
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell. Infect. Microbiol., 24 April 2023
Sec. Fungal Pathogenesis
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1195968
This article is part of the Research TopicProtein Metabolism in Fungal Infection and Diseases Volume IIView all articles
Cryptococcus species are opportunistic human fungal pathogens. Survival in a hostile environment, such as the elevated body temperatures of transmitting animals and humans, is crucial for Cryptococcus infection. Numerous intriguing investigations have shown that the Hsf family of thermotolerance transcription regulators plays a crucial role in the pathogen-host axis of Cryptococcus. Although Hsf1 is known to be a master regulator of the heat shock response through the activation of gene expression of heat shock proteins (Hsps). Hsf1 and other Hsfs are multifaceted transcription regulators that regulate the expression of genes involved in protein chaperones, metabolism, cell signal transduction, and the electron transfer chain. In Saccharomyces cerevisiae, a model organism, Hsf1’s working mechanism has been intensively examined. Nonetheless, the link between Hsfs and Cryptococcus pathogenicity remains poorly understood. This review will focus on the transcriptional regulation of Hsf function in Cryptococcus, as well as potential antifungal treatments targeting Hsf proteins.
Emerging and re-emerging fungal pathogens are one of the leading causes of human and animal illness and mortality (Akerfelt et al., 2010; Anckar and Sistonen, 2011; Brown et al., 2012; Fisher et al., 2012; Vos et al., 2012; Denning and Bromley, 2015; Gomez-Pastor et al., 2018). Cryptococcus species are encapsulated opportunistic pathogenic fungi that threaten human societies. Around 15% of AIDS-related deaths are caused by Cryptococcus infections, resulting in approximately 220,000 deaths per year (Idnurm et al., 2005; Kronstad et al., 2012; Erwig and Gow, 2016; Rajasingham et al., 2017). Recent researches have shown that infections caused by Cryptococcus species are a substantial public health concern in the Pacific Northwest of Northern America, Europe, Africa, and China (Byrnes et al., 2010; Byrnes et al., 2011; May et al., 2016; Liu et al., 2017). Nevertheless, antifungal medication options are limited, and the rapid development of drug resistance frequently impedes fungal treatment (Coste et al., 2004; Silver et al., 2004; Morschhauser et al., 2007; Dunkel et al., 2008; van der Linden et al., 2011; Kwon-Chung and Chang, 2012; Billmyre et al., 2020; Li et al., 2020; Priest et al., 2022).
Infections caused by Cryptococcus begin in the lung tissue and subsequently spread to the central nervous system, resulting in fatal meningitis (Hull and Heitman, 2002; Kronstad et al., 2011; Kronstad et al., 2013). Machineries, such as those involved in morphological alterations, capsule formation, nutrition acquisition, and thermotolerance, play an essential role in regulating the colonization, invasion, and replication of cells in host tissues (Idnurm et al., 2005; Lee et al., 2013; Gerwien et al., 2018; Verbancic et al., 2018; Gao et al., 2022). The ability of Cryptococcus to survive at human body temperature of 37 °C as well as in even harsher temperature conditions, such as those found in the transmission animal species, such as pigeons, whose body temperatures are 42 ± 1.3°C (Adams et al., 1999), is a key factor that contributes to its success as a fungal pathogen (Figure 1). The heat shock transcription factor (Hsf) regulation axis is one of the most extensively studied systems controlling thermotolerance in eukaryotic cells, involving both canonical and non-canonical transcription regulation patterns (Akerfelt et al., 2010; Anckar and Sistonen, 2011; Mendillo et al., 2012; Gomez-Pastor et al., 2018; Veri et al., 2018). Numerous outstanding reviews provide a comprehensive overview of the relationship between Hsf activity and fungal biology (Yamamoto et al., 2007; Morano et al., 2012; Veri et al., 2018). Hence, this review focuses on the function of the Hsf family in Cryptococcus and elucidates its function and control in Cryptococcus pathogenicity.
