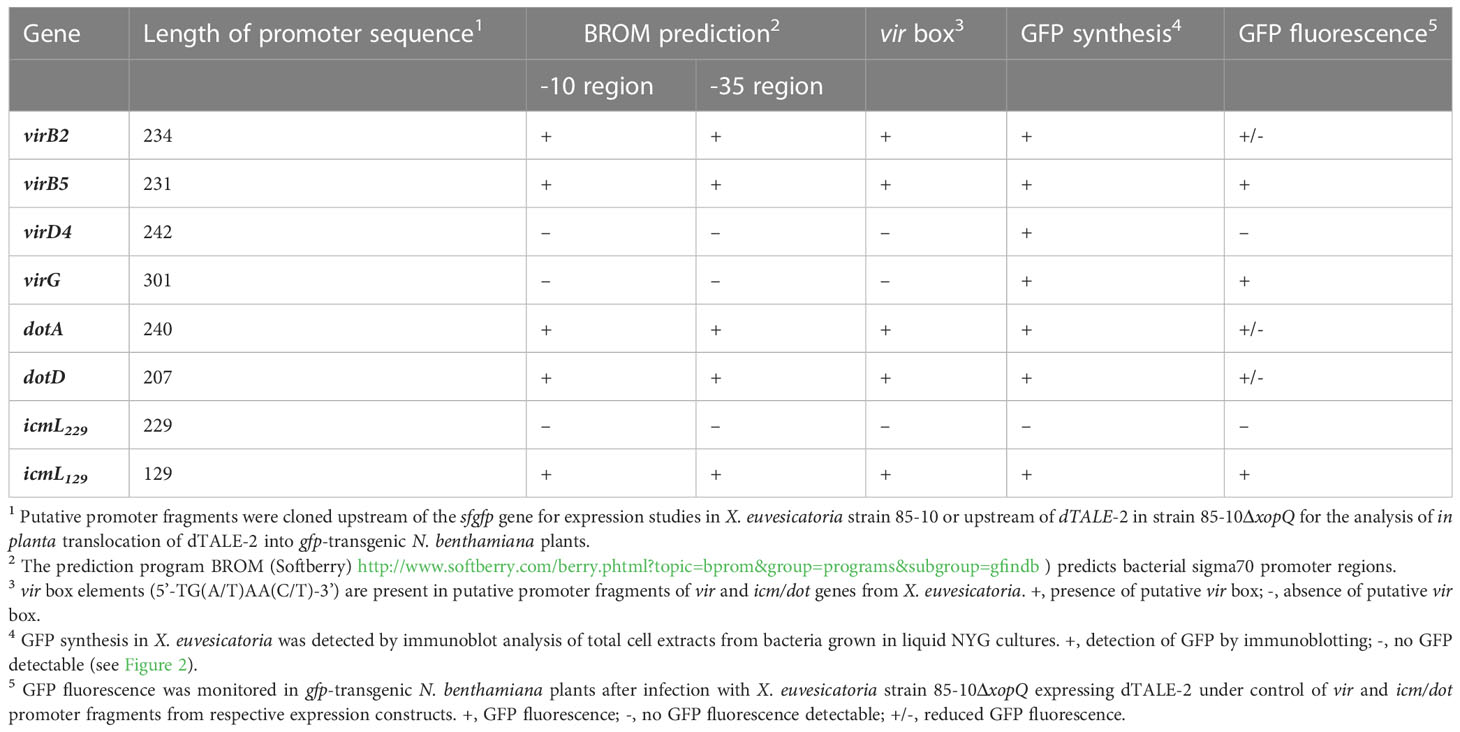
Table 1 Identification of promoters in upstream regions of vir and icm/dot genes from X. euvesicatoria strain 85-10.
In addition to chromosomal vir genes, X. euvesicatoria contains a second virB T4S gene cluster with virB1 – virB11 genes on plasmid pXCV38 (Figure 1B). The region adjacent to virB1 encodes predicted proteins with homology to TraI and TraG, which are involved in DNA transfer by T4S systems (Audette et al., 2007; Waksman, 2019). TraI belongs to the group of relaxases, which cut DNA prior to transfer at the origin of transfer (oriT) and remain attached to the DNA during T4S-dependent transport (Waksman, 2019; Costa et al., 2021). The plasmid-encoded Vir proteins from X. euvesicatoria share an overall amino acid identity between 32 – 98% with corresponding proteins encoded by the virB gene cluster located on plasmid pXAC64 of X. axonopodis pv. citri strain 306 (Figure 1). In contrast, the amino acid identity between chromosomally and plasmid-encoded Vir proteins is significantly lower in both X. euvesicatoria and X. axonopodis pv. citri and ranges between 20 – 50% (Figure 1). In both pathogens, VirD4 is encoded as a single copy gene on the chromosome.
Notably, X. euvesicatoria also contains a predicted icm/dot T4S gene cluster on plasmid pXCV183 (Thieme et al., 2005) (Figure 2A). The corresponding gene products are homologous to major structural components of Icm/Dot T4S systems and share between 22 – 46% amino acid identity with respective Icm/Dot proteins from L. pneumophila (Figure 2A, Table S5). The organization of the 16 putative icm/dot genes from X. euvesicatoria including icmL, K, E, G, C, D, J as well as dotD, C, B, O and L is similar to the order of the corresponding genes in L. pneumophila (Figure 2B). However, genes encoding predicted IcmF, H, N, M, Q, R, S, V and X proteins are missing in X. euvesicatoria.
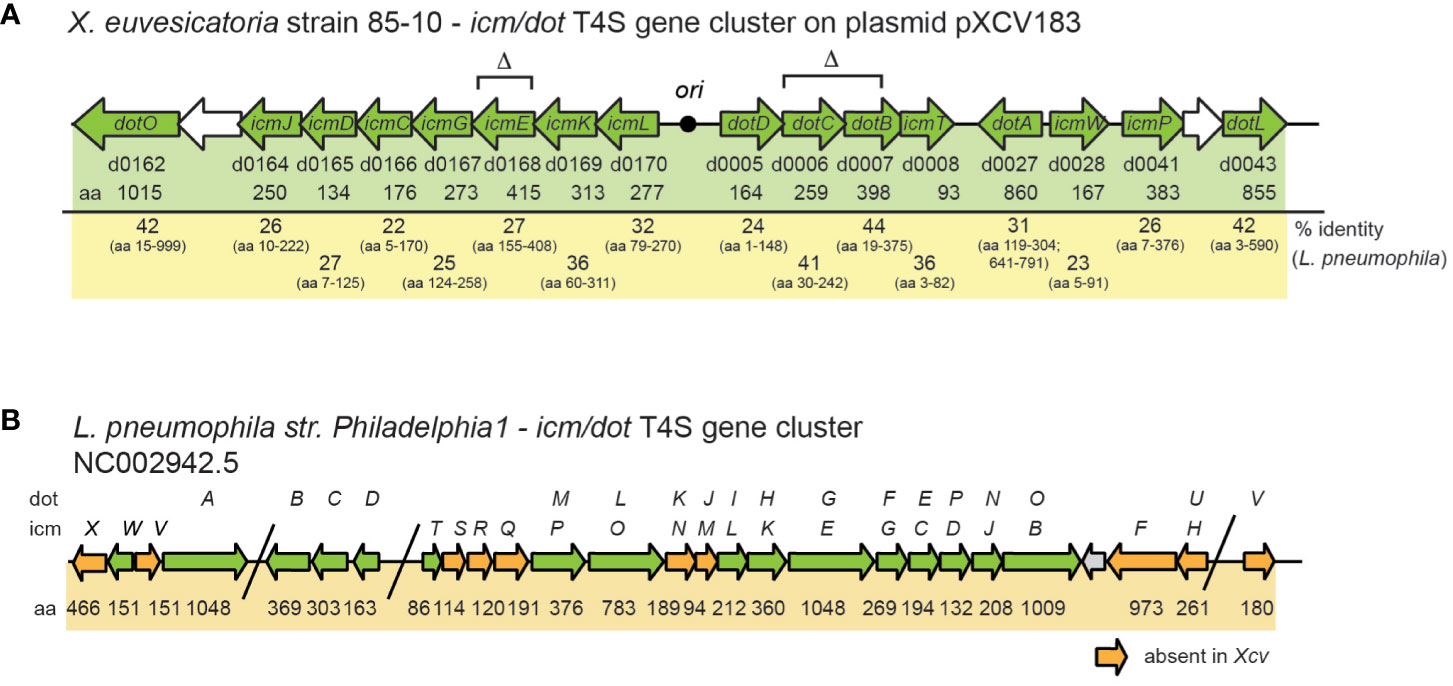
Figure 2 Schematic representation of icm/dot T4S gene clusters from X. euvesicatoria strain 85-10 and L. pneumophila strain Philadelphia 1. (A) icm/dot T4S gene cluster on plasmid pXCV183 from X. euvesicatoria strain 85-10. Genes, which are present in both L. pneumophila and X. euvesicatoria, are represented by green arrows. Gene numbers, the length of the gene products in amino acids (aa) and the amino acid sequence identity with corresponding proteins from L. pneumophila are indicated. The black circle refers to the origin of plasmid pXCV183. Δ refers to genes which were deleted in the present study. (B) Organization of the icm/dot T4S gene cluster from L. pneumophila strain Philadelphia 1. The nomenclature of genes as icm or dot genes is given above the arrows. Numbers refer to the size of the corresponding gene products in amino acids (aa). Orange arrows indicate genes, which are absent in X. euvesicatoria strain 85-10.
To investigate whether vir and icm/dot T4S genes from X. euvesicatoria are expressed, we generated reporter constructs containing sfgfp (superfolder green fluorescent protein) downstream of predicted promoters of virB2, virB5, virD4, virG, dotA, dotD and icmL (see above, Table 1). Constructs were transferred into X. euvesicatoria strains 85-10 and 85-10ΔvirG, which lacks the predicted regulatory gene virG. The lac promoter, which activates gene expression in X. euvesicatoria, served as positive control (Szczesny et al., 2010). As negative control, we cloned sfgfp downstream of the hrpB1 promoter, which is inactive in NYG medium (Wengelnik and Bonas, 1996). hrpB1 is a gene of the T3S gene cluster and is only activated in planta or when bacteria are cultivated in special minimal media (Wengelnik and Bonas, 1996). For the analysis of promoter activities, bacteria were cultivated in NYG medium and bacterial cell extracts were analyzed by immunoblotting using a GFP-specific antibody. As expected, sfGFP was detectable in cell extracts of cultures containing sfgfp under control of the lac but not of the hrpB1 promoter (Figure 3A). All tested vir promoters as well as the dotA and dotD promoters led to sfGFP synthesis. Similar sfGFP levels were obtained when constructs were analyzed in strains 85-10 and 85-10ΔvirG, suggesting that VirG is dispensable for the activation of gene expression (Figure 3A). Notably, the predicted promoter region spanning 229 bp upstream of the annotated start site of icmL did not lead to detectable sfgfp expression (Figures 3A, B).
