- Faculty of Science and Engineering, Southern Cross University, Lismore, NSW, Australia
Extracellular vesicles (EVs or exosomes) are well described for bacterial pathogens associated with our gastrointestinal system, and more recently as a novel mechanism for environmental persistence, dissemination and infection for human enteric viruses. However, the roles played by EVs in the ancient arms race that continues between amoebae and one of their prey, Legionella pneumophila, is poorly understood. At best we know of intracellular vesicles of amoebae containing a mix of bacterial prey species, which also provides an enhanced niche for bacteriophage infection/spread. Free-living amoeba-associated pathogens have recently been recognized to have enhanced resistance to disinfection and environmental stressors, adding to previously understood (but for relatively few species of) bacteria sequestered within amoebal cysts. However, the focus of the current work is to review the likely impacts of large numbers of respiratory-sized EVs containing numerous L. pneumophila cells studied in pure and biofilm systems with mixed prey species. These encapsulated pathogens are orders of magnitude more resistant to disinfection than free cells, and our engineered systems with residual disinfectants could promote evolution of resistance (including AMR), enhanced virulence and EV release. All these are key features for evolution within a dead-end human pathogen post lung infection. Traditional single-hit pathogen infection models used to estimate the probability of infection/disease and critical environmental concentrations via quantitative microbial risk assessments may also need to change. In short, recognizing that EV-packaged cells are highly virulent units for transmission of legionellae, which may also modulate/avoid human host immune responses. Key data gaps are raised and a previous conceptual model expanded upon to clarify where biofilm EVs could play a role promoting risk as well as inform a more wholistic management program to proactively control legionellosis.
1 Introduction
Water-based (saprozoic) pathogens cost the US over $2.39 billion per year (approximately $7m per head of population), some ten-fold the cost of gastrointestinal waterborne pathogens (Collier et al., 2021). These saprozoic pathogens predominantly grow within microeukaryotes, such as free-living amoebae and their analogues, macrophages, differentiating them from saprotrophic microorganisms that live off dead and decaying matter. In Europe, legionellosis (largely caused by three species of saprozoic Legionella from drinking water) was identified as the fifth most significant contributor to disability-adjusted life years (DALYs) from 31 selected diseases, following influenza, tuberculosis, human immunodeficiency virus (HIV) infection/AIDS and invasive pneumococcal disease (Cassini et al., 2019). However, unlike the top four infectious diseases, legionellosis is not spread person-to-person, but largely infect humans from water/moist soil/mulch habitats via aerosols to lung macrophages and alveolar epithelial cells (Ashbolt, 2015) – placing management in multiple hands, given the various jurisdictions addressing water provision (treatment, distribution and premise) and its other environmental exposures. Also, our aging population and various increasing vulnerable groups to these opportunistic pathogens are escalating the disease burden from water-based pathogens globally.
In the general absence of regulations requiring saprozoic pathogen monitoring near points of potential exposures (Bartram et al., 2007; CDC, 2017; HSE, 2017; Gamage et al., 2022), along with most cases being sporadic and rarely followed up (Abu Khweek and Amer, 2018; Clopper et al., 2021), there is significant under reporting of the true impact from these pathogens (Burillo et al., 2017; Vermeulen et al., 2020). What we know most is from Legionnaires’ Disease outbreaks from cooling tower aerosols, ornamental fountains and within hospitals, but these only account for some 15% of all cases, the bulk of the remainder being sporadic within the community, largely thought to be from premise plumbing systems (Falkinham et al., 2015; NAS, 2020).
Many saprozoic pathogens are transmitted to humans via aerosols, typically following distal disinfection processes (e.g., residual chlorine in drinking water, point-of-use UV irradiation and environmental desiccation). However, recent work has shown that extracellular vesicles (EVs) from amoebae, along with their trophozoites and cysts provide orders of magnitude resistance to disinfection for internalized pathogens compared to the freely suspended bacteria and human viruses (Storey et al., 2004; Folkins et al., 2020; Martin et al., 2020; Dey et al., 2022). From a control point of view this is important, as current guidance/regulation removal is only based on disinfection performance of the more readily inactivated freely suspended. Also relevant is L. pneumophila’s dual life phases (replicative & transmissive), with replicative cells (typical form used in disinfection studies) repressing, while the transmissive form induces virulence, motility and most importantly for persistence, various stress factors via activity of RNA-binding proteins, including CsrA and its thiamine pyrophosphate riboswitch (Sahr et al., 2017). Overall, this begs the question, has L. pneumophila, like other amoeba-resisting pathogens (Schmitz-Esser et al., 2010), evolved not only to grow and control host cells (using a large fraction of its genome to do so [(Burstein et al., 2016)]), but is it also adapted to persist within EVs or even generate host EVs to extend its transmissive phase? Recent transcriptomics is helping us to understand the differentially expressed Legionella genes within their infected amoebal hosts (Quan et al., 2020; Chauhan et al., 2023). Such techniques have yet to be applied to EVs production and within these vesicles.
Overall, identifying amoeba-associated pathogens may not only identify amplification niches in engineered systems (Thomas and Ashbolt, 2011; Thomas et al., 2014), but also pathogens more likely to be protected and deliver in an infectious dose (Shaheen and Ashbolt, 2018). No such routine amoeba-targeted monitoring is undertaken today but may well provide the proactive approach sought in water safety plans (WSPs) that are used around the world to manage waterborne enteric pathogens (WHO, 2017), but are in their infancy in being adapted to provide safe water for the management of saprozoic pathogens (Papadakis et al., 2018; De Giglio et al., 2021). Hence, the goal of this article is to summarize what is understood about the biology of L. pneumophila relevant to its in-situ biofilm hosts with the purpose of identifying promising targets to manage this critical saprozoic pathogen of engineered water systems.