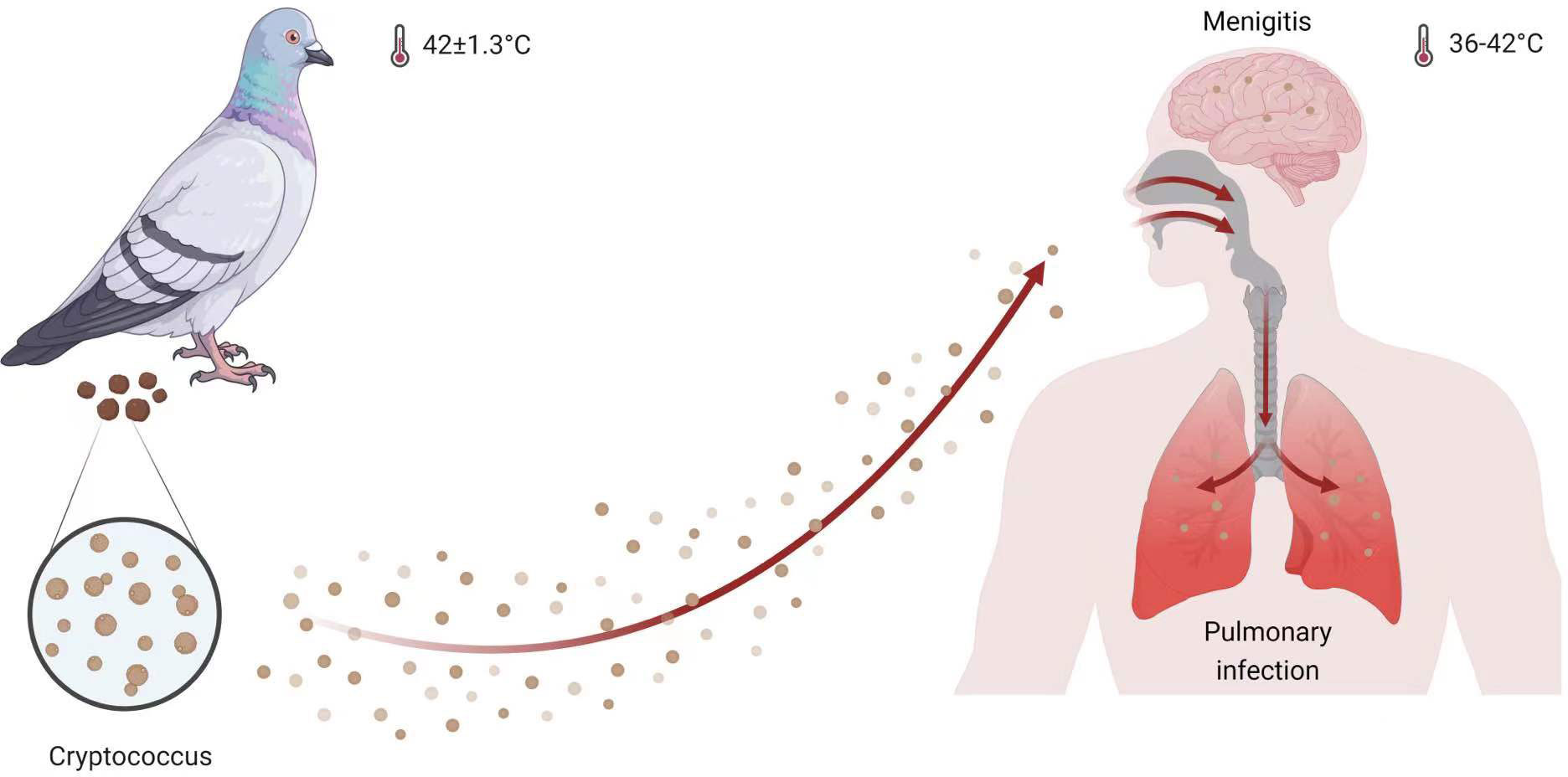
Figure 1 Cryptococcus infection model. Both humans and pigeons are hosts for Cryptococcus species, which specutilize thermotolerance to survive in pigeons with a body temperature of 42 ± 1.3°C and in humans with a temperature range of 36 to 42°C (Cramer et al., 2022). This figure was created with BioRender.com.
Heat shock transcription factors (Hsfs) are a family of highly conserved DNA-binding proteins that provide thermoprotection to cells by activating the expression of canonical target genes encoding heat shock proteins (Hsps) (Akerfelt et al., 2010; Anckar and Sistonen, 2011). The activation of Hsf1 is dependent on an increase in temperature, which dissociates the Hsf1 inhibitory complex in the cytosol compartment to produce the DNA-binding-competent homotrimer complex (Akerfelt et al., 2010; Anckar and Sistonen, 2011) (Figure 2). Hsf1 complex binds heat shock element sequences of its target genes, which then activates downstream gene expression in response to intracellular stressors such as protein misfolding and oxidative damage. We now know that the Hsf family and its regulatory mechanism are highly conserved between Saccharomyces cerevisiae (which contains one HSF gene) and human cells, which contain six HSF genes (HSF1, HSF2, HSF4, HSF5, HSFX and HSFY) (Gomez-Pastor et al., 2018). Hsf1 is the most extensively studied of these Hsf transcription factors. Studies have indicated that Hsf1 is a crucial element in the development of human diseases, although little is known about its regulation in the production of fungal virulence factors (Neef et al., 2010; Neef et al., 2011; Neef et al., 2014; Gomez-Pastor et al., 2018; Dong et al., 2019; Dong et al., 2020). In mammals, for instance, the inhibition or elevation of Hsf1 results in the development of neurological disorders and cancer, respectively.