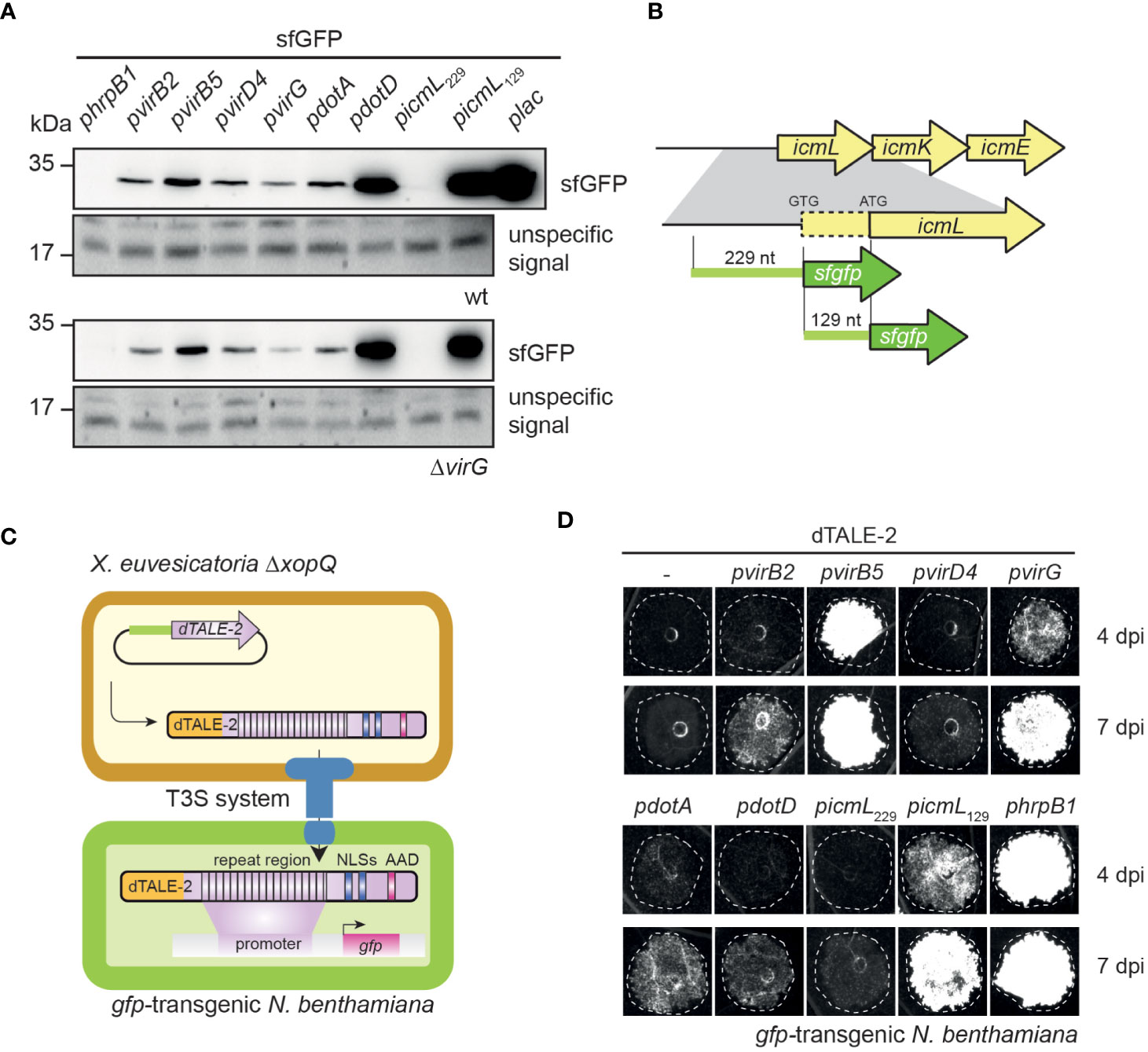
Figure 3 The analysis of promoter-reporter fusions reveals in vitro and in planta activities of vir and icm/dot promoters. (A) vir and icm/dot promoters activate expression of the sfgfp reporter gene in X. euvesicatoria. Strains 85-10 and 85-10ΔvirG containing sfgfp under control of predicted promoters of virB2, virB5, virD4, virG, dotA, dotD and icmL on corresponding expression constructs were grown in NYG medium, and total cell extracts were analyzed by immunoblotting using a GFP-specific antibody. Expression constructs containing sfgfp under control of the hrpB1 promoter, which is inactive in NYG medium, and the lac promoter, which is active in X. euvesciatoria, were used as negative and positive controls, respectively. The experiment was performed three times with similar results. Unspecific signals detected by the GFP-specific antibody are shown to demonstrate equal loading. Given the high expression level of gfp in strain 85-10 containing gfp under control of the plac promoter, only 0.5 µl of the protein extract were loaded. (B) Schematic representation of icmL and predicted promoter regions. icmL is located upstream of icmK and icmE in the icm/dot T4S gene cluster on plasmid pXCV183 from X. euvesicatoria strain 85-10. A putative ATG start codon is located 129 nucleotides (nt) downstream of the annotated GTG translation initiation start site. For the analysis of putative icmL promoter regions, 229 nt upstream of the annotated start site and 129 nt upstream of the putative ATG start codon, respectively, were fused to sfgfp as indicated. (C) Principle of the dTALE-2-based in vivo expression and translocation assay. Expression constructs encoding dTALE-2 downstream of the analyzed vir or icm/dot promoter regions were introduced into X. euvesicatoria strain 85-10ΔxopQ which is pathogenic on N. benthamiana plants (Adlung et al., 2016). For translocation assays, bacteria were infiltrated into gfp-transgenic N. benthamiana plants which contain a viral vector construct encoding GFP under control of a dTALE-2-responsive promoter. Expression of dTALE-2 leads to its translocation by the T3S system because dTALE-2 contains an N-terminal secretion and translocation signal (depicted in orange). The central repeat region binds to specific bases in the dTALE-2-responsive promoter. C-terminal nuclear localization signals (NLSs) and an acidic activation domain (AAD) are required for nuclear localization and activation of in planta gene expression, respectively. Activation of gfp expression by dTALE-2 leads to GFP fluorescence in the infected leaf areas. (D) In vivo reporter assay for the detection of dTALE-2 translocation. X. euvesicatoria strain 85-10ΔxopQ with expression constructs containing dTALE-2 under control of predicted vir and icm/dot promoters as indicated was infiltrated into leaves of gfp-transgenic N. benthamiana plants containing gfp on a viral vector construct under control of a dTALE-2-responsive promoter. In planta activation of vir and icm/dot promoters allowed the synthesis and translocation of dTALE-2 via the T3S system, thus inducing GFP fluorescence which was monitored 4 and 7 dpi using a chemiluminescence/fluorescence imager which displays GFP fluorescence in black and white. The experiment was performed three times with similar results.
In addition to the in vitro studies, we analyzed a potential in planta activity of vir and icm/dot promoters. For this, we generated expression constructs containing vir and icm/dot promoters upstream of an artificial TAL (transcription activator-like) effector gene (dTALE-2, designer TAL effector-2). dTALE-2 is a derivative of the type III effector protein AvrBs3 from X. euvesicatoria which is imported into the plant cell nucleus where it acts as a transcription factor and activates plant gene expression (Boch et al., 2014). TAL effectors bind to defined nucleotide sequences in the promoters of target genes via their central region, which consists of a variable number of amino acid repeats and allows the base-specific binding to DNA (Boch et al., 2009; Moscou and Bogdanove, 2009). dTALE-2 is used as a reporter for T3S-dependent protein translocation because it binds to a dTALE-2-responsive promoter which controls the expression of gfp on a stably integrated viral vector construct in transgenic Nicotiana benthamiana plants (Werner et al., 2011; Scheibner et al., 2016). vir and icm/dot promoter-driven expression in X. euvesicatoria, which leads to the translocation of dTALE-2 by the T3S system, therefore activates gfp expression in transgenic N. benthamiana plants (Figures 3C, D). For these assays, expression constructs encoding dTALE-2 under control of vir or icm/dot promoters were transferred into X. euvesicatoria strain 85-10ΔxopQ which lacks the T3E XopQ and induces disease symptoms in N. benthamiana plants (Adlung et al., 2016). Inoculation of gfp-transgenic N. benthamiana plants with xopQ deletion mutants containing dTALE-2 expression constructs led to GFP fluorescence 4 dpi when dTALE-2 was expressed under control of the virB5, virG or hrpB1 promoter (Figure 3D). Expression of dTALE-2 under control of the virB2, dotA and dotD promoters led to weaker fluorescence which was detectable 7 dpi (Figure 3D). No GFP fluorescence was observed when dTALE-2 was expressed under control of the virD4 or the icmL promoter (Figure 3D). The lack of detectable GFP fluorescence after virD4 promoter-driven expression of dTALE-2 is in contrast to the in vitro activity of the virD4 promoter and suggests that virD4 is not or only weakly expressed in planta (Figures 3A, D). In case of the predicted icmL promoter, we assume that the region 229 nucleotides upstream of the annotated GTG start codon of icmL does not activate in vitro or in planta reporter gene expression. Interestingly, DNA sequence analyses revealed the presence of an additional putative ATG translation initiation codon 129 nucleotides downstream of the predicted GTG start codon of icmL (Figure 3B). To investigate whether the icmL promoter is located between the annotated GTG and the alternative ATG start codon, we generated an additional expression construct containing dTALE-2 under control of a 129-bp DNA fragment upstream of the ATG codon. This led to detectable in vitro and in planta reporter gene expression, indicative of a promoter downstream of the annotated GTG translation start site (Figures 3A, D). In agreement with this finding, a promoter and a vir box was predicted in the region spanning 129 bp downstream of the annotated GTG start codon (Table 1 and Table S4).
To analyze a potential virulence function of T4S genes in X. euvesicatoria strain 85-10, we deleted single and multiple vir and icm/dot genes encoding predicted essential components of the T4S systems such as ATPases (VirD4, VirB4, VirB11 and DotB), the predicted structural components VirB6, VirB8, VirB9, VirB10, DotC and IcmE, and the putative regulators VirA and VirG (Figures 1, 2). For infection experiments, the wild-type X. euvesicatoria strain 85-10 and corresponding deletion mutants were infiltrated into leaves of susceptible ECW pepper plants and disease symptoms were inspected over a period of five to eight days. As expected, the wild-type strain 85-10 induced disease symptoms in form of water-soaked lesions that became necrotic five to seven dpi (Figure 4A). No reduction in symptom formation was detectable after infiltration of mutant strains deleted in single or multiple vir or icm/dot genes (Figure 4A). To investigate a potential redundant virulence function of both T4S systems, we also combined deletions in vir and icm/dot genes, thus generating strains 85-10ΔvirB4ΔicmE, 85-10ΔdotCBΔicmEΔvirB4 and 85-10ΔvirB6-10ΔicmEΔdotCB. However, no significant reduction in disease symptoms was observed (Figure 4A).
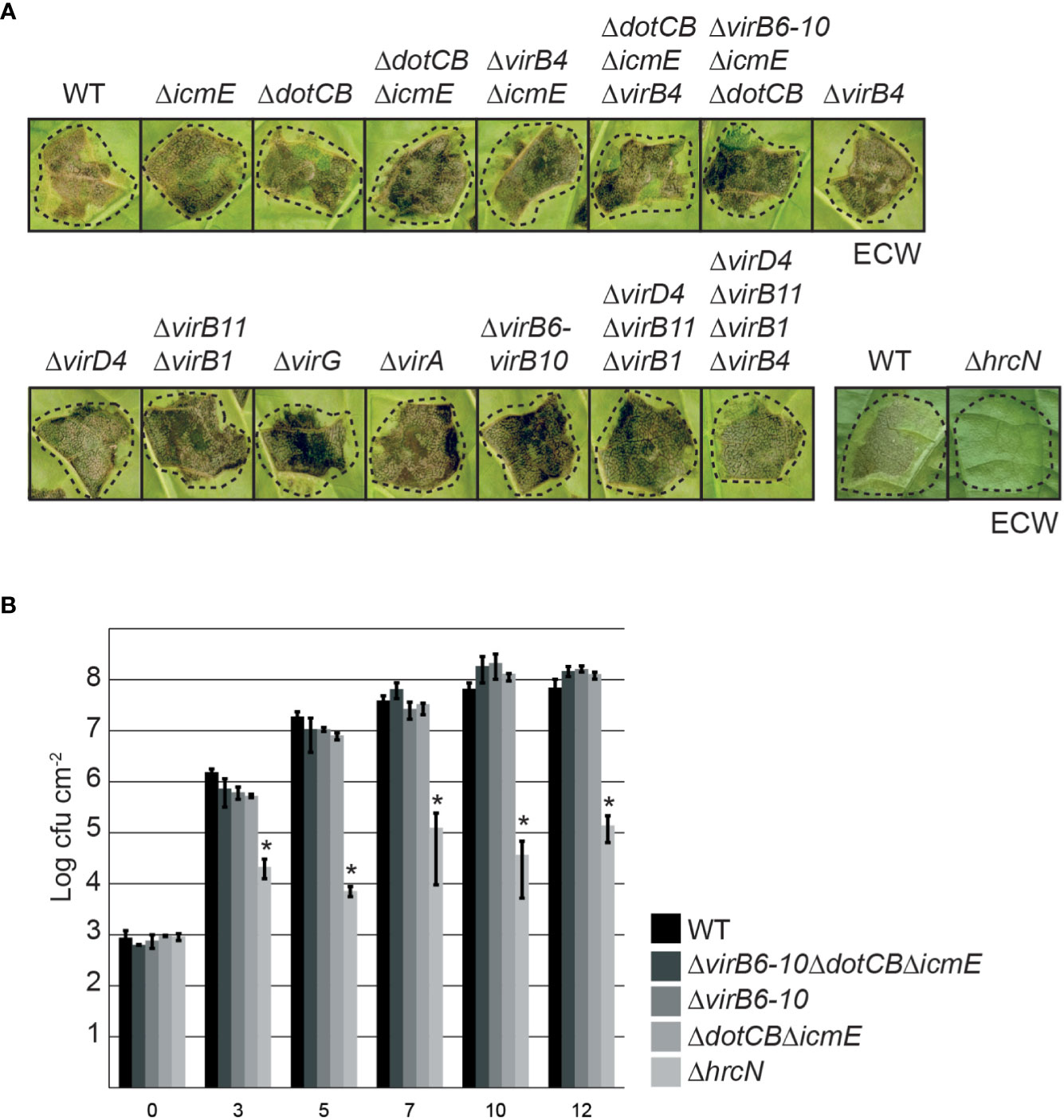
Figure 4 The putative Vir- and Icm/Dot-like T4S systems do not significantly contribute to virulence of X. euvesicatoria. (A) Infection studies with X. euvesicatoria wild-type, hrcN and T4S mutant derivatives. Strain 85-10 (wt) and deletion mutant derivatives thereof lacking the T3S ATPase gene hrcN or single or multiple vir and icm/dot genes as indicated were infiltrated at a density of 108 cfu ml-1 into leaves of susceptible ECW pepper plants. Disease symptoms were photographed 7 dpi. Dashed lines indicate the inoculated leaf areas. The experiment was performed three times with similar results. (B) In planta growth of wild-type and T4S mutant derivatives of X. euvesicatoria. Strain 85-10 (wt) and deletion mutant derivatives thereof lacking single or multiple vir and icm/dot genes as indicated were infiltrated into leaves of three susceptible ECW pepper plants. Bacterial in planta growth was monitored over a period of 12 days. The T3S-deficient and non-pathogenic strain 85-10ΔhrcN, which lacks the ATPase of the T3S system, was analyzed as control. Mean values of cfu/cm2 and standard deviations from one experiment are depicted in the diagram. The experiment was performed three times with similar results. According to a one-tailed paired t-test, the difference in growth of X. euvesicatoria wt and hrcN deletion mutant strains is significant (with a p value of 0.024; indicated by asterisks). Growth of T4S mutants is not significantly different from that of the wild-type strain (p values of 0.075 for strain 85-10ΔvirB6-10ΔdotCBΔicmE, 0.121 for strain 85-10ΔvirB6-10 and 0.134 for strain 85-10ΔdotCBΔicmE).