1.1 Monitoring and management options for L. pneumophila
Current culture-based methods for known saprozoic bacterial pathogens (Legionella spp., nontuberculous mycobacteria etc.) take 10-days to weeks for confirmation (e.g., (ISO 11731, 2017; Pfaller et al., 2021)) – far too long for a timely response. Further, cells of these pathogens may largely be present in viable but non-culturable (VBNC) states (hence missed by traditional culture methods) yet still infectious to humans (Dietersdorfer et al., 2018). Therefore, specialized amoeba co-culture is recommended for resuscitation (García et al., 2007; Dey et al., 2019a; Dey et al., 2019b; Dey et al., 2020). While still disputed by some, molecular methods are preferred for the management of water systems, such as qPCR (Lee et al., 2011). However, determining the fraction of infectious cells by qPCR is still problematic, given uncertainties in infectivity status and the variable residual chlorine levels likely present in piped water systems (Casini et al., 2018; Donohue et al., 2019; Donohue, 2021). Flow cytometry using immunocapture (Füchslin et al., 2010) or with cell-sorting in combination with qPCR shows promise to also identify VBNC cells, particularly those missed by conventional culture (e.g., ISO 11731:2017-05 pre-treatment procedure) but capable of infecting host cells (Nisar et al., 2023).
Nonetheless, live or dead, high concentrations (in excess of 103 cells/100 mL, (Hamilton et al., 2021)) in distal parts of water delivery systems infer growth in the system that needs to be addressed, else risk exposure to infectious aerosols at some stage. The major amplification site for L. pneumophila in water systems is within biofilm amoebae (NAS, 2020). Therefore, herein a prior conceptual model for legionellae growth within pipe biofilms (Shaheen and Ashbolt, 2021) is expanded upon to include EV and possible amoeba monitoring targets and identifies research gaps. Overall, targeting hosts prior to rapid, explosive growth of legionellae is hypothesized as a useful target within a system of checks (flushing, temp <25 or >50°C, disinfectant residual etc.) for a proactive early warning water management system.
2 Primary role of free-living amoebae supporting problematic L. pneumophila growth
2.1 Protist hosts for explosive growth of L. pneumophila
The primary hosts for Legionella within biofilms are various free-living protozoa, principally free-living amoebae (FLA) (Thomas et al., 2010) and ciliates (Tsao et al., 2019). While ciliates may excrete viable legionellae within fecal pellets for subsequent re-ingestion (Hojo et al., 2012; Berk and Garduño, 2013), L. pneumophila released following their lysis of amoebal trophozoites are more likely to lead to explosive growth cycles of phagocytosis-growth-release (Shaheen et al., 2019), given several hundred of cells that can grow per amoeba (Buse and Ashbolt, 2012).
A recent review of FLA is provided by Scheid (2019), in which he describes their diversity and the predatory heterotrophic feeding by trophozoites on biofilm microbiota and extracellular polymeric substances. Indeed, FLA are ubiquitous and instrumental to both biofilm formation and ecological successions. Important amoebal hosts supporting Legionella and other amoeba-resisting bacterial (ARB) pathogens include members of Acanthamoeba, Naegleria, Vermamoeba and Willaertia (Thomas et al., 2010). Although the term FLA has no relevance to amoebal taxonomy or phylogeny, it does separate them from the parasitic intestinal amoebae (e.g., Entamoeba histolytica) that have significantly reduced genomes due to their host’s providing most needs (Shabardina et al., 2018). Critical to FLA success and dispersal (air, water, soils & by wildlife) is the environmentally robust cyst form, that may also encase/protect beneficial and pathogenic bacteria and their viruses (Thomas and Greub, 2010; Schulz et al., 2020; Shi et al., 2021), many of whom may carry virus-encoded auxiliary metabolic genes (vAMGs) (Yuan and Ju, 2023).
Pertinent to engineered water systems has been the increasing recognition of the importance of suspended flocs/particulates in amoebal planktonic dynamics (Anderson, 2018). While not considered biofilms of fixed surfaces, these planktonic niches may represent sloughed biofilm material or biofilm-like growth on suspended particulates that contribute to microbial activity in piped water systems (Liu et al., 2016) and ultimately the bulk of internalized pathogens transmitted via aerosols (Hamilton et al., 2018; Shaheen and Ashbolt, 2021).
2.2 Extracellular vesicles of FLA
Nearly all living cells may excrete EVs, which are described based on their site of origin (i.e., exosomes, ectosomes, cytonemes & nanotubes) (Vermeulen et al., 2020.). However, only recently have EVs been recognized as participating in cell-to-cell communication processes, including between predator and prey (de Souza and Barrias, 2020), and carry specific virulence factors (Cruz Camacho et al., 2023) with known immunomodulatory properties (Costa et al., 2021). Exosomes are EVs that originate from the endocytic pathway of a cell (de Souza and Barrias, 2020; Cruz Camacho et al., 2023) and FLA-EVs are of focus here because of their likely multiple connections across the life cycle of L. pneumophila in biofilms of engineered environments (Figure 1).