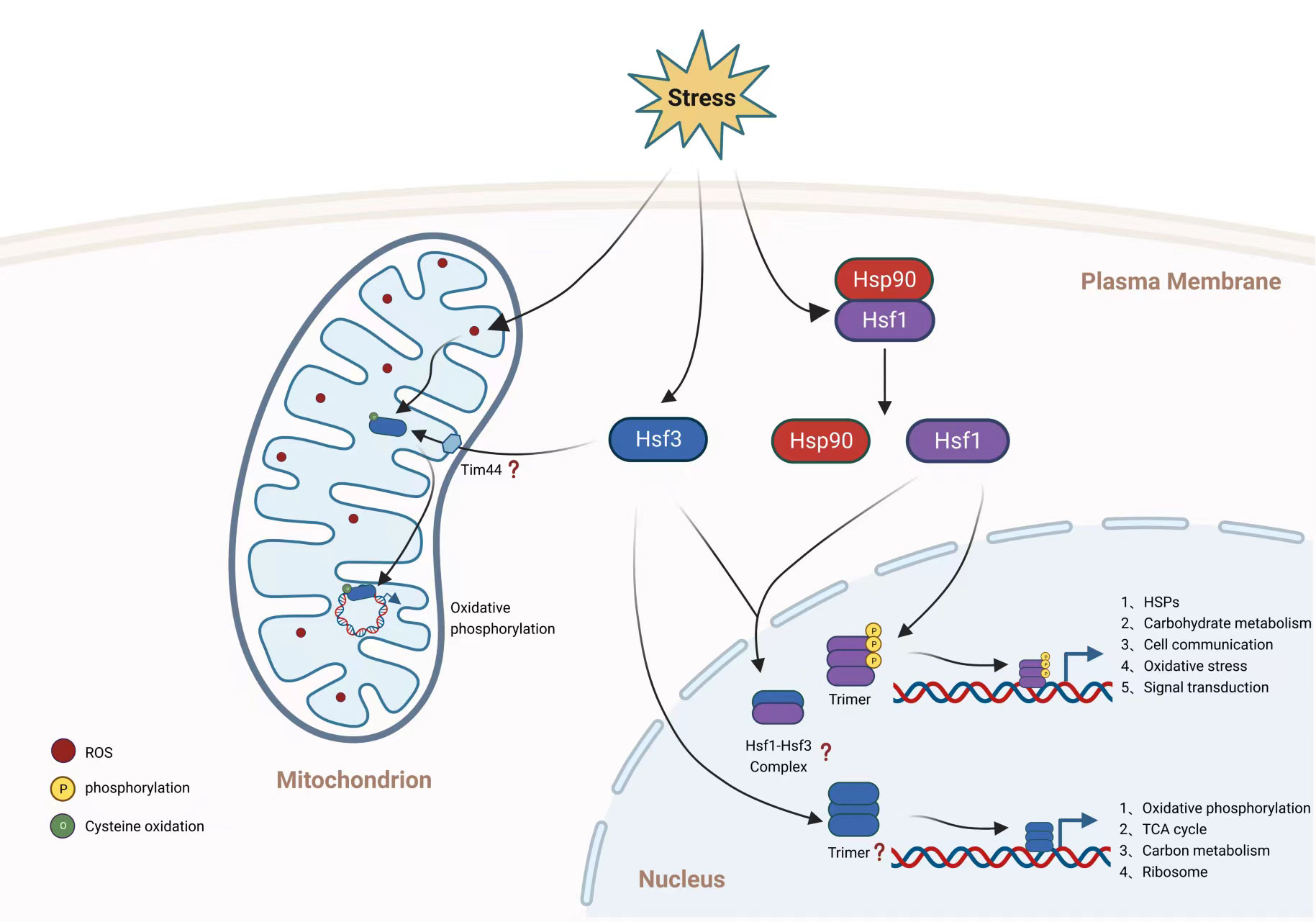
Figure 2 Hsf transcription regulation of C. neoformans. Hsf1 and Hsf3 participate in transcriptional regulation of gene expression, however the role of Hsf2 in C. neoformans is unknown. While Hsf1 and Hsf3 are transcription factors confined to the nucleus, Hsf3 is also translocated to the mitochondrion. As investigated in other organisms, Hsf1 in C. neoformans is likely repressed by Hsp90 under normal conditions and then translocated to the nucleus, where it is phosphorylated. The activated Hsf1 is subsequently trimerized to bind to gene promoter regions. Hsf1 of C. neoformans regulates the gene expression of canonical and noncanonical genes involved in glucose metabolism, cell communication, oxidative stress, and signal transduction. It is unknown if C. neoformans Hsf3 trimerizes in response to stimulation or if an Hsf1-Hsf3 complex is produced. Hsf3 in the nucleus of C. neoformans regulates the expression of genes involved in the TCA cycle, the electron transport chain, and the ribosome. ROS levels regulate the activity of mitochondrial Hsf3, which binds to the mitochondrial genome to promote the production of genes encoding the electron transport chain. This figure was created with BioRender.com.
C. neoformans HSF1, similar to that of S. cerevisiae, is an essential gene for cell growth under normal conditions, as the HSF1 endogenous promoter cannot be replaced with an inducible promoter (a galactose inducible promoter) without inhibiting cell development (Yang et al., 2017; Gao et al., 2022). Yang et al. studied the regulation mechanism of C. neoformans HSF1 for the first time in a thermotolerance transcriptome analysis using DNA microarrays. They observed an unanticipated down-regulation of HSF1 gene expression in response to elevated temperature. Another investigation of HSF1 further supported this observation in C. neoformans (Gao et al., 2022). HSP90 gene expression is strongly induced by temperature shifts despite the fact that HSP90 has a conventional Hsf1 binding motif and is a direct target of Hsf1. Curiously, HSF1 overexpression conferred tolerance to deadly temperature conditions (Yang et al., 2017). Why does C. neoformans inhibit the expression of the HSF1 gene if it is essential for thermotolerance? In one scenario, the dramatic increase of Hsp90 inhibits C. neoformans HSF1 gene expression in order to prevent its autoregulation of transcription (Gomez-Pastor et al., 2017). However, Hsf1 chromatin immunoprecipitation PCR analysis revealed no DNA amplification of its own promoter sequence, indicating that C. neoformans HSF1 is regulated differently than in other fungal species (Yang et al., 2017).
In addition to regulating protein chaperones, a DNA microarray study revealed that C. neoformans HSF1 acts as both a positive and negative transcriptional regulator in response to oxidative stress (Yang et al., 2017). In fact, a high-throughput ChIP sequencing investigation revealed that Hsf1 is a wide transcription regulator capable of DNA binding to noncanonical target genes involved in various biological processes, including glycolysis, cell communication, and signal transduction (Gao et al., 2022). The question of whether Hsf1 is a crucial virulence determinant remains unanswered, as the first experiment with an HSF1 overexpression strain exhibited the same pathogenicity as the wild-type strain. However, given that HSF1 is an essential gene, suppression of C. neoformans HSF1 is likely a promising avenue for the development of anti-cryptococcal treatment.