In addition to phenotypic analyses, we performed in planta growth curves with strains 85-10, 85-10ΔvirB6-10, 85-10ΔdotCBΔicmE and 85-10ΔvirB6-10ΔdotCBΔicmE. For this, bacteria were infiltrated into leaves of susceptible ECW pepper plants, and bacterial growth was analyzed over a period of twelve days. As control, we included the non-pathogenic strain 85-10ΔhrcN which lacks the ATPase HrcN of the T3S system (Lorenz and Büttner, 2009). The wild-type strain 85-10 multiplied up to 108 cfu ml-1 whereas a significant reduction in bacterial growth was observed for strain 85-10ΔhrcN as expected (Figure 4B). T4S mutant strains displayed a wild-type growth rate, suggesting that vir and icm/dot genes do not significantly contribute to bacterial multiplication in planta (Figure 4B).
The chromosomally encoded VirB/VirD4 T4S system from X. axonopodis pv. citri and the corresponding T4S system from the related pathogen S. maltophilia were previously shown to deliver toxins into Gram-negative bacteria such as E. coli, thus resulting in cell lysis (Souza et al., 2015; Bayer-Santos et al., 2019). To investigate a similar toxic effect of X. euvesicatoria on E. coli, we incubated X. euvesicatoria strain 85-10 and deletion mutants thereof lacking single or multiple vir or icm/dot genes in the presence of decreasing concentrations of E. coli strain BL21. Co-cultures were spotted on LB agar plates containing X-Gal which is a chromogenic artificial substrate of the β-galactosidase from E. coli and is converted into a blue pigment. The formation of blue colonies by E. coli indicates bacterial multiplication and was unaffected upon cocultivation with X. euvesicatoria (Figure 5A).
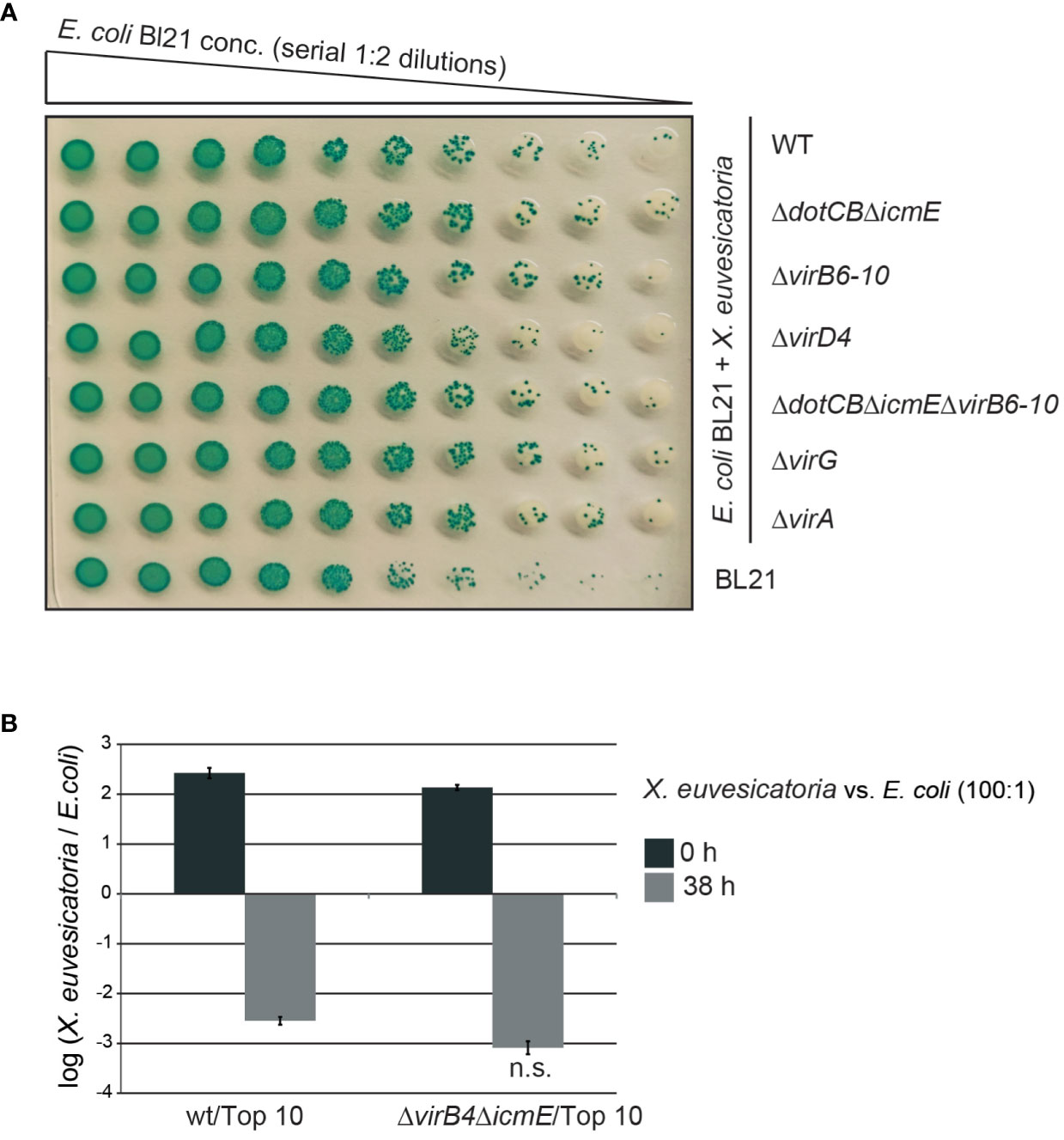
Figure 5 In vitro growth of E. coli is unaffected by X. euvesicatoria. (A) Cocultivation with X. euvesicatoria strains does not affect in vitro growth of E. coli. Serial dilutions of E. coli strain BL21, which naturally expresses the β-galactosidase, were incubated with X. euvesicatoria strain 85-10 (wt) or deletion mutant derivatives thereof lacking single or multiple vir and icm/dot genes as indicated. X. euvesicatoria strains were incubated at initial concentrations of 5 × 108 CFU/ml with E. coli BL21 at a concentration of 5 × 106 CFU/ml or serial 1:2 dilutions on LB agar plates containing X-gal and IPTG overnight. One representative image is shown. The experiment was performed three times with similar results. (B) Ratio of X. euvesicatoria and E. coli cells after cocultivation. X. euvesicatoria strain 85-10 (wt) or 85-10ΔicmEΔvirB4 and E. coli strain Top 10 were mixed at a ratio of 100: 1, spotted on an LB agar plate and serial dilutions were plated after 0 and 38 hours on NYG medium with rifampicin for X. euvesicatoria strains and on LB medium with streptomycin for growth of E. coli to count bacterial colonies. Mean values and standard deviations of the ratio of X. euvesicatoria and E. coli colonies from one representative experiment are shown. According to a t-test, the difference in growth of E. coli in the presence of the X. euvesicatoria wt or the T4S mutant is not significant (n.s.; p value of 0.2487). The experiment was performed three times with similar results.
In additional experiments, we incubated X. euvesicatoria wild-type and T4S mutant strains with E. coli strain Top10 in a ratio of 100:1 on solid LB medium and plated the bacteria 0 and 38 hours later for quantitative analysis. The X. euvesicatoria/E. coli ratio decreased 38 h after incubation, indicating a faster growth rate of E. coli compared to X. euvesicatoria (Figure 5B). However, no significant difference with X. euvesicatoria wild-type and T4S mutant strains was observed, suggesting that the T4S systems from X. euvesicatoria do not exert a negative effect on the growth of E. coli upon cocultivation under the tested conditions (Figure 5B). This is in contrast to the VirB/VirD4 T4S system from X. axonopodis pv. citri which exerts a toxic effect on E. coli (Souza et al., 2015).
The association of traI and traG genes with the plasmid-localized vir gene cluster from X. euvesicatoria prompted us to investigate the role of the predicted VirB/VirD4 T4S system in plasmid transfer. It was previously reported that X. euvesicatoria and X. citri pv. citri transfer plasmids and chromosomal DNA into other Xanthomonas strains (Basim et al., 1999; El Yacoubi et al., 2007). In the present study, we analyzed the transfer of a derivative of plasmid pXCV38 (hereafter referred to as pXCV3882-8::specR) which originates from strain 82-8 and contains a spectinomycin resistance gene inserted into the avrBs3 T3E gene (Bonas et al., 1989). Donor and recipient strains with plasmid pXCV3882-8::specR were, therefore, selected on medium containing spectinomycin. For conjugation experiments, strain 82-8 with plasmid pXCV3882-8::specR was incubated with a derivative of strain 85-10 containing a gentamicin resistance gene inserted into the genome in the hpaFG region adjacent to the hrp gene cluster. The hpaFG region is dispensable for virulence and serves as a landing platform for gene insertion (Lorenz et al., 2012). Cocultivation of strains 82-8 pXCV3882-8::specR and 85-10::gentR resulted in transconjugants which were gentamicin- and spectinomycin-resistant, suggesting that pXCV3882-8::specR had been transferred from strain 82-8 into strain 85-10::gentR (Figure 6A and Table S6). The presence of pXCV3882-8::specR in the recipient strains was confirmed by PCR amplification of the 5’ region of avrBs3 and by agarose gel electrophoresis of isolated plasmids (Figures 6B and C). pXCV3882-8::specR from strain 82-8, which has not yet been sequenced, is larger than pXCV38 from strain 85-10. Both plasmids could, therefore, be separated by agarose gel electrophoresis and were present in the resulting transconjugants (Figure 6C). Strain 82-8 thus likely contains a functional T4S system which acts as a conjugation machine and mediates transfer of plasmid pXCV3882-8::specR. To investigate whether strain 82-8 contains vir genes, we performed PCR analysis using primers specific for vir genes from strain 85-10. Notably, we did not amplify virB4, virB10 and virB11 from strain 82-8, suggesting that the primers did not efficiently bind to the template DNA (Figure 6D). In contrast, the chromosomal virD4 gene was amplified from strains 85-10 and 82-8 (Figure 6D). We assume that strain 82-8 contains a functional T4S system because it mediates transfer of plasmid pXCV3882-8::specR (see above).
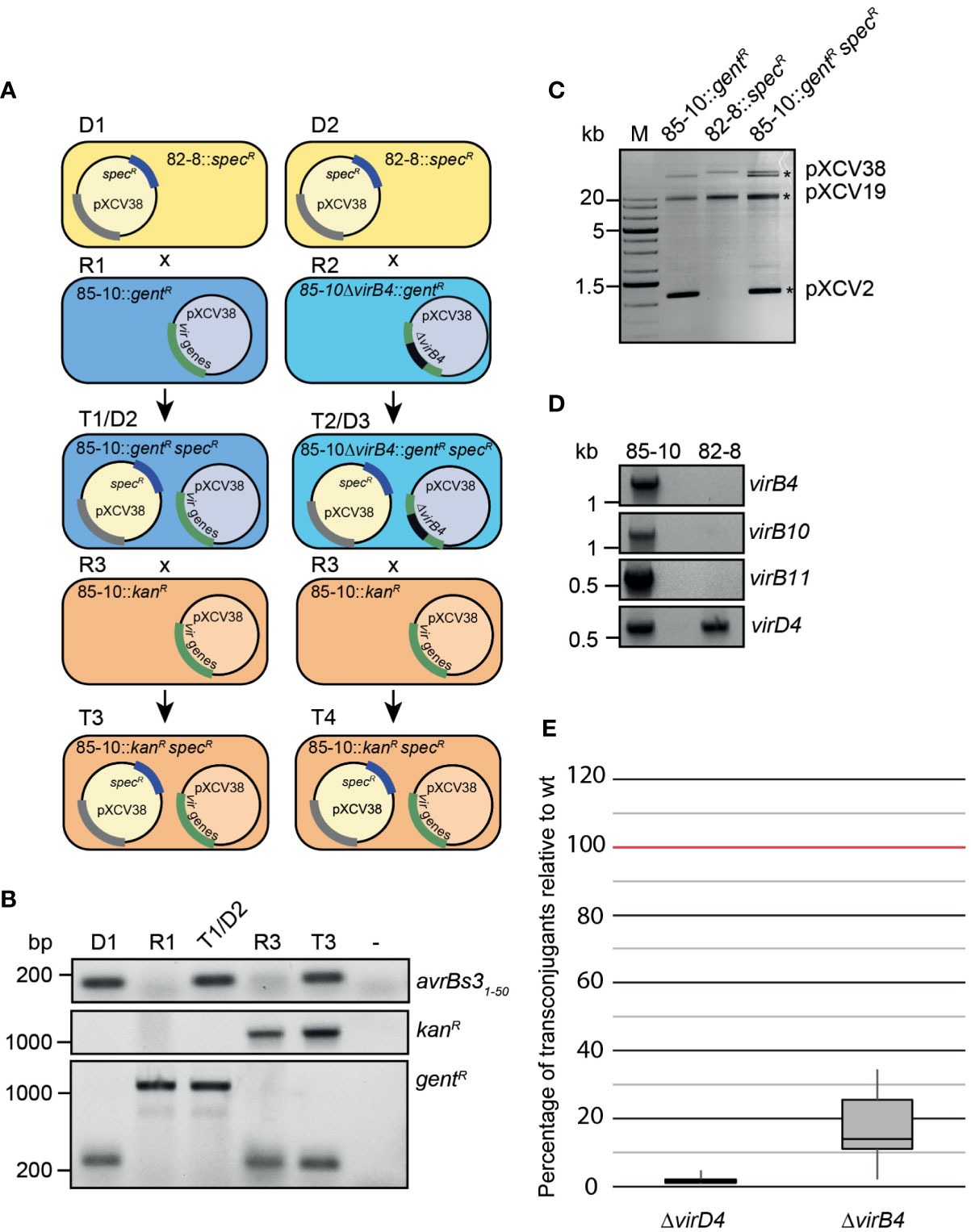
Figure 6 Plasmid transfer by X. euvesicatoria strain 85-10 is controlled by VirD4. (A) Schematic overview on conjugation experiments with X. euvesicatoria strains. Plasmid pXCV38 from strain 82-8 (donor 1, D1) containing a spectinomycin resistance gene (pXCV3882-8::specR) was transferred by conjugation to strain 85-10::gentR (recipient 1, R1) which contains the vir gene cluster on plasmid pXCV38. A grey bar indicates that the presence of a similar cluster on plasmid pXCV3882-8::specR is still unknown. The resulting transconjugant T1 (referred to as 85-10::gentR specR) was gentamicin- and spectinomycin-resistant and contained both plasmids. The numbers of transconjugants from three independent experiments resulting from the transfer of plasmid pXCV3882-8::specR is given in Table S6. Similarly, pXCV3882-8::specR was transferred to a derivative of strain 85-10::gentR with a deletion in virB4 (85-10ΔvirB4::gentR; recipient 2, R2), resulting in transconjugants T2. T1 and T2 served as donor strains (D2 and D3) in a second conjugation experiment using strain 85-10::kanR, which contains a kanamycin resistance gene, as recipient (R3). The resulting transconjugants (T3 and T4) grew on medium with kanamycin and spectinomycin and contained plasmid pXCV3882-8::specR. In addition to the virB4 deletion mutant, we used virD4 mutant strains for the conjugation experiments (see panel E). virD4 is located on the chromosome and absent from plasmid pXCV38. (B) Confirmation of plasmid transfer by PCR analysis of transconjugants. The first 50 codons of avrBs3, the kanamycin and the gentamicin resistance gene were amplified by PCR using donor, recipient and transconjugant strains as depicted in (A) as templates. Amplicons were analyzed by agarose gel electrophoresis and visualized by an UV imager. Primers used to amplify the gentamicin resistance gene annealed to the chromosomal hpaFG region, thus leading to a smaller PCR product in the absence of the gentamicin resistance gene. (C) Analysis of isolated plasmids from X. euvesicatoria by agarose gel electrophoresis. Plasmids were isolated from X. euvesicatoria strains 85-10::gentR, 82-8 pXCV3882-8::specR (82-8::specR) and 85-10::gentR pXCV3882-8::specR (85-10::gentR specR) and separated by agarose gel electrophoresis. Asterisks indicate the expected sizes of plasmids pXCV2 (1852 bp), pXCV19 (19146 bp) and pXCV38 (38116 bp) from X. euvesicatoria strain 85-10. Plasmid pXCV183 (182572 bp), which is also present in strain 85-10, presumably did not migrate into the agarose gel. (D) PCR amplification of vir genes from X. euvesicatoria strains 85-10 and 82-8. vir genes were amplified by PCR using specific primers for virB4, virB10, virB11 and virD4 from strain 85-10 and genomic DNA of strains 85-10 and 82-8 as template. PCR products were analyzed by agarose gel electrophoresis and visualized by an UV imager. (E) VirD4 and VirB4 are required for efficient plasmid transfer. Donor and recipient were mixed at a ratio of 1:1 and incubated overnight on a cellulose nitrate filter placed on solid NYG medium and covered with 1% water agar. Serial dilutions were plated on selective plates to count the transconjugants. The highest numbers of transconjugants were obtained when the T4S wild-type strain 85-10::gentR containing pXCV3882-8::specR was used as donor and were set to 100%. Reduced numbers of transconjugants obtained with strains 85-10ΔvirD4 and 85-10ΔvirB4 both containing pXCV3882-8::specR are depicted in a boxplot diagram. Values and standard deviations represent the percentage of transconjugants from six experiments with strain 85-10ΔvirD4 and from ten experiments with strain 85-10ΔvirB4.