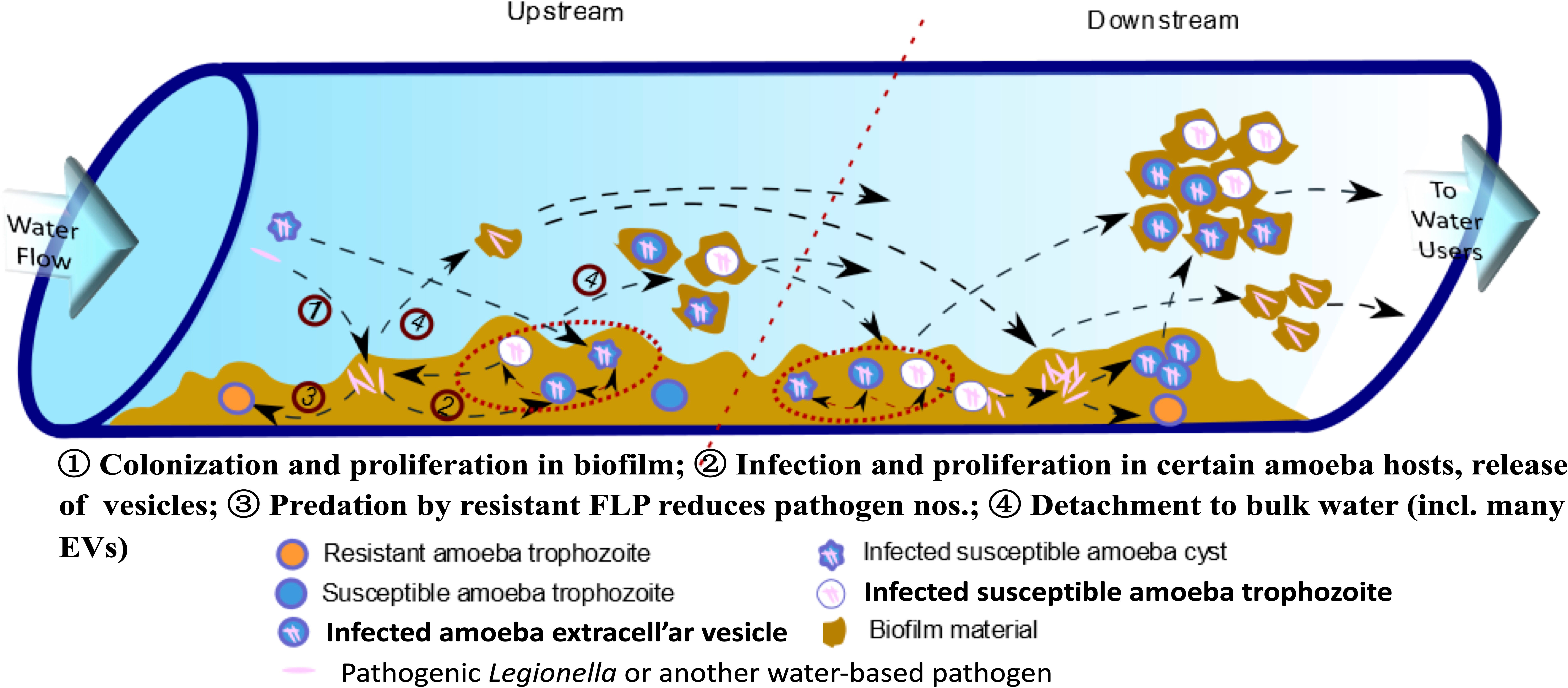
Figure 1 Conceptual model for explosive growth of Legionella in engineered water system biofilms. (Adapted from Shaheen et al. (2019), FLP - free-living protozoa [primarily amoebae and ciliates] grow on biofilm constituents, ultimately limited to feeding on less-preferred amoeba-resisting bacterial, such as L. pneumophila, leading to explosive growth and release in extracellular amoebal vesicles [EVs] that may enhance downstream amoebal predation over less preferred free bacteria – so increasing proliferation of pathogens.).
Various drinking water associated FLA have been shown to generate EVs containing tens to hundreds of L. pneumophila cells within the respiratory range (< 10 µm dia.) (Shaheen and Ashbolt, 2018). These EVs may provide packages of pathogens directly to the alveoli of our lungs, potentially influencing current quantitative microbial risk assessment (QMRA) models of Legionella risks and specifically the dose-response relationship (Hamilton et al., 2019). Hypothesized in this article are the environmental queues that may not only influence EV production but also their role leading to explosive growth of legionellae in their native biofilms (Figure 1).
While speculative for FLA, in bacterial predators, such as Myxococcus xanthus, outer membrane vesicles (OMVs) appear to specifically carry a ‘personalized’ subset of proteins that only in-part are dictated by the predator’s genome (Zwarycz et al., 2020). In other words, Zwarycz et al. (2020) hypothesize that the non-genome-derived proteome fraction is there to target its prey. Hence, is L. pneumophila really the predator not amoebae, utilizing a large fraction of its genome to not only manipulate the hosts into providing a growth niche in the form of Legionella-containing vacuoles (LCV) but potentially also EVs production to increase its persistence/transmission? Furthermore, as seen with other EVs, those containing legionellae may also modulate/avoid human host immune responses (e.g., triggered by phosphatidylserine lipids on the surface of EVs (Cole and Nizet, 2016)).
In bacteria, outer membrane vesicles (OMVs) have also been shown to provide a defense mechanism to phage infection (Reyes-Robles et al., 2018). Hence, next genomic then viruses specific to FLA are discussed and how they may impact on the evolution of these important hosts and ARB.
2.3 Genomic implications for L. pneumophila within engineered systems
As with their FLA hosts suited to the diversity of conditions in environmental biofilms, ARB maintain a larger genome than their relatives – so contradicting the expected genome reduction theory accepted for most intracellular pathogens (Moliner et al., 2010). This is likely due to the importance of gene transfer of pathogenicity factors, and the near 800 million year old arms-race between these predators and prey (Dupont et al., 2011; Shames, 2023) along with the role played by their respective viruses (Kaján et al., 2020). Of particular focus in this article is the remodeling of the Legionella-containing vesicle (LCV), with legionellae acquiring effector protein genes, such as Sar1/CopII that impact early secretory vesicle production genes (Robinson and Roy, 2006). As introduced in Figure 1, it is unknown how ARB may influence the release of EVs to enhance predator infection downstream, and provide a hot-spot for horizontal gene transfer (HGT). Recent work has pointed to FLA preferentially preying on non-ARB free bacteria over L. pneumophila (Shaheen and Ashbolt, 2021). What is unknown, but hypothesized here is that legionellae within EVs may not only avoid environmental stressors, but also overcome negative selective feeding on L. pneumophila by FLA. Indeed, are EVs preferential prey for FLA and an enhanced mechanism for prey to counter predators’ sensing of preferred food?