In other organisms, particularly mammals, posttranslational modifications (PTM) such as phosphorylation, acetylation, and SUMOylation have been demonstrated to regulate Hsf1 activity (Westerheide et al., 2009; Asano et al., 2016; Hendriks et al., 2017). Fungal Hsf1 has been found to be phosphorylated (Hoj and Jakobsen, 1994; Liu and Thiele, 1996; Hashikawa and Sakurai, 2004), although additional PTM are still unknown (Figure 2). Hsf1 may not be acetylated in C. neoformans, according to a recent comprehensive acetylome investigation, which failed to detect Hsf1 acetylation (Li et al., 2019). In S. cerevisiae, the transition from a basal level of Hsf1 phosphorylation to a hyperphosphorylation process was assumed to constitute a regulation of Hsf1 transcriptional activity levels. When C. neoformans was exposed to a higher temperature, temporary phosphorylation of Hsf1 was also detected, likely indicating the activation of Hsf1 activity (Yang et al., 2017). Although direct phosphorylation enzymes to C. neoformans Hsf1 have not yet been identified, it has been demonstrated that a protein kinase, Sch9, regulates HSF1 gene expression and protein level in a sophisticated way; that is, under basal conditions Sch9 regulates Hsf1 protein levels, whereas under heat shock conditions Sch9 represses HSF1 gene expression. However, the PTM analysis of C. neoformans Hsf1 is greatly hindered by the lack of a thorough study of Hsf1 PTM utilizing pan-antibody coupled mass spectrometry analysis and the inadequacy of the production of specialized antibodies for Hsf1 PTM sites. Recently generated phosphatase and kinase knockout libraries provide key tools to facilitate and accelerate the identification of upstream PTM enzymes of HSF1 and to map the regulation network of this essential transcription factor in C. neoformans (Lee et al., 2016; Jin et al., 2020).
Despite the fact that Hsf1 is the master transcription regulator of canonical HSP gene expression in response to temperature elevation, Hsf1 also regulates a large number of noncanonical target genes (Mendillo et al., 2012; Gao et al., 2022). For example, in mammals, Hsf1 directly coordinates in regulation of gene expression of malignancy, including cell cycle regulation, signaling, metabolism and translation. In addition, other mammalian Hsfs, including human Hsf2, have been shown to coordinate with Hsf1 to form heterotrimer complexes that regulate the expression of noncanonical target genes involved in tumor progression and neurodevelopment (Shinkawa et al., 2011; Mendillo et al., 2012; El Fatimy et al., 2014; Jaeger et al., 2016).
According to the traditional paradigm of fungal Hsfs, fungal species contain only one copy of Hsf protein (Gomez-Pastor et al., 2018; Veri et al., 2018; Gao et al., 2022), particularly in the model yeast S. cerevisiae and C. albicans. S. cerevisiae may have deceived our knowledge of the evolution and transcriptional regulation of the Hsf family in fungi. However, a recent study identified at least two Hsf orthologs in the genomes of substantially all fungal species, although their roles in transcription regulation are unknown. Except for S. cerevisiae and C. albicans, all fungal species contain multiple copies of HSF genes. Protein sequence comparisons revealed that the preponderance of fungal pathogen Hsf1s have a high degree of similarity, whereas C. neoformans Hsf3 and Hsfs from other fungal species have a moderate degree of divergence. (Gao et al., 2022). In C. neoformans, expression of all three Hsfs, namely Hsf1, Hsf2 and Hsf3, are all responsive to temperature shift, with reciprocal pattern in transcription regulation; Hsf1 is downregulated under temperature elevation conditions, as Hsf2 and Hsf3 are induced in expression upon heat treatment. Loss of C. neoformans HSF3 decreases the rate of cell survival under lethal temperature conditions and reduces fungal virulence and fitness in a mouse infection model (Gao et al., 2022). C. neoformans Hsf3 does not regulate gene expression of HSPs, instead binds directly to promoter regions of genes involved in carbohydrate metabolism (primarily genes involved in the tricarboxylic acid cycle, TCA) (Figure 2), and this resembles the noncanonical Hsf in mammals. However, C. neoformans Hsf1 and Hsf3 show high similarity in DNA-binding domain and share substantial overlap in target genes (mainly carbohydrate metabolic genes). Since it is now known that mammalian Hsf1 cooperates with other Hsfs, such as human Hsf2 (Jaeger et al., 2016; Gomez-Pastor et al., 2018), it is intriguing to examine whether C. neoformans Hsfs are regulated in a similar manner. Recent examination of mammalian Hsf2 revealed a transcription regulation in aerobic glycolysis (glycolysis) by interacting with euchromatic histone lysine methyltransferase 2 (EHMT2) to inhibit gene expression of fructose-bisphosphatase 1 (Fbp1) (Yang et al., 2019). Remarkably, C. neoformans promoter region of Fbp1 is concurrently bound by both C. neoformans Hsf1 and Hsf3. Five TCA intermediates, including citrate, isocitric acid, malate, fumarate, and alpha-ketoglutarate, are significantly induced when C. neoformans HSF3 is absent.