To investigate a possible contribution of the T4S systems to plasmid transfer in strain 85-10, pXCV3882-8::specR was transferred to derivatives of strain 85-10::gentR deleted in virD4 or virB4 (Figure 6A). The resulting T4S wild-type and mutant derivatives of strain 85-10::gentR containing pXCV3882-8::specR served as donor strains in additional conjugation experiments, using a kanamycin-resistant derivative of strain 85-10 (85-10::kanR) as recipient (Figure 6A). Transconjugants grew on medium containing kanamycin and spectinomycin but were sensitive to gentamicin, indicating a transfer of pXCV3882-8::specR from strain 85-10::gentR to strain 85-10::kanR (data not shown). PCR analyses confirmed the presence of the kanamycin resistance gene and the 5’ region of avrBs3 in the transconjugants (Figure 6B). pXCV3882-8::specR was most efficiently transferred by strain 85-10::gentR, whereas significantly less transformants were obtained when strain 85-10ΔvirD4::gentR was used as donor strain (Figure 6E). Residual plasmid transfer was observed with strain 85-10ΔvirB4::gentR, resulting in approximately 20% of the transconjugants obtained with the wild-type strain (Figure 6E). It is possible that the loss of VirB4 was partially compensated by functionally redundant proteins encoded in the genome of the donor strain or on plasmid pXCV3882-8::specR, which was used to analyze the plasmid transfer. However, given the severe reduction in plasmid transfer by the virD4 mutant, we assume that the VirB/VirD4 T4S system acts as a conjugation machine and mediates horizontal gene transfer.
Next, we investigated whether VirB/VirD4 and Icm/Dot proteins from X. euvesicatoria assemble into oligomeric complexes as would be expected from their predicted functions as components of T4S systems. For protein-protein interaction studies, we used the bacterial adenylate cyclase-based two-hybrid (BACTH) system which detects the interaction of proteins fused to the T18 or the T25 subdomain of the catalytic region of the adenylate cyclase (Cya). Interaction of T18 and T25 fusions leads to the reconstitution of Cya and thus to the production of cAMP. This activates the expression of the lac operon in E. coli reporter strains lacking the native cya gene (Karimova et al., 1998; Battesti and Bouveret, 2012). LacZ activity results in blue colonies, indicative of an interaction between both fusion proteins.
We performed BACTH assays with T18 and T25 fusions of the predicted ATPases VirD4, VirB4 and VirB11 as well as of the predicted core components of the VirB/VirD4 T4S system VirB7 and VirB10 (Figure 7A). Corresponding expression constructs were cotransformed into the E. coli BTH101 reporter strain and transformants were grown on plates containing X-Gal. We detected homo- and heterooligomerizations of the putative ATPases VirD4 and VirB11 as well as interactions between all tested putative ATPases with VirB10 (Figure 7B and Table 2). Furthermore, VirB7 interacted with itself and with VirB10 (Figure 7B and Table 2). The interactions were specific because the analyzed T4S system components did not interact with the T25 and T18 domains alone (Figure 7B). Immunoblot analysis of bacterial cell extracts showed that most tested T25 and T18 fusion proteins were stably synthesized (Figure S1). T25-VirB4 was not detectable and was, therefore, not included in the analyses.
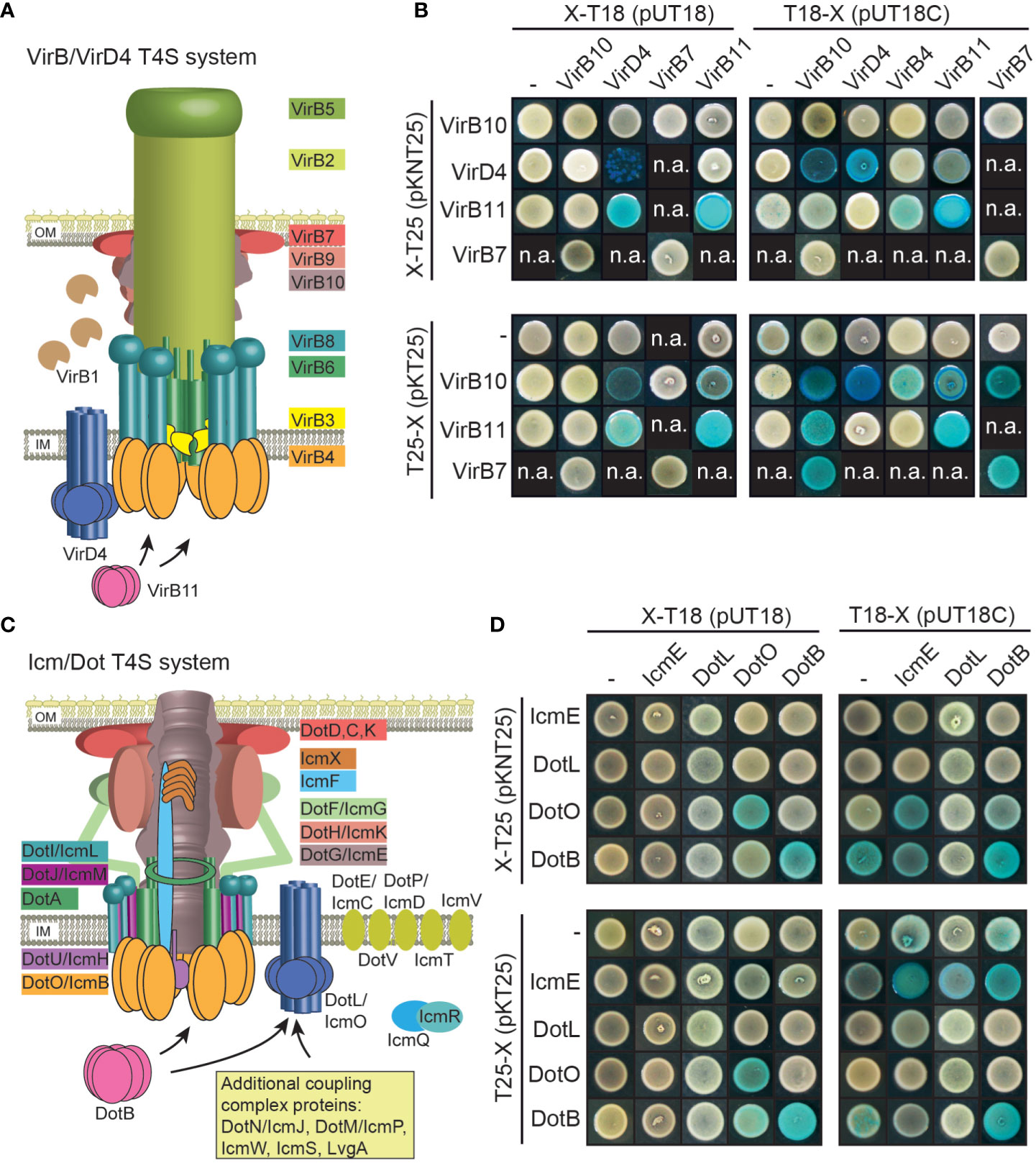
Figure 7 Vir and Icm/Dot proteins from X. euvesicatoria assemble into oligomeric complexes. (A) Schematic representation of the VirB/VirD4 T4S system. The model was adapted according to Mace et al., 2022. IM, inner membrane; OM, outer membrane. (B) Results of BACTH assays with Vir proteins from X. euvesicatoria. T18 and T25 fusions of the predicted structural components VirB7 and VirB10 as well as of the putative ATPases VirD4, VirB4 and VirB11 were analyzed in E. coli BTH101 cells as indicated and bacterial cultures were grown on indicator plates containing X-gal and IPTG. As control, fusion proteins were tested against the T18 or T25 domain alone as indicated. All proteins were stably synthesized as was shown by immunoblot analysis of bacterial cell extracts, using a FLAG epitope-specific antibody (see Figure S1). n.a., not analyzed. Interaction studies were performed at least three times with similar results. One representative colony is shown. Results are summarized in Table 2. (C) Schematic representation of the Icm/Dot-type T4S system. The model was adapted according to previous publications (Christie et al., 2017; Ghosal et al., 2019; Gomez-Valero et al., 2019; Sheedlo et al., 2022). IM, inner membrane; OM, outer membrane. (D) BACTH assays with Icm/Dot proteins from X. euvesicatoria. T18 and T25 fusions of the predicted structural component IcmE and the putative ATPases DotL, DotO and DotB were analyzed as described in (B). All proteins were stably synthesized as was shown by immunoblot analysis of bacterial cell extracts, using a FLAG epitope-specific antibody (see Figure S1). n.a., not analyzed. Interaction studies were performed at least three times with similar results. Results are summarized in Table 2.
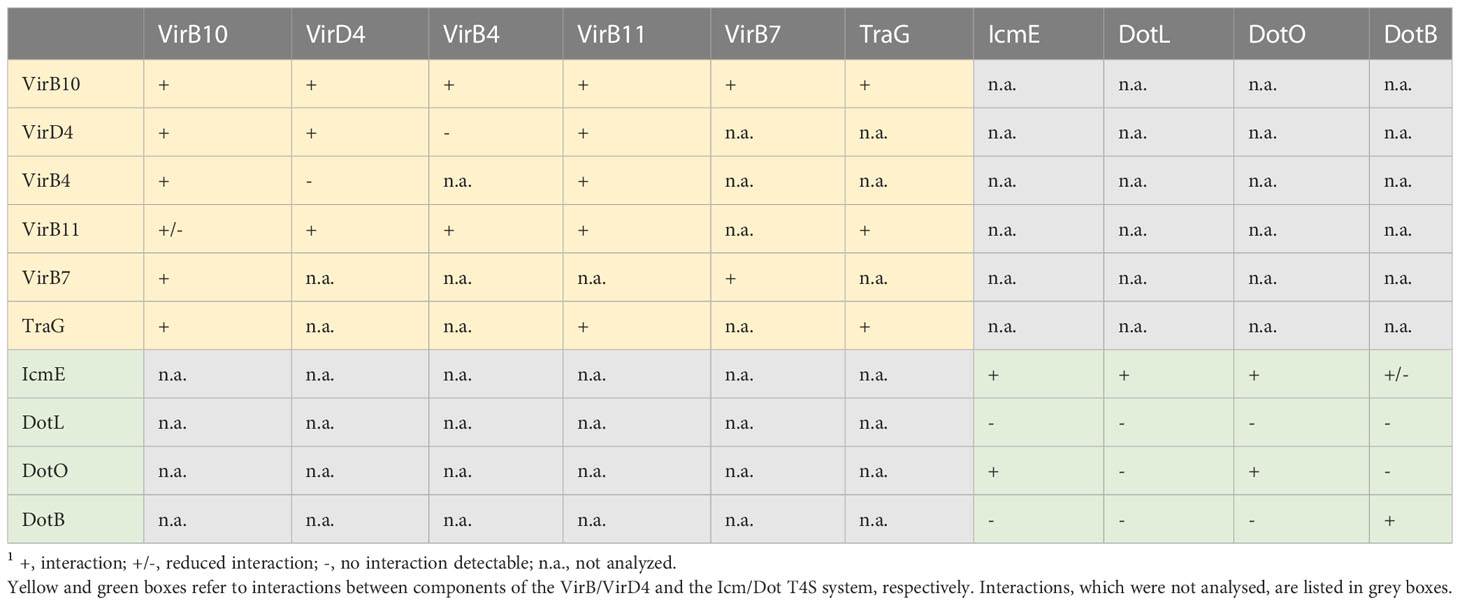
Table 2 Summary of the results of BACTH assays with components of VirB/VirD4 and Icm/Dot T4S substrates.1.