The role played by quorum sensing molecules (such as LAI-1 [3-hydroxypentadecane-4-one]) involved in legionellae changing from a reproductive to a transmissive phase (Schell et al., 2016), could also be part of the evolutionary arms race between predator and prey (Simon et al., 2015). It is currently unknown if LAI-like molecules are activated within EVs, as known to occur within trophozoites. Other potential changes within trophozoites and EVs that HGT may also impact could include environmental antimicrobial resistance (AMR) (Mortensen et al., 2021). AMR has generally not been viewed as important with pathogens that do not spread zoonotically or by person-to-person (such as with L. pneumophila). However, AMR within amoebal ecosystems increasingly impacted by AMR (e.g., cooling towers or irrigation systems receiving AMR-laden wastewater) could present a ‘perfect storm’ for enhance AMR within L. pneumophila-FLA ecosystems.
2.4 FLA evolution and their nucleocytoplasmic large DNA viruses
FLA evolved some 800-850 My ago (Cavalier-Smith, 2009), but have and continue to have exchange with bacteria/archaea and related viruses. A particular feature of FLA are their nucleocytoplasmic large DNA viruses (NCLDV), first described in 2003 (Abrahao et al., 2014) as Acanthamoeba polyphaga mimivirus (APMV) (now in the genus Mimivirus). In a recent whole genome analysis of A. polyphaga various NCLDVs (Marseillevirus, Mimivirus, Mollivirus, Pandoravirus, Pithovirus and a yet to be described family) were identified - inferring that NCLDV may have been domesticated during the evolution of amoebae (Schulz et al., 2020). Hence, though considered rather unusual, NCLDVs are not just a result of recent horizontal gene transfer (HGT) but in fact are associated with most major eukaryotic lineages, so impacting all ecosystems on earth (Schulz et al., 2020).
Another feature, potentially relevant to L. pneumophila’s human infectivity, has been described for human enteric viruses. A good exemplar is seen with human Norovirus, which change commensal gut bacteria’s EV production, likely impacting human host responses to infection (Mosby et al., 2022). While speculative for human infection by amoeba-legionellae material, it is intriguing that various enveloped and non-enveloped viruses generate a cytopathic effect in a range of amoebae and are released in amoebal EVs (Folkins et al., 2020; Dey et al., 2021). Hence, what roles may viruses play within amoebae (possibly in concert with internalized bacteria) regarding their expression of EVs and in modulating/avoiding human host immune responses? Important first insights to such mechanisms have been reported by Dey et al. (2022), who first described amoebic mitochondrial impacts by RNA viruses that appear to initiate and regulate apoptotic cell death. Overall this work points to research gaps that could be important to understand from an evolutionally perspective as well as to better describe human dose-response relationships for ARB pathogens.
Taking the above findings of NCLDV evolution, their likely (but not documented) presence within EVs and that specific genes from Legionella may be packaged within EVs – illustrates the is much to understand in the role that EVs may play in predator-prey interactions of ARB pathogens – and possibly much more to learn as to what actually leads to Legionella infection in human macrophages and alveolar epithelial cells.
3 Considerations for pro-active management of L. pneumophila
Managing emerging hazards has taken a more wholistic perspective in the last decade, and such an approach maybe particularly pertinent to legionellosis management. Stepping back to a broader view, we have an increasingly urbanized world population with its pollution becoming a dominant public health concern (via what is call the ‘pollutome’) (Pini et al., 2023). For chemical exposures there is also an increasing use of omics informed decisions about the exposome (Vlaanderen et al., 2021). Hence, it seems timely to also provide a One-Health lens to include microbial exosomes given their relevance to approximately one-third of community acquired (atypical) pneumonias (CAPs); which for those over 50 years of age is focused on healthcare settings, but recent work, reported for middle-eastern countries, has exposed CAP prevalent amongst young adults (20-40 years) within the broader community where legionellosis is also a major disease (Alhoufie et al., 2022).
As discussed above, molecular methods provide near real-time ways to target markers of pathogens, and in the current study the major vehicles for L. pneumophila amplification (FLA) and means for its dispersal and potential transmission (EVs). Clinical studies should also focus on the role amoeba trophozoites, EVs and fragments thereof may modulate human infection (including Pontiac fever), so such water monitoring targets could also be useful in clinical investigations. In the absence of being able to directly sample biofilm materials (such as though use of pipe wall coupons etc.), first flush tap samples are probably most informative of the last few meters where most problematic concentrations of ARB develop prior to aerosol exposures (Schoen and Ashbolt, 2011; Proctor et al., 2018). Hence, in concert with proactive management of flow conditions to keep cold waters below 25°C and hot water > 50°C, flushing of distal parts of water systems etc. (CDC, 2017), monitoring for amoebal aspects should provide the first hint of an increased potential for legionellae growth. Various qPCR protocols already exist for the major water-associated FLA and research should next focus on developing a more nuanced understanding of ARB growth conditions and targets.
4 Conclusions
While EVs seem to permeate all aspects of biology, surprisingly little is known about L. pneumophila and other amoeba-resisting pathogens’ role in driving EVs associated with FLA and engineered water systems. Seeking to provide proactive, rather than reactive management of legionellosis, by targeting management at the starting point for rapid amplification of L. pneumophila, i.e., a biomass of supportive amoebae, and promising EV telltales of their activity needs further examination. Molecular markers within EVs and of FLA would seem likely areas to first develop potential monitoring targets. Major data gaps identified during this study include: What role do internalized bacteria/viruses play in generating EVs and in modulating their behavior; Are amoebae induced to prey upon EVs; and, how FLA/EVs act in the gene pool leading to synchronized behavior within biofilm microbiomes and in human infection.
Author contributions
One author contributed to the article and approved the submitted version.
Funding
The work reported herein was previously funded by Alberta Innovates (grant #201300490), Alberta, and the Canadian Institute for Health Research (CIHR grant TGEHIPR 150713), Canada. Southern Cross University supported the salary for the author to prepare this article.