Despite the fact that Hsfs are thought to be nucleus-localized transcription factors, C. neoformans Hsf3 was unexpectedly found in the fungal mitochondrial organelles, showing ubiquitous binding of Hsf3 to the mitochondrial DNA (Gao et al., 2022) (Figure 2). The induction of HSF3 gene expression by mitochondrial stresses, such as heat and inhibitors of the mitochondrial complex, results in ROS overloads in C. neoformans mitochondria. Subsequently, intramitochondrial ROS oxidized Hsf3 to improve its ability to bind to DNA and activate the expression of downstream genes. How Hsf3 is recruited and translocated into the mitochondria of C. neoformans is unknown. It is probably via the universal mitochondrial translocase complex, TOM-TIM (Pfanner and Meijer, 1997). Protein co-IP followed by mass spectrometry identified the physical interaction between C. neoformans Hsf3 and Tim44 protein; however, further analysis is necessary to elucidate the mechanism of Hsf3 translocation. Results demonstrated that Hsf3 serves as a thermoprotector of C. neoformans via the regulation of gene expression of key components involved in electron transport chain, particularly the NDUFA5 subunit from complex I and Qcr9 subunit from complex III. Dampening Hsf3 protein level results in downregulation of ETC activity which promotes accumulation of intramitochondrial reactive oxygen species. The heat sensitive growth of the HSF3 deletion C. neoformans strain is readily rescued by overexpression mitochondrial specific superoxide dismutase (SOD2). Furthermore, C. neoformans Hsf3 acts as a redox sensing protein via the oxidation of the 130th cysteine residues. The oxidation by reactive oxygen species improves C. neoformans Hsf3’s mitochondrial DNA binding affinity. In a separate study, mammalian Hsf2 was suggested to be the redox sensor that determines cell fate via its regulation axis of HSF2-BTG2-SOD2, in which ROS activation of mammalian Hsf2 triggers the gene expression of BTG2 and SOD2 (Kanugovi Vijayavittal et al., 2022). The major disparity between two sensing mechanisms is that C. neoformans Hsf3 does not control gene expression of SOD2 (Gao et al., 2022). In addition, how does mammalian Hsf2 detect exclusive ROS? It is interesting to determine whether the cysteine residue at position 130 is conserved between two Hsfs.
Cryptococcosis has a significant death rate, yet treatment remains difficult due to the restricted number of antifungal drugs available (van der Linden et al., 2011; Billmyre et al., 2020; Li et al., 2020). In view of the therapeutic limitations, risks, and high costs associated with developing new antifungal medicines, the US Food and Drug Administration (FDA) has classified anti-cryptococcus medications as “orphan drugs,” offering regulatory support by decreasing the requirements for clinical research (Denning and Bromley, 2015). However, anticryptococcal medication resistance develops rapidly, surpassing the development of new therapeutic alternatives. Given that C. neoformans Hsf3 modulates virulence and Hsf1 is critical for regulating fungal growth (Yang et al., 2017), the Hsf family are prospective drug development targets. Given that the protein sequence of C. neoformans Hsf3 is similar to that of human Hsfs (Gao et al., 2022), it is necessary to develop inhibitors that target C. neoformans Hsfs but not human Hsfs (Gao et al., 2022). While the Hsf3 regulation axis may or may not be conserved in Aspergillus and Candida, Hsf1 is the highest priority option for therapeutical targets in the context of a larger understanding of fungal infections. Mammalian researches have offered new light on the production of HSF-targeting compounds for fungi (Neef et al., 2011; Neef et al., 2014; Dong et al., 2020). Mammalian Hsf1 activator and inhibitor have demonstrated efficacy in the treatment of mammalian ailments, such as neurological disorders and malignancies (Neef et al., 2011; Neef et al., 2014; Dong et al., 2020). For instance, a direct Hsf1 inhibitory chemical (Direct Targeted Hsf1 InhiBitor, DTHIB) that physically binds to human Hsf1 and induces its protein degradation suppresses the growth of tumors significantly (Dong et al., 2020). Given the significance of Hsf1 in Cryptococcus, it is intriguing to examine whether DTHIB can bind to and inhibit fungal proliferation in vivo.
Fungal Hsf1 has been found and thoroughly characterized during the past thirty years, but our understanding of the regulation of the Hsf family in fungal virulence lags far behind. Not only have recent intriguing investigations of Hsfs from mammals and fungi revealed new regulatory mechanisms and potential therapeutical targets for infectious disease, but they have also inspired the microbial community to address more biological questions. The coordination relationship between C. neoformans Hsf1 and Hsf3, as well as the unexplored function of Hsf2, remain to be clarified, and a comprehensive regulation map of the C. neoformans Hsf regulation network is urgently required. The upshot could be the development of strategies for promoting the prevention of cryptococcosis and other invasive fungal infections.
All authors contributed to the article and approved the submitted version. CS prepared the figure.