In addition to components of the predicted VirB/VirD4 T4S system, we performed BACTH assays with Icm/Dot proteins including the predicted ATPases DotO and DotB, which correspond to VirB4 and VirB11, respectively (Sheedlo et al., 2022) (Figure 7C). We also performed BACTH assays with the predicted coupling protein DotL as well as the putative major structural component IcmE which is also referred to as DotG and corresponds to VirB10. When analyzed as T25 and T18 fusions, DotO and DotB interacted with themselves as is expected for hexameric ATPases (Figure 7D and Table 2). Furthermore, a weak interaction between T18-IcmE and DotO-T25 was detected and IcmE interacted with itself (Figure 7D and Table 2). No self-interaction was observed for the predicted coupling protein DotL (Figure 7D). Yet, it cannot be excluded that N- or C-terminal T25 or T18 fusion partners interfere with the self-interaction of DotL. All tested fusion proteins were synthesized as was revealed by immunoblot analysis (Figure S1). Taken together, we conclude from the results of the BACTH assays that the predicted ATPases of both T4S systems self-interact as expected. Furthermore, VirD4, VirB4 and VirB11 interact with VirB10, which is the putative major structural component of the VirB/VirD4 T4S system and also associates with VirB7 (Table 2). Our results thus suggest that Vir and Icm/Dot proteins are involved in the assembly of heterooligomeric protein complexes, likely corresponding to T4S systems.
To identify possible substrates of the VirB/VirD4 T4S system from X. euvesicatoria, we searched for homologs of known type IV effectors from X. axonopodis pv. citri which are referred to as X-Tfes and were previously identified due to their interaction with the T4CP VirD4 (Alegria et al., 2005). In the present study, we focused on the X-Tfe homologs XCV0332, XCV1120 and XCV3571, which contain C-terminal motifs of XVIPCDs such as blocks of conserved amino acid sequences (GLxRIDHV and FAVQ GxxDPAHxRAHV; x refers to any residue) and a glutamine-rich C-terminal tail (Alegria et al., 2005) (Tables 3 and S7). Given that the XVIPCD provides the binding site for the AAD of VirD4 from X. axonopodis pv. citri (Alegria et al., 2005; Oka et al., 2022), we investigated possible interactions of the identified T4S candidate substrates with VirD4 from X. euvesicatoria. BACTH assays revealed interactions of T25 fusions of XCV3751, XCV1120 and XCV0332 with T18 fusions of VirD4 (Figure 8A and Table 2). The interactions were specific for VirD4, because the analyzed VirD4 interaction partners did not interact with the T18 domain alone (Figure 8A). Immunoblot analyses showed that most fusion proteins were stably synthesized (Figure S1). Exceptions were XCV0332-T25 and XCV3751-T25, which were, therefore, not included in the analyses (Figure S1). It was previously reported that the N-terminal region of the XVIPCD from X-Tfes adopts an ααβββ fold which interacts with the AAD of VirD4 (Oka et al., 2022). Interestingly, in silico modelling using the AlphaFold 2 algorithm (Jumper et al., 2021) revealed a similar fold of XVIPCDs in XCV0332, XCV1120 and XCV3751 which might associate with the AAD of VirD4 from X. euvesicatoria (Figures 8B-D). As additional candidate T4S substrate, we analyzed TraI which is a predicted relaxase and likely secreted by the VirB/VirD4 T4S system. TraI lacks an XVIPCD but also interacted with VirD4 when analyzed as T25 fusion (Figure 8A and Table 2).
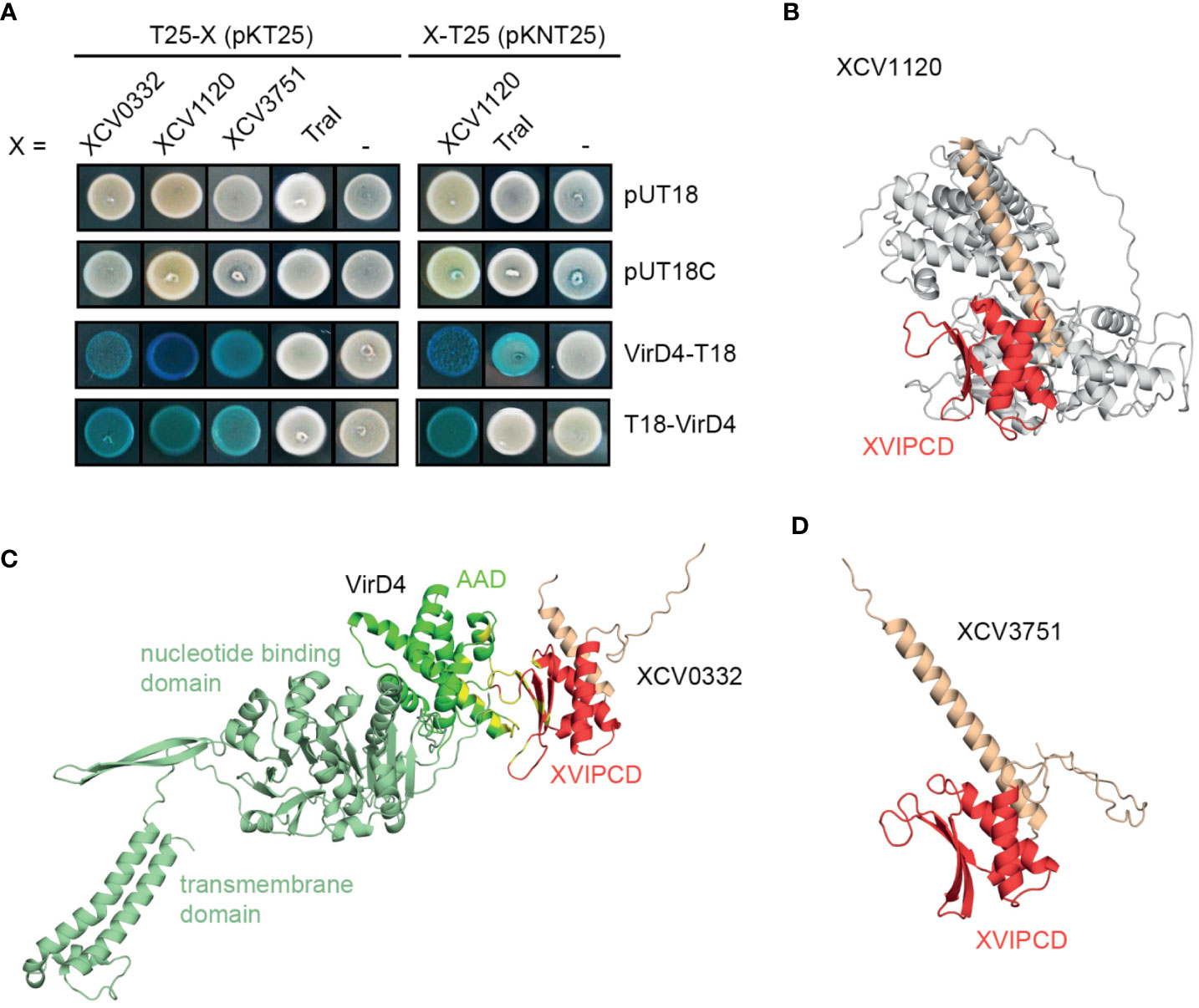
Figure 8 Candidate T4S substrates from X. euvesicatoria interact with VirD4 and contain a predicted ααβββ motif in the C-terminal XVIPCD. (A) Candidate T4S substrates from X. euvesicatoria interact with VirD4 in BACTH-based interaction studies. The interaction of T18 and T25 fusions of the candidate T4S substrates XCV0332, XCV1120, XCV3571 and TraI with VirD4 was analyzed in E. coli BTH101 cells as indicated and bacterial cultures were grown on indicator plates containing X-gal and IPTG. As control, fusion proteins were tested against the T18 or T25 domain alone as indicated. All tested proteins were stably synthesized as was shown by immunoblot analysis of bacterial cell extracts, using a FLAG epitope-specific antibody (see Figure S1). Interaction studies were performed at least three times with similar results. One representative colony is shown. (B) Structure prediction of VirD4 and in complex with the T4S substrate XCV0332 from X. euvesicatoria. Structures were predicted using the AlphaFold 2 algorithm and visualized using PyMOL (https://pymol.org). The AAD from VirD4 is shown in dark green, the ααβββ motif in the XVIPCDs of XCV0332 is shown in red. Residues predicted to be involved in the interaction between the AAD from VirD4 and the XVIPCD of XCV0332 are shown in yellow. (C) Structure prediction of XCV1120 from X. euvesicatoria. using the AlphaFold 2 algorithm. The ααβββ motif in the XVIPCDs is shown in red. (D) Structure prediction of XCV3751 from X. euvesicatoria. using the AlphaFold 2 algorithm. The ααβββ motif in the XVIPCDs is shown in red.
Given the results of our interaction and mutant studies, we assume that VirD4, which is encoded on the chromosome, acts as coupling protein for the VirB-like T4S system encoded on plasmid pXCV38. It can, however, not be excluded that substrate recognition is mediated by TraG which is encoded next to the virB T4S gene cluster on plasmid pXCV38 and shares 23% amino acid identity with VirD4 (Figure 1 and Figure S2). Since TraG-like proteins were shown to contribute to T4S (Schröder et al., 2002; Khara et al., 2021; Wang et al., 2022), we analyzed a possible interaction of TraG with components and putative T4S substrates of the VirB-like T4S system. When analyzed by BACTH assays, TraG interacted with itself, with the predicted ATPase VirB11 and the structural component VirB10 but not with the potential T4S substrate TraI (Figure S2). This is in contrast to VirD4 and suggests that VirD4 rather than TraG is involved in substrate recruitment.
To analyze a possible in vitro secretion of candidate T4S substrates from X. euvesicatoria, we generated expression constructs encoding XCV3751, XCV1120, XCV0332 and TraI fused to an N-terminal c-Myc epitope under control of the lac promoter. We assume that an N-terminal epitope does not significantly interfere with secretion because T4S signals are often located in the C-terminal protein region (Vergunst et al., 2000; Nagai et al., 2005). When analyzed in strain 85-10, epitope-tagged derivatives of XCV3751, XCV1120 and XCV0332 were stably synthesized. In contrast, c-Myc-TraI was not detected in cell extracts by immunoblotting, suggesting that it was unstable when expressed under control of the lac promoter (data not shown). traI was, therefore, expressed under control of a native promoter from X. euvesicatoria, which resulted in detectable protein synthesis (see below). For in vitro secretion assays, bacteria were cultivated in NYG medium containing BSA which likely stabilizes secreted proteins as was shown for substrates of the T3S system (Rossier et al., 1999). Total cell extracts and culture supernatants were separated by filtration and analyzed by immunoblotting. Notably, all tested proteins were detected in the culture supernatant of strain 85-10, suggesting that they were secreted (Figures 9A, B).