Acknowledgments
The author wishes to acknowledge the thoughtful comments of two reviewers, that much of the initial conceptual model was in collaboration with Dr. Md. Shaheen, and that initial research on amoebal vesicles protecting enteric viruses by Dr Rafik Dey and Masters students Nikki Atanasova and Melanie Folkins. The author is most thankful for their hard efforts in the laboratory and Dr. Dey’s broader vision of the significance of EVs in the biology of FLA in engineered water systems.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abrahao, J. S., Dornas, F. P., Silva, L. C., Almeida, G. M., Boratto, P. V., Colson, P., et al. (2014). Acanthamoeba polyphaga mimivirus and other giant viruses: an open field to outstanding discoveries. Virol. J. 11, 120. doi: 10.1186/1743-422X-11-120
Abu Khweek, A., Amer, A. O. (2018). Factors mediating environmental biofilm formation by Legionella pneumophila. Front. Cell. Infect. Microbiol. 8, 38. doi: 10.3389/fcimb.2018.00038
Alhoufie, S. T., Alhhazmi, A. A., Mahallawi, W. H., Alfarouk, K. O., Ibrahim, N. A. (2022). Serostatus and epidemiological characteristics for atypical pneumonia causative bacteria among healthy individuals in Medina, Saudi Arabia, a retrospective study. Healthc. (Basel) 10, 2316. doi: 10.3390/healthcare10112316
Anderson, O. R. (2018). A half-century of research on free-living amoebae, (1965-2017): review of biogeographic, ecological and physiological studies. Acta Protozologica 57, 1–28. doi: 10.4467/16890027ap.18.001.8395
Ashbolt, N. J. (2015). Environmental (saprozoic) pathogens of engineered water systems: understanding their ecology for risk assessment and management. Pathogens 4, 390–405. doi: 10.3390/pathogens4020390
Bartram, J., Chartier, Y., Lee, J. V., Pond, K., Surman-Lee, S. (2007). Legionella and the prevention of legionellosis (World Health Organization: Geneva).
Berk, S. G., Garduño, R. A. (2013). The tetrahymena and Acanthamoeba model systems. Methods Mol. Biol. 954, 393–416. doi: 10.1007/978-1-62703-161-5_25
Burillo, A., Pedro-Botet, M. L., Bouza, E. (2017). Microbiology and epidemiology of legionnaire's disease. Infect. Dis. Clin. North Am. 31, 7–27. doi: 10.1016/j.idc.2016.10.002
Burstein, D., Amaro, F., Zusman, T., Lifshitz, Z., Cohen, O., Gilbert, J. A., et al. (2016). Genomic analysis of 38 Legionella species identifies large and diverse effector repertoires. Nat. Genet. 48, 167–175. doi: 10.1038/ng.3481
Buse, H. Y., Ashbolt, N. J. (2012). Counting Legionella cells within single amoeba host cells. Appl. Environ. Microbiol. 78, 2070–2072. doi: 10.1128/AEM.07392-11
Casini, B., Baggiani, A., Totaro, M., Mansi, A., Costa, A. L., Aquino, F., et al. (2018). Detection of viable but non-culturable Legionella in hospital water network following monochloramine disinfection. J. Hosp. Infect. 98, 46–52. doi: 10.1016/j.jhin.2017.09.006
Cassini, A., Högberg, L. D., Plachouras, D., Quattrocchi, A., Hoxha, A., Simonsen, G. S., et al. (2019). Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European economic area in 2015: a population-level modelling analysis. Lancet Infect. Dis. 19, 56–66. doi: 10.1016/S1473-3099(18)30605-4
Cavalier-Smith, T. (2009). Predation and eukaryote cell origins: a coevolutionary perspective. Int. J. Biochem. Cell Biol. 41, 307–322. doi: 10.1016/j.biocel.2008.10.002
CDC (2017). Developing a water management program to reduce legionella growth and spread in buildings: a practical guide to implementing industry standards 13.2 (U.S. Department of Health and Human Services. Centers for Disease Control and Prevention). Available at: https://www.cdc.gov/legionella/downloads/toolkit.pdf.
Chauhan, D., Joseph, A. M., Shames, S. R. (2023). Intrabacterial regulation of a cytotoxic eefector by its cognate metaeffector promotes Legionella pneumophila virulence. mSphere 8, e0055222. doi: 10.1128/msphere.00552-22
Clopper, B. R., Kunz, J. M., Salandy, S. W., Smith, J. C., Hubbard, B. C., Sarisky, J. P. (2021). A methodology for classifying root causes of outbreaks of legionnaires' disease: deficiencies in environmental control and water management. Microorganisms 9, 89. doi: 10.3390/microorganisms9010089
Cole, J. N., Nizet, V. (2016). Bacterial evasion of host antimicrobial peptide defenses. Microbiol. Spectr. 4. doi: 10.1128/9781555819286.ch15
Collier, S. A., Deng, L., Adam, E. A., Benedict, K. M., Beshearse, E. M., Blackstock, A. J., et al. (2021). Estimate of burden and direct healthcare cost of infectious waterborne disease in the united states. Emerg. Infect. Dis. 27, 140–149. doi: 10.3201/eid2701.190676
Costa, A. O., Chagas, I. A. R., de Menezes-Neto, A., Rego, F. D., Nogueira, P. M., Torrecilhas, A. C., et al. (2021). Distinct immunomodulatory properties of extracellular vesicles released by different strains of Acanthamoeba. Cell Biol. Int. 45, 1060–1071. doi: 10.1002/cbin.11551
Cruz Camacho, A., Alfandari, D., Kozela, E., Regev-Rudzki, N. (2023). Biogenesis of extracellular vesicles in protozoan parasites: the ESCRT complex in the trafficking fast lane? PloS Pathog. 19, e1011140. doi: 10.1371/journal.ppat.1011140