This work was supported by the National Key Research and Development Program of China (2022YFC2303000). Funds for this program were also provided by the National Natural Science Foundation of China (32270205 to C.D.) and National High Level Hospital Clinical Research Funding (2022-PUMCH-A-264 to T.S.).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adams, N. J., Pinshow, B., Gannes, L. Z., Biebach, H. (1999). Body temperatures in free-flying pigeons. J. Comp. Physiol. B 169, 195–199. doi: 10.1007/s003600050211
Akerfelt, M., Morimoto, R. I., Sistonen, L. (2010). Heat shock factors: integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 11, 545–555. doi: 10.1038/nrm2938
Anckar, J., Sistonen, L. (2011). Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu. Rev. Biochem. 80, 1089–1115. doi: 10.1146/annurev-biochem-060809-095203
Asano, Y., Kawase, T., Okabe, A., Tsutsumi, S., Ichikawa, H., Tatebe, S., et al. (2016). IER5 generates a novel hypo-phosphorylated active form of HSF1 and contributes to tumorigenesis. Sci. Rep. 6, 19174. doi: 10.1038/srep19174
Billmyre, R. B., Applen Clancey, S., Li, L. X., Doering, T. L., Heitman, J. (2020). 5-fluorocytosine resistance is associated with hypermutation and alterations in capsule biosynthesis in cryptococcus. Nat. Commun. 11, 127. doi: 10.1038/s41467-019-13890-z
Brown, G. D., Denning, D. W., Gow, N. A., Levitz, S. M., Netea, M. G., White, T. C. (2012). Hidden killers: human fungal infections. Sci. Transl. Med. 4, 165rv113. doi: 10.1126/scitranslmed.3004404
Byrnes, E. J., 3rd, Li, W., Lewit, Y., Ma, H., Voelz, K., Ren, P., et al. (2010). Emergence and pathogenicity of highly virulent cryptococcus gattii genotypes in the northwest united states. PloS Pathog. 6, e1000850. doi: 10.1371/journal.ppat.1000850
Byrnes, E. J., 3rd, Li, W., Ren, P., Lewit, Y., Voelz, K., Fraser, J. A., et al. (2011). A diverse population of cryptococcus gattii molecular type VGIII in southern Californian HIV/AIDS patients. PloS Pathog. 7, e1002205. doi: 10.1371/journal.ppat.1002205
Coste, A. T., Karababa, M., Ischer, F., Bille, J., Sanglard, D. (2004). TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of candida albicans ABC transporters CDR1 and CDR2. Eukaryot Cell 3, 1639–1652. doi: 10.1128/EC.3.6.1639-1652.2004
Cramer, M. N., Gagnon, D., Laitano, O., Crandall, C. G. (2022). Human temperature regulation under heat stress in health, disease, and injury. Physiol. Rev. 102, 1907–1989. doi: 10.1152/physrev.00047.2021
Denning, D. W., Bromley, M. J. (2015). Infectious disease. how to bolster the antifungal pipeline. Science 347, 1414–1416. doi: 10.1126/science.aaa6097
Dong, B., Jaeger, A. M., Hughes, P. F., Loiselle, D. R., Hauck, J. S., Fu, Y., et al. (2020). Targeting therapy-resistant prostate cancer via a direct inhibitor of the human heat shock transcription factor 1. Sci. Transl. Med. 12 (574), eabb5647. doi: 10.1126/scitranslmed.abb5647
Dong, B., Jaeger, A. M., Thiele, D. J. (2019). Inhibiting heat shock factor 1 in cancer: a unique therapeutic opportunity. Trends Pharmacol. Sci. 40, 986–1005. doi: 10.1016/j.tips.2019.10.008
Dunkel, N., Blass, J., Rogers, P. D., Morschhauser, J. (2008). Mutations in the multi-drug resistance regulator MRR1, followed by loss of heterozygosity, are the main cause of MDR1 overexpression in fluconazole-resistant candida albicans strains. Mol. Microbiol. 69, 827–840. doi: 10.1111/j.1365-2958.2008.06309.x
El Fatimy, R., Miozzo, F., Le Mouel, A., Abane, R., Schwendimann, L., Saberan-Djoneidi, D., et al. (2014). Heat shock factor 2 is a stress-responsive mediator of neuronal migration defects in models of fetal alcohol syndrome. EMBO Mol. Med. 6, 1043–1061. doi: 10.15252/emmm.201303311
Erwig, L. P., Gow, N. A. (2016). Interactions of fungal pathogens with phagocytes. Nat. Rev. Microbiol. 14, 163–176. doi: 10.1038/nrmicro.2015.21
Fisher, M. C., Henk, D. A., Briggs, C. J., Brownstein, J. S., Madoff, L. C., McCraw, S. L., et al. (2012). Emerging fungal threats to animal, plant and ecosystem health. Nature 484, 186–194. doi: 10.1038/nature10947
Gao, X., Fu, Y., Sun, S., Gu, T., Li, Y., Sun, T., et al. (2022). Cryptococcal Hsf3 controls intramitochondrial ROS homeostasis by regulating the respiratory process. Nat. Commun. 13, 5407. doi: 10.1038/s41467-022-33168-1
Gerwien, F., Skrahina, V., Kasper, L., Hube, B., Brunke, S. (2018). Metals in fungal virulence. FEMS Microbiol. Rev. 42 (1), fux050. doi: 10.1093/femsre/fux050
Gomez-Pastor, R., Burchfiel, E. T., Neef, D. W., Jaeger, A. M., Cabiscol, E., McKinstry, S. U., et al. (2017). Abnormal degradation of the neuronal stress-protective transcription factor HSF1 in huntington's disease. Nat. Commun. 8, 14405. doi: 10.1038/ncomms14405