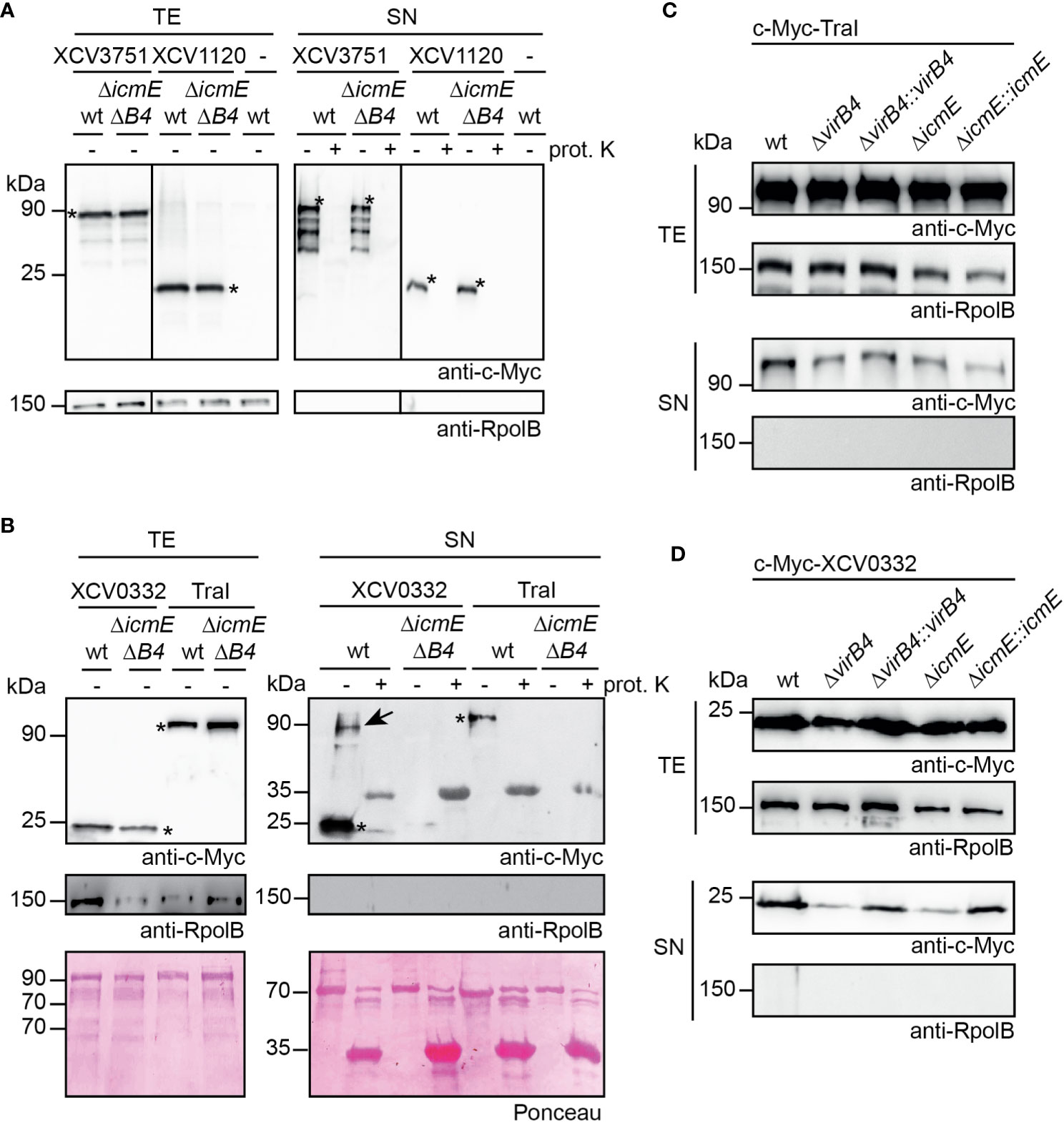
Figure 9 T4S systems from X. euvesicatoria secrete XCV0332 and TraI. (A) XCV3751 and XCV1120 are secreted independently of the T4S systems. X. euvesicatoria strains 85-10 (wt) and 85-10ΔicmEΔvirB4 (ΔicmEΔB4) with or without (-) expression constructs encoding c-Myc-XCV3751 and c-Myc-XCV1120 as indicated were incubated in NYG medium containing BSA. Total cell extracts (TE) and culture supernatants (SN) were analyzed by immunoblotting using a c-Myc epitope-specific antibody. To investigate whether proteins were secreted via OM vesicles, SN were incubated in the presence (+) or absence (-) of proteinase K at a final concentration of 0.1 mg/µl. Blots were reprobed with an antibody specific for the RNA polymerase B (RpolB) to analyze whether the presence of proteins in the culture supernatant was caused by cell lysis. Asterisks indicate signals corresponding to the expected sizes of c-Myc-XCV3751 or c-Myc-XCV1120, additional signals presumably represent degradation products. (B) In vitro secretion assays with XCV0332 and TraI. X. euvesicatoria strains 85-10 (wt) and 85-10ΔicmEΔvirB4 with expression constructs encoding c-Myc-XCV0332 or c-Myc-TraI as indicated were incubated in NYG medium containing BSA. TE and SN were analyzed by Ponceau staining and by immunoblotting using c-Myc- and RpolB-specific antibodies. SN were incubated in the presence (+) or absence (-) of proteinase K as described in (A). Asterisks indicate signals corresponding to the expected sizes of c-Myc-XCV0332 or c-Myc-TraI. The signal at a size of approximately 35 kDa results from unspecific binding of the antibodies to proteinase K. The signal at 90 kDa (indicated by an arrow) detected in the culture supernatant of strain 85-10 containing c-Myc-XCV0332 presumably corresponds to a XCV0332-containing protein complex. (C) The predicted relaxase TraI is secreted in icmE and virB4 mutants. For in vitro secretion assays, c-Myc-TraI was ectopically expressed in strain 85-10 (wt) and deletion mutant derivatives thereof lacking virB4 or icmE as indicated. For complementation studies, virB4 and icmE were reinserted into the genome of the corresponding deletion mutants (ΔvirB4::virB4 and ΔicmE::icmE). Bacteria were incubated in NYG medium in the presence of BSA, and TE and SN were analyzed by immunoblotting, using c-Myc epitope- and RpolB-specific antibodies. (D) VirB4 and IcmE contribute to the secretion of XCV0332. X. euvesicatoria strains as described in (C) and containing c-Myc-XCV0332 were incubated in NYG medium containing BSA, and TE and SN were analyzed by immunoblotting, using c-Myc epitope- and RpolB-specific antibodies. All secretion assays were performed at least three times with similar results.
To investigate whether secretion of the analyzed proteins was dependent on the T4S systems, we performed in vitro secretion assays with strain 85-10ΔicmEΔvirB4, which lacks predicted essential components of the VirB/VirD4 and Icm/Dot T4S system. Secretion of c-Myc-XCV1120 and c-Myc-XCV3751 was unaffected in strain 85-10ΔicmEΔvirB4, suggesting that the presence of the XVIPCD and the interaction with VirD4 is not sufficient to exclusively target both proteins to the T4S systems (Figure 9A). In contrast, significantly reduced amounts of c-Myc-XCV0332 and c-Myc-TraI were detected in the culture supernatant of strain 85-10ΔicmEΔvirB4, indicating a contribution of the T4S systems to secretion (Figure 9B). As a control, all supernatants were incubated in the presence of proteinase K to investigate a possible contribution of OM vesicles to secretion. OM vesicles were previously shown to mediate secretion of degradative enzymes from X. euvesicatoria (Solé et al., 2015). Detectable secretion of all tested candidate T4S substrates, however, was abolished upon treatment of the culture supernatants with proteinase K, suggesting that the secreted proteins were not present in OM vesicles (Figures 9A, B).
To further elucidate the contribution of the VirB/VirD4 or the Icm/Dot T4S system to secretion of c-Myc-XCV0332 and c-Myc-TraI, we introduced the corresponding expression constructs into virB4 and icmE single deletion mutants. Secretion of c-Myc-TraI was unaffected in both strains, suggesting that it is secreted by either the VirB/VirD4 or the Icm/Dot T4S system (Figure 9C). In contrast, secretion of c-Myc-XCV0332 was reduced but not completely abolished in strains 85-10ΔicmE and 85-10ΔvirB4 (Figure 9D). We, therefore, assume that the VirB/VirD4 and Icm/Dot T4S systems both contribute to the efficient secretion of XCV0332, which was abolished in the absence of both functional T4S systems (Figure 9B). Wild-type secretion levels of c-Myc-XCV0332 were restored when virB4 and icmE were reinserted into the genomes of strains 85-10ΔvirB4 and 85-10ΔicmE, respectively (Figure 9D). This suggests that reduced secretion of XCV0332 in the single mutants was specifically caused by the absence of VirB4 and IcmE, respectively, and not by a polar effect of the deletions on other genes. Our findings thus indicate that VirB4/VirD and Icm/Dot T4S systems are involved in in vitro protein secretion and that XCV0332 and TraI are targeted to both T4S systems. In case of TraI, the Icm/Dot T4S system appears to fully compensate for the loss of a functional VirB/VirD4 T4S system.
X. euvesciatoria is the only known plant-pathogenic bacterium which contains a predicted Icm/Dot T4S system in addition to the VirB/VirD4 T4S system. The corresponding T4S gene clusters are located on plasmids pXCV138 and pXCV38, respectively, in X. euvesicatoria strain 85-10 (Thieme et al., 2005). Additional virB genes as well as virD4 are present on the chromosome, however, a complete chromosomal virB/virD4 T4S gene cluster as described for X. axonopodis pv. citri is missing in X. euvesicatoria. Given that the role of T4S systems from X. euvesicatoria has not yet been investigated, we performed functional studies with T4S genes and analyzed secretion of candidate T4S substrates and their interaction with the T4S systems. The analysis of promoter-reporter fusions suggests that vir and icm/dot genes are expressed in vitro and in planta. In agreement with this finding, several Vir proteins including VirD4 and VirB proteins encoded on plasmid pXCV38 were previously identified in X. euvesicatoria cell extracts by mass spectrometry (Abendroth et al., 2017). Interestingly, the chromosomal virD4 gene, which encodes the predicted T4CP of the VirB/VirD4 T4S system, appears to be predominantly expressed in vitro. Given that VirD4 is an essential component of T4S systems, the reduced in planta promoter activity of virD4 indicates that T4S does not significantly contribute to the interaction of X. euvesicatoria with its host plants. This hypothesis is supported by the finding that deletion of predicted essential genes of both T4S systems did not interfere with disease symptom formation and in planta growth of X. euvesicatoria. In agreement with our observations, a virulence function or contribution to the host-pathogen interaction has not been attributed to any T4S system from Xanthomonas spp. (El Yacoubi et al., 2007; He et al., 2007; Souza et al., 2011; Jacob et al., 2014).
The chromosomally encoded VirB/VirD4 T4S system from X. axonopodis pv. citri was previously shown to deliver toxins into bacterial target cells and thus acts as a protein translocation machine (Souza et al., 2015). A similar function was reported for the VirB/VirD4 T4S system from the related opportunistic human pathogen S. maltophilia, suggesting a function of T4S systems as bacterial killing devices (Bayer-Santos et al., 2019). Components of the chromosomally encoded VirB/VirD4 T4S system from X. axonopodis pv. citri are homologous to corresponding proteins encoded on the chromosome of X. euvesicatoria. However, the chromosomal gene cluster in X. euvesicatoria is incomplete and might thus have resulted from chromosomal rearrangements, leading to the loss of function of the corresponding VirB/VirD4 T4S system. In agreement with this hypothesis, we did not detect a toxic effect of X. euvesicatoria T4S systems on E. coli strains.
Notably, X. axonopodis pv. citri and X. euvesicatoria contain a second virB gene cluster on plasmids pXAC64 and pXCV38, respectively. The association of the plasmid-localized T4S gene clusters with traI and traG genes suggests a role in conjugation which remains to be demonstrated in X. axonopodis pv. citri. The predicted functional differences of plasmid- and chromosomally encoded T4S systems as possible conjugation machines and toxin translocators, respectively, are reflected by the low sequence similarities of the corresponding VirB proteins. Interestingly, VirD4 is encoded as a single copy gene on the chromosome and might thus contribute to secretion by both VirB/VirD4 T4S systems in X. axonopodis pv. citri, suggesting that it interacts with a broad spectrum of substrates.
Plasmid transfer in Xanthomonas spp. was previously shown to occur in vitro and in planta, and leads to the exchange of virulence factors, thus potentially generating strains with altered host range or pathogenic potential (Basim et al., 1999; El Yacoubi et al., 2007; Wang et al., 2007). Mobilizable plasmids often carry the genes required for horizontal gene transfer such as plasmid pXcB from X. citri pv. citri, which encodes components of a VirB/VirD4 T4S system (El Yacoubi et al., 2007; Virolle et al., 2020). Mutation of virB4 affects self-transmission of pXcB from E. coli, thus confirming a role of the VirB/VirD4 T4S system in conjugation (El Yacoubi et al., 2007). When analyzed in X. citri, however, self-transmission of pXcB is unaffected in the absence of VirB4, suggesting the contribution of redundant plasmid transfer functions outside of pXcB which compensate for the loss of VirB4 (El Yacoubi et al., 2007). Similarly to pXcB, plasmid pXCV38 is transferable between different X. euvesicatoria strains as was demonstrated using a derivative of pXCV38 from strain 82-8 with a spectinomycin resistance gene. Deletion of the chromosomal virD4 gene in the donor strain significantly reduced plasmid transfer, indicating that the VirB/VirD4 T4S system from X. euvesicatoria acts as a conjugation machine. In strains lacking VirB4, however, residual plasmid transfer was observed, suggesting the contribution of functionally redundant proteins which partially compensate the loss of VirB4 as was described for X. citri pv. citri virB4 mutants (El Yacoubi et al., 2007). It is possible that a virB4 homolog on plasmid XCV3882-8 partially promotes plasmid transfer in virB4 mutants. In future studies, genome and plasmid sequence analyses are required to investigate the presence of T4S gene clusters in strain 82-8.
In agreement with their role in conjugation, VirB proteins assemble into heterooligomeric complexes corresponding to known subassemblies of T4S systems as was suggested by the results of protein-protein interaction studies. BACTH assays detected homo- and heterooligomerization of the predicted core components VirB7 and VirB10, which is in agreement with known assemblies of homologous proteins in T4S systems. As was shown for the conjugative T4S system encoded on plasmid pKM101 and the VirB/VirD4 T4S system from X. axonopodis pv. citri, a complex of 14 copies of VirB10 spans both bacterial membranes and associates in the OM layer with 14 copies of VirB9 and VirB7 (Sgro et al., 2018; Sheedlo et al., 2022). Interactions between VirB7 and VirB9 and the self-interaction of VirB7 are required for T4S system assembly and activity (Oliveira et al., 2016). VirB7 proteins are usually small lipoproteins of approximately 40 amino acids. In Xanthomonas spp., however, VirB7 proteins contain between 130 and 185 amino acids due to the presence of an additional globular C-terminal domain, designated N0 domain, which likely mediates VirB7 oligomerization and thus promotes the assembly of the T4S system (Sgro et al., 2019). Cryo-electron microscopy studies of a VirB7/VirB9/VirB10 complex from X. axonopodis pv. citri revealed that the N0 domain assembles as extra peripheral ring of the VirB7-VirB9-VirB10 OM complex (Souza et al., 2011; Sgro et al., 2019). Interestingly, however, in X. euvesicatoria and X. axonopodis pv. citri, chromosomally and plasmid-encoded VirB7 proteins share limited amino acid sequence similarities, which possibly reflects structural differences of translocation- and conjugation-associated X-T4S systems.