De Giglio, O., Diella, G., Lopuzzo, M., Triggiano, F., Calia, C., Pousis, C., et al. (2021). Management of microbiological contamination of the water network of a newly built hospital pavilion. Pathogens 10, 75. doi: 10.3390/pathogens10010075
de Souza, W., Barrias, E. S. (2020). Membrane-bound extracellular vesicles secreted by parasitic protozoa: cellular structures involved in the communication between cells. Parasitol. Res. 119, 2005–2023. doi: 10.1007/s00436-020-06691-7
Dey, R., Dlusskaya, E., Ashbolt, N. J. (2022). SARS-CoV-2 surrogate (Phi6) environmental persistence within free-living amoebae. J. Wat. Health 20, 83–91. doi: 10.2166/wh.2021.167
Dey, R., Folkins, M. A., Ashbolt, N. J. (2021). Extracellular amoebal-vesicles: potential transmission vehicles for respiratory viruses. NPJ Biofilms Microbiomes 7, 25. doi: 10.1038/s41522-021-00201-y
Dey, R., Mount, H., Ensminger, A. W., Tyrrell, G. J., Ward, L. P., Ashbolt, N. J. (2019a). Isolation of Legionella pneumophila by co-culture with local ameba, Canada. Emerg. Infect. Dis. 25, 2104–2107. doi: 10.3201/eid2511.190522
Dey, R., Rieger, A., Banting, G., Ashbolt, N. J. (2020). Role of amoebae for survival and recovery of 'non-culturable' Helicobacter pylori cells in aquatic environments. FEMS Microbiol. Ecol. 96. doi: 10.1093/femsec/fiaa182
Dey, R., Rieger, A. M., Stephens, C., Ashbolt, N. J. (2019b). Interactions of Pseudomonas aeruginosa with Acanthamoeba polyphaga observed by imaging flow cytometry. Cytometry Part A. J. Int. Soc. Anal. Cytol. 95A, 555–564. doi: 10.1002/cyto.a.23768
Dietersdorfer, E., Kirschner, A., Schrammel, B., Ohradanova-Repic, A., Stockinger, H., Sommer, R., et al. (2018). Starved viable but non-culturable (VBNC) Legionella strains can infect and replicate in amoebae and human macrophages. Water Res. 141, 428–438. doi: 10.1016/j.watres.2018.01.058
Donohue, M. J. (2021). Quantification of legionella pneumophila by qPCR and culture in tap water with different concentrations of residual disinfectants and heterotrophic bacteria. Sci. Total Environ. 774, 145142. doi: 10.1016/j.scitotenv.2021.145142
Donohue, M. J., Vesper, S., Mistry, J., Donohue, J. M. (2019). Impact of chlorine and chloramine on the detection and quantification of Legionella pneumophila and Mycobacterium species. Appl. Environ. Microbiol. 85, e01942-19. doi: 10.1128/AEM.01942-19
Dupont, C. L., Grass, G., Rensing, C. (2011). Copper toxicity and the origin of bacterial resistance–new insights and applications. Metallomics 3, 1109–1118. doi: 10.1039/c1mt00107h
Falkinham, J. O., III, Pruden, A., Edwards, M. (2015). Opportunistic premise plumbing pathogens: increasingly important pathogens in drinking water. Pathogens 4, 373–386. doi: 10.3390/pathogens4020373
Folkins, M. A., Dey, R., Ashbolt, N. J. (2020). Interactions between human reovirus and free-living amoebae: implications for enteric virus disinfection and aquatic persistence. ACS ES&T 54, 10201–10206. doi: 10.1021/acs.est.0c02896
Füchslin, H. P., Kötzsch, S., Keserue, H. A., Egli, T. (2010). Rapid and quantitative detection of Legionella pneumophila applying immunomagnetic separation and flow cytometry. Cytometry A. 77, 264–274. doi: 10.1002/cyto.a.20858
Gamage, S. D., Jinadatha, C., Coppin, J. D., Kralovic, S. M., Bender, A., Ambrose, M., et al. (2022). Factors that affect legionella positivity in healthcare building water systems from a Large, national environmental surveillance initiative. Environ. Sci. Technol. 56, 11363–11373. doi: 10.1021/acs.est.2c02194
García, M. T., Jones, S., Pelaz, C., Millar, R. D., Abu Kwaik, Y. (2007). Acanthamoeba polyphaga resuscitates viable non-culturable Legionella pneumophila after disinfection. Environ. Microbiol. 9, 1267–1277. doi: 10.1111/j.1462-2920.2007.01245.x
Hamilton, K. A., Hamilton, M. T., Johnson, W., Jjemba, P., Bukhari, Z., LeChevallier, M., et al. (2018). Health risks from exposure to Legionella in reclaimed water aerosols: toilet flushing, spray irrigation, and cooling towers. Water Res. 134, 261–279. doi: 10.1016/j.watres.2017.12.022
Hamilton, K. A., Hamilton, M. T., Johnson, W., Jjemba, P., Bukhari, Z., LeChevallier, M., et al. (2019). Risk-based critical concentrations of Legionella pneumophila for indoor residential water uses. Environ. Sci. Technol. 53, 4528–4541. doi: 10.1021/acs.est.8b03000
Hamilton, K. A., Kuppravalli, A., Heida, A., Joshi, S., Haas, C. N., Verhougstraete, M., et al. (2021). Legionnaires' disease in dental offices: quantifying aerosol risks to dental workers and patients. J. Occup. Environ. Hyg. 18, 378–393. doi: 10.1080/15459624.2021.1939878
Hojo, F., Sato, D., Matsuo, J., Miyake, M., Nakamura, S., Kunichika, M., et al. (2012). Ciliates expel environmental Legionella-laden pellets to stockpile food. Appl. Environ. Microbiol. 78, 5247–5257. doi: 10.1128/AEM.00421-12
HSE (2017). Control of legionella and other infectious agents in spa-pool systems. norwick (UK: Health and Safety Executive, TSO (The Stationery Office).
ISO 11731 (2017). “Water quality,” in Enumeration of legionella (Geneva: International Organization for Standardization (ISO). Available at: https://www.iso.org/standard/61782.html.