Gomez-Pastor, R., Burchfiel, E. T., Thiele, D. J. (2018). Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol. 19, 4–19. doi: 10.1038/nrm.2017.73
Hashikawa, N., Sakurai, H. (2004). Phosphorylation of the yeast heat shock transcription factor is implicated in gene-specific activation dependent on the architecture of the heat shock element. Mol. Cell Biol. 24, 3648–3659. doi: 10.1128/MCB.24.9.3648-3659.2004
Hendriks, I. A., Lyon, D., Young, C., Jensen, L. J., Vertegaal, A. C., Nielsen, M. L. (2017). Site-specific mapping of the human SUMO proteome reveals co-modification with phosphorylation. Nat. Struct. Mol. Biol. 24, 325–336. doi: 10.1038/nsmb.3366
Hoj, A., Jakobsen, B. K. (1994). A short element required for turning off heat shock transcription factor: evidence that phosphorylation enhances deactivation. EMBO J. 13, 2617–2624. doi: 10.1002/j.1460-2075.1994.tb06552.x
Hull, C. M., Heitman, J. (2002). Genetics of cryptococcus neoformans. Annu. Rev. Genet. 36, 557–615. doi: 10.1146/annurev.genet.36.052402.152652
Idnurm, A., Bahn, Y. S., Nielsen, K., Lin, X., Fraser, J. A., Heitman, J. (2005). Deciphering the model pathogenic fungus cryptococcus neoformans. Nat. Rev. Microbiol. 3, 753–764. doi: 10.1038/nrmicro1245
Jaeger, A. M., Pemble, C., Sistonen, L., Thiele, D. J. (2016). Structures of HSF2 reveal mechanisms for differential regulation of human heat-shock factors. Nat. Struct. Mol. Biol. 23, 147–154. doi: 10.1038/nsmb.3150
Jin, J. H., Lee, K. T., Hong, J., Lee, D., Jang, E. H., Kim, J. Y., et al. (2020). Genome-wide functional analysis of phosphatases in the pathogenic fungus cryptococcus neoformans. Nat. Commun. 11, 4212. doi: 10.1038/s41467-020-18028-0
Kanugovi Vijayavittal, A., Kumar, P., Sugunan, S., Joseph, C., Devaki, B., Paithankar, K., et al. (2022). Heat shock transcription factor HSF2 modulates the autophagy response through the BTG2-SOD2 axis. Biochem. Biophys. Res. Commun. 600, 44–50. doi: 10.1016/j.bbrc.2022.02.018
Kronstad, J. W., Attarian, R., Cadieux, B., Choi, J., D'Souza, C. A., Griffiths, E. J., et al. (2011). Expanding fungal pathogenesis: cryptococcus breaks out of the opportunistic box. Nat. Rev. Microbiol. 9, 193–203. doi: 10.1038/nrmicro2522
Kronstad, J. W., Hu, G., Jung, W. H. (2013). An encapsulation of iron homeostasis and virulence in cryptococcus neoformans. Trends Microbiol. 21, 457–465. doi: 10.1016/j.tim.2013.05.007
Kronstad, J., Saikia, S., Nielson, E. D., Kretschmer, M., Jung, W., Hu, G., et al. (2012). Adaptation of cryptococcus neoformans to mammalian hosts: integrated regulation of metabolism and virulence. Eukaryot. Cell 11, 109–118. doi: 10.1128/EC.05273-11
Kwon-Chung, K. J., Chang, Y. C. (2012). Aneuploidy and drug resistance in pathogenic fungi. PloS Pathog. 8, e1003022. doi: 10.1371/journal.ppat.1003022
Lee, I. R., Morrow, C. A., Fraser, J. A. (2013). Nitrogen regulation of virulence in clinically prevalent fungal pathogens. FEMS Microbiol. Lett. 345, 77–84. doi: 10.1111/1574-6968.12181
Lee, K. T., So, Y. S., Yang, D. H., Jung, K. W., Choi, J., Lee, D. G., et al. (2016). Systematic functional analysis of kinases in the fungal pathogen cryptococcus neoformans. Nat. Commun. 7, 12766. doi: 10.1038/ncomms12766
Li, Y., Li, H., Sui, M., Li, M., Wang, J., Meng, Y., et al. (2019). Fungal acetylome comparative analysis identifies an essential role of acetylation in human fungal pathogen virulence. Commun. Biol. 2, 154. doi: 10.1038/s42003-019-0419-1
Li, Y., Zhang, Y., Zhang, C., Wang, H., Wei, X., Chen, P., et al. (2020). Mitochondrial dysfunctions trigger the calcium signaling-dependent fungal multidrug resistance. Proc. Natl. Acad. Sci. U.S.A. 117, 1711–1721. doi: 10.1073/pnas.1911560116
Liu, Y., Kang, M., Wu, S. Y., Ma, Y., Chen, Z. X., Xie, Y., et al. (2017). Different characteristics of cryptococcal meningitis between HIV-infected and HIV-uninfected patients in the southwest of China. Med. Mycol 55, 255–261. doi: 10.1093/mmy/myw075
Liu, X. D., Thiele, D. J. (1996). Oxidative stress induced heat shock factor phosphorylation and HSF-dependent activation of yeast metallothionein gene transcription. Genes Dev. 10, 592–603. doi: 10.1101/gad.10.5.592
May, R. C., Stone, N. R., Wiesner, D. L., Bicanic, T., Nielsen, K. (2016). Cryptococcus: from environmental saprophyte to global pathogen. Nat. Rev. Microbiol. 14, 106–117. doi: 10.1038/nrmicro.2015.6
Mendillo, M. L., Santagata, S., Koeva, M., Bell, G. W., Hu, R., Tamimi, R. M., et al. (2012). HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell 150, 549–562. doi: 10.1016/j.cell.2012.06.031