In addition to the interactions between VirB7 and VirB10, BACTH assays revealed interactions between VirB10 and the three predicted ATPases VirD4, VirB4 and VirB11 which homo- and/or heterooligomerize as expected. Furthermore, IcmE, which corresponds to VirB10, interacts with all three predicted ATPases DotL, DotO and DotB of the predicted Icm/Dot T4S system. We, therefore, assume that Vir and Icm/Dot proteins assemble into heterooligomeric complexes, most likely corresponding to T4S systems. Notably, however, the icm/dot T4S gene cluster on plasmid pXCV183 only contains 16 genes which encode the predicted essential core components of the secretion apparatus whereas ten genes present in the icm/dot gene cluster from L. pneumophila are absent in X. euvesicatoria. The contribution of icm/dot genes to in vitro T4S in X. euvesicatoria, however, suggests that the icm/dot T4S gene cluster encodes a functional T4S system for protein delivery. Given that icm/dot genes do not significantly contribute to virulence of X. euvesicatoria and plasmid transfer (data not shown), a possible contribution to protein delivery into bacterial or eukaryotic cells remains to be investigated.
Substrates of T4S systems, so-called X-Tfes, were previously identified from X. axonopodis pv. citri based on their interactions with the T4CP VirD4 which was used as a bait in yeast two-hybrid screens (Alegria et al., 2005). The identified XVIPs contain a C-terminal domain, designated XVIPCD, of approximately 120 amino acids which provides the binding site for VirD4 (Oka et al., 2022). Additional XVIPCD-containing proteins were identified by bioinformatic analyses and often share similarities with peptidoglycan-modifying enzymes, suggesting a function in the bacterial periplasm (Sgro et al., 2019). X-Tfes are likely delivered into bacterial target cells where they presumably cause lysis as was shown for the X. axonopodis pv. citri X-Tfe XAC2609 (Souza et al., 2015). In the present study, we analyzed homologous proteins from X. euvesicatoria with C-terminal XVIPCDs as well as the predicted relaxase TraI, which is a known substrate of conjugation-associated VirB/VirD4 T4S systems and lacks an XVIPCD. All candidates including TraI interacted with VirD4, suggesting that substrate binding to VirD4 is not exclusively linked to the presence of an XVIPCD.
The N-terminal region of the XVIPCD adopts an ααβββ fold which interacts with the C-terminal AAD from VirD4 as was shown by previous solution NMR spectroscopy studies of the X-Tfe XAC2609 (Oka et al., 2022). In contrast, the C-terminal glutamine-rich tail of the XVIPCD is dispensable for the interaction with the AAD from VirD4 (Oka et al., 2022). Interestingly, structure predictions revealed similar ααβββ motifs in the C-terminal regions of the XVIPCD-containing candidate substrates XCV0332, XCV1120 and XCV3571 from X. euvesicatoria. Yet, a contribution of the XVIPCD of candidate X-Tfes from X. euvesicatoria to the interaction with VirD4 remains to be investigated. Notably, the analyzed candidate substrates XCV3571 and XCV1120 were secreted by wild-type and T4S mutant strains, suggesting that the presence of the XVIPCD and the interaction with VirD4 is not sufficient for the exclusive targeting of proteins to the T4S system. This is in agreement with the finding that the ααβββ motif in XAC2609 from X. axonopodis pv. citri is required for initial substrate recognition but is presumably not sufficient for T4S-dependent protein delivery (Oka et al., 2022). It was proposed that the C-terminal protein region outside the conserved XVIPCD promotes translocation, possibly by interacting with a yet unidentified adaptor protein of the T4S system (Oka et al., 2022). The presence of an additional translocation signal outside the XVIPCD in T4S substrates from X. euvesicatoria is supported by the finding that TraI is secreted by the T4S systems despite the lack of an XVIPCD.
Taken together, our in vitro secretion assays led to the identification of TraI and XCV0332 as T4S substrates from X. euvesicatoria. XCV0332 is a homolog of the X-Tfe XAC0323 and mainly consist of the XVIPCD. Both proteins might thus represent parts of ancient X-Tfes which result from mutations leading to the loss of the N-terminal protein region (Sgro et al., 2019). Secretion of XCV0332 was abolished in the absence of functional VirB/VirD4 and Icm/Dot T4S systems whereas icmE and virB4 single mutants led to reduced but still detectable secretion of XCV0332. We, therefore, assume that both T4S systems additively contribute to XCV0332 secretion, indicating that XVIPCD-containing proteins can be targeted to both T4S systems in X. euvesicatoria. A similar finding was observed for the secretion of TraI. In contrast to XCV0332, however, secretion of TraI was unaffected in icmE and virB4 single mutants. This suggests that the presence of either the VirB/VirD4 or the Icm/Dot T4S system is sufficient to promote TraI secretion at wild-type levels. The dual targeting of TraI was unexpected and points to a broad substrate specificity of both T4S systems. In future studies, we will, therefore, investigate possible interactions of TraI with components of the Icm/Dot T4S system as well as the biological significance of Icm/Dot-mediated protein delivery. Notably, in preliminary experiments, we did not detect any contribution of the Icm/Dot T4S system to plasmid transfer. However, given that mobilizable plasmids often contain T4S genes which are involved in plasmid transfer, a possible contribution of the Icm/Dot T4S system to the transfer of plasmid pXCV183, which contains the icm/dot T4S genes, needs to be analyzed in future experiments.
In summary, our study showed that X. euvesicatoria contains two functional T4S systems which contribute to protein secretion and presumably do not promote toxin delivery into E. coli. The VirB/VirD4 T4S system acts as a conjugation system for plasmid transfer between X. euvesicatoria strains and shares substrate specificity with the Icm/Dot system which serves as additional protein delivery system. The contribution of the Icm/Dot T4S system to the secretion of additional T4S effectors and a possible protein transport into plant cells remains to be investigated.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
SD, FS and DB conceived and designed the experiments. SD and FS performed the experiments and analyzed the data. SD and DB wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was funded by a grant from the Deutsche Forschungsgemeinschaft to DB (BU 2145/6-2).
We are grateful to M. Jordan for excellent technical assistance.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1203159/full#supplementary-material
Abendroth, U., Adlung, N., Otto, A., Grüneisen, B., Becher, D., Bonas, U. (2017). Identification of new protein-coding genes with a potential role in the virulence of the plant pathogen Xanthomonas euvesicatoria. BMC Genomics 18, 625. doi: 10.1186/s12864-017-4041-7
Adlung, N., Prochaska, H., Thieme, S., Banik, A., Blüher, D., John, P., et al. (2016). Non-host resistance induced by the Xanthomonas effector XopQ is widespread within the genus Nicotiana and functionally depends on EDS1. Front. Plant Sci. 7, 1796. doi: 10.3389/fpls.2016.01796
Alegria, M. C., Souza, D. P., Andrade, M. O., Docena, C., Khater, L., Ramos, C. H., et al. (2005). Identification of new protein-protein interactions involving the products of the chromosome- and plasmid-encoded type IV secretion loci of the phytopathogen Xanthomonas axonopodis pv. citri. J. Bacteriol 187, 2315–2325. doi: 10.1128/JB.187.7.2315-2325.2005
Alvarez-Martinez, C. E., Sgro, G. G., Araujo, G. G., Paiva, M. R. N., Matsuyama, B. Y., Guzzo, C. R., et al. (2021). Secrete or perish: the role of secretion systems in Xanthomonas biology. Comput. Struct. Biotechnol. J. 19, 279–302. doi: 10.1016/j.csbj.2020.12.020
Audette, G. F., Manchak, J., Beatty, P., Klimke, W. A., Frost, L. S. (2007). Entry exclusion in F-like plasmids requires intact TraG in the donor that recognizes its cognate TraS in the recipient. Microbiol 153, 442–451. doi: 10.1099/mic.0.2006/001917-0
Basim, H., Stall, R., Minsavage, G., Jones, J. (1999). Chromosomal gene transfer by conjugation in the plant pathogen Xanthomonas axonopodis pv. vesicatoria. Phytopathol 89, 1044–1049. doi: 10.1094/PHYTO.1999.89.11.1044
Bastedo, D. P., Lo, T., Laflamme, B., Desveaux, D., Guttman, D. S. (2020). Diversity and evolution of type III secreted effectors: a case study of three families. Curr. Top. Microbiol. Immunol. 427, 201–230. doi: 10.1007/82_2019_165
Battesti, A., Bouveret, E. (2012). The bacterial two-hybrid system based on adenylate cyclase reconstitution in Escherichia coli. Methods 58, 325–334. doi: 10.1016/j.ymeth.2012.07.018
Bayer-Santos, E., Cenens, W., Matsuyama, B. Y., Oka, G. U., Di Sessa, G., Mininel, I. D. V., et al. (2019). The opportunistic pathogen Stenotrophomonas maltophilia utilizes a type IV secretion system for interbacterial killing. PloS Pathog. 15, e1007651. doi: 10.1371/journal.ppat.1007651
Boch, J., Bonas, U., Lahaye, T. (2014). TAL effectors - pathogen strategies and plant resistance engineering. New Phytol. 204, 823–832. doi: 10.1111/nph.13015
Boch, J., Scholze, H., Schornack, S., Landgraf, A., Hahn, S., Kay, S., et al. (2009). Breaking the code of DNA-binding specificity of TAL-type III effectors. Science 326, 1509–1512. doi: 10.1126/science.1178811
Bonas, U., Schulte, R., Fenselau, S., Minsavage, G. V., Staskawicz, B. J., Stall, R. E. (1991). Isolation of a gene-cluster from Xanthomonas campestris pv. vesicatoria that determines pathogenicity and the hypersensitive response on pepper and tomato. Mol. Plant-Microbe Interact. 4, 81–88. doi: 10.1094/MPMI-4-081
Bonas, U., Stall, R. E., Staskawicz, B. (1989). Genetic and structural characterization of the avirulence gene avrBs3 from Xanthomonas campestris pv. vesicatoria. Mol. Gen. Genet. 218, 127–136. doi: 10.1007/BF00330575
Büttner, D. (2012). Protein export according to schedule – architecture, assembly and regulation of type III secretion systems from plant and animal pathogenic bacteria. Microbiol. Mol. Biol. Rev. 76, 262–310. doi: 10.1128/MMBR.05017-11
Büttner, D., Bonas, U. (2010). Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol. Rev. 34, 107–133. doi: 10.1111/j.1574-6976.2009.00192.x
Ceulemans, E., Ibrahim, H. M. M., De Coninck, B., Goossens, A. (2021). Pathogen effectors: exploiting the promiscuity of plant signaling hubs. Trends Plant Sci. 26, 780–795. doi: 10.1016/j.tplants.2021.01.005
Christie, P. J. (2016). The mosaic type IV secretion systems. EcoSal Plus 7. doi: 10.1128/ecosalplus.ESP-0020-2015
Christie, P. J., Gomez Valero, L., Buchrieser, C. (2017). Biological diversity and evolution of type IV secretion systems. Curr. Top. Microbiol. Immunol. 413, 1–30. doi: 10.1007/978-3-319-75241-9_1
Costa, T. R., Felisberto-Rodrigues, C., Meir, A., Prevost, M. S., Redzej, A., Trokter, M., et al. (2015). Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat. Rev. Microbiol. 13, 343–359. doi: 10.1038/nrmicro3456
Costa, T. R. D., Harb, L., Khara, P., Zeng, L., Hu, B., Christie, P. J. (2021). Type IV secretion systems: advances in structure, function, and activation. Mol. Microbiol. 115, 436–452. doi: 10.1111/mmi.14670
El Yacoubi, B., Brunings, A. M., Yuan, O., Shankar, S., Gabriel, D. W. (2007). In planta horizontal transfer of a major pathogenicity effector gene. Appl. Environ. Microbiol. 73, 1612–1621. doi: 10.1128/AEM.00261-06
Engler, C., Kandzia, R., Marillonnet, S. (2008). A one pot, one step, precision cloning method with high throughput capability. PloS One 3, e3647. doi: 10.1371/journal.pone.0003647
Galan, J. E., Waksman, G. (2018). Protein-injection machines in bacteria. Cell 172, 1306–1318. doi: 10.1016/j.cell.2018.01.034
Gelvin, S. B. (2003). Agrobacterium-mediated plant transformation: the biology behind the "gene-jockeying" tool. Microbiol. Mol. Biol. Rev. 67, 16–37. doi: 10.1128/MMBR.67.1.16-37.2003
Ghosal, D., Jeong, K. C., Chang, Y. W., Gyore, J., Teng, L., Gardner, A., et al. (2019). Molecular architecture, polar targeting and biogenesis of the Legionella Dot/Icm T4SS. Nat. Microbiol. 4, 1173–1182. doi: 10.1038/s41564-019-0427-4