Kaján, G. L., Doszpoly, A., Tarján, Z. L., Vidovszky, M. Z., Papp, T. (2020). Virus-host coevolution with a focus on animal and human DNA viruses. J. Mol. Evol. 88, 41–56. doi: 10.1007/s00239-019-09913-4
Lee, J. V., Lai, S., Exner, M., Lenz, J., Gaia, V., Casati, S., et al. (2011). An international trial of quantitative PCR for monitoring Legionella in artificial water systems. J. Appl. Microbiol. 110, 1032–1044. doi: 10.1111/j.1365-2672.2011.04957.x
Liu, G., Ling, F. Q., van der Mark, E. J., Zhang, X. D., Knezev, A., Verberk, J. Q., et al. (2016). Comparison of particle-associated bacteria from a drinking water treatment plant and distribution reservoirs with different water sources. Sci. Rep. 6, 20367. doi: 10.1038/srep20367
Martin, R. L., Harrison, K., Proctor, C. R., Martin, A., Williams, K., Pruden, A., et al. (2020). Chlorine disinfection of legionella spp., L. pneumophila, and Acanthamoeba under warm water premise plumbing conditions. Microorganisms, 8, 1452. doi: 10.3390/microorganisms8091452
Moliner, C., Fournier, P. E., Raoult, D. (2010). Genome analysis of microorganisms living in amoebae reveals a melting pot of evolution. FEMS Microbiol. Rev. 34, 281–294. doi: 10.1111/j.1574-6976.2009.00209.x
Mortensen, K., Lam, T. J., Ye, Y. (2021). Comparison of CRISPR-cas immune systems in healthcare-related pathogens. Front. Microbiol. 12, 758782. doi: 10.3389/fmicb.2021.758782
Mosby, C. A., Bhar, S., Phillips, M. B., Edelmann, M. J., Jones, M. K. (2022). Interaction with mammalian enteric viruses alters outer membrane vesicle production and content by commensal bacteria. J. Extracell Vesicles 11, e12172. doi: 10.1002/jev2.12172
NAS (2020). Management of legionella in water systems (Washington, DC: National Academies of Sciences, Engineering, and Medicine, The National Academies Press).
Nisar, M. A., Ross, K. E., Brown, M. H., Bentham, R., Best, G., Whiley, H. (2023). Detection and quantification of viable but non-culturable Legionella pneumophila from water samples using flow cytometry-cell sorting and quantitative PCR. Front. Microbiol. 14, 1094877. doi: 10.3389/fmicb.2023.1094877
Papadakis, A., Chochlakis, D., Sandalakis, V., Keramarou, M., Tselentis, Y., Psaroulaki, A. (2018). Legionella spp. risk assessment in recreational and garden areas of hotels. Int. J. Environ. Res. Public Health 15, 598. doi: 10.3390/ijerph15040598
Pfaller, S., King, D., Mistry, J. H., Alexander, M., Abulikemu, G., Pressman, J. G., et al. (2021). Chloramine concentrations within distribution systems and their effect on heterotrophic bacteria, mycobacterial species, and disinfection byproducts. Water Res. 205, 117689. doi: 10.1016/j.watres.2021.117689
Pini, L., Salvalaggio, A., Wennberg, A. M., Dimakou, A., Matteoli, M., Corbetta, M. (2023). The pollutome-connectome axis: a putative mechanism to explain pollution effects on neurodegeneration. Ageing Res. Rev. 86, 101867. doi: 10.1016/j.arr.2023.101867
Proctor, C. R., Reimann, M., Vriens, B., Hammes, F. (2018). Biofilms in shower hoses. Water Res. 131, 274–286. doi: 10.1016/j.watres.2017.12.027
Quan, F. S., Kong, H. H., Lee, H. A., Chu, K. B., Moon, E. K. (2020). Identification of differentially expressed Legionella genes during its intracellular growth in Acanthamoeba. Heliyon 6, e05238. doi: 10.1016/j.heliyon.2020.e05238
Reyes-Robles, T., Dillard, R. S., Cairns, L. S., Silva-Valenzuela, C. A., Housman, M., Ali, A., et al. (2018). Vibrio cholerae outer membrane vesicles inhibit bacteriophage infection. J. Bacteriol. 200, e00792-17. doi: 10.1128/JB.00792-17
Robinson, C. G., Roy, C. R. (2006). Attachment and fusion of endoplasmic reticulum with vacuoles containing Legionella pneumophila. Cell. Microbiol. 8, 793–805. doi: 10.1111/j.1462-5822.2005.00666.x
Sahr, T., Rusniok, C., Impens, F., Oliva, G., Sismeiro, O., Coppée, J. Y., et al. (2017). The Legionella pneumophila genome evolved to accommodate multiple regulatory mechanisms controlled by the CsrA-system. PloS Genet. 13, e1006629. doi: 10.1371/journal.pgen.1006629
Scheid, P. (2019). “Free-living amoebae in rivers and ponds and their multiple role in environmental health,” in Parasite and disease spread by major rivers on earth. Springer Nature Switzerland. Available at: https://www.sciencedirect.com/science/article/pii/B9780124095489109698?via%3Dihub.