Morano, K. A., Grant, C. M., Moye-Rowley, W. S. (2012). The response to heat shock and oxidative stress in saccharomyces cerevisiae. Genetics 190, 1157–1195. doi: 10.1534/genetics.111.128033
Morschhauser, J., Barker, K. S., Liu, T. T., Bla, B. W. J., Homayouni, R., Rogers, P. D. (2007). The transcription factor Mrr1p controls expression of the MDR1 efflux pump and mediates multidrug resistance in candida albicans. PloS Pathog. 3, e164. doi: 10.1371/journal.ppat.0030164
Neef, D. W., Jaeger, A. M., Gomez-Pastor, R., Willmund, F., Frydman, J., Thiele, D. J. (2014). A direct regulatory interaction between chaperonin TRiC and stress-responsive transcription factor HSF1. Cell Rep. 9, 955–966. doi: 10.1016/j.celrep.2014.09.056
Neef, D. W., Jaeger, A. M., Thiele, D. J. (2011). Heat shock transcription factor 1 as a therapeutic target in neurodegenerative diseases. Nat. Rev. Drug Discov. 10, 930–944. doi: 10.1038/nrd3453
Neef, D. W., Turski, M. L., Thiele, D. J. (2010). Modulation of heat shock transcription factor 1 as a therapeutic target for small molecule intervention in neurodegenerative disease. PloS Biol. 8, e1000291. doi: 10.1371/journal.pbio.1000291
Pfanner, N., Meijer, M. (1997). The tom and Tim machine. Curr. Biol. 7, R100–R103. doi: 10.1016/s0960-9822(06)00048-0
Priest, S. J., Yadav, V., Roth, C., Dahlmann, T. A., Kuck, U., Magwene, P. M., et al. (2022). Uncontrolled transposition following RNAi loss causes hypermutation and antifungal drug resistance in clinical isolates of cryptococcus neoformans. Nat. Microbiol. 7, 1239–1251. doi: 10.1038/s41564-022-01183-z
Rajasingham, R., Smith, R. M., Park, B. J., Jarvis, J. N., Govender, N. P., Chiller, T. M., et al. (2017). Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect. Dis. 17, 873–881. doi: 10.1016/S1473-3099(17)30243-8
Shinkawa, T., Tan, K., Fujimoto, M., Hayashida, N., Yamamoto, K., Takaki, E., et al. (2011). Heat shock factor 2 is required for maintaining proteostasis against febrile-range thermal stress and polyglutamine aggregation. Mol. Biol. Cell 22, 3571–3583. doi: 10.1091/mbc.E11-04-0330
Silver, P. M., Oliver, B. G., White, T. C. (2004). Role of candida albicans transcription factor Upc2p in drug resistance and sterol metabolism. Eukaryot. Cell 3, 1391–1397. doi: 10.1128/EC.3.6.1391-1397.2004
van der Linden, J. W., Snelders, E., Kampinga, G. A., Rijnders, B. J., Mattsson, E., Debets-Ossenkopp, Y. J., et al. (2011). Clinical implications of azole resistance in aspergillus fumigatus, the Netherlands 2007-2009. Emerg. Infect. Dis. 17, 1846–1854. doi: 10.3201/eid1710.110226
Verbancic, J., Lunn, J. E., Stitt, M., Persson, S. (2018). Carbon supply and the regulation of cell wall synthesis. Mol. Plant 11, 75–94. doi: 10.1016/j.molp.2017.10.004
Veri, A. O., Robbins, N., Cowen, L. E. (2018). Regulation of the heat shock transcription factor Hsf1 in fungi: implications for temperature-dependent virulence traits. FEMS Yeast Res. 18 (5), foy041. doi: 10.1093/femsyr/foy041
Vos, T., Flaxman, A. D., Naghavi, M., Lozano, R., Michaud, C., Ezzati, M., et al. (2012). Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990-2010: a systematic analysis for the global burden of disease study 2010. Lancet 380, 2163–2196. doi: 10.1016/S0140-6736(12)61729-2
Westerheide, S. D., Anckar, J., Stevens, S. M., Jr., Sistonen, L., Morimoto, R. I. (2009). Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science 323, 1063–1066. doi: 10.1126/science.1165946
Yamamoto, A., Ueda, J., Yamamoto, N., Hashikawa, N., Sakurai, H. (2007). Role of heat shock transcription factor in saccharomyces cerevisiae oxidative stress response. Eukaryot Cell 6, 1373–1379. doi: 10.1128/EC.00098-07
Yang, D. H., Jung, K. W., Bang, S., Lee, J. W., Song, M. H., Floyd-Averette, A., et al. (2017). Rewiring of signaling networks modulating thermotolerance in the human pathogen cryptococcus neoformans. Genetics 205, 201–219. doi: 10.1534/genetics.116.190595
Keywords: heat shock factor, protein chaperone, Cryptococcus, fungal infection, thermotolerance
Citation: Suo C, Gao Y, Ding C and Sun T (2023) The function and regulation of heat shock transcription factor in Cryptococcus. Front. Cell. Infect. Microbiol. 13:1195968. doi: 10.3389/fcimb.2023.1195968
Received: 29 March 2023; Accepted: 10 April 2023;
Published: 24 April 2023.
Edited by:
Tong-Bao Liu, Southwest University, ChinaReviewed by:
Ning-Ning Liu, Shanghai Jiao Tong University, ChinaCopyright © 2023 Suo, Gao, Ding and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tianshu Sun, c3VuX3RpYW5zaHVAMTYzLmNvbQ==; Chen Ding, ZGluZ2NoZW5AbWFpbC5uZXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.