Gomez-Valero, L., Chiner-Oms, A., Comas, I., Buchrieser, C. (2019). Evolutionary dissection of the Dot/Icm system based on comparative genomics of 58 Legionella species. Genome Biol. Evol. 11, 2619–2632. doi: 10.1093/gbe/evz186
He, Y. Q., Zhang, L., Jiang, B. L., Zhang, Z. C., Xu, R. Q., Tang, D. J., et al. (2007). Comparative and functional genomics reveals genetic diversity and determinants of host specificity among reference strains and a large collection of Chinese isolates of the phytopathogen Xanthomonas campestris pv. campestris. Genome Biol. 8, R218. doi: 10.1186/gb-2007-8-10-r218
Huguet, E., Hahn, K., Wengelnik, K., Bonas, U. (1998). ). hpaA mutants of Xanthomonas campestris pv. vesicatoria are affected in pathogenicity but retain the ability to induce host-specific hypersensitive reaction. Mol. Microbiol. 29, 1379–1390. doi: 10.1046/j.1365-2958.1998.01019.x
Jacob, T. R., de Laia, M. L., Moreira, L. M., Goncalves, J. F., Carvalho, F. M., Ferro, M. I., et al. (2014). Type IV secretion system Is not involved in infection process in citrus. Int. J. Microbiol. 2014, 763575. doi: 10.1155/2014/763575
Jin, S. G., Roitsch, T., Christie, P. J., Nester, E. W. (1990). The regulatory VirG protein specifically binds to a cis-acting regulatory sequence involved in transcriptional activation of Agrobacterium tumefaciens virulence genes. J. Bacteriol 172, 531–537. doi: 10.1128/jb.172.2.531-537.1990
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Karimova, G., Dautin, N., Ladant, D. (2005). Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J. Bacteriol 187, 2233–2243. doi: 10.1128/JB.187.7.2233-2243.2005
Karimova, G., Pidoux, J., Ullmann, A., Ladant, D. (1998). A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U.S.A. 95, 5752–5756. doi: 10.1073/pnas.95.10.5752
Kaur, A., Bansal, K., Kumar, S., Sonti, R. V., Patil, P. B. (2019). Complete genome dynamics of a dominant-lineage strain of Xanthomonas oryzae pv. oryzae harbouring a novel plasmid encoding a type IV secretion system. Access Microbiol. 1, e000063. doi: 10.1099/acmi.0.000063
Khara, P., Song, L., Christie, P. J., Hu, B. (2021). In situ visualization of the pKM101-encoded type IV secretion system reveals a highly symmetric ATPase energy center. mBio 12, e0246521. doi: 10.1128/mBio.02465-21
Kitao, T., Kubori, T., Nagai, H. (2022). Recent advances in structural studies of the Legionella pneumophila Dot/Icm type IV secretion system. Microbiol. Immunol. 66, 67–74. doi: 10.1111/1348-0421.12951
Kubori, T., Nagai, H. (2016). The type IVB secretion system: an enigmatic chimera. Curr. Opin. Microbiol. 29, 22–29. doi: 10.1016/j.mib.2015.10.001
Li, Y. G., Christie, P. J. (2018). The Agrobacterium VirB/VirD4 T4SS: mechanism and architecture defined through in vivo mutagenesis and chimeric systems. Curr. Top. Microbiol. Immunol. 418, 233–260. doi: 10.1007/82_2018_94
Li, Y. G., Hu, B., Christie, P. J. (2019). Biological and structural diversity of type IV secretion systems. Microbiol. Spectr. 7. doi: 10.1128/9781683670285.ch22
Liu, X., Shin, S. (2019). Viewing Legionella pneumophila pathogenesis through an immunological lens. J. Mol. Biol. 431, 4321–4344. doi: 10.1016/j.jmb.2019.07.028
Lorenz, C., Büttner, D. (2009). Functional characterization of the type III secretion ATPase HrcN from the plant pathogen Xanthomonas campestris pv. vesicatoria. J. Bacteriol 191, 1414–1428. doi: 10.1128/JB.01446-08
Lorenz, C., Hausner, J., Büttner, D. (2012). HrcQ provides a docking site for early and late type III secretion substrates from Xanthomonas. PloS One 7, e51063. doi: 10.1371/journal.pone.0051063
Mace, K., Vadakkepat, A. K., Redzej, A., Lukoyanova, N., Oomen, C., Braun, N., et al. (2022). Cryo-EM structure of a type IV secretion system. Nature 607, 191–196. doi: 10.1038/s41586-022-04859-y
Meir, A., Mace, K., Vegunta, Y., Williams, S. M., Waksman, G. (2023). Substrate recruitment mechanism by gram-negative type III, IV, and VI bacterial injectisomes. Trends Microbiol. doi: 10.1016/j.tim.2023.03.005
Moscou, M. J., Bogdanove, A. J. (2009). A simple cipher governs DNA recognition by TAL effectors. Science 326, 1501. doi: 10.1126/science.1178817
Nagai, H., Cambronne, E. D., Kagan, J. C., Amor, J. C., Kahn, R. A., Roy, C. R. (2005). A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc. Natl. Acad. Sci. U.S.A. 102, 826–831. doi: 10.1073/pnas.0406239101
Nagai, H., Kubori, T. (2011). Type IVB secretion systems of Legionella and other Gram-negative bacteria. Front. Microbiol. 2, 136. doi: 10.3389/fmicb.2011.00136
Oka, G. U., Souza, D. P., Cenens, W., Matsuyama, B. Y., Cardoso, M. V. C., Oliveira, L. C., et al. (2022). Structural basis for effector recognition by an antibacterial type IV secretion system. Proc. Natl. Acad. Sci. U.S.A. 119. doi: 10.1073/pnas.2112529119
Oliveira, L. C., Souza, D. P., Oka, G. U., Lima, F. D. S., Oliveira, R. J., Favaro, D. C., et al. (2016). VirB7 and VirB9 interactions are required for the assembly and antibacterial activity of a type IV secretion system. Structure 24, 1707–1718. doi: 10.1016/j.str.2016.07.015
Otten, C., Büttner, D. (2021). HrpB4 from Xanthomonas campestris pv. vesicatoria acts similarly to SctK proteins and promotes the docking of the predicted sorting platform to the type III secretion system. Cell Microbiol. 23, e13327. doi: 10.1111/cmi.13327
Rossier, O., Wengelnik, K., Hahn, K., Bonas, U. (1999). The Xanthomonas Hrp type III system secretes proteins from plant and mammalian pathogens. Proc. Natl. Acad. Sci. U.S.A. 96, 9368–9373. doi: 10.1073/pnas.96.16.9368
Scheibner, F., Schulz, S., Hausner, J., Marillonnet, S., Büttner, D. (2016). Type III-dependent translocation of HrpB2 by a non-pathogenic hpaABC mutant of the plant-pathogenic bacterium Xanthomonas campestris pv. vesicatoria. Appl. Environ. Microbiol. 82, 3331–3347. doi: 10.1128/AEM.00537-16
Schröder, G., Krause, S., Zechner, E. L., Traxler, B., Yeo, H. J., Lurz, R., et al. (2002). TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J. Bacteriol 184, 2767–2779. doi: 10.1128/JB.184.10.2767-2779.2002
Sgro, G. G., Costa, T. R. D., Cenens, W., Souza, D. P., Cassago, A., Coutinho de Oliveira, L., et al. (2018). Cryo-EM structure of the bacteria-killing type IV secretion system core complex from Xanthomonas citri. Nat. Microbiol. 3, 1429–1440. doi: 10.1038/s41564-018-0262-z
Sgro, G. G., Oka, G. U., Souza, D. P., Cenens, W., Bayer-Santos, E., Matsuyama, B. Y., et al. (2019). Bacteria-killing type IV secretion systems. Front. Microbiol. 10, 1078. doi: 10.3389/fmicb.2019.01078
Sheedlo, M. J., Ohi, M. D., Lacy, D. B., Cover, T. L. (2022). Molecular architecture of bacterial type IV secretion systems. PloS Pathog. 18, e1010720. doi: 10.1371/journal.ppat.1010720
Shen, Z., Tang, C. M., Liu, G. (2022). Towards a better understanding of antimicrobial resistance dissemination: what can be learnt from studying model conjugative plasmids? Mil Med. Res. 9, 3. doi: 10.1186/s40779-021-00362-z
Solé, M., Scheibner, F., Hoffmeister, A. K., Hartmann, N., Hause, G., Rother, A., et al. (2015). Xanthomonas campestris pv. vesicatoria secretes proteases and xylanases via the Xps type II secretion system and outer membrane vesicles. J. Bacteriol 197, 2879–2893. doi: 10.1128/JB.00322-15
Soucy, S. M., Huang, J., Gogarten, J. P. (2015). Horizontal gene transfer: building the web of life. Nat. Rev. Genet. 16, 472–482. doi: 10.1038/nrg3962
Souza, D. P., Andrade, M. O., Alvarez-Martinez, C. E., Arantes, G. M., Farah, C. S., Salinas, R. K. (2011). A component of the Xanthomonadaceae type IV secretion system combines a VirB7 motif with a N0 domain found in outer membrane transport proteins. PloS Pathog. 7, e1002031. doi: 10.1371/journal.ppat.1002031
Souza, D. P., Oka, G. U., Alvarez-Martinez, C. E., Bisson-Filho, A. W., Dunger, G., Hobeika, L., et al. (2015). Bacterial killing via a type IV secretion system. Nat. Commun. 6, 6453. doi: 10.1038/ncomms7453
Stall, R. E., Loschke, D. C., Jones, J. B. (1986). Linkage of copper resistance and avirulence loci on a self-transmissible plasmid in Xanthomonas campestris pv. vesicatoria. Phytopathol 76, 240–243. doi: 10.1094/Phyto-76-240
Szczesny, R., Jordan, M., Schramm, C., Schulz, S., Cogez, V., Bonas, U., et al. (2010). Functional characterization of the Xps and Xcs type II secretion systems from the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria. New Phytol. 187, 983–1002. doi: 10.1111/j.1469-8137.2010.03312.x
Tamamoto, S., Aoyama, T., Takanami, M., Oka, A. (1990). Binding of the regulatory protein VirG to the phased signal sequences upstream from virulence genes on the hairy-root-inducing plasmid. J. Mol. Biol. 215, 537–547. doi: 10.1016/S0022-2836(05)80166-4
Thieme, F., Koebnik, R., Bekel, T., Berger, C., Boch, J., Buttner, D., et al. (2005). Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J. Bacteriol 187, 7254–7266. doi: 10.1128/JB.187.21.7254-7266.2005
Vergunst, A. C., Schrammeijer, B., den Dulk-Ras, A., de Vlaam, C. M., Regensburg-Tuink, T. J., Hooykaas, P. J. (2000). VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290, 979–982. doi: 10.1126/science.290.5493.979
Virolle, C., Goldlust, K., Djermoun, S., Bigot, S., Lesterlin, C. (2020). Plasmid transfer by conjugation in Gram-negative bacteria: from the cellular to the community level. Genes 11, 1239. doi: 10.3390/genes11111239
Wagner, S., Grin, I., Malmsheimer, S., Singh, N., Torres-Vargas, C. E., Westerhausen, S. (2018). Bacterial type III secretion systems: a complex device for the delivery of bacterial effector proteins into eukaryotic host cells. FEMS Microbiol. Lett. 365, fny201. doi: 10.1093/femsle/fny201
Waksman, G. (2019). From conjugation to T4S systems in Gram-negative bacteria: a mechanistic biology perspective. EMBO Rep. 20. doi: 10.15252/embr.201847012
Wang, D., Wang, L., Bi, D., Song, J., Wang, G., Gao, Y., et al. (2022). Conjugative transfer of acute hepatopancreatic necrosis disease-causing pVA1-type plasmid is mediated by a novel self-encoded type IV secretion system. Microbiol. Spectr. 10, e0170222. doi: 10.1128/spectrum.01702-22
Wang, Y., Xiao, M., Geng, X., Liu, J., Chen, J. (2007). Horizontal transfer of genetic determinants for degradation of phenol between the bacteria living in plant and its rhizosphere. Appl. Microbiol. Biotechnol. 77, 733–739. doi: 10.1007/s00253-007-1187-2
Weber, E., Engler, C., Gruetzner, R., Werner, S., Marillonnet, S. (2011). A modular cloning system for standardized assembly of multigene constructs. PloS One 6, e16765. doi: 10.1371/journal.pone.0016765
Wengelnik, K., Bonas, U. (1996). HrpXv, an AraC-type regulator, activates expression of five of the six loci in the hrp cluster of Xanthomonas campestris pv. vesicatoria. J. Bacteriol 178, 3462–3469. doi: 10.1128/jb.178.12.3462-3469.1996
Werner, S., Breus, O., Symonenko, Y., Marillonnet, S., Gleba, Y. (2011). High-level recombinant protein expression in transgenic plants by using a double-inducible viral vector. Proc. Natl. Acad. Sci. U.S.A. 108, 14061–14066. doi: 10.1073/pnas.1102928108
Winans, S. C. (1990). Transcriptional induction of an Agrobacterium regulatory gene at tandem promoters by plant-released phenolic compounds, phosphate starvation, and acidic growth media. J. Bacteriol 172, 2433–2438. doi: 10.1128/jb.172.5.2433-2438.1990
Winans, S. C., Jin, S., Komari, T., Johnson, K. M., Nester, E. W. (1987). “The role of virulence regulatory loci in determining Agrobacterium host range,” in Plant molecular biology. Eds. Wettstein, D.v., Chua, N. H. (Plenum Press), 573–582.
Keywords: Type IV secretion, conjugation, Icm/Dot, VirD4, plant pathogen, effector, relaxase
Citation: Drehkopf S, Scheibner F and Büttner D (2023) Functional characterization of VirB/VirD4 and Icm/Dot type IV secretion systems from the plant-pathogenic bacterium Xanthomonas euvesicatoria. Front. Cell. Infect. Microbiol. 13:1203159. doi: 10.3389/fcimb.2023.1203159
Received: 10 April 2023; Accepted: 17 July 2023;
Published: 01 August 2023.
Edited by:
Yangyang Zhao, Jiangsu Academy of Agricultural Sciences (JAAS), ChinaReviewed by:
Jason Eugene Heindl, Rowan University, United StatesCopyright © 2023 Drehkopf, Scheibner and Büttner. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniela Büttner, ZGFuaWVsYS5idWV0dG5lckBnZW5ldGlrLnVuaS1oYWxsZS5kZQ==
†Present address: Felix Scheibner, Bristol Myers Squibb GmbH & Co. KGaA, Munich, Germany
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.