Schell, U., Simon, S., Hilbi, H. (2016). Inflammasome recognition and regulation of the legionella flagellum. Curr. Top. Microbiol. Immunol. 397, 161–181. doi: 10.1007/978-3-319-41171-2_8
Schmitz-Esser, S., Tischler, P., Arnold, R., Montanaro, J., Wagner, M., Rattei, T., et al. (2010). The genome of the amoeba symbiont "Candidatus amoebophilus asiaticus" reveals common mechanisms for host cell interaction among amoeba-associated bacteria. J. Bacteriol. 192, 1045–1057. doi: 10.1128/JB.01379-09
Schoen, M. E., Ashbolt, N. J. (2011). An in-premise model for Legionella exposure during showering events. Water Res. 45, 5826–5836. doi: 10.1016/j.watres.2011.08.031
Schulz, F., Roux, S., Paez-Espino, D., Jungbluth, S., Walsh, D. A., Denef, V. J., et al. (2020). Giant virus diversity and host interactions through global metagenomics. Nature 578, 432–436. doi: 10.1038/s41586-020-1957-x
Shabardina, V., Kischka, T., Kmita, H., Suzuki, Y., Makalowski, W. (2018). Environmental adaptation of Acanthamoeba castellanii and Entamoeba histolytica at genome level as seen by comparative genomic analysis. Int. J. Biol. Sci. 14, 306–320. doi: 10.7150/ijbs.23869
Shaheen, M., Ashbolt, N. J. (2018). Free-living amoebae supporting intracellular growth may produce vesicle-bound respirable doses of Legionella within drinking water systems. Exposure Health 10, 201–209. doi: 10.1007/s12403-017-0255-9
Shaheen, M., Ashbolt, N. J. (2021). Differential bacterial predation by free-living amoebae may result in blooms of Legionella in drinking water systems. Microorganisms 9, 174. doi: 10.3390/microorganisms9010174
Shaheen, M., Scott, C., Ashbolt, N. J. (2019). Long-term persistence of infectious Legionella with free-living amoebae in drinking water biofilms. Int. J. Hygiene Environ. Health 222, 678–686. doi: 10.1016/j.ijheh.2019.04.007
Shames, S. R. (2023). Eat or be eaten: strategies used by Legionella to acquire host-derived nutrients and evade lysosomal degradation. Infect. Immun. 91, e0044122. doi: 10.1128/iai.00441-22
Shi, Y., Queller, D. C., Tian, Y., Zhang, S., Yan, Q., He, Z., et al. (2021). The ecology and evolution of amoeba-bacterium interactions. Appl. Environ. Microbiol. 87, e01866-20. doi: 10.1128/AEM.01866-20
Simon, S., Schell, U., Heuer, N., Hager, D., Albers, M. F., Matthias, J., et al. (2015). Inter-kingdom signaling by the Legionella quorum sensing molecule LAI-1 modulates cell migration through an IQGAP1-Cdc42-ARHGEF9-dependent pathway. PloS Pathog. 11, e1005307. doi: 10.1371/journal.ppat.1005307
Storey, M. V., Winiecka-Krusnell, J., Ashbolt, N. J., Stenström, T. A. (2004). The efficacy of heat and chlorine treatment against thermotolerant acanthamoebae and legionellae. Scand. J. Infect. Dis. 36, 656–662. doi: 10.1080/00365540410020785
Thomas, J. M., Ashbolt, N. J. (2011). Do free-living amoebae in treated drinking water systems present an emerging health risk? Environ. Sci. Technol. 45, 860–869. doi: 10.1021/es102876y
Thomas, V., Greub, G. (2010). Amoeba/amoebal symbiont genetic transfers: lessons from giant virus neighbours. Intervirology 53, 254–267. doi: 10.1159/000312910
Thomas, V., McDonnel, G., Denyer, S. P., Maillard, J.-Y. (2010). Freeliving amoebae and their intracellular pathogenic microorganisms: risk for water quality. FEMS Microbiol. Rev. 34, 231–259. doi: 10.1111/j.1574-6976.2009.00190.x
Thomas, J. M., Thomas, T., Stuetz, R. M., Ashbolt, N. J. (2014). Your garden hose: a potential health risk due to legionella spp. growth facilitated by free-living amoebae. Environ. Sci. Technol. 48, 10456–10464. doi: 10.1021/es502652n
Tsao, H. F., Scheikl, U., Herbold, C., Indra, A., Walochnik, J., Horn, M. (2019). The cooling tower water microbiota: seasonal dynamics and co-occurrence of bacterial and protist phylotypes. Water Res. 159, 464–479. doi: 10.1016/j.watres.2019.04.028
Vermeulen, L. C., Brandsema, P. S., van de Kassteele, J., Bom, B. C. M., van den Berg, H. H. J. L., de Roda Husman, A. M. (2020). “Mogelijke luchtverspreiding van legionella door afvalwaterzuiveringsinstallaties: een patiënt-controle onderzoek,” in RIVM briefrapport 2019-0195 (Rijksinstituut voor Volksgezondheid en Milieu (RIVM).
Vlaanderen, J., de Hoogh, K., Hoek, G., Peters, A., Probst-Hensch, N., Scalbert, A., et al. (2021). Developing the building blocks to elucidate the impact of the urban exposome on cardiometabolic-pulmonary disease: the EU EXPANSE project. Environ. Epidemiol. 5, e162. doi: 10.1097/EE9.0000000000000162
WHO (2017). “Global status report on water safety plans,” in A review of proactive risk assessment and risk management practices to ensure the safety of drinking-water. WHO/FWC/WSH/17.03 (Geneva: World Health Organization).
Yuan, L., Ju, F. (2023). Potential auxiliary metabolic capabilities and activities reveal biochemical impacts of viruses in municipal wastewater treatment plants. Environ. Sci. Technol. 57, 5485–5498. doi: 10.1021/acs.est.2c07800
Keywords: engineered water systems, environmental persistence, disinfection resistance, QMRA, monitoring
Citation: Ashbolt NJ (2023) Conceptual model to inform Legionella–amoebae control, including the roles of extracellular vesicles in engineered water system infections. Front. Cell. Infect. Microbiol. 13:1200478. doi: 10.3389/fcimb.2023.1200478
Received: 05 April 2023; Accepted: 08 May 2023;
Published: 18 May 2023.
Edited by:
Kenta Watanabe, Yamaguchi University, JapanReviewed by:
Carolina Hurtado Marcos, CEU San Pablo University, SpainJulia Walochnik, Medical University of Vienna, Austria
Copyright © 2023 Ashbolt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nicholas John Ashbolt, Tmljay5Bc2hib2x0QFNDVS5lZHUuYXU=
 Nicholas John Ashbolt
Nicholas John Ashbolt